
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 28 July 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.953479
In industrial settings and processes, yeasts may face multiple adverse environmental conditions. These include exposure to non-optimal temperatures or pH, osmotic stress, and deleterious concentrations of diverse inhibitory compounds. These toxic chemicals may result from the desired accumulation of added-value bio-products, yeast metabolism, or be present or derive from the pre-treatment of feedstocks, as in lignocellulosic biomass hydrolysates. Adaptation and tolerance to industrially relevant stress factors involve highly complex and coordinated molecular mechanisms occurring in the yeast cell with repercussions on the performance and economy of bioprocesses, or on the microbiological stability and conservation of foods, beverages, and other goods. To sense, survive, and adapt to different stresses, yeasts rely on a network of signaling pathways to modulate the global transcriptional response and elicit coordinated changes in the cell. These pathways cooperate and tightly regulate the composition, organization and biophysical properties of the cell wall. The intricacy of the underlying regulatory networks reflects the major role of the cell wall as the first line of defense against a wide range of environmental stresses. However, the involvement of cell wall in the adaptation and tolerance of yeasts to multiple stresses of biotechnological relevance has not received the deserved attention. This article provides an overview of the molecular mechanisms involved in fine-tuning cell wall physicochemical properties during the stress response of Saccharomyces cerevisiae and their implication in stress tolerance. The available information for non-conventional yeast species is also included. These non-Saccharomyces species have recently been on the focus of very active research to better explore or control their biotechnological potential envisaging the transition to a sustainable circular bioeconomy.
Saccharomyces cerevisiae is an essential eukaryotic cell model that also has a plethora of industrial applications (Parapouli et al., 2020). Since early in human civilization, it has been extensively used in the production of fermented food and beverages, which nowadays include bread, chocolate, wine, beer, cider, sake, spirits (rum, vodka, whisky, brandy), and other alcoholic beverages arising from the fermentation of fruits, honey, and tea (Walker and Stewart, 2016; Parapouli et al., 2020). Nowadays, many enzymes, pharmaceuticals, nutraceuticals, and other added-value bioproducts can be produced in engineered yeast cell factories (Borodina and Nielsen, 2014; Runguphan and Keasling, 2014; Kwak and Jin, 2017; Nielsen, 2019; Fathi et al., 2021; Wang et al., 2021; Arnesen et al., 2022; Stovicek et al., 2022). The implementation of a sustainable circular bioeconomy requires the development of Advanced Yeast Biorefineries to produce biofuels (e.g., bioethanol and biodiesel), chemicals, materials, and other bioproducts from organic residues from agriculture, forestry, and industry residues (Madhavan et al., 2017; Mukherjee et al., 2017; Yamakawa et al., 2018; Panahi et al., 2019; Saini and Sharma, 2021; Usmani et al., 2021; Raj et al., 2022). For the transition to a sustainable biobased economy, the use of non-conventional yeasts is gaining momentum since strains of this heterogenous group of non-Saccharomyces species are advantageous alternatives to S. cerevisiae whenever they can natively express highly interesting metabolic pathways, assimilate a wider range of carbon sources, and/or exhibit higher tolerance to relevant bioprocess-related stresses (Thorwall et al., 2020). Non-conventional yeasts are also being explored to enhance the flavor profiles and reduce the ethanol content of alcoholic beverages (Gschaedler, 2017; Holt et al., 2018; Bourbon-Melo et al., 2021). Spoilage yeasts, as is the case of the osmophilic and highly weak acid tolerant yeast species of the Zygosaccharomyces genera, are also being studied due to their high tolerance to stresses associated to food preservation (Palma and Sá-Correia, 2019). However, the advanced genome-editing techniques and other genomic and bioinformatics information and tools available for S. cerevisiae, are arguably the major reason why this species is still considered the major yeast cell factory (Nandy and Srivastava, 2018; Nielsen, 2019).
Regardless of the yeast species or the specific application, industrial yeasts typically encounter stresses that trigger intricate cellular responses involving multiple players (Bauer and Pretorius, 2000; Gibson et al., 2007; Eardley and Timson, 2020). Among these players is the cell wall. The cell wall maintains cell shape and integrity and, together with the plasma membrane, is part of the cell envelope and the first line of defense against multiple adverse environmental conditions (Stewart, 2017). The cell wall is a highly dynamic organelle whose biochemical and biophysical properties can be finely tuned as yeast cells encounter different stresses throughout the course of industrial bioprocesses (Klis, 2002; Stewart, 2017). Considerable cell energy must be invested for maintaining such complex and energetically expensive response, considering that over 1200 genes are estimated to be involved, directly or indirectly, in cell wall synthesis and regulation (De Groot et al., 2001). A deeper mechanistic insight into how the cell enacts such changes at the level of the cell wall is expected to lead to a better understanding of yeast’s adaptation to stresses of industrial relevance that ultimately may lead to improved bioprocess productivity and product yield. This understanding is also instrumental to guide the improvement of food preservation practices, in particular those involving the use of weak acid food preservatives, temperature- and osmotic- induced-stresses for the microbiological stabilization and conservation of foods, beverages, and other goods.
The suggested role for the cell wall in yeast stress response and tolerance, mostly emerged from the datasets obtained from the numerous genome-wide expression analyses or chemogenomic analyses under stress reported in recent years. However, the cell wall has not received the attention it deserves as a tolerance determinant toward multiple stresses. The objective of this review paper is to gather the literature available on the topic and provide a critical opinion and a comprehensive view on the current knowledge on the role of the cell wall and related signaling pathways in yeast adaptation and survival under industrially relevant stresses. Attempts of stress tolerance improvement through the manipulation of cell wall biosynthetic pathway are also included. Most of the published knowledge pertains to the yeast model S. cerevisiae but the information available in the scientific literature for other yeast species of biotechnological relevance is also offered.
The cell wall of S. cerevisiae represents up to 30% of the cell’s dry weight (w/w) being almost entirely composed of polysaccharides (≈85%) and proteins (≈15%) (Nguyen et al., 1998) with a thickness of 115—120 nm, as determined by atomic force microscopy and ultrathin-sectioning electron microscopy (Dupres et al., 2010; Yamaguchi et al., 2011). It is a layered structure with two different layers (Figure 1), distinguishable by ultrathin-sectioning electron microscopy (Hagen et al., 2004; Yamaguchi et al., 2011; Orlean, 2012). The electron-dense outer layer mostly consists of mannoproteins whereas, in the more electron-transparent inner layer, glucans are the major component and chitin is in a lesser extent (Hagen et al., 2004; Yamaguchi et al., 2011; Orlean, 2012). Most of the cell wall mechanical strength derives from its inner layer, in which the β-linked glucans are the major components of the polysaccharide fraction, and alone represent 30–60% of the cell wall dry weight (Fleet, 1991). Approximately 30–60% of the cell wall dry biomass is composed by β-glucans, around 85% of these are 1,3-β-glucans and the remaining are 1,6-β-glucans (Aguilar-Uscanga and François, 2003; Orlean, 2012; Figure 1). The β1,3-glucan chains are composed of ≈1500 glucose units and are assembled in coiled spring-like structure that confer elasticity and tensile strength to the cell wall (Klis, 2002). The β1,6-glucans chains are shorter than the β1,3-glucan chains and, although quantitatively being a minor component of the wall, β1,6-glucans have a central role in cross-linking β1,3-glucans together (Kollár et al., 1997; Orlean, 2012). Additionally, β1,6-glucans can be connected to mannoproteins with a glycosylphosphatidylinositol (GPI) anchor and to chitin (Kollár et al., 1995; Orlean, 2012). The reducing ends of β1,3-glucans chains can be linked to a side-branching β1,6-glucan on β1,3-glucans chains, forming a fibrillar structure that serves as backbone and anchorage point for other constituents of the cell wall (Lesage and Bussey, 2006; Figure 1). The non-reducing ends of 1,3-β-glucans are linked to the reducing ends of chitin through a β-1,4 link (Kollár et al., 1995, 1997; Orlean, 2012).
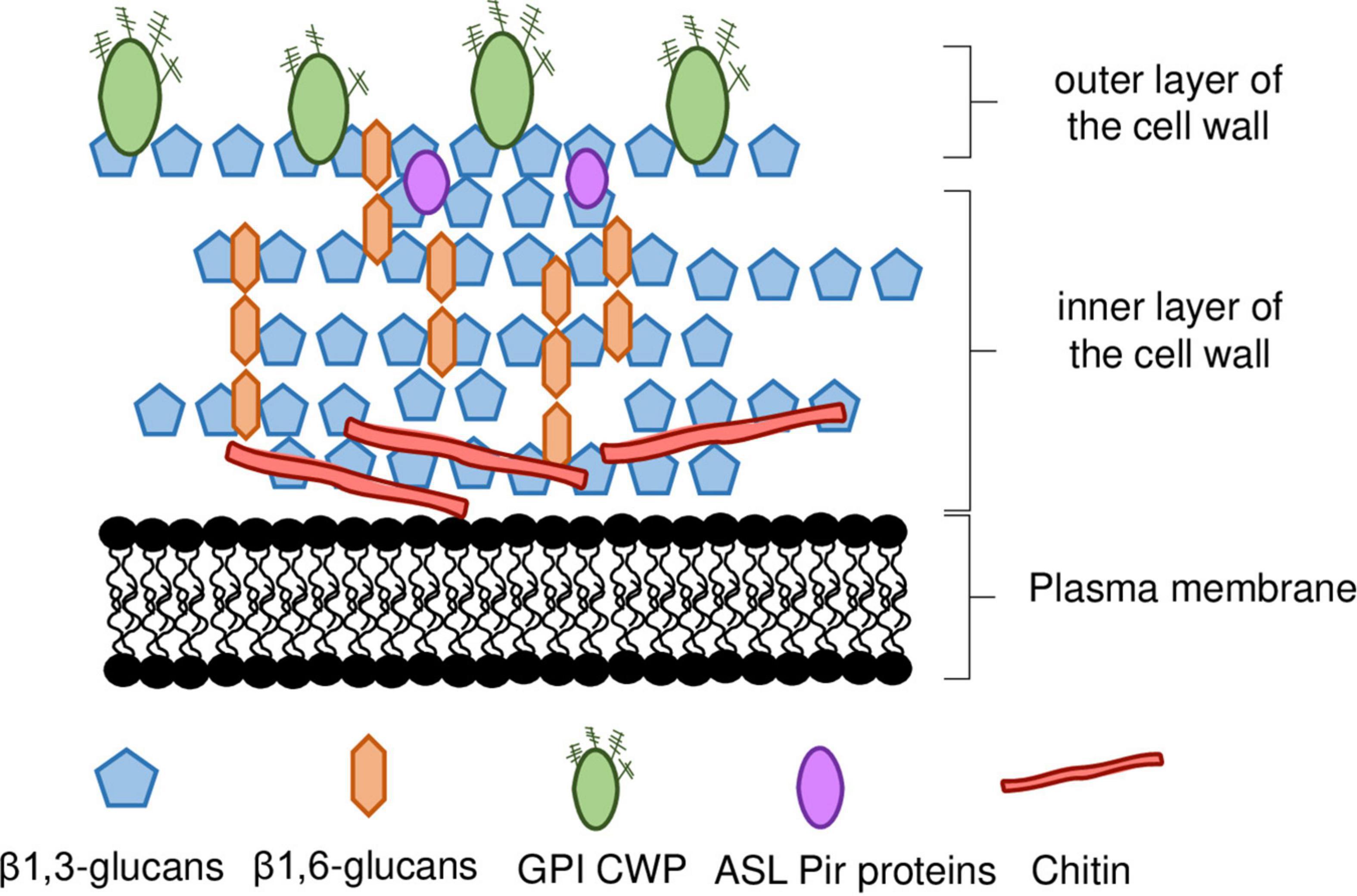
Figure 1. Schematic representation of S. cerevisiae cell wall. The inner layer of the cell wall is mostly composed of β1,3-glucan chains branched with β1,6-glucans, and chitin. The outer layer is composed of mannoproteins, most of which are linked to the inner wall by a GPI anchor, whereas ASL-Pir proteins seem to be uniformly distributed throughout the inner layer. ASL-CWP, alkali-sensitive linkage cell wall protein. GPI-CWP, glycosylphosphatidylinositol cell wall protein. Details in the main text.
Chitin is a linear polymer of β1,4-linked N-acetylglucosamine (GlcNAc), representing less than 2% of S. cerevisiae cell wall dry weight (Figure 1). Chitin can occur both in the free form, or bound to β-glucans (Kollár et al., 1995, 1997; Orlean, 2012). Chitin is normally concentrated as a ring between the mother cell and the emerging bud and in the lateral walls of the mother cell after septation (Cabib, 2009). In response to stress, chitin levels can increase to as much as 20% of the cell wall dry weight (Popolo et al., 1997; Dallies et al., 1998; Ram et al., 1998; Osmond et al., 1999; Valdivieso et al., 2000; Magnelli et al., 2002; Orlean, 2012). Changes occurring in cell wall nanomechanical properties, such as cell surface stiffness, are mostly dependent on the crosslinking between β-glucans and chitin (Dague et al., 2010) and occur in response to stresses of industrial relevance (Dague et al., 2010; Pillet et al., 2014; Schiavone et al., 2016; Ribeiro et al., 2021, 2022).
The outer layer is composed by cell wall mannoproteins (Figure 1). These mannoproteins are heavily glycosylated, modified with both N-and O-linked carbohydrates, commonly formed by mannose (Klis et al., 2006; Schiavone et al., 2014). The outer layer serves an important protective role in the cell by limiting the access of external aggressors, such as foreign enzymes, to the inner layer (Klis et al., 2006). The proteins that constitute this layer are involved in a wide range of functions often related to cell-to-cell interactions (e.g., mating, flocculation, biofilm formation, etc.) (Klis, 2002). Increased cell wall hydrophobicity influences flocculation leading to increased robustness to inhibitory chemical compounds (Kahar et al., 2022). This was associated with changes in the expression of MOT3 gene, encoding a transcription regulator that controls the expression of a cell wall protein (CWPs) encoding gene, YGP1, that influences cell wall hydrophobicity (Kahar et al., 2022).
There are two major classes of CWPs, namely the glycosylphosphatidylinositol CWPs (GPI-CWPs), and the alkali-sensitive linkage CWPs (ASL-CWPs), which include proteins of the internal repeats (Pir) family (Klis, 2002; Klis et al., 2006; Figure 1). While GPI-CWPs are typically linked to β1,6-chains by a GPI anchor, ASL-CWPs are directly linked to β1,3-glucans through an alkali-labile bond (Lesage and Bussey, 2006). They also differ in their distribution throughout the cell wall. While GPI-CWPs are found in the outer layer, Pir-CWPs seem to be uniformly distributed throughout the inner layer, which is consistent with their direct association with β1,3-glucans (Kapteyn et al., 1999b; Klis et al., 2006; Figure 1).
Cell wall biogenesis involves cell wall polysaccharide synthases and enzymes involved in cell wall remodeling, assembly and degradation (Klis et al., 2006; Free, 2013; Figure 2). The β1,3-glucans are synthetized as a linear polymer by the FKS family of genes (FKS1-3). The FKS1 and FKS2 genes code for β1,3-glucan synthases and differ mostly on the expression pattern, whereas FKS3 remains poorly characterized (Garrett-Engele et al., 1995; Mazur et al., 1995; Igual et al., 1996; Zhao et al., 1998; Lesage and Bussey, 2006). While FKS1 expression is prevalent under optimal growth, FKS2 expression is induced in response to different stresses such as glucose depletion, alternative carbon sources (e.g., acetate, glycerol, galactose), high extracellular Ca2+ or heat stress (Garrett-Engele et al., 1995; Mazur et al., 1995; Igual et al., 1996; Zhao et al., 1998). Several genes influence β-1,6-glucan levels, in particular the KRE6 and SKN1 genes, encoding proteins involved in β1,6-glucan synthesis, and the KRE9 and KHN1 encoding CWPs presumably involved in β1,6-glucan cross-link to other components of the cell wall (Roemer et al., 1994; Lesage and Bussey, 2006). Chitin is synthetized as a linear polymer by the chitin synthases encoded by the CHS1-3 gene family. The gene CHS3, encoding chitin synthase III, is by far responsible for the majority of chitin synthesis, whether it is during optimal growth conditions or during stress response when increased chitin deposition in the cell wall occurs (Roncero, 2002; Klis et al., 2006; Lesage and Bussey, 2006; Orlean, 2012). Also, CWPs mannosylation requires several genes encoding proteins with mannosyltransferase activity (Gonzalez et al., 2009a; Orlean, 2012). The GAS family (GAS1-5), encodes β1,3-glucanosyltransferases involved in β1,3-glucan branching and elongation, in which GAS1 has a major role and is also required for β1,6-glucan linkage with β1,3-glucan chains (Ram et al., 1998; Ragni et al., 2007; Orlean, 2012; Aimanianda et al., 2017). Shortening of glucan chains is also required for cell wall remodeling and involves BGL2 and EXG1-2 encoding a major endoglucanase and major exoglucanases, respectively (Larriba et al., 1995; Lesage and Bussey, 2006; Orlean, 2012; Aimanianda et al., 2017). The transglycosylases GPI proteins encoded by the CRH1-2 genes have a major role in the cross-linkage between chitin and glucans (Cabib et al., 2007, 2008; Cabib, 2009; Orlean, 2012; Blanco et al., 2015). Endochitinases, encoded by CTS1-2, are required for septation and cell separation (Kuranda and Robbins, 1991; Cabib et al., 1992; Orlean, 2012).
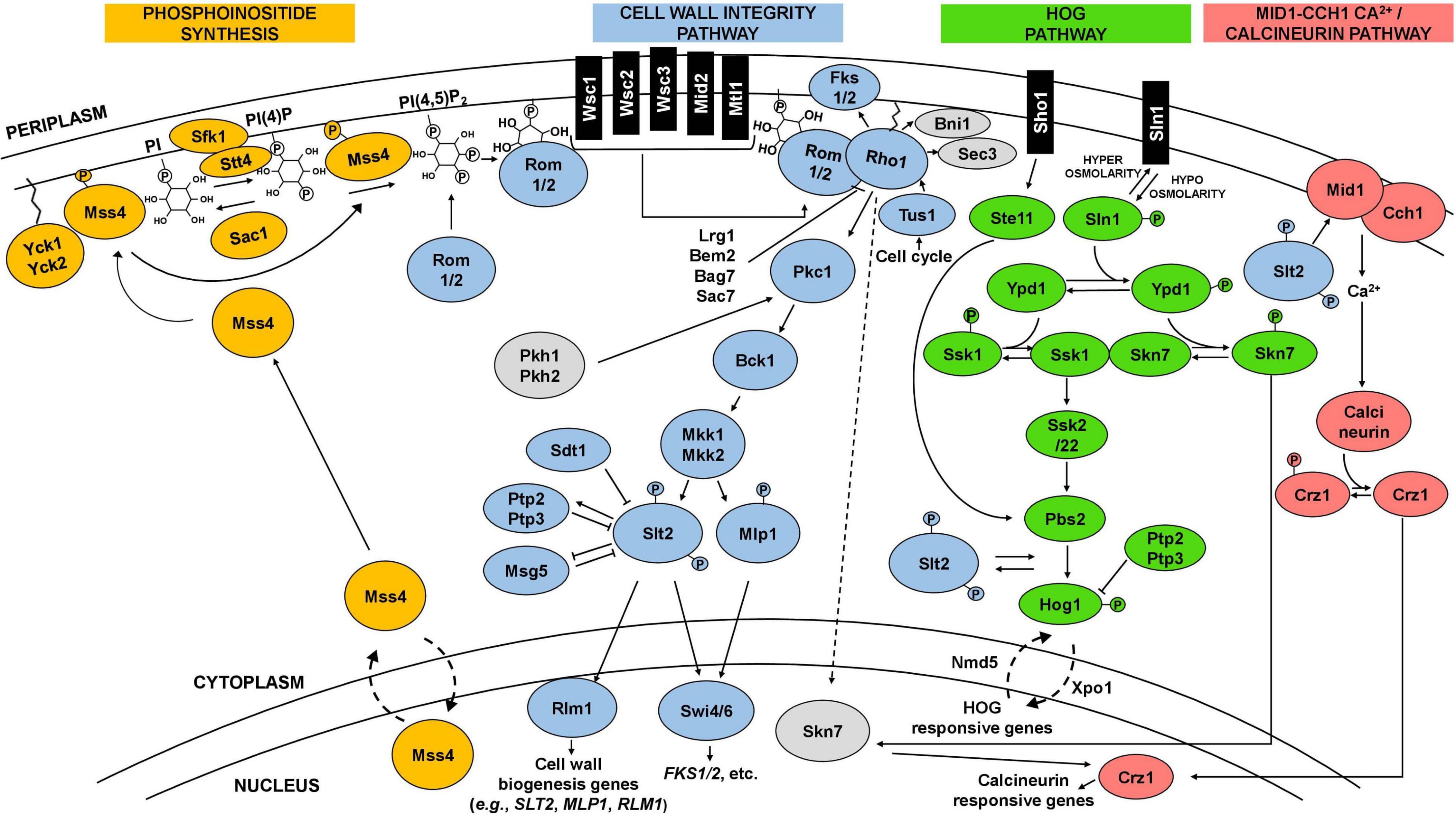
Figure 2. Schematic representation of the CWI, HOG, and calcineurin signaling pathways and interactions. The cell wall integrity pathway (CWI), in blue, is the main pathway involved in the maintenance of yeast cell wall integrity. It is frequently activated by a signal starting at plasma membrane sensors (Wsc1-3, Mid2, Mtl1), which then trigger a cascade culminating in the phosphorylation of Slt2. Phosphorylated Slt2 activates the transcription factor Rlm1 and the SBF complex (Swi4/6), triggering a transcriptional response that elicit changes at cell wall level. Phosphoinositide synthesis (in orange) plays an important role in the activation of the CWI pathway by recruiting Rom1/2 to the plasma membrane, which activates the main regulator of the pathway, the GTPase Rho1. The CWI pathway receives input and interplays with other pathways, including the high osmolarity glycerol (HOG) pathway (in green) and the calcineurin pathway (in pink), represented in the figure, but also with the protein kinase A (PKA) and TOR signaling pathways. Details in the text.
Saccharomyces cerevisiae cell wall appears to have many organizational similarities with the walls of other ascomycetous yeasts and even of basidiomycetous yeasts (Jaafar and Zueco, 2004; De Groot et al., 2005, 2008; Patel and Free, 2019; Garcia-Rubio et al., 2020). However, there are differences between the cell walls of S. cerevisiae and other fungi. For instance, the cell wall of S. cerevisiae does not contain α-glucans, chitosan, or melanin, and its chitin content is relatively low, especially when compared to other yeasts or filamentous fungi (e.g., Candida albicans, Aspergillus niger, Paracoccidioides brasiliensis, Blastomyces dermatitidis) (Kanetsuna et al., 1969; Garcia-Rubio et al., 2020; van Leeuwe et al., 2020).
Yeast cells sense and respond to environmental stresses through the induction and activity of different signaling pathways (Chen and Thorner, 2007). Depending on the type of stress, specific pathways can be triggered and directly, or indirectly, elicit changes in the composition and architecture of the cell wall (Levin, 2011; Figure 2). The CWI signaling pathway is one of the most well-described pathways, characterized for its role in cell wall maintenance and homeostasis, particularly in response to environmental external stimuli that may damage the cell wall (Gustin et al., 1998; Levin, 2011; Sanz et al., 2018; Figure 2). CWI pathway relies on a family of plasma membrane sensors, namely Wsc1-3, Mid2, and Mtl1. Stimuli that may impose a stress to the cell are detected by these sensors and triggers a phosphorylation cascade that elicit transcriptional changes to enable the cell to adapt to stress conditions (Kock et al., 2016; Sanz et al., 2018; Figure 2). For the Wsc1 sensor, the signaling transduction to the downstream components of the pathway is influenced by the spatial distribution of these sensors within the plasma membrane, requiring a functional extracellular cysteine-rich domain to form clusters in specific microdomains or rafts (Heinisch et al., 2010; Schöppner et al., 2022). Industrially relevant stresses reported to trigger a response by the referred CWI pathway membrane sensors are highlighted in Table 1. Together with phosphatidylinositol 4,5-biphosphate, PIP2, which recruits the guanine nucleotide exchange factors (GEFs) Rom1 and Rom2, the surface sensors activate the Rho1 GTPase (Levin, 2011; Sanz et al., 2018). Rho1 regulates both β1,3-glucan synthases encoded by FKS1 and FKS2, and β1,6-glucan synthase activities, and is considered a key regulator of CWI signaling (Levin, 2011). Rho1 activates Pkc1 and sets in motion a series of phosphorylation events which sequentially activate Bck1, Mkk1/2, and the pathway’s MAPK, Slt2 (Levin, 2011; Figure 2). Finally, Slt2 activates two transcription factors, Rlm1 and SBF (Swi4/Swi6 complex), that coordinate the CWI transcriptional response (Levin, 2011; Orlean, 2012; Figure 2). This response mostly involves the activation of cell wall biogenesis genes. The Rlm1 transcription factor is responsible for the expression of most of the genes induced in response to cell wall stress, such as the CHS3 gene, and also regulates the expression of FKS1 (Jung and Levin, 1999; Klis et al., 2006). Importantly, Rlm1 activates MLP1, coding for a Slt2 pseudo-kinase paralog, that, together with Slt2, activates the SBF complex for transcription of a subset of cell wall stress-activated genes. Among them are genes related with glucan synthesis (FKS1, FKS2) or glucan elongation and branching (GAS1) (Igual et al., 1996; Baetz et al., 2001; Kim et al., 2008; Levin, 2011; Harris et al., 2013)This brief description of the CWI pathway as a simple linear cascade of events can be deceiving. In fact, the CWI pathway receives lateral influences from cAMP-Protein Kinase A (PKA) signaling, TOR signaling, calcineurin signaling, the HOG pathway, and likely from others not yet clarified (Fuchs and Mylonakis, 2009; Rodríguez-Peña et al., 2010; García et al., 2017; Udom et al., 2019; Jiménez-Gutiérrez et al., 2020a,b; Figure 2). This complex network is what enables the CWI pathway to be activated by numerous types of stressors, ensuring an adequate response to each stress or group of stresses (Figure 2). The existence of a complex interplay between the CWI pathway and other signaling pathways is also being revealed for other yeast species (Donlin et al., 2014; Valiante et al., 2015; Madrid et al., 2016; Assis et al., 2018).
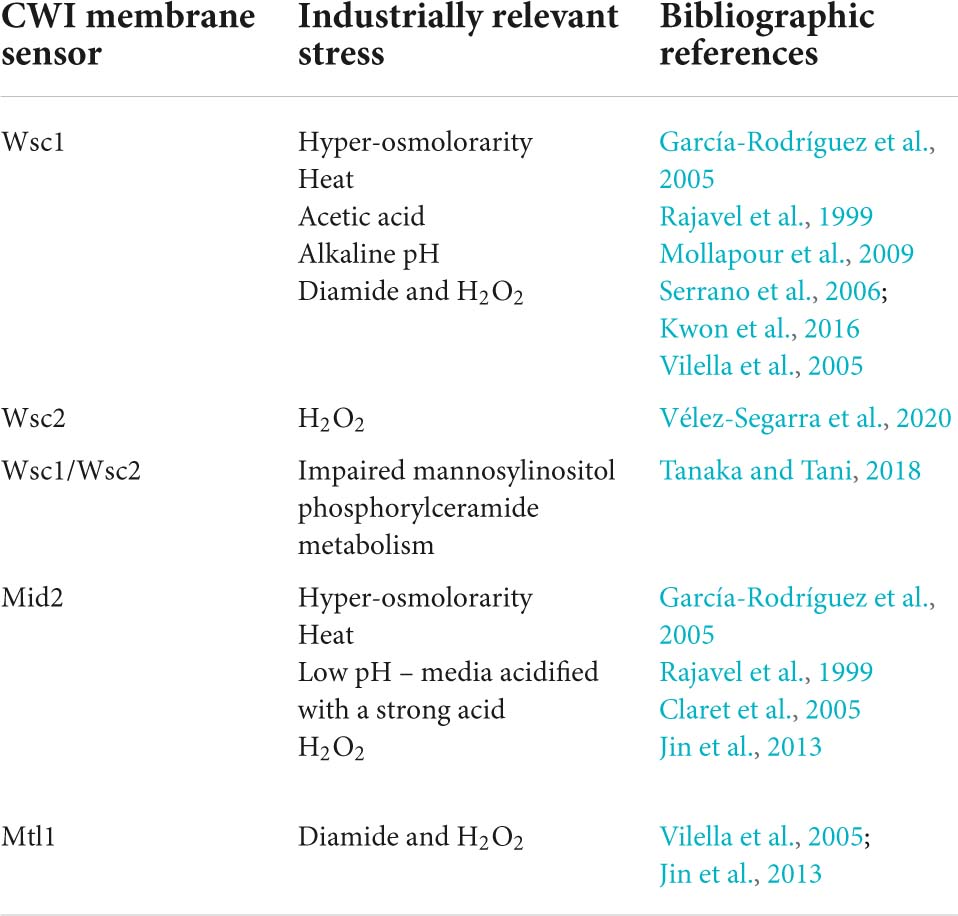
Table 1. Cell wall integrity membrane sensors implicated in the sensing of industrially relevant stresses.
Changes occurring at the level of plasma membrane can also elicit responses at the level of the cell wall. Plasma membrane stretching appears to be the main factor in activating the CWI pathway in response to a number of stresses, including high osmotic pressure and supra-optimal temperature (Kamada et al., 1995; Hohmann, 2002b). This is consistent with the CWI pathway-activating sensors being located at the membrane and the likely role of Wsc1 as a mechanosensor (Kock et al., 2015). Alterations of plasma membrane lipid composition have also been shown to impact the CWI pathway, namely, a defective biosynthesis of the complex sphingolipid mannosylinositol phosphorylceramide (MIPC) leads to increased abundance of the phosphorylated form of Slt2 and sensitivity to cell wall-perturbing agents (Morimoto and Tani, 2015). This phenotype is partly suppressed by the upregulation of ergosterol biosynthesis, suggesting that MIPC and ergosterol are coordinately involved in maintaining the integrity of the cell wall (Tanaka and Tani, 2018). Also, a reported crosstalk between plasma membrane ergosterol content (related with the level of expression of the plasma membrane ergosterol transporter Pdr18), and cell wall biophysical properties suggested a coordinated response to counteract acetic acid deleterious effects, reinforcing the notion that plasma membrane lipid composition influences cell wall integrity during stress (Ribeiro et al., 2022).
For the main and better studied stresses encountered during industrial bioprocesses, this review provides, whenever possible, an integrative view of the pathways and responses that cooperatively maintain cell wall integrity and, in so doing, helps yeasts to resist to multiple stresses.
All aerobically growing organisms suffer exposure to oxidative stress, caused by reactive oxygen species (ROS) capable of damaging cellular DNA, lipids, carbohydrates, and proteins, threatening cell integrity (Jiménez-Gutiérrez et al., 2020b). Consequently, mechanisms to protect cell components against ROS were evolved and the antioxidant defenses can be induced either by respiratory growth or in the presence of pro-oxidants (Moradas-Ferreira and Costa, 2000). Yeasts have several inducible adaptive stress responses to oxidants regulated at the transcriptional and posttranscriptional levels (Jamieson, 1998; Moradas-Ferreira and Costa, 2000). Stresses commonly arising during industrial fermentations, such as supra-optimal temperatures or presence of inhibitory concentrations of ethanol, acetic acid or lactic acid lead to oxidative stress that occurs when cellular defense mechanisms are unable to cope with existing ROS (Costa and Moradas-Ferreira, 2001; Abbott et al., 2009; Teixeira et al., 2009; Morano et al., 2012; Charoenbhakdi et al., 2016).
The cell wall or, more broadly, the cell envelope, has been associated with oxidative stress toxicity and tolerance. This involvement has been unveiled by studies focused on S. cerevisiae exposure to pro-oxidant agents such as hydrogen peroxide, lipid hyperoxides, diamine, catecholamines, and organic hydroperoxides, such as cumene hydroperoxide (CHP) and linoleic acid hydroperoxide (LoaOOH) (Staleva et al., 2004; Vilella et al., 2005; Petkova et al., 2010; Taymaz-Nikerel et al., 2016). Oxidative stress resulting from exposure to these agents induces distinct responses in S. cerevisiae. For instance, a quantitative proteomics study reported differences in the activation of the CWI pathway between hydrogen peroxide, CHP and diamide (Pandey et al., 2020). Decreased cell permeability, a property influenced by the thickness and composition of the cell wall and plasma membrane, results in increased resistance to pro-oxidant compounds by limiting their diffusion into the cell (Sousa-Lopes et al., 2004). Furthermore, membrane lipid composition is a determinant of oxidative stress resistance, with cells containing a higher level of saturated fatty acids being more resistant than cells with a higher level of polyunsaturated fatty acids (Jamieson, 1998).
Diamide induces the formation of disulfide bonds in the three-dimensional structure of CWPs, causing changes in the morphology of the cell outer layer and the increase of cell wall thickness (De Souza Pereira and Geibel, 1999; Vilella et al., 2005). CHP causes oxidative damage to the cell wall periphery leading to the upregulation of genes related to cell wall biogenesis (HSP150), CWI pathway regulation (RHO1, ROM2) and β1,6-glucan synthesis (KRE5, KRE6, KHN1) (Sha et al., 2013; Figure 3). The transcriptional reprogramming of yeast in response to oxidative stress is mainly regulated by two oxidative stress-responsive transcription factors, Yap1 and Skn7, and the general stress transcription factors, Msn2/Msn4 (Auesukaree, 2017). Notably, Skn7 has an important role in the regulation of genes involved in the maintenance of cell wall integrity, including genes coding for CWPs (Alloush et al., 2002; Li et al., 2002; Levin, 2011). The CWI pathway sensors Wsc1, Wsc2, Mtl1, and Mid2 play an important role in the sensing of oxidative stress induced by H2O2 (Vilella et al., 2005; Jin et al., 2013; Vélez-Segarra et al., 2020). Surviving and overcoming oxidative stress induced by H2O2, as for diamine, requires Pkc1 and the upstream element of the CWI pathway Rom2 (Vilella et al., 2005; Figure 3).
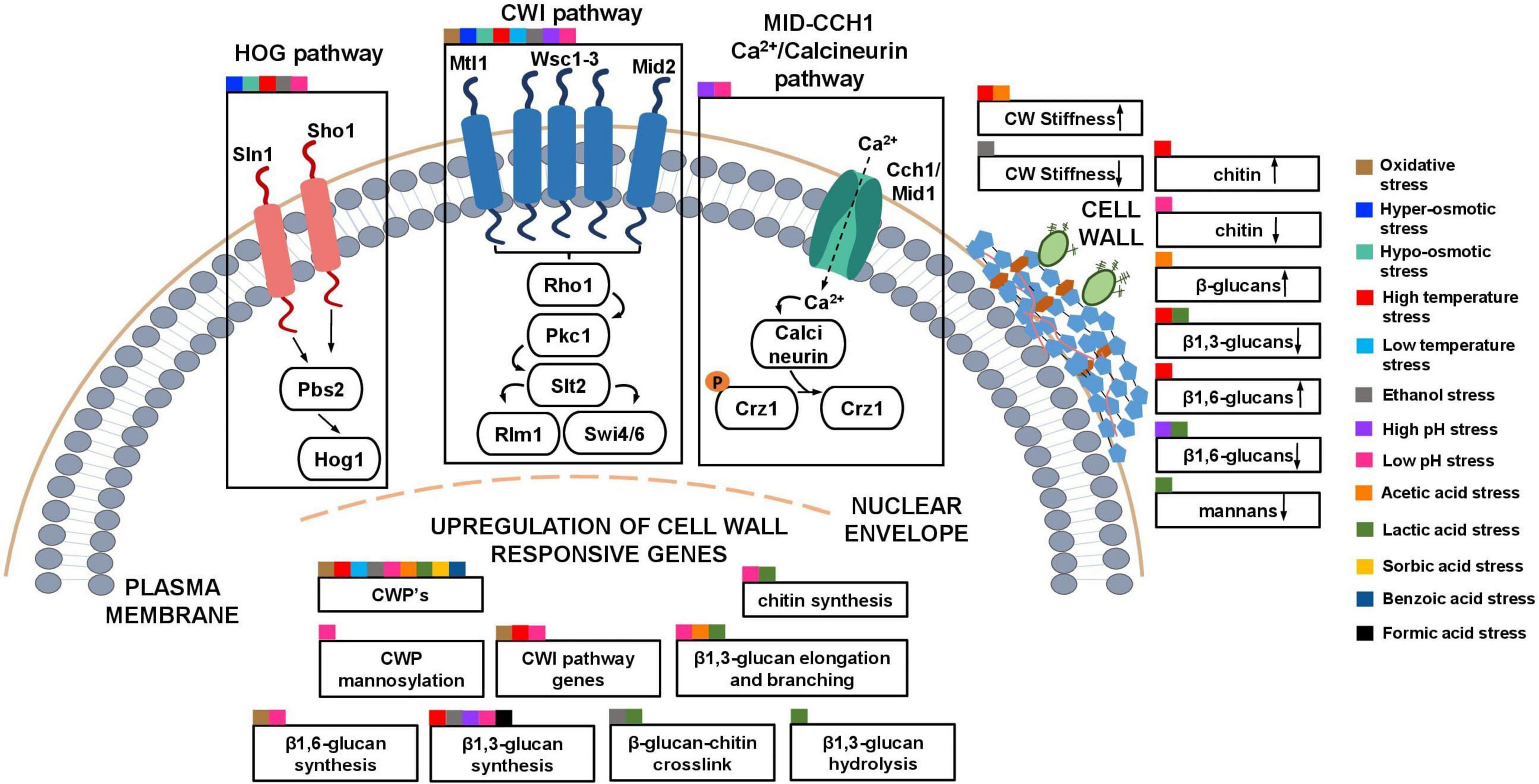
Figure 3. Schematic model of the described S. cerevisiae adaptive responses common to multiple industrial relevant stresses involving the cell wall. The cell wall integrity pathway (CWI) and other signaling pathways (HOG and Calcineurin pathway) that, in collaboration with the CWI pathway and the upregulation of cell wall genes, lead to changes in the composition and nanomechanical properties of the cell wall in response to different stresses are highlighted. Squares of different colors indicate the different industrial-relevant stresses for which data is available in the literature.
In Yarrowia lipolytica, a promising oleaginous yeast (Ledesma-Amaro and Nicaud, 2016), oxidative stress induced by H2O2 leads to changes in the morphology of the cell wall, with the formation of globular surface structures in the cell wall surface (Arinbasarova et al., 2018). When exposed to soluble complexes of UO2, the mRNA levels from genes involved in elongation of β1,3-glucan and chitin synthesis were reduced in a tropical marine strain of Y. lipolytica, able to immobilize in the cell surface uranium often associated with oxidative stress (Kolhe et al., 2020). These complexes, formed above pH 7.0, are prevalent in aquatic environments such as rivers or sea water, being this strain considered promising for the bioremediation of uranium-contaminated aquatic environments (Kolhe et al., 2020, 2021).
Hyper-osmotic stress is common in industrial bioprocesses (Mattanovich et al., 2004). Very high gravity fermentations are frequently used to enhance product titres in specific sectors, such as in first-generation bioethanol production by increasing initial sugar concentration and, consequently, osmotic pressure (Deparis et al., 2017). In winemaking and baking, yeasts also have to grow in hyper-osmotic environments and their performance depends on the ability to adapt and respond to this stress (Attfield and Kletsas, 2000; Noti et al., 2015). Also, the increase of osmotic pressure induced by the addition of stressful concentrations of sugars or salts is beneficial for food preservation (Gonzalez et al., 2016). In yeasts, the High Osmolarity Glycerol (HOG) and the CWI signaling pathways are central to osmotic stress-induced response and control of cellular turgor (Davenport et al., 1995; Hohmann, 2002a). Although there are many similarities in how the yeast cell responds to osmotic stress caused by different agents, thus allowing the description of a generalized stress response, there are also specific features for each osmolyte or salt stress (Gonzalez et al., 2016).
Sudden exposure to increased osmolarity leads to cell shrinkage due to water efflux and likely causes the release of plasma membrane material, as well as changes in cell wall structure (Morris et al., 1983; Claro et al., 2007; Saldaña et al., 2021). The decrease in cell volume is accompanied by changes in cell morphology and surface roughness (Adya et al., 2006; Saldaña et al., 2021). Intracellularly, a compensatory response is triggered, recruiting water from the vacuole into the cytoplasm (Mager and Siderius, 2002). These changes occur immediately after an osmotic upshift, likely caused by rapid biophysical forces (Ene et al., 2015). To survive such a shock, the properties of the cell wall have to be modified to have enough elasticity to respond to shifts in osmolarity, while maintaining enough rigidity to preserve cell morphology and integrity (Ene et al., 2015). The rapid changes of cellular volume and shape occurring in response to hyperosmotic shock that are fully reversible when cells are introduced in an isosmotic solution, are evidences of the remarkable plasticity of the cell wall (Saldaña et al., 2021).
The HOG pathway (Figures 2, 3) is a MAPK signal transduction system and the major pathway in the adaptation of yeast cells to increased osmolarity (Hohmann, 2002a). The pathway relies on two osmolarity sensors, Sln1 and Sho1. During an osmotic upshift, Sln1 is transiently inhibited, diminishing the levels of phosphorylated Ssk1. Unphosphorylated Ssk1 can then interact with Ssk2/Ssk22, which in turn phosphorylates Pbs2. The Sho1 osmosensor also transmits a signal to Pbs2 via the MAPKKK Ste11 (Hohmann, 2009). The phosphorylated Pbs2 activates Hog1 and modulates the expression of several genes (Hohmann, 2009), including the transcriptional activation of GPD1 and GPP2 genes, encoding a glycerol 3-phosphate dehydrogenase and a glycerol-1-phosphatase respectively, involved in glycerol production (Hohmann, 2009). In response to hyper-osmotic stress, Hog1 phosphorylates Rgc2, a regulator of the Fps1 glycerol channel, causing the dissociation of Rgc2 from Fps1 and the consequent glycerol channel closure (Lee et al., 2013). Together, the blockade of glycerol efflux by the closing of the Fps1 channel and the increase in glycerol biosynthetic capacity lead to glycerol accumulation in the cytoplasm and effectively counteract the loss of turgor pressure caused by an osmotic upshift (Jiménez-Gutiérrez et al., 2020b).
A coordinated interplay between the HOG pathway and the CWI pathway allows yeast cells to better adapt to hyper-osmotic stress conditions (García-Rodríguez et al., 2005; Vaz et al., 2020; Figure 3). The CWI pathway appears to be indirectly activated by an increase in turgor pressure caused by the accumulation of glycerol, leading to transient phosphorylation of Slt2 that depends on a functional HOG pathway (García-Rodríguez et al., 2005). Nevertheless, the CWI sensors Mid2 and Wsc1 seem to be involved in the sensing of Hog1-driven accumulation of glycerol (García-Rodríguez et al., 2005). Further evidences of the interplay between the HOG pathway and the CWI pathway were reported (Vaz et al., 2020). Chemogenomic studies have implicated genes related to β1,3-glucan synthesis (FKS1), β1,6-glucan synthesis (KRE6) and cell wall mannosylation (MNN10, ANP1) as determinants of tolerance to high-glucose and sucrose concentrations (Ando et al., 2006; Teixeira et al., 2010), known to induce hyper-osmotic stress (Deparis et al., 2017).
In Y. lipolytica, hyper-osmotic stress was also found to lead to cell wall remodeling (Kubiak-Szymendera et al., 2022). YlHOG1 deletion impacts filamentous growth, cytokinesis, and resistance to cell wall perturbing agents (Rzechonek et al., 2018). A quantitative proteomic analysis has shown that, among the upregulated proteins under hyper-osmotic stress is Pil1, involved in formation of membrane-associated protein complexes commonly referred as eisosomes and distributed across the cell surface periphery, and UTR2 coding for a CWP, involved in glucan-chitin crosslinking in S. cerevisiae (Kubiak-Szymendera et al., 2022). Other non-Saccharomyces species, such as Zygosaccharomyces rouxii, Debaryomyces hansenii, and Pichia sorbitophila are notable for their osmotolerance (Bubnová et al., 2014). The spoilage yeast of high-sugar or high-salt foods Z. rouxii appears to react to hyperosmolarity similarly to S. cerevisiae, accumulating intracellular glycerol through the increased expression of ZrGPS1 and decreased expression of ZrFPS1 (Pøibylová et al., 2007; Hou et al., 2013; Guo et al., 2020). Among Z. rouxii upregulated genes under high osmolarity stress are FKS1(encoding a β1,3-glucan synthase), UTR2 (encoding a cell wall transglycosylase), KRE9 (encoding a glycoprotein involved in β-glucan assembly), CHS1 (encoding a chitin synthase) and KAR2 (encoding an ATPase involved β1,6-glucan synthesis and also involved in the translocation of proteins into the endoplasmic reticulum) (Guo et al., 2020). The upregulation of the encoding genes helps to explain why the cell wall became thicker as the cell volume decreases, resulting in a smaller amplitude of cell size variation (Guo et al., 2020).
In a Zygosaccharomyces mellis strain, isolated from honey and tolerant to high-glucose concentrations (Liu et al., 2016, 2021), the genes KRE5 (involved in β1,6-glucan synthesis) and SLT2 (encoding a kinase of the CWI pathway), are upregulated under hyper-osmotic stress (Liu et al., 2021). This indicates that, in Z. mellis, the maintenance of cell wall integrity under this stress is also important. In D. hansenii, the cell wall was also shown to play a critical role in osmosensing and genes involved in CWP mannosylation (MNN1, PMT2, PSA1, and MNT1) are upregulated in response to hyper-osmotic stress (Thomé, 2007; Gonzalez et al., 2009b).
Another obstacle that can be faced by yeast cell factories is temperature stress, in particular heat stress. Heat stress is common during alcoholic fermentations to produce alcoholic beverages or bioethanol due to their exothermic nature. If the temperature is not controlled, a significant rise in temperature occurs (Parapouli et al., 2020; Walker and Basso, 2020). Given that in the presence of other stresses, in particular of ethanol or acetic acid stress, the optimum and the maximum temperatures for growth decrease as the stress level increase, even temperatures close to the optimal range of growth can become lethal temperatures, depending on the level of stress (van Uden, 1985; Godinho et al., 2021). Despite the importance of cold stress, the related literature is scarce in the context of this review, so this part of the article is essentially dedicated to heat stress.
At supra-optimal temperatures in the absence of any other stress (higher than 35–37°C), S. cerevisiae activates the heat shock response (HSR) and undergoes physiological changes which include membrane and cell wall restructuring (Verghese et al., 2012; Schiavone et al., 2014). During heat stress, β1,6-glucan and chitin content increases by 20 and 100% respectively, and β1,3-glucans levels decrease by 45% (Schiavone et al., 2014; Figure 3). The increased β1,6-glucan synthesis under heat stress is in accordance with a higher number of cross-linkages between this polymer and chitin, presumably compensating cell wall weakening during this stress (Kapteyn et al., 1999a; Schiavone et al., 2014). Heat stress also induces changes in the morphology of the yeast cell surface with the formation of circular structures in the surface of heat-stressed cells (Pillet et al., 2014). The emergence of these structures is, apparently, related to a dysfunction in the budding machinery, accompanied by a concomitant increase in chitin and cell wall stiffness, regulated by the CWI pathway (Pillet et al., 2014; Figure 3) activated in response to heat stress (Torres et al., 1991; Lee and Levin, 1992; Irie et al., 1993). Together with Msn2/4, Hsf1 is a transcription factor responsible for the bulk of the HSR (Imazu and Sakurai, 2005; Truman et al., 2007). Heat stress also promotes glycerol efflux by the opening of Fps1 channels regulated by the CWI pathway (Dunayevich et al., 2018). The resulting turgor loss stimulates the activation of the HOG pathway, promoting glycerol production and the re-establishment of turgor pressure (Dunayevich et al., 2018). Heat shock leads to the intracellular accumulation of trehalose in S. cerevisiae (Attfield, 1987; Hottiger et al., 1987). Since trehalose and thermotolerance are closely related, trehalose was suggested to act as a thermoprotectant (Hottiger et al., 1989). The intracellular accumulation of trehalose causes a decrease of the specific growth rate that may trigger the environmental stress response (ESR) and higher thermotolerance (Gibney et al., 2015). The accumulation of trehalose increases cytosolic osmolarity and turgor pressure, mimicking hypotonic stress, and causing plasma membrane stretching known to lead to activation of the CWI pathway (Kamada et al., 1995; Mensonides et al., 2005). Consistently, preventing trehalose synthesis by deletion of the TPS1 gene led to decreased activation of the CWI pathway upon exposure of yeast cells to heat stress (Mensonides et al., 2005). Interestingly, a recent study suggests that the UDP-Glucose Pyrophosphorylase Ugp1 is required for heat stress response by influencing trehalose and glucan content (Zhang and Xu, 2021).
Although the mechanism behind heat-induced activation of the CWI pathway is not clear, the Wsc1 and Mid2 CWI sensors appear to be involved in the activation of CWI pathway, reinforcing the notion that this stress is ultimately transmitted to the cell surface (Verna et al., 1997; Rajavel et al., 1999; Martín et al., 2000; Levin, 2011; Verghese et al., 2012). In fact, Wsc1 sensors form clusters in the plasma membrane upon heat stress (the so-called Wsc1 sensosomes) enhancing the CWI pathway signaling capability (Heinisch et al., 2010). The expression of the CWI pathway SLT2 gene, the HSP150 gene encoding a protein required for cell stability and the CWP-encoding gene YGP1 are induced in response to heat stress (Causton et al., 2001; Varol et al., 2018; Figure 3). Also, FKS2, encoding a β1,3-glucan synthase, is upregulated during heat stress, being its expression regulated by both CWI and Calcineurin pathways (Zhao et al., 1998; Figure 3).
The ability to withstand elevated temperatures while maintaining high growth rates and ethanol productivity can be beneficial for bioethanol production and could alleviate some of the costs associated with the cooling of the bioreactors often required to allow an adequate fermentative performance (Lehnen et al., 2019; Thorwall et al., 2020). Two prominent examples of thermotolerant yeast species, shown to be able to grow relatively well above 40°C, are Kluyveromyces marxianus and Ogataea polymorpha (formerly Hansenula polymorpha) (Lehnen et al., 2019; Thorwall et al., 2020). In K. marxianus, high temperatures were shown to lead to upregulation of genes associated with changes in plasma membrane composition (Fu et al., 2019). Very few is reported concerning changes occurring in K. marxianus cell wall under heat stress but a strain isolated from agave when grown at 42°C exhibit increased sensitivity to lyticase than when grown at 30°C, indicating that heat stress may affect cell wall integrity (Castillo-Plata et al., 2021). In Ogataea species, thermotolerance has been attributed to a structural predisposition of the cell envelope, related with membrane and cell wall composition, allowing a higher cell envelope stability (Lehnen et al., 2019). Among the candidate genes that contribute to heat tolerance in O. polymorpha is an ortholog of S. cerevisiae PSA1, coding for GPD-mannose pyrophosphorylase, involved in cell wall biosynthesis (Seike et al., 2021). In the genus Ogataea, Mpk1 appears to be involved in CWI signaling in response to heat stress, but, different Ogataea strains exhibit different growth phenotypes when MPK1 is absent (Kim et al., 2018). Compared to O. polymorpha, O. parapolymorpha has a thinner β-glucan and chitin layer with short mannan chains and the derived deletion mutant mpk1Δ exhibit more severe growth defects during heat stress and higher susceptibility to cell wall-perturbing agents, leading the authors to consider these differences related to cell wall structural differences (Kim et al., 2018).
The characterization of the yeast response to low temperatures is also important in industry-related bioprocesses (Liszkowska and Berlowska, 2021), in particular in many wine and beer fermentations, resulting in the retention of more volatile compounds that influence the sensory properties of the product (Liszkowska and Berlowska, 2021). Furthermore, cold-adapted spoilage yeasts can potentially impose health risks to the consumers and economic burden in the Food industry, being able to grow and proliferate at temperatures at which food products are refrigerated (Fleet, 2011). Among the genes that are upregulated in response to cold temperatures are genes encoding cell wall mannoproteins (TIR1-2, TIR4, TIP1, and DAN1) (Abramova et al., 2001; Sahara et al., 2002; Homma et al., 2003; Schade et al., 2004; Murata et al., 2006; Abe, 2007; Figure 3). Increased resistance to the cell wall-perturbing compound SDS in cold-shock-stress-induced cells has also been attributed, at least partially, to the upregulation of DAN1 (Abe, 2007). The CWI pathway WSC1, BCK1, and SLT2 genes encoding a CWI sensor, the MAPKKK Bck1 and the MAPK Slt2, respectively, have also been implicated in response and adaptation to cold temperature-induced-stress, being Slt2 phosphorylation partially dependent on the Wsc1 CWI sensor (Córcoles-Sáez et al., 2012; Figure 3). Psychrophilic yeasts, yeasts adapted to low temperatures, with an optimal growth performance at 15°C, are studied due to its biotechnological potential, in particular for the production of cold-active enzymes (Buzzini et al., 2012). Interestingly, the absence of the SWI4 or SWI6 genes, encoding transcriptional activators of the CWI pathway in Metschnikowia australis W7-5, leads to impaired growth at temperatures as low as 5°C, suggesting that these genes are determinants of tolerance to low temperature in this species (Wei et al., 2021).
Ethanol toxicity is arguably the major environmental stress limiting industrial titres and overall productivity in a wide range of industrial yeast fermentations (Gibson et al., 2007; Parapouli et al., 2020). Accumulation of high concentrations of ethanol, whether during bioethanol production, wine and brewing industries, or other alcoholic fermentations, often results in decreased fermentation yield or complete cessation of yeast metabolic activity and fermentation arrest (Walker and Basso, 2020).
Due to its liposolubility, ethanol disrupts plasma membrane lipid organization, increasing its fluidity, non-specific permeability, and compromising transmembrane electrochemical potential (van Uden, 1985). Among the adaptive responses is the alteration of plasma membrane composition (e.g., increase in ergosterol content and in unsaturated/saturated fatty acid ratio) leading to increased lipid order and counteracting plasma membrane fluidisation (Navarro-Tapia et al., 2018), together with and adaptive response at the level of the cell wall (Ding et al., 2009; Stanley et al., 2010). Remarkably, the impairment of plasma membrane integrity was shown to influence the nanomechanical properties of the cell wall during ethanol stress (Schiavone et al., 2016) (Figure 3). Specifically, ethanol-induced changes to the plasma membrane, compromising the proper delivery of plasma membrane-anchored GPI-CWPs to the membrane via the secretory pathway, affect their crosslinking to cell wall polysaccharides resulting in reduced cell wall stiffness (Schiavone et al., 2016; Figure 3).
Several chemogenomic studies have implicated genes related to cell wall biosynthesis, cell wall remodeling, and CWI pathway as determinants of ethanol tolerance (Kubota et al., 2004; Fujita et al., 2006; van Voorst et al., 2006; Auesukaree et al., 2009; Teixeira et al., 2009). Additionally, suitable supplementation of the fermentation medium with potassium, zinc or inositol was shown to improve tolerance to ethanol stress (Krause et al., 2007; Zhao et al., 2009; Xu et al., 2020). Genes reported as determinants of ethanol tolerance include MNN10, MNN11, ANP1, and HOC1, encoding four subunits of the polymerase complex responsible for the elongation of the mannose backbone present in CWPs, and LDB7, encoding a component of the chromatin structure remodeling complex, involved in the regulation of the mannosylphosphate content of mannoproteins (Kubota et al., 2004; Auesukaree et al., 2009; Teixeira et al., 2009). Moreover, the CWP-encoding genes PIR3, SED1, SPI1, and YGP1, as well as the GPI-CWPs-encoding genes of the TIR family (TIR1-3 and TIP1), were found to be transcriptionally responsive to ethanol stress (Ogawa et al., 2000; Rossignol et al., 2003; Wu et al., 2006; Udom et al., 2019; Figure 3). The overrepresentation of genes related to the mannoprotein-rich outer layer of the cell wall in response to ethanol-induced stress suggests that mannoproteins have a role in yeast adaptation to this stress, limiting ethanol’s access to the plasma membrane and thus counteracting ethanol-induced membrane permeabilization and subsequent deleterious effects. Due to its amphiphilic nature, ethanol can bind to the exposed proteins at the outer layer of the cell wall, likely altering its organization and increasing cell wall porosity (Zlotnik et al., 1984; De Nobel et al., 1990; Udom et al., 2019).
Genes related to chitin and glucan synthesis, namely CHS1 (chitin), FKS1 (β1,3-glucans), ACF2 (β1,3-glucans), and KRE6 (β1,6-glucans), as well as genes involved in cell wall integrity (PUN1) and cell wall organization (SIT4), were also found to be involved in ethanol tolerance (Kubota et al., 2004; Auesukaree et al., 2009; Teixeira et al., 2009; Mota et al., 2021). The upregulation of FKS1 and also of SED1 and SMI1 genes, involved in cell wall biosynthesis depends on the Znf1 transcription factor, recently implicated in adaptation to ethanol stress (Samakkarn et al., 2021). Consistent with a transcriptional response involving multiple components of the cell wall under ethanol stress, elements of the CWI signaling pathway, namely the membrane sensors Mid2 and Wsc1, the MAPKKK Bck1, and the MAPK Slt2, were found to be crucial for maximum tolerance to ethanol stress (Fujita et al., 2006; Auesukaree et al., 2009; Teixeira et al., 2009). Also, yeasts deficient in either one of the two Slt2-activated SBF subunits, Swi4 and Swi6, reveal higher sensitivity to ethanol (Udom et al., 2019). The SBF complex controls the expression of FKS2 and CRH1, coding for a β1,3-glucan synthase and a chitin transglycosylase involved in glucan-chitin crosslinking, respectively; these genes were shown to be upregulated during ethanol stress (Udom et al., 2019; Figure 3). Additionally, the HOG pathway also seems to contribute to the regulation of the transcriptional response of cell wall genes during ethanol stress, since the dysfunction of the HOG pathway leads to decreased expression of FKS2 and CRH1 (Udom et al., 2019). Furthermore, the wall of yeast cells exposed to ethanol exhibits higher resistance to lyticase and zymolyase treatment (Udom et al., 2019). In summary, the mechanism underlying ethanol tolerance involves a collaborative role of the CWI and HOG signaling pathway in the transcriptional regulation of cell wall genes leading to cell wall changes (Udom et al., 2019; Figure 3).
Not many non-conventional yeasts can compete with S. cerevisiae when it comes to ethanol production and tolerance, but some have other interesting features that could make their use in the bioethanol industry appealing. K. marxianus, known for its thermotolerance and ability to metabolize several carbon sources (Thorwall et al., 2020), has a fair number of studies dedicated to understanding the response to ethanol stress (Diniz et al., 2017; Alvim et al., 2019; Mo et al., 2019; da Silveira et al., 2020), although not much is known about ethanol-induced changes to its cell wall. However, the adaptive evolution under ethanol stress was found to lead to the improvement of multiple pathways, including cell wall biogenesis, suggesting that cell wall remodeling is part of a strategy to mitigate the toxic effects of ethanol, also in this species (Mo et al., 2019). The increase in cell wall thickness also occurs in Saccharomyces boulardii under ethanol stress (Ramírez-Cota et al., 2021). S. boulardii is known for its probiotic capacities as a biotherapeutic agent in infections and medical disorders (Hudson et al., 2016; Suvarna et al., 2018; Hossain et al., 2020) and is a promising species to be used in certain crafts of beer fermentation (Ramírez-Cota et al., 2021). In Issatchenkia orientalis, with potential to be used in winemaking, cell wall-related genes GAS4 (β-1,3-glucanosyltransferase involved in glucan elongation), FLO1 (CWP involved in adhesion events important for flocculation) and IFF6 (GPI-CWP involved in cell wall organization) were found to be up-regulated under ethanol stress (Miao et al., 2018).
A recent chemogenomic analysis reported that, in S. cerevisiae, methanol and ethanol, share genetic determinants of tolerance involved in cell wall maintenance (Mota et al., 2021). Methanol is a feedstock alternative to sugar-based raw substrates for biorefinery processes and a toxic compound commonly found in crude glycerol, a by-product of the biodiesel industry, and in hydrolysates from pectin-rich biomass residues (Yasokawa et al., 2010; Martins et al., 2020; Mota et al., 2021). Methanol and ethanol tolerance genes include FKS1 and SMI1 (β1,3-glucan synthesis), ROT2 (β1,6-glucan synthesis), MNN11 (mannosylation of CWPs), and WSC1 (CWI pathway membrane sensor). However, KRE6, CWH41 (β1,6-glucan synthesis) and GAS1 (β1,3-glucan chain elongation and branching) were found to be required for maximum tolerance to methanol but not for equivalent inhibitory concentrations of ethanol (Mota et al., 2021). This suggests that, despite the similarities of these alcohols, equivalent inhibitory concentrations might impact the cell wall differently and, as such, elicit not fully overlapping remodeling responses (Mota et al., 2021). The CWI pathway is implicated in methanol adaptation in Komagataella phaffii (formerly Pichia pastoris), involving the upregulation of the SLT2 homolog encoding gene in K. phaffii (Zhang et al., 2020). The homolog of Wsc1 and Wsc3 CWI sensors in the methylotrophic yeast K. phaffii were implicated in sensing methanol, interacting upstream with the K. phaffii homolog of Rom2, a GEF of S. cerevisiae CWI pathway for activation of methanol-inducible genes (Ohsawa et al., 2017). Additionally, differences in methanol metabolism, vector transformation efficiency, growth and heterologous protein production between different K. phaffii strains were related with cell wall integrity (Zhang et al., 2020), providing another example of the important role of cell wall.
Cell wall composition and architecture and the CWI pathway were also implicated in yeast adaptive response to acid and alkaline stress conditions (Claret et al., 2005; Serrano et al., 2006; De Lucena et al., 2012). However, the mechanisms underlying yeast response and tolerance to acidic conditions are complex and dependent not only on the pH value but also on the nature of the acid used to adjust low pH. Strong inorganic acids, such as sulphuric acid and hydrochloric acid, are fully dissociated at any external pH, while weak organic acids dissociation depends on medium pH and their pKa, the toxic form being the liposoluble non dissociated form (Carmelo et al., 1996, 1997; Booth and Stratford, 2003). Since the plasma membrane of unstressed cells is very poorly permeable to H+, the effect of strong acids relies, essentially, on the concentration of H+/medium pH (Carmelo et al., 1996, 1997; Lucena et al., 2020). For this reason, pH stress and stress induced by organic acid stress at low pH are discussed separately.
In bioethanol production, yeast biomass is reused after being washed between successive batches using inorganic acids. It is a common procedure, frequently carried out at a pH below 3.0 as a means of eliminating contaminant bacteria from pitching yeast. This disinfection treatment, together with the presence of toxic metabolites and other stressful conditions occurring during fermentation, may lead to the loss of cell viability and limit fermentation yield (Simpson and Hammond, 1989; De Melo et al., 2010; Lucena et al., 2020). A scalable and economic solution to control bacterial contamination during alcoholic fermentation is to run the fermentations at low pH (<4.0), at which growth and viability of most bacteria are drastically reduced (Narayanan et al., 2016). Thus, understanding how yeast strains tolerate low pH set up with strong acids may enable the improvement of ethanol yield and reduce production costs (Brosnan et al., 2000). It is likely that during fermentation, organic acids (weak acids) may also play a role since they are produced during yeast metabolism and may be already present in the fermentation medium (e.g., when lignocellulosic biomass hydrolysates are used).
In response to acid pH, the cell wall structure and composition suffers alteration (Cabib et al., 1989; Kapteyn et al., 2001) leading to deformation of surface morphology (De Lucena et al., 2015). Cell wall chitin levels decrease at growth pH values below 5.0, likely as a result of increased chitinase activity (Cabib et al., 1989; Figure 3). Many of the genes found to be upregulated at low pH are related to cell wall biogenesis, including FKS1 (β1,3-glucan synthase), GAS1 (β-1,3-glucanosyltransferase involved in cell wall remodeling-elongation of (1 → 3)-β-D-glucan chains and branching), CHS1 (chitin synthase), NCW2 (GPI-protein involved in chitin-glucan assembly), KRE6 (glucosyl hydrolase required for β1,6-glucan synthesis) and MNN9 (mannosyltransferase subunit involved in wall protein mannosylation) (De Melo et al., 2010; De Lucena et al., 2012; Figure 3). Interestingly, using QTL mapping to uncover the genetic basis of a bioethanol industrial strain Pedra-2 (PE-2) tolerance, a prevalent non-synonymous mutation (A631G) in GAS1 was identified during growth at low pH induced by sulfuric acid exposure, reinforcing the idea of the relevant role of this GPI-protein in yeast tolerance in acidic environments (Coradini et al., 2021). Together with the up-regulation of genes related with 1,3-β-glucan synthesis, elongation, and anchoring, low pH stress leads to increased cell wall resistance to compounds with β1,3-glucanase activity and to the establishment of more alkali-sensitive linkages between CWPs and the β1,3-glucan network (Kapteyn et al., 2001; De Melo et al., 2010; De Lucena et al., 2012, 2015; Lucena et al., 2020). This suggests that low pH established by a strong inorganic acid affects the β-glucan fraction of the cell wall.
As for other stresses, response to acid pH imposed by inorganic acids involves a crosstalk between different signaling pathways. The CWI signaling pathway is involved in response to low pH in S. cerevisiae and has been proposed as the main mechanism for tolerance to acid pH (Claret et al., 2005; De Lucena et al., 2012; Figure 3). Its activation is mainly mediated by Mid2, but also by Wsc1, with the latter appearing to have a more prominent role in activating the general response of the cell to this stress (Claret et al., 2005; De Lucena et al., 2012). The proposed model is that cell wall injury due to acid stress results in lower turgor and consequently mimics the effect of hyper-osmotic shock, justifying the activation of the HOG pathway (De Lucena et al., 2012; Figure 3). The HOG pathway then appears to have a dual role. First, Hog1 activates the protein complex Msn2/4, which induces the expression of ESR genes, including RGD1, a major regulator of yeast survival at low pH stress, that encodes a protein implicated in the activation of CWI pathway under acid stress (Claret et al., 2005). Second, the Hog1 kinase may help to establish a positive feedback loop at the downstream module of the CWI pathway by cooperating with the Slt2-activated Rlm1 transcription factor to increase the expression of SLT2 (Claret et al., 2005; De Lucena et al., 2012). Therefore, the HOG pathway can activate the CWI pathway while bypassing its membrane sensors. The Ca2+/calmodulin-dependent calcineurin pathway is also involved in the response to acid pH, and interacts with the CWI pathway by activating Cch1/Mid1 calcium channels by Slt2, the Crz1 transcription factor by Rho1-Skn7, and through the cooperation between Slt2 and Crz1 for the expression of FKS2 in response to cell wall injury (De Lucena et al., 2012; Figure 3). This Ca2+-dependent response is likely responsible for the increment of CWI protein trafficking and their localisation at cell surface to repair the structural changes caused by medium acidification (Lucena et al., 2020). Together, CWI, HOG, and calcineurin signaling pathways ensure the post-translational activation of the transcription factors needed to promote cell wall maintenance and regeneration to survive acidic pH stress.
The yeast S. cerevisiae proliferates better at acidic than at neutral or alkaline pH and medium alkalinisation has widespread effects in yeast physiology (Serra-Cardona et al., 2015). An increase of pH from 4.0 to 6.0 leads to the decrease of the relative proportion of β1,6-glucans in the β-glucan fraction of the cell wall (Aguilar-Uscanga and François, 2003) and cells grown at pH 6.0 are more susceptible to zymolyase treatment (Aguilar-Uscanga and François, 2003; Figure 3). The CWI pathway is necessary for tolerance to alkaline pH, as shown by the strong alkali-sensitive phenotype of the bck1Δ, slt2Δ, swi4Δ, and swi6Δ mutants (Serrano et al., 2006). The alkaline stress-mediated activation of Slt2 was also shown to depend on the CWI Wsc1 membrane sensor (Serrano et al., 2006; Kwon et al., 2016; Figure 3). Both FKS2 and GAS1 encoded proteins, involved in the synthesis and elongation of β1,3-glucans respectively, are required to resist alkaline stress, and likely play a major role in altering the ratio between different types of glucans in the cell wall (Serrano et al., 2006). The Ca2+-dependent calcineurin response has also been implicated in the regulation of cell wall synthesis during alkaline stress by the upregulation of FKS2 expression via the calcineurin-activated transcription factor Crz1 (Viladevall et al., 2004; Figure 3).
Changes in cell wall composition were also implicated in Y. lipolytica adaptation to high pH stress (Sekova et al., 2019). In particular, the structural mannoprotein YlPir1 is abundant in the cell wall in unstressed conditions but absent when Y. lipolytica cells are exposed to high pH stress (Sekova et al., 2019). This readjustment is consistent with the fact that mannans, unlike other main polysaccharides of the cell wall, are prone to alkaline hydrolysis, and therefore unstable at high pH (Sekova et al., 2019).
To summarize, there are major differences between the signaling responses elicited by acidic and alkaline pH stresses involving the cell wall. First, Mid2 appears to be the main sensor activating the CWI pathway in response to acidic pH, while Wsc1 is the main sensor in alkaline pH (Claret et al., 2005; Fuchs and Mylonakis, 2009; Serra-Cardona et al., 2015). Second, acidic pH stress mostly leads to the transcription of Rlm1-dependent genes, while alkaline pH stress favors transcription of SBF-dependent genes. Third, while the CWI pathway manages acidic stress in a Hog1-dependent manner, response to alkaline stress is Hog1-independent (Fuchs and Mylonakis, 2009).
The response and tolerance of the yeast cell to the various industrially relevant weak acids and the underlying toxicity mechanisms are not fully shared by all the acids, with specific mechanisms for a weak acid/group of weak acids (Mira et al., 2010c). In general, and broadly speaking, the higher the lipophilicity of each of a weak acid is, the higher its toxicity. The straight medium chain weak acids (e.g., butyric, hexanoic, octanoic, and decanoic acids) and sorbic and benzoic acids are more lipophilic and toxic than the short-chain volatile fatty acids (VFA) formic, acetic, and propionic acids (Mira et al., 2010c; Skoneczny, 2018).
Acetic acid is widely used as a food preservative in the food industry and is also a major inhibitory compound present in lignocellulosic biomass hydrolysates limiting the use of this low cost and abundant biomass (Palma et al., 2018; Cunha et al., 2019). Acetic acid is also produced by yeast metabolic activity and can lead, together with ethanol and other yeast toxic metabolites, to decreased ethanol yield and even fermentation arrest depending on the level of stress (Palma et al., 2018; Cunha et al., 2019; Palma and Sá-Correia, 2019). Elucidation of the mechanisms underlying yeast adaptation and tolerance to acetic acid is instrumental to pave way for strain and process optimisation in several important biotechnological and food industries.
When the external pH is below acetic acid pKa (below 4.75 at 25°C) (Lide, 2003), the undissociated form of the acid (CH3COOH) is able to passively diffuse through the plasma membrane lipid bilayer (Casal et al., 1996; Mira et al., 2010c; Palma et al., 2018; Palma and Sá-Correia, 2019). Once in the near-neutral cytosol, acetic acid dissociates into the negatively charged acetate counterion, CH3COO–, releasing protons, H+. Being unable to cross the hydrophobic lipid layer due to the electric charge, these ions accumulate in the cytosol, resulting in decreased intracellular pH, increased turgor pressure and oxidative stress, disrupting normal metabolism (Mira et al., 2010c; Palma et al., 2018; Palma and Sá-Correia, 2019; Ribeiro et al., 2022).
Several genome-wide studies have sought to shed light into the global mechanisms involved in the response and tolerance of S. cerevisiae to acetic acid (Kawahata et al., 2006; Abbott et al., 2007; Almeida et al., 2009; Mira et al., 2010a,b; Bajwa et al., 2013; Longo et al., 2015). Increased cell wall impermeabilization in adapted yeast cells can reduce the passive diffusion of the weak acids into the cytosol, in this way restraining the futile cycle associated with the re-entry of the liposoluble acid form after the active expulsion of its counter-ion from the cell interior (Ullah et al., 2013). Recently, it was reported a coordinate and comprehensive view on the time course of the alterations occurring at the level of the cell wall during adaptation of a yeast cell population to sudden exposure to a sub-lethal stress induced by acetic acid (Ribeiro et al., 2021). Yeast cell wall resistance to lyticase activity was found to increase during acetic acid-induced growth latency, corresponding to the period of yeast population adaptation to sudden exposure to acetic acid. This response was correlated with the increase of cell stiffness, assessed by atomic force microscopy (Ribeiro et al., 2021; Figure 3). The increased content of cell wall β-glucans, also assessed by fluorescence microscopy, and the slight increase of the transcription level of the GAS1 gene encoding a β-1,3-glucanosyltransferase that leads to elongation of β1,3-glucan chains, were also implicated (Ribeiro et al., 2021; Figure 3). These observations reinforce the notion that the adaptive yeast response to acetic acid stress involves a coordinate alteration of the cell wall at the biophysical and molecular levels, essential to limit the futile cycle associated to the re-entry of the toxic acid form after the active expulsion of acetate from the cell interior (Ribeiro et al., 2021).
The adaptive genomic response to acetic acid in S. cerevisiae is mainly regulated by the Haa1 transcription factor involved in the direct, or indirect, transcriptional activation of approximately 80% of acetic acid-responsive genes and likely involved in the response at the cell wall level (Mira et al., 2010a, 2011). Haa1 increased expression or Haa1 specific mutations lead to increased tolerance to acetic acid stress and to a lower intracellular accumulation of acetate (Tanaka et al., 2012; Palma et al., 2018). Overexpression of HAA1 improves cell wall robustness in response to this weak acid, as suggested by the decreased susceptibility of the cell wall to lyticase activity mediated disruption (Cunha et al., 2018). Also, YGP1 and SPI1, encoding CWPs, belong to the Haa1 regulon; they are upregulated under acetic acid stress and contribute to yeast tolerance (Simões et al., 2006; Sakihama et al., 2015; Figure 3). Increased mRNA levels from YGP1 were reported in cells overexpressing HAA1 (Tanaka et al., 2012). These results suggest that not only the activation of acetate expulsion through efflux pumps is involved in acetic acid tolerance, as proposed (Piper et al., 1998; Holyoak et al., 1999; Tenreiro et al., 2002; Fernandes et al., 2005; Kawahata et al., 2006; Mira et al., 2010b), but a more efficient restriction of the diffusional entry of acetic acid, partially dependent of the CWP Ygp1, can also be involved (Tanaka et al., 2012). The Znf1 transcription factor was also implicated in acetic acid tolerance and in the upregulation of the YGP1 gene (Songdech et al., 2020).
Genes involved in CWP mannosylation (MNN2, MNN9, MNN11, KTR4), chitin synthesis (CHS1, CHS5), β1,3-glucan synthesis (FKS1, ROM2), and β1,6-glucan synthesis (KRE6) were also reported as being required for maximum tolerance of S. cerevisiae to acetic acid stress (Mollapour et al., 2009; Mira et al., 2010b). However, the mRNA levels from RLM1, encoding a major transcriptional regulator of the CWI pathway, and from Rlm1-target genes, were found to decrease in cells exposed to acetic acid stress, suggesting that the CWI pathway is not the major key player in acetic acid stress response (Ribeiro et al., 2021).
Zygosaccharomyces bailii is a common food spoilage yeast capable of adapting and proliferating in the presence of remarkably high concentrations of acetic acid (Palma et al., 2018). During exposure to acetic acid, several genes involved in the modulation of plasma membrane composition and cell wall architecture were found to be differentially expressed (Antunes et al., 2018). Among those genes is the homologue of S. cerevisiae YGP1, encoding a cell wall-related glycoprotein, whose upregulation in the presence of acetic acid was shown to depend on ZbHaa1 (Palma et al., 2017; Antunes et al., 2018), showing a high degree of similarity between the responses involving the cell wall in both yeast species. In a K. marxianus strain isolated from agave, lyticase assays showed that the addition of KCl/KOH leads to the increase of cell wall robustness in cells grown in the presence of acetic acid (Castillo-Plata et al., 2021). However, further studies are necessary to elucidate the link between changes in cell wall during acetic acid adaptation and potassium homeostasis (Castillo-Plata et al., 2021). In I. orientalis, the expression of MPG1 gene, coding for a GDP-mannose pyrophosphorylase involved in cell wall synthesis, was also found to be upregulated under acetic acid stress (Li et al., 2021).
Lactic acid is another important weak acid in the food industry and also in the pharmaceutical and cosmetic industries. Its industrial production is currently carried out by lactic acid bacteria (Van Maris et al., 2004; Sauer et al., 2010) but bacteria are sensitive to low pH, requiring large amounts of neutralizing agents to counteract the acidification of the fermentation media, thus compromising the recovery yield of precipitated lactic acid (Altaf et al., 2007; Yen et al., 2010). Yeasts typically fare better than bacteria in acidic environments, which has motivated attempts to produce lactic acid through the heterologous expression of lactate dehydrogenases in yeast (Dequin and Barre, 1994; Porro et al., 1999; Pacheco et al., 2012). The success of such strategy and its industrial application require knowledge of the mechanisms behind yeast tolerance to lactic acid stress. Exposure to lactic acid leads to a decrease in cell wall glucan content and, to a lesser extent, of the mannan content of S. cerevisiae cell wall (Berterame et al., 2016; Figure 3). Several genes involved in the synthesis of cell wall polysaccharides, cell wall remodeling, and synthesis of mannoproteins are transcriptionally responsive and/or are determinants of tolerance to lactic acid stress (Kawahata et al., 2006; Abbott et al., 2007; Suzuki et al., 2012). Specifically, genes required for glucan remodeling (EXG1, GAS2, SCW10), cross-linking between β-glucans and chitin (CRH1), chitin synthesis (CHS1), and genes encoding mannoproteins required for cell wall stability (HSP150, PIR3, SED1), are up-regulated in response to lactic acid stress (Kawahata et al., 2006; Figure 3). Genes encoding elements of the MAPK module of the CWI pathway (BCK1, SLT2), as well as genes involved in glucan synthesis (KRE1, KRE11) and remodeling (GAS1), have also been implicated in lactic acid tolerance (Kawahata et al., 2006; Suzuki et al., 2012).
The Haa1 transcription factor also plays an important role in tolerance to lactic acid and is involved in the control of the expression of CWPs (Abbott et al., 2008; Mira et al., 2010c; Sugiyama et al., 2014). As found for acetic acid stress (Kim et al., 2019), exposure to lactic acid leads to Haa1 translocation from the cytoplasm to the nucleus where gene transcription, in particular of YGP1 and SPI1 occurs (Sugiyama et al., 2014). The overexpression of these two genes, encoding CWP, likely confers a stronger protective effect against lactic acid-induced toxicity (Sugiyama et al., 2014).
Due to the high tolerance of Zygosaccharomyces parabailii to high lactic acid concentrations at low pH, this species was proposed as a promising novel host for lactic acid production (Ortiz-Merino et al., 2018). In Z. parabailii, several cell wall-related genes were found to be down-regulated in the presence of lactic acid (Ortiz-Merino et al., 2018). Under the stress conditions tested, during exponential growth in the presence of lactic acid, a slight decrease in glucans was reported in S. cerevisiae and Z. bailii, and a slight decrease in mannans in S. cerevisiae (Berterame et al., 2016; Kuanyshev et al., 2016).
Propionic acid is commonly used to preserve baked goods and dairy (Suhr and Nielsen, 2004). Transcriptomic and chemogenomic studies have hinted at a role for the cell wall in yeast adaptation to this weak acid (Fernandes et al., 2005; Mira et al., 2009). Specifically, CWP1 (encoding a GPI-CWP), BAG7 (β1,3-glucan synthesis), and KNH1 (β1,6-glucan synthesis) are required to resist propionic acid stress, all of which are regulated by the transcription factor Rim101 (Mira et al., 2009), suggesting that cell wall remodeling during adaptation to propionic acid stress may be dependent on RIM101 expression.
Sorbic and benzoic acids are two other weak acids used to preserve foods and beverages. In both cases, the S. cerevisiae cell wall has been implicated in stress tolerance (De Nobel et al., 2001; Simões et al., 2006). Common to the responses to sorbic and benzoic acids is the induction of SPI1, a GPI-CWP that likely leads to the decrease of cell wall porosity and, in turn, limits the access to plasma membrane, thus reducing membrane damage, intracellular acidification, and viability loss (De Nobel et al., 2001; Simões et al., 2006; Figure 3). As previously mentioned, SPI1 is also activated in response to other weak acids and ethanol stress (Ogawa et al., 2000; Simões et al., 2006; Wu et al., 2006).
Formic acid in an inhibitory weak acid present in lignocellulosic hydrolysates negatively impacting lignocellulosic-based biorefining (Cunha et al., 2019).
Under formic acid stress, S. cerevisiae cells exhibited a deformed shape, with collapsed cell wall edges, indicating that the cell wall was damaged, and an FTIR analysis suggested that chitin structure was altered (Zeng et al., 2022). S. cerevisiae genes involved in β1,3-glucan synthesis (FKS1, ELO2), β1,6-glucan synthesis (TRS65), chitin synthesis (CHS5), CWP mannosylation (PMT2), cell wall integrity (PUN1), and CWI pathway regulation (ROM2) are important determinants of formic acid tolerance while FKS3, encoding an FKS1-2 homolog, is upregulated in response to this acid stress (Henriques et al., 2017; Zeng et al., 2022; Figure 3). Moreover, the upregulation of EXG2 encoding a major exoglucanase, and PKC1, encoding a CWI pathway kinase, was reported in an industrial S. cerevisiae strain (S6), engineered to ferment xylose, when grown in a medium with glucose and xylose supplemented with formic acid (Li et al., 2020).
Some of the responses involving the modification cell wall metabolism and cell wall physicochemical properties in yeasts are shared by relevant industrial stresses (Figure 3). For example, the CWI pathway is implicated in oxidative-, osmotic-, heat and cold-, ethanol- and low and high pH- induced stresses (Kamada et al., 1995; Hohmann, 2002a; Claret et al., 2005; García-Rodríguez et al., 2005; Mensonides et al., 2005; Vilella et al., 2005; Fujita et al., 2006; Serrano et al., 2006; Auesukaree et al., 2009; Teixeira et al., 2009; Córcoles-Sáez et al., 2012; De Lucena et al., 2012; Pillet et al., 2014; Kwon et al., 2016; Udom et al., 2019; Vaz et al., 2020). Also, the coordinated regulation involving the CWI and the HOG pathways occurs during osmotic, heat, ethanol and low pH stresses (Kamada et al., 1995; Hohmann, 2002a; García-Rodríguez et al., 2005; Mensonides et al., 2005; De Lucena et al., 2012; Dunayevich et al., 2018; Udom et al., 2019; Vaz et al., 2020) and the Calcineurin pathway interacts with the CWI pathway during high and low pH stresses (Viladevall et al., 2004; De Lucena et al., 2012). The reported upregulation of genes encoding CWPs was also observed under several stress induced conditions (e.g., under ethanol stress in S. cerevisiae and I. orientalis and under acetic acid stress in S. cerevisiae and Z. bailii (Ogawa et al., 2000; Rossignol et al., 2003; Simões et al., 2006; Wu et al., 2006; Sakihama et al., 2015; Palma et al., 2017; Antunes et al., 2018; Miao et al., 2018; Udom et al., 2019) and under oxidative, high and low temperature, low pH, lactic, sorbic and benzoic acids-induced stresses in S. cerevisiae) (Abramova et al., 2001; Causton et al., 2001; De Nobel et al., 2001; Sahara et al., 2002; Homma et al., 2003; Schade et al., 2004; Murata et al., 2006; Simões et al., 2006; Abe, 2007; Sha et al., 2013; Sugiyama et al., 2014; Varol et al., 2018). This common response is consistent with the important role of CWP in decreasing cell surface porosity and increasing cell wall stability when coping with stress. The upregulation of genes involved in β1,3-glucan synthesis was also reported to be shared by several stresses (heat, ethanol, low and high pH and formic acid stress) and during hyper-osmotic stress in Z. rouxii (Zhao et al., 1998; Viladevall et al., 2004; De Melo et al., 2010; De Lucena et al., 2012; Udom et al., 2019; Guo et al., 2020; Samakkarn et al., 2021; Zeng et al., 2022). Genes involved in β1,3-glucan elongation and branching were also found to be upregulated in response to low pH, acetic and lactic acids-induced stresses and under ethanol stress in I. orientalis (Kawahata et al., 2006; De Melo et al., 2010; De Lucena et al., 2012; Miao et al., 2018; Ribeiro et al., 2021). Genes involved in chitin synthesis found to be upregulated were also found to be shared by the response to low pH and lactic acid stress in S. cerevisiae, and to hyper-osmotic stress in Z. rouxii (Kawahata et al., 2006; De Melo et al., 2010; De Lucena et al., 2012; Guo et al., 2020). Genes involved in β-glucan-chitin crosslinking were also found to be upregulated under ethanol and lactic acid stresses in S. cerevisiae, and under hyper-osmotic stress in Z. rouxii (Kawahata et al., 2006; Udom et al., 2019; Guo et al., 2020). The upregulation of genes involved in β1,6-glucan synthesis are shared by oxidative and low pH stress in S. cerevisiae, and by hyper-osmotic stress in Z. rouxii and Z. mellis (De Melo et al., 2010; De Lucena et al., 2012; Sha et al., 2013; Guo et al., 2020; Liu et al., 2021). Genes involved in CWI pathway were found to be upregulated in oxidative-, heat- and low pH-induced stress in S. cerevisiae, under hyper-osmotic stress in Z. mellis, and under methanol stress in K. phaffii (Claret et al., 2005; De Lucena et al., 2012; Sha et al., 2013; Varol et al., 2018; Zhang et al., 2020; Liu et al., 2021). Genes involved in CWP mannosylation were found to be upregulated in S. cerevisiae under low pH stress and in D. hansenii under hyper-osmotic stress (Thomé, 2007; Gonzalez et al., 2009b; De Melo et al., 2010; De Lucena et al., 2012). Genes involved in β1,3-glucan hydrolysis were found to be upregulated under lactic acid stress in S. cerevisiae (Kawahata et al., 2006).
Of all the industrially relevant stresses herein described, exposure to several stresses were found to decrease, at least, one type of cell wall polysaccharide in S. cerevisiae. Specifically, a decrease in the β-glucan content was reported for heat- (β1,3-glucans), high pH- (β1,6-glucans), lactic acid- (β1,3 and β1,6-glucans) induced stresses, and a decrease in mannans was reported during lactic acid stress (Aguilar-Uscanga and François, 2003; Schiavone et al., 2014; Berterame et al., 2016). A slight decrease in the β-glucan content (β1,3 and β1,6-glucans) was also reported for Z. bailii during lactic acid stress (Kuanyshev et al., 2016). A decrease in the chitin content was reported for inorganic acid-induced low pH in S. cerevisiae (Cabib et al., 1989). Nevertheless, an increase of cell wall polysaccharide content was also reported, in particular an increase in chitin and β1,6-glucans under heat stress and an increase in the β-glucans content under acetic acid stress in S. cerevisiae (Schiavone et al., 2014; Ribeiro et al., 2021). It is noteworthy to take in consideration that the methods used for cell wall polysaccharides quantification were not the same in different articles and both the levels of stress and the adaptation phase of the cells examined (early response, cells adapted to the stress) could be different or not clearly reported.
A few successful examples of the alteration of the physicochemical properties of the cell wall either by the genetic engineering of the yeast cell or by the adaptive laboratory evolution (ALE) of yeast cells leading to the increase of tolerance to stress(es) have been reported in the literature.
As referred above, through chemogenomic analyses, it was demonstrated that the expression of genes related with chitin and glucan synthesis, namely CHS1 (chitin), FKS1 and ACF2 (β1,3-glucans), KRE6 (β1,6-glucans), and others involved in cell wall integrity (PUN1) and cell wall organization (SIT4), are required for maximum tolerance to ethanol in S. cerevisiae (Kubota et al., 2004; Auesukaree et al., 2009; Teixeira et al., 2009; Mota et al., 2021). Superior fermentation performance of lignocellulosic hydrolysates was reported for a recombinant S. cerevisiae WXY70 strain overexpressing the CCW12 gene, encoding a cell wall mannoprotein, compared to the control strain, and CCW12 expression was found to improve cell wall stability and tolerance to the growth inhibitors present (Kong et al., 2021). The deletion of GAL6, encoding a cysteine aminopeptidase with homocysteine-thiolactonase activity, was found to lead to improved growth and higher viability of S. cerevisiae, in the presence of ethanol stress (Yazawa et al., 2007). The gal6Δ cells showed increased resistance to zymolyase activity indicating the occurrence of structural changes in the cell wall (Yazawa et al., 2007). A marked increased tolerance to ethanol stress associated with transcription rewiring involving cell wall synthesis was also identified in the highly–ethanol tolerant strain K. marxianus FIM1 obtained by ALE in an ethanol supplemented medium (Mo et al., 2019). Among the identified changes at the transcriptional level were the alterations related with the upregulation of cell wall metabolism involving, in particular, chitin synthesis (CHS3), β1,6-glucan synthesis (KNH1), glucanosyltransferases activity involved in β1,3-glucan branching and elongation (GAS4) and the cell wall integrity pathway (BCK1, MID2, WSC3) (Mo et al., 2019).
Numerous genes encoding proteins required for cell wall polysaccharides synthesis (FKS1, ROM2, KRE6, CHS1, CHS5), cell wall remodeling (GAS1) and protein mannosylation (MNN2, MNN9, MNN11, KTR4) were found to be determinants of acetic acid stress tolerance by chemogenomic analyses (Kawahata et al., 2006; Mira et al., 2010b). The increased expression of the HAA1 gene, encoding a transcription factor and a major determinant of acetic acid tolerance in S. cerevisiae, led to the improvement of cell wall robustness under acetic acid stress, as suggested by the decreased susceptibility of the cell wall to lyticase activity mediated disruption (Cunha et al., 2018). Through Haa1 amino acid sequence engineering, a single amino acid exchange at position 135 (serine to phenylalanine) was found to lead to the upregulation of genes of the Haa1-regulon, increasing acetic acid tolerance, in particular YGP1 (Swinnen et al., 2017). The deletion of ATG22, encoding a vacuolar membrane protein that mediates the efflux of amino acids resulting from autophagic protein degradation, was found to delay programmed cell death (PDC) induced by acetic acid. The deletion of ATG22 contributes to the maintenance of cell wall integrity, by preventing the decrease in total cell wall polysaccharides induced by PDC caused by severe acetic acid stress, increasing the transcript levels of CWI pathway genes (Yang et al., 2006; Hu et al., 2019). This suggested ATG22 as a potential target for genetic engineering strategies to improve yeast cell wall robustness and tolerance to acetic acid and other industrial stresses. Enhanced tolerance to both acetic and formic acids at low pH (pH 2.4 or below) by expressing the GAS1 gene of Issatchenkia orientalis in S. cerevisiae was reported (Wada et al., 2020). Also, the overexpression of GAS1 in S. cerevisiae led to increased lactic acid productivity at low pH (Zhong et al., 2021), reinforcing the importance of GAS1, encoding a beta-1,3-glucanosyltransferase, in this context (Baek et al., 2017).
As previously referred, chemogenomic studies demonstrated that genes related with cell wall polysaccharide synthesis (FKS1, KRE6) and cell wall mannosylation (MNN10, ANP1) are determinants of tolerance to osmotic stress (high glucose concentrations) in S. cerevisiae (Ando et al., 2006; Teixeira et al., 2010). In Pichia pastoris (now Komagataella phaffii), the deletion of YPS7, encoding a putative GPI-linked aspartyl protease, led to increased osmotic tolerance and this gene was proposed as a promising molecular target for the engineering of yeast robustness (Guan et al., 2012). This species is used to produce heterologous proteins in the pharmaceutical and food industry.
In a laboratory adaptively evolved K. marxianus strain exhibiting a significantly improved tolerance to high temperatures, a single nucleotide polymorphism was found in the coding region of the exoglucanase gene EXGI, required for cell wall remodeling (Ai et al., 2022).
The important role played by the cell wall in the adaptation and tolerance of yeasts to different stresses of biotechnological interest emerges from this review article. Although several evidences support this idea, the truth is that these evidences largely come from genome-wide analyses of the response of yeasts to various stresses. Furthermore, the experimental conditions and levels of stress used are in general different and only part of the studies involves a time-course analysis covering the various stages of adaptation and growth under stress. Therefore, it is likely that the apparent divergences reported are due to this fact.
This review article also provides information on how the different signaling pathways coordinate to elicit changes at the level of the yeast cell wall in response to different relevant stresses. However, it is important to highlight that, in industrial bioprocesses, several stresses are often present simultaneously, which further complicates the goal of providing a comprehensive description of what happens during adaptation to adverse process conditions. It is noteworthy to take in consideration that yeast cells exposed to mild stress develop tolerance not only to higher doses of the same stress, but also to stress caused by other agents. This phenomenon, known as cross-protection, is based on the existence of integrating mechanisms that senses and responds to different forms of stress (Estruch, 2000). Concerning the cell wall, the physiological observations reported and the integrated molecular responses here described provide the basis for the involvement of this dynamic organelle in cross-stress protection.
Most of the review paper is dedicated to the model yeast and cell factory Saccharomyces cerevisiae. However, the scarce available information in the literature concerning non-conventional yeast species of biotechnological relevance is also mentioned throughout the text. A deeper understanding of the nature of the molecular response and the changes occurring at the cell envelope level would be valuable and allow the development of more rational strategies to construct superior yeasts for biotechnology and to control the activity of food spoiling yeasts. Genome-wide analyses are contributing to identify a wealth of cell wall-related promising targets for the improvement of yeast tolerance. However, more in depth molecular and cellular studies are instrumental to better understand the somewhat overlooked role of the cell wall in tolerance to multiple stresses in yeasts.
IS-C, RR, and NB-M conceived and designed the study. RR and NB-M performed the literature search, prepared the figures, and wrote the first manuscript draft. IS-C and RR completed and revised the manuscript drafts. All authors approved the final manuscript.
Work in the laboratory of IS-C was supported by ‘Fundação para a Ciência e a Tecnologia’ (FCT) projects: PTDC/BBB-BEP/0385/2014, ERA-IB-2/0003/2015, PTDC/FIS-NAN/6101/2014; and Ph.D. fellowships to RR (FCT-Ph.D. Programme AEM—Applied and Environmental Microbiology; PD/BD/135204/2017) and NB-M (FCT Ph.D. Programme BIOTECnico-Biotechnology and Biosciences; PD/BD/150339/2019). Funding received from FCT by iBB—Institute for Bioengineering and Biosciences (UIDB/04565/2020 and UIDP/04565/2020) and by i4HB (LA/P/0140/2020), is also acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbott, D. A., Knijnenburg, T. A., De Poorter, L. M. I., Reinders, M. J. T., Pronk, J. T., and Van Maris, A. J. A. (2007). Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae. FEMS Yeast Res. 7, 819–833. doi: 10.1111/j.1567-1364.2007.00242.x
Abbott, D., Suir, E., Duong, G., de Hulster, E., Pronk, J., and van Maris, A. (2009). Catalase overexpression reduces lactic acid-induced oxidative stress in Saccharomyces cerevisiae. Appl. Env. Microbiol. 75, 2320–2325. doi: 10.1128/AEM.00009-09
Abbott, D., Suir, E., van Maris, A., and Pronk, J. (2008). Physiological and transcriptional responses to high concentrations of lactic acid in anaerobic chemostat cultures of Saccharomyces cerevisiae. Appl. Env. Microbiol. 74, 5759–5768. doi: 10.1128/AEM.01030-08
Abe, F. (2007). Induction of DAN/TIR yeast cell wall mannoprotein genes in response to high hydrostatic pressure and low temperature. FEBS Lett. 581, 4993–4998. doi: 10.1016/j.febslet.2007.09.039
Abramova, N., Sertil, O., Mehta, S., and Lowry, C. V. (2001). Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol. 183, 2881–2887. doi: 10.1128/JB.183.9.2881-2887.2001
Adya, A. K., Canetta, E., and Walker, G. M. (2006). Atomic force microscopic study of the infuence of physical stresses on Saccharomyces cerevisiae and Schizosaccharomyces pombe. FEMS Yeast Res. 6, 120–128. doi: 10.1111/j.1567-1364.2005.00003.x
Aguilar-Uscanga, B., and François, J. M. (2003). A study of the yeast cell wall composition and structure in response to growth conditions and mode of cultivation. Lett. Appl. Microbiol. 37, 268–274. doi: 10.1046/j.1472-765X.2003.01394.x
Ai, Y., Luo, T., Yu, Y., Zhou, J., and Lu, H. (2022). Downregulation of ammonium uptake improves the growth and tolerance of Kluyveromyces marxianus at high temperature. Microbiol. Open 11, 1–13. doi: 10.1002/mbo3.1290
Aimanianda, V., Simenel, C., Garnaud, C., Clavaud, C., Tada, R., Barbin, L., et al. (2017). The dual activity responsible for the elongation and branching of β-(1,3)- glucan in the fungal cell wall. MBio 8, e619. doi: 10.1128/mBio.00619-17
Alloush, H. M., Edwards, T. A., Valle-Lisboa, V., and Wheals, A. E. (2002). Cdc4 is involved in the transcriptional control of OCH1, a gene encoding α-1,6-mannosyltransferase in Saccharomyces cerevisiae. Yeast 19, 69–77. doi: 10.1002/yea.801
Almeida, B., Ohlmeier, S., Almeida, A. J., Madeo, F., Leão, C., Rodrigues, F., et al. (2009). Yeast protein expression profile during acetic acid-induced apoptosis indicates causal involvement of the TOR pathway. Proteomics 9, 720–732. doi: 10.1002/pmic.200700816
Altaf, M., Venkateshwar, M., Srijana, M., and Reddy, G. (2007). An economic approach for L-(+) lactic acid fermentation by Lactobacillus amylophilus GV6 using inexpensive carbon and nitrogen sources. J. Appl. Microbiol. 103, 372–380. doi: 10.1111/j.1365-2672.2006.03254.x
Alvim, M. C. T., Vital, C. E., Barros, E., Vieira, N. M., da Silveira, F. A., Balbino, T. R., et al. (2019). Ethanol stress responses of Kluyveromyces marxianus CCT 7735 revealed by proteomic and metabolomic analyses. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 112, 827–845. doi: 10.1007/s10482-018-01214-y
Ando, A., Tanaka, F., Murata, Y., Takagi, H., and Shima, J. (2006). Identification and classification of genes required for tolerance to high-sucrose stress revealed by genome-wide screening of Saccharomyces cerevisiae. FEMS Yeast Res. 6, 249–267. doi: 10.1111/j.1567-1364.2006.00035.x
Antunes, M., Palma, M., and Sá-correia, I. (2018). Transcriptional profiling of Zygosaccharomyces bailii early response to acetic acid or copper stress mediated by ZbHaa1. Sci. Rep. 2, 1–14. doi: 10.1038/s41598-018-32266-9
Arinbasarova, A. Y., Machulin, A. V., Biryukova, E. N., Sorokin, V. V., Medentsev, A. G., and Suzina, N. E. (2018). Structural changes in the cell envelope of Yarrowia lipolytica yeast under stress conditions. Can. J. Microbiol. 64, 359–365. doi: 10.1139/cjm-2018-0034
Arnesen, J. A., Belmonte Del Ama, A., Jayachandran, S., Dahlin, J., Rago, D., Andersen, A. J. C., et al. (2022). Engineering of Yarrowia lipolytica for the production of plant triterpenoids: asiatic, madecassic, and arjunolic acids. Metab. Eng. Commun. 14:e00197. doi: 10.1016/j.mec.2022.e00197
Assis, L., Manfiolli, A., Mattos, E., Fabri, J., Malavazi, I., Jacobsen, I., et al. (2018). Protein kinase a and high-osmolarity glycerol response pathways cooperatively control cell wall carbohydrate mobilization in Aspergillus fumigatus. Mol. Biol. Physiol. 9, 1–15. doi: 10.1128/mBio.01952-18
Attfield, P. V. (1987). Trehalose accumulates in Saccharomyces cerevisiae during exposure to agents that induce heat shock response. FEBS Lett. 225, 259–263. doi: 10.1016/0014-5793(87)81170-5
Attfield, P. V., and Kletsas, S. (2000). Hyperosmotic stress response by strains of bakers’ yeasts in high sugar concentration medium. Lett. Appl. Microbiol. 31, 323–327. doi: 10.1046/j.1472-765X.2000.00825.x
Auesukaree, C. (2017). Molecular mechanisms of the yeast adaptive response and tolerance to stresses encountered during ethanol fermentation. J. Biosci. Bioeng. 124, 133–142. doi: 10.1016/j.jbiosc.2017.03.009
Auesukaree, C., Damnernsawad, A., Kruatrachue, M., Pokethitiyook, P., Boonchird, C., Kaneko, Y., et al. (2009). Genome-wide identification of genes involved in tolerance to various environmental stresses in Saccharomyces cerevisiae. J. Appl. Genet. 50, 301–310. doi: 10.1007/BF03195688
Baek, S.-H., Kwon, E. Y., Bae, S.-J., Cho, B.-R., Kim, S.-Y., and Hahn, J.-S. (2017). Improvement of d-lactic acid production in saccharomyces cerevisiae under acidic conditions by evolutionary and rational metabolic engineering. Biotechnol. J. 12:1700015. doi: 10.1002/biot.201700015
Baetz, K., Moffat, J., Haynes, J., Chang, M., and Andrews, B. (2001). Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol. Cell. Biol. 21, 6515–6528. doi: 10.1128/MCB.21.19.6515-6528.2001
Bajwa, P. K., Ho, C. Y., Chan, C. K., Martin, V. J. J., Trevors, J. T., and Lee, H. (2013). Transcriptional profiling of Saccharomyces cerevisiae T2 cells upon exposure to hardwood spent sulphite liquor: comparison to acetic acid, furfural and hydroxymethylfurfural. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 103, 1281–1295. doi: 10.1007/s10482-013-9909-1
Bauer, F. F., and Pretorius, I. S. (2000). Yeast stress response and fermentation efficiency: how to survive the making of wine - a review. South African J. Enol. Vitic. 21, 27–51. doi: 10.21548/21-1-3557
Berterame, N. M., Porro, D., Ami, D., and Branduardi, P. (2016). Protein aggregation and membrane lipid modifications under lactic acid stress in wild type and OPI1 deleted Saccharomyces cerevisiae strains. Microb. Cell Fact. 15, 1–12. doi: 10.1186/s12934-016-0438-2
Blanco, N., Sanz, A. B., Rodríguez-Peña, J. M., Nombela, C., Farkaš, V., Hurtado-Guerrero, R., et al. (2015). Structural and functional analysis of yeast Crh1 and Crh2 transglycosylases. FEBS J. 282, 715–731. doi: 10.1111/febs.13176
Booth, I. R., and Stratford, M. (2003). “Acidulants and low pH,” in Food Preservatives, eds N. J. Russell and G. W. Gould (Boston, MA: Springer), 25–47. doi: 10.1007/978-0-387-30042-9_3
Borodina, I., and Nielsen, J. (2014). Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals. Biotechnol. J. 9, 609–620. doi: 10.1002/biot.201300445
Bourbon-Melo, N., Palma, M., Rocha, M. P., Ferreira, A., Bronze, M. R., Elias, H., et al. (2021). Use of Hanseniaspora guilliermondii and Hanseniaspora opuntiae to enhance the aromatic profile of beer in mixed-culture fermentation with Saccharomyces cerevisiae. Food Microbiol. 95:103678. doi: 10.1016/j.fm.2020.103678
Brosnan, M. P., Donnelly, D., James, T. C., and Bond, U. (2000). The stress response is repressed during fermentation in brewery strains of yeast. J. Appl. Microbiol. 88, 746–755. doi: 10.1046/j.1365-2672.2000.01006.x
Bubnová, M., Zemanciková, J., and Sychrová, H. (2014). Osmotolerant yeast species differ in basic physiological parameters and in tolerance of non-osmotic stresses. Yeast 31, 309–321.
Buzzini, P., Branda, E., Goretti, M., and Turchetti, B. (2012). Psychrophilic yeasts from worldwide glacial habitats: diversity, adaptation strategies and biotechnological potential. FEMS Microbiol. Ecol. 82, 217–241. doi: 10.1111/j.1574-6941.2012.01348.x
Cabib, E. (2009). Two novel techniques for determination of polysaccharide cross-links show that Crh1p and Crh2p attach chitin to both β(1-6)- and β(1-3)glucan in the Saccharomyces cerevisiae cell wall. Eukaryot. Cell 8, 1626–1636. doi: 10.1128/EC.00228-09
Cabib, E., Blanco, N., Grau, C., Rodríguez-Peña, J. M., and Arroyo, J. (2007). Crh1p and Crh2p are required for the cross-linking of chitin to beta(1-6)glucan in the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 63, 921–935. doi: 10.1111/j.1365-2958.2006.05565.x
Cabib, E., Farkas, V., Kosík, O., Blanco, N., Arroyo, J., and McPhie, P. (2008). Assembly of the yeast cell wall. Crh1p and Crh2p act as transglycosylases in vivo and in vitro. J. Biol. Chem. 283, 29859–29872. doi: 10.1074/jbc.M804274200
Cabib, E., Silverman, S. J., and Shaw, J. A. (1992). Chitinase and chitin synthase 1: counterbalancing activities in cell separation of Saccharomyces cerevisiae. J. Gen. Microbiol. 138, 97–102. doi: 10.1099/00221287-138-1-97
Cabib, E., Sburlati, A., Bowers, B., and Silverman, S. J. (1989). Chitin synthase 1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 108, 1665–1672. doi: 10.1083/jcb.108.5.1665
Carmelo, V., Bogaerts, P., and Sá-Correia, I. (1996). Activity of plasma membrane H+-ATPase and expression of PMA1 and PMA2 genes in Saccharomyces cerevisiae cells grown at optimal and low pH. Arch. Microbiol. 166, 315–320. doi: 10.1007/s002030050389
Carmelo, V., Santos, H., and Sá-Correia, I. (1997). Effect of extracellular acidification on the activity of plasma membrane ATPase and on the cytosolic and vacuolar pH of Saccharomyces cerevisiae. Biochim. Biophys. Acta - Biomembr. 1, 63–70. doi: 10.1016/S0005-2736(96)00245-3
Casal, M., Cardoso, H., and Leão, C. (1996). Mechanisms regulating the transport of acetic acid in Saccharomyces cerevisiae. Microbiology 142, 1385–1390. doi: 10.1099/13500872-142-6-1385
Castillo-Plata, A. K., Sigala, J. C., Lappe-Oliveras, P., and Le Borgne, S. (2021). KCl/KOH supplementation improves acetic acid tolerance and ethanol production in a thermotolerant strain of Kluyveromyces marxianus isolated from henequen (agave fourcroydes). Rev. Mex. Ing. Química 21:Bio2567. doi: 10.24275/rmiq/Bio2567
Causton, H. C., Ren, B., Koh, S. S., Harbison, C. T., Kanin, E., Jennings, E. G., et al. (2001). Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12, 323–337. doi: 10.1091/mbc.12.2.323
Charoenbhakdi, S., Dokpikul, T., Burphan, T., Techo, T., and Auesukaree, C. (2016). Vacuolar H+-ATPase protects Saccharomyces cerevisiae cells against ethanolinduced oxidative and cell wall stresses. Appl. Environ. Microbiol. 82, 3121–3130. doi: 10.1128/AEM.00376-16
Chen, R. E., and Thorner, J. (2007). Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta Mol. Cell Res. 1773, 1311–1340. doi: 10.1016/j.bbamcr.2007.05.003
Claret, S., Gatti, X., Doignon, F., Thoraval, D., and Crouzet, M. (2005). The rgd1p rho GTPase-activating protein and the Mid2p cell wall sensor are required at low pH for protein kinase C pathway activation and cell survival in Saccharomyces cerevisiae. Eukaryot. Cell 4, 1375–1386. doi: 10.1128/EC.4.8.1375-1386.2005
Claro, F. B., Rijsbrack, K., and Soares, E. V. (2007). Flocculation onset in Saccharomyces cerevisiae: effect of ethanol, heat and osmotic stress. J. Appl. Microbiol. 102, 693–700. doi: 10.1111/j.1365-2672.2006.03130.x
Coradini, A. L. V., da Silveira Bezerra de Mello, F., Furlan, M., Maneira, C., Carazzolle, M. F., Pereira, G. A. G., et al. (2021). QTL mapping of a brazilian bioethanol strain links the cell wall protein-encoding gene GAS1 to low pH tolerance in S. cerevisiae. Biotechnol. Biofuels 14:239. doi: 10.1186/s13068-021-02079-6
Córcoles-Sáez, I., Ballester-Tomas, L., Torre-Ruiz, M. A., Prieto, J. A., and Randez-Gil, F. (2012). Low temperature highlights the functional role of the cell wall integrity pathway in the regulation of growth in Saccharomyces cerevisiae. Biochem. J. 446, 477–488. doi: 10.1042/BJ20120634
Costa, V., and Moradas-Ferreira, P. (2001). Oxidative stress and signal transduction in Saccharomyces cerevisiae: insights into ageing, apoptosis and diseases. Mol. Aspects Med. 22, 217–246. doi: 10.1016/S0098-2997(01)00012-7
Cunha, J. T., Costa, C. E., Ferraz, L., Romaní, A., Johansson, B., Sá-Correia, I., et al. (2018). HAA1 and PRS3 overexpression boosts yeast tolerance towards acetic acid improving xylose or glucose consumption: unravelling the underlying mechanisms. Appl. Microbiol. Biotechnol. 102, 4589–4600. doi: 10.1007/s00253-018-8955-z
Cunha, J. T., Romaní, A., Costa, C. E., Sá-Correia, I., and Domingues, L. (2019). Molecular and physiological basis of Saccharomyces cerevisiae tolerance to adverse lignocellulose-based process conditions. Appl. Microbiol. Biotechnol. 103, 159–175. doi: 10.1007/s00253-018-9478-3
da Silveira, F. A., de Oliveira Soares, D. L., Bang, K. W., Balbino, T. R., de Moura Ferreira, M. A., Diniz, R. H. S., et al. (2020). Assessment of ethanol tolerance of Kluyveromyces marxianus CCT 7735 selected by adaptive laboratory evolution. Appl. Microbiol. Biotechnol. 104, 7483–7494. doi: 10.1007/s00253-020-10768-9
Dague, E., Bitar, R., Ranchon, H., Durand, F., and François, J. M. (2010). An atomic force microscopy analysis of yeast mutants defective in cell wall architecture. Yeast 27, 673–684. doi: 10.1002/yea.1801
Dallies, N., François, J., and Paquet, V. (1998). A new method for quantitative determination of polysaccharides in the yeast cell wall. application to the cell wall defective mutants of Saccharomyces cerevisiae. Yeast 14, 1297–1306. doi: 10.1002/(SICI)1097-0061(1998100)14.0.CO;2-L
Davenport, K. R., Sohaskey, M., Kamada, Y., Levin, D. E., and Gustin, M. C. (1995). A second osmosensing signal transduction pathway in yeast. hypotonic shock activates the PKC1 protein kinase-regulated cell integrity pathway. J. Biol. Chem. 270, 30157–30161. doi: 10.1074/jbc.270.50.30157
De Groot, P. W. J., Kraneveld, E. A., Qing, Y. Y., Dekker, H. L., Groß, U., Crielaard, W., et al. (2008). The cell wall of the human pathogen candida glabrata: differential incorporation of novel adhesin-like wall proteins. Eukaryot. Cell 7, 1951–1964. doi: 10.1128/EC.00284-08
De Groot, P. W. J., Ram, A. F., and Klis, F. M. (2005). Features and functions of covalently linked proteins in fungal cell walls. Fungal. Genet. Biol. 42, 657–675. doi: 10.1016/j.fgb.2005.04.002
De Groot, P. W. J., Ruiz, C., de Aldana, C. R. V., Duenas, E., Cid, V. J., Del Rey, F., et al. (2001). A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae. Comput. Funct. Geno. 2, 124–142. doi: 10.1002/cfg.85
De Lucena, R. M., Elsztein, C., de Barros Pita, W., de Souza, R. B., de Sá Leitão Paiva Júnior, S., and de Morais Junior, M. A. (2015). Transcriptomic response of Saccharomyces cerevisiae for its adaptation to sulphuric acid-induced stress. Antonie Van Leeuwenhoek 108, 1147–1160. doi: 10.1007/s10482-015-0568-2
De Lucena, R. M., Elsztein, C., Simões, D. A., and De Morais, M. A. (2012). Participation of CWI, HOG and Calcineurin pathways in the tolerance of Saccharomyces cerevisiae to low pH by inorganic acid. J. Appl. Microbiol. 113, 629–640. doi: 10.1111/j.1365-2672.2012.05362.x
De Melo, H. F., Bonini, B. M., Thevelein, J., Simões, D. A., and Morais, M. A. (2010). Physiological and molecular analysis of the stress response of Saccharomyces cerevisiae imposed by strong inorganic acid with implication to industrial fermentations. J. Appl. Microbiol. 109, 116–127. doi: 10.1111/j.1365-2672.2009.04633.x
De Nobel, H., Lawrie, L., Brul, S., Klis, F., Davis, M., Alloush, H., et al. (2001). Parallel and comparative analysis of the proteome and transcriptome of sorbic acid-stressed Saccharomyces cerevisiae. Yeast 18, 1413–1428. doi: 10.1002/yea.793
De Nobel, J. G., Klis, F. M., Priem, J., Munnik, T., and Van Den Ende, H. (1990). The glucanase-soluble mannoproteins limit cell wall porosity in Saccharomyces cerevisiae. Yeast 6, 491–499. doi: 10.1002/yea.320060606
De Souza Pereira, R., and Geibel, J. (1999). Direct observation of oxidative stress on the cell wall of Saccharomyces cerevisiae strains with atomic force microscopy. Mol. Cell. Biochem. 201, 17–24. doi: 10.1023/a:1007007704657
Deparis, Q., Claes, A., Foulquié-Moreno, M. R., and Thevelein, J. M. (2017). Engineering tolerance to industrially relevant stress factors in yeast cell factories. FEMS Yeast Res. 17, 1–17. doi: 10.1093/femsyr/fox036
Dequin, S., and Barre, P. (1994). Mixed lactic acid–alcoholic fermentation by Saccharomyes cerevisiae expressing the Lactobacillus casei L(+)–LDH. Nat. Biotechnol. 12, 173–177. doi: 10.1038/nbt0294-173
Ding, J., Huang, X., Zhang, L., Zhao, N., Yang, D., and Zhang, K. (2009). Tolerance and stress response to ethanol in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 85, 253–263. doi: 10.1007/s00253-009-2223-1
Diniz, R. H. S., Villada, J. C., Alvim, M. C. T., Vidigal, P. M. P., Vieira, N. M., Lamas-Maceiras, M., et al. (2017). Transcriptome analysis of the thermotolerant yeast Kluyveromyces marxianus CCT 7735 under ethanol stress. Appl. Microbiol. Biotechnol. 101, 6969–6980. doi: 10.1007/s00253-017-8432-0
Donlin, M. J., Upadhya, R., Gerik, K. J., Lam, W., Vanarendonk, L. G., Specht, C. A., et al. (2014). Cross talk between the cell wall integrity and cyclic AMP/protein kinase a pathways in Cryptococcus neoformans. MBio 5, 1–10. doi: 10.1128/mBio.01573-14
Dunayevich, P., Baltanás, R., Clemente, J. A., Couto, A., Sapochnik, D., Vasen, G., et al. (2018). Heat-stress triggers MAPK crosstalk to turn on the hyperosmotic response pathway. Sci. Rep. 8:33203. doi: 10.1038/s41598-018-33203-6
Dupres, V., Dufrene, Y., and Heinisch, J. (2010). Measuring cell wall thickness in living yeast cells using single molecular rulers. ACS Nano 4, 5498–5504. doi: 10.1021/nn101598v
Eardley, J., and Timson, D. J. (2020). Yeast cellular stress: impacts on bioethanol production. Fermentation 6:109. doi: 10.3390/fermentation6040109
Ene, I. V., Walker, L. A., Schiavone, M., Lee, K. K., Martin-Yken, H., Dague, E., et al. (2015). Cell wall remodeling enzymes modulate fungal cell wall elasticity and osmotic stress resistance. MBio 6:e00986. doi: 10.1128/mBio.00986-15
Estruch, F. (2000). Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24, 469–486. doi: 10.1016/S0168-6445(00)00035-8
Fathi, Z., Tramontin, L. R. R., Ebrahimipour, G., Borodina, I., and Darvishi, F. (2021). Metabolic engineering of Saccharomyces cerevisiae for production of β-carotene from hydrophobic substrates. FEMS Yeast Res. 21:foaa068. doi: 10.1093/femsyr/foaa068
Fernandes, A. R., Mira, N. P., Vargas, R. C., Canelhas, I., and Sá-Correia, I. (2005). Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem. Biophys. Res. Commun. 337, 95–103. doi: 10.1016/j.bbrc.2005.09.010
Fleet, G. H. (1991). “Cell walls,” in The Yeasts, Vol. 4, ed. H. J. S. Rose (London: Academic Press), 199–277. doi: 10.1002/jobm.19720120411
Fleet, G. H. (2011). Yeast spoilage of foods and beverages. Yeasts 2011, 53–63. doi: 10.1016/B978-0-444-52149-1.00005-7
Free, S. J. (2013). “Chapter two - fungal cell wall organization and biosynthesis,” in Advances in Genetics, eds T. Friedmann and J. C. Dunlap (Academic Press), 33–82. doi: 10.1016/B978-0-12-407677-8.00002-6
Fu, X., Li, P., Zhang, L., and Li, S. (2019). Understanding the stress responses of Kluyveromyces marxianus after an arrest during high-temperature ethanol fermentation based on integration of RNA-Seq and metabolite data. Appl. Microbiol. Biotechnol. 103, 2715–2729. doi: 10.1007/s00253-019-09637-x
Fuchs, B. B., and Mylonakis, E. (2009). Our paths might cross: the role of the fungal cell wall integrity pathway in stress response and cross talk with other stress response pathways. Eukaryot. Cell 8, 1616–1625. doi: 10.1128/EC.00193-09
Fujita, K., Matsuyama, A., Kobayashi, Y., and Iwahashi, H. (2006). The genome-wide screening of yeast deletion mutants to identify the genes required for tolerance to ethanol and other alcohols. FEMS Yeast Res. 6, 744–750. doi: 10.1111/j.1567-1364.2006.00040.x
García-Rodríguez, L. J., Valle, R., Durán, Á, and Roncero, C. (2005). Cell integrity signaling activation in response to hyperosmotic shock in yeast. FEBS Lett. 579, 6186–6190. doi: 10.1016/j.febslet.2005.10.001
Garcia-Rubio, R., de Oliveira, H. C., Rivera, J., and Trevijano-Contador, N. (2020). The fungal cell wall: candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 10:1–13. doi: 10.3389/fmicb.2019.02993
García, R., Bravo, E., Diez-Muñiz, S., Nombela, C., Rodríguez-Peña, J. M., and Arroyo, J. (2017). A novel connection between the cell wall integrity and the PKA pathways regulates cell wall stress response in yeast. Sci. Rep. 7, 1–15. doi: 10.1038/s41598-017-06001-9
Garrett-Engele, P., Moilanen, B., and Cyert, M. S. (1995). Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H(+)-ATPase. Mol. Cell. Biol. 15, 4103–4114. doi: 10.1128/MCB.15.8.4103
Gibney, P. A., Schieler, A., Chen, J. C., Rabinowitz, J. D., and Botstein, D. (2015). Characterizing the in vivo role of trehalose in Saccharomyces cerevisiae using the AGT1 transporter. Proc. Natl. Acad. Sci. U.S.A. 112, 6116–6121. doi: 10.1073/pnas.1506289112
Gibson, B. R., Lawrence, S. J., Leclaire, J. P. R., Powell, C. D., and Smart, K. A. (2007). Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol. Rev. 31, 535–569. doi: 10.1111/j.1574-6976.2007.00076.x
Godinho, C. P., Costa, R., and Sá-Correia, I. (2021). The ABC transporter Pdr18 is required for yeast thermotolerance due to its role in ergosterol transport and plasma membrane properties. Environ. Microbiol. 23, 69–80. doi: 10.1111/1462-2920.15253
Gonzalez, M., Lipke, P. N., and Ovalle, R. B. (2009a). “Chapter 15 - GPI proteins in biogenesis and structure of yeast cell walls,” in Glycosylphosphatidylinositol (GPI) Anchoring of Proteins, eds A. K. Menon, T. Kinoshita, P. Orlean, and F. Tamanoi (New York, NY: Academic Press), 321–356. doi: 10.1016/S1874-6047(09)26015-X
Gonzalez, M, Vásquez, A., Zuazaga, H., Sen, A., Olvera, H., Ortiz, S., et al. (2009b). Genome-wide expression profiling of the osmoadaptation response of Debaryomyces hansenii. Yeast 26, 111–124.
Gonzalez, R., Morales, P., Tronchoni, J., Cordero-Bueso, G., Vaudano, E., Quirós, M., et al. (2016). New genes involved in osmotic stress tolerance in Saccharomyces cerevisiae. Front. Microbiol. 7:1–12. doi: 10.3389/fmicb.2016.01545
Gschaedler, A. (2017). Contribution of non-conventional yeasts in alcoholic beverages. Curr. Opin. Food Sci. 13, 73–77. doi: 10.1016/j.cofs.2017.02.004
Guan, B., Lei, J., Su, S., Chen, F., Duan, Z., Chen, Y., et al. (2012). Absence of Yps7p, a putative glycosylphosphatidylinositol-linked aspartyl protease in pichia pastoris, results in aberrant cell wall composition and increased osmotic stress resistance. FEMS Yeast Res. 12, 969–979. doi: 10.1111/1567-1364.12002
Guo, H., Qiu, Y., Wei, J., Niu, C., Zhang, Y., Yuan, Y., et al. (2020). Genomic insights into sugar adaptation in an extremophile yeast Zygosaccharomyces rouxii. Front. Microbiol. 10:1–10. doi: 10.3389/fmicb.2019.03157
Gustin, M. C., Albertyn, J., Alexander, M., and Davenport, K. (1998). MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264. doi: 10.1128/mmbr.62.4.1264-1300.1998
Hagen, I., Ecker, M., Lagorce, A., Francois, J. M., Sestak, S., Rachel, R., et al. (2004). Sed1p and Srl1p are required to compensate for cell wall instability in Saccharomyces cerevisiae mutants defective in multiple GPI-anchored mannoproteins. Mol. Microbiol. 52, 1413–1425. doi: 10.1111/j.1365-2958.2004.04064.x
Harris, M. R., Lee, D., Farmer, S., Lowndes, N. F., and de Bruin, R. A. M. (2013). Binding specificity of the G1/S transcriptional regulators in budding yeast. PLoS One 8:e61059. doi: 10.1371/journal.pone.0061059
Heinisch, J. J., Dupres, V., Wilk, S., Jendretzki, A., and Dufrêne, Y. F. (2010). Single-molecule atomic force microscopy reveals clustering of the yeast plasma-membrane sensor Wsc1. PLoS One 5:e11104. doi: 10.1371/journal.pone.0011104
Henriques, S. F., Mira, N. P., and Sá-Correia, I. (2017). Genome-wide search for candidate genes for yeast robustness improvement against formic acid reveals novel susceptibility (Trk1 and positive regulators) and resistance (Haa1-regulon) determinants. Biotechnol. Biofuels 10:96. doi: 10.1186/s13068-017-0781-5
Hohmann, S. (2002a). Osmotic adaptation in yeast-control of the yeast osmolyte system. Int. Rev. Cytol. 215, 149–187. doi: 10.1016/S0074-7696(02)15008-X
Hohmann, S. (2002b). Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66, 300–372. doi: 10.1128/mmbr.66.2.300-372.2002
Hohmann, S. (2009). Control of high osmolarity signalling in the yeast Saccharomyces cerevisiae. FEBS Lett. 583, 4025–4029. doi: 10.1016/j.febslet.2009.10.069
Holt, S., Mukherjee, V., Lievens, B., Verstrepen, K. J., and Thevelein, J. M. (2018). Bioflavoring by non-conventional yeasts in sequential beer fermentations. Food Microbiol. 72, 55–66. doi: 10.1016/j.fm.2017.11.008
Holyoak, C. D., Bracey, D., Piper, P. W., Kuchler, K., and Coote, P. J. (1999). The Saccharomyces cerevisiae weak acid-inducible ABC transporter Pdr12 transports fluorescein and preservative anions from the cytosol by an energy-dependent mechanism. J. Bacteriol. 181, 4644–4652. doi: 10.1128/JB.181.15.4644-4652.1999
Homma, T., Iwahashi, H., and Komatsu, Y. (2003). Yeast gene expression during growth at low temperature. Cryobiology 46, 230–237. doi: 10.1016/s0011-2240(03)00028-2
Hossain, M. N., Afrin, S., Humayun, S., Ahmed, M. M., and Saha, B. K. (2020). Identification and growth characterization of a novel strain of Saccharomyces boulardii isolated from soya paste. Front. Nutr. 7:27. doi: 10.3389/fnut.2020.00027
Hottiger, T., Boller, T., and Wiemken, A. (1989). Correlation of trenalose content and heat resistance in yeast mutants altered in the RAS/adenylate cyclase pathway: is trehalose a thermoprotectant? FEBS Lett. 255, 431–434. doi: 10.1016/0014-5793(89)81139-1
Hottiger, T., Schmutz, P., and Wiemken, A. (1987). Heat-induced accumulation and futile cycling of trehalose in Saccharomyces cerevisiae. J. Bacteriol. 169, 5518–5522. doi: 10.1128/jb.169.12.5518-5522.1987
Hou, L., Wang, M., Wang, C., Wang, C., and Wang, H. (2013). Analysis of salt-tolerance genes in Zygosaccharomyces rouxii. Appl. Biochem. Biotechnol. 170, 1417–1425. doi: 10.1007/s12010-013-0283-2
Hu, J., Dong, Y., Wang, W., Zhang, W., Lou, H., and Chen, Q. (2019). Deletion of Atg22 gene contributes to reduce programmed cell death induced by acetic acid stress in Saccharomyces cerevisiae. Biotechnol. Biofuels 12, 1–20. doi: 10.1186/s13068-019-1638-x
Hudson, L. E., McDermott, C. D., Stewart, T. P., Hudson, W. H., Rios, D., Fasken, M. B., et al. (2016). Characterization of the probiotic yeast Saccharomyces boulardii in the healthy mucosal immune system. PLoS One 11:e0153351. doi: 10.1371/journal.pone.0153351
Igual, J. C., Johnson, A. L., and Johnston, L. H. (1996). Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 34, 1049–1057. doi: 10.1002/j.1460-2075.1996.tb00880.x
Imazu, H., and Sakurai, H. (2005). Saccharomyces cerevisiae heat shock transcription factor regulates cell wall remodeling in response to heat shock. Eukaryot. Cell 4, 1050–1056. doi: 10.1128/EC.4.6.1050-1056.2005
Irie, K., Takase, M., Lee, K. S., Levin, D. E., Araki, H., Matsumoto, K., et al. (1993). MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol. Cell. Biol. 13, 3076–3083. doi: 10.1128/mcb.13.5.3076
Jaafar, L., and Zueco, J. (2004). Characterization of a glycosylphosphatidylinositol-bound cell-wall protein (GPI-CWP) in Yarrowia lipolytica. Microbiology 150, 53–60. doi: 10.1099/mic.0.26430-0
Jamieson, D. J. (1998). Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14, 1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:163.0.CO;2-S
Jiménez-Gutiérrez, E., Alegría-Carrasco, E., Alonso-Rodríguez, E., Fernández-Acero, T., Molina, M., and Martín, H. (2020a). Rewiring the yeast cell wall integrity (CWI) pathway through a synthetic positive feedback circuit unveils a novel role for the MAPKKK Ssk2 in CWI pathway activation. FEBS J. 22, 4881–4901. doi: 10.1111/febs.15288
Jiménez-Gutiérrez, E., Alegría-Carrasco, E., Sellers-Moya, Á, Molina, M., and Martín, H. (2020b). Not just the wall: the other ways to turn the yeast CWI pathway on. Int. Microbiol. 23, 107–119. doi: 10.1007/s10123-019-00092-2
Jin, C., Parshin, A. V., Daly, I., Strich, R., and Cooper, K. F. (2013). The cell wall sensors Mtl1, Wsc1, and Mid2 are required for stress-induced nuclear to cytoplasmic translocation of cyclin C and programmed cell death in yeast. Oxid. Med. Cell. Longev. 2013:320823. doi: 10.1155/2013/320823
Jung, U. S., and Levin, D. E. (1999). Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 34, 1049–1057. doi: 10.1046/j.1365-2958.1999.01667.x
Kahar, P., Itomi, A., Tsuboi, H., Ishizaki, M., Yasuda, M., Kihira, C., et al. (2022). The flocculant Saccharomyces cerevisiae strain gains robustness via alteration of the cell wall hydrophobicity. Metab. Eng. 72, 82–96. doi: 10.1016/j.ymben.2022.03.001
Kamada, Y., Jung, U. S., Piotrowski, J., and Levin, D. E. (1995). The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9, 1559–1571. doi: 10.1101/gad.9.13.1559
Kanetsuna, F., Carbonell, L. M., Moreno, R. E., and Rodriguez, J. (1969). Cell wall composition of the yeast and mycelial forms of Paracoccidioides brasiliensis. J. Bacteriol. 97, 1036–1041. doi: 10.1128/jb.97.3.1036-1041.1969
Kapteyn, J. C., Ter Riet, B., Vink, E., Blad, S., De Nobel, H., Van Den Ende, H., et al. (2001). Low external ph induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 39, 469–479. doi: 10.1046/j.1365-2958.2001.02242.x
Kapteyn, J. C., Van Den Ende, H., and Klis, F. M. (1999a). The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta Gen. Subj. 1426, 373–383. doi: 10.1016/s0304-4165(98)00137-8
Kapteyn, J. C., Van Egmond, P., Sievi, E., Van Den Ende, H., Makarow, M., and Klis, F. M. (1999b). The contribution of the O-glycosylated protein Pir2p/Hsp150 to the construction of the yeast cell wall in wild-type cells and beta 1,6-glucan-deficient mutants. Mol. Microbiol. 31, 1835–1844. doi: 10.1046/j.1365-2958.1999.01320.x
Kawahata, M., Masaki, K., Fujii, T., and Iefuji, H. (2006). Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res. 6, 924–936. doi: 10.1111/j.1567-1364.2006.00089.x
Kim, H., Thak, E. J., Yeon, J. Y., Sohn, M. J., Choo, J. H., Kim, J.-Y., et al. (2018). Functional analysis of Mpk1-mediated cell wall integrity signaling pathway in the thermotolerant methylotrophic yeast Hansenula polymorpha. J. Microbiol. 56, 72–82. doi: 10.1007/s12275-018-7508-6
Kim, K.-Y., Truman, A. W., and Levin, D. E. (2008). Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol. Cell. Biol. 28, 2579–2589. doi: 10.1128/mcb.01795-07
Kim, M. S., Cho, K. H., Park, K. H., Jang, J., and Hahn, J.-S. (2019). Activation of Haa1 and War1 transcription factors by differential binding of weak acid anions in Saccharomyces cerevisiae. Nucleic Acids Res. 47, 1211–1224. doi: 10.1093/nar/gky1188
Klis, F. M. (2002). Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26, 239–256. doi: 10.1111/j.1574-6976.2002.tb00613.x
Klis, F. M., Boorsma, A., and De Groot, P. W. J. (2006). Cell wall construction in Saccharomyces cerevisiae. Yeast 23, 185–202. doi: 10.1002/yea.1349
Kock, C., Arlt, H., Ungermann, C., and Heinisch, J. J. (2016). Yeast cell wall integrity sensors form specific plasma membrane microdomains important for signalling. Cell. Microbiol. 18, 1251–1267. doi: 10.1111/cmi.12635
Kock, C., Dufrêne, Y. F., and Heinisch, J. J. (2015). Up against the wall: is yeast cell wall integrity ensured by mechanosensing in plasma membrane microdomains? Appl. Environ. Microbiol. 81, 806–811. doi: 10.1128/AEM.03273-14
Kolhe, N., Kulkarni, A., Zinjarde, S., and Acharya, C. (2021). Transcriptome response of the tropical marine yeast Yarrowia lipolytica on exposure to uranium. Curr. Microbiol. 78, 2033–2043. doi: 10.1007/s00284-021-02459-z
Kolhe, N., Zinjarde, S., and Acharya, C. (2020). Impact of uranium exposure on marine yeast, Yarrowia lipolytica: insights into the yeast strategies to withstand uranium stress. J. Hazard. Mater. 381:121226. doi: 10.1016/j.jhazmat.2019.121226
Kollár, R., Petráková, E., Ashwell, G., Robbins, P. W., and Cabib, E. (1995). Architecture of the yeast cell wall. the linkage between chitin and β(1 → 3)-glucan. J. Biol. Chem. 270, 1170–1178. doi: 10.1074/jbc.270.3.1170
Kollár, R., Reinhold, B. B., Petráková, E., Yeh, H. J. C., Ashwell, G., Drgonová, J., et al. (1997). Architecture of the yeast cell wall. beta(1–>6)-glucan interconnects mannoprotein, beta(1–>)3-glucan, and chitin. J. Biol. Chem. 272, 17762–17775. doi: 10.1074/jbc.272.28.17762
Kong, M., Li, X., Li, T., Zhao, X., Jin, M., Zhou, X., et al. (2021). Overexpressing CCW12 in Saccharomyces cerevisiae enables highly efficient ethanol production from lignocellulose hydrolysates. Bioresour. Technol. 337:125487. doi: 10.1016/j.biortech.2021.125487
Krause, E. L., Villa-García, M. J., Henry, S. A., and Walker, L. P. (2007). Determining the effects of inositol supplementation and the opi1 mutation on ethanol tolerance of Saccharomyces cerevisiae. Ind. Biotechnol. (New Rochelle. N. Y). 3, 260–268. doi: 10.1089/ind.2007.3.260
Kuanyshev, N., Ami, D., Signori, L., Porro, D., Morrissey, J. P., and Branduardi, P. (2016). Assessing physio-macromolecular effects of lactic acid on Zygosaccharomyces bailii cells during microaerobic fermentation. FEMS Yeast Res. 16:fow058. doi: 10.1093/femsyr/fow058
Kubiak-Szymendera, M., Skupien-Rabian, B., Jankowska, U., and Celiñska, E. (2022). Hyperosmolarity adversely impacts recombinant protein synthesis by Yarrowia lipolytica-molecular background revealed by quantitative proteomics. Appl. Microbiol. Biotechnol. 106, 349–367. doi: 10.1007/s00253-021-11731-y
Kubota, S., Takeo, I., Kume, K., Kanai, M., Shitamukai, A., Mizunuma, M., et al. (2004). Effect of ethanol on cell growth of budding yeast: genes that are important for cell growth in the presence of ethanol. Biosci. Biotechnol. Biochem. 68, 968–972. doi: 10.1271/bbb.68.968
Kuranda, M. J., and Robbins, P. W. (1991). Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J. Biol. Chem. 266, 19758–19767.
Kwak, S., and Jin, Y. S. (2017). Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: a review and perspective. Microb. Cell Fact. 16, 1–15. doi: 10.1186/s12934-017-0694-9
Kwon, Y., Chiang, J., Tran, G., Giaever, G., Nislow, C., Hahn, B.-S., et al. (2016). Signaling pathways coordinating the alkaline pH response confer resistance to the hevein-type plant antimicrobial peptide Pn-AMP1 in Saccharomyces cerevisiae. Planta 244, 1229–1240. doi: 10.1007/s00425-016-2579-2
Larriba, G., Andaluz, E., Cueva, R., and Basco, R. D. (1995). Molecular biology of yeast exoglucanases. FEMS Microbiol. Lett. 125, 121–126. doi: 10.1111/j.1574-6968.1995.tb07347.x
Ledesma-Amaro, R., and Nicaud, J. M. (2016). Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Prog. Lipid Res. 61, 40–50. doi: 10.1016/j.plipres.2015.12.001
Lee, J., Reiter, W., Dohnal, I., Gregori, C., Beese-Sims, S., Kuchler, K., et al. (2013). MAPK Hog1 closes the S. cerevisiae glycerol channel Fps1 by phosphorylating and displacing its positive regulators. Genes Dev. 27, 2590–2601. doi: 10.1101/gad.229310.113
Lee, K. S., and Levin, D. E. (1992). Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol. Cell. Biol. 12, 172–182. doi: 10.1128/mcb.12.1.172
Lehnen, M., Ebert, B. E., and Blank, L. M. (2019). Elevated temperatures do not trigger a conserved metabolic network response among thermotolerant yeasts. BMC Microbiol. 19:1–11. doi: 10.1186/s12866-019-1453-3
Lesage, G., and Bussey, H. (2006). Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 317–343. doi: 10.1128/MMBR.00038-05
Levin, D. E. (2011). Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189, 1145–1175. doi: 10.1534/genetics.111.128264
Li, B., Xie, C. Y., Yang, B. X., Gou, M., Xia, Z. Y., Sun, Z. Y., et al. (2020). The response mechanisms of industrial Saccharomyces cerevisiae to acetic acid and formic acid during mixed glucose and xylose fermentation. Proc. Biochem. 91, 319–329. doi: 10.1016/j.procbio.2020.01.002
Li, S., Dean, S., Li, Z., Horecka, J., Deschenes, R. J., and Fassler, J. S. (2002). The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via the transcription factor, Skn7p. Mol. Biol. Cell 13, 412–424. doi: 10.1091/mbc.01-09-0434
Li, Y., Li, Y., Li, R., Liu, L., Miao, Y., Weng, P., et al. (2021). Metabolic changes of Issatchenkia orientalis under acetic acid stress by transcriptome profile using RNA-sequencing. Int. Microbiol. doi: 10.1007/s10123-021-00217-6 [Epub ahead of print].
Lide, D. R. (2003). “Dissociation constants of organic acids and bases,” in CRC Handbook of Chemistry and Physics, ed. D. R. Lide (Boca. Raton, FL: CRC Press), 8–46.
Liszkowska, W., and Berlowska, J. (2021). Yeast fermentation at low temperatures: adaptation to changing environmental conditions and formation of volatile compounds. Molecules 26:1035. doi: 10.3390/molecules26041035
Liu, G., Bi, X., Tao, C., Fei, Y., Gao, S., Liang, J., et al. (2021). Comparative transcriptomics analysis of Zygosaccharomyces mellis under high-glucose stress. Food Sci. Hum. Wellness 10, 54–62. doi: 10.1016/j.fshw.2020.05.006
Liu, G., Tao, C., Zhu, B., Bai, W., Zhang, L., Wang, Z., et al. (2016). Identification of Zygosaccharomyces mellis strains in stored honey and their stress tolerance. Food Sci. Biotechnol. 25, 1645–1650. doi: 10.1007/s10068-016-0253-x
Longo, V., Ždralević, M., Guaragnella, N., Giannattasio, S., Zolla, L., and Timperio, A. M. (2015). Proteome and metabolome profiling of wild-type and YCA1-knock-out yeast cells during acetic acid-induced programmed cell death. J. Proteomics 128, 173–188. doi: 10.1016/j.jprot.2015.08.003
Lucena, R. M., Dolz-Edo, L., Brul, S., de Morais, M. A., and Smits, G. (2020). Extreme low cytosolic pH is a signal for cell survival in acid stressed yeast. Genes 11:56. doi: 10.3390/genes11060656
Madhavan, A., Jose, A. A., Binod, P., Sindhu, R., Sukumaran, R. K., Pandey, A., et al. (2017). Synthetic biology and metabolic engineering approaches and its impact on non-conventional yeast and biofuel production. Front. Energy Res. 5:8. doi: 10.3389/fenrg.2017.00008
Madrid, M., Vázquez-Marín, B., Franco, A., Soto, T., Vicente-Soler, J., Gacto, M., et al. (2016). Multiple crosstalk between TOR and the cell integrity MAPK signaling pathway in fission yeast. Sci. Rep. 6, 1–17. doi: 10.1038/srep37515
Mager, W. H., and Siderius, M. (2002). Novel insights into the osmotic stress response of yeast. FEMS Yeast Res. 2, 251–257. doi: 10.1016/S1567-1356(02)00116-2
Magnelli, P., Cipollo, J. F., and Abeijon, C. (2002). A refined method for the determination of Saccharomyces cerevisiae cell wall composition and beta-1,6-glucan fine structure. Anal. Biochem. 301, 136–150. doi: 10.1006/abio.2001.5473
Martín, H., Rodríguez-Pachón, J. M., Ruiz, C., Nombela, C., and Molina, M. (2000). Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275, 1511–1519. doi: 10.1074/jbc.275.2.1511
Martins, L. C., Monteiro, C. C., Semedo, P. M., and Sá-Correia, I. (2020). Valorisation of pectin-rich agro-industrial residues by yeasts: potential and challenges. Appl. Microbiol. Biotechnol. 104, 6527–6547. doi: 10.1007/s00253-020-10697-7
Mattanovich, D., Gasser, B., Hohenblum, H., and Sauer, M. (2004). Stress in recombinant protein producing yeasts. Jounal 113, 121–135. doi: 10.1016/j.jbiotec.2004.04.035
Mazur, P., Morin, N., Baginsky, W., El-Sherbeini, M., Clemas, J. A., Nielsen, J. B., et al. (1995). Differential expression and function of two homologous subunits of yeast 1,3-beta-D-glucan synthase. Mol. Cell. Biol. 15, 5671–5681. doi: 10.1128/MCB.15.10.5671
Mensonides, F. I. C., Brul, S., Klis, F. M., Hellingwerf, K. J., and De Mattos, M. J. T. (2005). Activation of the protein kinase C1 pathway upon continuous heat stress in Saccharomyces cerevisiae is triggered by an intracellular increase in osmolarity due to trehalose accumulation. Appl. Environ. Microbiol. 71, 4531–4538. doi: 10.1128/AEM.71.8.4531-4538.2005
Miao, Y., Xiong, G., Li, R., Wu, Z., Zhang, X., and Weng, P. (2018). Transcriptome profiling of Issatchenkia orientalis under ethanol stress. AMB Exp. 8:39. doi: 10.1186/s13568-018-0568-5
Mira, N. P., Becker, J. D., and Sá-Correia, I. (2010a). Genomic expression program involving the haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. Omi. A J. Integr. Biol. 14, 587–601. doi: 10.1089/omi.2010.0048
Mira, N. P., Palma, M., Guerreiro, J. F., and Sá-Correia, I. (2010b). Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microb. Cell Fact. 9:79. doi: 10.1186/1475-2859-9-79
Mira, N. P., Teixeira, M. C., and Sá-Correia, I. (2010c). Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: a genome-wide view. OMICS 14, 525–540. doi: 10.1089/omi.2010.0072
Mira, N. P., Henriques, S. F., Keller, G., Teixeira, M. C., Matos, R. G., Arraiano, C. M., et al. (2011). Identification of a DNA-binding site for the transcription factor Haa1, required for Saccharomyces cerevisiae response to acetic acid stress. Nucleic Acids Res. 39, 6896–6907. doi: 10.1093/nar/gkr228
Mira, N. P., Lourenço, A. B., Fernandes, A. R., Becker, J. D., and Sá-Correia, I. (2009). The RIM101 pathway has a role in Saccharomyces cerevisiae adaptive response and resistance to propionic acid and other weak acids. FEMS Yeast Res. 9, 202–216. doi: 10.1111/j.1567-1364.2008.00473.x
Mo, W., Wang, M., Zhan, R., Yu, Y., He, Y., and Lu, H. (2019). Kluyveromyces marxianus developing ethanol tolerance during adaptive evolution with significant improvements of multiple pathways. Biotechnol. Biofuels 12, 1–15. doi: 10.1186/s13068-019-1393-z
Mollapour, M., Shepherd, A., and Piper, P. W. (2009). Presence of the Fps1p aquaglyceroporin channel is essential for Hog1p activation, but suppresses Slt2(Mpk1)p activation, with acetic acid stress of yeast. Microbiology 155, 3304–3311. doi: 10.1099/mic.0.030502-0
Moradas-Ferreira, P., and Costa, V. (2000). Adaptive response of the yeast Saccharomyces cerevisiae to reactive oxygen species: defences, damage and death. Redox Rep. 5, 277–285. doi: 10.1179/135100000101535816
Morano, K. A., Grant, C. M., and Moye-Rowley, W. S. (2012). The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190, 1157–1195. doi: 10.1534/genetics.111.128033
Morimoto, Y., and Tani, M. (2015). Synthesis of mannosylinositol phosphorylceramides is involved in maintenance of cell integrity of yeast Saccharomyces cerevisiae. Mol. Microbiol. 95, 706–722. doi: 10.1111/mmi.12896
Morris, G. J., Winters, L., Coulson, G. E., and Clarke, K. J. (1983). Effect of osmotic stress on the ultrastructure and viability of the yeast Saccharomyces cervisiae. J. Gen. Microbiol. 132, 2023–2034. doi: 10.1099/00221287-132-7-2023
Mota, M. N., Martins, L. C., and Sá-Correia, I. (2021). The identification of genetic determinants of methanol tolerance in yeast suggests differences in methanol and ethanol toxicity mechanisms and candidates for improved methanol tolerance engineering. J. Fungi 7, 1–28. doi: 10.3390/jof7020090
Mukherjee, V., Radecka, D., Aerts, G., Verstrepen, K. J., Lievens, B., and Thevelein, J. M. (2017). Phenotypic landscape of non-conventional yeast species for different stress tolerance traits desirable in bioethanol fermentation. Biotechnol. Biofuels 10, 1–19. doi: 10.1186/s13068-017-0899-5
Murata, Y., Homma, T., Kitagawa, E., Momose, Y., Sato, M. S., Odani, M., et al. (2006). Genome-wide expression analysis of yeast response during exposure to 4 degrees C. Extremophiles 10, 117–128. doi: 10.1007/s00792-005-0480-1
Nandy, S. K., and Srivastava, R. K. (2018). A review on sustainable yeast biotechnological processes and applications. Microbiol. Res. 207, 83–90. doi: 10.1016/j.micres.2017.11.013
Narayanan, V., Sànchez i Nogué, V., van Niel, E. W. J., and Gorwa-Grauslund, M. F. (2016). Adaptation to low pH and lignocellulosic inhibitors resulting in ethanolic fermentation and growth of Saccharomyces cerevisiae. AMB Exp. 6:8. doi: 10.1186/s13568-016-0234-8
Navarro-Tapia, E., Querol, A., and Pérez-Torrado, R. (2018). Membrane fluidification by ethanol stress activates unfolded protein response in yeasts. Microb. Biotechnol. 11, 465–475. doi: 10.1111/1751-7915.13032
Nguyen, T. H., Fleet, G. H., and Rogers, P. L. (1998). Composition of the cell walls of several yeast species. Appl. Microbiol. Biotechnol. 50, 206–212. doi: 10.1007/s002530051278
Nielsen, J. (2019). Yeast systems biology: model organism and cell factory. Biotechnol. J. 14, e1800421. doi: 10.1002/biot.201800421
Noti, O., Vaudano, E., Pessione, E., and Garcia-Moruno, E. (2015). Short-term response of different Saccharomyces cerevisiae strains to hyperosmotic stress caused by inoculation in grape must: RT-qPCR study and metabolite analysis. Food Microbiol. 52, 49–58. doi: 10.1016/j.fm.2015.06.011
Ogawa, Y., Nitta, A., Uchiyama, H., Imamura, T., Shimoi, H., and Ito, K. (2000). Tolerance mechanism of the ethanol-tolerant mutant of sake yeast. J. Biosci. Bioeng. 90, 313–320. doi: 10.1016/S1389-1723(00)80087-0
Ohsawa, S., Yurimoto, H., and Sakai, Y. (2017). Novel function of Wsc proteins as a methanol-sensing machinery in the yeast Pichia pastoris. Mol. Microbiol. 104, 349–363. doi: 10.1111/mmi.13631
Orlean, P. (2012). Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 192, 775–818. doi: 10.1534/genetics.112.144485
Ortiz-Merino, R. A., Kuanyshev, N., Byrne, K. P., Varela, J. A., Morrissey, J. P., Porro, D., et al. (2018). Transcriptional response to lactic acid stress in the hybrid yeast Zygosaccharomyces parabailii. Appl. Environ. Microbiol. 84, 1–17. doi: 10.1128/AEM.02294-17
Osmond, B. C., Specht, C. A., and Robbins, P. W. (1999). Chitin synthase III: synthetic lethal mutants and “stress related” chitin synthesis that bypasses the CSD3/CHS6 localization pathway. Proc. Natl. Acad. Sci. U.S.A. 96, 11206–11210. doi: 10.1073/pnas.96.20.11206
Pacheco, A., Talaia, G., Sá-Pessoa, J., Bessa, D., Gonçalves, M. J., Moreira, R., et al. (2012). Lactic acid production in Saccharomyces cerevisiae is modulated by expression of the monocarboxylate transporters Jen1 and Ady2. FEMS Yeast Res. 12, 375–381. doi: 10.1111/j.1567-1364.2012.00790.x
Palma, M., Dias, P. J., Roque, F., de, C., Luzia, L., Guerreiro, J. F., et al. (2017). The Zygosaccharomyces bailii transcription factor Haa1 is required for acetic acid and copper stress responses suggesting subfunctionalization of the ancestral bifunctional protein Haa1/Cup2. BMC Geno. 18:75. doi: 10.1186/s12864-016-3443-2
Palma, M., Guerreiro, J. F., and Sá-Correia, I. (2018). Adaptive response and tolerance to acetic acid in Saccharomyces cerevisiae and Zygosaccharomyces bailii: a physiological genomics perspective. Front. Microbiol. 9:274. doi: 10.3389/fmicb.2018.00274
Palma, M., and Sá-Correia, I. (2019). “Physiological genomics of the highly weak-acid-tolerant food spoilage yeasts of Zygosaccharomyces bailii sensu lato,” in Progress in Molecular and Subcellular Biology, ed. I. Sá-Correia (Cham: Springer International Publishing), doi: 10.1007/978-3-030-13035-0_4
Panahi, H., Dehhaghi, M., Kinder, J. E., and Ezeji, T. C. (2019). A review on green liquid fuels for the transportation sector: a prospect of microbial solutions to climate change. Biofuel Res. J. 6, 995–1024. doi: 10.18331/BRJ2019.6.3.2
Pandey, P., Zaman, K., Prokai, L., and Shulaev, V. (2020). Comparative proteomics analysis reveals unique early signaling response of Saccharomyces cerevisiae to oxidants with different mechanism of action. Int. J. Mol. Sci. 22:167. doi: 10.3390/ijms22010167
Parapouli, M., Vasileiadis, A., Afendra, A. S., and Hatziloukas, E. (2020). Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 6, 1–31. doi: 10.3934/microbiol.2020001
Patel, P. K., and Free, S. J. (2019). The genetics and biochemistry of cell wall structure and synthesis in Neurospora crassa, a model filamentous fungus. Front. Microbiol. 10:1–18. doi: 10.3389/fmicb.2019.02294
Petkova, M. I., Pujol-Carrion, N., Arroyo, J., García-Cantalejo, J., and De La Torre-Ruiz, M. A. (2010). Mtl1 is required to activate general stress response through TOR1 and RAS2 inhibition under conditions of glucose starvation and oxidative stress. J. Biol. Chem. 285, 19521–19531. doi: 10.1074/jbc.M109.085282
Pillet, F., Lemonier, S., Schiavone, M., Formosa, C., Martin-yken, H., Francois, J. M., et al. (2014). Uncovering by atomic force microscopy of an original circular structure at the yeast cell surface in response to heat shock. BMC Biol. 12:1–11. doi: 10.1186/1741-7007-12-6
Piper, P., Mahé, Y., Thompson, S., Pandjaitan, R., Holyoak, C., Egner, R., et al. (1998). The pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 17, 4257–4265. doi: 10.1093/emboj/17.15.4257
Popolo, L., Gilardelli, D., Bonfante, P., and Vai, M. (1997). Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J. Bacteriol. 179, 463–469. doi: 10.1128/jb.179.2.463-469.1997
Porro, D., Bianchi, M. M., Brambilla, L., Menghini, R., Bolzani, D., Carrera, V., et al. (1999). Replacement of a metabolic pathway for large-scale production of lactic acid from engineered yeasts. Appl. Environ. Microbiol. 65, 4211–4215. doi: 10.1128/aem.65.9.4211-4215.1999
Pøibylová, L., Montigny, J., and Sychrova, H. (2007). Osmoresistant yeast Zygosaccharomyces rouxii:the two most studied wild-type strains (ATCC 2623 and ATCC 42981) differ in osmotolerance and glycerol metabolism. Yeast 24, 171–180.
Ragni, E., Fontaine, T., Gissi, C., Latgè, J. P., and Popolo, L. (2007). The gas family of proteins of Saccharomyces cerevisiae: characterization and evolutionary analysis. Yeast 24, 297–308. doi: 10.1002/yea.1473
Raj, T., Chandrasekhar, K., Naresh Kumar, A., Rajesh Banu, J., Yoon, J.-J., Kant Bhatia, S., et al. (2022). Recent advances in commercial biorefineries for lignocellulosic ethanol production: current status, challenges and future perspectives. Bioresour. Technol. 344:126292. doi: 10.1016/j.biortech.2021.126292
Rajavel, M., Philip, B., Buehrer, B. M., Errede, B., and Levin, D. E. (1999). Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 3969–3976. doi: 10.1128/mcb.19.6.3969
Ram, A. F. J., Kapteyn, J. C., Montijn, R. C., Caro, L. H. P., Douwes, J. E., Baginsky, W., et al. (1998). Loss of the plasma membrane-bound protein gas1p in Saccharomyces cerevisiae results in the release of β1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J. Bacteriol. 180, 1418–1424. doi: 10.1128/jb.180.6.1418-1424.1998
Ramírez-Cota, G. Y., López-Villegas, E. O., Jiménez-Aparicio, A. R., and Hernández-Sánchez, H. (2021). Modeling the ethanol tolerance of the probiotic yeast Saccharomyces cerevisiae var. boulardii CNCM I-745 for its possible use in a functional beer. Probiot. Antimicrob. Proteins 13, 187–194. doi: 10.1007/s12602-020-09680-5
Ribeiro, R. A., Godinho, C. P., Vitorino, M. V., Robalo, T. T., Fernandes, F., Rodrigues, M. S., et al. (2022). Crosstalk between yeast cell plasma membrane ergosterol content and cell wall stiffness under acetic acid stress involving Pdr18. J. Fungi. 8:103. doi: 10.3390/jof8020103
Ribeiro, R. A., Vitorino, M. V., Godinho, C. P., Bourbon-Melo, N., Robalo, T. T., Fernandes, F., et al. (2021). Yeast adaptive response to acetic acid stress involves structural alterations and increased stiffness of the cell wall. Sci. Rep. 11:12652. doi: 10.1038/s41598-021-92069-3
Rodríguez-Peña, J. M., García, R., Nombela, C., and Arroyo, J. (2010). The high-osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: a yeast dialogue between MAPK routes. Yeast 27, 495–502. doi: 10.1002/yea
Roemer, T., Paravicini, G., Payton, M. A., and Bussey, H. (1994). Characterization of the yeast (1–>6)-beta-glucan biosynthetic components, Kre6p and Skn1p, and genetic interactions between the PKC1 pathway and extracellular matrix assembly. J. Cell Biol. 127, 567–579. doi: 10.1083/jcb.127.2.567
Roncero, C. (2002). The genetic complexity of chitin synthesis in fungi. Curr. Genet. 41, 367–378. doi: 10.1007/s00294-002-0318-7
Rossignol, T., Dulau, L., Julien, A., and Blondin, B. (2003). Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 20, 1369–1385. doi: 10.1002/yea.1046
Runguphan, W., and Keasling, J. D. (2014). Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 21, 103–113. doi: 10.1016/j.ymben.2013.07.003
Rzechonek, D. A., Day, A. M., Quinn, J., and Miroñczuk, A. M. (2018). Influence of ylHog1 MAPK kinase on Yarrowia lipolytica stress response and erythritol production. Sci. Rep. 8, 1–12. doi: 10.1038/s41598-018-33168-6
Sahara, T., Goda, T., and Ohgiya, S. (2002). Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature. J. Biol. Chem. 277, 50015–50021. doi: 10.1074/jbc.M209258200
Saini, S., and Sharma, K. K. (2021). Fungal lignocellulolytic enzymes and lignocellulose: a critical review on their contribution to multiproduct biorefinery and global biofuel research. Int. J. Biol. Macromol. 193, 2304–2319. doi: 10.1016/j.ijbiomac.2021.11.063
Sakihama, Y., Hasunuma, T., and Kondo, A. (2015). Improved ethanol production from xylose in the presence of acetic acid by the overexpression of the HAA1 gene in Saccharomyces cerevisiae. J. Biosci. Bioeng. 119, 297–302. doi: 10.1016/j.jbiosc.2014.09.004
Saldaña, C., Villava, C., Ramírez-Villarreal, J., Morales-Tlalpan, V., Campos-Guillen, J., Chávez-Servín, J., et al. (2021). Rapid and reversible cell volume changes in response to osmotic stress in yeast. Brazilian J. Microbiol. 52, 895–903. doi: 10.1007/s42770-021-00427-0
Samakkarn, W., Ratanakhanokchai, K., and Soontorngun, N. (2021). Reprogramming of the ethanol stress response in Saccharomyces cerevisiae by the transcription factor Znf1 and its effect on the biosynthesis of glycerol and ethanol. Appl. Environ. Microbiol. 87:e0058821. doi: 10.1128/AEM.00588-21
Sanz, A. B., García, R., Rodríguez-Peña, J. M., and Arroyo, J. (2018). The CWI pathway: regulation of the transcriptional adaptive response to cell wall stress in yeast. J. Fungi 4:1. doi: 10.3390/jof4010001
Sauer, M., Porro, D., Mattanovich, D., and Branduardi, P. (2010). 16 years research on lactic acid production with yeast–ready for the market? Biotechnol. Genet. Eng. Rev. 27, 229–256. doi: 10.1080/02648725.2010.10648152
Schade, B., Jansen, G., Whiteway, M., Entian, K. D., and Thomas, D. Y. (2004). Cold adaptation in budding yeast. Mol. Biol. Cell 15, 5492–5502. doi: 10.1091/mbc.e04-03-0167
Schiavone, M., Formosa-Dague, C., Elsztein, C., Teste, M. A., Martin-Yken, H., De Morais, M. A., et al. (2016). Evidence for a role for the plasma membrane in the nanomechanical properties of the cell wall as revealed by an atomic force microscopy study of the response of Saccharomyces cerevisiae to ethanol stress. Appl. Environ. Microbiol. 82, 4789–4801. doi: 10.1128/AEM.01213-16
Schiavone, M., Vax, A., Formosa, C., Martin-Yken, H., Dague, E., and François, J. M. (2014). A combined chemical and enzymatic method to determine quantitatively the polysaccharide components in the cell wall of yeasts. FEMS Yeast Res. 14, 933–947. doi: 10.1111/1567-1364.12182
Schöppner, P., Lutz, A. P., Lutterbach, B. J., Brückner, S., Essen, L.-O., and Mösch, H.-U. (2022). Structure of the yeast cell wall integrity sensor Wsc1 reveals an essential role of surface-exposed aromatic clusters. J. Fungi 8:379. doi: 10.3390/jof8040379
Seike, T., Narazaki, Y., Kaneko, Y., Shimizu, H., and Matsuda, F. (2021). Random transfer of Ogataea polymorpha genes into Saccharomyces cerevisiae reveals a complex background of heat tolerance. J. Fungi 7:302. doi: 10.3390/jof7040302
Sekova, V. Y., Dergacheva, D. I., Isakova, E. P., Gessler, N. N., Tereshina, V. M., and Deryabina, Y. I. (2019). Soluble sugar and lipid readjustments in the Yarrowia lipolytica yeast at various temperatures and pH. Metabolites 9:307. doi: 10.3390/metabo9120307
Serra-Cardona, A., Canadell, D., and Ariño, J. (2015). Coordinate responses to alkaline pH stress in budding yeast. Microb. Cell 2, 182–196. doi: 10.15698/mic2015.06.205
Serrano, R., Martín, H., Casamayor, A., and Ariño, J. (2006). Signaling alkaline pH stress in the yeast Saccharomyces cerevisiae through the Wsc1 cell surface sensor and the Slt2 MAPK pathway. J. Biol. Chem. 281, 39785–39795. doi: 10.1074/jbc.M604497200
Sha, W., Martins, A. M., Laubenbacher, R., Mendes, P., and Shulaev, V. (2013). The genome-wide early temporal response of saccharomyces cerevisiae to oxidative stress induced by cumene hydroperoxide. PLoS One 8:1–15. doi: 10.1371/journal.pone.0074939
Simões, T., Mira, N., Fernandes, A., and Sá-correia, I. (2006). The SPI1 gene, encoding a glycosylphosphatidylinositol-anchored cell wall protein, plays a prominent role in the development of yeast resistance to lipophilic weak-acid food preservatives. Appl. Environ. Microbiol. 18, 7168–7165. doi: 10.1128/AEM.01476-06
Simpson, W. J., and Hammond, J. R. M. (1989). The response of brewing yeasts to acid washing. J. Inst. Brew. 95, 347–354.
Skoneczny, M. (2018). Stress Response Mechanisms in Fungi: Theoretical and Practical Aspects. Cham: Springer, doi: 10.1007/978-3-030-00683-9
Songdech, P., Ruchala, J., Semkiv, M. V., Jensen, L. T., Sibirny, A., Ratanakhanokchai, K., et al. (2020). Overexpression of transcription factor ZNF1 of glycolysis improves bioethanol productivity under high glucose concentration and enhances acetic acid tolerance of Saccharomyces cerevisiae. Biotechnol. J. 15, 1–10. doi: 10.1002/biot.201900492
Sousa-Lopes, A., Antunes, F., Cyrne, L., and Marinho, H. S. (2004). Decreased cellular permeability to H2O2 protects Saccharomyces cerevisiae cells in stationary phase against oxidative stress. FEBS Lett. 578, 152–156. doi: 10.1016/j.febslet.2004.10.090
Staleva, L., Hall, A., and Orlow, S. J. (2004). Oxidative stress activates FUS1 and RLM1 transcription in the yeast Saccharomyces cerevisiae in an oxidant-dependent manner. Mol. Biol. Cell 15, 5574–5582. doi: 10.1091/mbc.E04-02-0142
Stanley, D., Bandara, A., Fraser, S., Chambers, P. J., and Stanley, G. A. (2010). The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J. Appl. Microbiol. 109, 13–24. doi: 10.1111/j.1365-2672.2009.04657.x
Stewart, G. G. (2017). “The structure and function of the yeast cell wall, plasma membrane and periplasm,” in Brewing and Distilling Yeasts, ed. G. G. Stewart (Cham: Springer), 55–75. doi: 10.1007/978-3-319-69126-8_5
Stovicek, V., Dato, L., Almqvist, H., Schöpping, M., Chekina, K., Pedersen, L. E., et al. (2022). Rational and evolutionary engineering of Saccharomyces cerevisiae for production of dicarboxylic acids from lignocellulosic biomass and exploring genetic mechanisms of the yeast tolerance to the biomass hydrolysate. Biotechnol. Biofuels Bioprod. 15:22. doi: 10.1186/s13068-022-02121-1
Sugiyama, M., Akase, S. P., Nakanishi, R., Horie, H., Kaneko, Y., and Harashima, S. (2014). Nuclear localization of Haa1, which is linked to its phosphorylation status, mediates lactic acid tolerance in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 80, 3488–3495. doi: 10.1128/AEM.04241-13
Suhr, K. I., and Nielsen, P. V. (2004). Effect of weak acid preservatives on growth of bakery product spoilage fungi at different water activities and pH values. Int. J. Food Microbiol. 95, 67–78. doi: 10.1016/j.ijfoodmicro.2004.02.004
Suvarna, S., Dsouza, J., Ragavan, M. L., and Das, N. (2018). Potential probiotic characterization and effect of encapsulation of probiotic yeast strains on survival in simulated gastrointestinal tract condition. Food Sci. Biotechnol. 27, 745–753. doi: 10.1007/s10068-018-0310-8
Suzuki, T., Sugiyama, M., Wakazono, K., Kaneko, Y., and Harashima, S. (2012). Lactic-acid stress causes vacuolar fragmentation and impairs intracellular amino-acid homeostasis in Saccharomyces cerevisiae. J. Biosci. Bioeng. 113, 421–430. doi: 10.1016/j.jbiosc.2011.11.010
Swinnen, S., Henriques, S. F., Shrestha, R., Ho, P. W., Correia, I. S., and Nevoigt, E. (2017). Improvement of yeast tolerance to acetic acid through Haa1 transcription factor engineering: towards the underlying mechanisms. Microb. Cell Fact. 16, 1–15. doi: 10.1186/s12934-016-0621-5
Tanaka, K., Ishii, Y., Ogawa, J., and Shima, J. (2012). Enhancement of acetic acid tolerance in Saccharomyces cerevisiae by overexpression of the Haa1 gene, encoding a transcriptional activator. Appl. Environ. Microbiol. 78, 8161–8163. doi: 10.1128/AEM.02356-12
Tanaka, S., and Tani, M. (2018). Mannosylinositol phosphorylceramides and ergosterol coodinately maintain cell wall integrity in the yeast Saccharomyces cerevisiae. FEBS J. 285, 2405–2427. doi: 10.1111/febs.14509
Taymaz-Nikerel, H., Cankorur-Cetinkaya, A., and Kirdar, B. (2016). Genome-wide transcriptional response of Saccharomyces cerevisiae to stress-induced perturbations. Front. Bioeng. Biotechnol. 4:17. doi: 10.3389/fbioe.2016.00017
Teixeira, M. C., Raposo, L. R., Mira, N. P., Lourenço, A. B., and Sá-Correia, I. (2009). Genome-wide identification of Saccharomyces cerevisiae genes required for maximal tolerance to ethanol. Appl. Environ. Microbiol. 75, 5761–5772. doi: 10.1128/AEM.00845-09
Teixeira, M. C., Raposo, L. R., Palma, M., and Sá-Correia, I. (2010). Identification of genes required for maximal tolerance to high-glucose concentrations, as those present in industrial alcoholic fermentation media, through a chemogenomics approach. OMICS 14, 201–210. doi: 10.1089/omi.2009.0149
Tenreiro, S., Nunes, P. A., Viegas, C. A., Neves, M. S., Teixeira, M. C., Cabral, M. G., et al. (2002). AQR1 gene (ORF YNL065w) encodes a plasma membrane transporter of the major facilitator superfamily that confers resistance to short-chain monocarboxylic acids and quinidine in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 292, 741–748. doi: 10.1006/bbrc.2002.6703
Thomé, P. E. (2007). Cell wall involvement in the glycerol response to high osmolarity in the halotolerant yeast Debaryomyces hansenii. antonie van leeuwenhoek. Int. J. Gen. Mol. Microbiol. 91, 229–235. doi: 10.1007/s10482-006-9112-8
Thorwall, S., Schwartz, C., Chartron, J. W., and Wheeldon, I. (2020). Stress-tolerant non-conventional microbes enable next-generation chemical biosynthesis. Nat. Chem. Biol. 16, 113–121. doi: 10.1038/s41589-019-0452-x
Torres, L., Martín, H., García-Saez, M. I., Arroyo, J., Molina, M., Sánchez, M., et al. (1991). A protein kinase gene complements the lytic phenotype of Saccharomyces cerevisiae lyt2 mutants. Mol. Microbiol. 11, 2845–2854. doi: 10.1111/j.1365-2958.1991.tb01993.x
Truman, A. W., Millson, S. H., Nuttall, J. M., Mollapour, M., Prodromou, C., and Piper, P. W. (2007). In the yeast heat shock response, Hsf1-directed induction of Hsp90 facilitates the activation of the Slt2 (Mpk1) mitogen-activated protein kinase required for cell integrity. Eukaryot. Cell 6, 744–752. doi: 10.1128/EC.00009-07
Udom, N., Chansongkrow, P., Charoensawan, V., and Auesukaree, C. (2019). Coordination of the cell wall integrity and highosmolarity glycerol pathways in response to ethanol stress in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 85, e551. doi: 10.1128/AEM.00551-19
Ullah, A., Chandrasekaran, G., Brul, S., and Smits, G. J. (2013). Yeast adaptation to weak acids prevents futile energy expenditure. Front. Microbiol. 4:142. doi: 10.3389/fmicb.2013.00142
Usmani, Z., Sharma, M., Awasthi, A. K., Lukk, T., Tuohy, M. G., Gong, L., et al. (2021). Lignocellulosic biorefineries: the current state of challenges and strategies for efficient commercialization. Renew. Sustain. Energy Rev. 148:111258. doi: 10.1016/j.rser.2021.111258
Valdivieso, M. H., Ferrario, L., Vai, M., Duran, A., and Popolo, L. (2000). Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J. Bacteriol. 182, 4752–4757. doi: 10.1128/JB.182.17.4752-4757.2000
Valiante, V., Macheleidt, J., Föge, M., and Brakhage, A. A. (2015). The Aspergillus fumigatus cell wall integrity signalling pathway: drug target, compensatory pathways and virulence. Front. Microbiol. 6:1–12. doi: 10.3389/fmicb.2015.00325
van Leeuwe, T. M., Arentshorst, M., Punt, P. J., and Ram, A. F. J. (2020). Interrogation of the cell wall integrity pathway in Aspergillus niger identifies a putative negative regulator of transcription involved in chitin deposition. Gene 5:100028. doi: 10.1016/j.gene.2020.100028
Van Maris, A. J. A., Winkler, A. A., Porro, D., Van Dijken, J. P., and Pronk, J. T. (2004). Homofermentative lactate production cannot sustain anaerobic growth of engineered Saccharomyces cerevisiae: possible consequence of energy-dependent lactate export. Appl. Environ. Microbiol. 70, 2898–2905. doi: 10.1128/AEM.70.5.2898-2905.2004
van Uden, N. (1985). Ethanol toxicity and ethanol tolerance in yeasts. Annu. Reports Ferment. Proc. 8, 11–58.
van Voorst, F., Houghton-Larson, J., Jønson, L., Kielland-Brandt, M. C., and Brandt, A. (2006). Genome-wide identification of genes required for growth of Saccharomyces cerevisiae under ethanol stress. Yeast 23, 351–359. doi: 10.1002/yea.1359
Varol, D., Purutçuoǧlu, V., and Yılmaz, R. (2018). Transcriptomic analysis of the heat stress response for a commercial baker’s yeast Saccharomyces cerevisiae. Genes Geno. 40, 137–150. doi: 10.1007/s13258-017-0616-6
Vaz, E., Lee, J., and Levin, D. (2020). Crosstalk between S. cerevisiae SAPKs Hog1 and Mpk1 is mediated by glycerol accumulation. Fungal Biol. 124, 361–367. doi: 10.1016/j.funbio.2019.10.002
Vélez-Segarra, V., González-Crespo, S., Santiago-Cartagena, E., Vázquez-Quiñones, L. E., Martínez-Matías, N., Otero, Y., et al. (2020). Protein interactions of the mechanosensory proteins Wsc2 and Wsc3 for stress resistance in Saccharomyces cerevisiae. G3 Gen. Geno. Genet. 10, 3121–3135. doi: 10.1534/g3.120.401468
Verghese, J., Abrams, J., Wang, Y., and Morano, K. A. (2012). Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol. Mol. Biol. Rev. 76, 115–158. doi: 10.1128/MMBR.05018-11
Verna, J., Lodder, A., Lee, K., Vagts, A., and Ballester, R. (1997). A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 94, 13804–13809. doi: 10.1073/pnas.94.25.13804
Viladevall, L., Serrano, R., Ruiz, A., Domenech, G., Giraldo, J., Barceló, A., et al. (2004). Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J. Biol. Chem. 279, 43614–43624. doi: 10.1074/jbc.M403606200
Vilella, F., Herrero, E., Torres, J., and De La Torre-Ruiz, M. A. (2005). Pkc1 and the upstream elements of the cell integrity pathway in Saccharomyces cerevisiae, Rom2 and Mtl1, are required for cellular responses to oxidative stress. J. Biol. Chem. 280, 9149–9159. doi: 10.1074/jbc.M411062200
Wada, K., Fujii, T., Akita, H., and Matsushika, A. (2020). IoGAS1, a GPI-anchored protein derived from Issatchenkia orientalis, confers tolerance of Saccharomyces cerevisiae to multiple acids. Appl. Biochem. Biotechnol. 190, 1349–1359. doi: 10.1007/s12010-019-03187-8
Walker, G. M., and Basso, T. O. (2020). Mitigating stress in industrial yeasts. Fungal Biol. 124, 387–397. doi: 10.1016/j.funbio.2019.10.010
Walker, G., and Stewart, G. (2016). Saccharomyces cerevisiae in the production of fermented beverages. Beverages 2:30. doi: 10.3390/beverages2040030
Wang, G., Kell, D. B., and Borodina, I. (2021). Harnessing the yeast Saccharomyces cerevisiae for the production of fungal secondary metabolites. Essays Biochem. 65, 277–291. doi: 10.1042/EBC20200137
Wei, X., Chi, Z., Liu, G.-L., Hu, Z., and Chi, Z.-M. (2021). The genome-wide mutation shows the importance of cell wall integrity in growth of the psychrophilic yeast metschnikowia australis W7-5 at different temperatures. Microb. Ecol. 81, 52–66. doi: 10.1007/s00248-020-01577-8
Wu, H., Zheng, X., Araki, Y., Sahara, H., Takagi, H., and Shimoi, H. (2006). Global gene expression analysis of yeast cells during sake brewing. Appl. Environ. Microbiol. 72, 7353–7358. doi: 10.1128/AEM.01097-06
Xu, Y., Yang, H., Brennan, C. S., Coldea, T. E., and Zhao, H. (2020). Cellular mechanism for the improvement of multiple stress tolerance in brewer’s yeast by potassium ion supplementation. Int. J. Food Sci. Technol. 55, 2419–2427. doi: 10.1111/ijfs.14491
Yamaguchi, M., Namiki, Y., Okada, H., Mori, Y., Furukawa, H., and Wang, J. (2011). Structome of Saccharomyces cerevisiae determined by freeze-substitution and serial ultrathin-sectioning electron microscopy. J. Electron Microsci. 60, 321–335. doi: 10.1093/jmicro/dfr052
Yamakawa, C. K., Qin, F., and Mussatto, S. I. (2018). Advances and opportunities in biomass conversion technologies and biorefineries for the development of a bio-based economy. Bio. Bioenergy 119, 54–60. doi: 10.1016/j.jbiotec.2018.07.041
Yang, Z., Huang, J., Geng, J., Nair, U., and Klionsky, J. (2006). Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell 17, 5094–5104. doi: 10.1091/mbc.E06
Yasokawa, D., Murata, S., Iwahashi, Y., Kitagawa, E., Nakagawa, R., Hashido, T., et al. (2010). Toxicity of methanol and formaldehyde towards Saccharomyces cerevisiae as assessed by DNA microarray analysis. Appl. Biochem. Biotechnol. 160, 1685–1698. doi: 10.1007/s12010-009-8684-y
Yazawa, H., Iwahashi, H., and Uemura, H. (2007). Disruption of URA7 and GAL6 improves the ethanol tolerance and fermentation capacity of Saccharomyces cerevisiae. Yeast 24, 551–560. doi: 10.1002/yea.1492
Yen, H. W., Chen, T. J., Pan, W. C., and Wu, H. J. (2010). Effects of neutralizing agents on lactic acid production by Rhizopus oryzae using sweet potato starch. World J. Microbiol. Biotechnol. 26, 437–441. doi: 10.1007/s11274-009-0186-0
Zeng, L., Huang, J., Feng, P., Zhao, X., Si, Z., Long, X., et al. (2022). Transcriptomic analysis of formic acid stress response in Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 38:34. doi: 10.1007/s11274-021-03222-z
Zhang, C., Ma, Y., Miao, H., Tang, X., Xu, B., Wu, Q., et al. (2020). Transcriptomic analysis of Pichia pastoris (Komagataella phaffii) GS115 during heterologous protein production using a high-cell-density fed-batch cultivation strategy. Front. Microbiol. 11:463. doi: 10.3389/fmicb.2020.00463
Zhang, Q., and Xu, Y. (2021). UDP-glucose pyrophosphorylase ugp1 is required in heat stress response of Saccharomyces cerevisiae. J. Bio. Mater. Bioenergy 15, 188–193. doi: 10.1016/j.bbrc.2015.10.090
Zhao, C., Jung, U. S., Garrett-Engele, P., Roe, T., Cyert, M. S., and Levin, D. E. (1998). Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol. Cell. Biol. 18, 1013–1022. doi: 10.1128/MCB.18.2.1013
Zhao, X. Q., Xue, C., Ge, X. M., Yuan, W. J., Wang, J. Y., and Bai, F. W. (2009). Impact of zinc supplementation on the improvement of ethanol tolerance and yield of self-flocculating yeast in continuous ethanol fermentation. J. Biotechnol. 139, 55–60. doi: 10.1016/j.jbiotec.2008.08.013
Zhong, W., Yang, M., Hao, X., Sharshar, M. M., Wang, Q., and Xing, J. (2021). Improvement of D-lactic acid production at low pH through expressing acid-resistant gene IoGAS1 in engineered Saccharomyces cerevisiae. J. Chem. Technol. Biotechnol. 96, 732–742. doi: 10.1002/jctb.6587
Keywords: cell wall, yeasts of biotechnological relevance, stress response, stress tolerance, signaling pathways
Citation: Ribeiro RA, Bourbon-Melo N and Sá-Correia I (2022) The cell wall and the response and tolerance to stresses of biotechnological relevance in yeasts. Front. Microbiol. 13:953479. doi: 10.3389/fmicb.2022.953479
Received: 26 May 2022; Accepted: 11 July 2022;
Published: 28 July 2022.
Edited by:
Markus Proft, Institute of Biomedicine of Valencia (CSIC), SpainReviewed by:
Javier Arroyo, Complutense University of Madrid, SpainCopyright © 2022 Ribeiro, Bourbon-Melo and Sá-Correia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isabel Sá-Correia, aXNhY29ycmVpYUB0ZWNuaWNvLnVsaXNib2EucHQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.