
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 07 July 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.952982
This article is part of the Research TopicNew Knowledge of Food Microbiology in Asia, Volume IIView all 15 articles
 Xiaoyun Qu1†
Xiaoyun Qu1† Jie Zhou1,2†
Jie Zhou1,2† Haoqi Huang1,2
Haoqi Huang1,2 Wen Wang2
Wen Wang2 Yingping Xiao2
Yingping Xiao2 Biao Tang2
Biao Tang2 Hanlin Liu1
Hanlin Liu1 Chenggang Xu1*
Chenggang Xu1* Xingning Xiao2*
Xingning Xiao2*Proteus mirabilis is a common opportunistic zoonotic pathogen, and its ongoing acquisition of antimicrobial resistance genes poses challenges to clinical treatments. Human-sourced whole genomic sequencing of human P. mirabilis isolates has been reported, but pig-sourced isolates have not been thoroughly investigated even though these animals can serve as reservoirs for human infections. In the current study, we report a molecular epidemiological investigation to unravel the antimicrobial and virulence gene risk factors for P. mirabilis contamination in 9 pig farms in 3 different cities in Zhejiang Province, China. We collected 541 swab samples from healthy pigs and 30 were confirmed as P. mirabilis. All 30 isolates were resistant to tetracyclines, macrolides, sulfonamides, β-lactams and chloramphenicol, and all were multiple drug-resistant and 27 were strong biofilm formers. Phylogenetic analyses indicated these 30 isolates clustered together in 2 major groups. Whole genome sequencing demonstrated that the isolates possessed 91 different antimicrobial resistance genes belonging to 30 antimicrobial classes including rmtB, sul1, qnrS1, AAC(6′) − Ib − cr, blaCTX − M − 65 and blaOXA − 1. All isolates contained mobile genetic elements including integrative conjugative elements (ICEs) and integrative and mobilizable elements (IMEs). Minimum inhibitory concentration (MIC) testing indicated direct correlates between cognate genes and antimicrobial resistance. We also identified 95 virulence factors, almost all isolates contained 20 fimbrial and flagellar operons, and this represents the greatest number of these operon types found in a single species among all sequenced bacterial genomes. These genes regulate biofilm formation and represent a confounding variable for treating P. mirabilis infections. Our P. mirabilis isolates were present in healthy animals, and multiple drug resistance in these isolates may serve as a reservoir for other intestinal and environmental Enterobacteriaceae members. This prompts us to more strictly regulate veterinary antibiotic use.
Proteus mirabilis is an Enterobacteriaceae member and a significant opportunistic and food-borne pathogen and second only to enteropathogenic Escherichia coli (EPEC) in prevalence for urinary tract infection. P. mirabilis can exist in a variety of environments including the intestinal tracts of humans and wild and domestic animals and can cause infections in immuno-compromised hosts resulting in diarrhea, urinary tract infection (UTI) and keratitis (Mobley and Belas, 1995; Drzewiecka, 2016; Gong et al., 2019). Importantly, antimicrobial resistance of P. mirabilis has become a serious impediment to clinical treatment and resistance to colistin, nitrofurans, tigecycline, tetracycline, and β-lactams have been documented (Stock, 2003).
China is the largest global antibiotic producer and consumer and more than half are used to treat food animals (Gilbert, 2012). Livestock farms are environments with high bacterial loads and high antimicrobial selective pressures; a combination favoring development of resistant bacteria (Klein et al., 2018; Qian et al., 2018). There is abundant direct and indirect evidence that the antibiotic use on farms correlates with the rise and spread of associated antibiotic resistance genes (ARG) in human pathogens and even the direct transfer of antibiotic-resistant bacteria from animals to humans (Marshall and Levy, 2011). Pigs are major meat source worldwide, especially in China and ARGs are increasingly detected in bacterial isolates from pig feces implicating pig husbandry in ARG development and spread (Larson, 2015; Shi et al., 2021). In particular, mobile genetic elements (MGE) including insertion sequences (IS), resistance plasmids, gene castle, transposons, integrative conjugative elements (ICE), and integrative and mobilizable elements (IME) allow ARG movement within or between DNA molecules. These MGE are now frequently detected in P. mirabilis isolates (Shelenkov et al., 2020; Wang et al., 2021).
P. mirabilis is also a biofilm former allowing its engraftment on living or abiotic surfaces and complicates disease treatment and enhances viability through antimicrobial and disinfectant resistance. Treatment of UTIs due to indwelling catheters can be complicated through biofilm formation that prolong bacterial presence and can lead to bladder and kidney infections that may progress to bacteremia or sepsis (Høiby et al., 2011). P. mirabilis contains numerous virulence factors related to adhesion, colonization, biofilm formation and pathogenicity. These include the MR/P and PMF fimbriae and the uroepithelial cell adhesin (UCA) and all can combine to enable urinary cell attachment and biofilm formation.
The emergence of multidrug-resistant (MDR) bacterial pathogens is now considered a public health risk and sources of these pathogens have been found in humans, livestock, wild animals and food. Importantly, molecular typing studies have identified animal strains that were transmitted to humans (Reich et al., 2013; Olonitola et al., 2015; Lei et al., 2016). Previously reported 7.07% P. mirabilis from broiler chicken samples (Li et al., 2022) and pig operations in China are currently experiencing large outbreaks of diarrheal disease in piglets (Li, 2021). As a pathogen that can cause diarrhea, P. mirabilis has not attracted enough attention. Therefore, the objectives were to investigated the prevalence, antimicrobial resistance and virulence genes for P. mirabilis to characterize risk factors of P. mirabilis contamination in Zhejiang Province, China. Whole genome sequencing and bioinformatics analyses are cost-effective methods for the investigation and characterization of zoonotic pathogens (Biswas et al., 2020; Liu et al., 2020, 2021). We conducted a study on 9 farms in 3 different cities in Zhejiang Province, China, which were rarely reported, to obtain P. mirabilis isolates from pig swab sources. The recovered strains were subjected to whole genome sequencing followed by in silico analysis to identify antimicrobial resistance and virulence genes. We also determined antibiotic MICs to correlate with ARG presence.
A total of 541 pig swab specimens were obtained from 9 farms located in 3 different cities in Zhejiang Province, China from May to December 2021. The strains isolated from Jinhua, Hangzhou, and Quzhou were designated PM 1 (n = 101), PM 2 (n = 240), and PM 3 (n = 200), respectively (Table 1). In brief, swabs were placed in sterile centrifuge tubes containing 3 ml Selenite Cystine Broth (SC; Hopebiol, Qingdao, China) and incubated at 37°C for 12 h. Loopfuls were then streaked on Salmonella-Shigella agar plates (SS; Becton Dickinson, Franklin Lakes, NJ, United States) and incubated as per above (Li et al., 2022). Suspected P. mirabilis colonies were streak-purified a second time. Presumptive P. mirabilis colonies displayed black centers with transparent edges on SS agar.
Suspected P. mirabilis were selected for genomic DNA extraction using a TIANamp bacteria DNA kit (Tiangen Biotech, Beijing, China) according to the instructions of the manufacturer and DNA was stored at −20°C. P. mirabilis identities were confirmed using PCR detection of the atpD gene (5′-AGAGTTTGATCCTGGCTCAG-3′5′-ACGGGCGGTGTGTRC-3′) as previously described (Bi et al., 2013). In addition, the 16S rDNA genes (5′-AGAGTTTGATCCTGGCTCAG-3′5′-ACGGGCGGTGTGTRC-3) from these isolates were sequenced. Pure cultures of identified strains were cryopreserved at −80°C in 30% glycerol.
Biofilm formation was assayed using the crystal violet (CV) staining method as previously described (Stepanović et al., 2007; Han et al., 2015; Lajhar et al., 2018). Briefly, bacterial suspension turbidities were adjusted to 0.5 McFarland standard (~108 CFU/ml) and 20 μl was then added to prepared microtiter plates containing 180 μl Luria Bertani (LB) broth per well; negative control wells contained only LB. The plates were then covered and incubated for 24 h at 37°C under static conditions. The liquid was then discarded and the wells were washed 3× with phosphate-buffered saline (PBS) and 200 μl 0.1% CV (Sigma, Aldrich, United States) was then added per well and the plates were allowed to stand for 30 min at room temperature. The CV was decanted and the bacteria were then heat-fixed by exposure to a stream of hot air at 60°C for 60 min. The dye bound to the cells was then resolubilized using three rinses with 200 μl of 95% ethanol per well. The optical density (OD) of combined washes was measured at 595 nm using a microtiter plate reader. The average OD595 values were calculated for triplicates and the tests were repeated three times.
Illumina pair-end sequencing of each strain utilized 1 μg genomic DNA for library construction. The qualified library was used for Illumina NovaSeq 6,000 sequencing (150 bp * 2) at Shanghai Biozeron. The raw paired end reads were trimmed and quality controlled by Trimmomatic version 0.36. ABySS1 was used for genome assembly with multiple-Kmer parameters for optimization. GapCloser2 was subsequently applied to fill remaining local inner gaps and correct single base polymorphisms for the final assembly results. Ab initio prediction methods were used to obtain gene models for P. mirabilis strains and were identified using GeneMark and then used for BLASTp searches against the non-redundant (NR) NCBI database. Potential virulence factors were assessed by searches against the Virulence Factors of Pathogenic Bacteria Database (VFDB) and BLAST results were considered significant at E < 1e−5 and is a generally accepted consensus cut-off (Chen et al., 2005). ARGs of the 30 P. mirabilis strains we sequenced were also examined using the Comprehensive Antibiotic Resistance Database (CARD), and ICEs were identified using ICEfinder (Liu et al., 2019). The public data of reference genome of P. mirabilis (HI4320) was obtained from NCBI3; gene annotation information including functional annotations and gene family information were also downloaded. The datasets generated for this study can be found in the NCBI Bioproject with the accession number no. PRJNA841796.
Broth dilution method was used for MIC test for a panel of 15 antimicrobial agents belonging to 9 classes as described previously (Kobylka, Kuth, Müller, Geertsma, & Pos, 2020). MIC results were interpreted according to the recommendations of the Clinical Laboratory Standard Institute guidelines (CLSI, 2017; Table 2). The antimicrobial agents were grouped in the following classes: tetracyclines (tetracycline, TET, 8–256 μg/ml; minocycline, MIN, 4–256 μg/ml), fluoroquinolones (ofloxacin, OFX, 2–64 μg/ml; nalidixic acid, NAL, 2–64 μg/ml; ciprofloxacin, CIP, 2–64 μg/ml), macrolides (azithromycin, AZ, 16–512 μg/ml; erythromycin, EM, 16–512 μg/ml), β-lactamase inhibitors (amoxicillin/clavulanic acid, AMC, 4/2–128/64 μg/ml), penicillins (ampicillin, AMP, 16–512 μg/ml), carbapenems (meropenem, MEM, 1–64 μg/ml), cephalosporins (ceftiofur, CEF, 16–512 μg/ml), aminoglycosides (amikacin, AMK, 2–64 μg/ml; gentamicin, GEN, 2–128 μg/ml), phenicols (chloramphenicol, CHL, 4–128 μg/ml; rifampicin, RIF, 1–64 μg/ml).
Proteus mirabilis CMCC 49005 was used for quality control.
GraphPad Prism 8 software (San Diego, CA, United States) was used for figure generation. ANOVA was performed using least squares techniques with IBM SPSS Statistics 20 software (SPSS, Chicago, IL, United States). A significant difference was established at p < 0.05. MIC results of intermediate susceptibility were merged with resistance and each test result (resistant or susceptible) was compared with the detection (presence or absence) of the corresponding ARG in silico.
Proteus mirabilis has been frequently incriminated in food-borne and urinary tract infections in humans. We first explored the prevalence and characteristics of P. mirabilis in pig farm isolates from Zhejiang province, China.
We isolated 30 P. mirabilis strains from 541 samples (5.55%) using atpD PCR detection, 16S rDNA and whole genome sequencing (Table 1). The PM1 group (Jinhua) was the most contaminated (8.91%), and this level was close to that previously reported (7.07%) from broiler chicken samples (Li et al., 2022) but lower than from human sources (Mirzaei et al., 2019; Tabatabaei et al., 2021). Phylogenetic analysis identified 2 major clusters including Group 1 that contained reference strain LR738973_1 from fecal samples of weaned piglets in Brazil and the smaller Group 2 that included model strain HI4320, MK758055.1 from human feces, AM231709.1 from fish bowels and OL629222.1 from raccoon feces in China (Figure 1A). The isolates were also clustered closely in the phylogenetic analysis of core SNPs especially between PM1 and PM3 (Figure 1B), and PCA analysis indicated PM 1 and PM 3 were closely linked (Figure 1C), which might because Jinhua and Quzhou are closer together.
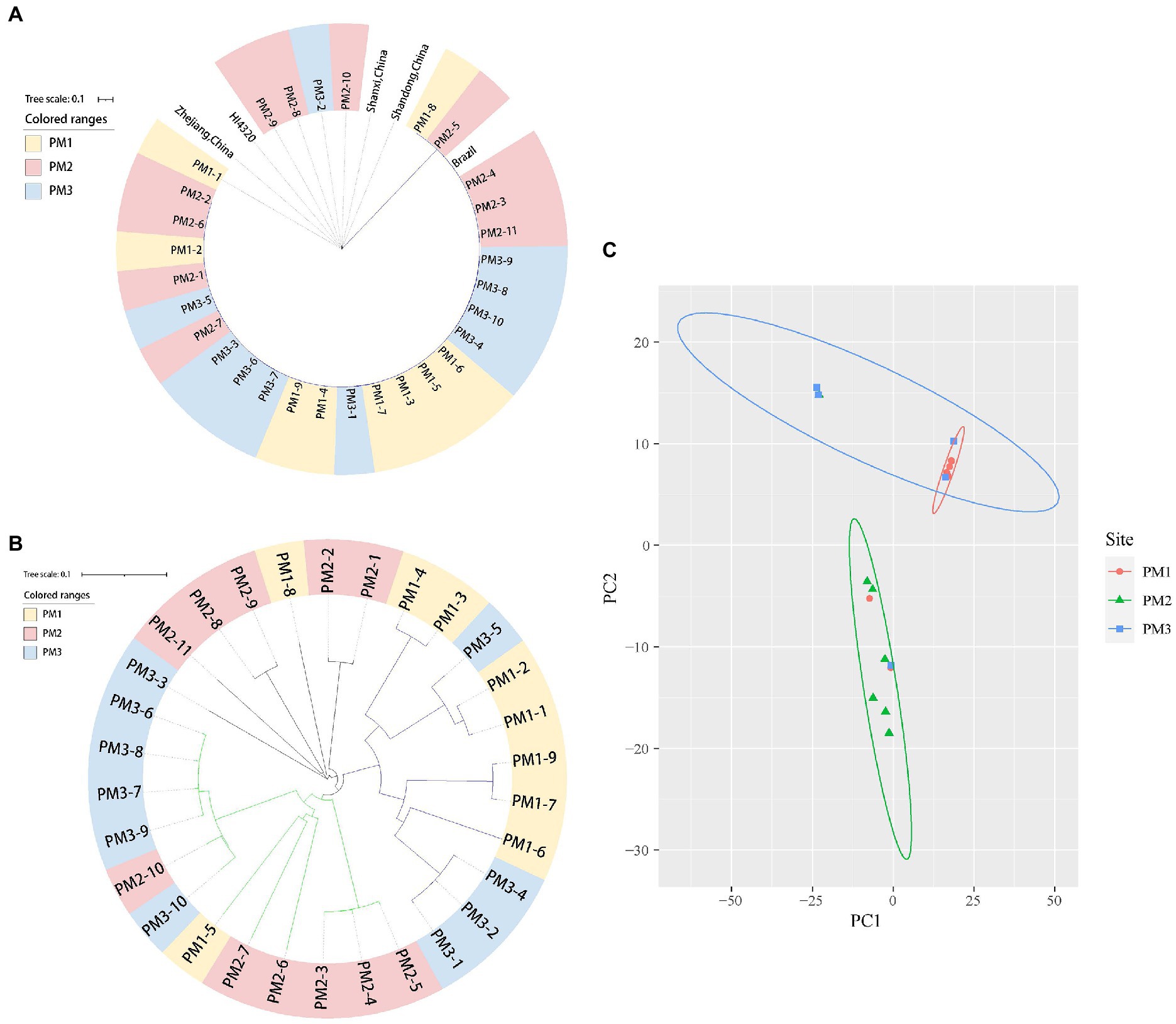
Figure 1. Phylogenetic tree of 31 P. mirabilis isolates in this study. (A) 16S rDNA tree. (B) SNP tree. (C) PCA analysis.
The 30 P. mirabilis isolates we identified were also biofilm formers (Figure 2). For example, isolates PM1-3, PM1-4 and PM1-6 displayed a moderate biofilm formation capacity (0.2314 ≤ OD595 ≤ 0.4628) while 27 isolates were strong biofilm formers (OD595 ≥ 0.4628). All PM2 and PM3 isolates were in this biofilm formation capacity category and significantly different from isolates from PM1 (p < 0.05). Bacterial biofilms are resistant to antibiotics, disinfectants and phagocytosis as well as other components of innate and adaptive inflammatory systems (Høiby et al., 2011). Compared with planktonic cells, biofilm cells are less sensitive to antimicrobial agents and limit antibiotic entry to low levels (Tseng et al., 2013). In this study, 90% (27/30) of our isolates were strong biofilm formers and these could be ranked as PM3 > PM2 > PM1. Interestingly, this ranking was consistent with the MIC results (see below) and intensity of biofilm formation has been previously linked to antibiotic resistance (Mirzaei et al., 2019) and MDR strains are often biofilm formers, similar to our findings (Sanchez et al., 2013).
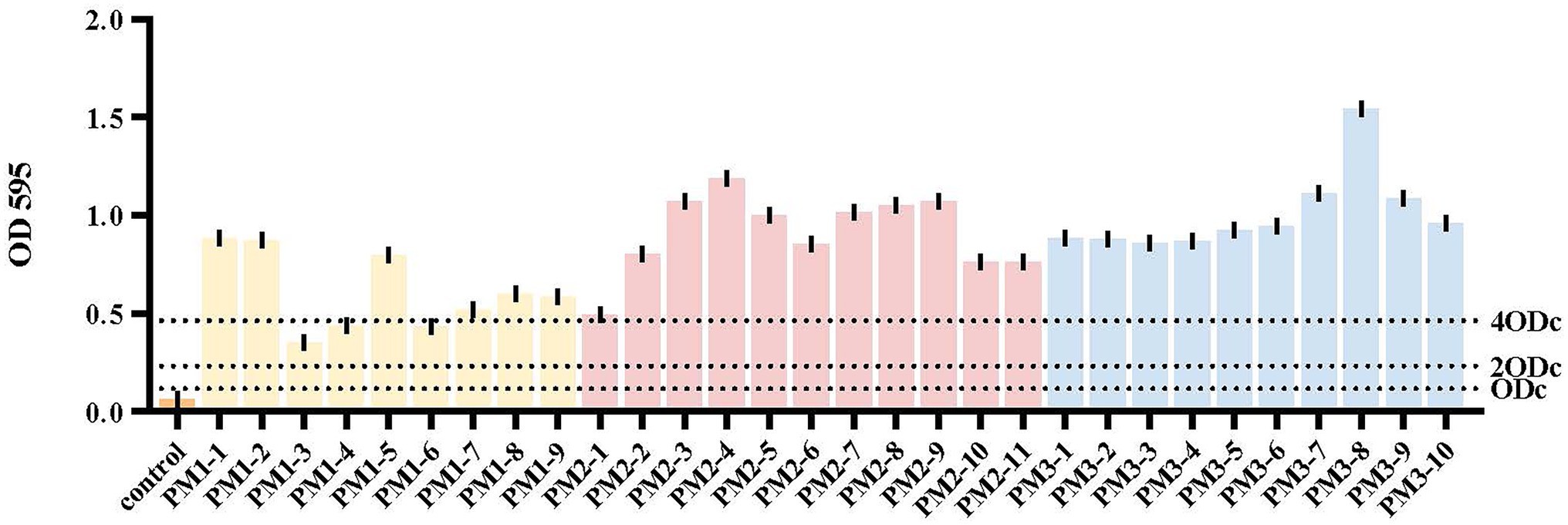
Figure 2. Biofilm formation. Yellow, PM1; pink, PM2; blue, PM3. The cut-off value (ODc) was defined as 3 SD above the mean OD595 of the negative control: ODc = average OD595 of negative control + (3 × SD of negative control). OD595 ≤ ODc = absence of biofilm; ODc ≤ OD595 ≤ 2 × ODc = weak biofilm producer; 2 × ODc ≤ OD595 ≤ 4 × ODc = moderate biofilm producer; 4 × ODc ≤ OD595 = strong biofilm producer.
MIC testing of our 30 isolates indicated different degrees of resistance to the 15 antimicrobials we analyzed. P. mirabilis is naturally resistant to tetracycline, tigecycline, and polymyxin, but sensitive to carbapenems and aminoglycosides although amikacin and meropenem resistance has been increasing and is linked with their abuse (Kang et al., 2021). Our results were consistent with these observations and most isolates were sensitive to the aminoglycoside amikacin (21/30) and the carbapenem meropenem (12/30). These levels were lower than those found in two recent hospital studies in Iran that documented 71% and 82% and 82% and 82%, respectively (Mirzaei et al., 2019; Tabatabaei et al., 2021), and higher than the 10.5% sensitive to meropenem in a Chinese hospital (Hu et al., 2012; Han et al., 2015). All our strains were resistant to the tetracyclines (tetracycline, minocycline), macrolides (azithromycin, erythromycin) and chloramphenicol (chloramphenicol) classes and to a lesser extent to amoxicillin/clavulanic acid (27/30), ampicillin (7/30), ciprofloxacin (23/30), and rifampicin (22/30; Table 2). Rifampicin is a broad-spectrum antibiotic used primarily in clinical treatment of tuberculosis (Chen et al., 2017). Chloramphenicol was forbidden for veterinary use in China in 2002 (The Ministry of Agriculture of the People’s Republic of China, 2002) so it is intriguing why we found such high levels of resistance to these drugs.
Our PM3 group displayed the highest prevalence for antibiotic resistance to amoxicillin/clavulanic acid, ampicillin, gentamycin and amikacin (Figure 4A). Moreover, all our isolates were resistant to at least 3 antimicrobial classes and were considered MDR (Figure 3). These data were similar to previous findings of resistance in animals in China (Li et al., 2022). These high levels of drug resistance and prevalence of MDR P. mirabilis on these farms pose a grave public health threat and would render clinical treatment protocols problematic.
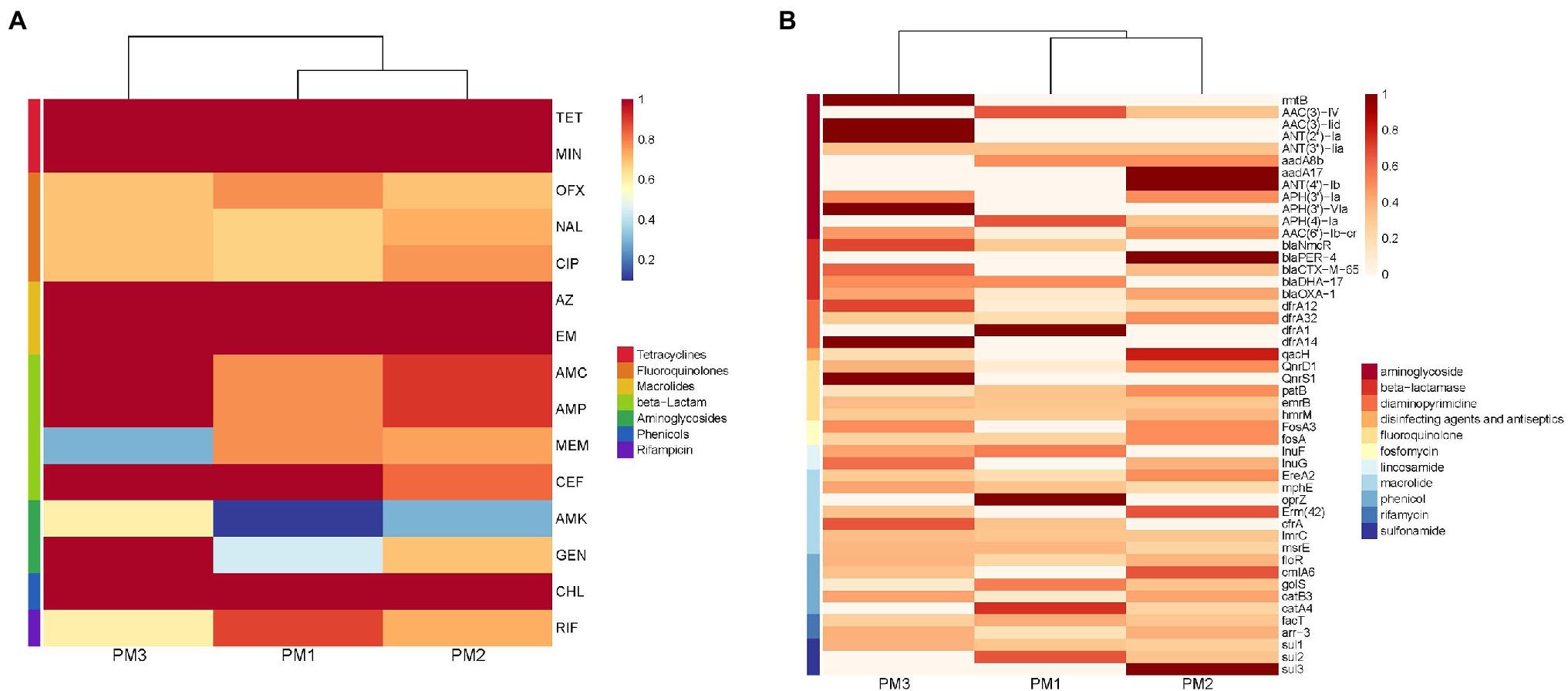
Figure 4. Heatmap of antimicrobial resistance of P. mirabilis isolates according to (A) phenotype and (B) genotype.
Genomic analysis identified the presence of 91 ARGs in our isolates that encoded resistance to 30 antimicrobial classes. The most prevalent (total 43 genes) resistance mechanisms were antibiotic efflux pumps (42.86%), antibiotic inactivation (27.47%), antibiotic target alteration (15.38%), antibiotic target protection (6.59%) and antibiotic target replacement (7.69%). Interestingly, 43 (47.25%) ARGs were present in all isolates. These ARGs included members of the ATP-binding cassette (ABC), major facilitator superfamily (MFS), pmr phosphoethanolamine transferase, resistance-nodulation-cell division (RND), glycopeptide resistance gene cluster, Acinetobacter mutant lpx gene conferring resistance to colistin and ABC-F ATP-binding cassette ribosomal protection protein MDR gene family members bcr-1, tet(J), tetA(48), msbA, mdtB, and mexA (Supplementary Table S1).
The remaining 48 ARGs were variably present among the isolates. These were associated with different antibiotic classes: blaCTX-M-65, blaDHA-17, blaNmcR, blaOXA-1, blaPER-4, and blaPER-4 for β-lactams; sul1, sul2, sul3 for sulfonamides; rmtB and AAC, ANT, APH groups for aminoglycosides, dfrA groups for trimethoprim, lnu for lincosamide, ereA2, mphE, emrB for macrolides; qnrD1, qnrS1 for quinolones; fos for fosfomycin; fact, arr-3 for rifamycin and cml, floR, cat for phenicols. In particular, >50% of these isolates contained hmrM, emrB, ImrC, floR, sul1, fact, arr-3, blaOXA-1, catB3 and AAC(6′)-Ib-cr that encode resistance to fluoroquinolones, macrolides, phenicols, sulfonamides, rifamycin, aminoglycosides, and β-lactams, respectively. We also detected the qacH gene in 5 isolates; this gene encodes resistance to disinfecting agents and antiseptics (Figure 5C).
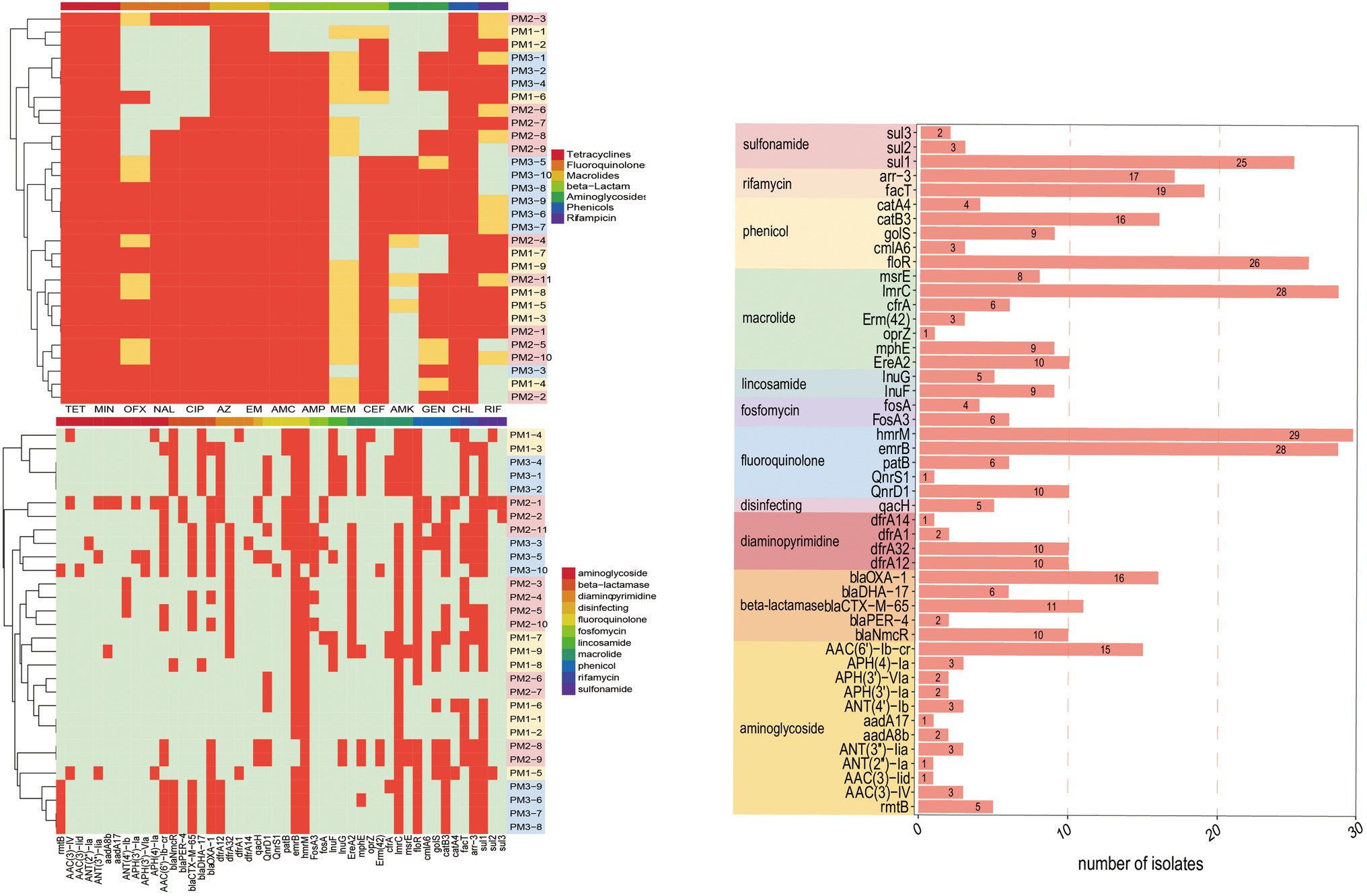
Figure 5. ARGs present in P. mirabilis genomes of study isolates. (A) Phenotype; PM 1–5, PM 2–12 were resistant to all the tested drugs. (B) Genotype, excepting ARGs that all strains contained. (C) Fluoroquinolone, macrolide, phenicol, and sulfonamide prevalence. Red, positive; green, negative.
The extended-spectrum β-lactamases (ESBL) were reported for the first time in 1983 (Knothe et al., 1983) and P. mirabilis is highly represented as an ESBL host. These representatives include the cephalosporinase (AmpC) and TEM and CTX-M type carbapenemases that can degrade penicillin, piperacillin, and the cephalosporins (Bontron et al., 2019). We found the presence of blaCTX-M in our P. mirabilis isolates that enables resistance to ceftiofur while piperacillin and cephalosporin resistance was linked to the presence of blaOXA-1 gene (Algammal et al., 2021). In addition, both blaOXA-1 and blaCTX-M genes synergistically enable P. mirabilis to resist β-lactams combination treatments (Schultz et al., 2015). In the current study, our isolates displayed a high prevalence of blaOXA-1 (53.33%), blaCTX-M-65 (36.67%), blaNmcR (33.33%), blaDHA-17 (20%), and blaPER-4 (13.33%) that are β-lactams resistance gene (Figure 5C). Remarkably, 10 isolates contained both blaOXA-1 and blaCTX-M-65 and 7 in PM3 that also displayed the highest phenotypic resistance to the β-lactams ceftiofur, ampicillin, and amoxicillin/clavulanic acid (Supplementary Material; Figure 3A). The primary aminoglycoside ARG carried by all isolates in our study was AAC (6′)-lb-cr (50%) and 5 rmtB 16S rRNA methyltransferases (rmt) were detected only in PM3. Resistance to amikacin was also prevalent in this group (Figure 4A). The rmtB gene is the most frequently detected ARG in Enterobacteriaceae from humans and animals and mediates a very high-level resistance to aminoglycosides including amikacin and is usually associated with MDR bacteria (Fang et al., 2019) and was consistent with our results. We also detected a large number of tetracycline, macrolide, peptide and fluoroquinolone resistance genes and PM3 displayed resistance to tetracycline, macrolide and phenicols (Figures 4A,B). Furthermore, we evaluated the relationship between phenotypic drug resistance and the presence or absence of corresponding ARGs. All the isolates were resistant to tetracycline, minocycline, azithromycin, erythromycin, and chloramphenicol ARG, and 76.67% resistant to ciprofloxacin, ceftiofur, and gentamicin, and 60% to meropenem and possessed the cognate ARGs. However, genotypic and phenotypic resistance are sometimes not directly linked although the drug-resistant phenotypes and genotypes of most bacteria were consistent (Table 3). Eight strains did not contain any β-lactamase resistance and only one (PM2-3) was sensitive to all the β-lactams, 6 were partially sensitive and PM1-9 displayed resistance similar to the other 22 isolates. For rifampicin, fluoroquinolone, and aminoglycoside resistance, we could not identify any significant correlation between genotype and phenotype and resistance may be mediated by efflux pump mechanisms (Supplementary Table S2).
Our results indicated that the distribution of drug-resistant phenotypes was correlated with genotypes for β-lactams and aminoglycosides although these differed by degree. These may due to the complex drug resistance mechanisms in agreement with a study indicating that the phenotypic test and not possession of a specific ARG is the gold standard for assessment of bacterial drug resistance (Wu et al., 2021).
The presence of MGEs in P. mirabilis are a great public health threat due to the capacity for autonomous ARG transfer (Partridge et al., 2018). ICEs are integrated DNA regions in the chromosome that are mobilizable by conjugation via type IV secretion systems that mediate cell to cell DNA transfer (Delavat et al., 2017). Tyrosine and serine recombinases can mediate att site-specific recombination between circular ICE and chromosomal targets (Johnson and Grossman, 2015). IMEs can be mobilized using ICE machinery or conjugative plasmids when ICEs erode and accumulate within the host chromosome leading to inactivation of their independent mobility (Partridge et al., 2018).
In our study, we identified 15 ICEs and 7 IMEs and 53.33% of our isolates contained ICEs or IMEs with 13 of 30 containing ICEs and 6 of 30 containing IMEs (Table 4). These results were higher than previously reported (23.53%) in P. mirabilis from the tree shrew (Gu et al., 2020). Most MGEs were detected in PM3 (90%) and 2 PM1 strains possessed the same ICE type (Table 5). This indicated that transconjugation was frequent for this group. Moreover, strains with ICEs from the same group carried similar resistance genes especially in PM1 and PM3. All of the PM1 strains with MGEs (PM1-7, PM1-8, PM1-9) were clustered. Strains with MGEs in PM3 gathered into 3 small groups and carried the same resistance gene (PM3-8, PM3-9 and PM3-1, PM3-2, PM3-4; Figure 5 and Table 5). Such results indicate the serious drug resistance of PM3 group may be associated with MGEs. The high MDR prevalence could be attributed to a combination of these factors. However, we did not perform a validation analysis of these detected MGE functions and further analyses are needed to understand the effect of these elements on drug resistance.
Most of the endemic strains in our study were MDR and the number of virulence genes identified would contribute to the establishment of persistent and drug-tolerant infections. In our 30 isolates, we identified 95 of 2,741 possible virulence factors composed of 10 categories: transporter, fimbriae, flagella, hemin, metal, enzyme, protein, regulator, type VI secretion system, and adhesin. Enzyme represented 22 virulence factors related to factors such as DP-heptose synthase, UDP-glucose 6-dehydrogenase, and decarboxylase. The flagella and fimbrial groups were also abundant and 20 different genes were represented. The least abundant were the type VI secretion system and adhesin (Figure 6).
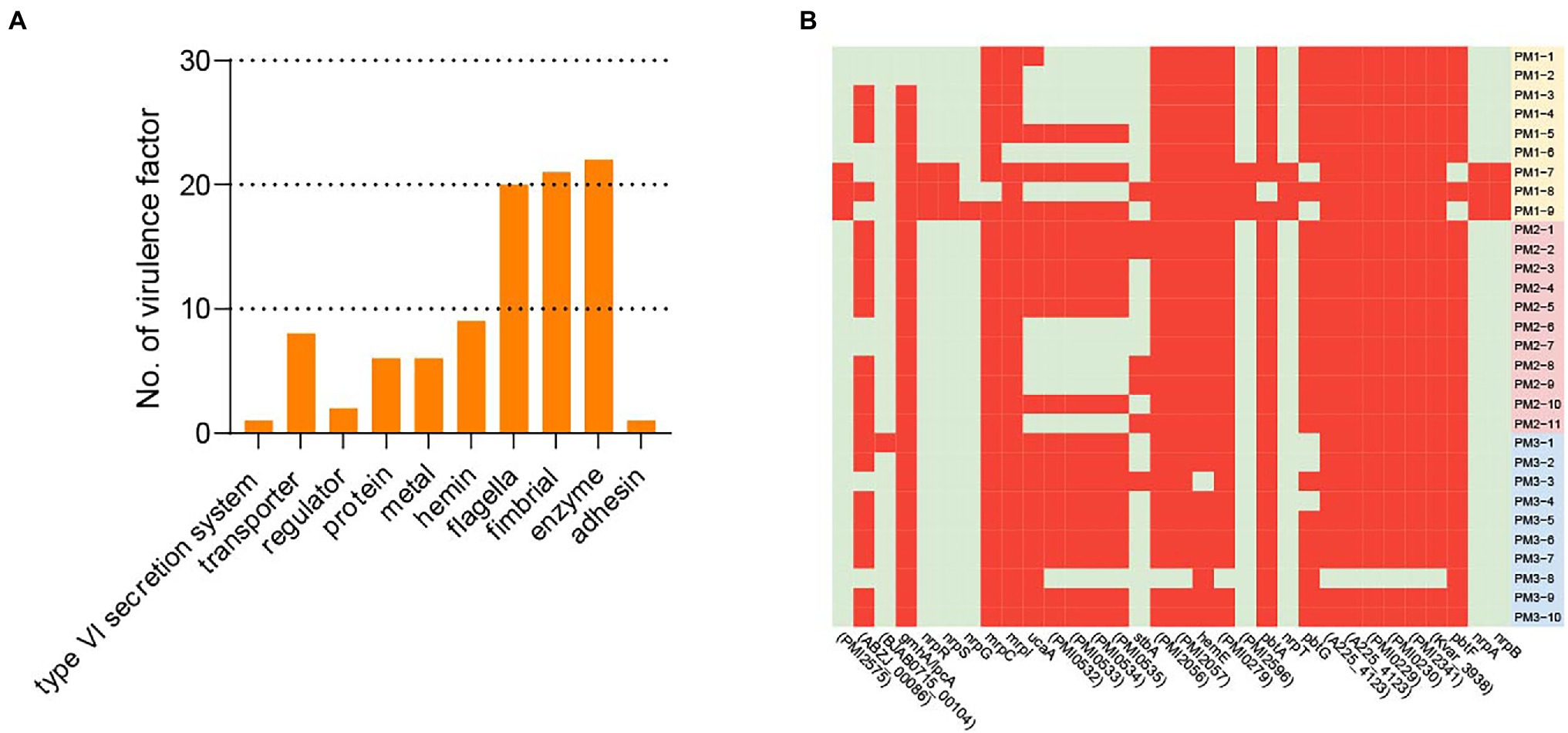
Figure 6. Virulence factors. (A) Virulence factor categories and the number represented in each category. (B) Virulence factors that were variably present among isolates. Red, positive; green, negative.
All isolates contained 63/95 virulence factors associated with biofilm formation, adhesin, carbon storage, hemin, metal and motility and 32/95 were variably present among the 30 isolates. All strains contained at least 8 of the 32 variable virulence genes that included the categories transporter, Proteus-like (MR/P) fimbriae, protein, adhesin, enzyme, fimbrial, metal, and regulator that included ABC transporter permease, hemolysin activator protein, fimbrial adhesin, IS4 family transposase ORF 2, siderophore biosynthesis protein, and non-ribosomal peptide synthase. The PM3 group contained the largest numbers of virulence factors while 10 isolates did not contain any virulence factors related to UCA in the PM1 and PM2 groups (Figure 6B, Supplementary Table S2).
The virulence factors for the MR/P and PMF fimbriae and UCA are considered essential in the initial phase of infection and contribute significantly to UTI development (Cook et al., 1995; Jansen et al., 2004; Govindarajan and Kandaswamy, 2022). Aggregation and initial biofilm formation are mediated by MR/P fimbriae that are products of the mrp operon (Wasfi et al., 2020). The UCA fimbriae are controlled via the ucaA gene and PMF confers bladder colonization. All of our isolates contained PMF and 20 contained ucaA and most contained 5 other UCA-related virulence genes (Supplementary Table S3). Moreover, we found 20 putative fimbrial operons and this is the largest number found in any sequenced bacterial species (Scavone et al., 2016).
P. mirabilis is highly mobile and swarm ability allows it to pass through catheters to the urinary tract. Flagella are important to swarmer cell differentiation and are necessary to swarming motility (Belas and Suvanasuthi, 2005). In our study, we found all the isolates carried 20 different flagellar virulence factors including flgI, film, and flgC. Most importantly, 29 isolates except PM3-8 contained the ZapA virulence factor (Belas et al., 2004) that enables the bacterium to survive in the urethra via degradation of host proteins using the Zap1 metalloproteinase (Supplementary Table S3). Our findings indicated the P. mirabilis isolated from healthy pigs is a potential threat to public health since its high prevalence and possession of virulence genes and ARGs can be transferred via MGEs.
In this study, we conducted the prevalence and characteristics of P. mirabilis from healthy farmed pigs in China. Phylogenetic analysis indicated the isolates were closely related and clustered into 2 major groups. Our 30 isolates harbored 91 ARGs and >50% contained MGEs as well as 95 different virulence factors that included the presence of 20 putative fimbrial operons. Most (90%) of the isolates were strong biofilm formers. This is the first study to report the genomic investigation and prevalence of P. mirabilis associated with pig farm operations in Zhejiang, China.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XX and CX: writing—review and editing. XQ and JZ: investigating and writing—original draft. HH and WW: data curation. YX, BT, and HL: resources. All authors contributed to manuscript revision, read, and approved the submitted version.
This research was supported by the Key Research and Development Program of Zhejiang Province (2022C02049), Ministry of Agriculture and Rural Affairs (14215033), and the Walmart Foundation (UA2020-152 and UA2021-247).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.952982/full#supplementary-material
1. ^http://www.bcgsc.ca/platform/bioinfo/software/abyss
2. ^https://sourceforge.net/projects/soapdenovo2/files/GapCloser/
Algammal, A. M., Hashem, H. R., Alfifi, K. J., Hetta, H. F., Sheraba, N. S., Ramadan, H., et al. (2021). atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci. Rep. 11:9476. doi: 10.1038/s41598-021-88861-w
Belas, R., Manos, J., and Suvanasuthi, R. (2004). Proteus mirabilis ZapA metalloprotease degrades a broad spectrum of substrates, including antimicrobial peptides. Infect. Immun. 72, 5159–5167. doi: 10.1128/iai.72.9.5159-5167.2004
Belas, R., and Suvanasuthi, R. (2005). The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J. Bacteriol. 187, 6789–6803. doi: 10.1128/jb.187.19.6789-6803.2005
Bi, S.-I., Tang, S.-z., Wu, X.-y., and Chen, S.-y. (2013). Quantitative detection of Proteus species by real-time polymerase chain reaction using SYBR green I. Ann. Microbiol. 63, 1205–1208. doi: 10.1007/s13213-012-0519-3
Biswas, S., Elbediwi, M., Gu, G., and Yue, M. (2020). Genomic characterization of new variant of hydrogen sulfide (H(2)S)-producing Escherichia coli with multidrug resistance properties carrying the mcr-1 gene in China †. Antibiotics (Basel) 9:80. doi: 10.3390/antibiotics9020080
Bontron, S., Poirel, L., Kieffer, N., Savov, E., Trifonova, A., Todorova, I., et al. (2019). Increased resistance to Carbapenems in Proteus mirabilis mediated by amplification of the Bla(VIM-1)-carrying and IS26-associated class 1 Integron. Microb. Drug Resist. 25, 663–667. doi: 10.1089/mdr.2018.0365
Chen, C.-M., Lai, C.-H., Wu, H.-J., and Wu, L.-T. (2017). Genetic characteristic of class 1 integrons in proteus mirabilis isolates from urine samples. Biomedicine 7:9. doi: 10.1051/bmdcn/2017070202
Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. doi: 10.1093/nar/gki008
CLSI (2017). Performance Standards for Antimicrobial Susceptibility Testing, 27th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Cook, S. W., Mody, N., Valle, J., and Hull, R. (1995). Molecular cloning of Proteus mirabilis uroepithelial cell adherence (uca) genes. Infect. Immun. 63, 2082–2086. doi: 10.1128/iai.63.5.2082-2086.1995
Delavat, F., Miyazaki, R., Carraro, N., Pradervand, N., and van der Meer, J. R. (2017). The hidden life of integrative and conjugative elements. FEMS Microbiol. Rev. 41, 512–537. doi: 10.1093/femsre/fux0008
Drzewiecka, D. (2016). Significance and roles of Proteus spp. bacteria in natural environments. Microbiol. Ecol. 72, 741–758. doi: 10.1007/s00248-015-0720-6
Fang, L. X., Deng, G. H., Jiang, Q., Cen, D. J., Yang, R. S., Feng, Y. Y., et al. (2019). Clonal expansion and horizontal transmission of epidemic F2:A1:B1 plasmids involved in co-spread of rmtB with qepA and blaCTX-M-27 in extensively drug-resistant Salmonella enterica serovar Indiana isolates. J. Antimicrob. Chemother. 74, 334–341. doi: 10.1093/jac/dky441
Gilbert, N. (2012). Rules tighten on use of antibiotics on farms. Nature 481:125. doi: 10.1038/481125a
Gong, Z., Shi, X., Bai, F., He, X., Zhang, H., Li, Y., et al. (2019). Characterization of a novel Diarrheagenic strain of Proteus mirabilis associated with food poisoning in China. Front. Microbiol. 10:2810. doi: 10.3389/fmicb.2019.02810
Govindarajan, D. K., and Kandaswamy, K. (2022). Virulence factors of uropathogens and their role in host pathogen interactions. Cell Surf. 8:100075. doi: 10.1016/j.tcsw.2022.100075
Gu, W., Wang, W., Tong, P., Liu, C., Jia, J., Lu, C., et al. (2020). Comparative genomic analysis of Proteus spp. isolated from tree shrews indicated unexpectedly high genetic diversity. PLoS One 15:e0229125.
Han, X., Liu, L., Fan, G., Zhang, Y., Xu, D., Zuo, J., et al. (2015). Riemerella anatipestifer lacks luxS, but can uptake exogenous autoinducer-2 to regulate biofilm formation. Res. Microbiol. 166, 486–493. doi: 10.1016/j.resmic.2015.06.004
Høiby, N., Ciofu, O., Johansen, H. K., Song, Z. J., Moser, C., Jensen, P., et al. (2011). The clinical impact of bacterial biofilms. Int. J. Oral Sci. 3, 55–65. doi: 10.4248/ijos11026
Hu, Y. Y., Cai, J. C., Zhang, R., Zhou, H. W., Sun, Q., and Chen, G. X. (2012). Emergence of Proteus mirabilis harboring blaKPC-2 and qnrD in a Chinese hospital. Antimicrob. Agents Chemother. 56, 2278–2282. doi: 10.1128/aac.05519-11
Jansen, A. M., Lockatell, V., Johnson, D. E., and Mobley, H. L. (2004). Mannose-resistant Proteus-like fimbriae are produced by most Proteus mirabilis strains infecting the urinary tract, dictate the in vivo localization of bacteria, and contribute to biofilm formation. Infect. Immun. 72, 7294–7305. doi: 10.1128/iai.72.12.7294-7305.2004
Johnson, C. M., and Grossman, A. D. (2015). Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet. 49, 577–601. doi: 10.1146/annurev-genet-112414-055018
Kang, Q., Wang, X., Zhao, J., Liu, Z., Ji, F., Chang, H., et al. (2021). Multidrug-resistant Proteus mirabilis isolates carrying bla(OXA-1) and bla(NDM-1) from wildlife in China: increasing public health risk. Integr. Zoo. 16, 798–809. doi: 10.1111/1749-4877.12510
Klein, E. Y., Van Boeckel, T. P., Martinez, E. M., Pant, S., Gandra, S., Levin, S. A., et al. (2018). Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. U. S. A. 115, e3463–e3470. doi: 10.1073/pnas.1717295115
Knothe, H., Shah, P., Krcmery, V., Antal, M., and Mitsuhashi, S. (1983). Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11, 315–317. doi: 10.1007/bf01641355
Lajhar, S. A., Brownlie, J., and Barlow, R. (2018). Characterization of biofilm-forming capacity and resistance to sanitizers of a range of E. coli O26 pathotypes from clinical cases and cattle in Australia. BMC Microbiol. 18:41. doi: 10.1186/s12866-018-1182-z
Larson, C. (2015). Pharmaceuticals. China’s lakes of pig manure spawn antibiotic resistance. Science 347:704. doi: 10.1126/science.347.6223.704
Lei, C. W., Zhang, A. Y., Wang, H. N., Liu, B. H., Yang, L. Q., and Yang, Y. Q. (2016). Characterization of SXT/R391 integrative and conjugative elements in Proteus mirabilis isolates from food-producing animals in China. Antimicrob. Agents Chemother. 60, 1935–1938. doi: 10.1128/aac.02852-15
Li, N. (2021). Isolation and identification of swine-sourced Proteus mirabilis, and detection of its drug resistance and virulence genes. Ani. Husb. Vet. Med. 53, 89–94.
Li, Z., Peng, C., Zhang, G., Shen, Y., Zhang, Y., Liu, C., et al. (2022). Prevalence and characteristics of multidrug-resistant Proteus mirabilis from broiler farms in Shandong Province, China. Poult. Sci. 101:101710. doi: 10.1016/j.psj.2022.101710
Liu, Q., Chen, W., Elbediwi, M., Pan, H., Wang, L., Zhou, C., et al. (2020). Characterization of Salmonella Resistome and Plasmidome in pork production system in Jiangsu, China. Front. Vet. Sci. 7:617. doi: 10.3389/fvets.2020.00617
Liu, Y., Jiang, J., Ed-Dra, A., Li, X., Peng, X., Xia, L., et al. (2021). Prevalence and genomic investigation of Salmonella isolates recovered from animal food-chain in Xinjiang, China. Food Res. Int. 142:110198. doi: 10.1016/j.foodres.2021.110198
Liu, M., Li, X., Xie, Y., Bi, D., Sun, J., Li, J., et al. (2019). ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47, d660–d665. doi: 10.1093/nar/gky1123
Marshall, B. M., and Levy, S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24, 718–733. doi: 10.1128/cmr.00002-11
Mirzaei, A., Habibi, M., Bouzari, S., and Karam, M. R. A. (2019). Characterization of antibiotic-susceptibility patterns, virulence factor profiles and clonal relatedness in Proteus mirabilis isolates from patients with urinary tract infection in Iran. Infect. Drug Resist. 12, 3967–3979. doi: 10.2147/idr.S230303
Mobley, H. L., and Belas, R. (1995). Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol. 3, 280–284. doi: 10.1016/s0966-842x(00)88945-3
Olonitola, O. S., Fahrenfeld, N., and Pruden, A. (2015). Antibiotic resistance profiles among mesophilic aerobic bacteria in Nigerian chicken litter and associated antibiotic resistance genes1. Poult. Sci. 94, 867–874. doi: 10.3382/ps/pev069
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-17. doi: 10.1128/cmr.00088-17
Qian, X., Gu, J., Sun, W., Wang, X. J., Su, J. Q., and Stedfeld, R. (2018). Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting. J. Hazard. Mater. 344, 716–722. doi: 10.1016/j.jhazmat.2017.11.020
Reich, F., Atanassova, V., and Klein, G. (2013). Extended-spectrum β-lactamase- and AmpC-producing enterobacteria in healthy broiler chickens, Germany. Emerg. Infect. Dis. 19, 1253–1259. doi: 10.3201/eid1908.120879
Sanchez, C. J. Jr., Mende, K., Beckius, M. L., Akers, K. S., Romano, D. R., Wenke, J. C., et al. (2013). Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect. Dis. 13:47. doi: 10.1186/1471-2334-13-47
Scavone, P., Iribarnegaray, V., Caetano, A. L., Schlapp, G., Härtel, S., and Zunino, P. (2016). Fimbriae have distinguishable roles in Proteus mirabilis biofilm formation. Pathog. Dis. 74:ftw033. doi: 10.1093/femspd/ftw033
Schultz, E., Haenni, M., Mereghetti, L., Siebor, E., Neuwirth, C., Madec, J. Y., et al. (2015). Survey of multidrug resistance integrative mobilizable elements SGI1 and PGI1 in Proteus mirabilis in humans and dogs in France, 2010-13. J. Antimicrob. Chemother. 70, 2543–2546. doi: 10.1093/jac/dkv154
Shelenkov, A., Petrova, L., Fomina, V., Zamyatin, M., Mikhaylova, Y., and Akimkin, V. (2020). Multidrug-resistant Proteus mirabilis strain with cointegrate plasmid. Microorganisms 8:1775. doi: 10.3390/microorganisms8111775
Shi, X., Li, Y., Yang, Y., Shen, Z., Cai, C., Wang, Y., et al. (2021). High prevalence and persistence of carbapenem and colistin resistance in livestock farm environments in China. J. Hazard. Mater. 406:124298. doi: 10.1016/j.jhazmat.2020.124298
Stepanović, S., Vuković, D., Hola, V., Di Bonaventura, G., Djukić, S., Cirković, I., et al. (2007). Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115, 891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x
Stock, I. (2003). Natural antibiotic susceptibility of Proteus spp., with special reference to P. mirabilis and P. penneri strains. J. Chemother. 15, 12–26. doi: 10.1179/joc.2003.15.1.12
Tabatabaei, A., Ahmadi, K., Shabestari, A. N., Khosravi, N., and Badamchi, A. (2021). Virulence genes and antimicrobial resistance pattern in Proteus mirabilis strains isolated from patients attended with urinary infections to tertiary hospitals, in Iran. Afr. Health Sci. 21, 1677–1684. doi: 10.4314/ahs.v21i4.22
The Ministry of Agriculture of the People’s Republic of China. (2002) Maximum residue limits for veterinary drugs in animal products. Available at: https://www.moa.gov.cn/gk/tzgg_1/gg/200302/t20030226_59300.htm (Accessed December 24, 2002).
Tseng, B. S., Zhang, W., Harrison, J. J., Quach, T. P., Song, J. L., Penterman, J., et al. (2013). The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ. Microbiol. 15, 2865–2878. doi: 10.1111/1462-2920.12155
Wang, Q., Peng, K., Liu, Y., Xiao, X., Wang, Z., and Li, R. (2021). Characterization of TMexCD3-TOprJ3, an RND-type efflux system conferring resistance to tigecycline in Proteus mirabilis, and its associated integrative conjugative element. Antimicrob. Agents Chemother. 65:e0271220. doi: 10.1128/aac.02712-20
Wasfi, R., Hamed, S. M., Amer, M. A., and Fahmy, L. I. (2020). Proteus mirabilis biofilm: development and therapeutic strategies. Front. Cell. Infect. Microbiol. 10:414. doi: 10.3389/fcimb.2020.00414
Keywords: antimicrobial resistance, biofilm information, whole genome sequencing, virulence factor, Proteus mirabilis
Citation: Qu X, Zhou J, Huang H, Wang W, Xiao Y, Tang B, Liu H, Xu C and Xiao X (2022) Genomic Investigation of Proteus mirabilis Isolates Recovered From Pig Farms in Zhejiang Province, China. Front. Microbiol. 13:952982. doi: 10.3389/fmicb.2022.952982
Received: 25 May 2022; Accepted: 21 June 2022;
Published: 07 July 2022.
Edited by:
Qingli Dong, University of Shanghai for Science and Technology, ChinaReviewed by:
Fei Jia, University of Arkansas, United StatesCopyright © 2022 Qu, Zhou, Huang, Wang, Xiao, Tang, Liu, Xu and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chenggang Xu, Y2hnYW5neHVAMTI2LmNvbQ==; Xingning Xiao, eGluZ25pbmd4aWFvQDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.