
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 August 2022
Sec. Microorganisms in Vertebrate Digestive Systems
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.950587
This article is part of the Research Topic Recent Advances and Perspectives on the Gastrointestinal Microbiota of Small Ruminants View all 31 articles
 Jia Zhou1
Jia Zhou1 Benchu Xue1
Benchu Xue1 Anhai Hu1
Anhai Hu1 Shuangming Yue2
Shuangming Yue2 Mei Wu1
Mei Wu1 Qionghua Hong3
Qionghua Hong3 Yuhan Wu1
Yuhan Wu1 Zhisheng Wang1
Zhisheng Wang1 Lizhi Wang1
Lizhi Wang1 Quanhui Peng1
Quanhui Peng1 Bai Xue1*
Bai Xue1*Physically effective neutral detergent fiber (peNDF) is a concept that accounts for the particle length of NDF in diets, sustaining the normal chewing behavior and rumen fermentation of ruminants. Specifically, peNDF>1.18 is the commonest one that is calculated from NDF and the percentage of feed dry matter left on the 1.18, 8.00, and 19.00 mm sieves. This study aimed to investigate the effects of different levels of peNDF>1.18 on the rumen microbiome and its correlation with nutrient digestibility and rumen fermentation in goats. A total of 30 Lezhi black goats were randomized and blocked to five dietary treatments (n = 6). All the diets were identical in composition but varied in hay lengths, leading to the different peNDF>1.18 content of the diets: 32.97, 29.93, 28.14, 26.48, and 24.75%. The results revealed that the nutrient digestibility increased when dietary peNDF>1.18 levels decreased from 32.97% to 28.14%, with the highest digestibility at 28.14% peNDF>1.18 treatment, after which nutrient digestibility decreased with the decreasing of dietary peNDF levels. Ruminal NH3-N concentrations in the 29.93% and 28.14% groups were higher than that in the 24.75% group (p < 0.05). Ruminal microbial protein concentration was the highest in the 32.97% group (p < 0.05). Daily CH4 production in the 32.97% and 24.75% peNDF>1.18 treatments was lower than that in the 26.48% group (p < 0.05) and no differences were observed among other groups. The relative abundance of rumen fungi at the phylum and genus levels and archaea at the species were affected by dietary peNDF>1.18 content. In conclusion, decreasing dietary peNDF>1.18 levels within a certain range can improve nutrient digestibility and change the rumen microbial community structure of goats. Dietary peNDF>1.18 level should be 28.14% (roughage length around 1 cm) among the five levels for 4 months Lezhi black goats with the purpose of optimal nutrient digestibility.
Forages comprise more than 40% of the diets for ruminants and are essential to maintain an appropriate rumen function and physiology (Bargo et al., 2002). The high fiber content is the major nutritional difference between forages and concentrates, resulting in a lower energy value of forages. Unlike monogastric animals, the rumen of ruminants is colonized with an abundance of microorganisms, which are capable of converting fiber in feed into available energy for the host. Meanwhile, fiber can stimulate chewing, salivation, rumination, and ruminal motility of ruminants. It also plays an important role in alleviating rumen acidosis, regulating dietary intake, synthesizing milk fat, and promoting the digestion of solid particles in the rumen (Adesogan et al., 2019). Consequently, evaluating and improving the utilization efficiency of dietary fiber is particularly important for the formulation of diets for ruminants.
In order to assess the adequacy of dietary fiber for ruminants, the concept of physically effective neutral detergent fiber (peNDF) was proposed by Mertens (Mertens, 1997) and attracted more and more attention. The peNDF is more efficient to indicate the physical effects of a diet because it incorporates information on particle size and chemical fiber content of the feedstuffs (Zebeli et al., 2012) and reflects the physical characteristics that influence masticatory activity and stimulate stratification of the rumen (Zhao et al., 2011). It seems to be an ideal and versatile method to calculate the peNDF by separating the diet or forage into pellets of various grain sizes using a manually operated Penn State Particle Separator (PSPS). The peNDF content in feedstuffs or forages could be determined by their neutral detergent fiber (NDF) content of them multiplied by the proportion of pellet retained on the 1.18-mm sieve or 8.00-mm sieve, defined as peNDF>1.18 and peNDF>8.00, respectively (Zebeli et al., 2012).
The rumen is a very important digestive organ of ruminants, hosting a large number of microorganisms that can effectively degrade proteins and carbohydrates through fermentation. The efficiency of rumen digestion mainly depends on microorganisms that may be affected by dietary peNDF (Li et al., 2014). It was well documented that increasing the dietary peNDF content resulted in increased time for ruminating and chewing (Yang and Beauchemin, 2007; Zhao et al., 2011; Li et al., 2014). However, the effects of dietary peNDF levels on nutrient digestibility are not consistent in the literature. Several recent studies revealed that the increase of dietary peNDF content could reduce the total tract apparent digestibility of dry matter (DM), crude protein (CP), and NDF in dairy cattle (Soita et al., 2002; Molavian et al., 2020) and lambs (de Paula Carlis et al., 2021). Other studies found that the nutrients digestibility was quadratically related to dietary peNDF content (Zhao et al., 2011; Wang et al., 2017), or not affected by peNDF (Oh et al., 2016; Jang et al., 2017; Kaeokliang et al., 2019; Li et al., 2020). In addition, nutrient digestibility is influenced by the rumen microbiome (Li and Guan, 2017). Previous studies have reported that dietary fiber is an important factor influencing rumen microbial composition, diversity, and microbial metabolites (Billenkamp et al., 2021; Yildirim et al., 2021). But till now, few studies have been conducted concerning the effects of dietary peNDF on rumen microbial community structure and the relationship between rumen microbiota and nutrient digestibility, especially fungi.
The unique rumen structure of ruminants allows goats to effectively utilize some unconventional feed resources such as straw and forage. To make more efficient use of feed resources such as straw, it is crucial to evaluate the nutritional value and its effect on various aspects including nutrient digestibility, rumen fermentation, and rumen microbial communities, which could dramatically influence the production performance and health condition of ruminant (Cao et al., 2021), thereby providing useful information and guidance for improving the production efficiency of goats. We hypothesized that variation in the dietary peNDF levels could affect nutrient digestibility, rumen fermentation, and rumen microbial diversity. The objective of this study was to investigate the effects of dietary peNDF levels on nutrient digestibility, rumen fermentation, and rumen microbial communities of Lezhi black goats, by analyzing the predicted functional differences of different ruminal microorganisms.
All experimental protocols were approved by the Animal Ethical and Welfare Committee of Animal Nutrition Institute, Sichuan Agricultural University (Approval No.201408-3).
This study was a part of a larger project which aimed to explore the effect of dietary peNDF level on the development of the digestive tract in growing goats. Animal feeding and management were described previously by Xue et al. (2022). In brief, a total of 30 male Lezhi black goats, aged 4.0 ± 0.1 months and weighed 21.42 ± 0.24 kg, were randomly blocked into five treatments, with six replicates per treatment and one goat per replicate. The diet composition and nutrient levels were the same for the five treatments except for the level of peNDF. The diets were formulated according to the nutrient requirements of 20 kg goats with a daily weight gain of 100 g by NRC (2007), and the ingredients and chemical composition as shown in Table 1.
Dietary peNDF levels were controlled by cutting the forage (Leymus chinensis, alfalfa, peanut vine, all of them are hays) at the following lengths with a forage cutter (FS60, Jining Nongfengli Machinery Equipment Co., Ltd., Jining, China): long (7 cm), medium (4 cm), short (1 cm), fine (5 mm sieve), very fine (1 mm sieve). Particle size distribution of forages and mixed diets was determined using the PSPS containing three sieves (19.00, 8.00, and 1.18 mm) and 1 pan (receive particles <1.18 mm), as previously described (Cao et al., 2013). Physically effective factors (pef) for forage and diets are calculated as the sum of DM proportion retained on the sieves, referring to the ability of a specific kind of feed to stimulate animals to chew, which range from 0 to 1.0 (Zebeli et al., 2012). For example, pef>1.18 and pef>8.00 equal to the sum of the DM proportion retained on sieves (19.00, 8.00, and 1.18 mm) and the sum of the DM proportion retained on two sieves (19.00 and 8.00 mm). The value of peNDF in the diet is calculated as follows: peNDF (% of diet) =NDF × pef, where NDF presents the percentage of NDF in the diet (Yang and Beauchemin, 2005). Based on this, peNDF>1.18 and peNDF>8.00 could be calculated as the dietary NDF content multiplied by pef>1.18 and pef>8.00, respectively.
In this study, we distinguished the five treated diets based on the difference in the content of peNDF>1.18, as fed <1.18 mm in length can pass through the rumen to the following digestive tract (Maulfair et al., 2011), while the feed particles retained on the 1.18 mm sieve have high resistance to passing through the rumen, resulting in increased chewing and rumination activities (Poppi et al., 1981). The concentrates and forages were mixed and stirred on a total mixed ration (TMR) machine (Shandong Shengshi Machinery Manufacturing Co., Ltd, Qufu, Shandong) for 2 min before feeding. TMR was supplied to the goats twice daily at 08: 00 and 20: 00 for ad libitum intake (at least 10% orts). Clean water was available ad libitum throughout the study.
All the animals were fed in a single cage for a 30-day formal feeding trial, after a 14-d adaptation period. Total collection of feces was conducted on day 7 of the formal period for six consecutive days till day 12. On days 13 to 28, three goats in each treatment were selected for methane (CH4) measurements in four separate open-circuit respiratory chambers. These goats stayed in chambers for four consecutive days with a 2-day adaptation period and a 2-day sampling and measurement period.
Before feeding on days 7 to 12, total fecal samples were collected by grab sampling two times daily, weighed, and recorded. A 10% aliquot of each fecal sample was removed and immediately stored in a freezer (−20°C) until further analysis. At the end of the feeding trial (day 30), 15 goats (three goats from each treatment) were slaughtered after electric shock and the others were continually kept in pens. Samples of rumen fluid and rumen chyme were collected from rumen immediately after slaughtering. Approximately 50 ml of rumen fluid was collected from each goat by filtering rumen content through four layers of gauze and then divided into two aliquots after pH determination using a PHS-3B acidometer (Shanghai INESA Scientific Instrument Co., Ltd., Shanghai, China). One aliquot was transferred into sterilized tubes and placed in liquid nitrogen as soon as possible and was taken to the laboratory and stored in a refrigerator at −80°C for DNA extraction. Another aliquot was aliquoted and transferred into sterilized tubes and stored at −80°C for chemical assay of volatile fatty acids (VFAs), ammonia nitrogen (N-NH3), and microbial crude protein (MCP) concentrations.
An open-circuit respiratory chamber was used for CH4 measurements, and the CH4 production was calculated as described previously (Aguilera and Prieto, 1986). Three goats in each treatment were selected for CH4 measurements, with one goat in each chamber. The total volume of 6 m3 (2.5 m long, 1.5 m wide, and 1.6 m high) of each chamber was ventilated by suction pumps set a range of 16–20 m3/h, allowing a slight negative pressure within the chambers. Temperature and relative humidity were set at 25 ± 1°C and 60 ± 10% respectively with air conditioning units. The CH4 concentration in the air entering and leaving each individual chamber was measured every 10 minutes by using an MGA3000 Multi-Gas Analyzer (ADC Gas Analysis Ltd., Hoddesdon, United Kingdom). The analyzer was calibrated weekly using oxygen-free nitrogen (zero gas) and a known quantity of CH4 (span gas). The flow measurement systems were checked before and immediately after the experiment by releasing analytical grade CH4 into the chambers, by determining the recovery of CH4. The purpose of the calibrations was to ensure a recovery rate of CH4 at a range of 97 to 103%. The concentration of CH4 was analyzed using gas chromatography. Each chamber contained a feed bin, drinking water container, and separate trays.
The contents of DM, organic matter (OM), and crude protein (CP), in feed samples and fecal samples, were analyzed according to AOAC (International, 1990), and the contents of neutral detergent fiber (NDF) and acid detergent fiber (ADF) in these samples were determined according to previously described (Van Soest, 1963). Subsequently, the apparent digestibility of DM, OM, CP, NDF, and ADF were calculated through the formula: apparent nutrient digestibility =100% × [(feed intake × nutrient content in the diet)—(fecal output × nutrient content in the feces)]/ feed intake × nutrient content in the diet (Almeida and Stein, 2010).
The concentration of VFAs in rumen fluid was analyzed with a Varian CP-3800 gas chromatograph (Agilent Technologies, Santa Clara, USA). The thawed rumen fluid samples were centrifuged at 500 × g for 15 min, 0.2 ml of the 25% metaphosphoric acid solution (w/v) and 23.3 μl of the 210 mmol/L crotonic acid solution were added to 1 mL of the supernatant. The mixed solution was incubated at 4°C for 30 min and then centrifuged at 16,000 × g for 10 min to obtain new supernatants. Subsequently, each supernatant sample (0.3 ml) was diluted 4-fold (v/v) with chromatographic grade methanol. In subsequent experiments, 1 μl of the rumen fluid sample was injected using a 50:1 split ratio at 220°C. The chromatographic column was programmed from 100 to 190°C at 20°C/min. Nitrogen was employed as the carrier gas at a constant flow of 1 ml/min. The concentration of NH3-N was measured using UV-3600i Plus spectrophotometrically (Shimadzu, Kyoto, Japan) according to the method described previously (Weatherburn, 1967). Briefly, the thawed samples of the rumen fluid were centrifuged (6,500 × g) at 4°C for 15 min, 2.5 ml of the prepared chromogenic solution (10 g of phenol and 50 mg of sodium nitroprusside to 1 L of solution) and 2 ml of the alkaline hypochlorite (5 g of sodium hydroxide with 50 ml of 5.25% sodium hypochlorite (w/v) per liter of solution) were added to 80 μl of the supernatant. The absorbance was read at the wavelength of 625 nm after the mixed solution was incubated at 37°C for 20 min to calculate the concentration of NH3-N. The microbial protein (MCP) in rumen fluid was obtained by repeated centrifugation as previously reported (Berthiaume et al., 2010) and the concentration of MCP was determined by using bicinchoninic acid (BCA) protein concentration kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). In brief, the thawed samples of the rumen fluid were centrifuged at 500 × g and 4°C for 10 min, then the supernatant was centrifuged at 20,000 × g and 4°C for 30 min. The precipitate was washed with deionized water and centrifuged at 20,000 × g and 4°C for 30 min, repeated 3 times. The final pellet was thoroughly diluted with deionized water, and 10 μl of the dilution was blended with 250 μl reaction fluid at 37°C for 30 min. The MCP content could be calculated by measuring the optical density value at 562 nm in a SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA, USA) according to the manufacturer's instructions.
Total DNA was extracted from each sample by using the FastPure® Microbiome DNA Isolation Kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the protocol provided by the manufacturer. The DNA was quantified by using the NanoDrop® ND-2000 micro-spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The integrity of DNA samples was assessed with 0.8% denatured agarose gel electrophoresis. The specific primers with barcode were used to amplify the V4-V5 region of the 16S ribosomal RNA (rRNA) gene of archaea, and the primer sequences were as follows: 519F (5′-CAGCMGCCGCGGTAA-3′) and 976R (5′-CCGGCGTTGAMTCCAATT-3′) (Teske and Sørensen, 2008). Similarly, the sequences of the primers used to amplify the fungal internal transcribed spacer (ITS) 2-rDNA region were as follows: ITS3_KYO2 (5′-GATGAAGAACGYAGYRAA-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Toju et al., 2012). The amplification was performed by GeneAmp® PCR System 9700 (Applied Biosystems, Inc., Foster City, CA, USA). The amplification conditions of archaea were 94°C for 1 min; 94°C for 20 s, 54°C for 30 s, 72°C for 30 s, repeated for 30 cycles; 72°C for 5 min. The amplification conditions of fungi were 94°C for 1 min; 94°C for 20 s, 48°C for 30 s, 72°C for 30 s, repeated for 30 cycles; 72°C for 5 min. The PCR product of each sample was detected by 2% agarose gel electrophoresis and sent to Rhonin Biotechnology Co., Ltd (Chengdu, China) for high-throughput sequencing analysis on the Illumina HiSeq 2500 (PE250) platform after fluorescent quantization.
The original offline data obtained by sequencing was spliced and filtered to obtain a high-quality target sequence for subsequent analysis. Subsequent bioinformatics operations were completed using Usearch and QIIME4 (Edgar, 2013). Statistics and mapping are mainly done using R5, python, and java. The main steps were as follows: stitching the sequences, distinguishing samples according to barcode, removing the low quality sequences, removing the chimera, clustering the operational taxonomic units (OTUs) at a similarity of 97%, selecting the representative sequence of OTUs, classifying the species using the SILVA ribosomal RNA database (http://www.arb-silva.de/) (Quast et al., 2012) and the UNITE database (https://unite.ut.ee/) (Kõljalg et al., 2013), aligning and filtering the representative sequences and then reconstructing the phylogenetic tree, filtering out the unwanted OTUs and resampling, calculating the abundance at each classification level, analyzing the community composition, analyzing the alpha diversity and beta diversity, differentiating the species, and analyzing the correlation between species and functions. For the functional classification prediction of fungal communities, we used FUNGuild (an open annotation tool for parsing fungal community datasets by ecological guild, https://github.com/UMNFuN/FUNGuild) to parse fungal OTUs into trophic modes and guilds (Toju et al., 2016).
The experimental data in this study were statistically analyzed using one-way analysis of variance (ANOVA) followed by Tukey's post hoc test or Kruskal–Wallis-test with SPSS version 26.0 (SPSS Inc., Chicago, IL, USA). The quadratic regression analysis was used to analyze the relationship between the nutrient digestibility and the level of dietary peNDF.Spearman correlation coefficients (r) and p value were analyzed using the OmicShare tools (a free online platform for data analysis, https://www.omicshare.com/tools) to show correlations between the feed intake, digestibility, rumen fermentation, methane production with the relative abundance of fungi genera and methanogenic species. p < 0.05 was accepted as statistically significant differences. Results are presented as mean and standard error.
As shown in Table 2, the values of pef>8.00 in five treatments could be calculated as 36.97, 22.95, 16.25, 6.27, and 3.12% according to the proportions of the particles retained on 19.00 mm and 8.00 mm sieves; the values of peNDF >8.00 in five treatments could be calculated as 15.86, 9.84, 6.97, 2.69, and 1.34% according to the values of pef>8.00 and the percentage of NDF in the diet (42.89%, Table 1). Similarly, the values of pef>1.18 in five treatments could be calculated as 76.88%, 69.78%, 65.61%, 61.73%, and 57.71% according to the proportions of the particles retained on 19.00, 8.00, and 1.18 mm sieves; the values of peNDF>1.18 in five treatments could be calculated as 32.97%, 29.93%, 28.14%, 26.48%, and 24.75%, respectively.
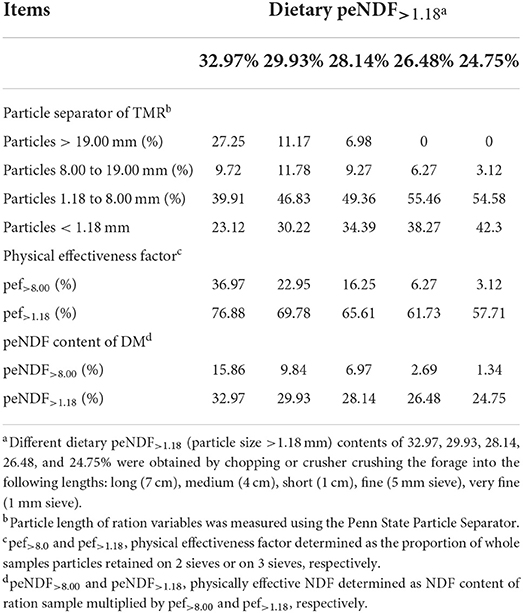
Table 2. Particle size distribution, physical effectiveness factors, and physically effective neutral detergent fiber content of the treatment rations fed to goats (DM basis).
Apparent digestibility of DM, OM, CP, NDF, and ADF varied greatly (p < 0.05) among the treatment groups (Table 3), the highest and the lowest digestibility of those nutrients were observed in the 28.14% and 24.75% peNDF>1.18 treatments, respectively. Regression analysis showed that the digestibility of DM, OM, NDF, and ADF was quadratically correlated with dietary peNDF>1.18 levels (Table 4). It could be calculated that the highest digestibility of DM, OM, NDF, and ADF appeared at dietary peNDF>1.18 levels of 30.25%, 30.18%, 30.36%, and 30.67%.
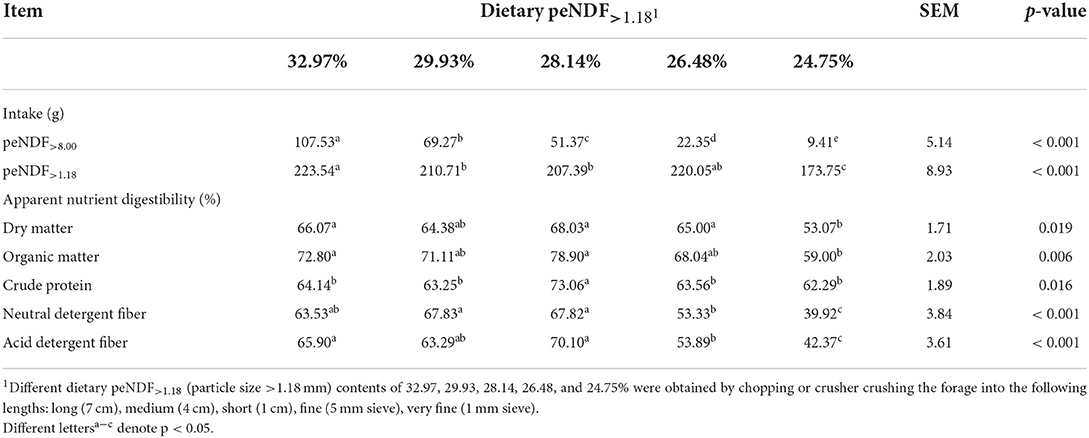
Table 3. Effects of different contents of peNDF>1.18 in diets on apparent nutrient digestibility in goats.
The effects of dietary peNDF>1.18 content on rumen fermentation parameters and methane production are presented in Table 5. Dietary peNDF>1.18 content did not affect ruminal pH and the concentration of VFAs. Ruminal NH3-N concentrations in the 29.93% and 28.14% groups were higher than that in the 24.75% group (p < 0.05). Ruminal MCP concentration was the highest in the 32.97% group (p < 0.05). Daily CH4 production in the 32.97% and 24.75% peNDF>1.18 treatments was lower than that in the 26.48% group (p < 0.05) and no differences were observed among other groups. CH4 production did not differ among the groups when expressed as per unit of DM intake.
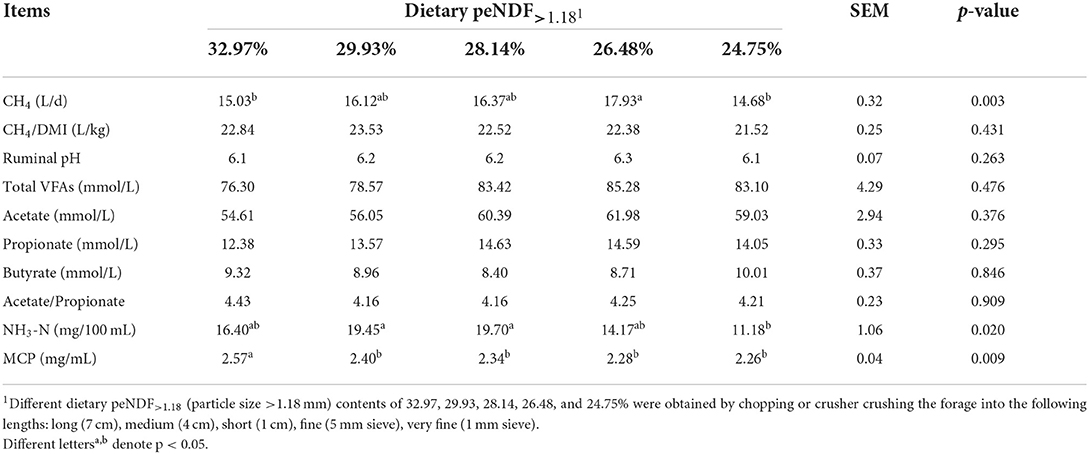
Table 5. Effects of different contents of peNDF>1.18 in diets on ruminal fermentation characteristics in goats.
A total of 482,535 effective sequences that met quality control were obtained from the 16S rRNA high-throughput sequencing analysis. Based on 97% sequence similarity from valid sequences, these sequences were completely clustered into 876 OTUs with a mean of 58.4 ± 17.8 (mean ± SD, n = 3) OTUs per sample. The average number of OTUs, Chao1 index, Shannon index, and Simpson index were the highest in the 32.97% peNDF>1.18 treatment (p < 0.05), and the Shannon index and Simpson index were the lowest in the 29.93, 28.14, and 26.48% peNDF>1.18 treatments (Table 6). A total of 44 OTUs were shared across the five treatments, and the number of sequences in shared OTUs accounted for 99.52% of the total effective sequences (Figure 1A).
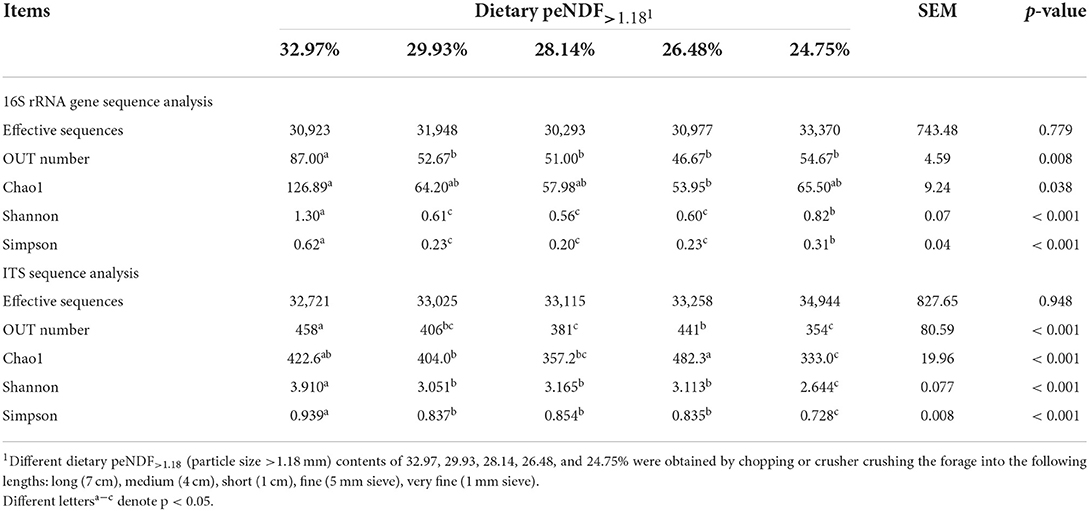
Table 6. Effects of different contents of peNDF>1.18 in diets on rumen microbial OTUs and alpha diversity of goats.
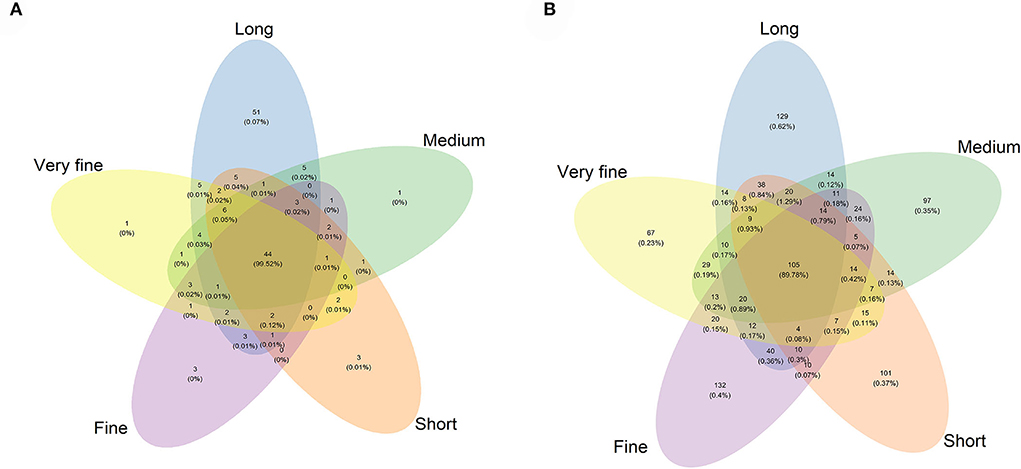
Figure 1. Venn diagram of the OTUs among the five treatments of archaea (A) and fungi (B). Long: 32.97% peNDF>1.18 treatment; Medium: 29.93% peNDF>1.18 treatment; Short: 28.14% peNDF>1.18 treatment; Fine: 26.48% peNDF>1.18 treatment; Very fine: 24.75% peNDF>1.18 treatment.
The principal co-ordinate analysis (PCoA) was conducted based on Bray-Curtis distance as shown in Figure 2A. Each treatment was evenly distributed among the clusters. All samples tended to cluster together in accordance with their own ratio treatment. These results suggested strong differences in the structure of archaea among the five treatments. Moreover, the samples of 26.48% and 24.75% peNDF>1.18 treatments were clustered in the same area, indicating that the archaeal community structure of the two treatments was more similar to that of other treatments. Similar results appeared in the 29.93% and 28.14% peNDF>1.18 treatments.
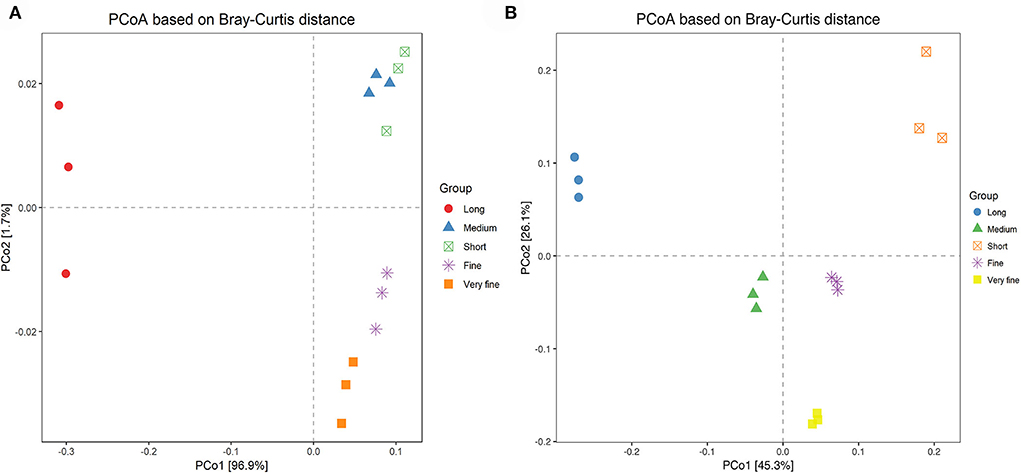
Figure 2. PCoA analysis based on Brary-Curtis distance among the five treatments of archaea (A) and fungi (B). Distances between the samples, based on similarity in OTU composition (OTU similarity ≥97%) calculated with Brary-Curtis distance, were visualized by principal coordinates analysis (PCoA) plots. A greater distance between two samples inferred a lower similarity. The percentage of variation explained by PC1 and PC2 were indicated in the axis.
Only the phylum Euryarchaeota of archaea was identified in all samples in this study. A total of four genera of archaea were identified in all samples, with the largest proportion of Methanobrevibacter accounting for 99.64%, followed by F_Methanomethylophilaceae|g_ Uncultured at 0.25%, Methanosphaera at 0.11% and Candidatus methanomethylophilus at 0.0034% (Figure 3A, Table 7). The relative abundance of Methanobrevibacter was higher in the 26.48% peNDF>1.18 treatment than that in the 24.75% peNDF>1.18 treatment (p < 0.05). The relative abundance of Methanosphaera was higher in the 32.97% and 24.75% peNDF>1.18 treatments than that in the 29.93%, 28.14%, and 26.48% peNDF>1.18 treatments (p < 0.05).
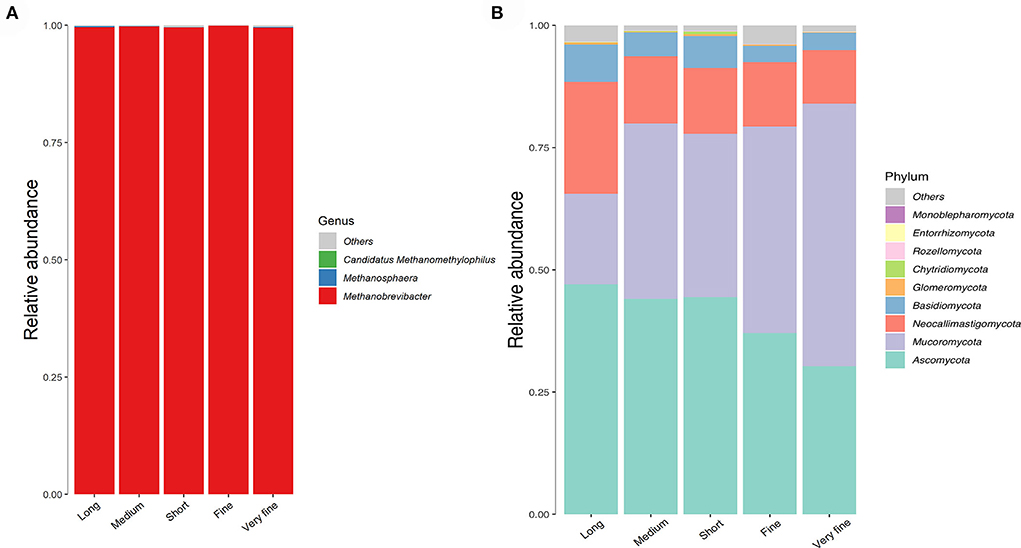
Figure 3. The histogram of the relative abundance of archaea at genus level (A) and fungi at phylum level (B). Long: 32.97% peNDF>1.18 treatment; Medium: 29.93% peNDF>1.18 treatment; Short: 28.14% peNDF>1.18 treatment; Fine: 26.48% peNDF>1.18 treatment; Very fine: 24.75% peNDF>1.18 treatment.
The archaea varied greatly with treatments at the species level (Figure 4). The dominant species for methanogenesis was Methanobrevibacter sp. YE315, followed by Methanobrevibacter boviskoreani JH1. The abundance and diversity of archaea are similar between the groups of consecutive peNDF levels, while the 32.97% peNDF>1.18 treatment was quite different from other treatments. The relative abundance of Methanobrevibacter sp. YE315 was lower in the 32.97% peNDF>1.18 treatment than that in other treatments (p < 0.05). The relative abundance of M. boviskoreani JH1 was higher in the 32.97% peNDF>1.18 treatment than that in other treatments (p < 0.05).
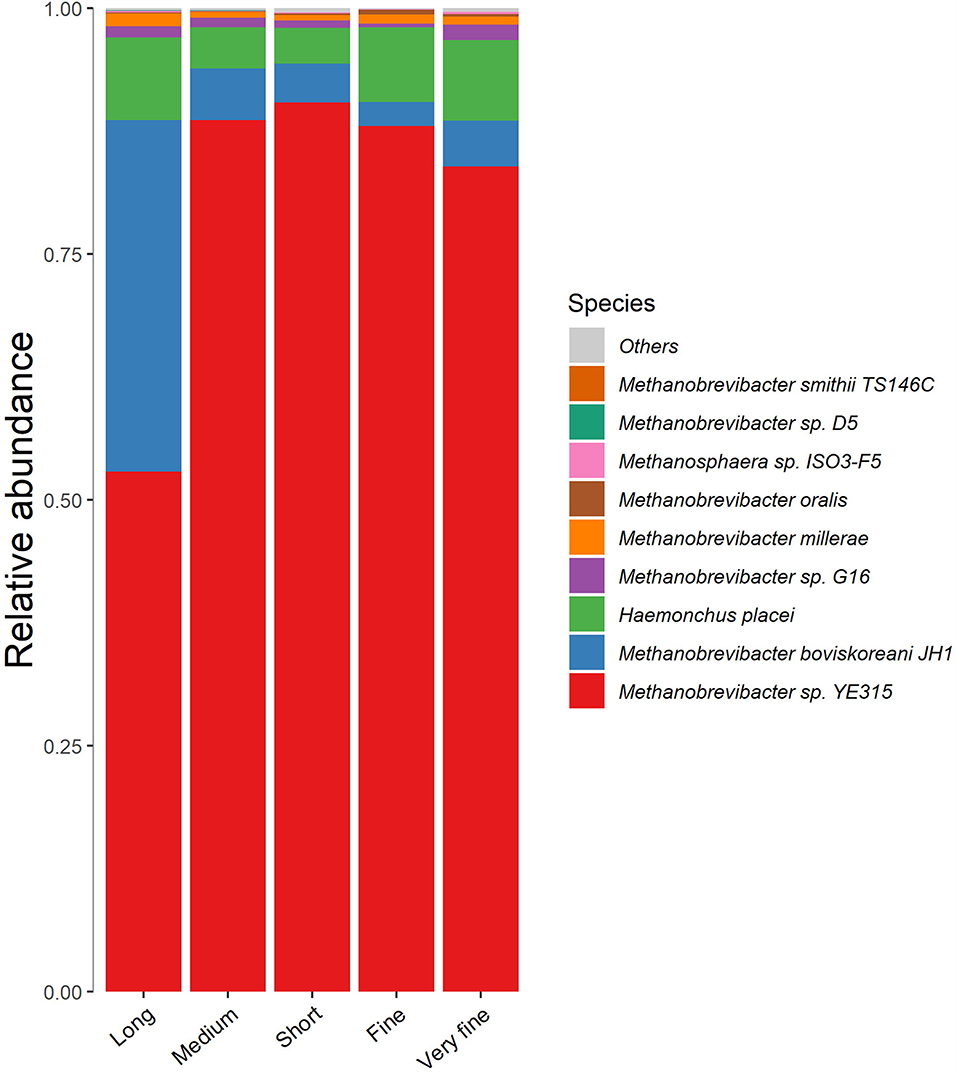
Figure 4. The cluster histogram of methanogen at species level. Long: 32.97% peNDF>1.18 treatment; Medium: 29.93% peNDF>1.18 treatment; Short: 28.14% peNDF>1.18 treatment; Fine: 26.48% peNDF>1.18 treatment; Very fine: 24.75% peNDF>1.18 treatment.
A total of 501,190 valid sequences that met quality control were obtained from the fungal ITS sequencing analysis. Based on 97% sequence similarity from valid sequences, these sequences were completely clustered into 1,013 OTUs with a mean of 301 ± 33.7 (mean ± SD, n = 3) OTUs per sample. The average number of ruminal fungal OTUs was the greatest in the 32.97% peNDF>1.18 treatment (p < 0.05), and the lowest value was observed in the 28.14% peNDF>1.18 and 24.75% peNDF>1.18 treatments (Table 6). Both the Shannon-Wiener index and Simpson index in the 32.97% peNDF>1.18 treatment were higher than that in other treatments, and in the 24.75 % peNDF>1.18 treatment was lower than that in other treatments (p < 0.05). A total of 105 OTUs were shared across the five treatments, and the number of sequences in shared OTUs accounted for 89.78% of the total effective sequences (Figure 1B).
The principal co-ordinate analysis (PCoA) was conducted based on the Euclidean distances between the different treatments, and the results are shown in Figure 2B. Each treatment was evenly distributed among the clusters. All samples tended to cluster together in accordance with their own ration treatment. These results suggested strong differences in the fungal structure among the treatments. Moreover, the samples of 24.75% and 26.48% peNDF>1.18 treatment were clustered in the same area, indicating that the fungal community structure of the two treatments was more similar to that of other treatments.
A total of 10 fungal phyla were identified in all samples, seven of which were shared (Figure 3B). The top four most abundant phyla ranked in descending order were Ascomycota, Mucoromycota, Neocallimastigomycota, and Basidiomycota, accounting for an average of 40.6, 36.8, 14.9, and 5.2% of the total fungi at the phyla level, respectively. In addition to Basidiomycota, the fungal community at the phylum level differed significantly among the treatments (p < 0.05). The relative abundance of Ascomycota was lowest in the 24.75% peNDF>1.18 treatment, followed by the 26.48% peNDF>1.18 treatment (p < 0.05), while the relative abundance of Mucoromycota was lowest in the 24.75% peNDF>1.18 treatment, followed by the 26.48% peNDF>1.18 treatment (p < 0.05). The relative abundance of Neocallimastigomycota and Glomeromycota were higher in the 32.97% peNDF>1.18 treatment than that in other treatments (p < 0.05).
At the genus level, a total of 186 fungal genera were detected, 14 of which were detected in all goats with an average abundance of >1%, including o_GS23|f_uncultured, f_Saccharomycetales_fam_Incertae_sedis|g_uncultured, Candida, f_Neocallimastigaceae|g_uncultured, Neocallimastix, Symbiotaphrina, Orpinomyces, o_Pleosporales|f_uncultured, o_Agaricostilbales|f_uncultured, o_Sebacinales|f_uncultured, f_Phaeosphaeriaceae|g_uncultured, Yarrowia, Aspergillus, p_Ascomycota|c_uncultured (Table 8, Supplementary Figure S1). The top 50 most abundant fungal genera were displayed in heatmap (Supplementary Figure S2). The dominant fungi at the genus level in the rumen of goats were Candida, Neocallimastix, and Orpinomyces, accounting for an average of 8.92% and 5.49% of the total fungi at the genus level, respectively. All the fungi at the genus level differed greatly among the treatments (p < 0.05). The relative abundance of Candida went up at first and then dropped with the decrease of dietary peNDF>1.18 content from 32.97 to 24.75%, and reached the maximum at the content of peNDF>1.18 was 28.1%. The relative abundance of Neocallimastigomycota Neocallimastix was the highest in the 29.93% peNDF>1.18 treatment, followed by the 32.97% peNDF>1.18 treatment.
The functional information of rumen archaea in each treatment based on the functional prediction of archaea by the Tax4Fun program and the SILVA database is summarized in Table 9. We found that the functional prediction of rumen archaea was significantly affected by dietary peNDF>1.18 levels. The main gene functions of rumen archaea were associated with amino acid metabolism, carbohydrate metabolism, membrane transport, and replication and repair. The functional classification prediction of rumen fungi in each treatment based on the OTU abundance is shown in Table 10. The rumen fungal functional guilds were significantly affected by dietary peNDF>1.18 levels. The trophic mode revealed that the most diverse fungal type was the pathotrophs. Meanwhile, the guild classification implied that the most diverse guilds were animal pathogens.
Correlations between microbiome composition and DM intake, digestibility, and rumen fermentation parameters are shown in Figure 5. The DM intake was negatively associated with the relative abundance of s_Methanobrevibacter sp. G16, s_M. boviskoreani JH1 and g_Methanosphaera (r < −0.6 and p < 0.05). The concentration of NH3-N was negatively associated with the relative abundance of s_Haemonchus placei (r < −0.7 and p < 0.05), and it was also positively correlated with the relative abundance of s_Methanobrevibacter sp. YE315 and p_Ascomycota (r > 0.5 and p < 0.05). The concentration of MCP was negatively associated with the relative abundance of p_Mucoromycota (r < −0.6 and p < 0.01), and it was also positively correlated with the relative abundance of p_Neocallimastigomycota and g_Neocallimastix (r > 0.6 and p < 0.05). The daily CH4 production was negatively associated with the relative abundance of s_M. boviskoreani JH1 and s_Methanobrevibacter sp. G16 (r < −0.6 and p < 0.01), and it was also positively correlated with the relative abundance of g_Candida and s_Methanobrevibacter sp. YE315 (r > 0.6 and p < 0.05). The concentration of total VFAs was negatively associated with the species s_M. boviskoreani JH1 (r < −0.5 and p < 0.05). The acetate concentration was negatively associated with the relative abundance of s_M. boviskoreani JH1 (r < −0.6 and p < 0.05). The propionate concentration was negatively associated with the relative abundance of s_M. boviskoreani JH1 and s_Methanobrevibacter sp. G16 (r < −0.5 and p < 0.05). The digestibility of OM was negatively associated with the relative abundance of p_Mucoromycota (r < −0.6 and p < 0.05), while positively associated with the relative abundance of g_Candida and p_Ascomycota (r > 0.6 and p < 0.05). The digestibility of NDF was negatively associated with the relative abundance of p_Mucoromycota (r < −0.6 and p < 0.05), while positively associated with the relative abundance of p_Ascomycota, p_Neocallimastigomycota, and g_Neocallimastix (r > 0.5 and p < 0.05). The digestibility of ADF was negatively associated with the relative abundance of p_Mucoromycota (r < −0.6 and p < 0.05), while positively associated with the relative abundance of p_Ascomycota, p_Neocallimastigomycota, g_Neocallimastix, and g_Candida (r > 0.5 and p < 0.05). The digestibility of DM was negatively associated with the relative abundance of p_Mucoromycota (r < −0.6 and p < 0.01), while positively associated with the relative abundance of p_Ascomycota and g_Candida (r > 0.5 and p < 0.05).
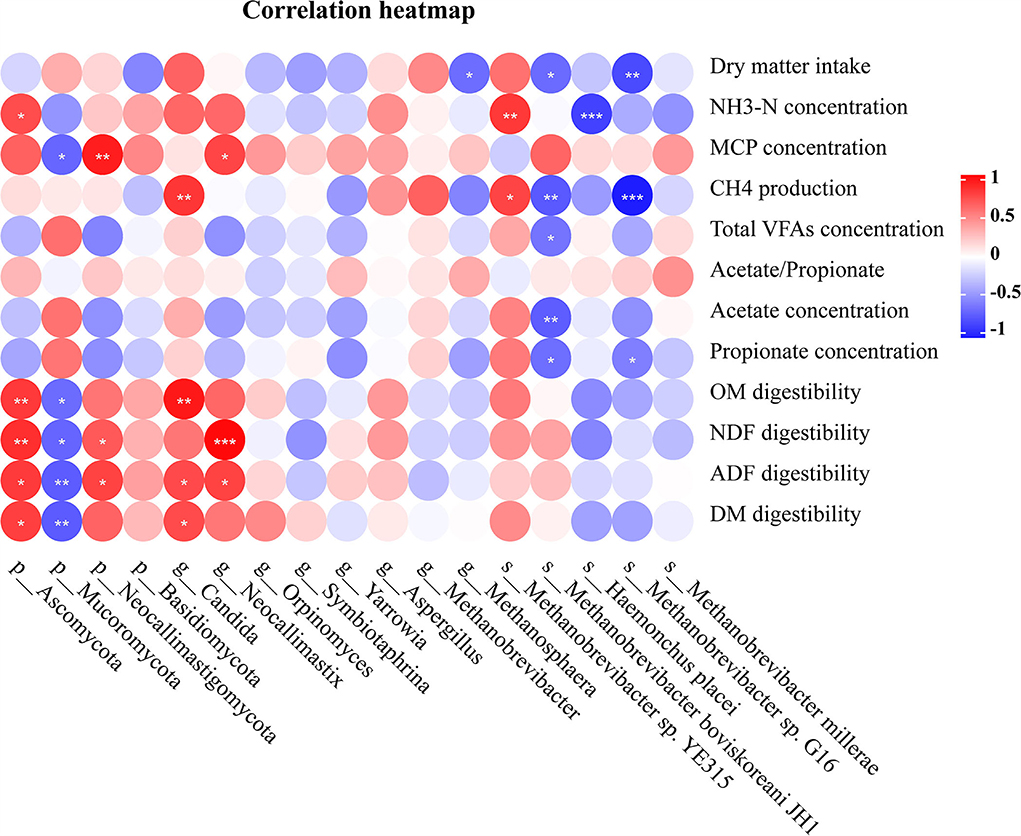
Figure 5. The correlation of dry matter intake, rumen fermentation parameters and nutrient digestibility with microbial abundance. The colors indicate positive (red, closer to 0.81) or negative (blue, closer to −0.90) correlations.
Due to the high forage content in ruminant feed, optimizing forage particle size is a significant feeding strategy to improve forage utilization for ruminants (Tafaj et al., 2005). A suitable particle size would lead to rapid digestion and passage of feed in the rumen, with less stimulation of masticatory activity and rumen contraction (Mertens, 1997). Excessively small or fine particle size would result in a decrease in rumen pH, negatively affecting fiber digestion, feed intake, and feed efficiency (Tafaj et al., 2005). In our recently published study, the feed intake of goats increased first and then decreased with the decrease of dietary peNDF>1.18 content from 33.0 to 24.8% (678 vs. 704 vs. 737 vs. 831 vs. 702 g/day), and reached the maximum at the content of peNDF>1.18 was 26.5% (Xue et al., 2022). The same trend appeared in the apparent digestibility of nutrients in this study, with the highest and the lowest digestibility of DM, OM, CP, NDF, and ADF observed in the 28.14% and 24.75% peNDF>1.18 treatment. It may be that more feed particles smaller than 1.88 mm have a faster passage rate from the rumen to escape rumen fermentation (Poppi et al., 1980), making against the digestion of nutrients, especially for the fiber. Jang et al. (2017) fed three diets with peNDF>1.18 content of 16.22, 23.85, and 21.71%, and found that the total rumen retention and transit time of feed increased with the escalating peNDF>1.18 content in the diet, providing more time for the digestion of nutrients. In addition, the particle size of ruminant feed also affected rumination and chewing behavior (Ramirez Ramirez et al., 2016; Jiang et al., 2019), determining the amount of saliva produced and secreted. Saliva is a buffer for maintaining ruminal pH, and acetate to propionate ratio, providing a stable environment for the fermentability and proper growth of rumen microbes (Banakar et al., 2018). Longer forage particles would be reduced in size during the prolonged chewing process, so as to partially eliminate the influence of particle length (Yang and Beauchemin, 2006c). Since 1.18 mm is considered the critical length for retention of chyme granules in the rumen, stimulating chewing and rumination behavior (Poppi et al., 1980, 1981), the apparent digestibility is poorly different in goats-fed diets with long (7 cm), medium (4 cm), and short (1 cm) forages in this study. Wang et al. (2017) investigated the effect of four dietary peNDF>1.18 levels (from 22.7% to 28.4%) on the nutrient digestibility of heifers and found that the digestibility increased when dietary peNDF>1.18 level increased from 22.7 to 25.9%, which was consistent with the results of this study. However, other studies reported that increasing dietary peNDF content reduced the apparent digestibility of DM, OM, NDF, and ADF (Soita et al., 2002; Yang and Beauchemin, 2006a) or did not affect the apparent digestibility (Park et al., 2014). The discrepancy in these findings may be due to the different ways of changing the content of peNDF and the type of concentrates. Wang et al. (2017) used the same corn grain-based diet and cut different forage lengths to control the peNDF content of the diet. Other studies, however, used barley as the main concentrate (Yang and Beauchemin, 2006a) or no concentrate (Soita et al., 2002) in the diet or adjusted the mixing time of TMR to control the content of dietary peNDF (Park et al., 2014). Fermentability of concentrate is also one of the important factors affecting the digestion of ruminants (Bekele et al., 2010). Compared with corn, the higher fermentability of the grain may be another factor affecting the digestion of nutrition (Yang and Beauchemin, 2006b).
Oh et al. (2016) found that the nutrient digestibility linearly increased with the dietary peNDF>1.18 content in heifers, in which the peNDF>1.18 levels ranged from 27.50 to 30.36%. The wide range of the peNDF>1.18 gradient (24.75–32.97%) in this study was useful in depicting the locus of nutrient digestibility with peNDF>1.18 levels. Regression analysis indicated that the digestibility of all nutrients reached the peak at the dietary peNDF>1.18 level of 30% (Table 4). The nutrient digestibility was quadratically correlated to dietary peNDF>1.18 content, and the inflection point falls in the range from 28.14 to 30.0%. Combining the results from Wang et al. (2017) and Oh et al. (2016), we deduced that the nutrient digestibility of ruminants kept on increasing when dietary peNDF>1.18 levels increased from 22.7 to 30.36%. It is well known that the increase in the residence time of feed in the gastrointestinal tract would improve its digestive efficiency (Shaver et al., 1988; Kaske and Engelhardt, 1990). In addition, the residence time in the gastrointestinal tract of 1.00 mm particles was 22.7 h less than that of 10.00 mm particles (Kaske and Engelhardt, 1990). Combined with the results of nutrient digestibility in this study, proper control of forage length (5.00–10.00 mm) was beneficial to the utilization of nutrients in goats.
Rumen pH reflects the balance between the production and absorption of organic acids in the rumen of ruminants. Physically effective fiber is the fraction of feed that stimulates chewing activity and salivary secretion (Mertens, 1997), which is beneficial to neutralizing excess hydrogen ions and therefore helps prevent the reduction of rumen pH. Our results suggested that dietary peNDF>1.18 content has no effect on rumen pH and VFAs concentration in the rumen. This may be due to the stable non-fibrous carbohydrate content across the diets. Although the content of peNDF in the diet is an important factor in maintaining the stability of the rumen environment (Allen, 1997), the rumen pH is more affected by the proportion of non-fiber carbohydrates in the diet, because they are more easily degraded in the rumen (Nasrollahi et al., 2012). Moreover, this study only measured the rumen pH at the time of slaughter before morning feeding and did not observe the dynamic change of rumen pH, all of this may result that no significant difference was observed between the groups.
However, our study found that the concentrates of NH3-N and MCP in the rumen were affected by the dietary peNDF>1.18 content. As the length of the forage particles decreased, the residence and fermentation time of the chyme in the rumen were shortened (Kaske and Engelhardt, 1990), which may be the reason for the low concentration of NH3-N and MCP in very fine treatment (peNDF>1.18 = 24.75%). Rumen NH3-N is an important nitrogen source for rumen microbial growth and microbial protein synthesis (Pisulewski et al., 2010). Wang et al. (2017) found that the ammonia concentration increased significantly with the increase of particle size and dietary peNDF>1.18 content, even though the peNDF>1.18 content in the diet was low (22.7 to 28.4%). In this study, the NH3-N concentration went up at first and then dropped with the decrease of the dietary peNDF>1.18 content from 33.0 to 24.8%, and reached the maximum at the content of peNDF>1.18 was 28.14%. This result is similar to the apparent digestibility of nutrients, with a quadratic linear relationship between rumen NH3-N and dietary peNDF>1.18 content.
Methane is the end-product of the degradation of dietary carbohydrates into VFAs in the rumen (Pinares-Patiño et al., 2007; Hylemon et al., 2018). Earlier studies have reported that CH4 production was positively correlated with the rate of feed degradation in vitro (Wang et al., 2014; Molina-Botero et al., 2020). The methane production in the 24.5% peNDF>1.18 treatment was the lowest in this study, consistent with the results of nutrient digestibility, this reduction might be related to the decrease in feed fermentation rate in the rumen. Furthermore, the variation in daily CH4 production when cattle are fed ad libitum is largely due to the differences in feed intake and feed nutritive value (Johnson and Johnson, 1995). The intake and nutritional value of feed determine the fermentation substrate of rumen microorganisms. A recent meta-analysis demonstrated a strong positive correlation between daily CH4 production and DMI in finishing beef cattle (Smith et al., 2021). Hence, we found that the variation trend of daily CH4 production was consistent with the DMI when goats were fed diets with different levels of peNDF (Xue et al., 2022), while the daily CH4 production per kg DMI was not affected by the dietary peNDF content.
Archaea only account for 2–4% of the total rumen microorganisms of ruminants, making it difficult to study the structure and composition of rumen archaea (Lin et al., 1997; Cruz et al., 2016). Furthermore, the strains identified by in vitro culture were short of representation due to the fact that these archaea are strictly anaerobic organisms (Paul et al., 2017). Much more identification efforts are awaited to describe the classification of major archaeal species (Paul et al., 2017). Nevertheless, ruminal archaea are responsible for converting hydrogen and carbon dioxide produced by other fermenting microorganisms to methane, thereby playing a non-negligible role in maintaining rumen metabolism and function (Patra et al., 2017). In the current study, all sequences were clustered into three genera with validly published names and an unknown genus, and nine species with validly published names and two unknown species. Moreover, we found that Methanobacter was the most dominant genus in goat rumen, which is consistent with the previously reported results in buffaloes (Kala et al., 2020), goats (Fliegerova et al., 2021), sheep (McLoughlin et al., 2020), and beef cattle (Paul et al., 2017). The most dominant archaeal species was Methanobrevibacter sp. YE315 and its relative abundance were significantly affected by dietary peNDF content. Unfortunately, we have not found any reports describing the role of Methanobrevibacter sp. YE315. The correlation analysis showed that the daily production of CH4 was positively correlated with the relative abundance of Methanobrevibacter sp. YE315, while negatively correlated with the relative abundances of both Methanobrevibacter sp. G16 and M. boviskoreani JH1. The relationship between methanogenesis and the abundance of methanogens in the rumen has not been unified in the literature. Previous studies suggested that methane production was related to specific methanogens species or to the overall abundance of methanogens (Wallace et al., 2014; Danielsson et al., 2017). However, other reports showed that there was no relationship between methane production and the abundance of methanogens (Shi et al., 2014; Casañas et al., 2015). The current study concluded that the low relative abundance of methanogens in the rumen compared to fungi and bacteria contributes to the low or no correlation with methane production (Janssen and Kirs, 2008).
Fungi account for 5–20% of the total microbial mass in the rumen of ruminants (Rezaeian et al., 2004). Fungi produce a large, comprehensive array of enzymes necessary for the digestion of plant material, such as cellulases, xylanases, and other hydrolases (Solomon et al., 2016). Due to the diversity and high activity of these enzymes, rumen fungi play a major role in lignocellulose fermentation (Orpin and Joblin, 1997). An earlier study showed that feeding a high proportion of enriched diets resulted in a significant reduction or even complete disappearance of fungal abundance in the rumen of 80% of lambs, while the fungal structure was restored to stability after feeding Alfalfa hay (Fonty et al., 1987). Another study also showed that increasing the proportion of concentrate in the diet would reduce the total number of rumen fungi in sheep (Faichney et al., 1997). These studies indicated that the composition and structure of rumen fungi were affected by the content of forage (fiber) in the diet. Rumen fungi are closely related to the number of plant substrates in the rumen due to their properties of colonizing fiber granules (Gordon and Phillips, 1998). In addition, it has been reported that fungi play an indispensable role in the degradation of cellulose in the rumen because they can penetrate deeply into plant tissues that are usually inaccessible to bacteria (Bauchop and Mountfort, 1981), which has been confirmed in controlling fungal activity to change the degradation rate of plant cell walls (Lee et al., 2000). Neocalimastigales family members play an important role in degrading the feed materials that has complex structures and are rich in cellulose, enabling the production of hydrogen gas as a substrate for other rumen microorganisms (Boots et al., 2013). The proportion of concentrate in the diet increased from 50 to 90% separated Neocallismastigales assemblies (Boots et al., 2013). In this study, we found that the relative abundance of f_Neocallimastigaceae|g_uncultured was the highest in the 32.97% peNDF>1.88 treatment and the lowest in the 28.14% peNDF>1.88 treatment, indicating that the content of dietary peNDF affected its abundance. Neocallimstixhas was observed to be the dominant fungal genus in the rumen of most ruminants (Kittelmann et al., 2013), which is consistent with the results of the current study. Neocallimastix produces lower amounts of cellulolytic and xylanolytic enzymes than other fungi (e.g., Piromyces) (Teunissen et al., 1992). The content of peNDF in the diet affected the abundance of Neocallimastix in this study. Overall, diet plays an important role in maintaining fungal diversity in the rumen of ruminants. The anaerobic rumen fungal population in the rumen of cattle fed the same growing stage forages was reduced compared with those fed mature forages (Kostyukovsky et al., 1991). Notably, microbes that comprise a larger proportion of the abundance contribute significantly to the function of the rumen microbial ecosystem, and a small subset of microbes in the rumen community may have important, yet unrecognized, ecological functions (Singh et al., 2015).
In this study, we inferred the function of microorganisms in the rumen of goats by Tax4Fun and FUNGuild. Many genes from rumen microbiota are closely related to nutrient metabolism, and therefore can be used to predict the function of microbial activity in nutrient metabolism (Noecker et al., 2016). The metabolic genes of archaea in the 24.75% peNDF>1.18 treatment is low, suggesting that low peNDF may not be beneficial to the metabolic activities of archaea. The production of methane is also associated with feed intake and rumen passage rate in addition to the abundance or activity of methanogens (Goopy et al., 2014; Fagundes et al., 2020). Both feed intake and rumen passage rate may affect the growth of rumen microbes (including methanogens) because they are related to the microbial energy flux and microbial production time (Satter, 1986; Martínez et al., 2009). The lower DM intake in the 32.97% peNDF>1.88 and 24.75% peNDF>1.88 treatments could result in a decrease in the relative abundance of Methanobrevibacter sp. YE315. The very fine forage particles in the 24.75% peNDF>1.88 treatment could increase the rumen passage rate, thus decreasing the relative abundance of Methanobrevibacter sp. YE315. The functional prediction value of carbohydrate metabolism was observed to be lower in the 32.97% peNDF>1.88 and 24.75% peNDF>1.88 treatments. These results were consistent with the daily production of CH4. On the other hand, archaea in rumen interact with fungi and bacteria (Patra et al., 2017). Although little is known about archaea related to fungi, rumen bacteria and archaea have been confirmed to interact through interspecific hydrogen transfer (Patra et al., 2017). A clear link between rumen fungal abundance and methane production has been demonstrated by experiments in vitro (Newbold et al., 1995; Qin et al., 2012) and in vivo (Schönhusen et al., 2003), implying that there was an association between fungi and archaea.
Based on the mode of nutrient acquisition (trophic mode), fungi can be classified into three main groups (Tedersoo et al., 2014): saprotroph, nutrients derived from degradation of dying host cells; symbiotroph, nutrients derived from exchanging resources with host cells; pathotroph, nutrients derived from harming host cells. Further, in these trophic modes, a total of 12 categories were included as guilds: animal pathogens, arbuscular mycorrhizal fungi, ectomycorrhizal fungi, ericoidmycorrhizal fungi, foliar endophytes, lichenicolous fungi, liche-nized fungi, mycoparasites, plant pathogens, undefined root en-dophytes, undefined saprotrophs, and wood saprotrophs. We found that the guild OUT richness of pathotroph was the lowest in the 28.14% peNDF>1.88 treatment while the highest in the 26.48% peNDF>1.88 treatment, indicating that the dietary peNDF level of 26.48% was unfavorable to the rumen health of the host.
To the best of our knowledge, this report is the first study to investigate the effect of dietary peNDF levels on rumen fungal diversity in goats. In this study, we reduced the content of peNDF>1.18 in diets by reducing the chop length of forages, and found that the community structures of rumen archaea and fungi varied with the levels of dietary peNDF>1.18. Without changing the composition of the diet, appropriately reducing the levels of peNDF>1.18 in the diet contributed to higher nutrient digestibility in goats, whereas excessive crushing resulted in negative effects. The apparent digestibility of nutrients was highest in the 28.14% PeNDF>1.18 treatment, indicating that the nutrient utilization was optimal at this level of peNDF>1.18. Therefore, we recommend that the content of peNDF>1.18 in goat diets should be 28.14% (roughage chopped to 1 cm) to get higher usage of feed. Moreover, whether the level of peNDF in the diets affected the production performance of ruminants, such as milk yield, growth performance, and meat quality, needs to be further studied.
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA865508.
The animal study was reviewed and approved by the Animal Ethical and Welfare Committee of Animal Nutrition Institute, Sichuan Agricultural University.
JZ and BeX wrote the original manuscript, performed data statistical analysis, and produced figures and tables. BeX and MW performed the experiments. AH and YW provided great help in data analysis. JZ, BaX, and QH participated in the design of the trial. SY, ZW, BaX, LW, and QP revised the manuscript. All authors have read and approved the final manuscript.
This research was funded by China Agriculture Research System of MOF and MARA (CARS-39).
We gratefully acknowledge the laboratory staff who provided assistance during the course of the study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.950587/full#supplementary-material
Adesogan, A., Arriola, K., Jiang, Y., Oyebade, A., Paula, E., Pech-Cervantes, A., et al. (2019). Symposium review: technologies for improving fiber utilization. J. Dairy Sci. 102, 5726–5755. doi: 10.3168/jds.2018-15334
Aguilera, J., and Prieto, C. (1986). Description and function of an open-circuit respiration plant for pigs and small ruminants and the techniques used to measure energy metabolism. Arch. Anim. Nutr. 36,1009–1018. doi: 10.1080/17450398609429522
Allen, M. S. (1997). Relationship between fermentation acid production in the rumen and the requirement for physically effective fiber. J.Dairy Sci. 80, 1447–1462. doi: 10.3168/jds.S0022-0302(97)76074-0
Almeida, F., and Stein, H. (2010). Performance and phosphorus balance of pigs fed diets formulated on the basis of values for standardized total tract digestibility of phosphorus. J. Anim. Sci. 88, 2968–2977. doi: 10.2527/jas.2009-2285
Banakar, P., Anand Kumar, N., and Shashank, C. (2018). Physically effective fibre in ruminant nutrition: a review. J. Pharmacogn. Phytochem. 7, 303–308
Bargo, F., Muller, L., Delahoy, J., and Cassidy, T. (2002). Performance of high producing dairy cows with three different feeding systems combining pasture and total mixed rations. J. Dairy Sci. 85, 2948–2963. doi: 10.3168/jds.S0022-0302(02)74381-6
Bauchop, T., and Mountfort, D. O. (1981). Cellulose fermentation by a rumen anaerobic fungus in both the absence and the presence of rumen methanogens. Appl. Environ. Microb. 42, 1103–1110. doi: 10.1007/BF00443249
Bekele, A. Z., Koike, S., and Kobayashi, Y. (2010). Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol. Lett. 305, 49–57. doi: 10.1111/j.1574-6968.2010.01911.x
Berthiaume, R., Benchaar, C., Chaves, A., Tremblay, G., Castonguay, Y., Bertrand, A., et al. (2010). Effects of nonstructural carbohydrate concentration in alfalfa on fermentation and microbial protein synthesis in continuous culture. J. Dairy Sci. 93, 693–700. doi: 10.3168/jds.2009-2399
Billenkamp, F., Schnabel, K., Hüther, L., Frahm, J., von Soosten, D., Meyer, U., et al. (2021). No hints at glyphosate-induced ruminal dysbiosis in cows. NPJ Biofilms. Microbi. 7, 1–13. doi: 10.1038/s41522-021-00198-4
Boots, B., Lillis, L., Clipson, N., Petrie, K., Kenny, D., Boland, T., et al. (2013). Responses of anaerobic rumen fungal diversity (phylum N eocallimastigomycota) to changes in bovine diet. J. Appl. Microbiol. 114, 626–635. doi: 10.1111/jam.12067
Cao, Y., Gao, Y., Xu, M., Liu, N., Zhao, X., Liu, C., et al. (2013). Effect of ADL to aNDF ratio and ryegrass particle length on chewing, ruminal fermentation, and in situ degradability in goats. Anim. Feed Sci. Tech. 186, 112–119. doi: 10.1016/j.anifeedsci.2013.08.010
Cao, Y., Wang, D., Wang, L., Wei, X., Li, X., Cai, C., et al. (2021). Physically effective neutral detergent fiber improves chewing activity, rumen fermentation, plasma metabolites, and milk production in lactating dairy cows fed a high-concentrate diet. J. Dairy Sci. 104, 5631–5642. doi: 10.3168/jds.2020-19012
Casañas, M. A., Rangkasenee, N., Krattenmacher, N., Thaller, G., Metges, C., Kuhla, B. J. J., et al. (2015). Methyl-coenzyme M reductase A as an indicator to estimate methane production from dairy cows. J. Dairy Sci. 98, 4074–4083. doi: 10.3168/jds.2015-9310
Cruz, G. D., Millen, D. D., and Rigueiro, A. L. N. (2016). “Rumen models,” in Rumenology, eds D. Millen, M. De Beni Arrigoni, and R. Lauritano Pacheco (Cham: Springer). doi: 10.1007/978-3-319-30533-2_10
Danielsson, R., Dicksved, J., Sun, L., Gonda, H., Müller, B., Schnürer, A., et al. (2017). Methane production in dairy cows correlates with rumen methanogenic and bacterial community structure. Front. Microbiol. 8, 226. doi: 10.3389/fmicb.2017.00226
de Paula Carlis, M. S., Sturion, T. U., da Silva, A. L. A., Eckermann, N. R., Polizel, D. M., de Assis, R. G., et al. (2021). Whole corn grain-based diet and levels of physically effective neutral detergent fiber from forage (pefNDF) for feedlot lambs: Digestibility, ruminal fermentation, nitrogen balance and ruminal pH. Small Ruminant. Res. 205, 106567. doi: 10.1016/j.smallrumres.2021.106567
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Fagundes, G. M., Benetel, G., Welter, K. C., Melo, F. A., Muir, J. P., Carriero, M. M., et al. (2020). Tannin as a natural rumen modifier to control methanogenesis in beef cattle in tropical systems: friend or foe to biogas energy production? Res. Vet. Sci. 132, 88–96. doi: 10.1016/j.rvsc.2020.05.010
Faichney, G., Poncet, C., Lassalas, B., Jouany, J., Millet, L., Dor,é, J., et al. (1997). Effect of concentrates in a hay diet on the contribution of anaerobic fungi, protozoa and bacteria to nitrogen in rumen and duodenal digesta in sheep. Anim. Feed Sci. Tech. 64, 193–213. doi: 10.1016/S0377-8401(96)01059-0
Fliegerova, K. O., Podmirseg, S. M., Vinzelj, J., Grilli, D. J., Kvasnov,á, S, Schierov,á, D., et al. (2021). The effect of a high-grain diet on the rumen microbiome of goats with a special focus on anaerobic fungi. Microorganisms 9, 157. doi: 10.3390/microorganisms9010157
Fonty, G., Gouet, P., Jouany, J.-P., and Senaud, J. (1987). Establishment of the microflora and anaerobic fungi in the rumen of lambs. Microbiology 133, 1835–1843. doi: 10.1099/00221287-133-7-1835
Goopy, J. P., Donaldson, A., Hegarty, R., Vercoe, P. E., Haynes, F., Barnett, M., et al. (2014). Low-methane yield sheep have smaller rumens and shorter rumen retention time. Br. J. Nutr. 111, 578–585. doi: 10.1017/S0007114513002936
Gordon, G. L., and Phillips, M. W. (1998). The role of anaerobic gut fungi in ruminants. Nutr. Res. Rev. 11, 133–168. doi: 10.1079/nrr19980009
Hylemon, P. B., Harris, S. C., and Ridlon, J. M. (2018). Metabolism of hydrogen gases and bile acids in the gut microbiome. FEBS Lett. 592, 2070–2082. doi: 10.1002/1873-3468.13064
International, A. (1990). Official methods of analysis. Gaithersburg, Washington, DC, USA: Aoac Intl.
Jang, S. Y., Kim, E. K., Park, J. H., Oh, M. R., Tang, Y. J., Ding, Y. L., et al. (2017). Effects of physically effective neutral detergent fiber content on dry matter intake, digestibility, and chewing activity in Korean native goats (Capra hircus coreanae) fed with total mixed ration. Asian. Austral. J. Anim. 30, 1405. doi: 10.5713/ajas.16.0868
Janssen, P. H., and Kirs, M. (2008). Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 74, 3619–3625. doi: 10.1128/AEM.02812-07
Jiang, F. G., Lin, X. Y., Yan, Z. G., Hu, Z. Y., Wang, Y., and Wang, Z. H. (2019). Effects of forage source and particle size on chewing activity, ruminal pH, and saliva secretion in lactating Holstein cows. Anim. Sci. J. 90, 382–392. doi: 10.1111/asj.13153
Johnson, K. A., and Johnson, D. E. (1995). Methane emissions from cattle. J. Anim. Sci. 73, 2483–2492. doi: 10.2527/1995.7382483x
Kaeokliang, O., Kawashima, T., Narmseelee, R., Butcha, P., Sunato, S., Thinowong, A., et al. (2019). Effects of physically effective fiber in diets based on rice straw and cassava pulp on chewing activity, ruminal fermentation, milk production, and digestibility in dairy cows. Anim. Sci. J. 90, 1193–1199. doi: 10.1111/asj.13271
Kala, A., Kamra, D., Agarwal, N., Chaudhary, L., and Joshi, C. (2020). Insights into metatranscriptome, and CAZymes of buffalo rumen supplemented with blend of essential oils. Indian J. Microbiol. 60, 485–493. doi: 10.1007/s12088-020-00894-3
Kaske, M., and Engelhardt, W. (1990). The effect of size and density on mean retention time of particles in the gastrointestinal tract of sheep. Brit. J. Nutr. 63, 457–465. doi: 10.1079/BJN19900133
Kittelmann, S., Seedorf, H., Walters, W. A., Clemente, J. C., Knight, R., Gordon, J. I., et al. (2013). Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PloS ONE. 8, e47879. doi: 10.1371/journal.pone.0047879
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Kostyukovsky, V. A., Okunev, O. N., and Tarakanov, B. V. (1991). Description of two anaerobic fungal strains from the bovine rumen and influence of diet on the fungal population in vivo. Microbiology. 137, 1759–1764. doi: 10.1099/00221287-137-7-1759
Lee, S., Ha, J., and Cheng, K.-J. (2000). Relative contributions of bacteria, protozoa, and fungi to in vitro degradation of orchard grass cell walls and their interactions. Appl. Environ. Microbiol. 66, 3807–3813. doi: 10.1128/AEM.66.9.3807-3813.2000
Li, C., Beauchemin, K. A., and Yang, W. (2020). Feeding diets varying in forage proportion and particle length to lactating dairy cows: I. effects on ruminal pH and fermentation, microbial protein synthesis, digestibility, and milk production. J. Dairy Sci. 103, 4340–4354. doi: 10.3168/jds.2019-17606
Li, F., and Guan, L. L. (2017). Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle. Appl. Environ. Microb. 83, e00061–e00017. doi: 10.1128/AEM.00061-17
Li, F., Li, Z., Li, S., d Ferguson, J., Cao, Y., Yao, J., et al. (2014). Effect of dietary physically effective fiber on ruminal fermentation and the fatty acid profile of milk in dairy goats. J. Dairy Sci. 97, 2281–2290. doi: 10.3168/jds.2013-6895
Lin, C., Raskin, L., and Stahl, D. A. (1997). Microbial community structure in gastrointestinal tracts of domestic animals: comparative analyses using rRNA-targeted oligonucleotide probes. FEMS Microbio. Ecol. 22, 281–294. doi: 10.1016/S0168-6496(97)00002-0
Martínez, M., Ranilla, M., Ramos, S., Tejido, M., and Carro, M. (2009). Effects of dilution rate and retention time of concentrate on efficiency of microbial growth, methane production, and ruminal fermentation in Rusitec fermenters. J. Dairy Sci. 92, 3930–3938. doi: 10.3168/jds.2008-1975
Maulfair, D., Fustini, M., and Heinrichs, A. (2011). Effect of varying total mixed ration particle size on rumen digesta and fecal particle size and digestibility in lactating dairy cows. J. Dairy Sci. 94, 3527–3536. doi: 10.3168/jds.2010-3718
McLoughlin, S., Spillane, C., Claffey, N., Smith, P. E., O'Rourke, T., Diskin, M. G., et al. (2020). Rumen microbiome composition is altered in sheep divergent in feed efficiency. Front. Microbiol. 11, 1981. doi: 10.3389/fmicb.2020.01981
Mertens, D. (1997). Creating a system for meeting the fiber requirements of dairy cows. J. Dairy Sci. 80, 1463–1481. doi: 10.3168/jds.S0022-0302(97)76075-2
Molavian, M., Ghorbani, G., Rafiee, H., and Beauchemin, K. (2020). Substitution of wheat straw with sugarcane bagasse in low-forage diets fed to mid-lactation dairy cows: Milk production, digestibility, and chewing behavior. J. Dairy Sci. 103, 8034–8047. doi: 10.3168/jds.2020-18499
Molina-Botero, I. C., Mazabel, J., Arceo-Castillo, J., Urrea-Benítez, J. L., Olivera-Castillo, L., Barahona-Rosales, R., et al. (2020). Effect of the addition of Enterolobium cyclocarpum pods and Gliricidia sepium forage to Brachiaria brizantha on dry matter degradation, volatile fatty acid concentration, and in vitro methane production. Trop. Anim. Health Pro. 52, 2787–2798. doi: 10.1007/s11250-020-02324-4
Nasrollahi, S., Khorvash, M., Ghorbani, G., Teimouri-Yansari, A., Zali, A., and Zebeli, Q. (2012). Grain source and marginal changes in forage particle size modulate digestive processes and nutrient intake of dairy cows. Animal 6, 1237–1245. doi: 10.1017/S1751731112000122
Newbold, C., Lassalas, B., and Jouany, J. (1995). The importance of methanogens associated with ciliate protozoa in ruminal methane production in vitro. Lett. Appl. Microbiol. 21, 230–234. doi: 10.1111/j.1472-765x.1995.tb01048.x
Noecker, C., Eng, A., Srinivasan, S., Theriot, C. M., Young, V. B., Jansson, J. K., et al. (2016). Metabolic model-based integration of microbiome taxonomic and metabolomic profiles elucidates mechanistic links between ecological and metabolic variation. MSystems 1, e00013–00015. doi: 10.1128/mSystems.00013-15
NRC. (2007). Nutrient requirements of small ruminants: sheep, goats, cervids, and new world camelids. The National Academies Press, Washington D.C. USA. doi: 10.17226/11654
Oh, M. R., Hong, H., Li, H. L., Jeon, B. T., Choi, C. H., Ding, Y. L., et al. (2016). Effects of Physically Effective Neutral Detergent Fiber Content on Intake, Digestibility, and Chewing Activity in Fattening Heifer Fed Total Mixed Ration. Asian. Austral. J. Anim. 29, 1719. doi: 10.5713/ajas.16.0344
Orpin, C., and Joblin, K. (1997). “The rumen anaerobic fungi,” in The rumen microbial ecosystem. p. 140–195. doi: 10.1016/0378-1097(87)90045-0
Park, J. H., Kim, K. H., Park, P. J., Jeon, B. T., Oh, M. R., Jang, S. Y., et al. (2014). Effects of physically effective neutral detergent fibre content on dry-matter intake, digestibility and chewing activity in beef cattle fed total mixed ration. Anim. Prod. Sci. 55, 166–169. doi: 10.1071/AN14241
Patra, A., Park, T., Kim, M., and Yu, Z. (2017). Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J. Anim. Sci. Biotechno. 8, 1–18. doi: 10.1186/s40104-017-0145-9
Paul, S. S., Dey, A., Baro, D., and Punia, B. S. (2017). Comparative community structure of archaea in rumen of buffaloes and cattle. J. Sci. Food Agr. 97, 3284–3293. doi: 10.1002/jsfa.8177
Pinares-Patiño, C., Waghorn, G., Machmüller, A., Vlaming, B., Molano, G., Cavanagh, A., et al. (2007). Methane emissions and digestive physiology of non-lactating dairy cows fed pasture forage. Can. J. Anim. Sci. 87, 601–613. doi: 10.4141/CJAS06023
Pisulewski, P. M., Okorie, A. U., Buttery, P. J., Haresign, W., and Lewis, D. (2010). Ammonia concentration and protein synthesis in the rumen. P. Nut. Soc. 32, 759–766. doi: 10.1002/jsfa.2740320803
Poppi, D., Minson, D., and Ternouth, J. (1981). Studies of cattle and sheep eating leaf and stem fractions of grasses. 1. The voluntary intake, digestibility and retention time in the reticulo-rumen. Aust. J. Agr. Res. 32, 99–108. doi: 10.1071/ar9810099
Poppi, D., Norton, B., Minson, D., and Hendricksen, R. (1980). The validity of the critical size theory for particles leaving the rumen. J. Agri. Sci. 94, 275–280. doi: 10.1017/s0021859600028859
Qin, W., Li, C., Kim, J., Ju, J., and Song, M.-K. (2012). Effects of defaunation on fermentation characteristics and methane production by rumen microbes in vitro when incubated with starchy feed sources. Asian-Australas. J. Anim. Sci. 25, 1381. doi: 10.5713/ajas.2012.12240
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramirez Ramirez, H., Harvatine, K., and Kononoff, P. (2016). Short communication: Forage particle size and fat intake affect rumen passage, the fatty acid profile of milk, and milk fat production in dairy cows consuming dried distillers grains with solubles. J. Dairy Sci. 99, 392–8. doi: 10.3168/jds.2015-10006
Rezaeian, M., Beakes, G. W., and Parker, D. S. (2004). Distribution and estimation of anaerobic zoosporic fungi along the digestive tracts of sheep. Nucleic Acids Res. 108, 1227–1233. doi: 10.1017/S0953756204000929
Satter, L. (1986). Protein and fiber digestion, passage and utilization in lactating cows. J. Dairy Sci. 69, 2734–2749. doi: 10.3168/jds.s0022-0302(87)80027-9
Schönhusen, U., Zitnan, R., Kuhla, S., Jentsch, W., Derno, M., and Voigt, J. (2003). Effects of protozoa on methane production in rumen and hindgut of calves around time of weaning. Arch. Anim. Nutr. 57, 279–295. doi: 10.1080/00039420310001594423
Shaver, R., Nytes, A., Satter, L., and Jorgensen, N. (1988). Influence of feed intake, forage physical form, and forage fiber content on particle size of masticated forage, ruminal digesta, and feces of dairy cows. J. Dairy Sci. 71, 1566–1572. doi: 10.3168/jds.S0022-0302(88)79720-9
Shi, W., Moon, C. D., Leahy, S. C., Kang, D., Froula, J., Kittelmann, S., et al. (2014). Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 24, 1517–1525. doi: 10.1101/gr.168245.113
Singh, K., Jisha, T., Reddy, B., Parmar, N., Patel, A., Patel, A., et al. (2015). Microbial profiles of liquid and solid fraction associated biomaterial in buffalo rumen fed green and dry roughage diets by tagged 16S rRNA gene pyrosequencing. Mol. Biol. Rep. 42, 95–103. doi: 10.1007/s11033-014-3746-9
Smith, P. E., Waters, S. M., Kenny, D. A., Kirwan, S. F., Conroy, S., and Kelly, A. K. (2021). Effect of divergence in residual methane emissions on feed intake and efficiency, growth and carcass performance, and indices of rumen fermentation and methane emissions in finishing beef cattle. J. Anim. Sci. 99, skab275. doi: 10.1093/jas/skab275
Soita, H., Christensen, D., McKinnon, J., and Mustafa, A. (2002). Effects of barley silage of different theoretical cut length on digestion kinetics in ruminants. Can. J. Anim. Sci. 82, 207–213. doi: 10.4141/A01-064
Solomon, K. V., Haitjema, C. H., Henske, J. K., Gilmore, S. P., Borges-Rivera, D., Lipzen, A., et al. (2016). Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science. 351, 1192–1195. doi: 10.1126/science.aad1431
Tafaj, M., Kolaneci, V., Junck, B., Maulbetsch, A., Steingass, H., and Drochner, W. (2005). Influence of fiber content and concentrate level on chewing activity, ruminal digestion, digesta passage rate and nutrient digestibility in dairy cows in late lactation. Asian. Austral. J. Anim. 18, 1116–1124. doi: 10.1080/17450390500217082
Tedersoo, L., Bahram, M., Põlme, S., Kõljalg, U., Yorou, N. S., Wijesundera, R., et al. (2014). Global diversity and geography of soil fungi. Science. 346, 1256688. doi: 10.1126/science.1256688
Teske, A., and Sørensen, K. B. (2008). Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME. J. 2, 3–18. doi: 10.1038/ismej.2007.90
Teunissen, M. J., de Kort, G. V., den Camp, H. J. O., and Huis, J. H. (1992). Production of cellulolytic and xylanolytic enzymes during growth of the anaerobic fungus Piromyces sp. on different substrates. Microbiology. 138, 1657–1664. doi: 10.1099/00221287-138-8-1657
Toju, H., Kishida, O., Katayama, N., and Takagi, K. (2016). Networks depicting the fine-scale co-occurrences of fungi in soil horizons. PloS ONE. 11, e0165987. doi: 10.1371/journal.pone.0165987
Toju, H., Tanabe, A. S., Yamamoto, S., and Sato, H. (2012). High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PloS ONE. 7, e40863. doi: 10.1371/journal.pone.0040863
Van Soest, P. (1963). A rapid method for the determination of fiber and lignin. J. Assoc. Off. Anal. Chem. 46, 829–835.
Wallace, R. J., Rooke, J. A., Duthie, C.-A., Hyslop, J. J., Ross, D. W., McKain, N., et al. (2014). Archaeal abundance in post-mortem ruminal digesta may help predict methane emissions from beef cattle. Sci. Rep. 4, 1–8. doi: 10.1038/srep05892
Wang, H., Chen, Q., Chen, L., Ge, R., Wang, M., Yu, L., et al. (2017). Effects of dietary physically effective neutral detergent fiber content on the feeding behavior, digestibility, and growth of 8-to 10-month-old Holstein replacement heifers. J. Dairy Sci. 100, 1161–1169. doi: 10.3168/jds.2016-10924
Wang, M., Sun, X., Janssen, P., Tang, S., and Tan, Z. (2014). Responses of methane production and fermentation pathways to the increased dissolved hydrogen concentration generated by eight substrates in in vitro ruminal cultures. Anim. Feed Sci. Tech. 194, 1–11. doi: 10.1016/j.anifeedsci.2014.04.012
Weatherburn, M. (1967). Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 39, 971–974. doi: 10.1021/ac60252a045
Xue, B., Wu, M., Yue, S., Hu, A., Li, X., Hong, Q., et al. (2022). Changes in Rumen Bacterial Community Induced by the Dietary Physically Effective Neutral Detergent Fiber Levels in Goat Diets. Front. Microbiol. 13, 820509–820509. doi: 10.3389/fmicb.2022.820509
Yang, W., and Beauchemin, K. (2005). Effects of physically effective fiber on digestion and milk production by dairy cows fed diets based on corn silage. J. Dairy Sci. 88, 1090–1098. doi: 10.3168/jds.S0022-0302(05)72776-4
Yang, W., and Beauchemin, K. (2006a). Increasing the physically effective fiber content of dairy cow diets may lower efficiency of feed use. J. Dairy Sci. 89, 2694–2704. doi: 10.3168/JDS.S0022-0302(06)72345-1
Yang, W., and Beauchemin, K. (2006b). Effects of physically effective fiber on chewing activity and ruminal pH of dairy cows fed diets based on barley silage. J. Dairy Sci. 89, 217–228. doi: 10.3168/jds.S0022-0302(06)72086-0
Yang, W., and Beauchemin, K. (2006c). Physically effective fiber: method of determination and effects on chewing, ruminal acidosis, and digestion by dairy cows. J. Dairy Sci. 89, 2618–2633. doi: 10.3168/jds.S0022-0302(06)72339-6
Yang, W., and Beauchemin, K. (2007). Altering physically effective fiber intake through forage proportion and particle length: Chewing and ruminal pH. J. Dairy Sci. 90, 2826–2838. doi: 10.3168/jds.2007-0032
Yildirim, E., Ilina, L., Laptev, G., Filippova, V., Brazhnik, E., Dunyashev, T., et al. (2021). The structure and functional profile of ruminal microbiota in young and adult reindeers (Rangifer tarandus) consuming natural winter-spring and summer-autumn seasonal diets. PeerJ 9, e12389. doi: 10.7717/peerj.12389
Zebeli, Q., Aschenbach, J., Tafaj, M., Boguhn, J., Ametaj, B., and Drochner, W. (2012). Invited review: Role of physically effective fiber and estimation of dietary fiber adequacy in high-producing dairy cattle. J. Dairy Sci. 95, 1041–1056. doi: 10.3168/jds.2011-4421
Keywords: goat, peNDF, nutrient digestibility, ruminal fermentation, microbial community
Citation: Zhou J, Xue B, Hu A, Yue S, Wu M, Hong Q, Wu Y, Wang Z, Wang L, Peng Q and Xue B (2022) Effect of dietary peNDF levels on digestibility and rumen fermentation, and microbial community in growing goats. Front. Microbiol. 13:950587. doi: 10.3389/fmicb.2022.950587
Received: 23 May 2022; Accepted: 28 July 2022;
Published: 25 August 2022.
Edited by:
Mukesh Kumar Awasthi, Northwest A&F University, ChinaReviewed by:
Alaa Emara Rabee, Desert Research Center, EgyptCopyright © 2022 Zhou, Xue, Hu, Yue, Wu, Hong, Wu, Wang, Wang, Peng and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bai Xue, MTM1OTJAc2ljYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.