
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 23 August 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.948657
This article is part of the Research TopicWomen in Evolutionary and Genomic Microbiology: 2022View all 4 articles
Colony growth is a common phenomenon of structured populations dispersed in nature; nevertheless, studies on the spatial distribution of colonies are largely insufficient. Here, we performed a systematic survey to address the questions of whether and how the spatial distribution of colonies was influenced by the genome and environment. Six Escherichia coli strains carrying either the wild-type or reduced genomes and eight media of varied nutritional richness were used to evaluate the genomic and environmental impacts, respectively. The genome size and nutritional variation contributed to the mean size and total area but not the variation and shape of size distribution of the colonies formed within the identical space and of equivalent spatial density. The spatial analysis by means of the Voronoi diagram found that the Voronoi correlation remained nearly constant in common, in comparison to the Voronoi response decreasing in correlation to genome reduction and nutritional enrichment. Growth analysis at the single colony level revealed positive correlations of the relative growth rate to both the maximal colony size and the Voronoi area, regardless of the genomic and nutritional variety. This result indicated fast growth for the large space assigned and supported homeostasis in the Voronoi correlation. Taken together, the spatial distribution of colonies might benefit efficient clonal growth. Although the mechanisms remain unclear, the findings provide quantitative insights into the genomic and environmental contributions to the growth and distribution of spatially or geographically isolated populations.
Studies on colony growth are important for understanding microbial ecology (Ernebjerg and Kishony, 2012; Skandamis and Jeanson, 2015), as microorganisms grown on solid surfaces often form colonies (Jeanson et al., 2015). The growth environment plays an important role in microbial growth; for example, both nitrogen and carbon can cause catabolite repression (Simpson-Lavy and Kupiec, 2019; Nair and Sarma, 2021). Studies on the growth of single colonies observed that colony growth was dependent on the nutritional condition, agar concentration and presence of other cells (Harcombe et al., 2014; Warren et al., 2019; Díaz-Pascual et al., 2021). The colony growth pattern and dynamics could be perturbed by spatial antibiotics in the growth environment (Frost et al., 2018; Sharma and Wood, 2021), which might be caused by physical and mechanical interactions (Persat et al., 2015; Tronnolone et al., 2018). Despite many studies linking environmental conditions to colony growth patterns (van Gestel et al., 2014; Cole et al., 2015; Xiong et al., 2020), the spatial distribution of colony size has rarely been reported. The landmark study first applied spatial analysis of Voronoi diagram to observe the size variation of the colonies grown on identical plates (Chacón et al., 2018). The correlation of the colony size to the spatial area assigned to the colony (i.e., Voronoi area) was observed (Xue et al., 2021). Whether such a correlation in the spatial distribution of colonies was affected by the environmental conditions was required to be addressed. In addition to the environment, genomic information is another crucial factor for colony growth. Previous studies reported that mutations in rpoS inhibited colony maturation (Ito et al., 2008, 2009) and that hemB was responsible for the formation of small mutant colonies (Bates et al., 2003). In addition, small RNAs (sRNAs) were found to be involved in biofilm formation (Van Puyvelde et al., 2013; Bak et al., 2015; Parker et al., 2017). These studies investigated the relationship between genetic information and colony growth in terms of the contribution of specific genes or sRNAs. Besides these genetic changes of defined mechanism and/or function, the large deficiency of genomic fragments was supposed to disturb colony growth significantly. Both systematic single-gene knockout (Baba et al., 2006; Joyce et al., 2006) and genome reduction (Kato and Hashimoto, 2007; Mizoguchi et al., 2008; Karcagi et al., 2016) were intensively reported. The genome-reduced strains were successfully applied to studies on metabolic engineering (Lee et al., 2009), experimental evolution (Choe et al., 2019), growth prediction (Ashino et al., 2019), origin of life (Lu et al., 2022), and so on. Whether and how genome reduction contributes to growth dynamics have been intensively studied in liquid media (Karcagi et al., 2016; Kurokawa et al., 2016), which has led to valuable insights into adaptive evolution and niche expansion (Nishimura et al., 2017; Choe et al., 2019; Kurokawa et al., 2022). Nevertheless, the contribution of genome reduction to colony growth remains unclear, as the growth dynamics in liquid media are somehow different from those on solid surfaces (Tsuchiya et al., 2018). Consequently, whether the correlation in the spatial distribution of colonies was affected by the genome reduction was unclear.
Taken together, the present study employed the model bacterium Escherichia coli (E. coli) to address the questions raised above, that is, of how genome reduction and nutritional variation effected the spatial distribution of colonies and whether genomic and environmental contributions are common. An assortment of genome-reduced E. coli strains was used to evaluate the contribution of genomic gradient to colony spatial distribution. The environmental contribution to colony spatial distribution was evaluated upon nutritionally varied media, which were adjusted by the medium composition of either the concentration of glucose in the minimal medium or the ratio of the nutritional rich medium and the minimal medium.
A total of six E. coli strains of varied genome sizes, i.e., the wild-type W3110 strain (N0) and its derivates of reduced genomes (N3, N10, N14, N20, and N28) (Supplementary Figure 1A), were employed to evaluate the effect of genome size on colony growth and spatial distribution. The concentration gradient of either glucose in the minimal medium or Luria-Bertani (LB) in the mixed medium were applied to evaluate the effect of the nutritional variation on the colony growth and spatial distribution. Multiple dilution rates and replicates of the plating were conducted for each condition, and the temporal changes in colony growth were recorded for the following analyses (Supplementary Figure 1B). The statistical parameters related to colony growth and spatial distribution, which are summarized in Table 1, were evaluated as follows. To capture the spatial distribution of the colonies grown on each plate, the number (Dk), the mean size (Mk), the total area (Tk), and the size variation (Vk) of all colonies on the plate, as well as the skewness (Zk), and kurtosis (Uk) of the distribution of colony size, which were introduced as the indicators of deviation from the normal distribution, were calculated (Figure 1 and Table 1). These parameters all described the size distribution of colonies located on the same solid space (agar plate).
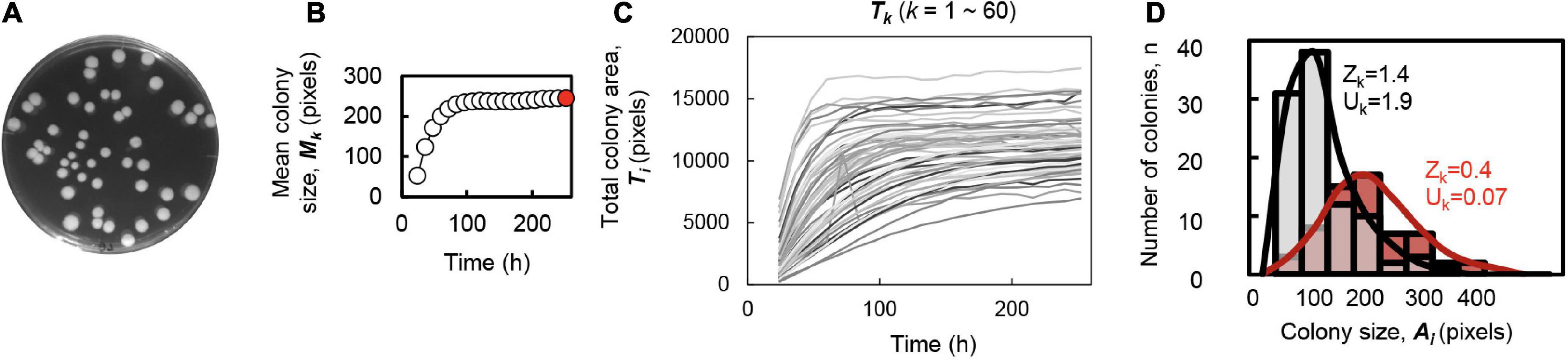
Figure 1. Parameters used for the colony spatial analyses. (A) Image of colonies on the agar plate. (B) Temporal changes in the averaged size of the colonies formed on a single plate. The steady state is indicated in red at the time point of 252 h, which is corresponded to the image shown in panel (A). (C) Temporal changes in the total area of all colonies formed on a single plate. The gradation in gray indicates a total of 60 colonies located on the identical plate. (D) Histogram of the colony size within a single plate. As representative examples, both the histograms and the fitted distributions of the areas of the colonies grown on the plates are shown. Color variation (gray and red) represents two different plates. The calculated values of skewness (Zk) and kurtosis (Uk) of the distributions are indicated.
In addition, the growth dynamics at the single colony level were evaluated with two parameters: the relative growth rate (ri) and the steady size (Ki). The spatial analysis according to the Voronoi diagram (Dale and Fortin, 2014; Baddeley et al., 2015) resulted in the parameters of the Voronoi area (Vori) for individual colonies, the Voronoi response (Resk) and the Voronoi correlation (Cork) for each plate (space). These parameters (Table 1) are further described in the corresponding results sections.
The spatial density of the colonies determined the colony size and variation within the same space, independent of the genomic and nutritional variations (Figure 2). The number of colonies grown on the agar plate (Dk) was significantly correlated with all five parameters representing the colony size and the size distribution (Mt, Tk, Vk, Uk, and Zk), regardless of genome reduction (Figure 2A), carbon abundance (Figure 2B), or nutritional richness (Figure 2C). As all these parameters described the colonies grown at steady state, the bias of the colony growth phase could be ignored. The common correlations demonstrated that the spatial distribution of colonies was largely influenced by the spatial density of colonies (i.e., the number of colonies per plate). Note that the colonies grown on the plates containing 0.2% glucose showed an enlarged Tk, which might be because the medium composition (e.g., glucose concentration) was occasionally the optimized condition for bacterial growth (Millard et al., 2017; Ashino et al., 2019).
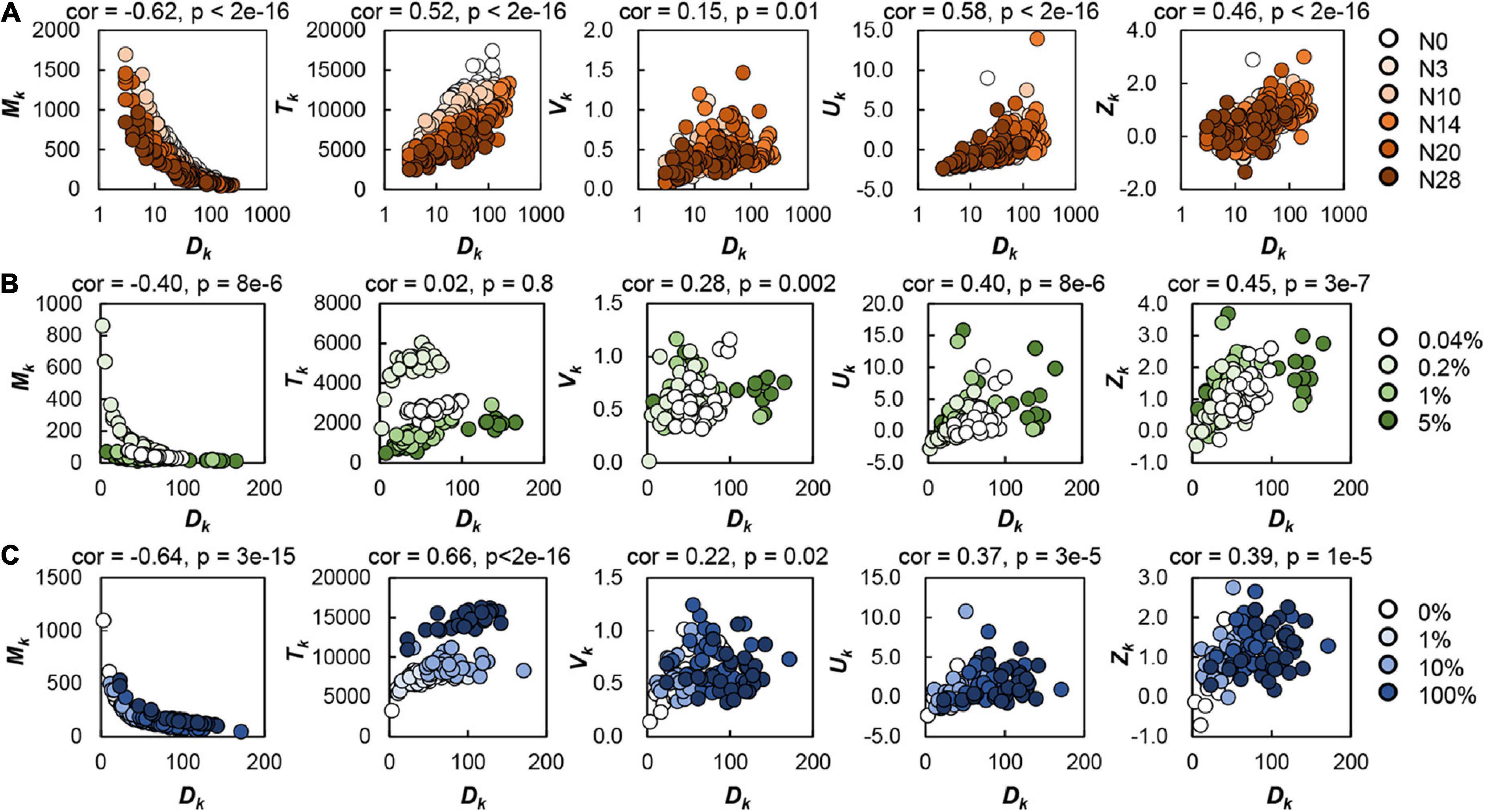
Figure 2. Effect of spatial density on colony formation. The number of colonies grown on the single plate (Dk) was plotted against the calculated parameters relating to the colony size and distribution on the identical plate, i.e., the mean size (Mk), total area (Tk) and variation (Vk) of the colonies, and the kurtosis (Uk) and skewness (Zk) of the distribution of colony size. Circles represent the plates. Orange gradation (N = 284) indicates the genome size from the wild type to the reduced ones (A). Green gradation (N = 117) represents the concentration gradient of glucose in the minimal medium M63 (B). Blue gradation (N = 119) represents the ratio gradient of LB in the mixed medium M63LB (C). Pearson’s correlation coefficients and the p-values are indicated.
The increase in the spatial density of colonies led to a decrease in the mean size of colonies, whereas it was associated with an increase in the total area of colonies (Figure 2, left two panels). A larger number of small colonies might be more efficient in resource utilization than a smaller number of large colonies. The variation in colonies within the same space changed in response to the increased spatial density (Figure 2, middle panels). The higher density of the colonies on the plate (Dk) resulted in the larger variation of colony size (Vk), indicating the larger differentiation in growth of the colonies located on the same space. This was supported by the positive correlations of Dk to both Zk and Uk (Figure 2, right two panels), as both the longer right tailed and the more sharped distributions, represented by larger Zk and Uk, respectively, indicated the biased growth of colonies.
Both the mean size and the total area of the colonies were significantly correlated with the genome size and nutritional variation; however, the variation and distribution of colony size remained constant (Figure 3 and Supplementary Figure 2). Note that only plates with a comparable number of colonies (25–75 colonies/plate) were used for the analyses to reduce the effect caused by the spatial density. As genome reduction was additively cumulative (Mizoguchi et al., 2008; Kurokawa et al., 2016), the negative correlation between colony size and the length of genomic deletion (Figure 3A) implied that it was the genome size but not the specific gene function that played a role in colony growth. The findings were consistent with the growth dynamics of genome-reduced E. coli strains in liquid media (Kurokawa et al., 2016; Nishimura et al., 2017).
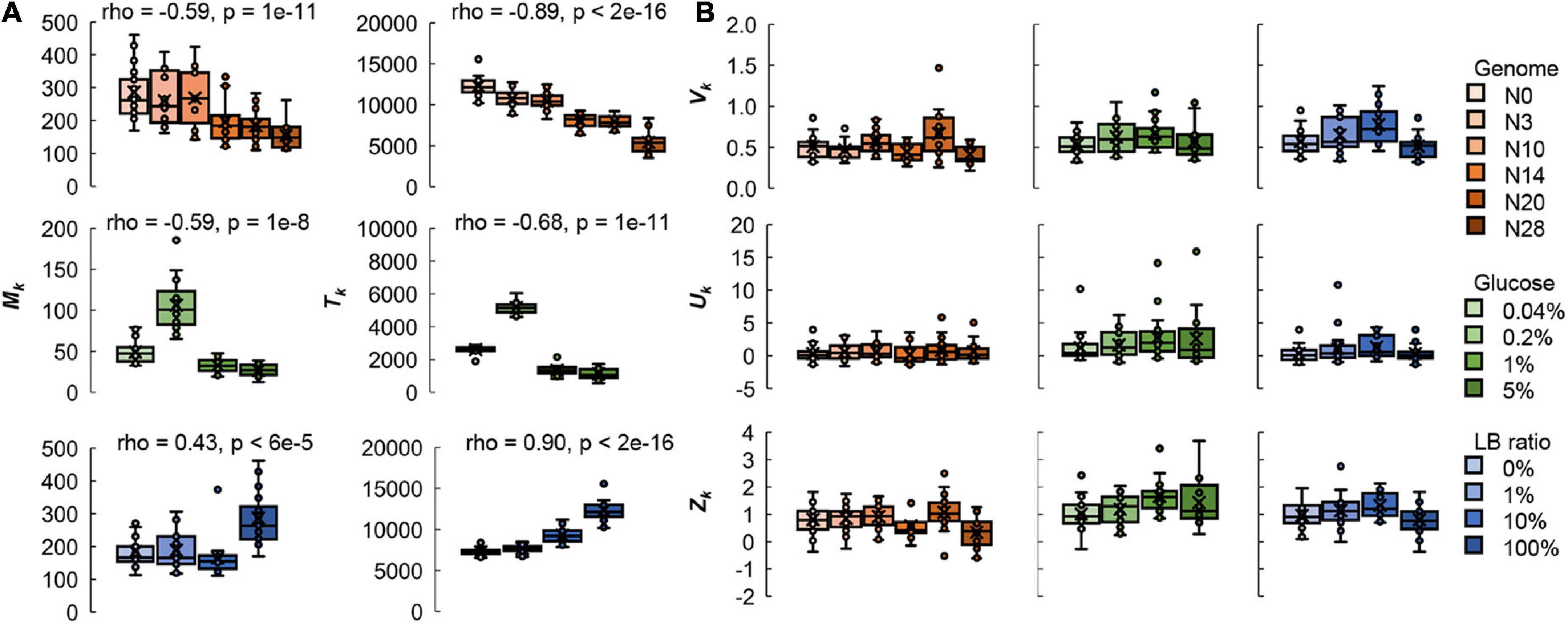
Figure 3. Boxplots of the global parameters relating to the size distribution of colonies. (A) Genomic and nutritional gradient-dependent parameters. The mean size (Mk) and total area (Tk) of the colonies on individual plates are shown. (B) Genomic and nutritional gradient independent parameters. The variation (Vk) of the colonies and the kurtosis (Uk) and skewness (Zk) of the distribution of colony size are shown. Tiny circles and crosses represent the parameters of individual plates of 25–75 colonies and their mean values, respectively. Gradations in orange, green, and blue indicate the genome size from the wild type to the reduced ones (N = 111), the concentration gradient of glucose in the minimal medium M63 (N = 78) and the ratio gradient of LB in the mixed medium M63LB (N = 80). The individual data points can be found in Supplementary Figure 3. Spearman’s correlation coefficients and the p-values, which were of statistical significance, are indicated.
The nutritional richness and the carbon abundance showed differentiated effects on colony growth. The mean size and total area of the colonies were negatively correlated with the concentration of glucose in the minimal medium, M63 (Figure 2B, left two panels), whereas they were positively correlated with the ratio of LB in the mixed medium, M63LB (Figure 2C, left two panels). The enrichment of the single resource of glucose and the overall nutritional richness (LB) in the media resulted in the reverse directional changes in the colony size. This result strongly suggested that the balance of the medium composition largely participated in colony growth, as similar as to population growth in liquid media (Ashino et al., 2019; Aida et al., 2022). In addition, the coefficient of variation of colony size (Vk), kurtosis (Uk), and skewness (Zk) of the size distribution of colonies all remained roughly comparable (p > 0.05), independent of either genome reduction or nutritional variation (Figure 3B). The results implied that the genetic and environmental variety disturbed the saturated colony size and/or the biomass abundance but not the size variation and distribution of colonies.
To evaluate the effect of the spatial distribution on the growth of colonies, the spatial analysis was performed using the Voronoi diagram, a practical method to divide the multiple distributed points into regions, as reported previously (Chacón et al., 2018; Xue et al., 2021). The parameter of the Voronoi area (Vori) was calculated, indicating the space region on the plate assigned to the corresponding colony (Figure 4A). To discover the relationship between the relative size of the colony (Ai) and the space in charge (Vori), the correlation coefficient (Cork) and the slope of linear regression (Resk) were evaluated (Figure 4B), as reported by previous studies (Chacón et al., 2018; Chacon et al., 2020). As Cork and Resk were generally employed in the studies of spatial analysis for the patterns in geography and ecology (Dale and Fortin, 2014), the explanation in microbiology (e.g., pattern in colony growth) were the fairness of space division for colony growth and the efficiency of resource utilizing of the colony in the assigned space area, respectively.
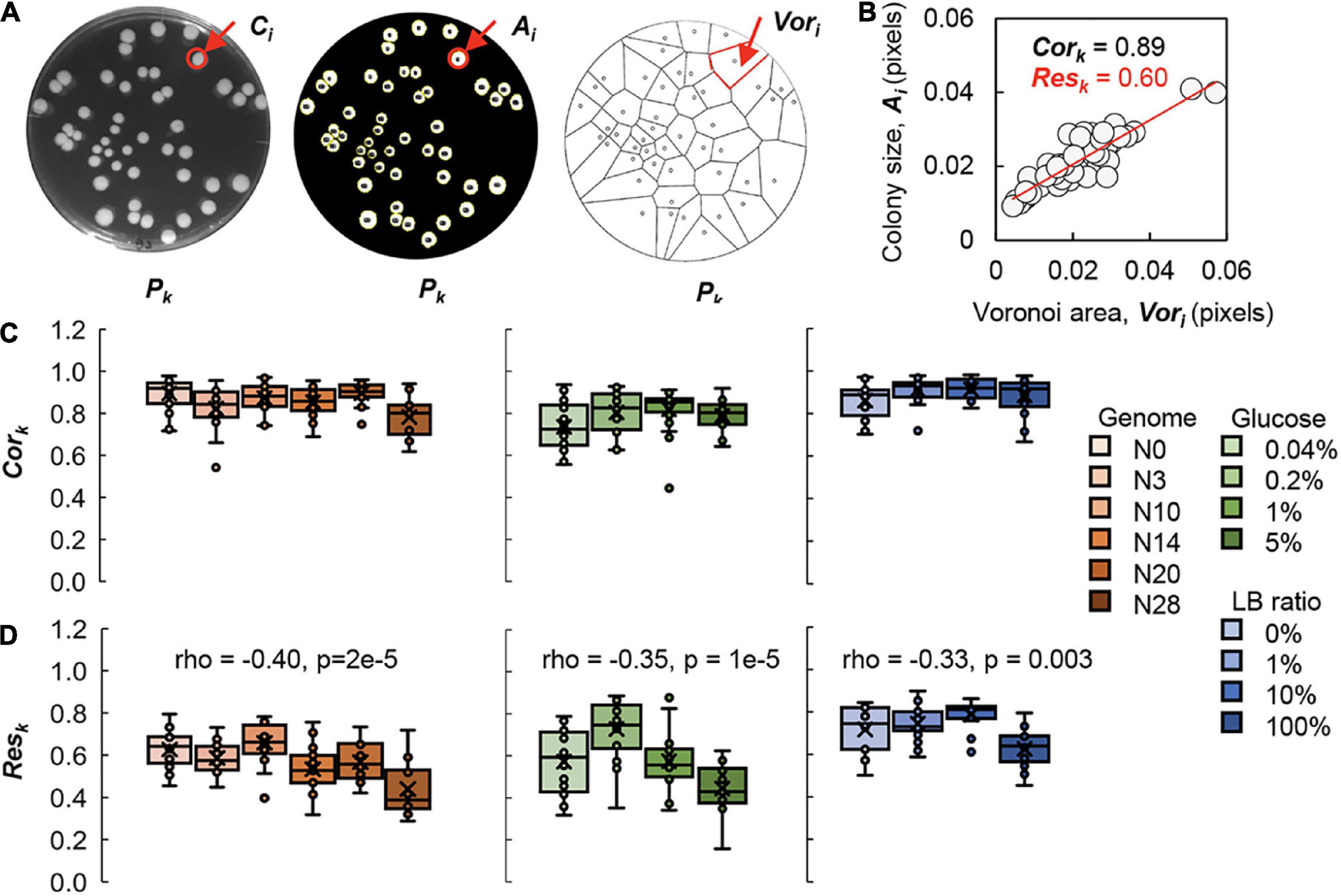
Figure 4. Voronoi diagram analysis. (A) Image analysis. The original image, the processed image and the Voronoi diagram of the agar plate (Pk) are shown from left to right as an example. The single colony (Ci), the corresponding colony size (Ai) and its Voronoi area (Vori) are indicated by arrows and highlighted in red. (B) Evaluation of Voronoi correlation and Voronoi response. The Voronoi correlation (Cork) and Voronoi response (Resk) are defined as the correlation coefficient and the slope of linear regression of the relative Voronoi area and the relative colony size, respectively. As an example, the values of Cork and Resk are indicated in black and red, respectively. Circles represent the colonies (N = 47). (C) Relationships of Cork to the genome size and nutritional gradient. Boxplots of Cork acquired from each plate are shown. (D) Correlations of Resk to the genome size and nutritional gradient. Boxplots of Resk acquired from each plate are shown. Tiny circles represent the Cork of individual plates of 25–75 colonies. The crosses indicate the average of Cork. Gradations in orange, green and blue indicate the genome size from the wild type to the reduced ones (N = 111), the concentration gradient of glucose in the minimal medium M63 (N = 78) and the ratio gradient of LB in the mixed medium M63LB (N = 80). Spearman’s correlation coefficients and the p-values are indicated.
The analytical results showed that Cork remained roughly constant, regardless of the changes in genome size or nutritional variety (Figure 4C). The value of Cork was close to 1, which revealed that the area charged by the colony was highly positively correlated with the growth capacity of the colony. The results clearly demonstrated the generality of the Voronoi correlation (Cork) in colony growth, which strongly supported our previous finding (Xue et al., 2021) and indicated the commonly fair division of the space area for colony growth.
In comparison to the conserved Voronoi correlation, the Voronoi response was changed in response to the genomic and nutritional differences, which was complementary to the previous report on the wild-type bacterium growing on different carbon sources (Chacón et al., 2018). Weak but significant correlations of Resk with both genome reduction and nutritional richness were detected (Figure 4D). The decrease in Resk indicated that the decrease in colony size was correlated with the genome size and nutritional variation, although the assigned space for the colony, i.e., Vori, remained equivalent. This strongly suggested the reduced efficiency of resource utilization associated with genome reduction and nutritional enrichment. Note that the findings were unbiased by the experimental (plating) conditions, as the Vori was independent of genome reduction or nutritional variation (Supplementary Figure 3). This might be the reason why the Voronoi correlation remained conserved. The spatial density of colonies (Dk) was slightly influenced by either Cork or Resk (Supplementary Figure 4); nevertheless, the conclusions were drawn from the plates of comparable Dk.
To understand the genomic and nutritional differences correlated with changes in the spatial distribution, the growth of a single colony was analyzed, as a previous study proposed that the Voronoi response might be attributed to the growth rate (Chacon et al., 2020). According to the temporal changes in the size of a single colony, the relative growth rate (r) and the maximal colony size (K) were calculated (Figure 5A). Analyzing a total of 80 single colonies, we observed a significant positive correlation between the relative growth rate and the maximal size at the single colony level (Figure 5B). Although the colonies were grown on the various plates, the faster the colonies grew, the larger the colonies formed. It seemed to be a general mechanism, regardless of genome reduction or nutritional variety. Furthermore, the spatial analysis found a positive correlation between the growth rate and the Voronoi area at the single colony level (Figure 5C). This result indicated that the fast-growing colonies tended to have a large spatial area for colony growth in the future. Taken together, the increased growth rate was beneficial to occupying (assigning) the broadened space for continuous growth to achieve the enlarged size of the final population/colony. The coordinated relationship among ri, Ki, and Vori at the single colony level well explained the universality of the Voronoi correlation at the spatial level (Figure 4C).
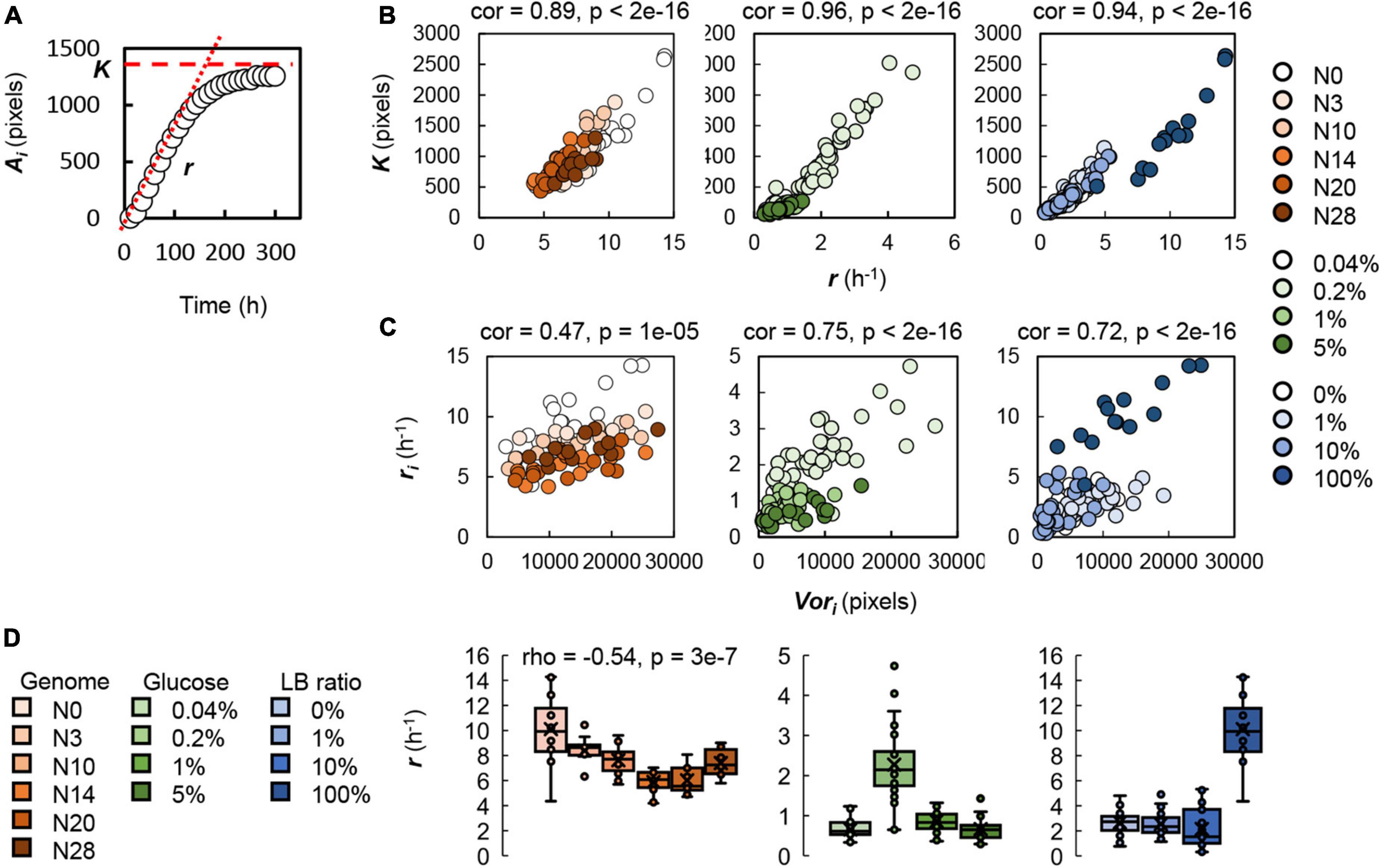
Figure 5. Single colony analysis. (A) Growth curve of a single colony. Circles indicate the temporal changes in the colony size. The relative growth rate (r) and the steady size of the colony (K) are indicated by red dashed lines. (B) Correlation between the relative growth rate and the steady size of the colonies. (C) Correlation between the Voronoi area and the relative growth rate. Circles represent the colonies. Gradations in orange, green and blue indicate the genome size from the wild type to the reduced ones (N = 80), the concentration gradient of glucose in the minimal medium M63 (N = 131) and the ratio gradient of LB in the mixed medium M63LB (N = 141). Pearson’s correlation coefficients and the statistical significance are indicated. (D) Relationships of genome reduction and nutritional richness with the growth rate. Boxplots of the relative growth rates acquired from each condition are shown. The tiny circles and the crosses represent the individual ri and the average ri, respectively. Color variation in orange, green and blue and the numbers of single colonies used for the analysis are described above. Spearman’s correlation coefficients and the p-value, which was of statistical significance, are indicated.
On the other hand, the relative growth rates of single colonies were correlated to genome reduction but not nutritional enrichment (Figure 5D). According to the coordination among ri, Ki, and Vori (Figures 5B,C), the genomic gradient-correlated decrease in the growth rate triggered the reduced Voronoi area. Consequently, the Voronoi response declined in association with genome reduction, that is, the decreased Vori for the equivalent size of colonies carrying the reduced genomes. The contribution of genome reduction to both the colony size at the spatial level (Figure 3A, upper panels) and the Voronoi response in spatial distribution (Figure 4D, left panel) could be explained by the growth rate at the single colony level. However, the single colony growth analysis failed to explain the nutritional enrichment-associated decrease in the Voronoi response in spatial distribution. Although both genome reduction and the medium variety led to changes in colony growth, the mechanisms of the consequent changes in the Voronoi response seemed to be differentiated. Genetic restriction and resource limitation must have stimulated the varied regulatory pathways to achieve the maximal population size, although both caused growth decline.
It was an intriguing finding that the Voronoi correlation remained homeostatic, whereas the Voronoi response was differentiated in response to genetic and environmental changes. The steady Cork indicated that the colonies were grown in proportion to the assigned space (Vori) regardless of the genomic and nutritional variety, which indicated that the resource space was distributed to the colonies according to their sizes. The fluctuation in Resk indicated that the magnitudes of the changes in colony size of the identical Vori were changed due to genomic and nutritional interruptions, which indicated that the efficiency of resource utilization was different depending on the genome and environment. Although Cork remained approximately steady, its slight change was somehow linked to the large fluctuation in Resk. A weak but significant correlation between Cork and Resk was observed (Figure 6). The spatial distribution of colonies was supposed to benefit the efficient utilization of space and/or nutritional resources. This finding well agreed with the natural relationship that the larger allocated region allowed more resources available for population increase. This relationship seemed to be true not only for the homogeneously growing populations in liquid media but also for the geographically isolated populations. The results quantitatively proved the ecological rule of the colony growth in response to its spatial distribution, which was likely common in E. coli independent of the genomic and nutritional variation.
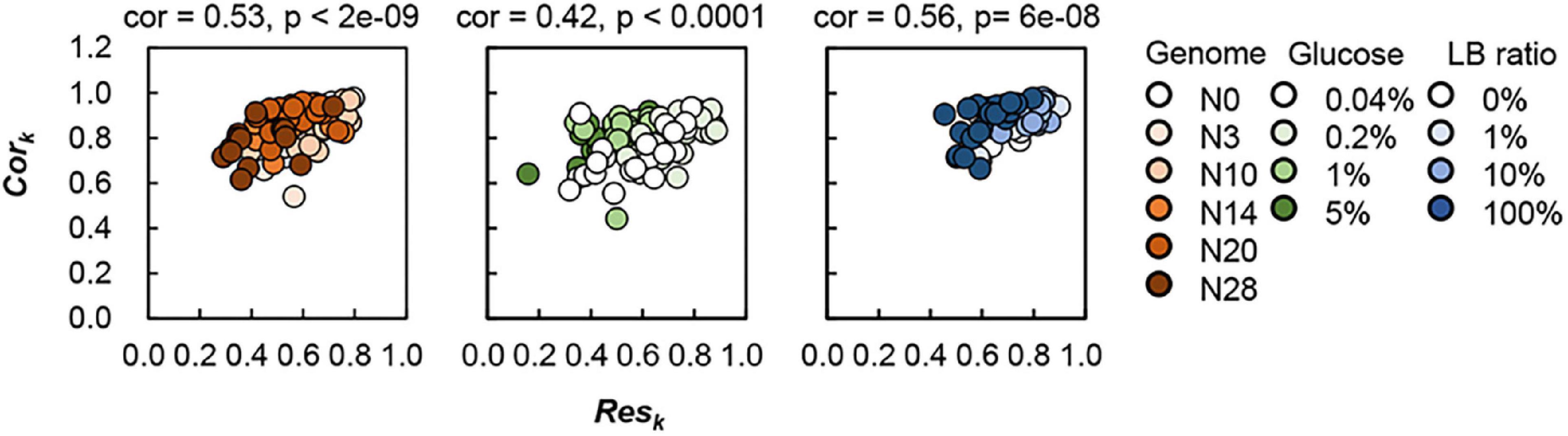
Figure 6. Relationship between the Voronoi response and Voronoi correlation. Circles represent the plates. Gradations in orange, green and blue indicate the genome size from the wild type to the reduced ones (N = 111), the concentration gradient of glucose in the minimal medium M63 (N = 78) and the ratio gradient of LB in the mixed medium M63LB (N = 80). Pearson’s correlation coefficients and the p-values are indicated.
The genetic and environmental dependent changes in Resk were supposed to be decided by the colony growth rate. As a consequence of ecological evolution (Sexton et al., 2017), the growth rate was supposed to be negatively correlated with the population size (Luckinbill, 1978; Engen et al., 2013), which is known as the trade-off mechanism (Cavalier-Smith, 1980; Weisse et al., 2015; Ferenci, 2016). However, instead of a trade-off, a positive correlation was observed in colony growth (Figure 5B). The decreased growth rate resulted in a small colony, which was assigned to the identical Vori. Consequently, the Voronoi response, the slope between colony size and Vori, decreased. The disappearance of the trade-off between colony growth rate and maximal colony size well explained the changes in the Voronoi response. As the trade-off mechanism was explained by the metabolic and enzymatic costs (Molenaar et al., 2009; Noor et al., 2014; Wortel et al., 2018), whether the positive correlation between growth rate and growth max was caused by any other alternative mechanism involved in metabolic and/or cellular pathways remains to be addressed in the future.
The systematic survey on colony growth evaluated the impacts of genome reduction and nutritional richness on the bacterial spatial distribution. The genetic and environmental dependent Voronoi response associated with the homeostatic Voronoi correlation was observed. The genome reduction-mediated growth decrease, which was observed in the uniformed populations grown in the liquid media (Kurokawa et al., 2016), was demonstrated to be general in the spatially separated colonies grown on the solid media. It seemed that it was the abundance but not the specificity of the genetic information that participated in the bacterial population propagation. In summary, the present study is the first to connect genomic and nutritional variety with the spatial distribution of bacterial colonies. These findings provide quantitative insights into the genomic and environmental contributions to the growth and distribution of spatially or geographically isolated populations. Since only an assortment of the laboratory engineered E. coli strains grown in the well-controlled conditions were examined in the present study, further investigation of different microbes is required to draw a universal conclusion on the fairness in the spatial distribution of colonies for efficient clonal growth. The spatial patterns and growth of colonies were supposed to be affected by multiple factors, such as the cross-sectional structure of colonies, the cellular interaction within a single colony, the interaction between neighboring colonies, and the nutritional diffusion in agar media. Clarifying these underground mechanisms is highly intriguing to connect bacterial morphology to colony life.
A total of six E. coli strains were used, including the wild-type E. coli strain K-12 W3110 and five of its derivative genome-reduced strains. These six strains were designated Nos. 0, 3, 10, 14, 20, and 28 (Supplementary Figure 1A), as described previously (Kurokawa et al., 2016; Nishimura et al., 2017). The lengths of deleted sequences are 88.7, 481.4, 709.5, 899.0, and 982.4 kb for strains Nos. 3, 10, 14, 20, and 28, respectively. The E. coli strains were from the KHK collection (Mizoguchi et al., 2008), distributed by the National BioResource Project, National Institute of Genetics, Shizuoka, Japan.
Both the rich medium LB (Sigma) and the minimal medium M63, of which the chemical composition was described in detail previously (Kurokawa and Ying, 2017), were used. Nutritional variation of the media was prepared either by diluting LB with M63 or by varying the amount of glucose in M63, which resulted in four different nutritional levels of 100, 10, 1, and 0% LB media and 5, 1, 0.2, and 0.04% glucose M63 media, respectively. The glycerol stocks of the E. coli strains, prepared beforehand as previously described (Kurokawa et al., 2016; Kurokawa and Ying, 2017), were diluted to a final concentration of ∼1,000 cells/ml. The diluted culture solutions were further diluted twofold, which led to different initial cell concentrations for plating (Supplementary Figure 1B). The media used for the dilution was equivalent to the media used for the agar plate for colony growth. For colony formation, every 100 μl of the diluted bacterial solution was plated on each plate (1.5% agar). The plates were incubated at 37°C in an incubator (THS030PA, ADVANTEC). More than 10 replicates were conducted for each dilution rate of each strain at each medium condition. Approximately 600 plates were inoculated and incubated for colony growth.
The plates were photographed with a high-sensitivity monochrome CCD camera of a gel imager (AE-6932GXES print graph, ATTO Co., Ltd.). Brightness, contrast, saturation, hue, sharpness, OSD time, and exposure time were set as previously described (Xue et al., 2021). Temporal changes in colony growth were captured at intervals of 12 h (Supplementary Figure 1B). The imaging of the plates (colony growth) was started at 24 h after inoculation and stopped at 300 h. The steady state of the colonies was determined at 252 h. The images were saved as TIF files and subjected to the computational analysis described in the following sections. A total of approximately 5,000 images of the temporal changes in colony growth were acquired from 520 plates, which were qualified for imaging and computational analysis.
Image data analysis was performed with Fiji, an open-source image processing package based on ImageJ.1 Background subtraction and binarization were performed on the acquired image data, and the edges of the plates were removed as noise. Only the images of high-resolution colonies were processed for the analysis. The coordinates of the center of each plate, the location of the colonies and the corresponding colony size Ai (i = 1∼n; n, number of colonies in the plate) were automatically calculated by the command Fiji. The number of colonies in each plate was defined as the colony density, Dk (k = 1∼N; N, number of plates). According to the calculated Ai, the total area of colonies in each plate Tk, the average colony size Mk, and the colony size variation Vk were calculated. Skewness (Zk) and kurtosis (Uk) were calculated using the “kurtosis” and “skewness” functions in the e1071 package of the R programming software. The calculations of these parameters were performed according to the following equations (Eqs. 1–6). These parameters are summarized in Table 1 and are shown in Figure 1.
The Voronoi region, Vori (i = 1∼n; n, number of colonies), which is the theoretical region assigned to each colony, was calculated according to the following formula, as previously described (Xue et al., 2021).
Here, V(cl) and d represent the Voronoi area of the colony cl and the function of the distance, respectively. l and m are natural numbers less than or equal to i, the number of colonies. x and y represent the X and Y coordinates of the colonies on the plates, which were used for the Voronoi division. The colonies and the agar plate were considered the genetic points (c1, c2, …, cn) and the metric space (C), respectively. The tessellation {V(c1), V(c2), …, V(ck)} refers to the Voronoi diagram. The Voronoi areas were calculated with the function “dirichlet” in the spatial statistics package “spatstat” in the software R.
The growth curves of a total of 80 colonies were analyzed in detail. To avoid the bias caused by the colony density, plates with an equivalent number of colonies (5–11 colonies/plate) were subjected to growth analysis. The growth rate of a single colony at a certain time point, t, was defined as the slope of two neighboring records at time points t and t-12 (Eq. 10.1).
Here, rt and A(t) represent the growth rate and the colony size at time point t, respectively. To avoid outliers, the maximal slope, rt, was averaged with its two neighboring slopes, r(t–12) and r(t+12). The advantages and methodology were described previously (Tsuchiya et al., 2018; Ashino et al., 2019). The resultant mean value was determined as the relative growth rate of colony i (Eq. 10.2).
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
B-WY conceived the research. KH and JW conducted the experiments. KH and B-WY analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the JSPS KAKENHI Grant-in-Aid for Scientific Research (B) (grant number: 19H03215) and partially by Grant-in-Aid for Challenging Exploratory Research (grant number: 21K19815).
We thank NBRP for providing the E. coli strains carrying the wild-type and reduced genomes.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.948657/full#supplementary-material
Aida, H., Hashizume, T., Ashino, K., and Ying, B.-W. (2022). Machine learning-assisted discovery of growth decision elements by relating bacterial population dynamics to environmental diversity. bioRxiv [Preprint]. doi: 10.1101/2022.02.10.479953
Ashino, K., Sugano, K., Amagasa, T., and Ying, B. W. (2019). Predicting the decision making chemicals used for bacterial growth. Sci. Rep. 9, 7251.
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2:2006.0008. doi: 10.1038/msb4100050
Baddeley, A., Rubak, E., and Turner, R. (2015). Spatial point patterns: Methodology and applications with R. Boca Raton, FL: Chapman and Hall/CRC.
Bak, G., Lee, J., Suk, S., Kim, D., Young Lee, J., Kim, K. S., et al. (2015). Identification of novel sRNAs involved in biofilm formation, motility, and fimbriae formation in Escherichia coli. Sci. Rep. 5:15287. doi: 10.1038/srep15287
Bates, D. M., von Eiff, C., McNamara, P. J., Peters, G., Yeaman, M. R., Bayer, A. S., et al. (2003). Staphylococcus aureus menD and hemB mutants are as infective as the parent strains, but the menadione biosynthetic mutant persists within the kidney. J. Infect. Dis. 187, 1654–1661. doi: 10.1086/374642
Cavalier-Smith, T. (1980). r- and K-tactics in the evolution of protist developmental systems: Cell and genome size, phenotype diversifying selection, and cell cycle patterns. Biosystems 12, 43–59. doi: 10.1016/0303-2647(80)90037-4
Chacón, J. M., Möbius, W., and Harcombe, W. R. (2018). The spatial and metabolic basis of colony size variation. ISME J. 12, 669–680. doi: 10.1038/s41396-017-0038-0
Chacon, J. M., Shaw, A. K., and Harcombe, W. R. (2020). Increasing growth rate slows adaptation when genotypes compete for diffusing resources. PLoS Computat. Biol. 16:e1007585. doi: 10.1371/journal.pcbi.1007585
Choe, D., Lee, J. H., Yoo, M., Hwang, S., Sung, B. H., Cho, S., et al. (2019). Adaptive laboratory evolution of a genome-reduced Escherichia coli. Nat. Commun. 10:935. doi: 10.1038/s41467-019-08888-6
Cole, J. A., Kohler, L., Hedhli, J., and Luthey-Schulten, Z. (2015). Spatially-resolved metabolic cooperativity within dense bacterial colonies. BMC Syst. Biol. 9:15. doi: 10.1186/s12918-015-0155-1
Dale, M. R., and Fortin, M.-J. (2014). Spatial analysis: A guide for ecologists. Cambridge: Cambridge University Press.
Díaz-Pascual, F., Lempp, M., Nosho, K., Jeckel, H., Jo, J. K., Neuhaus, K., et al. (2021). Spatial alanine metabolism determines local growth dynamics of Escherichia coli colonies. elife 10:e70794. doi: 10.7554/eLife.70794
Engen, S., Lande, R., and Saether, B. E. (2013). A quantitative genetic model of r- and K-selection in a fluctuating population. Am. Nat. 181, 725–736. doi: 10.1086/670257
Ernebjerg, M., and Kishony, R. (2012). Distinct growth strategies of soil bacteria as revealed by large-scale colony tracking. Appl. Environ. Microbiol. 78, 1345–1352. doi: 10.1128/AEM.06585-11
Ferenci, T. (2016). Trade-off mechanisms shaping the diversity of bacteria. Trends Microbiol. 24, 209–223.
Frost, I., Smith, W. P. J., Mitri, S., Millan, A. S., Davit, Y., Osborne, J. M., et al. (2018). Cooperation, competition and antibiotic resistance in bacterial colonies. ISME J. 12, 1582–1593. doi: 10.1038/s41396-018-0090-4
Harcombe, W. R., Riehl, W. J., Dukovski, I., Granger, B. R., Betts, A., Lang, A. H., et al. (2014). Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 7, 1104–1115. doi: 10.1016/j.celrep.2014.03.070
Ito, A., May, T., Kawata, K., and Okabe, S. (2008). Significance of rpoS during maturation of Escherichia coli biofilms. Biotechnol. Bioeng. 99, 1462–1471.
Ito, A., May, T., Taniuchi, A., Kawata, K., and Okabe, S. (2009). Localized expression profiles of rpoS in Escherichia coli biofilms. Biotechnol. Bioeng. 103, 975–983. doi: 10.1002/bit.22305
Jeanson, S., Floury, J., Gagnaire, V., Lortal, S., and Thierry, A. (2015). Bacterial colonies in solid media and foods: A review on their growth and interactions with the Micro-environment. Front. Microbiol. 6:1284. doi: 10.3389/fmicb.2015.01284
Joyce, A. R., Reed, J. L., White, A., Edwards, R., Osterman, A., Baba, T., et al. (2006). Experimental and computational assessment of conditionally essential genes in Escherichia coli. J. Bacteriol. 188, 8259–8271. doi: 10.1128/JB.00740-06
Karcagi, I., Draskovits, G., Umenhoffer, K., Fekete, G., Kovács, K., Méhi, O., et al. (2016). Indispensability of horizontally transferred genes and its impact on bacterial genome streamlining. Mol. Biol. Evol. 33, 1257–1269. doi: 10.1093/molbev/msw009
Kato, J., and Hashimoto, M. (2007). Construction of consecutive deletions of the Escherichia coli chromosome. Mol. Syst. Biol. 3:132. doi: 10.1038/msb4100174
Kurokawa, M., and Ying, B. W. (2017). Precise, high-throughput analysis of bacterial growth. J. Vis. Exp. 127:56197.
Kurokawa, M., Nishimura, I., and Ying, B. W. (2022). Experimental evolution expands the breadth of adaptation to an environmental gradient correlated with genome reduction. Front. Microbiol. 13:826894. doi: 10.3389/fmicb.2022.826894
Kurokawa, M., Seno, S., Matsuda, H., and Ying, B. W. (2016). Correlation between genome reduction and bacterial growth. DNA Res. 23, 517–525.
Lee, J. H., Sung, B. H., Kim, M. S., Blattner, F. R., Yoon, B. H., Kim, J. H., et al. (2009). Metabolic engineering of a reduced-genome strain of Escherichia coli for L-threonine production. Microb. Cell Fact. 8:2.
Lu, H., Aida, H., Kurokawa, M., Chen, F., Xia, Y., Xu, J., et al. (2022). Primordial mimicry induces morphological change in Escherichia coli. Commun. Biol. 5:24. doi: 10.1038/s42003-021-02954-w
Luckinbill, L. S. (1978). r and K selection in experimental populations of Escherichia coli. Science 202, 1201–1203.
Millard, P., Smallbone, K., and Mendes, P. (2017). Metabolic regulation is sufficient for global and robust coordination of glucose uptake, catabolism, energy production and growth in Escherichia coli. PLoS Comput. Biol. 13:e1005396. doi: 10.1371/journal.pcbi.1005396
Mizoguchi, H., Sawano, Y., Kato, J., and Mori, H. (2008). Superpositioning of deletions promotes growth of Escherichia coli with a reduced genome. DNA Res. 15, 277–284. doi: 10.1093/dnares/dsn019
Molenaar, D., van Berlo, R., de Ridder, D., and Teusink, B. (2009). Shifts in growth strategies reflect tradeoffs in cellular economics. Mol. Syst. Biol. 5:323. doi: 10.1038/msb.2009.82
Nair, A., and Sarma, S. J. (2021). The impact of carbon and nitrogen catabolite repression in microorganisms. Microb. Res. 251:126831. doi: 10.1016/j.micres.2021.126831
Nishimura, I., Kurokawa, M., Liu, L., and Ying, B. W. (2017). Coordinated changes in mutation and growth rates induced by genome reduction. mBio 8:e00676-17. doi: 10.1128/mBio.00676-17
Noor, E., Bar-Even, A., Flamholz, A., Reznik, E., Liebermeister, W., and Milo, R. (2014). Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLoS Comput. Biol. 10:e1003483. doi: 10.1371/journal.pcbi.1003483
Parker, A., Cureoglu, S., De Lay, N., Majdalani, N., and Gottesman, S. (2017). Alternative pathways for Escherichia coli biofilm formation revealed by sRNA overproduction. Mol. Microbiol. 105, 309–325. doi: 10.1111/mmi.13702
Persat, A., Nadell, C. D., Kim, M. K., Ingremeau, F., Siryaporn, A., Drescher, K., et al. (2015). The mechanical world of bacteria. Cell 161, 988–997.
Sexton, J. P., Montiel, J., Shay, J. E., Stephens, M. R., and Slatyer, R. A. (2017). Evolution of ecological niche breadth. Annu. Rev. Ecol. Evol. Syst. 48, 183–206.
Sharma, A., and Wood, K. B. (2021). Spatial segregation and cooperation in radially expanding microbial colonies under antibiotic stress. ISME J. 15, 3019–3033. doi: 10.1038/s41396-021-00982-2
Simpson-Lavy, K., and Kupiec, M. (2019). Carbon catabolite repression: Not only for glucose. Curr. Genet. 65, 1321–1323.
Skandamis, P. N., and Jeanson, S. (2015). Colonial vs. planktonic type of growth: Mathematical modeling of microbial dynamics on surfaces and in liquid, semi-liquid and solid foods. Front. Microbiol. 6:1178. doi: 10.3389/fmicb.2015.01178
Tronnolone, H., Tam, A., Szenczi, Z., Green, J. E. F., Balasuriya, S., Tek, E. L., et al. (2018). Diffusion-limited growth of microbial colonies. Sci. Rep. 8:5992.
Tsuchiya, K., Cao, Y. Y., Kurokawa, M., Ashino, K., Yomo, T., and Ying, B. W. (2018). A decay effect of the growth rate associated with genome reduction in Escherichia coli. BMC Microbiol. 18:101. doi: 10.1186/s12866-018-1242-4
van Gestel, J., Weissing, F. J., Kuipers, O. P., and Kovacs, A. T. (2014). Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME J. 8, 2069–2079. doi: 10.1038/ismej.2014.52
Van Puyvelde, S., Steenackers, H. P., and Vanderleyden, J. (2013). Small RNAs regulating biofilm formation and outer membrane homeostasis. RNA Biol. 10, 185–191.
Warren, M. R., Sun, H., Yan, Y., Cremer, J., Li, B., and Hwa, T. (2019). Spatiotemporal establishment of dense bacterial colonies growing on hard agar. eLife 8:e41093. doi: 10.7554/eLife.41093
Weisse, A. Y., Oyarzun, D. A., Danos, V., and Swain, P. S. (2015). Mechanistic links between cellular trade-offs, gene expression, and growth. Proc. Natl. Acad. Sci. U.S.A. 112, E1038–E1047. doi: 10.1073/pnas.1416533112
Wortel, M. T., Noor, E., Ferris, M., Bruggeman, F. J., and Liebermeister, W. (2018). Metabolic enzyme cost explains variable trade-offs between microbial growth rate and yield. PLoS Comput. Biol. 14:e1006010. doi: 10.1371/journal.pcbi.1006010
Xiong, L., Cao, Y., Cooper, R., Rappel, W. J., Hasty, J., and Tsimring, L. (2020). Flower-like patterns in multi-species bacterial colonies. eLife 9:48885. doi: 10.7554/eLife.48885
Keywords: spatial analysis, Voronoi diagram, genome reduction, colony size, single colony growth, agar plate, medium
Citation: Hitomi K, Weng J and Ying B-W (2022) Contribution of the genomic and nutritional differentiation to the spatial distribution of bacterial colonies. Front. Microbiol. 13:948657. doi: 10.3389/fmicb.2022.948657
Received: 20 May 2022; Accepted: 29 July 2022;
Published: 23 August 2022.
Edited by:
Ana M. S. Guimaraes, University of São Paulo, BrazilReviewed by:
Lev Tsimring, University of California, San Diego, United StatesCopyright © 2022 Hitomi, Weng and Ying. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bei-Wen Ying, eWluZy5iZWl3ZW4uZ2ZAdS50c3VrdWJhLmFjLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.