
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 08 August 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.948369
 Jonathan Lo*†
Jonathan Lo*† Jonathan R. Humphreys†
Jonathan R. Humphreys† Lauren Magnusson
Lauren Magnusson Benton Wachter
Benton Wachter Chris Urban
Chris Urban Skyler D. Hebdon
Skyler D. Hebdon Wei Xiong
Wei Xiong Katherine J. Chou
Katherine J. Chou Pin Ching Maness
Pin Ching Maness
3-Hydroxybutyrate (3HB) is a product of interest as it is a precursor to the commercially produced bioplastic polyhydroxybutyrate. It can also serve as a platform for fine chemicals, medicines, and biofuels, making it a value-added product and feedstock. Acetogens non-photosynthetically fix CO2 into acetyl-CoA and have been previously engineered to convert acetyl-CoA into 3HB. However, as acetogen metabolism is poorly understood, those engineering efforts have had varying levels of success. 3HB, using acetyl-CoA as a precursor, can be synthesized by a variety of different pathways. Here we systematically compare various pathways to produce 3HB in acetogens and discover a native (S)-3-hydroxybutyryl-CoA dehydrogenase, hbd2, responsible for endogenous 3HB production. In conjunction with the heterologous thiolase atoB and CoA transferase ctfAB, hbd2 overexpression improves yields of 3HB on both sugar and syngas (CO/H2/CO2), outperforming the other tested pathways. These results uncovered a previously unknown 3HB production pathway, inform data from prior metabolic engineering efforts, and have implications for future physiological and biotechnological anaerobic research.
The Wood Ljungdahl Pathway (WLP), used by acetogens, is considered the most energy efficient natural carbon fixation pathway (Bar-Even et al., 2012). This natural efficiency makes acetogens attractive for biotechnological purposes. Despite the promise of acetogens, the main products natively made are normally acetate and ethanol, which are generally lower value products with limited market sizes. 3-Hydroxybutyrate (3HB) is a chiral bioproduct of interest, with a variety of uses and applications. 3HB can be used for the synthesis of fine chemicals, medicines, biofuels, and bioplastics, especially polyhydroxybutyrate (PHB), which is a highly biodegradable bioplastic (Tokiwa and Ugwu, 2007). 3HB can also be co-polymerized with other biodegradable polymers to extend their use case and improve physical properties (Li et al., 2016; Cywar et al., 2021). With the environmental persistence of petroleum-based plastics and concerns over fossil fuels use and emissions, PHBs offer a potentially environmentally friendly alternative that is both biodegradable and reduces CO2 emissions. As a result of these attributes, PHBs are currently industrially produced by a number of companies, representing a growing market and active area of research (Chen, 2009).
As PHB/3HB prices are heavily influenced by substrates costs (i.e., sugar/oils), increased efficiency and use of marginal inexpensive substrates allows for cheaper production and reduced carbon emissions (Lee, 1996). Production of 3HB in acetogens is attractive for several reasons. Acetogens are potentially more carbon efficient at producing 3HB from sugars due to their ability to grow mixotrophically (Jones et al., 2016). Using the WLP, acetogens can recapture CO2 from pyruvate decarboxylation to generate 3 acetyl-CoA per glucose, versus the 2 acetyl-CoA normally produced by heterotrophic glycolysis. As such, 3HB production is theoretically 36% more efficient in acetogens on sugar alone and can theoretically result in complete carbon conversion with added H2 (Jones et al., 2016). Acetogens can also use syngas to generate acetyl-CoA, a precursor of 3HB biosynthesis, which means 3HB can be generated from carbon emitting sources directly, resulting in a carbon negative product. Recent technoeconomic analysis has shown that PHB from CO2 is potentially cost competitive at current prices (Huang et al., 2021). As a result, there is increasing environmental interest in engineering 3HB production in acetogens.
In literature, there are two main pathways proposed to generate 3HB in acetogens, with varying levels of success (Figure 1). These two pathways will be referred to by their critical genes as hbd1 ((S)-3-hydroxybutyryl-CoA dehydrogenase) (Figure 1A), and ctfAB (CoA transferase)/3hbdh (3-hydroxybutyrate dehydrogenase) (Figure 1B) (Jones et al., 2016; Woolston et al., 2018; de Souza Pinto Lemgruber et al., 2019; Flüchter et al., 2019; Karim et al., 2020; Jia et al., 2021). Both pathways require a thiolase to combine two acetyl-CoA molecules into acetoacetyl-CoA, however, from here the pathways diverge. After the formation of acetoacetyl-CoA, Hbd1 reduces acetoacetyl-CoA to (S)-3-hydroxybutyryl-CoA which is subsequently converted to 3HB via a thioesterase through removal of CoA. Conversely, the ctfAB/3hbdh pathway first uses the CtfAB to convert acetoacetyl-CoA to acetoacetate by transferring the CoA to acetate. From here, acetoacetate is then reduced by the 3Hbdh resulting in 3HB (Figure 1). The hbd1 pathway showed 3HB high titers from syngas (Karim et al., 2020), while ctfAB/3hbdh, relatively understudied, showed some ability to produce 3HB (Jones et al., 2016; Flüchter et al., 2019; Jia et al., 2021). The (R)-3-hydroxybutyryl-CoA dehydrogenase gene phaB, from Cupravidus necator, has also been utilized but showed poor 3HB production compared to the other pathways (Woolston et al., 2018; Flüchter et al., 2019). In this study, we investigated these promising 3HB production pathways in the acetogen Clostridium ljungdahlii, to compare their ability to make 3HB and explore methods to improve 3HB production.
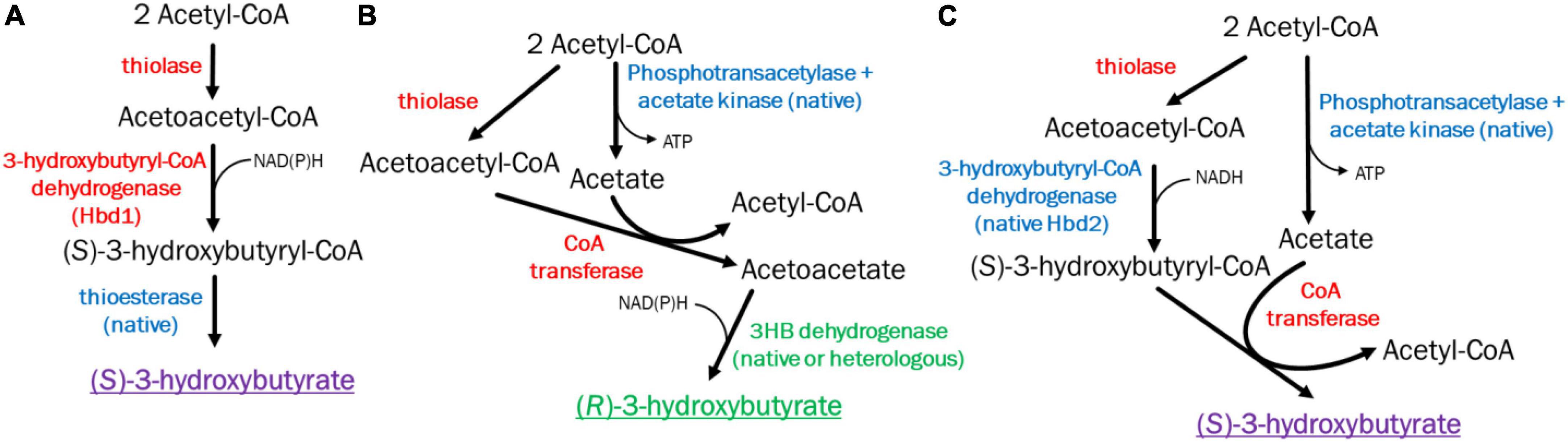
Figure 1. Pathways and enzymes for acetogenic 3HB production. (A) (S)-3-Hydroxybutyrate production pathway via Hbd1. (B) (R)-3-Hydroxybutyrate production pathway via CoA transferase/3Hbdh. (C) New (S)-3-Hydroxybutyrate production pathway via CoA transferase Hbd2. Heterologous enzymes in red, native enzymes in blue, native/heterologous R-3HB enzymes in green. S-3HB in purple.
Different 3HB pathways have different implications for productivity, specificity, cellular metabolism, ATP productivity, and redox (Bertsch and Müller, 2015; Woolston et al., 2018; Emerson et al., 2019). For instance, while hbd1 appears promising with regards to titer, its performance on syngas was inconsistent, with high and low 3HB production reported (Woolston et al., 2018; Karim et al., 2020). This may be because pathways using hbd1 enzymes do not produce ATP via substrate level phosphorylation (Figure 1A). As cell metabolism requires surplus ATP for growth and maintenance, acetate production may be required for ATP production. Thus, acetate production may “subsidize” 3HB production. In this pathway acetate is a terminal product and diverts acetyl-CoA from 3HB (Emerson et al., 2019).
In contrast, the ctfAB/3hbdh pathway relies on a CoA transferase of acetoacetyl-CoA to acetate, generating acetyl-CoA and acetoacetate, which is then reduced by 3HB dehydrogenase (3HBDH) to 3HB. The ctfAB/3hbdh pathway uses acetate as an intermediate and regenerates acetyl-CoA (Figure 1B). As a result, this pathway has a better ATP yield, which is important as growth on syngas is ATP limited (Bertsch and Müller, 2015; Emerson et al., 2019). Data on the ctfAB/3hbdh pathway showed promise but had inconsistent results. Jones et al suggested that acetoacetate can be natively converted to 3HB, as they saw significant levels of 3HB from their acetone generating strain which contained all genes required for the ctfAB/3hbdh pathway without 3hbdh itself (Jones et al., 2016). Data from Flüchter et al. (2019) suggests the ctfAB/3hbdh was highly active in Clostridium coskatii under heterotrophic conditions, but not autotrophically. This pathway was also tested in C. ljungdahlii but did not function under their conditions. While not optimizing for 3HB, Jia et al. (2021), also showed good titers (3 g/L) in C. ljungdahlii from this pathway, but low carbon yields (∼7%).
In investigating these pathways, we uncovered a pathway that performed substantially better in C. ljungdahlii, relying on ctfAB and an endogenous hbd2, referred to as ctfAB/hbd2 (Figure 1C). Annotated as (S)-3-hydroxybutyryl-CoA dehydrogenase (EC 1.1.1.57), little is known about hbd2. Hbd2 is an NADH specific enzyme, however, the functions and applications of this enzyme are not well known (Seedorf et al., 2008; Wang et al., 2010; Karim and Jewett, 2016). As far as we know, this work is the first time hbd2 has been validated for in vivo targeted product formation. We chose to investigate hbd2 further to both improve 3HB titers from syngas and expand our knowledge of the function and potential applications of this enzyme.
Clostridium ljungdahlii DSM 13528 and Clostridium kluyveri DSM 555 were from The Leibniz Institute DSMZ (Germany). Clostridium acetobutylicum ATCC 824 was from American Type Culture Collection (Manassas, VA, United States). C. ljungdahlii growth manipulations were based on previously reported techniques (Lo et al., 2020). Routine growth was performed at 37°C in modified YTF media (10 g/L yeast extract, 16 g/L Bacto tryptone, 4 g/L sodium chloride, 5 g/L fructose, 0.5 g/L cysteine, pH 6). YT media was the same as previous, omitting fructose as a carbon source. Bacterial manipulations were performed in a COY chamber (COY lab, Grass Lake, MI, United States) maintained anaerobic via palladium catalyst with 95% N2 and 5% H2 from Airgas (Randor, PA, United States). In general, chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO, United States) or Thermo Fisher (Waltham, MA, United States), unless otherwise indicated.
Standard molecular cloning and PCR techniques were used with enzymes from New England Biolabs (Ipswich, MA, United States). Routine PCR was performed using Phusion polymerase. For routine cloning and plasmid propagation, Escherichia coli strain NEB 10-Beta was utilized from New England Biolabs. The 1 kb Opti-DNA Marker ladder was from Applied Biological Materials (Vancouver, Canada). Primers and C. ljungdahlii optimized genes were generated from IDT (Coralville, IA, United States). Codon optimized genes atoB from E. coli, 3HBDH from Rhodobacter sphaeroides, the 3HBDH from Clostridium difficile were generated from the IDT algorithm using the Clostridium acetobutylicum option, which has a similar codon usage as C. ljungdahlii. Exact codon optimized sequences and primers are described in the Supplementary Materials. The pMTL80000 plasmids were from Chain Biotech (Nottingham, United Kingdom). Plasmids were generated using Gibson assembly from NEB. Confirmation of plasmids was performed by whole plasmid sequencing from the MGH DNA Core Facility (Cambridge, MA, United States).
Preparation of electrocompetent cells and transformation was performed based on previously reported protocols (Leang et al., 2013). Briefly, cells were grown overnight on YTF containing 40 mM DL-Threonine to an OD of 0.2–0.7, harvested, then washed with ice cold SMP buffer (270 mM sucrose, 1 mM MgCl2, 7 mM sodium phosphate, pH 6), then resuspended in SMP buffer with 10% (dimethyl sulfoxide) DMSO and frozen at –80°C until transformed. Cells mixed with 2–10 μg of DNA in a 1 mm cuvette, then transformed using a Bio-Rad Gene Pulser Xcell Electroporator (Hercules, CA, United States) with the following conditions: 625 kV, resistance at 600 Ω, capacitance of 25 μF. Cells were recovered overnight in YTF and plated the next day embedded in molten YTF 1.5% agar with 10 μg/mL thiamphenicol. Colonies appeared after 3 days. For generating the “3HB integration strain”, cells were grown in liquid YTF with thiamphenicol and 500 μg/mL 5-Fluoroorotic Acid (5FOA) based on previously reported protocols (Olson and Lynd, 2012; Minton et al., 2016). Cells were then single colony plated in YTF agar with thiamphenicol and 5FOA, picked, and colony screened using PCR. To cure the thiamphenicol resistant plasmid, PCR confirmed colonies were then passaged in YTF without thiamphenicol until thiamphenicol sensitivity was restored.
Liquid fermentation products were processed via previously described methods (Marcano-Velazquez et al., 2019). Briefly, samples were collected and filtered using Corning Costar Spin-X 0.45 μm (Corning, NY, United States) and routinely measured via HPLC, on a 1200 series Agilent (Santa Clara, CA, United States) Aminex HPX-87H column at 55°C with a 4 mM H2SO4 mobile phase. Enzymatic determination of R-3-Hydroxybutyrate was performed on HPLC filtered samples using the “D-3-Hydroxybutyric Acid (β-Hydroxybutyrate) Assay Kit” from Megazyme (Ireland), using the manufacturer’s instructions for the 96 well plate-based assay on a Tecan infinite M200 pro plate reader (Tecan Life Sciences, Switzerland). (R)-3-Hydroxybutyric acid and (S)-3-Hydroxybutyric acid were purchased from Sigma-Aldrich and used as standards from a 1 to 20 mM concentration. Optical density was measured using a Nanodrop (Thermofisher Scientific, Waltham, MA) at 600 nm. Carbon distribution was determined using only measured liquid components (i.e., Acetate, Ethanol, 3-Hydroxybutyrate, Fructose), measuring the mM and multiplying by the number of carbons in each product.
Heterotrophic growth of the strains was carried out in 15 mL Falcon tubes (Fisher scientific) using a 4 mL YTF medium with/without the addition of 10 μg/mL thiamphenicol at 37°C. Cells from an overnight seed culture were added in a 1:5 ratio (1 mL culture into 4 mL YTF) and left for 3 days before sampling. Autotrophic growth with CO, CO2, and H2 was carried out using 250 mL Duran Pressure Plus bottles (DWK Life Sciences, USA) containing 50 mL YT medium (YTF without fructose). 10 mL of an overnight culture was added to the bottles. Bottles were sealed and aseptically flushed with a CO, CO2, and H2 mixture (70%/20%/10% CO/CO2/H2) for 3 min. The same gas mixture was then added to 6 PSI of pressure within the bottles. YT within the bottles was supplemented with 10 μg/mL thiamphenicol for growth with plasmid bearing strains. Bottle growth was carried out at 37°C with 200 RPM shaking.
Autotrophic bioreactor growth was carried out using an Electrolab 2L bioreactor, containing 1.70 L YT with 10 μg/mL thiamphenicol. 300 mL of an autotrophic seed culture was added, and growth was carried out with a CO, CO2, and H2 mixture (70%/20%/10% CO/CO2/H2) at a flow rate of 300 standard cubic centimeter per min (sccm) with a fine steel diffusion stone. The pH was maintained at 5.2 using 3 M NaOH and the temperature was kept at 37°C using a heating wrap. Initial stirring was at 300 RPM which was increased to 500 RPM, followed by 900 RPM once cells began to grow based on OD600. OD600 and HPLC samples were taken daily.
Enzyme assays were performed based on previously reported procedures (Madan et al., 1973; Lo et al., 2020). Briefly, 50 mL of E. coli cells expressing C. kluyveri hbd1, C. ljungdahlii hbd2, and control vector pMTL83151 were harvested at mid-log phase and kept at –80°C until the day of enzyme assays. The lysis was performed using a bead-beating method. The 3-Hydroxybutyryl-CoA dehydrogenase assay was performed under the following conditions: 100 mM potassium phosphate buffer (pH 6.5), 25 mM potassium citrate, 75 μM NAD(P)H, and 125 μM acetoacetyl-CoA in 200 μL 96-well plate with a BioTek Synergy Neo2 plate reader (BioTek Instruments, United States) at 6 second intervals for 10 min. Oxidation of NAD(P)H at 340nm was used to follow enzyme activity, which is reported as μmol min–1 mg–1. To control for non-Hbd2 activity, we subtracted activity from E. coli cell free extract with pMTL83151, which was low (<0.01). Both hbd1 and hbd2 E. coli cell free extracts showed low activity without acetoacetyl-CoA (<0.01). Cell free extract protein was measured using a Bradford assay.
3HB production via acetoacetate is a straightforward pathway, requiring three steps: thiolase (Thl) to condense two acetyl-CoA to acetoacetyl-CoA, acetate:acetoacetyl CoA transferase (CtfAB) to transfer the CoA from acetoacetate-CoA to acetate, generating acetyl-CoA and acetoacetate, and 3Hbdh, which catalyzes the reduction of acetoacetate to 3HB (Figure 1B). We were initially interested in the relatively understudied ctfAB/3hbdh pathway due to previous literature showing substantial unintended 3HB production during C. ljungdahlii acetone/isopropanol production via Thl and CtfAB. First, we wanted to determine whether C. ljungdahlii contains an effective and native enzyme to efficiently reduce exogenous acetoacetate to 3HB as previously suggested (Jones et al., 2016; Jia et al., 2021). A similar study had been done by spiking in acetone, whereby a native reductase efficiently converted +90% acetone to isopropanol (Köpke et al., 2014), making it unnecessary to express a separate acetone reductase.
We therefore added 15 mM of acetoacetate to a growing culture of C. ljungdahlii to test endogenous reduction of acetoacetate. We measured ∼5 mM 3HB, demonstrating that acetoacetate can be natively reduced to 3HB. However, as only ∼5 mM of 3HB was detected, this suggests two thirds of the acetoacetate had been lost, and native acetoacetate conversion to 3HB is low.
Since it appears that acetoacetate reduction could be a limiting factor, we wanted to test various 3Hbdhs that could improve acetoacetate reduction. For the thiolase reaction, we chose the thiolase atoB from E. coli which showed good performance in C. acetobutylicum for butanol production (Nguyen et al., 2018). For the CoA transferase, we chose the ctfAB from C. acetobutylicum since this enzyme is well characterized and previously showed good functionality in C. ljungdahlii (Jones et al., 2016). We then tested putative 3HBDHs previously from literature: one from C. difficile (CDIF630_02933, native sequence and a codon optimized version), one from Rhodobacter sphaeroides (Rsph17025_1507), and one from C. ljungdahlii (CLJU_c23220) (Jones et al., 2016; Flüchter et al., 2019). We also note that the native C. ljungdahlii gene we overexpressed is not the gene responsible for 3HBDH activity in a recently published paper (Jia et al., 2021). Nevertheless, the overexpression of CLJU_c23220 serves as a “non-3hbdh” overexpression control.
We also wanted to compare the hbd1 pathway to our ctfAB/3hbdh pathway due to a report of high 3HB titers in C. autoethanogenum (Karim et al., 2020). The hbd1 pathway uses the C. acetobutylicum thiolase (Cac thl) and hbd1 from Clostridium kluyveri (Figure 1A). Expression of the pathways on each tested plasmid was driven by a C. ljungdahlii ferredoxin promoter (Pfdx) to allow for equal comparison. Plasmid bearing strains were grown in YTF medium containing fructose to observe heterotrophic production of 3HB. Final 3HB were determined after 3 days of growth (Figure 2).
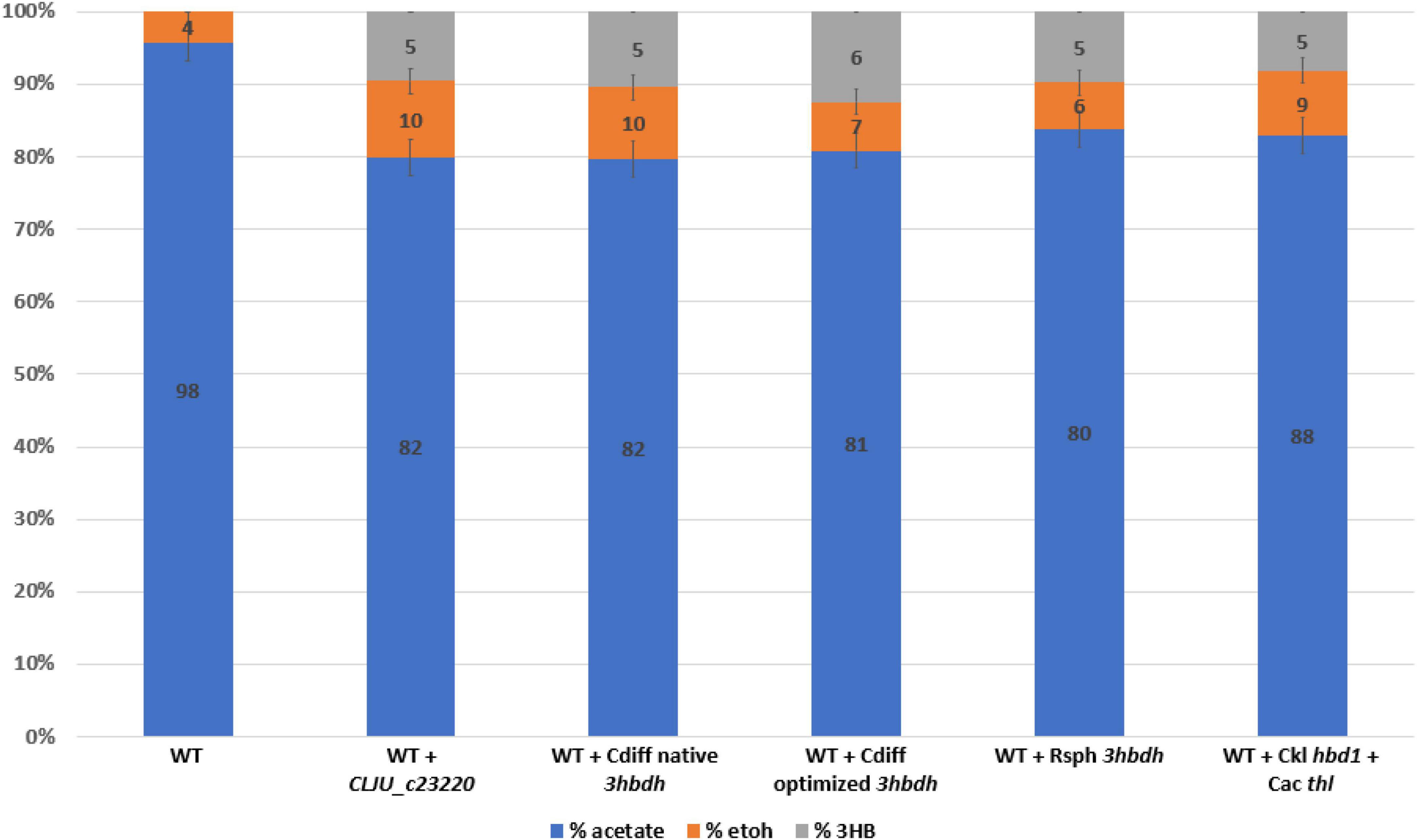
Figure 2. Heterotrophic 3HB production by Clostridium ljungdahlii strains expressing different 3HB pathways. Carbon distribution of fermentation products is indicated by the bar chart %, while the number label is the mM amount after 5 days of growth. In addition to the thiolase (thl) and CoA transferase, four genes were expressed on separate replicating plasmids. A codon optimized 3-Hydroxybutyrate dehydrogenase (3hbdh) from R. sphaeroides (Rsph), from C. difficile (Cdiff), native and codon optimized, and Clju_C23220 were expressed. Note that the C. ljungdahlii gene expressed is not a true 3HB Dehydrogenase (see text). A separate plasmid containing only C. kluyveri hbd1 and C. acetobutyulicum thiolase (thl) was also expressed. Error bars show standard error of the mean (SEM) for 3 biological replicates.
Results showed significant heterotrophic 3HB production via these both pathways. The transformed constructs demonstrated 5–6 mM of 3HB, which was comparable to the hbd1 based pathway (Figure 2). This is consistent with expectations reported by Jones et al and Flüchter et al that acetogenic heterotrophic 3HB production through this pathway is possible. Surprisingly, while Flüchter et al., 2019 specifically tried to express this pathway in both Clostridium coskatii and C. ljungdahlii, they were only able to demonstrate 3HB production in C. coskatii, as they did not detect 3HB production in C. ljungdahlii. Nevertheless, our data suggested that this pathway is in fact functional for 3HB production in C. ljungdahlii. Interestingly, explicit 3hbdh expression had only a marginal improvement on 3HB titers (2 mM difference between ‘Clj’ and ‘Cdiff’), despite our data showing native 3HBDH activity was poor.
To improve 3HB titers, we planned on coexpressing the ctfAB/3hbdh and hbd1 based pathways. We decided to integrate atoB and ctfAB into the pyrE locus to serve as base strain to allow us to test strategies for improving 3HB production with different expression constructs. Using 5-Fluoroorotic Acid (5FOA), homologous recombination can be used to place genes of interest into the pyrE locus, as pyrE+ cells are sensitive to 5FOA (Figure 3) (Minton et al., 2016). The genes atoB and ctfAB were successfully integrated into the genome, generating a strain that was 5FOA resistant. To cure the replicating plasmid, we subsequently passaged the strain on non-selective YTF media. We then isolated colonies that had become thiamphenicol sensitive, allowing for further transformations. PCR confirmation of integration was also carried out (Figure 3B). This is referred to as the “integrated 3HB strain” and was confirmed to make 3HB (Figure 4).
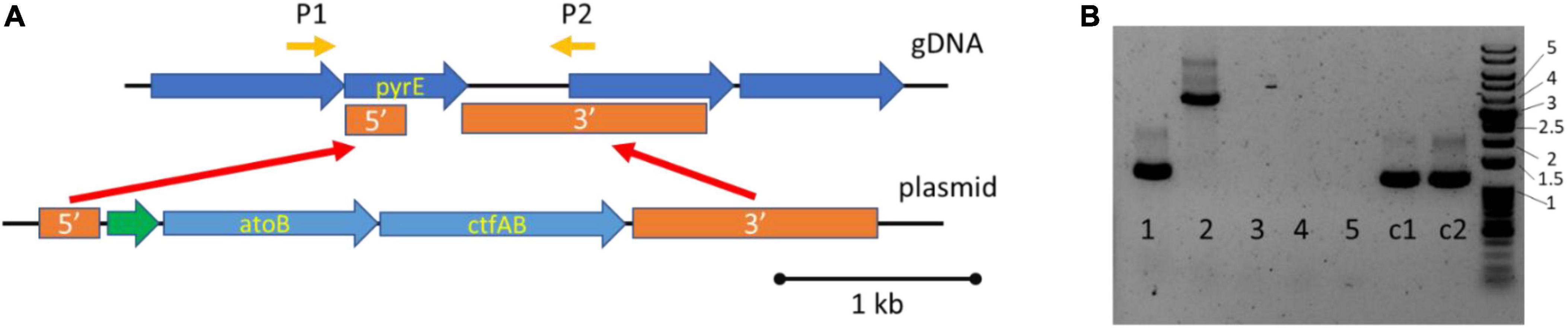
Figure 3. Integration of atoB and ctfAB into the pyrE locus of Clostridium ljungdahlii to generate the “integrated 3HB strain.” (A) Genome integration of atoB and ctfAB into pyrE locus, driven by Pfdx in green. P1 & P2 are primer binding sites for screening genomic pyrE locus. (B) PCR confirmation of integrated atoB and ctfAB using primers P1/P2 to amplify genomic locus. Lanes 1–5 contain screened 5FOA colonies, C1 and C2 are control gDNA from wild type DNA. A successful integration generates a size of 3.8 kb (Lane 2), whereas the wild type size is 1.2 kb (Lane 1, C1, and C2). The numbers next to the DNA ladder indicate kb size.
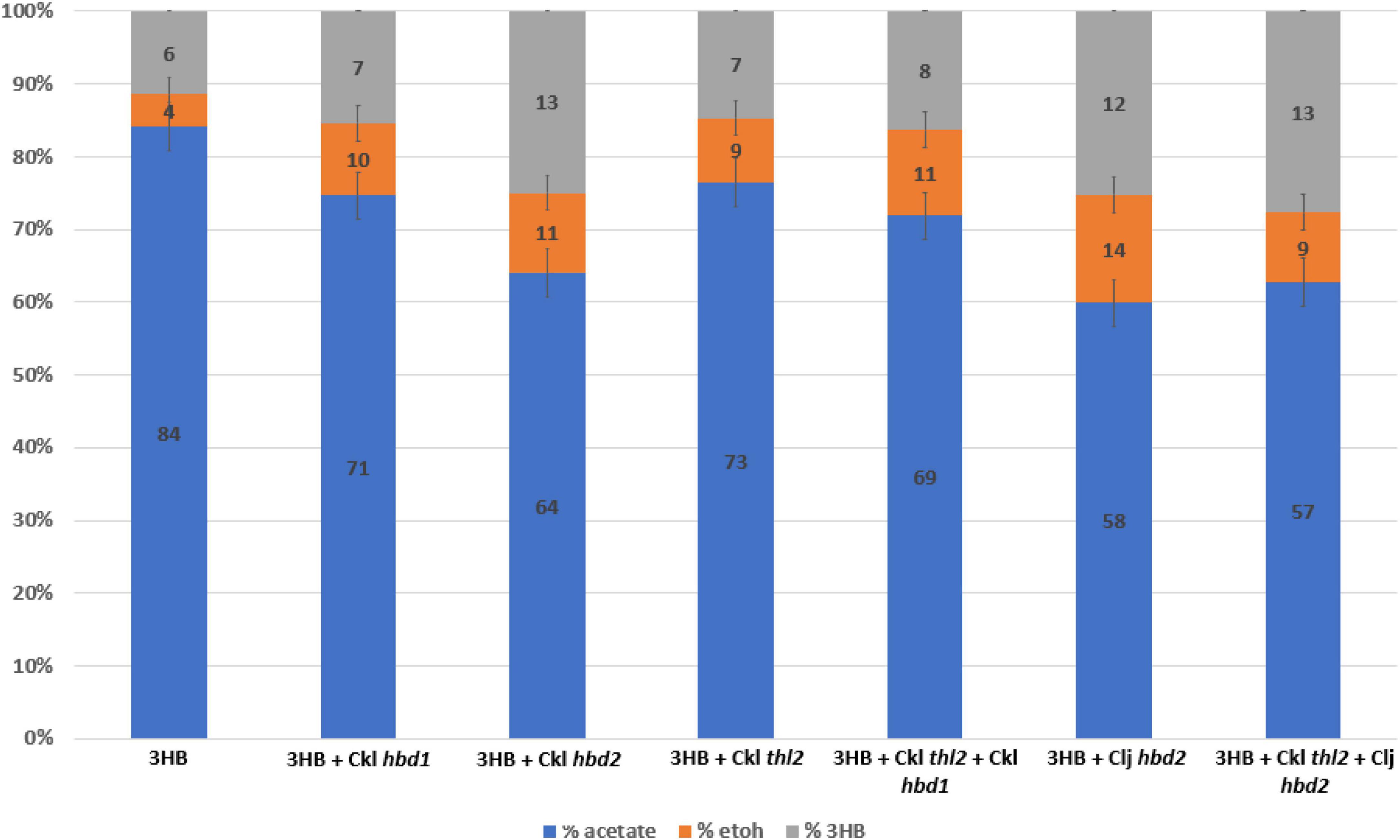
Figure 4. Heterotrophic fermentation product profile of Clostridium ljungdahlii strains expressing hbd1 vs. hbd2 in the 3HB strain background. Carbon distribution of fermentation products is indicated by the bar chart%, while the number label is the mM amount after 5 days of growth. The “integrated 3HB strain” contains the atoB from Escherichia coli and CoA transferase from C. acetobutylicum integrated into the genome. Replicating plasmids expressing a combination of thiolase, hbd1, and hbd2 were transformed into the integrated 3HB strain and measured for 3HB production. The expression of hbd2 results in superior 3HB production. Ckl for C. kluyveri, Clj for C. ljungdahlii. Carbon distribution to 3HB out of the total products is given as a percentage above bars. Error bars show standard error of the mean (SEM) for 3 biological replicates.
Introduction of a CoA-transferase based acetone/isopropanol pathway noted significant 3HB production, even though acetoacetate should be rapidly decarboxylated to acetone in those strains (Jones et al., 2016; Jia et al., 2021). Our experiment indicated that native acetoacetate conversion to 3HB was not robust, with only a third of the acetoacetate converted to 3HB. In light of this evidence, it seems suspicious that previously reported 3HB was derived from acetoacetate, given poor 3HBDH activity and robust decarboxylation to acetone. Explicit 3hbdh expression did improve 3HB titers (Figure 2), but not as much as we were expecting. While we were prospecting genes for overexpression, we noticed an annotated hbd2 in C. ljungdahlii and wondered whether it could be partially responsible for 3HB production. Transcriptomics evidence shows hbd2 (CLJU_c37300) is moderately expressed in C. ljungdahlii (∼300 FPKM), and we speculated this native gene could play a role in 3HB production (Nagarajan et al., 2013). In contrast, hbd1 (CLJU_c23560) was not expressed (>1 FPKM) and therefore not pursued.
We generated a hbd2 expression construct similar to the ones expressed in C. autoethanogenum (Figure 1A) (Karim et al., 2020). The constructs in Karim et al. only expressed thiolases and 3-hydroxybutyryl-CoA dehydrogenases but were very effective at generating 3HB in C. autoethanogenum. Our hbd2 construct used the ferredoxin promoter (Pfdx) to express a C. kluyveri thiolase (thl2) and the hbd2 from C. ljungdahlii. Since we were looking for further 3HB production enhancement, we first transformed the thl2 hbd2 construct into our “integrated 3HB strain.” Surprisingly, we found a 2.5-fold enhancement of 3HB production, from a 3HB carbon-yield of 11% (∼5 mM) to around 25% (∼12 mM) (Figure 4).
We wondered whether C. kluyveri thl2 and C. ljungdahlii hbd2 could be efficient alone at making 3HB as was seen in Karim et al, so we transformed them into the wild-type background without the integrated thiolase and CoA transferase. Interestingly, we saw no 3HB production in this strain (data not shown), suggesting that ctfAB is a critical gene, presumably to remove the CoA group from the 3HB-CoA generated by hbd2. A literature search showed a similar 3HB pathway in E. coli, using CoA transferase in conjunction with PhaB as the acetoacetyl-CoA reductase, where the CoA transferase was essential (Matsumoto et al., 2013). To compare C. kluyveri hbd1, we also transformed hbd1 into the “3HB integrated strain,” which showed improved 3HB production to 7.5 mM (∼2 mM over the 3HB parent) but was far inferior to the hbd2 strains (∼12 mM total). C. kluyveri also contains a hbd2 (CKL_2795), which we transformed as well, and performed similarly to the C. ljungdahlii hbd2 in 3HB titer (∼12 mM). This indicates that the hbd2 gene itself is important for increasing 3HB flux and is superior to the hbd1 gene in our system. In contrast, the plasmid expressing only thl2 in the “integrated 3HB strain” slightly improved 3HB to 7.5 mM, showing thiolase expression was not a major factor for 3HB production.
The C. kluyveri cofactor specificities of Hbd1 and Hbd2 are known from previous work (NADPH for Hbd1 and NADH for Hbd2) (Madan et al., 1973; Wang et al., 2010), but unknown for C. ljungdahlii Hbd2. Using cell free extract of E. coli expressing C. ljungdahlii hbd2, we were able to measure a specific activity of 0.07 μmol min–1 mg–1, and we confirmed NADH specificity and acetoacetyl-CoA-dependence. C. kluyveri hbd1 E. coli extracts were used as an NADPH-specific control and measured a specific activity of 0.22 μmol min–1 mg–1. We attempted to determine Km values for the C. ljungdahlii Hbd2 enzyme by varying either the NADH or the acetoacetyl-CoA concentrations. However, during this assay we noticed decreasing Hbd2 enzyme activity over several hours, inconsistent with the expected substrate loading conditions. We repeated the standard conditions of 75 μM NAD(P)H and 125 μM acetoacetyl-CoA with the hbd2 cell-free extracts, and we found markedly worse Hbd2 activity. In contrast, Hbd1 activity appeared to be intact, indicating that Hbd2 loses activity over time, even when kept on ice. We then lysed C. ljungdahlii cell-free extracts expressing hbd2 and found similar inconsistent activity results, suggesting that this was an Hbd2 related phenomena.
The stereoisomer of 3HB is important to determine, as the stereoisomer can determine its suitability for specific use cases and change bioplastics thermal/mechanical properties. Bioplastics physical properties can be driven and altered by the stereoisomer of the monomers (Cywar et al., 2021). Importantly, we wanted to note the relative contribution of the pathways in 3HB production, which can be determined by measuring the R or S form of 3HB. In the “integrated 3HB strain”, there are two possible pathways functioning to produce 3HB: the ctfAB/3hbdh and ctfAB/hbd2 pathway. 3hbdh pathway produces the R form (Figure 5A), while hbd2 produces the S form (Figure 5B), and thus their relative contribution can be determined. While we quantified total 3HB via HPLC, we could not resolve the composition of 3HB stereoisomers, so we needed an alternative method.
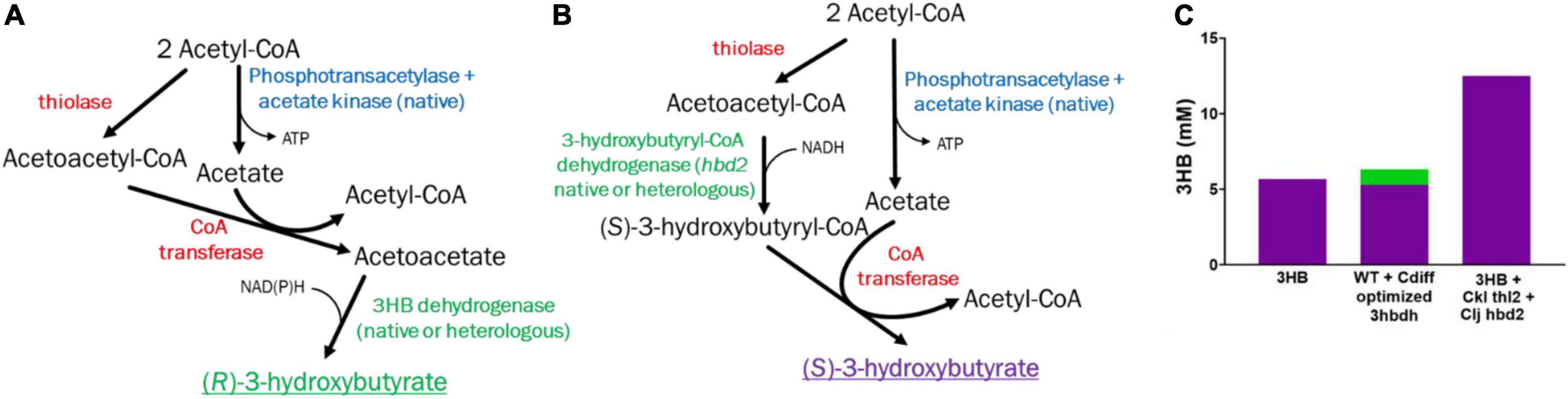
Figure 5. (R) vs. (S) 3HB Pathways and quantification. (A) The R-3HB pathway (B) The S-3HB pathway. Red indicates heterologous genes, blue native genes, green native or heterologous. (C) R (in green) vs. S (in purple) 3HB distribution from heterotrophic fermentation samples using HPLC and enzymatic analysis.
To confirm that ctfAB/hbd2 was the main cause of 3HB production, versus the ctfAB/3hbdh endogenous pathway, we used a commercial R-3HB enzyme assay kit (See Materials and Methods) to detect R-3HB from the heterotrophic samples from Figures 2, 4. We tested three strains: the “integrated 3HB strain”, 3HB + Ckl thl2 + Clj hbd2, and the wild type with ctfAB/3hbdh optimized from C. difficile. If the ctfAB/3hbdh pathway was the main pathway, we should get a detection from the enzymatic assay as it is specific for R-3HB, while S-3HB will not give a response. We did not detect any R-3HB in any of the “integrated 3HB strains” without 3hbdh overexpression <0.2 mM). In the wild type harboring the ctfAB/3hbdh with the optimized 3hbdh from Clostridium difficile, we detected around 1 mM R-3HB, which is around the magnitude of increase viewed in vivo with the added 3hbdh on the plasmid (Figure 5C). This shows that the 3HB produced is mainly S-3HB, via the ctfAB/hbd2 pathway, and not R-3HB via acetoacetate reduction with 3HBDH. As a control, we added known amounts of R and S-3HB and confirmed only R-3HB is detected via this assay.
Next, we tested the performance of select strains growing on syngas. Previous experiments with acetogens showed drastically different 3HB production on syngas versus on sugar. For instance, in two previous works, high C. ljungdahlii 3HB titers (∼20 mM) were generated on fructose. However, that did not translate to autotrophic 3HB production (>1 mM) (Woolston et al., 2018; Flüchter et al., 2019). Thus, we took promising heterotrophic strains and tested their autotrophic performance in bottles (Figure 6). Each strain was grown using a syngas mix of 70% CO, 20% CO2, and 10% H2 and product titers were analyzed after growth and gas consumption ceased. The hbd1 based pathway did not generate detectable 3HB. The “integrated 3HB strain” generated a small amount of 3HB, around 4 mM. The atoB, ctfAB, and 3hbdh strain generated the next most at ∼6 mM, while the “integrated 3HB strain” +thl2+ hbd2 strain generated the most at over 8 mM 3HB.
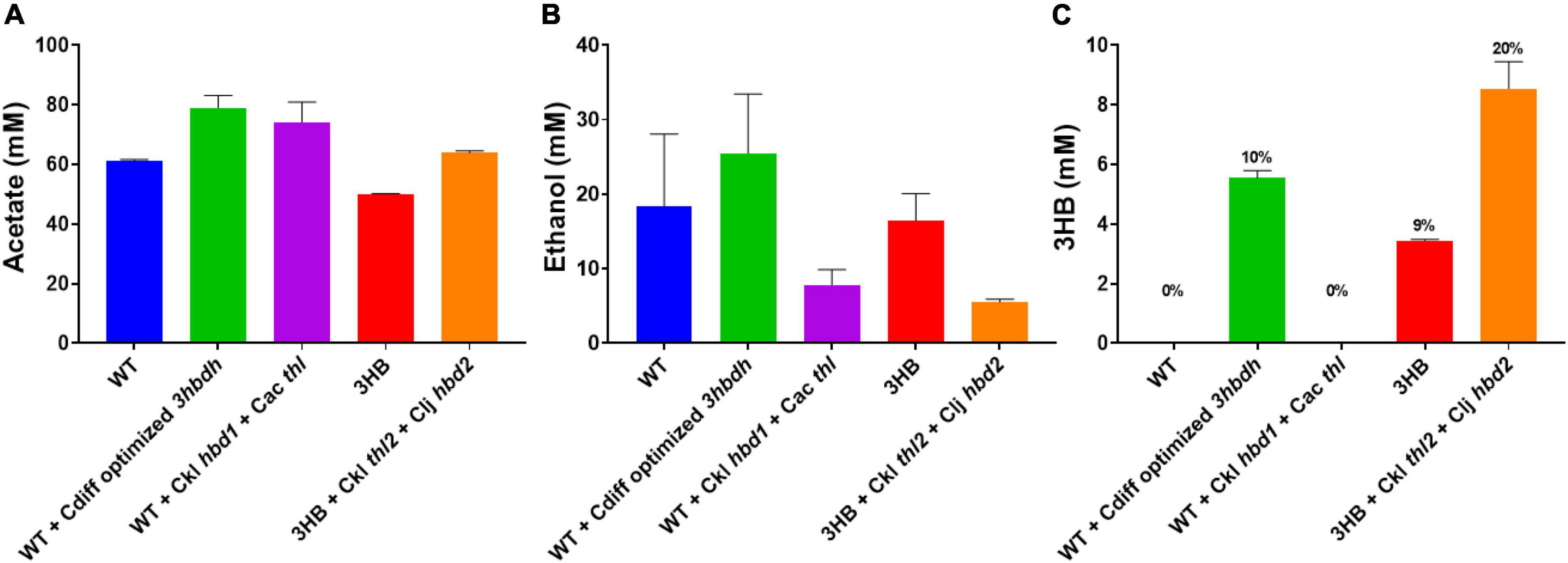
Figure 6. Autotrophic 3HB production in bottle fermentations. Strains were grown on syngas in 250 mL pressure bottles with 70% CO, 10% H2, and 20% CO2. (A) Acetate production, (B) ethanol production, and (C) 3HB production. Carbon distribution to 3HB out of the total products is given as a percentage above bars. Error bars show standard error of the mean (SEM) for 3 biological replicates.
We then tested the autotrophic performance of the 3HB + thl2+ hbd2 strain during bioreactor growth. We ran the strain under autotrophic conditions in a 2 L bioreactor, feeding 300 sccm of 70% CO, 10% H2, 20% CO2 maintained at pH 5.2 (Figure 7). The gas fermentation was run for 263 h after which growth ceased. By the end, the strain had produced 476 mM acetate, 53 mM ethanol, and 88 mM 3HB, with a carbon distribution of 67%, 8%, and 25%, respectively. The highest 24-h 3HB productivity rate (that is, the max 3HB rate over a 24-h period) was 0.083 g/L/hr. This shows that the improvement in 3HB production also translates to autotrophic fermentations on syngas. It is also higher than the previously highest titer (28 mM) and yield (∼7%) reported in C. ljungdahlii from syngas fed-batch fermentation, which had focused on enhancing 3HBDH to improve 3HB titers (Jia et al., 2021).
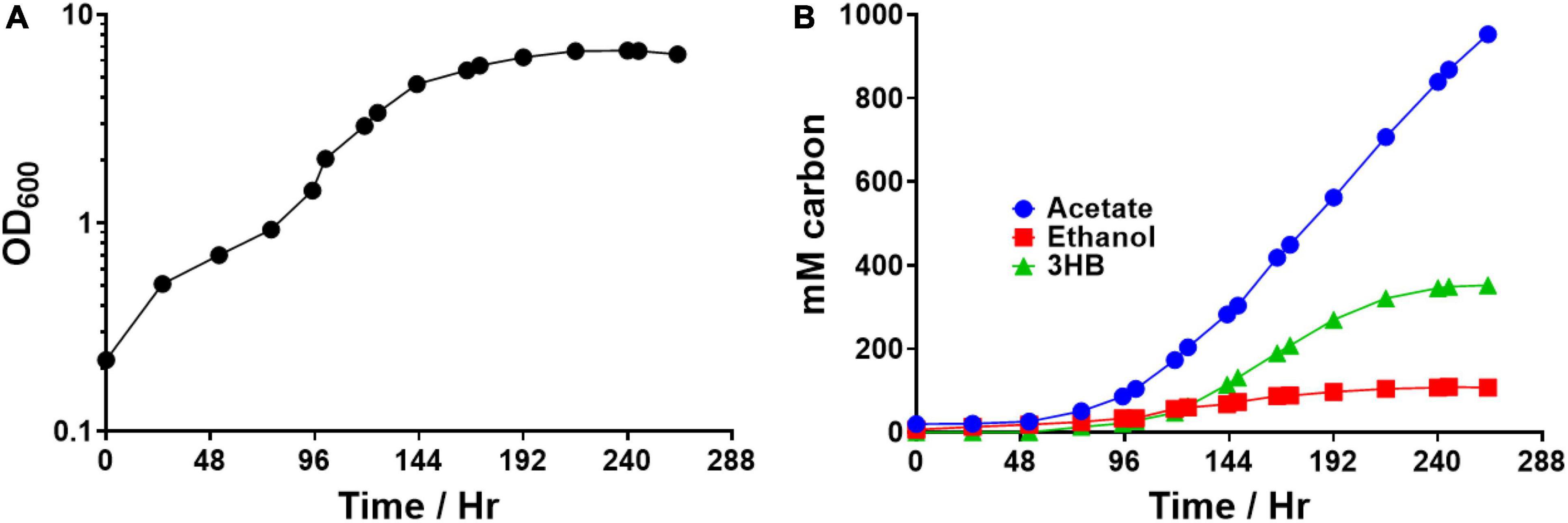
Figure 7. Autotrophic bioreactor fermentation product profile of Clostridium ljungdahlii integrated 3HB strain expressing additional thl and hbd2. The strain contains the atoB thiolase from Escherichia coli and CoA transferase from C. acetobutylicum integrated into the genome with an additional thl2 and hbd2 overexpressed on a replicating plasmid. (A) Optical density over time (B) Fermentation product profile over time in mM carbon.
Our data shows that among the different 3HB pathways, ctfAB/hbd2 expression outperformed other pathways we tested in C. ljungdahlii. Furthermore, ctfAB/hbd2 was probably responsible for 3HB detection in previous reports targeting isopropanol/acetone production (Jones et al., 2016; Jia et al., 2021). Despite C. ljungdahlii native 3HBDH activity, acetoacetate conversion is poor, and seems unlikely to outcompete acetoacetate decarboxylation to acetone in those strains. Rather, our data indicates that the native hbd2 was likely responsible for the 3HB in those strains, and targeted overexpression of hbd2 can enhance the yield of 3HB over other tested pathways.
What explains the improved 3HB production by this pathway? The most likely answer lies in Hbd2 (S)-3-hydroxybutyryl-CoA dehydrogenase. It has been previously shown in in vitro and in vivo systems that Hbd activity is key to driving high titers of both butanol and 3HB production, especially since thiolase condensation of acetyl-CoA is an endergonic reaction and downstream reactions are needed to “pull” the reaction forward (Karim and Jewett, 2016; Nguyen et al., 2018; Woolston et al., 2018; Karim et al., 2020). Interestingly, C. kluyveri hbd2 was tested for 3HB production and was found to underperform in a cell-free system compared to hbd1 (Karim et al., 2020). The cell-free system used in these experiments did not have an explicit CoA-transferase, which may explain its in vitro underperformance. The hbd2 from C. beijerinckii was used in vitro to generate butanol, with superior performance versus hbd1, but as far as we know this was the only application examined for hbd2 and was not performed in vivo (Karim and Jewett, 2016). The two in vitro studies provided conflicting evidence of the effectiveness of hbd2 vs. hbd1 and are the only biotechnical applications of hbd2 reported in literature.
Another explanation could be the presence of CoA-transferase in the 3HB strain. We note that previous work in acetogens found good 3HB production on heterotrophic conditions but poor results on autotrophic conditions (Woolston et al., 2018; Flüchter et al., 2019). It has been shown that acetyl-CoA levels can drastically change depending on heterotrophic versus autotrophic growth (Emerson et al., 2019), so autotrophic flux towards 3HB may be improved by regenerating acetyl-CoA for (S)-3-hydroxybutyryl-CoA dehydrogenase activity. In E. coli, Matsumoto et al., 2013 elucidated a CoA-transferase dependent 3HB pathway, where 3HB-CoA (generated by PhaB from Cupriavidus necator) would transfer the CoA to acetate, generating 3HB and acetyl-CoA. This pathway proved surprisingly efficient, generating a 3HB titer of 1 g/L. Relevant to acetogens, the 3HB titer was improved to 5.2 g/L with high acetate concentrations.
Little is known about hbd2 even though several Clostridia possess the gene. A BLAST search with C. ljungdahlii and C. kluyveri hbd2 indicates high identity to proteins in several Clostridia of scientific interest. Clostridium beijerinckii, Clostridium botulinum, Clostridium coskatii, Clostridium carboxidivorans and C. autoethanogenum generated high identity (>70%) hits with hbd2. Interestingly, C. acetobutylicum only has an hbd1, and not hbd2. In contrast, the identities between hbd2 vs. hbd1 from C. kluyveri have ∼40% amino acid identity to each other, suggesting that these enzymes are phylogenetically different. Butyryl-CoA synthesis genes often form an operon with hbd1, indicating a clear role in butyrate/butanol synthesis (Boynton et al., 1996; Hu et al., 2016). In contrast, as far as we can tell, in both C. kluyveri and C. ljungdahlii, hbd2 are co-localized to genes unrelated to butyrate/butanol synthesis.
The C. kluyveri Hbd2 is NADH-linked, but its functionality is unknown. It is speculated to be important for redox balancing and chain elongation in C. kluyveri, where hbd1 is notably NADPH-linked (Wang et al., 2010). Almost nothing is known about hbd2 in C. ljungdahlii. We confirmed that C. ljungdahlii Hbd2 is NADH specific, like C. kluyveri Hbd2, but its function remains a mystery. C. ljungdahlii does not natively produce 3HB, PHB, butyrate, or butanol, and its genomic context doesn’t appear to contain any obvious clues. Interestingly, it is moderately expressed in both heterotrophic and autotrophic conditions (FPKM 339 and 291, respectively) (Nagarajan et al., 2013), suggesting it could have an undetermined metabolic role.
This work put into context several interesting pieces of data, especially for the production of 3HB. C. ljunghdahlii has a number of functional genes that can natively catalyze 3HB production. It naturally converts acetoacetate to 3HB and has a highly functional Hbd2 that converts acetoacetyl-CoA to 3HB-CoA. 3HB has been produced in C. ljungdahlii and related acetogens, and although it was assumed that the heterologous expression of pathway components was responsible for 3HB production, native enzymes may also be playing a role in 3HB production. Published work expressing ctfAB in C. ljungdahlii assumed that 3HB was derived from acetoacetate reduction, but this 3HB could be from Hbd2 reducing acetoacetyl-CoA. One way to determine the relative contribution is through determination of the R vs. S stereoisomer via enzymatic assay analysis, which is a cheaper alternative than purchasing a chiral column. Furthermore, as different pathways have been expressed to produce 3HB in acetogens, 3HB titers could be increased by combining different pathway strategies into a single organism. We show that both hbd2 and 3hbdh can be functional, as we did detect both S and R-3HB, but the 3hbdh contribution was low compared to hbd2.
Finally, this ctfAB/hbd2 pathway may have other advantages vs. previously described pathways. In our experimental conditions, hbd2 expression appears to be superior to hbd1 and 3hbdh. As a native acetogen/Clostridia derived enzyme, the Hbd2 may function better in its native host than heterologous enzymes. For instance, phaB has been tested in C. ljungdahlii with poor results, possibly due to compromised expression, despite good results in E. coli (Matsumoto et al., 2013; Woolston et al., 2018; Flüchter et al., 2019). Furthermore, the CoA transferase from 3-hydroxybutyryl-CoA to acetate regenerates acetyl-CoA, which may be important for acetyl-CoA concentrations and addressing ATP limitation when growing on H2/CO2/CO. For instance, previously described Hbd1-based 3HB production does not involve substrate-level phosphorylation, whereas the ctfAB/hbd2 described pathway would.
While we have shown good production of 3HB with our new pathway using ctfAB/hbd2, we believe there is the opportunity for much further optimization and application. We have only tested only ctfAB from C. acetobutylicum and two hbd2, but BLAST screening shows a multitude of genes that could be tested. Greater 3HB yields could probably be gained by testing new genes and targeting acetyl-CoA related pathways, in particular acetate and ethanol (Woolston et al., 2018; Lo et al., 2020). In vitro testing has recently proven successful in screening multiple 3HB genes (Karim et al., 2020). Furthermore, different 3HB pathways could potentially be combined in a single strain, as they do not appear to be incompatible and may improve 3HB yield/rate/titer. For instance, the hbd1 pathway in C. autoethanogenum generates significant amounts of acetate, which could be reassimilated when combined with ctfAB/hbd2 (Karim et al., 2020). Additionally, while Karim et al. reported impressive titers of 3HB production based on the C. kluyveri Hbd1 in C. autoethanogenum, we were unable to repeat those results in C. ljungdahlii, suggesting there may be strain specific differences contributing to 3HB production. It is worth emphasizing that C. kluyveri Hbd2 is NADH-linked, while C. kluyveri Hbd1 is NADPH-linked (Wang et al., 2010). NAD(P)H redox differences may be a factor in 3HB production. Improved Clostridia product formation is often driven by changes to NAD(P)H metabolism (Lo et al., 2017, 2015; Nguyen et al., 2018; de Souza Pinto Lemgruber et al., 2019; Karim et al., 2020; Soucaille et al., 2020). Beyond 3HB, Hbd2 could be important for other related products including PHB and longer chain fatty acids/alcohols (C4-C6) (Phillips et al., 2015; Richter et al., 2016). Hbd2 has been drastically understudied in the context of both Clostridial physiology and engineering, and we have shown that it could have direct biotechnological uses.
To our knowledge, this is the first time hbd2 has been utilized in vivo for enhancing targeted product formation. Little is known about the native function of these genes, their biochemical characteristics/activity, and how they may be utilized to improve target product formation. The native activity of Hbd2 could be unknowingly contributing to efforts to engineer 3HB/PHB/fatty acid/alcohol production in Clostridia, as hbd2 is commonly found in many Clostridia studied for metabolic engineering. Our data shows that these Hbd2 enzymes may be a useful avenue of further interest for 3HB/PHB production. Additionally, Hbd2 catalyzes an important step in butanol/butyrate production, which are other value-added chemicals of interest that have been produced in acetogens. Furthermore, CoA-transferase based formation of 3HB appears to be a fruitful area of research for high 3HB production, especially considering that acetate formation is important for ATP synthesis in acetogens. This work sets the stage to explore the applications and diversity of hbd2 and CoA-transferases. We anticipate that higher titers could be reached by exploring metabolic engineering strategies with these enzymes.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
JL, JH, LM, SH, WX, KC, and PCM designed research. JL, JH, LM, CU, SH, and BW performed research. JL, JH, WX, KC, and PCM wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was authored by the National Renewable Energy Laboratory, operated by Alliance for Sustainable Energy, LLC, for the US Department of Energy (DOE) under Contract No. DE-AC36-08GO28308. Funding provided by DOE and Bioenergy Technology Office (BETO) under Contract No. DE-LC-000L079.
The views expressed in the article do not necessarily represent the views of the DOE or the US Government. The US Government retains and the publisher, by accepting the article for publication, acknowledges that the US Government retains a nonexclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this work, or allow others to do so, for US Government purposes.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.948369/full#supplementary-material
Bar-Even, A., Noor, E., and Milo, R. (2012). A survey of carbon fixation pathways through a quantitative lens. J. Exp. Bot. 63, 2325–2342. doi: 10.1093/jxb/err417
Bertsch, J., and Müller, V. (2015). Bioenergetic constraints for conversion of syngas to biofuels in acetogenic bacteria. Biotechnol. Biofuels 8:210. doi: 10.1186/s13068-015-0393-x
Boynton, Z. L., Bennet, G. N., and Rudolph, F. B. (1996). Cloning, sequencing, and expression of clustered genes encoding beta-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. J. Bacteriol. 178, 3015–3024. doi: 10.1128/jb.178.11.3015-3024.1996
Chen, G.-Q. (2009). A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem. Soc. Rev. 38, 2434–2446. doi: 10.1039/B812677C
Cywar, R. M., Rorrer, N. A., Hoyt, C. B., Beckham, G. T., and Chen, E. Y.-X. (2021). Bio-based polymers with performance-advantaged properties. Nat. Rev. Mater. 7, 83–103. doi: 10.1038/s41578-021-00363-3
de Souza Pinto Lemgruber, R., Valgepea, K., Tappel, R., Behrendorff, J. B., Palfreyman, R. W., Plan, M., et al. (2019). Systems-level engineering and characterisation of Clostridium autoethanogenum through heterologous production of poly-3-hydroxybutyrate (PHB). Metab. Eng. 53, 14–23. doi: 10.1016/j.ymben.2019.01.003
Emerson, D. F., Woolston, B. M., Liu, N., Donnelly, M., Currie, D. H., and Stephanopoulos, G. (2019). Enhancing hydrogen-dependent growth of and carbon dioxide fixation by Clostridium ljungdahlii through nitrate supplementation. Biotechnol. Bioeng. 116, 294–306. doi: 10.1002/bit.26847
Flüchter, S., Follonier, S., Schiel-Bengelsdorf, B., Bengelsdorf, F. R., Zinn, M., and Dürre, P. (2019). Anaerobic production of poly(3-hydroxybutyrate) and its precursor 3-hydroxybutyrate from synthesis gas by autotrophic clostridia. Biomacromolecules 20, 3271–3282. doi: 10.1021/acs.biomac.9b00342
Hu, L., Huang, H., Yuan, H., Tao, F., Xie, H., and Wang, S. (2016). Rex in Clostridium kluyveri is a global redox-sensing transcriptional regulator. J. Biotechnol. 233, 17–25. doi: 10.1016/j.jbiotec.2016.06.024
Huang, Z., Grim, R. G., Schaidle, J. A., and Tao, L. (2021). The economic outlook for converting CO2 and electrons to molecules. Energy Environ. Sci. 14, 3664–3678. doi: 10.1039/D0EE03525D
Jia, D., He, M., Tian, Y., Shen, S., Zhu, X., Wang, Y., et al. (2021). Metabolic engineering of gas-fermenting Clostridium ljungdahlii for efficient co-production of isopropanol, 3-hydroxybutyrate, and ethanol. ACS Synth. Biol. 10, 2628–2638. doi: 10.1021/acssynbio.1c00235
Jones, S. W., Fast, A. G., Carlson, E. D., Wiedel, C. A., Au, J., Antoniewicz, M. R., et al. (2016). CO2 fixation by anaerobic non-photosynthetic mixotrophy for improved carbon conversion. Nat. Commun. 7:12800. doi: 10.1038/ncomms12800
Karim, A. S., and Jewett, M. C. (2016). A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab. Eng. 36, 116–126. doi: 10.1016/j.ymben.2016.03.002
Karim, A. S., Dudley, Q. M., Juminaga, A., Yuan, Y., Crowe, S. A., Heggestad, J. T., et al. (2020). In vitro prototyping and rapid optimization of biosynthetic enzymes for cell design. Nat. Chem. Biol. 16, 912–919. doi: 10.1038/s41589-020-0559-0
Köpke, M., Gerth, M. L., Maddock, D. J., Mueller, A. P., Liew, F., Simpson, S. D., et al. (2014). Reconstruction of an acetogenic 2,3-butanediol pathway involving a novel nadph-dependent primary-secondary alcohol dehydrogenase. Appl. Environ. Microbiol. 80, 3394–3403. doi: 10.1128/AEM.00301-14
Leang, C., Ueki, T., Nevin, K. P., and Lovley, D. R. (2013). A genetic system for Clostridium ljungdahlii: a chassis for autotrophic production of biocommodities and a model homoacetogen. Appl. Environ. Microbiol. 79, 1102–1109. doi: 10.1128/AEM.02891-12
Lee, S. Y. (1996). Plastic bacteria? Progress and prospects for polyhydroxyalkanoate production in bacteria. Trends Biotechnol. 14, 431–438. doi: 10.1016/0167-7799(96)10061-5
Li, Z., Yang, J., and Loh, X. J. (2016). Polyhydroxyalkanoates: opening doors for a sustainable future. NPG Asia Mater. 8:e265. doi: 10.1038/am.2016.48
Lo, J., Humphreys, J. R., Jack, J., Urban, C., Magnusson, L., Xiong, W., et al. (2020). The metabolism of Clostridium ljungdahlii in phosphotransacetylase negative strains and development of an ethanologenic strain. Front. Bioeng. Biotechnol. 8:560726. doi: 10.3389/fbioe.2020.560726
Lo, J., Olson, D. G., Murphy, S. J.-L., Tian, L., Hon, S., Lanahan, A., et al. (2017). Engineering electron metabolism to increase ethanol production in Clostridium thermocellum. Metab. Eng. 39, 71–79. doi: 10.1016/j.ymben.2016.10.018
Lo, J., Zheng, T., Olson, D. G., Ruppertsberger, N., Tripathi, S. A., Guss, A. M., et al. (2015). Deletion of nfnAB in thermoanaerobacterium saccharolyticum and its effect on metabolism. J. Bacteriol. 197, 2920–2929. doi: 10.1128/JB.00347-15
Madan, V. K., Hillmer, P., and Gottschalk, G. (1973). Purification and properties of nadp-dependent l(+)-3-hydroxybutyryl-coa dehydrogenase from Clostridium kluyveri. Eur. J. Biochem. 32, 51–56. doi: 10.1111/j.1432-1033.1973.tb02577.x
Marcano-Velazquez, J. G., Lo, J., Nag, A., Maness, P.-C., and Chou, K. J. (2019). Developing riboswitch-mediated gene regulatory controls in thermophilic bacteria. ACS Synth. Biol. 8, 633–640. doi: 10.1021/acssynbio.8b00487
Matsumoto, K., Okei, T., Honma, I., Ooi, T., Aoki, H., and Taguchi, S. (2013). Efficient (R)-3-hydroxybutyrate production using acetyl CoA-regenerating pathway catalyzed by coenzyme A transferase. Appl. Microbiol. Biotechnol. 97, 205–210. doi: 10.1007/s00253-012-4104-2
Minton, N. P., Ehsaan, M., Humphreys, C. M., Little, G. T., Baker, J., Henstra, A. M., et al. (2016). A roadmap for gene system development in Clostridium. Anaerobe Clost Path 41, 104–112. doi: 10.1016/j.anaerobe.2016.05.011
Nagarajan, H., Sahin, M., Nogales, J., Latif, H., Lovley, D. R., Ebrahim, A., et al. (2013). Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii. Microb. Cell Fact. 12:118. doi: 10.1186/1475-2859-12-118
Nguyen, N.-P.-T., Raynaud, C., Meynial-Salles, I., and Soucaille, P. (2018). Reviving the Weizmann process for commercial n-butanol production. Nat. Commun. 9:3682. doi: 10.1038/s41467-018-05661-z
Olson, D. G., and Lynd, L. R. (2012). “Chapter seventeen – transformation of Clostridium thermocellum by electroporation,” in Methods in Enzymology, Cellulases, ed. H. J. Gilbert (Cambridge, MA: Academic Press), 317–330. doi: 10.1016/B978-0-12-415931-0.00017-3
Phillips, J. R., Atiyeh, H. K., Tanner, R. S., Torres, J. R., Saxena, J., Wilkins, M. R., et al. (2015). Butanol and hexanol production in Clostridium carboxidivorans syngas fermentation: medium development and culture techniques. Bioresour. Technol. 190, 114–121. doi: 10.1016/j.biortech.2015.04.043
Richter, H., Molitor, B., Diender, M., Sousa, D. Z., and Angenent, L. T. (2016). A narrow pH Range supports butanol, hexanol, and octanol production from syngas in a continuous co-culture of Clostridium ljungdahlii and Clostridium kluyveri with in-line product extraction. Front. Microbiol. 7:1773. doi: 10.3389/fmicb.2016.01773
Seedorf, H., Fricke, W. F., Veith, B., Brüggemann, H., Liesegang, H., Strittmatter, A., et al. (2008). The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc. Natl. Acad. Sci. U.S.A. 105, 2128–2133. doi: 10.1073/pnas.0711093105
Soucaille, P., Meynial-Salles, I., Foulquier, C., and Riviere, A. (2020). Polypeptide having ferredoxin-NADP+ reductase activity, polynucleotide encoding the same and uses thereof. US patent US10760100B2.
Tokiwa, Y., and Ugwu, C. U. (2007). Biotechnological production of (R)-3-hydroxybutyric acid monomer. J. Biotechnol. 132, 264–272. doi: 10.1016/j.jbiotec.2007.03.015
Wang, S., Huang, H., Moll, J., and Thauer, R. K. (2010). NADP+ reduction with reduced ferredoxin and NADP+ reduction with NADH are coupled via an electron-bifurcating enzyme complex in Clostridium kluyveri. J. Bacteriol. 192, 5115–5123. doi: 10.1128/JB.00612-10
Keywords: metabolic pathway discovery, acetogen, Clostridium ljungdahlii, 3-hydroxybutyrate, syngas utilization
Citation: Lo J, Humphreys JR, Magnusson L, Wachter B, Urban C, Hebdon SD, Xiong W, Chou KJ and Ching Maness P (2022) Acetogenic production of 3-Hydroxybutyrate using a native 3-Hydroxybutyryl-CoA Dehydrogenase. Front. Microbiol. 13:948369. doi: 10.3389/fmicb.2022.948369
Received: 19 May 2022; Accepted: 11 July 2022;
Published: 08 August 2022.
Edited by:
Ivan A Berg, University of Münster, GermanyReviewed by:
Wolfgang Buckel, University of Marburg, GermanyCopyright © 2022 Lo, Humphreys, Magnusson, Wachter, Urban, Hebdon, Xiong, Chou and Ching Maness. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonathan Lo, loj04@hotmail.com
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.