
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 27 June 2022
Sec. Microbiotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.945184
Cis-3-hydroxypipecolic acid (cis-3-HyPip), a key structural component of tetrapeptide antibiotic GE81112, which has attracted substantial attention for its broad antimicrobial properties and unique ability to inhibit bacterial translation initiation. In this study, a combined strategy to increase the productivity of cis-3-HyPip was investigated. First, combinatorial optimization of the ribosomal binding site (RBS) sequence was performed to tune the gene expression translation rates of the pathway enzymes. Next, in order to reduce the addition of the co-substrate α-ketoglutarate (2-OG), the major engineering strategy was to reconstitute the tricarboxylic acid (TCA) cycle of Escherichia coli to force the metabolic flux to go through GetF catalyzed reaction for 2-OG to succinate conversion, a series of engineered strains were constructed by the deletion of the relevant genes. In addition, the metabolic flux (gltA and icd) was improved and glucose concentrations were optimized to enhance the supply and catalytic efficiency of continuous 2-OG supply powered by glucose. Finally, under optimal conditions, the cis-3-HyPip titer of the best strain catalysis reached 33 mM, which was remarkably higher than previously reported.
Hydroxypipecolic acids (HyPips) are the core structure of many alkaloids and drugs, such as tetrazomine (Qi et al., 2021), non-ribosomal peptide GE81112 (Zwick et al., 2021), palinavir (Hibi et al., 2016), and relebactam (Miller et al., 2014). Currently, HyPips are predominantly produced using chemical methods, which have several disadvantages, including expensive substrates, complicated steps, negative environmental impacts, and challenges surrounding regio- and stereo-selectivity reactions that are inevitable during this process (Ahuja and Sudalai, 2015; Chavan et al., 2015; Zhang and Sun, 2020). Bioproduction of HyPips via microbial fermentation and enzymatic catalysis is an attractive alternative to chemical production methods, owing to its simplicity, stereoselectivity, mild reaction conditions and eco-friendly properties. L-pipecolic acid (L-Pip) is often used to produce HyPips through hydroxylation with Fe(II)/α-ketoglutarate (2-OG)-based oxygenase (Mori et al., 1997; Shibasaki et al., 1999; Hara and Kino, 2009; Mattay and Huettel, 2017; Lu et al., 2020). However, the high cost of substrate L-Pip restricts its practical application. To reduce the cost, we previously constructed a microbial cell factory that can convert economic L-lysine into cis-3-hydroxypipecolic acid (cis-3-HyPip) with whole-cell cascade catalysis (Hu et al., 2022; Scheme 1). First, L-lysine was converted to L-Pip through the cyclization deamination by lysine cyclodeaminase (SpLCD), which was further hydroxylated by Fe(II)/2-OG-based oxygenase GetF to form cis-3-HyPip. Although we used plasmids with different copy numbers to regulate the expression of SpLCD and GetF and systematically optimized the reaction parameters, the central problem in the co-expression system was always the imbalance caused by differences in a specific activity or expression level. The lowest reaction rate limits the overall reaction rate, resulting in L-Pip accumulation. Therefore, developing a tunable multi-enzyme coordinate expression system in Escherichia coli for cis-3-HyPip production remains a top priority.
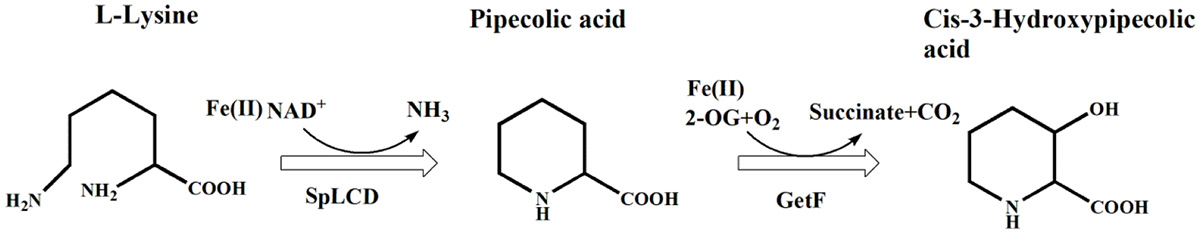
Scheme 1. Biotransformation pathway from L-lysine to cis-3-HyPip by the whole-cell biocatalytic system.
Precise control of the expression levels of multiple genes is critical in co-expression biocatalysis (Chen et al., 2017), and one strategy is to use different types of vectors or co-express multiple genes through one or more vectors to avoid low expression of rate-limiting enzymes and overexpression of non-rate-limiting enzymes (Liu et al., 2019a; Yang et al., 2021; Hu et al., 2022). Other strategies are generally possible to precisely control certain enzyme expression levels by using different promoters and ribosomal binding sites (RBSs) (Jiang and Fang, 2016; Zhang et al., 2019). For example, researchers have found that using the RBS-optimized strain, the conversion rate of L-aspartate biotransformed from maleate was nearly 100%, with no intermediates or byproducts (Liu et al., 2019b). To further avoid pyruvate overoxidation, the RBS sequences of UtCAT with various translation initiation rates were designed using a recently developed biophysical model of translation initiation, yielding 59.9 g/L pyruvate (Li et al., 2020).
Moreover, GetF from Streptomyces sp. L-49973 (Binz et al., 2010) was a Fe(II)/2-OG-based oxygenase and requires the co-substrate 2-OG to undergo oxidative decarboxylation and form succinate, which is another limiting factor. An adequate supply of 2-OG derived from inexpensive substrates is necessary for a whole-cell catalysis system. Previously, researchers have used L-glutamic acid oxidase to catalyze the production of 2-OG from glutamate, requiring additional overexpression of catalase to eliminate the effect of hydrogen peroxide toward enzymatic activity (Sun et al., 2019). Increased gene expression increases whole-cell pressure, which may affect the catalytic activity. Normally, 2-OG is metabolized via the tricarboxylic acid (TCA) cycle. Thus, little flux typically enters the synthesis pathway of the desired product. To address this problem, a smart strategy was developed by constructing a modified TCA cycle that changed the role of 2-OG from co-substrate to cofactor and that regenerated 2-OG (Lin et al., 2015). For example, E. coli cells expressing deacetoxycephalosporin-C synthase (DAOCS) were developed as a whole-cell biocatalyst to convert penicillin G to G-7-aminodeacetoxycephalosporanic acid, where the TCA cycle was engineered in vivo by blocking the normal TCA reaction from 2-OG to succinate, effectively coupling it with the DAOCS-catalyzed reaction to form a modified TCA cycle (Lin et al., 2015).
In this study, a combined strategy to increase the productivity of cis-3-HyPip was investigated. Firstly, GetF and SpLCD expression was fine-tuned via RBS engineering to improve cis-3-HyPip production. Subsequently, an efficient and sustainable 2-OG supply system was constructed, and the TCA cycle was disrupted at the 2-OG to the succinate step, forcing the cycle to incorporate a GetF catalyzed reaction. We also overexpressed gltA and icd to strengthen the artificial TCA cycle and increase the yield of cis-3-HyPip.
The strains used in this study were listed in Supplementary Table 1, E. coli DH5α served as the host for recombinant DNA manipulation and plasmid construction. For cis-3-HyPip production, E. coli BL21(DE3) and derived strains were used. The plasmid pEcgRNA carrying the ccdB gene was constructed and maintained in E. coli DB3.1. The DNA polymerases used for polymerase chain reaction (PCR) including 2 × Phanta Max Master Mix, 2 × Taq Master Mix and ClonExpress One Step Cloning Kit were purchased from Vazyme (Nanjing, China). Molecular biological reagents, such as T4 DNA ligase and DNA gel extraction kit, were obtained from TaKaRa (Dalian, China). Isopropyl β-D-1-thiogalactopyranoside (IPTG), ampicillin, spectinomycin, and kanamycin were provided from Sangon Biotech (Shanghai, China). All other chemicals were purchased from Sigma-Aldrich (Shanghai, China) or Sangon Biotech and were of analytical grade (Shanghai, China).
All strains were grown at 37°C and shaken at 220 rpm in lysogeny broth (LB). Based on plasmid maintenance requirements, ampicillin (100 μg/mL), kanamycin (50 μg/mL), or spectinomycin (50 μg/mL) were added to the medium. For recombinant strain expression, overnight cultures were inoculated into medium [12 g/L tryptone, 24 g/L yeast extract, 0.25% (w/v) glycerol, 2.3 g/L KH2PO4, and 12 g/L K2HPO4, 2.0 mM MgSO4, and trace metal mix containing 0.05 mM FeCl3, 0.02 mM CaCl2, 0.01 mM MnCl2, 0.01 mM ZnSO4, and 2 mM each of CoCl2, NiCl2, Na2MoO4, Na2SeO3, and H3BO3]. Cultures were shaken at 200 rpm and incubated at 37°C until the OD600 reached 0.4–0.6. Cells were then induced with 0.3 mM IPTG for 24 h at 20°C. After the end of the cultivation, cells were harvested by centrifugation and washed twice with 0.85% NaCl solution.
The strains and plasmids used in the present study are described in Supplementary Table 1. The primers used for gene cloning in this study were synthesized at General Biol (Chuzhou, China). Primer sequences are listed in Supplementary Tables 2–6. The sequences of all constructed plasmids were verified using DNA sequencing.
Ribosomal binding site sequences with different initial translation rates were used, and the sequences were derived from the International Genetically Engineered Machine Competition (iGEM1). The template was the pETDuet-RBS-getf-RBS-splcd plasmid, and the substitution was done by PCR of the whole plasmid with a forward primer (X-F) and a reverse primer (X-R). All successfully constructed plasmids were transformed into E. coli BL21(DE3) to express the genes.
According to a previously described method (Li et al., 2021), the pEcgRNA-ΔsucA and pEcgRNA-ΔaceA were constructed using the pEcgRNA plasmid. Positive clones were selected on LB plates supplemented with 40 μg/mL spectinomycin. Two homologous arms and the sequence to be inserted were separately amplified and then fused together by overlapping PCR to create donor DNA for integration. The PCR products were purified by gel extraction prior to electroporation.
The citrate synthase gene (gltA) was amplified from the genome of E. coli BL21(DE3), digested with EcoRI and HindIII, and ligated to the plasmid pCDFDuet-1-gltA. The isocitrate dehydrogenase gene (icd) fragment was digested with BglII and XhoI and inserted into the pCDFDuet-1-gltA digested with the same enzymes, resulting in the vector pCDFDuet-1-gltA-icd.
The recombinant plasmids were introduced to E. coli BL21 (DE3) and then the cells were induced with 0.3 mM IPTG for 24 h at 20°C and collected by centrifugation. Under the same reaction conditions, the ability of recombinant strains was evaluated in terms of product yield. The reaction system consisted of 50 mM L-lysine, 60 mM 2-OG, 10 mM Fe2+, 10 mM L-ascorbate (VC), 0.1% (wt/v) trition and cells with a final OD600 of 30 in PBS buffer (50 mM). The reaction was carried out in a 50 mL Erlenmeyer flask for 20 h at 30°C with shaking at 250 rpm and then terminated by boiling in a water bath for 3 min. All reactions were carried out in triplicate.
Electroporation of competent E. coli BL21(DE3) carrying pEcCas was carried out as described previously (Li et al., 2021). For λ-Red induction, L-arabinose (30 mM final concentration) was added to the culture. For genome editing, electroporation was performed using MicroPulser (Bio-Rad) with the default E. coli program (2 mm, 1.8 kV, and 6.1 ms). Each electroporation reaction included donor DNA and the plasmid pEcgRNA-ΔsucA. The electroporation mixture was suspended in 1 mL of LB medium right away. Cells were recovered by incubating for 2 h at 37°C before spreading on LB plates containing kanamycin (50 μg/mL) and spectinomycin (40 μg/mL) and incubating overnight at 37°C. Colonies were chosen at random and successful transformation was confirmed using colony PCR and DNA sequencing.
A colony of the edited clone containing both pEcCas and pEcgRNA-ΔsucA was inoculated into 2 mL LB medium containing rhamnose (20 mM) and kanamycin (50 μg/ml) to eliminate the pEcgRNA-ΔsucA and pEcCas series plasmids (Figure 1). The culture was shaken at 220 rpm for 8 h before being transferred into a liquid LB medium without antibiotics and then grown for another 8 h. The cells were spread on LB plates containing 10 μg/mL sucrose and incubated overnight at 37°C on LB plates with or without kanamycin (50 μg/mL). LB plates, LB plates with kanamycin, and LB plates with spectinomycin were used to screen for positive colonies. The pEcgRNA-ΔsucA-cured colonies were used in the next round of aceA gene editing. The methods used for aceA gene editing were identical to those used for sucA deletion.
For the biotransformation, cells were suspended in the reaction mixture containing 0.5% (wt/v) glucose, 50 mM L-lysine, 10 mM Fe2+, 10 mM VC, 0.1%(w/v) Triton X-100 and cells with a final OD600 of 30 in PBS buffer (50 mM). The bioconversion reactions were performed at 30°C and 250 rpm on a shaker. Aliquots were taken at various time points, centrifuged, and heated at 100°C for 5 min to terminate the reaction. At least three independent experiments were performed for each strain.
Cell growth was evaluated by measuring the absorbance at 600 nm using an UV spectrophotometer. For Fmoc derivatization, 50 μL of the culture supernatant was diluted with 250 μL of 100 mM borate buffer (pH 9.0). Diluted samples were derivatized with 10 mM Fmoc-Cl (TCI, Tokyo, Japan) in acetone (300 μL) and vigorously mixed and then reacted for 30 min. These mixtures were quenched with 600 μL of 25% acetonitrile/250 mM borate buffer (pH 5.5).
The amounts of L-lysine, L-Pip, and cis-3-HyPip were determined using a high-performance liquid chromatography system (Agilent 1260, Agilent Technologies, Santa Clara, CA, United States) with an HC-C18 column (Agilent Technologies, Santa Clara, CA, United States, 4.6 × 250 mm, 5 μm) and a flow rate of 1.0 mL/min at 40°C. The solvent gradient used was as follows: Buffer A [0.1% trifluoroacetic acid (TFA) water], B (0.1% TFA acetonitrile); 0–5 min, 30% B; 8–15 min, 40% B; 15–20 min, 40–50% B; 20–25 min, 50% B; 25–30 min, 50–90% B; 30–38 min, 90% B; 38–41 min, 90–30% B; and 41–45 min, 30% B.
In our previous study, a dual-enzyme system containing SpLCD and GetF was constructed and reaction parameters were systematically optimized to biosynthesize cis-3-HyPip from L-lysine (Hu et al., 2022). However, the development of this whole-cell cascade catalyst is challenged by difficulty in balancing the rates of the two main reaction steps, namely, eliminating intermediate pipecolic acid accumulation and shifting the equilibrium toward cis-3-HyPip production. Tuning translation rates by varying the strength of RBS sequences has been widely used as an effective approach to balance the expression of biosynthetic enzymes in metabolic pathways and optimize the synthesis of target metabolites due to convenience and high efficiency (Liu et al., 2019b; Li et al., 2020).
In the present study, the translation initiation rates of GetF and SpLCD were refined to enhance the titer of cis-3-HyPip. The expression levels of GetF and SpLCD were fine-tuned using RBS sequences with different translation initiation rates, and six RBS sequences were preliminarily selected in this study. T7 RBS originally existed in the Duet vectors, and five were from the iGEM Standard Parts and the RBS strength order was RBS 29 > 30 > RBS(T7) > 32 > 64 > 31 (Zhu et al., 2021). Using strain 1, which harbors pETduet-1-RBS(T7)-getf-RBS(T7)-splcd, as a starting strain, the GetF expression was combined with RBS sequences of various strengths to construct five strains (Figure 2A). As shown in Figure 2B, the efficiency of whole-cell catalysis was enhanced by increasing the strength of the RBS sequence. Finally, the highest activity was exhibited by the cells containing RBS 29 and the cis-3-HyPip productivity increased 30% through RBS optimization. Based on these results, the strain 2 harboring pETduet-1-29-getf-RBS(T7)-splcd as a starting strain, the SpLCD expression were combined with RBS sequences of various strengths to construct other five strains. As shown in Figure 2C, we found that the titers were much more impacted by the type of RBS used for GetF expression than that used for SpLCD expressions. Furthermore, it also showed that a moderate translational initiation rate of SpLCD is more suitable than the high or low rates, and that neither higher nor lower initiation rates not improved whole-cell activity. Similar results were also reported in previous studies, high-intensity RBS regulates the expression of non-rate-limiting enzymes, which may not result in a high yield of target products. Instead, medium-intensity RBS was observed to achieve a relatively highest yield (Alagesan et al., 2018; Sun et al., 2020). Finally, 32 mM cis-3-HyPip was obtained using strain 2 as the biocatalyst (Figure 2D). These results indicated that regulating gene expression by optimizing RBS strength could be utilized to solve the effect of rate-limiting steps and was useful for the target product yield improvement (Ge et al., 2022).
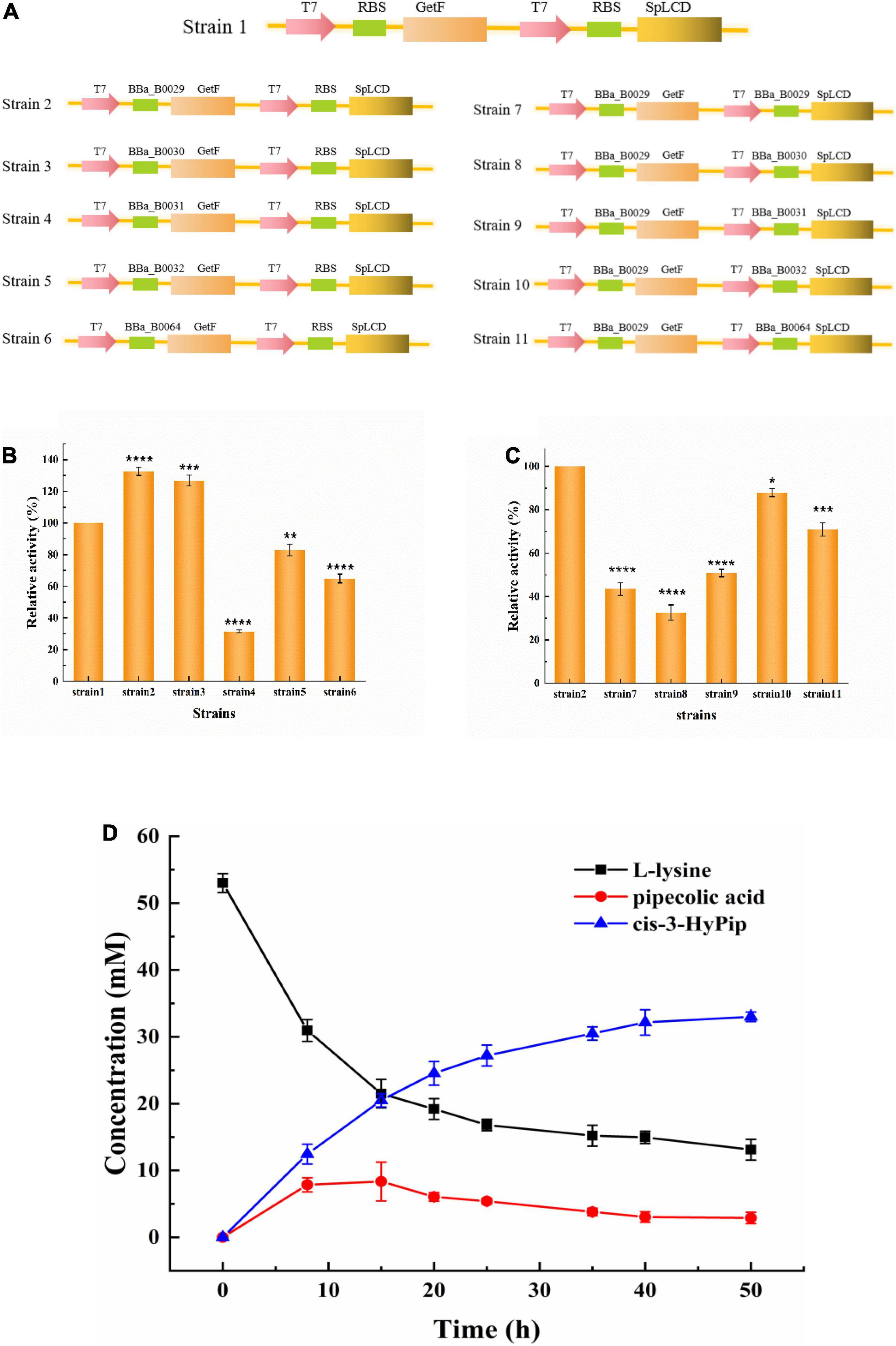
Figure 2. Whole-cell catalysis effects of strains with different RBS strengths. (A) Construction of different strains. (B) The effect of GetF RBS translation strength on catalysis. (C) The effect of SpLCD RBS translation strength on catalysis. (D) Time course of the bioconversion by the strain 2 (containing pETduet-1-29-getf-RBS-splcd). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
GetF catalyzes the conversion of L-Pip to cis-3-HyPip, which requires 2-OG as a co-substrate, resulting in a high cost. Several studies have linked glutamate to the 2-OG conversion reaction (Wu et al., 2018; Sun et al., 2019). This reaction also produces H2O2, which can impair enzymatic activity (Niu et al., 2014; Jing et al., 2020). In the TCA cycle, 2-OG is an important intermediate metabolite. To simplify the technological processes and lower the overall production costs, a whole-cell biocatalyst was reconstructed by using a metabolic engineering strategy in E. coli. This intervention could introduce a 2-OG accumulation system, eliminating the need for additional 2-OG supplementation (Figure 4A).
To engineer the metabolic flux of the TCA cycle, genes encoding sucA and aceA were separately knocked out in E. coli BL21(DE3). In this study, the sucA and aceA genes were knocked out in BL21(DE3) using the pEcCas/pEcgRNA system (Li et al., 2021). As shown in Figure 3A, the PCR amplification fragment was 4,563-bp (lane 2), indicated that sucA was not knocked out, while the PCR amplification fragment was 1,761-bp (lane 1), indicating that the sucA was knocked out. The PCR amplification fragment was 1,586-bp (lane 3), indicating that that it had been knocked out. By contrast, the 2,891-bp PCR amplification fragment (lane 4) indicated that aceA had not been knocked out (Figure 3B).
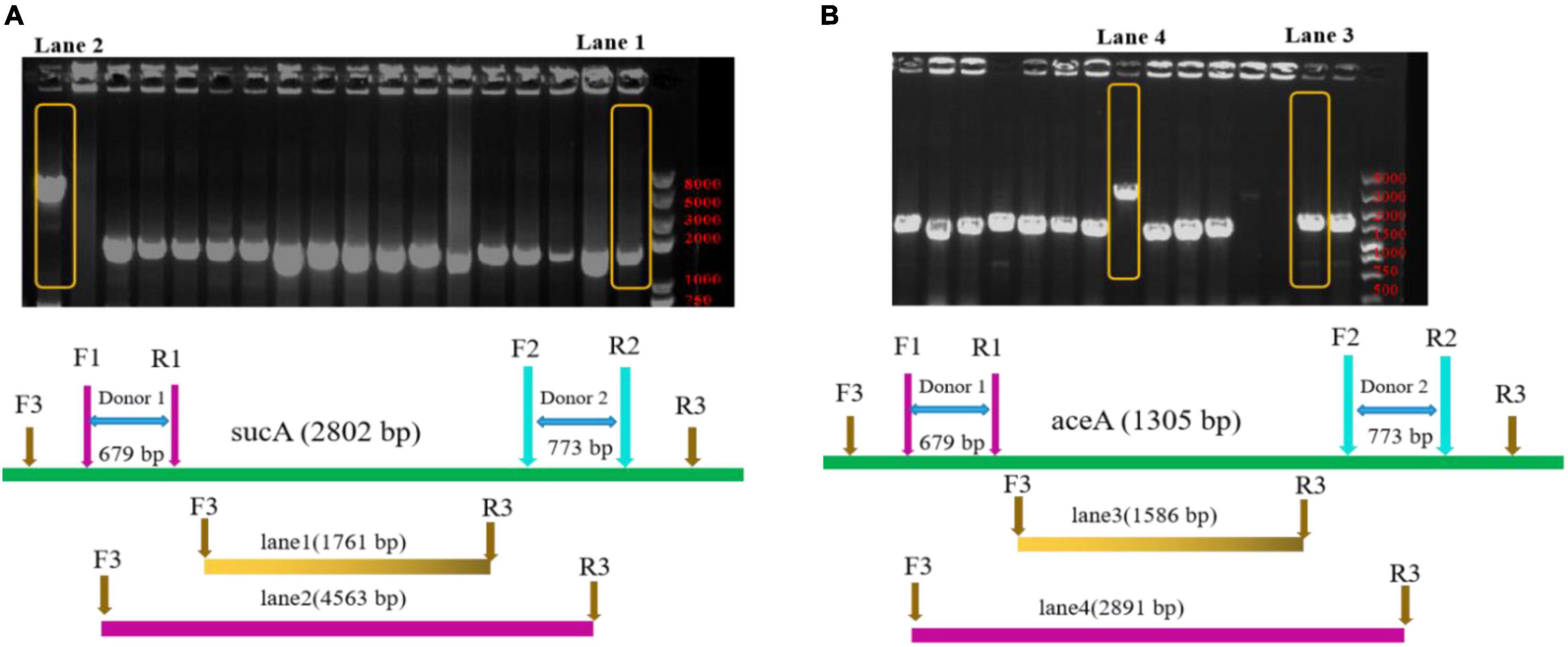
Figure 3. Polymerase chain reaction analysis of mutant strains. (A) Primers (F3/R3) were designed to target both ends of the sucA for the PCR detection of positive strains. The 1761-bp PCR-amplified fragment indicates that the sucA was knocked out, while the presence of a 4,563-bp fragment indicates that this gene was not knocked out. Lane 1: E. coli BL21(DE3) ΔsucA; lane 2: E. coli BL21(DE3). (B) Primers (F3/R3) were designed to target both ends of the aceA for the PCR detection of positive strains. The 1586-bp PCR-amplified fragment indicates that the aceA was knocked out, whereas the presence of a 2,891-bp fragment indicates that this gene was not knocked out. Lane 3: E. coli BL21(DE3) ΔaceA; Lane 4: E. coli BL21(DE3).
Three E. coli mutants were obtained through knockout, and the plasmid pETduet-029-getf-RBS-splcd was transformed into these engineered strains, respectively. In the resulting recombinant strains (strain 12, strain 13, and strain 14) showed higher production of cis-3-HyPip than the starting strain 2. As shown in Figure 4B, the production of cis-3-HyPip by strain 2, and strain 12(ΔsucA), strain 13(ΔaceA), and strain 14(ΔsucAΔaceA) was 3.1, 18, 4.4, and 20.1 mM. The strain 12(ΔsucA) showed the more obvious effect of increasing the yield of cis-3-HyPip. The deletion of aceA, the gene encoding the key enzyme for glyoxylate bypass, also caused a moderate increase in cis-3-HyPip yield, suggesting that aceA only played a minor role in 2-OG catabolism and only sucA mutation can completely block the conversion of 2-OG to succinate. These results indicate that using the redesigned TCA pathway to produce cis-3-HyPip in E. coli was effective. The TCA cycle can be engineered to adopt an alternative route without having significant impact on cell physiology (Lin et al., 2015).
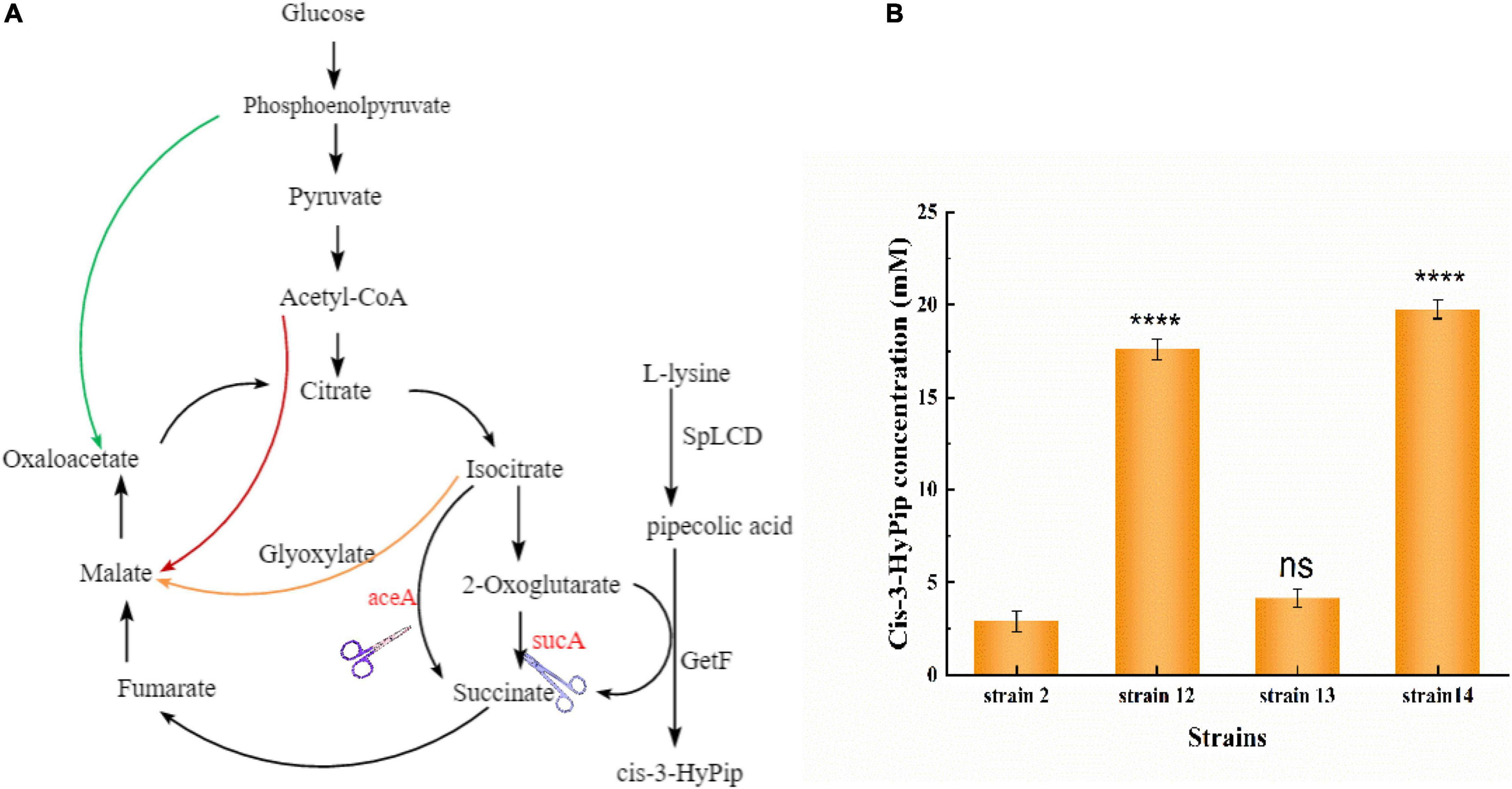
Figure 4. Reconstitution of the TCA cycle using GetF catalyzed reaction. (A) The manipulations to disrupt the TCA cycle and glyoxylate bypass. (B) Effect of sucA and aceA gene deletion on cis-3-HyPip production, strain 12 [E. coli BL21(DE3) ΔsucA pETduet-1-29-getf-RBS(T7)-splcd], strain 13 [E. coli BL21(DE3) ΔaceA pETduet-1-29-getf-RBS(T7)-splcd], strain 14 [E. coli BL21(DE3) ΔsucAΔaceA pETduet-1-29-getf-RBS(T7)-splcd]. ****P < 0.0001.
As previously reported, increasing the expression of genes encoding citrate synthase (gltA) and isocitrate dehydrogenase (icd) can increase the carbon flux to 2-OG (Zhang et al., 2018; Sun et al., 2019). Here, we used pCDFDuet-1, which contains the genes for citrate synthase (gltA) and isocitrate dehydrogenase (icd), to strengthen the TCA cycle (Figure 5A). Strain 15 [E. coli BL21(DE3) △sucA △aceA pETduet-1-29-getf-RBS(T7)-splcd &pCDF-glta-icd] was constructed on the basis of strain 14, and its produced slightly more than strain 14 (Figure 5B), indicating that overexpression of gltA and icd strengthen TCA cycle activity, thereby increasing the yield of cis-3-HyPip.
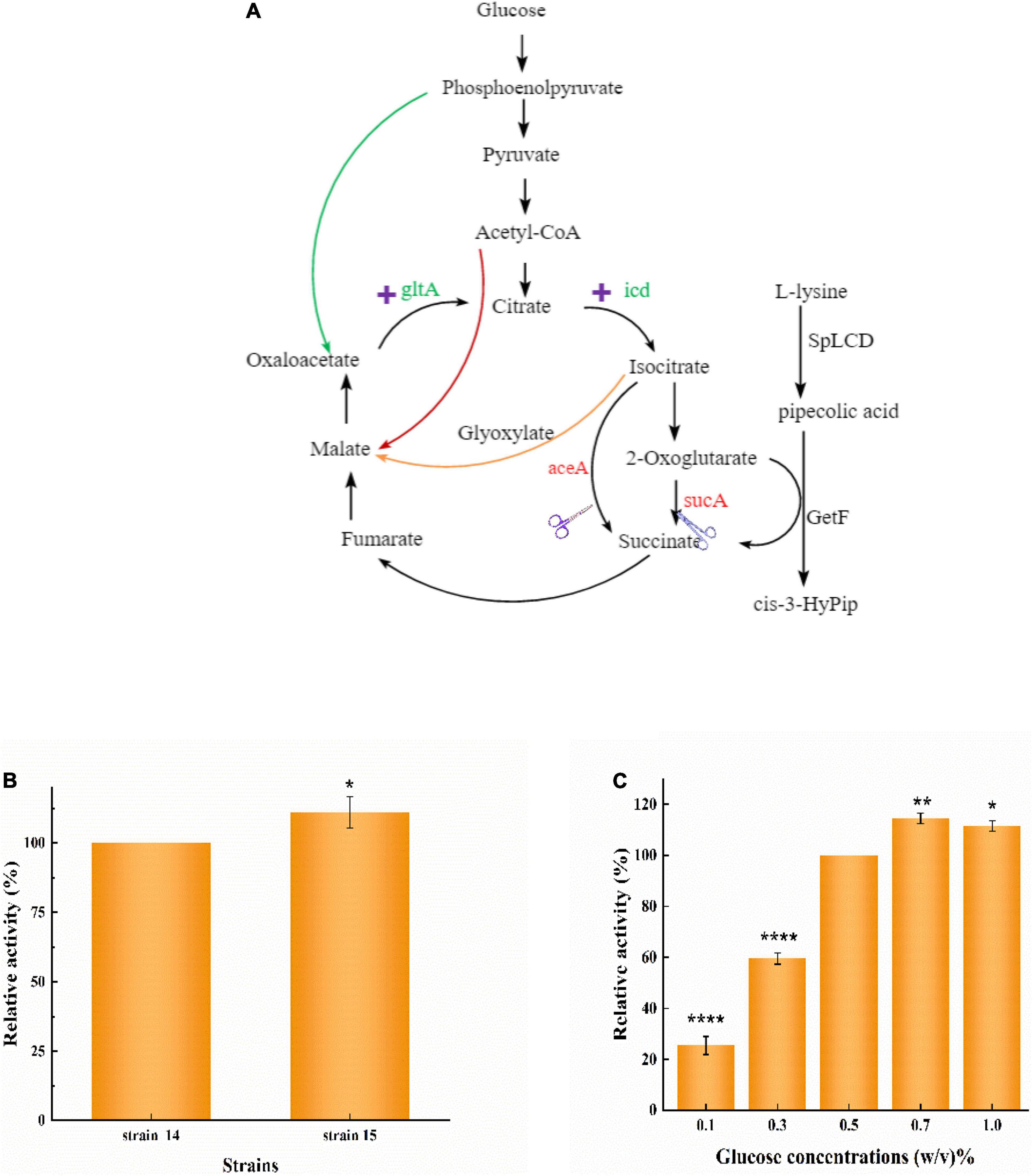
Figure 5. Increase whole-cell activity for cis-3-HyPip production by over-accumulation of 2-OG. (A) The combined manipulations to convert E. coli into an efficient whole-cell biocatalyst for cis-3-HyPip production. (B) Effect of overexpressing glta and icd on whole-cell catalysis. Strain 15 [E. coli BL21(DE3) △sucA △aceA pETduet-1-29-getf-RBS(T7)-splcd &pCDF-glta-icd], Strain 14 [E. coli BL21(DE3) △sucA △aceA pETduet-1-29-getf-RBS(T7)-splcd]. (C) Effects of glucose concentrations on whole-cell (strain 15) catalysis. *P < 0.05, **P < 0.01, ****P < 0.0001.
When glucose is taken up by E. coli, it is first broken down into pyruvate via glycolysis. Pyruvate is further converted to acetyl-CoA, which then enters the TCA cycle (Figure 5A). To increase the flux of the TCA cycle, different concentrations of glucose were also optimized. Our results showed that different glucose concentrations had a significant impact on cis-3-HyPip production in strain 15. As the glucose concentration increased from 0.1 to 0.7%, the production of cis-3-HyPip increased in concert (Figure 5C). At glucose concentrations higher than 0.7% (such as 0.7–1% w/v), no significant increase of cis-3-HyPip production was observed (Figure 5C). Such effects of glucose on cis-3-HyPip production were not observed in strain 2, which the TCA cycle was not coupled with the GetF catalyzed reaction. These results further demonstrated that the flux of the TCA cycle can be coupled with the catalyzed reaction (Ruan et al., 2019; An et al., 2020), and that the efficiency of the whole-cell biocatalysis for production can be improved by pushing the TCA cycle (Lin et al., 2015; Sun et al., 2019).
Under the optimized conditions described above, a curve for whole-cell biocatalyst mediated conversion of L-lysine into cis-3-HyPip was obtained. As shown in Figure 6, at 0–20 h, the yield of cis-3-HyPip accumulated rapidly up to 21 mM. Subsequently, the rate of product generation increased slightly and slowly after 20 h. The product yield stabilized during the final phase (>40 h), which may be due to the decrease of thermal stability of GetF was the reason for the loss of catalytic activity. Finally, the reaction reached equilibrium when 33 mM cis-3-HyPip was produced after bioconversion for 50 h. Compared to that in previous studies (Table 1), the engineered strain used in this study displays the highest yield reported to date via whole-cell biotransformation, and without the addition of 2-OG.
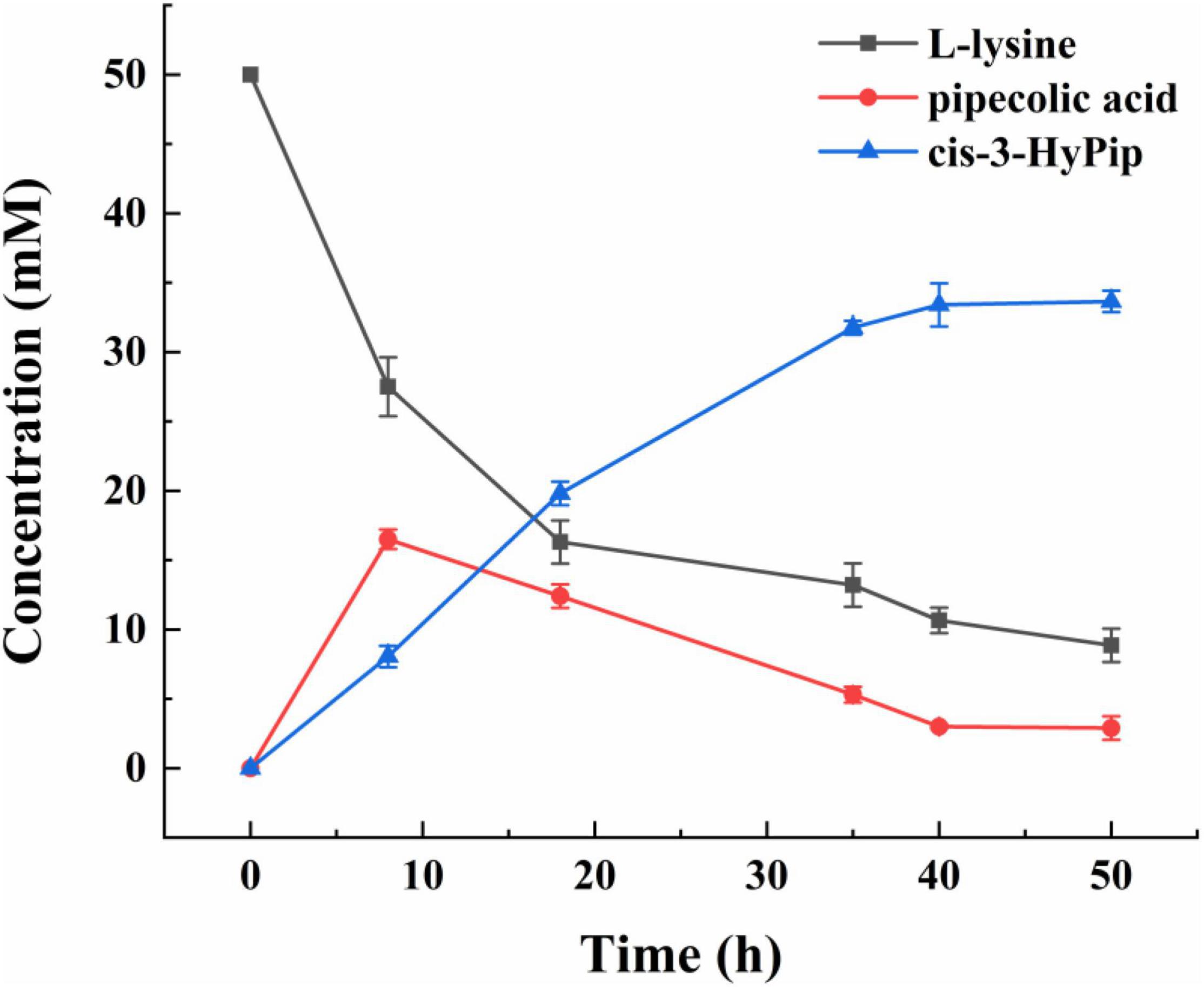
Figure 6. The time course of the bioconversion by strain 15 [E. coli BL21(DE3) △sucA △aceA pETduet-1-29-getf-RBS(T7)-splcd &pCDF-glta-icd].
In this study, we demonstrated synthetic biological strategies for enhancing cis-3-HyPip biosynthesis in E. coli. The translation rate was modulated by changing the strength of the RBS sequence to balance GetF and SpLCD expression and improve the titer of cis-3-HyPip. We also reconstituted the TCA cycle by knocking out the sucA and aceA genes, which reduced the addition of exogenous 2-OG and lowered the catalytic cost. The supply and catalytic efficiency of continuous 2-OG supplementation powered by glucose were improved by improving metabolic flux (gltA and icd) and optimizing glucose concentration. The maximum cis-3-HyPip titer was 33 mM. This is the first report of microbial biosynthesis of cis-3-HyPip that did not require the addition of 2-OG. Moreover, this strategy can also be applied to enzymes that catalyze 2-OG-coupled reactions and synthesize other functional compounds. It should be noted that there is still be much room for the improved production of cis-3-HyPip, for example, the directed evolution of GetF or high-throughput screening of GetF mutations can be studied in the future.
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
SH: methodology, data curation, writing – original draft, and investigation. YL: methodology. HL: supervision. KC: conceptualization, supervision, project administration, and funding acquisition. AZ: review and editing. PO: project administration and funding acquisition. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (2021YFC2104100), the National Science Foundation for Young Scientists of China (Grant No. 21908101), and China Postdoctoral Science Foundation (2018M642237 and 2020T130295).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.945184/full#supplementary-material
cis-3-HyPip, cis-3-hydroxypipecolic acid; 2-OG, α -ketoglutarate; HyPips, hydroxypipecolic acids; L-Pip, L-pipecolic acid; IPTG, isopropyl β-D -1-thiogalactopyranoside; PCR, polymerase chain reaction; RBS, ribosomal binding site; TCA, tricarboxylic acid cycle; VC, L-ascorbate.
Ahuja, B. B., and Sudalai, A. (2015). A concise enantioselective synthesis of (2S,3S)-3-hydroxypipecolic acid via proline catalyzed alpha-aminooxylation of aldehydes and Pd-catalyzed ether directed aza-Claisen rearrangements. Tetrahedron-Asymmetry 26, 24–28. doi: 10.1016/j.tetasy.2014.11.010
Alagesan, S., Hanko, E. K. R., Malys, N., Ehsaan, M., Winzer, K., and Minton, N. P. (2018). Functional genetic elements for controlling gene expression in Cupriavidus necator H16. Appl. Environ. Microbiol. 8, e00878–e918. doi: 10.1128/aem.00878-18
An, J. H., Zhang, W. L., Jing, X. R., Nie, Y., and Xu, Y. (2020). Reconstitution of TCA cycle involving l-isoleucine dioxygenase for hydroxylation of l-isoleucine in Escherichia coli using CRISPR-Cas9. 3 Biotech 10, 167. doi: 10.1007/s13205-020-2160-3
Binz, T. M., Maffioli, S. I., Sosio, M., Donadio, S., and Mueller, R. (2010). Insights into an unusual nonribosomal peptide synthetase biosynthesis identification and characterization of the GE81112 biosynthetic gene cluster. J. Biol. Chem. 285, 32710–32719. doi: 10.1074/jbc.M110.146803
Chavan, S. P., Khairnar, L. B., Pawar, K. P., Chavan, P. N., and Kawale, S. A. (2015). Enantioselective syntheses of (R)-pipecolic acid, (2R,3R)-3-hydroxypipecolic acid, beta-(+)-conhydrine and (-)-swainsonine using an aziridine derived common chiral synthon. RSC Adv. 5, 50580–50590. doi: 10.1039/c5ra06429e
Chen, H., Huang, R., and Zhang, Y. H. P. (2017). Systematic comparison of co-expression of multiple recombinant thermophilic enzymes in Escherichia coli BL21(DE3). Appl. Microbiol. Biotechnol. 101, 4481–4493. doi: 10.1007/s00253-017-8206-8
Ge, C., Yu, Z., Sheng, H. K., Shen, X. L., Sun, X. X., Zhang, Y. F., et al. (2022). Redesigning regulatory components of quorum-sensing system for diverse metabolic control. Nat. Commun. 13, 2182. doi: 10.1038/s41467-022-29933-x
Hara, R., and Kino, K. (2009). Characterization of novel 2-oxoglutarate dependent dioxygenases converting L-proline to cis-4-hydroxy-L-proline. Biochem. Biophys. Res. Commun. 379, 882–886. doi: 10.1016/j.bbrc.2008.12.158
Hibi, M., Mori, R., Miyake, R., Kawabata, H., Kozono, S., Takahashi, S., et al. (2016). Novel Enzyme Family Found in Filamentous Fungi Catalyzing trans-4-Hydroxylation of L-Pipecolic Acid. Appl. Environ. Microbiol. 82, 2070–2077. doi: 10.1128/aem.03764-15
Hu, S., Yang, P., Li, Y., Zhang, A., Chen, K., and Ouyang, P. (2022). Biosynthesis of cis-3-hydroxypipecolic acid from L-lysine using an in vivo dual-enzyme cascade. Enzyme Microb. Technol. 154, 109958–109958. doi: 10.1016/j.enzmictec.2021.109958
Jiang, W., and Fang, B. (2016). Construction of a tunable multi-enzyme-coordinate expression system for biosynthesis of chiral drug intermediates. Sci. Rep. 6, 30462. doi: 10.1038/srep30462
Jing, X. R., Liu, H., Nie, Y., and Xu, Y. (2020). 2-Ketoglutarate-generated in vitro enzymatic biosystem facilitates Fe(II)/2-Ketoglutarate-dependent dioxygenase-mediated C-H bond oxidation for (2s,3r,4s)-4-Hydroxyisoleucine synthesis. Int. J. Mol. Sci. 21, 5347. doi: 10.3390/ijms21155347
Klein, C., and Huettel, W. (2011). A simple procedure for selective hydroxylation of L-proline and L-pipecolic acid with recombinantly expressed proline hydroxylases. Adv. Synth. Catal. 353, 1375–1383. doi: 10.1002/adsc.201000863
Koketsu, K., Shomura, Y., Moriwaki, K., Hayashi, M., Mitsuhashi, S., Hara, R., et al. (2015). Refined regio- and stereoselective hydroxylation of L-pipecolic acid by protein engineering of L-proline cis-4-Hydroxylase based on the X-ray crystal structure. ACS Synth. Biol. 4, 383–392. doi: 10.1021/sb500247a
Li, G., Lian, J., Xue, H., Jiang, Y., Wu, M., Lin, J., et al. (2020). Enzymatic preparation of pyruvate by a whole-cell biocatalyst coexpressing L-lactate oxidase and catalase. Process Biochem. 96, 113–121. doi: 10.1016/j.procbio.2020.04.014
Li, Q., Sun, B., Chen, J., Zhang, Y., Jiang, Y., and Yang, S. (2021). A modified pCas/pTargetF system for CRISPR-Cas9-assisted genome editing in Escherichia coli. Acta Biochim. Biophys. Sin. 53, 620–627. doi: 10.1093/abbs/gmab036
Lin, B., Fan, K., Zhao, J., Ji, J., Wu, L., Yang, K., et al. (2015). Reconstitution of TCA cycle with DAOCS to engineer Escherichia coli into an efficient whole cell catalyst of penicillin G. Proc. Natl. Acad. Sci. U. S. A. 112, 9855–9859. doi: 10.1073/pnas.1502866112
Liu, S., Zhang, X., Liu, F., Xu, M., Yang, T., Long, M., et al. (2019a). Designing of a cofactor self-sufficient whole-cell biocatalyst system for production of 1,2-amino alcohols from epoxides. ACS Synth. Biol. 8, 734–743. doi: 10.1021/acssynbio.8b00364
Liu, Z., Yu, L., Zhou, L., and Zhou, Z. (2019b). One-pot biosynthesis of L-aspartate from maleate via an engineered strain containing a dual-Enzyme System. Appl. Environ. Microbiol. 85, 1001. doi: 10.1128/aem.01327-19
Lu, F., Chen, J., Ye, H., Wu, H. L., Sha, F., Huang, F. J., et al. (2020). Enzymatic hydroxylation of L-pipecolic acid by L-prolinecis-4-hydroxylases and isomers separation. Biotechnol. Lett. 42, 2607–2617. doi: 10.1007/s10529-020-03002-z
Mattay, J., and Huettel, W. (2017). Pipecolic Acid Hydroxylases: a monophyletic clade among cis-selective bacterial proline hydroxylases that discriminates L-proline. Chembiochem 18, 1523–1528. doi: 10.1002/cbic.201700187
Miller, S. P., Zhong, Y.-L., Liu, Z., Simeone, M., Yasuda, N., Limanto, J., et al. (2014). Practical and cost-effective manufacturing route for the synthesis of a beta-lactamase inhibitor. Org. Lett. 16, 174–177. doi: 10.1021/ol4031606
Mori, H., Shibasaki, T., Yano, K., and Ozaki, A. (1997). Purification and cloning of a proline 3-hydroxylase, a novel enzyme which hydroxylates free L-proline to cis-3-hydroxy-L-proline. J. Bacteriol. 179, 5677–5683. doi: 10.1128/jb.179.18.5677-5683.1997
Niu, P. Q., Dong, X. X., Wang, Y. C., and Liu, L. M. (2014). Enzymatic production of alpha-ketoglutaric acid from L-glutamic acid via L-glutamate oxidase. J. Bacteriol. 179, 56–62. doi: 10.1016/j.jbiotec.2014.03.021
Qi, W.-Y., Fang, S.-L., Xu, X.-T., Zhang, K., and Shi, B.-F. (2021). Asymmetric formal synthesis of (-)-tetrazomine. Org. Chem. Front. 8, 1802–1807. doi: 10.1039/d1qo00029b
Ruan, L. Y., Li, L., Zou, D., Jiang, C., Wen, Z. Y., Chen, S. W., et al. (2019). Metabolic engineering of Bacillus amyloliquefaciens for enhanced production of S-adenosylmethionine by coupling of an engineered S-adenosylmethionine pathway and the tricarboxylic acid cycle. Biotechnol. Biofuels 12, 211. doi: 10.1186/s13068-019-1554-0
Shibasaki, T., Mori, H., Chiba, S., and Ozaki, A. (1999). Microbial proline 4-hydroxylase screening and feme cloning. Appl. Environ. Microbiol. 65, 4028–4031. doi: 10.1128/AEM.65.9.4028-4031.1999
Sun, D., Gao, D., Liu, X., Zhu, M., Li, C., Chen, Y., et al. (2019). Redesign and engineering of a dioxygenase targeting biocatalytic synthesis of 5-hydroxyl leucine. Catal. Sci. Technol. 9, 1825–1834. doi: 10.1039/c9cy00110g
Sun, H., Yang, J., and Song, H. (2020). Engineering mycobacteria artificial promoters and ribosomal binding sites for enhanced sterol production. Biochem. Eng. J. 162, 107739. doi: 10.1016/j.bej.2020.107739
Wu, J., Fan, X., Liu, J., Luo, Q., Xu, J., and Chen, X. (2018). Promoter engineering of cascade biocatalysis for alpha-ketoglutaric acid production by coexpressing L-glutamate oxidase and catalase. Appl. Microbiol. Biotechnol. 102, 4755–4764. doi: 10.1007/s00253-018-8975-8
Yang, B., Zheng, P., Wu, D., and Chen, P. (2021). Efficient biosynthesis of raspberry ketone by engineered Escherichia coli coexpressing zingerone synthase and glucose dehydrogenase. J. Agricult. Food Chem. 69, 2549–2556. doi: 10.1021/acs.jafc.0c07697
Zhang, C., Li, Y., Ma, J., Liu, Y., He, J., Li, Y., et al. (2018). High production of 4-hydroxyisoleucine in corynebacterium glutamicum by multistep metabolic engineering. Metab. Eng. 49, 287–298. doi: 10.1016/j.ymben.2018.09.008
Zhang, J., Weng, H., Zhou, Z., Du, G., and Kang, Z. (2019). Engineering of multiple modular pathways for high-yield production of 5-aminolevulinic acid in Escherichia coli. Bioresour. Technol. 274, 353–360. doi: 10.1016/j.biortech.2018.12.004
Zhang, Z. D., and Sun, Z. H. (2020). A new synthesis of L-hydroxypipecolic acid. Synlett 31, 355–358. doi: 10.1055/s-0039-1690771
Zhu, Y., Wan, L., Meng, J., Luo, G., Chen, G., Wu, H., et al. (2021). Metabolic engineering of Escherichia coli for Lacto-N-triose II production with high productivity. J. Agricult. Food Chem. 69, 3702–3711. doi: 10.1021/acs.jafc.1c00246
Keywords: ribosome binding sites, tricarboxylic acid cycle, reconstitute, α-ketoglutarate, whole-cell catalysis
Citation: Hu S, Li Y, Zhang A, Li H, Chen K and Ouyang P (2022) Designing of an Efficient Whole-Cell Biocatalyst System for Converting L-Lysine Into Cis-3-Hydroxypipecolic Acid. Front. Microbiol. 13:945184. doi: 10.3389/fmicb.2022.945184
Received: 16 May 2022; Accepted: 03 June 2022;
Published: 27 June 2022.
Edited by:
Ang Li, Harbin Institute of Technology, ChinaReviewed by:
Xuwang Zhang, Dalian University of Technology, ChinaCopyright © 2022 Hu, Li, Zhang, Li, Chen and Ouyang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kequan Chen, a3FjaGVuQG5qdGVjaC5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.