- 1Department of Clinical Laboratory, Kunming Children’s Hospital, Kunming, China
- 2Yunnan Key Laboratory of Children’s Major Disease Research, Kunming Children’s Hospital, Kunming, China
- 3Yunnan Province Clinical Research Center for Children’s Health and Disease, Kunming Children’s Hospital, Kunming, China
- 4Yunnan Institute of Pediatrics, Kunming Children’s Hospital, Kunming, China
- 5Department of Laboratory, Chuxiong Higher College of Medicine, Chuxiong, China
Invasive Staphylococcus aureus (S. aureus) infection is associated with high rates of mortality in children. No studies have been reported on invasive S. aureus infection among children in Kunming, China, and it remains unknown whether the COVID-19 epidemic has affected S. aureus prevalence in this region. Thus, this study investigated the changes in molecular characteristics and antimicrobial resistance of invasive S. aureus strains isolated from children in Kunming during 2019–2021. In total, 66 invasive S. aureus strains isolated from children were typed by multilocus sequence typing (MLST), spa, and Staphylococcal cassette chromosome mec (SCCmec), and antimicrobial resistance and virulence genes were analyzed. A total of 19 ST types, 31 spa types and 3 SCCmec types were identified. Thirty nine (59.09%) strains were methicillin-sensitive S. aureus (MSSA) and 27 (40.91%) strains were methicillin-resistant S. aureus (MRSA). The most common molecular type was ST22-t309 (22.73%, 15/66), followed by ST59-t437 (13.64%, 9/66). In 2019 and 2021, the dominant molecular type was ST22-t309, while in 2020, it was ST59-t437. After 2019, the dominant molecular type of MRSA changed from ST338-t437 to ST59-t437. All strains were susceptible to tigecycline, ciprofloxacin, moxifloxacin, vancomycin, quinopudine-dafoputin, linezolid, levofloxacin, and rifampicin. From 2019 to 2021, the resistance to penicillin and sulfamethoxazole initially decreased and then increased, a trend that contrasted with the observed resistance to oxacillin, cefoxitin, erythromycin, clindamycin, and tetracycline. Sixteen antimicrobial resistance profiles were identified, with penicillin-tetracycline-erythromycin-clindamycin-oxacillin-cefoxitin being the most common, and the antimicrobial resistance profiles varied by year. The carrier rates of virulence genes, icaA, icaD, hla, fnbA, fnbB, clfA, clfB, and cna were 100.00%. Furthermore, sak, pvl, icaC, icaR, fib, lip, hlb, hysA, sea, seb, and tsst-1 had carrier rates of 96.97, 92.42, 87.88, 69.70, 84.85, 62.12, 56.06, 50, 37.87, 30.30, and 7.58%, respectively. Since COVID-19 epidemic, the annual number of invasive S. aureus strains isolated from children in Kunming remained stable, but the molecular characteristics and antimicrobial resistance profiles of prevalent S. aureus strains have changed significantly. Thus, COVID-19 prevention and control should be supplemented by surveillance of common clinical pathogens, particularly vigilance against the prevalence of multidrug-resistant and high-virulence strains.
Introduction
Staphylococcus aureus (S. aureus) has emerged as one of the most common bacteria in hospital and community-acquired infections, causing a wide range of infections including skin and soft tissue infections, cellulitis, and arthritis. It can also induce invasive infections involving deep tissues or organs, such as bacteremia, meningitis, osteomyelitis, and septicemia (Magill et al., 2018). S. aureus is more likely to cause severe invasive infections in children, whose immune systems, mucosa, and biological barriers are not yet fully developed (Ondusko and Nolt, 2018).
The incidence rate of invasive S. aureus infection has obvious regional and temporal differences. Williamson et al. (2014) found that the incidence rate of invasive S. aureus infection was approximately 52/100,000 among children in New Zealand. In the United States, the incidence rate of invasive community-associated methicillin-resistant S. aureus (CA-MRSA) infection among children rose from 1.10/100,000 in 2005 to 1.70/100,000 in 2010, with an annual growth rate of 10.20% (Iwamoto et al., 2013). The incidence rate of invasive MRSA infection in Chinese children increased from 3.75/10,000 in 2006 to 8.90/10,000 in 2011, with the incidence of invasive CA-MRSA infection increasing from zero to 2.43/10,000 (Qiao et al., 2013). These findings suggest that the incidence rate of invasive S. aureus infection in Chinese children is higher than that observed in developed countries.
More worrisome is the fact that invasive S. aureus infection in children is often accompanied by severe clinical signs and symptoms, which is even life-threatening. The mortality rate of children with invasive S. aureus infection has increased to 14.00% in the United States, of whom 30.00% die of sepsis despite appropriate treatment (Iwamoto et al., 2013). Li et al. (2015) showed that of 56 Chinese children with invasive S. aureus infection, two died, 35.72% had dyspnea, 28.57% were admitted to the ICU, and 37.5% had severe complications such as respiratory failure. These findings indicate that invasive S. aureus infection is a serious threat to the health and life of children and poses a significant challenge to clinical treatment.
The distribution characteristics of S. aureus differ considerably by geographic region. Studies indicate that ST8 and ST121 are the dominant S. aureus strains responsible for invasive infection in the United States, ST59-t437 and ST398-t571 in China, ST8-t190 in Australia, ST8-t008 and ST5-t777 in France, ST22-t032 in Germany, and ST22-t032 in the United Kingdom (Grundmann et al., 2010; O’Hara et al., 2012; Qiao et al., 2013). The molecular characteristics of prevalent S. aureus strains also vary over time. Wang et al. (2021a) found that the S. aureus strain responsible for bloodstream infection was ST398-t571 from 2016 to 2017 and ST59-t437 from 2018 to 2020 in Shandong Province, China. Thus, it is important to continuously monitor the prevalence of S. aureus in various regions. Unfortunately, no studies on invasive S. aureus infection among children in Kunming have been reported yet.
Since December 2019, COVID-19 has rapidly spread worldwide, and a variety of prevention and control measures have been employed in different regions and countries. The lifestyle of Chinese people has changed greatly since the COVID-19 pandemic, including lower population movement between different regions and provinces, wearing a mask, home quarantine, and COVID-19 vaccination. A study indicated that the positivity rates of viruses and atypical pathogens responsible for acute respiratory tract infection (ARI) in Chinese children have declined significantly since the COVID-19 pandemic and that the pathogenic spectrum has also changed (Wang et al., 2021b). Therefore, it is also worth considering and studying whether these changes have also affected the prevalence of common clinical bacterial pathogens.
To reveal the impact of the COVID-19 pandemic, this study aims to investigate the changes in molecular characteristics and antimicrobial resistance of invasive S. aureus strains isolated from children in Kunming during 2019–2021.
Materials and methods
Strain collection and identification
Invasive S. aureus infection is an infection in which S. aureus is isolated from a normally sterile part of the human body. Samples collected repeatedly from the same body part of the same patient should be eliminated. From January 2019 to December 2021, 66 invasive S. aureus strains (22, 24, and 20 in 2019, 2020, and 2021, respectively) were isolated from children < 18 years of age who were treated at Kunming Children’s Hospital, also called Yunnan Children’s Medical Center. This medical center is an affiliated hospital of Kunming Medical University, the largest tertiary hospital for children in Yunnan Province, and is responsible for the diagnosis and treatment of all refractory and critically ill children in the province.
Clinical samples include sterile body fluid specimens such as blood, cerebrospinal fluid, pleural fluid, bone marrow, and alveolar lavage fluid. The samples were inoculated onto blood agar plates and cultured in 5% CO2 at 37°C for 24 h. The strain colonies were subjected to morphological observation, Gram staining and microscopic examination, automatic bacterial identification using VITEK-2 Compact. MRSA was preliminarily determined based on a positive result of cefoxitin screening and confirmed by the presence of the mecA gene using PCR (Stegger et al., 2012).
Multilocus sequence typing
All stains were subjected to molecular typing by Multilocus sequence typing (MLST). Seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqiL) of MLST primer sequences and reaction conditions amplified by PCR were obtained from the MLST website.1 The sequences of the PCR products were compared to those of the existing alleles available from the MLST website, and the allelic number (sequence type, ST) was determined for each sequence.
Spa typing
All strains were typed by the spa typing according to published protocols, by amplifying the spa X region (Koreen et al., 2004). The PCR products were sequenced. Repeats and spa types were assigned using the spa database.2 The strains that could not be classified as any known spa type were defined as non-typable (NT).
Staphylococcal cassette chromosome mec typing
The SCCmec classification of all MRSA strains was performed as previously described (Zhang et al., 2005).
VITEK 2 Compact automated antimicrobial susceptibility test
The antimicrobial susceptibility test (AST) was performed according to the VITEK 2 Compact Automatic Bacterial Identification and Antimicrobial Susceptibility Analyzer user manual. In brief, a 0.5 MCF bacterial suspension was prepared. The instrument automatically adds the bacteria suspension to the Gram-positive bacteria AST card GP-639. After 18–24 h incubation, the Advanced Expert System™ system analyzes the susceptibility of bacteria to various antibacterial drugs. The interpretation of AST results was based on Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2019). The GP-639 card contains sixteen antimicrobials: penicillin (PEN), tigecycline (TJH), ciprofloxacin (CIP), moxifloxacin (MFX), tetracycline (TCY), vancomycin (VAN), quinopudine-dafoputin (QDA), clindamycin (CLI), linezolid (LNZ), levofloxacin (LVX), erythromycin (ERY), gentamicin (GEN), sulfamethoxazole (SXT), rifampicin (RIF), cefoxitin (FOX), and oxacillin (OXA). S. aureus ATCC 29213 is the quality control strain for the assay.
Virulence genes detection
Several virulence genes, biofilm-forming genes and genes encoding extracellular enzyme of S. aureus strains were detected by PCR, including fibronectin-binding proteins A and B (fnbA, fnbB), clumping factors A and B (clfA, clfB), fibrinogen binding protein (fib), collagen binding adhesin (cna), polysaccharide intercellular adhesions A, C, D, and R (icaA, icaC, icaD, icaR), Staphylococcal enterotoxins A and B (sea, seb), Panton–Valentine leucocidin (pvl), toxic shock syndrome1 (tsst-1), hemolysins a and β (hla, hlb), staphylokinase (sak), hyaluronate lyase (hysA), and lipase (lip). The primer sequences and PCR amplification reaction conditions were described previously (Zhang et al., 2014; He et al., 2015; Qu, 2019; Wu et al., 2019).
Statistical analysis
Statistical analysis and the plotting of experimental data were performed using graphPad Prism 8.0 software. The measurement data were tested for normal distribution using K-S normality. Those that conformed to a normal distribution were represented by the mean ± standard deviation. For the data of two groups with homoscedasticity, the independent sample t-test was used. For the data of two groups with heteroscedasticity, the t-test with a Welch’s correction was used. Measurement data that did not conform to a normal distribution are presented as the median (interquartile range), and data were compared between two groups using the Mann-Whitney U-test. Enumeration data were showed as numbers or percentages, and data were compared between groups using the Chi-square or Fisher exact test. P < 0.05 was considered statistically significant.
Ethics statement
The current study was approved by the Ethics Committee of Kunming Children’s Hospital (registration no. 2020-03-134-K01). Written informed consent was exempted, since this retrospective study mainly focused on bacteria and patient intervention was not required.
Results
Patient clinical characteristics
Sixty-six children (42 males and 26 females) with invasive S. aureus infection were admitted to Kunming Children’s Hospital from January 2019 to December 2021. The patients were 1 day to 14 years of age, of whom 30.30% (20/66) were ≤ 1 year of age. Twenty-seven (40.91%) and 39 (59.09%) patients were infected with MRSA and MSSA, respectively. Community-acquired infections accounted for 83.33% (55/66) of all cases. Patients were frequently admitted to the intensive care unit (28.79%, 19/66), the Department of Orthopedics (18.18%, 12/66), and the Department of Hematology (16.67%, 11/66). Blood was the most common type of clinical specimens, accounting for 62.12% (41/66). The invasive infections were common in patients with leukemia, osteomyelitis and pneumonia. In addition, 33.33% (22/66) of patients have multiple infections and 25.76% (17/66) complicated sepsis. The median length of hospital stay was 24.5 and 23 days in patients with MRSA and MSSA infection, respectively. Six patients had not yet recovered when their parents asked for them to be discharged from the hospital (Table 1).
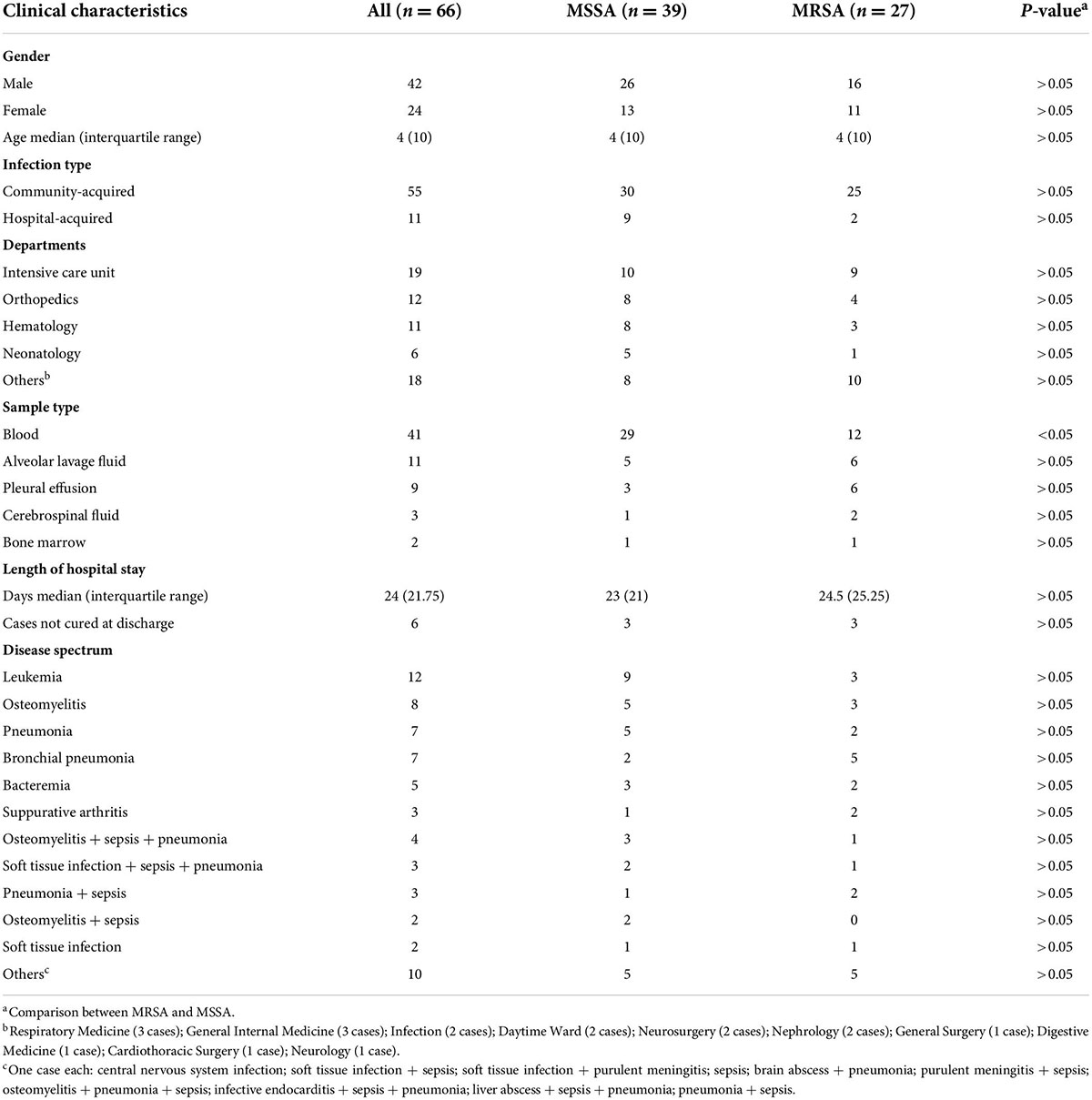
Table 1. Demographic and clinical characteristics of patients infected with methicillin-sensitive Staphylococcus aureus (MSSA) or methicillin-resistant Staphylococcus aureus (MRSA).
Multilocus sequence typing of strains
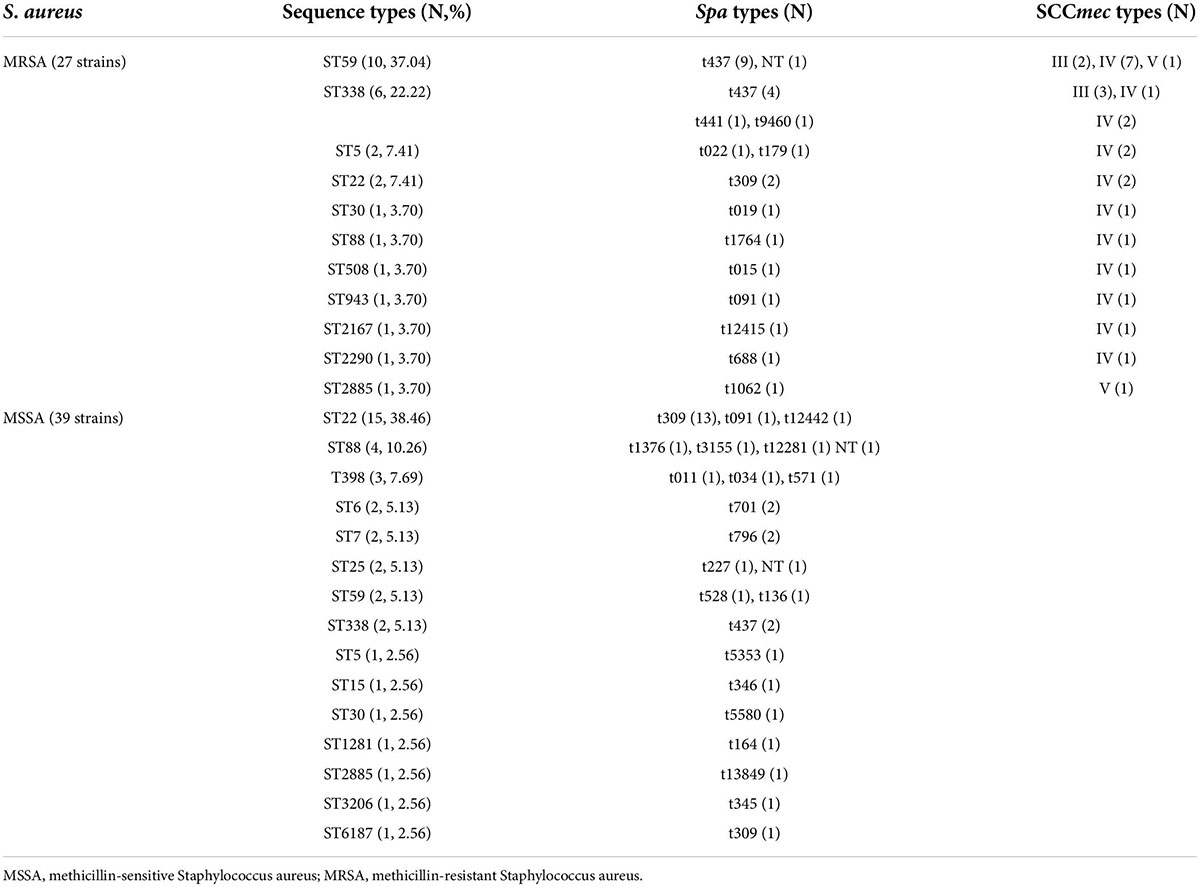
Nineteen ST types were identified among the 66 S. aureus strains, the most common of which was ST22 (25.76%, 17/66), followed by ST59 (18.18%, 12/66) and ST338 (12.12%, 8/66). ST22 was the dominant ST type among the MSSA strains (15/39, 38.46%), followed by ST88 (4/39, 10.26%). ST59 was the dominant ST type among the MRSA strains (37.04%, 10/27), followed by ST338 (6/27, 22.22%). In 2019 and 2021, the dominant ST type was ST22, while in 2020, it was ST59. After 2019, dominant ST type among MRSA was replaced from ST338 to ST59 (Figure 1).
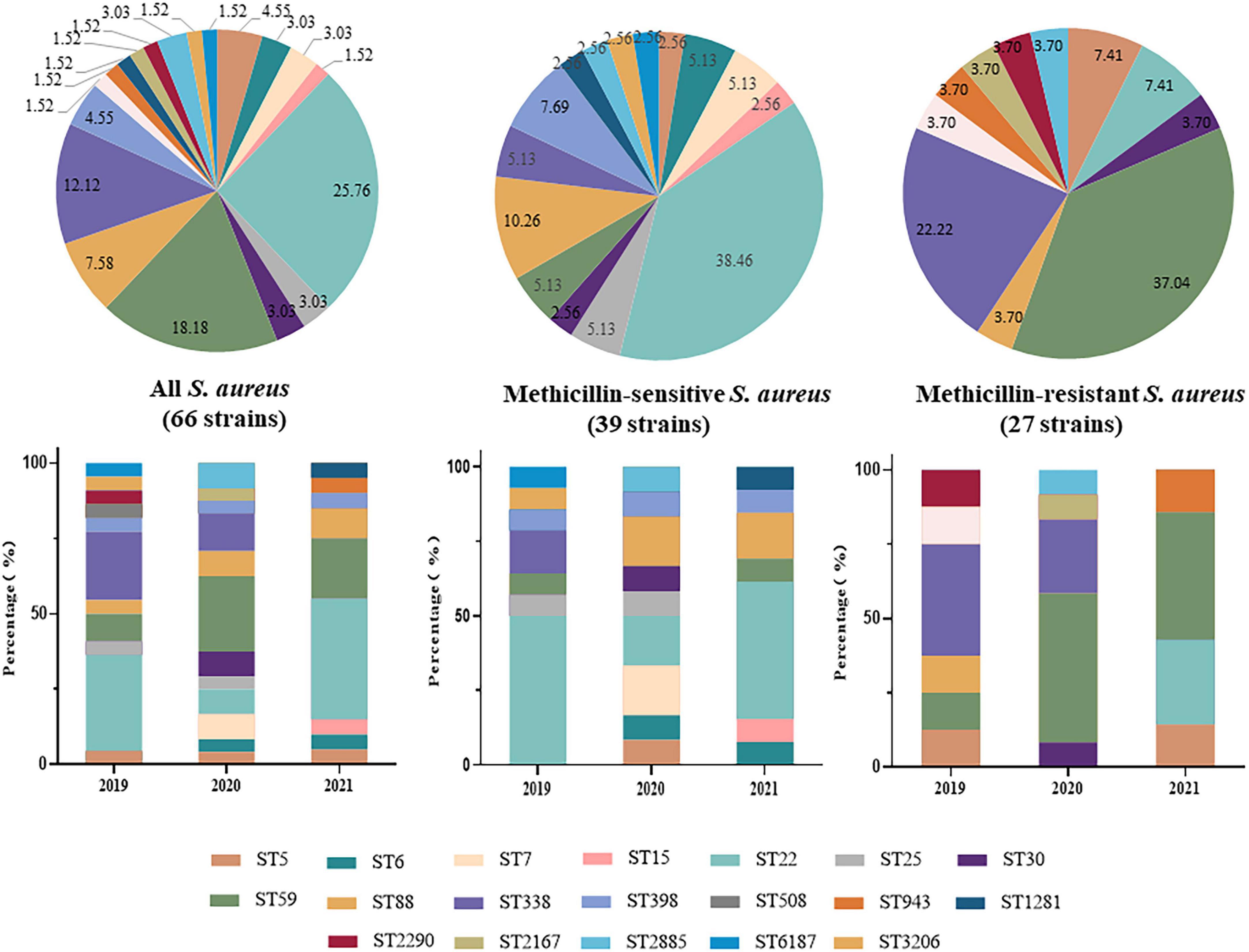
Figure 1. Distribution and changes in Sequence Types (STs) of 66 invasive Staphylococcus aureus strains from 2019 to 2021.
Spa typing of strains
Spa typing failed for three S. aureus strains and 31 spa types were identified among the remaining 63 S. aureus strains. Of these, t309 (24.24%, 16/66) was the dominant spa type, followed by t437 (22.73%, 15/66), and the remaining types were only found in one or two strains. Given the MLST results, a strong correlation was found between the dominant spa types and ST types, t309 correlated with ST22 (88.24%, 15/17), and t437 correlated with ST59 (75.00%, 9/12) and ST338 (75.00%, 6/8). A correlation was not identified between the remaining spa and ST types because of the small number of strains. Thus, the most common S. aureus type in this study was ST22-t309 (22.73%, 15/66), followed by ST59-t437 (13.64%, 9/66) and ST338-t437 (9.09%, 6/66). In MSSA strains, the dominant type was ST22-t309 (33.33%, 13/39), while in MRSA strains, the dominant types were ST59-t437 (33.33%, 9/27) and ST338-t437 (14.81%, 4/27) (Table 2).
Staphylococcal cassette chromosome mec typing of methicillin-resistant Staphylococcus aureus strains
Three SCCmec types were detected in MRSA strains, SCCmecIV (74.07%, 20/27), SCCmecIII (18.52%, 5/27) and SCCmecV (7.41%, 2/27). MLST, spa, and SCCmec typing showed that the major MRSA molecular types in present study were ST59-t437-SCCmecIV (25.93% 7/27) and ST338-t437-III (11.11%, 3/27) (Table 2).
Analysis of antimicrobial resistance
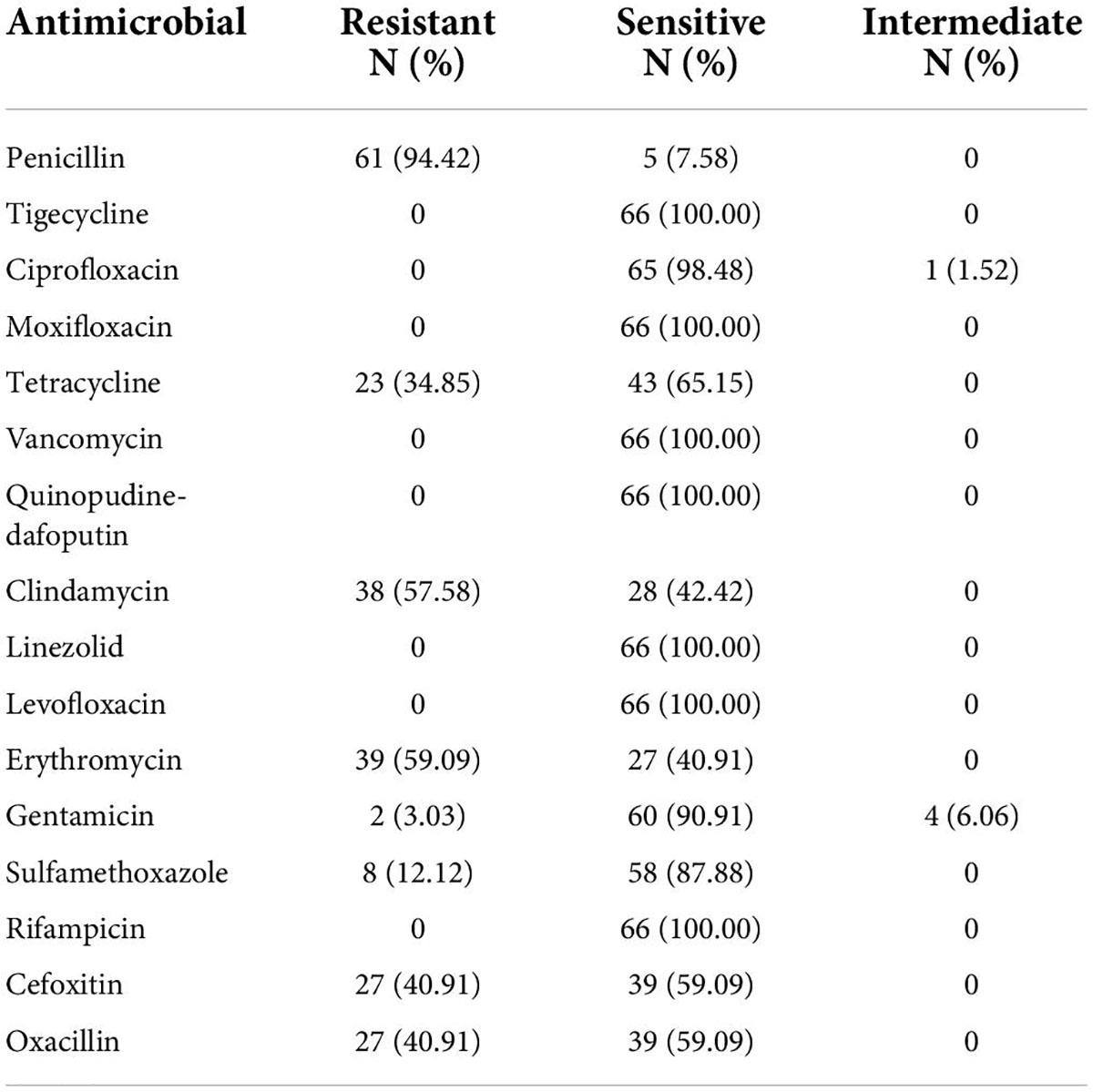
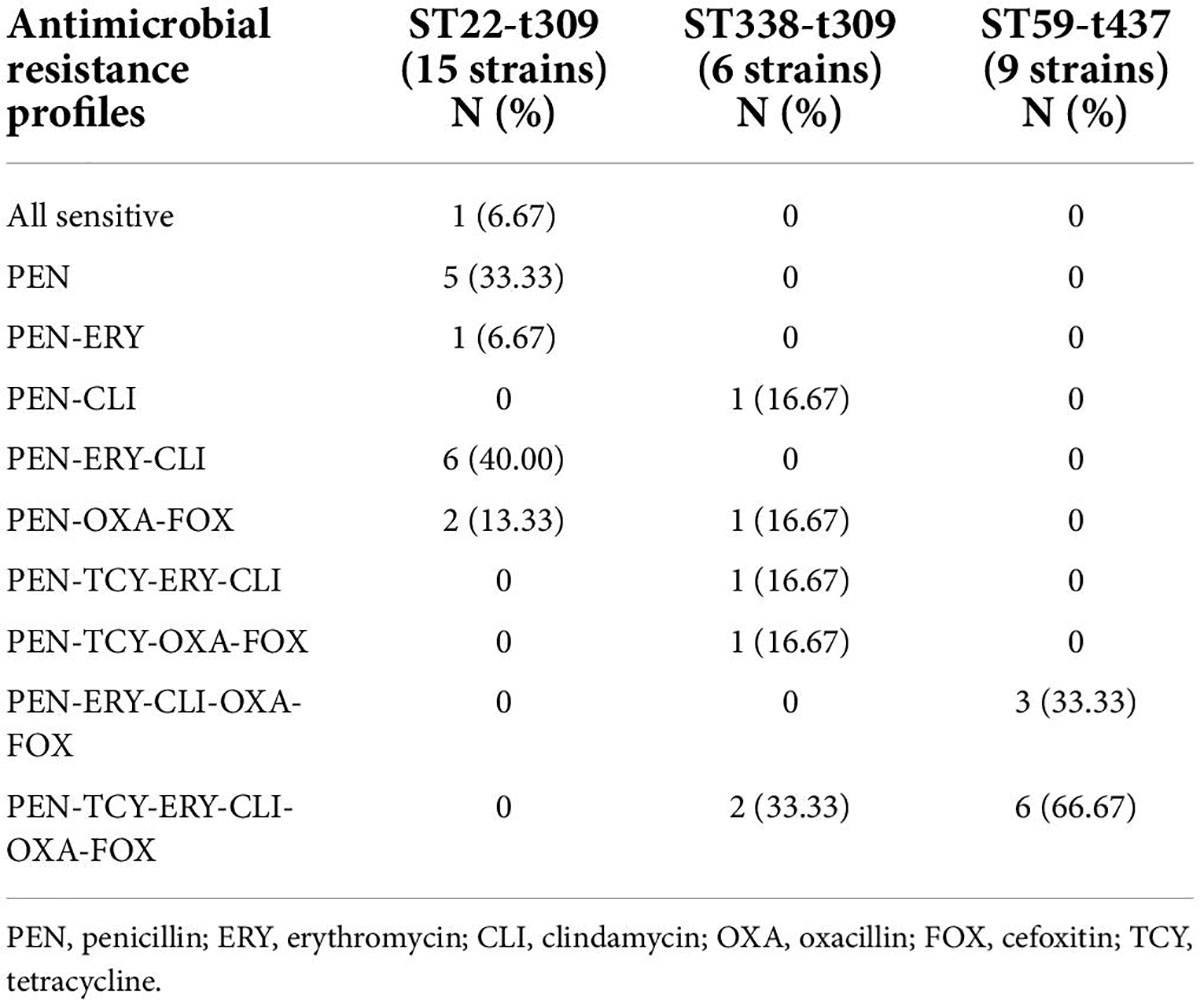
VITEK 2 Compact testing showed that 66 S. aureus strains were susceptible to TJH, CIP, MFX, VAN, QDA, LNZ, LVX, and RIF, and that the resistance rates to PEN, ERY, CLI, OXA, FOX, TCY, SXT, and GEN were 92.42, 59.09, 57.58, 40.91, 40.91, 34.85, 12.12, and 3.03%, respectively (Table 3). Further analysis showed that the resistance rates to PEN and SXT initially decreased and then increased from 2019 to 2021, a trend that contrasted with the resistance rates to OXA, FOX, ERY, CLI, and TCY (Figure 2). And there were differences in antimicrobial resistance rates among different ST-type strains, for example, ST59 was significantly more resistant to PEN, TCY, ERY, CLI, OXA, and FOX than ST22 and ST338 (Figure 3). In this study, S. aureus had 16 different antimicrobial resistance profiles, the most common being resistance to PEN-TCY-ERY-CLI-OXA-FOX (10/66, 15.15%), followed by resistance to PEN only and PEN-ERY-CLI (9/66, 13.64% for each profile) (Table 4). There were some differences in the antimicrobial resistance profiles by year. The most common antimicrobial resistance profiles in 2019 and 2021 were resistance to PEN only and PEN-ERY-CLI, while in 2020 it was resistance to PEN-TCY-ERY-CLI-OXA-FOX (Table 4).
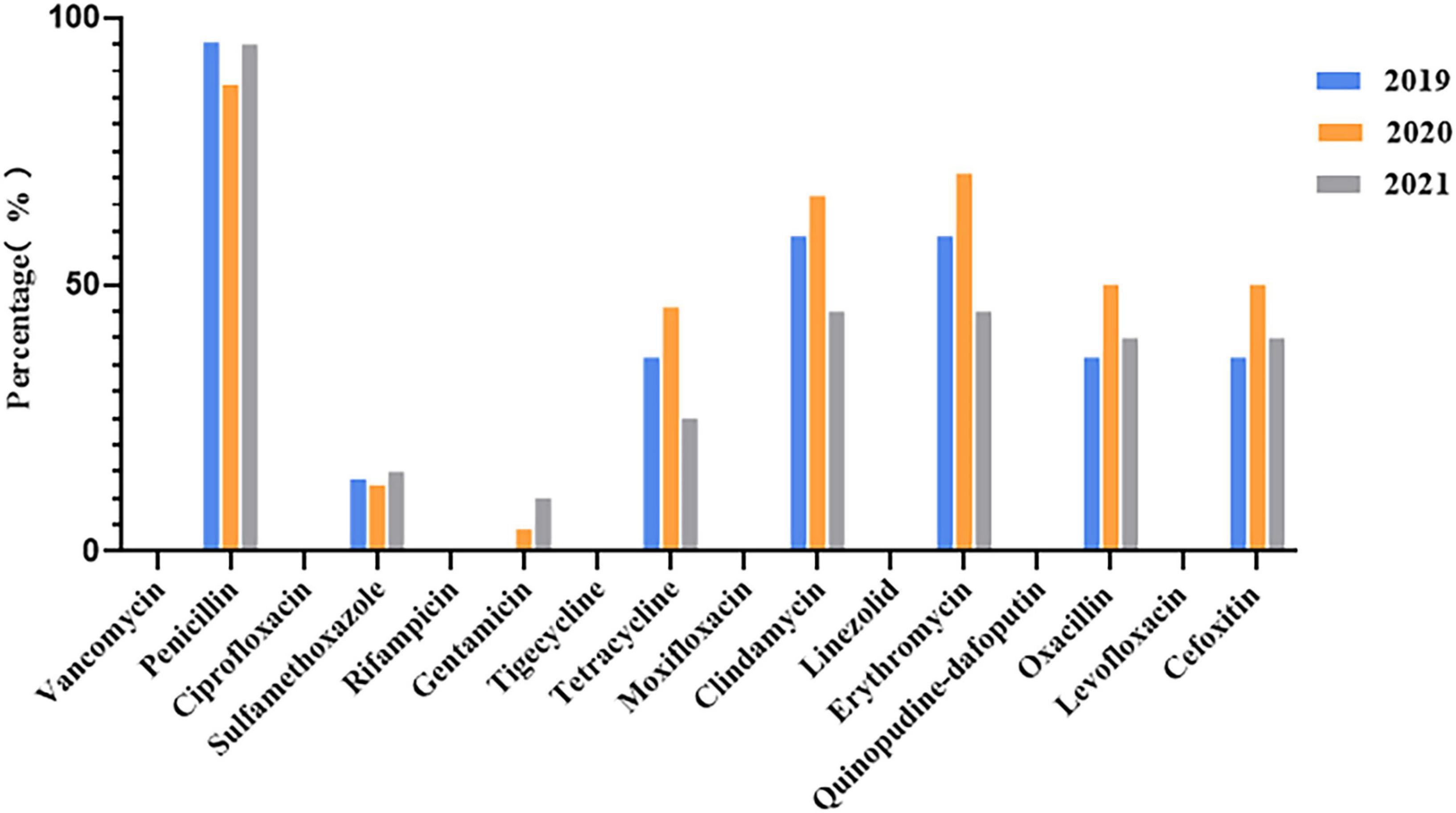
Figure 2. Changes in antimicrobial resistance rates of 66 invasive Staphylococcus aureus strains from 2019 to 2021.
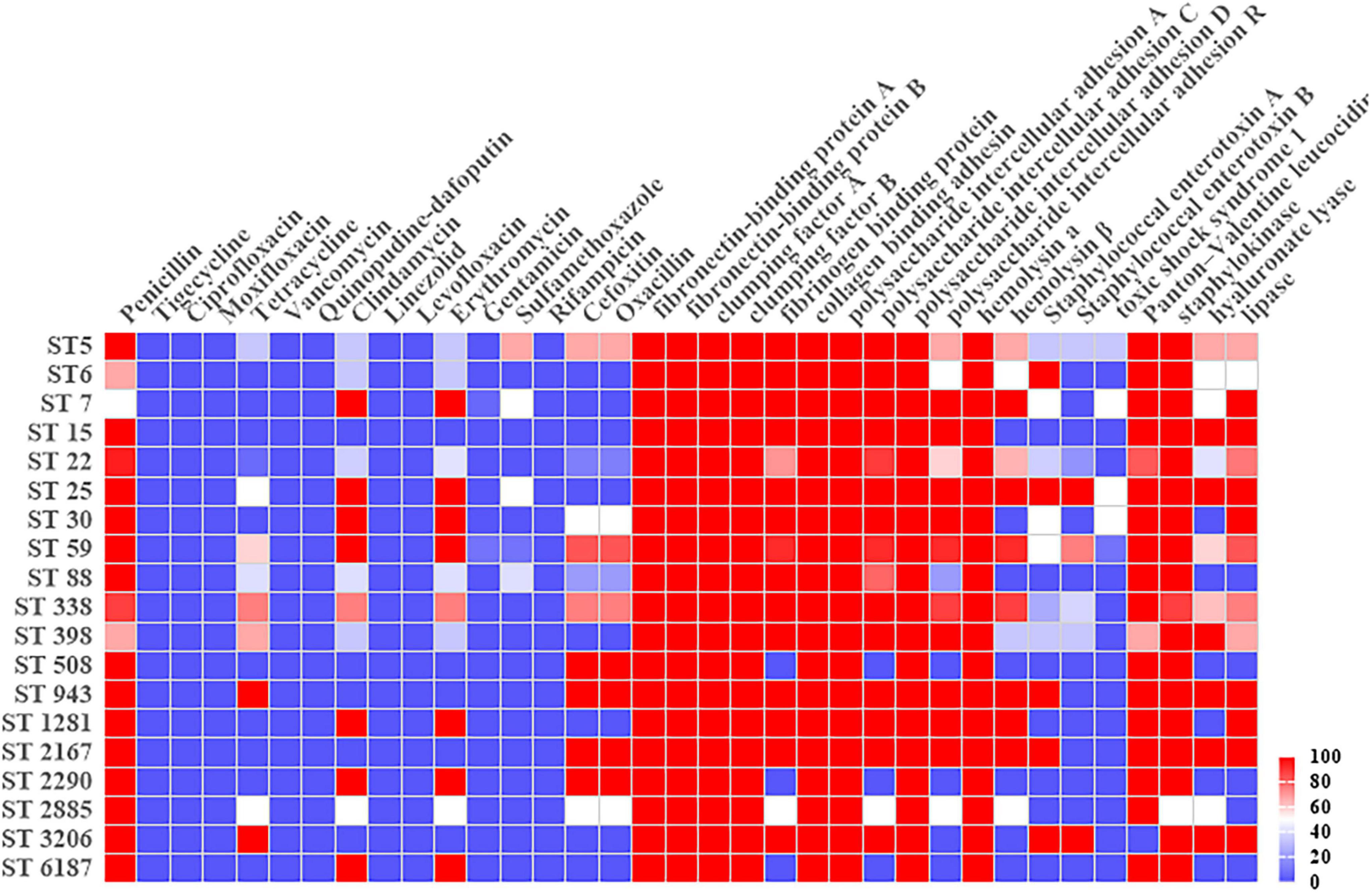
Figure 3. Heatmap of antimicrobial resistance rates and virulence gene carrier rates in different Sequence-Type Staphylococcus aureus strains.

Table 4. Antimicrobial resistance profiles of invasive Staphylococcus aureus strains from 2019 to 2021.
Detection of virulence genes
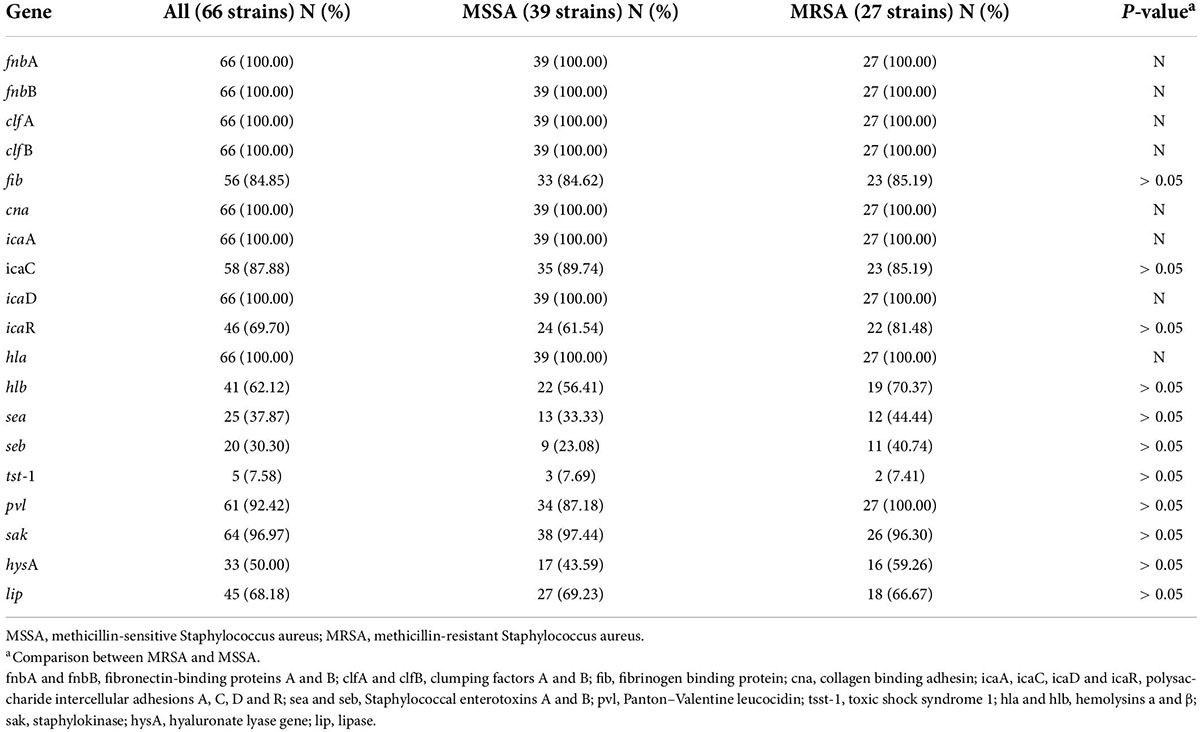
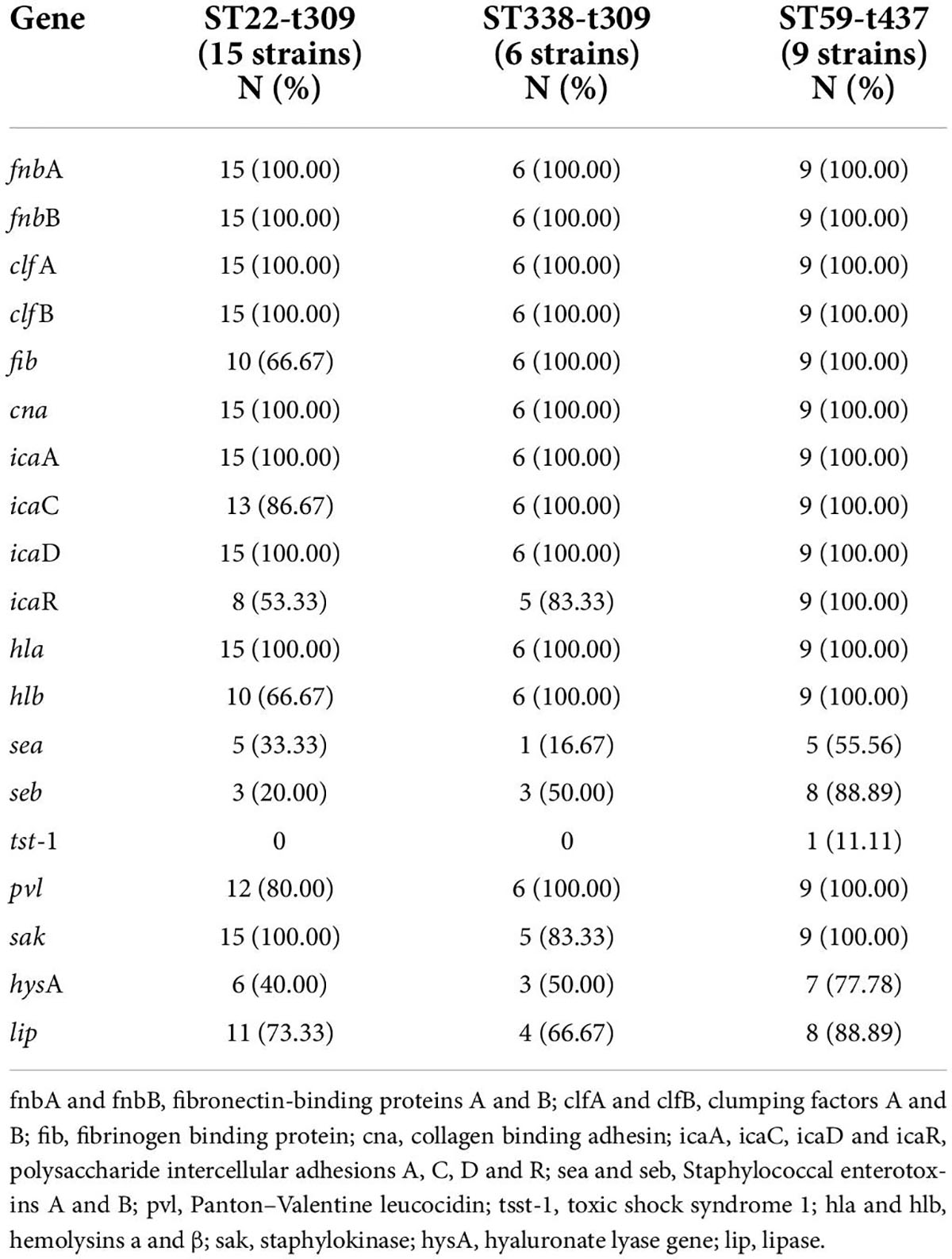
The carrier rates for common S. aureus virulence genes, icaA, icaD, fnbA, fnbB, clfA, clfB, hla, and cna, were 100.00%. sak, pvl, icaC, icaR, fib, lip, hlb, and hysA had carrier rates of 96.97, 92.42, 87.88, 69.70, 84.85, 62.12, 56.06, and 50%, respectively, while sea, seb, and tsst-1 had carrier rates of 37.87, 30.30, and 7.58%, respectively. The distribution of virulence genes in MRSA and MSSA strains did not differ significantly, but there were some differences in ST type strains, particularly for icaR, hlb, sea, seb, hysA, and lip (Table 5 and Figure 3).
Comparison of the characteristics in the dominant strains ST22-t309, ST338-t437, and ST59-t437
Antimicrobial resistance rates and profiles
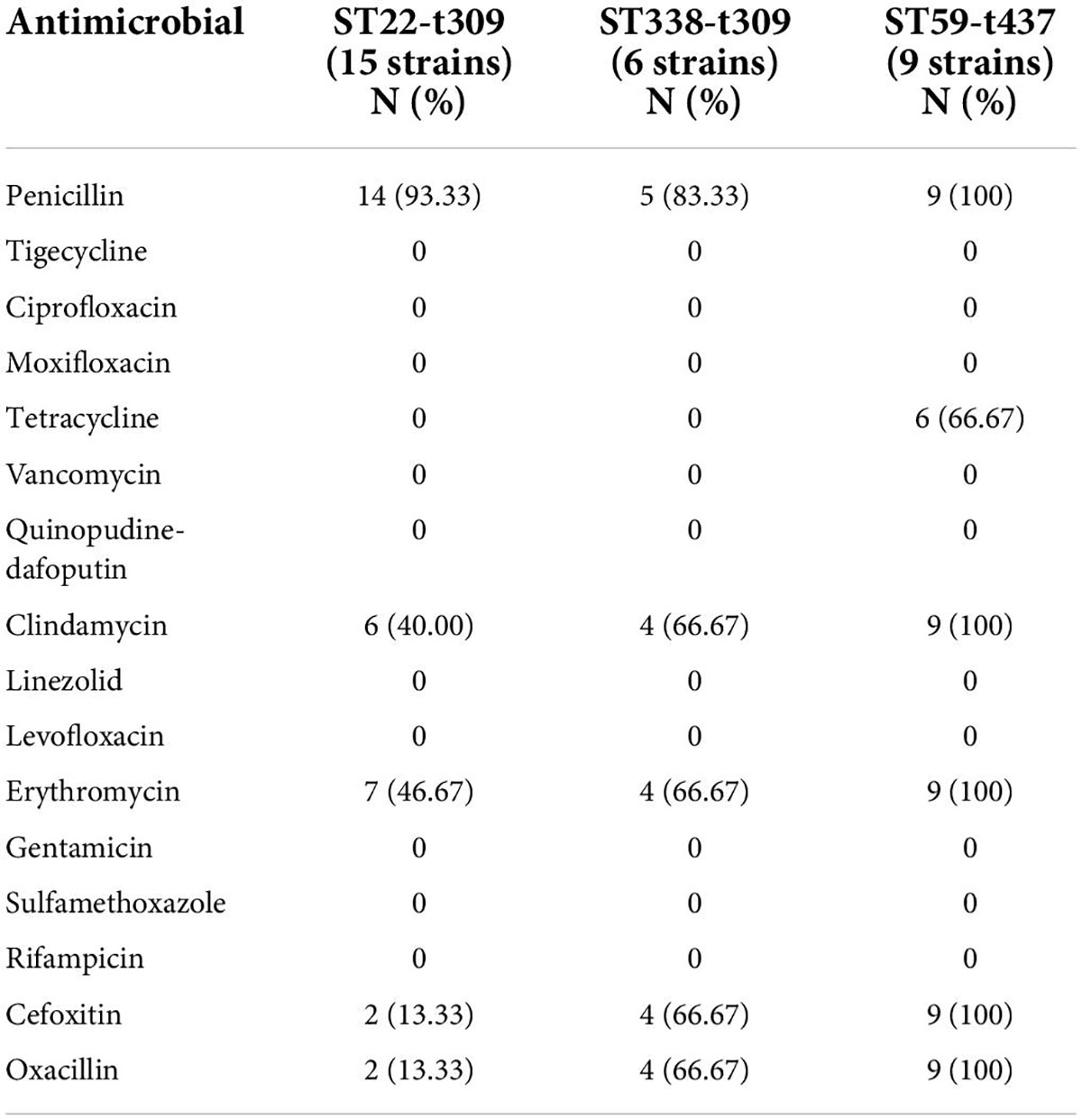
Except for PEN, ST338-t437 strains had higher resistance rates to CLI, ERY, OXA, and FOX than ST22-t309 strains. ST59-t437 strains were resistant to PEN, ERY, CLI, OXA, and FOX, with a TCY resistance rate of 66.67%. MRSA was found in only 13.33% of ST22-t309 strains, but in 66.67% of ST338-t437 strains and 100.00% of ST59-t437 strains, respectively (Table 6). Resistance to PEN-ERY-CLI and PEN alone was the most common resistance profile of the ST22-t309 strains, while resistance to PEN-TCY-ERY-CLI-OXA-FOX was the most common resistance profile of the ST338-t437 and ST59-t437 strains (Table 7).
Virulence genes
In the three dominant strains, fnbA, fnbB, cna, icaA, icaD, and hla had 100.00% carrier rates. While the carrier rates of fib, icaC, hlb, and pvl in the ST59-t437 and ST338-t437 strains were 100.00%, which were significantly greater than the carrier rates in the ST22-t309 strains. The carrier rates of icaR, seb and hysA were highest in the ST59-t437 strains, followed by the ST338-t437 and ST22-t309 strains (Table 8).
Discussion
This study collected 66 invasive S. aureus strains at Kunming Children’s Hospital from 2019 to 2021 and analyzed them for molecular typing, antimicrobial resistance, and virulence genes. The findings revealed a unique prevalence characteristic and change in invasive S. aureus strains found among children in this region, which differ from those reported from other countries and regions of China. Moreover, the molecular characteristics of prevalent strains in Kunming have changed significantly since the COVID-19 epidemic.
In this study, 66 children with invasive S. aureus infection were detected in three years (22, 24, and 20 in 2019, 2020, and 2021, respectively), showing little change in the annual number of cases. This suggests that the increased use of personal protection during the COVID-19 epidemic did not significantly reduce the incidence of this invasive disease, which is consistent with the findings of McNeil et al. (2021). Some studies have found that family members and household environment play a key role in S. aureus transmission (Mork et al., 2018; Mork et al., 2020). Therefore, increased time spent indoors and among household contacts may have contributed to the relatively stable incidence of invasive S. aureus infection during COVID-19 epidemic. The age of the children varied widely from 1 day to 14 years, however, 30.30% of the patients were < 1 year of age. In addition, invasive S. aureus infection was frequently seen among children with leukemia in present study. These findings suggest that such infections are likely to occur in infants and young children with underdeveloped immune systems or immunodeficiency. The patients in this study were primarily treated in the ICU and in the departments of orthopedics, and hematology, which is consistent with the characteristics of the diseases covered by these departments. Patients in the ICU and the department of orthopedics often receive invasive therapies such as surgery, invasive ventilation, gastric tube placement, and tracheal intubation or incision. Many children in the department of hematology are immunodeficient or are on tumor chemotherapy drugs so often have compromised immunity and may be more susceptible to invasive infections.
Data from the China Antimicrobial Surveillance Network3 indicates that the MRSA rate in China has dropped from 31.40% in 2019 to 30.00% in 2021, showing a decreasing annual trend. In this study, however, the average detection rate of MRSA was 40.91% from 2019 to 2021. And the detection rate of MRSA was also higher than the national average each year, which may be related to different regions or infection types. In addition, the MSSA rate (59.09%) in this study was much higher than that observed for MRSA (40.91%). This suggests that MSSA is the most common cause of invasive S. aureus infection among children in Kunming. While MRSA has gained considerable attention for its multi-drug resistance since its emergence in the 1980s, MSSA has been largely ignored, especially regarding invasive S. aureus infection. Some studies found that MSSA was more likely to cause bacteremia, endocarditis, and sepsis than MRSA and that there may be a “role reversal” between these two types of drug-resistant bacteria (Diep et al., 2011; Koeck et al., 2019). In China, current S. aureus-related infections are primarily caused by MSSA, so this bacterium should not be ignored in favor of focusing solely on MRSA. Resistance to TJH, VAN, QDA, LNZ, and RIF was not found in this study, so these antibiotics could be used as a clinical treatment for relevant infections.
In this study, the 66 S. aureus strains belonged to 19 ST types and 31 spa types, indicating that S. aureus strains in Kunming have a diverse genetic background. ST22, ST59, and ST338 were the top three ST types, accounting for up to 56.06% of all strains. ST22 and ST88 were the dominant MSSA ST types. ST59 and ST338 were the dominant MRSA ST types. Qiao et al. (2014) reported that ST88, ST25, and ST7 were the most common ST types of strains responsible for invasive CA-MSSA infection among Chinese children, and no ST22 strains were found, which differs substantially from the results of the current study. ST22, a highly transmissible strain originating from Europe, has caused several major outbreaks in Europe, India, Gaza Strip, and Singapore, and has become one of the main MRSA ST types worldwide (Tinelli et al., 2009; Manoharan et al., 2012; Hsu et al., 2015; Chang et al., 2018; Htun et al., 2018). In 2013, Li et al. (2013) first identified two ST22 strains in Shanghai, both of which belonged to MSSA. The detection rate of ST22 was low at the time, which may explain why this study’s results differed from Qiao’s. At present, ST22 becomes the dominant MSSA strain in several important Chinese cities such as Beijing, Chongqing, and Urumqi (Yang et al., 2017; Xiao et al., 2019; Yuan et al., 2019). According to the findings of this study, ST22 may have also become the predominant ST type of MSSA strains in Kunming. Unfortunately, current researches on invasive S. aureus infections in China have primarily focused on MRSA, and there have been no reports on the predominance of ST22. Many studies have found that ST59 is the predominant ST type among MRSA strains of invasive S. aureus infections in China (Qiao et al., 2013; Li et al., 2015), supporting the results of this study.
Previous studies have shown that virulence factors play an important role in severe S. aureus infections, and the types and numbers of S. aureus strains carrying virulence genes are closely related to their invasiveness and pathogenicity (Loughman et al., 2009; Jenkins et al., 2015). In this study, all strains carried the microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) genes, fnbA, fnbB, clfA, clfB, and fib. The proteins encoded by these genes play an important role in pathogen recognition and adhesion of host cells and extracellular matrix proteins, which are the first steps in disease development (Foster et al., 2014). In addition, the carrier rates of the key biofilm-forming genes, icaA, icaC, icaD, and icaR, were high in this study, suggesting that the strains were not only highly virulent but also had a strong ability to form biofilms. Biofilms formed by bacteria not only promote their survival in unfavorable environments but also allow them to acquire drug resistance (Omidi et al., 2020). This may explain the ability of these strains to survive and cause disease during the COVID-19 epidemic. Analysis of several other common S. aureus virulence genes showed that the hemolysin genes, hla and hlb, had the highest carrier rates (62.12–100.00%). These genes encode hemolysin, a factor that destroys multiple host cell types, promotes inflammation and causes S. aureus pneumonia and sepsis in clinical practice. It is also worth noting that the pvl gene carrier rate in this study was 92.42%. Several studies have shown that pvl has a high carrier rate in ST22 strains (Yang et al., 2017, Yuan et al., 2019), and in our study, it was as high as 82.35% in ST22 strains. Previous studies have shown the association between PVL and severe invasive infections (Yonezawa et al., 2015; Rajova et al., 2016). Furthermore, PVL is shown to play an important role in the development of pneumonia, and pvl gene expression level is positively correlated with pneumonia severity, which may explain why almost half of the patients in this study developed this complication. The three extracellular enzyme genes, sak, hysA, and lip, also had a higher carrying rates in this study (96.97, 50.00, and 68.18%, respectively). Therefore, invasive S. aureus infection results from the combination of multiple S. aureus toxins and extracellular enzymes. Further analysis showed no apparent differences in the carrier rates of these genes between MSSA and MRSA strains, suggesting that the virulence of MSSA and MRSA strains responsible for invasive infection in children is comparable.
The molecular types of S. aureus by geographical region vary over time, a finding also demonstrated in this study. The dominant S. aureus strain responsible for invasive infection in children from Kunming was ST22-t309 in 2019 and ST59-t437 in 2020. However, the dominant S. aureus strain returned to ST22-t309 in 2021. At the same time, dominant MRSA strain was replaced from ST338-t309 to ST59-t437. Thus, the dominant S. aureus and MRSA strains responsible for invasive infection in this region have changed over time. This could have resulted from (i) natural changes in prevalent strains or (ii) To prevent COVID-19 infection and transmission since the end of 2019, various prevention and control measures were taken. Local citizens were required to wear a mask, home quarantine was enforced, personal hygiene was stressed, and individual lifestyles changed dramatically. These alterations may have driven changes in the prevalent S. aureus strain type. Additional analysis of the antimicrobial resistance and virulence of the three dominant types showed that ST59-t437 had a stronger resistance to common antibiotics, a broader antimicrobial resistance profile, and higher carrying rates of various virulence genes than ST22-t309 and ST338-t437. These characteristics may have contributed to the survival and transmission of the ST59-t437 strain during the COVID-19 epidemic, especially when it was worst in 2020, allowing ST59-t437 to become the absolute dominant strain. In contrast, before the COVID-19 epidemic in 2019, and once the COVID-19 epidemic in China had been obviously mitigated and personal protection was reduced in 2021, ST22-t309 became the major prevalent strain.
There are limitations to this study which should be acknowledged. Foremost, our results are from a single center, which may affect the representativeness of the samples. In addition, the sample size was small and type II errors could not be overcome. Future multi-center, large-sample studies will be required to confirm the findings of the present study.
In summary, our results showed that the annual number of invasive S. aureus strains isolated from children in Kunming remained stable during 2019–2021. The dominant molecular types of strains were ST22-t309 and ST59-t437, and with diverse genetic backgrounds. The molecular characteristics and antimicrobial resistance profiles of prevalent S. aureus strains changed significantly during the COVID-19 epidemic. Thus, COVID-19 prevention and control should be supplemented by surveillance of common clinical pathogens, particularly vigilance against the prevalence of multidrug-resistant and high-virulence strains.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
MM and TD conceived and designed the experiments. YL and LT contributed to the molecular experiment and analyses of the data. XL, JL, and HW contributed to the identification of strains and antibiotic-susceptibility testing. HJ, JD, and DH contributed to collection of strains and data. All authors contributed to drafting the manuscript, critically revising the manuscript, and approved the final version of the manuscript.
Funding
This project was supported by the National Natural Science Foundation of China (No. 81960294), Science and Technology Planning Project of Yunnan province (202201AT070280 and 202101AY070001-216), “Spring City Plan” Special Young Talent Project, Top Experts training Project for the Academy and Technology in Yunnan province (202005AC160066), Experts training Project for the Academy and Technology in Kunming City (KMRCD-2021019), Top young Experts Project in Kunming (C201914013), and Health Science and Technology Personnel Training Foundation of Kunming City [2020-SW(reserve personnel)-109 and 2021-SW(reserve personnel)-72].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Chang, Q., Abuelaish, I., Biber, A., Jaber, H., Callendrello, A., Andam, C. P., et al. (2018). Genomic epidemiology of meticillin-resistant Staphylococcus aureus ST22 widespread in communities of the Gaza Strip, 2009. Euro Surveill. 23:1700592. doi: 10.2807/1560-7917.ES.2018.23.34.1700592
CLSI (2019). Performance Standards For Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standards Institute.
Diep, B. A., David, M. Z., Boyle-Vavra, S., Zychowski, D. L., and Daum, R. S. (2011). Methicillin-Susceptible Staphylococcus aureus as a Predominantly Healthcare-Associated Pathogen: A Possible Reversal of Roles? PLoS One 6:e18217. doi: 10.1371/journal.pone.0018217
Foster, T. J., Geoghegan, J. A., Ganesh, V. K., and Hook, M. (2014). Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62. doi: 10.1038/nrmicro3161
Grundmann, H., Aanensen, D. M., van den Wijngaard, C. C., Spratt, B. G., Harmsen, D., Friedrich, A. W., et al. (2010). Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: A molecular-epidemiological analysis. PLoS Med. 7:e1000215. doi: 10.1371/journal.pmed.1000215
He, J., Chen, H., Wang, G., Wang, Y., Wan, W., Guo, X., et al. (2015). Investigation of biofilm formation and biofilm related genes in Staphylococcus aureus. Jiangsu Agric. Sci. 43, 55–59.
Hsu, L. Y., Harris, S. R., Chlebowicz, M. A., Lindsay, J. A., Koh, T. H., Krishnan, P., et al. (2015). Evolutionary dynamics of methicillin-resistant Staphylococcus aureus within a healthcare system. Genome Biol. 16:81. doi: 10.1186/s13059-015-0643-z
Htun, H. L., Kyaw, W. M., de Sessions, P. F., Low, L., Hibberd, M. L., Chow, A., et al. (2018). Methicillin-resistant Staphylococcus aureus colonisation: Epidemiological and molecular characteristics in an acute-care tertiary hospital in Singapore. Epidemiol. Infect. 146, 1785–1792. doi: 10.1017/S0950268818001966
Iwamoto, M., Mu, Y., Lynfield, R., Bulens, S. N., Nadle, J., Aragon, D., et al. (2013). Trends in invasive methicillin-resistant Staphylococcus aureus infections. Pediatrics 132:e817–24. doi: 10.1542/peds.2013-1112
Jenkins, A., Diep, B. A., Mai, T. T., Vo, N. H., Warrener, P., Suzich, J., et al. (2015). Differential expression and roles of Staphylococcus aureus virulence determinants during colonization and disease. mBio 6:e2272–e2214. doi: 10.1128/mBio.02272-14
Koeck, M., Como-Sabetti, K., Boxrud, D., Dobbins, G., Glennen, A., Anacker, M., et al. (2019). Burdens of Invasive Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus Disease, Minnesota, USA. Emerg. Infect. Dis. 25, 171–174. doi: 10.3201/eid2501.181146
Koreen, L., Ramaswamy, S. V., Graviss, E. A., Naidich, S., Musser, J. M., and Kreiswirth, B. N. (2004). spa typing method for discriminating among Staphylococcus aureus isolates: Implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42, 792–799. doi: 10.1128/JCM.42.2.792-799.2004
Li, T., Song, Y., Zhu, Y., Du, X., and Li, M. (2013). Current status of Staphylococcus aureus infection in a central teaching hospital in Shanghai, China. BMC Microbiol. 13:153–163. doi: 10.1186/1471-2180-13-153
Li, W., Dong, Y., Ning, X., Zeng, T., Yu, S., Yao, K., et al. (2015). Clinical and molecular characteristics of invasive community-asociated Staphylococus aureus infections in Chinese neonates. Chin. J. Pract. Pediatr. 30, 1692–1696. doi: 10.3760/cma.j.issn.2095-428X.2015.22.004
Loughman, J. A., Fritz, S. A., Storch, G. A., and Hunstad, D. A. (2009). Virulence gene expression in human community-acquired Staphylococcus aureus infection. J. Infect. Dis. 199, 294–301. doi: 10.1086/595982
Magill, S. S., O’Leary, E., Janelle, S. J., Thompson, D. L., Dumyati, G., Nadle, J., et al. (2018). Changes in Prevalence of Health Care-Associated Infections in U.S. Hospitals. N. Engl. J. Med. 379, 1732–1744. doi: 10.1056/NEJMoa1801550
Manoharan, A., Zhang, L., Poojary, A., Bhandarkar, L., Koppikar, G., and Robinson, D. A. (2012). An outbreak of post-partum breast abscesses in Mumbai, India caused by ST22-MRSA-IV: Genetic characteristics and epidemiological implications. Epidemiol. Infect. 140, 1809–1812. doi: 10.1017/S0950268812000593
McNeil, J. C., Flores, A. R., Kaplan, S. L., and Hulten, K. G. (2021). The Indirect Impact of the SARS-CoV-2 Pandemic on Invasive Group a Streptococcus, Streptococcus Pneumoniae and Staphylococcus Aureus Infections in Houston Area Children. Pediatr. Infect. Dis. J. 40:e313–e316. doi: 10.1097/INF.0000000000003195
Mork, R. L., Hogan, P. G., Muenks, C. E., Boyle, M. G., Thompson, R. M., Morelli, J. J., et al. (2018). Comprehensive modeling reveals proximity, seasonality, and hygiene practices as key determinants of MRSA colonization in exposed households. Pediatr. Res. 84, 668–676. doi: 10.1038/s41390-018-0113-x
Mork, R. L., Hogan, P. G., Muenks, C. E., Boyle, M. G., Thompson, R. M., Sullivan, M. L., et al. (2020). Longitudinal, strain-specific Staphylococcus aureus introduction and transmission events in households of children with community-associated meticillin-resistant S aureus skin and soft tissue infection: A prospective cohort study. Lancet Infect. Dis. 20, 188–198. doi: 10.1016/s1473-3099(19)30570-5
O’Hara, F. P., Amrine-Madsen, H., Mera, R. M., Brown, M. L., Close, N. M., Suaya, J. A., et al. (2012). Molecular characterization of Staphylococcus aureus in the United States 2004-2008 reveals the rapid expansion of USA300 among inpatients and outpatients. Microb. Drug Resist. 18, 555–561. doi: 10.1089/mdr.2012.0056
Omidi, M., Firoozeh, F., Saffari, M., Sedaghat, H., Zibaei, M., and Khaledi, A. (2020). Ability of biofilm production and molecular analysis of spa and ica genes among clinical isolates of methicillin-resistant Staphylococcus aureus. BMC Res. Notes 13:19. doi: 10.1186/s13104-020-4885-9
Ondusko, D. S., and Nolt, D. (2018). Staphylococcus aureus. Pediatr. Rev. 39, 287–298. doi: 10.1542/pir.2017-0224
Qiao, Y., Dong, F., Song, W., Wang, L., Yang, Y., and Shen, X. (2013). Hospital- and community-associated methicillin-resistant Staphylococcus aureus: A 6-year surveillance study of invasive infections in Chinese children. Acta Paediatr. 102, 1081–1086. doi: 10.1111/apa.12386
Qiao, Y., Ning, X., Chen, Q., Zhao, R., Song, W., Zheng, Y., et al. (2014). Clinical and molecular characteristics of invasive community-acquired Staphylococcus aureus infections in Chinese children. BMC Infect. Dis. 14:582–591. doi: 10.1186/s12879-014-0582-4
Qu, W. (2019). Study On Drug Resistance And Virulence Factors Of Staphylococcu Aureus Isolated From Clinical Infected Patients. Tianjin: Tianjin Medical University.
Rajova, J., Pantucek, R., Petras, P., Varbanovova, I., Maslanova, I., and Benes, J. (2016). Necrotizing pneumonia due to clonally diverse Staphylococcus aureus strains producing Panton-Valentine leukocidin: The Czech experience. Epidemiol. Infect. 144, 507–515. doi: 10.1017/S0950268815001521
Stegger, M., Andersen, P. S., Kearns, A., Pichon, B., Holmes, M. A., Edwards, G., et al. (2012). Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251). Clin. Microbiol. Infect. 18, 395–400. doi: 10.1111/j.1469-0691.2011.03715.x
Tinelli, M., Monaco, M., Vimercati, M., Ceraminiello, A., and Pantosti, A. (2009). Methicillin-susceptible Staphylococcus aureus in skin and soft tissue infections, Northern Italy. Emerg. Infect. Dis. 15, 250–257. doi: 10.3201/eid1502.080010
Wang, X., Chen, Q., Chen, L., Pan, J., Ke, J., and Li, L. (2021b). Pathogenic spectrum of acute respiratory infections in children during the COVID-19 epidemic. Jiangxi Med. J. 56, 2250–2224. doi: 10.3969/j.issn.1006-2238.2021.12.032
Wang, X., Lin, D., Huang, Z., Zhang, J., Xie, W., Liu, P., et al. (2021a). Clonality, virulence genes, and antibiotic resistance of Staphylococcus aureus isolated from blood in Shandong, China. BMC Microbiol. 21:281. doi: 10.1186/s12866-021-02344-6
Williamson, D. A., Ritchie, S. R., Roberts, S. A., Coombs, G. W., Thomas, M. G., Hannaford, O., et al. (2014). Clinical and molecular epidemiology of community-onset invasive Staphylococcus aureus infection in New Zealand children. Epidemiol. Infect. 142, 1713–1721. doi: 10.1017/S0950268814000053
Wu, Y., Meng, Q., Qiao, J., Li, J., Cai, K., Wang, D., et al. (2019). Analysis of biofilm formation, related genes distribution and transcription level in Xinjiang isolates of Staphylococcus aureus from cows. J. South. Agric. 50, 1829–1835. doi: 10.3969/j.issn.2095-1191.2019.08.25
Xiao, N., Yang, J., Duan, N., Lu, B., and Wang, L. (2019). Community-associated Staphylococcus aureus PVL(+) ST22 predominates in skin and soft tissue infections in Beijing, China. Infect. Drug Resist. 12, 2495–2503. doi: 10.2147/IDR.S212358
Yang, Y., Hu, Z., Shang, W., Hu, Q., Zhu, J., Yang, J., et al. (2017). Molecular and Phenotypic Characterization Revealed High Prevalence of Multidrug-Resistant Methicillin-Susceptible Staphylococcus aureus in Chongqing, Southwestern China. Microb. Drug Resist. 23, 241–246. doi: 10.1089/mdr.2016.0078
Yonezawa, R., Kuwana, T., Kawamura, K., and Inamo, Y. (2015). Invasive Community-Acquired Methicillin-Resistant Staphylococcus aureus in a Japanese Girl with Disseminating Multiple Organ Infection: A Case Report and Review of Japanese Pediatric Cases. Case Rep. Pediatr. 2015:291025. doi: 10.1155/2015/291025
Yuan, W., Liu, J., Zhan, Y., Wang, L., Jiang, Y., Zhang, Y., et al. (2019). Molecular typing revealed the emergence of pvl-positive sequence type 22 methicillin-susceptible Staphylococcus aureus in Urumqi, Northwestern China. Infect. Drug Resist. 12, 1719–1728. doi: 10.2147/IDR.S202906
Zhang, K., McClure, J. A., Elsayed, S., Louie, T., and Conly, J. M. (2005). Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43, 5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005
Keywords: Staphylococcus aureus, children, invasive infection, COVID-19, molecular characteristic, antimicrobial
Citation: Ma M, Tao L, Li X, Liang Y, Li J, Wang H, Jiang H, Dong J, Han D and Du T (2022) Changes in molecular characteristics and antimicrobial resistance of invasive Staphylococcus aureus infection strains isolated from children in Kunming, China during the COVID-19 epidemic. Front. Microbiol. 13:944078. doi: 10.3389/fmicb.2022.944078
Received: 14 May 2022; Accepted: 27 July 2022;
Published: 11 August 2022.
Edited by:
Tomislav Mestrovic, University North – University center Varaždin, CroatiaReviewed by:
Majid Sharifi-Rad, Zabol University, IranSilvia Spoto, Policlinico Universitario Campus Bio-Medico, Italy
Copyright © 2022 Ma, Tao, Li, Liang, Li, Wang, Jiang, Dong, Han and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tingyi Du, ZHV0aW5neWlAZXR5eS5jbg==
†These authors have contributed equally to this work
 Mingbiao Ma
Mingbiao Ma Lvyan Tao2,3,4†
Lvyan Tao2,3,4† Xinyue Li
Xinyue Li