- School of Life Science and Engineering, Foshan University, Foshan, China
Bartonella spp. are gram-negative bacteria that can infect a wide spectrum of mammals. Rodents are considered to be the natural reservoir of many Bartonella species that are transmitted by various blood-sucking arthropods. The close contact between rodents and humans in urban areas increased the chance of transmitting rodent-borne Bartonella to humans. Investigation of the epidemiological characteristics of Bartonella infection in rodents is of great significance for the prevention and control of human Bartonellosis. In this study, rodents were captured to monitor the prevalence of Bartonella in urban areas of Guangzhou city. Six official or candidate species of Bartonella, including two confirmed zoonotic species, were detected with an overall prevalence of 6.4% in rodents captured herein. In addition, Rattus norvegicus was the predominant host species for Bartonella infection, and B. queenslandensis was the dominant species circulating in rodents in these areas. These results provide insights into the prevalence and genetic diversity of Bartonella species circulating in rodents in the urban areas of Guangzhou, and also urged the surveillance of rodent-associated Bartonella species in these areas.
Introduction
Bartonella spp. are gram-negative bacteria that belong to the genus of α-proteobacteria within the family Bartonellaceae (Anderson and Neuman, 1997). They can infect a wide range of mammals and are mainly transmitted by blood-sucking arthropods, such as fleas, lice, sandflies, biting flies, and ticks (Álvarez-Fernández et al., 2018). More than 50 validated species of Bartonella have been identified from domestic and wild animals, including horses, cattle, deer, sheep, rodents, and bats (Chang et al., 2000; Valentine et al., 2007; Okaro et al., 2017). As far as we know, at least 18 species of Bartonella, including Bartonella alsatica, B. ancashensis, B. bacilliformis, B. clarridgeiae, B. doshiae, B. elizabethae, B. grahamii, B. henselae, B. koehlerae, B. kosoyi, B. mayotimonensis, B. quintana, B. rattimassiliensis, B. rochalimae, B. tamiae, B. tribocorum, B. vinsonii, and B. washoensis, have been recognized to be associated with human diseases (Gundi et al., 2004; Kosoy et al., 2010; Kandelaki et al., 2016; Vayssier-Taussat et al., 2016; Okaro et al., 2017; Von Loewenich et al., 2019). Infection of different species of Bartonella can cause human diseases with different clinical manifestations, sometimes even fatal in immunocompromised patients (Mosepele et al., 2012).
Rodents represent the most abundant taxonomic order with some special ecological traits, such as wide geographic distribution and intimate interactions with humans and livestock. Thus, rodents play key roles in the transmission of a large variety of pathogenic agents of infectious diseases, including Bartonella (Meerburg et al., 2009). Among all known reservoir hosts of Bartonella species, rodents are considered the primary host, and Bartonella spp. in rodents have been reported in Asia (Li et al., 2015; Saengsawang et al., 2021; Liu et al., 2022), Africa (Hatyoka et al., 2019), Europe (Divari et al., 2020; Szewczyk et al., 2021), Americas (De Sousa et al., 2018; Müller et al., 2020), and Australia (Dybing et al., 2016; Egan et al., 2021). Approximately 90 species of rodents are known to be associated with more than 22 different species of Bartonella, ten of which have been confirmed to cause human infection (Daly et al., 1993; Welch et al., 1999; Birtles et al., 2002; Kosoy et al., 2003, 2008, 2010; Lin et al., 2008; Kandelaki et al., 2016; Vayssier-Taussat et al., 2016). Interestingly, many different rodent species have been reported to be infected with different Bartonella spp. in high prevalence globally (Gutiérrez et al., 2015). The high genetic diversity and infection rate of Bartonella in rodents emphasized the important roles that rodents played in the maintenance and transmission of Bartonella.
A previous study showed high infection rates, ranging from 4 to 50%, of Bartonella in rodents in China (Gutiérrez et al., 2015). Importantly, several studies reported the high prevalence of Bartonella species in rodents in the western (Rao et al., 2021), eastern (Qin et al., 2019), southern (Su et al., 2020), southeastern (Liu et al., 2022), southwest (Li et al., 2007), and northeastern China (Li et al., 2015), indicating the wide geographical distribution of Bartonella in rodents. However, most of the previous studies were conducted in the field environment. Information about the epidemiology of Bartonella spp. in urban areas where rodents have more chance of contact with humans is limited. Guangzhou, the provincial capital city of Guangdong province and the largest city in southern China, is located in the tropical and subtropical regions with a warm and moist climate (Wei et al., 2018), an ideal habitat for rodents. However, little information about the epidemiology of Bartonella in rodents in urban areas of Guangzhou is available. Therefore, in this study, we collected rodent samples in urban areas of Guangzhou to investigate the epidemiology of Bartonella spp. in rodents. The results of this study will provide insights into the genetic diversity and prevalence of Bartonella in Guangzhou, and also formulate the prevention and control strategies for Bartonella infections in this city.
Materials and Methods
Rodent Samples: Collection and Processing
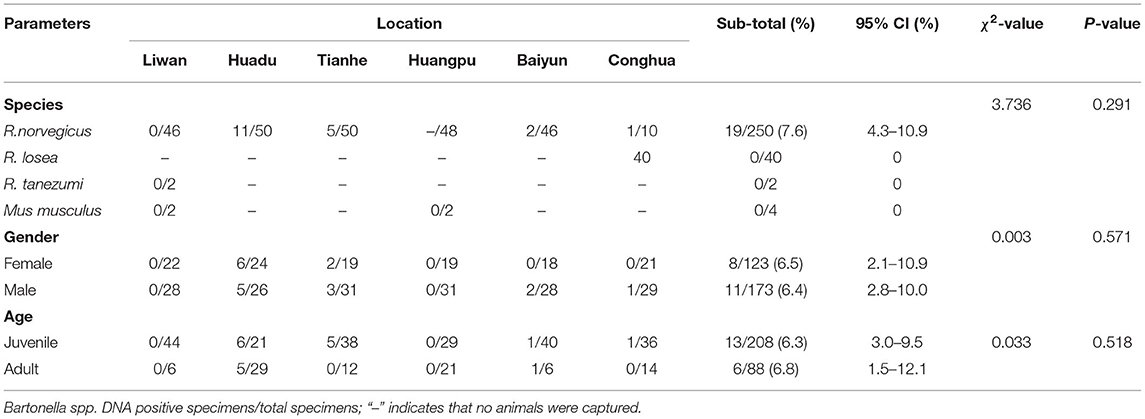
Rodents were captured using live-capture traps baited with cooked food in the urban areas of six districts (Liwan, Huadu, Tianhe, Huangpu, Baiyun, and Conghua) in Guangzhou city in 2020 (Figure 1). All rodents were euthanized, and the species, age, and sex were identified by trained field biologists, and the species were further confirmed by the sequence analysis of the mt-cyt b gene (Guo et al., 2013). Spleen samples were aseptically obtained immediately after euthanasia and stored at −80 °C until further use.
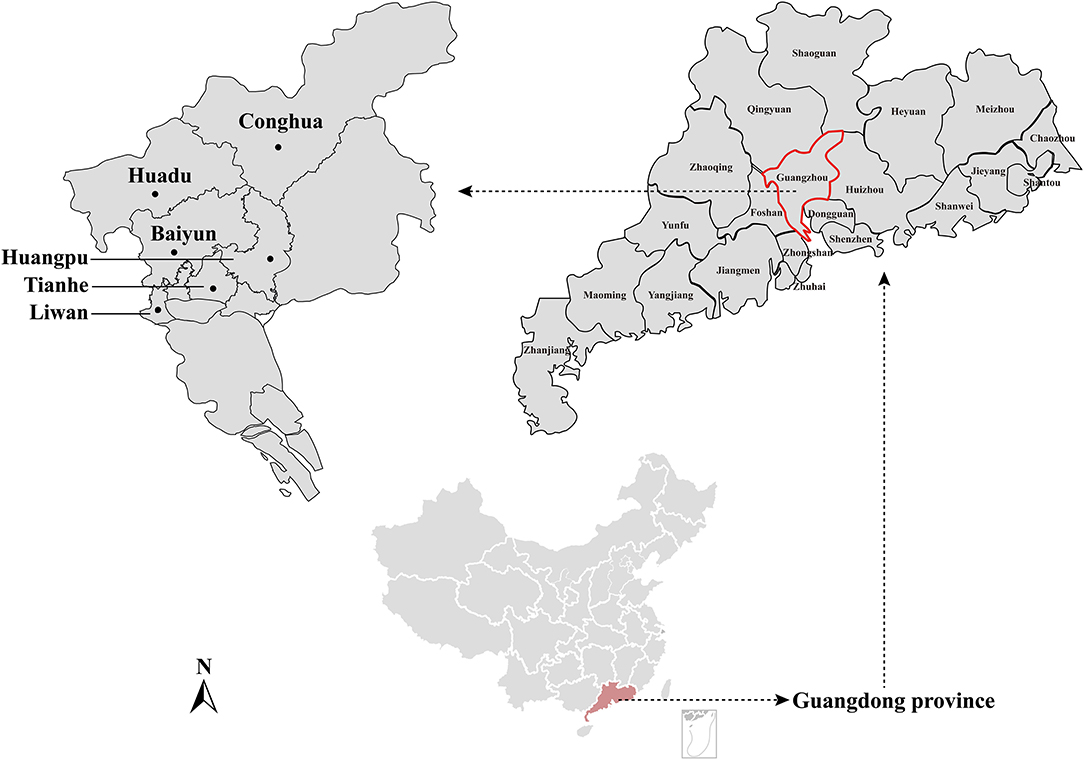
Figure 1. Geographic maps showing the location of sampling sites from where the rodents were captured in this study. This map was plotted by combination of Surfer software version-4 (Golden Software, USA) and Adobe illustrator version CC2017 (Adobe, USA). The black dots indicate the sampling regions in this study.
Approximately 20 mg of spleen samples were homogenized and further subjected to extract the total DNA using a DNA extraction kit (OMEGA, Doraville, CA, USA) as per the manufacturer's instructions. Then, all extracted DNA samples were used to detect the presence of Bartonella.
Detection and Molecular Characterization of Bartonella
Bartonella spp. were detected by nested-PCR using the primers targeting the conserved region of 16S rRNA (rrs) gene of Bartonella species as previously described (Zhang et al., 2019). The targeted region, about 1,100 bp fragment, was suspected to be a Bartonella-positive sample and was further confirmed by sequencing. To better determine and characterize the species of detected Bartonella strains, the citrate synthase gene (gltA) and the RNA polymerase beta-subunit gene (rpoB) were also amplified from the positive samples. All primer sequences used in the present study are listed in Table 1.
The PCR products with the expected size of each primer set were purified using a gel extraction kit (TaKaRa, Dalian, China) after electrophoresis. The purified DNA was cloned into a pMD19-T vector (TaKaRa, China), and the resulting plasmid was used to transform into competent E. coli cells. Positive inserts were confirmed by PCR, and five positive clones were sequenced by the Sangon Biotechnology Company (Shanghai, China). To prevent contamination, the procedures including extraction of total DNA, preparation of PCR mix, adding the template DNA, and agarose gel electrophoresis were performed in separate rooms using dedicated pipets and filtered tips. Distilled water was used as the negative control in all the PCR amplifications.
Sequence Comparison and Phylogenetic Analysis
Sequence assembly and manual editing were performed using the SeqMan program (DNASTAR, Madison, WI). The nucleotide (nt) sequence identities were calculated by the MegAlign program available within the Lasergene software package (version 7.1, DNAstar). All the sequences obtained in this study have been submitted to GenBank under the accession numbers ON413697–ON413715 for rrs gene and ON394008–ON394045 for gltA and rpoB genes, respectively.
The maximum-likelihood (ML) trees were constructed based on the general time-reversible (GTR) nucleotide substitution model and the optimized parameters of gamma (Γ)-distribution and proportion of invariable sites (i.e., GTR + Γ + I) with bootstrap support values calculated from 100 replicates implemented in MEGA X (Kumar et al., 2018).
Statistical Analysis
Statistical Package for Social Sciences (SPSS) Version 21.0 software was used in the statistical analyses of this study, and Fisher exact test was used to calculate the P-value to determine the differences in Bartonella positive rates between sampling sites and animal hosts. A P < 0.05 was considered statistically significant.
Ethics Statement
The authors confirmed that this study complies with the ethical policies of the journal, as specified in the journal's guidelines. All rodents were trapped by experienced field workers, and the sampling procedures and sample processing have been approved by the ethics committee of Foshan University.
Results
Rodent Trapping
A total of 296 rodents were captured in six districts of Guangzhou city in 2020 (Table 2). The captured rodents were identified into four species, including 250 Rattus norvegicus, 40 R. losea, 2 R. tanezumi, and 4 Mus musculus. R. norvegicus (250/296, 84.5%) was the most abundant species in urban environment in Guangzhou city. Among all rodents, the female and male were 123 and 173, respectively. In addition, the juvenile and adult rodents were 208 and 88, respectively.
Prevalence of Bartonella in Rodents
The total DNAs extracted from the rodents' spleen were subjected to PCR targeting the rrs gene of Bartonella spp., and PCR products with expected sizes were detected in 19 samples (Table 2). Sequencing and blast analysis of the PCR products confirmed that these 19 samples were Bartonella positive. Overall, the prevalence of Bartonella spp. in this study was 6.4% (19/296, 95% CI: 3.6–9.2%). Specifically, all Bartonella-positive samples were exclusively detected from R. norvegicus (7.6%, 95% CI: 4.3–10.9%). In addition, the infection rates of Bartonella in female and male rodents were 6.5% (95% CI: 2.1–10.9%) and 6.4% (95% CI: 2.8–10.0%), respectively. Moreover, 6.3% (95% CI: 3.0–9.5%) of the juvenile rodents and 6.8% (95% CI: 1.5–12.1%) of adult rodents were determined as Bartonella-positive. Notably, no significant difference was observed associated either with the rodent species or with the gender or age of the rodents.
Molecular Characterization of Bartonella
The rrs, gltA, and rpoB genes were recovered from positive samples to better characterize the species of detected Bartonella strains. Sequencing and blast analyses based on the rrs gene sequences (1,100 bp) revealed that Bartonella strains detected in this study were B. queenslandensis (n = 7), B. mastomydis (n = 5), B. tribocorum (n = 2), B. rattimassiliensis (n = 1), Bartonella sp. AA86HXZ (n = 3), and Bartonella sp. Fuji 12-1 (n = 1).
Sequence comparison analysis showed that all rrs gene sequences generated herein shared 98.2–100% nucleotide sequences identity with each other. The seven rrs gene sequences of B. queenslandensis obtained in the present study shared the highest sequence similarity with B. queenslandensis strain AUST/NH8 (EU111756), with 99.3–100% nucleotide sequences identity. Similarly, the rrs gene sequences of B. mastomydis, B. tribocorum, B. rattimassiliensis, Bartonella sp. AA86HXZ, and Bartonella sp. Fuji 12-1 obtained in the present study exhibited 99.5–99.9%, 99.8–100%, 100%, 99.3–99.6%, and 99.7% nucleotide sequences identical with the sequences of the corresponding species of Batonella under the GenBank accession number KY555064, NR074354, NR115255, KJ361602, and AB242293, respectively. Phylogenetic trees reconstructed based on the rrs gene sequences and the concatenated sequence of rrs, gltA, and rpoB genes showed a similar topology that these sequences determined in this study clustered together with the corresponding reference sequences, respectively (Figures 2, 3).
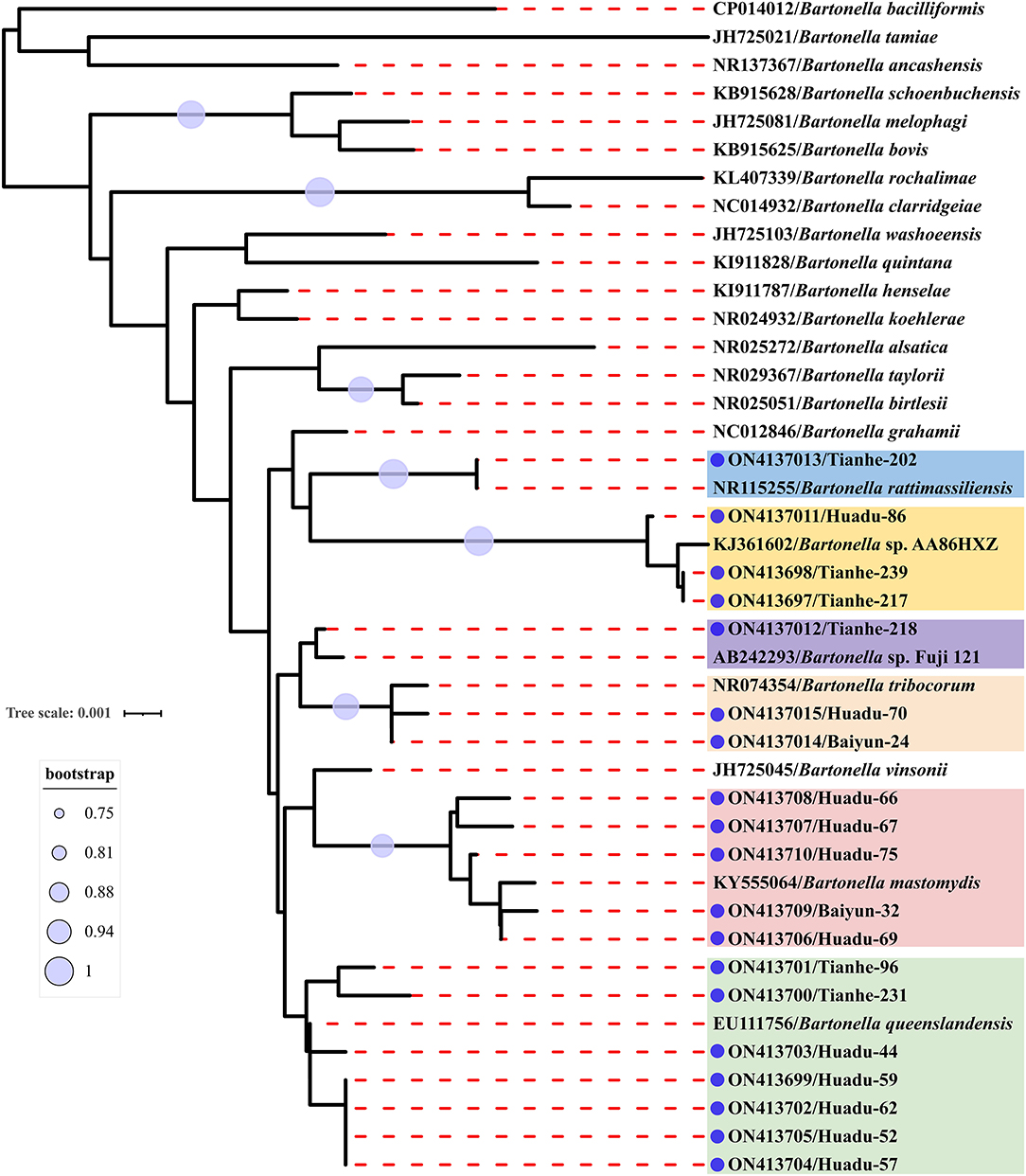
Figure 2. Maximum-likelihood phylogenetic tree reconstructed based on the nucleotide sequences of rrs gene of Bartonella. Bootstrap values were calculated with 100 replicates of the alignment, and only bootstrap values >70% are shown at appropriate nodes. Sequences of Bartonella determined in this study are marked with blue dot.
Figure 3. Maximum-likelihood phylogenetic tree reconstructed based on the concatenated nucleotide sequences of rrs, gltA, and rpoB genes of Bartonella. Bootstrap values were calculated with 100 replicates of the alignment, and only bootstrap values >70% are shown at appropriate nodes. Sequences of Bartonella determined in this study are marked with blue dot.
Discussion
A wide range of emerging and re-emerging infectious diseases have become major global threats to human health (Dong and Soong, 2021). Bartonella spp. are emerging vector-borne pathogens distributed worldwide and cause global public health concerns (Mogollon-Pasapera et al., 2009). Rodents are considered the major reservoirs that harbor the largest number and the greatest diversity of Bartonella species (Gutiérrez et al., 2015), and the presence of many pathogenic Bartonella species have been confirmed (Okaro et al., 2017). Therefore, investigation of the epidemiological characteristics of Bartonella infection in rodents is of great significance for the prevention and control of human Bartonellosis.
In the present study, rodents were captured in urban areas of Guangzhou city, and Bartonella was detected with an overall prevalence of 6.4%, which was lower than the results reported previously in the urban environments in Thailand (Saengsawang et al., 2021), Argentina (De Salvo et al., 2020), and Malaysia (Blasdell et al., 2019). In addition, the prevalence of Bartonella in our study was lower than those in other areas in China (Li et al., 2015; An et al., 2020; Rao et al., 2021; Liu et al., 2022). The observed differences in the prevalence of Bartonella in rodents may be related to rodent species, habitats, and arthropod vector populations (Meheretu et al., 2013). Herein, all Bartonella-positive were detected from R. norvegicus, which is the predominant species of rodents captured in our study and also a synanthropic rodent species mostly found in the urban environments (Gardner-Santana et al., 2009; Kosoy and Bai, 2019). Particularly, fleas have been shown to play an extremely important role in the transmission and acquisition of Bartonella species in rodents (Silaghi et al., 2016). Rodents inhabiting urban environments have less chance of flea-parasitic infestation, thus reducing the possibility of Bartonella infection. However, R. norvegicus closely cohabitates with humans living inside buildings and enhances the likelihood of human contact with rodents, suggesting that R. norvegicus may be an important source of zoonotic pathogens (Firth et al., 2014). Bartonella species detected in this study also suggest that much more attention should be focused on the increased risk of Bartonella transmission to humans in urban environments.
revious data indicated that the majority of known Bartonella spp. are carried by rodents, some of which have been implicated as the causative agents of human diseases (Gutiérrez et al., 2015; Okaro et al., 2017). In this study, six official or candidate species of Bartonella, including B. queenslandensis, B. mastomydis, B. tribocorum, B. rattimassiliensis, Bartonella sp. AA86HXZ, and Bartonella sp. Fuji 12-1 were detected in rodents, indicating the genetic diversity of Bartonella circulating in the rodent population in the urban areas of Guangzhou city. Notably, two confirmed human-pathogenic Bartonella species, i.e., B. tribocorum and B. rattimassiliensis, were detected in this study, indicating that infection risks exist in human populations in urban areas that originated from rodent-associated Bartonella, and also highlighting the importance of the surveillance of Bartonella infection in rodents.
In conclusion, genetically diversified Bartonella, including two confirmed zoonotic species, were detected in rodents in the urban areas of Guangzhou city with an overall prevalence of 6.4%. Moreover, R. norvegicus was the predominant host species for Bartonella infection, and B. queenslandensis was the dominant species circulating in rodents in these areas. These results provide a better understanding of the prevalence and genetic diversity of Bartonella species circulating in rodents in the urban areas of Guangzhou and also urged the necessity of the surveillance of rodent-associated Bartonella species in these areas.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Ethics Committee of Foshan University.
Author Contributions
J-WS conceived and designed the experiments and writing—review and editing. X-YY, HL, and JS performed the experiments and analyzed the data. Y-QZ, Z-HL, and X-LZ help to collect the samples. X-YY writing—original draft preparation. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Key Project of Agricultural and Social Development Science and Technology Projects of Guangzhou (No. 202103000008). The funder had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We wish to thank all the people who had collected samples in the field.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.942587/full#supplementary-material
References
Álvarez-Fernández, A., Breitschwerdt, E. B., and Solano-Gallego, L. (2018). Bartonella infections in cats and dogs including zoonotic aspects. Parasit Vectors 11, 624. doi: 10.1186/s13071-018-3152-6
An, C. H., Chen, B. B., Lyu, W., Nie, S. M., Li, S. Z., Fan, S. P., et al. (2020). Bartonella species investigated among rodents from Shaanxi Province of China. Biomed. Environ. Sci. 33, 201–205.
Anderson, B. E., and Neuman, M. A. (1997). Bartonella spp. as emerging human pathogens. Clin. Microbiol. Rev. 10, 203–219. doi: 10.1128/CMR.10.2.203
Birtles, R. J., Laycock, G., Kenny, M. J., Shaw, S. E., and Day, M. J. (2002). Prevalence of Bartonella species causing bacteraemia in domesticated and companion animals in the United Kingdom. Vet. Rec. 151, 225–229. doi: 10.1136/vr.151.8.225
Blasdell, K. R., Perera, D., and Firth, C. (2019). High prevalence of rodent-borne Bartonella spp. in urbanizing environments in sarawak, malaysian Borneo. Am. J. Trop. Med. Hyg. 100, 506–509. doi: 10.4269/ajtmh.18-0616
Chang, C. C., Chomel, B. B., Kasten, R. W., Heller, R. M., Kocan, K. M., Ueno, H., et al. (2000). Bartonella spp. isolated from wild and domestic ruminants in North America. Emerg. Infect. Dis. 6, 306–311. doi: 10.3201/eid0603.000313
Daly, J. S., Worthington, M. G., Brenner, D. J., Moss, C. W., Hollis, D. G., Weyant, R. S., et al. (1993). Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J. Clin. Microbiol. 31, 872–881. doi: 10.1128/jcm.31.4.872-881.1993
De Salvo, M. N., Hercolini, C., Arístegui, E., Bruno, A., Brambati, D. F., and Cicuttin, G. L. (2020). Bartonella spp. associated with rodents in an urban protected area, Buenos Aires (Argentina). Comp. Immunol. Microbiol. Infect. Dis. 72, 101515. doi: 10.1016/j.cimid.2020.101515
De Sousa, K. C. M., Do Amaral, R. B., Herrera, H. M., Santos, F. M., Macedo, G. C., De Andrade Pinto, P. C. E., et al. (2018). Genetic diversity of Bartonella spp. in wild mammals and ectoparasites in Brazilian Pantanal. Microb. Ecol. 76, 544–554. doi: 10.1007/s00248-017-1138-0
Divari, S., Pregel, P., Zanet, S., Ferroglio, E., Giannini, F., Scaglione, F. E., et al. (2020). Molecular evidence of Bartonella spp. in rodents: a study in Pianosa Island, Italy. Animals. 10, 2070. doi: 10.3390/ani10112070
Dong, X., and Soong, L. (2021). Emerging and re-emerging zoonoses are major and global challenges for public health. Zoonoses 1, 1. doi: 10.15212/ZOONOSES-2021-0001
Dybing, N. A., Jacobson, C., Irwin, P., Algar, D., and Adams, P. J. (2016). Bartonella species identified in rodent and feline hosts from island and mainland western Australia. Vector Borne. Zoonotic. Dis. 16, 238–244. doi: 10.1089/vbz.2015.1902
Egan, S. L., Taylor, C. L., Banks, P. B., Northover, A. S., Ahlstrom, L. A., Ryan, U. M., et al. (2021). The bacterial biome of ticks and their wildlife hosts at the urban-wildland interface. Microb. Genom. 7, 000730. doi: 10.1099/mgen.0.000730
Firth, C., Bhat, M., Firth, M. A., Williams, S. H., Frye, M. J., Simmonds, P., et al. (2014). Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. mBio 5, e01933–e01914. doi: 10.1128/mBio.01933-14
Gardner-Santana, L. C., Norris, D. E., Fornadel, C. M., Hinson, E. R., Klein, S. L., and Glass, G. E. (2009). Commensal ecology, urban landscapes, and their influence on the genetic characteristics of city-dwelling Norway rats (Rattus norvegicus). Mol. Ecol. 18, 2766–2778. doi: 10.1111/j.1365-294X.2009.04232.x
Gundi, V. A., Davoust, B., Khamis, A., Boni, M., Raoult, D., and La Scola, B. (2004). Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J. Clin. Microbiol. 42, 3816–3818. doi: 10.1128/JCM.42.8.3816-3818.2004
Guo, W. P., Lin, X. D., Wang, W., Tian, J. H., Cong, M. L., Zhang, H. L., et al. (2013). Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 9, e1003159. doi: 10.1371/journal.ppat.1003159
Gutiérrez, R., Krasnov, B., Morick, D., Gottlieb, Y., Khokhlova, I. S., and Harrus, S. (2015). Bartonella infection in rodents and their flea ectoparasites: an overview. Vector Borne. Zoonotic. Dis. 15, 27–39. doi: 10.1089/vbz.2014.1606
Hatyoka, L. M., Brettschneider, H., Bennett, N. C., Kleynhans, D. J., Muteka, S. P., and Bastos, A. D. S. (2019). Bartonella diversity and zoonotic potential in indigenous Tete Veld rats (Aethomys ineptus) from south Africa. Infect. Genet. Evol. 73, 44–48. doi: 10.1016/j.meegid.2019.04.012
Kandelaki, G., Malania, L., Bai, Y., Chakvetadze, N., Katsitadze, G., Imnadze, P., et al. (2016). Human Lymphadenopathy Caused by Ratborne Bartonella, Tbilisi, Georgia. Emerg. Infect. Dis. 22, 544–546. doi: 10.3201/eid2203.151823
Kosoy, M., and Bai, Y. (2019). Bartonella bacteria in urban rats: a movement from the jungles of southeast Asia to metropoles around the globe. Front. Ecol. Evol. 7, 88. doi: 10.3389/fevo.2019.00088
Kosoy, M., Bai, Y., Sheff, K., Morway, C., Baggett, H., Maloney, S. A., et al. (2010). Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. Am. J. Trop. Med. Hyg. 82, 1140–1145. doi: 10.4269/ajtmh.2010.09-0778
Kosoy, M., Morway, C., Sheff, K. W., Bai, Y., Colborn, J., Chalcraft, L., et al. (2008). Bartonella tamiae sp. nov., a newly recognized pathogen isolated from three human patients from Thailand. J. Clin. Microbiol. 46, 772–775. doi: 10.1128/JCM.02120-07
Kosoy, M., Murray, M., Gilmore, R. D. Jr., Bai, Y., and Gage, K. L. (2003). Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J. Clin. Microbiol. 41, 645–650. doi: 10.1128/JCM.41.2.645-650.2003
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, D. M., Hou, Y., Song, X. P., Fu, Y. Q., Li, G. C., Li, M., et al. (2015). High prevalence and genetic heterogeneity of rodent-borne Bartonella species on Heixiazi Island, China. Appl. Environ. Microbiol. 81, 7981–7992. doi: 10.1128/AEM.02041-15
Li, D. M., Liu, Q. Y., Yu, D. Z., Zhang, J. Z., Gong, Z. D., and Song, X. P. (2007). Phylogenetic analysis of Bartonella detected in rodent fleas in Yunnan, China. J. Wildl. Dis. 43, 609–617. doi: 10.7589/0090-3558-43.4.609
Lin, J. W., Chen, C. Y., Chen, W. C., Chomel, B. B., and Chang, C. C. (2008). Isolation of Bartonella species from rodents in Taiwan including a strain closely related to 'Bartonella rochalimae' from Rattus norvegicus. J. Med. Microbiol. 57, 1496–1501. doi: 10.1099/jmm.0.2008/004671-0
Liu, H., Han, T., Liu, W., Xu, G., Zheng, K., and Xiao, F. (2022). Epidemiological characteristics and genetic diversity of Bartonella species in rodents from southeastern China. Zoonoses Public Health 69, 224–234. doi: 10.1111/zph.12912
Meerburg, B. G., Singleton, G. R., and Kijlstra, A. (2009). Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 35, 221–270. doi: 10.1080/10408410902989837
Meheretu, Y., Leirs, H., Welegerima, K., Breno, M., Tomas, Z., Kidane, D., et al. (2013). Bartonella prevalence and genetic diversity in small mammals from Ethiopia. Vector Borne Zoonotic. Dis. 13, 164–175. doi: 10.1089/vbz.2012.1004
Mogollon-Pasapera, E., Otvos, L. Jr., Giordano, A., and Cassone, M. (2009). Bartonella: emerging pathogen or emerging awareness? Int. J. Infect. Dis. 13, 3–8. doi: 10.1016/j.ijid.2008.04.002
Mosepele, M., Mazo, D., and Cohn, J. (2012). Bartonella infection in immunocompromised hosts: immunology of vascular infection and vasoproliferation. Clin. Dev. Immunol. 2012, 612809. doi: 10.1155/2012/612809
Müller, A., Gutiérrez, R., Seguel, M., Monti, G., Otth, C., Bittencourt, P., et al. (2020). Molecular survey of Bartonella spp. in rodents and fleas from Chile. Acta Trop 212, 105672. doi: 10.1016/j.actatropica.2020.105672
Okaro, U., Addisu, A., Casanas, B., and Anderson, B. (2017). Bartonella species, an emerging cause of blood-culture-negative endocarditis. Clin. Microbiol. Rev. 30, 709–746. doi: 10.1128/CMR.00013-17
Qin, X. R., Liu, J. W., Yu, H., and Yu, X. J. (2019). Bartonella species detected in rodents from eastern China. Vector Borne Zoonotic Dis. 19, 810–814. doi: 10.1089/vbz.2018.2410
Rao, H., Li, S., Lu, L., Wang, R., Song, X., Sun, K., et al. (2021). Genetic diversity of Bartonella species in small mammals in the Qaidam Basin, western China. Sci. Rep. 11, 1735. doi: 10.1038/s41598-021-81508-w
Saengsawang, P., Morand, S., Desquesnes, M., Yangtara, S., and Inpankaew, T. (2021). Molecular detection of Bartonella Species in rodents residing in urban and suburban areas of central Thailand. Microorganisms 9, 2588. doi: 10.3390/microorganisms9122588
Silaghi, C., Pfeffer, M., Kiefer, D., Kiefer, M., and Obiegala, A. (2016). Bartonella, rodents, fleas and ticks: a molecular field study on host-vector-pathogen associations in Saxony, Eastern Germany. Microb. Ecol. 72, 965–974. doi: 10.1007/s00248-016-0787-8
Su, Q., Chen, Y., Wang, B., Huang, C., Han, S., Yuan, G., et al. (2020). Epidemiology and genetic diversity of zoonotic pathogens in urban rats (Rattus spp.) from a subtropical city, Guangzhou, southern China. Zoonoses Public Health 67, 534–545. doi: 10.1111/zph.12717
Szewczyk, T., Werszko, J., Slivinska, K., Laskowski, Z., and Karbowiak, G. (2021). Molecular detection of Bartonella spp. in rodents in Chernobyl Exclusion Zone, Ukraine. Acta Parasitol. 66, 222–227. doi: 10.1007/s11686-020-00276-1
Valentine, K. H., Harms, C. A., Cadenas, M. B., Birkenheuer, A. J., Marr, H. S., Braun-Mcneill, J., et al. (2007). Bartonella DNA in loggerhead sea turtles. Emerg. Infect. Dis. 13, 949–950. doi: 10.3201/eid1306.061551
Vayssier-Taussat, M., Moutailler, S., Féménia, F., Raymond, P., Croce, O., La Scola, B., et al. (2016). Identification of novel zoonotic activity of Bartonella spp., France. Emerg. Infect. Dis. 22, 457–462. doi: 10.3201/eid2203.150269
Von Loewenich, F. D., Seckert, C., Dauber, E., Kik, M. J. L., De Vries, A., Sprong, H., et al. (2019). Prosthetic valve endocarditis with Bartonella washoensis in a human european patient and its detection in red squirrels (Sciurus vulgaris). J. Clin. Microbiol. 58, e01404–e01419. doi: 10.1128/JCM.01404-19
Wei, Y., Wang, Y., Li, X., Qin, P., Lu, Y., Xu, J., et al. (2018). Meteorological factors and risk of hemorrhagic fever with renal syndrome in Guangzhou, southern China, 2006-2015. PLoS Negl. Trop. Dis. 12, e0006604. doi: 10.1371/journal.pntd.0006604
Welch, D. F., Carroll, K. C., Hofmeister, E. K., Persing, D. H., Robison, D. A., Steigerwalt, A. G., et al. (1999). Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J. Clin. Microbiol. 37, 2598–2601. doi: 10.1128/JCM.37.8.2598-2601.1999
Keywords: Bartonella, epidemiology, genetic diversity, rodents, urban areas
Citation: Yao X-Y, Liu H, Sun J, Zhang Y-Q, Lv Z-H, Zhang X-L and Shao J-W (2022) Epidemiology and Genetic Diversity of Bartonella in Rodents in Urban Areas of Guangzhou, Southern China. Front. Microbiol. 13:942587. doi: 10.3389/fmicb.2022.942587
Received: 12 May 2022; Accepted: 13 June 2022;
Published: 04 July 2022.
Edited by:
Wei Wang, Jiangsu Institute of Parasitic Diseases (JIPD), ChinaCopyright © 2022 Yao, Liu, Sun, Zhang, Lv, Zhang and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Wei Shao, andzaGFvMTk4OEAxNjMuY29t
†These authors have contributed equally to this work
 Xin-Yan Yao†
Xin-Yan Yao† Jing Sun
Jing Sun Jian-Wei Shao
Jian-Wei Shao