
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 04 August 2022
Sec. Phage Biology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.936267
This article is part of the Research Topic Bacteria-Phage Coevolution in Antimicrobial Resistance View all 6 articles
Bacteriophages (phages), the most abundant biological entities on Earth, have a significant effect on the composition and dynamics of microbial communities, biogeochemical cycles of global ecosystems, and bacterial evolution. A variety of antibiotic resistance genes (ARGs) have been identified in phage genomes in different soil samples. Phages can mediate the transfer of ARGs between bacteria via transduction. Recent studies have suggested that anthropogenic activities promote phage-mediated horizontal gene transfer events. Therefore, the role of phages in the dissemination of ARGs, which are a potential threat to human health, may be underestimated. However, the contribution of phages to the transfer of ARGs is still poorly understood. Considering the growing and wide concerns of antibiotic resistance, phages should be considered a research focus in the mobile resistome. This review aimed to provide an overview of phages as vehicles of ARGs in soil. Here, we summarized the current knowledge on the diversity and abundance of ARGs in soilborne phages and analyzed the contribution of phages to the horizontal transfer of ARGs. Finally, research deficiencies and future perspectives were discussed. This study provides a reference for preventing and controlling ARG pollution in agricultural systems.
With the extensive use of antibiotics in clinics and agriculture, the problem of antibiotic resistance has become increasingly prominent and has become a major public health risk faced by human society in the 21st century (Kavagutti et al., 2019; Pu et al., 2020). According to the latest report by the Lancet, there were an estimated 4.95 million deaths associated with bacterial antimicrobial resistance in 2019, including 1.27 million deaths attributable to bacterial antimicrobial resistance (Murray et al., 2022). If it is not effectively controlled, it is expected that this number will be as high as 10 million by 2050 (Willyard, 2017; Amarasiri et al., 2020). Recently, it has been shown that antibiotic resistance genes (ARGs) carried by clinically drug-resistant pathogens originate from the environment (Forsberg et al., 2012; Yang et al., 2013; Fang et al., 2015; Huijbers et al., 2019). Therefore, ARGs have been widely considered as a new type of environmental pollutant and have become a research hotspot worldwide (Forsberg et al., 2012; Zhang et al., 2021).
Microorganisms in the soil can exchange signals by secreting low concentrations of antibiotics, which provide selective pressure for bacteria to obtain resistance (Adriaenssens et al., 2017). However, anthropogenic activities, such as sewage run-off, manure application, and reclaimed water irrigation, cause soil to become one of the largest ARG pools in the environment (Calero-Cáceres et al., 2019; Pu et al., 2020). The long-term use of antibiotics in the breeding industry provides a selective pressure for animal intestinal microflora. Therefore, the diversity and abundance of ARGs are increasing in the gut microbiome, resulting in livestock manure becoming an important source of ARGs in the environment (Wright and Poinar, 2012). Livestock manure is applied to farmland soil as organic fertilizers and significantly increases soil antibiotic resistance. A previous study found that the abundance of 63 ARGs subtypes in soil treated with manure was 192–28,000 times higher than that in the control soil (Chen et al., 2017). ARGs in soil can migrate into the endophytic and phyllospheric microbiomes of plants (Zhang et al., 2019; Pu et al., 2020; Guo et al., 2021), and subsequently enter the food chain, posing an eventual potential threat to human health (Forsberg et al., 2012; Ye et al., 2019).
Horizontal gene transfer (HGT) is the main mechanism underlying the enrichment and spread of ARGs in the environment (Debroas and Siguret, 2019; Lerminiaux and Cameron, 2019). ARGs can be horizontally transferred in three ways: conjugation mediated by mobile genetic elements, transformation by cell-free DNA, and phage-mediated transduction (Brown-Jaque et al., 2015; Mishra et al., 2021). Among the three HGT mechanisms, conjugation is generally considered the predominant mode of ARG dissemination in environmental settings (von Wintersdorff et al., 2016). Recently, a variety of ARG sequences were found in soil phage genomes, suggesting that phages may play an important role in mobile antibiotic resistance. However, the contribution of phages as reservoirs to HGT of ARGs has not been extensively explored. In this study, we reviewed the occurrence and distribution of ARGs in soilborne phage genomes (pARGs), the mode and characteristics of phage-mediated horizontal transfer of ARGs (transduction), and the contribution of phages to the mobile antibiotic resistome. In addition, the current research gap and future prospects are discussed. This article provides a scientific foundation for further expansion of the knowledge of the soil antibiotic resistome and controlling the spread of phage-mediated ARGs.
Phages, as bacterial viruses, are the most abundant and diverse entities in the biosphere, with a total population of approximately 1031–33, which is 10 times that of bacteria (Balcazar, 2014; Dion et al., 2020). The complete biological structure of a phage consists of a nucleic acid core (DNA or RNA) surrounded by an outer shell of a protein coat (capsid) with an estimated size of 20–200 nm (Jebri et al., 2021). Additionally, most phages belong to the order Caudovirales, which includes five families: Myoviridae, Siphoviridae, Podoviridae, Ackermannviridae, and Herelleviridae (Adams et al., 2017; Walker et al., 2019; Jebri et al., 2021). Phages are divided into two categories according to their life cycle: temperate and virulent phages. Temperate phages can be inserted into the host genome as a prophage and replicate with the host; this process is known as the lysogenic cycle (Ghosh et al., 2008). When external conditions change (such as UV radiation and heavy metal exposure), the prophage enters the lytic cycle using host nutrients to synthesize the offspring phage and kill the host (Ghosh et al., 2008; Zhong et al., 2021). Phages have a significant effect on the composition and diversity of microbial communities, biogeochemical cycles of global ecosystems, and bacterial evolution (Maganha De Almeida Kumlien et al., 2021; Lobo and Faciola, 2021).
Phages exist in various environments, among which the soil is an important habitat (Williamson et al., 2007; Ghosh et al., 2008). It is estimated that the number of phage particles in soil accounts for 10% of the total number of viruses globally (Vos et al., 2009; Cobian et al., 2016). High biodiversity and rich soil nutrition provide a suitable living environment for phages (Gomez and Buckling, 2011). Phages have been detected in various types of soil samples, including farmland, forests, wetlands, croplands, sand dunes, and even extreme environments, such as deserts and Antarctic soil (Williamson et al., 2007; Gomez and Buckling, 2011; Wang, 2019; Bezuidt et al., 2020; Chen et al., 2021). The abundance of phages in the different soil types are summarized in Table 1. Among the various soil samples that have been investigated by epi-fluorescence microscopy, the abundances of phages in farmlands and forests are the highest, reaching 109/g, whereas the abundance of phages in deserts is nearly 3–6 orders of magnitude lower (Zablocki et al., 2016; Williamson et al., 2017; Trubl et al., 2018). With the continuous progress of metagenome sequencing and bioinformatics technology, the research of the “dark matter” of the microbial world—the Environmental virome (mainly composed of phages) has developed rapidly (Monaco and Kwon, 2017; Ofir and Sorek, 2018; Han et al., 2022). A viral metagenomic analysis of soil has suggested that viruses (phages) affect the microbial ecology (Ashelford et al., 2003; Dinsdale et al., 2008; Parsley et al., 2010; Cobian et al., 2016; Kavagutti et al., 2019). A recent study showed that phages play an important role in phosphorus metabolism, carbon metabolism, soil organic matter degradation, and polysaccharide binding after analyzing the permafrost thaw gradient (Emerson et al., 2018; Trubl et al., 2018; Emerson, 2019; Han et al., 2022). Additionally, soil properties and the bacterial community significantly affects the structure of the viral community (Chen et al., 2016, 2021). However, our understanding of soil phages is still relatively backward due to the high soil heterogeneity and the limitation of publicly available viral databases, especially regarding the contribution of phages to the soil antibiotic resistome (Larrañaga et al., 2018; Sun et al., 2018; Zhao et al., 2019).
Research on ARGs in soilborne phages began relatively late compared with that in the aquatic environment. Ross and Topp (2015) quantified a set of ARGs from soilborne phages using quantitative polymerase chain reaction (qPCR) technology. In the following years, the occurrence and abundance of soilborne pARGs from different regions were investigated using qPCR technology (Anand et al., 2016; Larrañaga et al., 2018; Sun et al., 2018; Zhao et al., 2019). Recently, metagenomics has become a powerful tool to study the diversity and abundance of pARGs in soil (Enault et al., 2017). The diversity and abundance of soilborne pARGs are summarized in Table 2.
The main methods for detecting pARGs are based on PCR technology, such as qPCR and droplet digital PCR (ddPCR), which can obtain absolute quantification information of specific ARG subtypes (Ross and Topp, 2015; Hu et al., 2022). Furthermore, recent metagenomic approaches provide a valuable tool for exploring the composition of the viral community, ARGs, and the correlations between them in different environmental settings (Chen et al., 2021). With the development of sequencing technology, more pARGs in soil will be unearthed, providing unprecedented opportunities to elucidate the mechanisms of mobile antibiotic resistance in soil (Chen et al., 2021). PCR technology and metagenomic sequencing have their own advantages. As a classical method for the absolute quantitative detection of ARGs in environmental samples and pure strains, PCR technology has the advantages of accuracy and rapidity (Hu et al., 2019a). In contrast, metagenomic sequencing does not depend on the microbial culture and screening process, and has the advantages of high specificity, sensitivity, and throughput (Hu et al., 2013; Hu et al., 2019a). Metagenomic sequencing facilitates the detection of more ARGs, which is conducive to the comprehensive understanding of the diversity of phage-mediated ARGs. The continuous innovation of sequencing technology can capture the phylogenetic and genetic diversity, fully understand the types, abundance, and transmission routes of phage-mediated ARGs, and predict the possible risk of antibiotic resistance and other potential threats to humans (Parsley et al., 2010; Bengtsson-Palme, 2017).
ARG subtypes that confer resistance to 13 antibiotics (β-lactams, tetracycline, quinolone, aminoglycosides, sulfonamide, streptomycin, chloramphenicol, trimethoprim, MLSB, vancomycin, rifamycin, pleuromutilin, and mupirocin) were identified in phage genomes from various soil sources (Table 2). Among these, tet genes are the main types of soilborne pARGs. Tetracyclines are one of the most commonly used antibiotics in the livestock breeding industry, and their resistance genes are ubiquitous in agricultural soil. For example, Sun et al. (2018) detected six genes conferring resistance to tetracyclines in the phage fraction of greenhouse soil, including tetC, tetE, tetG, tetM, tetO, and tetX. In addition, large amounts of the bla gene have been identified in soilborne phages, including blaTEM, blaCTX-M, and blaOXA-2. Hu et al. (2022) analyzed a total of 25 ARG subtypes using ddPCR technology and demonstrated that the detection rate of soilborne pARGs with the application of organic fertilizer was 76%, whereas the rates of soilborne pARGs with the application of non-fertilizer and chemical fertilizer were only 68 and 72%, respectively. Chen et al. (2021) analyzed the viral DNA of soil samples using the Illumina NovaSeq 6000 platform and detected 16 unique ARG subtypes, including catB, macB, vatB, dfrB2, dfrB6, and so on. Another study detected 144 ARGs in soil viruses, which were divided into 12 categories according to the types of antibiotic resistance (Wang, 2019).
Generally, the abundance of pARGs varies greatly between different soil types and ARG subtypes, ranging from 101 to 108 copies/g (Wang, 2019). A study of soilborne pARGs resistant to tetracycline found that the order of accumulative pARG abundance is as follows: efflux pump ARGs (tetC + tetE + tetG) > ribosome protection ARGs (tetM + tetO) > enzymatic modification ARG (tetX; Sun et al., 2018). As shown in Table 2, human activities, such as the application of organic fertilizer, affect the abundance of ARGs carried by soil phages. Another study also found that the relative abundance of pARGs in the soil receiving organic fertilizer treatment is significantly higher than that in soil receiving chemical fertilizer (p < 0.05; Wang, 2019). Hu et al. (2022) also found that soil receiving organic fertilizer has the highest total abundance of pARGs, reaching 1.6 × 103 copies/g, and soil treated with non-fertilizer has the lowest pARG abundance (6.0 × 102 copies/g).
The mechanisms of the horizontal transfer of ARGs are conjugation, natural transformation, and transduction (von Wintersdorff et al., 2016). Among them, plasmid-mediated conjugation has been identified as the main mechanism of HGT (Svara and Rankin, 2011). Some studies have found that ARGs resistant to amoxicillin and sulfanilamide are located in plasmids, with frequencies ranging from 10−6 to 10−4 (Musovic et al., 2010; Zhang et al., 2017). In addition, many plasmids have a wide-range of hosts, enabling ARGs to transfer among different biological species (Binh et al., 2008; Musovic et al., 2010). Transformation refers to the process in which the recipient bacteria directly obtain extracellular fragments of the donor bacteria to obtain new genetic traits (Hu et al., 2019a; Ding et al., 2021). However, the poor stability of extracellular DNA and low proportion of bacteria with natural transformation abilities make excludes transformation as the main mechanism of the horizontal transfer of ARGs (Nielsen et al., 2007). Recently, phage-mediated transduction has been considered as another mechanism for the horizontal transfer of ARGs (Subirats et al., 2016; Calero-Cáceres et al., 2019; Debroas and Siguret, 2019; Jebri et al., 2021).
Phage-mediated transduction of ARGs has been documented for several bacterial species, including Streptococcus pyogenes, Enterococci sp., Escherichia coli, and Salmonella sp., and the transduced ARGs are mainly resistant to erythromycin, tetracycline, gentamicin, and β-lactam (Ubukata et al., 1975; Hyder and Streitfeld, 1978; Schmieger and Schicklmaier, 1999; Mazaheri et al., 2011; Billard-Pomares et al., 2014). Transduction can be divided into generalized or specialized transduction according to classical microbiology (Figures 1A,B). Generalized transduction that achieves a genetic trait is carried by two types of phage particles (temperate and virulent phages) from a donor cell to a recipient cell during lytic pathways (Sander and Schmieger, 2001). Phages can transfer any gene from one bacterium to another during this process (Penadés et al., 2014). Transduction particles wrap host DNA (including ARGs) by mispackaging, and their frequency is very low (10−8–10−6; Maganha De Almeida Kumlien et al., 2021). Another model of genetic material mediated by phages is specialized transduction, which can only be carried out by temperate phages during the late lysogenic cycle (Debroas and Siguret, 2019). More specifically, specialized transduction particles carry fixed genes of the bacterial chromosome located adjacent to the prophage (including ARGs) attachment site with a low occurrence frequency (10−6; Balcazar, 2014). The frequency of gene transfer due to “misloading” of host DNA fragments is very low, whether by general transduction or specialized transduction (Poté et al., 2003; Muniesa et al., 2013). The third recently discovered transduction model, lateral transduction, may be the most effective way for bacteria to obtain ARGs (Figure 1C; Chen et al., 2018; Fillol-Salom et al., 2021). Kenzaka et al. (2010) accurately measured the transduction frequency between E. coli mediated by phages using cycling primed in situ amplification-fluorescent in situ hybridization (CPRINS-FISH). The results revealed that the frequency of DNA transfer is 10−4–10−3, indicating that the phage-mediated exchange of genes occurs at an unexpectedly high frequency (Kenzaka et al., 2007, 2010). Phage-mediated HGT has many distinct characteristics, which can be summarized into the following three aspects.
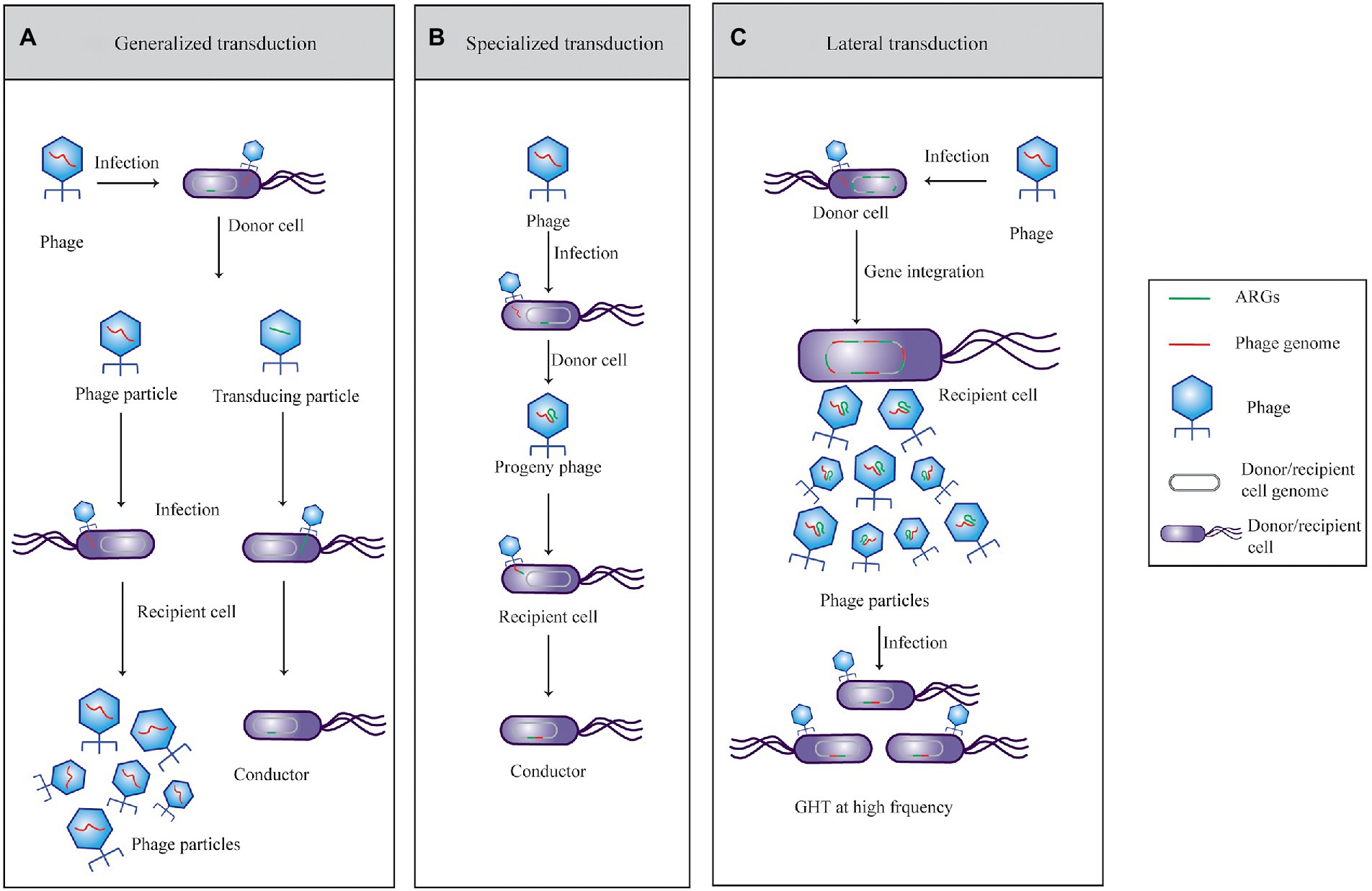
Figure 1. Horizontal transfer of phage-mediated antibiotic resistant genes through (A) general, (B) specialized, and (C) lateral transduction ARGs mediated by phages through transduction.
A previous study showed that the transduction frequency of phages in aquatic environments is several orders of magnitude higher than expected (nearly 1%; Muniesa et al., 2013). Recently, Chen et al. (2018) found that lateral transduction in the process of HGT is mediated by the temperate phage of Staphylococcus aureus, proving that phages can transfer genes with extremely high frequency. The mechanism of this newly discovered transduction is DNA packaging initiated in situ from integrated prophages. Several hundred kilobases of the S. aureus genome, are packaged in phage heads at very high frequencies. In situ replication before DNA packaging creates multiple prophage genomes, such that lateral-transducing particles form during normal phage maturation, transforming parts of the S. aureus chromosome into hypermobile regions of gene transfer (Chen et al., 2018). With the deepening of our understanding of phages and their transduction mechanisms, the frequency of phage-mediated HGT in the environment may be much higher than expected. Overall, phages act as powerful agents to transfer bacterial chromosomal DNA to another organism via genetic transduction (Fillol-Salom et al., 2021).
Conjugation has been considered to play a major role in HGT and the consequent spread of ARGs and requires cell-to-cell contact (Muniesa et al., 2013). However, phage transduction does not need to meet the above conditions. Additionally, ARGs can be transferred by phages at different time points (Muniesa et al., 2013). Researchers have found that viral DNA can be transferred from hot springs (−82°C) to ice (−0°C), which strongly supports this view (Sano et al., 2004). Paez-Espino et al. (2016) identified that the virus can infect organisms from different phyla by linking the virus to the microbial host through the CRISPR spacer and transfer RNA matches. Furthermore, phage-mediated HGT is not restricted to microorganisms of the same species, but occurs between different species, genera, or even phyla (Muniesa et al., 2013).
The genetic material of the phage is tightly surrounded by a protein coat called a capsid. Owing to their structure, phages thrive successfully in the environment and are resistant to natural and anthropogenic stressors such as enzymes, radiation, and antimicrobial substances (Muniesa et al., 2013; Göller et al., 2020). Durán et al. (2002) conducted an inactivation experiment of phages in rivers and showed that phages were more resistant than fecal coliforms and enterococci. As mentioned above, phage DNA will not be damaged in different extreme environments, and this conclusion is supported by the premise that the protein capsid is not broken (Sano et al., 2004). Overall, phages have stronger persistence and a broader time scale in the environment, making them more suitable as carriers for ARG transfer (Calero-Cáceres and Muniesa, 2016).
Phages act as vectors for genetic exchange, facilitate reproduction (short-term) or promote microbial evolution (long-term). When phages containing ARGs move between different bacteria, it leads to the horizontal spread of ARGs and even the emergence of drug-resistant bacteria in soil (Kenzaka et al., 2007). Understanding the horizontal transfer mechanism of pARGs is important when investigating the mobile resistome, which is conducive to controlling the spread of ARGs.
A summary of the research methods for phage-mediated transduction of ARGs is shown in Figure 2. Before the advent of new generation sequencing technology, phage-mediated ARG transduction was mainly studied through cultivation-based methods (Bowring et al., 2022). Firstly, a specific phage containing ARGs is used to infect antibiotic sensitive bacteria (such as E. coli). After co-culturing, the lysate is diluted and plated on drug-free and drug-containing plates under appropriate conditions. The frequency of transduction is calculated by dividing the number of colonies on the drug-containing plate by that on the drug-free plate (Zeph et al., 1988; Mazaheri et al., 2011; Figure 2A1). Another research method uses the phage community as a whole to conduct in vitro transduction experiments. First, the bacteria and phages are isolated from environmental samples. After co-culturing, the plate count of the antibiotic-resistant bacteria (ARB) is carried out to investigate the rate of ARG transfer (Modi et al., 2013; Figure 2A2). With the emergence of molecular biotechnology, research on the horizontal transfer of ARGs is more profound and rigorous (Ross and Topp, 2015). Bacteria and phages are obtained from environmental samples by means of centrifugal filtration, and their DNA is extracted. Then, the abundance of ARG is analyzed through PCR and fluorescent labeling technology (Harrington et al., 2012; Larrañaga et al., 2018; Watson et al., 2018; Zhao et al., 2019; Figure 2B1). In recent years, genome sequencing technology has overcome the limitations of bacterial culture and microbial loss caused by isolation. As shown in Figure 2B2, the transfer of ARGs is studied through the extraction of total environmental DNA, followed by the sequencing and analysis of the resistome and mobilome. The origin of AGRs and the role of phages in ARG transmission are explored through phylogenetic diversity and phage-host linkage analysis, respectively. In addition, the results of metagenomic or (virome) analysis can simultaneously reveal the microbial (viral) composition and function in soil, which contribute to the reliable risk assessment of ARG transmission through the source-tracking method and correlation analysis (Lekunberri et al., 2017; Chen et al., 2021).
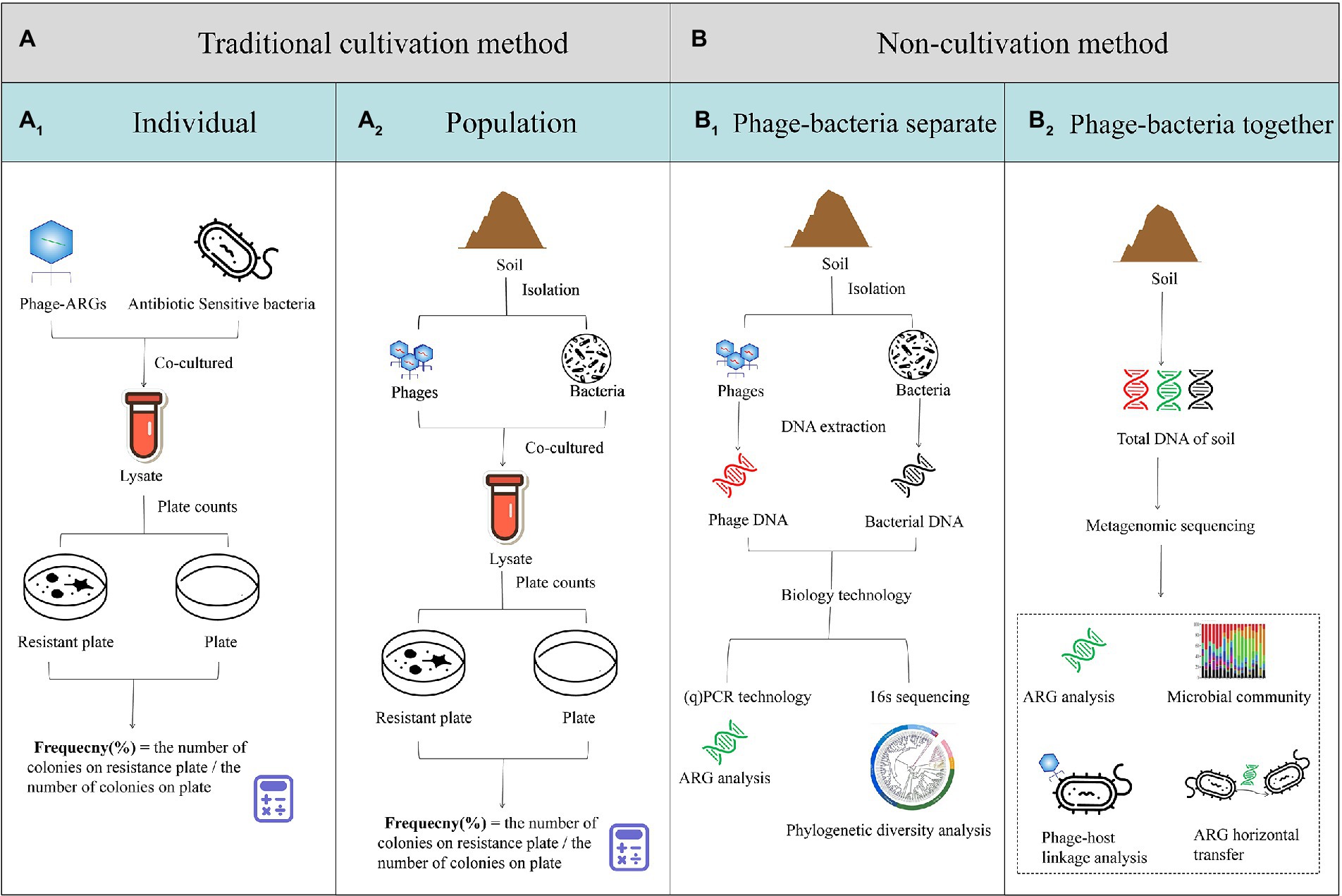
Figure 2. Research methods for phage-mediated transduction of antibiotic resistant genes (ARGs) in vitro. (A) Traditional cultivation methods using an individual phage containing ARGs co-cultured with antibiotic-sensitive bacteria (A1) or a soil sample from which the phage and bacteria populations were isolated and co-cultured (A2). (B) Non-cultivation methods using soil samples from which the phage and bacteria populations are isolated using centrifugal filtration, their DNA is extracted and ARGs and phylogenetic diversity is analyzed through PCR or 16 s sequencing, respectively (B1), or the total DNA of the soil is extracted analyzed through metagenomic sequencing (B2).
Many experimental studies have provided evidence that phages participate in the horizontal transfer of ARGs. Ross and Topp (2015) conducted transduction experiments using phages isolated from the soil to infect E. coli K-12. They found that a certain selective pressure (such as antibiotics) could promote the horizontal transfer of phage-mediated soilborne ARGs. Another study used phages-mediated ARGs to perform subculture experiments with E. coli as the host cells. By measuring the ARGs in the phage DNA before and after culture, it was proven that phages can reinfect the host and are important carriers for the transfer and transmission of ARGs in the environment (Larrañaga et al., 2018). Researchers have successfully transferred the bla gene in phage DNA into the E. coli genome using electroporation technology, which also suggests that phages are vehicles that transfer ARGs effectively (Colomer-Lluch et al., 2011).
However, our knowledge of phage-mediated HGT in soil lags far behind from what is known in other environmental media (e.g., water, animal gut) because of the high heterogeneity and rich biodiversity of the soil. A virome study showed that mouse intestinal phages were enriched with ARGs due to antibiotic treatment; these ARG-carrying phages infected intestinal bacteria, increasing the number of antibiotic-resistant bacteria (Modi et al., 2013). Shousha et al. (2015) found that phages isolated from chickens transduced the resistance of E. coli to various antibiotics (including ampicillin, tetracycline, and chloramphenicol). Another study demonstrated that phages released from S.aureus enable their hosts to acquire streptomycin-resistant genes from adjacent cells (Haaber et al., 2016).
Only a few studies were conducted to explore the relationship between antibiotics and the transfer of pARGs. A study on the gut virome showed that antibiotic treatment lead to the enrichment of pARGs. In addition, they assessed the proportion of resistant bacteria before and after phage infection with naive microbiota from mice gut as host by in vitro experiments, and found that the basal frequency (before infection) was lower than that microbiota infected with phages, especially in antibiotic-treated mice (Modi et al., 2013). These results demonstrated that antibiotic treatment enhanced the ability of phages to transmit resistance.
Numerous studies have demonstrated a positive correlation between the transfer of ARGs and the physicochemical characteristics of soil (Ji et al., 2012; Debroas and Siguret, 2019). Environmental factors, such as temperature, affected the dissemination of ARGs by exchange community boundaries (Iyer et al., 2013; Marti et al., 2014). Heavy metals could exert pressure on ARGs, thereby promoting the transfer of ARGs (He et al., 2020). Another study found that the concentration of Cu and Zn in soil was significantly positively correlated with the transduction of pARGs (Yang et al., 2021). Organic matter and heavy metals (Zn, Cr, and Cu) had a greater effect on pARGs than others factors, such as Hg, As, available phosphorus, available potassium, and total nitrogen (Hu et al., 2022). The concentrations of NH4-N and total phosphorus are positively correlated with two types of pARGs conferring resistant to tetracyclines and macrolide-lincosamide-streptogramin B (Yang et al., 2021). Random forest modeling and partial least squares path modeling results indicated that pH is a key factor affecting the occurrence of pARGs during the fermentation process (Xu et al., 2022).
Soil bacteria and phages are crucial reservoirs of ARGs in the natural environment (Sun et al., 2018; Guo et al., 2021). The ARGs in phages are generally positively correlated with that of their bacterial hosts (Yang et al., 2021). Virus-host linkage analyses revealed that the phage-mediated ARGs are closely related to five bacterial phyla, including Firmicutes, Bacteroidetes, Proteobacteria, Crenarchaeota, and Planctomycetes (Chen et al., 2021). Yang et al. (2021) investigated the association between bacterial communities and pARGs and found that the bacterial community contributes to 16.7% of the variation in the pARG profiles. The results showed that Terrisporobacter, Desulfovibrio, and Acinetobacter are the main drivers impacting pARGs (Yang et al., 2021). Several recent studies have confirmed that the bacterial community is the main driver impacting the pARG profiles. Using the variance partitioning analysis (VPA), our previous study also found that the bacterial community contributes the most to pARG variation (10.81%; Hu et al., 2022).
Recently, some studies have found that nano-metal oxide particles have an effect on the horizontal transfer of ARGs (Hu et al., 2019b; Otinov et al., 2020; Li and Zhang, 2022). Han et al. (2020) conducted an experiment using phage gM13 and E.coli exposed to nano-TiO2, and the results showed that nano-TiO2 increases the transduction frequency up to 4.5 fold compared to the control. TiO2 photoexcitation can drastically improve phage transduction efficiency 20.4-fold (Xiao et al., 2021). The transfer rate of ARGs mediated by nano-Al2O3 via a transduction-like pathway was 104 times higher than that of the control. (Ding et al., 2021). The possible mechanisms of these nanomaterials promoting transduction are presumed to the increase of phage attachment on host cell surface, and cell membrane permeability. In the contrast, excessive UV irradiation led to a decrease in transduction efficiency (Xiao et al., 2021). Additionally, ionic liquid, as an environmental friendly compound, facilitates the horizontal transfer of ARGs (Wang et al., 2015a,b).
Phages play an important role in the antibiotic resistome of soil, and their contributions to the spread of ARGs should not be underestimated. However, there are still many uncertainties regarding the distribution characteristics of ARGs in the phage genome and the mechanism of phage-mediated HGT at different environmental sites. Therefore, additional studies are required to elucidate this and to subsequently support the inclusion of phages in monitoring, evaluation, and surveillance programs aimed at the emergence and spread of ARGs (Balcázar, 2020). The following gaps in research should be addressed:
1. The influence mechanism of the physicochemical properties and biological communities of soil on ARG transduction via phages is unclear. Although phages can mediate the spread of ARGs in agricultural soil and increase the number of antibiotic resistant bacteria under selective pressures, whether they can improve the antibiotic resistance of other indigenous bacteria and related mechanisms needs to be elucidated.
2. The research methodology on soilborne pARGs needs to be improved. Existing studies have only focused on ARGs in free phages, ignoring prophages in bacterial genomes, which leads to the inevitable underestimation of the contribution of phages to the environmental antibiotic resistome. In addition, the removal of bacterial contamination during phage DNA extraction is a challenge in the quantitative study of phage metagenomes in the soil (Göller et al., 2020). Although the development of sequencing technologies will undoubtedly help solve these problems, these approaches should be standardized to avoid misleading and incorrect conclusions.
Phages are extremely abundant in soil and a large number of ARGs are present in their genomes. ARGs can be horizontally transferred through transduction at high frequencies, including specialized, generalized, and lateral transduction. Transduction can occur without contact between donor and recipient bacteria and is not limited by time and space. Moreover, there is increasing evidence that human activities, especially the application of organic fertilizers, lead to the enrichment and transmission of ARGs in soilborne phages. Therefore, the role of phages in the transfer of ARGs should not be ignored or underestimated. However, the contribution of phages to the transfer of ARGs has been relatively poorly studied because of the high heterogeneity and complexity of soils. With the advancement of sequencing technology, recent related research has been greatly developed. Additional studies are needed to elucidate the mechanisms and influencing factors contributing to the dissemination of ARGs via phages.
YZ: writing — original draft preparation, visualization, and conceptualization. YG: writing — original draft preparation and conceptualization. TQ: writing — review and editing and funding acquisition. MG: writing — review and editing. XW: supervision, project administration, writing — review and editing, and funding acquisition. All authors have approved this work for publication.
This work was supported by the National Natural Science Foundation of China (grant no. 32070089), Beijing Natural Science Foundation (grant no. 5222005), the Research Foundation of BAAFS (grant no. KJCX20200302), Beijing Postdoctoral Science Foundation (grant no. 202222104), and Postdoctoral Science Foundation of BAAFS (grant no. 2021ZZ006).
We would like to thank Editage (www.editage.cn) for English language editing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
HGT, Horizontal gene transfer; ARGs, Antibiotic resistance genes; AMGs, Auxiliary metabolic genes; qPCR, Quantitative polymerase chain reaction; SOM, Organic matter in soil; pARGs, Antibiotic resistance genes carried by phages.
Adams, M. J., Lefkowitz, E. J., King, A. M. Q., Harrach, B., Harrison, R. L., Knowles, N. J., et al. (2017). Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2017). Arch. Virol. 162, 2505–2538. doi: 10.1007/s00705-017-3358-5
Adriaenssens, E. M., Kramer, R., Van Goethem, M. W., Makhalanyane, T. P., Hogg, I., and Cowan, D. A. (2017). Environmental drivers of viral community composition in Antarctic soils identified by viromics. Microbiome. 5, 83. doi: 10.1186/s40168-017-0301-7
Amarasiri, M., Sano, D., and Suzuki, S. (2020). Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: current knowledge and questions to be answered. Crit. Rev. Env. Sci. Tec. 50, 2016–2059. doi: 10.1080/10643389.2019.1692611
Anand, T., Bera, B. C., Vaid, R. K., Barua, S., Riyesh, T., Virmani, N., et al. (2016). Abundance of antibiotic resistance genes in environmental bacteriophages. J. Gen. Virol. 97, 3458–3466. doi: 10.1099/jgv.0.000639
Ashelford, K. E., Day, M. J., and Fry, J. C. (2003). Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microb. 69, 285–289. doi: 10.1128/AEM.69.1.285
Balcazar, J. L. (2014). Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLoS Pathog. 10:e1004219. doi: 10.1371/journal.ppat.1004219
Balcázar, J. L. (2020). Implications of bacteriophages on the acquisition and spread of antibiotic resistance in the environment. Int. Microbiol. 23, 475–479. doi: 10.1007/s10123-020-00121-5
Bengtsson-Palme, J. (2017). Antibiotic resistance in the food supply chain: where can sequencing and metagenomics aid risk assessment? Curr. Opin. Food Sci. 14, 66–71. doi: 10.1016/j.cofs.2017.01.010
Bezuidt, O., Lebre, P. H., Pierneef, R., Leon-Sobrino, C., Adriaenssens, E. M., Cowan, D. A., et al. (2020). Phages actively challenge niche communities in antarctic soils. mSystems 5:e00234-20. doi: 10.1128/mSystems.00234-20
Billard-Pomares, T., Fouteau, S., Jacquet, M. E., Roche, D., Barbe, V., Castellanos, M., et al. (2014). Characterization of a P1-like bacteriophage carrying an SHV-2 extended-spectrum β-lactamase from an Escherichia coli strain. Antimicrob. Agents Chemother. 58, 6550–6557. doi: 10.1128/AAC.03183-14
Binh, C. T., Heuer, H., Kaupenjohann, M., and Smalla, K. (2008). Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol. Ecol. 66, 25–37. doi: 10.1111/j.1574-6941.2008.00526.x
Bowring, J. Z., Su, Y., Alsaadi, A., Svenningsen, S. L., Parkhill, J., Ingmer, H., et al. (2022). Screening for highly transduced genes in staphylococcus aureus revealed both lateral and specialized transduction. Microbiol. Spectrum. 10:e242321. doi: 10.1128/spectrum.02423-21
Brown-Jaque, M., Calero-Cáceres, W., and Muniesa, M. (2015). Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid 79, 1–7. doi: 10.1016/j.plasmid.2015.01.001
Calero-Cáceres, W., and Muniesa, M. (2016). Persistence of naturally occurring antibiotic resistance genes in the bacteria and bacteriophage fractions of wastewater. Water Res. 95, 11–18. doi: 10.1016/j.watres.2016.03.006
Calero-Cáceres, W., Ye, M., and Balcázar, J. L. (2019). Bacteriophages as environmental reservoirs of antibiotic resistance. Trends Microbiol. 27, 570–577. doi: 10.1016/j.tim.2019.02.008
Chen, Q., An, X., Li, H., Su, J., Ma, Y., and Zhu, Y. (2016). Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 92-93, 1–10. doi: 10.1016/j.envint.2016.03.026
Chen, M., An, X., Liao, H., Yang, K., Su, J., and Zhu, Y. (2021). Viral community and virus-associated antibiotic resistance genes in soils amended with organic fertilizers. Environ. Sci. Technol. 55, 13881–13890. doi: 10.1021/acs.est.1c03847
Chen, Q., An, X., Zhu, Y., Su, J., Gillings, M. R., Ye, Z., et al. (2017). Application of struvite alters the antibiotic resistome in soil, rhizosphere, and phyllosphere. Environ. Sci. Technol. 51, 8149–8157. doi: 10.1021/acs.est.7b01420
Chen, J., Quiles-Puchalt, N., Chiang, Y. N., Bacigalupe, R., Fillol-Salom, A., Chee, M., et al. (2018). Genome hypermobility by lateral transduction. Science 362, 207–212. doi: 10.1126/science.aat5867
Chen, L., Xun, W., Sun, L., Zhang, N., Shen, Q., and Zhang, R. (2014). Effect of different long-term fertilization regimes on the viral community in an agricultural soil of Southern China. Eur. J. Soil Biol. 62, 121–126. doi: 10.1016/j.ejsobi.2014.03.006
Cobian, G. A., Youle, M., Cantu, V. A., Felts, B., Nulton, J., and Rohwer, F. (2016). Viruses as winners in the game of life. Annu. Rev. Virol. 3, 197–214. doi: 10.1146/annurev-virology-100114-054952
Colomer-Lluch, M., Jofre, J., Muniesa, M., and Aziz, R. (2011). Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One 6:e17549. doi: 10.1371/journal.pone.0017549
Debroas, D., and Siguret, C. (2019). Viruses as key reservoirs of antibiotic resistance genes in the environment. ISME J. 13, 2856–2867. doi: 10.1038/s41396-019-0478-9
Ding, C., Jin, M., Ma, J., Chen, Z., Shen, Z., Yang, D., et al. (2021). Nano-Al2O3 can mediate transduction-like transformation of antibiotic resistance genes in water. J. Hazard. Mater. 405:124224. doi: 10.1016/j.jhazmat.2020.124224
Dinsdale, E. A., Edwards, R. A., Hall, D., Angly, F., Breitbart, M., Brulc, J. M., et al. (2008). Erratum: functional metagenomic profiling of nine biomes. Nature 455, 830. doi: 10.1038/nature07346
Dion, M. B., Oechslin, F., and Moineau, S. (2020). Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 18, 125–138. doi: 10.1038/s41579-019-0311-5
Durán, A. E., Muniesa, M., Méndez, X., Valero, F., Lucena, F., and Jofre, J. (2002). Removal and inactivation of indicator bacteriophages in fresh waters. J. Appl. Microbiol. 92, 338–347. doi: 10.1046/j.1365-2672.2002.01536.x
Emerson, J. B. (2019). Soil viruses: a new hope. mSystems. 4:e00120-19. doi: 10.1128/mSystems.00120-19
Emerson, J. B., Roux, S., Brum, J. R., Bolduc, B., Woodcroft, B. J., Jang, H. B., et al. (2018). Host-linked soil viral ecology along a permafrost thaw gradient. Nat. Microbiol. 3, 870–880. doi: 10.1038/s41564-018-0190-y
Enault, F., Briet, A., Bouteille, L., Roux, S., Sullivan, M. B., and Petit, M. A. (2017). Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 11, 237–247. doi: 10.1038/ismej.2016.90
Fang, H., Wang, H., Cai, L., and Yu, Y. (2015). Prevalence of antibiotic resistance genes and bacterial pathogens in long-term manured. Environ. Sci. Technol. 49, 1095–1104. doi: 10.1021/es504157v
Fillol-Salom, A., Bacigalupe, R., Humphrey, S., Chiang, Y. N., Chen, J., and Penadés, J. R. (2021). Lateral transduction is inherent to the life cycle of the archetypical Salmonella phage P22. Nat. Commun. 12, 6510. doi: 10.1038/s41467-021-26520-4
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O. A., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111. doi: 10.1126/science.1220761
Ghosh, D., Roy, K., Williamson, K. E., White, D. C., Wommack, K. E., Sublette, K. L., et al. (2008). Prevalence of lysogeny among soil bacteria and presence of 16S rRNA andtrzN genes in viral-community DNA. Appl. Environ. Microb. 74, 495–502. doi: 10.1128/AEM.01435-07
Göller, P. C., Haro-Moreno, J. M., Rodriguez-Valera, F., Loessner, M. J., and Gómez-Sanz, E. (2020). Uncovering a hidden diversity: optimized protocols for the extraction of dsDNA bacteriophages from soil. Microbiome 8, 17. doi: 10.1186/s40168-020-0795-2
Gomez, P., and Buckling, A. (2011). Bacteria-phage antagonistic coevolution in soil. Science 332, 106–109. doi: 10.1126/science.1198767
Gonzalez-Martin, C., Teigell-Perez, N., Lyles, M., Valladares, B., and Griffin, D. W. (2013). Epifluorescent direct counts of bacteria and viruses from topsoil of various desert dust storm regions. Res. Microbiol. 164, 17–21. doi: 10.1016/j.resmic.2012.08.009
Guo, Y., Qiu, T., Gao, M., Sun, Y., Cheng, S., Gao, H., et al. (2021). Diversity and abundance of antibiotic resistance genes in rhizosphere soil and endophytes of leafy vegetables: focusing on the effect of the vegetable species. J. Hazard. Mater. 415:125595. doi: 10.1016/j.jhazmat.2021.125595
Haaber, J., Leisner, J. J., Cohn, M. T., Catalan-Moreno, A., Nielsen, J. B., Westh, H., et al. (2016). Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat. Commun. 7:13333. doi: 10.1038/ncomms13333
Han, X., Lv, P., Wang, L., Long, F., Ma, X., Liu, C., et al. (2020). Impact of nano-TiO2 on horizontal transfer of resistance genes mediated by filamentous phage transduction. Environ. Sci. Nano 7, 1214–1224. doi: 10.1039/C9EN01279F
Han, L., Yu, D., Bi, L., Du, S., Silveira, C., Cobián Güemes, A. G., et al. (2022). Distribution of soil viruses across China and their potential role in phosphorous metabolism. Environm. Microbiome 17, 6. doi: 10.1186/s40793-022-00401-9
Harrington, C., Del Casale, A., Kennedy, J., Neve, H., Picton, B. E., Mooij, M. J., et al. (2012). Evidence of bacteriophage-mediated horizontal transfer of bacterial 16S rRNA genes in the viral metagenome of the marine sponge Hymeniacidon perlevis. Microbiology 158, 2789–2795. doi: 10.1099/mic.0.057943-0
He, Y., Yuan, Q., Mathieu, J., Stadler, L., Senehi, N., Sun, R., et al. (2020). Antibiotic resistance genes from livestock waste: occurrence, dissemination, and treatment. npj Clean Water 3, 1–11. doi: 10.1038/s41545-020-0051-0
Helsley, K. R., Brown, T. M., Furlong, K., and Williamson, K. E. (2014). Applications and limitations of tea extract as a virucidal agent to assess the role of phage predation in soils. Biol. Fert. Soils 50, 263–274. doi: 10.1007/s00374-013-0855-x
Hu, H. Y., Liu, H. M., Meng, L., Lan, T., Zheng, N., Cheng, J. B., et al. (2019a). Application of metagenomics in the detection of microbial antibiotic resistance genes. Microbiol. China, 3110–3213. doi: 10.13344/j.microbiol.china.190009
Hu, Y., Yang, X., Qin, J., Lu, N., Cheng, G., Wu, N., et al. (2013). Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 4, 2151. doi: 10.1038/ncomms3151
Hu, X., Yang, B., Zhang, W., Qin, C., Sheng, X., Oleszczuk, P., et al. (2019b). Plasmid binding to metal oxide nanoparticles inhibited lateral transfer of antibiotic resistance genes. Environ. Sci. Nano 6, 1310–1322. doi: 10.1039/c8en01447g
Hu, X. Y., Zhang, Y., Guo, Y. J., Chou, T. L., Gao, M., Sun, X. B., et al. (2022). Comparison in antibiotic resistance genes carried by bacteriophages and bacterial in farmland soil amended with different fertilizers. Biotechnol. Bull. 1985, 2021–1544.
Huijbers, P. M. C., Flach, C., and Larsson, D. G. J. (2019). A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ. Int. 130:104880. doi: 10.1016/j.envint.2019.05.074
Hyder, S. L., and Streitfeld, M. M. (1978). Transfer of erythromycin resistance from clinically isolated lysogenic strains of Streptococcus pyogenes via their endogenous phage. J. Infect. Dis. 138, 281–286. doi: 10.1093/infdis/138.3.281
Iyer, A., Barbour, E., Azhar, E., Qadri, I., Chaudhary, A., Abuzenadah, A., et al. (2013). Bacteriophages in Escherichia coli antimicrobial resistance. Adv. Biosci. Biotechnol. 4, 469–476. doi: 10.4236/abb.2013.43A062
Jebri, S., Rahmani, F., and Hmaied, F. (2021). Bacteriophages as antibiotic resistance genes carriers in agro-food systems. J. Appl. Microbiol. 130, 688–698. doi: 10.1111/jam.14851
Ji, X., Shen, Q., Liu, F., Ma, J., Xu, G., Wang, Y., et al. (2012). Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai. China. J. Hazard. Mater. 235-236, 178–185. doi: 10.1016/j.jhazmat.2012.07.040
Kavagutti, V. S., Andrei, A., Mehrshad, M., Salcher, M. M., and Ghai, R. (2019). Phage-centric ecological interactions in aquatic ecosystems revealed through ultra-deep metagenomics. Microbiome 7, 135. doi: 10.1186/s40168-019-0752-0
Kenzaka, T., Tani, K., and Nasu, M. (2010). High-frequency phage-mediated gene transfer in freshwater environments determined at single-cell level. ISME J. 4, 648–659. doi: 10.1038/ismej.2009.145
Kenzaka, T., Tani, K., Sakotani, A., Yamaguchi, N., and Nasu, M. (2007). High-frequency phage-mediated gene transfer among Escherichia coli cells, determined at the single-cell level. Appl. Environ. Microb. 73, 3291–3299. doi: 10.1128/AEM.02890-06
Larrañaga, O., Brown-Jaque, M., Quirós, P., Gómez-Gómez, C., Blanch, A. R., Rodríguez-Rubio, L., et al. (2018). Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil. Environ. Int. 115, 133–141. doi: 10.1016/j.envint.2018.03.019
Lekunberri, I., Subirats, J., Borrego, C. M., and Balcázar, J. L. (2017). Exploring the contribution of bacteriophages to antibiotic resistance. Environ. Pollut. 220, 981–984. doi: 10.1016/j.envpol.2016.11.059
Lerminiaux, N. A., and Cameron, A. D. S. (2019). Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65, 34–44. doi: 10.1139/cjm-2018-0275
Li, Y., Sun, H., Yang, W., Chen, G., and Xu, H. (2019). Dynamics of bacterial and viral communities in paddy soil with irrigation and urea application. Viruses 11, 347. doi: 10.3390/v11040347
Li, W., and Zhang, G. (2022). Detection and various environmental factors of antibiotic resistance gene horizontal transfer. Environ. Res. 212:113267. doi: 10.1016/j.envres.2022.113267
Lobo, R. R., and Faciola, A. P. (2021). Ruminal phages – a review. Front. Microbiol. 12:763416. doi: 10.3389/fmicb.2021.763416
Maganha De Almeida Kumlien, A. C., Borrego, C. M., and Balcázar, J. L. (2021). Antimicrobial resistance and bacteriophages: An overlooked intersection in water disinfection. Trends Microbiol. (Regular ed.) 29, 517–527. doi: 10.1016/j.tim.2020.12.011
Marti, E., Variatza, E., and Balcazar, J. L. (2014). The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 22, 36–41. doi: 10.1016/j.tim.2013.11.001
Mazaheri, N. F. R., Barton, M. D., and Heuzenroeder, M. W. (2011). Bacteriophage-mediated transduction of antibiotic resistance in enterococci. Lett. Appl. Microbiol. 52, 559–564. doi: 10.1111/j.1472-765X.2011.03043.x
Mishra, S., Klümper, U., Voolaid, V., Berendonk, T. U., and Kneis, D. (2021). Simultaneous estimation of parameters governing the vertical and horizontal transfer of antibiotic resistance genes. Sci. Total Environ. 798:149174. doi: 10.1016/j.scitotenv.2021.149174
Modi, S. R., Lee, H. H., Spina, C. S., and Collins, J. J. (2013). Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222. doi: 10.1038/nature12212
Monaco, C., and Kwon, D. (2017). Next-generation sequencing of the DNA virome from fecal samples. BIO-PROTOCOL. 7:e2159 doi: 10.21769/BioProtoc.2159
Muniesa, M., Colomer-Lluch, M., and Jofre, J. (2013). Potential impact of environmental bacteriophages in spreading antibiotic resistance genes. Future Microbiol. 8, 739–751. doi: 10.2217/fmb.13.32
Murray, C., Ikuta, K. S., Sharara, F., Swetschinski, L., and Aguilar, G. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Musovic, S., Dechesne, A., Sørensen, J., and Smets, B. F. (2010). Novel assay to assess permissiveness of a soil microbial community toward receipt of mobile genetic elements. Appl. Environ. Microb. 76, 4813–4818. doi: 10.1128/AEM.02713-09
Nielsen, K. M., Johnsen, P. J., Bensasson, D., and Daffonchio, D. (2007). Release and persistence of extracellular DNA in the environment. Environ. Biosaf. Res. 6, 37–53. doi: 10.1051/ebr:2007031
Ofir, G., and Sorek, R. (2018). Contemporary phage biology: from classic models to new insights. Cell 172, 1260–1270. doi: 10.1016/j.cell.2017.10.045
Otinov, G. D., Lokteva, A. V., Petrova, A. D., Zinchenko, I. V., Isaeva, M. V., Kovtunov, E. A., et al. (2020). Positive and negative effects of metal oxide nanoparticles on antibiotic resistance genes transfer. Antibiotics. 9, 742. doi: 10.3390/antibiotics9110742
Paez-Espino, D., Eloe-Fadrosh, E. A., Pavlopoulos, G. A., Thomas, A. D., Huntemann, M., Mikhailova, N., et al. (2016). Uncovering Earth’s virome. Nature 536, 425–430. doi: 10.1038/nature19094
Parsley, L. C., Consuegra, E. J., Kakirde, K. S., Land, A. M., Harper, W. F., and Liles, M. R. (2010). Identification of diverse antimicrobial resistance determinants carried on bacterial, plasmid, or viral metagenomes from an activated sludge microbial assemblage. Appl. Environ. Microb. 76, 3753–3757. doi: 10.1128/AEM.03080-09
Penadés, J. R., Chen, J., Quiles-Puchalt, N., Carpena, N., and Novick, R. P. (2014). Bacteriophage-mediated spread of bacterial virulence genes. Curr. Opin. Microbiol. 23, 171–178. doi: 10.1016/j.mib.2014.11.019
Poté, J., Ceccherini, M. T., Van, V. T., Rosselli, W., Wildi, W., Simonet, P., et al. (2003). Fate and transport of antibiotic resistance genes in saturated soil columns. Eur. J. Soil Biol. 39, 65–71. doi: 10.1016/S1164-5563(03)00003-7
Pu, Q., Zhao, L., Li, Y., and Su, J. (2020). Manure fertilization increase antibiotic resistance in soils from typical greenhouse vegetable production bases. China. J. Hazard. Mater. 391:122267. doi: 10.1016/j.jhazmat.2020.122267
Ross, J., and Topp, E. (2015). Abundance of antibiotic resistance genes in bacteriophage following soil fertilization with dairy manure or municipal biosolids, and evidence for potential transduction. Appl. Environ. Microb. 81, 7905–7913. doi: 10.1128/AEM.02363-15
Sander, M., and Schmieger, H. (2001). Method for host-independent detection of generalized transducing bacteriophages in natural habitats. Appl. Environ. Microbiol. 67, 1490–1493. doi: 10.1128/AEM.67.4.1490-1493.2001
Sano, E., Carlson, S., Wegley, L., and Rohwer, F. (2004). Movement of viruses between biomes. Appl. Environ. Microb. 70, 5842–5846. doi: 10.1128/AEM.70.10.5842-5846.2004
Schmieger, H., and Schicklmaier, P. (1999). Transduction of multiple drug resistance of Salmonella enterica serovar typhimurium DT104. FEMS Microbiol. Lett. 170, 251–256. doi: 10.1111/j.1574-6968.1999.tb13381.x
Shousha, A., Awaiwanont, N., Sofka, D., Smulders, F. J. M., Paulsen, P., Szostak, M. P., et al. (2015). Bacteriophages isolated from chicken meat and the horizontal transfer of antimicrobial resistance genes. Appl. Environ. Microb. 81, 4600–4606. doi: 10.1128/AEM.00872-15
Subirats, J., Sànchez-Melsió, A., Borrego, C. M., Balcázar, J. L., and Simonet, P. (2016). Metagenomic analysis reveals that bacteriophages are reservoirs of antibiotic resistance genes. Int. J. Antimicrob. Agents 48, 163–167. doi: 10.1016/j.ijantimicag.2016.04.028
Sun, M., Ye, M., Jiao, W., Feng, Y., Yu, P., Liu, M., et al. (2018). Changes in tetracycline partitioning and bacteria/phage-comediated ARGs in microplastic-contaminated greenhouse soil facilitated by sophorolipid. J. Hazard. Mater. 345, 131–139. doi: 10.1016/j.jhazmat.2017.11.036
Svara, F., and Rankin, D. J. (2011). The evolution of plasmid-carried antibiotic resistance. BMC Evol. Biol. 11, 130. doi: 10.1186/1471-2148-11-130
Trubl, G., Jang, H. B., Roux, S., Emerson, J. B., Solonenko, N., Vik, D. R., et al. (2018). Soil viruses are underexplored players in ecosystem carbon processing. mSystems. 3, e00076–18. doi: 10.1128/mSystems.00076-18
Ubukata, K., Konno, M., and Fujii, R. (1975). Transduction of drug resistance to tetracycline, chloramphenicol, macrolides, lincomycin and clindamycin with phages induced from Streptococcus pyogenes. J. Antibiot. 28, 681–688. doi: 10.7164/antibiotics.28.681
von Wintersdorff, C., Penders, J., van Niekerk, J., Mills, N. D., Majumder, S., van Alphen, L., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173. doi: 10.3389/fmicb.2016.00173
Vos, M., Birkett, P. J., Birch, E., Griffiths, R. I., and Buckling, A. (2009). Local adaptation of bacteriophages to their bacterial hosts in soil. Science 325, 833. doi: 10.1126/science.1174173
Walker, P. J., Siddell, S. G., Lefkowitz, E. J., Mushegian, A. R., Dempsey, D. M., Dutilh, B. E., et al. (2019). Changes to virus taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses. Arch. Virol. 164, 2417–2429. doi: 10.1007/s00705-019-04306-w
Wang, J. Y. (2019). Effect of Fertilization on the Antibiotic Resistance Genes in Soil Bacteria and Bacteriophages[D]. Beijing: University of Chinese Academy of Science. (in Chinese)
Wang, Q., Lu, Q., Mao, D., Cui, Y., and Luo, Y. (2015a). The horizontal transfer of antibiotic resistance genes is enhanced by ionic liquid with different structure of varying alkyl chain length. Front. Microbiol. 6:864. doi: 10.3389/fmicb.2015.00864
Wang, Q., Mao, D., Mu, Q., and Luo, Y. (2015b). Enhanced horizontal transfer of antibiotic resistance genes in freshwater microcosms induced by an ionic liquid. PLoS One 10:e126784. doi: 10.1371/journal.pone.0126784
Watson, B., Staals, R., and Fineran, P. C. (2018). CRISPR-Cas-mediated phage resistance enhances horizontal gene transfer by transduction. MBio 9, e02406-17. doi: 10.1128/mBio.02406-17
Williamson, K. E., Fuhrmann, J. J., Wommack, K. E., and Radosevich, M. (2017). Viruses in soil ecosystems: an unknown quantity within an unexplored territory. Annu. Rev. Virol. 4, 201–219. doi: 10.1146/annurev-virology-101416-041639
Williamson, K. E., Radosevich, M., Smith, D. W., and Wommack, K. E. (2007). Incidence of lysogeny within temperate and extreme soil environments. Environ. Microbiol. 9, 2563–2574. doi: 10.1111/j.1462-2920.2007.01374.x
Williamson, K. E., Radosevich, M., and Wommack, K. E. (2005). Abundance and diversity of viruses in six Delaware soils. Appl. Environ. Microb. 71, 3119–3125. doi: 10.1128/AEM.71.6.3119-3125.2005
Wright, G. D., and Poinar, H. (2012). Antibiotic resistance is ancient: implications for drug discovery. Trends Microbiol. 20, 157–159. doi: 10.1016/j.tim.2012.01.002
Xiao, X., Ma, X., Han, X., Wu, L., Liu, C., and Yu, H. (2021). TiO2 photoexcitation promoted horizontal transfer of resistance genes mediated by phage transduction. Sci. Total Environ. 760:144040. doi: 10.1016/j.scitotenv.2020.144040
Xu, L., Gu, J., Wang, X., Song, Z., Jiang, H., Li, N., et al. (2022). Risk of horizontal transfer of intracellular, extracellular, and bacteriophage antibiotic resistance genes during anaerobic digestion of cow manure. Bioresource Technol. 351:127007. doi: 10.1016/j.biortech.2022.127007
Yang, J., Wang, C., Shu, C., Liu, L., Geng, J., Hu, S., et al. (2013). Marine sediment bacteria harbor antibiotic resistance genes highly similar to those found in human pathogens. Microb. Ecol. 65, 975–981. doi: 10.1007/s00248-013-0187-2
Yang, Y., Xing, S., Chen, Y., Wu, R., Wu, Y., Wang, Y., et al. (2021). Profiles of bacteria/phage-comediated ARGs in pig farm wastewater treatment plants in China: association with mobile genetic elements, bacterial communities and environmental factors. J. Hazard. Mater. 404:124149. doi: 10.1016/j.jhazmat.2020.124149
Ye, M., Sun, M., Huang, D., Zhang, Z., Zhang, H., Zhang, S., et al. (2019). A review of bacteriophage therapy for pathogenic bacteria inactivation in the soil environment. Environ. Int. 129, 488–496. doi: 10.1016/j.envint.2019.05.062
Zablocki, O., Adriaenssens, E. M., and Cowan, D. (2016). Diversity and ecology of viruses in hyperarid desert soils. Appl. Environ. Microb. 82, 770–777. doi: 10.1128/AEM.02651-15
Zeph, L. R., Onaga, M. A., and Stotzky, G. (1988). Transduction of Escherichia coli by bacteriophage P1 in soil. Appl. Environ. Microb. 54, 1731–1737. doi: 10.1128/AEM.54.7.1731-1737.1988
Zhang, A., Gaston, J. M., Dai, C. L., Zhao, S., Poyet, M., Groussin, M., et al. (2021). An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nat. Commun. 12, 4765–4711. doi: 10.1038/s41467-021-25096-3
Zhang, Y., Gu, A. Z., He, M., Li, D., and Chen, J. (2017). Subinhibitory concentrations of disinfectants promote the horizontal transfer of multidrug resistance genes within and across genera. Environ. Sci. Technol. 51, 570–580. doi: 10.1021/acs.est.6b03132
Zhang, Y., Hu, H., Chen, Q., Singh, B. K., Yan, H., Chen, D., et al. (2019). Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 130:104912. doi: 10.1016/j.envint.2019.104912
Zhao, Y., Ye, M., Zhang, X., Sun, M., Zhang, Z., Chao, H., et al. (2019). Comparing polyvalent bacteriophage and bacteriophage cocktails for controlling antibiotic-resistant bacteria in soil-plant system. Sci. Total Environ. 657, 918–925. doi: 10.1016/j.scitotenv.2018.11.457
Keywords: bacteriophages, antibiotic resistance genes, mobile antibiotic resistome, horizontal gene transfer, soil
Citation: Zhang Y, Guo Y, Qiu T, Gao M and Wang X (2022) Bacteriophages: Underestimated vehicles of antibiotic resistance genes in the soil. Front. Microbiol. 13:936267. doi: 10.3389/fmicb.2022.936267
Received: 05 May 2022; Accepted: 04 July 2022;
Published: 04 August 2022.
Edited by:
Hanpeng Liao, Fujian Agriculture and Forestry University, ChinaReviewed by:
Yanan Wang, Henan Agricultural University, ChinaCopyright © 2022 Zhang, Guo, Qiu, Gao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuming Wang, d2FuZ3h1bWluZ0BiYWFmcy5uZXQuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.