- 1Department of Plant Protection, College of Horticulture and Plant Protection, Yangzhou University, Yangzhou, China
- 2Jiangsu Key Laboratory for Microbes and Functional Genomics, Jiangsu Engineering and Technology Research Center for Microbiology, College of Life Sciences, Nanjing Normal University, Nanjing, China
Phosphorylation is one of the most extensively investigated post-translational modifications that orchestrate a variety of cellular signal transduction processes. The phosphorylation of virus-encoded proteins plays an important regulatory role in the infection cycle of such viruses in plants. In recent years, molecular mechanisms underlying the phosphorylation of plant viral proteins have been widely studied. Based on recent publications, our study summarizes the phosphorylation analyses of plant viral proteins and categorizes their effects on biological functions according to the viral life cycle. This review provides a theoretical basis for elucidating the molecular mechanisms of viral infection. Furthermore, it deepens our understanding of the biological functions of phosphorylation in the interactions between plants and viruses.
Introduction
Rapid responses to internal and external cues are vital for complex life on the earth. Whether to maintain optimal conditions for growth or to avoid biotic/abiotic stresses, these responses must occur on a timescale that affords the organisms an obvious survival advantage. Cells have evolved sophisticated regulatory systems that can sense, transmit, sort, store, and interpret information that could enact a coordinated and timely response to the changes in environments. Post-translational modification (PTM) is an elegant transducer of the signals for adaptations of living organisms by the reversible and rapid nature, relatively small expenses, and profoundly function modulation of the target protein. Examples of PTM include phosphorylation (Ramazi and Zahiri, 2021), glycosylation (Vigerust and Shepherd, 2007), ubiquitination (Shen et al., 2016), acetylation (Diallo et al., 2019), lipidization (Chiang et al., 2013), small ubiquitin-like modifier (SUMOylation) (Cheng et al., 2017), and lysine succinylation (Zhang et al., 2011), butyrylation (Chen et al., 2007), or crotonylation (Wan et al., 2019). Phosphorylation is the most intensively investigated PTM and is involved in almost every cellular process (Cohen, 2002; Duan and Walther, 2015). Numerous metabolic enzymes, such as pyruvate dehydrogenase and glycogen synthase, occur in the phosphorylation PTM (Fischer and Krebs, 1955). The mechanisms by which phosphorylation controls protein functions include modulation of protein–protein interactions and changes in protein conformation. Phosphorylation often functions as a docking site for intra- or inter-molecular protein interactions via protein conformation changes, and directly modulates enzymatic activity, regulates subcellular localization, targets turnover, and affects signaling transduction by other PTMs (Figure 1). Overall, the phosphorylation by kinases and de-phosphorylation by phosphatases serve as low-cost and high-efficiency molecular regulators that can alter the behavior of the targets directly or indirectly.
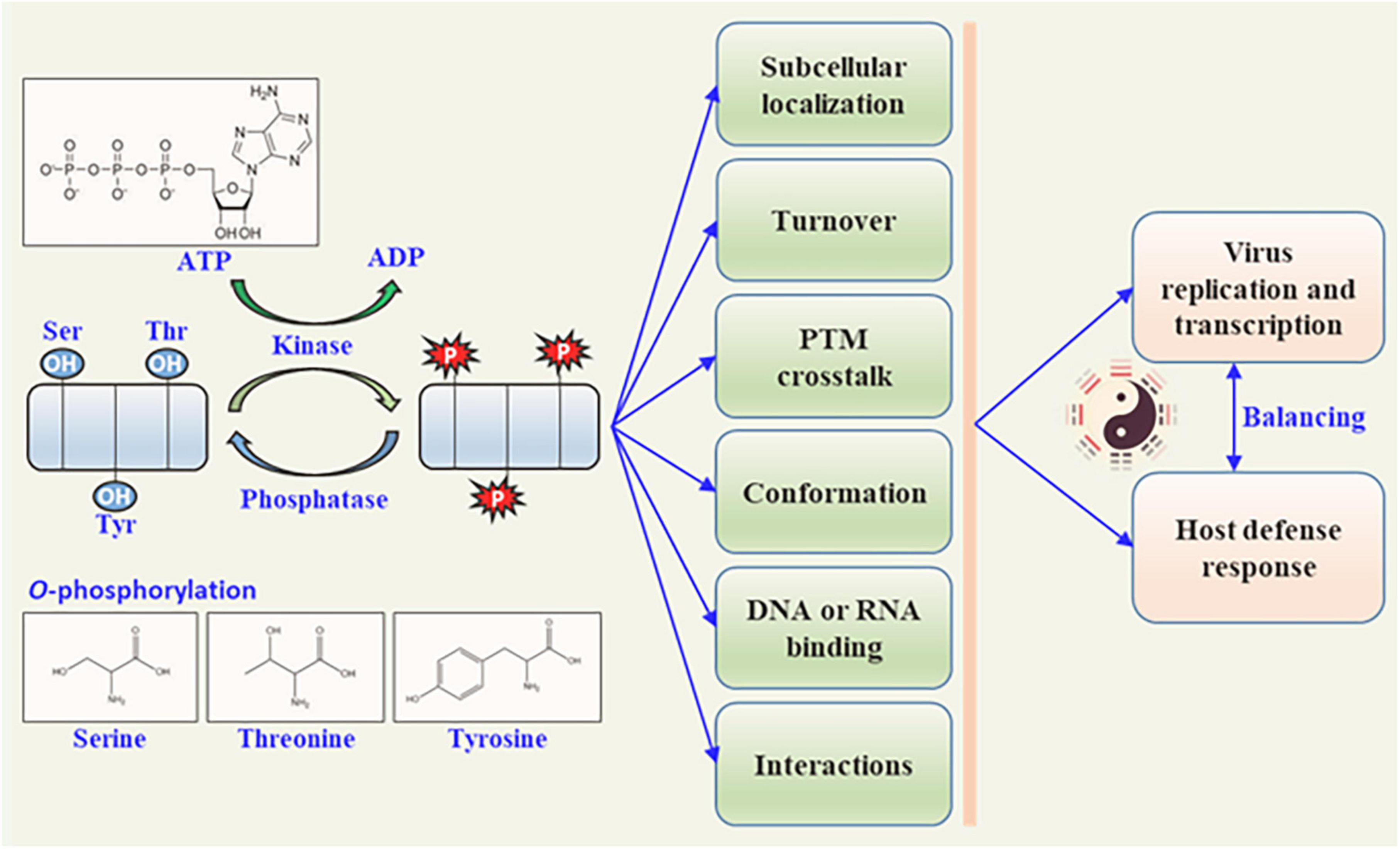
Figure 1. Biological function of plant viral protein phosphorylation. Protein kinases phosphorylate plant viral proteins to regulate the viral protein activity by phosphorylation and dephosphorylation (catalyzed by phosphatases). The functions of viral proteins are altered in different ways, including subcellular location, pathogenicity, intrinsic biological activity, DNA or RNA binding activity, self-interaction, and interaction with other proteins. These functional changes in plant viral proteins further affect viral replication or transcription, and the host defense responses.
There are four types of phosphorylations, the first being O-phosphorylation, which mainly occurs at the hydroxyl residues of serine (Ser), threonine (Thr), tyrosine (Tyr), and hydroxyproline (Emmer et al., 2013). The second is N-phosphorylation, which occurs on histidine and lysine amino residues (Ni et al., 2009). The third is S-phosphorylation, which occurs in the sulfhydryl group of cysteine (Buchowiecka, 2014). The last type is Acyl-phosphorylation, which is mainly observed in the acyl groups of aspartic acid and glutamate (Cheng et al., 1996). O-phosphorylation is the most common type of phosphorylation in eukaryotes (Panni, 2019). Heibeck et al. (2009) showed that the relative abundance ratio of three phosphorylated amino acids (AA) (pSer : pThr : pTyr) in a single cell is 1800 : 200 : 1, indicating that Ser phosphorylation is the most common modification (Heibeck et al., 2009). In eukaryotes, O-phosphorylation is catalyzed by protein kinases, which function by adding the γ-phosphate group of ATP or GTP to the hydroxyl side chain of Ser, Thr, Tyr, and hydroxyproline (Krupa et al., 2004). Different protein kinases can recognize and modify different sites of the target protein. Phosphorylated proteins can also be dephosphorylated by phosphatases, which reduce the phosphate group from the side chain of the target protein (Millar et al., 2019). These dynamic and reversible PTM changes are energy-consuming processes that require coordination between the protein kinase and phosphatase to regulate the folding style and conformation of the target protein (Jakubiec and Jupin, 2007). The reversible processes of phosphorylation and dephosphorylation act as an “ON-OFF SWITCH”-like modulator of their functions, such as regulating enzymatic activity, protein stability, protein virulence, subcellular localization of target proteins, signaling transduction and recognition, and interaction with other proteins (Friso and van Wijk, 2015).
In plant virology, studies on the phosphorylation modification of viral proteins have recently progressed. The regulatory function of phosphorylation is involved in most steps of the plant viral life cycle, including protein translation (Martínez-Turiño et al., 2018), replication of viral genomic materials (Gao et al., 2020), intercellular movement (Nemes et al., 2017), cell-to-cell movement (Hung et al., 2014), virion assembly (Nemes et al., 2019), and long-distance movement (Zhao et al., 2015). Although several viral proteins have been extensively studied, the molecular mechanisms of their action in viral infection have not been elucidated. A lack of unified, standard, and high-efficiency phosphorylation detection methods and detailed phosphorylation site identification still remains. In addition, the biological functions of phosphorylated viral proteins are distinct and independent for each plant virus studied. In this review, the functional phosphorylation analyses that were not within the scope of the plant virus life cycle and, accordingly, the relevant studies have not been included.
A clear understanding of the phosphorylation status of plant viral proteins in different environments would be helpful to further analyze the biological functions of phosphorylation. Hence, summarizing the methods for identifying the phosphorylation of plant viral proteins and the biological functions of phosphorylated plant viral proteins is urgently needed to gain a deeper understanding of the multiple functions of viral proteins during infection. This study aims to provide a reference for further study of the molecular mechanisms of plant viral protein phosphorylation and underlines the significance of the PTM in virus infection cycles.
Common assays for identification of phosphorylated plant viral proteins
The online services, such as Kinasephos1 and Netphos 3.12, can accurately predict the phosphorylation sites of plant virus proteins and the corresponding kinases involved (Zhang et al., 2018). Furthermore, various methods, including liquid chromatography-mass spectrometry (LC-MS/MS) (Gao et al., 2020) and radioisotope labeling ([γ-32P]ATP) experiments in vivo and in vitro, have been widely used for the identification of viral protein phosphorylation (Hu et al., 2015). Selective protein kinase inhibitor screening (Zhang et al., 2018), commercial pSer/pThr/pTyr phosphorylation-specific antibodies (Li et al., 2022), and commercial Phos-tag™ gel-based immunoblotting have been widely used for the identification of the phosphorylation of viral proteins. Here, we summarize the recently published phosphorylation identification assays for plant viral proteins, which include identification of the phosphorylation of target proteins and the detailed phosphorylation sites on target proteins, in order to provide a basis and reference for further studies.
Liquid chromatography-mass spectrometry-based identification of phosphorylated plant viral proteins
The continuous and rapid development of mass spectrometry and computer-based bioinformatics have improved the feasibility of research on viral protein PTMs and the interactions between plant viruses and hosts. Trypsinized protein samples are ionized and analyzed via mass spectrometry using the time-of-flight under an electric or magnetic field (Domon and Aebersold, 2006). The detector reports the relative intensity and mass-to-charge ratio of each peptide or signal. Based on the extra 79.966 KDa difference between the phosphorylated and unphosphorylated peptides under the condition of one potential phosphorylation site, AA sequences of the target peptide, and other fragmented ion signals from secondary mass spectrometry, phosphorylation can be easily identified with the bioinformatics software available nowadays (Zhang G. et al., 2014).
Various strategies have emerged for enriching phosphorylated peptides or proteins to improve the sensitivity and validity of phosphorylation assays. These approaches increase the abundance of phosphorylated peptides or proteins in LC-MS/MS assays. These enrichment methods include pSer/pThr/pTyr phosphorylation-specific antibody-based affinity capture (Stoevesandt and Taussig, 2013), chemical derivatization of phosphorylated AAs (Deng et al., 2007), metal ion-based affinity capture (Blacken et al., 2008), ion-exchange chromatography (Jungbauer and Hahn, 2009), and classical immunoprecipitation technology using phosphorylation-specific antibodies (Kurosaki et al., 2019). The enriched proteins are hydrolyzed to form peptides via trypsin digestion in vitro, and LC-MS/MS analyses are performed to detect the phosphorylated sites. Various phosphorylation sites can be identified using conventional LC-MS/MS analysis combined with the appropriate software. However, it is not sufficient to show that the target protein is phosphorylated, and additional phosphorylation analyses are necessary to further confirm the results.
Radioisotope (32P) labeling of plant viral proteins in vitro
Under kinase catalysis, the radioisotope 32P-labeled ATP that acts as an energy source (γ-position phosphate group) is transferred to the substrate protein by kinase (Lamberti et al., 2011). In the study by Lamberti et al. (2011), the corresponding proteins were separated via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) after in vitro catalytic reactions were completed. The gel was dried using a Model 583 Gel Dryer (Bio-Rad, Hercules, CA, United States), and the phosphorylated proteins were detected using X-autoradiography or a phosphorous storage screen (BAS-IP TR 2040 E Tritium Screen, GE Healthcare, Chicago, IL, United States). This method is the most direct protein phosphorylation detection assay in vitro. It has the advantages of high sensitivity and accuracy and has been widely applied in the identification of plant viral protein phosphorylation. Using this method, the phosphorylation of a single protein was proven in vitro, and the detailed AA sequences of the substrate protein and catalytic kinase were validated. However, this is a hazardous procedure for operators, especially for first-time and inexperienced researchers.
The specific steps for in vitro phosphorylation experiment are as follows. A 20 μL phosphorylation reaction system is prepared with 20 mmol.L–1 Tris-HCl buffer (pH = 7.5), 0.1 μg kinase/total protein extractions, 1.0 μg substrate (purified protein), 25 μmol.L–1 ATP, 1 μCi [γ-32P]-ATP (China Isotope & Radiation Corporation, Beijing, China), 5 mmol.L–1 MgCl2 (or MnCl2, CaCl2, etc.), and ultrapure water. After incubation at 30°C for 30 min, 5 μL of 2 × SDS loading buffer [100 mM Tris–HCl, pH = 6.8, 4% SDS (v/v), 20% glycerol (v/v), 0.2% bromophenol blue (v/v), and 5% β-mercaptoethanol (v/v)] are added. The reaction is terminated by denaturation at 95°C for 5 min. All proteins in the reaction system are then separated via 12.5% SDS-PAGE for 2 h at 80 V. The gel is placed on a filter paper (8 cm × 10 cm), and dried in a gel dryer for 40 min. The dried gel is exposed to X-rays and detected using a phosphorous storage screen or film (Kodak X-OMAT BT, Nanjing, China) (Hu et al., 2015). Specific antibodies or phosphorylated antibodies can also be used to detect the target substrate proteins via western blotting.
Phos-tag™ sodium dodecyl sulfate polyacrylamide gel electrophoresis for identification of phosphorylated plant viral protein
Phos-tag™ reagent can specifically bind to the phosphate group on the phosphorylated protein after binding with Mn2+ or Zn2+ to form a relatively stable compound (Kinoshita-Kikuta et al., 2014). When Mn2+ or Zn2+ and Phos-tag™ are added to a conventional SDS-polyacrylamide gel, the phosphorylated protein binds to the Phos-tag™. It forms a larger complex than the unphosphorylated protein, leading to a significant reduction in the electrophoretic migration rate of the phosphorylated protein in the gel under electrophoretic conditions (Kinoshita et al., 2006). Therefore, the Phos-tag™ reagent can specifically separate the phosphorylated proteins from non-phosphorylated proteins. Specific or phosphorylated antibodies are usually combined with a Phos-tag™ for western blotting.
This simple method can be used to detect all types of phosphorylation in vivo and in vitro. The Phos-tag™ reagent in SDS-polyacrylamide gel is generally twice the concentration used in most experiments, often at a concentration of 20–100 μM (Nagy et al., 2018). The optimal concentration of the Phos-tag™ reagent used for phosphorylation assays should be determined based on the actual characteristics of the specific protein in the experiment. Combined with the western blotting and LC-MS/MS, the Phos-tag™ reagent could be used for protein purification in some situations. However, it often has low accuracy in protein phosphorylation assays used for the preliminary determination of protein phosphorylation. Further identification of protein phosphorylation usually requires additional evidence, such as western blotting or LC-MS/MS.
Western blotting for identification of phosphorylated plant viral proteins
The target protein or system containing the target protein is electrophoresed via SDS-PAGE at 80 V for 2 h and then transferred onto a piece of nitrocellulose membranes (Bio-Rad, United States). Specific antibodies with high affinity for the target protein or phosphorylated Ser/Thr/Tyr are used to detect the target protein (Mei et al., 2018a). Secondary antibodies are cross-linked with the horseradish peroxidase or alkaline phosphate. Based on the specific binding activity of the antibody to the corresponding antigen, only the phosphorylated target protein can be detected, as evidenced by the strong bands or signals at specific positions on the membrane (Li et al., 2022).
The western blot assay is relatively easy to perform and has no equipment or location restrictions. Antigen–antibody recognition of the phosphorylation site is specific and has high resolution in western blotting. It is often used for the validation of LC-MS/MS results in phosphorylation assays. However, this method is semi-quantitative, it is difficult to identify new or multiple phosphorylation sites in a single protein because of the problems in preparing the corresponding specific phosphorylation antibody.
Common kinases that phosphorylate plant viral proteins
Phosphorylation/dephosphorylation is involved in multiple biological processes that regulate plant growth, development, and aging, such as plant biotic/abiotic stresses, hormone signal transduction, ion transportation, and material metabolism (Yin et al., 2018). Different kinases often mediate different types of phosphorylation in the target protein. An increasing number of studies indicate that phosphorylation plays an important role in regulating viral genome replication, viral intracellular/intercellular movement, viral gene transcription, virus infectivity to the host plant, defense and counter-defense of plant innate immunity, and virion assembly.
Understanding the recognition mechanisms and biological functions of kinases and their substrates is essential. The surrounding phosphorylated AAs (Tyr/Ser/Thr) can be specifically recognized by the modular phosphoprotein-binding domains of kinases, called p-motifs. van Wijk et al. (2014) have explored the certain kind of kinases that phosphoprotein of the p-motifs (van Wijk et al., 2014). Several algorithms, such as PTMphinder, Motif-x, and Motif-All, have been developed to predict the relatively conserved motifs in phosphopeptides, based on the concept of p-motifs. The obvious phosphorylation motifs were categorized into two major types: pS type and pT types. The pS type was clustered into seven clades, including glutamate-rich, glycine-rich, DS with acidic residues downstream or basic residues upstream, SP motif, pS with acid residues, and serine-rich pS motif. The pT types were also divided into seven clades, namely, aspartate-rich, glutamate-rich, proline-rich (2 X), arginine-rich (2 X), and lysine-rich pT (van Wijk et al., 2014). The acidic S-type motifs included S-[DE], S-E, S-X-[DE], and S-X-X-[DE], and the common basic S-type motifs comprised [RK]-X-X-S, R-S, and [RK]-X-S. T-P is the most common motif in the pT motif. Arginine was often present in the basic T-type motif ([RK]-X-X-T), while aspartate or glutamate was included in the acidic T-type motifs (X-T-X-[ED]). Annotation from the PhosPhAt 4.0 database and the known-target relationships were obtained for the p-motifs identified by Li et al. (2022) in common substrates of different kinase families. This analysis revealed that one-third of the identified p-motifs in the substrate of MAPK were T/S-P (Pitzschke, 2015; Rayapuram et al., 2018). CDKs are tightly associated substrates that contained the S-E, S-[DE], and S-X-[DE] motifs (Xi et al., 2021). The basic S-motif was over-represented in the substrates of SnRK1, SnRK2, SnRK3, and LRR-RK. In the T-motifs group, T-P was found in substrates of multiple kinase families, such as CKII, CDK, CDPK, SnRK1, and SnRK3 (Xi et al., 2021). The existing database and the corresponding relationship between substrates and kinases contribute to the prediction of target kinases that are capable of phosphorylating proteins containing the specific p-motifs.
Rapid and directed identification of hundreds of p-motifs via advanced mass-spectrometry and faster bioinformatics techniques has become feasible (Amanchy et al., 2007). Several phosphorylation prediction databases based on these accumulated datasets have based customized retrieval (Gao et al., 2009; Cheng et al., 2014; Schulze et al., 2015; Millar et al., 2019), thereby allowing the prediction of a possible optimal kinases type. In this study, we summarized the identification methods used in published studies. According to the operations, these methods are divided into three types. The first method involves comparative LC-MS/MS and the kinase-interaction assay, which can identify the corresponding kinase that can phosphorylate and directly interact with the target substrate (Figure 2A; Gil et al., 2019). The second method is high-throughput phosphoproteomics analyses, which can be applied where the corresponding kinase can phosphorylate but not interact with the target substrate (Figure 2B; Poss et al., 2016). The third method is the use of in vitro methods for validation of phosphorylation (Figure 2C; Lipton et al., 2015). Generally, the phosphorylation reaction in vitro (Figure 2C) is often combined with the top two analysis methods (Figures 2A,B) to further validate the substrate and the corresponding kinase. An in vitro phosphorylation reaction system or a cell lysis reaction should contain a phosphatase inhibitor.
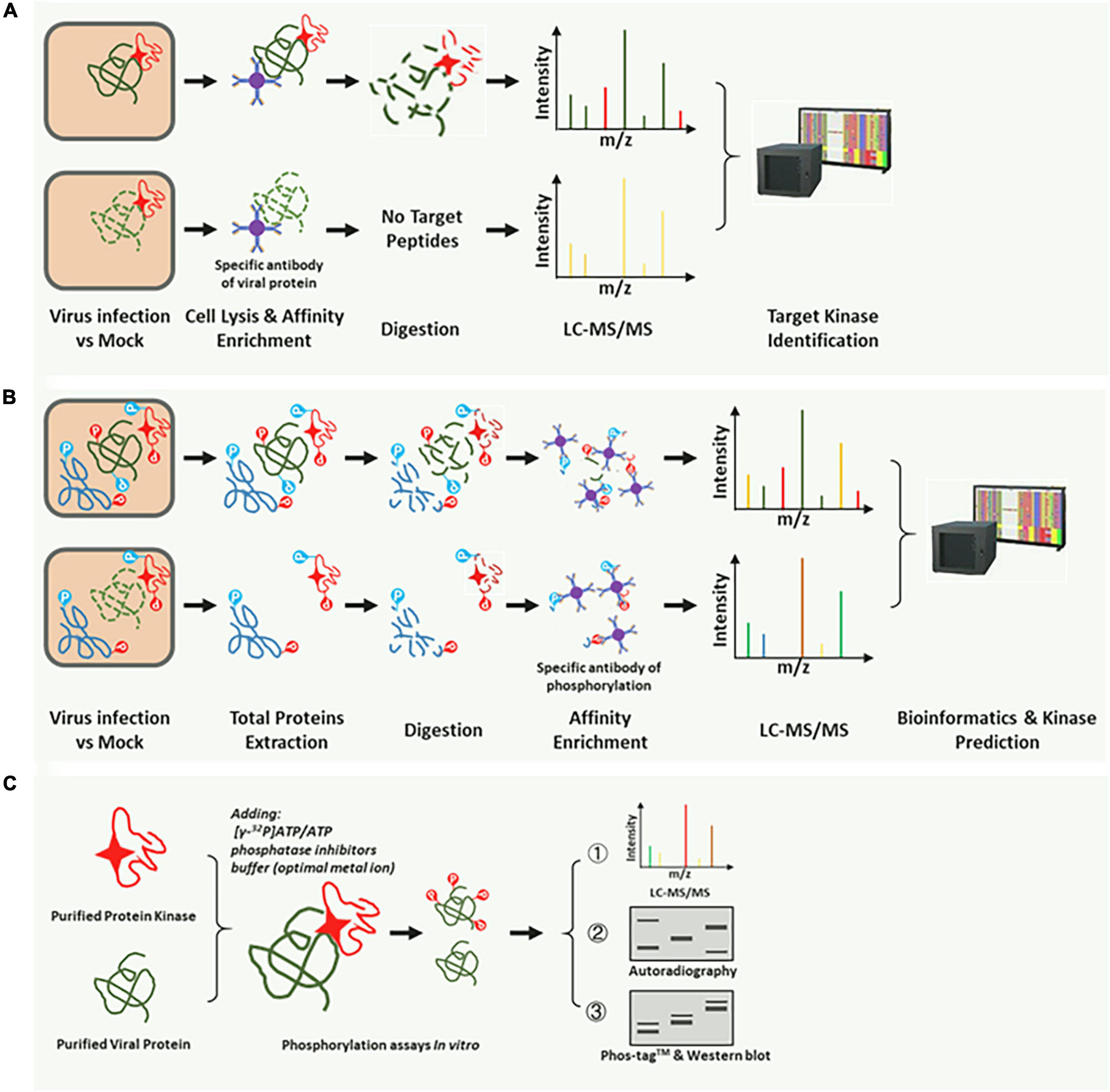
Figure 2. Common methods for identification of the corresponding kinase of the target substrate. (A) The target substrate interacts with and is phosphorylated by a specific kinase. A substrate-specific antibody was used for co-immunoprecipitation (Co-IP) of the corresponding kinase. The kinase could be identified by the LC-MS, and BLASTP was performed on the NCBI or the species-specific database. (B) High-throughput phosphoproteomic analysis based on the LC-MS/MS. The p-motifs could be determined using phosphor-peptides and interrogation of the corresponding database. Then, the kinase type could be identified using the already phosphorylation mode in the database. Using the third method (C), a specific kinase can be confirmed. (C) In vitro phosphorylation reaction system for further validation of the target kinase. Radioisotope (32P)-labeled ATP, Phos-tag™ SDS-PAGE, and pS/pT-specific antibodies were used in the reaction, and specific kinases were easily identified.
Previous investigations have shown that a variety of plant kinases can phosphorylate the proteins encoded by plant viruses, including the casein kinase family (CKs) (Martínez-Turiño et al., 2018), glycogen synthase kinase-3 (GSK-3) (Mei et al., 2018a), sucrose non-fermenting-1-related protein kinase (SnRK) (Shen et al., 2018), protein kinase A (PKA) (Makarov et al., 2012), and protein kinase C (PKC) (Samuilova et al., 2013). We have summarized these publications and listed the common motifs of phosphorylated substrates of these different protein kinases in Table 1. This summary will provide information to help identify common kinases in phosphorylation assays. In particular, by combining the in vitro and in vivo phosphorylation reaction experiments, the corresponding kinase or target phosphorylation site can be quickly confirmed.
Casein kinase family
Protein kinase 1 (CK1) is a highly conserved serine/threonine kinase in eukaryotes. Its N-terminal catalytic domain is relatively conserved and functions in the recognition of substrate proteins (Gross and Anderson, 1998). The C-terminal domain of CK1 has low conservation in different species and is responsible for the binding specificity of different substrate proteins (Graves et al., 1993; Graves and Roach, 1995; Cegielska et al., 1998). CK1 has been widely studied in both yeasts and animals (Knippschild et al., 2005). To date, seven CK1 types have been identified in mammals that are involved in the regulation of vesicle transport, growth, development, morphological changes, DNA repair, and cytokinesis. CK1 participates in signal transduction and hormone metabolism in rice (Liu et al., 2003; Dai and Xue, 2010). In Arabidopsis, CK1 regulates blue-light signaling and ethylene biosynthesis through phosphorylation (Tan and Xue, 2014). Additionally, the Arabidopsis genome encodes at least 14 CK1-like (CKL) protein kinases located in the cytoplasm, nucleus, endoplasmic reticulum, and vesicular granular structure and participates in the regulation of multiple biological processes (Lee, 2009).
Protein kinase 2 (CK2) phosphorylates various plant viral proteins and plays an important role in the regulation of viral infection and host defense. CK2 is a highly conserved serine/threonine-specific kinase with multiple physiological functions in eukaryotes (Mulekar and Huq, 2014). The CK2 holoenzyme comprises two α-catalytic subunits and two β-regulatory subunits that constitute heterotetramers in vivo (Litchfield, 2003). In plants, the core CK2α catalytic subunits can play an independent role in the phosphorylation of target proteins (Filhol et al., 2004). Most CK2 proteins belong to a multigene family in plants. CK2 homologous genes have been cloned from various plants, including Nicotiana attenuata, wheat, corn, rice, and barley (Mulekar and Huq, 2014). CK2 is involved in the regulation of plant growth, development, biotic/abiotic stresses, light signals, and circadian rhythms (Huq, 2006; Moreno-Romero et al., 2008; Nagel and Kay, 2012; Mulekar and Huq, 2014). CK2-mediated phosphorylation of substrate proteins usually leads to changes in protein quaternary structure and conformation, which results in changes in DNA binding, RNA-binding, dimerization, stability, protein–protein interactions, and subcellular localization (Mulekar and Huq, 2014).
Glycogen synthesis kinase-3
Glycogen synthesis kinase-3 (GSK3) is a serine/threonine phosphokinase that participates in all biological processes (Kim and Kimmel, 2000). In mammals, GSK3 regulates cell metabolism, signal transduction, embryonic development, and neuronal differentiation (Doble and Woodgett, 2003; Kim and Kimmel, 2006; Hur and Zhou, 2010; Wu and Pan, 2010). Most protein substrates of GSK3 contain a common phosphorylation motif (Ser/Thr-x-x-Ser/Thr) (Fiol et al., 1990; Pinna and Ruzzene, 1996; Table 1). Various GSK3-like kinases are also present in plants. Compared with animal GSK3, plant GSK genes are more diverse (Yoo et al., 2006). Phosphorylation of the C-terminal serine/threonine residue of GSK3 is catalyzed by another kinase, which promotes its catalytic activity in animals (Thomas et al., 1999). In plants, the GSK3-like kinases tend to interact directly with the substrate proteins and phosphorylate them. GSK3-like kinases play different roles in cell growth, root and stomatal cell development, flowering, and fruiting (Youn and Kim, 2015).
Protein kinase A and protein kinase C
Protein kinase A (PKA) is an enzyme family whose catalytic activity depends on the levels of cyclic adenylate (cAMP) (Gold, 2019). It is a serine/threonine phosphokinase that is involved in the regulation of multiple biological processes, including cell proliferation and glycogen, and lipid metabolism (Endicott et al., 2012). PKA is a tetramer comprising two regulatory and catalytic subunits (Zelada et al., 2002). In eukaryotic cells, cAMP activates PKA, which in turn phosphorylates the target substrate proteins. The activated PKA catalytic subunit can phosphorylate the serine or threonine residues of target proteins in cells, leading to changes in the activity, which further affects the expression of related genes (Yang, 2018). Although PKA has not been identified in plants thus far, plant virologists have found that the PKA-like kinases, which phosphorylate plant viral proteins and regulate their functions in the viral life cycle, are common in plants.
Protein kinase C is also a serine/threonine phosphokinase, composed of an N-terminal regulatory region (approximately 20—40 kDa) and a C-terminal catalytic region (approximately 45 kDa) (Newton, 1995). It regulates the functions of the target protein in vivo by phosphorylating the hydroxyl groups and adding a phosphate group to the serine/threonine side chains (Lim et al., 2015). PKC can be divided into three subtypes according to its structural motifs and activation conditions (Keenan et al., 1997). It plays an important role in apoptosis and immune gene expression in mammals (Newton, 2018).
Functions of phosphorylated plant viral protein
An increasing number of recent studies have revealed that plant viral proteins, including coat protein (CP), movement protein (MP), RNA-dependent RNA polymerase (RdRp), and viral suppressors of RNA silencing (VSRs), undergo phosphorylation during viral infection. The phosphorylation state of viral proteins is important for the stable infection of the virus in the host plant. In vivo, phosphorylation can regulate the interaction between large and small subunits of replicase, viral genomic RNA or DNA catalytic synthesis activity of replicase, stability of the viral replication complex (VRC), the interaction between viral proteins and host factors, and the binding of viral protein to RNA/DNA. This study summarizes the research on the biological functions of phosphorylated plant viral proteins (Figures 3, 4) from the perspective of the life cycle of plant viruses in plant viruses. Our research provides a reference for further plant viral protein phosphoproteomics research and provides a theoretical basis for plant viral disease prevention and control.
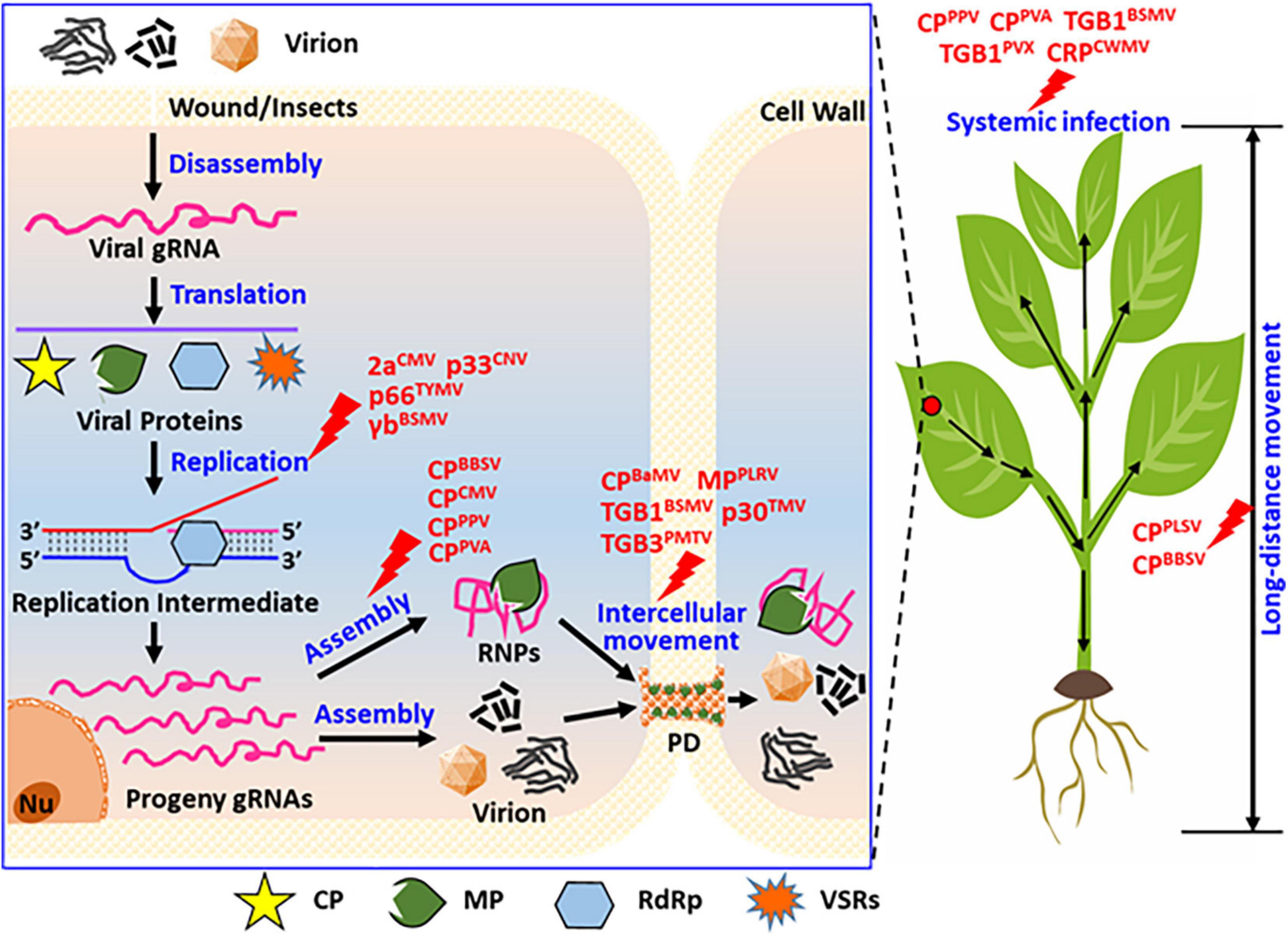
Figure 3. Summary of the functions of phosphorylated proteins encoded by positive single-strand RNA viruses during their life cycle. In view of the entire cell and whole plants, viral protein phosphorylation affects the replication, virion assembly, intracellular movement, long-distance movement, and systemic infection of the plants. As displayed, phosphorylation modification affected virus replication, for instance, the replicase of CMV (2a), CNV (p33), TYMV (p66), and the suppressor of BSMV (γb), these proteins not all are replicase, also contains virus-encoded RNA silencing suppressor. Phosphorylation modification of CP affected virion assemblies, such as the coat protein of BBSV, CMV, PPV, and PVA. Phosphorylation modification of viral proteins affected virus intercellular movement, such as movement protein of PLRV (MP), BSMV (TGB1), TMV (p30), PMTV (TGB3), also contains the coat protein of BaMV (CP). Phosphorylation modification of viral proteins affected virus long-distance movement, such as coat protein of PLSV (CPPLSV) and BBSV (CPBBSV). Phosphorylation modification of viral proteins affected virus systemic infection of host plant, such as coat protein of PPV (CPPPV), PVA (CPPVA), and movement protein of BSMV (TGB1BSMV), PVX (TGB1PVX), and the cysteine-rich protein of CWMV (CRPCWMV). These viruses display rod-shaped, filamentous, and spherical particles under electron-microscopy.
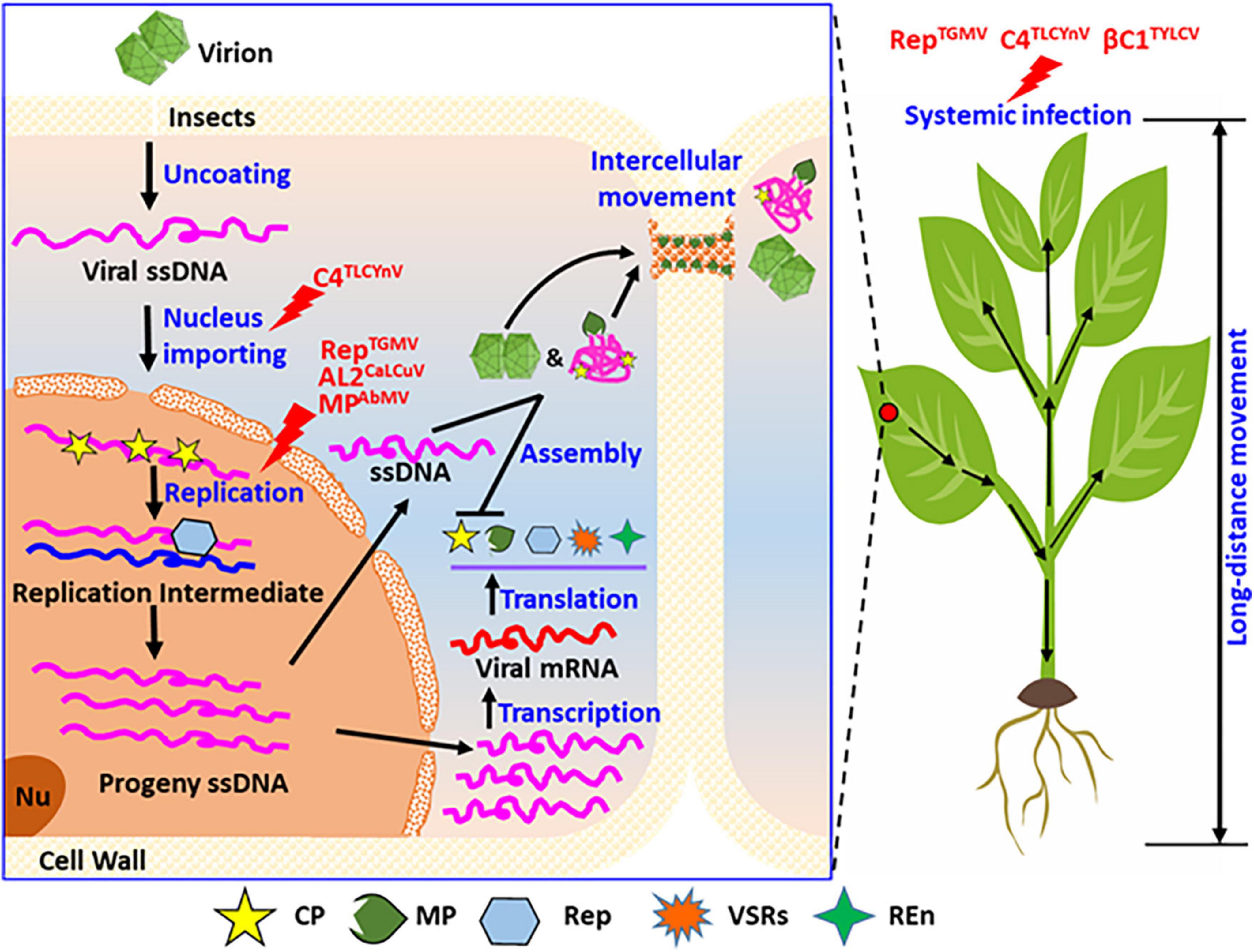
Figure 4. Summary of the phosphorylation functions of proteins encoded by geminiviruses during their life cycle. Considering the entire cell and whole plants, phosphorylation modification of the viral proteins regulates the different infection steps in the life cycle of geminiviruses in host plants, including the nuclear import, genomic DNA replication, and systemic infection. As shown, phosphorylation modification of C4 (C4TLCYnV) affected virus nucleocytoplasmic shuttling. Phosphorylation modification also affects the virus replication, for instance, replicase of TGMV (RepTGMV), AL2 of CaLCuV (AL2CaLCuV), and MP of AbMV (MPAbmV). Phosphorylation modification also affects virus systemic infection of host plant, such as replicase of TGMV (RepTGMV), C4 of TLCYnV (C4TLCYnV), and βC1 of TYLCV (βC1TYLCV). REn, replication enhancer; PD, plasmodesmata; Nu, nucleolus.
Effects of phosphorylation on viral replication and transcription
Cucumber necrosis virus (CNV) belongs to the genus Tombusvirus (Alam et al., 2021). The p33 protein of CNV (p33CNV) can be phosphorylated by membrane-related kinases and PKC. The phosphorylation sites of p33CNV are Thr-205, Ser-210, and Thr-211. The replicase cofactor function of p33CNV is completely lost in plant protoplasts and yeast when p33 is mutated to a phosphorylation mimic (T/S to D). Contrastingly, when p33 was mutated to the state of dephosphorylation (T/S to A), replication can occur in plant protoplasts and yeast could occur, but the viral infection efficiency is lower than that of the wild-type virus. These results indicate that in the process of viral infection, the alanine mutants of p33CNV in the viral genome affected the synthesis of subgenomic RNA and reduced the proportions of positive- and negative-strand RNA, ultimately affecting viral replication (Figure 3, Replication). Phosphorylated p33CNV attenuates the RNA-binding ability and RdRp activity in plants (Stork et al., 2005).
Turnip yellow mosaic virus (TYMV) is a member of Tymovirus (Ni et al., 2017). The ORF1 of TYMV (ORF1TYMV) encodes an RdRp with a mass of 206 kDa, which functions in viral RNA replication. In vivo, ORF1TYMV produces 140 kDa (p140) and 66 kDa (p66) replicase subunits via self-cleavage. The 66 kDa replicase subunits can be phosphorylated in plants, three main phosphorylation sites, thr-64, Ser-80, and Ser-326, have been identified via mass spectrometry. Dephosphorylation or phosphorylation-mimic mutants at the three phosphorylation sites of the 66 kDa replicase subunit have different effects on viral infectivity. The double phosphorylation-mimic mutant p66T64D/S80D and the single mutants p66S326A, p66S326D, and p66S326T have remarkable effects on viral infectivity, thereby significantly reducing the viral genomic RNA and CP protein accumulation levels. Phosphorylation of p66 does not affect its interaction with p140. In the viral infection cycle, the p66 phosphorylation is inhibited by the interactions between p140 and p66 subunits. These interactions mark the initiation of the viral replication complex assembly at the host cell membrane. Ser-326 site of p66 is the key to viral RNA synthesis regulation, and the phosphorylation/dephosphorylation of p66S326 affects the binding and depolymerization of viral genomic RNA and the replication complex. These results indicate that the phosphorylation of p66 is a molecular switch that regulates the speed of viral replication speed in a self-optimized manner and balances the interaction between the virus and the host plant. The speed of adaptive viral genomic RNA replication could guarantee the long-term coexistence of the virus in the host plant in terms of evolution (Jakubiec et al., 2006; Jakubiec and Jupin, 2007; Figure 3, Replication).
Barley stripe mosaic virus (BSMV) γb protein is a typical VSR and viral symptom determinant (Bragg et al., 2004). The γbBSMV can be phosphorylated by PKA-like protein kinase in N. benthamiana in vitro and in vivo. Ser-96 is the main phosphorylation site of γb in plants. The γbS96A mutant has reduced local and systemic VSR activities due to the compromised 21-bp dsRNA binding activity and causes necrosis of infected leaves. Overexpression of other VSRs in trans or cis cannot rescue the necrosis induced by the BSMV γbS96A mutant. These results demonstrated that suppression of cell death via γb phosphorylation at Ser-96 is functionally distinct from the RNA silencing suppressor activity. In addition, the serine/threonine/tyrosine kinase (NbSYT46) phosphorylates and interacts with γb. The antiviral role of NbSTY46 involves kinase activity, accordingly, the NbSTY46T436A mutant lacking kinase activity not only loses the ability to phosphorylate or interact with γb but also fails to sustain virus systemic infection in plants. Self-phosphorylated NbSYT46 directly interacts with and phosphorylates the γb protein, which negatively regulates the viral replication and inhibits viral infection (Zhang et al., 2018, 2021; Figure 3, Replication).
Barley yellow striate mosaic virus (BYSMV) is a member of the genus Cytorhabdovirus, which elicits chlorotic striate and mosaic symptoms in the leaves of cereal plant species (Cao et al., 2018). BYSMV genome is a long, negative, single-stranded RNA (Yan et al., 2015). In BYSMV infection cycle, the phosphoprotein (P) in barley has two forms, with different migration rates corresponding to 42 kDa (p42) and 44 kDa (p44). Mass-spectrometry identified five C-terminal serine-rich regions (SR) of the phosphoprotein (189SASRPSSIAS198) that were phosphorylated. The viral dephosphorylation mutant (BYSMV-PS5A) enhanced viral replication, whereas the transcription was facilitated by the phosphorylation-mimic mutant (BYSMV-PS5D). In vivo, the two forms of P proteins (PS5A and PS5D) were preferentially associated with the nucleocapsid protein-RNA template and large polymerase protein, providing optimal replication and transcription of BYSMV, respectively. Biochemical assays demonstrated that plant and insect CK1 proteins can phosphorylate the SR motif of the P protein. Superphosphorylation of SR induces conformational changes in proteins. Moreover, the overexpression of CK1 or a dominant-negative mutant impairs the phosphorylation/dephosphorylation balance between p42 and p44, thereby compromising viral infection in barley. These observations indicate that BYSMV recruits the conserved CK1 kinases to finely regulate the replication-transcription phase during the viral life cycle and achieve optimal cross-kingdom infection of plants and insect vectors (Gao et al., 2020).
Ding et al. (2022) demonstrated that the barley (Hordeum vulgare) MAPK-MPK3 (HvMPK3) and the planthopper ERK (LsERK) proteins can interact with the viral structural protein nucleoprotein and directly phosphorylate this N protein at Ser-290. Overexpression of HvMPK3 could inhibit BYSMV infection, and barley plants treated with the MAPK pathway inhibitor U0126 are more susceptible to BYSMV. The dsRNA was synthesized and microinjected into the planthopper to knock down the LsERK, then the virus infection was promoted. A phosphomimetic mutant of the nucleoprotein Ser-290 (S290D) completely abolished virus infection because of the impaired self-interaction of BYSMV N and the formation of unstable N–RNA complexes. These results demonstrated that the conserved MAPK and ERK directly phosphorylate the viral nucleoprotein to trigger immunity against cross-kingdom immunity to BYSMV in host plants and its insect vectors (Ding et al., 2022).
Effect of phosphorylation on the packaging of viral particles
Beet black scorch virus (BBSV) belongs to the genus Necrovirus in the family Tombusviridae (Xu et al., 2016). CPBBSV can be phosphorylated at Thr-41 by the cAMP-dependent protein kinase (PKA)-like kinases in vivo and in vitro. The non-phosphorylated state of CPT41 does not affect the initial viral RNA replication stage, however, viral genomic RNA accumulation is affected as the infection progresses. CPBBSV phosphorylation is inferred to be involved in the later stages of genomic RNA replication, effectively interfering with the host defense system. RNase-sensitivity assays revealed that the BBSVT41A and BBSVT41E mutant viruses are easily degraded by endogenous ribonucleases in Nicotiana benthamiana. Electron microscopy revealed that the BBSVT41A and BBSVT41E mutants are abnormal compared to the wild-type BBSV particle (Figure 3, Virion Assembly). These findings indicated that the CPT41 mutant could not package viral RNAs into intact viral particles to protect against genomic RNA degradation. The phosphorylation state of CPT41 can regulate the virus particles in plants, thereby finely regulating BBSV infection in plants (Zhao et al., 2015).
Effect of phosphorylation on viral movement
Bamboo mosaic virus (BaMV) is a member of the genus Potexvirus and possesses a positive single-stranded RNA genome (Chen et al., 2017). The CP is phosphorylated by CK2 in N. benthamiana (NbCK2). Studies revealed that the main phosphorylation site is Ser-241 and both CPBaMV and NbCK2α subunits are colocalized on the plasmodesmata (PD). These results suggest that phosphorylation of CPBaMV may be involved in the viral cell-to-cell movement. Although in vitro binding assays showed that the RNA-binding ability of CPS241A mutant was not impaired, the cell-to-cell movement ability of mutant BaMV-CPS241A was reduced. If the Ser-241 site is mutated to D (phosphorylation mimics, CP S241D), its RNA-binding ability is significantly weakened, and the cell-to-cell movement ability of the corresponding mutant virus is blocked. Further deletion of the AA from 240 to 242 in CPBaMV enhances the RNA-binding ability of CP, but the cell-to-cell movement ability of the corresponding mutant virus is affected. These results indicate that the NbCK2 regulates the RNA-binding ability of CPBaMV by phosphorylating the AA 241 (Hung et al., 2014). The p20 protein, encoded by BaMV satellite RNA (satBaMV), can also be phosphorylated in plants. In vitro experiments showed that p20 can be phosphorylated via a total intracellular protein extraction and the Ser-11 is the phosphorylation site. The p20S11D mutant almost completely loses its ability to bind to wild-type p20 and mutant p20S11A in vitro (Vijayapalani et al., 2012; Figure 3, Intercellular Movement).
Potato mop-top virus (PMTV), belongs to the genus of Pomovirus and the family Virgaviridae (Cowan et al., 2018). Genomic RNA3 of PMTV encodes TGB3, which is one of the three movement proteins. TGB3PMTV participates in viral intracellular movement in plants. Studies have found that the Tyr-87, Tyr-88, Tyr-89, and Tyr-120 are phosphorylated in TGB3. In vitro phosphorylation experiments have shown that the phosphorylation signal of TGB3 is absent when the tyrosine (Y) is mutated to alanine (A). These results demonstrated that the Tyr (87—89, 120) is the actual phosphorylation site of TGB3.
In addition, the phosphorylation/dephosphorylation state of TGB3 affects the intensity of its interaction with TGB2. Mutation of the tyrosine to alanine at positions 87–89 of TGB3PMTV enhances the interaction intensity between TGB3 and TGB2 in yeast. However, the intercellular movement ability of the corresponding viral mutant (PMTV-TGB3T87–89A) is weakened in N. benthamiana. Although the TGB3Y120A mutant does not affect the interaction with TGB2 protein, it cannot infect N. benthamiana. This demonstrates that tyrosine phosphorylation modification exists in plant viral proteins and is also a method of regulating plant virus movement and infectivity (Figure 3, Intercellular Movement; Samuilova et al., 2013).
Barley stripe mosaic virus (BSMV) is a representative species of hordeivirus (Wang et al., 2021). The triple gene blocks 1 (TGB1) protein is one of the three movement proteins encoded by BSMV. In vitro phosphorylation experiments showed that TGB1BSMV can be phosphorylated by CK2 kinase in the N. benthamiana and barley. Further research revealed that the Thr-401 is the main phosphorylation site, and Thr-395 is the anchor site of CK2 in vivo. Mutations in Thr-395 and Thr-401 mimic the dephosphorylation status (T395A, T395E, T401A, and T401E). The mutant viruses can systematically infect barley and wheat, however, the infection efficiency and viral accumulation levels decreased dramatically. In N. benthamiana, only the mutant virus BSMV-TGB1T395A can establish a systemic infection. Together, these results demonstrate that the phosphorylation/dephosphorylation status of TGB1 at Thr-395 and Thr-401 affects the viral systemic movement ability differently in monocot and dicot hosts. This may be a consequence of the architecture of host-specific vasculature. Furthermore, the phosphorylation of TGB1 promotes viral infection by enhancing the interaction intensity with TGB3 proteins, which is apt to form the viral ribonucleoprotein complex (BSMV intracellular movement complex). However, the phosphorylation modification of TGB1 is not sufficient to substantially alter the TGB localization patterns visualized via laser confocal microscopy (Figure 3, Intercellular Movement; Hu et al., 2015).
Effects of phosphorylation on viral infectivity and pathogenicity
Chinese wheat mosaic virus (CWMV), a member of the genus Furovirus and family of Virgaviridae, is an RNA virus with two positive-sense single-stranded genomic RNAs (Yang et al., 2020). Ser-162 and Ser-165 of the CWMV cysteine-rich protein (CRPCWMV) are the two phosphorylation sites catalyzed by wheat SAPK7 protein in vivo and in vitro. Mutational analyses have shown that the double-site phosphorylation-mimic mutant virus (CWMV-CRPS162/165D) enhances viral infectivity in plants because of the suppressed cell death activity and hydrogen peroxide production by the CRPS162/165D protein. Furthermore, the CWMV-CRPS162/165D interacts with the RNA-binding protein UBP1-associated protein 2C (TaUBA2C) in wheat, whereas the phosphorylation-deficient mutant virus (CWMV-CRPS162/165A) does not. Silencing of TaUBA2C promotes viral infection in wheat plants, while the overexpression of TaUBA2C inhibits viral infection. TaUBA2C recruits the pre-mRNAs of TaNPR1, TaPR1, and TaRBOHD to induce plant cell death and hydrogen peroxide production, ultimately inhibiting viral infection. This effect can be suppressed by phosphorylated CRPS162/165D through direct binding of TaUBA2C, resulting in changes in the chromatin-bound status and the attenuation of the RNA- or DNA-binding activity of TaUBA2C in plants (Figure 3, Systemic Infection; Li et al., 2022).
Tomato yellow leaf curl virus (TYLCV) is a member of Geminivirus with a single genomic component that is composed of a single-stranded circular DNA (DNA-A) and satellite DNA (DNA-B) (Prasad et al., 2020). The βC1 protein is encoded by satellite DNA-B, which functions as a VSR and a symptom determinant (Hu et al., 2019). βC1TYLCV interacts with a variety of host factors that interfere with multiple antiviral immune pathways in plants. An in vitro yeast two-hybrid assay (Y2H) and in vivo bimolecular fluorescence complementation (BiFC) revealed that βC1TYLCV interacts with tomato SNFI-related kinase (SnRK1). Phosphorylation experiments showed that βC1TYLCV is phosphorylated by SnRK1 kinase, and Ser-33, Thr-78, Tyr-5, and Tyr-110 are the main phosphorylation sites. The symptoms of phosphorylation-deficient mutant viruses (TYLCV-βC1S33A, -βC1T78A, -βC1S33A, and -βC1T78A) are aggravated, and the viral genomic DNA accumulation levels are significantly higher than those of the wild-type virus. The phosphorylation-mimic mutant viruses (TYLCV-βC1S33D, -βC1T78D, and -βC1S33D/T78D) delay infection, and the viral DNA accumulation levels are completely opposed to those in the phosphorylation-deficient mutant viruses. These results indicate that SnRK1 interacts with and phosphorylates βC1TYLCV protein to weaken viral infectivity in tomatoes. Compared with those of the double mutant virus TYLCV-βC1-2D infection, the symptoms of TYLCV-βC1-2D/2E (S33D/T78D/Y5E/Y110E) infection are further attenuated, whereas the VSR activity of βC1-2D/2E is significantly weakened. Furthermore, phosphorylation of the βC1 protein by SnRK1 greatly affects its interaction with N. benthamiana ASYMMETRIC LEAVES 1, and regulates the induction of symptoms (Figure 4, Systemic Infection; Shen et al., 2011; Zhong et al., 2017).
Yunnan tomato leaf curl virus (TLCYnV) is a member of the Geminivirus family with a single-component genomic DNA (Mei et al., 2020). The C4 protein of TLCYnV (C4TLCYnV) is a symptom determinant in plants (Mei et al., 2018b). In vivo and in vitro phosphorylation experiments demonstrated that C4TLCYnV is phosphorylated by the host NbSKη, and the Thr-51 is the corresponding phosphorylation site. Phosphorylation of C4TLCYnV plays a key role in plant viral pathogenicity. The pathogenicity and viral genomic DNA accumulation levels of the dephosphorylation mutant (TLCYnV-C4T51A) are significantly reduced. Furthermore, subcellular localization revealed that the C4T51A mutant confines the NbSKη protein to the cell membrane, and myristoylation of the C4 N-terminal Gly-2 position directly results in the cell membrane localization. The pathogenicity and viral genomic DNA accumulation levels of the mutant TLCYnV-C4G2A virus are significantly reduced, respectively. During TLCYnV-C4G2A virus infection, NbSKη cannot localize to the cell membrane. Chemical inhibition of N-myristoyltransferases or exportin-α enhances the nuclear retention of C4TLCYnV, and mutations in the putative phosphorylation (T51A) or myristoylation sites (G2A) of C4TLCYnV result in increased nuclear localization and reduced severity of the C4-induced developmental abnormalities. These results suggest that the nucleocytoplasmic shuttling of C4TLCYnV via protein modifications, including NbSKη-mediated protein phosphorylation, protein myristoylation, and the interaction with exportin-α, is critical for viral pathogenicity in plants (Figure 4, Systemic Infection; Mei et al., 2018a).
Table 2 shows several plant virus-encoded proteins that can be phosphorylated. Single viral protein phosphorylation often changes the biological characteristics of the protein itself and can affect the viral infection cycle in plants. The host CK2 is not only involved in plant growth and development but also regulates the function of a variety of viral proteins through phosphorylation. The virus often achieves maximum replication during co-evolution through an interaction balance between host defense and survival. Phosphorylation modifications often act as a molecular switch that guarantees optimal viral replication and prolonged viral existence in virus-specific hosts.
Conclusion and perspectives
As one of the most extensive PTMs, phosphorylation has now reached the stage where it is significant in almost every physiological event and plays important roles in regulating multiple biological activities, including cell signal transduction, protein–protein interactions, PTM-crosstalk, self-stability, and subcellular localization (Figure 1). Investigations of protein phosphorylation modification are important for revealing the molecular mechanisms underlying interactions between plant defense and viral counterdefense. Here, we summarized the current phosphorylation detection methods for plant viral proteins. Using the combination of in vivo and in vitro phosphorylation experiments, it is easy to determine the detailed phosphorylation sites of viral protein and host kinases. Various studies have revealed that viral proteins often act as molecular switches that display functions according to the two statuses, phosphorylation and dephosphorylation, promoting or preventing infection in a particular host. The different statuses of viral proteins are catalyzed by host kinases, including CK1, CK2, GSK3, PKA, and PKC. Table 2 summarizes the common phosphorylation motifs for the substrates of different kinases and provides a reference for identifying the phosphorylation sites of target viral proteins and their corresponding host kinases. CK2 can phosphorylate the CP and MP of different plant viruses, and the function of CK2 has been widely studied in relation to plant–virus interactions. These findings suggest that CK2 plays an essential role in the regulation of the viral infection cycle. However, it is unclear how the same kinase phosphorylates MP and CP, and how they coordinate with each other to benefit virus infection. Our study reviewed the methods for identifying phosphorylation and the biological functions of phosphorylated viral proteins reported to date. After phosphorylation in plant cells, the subcellular localization, pathogenicity, self-interactions, and interactions with other viral proteins are altered, which finally regulates virus replication and transcription during the infection process (Figure 1). The summaries provide a reference and direction for further research into how viral protein phosphorylation balances the continuous viral infection and the host’s moderate response and the defense or counter-defense role of plant kinases in the interactions between the plant virus and host.
In addition to phosphorylation, the side chain hydroxyl group on the serine and threonine residues of the target protein often undergo O-GlcNAcylation in eukaryote. The O-GlcNAcylation modification is catalyzed and removed by the N-acetylglucosamine transferase (OGT) and the O-linked N-acetylglucosamine hydrolase (OGA), respectively (Yang and Qian, 2017). The O-GlcNAcylation modification can regulate various basic cellular processes, such as transcription (Golks et al., 2007), epigenetic modification (Hanover et al., 2012; Dehennaut et al., 2014; Lewis and Hanover, 2014; Singh et al., 2015), intracellular signaling transduction (Vosseller et al., 2002; Zhang K. et al., 2014; Xu et al., 2019), and virus infection (Chen et al., 2005; Kim et al., 2011; Pérez et al., 2013; Song et al., 2019; Hu et al., 2021). The same serine and threonine target sites determine the interactions between O-GlcNAcylation and phosphorylation in vivo. Many studies have shown that in various proteins, O-GlcNAcylation often occurs with reciprocal or sequential phosphorylation of the same or neighboring residues (Hart et al., 2007, 2011). O-GlcNAcylation can temporally regulate the insulin signaling pathways in humans (Zhang K. et al., 2014). ATK is a type of serine/threonine kinases that are involved in cell survival and metabolism (Vosseller et al., 2002). The O-GlcNAcylation and phosphorylation sites of AKT were mapped to the same positions (Thr-308 and Ser-473) (Shi et al., 2015). These observations demonstrate that O-GlcNAcylation of AKT kinases directly suppresses its phosphorylation, which shows a “Yin-Yang” model with antagonistic effects on specific AAs of particular proteins (Butkinaree et al., 2010). In winter wheat verbalizations, the possible correlation between the modified sites and the computationally predicted structures of the 31 proteins containing both the O-GlcNAcylation and phosphorylation modifications were explored, and their result showed that they could be divided into two major patterns: competitive (Yin-Yang model) and coexisting (coordinately regulated) (Xu et al., 2019). In plant virology studies, CPPPV was found to be modified via O-GlcNAcylation during virus infection, which promoted the stability of the CP protein and affected the virus movement and replication (Chen et al., 2005; Kim et al., 2011; Pérez et al., 2013). Seven AA sites of the CPPPV, including the Thr-19, Thr-21, Thr-24, Thr-26, Thr-41, Thr-53, and Ser-65, show O-GlcNAcylation modifications during PPV infection (Pérez et al., 2013), whereas Ser-25, Ser-81, Ser-101, and Ser-118 were phosphorylated (Martínez-Turiño et al., 2018). The correlation between the Ser-25 phosphorylation and O-GlcNAcylation of Thr-19, Thr-21, Thr-24, and Thr-26 in the CPPPV needs further investigation. Exploration of the coexisting or competitive patterns of these two major modifications on neighboring residues (AA 19-26) of CPPPV has profound significance for a deeper understanding of the plant virus life cycle.
More than 20 viral proteins are phosphorylated during viral infection cycle (Figures 3, 4). Do other plant viral proteins also undergo phosphorylation modification with different regulatory effects on viral infection? A variety of kinases in plants have been identified in plants, with less than 10 types found to be directly involved in the phosphorylation. Nevertheless, the types and functions of kinases involved in the phosphorylation of viral proteins require further exploration. This study revealed that only the CK2 and PKA can phosphorylate various plant viral proteins. Are other viral proteins phosphorylated by these two kinases? Can different viral proteins encoded by the same virus be phosphorylated by the same host kinase? We identified numerous phosphorylation sites of plant viral proteins via mass spectrometry, especially in target proteins that can be phosphorylated via total protein extractions in vitro. The finding is not representative of the in vivo conditions because the total protein obtained from plant contained many kinases, and phosphorylation can occur between the kinase and substrate that would not normally be in contact in vivo. Different protein kinases can phosphorylate multiple sites of the same and different viral proteins encoded by the same virus, and these modifications have different regulatory roles in plant–virus interactions.
In plant virology, the core question is how to coordinate plant resistance with yield and quality. The phosphorylation studies discussed here are promising. Massive progress in the identification of the major substrates of each protein kinase and phosphatase has been achieved in recent years using the modern MS-based proteomics. However, major gaps in our knowledge still exist regarding the identities of the key substances and protein kinases, and how their phosphorylation contributes to changes in cell physiology in response to particular stimuli. A certain genome size determines a finite number of kinases and phosphatase in plants, and there are multiple potential phosphorylation sites within a single protein, which implies that the kinase and phosphatase isoforms may have overlapping specificities. These kinases are involved in the phosphorylation of several hundreds of substrates in vivo. In addition, the substrate of the target kinase is often cell-specific. Explaining the distinctive effects of various stimuli on different tissues requires further consideration. Hence, powerful methods, coupled with further exploitation of important methodological and technical advances, are needed to identify the substance protein and the corresponding target kinase. Of greater importance is research to control the outbreaks and devastation caused by plant viruses, develop tissue and phosphor-specific antibodies, certain cell-type-specific permeant inhibitors of protein kinase, and design new therapies via taking advantage of the distinct cell types not expressing a particular protein kinase or phosphatase.
In future plant–virus protein phosphorylation studies, the phosphorylation sites on viral proteins can be identified in detail by mass spectrometry combined with the in vitro phosphorylation experiments. The corresponding kinases can also be identified in the follow-up research using reverse genetics techniques. Furthermore, it would be beneficial to explore the impact of certain types of kinase and viral mechanism, including viral replication, viral transcription, and the host’s defense response. However, several proteins encoded by one certain plant virus can be phosphorylated by several types of kinases. The elucidation of specific kinases and their substrates and determination of the corresponding relationship are essential aspects that should be considered in future studies of biological function. If the common pathways involved in the broad-spectrum antiviral immunity conferred by kinases can be identified, we could cultivate new antiviral crop germplasm through transgenic or CRISPR-Cas9-mediated gene editing methods. Moreover, small-molecule activators or inhibitors could be designed according to the advanced structures of target kinases, which could be used as novel pesticides for spraying crop leaves in the field for virus control.
Author contributions
KZ and XZ contributed substantially to the conception and design of this review article and co-wrote the manuscript. KZ and ZH reviewed the manuscript before submission for its intellectual content. All authors gave final approval of the published version.
Funding
This work was supported by the National Natural Science Foundation of China (31801699 and 81903764), the Excellent Youth Fund of Jiangsu Natural Science Foundation (SBK2022030096), the Project of Science and Technology Development Plan for Traditional Chinese Medicine of Jiangsu Province (YB201993), and the Guangdong Provincial Key Laboratory for Plant Protection (Zhizhong2021-07).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AbMV, abutilon mosaic virus; BaMV, bamboo mosaic virus; BBSV, beet black scorch virus; BSMV, barley mosaic stripe virus; CaLCuV, cabbage leaf curl virus; CMV, cucumber mosaic virus; CNV, cucumber necrosis virus; CWMV, Chinese wheat mosaic virus; CP, coat protein; MP, movement protein; Nu, nucleolus; PD, plasmodesmata; PLRV, potato leaf roll virus; PMTV, potato mop-top virus; PPV, plum pox virus; PSLV, poa semilatent virus; PTM, post-translational modifications; PVA, potato virus A; PVX, potato virus X; TLCYnV, Yunnan tomato leaf curl virus; TGMV, tomato golden mosaic virus; CaLCuV, cabbage leaf curl virus; TYLCV, tomato yellow leaf curl virus; RdRp, RNA-dependent RNA polymerase; Ren, replication enhancer; Rep, replicase; RNPs, ribonucleoprotein; TMV, tobacco mosaic virus; TYMV, turnip yellow mosaic virus; VSR, virus-encoded RNA silencing suppressors; REn, replication enhancer.
Footnotes
References
Alam, S. B., Reade, R., Maghodia, A. B., Ghoshal, B., Theilmann, J., and Rochon, D. (2021). Targeting of cucumber necrosis virus coat protein to the chloroplast stroma attenuates host defense response. Virology 554, 106–119. doi: 10.1016/j.virol.2020.10.012
Amanchy, R., Periaswamy, B., Mathivanan, S., Reddy, R., Tattikota, S. G., and Pandey, A. (2007). A curated compendium of phosphorylation motifs. Nat. Biotechnol. 25, 285–286. doi: 10.1038/nbt0307-285
Atabekov, J. G., Rodionova, N. P., Karpova, O. V., Kozlovsky, S. V., Novikov, V. K., and Arkhipenko, M. V. (2001). Translational activation of encapsidated potato virus X RNA by coat protein phosphorylation. Virology 286, 466–474. doi: 10.1006/viro.2001.1013
Blacken, G. R., Sadílek, M., and Turecek, F. (2008). Gallium metal affinity capture tandem mass spectrometry for the selective detection of phosphopeptides in complex mixtures. J. Mass Spectrom. JMS 43, 1072–1080. doi: 10.1002/jms.1387
Bragg, J. N., Lawrence, D. M., and Jackson, A. O. (2004). The N-terminal 85 amino acids of the barley stripe mosaic virus gammab pathogenesis protein contain three zinc-binding motifs. J. Virol. 78, 7379–7391. doi: 10.1128/JVI.78.14.7379-7391.2004
Buchowiecka, A. K. (2014). Puzzling over protein cysteine phosphorylation–assessment of proteomic tools for S-phosphorylation profiling. The Analyst 139, 4118–4123. doi: 10.1039/c4an00724g
Butkinaree, C., Park, K., and Hart, G. W. (2010). O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 1800, 96–106. doi: 10.1016/j.bbagen.2009.07.018
Cao, Q., Xu, W.-Y., Gao, Q., Jiang, Z.-H., Liu, S.-Y., Fang, X.-D., et al. (2018). Transmission characteristics of barley yellow striate mosaic virus in its planthopper vector Laodelphax striatellus. Front. Microbiol. 9:1419. doi: 10.3389/fmicb.2018.01419
Cegielska, A., Gietzen, K. F., Rivers, A., and Virshup, D. M. (1998). Autoinhibition of casein kinase I epsilon (CKI epsilon) is relieved by protein phosphatases and limited proteolysis. J. Biol. Chem. 273, 1357–1364. doi: 10.1074/jbc.273.3.1357
Champagne, J., Laliberté-Gagné, M.-E., and Leclerc, D. (2007). Phosphorylation of the termini of cauliflower mosaic virus precapsid protein is important for productive infection. Mol. Plant-Microbe Interact. MPMI 20, 648–658. doi: 10.1094/MPMI-20-6-0648
Chapdelaine, Y., Kirk, D., Karsies, A., Hohn, T., and Leclerc, D. (2002). Mutation of capsid protein phosphorylation sites abolishes cauliflower mosaic virus infectivity. J. Virol. 76, 11748–11752. doi: 10.1128/jvi.76.22.11748-11752.2002
Chen, D., Juárez, S., Hartweck, L., Alamillo, J. M., Simón-Mateo, C., Pérez, J. J., et al. (2005). Identification of secret agent as the O-GlcNAc transferase that participates in Plum pox virus infection. J. Virol. 79, 9381–9387. doi: 10.1128/JVI.79.15.9381-9387.2005
Chen, I.-H., Huang, Y.-W., and Tsai, C.-H. (2017). The functional roles of the cis-acting elements in bamboo mosaic virus RNA genome. Front. Microbiol. 8:645. doi: 10.3389/fmicb.2017.00645
Chen, Y., Sprung, R., Tang, Y., Ball, H., Sangras, B., Kim, S. C., et al. (2007). Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics MCP 6, 812–819. doi: 10.1074/mcp.M700021-MCP200
Cheng, C. M., Tu, J., Yang, C. C., and Kuo, T. T. (1996). Rifampicin: an inhibitor of Xp12-specific protein phosphorylation in Xanthomonas oryzae pv. oryzae. FEMS Microbiol. Lett. 143, 141–149. doi: 10.1111/j.1574-6968.1996.tb08473.x
Cheng, H., Deng, W., Wang, Y., Ren, J., Liu, Z., and Xue, Y. (2014). dbPPT: a comprehensive database of protein phosphorylation in plants. Database J. Biol. Databases Curation 2014, bau121. doi: 10.1093/database/bau121
Cheng, X., Xiong, R., Li, Y., Li, F., Zhou, X., and Wang, A. (2017). Sumoylation of turnip mosaic virus rna polymerase promotes viral infection by counteracting the host NPR1-mediated immune response. Plant Cell 29, 508–525. doi: 10.1105/tpc.16.00774
Chiang, C.-Y., Pan, C.-H., Hsieh, C.-H., Tsai, J.-P., Chen, M.-Y., Liu, H.-H., et al. (2013). Lipidated dengue-2 envelope protein domain III independently stimulates long-lasting neutralizing antibodies and reduces the risk of antibody-dependent enhancement. PLoS Negl. Trop. Dis. 7:e2432. doi: 10.1371/journal.pntd.0002432
Cohen, P. (2002). The origins of protein phosphorylation. Nat. Cell Biol. 4, E127–E130. doi: 10.1038/ncb0502-e127
Cowan, G. H., Roberts, A. G., Jones, S., Kumar, P., Kalyandurg, P. B., Gil, J. F., et al. (2018). Potato mop-top virus co-opts the stress sensor HIPP26 for long-distance movement. Plant Physiol. 176, 2052–2070. doi: 10.1104/pp.17.01698
Dai, C., and Xue, H.-W. (2010). Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 29, 1916–1927. doi: 10.1038/emboj.2010.75
Dehennaut, V., Leprince, D., and Lefebvre, T. (2014). O-GlcNAcylation, an epigenetic mark. focus on the histone code, TET family proteins, and polycomb group proteins. Front. Endocrinol. 5:155. doi: 10.3389/fendo.2014.00155
Deng, Y.-H., Li, R.-J., Zhang, H.-S., Du, X.-L., and Wang, H. (2007). Liquid chromatographic analysis of phosphoamino acids at femtomole level using chemical derivatization with N-hydroxysuccinimidyl fluorescein-O-acetate. Anal. Chim. Acta 601, 118–124. doi: 10.1016/j.aca.2007.08.023
Diallo, I., Seve, M., Cunin, V., Minassian, F., Poisson, J.-F., Michelland, S., et al. (2019). Current trends in protein acetylation analysis. Expert Rev. Proteomics 16, 139–159. doi: 10.1080/14789450.2019.1559061
Ding, Z.-H., Gao, Q., Tong, X., Xu, W.-Y., Ma, L., Zhang, Z.-J., et al. (2022). MAPKs trigger antiviral immunity by directly phosphorylating a rhabdovirus nucleoprotein in plants and insect vectors. Plant Cell koac143. doi: 10.1093/plcell/koac143
Doble, B. W., and Woodgett, J. R. (2003). GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 116, 1175–1186. doi: 10.1242/jcs.00384
Domon, B., and Aebersold, R. (2006). Mass spectrometry and protein analysis. Science 312, 212–217. doi: 10.1126/science.1124619
Duan, G., and Walther, D. (2015). The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput. Biol. 11:e1004049. doi: 10.1371/journal.pcbi.1004049
Emmer, J., Vavrinská, A., Sychrovský, V., Benda, L., Kříž, Z., Koča, J., et al. (2013). Influence of the O-phosphorylation of serine, threonine and tyrosine in proteins on the amidic 15N chemical shielding anisotropy tensors. J. Biomol. NMR 55, 59–70. doi: 10.1007/s10858-012-9686-6
Endicott, J. A., Noble, M. E. M., and Johnson, L. N. (2012). The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 81, 587–613. doi: 10.1146/annurev-biochem-052410-090317
Filhol, O., Martiel, J.-L., and Cochet, C. (2004). Protein kinase CK2: a new view of an old molecular complex. EMBO Rep. 5, 351–355. doi: 10.1038/sj.embor.7400115
Fiol, C. J., Wang, A., Roeske, R. W., and Roach, P. J. (1990). Ordered multisite protein phosphorylation. Analysis of glycogen synthase kinase 3 action using model peptide substrates. J. Biol. Chem. 265, 6061–6065.
Fischer, E. H., and Krebs, E. G. (1955). Conversion of phosphorylase b to phosphorylase a in muscle extracts. J. Biol. Chem. 216, 121–132.
Flotow, H., Graves, P. R., Wang, A. Q., Fiol, C. J., Roeske, R. W., and Roach, P. J. (1990). Phosphate groups as substrate determinants for casein kinase I action. J. Biol. Chem. 265, 14264–14269.
Friso, G., and van Wijk, K. J. (2015). Posttranslational Protein Modifications in Plant Metabolism. Plant Physiol. 169, 1469–1487. doi: 10.1104/pp.15.01378
Gao, J., Agrawal, G. K., Thelen, J. J., and Xu, D. (2009). P3DB: a plant protein phosphorylation database. Nucleic Acids Res. 37, D960–D962. doi: 10.1093/nar/gkn733
Gao, Q., Yan, T., Zhang, Z.-J., Liu, S.-Y., Fang, X.-D., Gao, D.-M., et al. (2020). Casein Kinase 1 regulates cytorhabdovirus replication and transcription by phosphorylating a phosphoprotein serine-rich motif. Plant Cell 32, 2878–2897. doi: 10.1105/tpc.20.00369
Gao, Z., Zhang, D., Wang, X., Zhang, X., Wen, Z., Zhang, Q., et al. (2022). Coat proteins of necroviruses target 14-3-3a to subvert MAPKKKα-mediated antiviral immunity in plants. Nat. Commun. 13, 716. doi: 10.1038/s41467-022-28395-5
Gil, M., Lima, A., Rivera, B., Rossello, J., Urdániz, E., Cascioferro, A., et al. (2019). New substrates and interactors of the mycobacterial Serine/Threonine protein kinase PknG identified by a tailored interactomic approach. J. Proteomics 192, 321–333. doi: 10.1016/j.jprot.2018.09.013
Gold, M. G. (2019). Swimming regulations for protein kinase A catalytic subunit. Biochem. Soc. Trans. 47, 1355–1366. doi: 10.1042/BST20190230
Golks, A., Tran, T.-T. T., Goetschy, J. F., and Guerini, D. (2007). Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J. 26, 4368–4379. doi: 10.1038/sj.emboj.7601845
Graves, P. R., and Roach, P. J. (1995). Role of COOH-terminal phosphorylation in the regulation of casein kinase I delta. J. Biol. Chem. 270, 21689–21694. doi: 10.1074/jbc.270.37.21689
Graves, P. R., Haas, D. W., Hagedorn, C. H., DePaoli-Roach, A. A., and Roach, P. J. (1993). Molecular cloning, expression, and characterization of a 49-kilodalton casein kinase I isoform from rat testis. J. Biol. Chem. 268, 6394–6401.
Gross, S. D., and Anderson, R. A. (1998). Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell. Signal. 10, 699–711. doi: 10.1016/s0898-6568(98)00042-4
Hanover, J. A., Krause, M. W., and Love, D. C. (2012). Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 13, 312–321. doi: 10.1038/nrm3334
Hart, G. W., Housley, M. P., and Slawson, C. (2007). Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022. doi: 10.1038/nature05815
Hart, G. W., Slawson, C., Ramirez-Correa, G., and Lagerlof, O. (2011). Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858. doi: 10.1146/annurev-biochem-060608-102511
Heibeck, T. H., Ding, S.-J., Opresko, L. K., Zhao, R., Schepmoes, A. A., Yang, F., et al. (2009). An extensive survey of tyrosine phosphorylation revealing new sites in human mammary epithelial cells. J. Proteome Res. 8, 3852–3861. doi: 10.1021/pr900044c
Hu, J., Gao, Q., Yang, Y., Xia, J., Zhang, W., Chen, Y., et al. (2021). Hexosamine biosynthetic pathway promotes the antiviral activity of SAMHD1 by enhancing O-GlcNAc transferase-mediated protein O-GlcNAcylation. Theranostics 11, 805–823. doi: 10.7150/thno.50230
Hu, T., Huang, C., He, Y., Castillo-González, C., Gui, X., Wang, Y., et al. (2019). βC1 protein encoded in geminivirus satellite concertedly targets MKK2 and MPK4 to counter host defense. PLoS Pathog. 15:e1007728. doi: 10.1371/journal.ppat.1007728
Hu, Y., Li, Z., Yuan, C., Jin, X., Yan, L., Zhao, X., et al. (2015). Phosphorylation of TGB1 by protein kinase CK2 promotes barley stripe mosaic virus movement in monocots and dicots. J. Exp. Bot. 66, 4733–4747. doi: 10.1093/jxb/erv237
Hung, C.-J., Huang, Y.-W., Liou, M.-R., Lee, Y.-C., Lin, N.-S., Meng, M., et al. (2014). Phosphorylation of coat protein by protein kinase CK2 regulates cell-to-cell movement of bamboo mosaic virus through modulating RNA binding. Mol. Plant-Microbe Interact. MPMI 27, 1211–1225. doi: 10.1094/MPMI-04-14-0112-R
Huq, E. (2006). Degradation of negative regulators: a common theme in hormone and light signaling networks? Trends Plant Sci. 11, 4–7. doi: 10.1016/j.tplants.2005.11.005
Hur, E.-M., and Zhou, F.-Q. (2010). GSK3 signalling in neural development. Nat. Rev. Neurosci. 11, 539–551. doi: 10.1038/nrn2870
Hutti, J. E., Jarrell, E. T., Chang, J. D., Abbott, D. W., Storz, P., Toker, A., et al. (2004). A rapid method for determining protein kinase phosphorylation specificity. Nat. Methods 1, 27–29. doi: 10.1038/nmeth708
Ivanov, K. I., Puustinen, P., Gabrenaite, R., Vihinen, H., Rönnstrand, L., Valmu, L., et al. (2003). Phosphorylation of the potyvirus capsid protein by protein kinase CK2 and its relevance for virus infection. Plant Cell 15, 2124–2139. doi: 10.1105/tpc.012567
Ivanov, K. I., Puustinen, P., Merits, A., Saarma, M., and Mäkinen, K. (2001). Phosphorylation down-regulates the RNA binding function of the coat protein of potato virus A. J. Biol. Chem. 276, 13530–13540. doi: 10.1074/jbc.M009551200
Jakubiec, A., and Jupin, I. (2007). Regulation of positive-strand RNA virus replication: the emerging role of phosphorylation. Virus Res. 129, 73–79. doi: 10.1016/j.virusres.2007.07.012
Jakubiec, A., Tournier, V., Drugeon, G., Pflieger, S., Camborde, L., Vinh, J., et al. (2006). Phosphorylation of viral RNA-dependent RNA polymerase and its role in replication of a plus-strand RNA virus. J. Biol. Chem. 281, 21236–21249. doi: 10.1074/jbc.M600052200
Jungbauer, A., and Hahn, R. (2009). Ion-exchange chromatography. Methods Enzymol. 463, 349–371. doi: 10.1016/S0076-6879(09)63022-6
Kawakami, S., Padgett, H. S., Hosokawa, D., Okada, Y., Beachy, R. N., and Watanabe, Y. (1999). Phosphorylation and/or presence of serine 37 in the movement protein of tomato mosaic tobamovirus is essential for intracellular localization and stability in vivo. J. Virol. 73, 6831–6840. doi: 10.1128/JVI.73.8.6831-6840.1999
Keenan, C., Long, A., and Kelleher, D. (1997). Protein kinase C and T cell function. Biochim. Biophys. Acta 1358, 113–126. doi: 10.1016/s0167-4889(97)00080-3
Kim, L., and Kimmel, A. R. (2000). GSK3, a master switch regulating cell-fate specification and tumorigenesis. Curr. Opin. Genet. Dev. 10, 508–514. doi: 10.1016/s0959-437x(00)00120-9
Kim, L., and Kimmel, A. R. (2006). GSK3 at the edge: regulation of developmental specification and cell polarization. Curr. Drug Targets 7, 1411–1419. doi: 10.2174/1389450110607011411
Kim, S. H., Palukaitis, P., and Park, Y. I. (2002). Phosphorylation of cucumber mosaic virus RNA polymerase 2a protein inhibits formation of replicase complex. EMBO J. 21, 2292–2300. doi: 10.1093/emboj/21.9.2292
Kim, Y.-C., Udeshi, N. D., Balsbaugh, J. L., Shabanowitz, J., Hunt, D. F., and Olszewski, N. E. (2011). O-GlcNAcylation of the plum pox virus capsid protein catalyzed by SECRET AGENT: characterization of O-GlcNAc sites by electron transfer dissociation mass spectrometry. Amino Acids 40, 869–876. doi: 10.1007/s00726-010-0706-0
Kinoshita, E., Kinoshita-Kikuta, E., Takiyama, K., and Koike, T. (2006). Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics MCP 5, 749–757. doi: 10.1074/mcp.T500024-MCP200
Kinoshita-Kikuta, E., Kinoshita, E., Matsuda, A., and Koike, T. (2014). Tips on improving the efficiency of electrotransfer of target proteins from Phos-tag SDS-PAGE gel. Proteomics 14, 2437–2442. doi: 10.1002/pmic.201400380
Kleinow, T., Holeiter, G., Nischang, M., Stein, M., Karayavuz, M., Wege, C., et al. (2008). Post-translational modifications of abutilon mosaic virus movement protein (BC1) in fission yeast. Virus Res. 131, 86–94. doi: 10.1016/j.virusres.2007.08.011
Kleinow, T., Nischang, M., Beck, A., Kratzer, U., Tanwir, F., Preiss, W., et al. (2009). Three C-terminal phosphorylation sites in the abutilon mosaic virus movement protein affect symptom development and viral DNA accumulation. Virology 390, 89–101. doi: 10.1016/j.virol.2009.04.018
Knippschild, U., Gocht, A., Wolff, S., Huber, N., Löhler, J., and Stöter, M. (2005). The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell. Signal. 17, 675–689. doi: 10.1016/j.cellsig.2004.12.011
Krupa, A., Preethi, G., and Srinivasan, N. (2004). Structural modes of stabilization of permissive phosphorylation sites in protein kinases: distinct strategies in Ser/Thr and Tyr kinases. J. Mol. Biol. 339, 1025–1039. doi: 10.1016/j.jmb.2004.04.043
Kurosaki, T., Myers, J. R., and Maquat, L. E. (2019). Defining nonsense-mediated mRNA decay intermediates in human cells. Methods San Diego Calif 155, 68–76. doi: 10.1016/j.ymeth.2018.12.005
Lamberti, G., Gügel, I. L., Meurer, J., Soll, J., and Schwenkert, S. (2011). The cytosolic kinases STY8, STY17, and STY46 are involved in chloroplast differentiation in Arabidopsis. Plant Physiol. 157, 70–85. doi: 10.1104/pp.111.182774
Lee, J.-Y. (2009). Versatile casein kinase 1: multiple locations and functions. Plant Signal. Behav. 4, 652–654. doi: 10.1104/pp.108.129346
Lewis, B. A., and Hanover, J. A. (2014). O-GlcNAc and the epigenetic regulation of gene expression. J. Biol. Chem. 289, 34440–34448. doi: 10.1074/jbc.R114.595439
Li, J., Feng, H., Liu, S., Liu, P., Chen, X., Yang, J., et al. (2022). Phosphorylated viral protein evades plant immunity through interfering the function of RNA-binding protein. PLoS Pathog. 18:e1010412. doi: 10.1371/journal.ppat.1010412
Lim, P. S., Sutton, C. R., and Rao, S. (2015). Protein kinase C in the immune system: from signalling to chromatin regulation. Immunology 146, 508–522. doi: 10.1111/imm.12510
Link, K., Vogel, F., and Sonnewald, U. (2011). PD trafficking of potato leaf roll virus movement protein in Arabidopsis depends on site-specific protein phosphorylation. Front. Plant Sci. 2. Available online at: https://www.frontiersin.org/article/10.3389/fpls.2011.00018 (Accessed April 3, 2022).
Lipton, J. O., Yuan, E. D., Boyle, L. M., Ebrahimi-Fakhari, D., Kwiatkowski, E., Nathan, A., et al. (2015). The circadian protein bmal1 regulates translation in response to S6K1-mediated phosphorylation. Cell 161, 1138–1151. doi: 10.1016/j.cell.2015.04.002
Litchfield, D. W. (2003). Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 369, 1–15. doi: 10.1042/BJ20021469
Liu, W., Xu, Z.-H., Luo, D., and Xue, H.-W. (2003). Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J. Cell Mol. Biol. 36, 189–202. doi: 10.1046/j.1365-313x.2003.01866.x
Makarov, V. V., Iconnikova, A. Y., Guseinov, M. A., Vishnichenko, V. K., and Kalinina, N. O. (2012). In vitro phosphorylation of the N-terminal half of hordeivirus movement protein. Biochem. Biokhimiia 77, 1072–1081. doi: 10.1134/S0006297912090155
Martínez-Turiño, S., Pérez, J. D. J., Hervás, M., Navajas, R., Ciordia, S., Udeshi, N. D., et al. (2018). Phosphorylation coexists with O-GlcNAcylation in a plant virus protein and influences viral infection. Mol. Plant Pathol. 19, 1427–1443. doi: 10.1111/mpp.12626
Matsushita, Y., Ohshima, M., Yoshioka, K., Nishiguchi, M., and Nyunoya, H. (2003). The catalytic subunit of protein kinase CK2 phosphorylates in vitro the movement protein of tomato mosaic virus. J. Gen. Virol. 84, 497–505. doi: 10.1099/vir.0.18839-0
Meggio, F., and Pinna, L. A. (2003). One-thousand-and-one substrates of protein kinase CK2? FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 17, 349–368. doi: 10.1096/fj.02-0473rev
Mei, Y., Wang, Y., Hu, T., Yang, X., Lozano-Duran, R., Sunter, G., et al. (2018a). Nucleocytoplasmic shuttling of geminivirus C4 Protein mediated by phosphorylation and myristoylation is critical for viral pathogenicity. Mol. Plant 11, 1466–1481. doi: 10.1016/j.molp.2018.10.004
Mei, Y., Yang, X., Huang, C., Zhang, X., and Zhou, X. (2018b). Tomato leaf curl Yunnan virus-encoded C4 induces cell division through enhancing stability of Cyclin D 1.1 via impairing NbSKη -mediated phosphorylation in Nicotiana benthamiana. PLoS Pathog. 14:e1006789. doi: 10.1371/journal.ppat.1006789
Mei, Y., Wang, Y., Li, F., and Zhou, X. (2020). The C4 protein encoded by tomato leaf curl Yunnan virus reverses transcriptional gene silencing by interacting with NbDRM2 and impairing its DNA-binding ability. PLoS Pathog. 16:e1008829. doi: 10.1371/journal.ppat.1008829
Millar, A. H., Heazlewood, J. L., Giglione, C., Holdsworth, M. J., Bachmair, A., and Schulze, W. X. (2019). The scope, functions, and dynamics of posttranslational protein modifications. Annu. Rev. Plant Biol. 70, 119–151. doi: 10.1146/annurev-arplant-050718-100211
Módena, N. A., Zelada, A. M., Conte, F., and Mentaberry, A. (2008). Phosphorylation of the TGBp1 movement protein of potato virus X by a Nicotiana tabacum CK2-like activity. Virus Res. 137, 16–23. doi: 10.1016/j.virusres.2008.04.007
Moreno-Romero, J., Espunya, M. C., Platara, M., Ariño, J., and Martínez, M. C. (2008). A role for protein kinase CK2 in plant development: evidence obtained using a dominant-negative mutant. Plant J. Cell Mol. Biol. 55, 118–130. doi: 10.1111/j.1365-313X.2008.03494.x
Mulekar, J. J., and Huq, E. (2014). Expanding roles of protein kinase CK2 in regulating plant growth and development. J. Exp. Bot. 65, 2883–2893. doi: 10.1093/jxb/ert401
Nagel, D. H., and Kay, S. A. (2012). Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. CB 22, R648–R657. doi: 10.1016/j.cub.2012.07.025
Nagy, Z., Comer, S., and Smolenski, A. (2018). Analysis of Protein Phosphorylation Using Phos-Tag Gels. Curr. Protoc. Protein Sci. 93, e64. doi: 10.1002/cpps.64
Nemes, K., Gellért, Á, Almási, A., Vági, P., Sáray, R., Kádár, K., et al. (2017). Phosphorylation regulates the subcellular localization of cucumber mosaic virus 2b protein. Sci. Rep. 7, 13444. doi: 10.1038/s41598-017-13870-7
Nemes, K., Gellért, Á, Bóka, K., Vági, P., and Salánki, K. (2019). Symptom recovery is affected by cucumber mosaic virus coat protein phosphorylation. Virology 536, 68–77. doi: 10.1016/j.virol.2019.08.003
Newton, A. C. (1995). Protein kinase C: structure, function, and regulation. J. Biol. Chem. 270, 28495–28498. doi: 10.1074/jbc.270.48.28495
Newton, A. C. (2018). Protein kinase C: perfectly balanced. Crit. Rev. Biochem. Mol. Biol. 53, 208–230. doi: 10.1080/10409238.2018.1442408
Ni, F., Sun, S., Huang, C., and Zhao, Y. (2009). N-phosphorylation of amino acids by trimetaphosphate in aqueous solution—learning from prebiotic synthesis. Green Chem. 11, 569–573. doi: 10.1039/B817013D
Ni, F., Wu, L., Wang, Q., Hong, J., Qi, Y., and Zhou, X. (2017). Turnip yellow mosaic virus P69 interacts with and suppresses glk transcription factors to cause pale-green symptoms in Arabidopsis. Mol. Plant 10, 764–766. doi: 10.1016/j.molp.2016.12.003
Obata, T., Yaffe, M. B., Leparc, G. G., Piro, E. T., Maegawa, H., Kashiwagi, A., et al. (2000). Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J. Biol. Chem. 275, 36108–36115. doi: 10.1074/jbc.M005497200
Panni, S. (2019). Phospho-peptide binding domains in S. cerevisiae model organism. Biochimie 163, 117–127. doi: 10.1016/j.biochi.2019.06.005
Pérez, J., de, J., Udeshi, N. D., Shabanowitz, J., Ciordia, S., Juárez, S., et al. (2013). O-GlcNAc modification of the coat protein of the potyvirus Plum pox virus enhances viral infection. Virology 442, 122–131. doi: 10.1016/j.virol.2013.03.029
Pinna, L. A., and Ruzzene, M. (1996). How do protein kinases recognize their substrates? Biochim. Biophys. Acta 1314, 191–225. doi: 10.1016/s0167-4889(96)00083-3
Pitzschke, A. (2015). Modes of MAPK substrate recognition and control. Trends Plant Sci. 20, 49–55. doi: 10.1016/j.tplants.2014.09.006
Poss, Z. C., Ebmeier, C. C., Odell, A. T., Tangpeerachaikul, A., Lee, T., Pelish, H. E., et al. (2016). Identification of mediator kinase substrates in human cells using cortistatin A and quantitative phosphoproteomics. Cell Rep. 15, 436–450. doi: 10.1016/j.celrep.2016.03.030
Prasad, A., Sharma, N., Hari-Gowthem, G., Muthamilarasan, M., and Prasad, M. (2020). Tomato yellow leaf curl virus: impact, challenges, and management. Trends Plant Sci. 25, 897–911. doi: 10.1016/j.tplants.2020.03.015
Ramazi, S., and Zahiri, J. (2021). Post-translational modifications in proteins: resources, tools and prediction methods. Database J. Biol. Databases Curation 2021, baab012. doi: 10.1093/database/baab012
Rayapuram, N., Bigeard, J., Alhoraibi, H., Bonhomme, L., Hesse, A.-M., Vinh, J., et al. (2018). Quantitative phosphoproteomic analysis reveals shared and specific targets of Arabidopsis mitogen-activated protein kinases (MAPKs) MPK3, MPK4, and MPK6. Mol. Cell. Proteomics MCP 17, 61–80. doi: 10.1074/mcp.RA117.000135
Samuilova, O., Santala, J., and Valkonen, J. P. T. (2013). Tyrosine phosphorylation of the triple gene block protein 3 regulates cell-to-cell movement and protein interactions of Potato mop-top virus. J. Virol. 87, 4313–4321. doi: 10.1128/JVI.03388-12
Schulze, W. X., Yao, Q., and Xu, D. (2015). Databases for plant phosphoproteomics. Methods Mol. Biol. Clifton NJ 1306, 207–216. doi: 10.1007/978-1-4939-2648-0_16
Shen, Q., Hu, T., Bao, M., Cao, L., Zhang, H., Song, F., et al. (2016). Tobacco RING E3 ligase NtRFP1 mediates ubiquitination and proteasomal degradation of a geminivirus-encoded βC1. Mol. Plant 9, 911–925. doi: 10.1016/j.molp.2016.03.008
Shen, Q., Liu, Z., Song, F., Xie, Q., Hanley-Bowdoin, L., and Zhou, X. (2011). Tomato SlSnRK1 protein interacts with and phosphorylates βC1, a pathogenesis protein encoded by a geminivirus β-satellite. Plant Physiol. 157, 1394–1406. doi: 10.1104/pp.111.184648
Shen, W., Bobay, B. G., Greeley, L. A., Reyes, M. I., Rajabu, C. A., Blackburn, R. K., et al. (2018). Sucrose nonfermenting 1-related protein kinase 1 phosphorylates a geminivirus rep protein to impair viral replication and infection. Plant Physiol. 178, 372–389. doi: 10.1104/pp.18.00268
Shen, W., Dallas, M. B., Goshe, M. B., and Hanley-Bowdoin, L. (2014). SnRK1 phosphorylation of AL2 delays cabbage leaf curl virus infection in Arabidopsis. J. Virol. 88, 10598–10612. doi: 10.1128/JVI.00761-14
Shi, J., Gu, J., Dai, C., Gu, J., Jin, X., Sun, J., et al. (2015). O-GlcNAcylation regulates ischemia-induced neuronal apoptosis through AKT signaling. Sci. Rep. 5, 14500. doi: 10.1038/srep14500
Singh, J. P., Zhang, K., Wu, J., and Yang, X. (2015). O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 356, 244–250. doi: 10.1016/j.canlet.2014.04.014
Song, N., Qi, Q., Cao, R., Qin, B., Wang, B., Wang, Y., et al. (2019). MAVS O-GlcNAcylation Is essential for host antiviral immunity against Lethal RNA Viruses. Cell Rep. 28, 2386.e–2396.e. ***2386-2396.e5, doi: 10.1016/j.celrep.2019.07.085
Songyang, Z., Blechner, S., Hoagland, N., Hoekstra, M. F., Piwnica-Worms, H., and Cantley, L. C. (1994). Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. CB 4, 973–982. doi: 10.1016/s0960-9822(00)00221-9
Stoevesandt, O., and Taussig, M. J. (2013). Phospho-specific antibodies by design. Nat. Biotechnol. 31, 889–891. doi: 10.1038/nbt.2712
Stork, J., Panaviene, Z., and Nagy, P. D. (2005). Inhibition of in vitro RNA binding and replicase activity by phosphorylation of the p33 replication protein of cucumber necrosis tombusvirus. Virology 343, 79–92. doi: 10.1016/j.virol.2005.08.005
Tan, S.-T., and Xue, H.-W. (2014). Casein kinase 1 regulates ethylene synthesis by phosphorylating and promoting the turnover of ACS5. Cell Rep. 9, 1692–1702. doi: 10.1016/j.celrep.2014.10.047
Thomas, G. M., Frame, S., Goedert, M., Nathke, I., Polakis, P., and Cohen, P. (1999). A GSK3-binding peptide from FRAT1 selectively inhibits the GSK3-catalysed phosphorylation of Axin and β-catenin. FEBS Lett. 458, 247–251. doi: 10.1016/S0014-5793(99)01161-8
Trutnyeva, K., Bachmaier, R., and Waigmann, E. (2005). Mimicking carboxyterminal phosphorylation differentially effects subcellular distribution and cell-to-cell movement of Tobacco mosaic virus movement protein. Virology 332, 563–577. doi: 10.1016/j.virol.2004.11.040
van Wijk, K. J., Friso, G., Walther, D., and Schulze, W. X. (2014). Meta-Analysis of Arabidopsis thaliana phospho-proteomics data reveals compartmentalization of phosphorylation motifs. Plant Cell 26, 2367–2389. doi: 10.1105/tpc.114.125815
Vigerust, D. J., and Shepherd, V. L. (2007). Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 15, 211–218. doi: 10.1016/j.tim.2007.03.003
Vijayapalani, P., Chen, J. C.-F., Liou, M.-R., Chen, H.-C., Hsu, Y.-H., and Lin, N.-S. (2012). Phosphorylation of bamboo mosaic virus satellite RNA (satBaMV)-encoded protein P20 downregulates the formation of satBaMV-p20 ribonucleoprotein complex. Nucleic Acids Res. 40, 638–649. doi: 10.1093/nar/gkr705
Vosseller, K., Wells, L., Lane, M. D., and Hart, G. W. (2002). Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. U. S. A. 99, 5313–5318. doi: 10.1073/pnas.072072399
Waigmann, E., Chen, M. H., Bachmaier, R., Ghoshroy, S., and Citovsky, V. (2000). Regulation of plasmodesmal transport by phosphorylation of tobacco mosaic virus cell-to-cell movement protein. EMBO J. 19, 4875–4884. doi: 10.1093/emboj/19.18.4875
Wan, J., Liu, H., Chu, J., and Zhang, H. (2019). Functions and mechanisms of lysine crotonylation. J. Cell. Mol. Med. 23, 7163–7169. doi: 10.1111/jcmm.14650
Wang, X., Jiang, Z., Yue, N., Jin, X., Zhang, X., Li, Z., et al. (2021). Barley stripe mosaic virus γb protein disrupts chloroplast antioxidant defenses to optimize viral replication. EMBO J. 40, e107660. doi: 10.15252/embj.2021107660
Wu, D., and Pan, W. (2010). GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem. Sci. 35, 161–168. doi: 10.1016/j.tibs.2009.10.002
Xi, L., Zhang, Z., Herold, S., Kassem, S., Wu, X. N., and Schulze, W. X. (2021). Phosphorylation site motifs in plant protein kinases and their substrates. Methods Mol. Biol. Clifton NJ 2358, 1–16. doi: 10.1007/978-1-0716-1625-3_1
Xu, J., Liu, D., Zhang, Y., Wang, Y., Han, C., Li, D., et al. (2016). Improved pathogenicity of a beet black scorch virus variant by low temperature and co-infection with its satellite RNA. Front. Microbiol. 7:1771. doi: 10.3389/fmicb.2016.01771
Xu, S., Xiao, J., Yin, F., Guo, X., Xing, L., Xu, Y., et al. (2019). The protein modifications of o-glcnacylation and phosphorylation mediate vernalization response for flowering in winter wheat. Plant Physiol. 180, 1436–1449. doi: 10.1104/pp.19.00081
Yan, T., Zhu, J.-R., Di, D., Gao, Q., Zhang, Y., Zhang, A., et al. (2015). Characterization of the complete genome of barley yellow striate mosaic virus reveals a nested gene encoding a small hydrophobic protein. Virology 478, 112–122. doi: 10.1016/j.virol.2014.12.042
Yang, J., Zhang, T., Li, J., Wu, N., Wu, G., Yang, J., et al. (2020). Chinese wheat mosaic virus-derived vsiRNA-20 can regulate virus infection in wheat through inhibition of vacuolar- (H+)-PPase induced cell death. New Phytol. 226, 205–220. doi: 10.1111/nph.16358
Yang, L. (2018). Neuronal cAMP/PKA signaling and energy homeostasis. Adv. Exp. Med. Biol. 1090, 31–48. doi: 10.1007/978-981-13-1286-1_3
Yang, X., and Qian, K. (2017). Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 18, 452–465. doi: 10.1038/nrm.2017.22
Yin, X., Wang, X., and Komatsu, S. (2018). Phosphoproteomics: Protein phosphorylation in regulation of seed germination and plant growth. Curr. Protein Pept. Sci. 19, 401–412. doi: 10.2174/1389203718666170209151048
Yoo, M.-J., Albert, V. A., Soltis, P. S., and Soltis, D. E. (2006). Phylogenetic diversification of glycogen synthase kinase 3/SHAGGY-like kinase genes in plants. BMC Plant Biol. 6:3. doi: 10.1186/1471-2229-6-3
Yoshioka, K., Matsushita, Y., Kasahara, M., Konagaya, K., and Nyunoya, H. (2004). Interaction of tomato mosaic virus movement protein with tobacco RIO kinase. Mol. Cells 17, 223–229.
Youn, J.-H., and Kim, T.-W. (2015). Functional insights of plant GSK3-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol. Plant 8, 552–565. doi: 10.1016/j.molp.2014.12.006
Zelada, A., Castilla, R., Passeron, S., Giasson, L., and Cantore, M. L. (2002). Interactions between regulatory and catalytic subunits of the Candida albicans cAMP-dependent protein kinase are modulated by autophosphorylation of the regulatory subunit. Biochim. Biophys. Acta 1542, 73–81. doi: 10.1016/s0167-4889(01)00168-9
Zhang, G., Annan, R. S., Carr, S. A., and Neubert, T. A. (2014). Overview of peptide and protein analysis by mass spectrometry. Curr. Protoc. Mol. Biol. 108, 10.21.1–10.21.30. doi: 10.1002/0471142727.mb1021s108
Zhang, K., Yin, R., and Yang, X. (2014). O-GlcNAc: A bittersweet switch in liver. Front. Endocrinol. 5:221. doi: 10.3389/fendo.2014.00221
Zhang, X., Dong, K., Xu, K., Zhang, K., Jin, X., Yang, M., et al. (2018). Barley stripe mosaic virus infection requires PKA-mediated phosphorylation of γb for suppression of both RNA silencing and the host cell death response. New Phytol. 218, 1570–1585. doi: 10.1111/nph.15065
Zhang, X., Wang, X., Xu, K., Jiang, Z., Dong, K., Xie, X., et al. (2021). The serine/threonine/tyrosine kinase STY46 defends against hordeivirus infection by phosphorylating γb protein. Plant Physiol. 186, 715–730. doi: 10.1093/plphys/kiab056
Zhang, Z., Tan, M., Xie, Z., Dai, L., Chen, Y., and Zhao, Y. (2011). Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63. doi: 10.1038/nchembio.495
Zhao, X., Wang, X., Dong, K., Zhang, Y., Hu, Y., Zhang, X., et al. (2015). Phosphorylation of beet black scorch virus coat protein by PKA is required for assembly and stability of virus particles. Sci. Rep. 5, 11585. doi: 10.1038/srep11585
Keywords: phosphorylation, replication, intracellular movement, long-distance movement, infection cycle
Citation: Zhuang X, Guo X, Gu T, Xu X, Qin L, Xu K, He Z and Zhang K (2022) Phosphorylation of plant virus proteins: Analysis methods and biological functions. Front. Microbiol. 13:935735. doi: 10.3389/fmicb.2022.935735
Received: 04 May 2022; Accepted: 28 June 2022;
Published: 26 July 2022.
Edited by:
Beilei Wu, Institute of Plant Protection (CAAS), ChinaReviewed by:
Guanwei Wu, Ningbo University, ChinaXuefeng Yuan, Shandong Agricultural University, China
Copyright © 2022 Zhuang, Guo, Gu, Xu, Qin, Xu, He and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kun Zhang, emtAeXp1LmVkdS5jbg==; emhhbmdrdW44ODAzMjRAY2F1LmVkdS5jbg==
 Xinjian Zhuang
Xinjian Zhuang Xiao Guo
Xiao Guo Tianxiao Gu
Tianxiao Gu Xiaowei Xu
Xiaowei Xu Lang Qin1
Lang Qin1 Kai Xu
Kai Xu Zhen He
Zhen He Kun Zhang
Kun Zhang