- 1Department of Human Genetics, McGill University, Montréal, QC, Canada
- 2McGill University Research Centre on Complex Traits, Montréal, QC, Canada
- 3Swine and Poultry Infectious Diseases Research Center and Research Group on Infectious Diseases in Production Animals, Faculty of Veterinary Medicine, University of Montreal, Saint-Hyacinthe, QC, Canada
- 4Department of Microbiology and Immunology, McGill University, Montréal, QC, Canada
- 5CNRS, ImmunoConcEpT, UMR 5164, Université de Bordeaux, Bordeaux, France
- 6CHU de Bordeaux, FHU ACRONIM, Centre National de Référence des Maladies Auto-Immunes et Systémiques Rares Est/Sud-Ouest, Bordeaux, France
- 7Department of Biochemistry, McGill University, Montréal, QC, Canada
- 8Department of Pharmacology, McGill University, Montréal, QC, Canada
Since the end of 2019, the world has been challenged by the coronavirus disease 2019 (COVID-19) pandemic. With COVID-19 cases rising globally, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to evolve, resulting in the emergence of variants of interest (VOI) and of concern (VOC). Of the hundreds of millions infected, immunodeficient patients are one of the vulnerable cohorts that are most susceptible to this virus. These individuals include those with preexisting health conditions and/or those undergoing immunosuppressive treatment (secondary immunodeficiency). In these cases, several researchers have reported chronic infections in the presence of anti-COVID-19 treatments that may potentially lead to the evolution of the virus within the host. Such variations occurred in a variety of viral proteins, including key structural ones involved in pathogenesis such as spike proteins. Tracking and comparing such mutations with those arisen in the general population may provide information about functional sites within the SARS-CoV-2 genome. In this study, we reviewed the current literature regarding the specific features of SARS-CoV-2 evolution in immunocompromised patients and identified recurrent de novo amino acid changes in virus isolates of these patients that can potentially play an important role in SARS-CoV-2 pathogenesis and evolution.
Introduction
In late 2019, a new viral outbreak in Wuhan city, China (World Health Organization [WHO], 2020a), rapidly identified as the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), resulted in the coronavirus disease 2019 (COVID-19) pandemic (World Health Organization [WHO], 2020b), which still continues with the rise of novel variants of concern (VOCs) and of interest (VOIs).
Increased age is perhaps the strongest risk factor for severe COVID-19 (Bonanad et al., 2020); obesity, male gender, and various comorbidities such as hypertension, cardiovascular disease, and diabetes also contribute to an increased odds ratio of severe disease (Hu and Wang, 2021). However, among infected individuals, patients with secondary immunodeficiency, due to preexisting health conditions, and those undergoing immunosuppressive treatment are particularly susceptible to SARS-CoV-2 (Gao et al., 2020a; Liu and Hill, 2020; Hoffmann et al., 2021; Jones et al., 2021). Many research groups have reported chronic infections and the accumulation of viral protein-coding mutations in such individuals in the presence of anti-COVID-19 treatments, with potential relevance at both biological and epidemiological levels. We hypothesized that two main kinds of mutations could be observed in such immunodeficient setting, namely, (1) variations selected by antiviral treatment and (2) variations reflecting the adaptation of the virus to the human host, particularly in the context of an environment with reduced immune responses, allowing niches of selective pressure.
To gain insights into the mutational signatures of secondary immunodeficiency in SARS-CoV-2 genetic profiles, we have queried the literature to review SARS-CoV-2 genome data from 44 patients with secondary immunodeficiency who underwent treatment against COVID-19. We retrieved 148 full genomes from 21 patients and partial genomes for 24 patients. By analyzing the viral genomes detected in these patients in comparison with circulating variants, we identified numerous new protein-coding mutations and inspected their predicted structural or functional impact at the protein level.
SARS-CoV-2
SARS-CoV-2 is a betacoronavirus that shares 96% of its genomic identity with the RaTG13 bat coronavirus and is hypothesized to be of zoonotic origin (Zhou et al., 2020; Banerjee et al., 2021). It is a positive sense ribonucleic acid virus (RNA), with a genome spanning around 30 kilobases in length (Wu et al., 2020; V’kovski et al., 2021). Notably, two-thirds of its genome is composed of overlapping open reading frames (ORF) 1a and 1b, which together encode for an RNA-dependent RNA polymerase (RdRp) and other non-structural proteins important for viral replication and transcription (Figure 1A; Wang Q. et al., 2020; Wang Y. et al., 2020; Yan et al., 2021). The remainder of the viral genome is composed of ORFs 2–10 encoding for structural and accessory proteins (Figure 1A; Davidson et al., 2020; Jiang et al., 2020; Michel et al., 2020; Mohammad et al., 2020; Pancer et al., 2020).
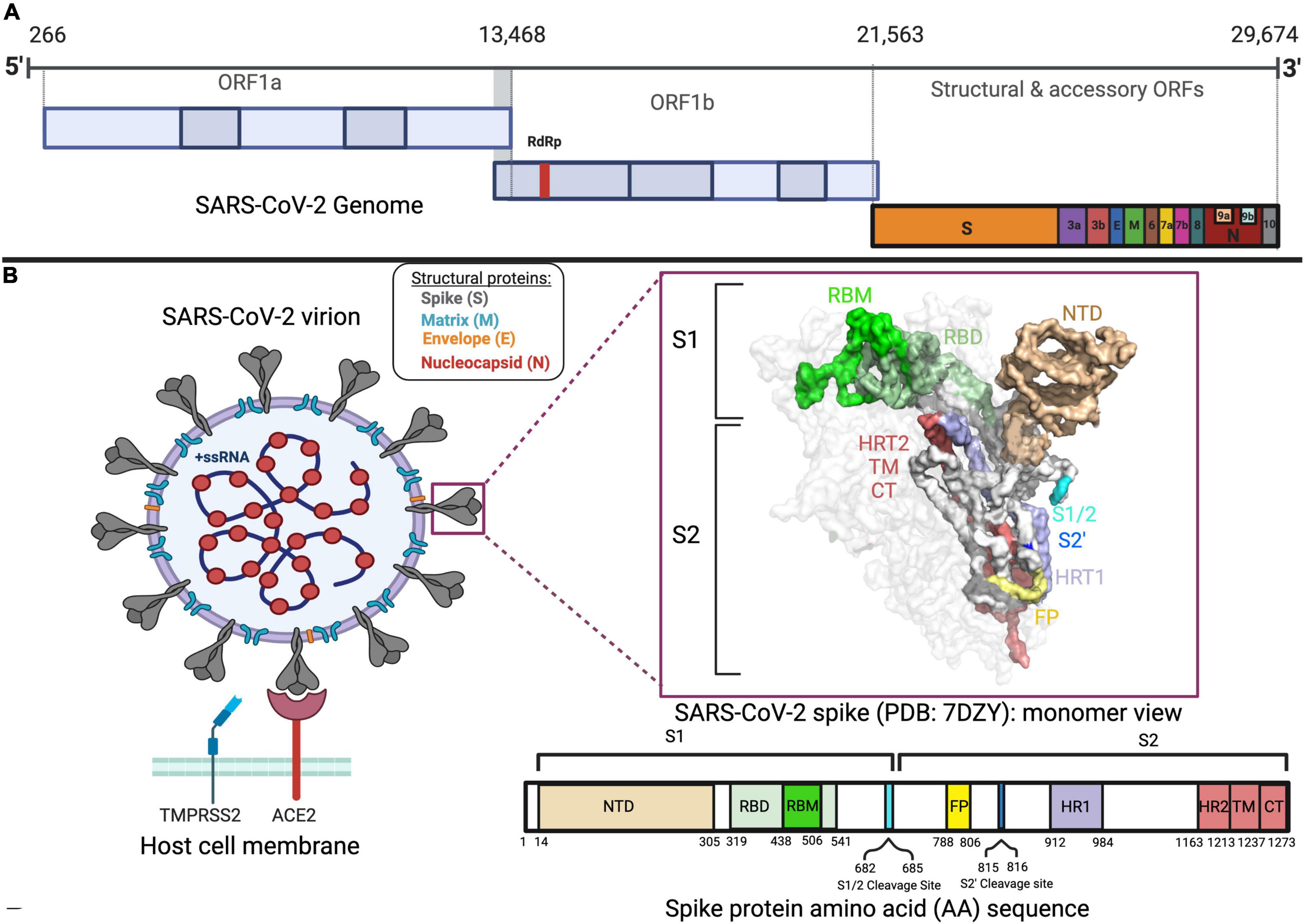
Figure 1. SARS-CoV-2 genome, spike, and virion. (A) The genomic sequence of SARS-CoV-2 with different open reading frames (ORFs) is displayed in different colors. (B) Representation of the SARS-CoV-2 virion structure, spike protein and amino acid sequence of spike protein, and its domains in different colors. The binding of SARS-CoV-2 virion to the ACE2 receptor adjacent to the TMPRSS2 protein is also shown.
Of the structural proteins, the spike is a large accessible homotrimeric protein of great importance in viral tropism and viral entry, making it a great target in therapeutic development (Conceicao et al., 2020). With a molecular weight of around 180 kDa, the spike protein is composed of 2 major subunits per monomer: the S1 (residues 14–685) and S2 (residues 686–1273) (Figure 1B; Huang et al., 2020; Martí et al., 2021). The former is the most variable part of the spike among coronaviruses and contains the amino (N)-terminal domain (NTD) and the receptor-binding domain (RBD) (Figure 1B; Huang et al., 2020; Martí et al., 2021). As for the S2, its domains, which are essential for viral fusion with the host cell membrane, are more conserved in structure and sequence (Figure 1B; Huang et al., 2020; Martí et al., 2021).
The main target of the spike is the angiotensin-converting enzyme (ACE2) (Li et al., 2020). The broad expression of ACE2 explains in part SARS-CoV-2 pathogenesis in a multitude of organs from respiratory, circulatory, urogenital, gastrointestinal, and nervous systems (Lopes-Pacheco et al., 2021).
Following cell entry, the replication of SARS-CoV-2 takes place in the cytoplasm with the help of the host ribosomal machinery, translating the ORF 1a and 1b genes into two large replicase polyproteins, namely, pp1a and pp1ab (V’kovski et al., 2021). Together, both pp1a and pp1ab polyproteins undergo proteolytic cleavages via the viral-encoded proteinases papain-like protease (PL-pro, Nsp3) and 3C-like protease (3CL-pro, Nsp5) to generate 16 mature non-structural proteins, i.e., Nsp1 to Nsp16 (Astuti and Ysrafil, 2020). Proteolysis is an essential step for viral replication, which is why antivirals targeting proteases are of interest (Dampalla et al., 2021; Roe et al., 2021). Later, the RNA-dependent RNA polymerase (RdRp and Nsp12), helicase (Nsp13), and Nsp7 to Nsp9 form the replication/transcription complex (RTC), allowing the synthesis of viral RNA in double-membrane vesicles (DMV) at the periphery of the endoplasmic reticulum (Brant et al., 2021).
Mutations in Emerging Variants
Like most RNA viruses, SARS-CoV-2 continues to mutate as it spreads, resulting in different variants, where the Pango numeric system assigns lineages with a number or letter such as B.1 (Oude Munnink et al., 2021). Among the variants circulating as of May 08 2022, five are known VOCs defined by the WHO based on their epidemiology and their association with disease severity or potential to escape available treatments or vaccines (Figure 2; Harvey et al., 2021; World Health Organization [WHO], 2021). Martin et al. (2021) reported that there has been a shift in the mutational landscape of some VOCs with the N501Y spike amino acid substitution (alpha, beta, and gamma) where there have been mutations arising independently and repeatedly in different viral lineages at 29 genome sites from 15 March 2021 to 1 June 2021. Such converging evolution in these sites could likely occur in variants of the same and different lineages (Martin et al., 2021). Variations in spike proteins that define VOCs are highlighted in Figure 2.
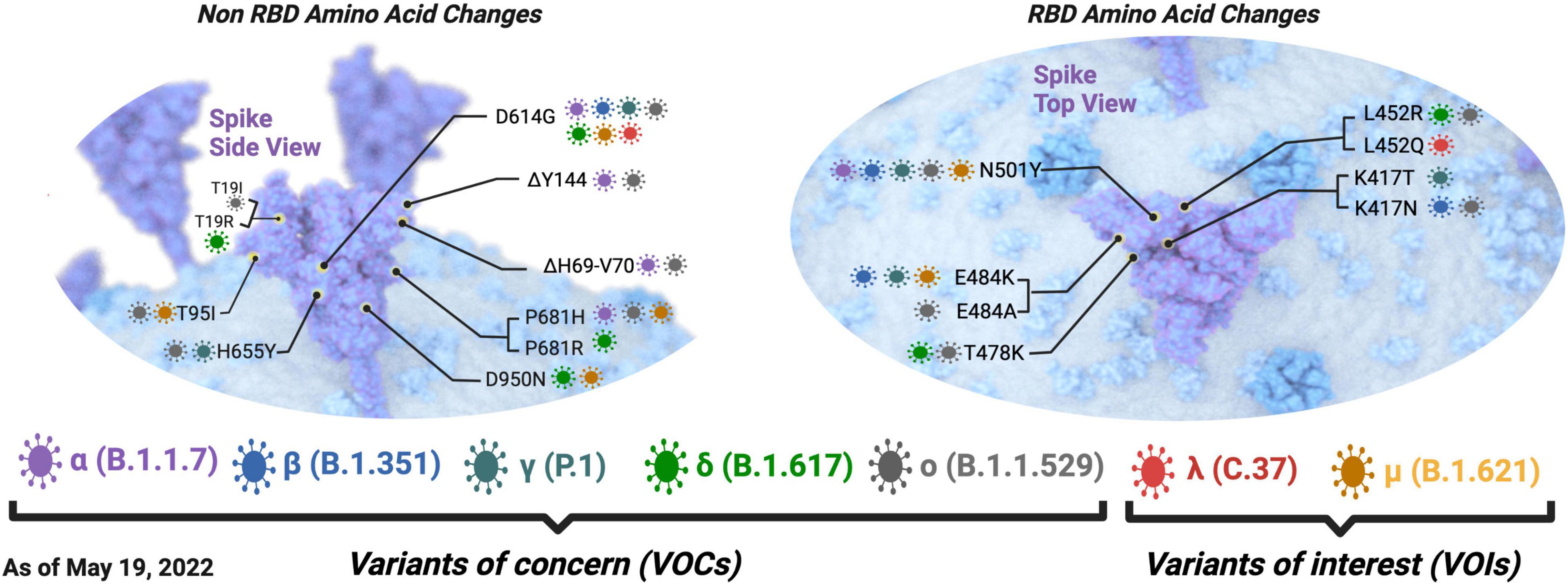
Figure 2. Defining spike amino acid changes in SARS-CoV-2 variant of concerns (VOCs) and interest (VOIs). The variants of concern are depicted in purple, blue, dark green, pale green, and pale gray for the alpha(α), beta (β), gamma (γ), delta (δ), and omicron (o) variants, respectively. The variants of interest are depicted in red and dark yellow for the lambda (λ) and mu (μ) variants, respectively. The envelope is shown in blue, and the spike protein is shown in purple. Common amino acid changes in different variants are also depicted. Non-RBD amino acid changes are shown on the left, and RBD amino acid changes are shown on the right. Defining amino acid changes are those appearing at the phylogenetic root of a variant (Hodcroft, 2021). Adapted from “The SARS-CoV-2 Variants of Concern,” by BioRender.com (2021).
As a nidovirus, SARS-CoV-2 encodes a unique proofreading enzyme 3′ to 5′ exonuclease (ExoN) involved in excising faulty nucleotides inserted by RNA polymerases, thus ensuring replication fidelity (Shannon et al., 2020; Gribble et al., 2021). Despite this proofreading mechanism, SARS-CoV-2 has shown a capacity to accumulate a wide range and high number of mutations (Chen et al., 2020). A study of samples from the first wave and second wave of COVID-19 in Japan noted a mutation rate of 1.16–1.87 × 10–3 base substitutions/site/year (Ko et al., 2021). This is relatively low compared with the human immunodeficiency virus (HIV) subtype B, which can have a nucleotide substitution rate ranging from 5.25 × 10–3 to 1.60 × 10–2 substitutions/site/year in gag and env-gp120 genes (Dapp et al., 2017).
However, there are more ways of generating genetic diversity; viral recombination, which is the generation of new progeny from two distinct strains of virus co-infecting a cell, is a way to generate viral genetic variation (Simon-Loriere and Holmes, 2011). In the case of SARS-CoV-2, Pollet et al. (2021) performed a recombination analysis of a variety of coronaviral sequences including 100,000 SARS-CoV-2 sequences. Through this analysis, they showed eight SARS-CoV-2 recombination events, two of them in the spike gene (Pollet et al., 2021). Earlier in 2021, a SARS-CoV-2 co-infection event of a single patient was reported with two strains with distinct lineages, which raises concern for the recombination of SARS-CoV-2 evolution (Francisco et al., 2021).
Furthermore, the genetic variability of viruses is shaped through the selection pressure of their host cell or environment. The host has multiple immune defense mechanisms at cellular, tissue, and systemic levels that can interfere with viral replication and spread. Letko et al. (2018) showed an example of MERS-CoV causing observable cytopathic effect due to the accumulation of amino acid variations in the spike protein after eight viral passages in BHK cells expressing the bat DPP4 receptor. Antiviral treatments that target specific viral proteins are another selective pressure that can result in the development of treatment-resistant mutants. For instance, the emergence of two mutations in the RdRp of murine hepatitis virus (MHV) conferred a 5.6-fold increased resistance to remdesivir (based on EC50 values) (Agostini Maria et al., 2018). The study of virus sequences that emerge in chronically infected patients could reveal regions of the virus genome that will be important as we prepare for and predict future variants.
Secondary Immunodeficiencies
The most common cause of immunodeficiency is acquired immunodeficiency, meaning impaired immune response secondary to a condition or its treatment. This review will focus on the four main types we have found to be associated with COVID-19, namely, cancer, organ transplantation, HIV infection/AIDS, and autoimmune diseases.
Indeed, it has been documented that immunosuppression leads to poorer prognosis in hospitalized patients (Ponsford et al., 2021), especially in cancer and organ-transplanted patients (Elkrief et al., 2020; Bhogal et al., 2021; Coll et al., 2021), as well as in HIV-infected patients (Suwanwongse and Shabarek, 2020; Kanwugu and Adadi, 2021). In cancer, this impaired immune response can result from the medical condition itself, for example, impaired humoral response in a chronic lymphocytic leukemia or bone marrow infiltration by an acute leukemia preventing the development of normal leukocytes. But immunosuppression can also be induced by the malignancy treatment: hypogammaglobulinemia induced by B cell depletion after rituximab use or by alkylating agent that impairs DNA from replicating cells (including cancer cells and leukocytes). The same kind of treatment-induced immune impairment happens in organ transplantation contexts, with the immunosuppressive regimen used for the prevention of graft rejection.
In HIV-affected patients, with incomplete or without antiretroviral treatment, HIV infection leads to low CD4 + T cells count, and the decrease in these cells gives rise to opportunistic diseases (Chinen and Shearer, 2010).
Autoimmune diseases are a heterogenous group of diseases characterized by loss of tolerance to self-antigens, leading to the development of autoantibodies and activation of the immune system, resulting in immune complex deposits, organ failure, and ultimately death in the most severe cases (Kaul et al., 2016; Denton and Khanna, 2017). Treatment options involve mainly the use of non-specific immunosuppressive agents such as high-dose corticosteroids or cyclophosphamide (alkylating agent), as well as targeted therapies such as rituximab (anti-CD20) and anti-TNFα (infliximab and adalimumab). The increased risk of infectious disease upon immunosuppressive therapies is well documented (Lode and Schmidt-Ioanas, 2005; Barber and Clarke, 2020; Mitratza et al., 2021), but the impact of autoimmune diseases and their therapies on COVID-19 disease course remains debated (Kastritis et al., 2020; Murtas et al., 2020; Pablos et al., 2020; Zen et al., 2020; Zhong et al., 2020; Liu et al., 2021b).
Study Population
Our literature review found 44 patients with prolonged SARS-CoV-2 infection affected with secondary immunodeficiency, summarized in Table 1. A more detailed version of Table 1 is attached in Supplementary Table 1. These patients were described in papers found using the two search engines: PubMed and Google Scholar with key phrases “SARS-CoV-2 chronic infection,” “SARS-CoV-2 evolution immunocompromised,” and “SARS-CoV-2 evolution immunodeficiency” queried until May 08 2022.
Among this population, 27 patients were affected with cancer, one with cholangiocarcinoma (P27) and 26 with hematopoietic malignancies. These encompass chronic lymphocytic leukemias (P1–6), acute leukemias (P7–12), lymphomas (P13–25), and multiple myeloma (P26). All these patients presented with a humoral deficiency, either due to the initial pathology or received treatments that combine anti-CD20 monoclonal antibodies that deplete B cells. This wide spectrum of antibody and chemotherapy regimens, including, for example, bendamustine or cyclophosphamide, has a broad effect on innate and adaptive immune responses. Four patients (P28–P31) were solid organ recipients with drug-induced immunosuppression designed to prevent graft rejection using a wide spectrum of immunosuppressors, such as mycophenolate mofetil, tacrolimus, cyclosporine, azathioprine, and steroids. Five patients (P32–P36) were HIV-infected individuals with CD4+ T cells impairment due to the viral infection. Two patients (P37 and P38) were affected with autoimmunity: one with antiphospholipid syndrome and one with ANCA-associated vasculitis. Finally, six patients (P39–P44) were immunocompromised by other associated comorbidities such as diabetes, chronic heart, or kidney disease. Of note, two of these patients (P40 and P44) were affected by autoimmune diseases (juvenile idiopathic arthritis and rheumatoid arthritis, respectively). (No information on an eventual immunosuppressive regimen was recorded for those two patients).
Among the 44 patients, 38 received treatment directly targeting the virus (associated or not to treatment for the cytokine storm or host-based therapies for immunomodulation). Notably, twenty-one (21) patients received anti-spike monoclonal antibodies (mAb) that target the receptor-binding domain (RBD) of the S protein (Kumar S. et al., 2021). Three patients (P7, P21, and P37) received an association of 2 mAb, casirivimab–imdevimab (Regeneron/Roche REGEN-COV/Ronapreve®), and 18 patients received bamlanivimab (16 in monotherapy, two in association with etesevimab, both from Lilly).
Of note, 21 patients received direct antiviral treatments; 21 patients were associated with remdesivir (an adenosine analog that directly inhibits the viral RNA polymerase, Gilead), and 3 patients were associated with lopinavir-ritonavir (HIV-1 protease inhibitors that bind to the catalytic site of the protease and impair virion production). Sixteen patients received convalescent plasma (CP), three patients in association with polyvalent immunoglobulins. Eight patients received polyvalent immunoglobulins intravenously [three patients with CP as mentioned, two patients with hyperimmune plasma (HP), and three patients in monotherapies]. Three patients received hyperimmune plasma (HP), two with IV immunoglobulins and one in monotherapy.
All these therapeutic data are summarized in Table 1, and previous immunosuppressive regimen, cytokine storm treatment, and host-based therapies are described in Supplementary Table 1.
Furthermore, viral genomes of patients were analyzed for predicted novel amino acid changes found in the different proteins. We defined novel amino acid changes as those detected as different from the earliest viral sequence obtained from the patient at admission. The underlying variations discussed are based on recurrence and/or structural significance, if reported in other studies. To have a common nomenclature for the corresponding amino acid changes, the ORF1ab polyprotein amino acid changes provided in some papers were converted to the corresponding Nsps. First, the amino acid sequence of the Nsp of interest was aligned with the sequence of polyprotein 1ab using BLAST. Second, the corresponding position of the amino acid change was determined and manually annotated. When only nucleotide sequences were available, the nucleotide changes in the different codons of different genes were manually determined. When our compiled studies showed amino acid changes with respect to the ORF1b gene only, we added nine amino acids to the position of the change to account for the ribosomal slippage in which the first nine amino acids are read in ORF1a and the following amino acids in the ORF1b (Bhatt et al., 2021).
In addition, in all patients examined (Table 1), the number of amino acid changes occurring in each SARS-CoV-2 protein reported by studies that performed full-length genome sequencing of SARS-CoV-2 was aggregated and divided by the number of amino acids per respective protein to generate a variation frequency and allow us to better visualize which proteins are the most changed (Figure 3). The full list of these variations is shown in Supplementary Table 2. To compare with the total number of SARS-CoV-2 sequenced viruses, the percentage of each amino acid change was calculated from the GISAID database as of May 18 2022 (using 10,900,892 as the total number of deposited sequences, and shown in Supplementary Table 3). The subsequent sections focus on the proteins with the most substitutions (in red-dark yellow heat map color on Figure 3) or substitutions with functional significance.
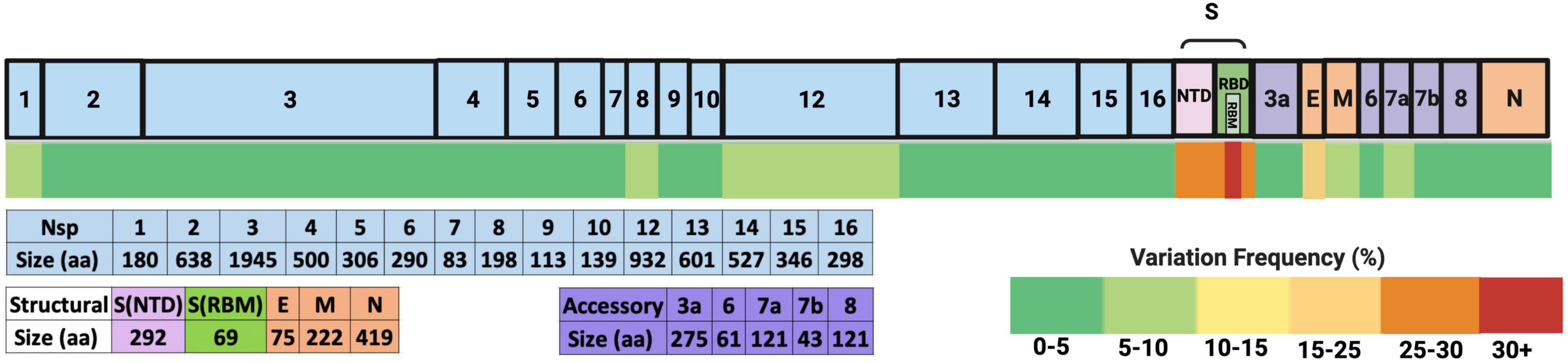
Figure 3. Frequency of amino acid variation in SARS-CoV-2 proteins from the analyzed secondary immunodeficient patients. The SARS-CoV-2 proteins are depicted on top, where the light blue boxes represent non-structural proteins (Nsps) generated from polyprotein 1ab (pp1ab). The NTD and RBM spike domains are shown as pink and green boxes, and other structural proteins are depicted in orange boxes (E, envelope; M, membrane; N, nucleocapsid). The accessory proteins are shown in purple. The total number of amino acids of each protein is depicted below. The heat map scale is shown on the bottom right. The changes shown were isolated from a total of 148 full-length SARS-CoV-2 genomic sequences from 21 patients (shown in S2).
From these data, it could be deduced that most variations occur in the spike protein, more particularly in the receptor-binding motif (RBM) and the NTD. Of note, the E protein has the second highest frequency, followed by Nsp12, Nsp1, Nsp8, ORF7a, and M. Variations in these proteins and others that are either recurrent and/or with functional significance are discussed in the following paragraphs.
Spike Protein Variations
Spike proteins had the highest number of amino acid changes, mainly in the RBM and in the NTD. Interestingly, the emergence of the E484K amino acid substitution (AAS) was observed in 43% of the patients studied (P3, P4, P12, P13, P28, P29, P32, P35, P37, P38, P39, P40, P41, P42, and P43). Secondary immunodeficient patients treated with bamlanivimab showcased a potential example of antiviral-induced selective pressure. Indeed, in April 2021, the emergency authorization use license of bamlanivimab monotherapy was revoked in the United States due to concerns about inefficiencies, as most circulating variants, especially the dominant delta variant, were resistant to neutralization (Gottlieb et al., 2021). Soon after, in January 2022, combined therapy of bamlanivimab and etesivimab, as well as casirivimab and imdevimab, was discouraged in the United States due to less neutralization activity against the now dominant omicron variant (Cavazzoni, 2022). All variations detected in SARS-CoV-2 spike proteins in the 44 patients are shown in Figure 4.
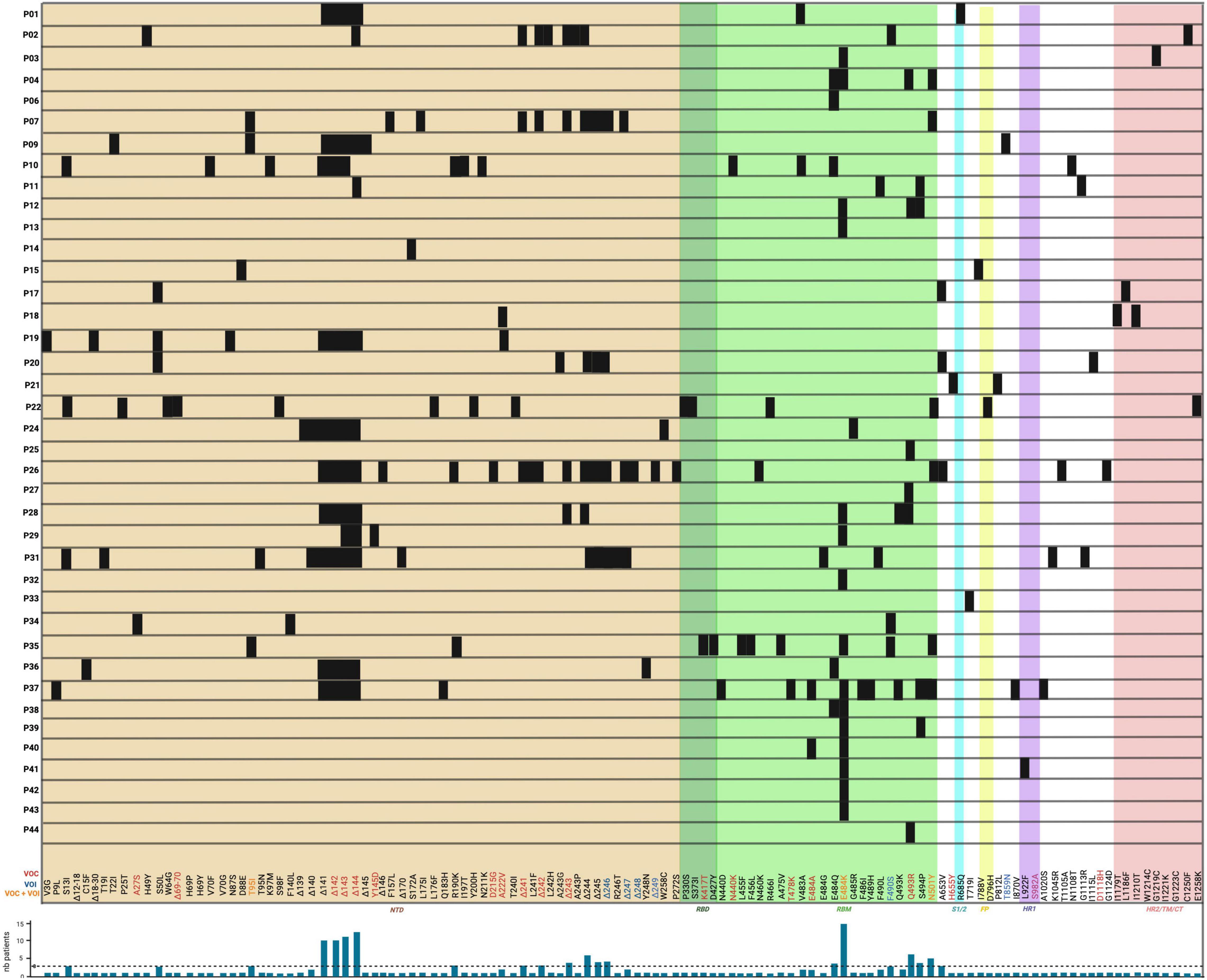
Figure 4. Location of novel SARS-CoV-2 amino acid changes in the spike protein emerging in some immunocompromised patients during chronic infection. The variations are shown as black boxes and represent either amino acid substitutions or deletion with their corresponding identity at the bottom. The different colored areas around the boxes are representative of the spike protein domains corresponding to those shown in Figure 1B: the brown area corresponds to the NTD; the dark green corresponds to the RBD; the pale green corresponds to the RBM of the RBD; the cyan corresponds to the S1/2; the yellow corresponds to the FP; the purple corresponds to the HR1, and the pale red corresponds to either HR2, TM, or CT domains. The amino acid change commonly found only in variants of concern (VOC) are in red font; those only in variants of interest (VOI) are in blue and those found in both VOC and VOI are in orange. Patients P5, P8, P16, P23, P30 did not present any novel amino acid changes in the spike protein and thus are not shown. A histogram depicts the occurrence of variations in the number of patients (shown on y-axis as “nb patients”) with the threshold of selection of three patients depicted by a dashed line.
Variations in the Receptor-Binding Motif
The spike RBM is a 69 amino acid motif (aa 438–506) involved in binding to the host cell receptor. From the selected patients, a total of 19 amino acid changes were found, with the most frequent change being the E484K substitution which was noted in 15 out of 44 patients (P3, P4, P12, P13, P28, P29, P32, P35, P37, P38, P39, P40, P41, P42, and P43). From these patients, 13 out of these 15 (29.5% of total patients) had received monoclonal antibody therapy, and of those, 12 (27% of total patients) were treated with bamlanivimab (Table 1). Furthermore, other variations at the same amino acid position (484) have also been reported. This includes the E484G, E484A, and E484Q substitutions that occurred in 1/44 (P31 – 2.3% of total patients), 2/44 (P37 and P40 – 4.5% of total patients), and 5/44 (P4, P6, P10, P36, and P38 – 11.4% of total patients) patients, respectively. For E484Q, 3 of 5 patients (6.8% of total patients) with this substitution received bamlanivimab, and both patients (4.5% of total patients) with E484A had received casirivimab and bamlanivimab, respectively, but P31 did not receive any monoclonal therapy. In recent studies, Jangra et al. (2021) showed that recombinant SARS-CoV-2 virus harboring the E484K AAS reduced in vitro antibody neutralization of human convalescent and post-vaccination sera relative to control virus without this variation. This result was also confirmed by Collier et al. (2021), showing loss of neutralizing activity by vaccine-elicited antibodies and monoclonal antibodies. In silico results by Wang et al. (2021) predicted that this AAS could result in favorable electrostatic interactions and tighter binding with the ACE2 receptor. In combination with another change not found in these patients (L452R), it has also been shown that a pseudotyped virus with the E484Q substitution resulted in a reduced neutralization of immune sera from vaccinated (against RBD) non-human primates, convalescent COVID-19 patients, as well as double-dose vaccinated individuals (also against RBD) (Li G. et al., 2021). This result was also confirmed by Ferreira et al. (2021) who also reported a decreased neutralization of pseudotyped virus with both E484Q and L452R alone or in combination using sera of vaccinated individuals. The AAS E484K is observed in beta, gamma, omicron, and mu variants, and E484A is found in the omicron variant (Hodcroft, 2021). Additionally, the 484 residue is in the proximity of suspected bamlanivimab- and casirivimab-binding sites (Figure 5), suggesting that these antibody therapies may have exerted selection pressure.
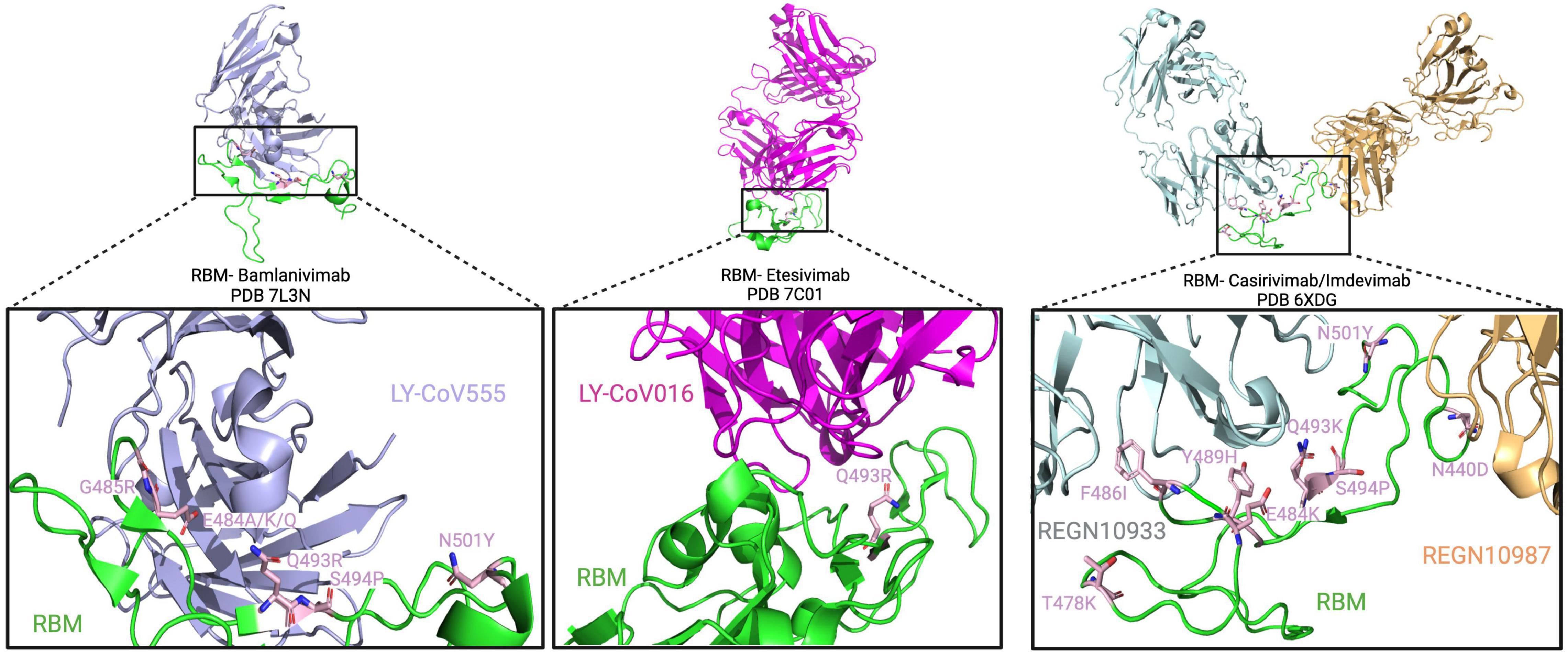
Figure 5. Spike RBM amino acid substitutions in patients treated with monoclonal antibodies. The interaction between the RBM and bamlanivimab, etesivimab, and casirivimab/imdevimab (light blue, pink, gray/yellow) is depicted on top from right to left, respectively. Below, a close-up view of the RBM/antibody interaction is shown with substitutions shown in light pink.
Another common variant observed in these secondary immunodeficient patients was the Q493R substitution, present in 6/44 (13.6% of total patients) patients (P4, P12, P25, P27, P28, and P44). All of them, except for P28, received bamlanivimab treatment. In combination with bamlanivimab, P25 and P27 also received etesivimab treatment, which binds the RBM in close proximity to this residue (Q493) (Figure 5). At the same position (493), the Q493K substitution was also reported in P28 as well as P37, where the latter had received casirivimab/imdevimab in combination (Choi et al., 2020). In terms of their abundance in the total number of SARS-CoV-2 sequences in GISAID, Q493R and Q493K have a frequency of 30.9 and 0.0079% respectively, suggesting an increased representation of the variation in secondary immunodeficient patients (Elbe and Buckland-Merrett, 2017). Clark et al. (2021) showed that SARS-CoV-2 spike pseudotyped viruses harboring the Q493K substitutions significantly decreased the neutralization of the REGN10933 (Casirivimab) and C1A-VH3-53 antibodies. This decreased neutralization was also noted in the case of Q493R pseudotyped virus (Clark et al., 2021). Similarly, a study by Starr Tyler et al. (2021) also investigated the potential of antibody escape mutations and demonstrated the escape of Q493K from the REGN10987 (imdevimab). Another nearby AAS is S494P, which has been observed in 4 patients (P11, P12, P37, and P39 – 9.1% of total patients), and is in 0.15% of all SARS-CoV-2 sequences in GISAID. This change has also led to the neutralization reduction in antibodies from convalescent and post-vaccinated sera, especially in combination with E484K and N501Y substitutions (Alenquer et al., 2021). These data suggest that Q493K and Q493R AAS could contribute to SARS-CoV-2 resistance to anti-spike monoclonal antibodies in immunodeficient patients.
Moreover, the N501Y AAS was also identified to emerge de novo in five patients (P4, P7, P22, P35, and P37), three of whom were treated with monoclonals (P4: bamlanivimab; P7 and P37: casirivimab/imdevimab). This AAS seems to be near the casirivimab-binding site to RBM (Figure 5). Furthermore, N501Y has also been determined to be present in alpha, beta, gamma, omicron, and mu variants. Having this AAS increases the binding affinity of spike proteins to human ACE2 as shown by Tian et al. (2021) and Liu et al. (2021a). Moreover, it has been shown that this substitution decreases neutralization by both H00S022 and 10F9 neutralizing monoclonal antibodies and increases the infectivity of pseudotyped virus by 5-fold compared with the 614G variant in HEK293T cells expressing mouse ACE2 (Li Q. et al., 2021). Interestingly, Niu et al. (2021) also noted that pseudotyped virus with the N501Y change effectively infected mouse-ACE2 expressing 293T cells and detected a successful infection of wild-type BALB/c with a SARS-CoV-2 strain bearing the substitution. A recent study by Liu et al. (2021a) reported that in vivo, this substitution enhanced viral fitness in intranasally infected hamsters and intra-cage transmissions. This suggests that N501Y may play a role in a potential viral spillover to mice (Huang et al., 2021).
Besides the aforementioned, other RBM changes were also found in the reviewed population, including the N440K (P10), T478K (P37), and F490S (P2, P34, and P35), which are found in the omicron, delta, and lambda variants, respectively. The N440K variant has been reported to evade the REGN10987 antibody (Starr Tyler et al., 2021), while F490S has been shown to allow resistance to vaccine-elicited sera (Kimura et al., 2021). Furthermore, among millions of GISAID sequences, the frequency of F490S, N440K, and T478K AASs is 0.17, 25.9, and 70.9%, respectively. Other AASs have also been identified in casirivimab/imdevimab receiving patients. In Figure 5, we showed the molecular structures and positions of the monoclonal antibodies and the AASs identified in the RBM of the spike. These findings suggest that either therapy (vaccine or antibody therapy) or convergent evolution explains their emergence.
(N)-Terminal Domain and other Spike Variations
Besides the RBM, spike amino acid changes were identified in other domains, especially in the 292 amino acid NTDs (aa 14–305). Among the NTD variations reported, the most frequent ones occurred in the range of residues between amino acid positions 139 and 146 and this occurred in 13 out of 44 patients (29.5% of the total patients, P1, P2, P9, P10, P11, P19, P24, P26, P28, P29, P31, P36, and P37). The second most common variant was a deletion occurring in a range of residues between amino acid positions 241 and 249 in 6 out of 44 patients (13.6% of total patients, P2, P7, P20, P26, P28, P31). In terms of substitutions, S50L (P17, P19, and P20), T95I (P7, P9, and P35), and R190K (P10, P26, and P35) were identified in 3 patients (6.8%). Moreover, other deletions were also reported at position 69–70 (Δ69–70) for P22, and both patients P19 and P37 had deletions from aa 18 to 30 (Δ18–30) and 12 to 18 (Δ12–18), respectively. Several of these amino acid changes were also present in VOCs and/or VOIs: a spike deletion at position 141 is present in the alpha variant, and the ones spanning from position 142–144 (Δ142–144) are in the omicron variant. Furthermore, the beta variant contains deletions from residue 241 to 243 (Δ241–243), whereas the lambda variant has a deletion from position 246 to 249 (Δ246–249). Both alpha and omicron variants contain the Δ69–70 deletion, and the lambda variant contains the T19I substitution (Hodcroft, 2021). Functionally, Mccarthy et al. (2021) showed that the combination of deletions (Δ69–70 and Δ 144/145), (Δ141–144, Δ144/145, and Δ146), and Δ243–244 all abolished binding to the 4A8 neutralizing antibody, indicating these regions in the NTD to be possible immunodominant epitopes for neutralization. Such an effect was also tested by Graham et al. (2021) where a significant reduction of neutralization by NTD targeting antibodies was also noted in pseudoviruses with the Δ141 spike deletion in combination with the D614G substitution. Furthermore, the researchers studying the chronic infection of P19 showed that double deletion Δ69–70, and another substitution reported in the spike fusion peptide D796H decreased the sensitivity to convalescent plasma in vitro (Kemp et al., 2021). It was also revealed that the Δ69–70 deletion had higher infectivity than a wild-type SARS-CoV-2 and that D796H was the main contributor to escaping neutralization while showing reduced infectivity (Kemp et al., 2021). The mutations seen in the NTD are functional mutations that overlap domains and have been observed in both VOCs and VOIs, and thus are worthy of significant focused interest for surveillance of future variants with altered biology. Besides the NTD and RBD, other amino acid changes occur in the spike such as the recurrent S13I in three patients (7.5% of total patients, P10, P22, and P31), the T859N (P9), and D1118H (P31) substitutions (2.5% of total patients). The T859N is also found in the lambda variant, and the D1118H is found in the alpha variant. The significance of these changes has not yet been determined. In sum, these results encompass the most recurrent amino acid changes observed in the spike protein of secondary immunodeficient patients described in the literature.
Variations in Non-Spike Proteins
Envelope
The SARS-CoV-2 envelope (E) is a 75 amino acid hydrophobic transmembrane protein that is crucial for infecting host cells (Boson et al., 2021). It is composed of three domains including the N-terminal domain (NTD; aa 1–8), transmembrane domain (TM; aa 9–38), and the C-terminal domain (CTD; aa 39–75) (Mandala et al., 2020). In other coronaviruses, it is thought that the TM acts as an ion channel and that the CTD interacts with other proteins like cellular adapters (Schoeman and Fielding, 2019). From our analysis of patients, all five AASs in the E protein were located in the TM and the CTD. From those, 9 out of 44 reviewed patients (20.4% of total patients) presented the T30I AAS (P10, P11, P15, P17, P18, P19, P20, P22, and P37) which is found in the transmembrane domain of this protein. A search of the 10,900,892 SARS-CoV-2 sequences recorded by GISAID, as of May 18 2022, indicated that this very rare variation is only found in 1,156 sequences (0.011%) (Elbe and Buckland-Merrett, 2017). Using FoldX, one study predicted that this change could be a stabilizing substitution (Rahman et al., 2021). To gain more insight on its structural effects, we modeled the T30I AAS into previously determined NMR structures of the SARS-CoV-1 (PDB: 5X29) and SARS-CoV-2 (PDB: 7K3G) envelope protein (Figure 6). Despite sequence similarity, there are notable differences between the structures, among them the positions of residue 30. In the 5X29 structure, Thr30 is in an interhelical position, whereas this residue is in a lipid-facing position in the 7K3G structure. It is unclear if these variations result from differing experimental techniques or simply plasticity of the protein complex. Due to the ambiguous position of Thr30, we additionally generated models using DeepMind AlphaFold 2. However, these models also suffered from inconsistent Thr30 positions, and thus, the precise position of this residue is uncertain. Nonetheless, in both the interhelical and lipid-facing positions, the T30I AAS increases the hydrophobicity of the transmembrane domain. The substitution from threonine, a hydrophilic amino acid, to isoleucine, a hydrophobic amino acid, would likely have a stabilizing effect, as the surrounding residues and lipid environment are also hydrophobic. Although the function of this precise change in SARS-CoV-2 is unknown, Nieto-Torres et al. (2014) have investigated the ion channel activity of the E protein in in vitro and in vivo pathogenesis of SARS-CoV; interestingly, they observed a lesser disease severity in mice infected with viruses lacking ion-channel (IC) activity, as opposed to those infected with viruses lacking IC activity with the T30I AAS, suggesting an impact on the presentation of the SARS-CoV-2 pathogenesis. The E protein can be sensed by TLR2-dependent host cell signaling to produce proinflammatory cytokines (Tasakis et al., 2021), suggesting that variations may have multiple effects on ion conductivity, pathogenesis, and inflammation.
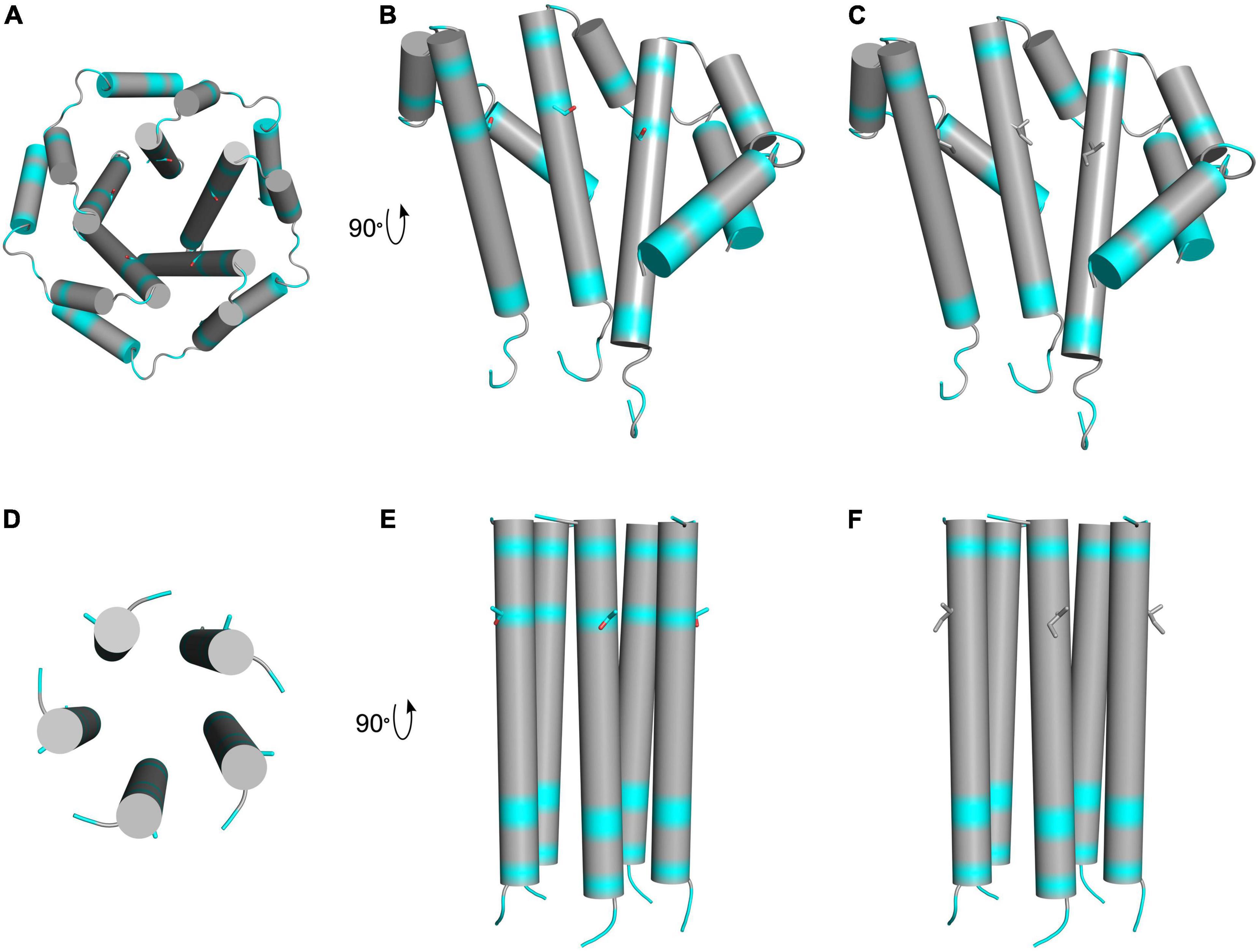
Figure 6. Schematic presentation of hydrophobic regions of T30I mutant E protein. The top row depicts LMPG micelle solution NMR structures of truncated SARS-CoV-1 E protein residues 8–65 in homopentameric channel complex (PDB: 5 × 29). Hydrophobic residues are colored in gray, and hydrophilic residues are colored in cyan. Oxygen atoms are colored in red. (A) Schematic view of the pore formed by the wild-type protein complex showing threonine 30 side chains. (B) Three subunits of the wild-type complex showing threonine 30 side chains. (C) Three subunits of the T30I mutant complex showing isoleucine 30 side chains. The bottom row depicts solid-state NMR structures of SARS-CoV-2 E protein transmembrane domain in homopentameric channel complex (PDB: 7K3G). Hydrophobic residues are colored in gray, and hydrophilic residues are colored in cyan. Oxygen atoms are colored in red. (D) View of the pore formed by the wild-type protein complex showing threonine 30 side chains. (E) Wild-type complex showing threonine 30 side chains. (F) T30I mutant complex showing isoleucine 30 side chains.
Besides T30I, five other AASs in the E protein were reported in the studied population, including the N48D and S50I (P10), T9I (P18), L19I (18), and L21F (P23) but as of yet, none has been reported to have an impact on the TM and CTD domains. Furthermore, as of May 18 2022, the only variation detected in the TM of E is the T9I substitution, with a variation frequency of 33.5% in 10,900,892 GISAID sequences, present in VOCs (Hodcroft, 2021). From our analysis, it could be speculated that the T30I AAS could perhaps be selected in immunodeficient settings, but more research on SARS-CoV-2 E AASs is needed to clarify whether this is the case.
Membrane Protein
The coronaviral membrane (M) is a 222 amino acid protein known to play a role in virion assembly and morphogenesis, among other processes (Liu et al., 2007; Jörrißen et al., 2021). In studied patients, the most common AAS identified was the H125Y, in five patients (11.4% – P10, P17, P20, P22, and P33). In addition, other AASs such as the A2S AAS were identified in 2 patients (4.5% – P1, P37). Yet, no functional impact has been reported from the literature, and the variation frequency of both H125Y and A2S in the total GISAID sequences is 0.11 and 0.24%, respectively. However, some defining AASs in the studied immunodeficient patients have been noted in VOCs, particularly in the delta (A82T) and omicron variants (D3G, Q19E, and 63T) (Hodcroft, 2021).
Nsp1
Nsp1 coronavirus proteins are known to shut down host protein translation to inhibit the expression of key genes involved in viral control (Nakagawa and Makino, 2021). As an 180 amino acid protein, it has been predicted to be made of an NTD (aa 1–128), a linker (aa 129–148), and a CTD domain (aa 149–180) (Schubert et al., 2020). The CTD has been shown to inhibit cellular gene expression by binding to the 40S ribosomal entry channel, whereas the NTD allows SARS-CoV-2 mRNA to escape this inhibition by binding to its leader sequence and stabilizing its interaction with the ribosome (Mendez et al., 2021).
In the analyzed secondary immunodeficient patients, several AASs and deletions were detected in the Nsp1 protein in patient viruses whose genomes were fully sequenced (P1, P2, P9, P10, P16, P17, P19, P21, P22, P28, and P31). From those, there were recurrent changes in the NTD including the amino acid deletion at position 85, which occurred in 2 (4.5%) patients (P1 and P31), and the AASs R124C (P10 and P22) and I114T (P2 and P19), which were also detected in two patients, respectively. The M85 deletion, R124C, and I114T substitutions are, respectively, present in 1.78, 0.14, and 0.05% of total GISAID sequences.
Structurally, the Nsp1 deletion at amino acid 85 has been previously shown to lead to a lower type I interferon response in infected Calu-3 cells, in contrast to wild-type Nsp1 (Lin et al., 2021). In the case of the AAS R124C, Mou et al. (2021) previously predicted in silico that this SNP has a destabilizing effect on the Nsp1 protein structure. Later, an in vitro study by Mendez et al. (2021) found that the R124A amino acid change at the same position along with the K125A AAS promotes host RNA decay, reduces host mRNA translation levels by destabilizing the binding to the 40S ribosomal subunit, and reduces the repression of SARS-CoV-2 leader containing transcripts. Moreover, Kim et al. (2021) also showed that the R124A/K125A changes did not have any effect on the levels of caspase-1 proteins in vitro, in contrast to the wild-type Nsp1, which significantly reduced caspase-1 levels and blocked its cleavage.
In summary, we identified recurrent Nsp1 variations in the NTD domain that could be involved in interfering with the host defenses. It would be of interest to investigate if there is a selection of Nsp1 NTD variations in immunodeficient individuals.
Nsp3
Nsp3 is the largest multi-domain coronaviral protein with a total of 1,945 amino acids (Lei et al., 2018; Gruca et al., 2021). It is involved in the proteolytic cleavage of polyproteins pp1a and pp1ab and the removal of K18-linked polyubiquitin and interferon-stimulated gene 15 (ISG15) from cellular proteins (Lei et al., 2018; Armstrong et al., 2021). From its many domains, its protease activity is conferred by the papain-like protease domain (aa 813–1076) (Armstrong et al., 2021). In this domain, two recurrent AASs were identified in the studied patients including T820I (4.5% of total patients, P11 and P23) and P822L (4.5%, P15 and P23). These occur with a frequency of 0.04 and 3.8% in the total GISAID sequences, and their impact on the role of Nsp3 is not yet studied. Other changes were also identified (S2), with the P1228L AAS (P7) being present in 35.6% of total GISAID sequences. This falls in the α-helical loop (aa 1,177–1,333), which is not yet well characterized. Therefore, it would be of great interest to investigate the role of the identified variations in Nsp3 and their possible impact on its catalytic activity.
Nsp6
Nsp6 is a transmembrane protein that is not very well characterized in SARS-CoV-2 infection (Kumar A. et al., 2021). Sun et al. (2022) showed that this protein can target the ATPase proton pump component involved in lysosomal acidification, ATP6AP1, to trigger NLRP3-dependent pyroptosis in lung epithelial cells. In the reviewed patients, the L37F protein-coding change was noted to emerge de novo in three different cases (11.4% of total patients, P10, P17, P20, P21, P31), and this AAS is part of around 2.0% of GISAID sequences (Lynch et al., 2021; Truong et al., 2021; Weigang et al., 2021). Recently, Benvenuto et al. (2020) predicted that such an amino acid change led to a lower stability of the Nsp6 protein structure and suggested a role of Nsp6 in binding with the ER. This AAS was also studied by Wang R. et al. (2020) who analyzed around 76,000 sequences in GISAID up to 19 October 2020, and correlated it with lower death ratios and transmission rates. Through bioinformatic analysis, it was also shown to be destabilizing and less functional compared with the wild-type (Wang R. et al., 2020). Furthermore, this same AAS has also been observed to weaken the interaction of Nsp6 with ATP6AP1, thus reducing lysosome acidification and pyroptosis induction (Sun et al., 2022). These observations stress the importance of the L37F AAS in both immunodeficient and immunocompetent individuals, given the relatively high variation rate in the reviewed patients and in GISAID, which might suggest an unappreciated fitness benefit conferred by this variation. The study of patients with chronic SARS-CoV-2 infection would be another source of data for predicting the transmissibility and lethality of SARS-CoV-2, especially in the context of immunodeficiency.
RdRp/Nsp12
The RNA-dependent RNA polymerase (RdRp) involved in SARS-CoV-2 genome replication and transcription of genes is composed of a catalytic subunit known as Nsp12 as well as two accessory subunits, Nsp8 and Nsp7 (Gao et al., 2020b). The Nsp12 domain resembles a right hand, comprising the fingers subdomain, which interact with the template strand RNA and direct it into the active site, and the palm domain, which forms the catalytic active center. The RdRp is the target for antiviral drugs such as remdesivir (RDV and GS-5734), which can incorporate itself in the nascent viral RNA chains, causing premature transcriptional termination (Warren et al., 2016). In the study population, many RdRp variations were identified in patients who were treated with remdesivir (nine patients: 20.4%, P2, P5, P9, P14, P16, P21, P22, P23, and P31). From these, a few AASs were identified in the RdRp palm domain including V792I (5%, P16 and P22), E796D (P9), C799R (P14), and E802D, respectively. In the whole GISAID database, these occur at a frequency of less than 0.0025%. Interestingly, an in vitro study by Szemiel et al. (2021) showed that a palm domain substitution in a conserved residue (E802D) of the RdRp decreases the sensitivity to remdesivir and viral fitness in a competition assay; this same amino acid change (E802D) was found in P21, decreased binding to remdesivir, and has a fitness cost (Gandhi et al., 2022). As this substitution was found to be close to residues involved in binding with nascent RNA (aa 813–815), it was suggested that the RdRp could have structural changes that could allow elongation of template RNA, even when remdesivir is incorporated (Szemiel et al., 2021). Indeed, this highlights those substitutions in the RdRp finger, and palm domain should be studied more carefully to determine if these play a role in conferring resistance to antivirals against the RdRp.
Nsp13
The helicase protein (Nsp13) plays a role in unwinding duplex RNA and DNA in a 5′ to 3′ direction (Vazquez et al., 2021). In the patients we studied, the D374E substitution identified in P22 is an AAS that occurs in one of the residues identified in the NTP hydrolysis active site, and its functional effect is yet to be determined (Jia et al., 2019). Although this AAS is very rare (9/10,900,892 GISAID sequences), since this Nsp13 is one of the proteins involved in the RTC formation, it would be of interest to investigate such changes and their role in viral replication.
ORF3a
The SARS-CoV-2 ORF3a is an integral membrane protein that has been shown to play a role in inducing apoptosis (Ren et al., 2020) in infected cells, in promoting lysosomal exocytosis (Chen D. et al., 2021), and in blocking the formation of autolysosomes (Miao et al., 2021; Qu et al., 2021). It has also been shown to inhibit STAT1 phosphorylation in vitro (Xia et al., 2020). In the described patients, nine variations were identified including the S171L (P9, 0.69% GISAID frequency). Chen D. et al. (2021) showed that the SARS-CoV-2 ORF3a S171E AAS abolished the production of trans-soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein receptor (SNARE) complex proteins involved in fusing the lysosome to plasma membranes; additionally, the amino acid substitution abolished the ability of ORF3a to increase the Ca2+ levels in the cytoplasm. Furthermore, the S171L substitution was predicted in silico to increase protein instability with a turn structure replaced by a coiled coil (Azad and Khan, 2021), suggesting that it could be a functional mutation selected in patients.
ORF7a
The ORF7a accessory protein is believed to play a role in modulating host immune responses (Redondo et al., 2021). In the case of the 44 patients described here, the AASs S81P occurred twice in P14 and P22 (4.5%), and the A105V, which occurred in P9, has been characterized before. In a 62-patient Romanian cohort, Lobiuc et al. (2021) showed that 27.5% were infected with the A105V AAS, and they experienced a twofold higher death rate than others without A105V. The researchers then did a bioinformatic analysis of this change and predicted an increased stability by allosteric effects (Lobiuc et al., 2021).
In summary, we have identified many recurrent SARS-CoV-2 amino acid changes to emerge de novo in immunodeficient patients in a variety of proteins that have previously been identified to have a structural effect. This could be the result of different host selection pressures as some proteins (E, Nsp1, M, and ORF7a) had a relatively higher frequency compared with others (Figure 3). More investigation and a bigger study population are needed to make a definitive conclusion.
Discussion
Prolonged infections resulting from weakened impaired immune responses allow the virus to persist, providing opportunities for increased viral replications and accumulations of mutations, some of which may be novel (Avanzato et al., 2020; Monrad et al., 2021). Thus, as the COVID-19 pandemic continues, it is crucial to track mutations arising in circulating and novel strains that can potentially become VOCs and VOIs to help predict their role in transmission and pathogenesis.
Case studies of chronically infected individuals with immunodeficiency could help gain insights into how the virus evolves in such settings. In this review, we highlighted 44 patients with secondary immunodeficiencies that were chronically infected with SARS-CoV-2 and received a variety of treatments, some of which may exert a selective pressure on the virus (De Vlaminck et al., 2013) (e.g., antiviral drugs targeting a specific protein site). Early studies showed that treatments with monoclonal antibodies should be used with great caution as they have been demonstrated to exert selective pressures on viruses. Focosi et al. (2021a) reviewed case series and reports and observed frequent emergence of single-nucleotide changes in the RBD regions of the spike gene when under the pressure of monoclonal antibodies; additionally, they noted that the mutational pressure from convalescent plasma was different in nature with deletions being more present, presumably due to the polyclonal nature of the antibodies. Alternatively, polyclonal antibodies recognizing different spike epitopes or combination therapy could be used to reduce selection pressure and treatment resistance. Efforts are being made to design broadly neutralizing SARS-CoV-2 and pan-coronavirus antibodies, some relying on the principle of targeting conserved regions that have a high fitness cost if altered (Cameroni et al., 2021; Martinez et al., 2021; Saunders et al., 2021; Shrestha et al., 2021; Tan et al., 2021). Such consideration and strategies when designing new therapies are needed to deliver therapeutics with longevity for the use of the current pandemic and future ones that will inevitably arise.
A careful approach is required in administering future antivirals targeting a specific site of SARS-CoV-2 proteins, especially as novel therapies such as molnupiravir, which targets the RdRp (Fischer et al., 2021), and paxlovid, which targets the SARS-CoV-2 main protease (Nsp5) by reacting reversibly with a cysteine residue at its active site (Pavan et al., 2021), are slated for approval. If these antivirals do not have a high genetic barrier for mutational escape, lengthy efforts and enormous resource commitments could be wasted on therapies that the world has already begun to hail as an end to the pandemic.
Besides the spike protein, it was interesting to find a recurrent AAS T30I in the E protein transmembrane motif in 9 out of 44 different patients (20.4% of total patients) who presented the T30I AAS (P10, P11, P15, P17, P18, P19, P20, P22, and P37). Like the latter, other variations were recurrent in the reviewed patients with a frequency greater than those found in the GISAID database. This was the case of the M protein A2S (4.5% of total patients, 0.24% of GISAID) and H125Y (11.4% of total patients, 0.11% of GISAID) AASs, as well as the Nsp1 R124C (4.5% of total patients, 0.14% of GISAID) and I114T (4.5% of total patients, 0.05% of GISAID), among others. One explanation for this difference could be that these mutations are specific to an immunodeficient environment, where certain immune selective pressures could be different, e.g., weakened. On the contrary, other AASs present in circulating VOCs were noted to emerge in the reviewed patients such as the Nsp3 P1228L (35.6% of GISAID), which could reflect the adaptation of the virus to the human host; however, the functional effect of such variations remains to be elucidated in further studies.
Although of interest, this literature review of SARS-CoV-2 variations in immunocompromised patients has limitations. Previous immunosuppressive regimens were not always known in detail, and this could lead to incomplete evaluation of the extent of immunodeficiency in the studied population. Another confounding factor is the variation in the standard of care for SARS-CoV-2 infection throughout the pandemic. Indeed, variation in treatment regimens over time makes the rise of mutations difficult to interpret, especially considering the variable time from treatment to the moment of infection. Moreover, virus sequencing is often from samples originating from the nasopharynx, which is part of the upper respiratory tract. These samples are not necessarily representative of the virus composition in the lower respiratory tract. Furthermore, the virus replicating in the lower respiratory tract may experience different selection pressures than the upper respiratory tract. In addition, the GISAID frequencies obtained are derived from uploaded consensus sequences in contrast to our patients’ sequencing data that have variable consensus agreement; therefore, the AASs we note may be more noisy and non-selective compared with the GISAID AASs. GISAID frequencies can also include immunocompromised individuals although we expect these to be in the minority. Of note, in the 44 patients that we compiled, all viruses were not sequenced at the beginning of SARS-CoV-2 infection or sequenced at the same timepoint during the disease, once again leading to comparison discrepancies. In addition, some of the papers focus on spike proteins only. Finally, although we chose to focus on secondary immunodeficiencies due to the larger number of patients with analyzed viral genomes, mutations in primary immunodeficient patients (Bucciol et al., 2021; Ciuffreda et al., 2021; Cabañero-Navalon et al., 2022) are slowly being characterized that could add to the topic of viral evolution in the context of immunodeficiencies at large.
Conclusion
In this review, several variations were found in the spike or Nsp12 proteins, which are important therapeutic targets. We also identified several recurrent variations in E, Nsp1, M, and ORF7a proteins that may play an important role in SARS-CoV-2 pathogenesis. Determining whether these variations emerged through selection in the immunodeficient patients or resulted from the adaptation of SARS-CoV-2 to the human host will require further study. However, the breadth and impact of mutations characterized in patients with secondary immunodeficiency highlight the relevance of monitoring the evolution of SARS-CoV-2 in immunocompromised individuals, not only to identify potentially adaptive novel mutations but also to mitigate the risk of introducing variants that may pose increased health threats to communities.
Author Contributions
SV and LA: conceptualization and supervision. NM, GG, DP, MH, PN, and AB: writing—original draft preparation. NM, GG, DP, MH, PN, AB, LA, and SV: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
NM and DP were supported by the Coronavirus Variants Rapid Response Network. SV was supported by the Canada Research Chair Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
All figures were created with Biorender.com, through a purchased license.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.933983/full#supplementary-material
References
Agostini Maria, L., Andres Erica, L., Sims Amy, C., Graham Rachel, L., Sheahan Timothy, P., Lu, X., et al. (2018). Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio 9:e00221-18. doi: 10.1128/mBio.00221-18
Alenquer, M., Ferreira, F., Lousa, D., Valério, M., Medina-Lopes, M., Bergman, M.-L., et al. (2021). Signatures in SARS-CoV-2 spike protein conferring escape to neutralizing antibodies. PLoS Pathog. 17:e1009772. doi: 10.1371/journal.ppat.1009772
Álvarez, H., Ruiz-Mateos, E., Juiz-González, P. M., Vitallé, J., Viéitez, I., Vázquez-Friol, M. D., et al. (2022). SARS-CoV-2 evolution and spike-specific CD4+ T-cell response in persistent COVID-19 with severe HIV immune suppression. Microorganisms 10:143. doi: 10.3390/microorganisms10010143
Armstrong, L. A., Lange, S. M., Dee Cesare, V., Matthews, S. P., Nirujogi, R. S., Cole, I., et al. (2021). Biochemical characterization of protease activity of Nsp3 from SARS-CoV-2 and its inhibition by nanobodies. PLoS One 16:e0253364. doi: 10.1371/journal.pone.0253364
Astuti, I., and Ysrafil. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. 14, 407–412. doi: 10.1016/j.dsx.2020.04.020
Avanzato, V. A., Matson, M. J., Seifert, S. N., Pryce, R., Williamson, B. N., Anzick, S. L., et al. (2020). Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 183, 1901–1912.e9. doi: 10.1016/j.cell.2020.10.049
Azad, G. K., and Khan, P. K. (2021). Variations in Orf3a protein of SARS-CoV-2 alter its structure and function. Biochem. Biophys. Rep. 26:100933. doi: 10.1016/j.bbrep.2021.100933
Baang, J. H., Smith, C., Mirabelli, C., Valesano, A. L., Manthei, D. M., Bachman, M. A., et al. (2021). Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J. Infect. Dis. 223, 23–27. doi: 10.1093/infdis/jiaa666
Bailly, B., Péré, H., Veyer, D., Berceanu, A., Daguindau, E., Roux, P., et al. (2021). Persistent COVID-19 in an immunocompromised host treated by SARS-CoV-2-specific monoclonal antibodies. Clin. Infect. Dis. 2021:ciab868. doi: 10.1093/cid/ciab868
Banerjee, A., Doxey, A. C., Mossman, K., and Irving, A. T. (2021). Unraveling the zoonotic origin and transmission of SARS-CoV-2. Trends Ecol. Evol. 36, 180–184. doi: 10.1016/j.tree.2020.12.002
Barber, M. R. W., and Clarke, A. E. (2020). Systemic lupus erythematosus and risk of infection. Expert Rev. Clin. Immunol. 16, 527–538. doi: 10.1080/1744666X.2020.1763793
Benvenuto, D., Angeletti, S., Giovanetti, M., Bianchi, M., Pascarella, S., Cauda, R., et al. (2020). Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. J. Infect. 81, e24–e27. doi: 10.1016/j.jinf.2020.03.058
Bhatt, P. R., Scaiola, A., Loughran, G., Leibundgut, M., Kratzel, A., Meurs, R., et al. (2021). Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science 372, 1306–1313. doi: 10.1126/science.abf3546
Bhogal, T., Khan, U. T., Lee, R., Stockdale, A., Hesford, C., Potti-Dhananjaya, V., et al. (2021). Haematological malignancy and nosocomial transmission are associated with an increased risk of death from COVID-19: results of a multi-center UK cohort. Leuk. Lymphoma 62, 1682–1691. doi: 10.1080/10428194.2021.1876865
Bonanad, C., García-Blas, S., Tarazona-Santabalbina, F., Sanchis, J., Bertomeu-González, V., Fácila, L., et al. (2020). The effect of age on mortality in patients with COVID-19: a meta-analysis with 611,583 subjects. J. Am. Med. Dir. Assoc. 21, 915–918. doi: 10.1016/j.jamda.2020.05.045
Borges, V., Isidro, J., Cunha, M., Cochicho, D., Martins, L., Banha, L., et al. (2021). Long-term evolution of SARS-CoV-2 in an immunocompromised patient with non-hodgkin lymphoma. mSphere 6:e00244-21. doi: 10.1128/mSphere.00244-21
Boson, B., Legros, V., Zhou, B., Siret, E., Mathieu, C., Cosset, F.-L., et al. (2021). The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 296:100111. doi: 10.1074/jbc.RA120.016175
Brant, A. C., Tian, W., Majerciak, V., Yang, W., and Zheng, Z.-M. (2021). SARS-CoV-2: from its discovery to genome structure, transcription, and replication. Cell Biosci. 11:136. doi: 10.1186/s13578-021-00643-z
Bronstein, Y., Adler, A., Katash, H., Halutz, O., Herishanu, Y., and Levytskyi, K. (2021). Evolution of spike mutations following antibody treatment in two immunocompromised patients with persistent COVID-19 infection. J. Med. Virol. 94, 1241–1245. doi: 10.1002/jmv.27445
Bucciol, G., Tangye, S. G., and Meyts, I. (2021). Coronavirus disease 2019 in patients with inborn errors of immunity: lessons learned. Curr. Opin. Pediatr. 33, 648–656. doi: 10.1097/MOP.0000000000001062
Cabañero-Navalon, M. D., Garcia-Bustos, V., Ruiz-Rodriguez, P., Comas, I., Coscollá, M., Martinez-Priego, L., et al. (2022). Persistent SARS-CoV-2 infection with repeated clinical recurrence in a patient with common variable immunodeficiency. Clin. Microbiol. Infect. 28, 308–310. doi: 10.1016/j.cmi.2021.10.021
Cameroni, E., Bowen, J. E., Rosen, L. E., Saliba, C., Zepeda, S. K., Culap, K., et al. (2021). Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 602, 664–670.
Cavazzoni, P. (2022). Coronavirus (COVID-19) Update: FDA Limits Use of Certain Monoclonal Antibodies to Treat COVID-19 Due to the Omicron Variant [Online]. Silver Spring, MD: U.S. Food & Drug Administration.
Cele, S., Karim, F., Lustig, G., San, J. E., Hermanus, T., Tegally, H., et al. (2022). SARS-CoV-2 prolonged infection during advanced HIV disease evolves extensive immune escape. Cell Host Microbe 30, 154–162.e5. doi: 10.1016/j.chom.2022.01.005
Chen, D., Zheng, Q., Sun, L., Ji, M., Li, Y., Deng, H., et al. (2021). ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress. Dev. Cell 56, 3250–3263.e5. doi: 10.1016/j.devcel.2021.10.006
Chen, L., Zody, M. C., Di Germanio, C., Martinelli, R., Mediavilla, J. R., Cunningham, M. H., et al. (2021). Emergence of multiple SARS-CoV-2 antibody escape variants in an immunocompromised host undergoing convalescent plasma treatment. mSphere 6:e0048021. doi: 10.1128/mSphere.00480-21
Chen, J., Wang, R., Wang, M., and Wei, G.-W. (2020). Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 432, 5212–5226. doi: 10.1016/j.jmb.2020.07.009
Chinen, J., and Shearer, W. T. (2010). Secondary immunodeficiencies, including HIV infection. J. Allergy Clin. Immunol. 125, S195–S203.
Choi, B., Choudhary, M. C., Regan, J., Sparks, J. A., Padera, R. F., Qiu, X., et al. (2020). Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 383, 2291–2293. doi: 10.1056/NEJMc2031364
Ciuffreda, L., Lorenzo-Salazar, J. M., Alcoba-Florez, J., Rodriguez-Pérez, H., Gil-Campesino, H., Íñigo-Campos, A., et al. (2021). Longitudinal study of a SARS-CoV-2 infection in an immunocompromised patient with X-linked agammaglobulinemia. J. Infect. 83, 607–635. doi: 10.1016/j.jinf.2021.07.028
Clark, S. A., Clark, L. E., Pan, J., Coscia, A., Mckay, L. G. A., Shankar, S., et al. (2021). SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell 184, 2605–2617.e18. doi: 10.1016/j.cell.2021.03.027
Coll, E., Fernández-Ruiz, M., Sánchez-Álvarez, J. E., Martínez-Fernández, J. R., Crespo, M., Gayoso, J., et al. (2021). COVID-19 in transplant recipients: the Spanish experience. Am. J. Transplant. 21, 1825–1837. doi: 10.1111/ajt.16369
Collier, D. A., De Marco, A., Ferreira, I. A. T. M., Meng, B., Datir, R. P., Walls, A. C., et al. (2021). Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 593, 136–141.
Conceicao, C., Thakur, N., Human, S., Kelly, J. T., Logan, L., Bialy, D., et al. (2020). The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol. 18:e3001016. doi: 10.1371/journal.pbio.3001016
Dampalla, C. S., Zheng, J., Perera, K. D., Wong, L.-Y. R., Meyerholz, D. K., Nguyen, H. N., et al. (2021). Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection. Proc. Natl. Acad. Sci. U.S.A. 118:e2101555118. doi: 10.1073/pnas.2101555118
Dapp, M. J., Kober, K. M., Chen, L., Westfall, D. H., Wong, K., Zhao, H., et al. (2017). Patterns and rates of viral evolution in HIV-1 subtype B infected females and males. PLoS One 12:e0182443. doi: 10.1371/journal.pone.0182443
Davidson, A. D., Williamson, M. K., Lewis, S., Shoemark, D., Carroll, M. W., Heesom, K. J., et al. (2020). Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 12:68. doi: 10.1186/s13073-020-00763-0
De Vlaminck, I., Khush, K. K., Strehl, C., Kohli, B., Luikart, H., Neff, N. F., et al. (2013). Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 155, 1178–1187. doi: 10.1016/j.cell.2013.10.034
Elbe, S., and Buckland-Merrett, G. (2017). Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 1, 33–46. doi: 10.1002/gch2.1018
Elkrief, A., Desilets, A., Papneja, N., Cvetkovic, L., Groleau, C., Lakehal, Y. A., et al. (2020). High mortality among hospital-acquired COVID-19 infection in patients with cancer: a multicentre observational cohort study. Eur. J. Cancer 139, 181–187. doi: 10.1016/j.ejca.2020.08.017
Ferreira, I. A. T. M., Kemp, S. A., Datir, R., Saito, A., Meng, B., Rakshit, P., et al. (2021). SARS-CoV-2 B.1.617 mutations L452R and E484Q Are Not synergistic for antibody evasion. J. Infect. Dis. 224, 989–994. doi: 10.1093/infdis/jiab368
Fischer, W., Eron, J. J., Holman, W., Cohen, M. S., Fang, L., Szewczyk, L. J., et al. (2021). Molnupiravir, an oral antiviral treatment for COVID-19. medRxiv [Preprint] doi: 10.1101/2021.06.17.21258639
Focosi, D., Maggi, F., Franchini, M., Mcconnell, S., and Casadevall, A. (2021a). Analysis of immune escape variants from antibody-based therapeutics against COVID-19: a systematic review. Int. J. Mol. Sci. 23:29. doi: 10.3390/ijms23010029
Focosi, D., Novazzi, F., Genoni, A., Dentali, F., Gasperina, D. D., Baj, A., et al. (2021b). Emergence of SARS-COV-2 spike protein escape mutation Q493R after treatment for COVID-19. Emerg. Infect. Dis. 27, 2728–2731. doi: 10.3201/eid2710.211538
Francisco, R. D. S. Jr., Benites, L. F., Lamarca, A. P., De Almeida, L. G. P., Hansen, A. W., Gularte, J. S., et al. (2021). Pervasive transmission of E484K and emergence of VUI-NP13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul, Brazil. Virus Res. 296:198345. doi: 10.1016/j.virusres.2021.198345
Gandhi, S., Klein, J., Robertson, A. J., Peña-Hernández, M. A., Lin, M. J., Roychoudhury, P., et al. (2022). De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report. Nat. Commun. 13;1547.
Gao, Y., Chen, Y., Liu, M., Shi, S., and Tian, J. (2020a). Impacts of immunosuppression and immunodeficiency on COVID-19: a systematic review and meta-analysis. J. Infect. 81, e93–e95. doi: 10.1016/j.jinf.2020.05.017
Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., et al. (2020b). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 368, 779–782. doi: 10.1126/science.abb7498
Gottlieb, R. L., Nirula, A., Chen, P., Boscia, J., Heller, B., Morris, J., et al. (2021). Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA 325, 632–644. doi: 10.1001/jama.2021.0202
Graham, C., Seow, J., Huettner, I., Khan, H., Kouphou, N., Acors, S., et al. (2021). Neutralization potency of monoclonal antibodies recognizing dominant and subdominant epitopes on SARS-CoV-2 Spike is impacted by the B.1.1.7 variant. Immunity 54, 1276–1289.e6. doi: 10.1016/j.immuni.2021.03.023
Gribble, J., Stevens, L. J., Agostini, M. L., Anderson-Daniels, J., Chappell, J. D., Lu, X., et al. (2021). The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 17:e1009226. doi: 10.1371/journal.ppat.1009226
Gruca, A., Ziemska-Legiecka, J., Jarnot, P., Sarnowska, E., Sarnowski, T. J., and Grynberg, M. (2021). Common low complexity regions for SARS-CoV-2 and human proteomes as potential multidirectional risk factor in vaccine development. BMC Bioinformatics 22:182. doi: 10.1186/s12859-021-04017-7
Guigon, A., Faure, E., Lemaire, C., Chopin, M.-C., Tinez, C., Assaf, A., et al. (2021). Emergence of Q493R mutation in SARS-CoV-2 spike protein during bamlanivimab/etesevimab treatment and resistance to viral clearance. J. Infect. 84, 248–288. doi: 10.1016/j.jinf.2021.08.033
Harvey, W. T., Carabelli, A. M., Jackson, B., Gupta, R. K., Thomson, E. C., Harrison, E. M., et al. (2021). SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19, 409–424.
Hensley, M. K., Bain, W. G., Jacobs, J., Nambulli, S., Parikh, U., Cillo, A., et al. (2021). Intractable coronavirus disease 2019 (COVID-19) and prolonged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication in a chimeric antigen receptor-modified T-cell therapy recipient: a case study. Clin. Infect. Dis. 73, e815–e821. doi: 10.1093/cid/ciab072
Hodcroft, E. B. (2021). CoVariants: SARS-CoV-2 Mutations and Variants of Interest. [Online]. Available online at: https://covariants.org/ (accessed May 18, 2022).
Hoffman, S. A., Costales, C., Sahoo, M. K., Palanisamy, S., Yamamoto, F., Huang, C., et al. (2021). SARS-CoV-2 neutralization resistance mutations in patient with HIV/AIDS, California, USA. Emerg. Infect. Dis. 27, 2720–2723. doi: 10.3201/eid2710.211461
Hoffmann, C., Casado, J. L., Härter, G., Vizcarra, P., Moreno, A., Cattaneo, D., et al. (2021). Immune deficiency is a risk factor for severe COVID-19 in people living with HIV. HIV Med. 22, 372–378. doi: 10.1111/hiv.13037
Hu, J., and Wang, Y. (2021). The clinical characteristics and risk factors of severe COVID-19. Gerontology 67, 255–266. doi: 10.1159/000513400
Huang, H., Zhu, Y., Niu, Z., Zhou, L., and Sun, Q. (2021). SARS-CoV-2 N501Y variants of concern and their potential transmission by mouse. Cell Death Differ. 28, 2840–2842. doi: 10.1038/s41418-021-00846-4
Huang, Y., Yang, C., Xu, X.-F., Xu, W., and Liu, S.-W. (2020). Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 41, 1141–1149. doi: 10.1038/s41401-020-0485-4
Jangra, S., Ye, C., Rathnasinghe, R., Stadlbauer, D., Alshammary, H., Amoako, A. A., et al. (2021). SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe 2, e283–e284. doi: 10.1016/S2666-5247(21)00068-9
Jensen, B., Luebke, N., Feldt, T., Keitel, V., Brandenburger, T., Kindgen-Milles, D., et al. (2021). Emergence of the E484K mutation in SARS-COV-2-infected immunocompromised patients treated with bamlanivimab in Germany. Lancet Reg. Health Eur. 8:100164. doi: 10.1016/j.lanepe.2021.100164
Jia, Z., Yan, L., Ren, Z., Wu, L., Wang, J., Guo, J., et al. (2019). Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 47, 6538–6550. doi: 10.1093/nar/gkz409
Jiang, H.-W., Zhang, H.-N., Meng, Q.-F., Xie, J., Li, Y., Chen, H., et al. (2020). SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 17, 998–1000. doi: 10.1038/s41423-020-0514-8
Jones, J. M., Faruqi, A. J., Sullivan, J. K., Calabrese, C., and Calabrese, L. H. (2021). COVID-19 outcomes in patients undergoing B cell depletion therapy and those with humoral immunodeficiency states: a scoping review. Pathog. Immun. 6, 76–103. doi: 10.20411/pai.v6i1.435
Jörrißen, P., Schütz, P., Weiand, M., Vollenberg, R., Schrempf, I. M., Ochs, K., et al. (2021). Antibody response to SARS-CoV-2 membrane protein in patients of the acute and convalescent phase of COVID-19. Front. Immunol. 12:679841. doi: 10.3389/fimmu.2021.679841
Kanwugu, O. N., and Adadi, P. (2021). HIV/SARS-CoV-2 coinfection: a global perspective. J. Med. Virol. 93, 726–732. doi: 10.1002/jmv.26321
Kastritis, E., Kitas, G. D., Vassilopoulos, D., Giannopoulos, G., Dimopoulos, M. A., and Sfikakis, P. P. (2020). Systemic autoimmune diseases, anti-rheumatic therapies, COVID-19 infection risk and patient outcomes. Rheumatol. Int. 40, 1353–1360. doi: 10.1007/s00296-020-04629-x
Kaul, A., Gordon, C., Crow, M. K., Touma, Z., Urowitz, M. B., Van Vollenhoven, R., et al. (2016). Systemic lupus erythematosus. Nat. Rev. Dis. Primers 2:16039.
Kemp, S. A., Collier, D. A., Datir, R. P., Ferreira, I. A. T. M., Gayed, S., Jahun, A., et al. (2021). SARS-CoV-2 evolution during treatment of chronic infection. Nature 592, 277–282. doi: 10.1038/s41586-021-03291-y
Kim, N.-E., Kim, D.-K., and Song, Y.-J. (2021). SARS-CoV-2 nonstructural proteins 1 and 13 suppress caspase-1 and the NLRP3 inflammasome activation. Microorganisms 9:494. doi: 10.3390/microorganisms9030494
Kimura, I., Kosugi, Y., Wu, J., Zahradnik, J., Yamasoba, D., Butlertanaka, E. P., et al. (2021). The SARS-CoV-2 Lambda variant exhibits enhanced infectivity and immune resistance. Cell Rep. 38:110218. doi: 10.1016/j.celrep.2021.110218
Ko, K., Nagashima, S., E, B., Ouoba, S., Akita, T., Sugiyama, A., et al. (2021). Molecular characterization and the mutation pattern of SARS-CoV-2 during first and second wave outbreaks in Hiroshima, Japan. PLoS One 16:e0246383. doi: 10.1371/journal.pone.0246383
Kumar, A., Kumar, P., Saumya, K. U., and Giri, R. (2021). Investigating the conformational dynamics of SARS-CoV-2 NSP6 protein with emphasis on non-transmembrane 91–112 & 231–290 regions. Microb. Pathog. 161:105236. doi: 10.1016/j.micpath.2021.105236
Kumar, S., Chandele, A., and Sharma, A. (2021). Current status of therapeutic monoclonal antibodies against SARS-CoV-2. PLoS Pathog. 17:e1009885. doi: 10.1371/journal.ppat.1009885
Lei, J., Kusov, Y., and Hilgenfeld, R. (2018). Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 149, 58–74. doi: 10.1016/j.antiviral.2017.11.001
Letko, M., Miazgowicz, K., Mcminn, R., Seifert, S. N., Sola, I., Enjuanes, L., et al. (2018). Adaptive evolution of MERS-CoV to species variation in DPP4. Cell Rep. 24, 1730–1737. doi: 10.1016/j.celrep.2018.07.045
Leung, W. F., Chorlton, S., Tyson, J., Al-Rawahi, G. N., Jassem, A. N., Prystajecky, N., et al. (2022). COVID-19 in an immunocompromised host: persistent shedding of viable SARS-CoV-2 and emergence of multiple mutations, a case report. Int. J. Infect. Dis. 114, 178–182. doi: 10.1016/j.ijid.2021.10.045
Li, G., Zhou, Z., Du, P., Yu, M., Li, N., Xiong, X., et al. (2021). The SARS-CoV-2 spike L452R-E484Q variant in the Indian B.1.617 strain showed significant reduction in the neutralization activity of immune sera. Precis. Clin. Med. 4, 149–154.
Li, Q., Nie, J., Wu, J., Zhang, L., Ding, R., Wang, H., et al. (2021). SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell 184, 2362–2371.e9. doi: 10.1016/j.cell.2021.02.042
Li, M.-Y., Li, L., Zhang, Y., and Wang, X.-S. (2020). Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty 9:45. doi: 10.1186/s40249-020-00662-x
Lin, J.-W., Tang, C., Wei, H.-C., Du, B., Chen, C., Wang, M., et al. (2021). Genomic monitoring of SARS-CoV-2 uncovers an Nsp1 deletion variant that modulates type I interferon response. Cell Host Microbe 29, 489–502.e8. doi: 10.1016/j.chom.2021.01.015
Liu, B. M., and Hill, H. R. (2020). Role of host immune and inflammatory responses in COVID-19 cases with underlying primary immunodeficiency: a review. J. Interferon Cytokine Res. 40, 549–554. doi: 10.1089/jir.2020.0210
Liu, D. X., Yuan, Q., and Liao, Y. (2007). Coronavirus envelope protein: a small membrane protein with multiple functions. Cell. Mol. Life Sci. 64, 2043–2048. doi: 10.1007/s00018-007-7103-1
Liu, Y., Liu, J., Plante, K. S., Plante, J. A., Xie, X., Zhang, X., et al. (2021a). The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 602, 294–299. doi: 10.1038/s41586-021-04245-0
Liu, Y., Sawalha, A. H., and Lu, Q. (2021b). COVID-19 and autoimmune diseases. Curr. Opin. Rheumatol. 33, 155–162.
Lobiuc, A., Şterbuleac, D., Sturdza, O., Dimian, M., and Covasa, M. (2021). A conservative replacement in the transmembrane domain of SARS-CoV-2 ORF7a as a putative risk factor in COVID-19. Biology 10:1276. doi: 10.3390/biology10121276
Lode, H. M., and Schmidt-Ioanas, M. (2005). Vasculitis and infection: effects of immunosuppressive therapy. Clin. Nephrol. 64, 475–479. doi: 10.5414/cnp64475
Lohr, B., Niemann, D., and Verheyen, J. (2021). Bamlanivimab treatment leads to rapid selection of immune escape variant carrying the E484K mutation in a B.1.1.7-infected and immunosuppressed patient. Clin. Infect. Dis. 73, 2144–2145. doi: 10.1093/cid/ciab392
Lopes-Pacheco, M., Silva, P. L., Cruz, F. F., Battaglini, D., Robba, C., Pelosi, P., et al. (2021). Pathogenesis of multiple organ injury in COVID-19 and potential therapeutic strategies. Front. Physiol. 12:593223. doi: 10.3389/fphys.2021.593223
Lynch, M., Macori, G., Fanning, S., O’regan, E., Hunt, E., O’callaghan, D., et al. (2021). Genomic evolution of SARS-CoV-2 Virus in immunocompromised patient, Ireland. Emerg. Infect. Dis. J. 27, 2499–2501. doi: 10.3201/eid2709.211159
Mancon, A., Rizzo, A., Mileto, D., Grosso, S., Foschi, A., Cutrera, M., et al. (2022). Viro-immunological evaluation in an immunocompromised patient with long-lasting SARS-CoV-2 infection. Emerg. Microb. Infect. 11, 786–789. doi: 10.1080/22221751.2022.2045877
Mandala, V. S., Mckay, M. J., Shcherbakov, A. A., Dregni, A. J., Kolocouris, A., and Hong, M. (2020). Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 27, 1202–1208. doi: 10.1038/s41594-020-00536-8
Martí, D., Torras, J., Bertran, O., Turon, P., and Alemán, C. (2021). Temperature effect on the SARS-CoV-2: a molecular dynamics study of the spike homotrimeric glycoprotein. Comput. Struct. Biotechnol. J. 19, 1848–1862. doi: 10.1016/j.csbj.2021.03.037
Martin, D. P., Weaver, S., Tegally, H., San, J. E., Shank, S. D., Wilkinson, E., et al. (2021). The emergence and ongoing convergent evolution of the SARS-CoV-2 N501Y lineages. Cell 184, 5189–5200.e7.
Martinez, D. R., Schaefer, A., Gobeil, S., Li, D., De La Cruz, G., Parks, R., et al. (2021). A broadly neutralizing antibody protects against SARS-CoV, pre-emergent bat CoVs, and SARS-CoV-2 variants in mice. bioRxiv [Preprint] doi: 10.1101/2021.04.27.441655
Martinot, M., Jary, A., Fafi-Kremer, S., Leducq, V., Delagreverie, H., Garnier, M., et al. (2021). Emerging RNA-dependent RNA polymerase mutation in a remdesivir-treated B-cell immunodeficient patient with protracted coronavirus disease 2019. Clin. Infect. Dis. 73, e1762–e1765. doi: 10.1093/cid/ciaa1474
Mccarthy, K. R., Rennick, L. J., Nambulli, S., Robinson-Mccarthy, L. R., Bain, W. G., Haidar, G., et al. (2021). Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 371, 1139–1142. doi: 10.1126/science.abf6950
Mendez, A. S., Ly, M., González-Sánchez, A. M., Hartenian, E., Ingolia, N. T., Cate, J. H., et al. (2021). The N-terminal domain of SARS-CoV-2 nsp1 plays key roles in suppression of cellular gene expression and preservation of viral gene expression. Cell Rep. 37:109841. doi: 10.1016/j.celrep.2021.109841
Miao, G., Zhao, H., Li, Y., Ji, M., Chen, Y., Shi, Y., et al. (2021). ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell 56, 427–442.e5. doi: 10.1016/j.devcel.2020.12.010
Michel, C. J., Mayer, C., Poch, O., and Thompson, J. D. (2020). Characterization of accessory genes in coronavirus genomes. Virol. J. 17:131. doi: 10.1186/s12985-020-01402-1
Mitratza, M., Klijs, B., Hak, A. E., Kardaun, J., and Kunst, A. E. (2021). Systemic autoimmune disease as a cause of death: mortality burden and comorbidities. Rheumatology 60, 1321–1330. doi: 10.1093/rheumatology/keaa537
Mohammad, S., Bouchama, A., Mohammad Alharbi, B., Rashid, M., Saleem Khatlani, T., Gaber, N. S., et al. (2020). SARS-CoV-2 ORF8 and SARS-CoV ORF8ab: genomic divergence and functional convergence. Pathogens 9:677. doi: 10.3390/pathogens9090677
Monrad, I., Sahlertz, S. R., Nielsen, S. S. F., Pedersen, L. Ø., Petersen, M. S., Kobel, C. M., et al. (2021). Persistent severe acute respiratory syndrome coronavirus 2 infection in immunocompromised host displaying treatment induced viral evolution. Open Forum Infect. Dis. 8:ofab295. doi: 10.1093/ofid/ofab295
Mou, K., Mukhtar, F., Khan, M. T., Darwish, D. B., Peng, S., Muhammad, S., et al. (2021). Emerging mutations in Nsp1 of SARS-CoV-2 and their effect on the structural stability. Pathogens 10:1285. doi: 10.3390/pathogens10101285
Murtas, R., Andreano, A., Gervasi, F., Guido, D., Consolazio, D., Tunesi, S., et al. (2020). Association between autoimmune diseases and COVID-19 as assessed in both a test-negative case-control and population case-control design. Auto Immun. Highlights 11:15. doi: 10.1186/s13317-020-00141-1
Nakagawa, K., and Makino, S. (2021). Mechanisms of coronavirus Nsp1-mediated control of host and viral gene expression. Cells 10:300. doi: 10.3390/cells10020300
Nieto-Torres, J. L., Dediego, M. L., Verdiá-Báguena, C., Jimenez-Guardeño, J. M., Regla-Nava, J. A., Fernandez-Delgado, R., et al. (2014). Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 10:e1004077. doi: 10.1371/journal.ppat.1004077
Niu, Z., Zhang, Z., Gao, X., Du, P., Lu, J., Yan, B., et al. (2021). N501Y mutation imparts cross-species transmission of SARS-CoV-2 to mice by enhancing receptor binding. Sig. Transduct. Target. Ther. 6:284. doi: 10.1038/s41392-021-00704-2
Nussenblatt, V., Roder, A. E., Das, S., De Wit, E., Youn, J.-H., Banakis, S., et al. (2022). Yearlong COVID-19 infection reveals within-host evolution of SARS-CoV-2 in a patient with B-cell depletion. J. Infect. Dis. 225, 1118–1123. doi: 10.1093/infdis/jiab622
Oude Munnink, B. B., Worp, N., Nieuwenhuijse, D. F., Sikkema, R. S., Haagmans, B., Fouchier, R. A. M., et al. (2021). The next phase of SARS-CoV-2 surveillance: real-time molecular epidemiology. Nat. Med. 27, 1518–1524.
Pablos, J. L., Galindo, M., Carmona, L., Lledó, A., Retuerto, M., Blanco, R., et al. (2020). Clinical outcomes of hospitalised patients with COVID-19 and chronic inflammatory and autoimmune rheumatic diseases: a multicentric matched cohort study. Ann. Rheum. Dis. 79, 1544–1549. doi: 10.1136/annrheumdis-2020-218296
Pancer, K., Milewska, A., Owczarek, K., Dabrowska, A., Kowalski, M., Łabaj, P. P., et al. (2020). The SARS-CoV-2 ORF10 is not essential in vitro or in vivo in humans. PLoS Pathog. 16:e1008959. doi: 10.1371/journal.ppat.1008959
Pavan, M., Bolcato, G., Bassani, D., Sturlese, M., and Moro, S. (2021). Supervised molecular dynamics (SuMD) insights into the mechanism of action of SARS-CoV-2 main protease inhibitor PF-07321332. J. Enzyme Inhib. Med. Chem. 36, 1646–1650. doi: 10.1080/14756366.2021.1954919
Peiffer-Smadja, N., Bridier-Nahmias, A., Ferré, V. M., Charpentier, C., Garé, M., Rioux, C., et al. (2021). Emergence of E484K mutation following bamlanivimab monotherapy among high-risk patients infected with the alpha variant of SARS-CoV-2. Viruses 13:1642. doi: 10.3390/v13081642
Pérez-Lago, L., Aldámiz-Echevarría, T., García-Martínez, R., Pérez-Latorre, L., Herranz, M., Sola-Campoy, P. J., et al. (2021). Different within-host viral evolution dynamics in severely immunosuppressed cases with persistent SARS-CoV-2. Biomedicines 9:808. doi: 10.3390/biomedicines9070808
Pollet, S., Conte, M. A., Sanborn, M., Jarman, R. G., Lidl, G. M., Modjarrad, K., et al. (2021). A comparative recombination analysis of human coronaviruses and implications for the SARS-CoV-2 pandemic. Sci. Rep. 11:17365. doi: 10.1038/s41598-021-96626-8
Ponsford, M. J., Ward, T. J. C., Stoneham, S. M., Dallimore, C. M., Sham, D., Osman, K., et al. (2021). A systematic review and meta-analysis of inpatient mortality associated with nosocomial and community COVID-19 exposes the vulnerability of immunosuppressed adults. Front. Immunol. 12:744696. doi: 10.3389/fimmu.2021.744696
Qu, Y., Wang, X., Zhu, Y., Wang, W., Wang, Y., Hu, G., et al. (2021). ORF3a-mediated incomplete autophagy facilitates severe acute respiratory syndrome coronavirus-2 replication. Front. Cell Dev. Biol. 9:716208. doi: 10.3389/fcell.2021.716208
Rahman, M. S., Hoque, M. N., Islam, M. R., Islam, I., Mishu, I. D., Rahaman, M. M., et al. (2021). Mutational insights into the envelope protein of SARS-CoV-2. Gene Rep. 22:100997. doi: 10.1016/j.genrep.2020.100997
Redondo, N., Zaldívar-López, S., Garrido, J. J., and Montoya, M. (2021). SARS-CoV-2 accessory proteins in viral pathogenesis: knowns and unknowns. Front. Immunol. 12:708264. doi: 10.3389/fimmu.2021.708264
Ren, Y., Shu, T., Wu, D., Mu, J., Wang, C., Huang, M., et al. (2020). The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 17, 881–883. doi: 10.1038/s41423-020-0485-9
Roe, M. K., Junod, N. A., Young, A. R., Beachboard, D. C., and Stobart, C. C. (2021). Targeting novel structural and functional features of coronavirus protease nsp5 (3CLpro, Mpro) in the age of COVID-19. J. Gen. Virol. 102:001558. doi: 10.1099/jgv.0.001558
Saunders, K. O., Lee, E., Parks, R., Martinez, D. R., Li, D., Chen, H., et al. (2021). Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature 594, 553–559. doi: 10.1038/s41586-021-03594-0
Schoeman, D., and Fielding, B. C. (2019). Coronavirus envelope protein: current knowledge. Virol. J. 16:69. doi: 10.1186/s12985-019-1182-0
Schubert, K., Karousis, E. D., Jomaa, A., Scaiola, A., Echeverria, B., Gurzeler, L.-A., et al. (2020). SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 27, 959–966.
Shannon, A., Le, N. T.-T., Selisko, B., Eydoux, C., Alvarez, K., Guillemot, J.-C., et al. (2020). Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 exonuclease active-sites. Antiviral Res. 178, 104793–104793. doi: 10.1016/j.antiviral.2020.104793
Shrestha, L. B., Tedla, N., and Bull, R. A. (2021). Broadly-neutralizing antibodies against emerging SARS-CoV-2 variants. Front. Immunol. 12:752003. doi: 10.3389/fimmu.2021.752003
Simon-Loriere, E., and Holmes, E. C. (2011). Why do RNA viruses recombine? Nat. Rev. Microbiol. 9, 617–626. doi: 10.1038/nrmicro2614
Starr Tyler, N., Greaney Allison, J., Addetia, A., Hannon William, W., Choudhary Manish, C., Dingens Adam, S., et al. (2021). Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 371, 850–854.
Sun, X., Liu, Y., Huang, Z., Xu, W., Hu, W., Yi, L., et al. (2022). SARS-CoV-2 non-structural protein 6 triggers NLRP3-dependent pyroptosis by targeting ATP6AP1. Cell Death Differ. [Epub ahead of print].
Suwanwongse, K., and Shabarek, N. (2020). Clinical features and outcome of HIV/SARS-CoV-2 coinfected patients in The Bronx, New York city. J. Med. Virol. 92, 2387–2389. doi: 10.1002/jmv.26077
Szemiel, A. M., Merits, A., Orton, R. J., Maclean, O. A., Pinto, R. M., Wickenhagen, A., et al. (2021). In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog. 17:e1009929. doi: 10.1371/journal.ppat.1009929
Tan, C.-W., Chia, W.-N., Young, B. E., Zhu, F., Lim, B.-L., Sia, W.-R., et al. (2021). Pan-sarbecovirus neutralizing antibodies in BNT162b2-immunized SARS-CoV-1 survivors. N. Engl. J. Med. 385, 1401–1406.
Tarhini, H., Recoing, A., Bridier-Nahmias, A., Rahi, M., Lambert, C., Martres, P., et al. (2021). Long-term severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectiousness among three immunocompromised patients: from prolonged viral shedding to SARS-CoV-2 superinfection. J. Infect. Dis. 223, 1522–1527. doi: 10.1093/infdis/jiab075
Tasakis, R. N., Samaras, G., Jamison, A., Lee, M., Paulus, A., Whitehouse, G., et al. (2021). SARS-CoV-2 variant evolution in the United States: high accumulation of viral mutations over time likely through serial founder events and mutational bursts. PLoS One 16:e0255169. doi: 10.1371/journal.pone.0255169
Tian, F., Tong, B., Sun, L., Shi, S., Zheng, B., Wang, Z., et al. (2021). N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. eLife 10:e69091. doi: 10.7554/eLife.69091
Truffot, A., Andréani, J., Le Maréchal, M., Caporossi, A., Epaulard, O., Germi, R., et al. (2021). SARS-CoV-2 variants in immunocompromised patient given antibody monotherapy. Emerg. Infect. Dis. J. 27, 2725–27280. doi: 10.3201/eid2710.211509
Truong, T. T., Ryutov, A., Pandey, U., Yee, R., Goldberg, L., Bhojwani, D., et al. (2021). Increased viral variants in children and young adults with impaired humoral immunity and persistent SARS-CoV-2 infection: a consecutive case series. EBioMedicine 67:103355. doi: 10.1016/j.ebiom.2021.103355
Vazquez, C., Swanson, S. E., Negatu, S. G., Dittmar, M., Miller, J., Ramage, H. R., et al. (2021). SARS-CoV-2 viral proteins NSP1 and NSP13 inhibit interferon activation through distinct mechanisms. PLoS One 16:e0253089. doi: 10.1371/journal.pone.0253089
V’kovski, P., Kratzel, A., Steiner, S., Stalder, H., and Thiel, V. (2021). Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 19, 155–170.
Wang, Q., Wu, J., Wang, H., Gao, Y., Liu, Q., Mu, A., et al. (2020). Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell 182, 417–428.e13. doi: 10.1016/j.cell.2020.05.034
Wang, R., Chen, J., Hozumi, Y., Yin, C., and Wei, G.-W. (2020). Decoding asymptomatic COVID-19 infection and transmission. J. Phys. Chem. Lett. 11, 10007–10015. doi: 10.1021/acs.jpclett.0c02765
Wang, Y., Mao, J.-M., Wang, G.-D., Luo, Z.-P., Yang, L., Yao, Q., et al. (2020). Human SARS-CoV-2 has evolved to reduce CG dinucleotide in its open reading frames. Sci. Rep. 10:12331.
Wang, W. B., Liang, Y., Jin, Y. Q., Zhang, J., Su, J. G., and Li, Q. M. (2021). E484K mutation in SARS-CoV-2 RBD enhances binding affinity with hACE2 but reduces interactions with neutralizing antibodies and nanobodies: binding free energy calculation studies. J. Mol. Graph. Model. 109, 108035–108035. doi: 10.1016/j.jmgm.2021.108035
Warren, T. K., Jordan, R., Lo, M. K., Ray, A. S., Mackman, R. L., Soloveva, V., et al. (2016). Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 531, 381–385. doi: 10.1038/nature17180
Weigang, S., Fuchs, J., Zimmer, G., Schnepf, D., Kern, L., Beer, J., et al. (2021). Within-host evolution of SARS-CoV-2 in an immunosuppressed COVID-19 patient as a source of immune escape variants. Nat. Commun. 12:6405. doi: 10.1038/s41467-021-26602-3
World Health Organization [WHO] (2020a). Pneumonia of unknown cause – China [Online]. Available Online at: https://www.who.int/emergencies/disease-outbreak-news/item/2020-DON229 [accessed October 25, 2021].
World Health Organization [WHO] (2020b). WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19 - 11 March 2020. Geneva: World Health Organization.
World Health Organization [WHO] (2021). Tracking SARS-CoV-2 Variants [Online]. Available Online at: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ [accessed November 14, 2021].
Wu, A., Peng, Y., Huang, B., Ding, X., Wang, X., Niu, P., et al. (2020). Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27, 325–328. doi: 10.1016/j.chom.2020.02.001
Xia, H., Cao, Z., Xie, X., Zhang, X., Chen, J. Y.-C., Wang, H., et al. (2020). Evasion of type I interferon by SARS-CoV-2. Cell Rep. 33:108234.
Yan, L., Ge, J., Zheng, L., Zhang, Y., Gao, Y., Wang, T., et al. (2021). Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell 184, 184–193.e10.
Zen, M., Fuzzi, E., Astorri, D., Saccon, F., Padoan, R., Ienna, L., et al. (2020). SARS-CoV-2 infection in patients with autoimmune rheumatic diseases in northeast Italy: a cross-sectional study on 916 patients. J. Autoimmun. 112:102502. doi: 10.1016/j.jaut.2020.102502
Zhong, J., Shen, G., Yang, H., Huang, A., Chen, X., Dong, L., et al. (2020). COVID-19 in patients with rheumatic disease in Hubei province, China: a multicentre retrospective observational study. Lancet Rheumatol. 2, e557–e564. doi: 10.1016/S2665-9913(20)30227-7
Keywords: SARS-CoV-2, viral evolution, secondary immunodeficiency, mutations, spike protein, COVID-19
Citation: Markarian NM, Galli G, Patel D, Hemmings M, Nagpal P, Berghuis AM, Abrahamyan L and Vidal SM (2022) Identifying Markers of Emerging SARS-CoV-2 Variants in Patients With Secondary Immunodeficiency. Front. Microbiol. 13:933983. doi: 10.3389/fmicb.2022.933983
Received: 01 May 2022; Accepted: 31 May 2022;
Published: 01 July 2022.
Edited by:
Axel Cloeckaert, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceReviewed by:
Barbara Schmidt, University of Regensburg, GermanyDaniele Focosi, Pisana University Hospital, Italy
Copyright © 2022 Markarian, Galli, Patel, Hemmings, Nagpal, Berghuis, Abrahamyan and Vidal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Levon Abrahamyan, bGV2b24uYWJyYWhhbXlhbkB1bW9udHJlYWwuY2E=; Silvia M. Vidal, c2lsdmlhLnZpZGFsQG1jZ2lsbC5jYQ==
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
 Nathan M. Markarian
Nathan M. Markarian Gaël Galli
Gaël Galli Dhanesh Patel
Dhanesh Patel Mark Hemmings
Mark Hemmings Priya Nagpal
Priya Nagpal Albert M. Berghuis
Albert M. Berghuis Levon Abrahamyan
Levon Abrahamyan Silvia M. Vidal
Silvia M. Vidal