
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 07 July 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.930658
This article is part of the Research Topic From Traditional to Modern: Progress of Molds and Yeasts in Fermented-food Production, Volume II View all 12 articles
Yeasts are often considered microorganisms for producing human therapeutic glycosylated end-products at an industrial scale. However, the products with non-humanized glycans limited their usage. Therefore, various methods to develop humanized glycosylated end-products have been widely reported in yeasts. To make full use of these methods, it is necessary to summarize the present research to find effective approaches to producing humanized products. The present research focuses on yeast species selection, glycosyltransferase deletion, expression of endoglycosidase, and expression of proteins with galactosylated and or sialylated glycans. Nevertheless, the yeasts will have growth defects with low bioactivity when the key enzymes are deleted. It is necessary to express the corresponding repairing protein. Compared with N-glycosylation, the function of yeast protein O-glycosylation is not well-understood. Yeast proteins have a wide variety of O-glycans in different species, and it is difficult to predict glycosylation sites, which limits the humanization of O-glycosylated yeast proteins. The future challenges include the following points: there are still many important potential yeasts that have never been tried to produce glycosylated therapeutic products. Their glycosylation pathway and related mechanisms for producing humanized glycosylated proteins have rarely been reported. On the other hand, the amounts of key enzymes on glycan pathways in human beings are significantly more than those in yeasts. Therefore, there is still a challenge to produce a large body of humanized therapeutic end-products in suitable yeast species, especially the protein with complex glycans. CRISPR-Cas9 system may provide a potential approach to address the important issue.
Engineering Escherichia coli has been widely used for the production of human advanced biopharmaceutical products (Arico et al., 2013). However, the recombinant proteins in non-glycosylated forms often suffer from short-term half-life, low activity, rapid clearance, and side effects. Pharmacokinetic studies in Wistar rats revealed 1.3-fold increase in plasma half-life for glycosylated IFN α2b compared to standard IFN α2b produced by E. coli (Baghban et al., 2018). Therefore, it is necessary to develop humanized glycosylated therapeutic products (Cheon et al., 2012). Yeasts are often firstly considered microorganisms for producing human therapeutic glycosylated end-products at an industrial scale. Long historical usage, vast data, and rich experiences have paved the way for Saccharomyces cerevisiae as an important expression platform and a microbial cell factory for producing human therapeutic products (Corbacho et al., 2005). In S. cerevisiae, the recombinant proteins are often typically hypermannosylated (Man>50GlcNAc2), which also affects the half-life, tissue distribution, and immunogenicity of the end-products. It is critical to developing the engineering of S. cerevisiae for producing the proteins with humanized glycosylation patterns by modulating mannosyltransferase and or other enzymes responsible for the chain biosynthesis of N-glycans (Crauwels et al., 2015).
Pichia pastoris expression system is one of the most popular tools for producing a recombinant protein with many advantages, including the low incidence of hyperglycosylated, appropriate folding in the endoplasmic reticulum (ER), and secretion expression with high purity (De Pourcq et al., 2012). Much effort has been paid to improving the N-glycosylation pathway of P. pastoris to mimic the human N-glycosylation pathway. Successful utilization of the CRISPR-Cas9 system has been used for the humanization of the glycosylation pathways in P. pastoris by disrupting och1 and alg3 (mannosyltransferase) genes (Dolashka-Angelova et al., 2010). Kluyveromyces lactis is a food-grade yeast commonly used for industrial applications. Gene modification has been applied to construct a humanized N-linked glycosylation pathway in K. lactis. Besides the above conventional yeast species, other established yeast expression systems include Hansenula polymorpha (Gündüz Ergün et al., 2019; Fukunaga et al., 2021a,b; Giorgetti et al., 2021; Gallo et al., 2022), Yarrowia lipolytica (Hamilton et al., 2013; Karbalaei et al., 2020; Jin et al., 2021), Schizosaccharomyces pombe (Kim et al., 2006; Katla et al., 2019; Lawton et al., 2021), Kluyveromyces marxianus (Lee et al., 2020), Candida albicans (Li et al., 2019), and Zygosaccharomyces cidri (Liu et al., 2009). The strategies to produce humanized glycosylated end-products were explored by using these yeast species.
The N-glycosylation has been widely studied in yeast species, indicating that the glycosylated signaling pathway is similar to that in mammalian cells. Man3GlcNAc2 is the common core of eukaryotic N-glycans, which further yields the Man8GlcNAc2 core via oligosaccharyltransferase. Man8GlcNAc2-Asn-protein is produced in the endoplasmic reticulum (ER) and conveyed to Golgi (Figure 1A). In Golgi, there is a significant difference in the glycosylation formation of proteins, Man8GlcNAc2 glycan is truncated into Man5GlcNAc2 via mannosidase in mammalian cells (Figure 1B) while Man8GlcNAc2 in yeasts is extended to form Man9GlcNAc2 and hypermannosylation via mannosyl transferase (Figure 1A), which will be cleared in human blood because of its non-human glycan (Ma et al., 2019). Genome-wide analysis for S. cerevisiae shows 19 different genes associated with mannoprotein-linked oligosaccharide generation, including CAX4 (dolichyl pyrophosphate (Dol-PP) phosphatase), OST4 (oligosaccharyltransferase), OCH1/LDB12 (Mannosyltransferase), MNN(Mannosyltransferase)2/LDB8, MNN4, MNN5, KTR6/MNN6, ANP1/MNN8, MNN9, MNN10, MNN11, GDA1 (Golgi GDPase), HOC1/LDB12 (glycosyltransferase), VAN1/LDB13 (glycosylation of secreted invertase), OST3, RHK1/ALG(mannosyltransferase)3, ALG5, ALG6, and ALG9, which mainly existed in nucleic ER and Golgi ER (Figure 1A; Narimatsu et al., 2019). Most yeast species produce the glycosylated protein with mannose units with long chains.
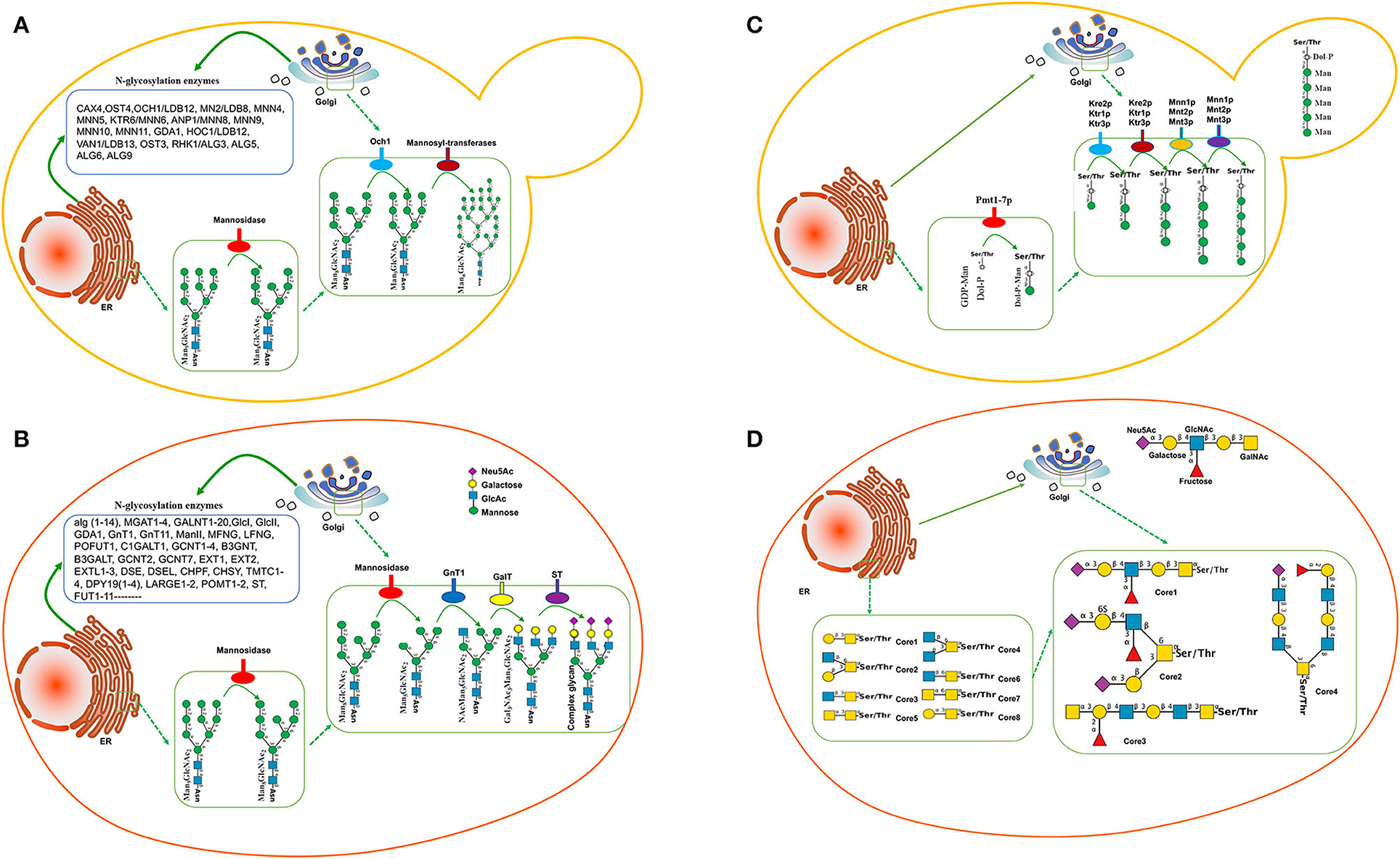
Figure 1. Glycosylation pathway between yeasts and mammalian cells. (A) N-glycosylation pathway in yeasts. (B) N-glycosylation pathway in mammalian cells. (C) O-glycosylation pathway in Saccharomyces cerevisiae. In the ER, the first mannose residue is added via protein-O-mannosyltransferases (Pmt1-7p). O-glycosylation is further elongated in the Golgi via several mannosyltransferases. (D) Mucin-type O-glycan synthesis in human cells.
Comparatively, the transferases in mammalian cells are significantly more than those in S. cerevisiae. More than 170 human glycosyltransferases have been found in the glycosylation pathway (Figure 1B; Oh et al., 2008). Most mammals produce the glycosylated protein with the mannose, galactose, and sialic acid N-glycolylneuraminic acid (Neu5Gc) units in the short-chain (Figure 1B). Therefore, the glycosylated proteins in yeast species are significantly longer than those in mammalian cells while the ingredients in mammalian cells are more complex than those in yeast species.
Compared with N-glycosylation, the function of yeast protein O-glycosylation is poorly understood. Yeast proteins have a wide variety of O-glycans in different species, and it is difficult to predict glycosylation sites, which limits the humanization of O-glycosylated yeast proteins. The first mannose linked to yeast protein is provided by polyterpenoid phosphate mannose (Dol-P-Man). Dol-P-Man is synthesized from guanosine diphosphate-mannose (GDP-Man) and polyterpenoid phosphate mannose (Dol-P) under the catalysis of Dol-P-Man synthase, and then, enters the endoplasmic reticulum to initiate protein O-glycosylation (Figure 1C). In contrast, mucin-type O-glycan synthesis in human cells starts with an N-acetylgalactosamine (GalNAc) residue attached with specific Ser/Thr residues with eight-type cores. The initiating residues are further elongated with a variety of monosaccharides. In the Golgi apparatus, the elongation of oligosaccharides is catalyzed by GalNAc transferases using UDP-GalNAc as a donor (Figure 1D). However, manufacturing the O-glycosylation pathway is not as progressive as N-glycosylation. Humanized mucin-type O-glycans had been identified in S. cerevisiae to provide a biosynthetic pathway for UDP-GalNAc and UDP-Gal/GalNAc-4-epimerase production (Luu et al., 2019).
Although yeasts have the potential to produce various types of industrial proteins, their failures to produce humanized glycoproteins limit their usage. Therefore, it is necessary to explore the strategies to regulate glycan forms of end-products in yeasts.
The glycan chains in yeast species are often longer than those in human beings. To fix the flaw, the key enzyme mannosyltransferase, which existed in N-glycosylation pathways, is often considered to be blocked. Och1(α-1,6-mannosyltransferase) and MNN1 (α-1,3-mannosyltransferase) produce mostly high-mannose glycans of the Man8GlcNAc2 (Figure 1A). Inactivation of the OCH1 gene is often an effective approach to eliminate hypermannosylation in H. polymorpha (Fukunaga et al., 2021b; Gallo et al., 2022), K. lactis (Park et al., 2011), P. pastoris (Ma et al., 2019), S. cerevisiae (Piirainen et al., 2022), and Y. lipolytica (Karbalaei et al., 2020), which results in the expressed proteins with short-chain glycan from Man3GlcNAc2 to Man14GlcNAc2 (Table 1). Especially in H. polymorpha, the deletion of OCH1 and alg3, and/or alg3 and alg11 can produce the protein with Man3GlcNAc2 (Gündüz Ergün et al., 2019; Fukunaga et al., 2021b). The expression of alg6 in S. pombe also produces the short-chain glycoprotein (Hamilton et al., 2013).
Protein glycosylation is often associated with cell wall integrity and its disruption may affect yeast bioactivity. The yeasts will have growth defects and low bioactivity when the key enzymes are deleted. Inactivation of the OCH1 gene will damage S. cerevisiae viability. The overexpression of glycosylphosphatidylinositol (GPI)-anchored protein can restore the bioactivity (Table 1; Prabhakar et al., 2019). Therefore, overexpression of repairing proteins may be an effective approach to improve the yields of humanized products.
The overexpression of endoglycosidase α-1,2-mannosidase can prevent the glycan chain from further extending in P. pastoris (Ma et al., 2019). ENGase isoform possesses great hydrolytic activities toward high-mannose N-glycans and short-chain glycan was produced after the ENGase was expressed in P. pastoris (Prill et al., 2005). EndoS2, an IgG-specific endoglycosidase, also has a similar function to ENGase and was expressed in S. cerevisiae to produce a single N-acetylglucosamine residue (Ryckaert et al., 2005).
In mammalian cells, GlcNAc sugars are attached to the paucimannose core with 2-4 GlcNac sugars, which can be further prolonged by adding galactose via GalTI (β-1,4-galactosyltransferase I). GalTI was expressed in H. polymorpha and increased the formation of galactosylated complex-type glycans (Giorgetti et al., 2021). A human galactosyltransferase fused protein with a UDP-glucose 4-epimerase domain from S. pombe was expressed in P. pastoris and also produced galactosylated proteins (Selas Castiñeiras et al., 2018).
The lack of sialylation on glycoprotein will reduce its efficacy as a therapeutic agent because of the rapid clearance of the protein from the human bloodstream. Human glycans are often capped terminally with sialic acid via sialyltransferase (ST, localized in Golgi, Figure 1B). A more effective biocatalyst system for producing sialylated glycoproteins was established in S. cerevisiae via the deletion of OCH1 and expression of ST more than one decade ago (Shenoy et al., 2021). Sialylated O-linked glycans were produced in P. pastoris via the expression of β-1,2-N-acetylglucosaminyltransferase 1 (AST; Song et al., 2010).
Among all the selected yeast species, H. polymorpha, K. marxianus, S. pombe, and Y. lipolytic tend to produce short-chain-glycan proteins than other yeast species (Table 1). Therefore, these yeast species may be more suitable to produce humanized end-products.
Over the past decades, engineering modification of N-glycosylation pathways in yeast species has developed significantly. Most yeast strains are suitable to produce the end-products with hybrid N-glycosylation, galactosylation, and sialyation complex glycans at high uniformity (Table 1). With the development of the yeast expression system, it is expected that yeast strains with the ability to generate fully humanized proteins or end-products will be attainable shortly. Nonetheless, there are still some challenges in exploring the yeast expressing system.
1) Arxula adeninivorans is an unconventional, non-pathogenic, and haploid yeast and maybe a new expression platform (Song et al., 2007). Brettanomyces bruxellensis, a wine yeast, mainly contributes to spontaneous beer fermentations associated with industrial fermentation ecosystems (Tanaka et al., 2021). Brettanomyces claussenii, a yeast often found on pineapple or other tropical fruits, expands the potential for the valorization of dairy by-products to functional beverages (Turakainen et al., 1994). Candida utilis is a commercial food additive and a potentially beneficial host for producing heterologous protein (Uccelletti et al., 2004). Hanseniaspora uvarum (anamorph Kloeckera apiculata) is an apiculate yeast species normally found on wine grapes and other fruits and has a strong influence on wine quality. Zygosaccharomyces bailii is a yeast species widely present in wine, tea, and vinegar fermentations (Uccelletti et al., 2006). However, these yeasts, and their N-glycosylation pathway and related mechanisms for producing humanized glycosylated proteins have rarely been reported.
2) There are more than 20 glycosyltransferases found in the glycosylation pathway in S. cerevisiae (Figure 1A; Narimatsu et al., 2019) and 170 human glycosyltransferases found in the glycosylation pathway (Figure 1B; Oh et al., 2008). Therefore, there is still a challenge to produce a large body of humanized therapeutic end-products with complex glycan in suitable yeasts. Making full use of the CRISPR-Cas9 technique may be a potential strategy to address the important issues (Ueda et al., 1993; Dolashka-Angelova et al., 2010).
XL, XC, LC, SW, and CC collected and analyzed all data. XQ, XL, XC, JS, KC, and HT contributed to the conception and designing the article and interpreted the relevant literature. XL and HT drafted and revised the article. The manuscript has been read and approved by all authors before submission.
The present work was supported by Guangzhou Zengcheng Qiaomengyuan Branch Project (Grant No. 20210300X).
XL, JS, XC, LC, SW, XQ, KC, CC, and HT were employed by Guangzhou Shenjingya Agricultural Technology Co., Ltd.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Arico, C., Bonnet, C., and Javaud, C. (2013). “N-glycosylation humanization for production of therapeutic recombinant glycoproteins in Saccharomyces cerevisiae,” in Glycosylation Engineering of Biopharmaceuticals, ed A. Beck (Berlin: Springer), 45–57. doi: 10.1007/978-1-62703-327-5_4
Baghban, R., Farajnia, S., Ghasemi, Y., Mortazavi, M., Zarghami, N., and Samadi, N. (2018). New developments in Pichia pastoris expression system, review and update. Curr. Pharmaceut. Biotechnol. 19, 451–467. doi: 10.2174/1389201019666180718093037
Cheon, S. A., Kim, H., Oh, D.-B., Kwon, O., and Kang, H. A. (2012). Remodeling of the glycosylation pathway in the methylotrophic yeast Hansenula polymorpha to produce human hybrid-type N-glycans. J. Microbiol. 50, 341–348. doi: 10.1007/s12275-012-2097-2
Corbacho, I., Olivero, I., and Hernández, L. M. (2005). A genome-wide screen for Saccharomyces cerevisiae non-essential genes involved in mannosyl phosphate transfer to mannoprotein-linked oligosaccharides. Fungal Genet. Biol. 42, 773–790. doi: 10.1016/j.fgb.2005.05.002
Crauwels, S., Steensels, J., Aerts, G., Willems, K., Verstrepen, K., and Lievens, B. (2015). Brettanomyces bruxellensis, essential contributor in spontaneous beer fermentations providing novel opportunities for the brewing industry. BrewingScience 68, 110–121.
De Pourcq, K., Tiels, P., Van Hecke, A., Geysens, S., Vervecken, W., and Callewaert, N. (2012). Engineering Yarrowia lipolytica to produce glycoproteins homogeneously modified with the universal Man3GlcNAc2 N-glycan core. PLoS ONE 7:e39976. doi: 10.1371/journal.pone.0039976
Dolashka-Angelova, P., Moshtanska, V., Kujumdzieva, A., Atanasov, B., Petrova, V., Voelter, W., et al. (2010). Structure of glycosylated Cu/Zn-superoxide dismutase from Kluyveromyces yeast NBIMCC 1984. J. Mol. Struct. 980, 18–23. doi: 10.1016/j.molstruc.2010.06.031
Fukunaga, T., Sakurai, Y., Ohashi, T., Higuchi, Y., Maekawa, H., and Takegawa, K. (2021a). Overexpression of cell-wall GPI-anchored proteins restores cell growth of N-glycosylation-defective och1 mutants in Schizosaccharomyces pombe. Appl. Microbiol. Biotechnol. 105, 8771–8781. doi: 10.1007/s00253-021-11649-5
Fukunaga, T., Tanaka, N., Furumoto, T., Nakakita, S., Ohashi, T., Higuchi, Y., et al. (2021b). Substrate specificities of α1, 2-and α1, 3-galactosyltransferases and characterization of Gmh1p and Otg1p in Schizosaccharomyces pombe. Glycobiology 31, 1037–1045. doi: 10.1093/glycob/cwab028
Gallo, G. L., Valko, A., Herrera Aguilar, N., Weisz, A. D., and D'alessio, C. (2022). A novel fission yeast platform to model N-glycosylation and the bases of congenital disorders of glycosylation type I. J. Cell Sci. 135:jcs259167. doi: 10.1242/jcs.259167
Giorgetti, S. I., Etcheverrigaray, M., Terry, F., Martin, W., De Groot, A. S., Ceaglio, N., et al. (2021). Development of highly stable and de-immunized versions of recombinant alpha interferon: promising candidates for the treatment of chronic and emerging viral diseases. Clin. Immunol. 233:108888. doi: 10.1016/j.clim.2021.108888
Gündüz Ergün, B., Hüccetogullari, D., Öztürk, S., Çelik, E., and Çalik, P. (2019). Established and upcoming yeast expression systems. Recombinant Protein Production Yeast 1, 1–74. doi: 10.1007/978-1-4939-9024-5_1
Hamilton, S. R., Cook, W. J., Gomathinayagam, S., Burnina, I., Bukowski, J., Hopkins, D., et al. (2013). Production of sialylated O-linked glycans in Pichia pastoris. Glycobiology 23, 1192–1203. doi: 10.1093/glycob/cwt056
Jin, Y., Yu, S., Liu, J.-J., Yun, E. J., Lee, J. W., Jin, Y.-S., et al. (2021). Production of neoagarooligosaccharides by probiotic yeast Saccharomyces cerevisiae var. boulardii engineered as a microbial cell factory. Microbial Cell Factor. 20, 1–10. doi: 10.1186/s12934-021-01644-w
Karbalaei, M., Rezaee, S. A., and Farsiani, H. (2020). Pichia pastoris: a highly successful expression system for optimal synthesis of heterologous proteins. J. Cell. Physiol. 235, 5867–5881. doi: 10.1002/jcp.29583
Katla, S., Yoganand, K., Hingane, S., Kumar, C. R., Anand, B., and Sivaprakasam, S. (2019). Novel glycosylated human interferon alpha 2b expressed in glycoengineered Pichia pastoris and its biological activity: N-linked glycoengineering approach. Enzyme Microbial Technol. 128, 49–58. doi: 10.1016/j.enzmictec.2019.05.007
Kim, M. W., Kim, E. J., Kim, J.-Y., Park, J.-S., Oh, D.-B., Shimma, Y.-I., et al. (2006). Functional characterization of the Hansenula polymorpha HOC1, OCH1, and OCR1 genes as members of the yeast OCH1 mannosyltransferase family involved in protein glycosylation. J. Biol. Chem. 281, 6261–6272. doi: 10.1074/jbc.M508507200
Lawton, M. R., Dana, L. D., and Alcaine, S. D. (2021). Lactose utilization by Brettanomyces claussenii expands potential for valorization of dairy by-products to functional beverages through fermentation. Curr. Opin. Food Sci. 42, 93–101. doi: 10.1016/j.cofs.2021.05.006
Lee, M.-H., Hsu, T.-L., Lin, J.-J., Lin, Y.-J., Kao, Y.-Y., Chang, J.-J., et al. (2020). Constructing a human complex type N-linked glycosylation pathway in Kluyveromyces marxianus. PLoS ONE 15:e0233492. doi: 10.1371/journal.pone.0233492
Li, S., Sun, P., Gong, X., Chang, S., Li, E., Xu, Y., et al. (2019). Engineering O-glycosylation in modified N-linked oligosaccharide (Man12GlcNAc2~ Man16GlcNAc2) Pichia pastoris strains. RSC Adv. 9, 8246–8252. doi: 10.1039/C8RA08121B
Liu, B., Gong, X., Chang, S., Yang, Y., Song, M., Duan, D., et al. (2009). Disruption of the OCH1 and MNN1 genes decrease N-glycosylation on glycoprotein expressed in Kluyveromyces lactis. J. Biotechnol. 143, 95–102. doi: 10.1016/j.jbiotec.2009.06.016
Luu, V.-T., Moon, H. Y., Yoo, S. J., Choo, J. H., Thak, E. J., and Kang, H. A. (2019). Development of conditional cell lysis mutants of Saccharomyces cerevisiae as production hosts by modulating OCH1 and CHS3 expression. Appl. Microbiol. Biotechnol. 103, 2277–2293. doi: 10.1007/s00253-019-09614-4
Ma, J., Li, Q., Tan, H., Jiang, H., Li, K., Zhang, L., et al. (2019). Unique N-glycosylation of a recombinant exo-inulinase from Kluyveromyces cicerisporus and its effect on enzymatic activity and thermostability. J. Biol. Eng. 13, 1–14. doi: 10.1186/s13036-019-0215-y
Narimatsu, Y., Joshi, H. J., Nason, R., Van Coillie, J., Karlsson, R., Sun, L., et al. (2019). An atlas of human glycosylation pathways enables display of the human glycome by gene engineered cells. Mol. Cell 75, 394–407.e395. doi: 10.1016/j.molcel.2019.05.017
Oh, D. B., Park, J. S., Kim, M. W., Cheon, S. A., Kim, E. J., Moon, H. Y., et al. (2008). Glycoengineering of the methylotrophic yeast Hansenula polymorpha for the production of glycoproteins with trimannosyl core N-glycan by blocking core oligosaccharide assembly. Biotechnol. J. 3, 659–668. doi: 10.1002/biot.200700252
Park, J.-N., Song, Y., Cheon, S. A., Kwon, O., Oh, D.-B., Jigami, Y., et al. (2011). Essential role of Yl MPO1, a novel Yarrowia lipolytica homologue of Saccharomyces cerevisiae MNN4, in mannosylphosphorylation of N-and O-linked glycans. Appl. Environ. Microbiol. 77, 1187–1195. doi: 10.1128/AEM.02323-10
Piirainen, M. A., Salminen, H., and Frey, A. D. (2022). Production of galactosylated complex-type N-glycans in glycoengineered Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 106, 301–315. doi: 10.1007/s00253-021-11727-8
Prabhakar, A., Vadaie, N., Krzystek, T., and Cullen, P. J. (2019). Proteins that interact with the mucin-type Glycoprotein Msb2p include a regulator of the actin cytoskeleton. Biochemistry 58, 4842–4856. doi: 10.1021/acs.biochem.9b00725
Prill, S. K. H., Klinkert, B., Timpel, C., Gale, C. A., Schröppel, K., and Ernst, J. F. (2005). PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 55, 546–560. doi: 10.1111/j.1365-2958.2004.04401.x
Ryckaert, S., Martens, V., De Vusser, K., and Contreras, R. (2005). Development of a S. cerevisiae whole cell biocatalyst for in vitro sialylation of oligosaccharides. J. Biotechnol. 119, 379–388. doi: 10.1016/j.jbiotec.2005.04.010
Selas Castiñeiras, T., Williams, S. G., Hitchcock, A. G., and Smith, D. C. (2018). E. coli strain engineering for the production of advanced biopharmaceutical products. FEMS Microbiol. Lett. 365:fny162. doi: 10.1093/femsle/fny162
Shenoy, A., Yalamanchili, S., Davis, A. R., and Barb, A. W. (2021). Expression and display of glycoengineered antibodies and antibody fragments with an engineered yeast strain. Antibodies 10:38. doi: 10.3390/antib10040038
Song, H., Qian, W., Wang, H., and Qiu, B. (2010). Identification and functional characterization of the HpALG11 and the HpRFT1 genes involved in N-linked glycosylation in the methylotrophic yeast Hansenula polymorpha. Glycobiology 20, 1665–1674. doi: 10.1093/glycob/cwq121
Song, Y., Choi, M. H., Park, J.-N., Kim, M. W., Kim, E. J., Kang, H. A., et al. (2007). Engineering of the yeast Yarrowia lipolytica for the production of glycoproteins lacking the outer-chain mannose residues of N-glycans. Appl. Environ. Microbiol. 73, 4446–4454. doi: 10.1128/AEM.02058-06
Tanaka, N., Kagami, A., Hirai, K., Suzuki, S., Matsuura, S., Fukunaga, T., et al. (2021). The fission yeast gmn2+ gene encodes an ERD1 homologue of Saccharomyces cerevisiae required for protein glycosylation and retention of luminal endoplasmic reticulum proteins. J. General Appl. Microbiol. 2020:2. doi: 10.2323/jgam.2020.07.002
Turakainen, H., Hankaanpää, M., Korhola, M., and Aho, S. (1994). Characterization of MEL genes in the genus Zygosaccharomyces. Yeast 10, 733–745. doi: 10.1002/yea.320100605
Uccelletti, D., Farina, F., Mancini, P., and Palleschi, C. (2004). KlPMR1 inactivation and calcium addition enhance secretion of non-hyperglycosylated heterologous proteins in Kluyveromyces lactis. J. Biotechnol. 109, 93–101. doi: 10.1016/j.jbiotec.2003.10.037
Uccelletti, D., Farina, F., Rufini, S., Magnelli, P., Abeijon, C., and Palleschi, C. (2006). The Kluyveromyces lactis α1, 6-mannosyltransferase KlOch1p is required for cell-wall organization and proper functioning of the secretory pathway. FEMS Yeast Res. 6, 449–457. doi: 10.1111/j.1567-1364.2006.00027.x
Ueda, K., Shimabuku, A. M., Konishi, H., Fujii, Y., Takebe, S., Nishi, K., et al. (1993). Functional expression of human P-glycoprotein in Schizosaccharomyces pombe. FEBS Lett. 330, 279–282. doi: 10.1016/0014-5793(93)80888-2
Vervecken, W., Kaigorodov, V., Callewaert, N., Geysens, S., De Vusser, K., and Contreras, R. (2004). In vivo synthesis of mammalian-like, hybrid-type N-glycans in Pichia pastoris. Appl. Environ. Microbiol. 70, 2639–2646. doi: 10.1128/AEM.70.5.2639-2646.2004
Wang, H., Song, H.-L., Wang, Q., and Qiu, B.-S. (2013). Expression of glycoproteins bearing complex human-like glycans with galactose terminal in Hansenula polymorpha. World J. Microbiol. Biotechnol. 29, 447–458. doi: 10.1007/s11274-012-1197-9
Wang, S., Rong, Y., Wang, Y., Kong, D., Wang, P. G., Chen, M., et al. (2020). Homogeneous production and characterization of recombinant N-GlcNAc-protein in Pichia pastoris. Microbial Cell Factor. 19, 1–11. doi: 10.1186/s12934-020-1280-0
Watanasrisin, W., Iwatani, S., Oura, T., Tomita, Y., Ikushima, S., Chindamporn, A., et al. (2016). Identification and characterization of Candida utilis multidrug efflux transporter CuCdr1p. FEMS Yeast Res. 16:fow042. doi: 10.1093/femsyr/fow042
Keywords: humanized glycosylation, yeasts, genetically engineering, end-products, galactosylated complex-type glycans
Citation: Li X, Shen J, Chen X, Chen L, Wan S, Qiu X, Chen K, Chen C and Tan H (2022) Humanization of Yeasts for Glycan-Type End-Products. Front. Microbiol. 13:930658. doi: 10.3389/fmicb.2022.930658
Received: 28 April 2022; Accepted: 31 May 2022;
Published: 07 July 2022.
Edited by:
Wanping Chen, University of Göttingen, GermanyReviewed by:
Kentaro Inokuma, Kobe University, JapanCopyright © 2022 Li, Shen, Chen, Chen, Wan, Qiu, Chen, Chen and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianlie Shen, c2psY2NtQDEyNi5jb20=; Haidong Tan, MTgwOTA1MDM1MUBxcS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.