- 1Singapore Centre for Environmental Life Sciences Engineering, Nanyang Technological University, Singapore, Singapore
- 2Institute of Materials Research and Engineering, Agency for Science, Technology and Research (A*STAR), Singapore, Singapore
- 3School of Bioscience and Technology, Vellore Institute of Technology, Vellore, India
- 4Australian Institute for Microbiology and Infection, The University of Technology Sydney, Sydney, NSW, Australia
- 5Department of Biological Sciences, National University of Singapore, Singapore, Singapore
- 6Agriculture and Food, Microbiomes for One Systems Health, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Canberra, ACT, Australia
Seawalls are important in protecting coastlines from currents, erosion, sea-level rise, and flooding. They are, however, associated with reduced biodiversity, due to their steep orientation, lack of microhabitats, and the materials used in their construction. Hence, there is considerable interest in modifying seawalls to enhance the settlement and diversity of marine organisms, as microbial biofilms play a critical role facilitating algal and invertebrate colonization. We assessed how different stone materials, ranging from aluminosilicates to limestone and concrete, affect biofilm formation. Metagenomic assessment of marine microbial communities indicated no significant impact of material on microbial diversity, irrespective of the diverse surface chemistry and topography. Based on KEGG pathway analysis, surface properties appeared to influence the community composition and function during the initial stages of biofilm development, but this effect disappeared by Day 31. We conclude that marine biofilms converged over time to a generic marine biofilm, rather than the underlying stone substrata type playing a significant role in driving community composition.
Introduction
A warming planet has led to thermal expansion of the oceans and melting of ice sheets, thus raising sea levels around the world (Warrick and Oerlemans, 1990; Douglas, 1991; Golledge et al., 2019) and accelerating coastal erosion and flooding. Low lying islands and coastal cities are particularly vulnerable (Bruun, 1962; Nicholls and Cazenave, 2010). As human populations continue to migrate toward the coast, the risk to property and life will increase (Temmerman et al., 2013). The installation of artificial defense structures such as seawalls has become the primary mitigation strategy for preventing some of the more extreme effects of sea level rise (Firth et al., 2014; Todd et al., 2019). These can be constructed from any material that can withstand the constant action of waves and currents, but concrete and natural stone materials are commonly used (Dugan et al., 2011). For many countries, large scale installations represent a significant amount of the coastline. For example, Korea’s artificial Saemangeum dyke is > 33 km in length (Lam et al., 2009), Japan is planning over 400 km of seawalls to protect from future tsunami disasters (Yamashita, 2020) and in Singapore, > 63% of the coast is some form of seawall, either vertical concrete or granite rip-rap (Lai et al., 2015). However, seawall proliferation comes at a considerable cost to coastal ecosystems. Heatherington and Bishop (2012) reported that seawalls affect the physical and hydrodynamic processes of a natural coastal region, such as sediment and larval transportation. Others have shown that they support lower species diversity and different communities (Ming et al., 2010; Lai et al., 2015) and can encourage invasive species (Simkanin et al., 2012). Critically, the pre-existing habitat is often obliterated by the installation of such large and intrusive infrastructure (Firth et al., 2014).
While there exists a strong ecological case for moving away from seawalls to ecosystem-based coastal defenses such as mangroves and saltmarshes, these require substantial shoreline space that is generally not available near cities (Temmerman et al., 2013). It is inevitable that seawalls will continue to be constructed. However, there remains the potential to improve their design to make them more viable as a habitat for marine life (Loke et al., 2015; Morris et al., 2019). One solution that is being extensively researched is to increase the topographical complexity of the seawall to provide niche availability for a wider range of species (Loke et al., 2019). It is thought that anthropogenic seawall installations are lacking in complexity and thus have a reduce niche environment (Lawrence et al., 2021). However, more recent investigations have highlighted that complexity alone is not universally positive with respect to increasing biodiversity, as this can be affected by local stressors, such as predation and should be seen only as an element in improving seawalls (Chee et al., 2021; Strain et al., 2021). Another, much less studied approach, is to identify construction materials that may encourage colonization by marine biota (Dodds et al., 2022; Hartanto et al., 2022). Before any invertebrates or other eukaryotic organisms settle on the seawall surface, it is known that microbial colonizers begin to form “conditioning” biofilms. These biofilms are a vital first step in the recruitment of other organisms (Freckelton et al., 2017), as the biofilms and species therein have been shown to have a role in regulating the settlement of sessile organisms onto the substratum (Hadfield, 2011; Freckelton et al., 2022). However, it is unclear whether manipulating the seawall construction material has any substantial impact on the biofilms that initially form on the seawall surface. Studies comparing seawall microbial biofilms to those found on natural rocky shorelines have shown differences in the relative abundance of biofilm inhabitants between the two environments (Tan et al., 2015). However, settlement control plates present at both sites (seawall and rocky shore) did not differ in biofilm settlement. This suggests that the locations are not driving any observed differences, but instead it is the chemical and/or physical properties of those sites that drive differences in the settled and formed microbial communities.
The current study selected natural stones comprising aluminosilicates and calcium carbonate, which present different surface chemistry. As substrates, they also differ in their resistance to erosion. Granite is an aluminosilicate that resists chemical erosion, which has led to its widespread implementation as a durable mechanical barrier on seawalls. In contrast, limestone is composed from calcium carbonate, which is more readily eroded in a mildly acidic environment (Revelle and Emery, 1957), thus giving rise to natural macrostructures with higher complexity (Higgins, 1980). Sandstone differs by offering a high degree of porosity and it has been reported to promote the growth of foliose algae (Green et al., 2012). Settlement on natural stones and the ensuing erosion generated by microbial species have been studied and shown to influence the microbial biofilms associated with the stones (Cockell et al., 2011; Summers et al., 2016). Differences have been observed between microorganisms weathering limestone, granite, and concrete (Coombes et al., 2011), with boring also being influenced by environmental factors (Cockell and Herrera, 2008). The surface chemistry of aluminosilicates presents silanol groups that have high affinity to water (Birch et al., 2008), while calcium carbonate substrates present moieties that are reported to bind to organic molecules (Suess, 1970; Carter, 1978; Zeller et al., 2020).
Based on these observations, we hypothesized that differing construction materials would recruit different microbial biofilm communities. If this hypothesis is correct, then construction material could be a key component not only for seawall strength and durability, but also for any biodiversity enhancement strategy, such as the addition of tiles or cladding. Therefore, to address the role of substrata in marine biofilm community composition, we examined several construction materials for the formation of natural tropical marine biofilms, to elucidate what impacts these varied materials may have on the initial colonization of an anthropogenic sea defense structures.
Materials and methods
Substrata used as proxies for potential seawall materials
To determine the effect that stone type has on marine biofilm formation on a tropical seawall, eight different substrata to monitor biofilm community formation were selected to represent salient potential construction materials, encompassing aluminosilicates and calcium carbonates, respectively. These were: granite (GRANITE), limestone (LIMESTN), marble (MARBLE), pink sandstone (SANDSP), violet sandstone (SANDSV), pH neutralized concrete (CONCTN), rough textured concrete (CONCRO), and smooth textured concrete (CONCTR). These were chosen to represent salient natural stone categories, with differing surface structure, mineral, and chemical compositions. They were selected with uniform surface appearance, on a cm-scale, purchased from a local stone mason (Hot Spring Stone Pte. Ltd., Singapore). The surfaces were saw-cut without polishing, thus mitigating surface roughness variations and avoiding alteration of their surface chemistry, respectively. Stone samples were used as 5 cm × 5 cm × 2.5 cm coupons and concrete coupons were prepared by pouring concrete premix into set molds of the same dimensions. Concrete grain composition was 80% sand and 20% cement, resulting in a mortar comparable to the standards employed in masonry. The intrinsic alkalinity of as-formed concrete, with pH ≈ 13, can be mitigated by carbonation. This was achieved by exposing the concrete coupons to a saturated CO2 environment, generated within a sealed vessel by sublimating dry ice, for 1 month (Hsiung et al., 2020). This process confers pH neutrality to the surface region, and progressively penetrates into the bulk volume of the concrete.
The stone surfaces were examined by scanning electron microscopy (SEM) using a JEOL JSM-7600F field emission scanning electron microscope operating at an accelerating voltage of 15 kV. Each was sputter coated with gold using a JEOL JFC-1200 fine coater and its surface morphology was imaged, together with an elemental cartography. Surface elemental and compositional analysis was performed with an Oxford Energy Dispersive X-Ray Analysis (EDX) system. Internal surface area, pore volume, and overall porosity measurements were obtained via Brunauer–Emmett–Teller (BET) nitrogen adsorption, using a Micromeritics ASAP 2020 system. Pore distribution was obtained from mercury intrusion, using a MicroActive Autopore V9600 mercury porosimeter. Average surface roughness (Ra) was measured over a 5 mm length using a TENCOR P-10 Surface Profiler at a scan speed of 50 μms–1, with a sampling rate of 100 Hz. Thermal conductivity was measured with a NETZSCH HFM 436 heat flow meter.
Natural biofilm generation— Amplicon sequencing experiment
To develop natural marine biofilms on the different stone types, each coupon was sterilized in an autoclave (121°C for 15 min), submerged vertically in one of two outdoor flow-through aquaria to mitigate sedimentation and were supplied with a constantly renewed supply of sand filtered natural seawater for 1 month. Each aquarium held 300 L of seawater and was fed at a flow rate of 15 L min–1, resulting in three water exchanges per hour. This exchange rate was chosen to mitigate any effects from leachates from the stone coupons during the incubation. Temperature and LUX intensity logs of the water and air (<5 cm above water line) were recorded each hour for 31 days, using Hobo data loggers (Onset, 8k data logger pendant).
To monitor biofilm development over the 1-month period, sacrificial coupons of all eight substrata types were removed at six time points (after 3 h and 1, 3, 7, 14, and 31 days) for examination. The experiment was divided over two aquaria, fed from the same seawater supply, with two replicates of each treatment were present in each tank, for a total of four replicates overall for each stone type at each time point (192 coupons in total). The position of each coupon within each aquarium was randomized, using the “sample” function in R base, to mitigate any positional bias within the aquaria with respect to biofilm growth. To determine the variability of the microbial community in the sand filtered seawater, 50 mL of seawater was collected at each time point to act as a background control. These aliquots were filtered onto a 0.22 μm polycarbonate filter and subsequently stored at −20°C.
Natural biofilm generation—Metagenome sequencing experiment
This experiment was repeated 12 months later using a similar experimental design to that employed for amplicon sequencing. The second run did not include concrete coupons and sacrificial sampling occurred weekly (weeks 1, 2, 3, and 4). One reason for omitting the concrete coupons was the emergence from our trials with chemically altered concretes that their differences appear to have little effect on biological diversity at the macro scale (Hsiung et al., 2020). Glass microscope slides were also included during this experiment to allow comparison with other studies which employ glass substrata as controls. All analyses involving the control glass slides were standardized to account for the difference in area between the glass and stone substrata. In addition, total nitrogen (TN), total organic carbon (TOC), and total carbon (TC) from the aquaria were measured each week for the duration of the experiment (TOC-L analyzer, Shimadzu, Singapore), from these data the total inorganic carbon (TIC) was calculated. The water chemistry was recorded each week using a YSI ProQuatro meter (YSI; Ohio, United States). The primary purpose of this second experiment was to enable metagenomic sequencing of the biofilms. All treatments were performed in triplicate for this experiment.
Biofilm collection and nucleic acid extraction
To assess the biodiversity of the biofilms across time, the stone coupons were removed from the aquaria at the time points indicated above. The stones were removed from the aquaria and the biofilm sampled from a single 5 × 5 cm face. A sterile cotton swab was used to collect the biofilm material formed on the surface. These were immediately labeled and frozen at −20°C, before being transported to the laboratory in a liquid nitrogen cooled dry shipper and then stored at −20°C until nucleic acid extraction was performed.
To extract and isolate the nucleic acids, individual cotton swab heads were removed and placed into a Lysis Matrix E tube, before extraction with the FastDNA soil extract kit (MP Biomedical, Singapore), which was implemented according to the manufacturer’s instructions. The resulting total nucleic acid extractions were quantified using a NanoDrop spectrophotometer, before being stored at −20°C for downstream analysis. These concentration data were employed as proxies for the general biomass of the biofilms and cannot distinguish between the main taxonomic groups (Bohórquez et al., 2017). For the background controls, the seawater was filtered and each filter was cut into half and placed in a sterile microcentrifuge tube, before being frozen in liquid N2 and then milled down to a powder, which was then subjected to the same process of extraction as the cotton swabs. All extractions were stored at −20°C, for downstream analysis.
Nucleic acid amplification and MiSeq amplicon library preparation
Two sets of primers were used to target the 16S rRNA (515f-806r; V4 region bacteria and archaea) (Caporaso et al., 2011) and the 18S rRNA (Euk 1392f—Euk Br; V9 region) (Medlin et al., 1988; Lane, 1991) to generate libraries for amplicon sequencing. For each reaction for the 16S rRNA gene, 0.5 μL of each primer (10 μM) was added to 10 μL of Jumpstart Taq Ready Mix (#P2893-100RXN, Merck, Singapore). Additionally, 1 μL of gDNA template was added to each reaction and then made up to 20 μL using sdH2O. For each reaction for the 18S rRNA gene, 1.0 μL of each primer (10 μM) was added to 12 μL of Jumpstart Taq Ready Mix. Additionally, 1 μL of gDNA template was added to each reaction and made up to 25 μL using sdH2O. The amplification condition for the 16S rRNA libraries were 94°C for 3 min initial denaturation, followed by 35 cycles of 94°C (45 s), 53°C (60 s), and 72°C (90 s); concluding with 72°C for 10 min. Amplification of the 18S rRNA libraries was achieved by 94°C for 3 min initial denaturation, followed by 35 cycles of 94°C (45 s), 57°C (60 s), and 72°C (90 s), concluding with 72°C for 10 min. Upon amplification, sequencing libraries were generated using the TruSeq kit v2.0 and paired-end MiSeq sequencing was conducted.
Amplicon sequence quality, assembly and identification
Quality filtration of the raw sequences was performed using the DaDa2 (Callahan et al., 2016) protocol, within an R environment (R Core Team, 2014). Briefly, all sequences were observed for QC Phred scores above 25 and bases trimmed from the start (10 bp) and end (50 bp) of the fragments. Subsequently, these sequences were assembled and processed using DaDa2 to obtain chimeric free amplicon single nucleotide variants (SNVs). Prokaryotic and eukaryotic sequences were identified through comparison to the Silva v.132 Ribosomal database. Each identified SNV required a minimum similarity of 80% in order to be classified as a particular taxon, otherwise it was labeled as unclassified. Rarefaction plots indicating sampling effort are available (Supplementary Figure 1).
Metagenome sequencing library
Total nucleic acid extractions from the second experiment were quantified using a Qubit HS (high sensitivity) dsDNA kit. Sequencing library preparation was achieved using the Swift Biosciences Accel-NGS 2S Plus DNA Kit, as per the manufacturer’s instructions. Using a Covaris S220 ultrasonicator, fragments were sheared to ∼450 bp, prior to adding barcodes using the Swift Biosciences 2S dual indexing kit. The subsequent libraries were checked for fragment length using a Bioanalyser DNA 7500 chip and quantification was performed using a picrogreen fluorescence assay. Libraries were then standardized to 4 nM as verified by qPCR (Kapa Biosystems Library Quantification kit, Applied Biosciences). An equimolar pool of all samples was generated and sequenced on Illumina HiSeq 2500 rapid runs (10–11 pM: V2 rapid sequencing chemistry), yielding reads of 251 bp paired-end sequences.
Metagenomic sequence analyses
Shotgun sequencing reads were assessed for quality and adapters were removed using the Trim Galore package (v0.6.4_dev). Low quality ends were removed from sequences with a phred score lower than 20. Paired reads were merged using Pear (v0.9.6), using default settings. The resulting sequences were trimmed to between 200 and 300 bases before being randomly sub-sampled, which resulted in 750,000 reads per sample. All sub-sampled reads were subjected to taxonomic identification by comparison to the Maxikraken2 database (Loman, 2020), using the Kraken2 package (v2.0.8_beta) and visualized as Sankey plots using R (Pavian Package). Independent of the taxonomic identification, the raw reads were de novo assembled using spades (v3.13.0), thus generating contigs. Gene annotation of the contigs was conducted using Prokka (1.13) and these were examined for functional identification using Microbeannotator (v2.0.5) which annotated the proteins against the KOfam and RefSeq databases, before being searches and compared again the KEGG database (release 551) using the KOfamscan package (Kanehisa et al., 2016; Kanehisa and Sato, 2019). From these metagenomic sequencing reads, all rRNA reads were identified and classified using sortmerna package (v4.3.4) and Kraken2 (v2.0.8_beta) package, both equipped with the Silva v.132 database used for the amplicon experiments. Rarefaction plots indicating sampling effort of all rRNAs are available (Supplementary Figure 3).
Statistical analysis of results
Data were processed and visualized in R Core Team (2014) using the vegan (Oksanen et al., 2011), complexheatmap (Gu et al., 2016), and ggplots (Wickham, 2016). Briefly, all absolute values of abundance were standardized to relative abundance for SNVs. Statistical analysis was performed using Permanova (Anderson, 2001) (adonis; vegan) to determine community differences and univariate differences between data were assessed using ANOVA or ANCOVA analyses (aov; vegan). Potential functional differences were assessed using ANOSIM to determine if identified complete KEGG pathways differed between treatment types. Post hoc analyses were conducted using the Tukey test (Tukey, 1949) of difference, to determine variables that differ significantly (p ≤ 0.05). Alpha-diversity was assessed using Shannon-Weiner indices (Shannon, 1948). All data was arcsine transformed prior to statistical assessment, to ensure the assumptions needed for parametric analyses were met where appropriate.
Data availability
Data generated during this study is available within the Supplementary Information. All sequences are available from the NCBI, Sequence Read Archive (BioProject: PRJNA698754).
Results
Biofilm physico-chemical environment
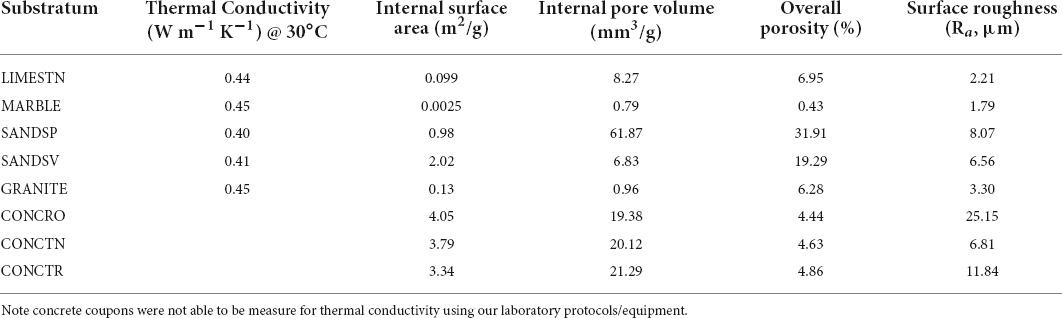
Three of the five natural stone substrata examined were classed as aluminosilicates (Al2SiO5; SANDSP, SANDSV, and GRANITE), while the remaining were calcium carbonates (LIMESTN and MARBLE). The elemental composition of LIMESTN was predominately Ca, C, and O, with trace amounts of Mg (0.02%) and Ni (0.1%) (Table 1). While broadly similar to LIMESTN, MARBLE contained a lower fraction of Ca, together with higher percentage (11.5%) of Mg and trace amounts of Co and Al. SANDSP, SANDSV, and GRANITE contained Si and Al, as anticipated for aluminosilicates, with higher fractions of Mg in SANDSV (3.64%) and GRANITE (1.72%). In GRANITE, Fe generally appeared to be co-localized with Mg. Al and Na were similarly co-located, but in complementary locations (Supplementary Figure 3). A similar trend was observed for Al and Na, in SANDSP. SANDSV presented comparatively small, localized regions of Ca, which are presumed to indicate the presence of incorporated calcium carbonate fragments. Si and O, associated with silica moieties, were exhibited uniformly over the natural stone aluminosilicates. In contrast, CONCRO samples showed Si in locations complementary to Ca.
While natural stone coupons were cut with identical tooling, their surfaces exhibited different roughness. This is attributed to differences in hardness and their exposed porosity. The structure of LIMESTN presents a fractional pore volume over 11% (Table 2), with a larger fraction in the sub-micron range ≈280–500 nm and a smaller proportion over ≈50–250 μm. This differs substantially from MARBLE, which presents a 10-fold lower overall porosity (≈ 1%) that occurs primarily around 250 nm and over a larger size range that is similar to LIMESTN ≈65–270 μm. These ranges were estimated from the width at half of the maximum of pore size (Supplementary Figure 2). SANDSP and SANDSV both present fractional pore volumes of the same order as limestone, with a significant proportion occurring in the range 150–500 nm. They contrast in their larger pore dimensions, with SANDSP exhibiting a narrow peak around 7.9 μm, while SANDSV offers a broad distribution, from ≈30 to 300 μm. GRANITE offers a comparatively closed structure, with a fractional pore volume that is similar to MARBLE. This appears as a broad distribution above 150 nm and larger size pores with a peak at 4.5 μm. Thermal conductivity was similar across the natural stones.
The environmental parameters of the aquaria hosting the stone coupons during biofilm growth showed some variability for the duration of the first experiment. Air and water data logs for temperature and LUX intensity indicated measurable diurnal fluctuations of both parameters, with light levels largely consistent and water temperature levels dropping over the course of the experiment (Supplementary Figure 5). For example, there was a temperature range of 5.68°C (max: 31.78°C, min: 26.10°C) in the air temperature over the course of the experiment, and the daily mean air temperature varied by 3.10°C. The water temperature showed an overall range of 2.39°C (max: 30.05°C, min: 27.67°C). The water temperature had a maximum diurnal variability of up to 1.20°C per day. However, the mean diurnal temperature change was 0.67°C. Over the course of the experiment the mean daily temperature of the water lowered by 1.85°C.
For the metagenomic experiment, the water samples tested throughout the experiment indicate some nutrient enrichment of inorganic carbon (Table 3). In addition, there was a reduction in TIC for weeks 2 and 3. TN did not follow the same pattern of fluctuations as the carbon.
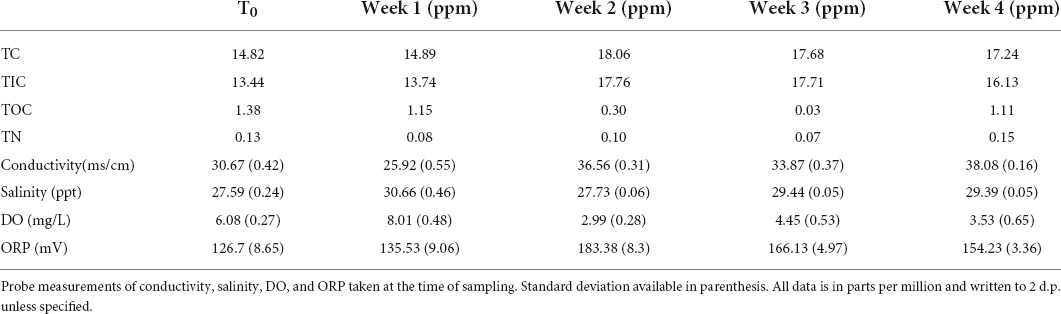
Table 3. Carbon and Nitrogen measure taken of the water present in the aquaria at the time of each sampling point.
Biofilm analyses
The amplicon experiment (with time points at 3 h and 1, 3, 7, 14, and 31 days) did not yield sufficient nucleic acids to provide a confident assessment of biomass. However, the metagenomic experiment yielded measurable nucleic acid concentrations at each of the four time points (Figure 1). Data indicate that there was no significant difference in biomass among substratum types [ANOVA, F(5,120) = 1.551, p = 0.179] or time points [ANOVA, F(3,120) = 0.551, p = 0.649]. However, while a visual inspection of these data suggested that the biomass on glass was lower than that of calcium carbonate-based substrata (MARBLE and LIMESTN), pairwise analyses revealed no significant differences (Tukey HSD, p = 0.121 and p = 0.251, respectively).
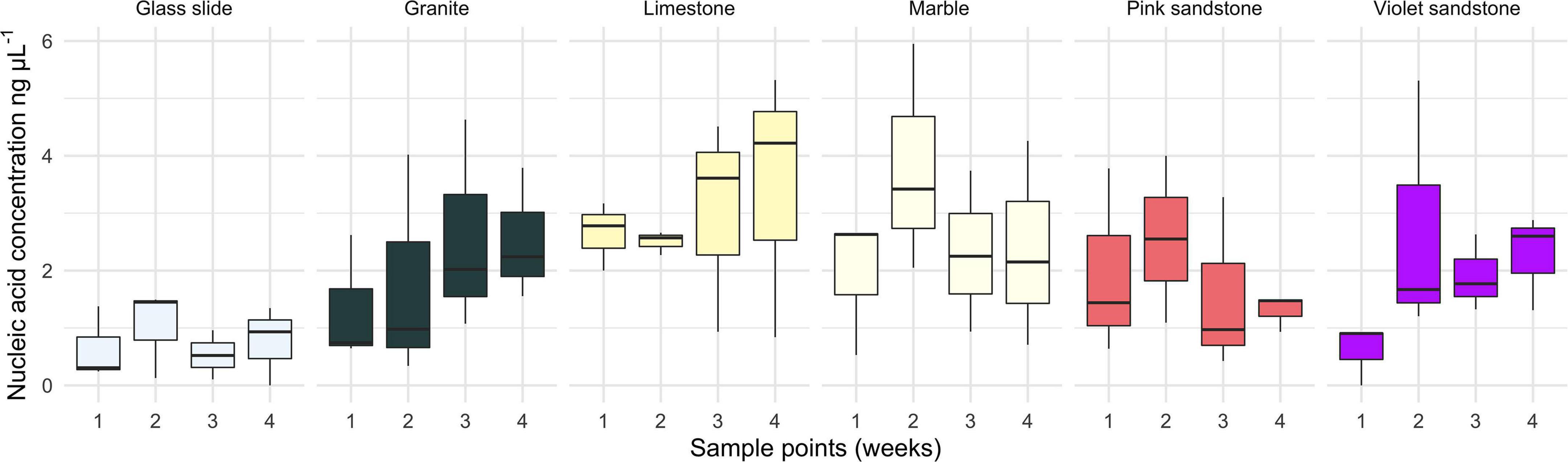
Figure 1. Box and whisker plot of the biofilm biomass estimates based on total nucleic acid concentration extracted from each time point grouped by substratum. Measured values were obtained using a Qubit fluorescence HS dsDNA kit.
Amplicon sequences
From the 192 samples processed, 190 produced suitable sequencing libraries, with two SANDSP coupons (days 3 and 31) failing to amplify. For the remaining 190 samples from the first experiment, there was no significant influence of substratum on the Prokaryotic composition based on 16S rRNA marker genes [PERMANOVA, F(7,181) = 1.203, p = 0.066; Figure 2). However, the age of the biofilm resulted in significant differences in bacterial composition [PERMANOVA, F(1,189) = 13.782, p = 0.001]. Pairwise investigation of these data indicates that no single substratum was significantly different in overall microbial community composition to any other substratum. Conversely, biofilm age had a significant effect on the microbial composition for all time point pairwise comparisons, with the exception of 3 h vs. 14 days and 7 days vs. 14 days which showed no statistical difference in microbial communities at these times.
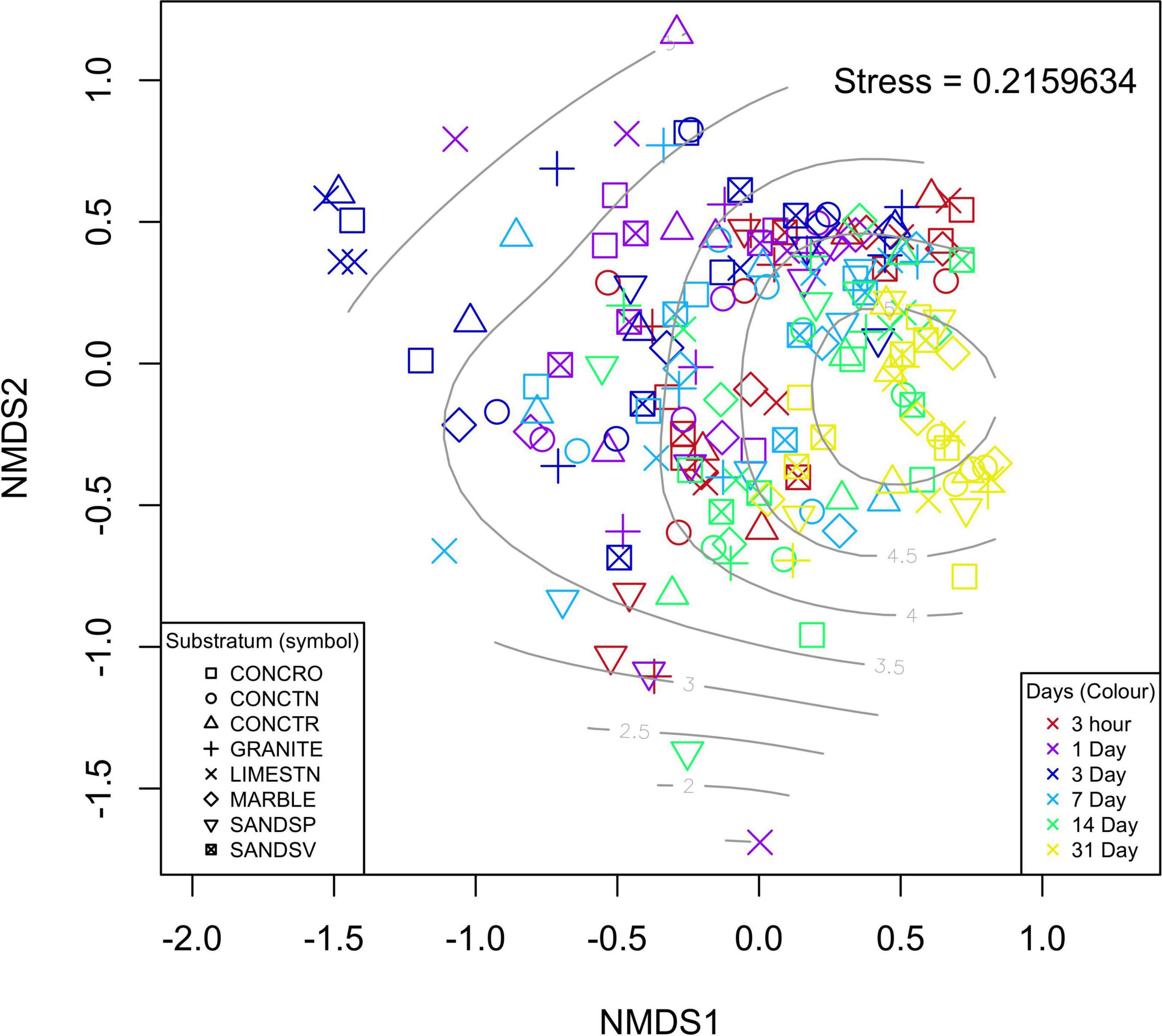
Figure 2. nMDS ordination of the individual samples used for 16S rRNA gene SNV analyses. Each substratum is indicated by a different symbol, whereas biofilm age is represented by color of the points on the plot. Light gray contour lines are indicative of Shannon-wiener diversity indices for each sample.
Univariate analysis of the effect of biofilm age and substratum on the biofilm’s α diversity also showed no significant influence of substratum [ANOVA, F(7,182) = 0.746, p = 0.634]. It was apparent that α diversity increased over time [ANOVA, F(1,188) = 31.87, p < 0.001], thought to result in the similarities in β diversity observed for all samples at the end of the experiment (31 days). Analyses of the seawater controls indicated that there was no change in Bray-Curtis variability over the course of the experiment and neither did microbial diversity differ [ANOVA, F(1,6) = 1.603, p = 0.29], indicating that no measurable outside influences impacted the experimental design.
The overall bacterial composition of the biofilms was dominated by a low abundance of single nucleotide variants (SNVs; Supplementary Figure 6). Most of the identified SNVs contributed < 1% of the overall relative abundance for any given sample. However, a few key taxa were dominant (high mean relative abundance) across all treatments. These included SNVs identified as Colwellia (5.3%), Escherichia-Shigella (4.3%), Neptuniibacter (3.2%), Thalassotalea (3.0%), and Rhodobacteraceae (2.9%); with Thalassotalea being the highest individual fraction (>80%) observed for one of the SANDSP replicates. While the overall bacterial community composition was not significantly different among substrata, 87 of the 3,631 individual SNVs identified in this study were significantly different among substratum type. These differences appeared to be primarily dominated by larger relative abundances of the SNVs on CONCRO (Supplementary Figure 7).
Examination of the 18S rRNA marker genes revealed no difference in Eukaryotic communities associated with the substratum material [PERMANOVA, F(9–95) = 0.9125, p = 0.691]. There was, however, a significant difference over time within the Eukaryotic communities [PERMANOVA, F(7–95) = 5.906, p = 0.001]. The 18S rRNA gene dataset yielded 4,419 specific SNVs across all treatments. From the 20 most abundant SNVs, it was clear that time was a contributing factor for their distribution (Supplementary Figure 8). From this subset (61.3% of total) of Eukaryotic SNVs, invertebrates contributed significantly to the SNVs, with Copepoda being responsible for > 27% of the total SNVs. Representatives of the Choradates, Vertebrata (4.53%), and Asciidians (4.38%) were also present. In addition, there was evidence of fungi (Basidiomycota; 3.48%), although the oligonucleotides used in this study were not optimized for fungal identification. In addition, the dominance of Copepoda 18S rRNA gene markers in the Eukaryotic fraction, is likely due to a high number of gene copy numbers within the genome his (Wyngaard et al., 1995).
Metagenomic sequence analyses
The metagenomic experiment revealed that the metagenome and amplicon data differed with respect to the dominant taxa in the Prokaryotic and Eukaryotic groups. This difference was likely due to amplification bias from the oligos used in amplicon sequencing or the gene copy number from the rRNA gene markers monitored. For example, amplicon analyses of the Prokaryotic fraction of the biofilm indicated a dominance of Colwellia spp., while the metagenomic data showed that Rhodobacterales was the most dominant of the contigs. This was also evident in the eukaryotic fraction, with Copeopoda being abundant in amplicon sequences, yet absent from the common metagenome sequences. All rRNA identified from the metagenome sequencing was compared to the rRNA data from the amplicon sequencing experiments. From these data we concluded that the most abundant organisms found in the amplicon experiments were also present in the metagenome experiment, with only relative abundances differing (Supplementary Figure 9). This abundance difference was expected due to different library preparation techniques employed for the two experiments. Assessment of the alpha diversity of these in silico obtained rRNA genes was performed by ANCOVA and we observed that the substrata had no measurable impact [ANCOVA, F(5) = 0.98, p > 0.05] on diversity, whereas the age of the biofilm did [ANCOVA, F(3) = 3.23, p = 0.028]. In addition to the differences between contigs and amplicon assessments, the use of contigs enabled us to determine the relative contribution of Eukaryotic and Prokaryotic fractions. While Eukaryotic contigs were observed, their contribution was minimal compared to Prokaryotic organisms (Bacteria ∼ 98%, Archaea ∼ 0.2%, Eukaryotes ∼ 0.8%, and Viruses ∼ 0.07%; Supplementary Data—taxa abundance tables).
Despite these subtle differences in biofilm community identifications, the metagenome experiment once again showed that biofilm age was a driving factor in biofilm composition [PERMANOVA, F(5–71) = 3.45, p < 0.001], as was previously observed in the amplicon experiment. In contrast to the amplicon experiment, we found a significant effect of substratum on biofilm communities. However, this was only between the glass control substratum and the other stone types—there were no significant differences amongst the experimental stone types and biofilm community (Figure 3).
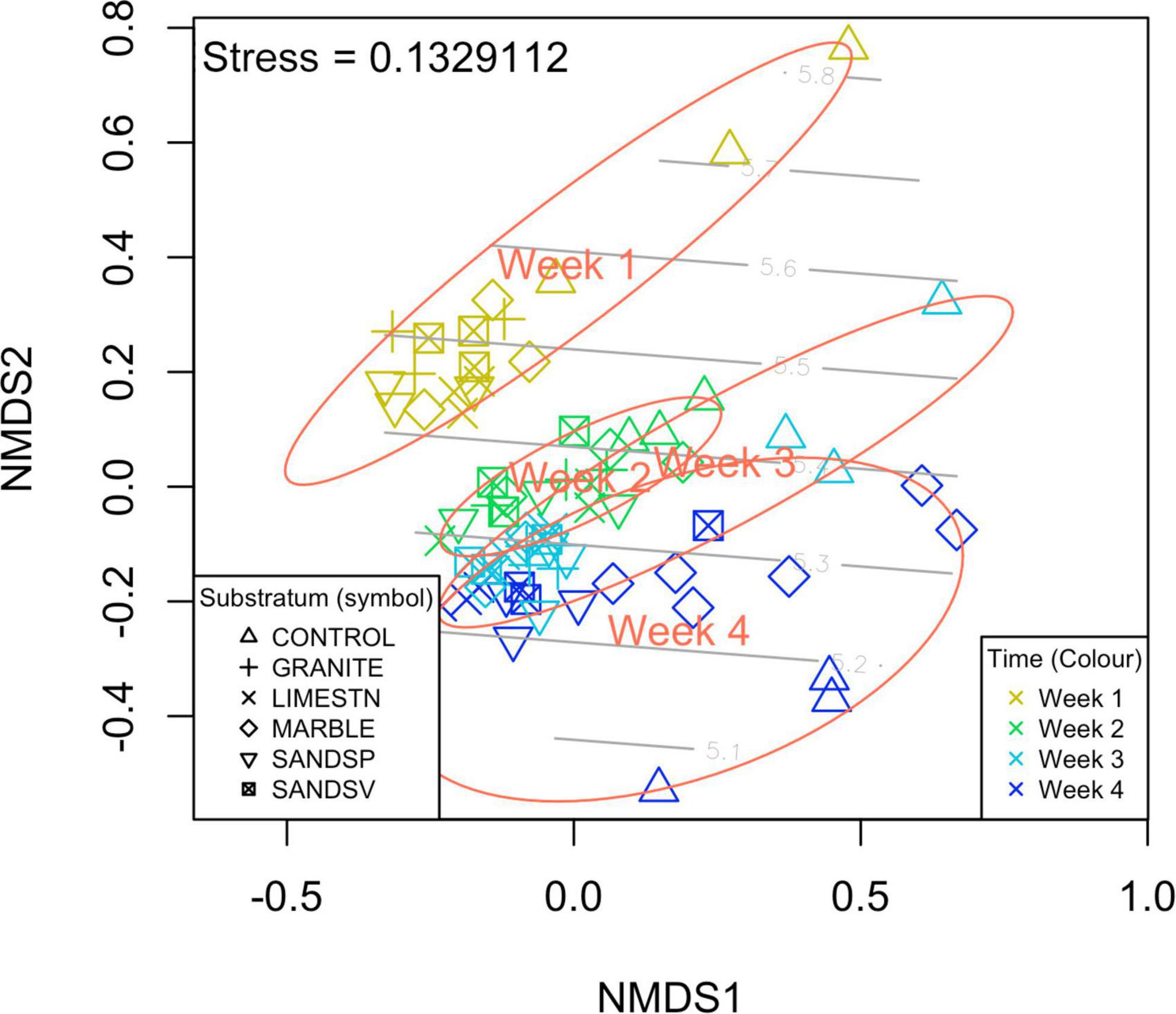
Figure 3. nMDS plot shows that that α and β diversity of the meta-contigs clearly separate over the y-axis suggesting that biofilm age is a factor of interest. Colors signify biofilm age: Gold = Week1, Green = Week2, light blue = Week 3, and dark blue = Week 4. Light gray contour lines indicate the Shannon Wiener diversity (H′).
Functional assessment of contigs
From the contigs there was a total of 3.9 M identified and hypothetical proteins annotated. When pooled for each treatment type across the four time points measured, these yielded 160 complete identified KEGG pathways. Of these, 81 KEGG pathways occurred within biofilms from all substrata. As with the taxonomic comparison, functional diversity also showed that the age of the biofilm had a statistically significant impact on the KEGG modules identified as complete (ANOSIM, R = 0.14, p = 0.01). However, substratum type had no significant effect (ANOSIM, R = 0.03, p > 0.05). Some KEGG modules which were common, but not shared across all samples, included Lipid A biosynthesis (M00866), Galactose degradation (M00632), and Thiosulfate oxidation (M00595). Some of the less common KEGG modules identified include Thyroid hormone biosynthesis (M00043), Glycosaminoglycan biosynthesis (M00058), and Lactosylceramide biosynthesis (M00066). As glass was the most dissimilar, significant differences in enrichment were observed for this substratum. Biofilms on glass slides were enriched for aromatic amino acid degradation while genes associated with assimilatory nitrate reduction (i.e., M00531) exhibited reduced abundance. As the biofilms increased in age, we also observed changes in the KEGG module enrichment, with initial (week 1) biofilms being enriched for salicylate (i.e., M00638). No enrichment of any KEGG module was observed for weeks 1 and 3. Four modules were enriched for weeks 1 and 4: homoprotocatechuate degradation, methane oxidation, ammonia nitrification, and comammox (i.e., M00533, M00174, M00528, and M00804, respectively). A full list of the identified modules, together with their level of completeness, can be found in Supplementary Figure 7.
Discussion
The construction of seawalls is often the go-to defense against erosion and flooding, especially in coastal cities, hence finding ways to mitigate their negative impacts on species’ biodiversity is critical to ecological functioning and sustainable development (Firth et al., 2014; Cole et al., 2015). The current study aims to determine whether seawall construction materials affect the initial biofilm formation, with a view to identifying stone types that can provide ecological uplift. Our results show that biofilms changed, in both taxonomic composition and functional potential, over the initial 4 weeks of growth. However, these were not significantly affected by the stone type in either of the two experiments; findings that align with a recent seawall study examining colonization by macrobiota on similar materials (Hartanto et al., 2022). In contrast, smooth glass supported a different biofilm community, as compared with the natural stones. It was also observed that some SNVs were significantly more abundant on rough concrete surfaces.
The calcium carbonate-based substrata supported a consistently high biomass throughout the experiment, while the glass slides induced growth of a consistently lower biomass biofilm. This is congruent with studies describing stronger adsorption of organic matter on calcium carbonate than on silica (Carter, 1978; Zeller et al., 2020). Moreover, glass slides had an intrinsic surface roughness approximately three orders of magnitude lower (Ra ∼ 3–5 nm) than that of the stones used in this study. This difference in roughness may limit the ability of biofilms to bind strongly onto the smoother surfaces (Encinas et al., 2020). This argument is supported by the increased relative abundance of some SNVs, as found on the rougher CONCRO samples (Supplementary Figure 2). Indeed, the latter substratum harbored many of the SNVs that differed significantly among substrata. However, some studies show that larger organisms such as diatoms have the opposite relationship with surface roughness, with rough surfaces yielding lower diversity and abundance (Sweat and Johnson, 2013). Previous research from other aquatic environments, such as rivers, have also shown that surface roughness is a driver of biofilm composition and diversity (Schneck et al., 2011), as well as augmenting the biomass of benthic algae (Murdock and Dodds, 2007). Furthermore, sandstone’s substantial fraction of micron-scale porosity is expected to facilitate the penetration of filaments and enhance water retention, leading to its propensity for supporting the growth of foliose algae (Green et al., 2012).
Porosity measurements indicate pore sizes that are compatible with promoting microbial adhesion, namely above 50 nm and up to several hundred nanometers (Feng et al., 2015). Granite presented almost no pores with sub-μm size and a narrow size distribution around 4.5 μm, together with a low (≈1%) fractional porosity. LIMESTN, SANDSP, and SANDSV incorporate significant porosity, ranging over sizes from μm to sub-mm. Their interior may thus potentially harbor micro-organisms, but these were not harvested and analyzed in the present study.
Biofilm diversity (α and β) did not expose significant differences for the tested substrata, thus implying the absence of surface-induced biofilm variations. While it is probable that surface roughness and the settling organism’s size and shape may contribute to driving the recruitment process of these initial colonizers (Lu et al., 2016; Huang et al., 2018), the homogeneity in marine microbial settlement contrasts with the diverse surface chemistry of stone surfaces, as broadly classified into calcium carbonate, aluminosilicates, and concrete (Table 1). Moreover, Huggett et al. (2009) found that marine biofilm taxonomy converged on similar substrata, but that invertebrate settlement was impacted by wettability (amongst other factors). Therefore, it is likely that surface roughness, surface chemistry, and sub-surface porosity in relation to the settling organism’s size and shape are key characteristics driving the recruitment process of these initial colonizers (Lu et al., 2016; Huang et al., 2018).
The overall diversity (α and β) of the biofilms appears not to have been driven by the stone materials, but rather regulated by the age of the biofilm. In total, we observed 863 16S SNVs (2.4% of total) that where significantly different the time periods sampled. Of the SNVs most enriched were candidates from Gemmatimonadetes, Calditrichaceae, Entotheonella, Microtrichales, Nitrsococcaceae, Lentisphaerae, and four uncultured bacteria. While the average water temperature did decline throughout the experiment, this was by only ∼1.84°C (mean daily average). There was no significant change in LUX over the duration of this experiment. Data showed (Figure 2) that, as the biofilms age, α diversity indices increased and the dissimilarly between biofilms on each substratum (β diversity) reduced, in the case of amplicon sequences. This increase in α diversity and decrease in β diversity is also observed in similar on-going field experiments in the region (data unpublished). However, the α diversity of the metagenome samples only showed minor fluctuations. This is most likely due to the lack of high frequency sampling in the first weeks, such as was employed for the amplicon experiments. This indicates a more homogenous biofilm with increased convergence in identified SNVs and identified contigs. For the contigs the α diversity was most similar at weeks 2 and 3, which matched the reduction in TOC measures. This suggests that recruitment from the water column may be a factor in regulating the α diversity. The β diversity changes on the glass controls (open triangles in Figure 3) were subtly different from other substrata of an equal age, supporting the observation that the (sub-nm) roughness of the glass may be more important than the chemical composition of the glass when compared to the silicate stones (SANDSP and SANDSV).
In addition to taxonomic diversity, we examined functional traits. Given the complexity in microbial systems, and that functional diversity does not have a global definition (Escalas et al., 2019), we opted for trait-based richness as a measure of functional potential. We showed that the age of the biofilm was a driver for changes in functional trait richness, although post hoc analyses show that it was only samples from weeks 1 to 3 that differed significantly. Both the functional and taxonomic differences observed could also be influenced by lower TOC levels at week 3, which may have altered recruitment. However, our data do not determine this conclusively (Supplementary Figure 10).
By observing enrichment of functional trait richness, the differences between the time points was associated with enrichment of specific KEGG modules only for weeks 1 and 4. This enrichment could have implications with respect to the function of the biofilm community and the efficacy by which invertebrate larvae settle on the biofilms. For example, the presence of methanotrophs in weeks 1 and 4 may reduce the settlement of some coral larvae as it has been shown that the presence of methanotrophs deter larval settlement (Sneed et al., 2015). Nevertheless, Petersen and Dubilier (2009) showed that methanotrophy can encourage symbiosis between invertebrates and bacteria, suggesting that not all invertebrates would be repelled by these bacteria. A similar scenario is also likely for coral settlement with regards to nitrogen fixation, as N2 is a major factor in coral health (Rädecker et al., 2015) and any changes in the nitrogen cycling ability of the biofilm may impact the settlement of coral larvae. Therefore, the enrichments observed for weeks 1 and 4, as well as the reduction observed on the glass substrata, could impact what types of invertebrate larvae choose to settle on these surface biofilms. Furthermore, the increase in biomass over time observed for some of the substrata may further enhance invertebrate settlement as it has been shown that increases in settlement occur with high bacterial cell numbers (Lema et al., 2019). However, it is unclear from these data whether this was an influence of biomass alone or a change in function that would likely result from a higher cell density within the biofilm.
Conclusion
The initial aim of this study was to determine what effect different seawall building material would have on the taxonomic and functional diversity of a typical marine biofilm during early settlement. We discovered, however, that differences in material surface chemistry and topography had no measurable lasting impact on the biofilms. Only glass, which was orders of magnitude smoother, and the rougher concrete substratum, had any significant influence on the biofilm composition, and only during the initial stages. The overall observation was that the marine biofilms converged over time to a generic marine biofilm, rather than the underlying stone substrata type playing a significant role in driving distinct communities.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
SS, YP, DM, PT, WB, and SR equally contributed to the design of this project. SS, YP, and DV generated all data and performed analyses. All authors contributed to the writing of this article.
Funding
This research was funded by the National Research Foundation, Prime Minister’s Office, Singapore under its Marine Science Research and Development Programme (Award No. MSRDP-P05). We acknowledge financial support from the Singapore Centre for Environmental Life Sciences Engineering, whose research is supported by the National Research Foundation Singapore, Ministry of Education, Nanyang Technological University and National University of Singapore, under its Research Centre of Excellence Programme. We also acknowledge financial support from the Institute of Materials Research and Engineering (IMRE) under the Agency of Science, Technology and Research (A*STAR).
Acknowledgments
We would like to thank the staff at St Johns Island National Marine Laboratory, Singapore for their help in facilitating these experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.928877/full#supplementary-material
References
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1046/j.1442-9993.2001.01070.x
Birch, W., Carré, A., and Mittal, K. L. (2008). “Wettability techniques to monitor the cleanliness of surfaces,” in Developments in Surface Contamination and Cleaning, eds R. Kohli and K. L. Mittal (Netherlands: Elsevier), 693–723. doi: 10.1016/B978-081551555-5.50015-0
Bohórquez, J., Mcgenity, T. J., Papaspyrou, S., García-Robledo, E., Corzo, A., and Underwood, G. J. (2017). Different types of diatom-derived extracellular polymeric substances drive changes in heterotrophic bacterial communities from intertidal sediments. Front. Microbiol. 8:245. doi: 10.3389/fmicb.2017.00245
Bruun, P. (1962). Sea-level rise as a cause of shore erosion. J. Waterways Harbors Division 88, 117–132. doi: 10.3390/antiox11020244
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13:581. doi: 10.1038/nmeth.3869
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Carter, P. W. (1978). Adsorption of amino acid-containing organic matter by calcite and quartz. Geochim. Cosmochim. Acta 42, 1239–1242. doi: 10.1016/0016-7037(78)90117-5
Chee, S. Y., Yee, J. C., Cheah, C. B., Evans, A. J., Firth, L. B., Hawkins, S. J., et al. (2021). Habitat complexity affects the structure but not the diversity of sessile communities on tropical coastal infrastructure. Front. Ecol. Evol. 9:673227. doi: 10.3389/fevo.2021.673227
Cockell, C. S., and Herrera, A. (2008). Why are some microorganisms boring? Trends Microbiol. 16, 101–106. doi: 10.1016/j.tim.2007.12.007
Cockell, C. S., Kelly, L. C., Summers, S., and Marteinsson, V. (2011). Following the kinetics: iron-oxidizing microbial mats in cold icelandic volcanic habitats and their rock-associated carbonaceous signature. Astrobiology 11, 679–694. doi: 10.1089/ast.2011.0606
Cole, M., Lindeque, P., Fileman, E., Halsband, C., and Galloway, T. S. (2015). The Impact of Polystyrene Microplastics on Feeding, Function and Fecundity in the Marine Copepod Calanus helgolandicus. Environ. Sci. Technol. 49, 1130–1137. doi: 10.1021/es504525u
Coombes, M. A., Naylor, L. A., Thompson, R. C., Roast, S. D., Gómez-Pujol, L., and Fairhurst, R. J. (2011). Colonization and weathering of engineering materials by marine microorganisms: an SEM study. Earth Surf. Process. Landf. 36, 582–593. doi: 10.1002/esp.2076
Dodds, K. C., Schaefer, N., Bishop, M. J., Nakagawa, S., Brooks, P. R., Knights, A. M., et al. (2022). Material type influences the abundance but not richness of colonising organisms on marine structures. J. Environ. Manag. 307:114549. doi: 10.1016/j.jenvman.2022.114549
Douglas, B. C. (1991). Global sea level rise. J. Geophys. Res. 96, 6981–6992. doi: 10.1029/91JC00064
Dugan, J., Airoldi, L., Chapman, M., Walker, S., Schlacher, T., Wolanski, E., et al. (2011). 8.02-Estuarine and coastal structures: environmental effects, a focus on shore and nearshore structures. Treat. Estuar. Coast. Sci. 8, 17–41. doi: 10.1016/B978-0-12-374711-2.00802-0
Encinas, N., Yang, C. Y., Geyer, F., Kaltbeitzel, A., Baumli, P., Reinholz, J., et al. (2020). Submicrometer-Sized Roughness Suppresses Bacteria Adhesion. ACS Appl. Mater. Interfaces 12, 21192–21200. doi: 10.1021/acsami.9b22621
Escalas, A., Hale, L., Voordeckers, J. W., Yang, Y., Firestone, M. K., Alvarez-Cohen, L., et al. (2019). Microbial functional diversity: from concepts to applications. Ecol. Evol. 9, 12000–12016. doi: 10.1002/ece3.5670
Feng, G., Cheng, Y., Wang, S. Y., Borca-Tasciuc, D. A., Worobo, R. W., and Moraru, C. I. (2015). Bacterial attachment and biofilm formation on surfaces are reduced by small-diameter nanoscale pores: how small is small enough? NPJ Biofilms Microbiomes 1:15022. doi: 10.1038/npjbiofilms.2015.22
Firth, L. B., Thompson, R. C., Bohn, K., Abbiati, M., Airoldi, L., Bouma, T. J., et al. (2014). Between a rock and a hard place: environmental and engineering considerations when designing coastal defence structures. Coast. Eng. 87, 122–135. doi: 10.1016/j.coastaleng.2013.10.015
Freckelton, M. L., Nedved, B. T., and Hadfield, M. G. (2017). Induction of Invertebrate Larval Settlement; Different Bacteria, Different Mechanisms? Sci. Rep. 7:42557. doi: 10.1038/srep42557
Freckelton, M. L., Nedved, B. T., Cai, Y. S., Cao, S., Turano, H., Alegado, R. A., et al. (2022). Bacterial lipopolysaccharide induces settlement and metamorphosis in a marine larva. Proc. Natl. Acad. Sci. U. S. A. 119:e2200795119. doi: 10.1073/pnas.2200795119
Golledge, N. R., Keller, E. D., Gomez, N., Naughten, K. A., Bernales, J., Trusel, L. D., et al. (2019). Global environmental consequences of twenty-first-century ice-sheet melt. Nature 566, 65–72. doi: 10.1038/s41586-019-0889-9
Green, D., Chapman, M., and Blockley, D. (2012). Ecological consequences of the type of rock used in the construction of artificial boulder-fields. Ecol. Eng. 46, 1–10. doi: 10.1016/j.ecoleng.2012.04.030
Gu, Z., Eils, R., and Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. doi: 10.1093/bioinformatics/btw313
Hadfield, M. G. (2011). Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Ann. Rev. Mar. Sci. 3, 453–470. doi: 10.1146/annurev-marine-120709-142753
Hartanto, R. S., Loke, L. H., Heery, E. C., Hsiung, A. R., Goh, M. W., Pek, Y. S., et al. (2022). Material type weakly affects algal colonisation but not macrofaunal community in an artificial intertidal habitat. Ecol. Eng. 176:106514. doi: 10.1016/j.ecoleng.2021.106514
Heatherington, C., and Bishop, M. J. (2012). Spatial variation in the structure of mangrove forests with respect to seawalls. Mar. Freshw. Res. 63, 926–933. doi: 10.1071/MF12119
Higgins, C. G. (1980). Nips, notches, and the solution of coastal limestone: an overview of the problem with examples from Greece. Estuar. Coast. Mar. Sci. 10, 15–30. doi: 10.1016/S0302-3524(80)80046-6
Hsiung, A. R., Tan, W. T., Loke, L. H., Firth, L. B., Heery, E. C., Ducker, J., et al. (2020). Little evidence that lowering the pH of concrete supports greater biodiversity on tropical and temperate seawalls. Mar. Ecol. Prog. Ser. 656, 193–205.
Huang, Y., Zheng, Y., Li, J., Liao, Q., Fu, Q., Xia, A., et al. (2018). Enhancing microalgae biofilm formation and growth by fabricating microgrooves onto the substrate surface. Bioresour. Technol. 261, 36–43. doi: 10.1016/j.biortech.2018.03.139
Huggett, M. J., Nedved, B. T., and Hadfield, M. G. (2009). Effects of initial surface wettability on biofilm formation and subsequent settlement of Hydroides elegans. Biofouling 25, 387–399. doi: 10.1080/08927010902823238
Kanehisa, M., and Sato, Y. (2019). KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 29, 28–35. doi: 10.1002/pro.3711
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Lai, S., Loke, L. H., Hilton, M. J., Bouma, T. J., and Todd, P. A. (2015). The effects of urbanisation on coastal habitats and the potential for ecological engineering: a Singapore case study. Ocean Coast. Manag. 103, 78–85. doi: 10.1016/j.ocecoaman.2014.11.006
Lam, N. W., Huang, R., and Chan, B. K. (2009). Variations in intertidal assemblages and zonation patterns between vertical artificial seawalls and natural rocky shores: a case study from Victoria Harbour, Hong Kong. Zool. Stud. 48, 184–195.
Lane, D. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (Chichester, UK: John Wiley and Sons), 115–175.
Lawrence, P. J., Evans, A. J., Jackson-Bué, T., Brooks, P. R., Crowe, T. P., Dozier, A. E., et al. (2021). Artificial shorelines lack natural structural complexity across scales. Proc. R. Soc. B 288:20210329. doi: 10.1098/rspb.2021.0329
Lema, K. A., Constancias, F., Rice, S. A., and Hadfield, M. G. (2019). High bacterial diversity in nearshore and oceanic biofilms and their influence on larval settlement by Hydroides elegans (Polychaeta). Environ. Microbiol. 21, 3472–3488. doi: 10.1111/1462-2920.14697
Loke, L. H., Heery, E. C., Lai, S., Bouma, T. J., and Todd, P. A. (2019). Area-independent effects of water-retaining features on intertidal biodiversity on eco-engineered seawalls in the tropics. Front. Mar. Sci. 6:16. doi: 10.3389/fmars.2019.00016
Loke, L. H., Ladle, R. J., Bouma, T. J., and Todd, P. A. (2015). Creating complex habitats for restoration and reconciliation. Ecol. Eng. 77, 307–313. doi: 10.1016/j.ecoleng.2015.01.037
Loman, N. (2020). MaxiKraken2_1903_140GB Mock Community Database. Available online at: https://lomanlab.github.io/mockcommunity/mc_databases.html (accessed April 2021).
Lu, N., Zhang, W., Weng, Y., Chen, X., Cheng, Y., and Zhou, P. (2016). Fabrication of PDMS surfaces with micro patterns and the effect of pattern sizes on bacteria adhesion. Food Control. 68, 344–351. doi: 10.1016/j.foodcont.2016.04.014
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Ming, C. L., Lionel, N. C. S., Jeremy, C. S. M., and Angie, S. L. (2010). Natural coral colonization of a marina seawall in Singapore. J. Coast. Dev. ISSN 14, 11–17.
Morris, R. L., Heery, E. C., Loke, L. H., Lau, E., Strain, E., Airoldi, L., et al. (2019). Design options, implementation issues and evaluating success of ecologically engineered shorelines. Oceanogr. Mar. Biol. 57, 169–228. doi: 10.1201/9780429026379-4
Murdock, J. N., and Dodds, W. K. (2007). Linking benthic algal biomass to stream substratum topography 1. J. Phycol. 43, 449–460. doi: 10.1111/j.1529-8817.2007.00357.x
Nicholls, R. J., and Cazenave, A. (2010). Sea-level rise and its impact on coastal zones. Science 328, 1517–1520. doi: 10.1126/science.1185782
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’hara, R., et al. (2011). Vegan: Community Ecology Package. R Package Version. Available online at: http://CRAN.Rproject.org/package=vegan (accessed April 2020).
Petersen, J. M., and Dubilier, N. (2009). Methanotrophic symbioses in marine invertebrates. Environ. Microbiol. Rep. 1, 319–335. doi: 10.1111/j.1758-2229.2009.00081.x
R Core Team (2014). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rädecker, N., Pogoreutz, C., Voolstra, C. R., Wiedenmann, J., and Wild, C. (2015). Nitrogen cycling in corals: the key to understanding holobiont functioning? Trends Microbiol. 23, 490–497. doi: 10.1016/j.tim.2015.03.008
Revelle, R., and Emery, K. O. (1957). Chemical Erosion of Beach Rock and Exposed Reef Rock: Bikini and Nearby Atolls. Marshall Islands: US Government Printing Office.
Schneck, F., Schwarzbold, A., and Melo, A. S. (2011). Substrate roughness affects stream benthic algal diversity, assemblage composition, and nestedness. J. North Am. Benthol. Soc. 30, 1049–1056. doi: 10.1899/11-044.1
Shannon, C. E. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Simkanin, C., Davidson, I. C., Dower, J. F., Jamieson, G., and Therriault, T. W. (2012). Anthropogenic structures and the infiltration of natural benthos by invasive ascidians. Mar. Ecol. 33, 499–511. doi: 10.1111/j.1439-0485.2012.00516.x
Sneed, J. M., Ritson-Williams, R., and Paul, V. J. (2015). Crustose coralline algal species host distinct bacterial assemblages on their surfaces. ISME J. 9, 2527–2536. doi: 10.1038/ismej.2015.67
Strain, E. M., Steinberg, P. D., Vozzo, M., Johnston, E. L., Abbiati, M., Aguilera, M. A., et al. (2021). A global analysis of complexity–biodiversity relationships on marine artificial structures. Glob. Ecol. Biogeogr. 30, 140–153. doi: 10.1111/geb.13202
Suess, E. (1970). Interaction of organic compounds with calcium carbonate—I. Association phenomena and geochemical implications. Geochim. Cosmochim. Acta 34, 157–168. doi: 10.1016/0016-7037(70)90003-7
Summers, S., Thomson, B. C., Whiteley, A. S., and Cockell, C. S. (2016). Mesophilic mineral-weathering bacteria inhabit the critical-zone of a perennially cold basaltic environment. Geomicrobiol. J. 33, 52–62. doi: 10.1080/01490451.2015.1039672
Sweat, L. H., and Johnson, K. B. (2013). The effects of fine-scale substratum roughness on diatom community structure in estuarine biofilms. Biofouling 29, 879–890. doi: 10.1080/08927014.2013.811492
Tan, E. L. Y., Mayer-Pinto, M., Johnston, E. L., and Dafforn, K. A. (2015). Differences in Intertidal Microbial Assemblages on Urban Structures and Natural Rocky Reef. Front. Microbiol. 6:1276. doi: 10.3389/fmicb.2015.01276
Temmerman, S., Meire, P., Bouma, T. J., Herman, P. M., Ysebaert, T., and De Vriend, H. J. (2013). Ecosystem-based coastal defence in the face of global change. Nature 504, 79–83. doi: 10.1038/nature12859
Todd, P. A., Heery, E. C., Loke, L. H., Thurstan, R. H., Kotze, D. J., and Swan, C. (2019). Towards an urban marine ecology: characterizing the drivers, patterns and processes of marine ecosystems in coastal cities. Oikos 128, 1215–1242. doi: 10.1111/oik.05946
Tukey, J. W. (1949). Comparing individual means in the analysis of variance. Biometrics 5, 99–114. doi: 10.2307/3001913
Warrick, R., and Oerlemans, J. (1990). “Sea Level Rise,” in Climate Change: The IPCC Scientific Assessment, eds J. T. Houghton, G. J. Jenkins, and J. J. Ephraums (Sweden: Intergovernmental Panel on Climate Change), 257–281.
Wickham, H. (2016). Ggplot2: Elegant Graphics for Data Analysis. New York: Springer. doi: 10.1007/978-3-319-24277-4
Wyngaard, G., Mclaren, I., White, M., and Sévigny, J.-M. (1995). Unusually high numbers of ribosomal RNA genes in copepods (Arthropoda: Crustacea) and their relationship to genome size. Genome 38, 97–104. doi: 10.1139/g95-012
Yamashita, H. (2020). Living together with seawalls: risks and reflexive modernization in Japan. Environ. Sociol. 6, 166–181. doi: 10.1080/23251042.2019.1709680
Keywords: seawall, biofilm, microbial diversity, marine microbes, coastal protection
Citation: Summers S, Pek YS, Vinod DP, McDougald D, Todd PA, Birch WR and Rice SA (2022) Bacterial biofilm colonization and succession in tropical marine waters are similar across different types of stone materials used in seawall construction. Front. Microbiol. 13:928877. doi: 10.3389/fmicb.2022.928877
Received: 26 April 2022; Accepted: 07 July 2022;
Published: 25 July 2022.
Edited by:
Hongbin Liu, Hong Kong University of Science and Technology, Hong Kong SAR, ChinaReviewed by:
Weipeng Zhang, Ocean University of China, ChinaRashmi Rathour, Jawaharlal Nehru University, India
Copyright © 2022 Summers, Pek, Vinod, McDougald, Todd, Birch and Rice. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen Summers, c3N1bW1lcnNAbnR1LmVkdS5zZw==; William R. Birch, dy1iaXJjaEBpbXJlLmEtc3Rhci5lZHUuc2c=
 Stephen Summers
Stephen Summers Y. Shona Pek
Y. Shona Pek Deepthi P. Vinod
Deepthi P. Vinod Diane McDougald
Diane McDougald Peter A. Todd
Peter A. Todd William R. Birch
William R. Birch Scott A. Rice
Scott A. Rice