
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 17 June 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.926710
Probiotics contribute a lot to human health and the occurrence of diseases. Correspondingly, probiotics’ safety evaluation and probiotic properties have received increasing attention in the food industry and disease treatment. Clostridium tyrobutyricum L319 is a short-chain fatty acid (SCFA)-producing strain isolated from Grana Padano cheese with a blowing defect. Our previous study has shown its safety at the genomic level. This study focused more on the safety evaluation and probiotic properties in vitro. According to the results, this strain has no potential virulence factors or the possibility of antibiotic resistance genes propagation. It also fulfilled several criteria to be used as a probiotic, including significant hydrophobicity under an acidic condition (pH 5.0) and resistance to simulate gastric juice and intestinal juice. Additionally, this strain was found to be tolerant to the harsh conditions of the external environment, including resistance to low (20°C) and high (50°C) temperatures, high salts (3% NaCl), and low pH (pH 5.0). Finally, we found that this strain could ferment prebiotics, such as chito-oligosaccharides, to produce SCFAs. It exhibited excellent growth performance whether using chito-oligosaccharide as a sole carbon source or combining glucose as the mixed carbon source. Furthermore, chito-oligosaccharide and glucose (1:1) mixed carbon sources were the optimal strategy for the production of SCFAs. Our findings demonstrated that this strain might be considered a promising candidate for future use as a probiotic to promote health benefits.
The gut microbiota is rich in diversity, including bacteria, viruses, fungi, and protozoa (Joukar et al., 2020). Among them, the number of bacteria alone is as high as 100 trillion; therefore, the gut microbiome is also called the second genome of human beings (Zhu et al., 2010). With the development of metagenomic sequencing and bioinformatics technology, various studies have shown an inseparable relationship between gut microbes and the host (Shreiner et al., 2015). The host’s balance of gut microbiota is mainly regulated through diet (Valdes et al., 2018). The gut microbiota obtains energy from the undecomposed substances in the gut for their growth and participates in the substance metabolism, nutrient absorption, and physiological and immune defense processes (Elzinga et al., 2019). Moreover, prebiotics is a class of substrates that can be selectively utilized by gut microbiota and are beneficial to health, including fructo-oligosaccharides, galacto-oligosaccharides, and inulin (Sonnenburg and Sonnenburg, 2019). They could enrich specific microorganisms and produce metabolites, such as short-chain fatty acids (SCFAs), that could contribute to human health (Bindels et al., 2015).
Short-chain fatty acids are the main products of intestinal microbial fermentation. Intestinal bacteria could produce SCFAs by using dietary fibers as substrates. The glycoside hydrolase produced by bacteria could convert dietary fibers into monosaccharides and then generate the main end-product SCFAs through anaerobic fermentation (Xu et al., 2020). SCFAs are a group of carboxylic acids containing six or fewer carbon molecules, particularly acetate, propionate, and butyrate. Approximately, 500–600 mmol of SCFAs in the gastrointestinal tract are produced per day, depending on dietary fiber content (Blaak et al., 2020). Moreover, acetate, propionate, and butyrate were produced in an overall molar proportion of ∼60:20:20, respectively, representing 95% of SCFAs in the human colon (Zhu et al., 2022). Numerous studies have shown that SCFAs are essential for maintaining the metabolic function of the host. They could provide energy for intestinal epithelial cells, protect the intestinal barrier, and regulate intestinal inflammation (Hu et al., 2018). Furthermore, among the SCFAs, butyrate has been investigated extensively and is the primary energy source for colon cells (Wang et al., 2020). It was found that the gut butyrate-producing bacteria mainly belong to phylum Firmicutes and can be further subdivided into the genus Clostridium (Xu et al., 2020).
At present, many studies are focusing on the health effects of Clostridium and its metabolite SCFAs. A mixture of human gut-derived 17 Clostridium strains was applied in a previous study to investigate the effects of preventing and treating multiple sclerosis and reveal the mediating role of their metabolite butyrate (Calvo-Barreiro et al., 2021). Also, Clostridium butyricum, a butyrate-producing probiotic, was found to increase SCFA levels, thereby inhibiting high-fat diet-induced intestinal tumor formation (Chen D. et al., 2020). Given the importance of Clostridium, it is necessary to evaluate its safety and probiotic properties. By now, only C. butyricum CBM588 was authorized as a novel probiotic for use by humans and food animals according to Regulation (EC) No. 258/97 (Isa et al., 2016). In addition, a novel Faecalibacterium prausnitzii strain was validated in vitro to identify its potential as a next-generation probiotic (Martín et al., 2017). Our previous study found that C. tyrobutyricum, a Gram-positive, acidogenic, anaerobic bacterium, could produce butyrate as the main fermentation product and acetate as co-products. It is the best strain reported for butyrate production (Jiang et al., 2018).
Meanwhile, C. tyrobutyricum can be used in the treatment and/or prevention of certain human and animal diseases. Recent studies have revealed that C. tyrobutyricum ATCC25755 could protect against lipopolysaccharide (LPS)-induced epithelial dysfunction and inflammation (Xiao et al., 2021b,c). Also, as a potential probiotic, it could alleviate Staphylococcus aureus-induced endometritis in mice by inhibiting endometrial barrier disruption and inflammatory response (Hu et al., 2019).
At present, there is an increasing interest in applying gut microbes as potential probiotic agents (Zhang et al., 2021). Due to safety concerns, lack of safety information, and legislation, only a limited number of Clostridium species are commercialized. Although little is known about the safety of C. tyrobutyricum, there is a clear promise for this species as a next-generation probiotic. Our previous study conducted a genome sequence analysis of C. tyrobutyricum L319 and evaluated its safety at the genomic level. The results indicated that this strain does not possess transferable resistance genes, invasive defensive pathogenicity factors, or harmful enzymes (Liu et al., 2020). In this study, as recommended (FAO/WHO) (Araya et al., 2002), phenotypic testing, prebiotics utilization, metabolite production, biochemical activity, and toxicity testing, in combination with genetic analysis, were performed to validate whether C. tyrobutyricum L319 is safe for use as a next-generation probiotic in the food industry and gut therapy.
Clostridium tyrobutyricum L319 used in this study was previously isolated from Grana Padano cheese with a blowing defect (Shanghai Rongyue Trading Co., Ltd., Shanghai, China). The genome was previously isolated drafted and analyzed in our previous study (Liu et al., 2020). C. tyrobutyricum was cultured at 37°C in reinforced clostridial medium (RCM), containing (per liter) 10 g tryptone, 10 g beef extract, 5 g glucose, 5 g NaCl, 3 g yeast extract, 3 g anhydrous sodium acetate, 1 g soluble starch, and 0.5 g L-cysteine, in serum bottles with a final concentration of 0.05% as an oxygen indicator under strictly anaerobic condition. The fermentation was carried out in a TGY medium containing (per liter) 30 g tryptone, 20 g glucose, 10 g yeast extract, and 1.3 g L-cysteine. In addition, the minimal medium used to evaluate the effects of different prebiotics containing (per liter) 1 g NH4Cl, 0.1 g KCl, 0.2 g MgSO4⋅7H2O, 0.8 g NaCl, 0.1 g KH2PO4, 20 mg CaCl2⋅2H2O, trace elements (per liter: 0.10 g ZnSO4⋅7H2O, 0.03 g MnCl2⋅4H2O, 0.30 g H3BO3, 0.20 g NiCl2⋅6H2O, 0.01 g CuCl22H2O, 0.02 g NiCl2⋅6H2O, 0.03 mg Na2MoO4⋅2H2O), and Wolfe’s vitamin solution (per liter: 20 mg biotin, 20 mg folate, 100 mg pyridoxine hydrochloride, 50 mg thiamine hydrochloride, 50 mg riboflavin, 50 mg nicotinic acid, 50 mg calcium pantothenate, 1 mg vitamin B12, 50 mg 4-aminobenzoic acid, 50 mg lipoic acid). All media were sterilized by autoclaving at 108°C for 30 min.
The neighbor-joining phylogenetic tree was constructed based on 16S rRNA gene sequences of the representative C. tyrobutyricum and its neighbors, C. butyricum. The average nucleotide identity (ANI) was calculated using OrthoANI (Lee et al., 2016). The evolutionary analysis of gene synteny and collinearity was conducted with MCScanX in the TBtools box (Wang et al., 2012; Chen C. et al., 2020).
Antibiotic resistance testing was performed using the minimum inhibitory concentration (MIC) method described in a previous study (Baccouri et al., 2019). The tested antibiotics were gentamicin, kanamycin, streptomycin, clindamycin, chloramphenicol, tetracycline, vancomycin, erythromycin, neomycin, ampicillin, linezolid, trimethoprim, rifampicin, and ciprofloxacin. The overnight cultures of bacteria were diluted with RCM medium to 105–106 CFU ml–1. Then, the antibiotic solutions and bacterial suspension were added to a 96-well plate from low to high concentrations, according to Supplementary Table 1. In addition, the one that contains the RCM medium and the bacterial suspension was considered the positive control. In contrast, others with RCM medium and phosphate-buffered saline (PBS) were negative control. MIC was determined by measuring optical density at 600 nm (OD600) with a microplate reader (SpectraMax, M2). The test was performed in triplicates. The MIC breakpoint is the concentration where no growth is exhibited. The sensitivity/resistance was confirmed regarding the microbiological cutoff values prescribed by the European Food Safety Authority (EFSA) in 2012 guidelines (ESFA, 2012). Strains exhibiting MIC values less than the breakpoints were considered sensitive. Otherwise, they were resistant.
According to the NCBI database, the neurotoxin genes of types A, B, E, and F from Clostridium botulinum [GenBank were A (EU429475.1), B (M81186.1), E (AB082519.1), F (M92906.1)] and alpha, beta, epsilon, iota from C. perfringens [alpha (L43546.1), beta (X83275.1), and epsilon (M95206.1), iota (X73562.1)] were selected to be detected on the genome of C. tyrobutyricum L319 by PCR reactions. Genes coding amino acid decarboxylases, HDC1 and HDC2 (both related to histidine decarboxylase), tyrosine decarboxylase (TDC), ornithine decarboxylase (ODC), and agmatine deaminase (AGDI) were also detected. The amplified products were separated by electrophoresis on 2% (w/v) agarose gels in 0.5 × TAE buffer. Gels were stained in TAE buffer containing 0.5 mg⋅ml–1 ethidium bromide. All the primers used are listed in Supplementary Table 2.
The overnight cultures of bacteria were diluted to 10–6 CFU with a 10-fold gradient of sterile PBS, and 100 μl of the bacterial suspension diluted to 10–6 was coated on the blood AGAR plate. Two parallel experiments were performed after anaerobic culture for 48 h, and two parallel experiments were performed to observe the hemolysis rings around the bacterial colonies on the blood agar plate.
To evaluate cell surface hydrophobicity at different pH values, the strain was collected after overnight cultivation by centrifugation (6,000 × g for 5 min at 4°C), washed twice with PBS (50 mM, pH 5, pH 6, pH 7) and then resuspended in the same buffer that was adjusted until A560 values (A0) around 1. In addition, N-hexadecane was then added five times the volume to the cell suspension (1:5) and adequately mixed for 120 s. The aqueous layer’s A560 value (A) was measured after 1 h at 37°C. Then, the hydrophobicity was calculated by the equation: %H = [(A0 − A)/A0] × 100.
The simulated intestinal juice contains 1% (w/v) pancreatin, 0.3% (w/v) bile salt, and was adjusted to pH 6.8, while the simulated gastric juice contains 3% (w/v) pepsin, 2% (w/v) NaCl, and was adjusted to pH 3.0. The strain was collected with the same method in hydrophobicity assay and 100 μl of the bacterial suspension was diluted to 10–6 coated on the RCM agar plate, considered as CFU0. Then, the remaining strains were divided into two groups (25 ml for each), separately growing in the intestinal juice for 4 h and gastric juice for 2 h (100 r/min, 37°C) with strains growing in PBS as control. The above steps were repeated to coat the bacterial suspension on the RCM agar plates. The colonies on the plates were counted after growing on the plates for 24–48 h, the CFUs were calculated, and the tolerance was evaluated by the equation: %T = [log10CFU/ml]/[log10CFU0/ml] × 100.
To assess the robustness of C. tyrobutyricum L319 at several temperatures, the overnight cultures were inoculated in 30 ml fresh RCM medium at 1% (v/v) with different temperatures (4, 20, 30, 37, 40, 45, and 50°C); NaCl (0, 1, 2, 3, and 4%); and pH (4.5, 5.0, and 6.5). The growth profiles were measured at OD600 at intervals. The expected growth conditions at 37°C, pH 6.5, and 0% NaCl were set as the control group. The growth experiments were performed in triplicate.
The overnight cultures were transferred to a 50 ml fresh TGY medium with a volume ratio of 5% at pH 7.0 and 5.0, respectively. The medium at pH 5.0 was adjusted with butyrate. The cell growth, butyrate, and acetate production were determined.
A total of eight prebiotics, including fructo-oligosaccharide, chitosan oligosaccharide, galacto-oligosaccharide, xylo-oligosaccharide, isomalto-oligosaccharide, inulin, raffinose, and lactitol, were used to determine the growth profiling of C. tyrobutyricum L319. Each prebiotic was the same C-mole as glucose in a minimal medium. The control group was the minimal medium adding 10 g⋅L–1 glucose. The other components in the medium were the same as the minimal medium. Strains were cultivated in serum bottles, while growth performances were determined at OD600. In addition, to investigate the growth profiling of C. tyrobutyricum L319 with glucose and prebiotics as mixed carbon sources, every prebiotic was the same C-mole as glucose (5 g⋅L–1 glucose) in the minimal medium that also contained 5 g⋅L–1 glucose. Strains were also cultivated in serum bottles, and the growth performances were determined at OD600. Lastly, the different ratios of chitosan oligosaccharide and glucose (1:1; 1:2; 1:3; 2:1, 3:1 C-mole) were added into the minimal medium, and the growth performances were determined.
Cell growth was monitored by measuring the optical density at 600 nm using a spectrophotometer (Shanghai Spectrum, China). The glucose, butyrate, and acetate concentrations were assessed using high-performance liquid chromatography (HPLC, LC-15C; Shimadzu) equipped with an organic acid analysis column (HPX-87H, Bio-Rad) at 65°C and with 0.005 M sulfuric acid as the mobile phase at 0.6 ml⋅min–1. HPLC also analyzed chito-oligosaccharide with a Carbonmix Ca-NP column (Sepax Technologies, Suzhou, China) kept at 85°C. Water was also used as the mobile phase with a 0.6 ml⋅min–1 flow rate.
The C. tyrobutyricum strain was isolated from Grana Padano cheese with a blowing defect. Moreover, C. butyricum was authorized as a novel ingredient according to Regulation (EC) No. 258/97. Therefore, in this study, we investigated the similarity of C. tyrobutyricum and the reference probiotic strain C. butyricum. The colony shapes of these two strains were relatively close, and it is not easy to find a significant difference (Figure 1A). Besides identifying colonies, a phylogenetic tree was constructed based on the 16S rRNA sequences from evolutionary distances by the neighbor-joining method. Analysis of the phylogenetic tree depicted the hierarchical relationship between the four strains and suggested that C. tyrobutyricum L319 was closely related to the potential probiotic C. tyrobutyricum ATCC25755 (Figure 1B). Whole-genome analysis using the ANI also exhibited that C. tyrobutyricum L319 is closer to C. tyrobutyricum ATCC25755 (ANI > 95%) in terms of genomic distance (Figure 1C). Finally, the overall genomic differences between C. tyrobutyricum L319 and C. tyrobutyricum ATCC25755 were analyzed by aligning the two genomes using MCScanX. The results suggested a high level of conservation along the chromosome and plasmid. Indeed, these two genomes were found to be organized in a very similar way (Figure 1D). These results indicated that C. tyrobutyricum L319 has a certain identity with the reference probiotic C. butyricum DSM10702 and has a high degree of homology with the potential probiotic C. tyrobutyricum ATCC25755 at the genome level.
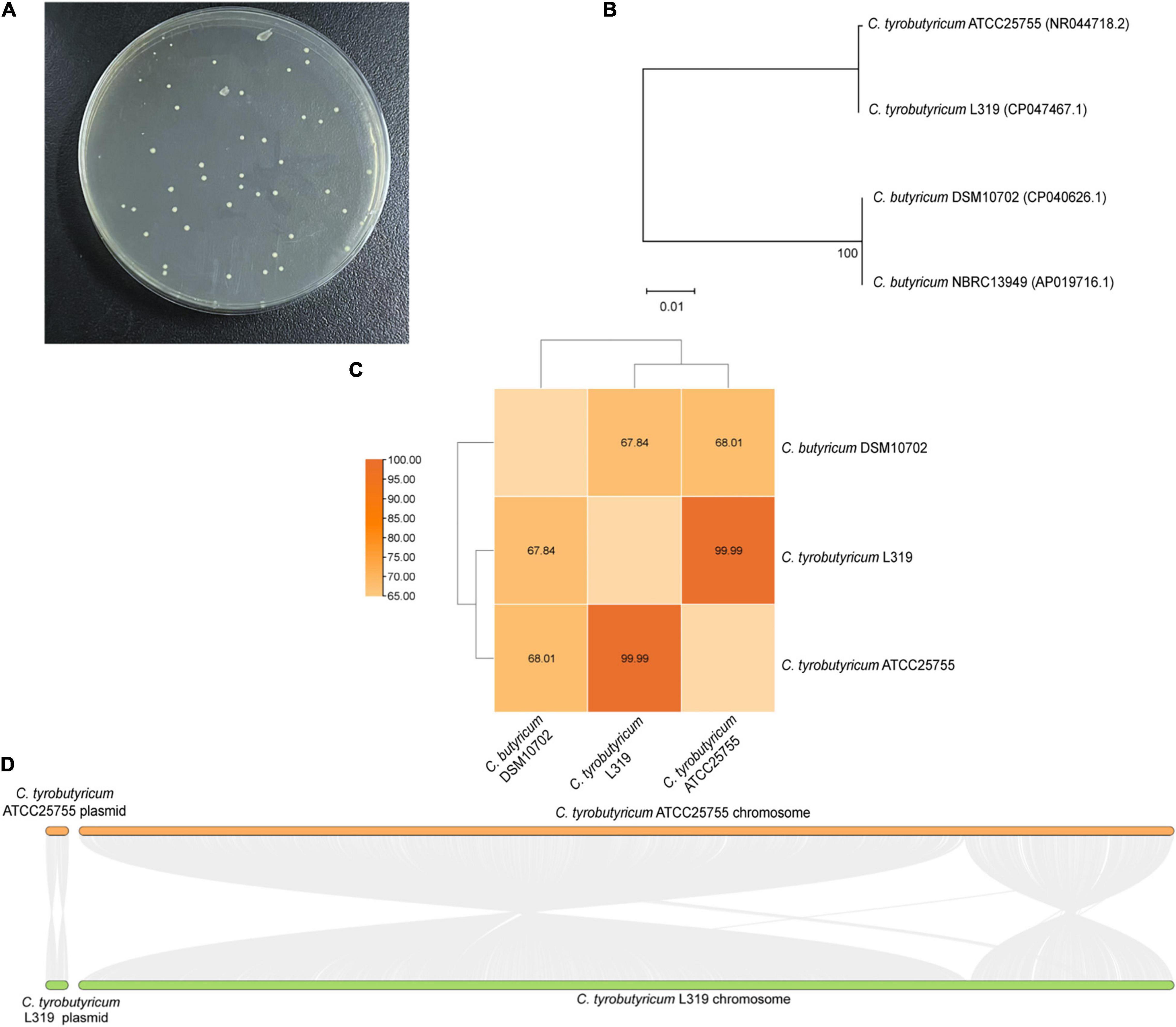
Figure 1. (A) The growth of C. tyrobutyricum L319 on RCM plates. (B) The neighbor-joining phylogenetic tree based on the 16S rRNA gene sequences. (C) Heatmap of ANI between C. tyrobutyricum L319, C. tyrobutyricum ATCC 25755, and C. butyricum DSM10702. ANI values (%) are indicated. (D) Pairwise alignment between the reference strain C. tyrobutyricum ATCC 25755 and the related strain C. tyrobutyricum L319.
The antibiotic resistance of C. tyrobutyricum L319 against 14 tested antibiotics is shown in Table 1. Generally, strains were considered resistant when the MIC values were higher than breakpoint values according to the EFSA. Overall, this strain showed the ability to resist the impact of aminoglycoside antibiotics (gentamicin, kanamycin, and streptomycin), clindamycin, and chloramphenicol. In contrast, this strain was susceptible to tetracycline, vancomycin, and erythromycin, which agrees with the antibiotic resistance analysis for C. butyricum CBM5881 (Isa et al., 2016). In addition, the breakpoints of other antibiotics, including neomycin, ampicillin, linezolid, trimethoprim, and rifampicin, were not established by the EFSA, the strain was also considered susceptible to neomycin (MIC = 0.5 μg⋅ml–1), ampicillin (MIC = 0.032 μg⋅ml–1), linezolid (MIC = 0.032 μg⋅ml–1), trimethoprim (MIC = 0.125 μg⋅ml–1), rifampicin (MIC = 0.125 μg⋅ml–1), and ciprofloxacin (MIC = 8 μg⋅ml–1) according to the results for antibiotic susceptibility of C. butyricum CBM5881 (Isa et al., 2016).
Identifying virulence genes in C. tyrobutyricum L319 by molecular and phenotypic procedures is necessary to avoid the risk of dissemination of virulence genes to other bacteria by genetic transfer. In our previous study, the genome of C. tyrobutyricum L319 was analyzed to determine the presence of neurotoxin genes (types A, B, E, and F) and toxin genes (alpha, beta, epsilon, and iota). Meanwhile, genes associated with biogenic amine biosynthesis, such as HDC1 and HDC2, TDC, ODC, and AGDI were also analyzed. Fortunately, these genes were not found in C. tyrobutyricum L319 by whole-genome sequencing (Liu et al., 2020). We checked these genes by PCR assays in this study, and the 16S rRNA gene was amplified as the control group (Supplementary Figure 1). The results showed that the neurotoxin and toxin genes were not detected in C. tyrobutyricum L319, and none of the genes associated with biogenic amine biosynthesis were detected.
To develop new beneficial microbes, their absence of hemolysis capacity should be evaluated. As mentioned above, hemolytic activity has been determined on the blood AGAR plate. Observation of the blood AGAR plates showed no α-hemolysis and β-hemolysis activities, while the strain showed γ-hemolysis activity, known as negative or no hemolysis activity (Figure 2A).
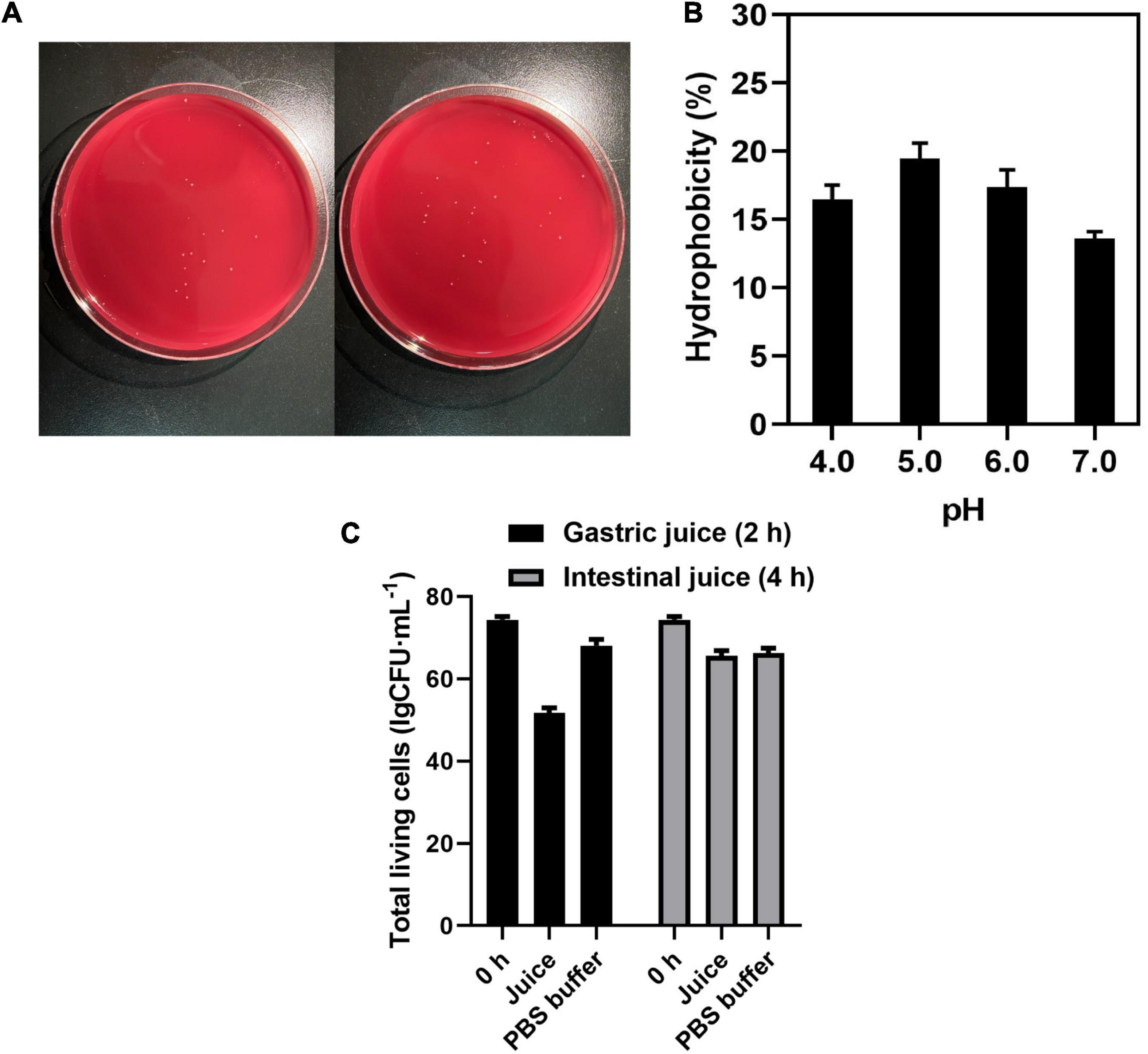
Figure 2. (A) Hemolysis of C. tyrobutyricum L319 on blood plate. (B) Hydrophobicity assay of C. tyrobutyricum L319 at pH 4.0, 5.0, 6.0, and 7.0. (C) The total living cells survive in the gastric juice and intestinal juice. Controls all refer to the strains suspended in PBS.
Cell surface hydrophobicity is a beneficial attribute for probiotics, and it is a non-specific interaction between microbial cells and the host. Therefore, the affinity of C. tyrobutyricum L319 toward the hydrophobic solvent N-hexadecane was investigated. The results of this experiment showed less than 20% hydrophobicity. However, it could be found that the strain presented the highest hydrophobicity at pH 5.0 (19.13%) when compared with that at pH 4.0 (16.23%), 6.0 (17.53%), and 7.0 (13.14%) (Figure 2B). It could be indicated that the strain exhibited better hydrophobicity under the pH conditions between 5.0 and 6.0.
The tolerance toward simulated intestinal juice and gastric juice is the essential characteristic for the strain to colonize the intestine. Our study indicated that C. tyrobutyricum L319 has a promising tolerance to gastric juice and intestinal juice with a survival ratio of 69.67% (2 h) and 88.27% (4 h) (Figure 2C), compared with other potential probiotic products (<56%) (Supplementary Table 3).
Probiotics are inevitably faced with various stresses from the external environment in the fermentation products and the human gastrointestinal tract, such as heat stress caused by high temperature and osmotic stress caused by water loss during the spray drying process of bacterial agents. Also, probiotics suffer gastric acid stress in the gastrointestinal tract. This stress seriously affected the growth, metabolism, and physiological functions of probiotics. Therefore, C. tyrobutyricum L319 was tested for its ability to survive in acidic conditions, low- and high-temperature conditions, and in the presence of NaCl. It could be found that C. tyrobutyricum L319 had different tolerance in different environments in this study (Figure 3). The growth profiling of C. tyrobutyricum L319 was slightly affected by temperature, and C. tyrobutyricum L319 displayed excellent tolerance to low and high temperatures, ranging from 20 to 50°C (Figure 3A). The maximum cell density at 30°C was 5.6; however, the lag period was longer than 37 and 40°C. The optimum growth temperature of 37°C was taken as the dividing line. With the culture temperature rising and falling, the maximum biomass was lower, and the lag period was longer. In addition, it can be found that the strain could not grow at 4°C but could still grow at 50°C, indicating that the strain has excellent tolerance to high temperatures.
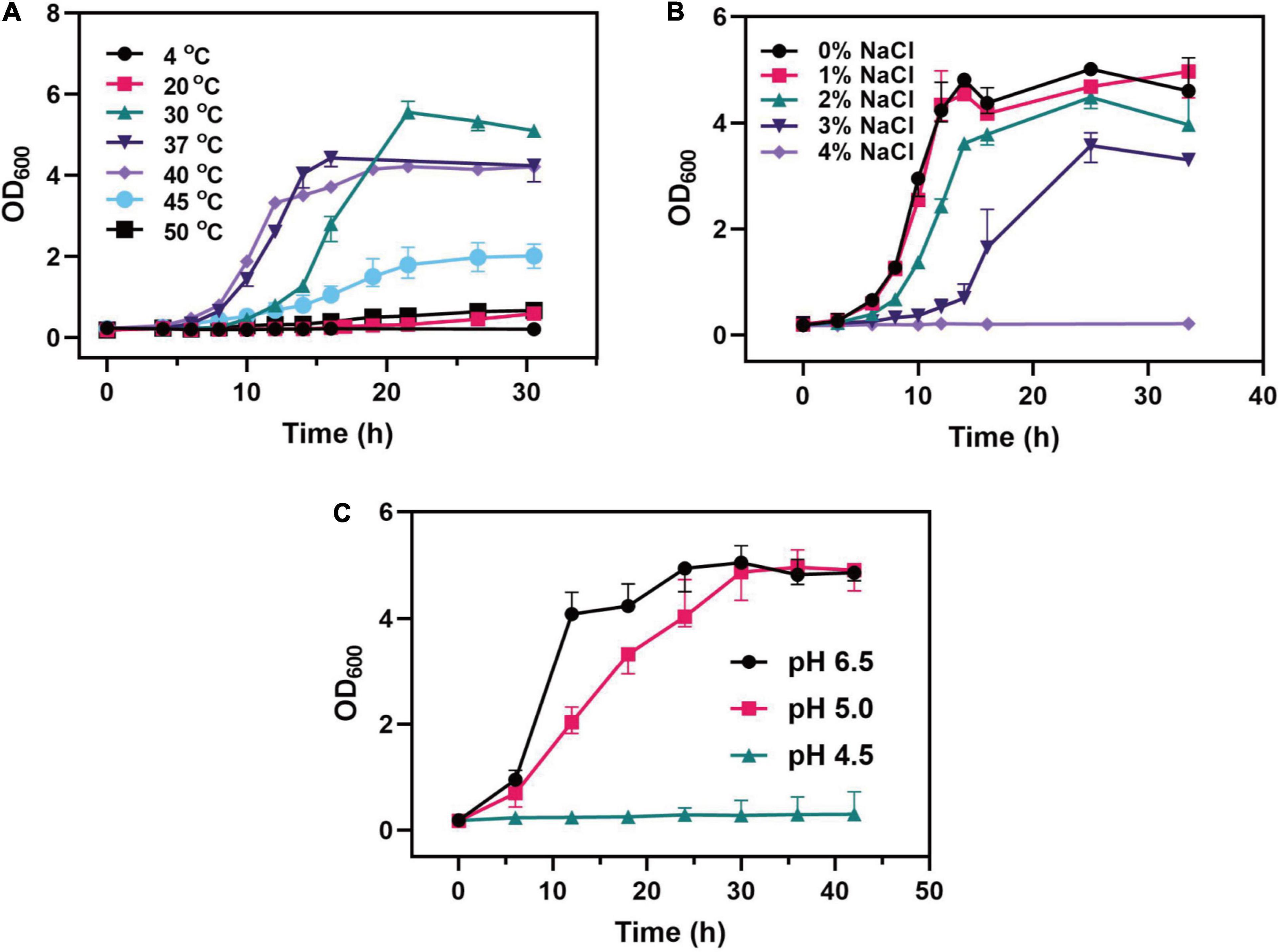
Figure 3. Effects of (A) different temperatures, (B) NaCl concentrations, and (C) pH values on the growth of the C. tyrobutyricum L319 strain.
Regarding the tolerance toward the osmotic pressure, C. tyrobutyricum L319 could survive under the 3% NaCl condition, and the maximum cell density reached about 31.5% of the density in the 0% NaCl condition. It was found that no significant variability was exhibited under the 1% NaCl condition. However, the strain could not grow under the 4% NaCl condition (Figure 3B). The viability of C. tyrobutyricum L319 was also assessed under the acidic condition, incubated in RCM broth supplemented with butyrate. Our findings indicated that the strain exhibited high tolerance to acidity after exposure to media pH 5.0. The maximum OD600 of C. tyrobutyricum L319 at pH 5.0 was close to the control at pH 6.5 (OD600max: 4.99 vs. 5.16). However, it exhibited no growth at pH 4.5.
In general, C. tyrobutyricum has already been considered a promising microbial host for the production of SCFAs, especially butyrate. Due to its unique butyrate synthetic pathway, C. tyrobutyricum mainly produces butyrate and acetate. Therefore, the batch fermentation results showed that the titer of butyrate was 6.29 g⋅L–1, when fed with 20 g⋅L–1 glucose, the yield was considerably achieved at 0.31 g⋅g–1. Furthermore, a much lower acetate titer was detected than butyrate (0.85 vs. 6.29 g⋅L–1) with a yield of 0.04 g⋅g–1 (Figure 4A). In addition, the fermentation of SCFAs at pH 5.0, which was close to the intestinal environment, was also investigated. Interestingly, the titer of butyrate was not significantly decreased (6.16 vs. 6.29 g⋅L–1) (Figure 4B). However, the proportion of butyrate/acetate came to 11, higher than that at pH 6.5 condition (BA/AA = 7.4). The results indicated that C. tyrobutyricum L319 has a great potential to be applied in the production of SCFA, which has already been recognized as a major product of intestinal microbial fermentation with beneficial effects on human health.
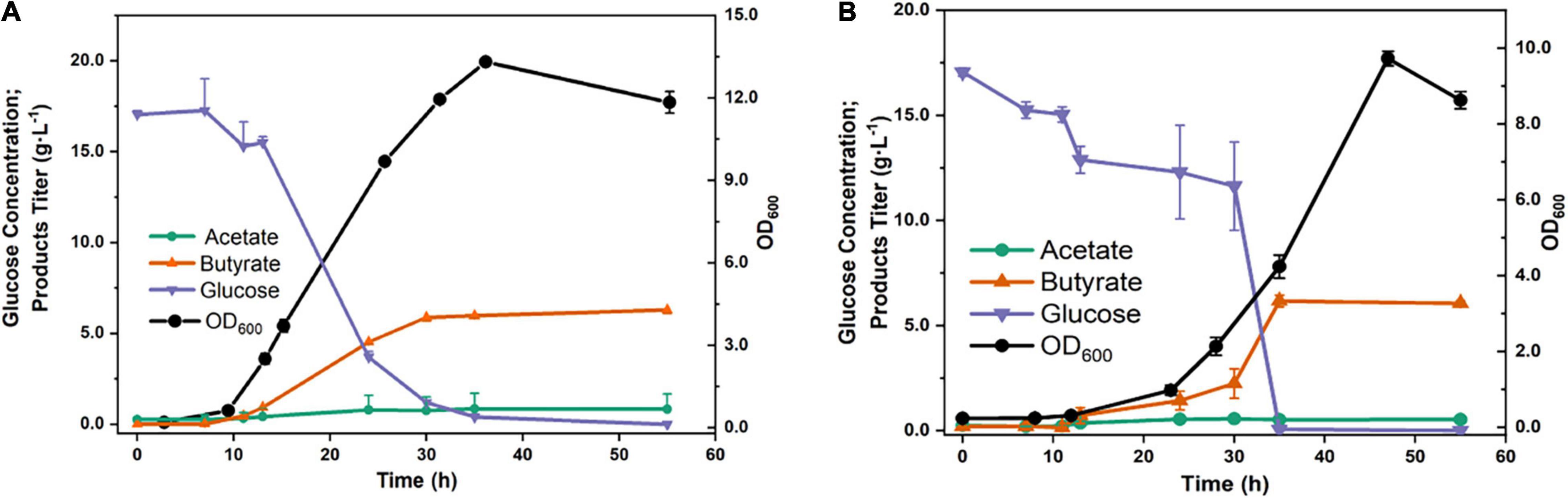
Figure 4. Fermentation products of strain under natural (pH 6.5) (A) and lower (pH 5.0) (B) pH values.
When the eight prebiotics were used separately as the sole carbon source, most of the strains did not exhibit growth compared with glucose as control, except for those media using galacto-oligosaccharide (OD600max: 1.29 vs. 2.05), xylo-oligosaccharide (OD600max: 1.04 vs. 2.05), and chito-oligosaccharide (OD600max: 1.26 vs. 2.05) (Figure 5A). Also, the strains growing in galacto-oligosaccharide showed a similar lag phase to those in glucose, while the strains in xylo-oligosaccharide and chito-oligosaccharide spent about four times longer adjusting to the conditions. In addition, prebiotics was also detected as complimentary carbon source separately added to an optimized medium containing 10 g⋅L–1 glucose with the same C-mole volume. The results (Figure 5B) showed that the strains growing in mixed carbon sources of glucose and chito-oligosaccharide exhibited outstanding biomass, even compared with glucose (3.88 vs. 1.94) as the sole carbon source. However, there are no significant differences found in other optimized media.
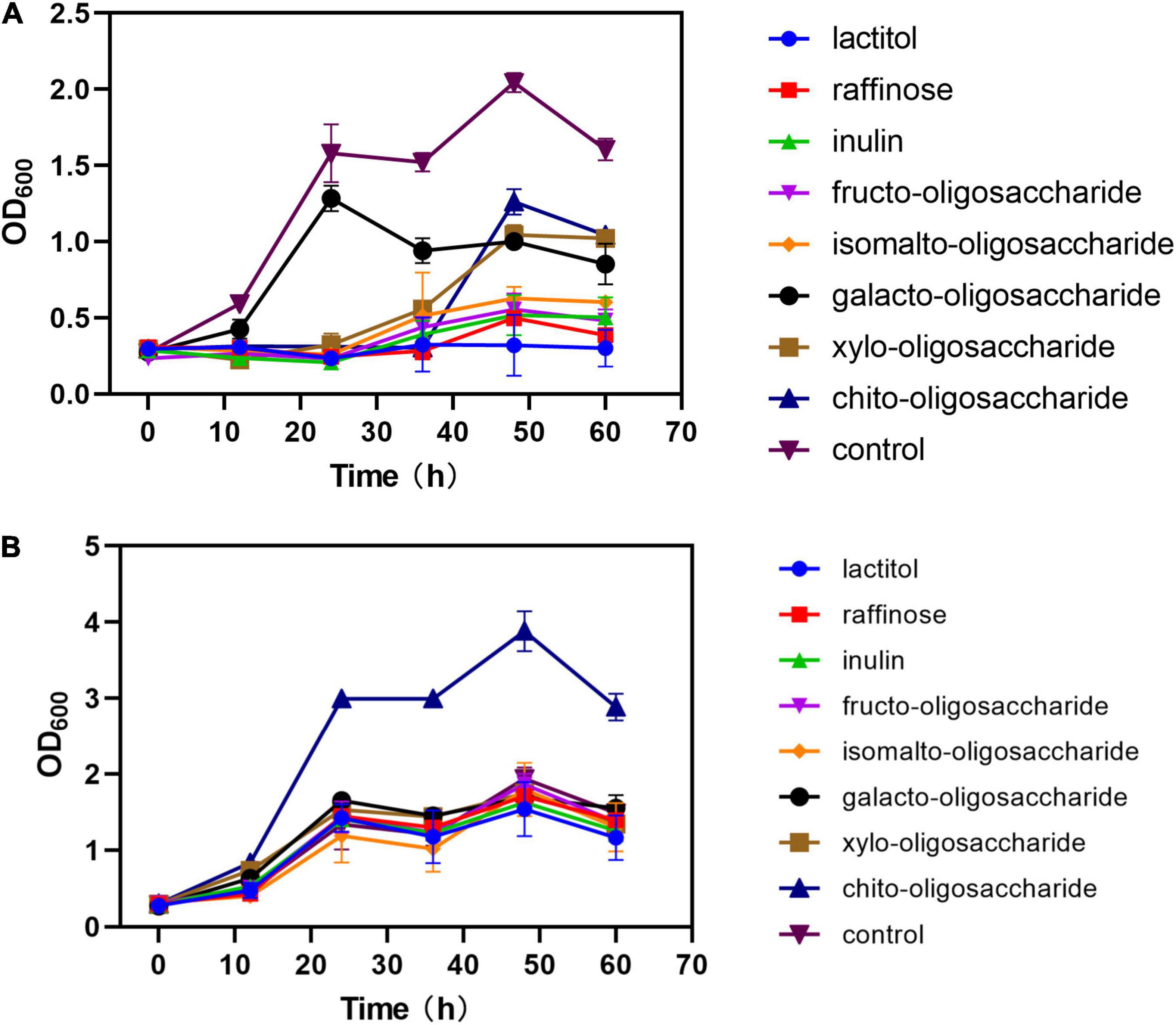
Figure 5. Effects of different prebiotics added on the growth of strain. (A) Prebiotics were added as sole carbon source in the same C-mole of 10 g⋅L– 1 glucose. (B) Prebiotics were added to the media with 5 g⋅L– 1 glucose (1:1 C-mole). Control in panels (A,B) refers to 10 g⋅L– 1 glucose as sole carbon source.
Further, the different ratios of chito-oligosaccharide and glucose as mixed carbon sources were investigated; the results (Figure 6A) showed that the strains growing in the mixed carbon sources of 3:1 C-mole reached the highest growth rate (μ = 0.06 h–1), followed by the ratio of 1 (μ = 0.05 h–1). Interestingly, when the same concentration of chito-oligosaccharide was added in the media. Commensurately, among the strains (Figure 6B), the strains in medium (3:1 C-mole) exhibited the highest glucose consumption (0.37 g⋅L–1⋅h–1) while the lowest (0.13 g⋅L–1⋅h–1) was obtained in the ratio of 1:3. In terms of the consumption of chito-oligosaccharide, strains in the medium of 3:1 C-mole reached the highest rate (0.15 g⋅L–1⋅h–1). However, the chito-oligosaccharides in all media were only consumed about 2 g⋅L–1 (Figure 6C).
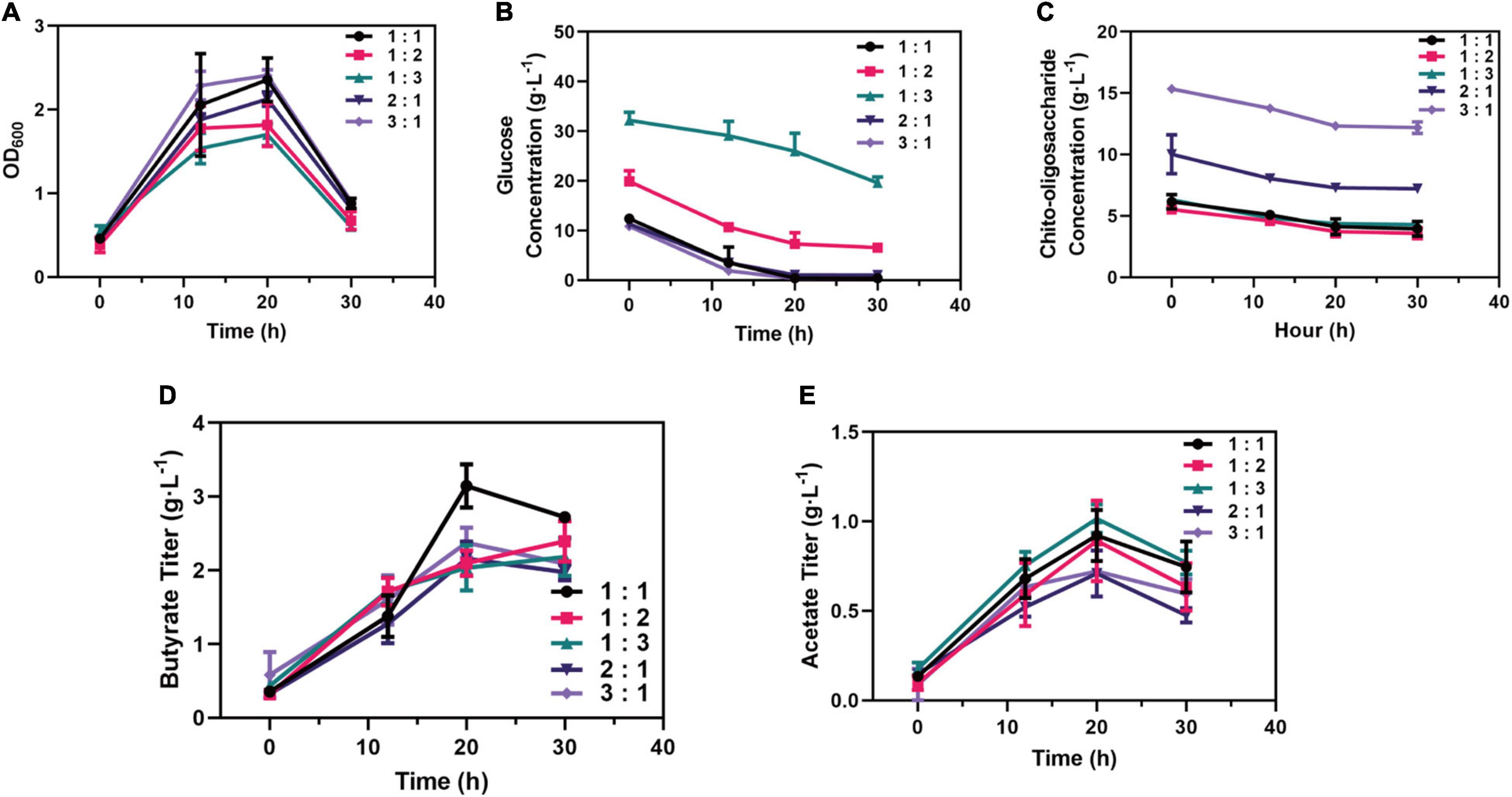
Figure 6. Optimizing added strategy of the proportion (C-mole) of chito-oligosaccharide and glucose. (A) Growth performance of strains in medium supplemented with different proportions of carbon sources. (B) Consumption of glucose in medium supplemented with different proportions of carbon sources. (C) Consumption of chito-oligosaccharide in medium supplemented with different proportions of carbon sources. (D) Production of butyrate supplemented with different proportions of carbon sources. (E) Production of acetate supplemented with different proportions of carbon sources.
In addition, the production of SCFAs in each medium was also investigated, which showed that the medium containing chito-oligosaccharide and glucose with 1:1 C-mole reached the outstanding titer of butyrate (3.15 g⋅L–1) and the yield of 0.31 g⋅g–1 from glucose and 0.50 g⋅g–1 from chito-oligosaccharide (Figure 6D). However, the highest titer of acetate (1.01 g⋅L–1, a yield of 0.03 g⋅g–1 from glucose consumed and 0.20 g⋅g–1 from chito-oligosaccharide) was obtained in the medium of 1:3 C-mole (Figure 6E), which might be related to the need to maintain the growth according to the growth profile in Figure 6A. Besides, the strains in the medium of 1:1 C-mole could also reach 0.92 g⋅L–1 in titer and yield of 0.09 g⋅g–1. Overall, the mixed carbon sources with 1:1 C-mole of chito-oligosaccharide and glucose were the optimal ratio for the production of SCFAs.
Probiotics are referred to as non-pathogenic live microorganisms and can confer health benefits on the host (Araya et al., 2002). Due to the potential for infection or toxin production, probiotics should meet health and safety requirements before commercial application. The primary responsibility for certifying a probiotic is determining its safety as any adverse effects should be anticipated ahead of time. Generally, biochemical characterization, risk assessment, and probiotic properties are critical components in evaluating a new probiotic (Oh and Jung, 2015). Previously, we demonstrated that the strain C. tyrobutyricum L319, isolated from the Grana Padano cheese with a blowing defect, is more competitive than isolates from other environments, exhibiting safety on the genome level (Liu et al., 2020). Although the health benefits of C. tyrobutyricum are well-acknowledged, considerable study remains to be done before this species can be certified as a probiotic. Therefore, we performed a more detailed characterization and systemic evaluation of C. tyrobutyricum L319 in this study, and we suggest that non-toxic C. tyrobutyricum is an excellent probiotic candidate.
Generally, C. butyricum was included in the list of microorganisms that have been developed as a probiotic for use in the food according to Regulation (EC) No. 258/97 (Isa et al., 2016). Meanwhile, previous studies have shown that a potential probiotic C. tyrobutyricum ATCC25755 could protect against LPS-induced colonic inflammation and epithelial dysfunction (Xiao et al., 2021a,c). Therefore, we investigated the similarity between the reference probiotic strain C. butyricum and C. tyrobutyricum, including identification of colonies, phylogenetic tree analysis, ANI analysis, and evolutionary analysis of gene synteny and collinearity. These results showed that C. tyrobutyricum L319 has a high degree of homology with the probiotic C. tyrobutyricum ATCC25755 at the genome level (Figure 1). Combined with the safety evaluation of C. tyrobutyricum at the genomic level in our previous study and its high homology and conservation with another C. tyrobutyricum, we can conclude that this bacterium exhibits a good safety profiling at the genomic level.
Except for the safety evaluation at the genomic level, antibiotic resistance, virulence-related assay, and hemolytic activity were all carried out. First, we verified the antibiotic resistance phenotype of C. tyrobutyricum L319 and found that it is resistant to gentamicin, kanamycin, streptomycin, clindamycin, and chloramphenicol but susceptible to tetracycline, vancomycin, erythromycin, neomycin, ampicillin, linezolid, trimethoprim, rifampicin, and ciprofloxacin (Table 1). The MICs of these antibiotics for C. tyrobutyricum L319 were also identified. Interestingly, we found that the vancomycin resistance exhibited discrepancies between the genotype and observed phenotype (Liu et al., 2020), which could be because potential antibiotic resistance genes were annotated based on 40% amino acid homology, which means that this gene may be not present in the genome of C. tyrobutyricum L319. Similarly, aminoglycoside (gentamicin, kanamycin, and streptomycin) resistance in C. tyrobutyricum L319 was also described as innate resistance in other C. butyricum strains, meaning that aminoglycoside resistance is not a specific trait of C. tyrobutyricum L319 (Isa et al., 2016). In addition, there is no safety hazard that all the genes associated with antibiotic resistance were found unable to be transferred into the environment according to our previous study (Liu et al., 2020). Furthermore, the absence of virulence-related genes is a desired feature for C. tyrobutyricum employed in the food industry.
Our previous study identified 30 potential virulence factors in the C. tyrobutyricum L319 genome. Fortunately, none of these genes need to be excluded according to EFSA guidelines (Liu et al., 2020). Meanwhile, the neurotoxin and toxin genes were not identified, and none of the genes associated with biogenic amines biosynthesis were also detected (Supplementary Figure 1). In addition, its hemolytic activity was confirmed by hemolysis tests, and the strain C. tyrobutyricum L319 exhibited no hemolytic activity (Figure 2A).
We also assessed the potential probiotic properties for the strain C. tyrobutyricum L319. Hydrophobicity is one of the suggestive parameters for cell surface properties of probiotics, and it is closely related to the adhesion ability of probiotics (Zuo et al., 2016). Previously, probiotics’ ability to attach to epithelial cells and promote health benefits increased as their hydrophobicity increased (Nami et al., 2019). In this study, C. tyrobutyricum L319 exhibited better hydrophobicity percentages under the conditions between pH 5.0 and 6.0 (Figure 2B) and an excellent tolerance toward simulated gastric juice and intestinal juice (Figure 2C), which exhibits a possibility for this strain to colonize the intestine.
In our results, C. tyrobutyricum L319 exhibited higher anti-stress tolerance (Figure 3). The growth of the strain was stimulated under different temperatures and NaCl concentrations. We found that the strain displayed excellent tolerance to low and high temperatures, ranging from 20 to 50°C, and it also could survive under 3% NaCl condition. These results indicated that we could optimize the fermentation conditions to promote better growth and faster enrichment of bacteria cells. Moreover, by simulating the gastrointestinal environment, we found that the strain exhibited excellent tolerance to acidity after exposure to media pH 5.0, and this indicated that C. tyrobutyricum L319 is highly active in the acidic gastrointestinal environment and has the potential to travel through the intestinal contents and colonize the intestinal location. Furthermore, we investigated the production of butyrate and acetate, the main components of SCFAs, under normal and acidic conditions (Figure 4). SCFAs play critical roles in regulating host metabolism, immune system, and cell proliferation. The highest concentrations of SCFAs are found in the cecum and proximal colon, where they are sources of energy for colon cells (primarily butyrate) (Koh et al., 2016). As a competitive strain for the production of SCFAs, we investigated the production of butyrate and acetate by C. tyrobutyricum L319, especially under acidic conditions. The results indicated that C. tyrobutyricum L319 has a great potential to be applied in the production of SCFAs, which could contribute to beneficial effects on human health (Figure 4; Zhu et al., 2022).
Prebiotics are substrates that are selectively utilized by the gut microbiota and promote human health (Cunningham et al., 2021). Previous studies have found that prebiotics can enhance the function of probiotics in gut microbiota and can delay the progression of immune diseases such as type 1 diabetes and acute pancreatitis by regulating the homeostasis of gut microbiota and the production of SCFAs (Fei et al., 2021). Studying the utilization of prebiotics by probiotics will contribute to the precise enrichment of target probiotics and improve their abundance. Therefore, we investigated the utilization of eight prebiotics by C. tyrobutyricum L319 and determined the production of butyrate and acetate. It was found that C. tyrobutyricum showed excellent growth performance whether using chito-oligosaccharide as a sole carbon source or using together with glucose as the mixed carbon source (Figure 5). Also, chito-oligosaccharide and glucose (1:1) mixed carbon sources were the optimal strategy for the production of SCFAs (Figure 6). Our results suggested that C. tyrobutyricum could ferment prebiotics, such as chito-oligosaccharides to produce SCFAs. This potential synergistic effect of prebiotics and probiotics could improve gut microbiota for health benefits.
This study confirmed that C. tyrobutyricum L319 has no potential virulence factors or the possibility of antibiotic resistance genes propagation. It also fulfilled several criteria for probiotics, including significant hydrophobicity under acidic conditions, resistance to low and high temperatures, high salts and low pH, availability of chito-oligosaccharides, and the production of SCFAs. To the best of our knowledge, this is the first study describing C. tyrobutyricum strains isolated from the Grana Padano cheese food sources, which could be a promising probiotic candidate for applications. Regarding the safety and probiotic properties, this strain is expected to be developed as the first probiotics representative of C. tyrobutyricum. Moreover, further studies about its safety and probiotic properties in vivo and the optimization of prebiotics utilization and SCFAs production will be necessary to establish its actual feasibility in being used as probiotics.
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
ZY performed the experiments, analyzed the data, and wrote the manuscript with the assistance of LY, LZ, YL, and ZZ. ZZ and LJ provided the experimental ideas and the design of this study. All authors edited, read, and approved the final version of the manuscript.
This study was supported by the National Natural Science Foundation of China (31922070, U2106228, and 22008114), the Natural Science Foundation of Jiangsu Province (BK20200684), the Jiangsu Synergetic Innovation Center for Advanced Bio-Manufacture (XTC2205), and the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (20KJB530017).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.926710/full#supplementary-material
Araya, M., Morelli, L., Reid, G., Sanders, M., Stanton, C., Pineiro, M., et al. (2002). Guidelines for the evaluation of probiotics in food. Rome: FAO, 1–11.
Baccouri, O., Boukerb, A. M., Farhat, L. B., Zébré, A., Zimmermann, K., Domann, E., et al. (2019). Probiotic potential and safety evaluation of Enterococcus faecalis OB14 and OB15, isolated from traditional tunisian testouri cheese and rigouta, using physiological and genomic analysis. Front. Microbiol. 10:881. doi: 10.3389/fmicb.2019.00881
Bindels, L. B., Delzenne, N. M., Cani, P. D., and Walter, J. (2015). Towards a more comprehensive concept for prebiotics. Nat. Rev. Gastro. Hepat. 12, 303–310. doi: 10.1038/nrgastro.2015.47
Blaak, E. E., Canfora, E. E., Theis, S., Frost, G., Groen, A. K., Mithieux, G., et al. (2020). Short chain fatty acids in human gut and metabolic health. Benef. Microb. 11, 411–455. doi: 10.3920/bm2020.0057
Calvo-Barreiro, L., Eixarch, H., Cornejo, T., Costa, C., Castillo, M., Mestre, L., et al. (2021). Selected Clostridia strains from the human microbiota and their metabolite, butyrate, improve experimental autoimmune encephalomyelitis. Neurotherapeutics 18, 920–937. doi: 10.1007/s13311-021-01016-7
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, D., Jin, D., Huang, S., Wu, J., Xu, M., Liu, T., et al. (2020). Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 469, 456–467. doi: 10.1016/j.canlet.2019.11.019
Cunningham, M., Azcarate-Peril, M. A., Barnard, A., Benoit, V., Grimaldi, R., Guyonnet, D., et al. (2021). Shaping the future of probiotics and prebiotics. Trends Microbiol. 29, 667–685. doi: 10.1016/j.tim.2021.01.003
Elzinga, J., Oost, J. V. D., Vos, W. M. D., and Smidt, H. (2019). The use of defined microbial communities to model host-microbe interactions in the human gut. Microbiol. Mol. Biol. R. 83, e18–e54. doi: 10.1128/MMBR.00054-18
ESFA (2012). Outcome of the public consultation on the draft Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. Parma: EFSA Supporting Publications. doi: 10.2903/sp.efsa.2012.EN-316
Fei, Y., Chen, Z., Han, S., Zhang, S., Zhang, T., Lu, Y., et al. (2021). Role of prebiotics in enhancing the function of next-generation probiotics in gut microbiota. Crit. Rev. Food Sci. Nutr. 2021, 1–18. doi: 10.1080/10408398.2021.1958744
Hu, J. M., Lin, S. L., Zheng, B. D., and Cheung, P. C. K. (2018). Short-chain fatty acids in control of energy metabolism. Crit. Rev. Food Sci. 58, 1243–1249. doi: 10.1080/10408398.2016.1245650
Hu, X., Guo, J., Xu, M., Jiang, P., Yuan, X., Zhao, C., et al. (2019). Clostridium tyrobutyricum alleviates Staphylococcus aureus-induced endometritis in mice by inhibiting endometrial barrier disruption and inflammatory response. Food Funct. 10, 6699–6710. doi: 10.1039/C9FO00654K
Isa, K., Oka, K., Beauchamp, N., Sato, M., Wada, K., Ohtani, K., et al. (2016). Safety assessment of the Clostridium butyricum MIYAIRI 588§probiotic strain including evaluation of antimicrobial sensitivity and presence of Clostridium toxin genes in vitro and teratogenicity in vivo. Hum. Exp. Toxicol. 35, 818–832. doi: 10.1177/0960327115607372
Jiang, L., Fu, H., Yang, H. K., Xu, W., Wang, J., and Yang, S.-T. (2018). Butyric acid: Applications and recent advances in its bioproduction. Biotechnol. Adv. 36, 2101–2117. doi: 10.1016/j.biotechadv.2018.09.005
Joukar, F., Mavaddati, S., Mansour-Ghanaei, F., and Samadani, A. A. (2020). Gut microbiota as a positive potential therapeutic factor in carcinogenesis: an overview of microbiota-targeted therapy. J. Gastr. Cancer 51, 363–378. doi: 10.1007/s12029-019-00237-6
Koh, A., De Vadder, F., Kovatcheva-Datchary, P., and Bäckhed, F. (2016). From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345. doi: 10.1016/j.cell.2016.05.041
Lee, I., Ouk Kim, Y., Park, S. C., and Chun, J. (2016). OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Liu, T., Zhu, L., Zhu, Z., and Jiang, L. (2020). Genome sequence analysis of Clostridium tyrobutyricum, a promising microbial host for human health and industrial applications. Curr. Microbiol. 77, 3685–3694. doi: 10.1007/s00284-020-02175-0
Martín, R., Miquel, S., Benevides, L., Bridonneau, C., Robert, V., Hudault, S., et al. (2017). Functional characterization of covel Faecalibacterium prausnitzii strains isolated from healthy volunteers: a step forward in the use of F. prausnitzii as a next-generation probiotic. Front. Microbiol. 8:1226. doi: 10.3389/fmicb.2017.01226
Nami, Y., Vaseghi Bakhshayesh, R., Mohammadzadeh Jalaly, H., Lotfi, H., Eslami, S., and Hejazi, M. A. (2019). Probiotic properties of Enterococcus isolated from artisanal dairy products. Front. Microbiol. 10:300. doi: 10.3389/fmicb.2019.00300
Oh, Y. J., and Jung, D. S. (2015). Evaluation of probiotic properties of Lactobacillus and Pediococcus strains isolated from Omegisool, a traditionally fermented millet alcoholic beverage in Korea. LWT-Food Sci. Technol. 63, 437–444. doi: 10.1016/j.lwt.2015.03.005
Shreiner, A. B., Kao, J. Y., and Young, V. B. (2015). The gut microbiome in health and in disease. Curr. Opin. Gastroen. 31, 69–75. doi: 10.1097/MOG.0000000000000139
Sonnenburg, J. L., and Sonnenburg, E. D. (2019). Vulnerability of the industrialized microbiota. Science 366:9255. doi: 10.1126/science.aaw9255
Valdes, A. M., Walter, J., Segal, E., and Spector, T. D. (2018). Role of the gut microbiota in nutrition and health. BMJ 361:k2179. doi: 10.1136/bmj.k2179
Wang, R. X., Lee, J. S., Campbell, E. L., and Colgan, S. P. (2020). Microbiota-derived butyrate dynamically regulates intestinal homeostasis through regulation of actin-associated protein synaptopodin. P. Natl. Acad. Sci. 117, 11648–11657. doi: 10.1073/pnas.1917597117
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Xiao, Z. P1., Liu, L. J., Jin, Y. Y., Pei, X., Sun, W. J., and Wang, M. Q. (2021a). Clostridium tyrobutyricum protects against LPS-induced colonic inflammation via IL-22 signaling in mice. Nutrients 13:215. doi: 10.3390/nu13010215
Xiao, Z. P., Liu, L. J., Jin, Y. Y., Pei, X., Sun, W. J., and Wang, M. Q. (2021b). A potential prophylactic probiotic for inflammatory bowel disease: The overall investigation of Clostridium tyrobutyricum ATCC25755 attenuates LPS-induced inflammation via regulating intestinal immune cells. Mol. Nutr. Food Res. 65:1213. doi: 10.1002/mnfr.202001213
Xiao, Z. P., Liu, L. J., Pei, X., Sun, W. J., Jin, Y. Y., Yang, S. T., et al. (2021c). A potential probiotic for diarrhea: Clostridium tyrobutyricum protects against LPS-induced epithelial dysfunction via IL-22 produced by Th17 cells in the Ileum. Front. Immunol. 12:227. doi: 10.3389/fimmu.2021.758227
Xu, Y., Zhu, Y., Li, X., and Sun, B. (2020). Dynamic balancing of intestinal short-chain fatty acids: the crucial role of bacterial metabolism. Trends Food Sci. Tech. 100, 118–130. doi: 10.1016/j.tifs.2020.02.026
Zhang, X., Cai, X., and Zheng, X. (2021). Gut microbiome-oriented therapy for metabolic diseases: challenges and opportunities towards clinical translation. Trends Pharmacol. Sci. 2021:3. doi: 10.1016/j.tips.2021.09.003
Zhu, B., Wang, X., and Li, L. (2010). Human gut microbiome: the second genome of human body. Protein Cell 1, 718–725. doi: 10.1007/s13238-010-0093-z
Zhu, Z., Zhu, L., and Jiang, L. (2022). Dynamic regulation of gut Clostridium-derived short-chain fatty acids. Trends Biotechnol. 40, 266–270. doi: 10.1016/j.tibtech.2021.10.005
Keywords: Clostridium tyrobutyricum, safety evaluation, probiotics, prebiotics, short-chain fatty acids
Citation: Yang Z, Amal FE, Yang L, Liu Y, Zhu L, Zhu Z and Jiang L (2022) Functional Characterization of Clostridium tyrobutyricum L319: A Promising Next-Generation Probiotic for Short-Chain Fatty Acid Production. Front. Microbiol. 13:926710. doi: 10.3389/fmicb.2022.926710
Received: 23 April 2022; Accepted: 25 May 2022;
Published: 17 June 2022.
Edited by:
Chongde Wu, Sichuan University, ChinaReviewed by:
Lixin Luo, South China University of Technology, ChinaCopyright © 2022 Yang, Amal, Yang, Liu, Zhu, Zhu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhengming Zhu, emh1em1Abmp0ZWNoLmVkdS5jbg==; Ling Jiang, amlhbmdsaW5nQG5qdGVjaC5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.