- 1Department of Environmental Science, V.B.S. Purvanchal University, Jaunpur, India
- 2Department of Biotechnology, M.H.P.G. College, Jaunpur, India
- 3Academy of Biology and Biotechnology, Southern Federal University, Rostov-on-Don, Russia
- 4Department of Agriculture and Veterinary Sciences, Mewar University, Chittorgarh, India
- 5Department of Molecular Biology and Biotechnology, Maharana Pratap University of Agriculture and Technology, Udaipur, India
- 6Cytogenetics Laboratory, Department of Zoology, Banaras Hindu University, Varanasi, India
- 7Department of Plant Physiology, Institute of Agricultural Sciences, Banaras Hindu University, Varanasi, India
- 8Department of Biochemistry, Faculty of Science, University of Allahabad, Allahabad, India
The breaking silence between the plant roots and microorganisms in the rhizosphere affects plant growth and physiology by impacting biochemical, molecular, nutritional, and edaphic factors. The components of the root exudates are associated with the microbial population, notably, plant growth-promoting rhizobacteria (PGPR). The information accessible to date demonstrates that PGPR is specific to the plant's roots. However, inadequate information is accessible for developing bio-inoculation/bio-fertilizers for the crop in concern, with satisfactory results at the field level. There is a need to explore the perfect candidate PGPR to meet the need for plant growth and yield. The functions of PGPR and their chemotaxis mobility toward the plant root are triggered by the cluster of genes induced by the components of root exudates. Some reports have indicated the benefit of root exudates in plant growth and productivity, yet a methodical examination of rhizosecretion and its consequences in phytoremediation have not been made. In the light of the afore-mentioned facts, in the present review, the mechanistic insight and recent updates on the specific PGPR recruitment to improve crop production at the field level are methodically addressed.
Introduction
According to the World Health Organization (WHO), the food shortage for sustaining the human population is on a steep upward trajectory, mainly owing to the quickly booming human population that is expected to cross the 10 billion mark by 2050 (DESA UN, 2015). Both WHO and the United Nations have proposed to intensify global food production by 50% in the near future. The agriculturally important microorganisms (AIMs) can play a pivotal role in realizing this colossal target considering the fact that fertile lands are sharply shrinking owing to urbanization and industrialization. AIMs not only improve plant growth and yield but provide sustained protection against a variety of phytopathogens (Bhattacharyya and Jha, 2012; Glick, 2012; Compant et al., 2019). The beneficial microbes of the rhizosphere zone interact positively with mutually guided components of root exudates, i.e., rhizodeposits (Hassan et al., 2019). During the rhizodeposition process, the plant roots secrete carbohydrates, fatty acids, essential amino acids, organic acids, hydrolytic enzymes, growth-regulating hormones, vitamins, nucleotides, flavonoids, polyphenols, sterols, and volatile organic compounds (Hartmann et al., 2009; Hu et al., 2018; Ankati and Podile, 2019).
In the last century, the word “rhizosphere” was introduced as a microbial hot spot in the area of the rootsystem (Hartmann et al., 2008). The rhizospheric region, a specific zone around the root and harbors various kinds of microorganisms, primarily bacteria, fungi, nematodes, insect larvae, mites, amoebas, and protozoa (Bonkowski et al., 2009). The bacterial colonies residing in the rhizospheric zone are called rhizobacteria (Hartmann et al., 2009). The rhizospheric zone supports the plant root system (Ahemad and Kibret, 2014) and modulates the physico-chemical and biological properties of the soil (Ahemad and Kibret, 2014; Zhalnina et al., 2018).
The rhizosphere zone provides a shelter for the exchange of biochemical components that establish inter-species relationships between the roots and microorganisms (Gupta et al., 2020). Plant roots release various types of enzymes/compounds in the soil that mediate the interaction between microorganisms and plants (Ankati and Podile, 2019). Factors influencing soil microbial population include soil quality, soil moisture, soil pH, and rhizospheric secretion (Bagyalakshmi et al., 2012; Upadhyay and Singh, 2015; Hu et al., 2018). There are various physical and chemical parameters of the rhizospheres that impact the function of microorganisms, which ultimately affect several mechanisms, such as the respiratory process, the secretion of organic acids by the roots, the breakdown of soil organic matter, nutrient uptake, symbiotic nitrogen fixation, etc. (Reinhold-Hurek et al., 2015; Mahmud et al., 2021).
The rhizosphere plays an important role in root excretion, microbial activity, genetic exchange, improving nutrient use efficiency, and gradient diffusion, which are jointly referred to as the rhizosphere effect (Badri and Vivanco, 2009; Ladygina and Hedlund, 2010; Mendes et al., 2013). Rhizobacteria associated with the plant root are often referred to as plant growth-promoting rhizobacteria (PGPR). The functions of plant growth-promoting rhizobacteria, such as direct and indirect mechanism, metabolism, chemotaxis, secretion, antibiotic production, etc., are mediated by its gene cluster that triggers host–PGPR interactions (Mark et al., 2005; Matilla et al., 2007; Ramachandran et al., 2011; Zhang et al., 2015; Bashir et al., 2021; South et al., 2021). Ultrastructure of the root cell wall mediated PGPR interaction, which was induced by the gene expression of the plant. Ryu et al. (2003) demonstrated that out of 38 genes, 30 genes of Bacillus subtilis-GB03 were associated with a change in the Arabidopsis root-ultrastructure and promote plant growth. Azospirillum irakense vitalized polygalacturonase gene (PG genes) in the roots of rice plant (Sekar et al., 2000). Among PG genes, PbrPG6 is responsible for fruit-soothe (Zhang et al., 2019). The root exudation and root exudates are relevant for the survivability of plants against various environmental conditions. The root exudates aid in the selection of microbial populations around the rhizosphere (Mendes et al., 2013; Zhang et al., 2015). In the purview to tackle this aspect, the review discusses the mechanisms of root exudation, the current updates on the selective plant growth-promoting rhizobacteria aggregation and their role in plant–microbe interface, and most importantly, the future developments in plant–PGPR interactions for sustainable agriculture.
Root Exudates and Plant Growth-Promoting Rhizobacteria
Plant root secretes 5–21% of photosynthetic matter such as carbohydrates, proteins, secondary metabolites, etc., into the rhizospheric soil environment, generally known as root exudates (Badri et al., 2013; Figure 1). The coping mechanism of plants under diverse environmental conditions mainly rests on the root acquisition of soil resources and their surroundings (Gupta et al., 2020). In the mid twentieth century, the world population increased quickly and posed various problems related to food, fiber, fuel, homeland, etc., which has consequences for hunger, poverty, water scarcity, and environmental degradation. The scarcity of food is a burgeoning challenge for humans that has been classified as goal number two of the Zero Hunger of the United Nations Sustainable Development Goals 2030.
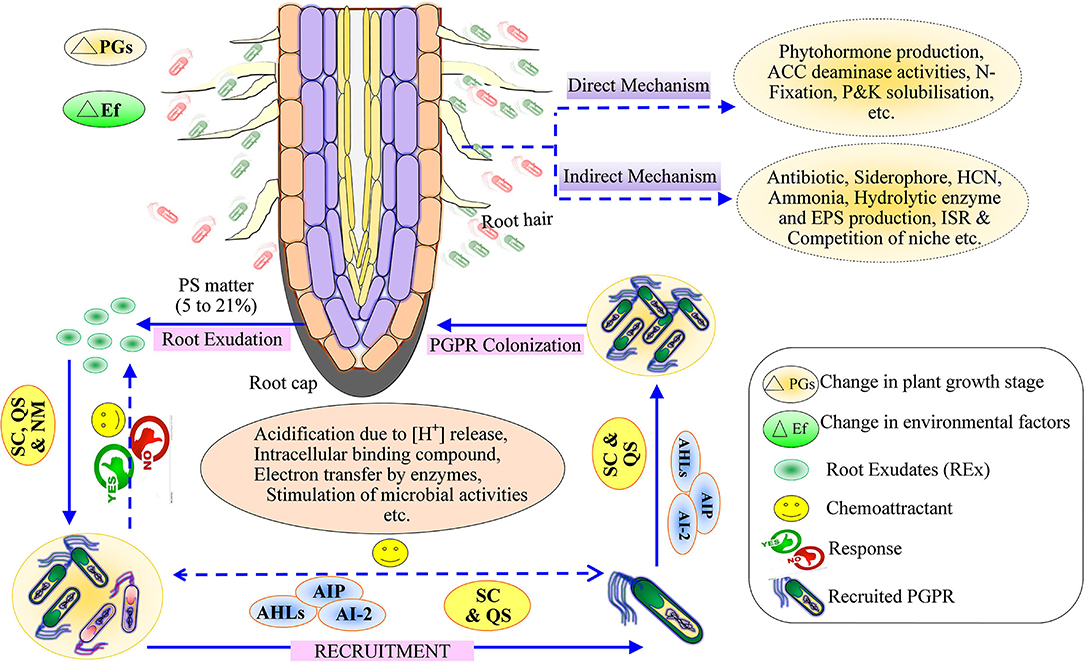
Figure 1. Schematic representation of the mechanism of root exudates for recruitment of plant growth-promoting rhizobacteria and plant growth-promoting mechanism (Direct and Indirect). SC, Selected compounds; QS, quorum sensing; NM, Nutrient management; AHL, Acyl Homoserine lactone; AIP, Autoinducing peptides; AI-2, Autoinducer; PS, Photosynthetic matter.
Recently, the researchers have introduced an eco-friendly concept based on free-living bacteria called PGPR (plant-growth promoting rhizobacteria). The plant growth-promoting rhizobacteria are soil-borne or root-colonizing rhizobacteria (Upadhyay et al., 2009, 2012a,b; Singh et al., 2017; Numana et al., 2018; Upadhyay and Chauhan, 2022), which play a functional role in plant growth through several mechanisms in terrestrial ecosystems. Plant-growth promoting rhizobacteria significantly reduce the dependence on chemical fertilizers and pesticides (Liu et al., 2017). Plant growth-promoting rhizobacteria promote plant growth through root-hair proliferation, enhancing root hair branching; increase in seedling emergence; early nodulation; nodule functioning; enhanced leaf surface area; improvement in vigor and biomass; increased indigenous plant hormones levels; and most importantly, by improving nutrient use efficiency (Vocciante et al., 2022). The plant growth-promoting rhizobacteria induce the accumulation of carbohydrates in plants and consequently the yield of various plant species (Bhattacharyya and Jha, 2012; Table 1). The most dominant endophytic plant growth-promoting rhizobacteria phyla are Proteobacteria and Actinobacteria, followed by Bacteroidetes and Firmicutes (Ray et al., 2017). Endophytic bacteria enter the plant tissues by the lateral root cracks, wounds, lenticels, germinating radicles, and other parts of the plant body (Chaturvedi et al., 2016). Endophytic-rhizospheric bacteria are involved in several functions such as internal protection of the environment (Santos et al., 2018), metabolism of carbon compounds, nitrogen fixation by nitrogenase (Santoyo et al., 2016), and capability for germination of nodes (Yousaf et al., 2017).
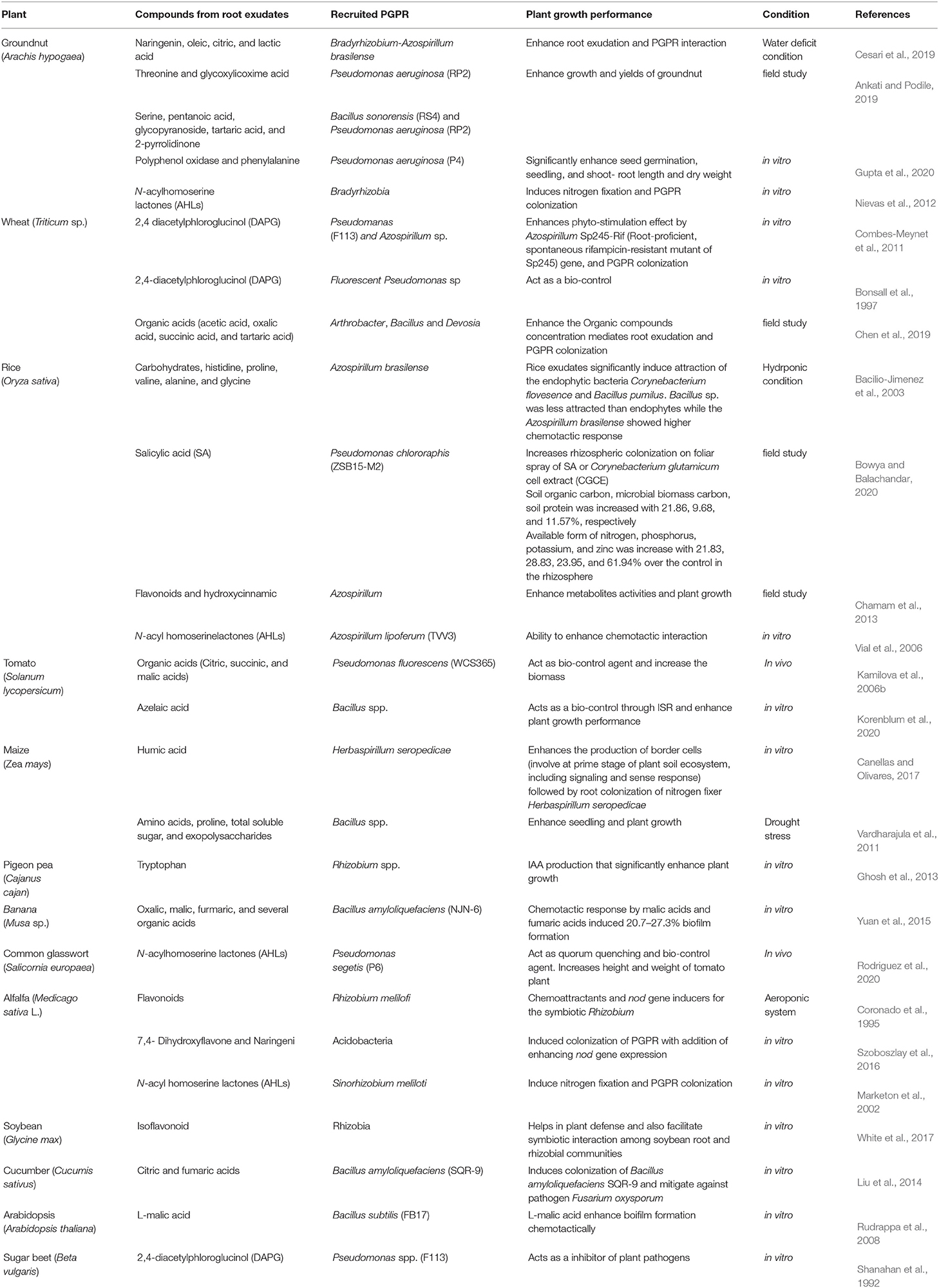
Table 1. Compounds from plant root exudates recruit perfect plant growth-promoting rhizobacteria (PGPR) improving plant growth performance.
The PGPR leads to increased soil fertility, plant growth promotion, and suppression of phytopathogens. These are involved in different functions of the soil ecosystem, nutrient availability, bioremediation of toxic heavy metals, degradation of pesticides, etc. (Chandler et al., 2008; Braud et al., 2009; Rajkumar et al., 2010; Paul et al., 2020; Bhojiya et al., 2021). The PGPR induces plant growth under varied environmental conditions, and the functional roles of bacteria vary with a specific plant (Table 1). The studies demonstrated that root exudates recruited microbial species that are more favorable for plant growth and productivity (Chowdhury et al., 2015; Zhang et al., 2015).
Plant–Microbe Interactions
Several researchers have reported that the population of microorganisms differs in the soil, for example, 12 × 108 bacteria/g dry soil, 12 × 105 fungi/g dry soil, 5 × 105 algae/g dry soil, and 46 × 106 actinomycetes/g dry soil; the bacterial population is highest followed by fungi, algae, and actinomycetes, as a general rule of the thumb (Yadav et al., 2015). Biotic interactions between the plants and microbes occur through communication that requires two essential conditions: one is the production of a specific signal, and another is the behavioral response generated from the signals (Keller and Surette, 2006). Plants communicate to rhizobacteria by secreting specific signaling molecules, viz. lectine enzymes, which are retraced by the bacterial species (Keller and Surette, 2006). B. subtilis detects only secondary metabolites as signals and produces a response against the secondary metabolites (Shank and Kolter, 2011; Singh et al., 2019). The behavior between the plants and the PGPR is mediated through root exudates, quorum sensing, cross-talk, electron-transfer mechanism, etc. (Tashiro et al., 2013; Singh et al., 2017; Keswani et al., 2020a). Positive plant–microbe interactions can be observed with PGPR, nitrogen-fixing bacteria, endo- and ecto-mycorrhizal fungi, whereas negative plant–microbe interactions are exhibited by pathogenic microbes (Haldar and Sengupta, 2015; Compant et al., 2019; Bashir et al., 2021; South et al., 2021). The legume rhizobia is an example of symbiotic interactions (Cai et al., 2009); the plant's secondary metabolite secretes flavonoids that activate a cascade of transcriptional events and mediates rhizobial nodulation signals commonly known as Nod-factors or lipo-chitooligosaccharides (Spaink, 2000). These factors trigger plant growth, leading to morphological changes in root hairs of legumes and the development of root nodules, while Nod-factors play a significant role in symbiotic nitrogen fixation (D'Haeze et al., 1998). The rhizospheric microbes act as biological control agents (BCA) that regulate plant pathogens. Thus, BCA ultimately increases plant productivity through the production of antimicrobial secondary metabolites (Weller, 2007; Singh et al., 2019), production of hydrolytic enzymes (Adesina et al., 2007), effectors (Rezzonico et al., 2005), and hyperparasitism (Harman et al., 2004). Plant growth-promoting microorganisms (PGPM) affect plant growth directly or indirectly through biofertilizers (Mahmud et al., 2021) and/or phytostimulators (Spaepen et al., 2007), as well as biocontrol activity (Figure 1).
The functional genes of B. amyloliquefaciens strains CAUB946, YAUB9601-Y2, and FZB42 are involved in the synthesis of phytohormones, and other gene clusters are involved in disease control (Chen et al., 2007; Borriss, 2011; Blom et al., 2012; Hao et al., 2012). The selection of the perfect candidate PGPR can be a remarkable development for biofertilizer technology. Zhang et al. (2015) demonstrated that B. amyloliquefaciens (SQR9) is an ideal and more efficient PGPR than other strains of Bacillus strains (Table 2). Thirteen unique mobile genomic islands (GIs) were observed for the SQR9 strain. These GIs were found to be involved in the synthesis of many known and unidentified novel compounds. A recent report also demonstrated that maize root exudates regulate 98 genes in SQR9 for carbohydrate and amino acid metabolism. The set of the genome in SQR9 performed several functions like extracellular matrix production and regulated gene expression (Table 2), which revealed a high density (1.8 × 106 CFU g−1 root) of SQR9 in the maize rhizosphere, and triggers plant growth (Zhang et al., 2015).

Table 2. Root exudates of maize plant recruit B. amyloliquefaciens strain. The whole genome sequence of B. amyloliquefaciens revealed that the group of genes induced by root exudates and its functional gene triggers rhizoadaptation, phytostimulation, bioferlilizer, and biocontrol activity.
Mechanistic Insight of Plant Growth-Promoting Rhizobacteria
Root Exudation
Narasimhan et al. (2003) reported two groups of root-exudates:(i) low molecular weight (LMW) such as sugars, amino acids, phenolics, secondary metabolites, and organic acids (citric, malic, oxalic, pyruvic, and succinic, etc.), and (ii) high molecular weight (HMW) that include proteins and complex carbohydrates. The nature and specificity of root exudates are dependent on the host species, plant developmental stages, physio-chemical nature of the soil, and surrounding microbial diversity (Hu et al., 2018; Singh et al., 2022). The maximum concentration of root exudates is found at the root tips and the lateral branching of the roots. Its amount also attenuates with increasing root surface (Compant et al., 2019), diffusion and degradation through sorption, deposition, or microbial consumption (Reinhold-Hurek et al., 2015). The microbial consumption contributes to the extravagance of root exudates owing to the valuable source of nutrition and energy for the rhizospheric microbes (Compant et al., 2019). The difference in the amount and nature of root exudates determines nutrient mobility, microbial population, and microbial diversity (Chamam et al., 2013; Bowya and Balachandar, 2020; Korenblum et al., 2020; Singh et al., 2022). Plant roots secrete root exudates in the rhizospheric region through passive (ion channels, vesicular transport, and diffusion) and active (secretion) mechanisms (Rohrbacher and St-Arnaud, 2016). LMW compounds are released through passive transport while HMW compounds through active transport mechanisms (Rohrbacher and St-Arnaud, 2016). The root exudates and solutes from cell membranes develop equilibrium between exterior and interior molecular transport (Weston et al., 2012; Cesari et al., 2019). In the passive mechanism, polar molecules and ions diffuse through the membrane using channels/permeases through a process called facilitated diffusion. These channels act as a passage for small ions like Na+, K+, Cl−, etc., and water, which aid in maintaining intra-cellular pH, membrane potential, osmotic status, and stabilized volume of the cell (Lee et al., 2007). The small polar and uncharged molecules can transport through direct passive diffusion depending on membrane permeability (Weston et al., 2012; Rohrbacher and St-Arnaud, 2016). The non-polar molecules pass through without using channels or transfer proteins (Weston et al., 2012). The electrochemical gradient arises owing to charged molecules or ions like amino acids, sugars, carboxylates ions, etc. (Rohrbacher and St-Arnaud, 2016). Passive transport across the membrane through channels is driven by an electrochemical gradient (Rohrbacher and St-Arnaud, 2016). Without any expense of energy, the movement based on an electrochemical gradient is called passive transport. The transport that requires energy from ATP for several ions or molecules against the concentration gradient or electrochemical gradient is called active transport (Rohrbacher and St-Arnaud, 2016). Plants have different coping mechanisms against the environment and secrete a large number of compounds that may require many transporters (Weston et al., 2012; Rohrbacher and St-Arnaud, 2016; Korenblum et al., 2020), and these transporters are capable of root exudation of aggregates into the rhizo-microbiome.
Weston et al. (2012) reported that the root exudates from root cells are transported by membrane transport proteins (MTPs). The ATP-binding cassette transporter helps in the phytochemical secretion from roots. Besides, Badri et al. (2013) have also described that out of 129 genes, 25 are significant for root exudation in Arabidopsis thaliana. A single gene mutation may influence the interaction among the microbial group of soil in A. thaliana (Badri et al., 2013). The MTPs include ABC transporter, multidrug and toxic compound extrusion (MATEs), major facilitator superfamily (MFS), and aluminum-activated malate transporter (ALMT). MATE transporter in rice root promotes exudation of polyphenolic compounds (Baetz and Martinoia, 2014). Recently, Wanga et al. (2018) reported that the aluminum exclusion from the root is facilitated by ALMT and citrate exudation through the MATE citrate transporter.
The Action of Root Exudates
Root exudates can mediate neutral, useful, or harmful interections between plant microbes and inter-species of microorganisms (Mendes et al., 2013; Hu et al., 2018). The secretion of root exudates rests on plant needs, and the rate of exudation is modified to cope with different biotic and abiotic stresses (Badri and Vivanco, 2009; Vardharajula et al., 2011). The root-driven changes in the microbial community observed by Donn et al. (2015) in the wheat rhizosphere demonstrated ten times more bacterial abundance than the bulk soil. Specific microbes like Burkholderiales, Sphingobacterium, and Xanthomonadales are dominant in the rhizospheres of Brachypodium distachyon in comparison to bulk soil (Kawasaki et al., 2016). Similar observations were recorded by Zhalnina et al. (2018) while studying the chemistry of root exudates of Avenabarbata where root-exudates were preferred as substrates for the specific bacterial community in the rhizospheres. The beneficial rhizobacterium Pseudomonas putida KT2440 is chemotactically attracted by 2-4-dihydroxy-7methoxy-1,4-benzoxazin-3-one from root exudates of Zea mays (Neal et al., 2012). The root exudate compounds like flavonoids act as signaling molecules, regulate nod-gene expression, activate nod-factors (lipochito-oligosaccharide), and trigger nodulation establishment in legumes (Abdel-Lateif et al., 2012; Figure 2). The flavonoids are released due to overcoming nitrogen deficiency in soil (Coronado et al., 1995). The flavonoid 7,4-dihydroxyflavone, from the root exudates of Medicago sativa, can mediate interaction with a diverse range of acid bacteria along with the induction of the nod-gene in the legumes (Szoboszlay et al., 2016). Strigolactone stimulates hyphal branching in mycorrhiza (Akiyama et al., 2005), and malic acid helps in the recruitment of plant growth-promoting rhizobacteria (Rudrappa et al., 2008). On the other hand, root-exudates have antimicrobial secondary metabolites such as benzoxazinoids (BXs), which can trim actinobacteria, proteobacteria, and pathogenic microbial populations in the maize rhizosphere (Hu et al., 2018). Root exudates influence the recruitment and make-up of microbiota in the plant rhizosphere (Hartmann et al., 2009; Ladyginaand Hedlund 2010; Reinhold-Hurek et al., 2015; Table 1). The ability of A. brasilense to modulate the plant root architecture was reported by Creus et al. (2005). Molina-Favero et al. (2008) observed that A. brasilense can synthesize nitric oxide (NO) aerobically, which mediates the IAA signaling pathway, leading to lateral and adventitious root formation in tomatoes. In Arabidopsis, under drought stress, root colonization of P. chlororaphis increases the expression of genes associated with ROS (reactive oxygen species) defense, auxin, jasmonic acid, and salicylic acid synthesis (Cho et al., 2013). P. chlororaphis also decreases the expression of ethylene and abscisic acid genes in Arabidopsis under drought stress (Cho et al., 2013; Figure 2). The roots of watermelon secrete more trans-chlorogenic acid and caffeic acid, followed by trans-cinnamic acid, which induced resistance against Fusarium oxysporum (Ling et al., 2013). Cai et al. (2009) reported that leguminous plant roots secrete canavanine, which recruits beneficial microorganisms. Canavanine favors the growth of selective rhizobia and also acts as an antimicrobial for pathogenic bacteria (Cai et al., 2009). Sugars and strigolactone, viz. 5-dexystrigal, components of non-legume root exudates mediate symbiotic association with mycorrhizal fungi (Fang and St Leger, 2010). Nguema-Ona et al., 2013 observed that AGPs (arabinogalactans protein) of root exudates also attract plant growth-promoting rhizobacteria through the chemo-attractant mechanism, and the maximum amount of AGPs was found at the root tip regions of the plants (Cannesan et al., 2012). AGPs induce a population of beneficial microbes in leguminous and non-leguminous plants (Xie et al., 2012; Vieira et al., 2020). The VOCs, myc-factors, nod-factors, exopolysaccharides, etc. are signaling components associated with rhizospheric microbes (Goh et al., 2013). VOCs (acetoin, 2-3-butanediol) mediate communication between plant microbes, induce ISR (induced systemic resistance) as bio-protestants (Ryu et al., 2004), and plant growth promotion. Ankati and Podile (2019) reported that threonine and glyoxylic oxime acid from root-exudates of groundnut influenced Pseudomonas aeruginosa (RP2), while serine, pentanoic acid, glucopyranoside, tartaric acid, and 2-pyrrolidinone influenced both P. aeruginosa (RP2) and B. sonorensis (RS4). These findings demonstrated that a specific component of root-exudates was responsible for selective PGPR interaction. Thus, the products of root exudates could be an effective agent for improving crop yield at the field level by enhancing PGPR colonization.
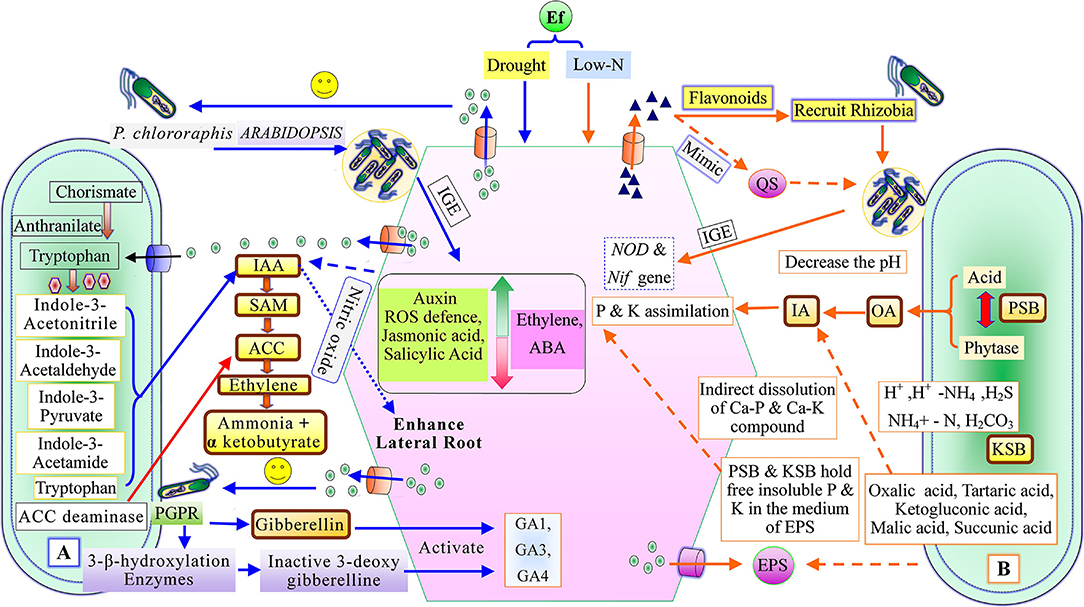
Figure 2. Plant growth-promoting mechanism of recruited plant growth-promoting rhizobacteria, (A) Phytostimulation, and (B) Biofertilizer activity [QS, Quorum sensing; IAA, indole-3-acetic acid; SAM, S-adenosyl methionine; ACC, 1-aminocyclopropane-1-carboxylate; IGE, Induce gene expression; ABA, Abscisic acid; GA1, GA2, and GA4, (Types of gibberellin); PSB, Phosphate solubilizing bacteria; KSB, Potassium solubilizing bacteria; EPS, exo-polysaccharides; OA, Organic acid; and IA, Inorganic acid].
Quorum sensing (QS) helps in establishing root microbe assemblage in the rhizosphere. The root exudates mimic QS signals of bacteria to repress QS-regulated responses of associated/adjacent bacteria. The root exudates have primary and secondary metabolites along with proteins (Korenblum et al., 2020). Some reports demonstrated that these proteins influenced the selective recruitment of useful bacteria (De-la-Pena et al., 2008). The QS compound in the root exudates of groundnut plants selects microbes and induces their population (Ankati and Podile, 2019). Bacteria communicate within the system through a density-dependent mechanism known as QS (Reinhold-Hurek et al., 2015). The QS regulates the metabolic as well as the behavioral activities of the bacterial community (Marketon et al., 2002; Nievas et al., 2012; Liu et al., 2014; Korenblum et al., 2020). This sort of interaction occurs through a dialect of chemical signals called autoinducers (AHLs: acyl homoserine lactones), autoinducing peptides (AIP), and autoinducer-2; furanone (AI-2), synthesized by bacteria (Figure 1). AHLs mediate signaling in gram-negative bacteria (Vial et al., 2006). AIP requires specialized membrane transport protein for signaling in the gram-positive bacteria, whereas AI-2 is required for both gram-positive and gram-negative bacteria (Abisado et al., 2018). Bacterial QS occurs through various complex pathways depending upon species diversity (Reinhold-Hurek et al., 2015). Therefore, the cognizance of the QS enables the regulation, thereby constraining bacterial communication (Figure 1). The inhibition strategies of QS are jointly called quorum-quenching, through which bacteria are ineffective in their interplay with each other. QS-mediates bacterial processes like growth, conjugation, bioluminescence, biofilm formation, siderophore production, and swarming (Barriuso et al., 2008). The threshold level of the initial plant growth-promoting rhizobacteria inoculum mediated by QS molecules strongly induces plant growth performance (Rodriguez et al., 2020). The plant rhizospheric region has a higher amount of AHL in comparison to the bulk soil, suggesting that these trigger bacterial colonization and establish a strong association between bacteria and plant roots (Vial et al., 2006).
Plant Growth-Promoting Rhizobacteria
Menendez and Garcia-Fraile (2017) classified plant growth-promoting rhizobacteria into extracellular plant growth-promoting rhizobacteria (e-PGPR) and intracellular plant growth-promoting rhizobacteria (i-PGPR). The plant growth-promoting rhizobacteria stimulate plant growth directly by the activity of phytostimulation and bio-fertilization, whereas indirectly through biopesticides or bio-control agents (Dwivedi and Dwivedi, 2002; Glick et al., 2007; Glick, 2012; Ngoma et al., 2012). The direct mechanism of plant growth-promoting rhizobacteria facilitates nutrient uptake or improvement in nutrient availability by nitrogen fixation (Cheng, 2008; Glick, 2012), solubilization of phosphorus and mineral nutrients, mineralizing organic compounds (Khan et al., 2010; Sharma et al., 2013), and phytohormones production including, IAA, ethylene, cytokinins, and gibberellins (Pliego et al., 2011; Upadhyay et al., 2016, 2019; South et al., 2021; Figure 2). On the basis of PGPR function, siderophore production may be considered as both a direct and an indirect mechanism (Ahmed and Holmstrom, 2014). The indirect mechanisms include antibiotic production (Sindhu et al., 2009), hydrolytic enzyme production (Dubey et al., 2014), induced systemic resistance (ISR), and exo-polysaccharides (EPS) production (Upadhyay et al., 2011; Figure 3).
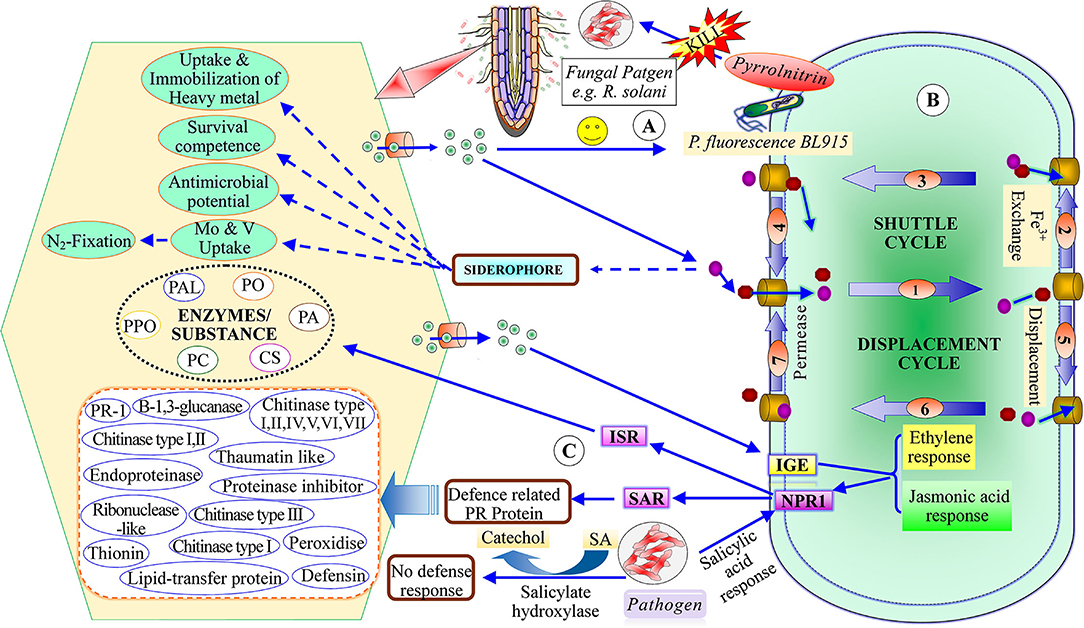
Figure 3. Indirect mechanism of plant growth-promoting rhizobacteria (A) recruited PGPR to produce antibiotic (B) siderophore production and plant growth-promotion, (C) mechanism of induced systematic resistance [ISR, induced systemic resistance; SAR, Systematic acquired resistance; IGE, Induce gene expression; NPRI, Non-expresser pathogenesis-related gene; SA, salicylic acid; PAL, phenylalanine amonislyase; PO, peroxidase; PPO, polyphenol oxidase; PC, poly-phenolic compounds; CS, chalcone synthase; PA, phytoalaxine.
Direct Mechanisms
IAA Production
IAA (indole-3-acetic acid), a product of the amino acid L-tryptophan (Yu et al., 2017), acts as a plant growth regulator. The IAA regulates the plant-growth through several cellular functions such as cell division, cell elongation and differentiation, increase in root length and root surface area, and the development of flowers (Gravel et al., 2007; Santner et al., 2009). About 80% of rhizobacteria produce auxins of microbial origin but with analogous functions, such as the auxins of plant origin (Patten and Glick, 1996; Ahemad and Kibret, 2014; Myresiotis et al., 2015; Keswani et al., 2020b. The IAA plays a crucial role in the interaction between plant and rhizobacteria and is synthesized by the associated plants and many microbes like plant growth-promoting rhizobacteria (Upadhyay et al., 2009). Several studies have demonstrated that the IAA production system was present in many bacterial species such as Agrobacterium tumefaciens, Alcaligenes faecalis, Agrobacterium tumefaciens (Costacurta and Vanderleyden, 1995), Pseudomonas syringae (Kosuge and Sanger, 1987), Streptomyces sp., B. subtilis spp. (Swain et al., 2007), Pseudomonas fluorescens (Oberhansli et al., 1991), and B. megaterium (Nghia et al., 2017).
Ethylene Production
Ethylene is a unique type of plant hormone secreted by plants and plant growth-promoting rhizobacteria (Babalola, 2010). Ethylene regulates plant physiological processes such as seed dormancy, enhances the formation of an adventitious root, leaf abscission, senescence of flower and leaf, and fruit ripening (Abeles et al., 1992). Different environmental conditions like salinity, drought, low temperature, pathogenic attack, and chemical exposure alter ethylene production and plant growth (Babalola, 2010). The optimum concentration of ethylene induces positive growth and development of plants (Saleem et al., 2007). Ethylene production directly depends on ACC production (Shaharoona et al., 2006).
Gibberellins and Cytokinin Production
Gibberellin (GA) is a tetracyclic di-terpenoid compound that acts as a plant hormone and is synthesized by many plant growth-promoting rhizobacteria. GA regulates many plant functions, such as fruit ripening, cell division, plant growth, etc. (Martinez et al., 2016; Plackett and Wilson, 2016). The endogenous GA concentration is raised by GA-producing plant growth-promoting rhizobacteria in the rhizospheric zone and it induces plant growth; for example, Leifsonia soli-SE134 and Enterococcus faecium-LKE12 in rice plant (Kang et al., 2014; Lee et al., 2015). Cytokinin is a plant hormone and is also synthesized by plant growth-promoting rhizobacteria. It is a member of adenine derivatives of the N-6 substituted group (Tsukanova et al., 2017), promotes cell cycle of the plant (Schaller et al., 2014), regulates plant growth and biosynthesis of chlorophyll (Cortleven and Schmülling, 2015). Plant growth-promoting rhizobacteria can produce Cytokinin and cause an increase in plant growth even under stressed conditions (O'Brien and Benkova, 2013). Tahir et al. (2017) demonstrated that B. subtilis-SYST2 induces the expression of cytokinin gene (s1CKX1) in tomato plants (Tahir et al., 2017). The plant growth-promoting rhizobacteria like Rhizobium sp., Azotobacter sp., Pantoea agglomerans, Pseudomonas fluorescens, Rhodospirillum rubrum, B. subtilis, and Paenibacillus polymyxa secrete cytokinins and/or gibberellins that promote plant growth (Kang et al., 2010).
Availability of Nutrients
The soil acts as a buzzword source of macro- and micro-nutrients for plant growth, while the compatible form of nutrients is a question of their availability to the plant. Most of the microorganisms can enhance the availability of nutrients in their compatible form to the plants (Upadhyay et al., 2019). Thus, soil fertility is one of the significant factors that governs diverse mechanisms of microorganisms. Nitrogen occurs in 78% of total atmospheric gases and cannot be utilized by higher plants directly. The nitrogen is utilized when it becomes fixed in the form of nitrogenous salt or ammonium ion. In nature, two kinds of N2 fixation occurs i.e., biological and non-biological (Raza et al., 2021). The biological nitrogen fixation (BNF) is carried out with the aid of bacteria, fungi, and algae, etc., which makes the atmospheric nitrogen available in the form of nitrogenous salt through the action of several plant growth-promoting rhizobacteria and blue-green algae (Dwivedi and Dwivedi, 2004). The BNF is borne out by two kinds of microbes, e.g., symbiotic and non-symbiotic (Tang et al., 2020; Raza et al., 2021). Symbiotic-BNF, a mutualistic link between the microbe and the plant, occurs in leguminous plants such as pea, chickpea, etc. (Cheng, 2008). In symbiotic-BNF, microbes enter the root and induce nodule formation (Cheng, 2008; Singh et al., 2022). Free nitrogen peroxide is converted into ammonia by nitrogen-fixing microorganisms and makes it available to the host (Ahemad and Kibret, 2014). This process involves a complex enzyme system known as nitrogenase (Gaby and Buckley, 2012), and the nif-gene, found in symbiotic as well as free-living bacterial systems (Reed et al., 2011). Nitrogen-fixing plant growth-promoting rhizobacteria such as Azospirillum (Montanez et al., 2009), Klebsiella (Arruda et al., 2013), Burkholderia (Chelius and Triplett, 2001), Bacillus (Zakry et al., 2012), and Pseudomonas (Piromyou et al., 2011) can significantly enhance crop productivity. Bacillus is associated with N2-fixing bacteria promoting plant growth and enhances the yield in non-leguminous cereals (Cakmakci et al., 2001; Ramirez and Mellado, 2005) such as maize (Pal, 1998), sugar beet, and barley (Sahin et al., 2004).
Mostly, phosphorus exists in the insoluble complex form like calcium phosphates in saline soil (Goldstein and Krishnaraj, 2007) and iron phosphates and aluminum phosphates in acidic soil (Mullen, 2005). Phosphorus is commonly present in soil in the range of 400–1,200 mg kg−1 (Fernández et al., 1988). Earth rocks are rich sources of phosphorus, in the form of primary apatites, and other primary minerals that previously existed in the geological age (Fernández et al., 1988). Indian soils are commonly deficient in phosphorus (Johri et al., 2003), and there are about 40 million tons of phosphorus deposits in India (Roychoudhury and Kaushik, 1989). Phosphate solubilizing bacteria (PSB), actinomycetes, and phosphate solubilizing fungi (PSF) solubilize the complex form of phosphates in the soil (Khan et al., 2007). The plant growth-promoting rhizobacteria induce the availability of phosphorus through mineralization and solubilization of the compounds of rock phosphates (organic and inorganic phosphorus; Nahas, 1996; Hilda and Fraga, 2000; Khiari and Parent, 2005). Bacterial species are superior phosphorus solubilizers than fungal species (Alam et al., 2002); PSB involves about 1–50% phosphorus solubilization followed by PSF, i.e., about 0.1–0.5% (Chen et al., 2006). Pseudomonas, Bacillus, Rhizobium, and Enterobacter, and fungal genera such as Penicillium and Aspergillus are the most efficient phosphate solubilizers (Kucey et al., 1989; Rodriguez and Fragal, 1990).
About 90% of potassium (K) in the soil exists in its complex form and is not available to the plants (Yadegari and Mosadeghzad, 2012; Zhang et al., 2013). Therefore, the solubilization of potassium is essential for K uptake by plants. K enhances seed germination, plant growth productivity, seedling vigor, and plantbiomass (Awasthi et al., 2011; Lynn et al., 2013; Meena et al., 2014; Zhang and Kong, 2014). The plant growth-promoting rhizobacteria can solubilize potassium rock (e.g., biotite, feldspar, illite, muscovite, orthoclase, and mica) into an available form of potassium for the plant. Potassium solubilizing plant growth-promoting rhizobacteria releases organic acids (e.g., oxalic acid, tartaric acids, gluconic acid, 2-ketogluconic acid, citric acid, malic acid, succinic acid, lactic acid, propionic acid, glycolic acid, malonic acid, fumaric acid, etc.) and inorganic acids (Awasthi et al., 2011; Etesami et al., 2017), which play an effective role in releasing K from K-bearing minerals (Hu et al., 2006; Liu et al., 2012; Keshavarz et al., 2013; Saiyad et al., 2015).
Different types of organic acids are involved in potassium solubilization, but the most prominent acids are tartaric acid, citric acid, succinic acid, α-ketogluconic acid, and oxalic acid released by KSB (Meena et al., 2014). Both aerobic and anaerobic plant growth-promoting rhizobacteria act as KSB, but most frequently aerobic bacteria that act as potassium solubilizers are Acidothiobacillus ferrooxidans, B. edaphicus, B. mucilaginosus, Burkholderia, Paenibacillus sp., and Pseudomonas (Etesami et al., 2017). Saprophytic bacteria, fungal strains, and actinomycetes also participate in K solubilization in a wide range (Gundala et al., 2013; Meena et al., 2014; Bakhshandeh et al., 2017). Thus, KSB–PGPR acts as a biofertilizer that improves soil fertility and plant growth. KSB is commonly found in different soil environments and can be isolated from rhizospheric and non-rhizospheric soil, including paddy soil (Bakhshandeh et al., 2017) and saline soil (Bhattacharya et al., 2016). KSB–PGPR is more effective for potassium solubilization (4.90 mg l−1) at a specific pH range of 6.5–8.0 (Badr et al., 2006). Similarly, Bacillus sp., Burkholderia sp., and Pseudomonas sp. can solubilize potassium at different temperatures and carbon sources from tea (Camellia sinensis; Bagyalakshmi et al., 2012).
Direct/Indirect Mechanism
Siderophore Production
Plant growth-promoting rhizobacteria secrete a low molecular weight (500–2,000 Da) iron-chelating compound called siderophore (Ahmed and Holmstrom, 2014). Siderophores are involved in the transportation and uptake of iron elements in the plant cells (Singh et al., 2022) and induce plant growth (Schwyn and Neilands, 1987; Hider and Kong, 2010; Ahmed and Holmstrom, 2014). Pseudomonas sp., Enterobacter genera, Bacillus, and Rhodococcus have a special capacity for binding iron through siderophores or siderochromes (Sah and Singh, 2015). Those microorganisms which cannot produce siderophores but use siderophores produced by other microorganisms are called xenosiderophores (Ahmed and Holmstrom, 2014). Production of siderophores occurs at specific conditions such as pH, temperature, and iron-concentration. The bacteria P.chlororaphis PCL1391 strain of rhizospheric roots of tomato plants can solubilize iron from the insoluble ferric oxides at neutral pH (Hernandez et al., 2004; Haas and Defago, 2005). Similarly, Sinha et al. (2018) reported that Psychrobacter piscatorii and Enterococcus casseliflavus from the Kerguelen Islands and B. cereus, Pseudoalteromonas tetraodonis, Psychrobacter pocilloporae, Pseudomonas weihenstephanensis, and Micrococcus aloeverae from the Prydz-Bay produced either hydroxamate-type siderophore or catecholate-type siderophores at 15–25°C with pH 8.5. Siderophores are produced by both aerobic and facultative anaerobic types of bacteria under the iron stress habitats (Neilands, 1995). Facultative aerobic bacterium such as Pseudomonas stutzeri CCUG 36651 produced siderophores under both aerobic and anaerobic conditions (Essen et al., 2007), Pseudomonas stutzeri CCUG 36651 produced four types of ferrioxamine siderophores under aerobic conditions but ferrioxamines siderophores under anaerobic conditions (Essen et al., 2007). Siderophore tightly binds with iron (Fe+3), then Fe+3-chelates move inside the cell through the cell membrane with the help of specific siderophore receptors (Haas and Defago, 2005). There are several types of siderophore binding proteins, such as permeases and ATPases involved in the transport of Fe+3 chelating compounds in the cell membrane, reported in gram-positive bacteria (Ahmed and Holmstrom, 2014). Gram-negative bacteria release many enzymes including periplasmic binding protein, outer membrane receptors, and cytoplasmic membrane protein, which help in the transport of iron-chelating compounds (Ahmed and Holmstrom, 2014).
Indirect Mechanisms of Plant Growth-Promoting Rhizobacteria
Indirect mechanisms involve antibiotics production, and hydrogen cyanide (HCN), ISR, and EPS production. The secretion of lytic enzymes of plant growth-promoting rhizobacteria induces plant growth (Maksimov et al., 2011; Upadhyay et al., 2016). The production of antibiotics is one of the most powerful bio-control tools for plant pathogens. Antibiotics are heterogeneous, low molecular weight, organic compounds secreted by microorganisms that help plant growth and metabolic activities (Duffy, 2003). The first antibiotic used as bio-control for plants was isolated from the bacterial species of fluorescent pseudomonads (Weller and Cook, 1983). Based on the mode of action, there are six classes of antibiotics, namely, phenazines, phloroglucinols, pyoluteorin, pyrrolnitrin, cyclic lipopeptides, and hydrogen cyanide (Haas and Defago, 2005). A large number of bacterial and fungal species secrete various types of antibiotics which induce plant growth by the suppression of phytopathogens (Maksimov et al., 2011). Pyrrolnitrin, an antibiotic isolated from P. fluorescens BL915 strain suppresses the growth of the fungal pathogen Rhizoctonia solani in cotton plants (Hill et al., 1994). Bacterial species of Pseudomonas secretes phenazine antibiotics that suppress various fungal pathogens including F. oxysporum and Gaeumannomyces graminis (Chin-A-Woeng et al., 2003). Bacillus sp. produces many types of antibiotics such as polymyxin, circulin, and colistin that suppress many plant diseases (Maksimov et al., 2011).
Hydrogen cyanide is a volatile secondary metabolite secreted by several gram-positive and gram-negative bacteria such as P.fluorescens, P. aeruginosa, Chromobacteria violaceum, etc. (Morrison and Askeland, 1983) that act as bio-control agents against soil-borne phytopathogens (Haas and Defago, 2005). HCN acts as a powerful inhibitor of various metallic enzymes including copper-bearing cytochrome C oxidase (Cho et al., 2013). HCN prevents many plant diseases like root-rot and black-rot diseases of tomato plants (Voisard et al., 1989) and also has nematicidal activities (Kang et al., 2010; Anderson and Kim, 2018). It is also very useful in agriculture and forestry due to the control of subterranean termites, Odontotermes obesus (Devi et al., 2007).
Induced systemic response (ISR) suppresses the disease of plants and animals that induces resistance against diseases (Van Loon et al., 1998). ISR induced by rhizobacteria shows resistance to several pathogens such as bacteria, fungi, and viruses (Korenblum et al., 2020). The plant growth-promoting rhizobacteria strain secretes salicylic acid that produces resistance to plant diseases, indicating that PGPR induces ISR (Krause et al., 2003; Idris et al., 2004). The treatment of tobacco plants with Bacillus rhizobacteria suppressed the impact of TMV (Tobacco Mosaic Virus) and also enhanced the height, weight, and yield of tobacco plants (Kloepper et al., 2004; Wang et al., 2009).
Exo-polysaccharides is a very active constituent of soil organic matter (Gouzou et al., 1993) produced by plant growth-promoting rhizobacteria under different soil environments like salinity (Upadhyay et al., 2011; Mohammed, 2018), drought, and normal conditions (Alami et al., 2000). EPS is the most important component of the extracellular matrix that shows two characters, slimy EPS and capsular EPS. EPS plays a significant role in several functions like biofilm formation (Bhaskar and Bhosle, 2005), bacterial cell protection (Mohammed, 2018), pollutants degradation (Fusconi and Godinho, 2002), plasma substituting capacity and bioremediation (Mohammed, 2018), maintenance of primary function of the cell, and antibacterial activity (Alami et al., 2000; Mohammed, 2018). EPS-producing PGPR influences root-adhering soils and establishes a balance between plant roots and microbial populations (Alami et al., 2000; Upadhyay et al., 2011).
Hydrolytic enzymes (HEs), mainly chitinase, glucanase, protease, and cellulase can hydrolyze chemical bonds of a wide variety of polymeric compounds including chitin, proteins, cellulose, hemicelluloses, and phytopathogenic DNA (Jadhav and Sayyed, 2016). HEs are capable of controlling phytopathogens through the hydrolysis of the cell wall, proteins, and DNA of pathogens. Thus, HEs play a major role in bio-control (Prathap and Ranjitha, 2015; Jadhav and Sayyed, 2016). The plant growth-promoting rhizobacteria act as an effective BCA through the lysis of phytopathogenic DNA (Garbeva et al., 2004). Microbial strains such as S. marcescens, B. cereus, B. subtilius, and B. thuringiensis can produce HEs that can control several phytopathogens namely R. solani, F. oxysporum, S. rolfsii, P. ultimum, etc. through different mechanisms (Someya et al., 2000).
Selective Recruitment of Plant Growth-Promoting Rhizobacteria
It is appealing to discuss what triggers the recruitment of microbiome in the rhizosphere. Microbiome in the rhizosphere affects plant growth and yield of the crop. There are two possibilities: (i) plant root creates an environment in the rhizosphere and attracts the useful microorganism/bacteria in the rhizosphere, (ii) soil already has a specified microbial population that allows the growth of selective plants. Here, we will discuss the first possibility which is more relevant to the root exudates and relevant to the remit of this review. Several previous reports demonstrated that the composition of root exudates varies with plant species, soil type, pH, and developmental stage (Berg and Smalla, 2009). The specific components of plant exudates promote the recruitment of specific microbiome/PGPR (Kamilova et al., 2006a). It is well-recognized that the microbial community in the rhizosphere is highly distinguished in different plant species (Edwards et al., 2015), the reason being the availability of different genotypes of the host plant. Based on the knowledge till date, the repertoire of the microbial community (especially the PGPR) around the plant root can be managed. It has been stated that some workers induced the recruitment of plant growth-promoting rhizobacteria by mutating the ABC transporter gene in plants (Badri et al., 2009). These results suggest the growth of a disease-resistant plant by influencing the root exudate components for the recruitment of plant growth-promoting rhizobacteria (Wei and Jousset, 2017). Natural disease-suppressive soil (term defined by Baker and Cook, 1974) can be achieved by manipulating the plant exudate resulting in the recruitment of the required PGPR (Exposito et al., 2017). Thus, differential components in plant exudates recruit microbial communities with a certain degree of specificity.
Future Perspectives
Many pieces of research demonstrated the diverse compounds of root exudates and their sensing toward beneficial microbes studied under confined and controlled laboratory conditions. Therefore, elucidation of the function of chemotaxis behavior of microbe-mediated compounds of root exudates is necessary for future research to make the success story at the field level. This will provide the structural foundation for a wide range of PGPR recognition by specific compounds (chemoattractants) of plant root exudates, respectively, and induce the growth of sustainable agriculture by chemotaxis to genetically modified plant growth-promoting rhizobacteria under degraded soil. Despite little knowledge of chemoattractant compounds of plant's root exudates, there are scopes for more researches for getting diverse advantages of root exudates through the application of emerging technology. Biotechnology is the utilization of biological resources for human welfare and industrial use. Plants have a well-developed system for the secretion of root exudates. Whether this secretion system can be utilized for biotechnological application is the central issue. Undoubtedly, there are several published reports, but the most fitting domains are (i) rhizosecretion and (ii) phytoremediation. Rhizosecretion is an alternative platform for manufacturing a large amount of pure target proteins (Drake et al., 2009). Borisjuk et al. (1999) demonstrated the production of recombinant proteins in plant root exudates. For this purpose, a genetically engineered plant with increased root mass can be used (Gaume et al., 2003). Rhizosecretion has been utilized for the production of antibodies. Madeira et al. (2016) have demonstrated that hyposecretion is an efficient and economical method for monoclonal antibody production. Catellani et al. (2020) recently evaluated the production of anti-fungal antibody scFvFc 2G8 using the root hair secretion system in Nicotiana benthamiana and Solanum lycopersicum. Another example of next-generation human therapeutic antibody production was demonstrated by Lonoce et al. (2016). They showed the production of tumor-targeting human-compatible monoclonalantibody H10 in hairy root plants (Lonoce et al., 2016).
Environmental pollution is the most devastating condition in the present ecological perspective. Phytoremediation is the process of removal of water and soil contaminants, especially by using the plant root system (Upadhyay and Edrisi, 2021). The role of root exudate in the removal of soil and water contaminants has been reported outstandingly (Gleba et al., 1999; Ma et al., 2016; Chen et al., 2020). Different components of root exudates play a specific role in the removal of certain contaminants from the soil and groundwater. Lu et al. (2017) reported that glucose present in plant exudates can remove the soil pyrene by promoting soil dehydrogenase activity. Palmitic acid present in plant exudates of tall fescue showed promising results in the removal of petroleum contaminants from the soil (Liu et al., 2015). Similarly, by manipulating the components of root exudates, it can be used for the removal of targeted pollutants from the soil and ground water. There are two main types of phytoremediation processes exploiting the plant exudates (i) rhizosphere biodegradation (plant root exudate recruited microbe mediated degradation of pollutants) and (ii) phytostabilization (plant exudate components immobilize the pollutants).
Concluding Remarks
Since the 1980s, plant growth-promoting rhizobacterial inoculants have been developed, but few of them revealed irregular performance at the field level. Although several researchers have developed the consortia of plant growth-promoting rhizobacteria, but with more or less similar outcomes in the farmer's field, the solution to these problems is somehow hidden in the root exudates and root microenvironment. Thus, the present review has concentrated on the remarkable views for future research to manage the challenges at the field level with PGPR inoculants. Several components of root exudates have functional interplay with PGPR either directly or through their gene expression. The recruitment of plant growth-promoting rhizobacteria through root exudates can enhance plant growth-promoting rhizobacteria root colonization, specifically, and induce close sustainable relationships between them for a long time. The hypothesis of specific recruitment would address the key gap for warranting the perfect plant growth-promoting rhizobacteria candidate and opening a new horizon of research in biofertilizer technology. It would be a promising technique for reducing the asymmetrical performance of plant growth-promoting rhizobacteria in the farmer's field.
Author Contributions
SU, AS, PC, PD, and AB contributed to conceptualization and visualization of the present review and writing the original draft. SU and AS designed the figures and tables. DJ, PD, and GC contributed to re-structuring the review. PD, BS, VDR, and TM contributed special remarks and edited the review for final submission. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This review work was supported by the Department of Environmental Science, V.B.S. Purvanchal University, Jaunpur, India. VDR and TM would like to recognize the support from the Ministry of Science and Higher Education of the Russian Federation (no. 220-5234-7520), laboratory “Soil Health” Southern Federal University.
References
Abdel-Lateif, K., Bogusz, D., and Hocher, V. (2012). The role of flavonoids in the establishment of plant roots endosymbioses with arbuscular mycorrhiza fungi, rhizobia and frankia bacteria. Plant Signal Behav. 7, 636–641. doi: 10.4161/psb.20039
Abeles, F. B., Morgan, P. W., and Saltveit, M. E. Jr. (1992). Ethylene in Plant Biology, 2nd Edn. San Diego, CA: Academic Press.
Abisado, R. G., Benomar, S., Klaus, J. R., Dandekar, A. A., and Chandler, J. R. (2018). Bacterial quorum sensing and microbioll community interactions. mBio 9, e01749–18. doi: 10.1128/mBio.01749-18
Adesina, M. F., Lembke, A., Costa, R., Speksnijder, A., and Smalla, K. (2007). Screening of bacterial isolates from various European soils for in vitro antagonistic activity towards Rhizoctonia solani and Fusarium oxysporum: site dependent composition and diversity revealed. Soil Biol. Biochem. 39, 2818–2828. doi: 10.1016/j.soilbio.2007.06.004
Ahemad, M., and Kibret, M. (2014). Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J. King Saud Univer. Sci. 26, 1–20. doi: 10.1016/j.jksus.2013.05.001
Ahmed, E., and Holmstrom, S. J. (2014). Siderophores in environmental research: roles and applications. Microbiol. Biotechnol. 7, 196–208. doi: 10.1111/1751-7915.12117
Akiyama, K., Matsuzaki, K., and Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827. doi: 10.1038/nature03608
Alam, S., Khalil, S., Ayub, N., and Rashid, M. (2002). in vitro solubilization of inorganic phosphate by phosphate solubilizing microorganism (PSM) from maize rhizosphere. Int. J. Agric. Biol. 4, 454–458.
Alami, Y., Achouk, W., Marol, C., and Heulin, T. (2000). Rhizosphere soil aggregation and plant growth promotion of sunflowers by an exopolysaccharide-producing Rhizobium sp. strain isolated from sunflower roots. Appl. Environ. Microbiol. 66, 3393–3398. doi: 10.1128/AEM.66.8.3393-3398.2000
Anderson, A. J., and Kim, Y. C. (2018). Biopesticides produced by plant-probiotic Pseudomonas chlororaphisisolates. Crop Prot. 105, 62–69. doi: 10.1016/j.cropro.2017.11.009
Ankati, S., and Podile, A. R. (2019). Metabolites in the root exudates of groundnut changes during interection with plant growth promoting rhizobacteria in a strain-specific manner. J. Plant Physiol. 24, 153057. doi: 10.1016/j.jplph.2019.153057
Arruda, L., Beneduzi, A., Martins, A., Lisboa, B., Lopes, C., and Bertolo, F. (2013). Screening of rhizobacteria from maize (Zea mays L.) in Rio Grande do Sul State (South Brazil) and analysis of their potential to improve plant growth. Appl. Soil Ecol. 63, 15–22. doi: 10.1016/j.apsoil.2012.09.001
Awasthi, R., Tewari, R., and Nayyar, H. (2011). Synergy between plants and P-solubilizing microbes in soils: effects on growth and physiology of crops. Int. Res. J. Microbiol. 2, 484–503.
Babalola, O. O. (2010). Ethylene quantification in three rhizobacterial isolates from Striga hermonthica-infested maize and sorghum Egypt. J. Biol. 12, 1–5.
Bacilio-Jimenez, M., Aguilar-Flores, S., Ventura-Zapata, E., Perez-Campos, E., Bouquelet, S., and Zenteno, E. (2003). Chemical characterization of root exudates from rice (Oryza sativa) and their effects on the chemotactic response of endophytic bacteria. Plant Soil 249, 271–277. doi: 10.1023/A:1022888900465
Badr, M. A., Shafei, A. M., and Sharaf El-Deen, S. H. (2006). The dissolution of K and P-bearing minerals by silicate dissolving bacteria and their effect on sorghum growth. Res. J. Agri. Biol. Sci. 2, 5–11.
Badri, D. V., Chaparro, J. M., Zhang, R., Shen, Q., and Vivanco, J. M. (2013). Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 288, 4502–4512. doi: 10.1074/jbc.M112.433300
Badri, D. V., Quintana, N., El Kassis, E. G., Kim, H. K., Choi, Y. H., Sugiyama, A., et al. (2009). An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 151, 2006–2017. doi: 10.1104/pp.109.147462
Badri, D. V., and Vivanco, J. M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32, 666–681. doi: 10.1111/j.1365-3040.2009.01926.x
Baetz, U., and Martinoia, E. (2014). Root exudates: the hidden part of plant defense. Trends Plant Sci. 2, 90–98. doi: 10.1016/j.tplants.2013.11.006
Bagyalakshmi, B., Ponmurugan, P., and Balamurugan, A. (2012). Impact of different temperature, carbon and nitrogen sources on solubilization efficiency of native potassium solubilizing bacteria from tea (Camellia sinensis). J. Biol. Res 3, 36–42.
Baker, K. F., and Cook, R. J. (1974). Biological Control of Plant Pathogens. San Fransisco, CA: Freeman and Company.
Bakhshandeh, E., Pirdashti, H., and Lendeh, K. S. (2017). Phosphate and potassium-solubilizing bacteria effect on the growth of rice. Ecol. Eng. 103, 164–169. doi: 10.1016/j.ecoleng.2017.03.008
Barriuso, J., Solano, B. R., Fray, R. G., Camara, M., Hartmann, A., and Gutierrez, M. F. J. (2008). Transgenic tomato plants alter quorum sensing in plant growth-promoting rhizobacteria. Plant Biotechnol. 6, 442–452. doi: 10.1111/j.1467-7652.2008.00331.x
Bashir, S., Iqbal, A., Hasnain, S., and White, J. F. (2021). Screening of sunfower associated bacteria as biocontrol agents for plant growth promotion. Arch. Microbiol. 203, 4901–4912. doi: 10.1007/s00203-021-02463-8
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Bhaskar, P. V., and Bhosle, N. B. (2005). Microbial extracellular polymeric substances in marine biogeochemical processes. Curr. Sci. 88, 45–53.
Bhattacharya, S., Bachani, P., Jain, D., Patidar, S. K., and Mishra, S. (2016). Extraction of potassium from K-feldspar through potassium solubilization in the halophilic acinetobacter soli (MTCC 5918) isolated from the experimental salt farm. Int. J. Min. Proc. 152, 53–57. doi: 10.1016/j.minpro.2016.05.003
Bhattacharyya, P. N., and Jha, D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350. doi: 10.1007/s11274-011-0979-9
Bhojiya, A. A., Joshi, H., Upadhyay, S. K., Srivastava, A. K., Pathak, V. V., Pandey, V. C., et al. (2021). Screening and optimization of zinc removal potential in Pseudomonas aeruginosa-HMR1 and its plant growth-promoting attributes. Bull. Environ. Contam. Toxicol. 108, 468–477. doi: 10.1007/s00128-021-03232-5
Blom, J., Rueckert, C., Niu, B., Wang, Q., and Borriss, R. (2012). The complete genome of Bacillus amyloliquefaciens sub sp. plantarum CAU B946 contains a gene cluster for non ribosomal synthesis of iturin A. J. Bacteriol. 194, 1845–1846. doi: 10.1128/JB.06762-11
Bonkowski, M., Villenave, C., and Griffiths, B. (2009). Rhizosphere fauna: the functional and structural diversity of intimate interactions of soil fauna with plant roots. Plant Soil. 321, 213–233. doi: 10.1007/s11104-009-0013-2
Bonsall, R. F., Weller, D. M., and Thomashow, L. S. (1997). Quantification of 2,4-diacetylphloroglucinol produced by fluorescent Pseudomonas spp. in vitro and in the rhizosphere of wheat. Appl. Environ. Microbiol. 63, 951–955. doi: 10.1128/aem.63.3.951-955.1997
Borisjuk, N. V., Borisjuk, L. G., Logendra, S., Petersen, F., Gleba, Y., and Raskin, I. (1999). Production of recombinant proteins in plant root exudates. Nat. Biotechnol. 17, 466–469. doi: 10.1038/8643
Borriss, R. (2011). “Use of plant-associated Bacillus strains as biofertilizers and biocontrol agents in agriculture,” in Bacteria in Agrobiology: Plant Growth Responces, ed D. K. Maheshwari (Berlin: Springer), 41–76. doi: 10.1007/978-3-642-20332-9_3
Bowya, T., and Balachandar, D. (2020). Rhizosphere engineering through exogenous growth-regulating small molecules improves the colonizing efficiency of a plant growth-promoting rhizobacterium in rice. 3 Biotech 10, 277. doi: 10.1007/s13205-020-02275-5
Braud, A., Jezequel, K., Bazot, S., and Lebeau, T. (2009). Enhanced phytoextraction of an agricultural Cr-, Hg- and Pb-contaminated soil by bio-augmentation with siderophore producing bacteria. Chemosphere 74, 280–286. doi: 10.1016/j.chemosphere.2008.09.013
Cai, T., Cai, W., Zhang, J., Zheng, H., Tsou, A. M., Xiao, L., et al. (2009). Host legume-exuded antimetabolites optimize the symbiotic rhizosphere. Mol. Microbiol. 73, 507–517. doi: 10.1111/j.1365-2958.2009.06790.x
Cakmakci, R., Kantar, F., and Sahin, F. (2001). Effect of N2-fixing bacterial inoculations on yield of sugar beet and barley. J. Plant Nutr. Soil Sci. 164, 527–531. doi: 10.1002/1522-2624(200110)164:5<527::AID-JPLN527>3.0.CO;2-1
Canellas, L. P., and Olivares, F. L. (2017). Production of border cells and colonization of maize root tips by Herbaspirillumseropedicae are modulated by humic acid. Plant Soil. 417, 403–413. doi: 10.1007/s11104-017-3267-0
Cannesan, M. A., Durand, C., Burel, C., Gangneux, C., Lerouge, P., Ishii, T., et al. (2012). Effect of arabinogalactan proteins from the root caps of pea and Brassica napus on Aphanomyces euteiches zoospore chemotaxis and germination. Plant Physiol. 159, 1658–1670. doi: 10.1104/pp.112.198507
Catellani, M., Lico, C., Cerasi, M., Massa, S., Bromuro, C., Torosantucci, A., et al. (2020). Optimised production of an anti-fungal antibody in Solanaceae hairy roots to develop new formulations against Candida albicans. BMC Biotechnol. 20, 15. doi: 10.1186/s12896-020-00607-0
Cesari, A., Paulucci, N., Lopez-Gomez, M., Hidalgo-Castellanos, J., Pla, C. L., and Dardanelli, M. S. (2019). Restrictive water condition modifies the root exudates composition during peanut-PGPR interaction and conditions early events, reversing the negative effects on plant growth Plant. Physiol. Biochem. 142, 519–527. doi: 10.1016/j.plaphy.2019.08.015
Chamam, A., Sanguin, H., Belllvert, F., Meiffren, G., Comte, G., Wisniewski-Dye, F., et al. (2013). Plant secondary metabolite profiling evidences strain-dependent effect in the Azospirillum- Oryza sativa association. Phytochemistry 87, 65–77. doi: 10.1016/j.phytochem.2012.11.009
Chandler, D., Davidson, G., Grant, W. P., Greaves, J., and Tatchell, G. M. (2008). Microbial biopesticides for integrated crop management: an assessment of environmental and regulatory sustainability. Trends Food Sci. Tech. 19, 275–283. doi: 10.1016/j.tifs.2007.12.009
Chaturvedi, H., Singh, V., and Gupta, G. (2016). Potential of bacterial endophytes as plant growth promoting factors. J. Plant Pathol. Microbiol. 7, 1–6. doi: 10.4172/2157-7471.1000376
Chelius, M. K., and Triplett, E. W. (2001). The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 3, 252–263. doi: 10.1007/s002480000087
Chen, C., Li, Z., Li, S., Deng, N., and Mei, P. (2020). Effects of root exudates on the activation and remediation of cadmium ion in contaminated soils. Environ. Sci. Pollut. Res. Int. 27, 2926–2934. doi: 10.1007/s11356-019-07263-8
Chen, S., Waghmode, T. R., Sun, R., Kuramae, E. E., Hu, C., and Liu, B. (2019). Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 7, 136. doi: 10.1186/s40168-019-0750-2
Chen, X. H., Koumoutsi, A., Scholz, R., Eisenreich, A., Schneider, K., Heinemeyer, I., et al. (2007). Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 25, 1007–1014. doi: 10.1038/nbt1325
Chen, Y. P., Rekha, P. D., Arunshen, A. B., Lai, W. A., and Young, C. C. (2006). Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl. Soil Ecol. 34, 33–41. doi: 10.1016/j.apsoil.2005.12.002
Cheng, Q. (2008). Perspectives in biological nitrogen fixation research. J. Integr. Plant Biol. 50, 786–798. doi: 10.1111/j.1744-7909.2008.00700.x
Chin-A-Woeng, T. F., Bloemberg, G. V., and Lugtenberg, B. J. (2003). Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol. 157, 503–523. doi: 10.1046/j.1469-8137.2003.00686.x
Cho, S. M., Kim, Y. H., Anderson, A. J., and Kim, Y. C. (2013). Nitric oxide and hydrogen peroxide production are involved in systemic drought tolerance induced by 2R, 3R-butanediol in Arabidopsis thaliana. Plant Pathol. 29, 427–434. doi: 10.5423/PPJ.OA.07.2013.0069
Chowdhury, S. P., Hartmann, A., Gao, X., and Borriss, R. (2015). Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 – a review. Front. Microbiol. 6, 780. doi: 10.3389/fmicb.2015.00780
Combes-Meynet, E., Potheir, J., Moenne-Loccoz, Y., and Prigent-Combaret, C. (2011). The pseudomonas secondary metabolite 2, 4-diacetylphloroglucinol is a signal inducing rhizoplane expression of azospirillum genes involve in plant-growth promotion. Mol. Plant Microbe Interact. 24, 271–284. doi: 10.1094/MPMI-07-10-0148
Compant, S., Samad, A., Faist, H., and Sessitsch, A. (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Coronado, C., Zuanazzi, J. A. S., Sallaud, C., Quirion, J. C., Esnault, R., Husson, H. P., et al. (1995). Alfalfa root flavonoid production 1s nitrogen regulated. Plant Physiol. 108, 533–542. doi: 10.1104/pp.108.2.533
Cortleven, A., and Schmülling, T. (2015). Regulation of chloroplast development and function by cytokinin. J. Exp. Bot. 66, 4999–5013. doi: 10.1093/jxb/erv132
Costacurta, A., and Vanderleyden, J. (1995). Synthesis of phytohormones by plant associated bacteria. Crit. Rev. Microbiol. 2, 1–18. doi: 10.3109/10408419509113531
Creus, M. C., Graziano, M., Casanovas, E. M., Pereyra, M. A., Simontacchi, M., Puntarulo, S., et al. (2005). Nitric oxide is involved in the Azospirillum brasilense-induced lateral root formation in tomato. Planta 221, 297–303. doi: 10.1007/s00425-005-1523-7
De-la-Pena, C., Lei, Z., Watson, B. S., Sumner, L. W., and Vivanco, J. M. (2008). Root-microbe communication through protein secretion. J. Biol. Chem. 283, 25247–25255. doi: 10.1074/jbc.M.801967200
DESA UN (2015). World Population Prospects: The 2015 Revision, Key Findings and Advance Tables. Working Paper No ESA/P/WP, 241. New York, NY: United Nations, Department of Economic and Social Affairs, Population Division.
Devi, K. K., Seth, N., Kothamasi, S., and Kothamasi, D. (2007). Hydrogen cyanide producing rhizobacteria killsubterranean termite Odontotermesobesus(Rambur) by cyanide poisoning under in Vitroconditions. Curr. Microbiol. 54, 74–78. doi: 10.1007/s00284-006-0473-z
D'Haeze, W., Gao, M., De, R., Rycke, Van, M., Montagu, E. G., and Holsters, M. (1998). Roles for azorhizobial nod factors and surface polysaccharides in intercellular invasion and nodule penetration, respectively. Mol. Plant Microbe Interact. 11, 999–1008. doi: 10.1094/MPMI.1998.11.10.999
Donn, S., Kirkegaard, J. A., Perera, G., Richardson, A. E., and Watt, M. (2015). Evolution of bacterial communities in the wheat crop rhizosphere. Environ. Microbiol. 17, 610–621. doi: 10.1111/1462-2920.12452
Drake, P. M. W., Barbi, T., Sexton, A., McGowan, E., Stadlmann, J., Navarre, C., et al. (2009). Development of rhizo secretion as a production system for recombinant proteins from hydroponic cultivated tobacco. FASEB J. 23:3581–3589. doi: 10.1096/fj.09-131771
Dubey, R. C., Khare, S., Kumar, P., and Maheshwari, D. K. (2014). Combined effect of chemical fertilisers and rhizosphere-competent Bacillus subtilis BSK17 on yield of Cicer arietinum. Arch. Phytopathol. Plant Protect. 47, 2305–2318. doi: 10.1080/03235408.2013.876744
Duffy, B. (2003). Pathogen self-defense: mechanisms to counteract microbial antagonism. Annu. Rev. Phytopathol. 41, 501–538. doi: 10.1146/annurev.phyto.41.052002.095606
Dwivedi, P., and Dwivedi, S. K. (2002). “Biopesticidal management of plant diseases,” in Microbial Diversity: Status and Potential, eds S. C. Tiwari, and G. D. Sharma (Guwahati: Scientific Book Publishers), 220–235.
Dwivedi, P., and Dwivedi, S. K. (2004). “Nitrogen fixation and biofertilizers,” in Environmental Microbiology and Biotechnology, eds D. P. Singh, and S. K. Dwivedi (New Delhi: New Age International Pvt Ltd), 126–148.
Edwards, J., Johnson, C., Santos-Medellin, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, 911–920. doi: 10.1073/pnas.1414592112
Essen, S. A., Johnsson, A., Bylund, D., Pedersen, K., and Lundstrom, U. S. (2007). Siderophore production by Pseudomonas stutzeri under aerobic and anaerobic conditions. Appl. Environ. Microbiol. 73, 5857–5864. doi: 10.1128/AEM.00072-07
Etesami, H., Emami, S., and Alikhani, H. A. (2017). Potassium solubilizing bacteria (KSB): mechanisms, promotion of plant growth, and future prospects - a review. J. Soil Sci. Plant Nutr. 17, 897–911. doi: 10.4067/S0718-95162017000400005
Exposito, R. G., de Bruijn, I., Postma, J., and Raaijmakers, J. M. (2017). Current insight into the role of rhizospheric bacteria in disease suppressive soil. Front. Microbiol. 8, 2529. doi: 10.3389/fmicb.2017.02529
Fang, W., and St Leger, R. J. (2010). Mrt a gene unique to fungi, encodes an oligosaccharide transporter and facilitates rhizosphere competency in Metarhiziumrobertsii. Plant Physiol. 154, 1549–1557. doi: 10.1104/pp.110.163014
Fernández, C., Novo, R., Microbiana, V., and Suelo, E. (1988). II. La habana: Editorial Puebloy Educación fibroblasts. Life Sci. 52, 1909–1915.
Fusconi, R., and Godinho, M. J. L. (2002). Screening for exopolysaccharide producing bacteria from sub- tropical polluted groundwater. Braz. J. Biol. 62, 363–369. doi: 10.1590/S1519-69842002000200020
Gaby, J. C., and Buckley, D. H. (2012). A comprehensive evaluation of PCR primers to amplify the nif H gene of nitrogenase. PLoS ONE 7, e42149. doi: 10.1371/journal.pone.0042149
Garbeva, P., van Veen, J. A., and Elas, J. D. (2004). Assessment of the diversity, and antagonism towards Rhizoctonia solani AG3, of Pseudomonas species in soil from different agricultural regimes. FEMS Microbiol. Ecol. 47, 51–64. doi: 10.1016/S0168-6496(03)00234-4
Gaume, A., Komarnytsky, S., Borisjuk, N., and Raskin, I. (2003). Rhizosecretion of recombinant proteins from plant hairy roots. Plant Cell Rep. 21, 1188–1193. doi: 10.1007/s00299-003-0660-3
Ghosh, P. K., Saha, P., Mayilraj, S., and Maiti, T. K. (2013). Role of IAA metabolizing enzymes on production of IAA in root, nodule of Cajanus cajanand its PGP Rhizobium sp. Biocatal. Agric. Biotechnol. 2, 234–239. doi: 10.1016/j.bcab.2013.04.002
Gleba, D., Borisjuk, N. V., Borisjuk, L. G., Kneer, R., Poulev, A., Skarzhinskaya, M., et al. (1999). Use of plant roots for phytoremediation and molecular farming. Proc. Natl. Acad. Sci. U.S.A. 96, 5973–5977. doi: 10.1073/pnas.96.11.5973
Glick, B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 12, 1–15. doi: 10.6064/2012/963401
Glick, B. R., Todorovic, B., Czarny, J., Cheng, Z., and Duan, J. (2007). Promotion of plant growth by bacterial ACC deaminase. Crit. Rev. Plant Sci. 26, 227–242. doi: 10.1080/07352680701572966
Goh, C. H., Veliz, V. D. F., Nicotra, A. B., and Mathesius, U. (2013). The impact of beneficial plant-associated microbes on plant phenotypic plasticity. J. Chem. Ecol. 7, 826–839. doi: 10.1007/s10886-013-0326-8
Goldstein, A. H., and Krishnaraj, P. U. (2007). “Phosphate solubilizing microorganisms vs. phosphate mobilizing microorganisms: what separates a phenotype from a trait?” in First International Meeting on Microbial Phosphate Solubilization, eds E. Velazquez, and C. Rodriguez-Barrueco (Dordrecht: Springer), 203–213. doi: 10.1007/978-1-4020-5765-6_31
Gouzou, L., Burtin, R., Philippy, R., Bartoli, F., and Heulin, T. (1993). Effect of inoculation with bacillus polymyxa on soil aggregation in the wheat rhizosphere: preliminary examination. Geoderma 56, 479–491. doi: 10.1016/0016-7061(93)90128-8
Gravel, V., Antoun, H., and Tweddell, R. J. (2007). Growth stimulation and fruit yield improvement of greenhouse tomato plants by inoculation with Pseudomonas putida or Trichoderma atroviride: Possible role of indole acetic acid (IAA) soil. Biol. Biochem. 39, 1968–1977. doi: 10.1016/j.soilbio.2007.02.015
Gundala, P. B., Chinthala, P., and Sreenivasulu, B. (2013). A new facultative alkaliphilic, potassium solubilizing, Bacillus Sp. SVUNM9 isolated from mica cores of Nellore District, Andhra Pradesh, India. Res. Rev. J. Microbiol. Biotechnol. 2, 1–7.
Gupta, V., Kumar, G. N., and Buch, A. (2020). Colonization by multi-potential Pseudomonas aeruginosa P4 stimulates (Arachis hypogaea L.) growth, defence physiology and root system functioning to benefit the root-rhizobacterial interface. J. Plant Physiol. 248, 153144. doi: 10.1016/j.jplph.2020.153144
Haas, D., and Defago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Haldar, S., and Sengupta, S. (2015). Plant-microbe cross-talk in the rhizosphere: insight and biotechnological potential. Open Microbiol. J. 9, 1–7. doi: 10.2174/1874285801509010001
Hao, K., He, P., Blom, J., Rueckert, C., Mao, Z., Wu, Y., et al. (2012). The genome of plant growth-promoting Bacillus amyloliquefaciens sub sp. plantarum strain YAU B9601-Y2 contains a gene cluster for mersacidin synthesis. J. Bacteriol. 194, 3264–3265. doi: 10.1128/JB.00545-12
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma species-opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Hartmann, A., Rothballer, M., Schmid, M., and Lorenz, H. (2008). A pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312, 7–14. doi: 10.1007/s11104-007-9514-z
Hartmann, A., Schmid, M., Tuinen, D., and Berg, G. (2009). Plant-driven selection of microbes. Plant Soil 321, 235–275. doi: 10.1007/s11104-008-9814-y
Hassan, M. K., McInroy, J. A., and Kloepper, J. W. (2019). The interactions of rhizodeposits with plant growth-promoting rhizobacteria in the rhizosphere: a review. Agriculture 9, 142. doi: 10.3390/agriculture9070142
Hernandez, M. E., Kappler, A., and Newman, D. K. (2004). Phenazines and other redox-active antibiotics promote microbial mineral reduction. Appl. Environ. Microbiol. 70, 921–928. doi: 10.1128/AEM.70.2.921-928.2004
Hider, R. C., and Kong, X. (2010). Chemistry and biology of siderophores. Nat. Product Rep. 27, 637–657. doi: 10.1039/b906679a
Hilda, R., and Fraga, R. (2000). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotech. Adv. 17, 319–359.
Hill, D. S., Stein, J. I., Torkewitz, N. R., Morse, A. M., Howell, C. R., Pachlatko, J. P., et al. (1994). Cloning of genes involved in the synthesis of pyrrolnitrin from Pseudomnas fluorescens and role of pyrrolnitrin synthesis in biological control of plant disease. Appl. Environ. Microbiol. 60, 78–85. doi: 10.1128/aem.60.1.78-85.1994
Hu, L., Robert, C. A. M., Cadot, S., Zhang, X., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9, 2738. doi: 10.1038/s41467-018-05122-7
Hu, X., Chen, J., and Guo, J. (2006). Two phosphate-and potassium-solubilizing bacteria isolated from Tianmu Mountain, Zhejiang, China. World J. Microbiol. Biotechnol. 22, 983–990. doi: 10.1007/s11274-006-9144-2
Idris, E. E. S., Bochow, H., Ross, H., and Borriss, R. (2004). Use of Bacillus subtilis as biocontrol agent. Phytohormone-like action of culture filtrates prepared from plant growth-promoting Bacillus amyloliquefaciens FZB24, FZB42, FZB45 and Bacillus subtilis FZB37. J. Plant Dis. Prot. 111, 583–597.
Jadhav, H. P., and Sayyed, R. Z. (2016). Hydrolytic enzymes of rhizospheric microbes in crop protection. MOJ Cell Sci. Rep. 3, 135–136. doi: 10.15406/mojcsr.2016.03.00070
Johri, B. N., Sharma, A., and Virdi, J. S. (2003). Rhizobacterial diversity in India and its in?uence on soil and plant health. Adv. Biochem. Eng. Biotechnol. 8, 49–89. doi: 10.1007/3-540-36488-9_2
Kamilova, F., Kravchenko, L. V., Shaposhnikov, A. I., Azarova, T., Makarova, N., and Lugtenberg, B. (2006b). Organic acids, sugars, and l-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere. Bacter. Mol. Plant Microbe Interact. 19, 250–256. doi: 10.1094/MPMI-19-0250
Kamilova, F., Kravchenko, L. V., Shaposhnikov, A. I., Makarova, N., and Lugtenberg, B. (2006a). Effects of the tomato pathogen Fusarium oxysporum f. sp. radicis-lycopersici and of the biocontrol bacterium Pseudomonas fluorescens WCS365 on the composition of organic acids and sugars in tomato root exudates. Mol. Plant Microbe Interact. 19, 1121–1126. doi: 10.1094/MPMI-19-1121
Kang, B. G., Kim, W. T., Yun, H. S., and Chang, S. C. (2010). Use of plant growth-promoting rhizobacteria to control stress responses of plant roots. Plant Biotechnol. Rep. 4, 179–183. doi: 10.1007/s11816-010-0136-1
Kang, S. M., Khan, A. L., You, Y. H., Kim, J. G., Kamran, M., and Lee, I. J. (2014). Gibberellin production by newly isolated strain Leifsonia soli SE134 and its potential to promote plant growth. J. Microbiol. Biotechnol. 24, 106–112. doi: 10.4014/jmb.1304.04015
Kawasaki, A., Donn, S., Ryan, P. R., Mathesius, U., Devilla, R., Jones, A., et al. (2016). Microbiome and exudates of the root and rhizosphere of Brachypodiumdistachyon, a model for wheat. PLoS ONE 11, e0164533. doi: 10.1371/journal.pone.0164533
Keller, L., and Surette, M. G. (2006). Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4, 249–258. doi: 10.1038/nrmicro1383
Keshavarz, Z. J., Aliasgharzad, N., Oustan, S., Emadi, M., and Ahmadi, A. (2013). Isolation and characterization of potassium solubilizing bacteria in some Iranian soils. Arch. Agron. Soil Sci. 59, 1713–1723. doi: 10.1080/03650340.2012.756977
Keswani, C., Singh, H. B., García-Estrada, C., Caradus, J., He, Y. W., Mezaache-Aichour, S., et al. (2020a). Antimicrobial secondary metabolites from agriculturally important bacteria as next-generation pesticides. Appl. Microbiol. Biotechnol. 104, 1013–1034. doi: 10.1007/s00253-019-10300-8
Keswani, C., Singh, S. P., Cueto, L., Garcia-Estrada, C., Mezaache-Aichour, S., Glare, T. R., et al. (2020b). Auxins of microbial origin and their use in agriculture. Appl. Microbiol. Biotechnol. 104, 8549–8565. doi: 10.1007/s00253-020-10890-8
Khan, M. S., Zaidi, A., Ahemad, M., Oves, M., and Wani, P. A. (2010). Plant growth promotion by phosphate solubilizing fungi-current perspective. Arch. Agron. Soil Sci. 56, 73–98. doi: 10.1080/03650340902806469
Khan, M. S., Zaidi, A., and Wani, P. A. (2007). Role of phosphate-solubilizing microorganisms in sustainable agriculture - a review. Agron. Sustain. Dev. 27, 29–43. doi: 10.1051/agro:2006011
Khiari, L., and Parent, L. E. (2005). Phosphorus transformations in acid light-textured soils treated with dry swine manure. Can. J. Soil Sci. 85, 75–87. doi: 10.4141/S03-049
Kloepper, J. W., Ryu, C. M., and Zhang, S. (2004). Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94, 1259–1266. doi: 10.1094/PHYTO.2004.94.11.1259
Korenblum, E., Donga, Y., Szymanski, J., Panda, S., Jozwiak, A., Massalha, H., et al. (2020). Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. PNAS 117, 3874–3883. doi: 10.1073/pnas.1912130117
Kosuge, T., and Sanger, M. (1987). Indole acetic acid, its synthesis and regulation: basis for tumorigen city in plant disease. Recent. Adv. Phytochem. 20, 147–161. doi: 10.1007/978-1-4684-8056-6_6
Krause, M. S., De Ceuster, T. J. J., Tiquia, S. M., Michel, J.r, F. C. Madden, L. V., et al. (2003). Isolation and characterization of rhizobacteria from composts that suppress the severity of bacterial leaf spot of radish. Phytopathology 93, 1292–1300. doi: 10.1094/PHYTO.2003.93.10.1292
Kucey, R. M. N., Janzen, H. H., and Legget, M. E. (1989). Microbial mediated increases in plant available phosphorus. Adv. Agron. 42, 199–228. doi: 10.1016/S0065-2113(08)60525-8
Ladygina, N., and Hedlund, K. (2010). Plant species influence microbial diversity and carbon allocation in the rhizosphere. Soil Biol. Biochem. 42, 162–168. doi: 10.1016/j.soilbio.2009.10.009
Lee, K. E., Radhakrishnan, R., Kang, S. M., You, Y. H., Joo, G. J., Lee, I. J., et al. (2015). Enterococcus faecium LKE12 cell-free extract accelerates host plant growth via gibberellin and indole-3-acetic acid secretion. J. Microbiol. Biotechnol. 25, 1467–1475. doi: 10.4014/jmb.1502.02011
Lee, Y. H., Foster, J., Chen, J., Voll, L. M., Weber, A. P. M., and Tegeder, M. (2007). App1 transports uncharged amino acids into roots of arabidopsis. Plant J. 50, 305–319. doi: 10.1111/j.1365-313X.2007.03045.x
Ling, N., Zhang, W., Wang, D., Mao, J., Huang, Q., Guo, S., et al. (2013). Root exudates from grafted-root watermelon showed a certain contribution in inhibiting Fusarium oxysporum f. sp. niveum. PLoS ONE 8, e63383. doi: 10.1371/journal.pone.0063383
Liu, D., Lian, B., and Dong, H. (2012). Isolation of Paenibacillus sp. and assessment of its potential for enhancing mineral weathering. Geomicrobiol J. 29, 413–421. doi: 10.1080/01490451.2011.576602
Liu, K., Newman, M., McInroy, J. A., Hu, C. H., and Kloepper, J. W. (2017). Selection and assessment of plant growth-promoting rhizobacteria (PGPR) for biological control of multiple plant diseases. Phytopathology 107, 928–936. doi: 10.1094/PHYTO-02-17-0051-R
Liu, W., Hou, J., Wang, Q., Yang, H., Luo, Y., and Christi, P. (2015). Collection and analysis of root exudates of Festuca arundinacea L. and their role in facilitating the phytoremediation of petroleum-contaminated soil. Plant Soil. 389, 109–119. doi: 10.1007/s11104-014-2345-9
Liu, Y., Zhang, N., Qiu, M., Feng, H., Vivanco, J. M., Shen, Q., et al. (2014). Enhanced rhizosphere colonization of beneficial Bacillus amyloliquefaciens SQR9 by pathogen infection. FEMS Microbiol. Lett. 353, 49–56. doi: 10.1111/1574-6968.12406
Lonoce, C., Salem, R., Marusic, C., Jutras, P. V., Scaloni, A., Salzano, A. M., et al. (2016). Production of a tumour-targeting antibody with a human-compatible glycosylation profile in N. benthamiana hairy root cultures. Biotechnol. J. 11, 1209–1220. doi: 10.1002/biot.201500628
Lu, H., Sun, J., and Zhu, L. (2017). The role of artificial root exudate components in facilitating the degradation of pyrene in soil. Sci. Rep. 7, 7130. doi: 10.1038/s41598-017-07413-3
Lynn, T. M., Win, H. S., Kyaw, E. P., Latt, Z. K., and Yu, S. S. (2013). Characterization of phosphate solubilizing and potassium decomposing strains and study on their effects on tomato cultivation. Int. J. Innov. Appl. Stud. 3, 959–966.
Ma, Y., Rajkumar, M., Zhang, C., and Freitas, H. (2016). Beneficial role of bacterial endophytes in heavy metal phytoremediation. J. Environ. Manage. 174, 14–25. doi: 10.1016/j.jenvman.2016.02.047
Madeira, L. M., Szeto, T. H., Henquet, M., Raven, N., Runions, J., Huddleston, J., et al. (2016). High-yield production of a human monoclonal IgG by rhizosecretion in hydroponic tobacco cultures. Plant Biotechnol J. 14, 615–624. doi: 10.1111/pbi.12407
Mahmud, A. A., Upadhyay, S. K., Srivastava, A. K., and Bhojiya, A. A. (2021). Biofertilizers: a nexus between soil fertility and crop productivity under abiotic stress. Curr. Res. Environ. Sust. 3, 100063. doi: 10.1016/j.crsust.2021.100063
Maksimov, I. V., Abizgildina, R. R., and Pusenkova, L. I. (2011). Plant growth promoting rhizobacteria as alternative to chemical crop protectors from pathogens (review). Appl. Biochem. Microbiol. 47, 333–345. doi: 10.1134/S0003683811040090
Mark, G. L., Dow, J. M., Kiely, P. D., Higgins, H., Haynes, J., Baysse, C., et al. (2005). Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc. Natl. Acad. Sci. U.S.A. 102, 17454–17459. doi: 10.1073/pnas.0506407102
Marketon, M. M., Gronquist, M. R., Eberhard, A., and Gonzalez, J. E. (2002). Characterization of the SinorhizobiummelilotisinR/sinI locus and the production of novel N-Acyl homoserine lactones. J. Bacteriol. 184, 5686–5695. doi: 10.1128/JB.184.20.5686-5695.2002
Martinez, C., Espinosa-Ruiz, A., and Prat, S. (2016). Gibberellins and plant vegetative growth. Ann. Plant Rev. 49, 285–322. doi: 10.1002/9781119210436.ch10
Matilla, M. A., Espinosa-Urgel, M., Rodriguez-Herva, J. J., Ramos, J. L., and Ramos-Gonzalez, M. L. (2007). Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol. 8, 179. doi: 10.1186/gb-2007-8-9-r179
Meena, V. S., Maurya, B. R., and Verma, J. P. (2014). Does a rhizospheric microorganism enhance K+ availability in agricultural soils. Microbiol. Res. 169, 337–347. doi: 10.1016/j.micres.2013.09.003
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Menendez, E., and Garcia-Fraile, P. (2017). Plant probiotic bacteria: solutions to feed the world. AIMS Microbiol. 3, 502–524. doi: 10.3934/microbiol.2017.3.502
Mohammed, A. F. (2018). Effectiveness of exopolysaccharides and biofilm forming plant growth promoting rhizobacteria on salinity tolerance of faba bean (Vicia faba L.). Afr. J. Microbiol. Res. 12, 399–404. doi: 10.5897/AJMR2018.8822
Molina-Favero, C., Creus, C. M., Simontacchi, M., Puntarulo, S., and Lamattina, L. (2008). Aerobic nitric oxide production by Azospirillumbrasilense Sp245 and its influence on root architecture in tomato. Mol. Plant Microbe Interact. 21, 1001–1009. doi: 10.1094/MPMI-21-7-1001
Montanez, A., Abreu, C., Gill, P. R., Hardarson, G., and Sicardi, M. (2009). Biological nitrogen fixation in maize (Zea mays L.) by N-15 isotope-dilution and identification of associated culturable diazotrophs. Biol. Fert. Soil 45, 253–263. doi: 10.1007/s00374-008-0322-2
Morrison, S. M., and Askeland, R. A. (1983). Cyanide production by Pseudomonas fluorescens and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 45, 1802–1807. doi: 10.1128/aem.45.6.1802-1807.1983
Mullen, M. D. (2005). “Phosphorus in soils: biological interactions,” in Encyclopedia of Soils in the Environment, eds D. Hillel, C. Rosenzweig, D. Powlson, K. Scow, M. Singer, and D. Sparks (Oxford: Academic Press, Elsevier, Ltd), 210–215. doi: 10.1016/B0-12-348530-4/00161-2
Myresiotis, C. K., Vryzas, Z., and Papadopoulou-Mourkidou, E. (2015). Effect of specific plant-growth-promoting rhizobacteria (PGPR) on growth and uptake of neonicotinoid insecticide thiamethoxam in corn (Zea mays L.) seedlings. Pest Manag. Sci. 71, 1258–1266. doi: 10.1002/ps.3919
Nahas, E. (1996). Factors determining rock phosphate solubilization by microorganism isolated from soil. World J. Microb. Biotechnol. 12, 18–23. doi: 10.1007/BF00327716
Narasimhan, K., Basheer, C., Bajic, V. B., and Swarup, S. (2003). Enhancement of plant-microbe interactions using a rhizosphere metabolomics-driven approach and its application in the removal of polychlorinated biphenyls. Plant Physiol. 132, 146–153. doi: 10.1104/pp.102.016295
Neal, A. L., Ahmad, S., Gordon-Weeks, R., and Ton, J. (2012). Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS ONE 7, e35498. doi: 10.1371/journal.pone.0035498
Neilands, J. B. (1995). Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270, 26723–26726. doi: 10.1074/jbc.270.45.26723
Nghia, N. K., Tien, T. T., Oanh, N. K., and Nuong, N. K. (2017). Isolation and characterization of indole acetic acid producing halophilic bacteria from salt affected soil of Rice-Shrimp farming system in the Mekong Delta, Vietnam. Agric. For. Fish. 6, 69–77. doi: 10.11648/j.aff.20170603.11
Ngoma, L., Babalola, O. O., and Ahmad, F. (2012). Ecophysiology of plant growth promoting bacteria. Sci. Res. Essays 7, 4003–4013. doi: 10.5897/SRE12.646
Nguema-Ona, E., Vicre-Gibouin, M., Cannesan, M. A., and Driouich, A. (2013). Arabinogalactan proteins in root-microbe interactions. Trends Plant Sci. 18, 440–449. doi: 10.1016/j.tplants.2013.03.006
Nievas, F., Bogino, P., Sorroche, F., and Giordano, W. (2012). Detection, characterization, and biological effect of quorum-sensing signaling molecules in peanut-nodulating bradyrhizobia. Sensors 12, 2851–2873. doi: 10.3390/s120302851
Numana, M., Bashir, S., Khan, Y., Mumtaz, R., Shinwari, Z. K., Khan, A. L., et al. (2018). Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: a review. Microbiol. Res. 209, 21–32. doi: 10.1016/j.micres.2018.02.003
Oberhansli, T., Defago, G., and Haas, D. (1991). Indole-3-acetic acid (IAA) synthesis in the biocontrol strain CHAO of Pseudomonas fluorescens: role of tryptophan side chain oxidase. J. Gen. Microbiol. 137, 2273–2279. doi: 10.1099/00221287-137-10-2273
O'Brien, J. A., and Benkova, E. (2013). Cytokinin cross-talking during biotic and abiotic stress responses. Front. Plant Sci. 4, 451. doi: 10.3389/fpls.2013.00451
Pal, S. S. (1998). Interaction of an acid tolerant strain of phosphate solubilizing bacteria with a few acid tolerant crops. Plant Soil 198, 169–177. doi: 10.1023/A:1004318814385
Patten, C. L., and Glick, B. R. (1996). Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 42, 207–220. doi: 10.1139/m96-032
Paul, S., Singh, V., Chauhan, P. K., Srivastava, A. K., and Upadhyay, S. K. (2020). Assessment of carrot growth performance with inoculation of AsT-PGPR under arsenic infested zone. G J. Environ. Sci. Technol. 7, 78–84.
Piromyou, P., Buranabanyat, B., Tantasawat, P., Tittabutr, P., Boonkerd, N., and Teaumroong, N. (2011). Effect of plant growth promoting rhizobacteria (PGPR) inoculation on microbial community structure in rhizosphere of forage corn cultivated in Thailand. Eur. J. Soil Biol. 47, 44–54. doi: 10.1016/j.ejsobi.2010.11.004
Plackett, A. R. G., and Wilson, Z. A. (2016). Gibberellins and plant reproduction. Ann. Plant Rev. 49, 323–358. doi: 10.1002/9781119210436.ch11
Pliego, C., Kamilova, F., and Lugtenberg, B. (2011). “Plant growth-promoting bacteria: fundamentals and exploitation,” in Bacteria in Agrobiology: Crop Ecosystems, ed D. K. Maheshwari (Berlin: Springer-Verlag), 295–343. doi: 10.1007/978-3-642-18357-7_11
Prathap, M., and Ranjitha, K. B. D. (2015). A critical review on plant growth promoting rhizobacteria. J. Plant Pathol. Microbiol. 6, 1–4. doi: 10.4172/2157-7471.1000266
Rajkumar, M., Ae, N., Prasad, M. N. V., and Freitas, H. (2010). Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 28, 142–149. doi: 10.1016/j.tibtech.2009.12.002
Ramachandran, V. K., East, A. K., Karunakaran, R., Downie, J. A., and Poole, P. S. (2011). Adaptation of Rhizobium leguminosarum to pea, alfalfa and sugar beet rhizospheres investigated by comparative transcriptomics. Genome Biol. 12, R106. doi: 10.1186/gb-2011-12-10-r106
Ramirez, L. E. F., and Mellado, J. C. (2005). “Bacterial biofertilizers,” in PGPR: Biocontrol and Biofertilization, ed Siddiqui ZA (Dordrecht: Springer), 143–172.
Ray, S., Singh, V., Bisen, K., Keswani, C., Singh, S., Singh, H. B., et al. (2017). “Endophytomicrobiont: a multifaceted beneficial interaction,” in H. B. Singh, B. K. Sarma, and C. Keswani. Advances in PGPR Research (Wallingford: CABI), 218–233. doi: 10.1079/9781786390325.0218
Raza, A., Ejaz, S., Saleem, M. S., Hejnak, V., Ahmad, F., Ahmed, M. A. A., et al. (2021). Plant growth promoting rhizobacteria improve growth and yield related attributes of chili under low nitrogen availability. PLoS ONE 16, e0261468. doi: 10.1371/journal.pone.0261468
Reed, S. C., Cleveland, C. C., and Townsend, A. R. (2011). Functional ecology of free-living nitrogen fixation: a contemporary perspective. Annu. Rev. Ecol. Evol. Syst. 42, 489–512. doi: 10.1146/annurev-ecolsys-102710-145034
Reinhold-Hurek, B., Bunger, W., Burbano, C. S., Sabale, M., and Hurek, T. (2015). Roots shaping their microbiome: global hotspots for microbial activity. Annu. Rev. Phytopathol. 53, 403–424. doi: 10.1146/annurev-phyto-082712-102342
Rezzonico, F., Binder, C., Defago, G., and Moenne-Loccoz, Y. (2005). The type III secretion system of biocontrol Pseudomonas fluorescens KD targets the phytopathogenic ChromistaPythiumultimum and promotes cucumber protection. Mol. Plant Microbe Interact. 18, 991–1001. doi: 10.1094/MPMI-18-0991
Rodriguez, H., and Fragal, R. (1990). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17, 319–339. doi: 10.1016/S0734-9750(99)00014-2
Rodriguez, M., Torres, M., Blanco, L., Bejar, V., Sampedro, I., and Llamas, I. (2020). Plant growth-promoting activity and quorum quenching mediated biocontrol of bacterial phytopathogens by Pseudomonas segetis strain P6. Sci. Rep. 10, 4121. doi: 10.1038/s41598-020-61084-1
Rohrbacher, F., and St-Arnaud, M. (2016). Root exudation: the ecological driver of hydrocarbon rhizoremediation. Agronomy 6, 19. doi: 10.3390/agronomy6010019
Roychoudhury, P., and Kaushik, B. D. (1989). Solubilization of mussorie rock phosphate by cyanobacteria. Curr. Sci. 58, 569–570.
Rudrappa, T., Czymmek, K. J., Pare, P. W., and Bais, H. P. (2008). Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 3, 1547–1556. doi: 10.1104/pp.108.127613
Ryu, C. M., Farag, M. A., Hu, C. H., Reddy, M. S., Kloepper, J. W., and Pare, P. W. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiology. 134, 1017–1026. doi: 10.1104/pp.103.026583
Ryu, C. M., Farag, M. A., Hu, C. H., Reddy, M. S., Wei, H. X., Pare, P. W., et al. (2003). Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 4927–4932. doi: 10.1073/pnas.0730845100
Sah, S., and Singh, R. (2015). Siderophore: structural and functional characterization – a comprehensive review. Agriculture 6, 97–114. doi: 10.1515/agri-2015-0015
Sahin, F. C., Akmakc, I. R., and Kantar, F. (2004). Sugar beet and barley yields in relation to inoculation with N2-fixing and phosphate solubilizing bacteria. Plant Soil 265, 123–129. doi: 10.1007/s11104-005-0334-8
Saiyad, S. A., Jhala, Y. K., and Vyas, R. V. (2015). Comparative efficiency of five potash and phosphate solubilizing bacteria and their key enzymes useful for enhancing and improvement of soil fertility. Int. J. Sci. Res. Public. 5, 1–6.
Saleem, M., Arshad, M., Hussain, S., and Bhatti, A. S. (2007). Perspective of plant growth promoting rhizobacteria (PGPR) containing ACC deaminase in stress agriculture. J. Indust. Microbiol. Biotechnol. 34, 635–648. doi: 10.1007/s10295-007-0240-6
Santner, A., Calderon-Villalobos, L. I. A., and Estelle, M. (2009). Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307. doi: 10.1038/nchembio.165
Santos, M. L. D., Berlitz, D. L., Wiest, S. L. F., Schunemann, R., Knaak, N., and Fiuza, L. M. (2018). Benefits associated with the interaction of endophytic bacteria and plants. Braz. Arch. Biol. Technol. 61, 1–11. doi: 10.1590/1678-4324-2018160431
Santoyo, G., Moreno-Hagelsieb, G., Orozco-Mosqueda, M. D. C., and Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Schaller, G. E., Street, I. H., and Kieber, J. J. (2014). Cytokinin and the cell cycle. Curr. Opin. Plant. Biol. 21, 7–15. doi: 10.1016/j.pbi.2014.05.015
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Sekar, C., Prasad, N. N., and Sundaram, M. D. (2000). Enhancement of polygalacturonase activity during auxin induced para nodulation and endorhizosphere colonization of Azospirillum in rice roots. Indian J. Exp. Biol. 38, 80–83.
Shaharoona, B., Arshad, M., and Zahir, Z. A. (2006). Effect of plant growth promoting rhizobacteria containing ACC-deaminase on maize (Zea mays L.) growth under axenic conditions and on nodulation in mung bean. Lett. Appl. Microbiol. 42, 155–159. doi: 10.1111/j.1472-765X.2005.01827.x
Shanahan, P., Osullivan, D. J., Simpson, P., Glennon, J. D., and O' Gara, F. (1992). Isolation of 2, 4-diacetylphloroglucinol from a Fluorescent Pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol. 58, 353–358. doi: 10.1128/aem.58.1.353-358.1992
Shank, E. A., and Kolter, R. (2011). Extracellular signaling and multicellularity in Bacillus subtilis. Curr. Opin. Microbiol. 14, 741–747. doi: 10.1016/j.mib.2011.09.016
Sharma, S. B., Sayyed, R. Z., Trivedi, M. H., and Gobi, T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2, 587. doi: 10.1186/2193-1801-2-587
Sindhu, S. S., Rakshiya, Y. S., and Sahu, G. (2009). Biological control of soilborne plant pathogens with rhizosphere. Bacter. Pest Technol. 3, 10–21.
Singh, H. B, Keswani, C., Reddy, M. S., Royano, E. S., and García-Estrada, C. (Eds.). (2019). Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms: Discovery and Applications. Singapore: Springer-Nature.
Singh, R., Singh, J., Deval, R., Upadhyay, S. K., and Kumar, D. (2017). The potentiality of selected strain of PGPR: azotobacter, for sustainable agriculture in India. G J. Environ. Sci. Technol. 4, 49–53.
Singh, R. K., Singh, P., Sharma, A., Guo, D.-J., Upadhyay, S. K., Song, Q.-Q., et al. (2022). Unraveling nitrogen fixing potential of endophytic diazotrophs of different saccharum species for sustainable sugarcane growth. Int. J. Mol. Sci. 23, 6242. doi: 10.3390/ijms23116242
Sinha, A. K., Venkateswaran, B. P., Tripathy, S. C., Sarkar, A., and Prabhakaran, S. (2018). Effects of growth conditions on siderophore producing bacteria and siderophore production from Indian ocean sector of Southern ocean. J. Basic Microbiol. 59, 412–424. doi: 10.1002/jobm.201800537
Someya, N., Kataoka, N., Komagata, T., Hirayae, K., Hibi, T., and Akutsu, K. (2000). Biological control of cyclamen soil borne diseases by Serratia marcescens strain B2. Plant Dis. 84, 334–340. doi: 10.1094/PDIS.2000.84.3.334
South, K. A., Nordstedt, N. P., and Jones, M. L. (2021). Identification of plant growth promoting rhizobacteria that improve the performance of greenhouse-grown petunias under low fertility conditions. Plants 10, 1410. doi: 10.3390/plants10071410
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Spaink, H. P. (2000). Root nodulation and infection factors produced by rhizobial bacteria. Annu. Rev. Microbiol. 54, 257–288. doi: 10.1146/annurev.micro.54.1.257
Swain, M. R., Naskar, S. K., and Ray, R. C. (2007). Indole 3-acetic acid production and effect on sprouting of yam (Dioscorearotundata L.) Minisetts by Bacillus subtilis isolated from culturable cowdung microflora. Pol. J. Microbiol. 56, 103–110.
Szoboszlay, M., White-Monsant, A., and Moe, L. A. (2016). The effect of root exudate 7,40- dihydroxyflavone and naringenin on soil bacterial community structure. PLoS ONE 11, e0146555. doi: 10.1371/journal.pone.0146555
Tahir, H. A. S., Gu, Q., Wu, H., Raza, W., Hanif, A., Wu, L., et al. (2017). Plant growth promotion by volatile organic compounds produced by Bacillus subtilis SYST2. Front. Microbiol. 8, 171. doi: 10.3389/fmicb.2017.00171
Tang, A., Haruna, A. O., Majid, N. M. A., and Jalloh, M. B. (2020). Potential PGPR properties of cellulolytic, nitrogen-fixing, phosphate-solubilizing bacteria in rehabilitated tropical forest soil. Microorganisms 8, 442. doi: 10.3390/microorganisms8030442
Tashiro, Y., Yawata, Y., Toyofuku, M., Uchiyama, H., and Nomura, N. (2013). Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ. 28, 13–24. doi: 10.1264/jsme2.ME12167
Tsukanova, K. A., Chebotar, V. K., Meyer, J. J. M., and Bibikova, T. N. (2017). Effect of plant growth-promoting rhizobacteria on plant hormone homeostasis. South Afr. J. Bot. 113, 91–102. doi: 10.1016/j.sajb.2017.07.007
Upadhyay, S. K., and Chauhan, P. K. (2022). Optimization of eco-friendly amendments as sustainable asset for salt-tolerant plant growth-promoting bacteria mediated maize (Zea Mays L.) plant growth, Na uptake reduction and saline soil restoration. Environ. Res. 211, 113081. doi: 10.1016/j.envres.2022.113081
Upadhyay, S. K., and Edrisi, S. A. (2021). Developing sustainable measures to restore fly ash contaminated lands: current challenges and future prospects. Land Degrad. Dev. 32, 4817–4831. doi: 10.1002/ldr.4090
Upadhyay, S. K., Maurya, S. K., and Singh, D. P. (2012a). Salinity tolerance in free living plant growth promoting rhizobacteria. Indian J. Sci. Res. 3, 73–78.
Upadhyay, S. K., Saxena, A. K., Singh, J. S, and Singh, D. P. (2019). Impact of native ST-PGPR (Bacillus pumilus; EU927414) on PGP traits, antioxidants activities, wheat plant growth and yield under salinity. Clim. Change Environ. Sust. 7, 157–168. doi: 10.5958/2320-642X.2019.00021.8
Upadhyay, S. K., and Singh, D. P. (2015). Effect of salt-tolerant plant growth-promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol. 17, 288–293. doi: 10.1111/plb.12173
Upadhyay, S. K., Singh, D. P., and Saikia, R. (2009). Genetic diversity of plant growth promoting rhizobacteria isolated from rhizospheric soil of wheat under saline condition. Curr. Microbiol. 59, 489–496. doi: 10.1007/s00284-009-9464-1
Upadhyay, S. K., Singh, G., and Singh, D. P. (2016). “Mechanism and understanding of PGPR: an approach for sustainable agriculture under a biotic stresses,” in Microbes and Environmental Management, eds J. S. Singh, and D. P. Singh (Stadum: Studium Press Pvt. Ltd), 225–254.
Upadhyay, S. K., Singh, J. S., Saxena, A. K., and Singh, D. P. (2012b). Impact of PGPR inoculation on growth and antioxidant status of wheat under saline conditions. Plant Biol. 14, 605–611. doi: 10.1111/j.1438-8677.2011.00533.x
Upadhyay, S. K., Singh, J. S., and Singh, D. P. (2011). Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere 21, 214–222. doi: 10.1016/S1002-0160(11)60120-3
Van Loon, L. C., Bakker, P. A., and Pieterse, M. J. (1998). Systemic induced resistance by rhizosphere bacteria. Annu. Rev. Phytopathol. 36, 453–483. doi: 10.1146/annurev.phyto.36.1.453
Vardharajula, S., Alia, S. Z., Grover, M., Reddy, G., and Bandi, V. (2011). Drought-tolerant plant growth promoting Bacillus spp. effect on growth, osmolytes and antioxidant status of maize under drought stress. J. Plant Interact. 6, 1–14. doi: 10.1080/17429145.2010.535178
Vial, L., Cuny, C., Gluchoff-Fiasson, K., Comte, G., Oger, P. M., Faure, D., et al. (2006). N-acyl-homoserine lactone-mediated quorum-sensing in Azospirillum: an exception rather than a rule. FEMS Microbiol. Ecol. 58, 155–168. doi: 10.1111/j.1574-6941.2006.00153.x
Vieira, S., Sikorski, J., Dietz, S., Herz, K., Schrumpf, M., Bruelheide, H., et al. (2020). Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J. 14, 463–475. doi: 10.1038/s41396-019-0543-4
Vocciante, M., Grifoni, M., Fusini, D., Petruzzelli, G., and Franchi, E. (2022). The role of plant growth-promoting rhizobacteria (PGPR) in mitigating plant's environmental stresses. Appl. Sci. 12, 1231. doi: 10.3390/app12031231
Voisard, C., Keel, C., Haas, D., and Defago, G. (1989). Cyanide production by Pseudomonas fluorescens suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 8, 351–358. doi: 10.1002/j.1460-2075.1989.tb03384.x
Wang, S., Huijun, W., Junqing, Q., Lingli, M., Jun, L., Yanfei, X., et al. (2009). Molecular mechanism of plant growth promotion and induced systemic resistance to tobacco mosaic virus by Bacillus spp. J. Microbiol. Biotechnol. 10, 1250–1258. doi: 10.4014/jmb.0901.008
Wanga, Y., Cai, Y., Cao, Y., Liu, Y., and Jiping, L. (2018). Aluminum-activated root malate and citrate exudation is independent of NIP1; 2-facilitated root-cell-wall aluminum removal in Arabidopsis. Plant Signal. Behav. 1, e1422469. doi: 10.1080/15592324.2017.1422469
Wei, Z., and Jousset, A. (2017). Plant breeding goes microbial. Trends Plant Sci. 22, 555–558. doi: 10.1016/j.tplants.2017.05.009
Weller, D. M. (2007). Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97, 250–256. doi: 10.1094/PHYTO-97-2-0250
Weller, D. M., and Cook, R. J. (1983). Suppression of take-all of wheat by seed-treatment with Fluorescent pseudomonads. Phytopathology 73, 463–469. doi: 10.1094/Phyto-73-463
Weston, L. A., Ryan, P. R., and Watt, M. (2012). Mechanisms for cellular transport and release of allelochemicals from plant roots into the rhizosphere. J. Exp. Bot. 63, 3445–3454. doi: 10.1093/jxb/ers054
White, L. J., Ge, X., Brozel, V. S., and Subramanian, S. (2017). Root isoflavonoids and hairy root transformation influence key bacterial taxa in the soybean rhizosphere. Environ. Microbiol 19, 1391–1406. doi: 10.1111/1462-2920.13602
Xie, F., Murray, J. D., Kim, J., Heckmann, A. B., Edwards, A., Oldroyd, G. E., et al. (2012). Legume pectate lyase required for root infection by rhizobia. Proc. Natl. Acad. Sci. U.S.A. 109, 633–638 doi: 10.1073/pnas.1113992109
Yadav, B. K., Akhtar, M. S., and Panwar, J. (2015). “Rhizospheric plant-microbe interactions: key factors to soil fertility and plant nutrition,” in Plant Microbes Symbiosis: Applied Facets, ed N. K. Arora (New Delhi: Springer), 127–145. doi: 10.1007/978-81-322-2068-8_6
Yadegari, M., and Mosadeghzad, Z. (2012). Biofertilizers effects on quantitative and qualitative yield of Thyme (Thymus vulgaris). Afr. J. Agri. Res. 7, 4716–4723. doi: 10.5897/AJAR11.2347
Yousaf, M., Rehman, Y., and Hasnain, S. (2017). High-yielding wheat varieties harbour superior plant growth promoting-bacterial endophytes. Appl. Food Biotech. 4, 143–154. doi: 10.22037/afb.v4i3.15014
Yu, X., Li, Y., Cui, Y., Liu, R., Li, Y., Chen, Q., et al. (2017). An indoleacetic acid-producing Ochrobactrumsp. MGJ11 counteracts cadmium effect on soybean by promoting plant growth. J. Appl Microbiol. 122, 987–996. doi: 10.1111/jam.13379
Yuan, J., Zhang, N., Huang, Q., Raza, W., Li, R., Vivanco, J. M., et al. (2015). Organic acids from root exudates of banana help root colonization of PGPR strain Bacillus amyloliquefaciens NJN-6. Sci. Rep. 5, 13438. doi: 10.1038/srep13438
Zakry, F. A. A., Shamsuddin, Z. H., Rahim, K. A., Zakaria, Z. Z., and Rahim, A. A. (2012). Inoculation of Bacillus sphaericus UPMB-10 to young oil palm and measurement of its uptake of fixed nitrogen using the 15N isotope dilution technique. Microbes Environ. 27, 257–262, doi: 10.1264/jsme2.ME11309
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., Rocha, U. N. D., Dhi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, A., Zhao, G., Gao, T., Wang, W., Li, J., Zhang, S., et al. (2013). Solubilization of insoluble potassium and phosphate by Paenibacilluskribensis CX-7: a soil microorganism with biological control potential. Afr. J. Microbiol. Res. 7, 41–47. doi: 10.5897/AJMR12.1485
Zhang, C., and Kong, F. (2014). Isolation and identification of potassium-solubilizing bacteria from tobacco rhizospheric soil and their effect on tobacco plants. Appl. Soil. Ecol. 82, 18–25. doi: 10.1016/j.apsoil.2014.05.002
Zhang, N., Yang, D., Wang, D., Miao, Y., Shao, J., Zhou, X., et al. (2015). Whole transcriptomic analysis of the plant-beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 during enhanced biofilm formation regulated by maize root exudates. BMC Genomics 16, 685. doi: 10.1186/s12864-015-1825-5
Keywords: chemoattractant, PGPR recruitment, plant-microbes interaction, root-exudate, rhizosphere
Citation: Upadhyay SK, Srivastava AK, Rajput VD, Chauhan PK, Bhojiya AA, Jain D, Chaubey G, Dwivedi P, Sharma B and Minkina T (2022) Root Exudates: Mechanistic Insight of Plant Growth Promoting Rhizobacteria for Sustainable Crop Production. Front. Microbiol. 13:916488. doi: 10.3389/fmicb.2022.916488
Received: 09 April 2022; Accepted: 09 June 2022;
Published: 14 July 2022.
Edited by:
Sandip Paul, JIS Institute of Advanced Studies and Research, IndiaReviewed by:
Divjot Kour, Eternal University, IndiaJosé David Flores Félix, Universidade da Beira Interior, Portugal
Copyright © 2022 Upadhyay, Srivastava, Rajput, Chauhan, Bhojiya, Jain, Chaubey, Dwivedi, Sharma and Minkina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sudhir K. Upadhyay, c2t1LmVudi5sa29AZ21haWwuY29torcid.org/0000-0002-2228-8063; Padmanabh Dwivedi, cGR3aXZlZGkyNUByZWRpZmZtYWlsLmNvbQ==
 Sudhir K. Upadhyay
Sudhir K. Upadhyay Abhishek K. Srivastava
Abhishek K. Srivastava Vishnu D. Rajput
Vishnu D. Rajput Prabhat K. Chauhan
Prabhat K. Chauhan Ali Asger Bhojiya
Ali Asger Bhojiya Devendra Jain
Devendra Jain Gyaneshwer Chaubey
Gyaneshwer Chaubey Padmanabh Dwivedi
Padmanabh Dwivedi Bechan Sharma
Bechan Sharma Tatiana Minkina
Tatiana Minkina