
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 June 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.913023
This article is part of the Research Topic Virome: Diversity, Function and Ecology View all 12 articles
Enterococcus faecalis is a common gram-positive non-spore-forming bacterium in nature and is found in the upper respiratory tract, intestine, and mouth of healthy people. E. faecalis is also one of the common pathogens causing nosocomial infections and is resistant to several antibiotics commonly used in practice. Thus, treating drug-resistant E. faecalis with antibiotics is challenging, and new approaches are needed. In this study, we isolated a bacteriophage named EFap02 that targets E. faecalis strain EFa02 from sewage at Southwest Hospital. Phage EFap02 belongs to the Siphoviridae family with a long tail of approximately 210 nm, and EFap02 can tolerate a strong acid and alkali environment and high temperature. Its receptor was identified as the capsular polysaccharide. Phage-resistant mutants had loss-of-function mutations in glycosyltransferase (gtr2), which is responsible for capsular polysaccharide biosynthesis, and this caused the loss of capsular polysaccharide and interruption of phage adsorption. Although phage-resistant mutants against EFap02 can be selected, such mutants are impaired in biofilm formation due to the loss of capsular polysaccharide, which compromises its virulence. Therefore, this study provided a detailed description of the E. faecalis EFap02 phage with the potential for treating E. faecalis infection.
Enterococcus faecalis is a common opportunistic pathogen that causes blood and urinary tract infections. It is one of the primary pathogens that infect root canals (Khalifa et al., 2016). E. faecalis is intrinsically resistant to several commonly used antibiotics, such as cephalosporin and aminoglycoside (García-Solache and Rice, 2019). Vancomycin-resistant Enterococcus (VRE) is resistant to vancomycin (Miller et al., 2016). With the emergence of multidrug-resistant strains of E. faecalis and its ability to form biofilms (Ch'ng et al., 2019), E. faecalis infection has caused great concern in practice (Fiore et al., 2019), and there is an urgent need to find new treatments.
In recent years, phage therapy has renewed interest as a promising alternative to antibiotics (Khalifa et al., 2016). Phage therapy has successfully treated patients with multidrug-resistant bacterial infections. In Belgium and France, a randomized, controlled, double-blind phase I/II trial (PhagoBurn) used a cocktail of 12 natural lytic phages to treat burn wounds infected with Pseudomonas aeruginosa. Compared to standard care, the bacterial load was reduced in the phage cocktail treatment group, indicating that phage therapy is promising, although the quality of the phage product needs improvement (Jault et al., 2019). Furthermore, in China, a 63-year-old woman who developed a recurrent urinary tract infection with extensively drug-resistant Klebsiella pneumoniae was cured by antibiotic and phage synergism (Bao et al., 2020). Only one human study described the treatment of chronic prostatitis associated with E. faecalis by phage therapy in 2007. Three patients who suffered chronic E. faecalis infection were not cured with antibiotics, autovaccines, and laser biostimulation. They were then treated with phage therapy twice daily for 1 month. The pathogen was eradicated, and clinical symptoms were relieved without recurrence (Letkiewicz et al., 2009).
In this study, we successfully isolated a lytic phage, EFap02, from the sewage of Southwest Hospital in China. Phage EFap02 belongs to the Siphoviridae family and has a potent lytic effect against E. faecalis. The capsular polysaccharide was identified as the EFap02 receptor. The phage-resistant mutant had loss-of-function mutations in glycosyltransferase Group 2 (gtr2), which is responsible for capsular polysaccharide biosynthesis. The mutations caused the loss of capsular polysaccharide and interruption of phage adsorption. The loss of capsular polysaccharide significantly decreased the biofilm formation capability compromising the EPap02 virulence. Overall, this study suggested that EFap02 could be a promising candidate for E. faecalis phage therapy.
The bacterial strains and phages are listed in Table 1. The EFa02 strain was isolated from the Daping Hospital Department of Clinical Laboratory Medicine. E. faecalis was cultured in brain heart infusion (BHI) medium at 37°C. When necessary, erythromycin (20 μg/ml) was added to the medium. The stock cultures were stored in medium supplemented with 20% glycerol at −80°C.
Isolation of bacteriophages was performed as previously described (Shen et al., 2018). Sewage (10 ml) from the Southwest Hospital was centrifuged at 10,000 × g for 10 min. The supernatant was filtered with a 0.45 μm aseptic filter, and 2 ml of the filtered supernatant was added to a 50 ml clean centrifuge tube. The logarithmic phase host bacteria EFa02 was added and then cultured in a 37°C shaking incubator overnight. The mixture was centrifuged at 21,000 × g for 1 min, and the supernatant was filtered using a 0.22 μm aseptic filter. Then, 10 μl supernatant was mixed with 100 μl host bacteria EFa02 in a 15 ml centrifuge tube. BHI soft agar (5 ml) was added, and the content was poured onto the surface of agar plates. The plates were incubated at 37°C overnight, and the plaques were observed on the top agar.
Phage morphology was observed by transmission electron microscopy (TEM; HT7700, Hitachi, Japan; Lee et al., 2019). The filtered phage lysate was dropped onto the prepared Formvar/carbon-coated copper grid and incubated for 1 min. The grid was negatively stained with 2% uranium acetate for 10 min and then observed using TEM at an acceleration voltage of 80 kV. Phages are classified according to the guidelines of the International Committee on the Taxonomy of Viruses (ICTV; Lefkowitz et al., 2018).
The optimal multiplicity of infection (MOI) is the ratio of bacteriophages to bacteria at the time of infection. The optimal MOI is the number of infections when the phage can achieve the best growth state (Lee et al., 2019). According to different MOIs, the phage (2 ml) and host bacteria (2 ml) were mixed. BHI medium was added to the mixture to a final volume of 10 ml. The mixture was incubated at 37°C with shaking at 220 rpm for 6 h. The 1 ml mixture was centrifuged at 12,000 × g for 1 min and filtered with a 0.45 μm filter. The phage titer was determined by the double-layer agar (DLA) method (Lee et al., 2019). The MOI with the highest phage titer was the optimal MOI. Three biological repeats were performed.
As previously described, one-step phage growth was performed (Lu et al., 2013). EFa02 was cultivated to the early logarithmic stage (OD600 = 0.5). The phage was mixed with EFa02 (108 CFU/ml) at an MOI of 0.01 and incubated at 37°C with shaking at 220 rpm. Samples were taken at time points 0, 10, 20, 30, 40, 50, 60, and 90 min. The phage titer was measured using the DLA method. Three biological replicates were performed.
The BHI medium was adjusted with HCl or NaOH to pH 2.0–14.0. Then, 990 μl of BHI medium was mixed with 10 μl phage stock solution (3 × 108 PFU/ml). After incubation at 37°C for 60 min, the phage titer was determined by the DLA method.
BHI medium (900 μl) was mixed with 100 μl phage stock solution (9 × 109 PFU/ml) and treated at 4, 25, 37, 50, 60, and 70°C. After 60 min, the samples were removed and cooled to room temperature. The phage titer was determined by the DLA method (Ding et al., 2020).
Phage EFap02 (1010 PFU/ml) was mixed with chloroform at ratios of 0, 10, 25, 50, 75, and 95%. The mixtures were incubated at 37°C with shaking at 220 rpm for 60 min. And the phage titer was determined using the DLA method (Oduor et al., 2020).
Isolating phage-resistant mutants was performed as described previously (Yang et al., 2020). Briefly, 10 μl of E. faecalis culture (OD600 = 0.2) was added to EFap02 (109 PFU), and the mixture was placed on BHI agar and incubated at 37°C for 24 h. Then, the phage resistance of the colonies was validated by the DLA assay.
The extraction of the phage genome was performed as previously described with a slight modification (Yuan et al., 2020). To remove contaminated DNA and RNA from the phage stocks, DNase I and RNase A were added to a final concentration of 1 μg/ml, and the phage stocks were incubated at 37°C for 60 min. EDTA (0.5 mol/l) was added to a final concentration of 0.02 mol/l. Proteinase K (20 g/l) and 10% SDS were added (final concentration 0.5%). The mixture was treated at 56°C for 60 min. An equal volume of balanced phenol (pH = 8.0) was added to extract nucleic acids, and then, the mixture was centrifuged at 10,000 × g for 5 min. The upper aqueous phase was transferred to a new 1.5 ml centrifuge tube. An equal volume of chloroform was added. The tube was gently mixed and centrifuged at 10,000 × g for 10 min. The supernatant was transferred to a new centrifuge tube, mixed with a 0.6 volume of isoamyl alcohol, and placed at −20°C for 1 h. The mixture was centrifuged at 12,000 × g for 20 min to collect the DNA pellet. The DNA pellet was washed with 75% ice-cold ethanol and centrifuged at 12,000 × g for 20 min. Finally, the pellet was dried at room temperature, dissolved in 50 μl of water, and stored at −20°C for future use.
The Bacterial Genomic DNA Kit (Tiangen-DP302) was used for DNA extraction. The phage genome was sequenced using the Illumina HiSeq 2,500 platform (~1 Gbp/sample) and assembled with Newbler (Version 2.9) under default parameters. The characteristics of the phage genes were predicted by RAST1 (Aziz et al., 2008) and FgeneSV.2 The homologous DNA sequences and proteins were searched and analyzed using BlastN and BlastP in the BLAST program3 (Altschul et al., 1997). Visualization of the phage circular genome was performed using the CGView server database (Stothard and Wishart, 2005). Phage virulence factors were analyzed by searching a virulence database (Underwood et al., 2005). A comparative analysis of the EFa02 genomes and phage-resistant mutants was performed by Breseq (Barrick et al., 2014). Analysis of phage DNA termini was performed using PhageTerm4 (Garneau et al., 2017), and the phylogenetic tree was constructed and displayed by MEGA 7.05 with the neighbor-joining method (Tamura et al., 2013).
EFa02 and EFap02 were cocultured for 72 h to explore the effect of phage on bacterial growth. The OD600 of the culture medium was measured at given time points. Three biological replicates were performed. The gtr2 gene was amplified by PCR using the primers listed in Table 2 to complement gtr2 in EFa02R. The PCR product was ligated into the plasmid pGM23 by Gibson assembly to generate pGM-gtr2, and pGM-gtr2 was electroporated into EFa02R, and the strain was named EFa02R::gtr2.
Phage adsorption tests were performed on different E. faecalis strains according to a previously reported protocol (Kęsik-Szeloch et al., 2013; Yang et al., 2020). The following steps were performed to verify the difference in the adsorption capacity of phage EFap02 to E. faecalis EFa02, EFa02R, and EFa02R::gtr2. The phage filtrate was diluted to 109 PFU/mL in sterile water to form the initial titer. The host bacteria (1 ml) and phage (105 PFU) were mixed and left in the EP tube for 5 min and centrifuged at 21,000 × g for 2 min. The phage titer was calculated by the DPA method (residual titer). The phage adsorption rate was calculated as [(initial titer −residual titer)/initial titer] × 100%. Three biological replicates were performed.
The phage was diluted in tenfold increments. Then, 100 μl of E. faecalis and 4 ml of BHI soft agar were mixed and poured onto a BHI agar plate. Then, 1 μl of the diluted phage was pipetted onto agar plates and incubated at 37°C overnight (Forti et al., 2018).
Bacterial was stained with crystal violet solution for 5 min and then stained with 20% copper sulfate solution. Then, the bacteria were observed under the microscope immediately.
Crystal violet staining was used to monitor biofilms (Forti et al., 2018). EFa02, EFa02R, and EFa02R::gtr2 were inoculated in 1 ml of BHI medium and cultured at 37°C overnight. The OD600 was measured, and the culture was diluted to an OD600 of 0.02. A diluted bacterial solution (1 ml) was added to a 24-well plate and incubated at 37°C for 24 h, 48 h, or 72 h. The medium was then gently removed, and the wells were washed three times with 1 ml of phosphate-buffered saline (PBS). Finally, the plate was stained with 1 ml of 0.1% crystal violet at room temperature for 20 min. The excess crystal violet was carefully removed, washed with sterile water three times, and air-dried for 3 h. Then, 1 ml of 95% ethanol was added for 10 min, and the OD570 of each well was measured.
Statistical analysis was performed using one-way ANOVA or Student’s t-test. A p < 0.05 was considered statistically significant.
Phage plaques were first discovered on the top agar lawn that contained a clinical strain of E. faecalis EFa02. Clear plaques, approximately 2 mm in diameter, were formed on the DLA plate (Figure 1A). The plaque was purified three times, and the isolated phage was named EFap02. Electron microscopy showed that phage EFap02 had an icosahedron head and a long tail of approximately 210 nm (Figure 1B). Phage EFap02 belonged to the Siphoviridae family according to the ICTV guidelines and was named vB_EFaS_EFap02.
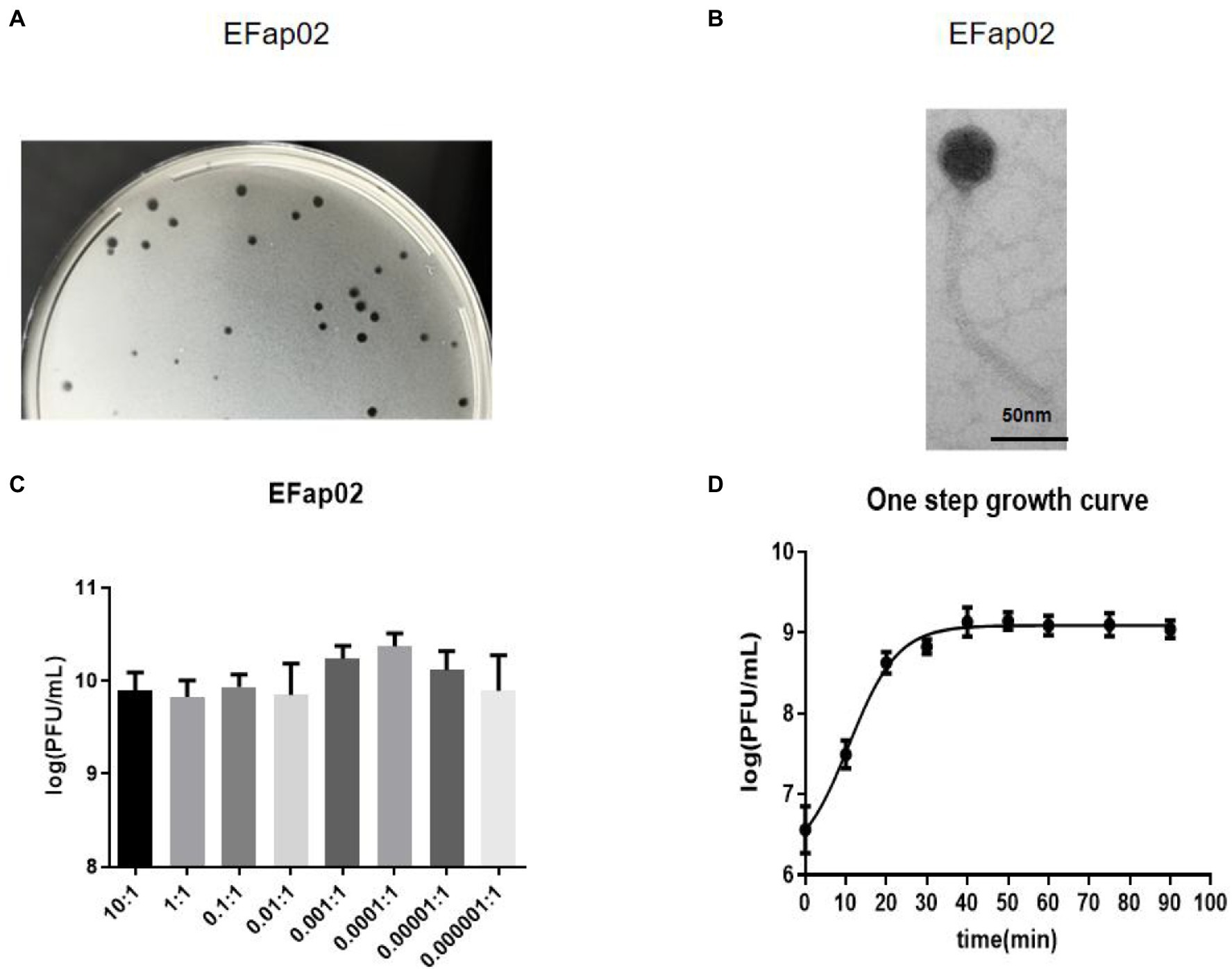
Figure 1. Phage EFap02 general biological characteristics. (A) Phage plaques on a double-layer agar plate of EFap02. (B) TEM image of phage EFap02. EFap02 has an icosahedral head (length 44 nm ± 5, width 40 nm ± 5) and a long tail (210 nm ± 5). The scale bar in the right corner is 50 nm. (C) Different multiplicities of infection (MOI) of EFap02. When the MOI was 0.0001, the EFap02 titer was the highest. (D) One-step growth curve of EFap02.
The highest phage EFap02 amount was 2.3 × 1010 PFU/ml when the MOI was 0.0001. Therefore, the optimal MOI of phage EFap02 is 0.0001 (Figure 1C). The one-step growth curve was measured to determine the latent period of phage EFap02 (Figure 1D). The phage titer increased 10 min after infection and peaked at 30 min, indicating a phage lysis time of approximately 30 min. These data suggest that EFap02 is effective at infecting EFa02.
The stability of EFap02 was tested under different pH, temperature, and chloroform treatments. EFap02 was viable when the pH was 4–11 (Figure 2A). Moreover, EFap02 was viable at 60°C and was rapidly inactivated at 70°C (Figure 2B). However, the EFap02 titer dropped to approximately 5 × 106 PFU/mL after chloroform treatment (Figure 2C). These data indicate that EFap02 can tolerate a strong acid and alkali environment and high temperature but is partially sensitive to chloroform.
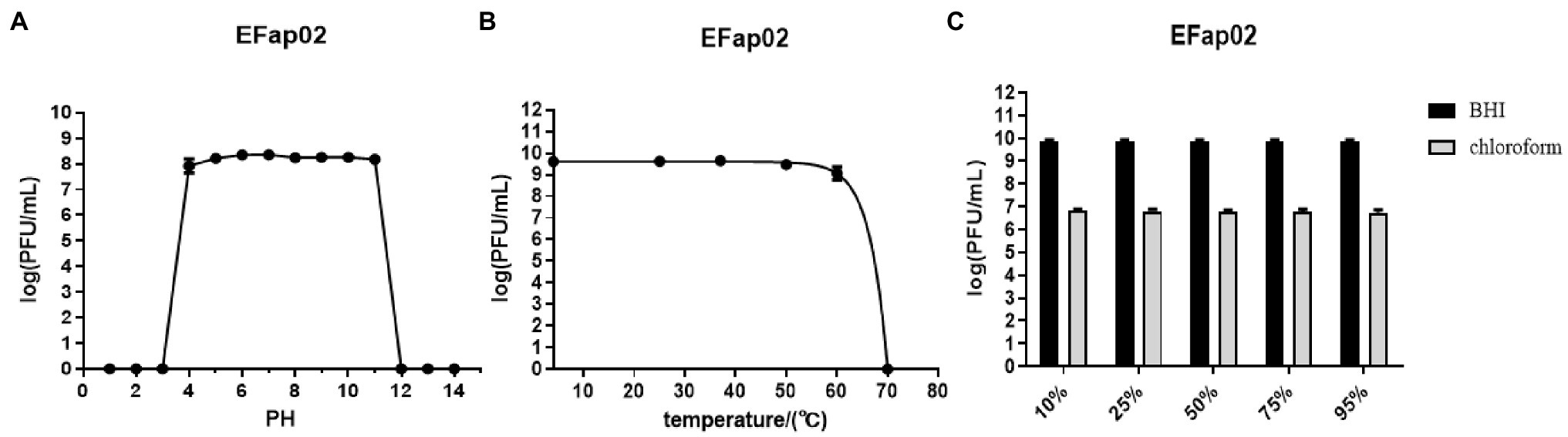
Figure 2. Stability of EFap02 under different conditions. (A) EFap02 is stable over a broad range of pH values (pH values from 4 to 11). (B) EFap02 is viable over a broad range of temperatures. (C) EFap02 is sensitive to chloroform, and the titer drops to approximately 5 × 106 PFU/ml after treatment.
The phage EFap02 genome structure is shown in Figure 3. The whole-genome sequencing results and phageTerm analysis results revealed that EFap02 had a linear double-stranded DNA, with a length of 39,776 bps and a (G + C) content of 35%. The complete genome sequence is available at GenBank (accession no. OL505084). The EFap02 genome sequence was searched as a query in the nucleotide database of the National Center for Biotechnology Information (NCBI). The results showed that the genome sequences of Enterococcus phage phiSHEF4 (GenBank accession no. NC_042022.1) and Enterococcus phage phiSHEF11 (GenBank accession no. OL799257.1) shared similar query coverage above 80% and identity above 91% with the EFap02 genome. The phylogenetic tree of EFap02 was constructed based on the nucleotide sequence terminase large subunit (ORF58), as shown in Figure 4. The result showed that EFap02 is closely related to the Enterococcus phage LY0323 (GenBank accession no. MH375074) genome, but was distantly related to the other phages included in the analyses. Based on these results, phage EFap02 could be considered as a novel phage. The EFap02 genome sequence encodes 60 predicted open reading frames (ORFs), 20 were functional genes, and 40 were hypothetical proteins (Table 3). No homologs of bacteriophage excisionases, repressors, integrases, or transposases were predicted in the EFap02 genome, supporting phage EFap02 as a lytic phage.
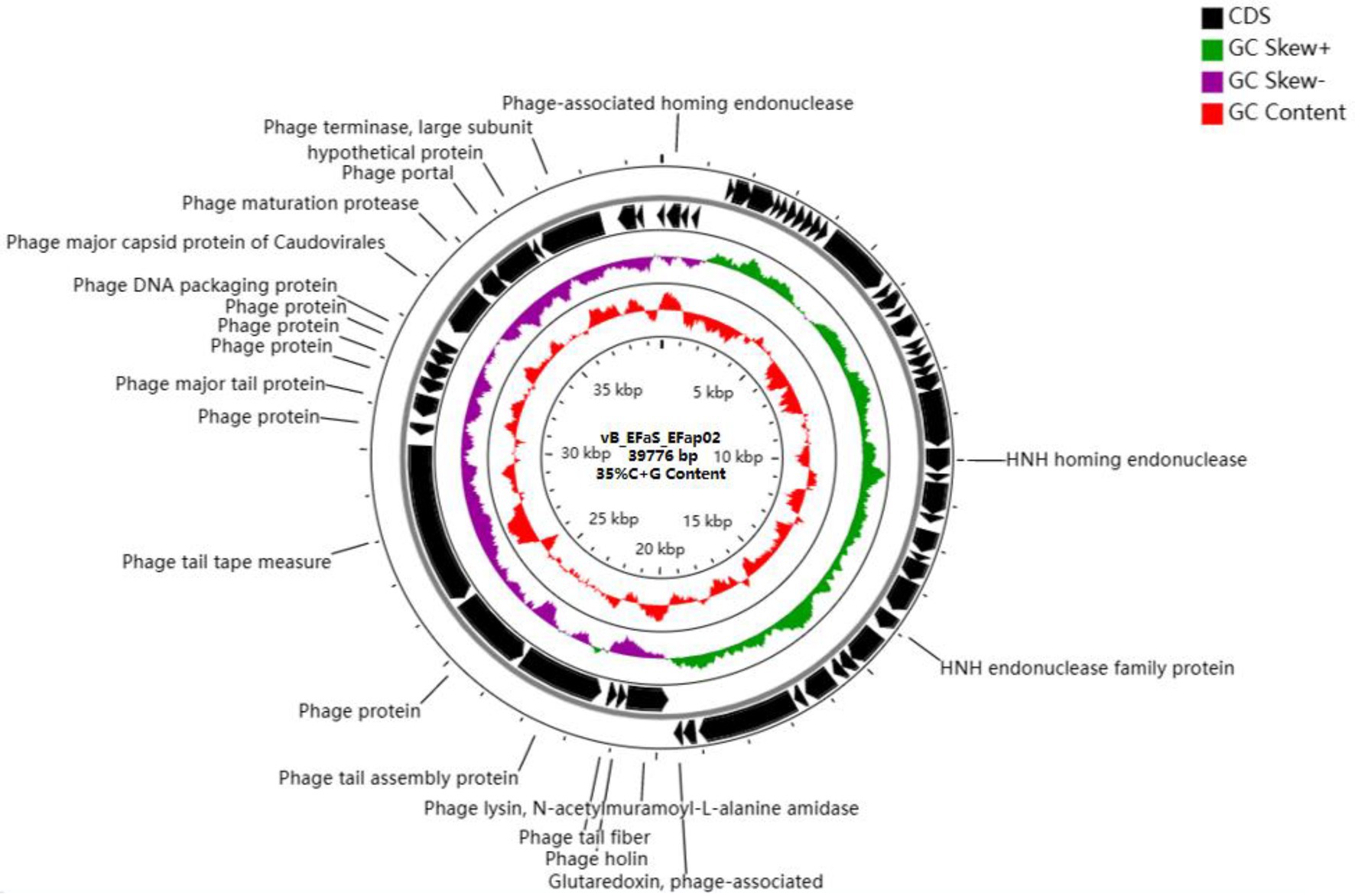
Figure 3. Genome characterization of phage EFap02. Genome map of phage EFap02 and genetic characteristics. Circular genome visualization of EFap02 was performed using the CGView server database. Annotation of the specific function of ORFs was conducted using RAST and the BLASTP database. The first inner circle with the red histogram indicates the GC content, while the second inner circle with the purple and green histograms indicates the GC skew. The outer black circle indicates the predicted ORFs of phage EFap02.
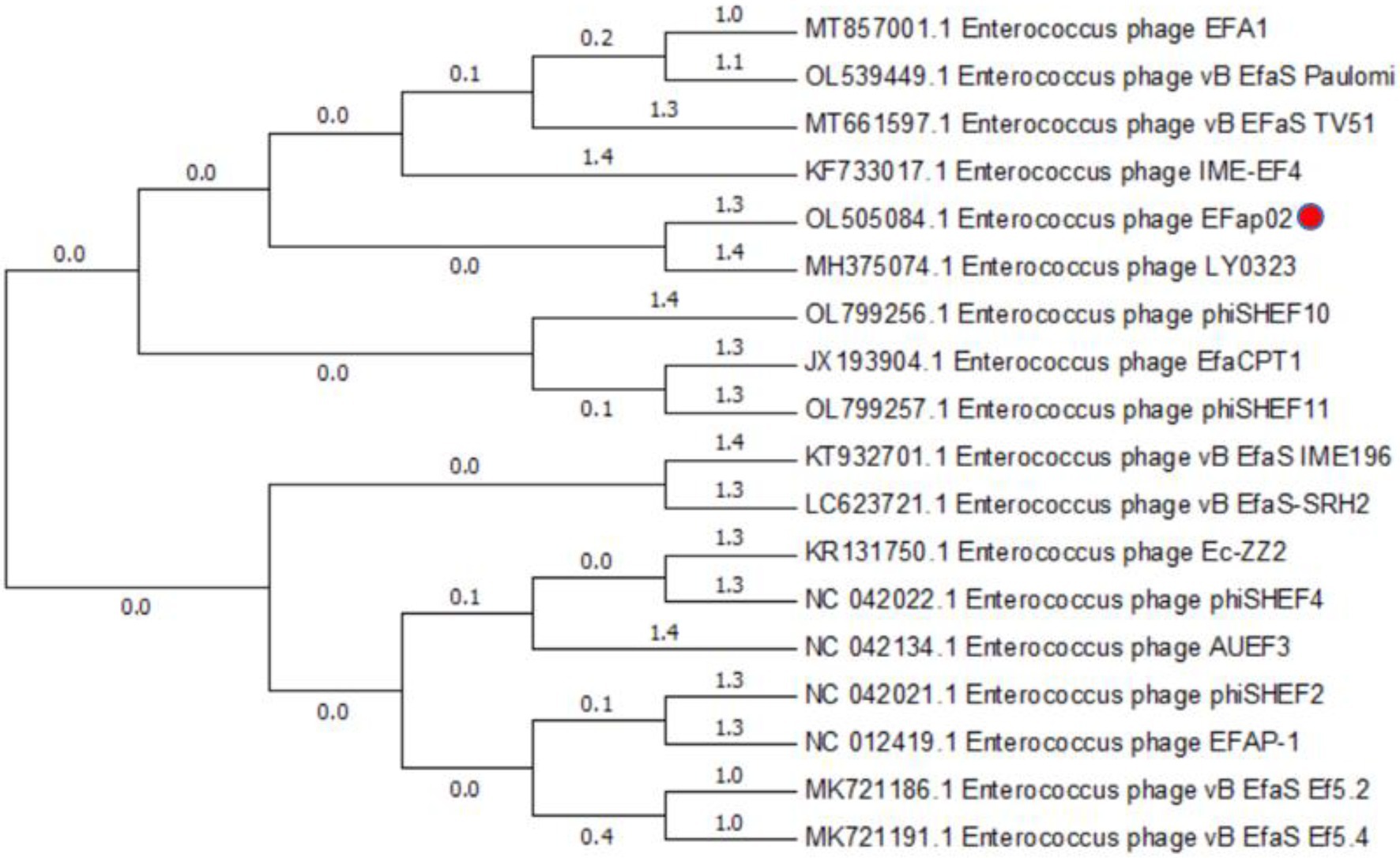
Figure 4. Phylogenetic relationships of phages infecting Enterococcus faecalis. Terminase large subunit and the neighbor-joining method were used to construct the phylogenetic tree.
The EFap02 phage genome includes DNA replication and modification, transcription regulation, phage packaging, structural protein, and host lysis protein modules. ORF53 encodes phage DNA packaging protein in the packaging module. ORF58 encodes a large terminase subunit, an enzyme that inserts a single viral genome into the viral procapsid. ORF56, a portal protein, injects DNA into the host cell through a pathway formed by portal protein. Among the structural proteins, ORF49 encodes the phage major tail protein. ORF44 encodes the phage tail fiber. ORF45 encodes the phage tail assembly protein, and ORF47 encodes the phage tail tape measure protein. These structural proteins are involved in binding to the host bacterium. For the lysis module, ORF42 is annotated as N-acetylmuramoyl-L-alanine amidase, and ORF43 is annotated as holin, which can form micron-scale holes in the inner membrane. The phage releases active endolysin (N-acetylmuramoyl-L-alanine amidase) into the periplasm to digest the peptidoglycan of E. faecalis. The Virulence Search Database shows that none of the ORFs encode virulence factors or antibiotic resistance genes, indicating that EFap02 can be safely used to treat E. faecalis-associated diseases.
In the bacterial growth curve, the OD600 of the phage–bacteria coculture increased 30 h after the phage was added (Figure 5A). The culture medium became turbid due to the growth of phage-resistant bacteria. We then isolated a phage-resistant mutant named EFa02R. Since most resistant bacteria exhibit phage resistance mediated by receptor mutation (Labrie et al., 2010), an adsorption assay was conducted. The results showed that the adsorption rate of EFa02R decreased to approximately 20% (Figure 5B). After sequencing, it was found that resistant bacteria contain a mutation in the gtr2 gene. The mutant site changed from AGC to ATC, and the amino acid changed from serine to isoleucine (Figure 5C).
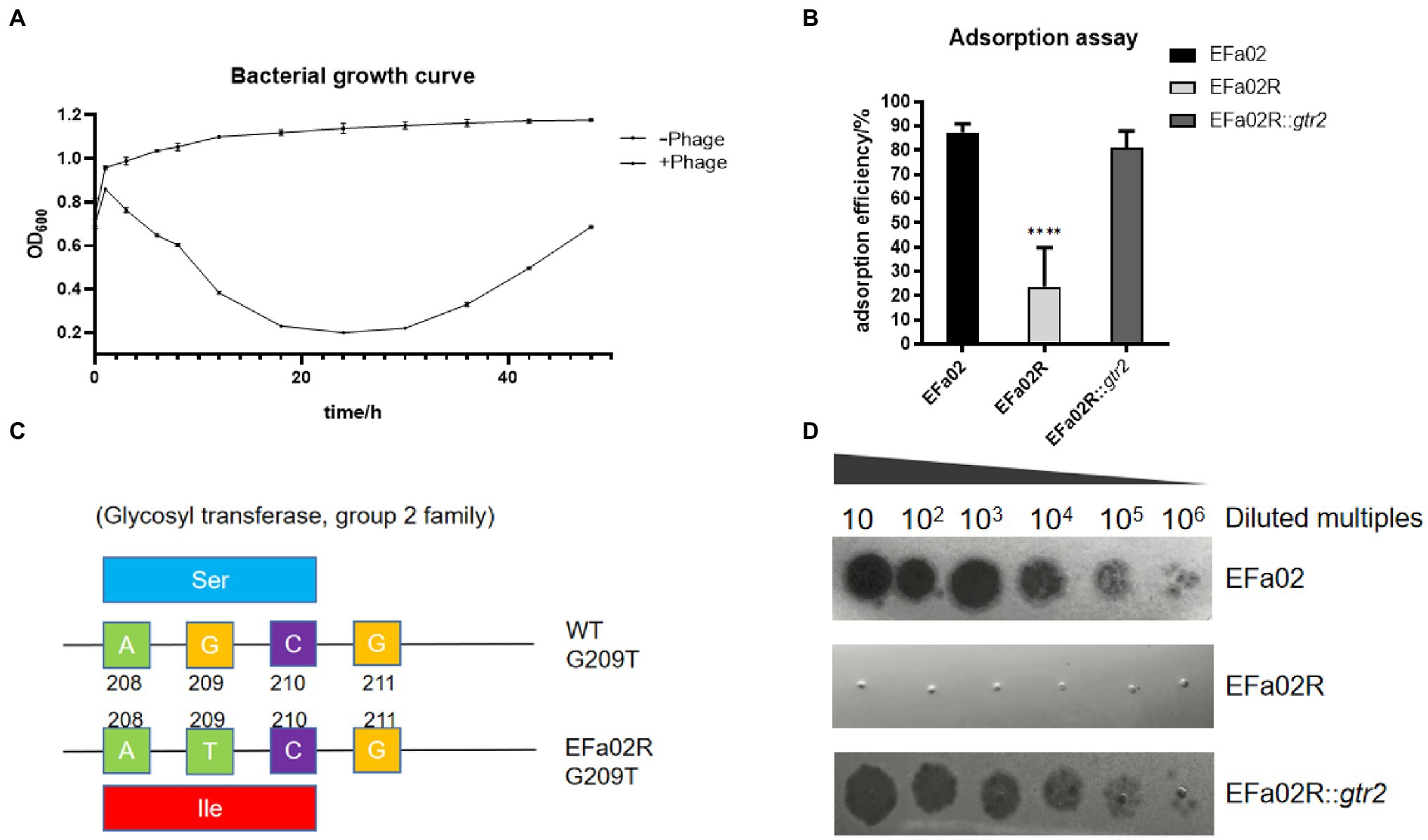
Figure 5. (A) Growth curve of the phage–bacteria coculture. Thirty hours after phage addition, the OD600 of the phage and bacteria coculture began to increase. (B) Adsorption rate of phage onto three strains. This result indicates that EFap02 can adsorb EFa02 and EFa02R::gtr2 but not EFa02R (**** p < 0.0001). (C) The mutation site of the glycosyltransferase Group 2 gene in EFa02R; the mutant site is G209T, which results in the amino acid change from serine to isoleucine. (D) EOP assay indicates that EFap02 can infect EFa02 and EFa02R::gtr2 but not EFa02R.
The gtr2 gene is related to the synthesis of capsular polysaccharide on the surface of Acinetobacter baumannii (Kenyon and Hall, 2013; Singh et al., 2018). We also performed capsule staining of EFa02, EFa02R and EFa02R::gtr2, which showed that EFa02 and EFa02R::gtr2 have the capsule, but EFa02R lost the capsule (Figure 6A). Furthermore, we performed a phage adsorption experiment to test whether capsular polysaccharide is the receptor for EFap02. We found that the adsorption capacity of EFa02R dropped dramatically to 23.9%, and the adsorption capacity recovered to 89% after complementing the gtr2 gene (Figure 5B). Additionally, EOP testing showed that EFa02R::gtr2 was resensitive to EFap02 (Figure 5D). These experiments suggest that the mutation of gtr2 leads to the loss of capsular polysaccharides and prevents EFap02 from adsorbing to EFa02R.
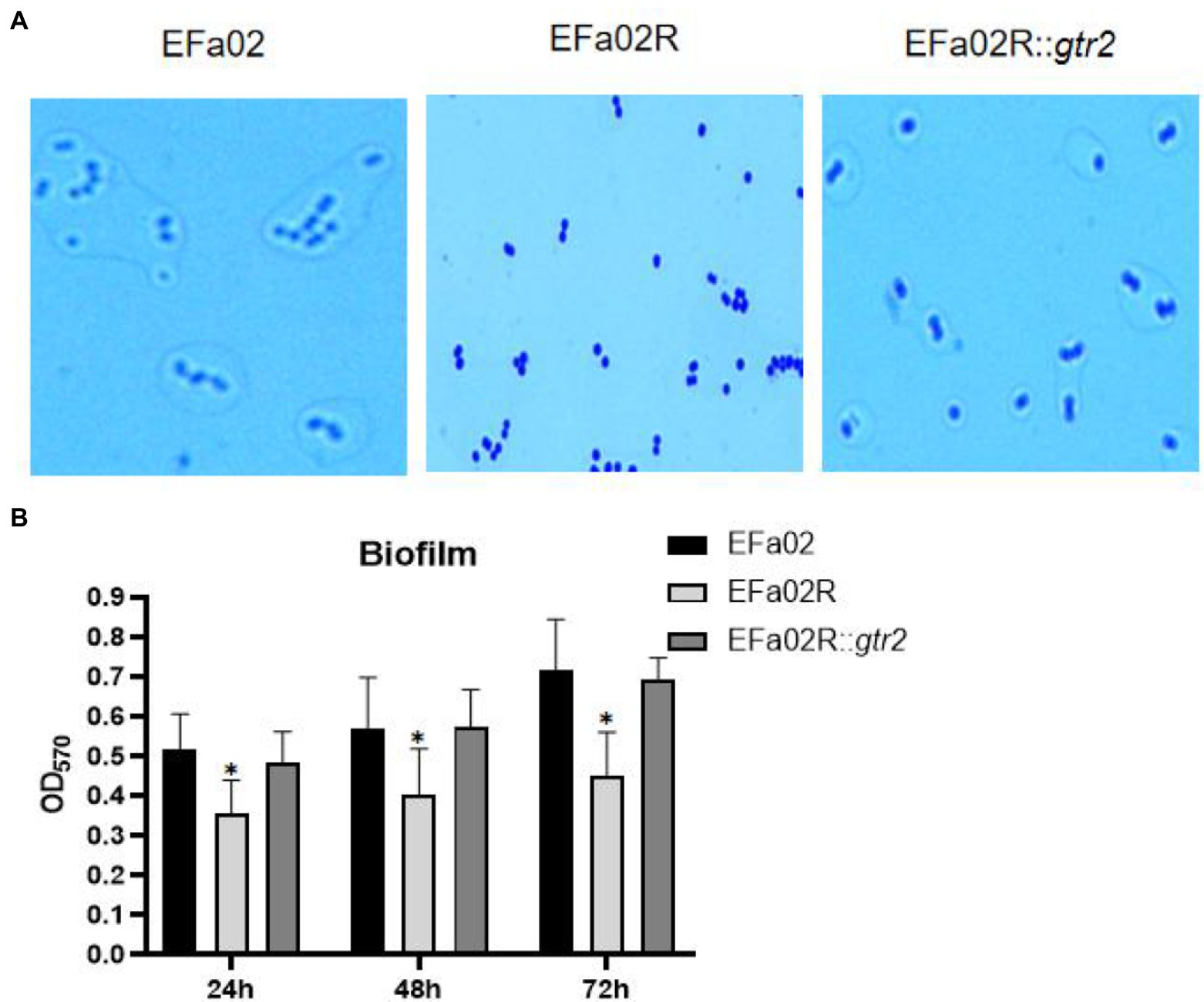
Figure 6. (A) Capsule staining of EFa02, EFa02R, and EFa02R::gtr2. Capsule staining showed that EFa02 and EFa02R::gtr2 have the capsule, but EFa02R lost the capsule. (B) The biofilm mass of EFa02R was less than those of EFa02 and EFa02R::gtr2 (*p < 0.05) at the given time points.
We measured the biofilm formation of these strains on the polystyrene surface after 24, 48, and 72 h of incubation. At each time point, the biofilm of the resistant mutant EFa02R was much less than that of the wild-type strain EFa02 and the complemented strain EFa02R::gtr2 (Figure 6B), indicating that the phage-resistant mutant had impaired biofilm formation.
Phage therapy has been used to treat E. faecalis infectious diseases (Letkiewicz et al., 2009; Bolocan et al., 2019), as well as many other diseases. For example, phages can treat alcoholic hepatitis by killing the E. faecalis that produces cytolysin which leads to hepatocyte lysis (Duan et al., 2019). Thus, the characterization of E. faecalis phages is important for treating E. faecalis-associated diseases.
Identification of the phage receptors is essential for the rational selection of phages for therapy. The E. faecalis phage receptors had been descried previously. Duerkop et al. found that phage infection of E. faecalis requires a predicted integral membrane protein named PIPEF (phage infection protein of E. faecalis). PIPEF is an integral membrane protein that spans the membrane six times (Duerkop et al., 2016). In 2018, Ho et al. showed that mutation of the glycosyltransferase epaR leads to the resistance to phage NPV1 by preventing phage adsorption (Ho et al., 2018). And these phage receptors are cell wall-associated structures that are important for cell viability under external stresses (Canfield and Duerkop, 2020).
In this study, the isolated EFap02 phage is stable under high temperatures and other conditions, making it convenient to store and transport for future clinical use. Although EFap02 has strong lytic activity, we observed the rapid emergence of phage-resistant mutants. Resistance is an issue in phage therapy, and its mechanisms are diverse. The receptor mutation is the most likely to occur, and it prevents phage adsorption (Labrie et al., 2010). In this study, the phage-resistant strain EFa02R had loss-of-function mutations in the glycosyltransferase gene Group 2 family responsible for the biosynthesis of capsular polysaccharides (Kenyon and Hall, 2013; Singh et al., 2018). We confirmed that the gtr2 mutation in EFa02 causes phage resistance and that the capsular polysaccharide is the phage receptor. Identifying phage receptors is essential for the rational development of phage cocktails (Gordillo Altamirano and Barr, 2021).
The loss of receptors prevents phage adsorption; however, there is always a trade-off for phage resistance. We observed that the capsular polysaccharide-loss phage-resistant mutant reduced the formation of biofilms. Previous studies found that the formation of biofilms increased the resistance to antibiotics (Ch'ng et al., 2019). Capsular polysaccharide loss may make bacteria resensitive to some antibiotics, which may allow for clearance of the bacteria by the body’s immune system or antibiotics (Gordillo Altamirano and Barr, 2021).
In summary, this study revealed the biological and genomic characteristics of a phage EFap02 and identified its receptor as the capsular polysaccharide. This study suggests EFap02 as a potential phage therapy agent.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, OL505084.
JL, QZ, KX, and JW conceived and designed the experiments. JL performed the experiments. JL, YZ, YLi, and YLu analyzed the data. JL, KX, QZ, and JW wrote the paper. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (NSFC, 32000120 to QZ) and Military Youth Training Program (2019XQN14).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bao, J., Wu, N., Zeng, Y., Chen, L., Li, L., Yang, L., et al. (2020). Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant. Emerg. Microbes Infect. 9, 771–774. doi: 10.1080/22221751.2020.1747950
Barrick, J. E., Colburn, G., Deatherage, D. E., Traverse, C. C., Strand, M. D., Borges, J. J., et al. (2014). Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics 15:1039. doi: 10.1186/1471-2164-15-1039
Bolocan, A. S., Upadrasta, A., Bettio, P. H. A., Clooney, A. G., Draper, L. A., Ross, R. P., et al. (2019). Evaluation of phage therapy in the context of Enterococcus faecalis and its associated diseases. Viruses 11:366. doi: 10.3390/v11040366
Canfield, G. S., and Duerkop, B. A. (2020). Molecular mechanisms of enterococcal-bacteriophage interactions and implications for human health. Curr. Opin. Microbiol. 56, 38–44. doi: 10.1016/j.mib.2020.06.003
Ch'ng, J. H., Chong, K. K. L., Lam, L. N., Wong, J. J., and Kline, K. A. (2019). Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 17, 82–94. doi: 10.1038/s41579-018-0107-z
Ding, T., Sun, H., Pan, Q., Zhao, F., Zhang, Z., and Ren, H. (2020). Isolation and characterization of Vibrio parahaemolyticus bacteriophage vB_VpaS_PG07. Virus Res. 286:198080. doi: 10.1016/j.virusres.2020.198080
Duan, Y., Llorente, C., Lang, S., Brandl, K., Chu, H., Jiang, L., et al. (2019). Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505–511. doi: 10.1038/s41586-019-1742-x
Duerkop, B. A., Huo, W., Bhardwaj, P., Palmer, K. L., and Hooper, L. V. (2016). Molecular basis for lytic bacteriophage resistance in enterococci. MBio 7:e01304-16. doi: 10.1128/mBio.01304-16
Fiore, E., Van Tyne, D., and Gilmore, M. S. (2019). Pathogenicity of enterococci. Microbiol. Spectr. 7, 4–7. doi: 10.1128/microbiolspec.GPP3-0053-2018
Forti, F., Roach, D. R., Cafora, M., Pasini, M. E., Horner, D. S., Fiscarelli, E. V., et al. (2018). Design of a broad-range bacteriophage cocktail That reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob. Agents Chemother. 62:e02573-17. doi: 10.1128/AAC.02573-17
García-Solache, M., and Rice, L. B. (2019). The Enterococcus: a model of adaptability to its environment. Clin. Microbiol. Rev. 32:00058-18. doi: 10.1128/CMR.00058-18
Garneau, J. R., Depardieu, F., Fortier, L.-C., Bikard, D., and Monot, M. (2017). PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 7:8292. doi: 10.1038/s41598-017-07910-5
Gordillo Altamirano, F. L., and Barr, J. J. (2021). Unlocking the next generation of phage therapy: the key is in the receptors. Curr. Opin. Biotechnol. 68, 115–123. doi: 10.1016/j.copbio.2020.10.002
Ho, K., Huo, W., Pas, S., Dao, R., and Palmer, K. L. (2018). Loss-of-function mutations in confer resistance to ϕNPV1 infection in Enterococcus faecalis OG1RF. Antimicrob. Agents Chemotherapy 62:e00758-18. doi: 10.1128/AAC.00758-18
Jault, P., Leclerc, T., Jennes, S., Pirnay, J. P., Que, Y.-A., Resch, G., et al. (2019). Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 19, 35–45. doi: 10.1016/S1473-3099(18)30482-1
Kenyon, J. J., and Hall, R. M. (2013). Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One 8:e62160. doi: 10.1371/journal.pone.0062160
Kęsik-Szeloch, A., Drulis-Kawa, Z., Weber-Dąbrowska, B., Kassner, J., Majkowska-Skrobek, G., Augustyniak, D., et al. (2013). Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol. J. 10:100. doi: 10.1186/1743-422X-10-100
Khalifa, L., Shlezinger, M., Beyth, S., Houri-Haddad, Y., Coppenhagen-Glazer, S., Beyth, N., et al. (2016). Phage therapy against Enterococcus faecalis in dental root canals. J. Oral Microbiol. 8:32157. doi: 10.3402/jom.v8.32157
Labrie, S. J., Samson, J. E., and Moineau, S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327. doi: 10.1038/nrmicro2315
Lee, D., Im, J., Na, H., Ryu, S., Yun, C. H., and Han, S. H. (2019). The novel Enterococcus phage vB_EfaS_HEf13 has broad lytic activity Against clinical isolates of Enterococcus faecalis. Front. Microbiol. 10:2877. doi: 10.3389/fmicb.2019.02877
Lefkowitz, E. J., Dempsey, D. M., Hendrickson, R. C., Orton, R. J., Siddell, S. G., and Smith, D. B. (2018). Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV). Nucleic Acids Res. 46, D708–D717. doi: 10.1093/nar/gkx932
Letkiewicz, S., Miedzybrodzki, R., Fortuna, W., Weber-Dabrowska, B., and Górski, A. (2009). Eradication of Enterococcus faecalis by phage therapy in chronic bacterial prostatitis — case report. Folia Microbiol. 54, 457–461. doi: 10.1007/s12223-009-0064-z
Lu, S., Le, S., Tan, Y., Zhu, J., Li, M., Rao, X., et al. (2013). Genomic and proteomic analyses of the terminally redundant genome of the Pseudomonas aeruginosa phage PaP1: establishment of genus PaP1-like phages. PLoS One 8:e62933. doi: 10.1371/journal.pone.0062933
Miller, W. R., Murray, B. E., Rice, L. B., and Arias, C. A. (2016). Vancomycin-resistant enterococci: therapeutic challenges in the 21st century. Infect. Dis. Clin. N. Am. 30, 415–439. doi: 10.1016/j.idc.2016.02.006
Oduor, J. M. O., Kadija, E., Nyachieo, A., Mureithi, M. W., and Skurnik, M. (2020). Bioprospecting Staphylococcus phages with therapeutic and bio-control potential. Viruses 12:133. doi: 10.3390/v12020133
Shen, M., Zhang, H., Shen, W., Zou, Z., Lu, S., Li, G., et al. (2018). Pseudomonas aeruginosa MutL promotes large chromosomal deletions through non-homologous end joining to prevent bacteriophage predation. Nucleic Acids Res. 46, 4505–4514. doi: 10.1093/nar/gky160
Singh, J. K., Adams, F. G., and Brown, M. H. (2018). Diversity and function of capsular polysaccharide in. Front. Microbiol. 9:3301. doi: 10.3389/fmicb.2018.03301
Stothard, P., and Wishart, D. S. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539. doi: 10.1093/bioinformatics/bti054
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Underwood, A. P., Mulder, A., Gharbia, S., and Green, J. (2005). Virulence searcher: a tool for searching raw genome sequences from bacterial genomes for putative virulence factors. Clin. Microbiol. Infect. 11, 770–772. doi: 10.1111/j.1469-0691.2005.01210.x
Yang, Y., Shen, W., Zhong, Q., Chen, Q., He, X., Baker, J. L., et al. (2020). Development of a bacteriophage cocktail to constrain the emergence of phage-resistant. Front. Microbiol. 11:327. doi: 10.3389/fmicb.2020.00327
Keywords: bacteriophage, Enterococcus faecalis, phage resistance, phage receptor, capsular polysaccharide, biofilm
Citation: Liu J, Zhu Y, Li Y, Lu Y, Xiong K, Zhong Q and Wang J (2022) Bacteriophage-Resistant Mutant of Enterococcus faecalis Is Impaired in Biofilm Formation. Front. Microbiol. 13:913023. doi: 10.3389/fmicb.2022.913023
Received: 05 April 2022; Accepted: 19 May 2022;
Published: 09 June 2022.
Edited by:
Mao Ye, Institute of Soil Science (CAS), ChinaReviewed by:
Mengzhe Li, Beijing University of Chemical Technology, ChinaCopyright © 2022 Liu, Zhu, Li, Lu, Xiong, Zhong and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kun Xiong, eGlvbmdrdW41MTVAc2luYS5jb20=; Qiu Zhong, NDEwODM1ODMwQHFxLmNvbQ==; Jing Wang, d2FuZ2ppbmcyMDA4QGFsaXl1bi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.