
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 13 September 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.910343
This article is part of the Research TopicAdvances in Emerging Coronavirus Identification and Tracing MethodsView all 21 articles
 Ting Wang1†
Ting Wang1† Jie Xu2,3†
Jie Xu2,3† Beibei Wang4
Beibei Wang4 Yulian Wang3
Yulian Wang3 Wei Zhao3
Wei Zhao3 Bin Xiang5
Bin Xiang5 Yuhua Xue6
Yuhua Xue6 Quan Yuan7
Quan Yuan7 Yiqiang Wang2,8*
Yiqiang Wang2,8*Background: The COVID-19 pandemic has killed over 6 million people worldwide. Despite the accumulation of knowledge about the causative pathogen severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the pathogenesis of this disease, cures remain to be discovered. We searched for certain peptides that might interfere with spike protein (S protein)-angiotensin-converting enzyme 2 (ACE2) interactions.
Methods: Phage display (PhD)-12 peptide library was screened against recombinant spike trimer (S-trimer) or receptor-binding domain (S-RBD) proteins. The resulting enriched peptide sequences were obtained, and their potential binding sites on S-trimer and S-RBD 3D structure models were searched. Synthetic peptides corresponding to these and other reference sequences were tested for their efficacy in blocking the binding of S-trimer protein onto recombinant ACE2 proteins or ACE2-overexpressing cells.
Results: After three rounds of phage selections, two peptide sequences (C2, DHAQRYGAGHSG; C6, HWKAVNWLKPWT) were enriched by S-RBD, but only C2 was present in S-trimer selected phages. When the 3D structures of static monomeric S-RBD (6M17) and S-trimer (6ZGE, 6ZGG, 7CAI, and 7CAK, each with different status of S-RBDs in the three monomer S proteins) were scanned for potential binding sites of C2 and C6 peptides, C6 opt to bind the saddle of S-RBD in both 6M17 and erected S-RBD in S-trimers, but C2 failed to cluster there in the S-trimers. In the competitive S-trimer-ACE2-binding experiments, synthetic C2 and C6 peptides inhibited S-trimer binding onto 293T-ACE2hR cells at high concentrations (50 μM) but not at lower concentrations (10 μM and below), neither for the settings of S-trimer binding onto recombinant ACE2 proteins.
Conclusion: Using PhD methodology, two peptides were generated bearing potentials to interfere with S protein-ACE2 interaction, which might be further exploited to produce peptidomimetics that block the attachment of SARS-CoV-2 virus onto host cells, hence diminishing the pathogenesis of COVID-19.
The COVID-19 pandemic had caused 6.35 million death worldwide as of 8 July 2022 and still poses a serious challenge in some nations. Worse was that over two more folds of “excess deaths” might have occurred due to indirect consequences of the pandemic, such as changes in “social, economic, and behavioral responses to the pandemic, including strict lockdowns” (Collaborators, 2022). While vaccinations and natural infections build herd immunity that helps to protect people from infection or prevent pandemic recurrence (Lipsitch and Dean, 2020; Mobarak et al., 2022), cures are still lacking for the infected individuals in most areas. Among the scientific efforts, various therapeutics have been tried, such as cells (Sang et al., 2021), engineered antibodies (Matthay and Luetkemeyer, 2021), natural products (Bhattacharya and Paul, 2021), synthetical biologicals (Robson, 2020), and small molecules (Tiwari et al., 2020). Intended targets included viral structural proteins (Sheward et al., 2022), host products [e.g., interleukin 6 (IL-6)] (Murthy and Lee, 2021), viral replication process (Schafer et al., 2022), or host-virus interactions (Gordon et al., 2020). The strategies aiming at the first step of virus-host interactions sound most attractive. The viral spike (S) protein trimers (S-trimers) are thought to be the main molecules mediating the affinity of exogenous severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus for angiotensin-converting enzyme 2 (ACE2) or other less-attended molecules, such as TMPRSS2 on host cells (Hoffmann et al., 2020). Since the structures of both S and ACE2 proteins are known, computation or computer-based methods are thought to be high for novel drug discovery (Cao et al., 2020; Jawad et al., 2021). However, though a few candidates had been proposed in these in silico studies, only part of them had been proven effective in functional experimental studies, highlighting the demand for more robust strategies that mimic the actual virus-host interactions more faithfully.
Phage display (PhD) methodology, as exemplified in other infectious diseases (Huang et al., 2012; Alfaleh et al., 2020; Roth et al., 2021), met this end and has been tried in the context of SARS-CoV-2 or COVID-19. In detail, PhD has been successful in producing antibodies for neutralizing or detection (Noy-Porat et al., 2020; Bertoglio et al., 2021) in identifying COVID-19-induced antibodies to the virus (Zhao et al., 2021) or in searching for viral epitopes responsible for virus escaping immune responses (Garrett et al., 2021). Based on our previous experience using PhD in studies of the host-pathogen interactions (Zhao et al., 2012; Shen et al., 2017), we performed PhD screening to search for peptides that would bind the receptor-binding domain (RBD) domain of SARS-CoV-2 S protein (S-RBD). Theoretically, if such peptides could bind the site(s) critical for S-RBD interaction with its receptors (e.g., ACE2 or other molecules), they should interfere with S-trimer-ACE2 interactions. Furthermore, such an S-protein Entrapped Affinity Ligand (SEAL) peptide should be able to block the binding of the viruses with their target cells. Here, we report that two SEAL peptides were obtained via phage displaying against S-RBD and S-trimer proteins, and preliminary functional studies demonstrated weak blocking effects at high concentrations. Encouragingly, while this project was ongoing, three groups reported their results obtained by protocols mainly relying on PhD (Petrenko et al., 2022; Sevenich et al., 2022; Yang et al., 2022). The promises and limitations of these studies were also discussed.
Recombinant SARS-CoV-2 S-trimer proteins were from the commercial resource (Cat# DRA49, MW 136.6 kDa; Novoprotein Company, Suzhou, China), and recombinant S-RBD products corresponding to aa319-541 of YP_009724390.1, MW 30.7 kDa (Lan et al., 2020) was a generous gift from Li (Tsinghua University, Beijing, China). PhD-12 Peptide PhD Library Kit (New England BioLabs, Beverly, MA, United States) was used for PhD screening against these two proteins. Briefly, S-trimer proteins were immobilized overnight at 4°C on the enzyme-linked immunosorbent assay (ELISA) plates at 100 μg/ml in 0.1 M NaHCO3, pH 8.6. The plates were then blocked with 0.5% bovine serum albumin (BSA) in 0.1 M NaHCO3 buffer (containing 0.02% NaN3) for 1 h. After six washes with tris base-buffered saline solution (TBST buffer containing 0.01% Tween-20), 2 × 1011 phages in TBST buffer were added for 45 min at room temperature. After ten washes, bound phages were recovered, amplified in Escherichia coli, harvested into TBS buffer (containing 0.02% NaN3), quantified with a plaque-forming assay, and used the product for the second round display. After two or three screening rounds, bound phages were harvested into elution buffer (0.2 M Glycine-HCl, 1 mg/ml BSA, pH 2.2) and neutralized with 1 M tris base-HCl buffer, pH 9.1. After dilution, the phage mix was applied onto bacterial plates to obtain blue plaques. Thirty (after the second round) or 25 (after the third round) isolated plaques were randomly picked for phage DNA sequencing using the primers in the kit. The resulting 12-amino acids peptides translated from phage DNA inserts were analyzed, and the most promising sequence was used for subsequent studies.
Enzyme-linked immunoassay (ELISA) was used to confirm the affinity of the resulting monoclonal phages for targeted proteins. ELISA plates (Corning, NY, United States) were coated with 10 μg/ml S-trimer or S-RBD proteins. With the starting original library phages (O virions) as control, all selected interest phages were amplified, titrated, and added to the plates at different concentrations (2.5 × 109, 1 × 1010, 4 × 1010, 1.6 × 1011, and 6.4 × 1011 phage virions in 100 μl) for 1 h at room temperature. The plates were washed six times with TBST washing buffer and then incubated with diluted horseradish peroxidase (HRP)-conjugated anti-M13 monoclonal antibody (GE Healthcare, Piscataway, NJ, United States) for 1 h. After six washes, 3,3′, 5,5-tetramethylbenzidine (TMB) solution (Beyotime Biotechnology, Shanghai, China) was added to the plate, and after 10 min of development, the reaction was stopped by adding 2 M H2SO4 solution. Optical absorbance was measured at 450 nm in a microplate reader (Synergy H1, BioTek Instruments, Inc., Winooski, VT, United States).
The surface of RBD or S-trimer proteins was scanned using the PEP-SiteFinder (Saladin et al., 2014). The 3D models of RBD to locate the potential docking site(s) of interest peptide(s) on S proteins (Yan et al., 2020) or S-trimers (Lv et al., 2020; Wrobel et al., 2020) were retrieved from Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB). The top 50 poses of each peptide in each protein model were checked to identify the most likely binding site(s). Cn3D (Wang et al., 2000) was also utilized for viewing these protein structures.
293T-ACE2hR cells, a cell line consistently expressing human ACE2 (hACE2) on the cell surface (Supplementary Figure 1; Zhang et al., 2021), or recombinant ACE2 proteins (Cat#10108-H02H, Novoprotein) were utilized to test the potential effect of interest peptides on S-trimer-ACE2 binding. In brief, two possible SEAL peptides derived from the above analysis (C2 and C6) and two reference peptides [spike-binding peptide 1 (SBP1) and spike-binding peptide 1 (SBP2)] (Zhang et al., 2020) were ordered from Biotech Bioscience and Technology (Shanghai, China) and dissolved in PBS. Their sequences were as follows: C2, DHAQRYGAGHSG; C6, HWKAVNWLKPWT; SBP1, IEEQAKTFLDKFNHEAEDLFYQSK; and SBP2, TFLDKFNHEAED. 293T-ACE2hR cells were grown in 96-well plates until confluent in the first measurement setting. S-trimer at 2 nM was mixed with equal volume (25 μl) of peptides at different concentrations (0, 0.16, 0.8, 4, 20, and 100 μM) and kept at room temperature for 1 h. After removing the culture medium from the cells, the mixture was added (50 μl/well) and kept at room temperature for 1 h. Unbound peptides and proteins were removed, and the cells were washed three times with PBS. HRP-conjugated Anti-6X His tag® antibody (diluted at 1:10,000; Abcam, Cambridge, MA, United States) was added to each well for 1 h at room temperature. After three washes with PBS, TMB Solution (Beyotime Biotechnology) was added to the plate, and the plate was read at OD370 nm in a Multiskan Go Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States).
Then, 293T-ACE2hR cells were substituted by recombinant ACE2 proteins and coated onto ELISA plates in the other measurement setting. In brief, recombinant ACE2 proteins (Novoprotein) were immobilized overnight at room temperature on ELISA plates at a 5 μg/ml concentration in 0.1 M NaHCO3, pH 9.6. The plates were then blocked with 10% fetal calf serum for 2 h and washed with tris-buffered saline solution containing 0.1% Tween-20 [phosphate-buffered saline (PBST) buffer]. Then a premixture of S-trimer proteins (final concentration 0.01 μM) with peptides (C2, C6, SBP1, and SBP2) of different concentrations (0.5, 1.65, and 5 μM) was added to each well (50 μl/well). The following procedures were described above for the 293T-ACE2hR cells setting.
Phage display has been widely used in identifying interacting partners of target molecules that were included in previous projects of this team (Zhao et al., 2012; Wang et al., 2019). In the current study, we applied PhD on S-trimers or S-RBD proteins, aiming to obtain peptides supposedly able to “seal” the potent binding site on their surface. After three rounds of panning PhD against recombinant S-RBD proteins, two phages with peptide sequences DHAQRYGAGHSG (C2) and HWKAVNWLKPWT (C6) were enriched in the elutes, each of them accounting for 10 clones in all 25 sequenced clones. Interestingly, C2 and C6 accounted for 7 and 6 clones in the elute after the second panning in all 30 sequenced clones. Therefore, we did not attempt more rounds of panning. When S-trimer proteins were used for panning, only the C2 sequence dominated the elutes, accounting for 20 of 30 clones after the second panning and 16 of 25 phages after the third panning, respectively. Next, using the starting library phages (O virions) as control, the ELISA assay demonstrated dose-dependent binding of monoclonal C2 and C6 phages to immobilized S-RBD proteins and C2 phages for S-trimer protein (Figure 1). An accurate comparison between affinity of C2 and C6 phages for the same target (e.g., S-RBD proteins) was not attempted, or between affinities of C2 phages for different target proteins (i.e., S-RBD or S-trimer). Measurement of affinity of C6 phages for S-trimer was not attempted either.
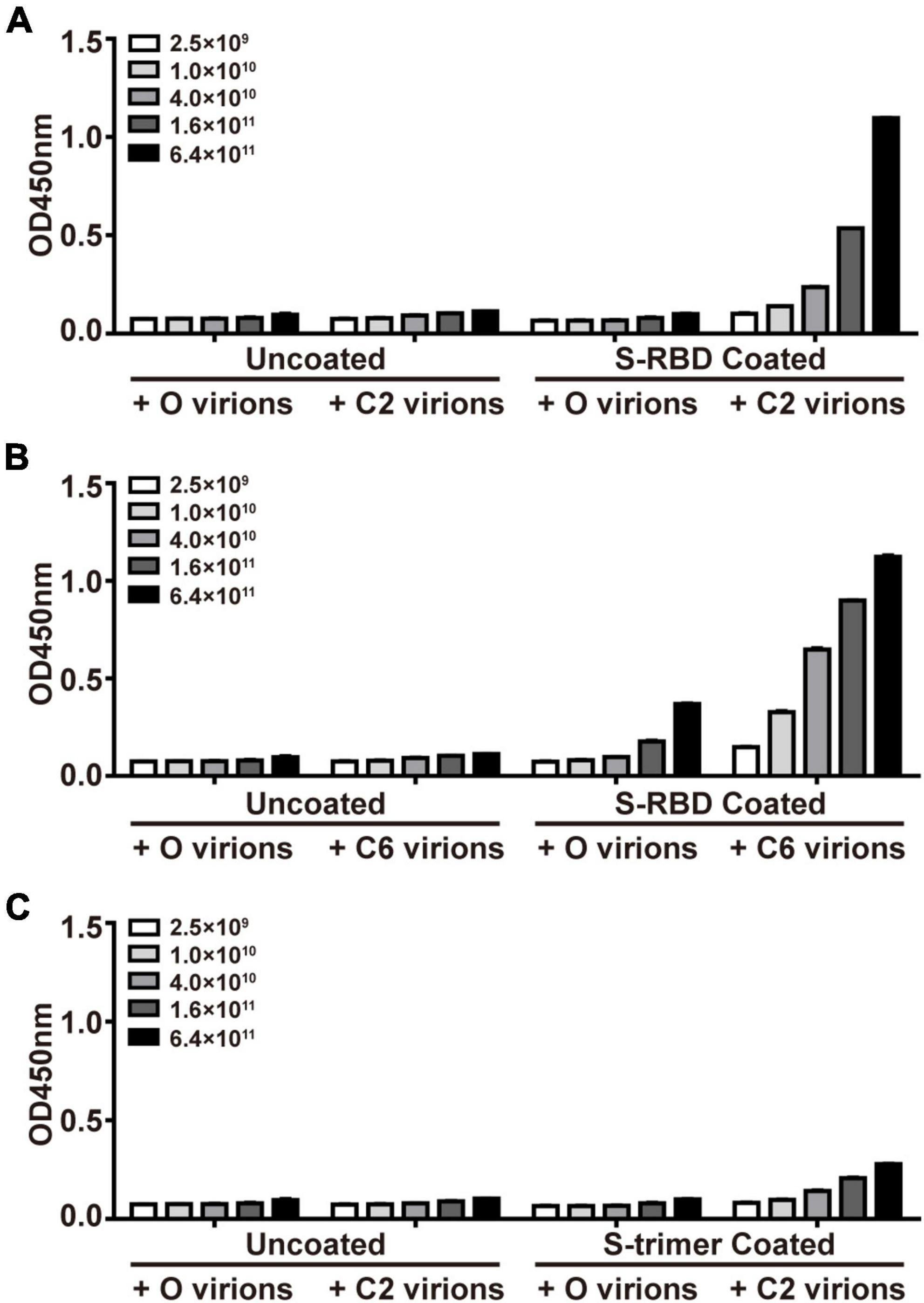
Figure 1. Confirmation of affinity binding of phages of S-protein Entrapped Affinity Ligand (SEAL) sequences with recombinant spike-receptor-binding domain (S-RBD) (A,B) or S-trimer proteins (C). The plates were coated with S-RBD (10 μg/ml, i.e., about 0.33 μM, A,B) or S-trimer proteins (10 μg/ml, i.e., about 0.073 μM, C). The interest phages (C2 and C6) and starting library phages (O) were serially diluted and tested for binding. The apparent less efficient C2 virions to bind S-trimer (C) than to bind S-RBD (A) might be due to these two targets’ difference in molar concentrations (by about 4.4-fold) at the same mass concentrations.
Previous crystal structural studies suggested that the resting S-trimers on the virus surface took a “closed” figuration and, upon contacting ACE2 (or other receptors) on host cells, went through the opening process and exposed the RBD (Wrobel et al., 2020). Surely, this opening process would also alter the configuration of the whole molecule. When the static model of single S-RBD protein (6M17) was used for predicting peptide binding sites, it was found that all fifty C6 peptide poses were docked onto the saddle of S-RBD, while a fraction of C2 peptide poses were docked onto other sites that were supposedly not to directly affect S-RBD’s receptor binding functions (Figure 2A). When S-trimer was used for modeling, binding sites for C2 poses were even more dispersed, and few of them would dock onto S-RBD saddlebacks, independent of S-trimers’ configuration (Figure 2B). For example, in the all-closing status of the S-trimer (6ZGE), 11 of 50 C2 poses were located in the spaces among the three S-RBDs, 6 between neighboring S-RBDs, and 8 in the middle between S-RBD and N-terminal domain (NTD). In the all-open configuration (7CAK), only 6 poses were related to S-RBD and not any poses were docked onto the saddle of open S-RBDs.
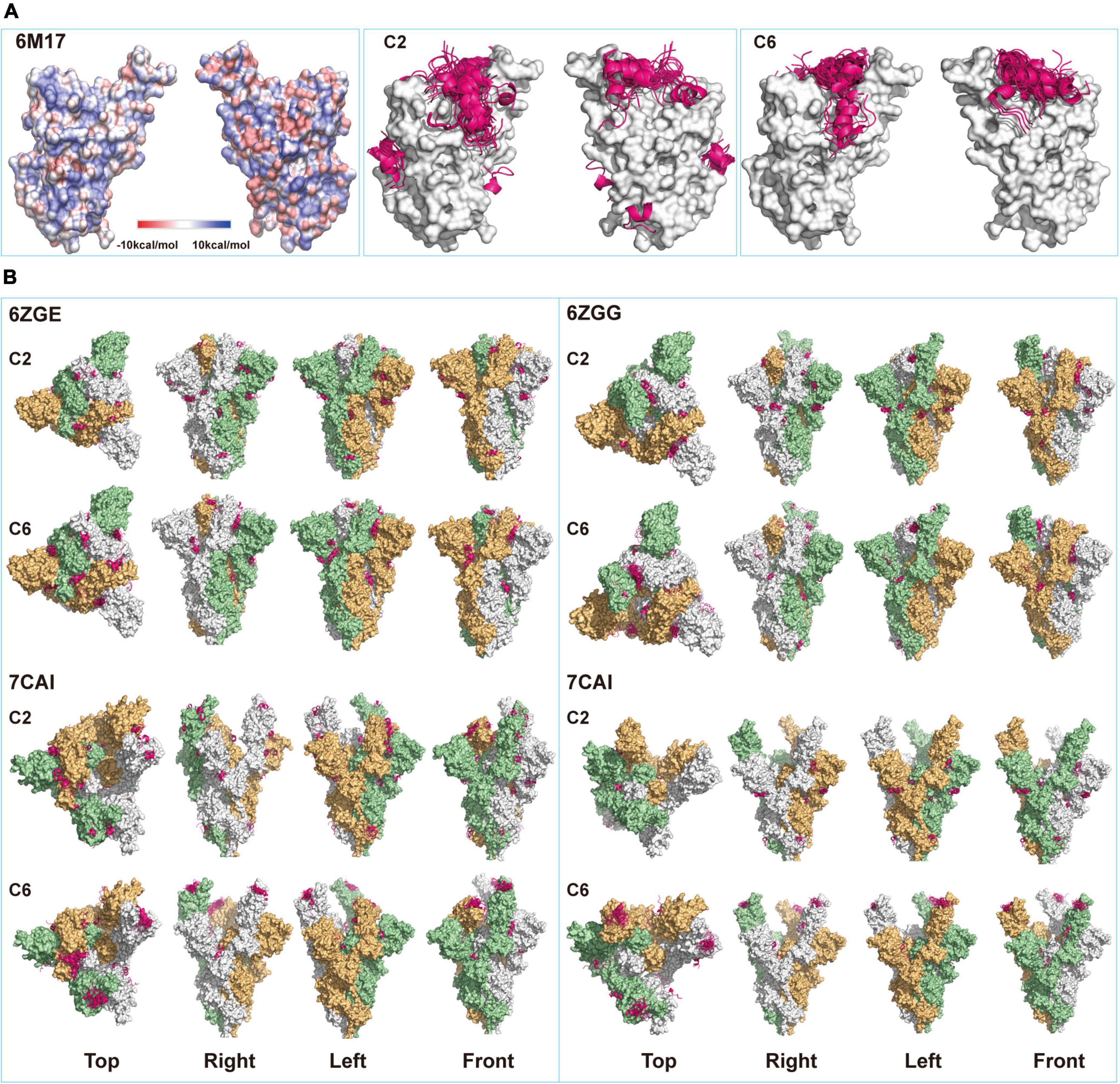
Figure 2. Structural modeling of peptides-spike interactions. (A) Comparing binding patterns for C2 and C6 peptides (pink belts) onto spike-receptor-binding domain (S-RBD). Shown are the fifty poses with the highest of each binding exclusively onto the saddle of RBD (6M17, open), some poses of C2 peptide bind onto other sites on the surface of RBD. (B) Four configurations corresponding to the different status of RBDs in the S-trimers (6ZGE, 6ZGG, 7CAI, and 7CAK for 0, 1, 2, and 3 RBDs open, respectively) were compared for their potential bindings with the two peptides. Each figuration was given a view from the top, right, left, and front sides.
On the contrary, once a single S-RBD was erected (6ZGG), several C6 poses were docked onto its saddle. When one or two more S-RBDs were erected (7CAI and 7CAK, respectively), almost all C6 poses were on their saddlebacks. We assumed that in an actual environment that contained both virus and host cells, if the C2 or C6 peptides were present when S-trimers were in the closed configuration just like in resting virus, their binding onto the surface of S-trimers might facilitate or hinder the opening or erection of S-RBD, which deserved in-depth investigation in future. However, once the S-trimer opening process was initiated, C6 peptides should be able to bind the saddle of S-RBDs. Since it has been well documented that the saddle section was critical for RBD functions, such as binding with ACE2 (Lan et al., 2020; Wang et al., 2020) and being immunogenic (Greaney et al., 2021), we proposed that C6 peptides might interfere with the interaction of S-trimer with ACE2 by SEALing the S-RBD saddle(s). The differential docking sites’ prediction observed for C2 and C6 might partially explain that C2 virions were enriched by both S-RBD and S-trimer proteins, while the C6 virions were enriched by S-RBD proteins only (Figure 1).
When the above hypothesis was tested on recombinant ACE2 proteins coated on a solid surface, no blocking effect was observed for any tested peptides even at a 500:1 (5 μM vs. 0.01 μM, peptide vs. S-trimer) ratio (Figure 3A). These peptides’ ineffectiveness in blocking S-trimer binding with ACE2 at equivalent concentrations was also observed when 293T-ACE2hR cells were used as the source of S-trimer targets in living cells. When 50,000-fold overdose of C2 or C6 peptides was present, namely, 50 μM peptides vs. 0.001 μM S-trimer proteins, a blocking effect was observed for C2 and C6, but still not for SBP1 or SBP2 peptides (Figure 3B).
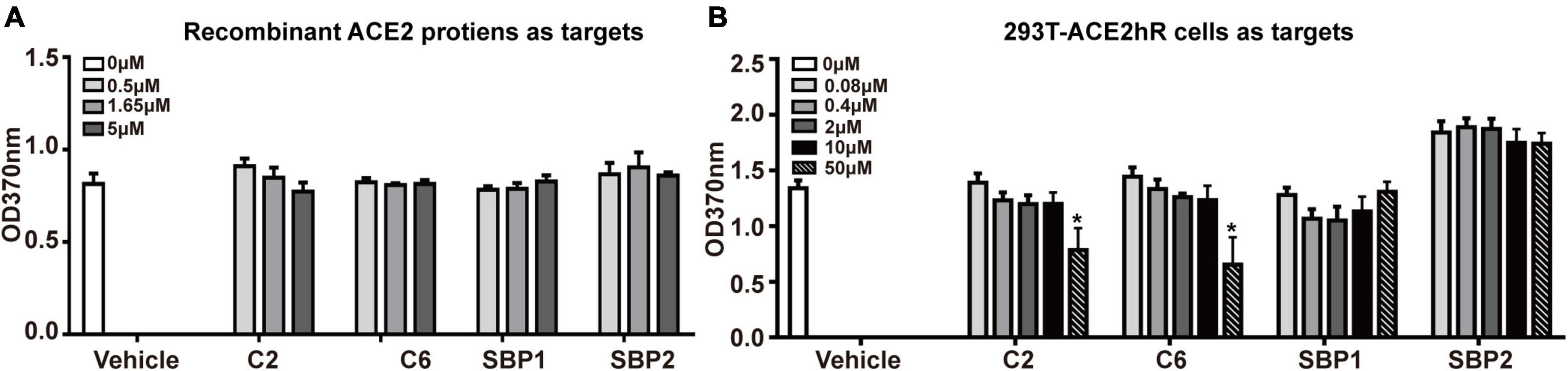
Figure 3. The potential blocking effect of peptides on the binding of S-trimer onto cellular or soluble angiotensin-converting enzyme 2 (ACE2). (A) The enzyme-linked immunoassay (ELISA) was applied to assess the blocking ability of synthetic peptides C2 and C6 at binding between ACE2 molecules and S-trimer protein, and the peptide SBP1 was used as positive control and peptide spike-binding peptide 2 (SBP2) as the negative control. (B) ELISA was applied to assess the blocking ability of synthetic peptides C2 and C6 at binding between 293T-ACE2hR cells and S-trimer protein. The peptide SBP1 was used as a positive and peptide SBP2 as the negative control. *p < 0.05, vs. vehicle, all by Student’s t-test.
Failure of 500-fold overdose of C2 or C6 peptides to block S-trimers binding with their receptors was disappointing, and we suggested two possible explanations for this failure, especially for C6. First, the occupation of the saddle by C6 did not cause enough stereotype blockade as expected, which might be due to the small size of this 12-aa peptide, especially when most poses lay on the saddle “seat” in a vertical-crossing orientation rather than paralleling along the saddle axis (Figure 2A). It has been demonstrated that the “cantle” and “pommel” contributed more than the “seat” (Supplementary Figure 2) to the overall affinity and configuration fitting between S-RBD and ACE2 (Lan et al., 2020; Wang et al., 2020). Second, the affinity of these peptides for S-RBD might be too low to constitute effective competitors when natural and intact receptors (i.e., ACE2 molecules) were present, which might be the case for the SBP1 and SBP2 peptides. SBP1 was from the N-terminal sequence of hACE2 (e.g., α-helix 1) that was supposed to contact S-RBD if it intact ACE2 (Supplementary Figure 2; Zhang et al., 2020). Though those investigators demonstrated the association of SBP1 peptides with S-RBD proteins at the level of micromolar scales using bio-layer interferometry, SBP1 peptides neither associated with cell surface S-RBD proteins nor did they outcompete ACE2 binding onto S-RBD proteins (Zhang et al., 2020). More rigorous studies should verify such explanations and determine the associated factors between C6 and C2 peptides for RBD or S-trimers.
Another issue deserving discussion was why C2 and C6 behaved similarly in the competition assay (Figure 3) though they manifested different enrichment patterns in PhD panning (Figure 1), as well as different binding properties in the 3D modeling assay (Figure 2). It was known that configuration changes of S-trimers when virus encountered host components were critical for higher affinity interactions between S-trimers and ACE2 molecules. It was understandable that such interactions would be unique in many aspects and depend on the dynamic minutiae of all parties, and we would arbitrarily assume that the factors causing differential binding patterns of C2 or C6 virions onto S-RBD/S-trimers in PhD assays did not contribute enough interference to the actual interactions between S-trimers and ACE2 in the competition context, which are reflected in Figure 3.
Lastly, as a useful tool for studying molecular interactions at both biophysical and functional levels, PhD has been successful in identifying peptides that might be directly utilized to block pathogen invasions in this lab (Zhao et al., 2012) or others (Hall et al., 2009; Wei et al., 2020). This study and several other efforts (Anand et al., 2021; Sokullu et al., 2021; Ballmann et al., 2022; Labriola et al., 2022; Petrenko et al., 2022; Sevenich et al., 2022; Yang et al., 2022) demonstrated that PhD was a plausible method for generating possible therapeutics to treat COVID-19 as well. For example, Petrenko used phage-displayed spike S1 protein mimotopes to search for “all” cellular receptors, including authentic and alternative ones. Interestingly, FGFR3 was identified as an alternative receptor to S proteins (Petrenko et al., 2022). Since FGFR3 manifested a distribution pattern different from that of ACE2, Petrenko’s work expanded the area of SARS-CoV-2 targets and might lead to the discovery of novel pathogenesis of SARS-CoV-2. Like us, Sevenich performed three rounds of screening on S-RBD proteins using a 16-aa phage library combined with high throughput sequencing. The five final sequences they obtained manifested variable affinity for S-RBD proteins in confirmation assays, with Kd from 1.3 to 89.4 μM (Sevenich et al., 2022). Others also demonstrated the stringent dependence of efficacy of intended therapeutics on their molecular compositions and configuration (Raghuvamsi et al., 2021). With the help of computation of RBD-ACE2 interactions, the Baker’s team generated mini-proteins of 56–64 residues with inhibitory concentrations of 24–35 nM for RBD-ACE2 binding (Cao et al., 2020). On the contrary, also based on analysis of motifs or amino acids involved in RBD-ACE2 interaction, Chitsike proposed six peptides (20–29 aa) mimicking S-RBD fragment or hACE2 fragment but found that their IC50 for inhibiting RBD-ACE2 binding in experiments varied from 27 to 363 μM (Chitsike et al., 2021). For us, the direction of our future study would be to use the current C6 SEAL peptides as the core sequences and to develop them into larger molecules (e.g., mini-protein) or other types of peptides (e.g., circular peptides) that would have a better chance to block RBD-ACE2 binding (Pomplun, 2020).
For the great endanger of the COVID-19 pandemic to the human, any strategies or approaches that might lead to the discovery of therapeutics or cures deserve a try (Singh et al., 2021; Vivekanandhan et al., 2021). We utilized PhD to generate two 12-aa peptides with the potentials to inhibit S-protein bindings onto cellular ACE2. Structural modeling revealed that one (C6) might take effect by binding onto the S-RBD-ACE2 interaction face. More efforts should be made to improve the binding affinity of the peptides for S proteins, such as by modifying or transforming them into other types of molecules to block S protein-ACE2 adherence more efficiently. Ultimately, such peptides or their derivatives might be developed into therapeutics that block virus-host attachment and hinder disease onset.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
YQW conceived and designed the study. TW, JX, YLW, and WZ performed the experiments. YX and QY supplied reagents. BW performed the 3D structural modeling. TW, JX, and YQW analyzed the data and prepared the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Zhejiang University Special Scientific Research Fund for COVID-19 Prevention and Control (2020XGZX049) and the Xiamen University Research Programs for COVID-19 (20720200006). YQW was partially supported by Xiang’an Hospital of Xiamen University (PM20205180001).
We thank Haitao Li at Tsinghua University for helpful discussion and for providing reagents.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.910343/full#supplementary-material
Alfaleh, M. A., Alsaab, H. O., Mahmoud, A. B., Alkayyal, A. A., Jones, M. L., Mahler, S. M., et al. (2020). Phage display derived monoclonal antibodies: From bench to bedside. Front. Immunol. 11:1986. doi: 10.3389/fimmu.2020.01986
Anand, T., Virmani, N., Bera, B. C., Vaid, R. K., Vashisth, M., Bardajatya, P., et al. (2021). Phage display technique as a tool for diagnosis and antibody selection for coronaviruses. Curr. Microbiol. 78, 1124–1134. doi: 10.1007/s00284-021-02398-9
Ballmann, R., Hotop, S. K., Bertoglio, F., Steinke, S., Heine, P. A., Chaudhry, M. Z., et al. (2022). ORFeome phage display reveals a major immunogenic epitope on the s2 subdomain of SARS-CoV-2 spike protein. Viruses 14:1326. doi: 10.3390/v14061326
Bertoglio, F., Meier, D., Langreder, N., Steinke, S., Rand, U., Simonelli, L., et al. (2021). SARS-CoV-2 neutralizing human recombinant antibodies selected from pre-pandemic healthy donors binding at RBD-ACE2 interface. Nat. Commun. 12:1577. doi: 10.1038/s41467-021-21609-2
Bhattacharya, S., and Paul, S. M. N. (2021). Efficacy of phytochemicals as immunomodulators in managing COVID-19: A comprehensive view. Virusdisease 32, 435–445. doi: 10.1007/s13337-021-00706-2
Cao, L., Goreshnik, I., Coventry, B., Case, J. B., Miller, L., Kozodoy, L., et al. (2020). De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 370, 426–431. doi: 10.1126/science.abd9909
Chitsike, L., Krstenansky, J., and Duerksen-Hughes, P. J. (2021). ACE2 : S1 RBD interaction-targeted peptides and small molecules as potential COVID-19 therapeutics. Adv. Pharmacol. Pharm. Sci. 2021:1828792. doi: 10.1155/2021/1828792
Collaborators, C.-E. M. (2022). Estimating excess mortality due to the COVID-19 pandemic: A systematic analysis of COVID-19-related mortality, 2020-21. Lancet 399, 1513–1536. doi: 10.1016/S0140-6736(21)02796-3
Garrett, M. E., Galloway, J., Chu, H. Y., Itell, H. L., Stoddard, C. I., Wolf, C. R., et al. (2021). High-resolution profiling of pathways of escape for SARS-CoV-2 spike-binding antibodies. Cell 184, 2927–2938.e11. doi: 10.1016/j.cell.2021.04.045
Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., et al. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. doi: 10.1038/s41586-020-2286-9
Greaney, A. J., Starr, T. N., Gilchuk, P., Zost, S. J., Binshtein, E., Loes, A. N., et al. (2021). Complete mapping of mutations to the SARS-CoV-2 Spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 29, 44–57.e9. doi: 10.1016/j.chom.2020.11.007
Hall, P. R., Hjelle, B., Njus, H., Ye, C., Bondu-Hawkins, V., Brown, D. C., et al. (2009). Phage display selection of cyclic peptides that inhibit Andes virus infection. J. Virol. 83, 8965–8969. doi: 10.1128/JVI.00606-09
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Kruger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and Is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8. doi: 10.1016/j.cell.2020.02.052
Huang, J. X., Bishop-Hurley, S. L., and Cooper, M. A. (2012). Development of anti-infectives using phage display: Biological agents against bacteria, viruses, and parasites. Antimicrob. Agents Chemother. 56, 4569–4582. doi: 10.1128/AAC.00567-12
Jawad, B., Adhikari, P., Podgornik, R., and Ching, W. Y. (2021). Key interacting residues between RBD of SARS-CoV-2 and ACE2 receptor: Combination of molecular dynamics simulation and density functional calculation. J. Chem. Inf. Model. 61, 4425–4441. doi: 10.1021/acs.jcim.1c00560
Labriola, J. M., Miersch, S., Chen, G., Chen, C., Pavlenco, A., Saberianfar, R., et al. (2022). Peptide-Antibody Fusions Engineered by Phage Display Exhibit an Ultrapotent and Broad Neutralization of SARS-CoV-2 Variants. ACS Chem. Biol. 17, 1978–1988. doi: 10.1021/acschembio.2c00411
Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., et al. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220. doi: 10.1038/s41586-020-2180-5
Lipsitch, M., and Dean, N. E. (2020). Understanding COVID-19 vaccine efficacy. Science 370, 763–765. doi: 10.1126/science.abe5938
Lv, Z., Deng, Y. Q., Ye, Q., Cao, L., Sun, C. Y., Fan, C., et al. (2020). Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 369, 1505–1509. doi: 10.1126/science.abc5881
Matthay, M. A., and Luetkemeyer, A. F. (2021). IL-6 receptor antagonist therapy for patients hospitalized for covid-19: Who, when, and how? JAMA 326, 483–485. doi: 10.1001/jama.2021.11121
Mobarak, A. M., Miguel, E., Abaluck, J., Ahuja, A., Alsan, M., Banerjee, A., et al. (2022). End COVID-19 in low- and middle-income countries. Science 375, 1105–1110. doi: 10.1126/science.abo4089
Murthy, S., and Lee, T. C. (2021). IL-6 blockade for COVID-19: A global scientific call to arms. Lancet Respir. Med. 9, 438–440. doi: 10.1016/S2213-2600(21)00127-2
Noy-Porat, T., Makdasi, E., Alcalay, R., Mechaly, A., Levy, Y., Bercovich-Kinori, A., et al. (2020). A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun. 11:4303. doi: 10.1038/s41467-020-18159-4
Petrenko, V. A., Gillespie, J. W., De Plano, L. M., and Shokhen, M. A. (2022). Phage-displayed mimotopes of SARS-CoV-2 spike protein targeted to authentic and alternative cellular receptors. Viruses 14:384. doi: 10.3390/v14020384
Pomplun, S. (2020). Targeting the SARS-CoV-2-spike protein: From antibodies to miniproteins and peptides. RSC Med. Chem. 12, 197–202. doi: 10.1039/d0md00385a
Raghuvamsi, P. V., Tulsian, N. K., Samsudin, F., Qian, X., Purushotorman, K., Yue, G., et al. (2021). SARS-CoV-2 S protein:ACE2 interaction reveals novel allosteric targets. Elife 10:e63646. doi: 10.7554/eLife.63646
Robson, B. (2020). Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Comput. Biol. Med. 119:103670. doi: 10.1016/j.compbiomed.2020.103670
Roth, K. D. R., Wenzel, E. V., Ruschig, M., Steinke, S., Langreder, N., Heine, P. A., et al. (2021). Developing recombinant antibodies by phage display against infectious diseases and toxins for diagnostics and therapy. Front. Cell. Infect. Microbiol. 11:697876. doi: 10.3389/fcimb.2021.697876
Saladin, A., Rey, J., Thevenet, P., Zacharias, M., Moroy, G., and Tuffery, P. (2014). PEP-SiteFinder: A tool for the blind identification of peptide binding sites on protein surfaces. Nucleic Acids Res. 42:W221–W226. doi: 10.1093/nar/gku404
Sang, L., Guo, X., Shi, J., Hou, S., Fan, H., and Lv, Q. (2021). Characteristics and developments in mesenchymal stem cell therapy for COVID-19: An update. Stem Cells Int. 2021:5593584. doi: 10.1155/2021/5593584
Schafer, A., Martinez, D. R., Won, J. J., Meganck, R. M., Moreira, F. R., Brown, A. J., et al. (2022). Therapeutic treatment with an oral prodrug of the remdesivir parental nucleoside is protective against SARS-CoV-2 pathogenesis in mice. Sci. Transl. Med. 14:eabm3410. doi: 10.1126/scitranslmed.abm3410
Sevenich, M., Thul, E., Lakomek, N. A., Klunemann, T., Schubert, M., Bertoglio, F., et al. (2022). Phage display-derived compounds displace hACE2 from Its complex with SARS-CoV-2 spike protein. Biomedicines 10:441. doi: 10.3390/biomedicines10020441
Shen, Y., Chen, L., Wang, M., Lin, D., Liang, Z., Song, P., et al. (2017). Flagellar hooks and hook protein FlgE participate in host microbe interactions at immunological level. Sci. Rep. 7:1433. doi: 10.1038/s41598-017-01619-1
Sheward, D. J., Kim, C., Ehling, R. A., Pankow, A., Castro Dopico, X., Dyrdak, R., et al. (2022). Neutralisation sensitivity of the SARS-CoV-2 omicron (B.1.1.529) variant: A cross-sectional study. Lancet Infect. Dis. 22, 813–820. doi: 10.1016/S1473-3099(22)00129-3
Singh, I. K., Kumari, P., Mittal, P., Kumar, A., Singal, B., Hasan, G. M., et al. (2021). Emerging Therapeutic Approaches to COVID-19. Curr. Pharm. Des. 27, 3370–3388. doi: 10.2174/1381612827666210125160703
Sokullu, E., Gauthier, M. S., and Coulombe, B. (2021). Discovery of Antivirals Using Phage Display. Viruses 13:1120. doi: 10.3390/v13061120
Tiwari, V., Beer, J. C., Sankaranarayanan, N. V., Swanson-Mungerson, M., and Desai, U. R. (2020). Discovering small-molecule therapeutics against SARS-CoV-2. Drug Discov. Today 25, 1535–1544. doi: 10.1016/j.drudis.2020.06.017
Vivekanandhan, K., Shanmugam, P., Barabadi, H., Arumugam, V., Daniel Raj, Daniel Paul Raj, D., et al. (2021). Emerging therapeutic approaches to combat COVID-19: Present status and future perspectives. Front. Mol. Biosci. 8:604447. doi: 10.3389/fmolb.2021.604447
Wang, Q., Zhang, Y., Wu, L., Niu, S., Song, C., Zhang, Z., et al. (2020). Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181, 894–904.e9. doi: 10.1016/j.cell.2020.03.045
Wang, T., Shen, Y., Li, Y., Wang, B., Wang, B., Wu, D., et al. (2019). Ectopic ATP synthase beta subunit proteins on human leukemia cell surface interact with platelets by binding glycoprotein IIb. Haematologica 104:e364–e368. doi: 10.3324/haematol.2019.216390
Wang, Y., Geer, L. Y., Chappey, C., Kans, J. A., and Bryant, S. H. (2000). Cn3D: Sequence and structure views for Entrez. Trends Biochem. Sci. 25, 300–302. doi: 10.1016/s0968-0004(00)01561-9
Wei, J., Hameed, M., Wang, X., Zhang, J., Guo, S., Anwar, M. N., et al. (2020). Antiviral activity of phage display-selected peptides against Japanese encephalitis virus infection in vitro and in vivo. Antiviral Res. 174:104673. doi: 10.1016/j.antiviral.2019.104673
Wrobel, A. G., Benton, D. J., Xu, P., Roustan, C., Martin, S. R., Rosenthal, P. B., et al. (2020). SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 27, 763–767.
Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., and Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448. doi: 10.1126/science.abb2762
Yang, F., Liu, L., Neuenschwander, P. F., Idell, S., Vankayalapati, R., Jain, K. G., et al. (2022). Phage display-derived peptide for the specific binding of SARS-CoV-2. ACS Omega 7, 3203–3211. doi: 10.1021/acsomega.1c04873
Zhang, G., Pomplun, S., Loftis, A. R., Tan, X., Loas, A., and Pentelute, B. L. (2020). Investigation of ACE2 N-terminal fragments binding to SARS-CoV-2 Spike RBD. bioRxiv [Preprint]. doi: 10.1101/2020.03.19.999318
Zhang, Y., Wang, S., Wu, Y., Hou, W., Yuan, L., Shen, C., et al. (2021). Virus-Free and live-cell visualizing SARS-CoV-2 cell entry for studies of neutralizing antibodies and compound inhibitors. Small Methods 5:2001031. doi: 10.1002/smtd.202001031
Zhao, G., Li, S., Zhao, W., He, K., Xi, H., Li, W., et al. (2012). Phage display against corneal epithelial cells produced bioactive peptides that inhibit Aspergillus adhesion to the corneas. PLoS One 7:e33578. doi: 10.1371/journal.pone.0033578
Keywords: SARS-CoV-2, spike protein, ACE2, RBD domain, phage display
Citation: Wang T, Xu J, Wang B, Wang Y, Zhao W, Xiang B, Xue Y, Yuan Q and Wang Y (2022) Receptor-binding domain-anchored peptides block binding of severe acute respiratory syndrome coronavirus 2 spike proteins with cell surface angiotensin-converting enzyme 2. Front. Microbiol. 13:910343. doi: 10.3389/fmicb.2022.910343
Received: 01 April 2022; Accepted: 18 August 2022;
Published: 13 September 2022.
Edited by:
Yi-Wei Tang, Cepheid, United StatesReviewed by:
Archana Singh, University of Delhi, IndiaCopyright © 2022 Wang, Xu, Wang, Wang, Zhao, Xiang, Xue, Yuan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiqiang Wang, WWlxaWFuZy5XYW5nQHhqdGx1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.