- 1Microbial Safety Division, National Institute of Agricultural Sciences, Rural Development Administration, Wanju, South Korea
- 2Functional Food and Nutrition Division, National Institute of Agricultural Sciences, Rural Development Administration, Wanju, South Korea
- 3Planning and Coordination Division, National Institute of Agricultural Sciences, Rural Development Administration, Wanju, South Korea
Lettuce wraps are popular in Korean cuisine for their high nutritional value and versatility as healthy additions to multiple dishes. Microbial contamination of lettuce is a major concern, as lettuce is consumed fresh without cooking. Among foodborne pathogens, the spore-forming, facultative anaerobic bacterium, Bacillus cereus is one of the frequently detected pathogen in lettuce in Korea. In this study, we investigated the prevalence and distribution of Bacillus cereus strains in lettuce production farms and further evaluated the enterotoxin gene profiles, antibiotic susceptibility, multidrug resistance pattern, and genetic differences among the B. cereus group isolates. Of the 140 samples isolated from 10 lettuce production farms, 30 samples (21.42%) were positive for B. cereus in which 19 (31.6%) and 10 (23.25%) were from soil and lettuce, respectively. The enterotoxin patterns A (hblCDA, nheABC, entFM, and cytK genes) and B (hblCDA, nheABC, and entFM genes) accounted for 50% and 20% of all the isolates, whereas the emetic gene cesB was not detected in any of the B. cereus group isolates. Antibiotic susceptibility testing of the B. cereus group isolates revealed that all the strains were predominantly resistant to β-lactam antibiotics except imipenem and generally susceptible to most of the non β-lactam antibiotics, including gentamycin, streptomycin, chloramphenicol, and tetracycline. ERIC-PCR and MLST analysis revealed high genetic diversity among the 30 B. cereus group isolates, which belonged to 26 different sequence types (STs) and seven new STs. Moreover, isolates with identical STs exhibited similar patterns of antibiotic resistance and enterotoxin profiles. Results of this study indicate a high prevalence of B. cereus group isolates in lettuce production farms in the Republic of Korea.
Introduction
Lettuce is one of the major commercial crops in Korea with a total domestic production of 86,128 tons from 3,387 ha cultivation area (Chang et al., 2020), recorded as the most cultivated leafy vegetable in Korea (Kim et al., 2018). Since lettuce is primarily consumed fresh without cooking as a wrap material or in salads, it could potentially be contaminated with pathogens across the food supply chain and may cause illness upon consumption (Bozkurt et al., 2021). The foodborne pathogens Escherichia coli O157:H7, Listeria monocytogenes, Salmonella, and Bacillus cereus are reported to be the major sources of lettuce-related outbreaks (Kim et al., 2012; Irvin et al., 2021; Qu et al., 2021). A total of 60 outbreaks related to consumption of leafy vegetables, mainly lettuce was reported to the Centers for Disease Control and Prevention (CDC) between 2014 and 2021 [Centers for Disease Control and Prevention (CDC), 2022]. Moreover, B. cereus was found to be the most frequently detected pathogen in fresh vegetables in Korea with a 37.5% contamination rate (Jo et al., 2011; Jung et al., 2016; Park et al., 2018). Hence, it is necessary to conduct microbial risk assessments in lettuce farms in Korea periodically and monitor the prevalence and characteristics of B. cereus group isolates.
The B. cereus group, also known as B. cereus sensu lato, constitutes numerous phylogenetically related species which may include B. cereus, B. anthracis, B. thuringiensis, B. cytotoxicus, B. toyonensis, B. mycoides, and B. pseudomycoides (Guinebretière et al., 2008; Liu et al., 2018; Carroll et al., 2021). These B. cereus group strains differ remarkably in their importance to medicine, public health and food safety (Mandic-Mulec et al., 2015). For example, some isolates of B. cereus and B. cytotoxicus strains cause foodborne illness (Naranjo et al., 2011; Guinebretière et al., 2013; Colaco et al., 2021). B. anthracis may cause different forms of anthrax diseases (cutaneous, gastrointestinal, and inhalational anthrax) to both animals and humans (Okinaka et al., 1999; Marston et al., 2005; Cote et al., 2015). (21)The B. thuringiensis strain which produces crystalline toxins is widely used in agriculture as pesticides (Bernhard et al., 1997). The psychrotolerant B. wiedmannii strain was reported to exhibit cytotoxic effects on vertebrate animals (Zhao et al., 2019). The probiotic strain B. toyonensis harbors chloramphenicol and tetracycline resistance genes catQ and tet(M) in their genome and may pose a risk for transmitting these antibiotic resistance genes to other species [EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP), 2012]. In addition, the B. toyonensis strain NCIMB 14858T may cause potential risks to humans when exposed, as this strain has the capacity to produce and release functional toxins [EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP), 2014]. B. pseudomycoides, which has been recognized as a non-pathogenic environmental strain, was shown to produce pore-forming toxins and exhibit cytotoxic effects to human cells (Miller et al., 2018). The B. weihenstephanensis and B. mycoides strains, which belong to group VI phylogeny based on phylogenetic classification by Guinebretière et al. (2010) and cytotoxicity studies by Miller et al. (2018) are generally considered as low risk pathogens. The 16S rRNA gene sequences of most of the B. cereus group strains share 99% sequence similarity (Sacchi et al., 2002) and seem to be members of a single species (Ceuppens et al., 2013). However, they are ecologically diverse from soil to gut (Guinebretière et al., 2008; Stenfors Arnesen et al., 2008), and the pathogenic potential of these B. cereus group strains varies between species (Guinebretière et al., 2010). Hence, it is necessary to discriminate the B. cereus isolates. Combining traditional methods with molecular typing methods. Such as enterobacterial repetitive intergenic consensus–polymerase chain reaction (ERIC-PCR) and multilocus sequence typing (MLST) is recommended to discriminate diverse B. cereus species (Maiden et al., 2013; Zhuang et al., 2019; Qu et al., 2021).
Bacillus cereus is a catalase-positive, toxin-producing, and endospore-forming facultative anaerobic gram-positive bacilli present ubiquitously in soil, water, vegetables, decaying matter, and food (Osimani et al., 2018). This mesophilic pathogen can quickly proliferate at room temperature with an ample amount of preformed toxins (McDowell et al., 2021), which upon ingestion may cause gastrointestinal illness. Clinical manifestations of B. cereus disease include two types of gastrointestinal syndromes, i.e., diarrheal syndrome and emetic syndrome (Stenfors Arnesen et al., 2008). The diarrheal syndrome is caused by B. cereus strains that produce one or several enterotoxins including two tripartite complexes (Fagerlund et al., 2010), namely non-hemolytic enterotoxin (NHE, encoded by nheA, nheB, and nheC), and hemolysin BL (HBL, encoded by hblA, hblC, and hblD; Sastalla et al., 2013) and single proteins cytotoxin K (CytK; Lund et al., 2000) and enterotoxin FM (EntFM; Ngamwongsatit et al., 2008). The emetic syndrome is caused by a heat-stable toxin cerulide (Ces) preformed in the food (Carroll et al., 2017).
Contamination of lettuce can occur at any stage from farming, harvesting, processing, packing, transporting, and storing the remains by consumers (Pang et al., 2017). Several factors including the spread of feces to soil via farm animals, contaminated irrigation water, cross-contamination by workers from one farm to another farm may lead to disease outbreaks. Information on the prevalence and characteristics of B. cereus in lettuce farms will help to understand transmission routes and develop intervention technologies to ensure food safety and public health.
This study provides information about the prevalence, distribution, enterotoxin profiles and antibiotic resistance patterns of B. cereus group isolates in lettuce production farms. In addition, genetic diversity of the B. cereus group isolates was also analyzed by ERIC-PCR and MLST. The results of this study may help to develop food safety measures to reduce the contamination and transmission of B. cereus in lettuce farms.
Materials and methods
Sampling
One hundred and forty samples including lettuce (n = 43), soil (n = 60), compost (n = 8), and irrigation water (n = 29) were collected from 10 different lettuce production farms located across Jeollabuk-do province in the Republic of Korea. All the samples were aseptically placed in separate sterile bags in a cool container and transported to the laboratory and analyzed within 24 h.
Analysis of coliforms, Escherichia coli, and Bacillus cereus
To analyze the prevalence of indicator bacteria such as aerobic bacteria, coliforms, and E. coli, 25 g of each sample (lettuce, soil, and compost) were homogenized in a sample bag containing 225 ml of 0.1% peptone water (PW; Oxoid, Basingstoke, United Kingdom) and homogenized in a BagMixer® (Interscience, Saint-Nom-la-Bretéche, France). The homogenized samples were tenfold serial diluted with 0.1% PW and aliquots of 1 ml were inoculated on 3 M™ Petrifilm™ Aerobic Count (AC)/Coliform Count (CC)/E. coli Count (EC) plates (3 M Microbiology, Minnesota, United States) and incubated at 37°C for 24 h. The irrigation water samples were evaluated for aerobic bacteria, coliforms, and E. coli using the Colilert-18 test kit (IDEXX laboratories, Westbrook, Maine, United States).
The prevalence of B. cereus was evaluated by mixing 25 g of lettuce, soil, or compost samples with 225 ml of buffered peptone water (BPW, Oxoid, Basingstoke, United Kingdom) in a stomacher bag using BagMixer® for 1 min. Tenfold serial dilutions of the homogenized samples were prepared and 0.1 ml of diluted samples were spread on Mannitol-egg yolk-polymyxin (MYP) agar plates (Oxoid, Basingstoke, United Kingdom). After incubation for 24 h at 30°C, typical pink colonies with turbid rings that generated lecithinase around them were presumptively identified as B. cereus and enumerated. For further confirmation, suspected colonies were sub-cultured on Tryptic Soy Agar (TSA; Difco, Sparks, MD, United States), plates, and the positive colonies were confirmed by PCR Detection Kit (Kogene Biotech, Korea).
Distribution of enterotoxin genes
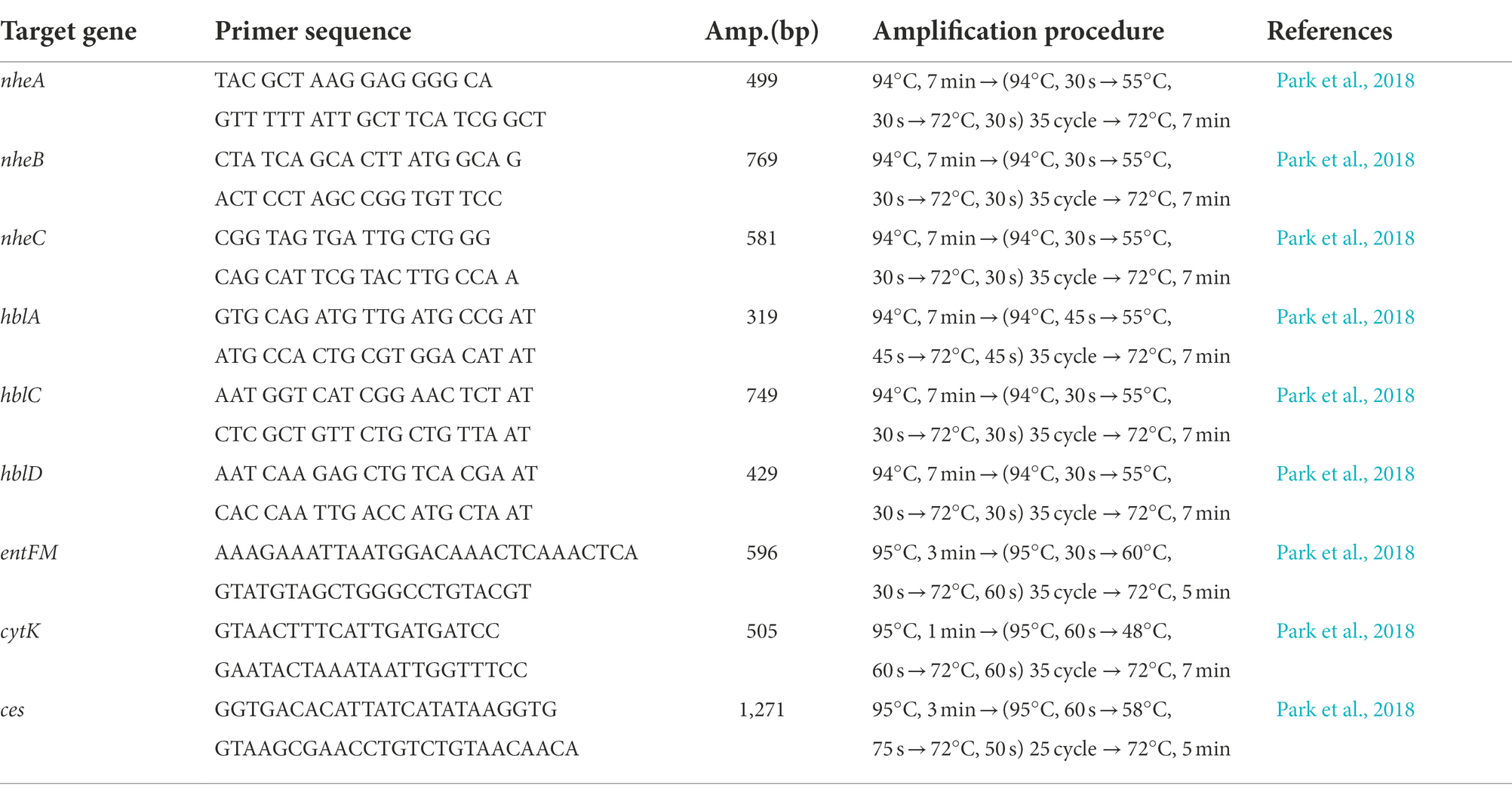
Genomic DNAs from B. cereus isolates were extracted using the G-spin™ bacterial genomic DNA extraction kit (iNtRON Biotechnology, Inc., Gyeonggi do, Korea) following the manufacturer’s protocol. The presence of diarrhea-causing enterotoxin genes (nheA, nheB, nheC, hblA, hblC, hblD, entFM, and cytK) and the emetic toxin cereulide synthesis (cesB) gene was determined by PCR. For PCR amplification, DNA templates of 50 ng and 10 pmol of each primer along with required distilled water (DW) were added to a PCR reaction mixture (20 μl final volume) consisting of AccuPower PCR premix (Bioneer, Daejeon, Korea) followed by amplification using the thermal cycler (C1000TM Thermal Cycler, BIORAD, CA, United States). The primer sequences and amplification procedures are described in Table 1.
Antimicrobial susceptibility testing
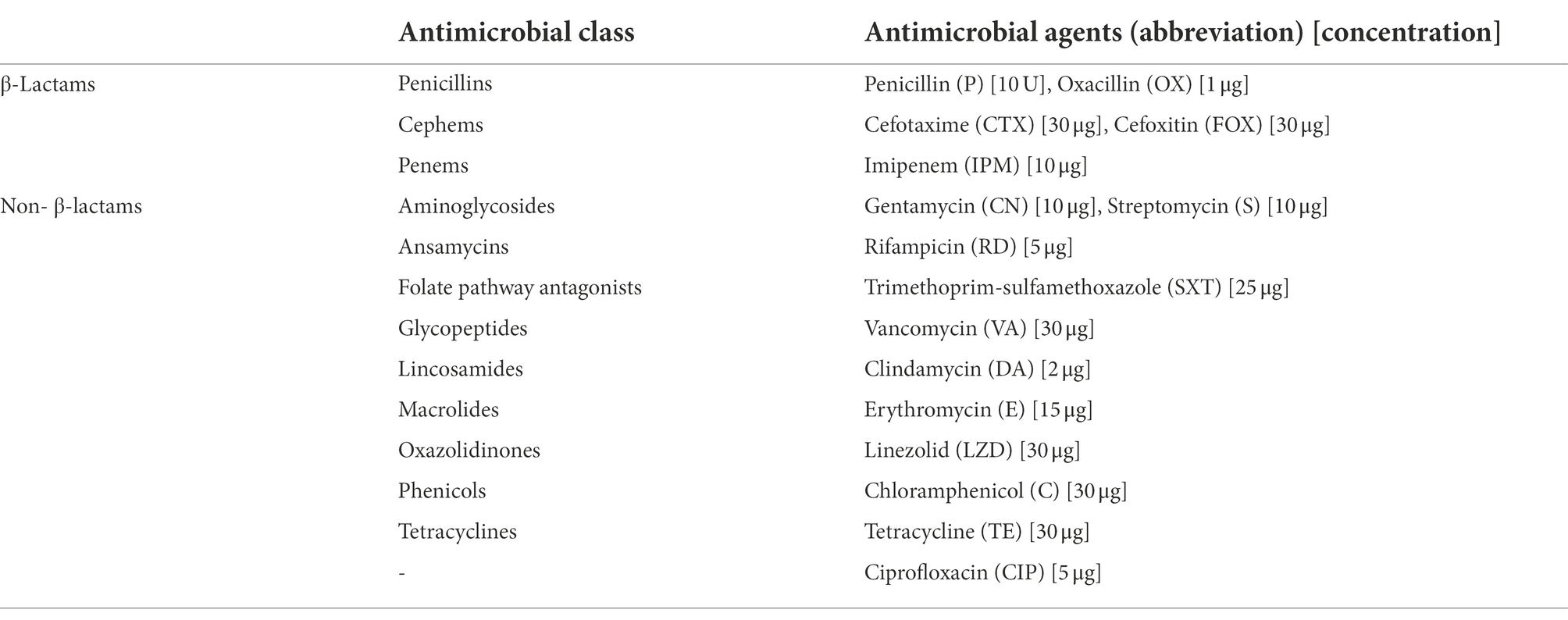
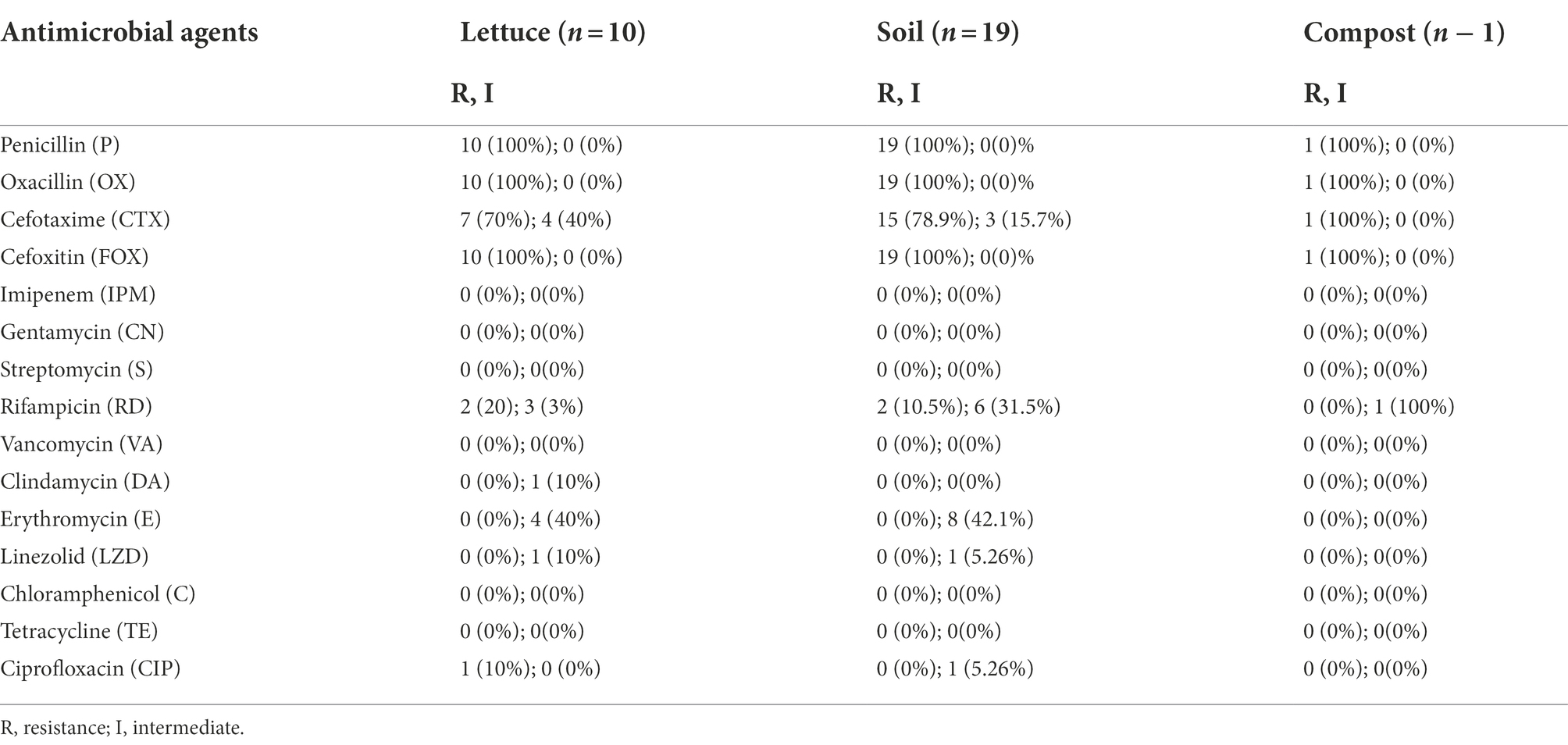
The antimicrobial susceptibility of B. cereus isolates was determined by the minimum inhibitory concentration (MIC) assay as reported by Manzulli et al. (2019) following the guidelines of the Clinical and Laboratory Standards Institute (CLSI). The antibiotics tested and concentrations used were as follows: penicillin (10 U), oxacillin (1 μg), cefotaxime (30 μg), cefoxitin (30 μg), imipenem (10 μg), gentamicin (10 μg), streptomycin (10 μg), rifampicin (5 μg), trimethoprim sulfamethoxazole (25 μg), vancomycin (30 μg), clindamycin (2 μg), erythromycin (15 μg), linezolid (30 μg), chloramphenicol (30 μg), tetracycline (30 μg), and ciprofloxacin (5 μg; Table 2). The antibiotic susceptibility of each B. cereus isolate was measured after incubation at 28°C for 16–18 h, and the results were interpreted following the guidelines provided by the CLSI document M100, 30th edition (CLSI, 2020). All the B. cereus isolates were classified as susceptible, intermediate susceptible, and resistant based on the diameter of inhibition zone and minimum inhibitory concentration (MIC) breakpoints for Staphylococcus spp.
Enterobacterial repetitive intergenic consensus–polymerase chain reaction
Genetic relatedness between the B. cereus isolates was determined by ERIC-PCR fingerprinting using the primer 5′-ATGTAAGCTCCTGGGGATTCAC-3′ designed by Versalovic et al. (1991). Amplification of repetitive ERIC sequences within the genomes of the isolates was performed with the following reaction conditions: initial denaturation at 95°C for 10 min followed by 4 cycles of 94°C for 5 min, 40°C for 5 min, 72°C for 5 min and 40 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 2 min and a final extension for 10 min at 72°C. The PCR products were evaluated by gel electrophoresis and with the Dice coefficient, the band patterns were used to generate a dendrogram by GelJ software (Heras et al., 2015) using the UPGMA (unweighted pair group method with arithmetic mean) at 1.5% optimization and tolerance level of 1.5% (Cheng et al., 2021).
Multilocus sequence typing
To analyze the genetic diversity among the B. cereus isolates, genomic DNAs of the isolates were extracted and gene fragments of seven housekeeping genes [glpF (glycerol uptake facilitator), gmk (guanylate kinase), ilvD (dihydroxy-acid dehydratase), pta (phosphate acetyltransferase), pur (phosphoribosylaminoimidazolecarboxamide), pycA (pyruvate carboxylase), and tpi (triosephosphate isomerase)] were amplified using the primers and conditions listed in the MLST database (Jolley et al., 2018).1 The resulting DNA fragments were purified and sequenced using an automated DNA analyzer (ABI 3730XL; Applied Biosystems, Foster City, CA) by SolGent Korea (SolGent, Daejeon, Korea). With the sequence information, allele numbers for each housekeeping gene were assigned based on the locus queries from the MLST database2 and the sequence types (STs) were determined based on the numeric allelic profile of the isolates. The genetic relationships among the B. cereus isolates were determined using the Global Optimal eBURST (goeBURST) algorithm (Francisco et al., 2009) implemented in the PHYLOViZ software (Ribeiro-Gonçalves et al., 2016; Liu et al., 2017).
Results and discussion
Prevalence of coliforms, Escherichia coli, and Bacillus cereus in lettuce production farms
The prevalence and distribution of aerobic bacteria, coliforms, E. coli, and B. cereus in the lettuce, soil, compost, and irrigation water samples collected from 10 different farms are presented in Table 3. Of the total 140 samples, aerobic bacteria and coliforms were more prevalent in soil (6.89 ± 0.19 log CFU/g; 3.07 ± 0.36 log CFU/g) and compost (5.44 ± 0.1 log CFU/g; ND) followed by lettuce (5.12 ± 0.37 log CFU/g; 2.43 ± 0.23 log CFU/g), while the irrigation water (2.18 ± 0.18 log CFU/ml; 0.32 ± 0.12 log CFU/ml) was least contaminated with indicator bacteria. E. coli was detected in lettuce samples from farms 2 and 3 and soil sample from farm 4, while E. coli was not detected in all the other samples.
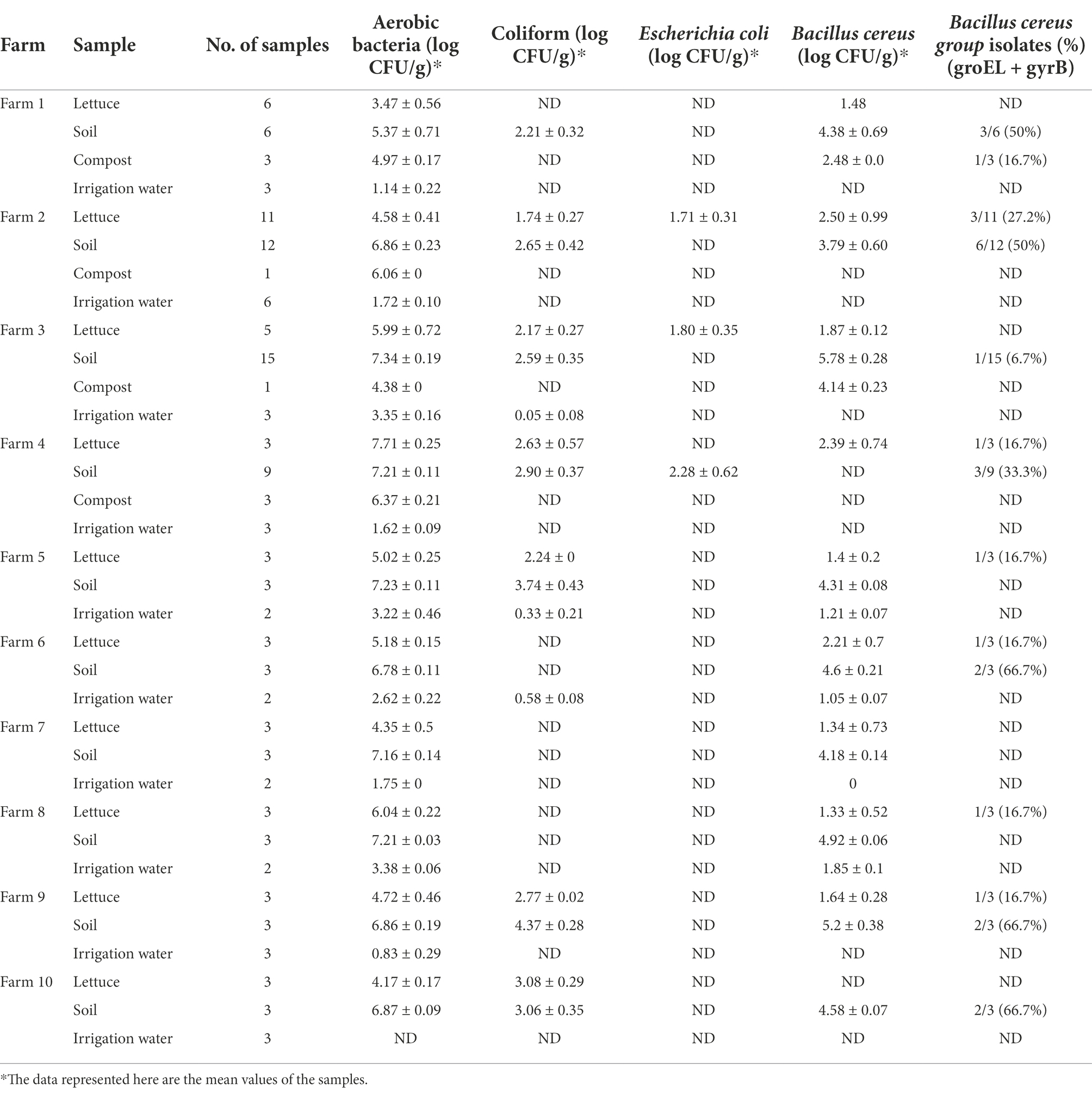
Table 3. Population density of indicator bacteria and Bacillus cereus in lettuce and its production environments.
Preliminary evaluation of B. cereus contamination analysis revealed that soil (4.63 ± 0.28 log CFU/g) and compost (3.31 ± 0.12 log CFU/g) was more prone to B. cereus contamination than lettuce (1.79 ± 0.64 log CFU/g) and irrigation water (1.03 ± 0.06 log CFU/ml). However, qualitative assessment results revealed that 28 out of 140 samples (20%) were contaminated with B. cereus group isolates, in which 19 isolates were from soil (31.6%), 8 from lettuce (18.60%), and 1 isolate from compost (12.50%). No B. cereus group strains were detected in irrigation water (Table 3).
Profiling of enterotoxin genes among Bacillus cereus isolates
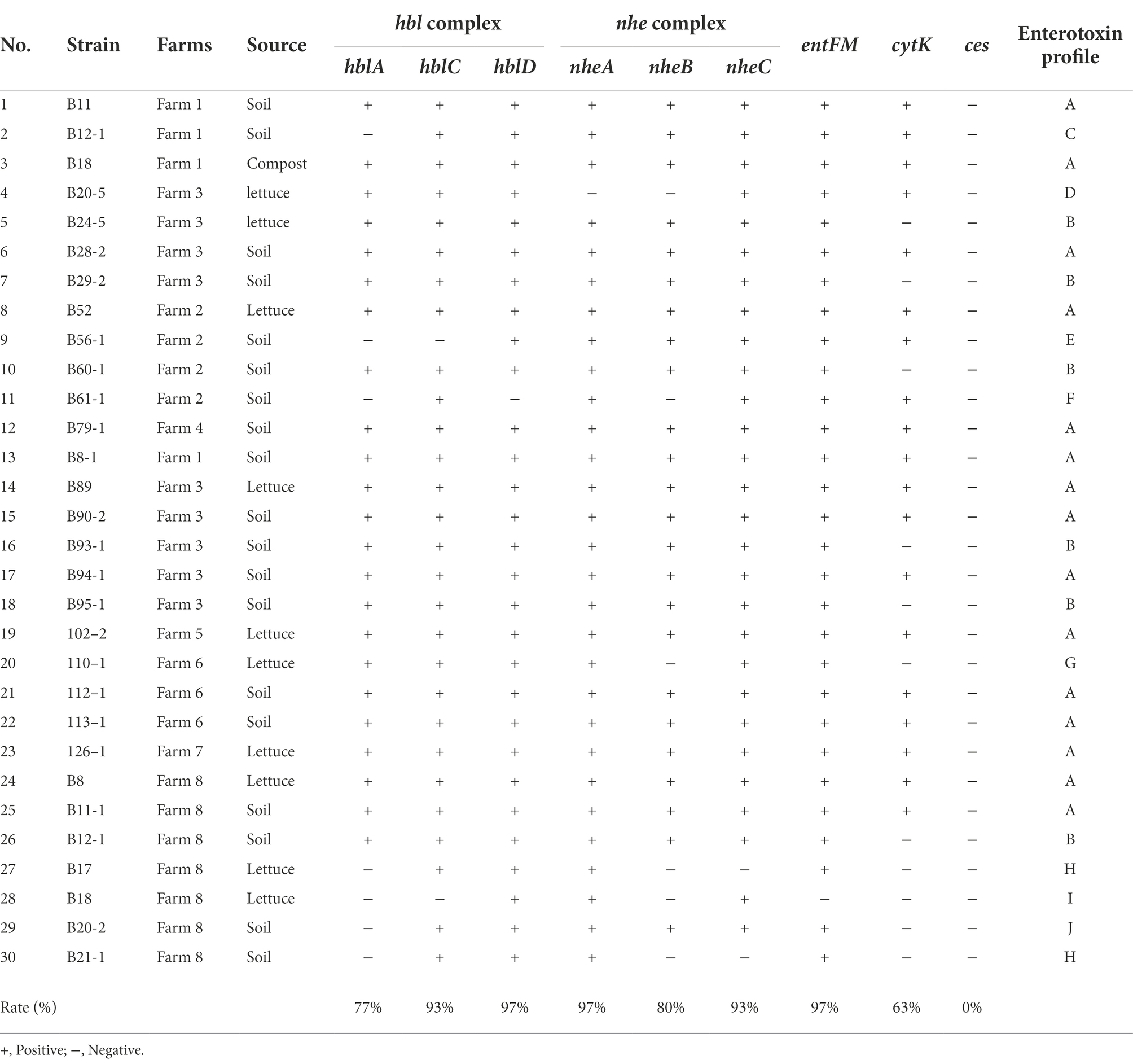
Identifying and profiling toxin genes of B. cereus group strains is an important step in characterizing the virulence potential of the B. cereus isolates mainly distinguishing the pathogenic potential of the strains. In this study, genomic DNA of 30 B. cereus isolates from lettuce, soil and compost samples from various farms were extracted, and the prevalence of enterotoxin genes nheA, nheB, nheC, hblA, hblC, hblD, entFM, and cytK and the emetic gene ces were determined by PCR. The distribution of enterotoxin genes among B. cereus isolates is summarized in Table 4. All the B. cereus isolates carried at least one gene encoding NHE toxin including nheA (29 out of 30 isolates, 96.6%), nheB (24 out of 30, 80%), nheC (29 out of 30, 97%), and at least one HBL toxin encoding gene including hblA (23 out of 30, 76.6%), hblC (28 out of 30, 93.3%), and hblD (29 out of 30, 97%). All the isolates except B18 possessed entFM (29 out of 30, 97%). The dermonecrotic cytK gene was detected in 19 out of 30 (63.3%) B. cereus group isolates (Table 4). However, the emetic gene ces was not detected in any of the B. cereus group isolates.
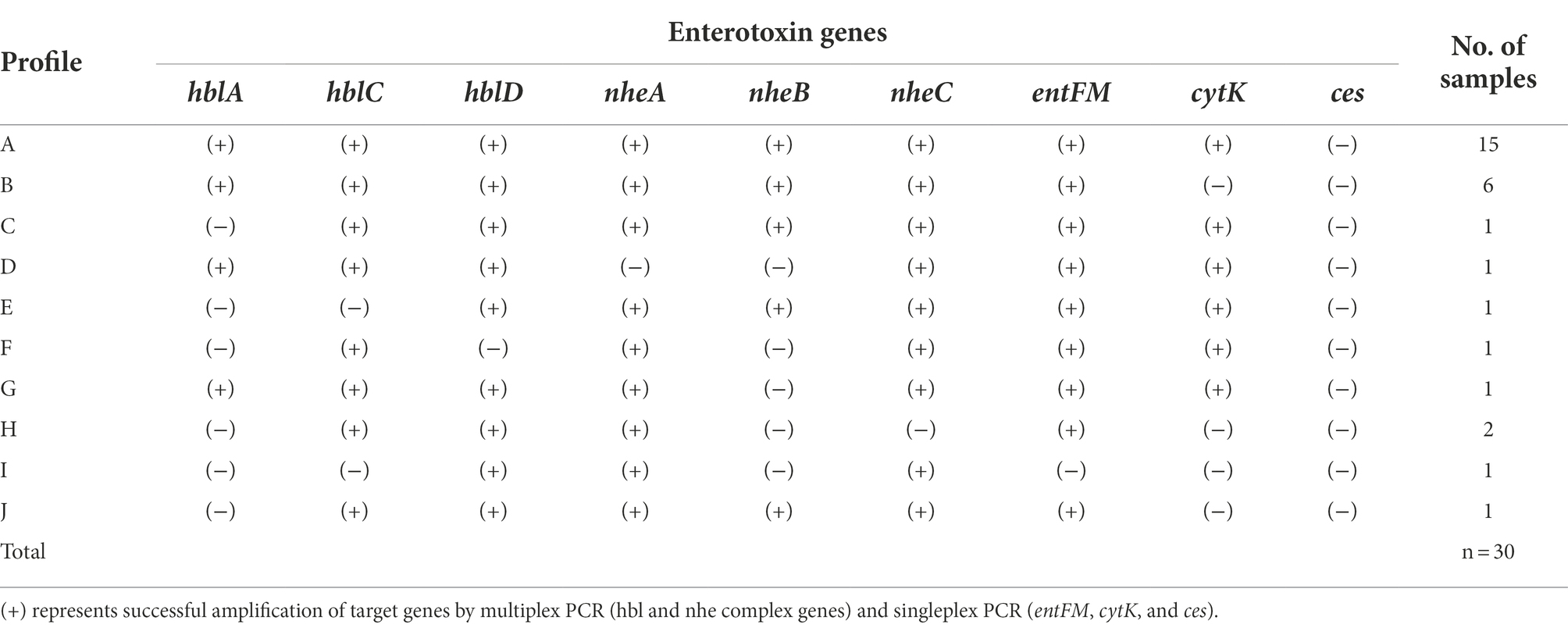
The B. cereus isolates were further classified into 11 groups (enterotoxin profile patterns A to K) based on the presence or absence of 8 enterotoxin genes and 1 emetic gene (Table 5). The enterotoxin patterns A (hblCDA, nheABC, entFM, and cytK gene) and B (hblCDA, nheABC, and entFM gene) accounted for 50% (15/30) and 20% (6/30) of all the isolates, while the remaining patterns, C to K were distributed evenly among the remaining 9 isolates, sharing 3.33% (1/30) each, i.e., 1 pattern per isolate. Corresponding to our observations, Kim et al. (2011) has also reported that the majority of their B. cereus isolates (79.2%) possessed enterotoxin profile patterns I and II which are patterns A and B in our study (Kim et al., 2011). Moreover, the B. cereus group strains with profile A containing all the enterotoxin genes are critically important strains among the environment, food, and clinical isolates (Hsu et al., 2021).
Since the major virulence factors hemolytic (HBL) and non-hemolytic (NHE) enterotoxins are three-component toxin complexes (Stenfors Arnesen et al., 2008), all the three genes should be present in the nheABC and hblCDA clusters to generate active toxins in both cases (Chaves et al., 2011; Sastalla et al., 2013). Based on these criteria, only 76% and 80% of the B. cereus isolates produce active form of HBL and NHE enterotoxins, respectively, and 70% of isolates which possess all the genes in both the clusters are highly pathogenic and may cause diarrhea (Table 4). The enterotoxin entFM, a cell wall peptidase necessary for bacterial morphology, motility, cell adhesion, and biofilm formation (Darrigo et al., 2016; Tran et al., 2020) is prevalent in the genome of almost all the members of the B. cereus group (Kim et al., 2011; Chon et al., 2015) which is consistent with our observation. The absence of emetic toxin gene ces in all strains indicates that the enterotoxin gene containing B. cereus strains are widespread in Korea (Kim et al., 2009) than the emetic strains as reported by Kim et al. (2011).
Antimicrobial susceptibility of Bacillus cereus isolates
Antibiotic resistance or susceptibility of B. cereus isolates to different antimicrobial agents is represented in Table 6. All the B. cereus isolates, regardless of the source (lettuce, soil, and compost) were resistant to penicillin (100%), oxacillin (100%), cefoxitin (100%), and cefotaxime (77% with 33% intermediate resistance). In contrast, all the B. cereus isolates were susceptible to imipenem (100%), gentamycin (100%), streptomycin (100%), trimethoprim-sulfamethoxazole (100%), ciprofloxacin (100%), chloramphenicol (100%), linezolid (100%), vancomycin (97%), erythromycin (93%) and tetracycline (93%). The ansamycin class antibiotic rifampicin inhibited the growth of 50% of the B. cereus isolates, while 33% showed intermediate resistance and the remaining 17% of isolates were resistant toward rifampicin. In the case of clindamycin, 60% of isolates were sensitive and 40% of strains showed intermediate resistance.
Pathogens develop antibiotic resistance through several mechanisms, including the acquisition of resistance determinants via horizontal gene transfer from soil and environmental bacteria (Munita and Arias, 2016; Peterson and Kaur, 2018) or by a random mutation generated by a consequence of antibiotic selection pressure, allowing them to survive and proliferate in the presence of the antimicrobial agent (Skalet et al., 2010). Since B. cereus group strains are clinically significant, determining their resistance/susceptibility toward antimicrobial agents is essential for treatment during disease outbreaks. Even though most infections caused by B. cereus group strains may get better without antimicrobial treatment, it is important to be vigilant as new B. cereus group infection strains may emerge in the future (Mills et al., 2022). Based on the antibiotic susceptibility profiles, the B. cereus group isolates were further classified into four different groups (I, II, III, and IV) according to the MDR patterns (Table 7).
Most of the B. cereus isolates (63.3%) were resistant to all the penicillins (P, OX) and cephems (CTX, FOX) class β-lactam antibiotics falling under pattern II while 23.3% of isolates were resistant to P, OX, and FOX (pattern I) but not to CTX. Generally, all the Bacillus cereus group strains are predominantly resistant to β-lactam antibiotics due to the abundant production of β-lactamase enzymes (Majiduddin et al., 2002; Owusu-Kwarteng et al., 2017). Correspondingly, all the B. cereus isolates in our study were resistant to β-lactam antibiotics except imipenem which showed 100% susceptibility toward B. cereus isolates (Table 7). This was due to the strong inhibition efficiency of the carbapenem antibiotic imipenem against β-lactamases (Vardakas et al., 2012). According to the results of our study, the B. cereus isolates were generally susceptible to most of the non-β-lactam antibiotics including gentamycin, streptomycin, trimethoprim-sulfamethoxazole, ciprofloxacin, chloramphenicol, linezolid, vancomycin, erythromycin, and tetracycline, which is consistent to the reports of other studies (Luna et al., 2007; Fiedler et al., 2019). However, the B. cereus isolates in our study showed an intermediate level of resistance toward rifampicin and clindamycin which has also been reported earlier in other studies (Park et al., 2009, 2018, 2020).
Genetic diversity analysis of Bacillus cereus group isolates by ERIC-PCR and MLST
Genomic fingerprinting by ERIC-PCR has been successfully used to analyze the genetic diversity of several bacteria and discriminate between closely related strains. The combined results of ERIC-PCR fingerprinting, MLST, enterotoxin profiles, and multidrug resistance patterns of B. cereus group isolates are summarized in Figure 1. ERIC-PCR fingerprinting of 30 B. cereus isolates from lettuce farms yielded 6 to 12 polymorphic bands ranging in size between 100 to over 3,000 base pairs (bp) with different band intensities. The most common molecular sizes of the bands in the ERIC-PCR gel image were around 300, 450, 600, and 900 bps. Based on the migration patterns of the PCR products, the B. cereus isolates were classified into 6 different groups (designated as I, II, III, IV, V, and VI) by UPGMA clustering. All the isolates from farms 1, 4, and 7 were within cluster I, while isolates from farms 6 and 8 were distributed within clusters I, II, and III, indicating that all these isolates are genetically related. The presence of B. cereus isolates from farms 2 and 3 in all the clusters shows their genetic diversity. However, the ERIC-PCR-based clustering did not correlate with the sources of the isolates or MDR patterns or enterotoxin profiles of the isolates (Figure 1).
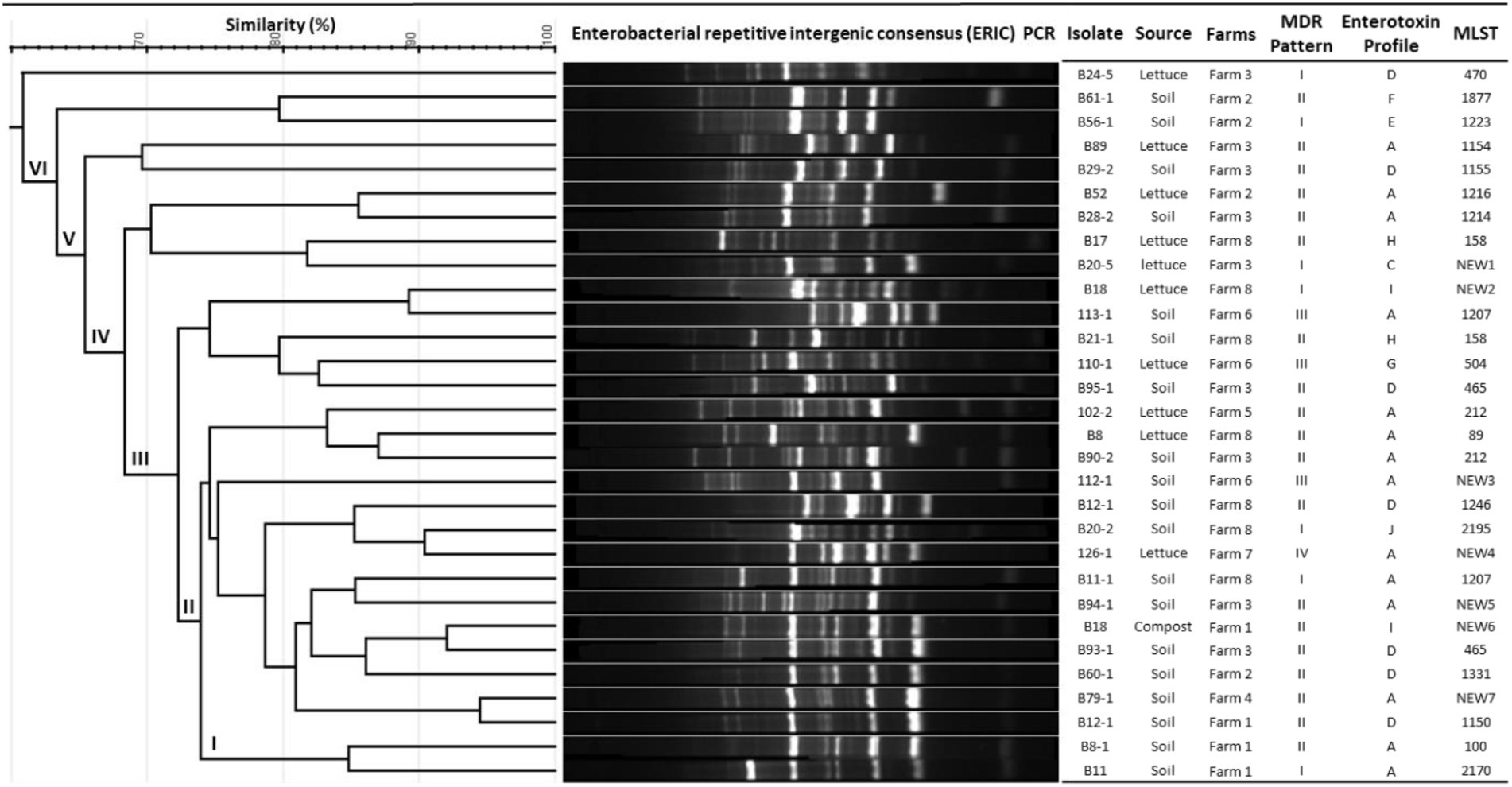
Figure 1. Genetic diversity analysis based on ERIC-PCR and MLST and their correlation with MDR patterns and enterotoxin profiles of Bacillus cereus isolates.
MLST-based genetic subtyping works by indexing the variation of nucleotide sequences in multiple housekeeping gene (loci) fragments distributed around the bacterial genome. Based on the nucleotide sequence variations, a numeric integer is designated to each locus and an allelic profile is created and with a unique combination of alleles, a sequence type (ST) number is assigned (Jolley and Maiden, 2014) with which the genetic diversity of the bacterial isolates can be characterized. In our study, MLST analysis of 30 B. cereus isolates resulted in identifying 26 STs from PubMLST database and 7 new STs.
Phylogenetic analysis helps us to better understand the evolutionary relationship between the organisms and thus helps us to characterize newly evolved isolates (Soltis and Soltis, 2003). MLST data were used to infer possible phylogenetic relationship between the isolates using the goeBURST algorithm (Francisco et al., 2009; Pérez-Losada et al., 2013). The goeBURST analysis revealed a close relationship between the soil isolate ST1150 and three other STs ST1246, ST1207, and a new ST New3 isolated from soil and between ST1207 and the novel ST New6 (Figure 2) isolated from lettuce. These STs pairs are single locus variants (SLVs) that differed from one another by one housekeeping gene allele.
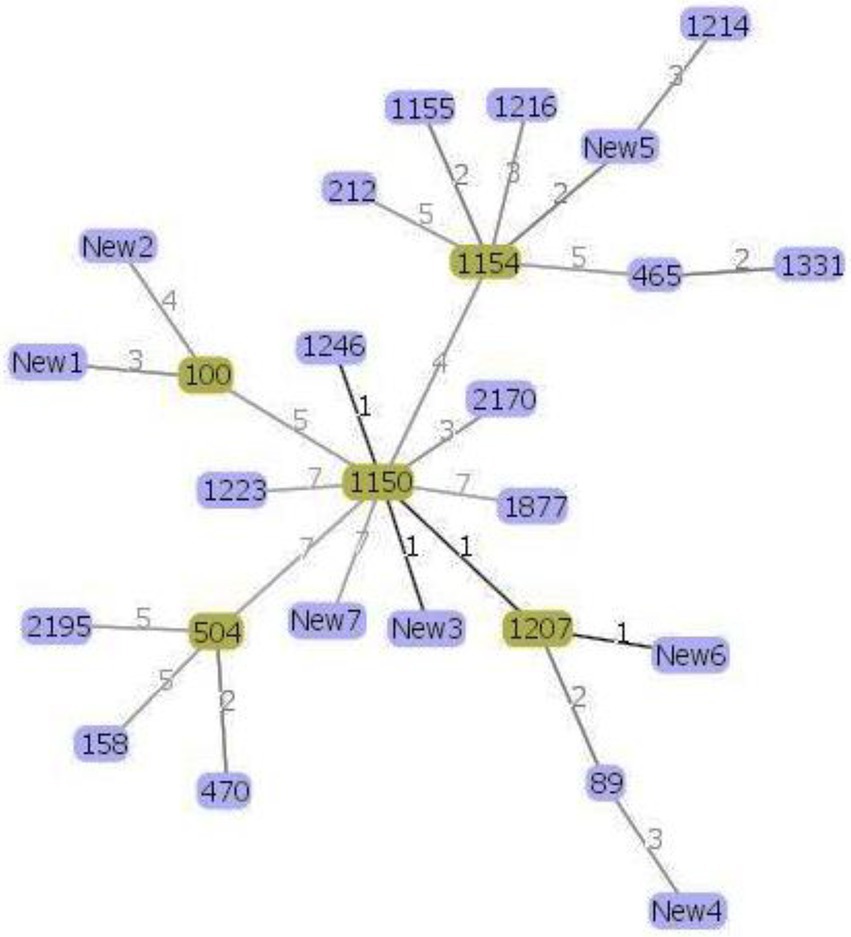
Figure 2. Genomic relatedness between the 30 Bacillus cereus isolates based on MLST data. The minimum spanning tree was constructed using the goeBURST algorithm implemented in the PHYLOViZ online platform. Green color represents the subgroup founders while single-locus variants are highlighted with darker links.
Moreover, according to the PubMLST database, the soil and lettuce isolates ST1214, ST1216, and ST1223 from our study were previously isolated from food packing materials in China; ST1150, ST1155, and ST1154 were previously isolated from soil samples from Japan; ST1207 and ST1246 were separated from feces samples from Korea and ST465 as previously isolated from Taiwan. The ST158 strain isolated from soil in this study was previously reported in isolates from Korea, Japan, and United States. ST100 was reported as blood isolate and milk product isolate from the USA and China, respectively. Moreover, other isolates from the USA included ST2195, ST89, ST504, and ST470. The soil isolates ST1223 and ST212 were identified in PubMLST database as isolates from unknown sources. Out of 7 novel STs, 1 isolate was from compost and 3 isolates each were from soil and lettuce samples. These results suggest that B. cereus isolates found in lettuce farms may be acquired through several sources. Furthermore, the results of MLST coincided with the patterns of antibiotic resistance and enterotoxin profiles of the isolates (Figure 1) and thus confirmed the genetic diversity within the B. cereus isolates.
Conclusion
In this study, we investigated the prevalence and distribution of B. cereus group isolates in 10 lettuce farms located in Korea and analyzed their toxin profiles, antibiotic resistance patterns, and genetic diversity among the isolates by ERIC-PCR and MLST. Thirty B. cereus group isolates were identified in 140 samples from lettuce farms. The enterotoxin patterns A (hblCDA, nheABC, entFM, and cytK gene) and B (hblCDA, nheABC, and entFM gene) accounted for 50% and 20% of all the isolates whereas the emetic gene ces was not detected in any of the B. cereus group isolates. The presence of enterotoxin genes in all the isolates indicates that the enterotoxin gene containing B. cereus strains are widespread in Korea than the emetic strains. Antibiotic susceptibility testing revealed that the B. cereus isolates in our study were predominantly resistant to β-lactam antibiotics except imipenem and generally susceptible to most of the non β-lactam antibiotics including gentamycin, streptomycin, trimethoprim-sulfamethoxazole, ciprofloxacin, chloramphenicol, linezolid, vancomycin, erythromycin, and tetracycline. Genetic fingerprinting by ERIC-PCR and MLST analysis revealed that high genetic diversity is prevalent among the B. cereus strains isolated from the lettuce farms in Korea. The results of this study provide information about the contamination levels and molecular characteristics of B. cereus group strains isolated from different lettuce farms which may help to improve the microbial safety standards in lettuce farms.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
NR performed some experiments, analyzed majority of the data, and wrote the whole manuscript. JJ, S-MS, and H-SJ prepared all the necessary materials and performed most of the experiments. B-EK and M-IJ contributed to the discussion of experimental results. J-GR, K-YR, and KO designed and supervised the experiments at different stages. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by “Research Program for Agricultural Science and Technology Development (project no. PJ015168022022),” National Institute of Agricultural Sciences, Rural Development Administration, South Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Bernhard, K., Jarrett, P., Meadows, M., Butt, J., Ellis, D., Roberts, G. M., et al. (1997). Natural isolates of bacillus thuringiensis: worldwide distribution, characterization, and activity against insect pests. J. Invertebr. Pathol. 70, 59–68. doi: 10.1006/jipa.1997.4669
Bozkurt, H., Bell, T., van Ogtrop, F., Phan-Thien, K. Y., and McConchie, R. (2021). Assessment of microbial risk during Australian industrial practices for Escherichia coli O157: H7 in fresh cut-cos lettuce: a stochastic quantitative approach. Food Microbiol. 95:103691. doi: 10.1016/j.fm.2020.103691
Carroll, L. M., Cheng, R. A., Wiedmann, M., and Kovac, J. (2021). Keeping up with the Bacillus cereus group: taxonomy through the genomics era and beyond. Crit. Rev. Food Sci. Nutr. 1–26. doi: 10.1080/10408398.2021.1916735
Carroll, L. M., Kovac, J., Miller, R. A., and Wiedmann, M. (2017). Rapid, high-throughput identification of anthrax-causing and emetic Bacillus cereus group genome assemblies via BTyper, a computational tool for virulence-based classification of Bacillus cereus group isolates by using nucleotide sequencing data. Appl. Environ. Microbiol. 83, e01096–e01017. doi: 10.1128/AEM.01096-17
Centers for Disease Control and Prevention (CDC) . Lettuce, other leafy greens, and food safety. (2022). Available at: https://www.cdc.gov/foodsafety/communication/leafy-greens.html#:~:text=CDC%20estimates%20that%20germs%20on,Salmonella%2C%20Listeria%2C%20and%20Cyclospora (Accessed 8 March 2022).
Ceuppens, S., Boon, N., and Uyttendaele, M. (2013). Diversity of Bacillus cereus group strains is reflected in their broad range of pathogenicity and diverse ecological lifestyles. FEMS Microbiol. Ecol. 84, 433–450. doi: 10.1111/1574-6941.12110
Chang, M. S., Kim, J. H., Lee, J. S., Park, M., Chang, E. H., and Hong, Y. (2020). Comparison of the shelf-life of commercial salad lettuces based on packaging types. Korean J. Food Preserv. 27, 255–260. doi: 10.11002/kjfp.2020.27.2.255
Chaves, J. Q., Pires, E. S., and Vivoni, A. M. (2011). Genetic diversity, antimicrobial resistance and toxigenic profiles of Bacillus cereus isolated from food in Brazil over three decades. Int. J. Food Microbiol. 147, 12–16. doi: 10.1016/j.ijfoodmicro.2011.02.029
Cheng, L. W., Rao, S., Poudyal, S., Wang, P. C., and Chen, S. C. (2021). Genotype and virulence gene analyses of Bacillus cereus group clinical isolates from the Chinese softshell turtle (Pelodiscus sinensis) in Taiwan. J. Fish Dis. 44, 1515–1529. doi: 10.1111/jfd.13473
Chon, J. W., Yim, J. H., Kim, H. S., Kim, D. H., Kim, H., Oh, D. H., et al. (2015). Quantitative prevalence and toxin gene profile of Bacillus cereus from ready-to-eat vegetables in South Korea. Foodborne Pathog. Dis. 12, 795–799. doi: 10.1089/fpd.2015.1977
CLSI . Performance Standards for Antimicrobial Susceptibility Testing, 30th edition CLSI supplement M100. Wayne, PA: Clinical Laboratory Standards Institute; (2020).
Colaco, C. M. G., Basile, K., Draper, J., and Ferguson, P. E. (2021). Fulminant Bacillus cereus food poisoning with fatal multi-organ failure. BMJ Case Rep. 14:e238716. doi: 10.1136/bcr-2020-238716
Cote, C. K., Heffron, J. D., Bozue, J. A., and Welkos, S. L. (2015). Bacillus anthracis and other bacillus species. Mol. Med. Microbiol. 3, 1789–1844. doi: 10.1016/B978-0-12-397169-2.00102-5
Darrigo, C., Guillemet, E., Dervyn, R., and Ramarao, N. (2016). The bacterial Mfd protein prevents DNA damage induced by the host nitrogen immune response in a NER-independent but RecBC-dependent pathway. PLoS One 11:e0163321. doi: 10.1371/journal.pone.0163321
EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) (2012). Scientific opinion on Toyocerin® (Bacillus cereus) as a feed additive for sows, piglets, pigs for fattening, cattle for fattening, calves for rearing, chickens for fattening and rabbits for fattening. EFSA J. 10:2924. doi: 10.2903/j.efsa.2012.2924
EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) (2014). Scientific opinion on the safety and efficacy of Toyocerin® (bacillus toyonensis) as a feed additive for chickens for fattening, weaned piglets, pigs for fattening, sows for reproduction, cattle for fattening and calves for rearing and for rabbits for fattening. EFSA J. 12:3766. doi: 10.2903/j.efsa.2014.3766
Fagerlund, A., Lindbäck, T., and Granum, P. E. (2010). Bacillus cereus cytotoxins Hbl, Nhe and CytK are secreted via the sec translocation pathway. BMC Microbiol. 10, 1–8. doi: 10.1186/1471-2180-10-304
Fiedler, G., Schneider, C., Igbinosa, E. O., Kabisch, J., Brinks, E., Becker, B., et al. (2019). Antibiotics resistance and toxin profiles of Bacillus cereus-group isolates from fresh vegetables from German retail markets. BMC Microbiol. 19, 1–13.
Francisco, A. P., Bugalho, M., Ramirez, M., and Carriço, J. A. (2009). Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinform. 10, 1–15. doi: 10.1186/1471-2105-10-152
Guinebretière, M. H., Auger, S., Galleron, N., Contzen, M., De Sarrau, B., De Buyser, M. L., et al. (2013). Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus group occasionally associated with food poisoning. Int. J. Syst. Evol. Microbiol. 63, 31–40. doi: 10.1099/ijs.0.030627-0
Guinebretière, M. H., Thompson, F. L., Sorokin, A., Normand, P., Dawyndt, P., Ehling-Schulz, M., et al. (2008). Ecological diversification in the Bacillus cereus group. Environ. Microbiol. 10, 851–865. doi: 10.1111/j.1462-2920.2007.01495.x
Guinebretière, M. H., Velge, P., Couvert, O., Carlin, F., Debuyser, M. L., and Nguyen-The, C. (2010). Ability of Bacillus cereus group strains to cause food poisoning varies according to phylogenetic affiliation (groups I to VII) rather than species affiliation. J. Clin. Microbiol. 48, 3388–3391. doi: 10.1128/JCM.00921-10
Heras, J., Domínguez, C., Mata, E., Pascual, V., Lozano, C., Torres, C., et al. (2015). BMC GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinform. 16, 1–8. doi: 10.1186/s12859-015-0703-0
Hsu, T. K., Tsai, H. C., Hsu, B. M., Yang, Y. Y., and Chen, J. S. (2021). Prevalence, enterotoxin-gene profiles, antimicrobial resistance, and genetic diversity of Bacillus cereus group in aquatic environments and shellfish. Sci. Total Environ. 758:143665. doi: 10.1016/j.scitotenv.2020.143665
Irvin, K., Viazis, S., Fields, A., Seelman, S., Blickenstaff, K., Gee, E., et al. (2021). An overview of traceback investigations and three case studies of recent outbreaks of Escherichia coli O157: H7 infections linked to romaine lettuce. J. Food Prot. 84, 1340–1356. doi: 10.4315/JFP-21-112
Jo, M. J., Jeong, A. R., Kim, H. J., Lee, N. R., Oh, S. W., Kim, Y. J., et al. (2011). Korean microbiological quality of fresh-cut produce and organic vegetables. J. Food Sci. Technol. 43, 91–97. doi: 10.9721/KJFST.2011.43.1.091
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Jolley, K. A., and Maiden, M. C. (2014). Using MLST to study bacterial variation: prospects in the genomic era. Future Microbiol. 9, 623–630. doi: 10.2217/fmb.14.24
Jung, K. H., Minseon, K., Daekeun, H., Choi, J. H., Min, K. S., and Se-Wook, O. (2016). Contamination patterns and molecular typing of Bacillus cereus in fresh-cut vegetable salad processing. Appl. Biol. Chem. 59, 573–577. doi: 10.1007/s13765-016-0198-z
Kim, T. S., Jang, Y. H., and Hwang, H. J. (2018). Strategies to increase domestic lettuce circulations through improving valuable end-user traits. J. Distribut. Sci. 16, 63–68. doi: 10.15722/jds.16.8.201808.63
Kim, W. I., Jung, H. M., Kim, S. R., Park, K. H., Kim, B. S., Yun, J. C., et al. (2012). Investigation of microbial contamination levels of leafy greens and its distributing conditions at different time-focused on perilla leaf and lettuce. J. Food Hygiene Saf. 27, 277–284. doi: 10.13103/JFHS.2012.27.3.277
Kim, J. B., Kim, J. M., Cho, S. H., Oh, H. S., Choi, N. J., and Oh, D. H. (2011). Toxin genes profiles and toxin production ability of Bacillus cereus isolated from clinical and food samples. J. Food Sci. 76, T25–T29. doi: 10.1111/j.1750-3841.2010.01958.x
Kim, S. K., Kim, K.-P., Jang, S. S., Shin, E. M., Kim, M.-J., Oh, S., et al. (2009). Prevalence and toxigenic profiles of Bacillus cereus isolated from dried red peppers, rice, and sunsik in Korea. J. Food Prot. 72, 578–582. doi: 10.4315/0362-028X-72.3.578
Liu, Y., Lai, Q., Du, J., and Shao, Z. (2017). Genetic diversity and population structure of the Bacillus cereus group bacteria from diverse marine environments. Sci. Rep. 7, 1–11. doi: 10.1038/s41598-017-00817-1
Liu, Y., Lai, Q., and Shao, Z. (2018). Genome analysis-based reclassification of bacillus weihenstephanensis as a later heterotypic synonym of bacillus mycoides. Int. J. Syst. Evol. Microbiol. 68, 106–112. doi: 10.1099/ijsem.0.002466
Luna, V. A., King, D. S., Gulledge, J., Cannons, A. C., Amuso, P. T., and Cattani, J. (2007). Susceptibility of bacillus anthracis, Bacillus cereus, bacillus mycoides, bacillus pseudomycoides and bacillus thuringiensis to 24 antimicrobials using Sensititre® automated microbroth dilution and Etest® agar gradient diffusion methods. J. Antimicrob. Chemother. 60, 555–567. doi: 10.1093/jac/dkm213
Lund, T., De Buyser, M. L., and Granum, P. E. (2000). A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 38, 254–261. doi: 10.1046/j.1365-2958.2000.02147.x
Maiden, M. C., Van Rensburg, M. J. J., Bray, J. E., Earle, S. G., Ford, S. A., Jolley, K. A., et al. (2013). MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 11, 728–736. doi: 10.1038/nrmicro3093
Majiduddin, F. K., Materon, I. C., and Palzkill, T. G. (2002). Molecular analysis of beta-lactamase structure and function. Int. J. Med. Microbiol. 292, 127–137. doi: 10.1078/1438-4221-00198
Mandic-Mulec, I., Stefanic, P., and Van Elsas, J. D. (2015). Ecology of bacillaceae. Microbiol. Spectr. 3:TBS-0017-2013. doi: 10.1128/microbiolspec.TBS-0017-2013
Manzulli, V., Fasanella, A., Parisi, A., Serrecchia, L., Donatiello, A., Rondinone, V., et al. (2019). Evaluation of in vitro antimicrobial susceptibility of bacillus anthracis strains isolated during anthrax outbreaks in Italy from 1984 to 2017. J. Vet. Sci. 20, 58–62. doi: 10.4142/jvs.2019.20.1.58
Marston, C. K., Hoffmaster, A. R., Wilson, K. E., Bragg, S. L., Plikaytis, B., Brachman, P., et al. (2005). Effects of long-term storage on plasmid stability in bacillus anthracis. Appl. Environ. Microbiol. 71, 7778–7780. doi: 10.1128/AEM.71.12.7778-7780.2005
McDowell, R. H., Sands, E. M., and Friedman, H. (2021). “Bacillus cereus,” in StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2022 Jan. Available from: https://www.ncbi.nlm.nih.gov/books/NBK459121/
Miller, R. A., Jian, J., Beno, S. M., Wiedmann, M., Kovac, J., and Björkroth, J. (2018). Intraclade variability in toxin production and cytotoxicity of Bacillus cereus group type strains and dairy-associated isolates. Appl. Environ. Microbiol. 84, e02479–e02417. doi: 10.1128/AEM.02479-17
Mills, E., Sullivan, E., Kovac, J., and Elkins, C. A. (2022). Comparative analysis of Bacillus cereus group isolates' resistance using disk diffusion and broth microdilution and the correlation between antimicrobial resistance phenotypes and genotypes. Appl. Environ. Microbiol. 88, e02302–e02321. doi: 10.1128/aem.02302-21
Munita, J. M., and Arias, C. A. (2016). Mechanisms of antibiotic resistance. Microbiol. Spectr. 4, 4–2. doi: 10.1128/microbiolspec.VMBF-0016-2015
Naranjo, M., Denayer, S., Botteldoorn, N., Delbrassinne, L., Veys, J., Waegenaere, J., et al. (2011). Sudden death of a young adult associated with Bacillus cereus food poisoning. J. Clin. Microbiol. 49, 4379–4381. doi: 10.1128/JCM.05129-11
Ngamwongsatit, P., Buasri, W., Pianariyanon, P., Pulsrikarn, C., Ohba, M., Assavanig, A., et al. (2008). Broad distribution of enterotoxin genes (hblCDA, nheABC, cytK, and entFM) among bacillus thuringiensis and Bacillus cereus as shown by novel primers. Int. J. Food Microbiol. 121, 352–356. doi: 10.1016/j.ijfoodmicro.2007.11.013
Okinaka, R. T., Cloud, K., Hampton, O., Hoffmaster, A. R., Hill, K. K., Keim, P., et al. (1999). Sequence and organization of pXO1, the large bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 181, 6509–6515. doi: 10.1128/JB.181.20.6509-6515.1999
Osimani, A., Aquilanti, L., and Clementi, F. (2018). Bacillus cereus foodborne outbreaks in mass catering. Int. J. Hosp. Manag. 72, 145–153. doi: 10.1016/j.ijhm.2018.01.013
Owusu-Kwarteng, J., Wuni, A., Akabanda, F., Tano-Debrah, K., and Jespersen, L. (2017). Prevalence, virulence factor genes and antibiotic resistance of Bacillus cereus sensu lato isolated from dairy farms and traditional dairy products. BMC Microbiol. 17, 1–8. doi: 10.1186/s12866-017-0975-9
Pang, H., Lambertini, E., Buchanan, R. L., Schaffner, D. W., and Pradhan, A. K. (2017). Quantitative microbial risk assessment for Escherichia coli O157: H7 in fresh-cut lettuce. J. Food Prot. 80, 302–311. doi: 10.4315/0362-028X.JFP-16-246
Park, K. M., Jeong, M., Park, K. J., and Koo, M. (2018). Prevalence, enterotoxin genes, and antibiotic resistance of Bacillus cereus isolated from raw vegetables in Korea. J. Food Prot. 81, 1590–1597. doi: 10.4315/0362-028X.JFP-18-205
Park, K. M., Kim, H. J., Jeong, M., and Koo, M. (2020). Enterotoxin genes, antibiotic susceptibility, and biofilm formation of low-temperature-tolerant Bacillus cereus isolated from green leaf lettuce in the cold chain. Foods 9:249. doi: 10.3390/foods9030249
Park, Y. B., Kim, J. B., Shin, S. W., Kim, J. C., Cho, S. H., Lee, B. K., et al. (2009). Prevalence, genetic diversity, and antibiotic susceptibility of Bacillus cereus strains isolated from rice and cereals collected in Korea. J. Food Prot. 72, 612–617. doi: 10.4315/0362-028X-72.3.612
Pérez-Losada, M., Cabezas, P., Castro-Nallar, E., and Crandall, K. A. (2013). Pathogen typing in the genomics era: MLST and the future of molecular epidemiology. Infect. Genet. Evol. 16, 38–53. doi: 10.1016/j.meegid.2013.01.009
Peterson, E., and Kaur, P. (2018). Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 9:2928. doi: 10.3389/fmicb.2018.02928
Qu, Y., Wei, C., Dai, X., Bai, Y., Zhao, X., Lan, Q., et al. (2021). The possible transmission and potential enterotoxicity of Bacillus cereus on lettuce farms in five Chinese provinces. Front. Microbiol. 12:746632. doi: 10.3389/fmicb.2021.746632
Ribeiro-Gonçalves, B., Francisco, A. P., Vaz, C., Ramirez, M., and Carriço, J. A. (2016). PHYLOViZ online: web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res. 44, W246–W251. doi: 10.1093/nar/gkw359
Sacchi, C. T., Whitney, A. M., Mayer, L. W., Morey, R., Steigerwalt, A., Boras, A., et al. (2002). Sequencing of 16S rRNA gene: a rapid tool for identification of bacillus anthracis. Emerg. Infect. Dis. 8, 1117–1123. doi: 10.3201/eid0810.020391
Sastalla, I., Fattah, R., Coppage, N., Nandy, P., Crown, D., Pomerantsev, A. P., et al. (2013). The Bacillus cereus Hbl and Nhe tripartite enterotoxin components assemble sequentially on the surface of target cells and are not interchangeable. PLoS One 8:e76955. doi: 10.1371/journal.pone.0076955
Skalet, A. H., Cevallos, V., Ayele, B., Gebre, T., Zhou, Z., Jorgensen, J. H., et al. (2010). Antibiotic selection pressure and macrolide resistance in nasopharyngeal Streptococcus pneumoniae: a cluster-randomized clinical trial. PLoS Med. 7:e1000377. doi: 10.1371/journal.pmed.1000377
Soltis, D. E., and Soltis, P. S. (2003). The role of phylogenetics in comparative genetics. Plant Physiol. 132, 1790–1800. doi: 10.1104/pp.103.022509
Stenfors Arnesen, L. P., Fagerlund, A., and Granum, P. E. (2008). From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32, 579–606. doi: 10.1111/j.1574-6976.2008.00112.x
Tran, S. L., Cormontagne, D., Vidic, J., André-Leroux, G., and Ramarao, N. (2020). Structural modeling of cell wall peptidase CwpFM (EntFM) reveals distinct intrinsically disordered extensions specific to pathogenic Bacillus cereus strains. Toxins 12:593. doi: 10.3390/toxins12090593
Vardakas, K. Z., Tansarli, G. S., Rafailidis, P. I., and Falagas, M. E. (2012). Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to Enterobacteriaceae producing extended-spectrum β-lactamases: a systematic review and meta-analysis. J. Antimicrob. Chemother. 67, 2793–2803. doi: 10.1093/jac/dks301
Versalovic, J., Koeuth, T., and Lupski, R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res. 19, 6823–6831. doi: 10.1093/nar/19.24.6823
Zhao, Y., Chen, C., Gu, H.-j., Zhang, J., and Sun, L. (2019). Characterization of the genome feature and toxic capacity of a bacillus wiedmannii isolate from the hydrothermal field in Okinawa trough. Front. Cell. Infect. Microbiol. 9:370. doi: 10.3389/fcimb.2019.00370
Keywords: Bacillus cereus, lettuce, toxin profiling, antimicrobial resistance, ERIC-PCR, MLST
Citation: Rajalingam N, Jung J, Seo S-M, Jin H-S, Kim B-E, Jeong M-I, Kim D, Ryu J-G, Ryu K-Y and Oh KK (2022) Prevalence, distribution, enterotoxin profiles, antimicrobial resistance, and genetic diversity of Bacillus cereus group isolates from lettuce farms in Korea. Front. Microbiol. 13:906040. doi: 10.3389/fmicb.2022.906040
Edited by:
Arun K. Bhunia, Purdue University, United StatesReviewed by:
Laura M. Carroll, European Molecular Biology Laboratory GermanyShuai Wei, Guangdong Ocean University, China
Copyright © 2022 Rajalingam, Jung, Seo, Jin, Kim, Jeong, Kim, Ryu, Ryu and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kwang Kyo Oh, b2hrd2FuZ0Brb3JlYS5rcg==
 Nagendran Rajalingam
Nagendran Rajalingam Jieun Jung2
Jieun Jung2 Kwang Kyo Oh
Kwang Kyo Oh