
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 09 June 2022
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.905826
This article is part of the Research Topic Microbial Associates of Blood-Sucking Arthropods and Other Animals: Relevance to Their Physiology, Ecology and Evolution View all 12 articles
Tsetse flies have socioeconomic significance as the obligate vector of multiple Trypanosoma parasites, the causative agents of Human and Animal African Trypanosomiases. Like many animals subsisting on a limited diet, microbial symbiosis is key to supplementing nutrient deficiencies necessary for metabolic, reproductive, and immune functions. Extensive studies on the microbiota in parallel to tsetse biology have unraveled the many dependencies partners have for one another. But far less is known mechanistically on how products are swapped between partners and how these metabolic exchanges are regulated, especially to address changing physiological needs. More specifically, how do metabolites contributed by one partner get to the right place at the right time and in the right amounts to the other partner? Epigenetics is the study of molecules and mechanisms that regulate the inheritance, gene activity and expression of traits that are not due to DNA sequence alone. The roles that epigenetics provide as a mechanistic link between host phenotype, metabolism and microbiota (both in composition and activity) is relatively unknown and represents a frontier of exploration. Here, we take a closer look at blood feeding insects with emphasis on the tsetse fly, to specifically propose roles for microRNAs (miRNA) and DNA methylation, in maintaining insect-microbiota functional homeostasis. We provide empirical details to addressing these hypotheses and advancing these studies. Deciphering how microbiota and host activity are harmonized may foster multiple applications toward manipulating host health, including identifying novel targets for innovative vector control strategies to counter insidious pests such as tsetse.
Tsetse flies are Dipterans belonging to the superfamily of exclusive blood feeders, Hippoboscoidea. Tsetse are exclusively grouped in the family Glossinidae, within the monophyletic genus Glossina, and are divided into four groups: morsitans, fusca, palpalis, and austeni (Krafsur, 2009). Tsetse flies are found only in sub-Saharan Africa with the different groups occupying distinct ecological terrains and blood meal preferences which effects the medical and agricultural significance of different species (Solano et al., 2010). Tsetse flies undergo adenotrophic viviparity (Benoit et al., 2015) meaning that a single larva develops in utero each gonotrophic cycle (Figure 1). Maternal secretions provide nutrition and seed larva with microbiota (Ma and Denlinger, 1974) through modified female accessory glands known as milk glands.
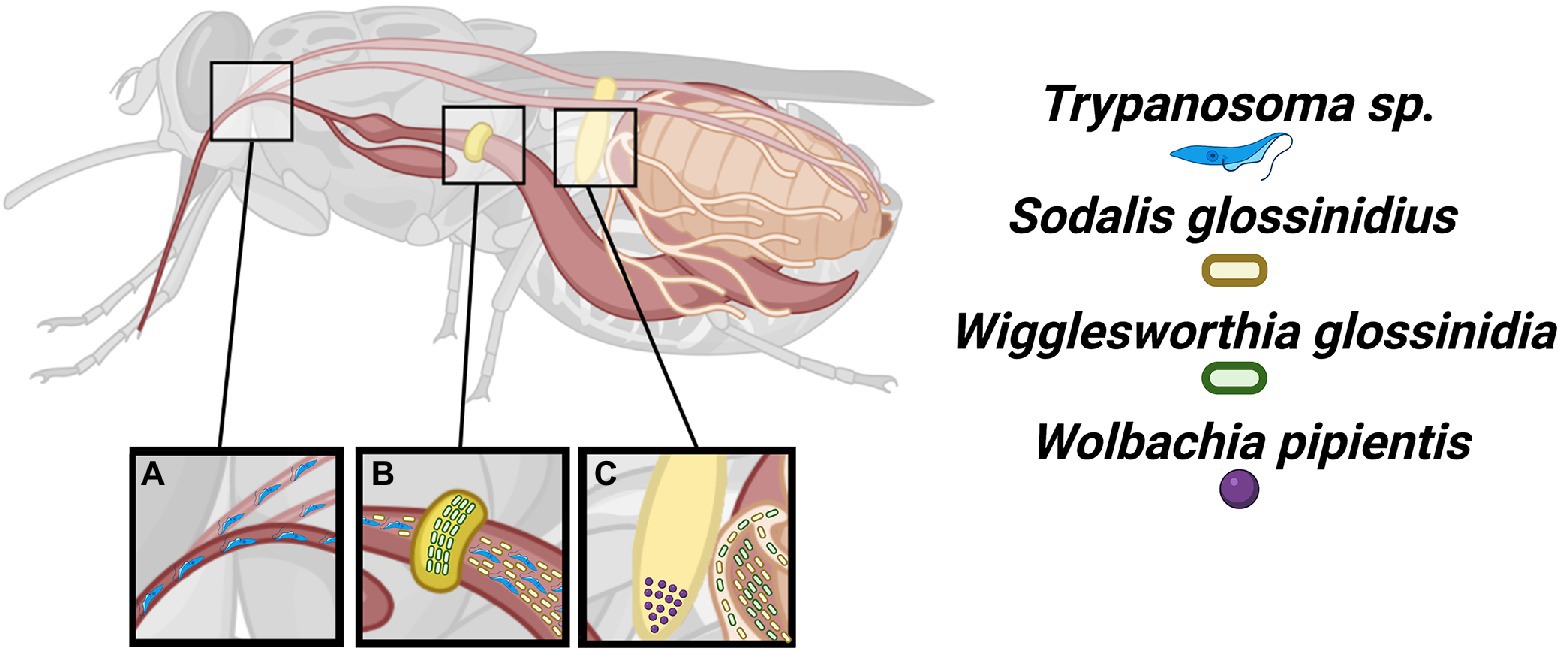
Figure 1. Localization of tsetse microbiota. The tsetse fly is the sole vector of most African trypanosomes. (A) These protozoan parasites are introduced into the tsetse fly by an infected blood meal where developmental differentiation, recombination, and migration to the salivary glands occur. (B) The Wigglesworthia and Sodalis symbionts may be found within the bacteriome and gut, respectively. (C) The Wigglesworthia, Sodalis, and Wolbachia symbionts are vertically transmitted. The Wigglesworthia and Sodalis bacteria specifically use milk gland infections while Wolbachia infects ovaries for transgenerational persistence.
A key feature in the evolution of eukaryotes has been the spatial and temporal partitioning of biochemical processes for the purpose of regulation (Chomicki et al., 1808; Martin, 2010; Gabaldón and Pittis, 2015). This partitioning reaches even greater complexity with the presence of microbiota and the necessity to coordinate their physiology with host biology particularly if they also rely on vertical transmission for their persistence as this additionally entails coordination with host reproductive biology. Tsetse flies possess a relatively simple core microbiota consisting of three different bacterial species (Wigglesworthia glossinidia, Sodalis glossinidius, and Wolbachia pipientis) varying in their occurrence and ranging in their impact toward host biology from parasitism to mutualism. The obligate mutualist, Wigglesworthia, is a focal point of this review and will be further discussed below. A commensal Sodalis (Dale and Maudlin, 1999) is not known to impact tsetse fitness but has emerged as a bacterium of interest as a target for paratransgenesis and introducing a trypanosome refractory phenotype (De Vooght et al., 2018). Lastly, W. pipientis (supergroup A) may be harbored by tsetse typically within reproductive tissues (O’Neill et al., 1993; Cheng et al., 2000; Balmand et al., 2013) which may result in cytoplasmic incompatibility between mating of differentially infected individuals (Alam et al., 2011).
Tsetse flies are the obligate vectors of most African Trypanosomes, Trypanosoma species, with an association dating back about 35 million years (Steverding, 2008). Trypanosome parasites are the causative agent of Human African Trypanosomiasis (HAT; T. brucei rhodesiense and T. b. gambiensis), a debilitating disease caused by the parasitic invasion of the central nervous system which is lethal if left untreated. The disease is endemic to 36 countries in sub-Saharan Africa. Animal African Trypanosomiasis (AAT; T. b. brucei, T. vivax, and T. congolense) is a wasting disease caused by trypanosome infections of domestic animals, contributing to food insecurity within impacted areas. Trypanosome infections of tsetse impose a reproductive burden on females (Hu et al., 2008) likely due to competition for resources with some of these provided by the microbiota (Michalkova et al., 2014; Rio et al., 2019).
Both sexes of tsetse feed exclusively on vertebrate blood and consequently have epidemiological significance toward trypanosome transmission. The blood, although rich in amino acids and iron, is particularly poor in B vitamins (Douglas, 2017), which are essential for animals. The provisioning of multiple B vitamins by the obligate mutualist W. glossinidia has enabled the restricted feeding ecology of the tsetse fly. The Wigglesworthia symbiont is the most predominant member of the tsetse microbiota (Aksoy, 1995; Chen et al., 1999; Aksoy et al., 2014; Tsagmo Ngoune et al., 2019) and inhabits the cytosol of specialized tsetse epithelial cells known as bacteriocytes that collectively form a bacteriome attached to the anterior midgut (Figure 1). Wigglesworthia cells are large, filamentous-like (Aksoy, 1995) and lie free in the cytoplasm, unabated from a host-generated membrane and likely also necessitating unique molecular transfer processes with tsetse.
The tsetse-Wigglesworthia association dates to around the incipient stages of species diversification of the Glossinidae family (Aksoy et al., 1995; Chen et al., 1999; Symula et al., 2011). This long interdependence has led to a profound impact on the evolutionary genomics of both species. Host adaptation has involved drastic Wigglesworthia genome size reduction (Akman et al., 2002; Rio et al., 2012) tailored to tsetse biology coupled with high genetic drift due to smaller population sizes arising from bottlenecks during vertical transfer. Despite its small size, the Wigglesworthia genome retains the potential to synthesize multiple B complex vitamins, namely, thiamine (B1), riboflavin (B2), nicotinamide (B3), pantothenic acid (B5), pyridoxine (B6), and folate (B9; Akman et al., 2002; Rio et al., 2012) believed to complement metabolic deficiencies in the blood feeding ecology of tsetse. To date, Wigglesworthia has not been cultured but with the availability of annotated genomes (Akman et al., 2002; Rio et al., 2012) and the advent of innovative culture technologies (Lagier et al., 2018; Cross et al., 2019) this may ultimately be achieved opening up a wide array of research questions. Additionally, the availability of an extracellular population of Wigglesworthia within maternal milk gland secretions (Ma and Denlinger, 1974; Attardo et al., 2008; Balmand et al., 2013) may also facilitate culturing.
As obligate mutualists, tsetse rely on Wigglesworthia for the optimal performance of several physiological processes involved in nutrition, digestion, immunological maturation and reproduction (and likely the connection between these; Wang et al., 2009; Snyder et al., 2010; Weiss et al., 2011, 2013; Michalkova et al., 2014; Snyder and Rio, 2015). In support of its specialization, the bacteriome is enriched in fly gene transcripts that belong to the transmembrane category (Bing et al., 2017; Medina Munoz et al., 2017, 2021), which includes amino acid transporters and multivitamin transporters, likely facilitating nutrient exchange between tsetse and Wigglesworthia. In turn, Wigglesworthia transcripts are enriched for the metabolism of cofactors and vitamins, supporting a complementary nutrient synthesis role for uptake by host transporters. Structural and functional examination of transporters, and how these may be regulated by epigenetics will help elucidate mechanisms used for interspecies metabolic regulation, likely involving some type of feedback network based on metabolites crucial for homeostasis, although this remains speculative.
Epigenetics controls gene expression and concomitant phenotype independent of gene sequence (Choudhuri, 2011), thereby enabling a relatively rapid adaptation independent of inheritance. Epigenetic mechanisms within insects include small RNA production (Asgari, 2013, 2015; Lucas and Raikhel, 2013; Zhang et al., 2014a; Lucas et al., 2015), histone post-translational modifications (Dickman et al., 2013; Glastad et al., 2015), chromatin remodeling (Rider et al., 2010; Riparbelli et al., 2012), and DNA methylation (Field et al., 2004). Epigenetics may be heritable but may also be erased and reestablished to address specific environmental cues (Waddington, 2012; Tammen et al., 2013; Bind et al., 2014; Deans and Maggert, 2015; Chatterjee et al., 2018; McCaw et al., 2020; Villagra and Frías-Lasserre, 2020). Our focus in this mini review will be specifically on the roles that microRNAs (miRNAs) and DNA methylation may have toward mediating the coordination of microbe-host interactions.
miRNAs are small (~22 nt) noncoding RNAs with a primary function in sequence-specific post-transcriptional gene regulation (Ibáñez-Ventoso et al., 2008; Fabian et al., 2010). Gene regulation (generally inhibitory) via miRNAs is highly conserved across eukaryotes (Bartel, 2018) with sequence conservation of seed regions (i.e., nucleotides present at positions 2–8 from the 5′ end) facilitating identification across often phylogenetically distant animals (Ligoxygakis et al., 2002; Marco et al., 2010). For example, more than 50% of the characterized Caenorhabditis elegans miRNAs are encoded in both human and Drosophila genomes (Ibáñez-Ventoso et al., 2008; Asgari, 2011). High conservation of miRNAs among Dipterans has also been observed in studies comparing mosquitoes to Drosophila (Lai et al., 2003; Li et al., 2009). Despite this conservation, miRNAs sharing high nucleotide identity may exhibit target variation in different species by undergoing “seed-shifting,” where slight changes in the 5′ end of a miRNA alters the seed region (Wheeler et al., 2009; Marco et al., 2010; Berezikov, 2011), consequentially generating a variety of new mRNA targets. Seed-shifting partnered with duplication is the primary evolutionary force for creating new miRNAs (Bartel, 2009; Berezikov, 2011). While miRNAs are known to be a significant source of regulators of endogenous genes (Carthew and Sontheimer, 2009), their potential role in modulating microbial homeostasis and in preventing dysbiosis has been comparatively understudied.
miRNAs are typically encoded within intergenic regions, in non-coding transcripts, or in rare cases within the coding region of genes (Slack, 2006; Asgari, 2011, 2013). To date, most miRNAs are produced through the canonical pathway, though a rare subset (known as non-canonical microRNAs) do not follow this pathway (Bartel, 2004; Abdelfattah et al., 2014). Canonical miRNA generation begins with transcription by RNA polymerase II of long RNA sequences known as polyadenylated primary transcripts (pri-miRNAs) in the nucleus (Asgari, 2011, 2013). The Drosha-Pasha/DGCR8 complex, also known as the Microprocessor complex, processes and cleaves the stem-loops of the pri-miRNA to form the hairpin precursor miRNA (pre-miRNA; Han et al., 2009; Asgari, 2011, 2013). The ~70 nt pre-miRNA is then transported into the cytoplasm by Exportin 5 and its terminal loop cleaved by Dicer and the loquacious protein (mammalian TRBP) forming a ~22 nt miRNA:miRNA* duplex (Asgari, 2011, 2013; Nguyen et al., 2015). Similar to Drosophila melanogaster (Tomari and Zamore, 2005), tsetse flies also encode two distinct Dicer proteins (Dicer-1 and Dicer-2) with Dicer-1 required for miRNA production (Lee et al., 2004). Argonaute (Ago) proteins then associate with the miRNA duplex using one of the strands as a guide strand forming a RNA-induced silencing complex (RISC; Kawamata and Tomari, 2010; Nguyen et al., 2015). The remaining strand, miRNA*, is known as the passenger miRNA and may play a regulatory role but is typically degraded (Asgari, 2011, 2013). Dicer cleavage also seems to selectively favor an arm of the precursor stem loop, though this preference can vary in different tissues in a context dependent manner (Griffiths-Jones et al., 2011; Chen et al., 2018; Kim et al., 2020). This variation leads to 3′ or 5′ (typical represented as miRNA -3p or -5p) isomiRs being present for miRNAs (Kim et al., 2020). This arm shifting is also responsible for generating a large portion of the diversity within miRNA families (Okamura et al., 2008; de Wit et al., 2009; Berezikov, 2011; Griffiths-Jones et al., 2011).
It is likely that miRNAs have a diversity of functions within the tsetse fly, as within the related Drosophila species a range of roles in development, endocrinology, viral immunity, and behavior have been described (Carthew et al., 2017). This is further supported by the conservation of many miRNAs homologs in the more distantly related mosquitos, although miRNA conservation does not necessarily suggest functional retention since some miRNAs are also predicted to have numerous targets (Lai et al., 2003; Friedman et al., 2009; Li et al., 2009). Of relevance, in Anopheles gambiae mosquitoes, an elevated abundance of miR-305 is known to increase susceptibility toward Plasmodium infections (Dennison et al., 2015), likely mediated by disrupting mRNAs involved in metabolic (Lampe and Levashina, 2018) and immunological processes (Dennison et al., 2015). Similarly, shed trypanosome VSG surface coat antigen when internalized by tsetse cardia cells, decreases miR-275 expression within the midgut (Aksoy et al., 2016; Vigneron et al., 2018). Consequently, the reduction of miR-275 results in compromising the synthesis of the peritrophic matrix (PM) by inhibiting peritrophin expression, the Wnt-signaling pathway and Iroquois/IRX family of transcription factors in the cardia thereby disrupting digestion and strengthening vector competence (Aksoy et al., 2016). As a proof of principle, paratransgenic S. glossinidius engineered to express tandem antagomir-275 repeats (3xant-miR275) phenocopies the compromised peritrophic matrix and offers an exciting (and economical) technological advancement toward studying the regulatory roles of other miRNAs. Lastly, tsetse with symptomatic Salivary Gland Hypertrophy Virus (SGHV) infections exhibit different tsetse miRNA and SGHV miRNA expression profiles upon comparison to asymptomatic flies. With symptomatic flies, the most highly expressed miRNAs are predicted to target immune-related mRNAs, including those encoded by fibrillin-1 (FBN1) and Ras-related protein-27 (Rab27), and others involved in reproduction such as apolipoprotein lipid transfer particle (Apoltp) and vitellogenin receptor (Vtgr; Meki et al., 2018). These genes are all downregulated within symptomatic flies contributing to viral immune evasion and associated ovarian aberrations and loss of reproductive fitness (Abd-Alla et al., 2010).
In previous insect research low or absent miRNA homology suggests novel biological or physiological functions of that miRNA (Marco et al., 2010). Using a custom pipeline of bioinformatic tools on publicly available tsetse Expressed Sequence Tags (ESTs), 10 miRNAs were found to be unique to tsetse flies with gmr-miR 619-5p and gmr-miR-2490-3p predicted to target genes impacted by trypanosome infection, including those encoding the thioester-containing protein (Tep-1) and heat shock protein 60A (Hsp60a; Yang et al., 2020), although experimental validation of molecular regulation remains to be shown.
miRNAs may also directly impact microbiota composition and activity (Ibáñez-Ventoso et al., 2008; Friedman et al., 2009). Besides pathogenic associations, miRNAs are also involved in the regulation of essential members of the microbiota. For example, with the symbiosis between aphids and their symbiont Buchnera (a similar ancient obligate nutritional mutualism to the tsetse-Wigglesworthia association; Douglas, 1998; Feng et al., 2019), 14 aphid-generated miRNAs are evolutionarily conserved among phylogenetically distant aphid species with significantly different expression of these within bacteriomes relative to symbiont-free tissue (Feng et al., 2018) strongly supporting roles in mediating symbiosis. Moreover, 84 mRNA targets with a predominant function in the principal functional role of the symbiosis, amino acid transport and metabolism (Feng et al., 2018), were identified as putative targets of these miRNAs. At least 10 of the 14 miRNAs have been identified to be of importance toward other host–microbe interaction studies (Skalsky et al., 2010; Jayachandran et al., 2013; Mehrabadi et al., 2013; Mayoral et al., 2014a; Zhang et al., 2014b; Jin et al., 2017; Qiang et al., 2017; Liu et al., 2019) suggesting a universal (and likely convergent) role in the regulation of symbioses. Compellingly, research in tsetse has indicated genes associated with both amino acid transport and metabolism (Wigglesworthia is auxotrophic for the majority of amino acids) have differential expression in aposymbiotic compared to wild-type flies, which may indicate a similar regulatory role toward these genes could be played by tsetse miRNAs (Medina Munoz et al., 2017).
A plethora of questions remain about whether animals can use miRNAs to impact gene expression in microbes. Previous research on miRNAs in insect microbial relations has focused on identifying miRNAs produced by the host and assumed to target host mRNAs involved in the symbiosis (Carthew et al., 2017; Feng et al., 2018, 2019). Encouraging research that suggests targeting of microbial (particularly bacteria) RNA may in fact be plausible comes from studies demonstrating miRNAs regulating mitochondrial mRNAs (Li et al., 2012; Duarte et al., 2014; Macgregor-Das and Das, 2018). Mitochondria, as remnants of an ancient Alphaproteobacterium endosymbiont rendered modern-day organelle, still retain a double membrane (Macgregor-Das and Das, 2018) similar to Wigglesworthia (Aksoy, 1995). If tsetse miRNAs interact with Wigglesworthia to coordinate gene expression, they are likely not alone. It is possible that other mutualists with significantly reduced genomes such as Wigglesworthia may also rely on these small RNAs as opposed to proteins for gene regulation, representing a novel avenue for experimental exploration to further our understanding of intracellular signaling (Hansen and Degnan, 2014). Lastly, a further compelling and reciprocal research focus is whether small RNAs encoded by bacterial mutualists may manipulate host genes, which is not unknown of within Gammaproteobacteria. For example, intracellular Salmonella produce a miRNA-like Sal-1 processed by human AGO2 proteins which enhances intracellular Salmonella survival (Gu et al., 2017). Further the production of a Wolbachia small noncoding RNA, WsnRNA-46A, enhances the transcription of Aedes aegypti Dynein heavy chain (Dhc) which facilitates Wolbachia association with microtubules enabling its transfer during mosquito oocyte or embryonic development (Mayoral et al., 2014b). Whether small noncoding RNAs produced by Wigglesworthia may impact tsetse metabolism or immunity remains to be seen.
DNA methylation is the addition of methyl (CH3) groups to cytosine residues (5mC) typically within 5′-cytosine-phosphate-guanine-3′ (CpG) dinucleotides (Lyko, 2018). Across insect taxa, genome methylation exhibits a patchy distribution and differs relative to those of vertebrates in regards to general localization (Head, 2014). For example, DNA methylation is prevalent in the promoter regions (creating CpG islands) of vertebrate genomes, with modifications altering the interactions of transcription factors and histones via steric effects (Moore et al., 2013). Within insect genomes, DNA methylation is pervasive within gene bodies (Cingolani et al., 2013; Takayama et al., 2014; Jeong et al., 2018; Huang et al., 2019), where it is involved in alternative gene splicing and the creation of isoforms (Lyko et al., 2010; Bonasio et al., 2012; Terrapon et al., 2014). Although CpG methylation is also present within insect genomes, methylation is more prevalent in the CpA and CpT dinucleotide contexts (Takayama et al., 2014). For example, splice junctions are enriched for non-CpG methylation (Cingolani et al., 2013) in bees and different splice variants of the same gene are associated with diverse methylation patterns (Lyko et al., 2010).
DNA methylation is among the most amenable epigenetic modifications to identify given its relative ease in identification. For example, commercially available antibodies detect the presence of methylated nucleotides within genomic DNA (Kunert et al., 2003) and may be used to enrich for methylated DNA prior to high-throughput sequencing (Glastad et al., 2014). Moreover, bisulfite sequencing and subsequent mapping (Ku et al., 2011), enables the characterization of nucleotide methylation across a reference genome of interest, permitting the discovery of preferential motifs (Takayama et al., 2014; Panikar et al., 2015). The generation of reference DNA methylomes for a variety of insects through developmental stages with validated ties to phenotypes will greatly facilitate our understanding of epigenetic modifications toward insect biology and fuel future research endeavors.
Although vitamins are essential to physiology, animals lack the ability to synthesize these de novo and must either obtain these critical nutrients through diet and/or microbiota provisioning (Brecher and Wigglesworth, 1944; Douglas, 2017). Folate (B9) is particularly deficient within blood (Brecher and Wigglesworth, 1944; Edwards et al., 1957; Pietrzik et al., 2010; Nikoh et al., 2014; Douglas, 2017; Duron et al., 2018), with symbiotic bacteria often provisioning this essential cofactor to strictly hematophagous animals (Duron and Gottlieb, 2020). A significant role for Wigglesworthia within their hosts is the production and provisioning of folate, which is critical for tsetse reproduction and larval development while also serving to enhance vector competence (Snyder and Rio, 2015; Rio et al., 2019).
Folate is necessary for DNA methylation because it is transformed into 5-methyltetrahydrofolate (5-methylTHF), needed for the formation of methionine from homocysteine (Crider et al., 2012). Once methionine has been synthesized, it is joined to ATP and converted into the universal methylation donor S-adenosyl methionine (SAM). SAM donates the methyl group during DNA methylation via the action of DNA methyltransferases (Crider et al., 2012; Shorter et al., 2015). Folate provisioning by Wigglesworthia may provide a means for connecting Wigglesworthia metabolism to tsetse genetic regulation via DNA methylation. In support of this connection, SAM abundance is significantly decreased in tsetse fly bacteriomes which have been cleared of their Wigglesworthia symbionts (Bing et al., 2017).
Due to the lack of DNMT-1 and -3 in the genome, tsetse has been predicted to lack DNA methylation (Bewick et al., 2017). However, due to its close evolutionary relation to D. melanogaster and the characterization of DNA methylation in the fruit fly genome (Takayama et al., 2014; Panikar et al., 2015; despite also lacking these DNMTs), we hypothesize the presence of methylation in the tsetse genomic DNA, particularly within Wigglesworthia harboring bacteriomes which may impact symbiosis activities. Symbiosis altering DNA methylation is not unprecedented in eukaryotes as previously reported in a wide array of organisms including plants (Vannier et al., 2015), anemones (Li et al., 2018), and mice (Warner et al., 1989; Yu et al., 2015), with concomitant changes in symbiosis phenotypes.
Metabolite provisioning is a fundamental role of host-associated microbiota, particularly of animals with limited diets such as the strictly blood feeding tsetse fly. The tsetse fly provides a valuable, and medically significant, model system to dissect regulatory mechanisms that coordinate host-microbiota activities, including nutrient exchange, immunological maturation and vector competence. Much has been gathered on the composition, functional contribution and evolutionary history of the tsetse microbiota, yet little is known regarding mechanisms coordinating microbial activity with host biology. Here we emphasize the investigation of epigenetics, specifically the role of miRNAs and DNA methylation, toward regulating interspecies activities as these may deliver rapid cues for the restoration and maintenance of homeostasis through tsetse development and following perturbations. We provide support for further investigations of these regulatory mechanisms and experimental guidance (Figure 2) for the simultaneous characterization of these epigenetic processes and assessing their impact toward the host-microbiota association. Besides providing the basis for a deeper understanding of ecological and organismal biology features and their evolution, the study of symbioses and its regulation, particularly in blood-feeding vectors is of significant consequence for epidemiological studies and the design of control strategies aimed at halting transmission of vector-borne diseases.
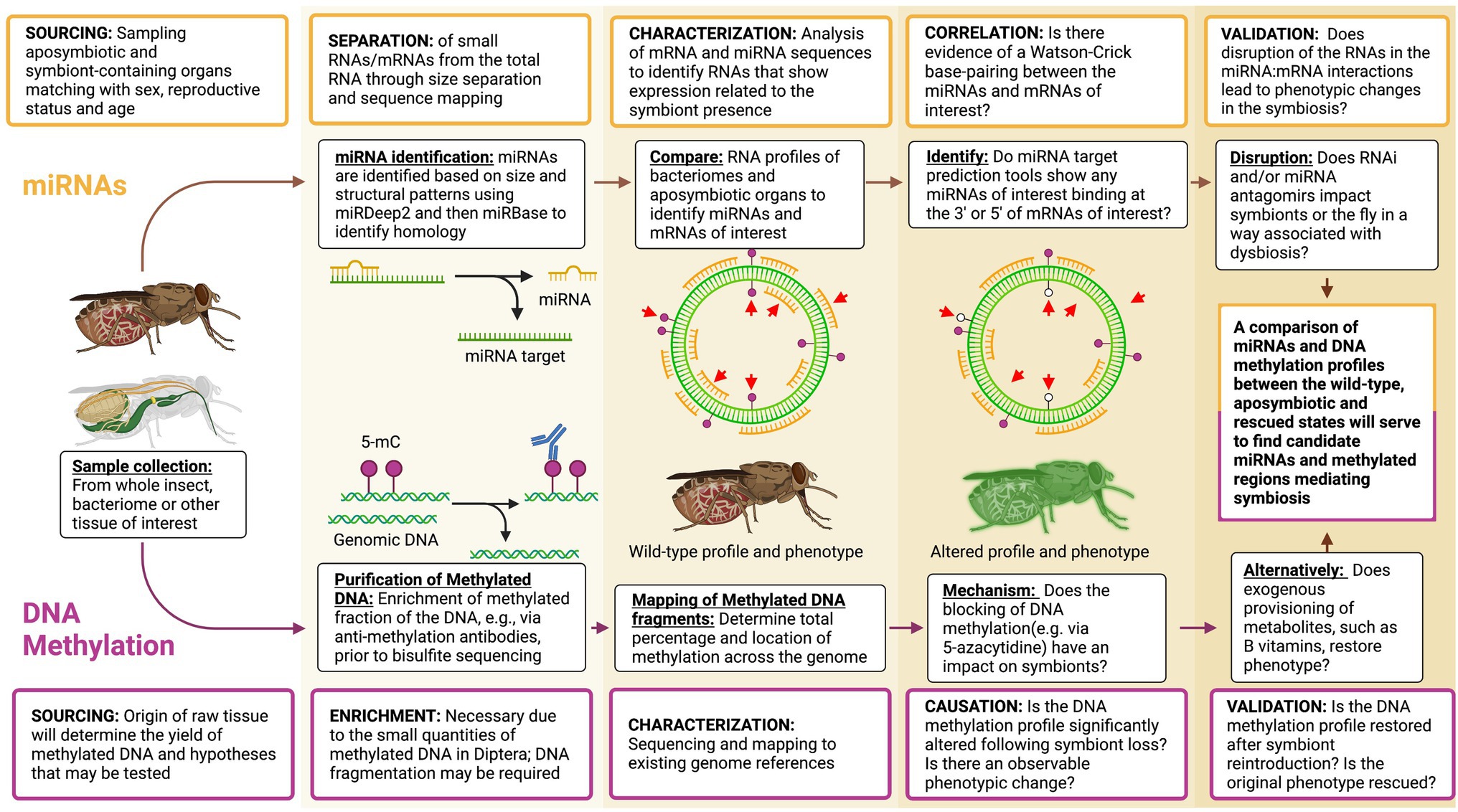
Figure 2. Flow chart for the empirical analyses of epigenetic mechanisms mediating the Wigglesworthia-tsetse fly symbiosis. Of primary importance is the establishment of wild-type miRNA and methylation profiles and phenotypes followed by the characterization of altered states. If altered states are fully or partially rescued by restoration of the epigenetic mechanism through either the reintroduction of the symbionts and/or their provisioned metabolites, then there is evidence in favor of epigenetics mediating symbiosis. Distinct branches highlight methodology for the detection of miRNAs and DNA methylation impact toward symbiosis, while smaller boxes feature comments on the relevant methodology. Red arrows indicate differences in nucleic acid profiles upon comparison of sample groups.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
We acknowledge the support for writing this review by a WVU Eberly College Faculty Development Grant. The tsetse miRNA work done in our laboratory is supported by NIH-NIAID R21AI145271 (RR). Figures were created through BioRender.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abd-Alla, A. M., Kariithi, H. M., Parker, A. G., Robinson, A. S., Kiflom, M., Bergoin, M., et al. (2010). Dynamics of the salivary gland hypertrophy virus in laboratory colonies of Glossina pallidipes (Diptera: Glossinidae). Virus Res. 150, 103–110. doi: 10.1016/j.virusres.2010.03.001
Abdelfattah, A. M., Park, C., and Choi, M. Y. (2014). Update on non-canonical microRNAs. Biomol. Concepts 5, 275–287. doi: 10.1515/bmc-2014-0012
Akman, L., Yamashita, A., Watanabe, H., Oshima, K., Shiba, T., Hattori, M., et al. (2002). Genome sequence of the endocellular obligate symbiont of tsetse, Wigglesworthia glossinidia. Nat. Genet. 32, 402–407. doi: 10.1038/ng986
Aksoy, S. (1995). Wigglesworthia gen nov. and Wigglesworthia glossinidia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbiont of tsetse flies. Int. J. Syst. Bacteriol. 45, 848–851. doi: 10.1099/00207713-45-4-848
Aksoy, S., Pourhosseini, A. A., and Chow, A. (1995). Mycetome endosymbionts of tsetse flies constitute a distinct lineage related to Enterobacteriaceae. Insect Mol. Biol. 4, 15–22. doi: 10.1111/j.1365-2583.1995.tb00003.x
Aksoy, E., Telleria, E. L., Echodu, R., Wu, Y., Okedi, L. M., Weiss, B. L., et al. (2014). Analysis of multiple tsetse fly populations in Uganda reveals limited diversity and species-specific gut microbiota. Appl. Environ. Microbiol. 80, 4301–4312. doi: 10.1128/AEM.00079-14
Aksoy, E., Vigneron, A., Bing, X. L., Zhao, X., O’Neill, M., Wu, Y. N., et al. (2016). Mammalian African trypanosome VSG coat enhances tsetse’s vector competence. Proc. Natl. Acad. Sci. U. S. A. 113, 6961–6966. doi: 10.1073/pnas.1600304113
Alam, U., Medlock, J., Brelsfoard, C., Pais, R., Lohs, C., Balmand, S., et al. (2011). Wolbachia symbiont infections induce strong cytoplasmic incompatibility in the tsetse fly Glossina morsitans. PLoS Pathog. 7:e1002415. doi: 10.1371/journal.ppat.1002415
Asgari, S. (2011). Role of microRNAs in insect host-microorganism interactions. Front. Physiol. 2:48. doi: 10.3389/fphys.2011.00048
Asgari, S. (2013). MicroRNA functions in insects. Insect Biochem. Mol. Biol. 43, 388–397. doi: 10.1016/j.ibmb.2012.10.005
Asgari, S. (2015). Regulatory role of cellular and viral microRNAs in insect-virus interactions. Curr. Opin. Insect Sci. 8, 104–110. doi: 10.1016/j.cois.2014.12.008
Attardo, G. M., Lohs, C., Heddi, A., Alam, U. H., Yildirim, S., and Aksoy, S. (2008). Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J. Insect Physiol. 54, 1236–1242. doi: 10.1016/j.jinsphys.2008.06.008
Balmand, S., Lohs, C., Aksoy, S., and Heddi, A. (2013). Tissue distribution and transmission routes for the tsetse fly endosymbionts. J. Invertebr. Pathol. 112, S116–S122. doi: 10.1016/j.jip.2012.04.002
Baradaran, E., Moharramipour, S., Asgari, S., and Mehrabadi, M. (2019). Induction of DNA methyltransferase genes in Helicoverpa armigera following injection of pathogenic bacteria modulates expression of antimicrobial peptides and affects bacterial proliferation. J. Insect Physiol. 118:103939. doi: 10.1016/j.jinsphys.2019.103939
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Benoit, J. B., Attardo, G. M., Baumann, A. A., Michalkova, V., and Aksoy, S. (2015). Adenotrophic viviparity in tsetse flies: potential for population control and as an insect model for lactation. Annu. Rev. Entomol. 60, 351–371. doi: 10.1146/annurev-ento-010814-020834
Berezikov, E. (2011). Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 12, 846–860. doi: 10.1038/nrg3079
Bewick, A. J., Vogel, K. J., Moore, A. J., and Schmitz, R. J. (2017). Evolution of DNA methylation across insects. Mol. Biol. Evol. 34, 654–665. doi: 10.1093/molbev/msw264
Bind, M. A., Zanobetti, A., Gasparrini, A., Peters, A., Coull, B., Baccarelli, A., et al. (2014). Effects of temperature and relative humidity on DNA methylation. Epidemiology 25, 561–569. doi: 10.1097/EDE.0000000000000120
Bing, X., Attardo, G. M., Vigneron, A., Aksoy, E., Scolari, F., Malacrida, A., et al. (2017). Unravelling the relationship between the tsetse fly and its obligate symbiont. Proc. Biol. Sci. 284:20170360. doi: 10.1098/rspb.2017.0360
Bonasio, R., Li, Q., Lian, J., Mutti, N. S., Jin, L., Zhao, H., et al. (2012). Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr. Biol. 22, 1755–1764. doi: 10.1016/j.cub.2012.07.042
Brecher, G., and Wigglesworth, V. B. (1944). The transmission of Actinomyces rhodnii Erkson in Rhodnius prolixus Stal (Hemiptera) and its influence on the growth of the host. Parasitology 35, 220–224. doi: 10.1017/S0031182000021648
Carthew, R. W., Agbu, P., and Giri, R. (2017). MicroRNA function in Drosophila melanogaster. Semin. Cell Dev. Biol. 65, 29–37. doi: 10.1016/j.semcdb.2016.03.015
Carthew, R. W., and Sontheimer, E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655. doi: 10.1016/j.cell.2009.01.035
Chatterjee, N., Gim, J., and Choi, J. (2018). Epigenetic profiling to environmental stressors in model and non-model organisms: ecotoxicology perspective. Environ. Health Toxicol. 33:e2018015. doi: 10.5620/eht.e2018015
Chen, X. A., Song, L., and Aksoy, S. (1999). Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossina and its bacteriome-asssociated endosymbiont Wigglesworthia glossinidia. J. Mol. Evol. 48, 49–58. doi: 10.1007/PL00006444
Chen, L., Sun, H., Wang, C., Yang, Y., Zhang, M., and Wong, G. (2018). miRNA arm switching identifies novel tumour biomarkers. EBioMedicine 38, 37–46. doi: 10.1016/j.ebiom.2018.11.003
Cheng, Q., Ruel, T., Zhou, W., Moloo, S., Majiwa, P., O’Neill, S., et al. (2000). Tissue distribution and prevalence of Wolbachia infections in tsetse flies, Glossina spp. Med. Vet. Entomol. 14, 44–50. doi: 10.1046/j.1365-2915.2000.00202.x
Chomicki, G., Werner, G. D. A., West, S. A., and Kiers, E. T. (1808). Compartmentalization drives the evolution of symbiotic cooperation. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2020:20190602. doi: 10.1098/rstb.2019.0602
Choudhuri, S. (2011). From Waddington’s epigenetic landscape to small noncoding RNA: some important milestones in the history of epigenetics research. Toxicol. Mech. Methods 21, 252–274. doi: 10.3109/15376516.2011.559695
Cingolani, P., Cao, X., Khetani, R. S., Chen, C. C., Coon, M., Sammak, A., et al. (2013). Intronic non-CG DNA hydroxymethylation and alternative mRNA splicing in honey bees. BMC Genomics 14:666. doi: 10.1186/1471-2164-14-666
Crider, K. S., Yang, T. P., Berry, R. J., and Bailey, L. B. (2012). Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Adv. Nutr. 3, 21–38. doi: 10.3945/an.111.000992
Cross, K. L., Campbell, J. H., Balachandran, M., Campbell, A. G., Cooper, C. J., Griffen, A., et al. (2019). Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat. Biotechnol. 37, 1314–1321. doi: 10.1038/s41587-019-0260-6
Dale, C., and Maudlin, I. (1999). Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int. J. Syst. Bacteriol. 49, 267–275.
De Vooght, L., Van Keer, S., and Van Den Abbeele, J. (2018). Towards improving tsetse fly paratransgenesis: stable colonization of Glossina morsitans morsitans with genetically modified Sodalis. BMC Microbiol. 18:165. doi: 10.1186/s12866-018-1282-9
de Wit, E., Linsen, S. E. V., Cuppen, E., and Berezikov, E. (2009). Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res. 19, 2064–2074. doi: 10.1101/gr.093781.109
Deans, C., and Maggert, K. A. (2015). What do you mean, “epigenetic”? Genetics 199, 887–896. doi: 10.1534/genetics.114.173492
Dennison, N. J., BenMarzouk-Hidalgo, O. J., and Dimopoulos, G. (2015). MicroRNA-regulation of Anopheles gambiae immunity to Plasmodium falciparum infection and midgut microbiota. Dev. Comp. Immunol. 49, 170–178. doi: 10.1016/j.dci.2014.10.016
Dickman, M. J., Kucharski, R., Maleszka, R., and Hurd, P. J. (2013). Extensive histone post-translational modification in honey bees. Insect Biochem. Mol. Biol. 43, 125–137. doi: 10.1016/j.ibmb.2012.11.003
Douglas, A. E. (1998). Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43, 17–37. doi: 10.1146/annurev.ento.43.1.17
Douglas, A. E. (2017). The B vitamin nutrition of insects: the contributions of diet, microbiome and horizontally acquired genes. Curr. Opin. Insect Sci. 23, 65–69. doi: 10.1016/j.cois.2017.07.012
Duarte, F. V., Palmeira, C. M., and Rolo, A. P. (2014). The role of microRNAs in mitochondria: small players acting wide. Genes 5, 865–886. doi: 10.3390/genes5040865
Duron, O., and Gottlieb, Y. (2020). Convergence of nutritional symbioses in obligate blood feeders. Trends Parasitol. 36, 816–825. doi: 10.1016/j.pt.2020.07.007
Duron, O., Morel, O., Noël, V., Buysse, M., Binetruy, F., Lancelot, R., et al. (2018). Tick-bacteria mutualism depends on B vitamin synthesis pathways. Curr. Biol. 28, 1896.e5–1902.e5. doi: 10.1016/j.cub.2018.04.038
Edwards, M. A., Kaufman, M. L., and Storvick, C. A. (1957). Microbiologic assay for the thiamine content of blood of various species of animals and man. Am. J. Clin. Nutr. 5, 51–55. doi: 10.1093/ajcn/5.1.51
Fabian, M. R., Sonenberg, N., and Filipowicz, W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379. doi: 10.1146/annurev-biochem-060308-103103
Feng, H., Park, J. S., Zhai, R. G., and Wilson, A. C. C. (2019). microRNA-92a regulates the expression of aphid bacteriocyte-specific secreted protein 1. BMC Res. Notes 12:638. doi: 10.1186/s13104-019-4665-6
Feng, H., Wang, L., Wuchty, S., and Wilson, A. C. C. (2018). microRNA regulation in an ancient obligate endosymbiosis. Mol. Ecol. 27, 1777–1793. doi: 10.1111/mec.14464
Field, L. M., Lyko, F., Mandrioli, M., and Prantera, G. (2004). DNA methylation in insects. Insect Mol. Biol. 13, 109–115. doi: 10.1111/j.0962-1075.2004.00470.x
Friedman, R. C., Farh, K. K. H., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi: 10.1101/gr.082701.108
Gabaldón, T., and Pittis, A. A. (2015). Origin and evolution of metabolic sub-cellular compartmentalization in eukaryotes. Biochimie 119, 262–268. doi: 10.1016/j.biochi.2015.03.021
Glastad, K. M., Hunt, B. G., and Goodisman, M. A. (2014). Evolutionary insights into DNA methylation in insects. Curr. Opin. Insect Sci. 1, 25–30. doi: 10.1016/j.cois.2014.04.001
Glastad, K. M., Hunt, B. G., and Goodisman, M. A. (2015). DNA methylation and chromatin organization in insects: insights from the ant Camponotus floridanus. Genome Biol. Evol. 7, 931–942. doi: 10.1093/gbe/evv039
Griffiths-Jones, S., Hui, J. H. L., Marco, A., and Ronshaugen, M. (2011). MicroRNA evolution by arm switching. EMBO Rep. 12, 172–177. doi: 10.1038/embor.2010.191
Gu, H., Zhao, C., Zhang, T., Liang, H., Wang, X. M., Pan, Y., et al. (2017). Salmonella produce microRNA-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival. Sci. Rep. 7:2392. doi: 10.1038/s41598-017-02669-1
Han, J., Pedersen, J. S., Kwon, S. C., Belair, C. D., Kim, Y. K., Yeom, K. H., et al. (2009). Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136, 75–84. doi: 10.1016/j.cell.2008.10.053
Hansen, A. K., and Degnan, P. H. (2014). Widespread expression of conserved small RNAs in small symbiont genomes. ISME J. 8, 2490–2502. doi: 10.1038/ismej.2014.121
Head, J. A. (2014). Patterns of DNA methylation in animals: an ecotoxicological perspective. Integr. Comp. Biol. 54, 77–86. doi: 10.1093/icb/icu025
Hu, C., Rio, R. V. M., Medlock, J., Haines, L. R., Nayduch, D., Savage, A. F., et al. (2008). Infections with immunogenic trypanosomes reduce tsetse reproductive fitness: potential impact of different parasite strains on vector population structure. PLoS Negl. Trop. Dis. 2:e192. doi: 10.1371/journal.pntd.0000192
Huang, H., Wu, P., Zhang, S., Shang, Q., Yin, H., Hou, Q., et al. (2019). DNA methylomes and transcriptomes analysis reveal implication of host DNA methylation machinery in BmNPV proliferation in Bombyx mori. BMC Genomics 20:736. doi: 10.1186/s12864-019-6146-7
Ibáñez-Ventoso, C., Vora, M., and Driscoll, M. (2008). Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS One 3:e2818. doi: 10.1371/journal.pone.0002818
Jayachandran, B., Hussain, M., and Asgari, S. (2013). Regulation of Helicoverpa armigera ecdysone receptor by miR-14 and its potential link to baculovirus infection. J. Invertebr. Pathol. 114, 151–157. doi: 10.1016/j.jip.2013.07.004
Jeong, H., Wu, X., Smith, B., and Yi, S. V. (2018). Genomic landscape of Methylation Islands in hymenopteran insects. Genome Biol. Evol. 10, 2766–2776. doi: 10.1093/gbe/evy203
Jin, P., Li, S., Sun, L., Lv, C., and Ma, F. (2017). Transcriptome-wide analysis of microRNAs in Branchiostoma belcheri upon Vibrio parahemolyticus infection. Dev. Comp. Immunol. 74, 243–252. doi: 10.1016/j.dci.2017.05.002
Kawamata, T., and Tomari, Y. (2010). Making RISC. Trends Biochem. Sci. 35, 368–376. doi: 10.1016/j.tibs.2010.03.009
Kim, H., Kim, J., Yu, S., Lee, Y. Y., Park, J., Choi, R. J., et al. (2020). A mechanism for microRNA arm switching regulated by Uridylation. Mol. Cell 78, 1224.e5–1236.e5. doi: 10.1016/j.molcel.2020.04.030
Krafsur, E. S. (2009). Tsetse flies: genetics, evolution, and role as vectors. Infect. Genet. Evol. 9, 124–141. doi: 10.1016/j.meegid.2008.09.010
Ku, C. S., Naidoo, N., Wu, M., and Soong, R. (2011). Studying the epigenome using next generation sequencing. J. Med. Genet. 48, 721–730. doi: 10.1136/jmedgenet-2011-100242
Kunert, N., Marhold, J., Stanke, J., Stach, D., and Lyko, F. (2003). A Dnmt2-like protein mediates DNA methylation in Drosophila. Development 130, 5083–5090. doi: 10.1242/dev.00716
Lagier, J. C., Dubourg, G., Million, M., Cadoret, F., Bilen, M., Fenollar, F., et al. (2018). Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 16, 540–550. doi: 10.1038/s41579-018-0041-0
Lai, E. C., Tomancak, P., Williams, R. W., and Rubin, G. M. (2003). Computational identification of Drosophila microRNA genes. Genome Biol. 4:R42. doi: 10.1186/gb-2003-4-7-r42
Lampe, L., and Levashina, E. A. (2018). MicroRNA tissue atlas of the malaria mosquito. G3 8, 185–193. doi: 10.1534/g3.117.300170
Lee, Y. S., Nakahara, K., Pham, J. W., Kim, K., He, Z., Sontheimer, E. J., et al. (2004). Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69–81. doi: 10.1016/S0092-8674(04)00261-2
LePage, D. P., Jernigan, K. K., and Bordenstein, S. R. (2014). The relative importance of DNA methylation and Dnmt2-mediated epigenetic regulation on Wolbachia densities and cytoplasmic incompatibility. PeerJ 2:e678. doi: 10.7717/peerj.678
Li, P., Jiao, J., Gao, G., and Prabhakar, B. S. (2012). Control of mitochondrial activity by miRNAs. J. Cell. Biochem. 113, 1104–1110. doi: 10.1002/jcb.24004
Li, Y., Liew, Y. J., Cui, G., Cziesielski, M. J., Zahran, N., Michell, C. T., et al. (2018). DNA methylation regulates transcriptional homeostasis of algal endosymbiosis in the coral model Aiptasia. Sci. Adv. 4:eaat2142. doi: 10.1126/sciadv.aat2142
Li, S., Mead, E. A., Liang, S., and Tu, Z. (2009). Direct sequencing and expression analysis of a large number of miRNAs in Aedes aegypti and a multi-species survey of novel mosquito miRNAs. BMC Genomics 10:581. doi: 10.1186/1471-2164-10-581
Ligoxygakis, P., Pelte, N., Ji, C., Leclerc, V., Duvic, B., Belvin, M., et al. (2002). A serpin mutant links toll activation to melanization in the host defence of Drosophila. EMBO J. 21, 6330–6337. doi: 10.1093/emboj/cdf661
Liu, L., Zhang, K. J., Rong, X., Li, Y. Y., and Liu, H. (2019). Identification of Wolbachia-responsive miRNAs in the small brown planthopper, Laodelphax striatellus. Front. Physiol. 10:928. doi: 10.3389/fphys.2019.00928
Lucas, K., and Raikhel, A. S. (2013). Insect microRNAs: biogenesis, expression profiling and biological functions. Insect Biochem. Mol. Biol. 43, 24–38. doi: 10.1016/j.ibmb.2012.10.009
Lucas, K. J., Zhao, B., Liu, S., and Raikhel, A. S. (2015). Regulation of physiological processes by microRNAs in insects. Curr. Opin. Insect Sci. 11, 1–7. doi: 10.1016/j.cois.2015.06.004
Lyko, F. (2018). The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92. doi: 10.1038/nrg.2017.80
Lyko, F., Foret, S., Kucharski, R., Wolf, S., Falckenhayn, C., and Maleszka, R. (2010). The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8:e1000506. doi: 10.1371/journal.pbio.1000506
Ma, W. C., and Denlinger, D. L. (1974). Secretory discharge and microflora of milk gland in tsetse flies. Nature 247, 301–303. doi: 10.1038/247301a0
Macgregor-Das, A. M., and Das, S. (2018). A microRNA’s journey to the center of the mitochondria. Am. J. Physiol. Heart Circ. Physiol. 315, H206–H215. doi: 10.1152/ajpheart.00714.2017
Marco, A., Hui, J. H. L., Ronshaugen, M., and Griffiths-Jones, S. (2010). Functional shifts in insect microRNA evolution. Genome Biol. Evol. 2, 686–696. doi: 10.1093/gbe/evq053
Martin, W. (2010). Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 365, 847–855. doi: 10.1098/rstb.2009.0252
Mayoral, J. G., Etebari, K., Hussain, M., Khromykh, A. A., and Asgari, S. (2014a). Wolbachia infection modifies the profile, shuttling and structure of microRNAs in a mosquito cell line. PLoS One 9:e96107. doi: 10.1371/journal.pone.0096107
Mayoral, J. G., Hussain, M., Joubert, D. A., Iturbe-Ormaetxe, I., O’Neill, S. L., and Asgari, S. (2014b). Wolbachia small noncoding RNAs and their role in cross-kingdom communications. Proc. Natl. Acad. Sci. U. S. A. 111, 18721–18726. doi: 10.1073/pnas.1420131112
McCaw, B. A., Stevenson, T. J., and Lancaster, L. T. (2020). Epigenetic responses to temperature and climate. Integr. Comp. Biol. 60, 1469–1480. doi: 10.1093/icb/icaa049
Medina Munoz, M., Brenner, C., Richmond, D., Spencer, N., and Rio, R. V. M. (2021). The holobiont transcriptome of teneral tsetse fly species of varying vector competence. BMC Genomics 22:400. doi: 10.1186/s12864-021-07729-5
Medina Munoz, M., Pollio, A. R., White, H. L., and Rio, R. V. M. (2017). Into the wild: parallel transcriptomics of the tsetse-Wigglesworthia mutualism within Kenyan populations. Genome Biol. Evol. 9, 2276–2291. doi: 10.1093/gbe/evx175
Mehrabadi, M., Hussain, M., and Asgari, S. (2013). MicroRNAome of Spodoptera frugiperda cells (Sf9) and its alteration following baculovirus infection. J. Gen. Virol. 94, 1385–1397. doi: 10.1099/vir.0.051060-0
Meki, I. K., İnce, İ. A., Kariithi, H. M., Boucias, D. G., Ozcan, O., Parker, A. G., et al. (2018). Expression profile of Glossina pallidipes microRNAs during symptomatic and asymptomatic infection with Glossina pallidipes salivary gland hypertrophy virus (Hytrosavirus). Front. Microbiol. 9:2037. doi: 10.3389/fmicb.2018.02037
Michalkova, V., Benoit, J. B., Weiss, B. L., Attardo, G. M., and Aksoy, S. (2014). Vitamin B6 generated by obligate symbionts is critical for maintaining proline homeostasis and fecundity in tsetse flies. Appl. Environ. Microbiol. 80, 5844–5853. doi: 10.1128/AEM.01150-14
Moore, L. D., Le, T., and Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38. doi: 10.1038/npp.2012.112
Negri, I., Franchini, A., Gonella, E., Daffonchio, D., Mazzoglio, P. J., Mandrioli, M., et al. (2009). Unravelling the Wolbachia evolutionary role: the reprogramming of the host genomic imprinting. Proc. Biol. Sci. 276, 2485–2491. doi: 10.1098/rspb.2009.0324
Nguyen, T. A., Jo, M. H., Choi, Y. G., Park, J., Kwon, S. C., Hohng, S., et al. (2015). Functional anatomy of the human microprocessor. Cell 161, 1374–1387. doi: 10.1016/j.cell.2015.05.010
Nikoh, N., Hosokawa, T., Moriyama, M., Oshima, K., Hattori, M., and Fukatsu, T. (2014). Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. U. S. A. 111, 10257–10262. doi: 10.1073/pnas.1409284111
O’Neill, S. L., Gooding, R. H., and Aksoy, S. (1993). Phylogenetically distant symbiotic microorganisms reside in Glossina midgut and ovary tissues. Med. Vet. Entomol. 7, 377–383. doi: 10.1111/j.1365-2915.1993.tb00709.x
Okamura, K., Phillips, M. D., Tyler, D. M., Duan, H., Chou, Y. T., and Lai, E. C. (2008). The regulatory activity of microRNA* species has substantial influence on microRNA and 3' UTR evolution. Nat. Struct. Mol. Biol. 15, 354–363. doi: 10.1038/nsmb.1409
Panikar, C. S., Rajpathak, S. N., Abhyankar, V., Deshmukh, S., and Deobagkar, D. D. (2015). Presence of DNA methyltransferase activity and CpC methylation in Drosophila melanogaster. Mol. Biol. Rep. 42, 1615–1621. doi: 10.1007/s11033-015-3931-5
Pietrzik, K., Bailey, L., and Shane, B. (2010). Folic acid and L-5-methyltetrahydrofolate: comparison of clinical pharmacokinetics and pharmacodynamics. Clin. Pharmacokinet. 49, 535–548. doi: 10.2165/11532990-000000000-00000
Qiang, J., Tao, Y. F., He, J., Li, H. X., Xu, P., Bao, J. W., et al. (2017). Inhibition of miR-92d-3p enhances inflammation responses in genetically improved farmed tilapia (GIFT, Oreochromis niloticus) with Streptococcus iniae infection by modulating complement C3. Fish Shellfish Immunol. 63, 367–375. doi: 10.1016/j.fsi.2017.02.036
Rider, S. D., Srinivasan, D. G., and Hilgarth, R. S. (2010). Chromatin-remodelling proteins of the pea aphid, Acyrthosiphon pisum (Harris). Insect Mol. Biol. 19, 201–214. doi: 10.1111/j.1365-2583.2009.00972.x
Rio, R. V. M., Jozwick, A. K. S., Savage, A. F., Sabet, A., Vigneron, A., Wu, Y., et al. (2019). Mutualist-provisioned resources impact vector competency. mBio 10:e00018-19. doi: 10.1128/mBio.00018-19
Rio, R. V., Symula, R. E., Wang, J., Lohs, C., Wu, Y. N., Snyder, A. K., et al. (2012). Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: glossinidae) obligate symbiont Wigglesworthia. mBio 3:e00240-11. doi: 10.1128/mBio.00240-11
Riparbelli, M. G., Giordano, R., Ueyama, M., and Callaini, G. (2012). Wolbachia-mediated male killing is associated with defective chromatin remodeling. PLoS One 7:e30045. doi: 10.1371/journal.pone.0030045
Shorter, K. R., Felder, M. R., and Vrana, P. B. (2015). Consequences of dietary methyl donor supplements: is more always better? Prog. Biophys. Mol. Biol. 118, 14–20. doi: 10.1016/j.pbiomolbio.2015.03.007
Skalsky, R. L., Vanlandingham, D. L., Scholle, F., Higgs, S., and Cullen, B. R. (2010). Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genomics 11:119. doi: 10.1186/1471-2164-11-119
Slack, F. J. (2006). Regulatory RNAs and the demise of ‘junk’ DNA. Genome Biol. 7:328. doi: 10.1186/gb-2006-7-9-328
Snyder, A. K., Deberry, J. W., Runyen-Janecky, L., and Rio, R. V. M. (2010). Nutrient provisioning facilitates homeostasis between tsetse fly (Diptera: Glossinidae) symbionts. Proc. Biol. Sci. 277, 2389–2397. doi: 10.1098/rspb.2010.0364
Snyder, A. K., and Rio, R. V. (2015). “Wigglesworthia morsitans” Folate (vitamin B9) biosynthesis contributes to tsetse host fitness. Appl. Environ. Microbiol. 81, 5375–5386. doi: 10.1128/AEM.00553-15
Solano, P., Ravel, S., and de Meeûs, T. (2010). How can tsetse population genetics contribute to African trypanosomiasis control? Trends Parasitol. 26, 255–263. doi: 10.1016/j.pt.2010.02.006
Steverding, D. (2008). The history of African trypanosomiasis. Parasit. Vectors 1:3. doi: 10.1186/1756-3305-1-3
Symula, R. E., Marpuri, I., Bjornson, R. D., Okedi, L., Beadell, J., Alam, U., et al. (2011). Influence of host phylogeographic patterns and incomplete lineage sorting on within-species genetic variability in Wigglesworthia species, obligate symbionts of tsetse flies. Appl. Environ. Microbiol. 77, 8400–8408. doi: 10.1128/AEM.05688-11
Takayama, S., Dhahbi, J., Roberts, A., Mao, G., Heo, S. J., Pachter, L., et al. (2014). Genome methylation in D. melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res. 24, 821–830. doi: 10.1101/gr.162412.113
Tammen, S. A., Friso, S., and Choi, S. W. (2013). Epigenetics: the link between nature and nurture. Mol. Asp. Med. 34, 753–764. doi: 10.1016/j.mam.2012.07.018
Terrapon, N., Li, C., Robertson, H. M., Ji, L., Meng, X., Booth, W., et al. (2014). Molecular traces of alternative social organization in a termite genome. Nat. Commun. 5:3636. doi: 10.1038/ncomms4636
Tomari, Y., and Zamore, P. D. (2005). Perspective: machines for RNAi. Genes Dev. 19, 517–529. doi: 10.1101/gad.1284105
Tsagmo Ngoune, J. M., Reveillaud, J., Sempere, G., Njiokou, F., Melachio, T. T., Abate, L., et al. (2019). The composition and abundance of bacterial communities residing in the gut of Glossina palpalis palpalis captured in two sites of southern Cameroon. Parasit. Vectors 12:151. doi: 10.1186/s13071-019-3402-2
Vannier, N., Mony, C., Bittebière, A.-K., and Vandenkoornhuyse, P. (2015). Epigenetic mechanisms and microbiota as a toolbox for plant phenotypic adjustment to environment. Front. Plant Sci. 6:1159. doi: 10.3389/fpls.2015.01159
Vigneron, A., Aksoy, E., Weiss, B. L., Bing, X., Zhao, X., Awuoche, E. O., et al. (2018). A fine-tuned vector-parasite dialogue in tsetse’s cardia determines peritrophic matrix integrity and trypanosome transmission success. PLoS Pathog. 14:e1006972. doi: 10.1371/journal.ppat.1006972
Villagra, C., and Frías-Lasserre, D. (2020). Epigenetic molecular mechanisms in insects. Neotrop. Entomol. 49, 615–642. doi: 10.1007/s13744-020-00777-8
Waddington, C. H. (2012). The epigenotype. 1942. Int. J. Epidemiol. 41, 10–13. doi: 10.1093/ije/dyr184
Wang, J., Wu, Y., Yang, G., and Aksoy, S. (2009). Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypansome transmission. Proc. Natl. Acad. Sci. U. S. A. 106, 12134–12138. doi: 10.1073/pnas.0901226106
Warner, R. E., van den Berg, A. M., Berland, K. M., Hinnefeld, J. D., Zhang, Z., Zhu, Y. T., et al. (1989). Total nuclear reaction probabilities and average cross sections for 27 to 92 MeV alpha particles in silicon. Phys. Rev. C Nucl. Phys. 40, 2473–2478. doi: 10.1103/PhysRevC.40.2473
Weiss, B. L., Wang, J., and Aksoy, S. (2011). Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol. 9:e1000619. doi: 10.1371/journal.pbio.1000619
Weiss, B. L., Wang, J., Maltz, M. A., Wu, Y., and Aksoy, S. (2013). Trypanosome infection establishment in the tsetse fly gut is influenced by microbiome-regulated host immune barriers. PLoS Pathog. 9:e1003318. doi: 10.1371/journal.ppat.1003318
Wheeler, B. M., Heimberg, A. M., Moy, V. N., Sperling, E. A., Holstein, T. W., Heber, S., et al. (2009). The deep evolution of metazoan microRNAs. Evol. Dev. 11, 50–68. doi: 10.1111/j.1525-142X.2008.00302.x
Yang, Z., Wang, M., Zeng, X., Wan, A. T. Y., and Tsui, S. K. W. (2020). In silico analysis of proteins and microRNAs related to human African trypanosomiasis in tsetse fly. Comput. Biol. Chem. 88:107347. doi: 10.1016/j.compbiolchem.2020.107347
Ye, Y. H., Woolfit, M., Huttley, G. A., Rancès, E., Caragata, E. P., Popovici, J., et al. (2013). Infection with a virulent strain of Wolbachia disrupts genome wide-patterns of cytosine methylation in the mosquito Aedes aegypti. PLoS One 8:e66482. doi: 10.1371/journal.pone.0066482
Yu, D. H., Gadkari, M., Zhou, Q., Yu, S., Gao, N., Guan, Y., et al. (2015). Postnatal epigenetic regulation of intestinal stem cells requires DNA methylation and is guided by the microbiome. Genome Biol. 16:211. doi: 10.1186/s13059-015-0763-5
Zhang, G., Hussain, M., and Asgari, S. (2014a). Regulation of arginine methyltransferase 3 by a Wolbachia-induced microRNA in Aedes aegypti and its effect on Wolbachia and dengue virus replication. Insect Biochem. Mol. Biol. 53, 81–88. doi: 10.1016/j.ibmb.2014.08.003
Zhang, P., Li, C., Shao, Y., Chen, X., Li, Y., Su, X., et al. (2014b). Identification and characterization of miR-92a and its targets modulating Vibrio splendidus challenged Apostichopus japonicus. Fish Shellfish Immunol. 38, 383–388. doi: 10.1016/j.fsi.2014.04.007
1. Wigglesworthia’s tight integration in tsetse physiology requires mechanisms for the regulatory control of population size and function. Intriguingly, Wigglesworthia lies free in the cytoplasm of bacteriocytes which may facilitate tsetse miRNAs to interact with these symbionts. Are tsetse miRNAs localized to the bacteriome acting on Wigglesworthia transcripts to control expression? Due to differences in nutrient demands during pregnancy and through aging, how may tsetse miRNA expression be impacted particularly toward the metabolic integration of Wigglesworthia symbionts?
2. Salmonella produce small RNAs which alter the host phenotype and lead to increased virulence. The DNA sequence which produces these small RNAs may also be found in many other Gamma-proteobacteria, making it plausible that these microRNA-like small RNAs have homologous roles in other bacteria. Do Wigglesworthia produce miRNA-like small RNAs to create favorable environments in the fly? Small bacterial RNAs represent an additional avenue for exploration toward advancing our understanding of interkingdom communication.
3. What types of epigenetic mechanisms may regulate the influx/efflux of substrates at symbiont and host transporters which lie at the interface of the association?
4. Pathogenic bacterial infections are associated with changes in the DNA methylation of several insects including members of Diptera (Ye et al., 2013; LePage et al., 2014), Lepidoptera (Baradaran et al., 2019), and Hemiptera (Negri et al., 2009). May beneficial symbionts also impact host DNA methylation? Establishing a cause-effect relationship and biochemical steps involved in these outcomes will be essential toward our understanding of the regulatory role exerted by bacteria on insect physiology.
5. What other epigenetic mechanisms may be affected by the metabolites of microbiota? For example, a role in epigenetics through biotin (B7) provisioning by symbionts may also occur given that histone biotinylation plays a role in transcriptional repression of genes and DNA repair.
Keywords: tsetse, Wigglesworthia, microbiota, insect, epigenetics
Citation: Lee MH, Medina Munoz M and Rio RVM (2022) The Tsetse Metabolic Gambit: Living on Blood by Relying on Symbionts Demands Synchronization. Front. Microbiol. 13:905826. doi: 10.3389/fmicb.2022.905826
Received: 27 March 2022; Accepted: 16 May 2022;
Published: 09 June 2022.
Edited by:
Takema Fukatsu, National Institute of Advanced Industrial Science and Technology (AIST), JapanReviewed by:
Aurélien Vigneron, Max Planck Institute for Chemical Ecology, GermanyCopyright © 2022 Lee, Medina Munoz and Rio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rita V. M. Rio, cml0YS5yaW9AbWFpbC53dnUuZWR1
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.