- 1Institute of Medical Microbiology and Hygiene, Ulm University Medical Center, Ulm, Germany
- 2Institute of Molecular Virology, Ulm University Medical Center, Ulm, Germany
Streptococcus anginosus produces the novel antimicrobial peptide Angicin, which inhibits Gram positive microorganisms and is classified as a group IId bacteriocin. Production of Angicin is regulated by the quorum sensing system Sil (Streptococcus invasion locus), which is located adjacent to the bacteriocin gene cluster. Within this genetic region a typical CAAX protease is encoded, which was designated SilX. Nelfinavir, a HIV protease inhibitor, led to a concentration dependent reduction in antimicrobial activity, presumably through the inhibition of SilX. Concentrations exceeding 25 μM Nelfinavir caused a complete abolishment of bacteriocin activity against Listeria monocytogenes. These results are supported by the observation, that a SilX deletion mutant of S. anginosus strain BSU 1211 no longer inhibits the growth of L. monocytogenes. Antimicrobial activity could be restored by addition of synthetically synthesized mature SilCR, implying that SilX may be involved in the export and processing of the signal peptide SilCR. Some CAAX proteases have been reported to provide immunity against bacteriocins. However, in a radial diffusion assay the deletion mutant S. anginosus BSU 1211ΔSilX showed no sensitivity toward Angicin arguing against a role of SilX in the immunity of S. anginosus. The putative processing of the signal peptide SilCR indicates a novel function of the CAAX protease SilX, in the context of S. anginosus bacteriocin production.
Introduction
Bacterial communication through quorum sensing (QS) represents a major regulatory mechanism for group behavior and bacteriocin production. Bacteriocins are small antimicrobial peptides of bacterial origin inhibiting the growth of other often closely related bacterial species. Producing bacteriocins provides a colonization advantage and influences the composition of microbial communities (Majeed et al., 2011; Thappeta et al., 2020; Vogel and Spellerberg, 2021). It is therefore critical to understand how bacteriocin production is regulated and which genes are required for bacteriocin production. CAAX proteases are often present in bacteriocin clusters. While they have repeatedly been implicated in immunity, their precise function in bacteriocin production is not clearly understood, for many species.
CAAX proteases are multipass, transmembrane proteins that contain two typical sequence motifs (EExxxR, FxxxH) that are necessary for the catalytic activity (Pei et al., 2011). The function of type II CAAX proteases in bacteria is ambiguous. CAAX proteases have been demonstrated to confer immunity to bacteriocins, process bacteriocins or function as a receptor for bacteriocins (Diep et al., 1996; Lux et al., 2007; Kjos et al., 2010; Biswas and Biswas, 2014; Maxson et al., 2015). For example, in the hemolysin gene cluster of Streptococcus pyogenes, SagE supposedly cleaves off the leader peptide of SagA, leading to the formation of the hemolysin StreptolysinS, the mature form of SagA (Maxson et al., 2015). SagE has previously been annotated as a bacteriocin self-immunity protein against Streptolysin, however, no clear evidence supporting this hypothesis has been found (Maxson et al., 2015). As another example, LsrS seems to have a receptor-like function for the two-peptide bacteriocin Smb produced by Streptococcus mutans (Biswas and Biswas, 2014). Overexpression of LsrS rendered cells more sensitive toward Smb, while inactivation led to Smb resistance. However, not the active CAAX domain is responsible for this phenotype since inactivation of this site had no effect on Smb sensitivity.
Besides their roles in bacteriocin production, CAAX proteases can interfere with QS systems. An interaction of CAAX proteases with histidine kinases has already been demonstrated. In these cases, the catalytically necessary motifs are not essential, e.g., SpdC, a CAAX protease of Staphylococcus aureus, interacts with various histidine kinases of this pathogen but lacks all conserved catalytic domains (Poupel et al., 2018). Furthermore, for Streptococcus agalactiae it was demonstrated that the CAAX protease Abx1 forms a signaling complex with histidine kinase CovS (Firon et al., 2013). There is a direct interaction between the transmembrane domain of CovS and Abx1, which is independent from its proteolytic activity. CovS forms together with CovR a two-component system, which regulates virulence and hemolysis in S. agalactiae (Lamy et al., 2004; Rosa-Fraile et al., 2014). The conserved catalytic motifs are present in Abx1 but are not necessary for its effect on CovS. In contrast to this, the conserved catalytic motifs are important for the activity of MroQ, a CAAX protease of Staphylococcus aureus. MroQ facilitates the maturation of the autoinducing peptide (AIP) of the accessory gene regulatory system (Agr) in S. aureus and thereby controls the expression of virulence factors (Le and Otto, 2015; Cosgriff et al., 2019).
A putative CAAX protease has also been identified in Streptococcus anginosus in the context of bacteriocin production (Vogel et al., 2021). S. anginosus is primarily a commensal of mucosal membranes. For a long time, the pathogenic potential of this species was underestimated, due to diagnostic difficulties in distinguishing the species from other viridans streptococci (Facklam, 2002; Asam and Spellerberg, 2014). As an opportunistic pathogen this species can cause severe invasive and pyogenic infection. Frequent isolation from abscesses, blood cultures and cystic fibrosis patients has been reported in more recent years (Siegman-Igra et al., 2012; Kobo et al., 2017; Jiang et al., 2020). We previously identified a novel bacteriocin of S. anginosus termed Angicin (Vogel et al., 2021). Angicin is a class IId bacteriocin and active against other streptococci as well as against Listeria spp. and enterococci. The QS system Sil (Streptococcal invasion locus) regulates bacteriocin production in S. anginosus and Streptococcus intermedius (Mendonca et al., 2016; Vogel et al., 2021). SilCR, the autoinducing peptide, is processed and exported by SilD and SilE (Belotserkovsky et al., 2009). Extracellular SilCR is then sensed by the histidine kinase SilB, which in turn phosphorylates the response regulator SilA. SilA induces gene expression of the sil genes as well as of the adjacent bacteriocin genes. A putative CAAX protease gene, designated silX, is located in the vicinity of the bacteriocin genes (blp3) and the sil genes of S. anginosus (Vogel et al., 2021). Whether this protease plays a part in self immunity, processing or as a bacteriocin receptor is unknown. To better understand the biosynthesis and regulation of Angicin production the role of SilX was investigated.
Materials and Methods
Bacterial Strains and Growth Conditions
Lysogeney Broth (LB-Miller) was used to cultivate Escherichia coli DH5α. Liquid E. coli cultures were incubated aerobically at 37°C on a shaker (180 rpm), whereas plates were kept at 37°C with 5% CO2. Two plasmid backbones, pAT28 and pAT18 were used in this study. 100 μg ml–1 spectinomycin (pAT28) (Sigma-Aldrich Chemie GmbH) or 400 μg mg–1 erythromycin (pAT18) (Serva) were supplemented for cultivation of E. coli strains carrying these plasmids. For cultivation on solid media, all bacterial stains were cultivated on sheep blood agar plates (Oxoid). For streptococci and listeria strains a liquid culture in THY medium [Todd-Hewitt Broth (Oxoid) supplemented with 0.5% yeast extract (BD)] was prepared. Liquid cultures as well as plates were incubated at 37°C and 5% CO2. Streptococcal mutants were incubated with 120 μg ml–1 spectinomycin or 10 μg ml–1 erythromycin if necessary. All strains and plasmids used in this study are summarized in Table 1.
General DNA Techniques
To isolate DNA (GeneEluteTM Bacterial Genomic DNA, Sigma-Aldrich; QIAamp® DNA Mini Kit, Qiagen) or plasmids (QIAprep® Spin Miniprep Kit, Qiagen) commercial kits were applied, following the manufacturer’s protocols. QIAamp DNA Mini Kit was used to screen clones for successful transformation, while GeneEluteTM Bacterial Genomic DNA was the standard DNA isolation kit. DNA and plasmid concentrations were measured using Quant-iT dsDNA Broad-Range (BR) Assay Kit (Invitrogen). Polymerase chain reactions (PCR) were conducted following standard protocols for Taq polymerase (Roche) or the Expand Long Template PCR system: DNA pol. Mix (Roche) with buffer 3. For Taq polymerase an initial denaturation at 95°C for 5 min was followed by 32 cycles of 1 min at 95°C, 30 s at 50°C and 1–3 min at 72°C. A final elongation of 7 min at 72°C was the last step. Elongation time was adjusted according to product length (1 min per 1ooo basepairs) Default settings for the Long Expand PCR were an initial denaturation for 5 min at 92°C followed by 11 cycles of 92°C for 10 s, 50°C for 20 s and 68°C for 3 min. This was followed by 25 cycles of 92°C for 15 s, 50°C for 20 s and 68°C for 3 min + 25 s per cycle. It was rounded up by a final elongation of 68°C for 5 min. Clean up of PCR products was performed using NucleoSpin Gel and PCR Clean-up (Machery-Nagel). All Primers used for this study are summarized in Table 2. Nucleotide sequencing was performed by Eurofins Genomics (Germany) and Microsynths laboratories (Switzerland).
Construction of Mutants
Construction of CAAX Deletion Mutants
For creating markerless deletion mutants the natural competence system of S. anginosus was exploited as described by Bauer et al. (2018). In a first step a splicing by overlap extension PCR (SOE-PCR) was performed. Flanking regions of the target gene, CAAX, were amplified with primers 1/2 for fragment 1 and 3/4 for fragment 2. Primers 2/3 introduced an overlap to either a lox66 or a lox71 sequence. Primers 5/6 were used to amplify a spectinomycin resistance gene from pGA14-spec and to simultaneously introduce a lox66 and a lox71 site adjacent to the spectinomycin gene. All three fragments were fused together by SOE-PCR. The fusion construct was loaded on an agarose gel (1%) to control for the right size. Subsequently, this linear construct was transformed into S. anginosus. S. anginosus BSU 1211 was incubated with 100 ng of competence-stimulating peptide 1 (CSP-1) to induce natural competence. After 40 min of incubation at 37°C the linear construct was added, and it was incubated for another 80 min at 37°C and subsequently plated on THY-spectinomycin agar. Colonies were picked after 24–48 h and screened for successful insertion with a colony-PCR. Therefore, streptococci were resuspended in 50 μl Aqua bidest and then incubated at 95°C for 30 min. Subsequently, the supernatant was used in a standard Taq-PCR (see “General DNA Techniques”). Primers 1/7 were used to amplify the DNA present in the supernatant. Positive clones were transformed with a Cre-recombinase harboring plasmid (pAT18-cre-rectufA) using the above-described method. In this case, transformed cells were plated on sheep blood agar plates supplemented with 10 μg ml–1 erythromycin The Cre-recombinase recombines the two lox sites to a singular lox72 site and thereby eliminates the spectinomycin resistance gene. Spectinomycin-sensitive but erythromycin resistant clones were incubated without antibiotics to induce plasmid loss resulting in a markerless deletion strain. The created deletion was amplified by PCR using primers 8/9 and checked by subsequent DNA sequencing.
The same method was used to create a markerless SilCR knockout strain of S. anginosus BSU 1211. Primers 10/11 (F1) and 12/13 (F2) were used to create the flanking fragments and primers 14/15 were used to verify a correct deletion.
EGFP Expressing Mutants
To analyze SilX expression a reporter construct with EGFP was created. The putative SilX promotor was amplified using primers 16/17, which introduced restriction cutting sites for BamHI and EcoRI. Both enzymes (New England Biolabs) were used to digest the PCR product as well as plasmid pBSU101. After purification both products were ligated and transformed into E. coli DH5α by heat shock transformation. Plasmid DNA was extracted and used for a transformation into S. anginosus strain BSU 1211 via inducing natural competence as described by Bauer et al. (2018). Correct plasmid construction was controlled by PCR using primers 18/19/20 and subsequent DNA sequencing.
Radial Diffusion Assay
Bacteriocin activity was quantified using a modified radial diffusion assay (RDA) (Vogel et al., 2021). Briefly, overnight cultures of putative target strains were washed and O.D.600 nm was determined. Per plate 2 × 107 bacterial cells were seeded. After washing with 10 mM Phosphatebuffer, overnight cultures of bacteriocin producing S. anginosus strains were adjusted to an O.D.600 nm of 0.5. With wide bore pipette tips (Ayxgen- A Corning Brand, Corning Inc., Salt Lake City, UT, United States) wells were placed into the agar plates of target strains, which were subsequently filled with the overnight cultures of bacteriocin producing strains. Following overnight incubation at 37°C and 5% CO2, inhibition zones were measured in cm.
In some assays 100 ng of the signal peptide SilCRSAG–C (GWLEDLFSPYFKKYKLGKLGQPDLG) were added simultaneously with the bacteria to induce bacteriocin production. The peptide was synthesized by the Core facility of functional peptidomics (UPEP, Ulm University, Ulm, Germany) based on the deduced sequence of SilCRSAG–C of S. anginosus strain BSU 1211.
In some experiments, the HIV protease inhibitors Saquinavir (Sigma-Aldrich) or Nelfinavir (Sigma-Aldrich) were administered in combination with bacteriocin producing bacteria. Dimethyl sulfoxid (DMSO, Sigma), in which both inhibitors were solved, was used as a control. Experiments were conducted in technical duplicates.
FACS Analysis
To assess the activity of the silX gene the silX promoter of S. anginosus strain BSU 1211 was cloned in front of egfp to construct plasmid pBSU 1202. The newly constructed plasmid was transformed into strain S. anginosus SK52. After overnight incubation it was freshly inoculated in 3 ml of THY supplemented with 120 μg ml–1 spectinomycin either in the presence or absence of 10 μg ml–1 SilCR. After 2 h and cells were harvested by centrifugation (3000 × g, 10 min), washed, and resuspended in Dulbecco’s phosphate buffered saline (DPBS, pH 7.4). The mean fluorescence of 10 000 events was determined by flow cytometer. The FACSCalibur (Becton Dickinson Immunocytometry Systems) was utilized with the following instrument settings: FSC: E00, SSC: 400, FL-1: 700. As a control egfp without promoter and egfp under the control of the cfb promoter was used. All experiments were performed with technical triplicates.
Bioinformatic and Statistical Analysis
The GenBank database1 was used as a source for nucleotide sequences. Homology searches were done with the Basic Local Alignment Search Tool (Altschul et al., 1990)2. Other genetic analysis was carried out using SnapGene 5.03. Phyre2 was used to analyze the SilX protein sequence (Kelley et al., 2015). An alignment of putative silA binding sites was constructed using CLC Main Workbench v7.7.34 with default settings (gap open cost value 10.0, gap extension cost value 1.0). Preparation of graphs and statistical analysis was performed by GraphPad Prism V6 (GraphPad Software Inc., La Jolla, CA, United States).
Results
In Silico Analysis of SilX in Streptococcus anginosus
SilX is located between the response regulator silA of the QS system Sil and the bacteriocin-like-peptide3 (blp3) region (Figure 1A), which encodes for the functional bacteriocin Angicin of S. anginosus (Vogel et al., 2021). The silX gene has the same orientation as silA and the deduced amino acid sequence displays homologies to immunity proteins and streptococcal metalloproteases, while its function has not been investigated. By Blast analysis silX homologs can only be found in the Streptococcus anginosus Group, while Streptococcus constellatus shows a higher percentage of sequence identity (98.74%) than Streptococcus intermedius (97.35%).
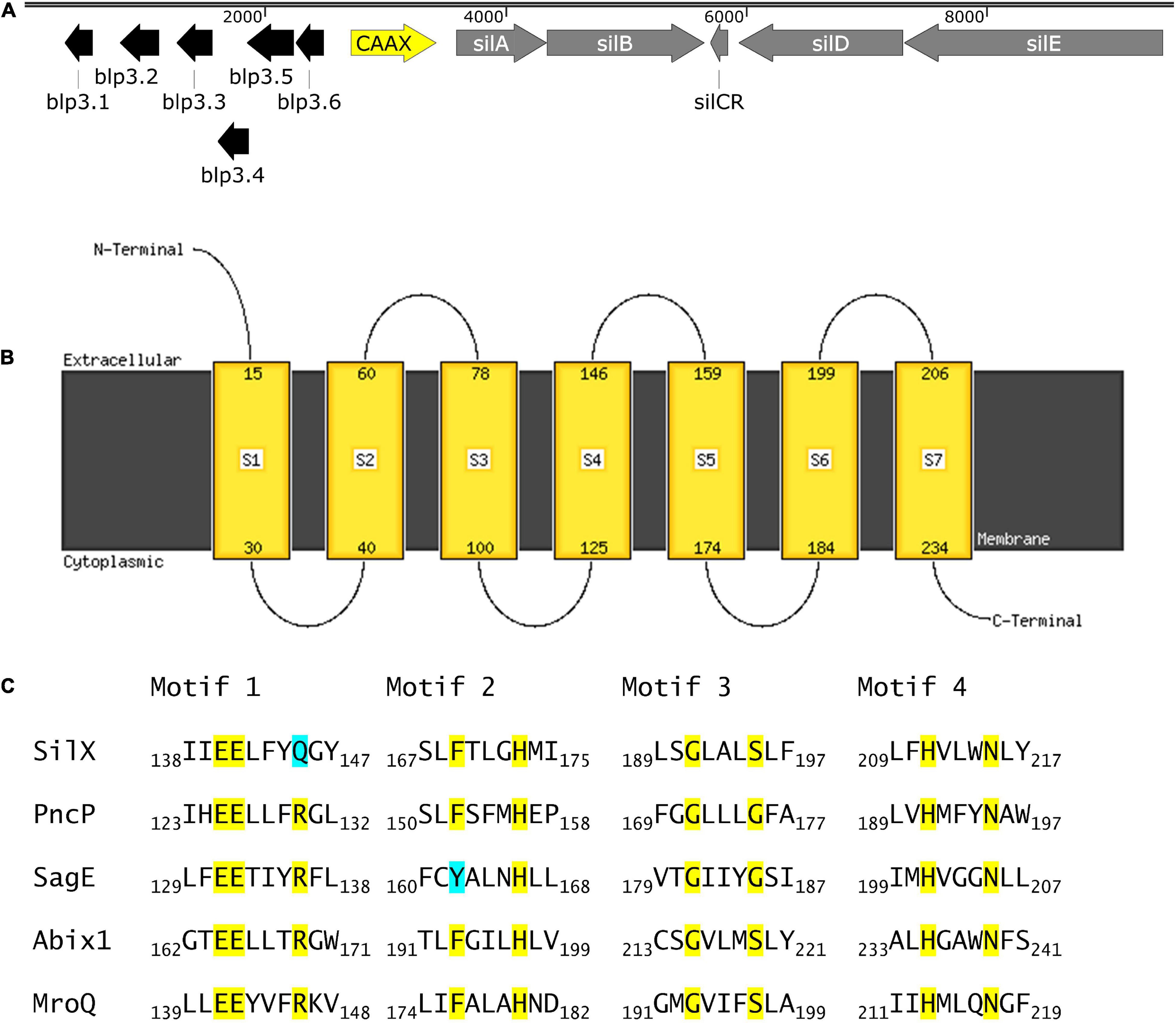
Figure 1. SilX is a transmembrane protein harboring conserved CAAX domains. (A) Genetic organization of the bacteriocin and quorum sensing locus in S. anginosus BSU 1211 (Accession: MZ766502). Modified from Vogel et al. (2021). (B) Prediction of protein transmembrane structure of SilX constructed using Phyre2. (C) SilX is a member of the CAAX proteins. Conserved domains of CAAX proteins are depicted with the important residues marked in yellow and differing residues marked in turquoise. SilX is compared to PncP of Streptococcus mitis (Acsession number: VTS34806.1), SagE of S. pyogenes (Accession number: QJC39319.1), Abix1 of Streptococcus agalactiae (Accession number: WP_001042316.1) and MroQ of S. aureus (Accession number: ABD20720.1).
To further characterize SilX its protein structure was predicted using Phyre2. This analysis revealed CAAX protease motifs with close homologies to other streptococcal and staphylococcal CAAX proteases. Seven transmembrane domains could be detected with the C-terminus of SilX being on the cytoplasmic site and the N-termiuns reaching into the extracellular lumen (Figure 1B). Most of the known motifs of CAAX proteases are conserved in SilX, however, in the diglutamate motif (EExxxR) the arginine residue is replaced by a glutamine (EExxxQ) (Figure 1C). The diglutamate motif (E140-E141) has been reported as necessary for the catalytic function of CAAX proteases (Pei et al., 2011). In summary, the in silico analysis supports a putative function of SilX as a CAAX protease.
The Role of SilX in Streptococcus anginosus Bacteriocin Production
It has previously been shown that HIV protease inhibitors are able to inhibit CAAX proteases, like SagE of Streptococcus pyogenes (Maxson et al., 2015). To explore, if a similar effect can be achieved for SilX we investigated the effect of HIV protease inhibitors Nelfinavir and Saquinavir on the bacteriocin production of S. anginosus. Bacteriocin activity of S. anginosus BSU 1211 against two highly susceptible target strains (Streptococcus constellatus, Listeria monocytogenes) was tested in a radial diffusion assay (RDA) either in the presence or absence of HIV protease inhibitors (Figure 2).
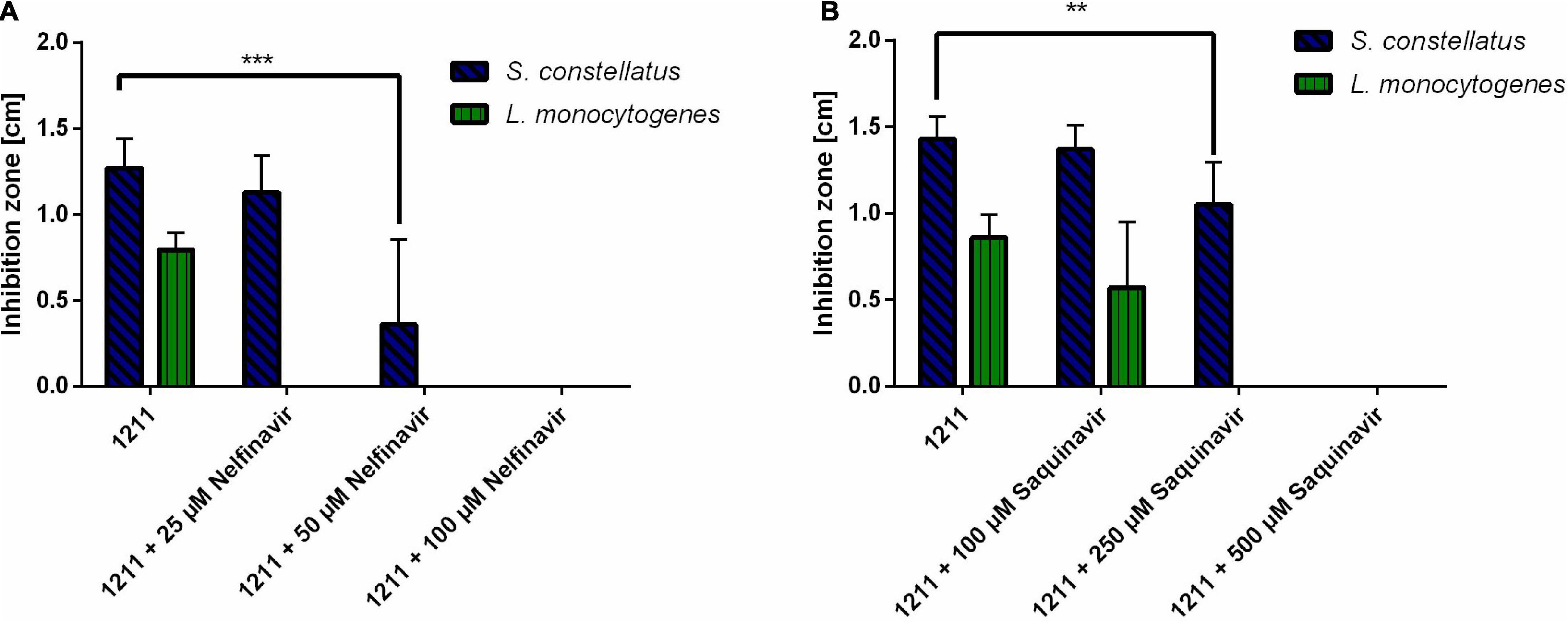
Figure 2. HIV protease inhibitors inhibit Angicin activity of S. anginosus. The antimicrobial activity of S. anginosus BSU 1211 against S. constellatus and L. monocytogenes was tested in an RDA. S. anginosus BSU 1211 was supplemented with (A) Nelfinavir concentrations ranging from 25 to 250 μM or (B) Saquinavir concentrations ranging from 100 to 500 μM. Depicted is inhibition zone diameter ± standard deviation of at least five independent experiments. Significance was calculated using Mann-Whitney U test with * presenting a p-value < 0.5, ** indicating p < 0.01 and *** illustrating p < 0.001.
The supplementation with Nelfinavir or Saquinavir caused a dose dependent reduction of bacteriocin activity. At concentrations of 250 μM Saquinavir S. anginosus BSU 1211 was no longer able to inhibit the growth of L. monocytogenes, while 500 μM Saquinavir were needed to completely abolish the antimicrobial effect against S. constellatus (Figure 2A). In the case of Nelfinavir, already 25 μM were sufficient to abolish antimicrobial activity against L. monocytogenes (Figure 2B). For a complete loss of inhibition against S. constellatus 100 μM of Nelfinavir are needed.
To investigate, if the gene silX is required for effective bacteriocin production of S. anginosus, silX of strain BSU1211 was deleted and the mutant strain was tested for antimicrobial activity in a RDA (Figure 3). S. anginosus BSU 1211ΔSilX demonstrated a complete loss of growth inhibition for the species L. monocytogenes and S. constellatus. This observation indicates an essential role of SilX in bacteriocin production. However, the antimicrobial activity of the deletion mutant could be reestablished to previous levels by supplementation with the signaling peptide SilCR at a concentration of 10 μg × ml–1.
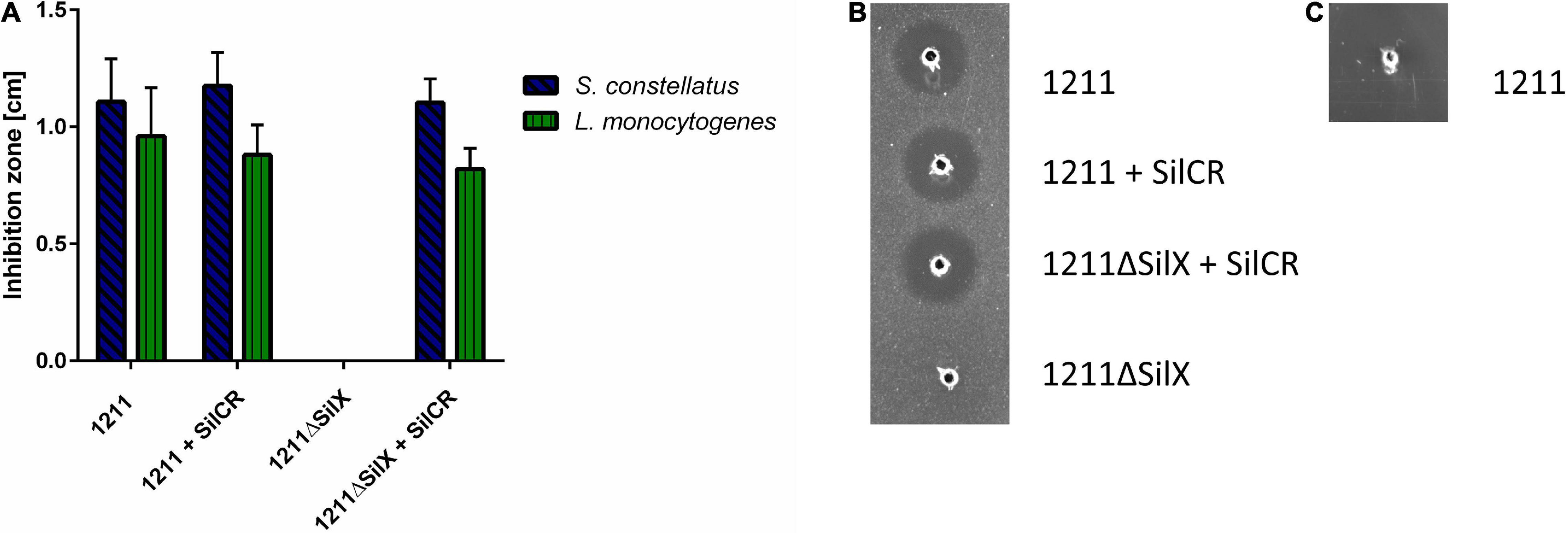
Figure 3. S. anginosus BSU 1211ΔSilX has no antimicrobial activity against S. constellatus and L. monocytogenes. A SilX deletion mutant of S. anginosus was tested in a RDA against S. constellatus and L. monocytogenes either alone or supplemented with 10 μg x ml– 1 SilCR. (A) Depicted is the mean of inhibition zone diameter ± standard deviation of at least 5 independent experiments. No significant differences were found with a Mann-Whitney-U test. (B) Depicted is a RDA of S. anginosus BSU 1211 and S. anginosusΔSilX with S. constellatus as target strain under supplementation with 10 μg x ml– 1 SilCR as indicated. (C) Depicted is a RDA of S. anginosus BSU1211ΔSilX as a target strain against S. anginosus BSU1211.
To investigated whether SilX may also serve as a immunity protein S. anginosus BSU 1211ΔSilX was checked for bacteriocin susceptibility toward the wildtype strain BSU 1211. The deletion mutant showed no sensitivity toward the wildtype strain arguing against an essential role in bacteriocin immunity. In conclusion our data support an essential role of the presumed protease function of SilX for bacteriocin activity, which can however be substituted by supplementation with the signal peptide SilCR.
SilX Promoter
The gene silX in S. anginosus is located adjacent to the streptococcal invasion locus (sil) (Figure 1A) and transcribed in the same orientation as the genes silA and silB. SilA encodes a response regulator belonging to the AlgR/AgrA/LytR family of transcription regulators that bind DNA via a LytTR-type domain (Nikolskaya, 2002; Belotserkovsky et al., 2009). Since these regulators form dimers and typically bind to direct repeats, we searched for this motif upstream of silX. A putative promoter could be located in this region, consisting of 11 bp long imperfect repeats interspaced with 10 bp. Similar putative promoters were identified in front of silCR and silE as well as upstream of the putative bacteriocin genes blp3.1 and blp3.6 (Figure 4A).
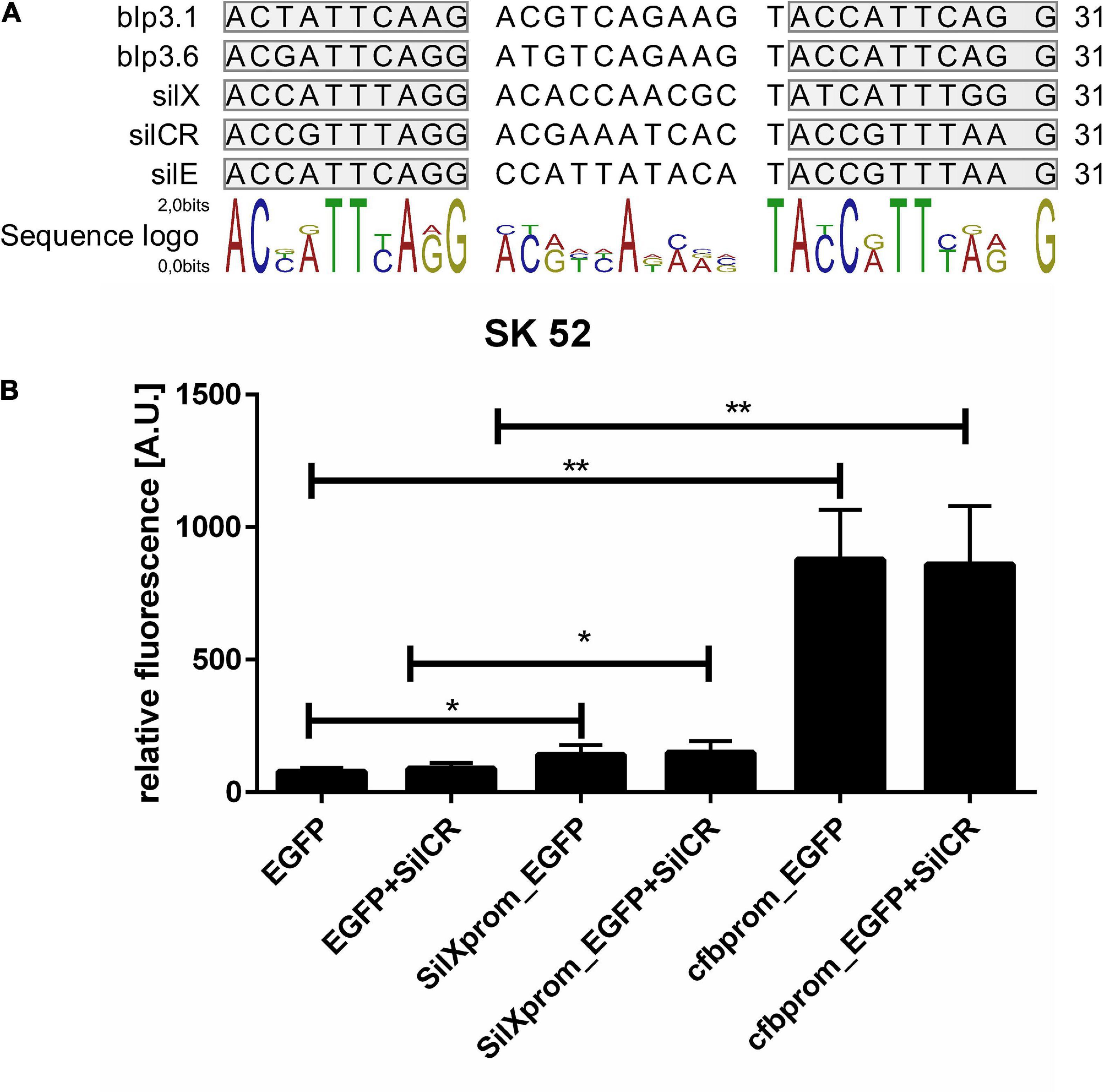
Figure 4. Identification and activity of the SilX promotor. (A) Predicted SilA binding sites. Overview was constructed using CLC main workbench 7. Imperfect repeats are marked in gray. (B) Depicted is the mean fluorescence of S. anginosus SK52 (B) transformed with egfp without promoter (negative control), with the SilX promoter or the cfb promoter (positive control) either with or without 10 mg/l synthetic SilCR. Depicted are means + standard deviation of five independent experiments. Significant differences were calculated using a Man-Whitney U test with * illustrating a p-value < 0.5 and ** indicating p < 0.01.
To assess the activity and functionality of the putative SilX promoter, it was cloned in front of an EGFP gene on the plasmid reporter gene construct pBSU 1202 and transformed into S. anginosus. As a control, the egfp gene alone (pBSU 100) or under the control of an overexpression promoter (pBSU 101) was investigated simultaneously. The activity of the S. anginosus BSU 1211 promoter was assessed in the S. anginosus type strain SK52 (Figure 4B). Promoter activity was surveyed after 2 h and was significantly higher when egfp was under the control of the SilX promoter than in the absence of putative promoter sequences. However, compared to the strong cfb promoter only low-grade activity could be observed. A supplementation with the signal peptide SilCR did not lead to a significantly increased promoter activity. Unfortunately, it was not possible to transform S. anginosus BSU 1211 with these constructs.
Discussion
Bacterial CAAX proteases are intramembrane metalloproteases with various functions that are commonly present in bacteriocin gene clusters (Lux et al., 2007; Denapaite et al., 2010). In the context of bacteriocin production a connection of CAAX proteases to bacteriocin immunity has frequently been reported (Kjos et al., 2010). In this report we investigated the function of SilX, a CAAX protease of S. anginosus, which is involved in bacteriocin activity and the regulation of antimicrobial activity through the QS system sil.
Analysis of the protein sequences demonstrated classical CAAX protease motifs (Figure 1). To determine if SilX displays the predicted protease activity, the effect of HIV protease inhibitors on bacteriocin production was investigated. HIV protease inhibitors, like Nelfinavir, have previously been shown to inhibit bacterial CAAX proteases e.g., SagE of S. pyogenes (Maxson et al., 2015), which is involved in processing the hemolytic SagA peptide of the streptolysin S gene cluster. We could show that by incubation with Nelfinavir a dose dependent inhibition of bacteriocin production occurred. Also, Saquinavir had a dose-dependent effect on bacteriocin inhibition, however the effect was not as pronounced as for Nelfinavir. This might be explained by the difference in molecular weight. Saquinavir has a molecular weight of 766.95 Dalton and Nelfinavir of 567.78 Dalton. The smaller Nelfinavir might more easily diffuse across the cytoplasmic membrane and therefore lower concentrations are able to sufficiently inhibit SilX. Furthermore, Nelfinavir is more lipophilic than Saquinavir, which might also lead to a better uptake of the drug (Maxson et al., 2015). Nelfinavir is associated with higher intracellular concentrations than Saquinavir (Hoggard, 2003).
The dose-dependency of the effect clearly shows that the reduction in antimicrobial activity is due to SilX. In line with this, a SilX deletion mutant lost antimicrobial activity against the target strains of Angicin, suggesting that SilX is essential for bacteriocin production. It has previously been noted that CAAX proteases play an important role in bacteriocin production and CAAX knock out mutants in Streptococcus pneumoniae no longer display antimicrobial activity (Lux et al., 2007). Interestingly, bacteriocin production and thereby inhibition of target strains could be reestablished by addition of the signaling peptide SilCR, which is encoded in the adjacent QS system. These results are consistent with the hypothesis that SilX is responsible for processing and cleavage of SilCR into its mature configuration. SilCR has previously been demonstrated to induce bacteriocin production of S. anginosus (Vogel et al., 2021). The catalytic activity of CAAX proteases depends on two distinct sequence motifs EExxxR and FxxxH (Pei et al., 2011). The FxxxH motif is conserved in S. anginosus BSU 1211, while in the diglutamate motif the arginine residue is replaced by glutamine. However, both of these amino acid residues are polar, thus representing a conservative switch, that may preserve the catalytic function. Leading to the conclusion that SilX could still be able to process SilCR. If SilX processes SilCR, the question arises what the function of SilD and SilE might be. It would be possible that this export system is responsible for the processing and export of Angicin. Further experiments should aim at elucidating this question.
A similar mechanism of peptide maturation is proposed for SagE of S. pyogenes (Maxson et al., 2015). SagE is part of the Streptolysin S gene cluster mediating ß-hemolysis of this species. The ß-hemolysin itself is produced as a peptide protoxin (SagA), which is subsequently modified to become hemolytic. The authors could demonstrate that after an inhibition with HIV protease inhibitors, SagE is no longer able to process the pro-toxin of Streptolysin S (Maxson et al., 2015).
For the CAAX protease MorQ of S. aureus it is suggested that this enzyme facilitates the optimal processing or the export of the AIP of the arg system (Cosgriff et al., 2019). However, the precise mechanism is not yet understood. Another possible mechanism of action of SilX is an interaction with the histidine kinase SilB. Other CAAX proteases have been shown to interact with histidine kinases either increasing or decreasing their activity (Firon et al., 2013; Cosgriff et al., 2019). In Staphylococcus aureus in particular the interaction of a CAAX protease with the histidine kinase of a QS system has been demonstrated. It’s possible that SilX promotes SilB activity leading to the expression of bacteriocins. Consequently, SilB would be inactive in the SilX knock out mutant. However, this effect can be reversed by high SilCR concentrations, which are achieved by exogenously adding synthetic SilCR.
In other bacterial species several studies on CAAX proteases revealed functions connected to bacteriocin self immunity (Lux et al., 2007; Kjos et al., 2010). Immunity proteins protect producer strains against the detrimental effects of their own bacteriocins (Fimland et al., 2002). We found however no evidence that SilX is important for bacteriocin self-immunity in S. anginosus. The CAAX knock-out mutant strain we generated was not sensitive toward the Angicin-producing S. anginosus strain BSU 1211. Moreover blp3.3 another gene of the blp3 gene cluster of S. anginosus has already been shown to provide immunity functions (Vogel et al., 2021). In a next step promoter functionality and activity was surveyed. The silX gene expression is regulated by the sil locus and dependent on a transcription activator SilA (Belotserkovsky et al., 2009; Vogel et al., 2021). SilA belongs to the LytR regulator family that bind DNA as dimers at direct repeats. For S. anginosus we predicted a binding site composed of two 10 bp repeats and a 11 bp spacer. The same basic composition is also found for S. pyogenes and S. intermedius (Belotserkovsky et al., 2009; Mendonca et al., 2016). The SilA binding site was cloned in front of the reporter gene egfp to assess its functionality. Due to unknown reasons, it was not possible to transform S. anginous BSU 1211 with this construct. Alternatively, it was transformed into the S. anginosus type strain (SK 52), where an increased fluorescence could be observed in the strains harboring the putative SilX promoter sequence (Figure 4). However, compared to cfb promoter, which has been shown to induce high level gene expression in various streptococci (Aymanns et al., 2011), the SilX promoter shows only a moderate activity in this background and a supplementation with the signal peptide SilCR did not induce promoter activity. This may be explained by the fact that in S. anginosus SK52 the silA and silB gene are not present and SilCR needs this two-component system to efficiently alter gene expression (Belotserkovsky et al., 2009). Alternatively, this data could also indicate that SilX has a constitutive low-level expression that is independent from the sil system. If SilX processes SilCR, this would ensure the maturation of SilCR as soon as SilCR production is turned on. However, since the sil locus is not intact in S. anginosus SK52, it is difficult to draw definite conclusions, without investigating this construct in a S. anginosus strain harboring a sil locus.
CAAX proteases are often part of bacteriocin operons in streptococci. For example two CAAX proteases are found adjacent to the biosynthetic operon of gallocin from Streptococcus gallolyticus (Harrington et al., 2021; Proutière et al., 2021). The function of these proteins has so far not been investigated. In S. mutans the CAAX protease LsrS is described as protein with receptor-like function (Biswas and Biswas, 2014). For S. pneumoniae it was shown that the knockout of a CAAX protease led to a loss of antimicrobial achtivity (Lux et al., 2007). To our knowledge this is the first report to indicate that a streptococcal CAAX protease may be involved in processing the signal peptide of a quorum sensing system. It thus links a bacteriocin associated CAAX protease to the regulation of bacteriocin production rather than to bacteriocin self-immunity.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
VV and BS designed this study. MF, MJ, AB, SM, and VV performed the experiments under the supervision of VV and BS. JM supported the experiments conducted with HIV inhibitors. VV wrote, and BS reviewed this manuscript. All authors contributed to the article and approved the submitted version.
Funding
MJ, JM, and BS were supported by the German Research Foundation within the CRC 1279. VV thanks the International Graduate School in Molecular Medicine Ulm for their support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Christian U. Riedel for providing microorganisms.
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ https://www.snapgene.com/
- ^ http://www.clcbio.com
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Asam, D., and Spellerberg, B. (2014). Molecular pathogenicity of Streptococcus anginosus. Mol. Oral Microbiol. 29, 145–155. doi: 10.1111/omi.12056
Aymanns, S., Mauerer, S., van Zandbergen, G., Wolz, C., and Spellerberg, B. (2011). High-Level Fluorescence Labeling of Gram-Positive Pathogens. PLoS One 6:e19822. doi: 10.1371/journal.pone.0019822
Bauer, R., Mauerer, S., Grempels, A., and Spellerberg, B. (2018). The competence system of Streptococcus anginosus and its use for genetic engineering. Mol. Oral Microbiol. 33, 194–202. doi: 10.1111/omi.12213
Bauer, R., Neffgen, N., Grempels, A., Furitsch, M., Mauerer, S., Barbaqadze, S., et al. (2020). Heterogeneity of Streptococcus anginosus ß-hemolysis in relation to CRISPR/Cas. Mol. Oral Microbiol. 35, 56–65. doi: 10.1111/omi.12278
Bécavin, C., Bouchier, C., Lechat, P., Archambaud, C., Creno, S., Gouin, E., et al. (2014). Comparison of Widely Used Listeria monocytogenes Strains EGD, 10403S, and EGD-e Highlights Genomic Differences Underlying Variations in Pathogenicity. mBio 5:e969–e914. doi: 10.1128/mBio.00969-14
Belotserkovsky, I., Baruch, M., Peer, A., Dov, E., Ravins, M., Mishalian, I., et al. (2009). Functional Analysis of the Quorum-Sensing Streptococcal Invasion Locus (sil). PLoS Pathog. 5:e1000651. doi: 10.1371/journal.ppat.1000651
Biswas, S., and Biswas, I. (2014). A Conserved Streptococcal Membrane Protein, LsrS, Exhibits a Receptor-Like Function for Lantibiotics. J. Bacteriol. 196, 1578–1587. doi: 10.1128/JB.00028-14
Cosgriff, C. J., White, C. R., Teoh, W. P., Grayczyk, J. P., and Alonzo, F. (2019). Control of Staphylococcus aureus Quorum Sensing by a Membrane-Embedded Peptidase. Infect. Immun 87:e00019–e19. doi: 10.1128/IAI.00019-19
Denapaite, D., Brückner, R., Nuhn, M., Reichmann, P., Henrich, B., Maurer, P., et al. (2010). The Genome of Streptococcus mitis B6 - What Is a Commensal? PLoS One 5:e9426. doi: 10.1371/journal.pone.0009426
Diep, D. B., Håvarstein, L. S., and Nes, I. F. (1996). Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178, 4472–4483. doi: 10.1128/jb.178.15.4472-4483.1996
Facklam, R. (2002). What Happened to the Streptococci: overview of Taxonomic and Nomenclature Changes. Clin. Microbiol. Rev. 15, 613–630. doi: 10.1128/CMR.15.4.613-630.2002
Fimland, G., Eijsink, V. G. H., and Nissen-Meyer, J. (2002). Comparative studies of immunity proteins of pediocin-like bacteriocins. Microbiology 148, 3661–3670. doi: 10.1099/00221287-148-11-3661
Firon, A., Tazi, A., Da Cunha, V., Brinster, S., Sauvage, E., Dramsi, S., et al. (2013). The Abi-domain Protein Abx1 Interacts with the CovS Histidine Kinase to Control Virulence Gene Expression in Group B Streptococcus. PLoS Pathog. 9:e1003179. doi: 10.1371/journal.ppat.1003179
Harrington, A., Proutière, A., Mull, R. W., du Merle, L., Dramsi, S., and Tal-Gan, Y. (2021). Secretion, Maturation, and Activity of a Quorum Sensing Peptide (GSP) Inducing Bacteriocin Transcription in Streptococcus gallolyticus. mBio 12, e3189–e3120. doi: 10.1128/mBio.03189-20
Hoggard, P. G. (2003). The mechanisms that control intracellular penetration of the HIV protease inhibitors. J. Antimicrob. Chemother. 51, 493–496. doi: 10.1093/jac/dkg137
Jiang, S., Li, M., Fu, T., Shan, F., Jiang, L., and Shao, Z. (2020). Clinical Characteristics of Infections Caused by Streptococcus Anginosus Group. Sci. Rep. 10:9032. doi: 10.1038/s41598-020-65977-z
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kjos, M., Snipen, L., Salehian, Z., Nes, I. F., and Diep, D. B. (2010). The Abi Proteins and Their Involvement in Bacteriocin Self-Immunity. J. Bacteriol. 192, 2068–2076. doi: 10.1128/JB.01553-09
Kobo, O., Nikola, S., Geffen, Y., and Paul, M. (2017). The pyogenic potential of the different Streptococcus anginosus group bacterial species: retrospective cohort study. Epidemiol. Infect. 145, 3065–3069. doi: 10.1017/S0950268817001807
Lamy, M.-C., Zouine, M., Fert, J., Vergassola, M., Couve, E., Pellegrini, E., et al. (2004). CovS/CovR of group B streptococcus: a two-component global regulatory system involved in virulence: the CovS/CovR regulatory system of Streptococcus agalactiae. Mol. Microbiol. 54, 1250–1268. doi: 10.1111/j.1365-2958.2004.04365.x
Le, K. Y., and Otto, M. (2015). Quorum-sensing regulation in staphylococci-an overview. Front. Microbiol. 6:1174. doi: 10.3389/fmicb.2015.01174
Lux, T., Nuhn, M., Hakenbeck, R., and Reichmann, P. (2007). Diversity of Bacteriocins and Activity Spectrum in Streptococcus pneumoniae. J. Bacteriol. 189, 7741–7751. doi: 10.1128/JB.00474-07
Majeed, H., Gillor, O., Kerr, B., and Riley, M. A. (2011). Competitive interactions in Escherichia coli populations: the role of bacteriocins. ISME J. 5, 71–81. doi: 10.1038/ismej.2010.90
Maxson, T., Deane, C. D., Molloy, E. M., Cox, C. L., Markley, A. L., Lee, S. W., et al. (2015). HIV protease inhibitors block streptolysin S production. ACS Chem. Biol. 10, 1217–1226. doi: 10.1021/cb500843r
Mendonca, M. L., Szamosi, J. C., Lacroix, A.-M., Fontes, M. E., Bowdish, D. M., and Surette, M. G. (2016). The sil Locus in Streptococcus Anginosus Group: interspecies Competition and a Hotspot of Genetic Diversity. Front. Microbiol. 7:2156. doi: 10.3389/fmicb.2016.02156
Nikolskaya, A. N. (2002). A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 30, 2453–2459. doi: 10.1093/nar/30.11.2453
Pei, J., Mitchell, D. A., Dixon, J. E., and Grishin, N. V. (2011). Expansion of Type II CAAX Proteases Reveals Evolutionary Origin of γ-Secretase Subunit APH-1. J. Mol. Biol. 410, 18–26. doi: 10.1016/j.jmb.2011.04.066
Poupel, O., Proux, C., Jagla, B., Msadek, T., and Dubrac, S. (2018). SpdC, a novel virulence factor, controls histidine kinase activity in Staphylococcus aureus. PLoS Pathog. 14:e1006917. doi: 10.1371/journal.ppat.1006917
Proutière, A., du Merle, L., Périchon, B., Varet, H., Gominet, M., Trieu-Cuot, P., et al. (2021). Characterization of a Four-Component Regulatory System Controlling Bacteriocin Production in Streptococcus gallolyticus. mBio 12:e3187–e3120. doi: 10.1128/mBio.03187-20
Rosa-Fraile, M., Dramsi, S., and Spellerberg, B. (2014). Group B streptococcal haemolysin and pigment, a tale of twins. FEMS Microbiol. Rev. 38, 932–946. doi: 10.1111/1574-6976.12071
Siegman-Igra, Y., Azmon, Y., and Schwartz, D. (2012). Milleri group streptococcus—a stepchild in the viridans family. Eur. J. Clin. Microbiol. Infect. Dis. 31, 2453–2459. doi: 10.1007/s10096-012-1589-7
Thappeta, K. R. V., Ciezki, K., Morales-Soto, N., Wesener, S., Goodrich-Blair, H., Stock, S. P., et al. (2020). R-type bacteriocins of Xenorhabdus bovienii determine the outcome of interspecies competition in a natural host environment. Microbiology 166, 1074–1087. doi: 10.1099/mic.0.000981
Trieu-Cuot, P., Carlier, C., Poyart-Salmeron, C., and Courvalin, P. (1990). A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in Gram-positive bacteria. Nucleic Acids Res. 18, 4296–4296. doi: 10.1093/nar/18.14.4296
Trieu-Cuot, P., Carlier, C., Poyart-Salmeron, C., and Courvalin, P. (1991). Shuttle vectors containing a multiple cloning site and a lacZα gene for conjugal transfer of DNA from Escherichia coli to Gram-positive bacteria. Gene 102, 99–104. doi: 10.1016/0378-1119(91)90546-N
Vogel, V., Bauer, R., Mauerer, S., Schiffelholz, N., Haupt, C., Seibold, G. M., et al. (2021). Angicin, a novel bacteriocin of Streptococcus anginosus. Sci. Rep. 11:24377. doi: 10.1038/s41598-021-03797-5
Keywords: Streptococcus anginosus, CAAX protease, bacteriocin, Angicin, streptococcal invasion locus, HIV protease inhibitor
Citation: Vogel V, Fuchs M, Jachmann M, Bitzer A, Mauerer S, Münch J and Spellerberg B (2022) The Role of SilX in Bacteriocin Production of Streptococcus anginosus. Front. Microbiol. 13:904318. doi: 10.3389/fmicb.2022.904318
Received: 25 March 2022; Accepted: 07 June 2022;
Published: 01 July 2022.
Edited by:
Marcus Fulde, Freie Universität Berlin, GermanyReviewed by:
Takeshi Zendo, Kyushu University, JapanKoshy Philip, University of Malaya, Malaysia
John Renye, Agricultural Research Service (USDA), United States
Hilario Mantovani, University of Wisconsin-Madison, United States
Copyright © 2022 Vogel, Fuchs, Jachmann, Bitzer, Mauerer, Münch and Spellerberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barbara Spellerberg, YmFyYmFyYS5zcGVsbGVyYmVyZ0B1bmlrbGluaWstdWxtLmRl
 Verena Vogel
Verena Vogel Miki Fuchs
Miki Fuchs Marie Jachmann1
Marie Jachmann1 Alina Bitzer
Alina Bitzer Jan Münch
Jan Münch Barbara Spellerberg
Barbara Spellerberg
