
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 14 July 2022
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.903136
This article is part of the Research TopicEnvironmental Influences on the Host-associated MicrobiomesView all 13 articles
Nematodes in the genus Phasmarhabditis can infect and kill slugs and snails, which are important agricultural pests. This useful trait has been commercialized by the corporation BASF after they mass produced a product labeled Nemaslug®. The product contains Phasmarhabditis hermaphrodita, which has been cultured with Moraxella osloensis, a bacterial strain that was originally thought to be responsible for causing mortality in slugs and snails. The exact mechanism leading to death in a Phasmarhabditis infected host is unknown but may involve contributions from nematode-associated bacteria. The naturally occurring microbial community of Phasmarhabditis is unexplored; the previous Phasmarhabditis microbial community studies have focused on laboratory grown or commercially reared nematodes, and in order to obtain a deeper understanding of the parasite and its host interactions, it is crucial to characterize the natural microbial communities associated with this organism in the wild. We sampled Phasmarhabditis californica, Phasmarhabditis hermaphrodita, and Phasmarhabditis papillosa directly from their habitats in Central and Southern California nurseries and garden centers and identified their native microbial community via 16S amplicon sequencing. We found that the Phasmarhabditis microbial community was influenced by species, location, and possibly gastropod host from which the nematode was collected. The predominant bacteria of the Phasmarhabditis isolates collected included Shewanella, Clostridium perfringens, Aeromonadaceae, Pseudomonadaceae, and Acinetobacter. Phasmarhabditis papillosa isolates exhibited an enrichment with species belonging to Acinetobacter or Pseudomonadaceae. However, further research must be performed to determine if this is due to the location of isolate collection or a species specific microbial community pattern. More work on the natural microbial community of Phasmarhabditis is needed to determine the role of bacteria in nematode virulence.
Nematodes are one of the most ecologically diverse groups of organisms on Earth. They exist on every continent, surviving in all climates where decomposition occurs (Ingham, 1994; Bongers and Bongers, 1998; De Ley, 2006; Schafer, 2016). Some exist as free-living organisms like Caenorhabditis elegans, and many have evolved to form a variety of parasitic relationships like Ascaris lumbricoides, or the entomopathogenic nematode (EPN) Steinernema feltiae that has been utilized for biological control against pestiferous insects (Kaya, 1993; Riddle et al., 1997; Marilyn, 2008). EPNs have evolved specific mutualistic relationships with bacterial species in their gut that helps to kill various insects (Grewal and Georgis, 1997). Recent metagenomic analyses have indicated that the commensal microbial community of EPNs, the gut microbial community, is more complex than originally thought, leading to the possibility of a native EPN pathobiome that assists with insect killing (Ogier et al., 2020).
One nematode that has been successfully used as a biological control agent is Phasmarhabditis hermaphrodita (Wilson et al., 1993b). All members of the genus are gastropod-specific facultative parasites. Phasmarhabditis hermaphrodita was discovered in Europe and has been commercialized for biological control in Europe under the name Nemaslug®. Phasmarhabditis nematodes are effective at killing pestiferous gastropods in laboratory and agricultural settings such as nurseries and a variety of crops (Wilson et al., 1994a,b; Rae et al., 2007b; McDonnell et al., 2020; Tandingan De Ley et al., 2020; Schurkman et al., 2022a) but are safe to tested non-gastropod organisms (Grewal and Grewal, 2003; Iglesias et al., 2003; Rae et al., 2007a; Nardo et al., 2010). However, due to the discovery of Phasmarhabditis in Europe, its use has not yet been permitted in the United States since invasive species are not permitted for biological control use in the country.
It was originally thought that Phasmarhabditis nematodes kill their hosts in a manner similar to EPNs, which employ mutualistic and pathogenic microbes to assist with insect killing (Wilson et al., 1993b; Tan and Grewal, 2001a). This hypothesis came about upon the discovery that P. hermaphrodita cultured with Moraxella osloensis was highly pathogenic to the grey field slug Deroceras reticulatum, more so than when it was cultured on other bacteria (Wilson et al., 1995a; Tan and Grewal, 2001b). The selection of bacteria to test came from isolates identified in P. hermaphrodita infective juveniles (IJs), dead D. reticulatum, and xenic foam chip cultures (Wilson et al., 1993a, 1995a). The species identified and tested included Aeromonas hydrophila, Aeromonas sp., Flavobacterium breve, Flavobacterium odoratum, M. osloensis, Providencia rettgeri, Pseudomonas fluorescens (isolate no. 140), P. fluorescens (isolate no. 141), and Serratia proteamaculans. When these bacteria were injected into D. reticulatum, A. hydrophila, and P. fluorescens (isolate no. 140) caused the most mortality. However, P. fluorescens (isolate no. 140) was able to facilitate better growth when culturing P. hermaphrodita, which may be indicative of this bacterial species serving as an optimum food source. However, nematodes which were grown on M. osloensis exhibited the highest pathogenicity while also allowing for good nematode growth. Another experiment showed that axenic P. hermaphrodita did not cause mortality in D. reticulatum while those reared on M. osloensis did (Tan and Grewal, 2001b). These experiments led to the assumption that P. hermaphrodita likely has a natural association with M. osloensis, similar to EPNs’ association with pathogenic bacteria (An et al., 2008; Wilson and Rae, 2015). After these experiments, it was generally accepted among the Phasmarhabditis community that these nematodes caused death in their gastropod hosts strictly through the utilization of the apparent commonly associated bacteria M. osloensis. However, the natural association of other bacteria with Phasmarhabditis in the wild and how this association contributes to nematode host virulence remained largely unexplored.
In 2010, it was shown that P. hermaphrodita associates with many bacterial species that do not affect its virulence (Rae et al., 2010), in contrast to the existing understanding of Phasmarhabditis virulence (Wilson et al., 1995a). Rae et al. (2010) suggested that P. hermaphrodita does not associate with specific bacteria due to the observation of inconsistent and varied bacterial assemblages with the nematode (Rae et al., 2010). However, the study was unable to identify key microbial species that regularly occur within Phasmarhabditis. This is because a limitation to PCR-DGGE is that specific bacterial species cannot be identified. Therefore, no conclusions were made on the presence of M. osloensis since that was not the goal of the study. The study only aimed to show that Phasmarhabditis associated with multiple bacterial assemblages. In another study, bacteria were analyzed from laboratory grown Phasmarhabditis, by allowing nematodes to crawl on LB agar plates and identifying some of the bacterial colonies that subsequently arose (Howe et al., 2020). Eight genera of bacteria were identified that were hypothesized to have come from the laboratory grown Phasmarhabditis. Pseudomonas was the only genus found in this most recent study that was also found in 1995 (Wilson et al., 1995a; Howe et al., 2020). Moraxella osloensis, which was expected to be identified, was not found. These mirror findings related to the native and naturally occurring microbial community of the model organism C. elegans (Dirksen et al., 2016), though prolonged in vitro growth in the laboratory raises the possibility of association with microbes not commonly found with Phasmarhabditis in the wild.
Describing the natural and infected microbial community of the host could help to distinguish whether microbes present within Phasmarhabditis originated from the host or from another source. Very little microbial community research has been done on D. reticulatum, the slug often used in Phasmarhabditis studies (Walker et al., 1999). However, gut microbial community metagenomic analyses have been performed on other slug species like Ambigolimax valentianus which identified a core microbial community of Citrobacter, Delftia, Erwinia, Arthrobacter, Stenotrophomonas, Pseudomonas, Rhodococcus, and Bacillus. Arion ater’s microbial community was also found to be influenced by the substrate they were on, while the soil microbial community itself could also be influenced by the introduction of the slug (Jackson, 2020; Jackson et al., 2021). A gut metagenomic analyses of the slug A. ater has also been performed. The most abundant bacterial genera in the gut of A. ater included Enterobacter, Citrobacter, Pseudomonas, and Escherichia (Joynson et al., 2017).
All microbial community studies that have taken place involving Phasmarhabditis have used laboratory cultured nematodes, and the microbial community changes upon introduction to a laboratory environment, especially when the nematodes are grown on monoxenic cultures (Dirksen et al., 2016). Recently, three species of Phasmarhabditis were discovered during gastropod surveys of California nurseries and garden centers (Tandingan De Ley et al., 2014, 2016). Between 2018 and 2021 additional surveys for Phasmarhabditis nematodes were performed to determine the distribution of these species (Schurkman et al., 2022b). Newly-isolated nematodes collected during these most recent surveys were used to identify the natural microbial communities of Phasmarhabditis isolates across the Central and Southern California regions. Similarities or differences between Phasmarhabditis isolates could help to further the understanding of the potential role that the microbial community plays between Phasmarhabditis nematodes and their hosts. In this study, we aimed to determine if the Phasmarhabditis microbial community is species specific, and if it is influenced by gastropod hosts, geographic locality, and collection methods.
Fourteen plant nurseries from Central California and five nurseries from Southern California were surveyed for gastropods infected with Phasmarhabditis as described in Schurkman et al. (2022b). In short, 1 person hour was spent searching for gastropods. Then, gastropods were sorted into containers by species and taken back to the laboratory at UC Riverside in coolers. Gastropods were decapitated and placed on 1% plain agar to create seed cultures (1 L: 10 g agar, 900 ml H2O) and their bodies were observed for the presence of nematodes under a dissecting microscope. Upon finding a nematode(s) which phenotypically resembled a member of the Phasmarhabditis genus (i.e., the significant presence of phasmids), up to five individuals were placed on Nematode Growth Medium [NGM; 1 L: 3 g NaCl, 20 g Agar, 2.5 g Peptone, 975 ml deionized H2O, and 10 ml Uracil (2 g/L) were added to a liter of deionized water, autoclaved, and let cool, to which were added 25 ml filtered KPO4, 1 ml filtered MgSO4, 1 ml CaCl2, and 1 ml Cholesterol (5 mg/ml)] to create axenic cultures. All nematodes on axenic culture plates were considered as identical species of the same strain with further verification after single nematode DNA sequencing. The axenic culture plates were stored at 17°C. Individual nematodes suspected to be Phasmarhabditis were picked from axenic culture plates and were prepared for PCR and DNA sequencing of the D2-D3 domains of the rDNA LSU, as described in Tandingan De Ley et al. (2014). Contigs were assembled and compared by BLAST with published sequences on Genbank using CodonCode Aligner (CodonCode Corp., 58 Beech Street, Dedham, MA, United States) to identify nematode species. A percent identity match near 99% was required in order to indicate a species identification using BLAST or CodonCode Aligner.
All nematodes suspected to be a member of the Phasmarhabditis genus via microscopy were prepared for microbial community analysis. This preparation was done for each nematode picked from gastropod cadavers and before axenic culture plates were created. To prepare, nematodes in seed cultures which phenotypically matched those that were used for axenic culture were subjected to a rinse. When possible, at least one nematode from each seed culture was washed in sterile M9 buffer (1 L: 3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, 1 ml 1 M MgSO4, and H2O to 1 L) thrice and then placed inside of 10 μl of sterile H2O in a 200 μl PCR tube, which was stored at −20°C for future use. These nematodes were labeled as washed nematodes. The washes were performed to rinse excess material from the cuticle of the nematode. When possible, at least one nematode was also not subjected to any treatment at all, and the nematode was immediately picked and placed inside of a 200 μl sterile microtube with 10 μl of sterile H2O and stored at −20°C. These nematodes were labeled as unwashed nematodes. Lastly, 10 μl of dead and partially decomposed gastropod tissue was pipetted into 10 μl of sterile H2O in a 200 μl PCR and was stored at −20°C. Comparisons to unwashed nematodes and decomposed slug tissue were done to observe whether the washes significantly altered the microbial community of Phasmarhabditis. Upon finding a Phasmarhabditis nematode via 28S sequencing, we used the corresponding nematode(s) previously frozen at −20°C for microbial community analysis.
Genomic DNA was extracted from all washed and unwashed nematodes, as well as from the decomposed dead gastropod tissue. The DNA extraction protocol included thawing samples on ice and breaking the individual nematodes into pieces within their PCR tube using a sterile 10 μl pipette tip. After breaking the nematodes, the total volume of all samples was brought up to 100 μl with sterile PCR grade water. An equal volume of phenol chloroform was then added to each sample. The contents of the small PCR tubes were then transferred to a 1.25-ml Eppendorf tube and were mixed via pipettor. The tubes were shaken by hand for 30 s and were then centrifuged at 12,000 rpm for 10 min at 4°C. After centrifugation, the aqueous phase of the solutions was removed and placed in a new 1.25-ml Eppendorf tube. The wash with phenol chloroform was repeated once more, and then 400 μl of isopropanol stored at −20°C was added to the solution. A 1:10 ratio of 3 M sodium acetate was then added to the solution and was mixed via pipetting up and down. The tubes were then shaken by hand for 30 s and to each tube, 1 μl of glycogen stored at −20°C was added before storing at −20°C for 24 h. The samples were then centrifuged at 13,000 rpm for 30 min at 4°C to form a pellet. All liquid was then removed from the tubes with a pipettor, being careful not to disturb the pellet at the base of the tube. Using a pipette, the pellet was carefully washed with 500 μl of ethanol which was immediately drawn from each tube. The tubes were then stored at 37°C in an incubator until no visible liquid was present. The pellet was then resuspended in 50 μl of sterile PCR grade water and the DNA was quantified using a Qubit 3 Fluorometer (Invitrogen by Thermo Fisher Scientific and life technologies, Waltham, MA, United States).
The bacterial 16S rDNA V4 region 515F-806R was amplified according to the earth microbial community project, 16S Illumina protocol (Thompson et al., 2017). Based on the concentrations of each single nematode DNA sample, 1–8 μl of the extracted DNA template, 10 μl Platinum Hot Start PCR Master Mix (ThermoFisher), and 0.5 μl of forward and reverse primers (10 μM) were added into the 25 μl PCR reaction system, with the barcode in the reverse primer. Thermocycler conditions were 4°C for 3 min, followed by 30 cycles (94°C for 45 s, 50°C for 60 s, 72°C for 90 s), and 72°C for 10 min. PCR products were pooled together and submitted to an Illumina MiSeq platform with 2 × 150 bp read lengths.
Raw reads were processed using the open-source software QIIME2 (Bolyen et al., 2019) version 2021.2. Samples that had >1,000 reads were retained and denoised using dada2 with default settings. Taxonomic classification was performed using classify-sklearn command against the Greengenes 13_8 99% OTUs from 515F/806R region of sequences (MD5: 9e82e8969303b3a86ac941ceafeeac86) set trimmed to 250 bp of the V4 hypervariable region (McDonald et al., 2012; Bokulich et al., 2018; Robeson et al., 2020). An amplicon sequence variant (ASV) was defined as a group of sequences with a similarity of 100%. Alpha and beta diversity analyses were calculated in QIIME2 with rarefied sample depth of 1,000, and results were plotted in GraphPad Prism 9. The heatmap was generated using pheatmap package in R program, samples and species clustered using hclust.
The statistical calculations used in QIIME2 were: Kruskal-Wallis test for alpha diversity comparisons, and permANOVA for beta diversity. Mann–Whitney U tests were performed in GraphPad Prism 9 for taxa comparisons.
Of the total 146 samples from three different nematode species from various gastropod hosts collected during surveys between 2019 and 2020, 26 were amplified and sequenced successfully (Supplementary Table 1). In total 475,226 raw reads were obtained from 26 samples. A total of 397,685 high-quality reads were clustered into 337 ASVs at 100% similarity level. Twenty-two samples with read depth > 1,000 remained for subsequent analyses (Supplementary Table 1).
Alpha diversity analysis showed that nematode species may be an important factor associated with the diversity of nematode microbial community’s. According to observed features, Shannon index, and Faith’s phylogenetic diversity (Faith pd), P. californica microbial community’s exhibited higher richness than those of P. hermaphrodita and P. papillosa (Figure 1A). Meanwhile, Central California samples had significantly higher observed features and Faith pd. index than Southern California. However, this may reflect the fact that all nematodes collected from Southern California were P. papillosa, which had the lowest diversity of observed features (Figure 1). The host of the nematode was not associated with differences in microbial richness or evenness (Figure 2A). No alpha diversity differences were noted across nematodes that were washed in M9 thrice, unwashed, or collected from decomposed gastropod tissue (Figure 2B).
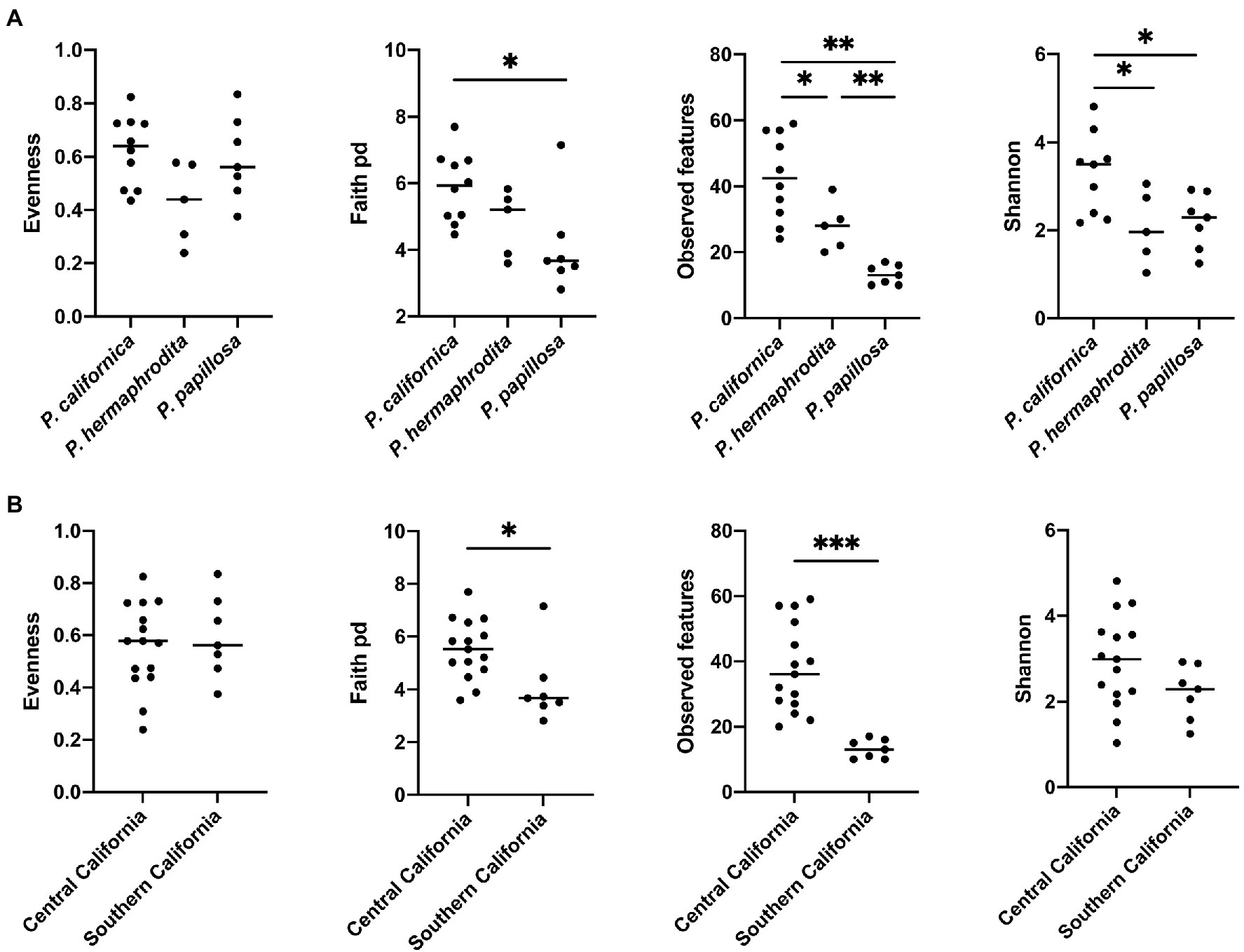
Figure 1. The comparison of microbial community alpha diversity of nematode-associated microbial communities. (A) Phasmarhabditis species and (B) location affect the richness of the microbial composition in nematode. Kruskal-Wallis test, *p < 0.05; **p < 0.01; and ***p < 0.001.
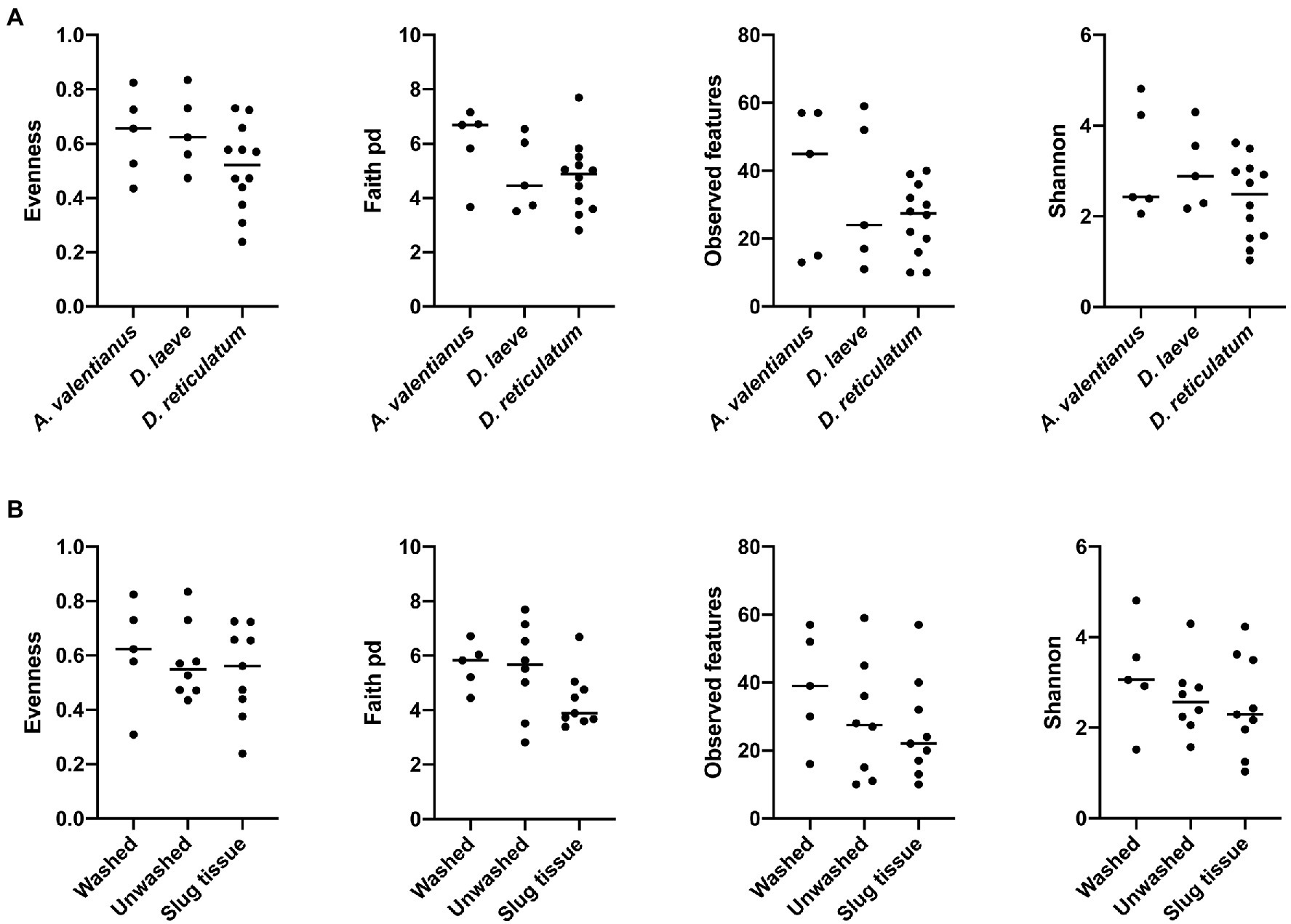
Figure 2. Comparison of microbial community alpha diversity across different host species or sample collection strategy. (A) Gastropod host and (B) collection strategy are not associated with differences in the diversity of the Phasmarhabditis microbial community.
The difference of overall microbial community structure showed similar trends with alpha diversity analyses. The PCoA plots based on Bray–Curtis distance between sample microbial community’s revealed a significant separation of bacterial composition depending on Phasmarhabditis species, especially between P. californica and P. papillosa (q value = 0.015), while the P. hermaphrodita microbial community overlapped with the other two species (Figure 3A), suggesting that P. hermaphrodita’s microbial community shared some of the same bacterial features with both P. californica and P. papillosa; these differences were statistically significant when tested using permANOVA (Table 1). Geographical location also contributed to differences in the nematode microbial community (Figure 3B), while treatment by washing with M9 did not (Figure 4B). The gastropod host also showed a slight overall association (p = 0.017) with the nematode microbial community (Figure 4A; Table 1), but no pairwise comparison of the hosts showed significantly separation. From these data, we conclude that nematode species and location may play an important role in shaping the native Phasmarhabditis microbial community and perhaps the gastropod microbial community.
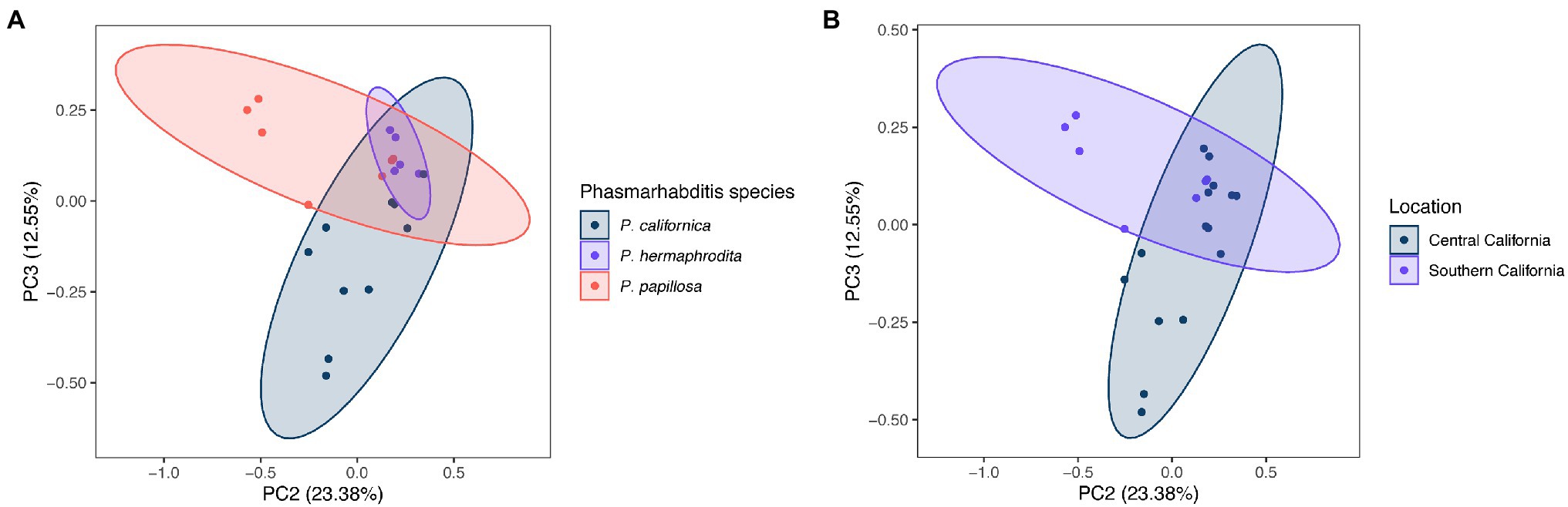
Figure 3. Principal coordinate analysis (PCoA) plots of nematode microbial community’s based on Bray Curtis distance. PCoA plots showing (A) Phasmarhabditis species and (B) location. Percent variance explained is shown in parentheses for each axis. Ellipses show 95% CI.
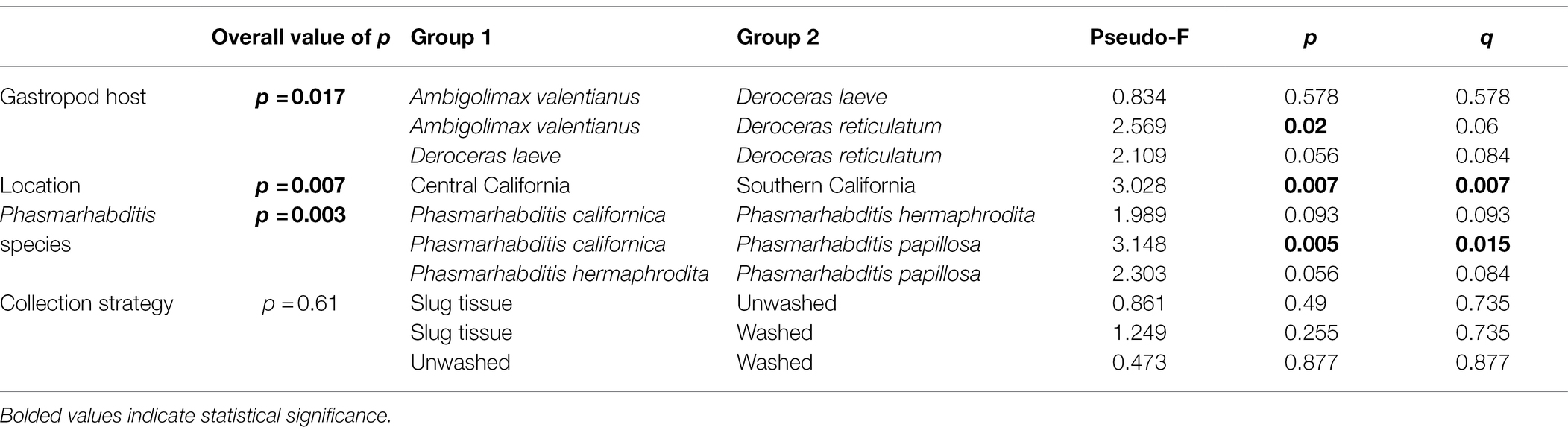
Table 1. permANOVA analysis reveals the microbial differences between gastropod hosts, locations, Phasmarhabditis species, or washed/unwashed/slug tissue.
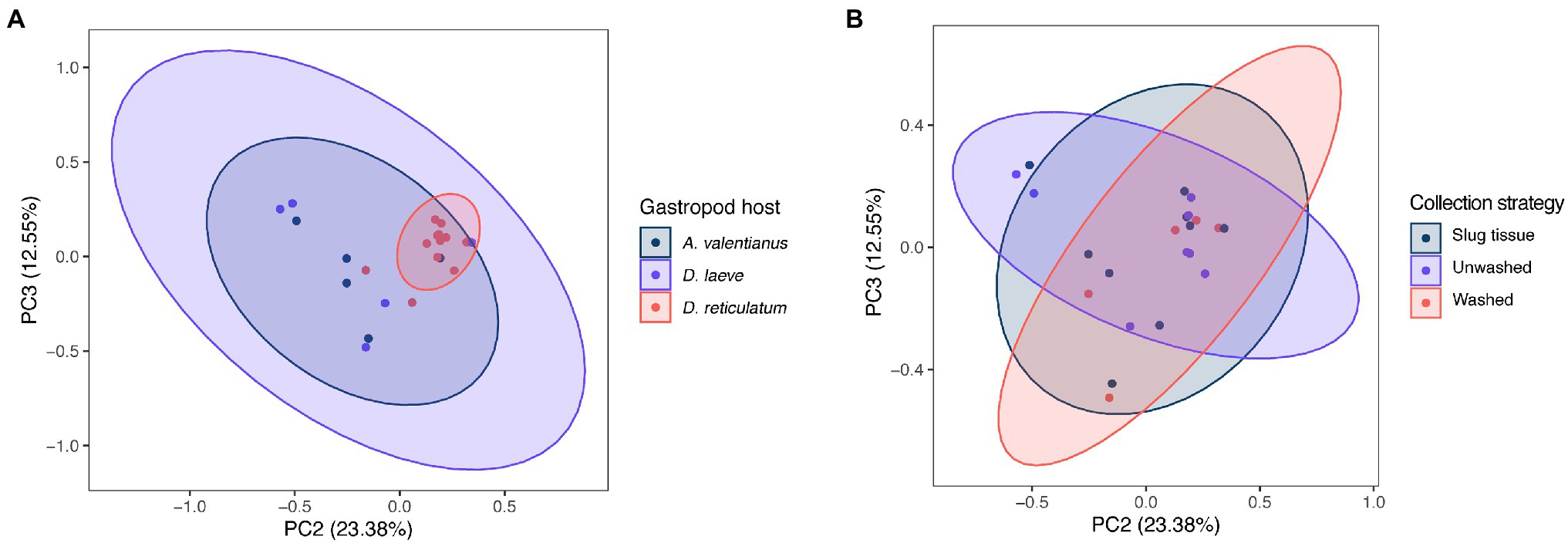
Figure 4. PCoA plots of nematode microbial community of different gastropod hosts, based on Bray Curtis distance. PCoA plots with samples clustered by (A) gastropod host and (B) collection strategy; % variance explained shown in parentheses for each axis. Ellipses show 95% CI.
The species-level bacterial community composition in the nematodes was analyzed using the unsupervised hierarchical cluster analysis. For this analysis, all the samples were divided into four groups (Figure 5). Cluster IV consisted of P. papillosa samples from Southern California and exhibited enrichment with species belonging to genus Acinetobacter or family Pseudomonadaceae. Cluster III consisted of P. californica from Central California. Cluster II consisted of a mixture of P. papillosa from Southern California and P. hermaphrodita from Central California, which were all collected from the same gastropod host, D. reticulatum; in these microbial communities, members of genus Shewanella and family Aeromonadaceae were the dominant microbial members. No members of the genus Moraxella were identified. Cluster I microbial samples were dominated by a high proportion of Clostridium perfringens, though these samples were collected from multiple nematodes and gastropod hosts from Central California. Among the most abundant species, Shewanella sp. was significantly increased in cluster II compared to the other clusters; samples in cluster I had 48%–86% of C. perfringens, which was not shown in any other clusters; species from family Pseudomonadaceae and genus Acinetobacter were significantly enriched in cluster IV; while family Aeromonadaceae was evenly distributed in all clusters (Figure 6).
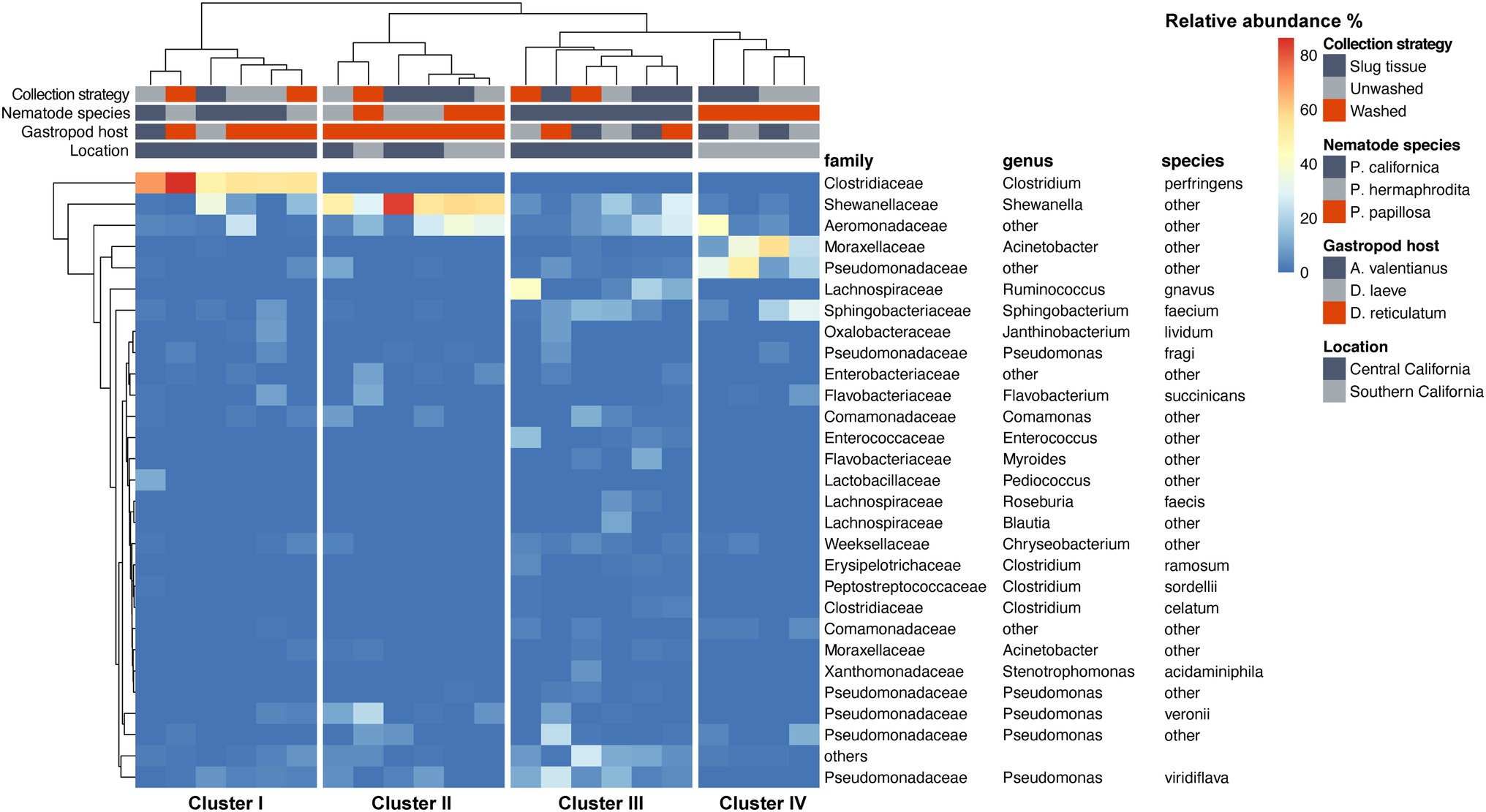
Figure 5. Heatmap of the nematode microbial community at species level. Species with relative abundance >5% across all samples are displayed.
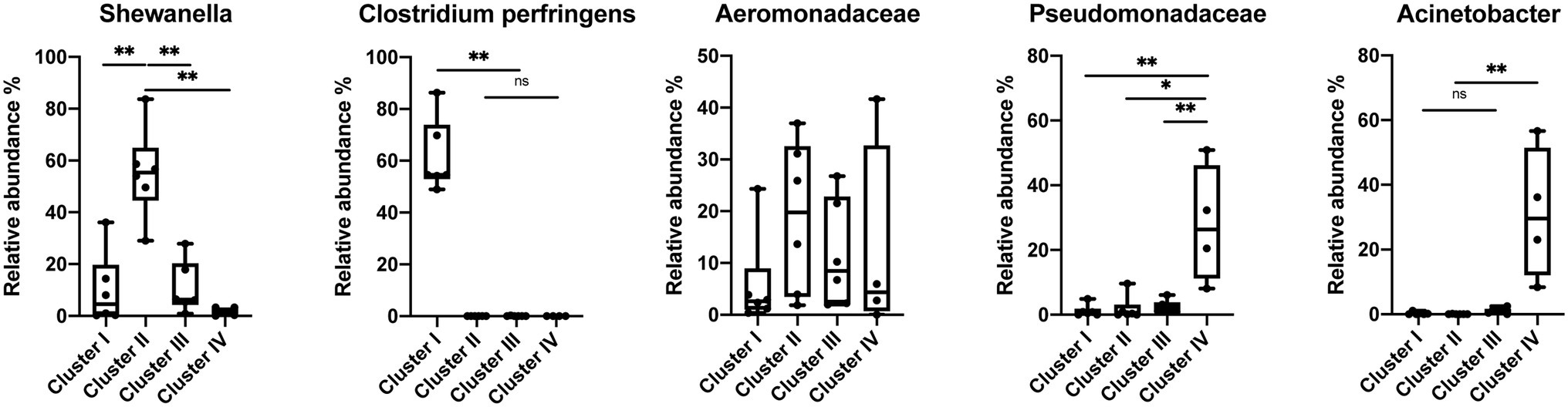
Figure 6. Relative abundance of the most abundant species in the Phasmarhabditis species. Mann–Whitney U-test, *p < 0.05; **p < 0.01. Boxplots show inter-quartile range, whiskers minimum to maximum.
This study is the first analysis of the native microbial community of Phasmarhabditis. It assessed the microbial community of Phasmarhabditis in multiple nursery and garden center habitats and aimed to help identify core microbial communities of Phasmarhabditis utilizing 16S metabarcoding analysis. The previous Phasmarhabditis microbial community work had been done using nematodes which had been kept in culture, leaving the possibility of the nematode’s microbial community being altered by laboratory conditions and frequent transfers (Wilson et al., 1993a, 1995b; Rae et al., 2010; Dirksen et al., 2016;Howe et al., 2020; Sheehy et al., 2022). Two other studies have used next generation sequencing techniques, however the studies did not use newly-recovered Phasmarhabditis, but instead utilized laboratory sub-cultured and maintained Phasmarhabditis (Howe et al., 2020; Sheehy et al., 2022). The most recent study by Sheehy et al. (2022) showed that Phasmarhabditis does not exclusively associate with a single bacterial species which it relies on for virulence. However, the study did not utilize native Phasmarhabditis and only utilized Phasmarhabditis nematodes which have been frequently cultured and transferred in laboratories (Sheehy et al., 2022). While some of the recent studies may suggest that Phasmarhabditis does not maintain a set core microbial community, understanding what microbes are naturally and commonly associated with Phasmarhabditis may help to solidify whether Phasmarhabditis utilizes any core microbial species which contributes to gastropod killing, similar to microbial contributors to EPN virulence. The findings may also reveal crucial bacterial species needed for Phasmarhabditis food consumption, survival, or host–parasite interactions. Our results suggest that the location, species, and possibly the gastropod host may affect the microbial diversity within the tested Phasmarhabditis.
Our findings are not entirely congruent with the previous Phasmarhabditis microbial community work. Similar to previous studies, we identified Acinetobacter and Pseudomonas spp. occurring on Phasmarhabditis, however, no previous studies identified predominant bacteria like Shewanella, Aeromonadaceae, or C. perfringens which were identified in this study (Wilson et al., 1995a; Rae et al., 2010; Howe et al., 2020; Sheehy et al., 2022). Pseudomonaceae and Acinetobacter species were enriched in some clusters of Phasmarhabditis nematodes, specifically cluster IV which consisted of P. papillosa (Figure 6). Acinetobacter and Pseudomonaceae bacteria are commonly found in the soil and have been discovered in multiple gastropod species (Ducklow et al., 1981; Wilson et al., 1993a; Ekperigin, 2007; Villena et al., 2010; Joynson et al., 2017; Howe et al., 2020; Jackson, 2020). It was previously found that unhealthy Biomphalaria glabrata snails had a core microbial community predominantly made up of Acinetobacter and Moraxella spp.; however, healthy snails had a microbial community predominantly made up of Pseudomonas spp. (Ducklow et al., 1981). It is possible that Phasmarhabditis and gastropods thrive with Pseudomonas spp., and the presence of other species like Acinetobacter or Moraxella spp. (the bacterial species used to mass produce Nemaslug®) in Phasmarhabditis cause increased pathogenicity. However, this hypothesis is disputed from a finding that showed that rearing Phasmarhabditis on Acinetobacter had no effect on its virulence (Nermut et al., 2014). The interesting pattern in which only P. papillosa (discovered only in Southern California) have both increased Pseudomonaceae and Acinetobacter needs more study. This pattern may be due to the bacterial diversity and population and potential dominance at the collection site, or a species-specific relationship with P. papillosa. However, another possibility is that Phasmarhabditis uses some of the predominant bacterial species as a major food source, and others as contributors toward virulence, or perhaps some bacteria are used as both food and a driver for pathogenesis. Since Acinetobacter and Pseudomonaceae are frequently found in soils and are not commonly known as highly virulent bacteria, it is possible that these predominant bacteria are used as a food source rather than enhance or contribute to pathogenicity. This hypothesis is further supported by the observation that Phasmarhabditis grew exceptionally well on agar cultured with P. fluorescens (isolate 141) or P. fluorescens PSG strain compared to other bacterial species. The Pseudomonas bacteria was still not selected for use in the commercial production of Nemaslug® possibly because it was not associated with the highest mortality rates, suggesting its role as a food source for the nematodes rather than a source of virulence (Wilson et al., 1995a,b).
Shewanella has been discovered in multiple gastropod species where it causes increased pathogenicity, however all studies which identified this were performed in aquatic environments (Cai et al., 2006; Wang et al., 2008). The finding of an association of Shewanella with Phasmarhabditis has not previously been reported. The bacteria were not detected in any P. californica isolates (Figure 5). This may have been due to a limited sample size throughout the study, or due to a random association of bacteria with Phasmarhabditis, as hypothesized in 2010 (Rae et al., 2010). However, P. californica had the largest representation throughout this study with a total of 10 utilized isolates (Supplementary Table 1). The occurrence of this predominant species may be indicative of it being used as a source of virulence toward the gastropod host; however further research is needed to assess this possibility.
Multiple gastropod species have been found associated with Clostridium bacteria (Charrier et al., 2006; Li, 2012). However, like Shewanella, the species C. perfringens had not previously been found in Phasmarhabditis or other nematodes. C. perfringens is most well known as a causative agent of food poisoning in mammals (Labbe, 1991). The species is frequently searched for and reported in foods for the sake of public health. There are over 1 million cases of poisoning from C. perfringens each year (Grass et al., 2013). A previous study demonstrated that C. perfringens enterotoxin could cause intestinal illness of mammals, and potentially fish and frogs (Robertson et al., 2010). However, it is not known how this bacterium affects gastropods and nematodes. Phasmarhabditis nematodes may serve as vectors for C. perfringens, using the bacteria as a weapon to kill their gastropod host. However, this seems less likely since it has been found that some gastropods can naturally harbor and vector the bacteria. It was previously thought that C. perfringens was only capable of reproducing in mammals and other endothermic organisms, and therefore only these organisms could vector the pathogenic bacteria. More recently it was found that ectotherms such as gastropods, frogs, and fish can also vector the bacteria and therefore these organisms should be monitored as sources of contamination (Frick et al., 2018). Our finding of these bacteria furthers the claim that ectotherms, specifically gastropod-associated nematodes, can act as vectors. However, it was only discovered in cluster I which consisted of P. californica and P. hermaphrodita. It is possible that only P. californica, and P. hermaphrodita use C. perfringens as a source of virulence. However, further study is needed.
The most predominant bacterial family found throughout all Phasmarhabditis species was Aeromonadaceae. This family was found in similar abundance across all species and served as the only predominant commonality within the genus. The family has not previously been found within Phasmarhabditis and is not common in many nematodes, but it has been discovered within multiple gastropod species (Villena et al., 2010; Li et al., 2019). It is possible that Phasmarhabditis largely assumes the microbial diversity of its gastropod host. However, this hypothesis needs further experimentation. Since Aeromonadaceae is not commonly known to be highly pathogenic to a variety of organisms and it was the most predominant species across all Phasmarhabditis nematodes, it can be hypothesized that Phasmarhabditis species utilize the bacteria as a major food source rather than a source of virulence. Phasmarhabditis hermaphrodita and other members of the genus are known to be bacterivorous (Tan and Grewal, 2001b). However, their food preferences at collection sites are unknown. Phasmarhabditis hermaphrodita has been found to grow well when reared on monoxenic cultures of P. fluorescens, but this does not prove its preferred bacterial food source in a native setting (Wilson et al., 1995b). Further experimentation may draw out explanations for the clustering of Aeromonadaceae observed in clusters III and IV within P. californica and P. papillosa (Figure 5).
Future work to assess the microbial diversity of Phasmarhabditis needs to utilize next generation sequencing technology and nematodes which have not been maintained in culture for a long period of time. Further microbial community work with the species from this study (P. californica, P. hermaphrodita, and P. papillosa) should be done in order to obtain more isolates for statistical power in identifying the microbial community of the species. Further study would also lead to the possibility of work with less discrepancies in read counts as we observed. Study of other Phasmarhabditis species microbial community should also be assessed in order to determine other species-specific microbial patterns. It is likely that the maintenance of nematodes in culture on specific media can influence the microbiota (Dirksen et al., 2016). It was recently found that C. elegans native microbial community differs from the previously described microbial community, and its microbial community has some consistencies across time at the genus level but can be influenced by various substrates and present bacteria. Interestingly, one of the consistent genera in C. elegans is Pseudomonas, which was one of the predominant bacteria we identified which may serve as a major food source for Phasmarhabditis (Dirksen et al., 2016; Johnke et al., 2020). To understand the natural relationships and mechanisms between Phasmarhabditis and their hosts, native isolates must be utilized. Next generation sequencing technologies allow for rapid sequencing and identification of these isolates and their microbial communities upon collection, allowing for easy assessment of the native microbial flora and their potential interactions.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accessionnumber(s) can be found in the article/Supplementary Material. The 16S sequence data has been deposited in NCBI under BioProject PRJNA816167.
JS, IL, AH, and AD: conceptualization; JS, RL, SA, and AH: methodology and software; JS, RL, AH, and AD: validation and original draft preparation; JS, RL, and SA: formal analysis and investigation; IL, AH, and AD: resources and funding acquisition; JS, RL, and AH: data curation; JS, RL, SA, IL, AH, and AD: writing; JS, RL, IL, AH, and AD: writing—review and editing; JS and RL: visualization; and AH and AD: supervision and project administration. All authors have read and agreed to the published version of the manuscript.
The work was supported by the United States Department of Agriculture Specialty Crop Multi-State Program (USDA-SCMP) in partnership with CDFA (2018–2022 grant #12509488), a generous gift (2019–2022) from CANERS Foundation through the California Association of Nurseries and Garden Centers (CANGC), Plant California Alliance (PCA), and National Institutes of Health GM124724.
IL is a co-inventor on a patent entitled Mollusk-killing Biopesticide (WO2017059342A1).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to acknowledge Rory McDonnell who has assisted in our project development as well as our identification of gastropods. We would also like to acknowledge Dee Denver for his assistance in project development, and Kyle Anesko for his assistance with gastropod maintenance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.903136/full#supplementary-material
An, R., Sreevatsan, S., and Grewal, P. S. (2008). Moraxella osloensis gene expression in the slug host Deroceras reticulatum. BMC Microbiol. 8:19. doi: 10.1186/1471-2180-8-19
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker gene amplicon sequences. PeerJ 6:p.e3208v2. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bongers, T., and Bongers, M. (1998). Functional diversity of nematodes. Appl. Soil Ecol. 10, 239–251. doi: 10.1016/S0929-1393(98)00123-1
Cai, J., Chen, H., Thompson, K. D., and Li, C. (2006). Isolation and identification of Shewanella alga and its pathogenic effects on post-larvae of abalone Haliotis diversicolor supertexta. J. Fish Dis. 29, 505–508. doi: 10.1111/j.1365-2761.2006.00732.x
Charrier, M. Y., Fonty, G., Gaillard-Martinie, B., Ainouche, K., and Andant, G. (2006). Isolation and characterization of cultivable fermentative bacteria from the intestine of two edible snails, Helixpomatia and Cornu aspersum (Gastropoda: Pulmonata). Biol. Res. 39, 669–681. doi: 10.4067/S0716-97602006000500010
De Ley, P. (2006). “A quick tour of nematode diverity and the backbone of nematode phylogeny,” in Wormbook: The Online Review of C. elegans Biology (Pasadena, CA: Wormbook), 2005–2018.
Dirksen, P., Marsh, S. A., Braker, I., Heitland, N., Wagner, S., Nakad, R., et al. (2016). The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 14, 1–16. doi: 10.1186/s12915-016-0258-1
Ducklow, H. W., Clausen, K., and Mitchell, R. (1981). Ecology of bacterial communities in the schistosomiasis vector snail Biomphalaria glabrata. Microb. Ecol. 7, 253–274. doi: 10.1007/BF02010308
Ekperigin, M. M. (2007). Preliminary studies of cellulase production by Acinetobacter anitratus and Branhamella sp. Afr. J. Biotechnol. 6, 634–649.
Frick, C., Vierhellig, J., Linke, R., Savio, D., Zornig, H., Antensteiner, R., et al. (2018). Poikilothermic animals as a previously unrecognized source of fecal indicator bacteria in a backwater ecosystem of a large river. Appl. Environ. Microbiol. 84, e00715–e00718. doi: 10.1128/AEM.00715-18
Grass, J. E., Gould, L. H., and Mahon, B. E. (2013). Epidemiology of foodborne disease outbreaks caused by Clostridium perfringens, United States, 1998-2010. Foodborne Pathog. Dis. 10, 131–136. doi: 10.1089/fpd.2012.1316
Grewal, P., and Georgis, R. (1997). “Entomopathogenic nematodes,” in Biopesticides: Use and Delivery. Vol. 5. eds. F. R. Hall and J. J. Menn (Totowa, NJ: Humana Press), 271–299.
Grewal, S. K., and Grewal, P. S. (2003). Survival of earthworms exposed to the slug-parasitic nematode Phasmarhabditis hermaphrodita. J. Invertebr. Pathol. 82, 72–74. doi: 10.1016/S0022-2011(02)00200-8
Howe, D. K., Ha, A. D., Colton, A., Tandingan De Ley, I., Rae, R. G., Ross, J., et al. (2020). Phylogenetic evidence for the invasion of a commercialized European Phasmarhabditis hermaphrodita lineage into North America and New Zealand. PLOS One 15:e0237249. doi: 10.1371/journal.pone.0237249
Iglesias, J., Castillejo, J., and Castro, R. (2003). The effects of repeated applications of the molluscicide metaldehyde and the biocontrol nematode Phasmarhabditis hermaphrodita on molluscs, earthworms, nematodes, acarids and collembolans: a two year study in north-west Spain. Pest Manag. Sci. 59, 1217–1224. doi: 10.1002/ps.758
Ingham, R. E. (1994). “Nematodes,” in Methods of Soil Analysis, Part 2, Microbiological and Biochemical Properties, eds. R. W. Weaver, J. S. Angle and P. S. Bottomley (Madison, WI: Soil Science Society of America), 459–490.
Jackson, D. (2020). Elucidating the Influences of an Invasive Slug on Soil Bacterial Communities. University of California, Riverside.
Jackson, D., Maltz, M. R., Freund, H. L., Borneman, J., and Aronson, E. (2021). Environment and diet influence the bacterial microbiome of Ambigolimax valentianus, and invasive slug in California. Insects 12:575. doi: 10.3390/insects12070575
Johnke, J., Dirksen, P., and Schulenburg, H. (2020). Community assembly of the native C. elegans microbiome is influenced by time, substrate, and individual bacterial taxa. Environ. Microbiol. 22, 1265–1279. doi: 10.1111/1462-2920.14932
Joynson, R., Pritchard, L., Osemwekha, E., and Ferry, N. (2017). Metagenomic analysis of the gut microbiome of the common black slug Arion ater in search of novel lignocellulose degrading enzymes. Front. Microbiol. 8:2181. doi: 10.3389/fmicb.2017.02181
Kaya, H. K. (1993). Entomopathogenic nematodes. Annu. Rev. Entomol. 38, 181–206. doi: 10.1146/annurev.en.38.010193.001145
Labbe, R. G. (1991). Clostridium perfringens. J. Assoc. Off. Anal. Chem. 74, 711–714. doi: 10.1093/jaoac/74.4.711
Li, K. (2012). Molecular analysis of intestinal bacterial communities in Cipangopaludina chinensis used in aquatic ecological restorations. Ecol. Eng. 39, 36–39. doi: 10.1016/j.ecoleng.2011.11.013
Li, L. H., Lv, S., Lu, Y., Bi, D. Q., Guo, Y. H., Wu, J. T., et al. (2019). Spatial structure of the microbiome in the gut of Pomacea canaliculata. BMC Microbiol. 19, 1–9. doi: 10.1186/s12866-019-1661-x
Marilyn, S. E. (2008). “Ascaris lumbricoides: a review of its epidemiology and relationship to other infections,” in Annales Nestle (English ed.), Vol. 66, 7–22. doi: 10.1159/000113305
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
McDonnell, R. J., Colton, A. J., Howe, D. K., and Denver, D. R. (2020). Lethality of four species of Phasmarhabditis (Nematoda: Rhabditidae) to the invasive slug, Deroceras reticulatum (Gastropoda: Agriolimacidae) in laboratory infectivity trials. Biol. Control 150:104349. doi: 10.1016/j.biocontrol.2020.104349
Nardo, E. A. B. d., Sindermann, A. B., Grewal, S. K., and Grewal, P. S. (2010). Non-susceptibility of earthworm Eisenia fetida to the Rhabditid nematode Phasmarhabditis hermaphrodita, a biocontrol agent of slugs. Biocontrol Sci. Tech. 14, 93–98. doi: 10.1080/0958315031000151693
Nermut, J., Puza, V., and Mracek, Z. (2014). The effect of different growing substrates on the development and quality of Phasmarhabditis hermaphrodita (Nematoda: Rhabditidae). Biocontrol Sci. Tech. 24, 1026–1038. doi: 10.1080/09583157.2014.915926
Ogier, J. C., Pages, S., Frayssinet, M., and Gaudriault, S. (2020). Entomopathogenic nematode-associated microbiota: from monoxenic paradigm to pathobiome. Microbiome 8, 1–17. doi: 10.1186/s40168-020-00800-5
Rae, R. G., Robertson, J., and Wilson, M. J. (2007a). Susceptibility of indigenous UK earthworms and an invasive pest flatworm to the slug parasitic nematode Phasmarhabditis hermaphrodita. Biocontrol Sci. Tech. 15, 623–626. doi: 10.1080/09583150500086870
Rae, R. G., Tourna, M., and Wilson, M. J. (2010). The slug parasitic nematode Phasmarhabditis hermaphrodita associates with complex and variable bacterial assemblages that do not affect its virulence. J. Invertebr. Pathol. 104, 222–226. doi: 10.1016/j.jip.2010.04.008
Rae, R., Verdun, C., Grewal, P. S., Robertson, J. F., and Wilson, M. J. (2007b). Biological control of terrestrial molluscs using Phasmarhabditis hermaphrodita progress and prospects. Pest Manag. Sci. 63, 1153–1164. doi: 10.1002/ps.1424
Riddle, D., Blumenthal, T., Meyer, B. J., and James, R. (1997). C. Elegans II. 2nd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Robertson, S. L., Smedley, J. G. III, and McClane, B. A. (2010). Identification of a claudin-4 residue important for mediating the host cell binding action of Clostridium perfringens enterotoxin. Infect. Immun. 78, 505–517. doi: 10.1128/IAI.00778-09
Robeson, M. S., O’Rourke, D. R., Kaehler, B. D., Ziemski, M., Dillon, M. R., Foster, J. T., et al. (2020). RESCRIPt: reproducible sequence taxonomy reference database management for the masses. bioRxiv [Preprint]. doi: 10.1101/2020.10.05.326504
Schafer, W. (2016). Nematode nervous systems. Curr. Biol. 26, R955–R959. doi: 10.1016/j.cub.2016.07.044
Schurkman, J., Dodge, C., McDonnell, R., de Ley, I. T., and Dillman, A. R. (2022a). Lethality of Phasmarhabditis spp. (P. hermaphrodita, P. californica, and P. papillosa) nematodes to the grey field slug Deroceras reticulatum on canna lilies in a lath house. Agronomy 12:20. doi: 10.3390/agronomy12010020
Schurkman, J., Tandingan De Ley, I., Anesko, K., Paine, T., McDonnell, R., and Dillman, A. R. (2022b). Distribution of Phasmarhabditis (Nematoda: Rhabditidae) and their gastropod hosts in California plant nurseries and garden centers. Review 13. doi: 10.3389/fpls.2022.856863
Sheehy, L., Cutler, J., Weedall, G. D., and Rae, R. (2022). Microbiome analysis of malacopathogenic nematodes suggests no evidence of a single bacterial symbiont responsible for gastropod mortality. Front. Immunol. 13:878783. doi: 10.3389/fimmu.2022.878783
Tan, L., and Grewal, P. S. (2001a). Infection behavior of the rhabditid nematode Phasmarhabditis hermaphrodita to the grey garden slug Deroceras reticulatum. J. Parasitol. 87, 1349–1354. doi: 10.1645/0022-3395(2001)087[1349:IBOTRN]2.0.CO;2
Tan, L., and Grewal, P. S. (2001b). Pathogenicity of Moraxella osloensis, a bacterium associated with the nematode Phasmarhabditis hermaphrodita, to the slug Deroceras reticulatum. Appl. Environ. Microbiol. 67, 5010–5016. doi: 10.1128/AEM.67.11.5010-5016.2001
Tandingan De Ley, I., Holovachov, O., McDonnell, R. J., Bert, W., Paine, T. D., and De Ley, P. (2016). Description of Phasmarhabditis californica n. sp. and first report of P. papillosa (Nematoda: Rhabditidae) from invasive slugs in the USA. Nematology 18, 175–193. doi: 10.1163/15685411-00002952
Tandingan De Ley, I., McDonnell, R., Lopez, S., Paine, T. D., and De Ley, P. (2014). Phasmarhabditis hermaphrodita (Nematoda: Rhabditidae), a potential biocontrol agent isolated for the first time from invasive slugs from North America. Nematology 16, 1129–1138. doi: 10.1163/15685411-00002838
Tandingan De Ley, I., Schurkman, J., Wilen, C., and Dillman, A. R. (2020). Mortality of the invasive white garden snail Theba pisana exposed to three US isolates of Phasmarhabditis spp (P. hermaphrodita, P. californica, and P. papillosa). PLOS One 15:e0228244. doi: 10.1371/journal.pone.0228244
Thompson, L. R., Sanders, J. G., McDonald, D., Amir, A., Ladau, J., Locey, K. J., et al. (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463. doi: 10.1038/nature24621
Villena, A. M., Morales, C. S., Soto, P. J., and Enciso, H. M. (2010). Bacterial flora in the digestive tract of Helix aspersa muller snails under two breeding systems. Rev. de Investig. Vet. del Peru. 21, 100–105.
Walker, A. J., Glen, D. M., and Shewry, P. R. (1999). Bacteria associated with the digestive system of the slug Deroceras reticulatum are not required for protein digestion. Soil Biol. Biochem. 31, 1387–1394. doi: 10.1016/S0038-0717(99)00054-1
Wang, X. J., Yu, R. C., Luo, X., Zhou, M. J., and Lin, X. T. (2008). Toxin-screening and identification of bacteria isolated from highly toxic marine gastropod Nassarius semiplicatus. Toxicon 52, 55–61. doi: 10.1016/j.toxicon.2008.04.170
Wilson, M. J., Glen, D. M., George, S. K., and Butler, R. C. (1993a). Mass cultivation and storage of the rhabditid nematode Phasmarhabditis hermaphrodita, a biocontrol agent for slugs. Biocontrol Sci. Tech. 3, 513–521. doi: 10.1080/09583159309355307
Wilson, M. J., Glen, D. M., George, S. K., Glen, D. M., and The, S. K. G. (1993b). The rhabditid nematode Phasmarhabditis hermaphrodita as a potential biological control agent for slugs. Biocontrol Sci. Tech. 3, 503–511. doi: 10.1080/09583159309355306
Wilson, M. J., Glen, D. M., George, S. K., and Pearce, J. D. (1995a). Selection of a bacterium for the mass production of Phasmarhabditis hermaphrodita (Nematoda: Rhabditidae) as a biocontrol agent for slugs. Fundam. Appl. Nematol. 18, 419–425.
Wilson, M. J., Glen, S. K., George, S. K., Pearce, J. D., and Wiltshire, C. W. (1994a). Biological control of slugs in winter wheat using the rhabditid nematode Phasmarhabditis hermaphrodita. Ann. Appl. Biol. 125, 377–390. doi: 10.1111/j.1744-7348.1994.tb04978.x
Wilson, M. J., Glen, D. M., Hughes, L. A., Pearce, J. D., and Rodgers, P. B. (1994b). Laboratory tests of the potential of entomopathogenic nematodes for the control of field slugs (Deroceras reticulatum). J. Invertebr. Pathol. 64, 182–187. doi: 10.1016/S0022-2011(94)90100-7
Wilson, M. J., Glen, D. M., Pearce, J. D., and Rodgers, P. B. (1995b). Monoxenic culture of the slug parasite Phasmarhabditis hermaphrodita (Nematoda: Rhabditidae) with different bacteria in liquid and solid phase. Fundam. Appl. Nematol. 18, 159–166.
Keywords: Phasmarhabditis californica, Phasmarhabditis hermaphrodita, Phasmarhabditis papillosa, microbiome, gastropods
Citation: Schurkman J, Liu R, Alavi S, Tandingan De Ley I, Hsiao A and Dillman AR (2022) The Native Microbial Community of Gastropod-Associated Phasmarhabditis Species Across Central and Southern California. Front. Microbiol. 13:903136. doi: 10.3389/fmicb.2022.903136
Received: 23 March 2022; Accepted: 17 May 2022;
Published: 14 July 2022.
Edited by:
Yi Luo, Nankai University, ChinaReviewed by:
Claudia S. L. Vicente, University of Évora, PortugalCopyright © 2022 Schurkman, Liu, Alavi, Tandingan De Ley, Hsiao and Dillman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adler R. Dillman, YWRsZXJkQHVjci5lZHU=; Ansel Hsiao, YW5zZWwuaHNpYW9AdWNyLmVkdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.