
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 15 June 2022
Sec. Aquatic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.900669
Low nucleic acid content (LNA) bacteria are ubiquitous and estimated to constitute 20%–90% of the total bacterial community in marine and freshwater environment. LNA bacteria with unique physiological characteristics, including small cell size and small genomes, can pass through 0.45-μm filtration. The researchers came up with different terminologies for low nucleic acid content bacteria based on different research backgrounds, such as: filterable bacteria, oligotrophic bacteria, and low-DNA bacteria. LNA bacteria have an extremely high level of genetic diversity and play an important role in material circulation in oligotrophic environment. However, the majority of LNA bacteria in the environment remain uncultivated. Thus, an important challenge now is to isolate more LNA bacteria from oligotrophic environments and gain insights into their unique metabolic mechanisms and ecological functions. Here, we reviewed LNA bacteria in aquatic environments, focusing on their characteristics, community structure and diversity, functions, and cultivation strategies. Exciting future prospects for LNA bacteria are also discussed.
Low nucleic acid content (LNA) bacteria exist widely in the natural environments, including oceans, lakes, rivers, springs, groundwater, drinking water, greenland glacier ice core, wetlands and sediments, ranging from 20% to 90% of the total bacterial community in marine and freshwater environment (Cottrell and Kirchman, 2000; Servais et al., 2003; Longnecker et al., 2005, 2006; Miteva and Brenchley, 2005; Suzina et al., 2011; Liu et al., 2017, 2018; Proctor et al., 2018; Eswaran and Khandeparker, 2021). Microbiologists have given different definitions and names to LNA bacteria based on their characteristics, such as filterable bacteria (Wang et al., 2007), low-DNA bacteria (Gasol et al., 1999). In the field of microbiology, filtration through 0.45-μm pore size filters is a common method to isolate and enrich LNA bacteria from aqueous samples (Wang et al., 2009). Here, we define the bacteria with the characters of fewer than 0.1 μm3 in volume, can pass through 0.45-μm microporous membrane, low DNA content showed in flow cytometry (FCM) as LNA bacteria. All bacteria with synonymous names that fall under the definition provided are considered LNA bacteria.
LNA bacteria were initially considered to be inactive, dead, fragment cells (Gasol et al., 1999) and high nucleic acid content (HNA) bacteria were assumed to be the most active group of heterotrophic bacteria in different marine ecosystems (Gasol and Del Giorgio, 2000). With the development of technology, investigators found that LNA bacteria may represent the most active and predominant bacterioplankton community in oligotrophic environments (Zubkov et al., 2001; Jochem et al., 2004; Longnecker et al., 2005). LNA bacteria with a simplified metabolism, efficient substrate uptake system, and special cellular membrane constitution (Liu et al., 2017), can respond quickly to various stressful environment conditions (Ramseier et al., 2011). Thus, it has been proposed that LNA bacteria can be used as bio-indicators to reflect the water quality (Sharuddin et al., 2018; Santos et al., 2019).
LNA bacteria, including species of free-living and parasitic bacteria, have an extremely high level of diversity. Phylogenetic analysis showed that the Alphaproteobacteria SAR11 clade (Mary et al., 2006; Schattenhofer et al., 2011), Gammaproteobacteria SAR86 clade (Zubkov and Sleigh, 1998; Zubkov et al., 2002), Betaproteobacteria, Firmicutes and Fibrobacter (Vila-Costa et al., 2012) were dominant in LNA bacteria populations in marine environments. In freshwater, Actinobacteria Gammaproteobacteria, and Deltaproteobacteria were the dominant phyla in LNA bacterial operational taxonomic units (OTUs; Proctor et al., 2018; Song et al., 2019a). Additionally, it was reported that many LNA bacteria attributed to candidate phyla radiation (CPR) in freshwater ecosystems (Proctor et al., 2018). High-throughput sequencing based on molecular markers and complex metagenomic methods have suggested that LNA bacteria play important roles in the biochemical cycles in marine and freshwater environment (Giovannoni, 2017; Henson et al., 2018; Song et al., 2019a). Whereas the majority of LNA bacteria remain uncultivated under laboratory conditions, the recognition of the physiological growth characteristics and ecological functions of LNA bacteria are still limited.
In this review, we provide an in-depth analysis on LNA bacteria focusing on the following aspects: (1) terminologies of LNA bacteria; (2) the characteristics of LNA bacteria; (3) the community structure and diversity of LNA bacteria; (4) the impact of environmental factors on LNA bacterial community; (5) the functions of LNA bacteria; (6) the cultivation strategy of LNA bacteria.
When considering LNA bacteria, it is important to note the importance of the terms used. Different terminologies and the precise definitions of the terms of small-sized have been widely discussed (Duda et al., 2012; Ghuneim et al., 2018; Nakai, 2020). From the point of view of bacterial species, terminologies can potentially overlap as a single strain can have one or more of these characteristics. Based on the descriptions used in the literature, we summarized the terms for LNA bacteria and their relationships, as shown in Table 1. The terminologies in Table 1 were defined mainly based on three different characteristics of LNA bacteria.
FCM, which is based on fluorescent nucleic acid-staining technologies and independent of microbial cultivability, has been rapidly applied in the field of aquatic microbiology as a useful tool for study LNA bacteria (Wang et al., 2009; Koch et al., 2014). Bacteria in the aquatic environment are generally divided into at least two groups by FCM analysis (Marie et al., 1997). The terms “Group I cells” and “Group II cells” were originally used to label the two groups (Li et al., 1995). Researchers later observed a correlation between fluorescence intensity and DNA/RNA content in the cells, and therefore renamed the two groups as “low-DNA (LDNA) bacteria” and “high-DNA (HDNA) bacteria,” respectively (Gasol et al., 1999). Lebaron et al. (2001) modified the designations of two groups again to “high nucleic acid-content (HNA) bacteria” and “low nucleic acid-content (LNA) bacteria” (Figure 1). Until now, the concept of LNA bacteria has remained confined to the field of FCM and it was a relative concept. Furthermore, scenarios have been proposed in which bacterial cells may switch between LNA and HNA bacteria (Bouvier et al., 2007).
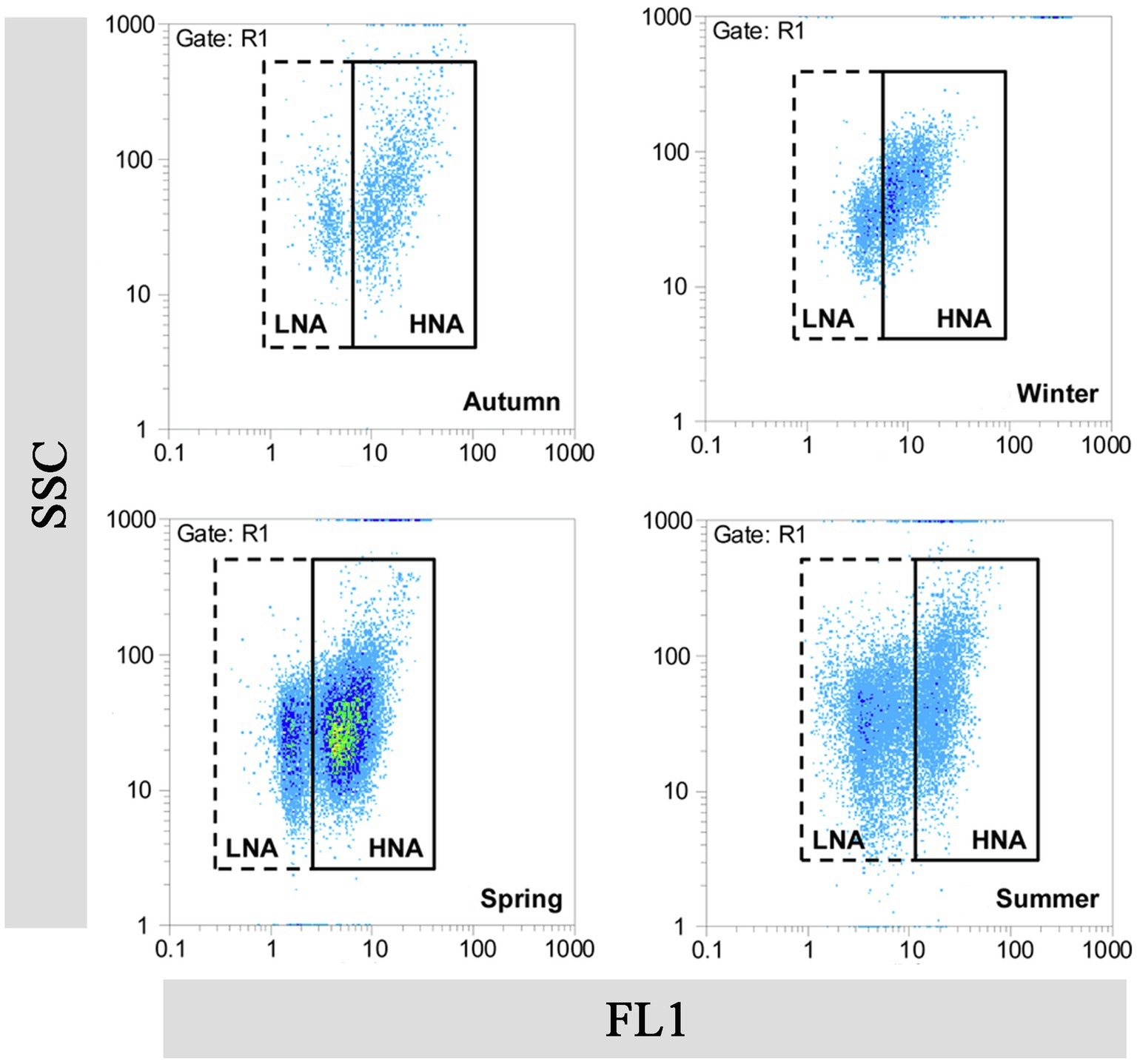
Figure 1. Flow cytogram fingerprint of LNA and HNA bacteria in the Haihe River. This figure is cited from the authors (Liu et al., 2016). FL1, fluorescence intensity; SSC, sideward scatter.
The characteristics of LNA bacteria were linked with the parameters in FCM. These links, including low fluorescence linked to low DNA content, low scatter linked to small cell size, filterability linked the small cell size, small cell sizes linked to small genome sizes and low DNA content, are supported by literature (Veldhuis et al., 1997; Gasol et al., 1999; Lebaron et al., 2002; Wang et al., 2009). When we link the concepts of LNA bacteria that can pass through 0.45-μm filter membrane to small-sized bacteria, such as: ultramicrobacteria, ultramicrocells, ultra-small bacteria, and nano-sized bacteria (Table 1), all these terms are within the scope of the definition of LNA bacteria. Accordingly, LNA bacteria is the most appropriate term to define such bacteria. Although all these names are vaguely defined based on isolated aspects of bacteria or different investigation techniques, they have the same properties, such as filterable (Proctor et al., 2018; Nakai, 2020). Additionally, from the perspective of cell volume, the cell volume of both LNA bacteria and small-sized bacteria is <0.1 μm3 (Table 2).
Wang et al. (2009) found that LNA bacteria can be effectively separated by 0.45-μm membrane filtration and widely exists in oligotrophic environments. Thus, LNA bacteria is also called oligotrophic bacteria and filterable bacteria. This will be described in detail in the following sections.
The most striking property of LNA bacteria is their small cell size. Previous studies have found that the cell sizes of LNA bacterial communities in marine environment ranged from 0.035 to 0.06 μm3 (Huete-Stauffer et al., 2016). The biovolumes of LNA bacteria were lower than that of HNA bacteria in marine environment (García et al., 2014). For example, Morán et al. (2015) reported that the cell size of HNA bacteria (0.032–0.115 μm3; n = 114) was higher than that of LNA bacteria (0.050–0.056 μm3; n = 114). Proctor et al. (2018) found that LNA bacteria had an average diameter of 0.18 μm, an average length of 0.57 μm, and an average volume of 0.016 μm3 (n = 12). Additionally, the small cellular volume (0.009 ± 0.002 μm3) of representative strains such as Candidatus Katanobacteria (WWE3), Candidatus Microgenomates (OP11), and Candidatus Parcubacteria (OD1) were detected by cryogenic transmission electron microscopy (Luef et al., 2015). Ghai et al. (2013) reported that the cellular volume of ultra-small marine Actinobacteria was about 0.013 μm3 as estimated by flow cytometry-fluorescence in situ hybridization (FISH) techniques. The cellular volume of isolated LNA bacteria was generally found to be less than 0.1 μm3 (Table 2).
LNA bacteria not only possess small cellular volumes, but also small genomes (Table 2). Studies found that the DNA content of HNA bacteria was about 3.5 times higher than that of LNA bacteria (Schattenhofer et al., 2011), which can be reflected by the genome sizes of closely-related cultured representatives of HNA clades (3–6 Mb) and LNA clades (1.3–1.5 Mb). It was also reported that the typical representative LNA bacteria have small genome sizes, such as Candidatus Pelagibacter ubique HTCC1062 (1.54 Mb; Giovannoni et al., 2005), Candidatus Fonsibacter ubiquis LSUCC0530 (1.16 Mb; Henson et al., 2018), Candidatus Katanobacteria (WWE3; 0.88 Mb) and Candidatus Parcubacteria (OD1; 0.69 Mb; Kantor et al., 2013; Luef et al., 2015), while the representatives HNA bacteria such as Roseobacter or Flavobacteria have genome sizes ranging from 4.2 to 4.7 Mb (Pinhassi et al., 2006; Kalhoefer et al., 2011). Additionally, Candidatus Dependentiae (TM6), suggested as an LNA bacterial taxa previously had a small genomes (0.5–1.5 Mb; Longnecker et al., 2005; Proctor et al., 2018). And the genomes of SAR86 was modestly sized at 1.25–1.7 Mb (Dupont et al., 2012).
Microporous filtration (0.1–0.45 μm) is commonly used as a disinfection method for thermal agents and experimental solvents, and as a physical method for the removal of microorganisms such as bacteria in drinking water and wastewater treatment (Wang et al., 2007; Lee et al., 2010). The bacteria small enough to pass through a 0.45-μm membrane filter was termed filterable bacteria (Tabor et al., 1981). In the field of microbiology, it is also used to isolate and enrich LNA bacteria (Ma et al., 2019; Song et al., 2019a). Studies found that most of LNA bacteria are able to pass through a 0.45-μm filter while about 90% of the HNA bacteria were retained (Wang et al., 2009). Song et al. (2019a) found that the fraction of LNA bacteria in lake water samples significantly increased after 0.45-μm filtration. Thus, LNA bacteria was also called filterable bacteria. Studies found that the relative abundance of LNA bacteria was increased after filtration (Proctor et al., 2018), 15.53% ± 5.17% and 2.78% ± 0.88% increases for Actinobacteria and Candidatus Parcubacteria in LNA bacteria compared with the total bacteria (Song et al., 2019a).
It is worth noting that laboratory filtration experiments on a variety of different forms of bacteria have shown that the morphology of bacteria (including its flexibility) is the decisive factor in determining the passage of bacteria through the microporous membrane (Wang et al., 2008). Other factors that influence the passage of bacteria through the porous membrane include cell flexibility, microporous filtration rate, and microporous materials. Notably, the bacteria that can pass through the membrane are not necessarily LNA bacteria. There are two possible explanations for this. First, the cell volume and cell morphology of bacteria can change when they are confronted with conditions of low solutes undergo certain changes, which can allow these bacteria to pass through small pores (Tabor et al., 1981; Torrella and Morita, 1981). Second, some groups such as Hylemonella gracilis, due to the absence of a rigid cell wall, can effectively squeeze through small pores (Ghuneim et al., 2018).
The main characteristics of oligotrophic environments are low energy flows and low absolute concentrations of nutrients (Egli, 2010). Nevertheless, such environments still contain 105–106 cells/ml bacteria (Morita, 1988). LNA bacteria are widespread in oligotrophic environments, including ocean, groundwater, tap water and alpine streams, and occupy a high proportion of the microbial communities (Wang et al., 2009). Studies reported that freshwater SAR11 bacteria (LD12) with the characteristics that slow but efficient uptake already at low substrate concentrations, enable them to adapt to oligotrophic habitats (Salcher et al., 2011). Consequently, LNA bacteria in such environments are considered to be “oligotrophic bacteria.” Two main physiological features of LNA bacteria enable them to survive in oligotrophic environments. One is their low growth rate. A previous study showed that the growth rate of Candidatus Pelagibacter ubique HTCC1062 was 0.40–0.58 cell divisions per day does not vary in response to nutrient addition (Giovannoni et al., 2005). Eguchi et al. (1996) also reported that the growth rate (0.13–0.16 h−1 at 23°C) of ultramicrobacteria (Sphingopyxis alaskensis RB2256) remains largely unchanged with carbon concentrations between 0.8 and 800 mg L−1, while the growth rate was 0.02–0.13 h−1 when cultured in glucose-limited media (Ostrowski et al., 2001). The second physiological feature is the adaptable surface-to-volume ratio. LNA bacteria can actively reduce their volume and increase the specific surface area to take up nutrients more efficiently in oligotrophic environments (Duda et al., 2012; Martínez-Cano et al., 2015; Liu et al., 2019).
LNA bacteria are widely distributed in various environments, including oceans, lakes, rivers, springs, groundwater, wetlands, and sediments (Servais et al., 2003; Longnecker et al., 2005, 2006; Suzina et al., 2011; Liu et al., 2017; Proctor et al., 2018; Song et al., 2019a; Eswaran and Khandeparker, 2021). The percentage of LNA bacteria in seawater and freshwater is summarized in Table 3. In marine environment, a relatively low proportion of LNA bacteria was reported in coastal seas, such as in NW Mediterranean coastal waters (42%–47%; Vila-Costa et al., 2012). In contrast, a higher proportion of LNA bacteria in open ocean was appeared than coastal or continental shelf communities (Servais et al., 2003; Longnecker et al., 2005, 2006), including the Southwest Atlantic Ocean (85%, Andrade et al., 2007), the North Atlantic Ocean (71%–85%, Schattenhofer et al., 2011). And in freshwater, the percentage of LNA bacteria ranges from 20% to 90% (generally >50%) of the total bacterial community, which varies with environmental conditions and ecosystem productivity (Bouvier et al., 2007; Wang et al., 2009; Proctor et al., 2018; Song et al., 2019a).
LNA bacteria are the most abundant group of planktonic bacteria. Actinobacteria, Gammaproteobacteria were the dominant phyla, hgcI_clade and pseudomonas were the most abundant bacterial genera in LNA bacterial OTUs (Song et al., 2019a). The rare taxa, such as: Alphaproteobacteria—LD12, SAR11, SAR86, Candidatus Katanobacteria (WWE3), and Candidatus Microgenomates (OP11) were also found in LNA bacteria in freshwater environment (Proctor et al., 2018). In addition, representative bacteria such as Candidatus Parcubacteria (OD1; Nelson and Stegen, 2015), Candidatus Gracilibacteria (GN02: Rinke et al., 2013), Candidatus Saccharibacteria (TM7; He et al., 2015), Candidatus Dependentiae (TM6; McLean et al., 2013), and Candidatus Omnitrophica (OP3; Glöckner et al., 2010) were obtained through gene sequencing in LNA bacteria and cannot yet be cultured in the laboratory (Proctor et al., 2018). In marine environment, LNA bacteria was dominated by the Alphaproteobacterial clade SAR11 (45%–74%), Betaproteobacteria (2%–4%) in the North Atlantic Ocean (Schattenhofer et al., 2011). SAR11 and SAR86 contributed largely to LNA bacteria in NW Mediterranean coastal waters (Vila-Costa et al., 2012). And most of the isolated LNA bacteria belong to Alphaproteobacteria, Betaproteobacteria, and Actinobacteria (Table 2).
So far, the community composition of LNA and HNA bacteria remains controversial. Studies reported that LNA bacteria share the same clades with HNA bacteria in the Mediterranean coast (Servais et al., 2003). However, other studies have claimed that the phylogenetic composition varied between LNA and HNA bacteria. For example, the HNA bacteria was comprised of members of Bacteroidetes (26%–32%), Alphaproteobacteria without SAR11 (19%–25%), and Gammaproteobacteria without SAR86 (12% and 8%) at stations (BPLR and ARCT) in the North Atlantic Ocean (Schattenhofer et al., 2011), whereas the Alphaproteobacterial SAR11 clade (62%–72%) dominated in LNA bacteria (Mary et al., 2006; Schattenhofer et al., 2011). Additionally, a high abundance (>10%) of the Gammaproteobacterial clade SAR86 was reported in LNA bacteria of prokaryotic picoplankton in coastal seas (Zubkov and Sleigh, 1998; Zubkov et al., 2002). Members of Betaproteobacteria, Firmicutes, and Fibrobacter were also found in LNA bacteria (Vila-Costa et al., 2012). Interestingly, researchers found that some bacteria can shift between LNA and HNA bacterial groups in accordance with changes in external conditions. For example, Gammaproteobacteria, Actinobacteria, and Betaproteobacteria were equally likely to appear in both HNA and LNA bacteria in NW Mediterranean coastal waters (Vila-Costa et al., 2012). A similar phenomenon was also reported in freshwater environments where Methylobacteriaceae, Pseudomonas, and Alteromonodaceae were identified as both LNA (Longnecker et al., 2005) and HNA bacteria (Proctor et al., 2018). Additionally, previous studies found that the diversity indices of LNA bacteria were relatively lower than in HNA bacteria in both freshwater and marine environments (Vila-Costa et al., 2012; Song et al., 2019a). This may be due to HNA bacteria being composed of versatile fast growing bacteria (Schattenhofer et al., 2011), whose larger genomes allow them to occupy more ecological niches.
LNA bacteria are sensitive to the living environment. Previous studies found that the distribution pattern of HNA and LNA bacteria in different marine and freshwater ecosystems was controlled by nutrients, substrate availability, pH, temperature, chlorophyll-a concentration and salinity (Joux et al., 2005; Nishimura et al., 2005; Bouvier et al., 2007; Belzile et al., 2008; Salcher et al., 2011; Liu et al., 2017; Song et al., 2019b). The net growth rates of LNA bacteria had a strong negative correlation with chlorophyll-a in temperate coastal waters (Huete-Stauffer and Morán, 2012). The decrease in cell size and nucleic acid content with temperature were also correlated, showing a common mean decrease of 0.4% per °C (Huete-Stauffer et al., 2016). The LNA/HNA ratios can shift with environmental conditions (Sharuddin et al., 2018), which lead to an exchange between LNA and HNA bacteria. The shifts in HNA and LNA bacterial fractions indicate the change in bacterial bulk activities as well as community composition. It was found that pH, total organic carbon (TOC), conductivity, and nitrogen-related concentrations played key roles in shaping LNA bacterial diversity (Song et al., 2019a). In addition, spatiotemporal changes also had significant effects on the cell granularity, DNA content, abundance, the distribution pattern and the community of LNA bacteria (Schattenhofer et al., 2011; Vila-Costa et al., 2012; Eswaran and Khandeparker, 2021).
The predicted metabolic functions of isolated LNA bacteria were investigated using the KEGG database and numerous proteins were annotated in pathways linked to carbohydrate metabolism, energy metabolism and lipid metabolism, which means that LNA bacteria can participate in material circulation in the environment (Figure 2; Giovannoni, 2017; Henson et al., 2018; Song et al., 2019a). In particular, enzymes found in carbohydrate and amino acid metabolism enable LNA bacteria to use small molecular weight compounds such as carboxyl groups and amino acids as a carbon/energy source for survival in oligotrophic environments (Tripp et al., 2008; Sun et al., 2011). Comparative genomic analysis from different LNA bacteria showed that some metabolic pathways missed in the strains (Figure 3). This phenomenon has been reported in previous studies. For example, LD12 lacks the glyoxylate shunt and some single carbon (C1) metabolism. However, the EMP pathway was contained in the genomes of LD12 and was not found in most marine SAR11 (Eiler et al., 2016). Additionally, a complete assimilatory sulfate reduction pathway is missing in Candidatus Fonsibacter ubiquis LSUCC0530 (Henson et al., 2018). In addition, LNA bacteria play an important role in the degradation of organic matter. They had the potential to degrade aromatic compounds in freshwater (Song et al., 2019a) and oxidize many labile, low-molecular-weight organic compounds, including some volatile organic compounds (VOCs) and methylated compounds (Giovannoni, 2017). Ma et al. (2016) found that the typical LNA bacteria, Curvibacter sp. PAE-UM, is capable of degrading phthalate esters under oligotrophic conditions. More functions discussed in this review were predicted by omics techniques, although the concerted effect of the entire LNA bacterial community in ecosystem functions remains unknown. To fully understand the functional characteristics of small-sized bacteria, more small-sized bacteria need to be isolated and cultured.
Figure 2. Analysis of the potential metabolic functions of LNA bacteria and ultramicrobacteria. The protein sequences of bacteria were obtained from NCBI database and annotated in KEGG database. The value in the diagram represents the number of proteins involved in the pathways. HTCC1062: Candidatus Pelagibacter ubique HTCC1062; LSUCC0530: Candidatus Fonsibacter ubiquis LSUCC0530; RB2256: Sphingopyxis alaskensis RB2256; KNCT: Aurantimicrobium minutum KNCT; 15G-AUS-rotT: Aquiluna borgnonia 15G-AUS-rotT; IMCC25003: Candidatus Planktophila rubra IMCC25003; MWH-Mo1T: Aurantimicrobium photophilum MWH-Mo1T.
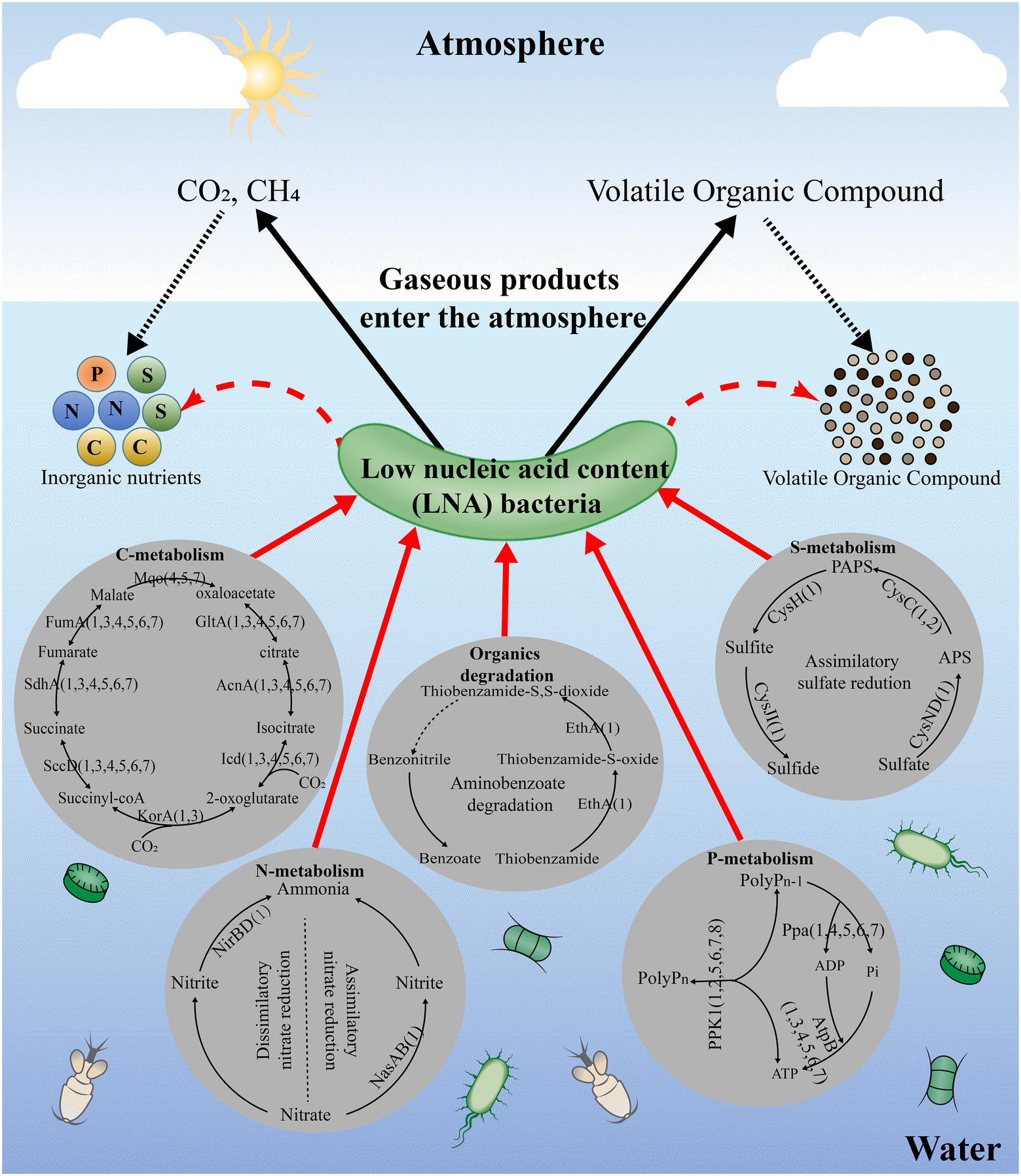
Figure 3. The potential metabolic functions of LNA bacteria and ultramicrobacteria in the aquatic environments. The enzymes in the figure means that can be annotated in pathways of the strains in KEGG and the number in the figure means the strains. All data used for the analysis were obtained from NCBI database; 1: Sphingopyxis alaskensis RB2256; 2: Candidatus Pelagibacter ubique HTCC1062; 3: Candidatus Fonsibacter ubiquis LSUCC0530; 4: Aurantimicrobium minutum KNCT; 5: Aquiluna borgnonia 15G-AUS-rotT; 6: Candidatus Planktophila rubra IMCC25003; 7: Aurantimicrobium photophilum MWH-Mo1T.
Although the unknown metabolic features of uncultured bacteria could be elucidated by metagenomics and single-cell genomics, the data are still difficult to interpret without fundamental knowledge of the underlying bacterial physiology. Without cultivation, many questions about the roles of uncultured bacteria in their natural settings remain unanswered (Lewis et al., 2020). To improve our understanding of the functions of uncultured bacteria, it is essential to increase our capacity for bringing the uncultured bacteria from the environment into culture.
Conventional cultivation methods are laborious, time consuming, and most importantly, selective and biased for the growth of specific microorganisms (Figure 4A; Zengler et al., 2002). Most cells obtained from nature and visualized by microscopy are viable, but the majority of the bacteria do not form visible colonies on plates. To cultivate the LNA bacteria from aquatic environments, many cultivation methods have been tested. First, dilution cultures. Dilution cultures were proposed by Button et al. (1993), who demonstrated that they have advantages for studying oligo-bacteria. This method relies on diluting samples in low nutrient media until the final dilution tube contains the most abundant members of the population which can grow in the media (Duda, 2011). The type strain of ultramicrobacteria named Sphingopyxis alaskensis RB2256 was isolated from Resurrection Bay in Alaska using dilution cultures (Schut et al., 1993). Second, filtration. Filtration was found to be a useful method for the isolation of LNA bacteria from the environments. Different ultramicrobacteria members also have been obtained through the filtration of environmental samples (Nakai, 2020). Additionally, to isolate more uncultured bacteria, researchers have tried to use cultivation media with a composition more suited to their natural environment (Connon and Giovannoni, 2002; Kaeberlein et al., 2002), such as filter-sterilized seawater supplemented with different concentrations of nutrients (Zengler et al., 2002). The advantage of this approach is that the conditions for bacterial growth are as natural as possible, even taking into account any alterations during experimental handling of seawater. Third, FCM. FCM is one approach that has been applied to the selection of non-cultivable microbes. The cell count in the marine and freshwater are barely detectable by conventional techniques. In such cases, FCM with the advantages, such as: being culture-independent, fast, sensitive, and accurate, are the most suitable quantitative techniques to study of non-cultivable microbes in aquatic environments (Wang et al., 2010). Hence, the combination of filtration for dilution cultures, cultivation media, and FCM facilitates the study of non-cultivable microbes in aquatic environments (Figure 4B). Such an approach was successfully applied in the isolation of LNA bacteria from freshwater (Wang et al., 2009), and in the cultivation of opportunistic pathogenic bacteria in oligotrophic aquatic environments (Vital et al., 2007).
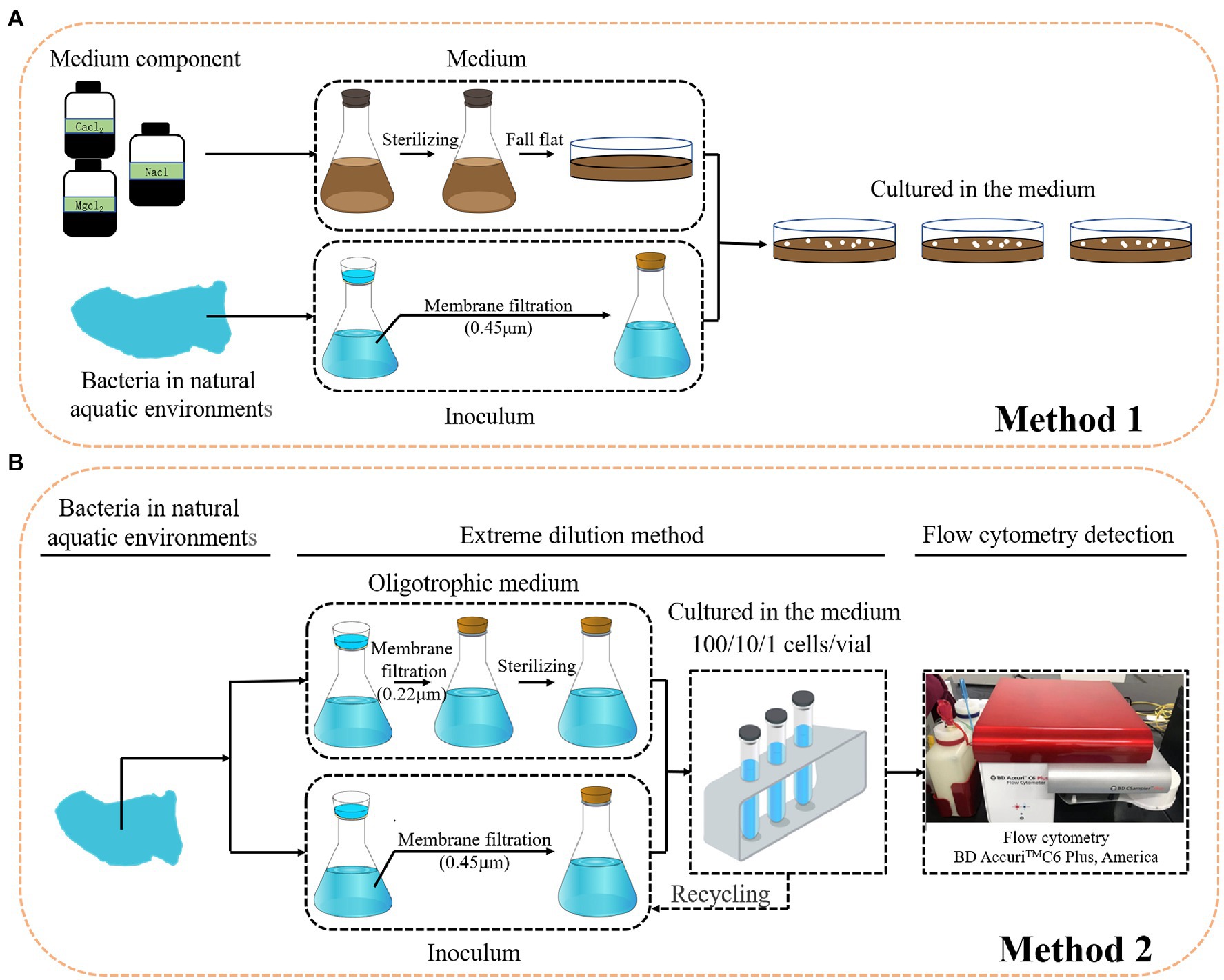
Figure 4. Cultivation strategies for LNA bacteria. (A): conventional cultivation; (B): dilution cultures, filtration and FCM.
A comprehensive understanding of the microbial world undoubtedly requires cultivation, yet the difficulties associated with the isolation and cultivation of oligotrophic bacteria are well documented (Schut et al., 1997). A fundamental problem that limits their cultivation is that obligately oligotrophic bacteria are innately sensitive to nutrients (Cavicchioli et al., 2003). In addition, certain physiological groups of bacteria require specific growth conditions, including resuscitation from dormancy; symbiotic interdependencies; physical contact or spatial proximity; and physicochemical environmental conditions (Lewis et al., 2020). While the key physiological traits for some microorganisms can be revealed through the metatranscriptome and metaproteome, accurately determining microbial features such as growth kinetics, metabolic mechanisms, and physiological properties is incredibly difficult, as these cannot be inferred from genome sequences alone. Thus, it is essential to increase our capacity for bringing microorganisms from the environment into pure culture.
LNA bacteria are an important part of marine microbial community, with the potential to be metabolically active and contributing toward mineralization in the oligotrophic marine environment (Cavicchioli et al., 2003). To some extent, bacteria with a genome size as low as <1 Mb have certain physiological and metabolic defects. For example, several carbon degradation pathways in SAR11 were lost in freshwater habitats (Eiler et al., 2016). Very limited information is available on the mechanism of adaptation by LNA bacteria, especially in oligotrophic environments. Cultivation of representative LNA bacteria is important to reveal their physiological and metabolic properties.
Groups that are highly abundant in an environment are likely to have pivotal ecological roles (Lewis et al., 2020). Many uncultivated microorganisms preserve a lot of new genes with unknown functions under extreme environmental conditions. Although the function of many microorganisms can be predicted by means of omics, these omics approaches cannot replace culture-based phylogenetic and functional studies, since functional prediction relies heavily on the availability of well-annotated genomes from cultures. Therefore, more in-depth investigations are needed to explore the functional properties of small-sized bacteria in the ecosystem.
In this work, the characteristics, biodiversity, and cultivation strategy of LNA bacteria were reviewed. The main conclusions are:
• Various terminologies have been used to define LNA bacteria, and all of these terms are within the scope of the definition of LNA bacteria.
• LNA bacteria not only have a small cell size and small genome, but also have filterable and oligotrophic characteristics.
• LNA bacteria are the main group of planktonic bacteria and are widely distributed in various environments. The distribution pattern and the composition of LNA bacteria can shift with environmental factors.
• Prediction of the metabolic functions of LNA bacteria revealed that LNA bacteria can participate in material cycling in the environment. However, to fully understand the functional characteristics of LNA bacteria, more LNA bacteria need to be isolated and cultured.
• For isolating pure LNA bacteria, a combination of filtration for dilution cultures, cultivation media, and FCM is considered to be effective.
WH: data curation and writing—original draft preparation. HZ: visualization. XL: data curation. RL: software. MB: writing—review and editing. YW: supervision, writing—review and editing, and funding acquisition. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (2020YFC1807000), the National Natural Science Foundation (31870485), the Tianjin Natural Science Foundation (19JCZDJC39600), and 111 program, Ministry of Education, China (T2017002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Andrade, L., Gonzalez, A. M., Rezende, C. E., Suzuki, M., Valentin, J. L., and Paranhos, R. (2007). Distribution of HNA and LNA bacterial groups in the Southwest Atlantic Ocean. Braz. J. Microbiol. 38, 330–336. doi: 10.1590/S1517-83822007000200028
Belzile, C., Brugel, S., Nozais, C., Gratton, Y., and Demers, S. (2008). Variations of the abundance and nucleic acid content of heterotrophic bacteria in Beaufort shelf waters during winter and spring. J. Mar. Syst. 74, 946–956. doi: 10.1016/j.jmarsys.2007.12.010
Bouvier, T., Del Giorgio, P. A., and Gasol, J. M. (2007). A comparative study of the cytometric characteristics of high and low nucleic-acid bacterioplankton cells from different aquatic ecosystems. Environ. Microbiol. 9, 2050–2066. doi: 10.1111/j.1462-2920.2007.01321.x
Button, D. K., Schut, F., Quang, P., Martin, R., and Robertson, B. R. (1993). Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59, 881–891. doi: 10.1128/aem.59.3.881-891.1993
Cavicchioli, R., Ostrowski, M., Fegatella, F., Goodchild, A., and Guixa-Boixereu, N. (2003). Life under nutrient limitation in oligotrophic marine environments: an eco/physiological perspective of Sphingopyxis alaskensis (formerly Sphingomonas alaskensis). Microb. Ecol. 45, 203–217. doi: 10.1007/s00248-002-3008-6
Connon, S. A., and Giovannoni, S. J. (2002). High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68, 3878–3885. doi: 10.1128/AEM.68.8.3878-3885.2002
Cottrell, M. T., and Kirchman, D. L. (2000). Natural assemblages of marine proteobacteria and members of the Cytophaga-flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66, 1692–1697. doi: 10.1128/AEM.66.4.1692-1697.2000
Duda, V. I., Suzina, N. E., Esikova, T. Z., Akimov, V. N., Oleinikov, R. R., Polivtseva, V. N., et al. (2009). A cytological characterization of the parasitic action of ultramicrobacteria NF1 and NF3 of the genus Kaistia on chemoorganotrophic and phototrophic bacteria. FEMS Microbiol. Ecol. 69, 180–193. doi: 10.1111/j.1574-6941.2009.00696.x
Duda, V. I., Suzina, N. E., Polivtseva, V. N., and Boronin, A. M. (2012). Ultramicrobacteria: formation of the concept and contribution of ultramicrobacteria to biology. Microbiology 81, 379–390. doi: 10.1134/S0026261712040054
Dupont, C. L., Rusch, D. B., Yooseph, S., Lombardo, M. J., Alexander Richter, R., Valas, R., et al. (2012). Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 6, 1186–1199. doi: 10.1038/ismej.2011.189
Egli, T. (2010). How to live at very low substrate concentration. Water Res. 44, 4826–4837. doi: 10.1016/j.watres.2010.07.023
Eguchi, M., Nishikawa, T., Macdonald, K., Cavicchioli, R., Gottschal, J. C., and Kjelleberg, S. (1996). Responses to stress and nutrient availability by the marine ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 62, 1287–1294. doi: 10.1128/aem.62.4.1287-1294.1996
Eiler, A., Mondav, R., Sinclair, L., Fernandez-Vidal, L., Scofield, D. G., Schwientek, P., et al. (2016). Tuning fresh: radiation through rewiring of central metabolism in streamlined bacteria. ISME J. 10, 1902–1914. doi: 10.1038/ismej.2015.260
Eswaran, R., and Khandeparker, L. (2021). Ecological relevance of high and low nucleic acid content bacteria in a monsoon influenced tropical estuary: implications on the β-glucosidase activity. Acta Oecol. 110:103697. doi: 10.1016/j.actao.2020.103697
García, F. C., López-Urrutia, Á., and Morán, X. A. G. (2014). Automated clustering of heterotrophic bacterioplankton in flow cytometry data. Aquat. Microb. Ecol. 72, 175–185. doi: 10.3354/ame01691
Gasol, J. M., and Del Giorgio, P. A. (2000). Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64, 197–224. doi: 10.3989/scimar.2000.64n2197
Gasol, J. M., Li Zweifel, U., Peters, F., Fuhrman, J. A., and Hagström, Å. (1999). Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl. Environ. Microbiol. 65, 4475–4483. doi: 10.1128/aem.65.10.4475-4483.1999
Ghai, R., Mizuno, C. M., Picazo, A., Camacho, A., and Rodriguez-Valera, F. (2013). Metagenomics uncovers a new group of low GC and ultra-small marine Actinobacteria. Sci. Rep. 3, 1–8. doi: 10.1038/srep02471
Ghuneim, L. A. J., Jones, D. L., Golyshin, P. N., and Golyshina, O. V. (2018). Nano-sized and filterable bacteria and archaea: biodiversity and function. Front. Microbiol. 9:1971. doi: 10.3389/fmicb.2018.01971
Giovannoni, S. J. (2017). SAR11 bacteria: The most abundant plankton in the oceans. Ann. Rev. Mar. Sci. 9, 231–255. doi: 10.1146/annurev-marine-010814-015934
Giovannoni, S. J., Tripp, H. J., Givan, S., Podar, M., Vergin, K. L., Baptista, D., et al. (2005). Genetics: genome streamlining in a cosmopolitan oceanic bacterium. Science 309, 1242–1245. doi: 10.1126/science.1114057
Glöckner, J., Kube, M., Shrestha, P. M., Weber, M., Glöckner, F. O., Reinhardt, R., et al. (2010). Phylogenetic diversity and metagenomics of candidate division OP3. Environ. Microbiol. 12, 1218–1229. doi: 10.1111/j.1462-2920.2010.02164.x
Hahn, M. W., Koll, U., Schmidt, J., Huymann, L. R., Karbon, G., and Lang, E. (2018). Polynucleobacter hirudinilacicola sp. Nov. and polynucleobacter campilacus sp. nov., both isolated from freshwater systems. Int. J. Syst. Evol. Microbiol. 68, 2593–2601. doi: 10.1099/ijsem.0.002880
Hahn, M. W., Lang, E., Brandt, U., Lu, H., and Victoria, L. (2010). Free-living planktonic bacteria inhabiting freshwater lakes and rivers. Int. J. Syst. Evol. Microbiol. 60, 166–173. doi: 10.1099/ijs.0.010595-0
Hahn, M. W., Lünsdorf, H., Wu, Q., Schauer, M., Höfle, M. G., Boenigk, J., et al. (2003). Isolation of novel ultramicrobacteria classified as Actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69, 1442–1451. doi: 10.1128/AEM.69.3.1442-1451.2003
Hahn, M. W., Pitt, A., Koll, U., Schmidt, J., Maresca, J. A., and Neumann-Schaal, M. (2021). Aurantimicrobium photophilum sp. Nov., a non-photosynthetic bacterium adjusting its metabolism to the diurnal light cycle and reclassification of cryobacterium mesophilum as terrimesophilobacter mesophilus gen. Nov., comb. nov. Int. J. Syst. Evol. Microbiol. 71:004975. doi: 10.1099/ijsem.0.004975
Hahn, M. W., Schmidt, J., Asiyo, G. S., Kyrpides, N. C., Woyke, T., and Whitman, W. B. (2017). Reclassification of a Polynucleobacter cosmopolitanus strain isolated from tropical Lake victoria as Polynucleobacter victoriensis sp. Nov. Int. J. Syst. Evol. Microbiol. 67, 5087–5093. doi: 10.1099/ijsem.0.002421
Hahn, M. W., Schmidt, J., Taipale, S. J., Ford Doolittle, W., and Koll, U. (2014). Rhodoluna lacicola gen. Nov., sp. nov., a planktonic freshwater bacterium with stream-lined genome. Int. J. Syst. Evol. Microbiol. 64, 3254–3263. doi: 10.1099/ijs.0.065292-0
He, X., McLean, J. S., Edlund, A., Yooseph, S., Hall, A. P., Liu, S. Y., et al. (2015). Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl. Acad. Sci. U. S. A. 112, 244–249. doi: 10.1073/pnas.1419038112
Henson, M. W., Lanclos, V. C., Faircloth, B. C., and Thrash, J. C. (2018). Cultivation and genomics of the first freshwater SAR11 (LD12) isolate. ISME J. 12, 1846–1860. doi: 10.1038/s41396-018-0092-2
Hoetzinger, M., Schmidt, J., Pitt, A., Koll, U., Lang, E., and Hahn, M. W. (2019). Polynucleobacter paneuropaeus sp. nov., characterized by six strains isolated from freshwater lakes located along a 3000 km north-south cross-section across Europe. Int. J. Syst. Evol. Microbiol. 69, 203–213. doi: 10.1099/ijsem.0.003130
Huete-Stauffer, T. M., Arandia-Gorostidi, N., Alonso-Sáez, L., and Morán, X. A. G. (2016). Experimental warming decreases the average size and nucleic acid content of marine bacterial communities. Front. Microbiol. 7:730. doi: 10.3389/fmicb.2016.00730
Huete-Stauffer, T. M., and Morán, X. A. G. (2012). Dynamics of heterotrophic bacteria in temperate coastal waters: similar net growth but different controls in low and high nucleic acid cells. Aquat. Microb. Ecol. 67, 211–223. doi: 10.3354/ame01590
Jochem, F. J., Lavrentyev, P. J., and First, M. R. (2004). Growth and grazing rates of bacteria groups with different apparent DNA content in the Gulf of Mexico. Mar. Biol. 145, 1213–1225. doi: 10.1007/s00227-004-1406-7
Joux, F., Servais, P., Naudin, J. J., Lebaron, P., Oriol, L., and Courties, C. (2005). Distribution of picophytoplankton and bacterioplankton along a river plume gradient in the Mediterranean Sea. Vie Milieu. 3, 301–311. doi: 10.1016/0923-5965(91)90036-2
Kaeberlein, T., Lewis, K., and Epstein, S. S. (2002). Isolating “uncultivabte” microorganisms in pure culture in a simulated natural environment. Science 296, 1127–1129. doi: 10.1126/science.1070633
Kalhoefer, D., Thole, S., Voget, S., Lehmann, R., and Brinkhoff, T. (2011). Comparative genome analysis and genome-guided physiological analysis of Roseobacter litoralis. BMC Genomics 12:324. doi: 10.1186/1471-2164-12-324
Kantor, R. S., Wrighton, K. C., Handley, K. M., Sharon, I., Hug, L. A., Castelle, C. J., et al. (2013). Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio 4, e00708–e00713. doi: 10.1128/mBio.00708-13
Kim, S., Kang, I., Seo, J. H., and Cho, J. C. (2019). Culturing the ubiquitous freshwater actinobacterial acI lineage by supplying a biochemical ‘helper’ catalase. ISME J. 13, 2252–2263. doi: 10.1038/s41396-019-0432-x
Koch, C., Harnisch, F., Schröder, U., and Müller, S. (2014). Cytometric fingerprints: evaluation of new tools for analyzing microbial community dynamics. Front. Microbiol. 5:273. doi: 10.3389/fmicb.2014.00273
Lebaron, P., Servais, P., Agogué, H., Courties, C., and Joux, F. (2001). Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active cells and inactive cells in aquatic systems? Appl. Environ. Microbiol. 67, 1775–1782. doi: 10.1128/AEM.67.4.1775-1782.2001
Lebaron, P., Servais, P., Baudoux, A. C., Bourrain, M., Courties, C., and Parthuisot, N. (2002). Variations of bacterial-specific activity with cell size and nucleic acid content assessed by flow cytometry. Aquat. Microb. Ecol. 28, 131–140. doi: 10.3354/ame028131
Lee, A., Mcvey, J., Faustino, P., Lute, S., Sweeney, N., Pawar, V., et al. (2010). Use of Hydrogenophaga pseudoflava penetration to quantitatively assess the impact of filtration parameters for 0.2-micrometer-pore-size filters. Appl. Environ. Microbiol. 76, 695–700. doi: 10.1128/AEM.01825-09
Lewis, W. H., Tahon, G., Geesink, P., Sousa, D. Z., and Ettema, T. J. G. (2020). Innovations to culturing the uncultured microbial majority. Nat. Rev. Microbiol. 19, 225–240. doi: 10.1038/s41579-020-00458-8
Li, W. K. W., Jellett, J. F., and Dickie, P. M. (1995). DNA distributions in planktonic bacteria stained with TOTO or TO-PRO. Limnol. Oceanogr. 40, 1485–1495. doi: 10.4319/lo.1995.40.8.1485
Liu, J., Hao, Z., Ma, L., Ji, Y., Bartlam, M., and Wang, Y. (2016). Spatio-temporal variations of high and low nucleic acid content bacteria in an exorheic river. PLoS One 11:e0153678. doi: 10.1371/journal.pone.0153678
Liu, J., Li, B., Wang, Y., Zhang, G., Jiang, X., and Li, X. (2019). Passage and community changes of filterable bacteria during microfiltration of a surface water supply. Environ. Int. 131:104998. doi: 10.1016/j.envint.2019.104998
Liu, J., Ma, D., Ma, L., Song, Y., Gao, G., and Wang, Y. (2017). Geographic distribution pattern of low and high nucleic acid content bacteria on a river-catchment scale. Mar. Freshw. Res. 68:1618. doi: 10.1071/mf16068
Liu, J., Zhao, R., Zhang, J., Zhang, G., Yu, K., Li, X., et al. (2018). Occurrence and fate of ultramicrobacteria in a full-scale drinking water treatment plant. Front. Microbiol. 9:2922. doi: 10.3389/fmicb.2018.02922
Longnecker, K., Sherr, B. F., and Sherr, E. B. (2005). Activity and phylogenetic diversity of bacterial cells with high and low nucleic acid content and electron transport system activity in an upwelling ecosystem. Appl. Environ. Microbiol. 71, 7737–7749. doi: 10.1128/AEM.71.12.7737-7749.2005
Longnecker, K., Sherr, B. F., and Sherr, E. B. (2006). Variation in cell-specific rates of leucine and thymidine incorporation by marine bacteria with high and with low nucleic acid content off the Oregon coast. Aquat. Microb. Ecol. 43, 113–125. doi: 10.3354/ame043113
Loveland-Curtze, J., Miteva, V., and Brenchley, J. (2010). Novel ultramicrobacterial isolates from a deep Greenland ice core represent a proposed new species, Chryseobacterium greenlandense sp. nov. Extremophiles 14, 61–69. doi: 10.1007/s00792-009-0287-6
Luef, B., Frischkorn, K. R., Wrighton, K. C., Holman, H. Y. N., Birarda, G., Thomas, B. C., et al. (2015). Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 6:6372. doi: 10.1038/ncomms7372
Ma, D., Hao, Z. Y., Sun, R., Bartlam, M., and Wang, Y. (2016). Genome sequence of a typical ultramicrobacterium, Curvibacter sp. strain PAE-UM, capable of phthalate ester degradation. Genome Announc. 4, 1–2. doi: 10.1128/genomeA.01510-15
Ma, L., Li, B., and Zhang, T. (2019). New insights into antibiotic resistome in drinking water and management perspectives: A metagenomic based study of small-sized microbes. Water Res. 152, 191–201. doi: 10.1016/j.watres.2018.12.069
Marie, D., Partensky, F., Jacquet, S., and Vaulot, D. (1997). Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR green I. Appl. Environ. Microbiol. 63, 186–193. doi: 10.1128/aem.63.1.186-193.1997
Martínez-Cano, D. J., Reyes-Prieto, M., Martínez-Romero, E., Partida-Martínez, L. P., Latorre, A., Moya, A., et al. (2015). Evolution of small prokaryotic genomes. Front. Microbiol. 5:742. doi: 10.3389/fmicb.2014.00742
Mary, I., Heywood, J. L., Fuchs, B. M., Amann, R., Tarran, G. A., Burkill, P. H., et al. (2006). SAR11 dominance among metabolically active low nucleic acid bacterioplankton in surface waters along an Atlantic meridional transect. Aquat. Microb. Ecol. 45, 107–113. doi: 10.3354/ame045107
McLean, J. S., Lombardo, M. J., Badger, J. H., Edlund, A., Novotny, M., Yee-Greenbaum, J., et al. (2013). Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc. Natl. Acad. Sci. U. S. A. 110, E2390–E2399. doi: 10.1073/pnas.1219809110
Miteva, V. I., and Brenchley, J. E. (2005). Detection and isolation of ultrasmall microorganisms from a 120,000-year-old Greenland glacier ice core. Appl. Environ. Microbiol. 71, 7806–7818. doi: 10.1128/AEM.71.12.7806-7818.2005
Morán, X. A. G., Alonso-Sáez, L., Nogueira, E., Ducklow, H. W., González, N., López-Urrutia, Á., et al. (2015). More, smaller bacteria in response to ocean’s warming? Proc. R. Soc. B Biol. Sci. 282:20150371. doi: 10.1098/rspb.2015.0371
Morita, R. Y. (1988). Bioavailability of energy and its relationship to growth and starvation survival in nature. Can. J. Microbiol. 34, 436–441. doi: 10.1139/m88-076
Nakai, R. (2020). Size matters: ultra-small and filterable microorganisms in the environment. Microbes Environ. 35, 1–11. doi: 10.1264/jsme2.ME20025
Nakai, R., Baba, T., Niki, H., Nishijima, M., and Naganuma, T. (2015). Aurantimicrobium minutum gen. Nov., sp. nov., a novel ultramicrobacterium of the family Microbacteriaceae, isolated from river water. Int. J. Syst. Evol. Microbiol. 65, 4072–4079. doi: 10.1099/ijsem.0.000541
Nakai, R., Fujisawa, T., Nakamura, Y., Nishide, H., Uchiyama, I., Baba, T., et al. (2016). Complete genome sequence of Aurantimicrobium minutum type strain KNCT, a planktonic ultramicrobacterium isolated from river water. Genome Announc. 4, 15–16. doi: 10.1128/genomeA.00616-16
Nelson, W. C., and Stegen, J. C. (2015). The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front. Microbiol. 6:713. doi: 10.3389/fmicb.2015.00713
Nishimura, Y., Kim, C., and Nagata, T. (2005). Vertical and seasonal variations of bacterioplankton subgroups with different nucleic acid contents: possible regulation by phosphorus. Appl. Environ. Microbiol. 71, 5828–5836. doi: 10.1128/AEM.71.10.5828-5836.2005
Ostrowski, M., Cavicchioli, R., Blaauw, M., and Gottschal, J. C. (2001). Specific growth rate plays a critical role in hydrogen peroxide resistance of the marine oligotrophic ultramicrobacterium Sphingomonas alaskensis strain RB2256. Appl. Environ. Microbiol. 67, 1292–1299. doi: 10.1128/AEM.67.3.1292-1299.2001
Pinhassi, J., Bowman, J. P., Nedashkovskaya, O. I., Lekunberri, I., Gomez-Consarnau, L., and Pedrós-Alió, C. (2006). Leeuwenhoekiella blandensis sp. nov., a genome-sequenced marine member of the family Flavobacteriaceae. Int. J. Syst. Evol. Microbiol. 56, 1489–1493. doi: 10.1099/ijs.0.64232-0
Pitt, A., Schmidt, J., Koll, U., and Hahn, M. W. (2019). Rhodoluna limnophila sp. Nov., a bacterium with 1.4 Mbp genome size isolated from freshwater habitats located in Salzburg, Austria. Int. J. Syst. Evol. Microbiol. 69, 3946–3954. doi: 10.1099/ijsem.0.003720
Pitt, A., Schmidt, J., Koll, U., and Hahn, M. W. (2021). Aquiluna borgnonia gen. Nov., sp. nov., a member of a Microbacteriaceae lineage of freshwater bacteria with small genome sizes. Int. J. Syst. Evol. Microbiol. 71:004825. doi: 10.1099/ijsem.0.004825
Proctor, C. R., Besmer, M. D., Langenegger, T., Beck, K., Walser, J. C., Ackermann, M., et al. (2018). Phylogenetic clustering of small low nucleic acid-content bacteria across diverse freshwater ecosystems. ISME J. 12, 1344–1359. doi: 10.1038/s41396-018-0070-8
Ramseier, M. K., von Gunten, U., Freihofer, P., and Hammes, F. (2011). Kinetics of membrane damage to high (HNA) and low (LNA) nucleic acid bacterial clusters in drinking water by ozone, chlorine, chlorine dioxide, monochloramine, ferrate(VI), and permanganate. Water Res. 45, 1490–1500. doi: 10.1016/j.watres.2010.11.016
Rinke, C., Schwientek, P., Sczyrba, A., Ivanova, N. N., Anderson, I. J., Cheng, J. F., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437. doi: 10.1038/nature12352
Sahin, N., Gonzalez, J. M., Iizuka, T., and Hill, J. E. (2010). Characterization of two aerobic ultramicrobacteria isolated from urban soil and a description of Oxalicibacterium solurbis sp. nov. FEMS Microbiol. Lett. 307, 25–29. doi: 10.1111/j.1574-6968.2010.01954.x
Salcher, M. M., Pernthaler, J., and Posch, T. (2011). Seasonal bloom dynamics and ecophysiology of the freshwater sister clade of SAR11 bacteria “that rule the waves” (LD12). ISME J. 5, 1242–1252. doi: 10.1038/ismej.2011.8
Santos, M., Oliveira, H., Pereira, J. L., Pereira, M. J., Gonçalves, F. J. M., and Vidal, T. (2019). Flow cytometry analysis of low/high DNA content (LNA/HNA) bacteria as bioindicator of water quality evaluation. Ecol. Indic. 103, 774–781. doi: 10.1016/j.ecolind.2019.03.033
Schattenhofer, M., Wulf, J., Kostadinov, I., Glöckner, F. O., Zubkov, M. V., and Fuchs, B. M. (2011). Phylogenetic characterisation of picoplanktonic populations with high and low nucleic acid content in the North Atlantic Ocean. Syst. Appl. Microbiol. 34, 470–475. doi: 10.1016/j.syapm.2011.01.008
Schut, F., Gottschal, J. C., and Prins, R. A. (1997). Isolation and characterisation of the marine ultramicrobacterium Sphingomonas sp. strain RB2256. FEMS Microbiol. Rev. 20, 363–369. doi: 10.1111/j.1574-6976.1997.tb00321.x
Schut, F., Vries, D. C. J., Gottschal, R. B., Robertson, B. R., Harder, W., Prins, R. A., et al. (1993). Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 111, 115–122. doi: 10.1016/0378-1097(93)90191-4
Servais, P., Casamayor, E. O., Courties, C., Catala, P., Parthuisot, N., and Lebaron, P. (2003). Activity and diversity of bacterial cells with high and low nucleic acid content. Aquat. Microb. Ecol. 33, 41–51. doi: 10.3354/ame033041
Sharuddin, S. S., Ramli, N., Mohd-Nor, D., Hassan, M. A., Maeda, T., Shirai, Y., et al. (2018). Shift of low to high nucleic acid bacteria as a potential bioindicator for the screening of anthropogenic effects in a receiving river due to palm oil mill effluent final discharge. Ecol. Indic. 85, 79–84. doi: 10.1016/j.ecolind.2017.10.020
Song, Y., Mao, G., Gao, G., Bartlam, M., and Wang, Y. (2019b). Structural and functional changes of groundwater bacterial community during temperature and pH disturbances. Microb. Ecol. 78, 428–445. doi: 10.1007/s00248-019-01333-7
Song, Y., Wang, Y., Mao, G., Gao, G., and Wang, Y. (2019a). Impact of planktonic low nucleic acid-content bacteria to bacterial community structure and associated ecological functions in a shallow lake. Sci. Total Environ. 658, 868–878. doi: 10.1016/j.scitotenv.2018.12.274
Sun, J., Steindler, L., Thrash, J. C., Halsey, K. H., Smith, D. P., Carter, A. E., et al. (2011). One carbon metabolism in SAR11 pelagic marine bacteria. PLoS One 6:e23973. doi: 10.1371/journal.pone.0023973
Suzina, N. E., Duda, V. I., Esikova, T. Z., Shorokhova, A. P., Gafarov, A. B., Oleinikov, R. R., et al. (2011). Novel ultramicrobacteria, strains NF4 and NF5, of the genus Chryseobacterium: facultative epibionts of bacillus subtilis. Microbiology 80, 535–548. doi: 10.1134/S0026261711040187
Tabor, P. S., Ohwada, K., and Colwell, R. R. (1981). Filterable marine bacteria found in the deep sea: distribution, taxonomy, and response to starvation. Microb. Ecol. 7, 67–83. doi: 10.1007/BF02010479
Torrella, F., and Morita, R. Y. (1981). Microcultural study of bacterial size changes and microcolony and ultramicrocolony formation by heterotropic bacteria in seawater. Appl. Environ. Microbiol. 41, 518–527. doi: 10.1128/aem.41.2.518-527.1981
Tripp, H. J., Kitner, J. B., Schwalbach, M. S., Dacey, J. W. H., Wilhelm, L. J., and Giovannoni, S. J. (2008). SAR11 marine bacteria require exogenous reduced Sulphur for growth. Nature 452, 741–744. doi: 10.1038/nature06776
Van Wambeke, F., Catala, P., Pujo-Pay, M., and Lebaron, P. (2011). Vertical and longitudinal gradients in HNA-LNA cell abundances and cytometric characteristics in the Mediterranean Sea. Biogeosciences 8, 1853–1863. doi: 10.5194/bg-8-1853-2011
Veldhuis, M. J. W., Cucci, T. L., and Sieracki, M. E. (1997). Cellular DNA content of marine phytoplankton using two new fluorochromes: taxonomic and ecological implications. J. Phycol. 33, 527–541. doi: 10.1111/j.0022-3646.1997.00527.x
Vila-Costa, M., Gasol, J. M., Sharma, S., and Moran, M. A. (2012). Community analysis of high- and low-nucleic acid-containing bacteria in NW Mediterranean coastal waters using 16S rDNA pyrosequencing. Environ. Microbiol. 14, 1390–1402. doi: 10.1111/j.1462-2920.2012.02720.x
Vital, M., Fuchslin, H. P., Hammes, F., and Egli, T. (2007). Growth of vibrio cholerae O1 ogawa eltor in freshwater. Microbiology 153, 1993–2001. doi: 10.1099/mic.0.2006/005173-0
Wang, Y., Hammes, F., Boon, N., Chami, M., and Egli, T. (2009). Isolation and characterization of low nucleic acid (LNA)-content bacteria. ISME J. 3, 889–902. doi: 10.1038/ismej.2009.46
Wang, Y., Hammes, F., Boon, N., and Egli, T. (2007). Quantification of the filterability of freshwater bacteria through 0.45, 0.22, and 0.1 μm pore size filters and shape-dependent enrichment of filterable bacterial communities. Environ. Sci. Technol. 41, 7080–7086. doi: 10.1021/es0707198
Wang, Y., Hammes, F., De Roy, K., Verstraete, W., and Boon, N. (2010). Past, present and future applications of flow cytometry in aquatic microbiology. Trends Biotechnol. 28, 416–424. doi: 10.1016/j.tibtech.2010.04.006
Wang, Y., Hammes, F., and Egli, T. (2008). The impact of industrial-scale cartridge filtration on the native microbial communities from groundwater. Water Res. 42, 4319–4326. doi: 10.1016/j.watres.2008.07.027
Zengler, K., Toledo, G., Rappé, M., Elkins, J., Mathur, E. J., Short, J. M., et al. (2002). Cultivating the uncultured. Proc. Natl. Acad. Sci. U. S. A. 99, 15681–15686. doi: 10.1073/pnas.252630999
Zubkov, M. V., Fuchs, B. M., Burkill, P. H., and Amann, R. (2001). Comparison of cellular and biomass specific activities of dominant bacterioplankton groups in stratified waters of the Celtic Sea. Appl. Environ. Microbiol. 67, 5210–5218. doi: 10.1128/aem.67.11.5210-5218.2001
Zubkov, M. V., Fuchs, B. M., Tarran, G. A., Burkill, P. H., and Amann, R. (2002). Mesoscale distribution of dominant bacterioplankton groups in the northern North Sea in early summer. Aquat. Microb. Ecol. 29, 135–144. doi: 10.3354/ame029135
Zubkov, M. V., and Sleigh, M. A. (1998). Picoplanktonic community structure on an Atlantic transect from 50 degrees N to 20 degrees S. Deep. Res. Part I Oceanogr. Res. Pap. 45:1339.
Keywords: physical characteristics, terminologies, diversity, functions, cultivation strategy, LNA bacteria
Citation: Hu W, Zhang H, Lin X, Liu R, Bartlam M and Wang Y (2022) Characteristics, Biodiversity, and Cultivation Strategy of Low Nucleic Acid Content Bacteria. Front. Microbiol. 13:900669. doi: 10.3389/fmicb.2022.900669
Received: 21 March 2022; Accepted: 24 May 2022;
Published: 15 June 2022.
Edited by:
Martin W. Hahn, University of Innsbruck, AustriaReviewed by:
Ryosuke Nakai, National Institute of Advanced Industrial Science and Technology (AIST), JapanCopyright © 2022 Hu, Zhang, Lin, Liu, Bartlam and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingying Wang, d2FuZ3l5QG5hbmthaS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.