
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 May 2022
Sec. Aquatic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.900234
The small protein B (SmpB), previously acting as a ribosome rescue factor for translation quality control, is required for cell viability in bacteria. Here, our study reveals that SmpB possesses new function which regulates the expression of outer membrane protein A (ompA) gene as a transcription factor in Aeromonas veronii. The deletion of SmpB caused the lower transcription expression of ompA by Quantitative Real-Time PCR (qPCR). Electrophoretic mobility shift assay (EMSA) and DNase I Footprinting verified that the SmpB bound at the regions of −46 to −28 bp, −18 to +4 bp, +21 to +31 bp, and +48 to +59 bp of the predicted ompA promoter (PompA). The key sites C52AT was further identified to interact with SmpB when PompA was fused with enhanced green fluorescent protein (EGFP) and co-transformed with SmpB expression vector for the fluorescence detection, and the result was further confirmed in microscale thermophoresis (MST) assays. Besides, the amino acid sites G11S, F26I, and K152 in SmpB were the key sites for binding to PompA. In order to further develop peptide antimicrobial agents, the peptide aptamer PA3 was screened from the peptide aptamer (PA) library by bacterial two-hybrid method. The drug sensitivity test showed that PA3 effectively inhibited the growth of A. veronii. In summary, these results demonstrated that OmpA was a good drug target for A. veronii, which was regulated by the SmpB protein and the selected peptide aptamer PA3 interacted with OmpA protein to disable SmpB-OmpA signal pathway and inhibited A. veronii, suggesting that it could be used as an antimicrobial agent for the prevention and treatment of pathogens.
Aeromonas veronii is a serious pathogen which is dispersed in different geographical locations. It causes not only human diseases such as meningitis, diarrhea, and trauma infection (Cui et al., 2007; Prediger et al., 2020; Zhang et al., 2020), but also aquaculture diseases such as rot skin disease, septicemia, and rotten tail disease (Dong et al., 2017; Lazado and Zilberg, 2018; Liu Z. G. et al., 2018; Ran et al., 2018). In addition, A. veronii is resistant to many antibiotics together with penicillin, ampicillin, oxacillin and piperacillin (Yang et al., 2022). So it’s intractable to prevent and treat, and it is urgent to study the regulation mechanism of its virulence.
Ribosome rescue systems are pivotal to recycle arrested ribosomes that stall at the 3′ end of mRNAs, subsequently releasing the tagged polypeptide for degradation by cellular proteases and eventually permitting disassembly and liberating ribosomal subunits for new rounds of protein synthesis (Himeno et al., 2015). It is widely believed that trans-translation is the most ubiquitous and effective ribosome rescue system in bacteria (Konno et al., 2007), which is required for cell viability, growth, and pathogenesis (Brunel and Charpentier, 2016), and its inactivation decreases the abilities to respond to and recover from stress conditions (Yang and Glover, 2009), indicating that it may be a promising target for the development of new antimicrobial agents. The molecule inhibitor KKL-10 or KKL-40 exhibited antimicrobial activity against Francisella Tularensis through intervening trans-translation (Goralski et al., 2016). The small-molecule inhibitor of trans-translation KKL-40 also interacts with the human antimicrobial peptide LL-37 to inhibit Staphylococcus aureus (Huang et al., 2019). Small protein B (SmpB) is conserved and found as the binding partner of tmRNA (Hong et al., 2007). However, the SmpB and tmRNA have been shown to play distinct roles. The SmpB mutant exhibits higher susceptibility to antibiotics and stresses than the ssrA (the gene encoding tmRNA) mutant in Escherichia coli (Li et al., 2013). According to the reports, the ssrA regulates S. aureus pigment synthesis through base pairing with crtMN mRNA as an antisense RNA (Liu et al., 2010). So, does SmpB have other functions as well?
Most researchers believe that the SmpB act through trans-translation in virulence, but our previous study demonstrated that the SmpB interacted with the promoter of bvgS gene and enhanced the transcriptional expression without the participation of tmRNA in A. veronii (Liu et al., 2015). Here, we detailed again that the SmpB increased the expression of downstream outer membrane protein A (OmpA) as a transcription factor (TF), an effect that was entirely distinct from the previous trans-translation mechanism. OmpA protein is a highly conserved outer-membrane protein in Gram-negative bacteria. In addition to its role as a structural protein for the maintenance of cell integrity and morphology (Krishnan and Prasadarao, 2012), OmpA serves as a receptor for adhesion and invasion associated with virulence (Smith et al., 2007). Peptide aptamers (PAs), a kind of small combinatorial proteins, bind to specific sites on the target molecules. Compared with antibodies, PAs have advantages such as small size, simple and stable structure, and soluble expression in cells (Reverdatto et al., 2015). Owing to these advantages, PAs today are widely applied in bioimaging, targeted drug delivery, and the drug selection of antitumor and antimicrobial agents (Blum et al., 2000; Wang and Farokhzad, 2014; Zhou and Rossi, 2014; Tan et al., 2021). To evaluate the validation of the SmpB-OmpA signal pathway, the particular PA was selected utilizing OmpA as a bait in the bacterial two-hybrid system (Liu et al., 2016), followed by the interaction with OmpA and the growth inhibition. Collectively, we uncovered that SmpB-activated OmpA expression as a TF. The interactions identified in our study deepened the insight into SmpB-mediated regulation, unveiled the outer membrane biogenesis and identified the drug targets for the development of new inhibitors in bacteria.
The strains, plasmids, and primers were defined in Supplementary Tables 1–3, respectively. The plasmids were constructed according to the conventional molecular biology techniques, such as PCR, DNA restriction, ligation, transformation, and positive colony selection (Aviv and Gal-Mor, 2018). Endonucleases and DNA polymerase were purchased from New England Biolabs (Ipswich, MA, United States). The DNA purification kit, oligonucleotide synthesis, and DNA sequencing were provided by Sangon Biotech (Shanghai, China). To construct the fluorescent reporter plasmid pDH116, both the promoter of the ompA gene and encoding region of enhanced green fluorescent protein (EGFP) were amplified by primers in Supplementary Table 3. The two DNA fragments were purified and combined to a fusion by overlap extension PCR and were subsequently inserted into the backbone of plasmid pDH113 (Liu et al., 2015).
Bacteria were cultured in LB broth or on LB agar plates at 37°C. A final concentration of 0.1 mM IPTG (isopropyl β-D-1-thiogalactopyranoside) was applied to induce the Lac or T7 promoter. Antibiotics were used at the following concentrations: 25 μg/ml of chloramphenicol, 100 μg/ml of ampicillin, 25 μg/ml of kanamycin, and 20 μg/ml of tetracycline. The culture was incubated at 37°C overnight with agitation and then inoculated in fresh medium until the exponential or stationary phase.
The upstream region from open reading frames (ORF) was uploaded and predicted by iPromoter-2L,1 BPROM,2 and Promoter Calculator.3 With multi-window-based PseKNC, iPromoter-2L is a two-layer predictor for identifying the sigma promoters in E. coli, such as σ24-promoter, σ28, σ32, σ38, σ70, and so on (Chen et al., 2014, 2015; Liu B. et al., 2018). BPROM is a bacterial sigma 70 promoter recognition program for annotating the upstream regions of ORFs (Solovyev and Salamov, 2011). Promoter Calculator is applied to predict promoter elements and transcriptional initiation rates across transcriptional start sites (TSSs) depending on free energy calculation.
Bacterial growth was monitored by taking 1 ml of culture to measure the optical density (OD) at 600 nm using a spectrophotometer (Shanghai Spectrum Instruments Co., Ltd., Shanghai, China) at appropriate times. The total fluorescence was determined by analyzing 200-μl aliquots of culture with a fluorescent plate reader by Infinite® 200 PRO instrument (Tecan, Shanghai, China), wherein the wavelengths of excitation and emission were programmed at 485 and 525 nm, respectively. Each treatment was performed in triplicate with separated bacterial cultures, and the fluorescence values were normalized to the OD.
Bacterial one-hybrid assay was described previously (Guo et al., 2009). Briefly, the E. coli XL1-Blue MRF′ strain was applied to the propagation of all pBX-cmT and pTRG recombinant plasmids. A pair of pBX-cmT and pTRG derivatives was co-transformed into the E. coli XL1-Blue MRF′ reporter strain for the growth test. The principle of DNA–protein interactions was dependent on the transcriptional activation of the HIS3 reporter gene, which allowed the growth in the presence of 3-amino-1,2,4-triazole (3-AT), a competitive inhibitor of His3 enzyme. Besides, the positives were verified by using the aadA gene, which conferred streptomycin resistance, as a secondary reporter. Collectively, the selective medium was appended with 3-AT, streptomycin, tetracycline (resistance of pTRG and its derivative plasmids), chloramphenicol (resistance of pBT and its derivative plasmids), and kanamycin (resistance of E. coli reporter strains) (Guo et al., 2009). Likewise, two-hybrid assay was performed except that the conjugated plasmids were pBT and pTRG derivatives (Strauch and Georgiou, 2007).
The strains were cultured in M9 minimal medium (standard). Briefly, total RNA was extracted from all strains, and the residual DNA was removed by DNase I treatment (Green and Sambrook, 2018). First-strand complementary DNA (cDNA) was synthesized employing the HiScript® IIQ RT SuperMix (Vazyme, Nanjing, China) and was subsequently diluted as the template for the quantitative real-time PCR (qPCR) analysis. The primers used in qPCR were listed in Supplementary Table 4. The qPCR reaction was conducted using the LightCycler® 96 instrument (Roche, Switzerland, Germany) for the fluorescence detection. The relative amounts of RNAs were calculated using the comparative Ct method by selecting Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the reference gene (Schmittgen and Livak, 2008).
To prepare the DNA probe, a 125-bp fragment of the predicted ompA promoter (PompA) region was amplified from the pDH116 plasmid with primers that had been labeled by 6-carboxyfluorescein (6-FAM) (Xu et al., 2017). To prepare the SmpB protein, the pET-28a-SmpB was transferred into E. coli BL21. The SmpB was purified as the calculated molecular weight of 19 kDa using NI-IDA column. For each assay, 40 ng of probe was incubated with increasing amounts of SmpB protein in a total volume of 20 μl. After incubation at 30°C for 30 min, the samples were loaded onto 5% polyacrylamide native gels for electrophoresis in 0.5 × TBE running buffer at 10 V/cm for approximately 1.5 h. The DNA fragment bound to SmpB protein was detected using the Image Quant LAS 4000 mini system (GE Healthcare, Piscataway, United States).
After the DNA probe and SmpB protein were mixed and incubated as previously described EMSA, 5 μl of solution including 0.015 units of DNase I (Promega, Madison, United States) and 100 nmol of CaCl2 were added, followed by further incubation for 1 min at 25°C. The reaction was terminated by adding 70 μl of DNase I stop solution. The samples were extracted with phenol/chloroform, precipitated with ethanol, and dissolved in 20 μl of Mini-Q water. To prepare the DNA ladders, the fmol DNA cycle sequencing system (Promega, Madison, United States) was applied. Twelve microliters of the sequencing reactions were performed using 15 ng of the PompA region as the template and 5 pmol of FAM-labeled oligonucleotides as the sequencing primer. Subsequently, 1 μl of digested DNA fragments was added to 8.5 μl of highly deionized (HiDi) formamide and 0.5 μl of GeneScan-LIZ600 size standards (Applied Biosystems, Foster City, United States), and then the mixture was analyzed using the 3130xl DNA analyzer and Peak Scanner software v1.0 (Applied Biosystems, Foster City, United States) (Cao et al., 2017).
The purified SmpB protein was labeled by the RED-tris-NTA 2nd Generation dye (NanoTemper Technologies, Munich, Germany) to obtain the final concentration of 50 nM fluorescent molecules. The DNA fragments including the native PompA (50 μM PompA C52AT) and the mutation (800 μM PompA T52GC) were used as the ligand molecules. The microscale thermophoresis (MST) reaction buffer was phosphate buffered saline-tween (PBS-T) buffer (2 mM KH2PO4, 8 mM Na2HPO4, 350 mM NaCl, 0.6 mM DTT, 0.05% Tween 20, and pH 7.4). Approximately, 10 μl of the serial diluted ligand molecules was mixed with the equal volume of fluorescent molecules. The reaction products were sampled in a standard capillary and placed in a MO NT.115 capillary tray (NanoTemper Technologies, Munich, Germany). The samples were measured at 60% LED/excitation power and medium MST power. The KD value was calculated by the MO.Affinity Analysis software (NanoTemper Technologies, Munich, Germany).
The peptide aptamer library (pTRG-SN-peptides) was constructed, and each PA comprised a scaffold protein S. aureus nuclease (SN) and a random peptide which exposed as a surface loop and bulged out on the surface of the scaffold. The plasmid pBT-ompA was applied as a bait to screen specific PA interacting with OmpA by the bacterial two-hybrid assay. The interactive key regions between OmpA and PA were identified as described previously (Liu et al., 2016).
The peptide aptamer PA3 with molecular weight of 21 kDa that interacted with OmpA was expressed and purified for further use in E. coli. The susceptibility test was performed using the broth dilution method. A. veronii was grown in LB broth overnight at 30°C, and 3 ml of culture was diluted until an OD600 of 0.05, in which the purified peptide aptamer PA3 was supplemented at a final concentration of 50 μg/ml. After incubation for 10 h at 30°C, the OD600 of A. veronii was measured. In the meantime, phosphate buffered saline (PBS) buffer and bovine serum albumin (BSA) and purified SN were chosen as the negative controls, and chloramphenicol (Cm) was chosen as the positive control. The purified peptide aptamer PA3 was serially diluted at a range of 3–50 μg/ml.
The result data were based on at least three independent experiments and were displayed as the means ± standard deviation. The statistical significance was determined using one-way ANOVA, where * and ** represented significant (p < 0.05) and extremely significant differences (p < 0.01), respectively.
In the previous study, the interference of PA on the function of SmpB resulted in the down-regulation of ompA transcriptional level in A. veronii (Liu et al., 2017). To analyze whether SmpB regulated OmpA directly, qPCR was performed in WT (wild type), ΔSmpB (SmpB deletion strain), and C-ΔSmpB (complementing smpB to ΔSmpB) at exponential phase and stationary phase, respectively. The transcription levels of ompA in ΔSmpB strain were lower at both phases compared to WT, while those of C-ΔSmpB strain had no difference at exponential phase and had significant increase at stationary phase (Figure 1A). We speculated that there might be an unknown TF that assisted SmpB protein to regulate the expression of OmpA and played unlike roles in different developmental stages of A. veronii. Whether this TF directly acted on OmpA or indirectly affected OmpA by regulating the expression of SmpB protein needed further study. Bacterial one-hybrid assays were performed to analyze the interaction between SmpB and the PompA. The results showed that the co-transformants conferring SmpB and the PompA were grown in the presence of 12 mM 3-AT media (Figure 1B and Supplementary Figure 1), indicating that SmpB specifically bound to the PompA region in vitro. However, the interaction of SmpB and OmpA could not be observed by the bacterial two-hybrid assay (Supplementary Figure 2).

Figure 1. Identification of the key region in the predicted ompA promoter (PompA) interacting with small protein B (SmpB). (A) Quantitative real-time PCR (qPCR) assays. The relative expression of ompA was analyzed in WT, ΔSmpB, and C-ΔSmpB in the exponential and stationary phase, respectively. The NS represents “not statistically significant” and * represents significant differences (p < 0.01). (B) Verification of SmpB interacting with the PompA by bacterial one-hybrid system. The interaction between SmpB and the PompA was monitored by the growth on 3-AT-free and 12 mM 3-AT medium. (C) Binding of SmpB to the PompA. Electrophoretic mobility shift assay (EMSA) was performed using the purified SmpB with the promoter regions of the ompA gene. The DNA segments were amplified from the promoter of the predicted ompA gene. (D) The DNA sequences of the protected regions were provided, and the sequences were underlined. Transcriptional start site was marked with an arrow. Nucleotide sequences were numbered considering the transcription initiation site as +1. (E) Identification of SmpB-protected DNA fragments in the promoter regions of ompA using DNase I footprinting assays. The regions protected by SmpB were marked with frames of dashed lines.
In addition, gel shift assay was performed to verify the binding between SmpB and the promoter of ompA. The 6 × His-tagged SmpB in the range of 0–3 μg was initially incubated with 40 ng of the labeled PompA (Figure 1C), in which DNA was completely bound with 3 μg of SmpB. When 4 μg of unlabeled probes was added to the reaction, the overloaded DNA was competitive to form the complex with SmpB, resulting in a surplus of labeled DNA and the disappearance of the labeled DNA–SmpB complex (Figure 1C). The upstream elements before ompA ORF of ompA gene were predicted by iPromoter-2L (Figure 1D), of which including a σ70 promoter sequence. In addition, −10 element, −35 element, and TSS were deduced by BPROM and Promoter Calculator (Supplementary Figure 3 and Supplementary Dataset 1). Since the average nucleotide number of paired mRNA: 16S rRNA was 6.3 in bacteria 33 (Schurr et al., 1993) and the sequence (AGGAGG) was confirmed as Shine–Dalgarno (SD) sequence for promoting translation (Vimberg et al., 2007), a similar SD sequence (AGAGGC) was exhibited at the PompA region in A. veronii. Subsequently, the SmpB-protected DNA sequences were precisely determined by the DNase I footprinting assay (Figure 1E). Four protected regions were located at −46 to −28 bp, −18 to +4 bp, +21 to +31 bp, and +48 to +59 bp, respectively, which were located in the upstream of the initiation codon of ompA (Figure 1D).
To further investigate the regulation of OmpA by SmpB, we monitored the fluorescence activities of the PompA and EGFP translational fusion in the presence or absence of SmpB expression. It has been reported that divalent metal ion Mg2+ or Ca2+ stabilized the molecule conformation and affected interactions through neutralizing the negative charges (Lusetti et al., 2003; Onoda et al., 2005; Ralec et al., 2017; Kim et al., 2018). Hence, divalent metal ion Mg2+ or Ca2+ was added and the results demonstrated all the cells maintained constant growth (Figure 2A), while the cells expressing SmpB showed the significant fluorescent enhancement when supplemented with 5 mM Mg2+ compared with the controls (Figure 2B), signifying Mg2+ may help to stabilize the binding of protein and DNA structure. To examine the contribution of each binding site in the PompA region, the point mutations were introduced to disrupt the binding of SmpB. Based on the experimental results of DNase I footprinting assay, the sites T–17GG, C25AC, and C52AT in the PompA were mutated to C–17AA, T25CT, and T52GC, respectively. After the native and mutated reporters were transformed separately, the fluorescent levels were measured to analyze the promoter activities at the stationary stage. The results revealed that C–17AA and T52GC mutations themselves had no effect on the promoter activities, while the promoter activity was changed by T25CT mutation, which was abnegated afterward (Figure 2C). When SmpB and the promoter were co-transformed, the cells containing native promoter or C–17AA mutation increased the reporter expression, while T52GC mutation did not, indicating that the site was required for SmpB regulation (Figure 2D). Next, SmpB was truncated and mutated to analyze the effects of the PompA. The results revealed that the G11S, F26I, and K152 sites of SmpB were vital to interact with the PompA (Figure 2E).
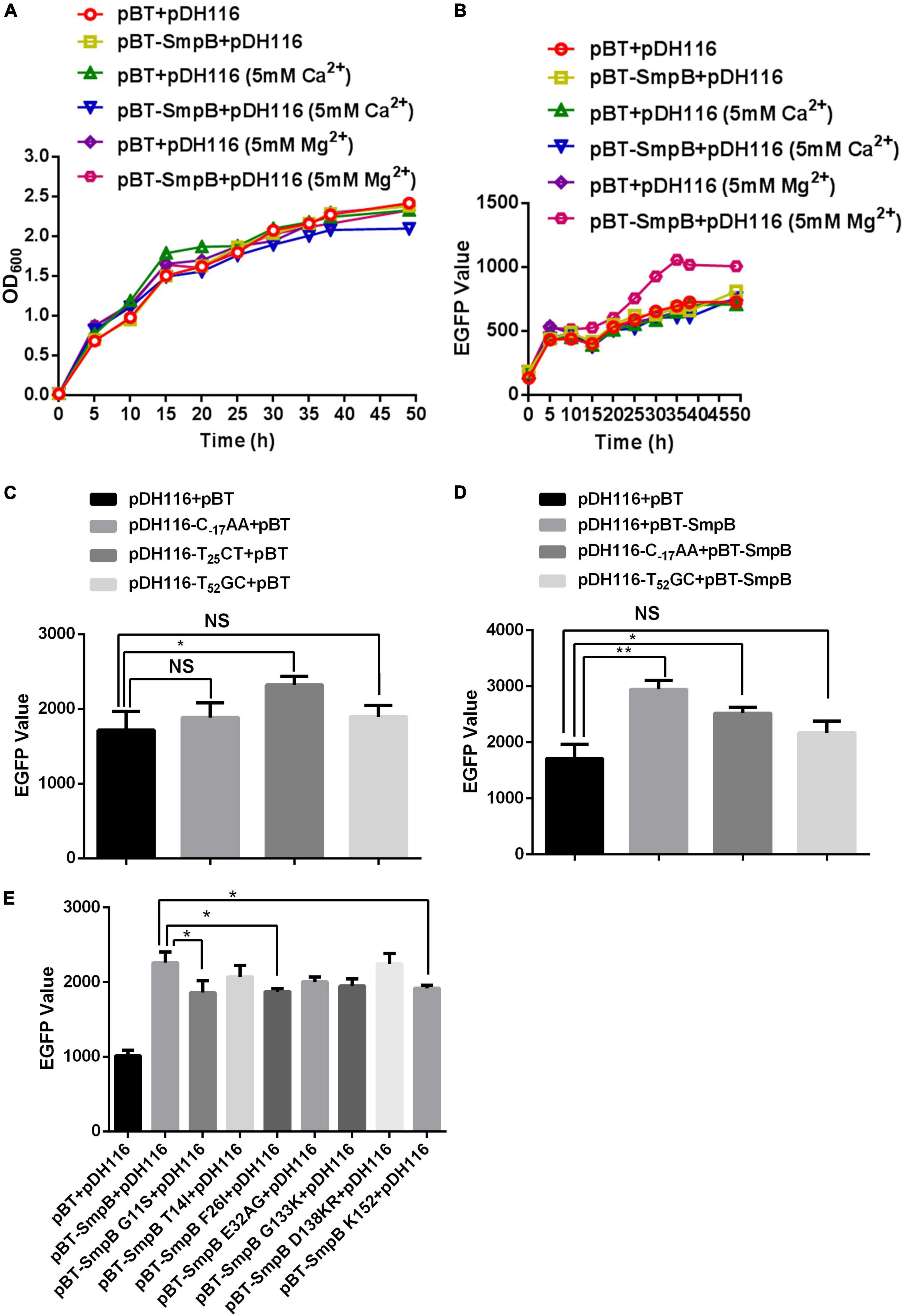
Figure 2. Small protein B regulates the transcription of PompA positively. (A) Growth measurement of the strain bearing pDH116 when co-transformed with either pBT-SmpB or empty vector pBT. The plasmid pDH116 is endowed with both the promoter of the ompA gene and encoding region of enhanced green fluorescent protein (EGFP). (B) Fluorescence measurement of the strain bearing pDH116 when co-transformed with either pBT-SmpB or empty vector pBT. (C) Promoter activity of the native and mutated pDH116 (pDH116-C–17AA, pDH116-T25CT, and pDH116-T52GC). (D) Effects of SmpB on the expression of the native and mutated pDH116. (E) Identifications of the key amino acid residues in SmpB interacting with pDH116. The “NS” represents “not statistically significant” and “***” is significant (p < 0.05) and extremely significant differences (p < 0.01), respectively.
The initial concentrations of the native ompA promoter (50 μM PompA C52AT) or the mutant (800 μM PompA T52GC) were serially diluted, and mixed in equal volume with the labeled SmpB probes (50 nM) to obtained the dose–response fit graph. The KD values were calculated using MO. Analysis Software. The KD value of PompA C52AT binding with SmpB was 0.93 ± 0.58 μM (Figure 3A), while that of the mutation PompA T52GC binding with SmpB was 126.41 ± 51.63 μM (Figure 3B). The affinity of the PompA with SmpB protein decreased significantly after C52AT was mutated to T52GC, demonstrating that C52AT site was the key site for SmpB binding to the PompA region.
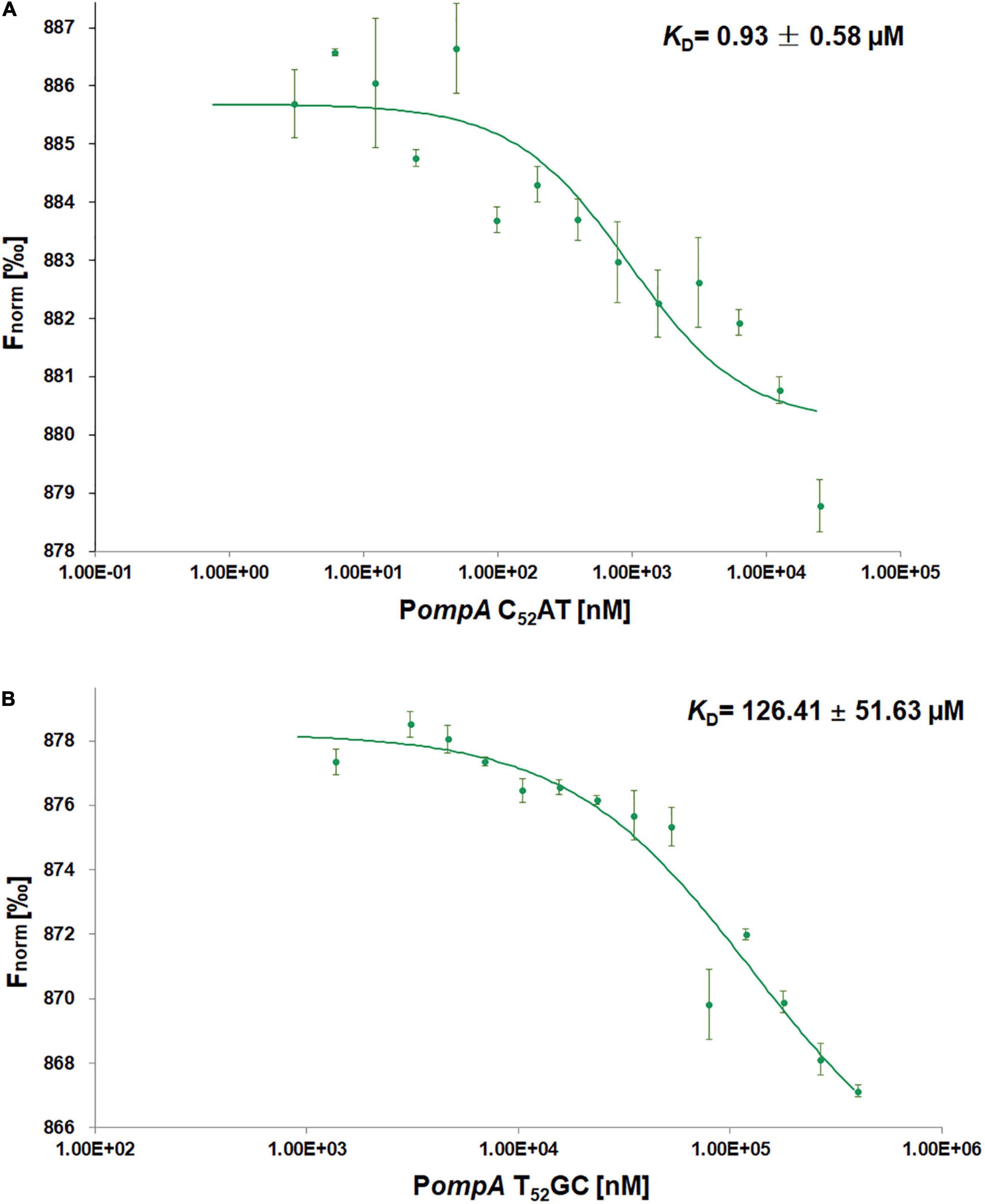
Figure 3. Small protein B protein interacts with the PompA by microscale thermophoresis (MST) assays. (A,B) Dose Response Fit of SmpB protein with PompA C52AT and PompA T52GC. Fnorm values of Y-axis in the dose response plot are calculated from the ratio of normalized fluorescence F0/F1, where F0 corresponds to the normalized fluorescence prior to MST activation. F1 is determined after an optimal MST power-dependent time interval which yields the best signal-to-noise ratio. The X-axis is the different concentration of PompA after successive dilution. The initial concentrations of PompA C52AT (native ompA promoter) and PompA T52GC (mutant) were, respectively, 25 and 400 μM in reaction buffer and the fluorescence molecules SmpB labeled by Monolith His-tag Labeling Kit RED-tris-NTA 2nd Generation was 25 nM in the buffer. MST experiments were performed in Phosphate buffered saline-Tween (PBS-T) buffer (2 mM KH2PO4, 8 mM Na2HPO4 350 mM NaCl, 0.6 mM DTT, 0.05% Tween 20, pH 7.4) with 40% MST-Power, 60% Excitation-Power).
The bacterial two-hybrid system was performed to identify PA that bound specifically to OmpA protein in vivo. Six clones that might interact with OmpA were isolated from 2 × 102 transformants (Table 1). The colony designated as PA3 exhibited the strongest interaction with OmpA (Figure 4A). To further explore the key region of OmpA that interacted with PA3, OmpA truncations including pBT-ΔN-OmpA and pBT-ΔC-OmpA, were co-transformed with pTRG-PA3. The results showed that C-terminal OmpA bound to PA3 specifically (Figure 4B).
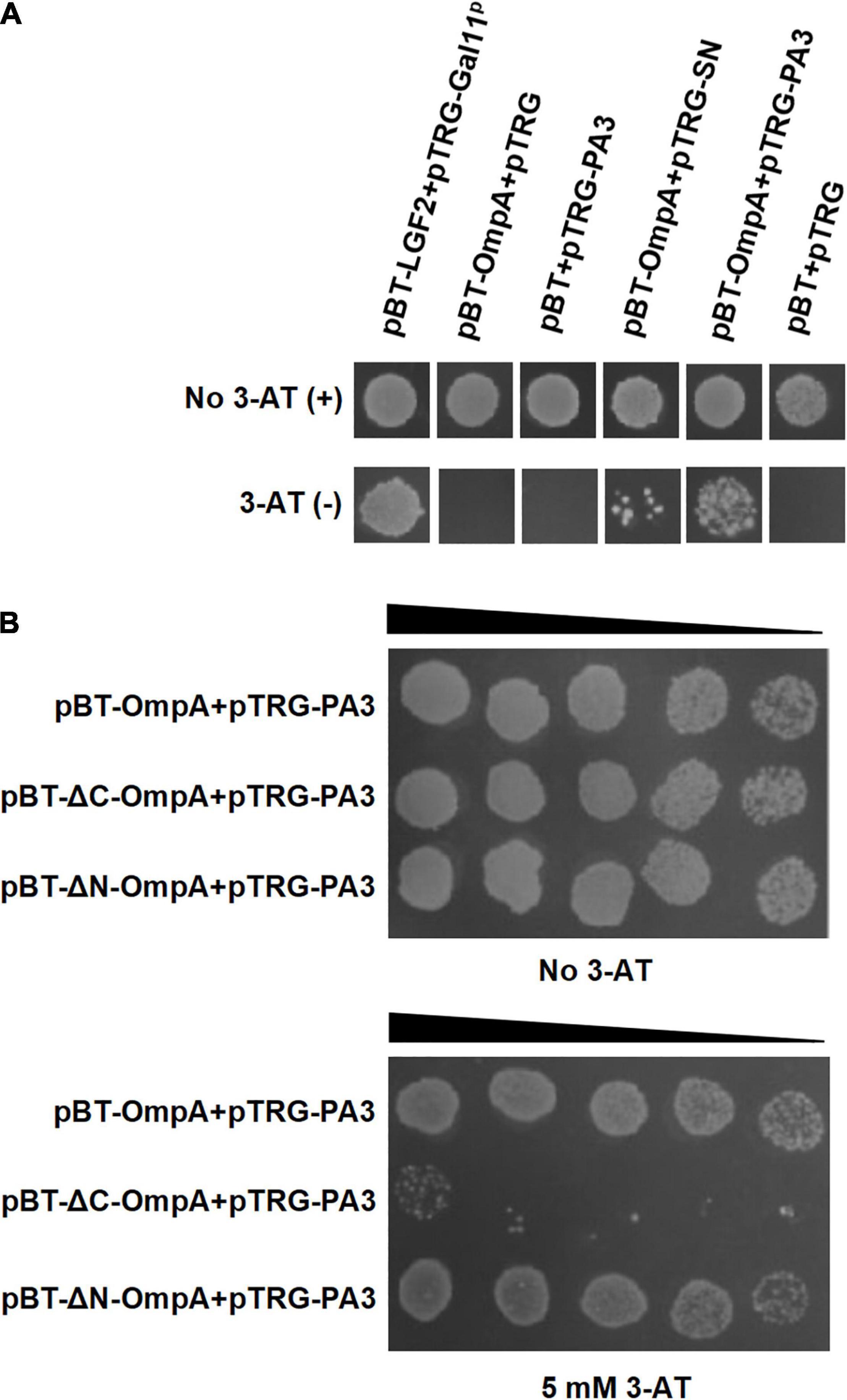
Figure 4. Interaction between OmpA and the specific peptide aptamer PA3. (A) The co-transformants were cultivated overnight and were spotted onto no 3-AT or 5-mM 3-AT media with 10 μl of 106 CFU/ml. (B) Identification of the key regions of OmpA interacting with PA3 by the bacterial two-hybrid system.
To confirm the antimicrobial activity of PA3, the growth assay was performed to test the inhibition of A. veronii. The results showed that the final concentration of purified PA3 was above 25 μg/ml; it could significantly inhibit the growth of A. veronii (Figure 5). These results demonstrated that PA3 could effectively inhibit the growth of A. veronii, suggesting PA3 possibly interfered the function of OmpA protein through the interaction with OmpA.
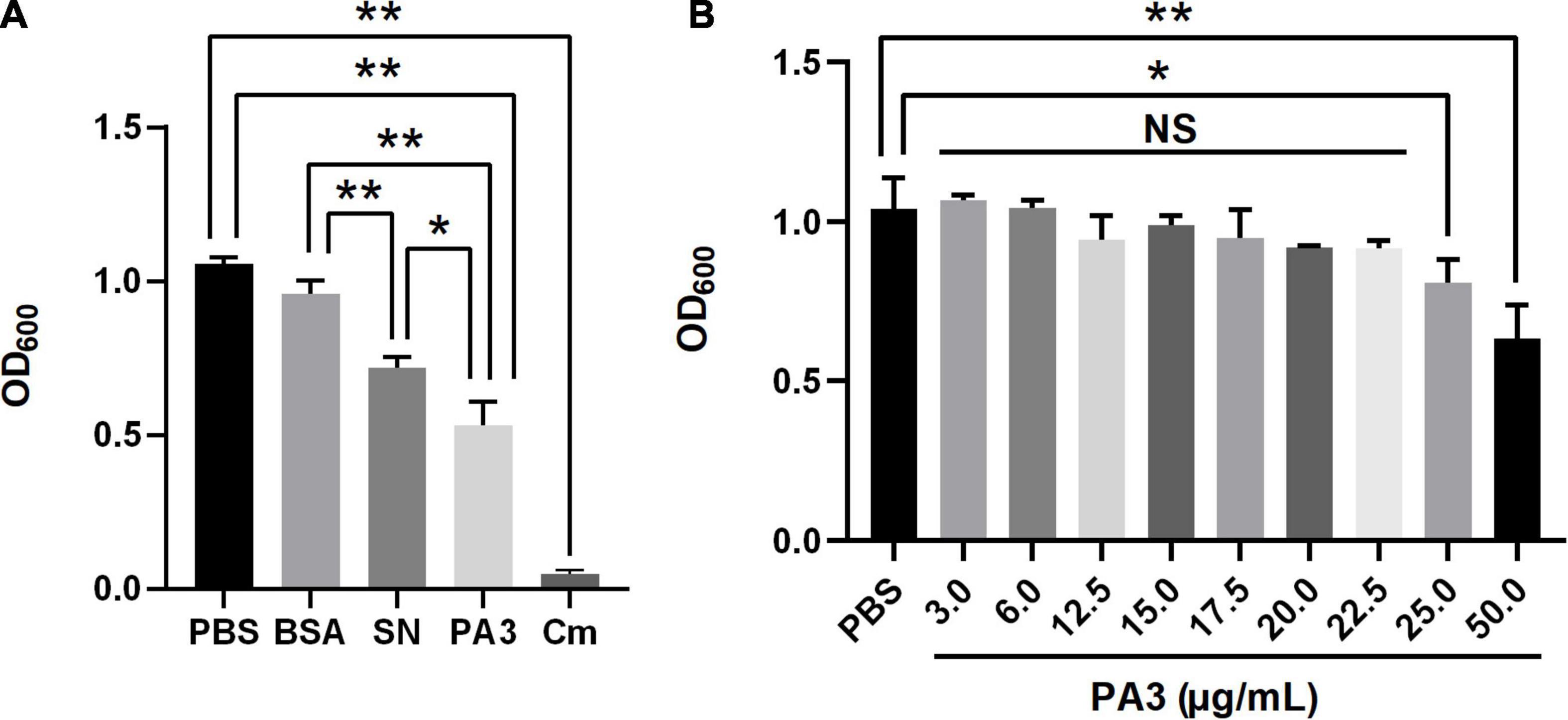
Figure 5. Characterization of peptide aptamer PA3. (A) Antibacterial activity of PA3 against Aeromonas veronii. Each of phosphate buffered saline (PBS), bovine serum albumin (BSA), SN, PA3, or Cm was added to the bacterial cultures, and OD600 measurements were carried out after 10 h of incubation. (B) Susceptibility test of PA3 in A. veronii. The serial concentrations of PA3 were diluted for supplementation in the bacterial cultures, and OD600 measurements were carried out after 10 h of incubation. The “NS” represents “not statistically significant” and “***” are significant (p < 0.05) and extremely significant differences (p < 0.01), respectively.
Outer membrane protein A maintains the structural integrity of the outer membrane together with cell wall lipoprotein and peptidoglycan in Gram-negative bacteria and contributes to nutrient absorption, pathogenicity, and multiple drug resistance (Vàzquez-Juárez et al., 2004; Confer and Ayalew, 2013). In addition, OmpA mediates the adhesion of pathogenic strains to leukocytes and macrophages, and the addition of exogenous OmpA antibody significantly reduces the adhesion to cells (Krishnan and Prasadarao, 2012). However, few studies are related to the regulation of OmpA.
Here, we reported a novel mechanism of SmpB participating in the regulation of outer membrane biogenesis of OmpA in A. veronii. SmpB is principally considered as a ribosome rescue factor, which works with tmRNA for quality control and dissociation of stalled ribosomes. The deficient SmpB attenuates growth, colonization, virulence, and antibiotic persistence (Mu et al., 2013; Liu et al., 2017; Mraheil et al., 2017). Apart from ribosome rescue, the action of SmpB alone was first shown to increase the bvgS expression as a TF without the participation of tmRNA (Liu et al., 2015). This work, building on our prior work, revealed that SmpB was required to bind to the promoter region of ompA (Figures 1, 2), which functioned in a manner similar to bvgS activation in A. veronii (Liu et al., 2015). The PompA C52AT of ompA promoter and the G11S, F26I, and K152 of SmpB were the key sites for association. However, the protein interaction of SmpB and OmpA could not be observed by the bacterial two-hybrid assay (Supplementary Figure 2). These results suggested that SmpB-regulated ompA at the gene level. Whether SmpB alters the PompA structure to make RNA polymerases or ribosomes more accessible still need to be further researched.
To clarify the importance of OmpA in A. veronii, a particular PA was identified to interact with OmpA for interfering with its function. PA exerts the intrinsic properties of antibody with small size, leading to convenient production and application (Blum et al., 2000; Gebauer and Skerra, 2009). PA3 screened from PA library exhibited strong interaction with OmpA. In the presence of 50 μg/ml of purified PA3, the growth of Acinetobacter baumannii was significantly inhibited compared with the control (Figure 5). The above results are similar to that the cyclic hexapeptide binding with OmpA inhibits its function and enhances the activity of colistin against A. baumannii (Parra-Millán et al., 2018). As the main component of Omp in Gram-negative bacteria and a key virulence factor of pathogenic bacteria, OmpA plays an important role in mediating bacterial biofilm formation, antibiotic resistance, and cell infection (Nie et al., 2020), which indicating that OmpA may serve as a potential drug target for combating A. veronii infection.
In summary, a possible mechanism for SmpB-mediated regulation of OmpA biogenesis in A. veronii was proposed based on multiple molecular biology techniques (Figure 6). Notably, the downstream node OmpA seemed terminal in the regulatory network because the inhibition of OmpA could hamper the pathway and seriously influence the growth by the PA (Table 1). We postulate that OmpA is a potential drug target for human and animal diseases.
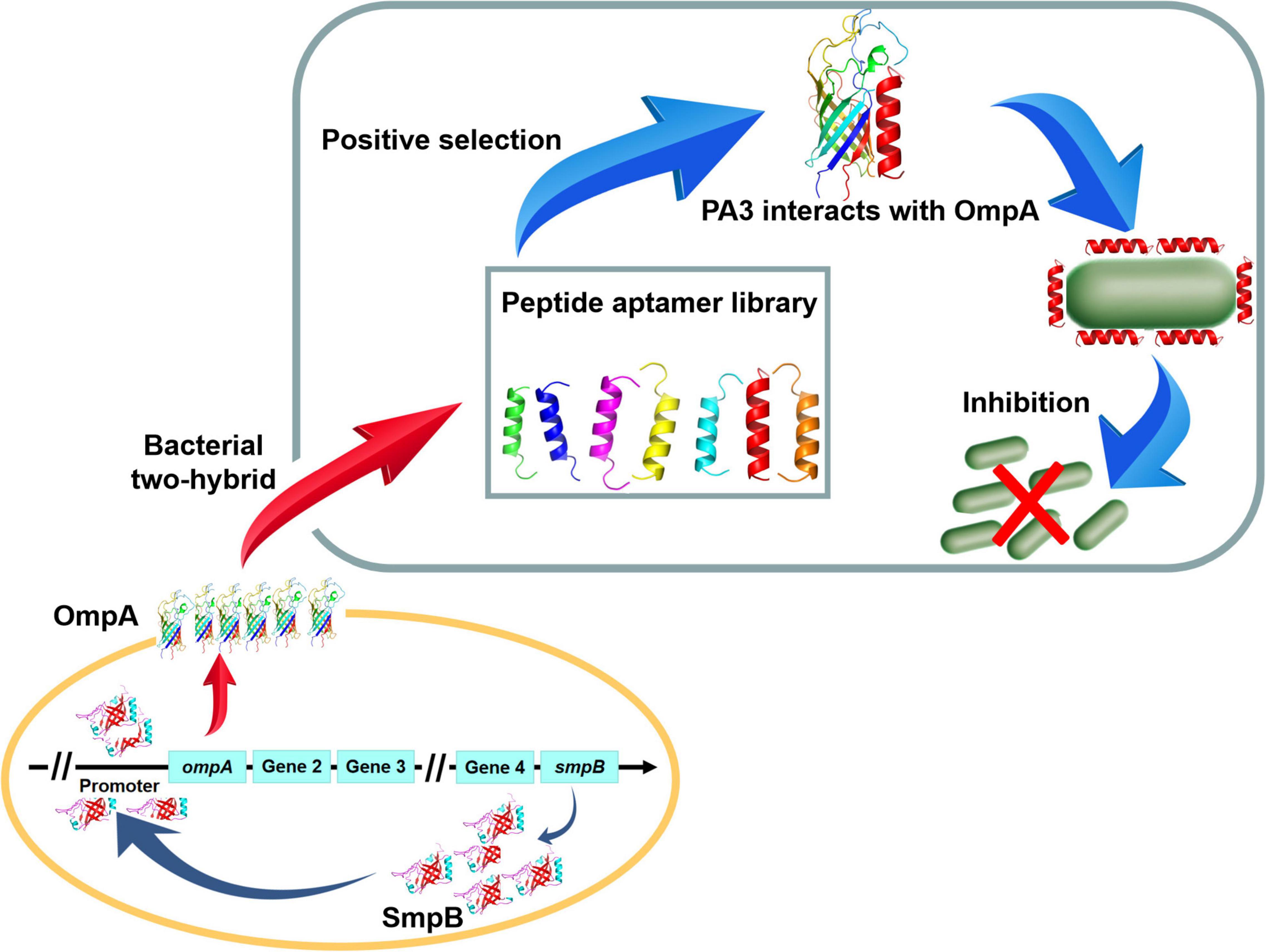
Figure 6. The schematic diagram of Peptide aptamer PA3 attenuates the viability of A. veronii by hindering of SmpB-OmpA signal pathway.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
PL and HC conceived the study, designed and performed the experiments, analyzed the data, and prepared the manuscript. QX, DW, and YT assisted in data analysis. XH and ML assisted in manuscript editing. ZL contributed to experimental design, data analysis, and preparation and editing of the manuscript. All authors have read and approved the manuscript.
This work was supported by the Key Research and Development Program of Hainan Province (No. ZDYF2021XDNY181 to ZL) and National Natural Science Foundation of China (Nos. 32160038 to ZL and 81902764 to PL).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are very thankful to the reviewers for the time and careful assessment of our manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.900234/full#supplementary-material
Aviv, G., and Gal-Mor, O. (2018). Western blotting against tagged virulence determinants to study bacterial pathogenicity. Methods Mol. Biol. 1734, 47–54. doi: 10.1007/978-1-4939-7604-1_6
Blum, J. H., Dove, S. L., Hochschild, A., and Mekalanos, J. J. (2000). Isolation of peptide aptamers that inhibit intracellular processes. Proc. Natl. Acad. Sci. U.S.A. 97, 2241–2246. doi: 10.1073/pnas.040573397
Brunel, R., and Charpentier, X. (2016). Trans-translation is essential in the human pathogen Legionella pneumophila. Sci. Rep. 6:37935. doi: 10.1038/srep37935
Cao, Y., Zheng, F., Wang, L., Zhao, G., Chen, G., Zhang, W., et al. (2017). Rce1, a novel transcriptional repressor, regulates cellulase gene expression by antagonizing the transactivator Xyr1 in Trichoderma reesei. Mol. Microbiol. 105, 65–83. doi: 10.1111/mmi.13685
Chen, W., Lei, T. Y., Jin, D. C., Lin, H., and Chou, K. C. (2014). PseKNC: a flexible web server for generating pseudo K-tuple nucleotide composition. Anal. Biochem. 456, 53–60. doi: 10.1016/j.ab.2014.04.001
Chen, W., Lin, H., and Chou, K. C. (2015). Pseudo nucleotide composition or PseKNC: an effective formulation for analyzing genomic sequences. Mol. Biosyst. 11, 2620–2634. doi: 10.1039/c5mb00155b
Confer, A. W., and Ayalew, S. (2013). The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet. Microbiol. 163, 207–222. doi: 10.1016/j.vetmic.2012.08.019
Cui, H., Hao, S., and Arous, E. (2007). A distinct cause of necrotizing fasciitis: Aeromonas veronii biovar sobria. Surg. Infect. 8, 523–528. doi: 10.1089/sur.2006.046
Dong, H. T., Techatanakitarnan, C., Jindakittikul, P., Thaiprayoon, A., Taengphu, S., Charoensapsri, W., et al. (2017). Aeromonas jandaei and Aeromonas veronii caused disease and mortality in Nile tilapia, Oreochromis niloticus (L.). J. Fish. Dis. 40, 1395–1403. doi: 10.1111/jfd.12617
Gebauer, M., and Skerra, A. (2009). Engineered protein scaffolds as next-generation antibody therapeutics. Curr. Opin. Struct. Biol. 13, 245–255. doi: 10.1016/j.cbpa.2009.04.627
Goralski, T. D., Dewan, K. K., Alumasa, J. N., Avanzato, V., Place, D. E., Markley, R. L., et al. (2016). Inhibitors of ribosome rescue arrest growth of Francisella tularensis at all stages of intracellular replication. Antimicrob. Agents Chemother. 60, 3276–3282. doi: 10.1128/AAC.03089-15
Green, M. R., and Sambrook, J. (2018). Quantification of RNA by Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR). Cold Spring Harb Protoc. 2018, 847–856. doi: 10.1101/pdb.prot095042
Guo, M., Feng, H., Zhang, J., Wang, W., Wang, Y., Li, Y., et al. (2009). Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 19, 1301–1308. doi: 10.1101/gr.086595.108
Himeno, H., Nameki, N., Kurita, D., Muto, A., and Abo, T. (2015). Ribosome rescue systems in bacteria. Biochimie. 114, 102–112. doi: 10.1016/j.biochi.2014.11.014
Hong, S. J., Lessner, F. H., Mahen, E. M., and Keiler, K. C. (2007). Proteomic identification of tmRNA substrates. Proc. Natl. Acad. Sci. U.S.A. 104, 17128–17133. doi: 10.1073/pnas.0707671104
Huang, Y., Alumasa, J. N., Callaghan, L. T., Baugh, R. S., Rae, C. D., Keiler, K. C., et al. (2019). A small-molecule inhibitor of trans-translation synergistically interacts with cathelicidin antimicrobial peptides to impair survival of Staphylococcus aureus. Antimicrob. Agents Chemother. 63:e02362-18. doi: 10.1128/AAC.02362-18
Kim, R., Kanamaru, S., Mikawa, T., Prévost, C., Ishii, K., Ito, K., et al. (2018). RecA requires two molecules of Mg2+ ions for its optimal strand exchange activity in vitro. Nucleic Acids Res. 46, 2548–2559. doi: 10.1093/nar/gky048
Konno, T., Kurita, D., Takada, K., Muto, A., and Himeno, H. (2007). A functional interaction of SmpB with tmRNA for determination of the resuming point of trans-translation. RNA. 13, 1723–1731. doi: 10.1261/rna.604907
Krishnan, S., and Prasadarao, N. V. (2012). Outer membrane protein A and OprF: versatile roles in Gram-negative bacterial infections. FEBS J. 279, 919–931. doi: 10.1111/j.1742-4658.2012.08482.x
Lazado, C. C., and Zilberg, D. (2018). Pathogenic characteristics of Aeromonas veronii isolated from the liver of a diseased guppy (Poecilia reticulata). Lett. Appl. Microbiol. 67, 476–483. doi: 10.1111/lam.13057
Li, J., Ji, L., Shi, W., Xie, J., and Zhang, Y. (2013). Trans-translation mediates tolerance to multiple antibiotics and stresses in Escherichia coli. J. Antimicrob. Chemother. 68, 2477–2481. doi: 10.1093/jac/dkt231
Liu, B., Yang, F., Huang, D. S., and Chou, K. C. (2018). iPromoter-2L: a two-layer predictor for identifying promoters and their types by multi-window-based PseKNC. Bioinformatics. 34, 33–40. doi: 10.1093/bioinformatics/btx579
Liu, P., Chen, Y., Wang, D., Tang, Y., Tang, H., Song, H., et al. (2016). Genetic selection of peptide aptamers that interact and inhibit both Small protein B and alternative ribosome-rescue factor A of Aeromonas veronii C4. Front. Microbiol. 7:1228. doi: 10.3389/fmicb.2016.01228
Liu, P., Huang, D., Hu, X., Tang, Y., Ma, X., Yan, R., et al. (2017). Targeting inhibition of SmpB by peptide aptamer attenuates the virulence to protect Zebrafish against Aeromonas veronii infection. Front. Microbiol. 8:1766. doi: 10.3389/fmicb.2017.01766
Liu, Y., Wu, N., Dong, J., Gao, Y., Zhang, X., Shao, N., et al. (2010). SsrA (tmRNA) acts as an antisense RNA to regulate Staphylococcus aureus pigment synthesis by base pairing with crtMN mRNA. FEBS. Lett. 584, 4325–4329. doi: 10.1016/j.febslet.2010.09.024
Liu, Z., Liu, P., Liu, S., Song, H., Tang, H., and Hu, X. (2015). Small protein B upregulates sensor kinase bvgS expression in Aeromonas veronii. Front. Microbiol. 6:579. doi: 10.3389/fmicb.2015.00579
Liu, Z. G., Zheng, A. F., Chen, M. M., Lian, Y. X., Zhang, X. K., Zhang, S. Z., et al. (2018). Isolation and identification of pathogenic Aeromonas veronii from a dead Yangtze finless porpoise. Dis. Aquat. Organ. 132, 13–22. doi: 10.3354/dao03288
Lusetti, S. L., Shaw, J. J., and Cox, M. M. (2003). Magnesium ion-dependent activation of the RecA protein involves the C terminus. J. Biol. Chem. 278, 16381–16388. doi: 10.1074/jbc.M212916200
Mraheil, M. A., Frantz, R., Teubner, L., Wendt, H., Linne, U., Wingerath, J., et al. (2017). Requirement of the RNA-binding protein SmpB during intracellular growth of Listeria monocytogenes. Int. J. Med. Microbiol. 307, 166–173. doi: 10.1016/j.ijmm.2017.01.007
Mu, X., Huan, H., Xu, H., Gao, Q., Xiong, L., Gao, R., et al. (2013). The transfer-messenger RNA-small protein B system plays a role in avian pathogenic Escherichia coli pathogenicity. J. Bacteriol. 195, 5064–5071. doi: 10.1128/JB.00628-13
Nie, D., Hu, Y., Chen, Z., Li, M., Hou, Z., Luo, X., et al. (2020). Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection. J. Biomed. Sci. 27:26. doi: 10.1186/s12929-020-0617-7
Onoda, A., Arai, N., Shimazu, N., Yamamoto, H., and Yamamura, T. (2005). Calcium ion responsive DNA binding in a zinc finger fusion protein. J. Am. Chem. Soc. 127, 16535–16540. doi: 10.1021/ja052477m
Parra-Millán, R., Vila-Farrés, X., Ayerbe-Algaba, R., Varese, M., Sánchez-Encinales, V., Bayó, N., et al. (2018). Synergistic activity of an OmpA inhibitor and colistin against colistin-resistant Acinetobacter baumannii: mechanistic analysis and in vivo efficacy. J. Antimicrob. Chemother. 73, 3405–3412. doi: 10.1093/jac/dky343
Prediger, K. C., Dallagassa, C. B., Moriel, B., Vizzotto, B. S., Volanski, W., Souza, E. M., et al. (2020). Virulence characteristics and antimicrobial resistance of Aeromonas veronii biovar sobria 312M, a clinical isolate. Braz. J. Microbiol. 51, 511–518. doi: 10.1007/s42770-019-00180-5
Ralec, C., Henry, E., Lemor, M., Killelea, T., and Henneke, G. (2017). Calcium-driven DNA synthesis by a high-fidelity DNA polymerase. Nucleic Acids Res. 45, 12425–12440. doi: 10.1093/nar/gkx927
Ran, C., Qin, C., Xie, M., Zhang, J., Li, J., Xie, Y., et al. (2018). Aeromonas veronii and aerolysin are important for the pathogenesis of motile aeromonad septicemia in cyprinid fish. Environ. Microbiol. 20, 3442–3456. doi: 10.1111/1462-2920.14390
Reverdatto, S., Burz, D. S., and Shekhtman, A. (2015). Peptide aptamers: development and applications. Curr. Top Med Chem. 15, 1082–1101. doi: 10.2174/1568026615666150413153143
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Schurr, T., Nadir, E., and Margalit, H. (1993). Identification and characterization of E. coli ribosomal binding sites by free energy computation. Nucleic Acids Res. 21, 4019–4023. doi: 10.1093/nar/21.17.4019
Smith, S. G. J., Mahon, V., Lambert, M. A., and Fagan, R. P. (2007). A molecular Swiss army knife: OmpA structure, function and expression. FEMS. Microbiol. Lett. 273, 1–11. doi: 10.1111/j.1574-6968.2007.00778.x
Solovyev, V., and Salamov, A. (2011). “Automatic annotation of microbial genomes and metagenomic sequences,” in Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies, ed. R. W. Li (New York, NY: Nova Science Publishers), 61–78.
Strauch, E. M., and Georgiou, G. (2007). A bacterial two-hybrid system based on the twin-arginine transporter pathway of E. coli. Protein. Sci. 16, 1001–1008. doi: 10.1110/ps.062687207
Tan, Y., Li, Y., Qu, Y. X., Su, Y., Peng, Y., Zhao, Z., et al. (2021). Aptamer-peptide conjugates as targeted chemosensitizers for breast cancer treatment. ACS. Appl. Mater Interfaces. 13, 9436–9444. doi: 10.1021/acsami.0c18282
Vàzquez-Juárez, R. C., Romero, M. J., and Ascencio, F. (2004). Adhesive properties of a LamB-like outer-membrane protein and its contribution to Aeromonas veronii adhesion. J. Appl. Microbiol. 96, 700–708. doi: 10.1111/j.1365-2672.2004.02177.x
Vimberg, V., Tats, A., Remm, M., and Tenson, T. (2007). Translation initiation region sequence preferences in Escherichia coli. BMC. Mol. Biol. 8:100. doi: 10.1186/1471-2199-8-100
Wang, A. Z., and Farokhzad, O. C. (2014). Current progress of aptamer-based molecular imaging. J. Nucl. Med. 55, 353–356. doi: 10.2967/jnumed.113.126144
Xu, T., Wu, Y., Lin, Z., Bertram, R., Götz, F., Zhang, Y., et al. (2017). Identification of Genes Controlled by the Essential YycFG Two-Component System Reveals a Role for Biofilm Modulation in Staphylococcus epidermidis. Front. Microbiol. 8:724. doi: 10.3389/fmicb.2017.00724
Yang, C., and Glover, J. R. (2009). The SmpB-tmRNA tagging system plays important roles in streptomyces coelicolor growth and development. PLoS. One. 4:e4459. doi: 10.1371/journal.pone.0004459
Yang, H., Zhang, M., Ji, T., Zhang, Y., Wei, W., and Liu, Q. (2022). Bacillus subtilis CK3 used as an aquatic additive probiotics enhanced the immune response of crayfish Procambarus clarkii against newly identified Aeromonas veronii pathogen. Aquac. Res. 53, 255–264. doi: 10.1111/are.15571
Zhang, H. P., Kang, Y. H., Kong, L. C., Ju, A. Q., Wang, Y. M., Muhammad, I., et al. (2020). Functional analysis of hisJ in Aeromonas veronii reveals a key role in virulence. Ann. N. Y. Acad. Sci. 1465, 146–160. doi: 10.1111/nyas.14265
Keywords: Aeromonas veronii, SmpB, OmpA, peptide aptamer, antimicrobial-agent
Citation: Liu P, Chang H, Xu Q, Wang D, Tang Y, Hu X, Lin M and Liu Z (2022) Peptide Aptamer PA3 Attenuates the Viability of Aeromonas veronii by Hindering of Small Protein B-Outer Membrane Protein A Signal Pathway. Front. Microbiol. 13:900234. doi: 10.3389/fmicb.2022.900234
Received: 20 March 2022; Accepted: 12 April 2022;
Published: 19 May 2022.
Edited by:
Jin Zhou, Tsinghua University, ChinaReviewed by:
Jianchun Guo, Chinese Academy of Tropical Agricultural Sciences, ChinaCopyright © 2022 Liu, Chang, Xu, Wang, Tang, Hu, Lin and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhu Liu, emh1bGl1QGhhaW5hbnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.