
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 June 2022
Sec. Aquatic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.894026
 James O’Brien1,2*
James O’Brien1,2* Erin L. McParland3
Erin L. McParland3 Anna R. Bramucci1
Anna R. Bramucci1 Martin Ostrowski1
Martin Ostrowski1 Nachshon Siboni1
Nachshon Siboni1 Timothy Ingleton4
Timothy Ingleton4 Mark V. Brown5
Mark V. Brown5 Naomi M. Levine6
Naomi M. Levine6 Bonnie Laverock1
Bonnie Laverock1 Katherina Petrou2
Katherina Petrou2 Justin Seymour1*
Justin Seymour1*The organic sulfur compounds dimethylsulfoniopropionate (DMSP) and dimethyl sulfoxide (DMSO) play major roles in the marine microbial food web and have substantial climatic importance as sources and sinks of dimethyl sulfide (DMS). Seasonal shifts in the abundance and diversity of the phytoplankton and bacteria that cycle DMSP are likely to impact marine DMS (O) (P) concentrations, but the dynamic nature of these microbial interactions is still poorly resolved. Here, we examined the relationships between microbial community dynamics with DMS (O) (P) concentrations during a 2-year oceanographic time series conducted on the east Australian coast. Heterogenous temporal patterns were apparent in chlorophyll a (chl a) and DMSP concentrations, but the relationship between these parameters varied over time, suggesting the phytoplankton and bacterial community composition were affecting the net DMSP concentrations through differential DMSP production and degradation. Significant increases in DMSP were regularly measured in spring blooms dominated by predicted high DMSP-producing lineages of phytoplankton (Heterocapsa, Prorocentrum, Alexandrium, and Micromonas), while spring blooms that were dominated by predicted low DMSP-producing phytoplankton (Thalassiosira) demonstrated negligible increases in DMSP concentrations. During elevated DMSP concentrations, a significant increase in the relative abundance of the key copiotrophic bacterial lineage Rhodobacterales was accompanied by a three-fold increase in the gene, encoding the first step of DMSP demethylation (dmdA). Significant temporal shifts in DMS concentrations were measured and were significantly correlated with both fractions (0.2–2 μm and >2 μm) of microbial DMSP lyase activity. Seasonal increases of the bacterial DMSP biosynthesis gene (dsyB) and the bacterial DMS oxidation gene (tmm) occurred during the spring-summer and coincided with peaks in DMSP and DMSO concentration, respectively. These findings, along with significant positive relationships between dsyB gene abundance and DMSP, and tmm gene abundance with DMSO, reinforce the significant role planktonic bacteria play in producing DMSP and DMSO in ocean surface waters. Our results highlight the highly dynamic nature and myriad of microbial interactions that govern sulfur cycling in coastal shelf waters and further underpin the importance of microbial ecology in mediating important marine biogeochemical processes.
Up to 10% of the carbon fixed by marine phytoplankton is used to synthesize a single organosulfur compound called dimethylsulfoniopropionate (DMSP) (Archer et al., 2001). For DMSP-producing phytoplankton, this molecule can play a number of important physiological roles, such as an intracellular osmolyte, cryoprotectant, and antioxidant (Karsten et al., 1996; Kirst, 1996; Sunda et al., 2002). Following the release of DMSP into the surrounding water column through phytoplankton cell exudation or lysis (Bratbak et al., 1995; Stefels, 2000), this dissolved a pool of DMSP represents a major source of carbon and sulfur for heterotrophic bacteria (up to 15% of carbon and 90% of sulfur demands) (Zubkov et al., 2001, 2002). DMSP also has substantial biogeochemical importance because it is the principal precursor to the volatile gas, dimethyl sulfide (DMS). This gas is a product of phytoplankton and bacterial DMSP degradation and represents the main vehicle for the efflux of sulfur from the ocean to the atmosphere (Kettle et al., 1999; Simó, 2001), where it can subsequently be converted into cloud condensation nuclei that increase albedo (Charlson et al., 1987). However, DMS efflux and its subsequent climatic importance can be limited by transformation of the volatile gas to dimethyl sulfoxide (DMSO) through photolysis and microbial DMS oxidation (Brimblecombe and Shooter, 1986; Kiene and Bates, 1990; Lidbury et al., 2016; Thume et al., 2018). Due to the physiological, ecological, and biogeochemical importance of DMS (O) (P), an improved understanding of the temporal dynamics of the microbial community that control DMSP cycling is crucial for understanding marine ecosystem function (Kiene et al., 2000; Kiene and Linn, 2000; Simo et al., 2002; Howard et al., 2006; Levine et al., 2012; Nowinski et al., 2019b).
The production of DMSP is widespread across marine eukaryotic species, including macroalgae, brackish plants, corals, and, most notably, marine phytoplankton (Van Diggelen et al., 1986; Keller, 1989; Van Alstyne and Puglisi, 2007; McParland and Levine, 2019). While DMSP biosynthesis is performed by a large diversity of phytoplankton, there is substantial variability in the amount of DMSP produced by different phytoplankton species (Keller et al., 1989; Caruana and Malin, 2014; McParland and Levine, 2019). Generally, it is recognized that prymnesiophytes and dinoflagellates are high DMSP producers (HiDPs), while diatoms and cyanobacteria are low DMSP producers (LoDPs) (Keller, 1989; McParland and Levine, 2019). Recent evidence has reported a significant correlation between previous measurements of high and low DMSP concentrations in marine phytoplankton isolates, with the presence of two recently identified DMSP biosynthesis genes, DSYB and TpMT2, respectively (Curson et al., 2018; Kageyama et al., 2018; McParland et al., 2021). Therefore, the expression of these separate DMSP biosynthesis genes may have an important role in determining differential DMSP production in marine phytoplankton.
A majority of DMSP produced by HiDP and LoDP phytoplankton is released into the marine environment where it is available for bacterial transformations (Simo et al., 2002). Heterotrophic bacteria can transform DMSP using two different DMSP degradation pathways (Curson et al., 2011). These include the DMSP lyase pathway, encoded by the ddd genes, which yields DMS (Curson et al., 2011; Sun et al., 2016; Li et al., 2021), and the DMSP demethylation pathway, which is encoded by the dmdA gene and allows for the assimilation of DMSP-derived carbon and sulfur, but, notably, does not produce DMS (Howard et al., 2008; Reisch et al., 2008; Dupont et al., 2012). DMSP degradation through these competing cleavage and demethylation pathways is widespread in marine bacterial communities, with diverse species able to mediate one or the other, or both pathways (Simó, 2001; Reisch et al., 2011; Moran et al., 2012; Varaljay et al., 2015; Nowinski et al., 2019a). The dmdA gene is the most abundant DMSP degradation gene (Landa et al., 2016) and is found in dominant bacterial lineages, including the SAR11, SAR86, SAR116, and Roseobacter clades (Howard et al., 2008; Reisch et al., 2008; Dupont et al., 2012). There are also eight known non-homologous ddd genes (Curson et al., 2011; Sun et al., 2016; Li et al., 2021). These genes have been identified in diverse lineages of bacteria, including members of the SAR116 and Roseobacter clade (Todd et al., 2011, 2012; Choi et al., 2015), and, most recently, with the discovery of dddK in the SAR11 clade (Sun et al., 2021). Of these genes, dddK has been reported as the most dominant in pelagic open water environments (Teng et al., 2021), while the most abundant in productive coastal waters is the Roseobacter and SAR116-associated dddP (Nowinski et al., 2019a).
Bacterial DMS oxidation transforms DMS to DMSO and is the primary process of DMS removal in marine surface waters (Kiene and Bates, 1990). DMS oxidation is performed by bacteria that possess one of three known enzymes, a multicomponent monooxygenase (DsoABCDEF), a DMS hydrogenase (DdhABC), and trimethylamine monooxygenase (Tmm) (Horinouchi et al., 1999; McDevitt et al., 2002; Chen et al., 2011). The most abundant gene encoding bacterial DMS oxidation is tmm (Teng et al., 2021), which requires methylamines to convert DMS to DMSO, is estimated to be found in 20% of all bacterial cells and is notably found in the SAR11 clade and Roseobacter group (Chen et al., 2011; Lidbury et al., 2016).
Somewhat intriguingly, it has recently been shown that some non-cyanobacterial marine bacteria, including the Alphaproteobacteria and Gammaproteobacteria (McParland and Levine, 2019), also have the capacity to synthesize DMSP (Curson et al., 2017). For example, the DMSP biosynthesis genes dsyB and mmtN have been identified in DMSP-producing isolates belonging to the marine Roseobacter clade and Actinobacteria (Curson et al., 2017; Williams et al., 2019; Liu et al., 2021; Sun et al., 2021). Bacteria that possess dsyB, which is more abundant in marine environments, have since been reported as important DMSP producers in coastal seawater, open ocean surface seawater, coastal sediments, the deep ocean, and the surface microlayer of the East China Sea (Williams et al., 2019; Sun et al., 2020, 2021; Zheng et al., 2020; Liu et al., 2021). These recent molecular insights have provided a transformative view of the role that bacteria play in the production and cycling of DMSP, but how the occurrence and ecological dynamics of these groups change seasonally is largely unestablished.
In coastal and open ocean environments, DMSP concentrations display marked seasonal variability (Kettle et al., 1999). Highest DMSP concentrations often occur in spring and are generally attributed to phytoplankton blooms (Stefels et al., 1995; Townsend and Keller, 1996; van Duyl et al., 1998; Simó and Dachs, 2002); however, DMSP concentrations are not always coupled to phytoplankton biomass (Townsend and Keller, 1996; Vila-Costa et al., 2008). Similarly, DMS concentrations also display seasonal trends, but, unlike DMSP, levels of DMS are often greatest in the summer (Simó and Dachs, 2002). Notably, there is often no clear linear relationship between DMSP and DMS concentrations, which has been attributed to differential DMS production among phytoplankton assemblages and bacterial degradation of DMSP (Levasseur et al., 1996). The abundance of bacterial genes encoding enzymes that catalyze DMS(P) degradation (e.g., dmdA, tmm, and dddP) exhibits wide geographical distributions and is detected in tropical to polar environments (Teng et al., 2021). These genes have also been shown to significantly vary in abundance over time (Nowinski et al., 2019a), although their seasonal dynamics, relationships with environmental DMS(P) levels, and links to bacterial assemblage structure have not been examined in detail.
Oceanographic time series have monitored DMSP and DMS concentrations in diverse open-ocean and coastal sites (North Sea, Atlantic Ocean, Pacific Ocean, Indian Ocean, Southern Ocean, Baltic Sea, and Mediterranean Sea) (Dacey et al., 1998; Shenoy and Patil, 2003; Vila-Costa et al., 2008; Levine et al., 2012; Varaljay et al., 2012, 2015; Zhao et al., 2021). Although, with a recently improved understanding of the molecular mechanisms underpinning DMSP production and degradation, a combination of biogeochemical and molecular ecology approaches applied to ocean time series will deliver an even greater capacity to elucidate how the microbial community influences variability in DMS (O) (P) over time. Here, we describe a 2-year DMSP time-series study, conducted at an oceanographic station located on the eastern Australian continental shelf. Measurements of DMS (O) (P) concentrations were combined with measurements of DMS production rates (DMSP lyase assays), quantification of bacterial DMSP cycling genes, and analysis of the diversity of the phytoplankton and bacterial communities involved in DMSP cycling. By integrating this diverse suite of measurements, we aimed to identify the ecological relationships involved in DMSP cycling, with a specific focus on the genetic potential for microbes to cycle DMSP.
Seawater samples were collected monthly (February 2017–January 2019) from the Australian Integrated Marine Observing System (IMOS) National Reference Station (NRS) located at Port Hacking (34° 07.06 S, 151° 13.09 E). This long-running oceanographic time-series site is situated 7 km offshore, near the city of Sydney, Australia (population, 4.3 million). Triplicate 1-L samples were collected in autoclaved and acid-washed 1-L polycarbonate bottles from 1 m below the surface, filled ensuring no headspace, and kept in the dark to avoid photo-oxidation in a cooler during transportation to the laboratory for analysis. These samples were used for characterization of dimethyl sulfide (DMS), dimethylsulfoniopropionate (DMSP), dimethyl sulfoxide (DMSO), DMSP lyase enzyme activity, and chlorophyll a levels. For microbial community analysis, an individual 2-L sample (n = 1) was collected from the same depth and immediately filtered onto a 0.22-μm polyethersulfone membrane filter (Millipore® Sterivex™) using a peristaltic pump (Watson-Marlow). For quantitative PCR (qPCR) of DMSP cycling genes, a set of triplicate 2-L samples (n = 3) was also collected from the same depth and filtered onto a 0.22-μm polycarbonate membrane filter (Millipore®) using the same methods. All filters were transported on ice in a cooler during transportation before being snap-frozen in liquid nitrogen within 4 h, and then stored at −80°C until processing.
Physicochemical data, including sea-surface temperature (°C) and salinity (PSU), dissolved oxygen (μmol L–1), and inorganic nutrients: nitrate/nitrites (μmol L–1), orthophosphate (μmol L–1), and silicate (μmol L–1), were retrieved from the IMOS curated Australian Ocean Data Network Portal1 (Supplementary Table 1).
Chlorophyll a (chl a) fluorescence (μg L–1) was measured using JGOFS protocols (Knap et al., 1996) with slight modification. Triplicate 300-ml surface water volumes were gently vacuum filtered onto a 0.7-μm glass fiber filter (GF/F, 25-mm diameter) to collect cells, which were submerged face down in a glass vial containing 3 ml of 90% acetone. The samples were vortexed and stored in the dark at −20°C for 24 h for extraction. One ml of a sample was loaded into a glass cuvette, and chl a concentration was measured using the chl a NA module in a Fluorometer (Turner Designs, Trilogy, Sunnyvale, CA, United States). A blank of 90% acetone was run to ensure background of fluorescence was at a minimum and chl a concentration was calculated using a standard curve (R2 = 0.99) made with chl a standard (Sigma-Aldrich C5753, St. Louis, MO, United States).
All dimethylated sulfur samples analyzed by gas chromatography were processed immediately after transportation from the vessel to the laboratory (within 4 h). The DMS samples were prepared by transferring 2 ml of unfiltered seawater into a 14-ml headspace vial that was capped (with a butyl rubber septum) and crimped (an aluminum crimp cap) before immediate headspace analysis. Dissolved DMSP (DMSPd) concentrations were prepared by gravity filtering no more than 3-ml seawater onto a 0.7 μm (nominal pore size) Whatman GF/F (25-mm diameter) to remove cells, while minimizing cell rupture (Kiene and Slezak, 2006). It should be noted the nominal pore size used to filter DMSPd does not exclude all bacteria and, as a result, could contain a small but likely negligible amount of bacterial particulate DMSPp (DMSPp). The first 2 ml of filtrate was collected in a 14-ml headspace vial before alkaline hydrolysis with 0.75-M NaOH. The sample was immediately capped, sealed, and left to rest for complete hydrolysis and equilibrium (at least 12 h) prior to analysis. To sample for total DMSP (DMSPt), 2 ml of unfiltered seawater was hydrolyzed with NaOH (0.75 M) before capping and sealing vials immediately, allowing sufficient time (at least 12 h) for total conversion of DMSPt to DMS and equilibrium before analysis. DMSPp was calculated as the difference between DMSPt and DMSPd.
Following DMSPt analysis, alkaline samples were uncapped and purged for 10 min with high purity nitrogen gas at a flow rate of 60 ml min–1 to remove any volatile sulfur compounds remaining from alkaline treatment. The samples were neutralized by adding 80 μL of 32% HCl, and DMSO was converted to DMS by adding 350 μL of 12% TiCl3 solution and immediately capped following previously described methods (Kiene and Gerard, 1994; Deschaseaux et al., 2014, 2019). Vials were immersed in a water bath at 50°C for 1 h and cooled to room temperature prior to purge-and-trap analysis on the GC-FPD as described below.
Analyses of sulfur compounds were performed on a gas chromatograph (GC-2010 Plus, Shimadzu, Japan), coupled with a flame photometric detector (FPD) set at 160°C with hydrogen and air flow rates at 40 and 60 ml min–1, respectively. A purge-and-trap methodology was used to analyze samples (Simo et al., 1993). Briefly, samples were sparged with high purity helium (He) to purge all volatile gas (including DMS) from the sample while trapping the DMS in a PTFE loop immersed in liquid nitrogen. After 4 min of cryotrapping, the loop was heated in warm water, allowing the DMS to desorb before injection into the GC. DMS was eluted onto a capillary column (30 m × 0.32 mm × 5 μm) heated to 130°C, using He as the carrier gas with a flow rate of 12 ml min–1 and a split ratio of five. Quantification of DMS was performed by integrating the peak area against a seven-point calibration curve of known DMS concentrations (1 pmol to 200 pmol).
The DMSP lyase activity (DLA) assay is used as a proxy for the activity of the phytoplankton and bacterial DMSP lyase pathway and provides a rate of DMS production from DMSP (Harada et al., 2004; Bell et al., 2007; Levine et al., 2012). DLA assays were performed via direct injection of 100 μL of headspace (column flow: 3.66 ml min–1) from two fractions (Levine et al., 2012): phytoplankton (>2.0 μm) and bacteria (0.22–2.0 μm) following the methods described in Harada et al. (2004). While these fractions are hereafter named phytoplankton and bacteria, it should be acknowledged that these are operational definitions only. The fractionation process does not guarantee the strict separation of both groups, whereby phytoplankton cells smaller than 2.0 μm could be collected in the bacteria fraction, and, alternatively, attached bacteria may be present in the phytoplankton fraction. Phytoplankton DLA (DLAp) assays were prepared by gentle vacuum filtration (<0.02 Pa) of 600-ml bulk seawater onto an autoclaved 2.0-μm polycarbonate filter (25-mm diameter). From the remaining filtrate, bacterial DLA assays were prepared by gentle filtration (<0.02 Pa) of 300 ml onto a 0.22-μm polycarbonate filter (25-mm diameter). All DLA samples were snap-frozen in liquid N2 and stored at −80°C until analysis. Prior to analysis, filters were thawed slowly on ice and then transferred facedown into a glass vial in 1 ml of a TRIS buffer (pH 8.0), capped with a rubber stopper, and vortexed for 10 s. After a 20-min incubation in a 20°C water bath, 20 μL of DMSP-HCl (Sigma-Aldrich, United States) was added to a final concentration of 5 mM, and the vial sealed and crimp capped following previously described methods (Harada et al., 2004; Sheehan and Petrou, 2020; Fernandez et al., 2021). While the addition of 5-mM DMSP is substantially higher than those observed in the environment (Galí and Simó, 2015), we have adopted the widely used methodology of Harada et al. (2004) to allow for inter-study comparison. DMSP lyase activity was then determined via direct injection of 100 μL of headspace (column flow: 3.66 ml min–1) as previously described (Harada et al., 2004; Sheehan and Petrou, 2020; Fernandez et al., 2021). All DLA assays were performed with a control (a TRIS buffer without DMSP addition) and a procedural control (a TRIS buffer with 5-mM DMSP addition). It should be noted that the procedural control of high substrate addition of DMSP (5 mM) and the alkaline TRIS buffer (pH 8.0) produced a measurable signal of DMS during the assays; these non-biological signals of DMS generation were deducted from the assay to calculate the microbial DLA for >2.0 μm and 0.22–2.0-μm fractions of seawater.
DNA was extracted from filters using a modified application of the PowerWater® DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, United States, now Qiagen) (Appleyard et al., 2013). Bacterial and eukaryotic assemblages were characterized using 16S rRNA and 18S rRNA gene sequencing, respectively. For 16S rRNA sequencing, the V1–V3 region of the bacterial 16S rRNA gene was amplified using the 27F (AGAGTTTGATCMTGGCTCAG) (Lane, 1991) and 519R (GWATTACCGCGGCKGCTG) primer pairing (Lane et al., 1985) under the following thermocycling conditions: 95°C for 10 min; 35 cycles of 94°C for 30 s, 55°C for 10 s, and 72°C, followed by a final extension at 72°C for 10 min. The V4 region of the 18S rRNA gene was amplified using the TAReuk454FWD1 (CCAGCASCYGCGGTAATTCC) and a modified TAReuk-Rev3 (ACTTTCGTTCTTGATYRATGA) primer (Piredda et al., 2017), designed to be less discriminant against Haptophytes than the original TAReuk-Rev3 primer (Stoeck et al., 2010). Amplification was performed using the following thermocycling conditions: 98°C for 30 s; 10 cycles of 98°C for 10 s, 44°C for 30 s, and 72°C for 15 s; 20 cycles of 98°C for 10 s, 62°C for 30 s, and 72°C, followed by a final extension at 72°C for 7 min. All 16S and 18S rRNA amplicons were subsequently sequenced using the Illumina MiSeq platform at the Ramaciotti Centre for Genomics at the University of New South Wales.
Raw paired end reads for bacterial 16S and eukaryotic 18S rRNA genes were downloaded from Australian Microbiome Initiative data portal2 in August 2020. Low-quality reads with “N” bases were removed, and then forward and reverse primers were removed from sequences using cutadapt (Martin, 2011), and reads were truncated to eliminate low-quality terminal bases (16S truncated at R1 = 255, R2 = 250, 18S truncated at R1 = 250, R2 = 228), dereplicated, denoised, and merged using pseudo pooling. Then chimera removal was performed using dada2 removeBimeraDenovo (Callahan et al., 2016), and identical sequences of differing lengths were combined using the dada2 collapse no-mismatch step (the full pipeline available here: https://github.com/martinostrowski/marinemicrobes/tree/master/dada2). Bacterial ASVs were taxonomically classified using the SILVA v132 database, with a 50% Bayesian probability cut-off (Wang et al., 2007; Yilmaz et al., 2014), and eukaryotic ASVs were classified using the Protist Ribosomal Reference Database (PR2) (Guillou et al., 2012). A summary of all accession numbers, data availability, and number of reads per sample are available in Supplementary Table 2.
Photosynthetic protists were identified as 18S rRNA gene sequences assigned as Chlorophyta, Dinophyta, Cryptophyta, Haptophyta, Ochrophyta, Cercozoa, Syndiniales, and Sarcomonadea by PR2 taxonomy (Vaulot et al., 2021) and were extracted from the Eukaryotic dataset, resulting in a subset of 10,875 18S ASVs. A curated bioinformatic pipeline was used to classify these ASVs as potential DMSP producers by incorporating previous measurements of cellular DMSP production in monocultures (58) to assign their putative ability to produce DMSP based on phylogenetic inference (the full pipeline available here: https://doi.org/10.5281/zenodo.5090864). First, full-length 18S sequences of isolates with previously reported intracellular DMSP concentrations (n = 107) were collected from NCBI, including diverse taxa from Chlorophyta, Dinophyta, Haptophyta, Ochrophyta, Pelagophyta, Rhizaria, and Rhodophyta. The 18S rRNA gene sequences were aligned with the eukaryotic small subunit ribosomal RNA Rfam (RF01960) using Infernal (v 1.1) (Nawrocki and Eddy, 2013) in order to build a reference phylogeny with RAxML (v 8.0) (Stamatakis, 2014) using the GTRGAMMA model. A second alignment of the 10,875 unique ASVs was created with Infernal, and then pplacer (Matsen et al., 2010) was used to place ASVs onto the reference phylogeny. The ASVs that had significant sequence similarity (posterior probability of 90%, likelihood <−4,000), with an isolate previously identified to produce DMSP, were assumed to be DMSP producers. These “DMSP-producing ASVs” were further categorized as low DMSP producers (LoDP) or high DMSP producers (HiDP) based on SILVA taxonomic assignment (at the genus level), matching isolates with previously measured intracellular DMSP concentrations of less than or greater than 50-mM DMSP, respectively (McParland and Levine, 2019). While these putative assignments are a good representation of our understanding of intracellular DMSP concentrations in marine phytoplankton, it should be noted that, currently, it is impossible to predict these assignments with absolute certainty, as there are exceptions of HiDP and LoDP genera, having less than or greater than 50-mM concentrations, respectively. If a DMSP-producing ASV was not of the same genus (based on SILVA taxonomy) as a known DMSP-producing isolate, then it was defined as a likely producer with unknown DMSP production potential. A summary of how many predicted HiDP and LoDP ASVs were per sample is available in Supplementary Table 2.
Quantitative PCR (qPCR) was used to determine the total abundance of bacterial 16S rRNA genes and genes involved in marine DMSP cycling. All qPCR analyses were performed using an epMotion 5075l automated Liquid Handling System on a Bio-Rad CFX Touch Real-Time PCR Detection System. All sample plates included a triplicate, six-point calibration curve constructed from a known amount of amplicon DNA measured by Qubit (according to the manufacturer’s instructions), followed by five successive 10-fold dilutions and negative controls of nuclease-free water. Absolute quantification of DMSP cycling genes encoding DMSP catabolism dddP (Levine et al., 2012) and dmdA (A/1 (Roseobacter), D/all (SAR11 clade), subclade (Varaljay et al., 2010), bacterial DMS oxidation (Lidbury et al., 2016), and the bacterial DMSP biosynthesis gene, dsyB (Williams et al., 2019) was performed using primers and annealing temperatures listed in Table 1. All assays incorporated technical triplicates of the following mixture: 2.5 μL of 2X SensiFAST SYBR Hi-ROX Master Mix, a 0.2-μL, 10-μM forward primer; a 0.2-μL, 10-μM reverse primer; 0.1-μL nuclease-free water, and 2 μL of neat DNA template. Quantification of DMSP cycling genes consisted of an initial denaturation step of 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, the specified annealing temperature for each gene in Table 1 for 30 s and 72°C for 30 s. To differentiate specific amplicons from non-specific products, a dissociation melt curve was generated after each reaction.
Quantification of the bacterial 16S rRNA gene was performed using an assay adopted from Suzuki et al. (2000). Each individual PCR reaction volume was 5 μL and contained 2.5 μL of iTaq Universal probes SMX (Bio-Rad), 0.1-μL TaqMan Probe Mix, TM1389F (5′–CTTGTACACACCGCCCGTC–3′), and 0.2-μL, 10-μM concentrations of 16S rRNA gene specific primers, BACT1369F (5′–CGGTGAATACGTTCYCGG–3′) and PROK1492R (5′–GGWTACCTTGTTACGACTT–3′). Quantitative PCR was performed with the following cycling conditions: 95°C for 3 min, followed by 39 cycles of 95°C for 30 s and 56°C for 60 s. The relative abundance of bacterial DMSP-degrading genes was acquired by normalizing their copy numbers to the copy number of the bacterial 16S rRNA gene, although it should be noted that some bacterial genomes have multiple copies of the 16S rRNA gene (Cui et al., 2015).
To test for differences in DMS (O) (P) concentrations over time and for differences in the abundance of bacterial DMSP cycling genes, the Kruskal–Wallis (KW) tests with the Bonferroni-corrected post hoc tests were performed using SPSS version 17.0 (SPSS Statistics, Inc., Chicago, IL, United States). Pearson’s correlations were performed to test for significant positive and negative correlations between biogeochemical measurements [DMS(P) and DLA], molecular measurements (16S/18S rRNA gene and qPCR) and environmental parameters. When comparing interdisciplinary variables, all data were log transformed before analyses to reduce error introduced by comparing variables with different units.
Environmental conditions at Port Hacking exhibited clear seasonal patterns throughout the time series, from February 2017 to January 2019. Sea surface temperature peaked in the austral autumn, specifically during April 2017 (23.9°C) and March 2018 (23.0°C) (Supplementary Table 1). Salinity ranged from 35.1 to 35.8 PSU and was consistently greater than 35.5 PSU between May and September in both 2017 and 2018, with the lowest salinity measured in March 2017 (Supplementary Table 1). Nutrient levels displayed clear temporal patterns, with highest levels of both nitrate/nitrite (NOx2–) and phosphate (PO34–), occurring between June and September in both years, with peak concentrations in September 2017 (3.2 μmol L–1 NOx2– and 0.3 μmol L–1 PO34–) (Supplementary Table 1). The highest concentrations of silicate (SiO32–) occurred between March and September in both years, with peak concentrations occurring in March 2017 (1.7 μmol L–1) (Supplementary Table 1). Average chlorophyll a (chl a) concentrations were 1.6 ± 0.29 μg L–1 (average ± SE), but levels of this proxy for phytoplankton biomass displayed significant shifts over time (KW test = 58.6, df = 20, p < 0.01), with highest levels occurring during three phytoplankton bloom events in March 2017 (4.3 ± 0.36 μg L–1), October 2017 (4.4 ± 0.96 μg L–1), and September 2018 (4.6 ± 0.16 μg L–1) (Supplementary Table 1, q < 0.05).
Clear seasonal shifts were apparent in the concentrations of total dimethylsulfoniopropionate (DMSPt, Figure 1A, KW test = 56.1, df = 20, p < 0.01), particulate DMSP (DMSPp, Figure 1B, KW test = 57.8 df = 20, p < 0.01), and dissolved DMSP (DMSPd, Figure 1C, KW test = 54.8 df = 20, p < 0.01). DMSPt concentrations averaged 25.7 ± 5.31 nM but increased significantly during the springtime (q < 0.05), whereby annual peak concentrations were measured in October 2017 (118 ± 11.8 nM) and November 2018 (64 ± 12.8 nM) (Figure 1A). Patterns in DMSPp and DMSPd concentrations were also characterized by significant seasonal increases (q < 0.05) in October 2017 (93 ± −14.9-nM DMSPp, 25 ± −5.6-nM DMSPd) and November 2018 (36 ± −6.0-nM DMSPp, 28 ± −6.4-nM DMSPd) (Figures 1B,C). Concentrations of the volatile sulfur compound, dimethyl sulfide (DMS), averaged 2.7 ± 0.30 nM throughout the time series (Figure 1D). Temporal patterns in DMS concentrations were not as clear as those observed for DMSP, and, notably, DMS peaks did not coincide with peaks of DMSP; instead, they occurred in the Austral summer months of February 2017 (5 ± 0.4 nM) and December 2018 (4 ± 0.2 nM) (Figure 1D). Concentrations of total dimethyl sulfoxide (DMSOt) averaged 15.1 ± 3.97 nM and demonstrated significant shifts over time (KW test = 59.2, df = 20, p < 0.01). Peak concentrations of DMSOt coincided with peak DMSPt in 2017, whereby DMSOt concentrations were as great as 74.8 ± 2.17 nM in October 2017 (Figure 1E).
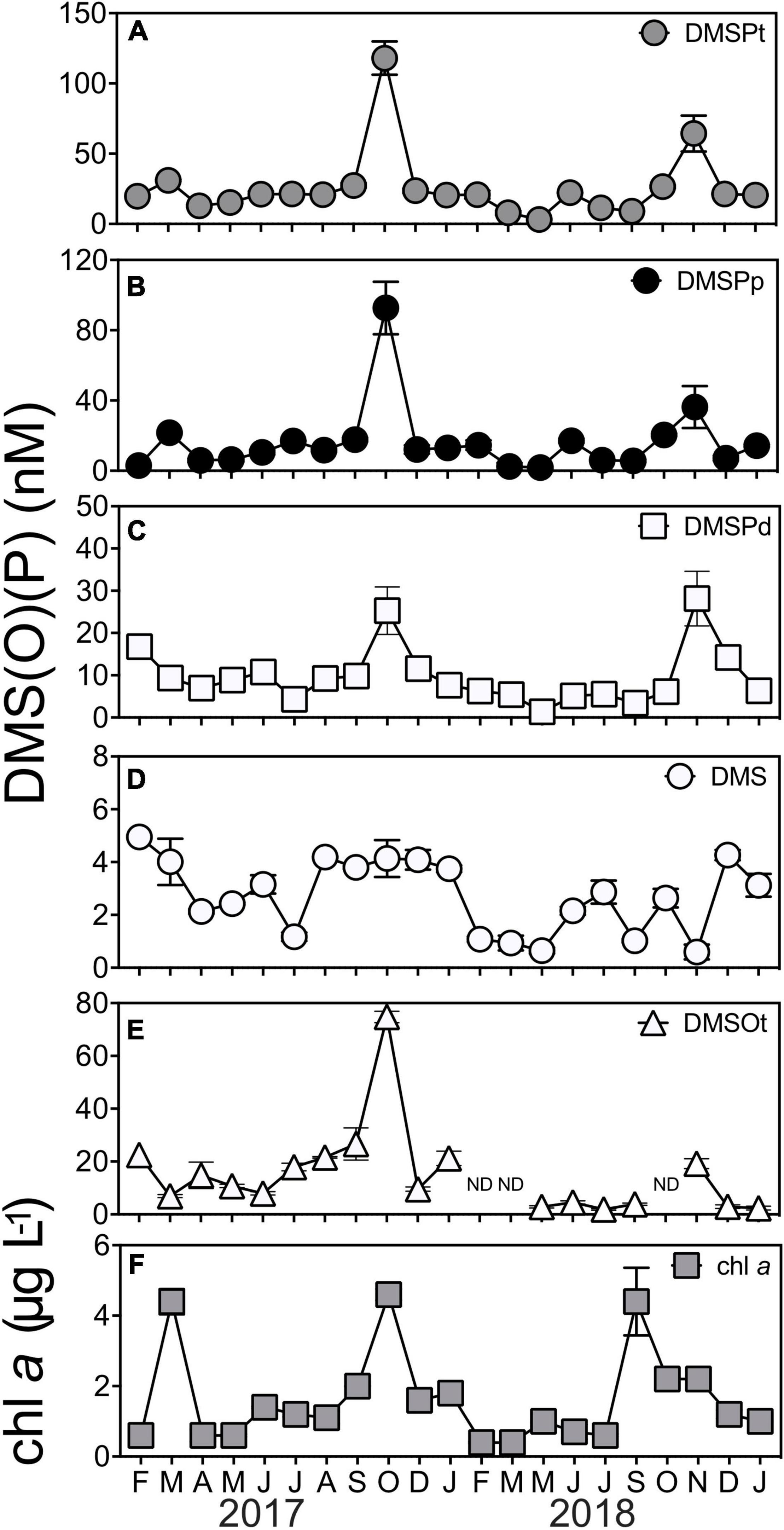
Figure 1. Concentrations of marine sulfur compounds (A) total DMSP (DMSPt) (B) particulate DMSP (DMSPp), (C) dissolved DMSP (DMSPd), (D) dimethyl sulfide (DMS), (E) total DMSO (DMSOt), and (F) chlorophyll a (chl a) at Port Hacking NRS between February 2017 and January 2019. DMS (P) data are means ± standard error (n = 3). ND, non-detected, indicating concentrations were below the detection limit of 1 pmol.
Overall, every fraction of DMSP (DMSPt, DMSPp, and DMSPd) was significantly correlated to each other (Supplementary Table 3). A correlation between chl a with these fractions (DMSP, DMSPp, and DMSPd) was also detected across the duration of the time series (Supplementary Table 3), emphasizing the importance of phytoplankton biomass to DMSP cycles in the time series. This was exemplified by a high chl a/high DMSP event in October 2017, where peak concentrations of chl a and DMSPt were recorded at 4.6 ± −0.16-μg L–1 chl a and 118 ± −11.8-nM DMSPt (Figure 1F). However, despite the significant correlation found between DMSPt and chl a (Pearson’s r = 0.48, p < 0.05, Supplementary Table 3), not all high chl a events (defined as those >4-μg L–1 chl a) were associated with DMSP concentrations as high as October 2017. Indeed, two high chl a/low DMSP events were captured in the time series in March 2017 and September 2018 when phytoplankton blooms occurred (Figure 1F, as indicated by high chl a concentration), but, at these times, DMSPt levels were only 31 ± −0.7-nM and 27 ± −1.1-nM DMSPt, respectively (Figure 1A). Notably, during November 2018, an event characterized by low chl a level (2 ± 0.1 μg L–1) was measured to have 2-fold higher concentrations of DMSP (64 ± −12.8-nM DMSPt) than those measured during the high chl a/low DMSP events seen in March 2017 and September 2018 (Figures 1A,F). As well as phytoplankton biomass (chl a), salinity was significantly correlated with concentrations of DMSPt (Pearson’s r = −0.45, p < 0.05, Supplementary Table 3) and DMSPp (Pearson’s r = −0.44, p < 0.05, Supplementary Table 3), whereby higher levels of DMSP were associated with lower salinity measured during the time series.
No significant correlations were detected between any of the fractions of DMSP with its degradation product, DMS, or with any environmental variables (temperature, salinity, NOx2–, PO34–, or SiO32–). Additionally, there was no relationship between DMS and chl a, or any other environmental variable (temperature, salinity, NOx2–, PO34–, or SiO32–) (Supplementary Table 3). DMSOt concentrations were significantly positively correlated with DMSPt and DMSPd, but no significant correlation was found between DMSOt with DMS, DMSPp or any environmental variable measured throughout the 2-year time series (Supplementary Table 3).
Given the significant correlations between DMSPp and chl a, we next examined the phytoplankton community dynamics, underpinning this relationship by using 18S rRNA gene amplicon sequencing. This analysis included a consideration of the relative occurrence of predicted high DMSP-producing (HiDP) and low DMSP-producing (LoDP) phytoplankton. After characterization of putative HiDP and LoDP sequences, no significant correlation between HiDP relative abundance and chl a was found, although a strong significant positive correlation existed between LoDP relative abundance and chl a concentration (Pearson’s r = 0.73, p < 0.01, n = 21). Notably, there was a significant positive correlation between the relative abundance of predicted HiDPs and DMSPt concentration (Figure 2A, Pearson’s r = 0.61, p < 0.01, n = 21), whereas no significant correlation was evident between predicted LoDP relative abundance with DMSPt concentration (Figure 2B, Pearson’s r = 0.265, p > 0.05, n = 21), suggesting LoDP phytoplankton has a smaller influence on bulk DMSP concentrations than HiDP phytoplankton.
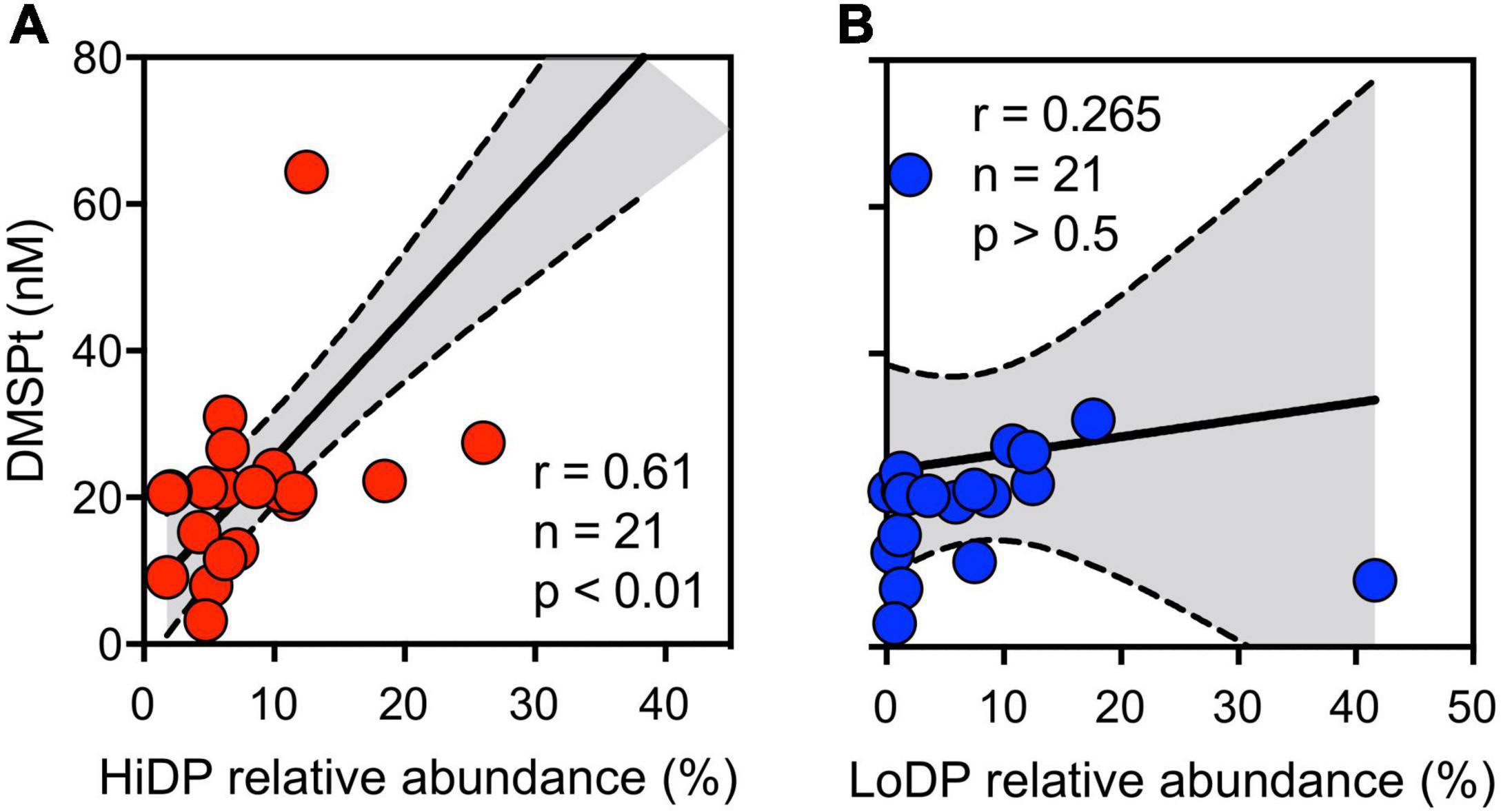
Figure 2. Concentration total DMSP (DMSPt), with the relative abundance of sequences putatively annotated as (A) high-DMSP-producing (red) and (B) low-DMSP-producing phytoplankton (blue) at Port Hacking NRS between February 2017 and January 2019. Pearson’s r indicates a significant correlation between variables.
The phytoplankton community associated with the high chl a/high DMSP event in October 2017 was dominated by HiDPs (48% of all 18S rRNA gene sequences) (Figure 3), with a much smaller contribution (13%) from LoDPs (Figure 3). More specifically, the phytoplankton community (the 18S phototrophic community) at this time was dominated by HiDP dinoflagellate genera, including Heterocapsa (24%), Prorocentrum (16%), Alexandrium (14%), and the picoeukaryote, Picochlorum (32%) (Figure 3).
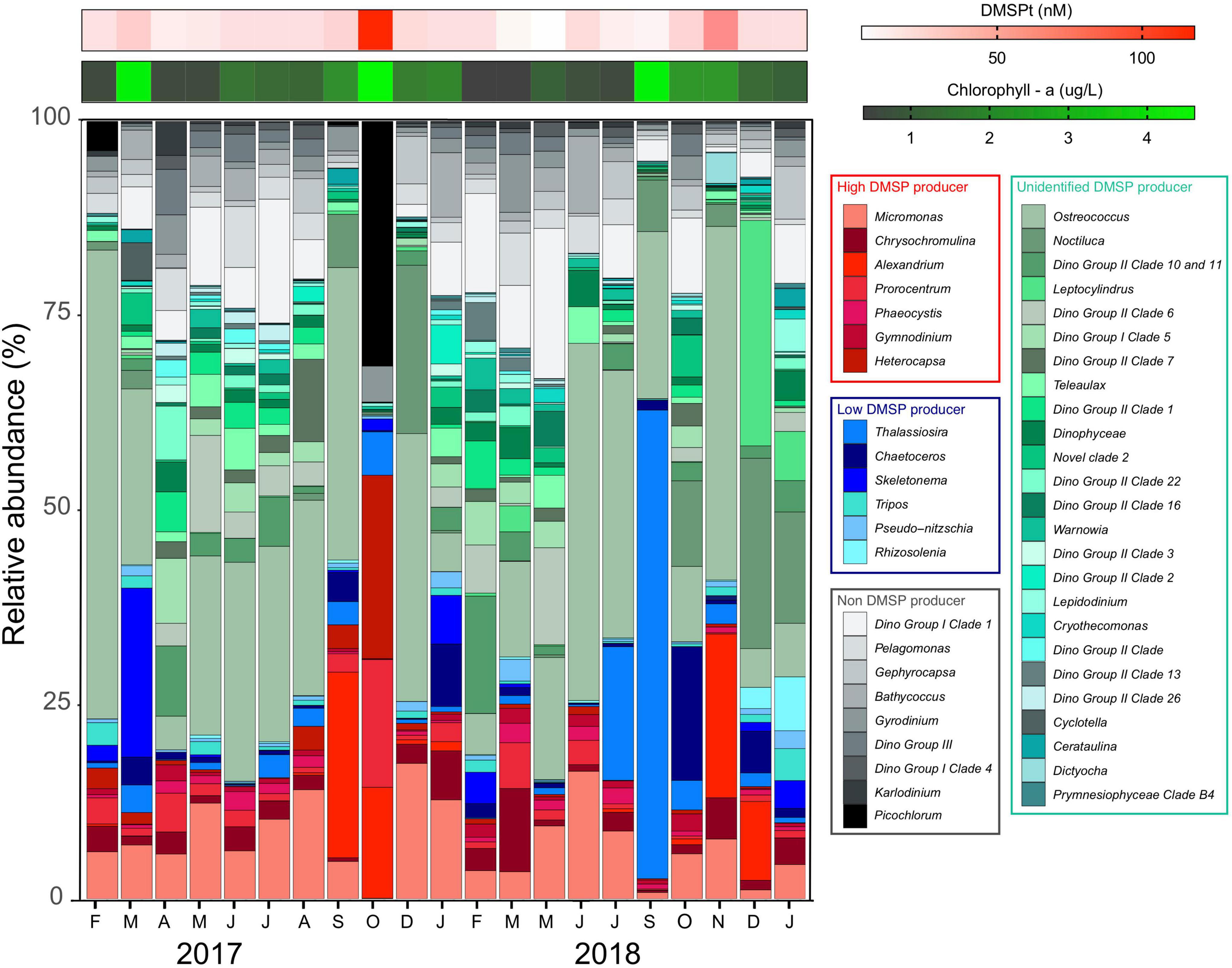
Figure 3. Relative abundance of eukaryotic phototrophs at Port Hacking NRS between February 2017 and January 2019. Phytoplankton taxa assigned as known high DMSP producers (red colors), known low DMSP producers (blue colors), likely producers with unknown DMSP production potential (green colors), and non-DMSP producers (gray-scale colors). Heatmaps represent concentration of total DMSP (red) and chl a (green).
In contrast to the patterns observed in October 2017, high chl a/low DMSP events in March 2017 and September 2018 were dominated by LoDPs (17 and 41% of all 18S rRNA gene sequences, respectively) (Figure 3), with a low relative abundance of HiDPs (6 and 2%, respectively) (Figure 3). The LoDP-dominated communities at these times were comprised of high relative abundances of diatoms, including Skeletonema (21% of 18S phototrophic sequences) in March 2017 and Thalassiosira (60% of 18S phototrophic sequences) in September 2018 (Figure 3).
Further evidence that suggests the abundance of predicted HiDP phytoplankton may be influencing DMSPt concentrations was demonstrated by the low chl a/high DMSP event in November 2018, whereby HiDPs and LoDPs were made up 12 and 2% of all 18S rRNA gene sequences, respectively (Figures 2A,B). The phytoplankton community during this event was dominated by Ostreococcus (46%) and a high relative abundance of the HiDP dinoflagellate, Alexandrium (21%) (Figure 3).
The DMSP lyase activity (DLA) assay is used as a proxy for the activity of the enzyme responsible for the cleavage of DMSP to DMS (Harada et al., 2004; Bell et al., 2007; Levine et al., 2012). Assays of operationally defined fractions of phytoplankton (DLAp, >1.2 μm) and bacteria (DLAb, 0.2–1.2 μm) revealed DLAp was significantly positively correlated with DMSPp; however, no significant correlations existed between DMSPd, DMSOt, chl a, and environmental variables (temperature, salinity, NOx2–, PO34–, or SiO32–) (Supplementary Table 4). Meanwhile, DLAb was not significantly correlated with DMSPp, DMSPd, DMSOt, chl a or environmental variables (temperature, salinity, NOx2–, PO34–, or SiO32–) (Supplementary Table 4). Notably, a significant positive correlation was detected between DLAp and DLAb with DMS (Supplementary Table 4), which demonstrates a direct link between the fractions of microbial DMSP lyase activity with its breakdown product.
Although no clear seasonal trend was evident in the rate of DLAp, significant differences over time were present (KW test = 53.1, df = 20, p < 0.01), with highest activity (2.5 ± −0.12-nM DMS min–1) measured during the high chl a/high DMSP event in October 2017 and high chl a/low DMSP event in March 2017 (1.9 ± −0.40-nM DMS min–1) (Figure 4A). Additionally, rates of DLAb significantly shifted during the time series (KW test = 48.9, df = 20, p < 0.01) (Figure 4B), where, in particular, the rates measured during high chl a/low DMSP event in March 2017 (0.05 ± −0.001-nM DMS min–1) significantly exceeded those recorded in the low chl a/high DMSP event in November 2018 (0.004 ± −0.0004-nM DMS min–1) (q = 0.38).
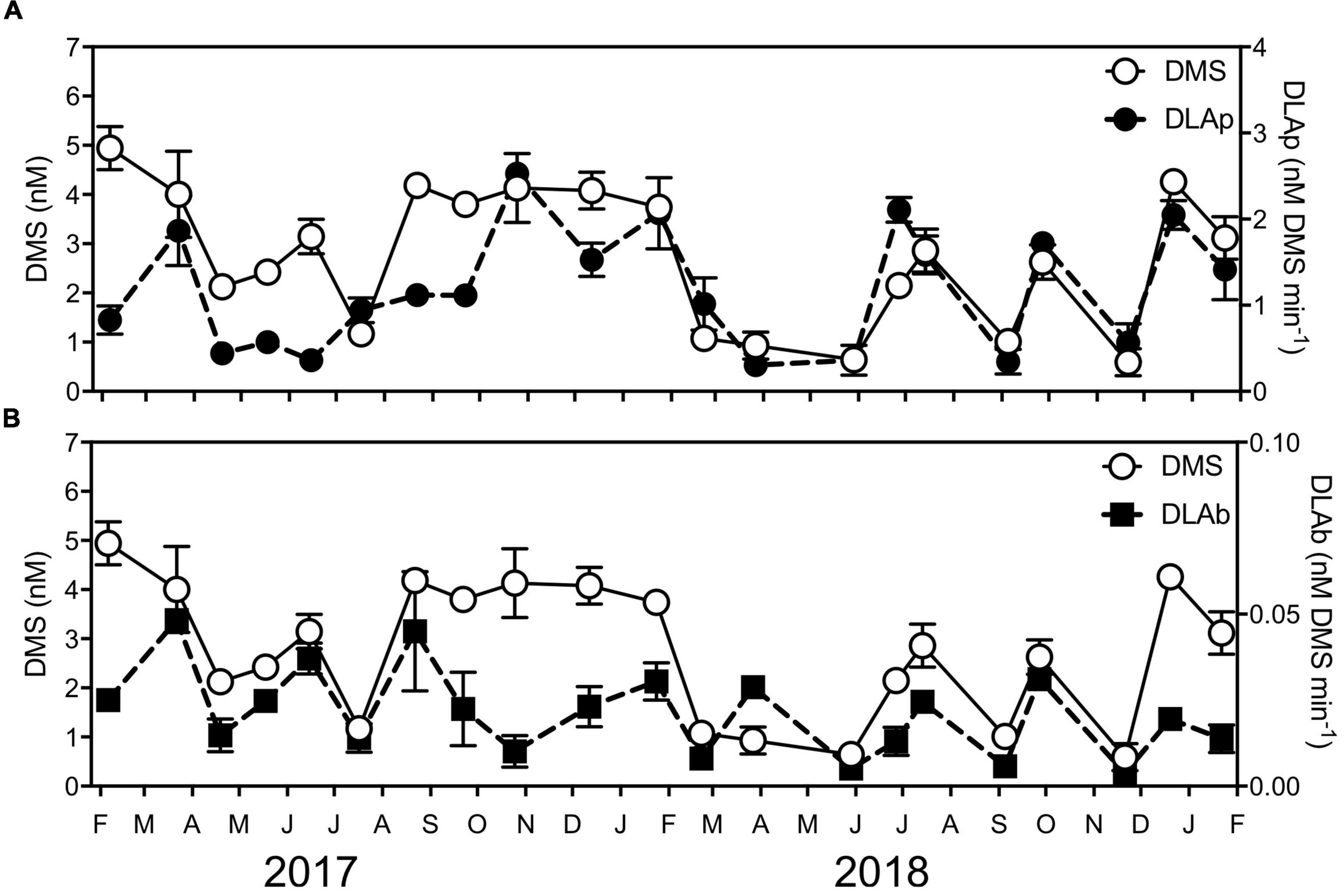
Figure 4. Concentrations of DMS and DMSP lyase activity (DLA) of (A) phytoplankton (DLAp) and (B) bacteria (DLAb) at Port Hacking NRS between February 2017 and January 2019. Data are means ± standard error (n = 3).
The dominant bacterial groups during the time series (indicated by relative abundance) were the SAR11 clade (18 ± 2.1%), Flavobacteriales (14 ± 2.1%), Synechococcales (8 ± 1.6%), Rhodobacterales (8 ± 1.2%), and the SAR86 clade (7 ± 1.2%) (Figure 5). Among the top 20 orders of bacteria, there were no significant positive relationships with any of the dimethylated sulfur compounds or chl a detected, with the notable exception of significant positive correlations between the relative abundance of Rhodobacterales and DMSPd (Pearson’s r = 0.57, p < 0.01, n = 21, Supplementary Table 5). Highest relative abundances of Rhodobacterales (∼10–27%) were evident in the austral spring to summer periods. In November 2018, during the low chl a/high DMSP event, Rhodobacterales dominated the bacterial community, comprising 28% of all 16S rRNA gene sequences (Figure 5). At this time, the relative abundance of Rhodobacterales was 6.6-fold, 2.8-fold, and 2.7-fold higher than during the high chl a/low DMSP event in March 2017, the high chl a/high DMSP event in October 2017, and the high chl a/low DMSP event in September 2018, respectively (Figure 5).
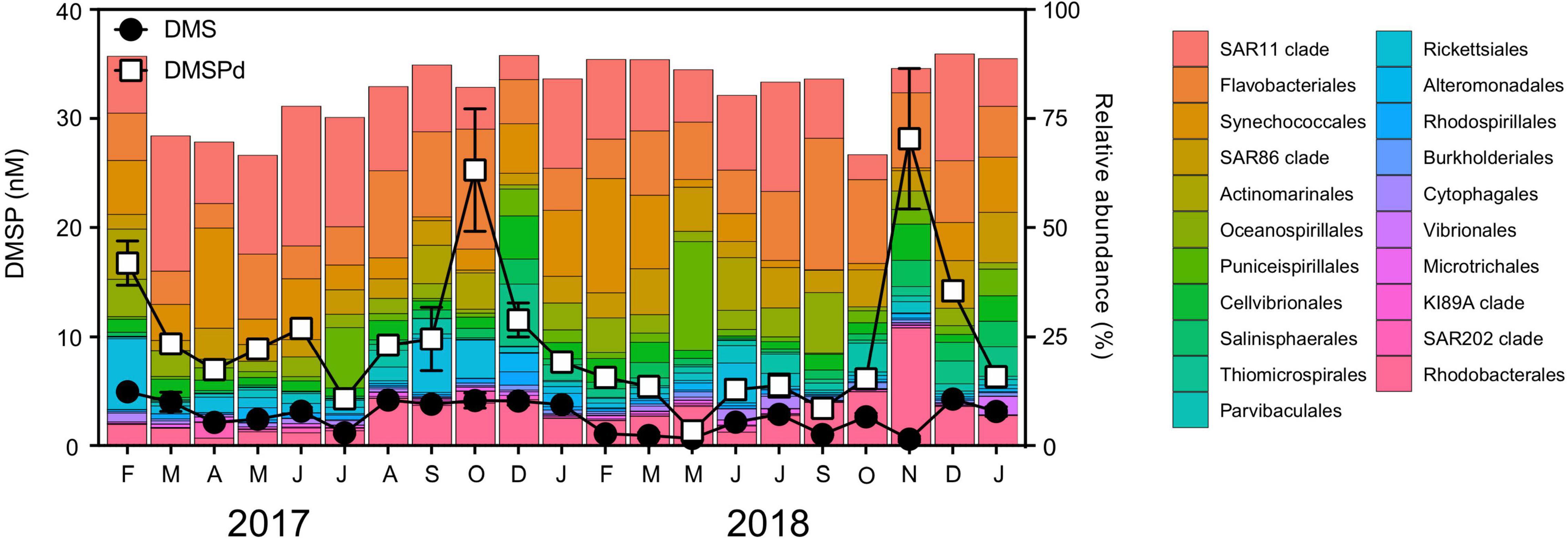
Figure 5. Relative abundance of the bacterial community at Port Hacking NRS between February 2017 and January 2019. Line scatter plots show nM concentrations of dissolved DMSP (DMSPd) and dimethyl sulfide (DMS) measured during the time series. DMS (P) data are means ± standard error (n = 3).
To further explore the significant increases in relative abundance of Rhodobacterales during the November 2018 DMSPd peak, we examined the dynamics of the 25 most dominant Rhodobacterales ASVs. Notably, just 5 Rhodobacterales ASVs collectively made up 21% of all 16S rRNA gene sequences in November 2018. These ASVs included HIMB11 sp. (Bc1000003) (7%), Ascidiaceihabitans sp. (Bc1000035, previously described as Ascidiaceihabitans sp. z2239) (6%), HIMB11 sp. (Bc1000116) (3%), Amylibacter sp. (Bc1000011, previously described as Amylibacter z3093) (3%), and Planktomarina sp. (Bc1000120, previously described as Planktomarina sp. z3603) (2%) (Supplementary Figure 1). Of the 5 dominant ASVs, none were significantly correlated with chl a throughout the time series; however, the relative abundance of Ascidiaceihabitans sp. (Bc1000035), HIMB11 sp. (Bc1000003), HIMB11 sp. (Bc1000116), and Planktomarina sp. (Bc1000120) was significantly positively correlated with DMSPd (Supplementary Figure 1). Three of these dominant ASVs [Ascidiaceihabitans sp. (Bc1000035), HIMB11 sp. (Bc1000116), and Planktomarina sp. (Bc1000120)] were greater than 10-fold more abundant in the low chl a/high DMSP event compared with their average abundance throughout the time series (Supplementary Figure 1).
Clear temporal patterns in the abundance of the genes encoding the DMSP demethylation pathway (dmdA subclade A/1 and D/all), the DMSP lyase pathway (dddP), and bacterial DMSP biosynthesis (dsyB) were apparent over the 21-month sampling period using quantitative polymerase chain reactions (qPCR) (Figure 6).
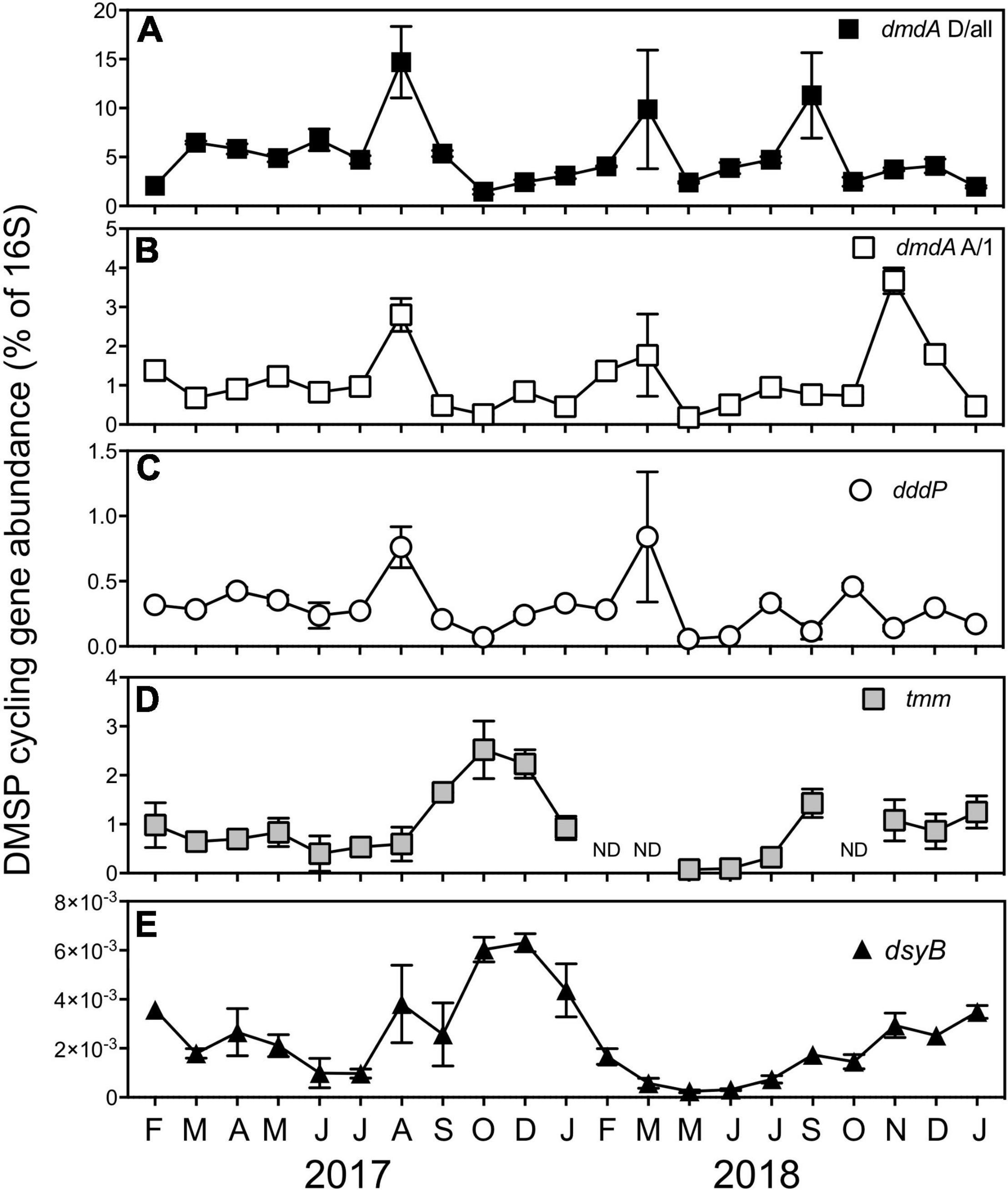
Figure 6. A relative proportion of DMSP cycling genes encoding the DMSP demethylation genes, (A) dmdA subclade D/all and (B) dmdA subclade A/1, (C) the DMSP lyase gene, dddP, (D) the bacterial DMS oxidation gene, tmm, and (E) the bacterial production gene dsyB at Port Hacking between February 2017 and January 2019. All data are means ± standard error (n = 3) and normalized to counts of the bacterial 16S rRNA gene. ND, non-detected.
The DMSP demethylase genes dmdA subclade A/1 and dmdA subclade D/all are recognized as genes that belong to members of the Roseobacter clade and SAR11 clade, respectively (Levine et al., 2012; Varaljay et al., 2012). Significant temporal shifts in the abundance of both dmdA subclade A/1 (KW test = 49.5, df = 20, p < 0.01) and dmdA subclade D/all (KW test = 51.1, df = 20, p < 0.01) were observed, including concurrent significant increases (q < 0.05) in August 2017 and March 2018 (Figure 6). Overall, there was a significant positive correlation between dmdA A/1 and dmdA D/all (Supplementary Table 6). However, the abundance of the DMSP demethylase genes was decoupled during the high chl a/low DMSP event in September 2018 (Figure 6), where the abundance of dmdA D/all displayed a significant 2.2-fold increase (q < 0.05, 11 ± 4.4 copies of D/all per 16S). In contrast, the abundance of dmdA A/1 recorded a significant 3.3-fold increase (q < 0.05, 4 ± 0.3 copies of A/1 per 16S) during the low chl a/high DMSP event in November 2018 (Figure 6). No correlation was found between dmdA D/all with any dimethylated sulfur compound, DMSP lyase activity (DLAp and DLAb) or an environmental variable (temperature, salinity, chl a, NOx2–, PO34–, or SiO32–) (Supplementary Table 6). Despite the dmdA subclade D targeting SAR11 genes, no significant correlation between dmdA D/all and the collective relative abundance of the SAR11 order was detected, nor was the gene correlated with the abundance of other dominant bacteria (Supplementary Table 6). The abundance of the Roseobacter-associated DMSP demethylase gene (dmdA A/1) was significantly positively correlated with the activity of the bacterial DMSP lyase pathway (DLAb), although was not correlated with dimethylated sulfur compounds or environmental factors (Supplementary Table 6). Correlations between dmdA A/1 and the relative abundance of dominant orders of bacteria revealed no relationships, with exception of a significant positive correlation between dmdA A/1 abundance and Rhodobacterales-relative abundance (Supplementary Table 7).
With the abundance of the DMSP lyase gene, dddP also shifted significantly over time (KW test = 51.1, df = 20, p < 0.01) and was significantly positively correlated with both dmdA genes (Supplementary Table 6), revealing similar patterns of significantly increased abundance (q < 0.05) in August 2017 and March 2018 (Figure 6). The only other parameter that dddP displayed a significant positive correlation was DLAb, which is important as it displays a direct link between the genetic potential of dddP to convert DMSP to DMS, with the activity of an enzyme responsible for DMSP cleavage (Supplementary Tables 5, 6).
Temporal variation was observed in the relative abundance of the gene encoding DMS oxidation via the trimethylamine monooxygenase enzyme, tmm (KW test = 31.8, df = 17, p = 0.01). The peak abundance of tmm coincided with peaks in DMSOt during the high chl a/high DMSP event in October 2017 (2.5 ± 0.59 copies of tmm per 16S) (Figure 6). A significant positive correlation existed between the relative abundance of tmm and DMSPt, DMSPd, and, notably, with DMSOt. These data give evidence to support the hypothesis that the gene tmm, under specific environmental conditions, may play a role in marine sulfur cycling (Supplementary Figure 2 and Supplementary Table 6). No relationship between tmm and environmental variables was identified during the time series (Supplementary Table 6); however, significant positive correlations were detected between tmm and the relative abundance of Cellvibrionales and Puniceispirillales (SAR116 clade) (Supplementary Table 7).
Seasonal patterns were observed in the abundance of the DMSP biosynthesis gene, dsyB. Abundances of dsyB showed significant increases in spring-summer months relative to winter months (q < 0.05), with peak abundances coinciding with the high chl a/high DMSP event experienced in October 2017 (0.006 ± 0.0005 copies of dsyB per 16S) and December 2017 (0.006 ± 0.0004 copies of dsyB per 16S) (Figure 6). A significant positive correlation was detected between the abundance of dsyB and concentrations of DMS, DMSPt, and DMSPd (Supplementary Figure 3 and Supplementary Table 6), suggesting a potential bacterial contribution to DMSP and DMS concentrations. No relationships between dsyB and environmental variables were found in the time-series data, although there was a significant positive correlation between dsyB and the relative abundance of three bacterial orders, specifically Cellvibrionales, Puniceispirillales (SAR116 clade), and Salinisphaerales (Supplementary Table 7).
Oceanic dimethylsulfoniopropionate (DMSP), dimethyl sulfoxide (DMSO), and dimethyl sulfide (DMS) concentrations are largely governed by ecological and metabolic interactions among marine phytoplankton and bacterial assemblages that synthesize and transform DMSP. Yet, how these interactions change over space and time and ultimately regulate the conversions among DMSP, DMSO, and DMS are poorly characterized. In this study, we coupled a suite of biological and chemical approaches to examine seasonal patterns in DMS (O) (P) cycling in coastal shelf waters in the south Pacific Ocean. Concentrations of these organosulfur compounds in our time series displayed clear seasonal trends that often share similarities with patterns observed in other coastal time series, specifically spring-time peaks of DMSP (Stefels et al., 1995; Townsend and Keller, 1996; van Duyl et al., 1998; Simó and Dachs, 2002) and summer-time peaks of DMS (Simó and Dachs, 2002). However, we revealed that, despite clear seasonal trends in DMS(P), the microbiology underpinning these dynamics is capricious.
Long-term observations of DMSP concentrations in eastern Australian coastal waters revealed heterogenous DMSP concentrations throughout the study. The concentrations of total DMSP (DMSPt) and particulate DMSP (DMSPp) did not display any significant correlations with temperature or any nutrient concentration (nitrate, phosphate, and silicate). No temporal relationship between DMSP with these variables is not uncommon and has previously been reported in other sulfur time series in the English channel (Archer et al., 2009), the Baltic Sea (Zhao et al., 2021), and the North Sea (Speeckaert et al., 2018). Conversely, concentrations of DMSPt and DMSPp demonstrated a significant negative correlation with salinity over the duration of the time series. This finding implies that changes in salinity play a major role in the production of DMSP by marine phytoplankton and corroborate previous research that has identified this relationship in coastal waters (Shenoy et al., 2000; Hu et al., 2005), possibly due to the hypothesized physiological role DMSP plays in marine phytoplankton as an osmolyte (Dickson and Kirst, 1986; Kirst, 1996). Shifts in DMSP observed in the time series were not just influenced by potential shifts in phytoplankton physiology but also phytoplankton biomass (chl a), which was significantly positively correlated with DMSPt, DMSPp, and DMSPd. Consistent with other time series (Stefels et al., 1995; Townsend and Keller, 1996; van Duyl et al., 1998; Simó and Dachs, 2002), significant annual increases of DMSP were recorded in the springtime of each year (specifically October 2017 and November 2018), which, generally, also peaked during the springtime (e.g., October 2017 and September 2018). However, our results demonstrate that elevated chl a levels did not always result in pulses of DMSP and that the determinants of DMSP concentrations in the study environment are more nuanced than simply the presence of phytoplankton blooms. This was clearly demonstrated by three distinctive springtime peak chl a and DMSP pulse events (Figure 7). The first of these events was characterized by significant peaks of both chl a and DMSP (a high chl a/high DMSP event, October 2017). The second event (September 2018) had equally high chl a concentrations, but much lower DMSP levels (the high chl a/low DMSP event), while the third event had the highest DMSP concentrations measured in 2018 (November 2018), but chl a levels that were half those of October 2017 (the low chl a/high DMSP event). These patterns indicate that springtime increases in DMSPp and DMSPd are not solely driven by phytoplankton biomass, a decoupling that might be attributed to shifts in the dominant representatives of the phytoplankton community and their DMSP-producing potential (Belviso et al., 2000; Sunda and Hardison, 2008; Archer et al., 2009).
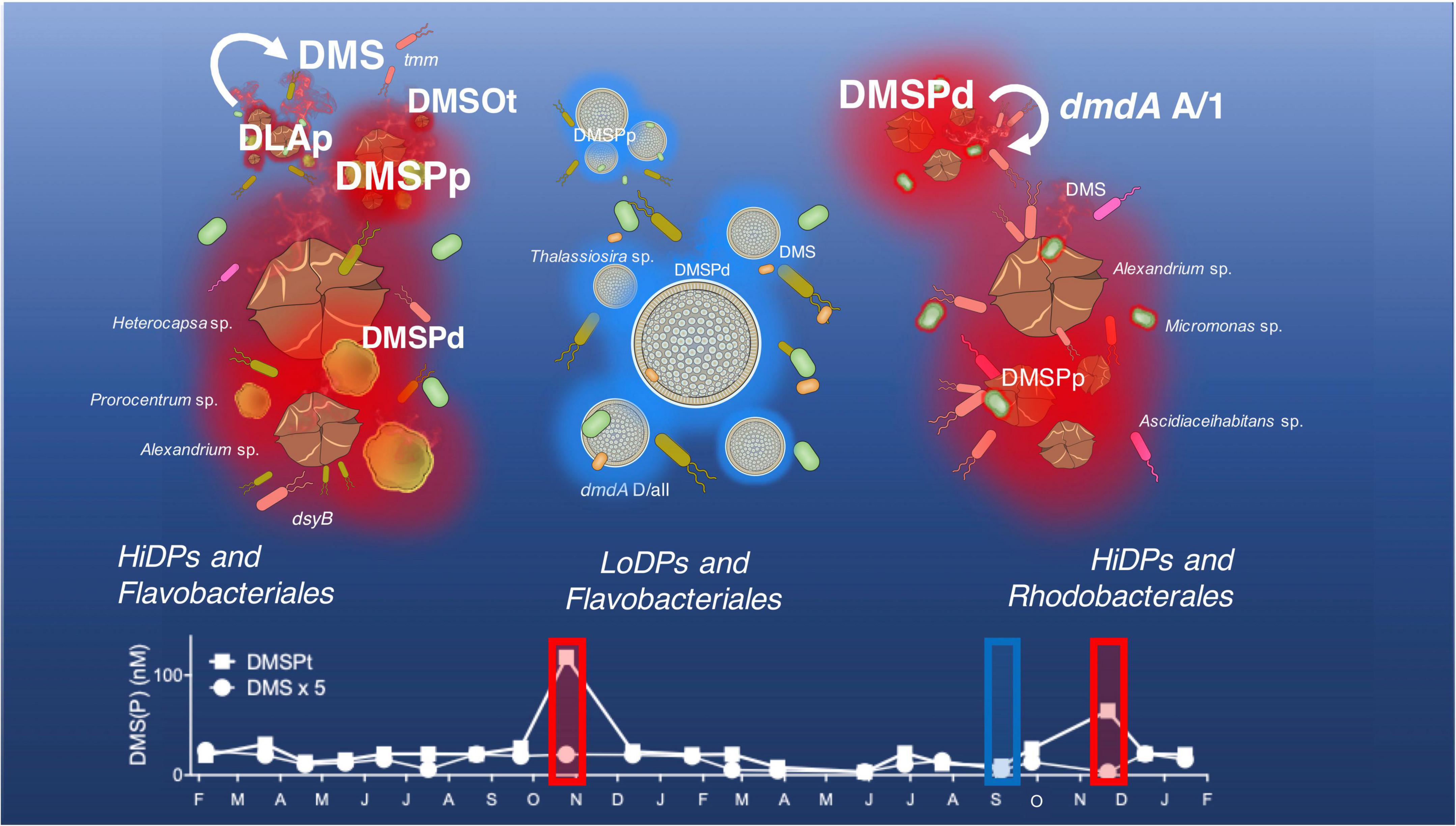
Figure 7. Dimethylsulfoniopropionate (DMSP)-producing phytoplankton and associated bacterial consortia influencing DMSP cycling at Port Hacking time series in October 2017 (the left panel), September 2018 (the middle panel) and November 2018 (the right panel). Concentrations of particulate DMSP (DMSPp) are driven by the composition and DMSP-producing potential of phytoplankton present. The low-DMSP-producing phenotype (LoDP) Thalassiosira sp. produces comparatively low-DMSPp to high-DMSP-producing phenotypes (HiDP), Heterocapsa sp., Prorocentrum sp., Alexandrium sp., and Micromonas sp. Concentrations of DMSP converted by HiDPs with active DMSP lyases enzymes (DLAp) contribute to high concentrations of the volatile sulfur emission dimethyl sulfide (DMS). High concentrations of DMS are accompanied by a high relative abundance of the bacterial DMS oxidation gene (tmm), which may contribute to the high dimethyl sulfoxide (DMSO) concentrations observed. The amount of DMS in the surface water at Port Hacking can be constrained by shifts in the genes encoding the DMSP demethylase pathway (dmdA A/1, dmdA D/all), including Roseobacter strains, like Ascidiaceihabitans sp.
To examine the influence of successions of phytoplankton influencing DMSP dynamics in the time series, we monitored temporal shifts in the phytoplankton community, with a specific focus on DMSP production, by measuring patterns in the relative abundance of putative high-DMSP-producing phytoplankton (HiDPs) and low-DMSP-producing phytoplankton (LoDPs) (McParland and Levine, 2020). Over the duration of the time series, a significant positive relationship was measured between the relative abundance of putative HiDPs and DMSPt concentration (Figure 2A, Pearson’s r = 0.61, p < 0.01, n = 21). Notably, this relationship was stronger than that observed between chl a and DMSPt. Conversely, no relationship was found between putative LoDP relative abundance and DMSPt. The relative abundance of HiDPs and LoDPs significantly shifted over time. Springtime pulses of high DMSP (i.e., the high chl a/high DMSP event and the low chl a/high DMSP event) were always accompanied by greater relative abundance of HiDPs. Meanwhile, peaks in chl a that corresponded with increased LoDP-relative abundance (the high chl a/low DMSP event in March 2017 and the high chl a/low DMSP event) demonstrated negligible increases in DMSP. Our results support previous predictions based on observations from the Southern Ocean and Sargasso Sea that propose HiDP biomass dominates DMSP production, even at times when they are not the dominant population within the phytoplankton assemblage (McParland and Levine, 2019).
The HiDP phytoplankton communities corresponding with pulses of DMSP in our time series were dominated by dinoflagellates. Blooms of dinoflagellates have elsewhere been linked to high DMSP concentrations (Varaljay et al., 2015; Kiene et al., 2019; Nowinski et al., 2019a). The high chl a/high DMSP event was dominated by a mixed dinoflagellate bloom of Heterocapsa, Prorocentrum, and Alexandrium strains that were closely related to known HiDP phytoplankton isolates (Supplementary Table 8). During the low chl a/high DMSP event, the HiDP phytoplankton community was comprised of dominant strains with closely related HiDP isolates, including the dinoflagellate Alexandrium and the picoeukaryote Micromonas (Supplementary Table 8). Conversely, the high chl a/low DMSP event was dominated by the LoDP diatom, Thalassiosira. The dominant Thalassiosira ASV (Eb1000094) was found to be related to a known LoDP Thalassiosira isolate (Supplementary Table 8). Overall, our data imply that seasonal increases of in situ DMSP concentration are a result of increased relative abundance of predicted HiDP phytoplankton lineages and that examples of decoupling between DMSP and chl a are a result of predicted LoDP phytoplankton blooms. This proposed relationship between HiDP-relative abundance and DMSPt concentration is consistent with a previous study, which considered HiDP and LoDP biomass in high-performance liquid chromatography samples (HPLC) from ocean waters (McParland and Levine, 2019). However, it should be acknowledged that both approaches are predictive and that an improved representation of subdominant phytoplankton communities is required to make accurate predictions of in situ DMSP concentrations. Nonetheless, these results point to the importance of considering phytoplankton community composition as a major biological factor governing in situ DMSP concentrations, which, in turn, may influence the composition of their bacterial consortia and DMSP breakdown products.
Temporal shifts in bacterial community composition observed during this study often corresponded with changes in chl a concentration and pulses of DMSP throughout the time series, providing evidence that phytoplankton-derived DMSP may play a critical role in shaping marine bacterioplankton assemblages. The strongest relationship between DMSP and any group of heterotrophic bacteria was between dissolved DMSP (DMSPd) and the Rhodobacterales. Members of the Rhodobacterales are often reported as a dominant feature of bacterial communities during blooms of DMSP-producing phytoplankton (González et al., 2000; Zubkov et al., 2002; Delmont et al., 2014). In our study, the greatest abundance of Rhodobacterales coincided with the low chl a/high DMSP Alexandrium and Micromonas bloom. During the bloom, inspection of Rhodobacterales at a higher taxonomic resolution found that five dominant amplicon sequence variants (ASVs) comprised over 20% of the entire bacterial population. Previously, we have demonstrated that two of these ASVs, Amylibacter sp. (Bc1000011) and Ascidiaceihabitans sp. (Bc1000035) display spatial and temporal relationships with the HiDP Micromonas at multiple time-series sites across subtropical and temperate environments (O’Brien et al., 2022). Importantly, the results of the present study demonstrate higher-than-average abundance of Amylibacter, Ascidiaceihabitans, and Micromonas occur during high-DMSP conditions.
Flavobacteriales were dominant members of the bacterial community coinciding with both spring high chl a phytoplankton blooms, although concentrations of DMSP greatly differed between the two blooms. Flavobacteria have been shown to grow while converting DMS to DMSO; however, no mechanism for this transformation has been discovered (Green et al., 2011). Instead, it is thought that the success of Flavobacteria during phytoplankton blooms is due to the breakdown and assimilation of high molecular weight carbohydrate polymers derived from phytoplankton (Kirchman, 2002; Avcı et al., 2020). In our time series, a positive correlation between the relative abundance of Flavobacteriales and chl a was found, but there were no significant correlations with any dimethylated sulfur compounds, indicating that the ecological links between the Flavobacteria and phytoplankton are not governed by DMS (O) (P). Overall, springtime increases in chl a and DMSP in the time series resulted in a shift from bacterial communities dominated by oligotrophic organisms, like the SAR11 clade (in low DMSP conditions) to copiotrophic specialists like Rhodobacterales.
Within the context of marine DMS(P) cycling processes, understanding patterns in the abundance of key groups of bacteria, including the Rhodobacterales and SAR11 clade, is crucial due to their ability to transform DMSP through both the DMSP lyase (ddd) and DMSP demethylation (dmdA) pathways (Todd et al., 2009; Curson et al., 2017). The total abundance of subclades of dmdA belonging to Roseobacter (Subclade A/1) and the SAR11 clade (Subclade D/all) displayed heterogenous patterns in abundance during the time series. Both Roseobacter dmdA and SAR11 dmdA exhibited significant positive correlations to each other across all samples. However, the abundance of Roseobacter-associated and SAR11-associated DMSP demethylation genes decoupled in springtime 2018, whereby SAR11 dmdA significantly increased during the high chl a/low DMSP phytoplankton bloom, while elevated levels of Roseobacter dmdA were associated with the low chl a/high DMSP event. Interestingly, the springtime increase in SAR11 dmdA was not concomitant with above-average SAR11-relative abundance, whereas the increase in Roseobacter dmdA was associated with more than a three-fold increase in Rhodobacterales-relative abundance. Contrasting patterns in the abundance of these demethylation genes emphasize the differential response to DMSP displayed by the SAR11 clade and the Roseobacter group. Indeed, members of the SAR11 clade have been shown to be reliant upon reduced forms of sulfur (including DMSP) for growth (Tripp et al., 2008), although transcriptomic analysis revealed that SAR11 members do not show substantial transcriptional responses to DMSP enrichments (Vila-Costa et al., 2010), albeit this transcriptional response may be dependent upon the concentration of DMSP enrichment (Gao et al., 2020). Conversely, DMSP additions to seawater have been demonstrated to stimulate upregulation of Roseobacter-like transcripts (Vila-Costa et al., 2010). This likely explains a significant increase in the abundance of Roseobacter dmdA, as members of Roseobacter transform abundant DMSPd concentrations into readily assimilated reduced sulfur (Howard et al., 2008; Reisch et al., 2008; Dupont et al., 2012).
The abundance of the Roseobacter-associated DMSP lyase gene (Levine et al., 2012), dddP, revealed temporal heterogeneity, but no clearly discernible seasonal patterns. Overall, dddP was significantly positively correlated with Roseobacter demethylation genes, but, unlike dmdA, showed no significant increase in abundance in Spring 2018. Despite dddP being involved in transformations of DMSP to DMS, no significant relationship was found between the abundance of dddP and DMS concentrations. This is understandable, while dddP has been reported as the most abundant bacterial gene encoding DMSP lyase in coastal ecosystems (Nowinski et al., 2019a), it is just one of eight identified bacterial DMSP lyase genes (Todd et al., 2011, 2012; Choi et al., 2015). Notably one of these genes, dddK, which is found in the dominant SAR11 clade (Sun et al., 2021), can, sometimes, be just as abundant as dddP in coastal waters (Nowinski et al., 2019a). It is possible that the SAR11 clade may also be significant contributors of DMS; however, we did not quantify SAR11 dmdA (dddK) in this study. Additionally, besides bacterial degradation of DMSP, a multitude of factors control the concentration of DMS in marine surface waters, including bacterial and photo-oxidation of DMS (Toole et al., 2003; Lidbury et al., 2016), or the activity of phytoplankton and bacterial DMSP lyase enzymes (Steinke et al., 2000; Alcolombri et al., 2015).
Temporal patterns in the abundance of the bacterial DMSP degradation genes measured were significantly correlated with one another and generally exhibited increases in the late winter to springtime. A decoupling between taxon-specific genes encoding the same DMSP transformation (DMSP demethylation) highlights the importance of considering diversity of the bacterial DMSP degraders as all responses to DMSP availability are not equal (Vila-Costa et al., 2010). Moreover, significant correlations between dominant Roseobacter ASVs and DMSP degradation genes found in our data support that strain-specific responses to DMSP availability exist in marine surface waters (Nowinski et al., 2019a) and should be considered to identify key DMS(P) cycling bacteria.
Variable concentrations of DMS were measured over the course of the time series. While no clear seasonal trend was evident over the 2-year study, concentrations of DMS were generally greatest in the late spring to summertime. Increased DMS concentrations in summer are a global phenomenon (Bates et al., 1987; Simó and Pedrós-Alió, 1999; Williams et al., 2019) and are often thought to be a result of high-DMSP-producing phytoplankton being situated in strongly sunlit, stratified, and nutrient-depleted waters (Stefels et al., 2007; Archer et al., 2009). However, our results do not indicate a direct relationship between DMS and HiDP-associated particulate DMSP (or other fractions of DMSP). Instead, our study reveals that DMSP lyase enzyme activity by marine microbes is a greater determinant of DMS, as indicated by significant positive correlations between DMS with size fractionated microbial community samples of >2.0 μm (hereafter referred to as DLAp) and 0.22 μm–2.0 μm (hereafter referred to as DLAb) with DMS concentrations during the time series (Supplementary Table 4). Levels of both DLAp and DLAb significantly varied over time. When comparing springtime chl a increases and DMSP pulses observed in the time series, both DMS concentrations and DLAp rates were greater during the high chl a/high DMSP bloom in spring 2017 compared to the following spring. These findings highlight the significant influence that microbial DMSP lyases can have on controlling marine DMS concentrations. Despite both DLAp and DLAb being significantly positively correlated with DMS concentrations, no relationship was found between DLAp and DLAb (Pearson’s r = 0.17, p > 0.05, n = 21). DLAp was significantly positively correlated with DMSPp in our time series; however, previous research has suggested physical (UV-A) stress is an equally or more important determinant of DLAp rates (Levine et al., 2012). DLAb rates have been previously linked to temperature (Levine et al., 2012), although our results indicate a stronger link between DLAb rates and the abundance of the Roseobacter-associated DMSP lyase gene (dddP). The variable nature of DMS concentrations throughout the time series could not be explained through environmental conditions alone, and our findings demonstrate the importance of monitoring microbial DMSP lyase enzyme activity as, potentially, the most important determinant of DMS in marine surface waters.
Dimethylsulfoniopropionate has substantial climatic importance as a precursor compound to DMS (Charlson et al., 1987). DMSO shares this role as a DMS precursor (via bacterial DMSO reduction) (Bray et al., 2001), but, notably, because photochemical and microbial DMS oxidation (of DMS to DMSO) represents a major sink for DMS in the marine environment (Kiene and Bates, 1990). Bacterial DMS oxidation is the primary process for removal of DMS in marine surface waters (Lidbury et al., 2016), and the most abundant DMS oxidation pathway in the marine environment is trimethylamine monooxygenase (Tmm) (Teng et al., 2021). Tmm is encoded by the gene, tmm, which has been estimated to be found in 20% of all bacteria (Chen et al., 2011; Chen, 2012). Our results show that tmm was less abundant compared to other environments, whereby it only occurred in approximately 2.5% of bacteria (normalized to 16S rRNA genes) at Port Hacking. The relative abundance of tmm revealed seasonal increases, including contemporaneous peaks in tmm with DMSP and DMSO. This finding is significant as DMSO is a product of Tmm activity (Chen et al., 2011), although it should be acknowledged that this activity is dependent upon the presence of methylamines (Lidbury et al., 2016). Overall, these findings highlight the significance of tmm-carrying bacteria in contributing to seasonal surface water concentrations of DMSO, its potential role as a DMS sink, and the need for this pathway of the marine sulfur cycle to be addressed more widely, including future measurements of in situ methylamine concentrations, transcription, and enzyme abundance and activity.
Bacteria possessing the DMSP biosynthesis gene, dsyB, have been recognized as potentially important producers of DMSP across a multitude of marine environments (Curson et al., 2017; Williams et al., 2019; Sun et al., 2020; Zheng et al., 2020). During this study, dsyB levels displayed seasonal increases over spring and summer and were significantly correlated with DMSPt, DMSPd, and DMS (Supplementary Figure 2 and Supplementary Table 6). This bacterial DMSP biosynthesis gene also demonstrated greater abundances during the high chl a/high DMSP event compared to other significant spring blooms. The relative abundance of dsyB was significantly correlated with the relative abundance of a number of bacterial orders, although the reason behind these correlations is currently unclear, as dsyB has not yet been identified in any members of Cellvibrionales, Puniceispirillales (SAR116 clade), and Salinisphaerales. Overall, our observations indicate that bacterial DMSP production may contribute to temporal variability in ocean surface DMSP concentrations. This contribution, however, is likely to play a subsidiary role compared to HiDP production, as some HiDP dinoflagellates are known to have 17-fold greater intracellular concentrations of DMSP compared to known bacterial isolates (McParland and Levine, 2019). Nonetheless, our work and other recent studies (Williams et al., 2019; Sun et al., 2020, 2021; Zheng et al., 2020; Liu et al., 2021) confirm a need to discard a singular focus on bacteria as DMSP degraders and highlight a need to quantify the role of DMSP production by marine bacteria in marine surface waters.
During the sulfur time series at Port Hacking, NSW, Australia, we revealed temporal shifts in the concentration of dimethylated sulfur compounds and important factors that have the potential to govern these cycles, including microbial community composition and DMSP cycling gene abundances. It is important to note that these measurements provide robust hypotheses but do not provide absolute certainty. In order to confirm these hypotheses, it is necessary to consider the behavior of these microbial communities and, importantly, the expression of the genes in relation to in situ DMS (O) (P) concentrations.
Identifying the ecological determinants of microbial DMSP cycling is a key to understanding their influence on climate and marine biogeochemical cycles. The present study shows that DMS (O) (P) cycling is highly complex and temporally dynamic within coastal shelf waters of the southern Pacific Ocean, with a diverse suite of processes governing surface water concentrations of DMS (O) (P) that shift in importance from one time to another. Our results emphasize that, among microbial communities, there is no “single story” behind DMSP cycling dynamics. This points to the importance of considering a wide and constantly expanding (Curson et al., 2017, 2018; McParland and Levine, 2019, 2020; McParland et al., 2021) range of biochemical processes and microbiological players involved in marine DMSP cycling.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JO’B, JS, and KP designed the project. JO’B, NS, and TI collected the samples. MO and AB designed the sequencing data pipeline. JO’B, NS, and BL prepared qPCR pipelines used. JO’B analyzed the dataset. EM and NL developed the DMSP-producer pipeline used in the study. JO’B, AB, and MO curated the data. JS and KP acquired the funding. JO’B, AB, MO, NS, BL, MB, KP, EM, NL, and JS performed writing, review, and editing of the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the Australian Research Council Grants FT130100218 and DP180100838 awarded to JS and DP140101045 awarded to JS and KP, as well as an Australian Government Research Training Program Scholarship awarded to JO’B.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to gratefully acknowledge the Australian Microbiome Initiative and the Integrated Marine Observing System (IMOS) for the generation of data used in this publication. The Australian Microbiome initiative was supported by funding from Bioplatforms Australia and IMOS through the Australian Government’s National Collaborative Research Infrastructure Strategy (NCRIS), Parks Australia, through the Bush Blitz program funded by the Australian Government and BHP, and the Commonwealth Scientific and Industrial Research Organisation (CSIRO). Oceanographic data were sourced from IMOS, which is enabled by NCRIS. We would also like to gratefully acknowledge Glynn Gorick for his original artwork, which was used in the final figure of this article.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.894026/full#supplementary-material
Alcolombri, U., Ben-Dor, S., Feldmesser, E., Levin, Y., Tawfik, D. S., and Vardi, A. (2015). Identification of the algal dimethyl sulfide–releasing enzyme: a missing link in the marine sulfur cycle. Science 348, 1466–9. doi: 10.1126/science.aab1586
Appleyard, S. A., Abell, G., and Watson, R. (2013). Tackling Microbial Related Issues in Cultured Shellfish Via Integrated Molecular and Water Chemistry Approaches. Hobart, Tas: CSIRO Marine and Atmospheric Research.
Archer, S. D., Cummings, D. G., Llewellyn, C. A., and Fishwick, J. R. (2009). Phytoplankton taxa, irradiance and nutrient availability determine the seasonal cycle of DMSP in temperate shelf seas. Mar. Ecol. Prog. Ser. 394, 111–24.
Archer, S. D., Widdicombe, C. E., Tarran, G. A., Rees, A. P., and Burkill, P. H. (2001). Production and turnover of particulate dimethylsulphoniopropionate during a coccolithophore bloom in the northern North Sea. Aquat. Microb. Ecol. 24, 225–41.
Avcı, B., Krüger, K., Fuchs, B. M., Teeling, H., and Amann, R. I. (2020). Polysaccharide niche partitioning of distinct Polaribacter clades during North Sea spring algal blooms. ISME J. 14, 1369–83. doi: 10.1038/s41396-020-0601-y
Bates, T. S., Charlson, R. J., and Gammon, R. H. (1987). Evidence for the climatic role of marine biogenic sulphur. Nature 329, 319–21. doi: 10.1126/science.1151109
Bell, T. G., Malin, G., Kim, Y.-N., and Steinke, M. (2007). Spatial variability in DMSP-lyase activity along an Atlantic meridional transect. Aquat. Sci. 69, 320–9.
Belviso, S., Christaki, U., Vidussi, F., Marty, J.-C., Vila, M., and Delgado, M. (2000). Diel variations of the Dmsp-to-chlorophyll a ratio in Northwestern Mediterranean surface waters. J. Mar. Syst. 25, 119–28.
Bratbak, G., Levasseur, M., Michaud, S., Cantin, G., Fernández, E., Heimdal, B. R., et al. (1995). Viral activity in relation to Emiliania huxleyi blooms: a mechanism of DMSP release? Mar. Ecol. Prog. Ser. 128, 133–42.
Bray, R. C., Adams, B., Smith, A. T., Richards, R. L., Lowe, D. J., and Bailey, S. (2001). Reactions of dimethylsulfoxide reductase in the presence of dimethyl sulfide and the structure of the dimethyl sulfide-modified enzyme. Biochemistry 40, 9810–20. doi: 10.1021/bi010559r
Brimblecombe, P., and Shooter, D. (1986). Photo-oxidation of dimethylsulphide in aqueous solution. Mar. Chem. 19, 343–53.
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). Dada2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–3. doi: 10.1038/nmeth.3869
Caruana, A. M., and Malin, G. (2014). The variability in DMSP content and DMSP lyase activity in marine dinoflagellates. Prog. Oceanogr. 120, 410–24.
Charlson, R. J., Lovelock, J. E., Andreae, M. O., and Warren, S. G. (1987). Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature 326, 655–61.
Chen, Y. (2012). Comparative genomics of methylated amine utilization by marine Roseobacter clade bacteria and development of functional gene markers (tmm, gmaS). Environ. Microbiol. 14, 2308–22. doi: 10.1111/j.1462-2920.2012.02765.x
Chen, Y., Patel, N. A., Crombie, A., Scrivens, J. H., and Murrell, J. C. (2011). Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 108, 17791–6. doi: 10.1073/pnas.1112928108
Choi, D. H., Park, K.-T., An, S. M., Lee, K., Cho, J.-C., Lee, J.-H., et al. (2015). Pyrosequencing revealed SAR116 clade as dominant dddP-containing bacteria in oligotrophic NW Pacific Ocean. PLoS One 10:e0116271. doi: 10.1371/journal.pone.0116271
Cui, Y., Suzuki, S., Omori, Y., Wong, S.-K., Ijichi, M., Kaneko, R., et al. (2015). Abundance and distribution of dimethylsulfoniopropionate degradation genes and the corresponding bacterial community structure at dimethyl sulfide hot spots in the tropical and subtropical pacific ocean. Appl. Environ. Microbiol. 81, 4184–94. doi: 10.1128/AEM.03873-14
Curson, A. R., Liu, J., Martínez, A. B., Green, R. T., Chan, Y., Carrión, O., et al. (2017). Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nat. Microbiol. 2:17009. doi: 10.1038/nmicrobiol.2017.9
Curson, A. R., Todd, J. D., Sullivan, M. J., and Johnston, A. W. (2011). Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat. Rev. Microbiol. 9, 849–59. doi: 10.1038/nrmicro2653
Curson, A. R., Williams, B. T., Pinchbeck, B. J., Sims, L. P., Martínez, A. B., Rivera, P. P. L., et al. (2018). DSYB catalyses the key step of dimethylsulfoniopropionate biosynthesis in many phytoplankton. Nat. Microbiol. 3, 430–9.
Dacey, J. W., Howse, F. A., Michaels, A. F., and Wakeham, S. G. (1998). Temporal variability of dimethylsulfide and dimethylsulfoniopropionate in the Sargasso Sea. Deep Sea Res. I Oceanogr. Res. Pap. 45, 2085–104.
Delmont, T. O., Hammar, K. M., Ducklow, H. W., Yager, P. L., and Post, A. F. (2014). Phaeocystis antarctica blooms strongly influence bacterial community structures in the Amundsen Sea polynya. Front. Microbiol. 5:646. doi: 10.3389/fmicb.2014.00646
Deschaseaux, E., Kiene, R., Jones, G. B., Deseo, M. A., Swan, H., Oswald, L., et al. (2014). Dimethylsulphoxide (DMSO) in biological samples: a comparison of the TiCl3 and NaBH4 reduction methods using headspace analysis. Mar. Chem. 164, 9–15.
Deschaseaux, E., O’brien, J., Siboni, N., Petrou, K., and Seymour, J. R. (2019). Shifts in dimethylated sulfur concentrations and microbiome composition in the red-tide causing dinoflagellate Alexandrium minutum during a simulated marine heatwave. Biogeosciences 16, 4377–91.
Dickson, D., and Kirst, G. (1986). The role of β-dimethylsulphoniopropionate, glycine betaine and homarine in the osmoacclimation of Platymonas subcordiformis. Planta 167, 536–43. doi: 10.1007/BF00391230
Dupont, C. L., Rusch, D. B., Yooseph, S., Lombardo, M.-J., Richter, R. A., Valas, R., et al. (2012). Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 6:1186. doi: 10.1038/ismej.2011.189
Fernandez, E., Ostrowski, M., Siboni, N., Seymour, J. R., and Petrou, K. (2021). Uptake of dimethylsulfoniopropionate (DMSP) by natural microbial communities of the great barrier reef (GBR), Australia. Microorganisms 9:1891. doi: 10.3390/microorganisms9091891
Galí, M., and Simó, R. (2015). A meta-analysis of oceanic DMS and DMSP cycling processes: disentangling the summer paradox. Global Biogeochem. Cycles 29, 496–515.
Gao, C., Fernandez, V. I., Lee, K. S., Fenizia, S., Pohnert, G., Seymour, J. R., et al. (2020). Single-cell bacterial transcription measurements reveal the importance of dimethylsulfoniopropionate (DMSP) hotspots in ocean sulfur cycling. Nat. Commun. 11:1942. doi: 10.1038/s41467-020-15693-z
González, J. M., Simó, R., Massana, R., Covert, J. S., Casamayor, E. O., Pedrós-Alió, C., et al. (2000). Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66, 4237–46. doi: 10.1128/AEM.66.10.4237-4246.2000
Green, D. H., Shenoy, D. M., Hart, M. C., and Hatton, A. D. (2011). Coupling of dimethylsulfide oxidation to biomass production by a marine flavobacterium. Appl. Environ. Microbiol. 77, 3137–40. doi: 10.1128/AEM.02675-10
Guillou, L., Bachar, D., Audic, S., Bass, D., Berney, C., Bittner, L., et al. (2012). The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–604. doi: 10.1093/nar/gks1160
Harada, H., Rouse, M.-A., Sunda, W., and Kiene, R. P. (2004). Latitudinal and vertical distributions of particle-associated dimethylsulfoniopropionate (DMSP) lyase activity in the western North Atlantic Ocean. Can. J. Fish. Aquat. Sci. 61, 700–11.
Horinouchi, M., Yoshida, T., Nojiri, H., Yamane, H., and Omori, T. (1999). Polypeptide requirement of multicomponent monooxygenase DsoABCDEF for dimethyl sulfide oxidizing activity. Biosci. Biotechnol. Biochem. 63, 1765–71.
Howard, E. C., Henriksen, J. R., Buchan, A., Reisch, C. R., Bürgmann, H., Welsh, R., et al. (2006). Bacterial taxa that limit sulfur flux from the ocean. Science 314, 649–52. doi: 10.1126/science.1130657
Howard, E. C., Sun, S., Biers, E. J., and Moran, M. A. (2008). Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ. Microbiol. 10, 2397–410. doi: 10.1111/j.1462-2920.2008.01665.x
Hu, M., Liu, L., Ma, Q., Zhu, T., Tian, X., and Dai, M. (2005). Spatial–temporal distribution of dimethylsulfide in the subtropical Pearl River estuary and adjacent waters. Continental Shelf Res. 25, 1996–2007.
Kageyama, H., Tanaka, Y., Shibata, A., Waditee-Sirisattha, R., and Takabe, T. (2018). Dimethylsulfoniopropionate biosynthesis in a diatom Thalassiosira pseudonana: identification of a gene encoding MTHB-methyltransferase. Arch. Biochem. Biophys. 645, 100–6. doi: 10.1016/j.abb.2018.03.019
Karsten, U., Kück, K., Vogt, C., and Kirst, G. (1996). “Dimethylsulfoniopropionate production in phototrophic organisms and its physiological functions as a cryoprotectant,” in Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds, eds R. P. Kiene, P. T. Visscher, M. D. Keller, and G. O. Kirst (Berlin: Springer), 143–53.
Keller, M. D. (1989). Dimethyl sulfide production and marine phytoplankton: the importance of species composition and cell size. Biol. Oceanogr. 6, 375–82.
Keller, M. D., Bellows, W. K., and Guillard, R. R. (1989). Dimethyl Sulfide Production in Marine Phytoplankton. Washington, DC: ACS Publications.
Kettle, A., Andreae, M., Amouroux, D., Andreae, T., Bates, T., Berresheim, H., et al. (1999). A global database of sea surface dimethylsulfide (DMS) measurements and a procedure to predict sea surface DMS as a function of latitude, longitude, and month. Global Biogeochem. Cycles 13, 399–444.
Kiene, R. P., and Bates, T. S. (1990). Biological removal of dimethyl sulphide from sea water. Nature 345, 702–5.
Kiene, R. P., and Gerard, G. (1994). Determination of trace levels of dimethylsulfoxide (DMSO) in seawater and rainwater. Mar. Chem. 47, 1–12.
Kiene, R. P., and Linn, L. J. (2000). Distribution and turnover of dissolved DMSP and its relationship with bacterial production and dimethylsulfide in the Gulf of Mexico. Limnol. Oceanogr. 45, 849–61.
Kiene, R. P., and Slezak, D. (2006). Low dissolved DMSP concentrations in seawater revealed by small-volume gravity filtration and dialysis sampling. Limnol. Oceanogr. Methods 4, 80–95.
Kiene, R. P., Linn, L. J., and Bruton, J. A. (2000). New and important roles for DMSP in marine microbial communities. J. Sea Res. 43, 209–24.
Kiene, R. P., Nowinski, B., Esson, K., Preston, C., Marin Iii, R., Birch, J., et al. (2019). Unprecedented DMSP concentrations in a massive dinoflagellate bloom in Monterey Bay, CA. Geophys. Res. Lett. 46, 12279–88.
Kirchman, D. L. (2002). The ecology of Cytophaga–Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39, 91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x
Kirst, G. (1996). “Osmotic adjustment in phytoplankton and macroalgae,” in Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds, eds R. P. Kiene, P. T. Visscher, M. D. Keller, and G. O. Kirst (Berlin: Springer), 121–9.
Knap, A., Michaels, A., Close, A., Ducklow, H., and Dickson, A. (1996). Protocols for the Joint Global Ocean Flux Study (JGOFS) Core Measurements. Paris: UNESCO.
Landa, M., Blain, S., Christaki, U., Monchy, S., and Obernosterer, I. (2016). Shifts in bacterial community composition associated with increased carbon cycling in a mosaic of phytoplankton blooms. ISME J. 10, 39–50. doi: 10.1038/ismej.2015.105
Lane, D. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackenbrandt and M. Goodfellow (Hoboken, NJ: John Wiley & Sons), 115–75.
Lane, D. J., Pace, B., Olsen, G. J., Stahl, D. A., Sogin, M. L., and Pace, N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U. S. A. 82, 6955–9. doi: 10.1073/pnas.82.20.6955
Levasseur, M., Michaud, S., Egge, J., Cantin, G., Nejstgaard, J., Sanders, R., et al. (1996). Production of DMSP and DMS during a mesocosm study of an Emiliania huxleyi bloom: influence of bacteria and Calanus finmarchicus grazing. Mar. Biol. 126, 609–18.
Levine, N. M., Varaljay, V. A., Toole, D. A., Dacey, J. W., Doney, S. C., and Moran, M. A. (2012). Environmental, biochemical and genetic drivers of DMSP degradation and DMS production in the Sargasso Sea. Environ. Microbiol. 14, 1210–23. doi: 10.1111/j.1462-2920.2012.02700.x
Li, C.-Y., Wang, X.-J., Chen, X.-L., Sheng, Q., Zhang, S., Wang, P., et al. (2021). A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA. Elife 10:e64045. doi: 10.7554/eLife.64045
Lidbury, I., Kröber, E., Zhang, Z., Zhu, Y., Murrell, J. C., Chen, Y., et al. (2016). A mechanism for bacterial transformation of dimethylsulfide to dimethylsulfoxide: a missing link in the marine organic sulfur cycle. Environ. Microbiol. 18, 2754–66. doi: 10.1111/1462-2920.13354
Liu, J., Zhang, Y., Liu, J., Zhong, H., Williams, B. T., Zheng, Y., et al. (2021). Bacterial dimethylsulfoniopropionate biosynthesis in the East China Sea. Microorganisms 9:657. doi: 10.3390/microorganisms9030657
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–2. doi: 10.1089/cmb.2017.0096
Matsen, F. A., Kodner, R. B., and Armbrust, E. V. (2010). pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11:538. doi: 10.1186/1471-2105-11-538
McDevitt, C. A., Hanson, G. R., Noble, C. J., Cheesman, M. R., and Mcewan, A. G. (2002). Characterization of the redox centers in dimethyl sulfide dehydrogenase from Rhodovulum sulfidophilum. Biochemistry 41, 15234–44. doi: 10.1021/bi026221u
McParland, E. L., and Levine, N. M. (2019). The role of differential DMSP production and community composition in predicting variability of global surface DMSP concentrations. Limnol. Oceanogr. 64, 757–73.
McParland, E. L., and Levine, N. M. (2020). Evidence for Two Independent Ecological Roles of Dimethylsulfoniopropionate (DMSP). Washington, DC: American Geophysical Union.
McParland, E. L., Lee, M. D., Webb, E. A., Alexander, H., and Levine, N. M. (2021). DMSP synthesis genes distinguish two types of DMSP producer phenotypes. Environ. Microbiol. 23, 1656–69. doi: 10.1111/1462-2920.15393
Moran, M. A., Reisch, C. R., Kiene, R. P., and Whitman, W. B. (2012). Genomic insights into bacterial DMSP transformations. Ann. Rev. Mar. Sci. 4, 523–42. doi: 10.1146/annurev-marine-120710-100827
Nawrocki, E. P., and Eddy, S. R. (2013). Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–5. doi: 10.1093/bioinformatics/btt509
Nowinski, B., Smith, C. B., Thomas, C. M., Esson, K., Marin, R., Preston, C. M., et al. (2019b). Microbial metagenomes and metatranscriptomes during a coastal phytoplankton bloom. Sci. Data 6:129. doi: 10.1038/s41597-019-0132-4
Nowinski, B., Motard-Côté, J., Landa, M., Preston, C. M., Scholin, C. A., Birch, J. M., et al. (2019a). Microdiversity and temporal dynamics of marine bacterial dimethylsulfoniopropionate genes. Environ. Microbiol. 21, 1687–701. doi: 10.1111/1462-2920.14560
O’Brien, J., Mcparland, E. L., Bramucci, A. R., Siboni, N., Ostrowski, M., Kahlke, T., et al. (2022). Biogeographical and seasonal dynamics of the marine Roseobacter community and ecological links to DMSP-producing phytoplankton. ISME Commun. 2:16.
Piredda, R., Tomasino, M., D’erchia, A., Manzari, C., Pesole, G., Montresor, M., et al. (2017). Diversity and temporal patterns of planktonic protist assemblages at a Mediterranean long term ecological research site. FEMS Microbiol. Ecol. 93:fiw200. doi: 10.1093/femsec/fiw200
Reisch, C. R., Moran, M. A., and Whitman, W. B. (2008). Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J. Bacteriol. 190, 8018–24. doi: 10.1128/JB.00770-08
Reisch, C. R., Stoudemayer, M. J., Varaljay, V. A., Amster, I. J., Moran, M. A., and Whitman, W. B. (2011). Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature 473, 208–11. doi: 10.1038/nature10078
Sheehan, C. E., and Petrou, K. (2020). Dimethylated sulfur production in batch cultures of Southern Ocean phytoplankton. Biogeochemistry 147, 53–69.
Shenoy, D., and Patil, J. S. (2003). Temporal variations in dimethylsulphoniopropionate and dimethyl sulphide in the Zuari estuary, Goa (India). Mar. Environ. Res. 56, 387–402. doi: 10.1016/S0141-1136(02)00337-9
Shenoy, D., Kumar, M. D., and Sarma, V. (2000). Controls of dimethyl sulphide in the Bay of Bengal during Bobmex-Pilot cruise 1998. J. Earth Syst. Sci. 109, 279–283.
Simó, R. (2001). Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 16, 287–94. doi: 10.1016/s0169-5347(01)02152-8
Simó, R., and Dachs, J. (2002). Global ocean emission of dimethylsulfide predicted from biogeophysical data. Global Biogeochem. Cycles 16:1078.
Simó, R., and Pedrós-Alió, C. (1999). Role of vertical mixing in controlling the oceanic production of dimethyl sulphide. Nature 402, 396–369.
Simo, R., Archer, S. D., Gilpin, L., and Stelfox-Widdicombe, C. E. (2002). Coupled dynamics of dimethylsulfoniopropionate and dimethylsulfide cycling and the microbial food web in surface waters of the North Atlantic. Limnol. Oceanogr. 47, 53–61.
Simo, R., Grimalt, J. O., and Albaiges, J. (1993). Field sampling and analysis of volatile reduced sulphur compounds in air, water and wet sediments by cryogenic trapping and gas chromatography. J. Chromatogr. A 655, 301–7.
Speeckaert, G., Borges, A. V., Champenois, W., Royer, C., and Gypens, N. (2018). Annual cycle of dimethylsulfoniopropionate (DMSP) and dimethylsulfoxide (DMSO) related to phytoplankton succession in the Southern North Sea. Sci. Total Environ. 622, 362–72. doi: 10.1016/j.scitotenv.2017.11.359
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–3. doi: 10.1093/bioinformatics/btu033
Stefels, J. (2000). Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J. Sea Res. 43, 183–97.
Stefels, J., Dijkhuizen, L., and Gieskes, W. (1995). DMSP-lyase activity in a spring phytoplankton bloom off the Dutch coast, related to Phaeocystis sp. abundance. Mar. Ecol. Prog. Ser. 123, 235–43.
Stefels, J., Steinke, M., Turner, S., Malin, G., and Belviso, S. (2007). Environmental constraints on the production and removal of the climatically active gas dimethylsulphide (DMS) and implications for ecosystem modelling. Biogeochemistry 83, 245–75.
Steinke, M., Malin, G., Turner, S., and Liss, P. (2000). Determinations of dimethylsulphoniopropionate (DMSP) lyase activity using headspace analysis of dimethylsulphide (DMS). J. Sea Res. 43, 233–44.
Stoeck, T., Bass, D., Nebel, M., Christen, R., Jones, M. D., Breiner, H. W., et al. (2010). Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19, 21–31. doi: 10.1111/j.1365-294X.2009.04480.x
Sun, H., Liu, J., Tan, S., Zheng, Y., Wang, X., Liang, J., et al. (2021). Spatiotemporal distribution of bacterial dimethylsulfoniopropionate producing and catabolic genes in the Changjiang Estuary. Environ. Microbiol. 23, 7073–92. doi: 10.1111/1462-2920.15813
Sun, H., Zhang, Y., Tan, S., Zheng, Y., Zhou, S., Ma, Q.-Y., et al. (2020). DMSP-producing bacteria are more abundant in the surface microlayer than subsurface seawater of the East China Sea. Microb. Ecol. 80, 350–365. doi: 10.1007/s00248-020-01507-8
Sun, J., Todd, J. D., Thrash, J. C., Qian, Y., Qian, M. C., Temperton, B., et al. (2016). The abundant marine bacterium Pelagibacter simultaneously catabolizes dimethylsulfoniopropionate to the gases dimethyl sulfide and methanethiol. Nat. Microbiol. 1:16065. doi: 10.1038/nmicrobiol.2016.65
Sunda, W. G., and Hardison, D. R. (2008). Contrasting seasonal patterns in dimethylsulfide, dimethylsulfoniopropionate, and chlorophyll a in a shallow North Carolina estuary and the Sargasso Sea. Aquat. Microb. Ecol. 53, 281–94.
Sunda, W., Kieber, D., Kiene, R., and Huntsman, S. (2002). An antioxidant function for DMSP and DMS in marine algae. Nature 418, 317–20. doi: 10.1038/nature00851
Suzuki, M. T., Taylor, L. T., and Delong, E. F. (2000). Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66, 4605–14. doi: 10.1128/AEM.66.11.4605-4614.2000
Teng, Z.-J., Qin, Q.-L., Zhang, W., Li, J., Fu, H.-H., Wang, P., et al. (2021). Biogeographic traits of dimethyl sulfide and dimethylsulfoniopropionate cycling in polar oceans. Microbiome 9:207.
Thume, K., Gebser, B., Chen, L., Meyer, N., Kieber, D. J., and Pohnert, G. (2018). The metabolite dimethylsulfoxonium propionate extends the marine organosulfur cycle. Nature 563, 412–5. doi: 10.1038/s41586-018-0675-0
Todd, J. D., Curson, A. R., Kirkwood, M., Sullivan, M. J., Green, R. T., and Johnston, A. W. (2011). DddQ, a novel, cupin-containing, dimethylsulfoniopropionate lyase in marine roseobacters and in uncultured marine bacteria. Environ. Microbiol. 13, 427–38. doi: 10.1111/j.1462-2920.2010.02348.x
Todd, J. D., Kirkwood, M., Newton-Payne, S., and Johnston, A. W. (2012). DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J. 6, 223–6. doi: 10.1038/ismej.2011.79
Todd, J., Curson, A., Dupont, C., Nicholson, P., and Johnston, A. (2009). The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some ascomycete fungi. Environ. Microbiol. 11, 1376–85. doi: 10.1111/j.1462-2920.2009.01864.x
Toole, D. A., Kieber, D. J., Kiene, R. P., Siegel, D. A., and Nelson, N. B. (2003). Photolysis and the dimethylsulfide (DMS) summer paradox in the Sargasso Sea. Limnol. Oceanogr. 48, 1088–100.
Townsend, D. W., and Keller, M. D. (1996). Dimethylsulfide (DMS) and dimethylsulfoniopropionate (DMSP) in relation to phytoplankton in the Gulf of Maine. Mar. Ecol. Prog. Ser. 137, 229–41.
Tripp, H. J., Kitner, J. B., Schwalbach, M. S., Dacey, J. W., Wilhelm, L. J., and Giovannoni, S. J. (2008). SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452, 741–4. doi: 10.1038/nature06776
Van Alstyne, K. L., and Puglisi, M. P. (2007). DMSP in marine macroalgae and macroinvertebrates: distribution, function, and ecological impacts. Aquat. Sci. 69, 394–402.
Van Diggelen, J., Rozema, J., Dickson, D., and Broekman, R. (1986). β-3-dimethylsulphoniopropionate, proline and quaternary ammonium compounds in Spartina anglica in relation to sodium chloride, nitrogen and sulphur. New Phytol. 103, 573–586.
van Duyl, F. C., Gieskes, W. W., Kop, A. J., and Lewis, W. E. (1998). Biological control of short-term variations in the concentration of DMSP and DMS during a Phaeocystis spring bloom. J. Sea Res. 40, 221–231.
Varaljay, V. A., Gifford, S. M., Wilson, S. T., Sharma, S., Karl, D. M., and Moran, M. A. (2012). Bacterial dimethylsulfoniopropionate degradation genes in the oligotrophic North Pacific subtropical gyre. Appl. Environ. Microbiol. 78, 2775–82. doi: 10.1128/AEM.07559-11
Varaljay, V. A., Howard, E. C., Sun, S., and Moran, M. A. (2010). Deep sequencing of a dimethylsulfoniopropionate-degrading gene (dmdA) by using PCR primer pairs designed on the basis of marine metagenomic data. Appl. Environ. Microbiol. 76, 609–17. doi: 10.1128/AEM.01258-09
Varaljay, V. A., Robidart, J., Preston, C. M., Gifford, S. M., Durham, B. P., Burns, A. S., et al. (2015). Single-taxon field measurements of bacterial gene regulation controlling DMSP fate. ISME J. 9, 1677–1686.
Vaulot, D., Geisen, S., Mahe, F., and Bass, D. (2021). pr2-primers: an 18S rRNA primer database for protists. bioRxiv [Preprint]. doi: 10.1101/2021.01.04.425170
Vila-Costa, M., Kiene, R. P., and Simí, R. (2008). Seasonal variability of the dynamics of dimethylated sulfur compounds in a coastal northwest Mediterranean site. Limnol. Oceanogr. 53, 198–211.
Vila-Costa, M., Rinta-Kanto, J. M., Sun, S., Sharma, S., Poretsky, R., and Moran, M. A. (2010). Transcriptomic analysis of a marine bacterial community enriched with dimethylsulfoniopropionate. ISME J. 4, 1410–20. doi: 10.1038/ismej.2010.62
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–7. doi: 10.1128/AEM.00062-07
Williams, B. T., Cowles, K., Martínez, A. B., Curson, A. R., Zheng, Y., Liu, J., et al. (2019). Bacteria are important dimethylsulfoniopropionate producers in coastal sediments. Nat. Microbiol. 4, 1815–25. doi: 10.1038/s41564-019-0527-1
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–8. doi: 10.1093/nar/gkt1209
Zhao, Y., Schlundt, C., Booge, D., and Bange, H. W. (2021). A decade of dimethyl sulfide (DMS), dimethylsulfoniopropionate (DMSP) and dimethyl sulfoxide (DMSO) measurements in the southwestern Baltic Sea. Biogeosciences 18, 2161–79.
Zheng, Y., Wang, J., Zhou, S., Zhang, Y., Liu, J., Xue, C.-X., et al. (2020). Bacteria are important dimethylsulfoniopropionate producers in marine aphotic and high-pressure environments. Nat. Commun. 11:4658. doi: 10.1038/s41467-020-18434-18444
Zubkov, M. V., Fuchs, B. M., Archer, S. D., Kiene, R. P., Amann, R., and Burkill, P. H. (2001). Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3, 304–11. doi: 10.1046/j.1462-2920.2001.00196.x
Keywords: DMSP, DMS, DLA, phytoplankton, bacteria, qPCR, 16S rRNA gene, 18S rRNA gene
Citation: O’Brien J, McParland EL, Bramucci AR, Ostrowski M, Siboni N, Ingleton T, Brown MV, Levine NM, Laverock B, Petrou K and Seymour J (2022) The Microbiological Drivers of Temporally Dynamic Dimethylsulfoniopropionate Cycling Processes in Australian Coastal Shelf Waters. Front. Microbiol. 13:894026. doi: 10.3389/fmicb.2022.894026
Received: 11 March 2022; Accepted: 20 May 2022;
Published: 15 June 2022.
Edited by:
Jose M. Gonzalez, University of La Laguna, SpainReviewed by:
Jonathan David Todd, University of East Anglia, United KingdomCopyright © 2022 O’Brien, McParland, Bramucci, Ostrowski, Siboni, Ingleton, Brown, Levine, Laverock, Petrou and Seymour. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James O’Brien, amFtZXMub2JyaWVuQHN0dWRlbnQudXRzLmVkdS5hdQ==; Justin Seymour, anVzdGluLnNleW1vdXJAdXRzLmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.