- 1Natural Science Research Institute, Gangneung-Wonju National University, Gangneung, South Korea
- 2Department of Zoology, Al Azhar University, Assiut, Egypt
- 3Department of Biology, Gangneung-Wonju National University, Gangneung, South Korea
The morphology and molecular phylogeny of two new ciliates, Monolamellophrya terricola gen. nov., sp. nov. and Trachelophyllum parapiculatum sp. nov., discovered in South Korea, were investigated. The two species belong to the suborder Trachelophyllina, which is characterized by the presence of a mucilaginous layer containing lepidosomes covering the cortex. Monolamellophrya terricola gen. nov., sp. nov. is characterized by the presence of a single layer of type II lepidosomes, representing a new genus. Trachelophyllum parapiculatum sp. nov. has only type I lepidosomes covering the cortex, a generic character of the genus Trachelophyllum, and is distinguished from other congeners by a combination of morphological features, including the 15–24 μm long rod-shaped extrusomes, the 9–13 ciliary rows, the 7–11 and 17–25 dikinetids in brush rows 1 and 2, respectively, and the bipolar brush row 3. Furthermore, the 18S rRNA gene sequences of the two new species were provided. The phylogenetic analyses show that the sequence of M. terricola gen. nov., sp. nov. clusters with two other trachelophyllid sequences, and the sequence of T. parapiculatum sp. nov. is placed at the base of these three sequences with full support. Furthermore, the four trachelophyllid sequences that are available so far form a monophyletic clade.
Introduction
Haptorid ciliates of the suborder Trachelophyllina Grain, 1994 are widely distributed and mostly found in terrestrial and semiterrestrial habitats but also found in freshwater and rarely in saline water (Kahl, 1930; Foissner, 1984, 1994, 2005, 2016; Foissner et al., 1995, 1999, 2002; Coats and Clamp, 2009; Telesh et al., 2009; Jang et al., 2015; Bourland, 2017). They are characterized by the presence of organic epicortical scales, of unknown function called lepidosomes, embedded in a mucilaginous layer covering the cortex. The shape of the lepidosomes is considered a genus-specific character within the Trachelophyllina and can only be revealed using a scanning electron microscope, thus identification of these taxa based only on live observation and/or silver impregnation is insufficient (Foissner et al., 2002; Foissner, 2005, 2016). So far, 12 types of lepidosomes were described in 19 well-characterized species and ten genera. Each genus is characterized by the presence of one, two, or three types of lepidosomes as follows: the genera Epitholiolus Foissner et al., 2002 and Trachelophyllum Claparéde and Lachmann, 1859 each has only a single type of lepidosomes; the genera Bilamellophrya Foissner et al., 2002, Cataphractes Foissner, 2016, Ileonema Stokes, 1884, Lingulothrix Foissner et al., 2002, Sleighophrys Foissner, 2005, and Spetazoon Foissner, 1994 each has two types of lepidosomes; and the genera Luporinophrys Foissner, 2005 and Trachelophyllides Foissner, 2016 each has three types of lepidosomes (Nicholls and Lynn, 1984; Foissner et al., 2002; Foissner, 2005, 2016; Bourland, 2017). Trachelophyllina genera belong to two different families mainly based on the dorsal brush rows organization. The family Lingulotrichidae Foissner, 2016 is characterized by the presence of more than three heteromorphic brush rows, while the family Trachelophyllidae Kent, 1881 is characterized by the presence of two dikinetidal and one monokinetidal brush rows (Foissner, 2016). Currently, the trachelophyllids are highly underrepresented in the phylogenetic trees, with only two 18S rRNA gene sequences available in the GenBank database. Unfortunately, the identification of these two sequences is doubtful because data on their lepidosomes are lacking (Vdačný et al., 2011; Jang et al., 2015, 2022; Huang et al., 2018).
In this study, we investigate the morphology of two species discovered in South Korea. The first species is unique within the Trachelophyllidae in having a single layer of type II lepidosomes and thus representing a new genus, Monolamellophrya gen. nov., while the second species has only type I lepidosomes as other members of the genus Trachelophyllum and is distinguishable from other congeners by a combination of features, i.e., the 15–24 μm long rod-shaped extrusomes, the 9–13 ciliary rows, the 7–11 and 17–25 dikinetids in brush rows 1 and 2, respectively, and the bipolar brush row 3. Moreover, the 18S rRNA gene sequences of the two species were analyzed to determine their phylogenetic position.
Materials and Methods
Sample Collection and Identification
Monolamellophrya terricola gen. nov., sp. nov. was discovered in a soil sample collected near Bongnae Falls, Ulleung Island, South Korea (N 37° 29′ 48.12″ E 130° 53′ 29.64″) in August 2018. The soil sample was air-dried for at least 2 weeks and stored in a plastic bag. The sample was rewetted in February 2019 with mineral water (Jeju Samdasoo, Jeju Province Development Co., South Korea) to induce excystment of ciliates using the non-flooded Petri dish method (Foissner et al., 2002). Trachelophyllum parapiculatum sp. nov. was discovered in a water sample including debris collected from a temporary pond on a lawn in the Gangneung-Wonju National University, Gangneung-si, South Korea (N 37°46′12.4″ E 128°52′16.5″) after heavy rainfall in July 2020. The water sample was kept in a plant culture dish at room temperature. Attempts to establish enriched cultures were unsuccessful for both species. Living specimens were investigated using a stereomicroscope (Olympus SZ61, Tokyo, Japan) and a light microscope (Olympus BX53) with differential interference contrast at magnifications of 50–1,000×. The infraciliature was revealed by protargol impregnation and scanning electron microscopy. Protargol powder was synthesized using the methods described by Pan et al. (2013) and Kim and Jung (2017), and the protargol impregnation technique is based on ‘procedure A' described by Foissner (2014). The SEM technique was conducted following the procedures described by Foissner (2014) and Moon et al. (2020). The terminology is according to Foissner et al. (2002) and Foissner (2016).
DNA Extraction, PCR Amplification, and Sequencing
Under the stereomicroscope, five cells were collected using a microcapillary from raw cultures of both species. The cells were transferred to habitat water filtered by a 0.2-μm syringe filter (Minisart® CA Syringe Filters; Sartorius, Aubagne, France), starved for at least 3 h, washed at least five times using the same filtered water to remove other eukaryotes, and then transferred to a 1.5-ml centrifuge tube each with a minimum volume of water. Genomic DNA was extracted using a RED-Extract-N-Amp Tissue PCR Kit (Sigma, St. Louis, MO, USA). The 18S rRNA gene was amplified using the primer New Euk A (5′-CTG GTT GAT YCT GCC AGT-3′) (Moon et al., 2017), which is a slightly modified version of the primer Euk A in Medlin et al. (1988), and the primer LSU rev4 (5′-GTT AGA CTY CTT GGT CCG TG-3′) (Sonnenberg et al., 2007) for M. terricola gen. nov., sp. nov. The primers New Euk A and Euk B (5′-TGA TCC TTC TGC AGG TTC ACC TAC-3′) were used to cover nearly the entire 18S rRNA gene of T. parapiculatum sp. nov. The PCR conditions for the primers New Euk A and LSU rev4 were as follows: denaturation at 94°C for 1 min 30 s, followed by 40 cycles of denaturation at 98°C for 10 s, annealing at 58.5°C for 30 s, and extension at 72°C for 3 min, and a final extension step at 72°C for 7 min. The PCR conditions for the primers New Euk A and Euk B were as follows: denaturation at 94°C for 90 s, followed by 40 cycles of denaturation at 98°C for 10 s, annealing at 58.5°C for 30 s, extension at 72°C for 2 min, and a final extension step at 72°C for 7 min. For purification of the PCR products, MEGAquickspin Total Fragment DNA Purification Kit (iNtRON Biotechnology, South Korea) was used. DNA sequencing was performed using an ABI 3700 sequencer (Applied Biosystems, Foster City, CA, USA). New Euk A, LSU rev4, and three internal primers [18SF790v2: 5′-AAA TTA KAG TGT TYM ARG CAG-3′, 18SR300: 5′-CAT GGT AGT CCA ATA CAC TAC-3′ (Park et al., 2017), and 18SF1470: 5′-TCT GTG ATG CCC TTA GAT GTC-3′ (Jung et al., 2018)] were used for the sequences of M. terricola gen. nov., sp. nov., and only New Euk A and Euk B primers were used for the sequences of T. parapiculatum sp. nov.
Phylogenetic Analyses
The 18S rRNA gene sequences of M. terricola gen. nov., sp. nov. and T. parapiculatum sp. nov. were assembled using Geneious 9.1.5 (Kearse et al., 2012). To determine the phylogenetic position of the two new species, 18S rRNA gene sequences of 77 ciliates were retrieved from the NCBI database, including three metopids as outgroup taxa: Clevelandella panesthiae (KC139719), Metopus palaeformis (AY007450), and Nyctotherus ovalis (AJ222678). The sequences were aligned using ClustalW (Thompson et al., 1994), and both ends were manually trimmed using BioEdit 7.0.9.0 (Hall, 1999). The length of the final alignment was 1609 bp. Using jModelTest 2.1.7 (Darriba et al., 2012), the best-fit evolutionary model TVM+I+G under the Akaike information criterion (AIC) was selected. The maximum likelihood (ML) tree was constructed using IQ-Tree 1.5.3 (Nguyen et al., 2015). The reliability of internal branches was assessed using a nonparametric bootstrap method with 100,000 replicates. The number of nucleotide differences and pairwise sequence similarity was calculated using MEGA 6.06 (Tamura et al., 2013). MrBayes 3.1.2 (Ronquist et al., 2012) was used for Bayesian inference (BI) analyses with Markov Chain Monte Carlo for 3,000,000 generations at a sampling frequency of every 100 generations, and the first 25% of trees were discarded as burn-in. Phylogenetic trees were visualized using the free software package FigTree version 1.4.3 by Rambaut (2006).
Results
ZooBank registration number of this study: urn:lsid:zoobank.org:pub:CE631A43-5895-4080-95EF-D43022C95A78
ZooBank registration number of Monolamellophrya gen. nov.: urn:lsid:zoobank.org:act:74B5B8AA-E968-47A7-AF9A-6F233605EE36
ZooBank registration number of M. terricola gen. nov., sp. nov.: urn:lsid:zoobank.org:act:2BAE8213-FC5D-4DD5-9472-03C79245E819
ZooBank registration number of T. parapiculatum sp. nov.: urn:lsid:zoobank.org:act:A49DCDEF-7A93-477D-AEC5-F6CAEBADF046
Taxonomy
Phylum Ciliophora Doflein, 1901
Subphylum Intramacronucleata Lynn, 1996
Class Litostomatea Small and Lynn, 1981
Subclass Haptoria Corliss, 1974
Order Spathidiida Foissner and Foissner, 1988
Suborder Trachelophyllina Grain, 1994
Family Trachelophyllidae Kent, 1881
Monolamellophrya gen. nov.
Diagnosis
Trachelophyllidae with only one layer of type II lepidosomes. Lepidosomes with conical superstructure composed of six or seven concave meridional arcs.
Etymology
Composite of the Greek numeral mono (one), the Latin noun lamella (thin plate), and the Greek noun ophrya (eyebrow, cilia, ciliate), referring to the single type of lepidosome. Feminine gender.
Type Species
Monolamellophrya terricola sp. nov.
Species Assignable
Only the type species.
Monolamellophrya terricola gen. nov., sp. nov.
Diagnosis
Size in vivo 100–160 × 15–25 μm; body slenderly fusiform and slightly flattened dorsoventrally. Two macronuclear nodules and two micronuclei. Extrusomes acicular, 7–9 μm long in oral bulge and cytoplasm. 11–13 meridional ciliary rows. Dorsal brush rows 1 and 2 dikinetidal and isostichad, each consisting of 11–18 and 12–18 dikinetids, respectively; brush row 3 monokinetidal, slightly longer than rows 1 and 2. Lepidosomes of type II, ~ 1.0 × 0.7 × 0.6 μm, with conical superstructure composed of six or seven arcs.
Etymology
The Latin species-group name terricola (living in soil) refers to the habitat in which the species was discovered.
Type Locality
Soil near Bongnae Falls, Ulleung Island, South Korea (N 37° 29′ 48.12″ E 130° 53′ 29.64″).
Type Material
The slide containing the holotype (Figures 1E,F, 3A; NNIBRPR21237) and one paratype slide (NNIBRPR21238) with protargol-impregnated specimens were deposited at the Nakdonggang National Institute of Biological Resources (NNIBR), Sangju, Korea.
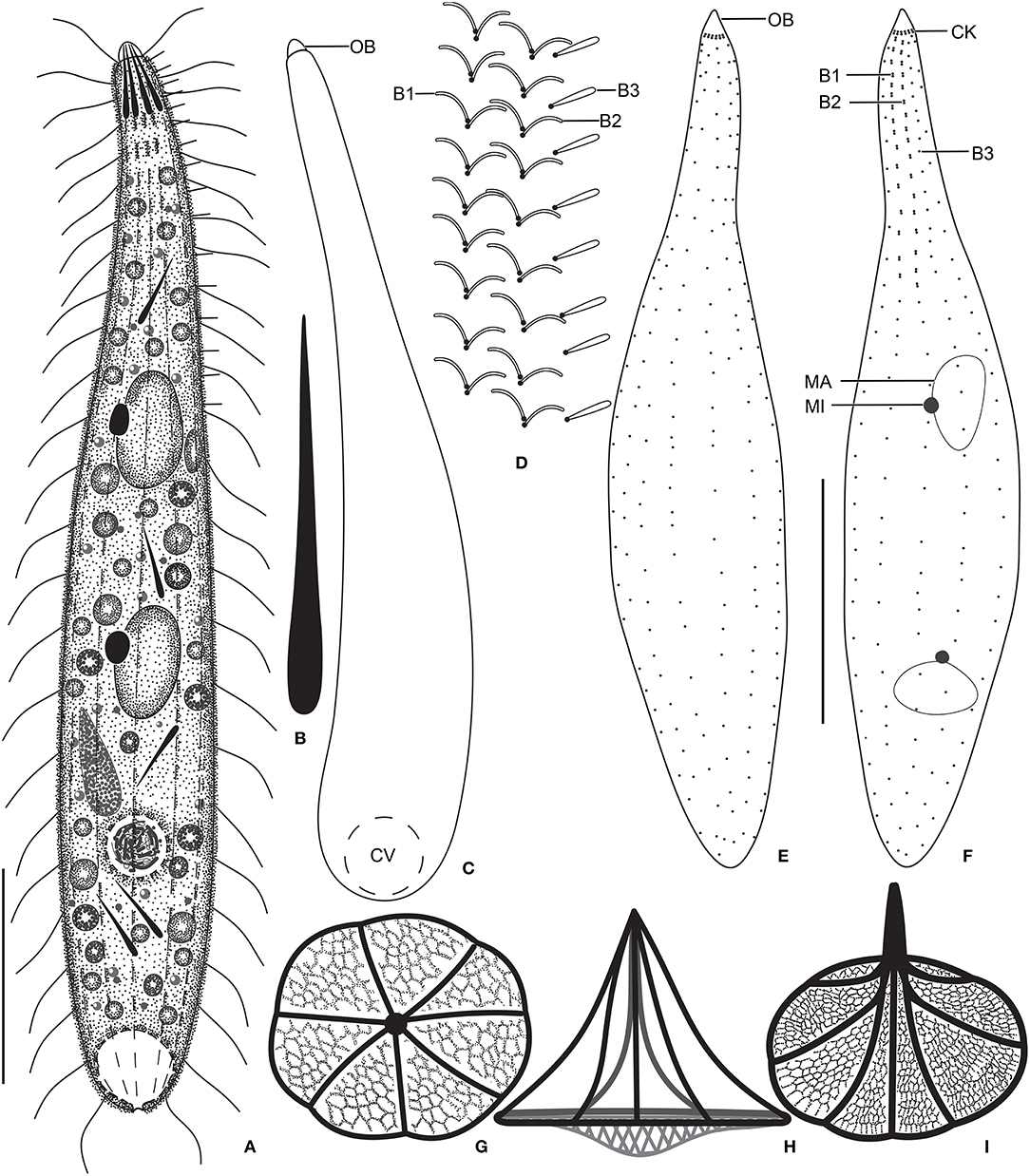
Figure 1. Monolamellophrya terricola gen. nov., sp. nov. from life (A–D), after protargol impregnation (E,F), and redrawn from scanning electron micrographs (G–I). (A) A representative specimen, showing the body shape and the acicular extrusomes in the oral region and cytoplasm. (B) Acicular extrusome. (C) A specimen with broadly rounded posterior end. (D) Ciliature of dorsal brush. (E,F) Ventral (E) and dorsal (F) views of the holotype specimen, showing the infraciliature and the nuclear apparatus. (G–I) Vertical (G), lateral (H), and oblique distal (I) views of the type II lepidosomes, showing the circular baseplate with convex middle portion, the fine arcs forming rosette-like structure in surface view (G), and a conspicuous cone structure in lateral view (H). B1–3, dorsal brush rows; CK, circumoral kinety; CV, contractile vacuole; MA, macronucleus; MI, micronucleus. Scale bars = 30 μm.
Morphological Description of Monolamellophrya terricola gen. nov., sp. nov.
Size 100–160 × 15–25 μm in vivo and 75–137 × 11–23 μm after protargol impregnation; length:width ratio ~ 6.7:1 in vivo and 3.9–11.4:1 (6.1:1 on average) after protargol impregnation. Body slenderly fusiform, slightly flattened dorsoventrally, with slightly (in contracted specimens) to distinctly (in extended specimens) narrow neck, ~ 5 μm wide in protargol preparations, slightly widened in the oral region, and gradually broadened posteriorly merging into the wider, slender trunk. The anterior end (oral bulge) narrow and conical, the posterior end usually narrowly rounded, rarely broadly rounded (Figures 1A,C,E,F, 2A–C, 3A–H). Cells very flexible and contractile by up to 30% of body length under cover glass; contracts and extends very slowly. The nuclear apparatus usually in the middle third of the cell, sometimes posterior macronuclear nodule displaced to a posterior quarter of the cell. Macronuclear nodules usually ellipsoidal to narrowly ellipsoidal and sometimes with irregular outline, each ~ 15 × 7 μm in vivo. Micronuclei near or attached to macronuclear nodules, globular to broadly ellipsoidal (Figures 1A,F, 2A–C, 3A). Contractile vacuole in the posterior body end with a single terminal excretory pore (Figures 1A,C, 2A–C). Extrusomes acicular, 7–9 μm long in vivo, form bundle in the oral bulge and scattered in the cytoplasm, and do not impregnate with protargol (Figures 1A,B, 2D,E). Cytoplasm colorless, contains lipid droplets 1–4 μm across and food vacuoles up to 10 μm across containing flagellates and small ciliates (Figures 1A, 2A–E, 3A–E). Usually glides slowly between soil particles and occasionally swims slowly by rotating about the main body axis.
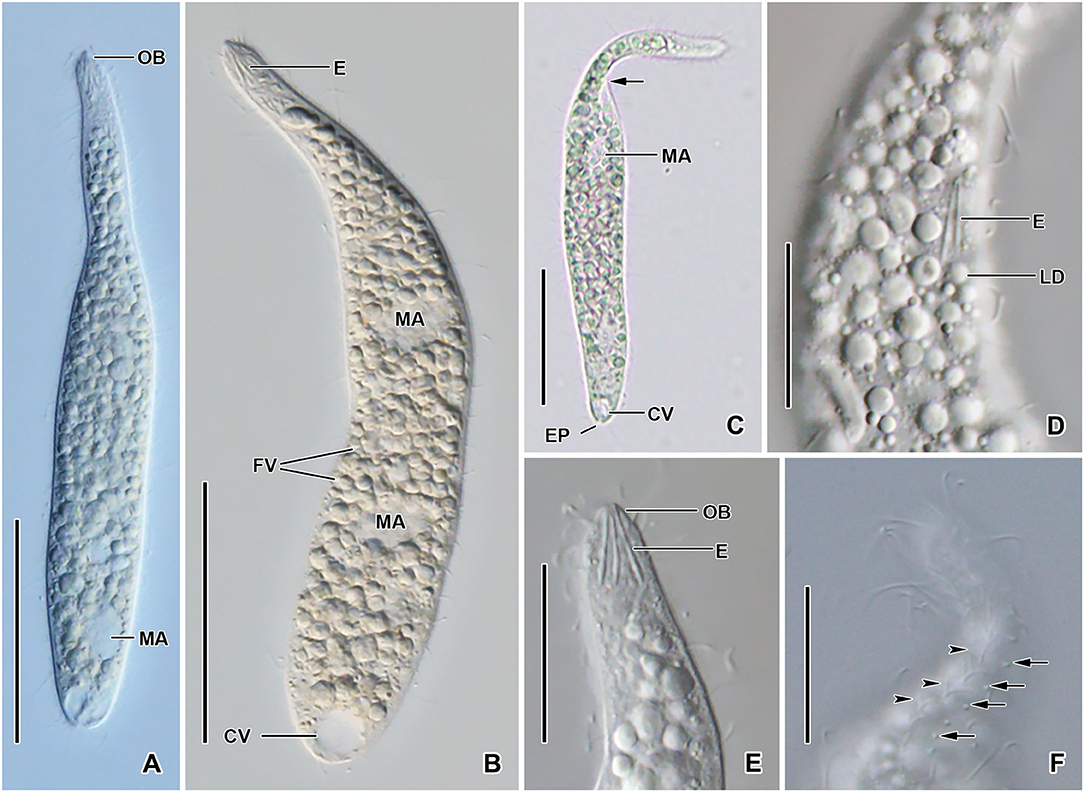
Figure 2. Monolamellophrya terricola gen. nov., sp. nov. from life. (A–C) Overviews showing the body shape, the narrow neck, the terminal contractile vacuole, the nuclear apparatus, and the cytoplasm studded with food vacuoles and lipid droplets. The arrow shows that the body is laterally flattened. (D,E) Optical sections showing the acicular extrusomes in cytoplasm (D) and oral bulge (E) and the lipid droplets. (F) The dorsal brush, showing the curved, V-like spread bristles of brush row 2 (arrowheads) and the acicular bristles of brush row 3 (arrows). B1–3, dorsal brush rows; CK, circumoral kinety; CV, contractile vacuole; E, extrusomes; EP, excretory pore; FV, food vacuoles; LD, lipid droplets; MA, macronucleus; MI, micronucleus; OB, oral bulge. Scale bars = 50 μm (A–C) and 20 μm (D–F).
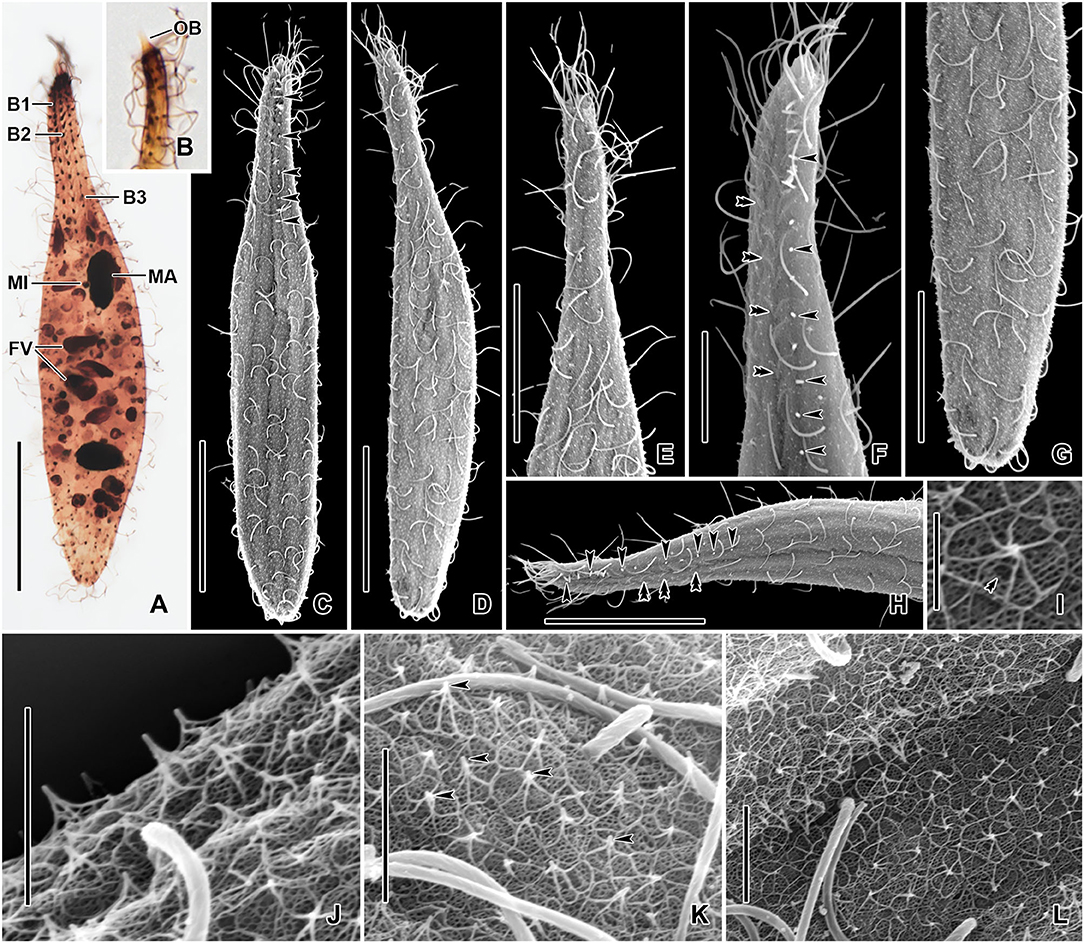
Figure 3. Monolamellophrya terricola gen. nov., sp. nov. after protargol impregnation (A,B) and in the scanning electron microscope (C–L). (A) Dorsal view of the holotype specimen showing the dorsal brush rows, the nuclear apparatus, and the cytoplasm packed with food vacuoles. (B) The narrow, conical oral bulge of a paratype specimen. (C–H) Dorsal (C,F,H) and ventral (D,E,G) views, showing the cortex covered with epicortical scales of type II indicated by the white dots, which represent the cone structure made by the fine arcs. Arrowheads mark the bristles of the dorsal brush row 3. Cilia of brush rows 1 and 2 V-like spread and covered by epicortical scales (double arrowheads). (I–L) Lepidosomes of type II showing the fine arcs fusing together and forming a rosette-like structure in the top view. The double arrowhead in (I) marks the depression in the middle area of the baseplate. B1–3, dorsal brush rows; FV, food vacuoles; MA, macronucleus; MI, micronucleus. Scale bars = 30 μm (A,C,D,H), 20 μm (E,G), 10 μm (F), 2 μm (J,K), and 1 μm (I).
Cortex thin and very flexible, covered with a very thin mucilaginous layer of lepidosomes, recognizable only in scanning electron micrographs. Mucilaginous layer composed of a single layer of tightly spaced type II lepidosomes (Figures 3C–L). Individual lepidosomes 1.0 × 0.7 × 0.6 μm on average in scanning electron micrographs, baseplate circular to broadly ellipsoidal, finely faceted, central area slightly to distinctly convex (Figures 1H, 3I), gives rise to six or seven fine, concave arcs, forming cone-shaped superstructure in lateral view and a rosette-like structure in top view (Figures 1G–I, 3I–L).
Cilia in vivo 8–10 μm in length. 11–13 meridional and equidistant ciliary rows composed of rather widely spaced monokinetids, three of them form dorsal brush rows anteriorly and continue posteriorly as ordinary somatic ciliary rows (Figures 1D–F, 3A–H). Brush rows 1 and 2 of similar length and structure (isostichad), extend to ~36% of body length and composed of rather widely spaced 11–18 and 12–18 dikinetids, respectively, dikinetids bear curved, immobile, V-like spread bristles, 3–4 μm long; bristles laying down and covered by mucilaginous layer and hardly recognizable both in vivo and in scanning electron micrographs (Figures 1D,F, 2F, 3H). Brush row 3 slightly longer than rows 1 and 2, composed of widely spaced monokinetids bearing 2–3 μm long, acicular and immobile bristles, piercing scale layer and recognizable both in vivo and in scanning electron micrographs (Figures 1D,E, 2F, 3A–C,F,H; Table 1).

Table 1. Morphometric data on Monolamellophrya terricola gen. nov., sp. nov. (Mt), and Trachylophyllum parapiculatum sp. nov. (Tp).
Oral bulge inconspicuous in vivo and after protargol impregnation because of hyaline and small size (2 × 2 μm in stained cells); conical and set off from body proper, contains extrusomes. Circumoral kinety at the base of the oral bulge, composed of ~12 dikinetids, each bearing a single cilium and inconspicuous nematodesmata extending posteriorly forming a pharyngeal basket (Figures 1A,C,E,F, 2A,B,E).
Trachelophyllum parapiculatum sp. nov.
Diagnosis
Size in vivo 140–200 × 15–25 μm; body slenderly clavate to fusiform and dorsoventrally flattened. Two macronuclear nodules and two micronuclei. Extrusomes rod-shaped, 15–24 μm in length, forming a bundle in the oral bulge and scattered or forming bundles in the cytoplasm. 9–13 meridional ciliary rows. Brush rows distinctly heterostichad, rows 1 and 2, each consists of 7–11 and 17–25 dikinetids, respectively, row 1 approximately half length of row 2; brush row 3 monokinetidal, extends to posterior body end. Lepidosomes of type I, ~1.4 × 1.1 × 0.4 μm, with hemispherical superstructure composed of ~11 polygons.
Etymology
Composite of the Greek prefix para- (besides, like, resembling) and the species-group name apiculatum, referring to the similarity to T. apiculatum (Perty, 1852) Claparéde and Lachmann, 1859.
Type Locality
Temporary pond on a lawn in the Gangneung-Wonju National University, Gangneung, South Korea (N 37°46′12.4″ E 128°52′16.5″).
Type Material
The slide containing the holotype (Figures 4B,C, 6A,B; NNIBRPR21239) and one paratype slide (NNIBRPR21240) with protargol-impregnated specimens were deposited at the Nakdonggang National Institute of Biological Resources (NNIBR), Sangju, Korea.
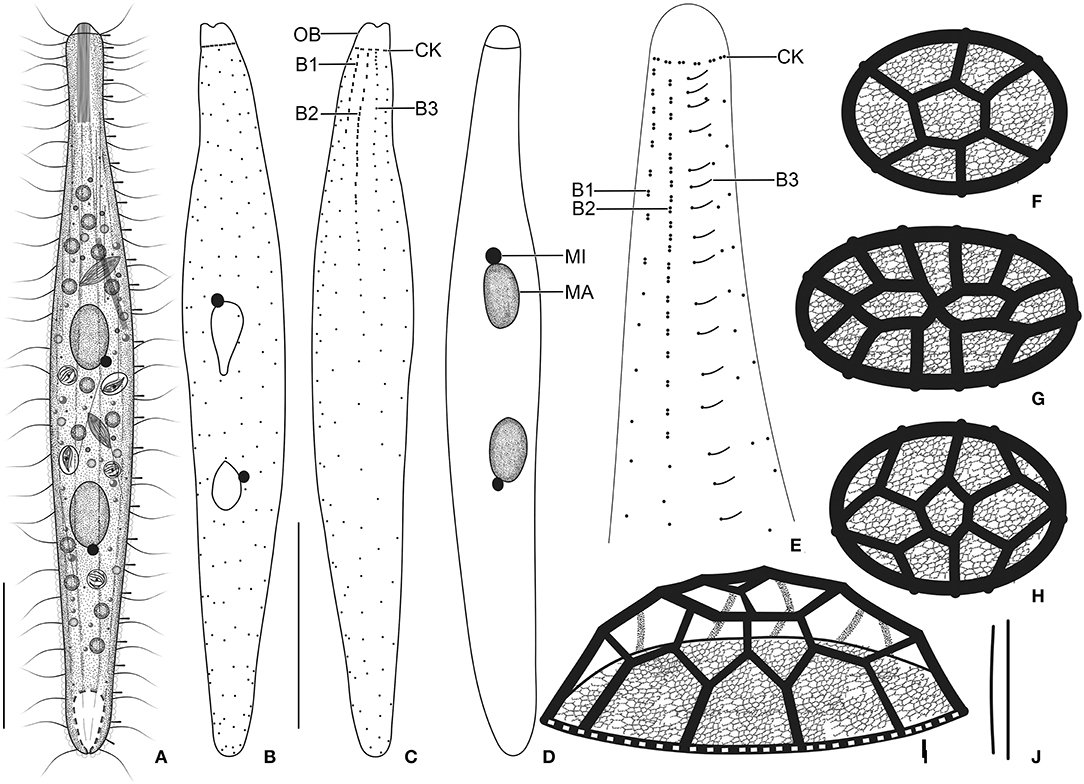
Figure 4. Trachelophyllum parapiculatum sp. nov. from life (A,J), after protargol impregnation (B–E), and redrawn from scanning electron micrographs (F–I). (A) A representative specimen showing the body shape and the long extrusomes in the oral bulge and cytoplasm, and the long brush row 3. (B–E) Ventral (B,D) and dorsal (C,E) views of the holotype (B,C) and two paratype (D,E) specimens, showing the somatic ciliature and the nuclear apparatus. Note the two dikinetidal brush rows, the single monokinetidal brush 3, and the dikinetidal circumoral kinety. (F–I) Surface (F–H) and lateral (I) views of lepidosomes type I. (J) Extrusomes. B1–3, dorsal brush rows; CK, circumoral kinety; MA, macronuclear nodules; MI, micronucleus; OB, oral bulge. Scale bars = 30 μm.
Morphological Description of Trachelophyllum parapiculatum sp. nov.
Size 140–200 × 15–25 μm in vivo and 107–150 × 9–20 μm after protargol impregnation; length:width ratio ~6.7:1 in vivo and 5.6–16.7:1 (on average 8.4:1) after protargol impregnation; body dorsoventrally flattened up to 2:1. Body slenderly clavate to fusiform with the neck slightly widened in the oral region and gradually broadened posteriorly merging into a wider trunk. The anterior end (oral bulge) narrow and cylindrical, posterior end narrowly rounded (Figures 4A–D, 5A–E,G,H, 6A,B,I). Cells very flexible and slightly contractile under cover glass; contracts and extends very slowly. The nuclear apparatus usually in the middle quarters of the cell. Macronuclear nodules usually ellipsoidal to narrowly ellipsoidal, each ~ 15 × 8 μm in vivo. Micronuclei near or attached to macronuclear nodules, spherical (Figures 4A,B,D, 5C,E, 6A,B). Contractile vacuole in posterior body end and connects with single terminal excretory pore by a distinct excretory canal (Figures 4A, 5C,E,F). Extrusomes straight or slightly curved rod-shaped, 15–24 μm long in vivo, form bundles in the oral region and extend into the oral bulge and scattered or form bundles in the cytoplasm, do not impregnate with protargol (Figures 4A,J, 5A,E,G). Cytoplasm colorless, contains 1–3 μm across lipid droplets and up to 10 μm across food vacuoles containing flagellates and small ciliates (Figures 4A, 5B,C,E). Usually swims slowly and rarely glides on the bottom of the culture dish.
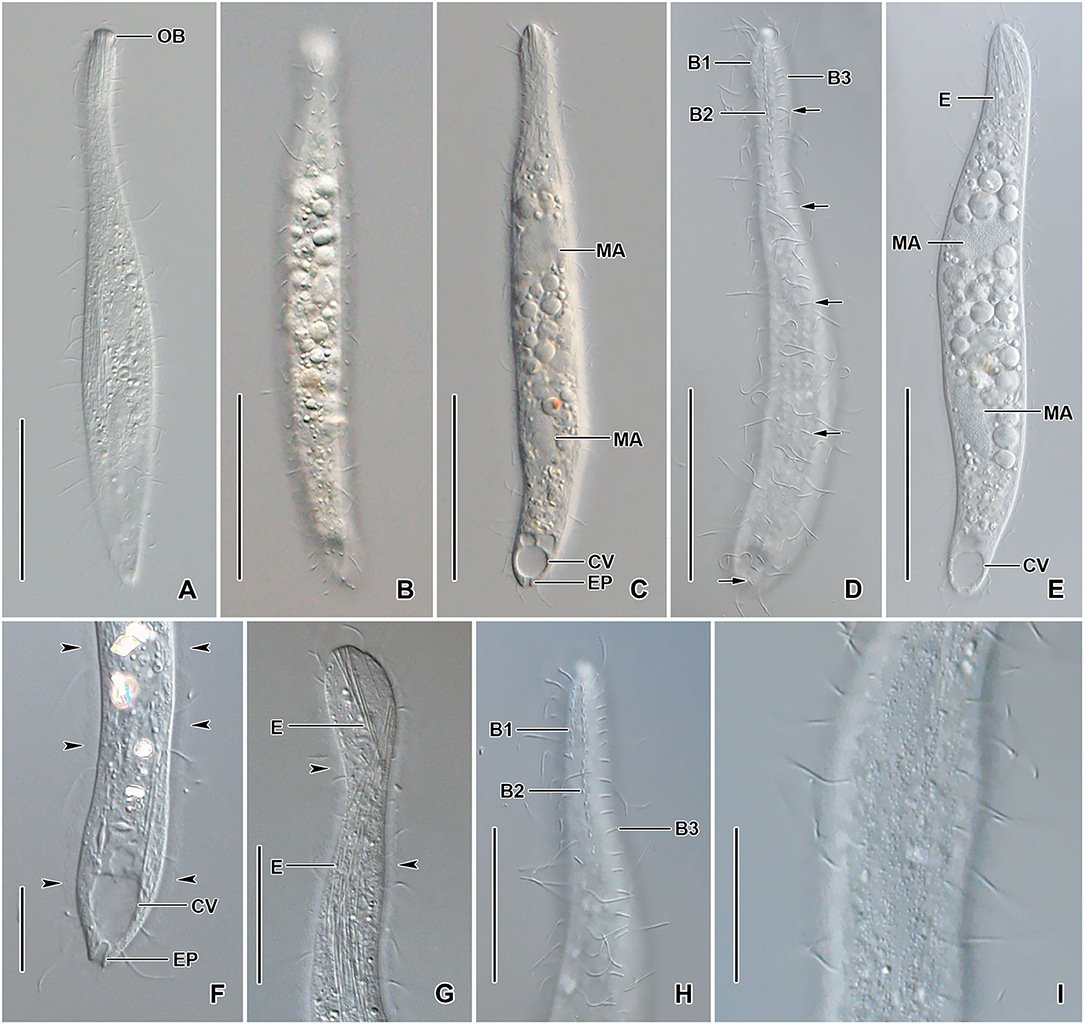
Figure 5. Trachelophyllum parapiculatum sp. nov. from life. (A–E) Overviews showing the body shape, the two widely spaced macronuclear nodules, the terminal contractile vacuole, the excretory pore, and the long rod-shaped extrusomes. Arrows denote the bipolar dorsal brush row 3. (F,G) Posterior (F) and anterior (G) body portion showing the contractile vacuole, the single excretory pore, and the extrusomes extending into the oral bulge. Arrowheads mark the mucilaginous layer. (H) Dorsal view of the anterior body portion showing the two dikinetidal and one monokinetidal brush rows. (I) Surface view showing the cortical granulation. B1–3, dorsal brush rows; CV, contractile vacuoles; E, extrusomes; EP, excretory pore; MA, macronuclear nodules; OB, oral bulge. Scale bars = 50 μm (A–E), 30 μm (H), 20 μm (G,I), and 15 μm (F).

Figure 6. Trachelophyllum parapiculatum sp. nov. after protargol impregnation (A–C) and in the scanning electron microscope (D–I). (A–C) Ventral (A) and dorsal (B,C) views of the holotype (A,B) and a paratype (C) specimen, showing the somatic ciliature, the dikinetidal dorsal brush rows 1 and 2, and the monokinetidal (arrowheads) brush row 3. Note the circumoral dikinetids each bearing a single cilium (C). (D,F) Anterior body portion, showing the oral bulge not covered with lepidosomes. Arrowheads show type I lepidosomes. (E) Lateral view of lepidosome type I. (G–I) Cortex covered with type I lepidosomes (arrowheads). (J) An extended specimen. B1–3, dorsal brush rows; MA, macronuclear nodules; MI, micronucleus; OB, oral bulge. Scale bars = 50 μm (J), 30 μm (A,B), 10 μm (C,D), and 5 μm (F–I).
Cortex thin and very flexible, contains small and irregularly arranged cortical granules (Figure 5I), covered by a mucilaginous layer of lepidosomes, recognizable in vivo and scanning electron micrographs. Mucilaginous layer composed of tightly spaced type I lepidosomes (Figures 4A, 5F,G, 6D–H). Individual scales 1.0–2.1 × 0.7–1.7 × 0.3–0.5 μm (1.4 × 1.1 × 0.4 μm on average; n = 12) in scanning electron microscopes, baseplate circular to elliptical finely faceted, perforations in baseplate margin hardly recognizable, superstructure hemispherical composed of an average of 11 polygons (Figures 4F–I, 6D–H).
Cilia in vivo ~8 μm in length, rather widely spaced. Ciliary rows meridional and equidistant; two of them (dorsal brush rows) form dikinetids anteriorly and continue posteriorly as ordinary somatic ciliary rows. Brush rows 1 and 2 composed of 7–11 and 17–25 dikinetids, respectively; row 1 approximately half the length of row 2, both bearing 3–4 μm long immobile, parallel bristles; bristles laying down and possibly covered by epicortical scale layer and hardly recognizable both in vivo and in scanning electron micrographs (Figures 4C,E). Brush row 3 extends to the posterior body end, composed of widely spaced monokinetids bearing 2–3 μm long immobile bristles, recognizable both in vivo and after protargol impregnation (Figures 4A, 5D,H, 6B,C,D; Table 1). In a few specimens, the row right of brush row 3 commences with shortened cilia as long as brush bristles and continues posteriorly as ordinary somatic ciliary rows.
Oral bulge conspicuous in vivo because of extrusomes, 4 × 5 μm after protargol impregnation; cylindrical and set off from body proper and not covered by lepidosomes. Circumoral kinety at the base of the oral bulge, composed of ~11 dikinetids, each bearing a single cilium and inconspicuous nematodesmata extending posteriorly, forming a pharyngeal basket (Figures 4C,E, 5A,D,G, 6C–E,I).
Phylogenetic Analyses
The 18S rRNA gene sequences of M. terricola gen. nov., sp. nov. and T. parapiculatum sp. nov. are 1,490 base pairs long each, having a GC content of 40.15% and 40.27%, and are available under GenBank accession numbers ON212658 and ON212659, respectively. The phylogenetic trees using ML and BI analyses show rather similar topologies, thus only the ML tree was used (Figure 7). In the phylogenetic tree, the trachelophyllid sequences form a monophyletic clade with full supports. The sequence of M. terricola gen. nov., sp. nov. exhibits a similarity of 99.86% (2 nucleotides difference) with both T. brachypharynx and an unidentified Trachelophyllum sp., and clusters with both sequences with high (93% ML) and low (0.78 BI) support forming a polytomy (Figures 7, 8; Table 2). The sequence of T. parapiculatum sp. nov. is placed as a sister branch to these three sequences with full support and shows a similarity of 99.39% (9 nucleotides difference) with both M. terricola gen. nov., sp. nov. and T. brachypharynx and shows a similarity of 99.46% (8 nucleotides difference) with Trachelophyllum sp. (Figure 8; Table 2).
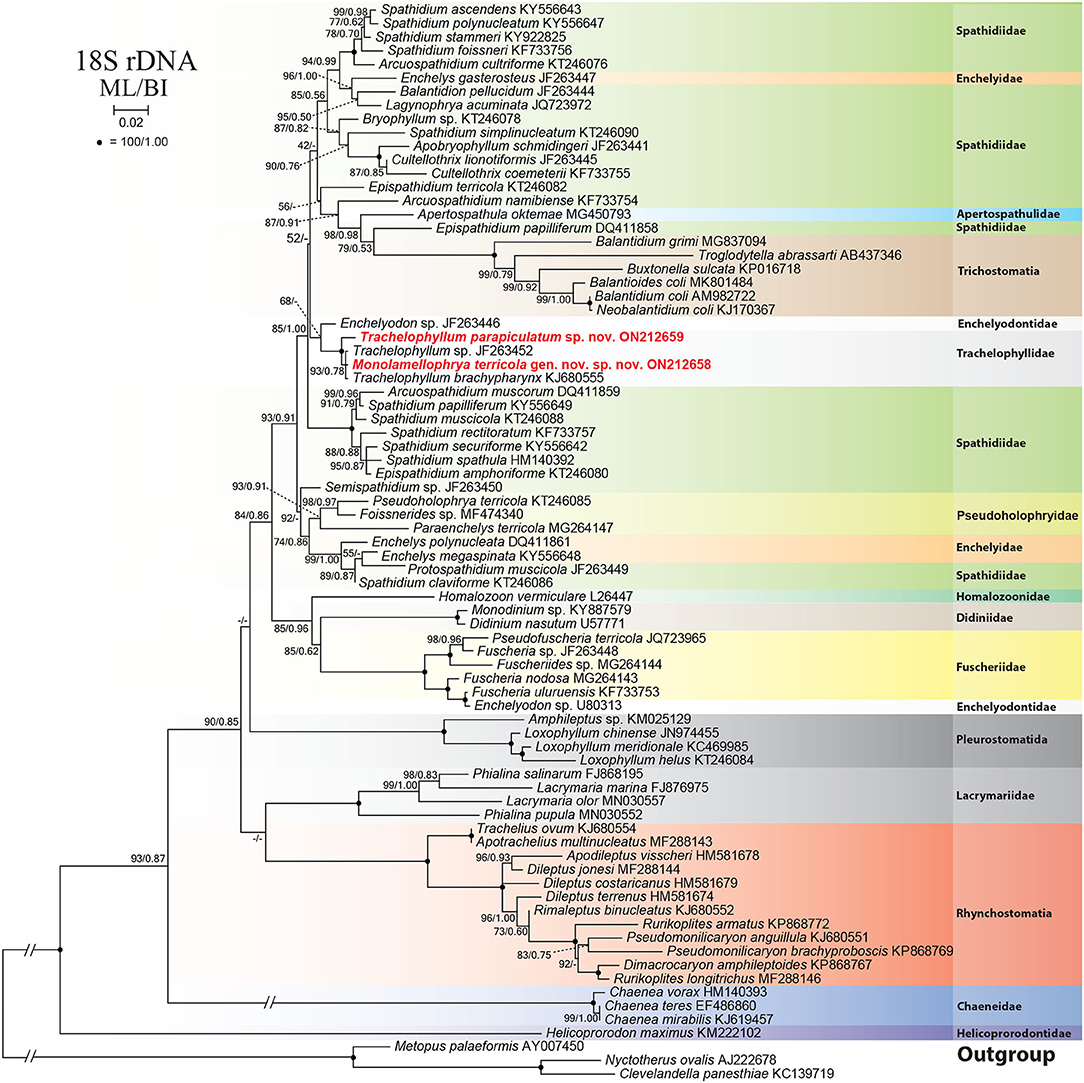
Figure 7. Maximum likelihood (ML) tree based on 18S rRNA gene sequences, showing the phylogenetic position of Monolamellophrya terricola gen. nov., sp. nov. and Trachelophyllum parapiculatum sp. nov. Newly obtained sequences are in bold. GenBank accession numbers follow species names. Numbers at the nodes represent the maximum likelihood (ML) bootstrap values and the Bayesian inference (BI) posterior probabilities. Dashes indicate bootstrap values < 50%, posterior probabilities < 0.5, or different topologies in BI and ML phylogenies. The scale bar represents two nucleotide substitutions per 100 nucleotides.
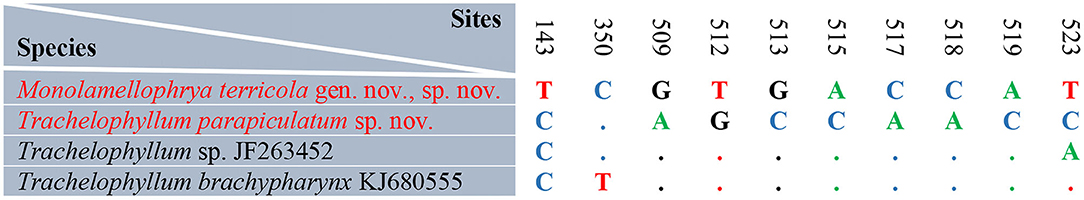
Figure 8. Sequence comparison of the 18S rRNA gene showing the unmatched nucleotides among the new species Monolamellophrya terricola gen. nov., sp. nov., Trachelophyllum parapiculatum sp. nov., and the other two available trachelophyllid species.
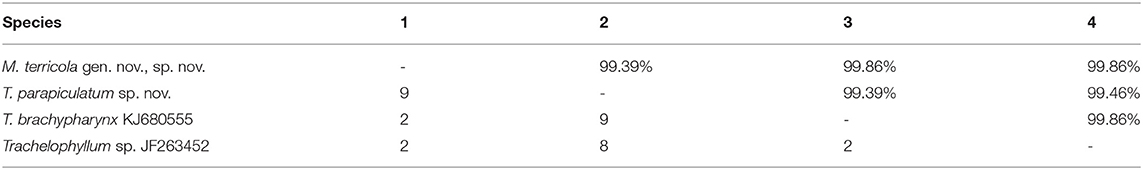
Table 2. Interspecific sequence similarity (above diagonal) and number of nucleotide differences (below diagonal) of the 18S rRNA gene sequences among members of the family Trachelophyllidae.
Discussion
Monolamellophrya gen. nov. as a New Genus
Members of the family Trachelophyllidae are characterized by the two dikinetidal and one monokinetidal dorsal brush rows and the presence of lepidosomes. Currently, 16 well-characterized trachelophyllid species belonging to eight genera are assigned to the family, mainly based on the types of the lepidosomes (Nicholls and Lynn, 1984; Foissner, 1994, 2005, 2016; Foissner et al., 2002; Bourland, 2017). Only two genera have a single type of lepidosomes each, namely, Epitholiolus with only type III lepidosomes and Trachelophyllum with only type I lepidosomes. Each of the other six genera has two or three types of lepidosomes. The new genus, Monolamellophrya gen. nov., is unique in having only one layer of type II lepidosomes. According to Foissner (2005, 2016) and Foissner et al. (2002), type II lepidosomes have four rather different shapes, and all of them are characterized basically by the cone-shaped superstructure and are found only in the genera Bilamellophrya and Luporinophrys. The first shape is found only in Bilamellophrya australiensis Foissner et al., 2002 and is characterized by the presence of ~10 concave, unevenly spaced arcs rising from the baseplate and fusing together at their ends forming a cone and connected to each other by three transverse rings. The second shape is found in B. hawaiiensis Foissner et al., 2002 and B. fraterculus Foissner, 2016 and is characterized by the presence of 12–16 scattered polygons in the cone-shaped superstructure. The third shape is found only in Luporinophrys micelae Foissner, 2005, and it has a larger size than other type II lepidosomes (4 × 4 μm) and its superstructure is formed by ~20 fine arcs (fibers) in two to five groups rising from the baseplate and fusing together at their ends forming a narrow, curved dome. The fourth shape is simple and consists of approximately seven arcs rising from the baseplate and fusing together forming a cone-shaped superstructure and found in B. etoschensis Foissner et al., 2002. The lepidosomes of M. terricola gen. nov., sp. nov. are similar to the fourth shape of type II in the shape of the superstructure but slightly differ in the thickness of the arcs (very thin vs. thick) and the shape of the baseplate (slightly to distinctly convex in middle portion vs. obconical). However, the convexity in the middle portion of the baseplate resembles the obconical baseplate described by Foissner et al. (2002) but is more shallow probably due to the preparation procedure. However, this variability in lepidosomes, especially type II, suggests that the trachelophyllids are an underestimated group and more studies are needed to reveal its true diversity.
Comparison of Monolamellophrya terricola gen. nov., sp. nov. With Closely Related Species
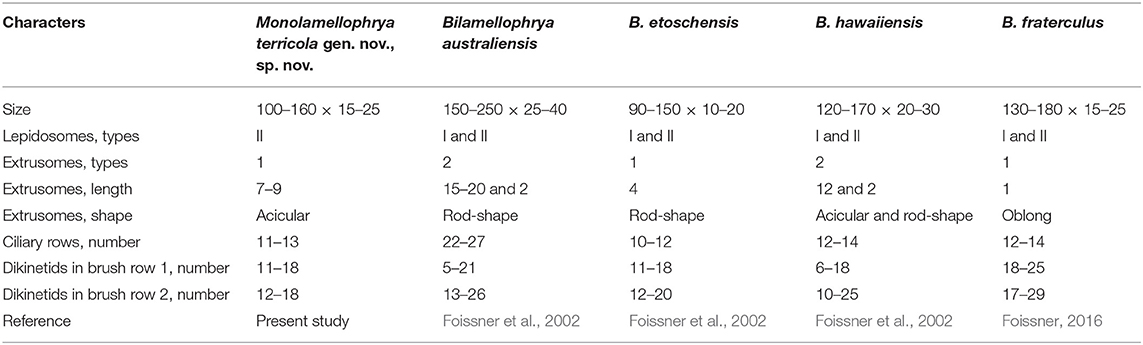
In vivo, M. terricola gen. nov., sp. nov. is hardly distinguishable from most trachelophyllid species because they have similar appearances and their sizes usually overlap. In addition, the mucilaginous layer of M. terricola gen. nov., sp. nov. is not recognizable in vivo and after protargol impregnation and thus could be misidentified as a non-trachelophyllid (without mucilaginous layer) haptorid. After protargol impregnation, many trachelophyllid species look similar and even have a similar number of ciliary rows and nuclear apparatus patterns. Thus, the scanning electron microscope is indispensable for the proper identification of such taxa. M. terricola gen. nov., sp. nov. is very similar to three Bilamellophrya species, namely, B. etoschensis, B. fraterculus, and B. hawaiiensis especially in body size and shape, the number of somatic kineties, and the overlapping numbers of dikinetids in dorsal brush rows (Table 3). However, M. terricola differs from these species mainly by the types of lepidosomes (one type vs. two types). Moreover, M. terricola gen. nov., sp. nov. is similar to B. hawaiiensis in having acicular extrusomes, but B. hawaiiensis has an extra type of extrusomes (rod-shaped). In addition, B. etoschensis and B. fraterculus each has a different type of extrusome, rod-shaped and oblong, respectively (Table 3) (Foissner et al., 2002; Foissner, 2016).
Comparison of Trachelophyllum parapiculatum sp. nov. With Closely Related Species
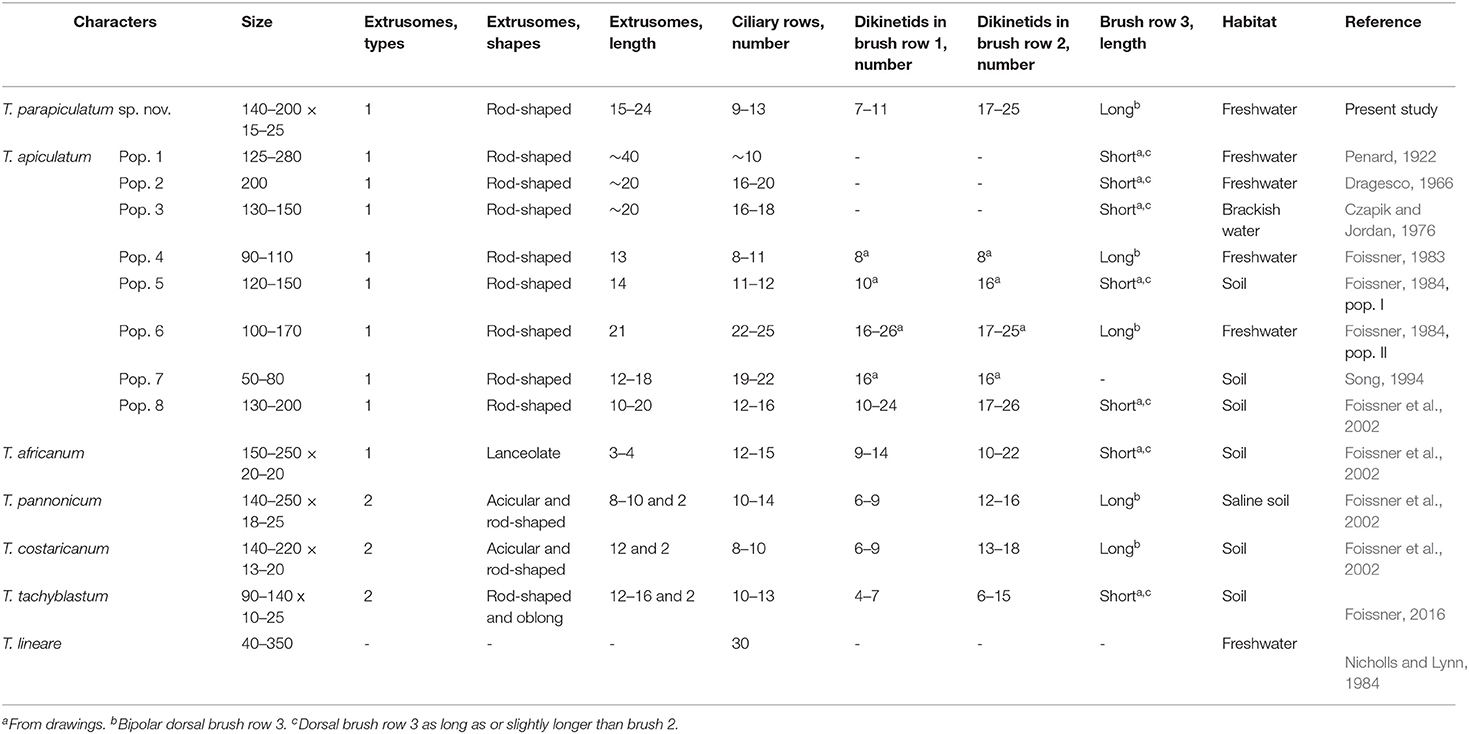
The trachelophyllid genus Trachelophyllum is characterized by having a mucilaginous layer composed only of type I lepidosomes (Foissner et al., 2002; Foissner, 2016). At present, ~31 species are assigned to the genus, most of which probably belong to different litostomatean families (Stokes, 1888; Levander, 1894; Penard, 1922; Dumas, 1930, 1937; Kahl, 1930; Gajewskaja, 1933; Lepsi, 1960; Tucolesco, 1962; Grolière, 1977; Foissner, 1983, 1984; Foissner et al., 2002). However, the lepidosomes of only six species, namely, T. apiculatum, T. africanum Foissner et al., 2002, T. costaricanum Foissner et al., 2002, T. lineare Lepsi, 1960, T. pannonicum Foissner et al., 2002, and T. tachyblastum Stokes, 1884, are characterized using the scanning electron microscope, and thus their assignment is doubtless. When comparing T. parapiculatum sp. nov. with other congeners, we recognized that the genus Trachelophyllum is divided into two groups based on the length of dorsal brush row 3: the first group consists of species with bipolar (extends to or near to posterior body end) brush row 3, namely, two populations of T. apiculatum, T. costaricanum, T. hyalinum Foissner, 1983, T. pannonicum, T. clavatum Stokes, 1886, and T. parapiculatum sp. nov.; and the second group consists of all other species and populations either with dorsal brush row 3 similar or slightly longer than row 2 or with brush row 3 of unknown morphology but with special morphological characters such as the coloration, the body shape, or the nuclear apparatus (Foissner et al., 1995, 2002; Foissner, 2016).
Several populations were described as T. apiculatum, but they differed from each other by at least a single important character (Table 4), suggesting that they should be classified as distinct species. The Venezuelan neotype population of T. apiculatum (population 8 in Table 4) designated by Foissner et al. (2002) has a short dorsal brush row 3, a character that was underestimated by the authors at that time, and differs also from T. parapiculatum sp. nov. by the number of dikinetids in dorsal brush row 1 (10–24 vs. 7–11). Trachelophyllum parapiculatum sp. nov. could be confused with two different populations described as T. apiculatum with long brush row 3: (1) the population described by Foissner (1983) (population 4 in Table 4) differs from T. parapiculatum sp. nov. in the extrusomes length (13 vs. 15–24 μm) and the number of dikinetids in dorsal brush row 2 (~8 vs. 17–25); and (2) population II in Foissner (1984) (population 6 in Table 4), which is distinguishable from T. parapiculatum sp. nov. by the number of somatic ciliary rows (22–25 vs. 9–13) and the number of dikinetids in dorsal brush row 1 (16–26 vs. 7–11) (Table 4). Trachelophyllum parapiculatum sp. nov. is also morphologically very similar to T. costaricanum and T. pannonicum and can be separated from each of them mainly by the types and shapes of the extrusomes, i.e., T. parapiculatum sp. nov. has only one type of rod-shaped extrusomes, while T. costaricanum and T. pannonicum each has two types of acicular and rod-shaped extrusomes. T. hyalinum, which was studied only in vivo and after silver impregnation and without morphometric data, differs from T. parapiculatum sp. nov. in having longer extrusomes (~30 μm vs. 15–24 μm long) that do not extend into the oral bulge. T. clavatum is also characterized by long brush row 3 (i.e., extends to two-thirds of the body length) and differs from other congeners by the presence of a single macronuclear nodule (Stokes, 1888; Foissner, 1983; Foissner et al., 2002).
Trachelophyllum tachyblastum resembles T. parapiculatum sp. nov. in body shape and size but differs mainly in the length of dorsal brush row 3 (short vs. bipolar), the types and shapes of the extrusomes (two types of rod-shaped and oblong vs. one type of rod-shaped extrusomes), and the numbers of dikinetids in brush rows 1 and 2 (4–7 and 6–15 vs. 7–11 and 17–25, respectively). T. lineare, which was described as Lepidotrachelophyllum fornicis by Nicholls and Lynn (1984) and Lynn and Nicholls (1985) and as L. lineare by Foissner (1994) and Foissner et al. (1999), has a large body size [up to 600 μm according to Lepsi (1960) and 40–350 μm according to Nicholls and Lynn (1984)] and a higher number of somatic ciliary rows (Table 4). T. brachypharynx Levander, 1894, which was studied recently by Jang et al. (2015), is a large species (330–445 × 35–45 μm) with filiform extrusomes ~30 μm long and 20–25 somatic ciliary rows. However, the lepidosomes of T. brachypharynx were observed only in vivo and described as hat-shaped. Thus, the assignment to the genus Trachelophyllum is questionable, as noted by Jang et al. (2015).
Phylogenetic Analyses
The new phylogenetic tree shows that the family Trachelophyllidae is monophyletic. The trachelophyllid clade comprises only four sequences: M. terricola gen. nov., sp. nov., T. parapiculatum sp. nov., T. brachypharynx, and an unidentified Trachelophyllum sp. However, like the unidentified Trachelophyllum sp., the assignment of T. brachypharynx is doubtful because the shape of the lepidosomes was not studied using a scanning electron microscope. According to Jang et al. (2015), the lepidosomes of T. brachypharynx in vivo look very similar to type V lepidosomes with their hat-shaped structure as described by Foissner (2005) and, up to date, found only in the genus Sleighophrys. Furthermore, the new phylogenetic tree shows a close relationship between the families Trachelophyllidae and Enchelyodontidae (Foissner et al., 2002) based on morphological similarities as discussed by Jang et al. (2015). Clearly, the two families are underrepresented in the phylogenetic tree and obtaining more trachelophyllid and enchelyodontid sequences would be of great help, especially from properly identified species (Foissner et al., 2002; Foissner, 2005, 2016; Jang et al., 2015; Bourland, 2017).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/ [GenBank accession numbers: ON212658 and ON212659].
Author Contributions
AO and J-HJ designed the study and revised the manuscript. AO performed morphological experiments, molecular experiments, data analyses, and wrote the manuscript. JHM collected the samples and helped in the scanning electron microscopy. All authors read and approved the final version of the manuscript.
Funding
This study was supported by a grant from the Nakdonggang National Institute of Biological Resources (NNIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NNIBR202201105).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to the Center for Research Facilities at Gangneung-Wonju National University for their assistance in the analysis of the cell structure (CPD, SEM, etc.).
References
Bourland, W. (2017). How far do ciliate flagships sail a proposed gondawanaland endemic species at anchor in idaho soils. Protist. 168, 352–361. doi: 10.1016/j.protis.2017.04.002
Coats, D. W., and Clamp, J. C. (2009). “Ciliated protists (Ciliophora) of the Gulf of Mexico.” In: Gulf of Mexico: Origin, Waters, and Biota, Vol. 1. Biodiversity, Eds D. L. Felder and D. K. Camp (Texas AandM University Press), 57–79.
Czapik, A., and Jordan, A. (1976). Les observations sur les cilies d'une mare. Acta Protozool. 15, 277–288.
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 9:772. doi: 10.1038/nmeth.2109
Dumas, E. (1930): Les microzoaires ou infusoires proprement dits. Faune du centre. 2e fascicule. Moulins (“Les imprimeries réunies”].
Dumas, E. (1937): Les microzoaires ou infusoires proprement dits. Faune du centre. 3e fascicule. Moulins (“Les imprimeries réunies”].
Foissner, W. (1983). Taxonomische Studien über die Ciliaten des Großglocknergebietes (Hohe Tauern, Österreich) I. Familien Holophryidae, Prorodontidae, Plagiocampidae, Colepidae, Enchelyidae und Lacrymariidae nov. fam. Annln Naturh. Mus. Wien B. 84, 49–85.
Foissner, W. (1984). Infraciliatur, Silberliniensystem und Biometrie einiger neuer und wenig bekannter terrestrischer, limnischer und mariner Ciliaten (Protozoa: Ciliophora) aus den Klassen Kinetofragminophora, Colpodea und Polyhymenophora. Stapfia. 12, 1–165.
Foissner, W. (1994). “Spetazoon australiense nov. gen., nov. spec., ein neues Wimpertier (Protozoa, Ciliophora) von Australien.” In: Die Urtiere, eine verborgene Welt, Eds E. Aescht Kataloge des ÖÖ. Landesmuseums (N.F.), 267–278.
Foissner, W. (2005). Two new “flagship” ciliates (Protozoa, Ciliophora) from Venezuela: Sleighophrys pustulata and Luporinophrys micelae. Eur. J. Protistol. 41, 99–117. doi: 10.1016/j.ejop.2004.10.002
Foissner, W. (2014). An update of ‘basic light and scanning electronmicroscopic methods for taxonomic studies of ciliated protozoa'. Int. J. Syst. Evol. Microbiol. 64, 271–292. doi: 10.1099/ijs.0.057893-0
Foissner, W. (2016). Terrestrial and semiterrestrial ciliates (Protozoa, Ciliophora) from Venezuela and Galápagos. Denisia. 35, 1–912.
Foissner, W., Agatha, S., and Berger, H. (2002). Soil ciliates (Protozoa, Ciliophora) from Namibia (Southwest Africa), with emphasis on two contrasting environments, the Etosha region and the Namib Desert. Denisia. 5, 1–1459.
Foissner, W., Berger, H., Blatterer, H., and Kohmann, F. (1995). Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems – Band IV: Gymnostomatea, Loxodes, Suctoria. Informationsberichte des Bayer. Landesamtes für Wasser-wirtschaft. 1/95, 1–540.
Foissner, W., Berger, H., and Schaumburg, J. (1999). Identification and ecology of limnetic plankton ciliates. Munich: Landesamt für Wasserwirtschaft.
Gajewskaja, N. (1933). Zur Oekologie, Morphologie und Systematik der Infusorien des Baikalsees. Zoologica, Stuttg. 32, l−298.
Grolière, C.-A. (1977). Contribution a l'étude des ciliés des sphaignes: II. Dynamique des populations. Protistologica. 13, 335–352.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nuc. Acid Symp. Ser. 41, 95–98.
Huang, J. B., Zhang, T., Zhang, Q., Li, Y., Warren, A., Pan, H., et al. (2018). Further insights into the highly derived haptorids (Ciliophora, Litostomatea): Phylogeny based on multigene data. Zool. Scr. 47, 231–242. doi: 10.1111/zsc.12269
Jang, S. W., Omar, A., Nam, S. W., and Jung, J.-H. (2022). Morphology and Molecular Phylogeny of Fuscheriides baugilensis sp. nov. (Protozoa, Ciliophora, Haptorida) from South Korea. Diversity. 14, 70. doi: 10.3390/d14020070
Jang, S. W., Vdačný, P., Shazib, S. U. A., and Shin, M. K. (2015). Morphology, ciliary pattern and molecular phylogeny of Trachelophyllum brachypharynx Levander, 1894 (Litostomatea, Haptoria, Spathidiida). Acta Protozool. 54, 123–135. doi: 10.4467/16890027AP.15.010.2735
Jung, J.-H., Moon, J. H., Park, K. M., Kim, S., Dolan, J. R., and Yang, E. J. (2018). Novel insights into the genetic diversity of Parafavella based on mitochondrial CO1 sequences. Zool. Scr. 47, 741–755. doi: 10.1111/zsc.12312
Kahl, A. (1930). Urtiere oder Protozoa I: Wimpertiere oder Ciliata (Infusoria) 1. Allgemeiner Teil und Prostomata. Tierwelt Dtl. 18, 1–180.
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kent, W. S. (1881). A Manual of the Infusoria: Including a Description of All Known Flagellate, Ciliate, and Tentaculiferous Protozoa British and Foreign, and An Account of the Organization and Affinities of the Sponges. Vol. II. London: David Bogue.
Kim, J. H., and Jung, J.-H. (2017). Cytological staining of protozoa: A case study on the impregnation of hypotrichs (Ciliophora: Spirotrichea) using laboratory-synthesized protargol. Anim. Cells Syst. 21, 412–418. doi: 10.1080/19768354.2017.1376707
Lepsi, I. (1960). Noi specii de infuzori (Neue Infusorienarten). Comunle Acad. Rep. pop. rom. 10, 1095–1101.
Levander, K. M. (1894). Materialien zur Kenntnis der Wasserfauna in der Umgebung von Helsingfors, mit besonderer Berücksichtigung der Meeresfauna. I. Protozoa. Acta Soc. Fauna Flora fenn. 12, 1–115.
Lynn, D. H., and Nicholls, K. H. (1985). Cortical microtubular structures of the ciliate Lepidotrachelophyllum fornicis Nicholls and Lynn, 1984 and phylogeny of the litostomate ciliates. Can. J. Zool. 63, 1835–1845. doi: 10.1139/z85-274
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Moon, J. H., Kim, J. H., and Jung, J.-H. (2017). Taxonomical reinvestigation of the colepid species Pinacocoleps pulcher (Spiegel, 1926) Foissner et al., 2008 (Ciliophora: Prorodontida: Colepidae). Acta Protozool. 56, 161–169. doi: 10.4467/16890027AP.17.014.7495
Moon, J. H., Kim, J. H., Quintela-Alonso, P., and Jung, J.-H. (2020). Morphology, morphogenesis, and molecular phylogeny of Neobakuella aenigmatica n. sp. (Ciliophora, Spirotrichea, Bakuellidae). J. Eukaryot. Microbiol. 67, 54–65. doi: 10.1111/jeu.12753
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nicholls, K. H., and Lynn, D. H. (1984). Lepidotrachelophyllum fornicis, n. g., n. sp., a ciliate with an external layer of organic scales (Ciliophora, Litostomatea, Haptoria). J. Protozool. 31, 413–419. doi: 10.1111/j.1550-7408.1984.tb02988.x
Pan, X., Bourland, W., and Song, W. (2013). Protargol synthesis: an in-house protocol. J. Eukaryot. Microbiol. 60, 609–614. doi: 10.1111/jeu.12067
Park, M. H., Moon, J. H., Kim, K. N., and Jung, J.-H. (2017). Morphology,morphogenesis, and molecular phylogeny of Pleurotricha oligocirrata nov. spec. (Ciliophora: Spirotrichea: Stylonychinae). Eur. J. Protistol. 59, 114–123. doi: 10.1016/j.ejop.2017.04.005
Penard, E. (1922). Études sur les Infusoires d'Eau Douce. Genève: Georg and Cie, 1–331. doi: 10.5962/bhl.title.122543
Perty, M. (1852). Zur Kenntniss kleinster Lebensformen nach Bau, Funktionen, Systematik, mit Specialverzeichniss der in der Schweiz beobachteten. Bern: Jent and Reinert. p. 228.
Rambaut, A. (2006). FigTree. Institute of Evolutionary Biology. Univ. of Edinburgh. Available online at: http://tree.bio.ed.ac.uk/software/figtree
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Song, W. (1994). Faunistical studies on some soil ciliates from Qingdao – I. Kinetofragminophora, Oligohymenophora, Colpodea. J. Ocean Univ. Qingdao. 24, 15–23.
Sonnenberg, R., Nolte, A. W., and Tautz, D. (2007). An evaluation of LSU rDNA D1–D2 sequences for their use in species identification. Front. Zool. 4, 6. doi: 10.1186/1742-9994-4-6
Stokes, A. C. (1888). A preliminary contribution towards a history of the fresh-water infusoria of the United States. J. Trenton nat. Hist. Soc. 1, 71–344.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA 6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2575–2579. doi: 10.1093/molbev/mst197
Telesh, I., Postel, L., Heerkloss, R., Mironova, E., and Skarlato, S. (2009). Zooplankton of the open Baltic Sea: extended atlas. Meereswiss. Ber., Warnemünde. 76, 1–290. doi: 10.12754/msr-2009-0076
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tucolesco, J. (1962). Études protozoologiques sur les eaux Roumaines I. Espèces nouvelles d'infusoires de la mer Noire et des bassins salés paramarins. Arch. Protistenk. 106, 1–36.
Keywords: dorsal brush, Korea, lepidosomes, SSU rRNA, Trachelophyllidae
Citation: Omar A, Moon JH and Jung J-H (2022) Morphology and Molecular Phylogeny of Two New Trachelophyllid Ciliates, Monolamellophrya terricola gen. nov., sp. nov. and Trachelophyllum parapiculatum sp. nov. (Litostomatea, Haptoria), From South Korea. Front. Microbiol. 13:893886. doi: 10.3389/fmicb.2022.893886
Received: 11 March 2022; Accepted: 26 April 2022;
Published: 06 June 2022.
Edited by:
Zhenzhen Yi, South China Normal University, ChinaReviewed by:
Hongbo Pan, Shanghai Ocean University, ChinaShahed Uddin Ahmed Shazib, Smith College, United States
Copyright © 2022 Omar, Moon and Jung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jae-Ho Jung, amhqdW5nQGd3bnUuYWMua3I=
 Atef Omar
Atef Omar Ji Hye Moon
Ji Hye Moon Jae-Ho Jung
Jae-Ho Jung