
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 July 2022
Sec. Microbiotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.888964
This article is part of the Research Topic Exploring the Role, Function, and Behavior of the Anaerobic Microbiota: Species, Communities, Networks, Biotechnological Potential, and Biomedical Aspects View all 5 articles
This study aims to obtain anaerobic fungi from the rumen and fecal samples and investigates their potential for lignocellulosic bioconversion. Multiple anaerobic strains were isolated from rumen contents (CR1–CR21) and fecal samples (CF1–CF10) of Bactrian camel using the Hungate roll tube technique. After screening for fiber degradability, strains from rumen contents (Oontomyces sp. CR2) and feces (Piromyces sp. CF9) were compared with Pecoramyces sp. F1 (earlier isolated from goat rumen, having high CAZymes of GHs) for various fermentation and digestion parameters. The cultures were fermented with different substrates (reed, alfalfa stalk, Broussonetia papyrifera leaves, and Melilotus officinalis) at 39°C for 96 h. The Oontomyces sp. CR2 had the highest total gas and hydrogen production from most substrates in the in vitro rumen fermentation system and also had the highest digestion of dry matter, neutral detergent fiber, acid detergent fiber, and cellulose present in most substrates used. The isolated strains provided higher amounts of metabolites such as lactate, formate, acetate, and ethanol in the in vitro rumen fermentation system for use in various industrial applications. The results illustrated that anaerobic fungi isolated from Bactrian camel rumen contents (Oontomyces sp. CR2) have the highest lignocellulosic bioconversion potential, suggesting that the Bactrian camel rumen could be a good source for the isolation of anaerobic fungi for industrial applications.
Lignocellulosic biomass refers to residues from agricultural and forestry industries, which are plentiful renewable resources with an annual yield of about 10 billion tons (Nguyen et al., 2019). Recent research has shown that lignocellulosic biomass is a large reserve of carbon, and it could be used as a substrate for bioenergy production (Yamamoto et al., 2014; Nanda et al., 2015; Sawatdeenarunat et al., 2015). However, considerable amount of lignocellulosic biomass is simply discarded or burnt in the field, leading to resource wastage and environmental pollution (Liang et al., 2021). Therefore, it has become increasingly important and urgent to rationally utilize lignocellulosic biomass. The complex chemical composition and structure (which includes cellulose, hemicellulose, and lignin, interlinked with each other) of lignocellulosic biomass limit its utilization as animal feed or as a substrate for biofuel production. High energy and corrosive chemicals are needed to break lignocellulosic biomass (Zabed et al., 2016).
Recently, some methods have achieved substantial degradation of lignocellulose. These methods use physical and/or chemical processes. Protein engineering has also been used to improve the performance of the existing lignocellulose-degrading enzymes (Taherzadeh and Karimi, 2008; Singh et al., 2015; Shi et al., 2019). Although these pretreatment methods have been used to enhance the efficiency of hydrolysis of lignocellulosic biomass, their use inevitably brings about secondary pollution and increased cost (Abraham et al., 2020; Mankar et al., 2021). Currently, the bioconversion strategy is regarded as the most efficient and environmental-friendly approach for the industrial utilization of lignocellulosic biomass. This strategy utilizes carbohydrate-active enzymes secreted by microorganisms to break down stubborn structural polymers into easily metabolized monosaccharides that are subsequently converted into products (Balan, 2014). Research on improving the bioconversion efficiency of lignocellulose has received increased attention in recent years.
Anaerobic fungi, belonging to the phylum Neocallimastigomycota, are few in number in the rumen but they perform one of the main functions of fiber degradation in ruminants (Trinci et al., 1994; Hess et al., 2020). Anaerobic fungi secrete comprehensive fiber-degrading enzymes (Solomon et al., 2016). Previous studies have shown that anaerobic fungi have high lignocellulosic bioconversion potential (Saye et al., 2021; Kazemi Shariat Panahi et al., 2022; Li et al., 2022). During the anaerobic digestion, the main metabolites formed by anaerobic fungi are formate, acetate, ethanol, lactate, hydrogen, and carbon dioxide (Boxma et al., 2004; Wilken et al., 2021). High fiber degradability and organic acid production are the advantages of using anaerobic fungi as inoculants for the bio-fermentation of feeds. The anaerobic fungi led to the breakdown of plant fibers, and an acidic environment during fermentation favors the growth of other microorganisms (Sugiharto et al., 2016; He et al., 2020). In addition, hydrogen and ethanol, as the main metabolites of anaerobic fungi, can be used for the development of clean energy (Cantarel et al., 2009). Although anaerobic fungi have a low ethanol production capacity, multiple techniques for increasing ethanol production and using anaerobic fungi have been reported (Stevenson and Weimer, 2002; Ali et al., 2013; Kazemi Shariat Panahi et al., 2022). The potential use of anaerobic fungi for the production of bioethanol and biohydrogen is worth exploring. Apart from this, other metabolites such as acetate and formate formed during the fermentation are suitable substrates for additional downstream industrial processes (Saye et al., 2021). Therefore, harnessing the bioconversion potential of anaerobic fungi to process lignocellulosic biomass is of great value.
Advancements in sequencing technology and morphological techniques over the past decade have led to the description of a total of 20 genera of anaerobic fungi from a few sources (Joshi et al., 2018; Hanafy et al., 2020a,2021; Stabel et al., 2020; Li Y. Q. et al., 2021). Previous research has shown differences in the extent of lignocellulose degradation among anaerobic fungi of different genera (Gruninger et al., 2014; Hanafy et al., 2020b; Li Y. Q. et al., 2021). Although studies have shown that the fungal community composition is similar in different sections of the ruminants’ gut (Davies et al., 1993a; Rabee et al., 2019), there are huge differences in their functions, and also their community structure and functions differ between samples of rumen and feces (Agustina et al., 2022). Therefore, we hypothesized that there were also significant differences between anaerobic fungi isolated from different rumen contents and feces.
In this study, a number of strains were isolated from rumen contents and fecal samples of Bactrian camel and subsequently compared with Pecoramyces sp. F1 (isolated from goat rumen liquid, having high glycoside hydrolases) (Jin et al., 2011) for lignocellulosic bioconversion using different substrates. The substrates included reed, alfalfa stalk, Broussonetia papyrifera leaves, and Melilotus officinalis, which have different lignocellulose contents. We expected differences in hydrogen production, lignocellulose degradation efficiency, and fermentation metabolite production to assess our hypothesis.
The complex medium used for isolation was according to the method of Cheng et al. (2009), which contained 1% wt/vol rice straw (hereafter listed as 1% rice straw) and antibiotics (1,600 IU/ml penicillin; 2,000 IU/ml streptomycin). The rice straw used was rinsed three times with distilled water, dried at 65°C in a blast drying oven (DHG-9245A, Shanghai Youyi Instrument Co., Ltd., Shanghai, China), pulverized, passed through a 3 mm sieve, sealed, and then stored at room temperature. Every 1,000 ml of the complex medium contained 150 ml of buffer solution A, 150 ml of buffer solution B, 150 ml of buffer solution C, 400 ml of basal medium (which contains 6 g NaHCO3, 10 g tryptone, and 2.5 g yeast extract), 150 ml of cell-free rumen liquid (the fresh rumen fluid centrifuged at 12,000 g for 5 min and the supernatant collected and stored at −20°C till use), 1 g of L-cysteine hydrochloride, and 1 ml of 0.1% (wt/vol) resazurin. Buffer solution A contained 0.3 g of K2HPO4 per 100 ml; buffer solution B contained 0.3 g of KH2PO4, 0.6 g of (NH4)2SO4, and 0.06 g of MgSO4⋅7H2O; and buffer solution C contained 0.6 g of NaCl and 0.06 g of CaCl2⋅2H2O per 100 ml. The complex medium (90 ml portions) was dispensed into 180 ml serum bottles under continuous flow of CO2. Thereafter, the serum bottles were sterilized by autoclaving at 121°C for 20 min.
Fresh rumen contents (5 g) and fresh feces (5 g) were collected from a Bactrian camel in Jimunai County (latitude 47°00′–47°59′ N and longitude 85°33′–87°09′ E), Xinjiang, China. After preheating 90 ml of the complex medium to be inoculated at 39°C for 2 h in advance, remove the aluminum cap and rubber stopper, and quickly transfer the collected samples to the complex medium. The cultures were incubated at 39°C in an incubator (HGPN-11-163, HENGZI, Shanghai, China) and transferred every 3 days. After three consecutive batch culture series, fungal growth (marked by rice straw floating in the complex medium) and methane production [detected by gas chromatography (GC)] were checked to ensure that mixed cultures of anaerobic fungi and methanogens were obtained (Cheng et al., 2009). An aliquot of 1 ml culture was inoculated into 9 ml anaerobic solution (complex medium without rumen liquid and basal medium) and diluted two times, and then 0.5 ml of the diluted culture was inoculated into 10 ml of the cellobiose medium supplemented with 2% agarose in the Hungate roll tube (Jin et al., 2011). After single fungal colonies were formed (in 3 days), they were picked up and transferred into the fresh complex medium with 1% rice straw (Ozkose et al., 2001). This process was repeated up to three times, until the fungal colonies appeared uniform under a microscope. After isolation, the cultures were maintained at 39°C in the complex medium with 1% rice straw and transferred every 3 days.
For the genetic characterization, biomass was collected from 10 ml of 3-day cultures after centrifugation at 12,000 g for 10 min. DNA was extracted from the precipitate using DNeasy PowerPlant Pro Kit (Qiagen, Germantown, Maryland) according to the manufacturer’s instructions. The region encompassing ITS1, 5.8S, ITS2, and D1/D2 of the LSU rRNA was targeted for amplification using the primers ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and NL4 (5′-TCAACATCCTAAGCGTAGGTA-3′) (Dagar et al., 2011). The PCR protocol consisted of an initial denaturation for 5 min at 95°C followed by 40 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, elongation at 72°C for 2 min, and a final extension of 72°C for 20 min (Dagar et al., 2011). The PCR products were Sanger sequenced at the Tsingke Biotechnology Co., Ltd., Nanjing, China. All raw sequences were deposited in the NCBI GenBank database under the accession number ON514405-ON514425, ON51427-ON514437.
The raw sequences were compared with the sequences available in the BLAST GenBank.1 The default settings for the neighbor-joining method in MEGA X were applied to construct the phylogenetic tree (Kumar et al., 2018).
A 10 ml portion of 3-day-old cultures, isolated from the camel rumen contents or feces, was inoculated into 90 ml fresh complex medium and incubated at 39°C for 96 h without shaking, with 1% of the rice straw (milled to pass through a 3 mm sieve) as a substrate. Each group had 4 replicates and a blank control to correct for the gas production. At 12, 24, 32, 40, 48, 60, 72, and 96 h of fermentation, the head-space gas was measured using the pressure transducer technique (PTT) and then vented to return the pressure back to ambient condition (Theodorou et al., 1994). At 96 h, pH was measured immediately after removing crimp seals and stoppers. The substrate after the fermentation was washed with deionized water and dried at 105°C in a blast drying oven (DHG-9245A, Shanghai Youyi Instrument Co., Ltd., Shanghai, China) to calculate dry matter digestibility (DMD). Anaerobic fungi strains that had the highest DMD and total gas production were taken to the next experiment. Two fungi (one each from the rumen contents and feces) were considered as having efficient dry matter degrading ability. Taxonomic features of these two fungi (in the medium with 1% rice straw) were examined by microscopy (DMi8, Leica, Germany) (Li Y. Q. et al., 2021).
Two strains isolated from the Bactrian camel rumen contents or feces, which had the highest DMD and Pecoramyces sp. F1. (Li et al., 2017), were inoculated separately into 90 ml media with several 1% wt/vol substrates (e.g., reed, alfalfa stalk, B. papyrifera leaves, and M. officinalis). The substrates were milled to pass through a 3 mm sieve before incubation. The experiment was set up with 12 different treatment groups (3 anaerobic fungi × 4 substrates), 4 replicates in each group, and a blank control to correct for gas production. The incubation was done at 39°C for 96 h without shaking. At 12, 24, 32, 40, 48, 60, 72, and 96 h of the fermentation, the head-space gas was measured using the PTT and then vented to return the pressure back to ambient condition. An airbag was connected to collect the gas for the determination of composition (Theodorou et al., 1994). Aliquots of 4 ml supernatant of culture were collected and stored at −20°C for the analysis of ethanol, formate, acetate, and lactate. In addition to this, for the analysis of two main fiber-degrading enzyme activities, aliquots of 2 ml supernatant of the culture were collected and stored in liquid nitrogen. The remaining fermentation medium was centrifuged at 12,000 g for 20 min to collect the residue, which was washed with deionized water and dried at 105°C in a blast drying oven to calculate DMD. Thereafter, it was used for the determination of the neutral detergent fiber (NDF), acid detergent fiber (ADF), neutral detergent solutes (NDS), cellulose, hemicellulose, and lignin, according to the methods reported in Zhu et al. (2001).
The relative abundance of hydrogen in the head-space gas in the bottle was measured by a GC system (Agilent 7890B, Agilent, California, United States). The gas sample was injected into a GC, and the gases were separated on packed GC columns (Fused Silica Capillary Column, Supelco, United States) at 80°C column temperature, 200°C injection temperature, 200°C TCD temperature, and air pressure and carrier gas (nitrogen) pressure of 0.05 MPa. The volume of hydrogen was calculated according to the method of Li et al. (2020).
The pH was measured at the end of fermentation immediately after removing crimp seals and stoppers from the serum vials (Seven2GO S2, METTLER TOLEDO, Switzerland). For the analyses of ethanol and acetate, 1 ml of supernatant was added to 1.5 ml centrifuge tubes with added 0.2 ml deproteinized metaphosphate solution containing crotonic acid. After centrifugation at 12,000 g for 10 min, the supernatant was filtered through a filter membrane (size 0.22 μm, SCAA-102; ANPEL, Shanghai, China); 1 μl of the filtered liquid was analyzed by GC (Agilent 7890B, Agilent, California, United States) at 40°C column temperature, 220°C injection temperature, and 230°C TCD temperature. The pressures of air, N2 carrier gas, and H2 were 0.05 MPa, according to the method of Li et al. (2016). Formate was measured by the Formate Assay Kit (K-FORM, Megazyme, Ireland), and lactate was measured with the Lactate Assay Kit (A019-2, Nanjing Jiancheng Biotechnology Institute, China) according to the manufacturer’s instructions (Zhu et al., 2001).
The activities of carboxymethyl cellulase (CMCase), a cellulose-degrading enzyme, and of xylanase, a hemicellulose-degrading enzyme, were measured according to the method of Li et al. (2020). One unit of CMCase and xylanase activity was defined as 1 μmol of glucose released per ml of supernatant per minute (U/ml/min) and as 1 μmol of xylose released per ml of supernatant per minute (U/ml/min), respectively.
A fiber analyzer (Ankom A200i; Ankom Technology, New York, United States) and a Muffle furnace (SX2-4-10N, Yiheng, Shanghai, China) were used for the determination of NDF, ADF, and lignin contents (Vansoest et al., 1991). Briefly, samples were treated in the neutral detergent solution in the fiber analyzer; the dissolved part was NDS, and the remaining part was NDF. The NDF was further treated using an acid detergent solution to dissolve hemicellulose and to obtain ADF. After treatment with 72% sulfuric acid, the ADF was digested to dissolve cellulose, and the residue, a mixture of lignin and silicate, was obtained. The residue was washed to remove lignin and to obtain silicate. The method of Niu et al. (2018) was used to calculate the contents of cell soluble or the soluble solutes in the neutral detergent solution, cellulose, and hemicellulose.
All experimental data were entered into Excel 2019 (Microsoft, Redmond, WA, United States) and preliminary processed, and then, Duncan’s analysis of one-way AVONA was used to calculate data at 95% level significance by using SPSS 26.0 (IBM SPSS Statistics, version 26.0; IBM Corp, Armonk, NY, United States). Data were presented as mean ± standard error of the mean.
A total of 21 and 10 cultures were isolated from the Bactrian camel rumen contents (CR1–CR21) and feces (CF1–CF10), respectively. After PCR amplification, we obtained 31 PCR products and subsequently obtained the raw sequence. After comparing with sequences available, 31 isolates were collected and identified as described in Supplementary Table 1. The results of ITS sequencing showed that the rumen-isolated anaerobic fungal strains were Oontomyces sp., and the fecal-isolated anaerobic fungal strains were Piromyces sp. The phylogenetic tree showed that the similarity between the rumen-isolated anaerobic fungi Oontomyces sp. CR1–CR22 and Oontomyces anksri strains SSD-CIB1 belonged to the same clade, and the similarity between fecal-isolated anaerobic fungi Piromyces sp. CF1–CF11 and Piromyces sp. Jen1 belonged to the same clade (Figure 1).
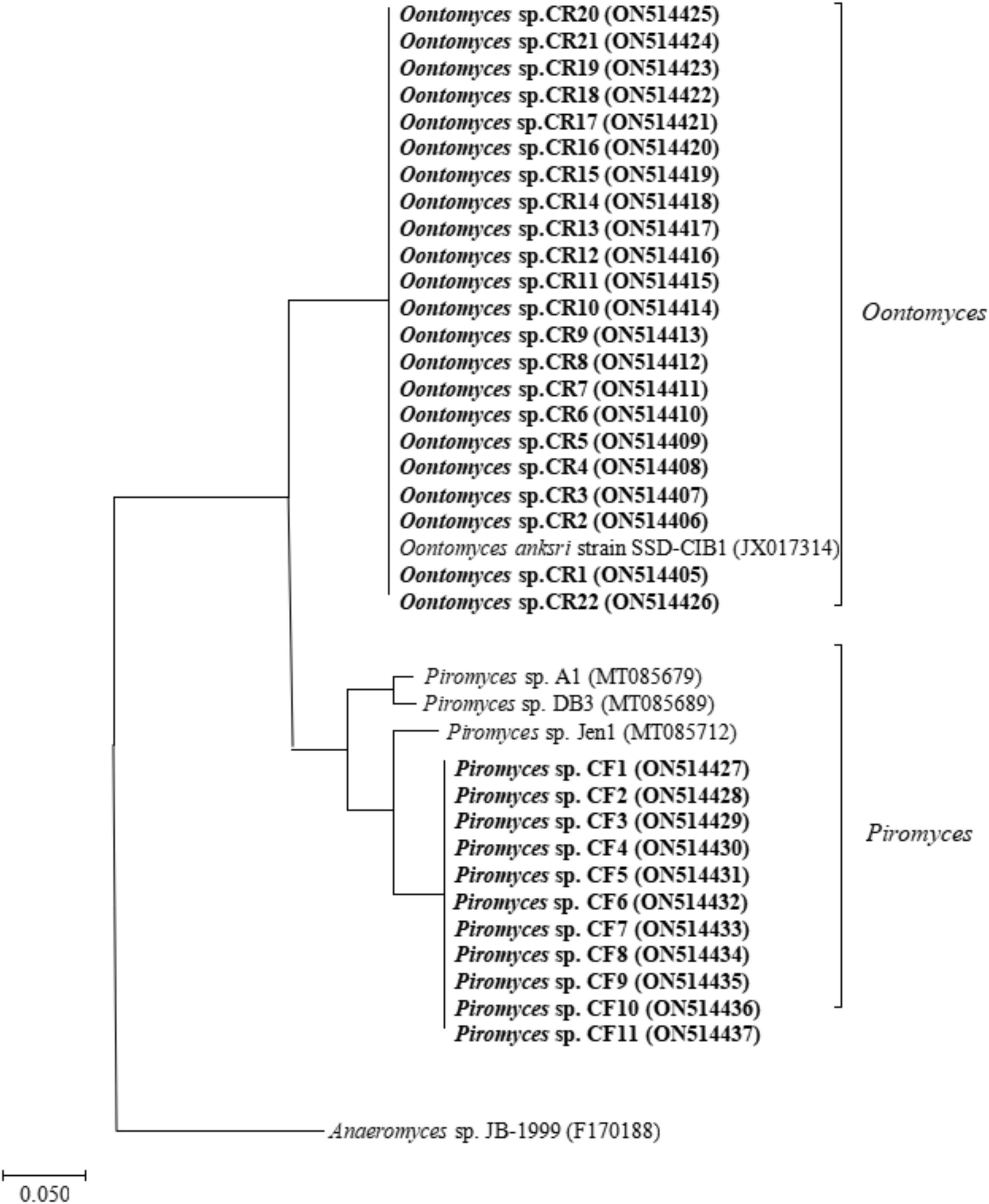
Figure 1. The phylogenetic tree of ITS1 sequences of anaerobic fungal strains isolated from the rumen and feces of Junggar Bactrian camels.
After 96 h of fermentation, the total gas production, DMD, and pH are presented in Supplementary Tables 2, 3. Anaerobic fungus Oontomyces sp. CR2 had the highest total gas production, DMD, and gas production per DM among CR1–CR21, with relatively lower pH than the other isolates from the Bactrian camel rumen contents (P < 0.05). For the isolates from fecal samples, the anaerobic fungus Piromyces sp. CF9 had the highest total gas production, DM loss, and gas production per DM among CF1–CF10, with relatively lower pH than the others from the Bactrian camel feces (P < 0.05) (Supplementary Table 3). Thus, Oontomyces sp. CR2 and Piromyces sp. CF9 were considered as the anaerobic fungi with efficient dry matter degradation ability and were used in further experiments.
After the selection of efficient fiber degrading fungi, Oontomyces sp. CR2 and Piromyces sp. CF9, they were identified morphologically. The taxonomic features are shown in Figure 2. Morphological characteristics showed that these fungi with monocentric thallus have spherical or oval sporangium with filamentous rhizoids. Zoospores are uniflagellate and spherical.
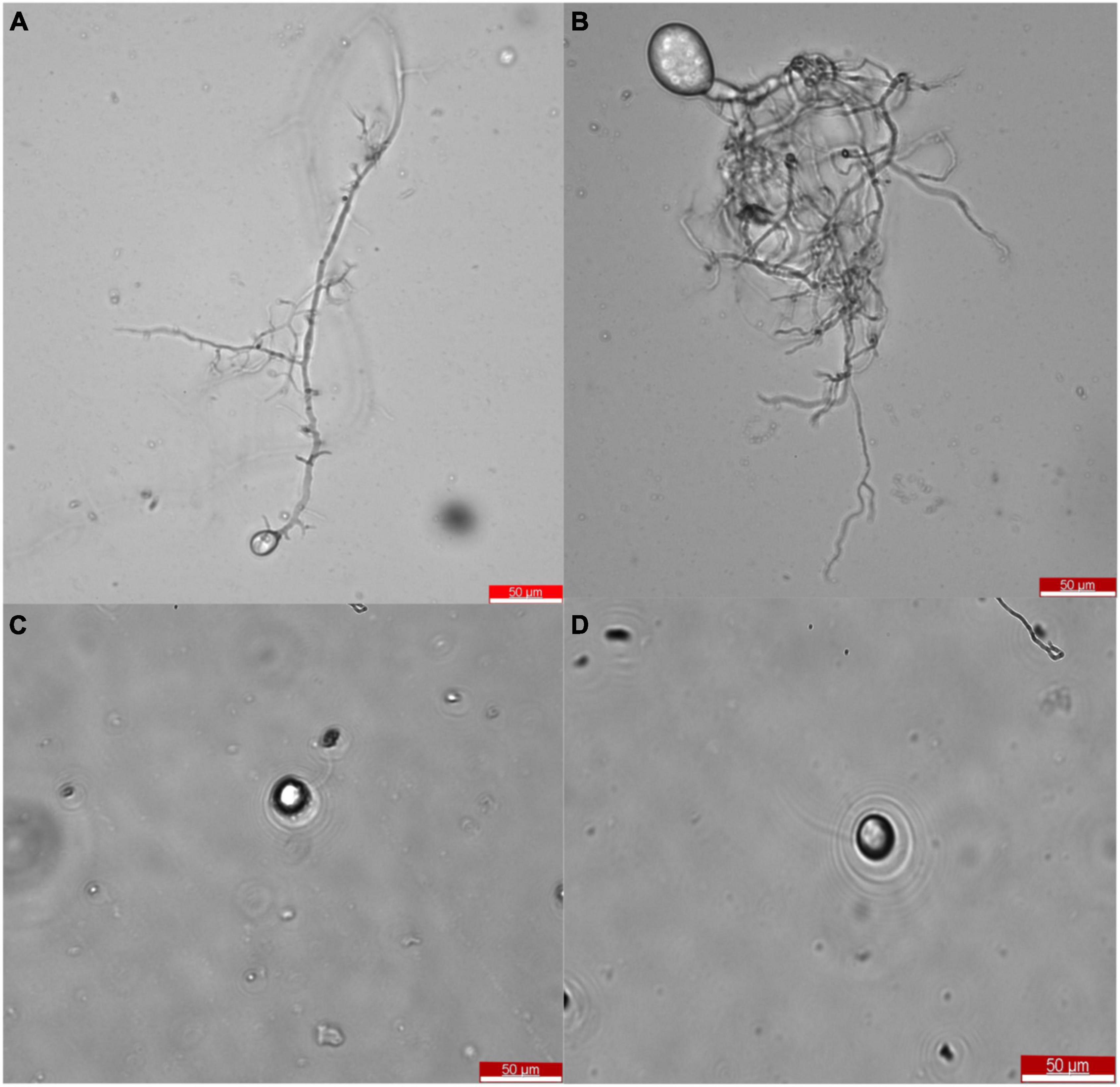
Figure 2. Morphology of Oontomyces sp. CR2 (A,C) and Piromyces sp. CF9 (B,D). Scale bar indicates 50 μm (A,B).
Figure 3A shows the cumulative gas production. Before 48 h of fermentation, the highest cumulative gas production was with B. papyrifera leaves using Oontomyces sp. CR2 as the inoculum. But after 48 h to the end of the fermentation, the highest cumulative gas production was with reed as the substrate. At the end of fermentation, Oontomyces sp. CR2 had the highest total hydrogen production (Figure 3D) and gas production with reed, B. papyrifera leaves, and alfalfa stalk (P < 0.05) as substrates; however, Pecoramyces sp. F1 produced the highest gas with M. officinalis (P < 0.05) (Figure 3C).
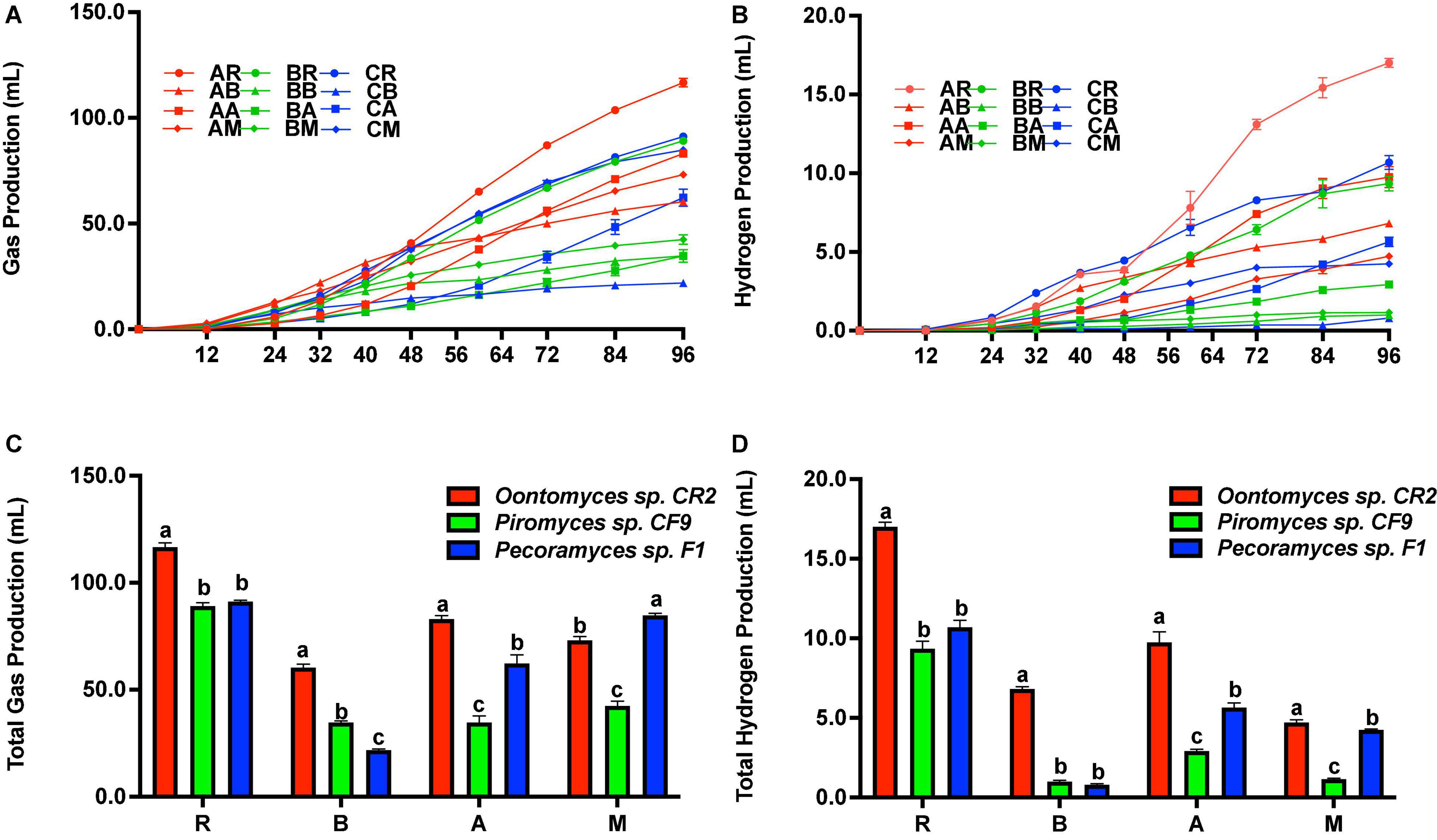
Figure 3. Cumulative gas and H2 production (A,B), and total gas and H2 production (C,D) after 96 h fermentation with Oontomyces sp. CR2, Piromyces sp. CF9, and Pecoramyces sp. F1 using different substrates (R, Reed; B, Broussonetia papyrifera leaves; A, Alfalfa stalk; M, Melilotus officinalis). The error bars represent the standard error of the mean (n = 4), and means with different letters in the same column are different at P < 0.05.
The cumulative hydrogen production is shown in Figure 3B. Before 50 h of fermentation, the highest cumulative hydrogen production was with reed as a substrate on using Pecoramyces sp. F1 as the inoculum. From 50 h onward, the highest cumulative hydrogen production was with reed on using Oontomyces sp. CR2 as the inoculum. At the end of the fermentation, Oontomyces sp. CR2 produced the highest total hydrogen in every substrate evaluated (P < 0.05) (Figure 2D).
Different substrates have different chemical compositions (Table 1). Reed has the lowest NDS content (P < 0.05) and high lignin content. On the contrary, B. papyrifera leaves have the highest NDS content (P < 0.05) and the lowest NDF, ADF, cellulose, and lignin contents (P < 0.05). There is not much difference in composition between alfalfa stalk and M. officinalis.
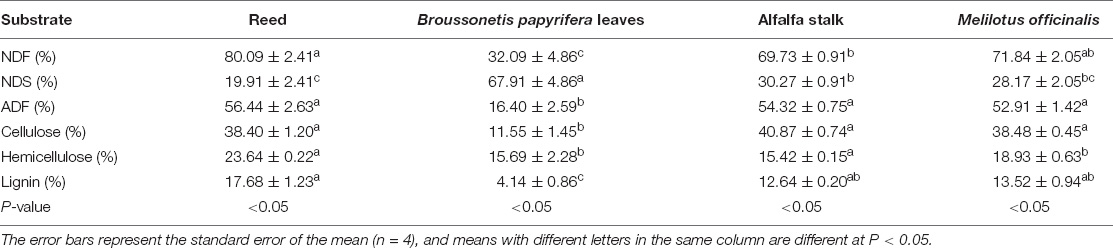
Table 1. Chemical composition of reed, alfalfa stalk, Broussonetia papyrifera leaves, and Melilotus officinalis.
The extent of degradation of different substrates after the fermentation is shown in Figure 4. By comparing the degradation characteristics of different anaerobic fungi on lignocellulose substrates, high-degrading anaerobic fungi were screened, thus laying a foundation for further exploration of their application potential (Dagar et al., 2015a,2018). The fermentation using different fungi and substrates shows a certain pattern in this research. The highest DMD for all the substrates was by Oontomyces sp. CR2, while this fungus had the highest NDFD for all the substrates except B. papyrifera leaves (P < 0.05). For all the substrates, the highest ADFD, cellulose digestion, and hemicellulose digestion were using Oontomyces sp. CR2 (P < 0.05). With Oontomyces sp. CR2, the highest lignin digestion was for all the substrates except alfalfa stalk.
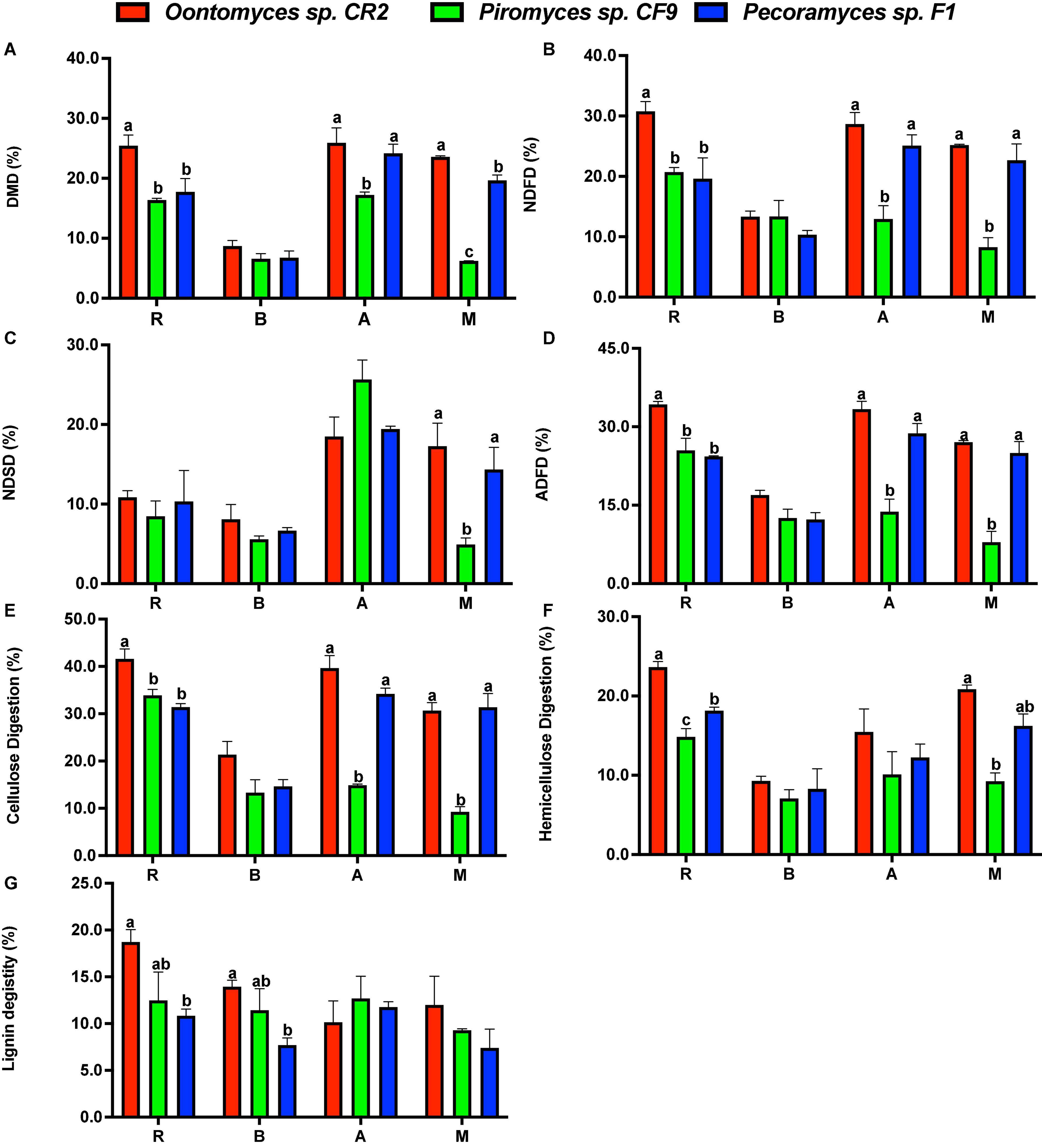
Figure 4. Degradation of reed, alfalfa stalk, Broussonetia papyrifera leaves, and Melilotus officinalis after fermentation. The figure includes DMD (A), NDF digestion (B), NDS digestion (C), ADF digestion (D), cellulose digestion (E), hemicellulose digestion (F) and lignin digestion (G) (R, Reed; B, Broussonetia papyrifera leaves; A, Alfalfa stalk; M, Melilotus officinalis). The error bars represent the standard error of the mean (n = 4), and means with different letters in the same column are different at P < 0.05.
Using the Pecoramyces sp. F1 culture, the pH value drop was lower on using reed and B. papyrifera leaves as substrates than using Oontomyces sp. CR2 or Piromyces sp. CF9, while using alfalfa and M. officinalis as substrates, Piromyces sp. CF9 had the least pH value drop, followed by Oontomyces sp. CR2 and Pecoramyces sp. F1 (Figure 5).
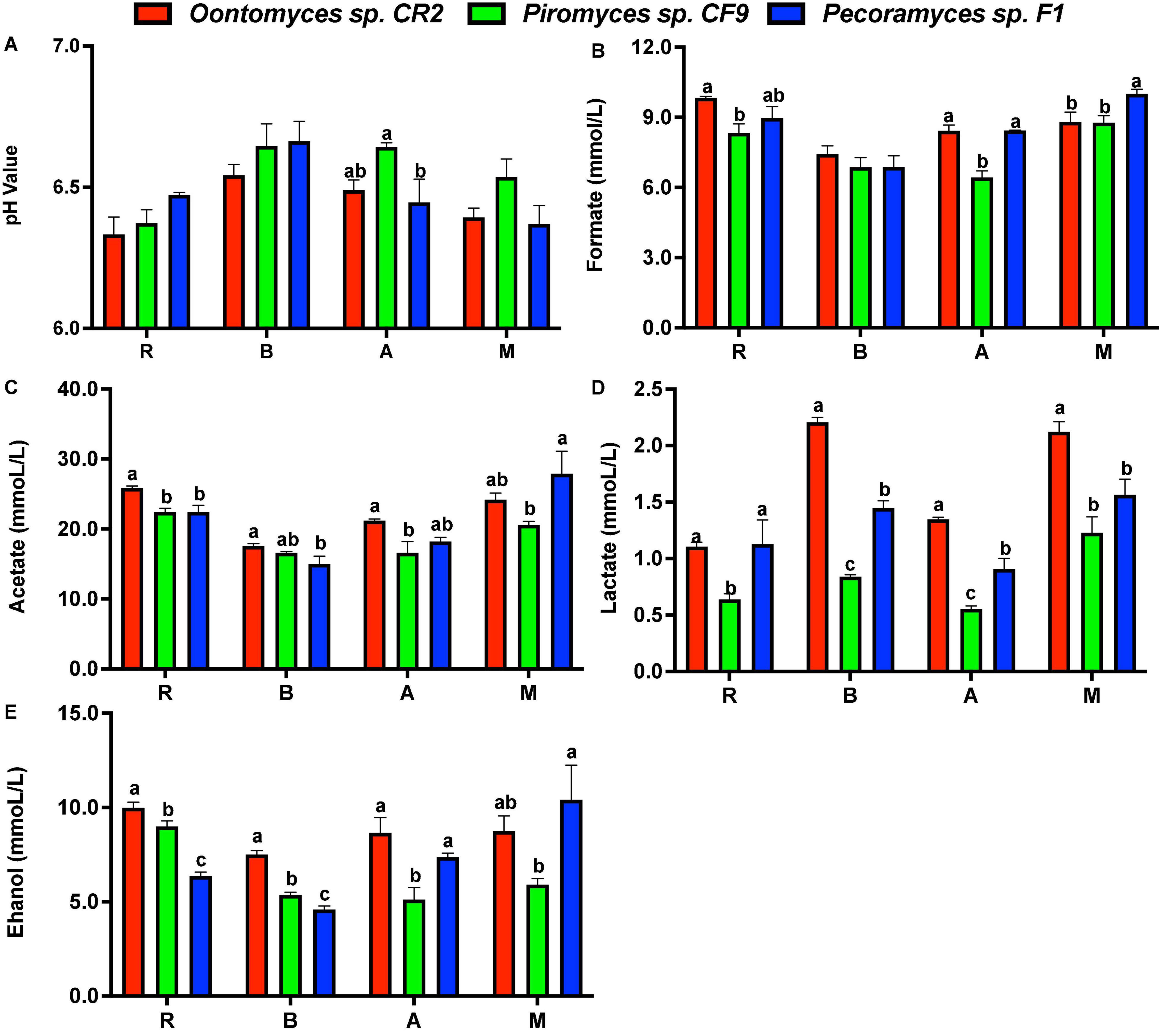
Figure 5. The pH value (A) and concentrations of formate (B), acetate (C), lactate (D), and ethanol (E) in the supernatant of different fermentations (R, Reed; B, Broussonetia papyrifera leaves; A, Alfalfa stalk; M, Melilotus officinalis). The error bars represent the standard error of the mean (n = 4), and means with different letters in the same column are different at P < 0.05.
Acetate was the main metabolite in the supernatant, followed by formate, ethanol, and lactate. Especially, Oontomyces sp. CR2 had the highest values of almost all metabolites. For example, the lactate content was highest except for reed (P < 0.05), and acetate and ethanol contents were highest except for M. officinalis (P < 0.05), while Pecoramyces sp. F1 had the highest formate content for M. officinalis (P < 0.05) (Figure 5).
The results of fiber-degrading enzyme activities are shown in Supplementary Figure 2. The results showed that Pecoramyces sp. F1, Oontomyces sp. CR2, and Piromyces sp. CF9 showed differences in enzyme activity. Former studies showed that different genera of anaerobic fungi exhibited significantly different enzyme activities, which were the same as our results (Dagar et al., 2018). For CMCase activity, Pecoramyces sp. F1 had higher activity than others for reed, alfalfa stalk, and M. officinalis, but Oontomyces sp. CR2 had the highest CMCase activity for B. papyrifera leaves (P < 0.05). For xylanase activity, Pecoramyces sp. F1 had the highest activity in every substrate.
The numbers of strains isolated from rumen were much higher than feces. Anaerobic fungi exist in all parts of the digestive tract, but due to limitations in accessing locations for sampling, anaerobic fungi have been isolated from rumen and feces in most cases (Voigt et al., 2021). The unique anaerobic environment of the rumen and suitable pH promote the growth of anaerobic fungi and make them degrade the feed that stays in the rumen for a long duration (Gruninger et al., 2014; Li M. M. et al., 2021). Previous studies have reported that almost 90% of the fungal population inhabited the rumen, illustrating that the total population of anaerobic fungi in the rumen is considerably larger than that in the omasum and hindgut organs (Davies et al., 1993b; Mao et al., 2016; Edwards et al., 2017). These results demonstrate that anaerobic fungi are isolated easily and in more numbers from the rumen, due to their abundance in this part of the digestive tract.
The isolated anaerobic fungus from the rumen contents, Oontomyces sp. CR2 had the highest total hydrogen production in every substrate (P < 0.05). Due to the absence of mitochondria in anaerobic fungi, the hydrogenosome is used to produce ATP (Borneman et al., 1989; Baykara, 2018). The hydrogenosomes produce hydrogen by using protons as electron acceptors during mixed-acid fermentation of monomeric sugars (predominantly glucose and xylose) derived from cellulose and hemicellulose (Saye et al., 2021). In the rumen environment, anaerobic fungi come into contact with a large number of substrates that have high cellulose and hemicellulose contents, which act as substrates for the production of hydrogen and an array of metabolites. The rumen is a relatively closed chamber having vast kinds of microorganisms. Due to the presence of methanogens, the accumulation of hydrogen is limited because it gets converted to methane (Hungate et al., 1970). Mizrahi et al. (2021) demonstrated that methanogenesis is likely to facilitate the activity of hydrogenase and the generation of hydrogen and ATP by anaerobic fungi (Hackstein et al., 2019). Anaerobic fungi in the rumen are exposed to higher concentrations of methane than anaerobic fungi in feces, which also explains higher hydrogen production by isolates from rumen contents. The rumen environment promotes the production of metabolites of hydrogenosome in anaerobic fungi, and this feature is preserved even in the anaerobic fungi isolated from the in vitro rumen fermentation system (Ma et al., 2022). In our study, although fungi from different sources share the same fermentation conditions (temperature and substrate types) in the in vitro system, strains isolated from rumen contents had higher hydrogen production than those from feces. These results further support the advantages of hydrogen metabolism in anaerobic fungi isolated from rumen contents. Also, feces get exposed to oxygen, and this could be the reason for lower abundance and activity of fungi isolated from feces.
Oontomyces sp. CR2 had the highest digestibility for most components of different substrates. Recently, Hanafy et al. (2020b) proposed the existence of a strong anaerobic fungi genus–host preference, which for some genera are encountered only in a specific animal. The anaerobic fungus, Oontomyces anksri gen.nov., sp. nov., was first isolated from the forestomach of the Indian camel (Camelus dromedarius) (Dagar et al., 2015b). Camel’s unique physical characteristics make it specially adapted to arid environments, and it has developed physiological mechanisms to face heat stress, dehydration, and shortage of nutrients (Guerouali and Wardeh, 1998). Meanwhile, the camel has a distinctive gastrointestinal morphology and is often described as a pseudoruminant. Its highly enlarged foregut comprises three distinct regions (Wilson, 1989). Due to the unique digestive system, the retention time of feed particles in camel is higher as compared with that in sheep and cattle, which increases the exposure of plant biomass to symbiotic microorganisms and helps in the efficient digestion of low-quality fibrous diets (Samsudin et al., 2011). The highest digestibility of Oontomyces sp. CR2, among the three anaerobic fungi investigated in our study, could be attributed to the unique digestive system and its unique environment. When anaerobic fungi with high capability to digest are used as an inoculant, the composition of the feed changes dramatically during fermentation. The plant cell wall of roughages gets degraded (de Vries and Visser, 2001), the contents of anti-nutritional factors are decreased (Abd El-Hack et al., 2018), and the palatability is improved (Yang et al., 2001; Oladosu et al., 2016). Our results showed that the anaerobic fungus isolated from the Bactrian camel rumen contents is a promising candidate for use as a feed inoculant in a bio-fermenter.
In addition to gas metabolites (hydrogen and carbon dioxide), anaerobic fungi also secrete a range of organic acids in their fermentation pathways (Breton et al., 1989; Boxma et al., 2004). These compounds have the potential for use as valuable substrates in various industrial processes. These substrates could have higher economic value than biogas as the final product (Silva et al., 2021). In our study, the fermentation products mainly included formate, acetate, lactate, and ethanol, which are consistent with the previous reports, and the different accumulation of metabolites can result in a variety of pH values (Li Y. Q. et al., 2021).
Moreover, Oontomyces sp. CR2, an anaerobic fungus isolated from the Bactrian camel rumen contents, showed significant differences in the production of a number of metabolites. Earlier research has shown that when the same anaerobic fungus is used to ferment different substrates, the types of metabolites do not change, but their proportions and contents change (Borneman et al., 1989). In the pathway of glucose metabolism, anaerobic fungi convert glucose to pyruvate in the cytosol or to malate and further to pyruvate in the hydrogenosome (Akhmanova et al., 1999; Li et al., 2016). After entering the hydrogenosome, malate is converted to CO2 and H2 or to pyruvate and further to format and acetate (Akhmanova et al., 1999). The higher levels of metabolite produced by Oontomyces sp. CR2 (isolated from the Bactrian camel rumen contents) may be attributed to two reasons. First, although different isolated anaerobic fungi fermented all the substrates in our study, Oontomyces sp. CR2 led to the degradation of higher proportion of the substrate to monosaccharides, which served as available substrates to facilitate the production of higher levels of metabolites. Second, higher levels of metabolite production created a lower pH acidic environment, which is more suitable for the growth of anaerobic fungi. This enhanced the metabolic capacity of cytosol and hydrogenosome (Saye et al., 2021).
Based on the observed high dry matter digestion and production of hydrogen and other fermentation metabolites, the anaerobic fungus (Oontomyces sp. CR2) isolated from the Bactrian camel rumen contents has the highest lignocellulosic bio-convention potential. The number of anaerobic fungi isolated from rumen contents was higher than that from feces, and the fungi isolated from the rumen contents had higher dry matter digestion and higher potential for various industrial applications. These results further highlight the advantages the rumen contents offer in the isolation of anaerobic fungi. The use of anaerobic fungi, isolated from the rumen, as an inoculum for various bio-conversion applications could be a good strategy for the future.
We used a single-substrate approach for the isolation of anaerobic fungi, which might have led to the isolation of fewer genera of anaerobic fungi. It would be interesting to use multi-substrates for the isolation. This might increase the number of genera of isolated anaerobic fungi having promising traits. Studies on these lines are suggested.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YX, YL, and RS conducted the experiment and analyzed the data. YX, XS, and ZS conceived and designed the work. YX, FL, XL, YC, and WZ wrote and revised the article. All authors contributed to the article and approved the submitted version.
This study was supported by the National Key R&D Program (2021YFD1300302), the Natural Science Foundation of China (32061143034 and 32161143028), and the Fundamental Research Funds for the Central Universities (KYYJ202003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.888964/full#supplementary-material
Abd El-Hack, M. E., Samak, D. H., Noreldin, A. E., Arif, M., Yaqoob, H. S., and Swelum, A. A. (2018). Towards saving freshwater: halophytes as unconventional feedstuffs in livestock feed: a review. Environ. Sci. Pollut. Res. 25, 14397–14406. doi: 10.1007/s11356-018-2052-9
Abraham, A., Mathew, A. K., Park, H., Choi, O., Sindhu, R., Parameswaran, B., et al. (2020). Pretreatment strategies for enhanced biogas production from lignocellulosic biomass. Bioresou. Technol. 301:122725. doi: 10.1016/j.biortech.2019.122725
Agustina, S., Wiryawan, K. G., Suharti, S., and Meryandini, A. (2022). The enrichment process and morphological identification of anaerobic fungi isolated from buffalo rumen. Biodiversitas J. Biol. Diversity 23, 469–477.
Akhmanova, A., Voncken, F. G., Hosea, K. M., Harhangi, H., Keltjens, J. T., Op Den Camp, H. J., et al. (1999). A hydrogenosome with pyruvate formate-lyase: anaerobic chytrid fungi use an alternative route for pyruvate catabolism. Mole. Microbiol. 32, 1103–1114. doi: 10.1046/j.1365-2958.1999.01434.x
Ali, S. S., Nugent, B., Mullins, E., and Doohan, F. M. (2013). Insights from the fungus fusarium oxysporum point to high affinity glucose transporters as targets for enhancing ethanol production from lignocellulose. Plos One 8:e54701. doi: 10.1371/journal.pone.0054701
Balan, V. (2014). Current challenges in commercially producing biofuels from lignocellulosic biomass. ISRN Biotechnol. 2014:463074. doi: 10.1155/2014/463074
Baykara, S. Z. (2018). Hydrogen: a brief overview on its sources, production and environmental impact. Int. J. Hydrogen Energy 43, 10605–10614.
Borneman, W. S., Akin, D. E., and Ljungdahl, L. G. (1989). Fermentation products and plant-cell wall-degrading enzymes produced by monocentric and polycentric anaerobic ruminal fungi. Appl. Environ. Microbiol. 55, 1066–1073. doi: 10.1128/Aem.55.5.1066-1073.1989
Boxma, B., Voncken, F., Jannink, S., van Alen, T., Akhmanova, A., van Weelden, S. W. H., et al. (2004). The anaerobic chytridiomycete fungus Piromyces sp E2 produces ethanol via pyruvate : formate lyase and an alcohol dehydrogenase E. Mole. Microbiol. 51, 1389–1399. doi: 10.1046/j.1365-2958.2003.03912.x
Breton, A., Bernalier, A., Bonnemoy, F., Fonty, G., Gaillard, B., and Gouet, P. (1989). Morphological and metabolic characterization of a new species of strictly anaerobic rumen fungus – Neocallimastix-joyonii. FEMS Microbiol. Lett. 58, 309–314. doi: 10.1111/j.1574-6968.1989.tb03065.x
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Cheng, Y. F., Edwards, J. E., Allison, G. G., Zhu, W. Y., and Theodorou, M. K. (2009). Diversity and activity of enriched ruminal cultures of anaerobic fungi and methanogens grown together on lignocellulose in consecutive batch culture. Bioresou. Technol. 100, 4821–4828. doi: 10.1016/j.biortech.2009.04.031
Dagar, S. S., Kumar, S., Griffith, G. W., Edwards, J. E., Callaghan, T. M., Singh, R., et al. (2015a). A new anaerobic fungus (Oontomyces anksri gen. nov., sp. nov.) from the digestive tract of the Indian camel (Camelus dromedarius). Fungal Biol. 119, 731–737. doi: 10.1016/j.funbio.2015.04.005
Dagar, S. S., Kumar, S., Mudgil, P., and Puniya, A. K. (2018). Comparative evaluation of lignocellulolytic activities of filamentous cultures of monocentric and polycentric anaerobic fungi. Anaerobe 50, 76–79. doi: 10.1016/j.anaerobe.2018.02.004
Dagar, S. S., Kumar, S., Mudgil, P., Singh, R., and Puniya, A. K. (2011). D1/D2 domain of large-subunit ribosomal DNA for differentiation of Orpinomyces spp. Appl. Environ. Microbiol. 77, 6722–6725. doi: 10.1128/AEM.05441-11
Dagar, S. S., Singh, N., Goel, N., Kumar, S., and Puniya, A. K. (2015b). Role of anaerobic fungi in wheat straw degradation and effects of plant feed additives on rumen fermentation parameters in vitro. Benefical Microbes 6, 353–360. doi: 10.3920/BM2014.0071
Davies, D. R., Theodorou, M. K., Brooks, A. E., and Trinci, A. P. (1993a). Influence of drying on the survival of anaerobic fungi in rumen digesta and faeces of cattle. FEMS Microbiol. Lett. 106, 59–63. doi: 10.1111/j.1574-6968.1993.tb05935.x
Davies, D. R., Theodorou, M. K., Lawrence, M. I., and Trinci, A. P. (1993b). Distribution of anaerobic fungi in the digestive tract of cattle and their survival in faeces. Microbiology 139, 1395–1400. doi: 10.1099/00221287-139-6-1395
de Vries, R. P., and Visser, J. (2001). Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mole. Biol. Rev. 65, 497–522.
Edwards, J. E., Forster, R. J., Callaghan, T. M., Dollhofer, V., Dagar, S. S., Cheng, Y. F., et al. (2017). PCR and Omics based techniques to study the diversity, ecology and biology of anaerobic fungi: insights, challenges and opportunities. Front. Microbiol. 8:1657. doi: 10.3389/fmicb.2017.01657
Gruninger, R. J., Puniya, A. K., Callaghan, T. M., Edwards, J. E., Youssef, N., Dagar, S. S., et al. (2014). Anaerobic fungi (phylum Neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential. FEMS Microbiol. Ecology 90, 1–17. doi: 10.1111/1574-6941.12383
Guerouali, A., and Wardeh, M. F. (1998). “Assessing nutrient requirements and limits to production of the camel under its simulated natural environment,” in Proceedings of the Third Annual Meeting for Animal Production under Arid Conditions, (Al-Ain: United Arab Emirates), 36–51.
Hackstein, J. H. P., Baker, S. E., van Hellemond, J. J., and Tielens, A. G. M. (2019). “Hydrogenosomes of anaerobic fungi: an alternative way to adapt to anaerobic environments,” in Hydrogenosomes and Mitosomes: Mitochondria of Anaerobic Eukaryotes, ed. J. Tachezy (Cham: Springer), 159–175.
Hanafy, R. A., Lanjekar, V. B., Dhakephalkar, P. K., Callaghan, T. M., Dagar, S. S., Griffith, G. W., et al. (2020a). Seven new Neocallimastigomycota genera from wild, zoo-housed, and domesticated herbivores greatly expand the taxonomic diversity of the phylum. Mycologia 112, 1212–1239. doi: 10.1080/00275514.2019.1696619
Hanafy, R. A., Johnson, B., Youssef, N. H., and Elshahed, M. S. (2020b). Assessing anaerobic gut fungal diversity in herbivores using D1/D2 large ribosomal subunit sequencing and multi-year isolation. Environ. Microbiol. 22, 3883–3908. doi: 10.1111/1462-2920.15164
Hanafy, R. A., Youssef, N. H., and Elshahed, M. S. (2021). Paucimyces polynucleatus gen. nov, sp. nov., a novel polycentric genus of anaerobic gut fungi from the faeces of a wild blackbuck antelope. Int. J. Syst. Evol. Microbiol. 71. doi: 10.1099/ijsem.0.004832
He, L. W., Zhou, W., Xing, Y. Q., Pian, R. Q., Chen, X. Y., and Zhang, Q. (2020). Improving the quality of rice straw silage with moringa oleifera leaves and propionic acid: fermentation, nutrition, aerobic stability and microbial communities. Bioresou. Technol. 299:122579. doi: 10.1016/j.biortech.2019.122579
Hess, M., Paul, S. S., Puniya, A. K., van der Giezen, M., Shaw, C., Edwards, J. E., et al. (2020). Anaerobic fungi: past, present, and future. Front. Microbiol. 11:584893. doi: 10.3389/fmicb.2020.584893
Hungate, R. E., Smith, W., Bauchop, T., Yu, I., and Rabinowitz, J. C. (1970). Formate as an intermediate in the bovine rumen fermentation. J. Bacteriol. 102, 389–397. doi: 10.1128/jb.102.2.389-397.1970
Jin, W., Cheng, Y. F., Mao, S. Y., and Zhu, W. Y. (2011). Isolation of natural cultures of anaerobic fungi and indigenously associated methanogens from herbivores and their bioconversion of lignocellulosic materials to methane. Bioresou. Technol. 102, 7925–7931. doi: 10.1016/j.biortech.2011.06.026
Joshi, A., Lanjekar, V. B., Dhakephalkar, P. K., Callaghan, T. M., Griffith, G. W., and Dagar, S. S. (2018). Liebetanzomycespolymorphus gen. et sp. nov., a new anaerobic fungus (Neocallimastigomycota) isolated from the rumen of a goat. MycoKeys 2018, 89–110. doi: 10.3897/mycokeys.40.28337
Kazemi Shariat Panahi, H. M., Dehhaghi, G. J., Guillemin, G., Gupta, V. K., Lam, S. S., Aghbashlo, M., et al. (2022). A comprehensive review on anaerobic fungi applications in biofuels production. Sci. Total Environ. 829:154521. doi: 10.1016/j.scitotenv.2022.154521
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mole. Biol. Evolu. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, M. M., White, R. R., Guan, L. L., Harthan, L., and Hanigan, M. D. (2021). Metatranscriptomic analyses reveal ruminal pH regulates fiber degradation and fermentation by shifting the microbial community and gene expression of carbohydrate-active enzymes. Animal Microbiome 3:32. doi: 10.1186/s42523-021-00092-6
Li, Y. F., Jin, W., Cheng, Y. F., and Zhu, W. Y. (2016). Effect of the associated methanogen Methanobrevibacter thaueri on the dynamic profile of end and intermediate metabolites of anaerobic fungus Piromyces sp F1. Curr. Microbiol. 73, 434–441. doi: 10.1007/s00284-016-1078-9
Li, Y. F., Jin, W., Mu, C. L., Cheng, Y. F., and Zhu, W. Y. (2017). Indigenously associated methanogens intensified the metabolism in hydrogenosomes of anaerobic fungi with xylose as substrate. J. Basic Microbiol. 57, 933–940. doi: 10.1002/jobm.201700132
Li, Y. Q., Hou, Z. S., Shi, Q. C., Cheng, Y. F., and Zhu, W. Y. (2020). Methane production from different parts of corn stover via a simple co-culture of an anaerobic fungus and methanogen. Front. Bioengineering Biotechnol. 8:314. doi: 10.3389/fbioe.2020.00314
Li, Y. Q., Meng, Z. X., Xu, Y., Shi, Q. C., Ma, Y. P., Aung, M., et al. (2021). Interactions between anaerobic fungi and methanogens in the rumen and their biotechnological potential in biogas production from lignocellulosic materials. Microorganisms 9:190. doi: 10.3390/microorganisms9010190
Li, Y. Q., Xu, Y., Xue, Y. H., Yang, S. H., Cheng, Y. F., and Zhu, W. Y. (2022). Ethanol production from lignocellulosic biomass by co-fermentation with Pecoramyces sp. F1 and Zymomonas mobilis ATCC 31821 in an integrated process. Biomass Bioenergy 161:106454.
Liang, J. S., Zhang, H. B., Zhang, P. Y., Zhang, G. M., Cai, Y. J., Wang, Q. Y., et al. (2021). Effect of substrate load on anaerobic fermentation of rice straw with rumen liquid as inoculum: hydrolysis and acidogenesis efficiency, enzymatic activities and rumen bacterial community structure. Waste Manag. 124, 235–243. doi: 10.1016/j.wasman.2021.02.017
Ma, J., Zhong, P., Li, Y., Sun, Z., Sun, X., Aung, M., et al. (2022). Hydrogenosome, pairing anaerobic fungi and H(2)-utilizing microorganisms based on metabolic ties to facilitate biomass utilization. J. Fungi 8:338. doi: 10.3390/jof8040338
Mankar, A. R., Pandey, A., Modak, A., and Pant, K. K. (2021). Pretreatment of lignocellulosic biomass: a review on recent advances. Bioresou. Technol. 334:125235.
Mao, S. Y., Huo, W. J., and Zhu, W. Y. (2016). Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 18, 525–541. doi: 10.1111/1462-2920.12724
Mizrahi, I., Wallace, R. J., and Morais, S. (2021). The rumen microbiome: balancing food security and environmental impacts. Nat. Rev. Microbiol. 19, 553–566. doi: 10.1038/s41579-021-00543-6
Nanda, S., Azargohar, R., Dalai, A. K., and Kozinski, J. A. (2015). An assessment on the sustainability of lignocellulosic biomass for biorefining. Renewable Sustainable Energy Rev. 50, 925–941.
Nguyen, L. N., Nguyen, A. Q., Johir, M. A., Guo, W. S., Ngo, H. H., Chaves, A. V., et al. (2019). Application of rumen and anaerobic sludge microbes for bio harvesting from lignocellulosic biomass. Chemosphere 228, 702–708. doi: 10.1016/j.chemosphere.2019.04.159
Niu, D. Z., Zuo, S. S., Jiang, D., Tian, P. J., Zheng, M. L., and Xu, C. C. (2018). Treatment using white rot fungi changed the chemical composition of wheat straw and enhanced digestion by rumen microbiota in vitro. Animal Feed Sci. Technol. 237, 46–54. doi: 10.1016/j.anifeedsci.2018.01.005
Oladosu, Y., Mohd, Y. R., Abdullah, N., Magaji, U., Ghazali, Hussin, Ramli, A., et al. (2016). Fermentation quality and additives: a case of rice straw silage. BioMed. Res. Int. 2016:7985167. doi: 10.1155/2016/7985167
Ozkose, E., Thomas, B. J., Davies, D. R., Griffith, G. W., and Theodorou, M. K. (2001). Cyllamyces aberensis gen.nov sp.nov., a new anaerobic gut fungus with branched sporangiophores isolated from cattle. Canadian J. Bot. Revue Canadienne De Botanique 79, 666–673. doi: 10.1139/b01-047
Rabee, A. E., Forster, R. J., Elekwachi, C. O., Kewan, K. Z., Sabra, E. A., Shawket, S. M., et al. (2019). Community structure and fibrolytic activities of anaerobic rumen fungi in dromedary camels. J. Basic Microbiol. 59, 101–110. doi: 10.1002/jobm.201800323
Samsudin, A. A., Evans, P. N., Wright, A. D., and Al Jassim, R. (2011). Molecular diversity of the foregut bacteria community in the dromedary camel (Camelus dromedarius). Environ. Microbiol. 13, 3024–3035. doi: 10.1111/j.1462-2920.2011.02579.x
Sawatdeenarunat, C., Surendra, K. C., Takara, D., Oechsner, H., and Khanal, S. K. (2015). Anaerobic digestion of lignocellulosic biomass: challenges and opportunities. Bioresou. Technol. 178, 178–186. doi: 10.1016/j.biortech.2014.09.103
Saye, L. M. G., Navaratna, T. A., Chong, J. P. J., O’Malley, M. A., Theodorou, M. K., and Reilly, M. (2021). The anaerobic fungi: challenges and opportunities for industrial lignocellulosic biofuel production. Microorganisms 9:694. doi: 10.3390/microorganisms9040694
Shi, Q. C., Li, Y. Q., Li, Y. F., Cheng, Y., and Zhu, W. (2019). Effects of steam explosion on lignocellulosic degradation of, and methane production from, corn stover by a co-cultured anaerobic fungus and methanogen. Bioresou. Technol. 290:121796. doi: 10.1016/j.biortech.2019.121796
Silva, A. F. R., Brasil, Y. L., Koch, K., and Amaral, M. C. S. (2021). Resource recovery from sugarcane vinasse by anaerobic digestion-A review. J. Environ. Manag. 295:113137. doi: 10.1016/j.jenvman.2021.113137
Singh, J., Suhag, M., and Dhaka, A. (2015). Augmented digestion of lignocellulose by steam explosion, acid and alkaline pretreatment methods: a review. Carbohydrate Polymers 117, 624–631. doi: 10.1016/j.carbpol.2014.10.012
Solomon, K. V., Haitjema, C. H., Henske, J. K., Gilmore, S. P., Borges-Rivera, D., Lipzen, A., et al. (2016). Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science 351, 1192–1195. doi: 10.1126/science.aad1431
Stabel, M., Hanafy, R. A., Schweitzer, T., Greif, M., Aliyu, H., Flad, V., et al. (2020). Aestipascuomyces dupliciliberans gen. nov, sp. nov., the first cultured representative of the uncultured SK4 clade from Aoudad sheep and alpaca. Microorganisms 8:1734. doi: 10.3390/microorganisms8111734
Stevenson, D. M., and Weimer, P. J. (2002). Isolation and characterization of a Trichoderma strain capable of fermenting cellulose to ethanol. Appl. Microbiol. Biotechnol. 59, 721–726. doi: 10.1007/s00253-002-1027-3
Sugiharto, S., Yudiarti, T., and Isroli, I. (2016). Performances and haematological profile of broilers fed fermented dried cassava (Manihot esculenta Crantz). Tropical Animal Health Production 48, 1337–1341. doi: 10.1007/s11250-016-1098-2
Taherzadeh, M. J., and Karimi, K. (2008). Pretreatment of lignocellulosic wastes to improve ethanol and biogas production: a review. Int. J. Mole. Sci. 9, 1621–1651. doi: 10.3390/ijms9091621
Theodorou, M. K., Williams, B. A., Dhanoa, M. S., Mcallan, A. B., and France, J. (1994). A simple gas-production method using a pressure transducer to determine the fermentation kinetics of ruminant feeds. Animal Feed Sci. Technol. 48, 185–197. doi: 10.1016/0377-8401(94)90171-6
Trinci, A. P. J., Davies, D. R., Gull, K., Lawrence, M. I., Nielsen, B. B., Rickers, A., et al. (1994). Anaerobic fungi in herbivorous animals. Mycological Res. 98, 129–152. doi: 10.1016/S0953-7562(09)80178-0
Vansoest, P. J., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Voigt, K., James, T. Y., Kirk, P. M., Santiago, A. L. C. M. D., Waldman, B., Griffith, G. W., et al. (2021). Early-diverging fungal phyla: taxonomy, species concept, ecology, distribution, anthropogenic impact, and novel phylogenetic proposals. Fungal Diversity 109, 59–98. doi: 10.1007/s13225-021-00480-y
Wilken, S. E., Monk, J. M., Leggieri, P. A., Lawson, C. E., Lankiewicz, T. S., Seppala, S., et al. (2021). Experimentally validated reconstruction and analysis of a genome-scale metabolic model of an anaerobic Neocallimastigomycota fungus. mSystems 6, e00002–e21. doi: 10.1128/mSystems.00002-21
Yamamoto, M., Iakovlev, M., Bankar, S., Tunc, M. S., and van Heiningen, A. (2014). Enzymatic hydrolysis of hardwood and softwood harvest residue fibers released by sulfur dioxide-ethanol-water fractionation. Bioresou. Technol. 167, 530–538. doi: 10.1016/j.biortech.2014.06.054
Yang, X. X., Chen, H. Z., Gao, H. L., and Li, Z. H. (2001). Bioconversion of corn straw by coupling ensiling and solid-state fermentation. Bioresou. Technol. 78, 277–280. doi: 10.1016/s0960-8524(01)00024-4
Zabed, H., Sahu, J. N., Boyce, A. N., and Faruq, G. (2016). Fuel ethanol production from lignocellulosic biomass: an overview on feedstocks and technological approaches. Renewable Sustainable Energy Rev. 66, 751–774.
Keywords: anaerobic fungi, bioconversion, strain isolation, hydrogenosome metabolism, lignocellulose degradation
Citation: Xue Y, Shen R, Li Y, Sun Z, Sun X, Li F, Li X, Cheng Y and Zhu W (2022) Anaerobic Fungi Isolated From Bactrian Camel Rumen Contents Have Strong Lignocellulosic Bioconversion Potential. Front. Microbiol. 13:888964. doi: 10.3389/fmicb.2022.888964
Received: 03 March 2022; Accepted: 26 May 2022;
Published: 19 July 2022.
Edited by:
Andreas Otto Wagner, University of Innsbruck, AustriaReviewed by:
Michelle Ann O’Malley, University of California, Santa Barbara, United StatesCopyright © 2022 Xue, Shen, Li, Sun, Sun, Li, Li, Cheng and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanfen Cheng, eWFuZmVuY2hlbmdAbmphdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.