
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 May 2022
Sec. Terrestrial Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.881450
 Daifa Su1,2
Daifa Su1,2 Shanyan Chen3
Shanyan Chen3 Wenxing Zhou3
Wenxing Zhou3 Junyu Yang1,2
Junyu Yang1,2 Zhiwei Luo3
Zhiwei Luo3 Zhenrong Zhang3
Zhenrong Zhang3 Yunxia Tian3
Yunxia Tian3 Qionge Dong3
Qionge Dong3 Xuemei Shen3
Xuemei Shen3 Shijie Wei3
Shijie Wei3 Jiangyun Tong3*
Jiangyun Tong3* Xiaolong Cui1,2*
Xiaolong Cui1,2*Anthracnose caused by Colletotrichum spp. was widespread in recent years and resulted in great damage to strawberry production. Soil microbial communities were key contributors to host nutrition, development, and immunity; however, the difference between the microbial communities of healthy and anthracnose-infected strawberry rhizosphere soils remains unclear. In this study, the Illumina sequencing technique was used to comparatively study the prokaryotic and fungal community compositions and structures between healthy and anthracnose-infected strawberry rhizosphere soils in Yuxi, Yunnan Province. Both microbial community diversities and richness of anthracnose-infected strawberry rhizosphere soils were higher than those of healthy strawberry rhizosphere soils. A total of 2,518 prokaryotic and 556 fungal operational taxonomic units (OTUs) were obtained at the 97% similarity threshold. Proteobacteria, Thaumarchaeota, and Acidobacteria were the dominant prokaryotic phyla; Ascomycota, unclassified_k__Fungi, and Mortierellomycota were the dominant fungal phyla. The relative abundances of beneficial bacterial phyla Actinobacteria and Firmicutes, genera Streptomyces, Azospirillum, and Bacillus were significantly reduced in anthracnose-infected strawberry rhizosphere soils; the relative abundance of beneficial fungal species Trichoderma asperellum shows a similar tendency with bacterial abundance. Besides Colletotrichum, 15 other potential fungal pathogen genera and seven fungal pathogen species were identified; among the potential pathogen genera and species, eight pathogen genera and Fusarium oxysporum showed significant differences between healthy and anthracnose-infected strawberry rhizosphere soils. The results suggested that strawberry planted in this area may be infected by other fungal pathogens except for Colletotrichum spp. Our present research will provide theoretical basis and data reference for the isolation and identification of strawberry pathogens and potential probiotics in future works.
Cultivated strawberry (Fragaria × ananassa Duch.) is one of the most popular fruits around the world (Cheng et al., 2014). Since it originated in the eighteenth century, cultivated strawberries have been broadly introduced all over the world (Su et al., 2018) and thousands of varieties have been developed for commercial purposes. Strawberry plants, like other commercial crops, can be damaged by environmental, genetic, and biological factors, either directly or by interactions between these factors (Garrido et al., 2011). In the field, strawberry plants can be infected by a large number of pathogens, including fungi, bacteria, viruses, nematodes, and arthropods, and these pathogens cause damage to the leaves, roots, crowns, and fruits (Husaini and Neri, 2016), resulting in the greatest fruit losses.
Colletotrichum spp. comprises a diverse range of important phytopathogenic fungi that cause pre- and postharvest crop losses all over the world. Many important economic crops, such as peach, chili, almond, citrus, apple, blueberry, tomato, dragon fruits, and mango, can be infected by Colletotrichum spp. (Peres et al., 2005; Phoulivong et al., 2010; Liao et al., 2012; Zakaria, 2021). Since Brooks (1931) first reported strawberry anthracnose in 1931, the disease has been reported all over the world; in China, strawberry anthracnose was first reported in 1990 by Hu (1990). The species C. acutatum, C. fragariae, and C. gloeosporioides have been reported as the major causal agents of strawberry anthracnose (Karimi et al., 2015; Husaini and Neri, 2016; Reddy, 2016). All three species can be found in all parts of the strawberry plant (Denoyes-Rothan et al., 2003), including fruits, flowers, leaves, runners, roots, and crowns; the typical symptoms are described as irregular and black leaf spots, flower blight, and fruit and crown rot (Freeman and Katan, 1997; Mertely and Legard, 2004). Other studies reported that species, including C. aenigma, C. boninense, C. changpingense, C. dematium, C. foriniae, C. fructicola, C. godetiae, C. kahawae, C. karstii, C. miaoliense, C. nymphaeae, C. salicis, C. siamense, C. simmondsii, C. theobromicola, C. truncatum, and C. viniferum (Mass, 1978; Singh et al., 2003; Bi et al., 2017; Adhikari et al., 2019; Dai et al., 2019; Hoseini et al., 2019; Liu et al., 2021), and Glomerella cingulate (Marian et al., 2020), can also cause strawberry anthracnose. The American Phytopathological Society (APS) also reported that Gloeosporium spp. is the pathogen of strawberry anthracnose (https://www.apsnet.org/edcenter/resources/commonnames/Pages/Strawberry.aspx).
The rhizosphere, the area surrounding plant roots (Li et al., 2020), consisting of soil, bacteria, archaea, eukaryotic microorganisms, and viruses, is one of the most complex microbial habitats on earth (Bowya and Balachandar, 2020; Li et al., 2021a), which plays a key role in nutrient acquisition, enhanced stress tolerance, protection against soil-borne pathogens, and host immune regulation (Pérez-Jaramillo et al., 2016; Pantigoso et al., 2020; Sun et al., 2020). Scientists have developed a wide range of methods to study microbial diversities and community structures in rhizosphere soils, including culture-dependent approaches (Garrido et al., 2008), Biolog microplates (Choi and Dobbs, 1999), DGGE (denaturing gradient gel electrophoresis) (Lu et al., 2012), and PLFA (phospholipid fatty acid) method (Yao et al., 2016). However, due to the complex components in soil samples, it is hard to isolate and cultivate the microorganisms from soil samples (approximately 1% can get the pure cultures) (Amann et al., 1995; Roose-Amsaleg et al., 2001). Developing new methods to study complex rhizosphere microbial diversities and community structures is urgent. With the advance of high-throughput sequencing techniques, making it possible to understand uncultured microorganisms in the rhizosphere soil of plants, it has been applied to study the complexity of microbial communities, and achievements have been made in the research of rhizosphere soil microorganisms using high-throughput sequencing technologies (Wang et al., 2019; Zhao et al., 2019; Yang et al., 2020b; Zhou et al., 2020).
China is the biggest strawberry planting country, and strawberry production has been ranked first in the world since 1994 (http://www.fao.org/faostat/zh/#data/QC/visualize). After Hu (1990) first reported strawberry anthracnose in China, Chinese researchers have done work on the strawberry anthracnose pathogens and found that C. acutatum is the first reported strawberry anthracnose pathogen (Dai et al., 2006), the main pathogens causing anthracnose of strawberry in China include C. acutatum, C. fragariae, and C. gloeosporioides, and the dominant species of strawberry anthracnose in different regions are varied (Jayawardena et al., 2016). Yunnan Province, located in the southwest of China, with diverse and complex landforms and climates as its main geographical advantages, is rich in abundant agricultural and sideline products. Strawberry cultivation started in the early 1980s in Yunnan Province, and now Yunnan is the only province in which strawberry cultivation can be carried out all year round in China with open-air cultivation. Until now, the microbial community differences between healthy and anthracnose-infected strawberry rhizosphere soils still remain unknown in Yunnan Province. In this study, a high-throughput sequencing technique was used to comparatively study the prokaryotic and fungal community differences between healthy and anthracnose-infected strawberry rhizosphere soils.
The experiment was carried out at an open field in Hongta District of Yuxi City (102.547871 E, 24.450693 N, 1651.0 ± 16.69 m), Yunnan Province, China. In this experiment, an open-field, strawberry cultivar Akihime has been cultivated using rotation with scallion (Allium fistulosum L. var. gigantum Makino) for 4 years. On November 16, 2018, samples were collected from the field in Hongta District using a five-point sampling method in the flowering and fruit ripening stages. Healthy (h) and disease-infected strawberry plants (d) were chosen, the whole plants were uprooted after removing the dead branches and leaves from the soil surface, and the rhizosphere soil samples were collected after strongly shaking the plants and brushing soil from root surfaces (Desgarennes et al., 2014; Edwards et al., 2015; Coleman-Derr et al., 2016; Cui et al., 2019). Each sample consisted of five subsamples, and healthy and anthracnose-infected strawberry plant samples were composed of three replicate samples, respectively. Healthy strawberry rhizosphere soil samples were marked as hRZ (hRZ 1–3, three replicates), and diseased strawberry rhizosphere soil samples were marked as dRZ (dRZ 1–3, three replicates). All samples were kept in ice bags, transported to laboratory, and immediately stored at −80°C for extraction of genomic DNA.
The total genomic DNA of soil samples was directly extracted using the PowerSoil DNA Isolation Kit (Mo Bio Laboratories, San Diego, CA, USA), following the manufacturer's protocol. The concentration and purity of the extracted DNA were measured using a NanoDrop 2000 spectrometer (Thermo Fisher Scientific, Wilmington, DE, USA).
The prokaryotic universal primer pairs 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′) were used to amplify the V3–V4 region of 16S rRNA gene (Yu et al., 2010; Yang et al., 2020a), and the fungi primer pairs ITS3F (5′-GCATCGATGAAGAACGCAGC-3′) and ITS4R (5′-TCCTCCGCTTATTGATATGC-3′) were used to amplify the ITS2 region (Tedersoo et al., 2015; Yang et al., 2020a). PCR amplification was conducted using TransGen AP221-02: TransStart FastPfu DNA Polymerase (TransGen Biotech, Beijing, China) and performed in a GeneAmp 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA). The reaction mixture included 4 μL 5 × FastPfu buffer, 2 μL 2.5 mM dNTPs, 0.4 μL FastPfu polymerase, 0.8 μL each primer (5 mM), 0.2 μL BSA, 10 ng template DNA, and ddH2O to a final volume of 20 μL. Thermal cycling conditions were as follows: 95°C for 3 min; V3–V4 region was followed by 27 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 45 s; ITS2 region was followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s, with a final extension at 72°C for 10 min. PCR amplification was detected using 2% agarose gel electrophoresis. The V3–V4 region and ITS2 region were sequenced on the Illumina MiSeq PE 300 platform at Shanghai Majorbio Bio-Pharm Technology Co., Ltd., following the manufacturer's protocols.
Since the data from the MiSeq sequencing platform were paired-end sequences, the paired reads were joined, quality controlled, and filtered to obtain high-quality reads using FLASH (v.1.2.11) (Magoč and Salzberg, 2011) and Trimmomatic (v.0.38) (Bolger et al., 2014) software. The obtained high-quality reads were processed to generate operational taxonomic units (OTUs) at a 97% sequence similarity threshold (Edgar, 2013). The taxonomic assignments were performed by the RDP classifier algorithm (http://rdp.cme.msu.edu/) against the Unite (v.8.0) ITS database (Abarenkov et al., 2010) and SILVA (SSU 132) 16S rRNA database (Quast et al., 2013) with a confidence threshold of 70%. All sequences taxonomically assigned to chloroplasts and mitochondria were removed in this study.
Bioinformatics analysis was performed on the free I-Sanger platform (Majorbio Bio-Pharm Technology Co. Ltd., Shanghai, China; www.i-sanger.com). Alpha diversity, which reflects the richness and diversity of microbial communities, including Sobs, Shannon, Simpson, Ace, Chao, and Coverage indexes, was calculated by Mothur (v.1.35.1; https://www.mothur.org/). Beta diversity was estimated as a representation of the compositional differences between communities, and NMDS (non-metric multidimensional scaling) was used to evaluate similarities across community structures using the Bray–Curtis distance metric. The potential functions of the prokaryotic and fungal communities between healthy and diseased rhizosphere soils were predicted by PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) (Douglas et al., 2020) and FunGuild (Fungi Functional Guild) (Nguyen et al., 2016) programs, respectively.
The data were collected using Microsoft Excel 2013, statistical analysis was performed using SPSS 17.0 (SPSS Inc., Chicago, IL, USA), the results were represented as the means ± SD (standard deviations), and P <0.05 was statistically significant.
In this study, six samples were collected and processed for high-throughput sequencing and analysis. A total of 227,697 and 302,804 raw reads were produced by the Illumina MiSeq PE 300 sequencing platform for prokaryotes and fungi; after low-quality reads were filtered, a total of 192,585 and 290,950 clean reads were obtained for prokaryotes and fungi, respectively (Supplementary Table 1). The reads in fungal samples were higher than in prokaryotic samples to obtain more informatics from samples. OTU numbers in healthy samples were significantly lower than in diseased samples for the prokaryotic community; for the fungal community, the tendency was similar to the prokaryotic community but did not show a significant difference (Supplementary Table 1). The good coverage in each sample was more than 98%, suggesting that the sequencing in each sample in this study was sufficient for subsequent analyses (Supplementary Table 1).
In this study, microbial community richness and diversity were assessed using alpha and beta diversity analyses. At the 97% threshold, the rarefaction curves of prokaryotic and fungal communities were nearly reached asymptotes, indicating that the sequence depth in each sample was sufficient, and most of the prokaryotic and fungal community diversities of the samples were captured (Supplementary Figure 1).
The community diversity was expressed by Shannon and Simpson indices; community richness was expressed by Ace, Chao, and Sobs indices. Shannon indices and community richness showed similar trends; the values in healthy samples were lower than in diseased samples (Table 1). Simpson index in this study showed an opposite tendency with the Shannon index (Table 1).
NMDS results indicated that the soil samples were divided into different groups for both prokaryotic and fungal communities and showed a good ordination (stress <0.1) (Figure 1).
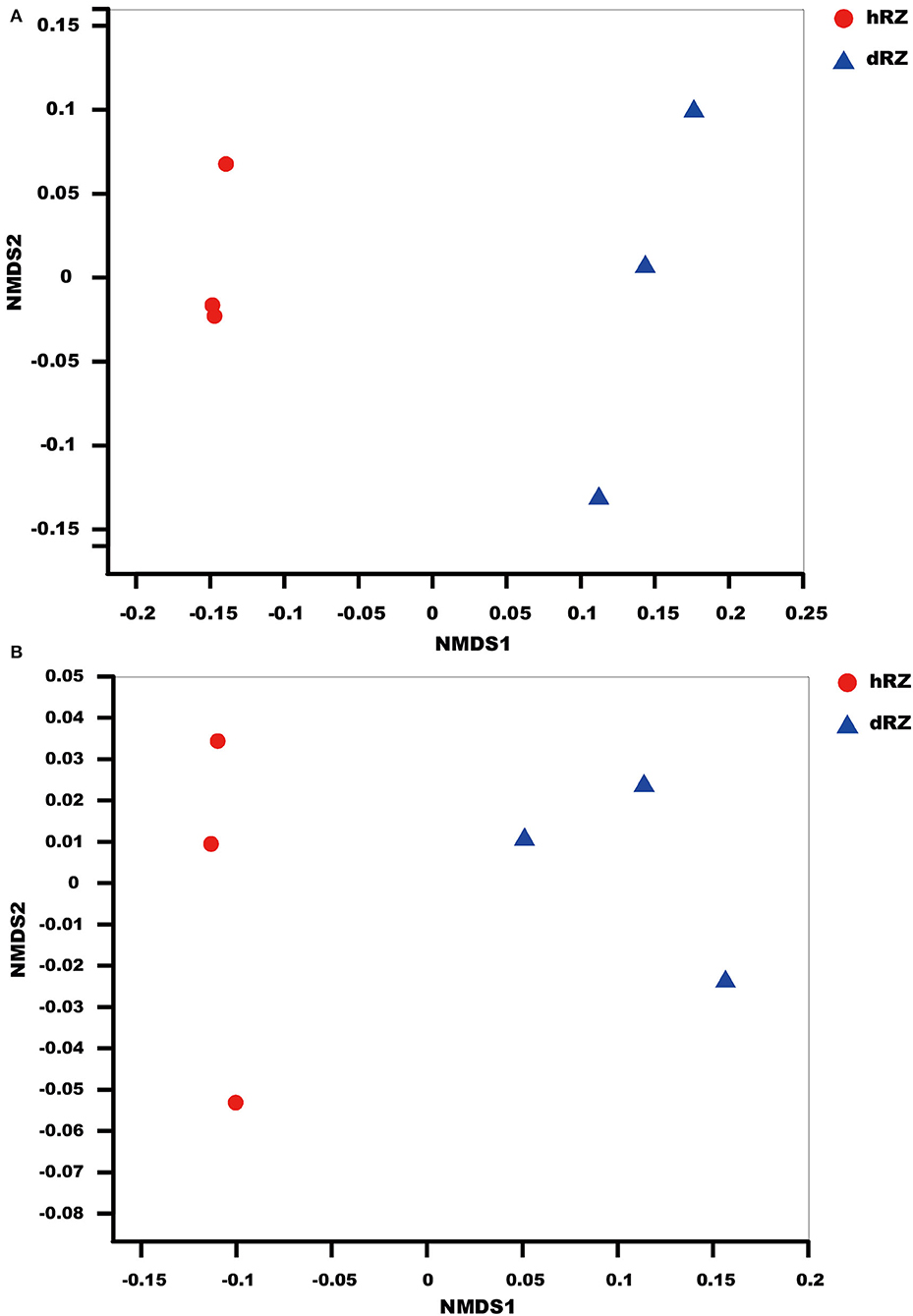
Figure 1. NMDS (non-metric multidimensional scaling) ordination based on Bray–Curtis similarities of prokaryotic (A) and fungal (B) OTU community structures among different samples. hRZ, healthy rhizosphere soils; dRZ, diseased rhizosphere soils (n = 3).
For the prokaryotic community, a total of 27 phyla, 62 classes, 180 orders, 309 families, 609 genera, and 2,518 OTUs were identified from six rhizosphere soil samples; 12 phyla, 30 classes, 63 orders, 122 families, 196 genera, and 556 OTUs were identified from six rhizosphere soil samples for the fungal community. At the phylum level, at least nine prokaryotic phyla showed their relative abundances >1%, and Proteobacteria was the most abundant phylum in both healthy and diseased rhizosphere soils (Figure 2A); for fungus, 10 phyla were identified from healthy rhizosphere soils, 12 phyla were identified from diseased rhizosphere soils, and Ascomycota was the most abundant phylum in both healthy and diseased rhizosphere soils (Figure 2C).
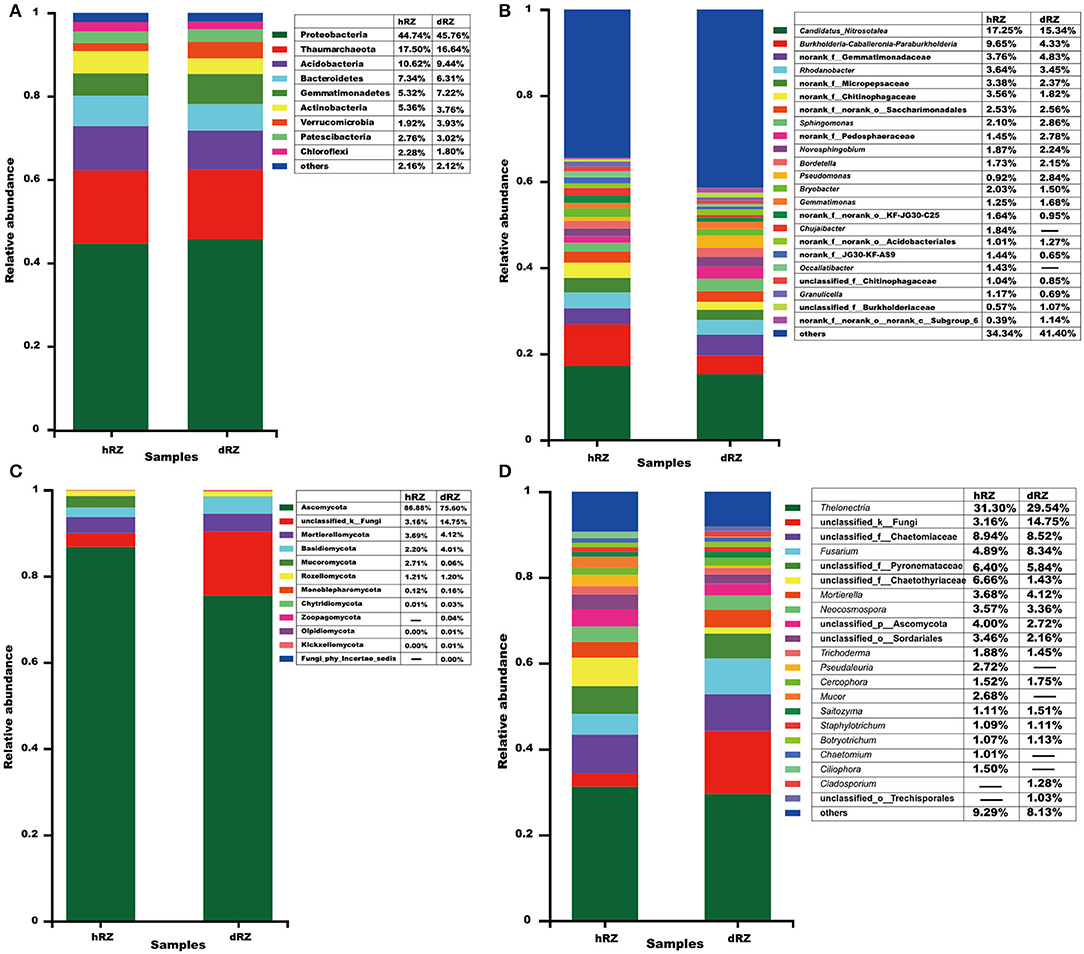
Figure 2. Relative abundances of prokaryotic (A,B) and fungal (C,D) community compositions at the phylum (A,C) and genus (B,D) levels are determined in different samples. The relative abundance <1% is combined into “others.” hRZ, healthy rhizosphere soils; dRZ, diseased rhizosphere soils (n = 3).
At the genus level, 23 prokaryotic genera were identified with relative abundance >1%, and the most abundant prokaryotic genera, including Candidatus_Nitrosotalea, Burkholderia–Caballeronia–Paraburkholderia, Rhodanobacter, Sphingomonas, Novosphingobium, Bordetella, Pseudomonas, Bryobacter, Gemmatimonas, Chujaibacter, Occallatibacter, Granulicella, and no rank and unclassified groups; hRZ and dRZ had 20 and 17 prokaryotic genera with a relative abundance >1%, respectively (Figure 2B). Twenty-one fungal genera showed that relative abundance > 1%, and the most abundant fungal genera, including Thelonectria, Fusarium, Mortierella, Neocosmospora, Trichoderma, Pseudaleuria, Cercophora, Mucor, Saitozyma, Staphylotrichum, Botryotrichum, Chaetomium, Ciliophora, Cladosporium, and unclassified groups; 19 and 17 fungal genera showed that relative abundance >1% in hRZ and dRZ, respectively (Figure 2D).
Common and unique taxonomic groups in two soil samples were visualized with a Venn diagram. At the phylum level, 24 prokaryotic phyla, namely, Proteobacteria, Diapherotrites, Actinobacteria, FCPU426, Firmicutes, Latescibacteria, Nitrospirae, Dependentiae, Acidobacteria, Planctomycetes, Euryarchaeota, Thaumarchaeota, Cyanobacteria, Armatimonadetes, Spirochaetes, Bacteroidetes, WPS-2, Gemmatimonadetes, Elusimicrobia, Verrucomicrobia, Chloroflexi, Rokubacteria, Patescibacteria, and unclassified_k__norank_d__Bacteria that were contained in both hRZ and dRZ; Epsilonbacteraeota, WS2, and Zixibacteria, were the prokaryotic phyla that were contained only in dRZ (Figure 3A and Supplementary Figure 2). Ten fungal phyla, namely, Mortierellomycota, Chytridiomycota, Ascomycota, Mucoromycota, Monoblepharomycota, Basidiomycota, Rozellomycota, Olpidiomycota, Kickxellomycota, and unclassified_k__Fungi, were contained in both hRZ and dRZ. Zoopagomycota and Fungi_phy_Incertae_sedis were the fungal phyla that were contained only in dRZ (Figures 2C, 3C and Supplementary Figure 3). At the genus level, 499 prokaryotic genera were contained in both hRZ and dRZ and 29 and 81 prokaryotic genera were contained only in hRZ and dRZ, respectively (Figure 3B and Supplementary Figure 4). One hundred and forty-eight fungal genera were contained in both hRZ and dRZ, and 14 and 34 fungal genera were contained only in hRZ and dRZ, respectively (Figure 3D and Supplementary Figure 5).
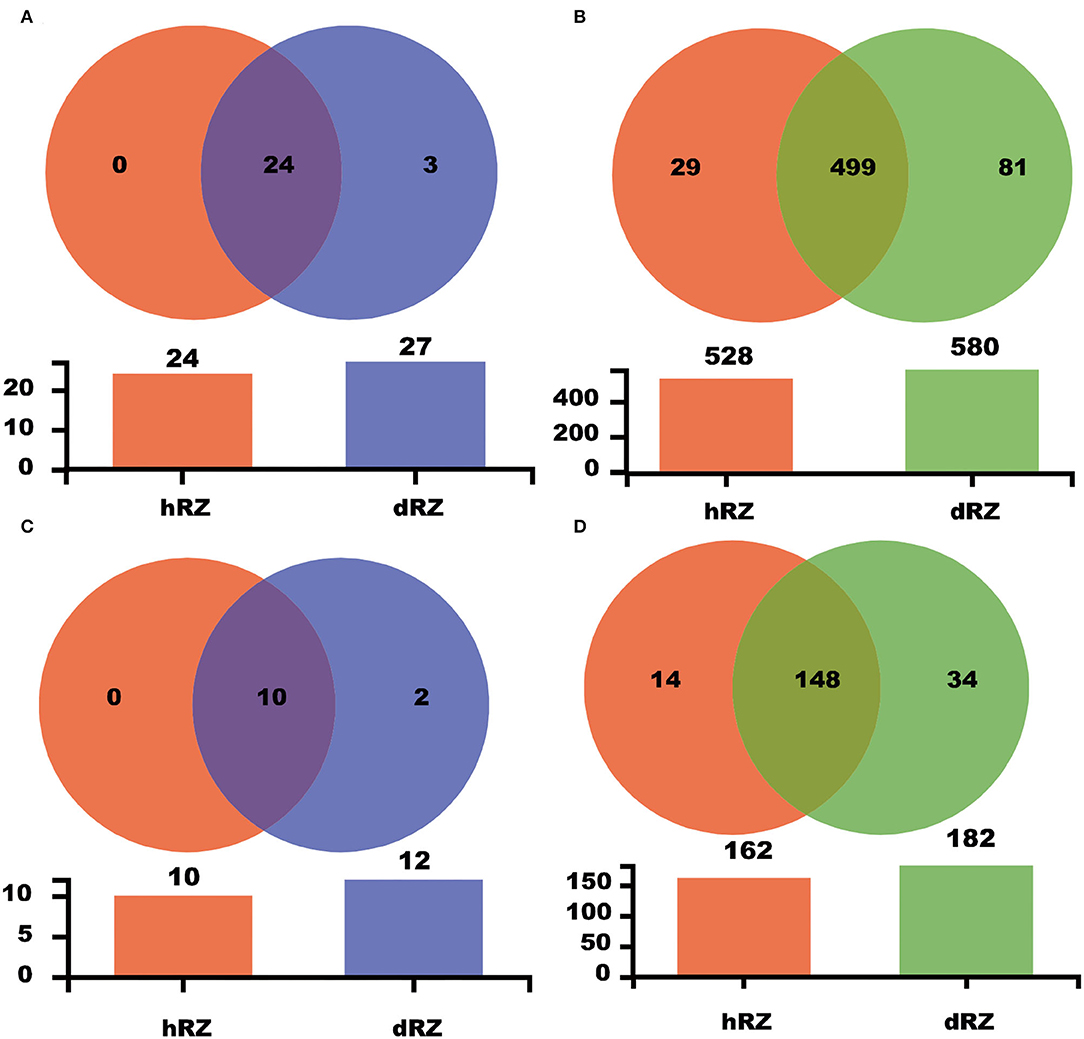
Figure 3. Venn diagram showing the common and unique OTUs among different samples at the phylum (A,C) and genus (B,D) levels for prokaryotic (A,B) and fungal (C,D) communities, respectively. hRZ, healthy rhizosphere soils; dRZ, diseased rhizosphere soils (n = 3).
Comparative analysis was used to study the prokaryotic and fungal community differences at the phylum and genus levels between healthy and diseased rhizosphere soils. Among 27 prokaryotic phyla, seven phyla were significantly different between healthy and diseased rhizosphere soils, the relative abundances of Rokubacteria, Cyanobacteria, Latescibacteria, Verrucomicrobia, and Gemmatimonadetes were significantly enriched in diseased rhizosphere soils compared with healthy rhizosphere soils, while the relative abundances of potential beneficial phyla Actinobacteria and Firmicutes in diseased rhizosphere soils were significantly reduced compared with healthy rhizosphere soils (Figure 4A). Among 12 fungal phyla, five phyla showed significant differences between healthy and diseased rhizosphere soils, the relative abundances of Ascomycota and Mucoromycota in healthy rhizosphere soils were significantly higher than in diseased rhizosphere soils, and the relative abundances of Basidiomycota and Zoopagomycota showed an opposite tendency with Ascomycota and Mucoromycota (Figure 4B).
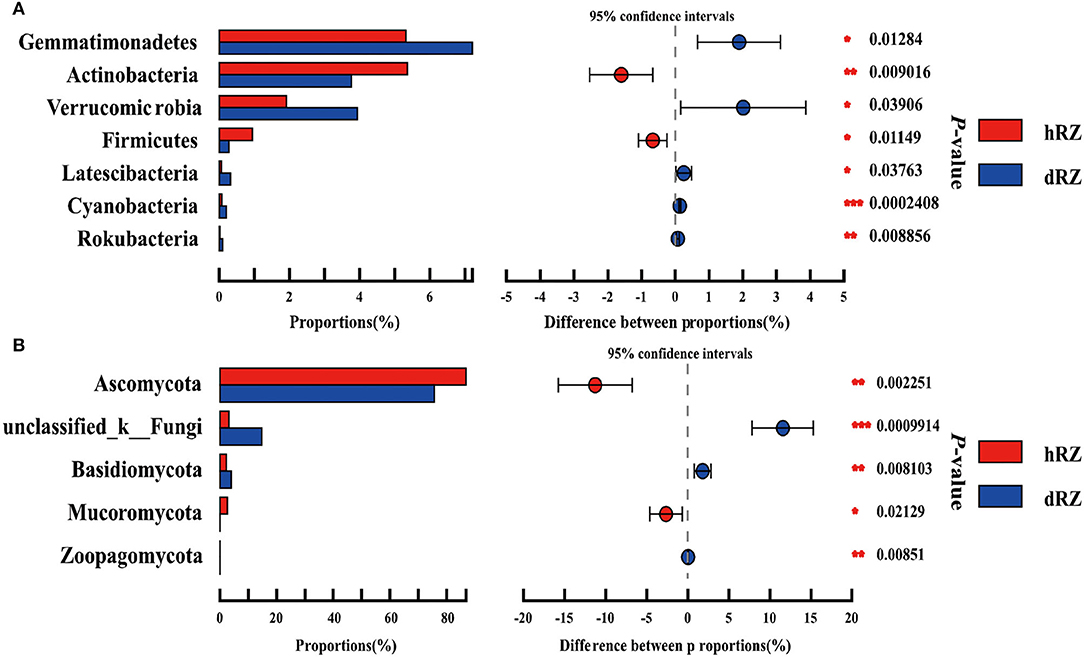
Figure 4. Relative abundances of prokaryotic (A) and fungal (B) phyla show significant differences between healthy and diseased rhizosphere soils. *Indicates significantly different values, *0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001. P-values are based on Student's t-tests. hRZ, healthy rhizosphere soils; dRZ, diseased rhizosphere soils (n = 3).
Among 609 identified prokaryotic genera in this study, 190 genera showed significant differences between healthy and diseased rhizosphere soils (data not shown). Among 190 genera with significant differences, 134 genera showed that the relative abundances in diseased rhizosphere soils were significantly higher than in healthy rhizosphere soils, while the other 56 genera showed an opposite tendency with the 134 genera. A total of 52 fungal genera showed significant differences between healthy and diseased rhizosphere soils, 26 genera showed that the relative abundances in healthy rhizosphere soils were significantly higher than in diseased rhizosphere soils, and the other 26 genera showed an opposite tendency (data not shown).
According to the APS, it is reported that bacterial pathogens, including Xanthomonas fragariae, Pseudomonas solanacearum, Rhodococcus fascians (= Corynebacterium fascians), and Aphelanchoides fragariae, can cause bacterial diseases in strawberries; more than 50 fungal pathogens can cause fungal diseases in strawberries (https://www.apsnet.org/edcenter/resources/commonnames/Pages/Strawberry.aspx). In this study, the bacterial pathogens were not annotated at the species level (data not shown); the relative abundances of genera Pseudomonas, Rhodococcus, and Xanthomonas in hRZ and dRZ were different; and the relative abundance of Pseudomonas in dRZ was significantly higher than in hRZ, while the relative abundances of Rhodococcus and Xanthomonas in hRZ were higher than in dRZ (Figure 5A). At the genus level, 15 potential fungal pathogen genera were identified and eight genera were significantly different between healthy and diseased rhizosphere soils (Figure 5B). Furthermore, seven fungal pathogens species, including Rhizoctonia fragariae, Plectosphaerella cucumerina, Neopestalotiopsis clavispora, Mucor hiemalis, Fusarium oxysporum, Dactylonectria torresensis, and Alternaria tenuissima, were annotated at the species level in two soil groups in this study, and the relative abundance of F. oxysporum was significantly different between hRZ and dRZ (Figure 5C).
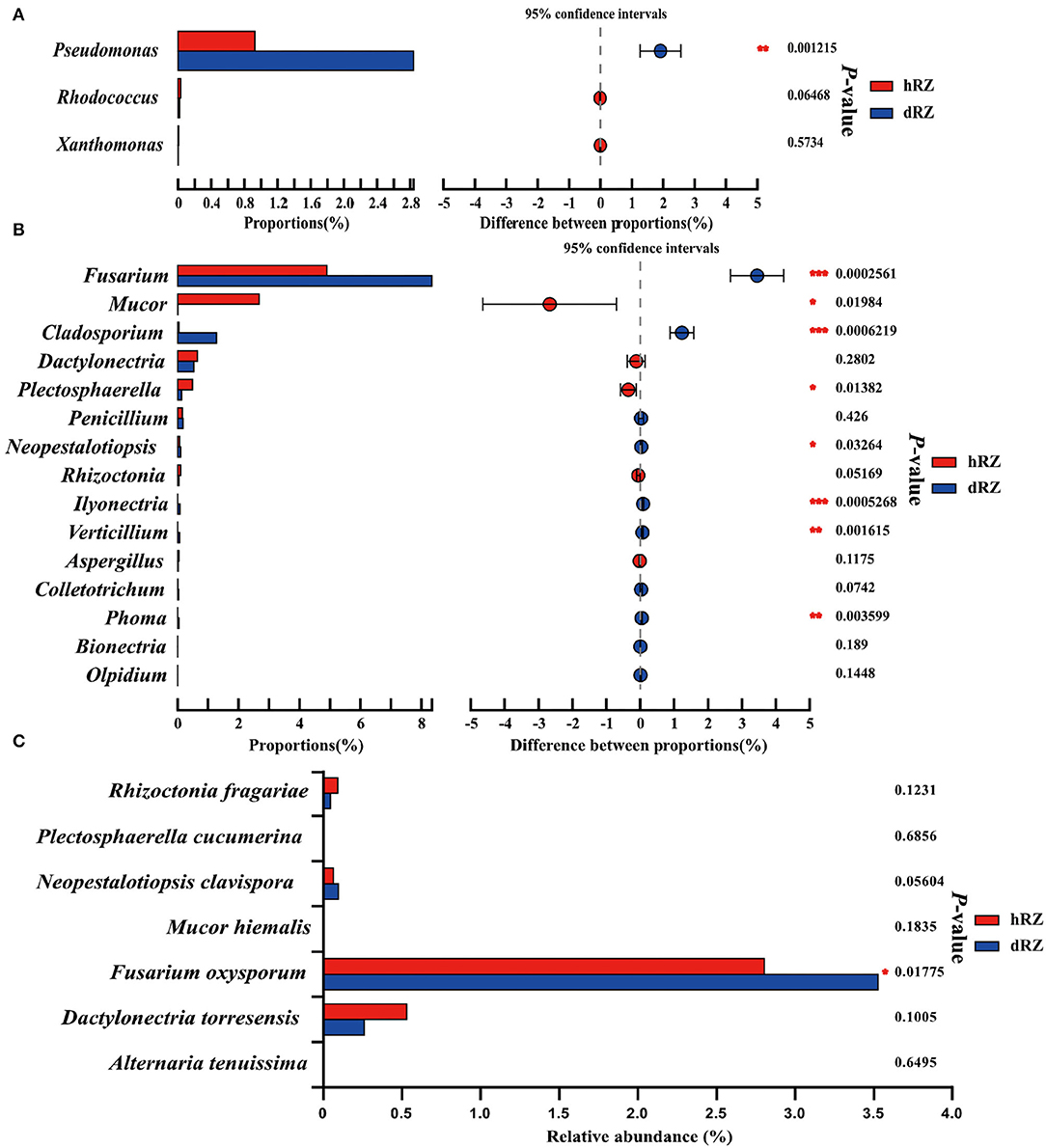
Figure 5. Relative abundances of bacterial (A) and fungal (B) pathogens between healthy and diseased rhizosphere soils at the genus level and (C) fungal pathogen species between healthy and diseased rhizosphere soils. *Indicates significantly different values, *0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001. P-values are based on Student's t-tests. hRZ, healthy rhizosphere soils; dRZ, diseased rhizosphere soils (n = 3).
As reported by some researchers, bacterial species within Streptomyces (Marian et al., 2020), Azospirillum (Tortora et al., 2011), and Bacillus (Mochizuki et al., 2012) can be used to biocontrol strawberry anthracnose. The relative abundances of Streptomyces and Bacillus in healthy rhizosphere soils were significantly higher than in diseased rhizosphere soils, and Azospirillum was only detected in healthy rhizosphere soils (Figure 6A). Bacillus, Paenibacillus, Streptomyces, Pseudomonas, Flavobacterium, Arthrobacter, Microbacterium, Azoarcus, Azospirillum, Caulobacter, Chromobacterium, Enterobacter, and Pantoea were usually seen as the source of beneficial microbes (Etcheverry et al., 2009; Ahemad and Kibret, 2014; Bruto et al., 2014; Jha and Saraf, 2015; He et al., 2020). In this study, the relative abundances of Azoarcus, Pantoea, Flavobacterium, and Pseudomonas were significantly increased when the plant was infected, and the relative abundances of Chromobacterium and Caulobacter also increased, but not significant; the relative abundances of Paenibacillus, Enterobacter, and Arthrobacter were significantly decreased after strawberry plant was diseased (Microbacterium excepted) (Figure 6A). Zhao et al. (2020) and Karimi et al. (2017) reported that Trichoderma asperellum can be used as a biocontrol fungus to control strawberry anthracnose caused by C. fragariae and C. nymphaeae, respectively. In this study, T. asperellum was detected between healthy and diseased samples in rhizosphere soils, and the relative abundance in hRZ was higher than in dRZ (Figure 6B).
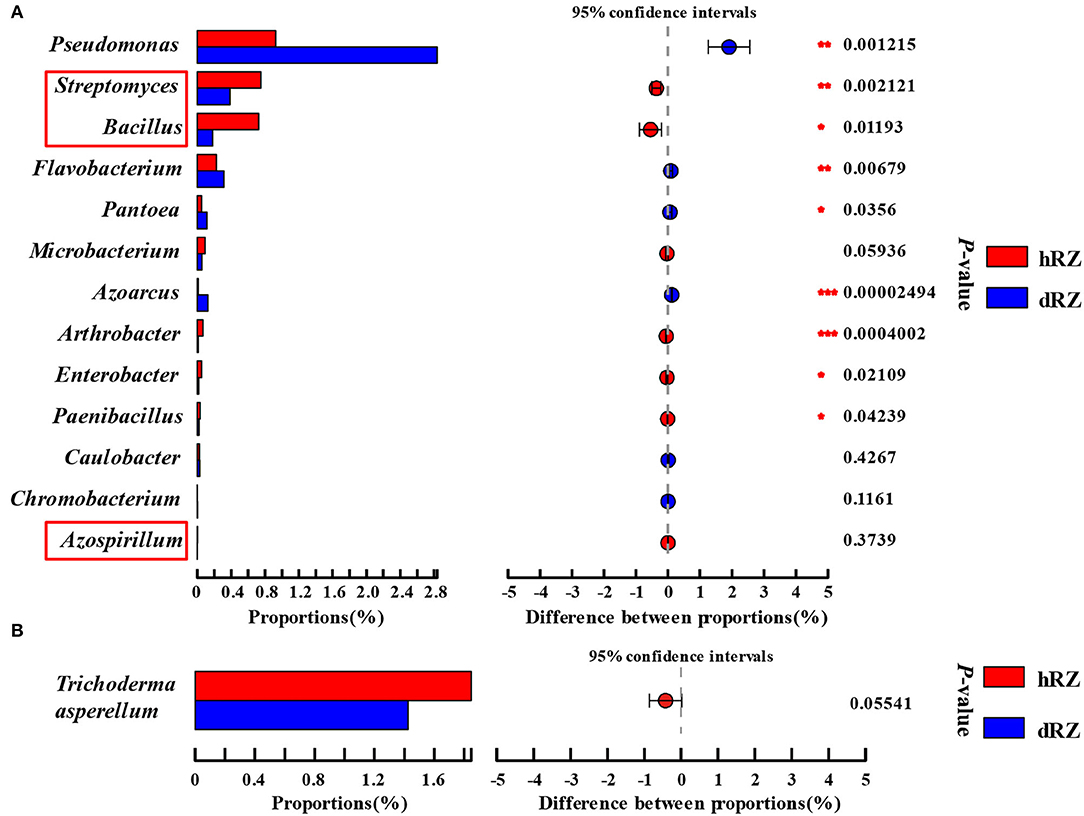
Figure 6. Relative abundances of beneficial bacterial genera (A) and fungal species Trichoderma asperellum (B) both in healthy and diseased rhizosphere soils. *Indicates significantly different values, *0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001. P-values are based on Student's t-tests. The red box indicates the antagonistic bacteria species used in strawberries. hRZ, healthy rhizosphere soils; dRZ, diseased rhizosphere soils (n = 3).
In this study, PICRUSt2 (Douglas et al., 2020) and FunGuild (Nguyen et al., 2016) programs were used to predict the potential functions of the prokaryotic and fungal communities within healthy and diseased rhizosphere soils, respectively. For prokaryotic communities, six level 1 KO groups containing 45 level 2 KO groups [metabolism (12/45), genetic information processing (4/45), organismal systems (10/45), cellular processes (4/45), human diseases (12/45), and environmental information processing (3/45)] were identified (Supplementary Tables 2, 3); two of the 45 level 2 KO groups showed different abundances (P <0.05) between the two soil groups (Supplementary Table 3 marked red). For fungal communities, the OTUs were annotated into 10 trophic modes, and four modes showed significant differences between healthy and diseased rhizosphere soils (Supplementary Table 4); 20 guilds were annotated, and six guilds showed significant differences between two soil groups (Supplementary Table 5 marked red).
In this study, the high-throughput sequencing technology was used to explore the rhizosphere soil microbial diversity and community structure between healthy and anthracnose-infected strawberry plants in an open field. The study provides new insight into the rhizosphere soil community differences in healthy and anthracnose-infected strawberry plants.
Like animal gut microbes, microbes growing in association with plant roots (in the rhizosphere) can affect the health of their host (Haney et al., 2015). The unbalance of microbial community will lead to plant disease, and the microbial community structure will also change after the plant was infected with pathogens (Hamel et al., 2005). Feng et al. (2018) study revealed that after Torreya grandis was infected by the F. oxysporum species complex, the growth and vitality of T. grandis, soil organic carbon content, and soil microbial abundance were decreased. Lee et al. (2021) investigated that the relative abundances of Actinobacteria and Firmicutes phyla were lower in diseased rhizosphere soil than in healthy rhizosphere soil, and disruption of two phyla caused the incidence of bacterial wilt disease. Chen et al. (2020) studied the endophytic phyllosphere microbiota between healthy and Pseudomonas syringae pv. tomato (Pst) DC3000-infected Arabidopsis quadruple mutants, and the results showed that the unbalance of relative abundance between Proteobacteria and Firmicutes was the main cause of the disease. In this study, the relative abundances of Actinobacteria and Firmicutes were significantly decreased in diseased rhizosphere soils compared with healthy samples, and this may be the main reason that strawberry plant was diseased.
It has been suggested that lower microbial diversity in the rhizosphere soils may lead to a higher occurrence of soil-borne disease (Bailey and Lazarovits, 2003), and most studies revealed that the rhizosphere soil microbial diversity in diseased samples was lower than that in healthy samples (Gorissen et al., 2004; Qiu et al., 2012; Deng et al., 2015; Lai et al., 2019; Yang et al., 2020a). Xiao et al. (2017) used high-throughput sequencing to comparatively study the rhizosphere soil bacterial communities of healthy strawberry and anthracnose-infected strawberry plants, and their results revealed that both OTU numbers and diversity index in infected rhizosphere soil were reduced when compared with the healthy samples. Other researchers' studies revealed that after crops, such as cotton (Liu et al., 2019), potato (Rosenzweig et al., 2012; Zhao et al., 2021), chili (Lai et al., 2019), Panax notoginseng (Wu et al., 2015), tobacco (Xiang et al., 2019), and monkshood (Li et al., 2021b), were diseased, the microbial diversity was decreased when compared with healthy samples. However, other researchers also found that when the microbial structure changes, plant roots may be more vulnerable to pathogen infection (Wu et al., 2016), and the microbial diversity in infected samples was higher than in healthy samples. Yang et al. (2021) studied the rhizosphere soil bacterial structures between healthy and root rot apples and found that the bacterial diversity in the rhizosphere soil of root rot apple plant was increased, and the balance of Actinobacteria and Acidobacteria might play an important role in keeping the health of rhizosphere soil. The study on the microbial differences between healthy and diseased crops, such as P. notoginseng (Wu et al., 2016), cherry trees (Ding et al., 2021), and banana (Fan et al., 2021), also revealed that diseased samples had higher microbial diversity than healthy samples. In this study, the results showed that both prokaryotic and fungal alpha diversities and OTU numbers in anthracnose-infected strawberry rhizosphere soils were higher than those of healthy strawberry rhizosphere soils, and our results coincide with other studies.
Rhizosphere microorganisms promote plant growth and protect plants from pathogen attack by a range of mechanisms, such as biofertilization, stimulation of root growth, rhizoremediation, control of abiotic stress, and disease control (Mendes et al., 2013). In this study, potentially beneficial bacteria in healthy strawberry rhizosphere soils were much more than in anthracnose-infected plants, and isolation of antagonistic bacteria from healthy samples may be much easier than from diseased samples. Species within Trichoderma were widely studied for biocontrol of strawberry fungal diseases, such as black root rot (Asad-Uz-Zaman et al., 2015), Armillaria root rot (Raziq and Fox, 1999, 2005; Pellegrini et al., 2014), root rot (El-Fiki and Ahmed, 2017), powdery mildew (Fiamingo et al., 2007), and strawberry anthracnose (Freeman et al., 2004; Porras et al., 2009; Karimi et al., 2017; Zhao et al., 2020). In this study, the relative abundances of Trichoderma in hRZ (1.881%) and dRZ (1.452%) were similar and did not show a significant difference (P > 0.05) (data not shown), but the beneficial fungal species T. asperellum showed a higher relative abundance in healthy strawberry rhizosphere soils. Wei et al. (2010) isolated a fungal strain MT-06 from anthracnose-infected strawberry tissues, and the strain showed an activity for biocontrol of strawberry anthracnose. In this study, species T. asperellum was identified in both healthy and diseased strawberry rhizosphere soils, that is, antagonists may be isolated not only from healthy samples but also from diseased samples.
For the sustainable development of the strawberry planting industry, disease biocontrol will be the main research direction in future. Our current research has laid a preliminary foundation for the isolation and identification of strawberry antagonists. Previous studies showed that species within Agrobacterium, Arthrobacter, Azoarcus, Azotobacter, Azospirillum, Bacillus, Burkholderia, Caulobacter, Chromobacterium, Enterobacter, Erwinia, Flavobacterium, Klebsiella, Micrococcous, Rhizobium, Pantoea, Pseudomonas, Microbacterium, Serratia, Paenibacillus, and Streptomyces had the potential roles in promoting plant growth and inhibiting different fungal pathogens (Etcheverry et al., 2009; Ahemad and Kibret, 2014; Bruto et al., 2014; Jha and Saraf, 2015; He et al., 2020). Patel et al. (2021) used the cultured method to select growth promotion isolates from peanut rhizosphere soil, and Burkholderia sp. RR18 was the most effective isolate; RR18 had activities, such as reduced collar rot disease incidence, increased germination rate and biomass of peanut seeds, and increased broad-spectrum antifungal activity. Li et al. (2021c) study revealed that after Astragalus mongholicus root infected by F. oxysporum, the plant roots can recruit some beneficial microbes, including Pseudomonas, Strenotrophomonas, Chryseobacterium, Achromobacter, and Flavobacterium. Wu et al. (2021) investigated the differences between Chinese wheat yellow mosaic resistant and susceptible wheat root endosphere and rhizosphere microbial, and the results revealed that beneficial rhizosphere microbes, such as Xanthomonadales, Actinomycetales, Sphingomonas, Rhizobium, Bacillaceae, Bacillus, Streptomycetaceae, Streptomyces, Nocardioides, Pseudonocardia, Bradyrhizobium, Pseudonocardiaceae, and Solibacteraceae, were enriched in the resistant wheat root. Some studies revealed that potential beneficial microbes, such as Bacillus, Paenibacillus, Streptomyces, Pseudomonas, Flavobacterium, Arthrobacter, Rhizobium, Microbacterium, Micrococcous, and Burkholderia, can establish disease suppression by a single strain or SynComs (synthetic microbial communities) via an antagonistic effect to protect the host from pathogens' invasion and attack (Weller et al., 2002; Lee et al., 2021; Li et al., 2021c; Marín et al., 2021). In this study, Streptomyces, Azospirillum, and Bacillus were identified as the potentially beneficial bacterial genera; some studies show that species within these genera can be used to biocontrol strawberry anthracnose; other beneficial microbes, such as Chromobacterium, Caulobacter, Paenibacillus, Enterobacter, Arthrobacter, Azoarcus, Microbacterium, Pantoea, Flavobacterium, and Pseudomonas, were also detected in rhizosphere soils, and their relative abundances were changed after plants were infected by pathogens. We speculated that when strawberry plants were invaded by pathogens, on the one hand, the rhizosphere soil microbial community was unbalanced and showed symptoms, on the other hand, plants will recruit probiotic groups to resist the further expansion of pathogens, so the relative abundances of beneficial microbes, such as Chromobacterium, Caulobacter, Azoarcus, Pantoea, Flavobacterium, and Pseudomonas, were higher in diseased samples than in healthy samples, and meanwhile, species within these groups may be the main source of probiotic bacteria. However, to determine whether these beneficial microbes can protect strawberry plants from pathogens infection, they need to be further isolated and tested in the laboratory.
When we collected strawberry rhizosphere soil samples, angular leaf spot-like symptoms were observed on plants from this strawberry-planted field. The angular leaf spot pathogen X. fragariae was not identified in this study, and the relative abundance of Xanthomonas was similar in the two soil samples; other bacterial pathogens, such as P. solanacearum, R. fascians, and A. fragariae, were still not identified. Some studies revealed that species within Colletotrichum could not only cause strawberry anthracnose but also cause other fungal diseases, such as crown and root rot (Han et al., 2014; Zhang et al., 2015). Referring to the APS, eight of 15 potential fungal pathogen genera were identified and showed significant differences between healthy and diseased strawberry rhizosphere soils in this study. Among seven potential fungal pathogen species, the relative abundance of F. oxysporum was significantly higher in diseased strawberry rhizosphere soils. The sampling field has planted strawberries for 4 years, soils maybe contained spores of pathogenic fungi, and the fungal pathogen species were identified in this study. Based on the above research, we can speculate that strawberries planted in this area were at risk of other diseases besides strawberry anthracnose.
In this study, Illumina sequencing was used to comparatively study the rhizosphere soil microbial communities between healthy and anthracnose-infected strawberry plants. Both rhizosphere soil community diversities and richness of the anthracnose-infected strawberry rhizosphere soils were higher than those of the healthy plants. When strawberry plants were infected, the diversities and compositions of rhizosphere soil microbial community were changed. Based on this, the potential probiotics and pathogens in the samples were comparatively analyzed, the potential probiotic abundances in healthy samples were higher than in diseased samples, and the changes of pathogens did not show a regularity. The results of this article will help researchers to carry out culture-dependent work and provide a basis for the isolation and identification of strawberry pathogens and probiotics.
Raw sequence data reported in this paper has been deposited to the National Center for Biotechnology Information (NCBI) under accession number PRJNA802716 (16S rRNA) and PRJNA802988 (ITS).
DS, SC, JY, JT, and XC planned and designed the research and performed the experiments. DS, SC, WZ, JY, ZL, ZZ, YT, QD, XS, and SW performed the experiments. DS, SC, WZ, and JY analyzed the data. DS, JT, and XC wrote the manuscript. SW, JT, and XC acquired the funds for the study. All authors read and improved the manuscript and agreed to the publication of the final manuscript.
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 31960220 and 31660089).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.881450/full#supplementary-material
Abarenkov, K., Nilsson, R. H., Larsson, K. H., Alexander, I. J., Eberhardt, U., Erland, S., et al. (2010). The UNITE database for molecular identification of fungi - recent updates and future perspectives. New Phytol. 186, 281–285. doi: 10.1111/j.1469-8137.2009.03160.x
Adhikari, T. B., Chacon, J. G., Fernandez, G. E., and Louws, G. J. (2019). First report of anthracnose causing both crown and fruit rot of strawberry by Colletotrichum siamense in North Carolina. Plant Dis. 103, 1775–1775. doi: 10.1094/PDIS-02-19-0314-PDN
Ahemad, M., and Kibret, M. (2014). Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J. King Saud Univ. Sci. 26, 1–20. doi: 10.1016/j.jksus.2013.05.001
Amann, R. I., Ludwig, W., and Schleifer, K. H. (1995). Phylogenetic identification of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169. doi: 10.1128/mr.59.1.143-169.1995
Asad-Uz-Zaman, M., Bhuiyan, M. R., Khan, M. A. I., Bhuiyan, M. K. I., and Latif, M. A. (2015). Integrated options for the management of black root rot of strawberry caused by Rhizoctonia solani Kuhn. C. R. Biol. 338, 112–120. doi: 10.1016/j.crvi.2014.11.006
Bailey, K. L., and Lazarovits, G. (2003). Suppressing soil-borne diseases with residue management and organic amendments. Soil Tillage Res. 72, 169–180. doi: 10.1016/S0167-1987(03)00086-2
Bi, Y., Guo, W., Zhang, G. J., Liu, S. C., and Chen, Y. (2017). First report of Colletotrichum truncatum causing anthracnose of strawberry in China. Plant Dis. 101, 832–832. doi: 10.1094/PDIS-07-16-1036-PDN
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinf. 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bowya, T., and Balachandar, D. (2020). Rhizosphere engineering through exogenous growth-regulating small molecules improves the colonizing efficiency of a plant growth-promoting rhizobacterium in rice. 3 Biotech 10:277. doi: 10.1007/s13205-020-02275-5
Brooks, A. N. (1931). Anthracnose of strawberry caused by Colletotrichum fragariae. Phytopathol. 21, 739–744.
Bruto, M., Prigent-Combaret, C., Muller, D., and Moënne-Loccoz, Y. (2014). Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related Proteobacteria. Sci. Rep. 4:6261. doi: 10.1038/srep06261
Chen, T., Nomura, K., Wang, X. L., Sohrabi, R., Xu, J., Xu, L. Y., et al. (2020). A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 580, 653–657. doi: 10.1038/s41586-020-2185-0
Cheng, X. F., Zhang, M., and Adhikari, B. (2014). Effect of ultrasonically induced nucleation on the drying kinetics and physical properties of freeze-dried strawberry. Drying Technol. 32, 1857–1864. doi: 10.1080/07373937.2014.952741
Choi, K. H., and Dobbs, F. C. (1999). Comparison of two kinds of biolog microplates (GN and ECO) in their ability to distinguish among aquatic microbial communities. J. Microbiol. Methods 36, 203–213. doi: 10.1016/S0167-7012(99)00034-2
Coleman-Derr, D., Desgarennes, D., Fonseca-Garcia, C., Gorss, S., Clingenpeel, S., Woyke, T., et al. (2016). Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 209, 798–811. doi: 10.1111/nph.13697
Cui, Y. X., Bing, H. J., Fang, L. C., Wu, Y. H., Yu, J. L., Shen, G. T., et al. (2019). Diversity patterns of the rhizosphere and bulk soil microbial communities along an altitudinal gradient in an alpine ecosystem of the eastern Tibetan Plateau. Geoderma 338, 118–127. doi: 10.1016/j.geoderma.2018.11.047
Dai, F. M., Ren, X. J., and Lu, J. P. (2006). First report of anthracnose fruit rot of strawberry caused by Colletotrichum acutatum in China. Plant Dis. 90, 1460–1460. doi: 10.1094/PD-90-1460A
Dai, T., Chang, X. N., Hu, Z. H., Liang, L., Sun, M. Y., Liu, P. F., et al. (2019). Untargeted metabolomics based on GC-MS and chemometrics: a new tool for the early diagnosis of strawberry anthracnose caused by Colletotrichum theobromicola. Plant Dis. 103, 2541–2547. doi: 10.1094/PDIS-01-19-0219-RE
Deng, X., Li, Q. F., Wu, C. Y., Li, Y., and Liu, J. K. (2015). Comparison of soil bacterial genetic diversity in root zone of banana (Musa paradisiaca) infected with Fusarium wilt and non-infected plants. Ecol. Environ. Sci. 24:402–408. doi: 10.16258/j.cnki.1674-5906.2015.03.006
Denoyes-Rothan, B., Guérin, G., Délye, C., Smith, B., Minz, D., Maymon, M., et al. (2003). Genetic diversity and pathogenic variability among isolates of Colletotrichum species from strawberry. Phytopathology. 93, 219–228. doi: 10.1094/PHYTO.2003.93.2.219
Desgarennes, D., Garrido, E., Torres-Gomez, M. J., Peña-Cabriales, J. J., and Partida-Martinez, L. P. (2014). Diazotrophic potential among bacterial communities associated with wild and cultivated Agave species. FEMS Microbiol. Ecol. 90, 844–857. doi: 10.1111/1574-6941.12438
Ding, L., Xu, L. J., Chu, X., Yang, L., Zhu, H. L., and Huang, J. X. (2021). Dissimilarity analysis of microbial communities in the rhizosphere and tissues of diseased and healthy cherry trees (Cerasus pseudocerasus). Can. J. Plant Pathol. 43, 612–621. doi: 10.1080/07060661.2020.1861101
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 112, E911–E920. doi: 10.1073/pnas.1414592112
El-Fiki, I. A. I., and Ahmed, M. F. A. (2017). Effect of biological control of root rot diseases of strawberry using Trichoderma spp. Res. J. Appl. Sci. 7, 482–492.
Etcheverry, M. G., Scandolara, A., Nesci, A., Ribeiro, M. S. V. B., Pereira, P., and Battilani, P. (2009). Biological interactions to select biocontrol agents against toxigenic strains of Aspergillus flavus and Fusarium verticillioides from Maize. Mycopathologia 167, 287–295. doi: 10.1007/s11046-008-9177-1
Fan, H. C., Wei, W., Zeng, L., Xu, S. T., Li, S., Guo, Z. X., et al. (2021). Comparative analysis on difference of bacterial community structure in rhizosphere soil between banana Fusarium wilt and healthy plants. Southwest China J. Agric. Sci. 24, 1885–1891. doi: 10.16213/j.cnki.scjas.2021.9.011
Feng, Y. X., Hu, Y. Y., Wu, J. S., Chen, J. H., Yrjälä, K., and Yu, W. W. (2018). Change in microbial communities, soil enzyme and metabolic activity in a Torreya grandis plantation in response to root rot disease. For. Ecol. Manage. 432, 932–941. doi: 10.1016/j.foreco.2018.10.028
Fiamingo, F., Elad, Y., and Pertot, I. (2007). Effect of application time of control agents on Podosphaera aphanis and side effect of fungicides on biocontrol agents survival on strawberry leaves. pp. 433–436.
Freeman, S., and Katan, T. (1997). Identification of Colletotrichum species responsible for anthracnose and root necrosis of strawberry in Israel. Phytopathol. 87, 516–521. doi: 10.1094/PHYTO.1997.87.5.516
Freeman, S., Minz, D., Kolesnik, I., Barbul, O., Zveibil, A., Maymon, M., et al. (2004). Trichoderma biocontrol of Colletotrichum acutatum and Botrytis cinerea and survival in strawberry. Eur. J. Plant Pathol. 110, 361–370. doi: 10.1023/B:EJPP.0000021057.93305.d9
Garrido, C., Carbú, M., Fernández-Acero, F. J., Budge, G. E., Vallejo, I., Colyer, A., et al. (2008). Isolation and pathogenicity of Colletotrichum spp. causing anthracnose of strawberry in south west Spain. Eur. J. Plant Pathol. 120, 409–415. doi: 10.1007/s10658-007-9224-7
Garrido, C., Carbú, M., Fernández-Acero, F. J., González-Rodríguez, V. E., and Cantoral, J. M. (2011). New insights in the study of strawberry fungal pathogens. Genes Genomes Genomics 5, 24–39.
Gorissen, A., van Overbeek, L. S., and van Elsas, J. D. (2004). Pig slurry reduces the survival of Ralstonia solanacearum biovar 2 in soil. Can. J. Microbiol. 50, 587–593. doi: 10.1139/w04-042
Hamel, C., Vujanovic, V., Jeannotte, R., Nakano-Hylander, A., and St-Arnaud, M. (2005). Negative feedback on a perennial crop: Fusarium crown and root rot of asparagus is related to changes in soil microbial community structure. Plant Soil 268, 75–87. doi: 10.1007/s11104-004-0228-1
Han, Y. C., Xiang, F. Y., Zeng, X. G., Zhang, P., and Gu, Y. C. (2014). Identification of pathogen causing crown and root rot on strawberry. Sci. Agric. Sin. 47, 53–60. doi: 10.3864/j.issn.0578-1752.2014.01.006
Haney, C. H., Samuel, B. S., Bush, J., and Ausubel, F. M. (2015). Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat. Plants 1:15051. doi: 10.1038/nplants.2015.51
He, Z. D., Gao, Y. F., Wang, Y., Li, C. X., Gao, X. Y., and Zhang, Z. H. (2020). Study on phosphate-solubilizing strain selection of plant growth promoting rhizobacteria and its effect on tomato growth promotion. Southwest China J. Agric. Sci. 33, 2891–2896. doi: 10.16213/j.cnki.scjas.2020.12.030
Hoseini, S., Amini, J., Nazemi-Rafie, J., and Khorshidi, J. (2019). Inhibitory effect of some plant essential oils against strawberry anthracnose caused by Colletotrichum nymphaeae under in vitro and in vivo conditions. Eur. J. Plant Pathol. 155, 1287–1302. doi: 10.1007/s10658-019-01856-2
Husaini, A. M., and Neri, D. (2016). Strawberry Growth, Development and Diseases. CAB International. doi: 10.1079/9781780646633.0000
Jayawardena, R. S., Huang, J. K., Jin, B. C., Yan, J. Y., Li, X. H., Hyde, K. D., et al. (2016). An account of Colletotrichum species associated with strawberry anthracnose in China based on morphology and molecular data. Mycosphere 7, 1147–1163. doi: 10.5943/mycosphere/si/2c/6
Jha, C. K., and Saraf, M. (2015). Plant growth promoting rhizobacteria (PGPR): a review. E3 J. Agric. Res. Dev. 5, 108–119. doi: 10.13140/RG.2.1.5171.2164
Karimi, K., Ahari, A. B., Arzanlou, M., Amini, J., and Pertot, I. (2017). Comparison of indigenous Trichoderma spp. strains to a foreign commercial strain in terms of biocontrol efficacy against Colletotrichum nymphaeae and related biological features. J. Plant Dis. Prot. 124, 453–466. doi: 10.1007/s41348-017-0088-6
Karimi, K., Babai-Ahari, A., and Arzanlou, M. (2015). Strawberry anthracnose disease. Plant Pathol. Sci. 4, 26–40.
Lai, B. C., Dai, R. Q., Wu, Z. Q., Li, F., Lin, D. F., and Wang, J. R. (2019). Bacterial diversities in rhizosphere soils at sites of healthy and Fusarium wilt infected chili plants. Fujian J. Agric. Sci. 34, 1073–1080. doi: 10.19303/j.issn.1008-0384.2019.09.012
Lee, S. M., Kong, H. G., Song, G. C., and Ryu, C. M. (2021). Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. ISME J. 15, 330–347. doi: 10.1038/s41396-020-00785-x
Li, J. Y., Zhao, Q. Q., Wuriyanghan, H. D., and Yang, C. (2021a). Biocontrol bacteria strains Y4 and Y8 alleviate tobacco bacterial wilt disease by altering their rhizosphere soil bacteria community. Rhizosphere 19:100390. doi: 10.1016/j.rhisph.2021.100390
Li, X. G., Panke-Buisse, K., Yao, X. D., Coleman-Derr, D., Ding, C. F., Wang, X. X., et al. (2020). Peanut plant growth was altered by monocropping-associated microbial enrichment of rhizosphere microbiome. Plant Soil 446, 655–669. doi: 10.1007/s11104-019-04379-1
Li, Y. L., He, F., Guo, Q., Feng, Z. Y., Zhang, M., Ji, C. L., et al. (2021b). Compositional and functional comparison on the rhizosphere microbial community between healthy and Sclerotium rolfsii-infected monkshood (Aconitum carmichaelii) revealed the biocontrol potential of healthy monkshood rhizosphere microorganisms. Biol. Control 165:104790. doi: 10.1016/j.biocontrol.2021.104790
Li, Z. F., Bai, X. L., Jiao, S., Li, Y. M., Li, P. R., Yang, Y., et al. (2021c). A simplified synthetic community rescues Astragalus mongholicus from root rot disease by activating plant-induced systemic resistance. Microbiome 9:217. doi: 10.1186/s40168-021-01169-9
Liao, C. Y., Chen, M. Y., Chen, Y. K., Wang, T. C., Sheu, Z. M., Kuo, K. C., et al. (2012). Characterization of three Colletotrichum acutatum isolates from Capsicum spp. Eur. J. Plant Pathol. 133, 599–608. doi: 10.1007/s10658-011-9935-7
Liu, H. Y., Wang, W., Zhang, R. F., Abdurahman, R., and Yao, J. (2019). Fungal community structure of cotton-field soil under different incidences of cotton Verticillium wilt. Sci. Agric. Sin. 52, 455–465. doi: 10.3864/j.issn.0578-1752.2019.03.006
Liu, Y., Ji, Y., Han, Y. C., Song, L. L., Zhang, L. Q., Ning, Z. Y., et al. (2021). Loop-mediated isothermal amplification and PCR combined assay to detect and distinguish latent Colletotrichum spp. infection on strawberry. J. Plant Pathol. 103, 887–99. doi: 10.1007/s42161-021-00873-7
Lu, X., Chen, L. H., and Li, Y. R. (2012). Effect of DNA extraction on DGGE analysis of microbial community in soil. Fujian J. Agric. Sci. 27, 367–372. doi: 10.19303/j.issn.1008-0384.2012.04.011
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Marian, M., Ohno, T., Suzuki, H., Kitamura, H., Kuroda, K., and Shimizu, M. (2020). A novel strain of endophytic Streptomyces for the biocontrol of strawberry anthracnose caused by Glomerella cingulate. Microbiol. Res. 234:126428. doi: 10.1016/j.micres.2020.126428
Marín, O., González, B., and Poupin, M. J. (2021). From microbial dynamics to functionality in the rhizosphere: a systematic review of the opportunities with synthetic microbial communities. Front. Plant Sci. 12:650609. doi: 10.3389/fpls.2021.650609
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Mertely, J. C., and Legard, D. E. (2004). Detection, isolation, and pathogenicity of Colletotrichum spp. from strawberry petioles. Plant Dis. 88, 407–418. doi: 10.1094/PDIS.2004.88.4.407
Mochizuki, M., Yamamoto, S., Aoki, Y., and Suzuki, S. (2012). Isolation and characterisation of Bacillus amyloliquefaciens S13-3 as a biological control agent for anthracnose caused by Colletotrichum gloeosporioides. Biocontrol Sci. Technol. 22, 697–709. doi: 10.1080/09583157.2012.679644
Nguyen, N. H., Song, Z. W., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Pantigoso, H. A., Manter, D. K., and Vivanco, J. M. (2020). Differential effects of phosphorus fertilization on plant uptake and rhizosphere microbiome of cultivated and non-cultivated potatoes. Microb. Ecol. 80, 169–180. doi: 10.1007/s00248-020-01486-w
Patel, R. R., Patel, D. D., Bhatt, J., Thakor, P., Triplett, L. R., and Thakkar, V. R. (2021). Induction of pre-chorismate, jasmonate, and salicylate pathways by Burkholderia sp. RR18 in peanut seedlings. J. Appl. Microbiol. 131, 1417–1430. doi: 10.1111/jam.15019
Pellegrini, A., Prodorutti, D., and Pertot, I. (2014). Use of bark mulch pre-inoculated with Trichoderma atroviride to control Armillaria root rot. Crop Prot. 64, 104–109. doi: 10.1016/j.cropro.2014.06.007
Peres, N. A., Timmer, L. W., Adaskaveg, J. E., and Correll, J. C. (2005). Lifestyles of Colletotrichum acutatum. Plant Dis. 89, 784–796. doi: 10.1094/PD-89-0784
Pérez-Jaramillo, J. E., Mendes, R., and Raaijmakers, J. M. (2016). Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 90, 635–644. doi: 10.1007/s11103-015-0337-7
Phoulivong, S., Cai, L., Chen, H., McKenzie, E. H. C., Abdelsalam, K., Chukeatirote, E., et al. (2010). Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Divers. 44, 33–43. doi: 10.1007/s13225-010-0046-0
Porras, M., Barrau, C., and Romero, F. (2009). Biological control of anthracnose with Trichoderma in strawberry fields. Acta Hortic. 842, 351–354. doi: 10.17660/ActaHortic.2009.842.66
Qiu, M. H., Zhang, R. F., Xue, C., Zhang, S. S., Li, S. Q., Zhang, N., et al. (2012). Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol. Fertil. Soils 48, 807–816. doi: 10.1007/s00374-012-0675-4
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Raziq, F., and Fox, R. T. V. (1999). Biological control of Armillaria root rot. Acta Hortic. 496, 115–126. doi: 10.17660/ActaHortic.1999.496.14
Raziq, F., and Fox, R. T. V. (2005). Combinations of fungal antagonists for biological control of Armillaria root rot of strawberry plants. Biol. Agric. Hortic. 23, 45–57. doi: 10.1080/01448765.2005.9755307
Reddy, P. P. (2016). Sustainable crop protection under protected cultivation. Springer. doi: 10.1007/978-981-287-952-3
Roose-Amsaleg, C. L., Garnier-Sillam, E., and Harry, M. (2001). Extraction and purification of microbial DNA from soil and sediment samples. Appl. Soil. Ecol. 18, 47–60. doi: 10.1016/S0929-1393(01)00149-4
Rosenzweig, N., Tiedje, J. M., Quensen, J. F., Meng, Q. X., and Hao, J. J. J. (2012). Microbial communities associated with potato common scab-suppressive soil determined by pyrosequencing analyses. Plant Dis. 96, 718–725. doi: 10.1094/PDIS-07-11-0571
Singh, B., Singh, S. K., Agarwal, P. C., Rani, I., and Khetarpal, R. K. (2003). Colletotrichum dematium causing anthracnose in hybrid strawberry (Fragaria × ananassa)-a new host record for India. Indian J. Agric. Sci. 73, 238–239.
Su, D. F., Tong, J. Y., Yang, J. Y., Chen, S. Y., Luo, Z. W., Shen, X. M., et al. (2018). Advances in research, exploitation and utilization of Fragaria spp. germplasm resources in China. J. Yunnan Univ. 40, 1261–1276. 10.7540/j.ynu.20180613
Sun, B., Liu, J., Meng, B., and Bao, K. (2020). Structural variability and co-occurrence pattern differentiation in rhizosphere microbiomes of the native invasive plant Echinochloa caudate in Momoge National Nature Reserve, China. Wetlands 40, 587–597. doi: 10.1007/s13157-019-01209-z
Tedersoo, L., Anslan, S., Bahram, M., Põlme, S., Riit, T., Liiv, I., et al. (2015). Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 10, 1–43. doi: 10.3897/mycokeys.10.4852
Tortora, M. L., Díaz-Ricci, J. C., and Pedraza, R. O. (2011). Azospirillum brasilense siderophores with antifungal activity against Colletotrichum acutatum. Arch. Microbiol. 193, 275–286. doi: 10.1007/s00203-010-0672-7
Wang, Y. T., Fu, L. N., Ji, G. H., Yang, J., Wang, X., Zhang, J. H., et al. (2019). A study of the microbial community diversity of corn rhizosphere in Yunnan province based on high-throughput sequencing technique. Acta Agric. Univ. Jiangxiensis 41, 491–500. doi: 10.13836/j.jjau.2019058
Wei, C. Y., Mao, X. Q., Cai, R. Y., Wang, Y. L., Wang, J. Y., Zhang, Z., et al. (2010). Identification and characterization of the fungal strain MT-06 for biocontrol of strawberry anthracnose. Mycosystema 29, 481–487. doi: 10.13346/j.mycosystema.2010.04.017
Weller, D. M., Raaijmakers, J. M., Gardener, B. B. M., and Thomashow, L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40, 309–348. doi: 10.1146/annurev.phyto.40.030402.110010
Wu, C. F., Wang, F. Y., Zhang, H. Q., Chen, G. X., Deng, Y. W., Chen, J. P., et al. (2021). Enrichment of beneficial rhizosphere microbes in Chinese wheat yellow mosaic virus-resistant cultivars. Appl. Microbiol. Biotechnol. 105, 9371–9383. doi: 10.1007/s00253-021-11666-4
Wu, Z. X., Hao, Z. P., Sun, Y. Q., Guo, L. P., Huang, L. Q., Zeng, Y., et al. (2016). Comparison on the structure and function of the rhizosphere microbial community between healthy and root-rot Panax notoginseng. Appl. soil Ecol. 107, 99–107. doi: 10.1016/j.apsoil.2016.05.017
Wu, Z. X., Hao, Z. P., Zeng, Y., Guo, L. P., Huang, L. Q., and Chen, B. D. (2015). Molecular characterization of microbial communities in the rhizosphere soils and roots of diseased and healthy Panax notoginseng. Antonie van Leeuwenhoek 108, 1059–1074. doi: 10.1007/s10482-015-0560-x
Xiang, L. G., Wang, H. C., Guo, Z. N., Xie, H. L., Cai, L. T., and Yu, Z. H. (2019). The influence of black shank disease infection on fungal community structure of rhizosphere soil and stem of tobacco plants. Mycosystema 38, 2099–2111. doi: 10.13346/j.mycosystema.190362
Xiao, R., Cao, Q. F., Nie, Y. J., Zhang, C. F., Deng, S., Sun, H. F., et al. (2017). A comparative study on rhizosphere soil bacterial communities of healthy strawberry and infected strawberry with anthracnose by high-throughput sequencing. Chin. Agric. Sci. Bull. 33, 14–20.
Yang, G. Z., Huang, W. J., Zheng, L. P., He, Y. Y., Zhang, Y., Kong, B. H., et al. (2021). Bacterial community structure and diversity of rhizosphere soil of healthy and root rot apple based on high-throughput sequencing. Southwest China J. Agric. Sci. 34, 1865–1869. doi: 10.16213/j.cnki.scjas.2021.9.008
Yang, J. Y., Wei, S. J., Su, D. F., Zhang, Z. R., Chen, S. Y., Luo, Z. W., et al. (2020a). Comparison of the rhizosphere soil microbial community structure and diversity between powdery mildew-infected and noninfected strawberry plants in a greenhouse by high-throughput sequencing technology. Curr. Microbiol. 77, 1724–1736. doi: 10.1007/s00284-020-01948-x
Yang, J. Y., Wei, S. J., Su, D. F., Zhang, Z. R., Chen, S. Y., Luo, Z. W., et al. (2020b). Study on prokaryotic communities in rhizospheres of powdery mildew-infected and non-infected strawberry in greenhouse by high-through sequencing technology. Southwest China J. Agric. Sci. 33, 85–91. doi: 10.16213/j.cnki.scjas.2020.1.014
Yao, X. D., Wang, W., and Zeng, H. (2016). Application of phospholipid fatty acid method in analyzing soil microbial community. Microbiol. China 43, 2086–2095. doi: 10.13344/j.microbiol.china.160014
Yu, Y., Lee, C., Kim, J., and Hwang, S. (2010). Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89, 670–679. doi: 10.1002/bit.20347
Zakaria, L. (2021). Diversity of Colletotrichum species associated with anthracnose disease in tropical fruit crops-a review. Agric. 11:297. doi: 10.3390/agriculture11040297
Zhang, Y., Liu, Z. P., Wei, Y. M., Shang, Q. X., Li, Y. Q., and Zhao, X. Y. (2015). Identification of the strawberry root rot pathogen in Changping District Beijing. Chin. Agric. Sci. Bull. 31, 278–284.
Zhao, D. L., He, H. Y., Wu, S. P., Chen, X. J., Tan, Q. Q., and Yang, X. H. (2020). Biocontrol mechanisms and control effects of Trichoderma asperellum GYSW-6m1 on strawberry anthracnose and growth-promoting effects on strawberry. Chin. J. Biol. Control 36, 587–595. doi: 10.16409/j.cnki.2095-039x.2020.04.020
Zhao, F., Zhao, M. Z., Wang, Y., Guan, L., and Pang, F. H. (2019). Microbial community structures and diversities in strawberry rhizosphere soils based on high-throughput sequencing. Soils 51, 51–60. doi: 10.13758/j.cnki.tr.2019.01.008
Zhao, W. S., Guo, Q. G., Su, Z. H., Wang, P. P., Dong, L. H., Hu, Q., et al. (2021). Characterization of fungal community structure in the rhizosphere soil of healthy and diseased-Verticillium wilt potato plants and carbon source utilization. Sci. Agric. Sin. 54, 296–309. doi: 10.3864/j.issn.0578-1752.2021.02.006
Zhou, L. T., Li, J. J., Zhao, Y. L., Luo, Y., Bai, Y., Chen, J., et al. (2020). Variation of bacterial communities in the rhizosphere soils of successive rotations Casuarina equisetifolia plantations based on high-throughput sequencing analysis. Acta. Ecol. Sin. 40, 2670–2679. doi: 10.5846/stxb201902250348
Keywords: strawberry anthracnose, rhizosphere soil, microbial community, high-throughput sequencing technology, cultivated strawberry
Citation: Su D, Chen S, Zhou W, Yang J, Luo Z, Zhang Z, Tian Y, Dong Q, Shen X, Wei S, Tong J and Cui X (2022) Comparative Analysis of the Microbial Community Structures Between Healthy and Anthracnose-Infected Strawberry Rhizosphere Soils Using Illumina Sequencing Technology in Yunnan Province, Southwest of China. Front. Microbiol. 13:881450. doi: 10.3389/fmicb.2022.881450
Received: 22 February 2022; Accepted: 28 March 2022;
Published: 16 May 2022.
Edited by:
Kauser Abdulla Malik, Forman Christian College, PakistanReviewed by:
Salma Mukhtar, Connecticut Agricultural Experiment Station, United StatesCopyright © 2022 Su, Chen, Zhou, Yang, Luo, Zhang, Tian, Dong, Shen, Wei, Tong and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiangyun Tong, dGp5NjcxMjA3QDEyNi5jb20=; Xiaolong Cui, eGxjdWlAeW51LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.