- 1Hubei International Scientific and Technological Cooperation Base of Traditional Fermented Foods, Huazhong Agricultural University, Wuhan, China
- 2College of Food Science and Technology, Huazhong Agricultural University, Wuhan, China
- 3Department of Microbiology, Technical University of Munich, Freising, Germany
- 4Department of Bioscience and Biotechnology, Faculty of Agriculture, University of the Ryukyus, Okinawa, Japan
Acetic acid bacteria (AAB) are a group of Gram-negative, strictly aerobic bacteria, including 19 reported genera until 2021, which are widely found on the surface of flowers and fruits, or in traditionally fermented products. Many AAB strains have the great abilities to incompletely oxidize a large variety of carbohydrates, alcohols and related compounds to the corresponding products mainly including acetic acid, gluconic acid, gulonic acid, galactonic acid, sorbose, dihydroxyacetone and miglitol via the membrane-binding dehydrogenases, which is termed as AAB oxidative fermentation (AOF). Up to now, at least 86 AOF products have been reported in the literatures, but no any monograph or review of them has been published. In this review, at first, we briefly introduce the classification progress of AAB due to the rapid changes of AAB classification in recent years, then systematically describe the enzymes involved in AOF and classify the AOF products. Finally, we summarize the application of molecular biology technologies in AOF researches.
Introduction
Acetic acid bacteria (AAB) are a group of Gram-negative, strictly aerobic bacteria within the family Acetobacteraceae (Saichana et al., 2015), which habitat in a large variety of different sources, such as flowers or fruits (Trček and Barja, 2015), guts of some insects (Crotti et al., 2016), and various traditionally fermented foods including vinegar, lambic beer, kefir, kombucha and so on (De Roos and De Vuyst, 2018). AAB may be named after their abilities to produce acetic acid via ethanol oxidation (Nanda et al., 2001), but actually some AAB strains are unable to produce acetic acid from ethanol, such as some strains within AAB genera of Asaia (As.) and Saccharibacter (Sa.; Moore et al., 2002; Jojima et al., 2004). Meanwhile, some AAB strains can fix nitrogen (Fuentes-Ramírez et al., 2001), produce pigment (Malimas et al., 2009), or exopolysaccharide (EPS; Gullo et al., 2017; La China et al., 2018). Very importantly, many AAB strains can incompletely oxidize various carbohydrates, alcohols and related compounds to yield the corresponding industrial products such as acetic acid, gluconic acid (GA), galactonic acid, 2-keto-L-gulonic acid (2-KGA), dihydroxyacetone (DHA), miglitol and so on (Mamlouk and Gullo, 2013), which have been successfully used in foods, cosmetics, medicines and other fields (Gullo et al., 2014; Saichana et al., 2015). These partial oxidation processes of AAB are named as AAB oxidative fermentation (AOF).
In AOF, the membrane-binding dehydrogenases (mDH) localized on the periplasmic side of the cytoplasmic membrane of AAB can deprive electrons from the substrates, followed by transferring them to ubiquinone (UQ, also called as coenzyme Q), which is then reduced to ubiquinol (UQH2), and eventually, the terminal oxidases (TO) transfer the electrons from UQH2 to oxygen to produce UQ, H2O, and the energy (ATP; Adachi et al., 2003; Matsushita et al., 2016; Zhang and Chen, 2022). Therefore, besides the common electronic respiratory chain which is located in the bacterial cell membrane, another special respiratory chain (hereinafter referred to as AOF respiratory chain) also co-exist in AAB cells, and make them rapidly get the energy released from AOF (Matsushita et al., 1994, 2004; Yakushi and Matsushita, 2010). The AAB strains with oxidative fermentation capacity are called as oxidative bacteria. It is worth mentioning that the oxidative fermentation and its corresponding respiratory chain not only exist in AAB cells, but also in other aerobic bacteria such as Pseudomonas spp. and Enterobacter spp. (Matsushita et al., 2016). In Figure 1 the two respiratory chains of alcohol (ethanol) in AAB are showed.
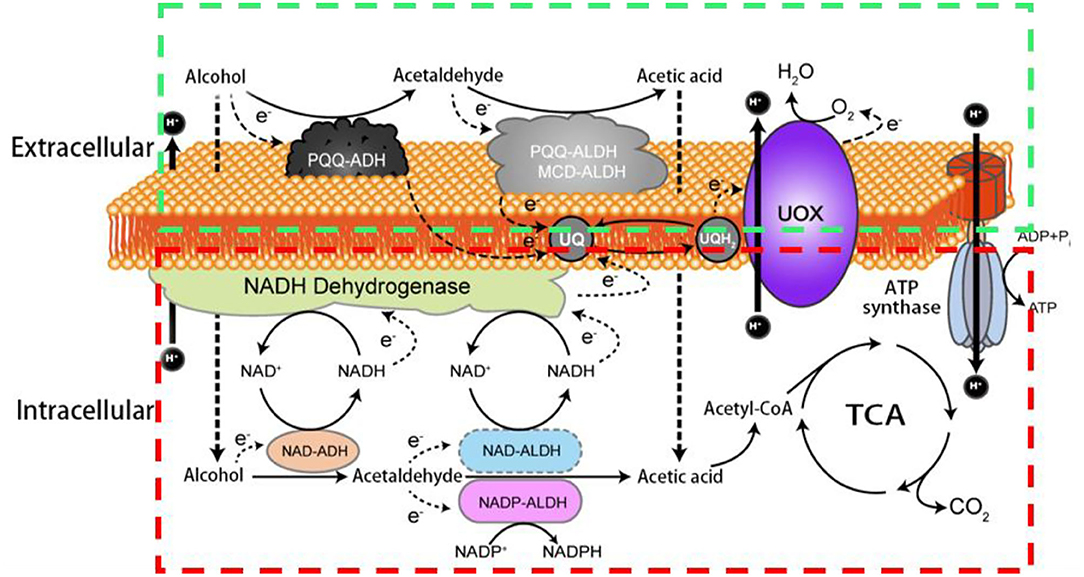
Figure 1. Schematic diagram of two respiratory chains of alcohol in acetic acid bacteria. PQQ-ADH: pyrroloquinoline quinone dependent alcohol dehydrogenase; PQQ-ALDH: pyrroloquinoline quinone dependent acetaldehyde dehydrogenase; MCD-ALDH: molybdenum-molybdopterin cytosine dinucleotide dependent acetaldehyde dehydrogenase; NAD-ADH: nicotinamide adenine dinucleotide dependent alcohol dehydrogenase; NAD- ALDH: nicotinamide adenine dinucleotide dependent acetaldehyde dehydrogenase; NADP-ALDH: nicotinamide adenine dinucleotide phosphate dependent acetaldehyde dehydrogenase; UQ: ubiquinone; UQH2: ubiquinol; UOX: ubiquinol oxidase; ATP: the energy; TCA: tricarboxylic acid cycle; The green box shows the AOF respiratory chain including PQQ-ADH, PQQ- ALDH (MCD-ALDH), UQ, UQH2; UOX and ATP synthase; the red box indicates the common respiratory chain including NAD-ADH, NAD-ALDH (NADP-ALDH), UQ, UQH2; UOX and ATP synthase.
Since AOF takes place on the periplasmic side of the cytoplasmic membrane of AAB, its products are directly released extracellular, and avoid the transport limitation from intracellular to extracellular, resulting that AAB cells are considered as very ideal biocatalysts to produce corresponding products (Matsushita et al., 2016; Sengun, 2017). In this review, we have collected 86 AOF products which have been published, and introduced the relative enzymes with AOF. In addition, we have summarized the advance of the AAB classification and molecular biotechnology.
AAB Classification Advance
The first AAB genus, Acetobacter (A.) was proposed by Beijerinck in 1898 (Wang and Chen, 2014). It was not until 37 years later (1935) that the second AAB genus, Gluconobacter (G.) was described by Asai (1968) and Yamada and Yukphan (2008). By the year of 1989, only 3 genera and 10 species of AAB were generally recognized (Wang and Chen, 2014). Since then the discovery and identification of AAB genera and species have been achieving rapid progress thanks to the development of molecular biology techniques (Wang and Chen, 2014). At the end of 2021, 19 genera and 110 species of AAB have been reported (https://lpsn.dsmz.de/; Supplementary Table 1).
The early classification and identification of AAB were mainly based on the phenotypic traits such as colony and microscopic morphologies, catalase test, Gram staining, chemotaxonomical characteristics mainly including UQ types (UQ9 or UQ10) and the profile of fatty methyl esters in AAB cells. For example, in Yamada and Kondo (1984) divided the genus of Acetobacter into two subgenera: Acetobacter subgenus with UQ9 and Gluconoacetobacter (Ga.) subgenus with UQ10, and Urakami et al. (1989) combined phenotypic characteristics, UQ types and the profile of fatty methyl esters to establish a new AAB genus—Acidomonas (Ac.), which was the third AAB genus recognized at that time.
In recent decades, with the development of molecular biology technologies, especially those related to rRNA, the AAB classification has developed very quickly, leading that some former AAB species or genera have excluded of the AAB group, meanwhile some new species and genera of AAB are proposed or independent from the former AAB (sub) species or (sub)genera. For instance, based on the phylogenic tree of 16S rRNA gene sequences, the genus Gluconacetobacter, which once belonged to the subgenus of Acetobacter, was elevated to the genus level (Yamada et al., 1997). Later, Yamada and his colleagues discovered that the 16S rRNA gene phylogenetic tree indicated two subclusters within the genus Gluconacetobacter, resulting that a new AAB genus Komagataeibacter (K.) was independent from the genus Gluconacetobacter (Yamada and Yukphan, 2008; Yamada et al., 2012a,b), and Ga. kakiaceti, Ga. Medellinesis, and Ga. maltaceti were re-classified into K. kakiaceti, K. medellinesis, and K. maltaceti, respectively (Yamada, 2014).
In Table 1, we have summarized the classification changes of AAB genera and species in recent decades.
However, only adopting the molecular biology methods related to rRNA may bring an error in AAB classification and identification. For example, according to the 16S rRNA phylogenic tree, the AAB genera of Asaia (As.), Kozakia (Ka.), Swaminathania (Sa.), and Neoasaia (N.) (Supplementary Table 1) could be categorized as one single genus, but their phenotypes warranted their distinction on the different genus level (Kersters et al., 2006). Therefore, internal transcribed spacer (ITS) of 16S-23S rRNA gene, restriction fragment length polymorphisms of genomic DNA and DNA-DNA hybridization (Janda and Abbott, 2007; Trček and Barja, 2015; Yamada, 2016) were also applied in AAB taxonomic studies. Moreover, the sequences of some genes were applied to identify AAB. For example, Trcek et al. (2006) used the nucleotide sequence of gene adhA for AAB identification, while genes of nifD and nifH were applied to identify nitrogen-fixing AAB species (Loganathan and Nair, 2004; Dutta and Gachhui, 2006). And multilocus sequence analysis of the three genes (dnaK, groEL, and rpoB) was performed to differentiate AAB species (Cleenwerck et al., 2010).
In the future, in order to obtain more objective, precise and reliable AAB classification, there is no doubt that a multidimensional method of the morphological classification combined with multiple molecular biological methods such as different genes or/ and the complete genome comparison, should be utilized for the classification and identification of AAB strains (Wang and Chen, 2014; Yamada, 2016).
AAB Oxidative Fermentation
Due to their long existence in sugar-rich environments such as fruits and flowers, some AAB strains are adaptively evolved their abilities to rapidly incompletely oxidize sugars, sugar alcohols, or/and alcohols by mDH to produce corresponding products like aldehydes, ketones, acids, and other products, and yield ATP via the AOF respiratory chain (Figure 1; Matsushita et al., 1994, 2002; Saichana et al., 2015).
In addition to AOF, AAB can also completely oxidize sugars, sugar alcohols, alcohols, organic acids, and other substances to CO2 and H2O through the Embden–Meyerhof–Parnas pathway, the tricarboxylic acid cycle, the pentose phosphate pathway, the Entner–Doudoroff pathway, and/or the glyoxylate pathway, and produce intermediates for cell growth and ATP through the common respiratory chain (Figure 1). Although both complete and incomplete oxidation systems simultaneously exist in AAB cells, usually two systems rarely show similar activities in the same growth period of AAB strains. In an environment of a high concentration of sugars, alcohols, and/or acids, AAB cells mainly carry out AOF in the early growth period and complete oxidation in the late growth period. Aldehydes, ketones, or/and acids accumulated via AOF can inhibit the growth of other microorganisms, subsequently, when substrates are almost consumed by AOF, AAB can utilize these AOF products to continue to grow through the complete oxidation, resulting in AAB cells possessing the good growth and survival competitiveness. From the view of ecological evolution, AOF should be considered as a unique characteristic for AAB to adapt to the growth and survival environments, so it leading the carbon sources used by AAB is very complex, especially when multiple carbon sources such as sugars and alcohols are simultaneously present in the media (Gupta et al., 2001; Deppenmeier et al., 2002; Adachi et al., 2003; Raspor and Goranovič, 2008; Mamlouk and Gullo, 2013; Nishikura-Imamura et al., 2014; Saichana et al., 2015). The strategies of AAB to cope with changes in their growth and survival environments (culture conditions) by adjusting AOF were summarized in the reference (Qin et al., 2022).
Since the dehydrogenases (DHs) involved in AOF are located in the cell membrane, they are called as mDHs, while DHs in the cytoplasm is called as cytoplasmic DHs (cDHs). When AAB are exposed to a high concentration of substrate such as sugars and/or alcohols, the DH activity in AAB cells is mainly reflected by mDH, whereas cDH is almost inactive, but with the decrease of concentration of substrates, the cDH activity gradually increases, while the mDH activity decreases or hardly functions (Baldrian, 2006; Hölscher and Görisch, 2006). There are great differences in the coenzymes between mDHs and cDHs, mDHs ones are quite diverse, mainly including pyrroloquinoline quinone (PQQ), molybdenum-molybdopterin cytosine dinucleotide (MCD), flavin adenine dinucleotide (FAD), nicotinamide adenine dinucleotide (NAD), or/and nicotinamide adenine dinucleotide phosphate (NADP; Table 2), while cDHs ones mainly include NAD or/and NADP (Figure 1; Matsushita et al., 1994). The electrons and protons (H+) from the substrate are deprived by both mDHs and cDHs, and transferred to UQ to produce UQH2, which is then oxidized to produce a proton potential between intracellular and extracellular by terminal oxidase (TO), thereby driving ATP synthase to yield ATP (Figure 1).
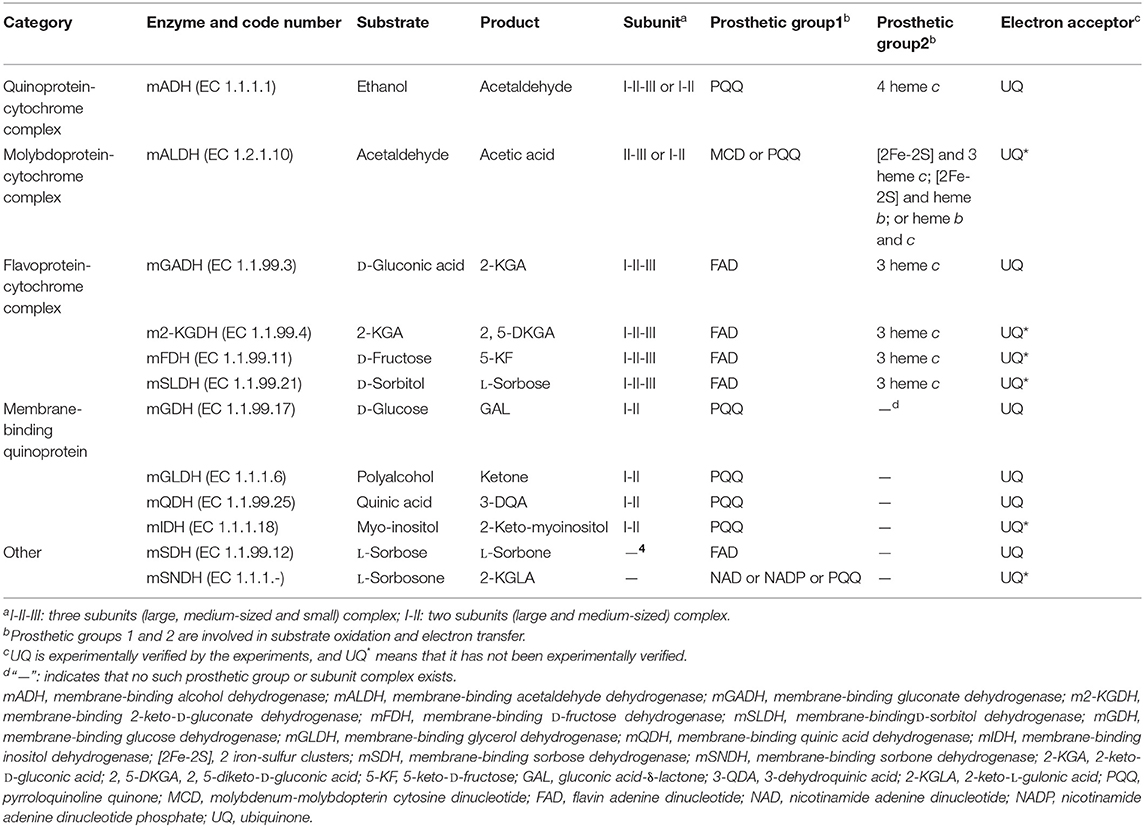
Table 2. The membrane-binding dehydrogenases in acetic acid bacteria based on their coenzyme differences.
Based on the coenzyme difference, the AAB mDH can be divided into five categories: quinoprotein-cytochrome complex, molybdoprotein-cytochrome complex, flavoprotein– cytochrome complex, quinoprotein, and others (Table 2). The mDH's types and characteristics vary in different genera, species, or strains of AAB. For example, membrane-binding alcohol dehydrogenase (mADH) from the genus Gluconobacter can oxidize ethanol into acetic acid, and can also oxidize D-glucose, GA, D-sorbitol, and glycerol into the corresponding products. In contrast, mADH from the genus Acetobacter or Komagataeibacter can only oxidize ethanol, almost impossible to oxidize other substrates (Matsushita et al., 2016).
TO is another key enzyme in the AOF respiratory chain, which can transfer electrons and protons from UQH2 to O2 to generate H2O2 or H2O. The TOs from AAB and other aerobic bacteria can be divided into heme-copper oxidase (HCO) and heme bd type oxidase (HBD-O). Among them, HCO includes cytochrome c oxidase (COX) which can accept electrons from cytochrome c, and ubiquinol oxidase (UOX) which can accept electrons from UQH2, and have binuclear O2-reducing sites consisting of heme a, o or/and b and one copper atom (Matsutani et al., 2014). Bacteria with COX are called “oxidase-positive” bacteria, such as strains from the genera of Paracoccus and Pseudomonas, while bacteria with UOX are called “oxidase-negative” bacteria, such as Escherichia coli and AAB strains. In addition, HBD-O of AAB is also one kind of UOX that can accept electrons from UQH2, but its oxygen reduction site contains heme b and d, which has a strong affinity for oxygen and can perform aerobic respiration under low oxygen conditions. Moreover, HBD-O has no proton pump function but has a certain proton release capacity that can produce part of the proton potential. In a word, the TOs of AAB mainly include UOX and HBD-O (Qin et al., 2022).
Membrane-Binding Dehydrogenase (mDH) in AOF
mDHs involved in the AOF drive functions are usually present in the form of heterotrimer or heterodimer. The structures and functions for some mDHs from AAB cells that have been clearly studied at present are shown in Figure 2.
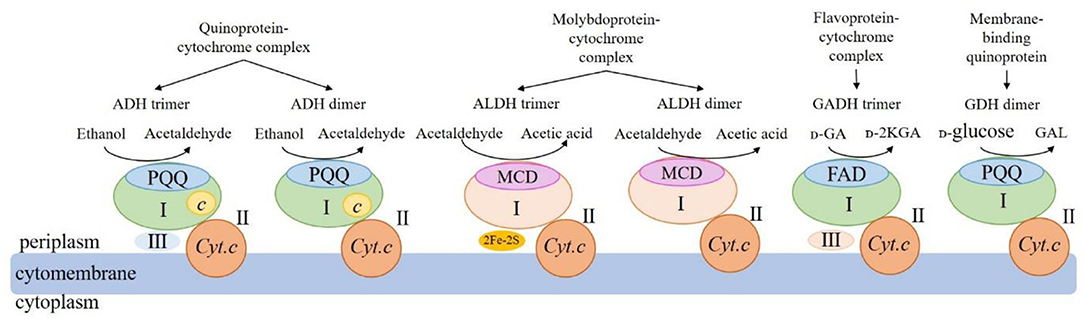
Figure 2. Schematic of structures and functions of the four main membrane-binding dehydrogenases from acetic acid bacteria. ADH: membrane-binding alcohol dehydrogenase; ALDH: membrane-binding acetaldehyde dehydrogenase; GADH: membrane-binding gluconate dehydrogenase; GDH: membrane-binding glucose dehydrogenase; PQQ: pyrroloquinoline quinone; MCD: molybdenum-molybdopterin cytosine dinucleotide; FAD: flavin adenine dinucleotide; c: heme c; Cyt.c: cytochrome c; UQ: ubiquinone; UQH2: reduced ubiquinone; 2Fe-2S: 2 iron-sulfur clusters; I: large subunits; II: medium-size subunits; III: small subunits; GA: gluconic acid; 2-KGA: 2-keto-D-gluconic acid; GAL: gluconic acid-ä-lactone.
Quinoprotein–Cytochrome Complex
The mDH of quinone protein-cytochrome complex clearly studied at present is mADH [EC.1.1.1.1], which is a constitutive ethanol ubiquinone oxidoreductase, and can catalyze the oxidation of ethanol to aldehyde and the reduction of UQ to UQH2 (Table 2). The mADH isolated from the cell membrane of Gluconobacter spp. consists of three subunits: the large subunit (I) containing one PQQ and a heme c binding-sites, respectively, can oxidize ethanol to acetaldehyde; the cytochrome c subunit (II) including three heme c binding sites, can reduce UQ to UQH2; and the small subunit (III) without any coenzyme binding site may be involved in cell membrane binding (Adachi et al., 2007; Masud et al., 2010). However, mADHs from K. europaeus and A. peroxydans (currently A. pasteurianus) contain only subunits I and II, no subunit III (Tayama et al., 1989; Trcek et al., 2006). When the dissociation of subunits I and II, the mADH enzyme activity of subunit I decrease significantly, but its enzyme activity is recovered after it re-combinates with subunit II, indicating that the complex of subunits I and II is necessary to maintain the mADH activity. The mADH contains a high-affinity UQ binding site and a catalytic site for the UQ oxidation-reduction enzyme activity, and UQ is involved in electron transfer among heme c, UQ, and UQH2. In summary, based on the current research results, the AAB mADH is a heterotrimer (I-II-III) or a dimer (I-II) membrane-binding quinoprotein-cytochrome complex and includes prosthetic groups: PQQ and heme c (Table 2 and Figure 2).
The substrate specificity of AAB mADH usually is very poor. Except for methanol, short-chain alcohols such as ethanol, 1-propanol, 1-butanol, 1-pentanol, and 1-hexanol can be utilized as its substrates (Shinagawa et al., 2006). In addition, mADH can also produce glyceraldehyde from glycerol, although it only has a low affinity for glycerol. Especially when the glycerol concentration is greater than 10% (W/V), its ability to oxidize glycerol is significantly improved (Habe et al., 2009). Moreover, aldehydes can be applied as substrates of mADH, too, and their oxidation rates by mADHs are almost same as those of the corresponding alcohols (Gómez-Manzo et al., 2008). Therefore, mADH alone can carry out the entire oxidation process from ethanol to acetaldehyde to acetic acid without the involvement of membrane-binding acetaldehyde dehydrogenase (mALDH) [EC 1.2.1.10] (Gómez-Manzo et al., 2015). In addition, although mADH cannot oxidize methanol, it can oxidize formaldehyde so that mADH can be developed as a formaldehyde scavenger (Shinagawa et al., 2006).
Molybdoprotein–Cytochrome Complex
The well-studied molybdoprotein-cytochrome complex mDH at present is mALDH, which is an acetaldehyde ubiquinone oxidoreductase. It is generally considered to be a heterotrimer (I-II-III) consisting of a large subunit (I) with MCD as one coenzyme, a medium-size subunit (II) with three heme c as prosthetic groups, and a small subunit (III) with two iron-sulfur clusters [2Fe-2S] as prosthetic groups (Table 2 and Figure 2). However, mALDHs are very different in various AAB strains. For instance, in term of the subunit composition, mALDHs from both A. aceti and K. europaeus are heterotrimers (I-II-III), while the mALDH from A. peroxydans (currently A.pasteurianus) is a heterodimer (I-II; Gómez-Manzo et al., 2010). In term of the prosthetic groups, the mALDH ones from K. europaeus include heme b, [2Fe-2S] cluster and MCD, while the mALDH ones from Ga. diazotrophicus include PQQ, heme b and c (Thurner et al., 1997; Gómez-Manzo et al., 2010).
As with mADH, mALDH possesses a poor substrate specificity, which can oxidize acetaldehyde, 1-propionaldehyde, 1-butyraldehyde, isobutyraldehyde, glutaraldehyde, and other major short-chain aldehydes except for formaldehyde (Toyama et al., 2007).
Flavoprotein–Cytochrome Complex
The flavoprotein-cytochrome complex mDH of AAB mainly includes membrane-binding gluconate dehydrogenase (mGADH) [EC 1.1.99.3], 2-keto-D-gluconate dehydrogenase (m2-KGDH) [EC 1.1.99.4], D-fructose dehydrogenase (mFDH) [EC 1.1.99.11], and D-sorbitol dehydrogenase (mSLDH) [EC 1.1.99.21] (Toyama et al., 2005; Kawai et al., 2013; Kataoka et al., 2015). These mDHs generally consist of a large subunit (I) with FAD as a coenzyme, a medium-size subunit (II) containing three heme c prosthetic groups, and a small subunit (III) with unknown function (Table 2 and Figure 2; Toyama et al., 2007).
mGADH, m2-KGDH, mFDH, and mSLDH are all UQ oxidoreductases with high substrate specificity. mGADH is also called GA 2-DH because it can oxidize the C-2 hydroxyl group of GA to produce 2-KGA. mGADH can only oxidize GA, and m2-KGDH can only oxidize 2-KGA to produce 2, 5-DKGA. mFDH can just oxidize fructose to produce 5-KF, which can be used as a biometric recognition molecule of the fructose biosensor. mSLDH can oxidize D-sorbitol to L-sorbose, and also weakly oxidize D-mannitol, whereas pentitol and erythritol are not oxidized by mSLDH (Shinagawa et al., 1982, 1984).
Membrane-Binding Quinoprotein
The membrane-binding quinoprotein AAB mDHs mainly include glucose dehydrogenase (mGDH) [EC 1.1.99.17], glycerol dehydrogenase (mGLDH) [EC 1.1.1.6], quinic acid dehydrogenase (mQDH) [EC 1.1.99.25], and inositol dehydrogenase (mIDH) [EC 1.1.1.18], which are composed of an N-terminal transmembrane domain and a C-terminal catalytic domain, including a large subunit (I) and a medium-size subunit (II). The coenzyme of the large subunit is PQQ, which plays a catalytic role, while the medium-size subunit does not contain any coenzyme and is only responsible for binding to the cell membrane (Table 2 and Figure 2).
mGDH is a D-glucose ubiquinone oxidoreductase, which can oxidize the C-1 hydroxyl group of D-glucopyranose to gluconic acid-δ-lactone (GAL; Ameyama et al., 1981), and GAL can be transformed into GA spontaneously or by one of glucolactonases on the cell membrane. mGDH has been developed as a glucose sensor because of its high substrate specificity, which can oxidize only glucose but not hexose and pentose. mGLDH is a glycerol ubiquinone oxidoreductase with the poor substrate specificity, which can oxidize glycerol to dihydroxyacetone, arabitol, sorbitol, mannitol, erytritol, ribiol, and other polyols to the corresponding ketones (Sugisawa and Hoshino, 2002), and GA to 5-KGA (Matsushita et al., 2003). Industrially, mGLDH from Glucobacter spp. has been used to produce L-sorbate, DHA, erythrose, and 5-KGA (Shinjoh et al., 2002; Sugisawa and Hoshino, 2002; Hoshino et al., 2003). mQDH is a quinic acid ubiquinone oxidoreductase, which can oxidize the C3 hydroxyl group of quinic acid to 3-dehydroquinic acid (3-DQA), then 3-DQA is transformed to shikimic acid and protocatechuic acid successively by mQDH, of which activity is only one fourth of that of quinic acid (Vangnai et al., 2010). mIDH is an inositol ubiquinone oxidoreductase, which can oxidize the C2 hydroxyl group of inositol to 2-keto-inositol (Holscher et al., 2007).
Other Membrane-Binding Dehydrogenases (mDH)
Other types of AAB mDHs include membrane-binding sorbose dehydrogenase (mSDH) [EC 1.1.99.12] and sorbosone dehydrogenase (SNDH) [EC 1.1.1.-] (Table 2). mSDH is an L-sorbose ubiquinone oxidoreductase with FAD as a coenzyme, which can oxidize the C1 hydroxyl group of sorbose to L-sorbosone (Sugisawa et al., 1991). mSDH has a high substrate specificity, which can only oxidize L-sorbose, not other sugars and alcohols (Pappenberger and Hohmann, 2013). There are 2 classes of SNDH, the first class of SNDH is a L-sorbosone ubiquinone oxidoreductase with NAD or NADP as a coenzyme, and can oxidize the C1 hydroxyl group of L-sorbosone to produce 2-keto-L-gulonic acid (2-KGLA; Pappenberger and Hohmann, 2013), present in the cytosol (Sugisawa et al., 1991; Shinjoh and Hoshino, 1995); the other class exists on the plasma membrane(mSNDH), are PQQ-dependent enzymes with L-sorbosone oxidation activity (Yakushi et al., 2020). Recently, it has been suggested that the substrate of mSNDH is the hemiacetal of L-sprbitol-1,5-pyranose (an isoform of L-sorbitol), that oxidized to 2-keto-L-gulone-1,5-pyranose(2-KGLL), and 2-KGLL will spontaneously be hydrolyzed into 2-KGLA under neutral pH conditions, but the substrate specificity, dynamics and structure of such mSNDH still need further investigation (Yakushi et al., 2020).
Terminal Oxidase (TO) in AOF
The TOs involved in the AOF respiratory chain mainly include UOX and HBD-O. Up to now, the TOs from G. oxydans and A. aceti have been intensively investigated.
Terminal Oxidases (TOs) in G. oxydans
The TOs in G. oxydans include cytochrome bo3-UOX and HBD-O (Prust et al., 2005; Miura et al., 2013; Richhardt et al., 2013; Matsushita et al., 2016). When bo3-UOX is absent, the early growth of G. oxydans is severely affected, whereas when HBD-O is absent, the early growth of cells is not affected (Matsushita et al., 1984). Further studies have indicated that at the early growth stage of G. oxydans, when the media pH is neutral, bo3-UOX is the major TO, and with the accumulation of acid and other products, when the media pH decrease to acid, HBD-O can replace bo3-UOX as the major TO, and act synergistically with cDH in the cytoplasm, leading that G. oxydans cells in acidic conditions continue to utilize organic acids and other products to grow (Miura et al., 2013; Richhardt et al., 2013).
Terminal Oxidases (TOs) in A. aceti
The TOs in A. aceti includes ba3/bo3-UOX, HBD-O, and two homologs of HBD-O, cyanide insensitive oxidase (CIO), CIO1, and CIO2, four TOs in total. Among them, ba3/bo3-UOX, CIO1 and CIO2 have a low affinity with oxygen, but a high turnover rate (efficiency), whereas HBD-O has a high affinity with oxygen, and can perform aerobic respiration under hypoxic conditions (Cunningham et al., 1997). Compared with HBD-O, CIO1 and CIO2, ba3/bo3-UOX have the stronger ability to form transmembrane proton potential and produce ATP, leading that ba3/bo3-UOX is the major TO in A. aceti (Matsutani et al., 2014).
Natural Products Yielded by AOF
Through various mDHs in AOF, AAB can oxidize various alcohols, sugars, sugar alcohols, acids and so on to the corresponding products such as acetic acid, GA, galactonic acid, 2-KGA, DHA, miglitol and so on (Mamlouk and Gullo, 2013), which have been successfully used in foods, cosmetics, medicines and other fields (Gullo et al., 2014; Saichana et al., 2015). The schematic diagram of typical mDHs from AAB strains and their main AOF products is shown in Figure 3. Up to now, 86 AOF products have been reported, and each AOF product is given a bold Arabic numeral (Supplementary Table 2). Based on the numbers in square brackets after each AOF product, the molecular and structural formula of the corresponding AOF compound can be found in Supplementary Table 2.
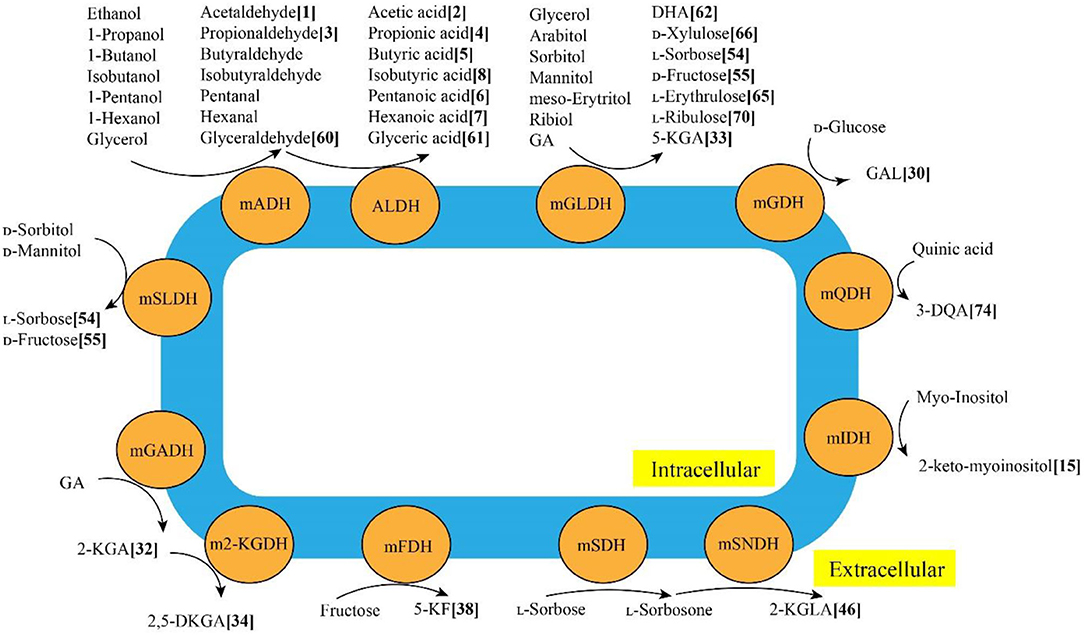
Figure 3. Typical membrane-binding dehydrogenases in acetic acid bacteria and their main AOF products. mADH: membrane-binding alcohol dehydrogenase; mALDH: membrane-binding acetaldehyde dehydrogenase; mGADH: membrane-binding gluconate dehydrogenase; m2-KGDH: membrane-binding 2-keto-D-gluconate dehydrogenase; mFDH: membrane-binding D-fructose dehydrogenase; mSLDH: membrane-binding D-sorbitol dehydrogenase; mGDH: membrane-binding glucose dehydrogenase; mGLDH: membrane-binding glycerol dehydrogenase; mQDH: membrane-binding quinic acid dehydrogenasw; mIDH: membranebinding inositol dehydrogenase; mSDH: membrane-binding sorbose dehydrogenase; mSNDH: membrane-binding sorbone dehydrogenase; GA: gluconic acid; 2-KGA: 2-keto-Dgluconic acid; 2, 5-DKGA: 2, 5-diketo-D-gluconic acid; 5-KF: 5-keto-D-fructose; DHA: dihydroxyacetone; 3-QDA: 3-dehydroquinic acid; 2-KGLA: 2-keto-l-gulonic acid.
AOF Products From Alcohols
AAB can partly oxidize a great number of primary, secondary and diol alcohols to yield the corresponding products, which have been utilized in foods, chemicals, and medicines.
Products From Primary Alcohols
The oxidation of ethanol to acetic acid may be the first-known AOF process from the AAB genus Acetobacter (Atkinson, 1956). The ethanol oxidation into acetic acid is a typical AOF process, which is divided into two steps (Matsushita et al., 1994). Ethanol is first oxidized to acetaldehyde [1] by PQQ-mADH, then converted to acetic acid [2] by PQQ-mALDH or MCD-mALDH. Besides the genus Acetobacter, the strains from Komagataeibacter spp. have very strong abilities to convert ethanol to acetic acid. Moreover, both of them have a high tolerance to ethanol and acetic acid, therefore they are the main species and strains in the vinegar production in the world (Adachi et al., 1978; Kanchanarach et al., 2010).
Except for methanol, mADH and mALDH can incompletely oxidize primary aliphatic normal alcohols with carbon chain length ≤ 6 to corresponding aldehydes and/or acids due to their poor substrate specificities (Adachi et al., 1978; Toyama et al., 2007), resulting in propionaldehyde [3] to propionic acid [4] from propanol, and butyric acid [5], pentanoic acid [6], hexanoic acid [7], isobutyric acid [8], and isovaleraldehyde [9] from butanol, pentanol, hexanol, isobutanol, and isoamyl alcohol, respectively (Švitel and Kutnik, 1995; Švitel and Šturdík, 1995; Molinari et al., 1996; Noyori, 2002).
Moreover, mADH and mALDH can also covert aromatic and other primary alcohols to the respective aldehydes and/or acids including phenylacetaldehyde [10], phenylacetic acid [11], 2-chloropropionic acid [12], (S)-2-phenyl-1-propionic acid [13], 2-methylbutanoic acid [14] and 2-keto-myoinositol [15] from 2-phenyl-ethanol, 2-chloropropanol, race-2-phenyl-1- propanol, 2-methylbutanol and myo-inositol, respectively (Molinari et al., 1996, 1999; Romano et al., 2002; Gandolfi et al., 2004; Hölscher and Görisch, 2006; Keliang and Dongzhi, 2006). Wei et al. (2016) found that racemic 1-(4-methoxyphenyl) ethanol (race-MOPE) could be oxidized to enantiopure (S)-MOPE [16] and 4-methoxyacetophenone [17] by mADH from Acetobacter sp. CCTCC M209061.
Products From Secondary Alcohols and Diols
Švitel and Kutnik (1995) found that G. oxydans CCM1783 could oxidize isopropanol and 2-butanol to acetone [18] and 2-buranone [19], respectively. Some AAB strains can also stereoselectively oxidize 2-methy-1,3-propanediol to (R)-β-hydroxyisobutyric acid [20] which is an important chiral building block in the synthesis of drugs (León et al., 2009).
The diols can be oxidized by some AAB strains, too. For example, 1,3-butandiol, 2,3- butandiol, (2R,3R)-2,3-butandiol, N-2-1,4-nonanodiol, 1,2-propanediol, ethanediol (ethylene glycol), racemical-1,2-butanediol, 1,3-propanediol and (R)-1-phenyl-1,2-ethanediol are oxidized to 3-hydroxybutyric acid [21] (Romano et al., 2002), (S)-acetoin [22] (Romano et al., 2002; Wang et al., 2013; Zhou et al., 2018), diacetyl [23], γ-nonanoic lactone [24] (Romano et al., 2002), (R)-2-hydroxy-propionic acid (D-(-)-lactic acid) [25] (Su et al., 2004), glycolic acid [26] (Wei et al., 2009), (R)-2-hydroxybutyric acid [27] (Gao et al., 2012), 3-hydroxypropionic acid [28] (Dishisha et al., 2015; Zhu et al., 2018), and (R)-mandelic acid [29] (Li et al., 2014), respectively.
Products From Sugars and Disaccharides via AOF
Through AOF, AAB can partially oxidize various sugars and disaccharides like glucose, fructose, arabinose, ribose, xylose, sorbose, lactose, isomaltose, gentiobiose, and melibiose to the corresponding products.
Products From Glucose
During AOF, glucose is first converted into gluconic acid-δ-lactone [30] (Shinagawa et al., 2009), then changed into GA [31] spontaneously or by membrane-binding gluconic acid-δ- lactonase (Shinagawa et al., 2009). GA can further be oxidized to 2-KGA [32] by GA dehydrogenase, or to 5-KGA [33] by PQQ dependent glycerol dehydrogenase (PQQ-GLDH); 2-KGA can be catalyzed to 2,5-DKGA [34] by FAD-dependent 2-keto-D-gluconic acid dehydrogenase (FAD-GADH; Matsushita et al., 2003; Shinagawa et al., 2009). 2,5-DKGA can be transferred to 4-keto-D-arabinose [35], which is further catalyzed to 4-keto-D-arabonate [36] by 4-keto-D-aldopentose-1-dehydrogenase (Adachi et al., 2011a). 2,5-DKGA can be decarboxylated to form D-lyxuronic acid [37], too (Kondô and Ameyama, 1958).
Glucose oxidative fermentation of AAB is not only closely related to glucose concentration and reaction pH (Qazi et al., 1991) but also to the sugars of the culture medium. When glucose in the medium is depleted, small amounts of 2-KGA and 5-KGA secreted in the medium can be transported to AAB cells by transporters and then reduced to GA by 2-KGA reductase (2-KGAR) or 5-KGA reductase (5-KGAR) in the cytoplasm, and utilized by AAB through the pentose phosphate pathway (PPP), allowing AAB to reproduce again and present a secondary growth curve (Saichana et al., 2015). The kinds of products obtained from glucose oxidation by AAB are also affected by pH, for example, when the pH of the culture medium is at 3.4-4.0, AAB oxidized glucose produces only 5-KGA. Therefore, when producing GA, 2-KGA, and 5-KGA using AAB species, to prevent them from being further consumed by AAB utilization, culture conditions with high glucose concentration, low pH and high O2 must be applied and fermentation terminated before the appearance of the secondary growth curve (Mamlouk and Gullo, 2013).
Products From Fructose, Arabinose, Ribose, Xylose, Sorbose, and Galactose
D-fructose can be oxidized to 5-keto-D-fructose [38] via mFDH and D-pentonate-4- dehydrogenase (P-4-DH) from AAB cells (Adachi et al., 2011b; Ano et al., 2017). mFDH possesses a high substrate specificity, whereas P-4-DH does not, which can oxidize D-psicose to 5-keto-D-psicose [39] (Ano et al., 2017), and shows mGLDH activity, too. In addition, fructose turns to glucosone [40] via a series of oxidized directly by G. roseus (Takahashi and Asai, 1932; Ikeda, 1955).
AAB strains can also transform D-arabinose to 4-keto-D-arabinose [35] and 4-keto-D-arabonate [36] (Adachi et al., 2010); D-ribose to 4-keto-D-ribose [41] and 4-keto-D-ribonate [42] (Adachi et al., 2011a); xylose to xylonic acid [43] (Buchert et al., 1988; Hahn et al., 2020), D-xylulose to xylitol [44] (Qi et al., 2016), sorbose to sorbitol [45] and 2-keto-D-gulonic acid [46] (Sugisawa et al., 1991; Adachi et al., 1999a), and D-galactose to D-galactonic acid [47] (Švitel and Šturdik, 1994). Adachi et al. (2013) also found that AAB cells could use 2-deoxy-D-ribose as a substrate to produce 2-deoxy-4-keto-D-ribose [48] and 2-deoxy-4-keto-D-ribonate [49], which are under the action of membrane-binding D-aldopentose 4-dehydrogenase and 4-keto-D-1-dehydrogenase, respectively (Adachi et al., 2013).
Products From Disaccharides
The strain A. orientalis KYG 22 can oxidize lactose to lactose acid [50] with the aid of D-glucose by mGDH (Kiryu et al., 2014). mGDH from some strains of Gluconobacter, Ga. hansenii NBRC 14816 and K. medellinensis NBRC 3288 can also oxidize isomaltose, gentiobiose and melibiose to isomaltobionic acid [51], gentiobionic acid, [52], and melibionic acid [53], respectively (Kiryu et al., 2020).
Products From Sugar Alcohols Through AOF
The sugar alcohols such as D-sorbitol, glycerol, D-mannitol, D-arabinol, D-erythritol and D-ribitol are transferred by AOF into L-sorbose, DHA, L-erythrose, D-xylulose, D-fructose, and L-ribulose, respectively (Cummins et al., 1957; Sugisawa and Hoshino, 2002; Prust et al., 2005; Adachi et al., 2007; Mamlouk and Gullo, 2013).
Products From Sorbitol
Sorbitol may be sequentially oxidized by two mDHs, sorbitol dehydrogenase and sorbose dehydrogenase from AAB cells, to L-sorbose [54] and D-fructose [55] (Cummins et al., 1957; Sato et al., 1967). Then sorbose is changed to 5-keto-D-fructose [56], which will be converted into three γ-pyrone compounds including kojic acid [57], 3-oxykojic acid, [58], and 5-oxymaltol [59] (Sato et al., 1967). In addition, L-sorbosone can enter AAB cells, then transformed to 2-keto-L-gulonic acid [46], which is secreted extracellularly as an intermediate of vitamin C synthesis (Sugisawa and Hoshino, 2002; Matsushita et al., 2003; Adachi et al., 2007).
Products From Glycerol
The G. frateurii NBRC3262 strain can sequentially convert glycerol to glyceraldehyde [60] to D-glyceric acid [61] by mADH (Habe et al., 2009). Some G. oxydans strains can oxidize glycerol to DHA [62] via mGLDH (Habe et al., 2009; Hu et al., 2010). DHA is widely used in cosmetics as an active sunscreen ingredient, as well as utilized in weight loss, antioxidant, vitiligo treatment, and so on (Levy, 1992). Some Acetobacter strains can convert racemic glycidol to glycidic acid [63] (Geerlof et al., 1994; Švitel and Kutnik, 1995).
Products From Other Sugar Alcohols
A. suboxydans strains can make L-fucitol convert to L-duco-4-ketose [64] (Richtmyer et al., 1950). Meso-erythritol is changed to L-erythrulose [65] by AAB cells (Richtmyer et al., 1950). D-arabitol (the 2-epimer of eylitol) was firstly oxidized to D-xylulose [66] by D-arabitol dehydrogenase (AraDH), then reduced to xylitol [67] by NAD-dependent xylitol dehydrogenase (Suzuki et al., 2002; Liu et al., 2019). Xylitol, a natural pentahydroxy sugar alcohol, has a sweet compound similar to sucrose and serves as a substitute for natural sweeteners. It also plays a role in preventing tooth decay (Suzuki et al., 2002). G. suboxydans can produce D-fructose [55] from D-mannitol (Adachi et al., 1999b). Ac. oxydans and could oxidize allitol to L-allulose (L-ribo-hexulose or L-psicose) [68] (Carr et al., 1968; Takeshita et al., 1996), which is rare and not natural ketohexose. AAB cells can also oxidize ribitol to L-ribulose [60] by membrane-binding NAD(P) independent ribitol dehydrogenase (Adachi et al., 2001). Recently, Xu et al. (2021) reported that polyol galactitol can be oxidized to two rare sugars, D-tagatose [70] and L-xylo-3-hexulose [71] by PQQ-dependent L-arabinitol 4-dehydrogenase (PQQ-LAD) from Acetobacter sp. and Gluconobacter sp. strains.
Products Converted From Organic Acids
Products From Quinate
Whiting and Coggins (1967) firstly reported that quinate oxidase from Ac. oxydans, quinate-cytochrome 555 oxidoreductase, could oxidize D-dihydroshikimic acid into 3,4- dihydroxy-5-oxocyclohexane-1-carboxylic acid [72]. Quinate is oxidized to 3-dehydroquinate (DQA) [73] by NAD(P) independent quinate dehydrogenase (QDH, EC 1.1.99.25), then to 3-dehydroshikimate (DSA) [74] via DQA dehydratase (EC. 4.2.1.10), and DSA can be converted to protocatechuate [75] by DSA dehydratase (Adachi et al., 2006).
Products From Other Organic Acids
Benziman and Perez (1965) proved that malate was converted to oxaloacetate [76] by a FAD-protein from A. xylinum (currently K. xylinus). Dosoretz et al. (1992) reported that the pure cultures of A. pasteurianus, G. cerinus and G. oxydans could oxidize calcium magnesium (Ca-Mg) lactate to Ca-Mg acetate (CMA) [77], which is considered to be a potentially noncorrosive and biodegradable deicing chemical. In 2015, Sato et al. discovered that some strains of Acetobacter spp. could partially oxidize L/D-lactate into pyruvate [78] by lactate dehydrogenase (Sato et al., 2015).
Products Converted From Other Substrates via AOF
Products From Hexosamine
In 1960, Takahashi and Kayamori discovered that A. melanogenum (currently G. oxydans) could transform glucosamine into glucosaminic acid [79], which is used as a commercial chemical in daily life (Takahashi and Kayamori, 1960). In 2004, Moonmangmee et al. found that D-mannosamine and D-galactosamine were changed into D-mannosaminate [80] and D-galatosaminate [81] by Gluconobacter sp. IFO 3264, respectively (Moonmangmee et al., 2004).
Products From Furfural
Zhou et al. (2017) prove that G. oxydans ATCC 621H could change furfural or furfuryl alcohol into 2-furoic acid [82], which is the raw material to produce many furoate esters and its derivatives widely used in the synthesis of pharmaceutical, agricultural and industrial chemicals (Zhou et al., 2017). Sayed et al. (2019) found that the resting cells of G. oxydans DSM 50049 were capable of highly selective production of 5-hydroxymethyl-2- furan-carboxylic acid [83], which is used as a monomer of a variety of polymers with antibacterial and antitumor activities (Sayed et al., 2019).
Products From Others
Sugisawa et al. (2005) first found that G. oxydans DSM 4025 could produce L-ascorbic acid [84] from L-gulono-γ-lactone with L-gulonate-γ-lactone dehydrogenase. Landis et al. (2002) prove that the strains of G. oxydans could also selectively region oxidize the N- butylglucamine into 6-deoxy-6-butylaminosorbose [85] (Landis et al., 2002). Some AAB strains can yield 6-(2-hydroxy-ethyl) amino-6-deoxy-α-L-sorbofuranose [86] using N-2-hydroxyethyl glucamine as substrate, which is an important precursor of miglitol, a α-glucosidase inhibitor, which was approved by the Food and Drug Administration of United States for the treatment of type II diabetes in December 1996 (Keliang and Dongzhi, 2006).
Molecular Biological Methods for AOF
In recent decades, a lot of molecular biology techniques have been applied to investigate AOF, especially its mDHs. Hereinafter, we just make a brief introduction about the application of molecular biology technologies in AOF, especially the establishment of a marker-less gene deletion system, for more detailed information, please read our recent review article (Yang et al., 2022).
Establishment of Marker-Less Gene Deletion System and Its Application in AOF
To investigate the function of mDH's genes from AAB, gene deletion is doubtlessly a direct and powerful tool. In this context, using a marker-less strategy is preferable to other molecular biology methods due to the following reasons (Peters et al., 2013): (1) the number of available markers such as antibiotic-resistant gene markers is so limited that the construction of the multi-gene knockout strain is sometimes impossible with marker genes; (2) as about 50% of genes in prokaryotes are located in the operons, where genes are co-transcribed into a single polycistronic mRNA (Osbourn and Field, 2009), the insertion of marker genes into the operon may exert polar effects and suppress the expression of downstream genes in the same operon.
Marker-less gene deletion techniques are often carried out in a two-step procedure using a plasmid vector carrying an antibiotic resistance selection marker, a counter selection marker, as well as the fused flanking fragments of the target gene (Gao et al., 2006). In the first step, the non-replicating plasmid vector is introduced into the host cell. Subsequently, the clones with the vector integrated at either the upstream or downstream site of the target gene by the homologous recombination are screened by antibiotic resistance and will be used for the second homologous recombination, in which the clones without the vector will be obtained through the recombination of the other homologous flanking region and the counter selection marker. The second homologous recombination will yield either the wild type strain or the desired gene deletion mutant with the theoretical ratio of 1:1 (Yang et al., 2022).
Peters et al. (2013) reported such a gene deletion system for AAB. The system involves a kanamycin resistance gene to select for AAB cells harboring the vector after its integration at the target site of the genome, together with gene upp, encoding uracil phosphoribosyl- transferase for the subsequent counter-selection to lose the vector from the genome, which could result in the desired deletion mutant without any marker sequence. In this case, uracil phosphoribosyltransferase converts the counter selection agent 5-fluorouracil (FU) to toxic 5-fluorouridinemonophosphate (F-UMP) to kill the wild type. However, most AAB species, including G. oxydans, possess the upp gene in their genomes. Therefore, deletion of the upp gene is required prior to application of this deletion method.
In order to avoid the deletion of upp before knocking out other genes, an improved method was established by Kostner et al. (2013) using the codA gene from Echerichia coli as the counter selection marker. This gene codes for cytosine deaminase, converting the nontoxic counters election agent 5-fluorocytosine (FC) to FU, which is subsequently converted to toxic F-UMP by uracil phosphoribosyltransferase encoded by gene upp. In addition, they found that the co-expression of codA with codB, coding for cytosine permease that facilitate the uptake of FC, can significantly increase the efficiency of the deletion method. Thus, codB was introduced to the deletion vector and also became a part of counter selection marker together with codA in this newly developed system.
Using the upp marker-based counter selection approach, Peters et al. (2013) knocked out all mDH-coding genes in G. oxydans 621H in a sequential manner, creating a series of mutants lacking one or more mDH (s), including G. oxydans BP.9 without all mDH genes and G. oxydans BP.8 only with the gene of a polyol dehydrogenase. Subsequently, by adopting a whole-cell 2,6-dichlorophenolindophenol (DCPIP) activity assay, the substrate specificities of the mDHs were clarified. Mientus et al. (2017) analyzed the substrate spectra of the mDHs of G. oxydans by applying a shuttle vector to express each individual enzyme in G. oxydans BP.9, and the substrate spectra of every one of the mDHs were defined by the whole-cell DCPIP assay. In addition, Peters et al. (2013) used G. oxydans BP.9 to express genes encoding mDHs from the metagenome of a mother of vinegar sample including many non-culturable AAB and other microorganisms. In 2019, Burger et al. applied G. oxydans BP.8 to investigate and optimize the production of L-erythrulose (Burger et al., 2019).
Molecular Biology Studies Regarding AOF Other Than Marker-Less Deletion
Besides the gene marker-less deletion method, a number of other molecular biological technologies have also been applied for AOF research. For example, PQQ-GLDH was once conferred different names owning to the diversity of its substrates, including D-gluconate, D-arabitol, and D-sorbitol (Shinagawa et al., 1999; Adachi et al., 2001; Sugisawa and Hoshino, 2002). It was not until the gene disruption experiments that people finally realized that these enzymes are actually the same (Miyazaki et al., 2002; Shinjoh et al., 2002). Later, the name PQQ-GLDH was confirmed by Matsushita et al. (2003).
The molecular techniques are also exploited with the aim of promoting AOF production. A typical example is the oxidation of D-glucose, which is the most common substrate for G. oxydans and can be transferred into a variety of different products, such as GA, 5-KGA, 2-KGA, and 2, 5-DKGA (Saichana et al., 2015). Therefore, in terms of GA production, further oxidation of GA to ketogluconate is unexpected, suggesting that promotion of GA could be achieved by suppressing the mDHs which can further oxidize GA (La China et al., 2018). On the other hand, an increase in the production of GA, 2-KGA, and 5-KGA could be achieved by over-expression of genes encoding FAD-dependent D-gluconate dehydrogenase (Shi et al., 2014), PQQ-GDH and PQQ-GLDH (Merfort et al., 2006), respectively. In addition, either enhancing the abundance of mRNA transcribed from PQQ-GLDH-coding genes through adding an A/T tail (Xu et al., 2014) or over-expression of genes responsible for PQQ biosynthesis (Wang et al., 2016) can lead to the improved production of L-sorbose, which could be used as the substrate for producing 2-keto-L-gulonic acid, a direct precursor of vitamin C (La China et al., 2018). Furthermore, Gao et al. (2014) heterologously expressed a different combination of genes encoding five L-sorbose mDHs and two L-sorbosone mDHs from Ketogulonicigenium vulgare in G. oxydans WSH-003, and screened the best recombinant strain G. oxydans/pGUC-k0203-GS-k0095 with the highest yield, achieving one-step production of 2-keto-L-gulonic acid from D-sorbitol, which was produced via a two-step process in the past.
Conclusion and Discussion
AAB is a large group of Gram-negative, strictly aerobic bacteria, some of which have the great ability to yield a number of products with commercial or potential commercial values by their unique oxidative fermentation (AOF). In this review, we first summarize the AAB classification progress, then systematically describe and classify AOF products and the relative enzymes. The application of molecular biology technologies in AOF research is also briefly introduced.
Although many research works have been carried out and remarkable progress has been made on AOF, some AOF products such as acetic acid (vinegar), bacterial cellulose, DHA and so on (La China et al., 2018), have been industrialized, there are still a lot of issues about AOF which need to be further investigated. For example, although we are pleased to witness the promising progress achieved in the research on the functions of AAB mDHs, as well as in the AOF application, 21 'orphan' mDHs with unknown substrate spectra in G. oxydans are still unexplored (Adachi and Yakushi, 2016), and few crystal structures of AAB mDHs have been published (Qin et al., 2022), which greatly hinders the understanding of substrate space- and enantio- specificities of AAB mDHs. Moreover, the relevant modern technologies, especially modern molecular biotechnology including omics, systems biology, combinatorial biology, synthetic biology, and metabolic engineering, should be utilized for constructing relevant engineering strains for efficient production of the target AOF products.
Author Contributions
YH contribute to the data collection, collation, and writing of the first draft. ZX and HZ responsible for the data collection and collation. WL and HT provide brief article ideas and language modifications. FC propose the overall idea for this article, reviewed and revised the first draft, and provided the corresponding financial support. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (No. 2662019PY015), the Major Special Projects of Technological Innovation of Hubei Province, China (No. 2018ABA075), the Major Science and Technology Project in Zhenjiang City, Jiangsu Province, China (No. ZD2019001), Programs of the International S&T Cooperation, Ministry of Science and Technology, China (No. 2014DFG32380), and China Scholarship Council (202006760070).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.879246/full#supplementary-material
References
Adachi, O., Ano, Y., Moonmangmee, D., Shinagawa, E., Toyama, H., Theeragool, G., et al. (1999a). Crystallization and properties of NADPH-dependent L-sorbose reductase from Gluconobacter melanogenus IFO 3294. Biosci. Biotechnol. Biochem. 63, 2137–2143. doi: 10.1271/bbb.63.2137
Adachi, O., Ano, Y., Toyama, H., and Matsushita, K. (2006). High shikimate production from quinate with two enzymatic systems of acetic acid bacteria. Biosci. Biotechnol. Biochem. 70, 2579–2582. doi: 10.1271/bbb.60259
Adachi, O., Ano, Y., Toyama, H., Matsushita, K., and KGaA, C. (2007). Biooxidation With PQQ-and FAD- Dependent Dehydrogenases. Modern Biooxidation: Enzymes, Reactions Applications. Weinheim: Wiley-VCH Verlag GmbH.
Adachi, O., Fujii, Y., Ano, Y., Moonmangmee, D., Toyama, H., Shinagawa, E., et al. (2001). Membrane-bound sugar alcohol dehydrogenase in acetic acid bacteria catalyzes L-ribulose formation and NAD-dependent ribitol dehydrogenase is independent of the oxidative fermentation. Biosci. Biotechnol. Biochem. 65, 115–125. doi: 10.1271/bbb.65.115
Adachi, O., Hours, R. A., Akakabe, Y., Shinagawa, E., Ano, Y., Yakushi, T., et al. (2013). Pentose oxidation by acetic acid bacteria led to a finding of membrane-bound purine nucleosidase. Biosci. Biotechnol. Biochem. 77, 1131–1133. doi: 10.1271/bbb.130066
Adachi, O., Hours, R. A., Akakabe, Y., Tanasupawat, S., Yukphan, P., Shinagawa, E., et al. (2010). Production of 4-keto-D-arabonate by oxidative fermentation with newly isolated Gluconacetobacter liquefaciens. Biosci. Biotechnol. Biochem. 74, 2555–2558. doi: 10.1271/bbb.100698
Adachi, O., Hours, R. A., Shinagawa, E., Akakabe, Y., Yakushi, T., and Matsushita, K. (2011a). Enzymatic synthesis of 4-pentulosonate (4-keto-D-pentonate) from D-aldopentose and D-pentonate by two different pathways using membrane enzymes of acetic acid bacteria. Biosci. Biotechnol. Biochem. 75, 2418–2420. doi: 10.1271/bbb.110575
Adachi, O., Hours, R. A., Shinagawa, E., Akakabe, Y., Yakushi, T., and Matsushita, K. (2011b). Formation of 4-keto-D-aldopentoses and 4-pentulosonates (4-keto-D-pentonates) with unidentified membrane-bound enzymes from acetic acid bacteria. Biosci. Biotechnol. Biochem. 75, 1801–1806. doi: 10.1271/bbb.110339
Adachi, O., Moonmangmee, D., Toyama, H., Yamada, M., Shinagawa, E., and Matsushita, K. (2003). New developments in oxidative fermentation. Appl. Microbiol. Biotechnol. 60, 643–653. doi: 10.1007/s00253-002-1155-9
Adachi, O., Tayama, K., Shinagawa, E., Matsushita, K., and Ameyama, M. (1978). Purification and characterization of particulate alcohol dehydrogenase from Gluconobacter suboxydans. Agri. Biol. Chem. 42, 2045–2056. doi: 10.1271/bbb1961.42.2045
Adachi, O., Toyama, H., and Matsushita, K. (1999b). Crystalline NADP-dependent D-mannitol dehydrogenase from Gluconobacter suboxydans. Biosci. Biotechnol. Biochem. 63, 402–407. doi: 10.1271/bbb.63.402
Adachi, O., and Yakushi, T. (2016). “Membrane-bound dehydrogenases of acetic acid bacteria,” in Acetic Acid bacteria: Ecology and Physiology, eds K. Matsushita, H., Toyama, N., Tonouchi, A., Okamoto-Kainuma (Tokyo: Springer Japan), 273–297. doi: 10.1007/978-4-431-55933-7_13
Ameyama, M., Shinagawa, E., Matsushita, K., and Adachi, O. (1981). D-Glucose dehydrogenase of Gluconobacter suboxydans: solubilization, purification and characterization. Agri. Biol. Chem. 45, 851–861. doi: 10.1271/bbb1961.45.851
Ano, Y., Hours, R. A., Akakabe, Y., Kataoka, N., Yakushi, T., Matsushita, K., et al. (2017). Membrane- bound glycerol dehydrogenase catalyzes oxidation of D-pentonates to 4-keto-D-pentonates, D-fructose to 5-keto-D-fructose, and D-psicose to 5-keto-D-psicose. Biosci. Biotechnol. Biochem. 81, 411–418. doi: 10.1080/09168451.2016.1254535
Asai, T. (1968). Acetic Acid Bacteria; Classification and Biochemical Activities. Tokyo: University of Tokyo Press.
Atkinson, D. E. (1956). The oxidation of ethanol and tricarboxylic acid cycle intermediates by Acetobacter peroxydans. J. Bacteriol. 72, 195–198. doi: 10.1128/jb.72.2.195-198.1956
Baldrian, P. (2006). Fungal laccases–occurrence and properties. FEMS Microbiol. Rev. 30, 215–242. doi: 10.1111/j.1574-4976.2005.00010.x
Benziman, M., and Perez, L. (1965). The participation of vitamin K in malate oxidation by Acetobacter xylinum. Biochem. Biophys. Res. Commun. 19, 127–132. doi: 10.1016/0006-291X(65)90130-0
Buchert, J., Puls, J., and Poutanen, K. (1988). Comparison of Pseudomonas fragi and Gluconobacter oxydans for production of xylonic acid from hemicellulose hydrolyzates. Appl. Microbiol. Biotechnol. 28, 367–372. doi: 10.1007/BF00268197
Burger, C., Kessler, C., Gruber, S., Ehrenreich, A., Liebl, W., and Weuster-Botz, D. (2019). L-Erythrulose production with a multideletion strain of Gluconobacter oxydans. Appl. Microbiol. Biotechnol. 103, 4393-4404. doi: 10.1007/s00253-019-09824-w
Carr, J., Coggins, R., Hough, L., Stacey, B., and Whiting, G. (1968). Micro-biological oxidation of allitol to l-ribo-hexulose by Acetomonas oxydans. Phytochemistry. 7, 1–4. doi: 10.1016/S0031-9422(00)88197-2
Cleenwerck, I., De Vos, P., and De Vuyst, L. (2010). Phylogeny and differentiation of species of the genus Gluconacetobacter and related taxa based on multilocus sequence analyses of housekeeping genes and reclassification of Acetobacter xylinus subsp. sucrofermentans as Gluconacetobacter sucrofermentans (Toyosaki et al. 1996) sp. nov., comb. nov. Int. J. Systemat. Evol. Microbiol. 60, 2277–2283. doi: 10.1099/ijs.0.018465-0
Crotti, E., Chouaia, B., Alma, A., Favia, G., Bandi, C., Bourtzis, K., et al. (2016). “Acetic acid bacteria as symbionts of insects,” in Acetic Acid Bacteria: Ecology and Physiology, eds K. Matsushita, H. Toyama, N. Tonouchi and A. Okamoto-Kainuma. (Tokyo: Springer Japan), 121–142. doi: 10.1007/978-4-431-55933-7_5
Cummins, J. T., King, T. E., and Cheldelin, V. H. (1957). The biological oxidation of sorbitol. J. Biol. Chem. 224, 323–329. doi: 10.1016/S0021-9258(18)65031-8
Cunningham, L., Pitt, M., and Williams, H. D. (1997). The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol. Microbiol. 24, 579–591. doi: 10.1046/j.1365-2958.1997.3561728.x
De Roos, J., and De Vuyst, L. (2018). Acetic acid bacteria in fermented foods and beverages. Curr. Opin. Biotechnol. 49, 115–119. doi: 10.1016/j.copbio.2017.08.007
Deppenmeier, U., Hoffmeister, M., and Prust, C. (2002). Biochemistry and biotechnological applications of Gluconobacter strains. Appl. Microbiol. Biotechnol. Adv. 60, 233–242. doi: 10.1007/s00253-002-1114-5
Dishisha, T., Pyo, S. H., and Hatti-Kaul, R. (2015). Bio-based 3-hydroxypropionic- and acrylic acid production from biodiesel glycerol via integrated microbial and chemical catalysis. Microb Cell Fact. 14:200. doi: 10.1186/s12934-015-0388-0
Dosoretz, C. G., Jain, M. K., and Grethlein, H. E. (1992). Oxidative fermentation of calcium-magnesium lactate to calcium-magnesium acetate deicing salt. Biotechnol. Lett. 14, 613–618. doi: 10.1007/BF01023951
Dutta, D., and Gachhui, R. (2006). Novel nitrogen-fixing Acetobacter nitrogenifigens sp. nov., isolated from Kombucha tea. Int. J. Systemat. Evol. Microbiol. 56, 1899–1903. doi: 10.1099/ijs.0.64101-0
Fuentes-Ramírez, L. E., Bustillos-Cristales, R., Tapia-Hernández, A., Jiménez-Salgado, T., Wang, E. T., Martínez-Romero, E., et al. (2001). Novel nitrogen-fixing acetic acid bacteria, Gluconacetobacter johannae sp. nov. and Gluconacetobacter azotocaptans sp. nov., associated with coffee plants. Int. J. Systemat. Evol. Microbiol. 51, 1305–1314. doi: 10.1099/00207713-51-4-1305
Gandolfi, R., Cavenago, K., Gualandris, R., Sinisterra Gago, J. V., and Molinari, F. (2004). Production of 2-phenylacetic acid and phenylacetaldehyde by oxidation of 2-phenylethanol with free immobilized cells of Acetobacter aceti. Process Biochem. 39, 749–753. doi: 10.1016/S0032-9592(03)00185-7
Gao, C., Zhang, W., Huang, Y., Ma, C., and Xu, P. (2012). Efficient conversion of 1,2-butanediol to (R)-2-hydroxybutyric acid using whole cells of Gluconobacter oxydans. Bioresour. Technol. 115, 75–78. doi: 10.1016/j.biortech.2011.11.009
Gao, L., Hu, Y., Liu, J., Du, G., Zhou, J., and Chen, J. (2014). Stepwise metabolic engineering of Gluconobacter oxydans WSH-003 for the direct production of 2-keto-L-gulonic acid from D-sorbitol. Metabol. Eng. 24, 30–37. doi: 10.1016/j.ymben.2014.04.003
Gao, W., Liu, Y., Giometti, C. S., Tollaksen, S. L., Khare, T., Wu, L., et al. (2006). Knock-out of SO1377 gene, which encodes the member of a conserved hypothetical bacterial protein family COG2268, results in alteration of iron metabolism, increased spontaneous mutation and hydrogen peroxide sensitivity in Shewanella oneidensis MR-1. BMC Genom. 7, 1–13. doi: 10.1186/1471-2164-7-76
Geerlof, A., Jongejan, J. A., van Dooren, T. J., Raemakers-Franken, P. C., van den Tweel, W. J., and Duine, J.A. (1994). Factors relevant to the production of (R)-(+)-glycidol (2, 3-epoxy-1-propanol) from racemic glycidol by enantioselective oxidation with Acetobacter pasteurianus ATCC 12874. Enzyme Microbial. Technol. 16, 1059–1063. doi: 10.1016/0141-0229(94)90143-0
Gillis, M., Kersters, K., Hoste, B., Janssens, D., Kroppenstedt, R., Stephan, M., et al. (1989). Acetobacter diazotrophicus sp. nov., a nitrogen-fixing acetic acid bacterium associated with sugarcane. Int. J. Systemat. Evol. Microbiol. 39, 361–364. doi: 10.1099/00207713-39-3-361
Gómez-Manzo, S., Chavez-Pacheco, J., Contreras-Zentella, M., Sosa-Torres, M., Arreguín-Espinosa, R., Perez De La Mora, M., et al. (2010). Molecular and catalytic properties of the aldehyde dehydrogenase of Gluconacetobacter diazotrophicus, a quinoheme protein containing pyrroloquinoline quinone, cytochrome b, and cytochrome c. J. Bacteriol. 192, 5718–5724. doi: 10.1128/JB.00589-10
Gómez-Manzo, S., Contreras-Zentella, M., González-Valdez, A., Sosa-Torres, M., Arreguín-Espinoza, R., and Escamilla-Marván, E. (2008). The PQQ-alcohol dehydrogenase of Gluconacetobacter diazotrophicus. Int. J. Food Microbiol. 125, 71–78. doi: 10.1016/j.ijfoodmicro.2007.10.015
Gómez-Manzo, S., Escamilla, J. E., González-Valdez, A., López-Velázquez, G., Vanoye-Carlo, A., Marcial-Quino, J., et al. (2015). The oxidative fermentation of ethanol in Gluconacetobacter diazotrophicus is a two-step pathway catalyzed by a single enzyme: alcohol-aldehyde dehydrogenase (ADHa). Int. J. Mol. Sci. 16, 1293–1311. doi: 10.3390/ijms16011293
Gosselé, F., Swings, J., Kersters, K., and De Ley, J. (1983). Numerical analysis of phenotypic features and protein gel electropherograms of Gluconobacter Asai 1935 emend. mut. char. Asai, Iizuka, and Komagata 1964. Int. J. Systemat. Evol. Microbiol. 33, 65-81. doi: 10.1099/00207713-33-1-65
Gullo, M., Sola, A., Zanichelli, G., Montorsi, M., Messori, M., and Giudici, P. (2017). Increased production of bacterial cellulose as starting point for scaled-up applications. Appl. Microbiol. Biotechnol. 101, 8115–8127. doi: 10.1007/s00253-017-8539-3
Gullo, M., Verzelloni, E., and Canonico, M. (2014). Aerobic submerged fermentation by acetic acid bacteria for vinegar production: process and biotechnological aspects. Process Biochem. 49, 1571–1579. doi: 10.1016/j.procbio.2014.07.003
Gupta, A., Singh, V. K., Qazi, G., and Kumar, A. (2001).Gluconobacter oxydans: its biotechnological applications. J. Mol. Microbiol. Biotechnol. Adv. 3, 445–456.
Habe, H., Shimada, Y., Yakushi, T., Hattori, H., Ano, Y., Fukuoka, T., et al. (2009). Microbial production of glyceric acid, an organic acid that can be mass produced from glycerol. Appl. Environ. Microbiol. 75, 7760–7766. doi: 10.1128/AEM.01535-09
Hahn, T., Torkler, S., van Der Bolt, R., Gammel, N., Hesse, M., Möller, A., et al. (2020). Determining different impact factors on the xylonic acid production using Gluconobacter oxydans DSM 2343. Process Biochem. 94, 172–179. doi: 10.1016/j.procbio.2020.04.011
Hölscher, T., and Görisch, H. (2006). Knockout and overexpression of pyrroloquinoline quinone biosynthetic genes in Gluconobacter oxydans 621H. J. Bacteriol. 188, 7668–7676. doi: 10.1128/JB.01009-06
Holscher, T., Weinert-Sepalage, D., and Gorisch, H. (2007). Identification of membrane-bound quinoprotein inositol dehydrogenase in Gluconobacter oxydans ATCC 621H. Microbiology153, 499–506. doi: 10.1099/mic.0.2006/002196-0
Hoshino, T., Sugisawa, T., Shinjoh, M., Tomiyama, N., and Miyazaki, T. (2003). Membrane-bound D-sorbitol dehydrogenase of Gluconobacter suboxydans IFO 3255—enzymatic and genetic characterization. Biochim. Biophys. Acta 1647, 278–288. doi: 10.1016/S1570-9639(03)00071-2
Hu, Z. C., Liu, Z. Q., Zheng, Y. G., and Shen, Y. C. (2010). Production of 1, 3-dihydroxyacetone from glycerol by Gluconobacter oxydans ZJB09112. J. Microbiol. Biotechnol. Lett. 20, 340–345. doi: 10.4014/jmb.0907.07011
Ikeda, Y. (1955). A presumable pathway of kojic acid formation formation from fructose by Gluconoeacter. J. General Appl. Microbiol. Biotechnol. 1, 152–163. doi: 10.2323/jgam.1.152
Janda, J. M., and Abbott, S. L. (2007). 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45, 2761–2764. doi: 10.1128/JCM.01228-07
Jojima, Y., Mihara, Y., Suzuki, S., Yokozeki, K., Yamanaka, S., and Fudou, R. (2004). Saccharibacter floricola gen. nov., sp. nov., a novel osmophilic acetic acid bacterium isolated from pollen. Int. J. Systemat. Evol. Microbiol. 54, 2263–2267. doi: 10.1099/ijs.0.02911-0
Kanchanarach, W., Theeragool, G., Inoue, T., Yakushi, T., Adachi, O., and Matsushita, K. (2010). Acetic acid fermentation of Acetobacter pasteurianus: relationship between acetic acid resistance and pellicle polysaccharide formation. Biosci. Biotechnol. Biochem. 2010:1007022033. doi: 10.1271/bbb.100183
Kataoka, N., Matsutani, M., Yakushi, T., and Matsushita, K. (2015). Efficient production of 2, 5-diketo-D-gluconate via heterologous expression of 2-ketogluconate dehydrogenase in Gluconobacter japonicus. Appl. Environ. Microbiol. 81, 3552–3560. doi: 10.1128/AEM.04176-14
Kawai, S., Goda-Tsutsumi, M., Yakushi, T., Kano, K., and Matsushita, K. (2013). Heterologous overexpression and characterization of a flavoprotein-cytochrome c complex fructose dehydrogenase of Gluconobacter japonicus NBRC3260. Appl. Environ. Microbiol. 79, 1654–1660. doi: 10.1128/AEM.03152-12
Keliang, G., and Dongzhi, W. (2006). Asymmetric oxidation by Gluconobacter oxydans. Appl. Microbiol Biotechnol. 70:135–139. doi: 10.1007/s00253-005-0307-0
Kersters, K., Lisdiyanti, P., Komagata, K., and Swings, J. (2006). The family Acetobacteraceae: the genera Acetobacter, Acidomonas, Asaia, Gluconacetobacter, Gluconobacter, and Kozakia. Prokaryotes 5, 163–200. doi: 10.1007/0-387-30745-1_9
Kiryu, T., Kiso, T., Sato, H., and Murakami, H. (2020). Oxidation of isomaltose, gentiobiose, and melibiose by membrane-bound quinoprotein glucose dehydrogenase from acetic acid bacteria. Biosci. Biotechnol. Biochem. 84, 507–517. doi: 10.1080/09168451.2019.1689095
Kiryu, T., Yamauchi, K., Masuyama, A., Ooe, K., Kimura, T., Kiso, T., et al. (2014). Optimization of lactobionic acid production by Acetobacter orientalis isolated from Caucasian Fermented Milk, “Caspian Sea Yogurt”. Biosci. Biotechnol. Biochem. 76, 361–363. doi: 10.1271/bbb.110608
Kondô, K., and Ameyama, M. (1958). Carbohydrate metabolism by acetobacter species: part I. oxidative activity for various carbohydrates. Part II. Ketogenic metabolism of glucose. Part III. Isolation and identification of d-lyxuronic acid on glucose oxidation by A. melanogenum. J. Agri. Chem. Soc. Jap. 22, 369–386. doi: 10.1080/03758397.1958.10857508
Kostner, D., Peters, B., Mientus, M., Liebl, W., and Ehrenreich, A. (2013). Importance of codB for new codA- based markerless gene deletion in Gluconobacter strains. Appl. Microbiol. Biotechnol. 97, 8341-8349. doi: 10.1007/s00253-013-5164-7
La China, S., Zanichelli, G., De Vero, L., and Gullo, M. (2018). Oxidative fermentations and exopolysaccharides production by acetic acid bacteria: a mini review. Biotechnol. Lett. 40, 1289–1302. doi: 10.1007/s10529-018-2591-7
Landis, B. H., McLaughlin, J. K., Heeren, R., Grabner, R. W., and Wang, P. T. (2002). Bioconversion of N-Butylglucamine to 6-Deoxy-6-butylamino Sorbose by Gluconobacter o xydans. Org. Process Res. Dev. 6, 547–552. doi: 10.1021/op0255128
León, R., Prazeres, D. M. F., Molinari, F., and Cabral, J. M. S. (2009). Microbial stereoselective oxidation of 2-methyl-1,3-propanediol to (R)-β-hydroxyisobutyric acid in aqueous/organic biphasic systems. Biocatal. Biotransform. 20, 201–207. doi: 10.1080/10242420290020723
Levy, S. B. (1992). Dihydroxyacetone-containing sunless or self-tanning lotions. J. Am. Acad. Dermatol. 27, 989–993. doi: 10.1016/0190-9622(92)70300-5
Li, D. H., Lin, J. P., and Wei, D. Z. (2014). Improving Gluconobacter oxydans performance in the in situ removal of the inhibitor for asymmetric resolution of racemic 1-phenyl-1,2-ethanediol. Bioresour. Technol. 159, 327–333. doi: 10.1016/j.biortech.2014.02.104
Li, L., Cleenwerck, I., De Vuyst, L., and Vandamme, P. (2017). Identification of acetic acid bacteria through matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and report of Gluconobacter nephelii Kommanee et al. 2011 and Gluconobacter uchimurae Tanasupawat et al. 2012 as later heterotypic synonyms of Gluconobacter japonicus Malimas et al. 2009 and Gluconobacter oxydans (Henneberg 1897) De Ley 1961 (Approved Lists 1980) emend. Gosselé et al. 1983, respectively. Systemat. Appl. Microbiol. 40, 123–134. doi: 10.1016/j.syapm.2017.01.003
Liu, L., Zeng, W., Du, G., Chen, J., and Zhou, J. (2019). Identification of NAD-dependent xylitol dehydrogenase from Gluconobacter oxydans WSH-003. ACS Omega 4, 15074–15080. doi: 10.1021/acsomega.9b01867
Loganathan, P., and Nair, S. (2004). Swaminathania salitolerans gen. nov., sp. nov., a salt-tolerant, nitrogen-fixing and phosphate-solubilizing bacterium from wild rice (Porteresia coarctata Tateoka). Int. J. Systemat. Evol. Microbiol. 54, 1185–1190. doi: 10.1099/ijs.0.02817-0
Malimas, T., Yukphan, P., Takahashi, M., Muramatsu, Y., Kaneyasu, M., Potacharoen, W., et al. (2008). Gluconobacter sphaericus (Ameyama 1975) comb. nov., a brown pigment-producing acetic acid bacterium in the Alphaproteobacteria. J. General Appl. Microbiol. 54, 211–220. doi: 10.2323/jgam.54.211
Malimas, T., Yukphan, P., Takahashi, M., Muramatsu, Y., Kaneyasu, M., Potacharoen, W., et al. (2009). Gluconobacter japonicus sp. nov., an acetic acid bacterium in the Alphaproteobacteria. Int. J. Systemat. Evol. Microbiol. 59, 466–471. doi: 10.1099/ijs.0.65740-0
Mamlouk, D., and Gullo, M. (2013). Acetic acid bacteria: physiology and carbon sources oxidation. Indian J. Microbiol. 53, 377–384. doi: 10.1007/s12088-013-0414-z
Marič, L., Cleenwerck, I., Accetto, T., Vandamme, P., and Trček, J. (2020). Description of Komagataeibacter melaceti sp. nov. and Komagataeibacter melomenusus sp. nov. isolated from apple cider vinegar. Microorganisms 8:1178. doi: 10.3390/microorganisms8081178
Masud, U., Matsushita, K., and Theeragool, G. (2010). Cloning and functional analysis of adhS gene encoding quinoprotein alcohol dehydrogenase subunit III from Acetobacter pasteurianus SKU1108. Int. J. Food Microbiol. 138, 39–49. doi: 10.1016/j.ijfoodmicro.2009.12.027
Matsushita, K., Fujii, Y., Ano, Y., Toyama, H., Shinjoh, M., Tomiyama, N., et al. (2003). 5-keto-D-gluconate production is catalyzed by a quinoprotein glycerol dehydrogenase, major polyol dehydrogenase, in Gluconobacter species. Appl. Environ. Microbiol. 69, 1959–1966. doi: 10.1128/AEM.69.4.1959-1966.2003
Matsushita, K., Patel, L., and Kaback, H. R. (1984). Cytochrome o oxidase from Escherichia coli. Characterization of the enzyme and mechanism of electrochemical proton gradient generation. Biochemistry 23, 4703–4714. doi: 10.1021/bi00315a028
Matsushita, K., Toyama, H., and Adachi, O. (2004). Respiratory Chains in Acetic Acid Bacteria: Membranebound Periplasmic Sugar and Alcohol Respirations. Respiration in Archaea and Bacteria. Dordrecht: Springer, 81–99. doi: 10.1007/978-1-4020-3163-2_4
Matsushita, K., Toyama, H., and Adachi, O. (1994). Respiratory chains and bioenergetics of acetic acid bacteria. Adv. Microbial. Physiol. 36, 247–301. doi: 10.1016/S0065-2911(08)60181-2
Matsushita, K., Toyama, H., Tonouchi, N., and Okamoto-Kainuma, A. (2016). Acetic Acid Bacteria—— Ecology and Physiology. Tokyo: Springer, Japan. doi: 10.1007/978-4-431-55933-7
Matsushita, K., Toyama, H., Yamada, M., and Adachi, O. (2002). Biotechnology quinoproteins: structure, function, and biotechnological applications. Appl. Microbiol. Biotechnol. 58, 13–22. doi: 10.1007/s00253-001-0851-1
Matsutani, M., Fukushima, K., Kayama, C., Arimitsu, M., Hirakawa, H., Toyama, H., et al. (2014). Replacement of a terminal cytochrome c oxidase by ubiquinol oxidase during the evolution of acetic acid bacteria. Biochim. Biophys. Acta 1837, 1810–1820. doi: 10.1016/j.bbabio.2014.05.355
Merfort, M., Herrmann, U., Bringer-Meyer, S., and Sahm, H. (2006). High-yield 5-keto-D-gluconic acid formation is mediated by soluble and membrane-bound gluconate-5-dehydrogenases of Gluconobacter oxydans. Appl. Microbiol. Biotechnol. 73, 443–451. doi: 10.1007/s00253-006-0467-6
Mientus, M., Kostner, D., Peters, B., Liebl, W., and Ehrenreich, A. (2017). Characterization of membrane-bound dehydrogenases of Gluconobacter oxydans 621H using a new system for their functional expression. Appl. Microbiol. Biotechnol. 101, 3189–3200. doi: 10.1007/s00253-016-8069-4
Miura, H., Mogi, T., Ano, Y., Migita, C. T., Matsutani, M., Yakushi, T., et al. (2013). Cyanide-insensitive quinol oxidase (CIO) from Gluconobacter oxydans is a unique terminal oxidase subfamily of cytochrome bd. J. Biochem. 153, 535–545. doi: 10.1093/jb/mvt019
Miyazaki, T., Tomiyama, N., Shinjoh, M., and Hoshino, T. (2002). Molecular cloning and functional expression of d-sorbitol dehydrogenase from Gluconobacter suboxydans IFO3255, which requires pyrroloquinoline quinone and hydrophobic protein SldB for activity development in E. coli. Biosci. Biotechnol. Biochem. 66, 262–270. doi: 10.1271/bbb.66.262
Molinari, F., Aragozzini, F., Cabral, J., and Prazeres, D. (1996). Continuous production and in situ extraction of isovaleraldehyde in a membrane bioreactor. Enzyme Microb. Technol. 20, 604–611. doi: 10.1016/S0921-0423(96)80074-8
Molinari, F., Gandolfi, R., Aragozzini, F., Leon, R., and Prazeres, D. M. (1999). Biotransformations in two-liquid-phase systems: production of phenylacetaldehyde by oxidation of 2-phenylethanol with acetic acid bacteria. Enzyme Microb. Technol. 25, 729–735. doi: 10.1016/S0141-0229(99)00107-6
Moonmangmee, D., Adachi, O., Toyama, H., and Matsushita, K. (2004). D-hexosaminate production by oxidative fermentation. Appl. Microbiol Biotechnol. 66, 253–258. doi: 10.1007/s00253-004-1707-2
Moore, J. E., McCalmont, M., Xu, J., Millar, B. C., and Heaney, N. (2002). Asaia sp., an unusual spoilage organism of fruit-flavored bottled water. Appl. Environ. Microbiol. 68, 4130–4131. doi: 10.1128/AEM.68.8.4130-4131.2002
Nanda, K., Taniguchi, M., Ujike, S., Ishihara, N., Mori, H., Ono, H., et al. (2001). Characterization of acetic acid bacteria in traditional acetic acid fermentation of rice vinegar (komesu) and unpolished rice vinegar (kurosu) produced in Japan. Appl. Environ. Microbiol. 67, 986–990. doi: 10.1128/AEM.67.2.986-990.2001
Nishikura-Imamura, S., Matsutani, M., Insomphun, C., Vangnai, A. S., Toyama, H., Yakushi, T., et al. (2014). Overexpression of a type II 3-dehydroquinate dehydratase enhances the biotransformation of quinate to 3-dehydroshikimate in Gluconobacter oxydans. Appl. Microbiol. Biotechnol. Adv. 98, 2955–2963. doi: 10.1007/s00253-013-5439-z
Noyori, R. (2002). Asymmetric catalysis: science and opportunities (Nobel lecture). Angewandte Chemie Int. Ed. 41, 2008-2022. doi: 10.1002/1521-3773(20020617)41:12<2008::AID-ANIE2008>3.0.CO;2-4
Osbourn, A. E., and Field, B. (2009). Operons. Cell. Mol. Life Sci. 66, 3755–3775. doi: 10.1007/s00018-009-0114-3
Pappenberger, G., and Hohmann, H. P. (2013). Industrial production of L-ascorbic acid (vitamin C) and D-isoascorbic acid. Biotechnol. Food Feed Additiv. 143–188. doi: 10.1007/10_2013_243
Peters, B., Junker, A., Brauer, K., Mühlthaler, B., Kostner, D., Mientus, M., et al. (2013). Deletion of pyruvate decarboxylase by a new method for efficient markerless gene deletions in Gluconobacter oxydans. Appl. Microbiol. Biotechnol. 97, 2521–2530. doi: 10.1007/s00253-012-4354-z
Prust, C., Hoffmeister, M., Liesegang, H., Wiezer, A., Fricke, W. F., Ehrenreich, A., et al. (2005). Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat. Biotechnol. 23, 195–200. doi: 10.1038/nbt1062
Qazi, G., Parshad, R., Verma, V., Chopra, C., Buse, R., Träger, M., et al. (1991). Diketo-gluconate fermentation by Gluconobacter oxydans. Enzyme Microb. Technol. 13, 504–507. doi: 10.1016/0141-0229(91)90010-8
Qi, X. H., Zhu, J. F., Yun, J. H., Lin, J., Qi, Y. L., Guo, Q., et al. (2016). Enhanced xylitol production: expression of xylitol dehydrogenase from Gluconobacter oxydans and mixed culture of resting cell. J. Biosci. Bioeng. 122, 257–262. doi: 10.1016/j.jbiosc.2016.02.009
Qin, Z., Yu, S., Chen, J., and Zhou, J. (2022). Dehydrogenases of acetic acid bacteria. Biotechnol. Adv. 54:107863. doi: 10.1016/j.biotechadv.2021.107863
Raspor, P., and Goranovič, D. (2008). Biotechnological applications of acetic acid bacteria. Crit. Rev. Biotechnol. 28, 101–124. doi: 10.1080/07388550802046749
Richhardt, J., Luchterhand, B., Bringer, S., Büchs, J., and Bott, M. (2013). Evidence for a key role of cytochrome bo 3 oxidase in respiratory energy metabolism of Gluconobacter oxydans. J. Bacteriol. 195, 4210–4220. doi: 10.1128/JB.00470-13
Richtmyer, N. K., Stewart, L. C., and Hudson, C. (1950). L-fuco-4-ketose, a new sugar produced by the action of Acetobacter suboxydans on L-Fucitol1. J. Am. Chem. Soc. 72, 4934–4937. doi: 10.1021/ja01167a025
Romano, A., Gandolfi, R., Nitti, P., Rollini, M., and Molinari, F. (2002). Acetic acid bacteria as enantioselective biocatalysts. J. Mol. Catalysis B 17, 235–240. doi: 10.1016/S1381-1177(02)00013-9
Saichana, N., Matsushita, K., Adachi, O., Frébort, I., and Frebortova, J. (2015). Acetic acid bacteria: a group of bacteria with versatile biotechnological applications. Biotechnol. Adv. 33, 1260–1271. doi: 10.1016/j.biotechadv.2014.12.001
Sato, J., Wakayama, M., and Takagi, K. (2015). Lactate dehydrogenase involved in lactate metabolism of Acetobacter Pasteurianus. Proc. Environ. Sci. 28, 67–71. doi: 10.1016/j.proenv.2015.07.010
Sato, K., Yamada, Y., Aida, K., and Uemura, T. (1967). On the formation of 5-keto-d-fructose and three γ-pyrone compounds from d-sorbitol by Acetobacter suboxydans. Agri. Biol. Chem. 31, 877–879. doi: 10.1080/00021369.1967.10858894
Sayed, M., Pyo, S.-H., Rehnberg, N., and Hatti-Kaul, R. (2019). Selective oxidation of 5-hydroxymethylfurfural to 5-hydroxymethyl-2-furancarboxylic acid using Gluconobacter oxydans. ACS Sustain. Chem. Eng. 7, 4406–4413. doi: 10.1021/acssuschemeng.8b06327
Sengun, I. Y. (2017). Acetic Acid Bacteria: Fundamentals and Food Applications. Oxfordshire: Taylor & Francis Group. doi: 10.1201/9781315153490
Shi, L., Li, K., Zhang, H., Liu, X., Lin, J., and Wei, D. (2014). Identification of a novel promoter gHp0169 for gene expression in Gluconobacter oxydans. J. Biotechnol. 175, 69–74. doi: 10.1016/j.jbiotec.2014.01.035
Shinagawa, E., Ano, Y., Yakushi, T., Adachi, O., and Matsushita, K. (2009). Solubilization, purification, and properties of membrane-bound D-glucono-delta-lactone hydrolase from Gluconobacter oxydans. Biosci. Biotechnol. Biochem. 73, 241–244. doi: 10.1271/bbb.80554
Shinagawa, E., Matsushita, K., Adachi, O., and Ameyama, M. (1982). Purification and characterization of D-sorbitol dehydrogenase from membrane of Gluconobacter suboxydans var. Agri. Biol. Chem. 46, 135–141. doi: 10.1271/bbb1961.46.135
Shinagawa, E., Matsushita, K., Adachi, O., and Ameyama, M. (1984). D-Gluconate dehydrogenase, 2-keto-D-gluconate yielding, from Gluconobacter dioxyacetonicus: purification and characterization. Agri. Biol. Chem. 48, 1517–1522. doi: 10.1271/bbb1961.48.1517
Shinagawa, E., Matsushita, K., Toyama, H., and Adachi, O. (1999). Production of 5-keto-D-gluconate by acetic acid bacteria is catalyzed by pyrroloquinoline quinone (PQQ)-dependent membrane-bound D-gluconate dehydrogenase. J. Mol. Catal. B. Enzym. 6, 341–350.
Shinagawa, E., Toyama, H., Matsushita, K., Tuitemwong, P., Theeragool, G., and Adachi, O. (2006). A novel type of formaldehyde-oxidizing enzyme from the membrane of Acetobacter sp. SKU 14. Biosci. Biotechnol. Biochem. 70, 850–857. doi: 10.1271/bbb.70.850
Shinjoh, M., and Hoshino, T. (1995). Development of a stable shuttle vector and a conjugative transfer system for Gluconobacter oxydans. J. Ferment. Bioeng. 79, 95–99. doi: 10.1016/0922-338X(95)94074-2
Shinjoh, M., Tomiyama, N., Miyazaki, T., and Hoshino, T. (2002). Main polyol dehydrogenase of Gluconobacter suboxydans IFO 3255, membrane-bound D-sorbitol dehydrogenase, that needs product of upstream gene, sldB, for activity. Biosci., Biotechnol. Biochem. 66, 2314–2322. doi: 10.1271/bbb.66.2314
Su, W., Chang, Z., Gao, K., and Wei, D. (2004). Enantioselective oxidation of racemic 1,2-propanediol to D-(–)-lactic acid by Gluconobacter oxydans. Tetrahedron 15, 1275–1277. doi: 10.1016/j.tetasy.2004.03.009
Sugisawa, T., and Hoshino, T. (2002). Purification and properties of membrane-bound D-sorbitol dehydrogenase from Gluconobacter suboxydans IFO 3255. Biosci. Biotechnol. Biochem. 66, 57–64. doi: 10.1271/bbb.66.57
Sugisawa, T., Hoshino, T., Nomura, S., and Fujiwara, A. (1991). Isolation and characterization of membrane-bound L-sorbose dehydrogenase from Gluconobacter melanogenus UV10. Agri. Biol. Chem. 55, 363–370. doi: 10.1271/bbb1961.55.363
Sugisawa, T., Miyazaki, T., and Hoshino, T. (2005). Microbial production of L-ascorbic acid from D-sorbitol, L-sorbose, L-gulose, and L-sorbosone by Ketogulonicigenium vulgare DSM 4025. Biosci. Biotechnol. Biochem. 69, 659–662. doi: 10.1271/bbb.69.659
Suzuki, S., Sugiyama, M., Mihara, Y., Hashiguchi, K., and Yokozeki, K. (2002). Novel enzymatic method for the production of xylitol from D-arabitol by Gluconobacter oxydans. Biosci. Biotechnol. Biochem. 66, 2614–2620. doi: 10.1271/bbb.66.2614
Švitel, J., and Kutnik, P. (1995). Potential of acetic acid bacteria for oxidation of low-molecular monoalcohols. Lett. Appl. Microbiol. 20, 365–368. doi: 10.1111/j.1472-765X.1995.tb01322.x
Švitel, J., and Šturdik, E. (1994). D-Galactose transformation to D-Galactonic acid by Gluconobacter oxydans. J. Biotechnol. 37, 85–88. doi: 10.1016/0168-1656(94)90206-2
Švitel, J., and Šturdík, E. (1995). n-Propanol conversion to propionic acid by Gluconobacter oxydans. Enzyme Microb. Technol. 17, 546–550. doi: 10.1016/0141-0229(94)00088-9
Swings, J., Gillis, M., Kersters, K., De Vos, P., Gosselé, F., and De Ley, J. (1980). Frateuria, a new genus for “Acetobacter aurantius”. Int. J. Systemat. Evol. Microbiol. 30, 547–556. doi: 10.1099/00207713-30-3-547
Takahashi, T., and Asai, T. (1932). On the formation of fructose and kojic acid by acetic acid bacteria. Proc. Imperial Acad. 8, 364–366. doi: 10.2183/pjab1912.8.364
Takahashi, T., and Kayamori, H. (1960). Studies on the formation of glucosamine acid by Acetobacter melanogenum Beijerinck and Pseudomonas fluorescens liquefaciens. J. Agri. Chem. Soc. Jap. 24, 231–234. doi: 10.1080/03758397.1960.10857663
Takeshita, K., Shimonishi, T., and Izumori, K. (1996). Production of L-psicose from allitol by Gluconobacter frateurii IFO 3254. J. Ferment. Bioeng. 81, 212–215. doi: 10.1016/0922-338X(96)82210-0
Tanasupawat, S., Thawai, C., Yukphan, P., Moonmangmee, D., Itoh, T., Adachi, O., et al. (2004). Gluconobacter thailandicus sp. nov., an acetic acid bacterium in the α-Proteobacteria. J. General Appl. Microbiol. 50, 159–167. doi: 10.2323/jgam.50.159
Tayama, K., Fukaya, M., Okumura, H., Kawamura, Y., and Beppu, T. (1989). Purification and characterization of membrane-bound alcohol dehydrogenase from Acetobacter polyoxogenes sp. nov. Appl. Microbiol. Biotechnol. Adv. 32, 181–185. doi: 10.1007/BF00165885
Thurner, C., Vela, C., Thöny-Meyer, L., Meile, L., and Teuber, M. (1997). Biochemical and genetic characterization of the acetaldehyde dehydrogenase complex from Acetobacter europaeus. Archiv. Microbiol. 168, 81–91. doi: 10.1007/s002030050473
Toyama, H., Furuya, N., Saichana, I., Ano, Y., Adachi, O., and Matsushita, K. (2007). Membrane-bound, 2-Keto-D-Gluconate-yielding D-gluconate dehydrogenase from Gluconobacter dioxyacetonicus IFO 3271: molecular properties and gene disruption. Appl. Environ. Microbiol. 73, 6551–6556. doi: 10.1128/AEM.00493-07
Toyama, H., Soemphol, W., Moonmangmee, D., Adachi, O., and Matsushita, K. (2005). Molecular properties of membrane-bound FAD-containing D-sorbitol dehydrogenase from thermotolerant Gluconobacter frateurii isolated from Thailand. Biosci. Biotechnol. Biochem. 69, 1120–1129. doi: 10.1271/bbb.69.1120
Trček, J., and Barja, F. (2015). Updates on quick identification of acetic acid bacteria with a focus on the 16S−23S rRNA gene internal transcribed spacer and the analysis of cell proteins by MALDI-TOF mass spectrometry. Int. J. Food Microbiol. 196, 137–144. doi: 10.1016/j.ijfoodmicro.2014.12.003
Trcek, J., Toyama, H., Czuba, J., Misiewicz, A., and Matsushita, K. (2006). Correlation between acetic acid resistance and characteristics of PQQ-dependent ADH in acetic acid bacteria. Appl. Microbiol. Biotechnol. Adv. 70, 366–373. doi: 10.1007/s00253-005-0073-z
Urakami, T., Tamaoka, J., Suzuki, K. I., and Komagata, K. (1989). Acidomonas gen. nov., incorporating Acetobacter methanolicus as Acidomonas methanolica comb. nov. Int. J. Systemat. Evol. Microbiol. 39, 50–55. doi: 10.1099/00207713-39-1-50
Urbance, J., Bratina, B., Stoddard, S., and Schmidt, T. (2001). Taxonomic characterization of Ketogulonigenium vulgare gen. nov., sp. nov. and Ketogulonigenium robustum sp. nov., which oxidize L-sorbose to 2-keto-L-gulonic acid. Int. J. Systemat. Evol. Microbiol. 51, 1059–1070. doi: 10.1099/00207713-51-3-1059
Vangnai, A., Promden, W., De-Eknamkul, W., Matsushita, K., and Toyama, H. (2010). Molecular characterization and heterologous expression of quinate dehydrogenase gene from Gluconobacter oxydans IFO3244. Biochemistry 75, 452–459. doi: 10.1134/S0006297910040085
Wang, B., and Chen, F. (2014). Taxonomy progress of acetic acid bacteria. China Brewing 33, 1–10. doi: 10.11882/j.issn.0254-5071.2014.12.001
Wang, P., Xia, Y., Li, J., Kang, Z., Zhou, J., and Chen, J. (2016). Overexpression of pyrroloquinoline quinone biosynthetic genes affects L-sorbose production in Gluconobacter oxydans WSH-003. Biochem. Eng. J.112, 70–77. doi: 10.1016/j.bej.2016.04.011
Wang, X., Lv, M., Zhang, L., Li, K., Gao, C., Ma, C., et al. (2013). Efficient bioconversion of 2, 3-butanediol into acetoin using Gluconobacter oxydans DSM 2003. Biotechnol. Biofuels 6, 1–9. doi: 10.1186/1754-6834-6-155
Wei, G., Yang, X., Zhou, W., Lin, J., and Wei, D. (2009). Adsorptive bioconversion of ethylene glycol to glycolic acid by Gluconobacter oxydans DSM 2003. Biochem. Eng. J. 47, 127–131. doi: 10.1016/j.bej.2009.07.016
Wei, P., Liang, J., Cheng, J., Zong, M. H., and Lou, W. Y. (2016). Markedly improving asymmetric oxidation of 1-(4-methoxyphenyl) ethanol with Acetobacter sp. CCTCC M209061 cells by adding deep eutectic solvent in a two-phase system. Microb. Cell Fact. 15:5. doi: 10.1186/s12934-015-0407-1
Whiting, G., and Coggins, R. J. (1967). The oxidation of D-quinate and related acids by Acetomonas oxydans. Biochem. J. 102, 283–293. doi: 10.1042/bj1020283
Xu, S., Wang, X., Du, G., Zhou, J., and Chen, J. (2014). Enhanced production of L-sorbose from D-sorbitol by improving the mRNA abundance of sorbitol dehydrogenase in Gluconobacter oxydans WSH-003. Microb. Cell Factor. 13, 1–7. doi: 10.1186/s12934-014-0146-8
Xu, Y., Chi, P., Lv, J., Bilal, M., and Cheng, H. (2021). L-xylo-3-hexulose, a new rare sugar produced by the action of acetic acid bacteria on galactitol, an exception to Bertrand Hudson's rule. Biochim. Biophys. Acta Gen Subj. 1865:129740. doi: 10.1016/j.bbagen.2020.129740
Yakushi, T., and Matsushita, K. (2010). Alcohol dehydrogenase of acetic acid bacteria: structure, mode of action, and applications in biotechnology. Appl. Microbiol. Biotechnol. 86, 1257–1265. doi: 10.1007/s00253-010-2529-z
Yakushi, T., Takahashi, R., Matsutani, M., Kataoka, N., Hours, R. A., Ano, Y., et al. (2020). The membrane-bound sorbosone dehydrogenase of Gluconacetobacter liquefaciens is a pyrroloquinoline quinone-dependent enzyme. Enzyme Microb. Technol. 137:109511. doi: 10.1016/j.enzmictec.2020.109511
Yamada, Y. (2000). Transfer of Acetobacter oboediens Sokollek et al 1998 and Acetobacter intermedius Boesch et al. 1998 to the genus Gluconacetobacter as Gluconacetobacter oboediens comb. nov. and Gluconacetobacter intermedius comb. nov. Int. J. Systemat. Evol. Microbiol. 50, 2225–2227. doi: 10.1099/00207713-50-6-2225
Yamada, Y. (2014). Transfer of Gluconacetobacter kakiaceti, Gluconacetobacter medellinensis and Gluconacetobacter maltaceti to the genus Komagataeibacter as Komagataeibacter kakiaceti comb. nov., Komagataeibacter medellinensis comb. nov. and Komagataeibacter maltaceti comb. nov. Int. J. Systemat. Evol. Microbiol. 64, 1670–1672. doi: 10.1099/ijs.0.054494-0
Yamada, Y. (2016). “Systematics of acetic acid bacteria,” in Acetic Acid Bacteria: Ecology and Physiology, eds. K. Matsushita, H. Toyama, N. Tonouchi and A. Okamoto-Kainuma (Tokyo: Springer Japan), 1–50. doi: 10.1007/978-4-431-55933-7_1
Yamada, Y., Hoshino, K. I., and Ishikawa, T. (1997). The phylogeny of acetic acid bacteria based on the partial sequences of 16S ribosomal RNA: the elevation of the subgenus Gluconoacetobacter to the generic level. Biosci. Biotechnol. Biochem. 61, 1244–1251. doi: 10.1271/bbb.61.1244
Yamada, Y., and Kondo, K. (1984). Gluconoacetobacter, a new subgenus comprising the acetate-oxidizing acetic acid bacteria with ubiquinone-10 in the genus Acetobacter. J. General Appl. Microbiol. 30, 297–303. doi: 10.2323/jgam.30.297
Yamada, Y., and Yukphan, P. (2008). Genera and species in acetic acid bacteria. Int. J. Food Microbiol. 125, 15–24. doi: 10.1016/j.ijfoodmicro.2007.11.077
Yamada, Y., Yukphan, P., Vu, H. T. L., Muramatsu, Y., Ochaikul, D., and Nakagawa, Y. (2012b). Subdivision of the genus Gluconacetobacter Yamada, Hoshino and Ishikawa 1998: the proposal of Komagatabacter gen. nov., for strains accommodated to the Gluconacetobacter xylinus group in the α-Proteobacteria. Ann. Microbiol. 62, 849–859. doi: 10.1007/s13213-011-0288-4
Yamada, Y., Yukphan, P., Vu, H. T. L., Muramatsu, Y., Ochaikul, D., Tanasupawat, S., et al. (2012a). Description of Komagataeibacter gen. nov., with proposals of new combinations (Acetobacteraceae). The Journal of general and applied microbiology. 58, 397–404. doi: 10.2323/jgam.58.397
Yang, H., Chen, T., Wang, M., Zhou, J., Liebl, W., Barja, F., et al. (2022). Molecular biology: fantastic toolkits to improve knowledge and application of acetic acid bacteria. Biotechnol. Adv. 2022:107911. doi: 10.1016/j.biotechadv.2022.107911
Zhang, X. Y., and Chen, F. S. (2022). Acetic Acid Bacteria and Their Application. Beijing: Chemical Industry Press.
Zhou, X., Zhou, X., Xu, Y., and Chen, R. R. (2017). Gluconobacter oxydans (ATCC 621H) catalyzed oxidation of furfural for detoxification of furfural and bioproduction of furoic acid. J. Chem. Technol. Biotechnol. 92, 1285–1289. doi: 10.1002/jctb.5122
Zhou, X., Zhou, X., Zhang, H., Cao, R., and Xu, Y. (2018). Improving the performance of cell biocatalysis and the productivity of acetoin from 2, 3-butanediol using a compressed oxygen supply. Process Biochem. 64, 46–50. doi: 10.1016/j.procbio.2017.09.027
Zhu, J., Xie, J., Wei, L., Lin, J., Zhao, L., and Wei, D. (2018). Identification of the enzymes responsible for 3-hydroxypropionic acid formation and their use in improving 3-hydroxypropionic acid production in Gluconobacter oxydans DSM. Bioresour. Technol. 265, 328–333. doi: 10.1016/j.biortech.2018.06.001
Keywords: acetic acid bacteria, classification, membrane-bound dehydrogenase, oxidative fermentation, molecular biology
Citation: He Y, Xie Z, Zhang H, Liebl W, Toyama H and Chen F (2022) Oxidative Fermentation of Acetic Acid Bacteria and Its Products. Front. Microbiol. 13:879246. doi: 10.3389/fmicb.2022.879246
Received: 19 February 2022; Accepted: 25 April 2022;
Published: 24 May 2022.
Edited by:
Albert Bordons, University of Rovira i Virgili, SpainReviewed by:
Maria J. Valera, Universidad de la República, UruguayCarla Jara, University of Chile, Chile
Montserrat Poblet, Rovira i Virgili University, Spain
Eveline Bartowsky, Lallemand, Canada
Copyright © 2022 He, Xie, Zhang, Liebl, Toyama and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fusheng Chen, Y2hlbmZzQG1haWwuaHphdS5lZHUuY24=
 Yating He
Yating He Zhenzhen Xie
Zhenzhen Xie Huan Zhang1,2
Huan Zhang1,2 Wolfgang Liebl
Wolfgang Liebl Fusheng Chen
Fusheng Chen