- 1CAS Key Laboratory of Biofuels, Shandong Provincial Key Laboratory of Synthetic Biology, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao, China
- 2Shandong Energy Institute, Qingdao, China
- 3Qingdao New Energy Shandong Laboratory, Qingdao, China
- 4Centre of Excellence in Integrated Omics and Computational Biology, Utkal University, Bhubaneswar, India
- 5College of Life Science, University of Chinese Academy of Sciences, Beijing, China
- 6Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Cyanobacteria from the representative hot springs of Yunnan Province, China are explored for their diversity and community composition following an integrative approach of cultivation-independent and -dependent studies and further isolation of potential taxa for future biotechnological perspective. 16S rRNA amplicon sequencing of microbial mats in these hot springs with temperature ranging from 38 to 90°C revealed Cyanobacteria and Proteobacteria constituting a bounteous portion of the bacterial community. The combined approach of 16S rRNA amplicon sequencing and phenotypic analysis revealed the diversity of cyanobacteria (a total of 45 genera). Out of these, a total of 19 cyanobacterial taxa belonging to 6 genera and 10 species were isolated as individuals with the possibility of biotechnological utilization. These isolates were subjected to a thorough morphological study and molecular characterization using 16S rRNA gene sequencing for identification and understanding their phylogeny. The identity and phenotypic and genotypic characteristics of 7 cyanobacterial isolates are not identical to any known cyanobacterial species, generating scope for future taxonomic novelties. Preliminary experiments based on high-temperature (50°C) cultivation showed that most of the isolates were thermotolerant and suggested for their high biotechnological usage potential.
Introduction
Hot springs or thermal springs are natural geological phenomena related to the expulsion of hot water (at or above 36.7°C) from the earth (Pentecost et al., 2003). These are stabilized ecosystems with highly constant abiotic components like temperature, pH, and ionic composition, which also inhabit consistent biotic resources. This consortium of microorganisms not only is adaptive to thermal extremity but also persisted through a long period of time, which can provide an insight into microbial evolution (McGregor and Rasmussen, 2008). The micro-vegetation in the thermal springs is dominated by cyanobacteria when the pH value is beyond 6 and temperature is below 74°C (Brock, 1967). There were several documentations of cyanobacteria that can survive at higher temperatures (Castenholz, 1969; Singh et al., 2018). Hot springs harbor potential thermotolerant strains for biotechnological applications. Mat-forming thermal cyanobacteria have the ability to produce a wide range of secondary metabolites with bioactive properties (Brito et al., 2015; Derikvand et al., 2017), which could be the rationality behind the isolation, and to enrich the culture collection of thermophilic cyanobacteria.
Hot springs of Yunnan Province are located in the Indo-Burma range, which is well known for widespread geothermal activity as a result of the complex geological history of the region (Hedlund et al., 2012; Hou et al., 2013). Extensive volcanism resulted in geothermal areas throughout Tengchong town, leading to many active hot springs (Du et al., 2005). Hot springs of Tengchong can be considered as biodiversity hotspots and compared with microbial diversity inhabiting other geothermal areas around the world, such as Yellowstone National Park, United States (Fournier, 1989), Japan (Yoshida, 1991), and Kamchatka, Russia (Kyle et al., 2007). Both cultivation-dependent and -independent examinations as well as biotechnological potential of thermophilic microbes (bacteria and archaea) from the hot springs of Yunnan Province were studied (Hedlund et al., 2012). However, a comprehensive census of the cyanobacterial communities, especially through next-generation sequencing (NGS) of hyper-variable regions of the 16S rRNA gene and isolation of potential strains for biotechnological applications, is still lacking.
Cyanobacteria were traditionally divided into five subsections based on their morphological complexity. Cyanobacteria of subsections I and II are unicellular while those of subsections III–V form various filamentous morphologies. Cells of subsection I divide by binary fission, whereas those of subsection II can also reproduce through multiple fissions in three planes to generate baeocytes (small, easily dispersed cells). Strains in subsection III have only vegetative cells, but in subsections IV and V, vegetative cells can differentiate into heterocysts (specialized nitrogen-fixing cells) or akinetes (resting cells that survive environmental stresses). In addition, strains in subsection V are further distinguished by branching patterns of their filaments (Tomitani et al., 2006; Shih et al., 2013).
To enrich the current state of the knowledge, the cyanobacterial diversity in the biofilms of different hot springs of Yunnan Province was explored for the first time. The objective of the study addresses four main questions: (1) What is the cyanobacterial composition in these thermal microbial mats? (2) How is the morphological diversity related to the meta-genomic diversity? (3) Could combined phenotypic and meta-genomic approach help for better understanding of cyanobacterial diversity? (4) Could the hot spring cyanobacterial isolates truly be able to endure heat stress under laboratory conditions? We have conducted both cultivation-independent (meta-genomic and traditional microscopic study together) and -dependent studies that offer a comprehensive assessment of microbial composition and isolation of novel cyanobacteria to explore their properties, respectively. Thus, the findings will not only expand our current knowledge of thermal microbiome in China but also provide clues and cyanobacterial strain resources for their biotechnological prospective.
Materials and Methods
Study Sites and Sampling
The sampling sites are under the Indo-Burma biodiversity hotspot, located near the China-Myanmar border (Figure 1) in Tengchong town (Dongshan, Huangpo, Rehai and Qinghai) and Longling town (Banglazhang), Yunnan Province, China. Sampling was conducted from 16 different sites including water, wet sediment, and microbial mats during August 2017, targeting especially cyanobacterial population and their isolation (Figure 1 and Table 1). Limnological features like pH, temperature (°C), and conductivity (μS/cm) were recorded using a portable pH meter equipped with a pH probe, a temperature probe, and a conductivity probe (Thermo Orion 420C-01A), respectively, on the spot. Microbial biofilms from various substrates were scrapped using sterile forceps and spatula into 50-ml Falcon tubes. Water samples were collected in pre-sterilized 1-L polyethylene bottles for chemical analysis. These samples were immediately stored on dry ice for transportation and later kept at a −80°C deep freezer in the laboratory until further analysis. Limnological parameters including concentration of cations (calcium, magnesium, potassium, ammonium, sodium, and lithium) and anions (nitrite, nitrate, phosphate, sulfate, bromide, chloride, and fluoride) were analyzed using Dionex ICS-5000 + ion-exchange chromatography system equipped with Dionex IonPac CS12A and Dionex IonPac CS18 column (250 × 4 mm, 5 μm, ThermoFisher, Waltham, MA, United States), respectively.
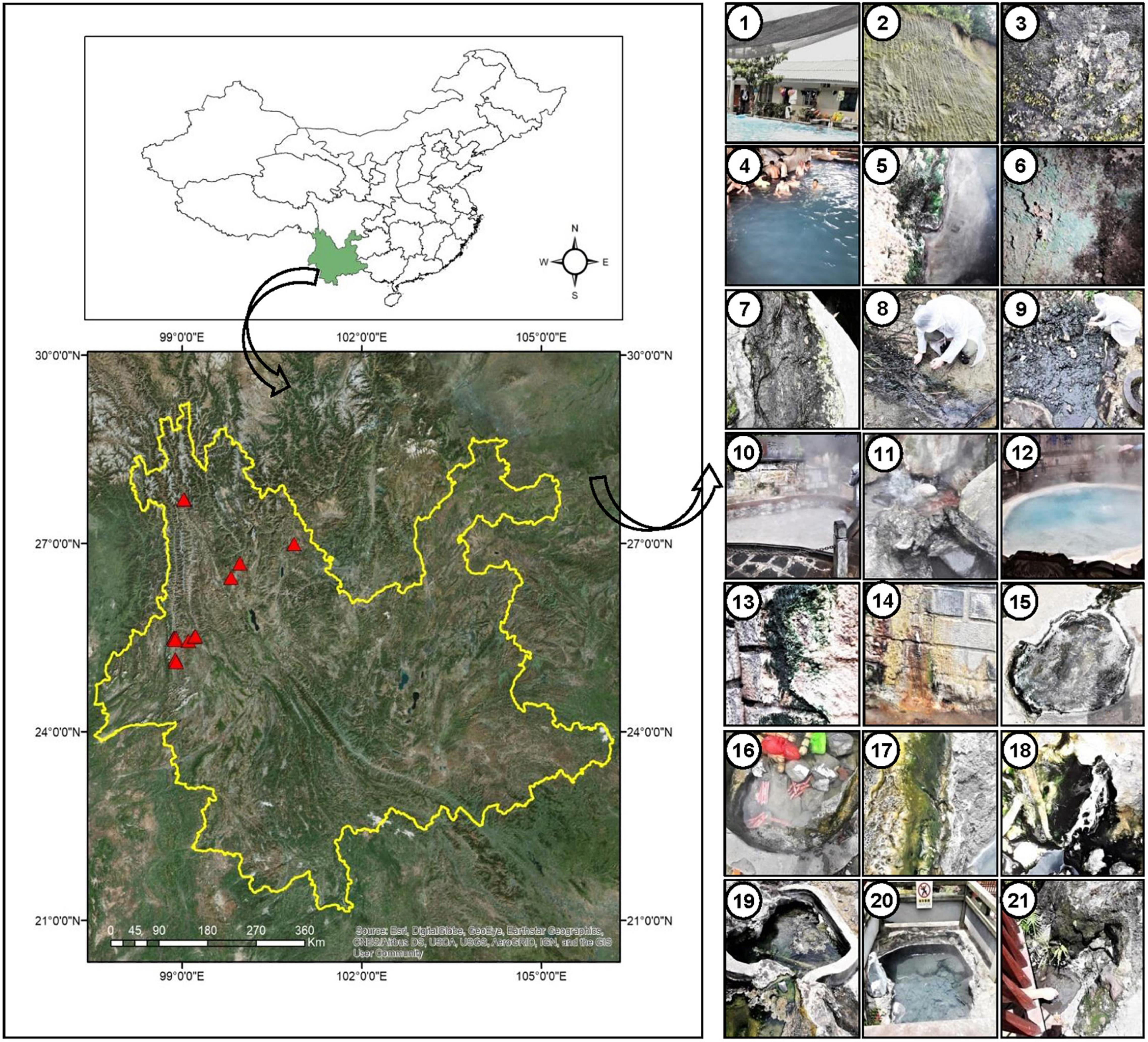
Figure 1. Location of sampling sites (marked with red triangles) in Yunnan Province, China. Geographic coordinates were determined using a Garmin Etrex GPS. Digital Elevation Model (DEM) data information was downloaded from Resource and Environment data cloud Platform of China (http://www.resdc.cn/). Pictures in the right panel indicate the surrounding sampling environment of the cyanobacterial biofilms in different hot springs, 1: Dongshan-001, 2–3: Huangpo-002 (soil), 4–7: Huangpo-003 (4 samples), 8: Rehai (Hamazui)-004, 9: Rehai (Hamazui)-005, 10: Rehai (Yanjingquan)-006, 11: Rehai (Zhenzhuquan)-007, 12: Rehai (Dagunguo)-008. 13–14: Rehai (Dagunguo)-009, 15: Banglazhang-011, 16–17: Banglazhang-012, 18: Banglazhang-013, 19: Banglazhang-014, 20: Banglazhang-015, 21: Banglazhang-016.
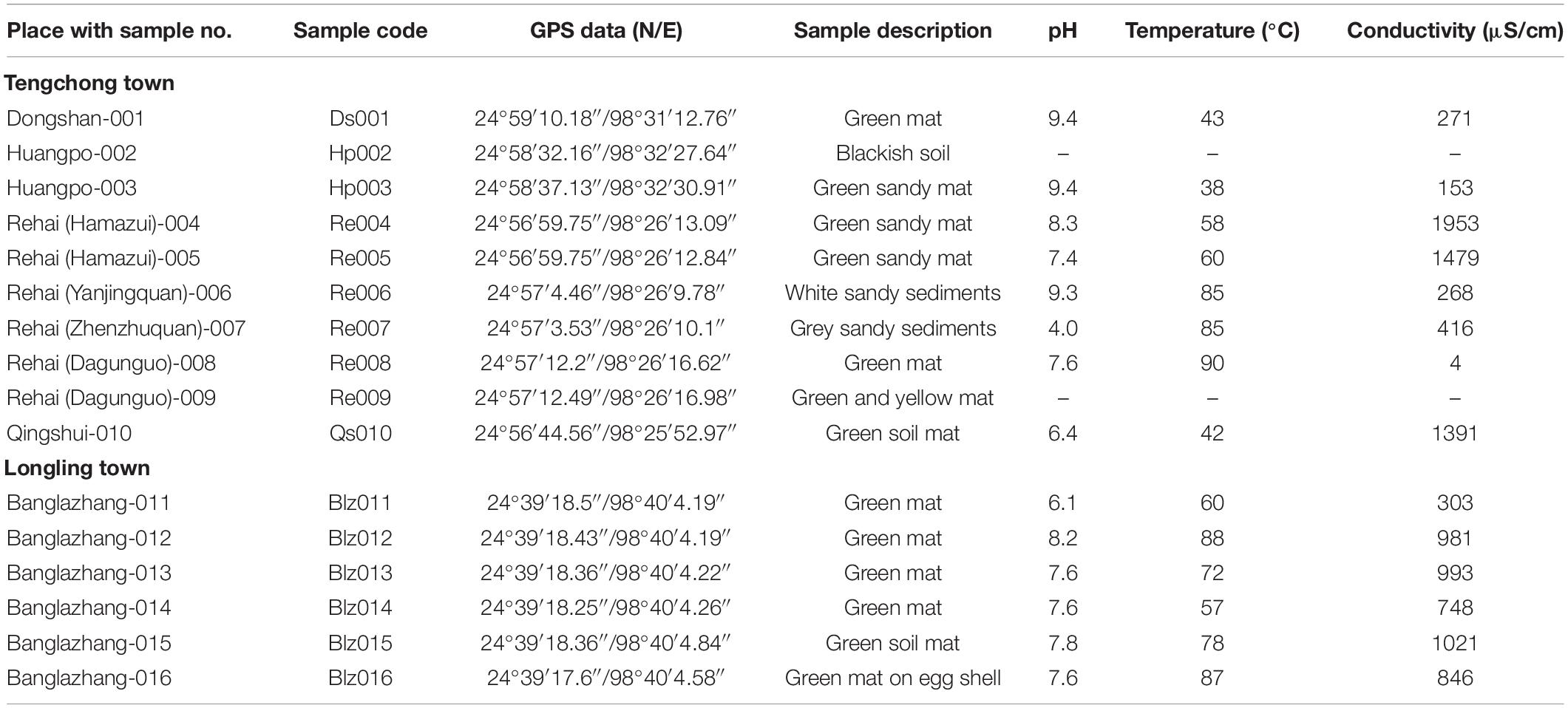
Table 1. Description of the sampling sites, hot spring samples, and environmental parameters (Yunnan Province).
16S rRNA Amplicon Sequencing and Data Analysis
The next-generation sequencing library preparations and Illumina MiSeq sequencing were conducted at GENEWIZ, Inc. (Suzhou, China). DNA samples were quantified using a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, United States). DNA (30–50 ng) was used to generate amplicons using a MetaVx™ Library Preparation kit (GENEWIZ, Inc., South Plainfield, NJ, United States). Primers targeting V3–V4 hypervariable regions of the 16S rRNA gene were proved to be able to capture the bacterial diversity in various environments (Klindworth et al., 2013). Thus, these regions were selected to generate amplicons and follow taxonomy analysis. These regions were amplified using primer pairs V3V4-F (CCTACGGRRBGCASCAGKVRVGAAT) and V3V4-R (GGACTACNVGGGTWTCTAATCC). First-round polymerase chain reaction (PCR) products were used as templates for second-round amplicon enrichment PCR. At the same time, indexed adapters were added to the ends of the 16S rRNA amplicons to generate indexed libraries ready for downstream NGS sequencing on Illumina Miseq. DNA libraries were validated by Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States), and quantified by Qubit 2.0 Fluorometer. DNA libraries were multiplexed and loaded on an Illumina MiSeq instrument according to the manufacturer’s instructions (Illumina, San Diego, CA, United States). Sequencing was performed using a 2 × 300 paired-end (PE) configuration; image analysis and base calling were conducted by the MiSeq Control Software (MCS) embedded in the MiSeq instrument. The QIIME data analysis package was used for 16S rRNA data analysis. The forward and reverse reads were joined and assigned to samples based on barcode and truncated by cutting off the barcode and primer sequences. Quality filtering on joined sequences was performed and sequences with length < 200 bp, ambiguous bases, and mean quality score ≥ 20 were discarded. Then, the sequences were compared with the reference database (RDP Gold database) using the UCHIME algorithm to detect chimeric sequences, and then the chimeric sequences were removed. The effective sequences were used in the final analysis. Sequences were grouped into operational taxonomic units (OTUs) using the clustering program VSEARCH (1.9.6) against the Silva 119 database pre-clustered at 97% sequence identity. The Ribosomal Database Program (RDP) classifier was used to assign taxonomic category to all OTUs at a confidence threshold of 0.8. The RDP classifier uses the Silva 123 database, which has taxonomic categories predicted to the species level.
Isolation and Culturing of Cyanobacteria Coupled With Temperature Stress Experiments
The biological mats were washed thoroughly by double-distilled water to remove inorganic debris, followed by homogenization by a glass homogenizer (1 ml) and then inoculated into Petri plates containing solidified BG11 medium (Rippka et al., 1979) at a slightly higher temperature of up to 40 ± 2°C under 30 μmol photon m–2 s–1 continuous light intensity in a temperature-controlled culture room. Axenic cultures of cyanobacteria were obtained by repeated sub-culturing on Petri plates.
To verify the thermotolerant features of these isolated cyanobacterial species, they were incubated in 10-ml test tubes at high temperature (50°C) with initial OD730 ∼0.06 under a continuous light intensity of 30 μmol photon m–2 s–1 without shaking.
Microscopy
Morphological features of the cyanobacteria were studied extensively under an Olympus CX31 microscope equipped with an Olympus DP-72 digital camera (Olympus, Tokyo, Japan) and software DP2-BSW (version 2.2) at 20 ms exposure time. The enumeration of phenotypic features was later considered for the identification of the taxa.
DNA Extraction and Polymerase Chain Reaction Amplification of 16S rRNA Gene of Isolated Cyanobacteria
Genomic DNA of all the isolates was extracted by using HiPurA™ plant genomic DNA Miniprep Purification Spin Kit. 16S rRNA gene from the isolated DNA was amplified using cyanobacteria-specific primers CYA106F and CYA781R (Nübel et al., 1997). PCR amplification consisted of an initial denaturation at 95°C for 5 min, 32 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 1 min, and extension at 72°C for 1 min, and a final extension at 72°C for 10 min. Individual reagents and their concentrations were as follows: 1 × PCR buffer with 1.5 mM MgCl2, dNTPs (100 μM each), 0.25 μM each primer, 2.5 U of DNA polymerase (Ex-Taq) (TaKaRa, Japan), and ∼50 ng of total DNA. PCR products were purified using an E.Z.N.A. Gel Extraction Kit (Omega Bio-Tek, United States) according to the manufacturer’s instructions. The obtained sequences of 16S rRNA gene were sequenced and the ambiguities were removed by visual editing. The sequences were compared with the NCBI GenBank database using the BLASTn1 and sequences of uncultured organisms were excluded.
Nucleotide Sequence Data Deposition
The paired-end nucleotide sequences have been submitted in NCBI SRA database with the accession IDs SRR11796781–SRR11796799 and SAMN14918520–SAMN14918537 for BioSample accessions under the BioProject PRJNA628565. Sequences of 16S rRNA genes obtained during the present study were deposited in GenBank with accession numbers (MK625304–MK625325).
Phylogenetic Analyses
All OTUs belonging to cyanobacteria and 16S rRNA gene sequence of cyanobacterial isolates were compared with the NCBI (National Center for Biotechnology Information) database using BLAST. An initial BLAST search of the NCBI GenBank database provided candidate sequences for comparing relationships of the cyanobacterial isolates with previously characterized cyanobacterial species. Different sets of sequences were combined together to construct 3 types of phylogenetic trees in the present study, which are as follows: (1) the sequences of major OTUs of cyanobacteria/bacteria of corresponding sampling spots and 16S rRNA gene sequence of cyanobacterial isolates; (2) the sequences of all reported OTUs of cyanobacteria and 16S rRNA gene sequence of cyanobacterial isolates; and (3) the 16S rRNA gene sequences of other hot spring cyanobacteria reported worldwide and 16S rRNA gene sequence of cyanobacterial isolates of the present study. Multiple alignments were created with reference to the selected GenBank sequences and Neighbor-joining phylogenetic trees were constructed from dissimilar distance and pairwise comparisons with the Jukes–Cantor distance model using the MEGA (molecular evolutionary genetics analysis) program, version 5 (Tamura et al., 2011). Bootstrap replications of 1,000 were assessed.
Results
Microbial Mat, Physicochemical Parameters, and Mineral Composition of Hot Spring Water
Microbial mats in the sampled thermal springs were both submerged and floating on the surface (Figure 1). Sometimes they appear as gelatinous films blanketed over sandy bottoms of a few hot springs. The textures of the mats were thin slimy gelatinous films to thick leathery mats with color ranging from bluish to yellowish green. The hot springs displayed a wide range of limnological characteristics, i.e., moderate to high temperature (38–90°C), acidic to slightly alkaline pH (4.0–9.4), and a variable conductivity (4–1,953 μS/cm), though the springs are in close proximity (Table 1). Based on the water temperature, the sampling sites were segregated into four groups representing different temperature gradients, i.e., Groups 1 (30–50°C), 2 (50–70°C), 3 (70–80°C), and 4 (80–90°C). Thermal springs Ds001, Hp003, and Qs010 (Group 1) were marked with the lowest temperature range (30–50°C), while the highest temperature range (80–90°C) was documented for the springs Re006, Re007, Re008, Blz012, and Blz016 (Group 4). Springs with temperature range 50–70°C were listed for Re004, Re005, Blz011, and Blz014 (Group 2). Only two springs Blz013 and Blz015 (Group 3) had a temperature range 70–80°C.
A comparative ionic composition and conductivity of water samples are presented in supporting information (Supplementary Figure 1). The abundance of sodium ion (Na+), among the cations in all the water samples, was distinctly observed followed by the concentration of potassium (K+). Similarly, chloride and sulfate were the dominant anions. The variation in ionic concentrations and conductivity in Longling town hot springs was negligible, but significant in the hot springs of Tengchong town. Total ion analysis (cations and anions) showed its richness in water sample of Re008 and minimum in Hp003. It was also revealed that the conductivity was directly proportional to the concentration of ions present in water samples. However, the derivation of relatedness was not made for the sample Re008, due to insufficient water sampling.
Overview of Meta-16S rRNA Gene Sequencing Data (Total Microorganisms)
A total of 584,226 reads (1787 OTUs) were identified from these samples. Operational taxonomic unit Venn or Petal diagram was also generated to illustrate the distribution of OTUs among these four groups (Supplementary Figure 2). (1) Group 1/30–50°C (1,186 OTUs): Ds001, Hp003, and Qs010; (2) Group 2/50–70°C (761 OTUs): Re004, Re005, Blz011, and Blz014; (3) Group 3/70–80°C (322 OTUs): Blz013 and Blz015, and (4) Group 4/80–90°C (713 OTUs): Re006, Re007, Re008, Blz012, and Blz016. The OTUs restricted to any individual group were 833, 103, 15, and 60 for Group 1, Group 2, Group 3, and Group 4, respectively. It also showed the information on the distribution of shared OTUs across these four groups, which were 260, 80, 253, 261, 541, and 268 between Groups 1 and 2, Groups 1 and 3, Groups 1 and 4, Groups 2 and 3, Groups 2 and 4, and Groups 3 and 4, respectively. Larger proportions of OTUs (179) were shared between Groups 2 and 4. There was relatively lower overlap between Groups 1 and 3. Common OTUs shared by all the groups was 58. Chao1 was used to compare species richness between different groups (Supplementary Figure 3), which supported that OTU estimate was significantly higher in Group 1, i.e., the lowest temperature gradient in all gradients.
Thirty major OTUs were represented as clustering heat maps (Supplementary Figure 4), which showed the color of different shades based on the relative abundance of each OTU. Among these, OTUs 1, 2, 5, 7, 12, and 21 belong to cyanobacteria, where OTUs 2 and 5 were related to morphological subsection III. Microbial community composition based on pyrosequencing of 16S rRNA gene (Supplementary Figure 5) revealed Cyanobacteria and Proteobacteria representing a larger portion of bacterial community in the hot spring mats of Yunnan Province. Other most common bacterial genera were Firmicutes, Chloroflexi, Bacteroidetes, Acidobacteria, and Deinococcus-thermus. Abundance of cyanobacteria was reported in all the sampling sites except RH007, which was rich with Proteobacteria (Supplementary Figure 5A). Furthermore, it can also be inferred that an extremely high temperature of 85°C and the acidic water (pH 4.0) of RH007 favored the growth of Proteobacteria, Bacteroidetes, and Actinobacteria, but inhibited the growth of cyanobacteria. Another site with the hottest (85°C) but alkaline water (pH 9.3), i.e., RH006, supported the growth of the members of phylum Thermodesulfobacteria and Aquificae rather than Cyanobacteria and Proteobacteria. Our result showed that the bacterial phylum Cyanobacteria and Proteobacteria were dominant in all the studied hot springs. The dominating organism in microbial mat depended on the temperature, pH, and geochemistry of sampling sites (Hou et al., 2013). Species richness of cyanobacteria was higher in the springs HP003, RH009, BLZ014, and BLZ015 than any other spots (Supplementary Figure 5A). The lowest number of OTUs was observed in Group 3, i.e., temperature range of 70–80°C, but interestingly with the highest proportion belonging to cyanobacteria (Supplementary Figure 5B).
Overview of Meta-16S rRNA Gene Sequencing Data (Cyanobacteria)
This study on the microbial composition in these hot springs was intended to explore cyanobacterial diversity in more detail rather than dealing with genera of other microorganisms precisely. Out of the total OTUs, 100 OTUs belonging to cyanobacteria were identified. Ninety-four and 36 cyanobacterial OTUs were found in Tengchong and Longling town, respectively. The OTU Venn diagram was illustrated based on the distribution of cyanobacterial OTUs among the four groups of temperature range and three groups of pH range (Figure 2). It is quite clear that there were diversified cyanobacteria in hot springs with temperature 30–50°C (Figure 2A) and pH > 9 (Figure 2B). This was also supported by the Shannon–Weaver (Figure 3A) and Simpson index (Figure 3B) analysis. When grouped by region, the results show that the diversity of cyanobacteria in Tengchong has changed greatly, while the diversity in Longling town is relatively small (Figures 3C,D).
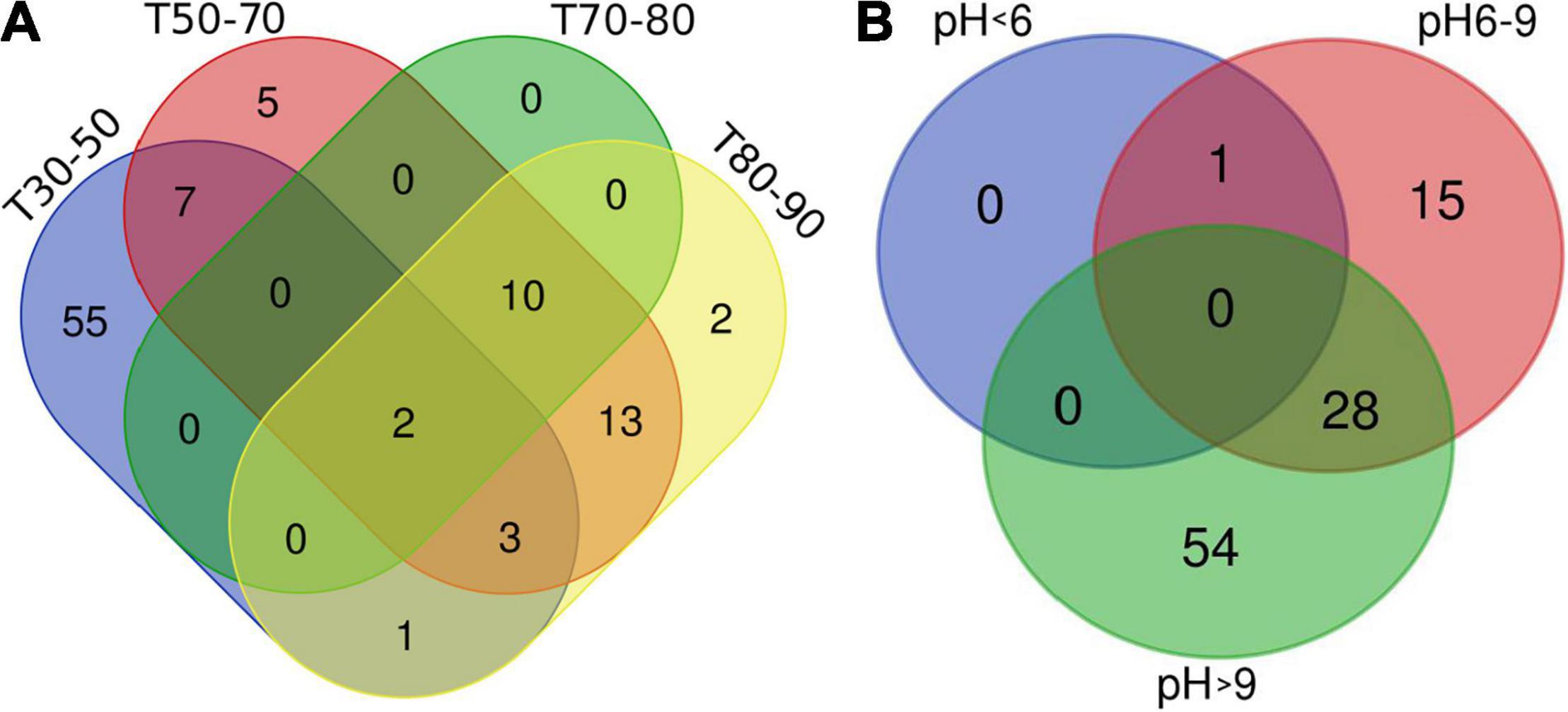
Figure 2. OTU Venn diagram illustrating distribution of cyanobacterial OTUs among the four groups of temperature range (A) and three groups of pH range (B). T indicates the temperatures of the water samples or the hot spring water temperatures of other sample types. T30–50 (Group 1: 30–50°C), T50–70 (Group 2: 50–70°C), T70–80 (Group 3: 70–80°C), and T80–90 (Group 4: 80–90°C). The Venn diagram was generated using the OmicStudio tools (https://www.omicstudio.cn/tool).
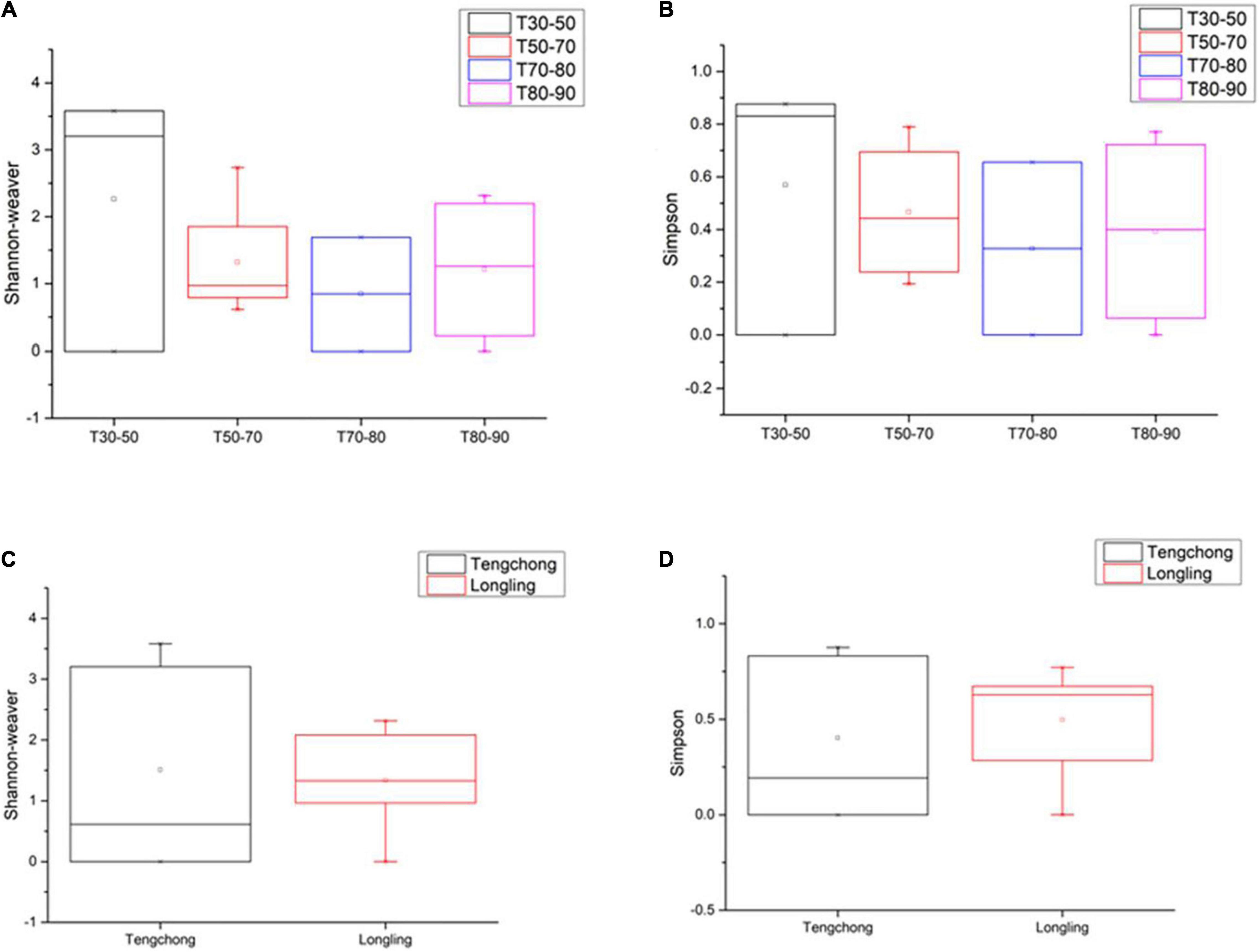
Figure 3. The Shannon–Weaver and Simpson indices of cyanobacterial OTUs in thermal springs based on temperature range (A,B) and location (C,D). The Box plot was generated using the Origin tools. Median values and interquartile ranges have been indicated in the plots. T indicates the temperatures of the water samples or the hot spring water temperatures of other sample types. T30–50 (Group 1: 30–50°C), T50–70 (Group 2: 50–70°C), T70–80 (Group 3: 70–80°C), and T80–90 (Group 4: 80–90°C).
According to the abundance of cyanobacteria OTUs at each sample point, 30 OTU species with higher abundance were selected to make a heat map (Figure 4). The abundance of different cyanobacteria in each sampling point (Figure 4A) as well as in different temperature ranges was depicted (Figure 4B), which showed that the types of cyanobacteria were quite different. In particular, in the T30–50 group, the cyanobacteria were significantly different from the other three groups, and the abundance of cyanobacteria was higher. The other three groups of cyanobacteria had relatively similar types and lower abundance, which may be due to high water temperature and insufficient nutrients in the water.
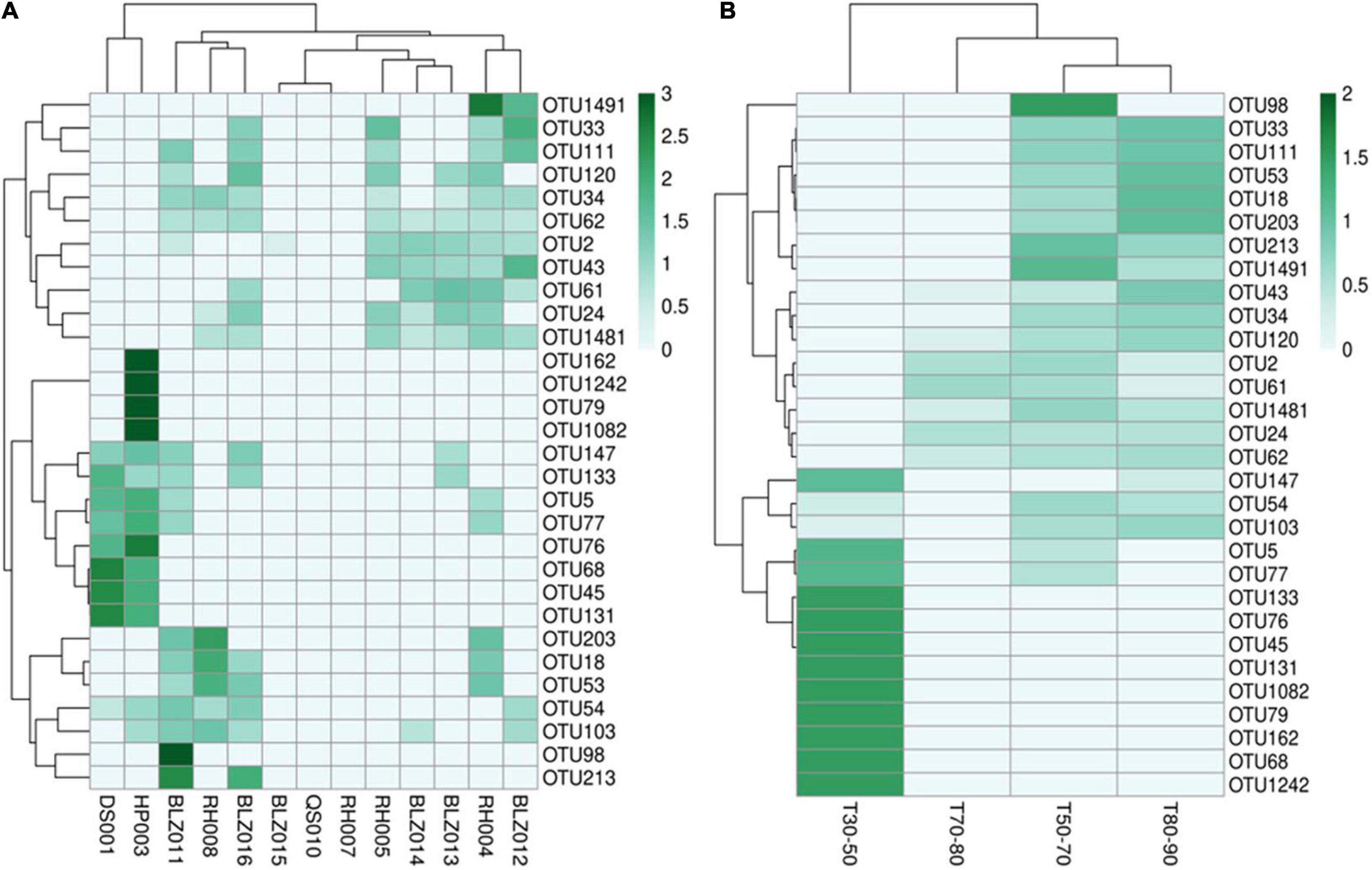
Figure 4. The cyanobacterial OTU abundance clustering heat map. The rows and columns represent the OTU IDs and the sampling sites (A) temperature groups (B), respectively. The OTU clustering tree is in the left. The value of each square color of the middle heat map corresponds to the relative abundance of each row of OTU. The heat map was generated using the OmicStudio tools (https://www.omicstudio.cn/tool).
The cyanobacterial OTUs found in the present study were assigned to 34 genera and 3 cyanobacterial groups (Synechococcus, Gloeocapsa, Aphanothece, Chroococcus, Cyanobium, Chondrocystis, Thermosynechococcus, Chroococcidiopsis, Pleurocapsa, Xenococcus, Gloeocapsopsis, Uncultured Chroococcales, Leptolyngbya, Oscillatoriales, Phormidium, Microcoleus, Leptolyngbyaceae cyanobacterium, Wilmottia, Arthronema, Pseudanabaena, Pseudanabaenaceae, Planktothricoides, Nodosilinea, Ancylothrix, Jaaginema, Oscillatoria, Microseira, Tolypothrix, Scytonema, Calothrix, Nostoc, Nodosilinea, Chlorogloeopsis, Stigonema, Mastigocladus, Iphinoe, and filamentous thermophilic cyanobacterium) approximately. More than 59% of total morphological subsection was represented by subsection III (59% of total subsections), especially the members of the genus Leptolyngbya. The second most abundant cyanobacterial group belonged to subsection II (18%) followed by subsections I (12%), V (6%), and IV (5%) (Figure 5A).
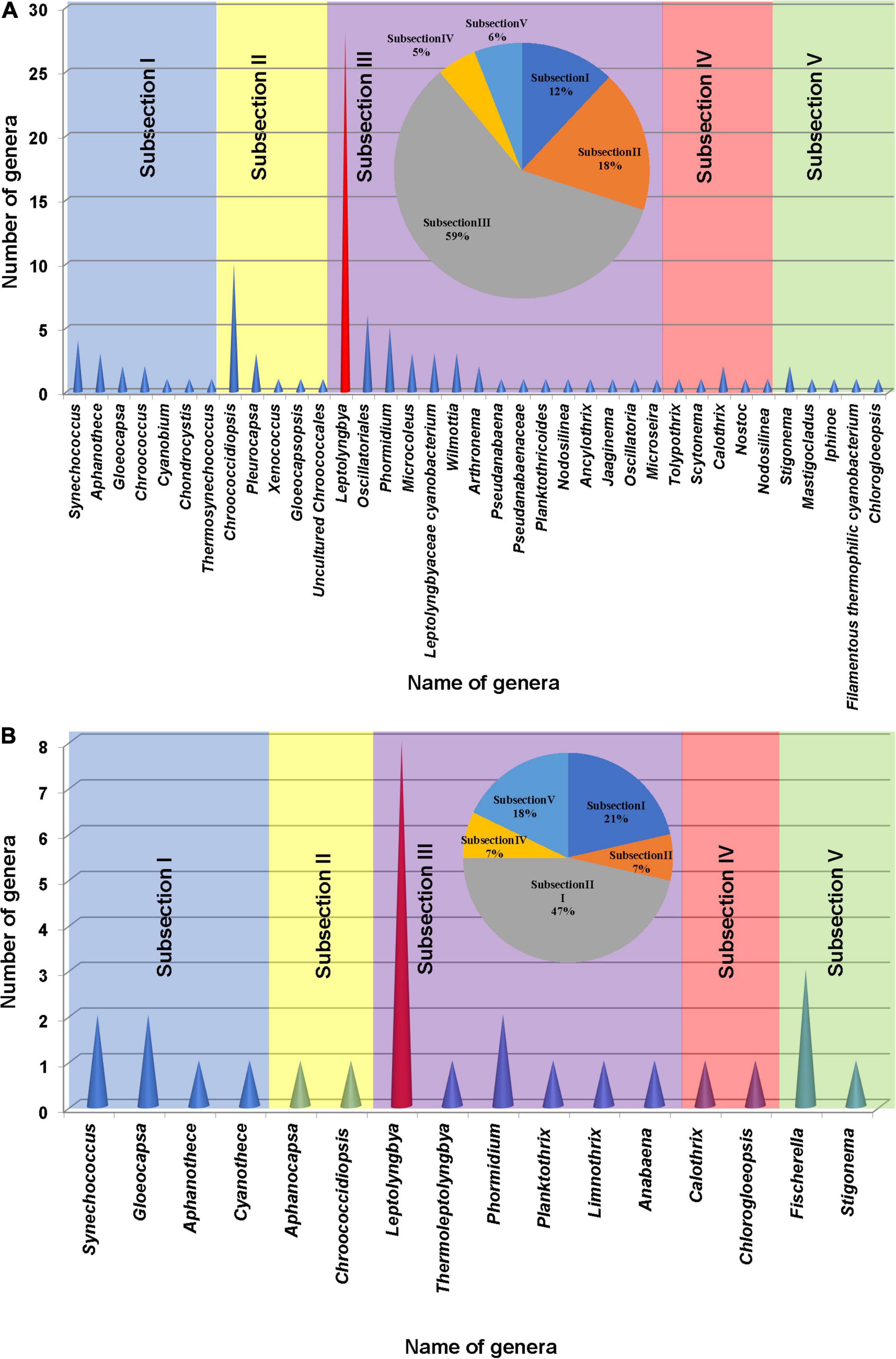
Figure 5. Cyanobacterial genera found based on the 16S rRNA amplicon sequencing analysis (A) and microscopic observation (B) of the samples collected from different studied sites of hot springs/hot water source of China. A pie chart was included in each histogram to display the percentage of cyanobacterial strains based on morphological subsections.
Phenotypic Characterization and Cyanobacterial Community Compositions
Floristic assessment of the mat-forming cyanobacteria and other associated algae through microscopy identified altogether 39 taxa from these thermal habitats of Yunnan Province (Supplementary Table 1). As stated, the thermal springs of Yunnan provide a wide range of habitats with diverse physicochemical parameters. The cyanobacteria in these extreme habitats thus get ample opportunity to adapt various environmental circumstances. To understand this, we tried to index the ecological specifications of the cyanobacterial species distributed in these thermal habitats. Cyanobacteria reported from Yunnan hot springs were categorized to adapt to various groups, i.e., moderate acidophilic, moderate alkaliphilic, mesothermophilic, thermophilic, and hyperthermophilic, depending on the pH and temperature of the hot springs from where they have reported/isolated (Table 2). Cyanobacteria were identified through extensive phenotypic characterizations (Figure 6 and Supplementary Figure 6). They displayed similar diversities at both species and generic levels. Leptolyngbya was the most diverse genus (8 taxa), followed by Fischerella (3 taxa). For more clarity in understanding the diversity, the taxa were grouped into different sub-sections to estimate their percentage of contribution to the biome (Figure 5B). It was revealed that the cyanobacterial species belonging to subsection III (47% of total cyanobacterial subsection diversity) were the major contributor in mat formation, which included taxa of Leptolyngbya, Thermoleptolyngbya, Phormidium, and Planktothrix, which were observed in 10 sampling sites out of the 16. Leptolyngbya dominated the hot springs of DS001, HP003–HP006, BLZ011, BLZ013, and BLZ014, whereas Thermoleptolyngbya dominated in HP006, HP007, and BLZ011. Likewise, Fischerella colonized the mats in QS010, BLZ012, and BLZ016. Synechococcus, Gloeocapsa, Chlorogloeopsis, and Anabaena were considered as the other associated mat-forming cyanobacterial species. It is noteworthy to mention that the sites with temperature > 80°C had only a few traces of cyanobacterial populations that were detected after extensive microscopy of the natural samples.
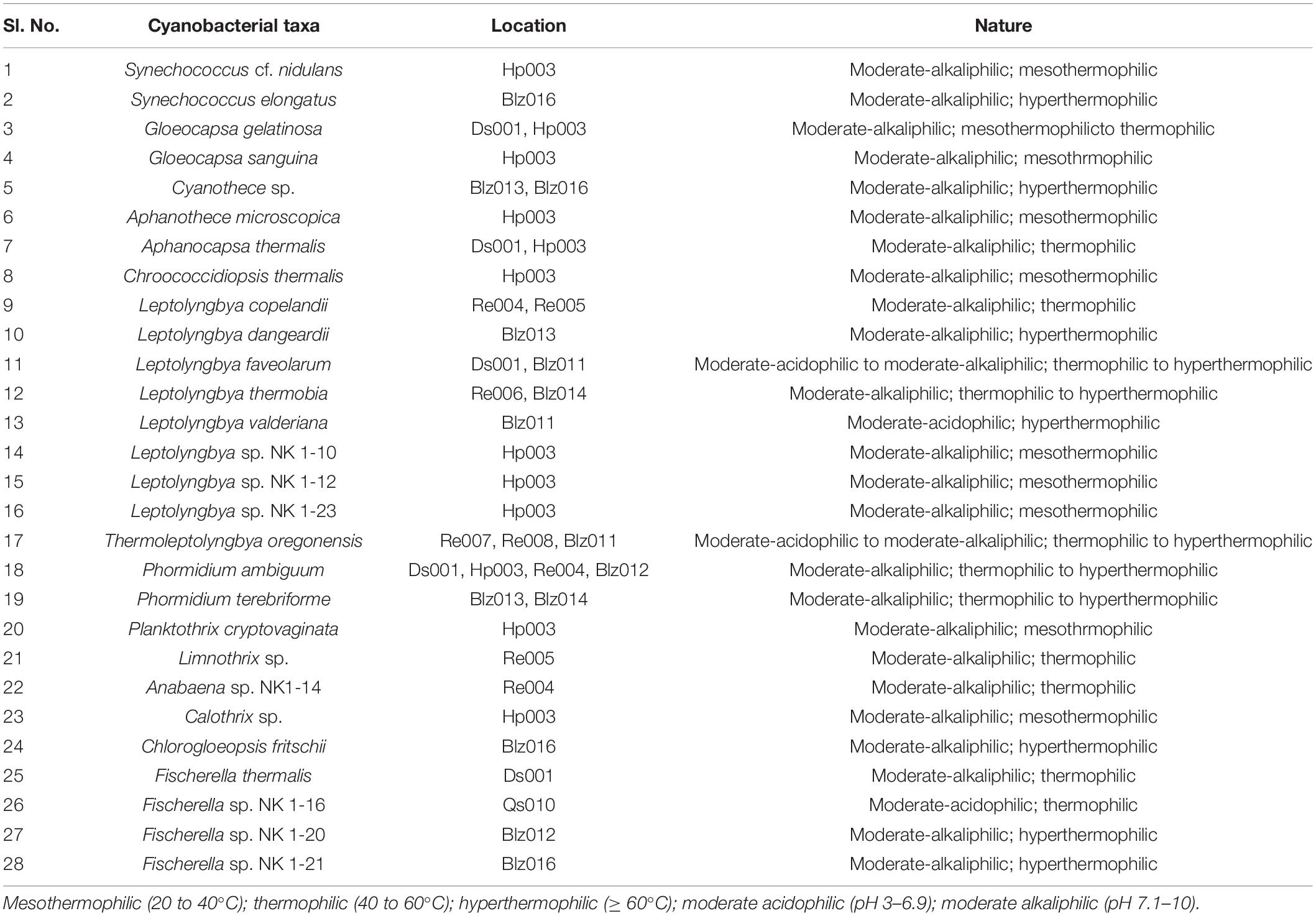
Table 2. Nature of reported cyanobacteria from Yunnan hot springs based on the range of pH and temperature of sampling sites.
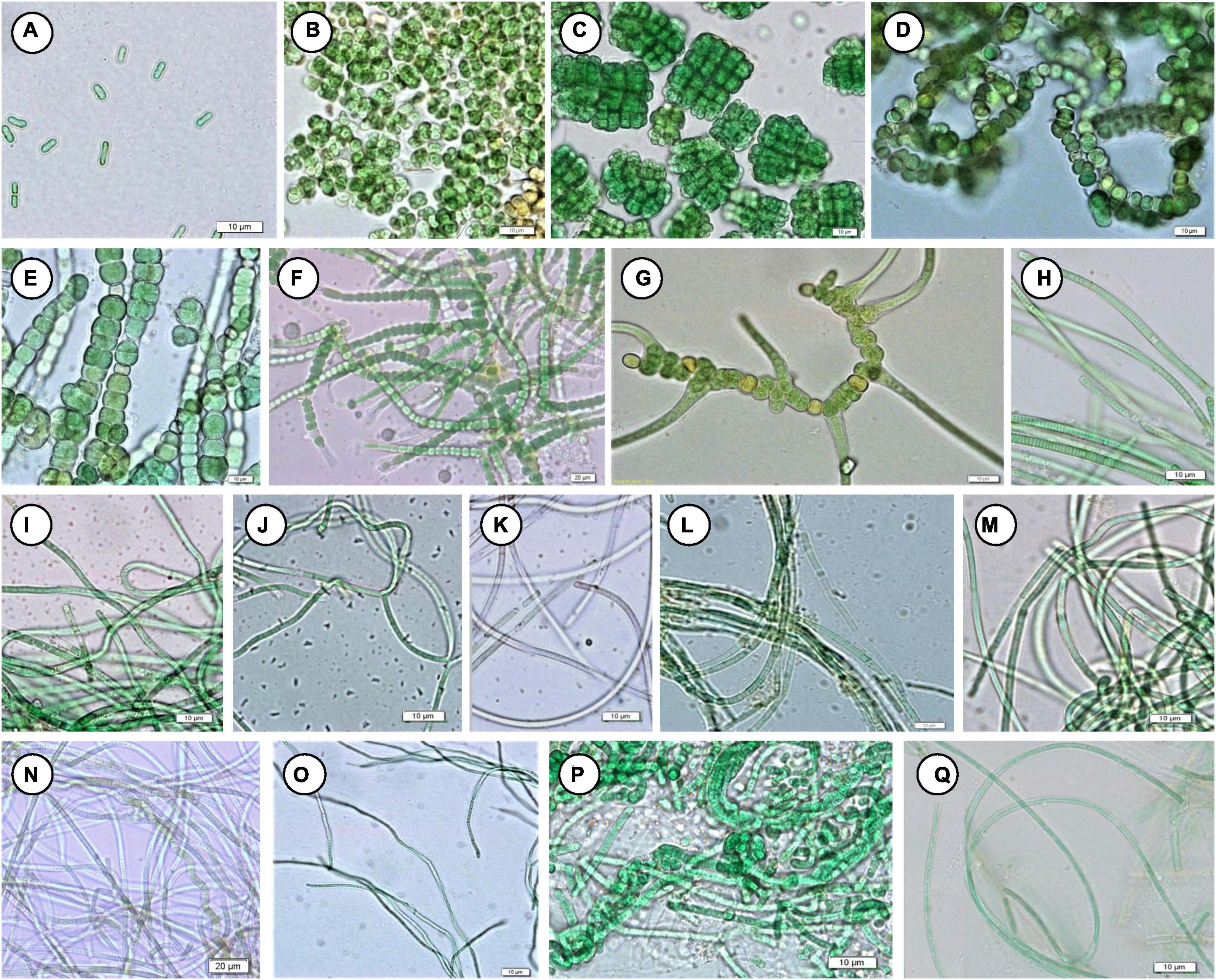
Figure 6. Mat-forming cyanobacteria identified in this study. (A) Synechococcus cf. nidulans, (B) Gloeocapsa gelatinosa, (C) Chlorogloeopsis fritschii, (D) Anabaena sp. NK1-14, (E) Fischerella sp. NK 1-16, (F) Fischerella sp. NK 1-20, (G) Fischerella thermalis, (H) Planktothrix cryptovaginata, (I) Leptolyngbya copelandii, (J) L. dangeardii, (K) L. faveolarum, (L) Leptolyngbya sp. NK 1-10, (M) Leptolyngbya sp. NK 1-23, (N) Leptolyngbya sp. NK 1-12, (O) Thermoleptolyngbya oregonensis, (P) Leptolyngbya thermobia, (Q) L. valderiana.
Comparison Between 16S rRNA Amplicon Sequencing and Phenotypic Observations
Operational taxonomic units (OTUs) belonging to genera Cyanothece, Aphanocapsa, Thermoleptolyngbya, Planktothrix, Limnothrix, Anabaena, and Fischerella were not identified in 16S rRNA sequencing, which may be due to insufficient samples/DNA quantity for high-throughput sequencing study as well as insufficient data in GenBank. The common genera in both 16S rRNA sequencing and phenotypic analysis were Synechococcus, Gloeocapsa, Aphanothece, Chroococcidiopsis, Leptolyngbya, Phormidium, Calothrix, and Chlorogloeopsis. Other cyanobacterial taxa reported in 16S rRNA sequencing analysis were not identified through microscopic observations because of their rarity in occurrence. We believe that the phenotypic characterization of any particular group of microorganisms, especially cyanobacteria, will impart more authenticity along with the 16S rRNA sequencing analysis in understanding the infrastructure of any microbial ecosystem. Further, the available genetic resource databases lack thorough taxonomic information including the identity of a species. Thus, we have adopted both methods together to reveal true cyanobacterial diversity in the hot springs of Yunnan Province, which was represented by a total of 45 cyanobacterial genera. The 16S rRNA sequencing analysis depicted the whole cyanobacterial diversity of the habitats with 100 different types of taxa, but their population and pattern of colonization were deeply elaborated after the phenotypic characterization, where the dominant mat-forming and associated taxa were segregated.
Isolation of Hot Spring Cyanobacteria and Verification of Their Thermotolerances
A total of 19 cyanobacterial strains belonging to 10 species and 6 genera were isolated to axenic cultures. These isolates will create the chance to understand the physiology of heat-tolerant cyanobacteria, screen for value-added products, and develop them as model microorganisms. They were closely observed for morphological alterations during their various growth phases, to facilitate more authentic identification. Their closest relatives in database (percentage similarity) and taxonomic assignment are presented in Table 3. Most of the isolates were identified up to species level except Leptolyngbya sp. NK 1-10, Leptolyngbya sp. NK 1-12, Anabaena sp. NK1-14, Fischerella sp. NK 1-16, Fischerella sp. NK 1-20, Fischerella sp. NK 1-21, and Leptolyngbya sp. NK 1-23. All these taxa were not identified up to species level, which could open the scope for identifying them as novel taxa with biotechnological potential in the near future.
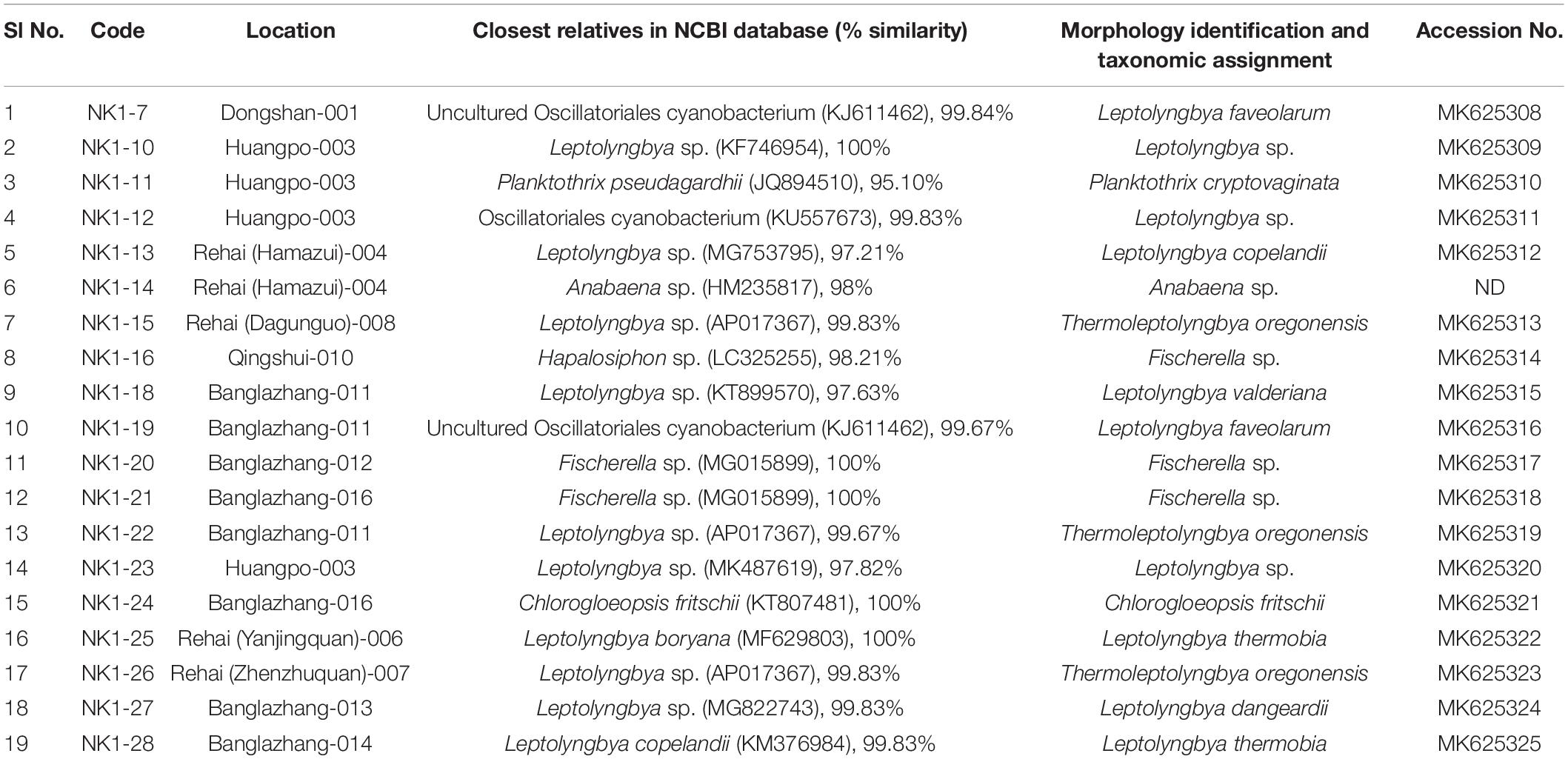
Table 3. List of identified cyanobacterial isolates from representative hot springs of Yunnan Province, China.
With the aim of confirming the thermotolerant feature of these isolated strains and evaluating their survival capacity at higher temperature, they were incubated at OD730 ∼0.06 in test tubes and cultivated statically at 50°C under 30 μmol photon m–2 s–1 light intensity for 10 days. Compared to their initial OD730, most of the cultures, except for NK1-10 and NK1-25, exhibited increased cell densities under these rigorous laboratory culture conditions (Figure 7). The results indicated the survivability of our isolates even at very low cell density under high temperature and further highlighted the significance of hot spring cyanobacterial strain mining using integrative approach.
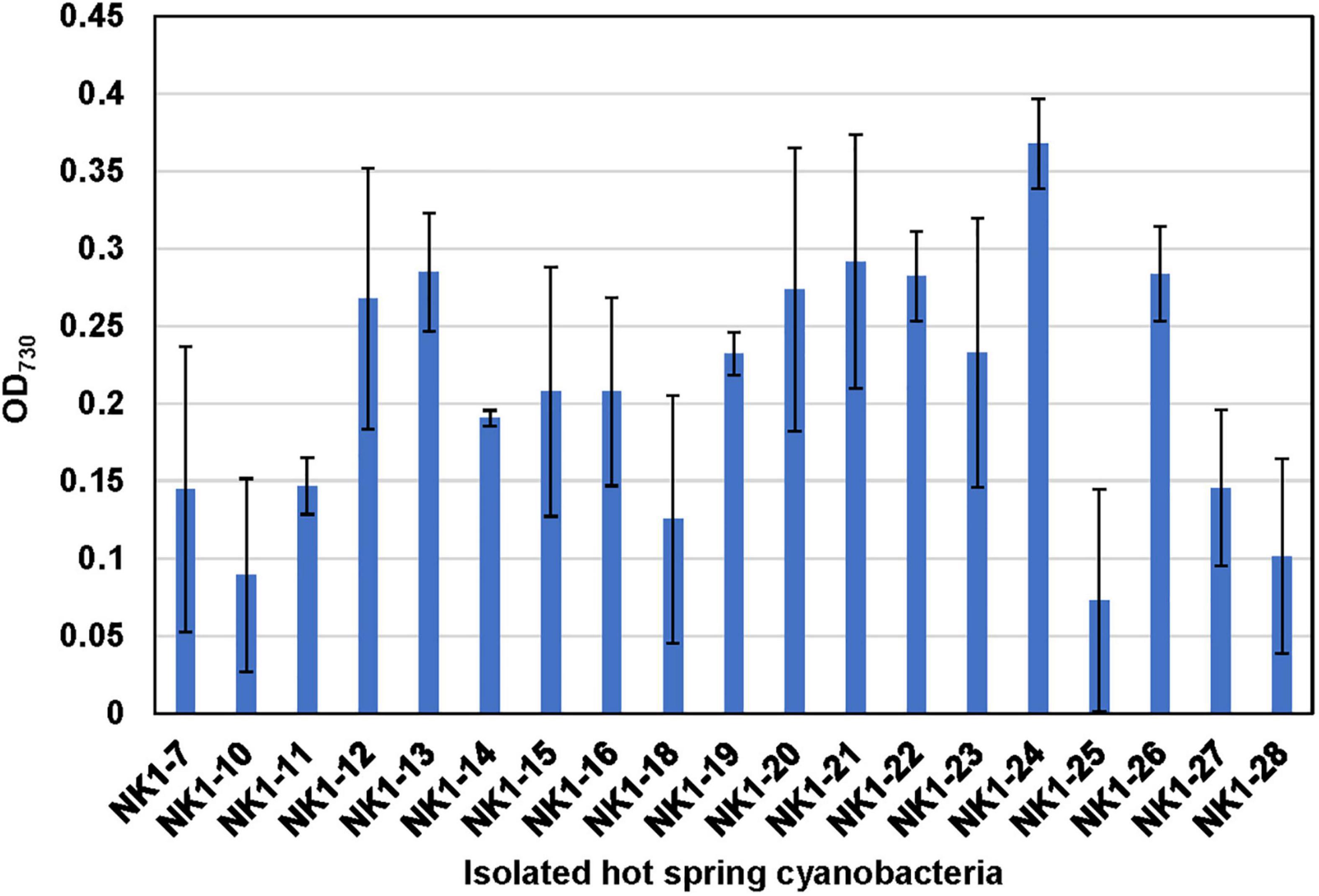
Figure 7. Verification of the thermotolerant feature of the hot spring cyanobacterial isolates. Note that the isolated cyanobacterial species was grown statically in 10-ml test tubes at 50°C under 30 μmol photon m–2 s–1 light intensity for 10 days.
Phylogenetic Analysis of Cyanobacteria
Phylogenetic relationships among the 16S rRNA gene sequences of isolated cyanobacterial species were analyzed along with the OTUs reported from our next-generation sequencing as well as other 16S rRNA gene sequences of related taxa. The OTUs obtained through next-generation sequencing were analyzed by NCBI BLAST search (one by one) and classified into different subsections (Supplementary Table 2). A total of 100 cyanobacterial OTUs were confirmed, which mainly belonged to subsection III and Leptolyngbya genera. Major OTUs of corresponding sampling spots and 16S rRNA gene sequences of isolated cyanobacteria species from the hot springs of Yunnan Province were compared, which showed that isolated cyanobacterial species belonged to same subsection as major cyanobacterial OTUs of the corresponding sampling site (Supplementary Table 3).
Different dendrograms of isolated cyanobacterial species were constructed using the following information: (1) major OTUs of cyanobacteria and microorganisms reported from 16S rRNA sequencing analysis (Supplementary Figure 7); (2) all OTUs of cyanobacteria reported in 16S rRNA sequencing (Supplementary Figure 8); and (3) already reported cyanobacteria isolated from hot springs (Figure 8).
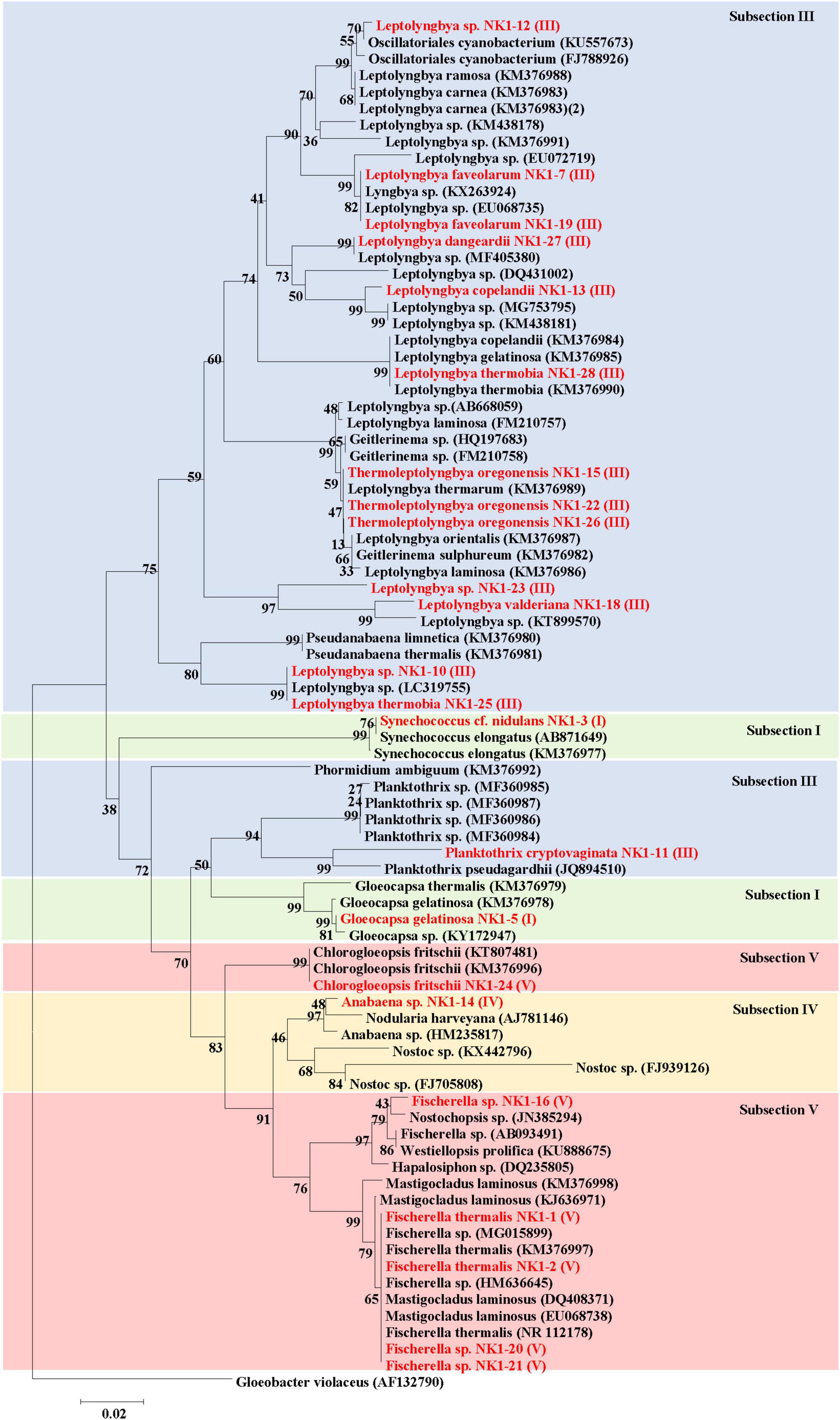
Figure 8. Phylogenetic relationships (Neighbor-Joining) between the isolated cyanobacteria species of hot springs of Yunnan Province, China and other closest relative hot spring cyanobacteria based on 16S rRNA gene sequences. Isolated cyanobacteria were indicated in red color. The number near the node represents the bootstrap value. Subsection was mentioned in brackets. Gloeobacter violaceus PCC 7421 (AF132790) was used as an out-group.
The sequences of major OTUs of corresponding sampling spots and 16S rRNA gene sequences of isolated cyanobacteria species from the hot springs of Yunnan Province were combined and Neighbor-joining phylogenetic trees were constructed (Supplementary Figure 7). The result showed that OTUs from 16S rRNA sequencing and isolated cyanobacterial species, belonging to the same subsection, clustered together. A major clade belonged to subsection III followed by subsection V. The OTUs 18, 33, 54, and 722 belonged to subsection III and clustered with different species of Leptolyngbya, Thermoleptolyngbya, and Planktothrix (isolated organism of the present study). Chlorogloeopsis fritschii NK1-24 was clustered with OTU1619, which showed the BLAST search similarity with C. fritschii (96%). Our isolated Fischerella sp. NK1-16, 20, and 21 clustered together with OTU298 under subsection V. OTUs 2 and 5 were the dominating cyanobacterial OTUs, which were classified as Uncultured Oscillatoriales and Phormidium autumnale based on the NCBI BLAST search, respectively. They clustered together and separated from the main subsection III clade. Other major OTUs belonging to bacterial taxa (OTU3, OTU4, OTU7, OTU8, OTU20, and OTU27) made a separate clade in the dendrogram.
Phylogenetic relationships between the isolated cyanobacterial species and all OTUs (100 OTUs) obtained through 16S rRNA sequencing of hot springs of Yunnan Province, China showed the clustering of isolated cyanobacteria in two major clades (Supplementary Figure 8). These two clades belong to subsections III, IV, and V. There were many other clades belonging to subsections I, II, and III, but showed separate clustering from our isolated organisms.
The 16S rRNA gene sequences of the isolated cyanobacteria were from BLAST in NCBI and sequences of other related taxa reported from hot springs were retrieved for the construction of a dendrogram (Figure 8). Phylogenetic analyses revealed that subsection III cyanobacteria were clustered in two clades. The first major clade of subsection III comprised different species of genera Leptolyngbya, Oscillatoriales cyanobacterium, Geitlerinema, Thermoleptolyngbya, and Pseudanabaena, while the second clade of subsection III included only Planktothrix and Phormidium. Cyanobacteria under Subsection V, i.e., Fischerella, Mastigocladus, Hapalosiphon, Westiellopsis, and Nostochopsis, were clustered together, while C. fritschii made a separate clade. Cyanobacterial members of subsection IV formed monophyletic clade including Anabaena and Nostoc. Subsection I clustered in different clades comprising mainly Gloeocapsa, Nodularia, and other hot spring cyanobacteria.
Discussion
Thermal habitats of China, specifically from the Tibetan plateau (Hongmei et al., 2005; Lau et al., 2009; Song et al., 2013), Yunnan Province (Xue et al., 2001; Lin et al., 2002; Xiang et al., 2003; Du et al., 2005; Song et al., 2009, 2010, 2013; Jiang et al., 2010; Hedlund et al., 2012, 2015; Pagaling et al., 2012; Hou et al., 2013; Li et al., 2015a), and Sichuan Province (Tang et al., 2018), were studied extensively for the last two decades because of their microbial diversity, taxonomy, basic physiology, cultivation, and biotechnological potential (Li et al., 2015b). While all previous studies were confined to various groups of bacteria (except cyanobacteria) and archaea, the present endeavor documented the cyanobacterial diversity in microbial mats from the hot springs of Yunnan Province, following both traditional microscopic methods as well as meta-genomic analysis of natural samples, followed by isolation of thermotolerant strains.
A wide physicochemical diversity of these springs (ambient to 97°C; pH from ≤1.8 to ≥9.3) provides a multitude of niches for extremophilic microorganisms and mainly limits them to bacteria, cyanobacteria, and archaea by restricting the other forms of life. Consistency in physicochemical properties in these hot springs can easily be observed by comparing our records with that of an earlier investigation that happened about a decade ago (Hedlund et al., 2012). This unchanged limnology provides a suitable ambience for thermotolerant species to adapt and thrive in these ecosystems. Species richness of microbes in thermal springs is dependent on temperature, pH, conductivity, and geochemistry of sampling sites (Hou et al., 2013). Sometimes, a single factor can regulate the richness of a particular species, which can be evidenced from the thermal habitats of Copahue ponds (Urbieta et al., 2014) and Garbanabra hot springs in Eritrea (Ghilamicael et al., 2017). The similar pattern was also observed for the thermal springs of Yunnan Province, where the total microbial richness was the lowest in Group 3 (70–80°C) springs, while, the highest richness was observed in Group 1 (30–50°C) springs. This supports the hypothesis by Miller et al. (2009) on the temperature regulation of the diversity of microorganisms in hot spring mats.
The hot springs of Yellowstone National Park have been used as the hot spot to explore the microbial community structure for many decades, which were dominated by Cyanobacteria and Chloroflexi (Ward et al., 1998, 2006; Klatt et al., 2011, 2013). The major phylotypes, including Cyanobacteria, Proteobacteria, Firmicutes, Chloroflexi, Acidobacteria, and Deinococcus-Thermus, have also been reported in the other hot springs at various geographical locations, i.e., China (Pagaling et al., 2012; Song et al., 2013), India (Mehetre et al., 2016; Saxena et al., 2016; Singh and Subudhi, 2016), Africa (Tekere et al., 2011; Ghilamicael et al., 2017), Malaysia (Chan et al., 2015), Kenya (Kambura et al., 2016), and Argentina (Urbieta et al., 2014). The combination of thermophilic cyanobacteria and anoxygenic photosynthetic bacteria seems to develop the photosynthetic microbial mats in diversified hot springs (Bateson et al., 1989; Ward et al., 1998).
Cyanobacteria in the hot springs of Yunnan mainly belonged to subsection III (filamentous form), especially Leptolyngbya as the most dominating genus. Therefore, it is believed to be the dominant and pioneer component of microbial mats in terms of biomass which governs the colonization of these mats and flow of nutrients within. Cyanobacteria need light, CO2, and N2 to fulfill their major requirements for growth, which makes them float. Hence, most of the cyanobacterial mats in hot springs of the present study were observed as floating on the surface of the water. The coccal Synechococcus and the filamentous heterocystous branched Cyanobacteria, like Mastigocladus and Fischerella, were the dominant mat-forming species around the globe (Castenholz, 1969; Ferris and Ward, 1997; Sompong et al., 2005; Miller et al., 2006, 2009; Ward et al., 2012). However, the Leptolyngbya dominated mats were also observed in several thermal springs (McGregor and Rasmussen, 2008; Moro et al., 2010; Coman et al., 2013; Mackenzie et al., 2013; Amarouche-Yala et al., 2014; Roy et al., 2015; Singh et al., 2018).
The study based on 16S rRNA amplicon sequencing and microscopic analysis together gave a clearer information on the cyanobacterial diversity. A total of 100 cyanobacterial OTUs comprising 37 genera were confirmed based on 16S rRNA gene pyrosequencing. Microscopic analysis also revealed cyanobacterial morphotypes and represented 27 cyanobacterial taxa belonging to 16 genera. The combined approach of morphological and high-throughput sequencing identified a wider cyanobacterial generic diversity (45 genera) in the hot springs of Yunnan Province, which were comparatively higher than the cyanobacterial diversity reported from the hot springs of other areas like Australia (McGregor and Rasmussen, 2008), Thailand (Sompong et al., 2005), and Bulgaria (Lukavský et al., 2011). However, 16S rRNA amplicon sequencing has revealed a much broader diversity of cyanobacteria than represented in culture, some taxa (of genera Cyanothece, Aphanocapsa, Thermoleptolyngbya, Planktothrix, Limnothrix, Anabaena and Fischerella), identified by microscopy, in 16S rRNA sequencing. Other cyanobacterial taxa reported in 16S rRNA amplicon sequencing were not reported in microscopic observation. It is evidenced that a cyanobacterial diversity study based on 16S rRNA amplicon sequencing alone often fails to recognize some taxa due to insufficient samples/DNA quantity as well as insufficient information in GenBank. Conversely, microscopic studies may fail to detect rare taxa due to limited resolution and a small part of microscopic samples when conducting analyses of cyanobacterial diversity of natural populations. Synechococcus, Gloeocapsa, Aphanothece, Chroococcidiopsis, Leptolyngbya, Phormidium, Calothrix, and Chlorogloeopsis were common genera in both 16S rRNA amplicon sequencing and microscopic analysis.
Almost all the cyanobacteria were moderately alkaliphilic (pH 7.1–10) except Leptolyngbya faveolarum and Thermoleptolyngbya oregonensis, which grew in moderately acidic springs (pH 4.0–6.1). Fischerella species generally occur in thermal springs of pH value > 5.0. In our study sites, Fischerella sp. NK 1-16 was found growing at a slightly acidic pH of 6.1, and the other three taxa of the genus, i.e., Fischerella sp. NK 1-20, Fischerella sp. NK 1-21, and F. thermalis, occurred in alkaline springs. About one-third of the cyanobacterial taxa preferred low temperature (20–40°C), specifically the members of the genera, Gloeocapsa, Aphanothece, Chroococcidiopsis, Planktothrix, and Calothrix. All the unidentified Leptolyngbya species were found to be mesothermophilic. However, Gloeocapsa gelatinosa was also recorded growing at a temperature slightly above 40°C in the hot springs of Dongshan in association with Aphanocapsa thermalis and Fischerella thermalis. Synechococcus, in general considered to be extremely thermotolerant, was reported to grow even at temperatures > 100°C around the globe. In the thermal springs of Yunnan Province, Synechococcus cf. nidulans was collected from Huangpo springs at 38°C and S. elongatus was found at 87°C in Banglazhang hot springs. Cyanothece species was also found to be hyperthermophilic. The most diverse genus in all these thermal springs, i.e., Leptolyngbya, was highly thermoadaptive, occurring in a wide temperature range of 38–85°C. The most adaptive ones were L. faveolarum and L. thermobia. Likewise, T. oregonensis was also found to be highly adaptive, with its distribution in a wide range of temperature (60–90°C) and pH (4.0–7.6). Leptolyngbya and its alied genera were also found dominating in the thermal habitats of Sichuan Province, China (Tang et al., 2018). Both the Phormidium species in these springs, despite preferring slightly alkaline habitats, seem to be thermoadaptive (38–88°C). C. fritschii, Fischerella sp. NK 1-20, and Fischerella sp. NK 1-21 were recorded from high temperatures (87–88°C) in the hot springs of Banglazhang. The springs of Rehai (Re008) have the highest chloride ion concentration and temperature (90°C) with pH 7.6, which favored the growth of only Thermoleptolyngbya, even on the adjacent soils. Rehai spring Re007 with a low pH (pH 4) and a high temperature (85°C) coupled with higher sulfur contents also favored the growth of only Thermoleptolyngbya species. It was also reported that the members of this genus inhabited the hot springs with pH 6.1 and temperature 60°C. However, in Yunnan Province, it was less distributed in other hot springs, possibly due to insufficient time for their dispersal and being outcompeted by predominant cyanobacterial strains (Leptolyngbya) therein. Microbiological studies on the Rehai hot springs also presented very low diversity, exclusively for Sulfolobales and their viruses at higher temperature (Hedlund et al., 2012). Lower microbial diversity in the hot springs of Tengchong, Yunnan Province was also concluded by Hou et al. (2013) along with non-significant correlation of diversity with geochemistry. Another important aspect that was observed in the study is that the adaptability of several cyanobacterial taxa in varied temperatures may also be inhibited by the increase in acidity of the springs. A similar pattern was also observed in the hot springs of Kenya, East Africa (Dadheech et al., 2013). However, there is no clear relation between distribution of a particular strain and the potential niche-determining parameters that explain the geographical distribution of these strains. Declined relative abundances of Deinococcus-Thermus, Cyanobacteria, and Chloroflexi were previously observed with increasing temperature in Tibetan hot springs, except for Aquificae, which remained constant throughout (Wang et al., 2013). The thermal springs with temperature higher than 75°C were reported for only few cyanobacterial strains in the present study. However, cyanobacterial diversity in the hot springs of Dongshan (Ds001), Huangpo (Hp003), and Rehai (Re004) with temperature range 38–58°C was the highest, which may provide optimal conditions for speciation of these strains. If pH is also considered with temperature range, it showed that hot springs Ds001 and Hp003 with pH 9.4 coupled with a moderate temperature range (38–43°C) had higher cyanbacterial report (14 taxa) than Re006 with pH 9.3 and temperature 85°C (only one species).
The occurrence of different cyanobacterial strains in these habitats was still not very clear based on environmental parameters and was likely to be caused by the combination of varied temperature, pH, and mineralogy of water and nearby soil/sediments. It is challenging to understand the community composition of diversified cyanobacterial strains of these hot springs using trait-based approaches because measured parameters might not be sufficient enough to analyze its role in shaping the community structure or important factors may go unmeasured. It was observed that temporal variation in physicochemical parameters may trigger the microbial community structure and diversity in a particular habitat (Wang et al., 2014). One study also suggests that the biotic interactions may be strong niche determinants, rather than relevant abiotic factors (nutrients, temperature, etc.), which are either unknown or cannot be precisely determined (Tromas et al., 2020). Moreover, the current findings significantly expand the knowledge of cyanobacterial distribution in diversified Yunnan hot springs. It was also observed that not all cyanobacterial species can be cultured in lab conditions. The trail for the isolation of many uncultured strains always opens the scope for research on these extreme habitats.
Hot springs are generally considered as “island-like habitats” for extremophilic organisms leading to geographical isolation (Ramette and Tiedje, 2007). Many potential and new cyanobacterial novelties were studied from China recently based on a culture-dependent approach (Tang et al., 2018). Therefore, the culture-dependent strategy was also implemented in the present study to explore culturable cyanobacterial taxa and their isolation from hot springs, in addition to a culture-independent strategy (16S rRNA amplicon sequencing and microscopic observation of natural samples). In a total of 19 cyanobacterial isolates, 6 genera were cultured, which were also characterized by 16S rRNA gene sequencing for confirmation and showed the congruency with corresponding morphological features. However, Leptolyngbya sp. NK 1-10, Leptolyngbya sp. NK 1-12, Fischerella sp. NK 1-16, Fischerella sp. NK 1-20, Fischerella sp. NK 1-21, and Leptolyngbya sp. NK 1-23 were not identified up to species level, suggesting the possibility of new species/genera. Leptolyngbya sensu lato has recently been the focus of several researchers to identify many “cryptogenus” as they have rarely distinctive phenotypic and ecological characteristics, yet can be segregated genotypically (Sciuto and Moro, 2016). The unidentified Leptolyngbya members from Yunnan hot springs also provided similar scopes for taxonomic novelties. Therefore, it is nevertheless to state that many new potential cyanobacteria can be described from these habitats in the near future. Additionally, these geographically isolated habitats harbor the indigenous thermophilic cyanobacteria (Castenholz, 1969; Miller et al., 2009), which confirmed our findings on the survival of most of the isolates at high temperatures. In the study of phylogeny of cyanobacterial isolates in the present work, the clustering of isolated cyanobacterial species with major OTUs of cyanobacteria under the same subsection reported from 16S rRNA amplicon sequencing analysis of Yunnan hot springs supports the possibility for isolation of major mat-forming strains in culture conditions. However, phylogenetic relationships between all isolated cyanobacteria species and their OTUs showed that many strains could not be isolated in culture conditions. Most of the cyanobacterial strains (especially Leptolyngbya) from Yunnan hot springs showed some phylogenetic relationship to thermophilic cyanobacteria in other biogeographically regions, which confirms the presence of cosmopolitan species in the Yunnan hot springs, which were supported by previous findings (Dadheech et al., 2013). It is unlikely that clear patterns of phylogeny and separate clades were observed for Synechococcus from hot springs of different countries (Papke et al., 2003). These different patterns of phylogeny and occurrence of endemic cyanobacterial strains in hot springs were supported by the hypothesis given by Papke et al. (2003) that either insufficient time for divergence at the genetic level or the restriction by the presence of dominating cyanobacterial strains in various local niches for evolutionary radiation is responsible. Overall, these findings suggest that geographical isolation of hot springs all over the world with distinct environmental parameters is the main reason for the divergence and endemism of thermophilic cyanobacteria.
In a nutshell, the current work is mainly focused on cyanobacterial census and thermophilic isolates from hot springs. Cyanobacteria are photosynthetic prokaryotes and one of the potential bio-resources for biotechnological applications. They represent a large and diverse group with only a limited history of characterization and exploitation. The application of thermophilic cyanobacteria is still relatively untapped due to the unavailability of novel isolates (unialgal/axenic strains). The isolation, characterization, and bio-prospection of thermophilic cyanobacteria have their own importance because they are the source of many useful temperature-tolerant enzymes and bioactive molecules (Forlani et al., 2008; Nozzi et al., 2013; Ibrahim et al., 2014; Li et al., 2015a). The present study also offers many potential thermophilic cyanobacteria for a detailed study and future biotechnological utilization.
Conclusion
The study of biodiversity and isolation of potential strains from extreme habitats is a key step for modern research, which not merely describes diversity, but offers the potential taxa for biotechnological exploration. The hot springs located in Tengchong of central-western Yunnan Province is the largest and most intensively studied geothermal field in China. Microbial diversity emphasizing cyanobacterial community structure with isolation of potential taxa is studied using 16S rRNA amplicon sequencing and microscopic approach as well as cultivation-dependent and -independent strategies. The taxonomy and isolation of thermophilic cyanobacteria from hot springs of Yunnan Province open a platform for increasing our understanding of cyanobacterial diversity, their potential, and novelty, which have never been cultivated under laboratory conditions. These strains can be further explored for natural products and temperature tolerance. Overall, the current findings are solid evidence for taxonomy and future exploration of thermotolerant cyanobacteria for biotechnological utilization.
Data Availability Statement
The data presented in the study are deposited in the NCBI repository, accession numbers SRR11796781–SRR11796799 and MK625304–MK625325.
Author Contributions
NK, TZ, and XL contributed to the sampling. NK and TZ performed the experiments and drafted the article with contributions from all authors. NK, YZ, SD, and TZ analyzed the data. TZ and XL designed the research and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (Grant number 2021YFA0909700), the National Natural Science Foundation of China (Grant numbers 31525002, 31570068, 31761133008, and M-0214), a grant from QIBEBT and Dalian National Laboratory for Clean Energy (DNL), CAS (grant number QIBEBT I201904), the Shandong Taishan Scholarship to XL, and the Talented Young Scientist Program of Ministry of Science and Technology of China (Grant IND-16-001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are thankful to the authorities of CoE-IOCB, Utkal University for assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.872598/full#supplementary-material
Footnotes
References
Amarouche-Yala, S., Benouadah, A., Bentabet, A. E. O., and López-García, P. (2014). Morphological and phylogenetic diversity of thermophilic cyanobacteria in Algerian hot springs. Extremophiles 18, 1035–1047. doi: 10.1007/s00792-014-0680-7
Bateson, M. M., Wiegel, J., and Ward, D. M. (1989). Comparative-analysis of 16s ribosomal-RNA sequences of thermophilic fermentative bacteria isolated from hot-spring cyanobacterial mats. Syst. Appl. Microbiol. 12, 1–7. doi: 10.1016/S0723-2020(89)80034-7
Brito, Â, Gaifem, J., Ramos, V., Glukhov, E., Dorrestein, P. C., Gerwick, W. H., et al. (2015). Bioprospecting Portuguese Atlantic coast cyanobacteria for bioactive secondary metabolites reveals untapped chemodiversity. Algal Res. 9, 218–226. doi: 10.1016/j.algal.2015.03.016
Brock, T. D. (1967). Life at high temperatures. Science 158, 1012–1019. doi: 10.1126/science.158.3804.1012
Castenholz, R. W. (1969). Thermophilic blue-green algae and the thermal environment. Bacteriol. Rev. 33, 476–504. doi: 10.1128/br.33.4.476-504.1969
Chan, C. S., Chan, K. G., Tay, Y. L., Chua, Y. H., and Goh, K. M. (2015). Diversity of thermophiles in a Malaysian hot spring determined using 16S rRNA and shotgun metagenome sequencing. Front. Microbiol. 6:177. doi: 10.3389/fmicb.2015.00177
Coman, C., Druga, B., Hegedus, A., Sicora, C., and Dragos, N. (2013). Archaeal and bacterial diversity in two hot spring microbial mats from a geothermal region in Romania. Extremophiles 17, 523–534. doi: 10.1007/s00792-013-0537-5
Dadheech, P. K., Glöckner, G., Casper, P., Kotut, K., Mazzoni, C. J., Mbedi, S., et al. (2013). Cyanobacterial diversity in the hot spring, pelagic and benthic habitats of a tropical soda lake. FEMS Microbiol. Ecol. 85, 389–401. doi: 10.1111/1574-6941.12128
Derikvand, P., Llewellyn, C. A., and Purton, S. (2017). Cyanobacterial metabolites as a source of sunscreens and moisturizers: a comparison with current synthetic compounds. Eur. J. Phycol. 52, 43–56. doi: 10.1080/09670262.2016.1214882
Du, J., Liu, C., Fu, B., Ninomiya, Y., Zhang, Y., Wang, C., et al. (2005). Variations of geothermometry and chemical-isotopic compositions of hot spring fluids in the Rehai geothermal field, southwestern China. J. Volcan. Geotherm. Res. 142, 243–261. doi: 10.1016/j.jvolgeores.2004.11.009
Ferris, M. J., and Ward, D. M. (1997). Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 63, 1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997
Forlani, G., Pavan, M., Gramek, M., Kafarski, P., and Lipok, J. (2008). Biochemical bases for a widespread tolerance of cyanobacteria to the phosphonate herbicide glyphosate. Plant Cell Physiol. 49, 443–456. doi: 10.1093/pcp/pcn021
Fournier, R. O. (1989). Geochemistry and dynamics of the yellowstone national park hydrothermal system. Annu. Rev. Earth Planet Sci. 17, 13–53. doi: 10.1146/annurev.ea.17.050189.000305
Ghilamicael, A. M., Budambula, N. L. M., Anami, S. E., Mehari, T., and Boga, H. I. (2017). Evaluation of prokaryotic diversity of five hot springs in Eritrea. BMC Microbiol. 17:203. doi: 10.1186/s12866-017-1113-4
Hedlund, B. P., Cole, J. K., Williams, A. J., Hou, W., Zhou, E., Li, W., et al. (2012). A review of the microbiology of the Rehai geothermal field in Tengchong, Yunnan Province, China. Geosci. Front. 3, 273–288. doi: 10.1016/j.gsf.2011.12.006
Hedlund, B. P., Reysenbach, A., Huang, L., Ong, J. C., Liu, Z., Dodsworth, J. A., et al. (2015). Isolation of diverse members of the Aquificalesfrom geothermal springs in Tengchong, China. Front. Microbiol. 6:157. doi: 10.3389/fmicb.2015.00157
Hongmei, J., Aitchison, J. C., Lacap, D. C., Peerapornpeesal, Y., Sompong, U., and Pointing, S. B. (2005). Community phylogenetic analysis of moderately thermophilic cyanobacterial mats from China, the Philipines and Thailand. Extremophiles 9, 325–332. doi: 10.1007/s00792-005-0456-1
Hou, W., Wang, S., Dong, H., Jiang, H., Briggs, B. R., Peacock, J. P., et al. (2013). A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province China using 16S rRNA gene pyrosequencing. PLoS One 8:e53350. doi: 10.1371/journal.pone.0053350
Ibrahim, W. M., Karam, M. A., Elshahat, R. M., and Adway, A. A. (2014). Biodegradation and utilization of organophosphorus pesticide malathion by cyanobacteria. Biomed. Res. Int. 2014:392682. doi: 10.1155/2014/392682
Jiang, H., Huang, Q., Dong, H., Wang, P., Wang, F., Li, W., et al. (2010). RNA-based investigation of ammonia-oxidizing archaea in hot springs of Yunnan Province, China. Appl. Environ. Microbiol. 76, 4538–4541. doi: 10.1128/AEM.00143-10
Kambura, A. K., Mwirichia, R. K., Kasili, R. W., Karanja, E. N., Makonde, H. M., and Boga, H. I. (2016). Bacteria and archaea diversity within the hot springs of lake magadi and little magadi in Kenya. BMC Microbiol. 16:136. doi: 10.1186/s12866-016-0748-x
Klatt, C. G., Liu, Z., Ludwig, M., Kühl, M., Jensen, S. I., Bryant, D. A., et al. (2013). Temporal metatranscriptomic patterning in phototrophic Chloroflexi inhabiting a microbial mat in a geothermal spring. ISME J. 7, 1775–1789. doi: 10.1038/ismej.2013.52
Klatt, C. G., Wood, J. M., Rusch, D. B., Bateson, M. M., Hamamura, N., Heidelberg, J. F., et al. (2011). Community ecology of hot spring cyanobacterial mats: predominant populations and their functional potential. ISME J. 5, 1262–1278. doi: 10.1038/ismej.2011.73
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks808
Kyle, J. E., Schroeder, P. A., and Wiegel, J. (2007). Microbial silicification in sinters from two terrestrial hot springs in the Uzon Caldera, Kamchatka, Russia. Geomicrobiol. J. 24, 627–641. doi: 10.1080/01490450701672158
Lau, M. C. Y., Aitchison, J. C., and Pointing, S. B. (2009). Bacterial community composition in thermophilic microbial mats from five hot springs in central Tibet. Extremophiles 13, 139–149. doi: 10.1007/s00792-008-0205-3
Li, H., Yang, Q., Li, J., Gao, H., Li, P., and Zhou, H. (2015a). The impact of temperature on microbial diversity and AOA activity in the Tengchong Geothermal Field, China. Sci. Rep. 5:17056. doi: 10.1038/srep17056
Li, J. J., Liu, Y., Cheng, J. J., Mos, M., and Daroch, M. (2015b). Biological potential of microalgae in China for biorefinery-based production of biofuels and high value compounds. New Biotechnol. 32, 588–596. doi: 10.1016/j.nbt.2015.02.001
Lin, L., Chen, C., Peng, Q., Ben, K., and Zhou, Z. (2002). Thermus rehai sp. nov., isolated from Rehai of Tengchong, Yunnan Province, China. J. Basic Microbiol. 42, 337–344. doi: 10.1002/1521-4028(200210)42:5<337:AID-JOBM337<3.0.CO;2-B
Lukavský, J., Furnadzhieva, S., and Pilarski, P. (2011). Cyanobacteria of the thermal spring at Pancharevo, Sofia, Bulgaria. Acta Bot. Croat. 70, 191–208. doi: 10.2478/v10184-010-0015-4
Mackenzie, R., Pedrós-Alió, C., and Díez, B. (2013). Bacterial composition of microbial mats in hot springs in Northern Patagonia: variations with seasons and temperature. Extremophiles 17, 123–136. doi: 10.1007/s00792-012-0499-z
McGregor, G. B., and Rasmussen, J. P. (2008). Cyanobacterial composition of microbial mats from an Australian thermal spring: a polyphasic evaluation. FEMS Microbiol. Ecol. 63, 23–35. doi: 10.1111/j.1574-6941.2007.00405.x
Mehetre, G. T., Paranjpe, A., Dastager, A. G., and Dharne, M. S. (2016). Investigation of microbial diversity in geothermal hot springs in Unkeshwar, India, based on 16S rRNA amplicon metagenome sequencing. Genome Announc. 4:e01766–15. doi: 10.1128/genomeA.01766-15
Miller, S. R., Purugganan, M. D., and Curtis, S. E. (2006). Molecular population genetics and phenotypic diversification of two populations of the thermophilic cyanobacterium Mastigocladus laminosus. Appl. Environ. Microbiol. 72, 2793–2800. doi: 10.1128/AEM.72.4.2793-2800.2006
Miller, S. R., Williams, C., Strong, A. L., and Carvey, D. (2009). Ecological specialization in a spatially structured population of the thermophilic cyanobacterium Mastigocladus laminosus. Appl. Environ. Microbiol. 75, 729–734. doi: 10.1128/AEM.01901-08
Moro, I., Rascio, N., La Rocca, N., Sciuto, K., Albertano, P., Bruno, L., et al. (2010). Polyphasic characterization of a thermo-tolerant filamentous cyanobacterium isolated from the Euganean thermal muds (Padova, Italy). Eur. J. Phycol. 45, 143–154. doi: 10.1080/09670260903564391
Nozzi, N. E., Oliver, J. W. K., and Atsumi, S. (2013). Cyanobacteria as a platform for biofuel production. Front. Bioeng. Biotechnol. 1:7. doi: 10.3389/fbioe.2013.00007
Nübel, U., Garcia-Pichel, F., and Muyzer, G. (1997). PCR primers to amplify 16s rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63, 3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997
Pagaling, E., Grant, W. D., Cowan, D. A., Jones, B. E., Ma, Y., Ventosa, A., et al. (2012). Bacterial and archaeal diversity in two hot spring microbial mats from the geothermal region of Tengchong, China. Extremophiles 16, 607–618. doi: 10.1007/s00792-012-0460-1
Papke, R. T., Ramsing, N. B., Bateson, M. M., and Ward, D. M. (2003). Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 5, 650–659. doi: 10.1046/j.1462-2920.2003.00460.x
Pentecost, A., Jones, B., and Renaut, R. W. (2003). What is a hot spring? Canad. J. Earth Sci. 40, 1443–1446. doi: 10.1139/e03-083
Ramette, A., and Tiedje, J. M. (2007). Biogeography. an emerging cornerstone for understanding prokaryotic diversity, ecology and evolution. Microb. Ecol. 53, 197–207. doi: 10.1007/s00248-005-5010-2
Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M., and Stanier, R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111, 1–61. doi: 10.1099/00221287-111-1-1
Roy, S., Bhattacharya, S., Debnath, M., and Ray, S. (2015). Diversity of cyanobacterial flora of Bakreswar geothermal spring, West Bengal, India – II. Algol. Stud. 147, 29–44. doi: 10.1127/1864-1318/2014/0178
Saxena, R., Dhakan, D. B., Mittal, P., Waiker, P., Chowdhury, A., Ghatak, A., et al. (2016). Metagenomic analysis of hot springs in central india reveals hydrocarbon degrading thermophiles and pathways essential for survival in extreme environments. Front. Microbiol. 7:2123. doi: 10.3389/fmicb.2016.02123
Sciuto, K., and Moro, I. (2016). Detection of new cosmopolitan genus Thermoleptolyngbya (Cyanobacteria, Leptolyngbyaceae) using the 16S rRNA gene and 16S-23S ITS region. Mol. Phylogenet. Evol. 105, 15–35. doi: 10.1016/j.ympev.2016.08.010
Shih, P. M., Wu, D. Y., Latifi, A., Axen, S. D., Fewer, D. P., Talla, E., et al. (2013). Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U.S.A 110, 1053–1058. doi: 10.1073/pnas.1217107110
Singh, A., and Subudhi, E. (2016). Structural insights of microbial community of Deulajhari (India) hot spring using 16s-rRNA based metagenomic sequencing. Genome Data 7, 101–102. doi: 10.1016/j.gdata.2015.12.004
Singh, Y., Gulati, A., Singh, D. P., and Khattar, J. I. S. (2018). Cyanobacterial community structure in hot water springs of Indian north-western himalayas: a morphological, molecular and ecological approach. Algal Res. 29, 179–192. doi: 10.1016/j.algal.2017.11.023
Sompong, U., Hawkins, P. R., Besley, C., and Peerapornpisal, Y. (2005). The distribution of cyanobacteria across physical and chemical gradients in hot springs in northern Thailand. FEMS Microbiol. Ecol. 52, 365–376. doi: 10.1016/j.femsec.2004.12.007
Song, Z., Wang, F., Zhi, X., Chen, J., Zhou, E., Liang, F., et al. (2013). Bacterial and archaeal diversities in Yunnan and Tibetan hot springs, China. Environ. Microbiol. 15, 1160–1175. doi: 10.1111/1462-2920.12025
Song, Z., Zhi, X., Li, W., Jiang, H., Zhang, C. L., and Dong, H. (2009). Actinobacterial diversity in hot springs in Tengchong (China), Kamchatka (Russia) and Nevada (USA). Geomicrobiol. J. 26, 256–263. doi: 10.1080/01490450902892373
Song, Z. Q., Chen, J. Q., Jiang, H. C., Zhou, E. M., Tang, S. K., Zhi, X. Y., et al. (2010). Diversity of Crenarchaeota in terrestrial hot springs in Tengchong, China. Extremophiles 14, 287–296. doi: 10.1007/s00792-010-0307-6
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tang, J., Jiang, D., Luo, Y., Liang, Y., Li, L., Shah, M. M. R., et al. (2018). Potential new genera of cyanobacterial strains isolated from thermal springs of western Sichuan, China. Algal Res. 31, 14–20. doi: 10.1016/j.algal.2018.01.008
Tekere, M., Lötter, A., Olivier, J., Jonker, N., and Venter, S. (2011). Metagenomic analysis of bacterial diversity of Siloam hot water spring, Limpopo, South Africa. African J. Biotechnol. 10, 18005–18012. doi: 10.5897/AJB11.899
Tomitani, A., Knoll, A. H., Cavanaugh, C. M., and Ohno, T. (2006). The evolutionary diversification of cyanobacteria: molecular-phylogenetic and paleontological perspectives. Proc. Natl. Acad. Sci. U.S.A 103, 5442–5447. doi: 10.1073/pnas.0600999103
Tromas, N., Taranu, Z. E., Castelli, M., Pimentel, J. S. M., Pereira, D. A., Marcoz, R., et al. (2020). The evolution of realized niches within freshwater Synechococcus. Environ. Microbiol. 22, 1238–1250. doi: 10.1111/1462-2920.14930
Urbieta, M. S., Toril, E. G., Giaveno, M. A., Bazan, A. A., and Donati, E. A. (2014). Archaeal and bacterial diversity in five different hydrothermal ponds in the Copahue region in Argentina. Syst. Appl. Microbiol. 37, 429–441. doi: 10.1016/j.syapm.2014.05.012
Wang, S., Dong, H., Hou, W., Jiang, H., Huang, Q., Briggs, B. R., et al. (2014). Greater temporal changes of sediment microbial community than its waterborne counterpart in Tengchong hot springs, Yunnan Province, China. Sci. Rep. 4:7479. doi: 10.1038/srep07479
Wang, S., Hou, W., Dong, H., Jiang, H., Huang, L., Wu, G., et al. (2013). Control of temperature on microbial community structure in hot springs of the Tibetan Plateau. PLoS One 8:e62901. doi: 10.1371/journal.pone.0062901
Ward, D. M., Bateson, M. M., Ferris, M. J., Kühl, M., Wieland, A., Koeppel, A., et al. (2006). Cyanobacterial ecotypes in the microbial mat community of mushroom spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function. Philos. Trans. R. Soc. B Biol. Sci. 361, 1997–2008. doi: 10.1098/rstb.2006.1919
Ward, D. M., Castenholz, R. W., and Miller, S. R. (2012). “Cyanobacteria in geothermal habitats,” in Ecology of Cyanobacteria II: Their Diversity in Space and Time, ed. B. A. Whitton (Berlin: Springer Science). 39–63. doi: 10.1007/978-94-007-3855-3_3
Ward, D. M., Ferris, M. J., Nold, S. C., and Bateson, M. M. (1998). A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62, 1353–1370. doi: 10.1128/MMBR.62.4.1353-1370.1998
Xiang, X., Dong, X., and Huang, L. (2003). Sulfolobus tengchongensis sp. nov., a novel thermoacidophilic archaeon isolated from a hot spring in Tengchong, China. Extremophiles 7, 493–498. doi: 10.1007/s00792-003-0355-2
Xue, Y., Xu, Y., Liu, Y., Ma, Y., and Zhou, P. (2001). Thermoanaerobacter tengcongensis sp. nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int. J. Syst. Evol. Microbiol. 51, 1335–1341. doi: 10.1099/00207713-51-4-1335
Keywords: hot springs, cyanobacteria, 16S rRNA amplicon sequencing, phenotype, phylogenetics
Citation: Keshari N, Zhao Y, Das SK, Zhu T and Lu X (2022) Cyanobacterial Community Structure and Isolates From Representative Hot Springs of Yunnan Province, China Using an Integrative Approach. Front. Microbiol. 13:872598. doi: 10.3389/fmicb.2022.872598
Received: 09 February 2022; Accepted: 28 February 2022;
Published: 25 April 2022.
Edited by:
Lei Chen, Tianjin University, ChinaReviewed by:
Jun Ni, Shanghai Jiao Tong University, ChinaHaojie Jin, Beijing Forestry University, China
Copyright © 2022 Keshari, Zhao, Das, Zhu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Zhu, emh1dGFvQHFpYmVidC5hYy5jbg==; Xuefeng Lu, bHZ4ZkBxaWJlYnQuYWMuY24=
 Nitin Keshari
Nitin Keshari Yang Zhao
Yang Zhao Sudipta Kumar Das
Sudipta Kumar Das Tao Zhu
Tao Zhu Xuefeng Lu
Xuefeng Lu