
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 July 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.870821
This article is part of the Research Topic Plant Microbiome: Diversity, Functions, and Applications View all 13 articles
Due to increased antimicrobial resistance against current drugs, new alternatives are sought. Endophytic bacteria associated with medicinal plants are recognized as valuable sources of novel secondary metabolites possessing antimicrobial, antitumor, insecticidal, and antiviral activities. In this study, five bacterial endophytes were isolated and identified from the medicinal plant, Alectra sessiliflora, and their antibacterial and antitumor activities were investigated. In addition, the crude extracts of the endophytes were analyzed using gas chromatography (GC) coupled with time-of-flight mass spectrometry (TOF-MS). The identified bacterial endophytes belong to three genera viz Lysinibacillus, Peribacillus, and Bacillus, with the latter as the dominant genus with three species. Ethyl acetate extracts from the endophytes were used for antimicrobial activity against eleven pathogenic strains through minimum inhibitory concentration (MIC). The antitumor activity against the Hela cervical, Hek 293 kidney, and A549 lung carcinoma cells was determined by the MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxy-phenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay. Lysinibacillus sp. strain AS_1 exhibited broad antibacterial activity against the pathogenic strains with MIC values ranging from 4 to 8 mg/ml, while Bacillus sp. strain AS_3 displayed MIC of 0.25 mg/ml. Crude extracts of Lysinibacillus sp. strain AS_1, Peribacillus sp. strain AS_2, and Bacillus sp. strain AS_3 showed growth inhibition of more than 90% against all the cancer cell lines at a concentration of 1,000 μg/ml. Untargeted secondary metabolite profiling of the crude extracts revealed the presence of compounds with reported biological activity, such as antimicrobial, antioxidant, anti-inflammatory, antitumor, and antidiabetic properties. This study reported for the first time, bacterial endophytes associated with A. sessiliflora with antibacterial and antitumor activities.
The frequency of infections caused by pathogenic bacteria has increased exponentially in the previous decades (Roca et al., 2015; Attia et al., 2020). In addition, the abuse and misuse of antimicrobial drugs, some of which are available over the counter without a prescription, has turned into a global health concern (Ayukekbong et al., 2017). All these compounded by the lack of new effective antimicrobial agents are contributing to the rise in antimicrobial resistance, and bacteria and fungi have developed resistance through a variety of mechanisms, including enzyme activation, altered target sites, decreased cell permeability, and increased efflux due to over-expression, among others (Baptista et al., 2018). This has resulted in a continual decrease in the development of new antimicrobial drugs; it is, therefore, necessary to discover and develop novel antimicrobial drugs from natural products (Sciarretta et al., 2016; Farhat et al., 2019).
To tackle antimicrobial resistance, recent breakthroughs in microbial ecology have led researchers to focus on studying ground-breaking and promising antimicrobial compounds from natural sources, such as medicinal plants. Medicinal plants have long been used to cure a variety of ailments, including skin conditions, coughs, microbiological infections, diabetes, colds, urinary issues, and inflammations (Aswani et al., 2020; Alotaibi et al., 2021). Medicinal plants have been recognized as good sources of bioactive substances that are vital for good health and are reservoirs for various microorganisms categorized as endophytes, such as bacteria, fungi, and actinomycetes (Petrini et al., 1993; Duhan et al., 2020).
Endophytes are microorganisms like fungi, bacteria, and actinomycetes, which have a mutual relationship with the host plant and inhabit the host tissues without causing detrimental symptoms (Gunatilaka, 2006). Bacterial endophytes have been identified as the prospective source of natural metabolites such as alkaloids, steroids, phenols, terpenoids, flavonoids, isocoumarins, and quinones, which have agricultural, industrial (Zinniel et al., 2002), and pharmaceutical applications (Palanichamy et al., 2018; Attia et al., 2020). Bacterial endophytes benefit the host plants by helping them survive abiotic and biotic conditions, solubilize minerals, nutrient acquisition, and protection against pathogens and parasitic nematodes (Dutta et al., 2014). Bacterial endophytes are diverse and range from Gram-positive to Gram-negative species of various genera such as Pantoea, Achromobacter, Acinetobacter, Xanthomonas, Bacillus, Agrobacterium, etc. (Sun et al., 2013). Bioactive compounds produced by various bacterial endophytes have antimicrobial and anticancer compounds that may be used for various diseases (Gouda et al., 2016). Furthermore, bioactive compounds which have been extracted from endophytic microorganisms exhibit antidiabetic, antifungal, immunosuppressant, and anti-inflammatory properties, thus they have received attention in drug discovery research (Egamberdieva et al., 2017; Panigrahi and Rath, 2021; Singh et al., 2021).
Research on medicinal plants and their associated endophytes, and their potential to synthesize distinct bioactive compounds, have opened the possibility of looking into more medicinal plants as well to explore their diverse endophytic bacteria (Duhan et al., 2020). Alectra sessiliflora is a medicinal plant that grows throughout Sub-Saharan Africa, China, India, and the Philippines (Morawetz and Wolfe, 2011; Gasa, 2015; Katembo et al., 2021). The eastern and southwestern provinces of South Africa, which include the Eastern Cape, Free State, Gauteng, KwaZulu-Natal, Limpopo, Mpumalanga, North-West, and Western Cape provinces, are home to A. sessiliflora (Morawetz and Wolfe, 2011). A. sessiliflora has been used in traditional medicine to treat toothaches, diarrhea, scabies, gastrointestinal illnesses, and oral thrush (Gasa, 2015; Katembo et al., 2021). It is used to treat tuberculosis in several African countries, such as Nigeria, and its leaves are used as a galactogen by pregnant women in Central Africa (Ogbole and Ajaiyeoba, 2010; Oosthuizen et al., 2019). The phytochemistry of A. sessiliflora has received significant attention, however, there is no information on its endophytes (Ogbole and Ajaiyeoba, 2010; Oosthuizen et al., 2019). The goal of this study was to isolate and identify bacterial endophytes from A. sessiliflora collected in Limpopo province, South Africa. In addition, the antibacterial and antitumor activities of the endophyte’s secondary metabolite crude extracts were investigated and further identified using gas chromatography high-resolution time-of-flight mass spectrometry (GC-TOF-MS).
The whole plant with a height up to 25 cm from the ground was collected from Eisleben, Botlokwa (23°31′49.5″S 29°49′27.1″E) in Limpopo province, South Africa. The whole plant including roots was placed in sterile polyethylene bags and transported to the laboratory at 4°C. The plant material was collected in March 2017 from a site with grassland. The identification of the plant material was carried out at the University of Johannesburg Herbarium (JRAU). A sample specimen of the plant material was deposited in the University of Johannesburg Herbarium (JRAU) with voucher specimen number Serepa-Dlamini 205 and species name A. sessiliflora. The remaining plant material was immediately processed in the laboratory.
The bacterial endophytes were isolated from fresh leaves (approximately 10–15 leaves) of 1 whole plant following the method by Ding and Melcher (2016). Following isolation of pure colonies of the bacterial endophytes, 35% glycerol (glycerol diluted in sterile distilled water) stock cultures were prepared and stored at −80°C for future use. Stock cultures of five bacterial isolates were retrieved from long-term storage and sub-cultured on fresh nutrient agar (NA) media followed by incubation for 2–7 days at 30°C. Sub-culturing of each bacterial isolate was repeated several times until pure colonies were obtained.
The bacterial strains were grown on NA for 24 h at 30°C and genomic DNA was extracted using the Zymo Research Fungal/Bacterial DNA MiniPrep Kit (Zymo Research, United States) as per the manufacturer’s protocol. The concentration of each endophyte DNA was quantified using the Nanodrop Spectrophotometer (Thermo Fisher Scientific, United States).
The 16S rRNA genes of each bacterial strain were amplified by polymerase chain reaction (PCR) using 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′CGGTTA CCTTGTTACGACTT-3′) primers (Yeates et al., 1997). The 25 μl PCR reactions contained 12.5 μl 2X PCR Master mix with standard buffer (20 mM Tris-HCI, 1.8 mM MgCl2, 22 mM NH4Cl, 22 mM KCl, 0.2 mM dNTPs, 5% glycerol, 0.06% IGEPAL® CA-630, 0.05% Tween® 20, 25 units/ml One Taq® DNA polymerase), 2.5 μl of each primer (10 μM), 2.5 μl nuclease-free water, and 5 μl of each DNA (>50 ng/μl) template. A negative control containing all the PCR mix without any DNA was included in the PCR experiment. The amplification was carried out on a MyCycler™ Thermal Cycler (Bio-Rad, United States). The PCR reaction conditions were initial denaturation at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min and extension at 72°C for 2 min, and a final extension of 72°C for 10 min. The amplicons were purified with ExoSAP-it™ (Thermo Fisher Scientific, United States) after which they were sent to a commercial service provider, Inqaba Biotechnical Industries (Pty) Ltd., Pretoria, South Africa for sequencing.
Raw sequence data of the 16S rRNA genes for each endophytic bacteria were used to create consensus sequences using the BioEdit Sequence Alignment Editor v.7.2.6 (Hall, 1999). The consensus sequences were subjected to BLAST analysis at the National Center for Biotechnology Information (NCBI) against the prokaryotic rRNA sequence database (Bacteria and Archaea), from which closely related bacterial species were identified (Altschul et al., 1990), and the type strains from the EzBioCloud database1 were also included. All phylogenetic analyses post-BLAST were performed using molecular evolutionary genetics analysis version (MEGA) v.7.27 software (Kumar et al., 2016). The sequences were aligned using multiple sequence comparison by log-expectation (MUSCLE) with default settings (Edgar, 2004). Phylogenetic trees were constructed using maximum likelihood (ML) following the Jukes-Cantor model (Jukes and Cantor, 1969). A total of 1,000 replications were used for the statistical confidence of the nodes. For Bayesian inference, a Markov Chain Monte Carlo (MCMC) method was used to reconstruct the phylogenetic trees using BEAST v.1.10.4 (Suchard et al., 2018). The resulting trees were visualized in FigTree v.1.4.4 (Rambaut, 2018).
Secondary metabolites of each bacterial isolate were extracted using the method described by Maloney et al. (2009) with slight modifications. Briefly, the endophytic bacteria isolated from A. sessiliflora were cultured in 1 L Luria Bertani (LB) broth and agitated at 200 rpm at 28°C for 7 days. After cultivation, 20 g/L of the Amberlite® XAD7HP 20–60 mesh (Sigma-Aldrich, Darmstadt, Germany) was added to each flask to absorb the secondary metabolites and was further agitated at 180 rpm for 2 h. A cheesecloth was used to filter the resin after which it was washed three times with 200 ml acetone. The acetone was concentrated using a rotary vapor (Lab Tech, Nantong, Jiangsu, China) at 5°C until a dark brown viscous extract was obtained. The residual water containing the crude extracts was transferred into a measuring cylinder and an equal volume of ethyl acetate (1:1 [v/v]) was added. The mixture was agitated vigorously for 5–10 min after which it was separated using a funnel. This process was repeated three times, and subsequently the ethyl acetate fraction was evaporated using a rotary vapor. The crude extracts were transferred into sterile beakers and covered with foil, then left at room temperature to dry.
In this study, the minimum inhibitory concentration (MIC) method described by Andrews (2001), was used to determine the antibacterial activities of the crude extracts from the bacterial endophytes with slight modifications. The test bacterial species included human clinical pathogens, and a number of the strains have previously exhibited antibiotic resistance to various antibiotics such as penicillin, ampicillin, quinolone, carbenicillin, cefalotin, cefotaxime, trimethoprim-sulfamethoxazole, clindamycin, dicloxacillin, and cetyltrimethylammonium bromide (Wagenlehner et al., 2003; Zhang et al., 2016; Hernández et al., 2021). Care was taken to include methicillin-resistant Staphylococcus saprophyticus (Higashide et al., 2008), members of the ESKAPE group (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) (Flores-Paredes et al., 2021), with the exception of Acinetobacter baumannii and Enterobacter spp.; and methicillin-susceptible S. aureus (MSSA) (Ham et al., 2010): The test strains included, Bacillus cereus (ATCC 10876), Escherichia coli (ATCC 10536), Klebsiella pneumoniae (ATCC 10031), Klebsiella oxytoca (ATCC 13182), Mycobacterium smegmatis (ATCC 21293), Pseudomonas aeruginosa (NCTC 10662), Staphylococcus aureus (ATCC 25923), S. saprophyticus (ATCC 15305), Staphylococcus epidermidis (ATCC 14990), Veillonella parvula (ATCC 10790), and Enterococcus faecium (ATCC 13048). Briefly, stock solutions of the crude endophyte extracts were prepared by dissolving 0.19 g in 1 ml dimethyl sulfoxide (DMSO) to make a stock solution of 32 mg/ml. This was then serially diluted to concentrations of 16 mg/L down to 0.25 mg/ml using Mueller-Hinton broth (MHB). Using McFarland 0.5 standard, 10 μl of each pathogenic strain was inoculated in 20 ml MHB and incubated at 30°C for 24 h. Using sterile 96 well microtiter plates, 100 μl of each pathogenic strain was added horizontally while 100 μl of the diluted crude extracts were added vertically starting from 16 mg/ml down to 0.25 mg/ml. The antibiotic Streptomycin with a concentration of 1 mg/ml (Sigma-Aldrich, Switzerland) was used as positive control while DMSO was used as a negative control. The MIC was conducted in triplicates. The plates were incubated at 37°C for 24 h after which 10 μl resazurin salt solution [0.02% (w/v)] was added to the wells as an indicator of microbial growth and incubated for an additional 2 h. The color change from blue to pink to clear indicated reduction had taken place as oxygen becomes limited within the medium, indicating that metabolism has taken place. The wells in which no color change occurred indicated no bacterial growth while the wells with a pink or clear color indicated bacterial growth. The MIC with the lowest concentration was visually inspected for color change.
The effect of bacterial endophyte’s crude extracts on the survival and growth of human cancer cell lines A549 lung carcinoma, Hek 293 kidney adenocarcinoma, and HeLa cervical adenocarcinoma cells was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT) in vitro cytotoxic assay. The crude extracts were prepared as described above and different concentrations of each extract (31.30, 62.60, 125, 250, 500, and 1,000 μg/ml) were prepared. Briefly, the A549, HeLa, and Hek 293 cells were grown using normal tissue culture techniques with the addition of 10% fetal bovine serum (FBS). The cells (1 × 106 cells/ml) were incubated in 96 well microtiter plates at 37°C for 24 h. Following incubation, the media was removed and 100 μl of fresh media was added to all the wells along with 100 μl of the diluted extracts from high (1,000 μg/ml) to low (31.3 μg/ml) concentrations. The cells were incubated for 72 h, after which 5 μl [3- (4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] (MTS) was added to the cells. The absorbance values were measured at 490 nm at 0, 1, 2, 3, and 4 h incubation periods using the Molecular Devices SpectraMax ABS Plus, and data were acquired with SoftMax Pro 7.1 Data Acquisition and Analysis Software. Auranofin was used as a positive control and DMSO was used as a negative control. In the MTT assay, the MTS compound is metabolized by viable cells from yellow to purple formazan by the mitochondria of viable cells which is detected at 490 nm. The cytotoxicity tests of the crude extracts were analyzed in duplicates across three plates (n = 6) and the absorbance value was reported. The results were expressed as growth inhibition and IC50 values were determined using the AAT Bioquest IC50 calculator [AAT Bioquest (2022), Sunnyvale, CA, United States] available at https://www.aatbio.com/tools/ic50-calculator. The IC50 is the half-maximal inhibitory concentration, which measures the effectiveness of a crude extract in inhibiting a given biological sample or process by half, in this study the inhibition of human cancer cell lines.
Metabolite profiling of the endophyte extracts was carried out on a GC-TOF-MS system (LECO Corporation St. Joseph, MI, United States) using the following conditions: primary column and a Rxi-5Sil MS (30 m, 250 μm i.d., 0.25 μm df) (Restek, Pennsylvania, United States) and a Rxi-17Sil MS (2 m, 250 μm i.d., 0.25 μm df) (Restek, Bellefonte, PA, United States) secondary column. In brief, samples were first prepared by adding 1 ml HPLC grade methanol (Sigma-Aldrich, Aston Manor, South Africa) to the extracts, 1 μl of each sample was injected, and Helium was used as a carrier gas with a flow rate of 1 ml/min. The oven temperature was maintained at 60°C for 1 min and then programmed at 10°C/min increment to 330°C, then 5°C/min to 280°C. The inlet temperature was 250°C. Mass spectra (MS) were optimized at an electron energy of −70 eV with an ion source at 250°C. The mass fragments used were from 40–660 m/z with an acquisition rate of 10 spectra/second. The interpretation of GC-MS mass-spectra was analyzed using the ChromaTOF software (LECO Corporation, St. Joseph, MI, United States). The functional groups and biological activities of the compounds were analyzed using the NCBI PubChem and PASS online databases available at https://pubchem.ncbi.nlm.nih.gov and http://www.way2drug.com/passonline, respectively.
Unless otherwise stated all experiments were carried out in triplicates. The mean values were calculated using the Microsoft Excel program version 2010. The t-test was performed to determine the significance of the difference between the mean values. One-way ANOVA was performed at p ≤ 0.05 significant levels to determine the variance.
In this study, a total of five bacterial endophytes all belonging to the Firmicute phylum were isolated and identified through 16S rRNA gene sequencing as shown in Table 1. The 16S rRNA sequences were deposited in GenBank with accession numbers from MZ976846—MZ976850. The 16S rRNA gene sequences of each strain were compared with other bacterial species available on the GenBank-NCBI database. The NCBI database confirmed the identity of the bacterial endophytes belonging to three genera Lysinibacillus, Peribacillus, and Bacillus with three isolates as shown in Table 1. All the isolates showed 94–99% similarities with other closely related strains retrieved from the NCBI database as indicated in Supplementary Table 1.
The evolutionary relationships between all the endophytic bacteria isolated from A. sessiliflora with other closely related species were constructed using the ML and Bayesian MCMC methods. Each species was delineated with closely related species in separate phylogenetic trees (Figures 1–3). Lysinibacillus sp. Strain AS_1 formed a polytomy clade with other Lysinibacillus macroides and two L. fusiformis strains, supported by a 95% bootstrap value (Figure 1). In the Bayesian phylogenetic tree (Supplementary Figure 1), Lysinibacillus sp. Strain AS_1 formed a paraphyletic group with L. fusiformis and L. endophyticus.
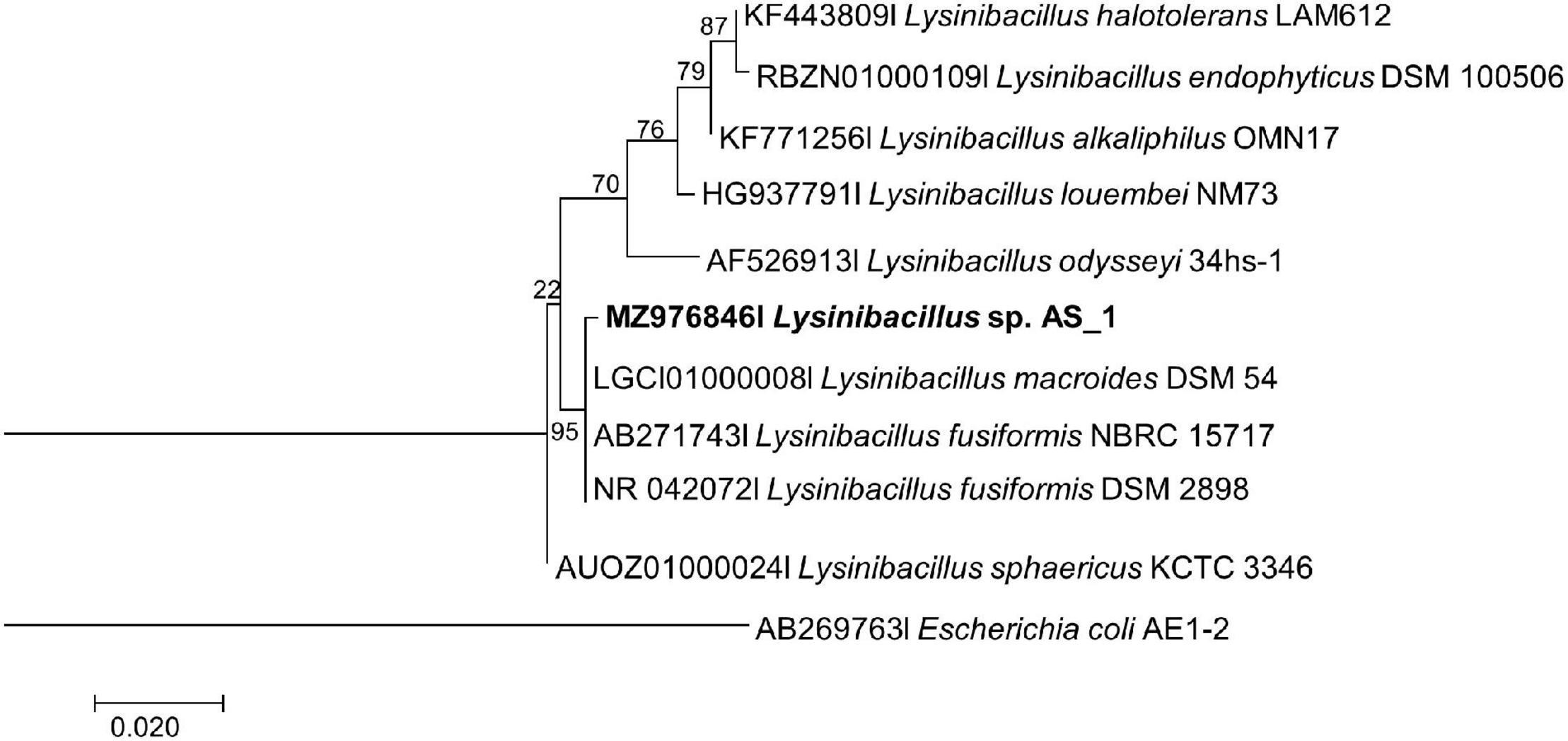
Figure 1. Maximum likelihood phylogenetic tree based on analysis of partial 16S rDNA nucleotide sequence of Lysinibacillus sp. Strain AS_1 with related strains from the Lysinibacillus genus. Numbers above or below the nodes indicate bootstrap values generated after 1,000 replications. Escherichia coli AE-1 (AB269763) was used as an outgroup.
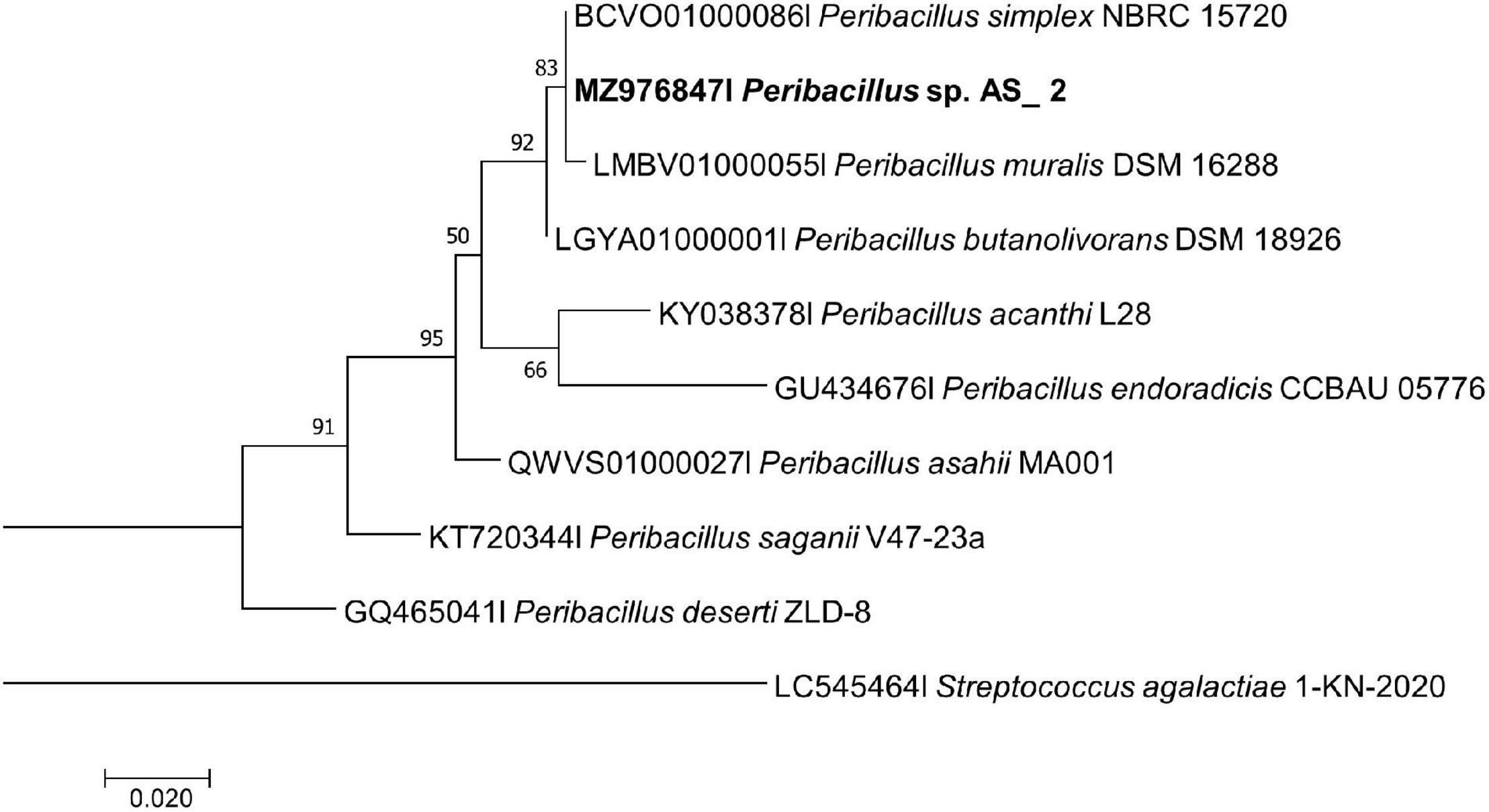
Figure 2. Maximum likelihood phylogenetic tree based on analysis of partial 16S rDNA nucleotide sequence of Peribacillus sp. Strain AS_2 with related strains from the Peribacillus genus. Numbers above or below the nodes indicate bootstrap values generated after 1,000 replications. Streptococcus agalactiae AE-1 (LC545464) was used as an outgroup.
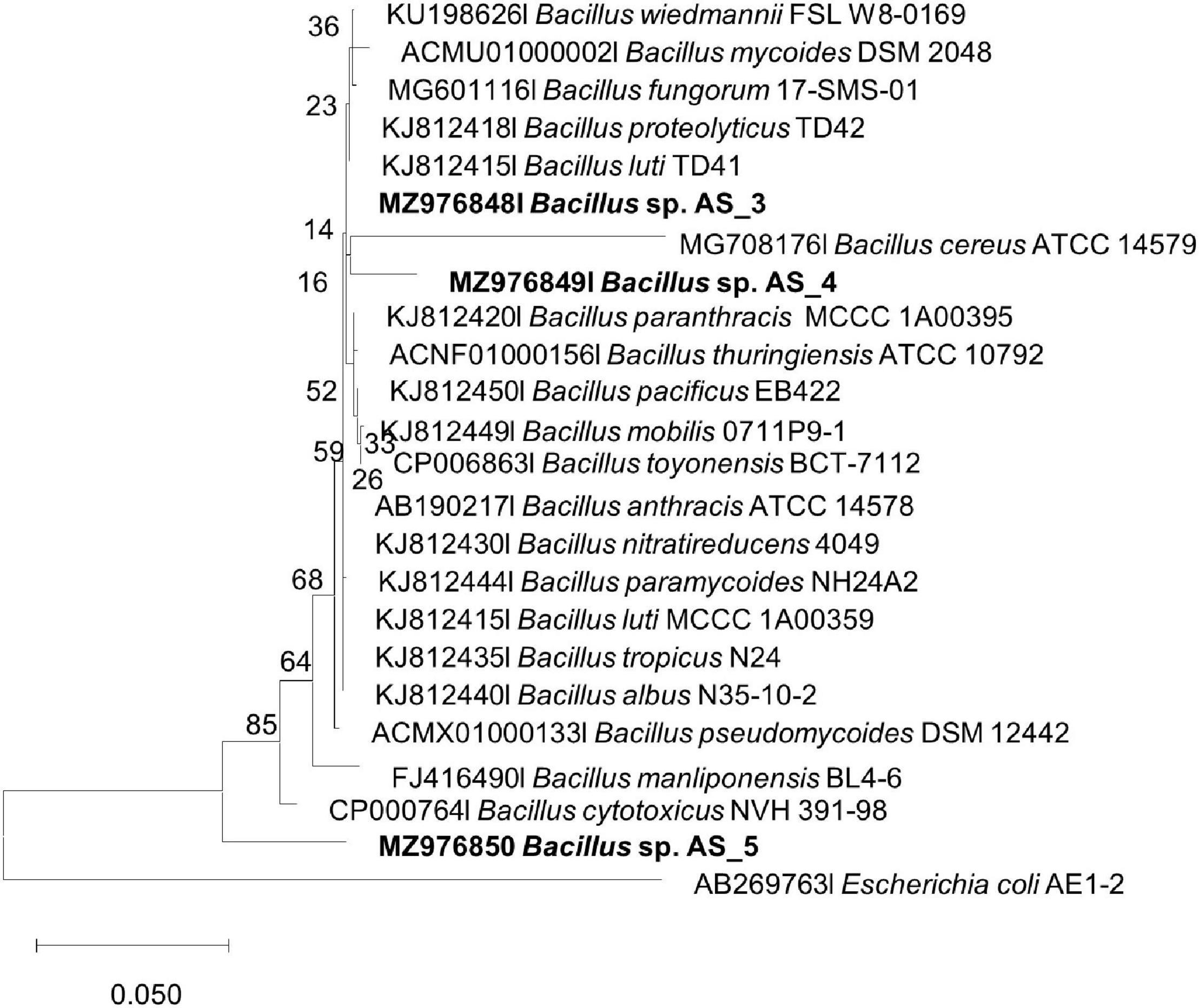
Figure 3. Maximum likelihood phylogenetic tree based on analysis of partial 16S rDNA nucleotide sequences of Bacillus sp. strain AS_3, Bacillus sp. AS_4 and Bacillus sp. AS_5 with related strains from the Bacillus genus. Numbers above or below the nodes indicate bootstrap values generated after 1,000 replications. Escherichia coli AE1-2 was used as an outgroup.
Peribacillus strain AS_2 showed a polytomy relationship with Peribacillus simplex and P. muralis supported by an 83% bootstrap value (Figure 2). In the Bayesian phylogenetic tree (Supplementary Figure 2) Peribacillus strain AS_2 formed a sister clade with Peribacillus soganii. Bacillus sp. Strain AS_3 had a polytomy clade with Bacillus luti and B. proteolyticus with a 23% bootstrap value (Figure 3). Strain AS_4 had a sister clade with B. cereus, and strain AS_5 did not cluster with any of the species (Figure 3). In the Bayesian phylogenetic tree (Supplementary Figure 3), strains AS_3 and AS_5 had sister clades with B. albus and B. pacificus, respectively. Strain AS_4 did not cluster with any species, although there was no congruency between the two methods, both indicate that strains in this study belong to Lysinibacillus, Peribacillus, and Bacillus genera. Strains AS_4 and AS_5 could represent new species and further studies are recommended.
The five isolated endophytic bacteria from medicinal plant A. sessiliflora were tested against 11 pathogenic strains for antibacterial activity as shown in Table 2. The minimum inhibitory concentration of extracted secondary metabolites ranged from 8 to 0.25 mg/ml. The crude extracts of Bacillus sp. strain AS_3 and Bacillus sp. strain AS_5 showed no inhibition against all the indicator strains. The lowest MIC value was recorded against K. pneumoniae (0.25 mg/ml), B. cereus (2 mg/ml), and S. saprophyticus (2 mg/ml) from strains AS_2 and AS_4. The highest concentration was recorded against M. smegmatis, E. coli, and V. parvula with MIC values ranging from 8 to 16 mg/ml from strains AS_1 and AS_2. Statistical analysis showed that only two of the endophytes’ extracts, strains AS_2 and AS_4 had significant (p < 0.05) inhibition values with the lowest MIC values of 0.25 and 2 mg/ml.
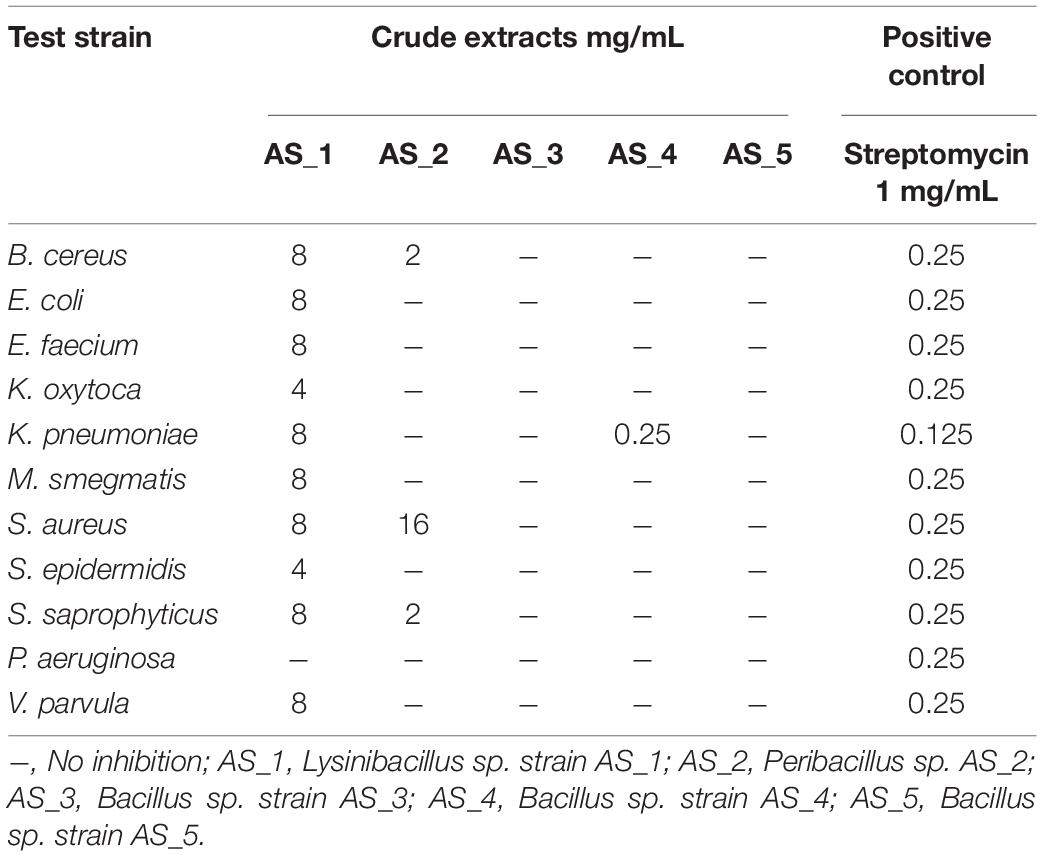
Table 2. Minimum inhibitory concentrations of crude extracts of bacterial endophytes associated with Alectra sessiliflora.
Different concentrations of crude ethyl acetate extracts of Lysinibacillus sp. strain AS_1, Peribacillus sp. strain AS_2, Bacillus sp. strain AS_3, Bacillus sp. strain AS_4, and Bacillus sp. strain AS_5 were used to determine antitumor activity against three cancer cell lines (A549, Hela and Hek 293). Dimethyl sulfoxide was used as a negative control, while auranofin was used as a positive control as it is known to kill most cancer cells by inhibiting thioredoxin reductase and the ubiquitin-proteasome system (McCauley et al., 2013; Li et al., 2016). Endophytic crude extracts showed varying activities against Hela cervical carcinoma cells with AS_1 and AS_2 showing 99% reduction at a concentration of 1,000–500 μg/ml (Figure 4). A cell reduction of 61% was observed for AS_3 at a concentration of 1,000 μg/ml. An increase in cell viability > 100% was noted for AS_4 and AS_5. Overall, only three strains AS_1, AS_2, and AS_3 had significant inhibition values (p < 0.05) except for strains AS_4 and AS_5 for Hela and Hek 293 cancer cell lines. Only strain AS_2 showed the most significant inhibition at higher concentrations (500 and 1,000 μg/ml) for A549 adenocarcinoma cells. The cell line viability was in the following order: Hela > Hek 293 > A459.
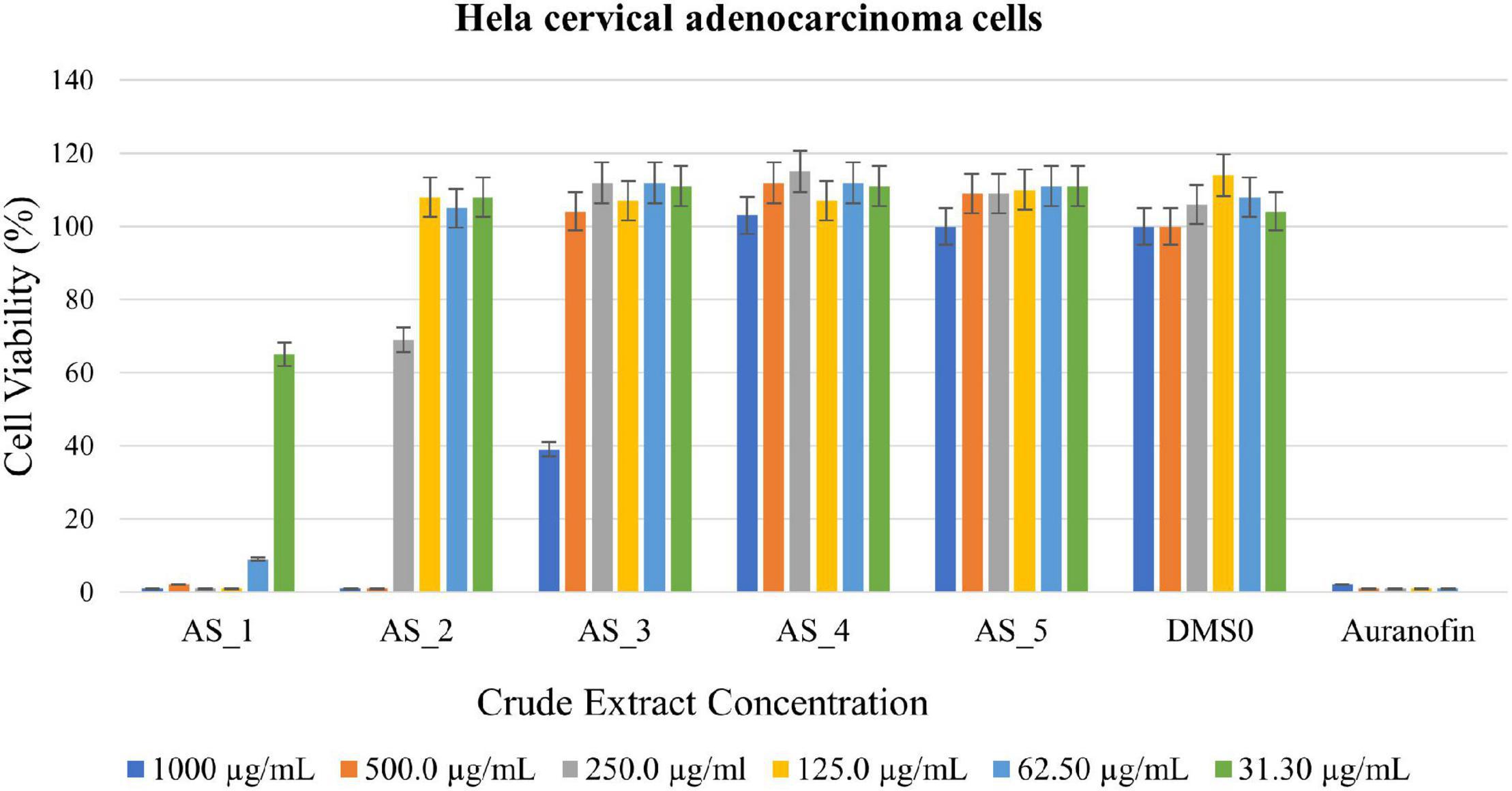
Figure 4. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT) cytotoxic assay of endophyte-derived secondary metabolites on Hela cervical adenocarcinoma cells tested at different concentrations ranging from 1,000 to 31.30 μg/ml. Auranofin was used as a positive control. AS_1, Lysinibacillus sp. strain AS_1; AS_2, Peribacillus sp. AS_2; AS_3, Bacillus sp. strain AS_3; AS_4, Bacillus sp. strain AS_4; AS_5, Bacillus sp. strain AS_5.
Strain AS_2 showed the highest cell reduction of 96% against Hek 293 kidney cells at a concentration of 500 μg/ml (Figure 5) and strain AS_3 showed a cell reduction of 92% (1,000 μg/ml), 83% (500 μg/ml), and 75% (250 μg/ml). A reduction of 62% was noted for strain AS_1 at a concentration of 1,000 μg/ml. No notable reduction was observed for strains AS_4 and AS_5 there was an increase in cell viability.
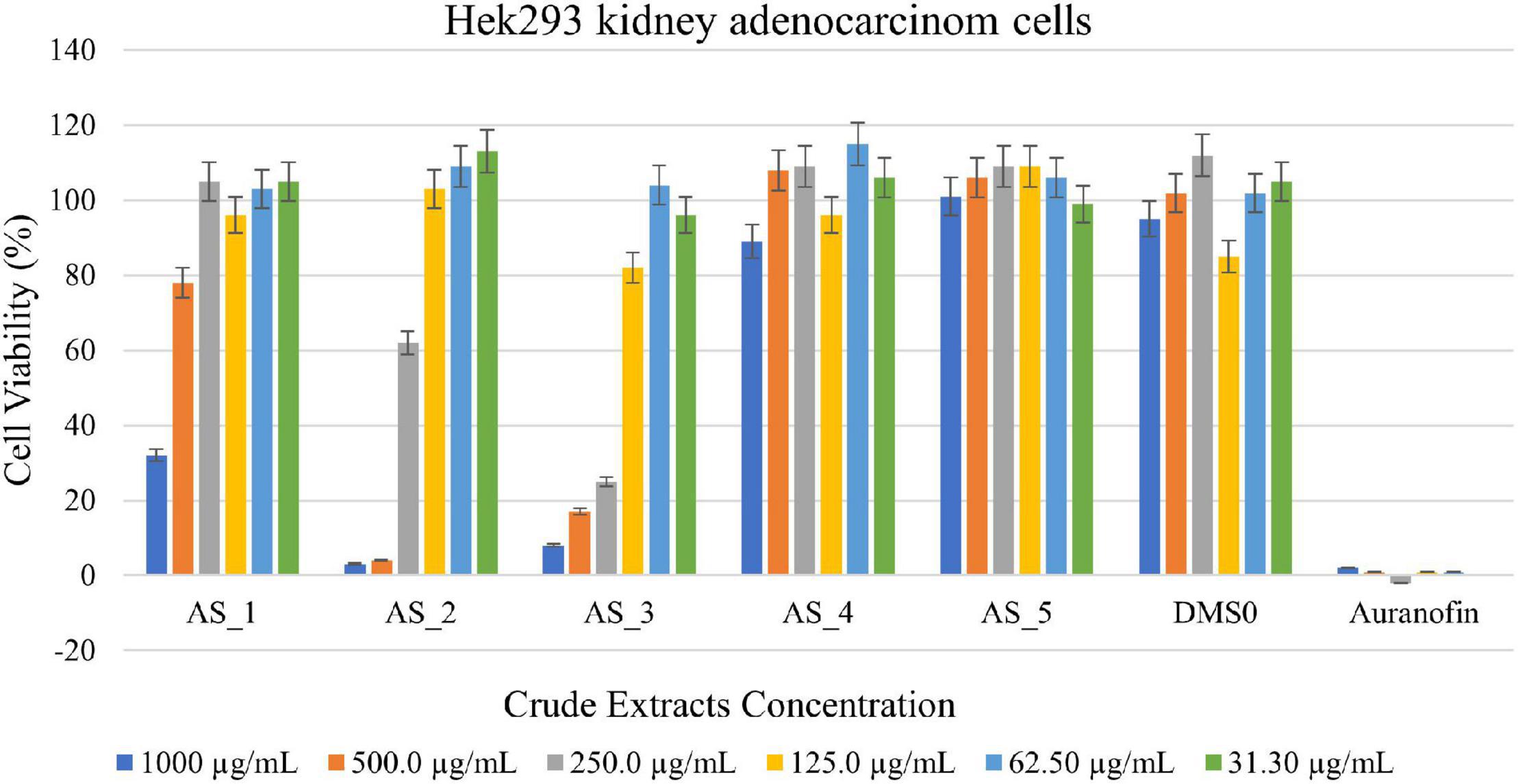
Figure 5. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT) cytotoxic assay of endophyte-derived secondary metabolites on Hek293 kidney adenocarcinoma cells tested at different concentrations ranging from 1,000 to 31.3 500 μg/ml. Auranofin was used as a positive control. AS_1, Lysinibacillus sp. strain AS_1; AS_2, Peribacillus sp. AS_2; AS_3, Bacillus sp. strain AS_3; AS_4, Bacillus sp. strain AS_4; AS_5, Bacillus sp. strain AS_5.
Bacillus sp. strain AS_3 crude extracts were able to kill all the A549 lung cells at a concentration of 1,000 μg/ml having cell viability of 0% (Figure 6) and strain AS_2 extracts showed a cell reduction of 99% at concentrations of 1,000–500 μg/ml respectively. Strain AS_1 showed a cell reduction of less than 50% at a concentration of 125 and 62.5 μg/ml respectively. An increase in cell viability was observed for strains AS_4 and AS_5.
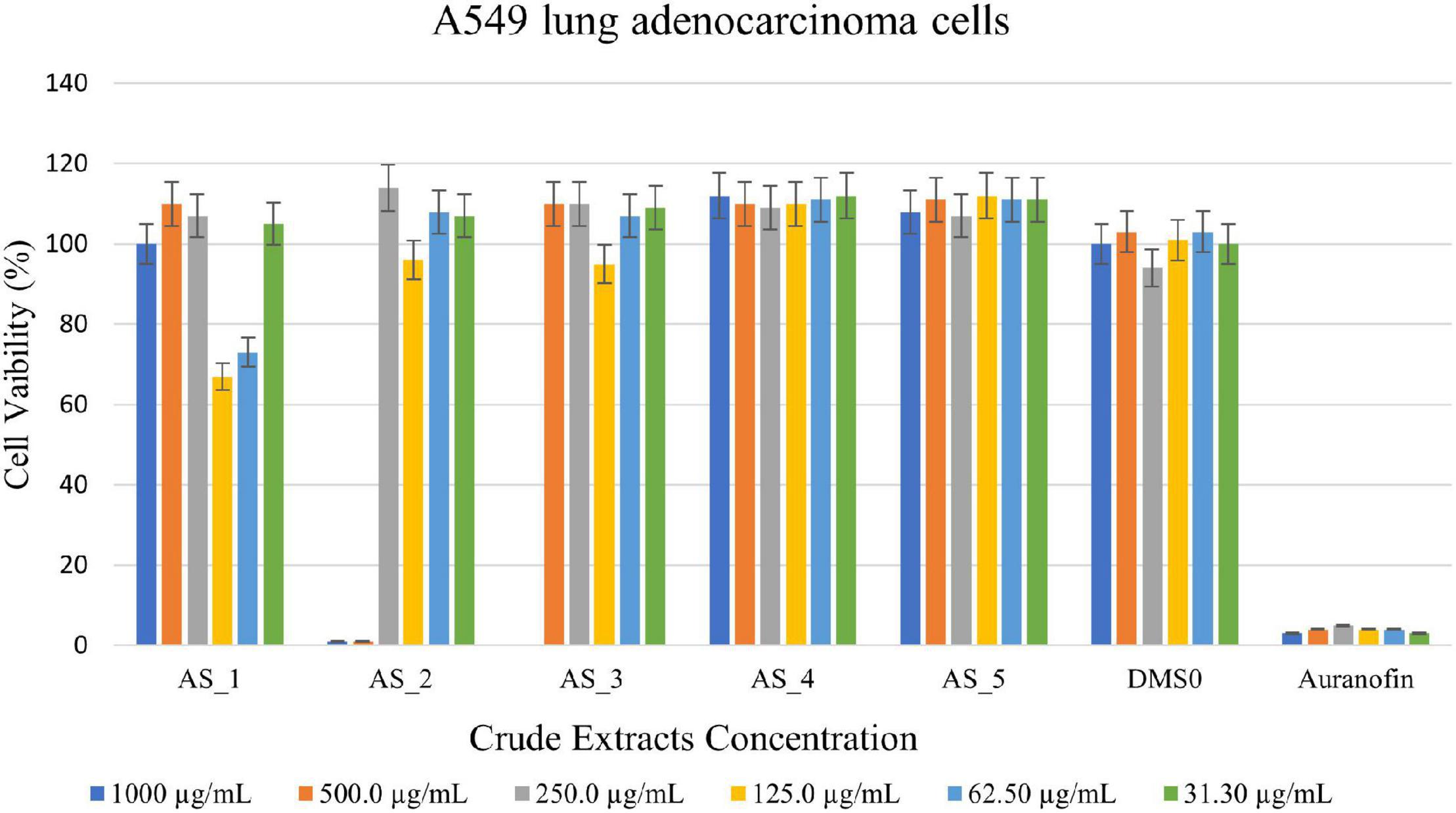
Figure 6. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT) cytotoxic assay of endophyte-derived secondary metabolites on A549 lung adenocarcinoma cells tested at different concentrations ranging from 1,000 to 31.3 500 μg/ml. Auranofin was used as a positive control. AS_1, Lysinibacillus sp. strain AS_1; AS_2, Peribacillus sp. AS_2; AS_3, Bacillus sp. strain AS_3; AS_4, Bacillus sp. strain AS_4; AS_5, Bacillus sp. strain AS_5.
The IC50 values were determined on all three cancer cell lines. From Table 3 it can be observed that the IC50 for Hela was 52.78, 262, and 700.7 μg/ml for the Hela adenocarcinoma cells for strains AS_1, AS_2, and AS_3, respectively. For Hek 293 adenocarcinoma cells, 50% inhibition was observed at 523.8 μg/ml (strain AS_1), 262.2 μg/ml (strain AS_2), 169.4 μg/ml (strain AS_3), and 18.31 μg/ml (strain AS_5). For A549 cells, 50% inhibition was observed at concentrations ranging from 190.9, 380.6, 753.3, and 165.4 μg/ml for strains AS_1, AS_2, AS_3, and AS_5, respectively. No notable inhibition was observed for strain AS_4 for all the cancer cell lines and AS_5 for Hela cells.
Metabolite profiling of the endophyte’s crude extracts from A. sessiliflora was subjected to GC-MS analysis. The bioactive compounds were identified and tabulated (Table 4). Table 4 shows the metabolite profiles for the ethyl acetate extracts. Only compounds having a retention time (RT) of ≥3 min were recorded. The gas chromatography results of the bacterial crude extracts identified a total of 80 secondary metabolites (Table 4, Supplementary Table 2, and Supplementary Figures 4–8), with AS_1 (Supplementary Figure 4) having the most identified compounds. The compounds prevalent in all the extracts were tridecane (C13H28), hexadecane (C16H34), tetracosane (C24H50), and ergotaman-3′,6′,18-trione,9,10-dihydro-12′-hydroxy-2′-methyl-5′-(phenylmethyl)-, (5′a,10a) (C33H37N5O5). Other interesting metabolites included benzyl benzoate (C9H16N2O2), benzene-acetamide (C8H9NO), 2-coumaranone (C8H6O2), and octacosane (C28H58).
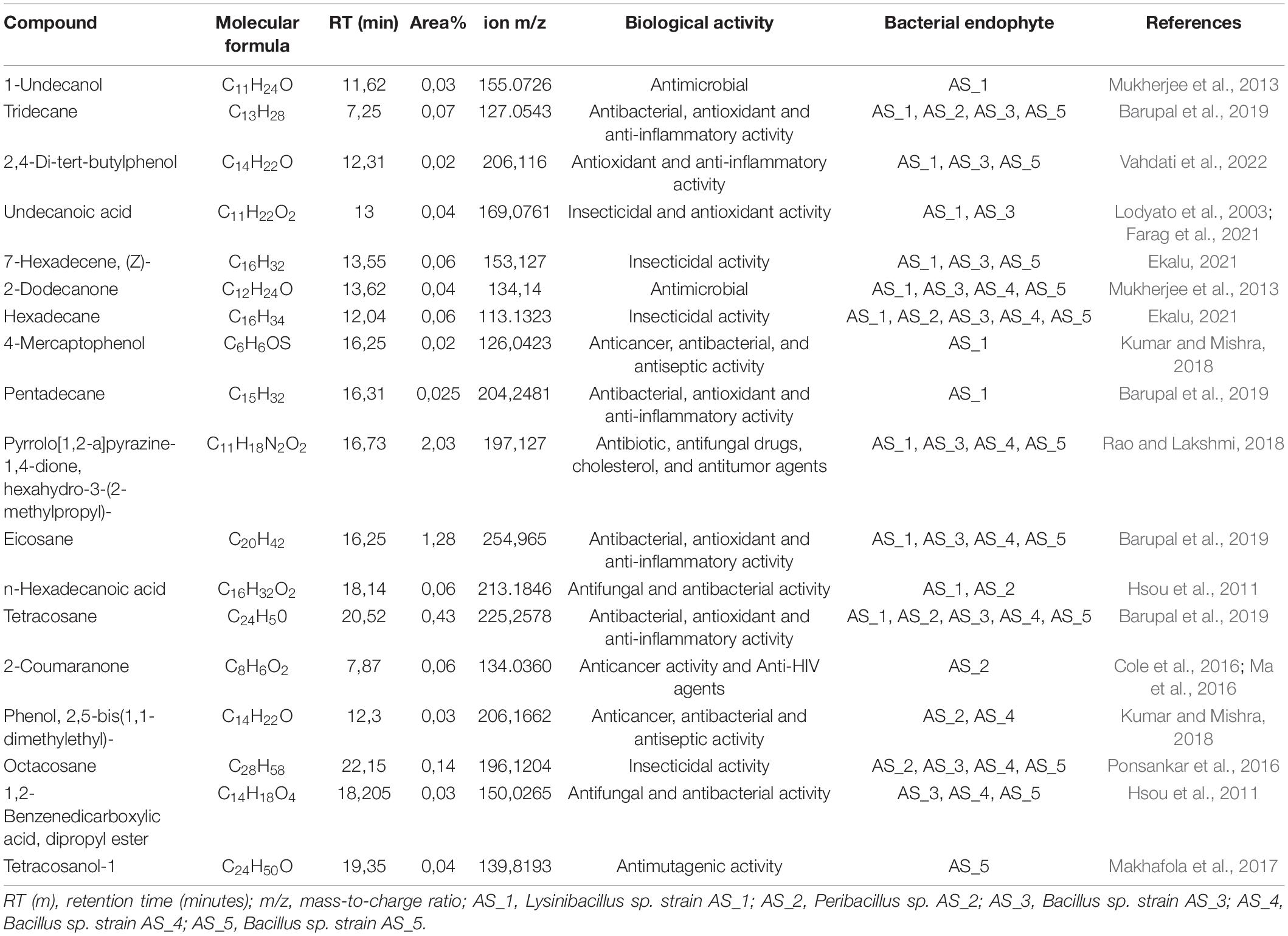
Table 4. GC-HRTOFMS analysis of bacterial endophyte’s crude extracts associated with Alectra sessiliflora.
Endophytic bacteria isolated from medicinal plants have gained much interest from researchers as they have been shown to possess antibacterial and antifungal activities (Duhan et al., 2020). They are also known to produce a wide range of secondary metabolites with various biological activities including antioxidant, antimalarial, antidiabetic, antimicrobial, anti-inflammatory, and cytotoxic (Abdalla et al., 2020). Due to the increasing number of deaths from infections caused by drug-resistant bacteria and cancer, there is an urgent need to search for new sources of drugs (Kusari et al., 2013; Prakash et al., 2020). Alectra sessiliflora is a medicinal plant with a limited history in ethnobotanical applications, but studies on the endophytic bacteria associated with it are scarce.
Based on the NCBI-BLAST database, the isolated strains had a 93–99% similarity to Lysinibacillus fusiformis strain POB29, Peribacillus simplex strain TP 141-1, Bacillus cereus strain NCIM 2158, Bacillus proteolyticus strain KLR12, and Bacillus safensis strain MF-86-1 (Supplementary Table 1). Although five bacterial endophytes were isolated in this study, the data adds to the minimally reported phyllosphere bacterial endophytes (Sivakumar et al., 2020). Li et al. (2021), reported a minimal number of bacterial endophytes from halophytes, with five bacterial endophytes isolated each from Reaumuria soongorica (PalL Maxim.) and Peganum harmala L., and three bacterial endophytes each from Artemisia carvifolia (Buch-Ham. ex Roxb. Hort. Beng.) and Suaeda dendroides (C. A. Mey. Moq.). The current study is the first to isolate and identify bacterial endophytes from A. sessiliflora, we strongly believe that more bacterial endophytes are associated with this plant host, thus necessitating further identification of its endophytes. The phyllosphere bacterial endophyte community is known to be affected by plant genotype, immune system and species, soil type, climatic conditions, and geographic location (Copeland et al., 2015). This could explain the low number of endophytes isolated in the current study. Similarly, previous studies reported the occurrence of Bacillus endophyte species within the phyllosphere of lettuce (Rastogi et al., 2012), and grapevine (Kembel et al., 2014). Peribacillus spp. and Lysinibacillus spp. have not been reported as phyllosphere endophytes, nonetheless, Lysinibacillus spp. have previously been isolated from tomato roots (Zhu et al., 2021), rice roots (Khaskheli et al., 2020; Shabanamol et al., 2021); and Peribacillus spp. was previously isolated from P. harmala (Li et al., 2021) and canola crop roots (Martínez-Hidalgo et al., 2021), making our study one of the few to isolate and report Peribacillus sp. as an endophyte. The 16S rRNA approach remains the gold standard for the initial identification of bacterial species, however, this approach cannot differentiate closely related bacterial species as is indicated by the formation of polytomy relationships in Figures 1–3 (Supplementary Figures 1–3; Kitahara and Miyazaki, 2013). The 16S rRNA gene has identified the endophytes to genus level, further studies like whole genome sequencing and multilocus sequence analysis (MLSA) are required for species delineation.
New and effective therapeutic drugs are required to combat microbial drug resistance (World Health Organization [WHO], 2021), and increasing incidence of cancers some of which have drug resistance (Housman et al., 2014). The use of medicinal plants as a source of bioactive compounds has paved the way for the discovery of novel drugs against microbial and cancer infections (Gagana et al., 2020). However, one setback of using medicinal plants is that several factors including the chemical composition of the plant, season and geographical specificity, cultivation requirements, and random use of the plant may limit their potential use (Katiyar et al., 2012). Moreover, their overuse can ultimately lead to plant extinction. Several studies suggest that endophytic bacteria isolated from medicinal plants can produce the same or similar bioactive compounds as their host plant including novel compounds (Mehanni and Safwat, 2009; Alvin et al., 2014; Gouda et al., 2016). These findings have attracted the interest of researchers as this indicates that endophytes can act as substitutes for plants when searching for novel bioactive compounds without causing major impacts on the environment.
A previous study has shown that plant extracts of A. sessiliflora exhibited antibacterial activity against selected pathogens including S. aureus, P. aeruginosa, E. coli, B. pumilus, and Shigella dysenteriae at MIC values ranging from 3.13–25 mg/ml (Mariita et al., 2010). In the current study, the antimicrobial activity of bacterial endophytes associated with A. sessiliflora was investigated against 11 pathogenic strains. A plant extract with a MIC value of ≤8 mg/ml is considered to possess some antimicrobial activity while those with a MIC value of ≤1 mg/ml are considered to have significant antibacterial activity (Fabry et al., 1998; Van Vuuren, 2008). Uche-Okereafor et al. (2019), reported antibacterial activities of bacterial endophytes isolated from Solanum mauritianum against pathogenic bacteria such as E. coli, S. aureus, K. pneumoniae, and P. aeruginosa, and the results indicated antimicrobial activity with MIC concentrations ranging from 0.0625 to 8 mg/ml. In a similar study by Tapfuma et al. (2020) bacterial endophytes isolated from Celtis africana had an antibacterial activity with MIC concentrations ranging from 4 to 8 mg/ml against B. cereus, E. coli, and S. aureus. Among the five bacterial endophytes, Lysinibacillus strain AS_1 extract had antibacterial activity against 10 test strains with antibacterial activity ranging from 4 and 8 mg/ml (Table 2), with the exception of P. aeruginosa. The most significant MIC value of 4 mg/ml was noted on the K. oxytoca and S. epidermidis. The previous studies demonstrated that the genus Lysinibacillus produced secondary metabolites such as antibiotics, hydrolytic enzymes, and bacteriocins with strong antibacterial activity against selected pathogens such as K. pneumoniae, S. aureus, and P. aeruginosa (Naureen et al., 2017). Peribacillus sp. strain AS_2 extracts had an MIC of 2 and 16 mg/ml, with S. saprophyticus and B. cereus being the most susceptible with an MIC value of 2 mg/ml except for S. aureus which had a higher MIC value of 16 mg/ml. All the test bacterial species were resistant to the Peribacillus crude extracts. To the best of our knowledge, this is the study on the antibacterial activity of bioactive compounds from Peribacillus sp. Bacillus sp. strain AS_4 had an MIC of 0.25 mg/ml against K. pneumoniae, which was the lowest MIC recorded in the study. A study by Akpor et al. (2021), determined the antibacterial potential of metabolites produced by B. proteolyticus, B. thuringiensis, B. cereus, and B. subtilis, and all the extracts showed antimicrobial activity against test pathogens at an MIC of 200 mg/ml. In a similar study by Makuwa and Serepa-Dlamini (2021), bioactive metabolites of Bacillus species isolated from a medicinal plant, Dicoma anomala were found to be effective against selected pathogens such as E. coli, K. oxytoca, and S. aureus with MIC values ranging from 0.625 to 10 mg/ml. According to Borriss et al. (2019), Bacillus species produce antibacterial agents such as surfactin and bacteriocins which may be responsible for their antibacterial activities (Huo et al., 2019). Interestingly, no antibacterial activities were reported for the crude extracts of Bacillus sp. strain AS_3 and Bacillus sp. strain AS_5 for all the pathogenic strains (Table 2). Strain AS_1 had MIC activities against most test strains, and we thus recommend its test against multi-drug resistant bacteria.
The antitumor activity of the bacterial endophyte’s crude extracts from A. sessiliflora was evaluated against three human cancer cells, Hela cervical, Hek 293 kidney, and A549 lung adenocarcinoma cells. Oosthuizen et al. (2019), conducted a study to determine the cytotoxic effects of A. sessiliflora plant extracts against U937 human macrophage cells, and the results showed an increase in cell viability of the U937 cells. In this study, crude secondary metabolites from A. sessiliflora bacterial endophytes showed the best antitumor activities showing cytotoxic effects against the three cancer cell lines. To the best of our knowledge, this is the first report on the antitumor cytotoxic activity of bacterial endophyte’s crude extracts from Lysinibacillus sp. strain AS_1, Peribacillus sp. strain AS_2, Bacillus sp. stain AS_3; Bacillus sp. strain AS_4, and Bacillus sp. strain AS_5, all isolated from A. sessiliflora.
Lysinibacillus sp. strain AS_1 crude extract showed antitumor activity against Hela cervical cells, with growth inhibition of more than 90% at concentrations ranging from 1,000 to 62.50 μg/ml (Figure 4). The minimum concentration of 31.3 μg/ml showed growth inhibition of 35%. For Hek 293 kidney cells (Figure 5), cell growth inhibition reduction of 68% was observed at a concentration of 1,000 μg/ml, while at concentrations of 500 and 125 a growth inhibition of less than 50% was recorded. Similar results were observed for A549 lung cells as no growth inhibition of more than 50% was noted (Figure 6). However, there was an increase in cell growth in the A549 lung cells. Peribacillus sp. strain AS_2 crude endophyte extract showed to have antitumor activity against all the cancer cell lines, with growth inhibition of more than 95% with concentrations ranging from 1,000 to 500 μg/ml. Growth inhibitions of 31 and 38% were observed at a concentration of 250 μg/ml for Hela cervical and Hek293 kidney cells, respectively. An increase in cell growth was observed at concentrations ranging from 125 to 31.3 μg/ml for Hela and Hek293, respectively, with the exception of A549 cells which showed less activity with a 4% reduction.
For Bacillus species, only Bacillus sp. strain AS_3 showed notable antitumor activity against all the cancer cells at a concentration of 1,000 μg/ml. Growth inhibition of 61% was observed for Hela cells at a concentration of 1,000 μg/ml. Growth inhibition of 92.83 and 75% was observed for Hek 293 kidney cells with a concentration ranging from 1,000 to 250 μg/ml, respectively (Figures 4–6). A 100% growth inhibition was achieved for A459 lung cells at a concentration of 1,000 μg/ml. No notable activity was observed for strains AS_4 and AS_5 for all the cancer cells, instead, there was an increase in cell growth. Overall, strains AS_1, AS_2, and AS_3, showed a significant effect on the growth inhibition of cells by decreasing the three cancer cells as compared to strains AS_4 and AS_5. Bacillus species are known to produce bioactive metabolites with antitumor and antibacterial species (Shao et al., 2021). Sebola et al. (2020), conducted a study to evaluate the antitumor activity of crude extracts from the medicinal plant Crinum macowanii Baker, and the results showed that the crude extracts of B. safensis had growth inhibition of 50% against A549 cells at a concentration of 100 μg/ml. Ramasubburayan et al. (2015), conducted a similar study in which the anticancer activity of the crude extract of B. subtilis subsp. subtilis RG was tested against MCF-7 human breast adenocarcinoma cells, and the results indicated growth of 37% at a concentration of 25 μg/ml.
The IC50 values were further determined to show the concentration at which 50% inhibition of the tumor or cancerous cells occurred. It has been proposed that extracts with IC50 values <20 μg/ml are significant when tested against cancer cell lines, whereas IC50 values <50 μg/ml are moderate, low when IC50 values are <200 μg/ml and non-toxic when IC50 > 200 μg/ml (Kuete and Efferth, 2015). For Hela cervical adenocarcinoma cells, strain AS_1 showed a low IC50 of 52.78 μg/ml. Strains AS_4 and AS_5 were found to be non-toxic (IC50 > 200 μg/ml) to the Hela cancer cells. For Hek 293 kidney adenocarcinoma cells, stain AS_5 showed a significant IC50 of 18.31 μg/ml, whereas strain AS_3 showed a low IC50 value of 169.4 μg/ml. For A549 lung cells, strain AS_5 showed low IC50 at a concentration of 165.4 μg/ml. Strains AS_1, AS_2, and AS_3 showed strong cytotoxic activities while the other extracts had poor activity. Bostanci et al. (2022), investigated the anticancer activity of the Salvia marashica plant in two cancer cell lines, breast cancer cells (MF-7) and healthy endothelial cell line (HUVEC), and the plant showed IC50 values at concentrations of 125 and 1,650 μg/ml, respectively. A similar study by Abdel-Fatah et al. (2021), showed that extracts isolated from Gink biloba showed anticancer activity of IC50 values of 4.06 and 6.07 μM for cancer cell lines HEPG2 (liver) and MCF7 (breast) cells, respectively.
The chemical composition of Alectra sessiliflora bacterial endophyte’s crude extracts was further analyzed using gas chromatography. The analysis identified 80 compounds (Table 4, Supplementary Table 2, and Supplementary Figures 4–8) belonging to different chemical groups of acids, alcohols, amino acids, aldehydes, amines, amides, ethers, esters, hydrocarbons, ketones, and carbohydrates. Several major compounds were identified in all the extracts including pyrrolo[1,2-a] pyrazine-1,4-dione, hexahydro-3-(2-methylpropyl), tridecane, eicosane, tetracosane, and hexadecane as shown in Table 4. Some alkane compounds or derivatives such as octacosane, tetracosane, and eicosane secreted by Bacillus sp. AS_3 have been reported to be a potential inhibitor against different cancer cells including cervical carcinoma, breast carcinoma, and, human embryonic lung cells W1-38 (Strobykina et al., 2019).
Phenol and phenol derivatives were identified in some bacterial extracts such as 4-mercaptophenol (AS_1), phenol,2,2-bis(1,1dimethyl) (AS_2 and AS_4), and 2,4-di-tertbutylphenol (AS_1, AS_3 and AS_5). These compounds are well-known to possess vital therapeutic properties including anticancer, antibacterial, antiseptic, and anti-inflammatory (Kumar and Mishra, 2018). The compound n-tetracosanol was isolated from leave extracts of Combretum microphyllum and it was found to have antimutagenic activity against Salmonella typhimurium TA98 at a low concentration of 5 μg/ml, and because mutations play a role in the pathogenesis and development of cancerous cells, it thus prevents the pathological process of cancer which can be caused by mutations (Makhafola et al., 2017). Vergara et al. (2015) conducted a study on the antiproliferative evaluation of tetracosanol over Chinese hamster ovary cells K1 (CHO-K1) and human melanoma cells and it was found that tetracosanol had no cytotoxic effect on the growth of CHO-K1 cells whereas, for human melanoma cells, it affected the cell density and inhibited growth by 58%. In this study, only Bacillus sp. strain AS_5 secreted this metabolite. Most of the compounds identified in this study have antibacterial and antitumor activity, which explains the results obtained from the crude extracts, especially for strains AS_1 and AS_2. The use of organic solvents such as ethyl acetate to extract bioactive metabolites from medicinal plants has been reported to yield a high number of metabolites with higher purity compared to water-based methods (Jose et al., 2002; Pintać et al., 2018). The culturing of bacterial endophytes for 7 days was sufficient to yield the expected metabolites. In a similar study by El-Naggar et al. (2017), 22 antimicrobial metabolites were achieved using the solvent ethyl acetate. Chakraborty et al. (2021), conducted a study in which 42 compounds from Streptomyces levis strain KS46 were found to possess antibacterial, antioxidant, antifungal, and antiproliferative activities. In this study, Lysinibacillus sp. strain AS_1 crude extract exhibited a high number of bioactive compounds which explains the significant antibacterial and antitumor activities as compared to the other bacterial endophyte extracts. In general, the presence of bioactive compounds in all the crude extracts of bacterial endophytes especially those with antibacterial and antitumor activities should be investigated further as they have shown potential to inhibit pathogenic bacteria and cancer cells and this further necessitates their investigation and use for drug development. To our knowledge, this is the first study to report on the bacterial endophytes associated with A. sessiliflora, with antibacterial and antitumor activity against bacterial pathogens and cancer cells, respectively.
With more studies conducted on plant-associated bacterial endophytes, more evidence has demonstrated that endophytes provide significant benefits to various sectors, such as pharmaceuticals, industry, and agriculture. The current results in this study showed that A. sessiliflora does harbor bacterial endophytes, some with notable antibacterial and antitumor activities, which are attributed to their bioactive constituents, we thus recommend further studies to be conducted for the isolation of more diverse endophytes from A. sessiliflora. The antibacterial and antitumor activities of the crude extracts show the potential use of endophytic bacteria and thus should be considered as a novel source for the isolation and production of pure bioactive compounds. Moreover, further research needs to be conducted on the specific compounds responsible for the antibacterial and antitumor activities of the endophytes as this would be useful in developing new antimicrobial drugs and understanding the mechanism of action of these compounds on the studied cancer cells.
The 16S rRNA gene sequences of this study are available from the corresponding author upon request. The isolated bacterial endophytes sequences in this study have been deposited in GenBank with the following accession numbers: MZ976846–MZ976850.
MM contributed to the experimental work. HW contributed to materials used for the antitumor studies. MS-D conceptualized the study, provided the materials used for the isolation, identification, and antimicrobial studies, and was the main supervisor of the project. All authors were involved in the writing of this manuscript and approved the submitted version.
This research was funded by the South African National Research Foundation Thuthuka grant (TTK210216586709). MM received the UJ Faculty of Science Bursary.
HW was employed by Mintek.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to acknowledge the South African National Research Foundation (NRF), Thuthuka, for the financial support for this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.870821/full#supplementary-material
Abdalla, M. A., Aro, A. O., Gado, D., Passari, A. K., Mishra, V. K., Singh, B. P., et al. (2020). Isolation of endophytic fungi from South African plants, and screening for their antimicrobial and extracellular enzymatic activities and presence of type I polyketide synthases. S. Afr. J. Bot. 134, 336–342. doi: 10.1016/j.sajb.2020.03.021
Abdel-Fatah, S. S., El-Batal, A. I., El-Sherbiny, G. M., Khalaf, M. A., and El-Sayed, A. S. (2021). Production, bioprocess optimization and γ-irradiation of Penicillium polonicum, as a new taxol producing endophyte from Ginko biloba. Biotechnol. Rep. 30:e00623. doi: 10.1016/j.btre.2021.e00623
Akpor, O. B., Akinwusi, O. D., and Ogunnusi, T. A. (2021). Production, characterization and pesticidal potential of Bacillus species metabolites against sugar ant (Camponotus consobrinus). Heliyon 7:08447. doi: 10.1016/j.heliyon.2021.e08447
Alotaibi, S. S., Alshoaibi, D., Alamari, H., Albogami, S., Khan, E., Alshanbari, A., et al. (2021). Potential significance of medicinal plants in forensic analysis: a review. Saudi J Biol Sci. 28, 3929–3935. doi: 10.1016/j.sjbs.2021.03.071
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Alvin, A., Miller, K. I., and Neilan, B. A. (2014). Exploring the potential of endophytes from medicinal plants as sources of antimycobacterial compounds. Microbiol. Res. 169, 483–495. doi: 10.1016/j.micres.2013.12.009
Andrews, J. M. (2001). Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48, 5–16. doi: 10.1093/jac/48.suppl_1.5
Aswani, R., Jishma, P., and Radhakrishnan, E. K. (2020). “Endophytic bacteria from the medicinal plants and their potential applications,” in Microbial Endophytes, eds A. Kumar and V. K. Singh (Sawston: Woodhead Publishing), 15–36. doi: 10.1016/B978-0-12-818734-0.00002-4
Attia, E. Z., Farouk, H. M., Abdelmohsen, U. R., and Mo’men, H. (2020). Antimicrobial and extracellular oxidative enzyme activities of endophytic fungi isolated from alfalfa (Medicago sativa) assisted by metabolic profiling. S. Afr. J. Bot. 134, 156–162. doi: 10.1016/j.sajb.2019.12.003
Ayukekbong, J. A., Ntemgwa, M., and Atabe, A. N. (2017). The threat of antimicrobial resistance in developing countries causes and control strategies. Antimicrob. Resist. Infect. Control. 6, 1–8. doi: 10.1186/s13756-017-0208-x
Baptista, P. V., McCusker, M. P., Carvalho, A., Ferreira, D. A., Mohan, N. M., Martins, M., et al. (2018). Nano-strategies to fight multidrug resistant bacteria “A Battle of the Titans”. Front. Microbiol. 9:1441. doi: 10.3389/fmicb.2018.01441
Barupal, T., Meena, M., and Sharma, K. (2019). Inhibitory effects of leaf extract of Lawsonia inermis on Curvularia lunata and characterization of novel inhibitory compounds by GC–MS analysis. Biotechnol. Rep. 23:e00335. doi: 10.1016/j.btre.2019.e00335
Borriss, R., Wu, H., and Gao, X. (2019). “Secondary metabolites of the plant growth promoting model Rhizobacterium Bacillus velezensis FZB42 are involved in direct suppression of plant pathogens and in stimulation of plant-induced systemic resistance,” in Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms, eds H. B. Singh, C. Keswani, M. S. Reddy, E. S. Royano, and C. García-Estrada (Singapore: Springer), 147–168.
Bostanci, M. T., Bulbul, A. S., Celik, I. S., Kocabas, Y. Z., Burhan, H., Bayat, R., et al. (2022). Investigation of antibacterial, antifungal, antibiofilm, antioxidant and anticancer properties of methanol extracts of Salvia marashica İlçim, Celep and Doğan and Salvia caespitosa Montbret and Aucher ex Benth plants with medicinal importance. Chemosphere 288:132602. doi: 10.1016/j.chemosphere.2021.132602
Chakraborty, B., Kumar, R. S., Almansour, A. I., Gunasekaran, P., and Nayaka, S. (2021). Bioprospection and secondary metabolites profiling of marine Streptomyces levis strain KS46. Saudi. J. Biol. Sci. 29, 667–679. doi: 10.1016/j.sjbs.2021.11.055
Cole, A. L., Hossain, S., Cole, A. M., and Phanstiel, O. IV (2016). Synthesis and bioevaluation of substituted chalcones, coumaranones and other flavonoids as anti-HIV agents. Bioorganic Med. Chem. 24, 2768–2776. doi: 10.1016/j.bmc.2016.04.045
Copeland, J. K., Yuan, L., Layeghifard, M., Wang, P. W., and Guttman, D. S. (2015). Seasonal community succession of the phyllosphere microbiome. Mol. Plant Microbe Interact. 28, 274–285. doi: 10.1094/MPMI-10-14-0331-FI
Ding, T., and Melcher, U. (2016). Influences of plant species, season and location on leaf endophytic bacterial communities of non-cultivated plants. PLoS Pathog. 11:e0150895. doi: 10.1371/journal.pone.0150895
Duhan, P., Bansal, P., and Rani, S. (2020). Isolation, identification and characterization of endophytic bacteria from medicinal plant Tinospora cordifolia. S. Afr. J. Bot. 134, 43–49. doi: 10.1016/j.sajb.2020.01.047
Dutta, D., Puzari, K. C., Gogoi, R., and Dutta, P. (2014). Endophytes: exploitation as a tool in plant protection. Braz. Arch. Biol. Technol. 57, 621–629. doi: 10.1590/S1516-8913201402043
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5:113. doi: 10.1186/1471-2105-5-113
Egamberdieva, D., Wirth, S., Behrendt, U., Ahmad, P., and Berg, G. (2017). Antimicrobial activity of medicinal plants correlates with the proportion of antagonistic endophytes. Front. Microbiol. 8:199. doi: 10.3389/fmicb.2017.00199
Ekalu, A. (2021). Medicinal uses, phytochemistry, and pharmacological activities of Mitracarpus species (Rubiaceae): a review. Sci. Afr. 11:e00692. doi: 10.1016/j.sciaf.2020.e00692
El-Naggar, N. E. A., El-Bindary, A. A. A., Abdel-Mogib, M., and Nour, N. S. (2017). In vitro activity, extraction, separation and structure elucidation of antibiotic produced by Streptomyces anulatus NEAE-94 active against multidrug-resistant Staphylococcus aureus. Biotechnol. Equip. 31, 418–430. doi: 10.1080/13102818.2016.1276412
Fabry, W., Okemo, P. O., and Ansorg, R. (1998). Antibacterial activity of East African medicinal plants. J. Ethnopharmacol. 60, 79–84. doi: 10.1016/S0378-8741(97)00128-1
Farag, S. M., Essa, E. E., Alharbi, S. A., Alfarraj, S., and El-Hassan, G. M. A. (2021). Agro-waste derived compounds (flax and black seed peels): toxicological effect against the West Nile virus vector, Culex pipiens L. with special reference to GC-MS analysis. Saudi. J. Biol. Sci. 28, 5261–5267. doi: 10.1016/j.sjbs.2021.05.038
Farhat, H., Urooj, F., Tariq, A., Sultana, V., Ansari, M., Ahmad, V. U., et al. (2019). Evaluation of antimicrobial potential of endophytic fungi associated with healthy plants and characterization of compounds produced by endophytic Cephalosporium and Fusarium solani. Biocatal. Agric. Biotechnol. 18:101043. doi: 10.1016/j.bcab.2019.101043
Flores-Paredes, W., Luque, N., Albornoz, R., Rojas, N., Espinoza, M., Pons, M. J., et al. (2021). Evolution of antimicrobial resistance levels of eskape microorganisms in a peruvian iv-level hospital. J. Infect. Chemother. 53:449. doi: 10.3947/ic.2021.0015
Gagana, S. L., Kumaraswamy, B. E., and Shivanna, M. B. (2020). Diversity, antibacterial and antioxidant activities of the fungal endophytes associated with Schleichera oleosa (Lour.) Merr. S. Afr. J. Bot. 134, 369–381. doi: 10.1016/j.sajb.2020.06.012
Gasa, N. (2015). Antibiofilm Activity of South African plant Extracts Against Mycobacterium spp. and their Mechanism of action Using Mycothiol reductase. Pretoria: University of Pretoria.
Gouda, S., Das, G., Sen, S. K., Shin, H. S., and Patra, J. K. (2016). Endophytes: a treasure house of bioactive compounds of medicinal importance. Front. Microbiol. 7:1538. doi: 10.3389/fmicb.2016.01538
Gunatilaka, A. L. (2006). Natural products from plant-associated microorganisms: distribution, structural diversity, bioactivity, and implications of their occurrence. J. Nat. Prod. 69, 509–526. doi: 10.1021/np058128n
Hall, T. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. In Nucleic Acids Symp. Ser. 41, 95–98.
Ham, J. S., Lee, S. G., Jeong, S. G., Oh, M. H., Kim, D. H., Lee, T., et al. (2010). Powerful usage of phylogenetically diverse Staphylococcus aureus control strains for detecting multidrug resistance genes in transcriptomics studies. Mol. Cells. 30, 71–76. doi: 10.1007/s10059-010-0090-3
Hernández, A. G. C., Ortiz, V. G., Gómez, J. L. A., López, M. ÁR., Morales, J. A. R., Macías, A. F., et al. (2021). Detection of Bacillus cereus sensu lato isolates posing potential health risks in Mexican chili powder. Microorganisms 9:2226. doi: 10.3390/microorganisms9112226
Higashide, M., Kuroda, M., Omura, C. T. N., Kumano, M., Ohkawa, S., Ichimura, S., et al. (2008). Methicillin-resistant Staphylococcus saprophyticus isolates carrying staphylococcal cassette chromosome mec have emerged in urogenital tract infections. Antimicrob. Agents Chemother. 52, 2061–2068. doi: 10.1128/AAC.01150-07
Housman, G., Byler, S., Heerboth, S., Lapinska, K., Longacre, M., Snyder, N., et al. (2014). Drug resistance in cancer: an overview. Cancers 6, 1769–1792. doi: 10.3390/cancers6031769
Hsou, A. B., Trigui, M., Mansour, R. B., Jarraya, R. M., Damak, M., and Jaoua, S. (2011). Chemical composition, cytotoxicity effect and antimicrobial activity of Ceratonia siliqua essential oil with preservative effects against listeria inoculated in minced beef meat. Int. J. Food Microbiol. 148, 66–72. doi: 10.1016/j.ijfoodmicro.2011.04.028
Huo, L., Hug, J. J., Fu, C., Bian, X., Zhang, Y., and Müller, R. (2019). Heterologous expression of bacterial natural product biosynthetic pathways. Nat. Prod. Rep. 36, 1412–1436. doi: 10.1039/c8np00091c
Jose, V. M. L. F., Ana, F. F. U., Sissi, M. F., and Vania, M. M. M. (2002). Antibacterial activity of extracts of six macroalgae from the northeastern brazilian coast. Braz. J. Microbiol. 33, 311–313. doi: 10.1590/S1517-83822002000400006
Jukes, T. H., and Cantor, C. R. (1969). “Evolution of protein molecules,” in Mammalian Protein Metabolism, ed. H. N. Munro (New York, NY: Academic Press), 21–132. doi: 10.1016/B978-1-4832-3211-9.50009-7
Katembo, S. P., Mukatakamba, G. K., Charles, V., and Ngulusansi, A. (2021). Clinical trial of Alectra sessiliflora (VAHL.) Kunze powder in the treatment of sheep’s foot rot in Lubero territory (North-Kivu/DR Congo). J. Anim. Plant. Sci. 48, 8722–8728. doi: 10.35759/JAnmPlSci.v48-3.3
Katiyar, C., Gupta, A., Kanjilal, S., and Katiyar, S. (2012). Drug discovery from plant sources: an integrated approach. Ayu 33:10. doi: 10.4103/0974-8520.100295
Kembel, S. W., O’Connor, T. K., Arnold, H. K., Hubbell, S. P., Wright, S. J., and Green, J. L. (2014). Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc. Natl. Acad. Sci. 111, 13715–13720. doi: 10.1073/pnas.1216057111
Khaskheli, M. A., Wu, L., Chen, G., Chen, L., Hussain, S., Song, D., et al. (2020). Isolation and characterization of root-associated bacterial endophytes and their biocontrol potential against major fungal phytopathogens of rice (Oryza sativa L.). Pathogens 9:172. doi: 10.3390/pathogens9030172
Kitahara, K., and Miyazaki, K. (2013). Revisiting bacterial phylogeny: natural and experimental evidence for horizontal gene transfer of 16S rRNA. Mob. Gent. Elements 3:e24210. doi: 10.4161/mge.24210
Kuete, V., and Efferth, T. (2015). African flora has the potential to fight multidrug resistance of cancer. Biomed. Res. Int. 2015:914813. doi: 10.1155/2015/914813
Kumar, A., and Mishra, A. K. (2018). Biological importance of phenol derivatives as potent bioactive compound: a review. Lett. Org. Chem. 15, 251–264. doi: 10.2174/1570178614666171130155539
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kusari, S., Pandey, S. P., and Spiteller, M. (2013). Untapped mutualistic paradigms linking host plant and endophytic fungal production of similar bioactive secondary metabolites. Phytochemistry 91, 81–87. doi: 10.1016/j.phytochem.2012.07.021
Li, H., Hu, J., Wu, S., Wang, L., Cao, X., Zhang, X., et al. (2016). Auranofin-mediated inhibition of PI3K/AKT/mTOR axis and anticancer activity in non-small cell lung cancer cells. Oncotarget 7:3548. doi: 10.18632/oncotarget.6516
Li, L., Gao, L., Liu, Y., Fang, B., Huang, Y., Mohamad, O. A., et al. (2021). Diversity of cultivable endophytic bacteria associated with halophytes in Xinjiang of China and their plant beneficial traits. J. Arid Land. 13, 790–800. doi: 10.1007/s40333-021-0016-2
Lodyato, V. I., Yurkova, I. L., Sorokin, V. L., Shadyro, O. I., Dolgopalets, V. I., and Kisel, M. A. (2003). Synthesis and properties of 11-(3, 5-Di-tert-butyl-2-hydroxyphenylcarbamoyl) undecanoic acid, a new amphiphilic antioxidant. Bioorg. Med. Chem. Lett. 13, 1179–1182. doi: 10.1016/S0960-894X(03)00041-6
Ma, Q., Liu, Y., Zhan, R., and Chen, Y. (2016). A new isoflavanone from the trunk of Horsfieldia pandurifolia. Nat. Prod. Res. 30, 131–137. doi: 10.1080/14786419.2015.1043554
Makhafola, T. J., Elgorashi, E. E., McGaw, L. J., Awouafack, M. D., Verschaeve, L., and Eloff, J. N. (2017). Isolation and characterization of the compounds responsible for the antimutagenic activity of Combretum microphyllum (Combretaceae) leaf extracts. BMC Complement. Altern. Med. 17:446. doi: 10.1186/s12906-017-1935-5
Makuwa, S. C., and Serepa-Dlamini, M. H. (2021). The antibacterial activity of crude extracts of secondary metabolites from bacterial endophytes associated with Dicoma anomala. Int. J. Microbiol. 2021:8812043. doi: 10.1155/2021/8812043
Maloney, K. N., MacMillan, J. B., Kauffman, C. A., Jensen, P. R., DiPasquale, A. G., Rheingold, A. L., et al. (2009). Lodopyridone, a structurally unprecedented alkaloid from a marine actinomycete. Org. Lett. 11, 5422–5424. doi: 10.1021/ol901997k
Mariita, R., Ogol, C. K. P. O., Oguge, N., and Okemo, P. (2010). Antitubercular and phytochemical investigation of methanol extracts of medicinal plants used by the samburu community in Kenya. Trop. J. Pharm. Res. 9, 379–385. doi: 10.4314/tjpr.v9i4.58935
Martínez-Hidalgo, P., Flores-Félix, J. D., Sánchez-Juanes, F., Rivas, R., Mateos, P. F., Santa Regina, I., et al. (2021). Identification of canola roots endophytic bacteria and analysis of their potential as biofertilizers for canola crops with special emphasis on sporulating bacteria. J. Agron. 11:1796. doi: 10.3390/agronomy11091796
McCauley, J., Zivanovic, A., and Skropeta, D. (2013). “Bioassays for anticancer activities,” in Metabolomics Tools for Natural Product Discovery, eds U. Roessner and D. Dias (Totowa, NJ: Humana Press), 191–205.
Mehanni, M. M., and Safwat, M. S. A. (2009). Endophytes of medicinal plants. Acta Hortic. 854, 31–39. doi: 10.17660/ActaHortic.2010.854.3
Morawetz, J. J., and Wolfe, A. D. (2011). Taxonomic revision of the Alectra sessiliflora complex (Orobanchaceae). Syst. Bot. 36, 141–152. doi: 10.1600/036364411X553234
Mukherjee, K., Tribedi, P., Mukhopadhyay, B., and Sil, A. K. (2013). Antibacterial activity of long-chain fatty alcohols against mycobacteria. FEMS Microbiol. Lett. 338, 177–183. doi: 10.1111/1574-6968.12043
Naureen, Z., Rehman, N. U., Hussain, H., Hussain, J., Gilani, S. A., Al Housni, S. K., et al. (2017). Exploring the potentials of Lysinibacillus sphaericus ZA9 for plant growth promotion and biocontrol activities against phytopathogenic fungi. Front. Microbiol. 8:1477. doi: 10.3389/fmicb.2017.01477
Ogbole, O., and Ajaiyeoba, E. (2010). Traditional management of tuberculosis in Ogun State of Nigeria: the practice and ethnobotanical survey. Afr. J. Tradit. Complement. Altern. Med. 7, 79–84. doi: 10.4314/ajtcam.v7i1.57270
Oosthuizen, C. B., Gasa, N., Hamilton, C. J., and Lall, N. (2019). Inhibition of mycothione disulphide reductase and mycobacterial biofilm by selected South African plants. S. Afr. J. Bot. 120, 291–297. doi: 10.1016/j.sajb.2018.09.015
Palanichamy, P., Krishnamoorthy, G., Kannan, S., and Marudhamuthu, M. (2018). Bioactive potential of secondary metabolites derived from medicinal plant endophytes. Egypt. J. Basic. Appl. Sci. 5, 303–312. doi: 10.1016/j.ejbas.2018.07.002
Panigrahi, S., and Rath, C. C. (2021). In vitro characterization of antimicrobial activity of an endophytic bacterium Enterobacter cloaca (MG001451) isolated from Ocimum sanctum. S. Afr. J. Bot. 143, 90–96. doi: 10.1016/j.sajb.2021.07.044
Petrini, O., Sieber, T. N., Toti, L., and Viret, O. (1993). Ecology, metabolite production, and substrate utilization in endophytic fungi. Nat. Toxins. 1, 185–196. doi: 10.1002/nt.2620010306
Pintać, D., Majkić, T., Torović, L., Orčić, D., Beara, I., Simin, N., et al. (2018). Solvent selection for efficient extraction of bioactive compounds from grape pomace. Ind. Crops. Prod. 111, 379–390. doi: 10.1016/j.indcrop.2017.10.038
Ponsankar, A., Vasantha-Srinivasan, P., Senthil-Nathan, S., Thanigaivel, A., Edwin, E. S., Selin-Rani, S., et al. (2016). Target and non-target toxicity of botanical insecticide derived from Couroupita guianensis L. flower against generalist herbivore, Spodoptera litura Fab. and an earthworm, Eisenia foetida Savigny. Ecotoxicol. Environ. Saf. 133, 260–270. doi: 10.1016/j.ecoenv.2016.06.043
Prakash, S., Elavarasan, N., Subashini, K., Kanaga, S., Dhandapani, R., Sivanandam, M., et al. (2020). Isolation of hesperetin-A flavonoid from Cordia sebestena flower extract through antioxidant assay guided method and its antibacterial, anticancer effect on cervical cancer via in vitro and in silico molecular docking studies. J. Mol. Struct. 1207:127751. doi: 10.1016/j.molstruc.2020.127751
Ramasubburayan, R., Sumathi, S., Bercy, D. M., Immanuel, G., and Palavesam, A. (2015). Antimicrobial, antioxidant and anticancer activities of mangrove associated bacterium Bacillus subtilis subsp. subtilis RG. Biocatal. Agric. Biotechnol. 4, 158–165. doi: 10.1016/j.bcab.2015.01.004
Rambaut, A. (2018). FigTree v.1.4.4. Available online at: https://github.com/rambaut/figtree/releases (accessed March 18, 2022).
Rao, M. R. K., and Lakshmi, N. V. (2018). Preliminary phytochemical and GC MS analysis of different extracts of Sphaeranthus indicus leaves. Indo Am. J. Pharm. 5, 1511–1520. doi: 10.5281/zenodo.1204485.svg
Rastogi, G., Sbodio, A., Tech, J. J., Suslow, T. V., Coaker, G. L., and Leveau, J. H. (2012). Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 6, 1812–1822. doi: 10.1038/ismej.2012.32
Roca, I., Akova, M., Baquero, F., Carlet, J., Cavaleri, M., Coenen, S., et al. (2015). The global threat of antimicrobial resistance: science for intervention. New. Microbes. New. Infect. 6, 22–29. doi: 10.1016/j.nmni.2015.02.007
Sciarretta, K., Røttingen, J. A., Opalska, A., Van Hengel, A. J., and Larsen, J. (2016). Economic incentives for antibacterial drug development: literature review and considerations from the transatlantic task force on antimicrobial resistance. Clin. Infect. Dis. 63, 1470–1474. doi: 10.1093/cid/ciw593
Sebola, T. E., Uche-Okereafor, N. C., Mekuto, L., Makatini, M. M., Green, E., and Mavumengwana, V. (2020). Antibacterial and anticancer activity and untargeted secondary metabolite profiling of crude bacterial endophyte extracts from Crinum macowanii baker leaves. Int. J. Microbiol. 2020:8839490. doi: 10.1155/2020/8839490
Shabanamol, S., Thampi, M., Sajana, P., Varghese, S., Karthika, S., George, T. K., et al. (2021). Characterization of the major antifungal extrolite from rice endophyte Lysinibacillus sphaericus against Rhizoctonia solani. Arch. Microbiol. 203, 2605–2613. doi: 10.1007/s00203-021-02229-2
Shao, Y., Wang, X. Y., Qiu, X., Niu, L. L., and Ma, Z. L. (2021). Isolation and purification of a new Bacillus subtilis Strain from deer dung with anti-microbial and anti-cancer activities. Curr. Med. Sci. 41, 832–840. doi: 10.1007/s11596-021-2383-5
Singh, A., Kumar, J., Sharma, V. K., Singh, D. K., Kumari, P., Nishad, J. H., et al. (2021). Phytochemical analysis and antimicrobial activity of an endophytic Fusarium proliferatum (ACQR8), isolated from a folk medicinal plant Cissus quadrangularis L. S. Afr. J. Bot. 140, 87–94. doi: 10.1016/j.sajb.2021.03.004
Strobykina, I. Y., Nemtarev, A. V., Garifullin, B. F., Voloshina, A. D., Sapunova, A. S., and Kataev, V. E. (2019). Synthesis and biological activity of alkane-1, 1-diylbis (phosphonates) of diterpenoid isosteviol. Russ. J. Org. Chem. 55, 17–24. doi: 10.1134/S1070428019010044
Sivakumar, N., Sathishkumar, R., Selvakumar, G., Shyamkumar, R., and Arjunekumar, K. (2020). “Phyllospheric microbiomes: diversity, ecological significance, and biotechnological applications,” in Plant Microbiomes for Sustainable Agriculture, eds A. Yadav, J. Singh, A. Rastegari, and N. Yadav (Cham: Springer), 113–172. doi: 10.1007/978-3-030-38453-1_5
Suchard, M. A., Lemey, P., Baele, G., Ayres, D. L., Drummond, A. J., and Rambaut, A. (2018). Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4:vey016. doi: 10.1093/ve/vey016
Sun, H., He, Y., Xiao, Q., Ye, R., and Tian, Y. (2013). Isolation, characterization, and antimicrobial activity of endophytic bacteria from Polygonum cuspidatum. Afr. J. Microbiol. Res. 7, 1496–1504. doi: 10.5897/AJMR12.899
Tapfuma, K. I., Nchabeleng, E. K., Adebo, O. A., Hussan, R., Williams, R. D., Ravuluvulu, A. B., et al. (2020). Antibacterial activity and gas chromatography mass spectrometry (GC–MS)-based metabolite profiles of Celtis africana and its endophytic extracts. Ind. Crops. Prod. 157:112933. doi: 10.1016/j.indcrop.2020.112933
Uche-Okereafor, N., Sebola, T., Tapfuma, K., Mekuto, L., Green, E., and Mavumengwana, V. (2019). Antibacterial activities of crude secondary metabolite extracts from Pantoea species obtained from the stem of Solanum mauritianum and their effects on two cancer cell lines. Int. J. Environ. Res. Public Health 16:602. doi: 10.3390/ijerph16040602
Vahdati, S. N., Lashkari, A., Navasatli, S. A., Ardestani, S. K., and Safavi, M. (2022). Butylated hydroxyl-toluene, 2, 4-Di-tert-butylphenol, and phytol of Chlorella sp. protect the PC12 cell line against H2O2-induced neurotoxicity. Biomed. Pharmacother. 145:112415. doi: 10.1016/j.biopha.2021.112415
Van Vuuren, S. F. (2008). Antimicrobial activity of South African medicinal plants. J. Ethnopharmacol. 119, 462–472. doi: 10.1016/j.jep.2008.05.038
Vergara, M., Olivares, A., and Altamirano, C. (2015). Antiproliferative evaluation of tall-oil docosanol and tetracosanol over CHO-K1 and human melanoma cells. Electron. J. Biotechnol. 18, 291–294. doi: 10.1016/j.ejbt.2015.05.004
Wagenlehner, F. M. E., Heisig, P., Irtenkauf, C., Notka, F., Decker, J., Lehn, N., et al. (2003). Clinically significant borderline resistance of sequential clinical isolates of Klebsiella pneumoniae. Int. J. Antimicrob. Agents 22, 367–373. doi: 10.1016/S0924-8579(03)00126-2
World Health Organization [WHO] (2021). Antimicrobial Resistance. Geneva: World Health Organization.
Yeates, C., Gillings, M. R., Davison, A. D., Altavilla, N., and Veal, D. A. (1997). PCR amplification of crude microbial DNA extracted from soil. Lett. Appl. Microbiol. 25, 303–307. doi: 10.1046/j.1472-765X.1997.00232.x
Zhang, A., He, X., Meng, Y., Guo, L., Long, M., Yu, H., et al. (2016). Antibiotic and disinfectant resistance of Escherichia coli isolated from retail meats in Sichuan, China. Microb. Drug Resist. 22, 80–87. doi: 10.1089/mdr.2015.0061
Zhu, L., Guo, J., Sun, Y., Wang, S., and Zhou, C. (2021). Acetic acid-producing endophyte Lysinibacillus fusiformis orchestrates jasmonic acid signaling and contributes to repression of cadmium uptake in tomato plants. Front. Plant Sci. 12:1041. doi: 10.3389/fpls.2021.670216
Keywords: Alectra sessiliflora, antibacterial activity, bioactive compounds, bacterial endophytes, antitumor activity
Citation: Maela MP, van der Walt H and Serepa-Dlamini MH (2022) The Antibacterial, Antitumor Activities, and Bioactive Constituents’ Identification of Alectra sessiliflora Bacterial Endophytes. Front. Microbiol. 13:870821. doi: 10.3389/fmicb.2022.870821
Received: 07 February 2022; Accepted: 02 June 2022;
Published: 05 July 2022.
Edited by:
Khondoker M. G. Dastogeer, Bangladesh Agricultural University, BangladeshReviewed by:
Weaam Ebrahim, Mansoura University, EgyptCopyright © 2022 Maela, van der Walt and Serepa-Dlamini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mahloro Hope Serepa-Dlamini, aG9wZXNAdWouYWMuemE=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.