
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 April 2022
Sec. Terrestrial Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.870667
 Chaoqun Wang1,2,3
Chaoqun Wang1,2,3 Ruzhen Jiao1,2,3*
Ruzhen Jiao1,2,3*Ecological stoichiometry is increasingly acknowledged as one of the main control factors for microbial activity and diversity. Soil carbon/nitrogen (C/N) and carbon/phosphorus (C/P) ratios are usually much higher than microbial nutrient requirements and vary with planting density and stand age in forestlands. However, how microorganisms cope with such stoichiometric imbalances and how they regulate nutrient cycling remain unclear. Here, 5- and 35-year-old experimental Cunninghamia lanceolata [Lamb.] Hook plantations with five planting densities (1,667, 3,333, 5,000, 6,667, and 10,000 stems ha−1) were used to explore the underlying mechanism of the response of microorganisms to stoichiometric imbalances. We found that (i) enzyme activity and microbial biomass and diversity increased with planting density at age 5 but decreased at age 35; (ii) soil microorganisms were P-limited at age 5, but gradually shifted from P- to N-limitation during the development of plantations from 5 to 35 years; and (iii) significantly negative relationships between microbial biomass stoichiometry and enzymatic stoichiometry were observed. The adaptive pathways of soil microorganisms to cope with stoichiometric imbalances include (i) adjusting the stoichiometry of microorganisms and enzymes; (ii) changing the relative abundance of the dominant microbial phyla; and (iii) increasing the ratio of fungal to bacterial diversity. These results highlight how to use the ecological stoichiometry method to identify soil microbial nutrient limitations with planting density during the development of plantations. By underlining the important role of stoichiometry on microbial growth and activity, these findings furthermore emphasize the dependency of organic matter transformation and nutrient cycling on the microbial community.
Ecological stoichiometry theory can be used to predict how living organisms will absorb exterior environmental resources [usually carbon (C), nitrogen (N), and phosphorus (P)] at ecosystem scales (Hillebrand and Kahler, 2005; Marichal et al., 2011; Xu et al., 2017; Yang et al., 2020). For microorganisms, its biomass C/P and N/P ratios are commonly used as indicators of the bioavailability of P (Güsewell and Freeman, 2003; Tessier and Raynal, 2003). However, the stoichiometry of soil microbial biomass and its available C, N, and P are more restricted to a fairly narrow range at the global scale (Bell et al., 2013), and there are large differences between communities (Xu et al., 2013). Therefore, it is difficult to understand the non-equilibrium flow mechanism of ecosystem nutrients only by evaluating the stoichiometry of microbial biomass. Ecological enzyme (enzymes not contained by membranes of living cells) activity and their stoichiometry are indicators of microbial nutrient requirements since enzymes are regulated by the availability of nutrients (Sinsabaugh et al., 2009). For example, lower β-glucosidase/acid phosphatase ratio and β-1,4-N-acetylglucosaminidase/acid phosphatase ratio in forests indicate greater microbial P demand relative to P availability (Waring et al., 2014). Nonetheless, large uncertainties still exist due to the different responses of soil enzymatic activity and stoichiometry to nutrient stoichiometry (Cleveland and Liptzin, 2007; Burns et al., 2013). Therefore, studies on the stoichiometry of soil hydrolytic enzymes that are responsible for the mineralization of C, N, and P will provide deeper insights into the mechanism of soil organic matter transformation and nutrient cycling.
In forestlands, root exudates strongly affect the soil organic C availability and ultimately alter soil nutrient stoichiometry (Phillips et al., 2011). Moreover, soil C/N/P stoichiometry is closely related to the composition of understory vegetation, because the stoichiometry of terrestrial plant organs varies widely (Bell et al., 2013). For example, C/P and N/P of plant tissues vary in the range of 230–1,100 and 20–50, respectively (Bell et al., 2013). Therefore, changes in understory vegetation communities with plantation development will input organic matter sources with different C/N/P stoichiometry into the soil. This eventually causes imbalances in microbial nutrient requirements (Cleveland and Liptzin, 2007). Furthermore, root exudates and understory communities widely vary with planting density during the development of plantations due to the change in the intensity of competition for environmental resources such as light, water, and nutrients (Bremer and Farley, 2010). For instance, decreased planting density supported better maintenance of understory vegetation in Pinus radiata plantations (Brockerhoff et al., 2003). The total number of understory plant species, density, and diversity gradually increased in 5–10 years after the establishment of Eucalyptus grandis plantations (Zhang et al., 2014). Therefore, large uncertainties concerning the effects of planting density during the development of plantations on ecological stoichiometry are huge challenges for predicting soil nutrient cycling and sustainable development of afforestation ecosystems.
Various responses of soil bacterial and fungal community diversity and composition to stoichiometric imbalances following afforestation were observed in different plantations (Deng et al., 2016; Zhang et al., 2016; Ren et al., 2017; Wang et al., 2020). This is mainly because bacteria and fungi have different C/N/P stoichiometries and target to decompose organic matters according to the theory of ecological stoichiometry (Mooshammer et al., 2014). For example, fast-growing bacteria with lower C/P or N/P ratios mainly decompose labile and nutrient-rich organic matter to meet their elemental requirements (Ganie et al., 2016), whereas fungi are the main controllers of N and P mobilization rates from root exudates (Li et al., 2015). Therefore, the fungal-to-bacterial ratio can be considered an indicator of nutrient availability (Vries et al., 2006). Changes in soil C/N/P stoichiometries affect microbial community compositions, diversity, and functions (Elser et al., 2000; Aanderud et al., 2018). For instance, the N/P ratio is the predominant factor for the abundances of Proteobacteria, Bacteroidetes, Cyanobacteria, and Nitrospirae in Robinia pseudoacacia L. forests (Ren et al., 2017). The N addition strongly decreased microbial respiration in Pinus koraiensis plantation soils (Zhou et al., 2017). Despite recent improvements in understanding the underlying mechanisms for the response of the microbial community to afforestation, fundamental questions regarding how microorganisms cope with stoichiometric imbalance following afforestation are still unclear. Therefore, it is crucial to investigate microbial distribution and composition and the relationship among microorganisms and soil biotic and abiotic properties following afforestation.
In this study, we evaluated enzyme activities and microbial biomass, diversity, and composition with planting density in 5- and 35-year-old Cunninghamia lanceolata [Lamb.] Hook (Chinese fir) plantations, which account for 5% of all plantation areas worldwide (Ma et al., 2007). We examined the stoichiometry of soil, microbial biomass C (MBC), N (MBN), and P (MBP), and C (β-glucosidase, BG)-, N (leucine aminopeptidase, LAP, and β-1,4-N-acetylglucosaminidase, NAG)-, and P (acid phosphatase, AP)-acquiring enzymes to address two fundamental questions: How does planting density affect C/N/P stoichiometry with the development of plantations? And, how do microbial communities cope with stoichiometric imbalances? We hypothesized (i) that there may be stoichiometric imbalances between soil microorganisms and their demand for nutrients and that microorganisms would be restricted by P due to increasing depletion of P and competition intensity with planting density during the development from 5 to 35 years (Wang et al., 2021b); (ii) that microbial biomass and diversity would decrease with planting density due to increasing competitiveness of plants, and thus enzyme activity would also decrease; and (iii) that the stoichiometry of microbial biomass would negatively correlate with enzymatic stoichiometry since microorganisms optimize growth by allocating more reserves to acquire the most limited nutrient (Treseder and Vitousek, 2001).
This study was carried out in 5-year-old (5a) and 35-year-old (35a) Chinese fir plantations, located in Shanxi Forest Farm (27°44′ N, 114°39′ E) in Jiangxi Province and Weimin Forest Farm (27°05′ N, 117°43′ E) in Fujian Province, China, respectively (Supplementary Figure 1). The two study sites are located in the mid-subtropical region of China. The annual average temperature of the region is 16.8–19.4°C, and the annual average precipitation is 1,590–1,712 mm. The soil is typical Ferrosols developed from highly weathered granites (Huang et al., 2017). The site index, the average tree height of 20-year-old Chinese fir, of the two study sites was 16 and was recorded by the farms. This allows us to compare the differences in soil parameters of Chinese fir plantations between two stand ages.
The 5- and 35-year-old plantations were established with 1-year-old seedlings in a completely randomized block design with three blocks in 2013 and 1982, respectively. Each block consisted of five planting densities: 1,667 (D1), 3,333 (D2), 5,000 (D3), 6,667 (D4), and 10,000 stems ha−1 (D5), resulting in 15 plots at each age (30 plots in total). The area of each plot is 600 m2. The stand density, mean tree height, diameter at breast height (DBH), fine root biomass, and standing litter in each plot were measured in May 2017 (Table 1). The stand density of each plot is lower than the planting density because of the self-thinning rule rather than the artificially thinning.
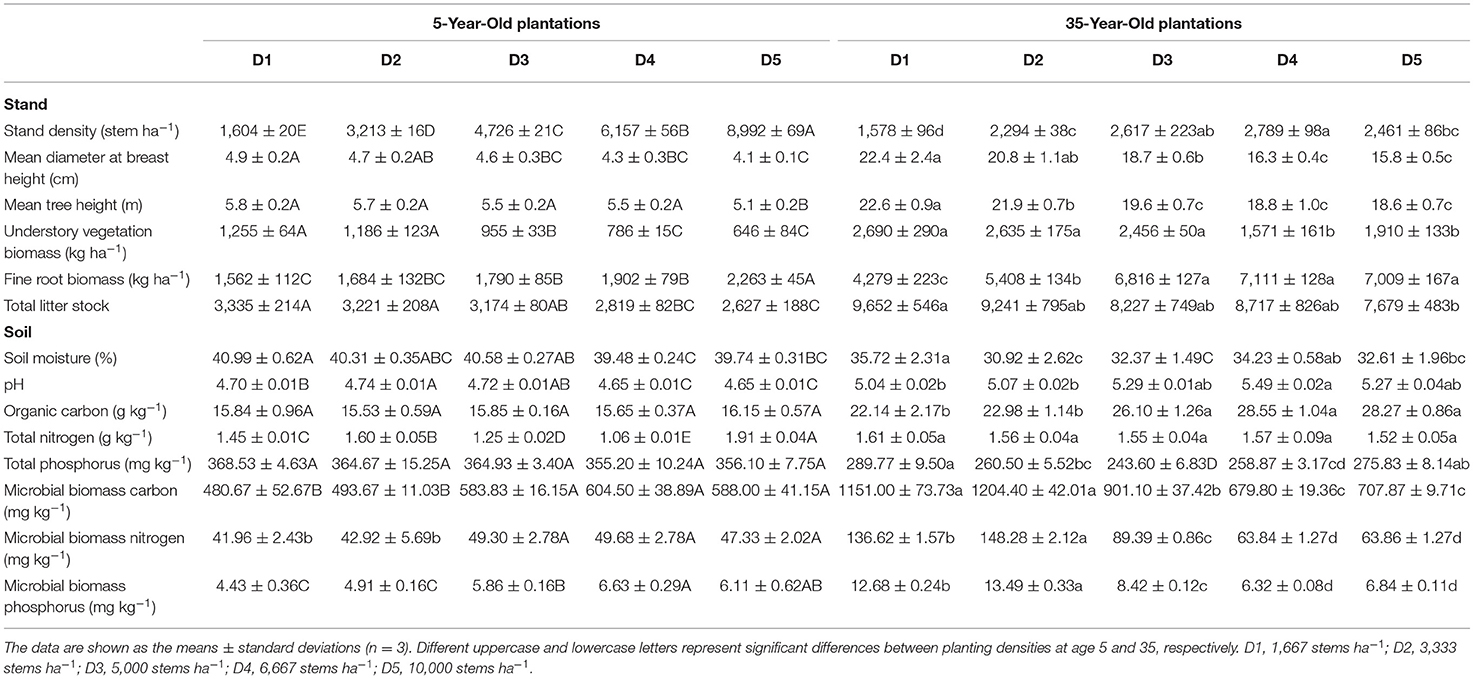
Table 1. Characteristics of the plots of different planting densities in 5- and 35-year-old plantations.
In May 2017, eight soil samples with a diameter of 5 cm were collected from 0 to 20 cm by the “S-sampling” method (Li, 2016) in each plot, combined, sieved through a 2-mm mesh, air-dried, and divided into three subsamples. One subsample was stored at −80°C before the analysis of DNA; one was kept at 4°C for the analysis of soil microbial biomass carbon (MBC), nitrogen (MBN), phosphorus (MBP), the activities of β-glucosidase (BG), leucine aminopeptidase (LAP), β-1,4-N-acetylglucosaminidase (NAG), and acid phosphatase (AP) and another was air-dried and ground to measure soil organic carbon (SOC), total nitrogen (TN), and total phosphorus (TP).
Soil moisture (SM) was measured by oven-drying samples at 105°C. Soil pH was measured in soil/water suspension (1:5). The SOC and TN contents were measured using the K2Cr2O7-H2SO4 oxidation method (Lefroy et al., 1993) and the Kjeldahl method (Bremner and Mulvaney, 1982), respectively. The TP content was measured after digestion with ammonium persulfate and sulfuric acid in an autoclave at 121°C on an ICP (Spectro Analytical Instruments, Spectro Arcos ICP, Kleve, Germany).
The MBC, MBN, and MBP contents were measured using the fumigation–extraction method (Brookes et al., 1984; Vance et al., 1987). The MBC and MBN contents were calculated according to the equations MBC = EC/kEC and MBN = EN/kEN, where kEC and kEN are equal to 0.45 and EC and EN are the difference between the content of organic C and N extracted from the fumigated and non-fumigated soils, respectively. The MBP content was calculated using a conversion factor of 0.40 and the recovery of an inorganic P spike by measuring the difference between the content of extractable P extracted from the fumigated and non-fumigated soils.
The BG and AP activities were assayed using a substrate of pNPG (4-nitrophenyl-β-D-glucopyranoside) (Geng et al., 2012) and p-nitrophenyl phosphate (Tabatabai, 1994), respectively. Further details of the procedure for BG and AP assays can be found in Wang et al. (2021a,b). The activities of LAP and NAG were measured by the method of Saiya-Cork et al. (2002). In brief, 1 g of soil was added to 9 ml of 50 mM sodium acetate buffer. The mixed solution was shaken well and then centrifuged at 4°C for 20 min (2,000 rpm). Finally, the supernatant was collected. Then, 10 μl of the supernatant was distributed to each well of a 96-well black microplate, and the reagents were added according to the kit instructions (Qingdao Jieshikang Biotechnology Co., Ltd., Qingdao, China). Microplates were incubated at 37°C for 60 min. Fluorescence at 450 nm was determined on a SpectraMax Paradigm Multi-Mode detection platform (Molecular Devices, San Jose, CA, USA). Soil C/N, C/P, and N/P acquisition ratios were calculated using BG/(LAP+NAG), BG/AP, and (LAP+NAG)/AP, respectively.
The total soil microbial genomic DNA was extracted by following the manufacturer's protocols of the Power Soil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA). The purity and quality of the extracted genomic DNA were checked by electrophoresis on 1% agarose gels. The PCR amplification of the bacterial V3–V4 region was conducted by using primers 338F (5′-GTACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GTGGACTACHVGGGTWTCTAAT-3′) (Zhang et al., 2015). The 5′ end of the forward and reverse primers each contained a 10-digit barcode sequence (provided by Allwegene Company, Beijing, China). The PCRs were carried out in 25 μl reaction volumes with 12.5 μl KAPA 2G Robust Hot Start Ready Mix, 1 μl forward primer (5 μM), 1 μl reverse primer (5 μM), 5 μl template DNA (30 ng in quality), and 5.5 μl H2O. Cycling parameters were 95°C for 5 min, followed by 28 cycles of 95°C for 45 s, 55°C for 50 s, and 72°C for 45 s with a final extension at 72°C for 10 min. Three PCR products per sample were pooled to mitigate reaction-level PCR biases. The fungal ITS-1 region was amplified by using primers ITS1F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS2 (5′-ATCCTCCGCTTATTGATATGC-3′) (White et al., 1990). The PCRs were conducted as follows: 4 μl 5 × FastPfu buffer, 1 μl forward primer (5 μM), 1 μl reverse primer (5 μM), 2 μl dNTP mixture (2.5 mM), 5 μl template DNA (30 ng in quality), and 10 μl H2O. Cycling parameters were 95°C for 2 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s with a final extension at 72°C for 5 min. Three separate PCR products per sample were pooled to mitigate reaction-level PCR biases.
The PCR products were purified using a QIAquick Gel Extraction Kit (QIAGEN, Germany), quantified by a real-time PCR, and sequenced on an Illumina MiSeq PE300 platform at Beijing Allwegene Tech, Ltd (Beijing, China). After the run, image analysis, base calling, and error estimation were performed using Illumina Analysis Pipeline Version 2.6 (Zhao et al., 2017). The erroneous and chimeric sequences were filtered out, and pair-end reads were initially trimmed using Illumina Analysis Pipeline Version 2.6 to remove low-quality sequences with a score below 20. Then, high-quality sequences were aligned to the Ribosomal Database Project (Cole et al., 2009) using QIIME (V1.7.0) (Caporaso et al., 2010) and were clustered into operational taxonomic units (OTUs) with a 97% similarity level using Uclust (Edgar, 2010). The taxonomy of each 16S rRNA and ITS gene sequence was analyzed by UCLUST against the Silva119 16S rRNA database using a confidence threshold of 90%. The initial sequencing data were submitted to the NCBI Sequence Read Archive (SRA) with the Bioproject of PRJNA811461. The Chao1 and Shannon diversity indexes were calculated using the QIIME in Python (v.1.8.0) (Sanner, 1999).
A one-way ANOVA, followed by the Tukey's HSD post-hoc test, was conducted to test the differences in enzyme activity, bacterial and fungal sequence and OUT numbers, microbial alpha diversity, the relative abundance of dominant microbial phyla, and soil, microbial, and enzymatic C/N/P stoichiometry among planting densities. The unary linear regression was conducted to identify the relationships among stoichiometry of soil, microbial, and enzymatic components. All statistical tests were conducted using SPSS (version 21, IBM, Armonk, NY, USA). Spearman's correlation analysis was performed to evaluate the relationships between enzyme activity and microbial alpha diversity (Chao1 and Shannon) and between microbial dominant phyla abundances and enzyme activities and soil physicochemical properties via one-way ANOVA in R (v.4.0.4). Redundancy analysis (RDA) was conducted to test the relative importance of soil physicochemical factors, microbial biomass C, N, and P, and soil and microbial C/N/P stoichiometry in explaining variations in microbial diversity using the “vegan” package in R. To eliminate multicollinearity, we selected explanatory variables using the variance inflation factor (VIF) lower than 10 (Tamura et al., 2019). Statistical analyses were tested using PERMANOVA based on 999 permutations in R.
At age 5, MBC, MBN, and MBP in high planting densities (>3,333 stem ha−1, in D3, D4, and D5) were higher by 10% to 50% than in low planting densities (<3,333 stem ha−1, in D1–D2) (Table 1). However, the MBC, MBN, and MBP were lower by 22 to 57% in high vs. low planting densities at age 35 (Table 1). The activities of four tested enzymes increased with planting density at age 5, while the opposite pattern was observed at age 35 (Figure 1).
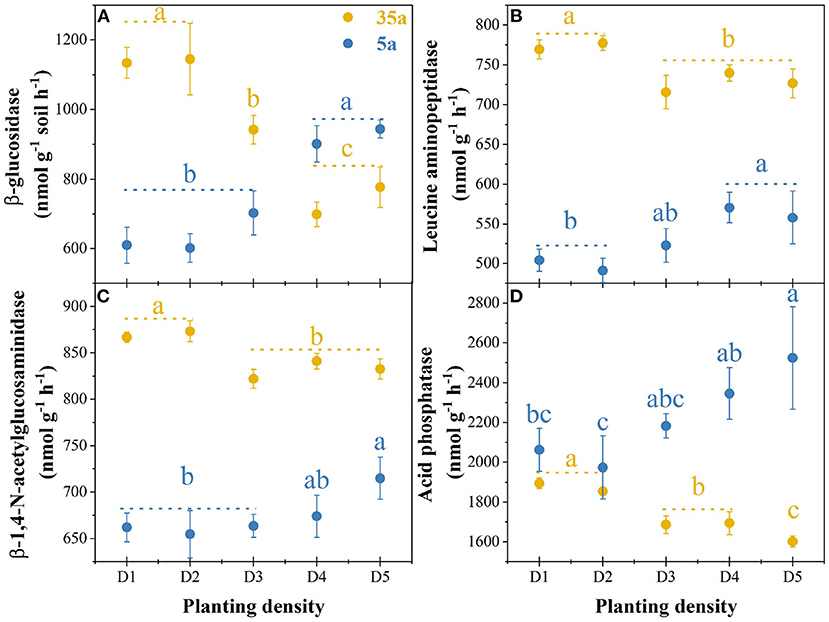
Figure 1. Enzyme activities in 5-year-old (5a, blue) and 35-year-old (35a, yellow) plantations. Different lowercase letters represent significant differences between different planting densities (p < 0.05). The data are shown as the mean ± standard deviations (n = 3). D1, 1,667 stems ha−1; D2, 3,333 stems ha−1; D3, 5,000 stems ha−1; D4, 6,667 stems ha−1; D5, 10,000 stems ha−1.
The SOC/TN ratio was higher in high vs. low planting densities at ages 5 and 35 (Figure 2A). The TN/TP ratio was higher in D1 and D2 than in D4 and D5 at age 5 (Figure 2C). The TN/TP ratio had the highest value in D3 and the lowest ones in D1 and D5 at age 35 (Figure 2C). At age 35, the SOC/TP ratio was higher in high vs. low planting densities (Figure 2B). The MBC/MBN ratio slightly increased with planting density at age 5, while MBC/MBP and MBN/MBP ratios were higher in D1, D2, and D3 vs. D4 and D5 (Figures 2D–F). At age 35, MBC/MBN and MBC/MBP ratios were higher in high vs. low planting densities, while MBN/MBP ratio was the opposite (Figures 2D–F). Finally, the opposite patterns for enzymatic C/N/P at ages 5 and 35 were observed (Figures 2G–I). At age 5, enzymatic C/N and C/P increased with planting density, while enzymatic N/P ratio decreased (Figures 2G–I).
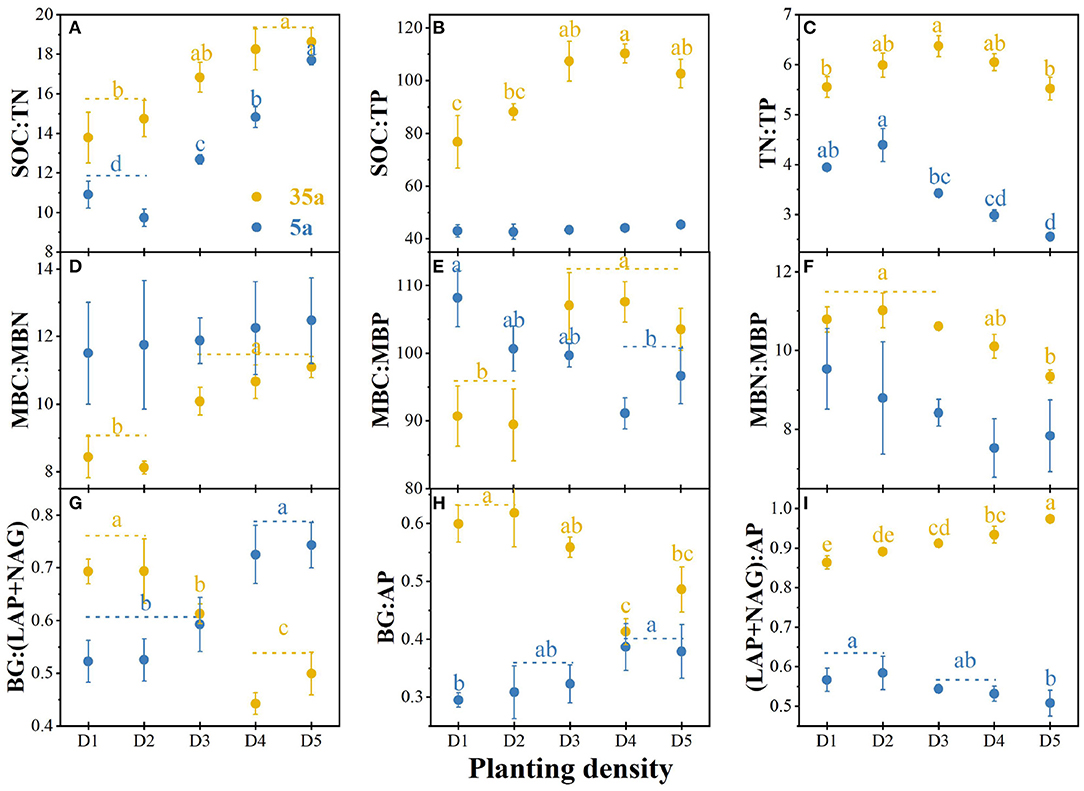
Figure 2. Soil carbon (C)/nitrogen (N)/phosphorus (P) stoichiometry, microbial biomass C/N/P stoichiometry, and enzymatic C/N/P stoichiometry in 5-year-old (5a, blue) and 35-year-old (35a, yellow) plantations. C acquisition is represented by β-glucosidase (BG); N acquisition is measured by the potential activities of leucine aminopeptidase (LAP) and β-1,4-N-acetylglucosaminidase (NAG); P acquisition is measured as acid phosphatase (AP) activity; MBC, microbial biomass carbon; MBN, microbial biomass nitrogen; MBP, microbial biomass phosphorus; SM, soil moisture; SOC, soil organic carbon; TN, total nitrogen; TP, total phosphorus. Different lowercase letters represent significant differences between different planting densities (p < 0.05). The data are shown as the mean ± standard deviations (n = 3). D1, 1,667 stems ha−1; D2, 3,333 stems ha−1; D3, 5,000 stems ha−1; D4, 6,667 stems ha−1; D5, 10,000 stems ha−1.
There were no differences in bacterial OTU number and Chao1 index among planting densities either in 5a or in 35a (Table 2, Supplementary Table 1). Fungal Shannon index and the ratio of fungal to bacterial Shannon increased with planting density at age 5 (Table 2). At age 35, fungal Chao1 and Shannon indexes decreased with planting density; moreover, the fungal-to-bacterial ratios of either Chao1 or Shannon index were higher in high vs. low planting densities (Table 2).
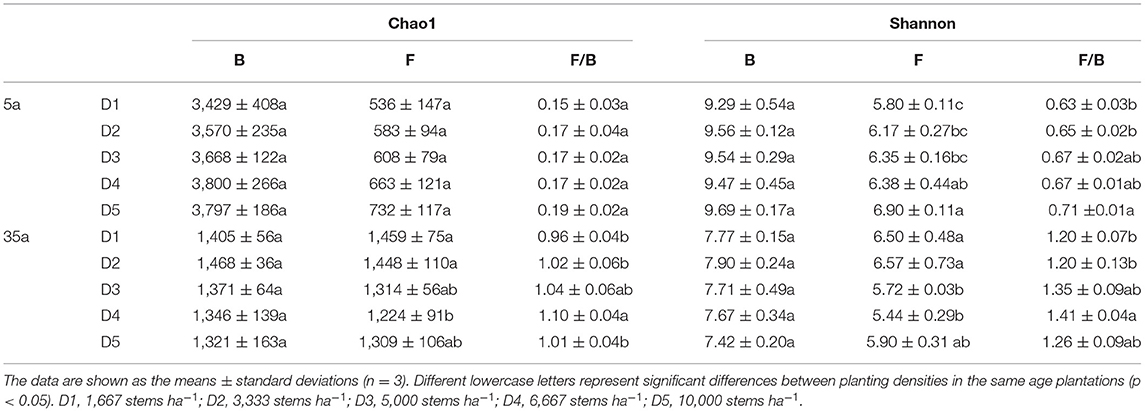
Table 2. Richness and diversity indices of bacterial (B) and fungal communities (F) in 5-year-old (5a) and 35-year-old (35a) Chinese fir plantations of different stand densities.
The dominant bacterial phyla (relative abundance > 1%) were as follows: Acidobacteria, Proteobacteria, Chloroflexi, Actinobacteria, Firmicutes, Gemmatimonadetes, Planctomycetes, and Verrucomicrobia at age 5 and Acidobacteria, Proteobacteria, Chloroflexi, Verrucomicrobia, Actinobacteria, Planctomycetes, Gemmatimonadetes, Nitrospirae, Elusimicrobia, and Bacteroidetes at age 35 (Figure 3A). The relative abundance of Acidobacteria, Firmicutes, and Gemmatimonadetes decreased with planting density at age 5, while Chloroflexi and Actinobacteria increased (Supplementary Table 2). No differences in the relative abundance of these phyla among five planting densities were observed at age 35.
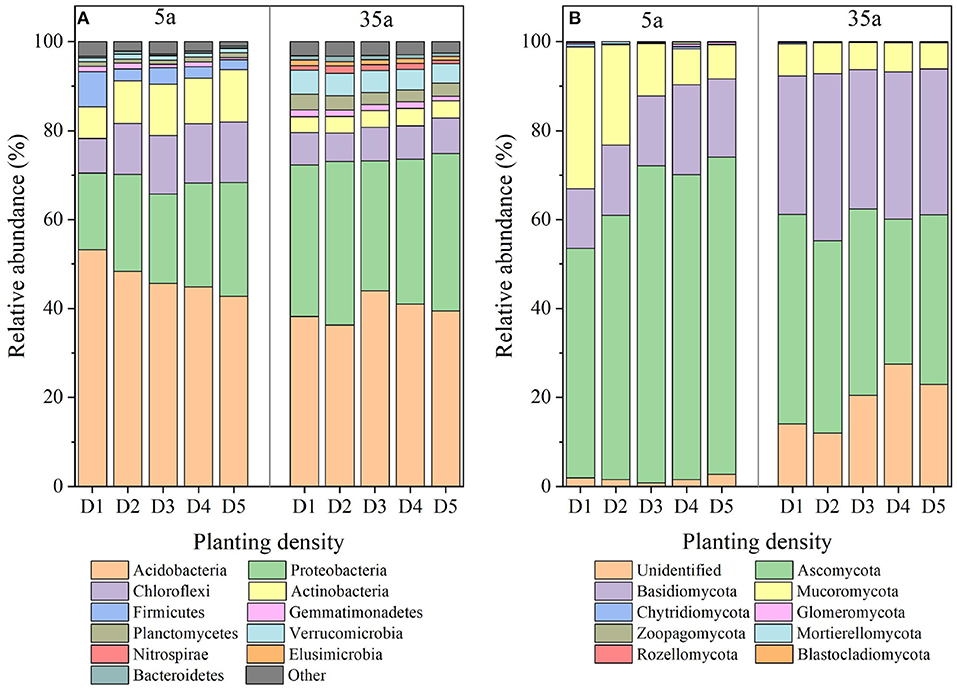
Figure 3. Bacterial (A) and fungal (B) community patterns in 5-year-old (left side) and 35-year-old (right side) plantations. D1, 1,667 stems ha−1; D2, 3,333 stems ha−1; D3, 5,000 stems ha−1; D4, 6,667 stems ha−1; D5, 10,000 stems ha−1.
The dominant fungal phyla at age 5 included Ascomycota, Basidiomycota, and Mucoromycota, with average contributions of 51.7–71.4, 13.3–20.1, and 7.6–31.9%, respectively (Figure 3B, left side). The relative abundance of Ascomycota and Basidiomycota increased with planting density at age 5 while Mucoromycota decreased (Supplementary Table 3). At age 35, the dominant fungal phyla included Ascomycota, Basidiomycota, and Mucoromycota with average contributions of 32.6–47.1, 31.1–37.5, and 5.9–7.2%, respectively (Figure 3B, right side). The relative abundance of Ascomycota in D4 was lower than that in D1, D2, and D3, and it was higher in D1 and D2 than in D5 (Supplementary Table 3). The relative abundance of Basidiomycota in D2 was higher than that in the other four planting densities (Supplementary Table 3).
At age 5, significant correlations were observed between TN/TP and MBN/MBP, between SOC/TN and BG/(LAP+NAG), between TN/TP and (LAP+NAG)/AP, between MBC/MBN and BG/(LAP+NAG), and between MBC/MBP and BG/AP (Figures 4C,D,F–H). The activities of BG, AP, and NAG were positively related to the ratio of fungal to bacterial Shannon index; moreover, positive relationships between AP and fungal Shannon index were observed at age 5 (Figure 5). At age 35, positive relationships between SOC/TN and MBC/MBN and between SOC/TP and MBC/MBP were found (Figures 4A,B). Negative relationships were found between SOC/TN and BG/(LAP+NAG), between SOC/TP and BG/AP, between MBC/MBN and BG/(LAP+NAG), between MBC/MBP and BG/AP, and between MBN/MBP and (LAP+NAG)/AP (Figures 4D,E,G–I). Moreover, BG, AP, LAP, and NAG were positively related to the fungal Chao1 index (Figure 5).
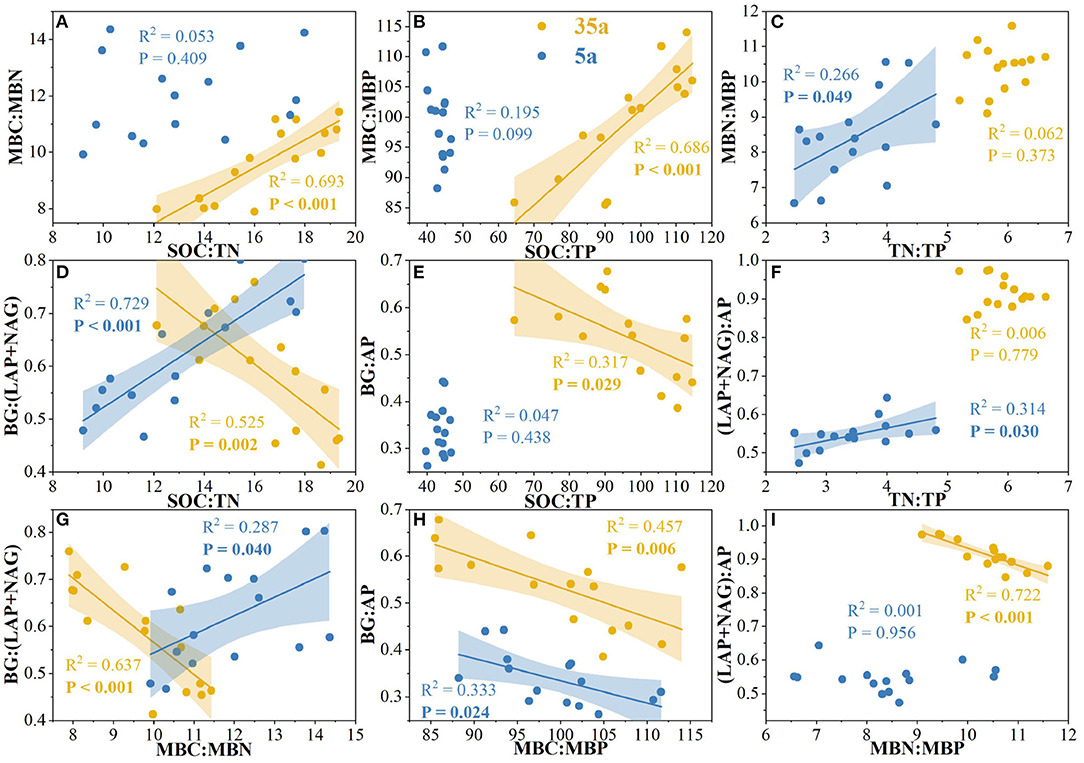
Figure 4. Relationships among soil carbon (C)/nitrogen (N)/phosphorus (P) stoichiometry, microbial biomass C/N/P stoichiometry, and enzymatic C/N/P stoichiometry in 5-year-old (5a, blue) and 35-year-old (35a, yellow) plantations. C acquisition is represented by β-glucosidase (BG); N acquisition is measured by the potential activities of leucine aminopeptidase (LAP) and β-1,4-N-acetylglucosaminidase (NAG); P acquisition is measured as acid phosphatase (AP) activity.
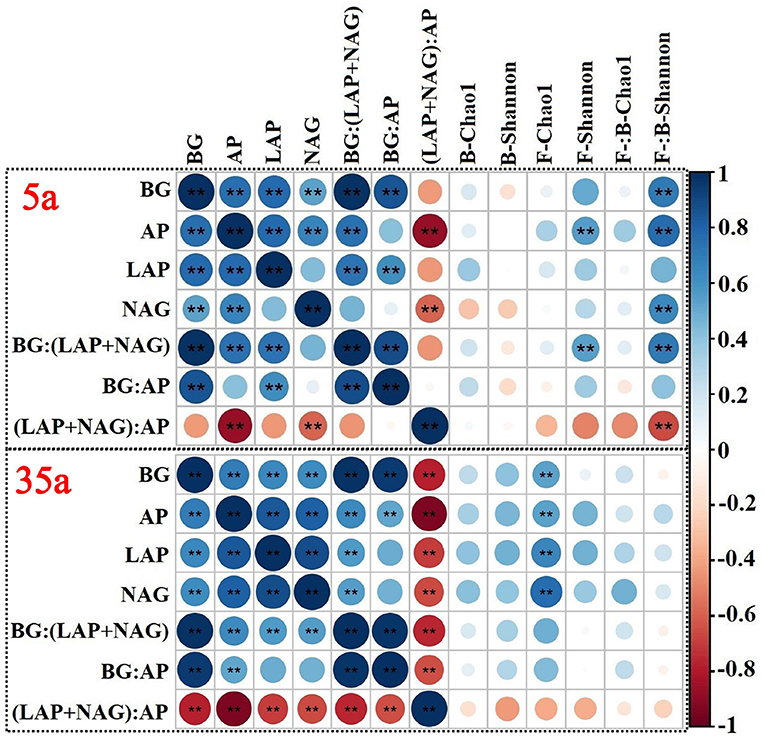
Figure 5. Spearman's rank correlation coefficients between enzyme activity and microbial alpha diversity (Chao1 and Shannon) in 5-year-old (top side) and 35-year-old (bottom side) plantations. BG, β-glucosidase; LAP, leucine aminopeptidase, NAG, β-1,4-N-acetylglucosaminidase; AP, acid phosphatase; B, bacteria; F, fungi.
At age 5, microbial dominant phyla abundances were mainly affected by SM, TN, TP, SOC/TN, and TN/TP; moreover, enzyme activities were strongly affected by the abundance of bacterial Acidobacteria, Chloroflexi, and Gemmatimonadetes and of fungal Ascomycota and Mucoromycota (Supplementary Figure 2, left side). In contrast, microbial dominant phyla abundances were mainly affected by pH, SOC, and SOC/TP; moreover, enzyme activities were strongly affected by the abundance of bacterial Chloroflexi and Planctomycetes and fungal Ascomycota (Supplementary Figure 2, right side).
Redundancy analysis (RDA) demonstrated that soil biotic and abiotic factors together accounted for 91.33 and 63.59% of the variation in bacterial (Figure 6A) and fungal communities (Figure 6B), respectively. RDA results revealed that strong positive relationships between microbial communities and SM, TP, and MBC/MBN and negative relationships between microbial communities and pH, TN, SOC/TN, and MBC were observed at age 5, while the opposite patterns were observed at age 35 (Figure 6, Supplementary Table 4).
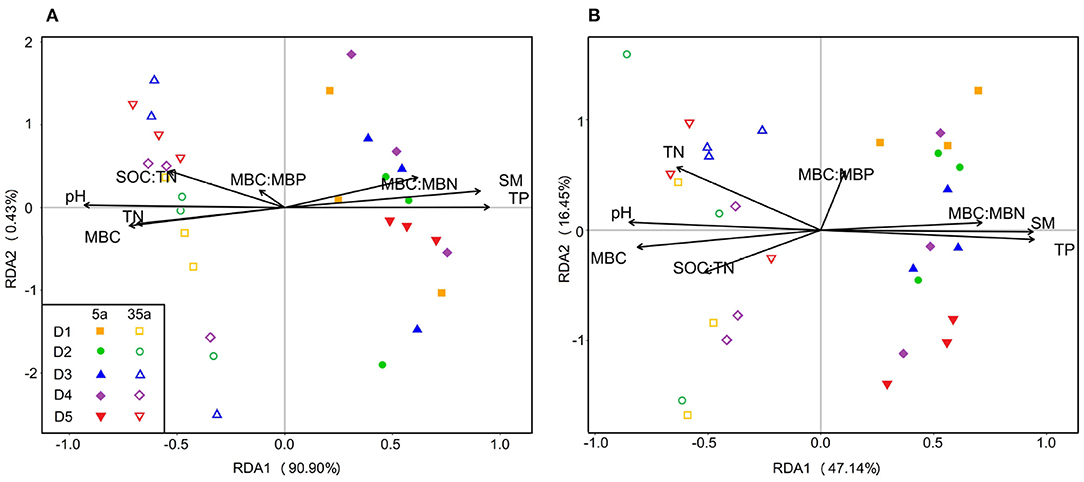
Figure 6. Ordination plots of the results from the redundancy analysis (RDA) to identify the relationships between bacterial (A) or fungal (B) communities and microbial biomass and soil physicochemical properties (black arrows) in 5-year-old (closed symbols) and 35-year-old (open symbols) plantations. BG, β-glucosidase; LAP, leucine aminopeptidase, NAG, β-1,4-N-acetylglucosaminidase; AP, acid phosphatase; MBC, microbial biomass carbon; MBN, microbial biomass nitrogen; MBP, microbial biomass phosphorus; SM, soil moisture; SOC, soil organic carbon; TN, total nitrogen; TP, total phosphorus; B, bacteria; F, fungi.
Our results demonstrated that soil, microbial, and enzymatic C/N/P stoichiometry differed among planting densities with the development of plantations from 5 to 35 years (Figure 2), thereby supporting our first hypothesis that there are stoichiometric imbalances between microbial biomass and their demand for nutrients with planting density during the development of plantations from 5 to 35 years. Enzymatic C/N and C/P ratios increased with planting density at age 5; moreover, enzymatic N/P ratio decreased with planting (Figures 2G–I). The opposite pattern was observed at age 35. This demonstrates that soil microorganisms provide more resources and energy for soil N-acquiring enzyme production other than P-acquiring enzymes with planting density during the development of plantations, indicating the shift from P- to N-limitation for microorganisms from 5 to 35 years. This was supported by the N-limitation of Chinese fir plantations after 12–22 years of development (Tong et al., 2020). Therefore, our first hypothesis regarding the P-limitation on microorganisms with planting density and stand age was only supported at age 5.
Soil eco-enzymatic stoichiometry was affected by plant residues since variations in plant tissue stoichiometry (Martiny et al., 2006). Therefore, the N-limitation could be attributed to increased input of coniferous litters with a higher C/N ratio stocks in high-density plantations (Table 1; Shao et al., 2017; Tong et al., 2021). The microbial growth rate is generally higher in soils with a lower soil C/N ratio, and microorganisms would release less C through respiration (Zhao et al., 2018). Conversely, microorganisms would convert more C to their biomass in soils with a higher C/N ratio (Schimel, 2016; Shoemaker et al., 2017). Importantly, this is supported by the negative correlation between MBC/MBN and BG/(LAP+NAG) with a planting density at age 35 (Figure 4). Another factor of N-limitation of microorganisms is that enzyme production itself requires investment N (Schimel and Weintraub, 2003). Moreover, the C/N ratio of proteins is usually much lower than the C/N of microbial biomass (Schimel and Weintraub, 2003). In addition, the greater amounts of fine root biomass in high-density plantations (Table 1) likely provided more resources to microorganisms, by increasing soil C contents (Creamer et al., 2014; Zhao et al., 2018). Accordingly, to acclimate to the N-limitation, microorganisms tend to secrete more N-acquiring enzymes than C-acquiring enzymes (Sinsabaugh et al., 2013; Rosinger et al., 2019; Li et al., 2020), thereby resulting in a decrease in enzymatic C/N and an increase in enzymatic N/P with planting density during the development of plantations from 5 to 35 years (Figure 2).
Microbial biomass and enzyme activity increased with planting density in 5-year-old plantations, while the opposite pattern was observed in 35-year-old plantations (Figure 1, Table 1). Therefore, the second hypothesis that microbial biomass and enzyme activity would decrease with planting density was only supported at age 35. These results are in line with the findings reported from C. lanceolata plantations (Wu et al., 2011; Wang et al., 2018, 2019) and Platycladus orientalis plantations (Wang and Shi, 2016). Therefore, microbial growth and activity were limited by increasing planting density with the development of plantations. The microbial metabolism theory (Sinsabaugh et al., 2008) proposed that soil microorganisms adopted a survival strategy that is increasing enzyme activity in the sufficient resource and decreasing enzyme activity in the deficient resource. At age 5, the intensity of competition between microorganisms and plants was weak, and the bioavailability of soil nutrients to microorganisms was relatively high. Moreover, fine root biomass increased with planting density (Table 1). Accordingly, the microbial enzymatic synthesis capacity was strong, which ultimately promoted soil enzyme activity (Li et al., 2020). By contrast, at age 35, plants grew with large roots and occupied an absolute competitive advantage (Yang et al., 2020), and the depletion of nutrients (especially N and P) increased with planting density (Wang et al., 2021b). Therefore, the availability of nutrients cannot meet microbial demands, which ultimately limits soil enzyme activity.
Soil enzyme activity is mainly affected by microorganisms and plant roots (Gu et al., 2009). Increased root biomass is expected to stimulate root exudates into soils, promoting microbial activity (Castorena et al., 2016; Fraser et al., 2017). If so, high planting densities with higher fine root biomass would increase enzyme activity. Interestingly, the result was the opposite in 35-year-old plantations, suggesting that soil microorganisms contributed much more to enzyme activity than plant roots. The decrease in enzyme activity with planting density in 35-year-old plantations could be partially by decreasing biomass of understory vegetation (Table 1; Rodriguez-Loinaz et al., 2008; Wang et al., 2021a). As the main driving factor for plant litter decomposition, soil enzymes produced by microorganisms strongly depend on the species and stoichiometry of the plant litter (Cleveland and Liptzin, 2007; Baker and Allison, 2017; Rosinger et al., 2019). From this perspective, soil microbial biomass and enzyme activity should be higher in plantations with a higher amount and diversity of understory vegetation. Importantly, such a pattern was observed in 35-year-old plantations.
Negative relationships between MBC/MBN and BG/(LAP+NAG), between MBC/MBP and BG/AP, and between MBN/MBP and (LAP+NAG)/AP (Figure 4) supported the third hypothesis. Soil C/N and C/P ratios were positively correlated with MBC/MBN and MBC/MBP ratios at age 35 (Figure 4), indicating that microorganisms adjusted their biomass C/N/P ratios to meet the nutrient stoichiometry of their substrates with the development of plantations (Mooshammer et al., 2014). This can be explained by the central concept of stoichiometric invariance (homeostasis) in ecological stoichiometry (Sterner and Elser, 2002). This concept proposed that non-homeostatic organisms would change their biomass C/N/P in response to the variation in soil C/N/P ratios with two potential mechanisms: (i) microbial storage of excess elements, resulting in a convergence between the biomass and resource stoichiometries, and (ii) changes in microbial community structure, resulting in shifts in biomass stoichiometry (Mooshammer et al., 2014). Essentially, the first mechanism seems to be confirmed by positive relationships between SOC/TN and MBC/MBN and between SOC/TP and MBC/MBP at age 35 (Figure 4), indicating a physiological adjustment of non-homeostatic microorganisms' stoichiometry. Moreover, our studies demonstrated changes in microbial community composition, and the ratio of fungal to bacterial diversity increased with planting density and stand age (Figure 3, Table 2). Accordingly, microorganisms may adopt non-homeostatic behaviors to reduce the stoichiometric imbalance to their resources with planting density during the development of plantations from 5 to 35 years.
We found that changes in soil bacterial and fungal compositions were mainly dependent on limited nutrients as observed in microbial community abundances positively related to TP at age 5 but related to TN and SOC/TN at age 35 (Supplementary Table 4). This again demonstrates the shift from P- to N-limitation for microorganisms from 5 to 35 years. To cope with the shift from P- to N-limitation, microorganisms changed the relative abundance of dominant phyla. For example, Proteobacteria, fast-growing and dominant bacteria, require higher P to synthesize a large amount of RNA to achieve a high rate of cell division (Elser et al., 2003; El Zahar et al., 2008). Moreover, the compositions of Proteobacteria, such as Rhizobiales, are considered to be rhizosphere plant-promoting bacteria (Jorquera et al., 2014) and can fix atmospheric nitrogen in symbiosis with plants (Van der Heijden et al., 2008). The same pattern was observed for Nitrospirae that can promote nitrogen accumulation (DeLuca et al., 2002). Thus, such an increase in the abundance of Proteobacteria and Nitrospirae should be an effective way to cope with the shift from P- to N-limitation of microorganisms with stand age. Remarkably, the fungal-to-bacterial ratios of either Chao1 (0.15–0.19 and 0.96–1.10 at age 5 and 35, respectively), or Shannon (0.63–0.71 and 1.20–1.41) index increased with stand age (Table 2). This was supported by the observed decreased fungal-to-bacterial ratio with inorganic nitrogen fertilization (Bloem and Vos, 2004). As compared to bacteria, the ability of fungi to degrade more recalcitrant litter, such as cellulose and lignin, in coniferous forests is stronger (Lauber et al., 2008). Moreover, the coniferous litter with a higher C/N ratio was increased with planting density and stand age (Table 1). Thus, an increased fungal-to-bacterial ratio may be another important pathway to cope with the shift from P- to N-limitation of microorganisms.
Finally, no significant differences in the relative abundance of the predominant bacteria were found among different planting densities at age 35, suggesting that bacterial communities strongly resist the change in planting density. This is supported by the fact that bacterial communities do not respond to plant–soil feedback, while the responses of fungal communities to plant–soil feedback are more remarkable ((Kardol et al., 2007; Lorenzo et al., 2010)). This is mainly because bacterial communities have a small-scale niche in the soil and are less symbiotic with the plant as compared to fungal communities (Sun et al., 2017). Therefore, the responses of bacterial communities to soil property changes caused directly or indirectly by planting density are weaker.
Microbial biomass, microbial diversity, and C-, N-, and P-acquiring enzyme activities increased with planting density at age 5, but the opposite pattern was observed at age 35. Contrasting patterns of stoichiometric imbalances between soil microorganisms and their demand for nutrients with planting density at ages 5 and 35 indicated changes in microbial nutrient limitation. Moreover, our study demonstrated that soil microbial nutrient limitation shifted from P- to N-limitation with planting density during the development of plantations from 5 to 35 years. Soil microorganisms may cope with the imbalance of stoichiometry by: (i) coordinating the adjustment of the stoichiometry of microorganisms and enzymes; (ii) changing the relative abundance of dominant microbial phyla; and (iii) increasing the fungal-to-bacterial diversity ratio. These findings support the ecological theory that microorganisms optimally allocate their resources when resources are limited. Altogether, our findings highlight that (i) stoichiometric imbalances are the main driver in shaping soil microbial communities and (ii) enzymatic stoichiometry can identify and explain microbial nutrient limitations and provide deeper insights for predicting nutrient bioavailability to microorganisms and modeling microbially mediated nutrient cycling in terrestrial ecosystems.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI GenBank; PRJNA811461.
CW: conceptualization, methodology, validation, formal analysis, data curation, writing-original draft preparation, writing-review and editing, and visualization. RJ: conceptualization, methodology, validation, formal analysis, resources, writing-review and editing, supervision, and funding acquisition. Both authors contributed to the article and approved the submitted version.
This study was supported by the State Key Research Development Program of China (2016YFD0600302).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.870667/full#supplementary-material
Aanderud, Z. T., Sabrina, S., Ball, B. A., Wall, D. H., Barrett, J. E., and Muscarella, M. E. (2018). Stoichiometric shifts in soil C:N:P promote bacterial taxa dominance, maintain biodiversity, and deconstruct community assemblages. Front. Microbiol. 9, 1401. doi: 10.3389/fmicb.2018.01401
Baker, N. R., and Allison, S. D. (2017). Extracellular enzyme kinetics and thermodynamics along a climate gradient in southern California. Soil Biol. Biochem. 114, 82–92. doi: 10.1016/j.soilbio.2017.07.005
Bell, C., Carrillo, Y., Boot, C. M., Rocca, J. D., Pendall, E., and Wallenstein, M. D. (2013). Rhizosphere stoichiometry: are C:N:P ratios of plants, soils, and enzymes conserved at the plant species-level? New Phytol. 201, 505–517. doi: 10.1111/nph.12531
Bloem, J., and Vos, A. (2004). “Fluorescent staining of microbes for total direct counts,” in Microbial Ecology Manual, ed Kowalchuk, G (Dordrecht: Kluwer Academic Publishers).
Bremer, L. L., and Farley, K. A. (2010). Does plantation forestry restore biodiversity or create green deserts? A synthesis of the effects of land-use transitions on plant species richness. Biodivers. Conserv. 19, 3893–3915. doi: 10.1007/s10531-010-9936-4
Bremner, J. M., and Mulvaney, C. S. (1982). “Nitrogen—total 1,” in Methods of soil analysis. Part 2. Chemical and Microbiological Properties, 595–624. doi: 10.2134/agronmonogr9.2.2ed.c31
Brockerhoff, E. G., Ecroyd, C. E., Leckie, A. C., and Kimberley, M. O. (2003). Diversity and succession of adventive and indigenous vascular understorey plants in Pinus radiata plantation forests in New Zealand. For. Ecol. Manag. 185, 307–326. doi: 10.1016/S0378-1127(03)00227-5
Brookes, P. C., Powlson, D. S., and Jenkinson, D. S. (1984). Phosphorus in the soil microbial biomass. Soil Biol. Biochem. 16, 169–175. doi: 10.1016/0038-0717(84)90108-1
Burns, R. G., DeForest, J. L., Marxsen, J., Sinsabaugh, R. L., Stromberger, M. E., Wallenstein, M. D., et al. (2013). Soil enzymes in a changing environment: current knowledge and future directions. Soil Biol. Biochem. 58, 216–234. doi: 10.1016/j.soilbio.2012.11.009
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., and Costello, E. K. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Castorena, E. V. G., Gutierrez-Castorena, M. D., Vargas, T. G., Bontemps, L. C., Martinez, J. D., Mendez, E. S., et al. (2016). Micromapping of microbial hotspots and biofilms from different crops using digital image mosaics of soil thin sections. Geoderma 279, 11–21. doi: 10.1016/j.geoderma.2016.05.017
Cleveland, C., and Liptzin, D. (2007). C:N:P stoichiometry in soil: is there a “redfield ratio” for the microbial biomass? Biogeochemistry 85, 235–252. doi: 10.1007/s10533-007-9132-0
Cole, S. E., LaRiviere, F. J., Merrikh, C. N., and Moore, M. J. (2009). A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol. Cell 34, 440–450. doi: 10.1016/j.molcel.2009.04.017
Creamer, C. A., Jones, D. L., Baldock, J. A., and Farrell, M. (2014). Stoichiometric controls upon low molecular weight carbon decomposition. Soil Biol. Biochem. 79, 50–56. doi: 10.1016/j.soilbio.2014.08.019
DeLuca, T. H., Zackrisson, O., Nilsson, M. C., and Sellstedt, A. (2002). Quantifying nitrogen-fixation in feather moss carpets of boreal forests. Nature 419, 917–920. doi: 10.1038/nature01051
Deng, Q., Cheng, X. X., Hui, D. F., Zhang, Q., Li, M., and Zhang, Q. F. (2016). Soil microbial community and its interaction with soil carbon and nitrogen dynamics following afforestation in central China. Sci. Total Environ. 541, 230–237. doi: 10.1016/j.scitotenv.2015.09.080
Edgar, R. C.. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
El Zahar, H. F., Marol, C., Berge, O., Rangel-Castro, J. I., Prosser, J. I., Balesdent, J., et al. (2008). Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2, 1221–1230. doi: 10.1038/ismej.2008.80
Elser, J. J., Acharya, K., Kyle, M., Cotner, J., Makino, W., Markow, T., et al. (2003). Growth rate-stoichiometry couplings in diverse biota. Ecol. Lett. 6, 936–943. doi: 10.1046/j.1461-0248.2003.00518.x
Elser, J. J., Sterner, R. W., Gorokhova, E., Fagan, W. F., Markow, T. A., and Cotner, J. B. (2000). Biological stoichiometry from genes to ecosystems. Ecol. Lett. 3, 540–550. doi: 10.1046/j.1461-0248.2000.00185.x
Fraser, T. D., Lynch, D. H., Gaiero, J., Khosla, K., and Dunfield, K. E. (2017). Quantification of bacterial non-specific acid (phoC) and alkaline (phoD) phosphatase genes in bulk and rhizosphere soil from organically managed soybean fields. Appl. Soil Ecol. 111, 48–56. doi: 10.1016/j.apsoil.2016.11.013
Ganie, M. A., Mukhtar, M., Dar, M. A., and Ramzan, S. (2016). Soil microbiological activity and carbon dynamics in the current climate change scenarios: a review. Pedosphere 26, 577–591. doi: 10.1016/S1002-0160(15)60068-6
Geng, Y., Dighton, J., and Gray, D. (2012). The effects of thinning and soil disturbance on enzyme activities under pitch pine soil in New Jersey Pinelands. Appl. Soil Ecol. 62, 1–7. doi: 10.1016/j.apsoil.2012.07.001
Gu, Y., Wang, P., and Kong, C. H. (2009). Urease, invertase, dehydrogenase and poly-phenoloxidase activities in paddy soil influenced by allelopathic rice variety. Eur. J. Soil Biol. 45, 436–441. doi: 10.1016/j.ejsobi.2009.06.003
Güsewell, S., and Freeman, C. (2003). Enzyme activity during N- and P-limited decomposition of wetland plant litter. Bull. Geobotan. Ins. ETH 69, 95–106.
Hillebrand, H., and Kahler, M. (2005). Low algal carbon content and its effect on the C:P stoichiometry. Freshw. Biol. 50, 1800–1807. doi: 10.1111/j.1365-2427.2005.01449.x
Huang, J., Ebach, M. C., and Triantafilis, J. (2017). Cladistic analysis of Chinese soil taxonomy. Geoderma Reg. 10, 11–20. doi: 10.1016/j.geodrs.2017.03.001
Jorquera, M. A., Inostroza, N. G., Lagos, L. M., Barra, P. J., Marileo, L. G., Rilling, J. I., et al. (2014). Bacterial community structure and detection of putative plant growth-promoting rhizobacteria associated with plants grown in Chilean agro-ecosystems and undisturbed ecosystems. Biol. Fert. Soils 50, 1141–1153. doi: 10.1007/s00374-014-0935-6
Kardol, P., Cornips, N. J., van Kempen, M. M. L., Bakx-Schotman, J. M. T., and van der Putten, W. H. (2007). Microbe-mediated plant-soil feedback causes historical contingency effects in plant community assembly. Ecol. Monog. 77, 147–162.
Lauber, C. L., Strickland, M. S., Bradford, M. A., and Fierer, N. (2008). The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 20, 2407–2415. doi: 10.1016/j.soilbio.2008.05.021
Lefroy, R. D. B., Blair, G. J., and Strong, W. M. (1993). Changes in soil organic matter with cropping as measured by organic carbon fractions and 13C natural isotope abundance. Plant Soil 155–156, 399–402. doi: 10.1007/BF00025067
Li, A., Fahey, T. J., Pawlowska, T. E., Fisk, M. C., and Burtis, J. (2015). Fine root decomposition, nutrient mobilization and fungal communities in a pine forest ecosystem. Soil Biol. Biochem. 83, 76–83. doi: 10.1016/j.soilbio.2015.01.019
Li, Q., Liu, Y., Gu, Y., Guo, L., Huang, Y., Zhang, J., et al. (2020). Ecoenzymatic stoichiometry and microbial nutrient limitations in rhizosphere soil along the Hailuogou Glacier forefield chronosequence. Sci. Total Environ. 704, 135413. doi: 10.1016/j.scitotenv.2019.135413
Li, Z. J.. (2016). Study on Soil Spatial Heterogeneity and Sampling Methods in Karst Region. Kunming: Guizhou University.
Lorenzo, P., Rodriguez-Echeverria, S., Gonzalez, L., and Freitas, H. (2010). Effect of invasive Acacia dealbata Link on soil microorganisms as determined by PCR-DGGE. Appl. Soil Ecol. 44, 245–251.
Ma, X., Heal, K. V., Liu, A., and Jarvis, P. G. (2007). Nutrient cycling and distribution in different-aged plantations of Chinese fir in southern China. For. Ecol. Manag. 243, 61–74. doi: 10.1016/j.foreco.2007.02.018
Marichal, R., Mathieu, J., Couteaux, M. M., Mora, P., Roy, J., and Lavelle, P. (2011). Earthworm and microbe response to litter and soils of tropical forest plantations with contrasting C:N:P stoichiometric ratios. Soil Biol. Biochem. 43, 1528–1535. doi: 10.1016/j.soilbio.2011.04.001
Martiny, J. B., Bohannan, B. J., Brown, J. H., Colwell, R. K., Fuhrman, J. A., Green, J. L., et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112. doi: 10.1038/nrmicro1341
Mooshammer, M., Wanek, W., Zechmeister-Boltenstern, S., and Richter, A. (2014). Stoichiometric imbalances between terrestrial decomposer communities and their resources: mechanisms and implications of microbial adaptations to their resources. Front. Microbiol. 5, 22. doi: 10.3389/fmicb.2014.00022
Phillips, R. P., Finzi, A. C., and Bernhardt, E. S. (2011). Enhanced root exudation induces microbial feedbacks to N cycling in a pine forest under long-term CO2 fumigation. Ecol. Lett. 14, 187–194. doi: 10.1111/j.1461-0248.2010.01570.x
Ren, C. J., Chen, J., Deng, J., Zhao, F. Z., Han, X. H., Yang, G. H., et al. (2017). Response of microbial diversity to C:N:P stoichiometry in fine root and microbial biomass following afforestation. Biol. Fert. Soils 53, 1–12. doi: 10.1007/s00374-017-1197-x
Rodriguez-Loinaz, G., Onaindia, M., Amezaga, I., Mijangos, I., and Garbisu, C. (2008). Relationship between vegetation diversity and soil functional diversity in native mixed-oak forests. Soil Biol. Biochem. 40, 49–60. doi: 10.1016/j.soilbio.2007.04.015
Rosinger, C., Rousk, J., and Sandén, H. (2019). Can enzymatic stoichiometry be used to determine growth-limiting nutrients for microorganisms?-A critical assessment in two subtropical soils. Soil Biol. Biochem. 128, 115–126. doi: 10.1016/j.soilbio.2018.10.011
Saiya-Cork, K. R., Sinsabaugh, R. L., and Zakb, D. R. (2002). The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol. Biochem. 34, 1309–1315. doi: 10.1016/S0038-0717(02)00074-3
Sanner, M. F.. (1999). Python: a programming language for software integration and development. J. Mol. Graph. Model. 17, 57–61.
Schimel, J.. (2016). Microbial ecology: linking omics to biogeochemistry. Nat. Microbiol. 1, 15028. doi: 10.1038/nmicrobiol.2015.28
Schimel, J. P., and Weintraub, M. N. (2003). The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: a theoretical model. Soil Biol. Biochem. 35, 549–563. doi: 10.1016/S0038-0717(03)00015-4
Shao, Y. N., Liu, Y. K., Li, Y. H., Chen, Y., and Tian, S. Y. (2017). Soil nutrient characteristics in Larix olgensis plantation with different stand densities. J. Cent. South Uni. For. Technol. 37, 27–31.
Shoemaker, W. R., Locey, K. J., and Lennon, J. T. (2017). A macroecological theory of microbial biodiversity. Nat. Ecol. Evol. 1, 1–6. doi: 10.1038/s41559-017-0107
Sinsabaugh, R. L., Hill, B. H., and Follstad Shah, J. J. (2009). Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature 462, 795–798. doi: 10.1038/nature08632
Sinsabaugh, R. L., Lauber, C. L., Weintraub, M. N., Ahmed, B., Allison, S. D., Crenshaw, C., et al. (2008). Stoichiometry of soil enzyme activity at global scale. Ecol. Lett. 11, 1252–1264. doi: 10.1111/j.1461-0248.2008.01245.x
Sinsabaugh, R. L., Manzoni, S., Moorhead, D. L., and Richter, A. (2013). Carbon use efficiency of microbial communities: stoichiometry, methodology and modelling. Ecol. Lett. 16, 930–939. doi: 10.1111/ele.12113
Sterner, R. W., and Elser, J. J. (2002). Ecological Stoichiometry: The Biology of Elements From Molecules to the Biosphere. Princeton, NJ: Princeton University Press. doi: 10.1515/9781400885695
Sun, S., Li, S., Avera, B. N., Strahm, B. D., and Badgley, B. D. (2017). Soil bacterial and fungal communities show distinct recovery patterns during forest ecosystem restoration. Appl. Environ. Microb. 83, e00966–e1017.
Tabatabai, M.. (1994). “Soil enzymes,” in Methods of Soil Analysis, ed J. Bigham (Madison, WI: SSSA), 775–834. doi: 10.2136/sssabookser5.2.c37
Tamura, R., Kobayashi, K., Takano, Y., Miyashiro, R., Nakata, K., and Matsui, T. (2019). Mixed integer quadratic optimization formulations for eliminating multicollinearity based on variance inflation factor. J. Glob. Optim. 73, 431–446. doi: 10.1007/s10898-018-0713-3
Tessier, J. T., and Raynal, D. J. (2003). Use of nitrogen to phosphorus ratios in plant tissue as an indicator of nutrient limitation and nitrogen saturation. J. Appl. Ecol. 40, 523–534. doi: 10.1046/j.1365-2664.2003.00820.x
Tong, R., Zhou, B., Jiang, L., Ge, X., and Cao, Y. (2020). The growth of Chinese fir is limited by nitrogen: evidences from N:P ratio, N or P variability and NuRE based on a regional investigation. For. Ecol. Manag. 460, 117905. doi: 10.1016/j.foreco.2020.117905
Tong, R., Zhou, B., Jiang, L., Ge, X., and Shi, J. (2021). Leaf litter carbon, nitrogen and phosphorus stoichiometry of Chinese fir (Cunninghamia lanceolata) across china. Glob. Ecol. Conserv. 27, e01542. doi: 10.1016/j.gecco.2021.e01542
Treseder, K. K., and Vitousek, P. M. (2001). Effects of soil nutrient availability on investment in acquisition of N and P in Hawaiian rain forests. Ecology 82, 946–954. doi: 10.1890/0012-9658(2001)082[0946:EOSNAO]2.0.CO;2
Van der Heijden, M. G. A., Bardgett, R. D., and van Straalen, N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
Vance, E., Brookes, P., and Jenkinson, D. (1987). An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 19, 703–707. doi: 10.1016/0038-0717(87)90052-6
Vries, F., Hoffland, E., Eekeren, N. V., Brussaard, L., and Bloem, J. (2006). Fungal/bacterial ratios in grasslands with contrasting nitrogen management. Soil Biol. Biochem. 38, 2092–2103. doi: 10.1016/j.soilbio.2006.01.008
Wang, C. Q., Xue, L., Dong, Y. H., Hou, L. Y., Wei, Y. H., and Jiao, R. Z. (2019). Responses of soil microbial community structure to stand densities of Chinese fir plantations. J. For. Res. 24, 162–167. doi: 10.1080/13416979.2019.1601652
Wang, C. Q., Xue, L., Dong, Y. H., and Jiao, R. Z. (2020). Effects of stand density on soil microbial community composition and enzyme activities in subtropical Cunninghamia lanceolate (Lamb.) hook plantations. For. Ecol. Manag. 479, 118559. doi: 10.1016/j.foreco.2020.118559
Wang, C. Q., Xue, L., Dong, Y. H., and Jiao, R. Z. (2021a). Soil organic carbon fractions, C-cycling hydrolytic enzymes, and microbial carbon metabolism in Chinese fir plantations. Sci. Total Environ. 758, 143695. doi: 10.1016/j.scitotenv.2020.143695
Wang, C. Q., Xue, L., Dong, Y. H., Wei, Y. H., and Jiao, R. Z. (2018). Unravelling the functional diversity of the soil microbial community of Chinese fir plantations of different densities. Forests 9, 532. doi: 10.3390/f9090532
Wang, C. Q., Xue, L., and Jiao, R. Z. (2021b). Soil phosphorus fractions, phosphatase activity, and the abundance of phoC and phoD genes vary with planting density in subtropical Chinese fir plantations. Soil Till. Res. 209, 104946. doi: 10.1016/j.still.2021.104946
Wang, W., and Shi, J. M. (2016). Characteristics of soil microbial carbon and water soluble carbon of Platycladus orientalis plantations with different stand density. Mod. Gard. 20, 19–20.
Waring, B. G., Weintraub, S. R., and Sinsabaugh, R. L. (2014). Ecoenzymatic stoichiometry of microbial nutrient acquisition in tropical soils. Biogeochemistry 117, 101–113. doi: 10.1007/s10533-013-9849-x
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, Vol. 18, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wu, Y. L., Wang, B., Zhao, C., Dai, W., and Li, P. (2011). Comprehensive evaluation of soil fertility in different developing stages of Chinese fir plantations. Chinese J. Northwest Agr. For. Uni. 39, 69–75.
Xu, X. F., Thornton, P. E., and Post, W. M. (2013). A global analysis of soil microbial biomass carbon, nitrogen and phosphorus in terrestrial ecosystems. Glob. Ecol. Biogeogr. 22, 737–749. doi: 10.1111/geb.12029
Xu, Z. W., Yu, G. R., Zhang, X. Y., He, N. P., Wang, Q. F., Wang, S. Z., et al. (2017). Soil enzyme activity and stoichiometry in forest ecosystems along the north-south transect in eastern china (NSTEC). Soil Biol. Biochem. 104, 152–163. doi: 10.1016/j.soilbio.2016.10.020
Yang, Y., Liang, C., Li, T., Cheng, H., and An, S. (2020). Soil extracellular enzyme stoichiometry reflects the shift from P- to N-limitation of microorganisms with grassland restoration. Soil Biol. Biochem. 149, 107928. doi: 10.1016/j.soilbio.2020.107928
Zhang, C., Liu, G. B., Xue, S., and Wang, G. (2016). Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the Loess Plateau. Soil Biol. Biochem. 97, 40–49. doi: 10.1016/j.soilbio.2016.02.013
Zhang, D. J., Zhang, J., Yang, W. Q., Wu, F. Z., and Huang, Y. M. (2014). Plant and soil seed bank diversity across a range of ages of Eucalyptus grandis plantations afforested on arable lands. Plant Soil 376, 307–325. doi: 10.1007/s11104-013-1954-z
Zhang, X., Tian, X., Ma, L., Feng, B., Liu, Q., and Yuan, L. (2015). Biodiversity of the symbiotic bacteria associated with toxic marine dinoflagellate Alexandrium tamarense. J. Bios. 3, 23–28. doi: 10.4236/jbm.2015.36004
Zhao, F. Z., Ren, C. J., Han, X. H., Yang, G. H., Wang, J., and Doughty, R. (2018). Changes of soil microbial and enzyme activities are linked to soil C, N and P stoichiometry in afforested ecosystems. For. Ecol. Manag. 427, 289–295. doi: 10.1016/j.foreco.2018.06.011
Zhao, Y., Wu, J., Yuan, X., Zhu, W., Wang, X., Cheng, X., et al. (2017). The effect of mixing intensity on the performance and microbial dynamics of a single vertical reactor integrating acidogenic and methanogenic phases in lignocellulosic biomass digestion. Bioresour. Technol. 238, 542–551. doi: 10.1016/j.biortech.2017.04.080
Keywords: C:N:P ratio, stoichiometric imbalances, ecological stoichiometry, microbial community, enzymatic activity, microbial nutrient limitation
Citation: Wang C and Jiao R (2022) Adaptive Pathways of Microorganisms to Cope With the Shift From P- to N-Limitation in Subtropical Plantations. Front. Microbiol. 13:870667. doi: 10.3389/fmicb.2022.870667
Received: 07 February 2022; Accepted: 30 March 2022;
Published: 29 April 2022.
Edited by:
Yurong Liu, Huazhong Agricultural University, ChinaCopyright © 2022 Wang and Jiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruzhen Jiao, amlhb3J6aEBjYWYuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.