
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 22 April 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.870658
This article is part of the Research TopicBasidiomycete Fungi: From Biosystematics and Biodiversity to BiotechnologyView all 21 articles
 Nan Tao1,2,3†
Nan Tao1,2,3† Bopu Cheng1,4†
Bopu Cheng1,4† Hongmei Chai1,2,3
Hongmei Chai1,2,3 Xianghua Cui1,2,3
Xianghua Cui1,2,3 Yuanhao Ma1,2,3
Yuanhao Ma1,2,3 Jinping Yan5
Jinping Yan5 Yongchang Zhao1,2,3*
Yongchang Zhao1,2,3* Weimin Chen1,2,3*
Weimin Chen1,2,3*
Guanosine triphosphate (GTP) cyclohydrolase I (GCH1) is the limiting enzyme of the tetrahydrobiopterin (BH4) synthesis pathway. The disruption of gch1 gene may cause conditional lethality due to folic acid auxotrophy in microorganisms, although the function of gch1 in basidiomycetes has not been deciphered so far. In the present study, gch1 expression in Cyclocybe aegerita (cagch1) was downregulated using the RNAi method, which resulted in growth retardation in both solid and liquid medium, with the hyphal tips exhibiting increased branching compared to that in the wild strain. The development of fruiting bodies in the mutant strains was significantly blocked, and there were short and bottle-shaped stipes. The transcriptional profile revealed that the genes of the MAPK pathway may be involved in the regulation of these effects caused by cagch1 knockdown, which provided an opportunity to study the role of gch1 in the development process of basidiomycetes.
Guanosine triphosphate (GTP) cyclohydrolase I (GCH1; EC 3.5.4.16) is the first enzyme to act in the biosynthesis of tetrahydrobiopterin (BH4). GCH1 catalyzes the conversion of GTP into dihydroneopterin triphosphate and formic acid (Nardese et al., 1996), and this reaction is the rate-limiting step in the de novo synthesis of BH4 in plants and microorganisms (Burg and Brown, 1968). Although bacteria, fungi, and plants can synthesize tetrahydrofolic acid; mammals consume folates in the form of vitamins in food. BH4 is crucial to invertebrates as it serves as a coenzyme of aromatic amino acid hydroxylase (Werner et al., 2011) and a cofactor of nitric oxide synthase (NOS) (Gorren and Mayer, 2002), in addition to being an essential regulatory factor of melanin biosynthesis (Chen et al., 2015) and a component of the synthesis process of various hormones and neurotransmitters (Werner et al., 2011). BH4 also serves as a cofactor for a variety of inflammatory factors and neurotransmitters (Latremoliere et al., 2015). In addition to being crucial for BH4 synthesis, GCH1 is reported to play an important role in the pathogenesis of neuropathological pain (Kim et al., 2009), Parkinson’s disease (Xu et al., 2017), diabetes (Wang et al., 2020), and cardiovascular diseases (Chuaiphichai et al., 2014).
Guanosine triphosphate cyclohydrolase I GCH1 is reported in various microorganisms, including Escherichia coli (Katzenmeier et al., 1990), Saccharomyces cerevisiae (Nardese et al., 1996), Nocardia sp. (He et al., 2004), and Rickettsia monacensis (Bodnar et al., 2020). The comparison of sequences from different organisms has revealed that the enzyme was highly conserved throughout the evolutionary process. The functional analysis of GCH1 has been focused mostly on the relationship between the enzyme and the folate content in organisms, such as folate enrichment by expressing the GCH1 gene in plants (Hossain et al., 2004; Storozhenko et al., 2007; Naqvi et al., 2009; Nunes et al., 2009; Blancquaert et al., 2015). So far, no one has described the effect of this enzyme on the development of the host organism. Furthermore, little is known about the function of GCHI in fungi. According to a report, the disruption of GCH1 in S. cerevisiae leads to conditional lethality due to folic acid auxotrophy (Nardese et al., 1996).
Cyclocybe aegerita is a delicious species cultivated in Asia (Zhang et al., 2015). The species has a short life cycle and complex development modes, such as monokaryon fruiting, which renders this species important as a model organism in genetics research (Labarère and Noël, 1992). In the present study, a gene encoding GCH1 named cagch1 was isolated from C. aegerita YSG, and its genome sequence was cloned, followed by comparing the deduced protein sequences with those of the other species for homology analysis. Using the RNAi method, the morphologies of the mycelium, as well as the young and mature fruiting bodies, were compared between the wild stain and the GCH1-mutant strain at various developmental stages. The findings of the current study would provide novel insights into the role of GCH1 in basidiomycetes development.
Cyclocybe aegerita dikaryotic strain YSG deposited in our laboratory was used in the transformation experiments. The mycelia were cultured at 25°C on a yeast extract peptone dextrose (YPD) medium plate (0.2% yeast extract, 0.2% peptone, 2% dextrose, and 1.5% agar) followed by culturing in the sawdust medium (64% hardwood sawdust, 15% wheat bran, 1% gypsum, and 20% cottonseed shells in 50% water) to produce fruiting bodies. The protoplast regeneration medium contained 205 g of sucrose, 2 g of tryptone, 2 g of yeast extract, and 10 g of agar per liter of dH2O. The DH5α strain of E. coli, to be used for plasmid amplification, was cultured at 37°C in Luria–Bertani (LB) medium containing 100 μg/ml of ampicillin. In order to conduct the growth rate test, equal amounts of mycelia were inoculated in both solid and liquid YPD media for the evaluation of colony diameter and biomass, respectively.
Cyclocybe aegerita YSG strain was cultured in YPD medium at 25°C in the dark for 8 days. The genomic DNA was extracted from the mycelia obtained from this culture of C. aegerita using the Fungal gDNA Isolation Kit (Biomiga, United States). Total RNA was extracted from the fungus using the RNAiso Plus in accordance with the manufacturer’s instruction (TaKaRa, Dalian, China). The complete genomic DNA and cDNA products of the cagch1 gene were amplified in a polymerase chain reaction (PCR) using the primer pair gch1F and gch1R based on the genomic information of C. aegerita monokaryotic strain YSG. The amplified products/fragments were then cloned into the pMD19-T vector (TaKaRa, Dalian, China) and sent for sequencing (Tsingke Biotechnology Co., Ltd.).
The transformation vector pAGH (given by Dr. Zhang, Kunming University of Science and Technology) was used for constructing the RNAi vector using the method described in a previous report (Zhang et al., 2021). The plasmid pAGH comprised the C. aegerita actin promoter, the glyceraldehyde-3-phosphate dehydrogenase (gpd) gene promoter, the CaMV35S terminator, the hygromycin B phosphotransferase gene, and the gpd promoter from Ustilago maydis. A 487-bp antisense fragment, amplified using primer Gch1-1F and Gch1-1R (Supplementary Table 1), of the cagch1 cDNA was also inserted into the EcoRV enzyme site. All fragments were sequentially ligated into the pAGH vector using the ClonExpress MultiS One Step Cloning Kit (Vazyme, Nanjing, China) in accordance with the manufacturer’s instructions.
The transformation was performed by following the PEG-CaCl2 protocol described in previous reports (Tao et al., 2020) with a few modifications. In brief, the protoplasts were prepared by digesting the C. aegerita mycelia from the liquid culture with a lysing enzyme solution containing 1% cellulase R-10 (Japan), 1% Lywallzyme, and 1% lysing enzymes from Trichoderma harzianum (Sigma-Aldrich) in 0.6 M mannitol buffer as the osmotic stabilizer. The obtained protoplasts were washed in the STC solution [1.2 M sorbitol, 10 mM CaCl2, and 50 mM Tris–HCl (pH 7.0)] and adjusted to a final concentration of 1 × 108 protoplasts ml–1. Subsequently, a volume of 100 μl of protoplasts was mixed with 1 μg of plasmid DNA digested with the KpnI enzyme and the reaction was allowed to occur for 30 min on ice. Afterward, 1 ml of the PTC solution (40% w/v polyethylene glycol 4,000, 60 mM CaCl2, and 1.2 M sorbitol) was added to the mixture, followed by incubation for 30 min at room temperature. Finally, the cells were added to 10 ml of molten YPD regeneration medium and then poured on plates. After 5 days of incubation at 25°C, 10 mL of molten YPD medium containing 150 μg/ml hygromycin B was poured over the plates for the screening of transformants.
The colonies resistant to hygromycin B were verified using the primer pair adhF and adhR targeted to the flanking region of the inserted fragment. Quantitative PCR (qPCR) was performed in the CFX96 Real-Time PCR detection system (Bio-Rad, Hercules, CA, United States). The PCR reaction was performed in a total volume of 20 μL using the iTaq™ Universal SYBR Green Supermix in accordance with the manufacturer’s instructions (Bio-Rad). The thermal cycling parameters for the PCR reactions were as follows: 20 s at 95°C for initial polymerase activation, followed by 40 cycles of 5 s at 95°C, and 30 s at 60°C. The primers used are listed in Supplementary Table 1. Amplification of each sample was performed in triplicate. Glyceraldehyde-3-phosphate dehydrogenase (GPD) was used as the reference gene for the normalization of the qPCR data. The relative expression levels were calculated using the 2–ΔΔCT method.
Total RNA was isolated from the mature YSG fruiting bodies as well as the transformants T5, T10, and T11. The corresponding transcriptomic data were obtained from six cDNA libraries sequenced on an Illumina sequencing platform at the Beijing Genome Institute (BGI, Shenzhen, China). The differentially expressed genes (DEGs) were classified into GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
On the basis of the genome annotation and sequence analysis, gDNA and cDNA were employed as templates for the amplification of the gene encoding CaGCH1 (the cagch1 gene), and the amplification products were sequenced and analyzed. The gDNA sequence of cagch1 was 1,067 bp in length and contained 3 introns and 4 exons (Accession Number OL303985). The cDNA of cagch1 was 894 bp in length and contained 297 amino acids (Figure 1A).
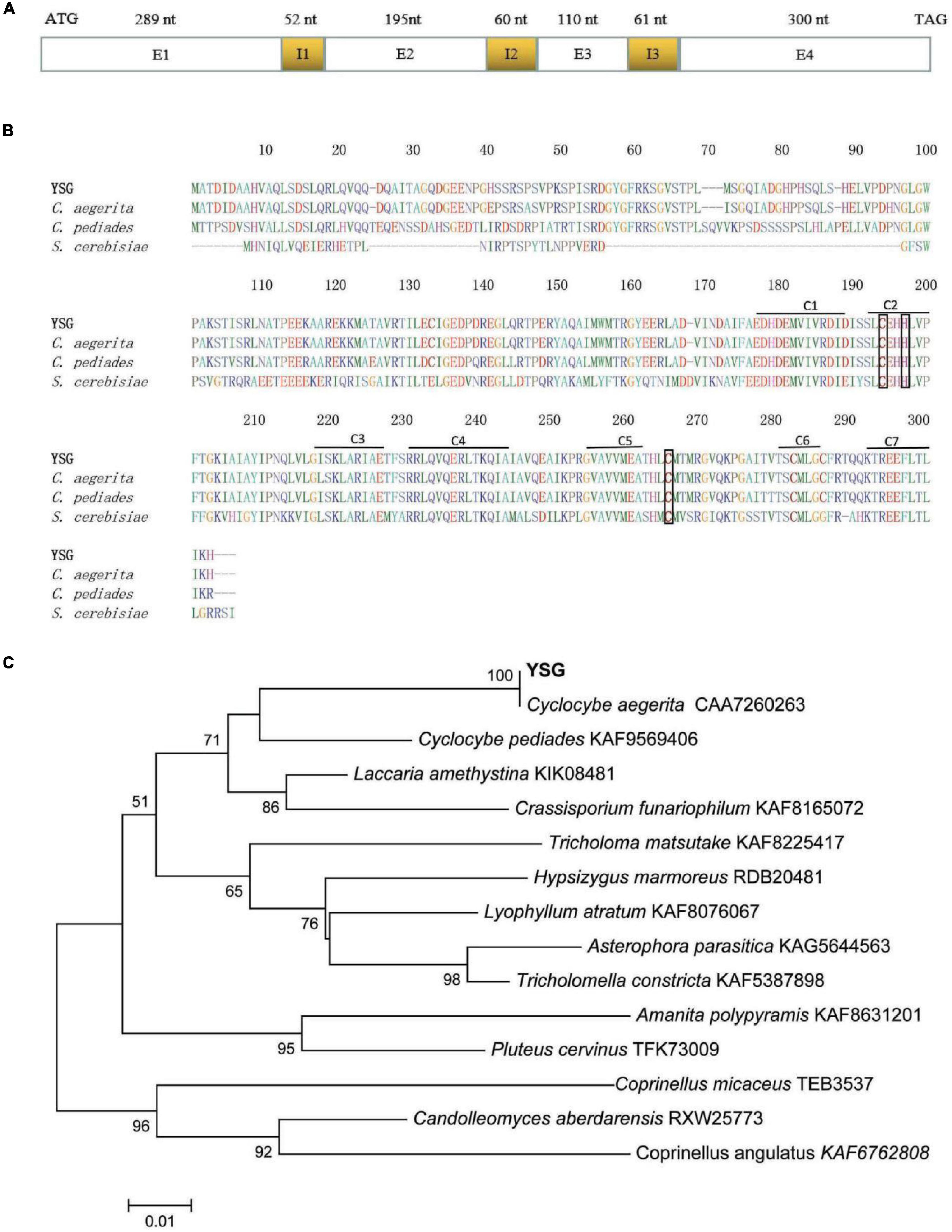
Figure 1. (A) Sequence structure of cagch1. (B) The characterization of the sequence of CaGCH1 (YSG) and its comparison with the sequences from other strains including C. aegerita (Accession Number CAA7260263), C. pediades (Accession Number KAF9569406), and S. cerevisiae (Accession Number P51601). (C) The phylogenetic tree for CaGCH1 with other GCH1 from basidiomycetes.
The protein sequences were aligned using ClustalW (soft package included in Mega 6) (Tamura et al., 2013) and the result revealed that the CaGCH1 protein shared a high level of similarity with the GCH1 proteins of C. aegerita (Accession Number CAA7260263), Cyclocybe pediades (Accession Number KAF9569406), and S. cerevisiae (Accession Number P51601). All four proteins contained seven conserved regions (C1–7) and three Zn2+ binding residues (Figure 1B). The phylogenetic analysis of the GCH1 protein sequences was performed with MEGA6 software using neighbor-joining, minimum evolution (Tamura et al., 2013), which indicated that CaGCH1 was the closest to that of the Cyclocybe species (previously used genus name Agrocybe) (Figure 1C).
In order to study the effect of cagch1 on the developmental process, the silence transformants were obtained by constructing a dual RNAi plasmid (Figure 2A). After the screening of transformants on the YPD medium containing hygromycin B, the specific primer pair adhF and adhR targeted to the actin and gpd promoter regions was used for confirming the positive transformants through PCR amplification (Figure 2B). Finally, correct PCR products were detected in three strains T5, T10, and T11 (Figure 2C).
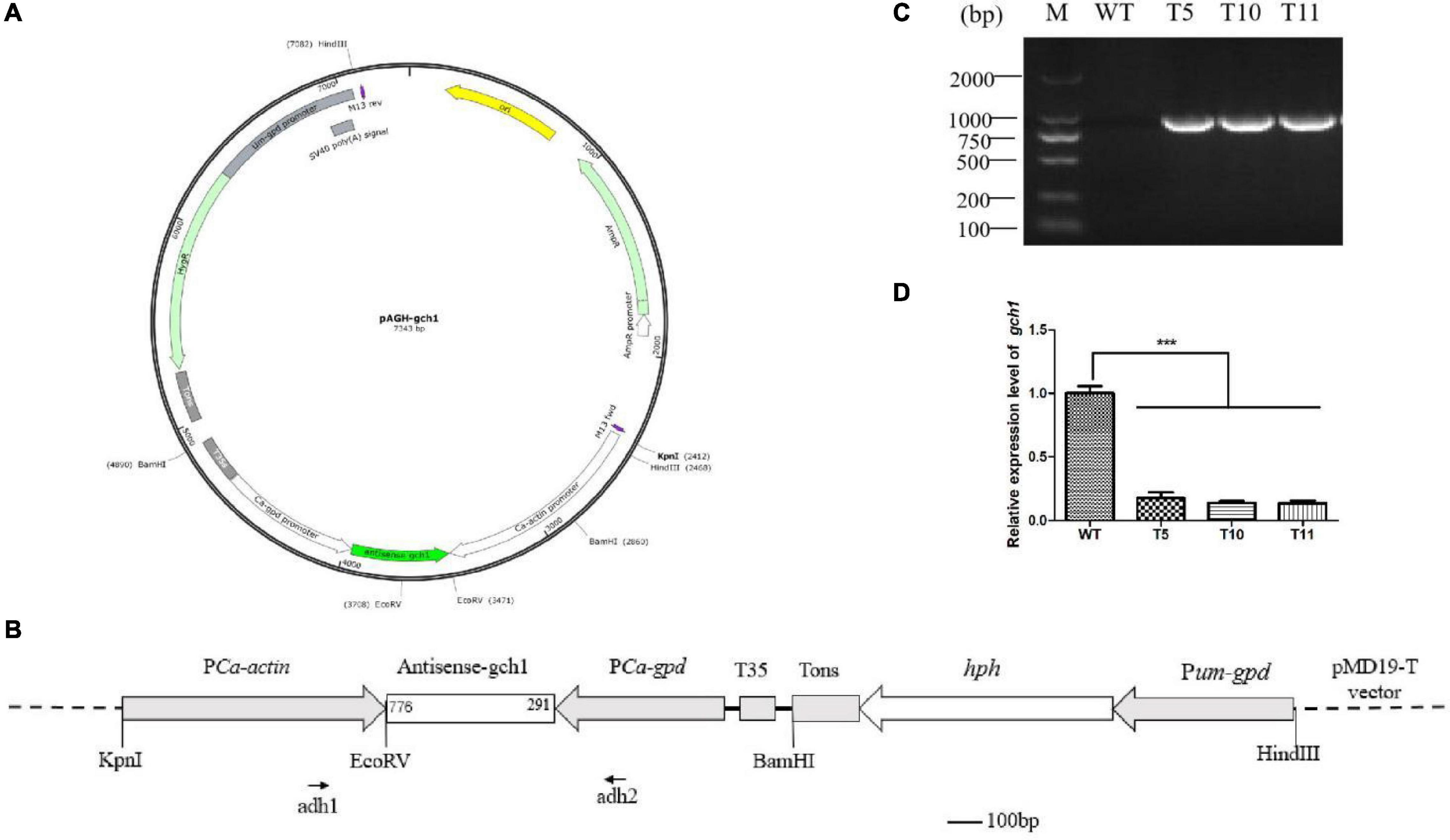
Figure 2. (A) Structure of the pAGH-gch1 plasmid. (B) Diagram of the RNAi vector construction. The antisense fragment of cagch1 was inserted in the site between the promoters actin and gpd. The primer pair of adh1 and adh2 was used for the verification of the transformants. (C) The verification of transformants. (D) The expression analysis of cagch1. Asterisks (***) indicate the data that differed significantly based on p < 0.0001 (t-test) as the significance threshold.
Mycelia were collected from WT, T5, T10, and T11 and subjected to total RNA extraction. The cDNA obtained after the reverse transcription of the extracted RNA was used for quantifying the expression of cagch1. It was observed that the expression level of cagch1 in all three transformants had decreased to a low level – approximately 17.63% in T5, 13.87% in T10, and 13.53% in T11 compared to that in the WT strain (Figure 2D).
After 7 days of culture on YPD plates, the transformants colonies appeared denser with smooth edges than WT colonies (Figure 3A). The colony diameter was measured for growth rate analysis, and it was observed that the WT strains spread faster than the transformants (Figure 3D). The microscopic observation revealed that transformants exhibited relatively greater branching in the hyphal tips growing on the cover glass (Figure 3B).
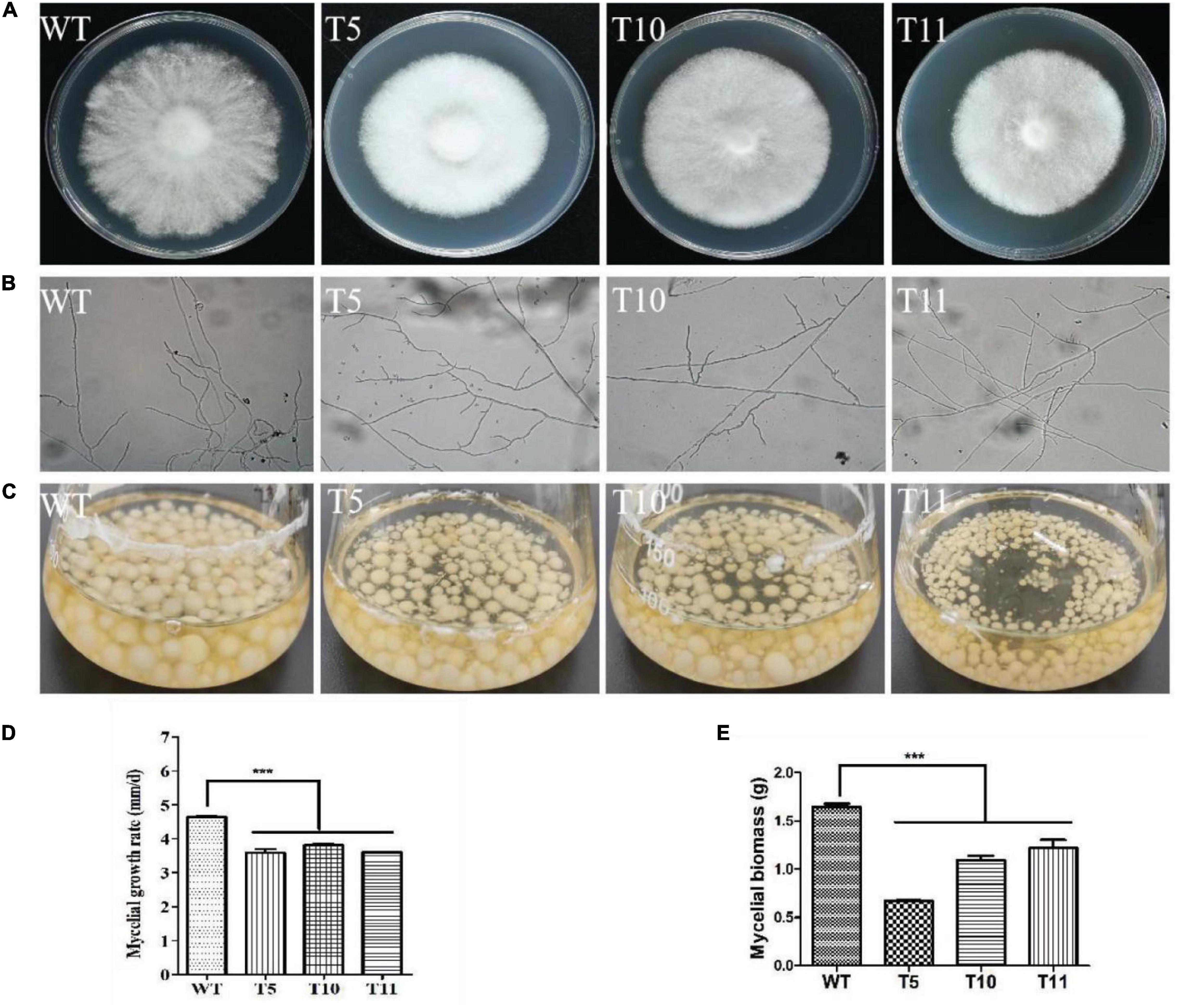
Figure 3. Morphology and growth characteristics of WT, T5, T10, and T11 strains. (A) Colony morphology. (B) Hyphal branch. (C) Culture in liquid. (D) Mycelial growth. (E) Mycelial biomass. Asterisks (***) indicate the data that differed significantly based on p < 0.0001 (t-test) as the significance threshold.
The equal mycelium block was cultured in the liquid YPD medium under shaking conditions of 180 rpm/min for 7 days (Figure 3C). It was observed that the mycelial biomass of all three transformants was decreased significantly compared to the WT strain, with the mycelium weight of the T5 strain approximately one-half that of the WT strain (Figure 3E).
The WT strain and the three mutant strains were cultured in sawdust bags at 25°C for 40 days to investigate the potential effect of cagch1 on the fruiting bodies of C. aegerita. The development of the transformant strains was observed to be blocked, and there were bottle-shaped stipes (Figure 4A). The longitudinal section of young fruiting bodies revealed that the stipes of the mutant strains comprised flocculent flesh relative to the compact flesh of the WT strain (Figure 4B). At the late stage, the stipe elongation in the mutant strain was significantly inhibited, and the length and the weight of fresh fruiting bodies were approximately one-sixth and one-fifteenth that of the wild type, respectively (Figures 4C–E). Moreover, the morphologies of all strains were stable in the subsequent two culture processes.
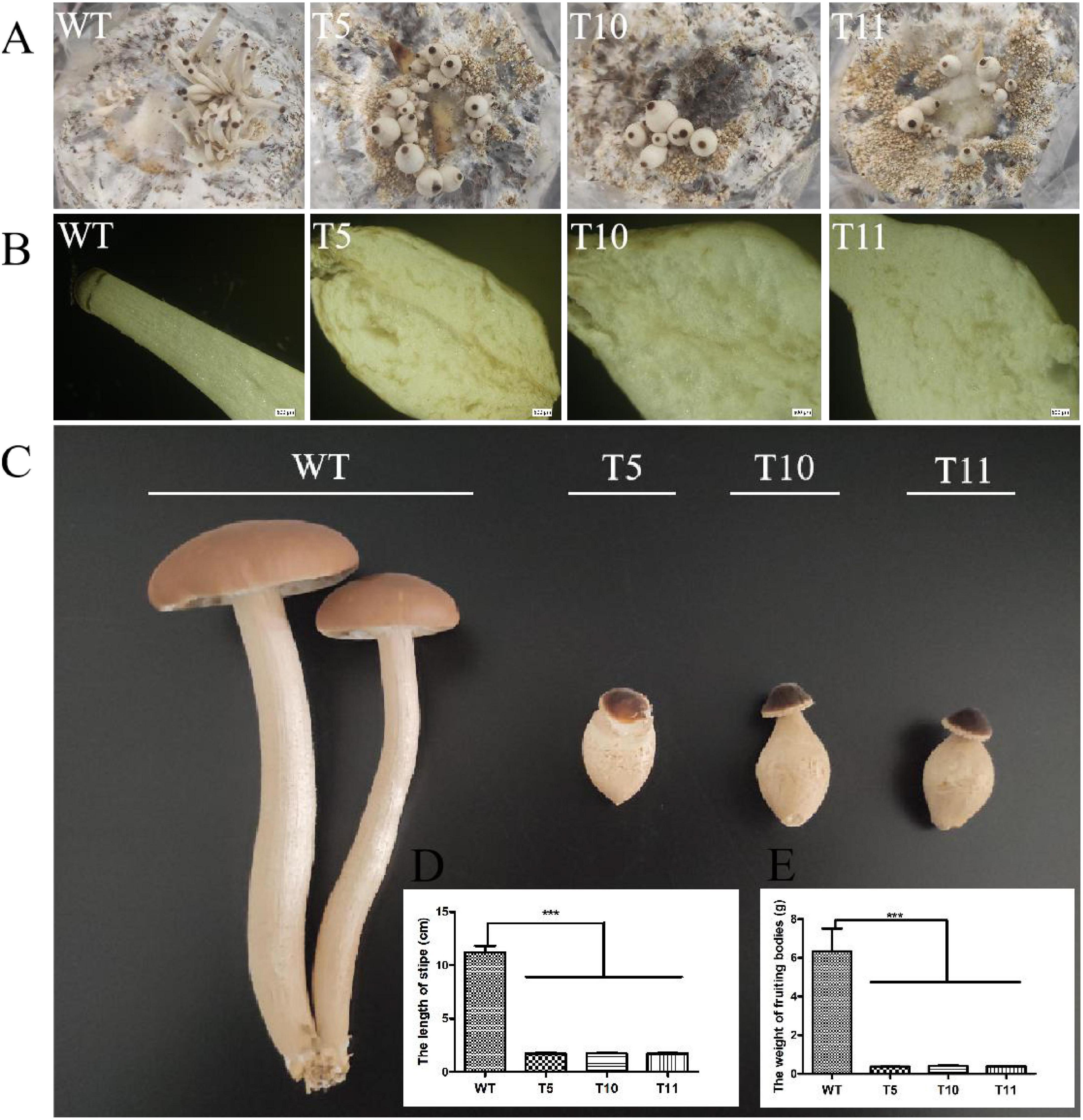
Figure 4. Morphology of the fruiting bodies in WT, T5, T10, and T11. (A) Young fruiting bodies. (B) Longitudinal section image of stipes. Bar, 500 μm. (C) Mature fruiting bodies. (D) The length of stipes. (E) The weight of fruiting bodies. Asterisks (***) indicate the data that differed significantly based on p < 0.0001 (t-test) as the significance threshold.
Furthermore, the expression level of cagch1 in the mature fruiting bodies was evaluated using the qPCR method. As depicted in Figure 5, the expression level of cagch1 in the three transformants exhibited different degrees of decrease—approximately 21.83% in T5, 23% in T10, and 48% in T11 compared to that in the WT strain (Figure 5).
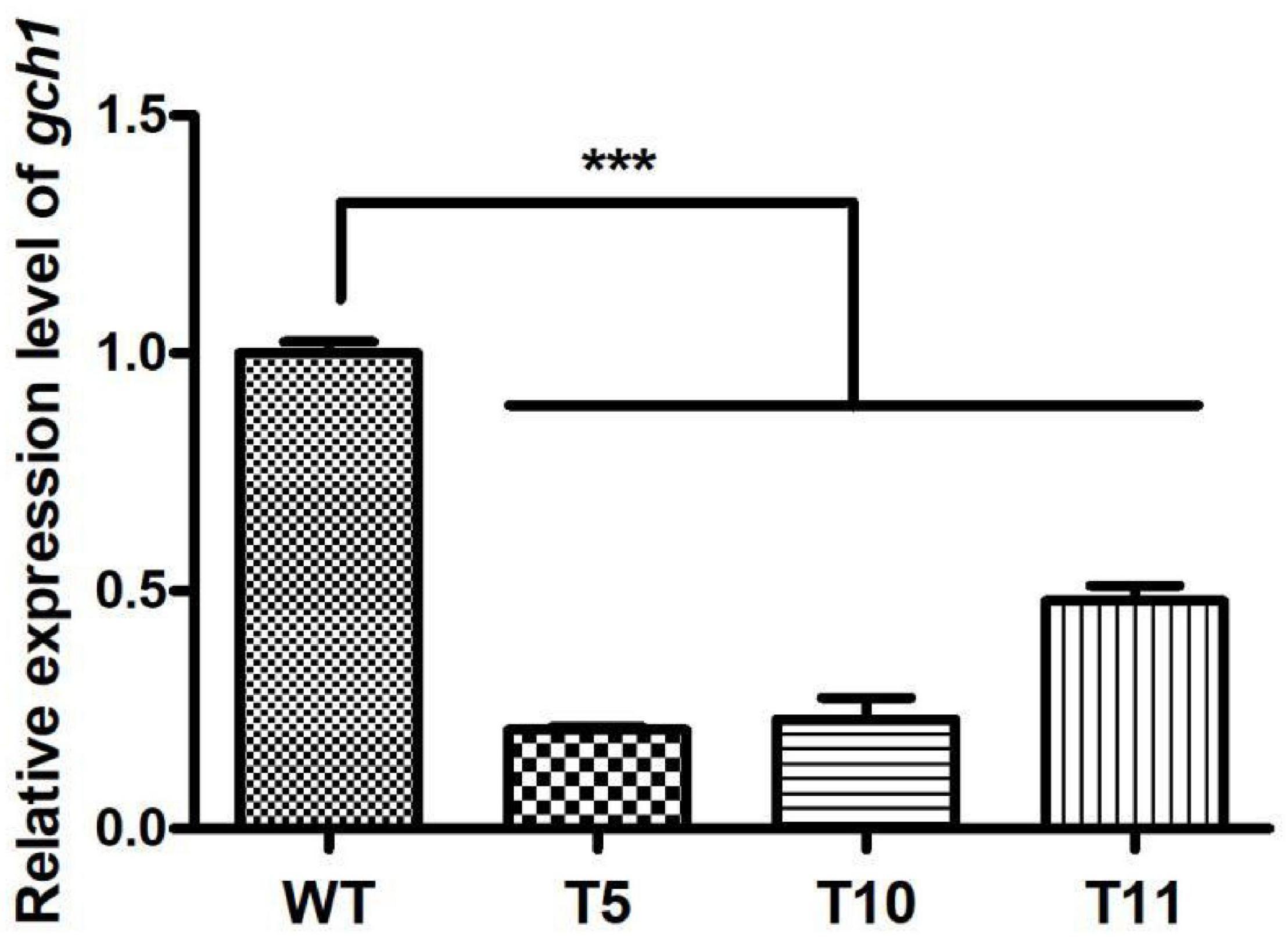
Figure 5. The expression level of cagch1 in mature fruiting bodies. Asterisks (***) indicate the data that differed significantly based on p < 0.0001 (t-test) as the significance threshold.
In order to evaluate the effect of cagch1 knockdown on the expression levels of different genes, the expression difference in the mating bodies of WT was compared to those of each of the three transgenic strains using the transcriptional profile analysis (PRJNA813015).
A total of 11,723 differentially expressed unigenes (DEGs) were identified, including 211 and 218 special genes in the WT fruiting bodies and in the mutant fruiting bodies, respectively (Figure 6A). Furthermore, the 544 DEGs involved in the KEGG metabolic pathway were categorized into five major types comprising 20 categories, among which a few pathways were associated with mycelial growth and development, such as cell wall remodeling in signal transduction pathway and polysaccharide anabolism pathway (Figure 6B). Among these 544 genes, most annotated in the MAPK pathway (Figure 6C) and upregulated in the mutant strains were possibly involved in cell wall remodeling and the mating process (Figure 6D).
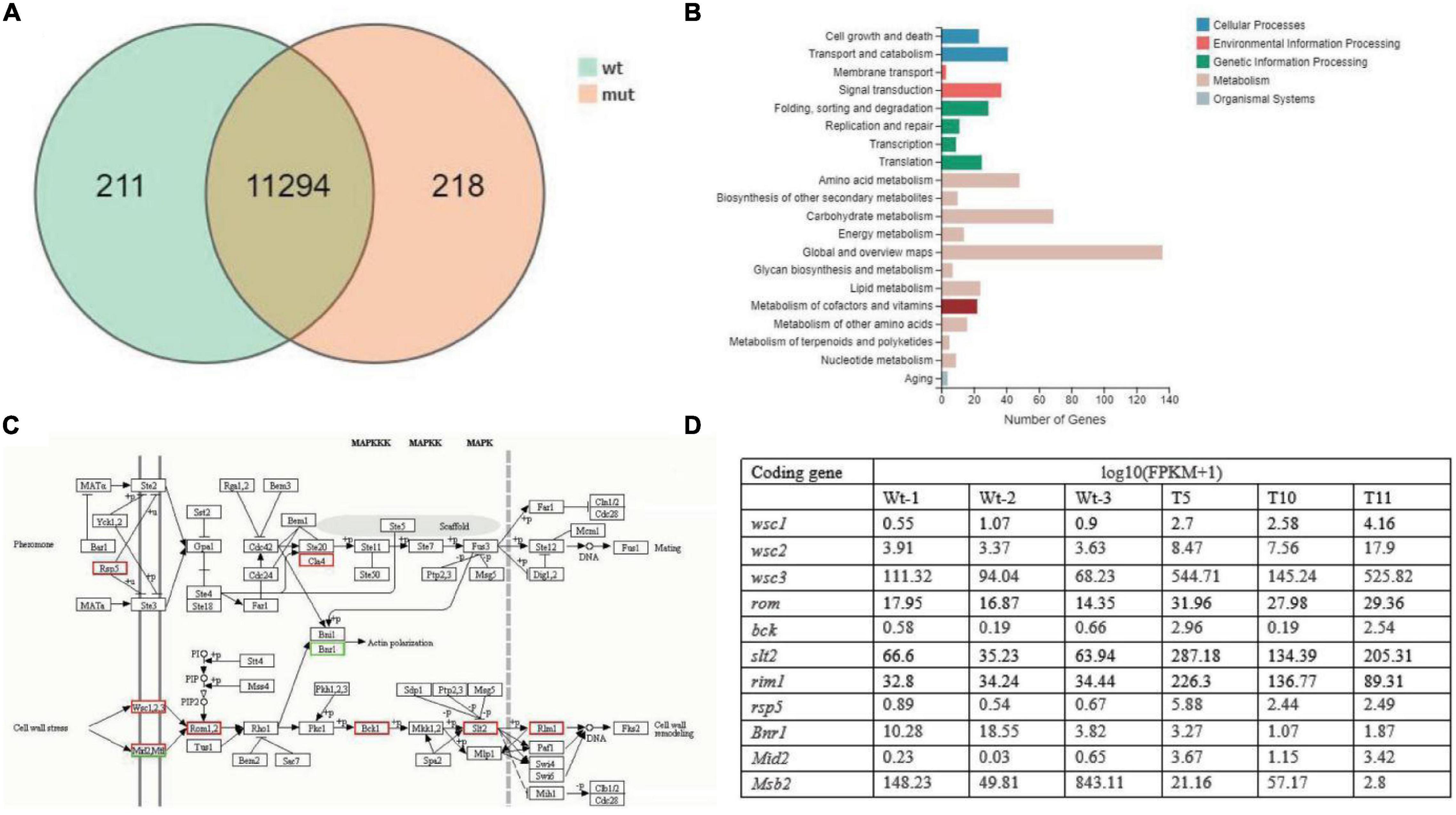
Figure 6. The number of differentially expressed genes (DEGs). (A) Venn diagram depicting the number of DEGs between WT and transformant strains (mut). (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) classification of the identified DEGs. (C) The putative pathway of signal transduction deduced in the KEGG analysis. (D) The expression levels of genes in the MAPK pathway.
Guanosine triphosphate cyclohydrolase I plays an important role in the biosynthetic pathway of various pteridines, including folic acid derivatives and tetrahydrobiopterin, where it serves as the enzyme cofactor for the one-carbon transfer reaction and the hydroxylation reaction of aromatic amino acids, respectively (Nardese et al., 1996). GCH1 is also involved in the production of nitric acid and, therefore, regulates several physiological processes via various molecular pathways (Tayeh and Marletta, 1989). In order to investigate the potential function of GCH1 in C. aegerita, the cagch1 gene was downregulated by constructing an RNAi system for cagch1. The results revealed that the transformants exhibited variable morphology during different developmental stages.
As an important functional enzyme, GCH1 contains amino acid sequences that are conserved in Homo sapiens, plants, and microorganisms (He et al., 2004; Liang et al., 2019; Bodnar et al., 2020). In particular, certain key residues for binding GTP or coordinating with the zinc ion, such as HH, LSK, and QE, present in GCH1 have been identified in CaGCH1 motifs C2, C3, and C4. Similar to the results reported in previous studies (He et al., 2004; Liang et al., 2019; Bodnar et al., 2020), in the present study, the amino acid residues of CaGCH1 were observed to be highly conserved in the downstream region compared to those of other species. These sequence characteristics provided a basis for the identification of GCH1, which is usually not annotated in the genome database.
The RNAi system was successfully constructed in C. aegerita using a dual promoter. The targeted gene cagch1 was evidently downregulated during the development of transformants. In a previous study on Ganoderma lucidum, four gene silencing constructs, namely, hairpin, sense, antisense, and dual promoter constructs, were introduced into the strain to be evaluated, with the dual promoter silencing vector demonstrating the highest rates of target gene silencing (Mu et al., 2012). Although the hairpin method has been used frequently for gene silencing in previous studies, the dual promoter method is simpler and more efficient in constructing the RNAi system in basidiomycetes.
The studies investigating the function of GCH1 in microorganisms are scarce. A study reported that the disruption of GCH1 in S. cerevisiae leads to conditional lethality due to the obstruction of folinic acid synthesis (Nardese et al., 1996). Another study reported that the important role of GCH1 in various physiological processes renders it suitable as a potential anti-infective drug target (Schüssler et al., 2019), which was confirmed when the inhibitors of tetrahydrofolate biosynthesis were successfully able to control a wide variety of microbial and protozoan infections (Swarbrick et al., 2008). However, the function of GCH1 in basidiomycetes has not been investigated so far. In the present study, the expression of cagch1 in C. aegerita was downregulated, and it was observed that the transformants differed from the wild-type strains in terms of mycelial branching, biomass, and fruiting bodies. The growth of the transformants was evidently inhibited in both solid and liquid media and the mycelium exhibited greater branching. In a previous study on G. lucidum, the gene encoding nicotinamide adenine dinucleotide phosphate oxidases (Nox) was reported to play an important role in hyphal branching and fruiting body development through the regulation of the Ca2+ signal pathway (Mu et al., 2014). Another study indicated that a cross-talk between Ca2+ and reactive oxygen species (ROS) regulates hyphal branching (Gao et al., 2018).
As cofactors of NOS, NADPH, FAD, BH4, and FMN are involved in the production of nitric oxide (NO) (Song et al., 2000). In fungi, NO is involved in the regulation of various developmental processes, including sporulation, spore germination (Maier et al., 2001; Gong et al., 2007), and fruiting body production (Song et al., 2000). When the underlying mechanism was investigated, the involvement of cGMP, MAPK, and light signaling pathways was revealed (Barhoom and Sharon, 2004; Vieira et al., 2009). In the present study, most of the genes that had been annotated in the mating process and cell wall remodeling in the MAPK pathway were upregulated in the mutant strains, which suggested that these genes could be induced by the products associated with CaGCH1 for participating in the developmental process.
A putative gene encoding the GCH1 protein, named CaGCH1, was cloned and identified in the basidiomycetes C. aegerita. The silencing of cagch1 resulted in the retardation of development, including the growth and the following morphological characteristics: (1) the growth rate of the mutants was decreased; (2) the hyphal tips of the mutants exhibited greater branching; and (3) the fruiting bodies in the mutants had short and bottle-shaped stipes. The transcriptomic analysis revealed that the regulation of the MAPK pathway by the knockdown of cagch1 could be involved in the development of fruiting bodies. Further comprehensive analysis of the function of cagch1 would improve the understanding of its role in the development process in basidiomycetes.
The data presented in the study are deposited in the NCBI repository, accession number PRJNA813015.
WC and YZ conceived and designed the experiments. NT, BC, and JY carried out the transformation and screening. NT, BC, and YM investigated the growth and microscopic observation. HC, BC, and XC performed the RT-PCR experiments and transcriptome analysis. WC, NT, and YZ analyzed all data. WC and NT wrote original draft manuscript. All authors have read and agreed to the final version of the manuscript.
This research was funded by National Natural Science Foundation of China (No. 31960017), Scientific Talents and Platform Plan of Yunnan province (202105AC160086), and China Agriculture Research System (CARS-20).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Yu Zhang (Kunming University of Science and Technology) is thanked for his helpful suggestions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.870658/full#supplementary-material
Barhoom, S., and Sharon, A. (2004). cAMP regulation of “pathogenic” and “saprophytic” fungal spore germination. Fungal Genet. Biol. 41, 317–326. doi: 10.1016/j.fgb.2003.11.011
Blancquaert, D., Van Daele, J., Strobbe, S., Kiekens, F., Storozhenko, S., De Steur, H., et al. (2015). Improving folate (vitamin B9) stability in biofortified rice through metabolic engineering. Nat. Biotechnol. 33, 1076–1078. doi: 10.1038/nbt.3358
Bodnar, J., Fitch, S., Sanchez, J., Lesser, M., Baston, D. S., and Zhong, J. (2020). GTP cyclohydrolase I activity from Rickettsia monacensis strain Humboldt, a Rickettsial endosymbiont of Ixodes pacificus. Tick Borne. Dis. 11:101434. doi: 10.1016/ttbdis.2020.101434
Burg, A. W., and Brown, G. M. (1968). The biosynthesis of folic acid. 8. Purification and properties of the enzyme that catalyzes the production of formate from carbon atom 8 of guanosine triphosphate. J. Biol. Chem. 243, 2349–2358. doi: 10.1016/S0021-9258(18)93481-2
Chen, P., Wang, J., Li, H., Li, Y., Chen, P., Li, T., et al. (2015). Role of GTP-CHI links PAH and TH in melanin synthesis in silkworm. Bombyx mori. Gene. 567, 138–145. doi: 10.1016/j.gene.2015.03.043
Chuaiphichai, S., McNeill, E., Douglas, G., Crabtree, M. J., Bendall, J. K., Hale, A. B., et al. (2014). Cell-autonomous role of endothelial GTP cyclohydrolase 1 and tetrahydrobiopterin in blood pressure regulation. Hypertension 64, 530–540. doi: 10.1161/HYPERTENSIONAHA.114.03089
Gao, T., Shi, L., Zhang, T., Ren, A., Jiang, A., Yu, H., et al. (2018). Cross talk between calcium and reactive oxygen species regulates hyphal branching and ganoderic acid biosynthesis in Ganoderma lucidum under copper stress. Appl. Environ. Microbiol. 84:e00438-18. doi: 10.1128/AEM.00438-18
Gong, X., Fu, Y., Jiang, D., Li, G., Yi, X., and Peng, Y. (2007). L-arginine is essential for conidiation in the filamentous fungus Coniothyrium minitans. Fungal Genet. Biol. 44, 1368–1379. doi: 10.1016/j.fgb.2007.07.007
Gorren, A. C., and Mayer, B. (2002). Tetrahydrobiopterin in nitric oxide synthesis: a novel biological role for pteridines. Curr. Drug. Metab. 3, 133–157. doi: 10.2174/1389200024605154
He, A., Simpson, D. R., Daniels, L., and Rosazza, J. P. (2004). Cloning, expression, purification, and characterization of Nocardia sp. GTP cyclohydrolase I. Protein. Expr. Purif. 35, 171–180. doi: 10.1016/j.pep.2004.02.008
Hossain, T., Rosenberg, I., Selhub, J., Kishore, G., Beachy, R., and Schubert, K. (2004). Enhancement of folates in plants through metabolic engineering. Proc. Natl. Acad. Sci. U.S.A. 101, 5158–5163. doi: 10.1073/pnas.0401342101
Katzenmeier, G., Schmid, C., and Bacher, A. (1990). Cloning and expression of the putative gene coding for GTP cyclohydrolase I from Escherichia coli. FEMS. Microbiol. Lett. 54, 231–234. doi: 10.1016/0378-1097(90)90288-2
Kim, S. J., Lee, W. I., Lee, Y. S., Kim, D. H., Chang, J. W., Kim, S. W., et al. (2009). Effective relief of neuropathic pain by adeno-associated virus-mediated expression of a small hairpin RNA against GTP cyclohydrolase 1. Mol. Pain. 5:67. doi: 10.1186/1744-8069-5-67
Labarère, J., and Noël, T. (1992). Mating type switching in the tetrapolar basidiomycete Agrocybe aegerita. Genetics 131, 307–319. doi: 10.1093/genetics/131.2.307
Latremoliere, A., Latini, A., Andrews, N., Cronin, S. J., Fujita, M., Gorska, K., et al. (2015). Reduction of neuropathic and inflammatory pain through inhibition of the tetrahydrobiopterin pathway. Neuron 86, 1393–1406. doi: 10.1016/j.neuron.2015.05.033
Liang, Q., Wang, K., Liu, X., Riaz, B., Jiang, L., Wan, X., et al. (2019). Improved folate accumulation in genetically modified maize and wheat. J. Exp. Bot. 70, 1539–1551. doi: 10.1093/jxb/ery453
Maier, J., Hecker, R., Rockel, P., and Ninnemann, H. (2001). Role of nitric oxide synthase in the light-induced development of sporangiophores in Phycomyces blakesleeanus. Plant Physiol. 126, 1323–1330. doi: 10.1104/pp.126.3.1323
Mu, D., Li, C., Zhang, X., Li, X., Shi, L., Ren, A., et al. (2014). Functions of the nicotinamide adenine dinucleotide phosphate oxidase family in Ganoderma lucidum: an essential role in ganoderic acid biosynthesis regulation, hyphal branching, fruiting body development, and oxidative-stress resistance. Environ. Microbiol. 16, 1709–1728. doi: 10.1111/1462-2920.12326
Mu, D., Shi, L., Ren, A., Li, M., Wu, F., and Jiang, A. (2012). The development and application of a multiple gene co-silencing system using endogenous URA3 as a reporter gene in Ganoderma lucidum. PLoS One 7:e43737. doi: 10.1371/journal.pone.0043737
Naqvi, S., Zhu, C., Farre, G., Ramessar, K., Bassie, L., Breitenbach, J., et al. (2009). Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc. Natl. Acad. Sci. U.S.A. 106, 7762–7767. doi: 10.1073/pnas.0901412106
Nardese, V., Gütlich, M., Brambilla, A., and Carbone, M. L. (1996). Disruption of the GTP-cyclohydrolase I gene in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 218, 273–279. doi: 10.1006/bbrc.1996.0048
Nunes, A. C., Kalkmann, D. C., and Aragão, F. J. (2009). Folate biofortification of lettuce by expression of a codon optimized chicken GTP cyclohydrolase I gene. Transgenic. Res. 18, 661–667. doi: 10.1007/s11248-009-9256-1
Schüssler, S., Haase, I., Perbandt, M., Illarionov, B., Siemens, A., Richter, K., et al. (2019). Structure of GTP cyclohydrolase I from Listeria monocytogenes, a potential anti-infective drug target. Acta. Crystallogr. F. Struct. Biol. Commun. 75, 586–592. doi: 10.1107/S2053230X19010902
Song, N. K., Jeong, C. S., and Choi, H. S. (2000). Identification of nitric oxide synthase in Flammulina velutipe. Mycologia 92, 1027–1032. doi: 10.2307/3761467
Storozhenko, S., Navarrete, O., Ravanel, S., De Brouwer, V., Chaerle, P., Zhang, G. F., et al. (2007). Cytosolic hydroxymethyldihydropterin pyrophosphokinase/dihydropteroate synthase from Arabidopsis thaliana: a specific role in early development and stress response. J. Biol. Chem. 282, 10749–10761. doi: 10.1074/jbc.M701158200
Swarbrick, J., Iliades, P., Simpson, J. S., and Macreadie, I. (2008). Folate biosynthesis – reappraisal of old and novel targets in the search for new antimicrobials. Open Enzym. Inhib. J. 1, 12–33. doi: 10.2174/1874940200801010012
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tao, N., Cui, X. H., Chai, H. M., Ma, Y. H., Zhao, Y. C., and Chen, W. M. (2020). Establishment of genetic transformation system of Agrocybe aegerita using PEG-mediated method. Mycosystema 39, 1100–1108. doi: 10.13346/j.mycosystema.200068
Tayeh, M. A., and Marletta, M. A. (1989). Macrophage oxidation of L-arginine to nitric oxide, nitrite, and nitrate. Tetrahydrobiopterin is required as a cofactor. J. Biol. Chem. 264, 19654–19658. doi: 10.1016/S0021-9258(19)47163-9
Vieira, A. L., Linares, E., Augusto, O., and Gomes, S. L. (2009). Evidence of a Ca(2+)-(*)NO-cGMP signaling pathway controlling zoospore biogenesis in the aquatic fungus Blastocladiella emersonii. Fungal. Genet. Biol. 46, 575–584. doi: 10.1016/j.fgb.2009.04.002
Wang, C., Chao, Y., Xu, W., Liang, M., Deng, S., Zhang, D., et al. (2020). CTRP13 Preserves endothelial function by targeting GTP cyclohydrolase 1 in diabetes. Diabetes 69, 99–111. doi: 10.2337/db19-0635
Werner, E. R., Blau, N., and Thöny, B. (2011). Tetrahydrobiopterin: biochemistry and pathophysiology. Biochem. J. 438, 397–414. doi: 10.1042/BJ20110293
Xu, Q., Li, K., Sun, Q., Ding, D., Zhao, Y., Yang, N., et al. (2017). Rare GCH1 heterozygous variants contributing to Parkinson’s disease. Brain 140:e41. doi: 10.1093/brain/awx110
Zhang, J. X., Chen, Q., Huang, C. Y., Gao, W., and Qu, J. B. (2015). History, current situation and trend of edible mushroom industry development. Mycosystema 34, 524–540.
Keywords: GCH1, fruiting body development, Cyclocybe aegerita, RNAi, hyphal branch
Citation: Tao N, Cheng B, Chai H, Cui X, Ma Y, Yan J, Zhao Y and Chen W (2022) A Putative Guanosine Triphosphate Cyclohydrolase I Named CaGCH1 Is Involved in Hyphal Branching and Fruiting Development in Cyclocybe aegerita. Front. Microbiol. 13:870658. doi: 10.3389/fmicb.2022.870658
Received: 07 February 2022; Accepted: 15 March 2022;
Published: 22 April 2022.
Edited by:
Jing Si, Beijing Forestry University, ChinaReviewed by:
Ryo Nasuno, Nara Institute of Science and Technology (NAIST), JapanCopyright © 2022 Tao, Cheng, Chai, Cui, Ma, Yan, Zhao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongchang Zhao, eWFhc211c2hyb29tQGFsaXl1bi5jb20=; Weimin Chen, Y2h3bWttQGFsaXl1bi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.