
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 06 April 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.869278
Pseudomonas plecoglossicida is a facultative fish pathogen that possesses three distinct type VI secretion systems (named T6SS-1, T6SS-2, and T6SS-3). Our previous work indicated that only T6SS-2 of P. plecoglossicida mediates interbacterial competition. However, the antibacterial T6SS effectors and their functions are unclear. Here, we reported two T6SS effectors that mediate antibacterial activity. We first identified four putative antibacterial effectors (denoted as Txe1, Txe2, Txe3, and Txe4) and their cognate immunity proteins encoded in P. plecoglossicida strain XSDHY-P by analyzing the regions downstream of three vgrG genes. We showed that the growth of Escherichia coli cells expressing Txe1, Txe2, and Txe4 was inhibited, and these three effectors exhibited nuclease activity in vivo. The interbacterial competition assays with single- or multi-effector deletion mutants as attackers revealed that Txe1 was the predominant T6SS toxin of P. plecoglossicida strain XSDHY-P mediating the interbacterial killing. This work contributes to our understanding of bacterial effectors involved in the interbacterial competition.
To survive and reproduce, bacteria need to compete with other microorganisms for nutrients and space. Many Gram-negative bacteria can utilize toxic proteins, known as “effectors” translocated by the type VI secretion system (T6SS) to eliminate their competitors (Basler et al., 2013; Cianfanelli et al., 2016; Hernandez et al., 2020). The T6SS is a contractile puncturing device with a needle-like structure, consisting of contractile sheath, an inner tube (composed of stacked rings of Hcp hexamers), and a transmembrane-baseplate complex (Mougous et al., 2006; Leiman et al., 2009). The Hcp tube is packed in the sheath. On top of the Hcp tube is a trimeric VgrG spike on which sits a cone-shaped proline–alanine–alanine–arginine repeat family (PAAR) domain-containing protein sharpening the tip (Cascales and Cambillau, 2012; Shneider et al., 2013). In response to unclear extracellular signals, the sheath contracts causing the needle structure to be expelled and penetrate the target membrane, thereby translocating effectors into neighboring bacterial or eukaryotic cells (Cascales and Cambillau, 2012; Jurėnas and Journet, 2021).
The T6SS needle structure carries effectors occurring in two modes. One class of effectors comprises covalent extensions of one of the components of the needle structure (Hcp, VgrG, or PAAR), and these are coined as “specialized” or “evolved” effectors. In contrast, “cargo” effectors non-covalently interact with Hcp, PAAR, or VgrG proteins. Often, adaptor or chaperone proteins are essential for stabilization and/or recruitment of effectors during assembly (Unterweger et al., 2017; Manera et al., 2022). Some of the T6SS adaptor proteins are those belonging to the DUF1795, DUF2169, and DUF4123 families. DUF1795 family proteins, termed Eag (effector-associated gene) adaptors, bind to the N-terminal half of PAAR-containing T6SS effectors, acting as a chaperone to stabilize the transmembrane domains of “specialized” effectors during the translocation across bacterial cells (Diniz et al., 2015; Whitney et al., 2015; Jurėnas et al., 2021). DUF2169 and DUF4123 family proteins seem to be less prevalent and were shown to assist the loading of “cargo” effectors onto the VgrG spike (Unterweger et al., 2015; Bondage et al., 2016; Pei et al., 2020).
Antimicrobial properties exist in many T6SS effectors, including those toxins with activities of nucleases, phospholipases, and peptidoglycan hydrolases (Durand et al., 2014). These effectors are toxic to essential cellular targets and cause either lysis or death of the attacked bacterial cells. Antibacterial effectors are often accompanied by cognate immunity proteins that confer self-protection (Russell et al., 2011) while preventing the bacteria from being damaged by sibling cells (MacIntyre et al., 2010). The antibacterial effector genes are often found immediately upstream of cognate immunity genes (Lewis et al., 2019).
Recently, we reported that a fish pathogenic Pseudomonas plecoglossicida strain carries three T6SS clusters (T6SS-1, T6SS-2, and T6SS-3), and each encodes a full set of core T6SS components (Tao et al., 2020). Our study specifically indicated that T6SS-2 participates in bacterial killing and is conserved across strains from either fish host or environmental sources. The T6SS-2 activity lends a competitive advantage to P. plecoglossicida over bacteria such as Photobacterium damselae subsp. damselae that is probably a competitor for host fish niches (Tao et al., 2020). It is unclear which effectors are deployed by P. plecoglossicida T6SS-2 to eliminate bacterial competitors from similar environments. This work aims to identify and characterize T6SS-2-dependent antibacterial effectors used by a fish pathogenic P. plecoglossicida strain XSDHY-P.
Bacterial strains used are listed in Supplementary Table S1. Pseudomonas plecoglossicida strain XSDHY-P (Zhang et al., 2014) and its derivatives were routinely grown in Luria–Bertani (LB) broth or solid medium (supplemented with 1.5% agar) at 28°C. Escherichia coli strain DH5α was used for plasmid maintenance and amplification. Escherichia coli strain BL21 (DE3) was used in growth inhibition assays. Escherichia coli strain XL10 (purchased from Vazyme Biotech, Nanjing, China) was used as the prey strain in interbacterial competition assays. The E. coli strains were cultured at 37°C and were grown in/on LB broth or agar plates at 28°C. Media were supplemented with the following antibiotics as required: ampicillin at 100 μg/ml, chloramphenicol at 25 μg/ml, kanamycin at 50 μg/ml, and tetracycline at 15 μg/ml.
The known antibacterial T6SS effectors are often encoded in the vicinity of or fused to vgrG genes (Hachani et al., 2014; Bernal et al., 2017; Wettstadt et al., 2019; Wood et al., 2019). Consequently, to identify putative effector and immunity proteins, we first searched for all putative vgrG genes encoded in P. plecoglossicida XSDHY-P. To this end, we used amino acid sequences of previously identified VgrG-1 (DVB73_RS06775), VgrG-2 (DVB73_RS03075), and VgrG-3 (DVB73_RS25130) to perform TBLASTN (Altschul et al., 1997) queries against a database of translated protein sequences belonging to the single strain XSDHY-P at www.pseudomonas.com with default settings (Winsor et al., 2016). Open-reading frames (ORFs) presented in vgrG gene neighborhoods were manually inspected using BLASTp searches against the NCBI “nr” database and analyzed for conserved domains using the NCBI Conserved Domain Database (Marchler-Bauer et al., 2017) tool for conserved effector domains. The SecreT6 webtool for T6SS-related protein prediction was used to search putative effector and immunity proteins against the database using an E-value threshold of e-4 (Li et al., 2015). Only those putative effectors encoded with downstream immunity proteins were included for further analysis. A putative immunity protein was identified using the following criteria: (a) the protein was encoded downstream (within 50 bp) of the cognate effector gene and (b) the length of the protein was at least 50 amino acids and was smaller than its cognate effector. PSORTb (v3.0.2) was used to predict subcellular localization of putative immunity proteins (Yu et al., 2010), thereby inferring the location of their cognate effectors, since immunity proteins are required in the correct cellular compartment of effector-producing cells to neutralize effector toxicity for self-protection. TMHMM (v.2.0) was used to predict transmembrane domains (Krogh et al., 2001), and SignalP 5.0 was used to predict signal peptides (Almagro Armenteros et al., 2019). BLASTp (Altschul et al., 1997) was used to calculate the identities between effector proteins. The deep-learning approach, AlphaFold2, was used to predict protein tertiary structures (Jumper et al., 2021). ESPript was used to visualize the sequence alignment (Robert and Gouet, 2014).
The amino acid sequences of analyzed proteins were multi-aligned with ClustalOmega (Sievers et al., 2011). The maximum likelihood (ML) phylogenetic tree was constructed using IQ-TREE (Trifinopoulos et al., 2016) under the LG + F + I + Gamma model, with 1,000 ultrafast bootstrap replicates (Minh et al., 2013). The phylogenetic tree was visualized in iTOL (Letunic and Bork, 2016).
Plasmids used are listed in Supplementary Table S1. To analyze the effector/immunity activity in E. coli, coding regions for putative effectors with or without cognate immunity genes were amplified from P. plecoglossicida XSDHY-P genomic DNA using Phanta® Max Super-Fidelity DNA Polymerase (Vazyme Biotech, China). The amplified fragments were cloned into the multiple cloning site (MCS) of the L-arabinose inducible vector, pBAD33.1 (Addgne#36267) either by Gibson assembly (Gibson et al., 2009) or by restriction cloning. For truncation analysis of effectors, the C-terminal domain of each effector coding sequence was inserted into pBAD33.1. Plasmid constructs were transformed into E. coli BL21(DE3) and confirmed by DNA sequencing. All primers used are listed in Supplementary Table S2.
In-frame deletion mutants of P. plecoglossicida were generated by two-step allelic exchange based on the suicide vector pK18mobSacB (Kvitko et al., 2009) as previously described (Tao et al., 2020). Briefly, 500–1,000 bp upstream and downstream of the gene to be deleted were amplified and then fused by overlapping PCR. The fused fragments were cloned into the suicide vector to construct the mutation plasmids (Supplementary Table S1). Mutation plasmids were transferred to P. plecoglossicida by conjugation. All mutants were verified by PCR. Mutants with multiple deletions were generated stepwisely. For multiple-deletion mutants, target genes were knocked out in a stepwise manner. The double-mutant Δtxe1Δtxe4 was generated by deleting txe4 on the Δtxe1 genetic background. To obtain the triple-mutant Δtxe1Δtxe2Δtxe4, txe2 was knocked out from the Δtxe1Δtxe4 mutant.
A bacterial toxicity assay was performed as previously described with several modifications (Jana et al., 2019). To test the toxicity of putative antibacterial effectors, E. coli BL21(DE3) with recombinant plasmids or an empty pBAD33.1 vector was streaked on LB agar containing either 0.2% glucose (repressing conditions) or 0.2% L-arabinose (inducing conditions) and chloramphenicol. The agar plates were incubated at 37°C. Images were acquired after 24 h.
To verify the protection conferred by immunity proteins, pBAD33.1 carrying both predicted effector and downstream immunity genes was transformed into E. coli BL21(DE3). Overnight cultures of E. coli cells were diluted 1:100 into fresh LB medium containing chloramphenicol and grown for 3 h at 37°C, 150 rpm. Each culture was normalized to an OD of 1.0 and diluted 100-fold in fresh LB with same antibiotics and divided into three sterile flasks as biological replicates. The initial OD600 was measured, and the flasks were incubated at 37°C, 150 rpm. After a 1.5-h incubation, E. coli cells were induced by adding 20% L-arabinose to a final concentration of 0.2% (wt/vol). The cultures continued to be grown under same conditions up to 9 h, and the OD600 was measured each hour since 30 min post-induction. A growth curve was plotted as the optical density (OD600) vs. the culture time. In addition, 10-fold dilutions were made from one replicate of each culture after incubation. Five-microliter aliquots of the dilutions was dropped onto LB agar to estimate the number of viable cells.
The plasmid DNA degradation analysis in E. coli cells was carried out as described previously (Ma et al., 2014). In brief, E. coli BL21(DE3) with recombinant or empty pBAD 33.1 have grown in LB containing chloramphenicol to an OD600 of 0.6–0.8. The expression of putative effector alone (or with cognate immunity) was then induced by 0.2% L-arabinose for 3 h. Equivalents of 1 ml of cells at OD600 = 1.0 from each culture were pelleted for plasmid extraction. Plasmid DNA was extracted using an E.Z.N.A. Plasmid DNA Mini Kit (Omega Bio-Tek, Inc., GA, United States). Plasmid DNA samples were analyzed by gel electrophoresis on a 1% agarose gel containing 1× GelRed (Biotium, Hayward, CA, United States) and run at 10 V/cm for 30 min. DNA was visualized under UV light transillumination using a Gel Doc system (BioRad, La Jolla, CA, United States).
Interbacterial competition assays were performed as previously described (Tao et al., 2020). In brief, wild-type P. plecoglossicida XSDHY-P and indicated mutants (sensitive to tetracycline) were used as attackers, and E. coli XL10 (resistant to tetracycline), which is susceptible to P. plecoglossicida T6SS-2-dependent killing (Tao et al., 2020), was used as the prey. Overnight cultures of bacterial strains were subcultured (1:50) in fresh LB for 3 h. The subcultures were adjusted to an OD600 of 0.5 in PBS and mixed at a 10:1 ratio of attacker to prey. Mixtures of 25 μl were spotted in triplicate onto 0.45-μm filter membranes that were placed on LB agar. The plates were incubated for 4 h at 30°C. Bacterial cells were dislodged from the membranes into 1 ml LB in 15-ml tubes by vortexing. The prey cells were enumerated by counting tetracycline-resistant colony-forming units (CFUs). The fold change in prey CFUs was calculated by dividing CFUs of the input (at 0 h) by the output (at 4 h). The prey growth index (PGI) was defined as log2 fold change in prey CFUs, which is a measure of how efficient an attacker strain is at prey-strain killing. A higher PGI indicates a lower ability of the attacker to kill the prey strain. The interbacterial competition assay was performed in triplicate.
Statistical analyses and figure production were performed using GraphPad Prism (version 8). One-way ANOVA, followed by Fisher’s least significant difference (LSD) test, was used to compare the PGI values between each effector mutant strain and wild-type strain or T6SS-2 inactivate mutant in the interbacterial competition assay. The differences were considered significant at p < 0.05.
T6SS effectors and cognate immunity proteins are frequently found to be genetically linked to vgrG genes (Hachani et al., 2011, 2014; Durand et al., 2014; Bondage et al., 2016; Ma et al., 2018; Lopez et al., 2020). We did TBLASTN searches using three known VgrGs as queries to identify all vgrG genes in strain XSDHY-P. We found two additional vgrG genes that were located remotely from three T6SS loci (Figure 1A). These two newly identified vgrG genes were arbitrarily named as vgrG-4 (DVB73_RS04055) and vgrG-5 (DVB73_RS08345). A pairwise analysis of amino acid identity showed that five VgrG proteins encoded by XSDHY-P shared 20.4%–71.6% identity; VgrG-5 (71.6%) had the highest sequence identity with VgrG-2, followed by VgrG-4 (58.6%; Figure 1B; Supplementary Figure S1). ML phylogenetic analysis of VgrGs from XSDHY-P showed that VgrG-5 and VgrG-2 were sister groups, together forming a cluster with VgrG-4, and they were separated from VgrG-1 or VgrG-3 (Figure 1C).
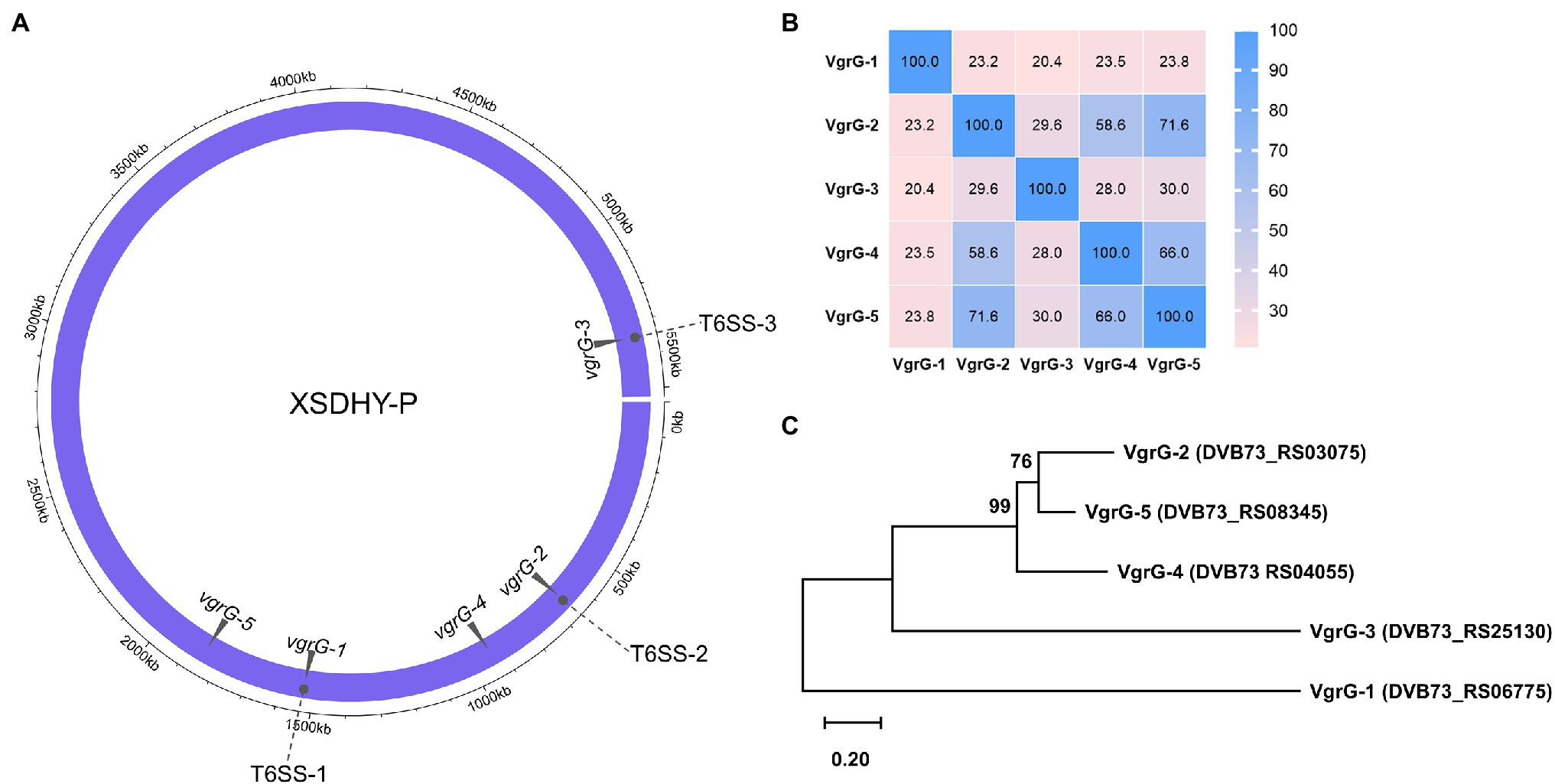
Figure 1. Distribution of the T6SS-1, T6SS-2, and T6SS-3 clusters, and the vgrG genes in the Pseudomonas plecoglossicida strain XSDHY-P (A). Heatmap showing identity comparison matrix of sequence alignment of VgrG proteins (B). A color key indicates the correspondence between pairwise identities and the colors displayed in the matrix. Phylogeny of VgrG proteins (C). Maximum likelihood tree showing the phylogenetic relationship of VgrG proteins in P. plecoglossicida XSDHY-P strain. Bootstrap n = 1,000 constructed based on ClustalOmega alignment of VgrG amino acid sequences.
We inspected ORFs within vgrG-2, vgrG-4, and vgrG-5 clusters for the presence of potential antibacterial E/I pairs. The vgrG-1 and vgrG-3 clusters were not analyzed, since T6SS-1 and T6SS-3 are not involved in antibacterial activities (Tao et al., 2020). A closer inspection of genes from relevant vgrG clusters allowed us to identify two putative E/I pairs within vgrG-2 cluster (Figure 2A) and one E/I pair within vgrG-4 and vgrG-5 clusters each (Figures 2B,C) according to the aforementioned criteria. These putative Type six XSDHY-P effector and immunity proteins were named Txe1 to 4 and Txi1 to 4, respectively. Notably, gene txe2, DVB73_RS03050 was previously annotated as a pseudogene, yet manual inspection of the ORF revealed an intact gene (positions 14,787–11,465 in reference sequence NZ_CP031146.1). Between vgrG and effector genes, one or two EagR adaptor (DUF1795 domain-containing protein) genes were identified. There was a striking similarity between vgrG-2 and vgrG-5 clusters in terms of genetic organization, as well as amino acid sequences of protein counterparts encoded between the two clusters (Figure 1B; Supplementary Figure S1). For example, EagR adaptor proteins encoded between vgrG-2 and vgrG-5 clusters were almost identical that EgaR-2a and EgaR-5a shared 96% amino acid identity, and EagR-2b and EagR-5b were identical (Supplementary Figure S2). In contrast, the homology of EagR-2a, EagR-2b, EagR-5a, and EagR-5b to EagR-4 from vgrG-4 cluster was low (22%–27% sequence identity). Considering the interaction between VgrG and Hcp proteins during T6SS assembly (Silverman et al., 2012) and the role of EagR in effector loading (Jurėnas and Journet, 2021), these data suggest that vgrG-2- and vgrG-5-associated effectors are likely linked to the same T6SS apparatus (i.e., T6SS-2) in strain XSDHYP.
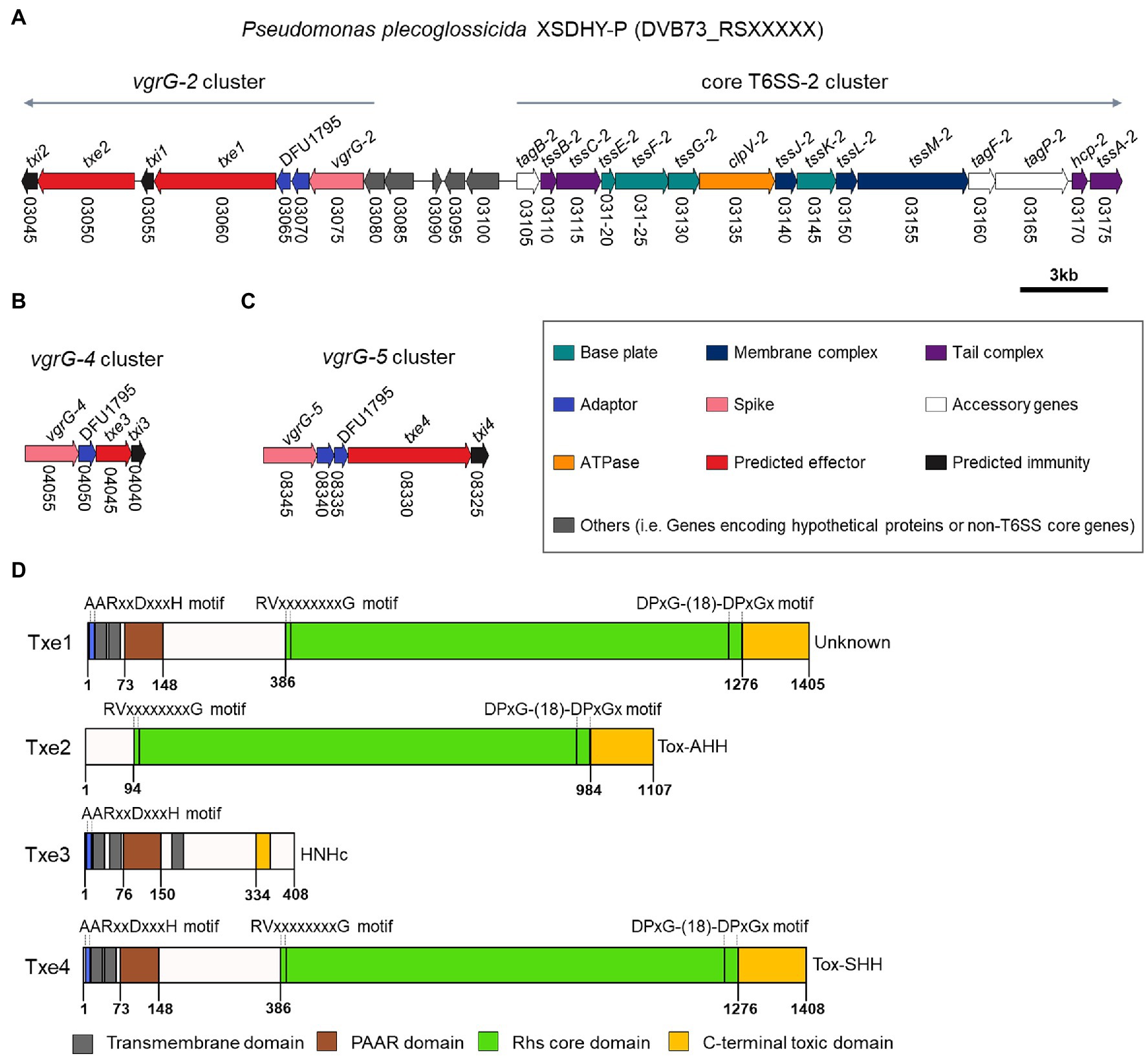
Figure 2. Genomic organization of T6SS-2 main cluster and vgrG-2 cluster (A), vgrG-4 cluster (B), and vgrG-5 cluster (C) from P. plecoglossicida XSDHY-P. Genes are represented as arrows and the direction represents the direction of transcription. Genes are color-coded to highlight their predicted functions as described in the legend; colors specific to each gene are used to indicate genes that encode similar proteins in other clusters. Gene nomenclature is indicated at the top, and locus numbers are shown at the bottom. Predicted domain composition of four putative antibacterial T6SS effectors identified from P. plecoglossicida XSDHY-P (D). Prediction was determined using the NCBI Conserved Domain Database tool (Marchler-Bauer et al., 2017). Schematic diagrams show the linear organization of relevant domains of putative effectors Txe1, Txe2, Txe3, and Txe4. The domains shown are: prePAAR motif (blue); transmembrane helices (dark grey); PAAR domain (brown); Rhs core domain (green); and C-terminal toxic domain (orange) as described in the legend. The lengths of the effector proteins and different domains are drawn to scale.
The immunity protein is required in the correct cellular compartment for protection against self- and/or sibling damage by its cognate effector (Lewis et al., 2019). Consequently, to infer the subcellular target locations of the putative effectors, we predicted the subcellular locations of potential immunity proteins, Txi1, Txi2, Txi3, and Txi4 using PSORTb and SOSUI GramN tools. All four putative immunity proteins were predicted to be cytoplasmic by at least one tool (Supplementary Table S3). Signal peptide analysis using the SignalP 5.0 program revealed no signal peptides present in any of immunity proteins (Supplementary Table S3). Moreover, none of the immunity protein contained transmembrane domains (Supplementary Table S3). These results suggest that the effectors Txe1, Txe2, Txe3, and Txe4 are likely to be toxic to bacterial cells in their cytoplasm.
Our analyses using the SecReT6 webtool (Li et al., 2015) revealed that Tke2 (an Rhs effector with an N-terminal PAAR domain and C-terminal HNH nuclease domain) from P. putida KT2440 (Bernal et al., 2017) was the best hit for Txe1, Txe2, and Txe4 with identity ranging from 87.4% to 90.8%. A sequence alignment analysis revealed that the four effectors harbored distinct CT domains (Supplementary Figure S3). To further predict the putative functions, we used sequence analysis to determine the domain structures of the four putative effectors. The domain architectures of each effector are shown in Figure 2D and Supplementary Table S3. Txe1 contains an N-terminal (NT) PAAR domain, a canonical Rhs (after “Rearrangement hot spots”) core domain [delimited by specific motifs matched to previously identified RVxxxxxxxxG and DPxG-(18)-DPxGx peptide motifs; Jackson et al., 2009; Hachani et al., 2014], and a C-terminal (CT) extension of 130 amino acids. The CT extension of Txe1 showed no similarity to any known domains in the CDD database. Txe2 contains a conserved Rhs core domain and a Tox-AHH toxin (pfam14412; HNH/ENDO VII nuclease superfamily). Txe3 contains an NT PAAR and a putative HNHc endonuclease (pfam01844). Txe4 has a similar domain architecture with Txe1, carrying a Tox-SHH toxin (pfam15652; HNH/Endo VII superfamily toxin with an SHH signature) at its C-terminus. In addition, Txe1, Txe3, and Txe4 encode prePAAR motif and TMDs (Figure 2D; Supplementary Figure S4). In addition, Txe1, Txe3, and Txe4 each contain a prePAAR motif and TMDs (Figure 2D; Supplementary Figure S4). It has been reported that prePAAR motifs are found in TMD-containing effectors, directing recruitment of cognate Eag proteins to facilitate folding and maintenance of the hydrophobic helices (Ahmad et al., 2020). According to the different number of TMDs, the effectors containing TMDs were proposed to be classified as Class I and Class II effectors (Ahmad et al., 2020). This proposed classification scheme assigns Txe1 and Txe4 to Class I and Txe3 to Class II. In addition, the potential toxic CT domains of Txe1, Txe3, and Txe4 are fused to each NT PAAR domain, so these three toxins are classified as “specialized” effectors (Durand et al., 2014).
To validate the antibacterial activity of putative T6SS effectors, we cloned each Txe protein individually into the expression plasmid pBAD33.1 under the control of a PBAD promoter (induced by L-arabinose and repressed by D-glucose) in E. coli BL21 (DE3). Escherichia coli cells were induced to express the four Txe proteins only for cytoplasmic localization, because the potential immunity proteins were predicted to be cytoplasmic. The results showed that induced expression of txe1, txe2, and txe4 within the cytoplasm of E. coli was detrimental (Figure 3A), confirming that Txe1, Txe2, and Txe4 are cytoplasmic toxins. Unexpectedly, the expression of txe3 did not arrest the growth of E. coli, though an HNHc endonuclease motif was predicted in Txe3. Truncation analysis demonstrated that heterologous expression of only the CT regions of Txe1 (residues 1,276–1,405), Txe2 (residues 984–1,107), and Txe4 (residues 1,276–1,408) was adequate to produce the observed toxicity (Figure 3B).
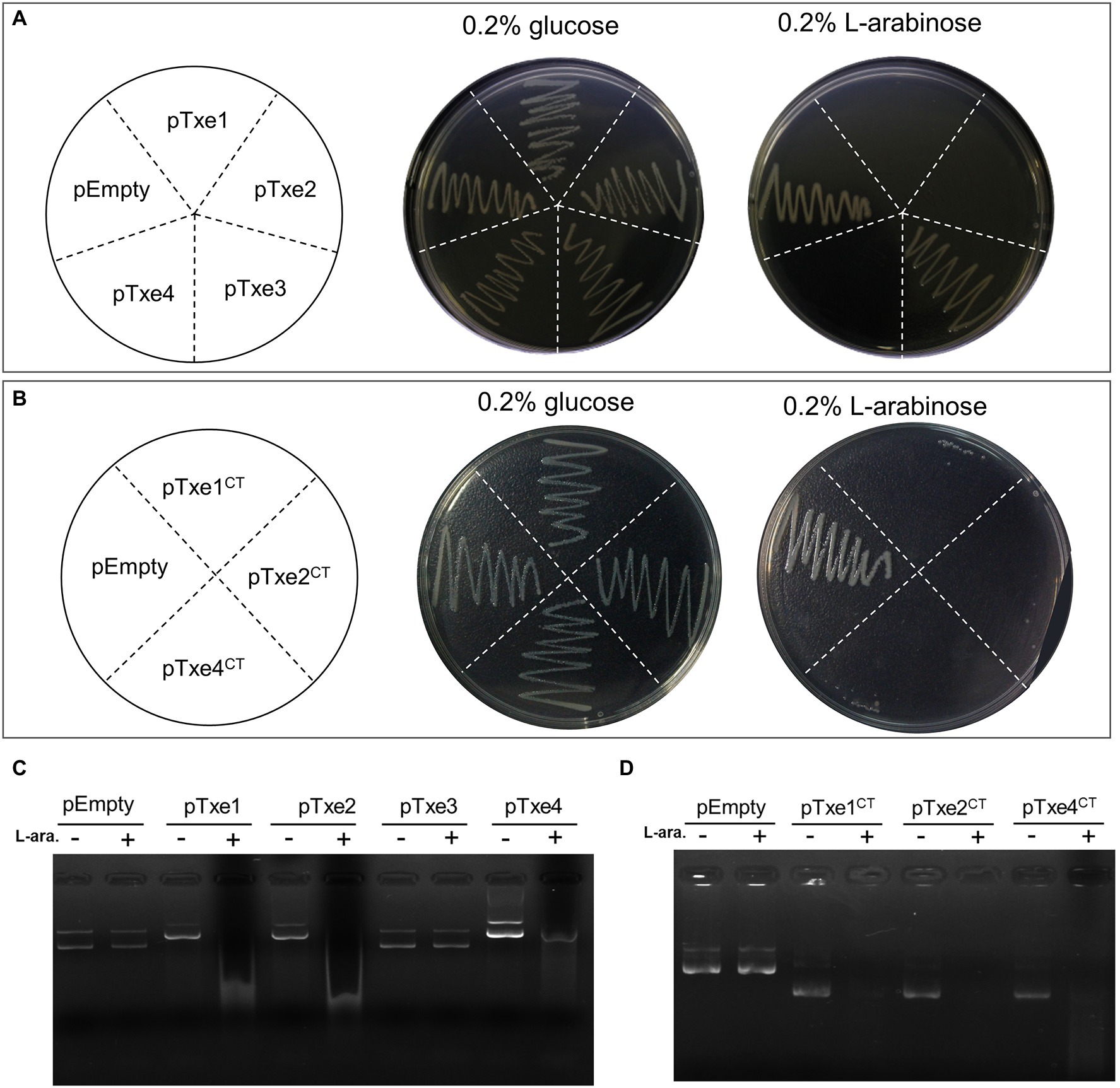
Figure 3. Txe1, Txe2, and Txe4 are three DNase toxins. Txe1, Txe2, or Txe4 were expressed on arabinose-inducible expression plasmid pBAD33.1 vector in Escherichia coli BL21(DE3), and the growth of E. coli was tested by streaking on LB agar containing 0.2% L-arabinose (induction) and/or 0.2% glucose (repression). The E. coli toxicity was not observed for expressed Txe3 (A). Toxicity of CT domains of Txe1 (residues 1,276–1,405), Txe2 (residues 984–1,107), and Txe4 (residues 1,276–1,408) expressed in E. coli BL21(DE3) from arabinose-inducible expression plasmids (B). Induced expression of full-length (C) or C-terminal extension (D) of Txe1, Txe2, or Txe4 for 3 h resulted in plasmid DNA degradation in vivo. Degradation was not observed for expressed Txe3. + And − symbols indicate addition or not of L-arabinose, respectively. The pBAD33.1 was transformed into E. coli BL21(DE3) to act as an empty vector control (pEmpty) for all assays. Representative data from three independent experiments are shown.
To assess the nuclease activity of the predicted effectors, we examined the degradation of the plasmid DNA extracted from E. coli strains after arabinose induction for 2 h. Expression of either txe1, txe2, or txe4 for 3 h resulted in the degradation of plasmid DNA in vivo (Figure 3C) as well for their respective CT domains (Figure 3D). Consistent with the results of the toxicity assay, expression of txe3 did not cause plasmid DNA degradation (Figure 3C). These results confirmed that Txe2 and Txe4 are DNase toxins as predicted from sequence analysis and demonstrated that Txe1 also has nuclease activity.
By using a query of Txe1 CT domain sequence, the BLASTp search revealed that homologs of this CT nuclease domain were also carried by some Rhs-repeats containing proteins from other bacterial species (data not shown). Partial sequence alignment of CT region of Txe1 with representative BLASTp hits showed that these proteins have a conserved dipeptide HH motif (H1362 and H1363; Figure 4A). These two conserved histidine residues are part of the loop (loop 4) connecting helix α6 and sheet β3 in a Txe1-CT domain structure model predicted by AlphaFold2 (Figure 4B). In this predicted structure model, sheets β3 and β4 and helices α6 and α7 together form the tertiary structure similar to the His-Me finger from caspase-activated DNase (PDB ID: 1v0d; Jablonska et al., 2017). Probably, the conserved dual histidine motif plays an indispensable role in the enzyme activity of Txe1. Here, we targeted H1362 and probed if it is import for the nuclease activity of Txe1. The histidine 1,362 was changed to alanine by site-directed mutagenesis (Figure 4C). As a result, the substitution of H1362 (H1362A) abolished the nuclease activity of Txe1 CT domain (Figure 4D) and its toxicity to host E. coli cells (Figure 4E). Hence, the residue H1362 is crucial for the DNase activity of Txe1.
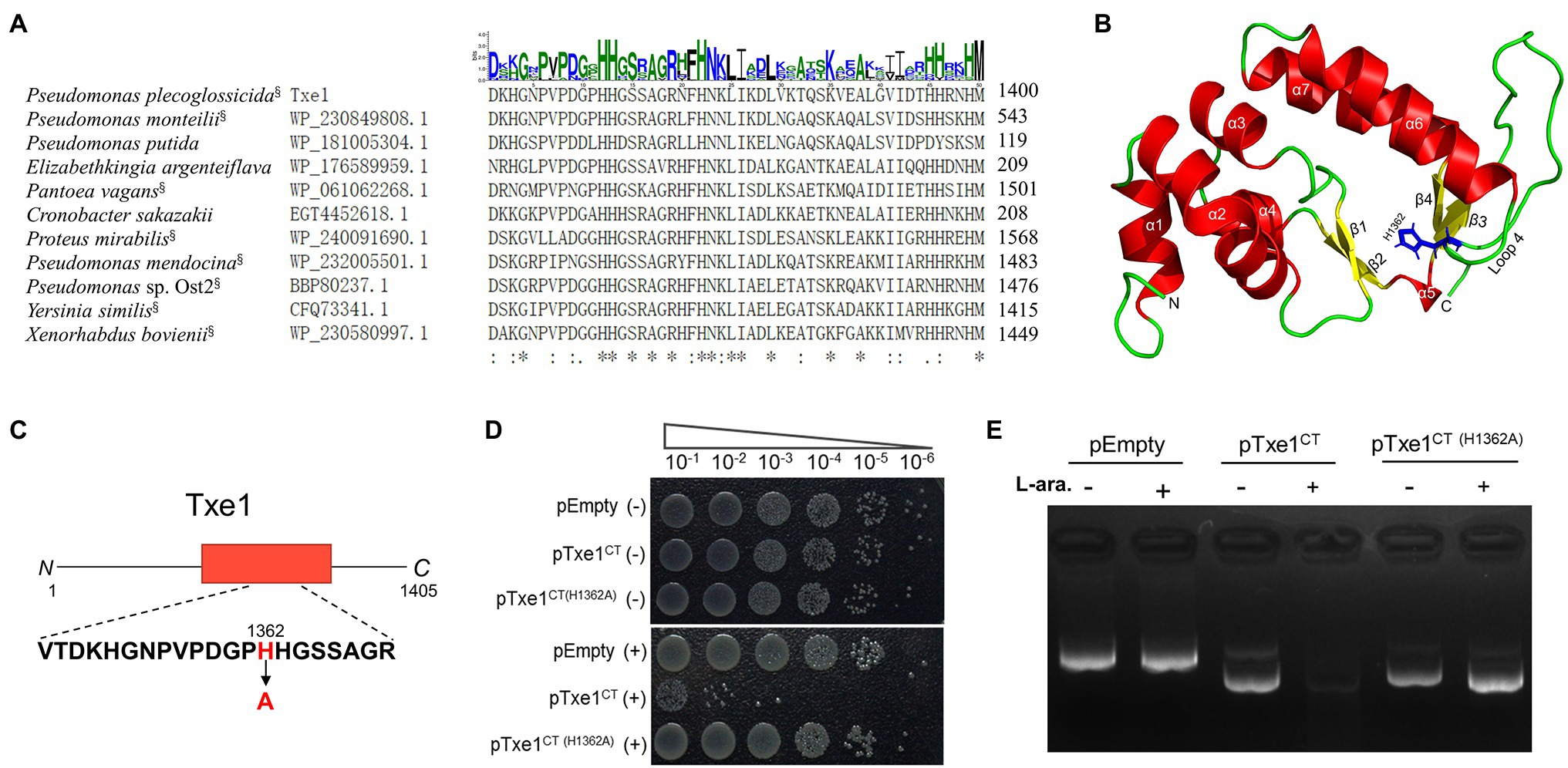
Figure 4. Mutation of H1362 abolishes toxicity of Txe1. Partial sequence alignment of Txe1 CT region with representative hits of BLASTp analysis (A). In each case, the species name and accession number are on the left, and the numbering for the last residue is on the right. CT domains that are linked to N terminal Rhs domain are indicted with a superscript “§” behind the species name. On the top is the sequence logo representation of the aligned region of 50 residues (this region contains all the identical histidine residues in the protein sequence alignment of Txe1 CT domain with the 100 best BlastP hits.). Consensus sequences were plotted with WebLogo web-based tool (http://weblogo.threeplusone.com/create.cgi). Height of letter indicates degree of conservation. Residues identical in all species are indicated by “*” on the bottom. AlphaFold2-generated tertiary structure models for the CT domain of Txe1 (B). Txe1 CT domain is colored by secondary structure: helices (red), sheets (yellow), and loop (green). A tertiary structure similar to the His-Me finger from caspase-activated DNase (PDB ID: 1v0d) is formed by sheets β3 and β4 and helices α6 and α7. Schematic representation indicating the mutation site of Txe1 (C). Mutation of histidine 1,362 to alanine (H1362A) in Txe1 abolishes toxicity (D). Growth of E. coli BL21(DE3) carrying empty pBAD33.1 (pEmpty) or plasmids directing the expression of the Txe1 CT domain or mutant on LB agar (E). + And − symbols indicate addition or not of 0.2% L-arabinose, respectively. The experiment was repeated twice with similar results, and the results from one representative experiment are shown.
Antibacterial T6SS toxins are usually encoded along with a cognate immunity gene (Dong et al., 2013). To examine whether those proteins (i.e., Txi1, Txi2, and Txi4) encoded immediately downstream of txe1, txe2, and txe4 are immunity proteins, we also cloned DNA fragments encoding each predicted E/I pair into the vector pBAD33.1. When induced with L-arabinose, Txe1, Txe2, and Txe4 were expressed in E. coli cytoplasm and exerted a bacteriostatic effect as measured by OD600 (Figures 5A–C). The growth inhibition of each effector was strikingly neutralized by the co-expression of the predicted immunity genes (i.e., txi1, txi2, and txi4; Figures 5A–C). Results of serial dilution spotting at the end of the experiment showed that viable cells of E. coli strains that co-expressed effector and immunity genes were 100–1,000 times higher than those that only expressed effector genes (Figures 5D–F). Moreover, the plasmid degradation analysis showed that co-production of Txi1, Txi2, and Txi4 partially suppressed the nuclease activity of cognate effectors (Supplementary Figure S5; Figure 3C). These results suggest that Txi1, Txi2, and Txi4 function as immunity proteins for Txe1, Txe2, and Txe4, respectively.
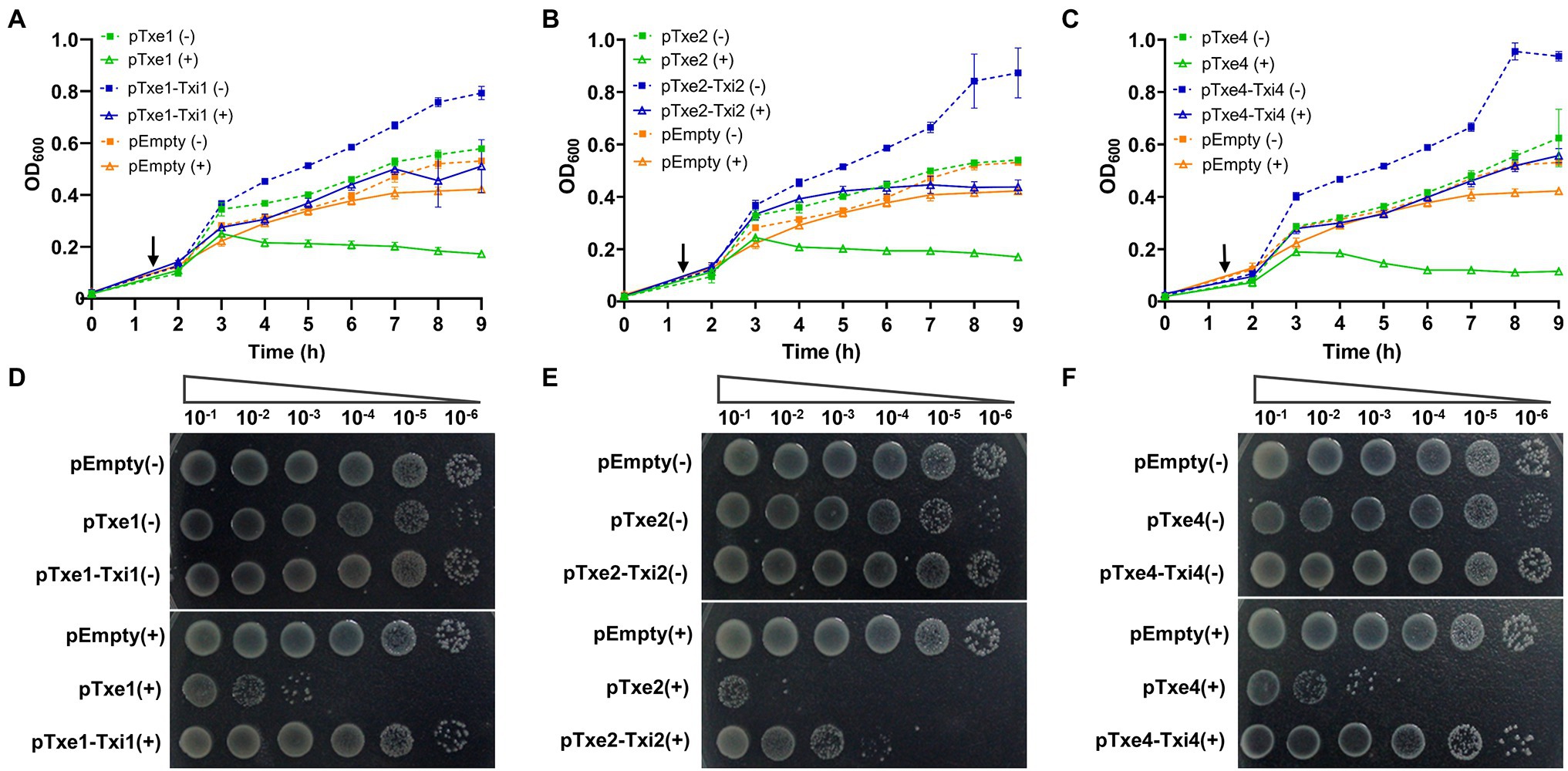
Figure 5. Txe1-Txei1, Txe2-Txei2, and Txe4-Txei4 are three effector–immunity pairs. Growth curves of E. coli BL21(DE3) harboring indicated plasmids were obtained by measuring the OD600 at 1-h intervals (A–C). Data are presented as the mean ± SD of three independent experiments. Levels of survival of cells were compared by 10-fold serial dilution on LB medium containing arabinose or not (D–F). The arrow indicates the time of induction with 0.2% L-arabinose, and (+) and (−) symbols indicate addition or not of L-arabinose, respectively. The experiment was repeated twice with similar results, and the results from a representative experiment are shown.
To probe the role of four Txe effectors in XSDHY-P T6SS-2-related antibacterial activity, we generated a series of single- or multiple-effector deletion mutants. For all these mutants, only the indicated effector gene(s) were deleted, and the cognate immunity gene(s) were kept in the genome. We first assessed the contribution of each individual effector in interspecies competition. To do so, we performed interbacterial competition assays between Δtxe1, Δtxe2, Δtxe3, or Δtxe4 (attacker strains) and E. coli strain XL10 (prey strain carrying tetracycline resistance). To measure the contribution of given effector(s) to T6SS killing, we determined the PGI, which reflects the survival of prey in co-culture with attacked strains on solid media. A high PGI indicates a higher survival rate for prey; a positive PGI value means a greater number of viable prey cells than baseline hour zero. Compared with the WT strain, the PGI was significantly higher in mutants deletion of txe1 alone or in combination with txe4 (p < 0.05), indicating that these two effectors contribute to the bacterial killing activity. Interestingly, no significant difference in PGI was observed between the WT strain and Δtxe2 or Δtxe3 (Figure 6A), although Txe2 encodes the toxic CT domain. Among all the single-deletion mutants, it was only when Δtxe1 was used as the attacker that the PGI value was positive, indicating that the deletion of txe1 attenuated T6SS killing the most (Figure 6A). The PGIs of Δtxe1 and Δtxe4 were all significantly lower than that of the T6SS-2-inactivated strain, ΔT6SS-2 (Figure 6A), indicating that mutants Δtxe1 and Δtxe4 possessed higher ability of bacterial killing than mutant ΔT6SS-2. Given that T6SS-2 is the sole T6SS present in P. plecoglossicida strain XSDHY-P involved in bacterial competition under the same experimental condition (Tao et al., 2020), these data suggest that the deletion of txe1 or txe4 alone did not completely abolish T6SS-2 killing.
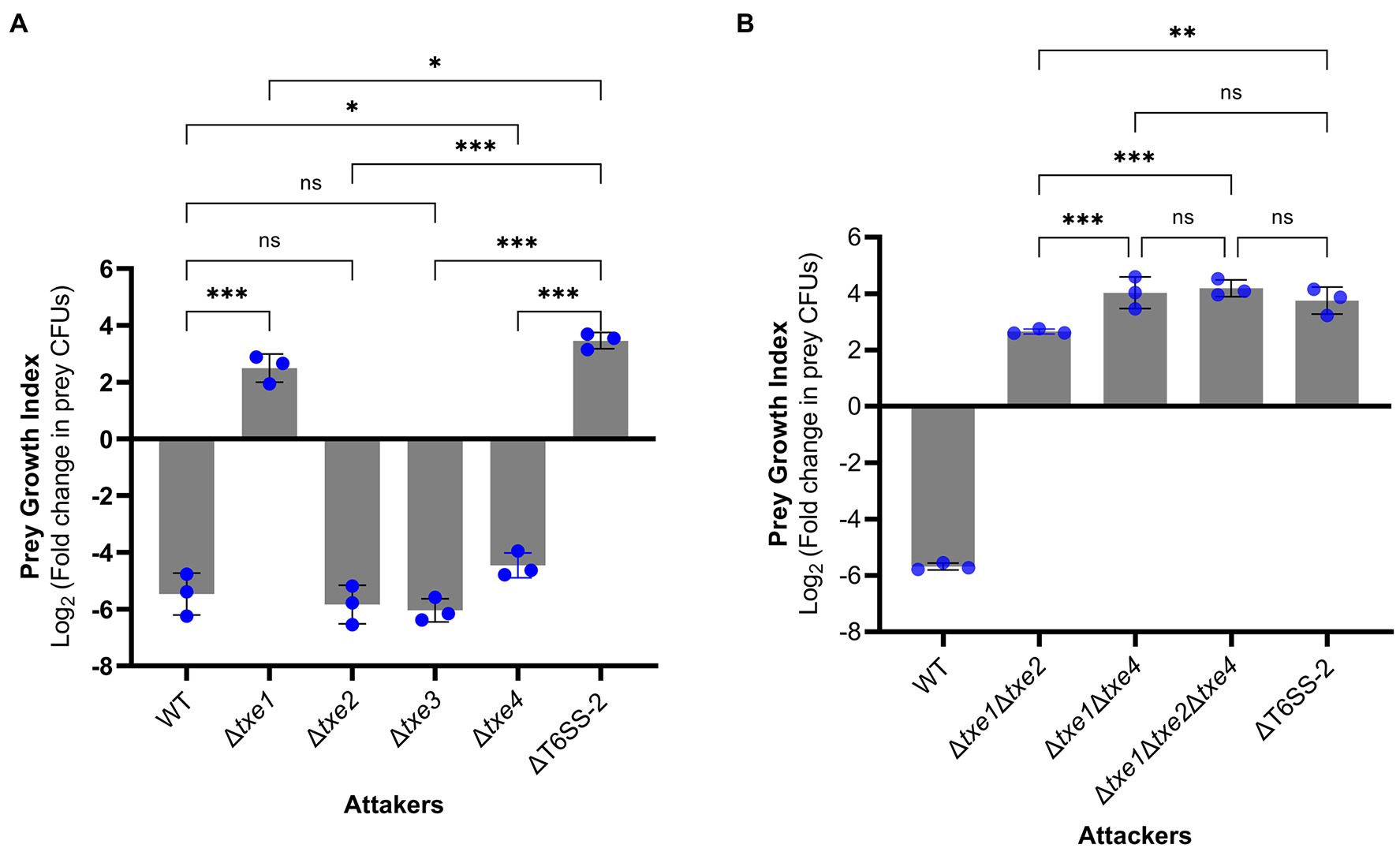
Figure 6. Interbacterial competition assay with P. plecoglossicida wild-type (WT) and deletion mutants as attackers. The prey growth index (PGI) was calculated from the prey strain E. coli XL10 when cocultured with P. plecoglossicida T6SS effector mutants or wild-type strain for 4 h. The PGI was defined as log2 fold change in prey CFUs (ratio of 4–0 h). A higher PGI indicates a lower ability of the attacker strain to kill the prey strain. PGI of single-effector deletion mutants vs. WT strain and ΔT6SS-2 (A). PGI of a triple-effector deletion mutant vs. WT strain and ΔT6SS-2 (B). Bars show mean ± SD of three biological replicates, with individual data points superimposed. Each data point represents the mean value of three technical replicates. The experiment was repeated twice with similar results, and the results from a representative experiment are shown. Comparison of PGI values between groups was performed using a one-way ANOVA and Fisher’s LSD test. ns, not significant (p > 0.05, *p ≤ 0.05, **p < 0.001, ***p < 0.001, and Student’s t-test).
Next, we tested whether txe1 and txe4 work synergically to mediate the observed bacterial killing in strain XSDHY-P. To this end, we created three multiple-effector deletion mutants including Δtxe1Δtxe4 and Δtxe1Δtxe2Δtxe4. Then, we compared the PGI values of these mutants with that of ΔT6SS-2 in another interbacterial competition assay. The PGI of Δtxe1Δtxe4 was not significantly different from those of ΔT6SS-2 or Δtxe1Δtxe2Δtxe4 (Figure 6B), indicating that Δtxe1Δtxe4 was attenuated as ΔT6SS-2, and Txe2 did not contribute to T6SS-2-mediated killing. Taken together, these results suggest that both Txe1 and Txe4 are used in the interbacterial competition by P. plecoglossicida strain XSDHY-P and that Txe1 is the predominant toxin responsible for killing E. coli in this assay.
Our previous work revealed that the fish pathogen, P. plecoglossicida strain XSDHY-P, encodes three T6SSs and T6SS-2 mediates interbacterial antagonism (Tao et al., 2020). In this work, we identified four putative antibacterial T6SS effectors that were named Txe1, Txe2, Txe3, and Txe4. The results presented here show that Txe1, Txe2, and Txe4 can effectively kill E. coli by exerting DNase activity when expressed in the cytosol (Figures 3A–C), suggesting that they are cytoplasmic-acting toxins. Although no conserved motif was identified in Txe1 CT domain based on the CDD search, a BLASTp search indicates that this nuclease might be commonly encoded with Rhs domain and in species belonging to the order Enterobacterales. Our site-directed mutagenesis analysis showed that one histidine residue (H1362) is essential for the toxicity of Txe1. However, more detailed analyses will be required to understand the exact role of His1362 in Txe1 and the corresponding residue in its homologs. Txe2 and Txe4 are AHH (pfam14412) and SHH (pfam15652) nuclease-domain containing effectors, respectively. Interbacterial competition assays show that txe1 and txe4 are necessary for the killing of E. coli in P. plecoglossicida strain XSDHY-P (Figures 6A,B). Although the data presented here largely confirm that Txe3 is not toxic to the E. coli strains used this study (Figure 5), we did not rule out the possibility that these particular E. coli strains have some T6SS-independent defense mechanisms that provide protection against activity of Txe3.
Txe1-Txi1, Txe2-Txi2, and Txe4-Txi4 are functional effector–immunity pairs. Our bioinformatic analysis revealed that these three effectors all contain a highly conserved Rhs core domain, but only Txe1 and Txe4 have a canonical Rhs-effector domain organization including a PAAR motif at the N-terminus, a domain of conserved Rhs-repeats in the central region, and a toxic domain at the C-terminus (Figures 3, 4). In addition, a conserved prePAAR motif and two transmembrane helices were identified in the region upstream of the respective PAAR domains of Txe1 and Txe4, a pattern that has also seen in other T6SS effectors. The prePAAR motif has been proposed to mediate effector–VgrG spike interactions (Ahmad et al., 2020), while the TMDs could facilitate the toxin entering into the inner membranes of target cells (Quentin et al., 2018). In contrast, Txe2 lacks the prePAAR motif and PAAR domain at its N terminus. It is still unclear whether the lack of PAAR domain influences the delivery of Txe2 in P. plecoglossicida and thereby restricts its role in the interbacterial competition, since PAAR is required for T6SS assembly in some species (Zheng et al., 2021). Our results regarding the function of these three toxins show that Txe1, Txe2, and Txe4 exhibit DNase activity and inhibit E. coli growth. Txe2 appears to be more toxic than Txe1 and Txe4, possibly due to differences in their mechanisms. In this study, the predicted immunity genes (txi1, txi2, and txi4) were co-expressed with cognate toxic effector genes in E. coli to prove that they are bona fide effector–immunity pairs. The putative immunity genes, when co-expressed, did not completely suppress plasmid DNA degradation by their cognate effectors in the cytoplasm; however, the toxicity of Txe1, Txe2, and Txe4 can be strikingly nullified based on growth curves and viable cell counts of E. coli strains (Figures 5A–F), confirming that Txe1-Txi1, Txe2-Txi2, and Txe4-Txi4 are functional effector–immunity pairs.
Txe1 is genetically linked to the vgrG-2 cluster (encoded in the main T6SS-2 structural module), and Txe4 is encoded within vgrG-5 cluster, which is not genetically linked to main T6SS-2 cluster. Two vgrG clusters have identical organization (one vgrG, two eagR genes, and downstream Rhs-toxin gene). In addition, VgrG, EagR, and Rhs effector counterparts encoded between the two gene clusters are also highly homologous (Supplementary Figures S1–S3), indicating that the two vgrG clusters are likely the result of a gene duplication event or are acquired from close relative by gene transfer. Together, these findings suggest that the proteins encoded in the vgrG-2 and vgrG-5 clusters may form VgrG-EagR-Rhs complexes of sufficient structural similarity to interact with the tube and baseplate of the same T6SS system (namely, T6SS-2) for toxin delivery (Jurėnas et al., 2021).
Txe1, Txe2, and Txe4 were identified to be homologous to Tke2 of P. putida strain KT2240 based on the analysis provided by the SecreT6 webtool (Li et al., 2015). Sequence alignment of the four effectors revealed that the sequence homology between Tke2 and Txe1, Txe2 or Txe4 was only found in the Rhs domains and not in the CT domains (Figure 2D). Tke2 is an effector deployed by K1-T6SS in P. putida KT2440 (Bernal et al., 2017). We analyzed the protein sequences of core structural components of T6SS-2 and K1-T6SS and found that they shared >90% amino acid sequence identity (Supplementary Table S4). We postulate that maintaining a conservative antibacterial T6SS across evolution confers a fitness advantage to these two Pseudomonas species, since the T6SS is energetically costly (Kudryashev et al., 2015). Nonetheless, P. putida and P. plecoglossicida have evolved their own distinctive toxin effectors and cognate immunity genes, and this may cause social incompatibilities and in turn lead to further genetic isolation (Vassallo et al., 2020).
In summary, we identified three type VI secretion DNase toxin and immunity pairs (Txe1-Txi1, Txe2-Txi2 and Txe4-Txi4) in P. plecoglossicida strain XSDHY-P, and they are encoded in two vgrG clusters. Txe1, Txe2, or Txe4 expressed in the cytoplasm of E. coli displayed comparable toxicity against host E. coli cells, while only Txe1 and Txe4 were shown to mediate the interspecies antagonism of strain XSDHY-P during the competition with an E. coli strain. The findings of this study contribute to our understanding of the bacterial effectors involved in competition between bacteria.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
ZT and XY conceived and designed the experiments. YL performed the experiments. ZT and YL analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Zhejiang Key Science and Technology Project (grant no. 2020C02004) and also supported by the Fundamental Research Funds for Zhejiang Provincial Universities and Research Institutes (grant no. 2021J012). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thanks Kequan Wang and Mingming Zhang for help with interbacterial competition assays. We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript. We thank two reviewers for insightful comments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.869278/full#supplementary-material
Ahmad, S., Tsang, K. K., Sachar, K., Quentin, D., Tashin, T. M., Bullen, N. P., et al. (2020). Structural basis for effector transmembrane domain recognition by type VI secretion system chaperones. elife 9:e62816. doi: 10.7554/eLife.62816
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Basler, M., Ho, B., and Mekalanos, J. (2013). Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894. doi: 10.1016/j.cell.2013.01.042
Bernal, P., Allsopp, L. P., Filloux, A., and Llamas, M. A. (2017). The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 11, 972–987. doi: 10.1038/ismej.2016.169
Bondage, D. D., Lin, J.-S., Ma, L.-S., Kuo, C.-H., and Lai, E.-M. (2016). VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor–effector complex. Proc. Natl. Acad. Sci. 113:E3931. doi: 10.1073/pnas.1600428113
Cascales, E., and Cambillau, C. (2012). Structural biology of type VI secretion systems. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 367, 1102–1111. doi: 10.1098/rstb.2011.0209
Cianfanelli, F. R., Monlezun, L., and Coulthurst, S. J. (2016). Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24, 51–62. doi: 10.1016/j.tim.2015.10.005
Diniz, J. A., Coulthurst, S. J., and Christie, P. J. (2015). Intraspecies competition in Serratia marcescens is mediated by type VI-secreted Rhs effectors and a conserved effector-associated accessory protein. J. Bacteriol. 197, 2350–2360. doi: 10.1128/JB.00199-15
Dong, T. G., Ho, B. T., Yoder-Himes, D. R., and Mekalanos, J. J. (2013). Identification of T6SS-dependent effector and immunity proteins by Tn-seq in vibrio cholerae. Proc. Natl. Acad. Sci. 110, 2623–2628. doi: 10.1073/pnas.1222783110
Durand, E., Cambillau, C., Cascales, E., and Journet, L. (2014). VgrG, tae, Tle, and beyond: the versatile arsenal of type VI secretion effectors. Trends Microbiol. 22, 498–507. doi: 10.1016/j.tim.2014.06.004
Gibson, D., Young, L., Chuang, R., Venter, J., Hutchison, C., and Smith, H. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6:343. doi: 10.1038/nmeth.1318
Hachani, A., Allsopp, L. P., Oduko, Y., and Filloux, A. (2014). The VgrG proteins are “à la carte” delivery systems for bacterial type VI effectors. J. Biol. Chem. 289, 17872–17884. doi: 10.1074/jbc.M114.563429
Hachani, A., Lossi, N. S., Hamilton, A., Jones, C., Bleves, S., Albesa-Jové, D., et al. (2011). Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J. Biol. Chem. 286, 12317–12327. doi: 10.1074/jbc.M110.193045
Hernandez, R. E., Gallegos-Monterrosa, R., and Coulthurst, S. J. (2020). Type VI secretion system effector proteins: effective weapons for bacterial competitiveness. Cell. Microbiol. 22:e13241. doi: 10.1111/cmi.13241
Jablonska, J., Matelska, D., Steczkiewicz, K., and Ginalski, K. (2017). Systematic classification of the his-me finger superfamily. Nucleic Acids Res. 45, 11479–11494. doi: 10.1093/nar/gkx924
Jackson, A. P., Thomas, G. H., Parkhill, J., and Thomson, N. R. (2009). Evolutionary diversification of an ancient gene family (rhs) through C-terminal displacement. BMC Genomics 10:584. doi: 10.1186/1471-2164-10-584
Jana, B., Fridman, C. M., Bosis, E., and Salomon, D. (2019). A modular effector with a DNase domain and a marker for T6SS substrates. Nat. Commun. 10:3595. doi: 10.1038/s41467-019-11546-6
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Jurėnas, D., and Journet, L. (2021). Activity, delivery, and diversity of type VI secretion effectors. Mol. Microbiol. 115, 383–394. doi: 10.1111/mmi.14648
Jurėnas, D., Rosa, L. T., Rey, M., Chamot-Rooke, J., Fronzes, R., and Cascales, E. (2021). Mounting, structure and autocleavage of a type VI secretion-associated Rhs polymorphic toxin. Nat. Commun. 12:6998. doi: 10.1038/s41467-021-27388-0
Krogh, A., Larsson, B., Von, H. G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kudryashev, M., Wang, R. Y. R., Brackmann, M., Scherer, S., Maier, T., Baker, D., et al. (2015). Structure of the type VI secretion system contractile sheath. Cell 160, 952–962. doi: 10.1016/j.cell.2015.01.037
Kvitko, B. H., Park, D. H., Velásquez, A. C., Wei, C.-F., Russell, A. B., Martin, G. B., et al. (2009). Deletions in the repertoire of pseudomonas syringae pv. Tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5:e1000388. doi: 10.1371/journal.ppat.1000388
Leiman, P. G., Marek, B., Ramagopal, U. A., Bonanno, J. B., Michael, J., Stefan, S., et al. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. 106, 4154–4159. doi: 10.1073/pnas.0813360106
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Lewis, J. M., Deveson Lucas, D., Harper, M., and Boyce, J. D. (2019). Systematic identification and analysis of Acinetobacter baumannii type VI secretion system effector and immunity components. Front. Microbiol. 10:2440. doi: 10.3389/fmicb.2019.02440
Li, J., Yao, Y., Xu, H. H., Hao, L., Deng, Z., Rajakumar, K., et al. (2015). SecReT6: a web-based resource for type VI secretion systems found in bacteria. Environ. Microbiol. 17, 2196–2202. doi: 10.1111/1462-2920.12794
Lopez, J., Ly, P. M., and Feldman, M. F. (2020). The tip of the VgrG spike is essential to functional type VI secretion system assembly in Acinetobacter baumannii. MBio 11, e02761–e02819. doi: 10.1128/mBio.02761-19
Ma, L. S., Hachani, A., Lin, J. S., Filloux, A., and Lai, E. M. (2014). Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16, 94–104. doi: 10.1016/j.chom.2014.06.002
Ma, J., Sun, M., Pan, Z., Lu, C., and Yao, H. (2018). Diverse toxic effectors are harbored by vgrG islands for interbacterial antagonism in type VI secretion system. Biochim. Biophys. Acta Gen. Subj. 1862, 1635–1643. doi: 10.1016/j.bbagen.2018.04.010
MacIntyre, D. L., Miyata, S. T., Kitaoka, M., and Pukatzki, S. (2010). The vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. 107, 19520–19524. doi: 10.1073/pnas.1012931107
Manera, K., Kamal, F., Burkinshaw, B., and Dong, T. G. (2022). Essential functions of chaperones and adaptors of protein secretion systems in Gram-negative bacteria. FEBS J. doi: 10.1111/febs.16056
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Minh, B. Q., Nguyen, M. A. T., and Haeseler, A. V. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Pei, T.-T., Li, H., Liang, X., Wang, Z.-H., Liu, G., Wu, L.-L., et al. (2020). Intramolecular chaperone-mediated secretion of an Rhs effector toxin by a type VI secretion system. Nat. Commun. 11:1865. doi: 10.1038/s41467-020-15774-z
Quentin, D., Ahmad, S., Shanthamoorthy, P., Mougous, J. D., Whitney, J. C., and Raunser, S. (2018). Mechanism of loading and translocation of type VI secretion system effector Tse6. Nat. Microbiol. 3, 1142–1152. doi: 10.1038/s41564-018-0238-z
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Russell, A. B., Hood, R. D., Bui, N. K., Leroux, M., Vollmer, W., and Mougous, J. D. (2011). Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347. doi: 10.1038/nature10244
Shneider, M. M., Buth, S. A., Ho, B. T., Basler, M., Mekalanos, J. J., and Leiman, P. G. (2013). PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353. doi: 10.1038/nature12453
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Silverman, J. M., Brunet, Y. R., Cascales, E., and Mougous, J. D. (2012). Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 66, 453–472. doi: 10.1146/annurev-micro-121809-151619
Tao, Z., Xu, Y., Zhou, S., Qian, D., Liu, M., Li, W., et al. (2020). Acquisition of a type VI secretion system is critical for pseudomonas plecoglossicida induced granulomas in fish internal organs. Aquaculture 516:734629. doi: 10.1016/j.aquaculture.2019.734629
Trifinopoulos, J., Nguyen, L.-T., Minh, B. Q., and Von Haeseler, A. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Unterweger, D., Kostiuk, B., Ötjengerdes, R., Wilton, A., Diaz-Satizabal, L., and Pukatzki, S. (2015). Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J. 34, 2198–2210. doi: 10.15252/embj.201591163
Unterweger, D., Kostiuk, B., and Pukatzki, S. (2017). Adaptor proteins of type VI secretion system effectors. Trends Microbiol. 25, 8–10. doi: 10.1016/j.tim.2016.10.003
Vassallo, C. N., Troselj, V., Weltzer, M. L., and Wall, D. (2020). Rapid diversification of wild social groups driven by toxin-immunity loci on mobile genetic elements. ISME J. 14, 2474–2487. doi: 10.1038/s41396-020-0699-y
Wettstadt, S., Wood, T. E., Fecht, S., and Filloux, A. (2019). Delivery of the Pseudomonas aeruginosa phospholipase effectors PldA and PldB in a VgrG- and H2-T6SS-dependent manner. Front. Microbiol. 10:1718. doi: 10.3389/fmicb.2019.01718
Whitney, J. C., Quentin, D., Sawai, S., LeRoux, M., Harding, B. N., Ledvina, H. E., et al. (2015). An interbacterial NAD(P)+ glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell 163, 607–619. doi: 10.1016/j.cell.2015.09.027
Winsor, G. L., Griffiths, E. J., Lo, R., Dhillon, B. K., Shay, J. A., and Brinkman, F. S. L. (2016). Enhanced annotations and features for comparing thousands of pseudomonas genomes in the pseudomonas genome database. Nucleic Acids Res. 44, D646–D653. doi: 10.1093/nar/gkv1227
Wood, T. E., Howard, S. A., Förster, A., Nolan, L. M., Manoli, E., Bullen, N. P., et al. (2019). The Pseudomonas aeruginosa T6SS delivers a periplasmic toxin that disrupts bacterial cell morphology. Cell Rep. 29, 187.e7–201.e7. doi: 10.1016/j.celrep.2019.08.094
Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615. doi: 10.1093/bioinformatics/btq249
Zhang, J. T., Zhou, S. M., An, S. W., Chen, L., and Wang, G. L. (2014). Visceral granulomas in farmed large yellow croaker, Larimichthys crocea (Richardson), caused by a bacterial pathogen, pseudomonas plecoglossicida. J. Fish Dis. 37, 113–121. doi: 10.1111/jfd.12075
Keywords: Pseudomonas plecoglossicida, type VI secretion system, VgrG, DNase, antibacterial activity, interbacterial competition, T6SS
Citation: Li Y, Yan X and Tao Z (2022) Two Type VI Secretion DNase Effectors are Utilized for Interbacterial Competition in the Fish Pathogen Pseudomonas plecoglossicida. Front. Microbiol. 13:869278. doi: 10.3389/fmicb.2022.869278
Received: 04 February 2022; Accepted: 17 March 2022;
Published: 06 April 2022.
Edited by:
Chih-Horng Kuo, Academia Sinica, TaiwanReviewed by:
Mary Nia Makimkim Santos, National Fisheries Research and Development Institute, PhilippinesCopyright © 2022 Li, Yan and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen Tao, dGFvemhlbjEyM0B6am91LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.