- 1Centre for Ecology and Evolution in Microbial Model Systems, Linnaeus University, Kalmar, Sweden
- 2Departamento de Ecoloxía e Bioloxía Animal, Centro de Investigación Mariña, Universidade de Vigo, Vigo, Spain
- 3CNRS, Laboratoire d’Ecogéochimie des Environnements Benthiques (LECOB), Sorbonne Université, Banyuls-sur-Mer, France
- 4CNRS, Laboratoire d’Océanographie Microbienne (LOMIC), Sorbonne Université, Banyuls-sur-Mer, France
- 5Earlham Institute, Norwich, United Kingdom
- 6IDAEA-CSIC, Barcelona, Spain
- 7Institut de Ciències del Mar-CSIC, Barcelona, Spain
- 8Department of Gene Technology, Science for Life Laboratory, KTH Royal Institute of Technology, Stockholm, Sweden
- 9Leibniz-Institute for Baltic Sea Research, Rostock, Germany
- 10Marine Biology Section, Department of Biology, University of Copenhagen, Helsingør, Denmark
Coastal ecosystems deteriorate globally due to human-induced stress factors, like nutrient loading and pollution. Bacteria are critical to marine ecosystems, e.g., by regulating nutrient cycles, synthesizing vitamins, or degrading pollutants, thereby providing essential ecosystem services ultimately affecting economic activities. Yet, until now bacteria are overlooked both as mediators and indicators of ecosystem health, mainly due to methodological limitations in assessing bacterial ecosystem functions. However, these limitations are largely overcome by the advances in molecular biology and bioinformatics methods for characterizing the genetics that underlie functional traits of key bacterial populations – “key” in providing important ecosystem services, being abundant, or by possessing high metabolic rates. It is therefore timely to analyze and define the functional responses of bacteria to human-induced effects on coastal ecosystem health. We posit that categorizing the responses of key marine bacterial populations to changes in environmental conditions through modern microbial oceanography methods will allow establishing the nascent field of genetic counselling for our coastal waters. This requires systematic field studies of linkages between functional traits of key bacterial populations and their ecosystem functions in coastal seas, complemented with systematic experimental analyses of the responses to different stressors. Research and training in environmental management along with dissemination of results and dialogue with societal actors are equally important to ensure the role of bacteria is understood as fundamentally important for coastal ecosystems. Using the responses of microorganisms as a tool to develop genetic counselling for coastal ecosystems can ultimately allow for integrating bacteria as indicators of environmental change.
Introduction
Earth is under tremendous pressure from human activities and the resulting global change. Coastal waters are productive ecosystems of high importance to society that suffer greatly from human activities causing water pollution, eutrophication, disrupted nutrient cycles, and loss of biodiversity (Rockström et al., 2009; Nash et al., 2017; Halpern et al., 2019). Legislative regulations for coastal management differ substantially between countries and continents, which represents a challenge for international agreements to promote coastal ecosystem health. As an example from the continent where we have our affiliations, it can be noted that the overarching aim of the Marine Strategy Framework Directive developed by the European Commission is to attain good environmental status across Europe’s marine environment. We think that achieving good environmental status in coastal marine waters requires actions based on due knowledge of the major ecosystem components – and an essential organism group that is left out of the equation is the bacteria (here denoting prokaryotes; Bacteria and Archaea).
So, why consider bacteria? Following the ground-breaking discovery in the early 1980s that planktonic bacteria in the oceans are actively growing and extremely abundant (109 cells/L), blossoming research on the metabolism, biodiversity and ecology of marine bacteria rapidly established their paramount role in the degradation of dissolved organic carbon (DOC). Moreover, bacteria regulate biogeochemical element cycles and influence overall marine productivity, processing more than 50% of the carbon fixed by photosynthesis and mediating most transformations of, e.g., nitrogen, phosphorus, and trace metals (Cole et al., 1988; Azam, 1998; Falkowski et al., 2008; Figure 1A). Recent advances in microbial oceanography have revealed that bacteria also provide a number of unexpected goods and services that influence ecosystem dynamics and productivity up to the level of fish, including the degradation of pollutants, production of vitamins, degradation of reactive oxygen species and the production of growth hormones (Amin et al., 2015; Durham et al., 2015; Cavicchioli et al., 2019). Moreover, because of their fast growth rates, bacteria react sensitively and rapidly to changes in environmental conditions. Nevertheless, bacteria have so far essentially been overlooked as indicators of ecosystem health, which has recently been highlighted (Zhang et al., 2020; Cordier et al., 2021; Alonso et al., 2022; Orel et al., 2022), and are currently not integrated in the development of policy making and management strategies to meet and mitigate the challenges imposed by human activity and global change on coastal waters.
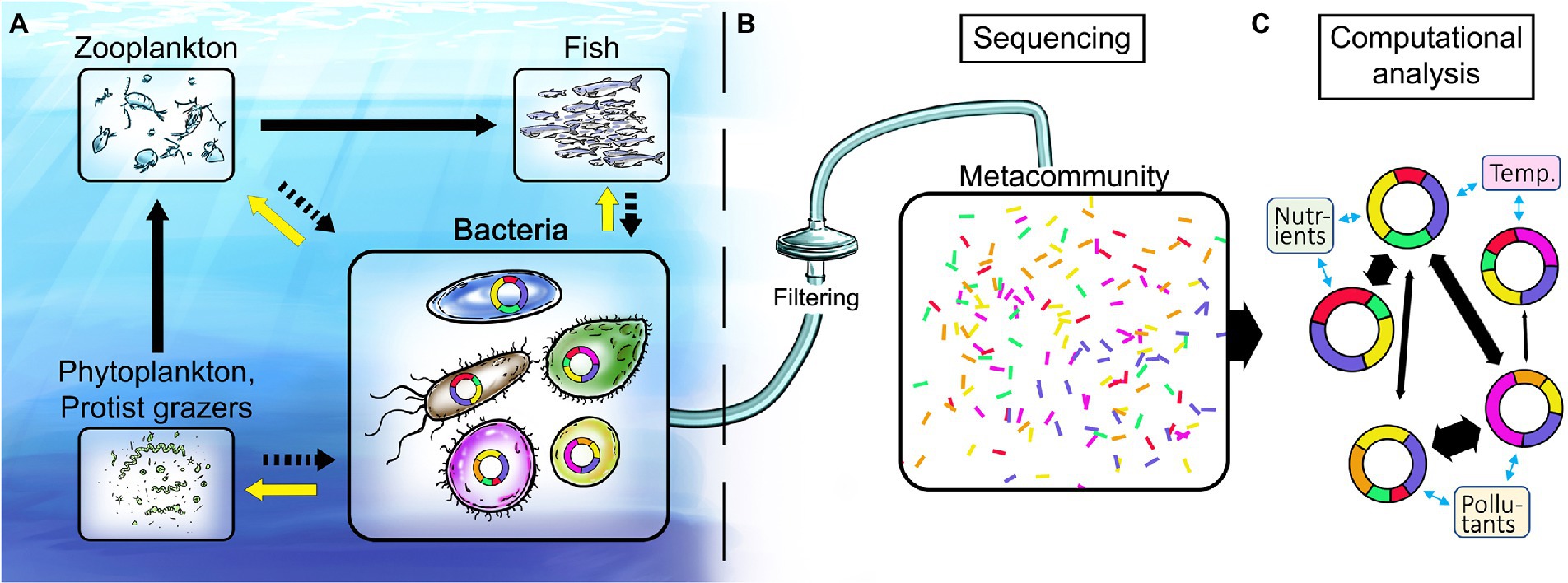
Figure 1. Illustration of the marine food web and proposed analysis workflow for disentangling linkages between environmental stressors and genomic structure and content of key marine bacteria. (A) Bacteria are fundamental in recycling of nutrients including dissolved organic carbon (black arrows to bacteria) and in providing ecosystem-wide services to higher trophic levels (yellow arrows). (B) Current state of the art methodology is based on sampling of the total nucleic acid (DNA and/or RNA) content of marine environments. (C) Computational analyses provide metagenome-assembled genomes (MAGs) that give unprecedented insights into both the identity of key bacterial populations and the genes underlying their different functional traits. Doughnut shapes represent genomes of such key bacterial populations and the colored doughnut sections denote traits. Co-occurrence network analysis of bacterial populations and their traits (black arrows between genomes) allows for identification of traits responsive to global change variables (blue arrows) and ultimately the ecosystem services supplied by key bacteria.
An analogy between human health and ecosystem health is warranted to grasp the potential of the mapping of the genetic basis for bacterial responses to environmental change. In medical practice, obtaining information about genetic disorders to make individually informed choices on medication and risk management – based on advice from genetics counsellors and medical doctors – is a rapidly spreading practice at hospitals worldwide, thanks to the large amount of affordable information gained by genome sequencing of patient material (be it cancer cells or gut microbiomes). In medicine, knowledge sources ranging from patient records to results from animal disease-model experiments (just to mention a few) have been essential to develop the understanding necessary to develop efficient treatments. In marine research, knowledge from time series measurements in the sea and controlled experiments with bacterial assemblages and isolates constitute a corresponding knowledge base (McCarren et al., 2010; Palovaara et al., 2014; Fuhrman et al., 2015; Bunse and Pinhassi, 2017; Landa et al., 2017). We find that it is now feasible and pertinent to build the knowledge for genetic counselling for the environment – effectively to interpret and predict alterations in microbial ecosystem functions in response to global change (Figure 1).
This is the background against which we call for systematically deploying state-of-the-art methods for genetic and genomic analyses of coastal ecosystems. This would permit identifying and characterizing functional traits of key bacterial populations, so that ultimately – through a dialogue between researchers and policy makers – bacteria can be implemented in environmental monitoring and used to improve the management of our world’s coastal waters.
Interrogating linkages between bacterial traits and ecosystem functions
Why are bacteria not yet explicitly considered in policy making, marine management practices, or ecosystem models? One central explanation is the lack of morphological traits that distinguish components of marine bacterioplankton – e.g. families, species or populations – which in turn severely complicates the study of their ecology. In this context, it is enlightening to make a comparison with another central plankton group, the photosynthetic eukaryotic and prokaryotic microorganisms referred to as phytoplankton. Importantly, the pronounced (and captivating) morphological differences between several phytoplankton groups allowed aquatic ecologists already in the early 1900s to establish a thorough understanding of functional traits of key taxa in a quantitative manner. Further, the spatiotemporal distribution of key phytoplankton taxa could be shown to vary predictably in relation to environmental conditions (Falkowski and Oliver, 2007). Therefore, major phytoplankton taxa, differing for example in size, motility or pigmentation, are represented in contemporary ecosystem models used for providing guidelines on sustainable management practices of aquatic environments around the globe (e.g., the BALTSEM model for the Baltic Sea). Conceptual and methodological advances in microbial ecology now allow for genetic and genomic characterization of the molecular mechanisms that determine the ecological niches of bacterial taxa (Figures 1B,C). We therefore posit that it is now both feasible and pertinent to develop corresponding knowledge of the metabolic and physiological characteristics representing functional traits that distinguish key bacterial taxa (Martiny et al., 2015; Coles et al., 2017; Baltar et al., 2019). Identification of such functional traits will be the first step to allow for a future inclusion of bacteria in ecosystem models.
Bacterial traits and ecosystem function
To obtain a thorough and mechanistic understanding of ecosystem functioning we reason that it is important to maneuver aquatic microbial ecology into the territory of trait-based ecology (Raes et al., 2011; Wallenstein and Hall, 2012; Brown et al., 2014; Krause et al., 2014). This includes defining what traits are and how they can be understood. One such example relates to microbial vitamin metabolism. The entire traditional food chain up to fish and birds depends on vitamin B1 produced by microbes. For this, it is possible to determine which bacteria contain the genes encoding vitamin B1 synthesis pathways and assign them to the trait “vitamin B1 synthesis,” and thus identifying the main providers of the ecosystem service “vitamin provision” (Paerl et al., 2018). In a similar manner, one can explore the role of bacteria in carbon cycling through analysis of traits like polysaccharide metabolism (conferred by, e.g., glycosyl hydrolases) and hydrocarbon metabolism (conferred by, e.g., aromatic-ring- hydroxylating dioxygenase genes; this at the same time addresses pollutant degradation traits (González-Gaya et al., 2019)). Also a variety of nitrogen and phosphorus cycling-related traits are pertinent to characterize, such as those for uptake or metabolism of different classes of dissolved organic or inorganic nitrogen or phosphorus. This could include genes for aminopeptidases versus phosphatases, which cross talk with the carbon cycle, or more specific traits like polyphosphate metabolism (conferred by purine nucleoside phosphorylases, PNPs), and genes involved in nitrification, denitrification or nitrogen fixation (Happel et al., 2019; Sosa et al., 2019). We recognize the challenge associated with defining traits, at an appropriately specific level, to obtain ecologically meaningful interpretations of patterns of trait distribution in nature. Nevertheless, we find that even traits carried by diverse taxa, e.g., nitrogen fixation (Zehr et al., 2003; Hallstrøm et al., 2022), provide pivotal insights into the interactions between microbes and the surrounding environment.
The potential to characterize linkages between bacteria and ecosystem services
A primary requisite to identify the genetics underlying particular traits is the access to detailed data on the genomes of key bacterial populations, which circumvents the dependence on difficult-to-assess differences in morphological or other phenotypic distinctions between taxa. In fact, standards are being developed so that new taxa and their names can be proposed based entirely on genomic information (Hedlund et al., 2022). Due to increasingly high-throughput and cost-effective sequencing methodologies, knowledge is accumulating on the spatiotemporal distribution of microbial groups and populations. In recent years, amplicon sequencing of phylogenetic marker genes (e.g., 16S rRNA) has been complemented by metagenomic and metatranscriptomic (−omics) studies of natural marine communities. Long-term field sampling provides important knowledge on the seasonal dynamics of key bacterial populations (Fuhrman et al., 2006; Teeling et al., 2012; Cram et al., 2015; Lambert et al., 2019, 2021; Auladell et al., 2022), and it has become evident that several ecosystem services are directly dependent on the diversity of bacterial communities (Galand et al., 2015, 2018; Garcia-Garcia et al., 2019). This complements theoretical considerations showing that biodiversity is fundamental for the capacity of natural communities to adapt to environmental change, i.e., the insurance effects of biodiversity (Yachi and Loreau, 1999). However, there is a critical lack of knowledge as to why different bacterial populations become dominant at different times and places or, even more importantly, how different populations influence their environment (i.e., what ecosystem functions they carry out). The answers to these questions are now tractable through molecular studies of the regulation of functional traits encoded in the genomes of key bacterial populations in response to environmental factors (Krause et al., 2014). Hereby we emphasize the use of -omics analyses in combination with the study of physiological and ecological responses. Moreover, the -omics approaches have the potential to resolve evolutionary long term adjustments in microbial communities.
The potential of linking -omics and environmental/ecological data has only recently been explored (Garcia et al., 2020; VanInsberghe et al., 2020), in part due to a lack of adequate computational methods (bioinformatics). Pioneering advances forming the basis for genetic counselling for the environment span from: 1) mapping bacterial taxa and their dynamics through 16S rRNA gene amplicon sequencing (Andersson et al., 2010; Edgar et al., 2011; Herlemann et al., 2011), via 2) the development of in situ instrumentation as a basis for non-biased -omics analyses (Ottesen et al., 2011; Bochdansky et al., 2017; Charvet et al., 2019), and 3) metagenomics and metatranscriptomics whereby functional genes are identified and their expression quantified (Gifford et al., 2013; Bunse et al., 2016; Rognes et al., 2016; Markussen et al., 2018; Alneberg et al., 2018b; Oberbeckmann et al., 2021), to 4) the present frontier, where it is possible to define bacterial populations and obtain genomes for key populations from metagenomic datasets, i.e., metagenome-assembled genomes (MAGs; Albertsen et al., 2013; Alneberg et al., 2014; Hugerth et al., 2015; Alneberg et al., 2018a; Figure 1C ). Moreover, from the content of functional genes in MAGs, their ecological niches across gradients of environmental conditions can be predicted, e.g., temperature, salinity, (in) organic nutrients and pollutants (Delmont et al., 2019; Alneberg et al., 2020; Cerro-Galvez et al., 2021; Pereira et al., 2021; Sjöqvist et al., 2021; Sun et al., 2021). Ultimately, this will move the field forward by determining how ecosystem services are distributed among key bacterial populations, rather than at community level as done before.
Adoption of MAG-assembly combined with long read sequencing approaches have recently given insights into the distribution of bacterial populations and their traits across the vastness of the open ocean (Ibarbalz et al., 2019), while leaving major blank spaces for the coastal seas. MAGs combined with metagenomic data sampled across environmental gradients can even reveal intra-species population structures and genes under environment-specific selection (Delmont et al., 2019; Sjöqvist et al., 2021). The finding of such strong links between bacterial genomes and the ecological niche provides a solid conceptual framework for predictive ecology based on genomic data from the environment. This allows, for the first time, to resolve some long standing yet urgent tasks: to determine how ecosystem services are distributed among marine bacteria (Ducklow, 2008), how key bacterial populations are affected by human impacts on coastal waters (Hutchins and Fu, 2017), and, in turn, how changes in bacterial abundances and activities influence the ecosystem, and vice versa (Azam and Malfatti, 2007). Now it is time to apply this knowledge to assess how human pressures on coastal waters influence bacterial communities and their ecosystem services. This has the potential to contribute evidence-based knowledge in both academic and non-academic settings where policies are shaped.
Research strategy into the future
We propose that systematic study of the microbial oceanography in coastal seas through the lenses of molecular biosciences could pave the way for the integration of bacteria into environmental management. In practice, we envision the following “road map”:
1. Compilation of prokaryotic biodiversity data (16S rRNA gene amplicons) from coastal locations, i.e., compilation and meta-analysis of existing data sets by taking advantage of existing projects and tools.
2. Establishment of MAG libraries from metagenomes across coastal locations of interest; through field sampling to identify key bacterial populations and their functional potentials; i.e. putative traits.
3. Identification of expressed traits from metatranscriptomes; field study catalogue of expressed genes from prokaryotic communities in general, and key bacterial populations (MAGs) in particular, from coastal locations.
4. Experimental testing of the regulation of key traits; environmental factors and anthropogenic stressors as determinants of trait responses of bacterial assemblages and key bacterial populations (MAGs).
5. Experimental testing of anthropogenic stressor effects on functional responses and traits in single isolates or mixtures of isolates representative of the major taxa of coastal marine bacterial communities.
6. Cross validation of trait responses to individual stressors in key bacterial populations of different coastal seas (international meta-analysis of linkages between environmental stressors and particular bacterial functional traits and taxa).
7. Establishment of national or regional “meeting points” for dialogue and collaborations with society – national environmental protection agencies, and local and regional authorities and industry.
8. Establishment of an international collaborative group to generate efficient layman descriptions/summaries of research findings – aimed at general public/education and authorities – to outline the utility of genetic counselling for the environment.
We think that systematically following such a road map would decipher the functionality and regulation of key bacterial populations in coastal waters. To reach this goal, it would be necessary to consolidate and develop existing computational methods for linking genes and populations to ecosystem services (Figures 1B,C, 2, “Bioinformatics”). An ambitious goal is then to delineate and ultimately predict the functionality of key bacterioplankton organisms via approaches ranging in complexity from time-resolved in situ studies (Figure 2, “Field studies”) to hypothesis-driven experiments with natural assemblages and/or experiments with single and multiple bacterial strains (Figure 2, “Experiments”). Such experiments could be done in both microcosms (up to some liters) and mesocosms (hundreds of liters) to address scale-dependent ecological processes. Together, these approaches are suited to address the overarching aim of resolving how traits and ecosystem functions are distributed among key bacterial populations present in coastal ecosystems, and how the abundance and activity of these bacteria – and thus their ecological traits – are affected by environmental factors in general, and environmental factors directly affected by global change in particular. In this framework, marine stations along the coasts of Europe are used as an example (Figure 2), but it can be applied and implemented globally. In parallel with these advances, it is necessary to engage in outreach and dissemination of research findings to a broad public along with training to bridge knowledge of microbial ecology and microbiome research with understanding of practices in management and governance (Figure 2, “Coastal Management”).
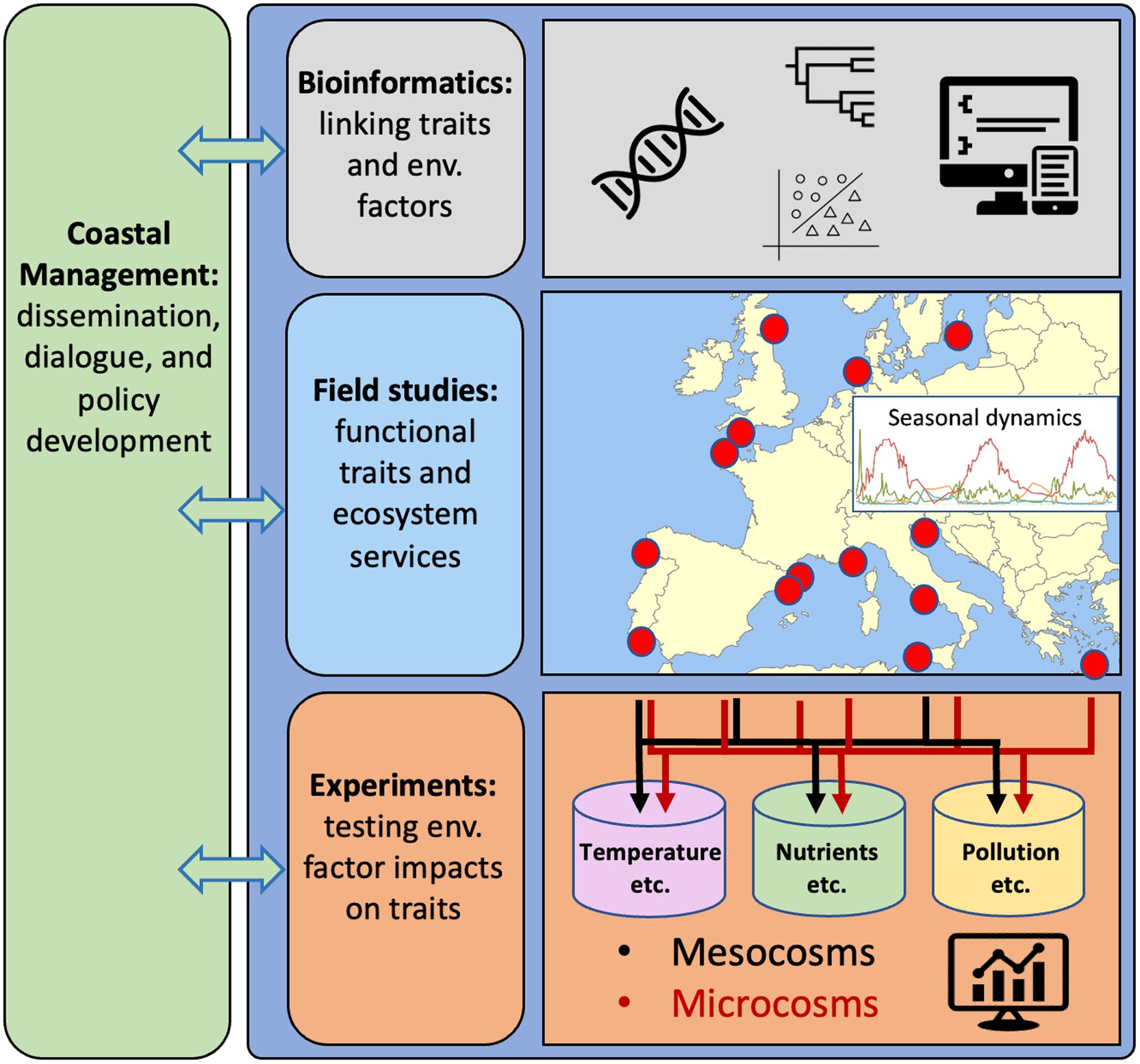
Figure 2. Research strategy to obtain comprehensive understanding of the genetic responses of key marine bacteria to environmental stressors in coastal waters, using European waters as an example. Bioinformatics for developing computational tools and workflows to effectively interpret linkages between functional traits and environmental factors. Field studies to generate and evaluate data on traits of key bacterial populations (from genomes and transcriptomes) and their distributions in time and space in relation to variability in environmental conditions. Experiments to identify and confirm relationships between environmental factors and specific traits and bacterial populations. Synergies between computational analyses of genomics, distributions and measured variability in environmental variables from field studies and experiments are essential to identify how bacterial functional traits can form the basis for genetic counselling of coastal seas. Dialogs with societal actors through all steps in the research strategy will be needed for reaching the goal of using bacteria to improve the quality of actions in coastal management.
Discussion
A fundamental incentive for this Perspective is the unifying framework of molecular biosciences, which daily brings important new knowledge to humanity, be it on human evolution, history, forensics, or medical sciences. Microbial oceanography pioneered the study of bacterial ecology and diversity using molecular methods (Giovannoni et al., 1990; Fuhrman et al., 1992; Béjà et al., 2000; Venter et al., 2004), setting the stage for the ongoing quest in environmental and medical sciences alike, to understand the interdependencies between microbiomes (i.e., microbial community composition and dynamics) and their “environment,” using the “omics” approaches (genomics, transcriptomics and proteomics; applied to tissues, cell lines, model bacteria and natural communities [termed “meta-omics” when applied to the latter]).
Coastal waters are highly valuable ecosystems with regard to the services they provide; e.g. fish and shellfish production, recreation, and waste assimilation. Understanding the factors that influence the productivity and stability of coastal ecosystems is therefore of utmost importance for a sustainable management of these regions. Working toward implementing genetic counselling for the marine environment is pertinent because increasing anthropogenically induced environmental disturbances, such as eutrophication and climate change, are placing numerous stressors on life in the oceans. It is also timely because, for the first time, we can decipher the functional role of key bacterial populations. Via advanced molecular techniques, the genes maintained and expressed by microbes can now be identified – revealing the functionality of these organisms and their linkages to prevailing environmental conditions and nutrient biogeochemistry. This facilitates the prediction of ecosystem services provided by global coastal waters in the future – a prerequisite for early mitigation of human impact and a critical step for identifying the societal actions required to ensure sustainable management of these sensitive regions in the face of global change.
We think it is timely to incorporate the emerging knowledge of genetics and ecology of bacteria – understood as genetics counselling for the environment – into developing increasingly sustainable management strategies and evaluation processes (Cordier et al., 2021). Beyond the systematic exploration of the relation between functional traits of key bacterial populations and environmental conditions, this requires education of the next generation of researchers with special emphasis on bridging bioinformatics, molecular biology, marine ecology, and ecosystem management. A prerequisite for success in this direction is interdisciplinary research involving modern environmental microbiology – computational and molecular biology, physiology, microbial oceanography and ecology – along with involvement of stakeholders in coastal management. Dissemination of results is an important component, as nicely exemplified through the TARA oceans project.1
In the outlined research strategy, continued use of and design of new coastal time-series stations will be critical for cross-system identification of environmental drivers of bacterial functionality – linkages between the environment and the processes carried out by bacteria. Time-series meta-omics datasets of at least 5–10 years are important because nutrient levels and biological activities are dynamic and seasonal features of temperate coastal ecosystems are expected to be affected by climate change. In turn, the established links furnish an ecophysiological understanding of key bacterial populations in natural waters, and this will be a precondition for the successful accomplishment of points 6 to 8 above. Research communication and dialogue with society has recently been emphasized in policy documents from, e.g., UNESCO, Global Research Council and Science Europe. Such closer collaboration between researchers and societal actors can be partially supported by national funding sources, but for example the efforts to reach a shared understanding across borders (emphasized in the last three points) would benefit from international funding. Certainly, researcher engagement in defining future (inter)national funding schemes would be warranted for this. The proposed actions have the potential to ensure that the role of bacteria is recognized as being of fundamental importance for the coastal marine ecosystem, and that this knowledge reaches decision makers involved in marine management. This will be invaluable for the continued refinement of sustainable management strategies for coastal zones.
Conclusion
We envision that microbes are sensitive sentinels (allowing detection of for example pathogens, nutrient loadings, pollution, and also in the long run as indicators of changes in food web structure in response to, e.g., over fishing) for the identification of undesired changes in the environment. With this view, the use of coastal microorganisms in genetic counselling for the environment could provide guidance for actions to reduce anthropogenic impact on the environment, which involves, e.g., the creation of marine reserves/protected areas or actual changes in legislation for the use of, e.g., fertilizers and pesticides. We posit that the actions presented in our road map for systematically exploring the responsiveness of key bacterial populations to environmental change – in time and space – will become critically important for laying the foundations for future assessment of environmental quality, optimization of coastal management, and policy. This is an urgent issue given the very rapid conceptual and methodological developments in using genetic and genomic analyses to obtain mechanistic understanding of ecological processes – paralleling and interacting with the corresponding developments in, e.g., medical sciences, forensics, biotechnology, and other industries. We believe that informed decisions based on environmental genetic counselling will ultimately aid mitigation of threats to coastal ecosystems posed by human activity, such as global change.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Acknowledgments
We thank for the many inspiring discussions with Ulla Li Zweifel and Åke Hagström on taking advantage of microbial genetic blueprints for informing on the health status of natural waters. We gratefully acknowledge Martin Brusin for contributing drawings of Figure 1. Research on this subject was supported by the marine strategic research program EcoChange to J. P and the BONUS BLUEPRINT project, which has received funding from BONUS, the Joint Baltic Sea Research and Development Program (Art 185), and Swedish, German and Danish research councils to JP, AA, ML and LR.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Albertsen, M., Hugenholtz, P., Skarshewski, A., Nielsen, K. L., Tyson, G. W., and Nielsen, P. H. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538. doi: 10.1038/nbt.2579
Alneberg, J., Bennke, C., Beier, S., Bunse, C., Quince, C., Ininbergs, K., et al. (2020). Ecosystem-wide metagenomic binning enables prediction of ecological niches from genomes. Commun. Biol. 3:119. doi: 10.1038/s42003-020-0856-x
Alneberg, J., Bjarnason, B. S., De Bruijn, I., Schirmer, M., Quick, J., Ijaz, U. Z., et al. (2014). Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146. doi: 10.1038/nmeth.3103
Alneberg, J., Karlsson, C. M. G., Divne, A. M., Bergin, C., Homa, F., Lindh, M. V., et al. (2018a). Genomes from uncultivated prokaryotes: a comparison of metagenome-assembled and single-amplified genomes. Microbiome 6:173. doi: 10.1186/s40168-018-0550-0
Alneberg, J., Sundh, J., Bennke, C., Beier, S., Lundin, D., Hugerth, L. W., et al. (2018b). BARM and BalticMicrobeDB, a reference metagenome and interface to meta-omic data for the Baltic Sea. Sci. Data 5:180146. doi: 10.1038/sdata.2018.146
Alonso, C., Pereira, E., Bertoglio, F., De Cáceres, M., and Amann, R. (2022). Bacterioplankton composition as an indicator of environmental status: proof of principle using indicator value analysis of estuarine communities. Aquat. Microb. Ecol. 88, 1–18. doi: 10.3354/ame01979
Amin, S. A., Hmelo, L. R., Van Tol, H. M., Durham, B. P., Carlson, L. T., Heal, K. R., et al. (2015). Interaction and signaling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101. doi: 10.1038/nature14488
Andersson, A. F., Riemann, L., and Bertilsson, S. (2010). Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 4, 171–181. doi: 10.1038/ismej.2009.108
Auladell, A., Barberan, A., Logares, R., Garces, E., Gasol, J. M., and Ferrera, I. (2022). Seasonal niche differentiation among closely related marine bacteria. ISME J. 16, 178–189. doi: 10.1038/s41396-021-01053-2
Azam, F. (1998). Microbial control of oceanic carbon flux: the plot thickens. Science 280, 694–696. doi: 10.1126/science.280.5364.694
Azam, F., and Malfatti, F. (2007). Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791. doi: 10.1038/nrmicro1747
Baltar, F., Bayer, B., Bednarsek, N., Deppeler, S., Escribano, R., Gonzalez, C. E., et al. (2019). Toward integrating evolution, metabolism, and climate change studies of marine ecosystems. Trends Ecol. Evol. 34, 1022–1033. doi: 10.1016/j.tree.2019.07.003
Béjà, O., Aravind, L., Koonin, E. V., Suzuki, M. T., Hadd, A., Nguyen, L. P., et al. (2000). Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289, 1902–1906. doi: 10.1126/science.289.5486.1902
Bochdansky, A. B., Clouse, M. A., and Herndl, G. J. (2017). Eukaryotic microbes, principally fungi and labyrinthulomycetes, dominate biomass on bathypelagic marine snow. ISME J. 11, 362–373. doi: 10.1038/ismej.2016.113
Brown, M. V., Ostrowski, M., Grzymski, J. J., and Lauro, F. M. (2014). A trait based perspective on the biogeography of common and abundant marine bacterioplankton clades. Mar. Genomics 15, 17–28. doi: 10.1016/j.margen.2014.03.002
Bunse, C., Lundin, D., Karlsson, C. M. G., Akram, N., Vila-Costa, M., Palovaara, J., et al. (2016). Response of marine bacterioplankton pH homeostasis gene expression to elevated CO2. Nat. Clim. Chang. 6, 483–487. doi: 10.1038/nclimate2914
Bunse, C., and Pinhassi, J. (2017). Marine bacterioplankton seasonal succession dynamics. Trends Microbiol. 25, 494–505. doi: 10.1016/j.tim.2016.12.013
Cavicchioli, R., Ripple, W. J., Timmis, K. N., Azam, F., Bakken, L. R., Baylis, M., et al. (2019). Scientists' warning to humanity: microorganisms and climate change. Nat. Rev. Microbiol. 17, 569–586. doi: 10.1038/s41579-019-0222-5
Cerro-Galvez, E., Dachs, J., Lundin, D., Fernandez-Pinos, M. C., Sebastian, M., and Vila-Costa, M. (2021). Responses of coastal marine microbiomes exposed to anthropogenic dissolved organic carbon. Environ. Sci. Technol. 55, 9609–9621. doi: 10.1021/acs.est.0c07262
Charvet, S., Riemann, L., Alneberg, J., Andersson, A. F., Von Borries, J., Fischer, U., et al. (2019). AFISsys-an autonomous instrument for the preservation of brackish water samples for microbial metatranscriptome analysis. Water Res. 149, 351–361. doi: 10.1016/j.watres.2018.11.017
Cole, J. J., Findlay, S., and Pace, M. L. (1988). Bacterioplankton production in fresh and saltwater ecosystems: a cross-system overview. Mar. Ecol. Prog. Ser. 43, 1–10. doi: 10.3354/meps043001
Coles, V. J., Stukel, M. R., Brooks, M. T., Burd, A., Crump, B. C., Moran, M. A., et al. (2017). Ocean biogeochemistry modeled with emergent trait-based genomics. Science 358, 1149–1154. doi: 10.1126/science.aan5712
Cordier, T., Alonso-Sáez, L., Apotheloz-Perret-Gentil, L., Aylagas, E., Bohan, D. A., Bouchez, A., et al. (2021). Ecosystems monitoring powered by environmental genomics: a review of current strategies with an implementation roadmap. Mol. Ecol. 30, 2937–2958. doi: 10.1111/mec.15472
Cram, J. A., Chow, C. E. T., Sachdeva, R., Needham, D. M., Parada, A. E., Steele, J. A., et al. (2015). Seasonal and interannual variability of the marine bacterioplankton community throughout the water column over ten years. ISME J. 9, 563–580. doi: 10.1038/ismej.2014.153
Delmont, T. O., Kiefl, E., Kilinc, O., Esen, O. C., Uysal, I., Rappe, M. S., et al. (2019). Single-amino acid variants reveal evolutionary processes that shape the biogeography of a global SAR11 subclade. elife 8:e46497. doi: 10.7554/eLife.46497
Ducklow, H. W. (2008). Microbial services: challenges for microbial ecologists in a changing world. Aquat. Microb. Ecol. 53, 13–19. doi: 10.3354/AME01220
Durham, B. P., Sharma, S., Luo, H., Smith, C. B., Amin, S. A., Bender, S. J., et al. (2015). Cryptic carbon and sulfur cycling between surface ocean plankton. Proc. Natl. Acad. Sci. U. S. A. 112, 453–457. doi: 10.1073/pnas.1413137112
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive Earth's biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Falkowski, P. G., and Oliver, M. J. (2007). Mix and match: how climate selects phytoplankton. Nat. Rev. Microbiol. 5, 813–819. doi: 10.1038/nrmicro1751
Fuhrman, J. A., Cram, J. A., and Needham, D. M. (2015). Marine microbial community dynamics and their ecological interpretation. Nat. Rev. Microbiol. 13, 133–146. doi: 10.1038/nrmicro3417
Fuhrman, J. A., Hewson, I., Schwalbach, M. S., Steele, J. A., Brown, M. V., and Naeem, S. (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proc. Natl. Acad. Sci. U. S. A. 103, 13104–13109. doi: 10.1073/pnas.0602399103
Fuhrman, J. A., Mccallum, K., and Davis, A. A. (1992). Novel major archaebacterial group from marine plankton. Nature 356, 148–149. doi: 10.1038/356148a0
Galand, P. E., Pereira, O., Hochart, C., Auguet, J. C., and Debroas, D. (2018). A strong link between marine microbial community composition and function challenges the idea of functional redundancy. ISME J. 12, 2470–2478. doi: 10.1038/s41396-018-0158-1
Galand, P. E., Salter, I., and Kalenitchenko, D. (2015). Ecosystem productivity is associated with bacterial phylogenetic distance in surface marine waters. Mol. Ecol. 24, 5785–5795. doi: 10.1111/mec.13347
Garcia, C. A., Hagstrom, G. I., Larkin, A. A., Ustick, L. J., Levin, S. A., Lomas, M. W., et al. (2020). Linking regional shifts in microbial genome adaptation with surface ocean biogeochemistry. Phil. Trans. R. Soc. B 375:20190254. doi: 10.1098/rstb.2019.0254
Garcia-Garcia, N., Tamames, J., Linz, A. M., Pedrós-Alió, C., and Puente-Sanchez, F. (2019). Microdiversity ensures the maintenance of functional microbial communities under changing environmental conditions. ISME J. 13, 2969–2983. doi: 10.1038/s41396-019-0487-8
Gifford, S., Sharma, A. K., Booth, M., and Moran, M. A. (2013). Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J. 7, 281–298. doi: 10.1038/ismej.2012.96
Giovannoni, S. J., Britschgi, T. B., Moyer, C. L., and Field, K. J. (1990). Genetic diversity in Sargasso Sea bacterioplankton. Nature 345, 60–63. doi: 10.1038/345060a0
González-Gaya, B., Martinez-Varela, A., Vila-Costa, M., Casal, P., Cerro-Gálvez, E., Berrojalbiz, N., et al. (2019). Biodegradation as an important sink of aromatic hydrocarbons in the oceans. Nat. Geosci. 12, 119–125. doi: 10.1038/s41561-018-0285-3
Hallstrøm, S., Raina, J. B., Ostrowski, M., Parks, D. H., Tyson, G. W., Hugenholtz, P., et al. (2022). Chemotaxis may assist marine heterotrophic bacterial diazotrophs to find microzones suitable for N-2 fixation in the pelagic ocean. ISME J. 16, 2525–2534. doi: 10.1038/s41396-022-01299-4
Halpern, B. S., Frazier, M., Afflerbach, J., Lowndes, J. S., Micheli, F., O'hara, C., et al. (2019). Recent pace of change in human impact on the world's ocean. Sci. Rep. 9:11609. doi: 10.1038/s41598-019-47201-9
Happel, E. M., Markussen, T., Teikari, J. E., Huchaiah, V., Alneberg, J., Andersson, A. F., et al. (2019). Effects of allochthonous dissolved organic matter input on microbial composition and nitrogen-cycling genes at two contrasting estuarine sites. FEMS Microbiol. Ecol. 95:fiz123. doi: 10.1093/femsec/fiz123
Hedlund, B. P., Chuvochina, M., Hugenholtz, P., Konstantinidis, K. T., Murray, A. E., Palmer, M., et al. (2022). SeqCode: a nomenclatural code for prokaryotes described from sequence data. Nat. Microbiol. 7, 1702–1708. doi: 10.1038/s41564-022-01214-9
Herlemann, D. P. R., Labrenz, M., Jurgens, K., Bertilsson, S., Waniek, J. J., and Andersson, A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579. doi: 10.1038/ismej.2011.41
Hugerth, L. W., Larsson, J., Alneberg, J., Lindh, M. V., Legrand, C., Pinhassi, J., et al. (2015). Metagenome-assembled genomes uncover a global brackish microbiome. Genome Biol. 16:279. doi: 10.1186/s13059-015-0834-7
Hutchins, D. A., and Fu, F. (2017). Microorganisms and ocean global change. Nat. Microbiol. 2:17058. doi: 10.1038/nmicrobiol.2017.58
Ibarbalz, F. M., Henry, N., Brandao, M. C., Martini, S., Busseni, G., Byrne, H., et al. (2019). Global trends in marine plankton diversity across kingdoms of life. Cell 179, 1084–1097.e21. doi: 10.1016/j.cell.2019.10.008
Krause, S., Le Roux, X., Niklaus, P. A., Van Bodegom, P. M., Lennon, J. T., Bertilsson, S., et al. (2014). Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front. Microbiol. 5:251. doi: 10.3389/fmicb.2014.00251
Lambert, S., Lozano, J. C., Bouget, F. Y., and Galand, P. E. (2021). Seasonal marine microorganisms change neighbors under contrasting environmental conditions. Environ. Microbiol. 23, 2592–2604. doi: 10.1111/1462-2920.15482
Lambert, S., Tragin, M., Lozano, J. C., Ghiglione, J. F., Vaulot, D., Bouget, F. Y., et al. (2019). Rhythmicity of coastal marine picoeukaryotes, bacteria and archaea despite irregular environmental perturbations. ISME J. 13, 388–401. doi: 10.1038/s41396-018-0281-z
Landa, M., Burns, A. S., Roth, S. J., and Moran, M. A. (2017). Bacterial transcriptome remodeling during sequential co-culture with a marine dinoflagellate and diatom. ISME J. 11, 2677–2690. doi: 10.1038/ismej.2017.117
Markussen, T., Happel, E. M., Teikari, J. E., Huchaiah, V., Alneberg, J., Andersson, A. F., et al. (2018). Coupling biogeochemical process rates and metagenomic blueprints of coastal bacterial assemblages in the context of environmental change. Environ. Microbiol. 20, 3083–3099. doi: 10.1111/1462-2920.14371
Martiny, J. B. H., Jones, S. E., Lennon, J. T., and Martiny, A. C. (2015). Microbiomes in light of traits: a phylogenetic perspective. Science 350:aac9323. doi: 10.1126/science.aac9323
Mccarren, J., Becker, J. W., Repeta, D. J., Shi, Y. M., Young, C. R., Malmstrom, R. R., et al. (2010). Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc. Natl. Acad. Sci. U. S. A. 107, 16420–16427. doi: 10.1073/pnas.1010732107
Nash, K. L., Cvitanovic, C., Fulton, E. A., Halpern, B. S., Milner-Gulland, E. J., Watson, R. A., et al. (2017). Planetary boundaries for a blue planet. Nat. Ecol. Evol. 1, 1625–1634. doi: 10.1038/s41559-017-0319-z
Oberbeckmann, S., Bartosik, D., Huang, S., Werner, J., Hirschfeld, C., Wibberg, D., et al. (2021). Genomic and proteomic profiles of biofilms on microplastics are decoupled from artificial surface properties. Environ. Microbiol. 23, 3099–3115. doi: 10.1111/1462-2920.15531
Orel, N., Fadeev, E., Klun, K., Ličer, M., Tinta, T., and Turk, V. (2022). Bacterial indicators are ubiquitous members of pelagic microbiome in anthropogenically impacted coastal ecosystem. Front. Microbiol. 12:765091. doi: 10.3389/fmicb.2021.765091
Ottesen, E. A., Marin, R., Preston, C. M., Young, C. R., Ryan, J. P., Scholin, C. A., et al. (2011). Metatranscriptomic analysis of autonomously collected and preserved marine bacterioplankton. ISME J. 5, 1881–1895. doi: 10.1038/Ismej.2011.70
Paerl, R. W., Sundh, J., Tan, D., Svenningsen, S. L., Hylander, S., Pinhassi, J., et al. (2018). Prevalent reliance of bacterioplankton on exogenous vitamin B1 and precursor availability. Proc. Natl. Acad. Sci. U. S. A. 115, E10447–E10456. doi: 10.1073/pnas.1806425115
Palovaara, J., Akram, N., Baltar, F., Bunse, C., Forsberg, J., Pedros-Alio, C., et al. (2014). Stimulation of growth by proteorhodopsin phototrophy involves regulation of central metabolic pathways in marine planktonic bacteria. Proc. Natl. Acad. Sci. U. S. A. 111, E3650–E3658. doi: 10.1073/pnas.1402617111
Pereira, O., Hochart, C., Boeuf, D., Auguet, J. C., Debroas, D., and Galand, P. E. (2021). Seasonality of archaeal proteorhodopsin and associated marine group IIb ecotypes (Ca. Poseidoniales) in the North Western Mediterranean Sea. ISME J. 15, 1302–1316. doi: 10.1038/s41396-020-00851-4
Raes, J., Letunic, I., Yamada, T., Jensen, L. J., and Bork, P. (2011). Toward molecular trait-based ecology through integration of biogeochemical, geographical and metagenomic data. Mol. Syst. Biol. 7:473. doi: 10.1038/msb.2011.6
Rockström, J., Steffen, W., Noone, K., Persson, A., Chapin, F. S. 3rd, Lambin, E. F., et al. (2009). A safe operating space for humanity. Nature 461, 472–475. doi: 10.1038/461472a
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahe, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Sjöqvist, C., Delgado, L. F., Alneberg, J., and Andersson, A. F. (2021). Ecologically coherent population structure of uncultivated bacterioplankton. ISME J. 15, 3034–3049. doi: 10.1038/s41396-021-00985-z
Sosa, O. A., Repeta, D. J., Delong, E. F., Ashkezari, M. D., and Karl, D. M. (2019). Phosphate-Limited Ocean regions select for bacterial populations enriched in the carbon-phosphorus lyase pathway for phosphonate degradation. Environ. Microbiol. 21, 2402–2414. doi: 10.1111/1462-2920.14628
Sun, Y., Debeljak, P., and Obernosterer, I. (2021). Microbial iron and carbon metabolism as revealed by taxonomy-specific functional diversity in the Southern Ocean. ISME J. 15, 2933–2946. doi: 10.1038/s41396-021-00973-3
Teeling, H., Fuchs, B. M., Becher, D., Klockow, C., Gardebrecht, A., Bennke, C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/Science.1218344
Vaninsberghe, D., Arevalo, P., Chien, D., and Polz, M. F. (2020). How can microbial population genomics inform community ecology? Phil. Trans. R. Soc. B 375:20190253. doi: 10.1098/rstb.2019.0253
Venter, J. C., Remington, K., Heidelberg, J. F., Halpern, A. L., Rusch, D., Eisen, J. A., et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74. doi: 10.1126/science.1093857
Wallenstein, M. D., and Hall, E. K. (2012). A trait-based framework for predicting when and where microbial adaptation to climate change will affect ecosystem functioning. Biogeochemistry 109, 35–47. doi: 10.1007/s10533-011-9641-8
Yachi, S., and Loreau, M. (1999). Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc. Natl. Acad. Sci. U. S. A. 96, 1463–1468. doi: 10.1073/pnas.96.4.1463
Zehr, J. P., Jenkins, B. D., Short, S. M., and Steward, G. F. (2003). Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5, 539–554. doi: 10.1046/j.1462-2920.2003.00451.x
Keywords: marine bacteria, ecosystem service, water quality, management, traits
Citation: Pinhassi J, Farnelid H, García SM, Teira E, Galand PE, Obernosterer I, Quince C, Vila-Costa M, Gasol JM, Lundin D, Andersson AF, Labrenz M and Riemann L (2022) Functional responses of key marine bacteria to environmental change – toward genetic counselling for coastal waters. Front. Microbiol. 13:869093. doi: 10.3389/fmicb.2022.869093
Edited by:
Chris Dupont, J. Craig Venter Institute, United StatesReviewed by:
Kerstin Johannesson, University of Gothenburg, SwedenCopyright © 2022 Pinhassi, Farnelid, García, Teira, Galand, Obernosterer, Quince, Vila-Costa, Gasol, Lundin, Andersson, Labrenz and Riemann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jarone Pinhassi, amFyb25lLnBpbmhhc3NpQGxudS5zZQ==
 Jarone Pinhassi
Jarone Pinhassi Hanna Farnelid
Hanna Farnelid Sandra Martínez García
Sandra Martínez García Eva Teira
Eva Teira Pierre E. Galand
Pierre E. Galand Ingrid Obernosterer
Ingrid Obernosterer Christopher Quince5
Christopher Quince5 Maria Vila-Costa
Maria Vila-Costa Josep M. Gasol
Josep M. Gasol Daniel Lundin
Daniel Lundin Anders F. Andersson
Anders F. Andersson Matthias Labrenz
Matthias Labrenz Lasse Riemann
Lasse Riemann