
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 April 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.867770
This article is part of the Research Topic Mechanisms of Biofilm Development and Antibiofilm Strategies View all 16 articles
 Ruifu Yang1†
Ruifu Yang1† Bipeng Lai1†
Bipeng Lai1† Kang Liao2
Kang Liao2 Baomo Liu1
Baomo Liu1 Lixia Huang1
Lixia Huang1 Shaoli Li1
Shaoli Li1 Jincui Gu1
Jincui Gu1 Ziying Lin1
Ziying Lin1 Yili Chen2
Yili Chen2 Shuaishuai Wang1
Shuaishuai Wang1 Yanli Qiu1
Yanli Qiu1 Jiating Deng1
Jiating Deng1 Simin Chen1
Simin Chen1 Chao Zhuo3*
Chao Zhuo3* Yanbin Zhou1*
Yanbin Zhou1*Acinetobacter baumannii, a strictly aerobic, non-lactose fermented Gram-negative bacteria, is one of the important pathogens of nosocomial infection. Major facilitator superfamily (MFS) transporter membrane proteins are a class of proteins that widely exists in microbial genomes and have been revealed to be related to biofilm formation in a variety of microorganisms. However, as one of the MFS transporter membrane proteins, little is known about the role of BIT33_RS14560 in A. baumannii. To explore the effects of BIT33_RS14560 on biofilm formation of A. baumannii, the biofilm formation abilities of 62 isolates were firstly investigated and compared with their transcript levels of BIT33_RS14560. Then, this specific gene was over-expressed in a standard A. baumannii strain (ATCC 19606) and two isolates of extensively drug-resistant A. baumannii (XDR-Ab). Bacterial virulence was observed using a Galleria mellonella infection model. High-throughput transcriptome sequencing (RNA seq) was performed on ATCC 19606 over-expressed strain and its corresponding empty plasmid control strain. Spearman’s correlation analysis indicated a significant negative correlation (R = −0.569, p = 0.000) between the △CT levels of BIT33_RS1456 and biofilm grading of A. baumannii isolates. The amount of A. baumannii biofilm was relatively high within 12–48 h. Regardless of standard or clinical strains; the biofilm biomass in the BIT33_RS14560 overexpression group was significantly higher than that in the control group ( p < 0.0001). Kaplan–Meier survival curve analysis showed that the mortality of G. mellonella was significantly higher when infected with the BIT33_RS14560 overexpression strain (χ2 = 8.462, p = 0.004). RNA-Seq showed that the mRNA expression levels of three genes annotated as OprD family outer membrane porin, glycosyltransferase family 39 protein, and glycosyltransferase family 2 protein, which were related to bacterial adhesion, biofilm formation, and virulence, were significantly upregulated when BIT33_RS14560 was over-expressed. Our findings provided new insights in identifying potential drug targets for the inhibition of biofilm formation. We also developed a practical method to construct an over-expressed vector that can stably replicate in XDR-Ab isolates.
Acinetobacter baumannii, a Gram-negative bacterium that exists widely in nature, is one of the most important opportunistic pathogens responsible for nosocomial infection. Immunocompromised or severe patients in the intensive care unit are more vulnerable to this pathogen, causing a wide range of infectious diseases, including pneumonia, meningitis, peritonitis, infections of the urinary tract and skin, and posing a great threat to severe patients (Ayoub Moubareck and Hammoudi Halat, 2020). The treatment of A. baumannii infection has always been a challenging problem in clinical practice due to its increased resistance to commonly-used antibacterial drugs, leading to a global epidemic of multidrug-resistant A. baumannii (MDR-Ab; Ibrahim et al., 2021) and extensively drug-resistant A. baumannii (XDR-Ab; Kengkla et al., 2018).
In addition to β-lactamase and aminoglycoside modification enzymes production, active efflux, modification of target sites, and changes in outer membrane permeability (Lee et al., 2017), the drug resistance mechanism of A. baumannii was also found to be closely related to the formation of biofilms (Yang et al., 2019; Saipriya et al., 2020). Biofilm refers to the complex, sessile communities of microbes found either attached to a surface or buried firmly in an extracellular matrix as aggregates (Roy et al., 2018). The biofilm formation of A. baumannii is a complex process regulated by many factors, including the pili assembly system (Tomaras et al., 2003), BfmRS two-component regulatory systems (Gaddy and Actis, 2009), bacterial quorum-sensing system (Zhang et al., 2020), biofilm-related proteins (Bap; Fattahian et al., 2011) and outer membrane protein A (OmpA; Gaddy et al., 2009), and so on. Most clinically isolated A. baumannii strains have been observed to have a strong ability to form biofilms, which results in significantly low sensitivity to antimicrobial agents.
Major facilitator superfamily (MFS), a class of transporter membrane proteins, belongs to the efflux pump families and has a strong substrate specificity, which is reflected in the transportation of secondary metabolites by an ion concentration gradient. Some MFS superfamily members have been confirmed to be closely related to biofilm formation and drug resistance. For example, in a pathogenic strain A. baumannii AIIMS 7, pmt, a putative MFS transporter-like ORF of 453 bp, was identified and found to be associated with adherence, biofilm formation, and probable extracellular DNA release (Sahu et al., 2012). The efflux gene A1S_1117, an MFS superfamily vanillate transporter, was reported to be expressed only in biofilm cells but inhibited in planktonic cells (Rumbo-Feal et al., 2013). AbaF, another member of the MFS superfamily, was proved to mediate the efflux of intracellular fosfomycin and promote the ability to form biofilms in A. baumannii (Sharma et al., 2017). Therefore, the in-depth study of MFS family members is of great significance to elucidate the mechanisms of biofilm formation in A. baumannii and control chronic infections.
Here, we focus on a new gene, which is labeled with a locus tag of BIT33_RS14560 in National Center of Biotechnology Information (NCBI) and presumed as a member of the MFS family. In this work, we successfully constructed two recombinant plasmids of BIT33_RS14560 over-expression, with one aimed at the standard A. baumannii strain (ATCC 19606) and the other on account of two XDR-Ab isolates. We transformed them into the target strains and managed to overexpress this target gene. We found that when BIT33_RS14560 was over-expressed, the abilities to form biofilm were significantly promoted in both standard and clinical strains of A. baumannii, which was also confirmed by quantitative real-time PCR (qRT-PCR) assays of 62 clinical isolates. Compared with wild-type ATCC 19606 strain, BIT33_RS14560 overexpression strain displayed higher mortality of Galleria mellonella (χ2 = 8.462, p = 0.004). We also presented RNA sequencing evidence to elucidate possible mechanisms for these phenotypic changes.
DNA or amino acid sequences of BIT33_RS14560 gene or protein (corresponding Accession Number: CP058289.1 or WP_002047564.1) and its homologs were searched using BLAST at the (NCBI) website1 and downloaded in a separate FASTA format. These sequences were analyzed to construct phylogenetic trees to determine the phylogenetic relationships between the selected strains (Supplementary Tables S1, S2). The best DNA or protein model (GTR + G + I) with the highest parameter was determined and chosen before the construction of phylogenetic trees. Dendrograms were generated by Maximum Likelihood (ML) with bootstrap values corresponding to 1,000 replications using the (MEGA) 11.0 software (Tamura et al., 2021).
Bacterial strains and plasmids used in this study are listed in Table 1. Of 62 A. baumannii isolates, 43 strains (69.4%) including two XDR-Ab isolates, XAb53 (Strain No. A53; Stored on June 2, 2021) and XAb50 (Strain No. A50; Stored on May 19th, 2020), were isolated from the First Affiliated Hospital of Sun Yat-sen University, the other 19 strains (30.6%) were obtained from the Microbiology Laboratory of State Key Laboratory of Respiratory Diseases, Guangzhou Medical University. Sources and proportion of 62 A. baumanii isolates used in this study were presented in Supplementary Table S3. Escherichia coli DH5α competent cells (Cat. No. 9057; TaKaRa) were purchased from Guangzhou Ruizhen Biotechnology Co., Ltd. The suicide plasmid pMo130-telR (Cat. No. P4951-ea; Wonder Biotech) was purchased from Guangzhou Qunlan Biotechnology Co., Ltd. Bacterial culturing was performed using Luria-Bertani (LB) agar medium (Cat. No. CM0337B; OXOID) or LB broth medium (Cat. No. CM0405; OXOID).
Overnight culture of A. baumannii strain was inoculated into fresh LB broth until the mid-log phase (OD = 0.6–0.8) was reached. Total RNA was extracted using the E.Z.N.A.® Bacterial RNA Kit (Cat. No. R6950-01; Omega). The quality and concentration of RNA were determined via spectrophotometry (NanoDrop 2000c; Fisher Scientific, United States). For reverse transcription PCR (RT-PCR), reverse transcription was performed using ProFlex PCR System cycler (Applied Biosystems, United States). The purified RNA (1 μg/ml) was reverse-transcribed into cDNA by the Evo M-MLV RT Premix for qPCR kit (Cat. No. AG11706; Accurate Biology), in a 20 μl reaction mixture containing 4 μl of Evo M-MLVRT Master Mix and 15 μl of RNAase free water, which was incubated for 15 min at 37°C and 5 s at 85°C. Then, the qRT-PCR assay was performed on a Roche LightCycler® 960 real-time PCR system. Here, SYBR Green Premix Pro Taq HS qPCR kit (Cat. No. 78 AG11702; Accurate Biology) was employed. Around 2 μl of the resulting cDNA products were put into a 20 μl reaction mixture containing 10 μl of SYBR® Green Pro Taq HS Premix II, 0.8 μl (or 10 μM) of each primer and 6.4 μl RNase free water. The parameters of PCR were set as followed: initial heat activation at 95°C for 30 s, followed by 35 cycles of the two-step cycling condition of denaturation at 95°C for 10 s and then annealing at 60°C for 30 s. The melting data of PCR products were measured and collected following the completion of PCR cycling. The rpoB gene was used as an internal reference for normalization. △CT value, defined as the CT value of the target gene minus that of the reference rpoB, was used to calculate the BIT33_RS14560 mRNA levels of A. baumannii. For each sample, three technical replicates were performed and the gene transcript levels were determined using the 2-△△Ct method. Primers used in this part are listed in Table 2.
All primers used in this study are listed in Table 2. PCR amplification was achieved using DNA polymerase (Cat. No. AG12202; Accurate 79 Biology). A plasmid DNA extraction kit (Cat. No. D2156-01; Omega), a DNA fragment purification kit (Cat. No. 9761; 77 TaKaRa), and a DNA Ligation kit (Cat. No. 6023; Accurate Biology) were also employed. The novel BIT33_RS14560 gene was PCR-amplified from the genomes of A. baumannii ATCC 19606 using specific primers RS14560-EcoRI-F and RS14560-6his-BamHI-R with the following conditions—initial hold at 94°C for 4 min, followed by 32 cycles of 98°C for 10 s, 55°C for 5 s, 72°C for 8 s, and final extension at 72°C for 7 min. Purified PCR products and pWH1266 plasmid were digested with BamHI and EcoRI, then, according to the manufacturer’s instructions, ligated to make a recombinant plasmid of overexpression, named as p-RS14560. After digesting with EcoRI and BamHI, blunt ends to the large fragments of pWH1266 plasmid were created with high-fidelity DNA polymerase and then self-ligated to make into an empty plasmid, known as p. Each plasmid was transformed into E. coli DH5α competent cells and then introduced into the A. baumannii ATCC 19606 by electroporation (1.75 kV). After overnight culture (16–18 h) at 37°C, monoclonal colony PCR was performed with Pwh1266-F and Pwh1266-R primers to select positive transformants on LB agar supplemented with 50 μg/ml Carbenicillin.
For the overexpression of XDR-Ab, based on the above work, we reconstructed a specific recombinant plasmid by replacing the Ampicillin resistance (AmpR) cassette in the p-RS plasmid with the Tellurite resistance (TelR) cassette. Briefly, the TelR cassette was amplified from the plasmid pMo130-telR using telR-PstI-F/telR-PvuI-R primers. After purification, these PCR products and p-RS plasmid were digested with PstI and PvuI then ligated to make a new plasmid p-telR-RS14560. Similarly, a corresponding empty plasmid p-telR was constructed according to the above-mentioned method. Each plasmid was transformed into E. coli DH5α competent cells and then electroporated into A. baumannii XAb53 and XAb50 isolates at the same voltage of 1.75 kV. Of particular note, when selecting positive transformants with telR-F/telR-R primers, the concentration of tellurite added into the LB agar should not be completely fixed and rigid, that is, 25 μg/ml for E. coli DH5α and 80 μg/ml for XAb53 or XAb50 isolates. Of note, these two XDRAB clinical isolates were randomly selected according to their results of multilocus sequence typing, that was, XAb50 was assigned to ST1145 (gltA-1, gyrB-3, gdhB-3, recA-102, cpn60-2, gpi-97, and rpoD-3), an ST recently prevalent at the First Affiliated Hospital of Sun Yat-sen University, while XAb53 was considered a new ST (gltA-1, gyrB-3, gdhB-3, recA-102, cpn60-1, gpi-103, and rpoD-26). The media should be cultured overnight for 24–30 h at 37°C.
We performed a microtiter-plate test to evaluate the biofilm formation ability of A. baumannii according to the method described previously (Rodríguez-Baño et al., 2008), but with certain modifications. Prior to inoculation, all strains were transferred from the stock cultures onto LB agar plates and incubated aerobically at 37°C for 18 h. Colonies from these plates were suspended in phosphate-buffered saline (PBS). After a 0.5 McFarland turbidity standard suspension (corresponding to 1.5 × 108 CFU/ml) of each isolate was determined, a 100-fold dilution with sterile LB broth was prepared. Then, 200 μl of each bacterial suspension were filled into no less than six wells of a sterile 96-well flat-bottomed plastic tissue culture plate with a lid. Negative and positive control wells contained sterile LB broth and standardized suspension of P. aeruginosa strain ATCC 27853, respectively. The plates were covered and incubated aerobically at 37°C for 0, 6, 12, 18, 24, 30, 36, 42, 48, 60, and 72 h, respectively. Then, the content of each well was gently aspirated, and each well was washed three times with 250 ml of ultrapure deionized water and left to dry. Each well was stained for 20 min at room temperature with 200 μl of 10 g/L crystal violet. Excess stain was rinsed off by sterile water. After the plates were air-dried, each well was added with 200 μl of 33.3% glacial acetic acid and resolubilized for 20 min. Absorbance at 570 nm (OD570) of each well was measured by Epoch 2 microplate reader (BioTek). A cut-off OD (ODc) was defined as three standard deviations (SDs) above the mean OD of the negative control. All strains were classified into the following categories (Stepanović et al., 2007): non-adherent (OD ≤ ODc, 0), weakly adherent (ODc < OD ≤ 2 × ODc, +), moderately adherent (2 × ODc < OD ≤ 4 × ODc, ++), or strongly adherent (4 × ODc < OD, +++). The results were averaged after all tests were carried out three times.
Bacterial cell suspensions at a concentration equivalent to a 0.5 McFarland Standard were prepared and followed by a 100-fold dilution with sterile LB broth. To make an infection model, the last instar healthy larvae with 2–3 cm long and 180–250 mg in weight were selected and randomly divided into groups with 10 individuals in each group. Each insect was carefully given an injection with 20 μl of bacterial solution (1.5 × 106 CFU/ml) and put in a Petri dish of 8 cm in diameter. Infect at least 30 larvae for each strain. Then place the dishes at 37°C in an incubator and check larval mortality regularly over a 6 h period for 3 days. A. baumannii 5075, a strain with high virulence, was served as the positive control while PBS served as the negative. Mortality data were analyzed using the Kaplan–Meier method.
Four single colonies, two derived from the overexpressed strain constructed by ATCC 19606, and two from the corresponding empty plasmid strain, were picked into 3 ml LB medium supplemented with 50 μg/ml Carbenicillin and inoculated, respectively. These cultures were grown for 18 h at 37°C and cells were then collected by centrifugation at 6,000 rpm for 10 min and washed three times with RNase free water. Total RNA extraction and the subsequent transcriptome sequencing were performed by Shanghai Majorbio Bio-pharm Technology Co., Ltd. After the quality analysis, clean data were obtained and analyzed on the online platform of Majorbio Cloud Platform.2 Basic functional annotation analysis was performed on the provided reference genome (Organism: A. baumannii; Genome ID: GCF_008632635.1). The sequences were then annotated with the following databases including NCBI Nr, NCBI Nt, Pfam,3 eggNOG4, SwissProt5, GO6, and KEGG.7 Differentially expressed genes (DEGs) were identified by DESeq2 software, Version 1.24.0.8 Those mRNAs with absolute value log2[Fold Change (FC)] ≥ 1 and p-adjust < 0.05 were considered as significant.
The IBM SPSS 26.0 statistical software was used for statistical analysis. The t test was applied to compare the difference of measurement data, which were presented as mean ± SD. Spearman’s rank correlation was conducted to analyze the correlations between biofilm formation and gene expression levels. The Kaplan–Meier method was used to construct a survival curve, with a calculation of p value by the log-rank test. p < 0.05 was considered to be statistically significant.
In the case of the DNA sequences, BIT33_RS14560 gene widely existed in all 14 A. baumannii strains with different identities and two clades were identified in the phylogenetic tree (Supplementary Figure S1). In the monophyletic clade I, the BIT33_RS14560 gene was found to most closely related to A. baumannii ATCC 17961 and they both clustered into a subgroup with other five A. baumannii isolates. Interestingly, the pattern strain A. baumannii ATCC 17978 was subgrouped with another six isolates into clade II, indicating a distant relationship with A. baumannii ATCC 19606.
When it came to the amino acid sequences, the phylogenetic tree showed that the BIT33_RS14560 membrane transporter was most closely related to A. baumannii WP_0657191181 (Supplementary Figure S2). In addition, this protein of interest was clustered into a subgroup with its homologous proteins A. baumannii HAV5588984.1, A. baumannii HAV6132876.1, A. baumannii HAV4460275.1, and Klebsiella pneumoniae SSW87290.1. What is more, the target protein was constituted into an evolutionary clade together with Acinetobacter nosocomialis WP_004709884.1, A. nosocomialis HAB71326.1, A. nosocomialis WP_207694075.1 as well as A. baumannii HAV5002142.1.
In this study, when biofilm biomass was measured at 72 h by crystal violet staining, 62 A. baumannii isolates showed a various range of OD570 from 0.09 to 2.29 nm, indicating varying abilities of biofilm formation. Among all the isolates, 9 (14.5%), 28 (45.2%), and 17 (27.4%) were strong, moderate, and weak biofilm producers, respectively, while no biofilm was observed in eight (12.9%) isolates. Interestingly, we found that these A. baumannii isolates also differed considerably in their mRNA levels of BIT33_RS14560, with the △CT ranging from 0.47 to 19.51 (Table 3). Spearman’s rank correlation analysis indicated that the △CT value of BIT33_RS14560 was negatively correlated with the biofilm rank of those A. baumannii isolates (R = − 0.569, p = 0.000), which suggested that a stronger biofilm-forming ability could be followed by a relative higher the expression level of BIT33_RS14560 (Supplementary Table S4).
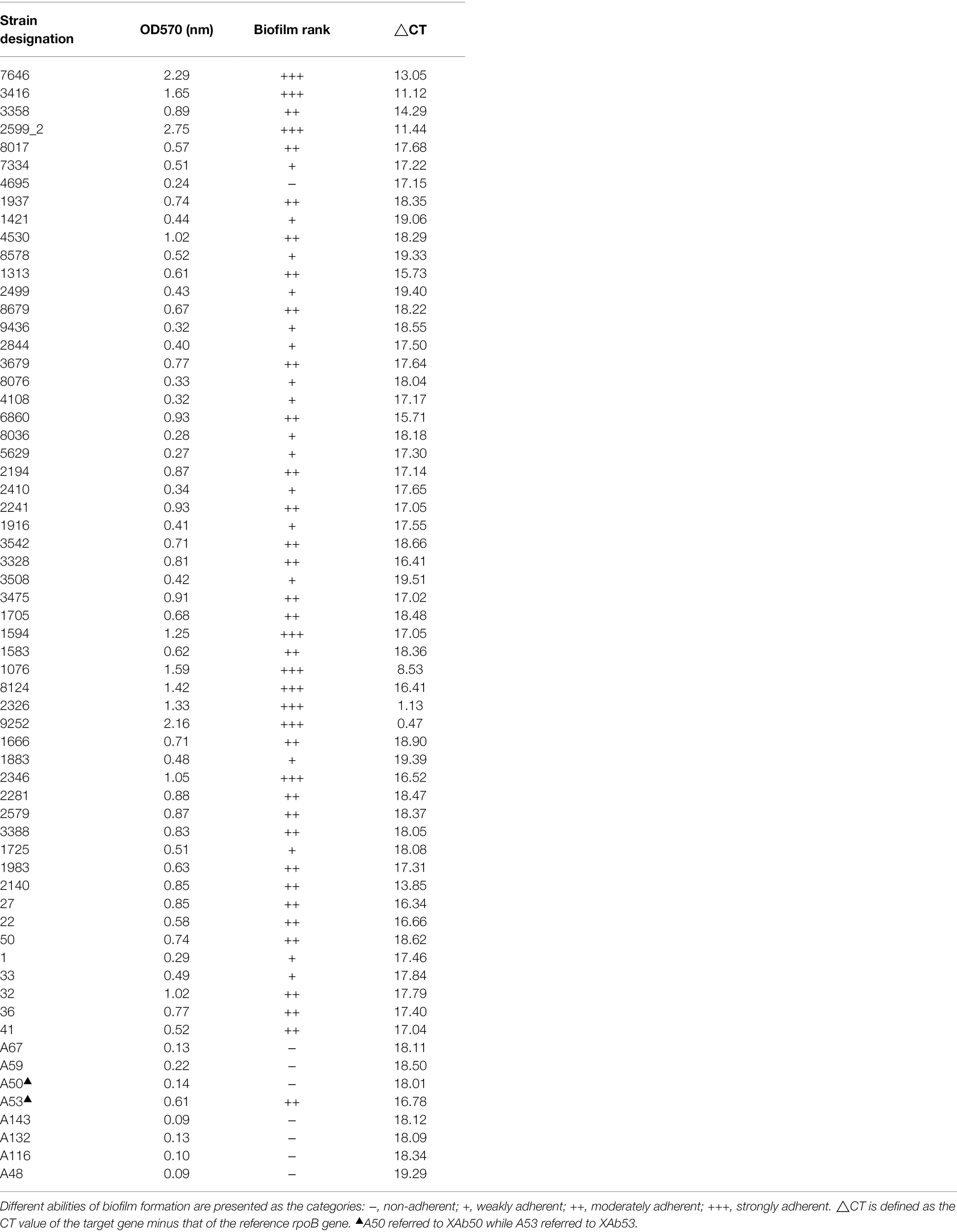
Table 3. The correlation between the biofilm biomass of 62 A. baumannii and their mRNA levels of BIT33_RS14560.
Fragment of BIT33_RS14560 coding sequence (CDS) plus its preceding promoter was cloned from the ATCC 19606 genome and 1% agarose gel electrophoresis of the PCR products yielded an expected band, 1,632 bp in size. The purified PCR products were inserted into pWH1266 to construct a vector p-RS14560 for the overexpression of BIT33_RS14560 in ATCC 19606. PCR analysis of the cloned plasmid followed by gene sequencing showed that the target fragment was successfully and correctly cloned into pWH1266 plasmid and the cloned gene was the same as BIT33_RS14560 (NCBI Reference Sequence: NZ_MJHA01000008.1) in the GenBank. These findings indicated that the expression vector p-RS14560 was successfully constructed (Supplementary Figures S3, S4). A 3,075 bp TelR cassette was successfully cloned from the suicide vector pMo130telR and inserted into the p-RS14560 plasmid to construct a new vector p-telR-RS14560. These were confirmed by obtaining two expected bands, 3,075 and 3,526 bp in size, respectively (Figure 1). Surprisingly, we managed to overexpress the target gene BIT33_RS14560 in two XDR-Ab isolates with this recombinant plasmid (see this article below).
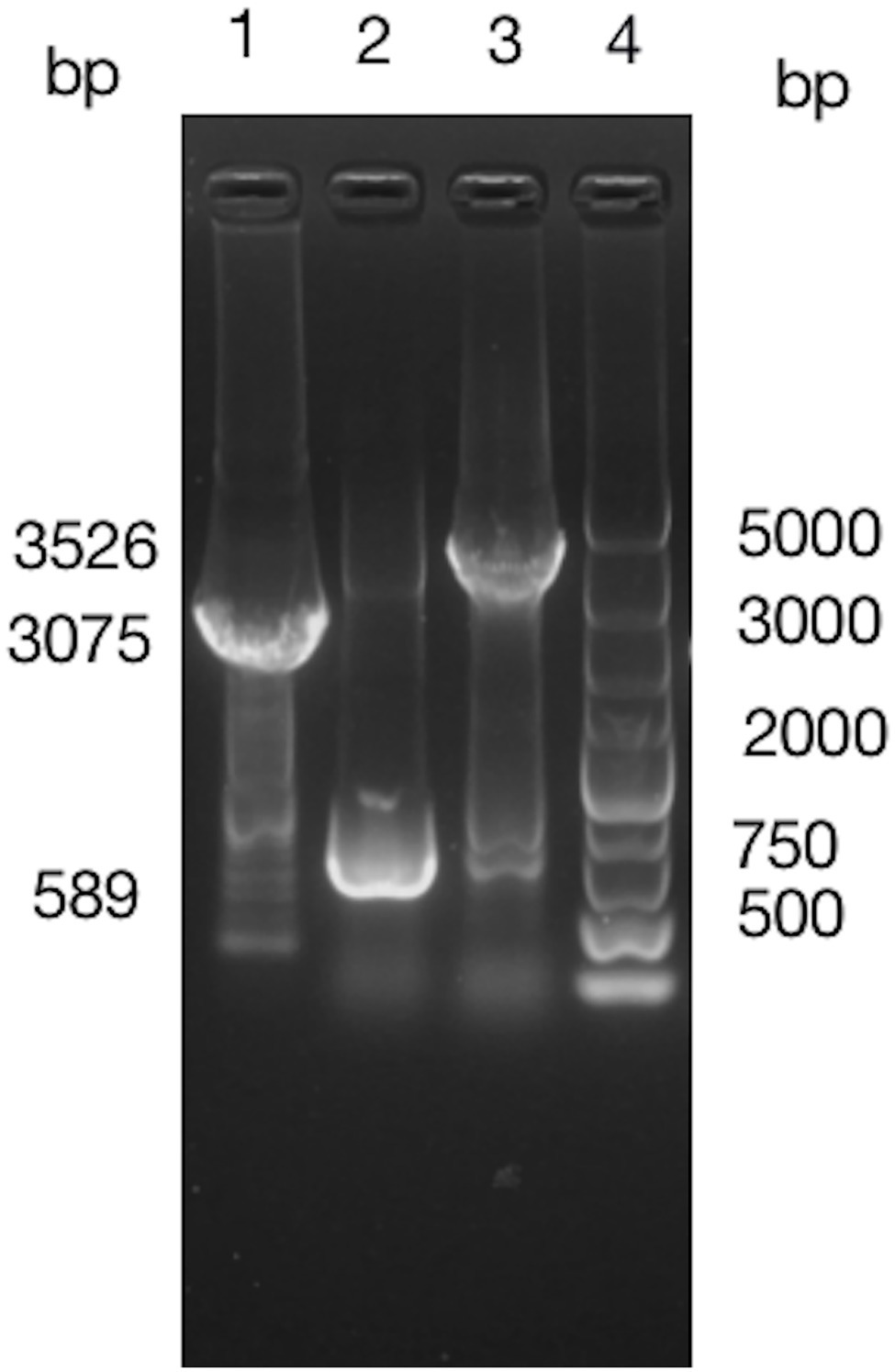
Figure 1. Around 1% agarose gel electrophoresis of PCR products indicates a successful construction of p-telR-RS14560 vector. The AmpR fragment in the p-RS14560 plasmid is replaced by telR cassette amplified from the pMo130telR vector. Lane 1: PCR products of telR cassette; lane 2: PCR products of p-RS14560 plasmid as negative control, lane 3: PCR products of positive clone, and lane 4: DNA marker 5,000 bp.
As mentioned above, those clinical A. baumannii isolates with higher BIT33_RS14560 mRNA levels tended to possess a stronger capacity of biofilm formation. To further investigate this relationship, a standard A. baumannii strain ATCC 19606, together with two XDR-Ab isolates, were transformed with the overexpression plasmids, p-RS14560 and p-telR-RS14560, respectively. To confirm if over-expression of BIT33_RS14560 occurred in the ATCC 19606 and these two XDR-Ab isolates, relative quantification of the BIT33_RS14560 transcript levels were determined by qRT-PCR with the corresponding strains transformed with empty plasmids as the control. In ATCC 19606, there was no significant difference between empty plasmid group and wild-type group, indicating that the transformation of two recombinant plasmids had little effect on the expression of BIT33_RS14560 gene (p > 0.05). For ATCC 19606 strain with the plasmid overexpressing the gene of interest, when compared with its control group, the expression level of the BIT33_RS14560 was significantly increased at 311.5 folds (p < 0.0001). For XAb53 strain, the relative expression levels of BIT33_RS14560 in the overexpression group was surprisingly higher at about 586 1000-fold than that of its control group (p < 0.0001). Similarly, for the other XDR-Ab strain, XAb50, the overexpression group significantly displayed approximately 619 1000-fold higher BIT33_RS14560 mRNA levels in comparison with its empty plasmid group (p < 0.0001). These results indicated the success of overexpression of BIT33_RS14560 in the three A. baumannii strains (Figure 2). The breathtaking multiples of overexpression in two XDR-Ab isolates may probably come from their very low transcript levels (or relative high △CT values) of this target gene, as shown in Table 3.
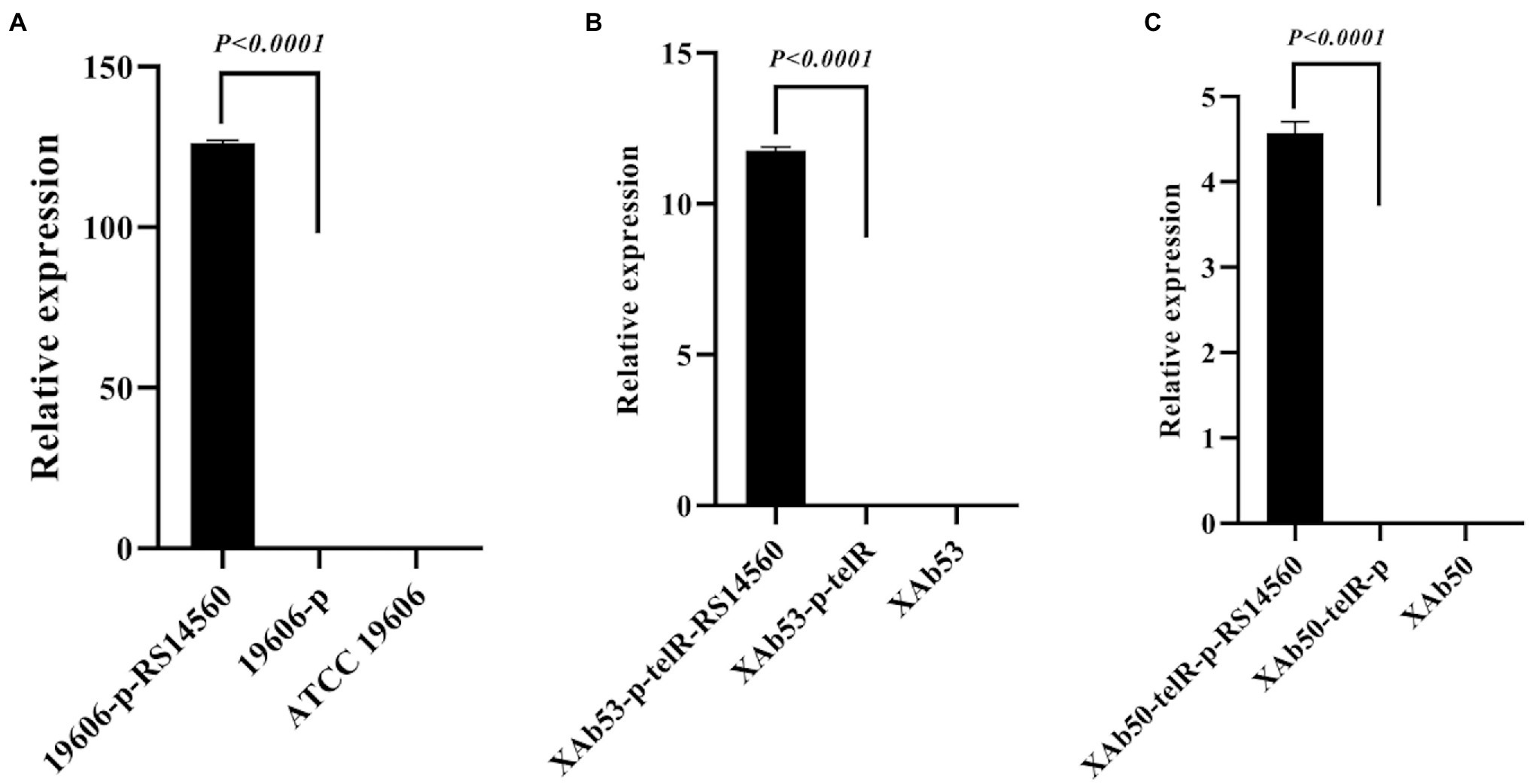
Figure 2. Relative expression of BIT33_RS14560 by qRT-PCR. The x-axis represents the A. baumannii strains, and the y-axis represents the relative mRNA levels of BIT33_RS14560 (2–△CT). p < 0.05 indicates a statistical significance. (A) Overexpression of ATCC 19606 strain; (B) Overexpression of XAb53 isolate; and (C) Overexpression of XAb50 isolate.
To verify the effect of BIT33_RS14560 overexpression on the biofilm formation of three A. baumannii strains, we evaluated the biofilm biomass with the microtiter-plate test at different points of time. We found that these three wild-type A. baumannii strains, including ATCC 19606 and other two XDR-Ab isolates, differed in their peak time of biofilm formation, but normally displayed relatively high levels of production within 12–48 h. When compared with each control group transformed with empty plasmids, we observed significantly higher biofilm biomass in the experimental groups of BIT33_RS14560-overexpression, whether in the standard strain ATCC 19606 or the two isolates XAb53 and XAb50 (Figure 3).
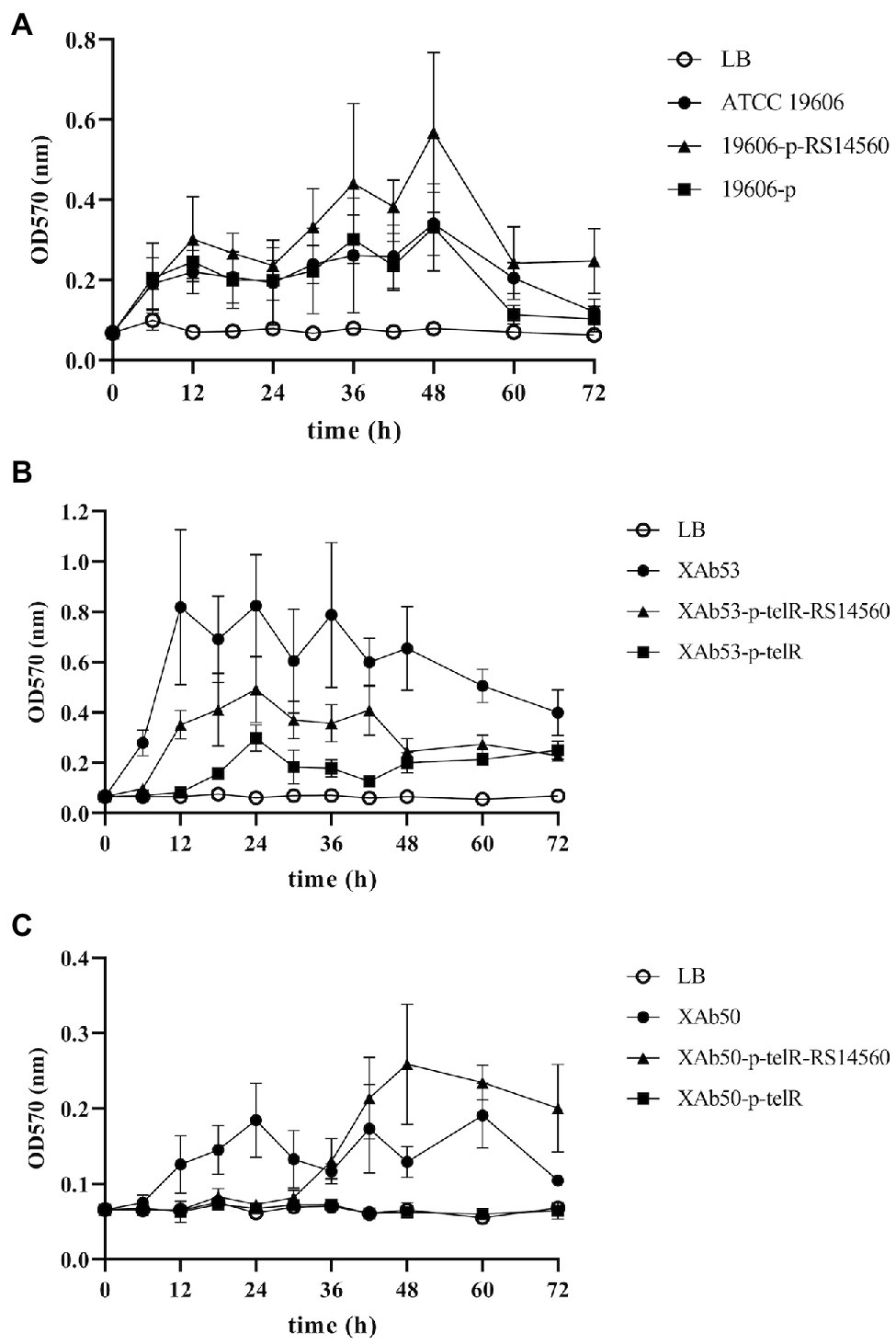
Figure 3. The dynamic changes of biofilm biomass produced by the nine A. baumannii strains at different time points. ATCC 19606, XAb53, and XAb50 are wild-type strains. 19,606-p-RS14560, XAb53-p-telR-RS14560, and XAb50-p-telR-RS14560 are BIT33_RS14560-overexpressed strains. 19,606-p, XAb53-p-telR, and XAb50-p-telR are empty plasmid control strains. The overexpression of BIT33_RS14560 exhibits enhancing effects, but with various time patterns, on the biofilm formation of strains in the experimental groups. Values are means ± SD of over three independent experiments. (A) Wild-type and recombinant strains of ATCC 19606; (B) Wild-type and recombinant strains of XAb53; and (C) Wild-type and recombinant strains of XAb50.
We used the G. mellonella infection model to investigate the effect on the virulence of the standard strain ATCC 19606 when BIT33_RS14560 was overexpressed. None of the G. mellonella died within 72 h when given with the equivalent PBS. By contrast, all G. mellonella died within 12 h following infection with the high-virulent strain A. baumannii 5075, which served as the positive control. When infected with the ATCC 19606 wild-type strain, G. mellonella mortality was 40.0% within 12 h and then rose to 70.0% at 24 h. In contrast, G. mellonella mortality following infection with the BIT33_RS14560 overexpression strain was 60.0% at 12 h and reached 90% within 24 h. No significant difference was observed in the survival rates between the wild-type strain ATCC 19606 and its empty plasmid strain 19,606-p (p > 0.05). As shown in Figure 4, Kaplan–Meier survival curve analysis showed that the survival rate of G. mellonella was significantly lower when infected with the BIT33_RS14560 overexpression strain (χ2 = 8.462, p = 0.004).
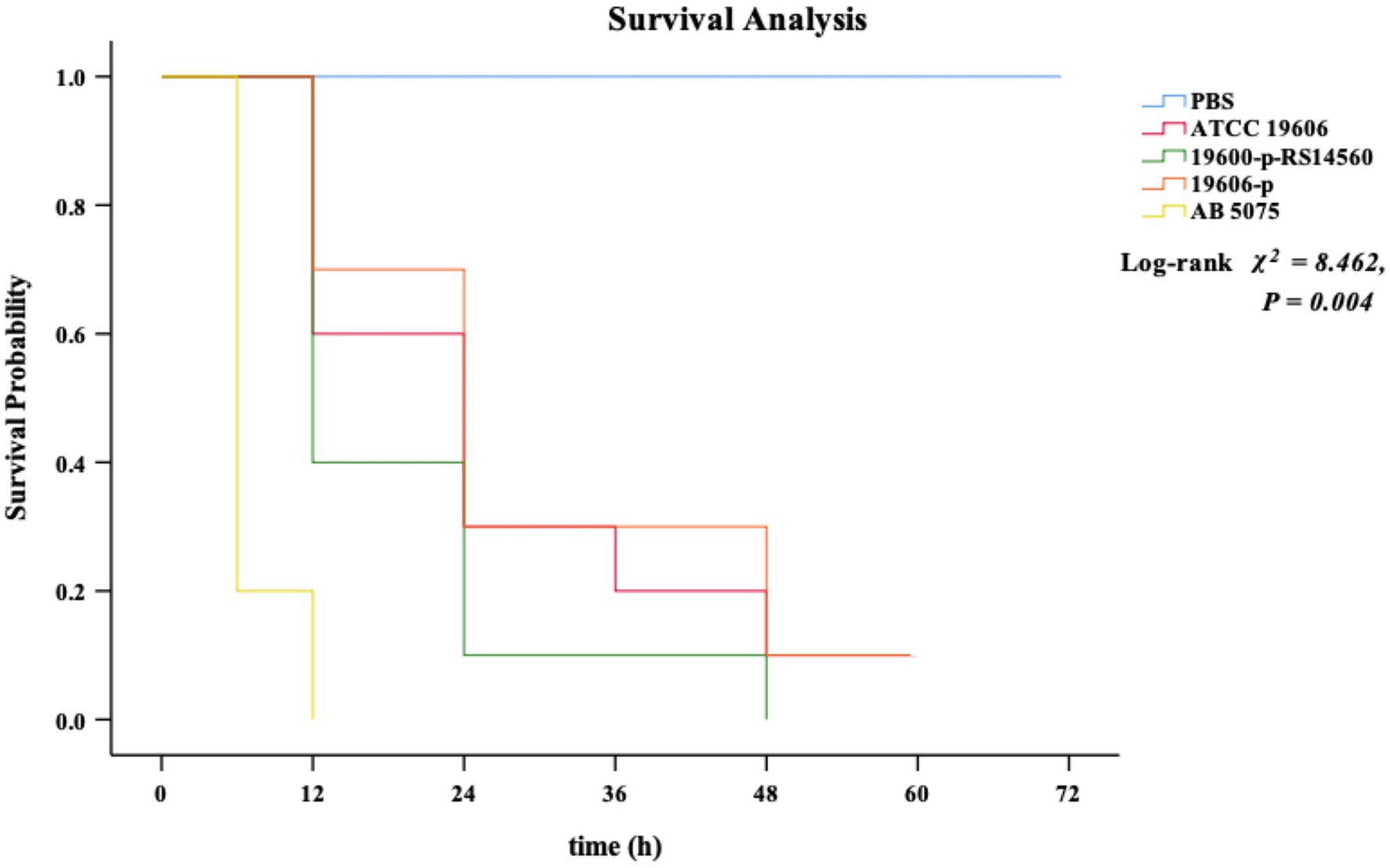
Figure 4. Kaplan–Meier curve analysis of the survival of G. mellonella after infection. PBS is the negative control group, ATCC 19606 is the wild-type strain, 19,606-p-RS14560 is the overexpression strain, 19,606-p is the empty plasmid control group, and A. baumannii 5075 is the positive control group. PBS, phosphate buffered saline. p < 0.05 indicates a statistical significance.
To better understand the mechanisms of the relationship between BIT33_RS14560 overexpression and A. baumannii biofilm formation, in the current study, we conducted a comparative transcriptomic analysis of four samples, including the treated group (two samples derived from ATCC 19606 with target gene overexpression) and the control group (two samples derived from ATCC 19606 with empty plasmid). A total of over 7.0 Gigabyte raw data were obtained, and the percentage of Q30 (an important index for the assessment of RNA-Seq quality, a higher value usually indicates a lower probability of base error) reached above 94.65%. Totally, 3,232 expressed genes were detected in this analysis, including 3,208 mRNAs and 24 small RNAs (sRNAs). Under the threshold of |log2(FC)| ≥ 1 and p-adjust < 0.05, a total of 12 DEGs (seven upregulated and five downregulated) were screened for subsequent analysis. The volcano plot and heatmap of these DEGs were drawn in Figures 5, 6, respectively. To further explore the biological functions of these DEGs, we performed GO and KEGG enrichment analysis. As presented in Figure 7, none of these DEGs were involved in GO biological process or cellular component category. However, in molecular function(MF)category, they significantly enriched in “magnesium transmembrane transporter activity, phosphorylative mechanism” (GO:0015444), “3-oxoacid CoA-transferase activity” (GO:0008260), “acetate CoA-transferase activity” (GO:0008775), “ATPase-coupled cation transmembrane transporter activity” (GO:0019829), and “transferase activity, transferring glycosyl groups” (GO:0016757; all FDR<0.05). In terms of the KEGG pathway, “aminobenzoate degradation” (map00627) and “valine, leucine, and isoleucine degradation” (map00280) were involved (FDR < 0.05; Figure 8).
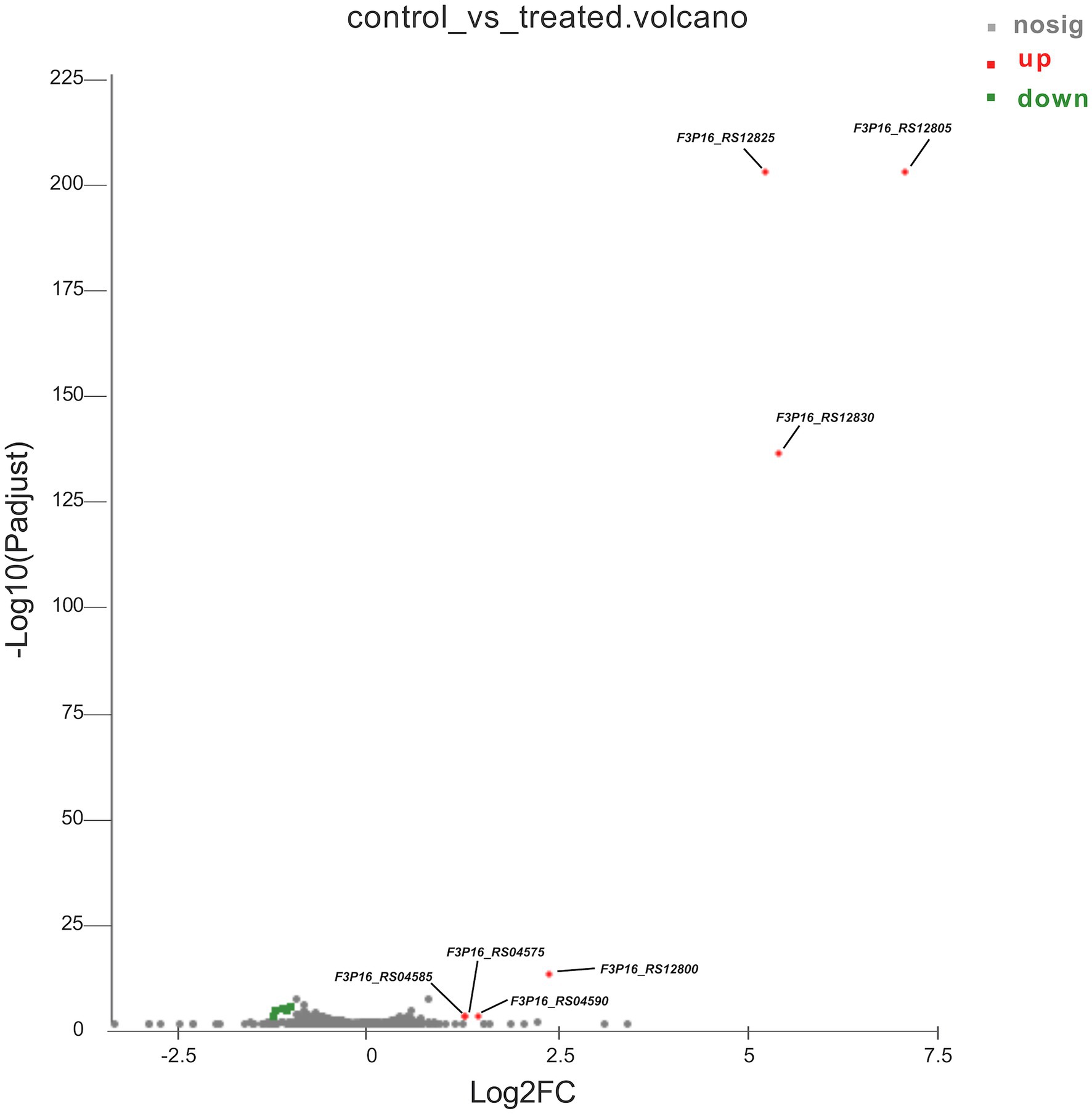
Figure 5. The volcano plot of differentially expressed genes identified by RNA-Seq. The x-axis represents the log2(FC), and the y-axis represents –log10 (p-adjust) calculated by the Student’s t-test. The red dots represent the upregulated genes with statistical significance ( p-adjust < 0.05 and |log2(FC)| ≥ 1). The green dots represent genes with down-regulated expression, while the gray ones represent genes with no statistical significance. Those upregulated DEGs are indicated. FC, fold change.
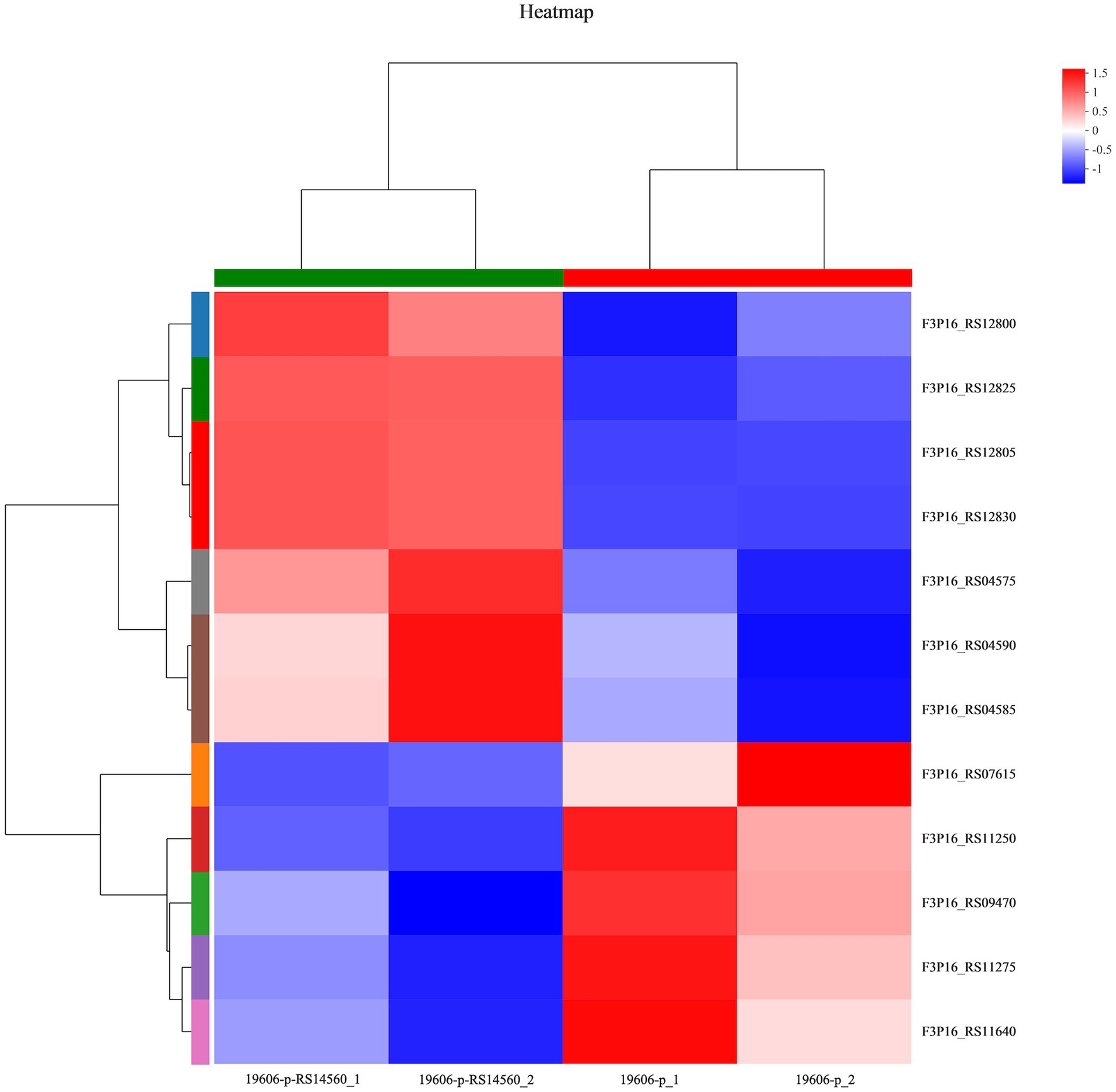
Figure 6. Heatmap of differentially expressed genes. 19,606-p-RS14,560_1 and 19,606-p-RS14,560_2 represent the treated groups while 19,606-p_1 and 19,606-p_2 represent the control groups. FC, fold change.
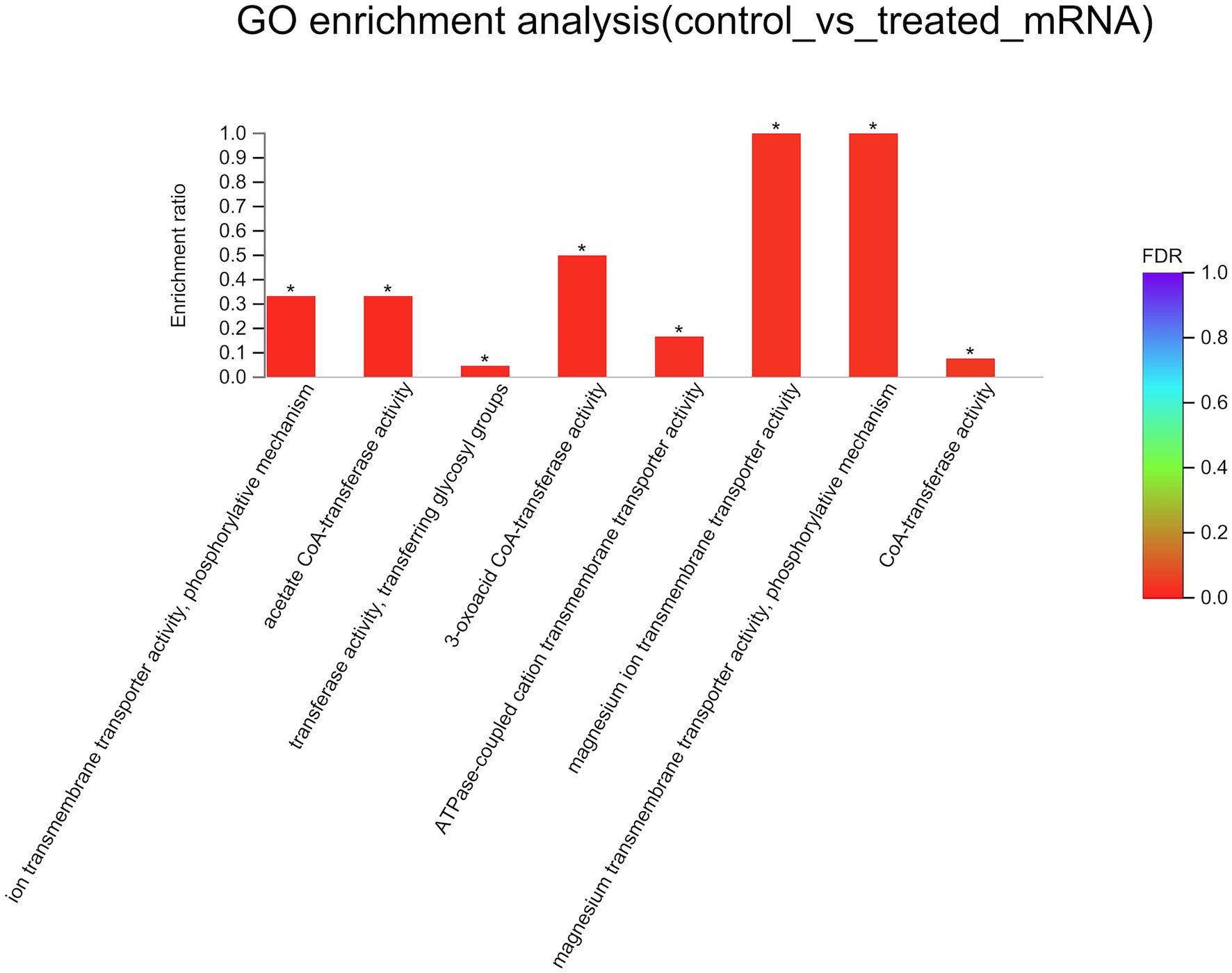
Figure 7. GO enrichment analysis of differentially expressed genes. The x-axis represents GO terms, and the y-axis represents the enrichment ratio. A higher ratio means a greater degree of enrichment. GO, Gene Ontology; FDR, false discovery rate. *FDR < 0.05.
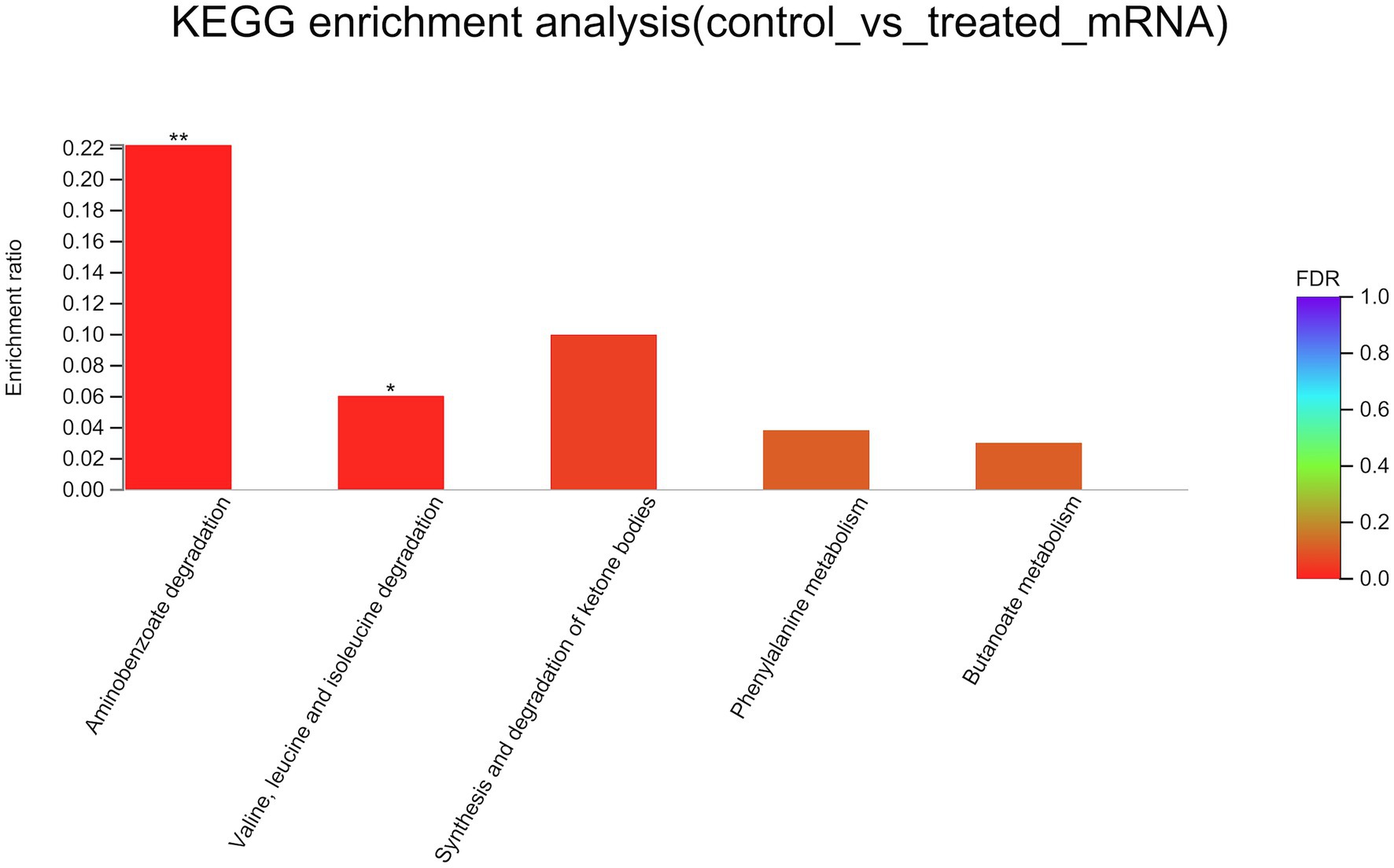
Figure 8. KEGG enrichment analysis of differentially expressed genes. The x-axis represents KEGG terms, and the y-axis represents the enrichment ratio. A higher ratio means a greater degree of enrichment. KEGG, Kyoto Encyclopedia of Genes and Genome. FDR, false discovery rate. *FDR < 0.05, **FDR < 0.01.
To investigate the distribution of the BIT33_RS134560 gene and its encoding protein in A. baumannii strains as well as their relationship, we conducted phylogenetic tree analysis. Our results showed that this gene existed in all the 14 A. baumannii strains of different genomic homology (Supplementary Figure S1). Similarly, the BIT33_RS14560 protein and its homologous ones could be found in many other strains of A. baumannii, even in other Acinetobacter spp. and K. pneumoniae (Supplementary Figure S2). These results indicated a wide distribution of the BIT33_RS134560 gene and its protein in A. baumannii strains. It was interesting to note that this target gene in ATCC19606 was very closely related to ATCC17961, but not to ATCC17978, although they were all pattern strains of A. baumannii. We have not figured out why and how this gene evolved the way it did. Further study of the difference in the expression of the BIT33_RS14560 gene between these two clades may shed light on the causes of this phenomenon.
In addition, that the BIT33_RS14560 protein was closely related to the MFS homologs of A. nosocomiae and K. pneumoniae indicated potential pathogenicity of this protein due to the fact that the latter two bacteria were also important pathogens of nosocomial infections (Nho et al., 2015; Wang et al., 2020). Therefore, we believed this gene of interest needed to be noticed.
The E. coli/Acinetobacter shuttle vector pWH1266 containing AmpR and TetR was initially generated for the cloning experiments of Acinetobacter calcoaceticus and E. coli (Hunger et al., 1990) and has been successfully applied in the overexpression construction of A. baumannii (Zander et al., 2013; Liou et al., 2014). However, for XDR-Ab, it should be mentioned that due to its extreme drug resistance, the number of antibiotic selection markers for transformants was greatly limited. One solution to this problem was that the antibiotic concentration should be increased as much as possible within the tolerant range of the vector’s resistance. In this case, we made our attempt to transform the recombinant plasmid p-RS14560, which contained only AmpR, into an XDR-Ab isolate by increasing the concentration of Carbenicillin from 50 to 1,700 μg/ml. However, we failed because the XDR-Ab was still able to grow at such a high concentration of Carbenicillin. To achieve this goal of overexpression in XDR-Ab, a replacement of AmpR by other resistant genes was the key to the deal.
When we made our attempts to knock out this target gene using a marker-less method (Amin et al., 2013), the TelR cassette of pMo130telR vector caught our attention. This suicide vector was derived from the modification of the pMo130 vector (Hamad et al., 2009) by carrying a TelR cassette (Sanchez-Romero et al., 1998). Researchers had tested A. baumannii isolates, including MDR-Ab isolates, and found that they were susceptible to tellurite (Amin et al., 2013). We also tested several XDR-Ab isolates to confirm their susceptibilities to tellurite and found that they could be counter-selected in LB agar containing tellurite of 30–100 μg/ml (data not shown). Fortunately, we managed to overexpress the target gene BIT33_RS14560 in two XDR-Ab isolates by replacing the AmpR with the TelR. The transformation of two recombinant vectors, p-telR and p-telR-RS14560, might to some extent make an impact on the transcript expression of BIT33_RS14560. This could be explained by the fitness cost of bacteria (San Millan and MacLean, 2017; Nang et al., 2018). Large plasmids could impose a metabolic burden for host bacterial strains (Ma et al., 2018). However, to our knowledge, this was the first time to achieve stable gene overexpression in XDR-Ab by the replacement of AmpR with telR, which would provide guidance for further investigations on XDR-Ab strains.
Like many other bacteria, A. baumannii displays a robust biofilm formation on many abiotic surfaces in hospital environments, raising the risk of chronic infections. Previous studies have shown that the degree of biofilm formation varied considerably depending on the A. baumannii isolates (Rodríguez-Baño et al., 2008). Our study also showed different abilities of biofilm formation among 62 A. baumannii isolates when measured at 72 h. Those with robust abilities of biofilm formation were more likely to survive under desiccation and nutrition-limiting conditions, which explained why some clinical isolates possessed more tenacious vitality in hospitals than others. As shown in Supplementary Table S4, Spearman’s rank correlation analysis suggested that a stronger biofilm-forming ability could be followed by a relatively higher expression level of BIT33_RS14560 (the △CT value vs. biofilm rank, R = −0.569, p = 0.000). This was validated by overexpression of the BIT33_RS14560 gene in the standard strain ATCC 19606 and the two isolates XAb53 and XAb50 (Figure 3).
Here, three wild-type A. baumannii strains used for the construction of BIT33-RS14560 overexpression normally displayed relatively high levels of production within 12–48 h, which was in accordance with the previous study (Rodríguez-Baño et al., 2008). However, the enhancing effects of BIT33_RS14560 on biofilm formation appeared to be variable in these three A. baumannii strains (Figure 3). Within 48 h, for the ATCC 19606 strain, overexpression of BIT33_RS14560 exhibited a gradual upward trend of facilitating biofilm formation, while in the XAb53 isolate; this effect seemed to be more stable, keeping it in fairly high levels of biofilm biomass. However, when it came to XAb50 isolate, the amount of biofilm remained relatively unchanged over the past 30 h, then, rose sharply into about 3.24-fold (data not shown) at 48 h. Further investigation is warranted to elucidate the mechanisms for this phenomenon.
Currently, A. baumannii biofilm formation, of which the mechanisms are still not fully elucidated, is still a tough issue in the treatment of refractory and chronic infections. As one of the largest superfamily of secondary carriers known to date, MFS family membrane transporters exist widely in the whole biological world (Reddy et al., 2012) and they are considered to be closely related to a wide variety of life phenomena due to a basic function of assisting in transmembrane transport of certain substances, including monosaccharides, oligosaccharides, amino acids, enzyme cofactors, drugs, and so on (Lorca et al., 2007; Chen et al., 2008; Yen et al., 2010). In bacteria, MFS superfamily proteins have been found not only to play an important role in the transport of many substances but also to be a member of the efflux pumps large families (Pasqua et al., 2019), which are involved in biofilm formation and drug resistance (Bay et al., 2017; Saranathan et al., 2017; Poudyal and Sauer, 2018; Tang et al., 2020).
BIT33_RS14560, although considered to be categorized into the MFS family, is still lacking reports about its effects on the biofilm formation of A. baumannii. In this study, we conducted a comparative transcriptomic analysis to investigate the possible mechanisms by which BIT33_RS14560 overexpression had an impact on the biofilm formation and virulence of A. baumannii ATCC19606. As shown in Table 4, we found that three DEGs, including F3P16_RS12800, F3P16_RS04575, and F3P16_RS04585, although displayed different up or down multiples, were hypothesized to be related more or less to bacterial biofilm formation or virulence.
F3P16_RS12800 was annotated as an OprD family outer membrane porin. The OprD family was described first for P. aeruginosa, but ever since that time, it has been found in many metabolically versatile soil bacteria and comprises over 100 members (Tamber et al., 2006). In the current study, we found that the overexpression of BIT33_RS14560 led to a significant upregulation of F3P16_RS12800, suggesting that in A. baumannii, BIT33_RS14560 may enhance biofilm formation by upregulating the expression of this outer membrane porin. This was supported by a previous comparative proteomics study (Cabral et al., 2011) revealing that some membrane proteins including OprD-like protein were involved in the adhesion and biofilm formation of A. baumannii.
Most importantly, we got F3P16_RS04575 and F3P16_RS04585, both encoded proteins belonging to the glycosyltransferase (GT) family and were upregulated when BIT33_RS14560 was overexpressed. GTs mainly participate in the transfer process of glycosyl from an activated donor to a receptor, such as sugar, lipid, protein, and nucleic acid and play an important role in pathogenic adhesion, biofilm formation, and virulence (Breton et al., 2006; Ribet and Cossart, 2010a). Structural analysis of three-dimensional (3D) protein fold has shown that the catalytic domains of GTs are categorized into GT-A, GT-B, and GT-C three types, the most common of which are GT-A and GT-B (Moremen and Haltiwanger, 2019). GT-B or GT-C is believed to play a role in adhesion, biofilm formation, and virulence of pathogenic bacteria while GT-A enters the host cell and interferes it with post-translational modifications thus affecting its signal transduction, protein translation, and immune response. It was also important to note that the protein glycosylation of GT was closely related to the virulence of pathogenic bacteria (Ribet and Cossart, 2010b; Yakovlieva and Walvoort, 2020). Many bacterial pathogens were found to bear toxic GTs that interfere with host post-translational modifications to promote their own survival and replication (Lu et al., 2015). Interestingly, when we further analyzed with the Carbohydrate-Active Enzymes (CAZy) database,9 we found that the proteins encoded by F3P16_RS04575 and F3P16_RS04585 belonged to GT-C and GT-A, respectively. This might explain why ATCC 19606 became more virulent when BIT33_RS14560 was overexpressed (Figure 4).
It was reported that proteins and carbohydrates were the main components of biofilm formation (Costerton et al., 1995). Amino acid metabolism, glycerolipid, and tricarboxylic acid cycle were also observed to be mostly affected during bacterial biofilm formation (Lu et al., 2019). Our results of GO and KEGG analysis (Figures 7, 8) also indicated that most DEGs participated in complicated metabolic processes including amino acid, carbohydrate metabolism, which also suggested the complexity of bacterial biofilm formation. However, the specific effects of these DEGs on A. baumannii biofilm formation and what metabolic pathways these DEGs may be involved in remained further investigations.
In this study, we demonstrated that when BIT33_RS14560, a member of the MFS family, was over-expressed, the abilities to form biofilm and the virulence of A. baumannii were significantly enhanced, no matter in standard or clinically isolated strains. This gene of interest existed widely in A. baumannii strains and its protein was closely related to the MFS homologs of A. nosocomiae and K. pneumoniae. Three DEGs including F3P16_RS12800, F3P16_RS04575, and F3P16_RS04585, annotated as OprD family outer membrane porin, glycosyltransferase family 39 protein and glycosyltransferase family 2 protein, respectively, were identified by RNA-Seq and speculated to be related to the biofilm formation and virulence of A. baumannii, which needs further studies. These findings provided new insights in identifying potential drug targets for the inhibition of biofilm formation. Besides, we developed a method for the construction of XDR-Ab overexpression, which may promote further microbiological research on their genotypes and phenotypes.
The data presented in the study are deposited in the Sequence Read Archive (SRA) repository, accession number at: https://www.ncbi.nlm.nih.gov/sra/PRJNA821219.
RY and BLa contributed to the conception and design of this study. RY, BLa, and BLi performed the experiments. KL, YC, and CZ provided the study materials. RY, LH, SL, JG, ZL, and SW analyzed and interpreted the data. YZ, CZ, and KL gave administrative supports. All authors contributed to the article and approved the submitted version.
This work is supported by grants from the National Natural Science Foundation of China (81570008, YZ) and the Natural Science Foundation of Guangdong Province of China (2021A1515010480, YZ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the Microbiology Laboratory of State Key Laboratory of Respiratory Diseases, Guangzhou Medical University for scientific research. We are grateful to Shanghai Majorbio Bio-pharm Technology Co., Ltd. for the analysis of RNA-Seq data.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.867770/full#supplementary-material
Supplementary Figure S1 | Phylogenetic analysis of BIT33_RS14560 gene. Bootstrap values >50 are displayed on the branches. A higher bootstrap value indicates a better reliability.
Supplementary Figure S2 | Phylogenetic analysis of BIT33_RS14560 protein. Bootstrap values >50 are displayed on the branches. A higher bootstrap value indicates a better reliability.
Supplementary Figure S3 | The construction of p-RS14560 overexpressed vector. (A) The amplification of BIT33_RS14560 fragment. Lane 1: DNA marker 2,000 bp, lane 2: sample; (B) PCR products of pWH1266 plasmid as negative control. Lane 1: DNA marker 2,000 bp, lane 2: PCR products of pWH1266 plasmid as negative control (C) Recombinant plasmid is successively transformed into competent E. coli DH5α and ATCC 19606 strains, with the Pwh1266-F/Pwh1266-R primers being used to ensure the success of directional linkage. Lane 1: DNA marker 2,000 bp, lane 2: PCR products of p-RS14560 plasmid as positive control, lane 3: PCR products of positive clone.
Supplementary Figure S4 | A representative part of the sequence alignment between the target gene in the p-RS14560 vector and the comparative analysis to the BIT33_RS14560 gene.
1. ^http://www.ncbi.nlm.nih.gov/BLAST/
6. ^http://www.geneontology.org/
7. ^http://www.genome.jp/kegg/
Amin, I. M., Richmond, G. E., Sen, P., Koh, T. H., Piddock, L. J., and Chua, K. L. (2013). A method for generating marker-less gene deletions in multidrug-resistant Acinetobacter baumannii. BMC Microbiol. 13:158. doi: 10.1186/1471-2180-13-158
Ayoub Moubareck, C., and Hammoudi Halat, D. (2020). Insights into Acinetobacter baumannii: a review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics 9:119. doi: 10.3390/antibiotics9030119
Bay, D. C., Stremick, C. A., Slipski, C. J., and Turner, R. J. (2017). Secondary multidrug efflux pump mutants alter Escherichia coli biofilm growth in the presence of cationic antimicrobial compounds. Res. Microbiol. 168, 208–221. doi: 10.1016/j.resmic.2016.11.003
Breton, C., Snajdrová, L., Jeanneau, C., Koca, J., and Imberty, A. (2006). Structures and mechanisms of glycosyltransferases. Glycobiology 16, 29R–37R. doi: 10.1093/glycob/cwj016
Cabral, M. P., Soares, N. C., Aranda, J., Parreira, J. R., Rumbo, C., Poza, M., et al. (2011). Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J. Proteome Res. 10, 3399–3417. doi: 10.1021/pr101299j
Chen, D. E., Podell, S., Sauer, J. D., Swanson, M. S., and Saier, M. H. Jr. (2008). The phagosomal nutrient transporter (Pht) family. Microbiology 154, 42–53. doi: 10.1099/mic.0.2007/010611-0
Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R., and Lappin-Scott, H. M. (1995). Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745. doi: 10.1146/annurev.mi.49.100195.003431
Fattahian, Y., Rasooli, I., Mousavi Gargari, S. L., Rahbar, M. R., Darvish Alipour Astaneh, S., and Amani, J. (2011). Protection against Acinetobacter baumannii infection via its functional deprivation of biofilm associated protein (bap). Microb. Pathog. 51, 402–406. doi: 10.1016/j.micpath.2011.09.004
Gaddy, J. A., and Actis, L. A. (2009). Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol. 4, 273–278. doi: 10.2217/fmb.09.5
Gaddy, J. A., Tomaras, A. P., and Actis, L. A. (2009). The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77, 3150–3160. doi: 10.1128/IAI.00096-09
Hamad, M. A., Zajdowicz, S. L., Holmes, R. K., and Voskuil, M. I. (2009). An allelic exchange system for compliant genetic manipulation of the select agents Burkholderia pseudomallei and Burkholderia mallei. Gene 430, 123–131. doi: 10.1016/j.gene.2008.10.011
Hunger, M., Schmucker, R., Kishan, V., and Hillen, W. (1990). Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87, 45–51. doi: 10.1016/0378-1119(90)90494-c
Ibrahim, S., Al-Saryi, N., Al-Kadmy, I. M. S., and Aziz, S. N. (2021). Multidrug-resistant Acinetobacter baumannii as an emerging concern in hospitals. Mol. Biol. Rep. 48, 6987–6998. doi: 10.1007/s11033-021-06690-6
Kengkla, K., Kongpakwattana, K., Saokaew, S., Apisarnthanarak, A., and Chaiyakunapruk, N. (2018). Comparative efficacy and safety of treatment options for MDR and XDR Acinetobacter baumannii infections: a systematic review and network meta-analysis. J. Antimicrob. Chemother. 73, 22–32. doi: 10.1093/jac/dkx368
Lee, C. R., Lee, J. H., Park, M., Park, K. S., Bae, I. K., Kim, Y. B., et al. (2017). Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol. 7:55. doi: 10.3389/fcimb.2017.00055
Liou, M. L., Soo, P. C., Ling, S. R., Kuo, H. Y., Tang, C. Y., and Chang, K. C. (2014). The sensor kinase BfmS mediates virulence in Acinetobacter baumannii. Microbiol. Immunol. Infect. 47, 275–281. doi: 10.1016/j.jmii.2012.12.004
Lorca, G. L., Barabote, R. D., Zlotopolski, V., Tran, C., Winnen, B., Hvorup, R. N., et al. (2007). Transport capabilities of eleven gram-positive bacteria: comparative genomic analyses. Biochim. Biophys. Acta 1768, 1342–1366. doi: 10.1016/j.bbamem.2007.02.007
Lu, Q., Li, S., and Shao, F. (2015). Sweet talk: protein glycosylation in bacterial interaction with the host. Trends Microbiol. 23, 630–641. doi: 10.1016/j.tim.2015.07.003
Lu, H., Que, Y., Wu, X., Guan, T., and Guo, H. (2019). Metabolomics deciphered metabolic reprogramming required for biofilm formation. Sci. Rep. 9:13160. doi: 10.1038/s41598-019-49603-1
Ma, K., Feng, Y., and Zong, Z. (2018). Fitness cost of a mcr-1-carrying IncHI2 plasmid. PLoS One 13:e0209706. doi: 10.1371/journal.pone.0209706
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug resistant, extensively drug resistant and pan drug resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Moremen, K. W., and Haltiwanger, R. S. (2019). Emerging structural insights into glycosyltransferase-mediated synthesis of glycans. Nat. Chem. Biol. 15, 853–864. doi: 10.1038/s41589-019-0350-2
Nang, S. C., Morris, F. C., McDonald, M. J., Han, M. L., Wang, J., Strugnell, R. A., et al. (2018). Fitness cost of mcr-1-mediated polymyxin resistance in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1604–1610. doi: 10.1093/jac/dky061
Nho, J. S., Jun, S. H., Oh, M. H., Park, T. I., Choi, C. W., Kim, S. I., et al. (2015). Acinetobacter nosocomialis secretes outer membrane vesicles that induce epithelial cell death and host inflammatory responses. Microb. Pathog. 81, 39–45. doi: 10.1016/j.micpath.2015.03.012
Pasqua, M., Grossi, M., Zennaro, A., Fanelli, G., Micheli, G., Barras, F., et al. (2019). The varied role of efflux pumps of the MFS family in the interplay of bacteria with animal and plant cells. Microorganisms 7:285. doi: 10.3390/microorganisms7090285
Poudyal, B., and Sauer, K. (2018). The ABC of biofilm drug tolerance: the MerR-like regulator BrlR is an activator of ABC transport systems, with PA1874-77 contributing to the tolerance of Pseudomonas aeruginosa biofilms to tobramycin. Antimicrob. Agents Chemother. 62, e01981–e01917. doi: 10.1128/AAC.01981-17
Reddy, V. S., Shlykov, M. A., Castillo, R., Sun, E. I., and Saier, M. H. Jr. (2012). The major facilitator superfamily (MFS) revisited. FEBS J. 279, 2022–2035. doi: 10.1111/j.1742-4658.2012.08588.x
Ribet, D., and Cossart, P. (2010a). Pathogen-mediated posttranslational modifications: a re-emerging field. Cell 143, 694–702. doi: 10.1016/j.cell.2010.11.019
Ribet, D., and Cossart, P. (2010b). Post-translational modifications in host cells during bacterial infection. FEBS Lett. 584, 2748–2758. doi: 10.1016/j.febslet.2010.05.012
Rodríguez-Baño, J., Martí, S., Soto, S., Fernández-Cuenca, F., Cisneros, J. M., Pachón, J., et al. (2008). Biofilm formation in Acinetobacter baumannii: associated features and clinical implications. Clin. Microbiol. Infect. 14, 276–278. doi: 10.1111/j.1469-0691.2007.01916.x
Roy, R., Tiwari, M., Donelli, G., and Tiwari, V. (2018). Strategies for combating bacterial biofilms: a focus on anti-biofilm agents and their mechanisms of action. Virulence 9, 522–554. doi: 10.1080/21505594.2017.1313372
Rumbo-Feal, S., Gómez, M. J., Gayoso, C., Álvarez-Fraga, L., Cabral, M. P., Aransay, A. M., et al. (2013). Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-Sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS One 8:e72968. doi: 10.1371/journal.pone.0072968
Sahu, P. K., Iyer, P. S., Gaikwad, M. B., Talreja, S. C., Pardesi, K. R., and Chopade, B. A. (2012). An MFS transporter-like ORF from MDR Acinetobacter baumannii AIIMS 7 is associated with adherence and biofilm formation on biotic/abiotic surface. Int. J. Microbiol. 2012:490647. doi: 10.1155/2012/490647
Saipriya, K., Swathi, C. H., Ratnakar, K. S., and Sritharan, V. (2020). Quorum-sensing system in Acinetobacter baumannii: a potential target for new drug development. J. Appl. Microbiol. 128, 15–27. doi: 10.1111/jam.14330
San Millan, A., and MacLean, R. C. (2017). Fitness costs of plasmids: a limit to plasmid transmission. Microbiol. Spectr. 5, 1–12. doi: 10.1128/microbiolspec.MTBP-0016-2017
Sanchez-Romero, J. M., Diaz-Orejas, R., and De Lorenzo, V. (1998). Resistance to tellurite as a selection marker for genetic manipulations of pseudomonas strains. Appl. Environ. Microbiol. 64, 4040–4046. doi: 10.1128/AEM.64.10.4040-4046.1998
Saranathan, R., Pagal, S., Sawant, A. R., Tomar, A., Madhangi, M., Sah, S., et al. (2017). Disruption of tetR type regulator adeN by mobile genetic element confers elevated virulence in Acinetobacter baumannii. Virulence 8, 1316–1334. doi: 10.1080/21505594.2017.1322240
Sharma, A., Sharma, R., Bhattacharyya, T., Bhando, T., and Pathania, R. (2017). Fosfomycin resistance in Acinetobacter baumannii is mediated by efflux through a major facilitator superfamily (MFS) transporter-AbaF. J. Antimicrob. Chemother. 72, 68–74. doi: 10.1093/jac/dkw382
Stepanović, S., Vuković, D., Hola, V., Di Bonaventura, G., Djukić, S., Cirković, I., et al. (2007). Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115, 891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x
Tamber, S., Ochs, M. M., and Hancock, R. E. (2006). Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa. J. Bacteriol. 188, 45–54. doi: 10.1128/JB.188.1.45-54.2006
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tang, M., Wei, X., Wan, X., Ding, Z., Ding, Y., and Liu, J. (2020). The role and relationship with efflux pump of biofilm formation in Klebsiella pneumoniae. Microb. Pathog. 147:104244. doi: 10.1016/j.micpath.2020.104244
Tomaras, A. P., Dorsey, C. W., Edelmann, R. E., and Actis, L. A. (2003). Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149, 3473–3484. doi: 10.1099/mic.0.26541-0
Wang, G., Zhao, G., Chao, X., Xie, L., and Wang, H. (2020). The characteristic of virulence, biofilm and antibiotic resistance of Klebsiella pneumoniae. Int. J. Environ. Res. Public Health 17:6278. doi: 10.3390/ijerph17176278
Yakovlieva, L., and Walvoort, M. T. C. (2020). Processivity in bacterial Glycosyltrans-ferases. ACS Chem. Biol. 15, 3–16. doi: 10.1021/acschembio.9b00619
Yang, C. H., Su, P. W., Moi, S. H., and Chuang, L. Y. (2019). Biofilm formation in Acinetobacter baumannii: genotype-phenotype correlation. Molecules 24:1849. doi: 10.3390/molecules24101849
Yen, M. R., Chen, J. S., Marquez, J. L., Sun, E. I., and Saier, M. H. (2010). Multidrug resistance: phylogenetic characterization of superfamilies of secondary carriers that include drug exporters. Methods Mol. Biol. 637, 47–64. doi: 10.1007/978-1-60761-700-6_3
Zander, E., Chmielarczyk, A., Heczko, P., Seifert, H., and Higgins, P. G. (2013). Conversion of OXA-66 into OXA-82 in clinical Acinetobacter baumannii isolates and association with altered carbapenem susceptibility. J. Antimicrob. Chemother. 68, 308–311. doi: 10.1093/jac/dks382
Keywords: Acinetobacter baumannii, BIT33_RS14560, biofilm, overexpression, RNA sequencing, extensively drug-resistant
Citation: Yang R, Lai B, Liao K, Liu B, Huang L, Li S, Gu J, Lin Z, Chen Y, Wang S, Qiu Y, Deng J, Chen S, Zhuo C and Zhou Y (2022) Overexpression of BIT33_RS14560 Enhances the Biofilm Formation and Virulence of Acinetobacter baumannii. Front. Microbiol. 13:867770. doi: 10.3389/fmicb.2022.867770
Received: 01 February 2022; Accepted: 22 March 2022;
Published: 25 April 2022.
Edited by:
Huancai Lin, Sun Yat-sen University, ChinaReviewed by:
Juan Carlos Vázquez Ucha, Institute of Biomedical Research of A Coruña (INIBIC), SpainCopyright © 2022 Yang, Lai, Liao, Liu, Huang, Li, Gu, Lin, Chen, Wang, Qiu, Deng, Chen, Zhuo and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanbin Zhou, emhvdXliQG1haWwuc3lzdS5lZHUuY24=; Chao Zhuo, Y2hhb3NoZWVwQHNpbmEuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.