- 1Department of Food and Experimental Nutrition, Faculty of Pharmaceutical Sciences, Food Research Center, University of São Paulo, São Paulo, Brazil
- 2Laboratory of Avian Pathology, Department of Pathology, Theriogenology, and One Health, São Paulo State University (FCAV-Unesp), São Paulo, Brazil
- 3Genomic Sciences Graduate Program, North Carolina State University, Raleigh, NC, United States
- 4Department of Food, Bioprocessing and Nutrition Sciences, North Carolina State University, Raleigh, NC, United States
- 5Department of Population Health and Pathobiology, College of Veterinary Medicine, North Carolina State University, Raleigh, NC, United States
- 6Department of Microbiology, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil
- 7Department of Clinical Analysis, Faculty of Pharmaceutical Sciences, University of São Paulo, São Paulo, Brazil
Salmonella enterica subsp. enterica serovar Heidelberg has been associated with a broad host range, such as poultry, dairy calves, swine, wild birds, environment, and humans. The continuous evolution of S. Heidelberg raises a public health concern since there is a global dispersal of lineages harboring a wide resistome and virulome on a global scale. Here, we characterized the resistome, phylogenetic structure and clustered regularly interspaced short palindromic repeats (CRISPR) array composition of 81 S. Heidelberg strains isolated from broiler farms (n = 16), transport and lairage (n = 5), slaughterhouse (n = 22), and retail market (n = 38) of the poultry production chain in Brazil, between 2015 and 2016 using high-resolution approaches including whole-genome sequencing (WGS) and WGS-derived CRISPR genotyping. More than 91% of the S. Heidelberg strains were multidrug-resistant. The total antimicrobial resistance (AMR) gene abundances did not vary significantly across regions and sources suggesting the widespread distribution of antibiotic-resistant strains from farm to market. The highest AMR gene abundance was observed for fosA7, aac(6′)-Iaa, sul2, tet(A), gyrA, and parC for 100% of the isolates, followed by 88.8% for blaCMY–2. The β-lactam resistance was essentially driven by the presence of the plasmid-mediated AmpC (pAmpC) blaCMY–2 gene, given the isolates which did not carry this gene were susceptible to cefoxitin (FOX). Most S. Heidelberg strains were classified within international lineages, which were phylogenetically nested with Salmonella strains from European countries; while CRISPR genotyping analysis revealed that the spacer content was overall highly conserved, but distributed into 13 distinct groups. In summary, our findings underscore the potential role of S. Heidelberg as a key pathogen disseminated from farm to fork in Brazil and reinforce the importance of CRISPR-based genotyping for salmonellae. Hence, we emphasized the need for continuous mitigation programs to monitor the dissemination of this high-priority pathogen.
Introduction
Salmonella enterica subsp. enterica serovar Heidelberg is most often associated with eggs and poultry (Hennessy et al., 2004; Chittick et al., 2006; Foley et al., 2008; Folster et al., 2012; Jackson et al., 2013). However, this scenario has changed since Salmonella Heidelberg has been associated with other sources from a broad host range, such as dairy calves (Centers for Disease Control and Prevention [CDC], 2017), swine (Cabral et al., 2017), wild birds (Liakopoulos et al., 2016a), environmental sources (Antony et al., 2018), human-derived clinical specimens (Abdullah et al., 2021), and outbreaks (Antony et al., 2018), which denotes their importance as a high-priority pathogen.
One of the most important risk factors surrounding foodborne illness is the international food trade that has been circumstantially accompanied by Salmonellae dispersal beyond borders. In this context, there is a global dispersal of multidrug-resistant lineages of serovar S. Heidelberg, reaching various countries in North America (Andrysiak et al., 2008; Centers for Disease Control and Prevention, 2014; Public Health Agency of Canada, 2014; Deblais et al., 2018; Cox et al., 2021), South America (Kipper et al., 2021), Europe (Liakopoulos et al., 2016b; Campos et al., 2018), and Asia (Wu et al., 2013). Therefore, the simultaneous increase and extended protraction of S. Heidelberg in many parts of the world have favored their genetic acquisition of virulence and antimicrobial resistance (AMR) genes through horizontal gene transfer (HGT), which has ultimately led to one of the most pressing global concerns.
Owing to their importance as a key poultry producer globally, Brazil quickly became the hotspot of S. Heidelberg and urgent actions were needed from the food safety authorities to mitigate this pathogen in order to reduce the economic losses in the poultry sector. In this context, most investigations to detect Salmonella enterica serovars in the poultry sector still had important methodological gaps, since the food industry focuses especially on Salmonella Typhimurium and Salmonella Enteritidis, which demonstrate the need for a combined approach between classical microbiology and high-resolution methods such as whole-genome sequencing (WGS) and clustered regularly interspaced short palindromic repeats (CRISPR) genotyping (Barrangou and Dudley, 2016; Thompson et al., 2018; Yousfi et al., 2020). Indeed, the use of these high-throughput sequencing analyses exemplifies a useful means, not only for identifying Salmonella serovars but also to trace back the origin of the contamination conferring a substantial aid in decision-making to the poultry sector. In this regard, we demonstrated the usefulness of WGS-based identification in our previous study for genotyping rare Salmonella enterica serovars isolated from food and related sources (Monte et al., 2021). This previous survey demonstrated that the CRISPR arrays were highly conserved, and this genomic inspection provides high-resolution genotyping of Salmonella serovars. Hence, we performed a genomic study by combining WGS and CRISPR genotyping to characterize S. Heidelberg isolates from different sources at broiler farms, slaughterhouses, transport, lairages, and retail markets in Brazil.
Materials and Methods
Salmonella Heidelberg Strains and Antimicrobial Susceptibility Testing
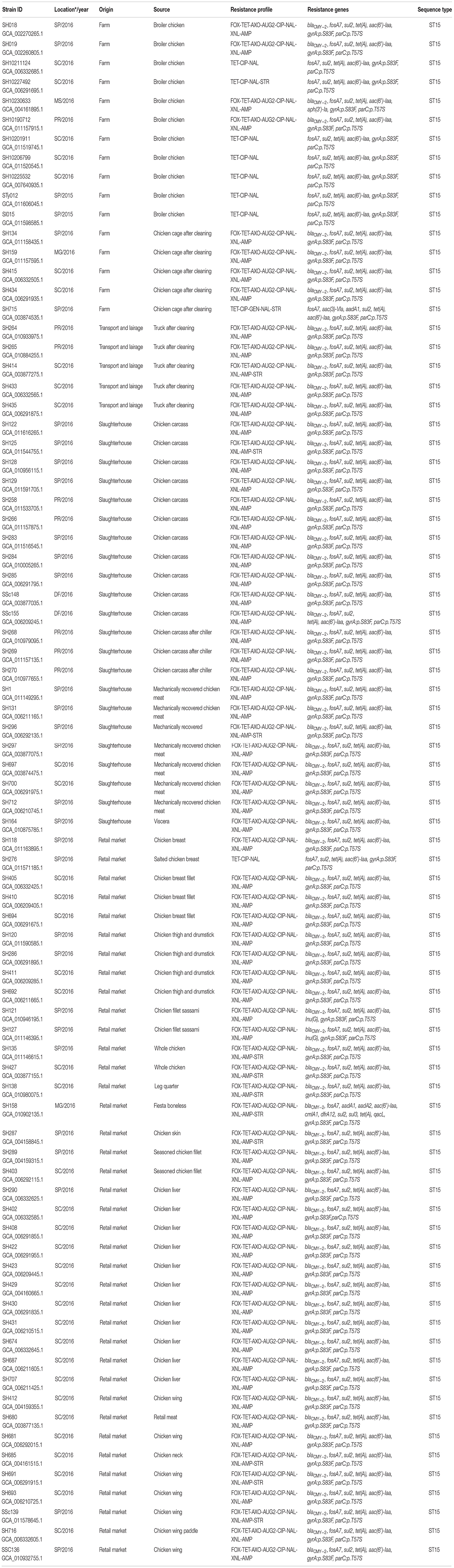
A total of 79 non-duplicate Salmonella enterica subsp. enterica serovar Heidelberg from our collection that included isolates obtained from broiler farms (n = 16), transport and lairage (n = 5), slaughterhouses (n = 22), and retail markets (n = 38) in Brazil between 2015 and 2016 were used in this study (refer Table 1). We also included two S. Heidelberg strains (SH159 and SSc139) from our previous work (Monte et al., 2019) for comparative purposes, totalizing eighty-one isolates. The Salmonella isolation was performed according to the International Organization for Standardization (Anonymous, 2007, 2017). These isolates were serotyped on the basis of somatic O, phase 1, and phase 2 of H flagellar antigens by agglutination tests with antisera as specified in the Kauffmann–White–Le Minor scheme (Grimont and Weil, 2007; Guibourdenche et al., 2010).
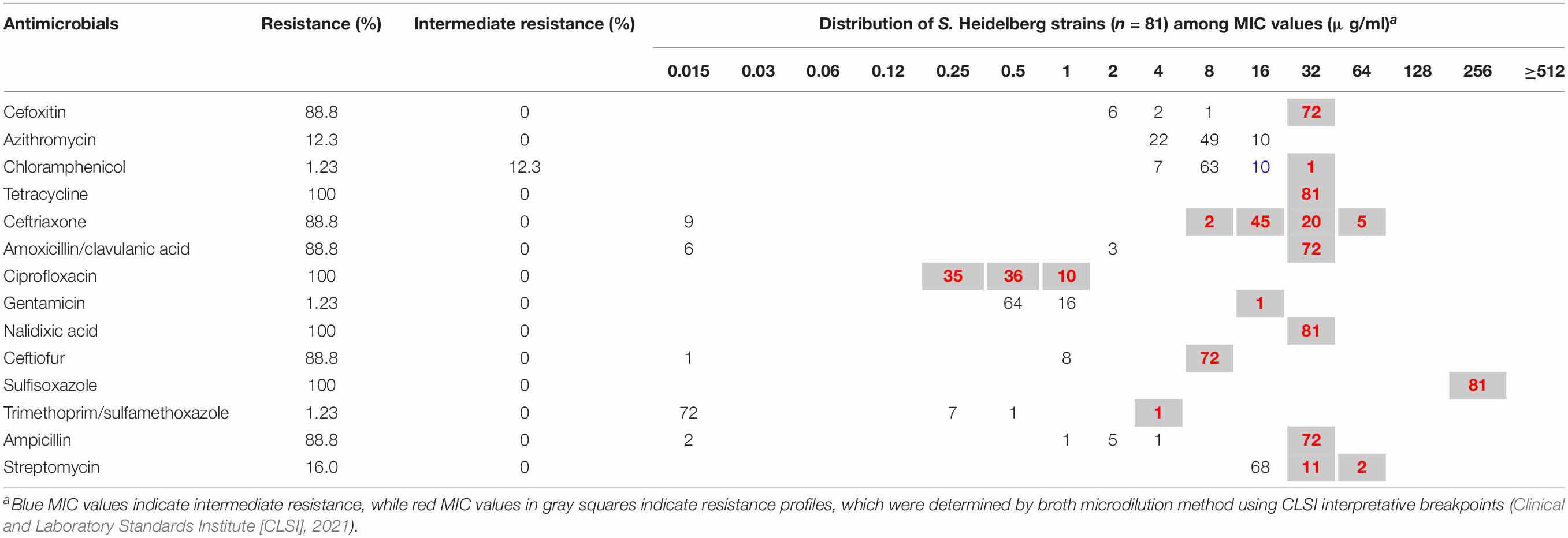
Minimum inhibitory concentrations (MICs) were determined by broth microdilution using Sensititre® Gram-Negative Plates (Trek Diagnostic Systems, OH), such as 14 antimicrobials: cefoxitin (FOX), ceftriaxone (AXO), amoxicillin/clavulanic acid 2:1 ratio (AUG2), ceftiofur (XNL), ampicillin (AMP), nalidixic acid (NAL), ciprofloxacin (CIP), chloramphenicol (CHL), tetracycline (TET), gentamicin (GEN), sulfisoxazole (FIS), trimethoprim/sulfamethoxazole (SXT), streptomycin (STR), and azithromycin (AZI). MIC values were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (Clinical and Laboratory Standards Institute [CLSI], 2021) and the National Antimicrobial Resistance Monitoring System (US Food and Drug Administration [FDA], 2015). Multidrug resistance was defined as resistant to three or more classes of antimicrobials (Magiorakos et al., 2012).
Genomic Analysis
All S. Heidelberg isolates (n = 81) underwent DNA extraction performed by using a commercial kit (QiAmp tissue, Qiagen, Germany) per manufacturer’s guidelines. Genomic DNA of eighty-one Salmonella isolates was sequenced at a 300-bp paired-end-read using the Nextera XT library preparation kit at the MiSeq platform (Illumina, San Diego, CA, United States).
Resulted raw sequence reads underwent strict quality control by using default settings in CLC workbench 10.1.1 (Qiagen) as per Monte et al. (2019), while assemblies were annotated with PROKKA version 1.14-dev (Seemann, 2014). A core genome phylogeny was constructed with an alignment of the core genes determined by the software version 3.11.2; the BlastP threshold was set to 95% (Page et al., 2015). A pan-genome genes presence–absence information from Roary was visualized with Phandango (Hadfield et al., 2018). The single nucleotide polymorphisms were extracted from the alignment using SNP-sites version 2.3.3 (Page et al., 2016). The phylogeny was reconstructed using RAxML version 8.2.12, using a General Time Reversible Model and Gamma distribution for rate heterogeneity (Stamatakis, 2014). The resulting phylogeny was tested against 1,000 bootstrap replications, as determined by implementing the majority rule, autoMR convergence criteria in the RAxML software (Pattengale et al., 2010). The phylogeny was visualized and annotated using iTol version 3 (Letunic and Bork, 2016).
Lastly, the assemblies were analyzed for acquired AMR genes and chromosomal point mutations using default settings of ResFinder 4.1 database available at the Center for Genome Epidemiology.1 In addition, we used MLST 2.0 to detect multilocus sequence typing (MLST), and the PlasmidFinder software version 2.0.1 was run with database version 2018-11-20 (Carattoli et al., 2014). A minimum identity threshold of 95% was used as a filter for identification.
Clustered Regularly Interspaced Short Palindromic Repeats Genotyping and Phylogenetic Analysis
An automated high-throughput processing pipeline previously described by Nethery and Barrangou (2019) was used to identify the CRISPR loci within each strain. Using CRISPR Visualizer, we extracted and imported CRISPR loci into the web interface for visualization and alignment of all CRISPR spacer and repeat sequences.2
Results
Salmonella Heidelberg Strains Harbored a Wide Resistome Against Critically Important Antimicrobials
A total of 81 (100%) S. Heidelberg strains were both phenotypically and genotypically resistant, whereas 91.3% (n = 74) were multidrug-resistant, defined as resistant to three or more classes of antimicrobial compounds (Magiorakos et al., 2012; Table 1). Results of the antimicrobial susceptibility testing are presented in Table 2. MICs vary among S. Heidelberg strains. All S. Heidelberg strains were resistant to TET, NAL, CIP, and FIS with MIC values ranging from 0.25 to ≥ 256 μg/ml (Table 2). The high MIC values observed in this study for β-lactams (AMP, amoxicillin/clavulanic acid, AXO, XNL, and FOX), TET, FIS, NAL, and STR (Table 2), confirm the high frequency of AMR genes and mutations predicted by genomic analysis. Yet, based on the MIC distribution, all S. Heidelberg strains displayed susceptibility to AZI (Table 2).
The total AMR gene abundances did not vary significantly across regions and sources suggesting pervasive distribution of antibiotic resistant strains from farm to market in six different States of Brazil (Figure 1). The highest AMR gene abundances were observed for fosfomycin (fosA7; 100%), sulfonamide (sul2; 100%), tetracycline [tet(A); 100%], and aminoglycoside [aac(6′)-Iaa; 100%]. Seventy-two (88.8%) S. Heidelberg strains harbored the plasmid-mediated AmpC β-lactamase (blaCMY–2), encoding resistance to third-generation cephalosporin (3GC). Unlike, Inu(G) (n = 2), aadA1 (n = 2), aph(3′)-Ia (n = 1), aac(3)-Via (n = 1), aadA2 (n = 1), cmlA1 (n = 1), dfrA12 (n = 1), sul3 (n = 1), and qacL (n = 1) AMR genes were detected at very low levels (Table 1). On the other hand, chromosomal point mutations in gyrA [p. Ser83Phe (tcc → ttc)] and parC [p. Thr57Ser (acc → agc)] were identified in 100% of the strains. This quinolone resistance-determining region (QRDR) among S. Heidelberg strains was sufficient to promote high-level resistance at > 32 μg/ml for NAL.
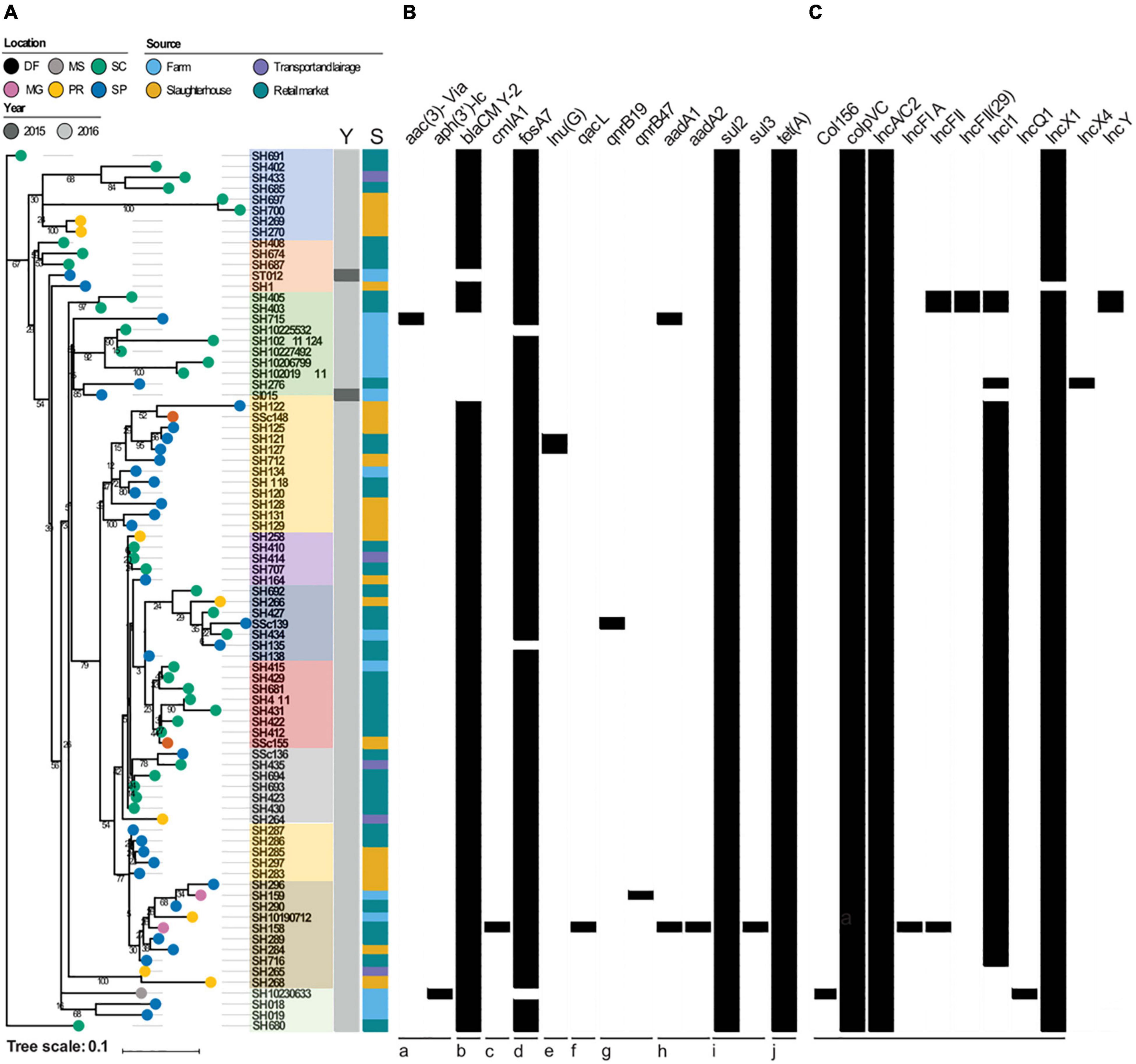
Figure 1. (A) Reconstructed phylogeny based on the core genome (4,139 genes) of the 81 S. Heidelberg strains. The percentage of bootstrap samples in which nodes appeared is shown. The location of isolation of each strain is labeled on its respective branch. Color strips depict the year (Y) and source (S) of isolation, respectively. (B) The presence and absence of selected antimicrobial resistance genes are shown, with black indicating presence. The drug classes impacted by these genes are: (a) aminoglycosides, (b) beta-lactams, (c) chloramphenicol, (d) fosfomycin, (e) lincosamide, (f) quaternary ammonium compounds, (g) quinolones, (h) streptomycin, (i) sulfonamides, (j) tetracylines. (C) Presence and absence of plasmid incompatibility groups, with black indicating presence. Brazilian States: PR, Paraná; SC, Santa Catarina; SP, São Paulo; MG, Minas Gerais; DF, Distrito Federal; MS, Mato Grosso do Sul.
All 81 Salmonella genomes were analyzed for the content of plasmid replicons by using the Center for Genomic Epidemiology (CGE) web-tool PlasmidFinder 2.1, with 100% of the genomes containing at least two replicons, like ColpVC and IncA/C2. The remaining plasmids replicons such as IncX1 (n = 80; 98.7%), Incl1 (n = 56; 69.1%), IncFII (n = 3; 3.7%), IncFII(29) (n = 2; 2.4%), IncY (n = 2; 2.4%), Col156 (n = 1; 1.2%), IncFIA (n = 1; 1.2%), IncQ1(n = 1; 1.2%), and IncX4 (n = 1; 1.2%) were identified within S. Heidelberg genomes (Figure 1).
Spacer Composition and Sequence Type Were Highly Conserved Within Salmonella Heidelberg Strains
Next, we visualized CRISPR loci extracted from WGS data to analyze the pattern of repeats and spacers distributed among S. Heidelberg strains (n = 81). In doing so, we observed 13 unique CRISPR array patterns [P1 (n = 13), P2 (n = 3), P3 (n = 1), P4 (n = 3), P5 (n = 26), P6 (n = 1), P7 (n = 16), P8 (n = 1), P9 (n = 1), P10 (n = 8), P11 (n = 5), P12 (n = 2), and P13 (n = 1)] as shown in Table 3. Overall, we observed a maximum of 44 spacers across S. Heidelberg strains (P7), spread across two loci. SH265 and SH268, belonging to profile P12, contained 37 spacers, the lowest number presented here. Spacer composition was highly conserved across strains, which shared 43 (P1, P2, P3, P4, P5, and P6), 42 (P7 and P8), 40 (P9 and P10), 39 (P11), and 36 (P12) identical spacers, reflecting a common ancestral origin (Figure 2). Next, we performed a comparative analysis of the architecture of the type I-E CRISPR-Cas system present in these strains and observed 100% amino acid identity across all strains—further evidence of shared ancestral origin (Figure 3). We further evaluated the multi-locus sequence typing by in silico prediction, which revealed that all S. Heidelberg strains matched the international sequence type (ST15) (Table 1).
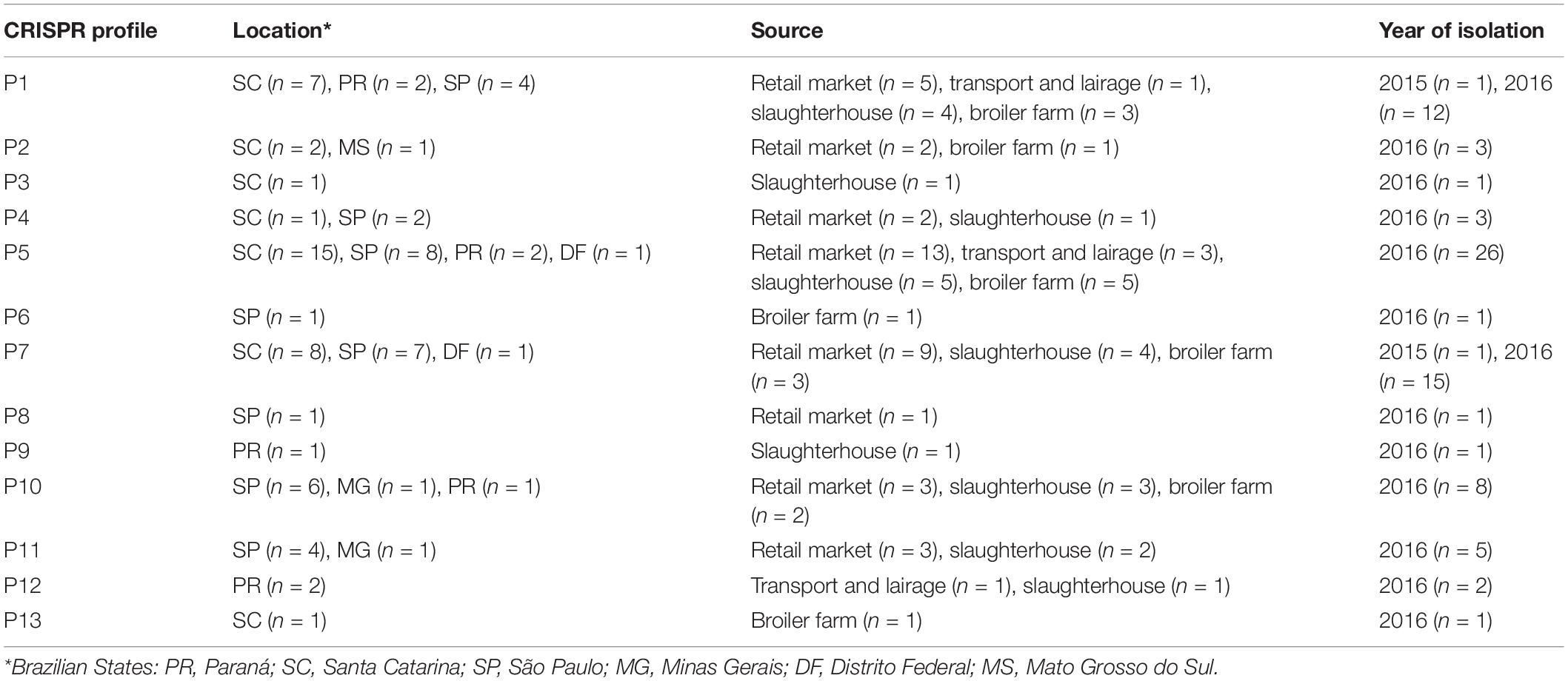
Table 3. Clustered regularly interspaced short palindromic repeats (CRISPR) patterns obtained from 81 Salmonella Heidelberg strains.
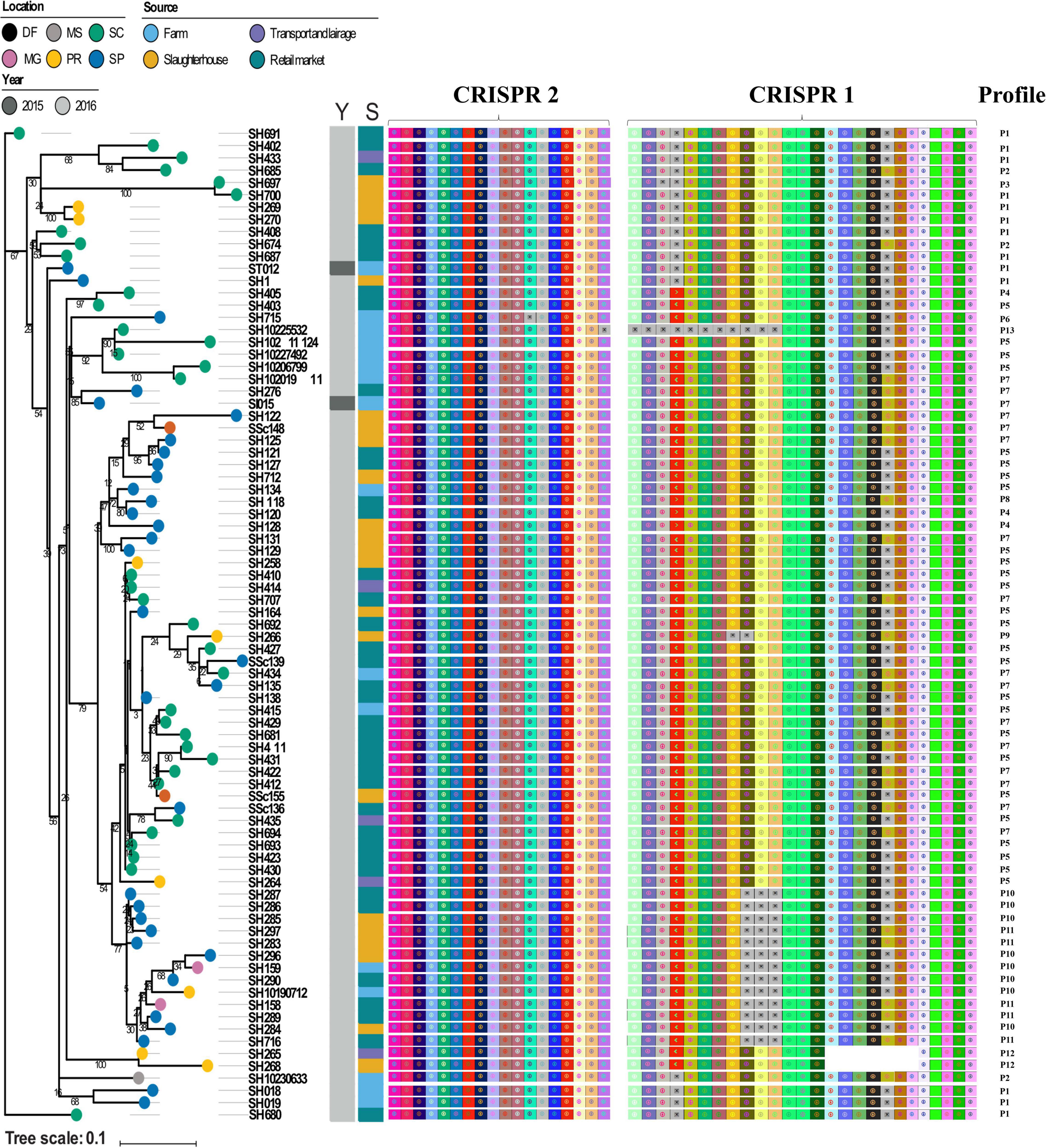
Figure 2. Reconstructed phylogeny based on the core genome, distribution of spacers composition, clustered regularly interspaced short palindromic repeats (CRISPR) loci, and CRISPR profiles among Salmonella Heidelberg strains. The location of isolation of each strain is labeled on its respective branch. Color strips depict the year (Y) and source (S) of isolation, respectively. Brazilian States: PR, Paraná; SC, Santa Catarina; SP, São Paulo; MG, Minas Gerais; DF, Distrito Federal; MS, Mato Grosso do Sul.

Figure 3. Salmonella enterica subsp. enterica serovar Heidelberg type I-E CRISPR locus architecture. This system contains two distinct CRISPR arrays—one associated with the cas genes and one disparate locus upstream (5′) from the cas genes.
Salmonella Heidelberg Strains Isolated From Brazil Are Genetically Related to South American, European, and Asian Isolates
While assessing the phylogenetics of S. Heidelberg strains (n = 81) sequenced in this study, we noticed that the core genome, calculated from WGS data, represented 74% of the pan-genome (4,139 out of 5,582 total genes). Little genomic variation was present among the core genome, as only 704 SNP sites were detected. Bootstrap values varied across the phylogeny, likely attributed to the small genomic variation among strains. S. Heidelberg strains did not cluster by year, source, or geographic location across the phylogeny suggesting the widespread distribution [regions (n = 6), sources (n = 4), years of isolation (n = 2)] and persistence of Salmonella strains in Brazil (Figure 1), which validate the previous surveys (Monte et al., 2019). Furthermore, 11 different clusters were identified as shown in Figure 1. Of these, 14 strains appear to be from independent lineages, given that they nested out of the main branches.
We also observed through SNP clustering (PDS000037185.127; n = 765 isolates; NCBI pathogen detection tool), cases of international clustering of S. Heidelberg from our collection (n = 77) with strains isolated from a variety of sources (food, human, and environment) from Brazil (n = 201), Chile (n = 9), the United Kingdom (n = 444), Germany (n = 1), the Netherlands (n = 2), South Korea (n = 1), and China (n = 1), which suggest a common ancestor origin (Supplementary Figure 1). Another two strains (SH265 and SH268) from this study nested in the SNP cluster (PDS000029160.10) with strains isolated from Brazil (n = 11), the United Kingdom (n = 4), and Chile (n = 1) (Supplementary Figure 2).
Discussion
There has been a great interest in surveying the adaptation of Salmonella serovars to the poultry production chain because of their extensive persistence in the past, notably with S. Typhimurium and S. Enteritidis, which have caused significant economic losses to this sector. Furthermore, the prevalence of S. Heidelberg shown in this study is not the only issue, but the fact that highly drug-resistant and/or MDR isolates are being recovered in most steps of the poultry production chain, particularly in Brazil could be considered a public health threat, as there is a risk of it becoming globalized.
Based on AMR results, the β-lactam resistance was essentially driven by the presence of plasmid-mediated AmpC (pAmpC) blaCMY–2 gene, given the isolates which did not carry this gene were susceptible to FOX, while QRDR such as gyrA and parC genes drove quinolone resistance (Table 1). Indeed, the presence of strains harboring blaCMY–2 gene could have implications on a one health interface, since this plasmid is more likely to persist (Teunis et al., 2018). Besides that, all strains harbored chromosomal mutations in gyrA and parC genes promoting high-level resistance against quinolones that could have implications on human health as treatment options become limited. Disturbingly, this result corroborates the findings by van den Berg et al. (2019) that found 98.4% of the S. Heidelberg isolates resistant to fluoroquinolones. On the other hand, all S. Heidelberg strains from our collection, displayed susceptibility to azithromycin, which could be considered a promising agent against Salmonellae infections (Crump et al., 2015; Wen et al., 2017). Azithromycin has been used as an alternative treatment option for enteric fever even when the guidelines on susceptibility testing were not available. Like fluoroquinolones, azithromycin is an antimicrobial agent with efficient intracellular penetration (Crump et al., 2015; Wen et al., 2017).
The total AMR load also included encoding resistance genes for aminoglycoside [aac(3)-VIa, aph(3′)-Ic, aadA1, aadA2], chloramphenicol (cmlA1), macrolides [Inu(G)], trimethoprim (dfrA12), and ammonium quaternary compounds (qacL) (Figure 1). Interestingly, the unique strain (SSc139) that carried the qacL gene was isolated from the retail market, which makes large use of such compounds to disinfect surfaces. Moreover, this strain nested in the same cluster with five strains isolated from broiler farms and slaughterhouse sources, in different states such as São Paulo, Santa Catarina, and Parana, denoting the successful establishment of this lineage (ST15) in the Southern and South regions of Brazil. In this regard, we visualized the co-occurrence of AMR genes in all S. Heidelberg strains.
The presence of AMR genes and transmissible plasmids demonstrated little variation across the strains. The broad distribution and abundance of S. Heidelberg in broiler farms, slaughterhouses, transport, lairages, and retail markets suggest the high adaptability of this serovar in the poultry production chain in Brazil. Similarly, a study by Edirmanasinghe et al. (2017) examining FOX-resistant S. Heidelberg strains isolated from human, abattoir poultry, and retail poultry sources in Canada revealed a potential common source among strains, which suggest the simultaneous dispersal of S. Heidelberg strains carrying CMY-2 gene in several sources and different geographical locations. In convergence with our results, another study revealed a high occurrence of S. Heidelberg in imported poultry meat in the Netherlands containing blaCMY–2 gene (van den Berg et al., 2019). Although we could not find colistin-resistant strains, it is worthwhile to note the presence of the IncX4 plasmid since it is a promiscuous plasmid with a high capacity of self-transmissibility that is commonly associated with the presence of plasmid-mediated colistin-resistance (mcr-1) in Brazil (Moreno et al., 2019).
Consistent with observations obtained in our study, recent surveys strongly support that S. Heidelberg may originate from a common ancestor, which circulates and persists in the Brazilian poultry production chain since at least 2004 (Kipper et al., 2021). More importantly, these strains are also nested with strains isolated from several sources and countries around the world supporting the hypotheses of intercontinental spread, which demonstrate that probably the common ancestor underwent diversification through genetic changes over time (Supplementary Material).
Protracted dissemination of S. Heidelberg via poultry might be a risk for a globalized food trade era. The introduction and clonal expansion of Salmonella strains across borders remain challenging due to the difficulties of identifying the origins of contamination. In light of this, the continued need for combined approaches between classical microbiology and high-resolution methods such as WGS and CRISPR genotyping truly illustrate to us what is hidden in plain sight.
For the purpose of discussion, other studies provide compelling validation data to support the usefulness of high-resolution methods for genotyping rare Salmonella enterica serovars (Monte et al., 2021) and/or to resolve S. Heidelberg isolates involved in foodborne outbreaks (Vincent et al., 2018). Of the latter, while assessing the CRISPR array of 145 S. Heidelberg isolates, Vincent et al. (2018) found 15 different CRISPR profiles endorsing our results.
This study further illustrates the potential of CRISPR for the tracking of variable genotypes in diverse Salmonella strains, as previously determined (DiMarzio et al., 2013; Shariat et al., 2013a,b, 2015; Monte et al., 2021), with noteworthy methodological convenience. Indeed, CRISPR-based analyses have proven relevant for subtyping of Salmonella enterica serovars Typhimurium and Heidelberg strains involved in outbreaks (Shariat et al., 2013b) and occasionally associated with antibiotic resistance (DiMarzio et al., 2013). Our findings underscore the potential role of S. Heidelberg as a key pathogen in the poultry production chain, particularly in Brazil.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
DM, RB, MN, PF-C, and ML designed the study. RB, PF-C, NL, and ML supervised the work. DM, MN, HB, SK, NL, PF-C, RB, and ML participated, coordinated, and analyzed the data. DM, MN, and RB wrote the original draft. All authors approved the final manuscript.
Funding
This study was granted by the North Carolina State University and Fundação de Amparo à Pesquisa do Estado de São Paulo-FAPESP [Food Research Center (FoRC-2013/07914-8) and 2016/03044-7]. The project has been partially developed during DM’s time as visiting scholar at the North Carolina State University under a fellowship grant from FAPESP (2017/15967-5).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.867278/full#supplementary-material
Supplementary Figures 1, 2 | SNP-based phylogram of S. Heidelberg isolates from various isolation sources and locations. Strain ID, isolate source, location, SNP cluster, and collection date were retrieved from Genbank. Red color indicates isolates from this study.
Footnotes
References
Abdullah, K., Iwen, P. C., and Abdalhamid, B. (2021). Identification of an mcr-9-Carrying Salmonella enterica serotype heidelberg strain isolated from blood. Microbiol. Resour. Announc. 10:e00676-21. doi: 10.1128/MRA.00676-21
Andrysiak, A. K., Olson, A. B., Tracz, D. M., Dore, K., Irwin, R., Ng, L. K., et al. (2008). Genetic characterization of clinical and agri-food isolates of multi drug resistant Salmonella enterica serovar Heidelberg from Canada. BMC Microbiol. 8:89. doi: 10.1186/1471-2180-8-89
Anonymous (2007). ISO 6579:2002 Amd. 1:2007 Microbiology of Food and Animal Feeding Stuffs–Horizontal Method for the Detection of Salmonella spp. – Annex D: Detection of Salmonella spp. in Animal Faeces and in Environmental Samples from the Primary Production Stage. Geneva: International Organization for Standardization.
Anonymous (2017). ISO 6579–1 Microbiology of the Food Chain–Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella–Part 1: Detection of Salmonella spp. Geneva: International Organization for Standardization.
Antony, L., Behr, M., Sockett, D., Miskimins, D., Aulik, N., Christopher-Hennings, J., et al. (2018). Genome divergence and increased virulence of outbreak associated Salmonella enterica subspecies enterica serovar Heidelberg. Gut Pathog. 10:53. doi: 10.1186/s13099-018-0279-0
Barrangou, R., and Dudley, E. G. (2016). CRISPR-based typing and next-generation tracking technologies. Annu. Rev. Food Sci. Technol. 7, 395–411. doi: 10.1146/annurev-food-022814-015729
Cabral, C. C., Panzenhagen, P., Delgado, K. F., Silva, G., Rodrigues, D., Franco, R. M., et al. (2017). Contamination of carcasses and utensils in small swine slaughterhouses by Salmonella in the Northwestern Region of the State of Rio de Janeiro, Brazil. J. Food Prot. 80, 1128–1132. doi: 10.4315/0362-028X.JFP-16-387
Campos, J., Mourão, J., Silveira, L., Saraiva, M., Correia, C. B., Maçãs, A. P., et al. (2018). Imported poultry meat as a source of extended-spectrum cephalosporin-resistant CMY-2-producing Salmonella Heidelberg and Salmonella Minnesota in the European Union, 2014-2015. Int. J. Antimicrob. Agents 51, 151–154. doi: 10.1016/j.ijantimicag.2017.09.006
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother 58, 3895–3903. doi: 10.1128/AAC.02412-14
Centers for Disease Control and Prevention (2014). National Antimicrobial Resistance Monitoring System (NARMS): 2014 Human Isolates Surveillance Report. Atlanta, GA: U.S. Department of Health and Human Services, 7.
Centers for Disease Control and Prevention [CDC] (2017). Multistate Outbreak of Multidrug-Resistant Salmonella Heidelberg Infections Linked to Contact with Dairy Calves. Atlanta, GA: Centers for Disease Control and Prevention.
Chittick, P., Sulka, A., Tauxe, R. V., and Fry, A. M. (2006). A summary of national reports of foodborne outbreaks of Salmonella Heidelberg infections in the United States: clues for disease prevention. J. Food Prot. 69, 1150–1153. doi: 10.4315/0362-028x-69.5.1150
Clinical and Laboratory Standards Institute [CLSI] (2021). Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Cox, G. W., Parmley, E. J., Avery, B. P., Irwin, R. J., Reid-Smith, R. J., Deckert, A. E., et al. (2021). A one-health genomic investigation of gentamicin resistance in Salmonella from Human and Chicken Sources in Canada, 2014 to 2017. Antimicrob. Agents Chemother. 65:e00966-21. doi: 10.1128/AAC.00966-21
Crump, J. A., Sjölund-Karlsson, M., Gordon, M. A., and Parry, C. M. (2015). Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 28, 901–937. doi: 10.1128/CMR.00002-15
Deblais, L., Lorentz, B., Scaria, J., Nagaraja, K. V., Nisar, M., Lauer, D., et al. (2018). Comparative genomic studies of Salmonella Heidelberg Isolated From Chicken- and Turkey-Associated Farm Environmental Samples. Front. Microbiol. 9:1841. doi: 10.3389/fmicb.2018.01841
DiMarzio, M., Shariat, N., Kariyawasam, S., Barrangou, R., and Dudley, E. G. (2013). Antibiotic Resistance in Salmonella enterica Serovar Typhimurium Associates with CRISPR Sequence Type. Antimicrob. Agents Chemother. 57, 4282–4289. doi: 10.1128/AAC.00913-13
Edirmanasinghe, R., Finley, R., Parmley, E. J., Avery, B. P., Carson, C., Bekal, S., et al. (2017). A whole-genome sequencing approach to study Cefoxitin-Resistant Salmonella enterica Serovar Heidelberg Isolates from Various Sources. Antimicrob. Agents Chemother. 61:e01919-16. doi: 10.1128/AAC.01919-16
Foley, S. L., Lynne, A. M., and Nayak, R. (2008). Salmonella challenges: prevalence in swine and poultry and potential pathogenicity of such isolates. J. Anim. Sci. 86, (Suppl. 14), E149–E162. doi: 10.2527/jas.2007-0464
Folster, J. P., Pecic, G., Rickert, R., Taylor, J., Zhao, S., Fedorka-Cray, P. J., et al. (2012). Characterization of multidrug-resistant Salmonella enterica serovar Heidelberg from a ground turkey-associated outbreak in the United States in 2011. Antimicrob. Agents Chemother. 56, 3465–3466. doi: 10.1128/AAC.00201-12
Grimont, P. A. D., and Weil, F. X. (2007). Antigenic Formulae of the Salmonella serovars, 9th Edn. (Paris: WHO Collaborating Centre for Reference and Research on Salmonella), 166.
Guibourdenche, M., Roggentin, P., Mikoleit, M., Fields, P. I., Bockemühl, J., Grimont, P. A., et al. (2010). Supplement 2003-2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 161, 26–29. doi: 10.1016/j.resmic.2009.10.002
Hadfield, J., Croucher, N. J., Goater, R. J., Abudahab, K., Aanensen, D. M., and Harris, S. R. (2018). Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34, 292–293. doi: 10.1093/bioinformatics/btx610
Hennessy, T. W., Cheng, L. H., Kassenborg, H., Ahuja, S. D., Mohle-Boetani, J., Marcus, R., et al. (2004). Egg consumption is the principal risk factor for sporadic Salmonella serotype Heidelberg infections: a case-control study in FoodNet sites. Clin. Infect. Dis. 38, (Suppl. 3), S237–S243. doi: 10.1086/381593
Jackson, B. R., Griffin, P. M., Cole, D., Walsh, K. A., and Chai, S. J. (2013). Outbreak-associated Salmonella enterica serotypes and food Commodities, United States, 1998-2008. Emerg. Infect. Dis. 19, 1239–1244. doi: 10.3201/eid1908.121511
Kipper, D., Orsi, R. H., Carroll, L. M., Mascitti, A. K., Streck, A. F., Fonseca, A., et al. (2021). Recent evolution and genomic profile of Salmonella enterica Serovar Heidelberg Isolates from Poultry Flocks in Brazil. Appl. Environ. Microbiol. 87:e01036-21. doi: 10.1128/AEM.01036-21
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Liakopoulos, A., Olsen, B., Geurts, Y., Artursson, K., Berg, C., Mevius, D. J., et al. (2016a). Molecular Characterization of Extended-Spectrum-Cephalosporin-Resistant Enterobacteriaceae from Wild Kelp Gulls in South America. Antimicrob. Agents Chemother. 60, 6924–6927. doi: 10.1128/AAC.01120-16
Liakopoulos, A., Geurts, Y., Dierikx, C. M., Brouwer, M. S., Kant, A., Wit, B., et al. (2016b). Extended-Spectrum Cephalosporin-Resistant Salmonella enterica serovar Heidelberg Strains, the Netherlands(1). Emerg. Infect. Dis. 22, 1257–1261. doi: 10.3201/eid2207.151377
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Monte, D., Nethery, M. A., Barrangou, R., Landgraf, M., and Fedorka-Cray, P. J. (2021). Whole-genome sequencing analysis and CRISPR genotyping of rare antibiotic-resistant Salmonella enterica serovars isolated from food and related sources. Food Microbiol. 93:103601. doi: 10.1016/j.fm.2020.103601
Monte, D. F., Lincopan, N., Berman, H., Cerdeira, L., Keelara, S., Thakur, S., et al. (2019). Genomic Features of High-Priority Salmonella enterica Serovars Circulating in the Food Production Chain, Brazil, 2000-2016. Sci. Rep. 9:11058. doi: 10.1038/s41598-019-45838-0
Moreno, L. Z., Gomes, V., Moreira, J., de Oliveira, C. H., Peres, B. P., Silva, A., et al. (2019). First report of mcr-1-harboring Salmonella enterica serovar Schwarzengrund isolated from poultry meat in Brazil. Diagn. Microbiol. Infect. Dis. 93, 376–379. doi: 10.1016/j.diagmicrobio.2018.10.016
Nethery, M. A., and Barrangou, R. (2019). CRISPR Visualizer: rapid identification and visualization of CRISPR loci via an automated high-throughput processing pipeline. RNA Biol. 16, 577–584. doi: 10.1080/15476286.2018.1493332
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Page, A. J., Taylor, B., Delaney, A. J., Soares, J., Seemann, T., Keane, J. A., et al. (2016). SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2:e000056. doi: 10.1099/mgen.0.000056
Pattengale, N. D., Alipour, M., Bininda-Emonds, O. R., Moret, B. M., and Stamatakis, A. (2010). How many bootstrap replicates are necessary? J Comput Biol. 17, 337–354. doi: 10.1089/cmb.2009.0179
Public Health Agency of Canada, (2014). National Enteric Surveillance Program (NESP) Annual Report. http://publicatio ns.gc.ca/colle ction s/colle ction _2016/aspc-phac/HP37-15-2014-eng.pdf (accessed April 2019).
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shariat, N., DiMarzio, M. J., Yin, S., Dettinger, L., Sandt, C. H., Lute, J. R., et al. (2013a). The combination of CRISPR-MVLST and PFGE provides increased discriminatory power for differentiating human clinical isolates of Salmonella enterica subsp. enterica serovar Enteritidis. Food Microbiol. 34, 164–173. doi: 10.1016/j.fm.2012.11.012
Shariat, N., Sandt, C. H., DiMarzio, M. J., Barrangou, R., and Dudley, E. G. (2013b). CRISPR-MVLST subtyping of Salmonella enterica subsp. enterica serovars Typhimurium and Heidelberg and application in identifying outbreak isolates. BMC Microbiol. 13:254. doi: 10.1186/1471-2180-13-254
Shariat, N., Timme, R. E., Pettengill, J. B., Barrangou, R., and Dudley, E. G. (2015). Characterization and evolution of Salmonella CRISPR-Cas systems. Microbiology 161, 374–386. doi: 10.1099/mic.0.000005
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Teunis, P., Evers, E. G., Hengeveld, P. D., Dierikx, C. M., Wielders, C., and van Duijkeren, E. (2018). Time to acquire and lose carriership of ESBL/pAmpC producing E. coli in humans in the Netherlands. PLoS One 13:e0193834. doi: 10.1371/journal.pone.0193834
Thompson, C. P., Doak, A. N., Amirani, N., Schroeder, E. A., Wright, J., Kariyawasam, S., et al. (2018). High-Resolution Identification of Multiple Salmonella Serovars in a Single Sample by Using CRISPR-SeroSeq. Appl. Environ. Microbiol. 84:e01859-18. doi: 10.1128/AEM.01859-18
US Food and Drug Administration [FDA] (2015). The National Antimicrobial Resistance Monitoring System Manual of Laboratory Methods. Silver Spring, MD: US Food and Drug Administration.
van den Berg, R. R., Dissel, S., Rapallini, M., van der Weijden, C. C., Wit, B., and Heymans, R. (2019). Characterization and whole genome sequencing of closely related multidrug-resistant Salmonella enterica serovar Heidelberg isolates from imported poultry meat in the Netherlands. PLoS One 14:e0219795. doi: 10.1371/journal.pone.0219795
Vincent, C., Usongo, V., Berry, C., Tremblay, D. M., Moineau, S., Yousfi, K., et al. (2018). Comparison of advanced whole genome sequence-based methods to distinguish strains of Salmonella enterica serovar Heidelberg involved in foodborne outbreaks in Québec. Food Microbiol. 73, 99–110. doi: 10.1016/j.fm.2018.01.004
Wen, S. C., Best, E., and Nourse, C. (2017). Non-typhoidal Salmonella infections in children: review of literature and recommendations for management. J. Paediatr. Child Health 53, 936–941. doi: 10.1111/jpc.13585
Wu, H., Xia, X., Cui, Y., Hu, Y., Xi, M., Wang, X., et al. (2013). Prevalence of extended-spectrum b-lactamase-producing Salmonella on retail chicken in six provinces and two national cities in the People’s Republic of China. J. Food Prot. 76, 2040–2044. doi: 10.4315/0362-028X.JFP-13-224
Yousfi, K., Usongo, V., Berry, C., Khan, R. H., Tremblay, D. M., Moineau, S., et al. (2020). Source Tracking Based on Core Genome SNV and CRISPR Typing of Salmonella enterica Serovar Heidelberg Isolates Involved in Foodborne Outbreaks in Québec, 2012. Front. Microbiol. 11:1317. doi: 10.3389/fmicb.2020.01317
Keywords: antibiotic resistance, CRISPR, phylogeny, Salmonella Heidelberg, foodborne disease, WGS
Citation: Monte DFM, Nethery MA, Berman H, Keelara S, Lincopan N, Fedorka-Cray PJ, Barrangou R and Landgraf M (2022) Clustered Regularly Interspaced Short Palindromic Repeats Genotyping of Multidrug-Resistant Salmonella Heidelberg Strains Isolated From the Poultry Production Chain Across Brazil. Front. Microbiol. 13:867278. doi: 10.3389/fmicb.2022.867278
Received: 31 January 2022; Accepted: 25 April 2022;
Published: 17 June 2022.
Edited by:
Laurent Dufossé, Université de La Réunion, FranceReviewed by:
Nikki W. Shariat, University of Georgia, United StatesAmal Awad, Mansoura University, Egypt
Copyright © 2022 Monte, Nethery, Berman, Keelara, Lincopan, Fedorka-Cray, Barrangou and Landgraf. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel F. M. Monte, bW9udGVfZGZtQGFsdW1uaS51c3AuYnI=; Mariza Landgraf, bGFuZGdyYWZAdXNwLmJy
 Daniel F. M. Monte
Daniel F. M. Monte Matthew A. Nethery
Matthew A. Nethery Hanna Berman
Hanna Berman Shivaramu Keelara
Shivaramu Keelara Nilton Lincopan
Nilton Lincopan Paula J. Fedorka-Cray5
Paula J. Fedorka-Cray5 Rodolphe Barrangou
Rodolphe Barrangou Mariza Landgraf
Mariza Landgraf