
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 31 March 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.866382
This article is part of the Research Topic Virome: Diversity, Function and Ecology View all 12 articles
Enterococcus faecalis is a Gram-positive opportunistic pathogen that causes nosocomial infections in humans. Due to the growing threat of antibiotic resistance of E. faecalis, bacteriophage therapy is a promising option for treating of E. faecalis infection. Here, we characterized a lytic E. faecalis bacteriophage vB_EfaS_efap05-1 with a dsDNA genome of 56,563 bp. Phage vB_EfaS_efap05-1 had a prolate head and a tail, and belongs to Saphexavirus which is a member of Siphoviridae. Efap05-1 uses either surface polysaccharide or membrane protein ComEA as the receptor because the mutation of both genes (ComEA and UDP-glucose 4-epimerase galE) prevents phage adsorption and leads to phage resistance, and complementation of ComEA or galE could recover its phage sensitivity. Our results provided a comprehensive analysis of a new E. faecalis phage and suggest efap05-1 as a potential antimicrobial agent.
Enterococcus faecalis is a gram-positive bacterium that could cause intestinal dysbiosis or infections in humans, such as nosocomial sepsis, urinary tract, and surgical site infections (Hayakawa et al., 2013; Jahansepas et al., 2018). In addition, the cytolysin-positive E. faecalis strains are correlated with mortality in patients with alcoholic hepatitis (Duan et al., 2019). However, the emergence of antibiotic resistance, especially vancomycin and daptomycin resistance, is especially troubling (Hu et al., 2018). Thus, new therapeutic approaches are needed to treat E. faecalis infections.
Bacteriophages are viruses that infect bacteria and are promising agents for antimicrobial treatment (Kortright et al., 2019; Wu et al., 2021). Recently, phage therapy clinical trials are initiated in many countries, and the number of case reports describing patients being treated increased significantly word-wide. For example, COVID-19 patients with carbapenem-resistant Acinetobacter baumannii infection were treated with a pre-optimized 2-phage cocktail and the infection was significantly relieved (Wu et al., 2021). However, phage resistance is a potential barrier to successful phage therapy (Egido et al., 2021). To confront this issue, the molecular mechanisms of phage resistance, as well as the genomic and biological characteristics of a phage, should be studied to provide the foundation for rational selection of the phages for therapy (Pires et al., 2020).
In this study, we isolated a new E. faecalis bacteriophage vB_EfaS_efap05-1 with a dsDNA genome of 56,563 bp. It belongs to Saphexavirus which is a member of Siphoviridae, and efap05-1 could use either surface polysaccharide or membrane protein ComEA as the receptor to adsorb to the bacterial surface. Thus, phage-resistant mutants had both mutations of ComEA and galE. In summary, this study provided a detailed characterization of an E. faecalis bacteriophage and suggests efap05-1 as a potential antimicrobial agent.
Enterococcus strains were collected from the Department of Clinical Laboratory Medicine and were grown aerobically on Brain-Heart Infusion Broth (BHI) broth at 37°C with shaking.
Bacteriophage was isolated from hospital sewage as previously described (Yang et al., 2016). Briefly, the sewage was pelleted, and the supernatant was filtered through a 0.22 μm-pore-size filter. Then, 500 μl of the sample was immediately mixed with 200 μl of bacterial culture, and 4 ml of molten BHI soft agar (0.4%) was added and poured onto BHI agar plates. After overnight culture, the formed plaque was picked, deposited in 1 ml of BHI, followed by a 10-fold dilution and double-layer agar assay to purify the phage. Then, one plaque from the third round of the purification process was picked for this study.
Phage particles were dropped on carbon-coated copper grids for 10 min. Then, the grids were stained with phosphotungstic acid (pH 7.0) for 15 s. The sample was examined under a Philips EM 300 electron microscope. The sizes of the phage were measured using AxioVision LE based on five randomly selected images.
The double-layer agar plate assay was used to calculate the phage titer. Briefly, 10-fold dilutions of phage solution were mixed with 200 μl of host bacteria, then mixed with 4 ml of molten BHI broth with 0.4% agar. Then, pour the mixture on a 1.5% agar plate. After overnight incubation at 37°C, the number of plaques was calculated as a plaque-forming unit (pfu). MOI experiments were performed by mixing log-phase bacteria (OD600 = 0.6) with a different number of phages, and the titer in the coculture was calculated using a double-layer agar plate assay after 5 h.
The one-step growth curve of efap05-1 was determined as described (Zhong et al., 2020). Briefly, 1 ml of log-phase bacteria and 1 ml of efap05-1 were mixed at an MOI of 10 and incubated at 37°C for 10 min. Then, the mixture was centrifuged for 1 min at a speed of 10,000 × g, and the pellet was resuspended in 6 ml of BHI medium. And samples were taken at the given time points, which are immediately pelleted and phage titer in the supernatant was measured immediately.
Bacteriophage adsorption assay with various time points was performed as previously described (Al-Zubidi et al., 2019). Briefly, the log phase bacterial cultures were pelleted and resuspended in medium to a final concentration of 3 × 108 CFU/ml. Then, phage was added to a final titer of 3 × 106 pfu/ml. The samples were cultured at 37°C for 10 min, and the phage titer in the supernatant were measured using the double-agar plating assays. The adsorption rate was calculated as (the original phage titer—the remaining phage titer)/the original phage titer.
Ten E. faecalis strains were selected to test the host range of efap05-1 through spot testing by dropping 1 μl of phage onto the double-layer soft agar premixed with the bacterial and cultured at 37°C for overnight. The formation of a clear plaque is considered as sensitive to phage efap05-1 infection.
Two microliter of serial 10-fold dilutions of phage efap05-1 were spotted on double layer agar plates containing a bacterial host. The number of plaques observed after overnight incubation were compared to the number obtained on the strain efa05.
The phage DNA extraction is performed as previously described (Khan et al., 2021). Then, phage genomic DNA was sequenced using an Illumina Hiseq 2,500 platform (~1 Gbp/sample). Fastp (Chen et al., 2018) was used for adapter trimming and filtering the raw reads. The data were assembled using the de novo assembly algorithm Newbler Version2.9 with default parameters, and the assembled genome was annotated using RAST (Overbeek et al., 2014). The DNA and protein sequences were checked for homologs with BLAST manually. The genome map was drawn by SnapGene 4.1.8. The sequence data is available in the NCBI under accession number OL505085.
The stability of phage under various conditions was tested by treating 109 pfu of phage under different pH (pH 2–13), temperature (0°C, 30°C, 40°C, 50°C, and 60°C), or chloroform concentration (10%, 25%, 50%, 75%, and 95%) for 60 min, the then the titer of the phage was determined by double-layer agar assay.
The phage resistant mutants were selected as previously described (Shen et al., 2018). The log phase bacteria were mixed with phage efap05-1 and cultured until the bacteria was lysed. Then, the lysate was inoculated onto the BHI agar. After overnight incubation, the single colonies were checked for its resistance against phage using the double-layer agar assay, which confirmed that all the colonies on the plates are resistant to phage infection.
The wide type strain efa05 and phage resistant mutant strain efa05R were selected for sequencing. Bacterial genomic DNA was extracted using UNlQ-10 Column Bacterial Genomic DNA Isolation Kit (sangon bitotec: SK1202), and then sent to Novogene Corporation for sequencing using the Illumina Hiseq 2,500 platform. Trimmomatic was used to remove adapter sequences and low-quality bases (Bolger et al., 2014). BWA was used to map clean reads to the reference genome sequence of efa05. Samtools (Li et al., 2009) was then used to prepare the data for use with the Integrative Genomics Viewer (IGV). DNA mutation locations were manually checked with IGV and SeqKit (Shen et al., 2016).
The ComEA gene and plasmid pMGP23:mCherry were amplified by PCR (the primers for the amplification of ComEA gene and the plasmid pMGP23:mCherry are listed in Table 1), and the PCR products were purified. The ComEA and plasmid were ligated by Gibson assembly to generate pMG-ComEA, and the constructed plasmid was comfired by sanger sequencing. The efa05R complementation strain was generated by electroporation of pMG-ComEA into strain efa05R followed by selection on BHI agar erythromycin (20ug/ml). The galE gene was complemented with the same protocol.
All the experiments were performed three times. The statistical analysis was performed using One-way ANOVA or t-test, and statistical significance was assumed if the value of p was <0.05.
An E. faecalis phage was isolated using plaque assay. It forms a clear plaque on the host strain efa05 in the double layer agar plates (Figure 1A). The phage particle was observed by transmission electron microscopy. It is a non-enveloped, head-tail structural particle. The prolate head is approximately 100 nm in length and 40 nm in width (Figure 1B). Thus, based on the morphology, phage vB_EfaS_efap05-1 belongs to Saphexavirus which is a member of Siphoviridae. The optimal multiplicity of infection (MOI) was 0.001, and the phage titer could reach approximately 8*1010 pfu/ml (Figure 1C). The one-step growth curve of efap05-1 indicates that this phage replicates quickly with a lysis period of about 20 min, and the phage titer approached plateau after 20 min (Figure 1D), and the burst size was estimated as about 20 pfu per bacterium.
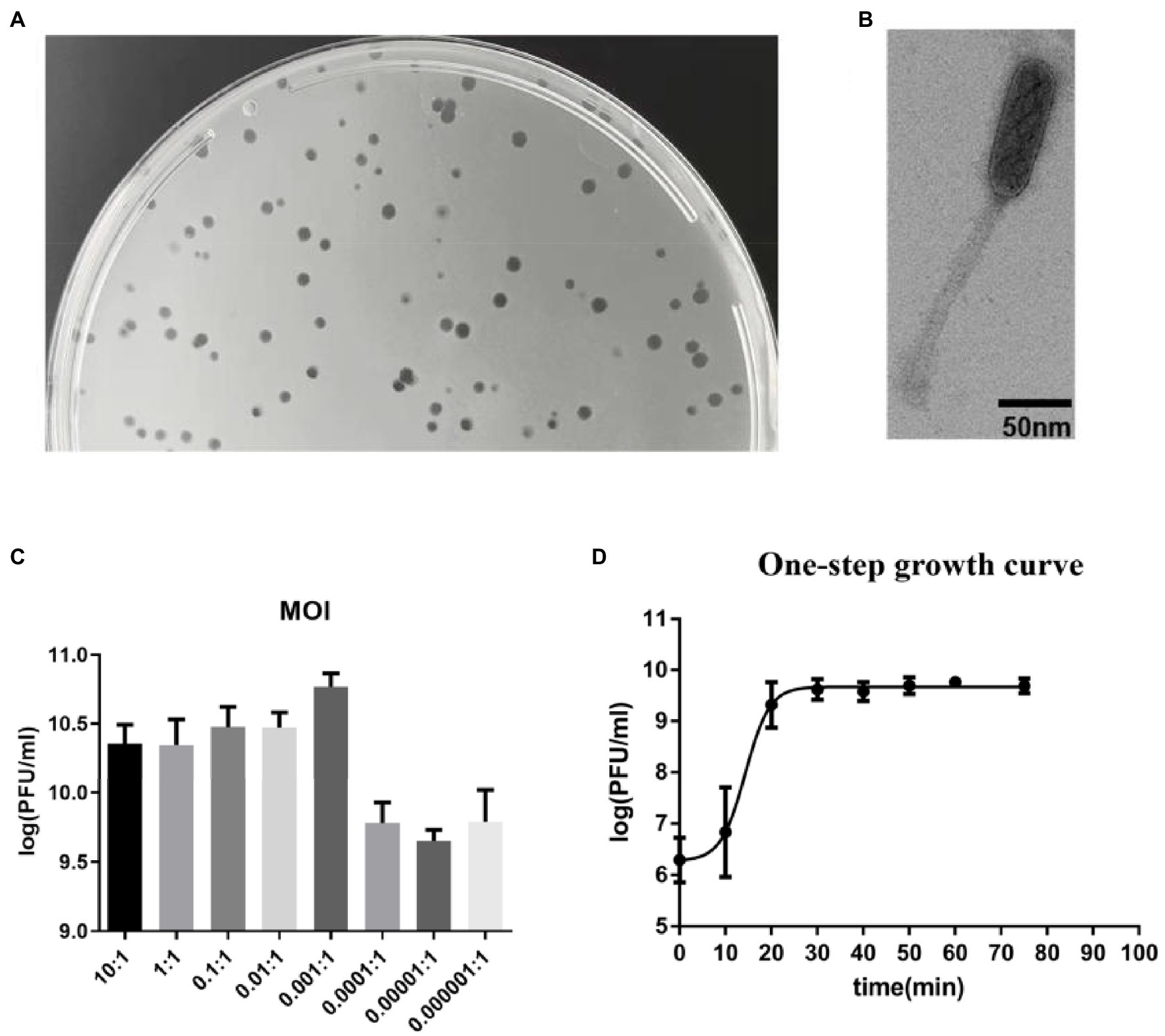
Figure 1. Biological characterization of E. faecalis phage vB_EfaS_efap05-1. (A) Phage efap05-1 forms clear plaques on the agar plate. (B) The transmission electron micrograph reveals that efap05-1 is a Saphexavirus which is a member of Siphoviridae. (C) The optimal MOI of phage efap05-1 is 0.001. (D) The one-step growth curve of efap05-1.
The host range of efap05-1 was estimated by EOP (efficiency of plating) assays. Ten clinically isolated E. faecalis strains were tested and five strains could be lysed by efap05-1, indicating a modest host range (Table 2).
The stability of efap05-1 under various conditions was tested. It could maintain stability under pH 5–10, and other pH solutions could impair the viability of efap05-1 (Figure 2A). And efap05-1 is stable under 50°C because its titer was not changed after 60 min incubation at 50°C (Figure 2B), and is completely inactivated over 70°C. Besides, chloroform treatment did not affect the viability of phage efap05-1, indicating that it is a non-enveloped phage (Figure 2C).
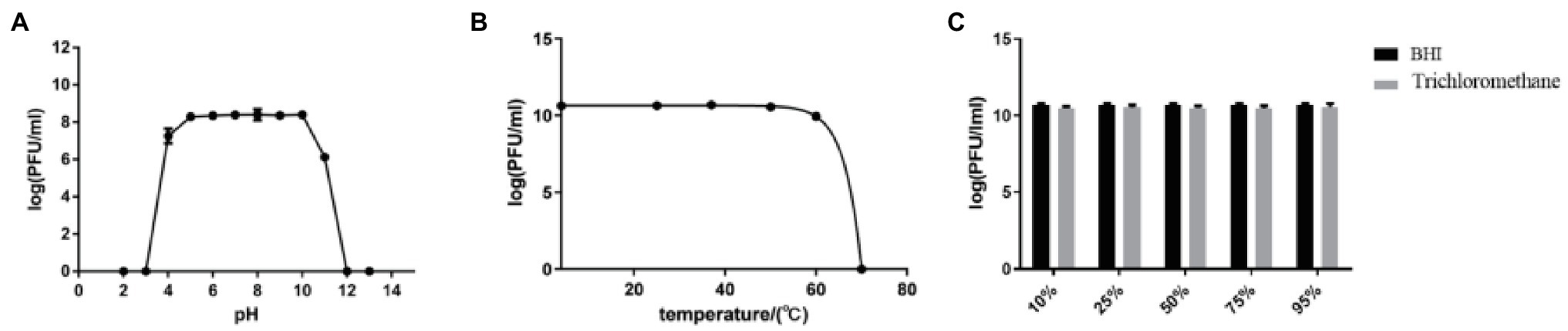
Figure 2. Stability of efap05-1: (A) Phage efap05-1 is stable under pH5 ~ 10, and is completely inactived under pH3. (B) Phage efap05-1 is inactivated by 70°C treatment. (C) Phage efap05-1 is resistant to chloroform because the titer was stable after chloroform treatment.
Phage efap05-1 is a double-stranded (ds) DNA phage with a linear genome of 56,564 base pairs (bp). Its G + C content is 40% and encodes 99 ORFs and one tRNA (Figure 3), which are predicted by RAST (Overbeek et al., 2014) and visualized by SnapGene.
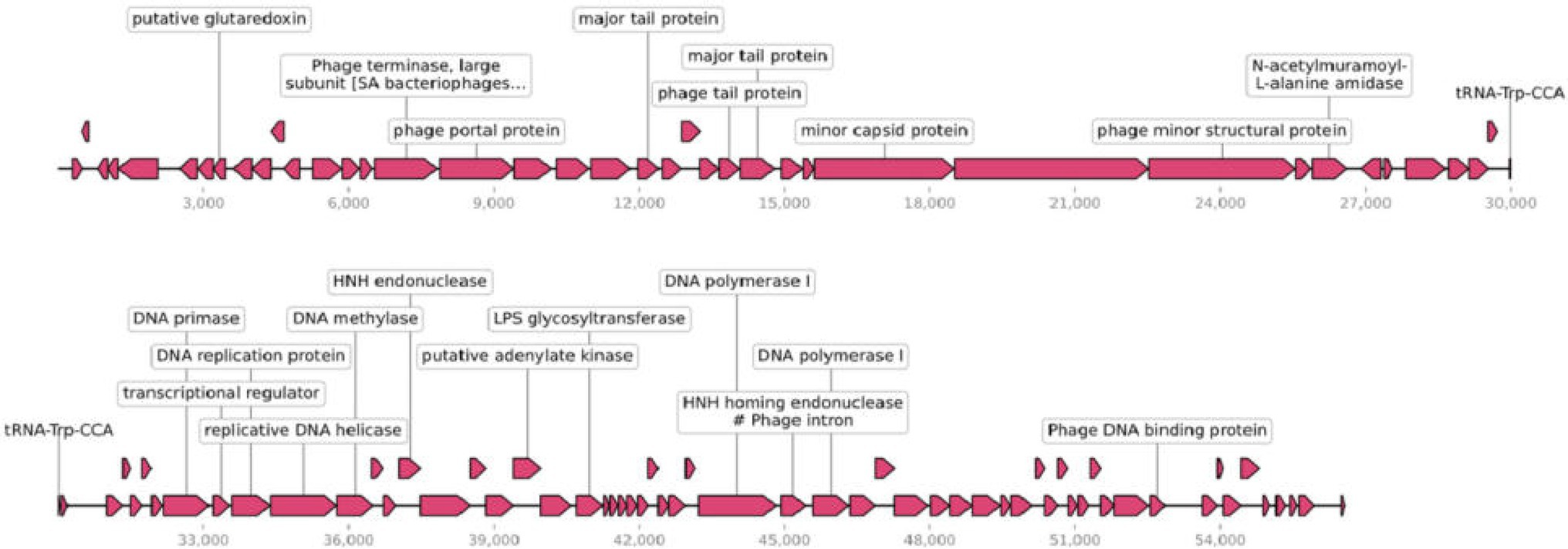
Figure 3. Genomic characterization of efap05-1. Phage efap05-1 is a dsDNA phage that encodes 99 predicted proteins and one tRNA.
Most of the ORFs are functionally unknown, and 22 ORFs are functionally annotated, which can be categorized into several functional modules, including phage DNA replications, lysis, phage structural protein (Figure 3). However, efap05-1 did not encode any antibiotic-resistant gene or virulence gene, indicating that it is a safe candidate for phage therapy.
To study the phage resistant mechanism of E. faecalis efa05 against phage efap05-1, we mixed phage with host and cultured until the bacteria are lysed. And then inoculate the lysate on BHI agar plate. One phage-resistant mutant efa05R was selected, and its resistance against efap05-1 was confirmed by EOP experiment (Figure 4B).
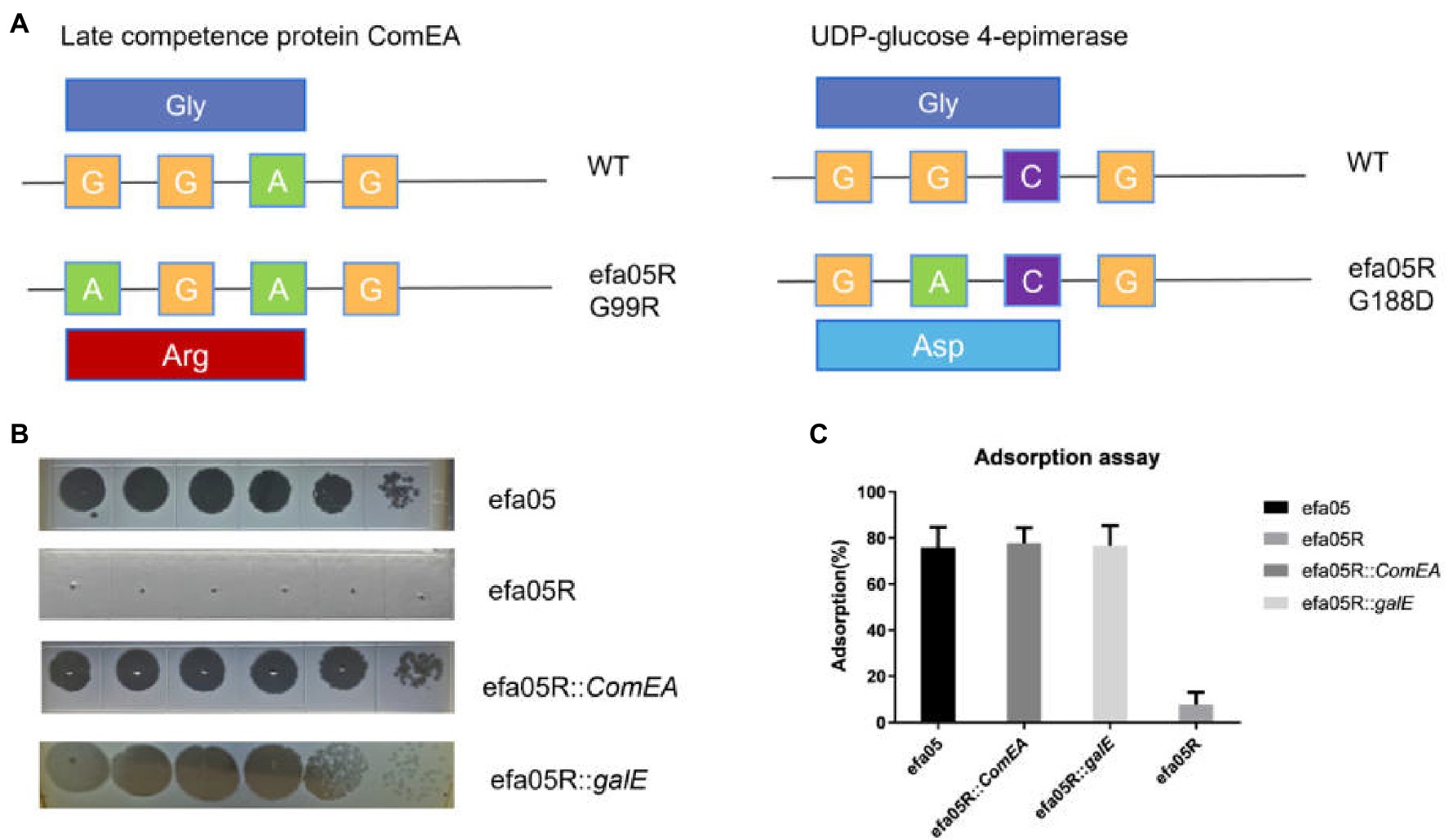
Figure 4. Characterization of the phage-resistant mutant. (A) The mutation site in efa05R was detected as G99R in ComEA and G188D in galE. (B) The EOP experiment of phage against wild type strain, phage resistant efa05R, and the complemented strains. (C) Adsorption assay of phage onto each strain.
Then, efa05R and wild-type strain efa05 was sent for whole-genome sequencing, and the mutation sites of these two strains were detected as Late competence protein ComEA and UDP-glucose 4-epimerase galE (Figure 4A). ComEA is a cell membrane protein that binds to the double-stranded DNA and initiates the DNA uptake process (Burghard-Schrod et al., 2021). And UDP-glucose 4-epimerase is involved in the biosynthesis of cell wall polysaccharides (Boels et al., 2001; Lee et al., 2014).
Usually, phage resistance is selected with one key mutation site (Li et al., 2018), the mutation of both genes detected in phage resistant mutant efa05R indicates that both genes are required for phage infection. As expected, the complementation of either ComEA or galE in efa05R could recover the phage sensitivity through enabling phage adsorption (Figures 4B,C). Thus, these data indicate that phage could bind to either polysaccharides or protein ComEA to initiate the phage infection cycle.
Enterococcus. faecalis have developed resistance to antibiotics, including vancomycin and daptomycin (Hayakawa et al., 2013; Jahansepas et al., 2018). Thus, phage therapy is a renewed interest to treat multidrug-resistant E. faecalis infection. The biological and genomic characterization of a phage is essential before applications in phage therapy (Barbu et al., 2016; El Haddad et al., 2019; Pires et al., 2020). In this study, we isolated an E. faecalis bacteriophage vB_EfaS_efap05-1 with a prolate head. It is a completely lytic phage without the antibiotic-resistant genes or virulence genes, indicating it as a potential candidate for phage therapy.
The identification of phage resistance mechanisms is important for rational select phage or designing a phage cocktail for therapy (Labrie et al., 2010; Duerkop et al., 2016). Phage resistance is quite common and is important for phage therapy because it could lead to treatment failure. Selecting different phages that target different receptors is a rational approach in selecting phages for therapy (Yang et al., 2020). And in vitro, most phage resistance is selected through modifications of the receptors (Castillo et al., 2015; Duerkop et al., 2016). For example, Pseudomonas aeruginosa phage resistant mutants are O-antigen deficient to prevent phage adsorption (Shen et al., 2018). In this study, phage resistance mutants are selected with mutations in two genes. And the complementation of each gene could restore the phage sensitivity as well as the phage adsorption. Thus, it is reasonable to infer that phage efap05-1 uses either polysaccharides or protein ComEA as the receptors. Polysaccharides are common phage receptors for a lot of E. faecalis phages (Duerkop et al., 2016; Chatterjee et al., 2020).
ComEA is a membrane protein, and the loss of the ComEA decreases the binding of DNA to the competent cell surface and the internalization of DNA and impairs DNA transformability (Inamine and Dubnau, 1995). However, protein ComEA, to our knowledge, is the first report to serve as a phage receptor, which is an interesting biological phenomenon.
The limitation of this study is the lack of identification of the receptor binding proteins in the phages. Most phages use either polysaccharides or protein as receptors (Lim et al., 2021). However, it is not common that phage uses two different receptors of polysaccharides and membrane protein, because the phage tail fiber is very specifically targeting the receptors, and usually one phage tail fiber could not adsorb to two different structural receptors. And the E.coli phage phi92 could adsorb to both encapsulated and nonencapsulated bacteria due to the presence of four different types of tail fibers and tail spikes in the viral particles, which enable the phage to use attachment different sites on the host cell surface (Schwarzer et al., 2012). And staphylococcal Twort-like phage ΦSA012 possesses two receptor binding proteins to expand its host range (Takeuchi et al., 2016). In our study, the current data suggest that phage efap05-1 might also encode different receptor binding proteins that enable it to adsorb to both polysaccharides and membrane proteins. Three tail fiber proteins are annotated in the genome of efap05-1 (Figure 3), which needs further study to demonstrate the function of each tail fiber protein.
In conclusion, we isolated and characterized an E. faecalis phage efap05-1, which is a candidate for the development of phage cocktails or phage-antibiotic combinations treatment for E. faecalis infections. The characterization of the phage-resistant mutant bacterium could help to develop a cocktail to avoid phage resistance.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
PW and FS designed the research. LH, WG, JL, and WP performed the laboratory work and collected the data. LH, WG, PW, and FS wrote the first draft of the manuscript and prepared figures. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (NSFC, 31660043 to WP) and Yunnan Medical leading talent project (L-2019001 to WP).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Al-Zubidi, M., Widziolek, M., Court, E. K., Gains, A. F., Smith, R. E., Ansbro, K., et al. (2019). Identification of novel bacteriophages with therapeutic potential that target Enterococcus faecalis. Infect. Immun. 87, e00512–e00519. doi: 10.1128/IAI.00512-19
Barbu, E. M., Cady, K. C., and Hubby, B. (2016). Phage therapy in the era of synthetic biology. Cold Spring Harb. Perspect. Biol. 8:a023879. doi: 10.1101/cshperspect.a023879
Boels, I. C., Ramos, A., Kleerebezem, M., and de Vos, W. M. (2001). Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microbiol. 67, 3033–3040. doi: 10.1128/AEM.67.7.3033-3040.2001
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Burghard-Schrod, M., Kilb, A., Kramer, K., and Graumann, P. L. (2021). Single molecule dynamics of DNA receptor ComEA, membrane permease ComEC and taken up DNA in competent Bacillus subtilis cells. J. Bacteriol. doi: 10.1128/jb.00572-21 [Epub ahead of print].
Castillo, D., Christiansen, R. H., Dalsgaard, I., Madsen, L., and Middelboe, M. (2015). Bacteriophage resistance mechanisms in the fish pathogen Flavobacterium psychrophilum: linking genomic mutations to changes in bacterial virulence factors. Appl. Environ. Microbiol. 81, 1157–1167. doi: 10.1128/AEM.03699-14
Chatterjee, A., Willett, J. L. E., Nguyen, U. T., Monogue, B., Palmer, K. L., Dunny, G. M., et al. (2020). Parallel genomics uncover novel enterococcal-bacteriophage interactions. mBio 11, e03120–e031219. doi: 10.1128/mBio.03120-19
Chen, S., Zhou, Y., Chen, Y., and Jia, G. (2018). Fastp: an ultra-fast all-in-one fastq preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Duan, Y., Llorente, C., Lang, S., Brandl, K., Chu, H., and Jiang, L., 2019., et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505–511, doi: 10.1038/s41586-019-1742-x
Duerkop, B. A., Huo, W., Bhardwaj, P., Palmer, K. L., and Hooper, L. V. (2016). Molecular basis for lytic bacteriophage resistance in Enterococci. mBio 7, e01304–e01316. doi: 10.1128/mBio.01304-16
Egido, J. E., Costa, A. R., Aparicio-Maldonado, C., Haas, P. J., and Brouns, S. J. J. (2021). Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol. Rev. 46:fuab048. doi: 10.1093/femsre/fuab048
El Haddad, L., Harb, C. P., Gebara, M. A., Stibich, M. A., and Chemaly, R. F. (2019). A systematic and critical review of bacteriophage therapy against multidrug-resistant ESKAPE organisms in humans. Clin. Infect. Dis. 69, 167–178. doi: 10.1093/cid/ciy947
Hayakawa, K., Marchaim, D., Palla, M., Gudur, U. M., Pulluru, H., Bathina, P., et al. (2013). Epidemiology of vancomycin-resistant Enterococcus faecalis: a case-case-control study. Antimicrob. Agents Chemother. 57, 49–55. doi: 10.1128/AAC.01271-12
Hu, F., Zhu, D., Wang, F., and Wang, M. (2018). Current status and trends of antibacterial resistance in China. Clin. Infect. Dis. 67, S128–S134. doi: 10.1093/cid/ciy657
Inamine, G. S., and Dubnau, D. (1995). ComEA, a Bacillus subtilis integral membrane protein required for genetic transformation, is needed for both DNA binding and transport. J. Bacteriol. 177, 3045–3051. doi: 10.1128/jb.177.11.3045-3051.1995
Jahansepas, A., Ahangarzadeh Rezaee, M., Hasani, A., Sharifi, Y., Rahnamaye Farzami, M., Dolatyar, A., et al. (2018). Molecular epidemiology of Vancomycin-resistant enterococcus faecalis and enterococcus faecium isolated from clinical specimens in the northwest of Iran. Microb. Drug Resist. 24, 1165–1173. doi: 10.1089/mdr.2017.0380
Khan, F. M., Gondil, V. S., Li, C., Jiang, M., Li, J., Yu, J., et al. (2021). A novel Acinetobacter baumannii bacteriophage Endolysin LysAB54 With high antibacterial activity Against multiple gram-negative microbes. Front. Cell. Infect. Microbiol. 11:637313. doi: 10.3389/fcimb.2021.637313
Kortright, K. E., Chan, B. K., Koff, J. L., and Turner, P. E. (2019). Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25, 219–232. doi: 10.1016/j.chom.2019.01.014
Labrie, S. J., Samson, J. E., and Moineau, S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327. doi: 10.1038/nrmicro2315
Lee, M. J., Gravelat, F. N., Cerone, R. P., Baptista, S. D., Campoli, P. V., Choe, S. I., et al. (2014). Overlapping and distinct roles of Aspergillus fumigatus UDP-glucose 4-epimerases in galactose metabolism and the synthesis of galactose-containing cell wall polysaccharides. J. Biol. Chem. 289, 1243–1256. doi: 10.1074/jbc.M113.522516
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, G., Shen, M., Yang, Y., Le, S., Li, M., Wang, J., et al. (2018). Adaptation of Pseudomonas aeruginosa to phage PaP1 predation via O-antigen polymerase mutation. Front. Microbiol. 9:1170. doi: 10.3389/fmicb.2018.01170
Lim, A. N. W., Yen, M., Seed, K. D., Lazinski, D. W., and Camilli, A. (2021). A tail fiber protein and a receptor-binding protein mediate ICP2 bacteriophage interactions with vibrio cholerae OmpU. J. Bacteriol. 203:e0014121. doi: 10.1128/JB.00141-21
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Pires, D. P., Costa, A. R., Pinto, G., Meneses, L., and Azeredo, J. (2020). Current challenges and future opportunities of phage therapy. FEMS Microbiol. Rev. 44, 684–700. doi: 10.1093/femsre/fuaa017
Schwarzer, D., Buettner, F. F., Browning, C., Nazarov, S., Rabsch, W., Bethe, A., et al. (2012). A multivalent adsorption apparatus explains the broad host range of phage phi92: a comprehensive genomic and structural analysis. J. Virol. 86, 10384–10398. doi: 10.1128/JVI.00801-12
Shen, W., Le, S., Li, Y., and Hu, F. Q. (2016). SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One 11:e0163962. doi: 10.1371/journal.pone.0163962
Shen, M., Zhang, H., Shen, W., Zou, Z., Lu, S., Li, G., et al. (2018). Pseudomonas aeruginosa MutL promotes large chromosomal deletions through non-homologous end joining to prevent bacteriophage predation. Nucleic Acids Res. 46, 4505–4514. doi: 10.1093/nar/gky160
Takeuchi, I., Osada, K., Azam, A. H., Asakawa, H., Miyanaga, K., and Tanji, Y. (2016). The presence of two receptor-binding proteins contributes to the wide host range of staphylococcal Twort-Like phages. Appl. Environ. Microbiol. 82, 5763–5774. doi: 10.1128/AEM.01385-16
Wu, N., Dai, J., Guo, M., Li, J., Zhou, X., and Li, F. (2021). Pre-optimized phage therapy on secondary Acinetobacter baumannii infection in four critical COVID-19 patients. Emerg. Microb. Infect. 10, 612–618. doi: 10.1080/22221751.2021.1902754
Yang, Y. H., Lu, S. G., Shen, W., Zhao, X., Shen, M. Y., et al. (2016). Characterization of the first double-stranded RNA bacteriophage infecting Pseudomonas aeruginosa. Sci. Rep. 6:38795. doi: 10.1038/srep38795
Yang, Y., Shen, W., Zhong, Q., Chen, Q., He, X., Baker, J. L., et al. (2020). Development of a bacteriophage cocktail to constrain the emergence of phage-resistant Pseudomonas aeruginosa. Front. Microbiol. 11:327. doi: 10.3389/fmicb.2020.00327
Keywords: bacteriophage, phage receptor, Enterococcus faecalis, exopolysaccharide, ComEA protein
Citation: Huang L, Guo W, Lu J, Pan W, Song F and Wang P (2022) Enterococcus faecalis Bacteriophage vB_EfaS_efap05-1 Targets the Surface Polysaccharide and ComEA Protein as the Receptors. Front. Microbiol. 13:866382. doi: 10.3389/fmicb.2022.866382
Received: 31 January 2022; Accepted: 08 March 2022;
Published: 31 March 2022.
Edited by:
Mao Ye, Institute of Soil Science (CAS), ChinaReviewed by:
Mengzhe Li, Beijing University of Chemical Technology, ChinaCopyright © 2022 Huang, Guo, Lu, Pan, Song and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuqiang Song, MzQ5NjU3MTYxQHFxLmNvbQ==; Peng Wang, d3AwMzA4MDFAMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.