
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 21 April 2022
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.865227
Wolbachia is a maternally inherited bacterium that is widely distributed among arthropods, in which it manipulates the reproduction of its hosts. Phage WO is the only bacteriophage known to infect Wolbachia, and may provide benefit to its host or arthropods. We screened for the presence of phage WO in Wolbachia-infected butterfly species for the first time, to investigate their diversity and evolutionary dynamics. All Wolbachia-infected butterfly species, including members of the families Hesperiidae, Lycaenidae, Nymphalidae, Papilionidae, and Pieridae, were found to harbor phage WO. Interestingly, 84% of 19 butterfly species, which were infected with a single Wolbachia strain harbored high levels of multiple phage types (ranging from 3 to 17 types), another three species harbored one or two phage types. For Wolbachia strains (ST-41, ST-19, ST-125 and ST-374) shared among various butterfly species, their host insects all harbored multiple phage types, while two Wolbachia strains (ST-297 and ST-wPcau) were found to infect one butterfly species, whose insect hosts harbored a single phage type, suggesting that horizontal transfer of Wolbachia between insects increased the likelihood of exposure to phages, resulting in increased phage genetic diversity. Twelve horizontal transmission events of phage WO were found, which shared common phage WO types among different Wolbachia strains associated with butterflies. Most horizontal transfer events involved different Wolbachia supergroups (A and B). Horizontal acquisition of phage WO might also occur between eukaryotes without Wolbachia transfer. Furthermore, 22 putative recombination events were identified in 13 of 16 butterfly species which harbored multiple phage types. These results showed that horizontal transfer of Wolbachia caused it to be exposed to the phage gene pool, and that horizontal transmission of phage WO, as well as intragenic recombination were important dynamics for phage WO genome evolution, which effectively promoted the high level of phage WO diversity associated with butterflies.
Wolbachia (Anaplasmataceae) are maternally inherited endosymbiotic bacteria that infect arthropods and filarial nematodes (Werren et al., 2008; Engelstädter and Hurst, 2009). They are extremely widespread and probably occur in 40–65% of arthropod species (Hilgenboecker et al., 2008; Zug and Hammerstein, 2012; Weinert et al., 2015). Although vertical transmission of Wolbachia from mother to offspring predominates within species, horizontal transmission between species often occurs in arthropods (Sintupachee et al., 2006; Yang et al., 2013; Johannesen, 2017; Cooper et al., 2019; Hou et al., 2020; Sanaei et al., 2021). Wolbachia manipulates its host’s reproduction by inducing several phenotypes, such as cytoplasmic incompatibility (CI), parthenogenesis, feminization of genetic males, and male-killing (Werren et al., 2008).
Bacterial viruses (bacteriophages or phages) are the most abundant organisms on earth and constitute a significant force in bacterial genome evolution (Hendrix et al., 1999; Bordenstein and Wernegreen, 2004). As a consequence of reductive evolution, mobile DNA elements were thought to be rare or absent in obligate intracellular bacteria given their isolated niche, but increasing reports have shown that the ecology of bacterial endosymbionts significantly influenced the amount of their genome populated by mobile elements. Endosymbionts that are strictly vertically transmitted from parents to offspring often lack mobile genetic elements, while intracellular bacteria that may be horizontally transmitted often retain a large amount of mobile DNA, including phages (Moran and Plague, 2004; Bordenstein and Reznikoff, 2005; Newton and Bordenstein, 2011; Metcalf and Bordenstein, 2012). Wolbachia phage, a λ phage-like temperate dsDNA phage, named phage WO, was first characterized from the Wolbachia strain wTai, infecting Teleogryllus taiwanemma, and can be either lysogenic and integrated into the Wolbachia chromosome, or lytic and free in the cytoplasm (Masui et al., 2000, 2001). Phage WO is estimated to infect about 90% of supergroups A and B of Wolbachia from various arthropod groups, but is absent from the mutualistic C and D supergroups commonly found in filarial nematodes (Bordenstein and Wernegreen, 2004; Gavotte et al., 2007; Gerth et al., 2014). Nearly all sequenced Wolbachia genomes, except those acting as obligate mutualists, harbored phage WO (Gavotte et al., 2007; Kent and Bordenstein, 2010; Metcalf and Bordenstein, 2012). Moreover, Wolbachia not currently infected by phages often show evidence of past infections (Foster et al., 2005; Kent and Bordenstein, 2010). An orf7-like phage pseudogene has even been found in a newly described Wolbachia-free gall wasp species, Latuspina jinzhaiensis, suggesting that vestiges of prophage DNA remain in the chromosomes of the host insect after a previous lateral gene transfer event (Abe et al., 2021; Zhu et al., 2021). Additionally, most phage-infected Wolbachia strains display low numbers of phage types, with 85% showing only one or two different phage types (Gavotte et al., 2007; Tanaka et al., 2009). On the other hand, multiple phage infections, where a Wolbachia strain displays more than two phage types, have been reported in several Wolbachia strains (Chauvatcharin et al., 2006; Gavotte et al., 2007). The results of Zhu et al. (2021) confirmed that a gall wasp, Andricus hakonensis, harbored 27 phage WO types. Thus, considering the wide distribution of Wolbachia, phage WO may be one of the most abundant phage lineages in arthropods (Zhu et al., 2021).
The persistence of the phage despite its documented lytic activity has led to the hypothesis that phage WO provides benefit to its Wolbachia or arthropod host (Kent and Bordenstein, 2010). As in other prokaryotes, the integration and transformation of prophages are considered major sources of Wolbachia lateral gene acquisition (Bordenstein et al., 2006). Phage WO may mediate lateral gene transfer between Wolbachia strains (Ishmael et al., 2009; Wang G. H. et al., 2016) and regulate Wolbachia density by inhibiting their replication or inducing cell lysis (Bordenstein et al., 2006). Recently, the factors underlying CI were identified as two genes, cifA and cifB, located adjacent to one another within WO prophage regions in the Wolbachia genome, with homologs in all known CI-inducing Wolbachia strains (Beckmann et al., 2017; LePage et al., 2017; Chen et al., 2019; Shropshire and Bordenstein, 2019). Mutation, recombination, and genome segment reassortment during replication may mediate genetic changes in viruses (Domingo, 2010). A phage genome can be divided into functional units (modules; each one responsible for head or tail formation, lysis, lysogeny, and so forth). It is generally believed that mixing through fragment rearrangement with other phages in the common gene pool comprise the main evolutionary dynamics of dsDNA phages (Hatfull, 2008). However, although phage WO is modular, it does not evolve according to modular gene exchange, but rather through point mutation, intragenic recombination, deletion, and purifying selection, given its intracellular habitat (Kent et al., 2011). The nucleotide sequence of the minor capsid gene orf7 from the wKueA1 strain of Wolbachia, infecting Ephestia kuehniella, is chimeric, and population genetic analysis has confirmed the occurrence of intragenic recombination events (Bordenstein and Wernegreen, 2004). Zhu et al. (2021) provided empirical molecular evidence of orf7 gene recombination, suggesting that intragenic recombination was the important evolutionary force, which effectively promoted the diversity of phage WO associated with gall wasps which harbor multiple phage WO types. Furthermore, base deletions during replication also significantly promoted the evolution of the phage genome, resulting in the diversity of phage WO associated with A. hakonensis (Su et al., 2021).
The order Lepidoptera (moths and butterflies) contains about 158,000 described species and it is one of the most widespread and diverse insect orders (van Nieukerken et al., 2011). Given its economic and ecological importance, the order Lepidoptera has been investigated extensively by entomologists and ecologists. The relationships between Lepidoptera and their heritable endosymbionts have received increasing attention. Wolbachia infections have been detected in many Lepidoptera, and in diverse butterfly species from various regions. They were present in 16% of 43 Lepidoptera species tested in Panama (Werren et al., 1995), in 58% of 120 Lepidoptera species in West Siberia, in particular, in 59% of 108 butterfly species (Ilinsky and Kosterin, 2017), in 45% of 49 butterfly species in Japan (Tagami and Miura, 2004), in 50% of 56 butterfly species in India (Salunke et al., 2012), in 35% of 52 butterfly species in southern China (Zhu and Gao, 2021), and in 43% of 53 Mylothris spp. and in 46% of 21 Bicyclus spp. (Duplouy et al., 2020), and in 29% of 24 Acraea spp. (Jiggins et al., 2001) in Africa. Many Wolbachia strains that infect Lepidoptera have evolved the ability to alter host reproduction; key mechanisms are inducing CI or shifting the sex ratio toward female prevalence in populations by male killing or feminization (Jiggins et al., 2001; Narita et al., 2011; Duplouy and Hornett, 2018). Lepidoptera are considered one of model systems for studying the manipulation of host reproduction by endosymbionts (Duplouy and Hornett, 2018). Given its potential impact on host genome evolution and reproductive regulation in insect hosts infected by Wolbachia, phage WO has received heightened interest. Nevertheless, only a minimal part of the phage biodiversity harbored in Wolbachia-infected Lepidoptera has been described (Tanaka et al., 2009; Batista et al., 2010; Furukawa et al., 2012; Wang et al., 2020). In this study, the presence of phage WO in 19 Wolbachia-infected butterfly species collected from China was detected by employing a PCR-based method with phage WO-specific gene markers in order to understand the prevalence patterns. To explore the evolutionary dynamics of phage WO diversity, we also analyzed intragenic recombination using the recombination detection procedure (RDP5) package and investigated horizontal transmission by comparative phylogenetic analysis of phages and their hosts.
Adult butterflies were collected from Liaoning, Hunan and Guangdong Provinces, China, from June 2019 to September 2020. In total, 148 individuals of 56 species were obtained from five families (Supplementary Table 1). The adult butterflies were taken back to the laboratory where their abdomens were removed with sterile scissors and stored in 100% ethanol at -80°C for subsequent DNA extraction.
The penultimate abdominal segments of all collected butterfly individuals were used for DNA extraction. The samples were soaked and cleaned twice in ultrapure water before DNA extraction. Total DNA was extracted from each individual using SDS/proteinase K digestion and a phenol-chloroform extraction method, as described previously (Zhu et al., 2007). The quality of all the DNA extracts was confirmed by PCR amplification of the COI gene with universal insect primer pairs (Folmer et al., 1994). Wolbachia infection screening were performed according to the methods of Zhu and Gao (2021). Wolbachia multi-locus sequence type (MLST) genes (gatB, coxA, hcpA, fbpA, and ftsZ) were amplified in Wolbachia-positive samples using the respective primers reported by Baldo et al. (2006).
Phage WO infections were screened by PCR using the primers WO-F (5’-CCCACATGAGCCAATGACGTCTG-3’) and WO-R (5’- CGTTCGCTCTGCAAGTAACTCCATTAAAAC-3’) to amplify a portion of the capsid protein gene orf7 (Masui et al., 2000). PCR amplification was conducted using a C1000 Touch thermal cycler (Bio-Rad, Hercules, CA, United States) in a 25-μL reaction volume comprising 2.5 μL 10 × PCR buffer, 0.125 μL Taq polymerase (Takara, Dalian, China), 2 μL dNTPs (10 mmol each), 1 μL forward and reverse primer (10 μM), 1 μL extracted DNA, and 18.4 μL ddH2O. The cycling conditions were 95°C for 3 min, 35 cycles of 95°C for 30 s, 57°C for 40 s, and 72°C for 40 s. All reactions were followed by a final extension step for 5 min at 72°C.
One to three individuals from each butterfly species were used to sequence the orf7 fragment. The PCR products were subsequently purified using a TaKaRa MiniBEST Agarose Gel DNA Extraction Kit Ver. 4.0 (Takara Biomedical Technology Co.), and the orf7 gene fragments were directly sequenced from purified PCR products using PCR primers. The appearance of multiple peaks in a sample at initial sequencing was taken as an indication of multiple infections. The PCR products were then purified using a DNA gene gel extraction kit and ligated directly into the vector, following the manufacturer’s protocols. For each sample, 10–20 independent positive colonies were isolated and cultured in a lysogeny broth medium fortified with ampicillin. Plasmids were extracted and partially sequenced in both directions using an ABI 3730XLDNA sequencer (Applied Biosystems, Foster City, CA, United States) with M13F/R at Wuhan Icongene Co., Ltd.
Sequence homology analysis was first performed using the BLAST1 program online. Genetic distances between all sequence pairs were calculated using the Kimura 2-parameter distance model in MEGA 7. Sequences having greater than 1.5% nucleotide diversity in the orf7 gene were defined as different haplotypes (Chafee et al., 2010; Zhu et al., 2021). The sequences have been deposited in GenBank under the following accession numbers: OM472181– OM472424.
The sequences obtained for the phage WO orf7 gene and Wolbachia multi-locus sequence typing (MLST) genes were aligned using SEQMAN PRO v.11.2 (DNASTAR, Madison, WI, United States). The data sets were then analyzed using a maximum likelihood (ML) method implemented in PAUP v.4.0b (Swofford, 2003). The best evolutionary model was selected according to the corrected Akaike information criterion, implemented in MEGA v.7.0. The ML trees were analyzed for 1,000 bootstrap replicates to assess branch support.
The individual segment alignments were analyzed using different methods described in the Recombination Detection Program (RDP5) package to detect evidence of intragenic recombination (Heath et al., 2006). The six recombination detection methods implemented in the RDP5 program for the identification of recombinant sequences and breakpoints were as follows: 3Seq (Martin and Rybicki, 2000), BootScan/rescan recombination test (Martin et al., 2005), GENECONV (Padidam et al., 1999), MaxChi (Smith, 1992), Chimaera (Posada and Crandall, 2001), and the Siscan method (Gibbs et al., 2000). The default settings were used for all methods, and the highest acceptable P-value cutoff was set to 0.05.
We collected 56 butterfly species belonging to five families from China. Among these, 19 species were infected with Wolbachia; two species, Limenitis doerriesi Staudinger and Parnassius stubbendorfii Menetries, were found to be infected with Wolbachia in this study, and the other 17 species detected by Zhu and Gao (2021; Supplementary Table 1). Following the method previously described (Zhu and Gao, 2021), the complete MLST and WSP profiles of the newly found Wolbachia strains were identified, and one unique ST and two wsp alleles were obtained (Table 1). Using the diagnostic PCR approach with the phage minor capsid protein gene (orf7)-specific primers, Wolbachia-infected species were screened for phage WO infection. Phage WO was found to be harbored in all Wolbachia-infected butterfly species (Table 1).
In total, 259 phage WO orf7 sequences were gained from 19 butterfly species infected with Wolbachia. Among these, 15 orf7-like sequences contained abnormal stop codons without being transcribed, which were treated as pseudogenes (accession no.: OM663002– OM663016). They were excluded from subsequent analysis. Phage WO types with similarity of orf7 DNA sequences greater than 98.5% were defined as identical types in accordance with a previous study (Chafee et al., 2010; Zhu et al., 2021). We named phage types as WO followed by the abbreviation of the insect name and haplotype numbers. L. doerriesi and Polygonia c-aureum L. were found to harbor a single phage type, WOLdo and WOPca, respectively, and Stichophthalma sp. harbored two phage types, WOSsp-1 and WOSsp-2. Unexpectedly, the other 16 Wolbachia-infected butterfly species (84% of 19 species tested) harbored multiple phage types: Ypthima praenubila Leech and Pseudozizeeria maha (Kollar) harbored 10 and 17 types, and the other species carried 3–9 types, respectively, while their Wolbachia infections were all of a single strain (Table 1 and Supplementary Figures 1–4).
As shown in Table 1, four Wolbachia strains, that is, ST-41, ST-19, ST-125 and ST-374, were shared among various butterfly species. To clarify the phage WO infections within these Wolbachia strains, based on the orf7 sequences, phylogenetic trees were constructed of the phage WO types infecting these Wolbachia strains from various insects, respectively (Figure 1).
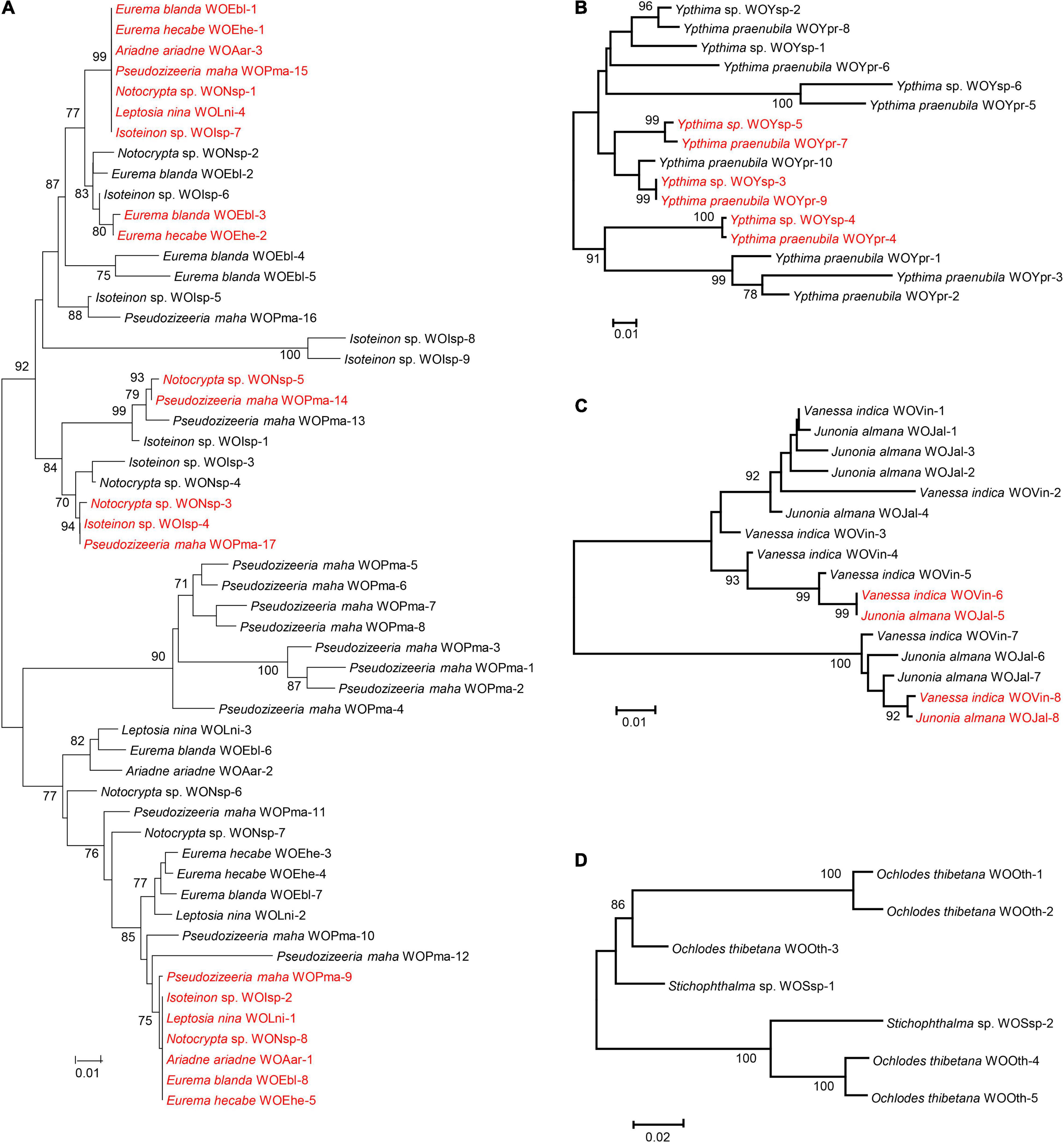
Figure 1. Maximum likelihood phylogenetic tree of the phage WO orf7 nucleotide sequences from Wolbachia strains ST-41 (A), ST-19 (B), ST-125 (C) and ST-374 (D). Numbers above branches are bootstrap values computed from 1,000 replications. WOEbl-1 refers to phage WO type. Red font indicates identical orf7 sequences or those with similarity of orf7 sequences greater than 98.5% among their respective accomplices.
Wolbachia strain ST-41 was found in seven butterfly species from four families (Hesperiidae, Lycaenidae, Nymphalidae, and Pieridae). These butterfly species harbored two common phage WO types; sequences of WOEbl-1, WOEhe-1, WOAar-3, WOPma-15, WONsp-1, WOLni-4 and WOIsp-7 were identical, and WOPma-9, WOIsp-2, WOLni-1, WONsp-8, WOAar-1, WOEbl-8 and WOEhe-5 were identical orf7 sequences or had one base substitution. Similarly, Eurema blanda (Boisduval) and E. hecabe (L.); Notocrypta sp. and P. maha; Notocrypta sp., Isoteinon sp. and P. maha harbored a common phage WO type, respectively. However, the remaining 33 phage WO types were carried by only one butterfly species (Figure 1A).
Two Ypthima species infected with Wolbachia strain ST-19 and Vanessa indica Herbst and Junonia almana (L.) infected with Wolbachia strain ST-125 harbored three and two common phage WO types, respectively. Meanwhile, these insects all carried their own unique phage WO types (Figures 1B,C). However, no common phage WO types were detected between Ochlodes thibetana Oberthür and Stichophthalma sp., although they were infected with the same Wolbachia strain (ST-374; Figure 1D).
Phylogenetic reconstruction of phage WO orf7 sequences and concatenated sequences of five Wolbachia MLST genes from butterflies was performed using ML methods (Figure 2). Obviously, there was no congruence between phage WO and its host Wolbachia phylogenies. Moreover, direct evidence was found for 12 horizontal transmission events of phage WO from butterflies, two involving three Wolbachia strains and 10 involving two Wolbachia strains, which shared common phage WO types among different Wolbachia strains. For example, WOIsp-4 (from Isoteinon sp.), WONsp-3 (from Notocrypta sp.), WOCcr-5 [from Colias croceus (Fourcroy)], WOMfr-3 (from Mycalesis francisca Cramer), WOPst-1 (from P. stubbendorfii), WOPma-17 (from P. maha), WOYpr-9 (from Y. praenubila), and WOYsp-3 (from Ypthima sp.) had identical orf7 sequences or one base substitution, thus belonging to the same phage WO type. They infected at least three Wolbachia strains, namely ST-41, ST-141, and ST-19. The Wolbachia strains of M. francisca and P. stubbendorfii could not be characterized because of failure to amplify some MLST loci. Among 12 horizontal transmission events, 10 involved different Wolbachia supergroups, occurring between supergroup A and supergroup B.
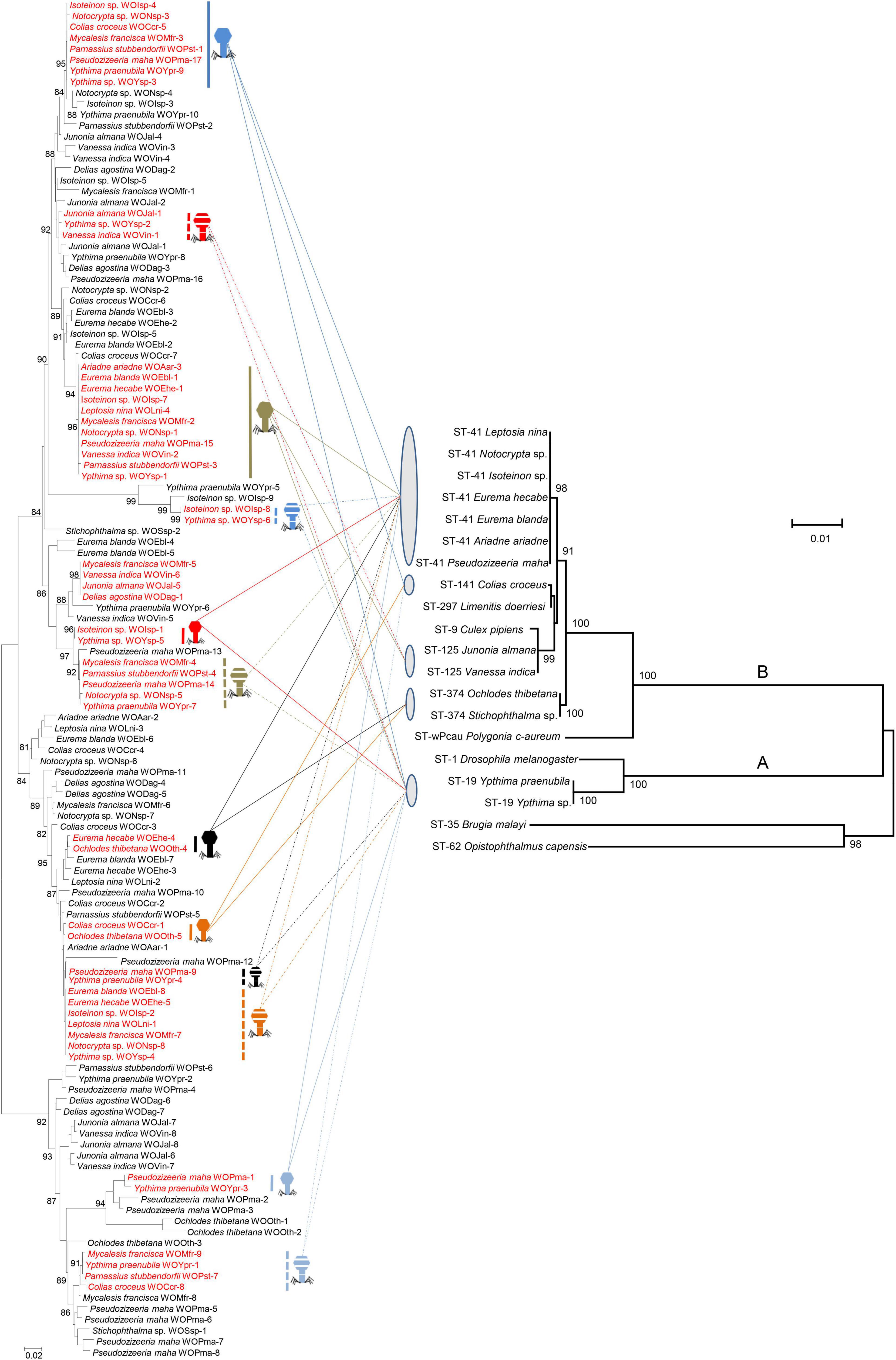
Figure 2. Comparison among phylogenies of phage WO based on orf7 nucleotide sequences (left) and Wolbachia based on concatenated sequences of multi-locus sequence type (MLST) genes (right). Numbers above branches are bootstrap values computed from 1,000 replications. Red font indicates identical orf7 sequences or those with similarity of orf7 sequences greater than 98.5% among their respective accomplices. These accomplices are shown with icons of different colors and shapes. The capital letters on the right indicate the Wolbachia supergroups.
Multiple phage types are carried by one butterfly species with a single Wolbachia strain infection, therefore genetic material might be exchanged for reassortment and intragenic recombination. To obtain direct evidence of intragenic recombination, we performed recombination analysis of the aligned orf7 sequences obtained from the same butterfly species using RDP5 programs. Regarding the results of recombination analysis, this study only provided the recombination events detected by more than three detection methods implemented in RDP5 programs.
A total of 22 putative recombination events were identified, resulting in new phage types, from 13 to 16 butterfly species which harbored diverse phage WOs (Table 2, Figure 3 and Supplementary Figures 5–11). These recombination events include two types; one major parent and one minor parent or one major parent and two to four minor parents were recombined with the same breakpoint into a new phage WO lineage (Table 2). For example, Phage WO type WOYpr-9 from Y. praenubila was detected as a recombinant by four of the six methods used: 3Seq (P < 10–9), BootScan (P < 10–8), GENECONV (P < 10–5), and MaxChi (P < 10–8). The major and minor parents were WOYpr-8 and WOYpr-10, and the beginning breakpoints were 141 and 360 bp (Figure 3A). Type WOVin-7 from V. indica was detected as a recombinant by three of the six methods used: 3Seq (P < 10–7), BootScan (P < 10–5–10–6), and GENECONV (P < 10–4–10–5). The major parent was WOVin-8, and minor parents may be WOVin-3 or WOVin-4, WOVin-5, or WOVin-6; the beginning breakpoint was 131 bp (Figure 3B). Furthermore, WOPma-2, WOVin-4, WOYpr-9, WOYpr-10, WOPst-1, WOpst-2, WOCcr-3, and WOCcr-4, which were obtained by recombination, could also be used as parents for recombination to contribute to the diversity of phages (Table 2). These results suggested that intragenic recombination occurred widely and frequently in phage WO harbored in butterflies.
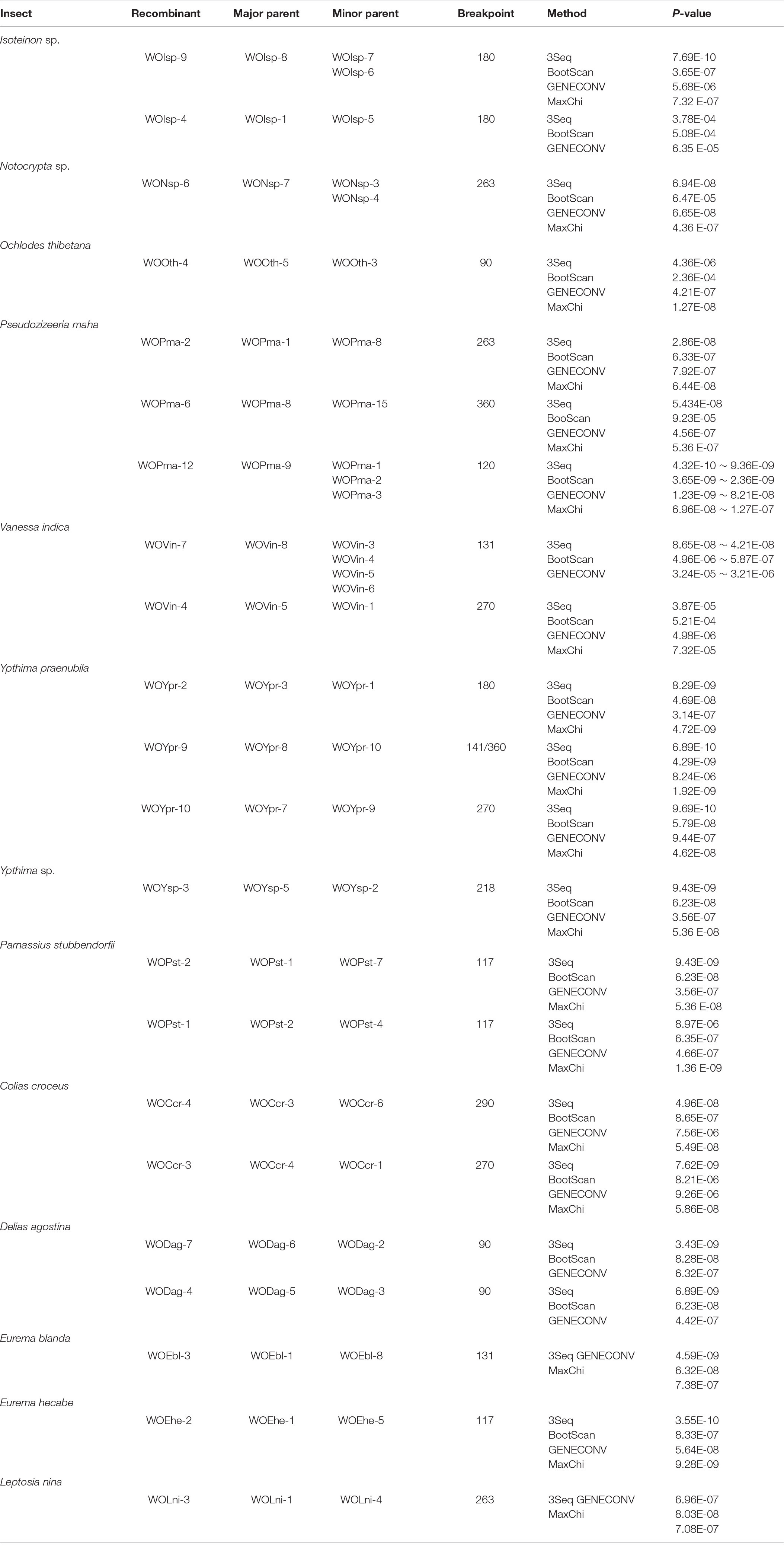
Table 2. Recombination analysis of the phage WO orf7 gene in butterflies using six methods implemented in the RDP package.

Figure 3. Recombination events of the orf7 gene between WOYpr-8 and WOYpr-10 resulting in recombinant WOYpr-9 (A) and among WOVin-8 and WOVin-3, WOVin-4, WOVin-5, and WOVin-6 resulting in recombinant WOVin-7 (B).
The prevalence of Wolbachia has been widely reported, and the bacterium has a high infection rate in butterflies (Jiggins et al., 2001; Tagami and Miura, 2004; Salunke et al., 2012; Ilinsky and Kosterin, 2017; Duplouy et al., 2020; Zhu and Gao, 2021). However, little information is available about phage WO associated with Wolbachia-infected butterflies. In this study, we detected the presence of phage WO infections associated with butterflies for the first time, and demonstrated that all 19 Wolbachia-infected species, belonging to Hesperiidae, Lycaenidae, Nymphalidae, Papilionidae, and Pieridae, harbor phage WO. Although multiple phage WO infections have been reported in several Wolbachia strains (Chauvatcharin et al., 2006; Gavotte et al., 2007), most phage-infected Wolbachia strains display low numbers of phage types, with 85% showing only one or two different phage types (Gavotte et al., 2007; Tanaka et al., 2009). Zhu et al. (2021) found that six gall wasp species with double or multiple Wolbachia infections harbored a high level of phage WO diversity. However, in that system, the number of phage types within a single Wolbachia strain remains unclear. Interestingly, in this study, the results indicated that 84% of 19 butterfly species which were infected with a single Wolbachia strain harbored high levels of multiple phage types: Y. praenubila and P. maha harbored 10 and 17 types, and the other 14 species carried 3–9 types, respectively. Although the number of positive clones selected was increased to 20 for samples with more than three types, the diversity of phage WO might still be underestimated in this study. The Wolbachia strain wRi, infecting Drosophila simulans, showed a single WO haplotype according to the initial PCR detection using phage WO-specific gene markers, but the genome sequence confirmed four protophage copies in the genome, three of which were unique (Gavotte et al., 2007; Klasson et al., 2009). In fact, primers used to screen for the presence or absence of the orf7 gene were not degraded enough to detect all orf7 variants (Kent and Bordenstein, 2010; Su et al., 2021). Su et al. (2021) designed specific detection primers, and may have significantly improved the detection efficiency of orf7 variants. Thus, the diversity of phage WO in Wolbachia-infected arthropods might be greater than current estimates.
The Wolbachia strains characterized in butterflies are often shared among various insect species, families and even orders (Ahmed et al., 2016; Ilinsky and Kosterin, 2017; Duplouy et al., 2020; Zhu and Gao, 2021). For example, strain ST-41, a core Wolbachia strain in butterflies, was shared among 30 butterfly species from four families, and it has also been found in four moth species from four families and one Diptera species (Ahmed et al., 2016; Ilinsky and Kosterin, 2017; Zhu and Gao, 2021). The occurrence of the similar Wolbachia strains in different butterfly species suggests that they may shift between host species via horizontal transfer, although the transfer mechanism is still not completely clear (Ahmed et al., 2016; Duplouy et al., 2020; Zhu and Gao, 2021). Horizontal transmission can increase the exposure of intracellular bacteria to the phage gene pool, thereby increasing the probability of being infected by phages (Moran and Plague, 2004; Bordenstein and Reznikoff, 2005; Newton and Bordenstein, 2011; Metcalf and Bordenstein, 2012). In this study, Wolbachia strains ST-41, ST-19, ST-125 and ST-374 were shared among two to seven butterfly species, respectively, and their host insects all harbored multiple phage types. In contrast, ST-297 and ST-wPcau were found to infect one butterfly species, whose insect hosts harbored a single phage type. Therefore, it might be considered that the horizontal transfer of Wolbachia between insect hosts increases the likelihood of exposure to phages, which is one of the reasons why butterflies harbor multiple types of phage WO.
Phage WO spreads among hosts through both vertical and horizontal transmission. Horizontal transfer of phage WO can occur between different Wolbachia strains, or it can be transmitted paternally by sperm from an infected male to the egg of a female carrying a phage-free Wolbachia (Gavotte et al., 2004; Bordenstein et al., 2006). The absence of an evolutionary correlation between WO and Wolbachia phylogenies (Bordenstein and Wernegreen, 2004; Gavotte et al., 2004; Wang N. X. et al., 2016) and divergent Wolbachia strains that infect the same (Gavotte et al., 2004; Chauvatcharin et al., 2006) or different hosts (Wang N. X. et al., 2016; Zhu et al., 2021) that have identical orf7 sequences indicated that many horizontal phage WO transfers occurred between different Wolbachia endosymbionts. In this study, 12 horizontal transmission events of phage WO were found, which shared common phage WO types among different Wolbachia strains associated with butterflies. Most horizontal transfer events involved different Wolbachia supergroups (A and B). Prophages undergo a lytic phase capable of rupturing bacterial and eukaryotic cell membranes, and phage WO occurs in the extracellular matrix of arthropods. Thus, they might pass through the eukaryote cell wall and then initiate new infections (Masui et al., 2001; Bordenstein et al., 2006; Gavotte et al., 2007). Wolbachia strains ST-41, ST-19, ST-125 and ST-374 were shared among various butterfly species. In addition to the common types, these butterfly species all harbored a variety of species-specific phage WO types. These results strongly suggested that Wolbachia may obtain phage WO types in butterflies through horizontal transfer of virus between eukaryotes, without Wolbachia. Therefore, the horizontal transmission of phage WO, with or without Wolbachia, was an important route for butterfly infecting Wolbachia strains to obtain phage diversity.
Reassortment and recombination have been confirmed in RNA and DNA viruses as a mechanism to adapt to the changing environment, expand host range and increase virulence, resulting in the genetic diversity of viruses (Domingo, 2010; Franzo et al., 2016; Chen et al., 2018). Bordenstein and Wernegreen (2004) confirmed the recombinogenic nature of phage WO, and, in the case of the capsid protein gene orf7, the recombination rate was the fastest reported for the Wolbachia genome. Zhu et al. (2021) provided practical molecular evidence supporting orf7 gene recombination in phage WO associated with gall wasps. Butterflies infected with a single Wolbachia strain carrying high levels of multiple phages provide an ideal model for detecting intragenic recombination. Twenty-two recombination events were identified in 13 of 16 butterfly species, which harbored diverse phage WO types, and some phage WO lineages derived from recombination could also be used as parents to form new phage WO types, displaying frequent and active recombination. These results suggested that intragenic recombination is an important evolutionary force, which effectively promotes the diversity of phage WO associated with butterflies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/; OM472181 – OM472424, and OM663002–OM663016.
D-HZ and SG designed the study. SG, Y-SR, and C-YS performed experiments and analyses. D-HZ and Y-SR wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the National Key Research and Development Program of China (no. 2018YFE0127100).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.865227/full#supplementary-material
Abe, Y., Ide, T., Su, C. Y., and Zhu, D. H. (2021). Leaf galls with the same morphology induced on the same plant species by two species of Latuspina (Hymenoptera: Cynipidae), with a description of a new species. Proc. Entomol. Soc. Wash. 123, 465–473. doi: 10.4289/0013-8797.123.3.465
Ahmed, M. Z., Breinholt, J. W., and Kawahara, A. Y. (2016). Evidence for common horizontal transmission of Wolbachia among butterflies and moths. BMC Evol. Biol. 16:118. doi: 10.1186/s12862-016-0660-x
Baldo, L., Bordenstein, S., Wernegreen, J. J., and Werren, J. H. (2006). Widespread recombination throughout Wolbachia genomes. Mol. Bio. Evol. 23, 437–449. doi: 10.1093/molbev/msj049
Batista, P. D., Keddie, B. A., Dosdall, L. M., and Harris, H. L. (2010). Phylogenetic placement and evidence for horizontal transfer of Wolbachia in Plutella xylostella (Lepidoptera: Plutellidae) and its parasitoid, Diadegma insulare (Hymenoptera: Ichneumonidae). Can. Entomol. 142, 57–64. doi: 10.4039/n09-050
Beckmann, J. F., Ronau, J. A., and Hochstrasser, M. (2017). A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2:17007. doi: 10.1038/nmicrobiol.2017.7
Bordenstein, S. R., Marshall, M. L., Fry, A. J., Kim, U., and Wernegreen, J. J. (2006). The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog. 2:e43. doi: 10.1371/journal.ppat.0020043
Bordenstein, S. R., and Reznikoff, W. S. (2005). Mobile DNA in obligate intracellular bacteria. Nat. Rev. Microbiol. 3, 688–699. doi: 10.1038/nrmicro1233
Bordenstein, S. R., and Wernegreen, J. J. (2004). Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol. Biol. Evol. 21, 1981–1991. doi: 10.1093/molbev/msh211
Chafee, M. E., Funk, D. J., Harrison, R. G., and Bordenstein, S. R. (2010). Lateral phage transfer in obligate intracellular bacteria (Wolbachia): verification from natural populations. Mol. Biol. Evol. 27, 501–505. doi: 10.1093/molbev/msp275
Chauvatcharin, N., Ahantarig, A., Baimai, V., and Kittayapong, P. (2006). Bacteriophage WO-B and Wolbachia in natural mosquito hosts: infection incidence, transmission mode and relative density. Mol. Ecol. 15, 2451–2461. doi: 10.1111/j.1365-294X.2006.02947.x
Chen, H., Ronau, J. A., Beckmann, J., and Hochstrasser, M. (2019). A Wolbachia nuclease and its binding partner provide a distinct mechanism for cytoplasmic incompatibility. Proc. Natl. Acad. Sci. U.S.A. 116, 22314–22321. doi: 10.1073/pnas.1914571116
Chen, Y., Trovao, N. S., Wang, G., Zhao, W., He, P., Zhou, H., et al. (2018). Emergence and evolution of novel Reassortant influenza a viruses in canines in southern China. mBio 9:e00909–18. doi: 10.1128/mBio.00909-18
Cooper, B. S., Vanderpool, D., Conner, W. R., Matute, D. R., and Turelli, M. (2019). Wolbachia acquisition by Drosophila yakuba-clade hosts and transfer of incompatibility loci between distantly related Wolbachia. Genetics 212, 1399–1419. doi: 10.1534/genetics.119.302349
Duplouy, A., and Hornett, E. A. (2018). Uncovering the hidden players in Lepidoptera biology: the heritable microbial endosymbionts. PeerJ 6:e4629. doi: 10.7287/peerj.preprints.26768v1
Duplouy, A., Pranter, R., Warren-Gash, H., Tropek, R., and Wahlberg, N. (2020). Towards unravelling Wolbachia global exchange: a contribution from the Bicyclus and Mylothris butterflies in the Afrotropics. BMC Microbiol. 20:319. doi: 10.1186/s12866-020-02011-2
Engelstädter, J., and Hurst, G. D. D. (2009). The ecology and evolution of microbes that manipulate host reproduction. Annu. Rev. Ecol. Evol. Syst. 40, 127–149. doi: 10.1146/annurev.ecolsys.110308.120206
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 3, 294–299.
Foster, J., Ganatra, M., Kamal, I., Ware, J., Makarova, K., Ivanova, N., et al. (2005). The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3:e121. doi: 10.1371/journal.pbio.0030121
Franzo, G., Cortey, M., Segalés, J., Hughes, J., and Drigo, M. (2016). Phylodynamic analysis of porcine circovirus type 2 reveals global waves of emerging genotypes and the circulation of recombinant forms. Mol. Phylogenet. Evol. 100, 269–280. doi: 10.1016/j.ympev.2016.04.028
Furukawa, S., Tanaka, K., Ikeda, T., Fukatsu, T., and Sasaki, T. (2012). Quantitative analysis of the lytic cycle of WO phages infecting Wolbachia. Appl. Entomol. Zool. 47, 449–456. doi: 10.1007/s13355-012-0142-6
Gavotte, L., Henri, H., Stouthamer, R., Charif, D., Charlat, S., Boulétreau, M., et al. (2007). A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol. Biol. Evol. 24, 427–435. doi: 10.1093/molbev/msl171
Gavotte, L., Vavre, F., Henri, H., Ravallec, M., Stouthamer, R., and Boulétreau, M. (2004). Diversity, distribution and specificity of WO phage infection in Wolbachia of four insect species. Insect Mol. Biol. 13, 147–153. doi: 10.1111/j.0962-1075.2004.00471.x
Gerth, M., Gansauge, M. T., Weigert, A., and Bleidorn, C. (2014). Phylogenomic analyses uncover origin and spread of the Wolbachia pandemic. Nat. Commun. 5:5117. doi: 10.1038/ncomms6117
Gibbs, M. J., Armstrong, J. S., and Gibbs, A. J. (2000). Sister-Scanning: a monte carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16, 573–582. doi: 10.1093/bioinformatics/16.7.573
Hatfull, G. F. (2008). Bacteriophage genomics. Curr. Opin. Microbiol. 11, 447–453. doi: 10.1016/j.mib.2008.09.004
Heath, L., Van Walt, E., Varsani, A., and Martin, D. P. (2006). Recombination patterns in aphthoviruses mirror those found in other picornaviruses. J. Virol. 80, 11827–11832. doi: 10.1128/JVI.01100-06
Hendrix, R. W., Smith, M. C., Burns, R. N., Ford, M. E., and Hatfull, G. F. (1999). Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc. Natl. Acad. Sci. U.S.A. 96, 2192–2197. doi: 10.1073/pnas.96.5.2192
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A., and Werren, J. H. (2008). How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol. Lett. 281, 215–220. doi: 10.1111/j.1574-6968.2008.01110.x
Hou, H. Q., Zhao, G. Z., Su, C. Y., and Zhu, D. H. (2020). Wolbachia prevalence patterns: horizontal transmission, recombination, and multiple infections in chestnut gall wasp-parasitoid communities. Entomol. Exp. Appl. 168, 752–765. doi: 10.1111/eea.12962
Ilinsky, Y., and Kosterin, O. (2017). Molecular diversity of Wolbachia in Lepidoptera: prevalent allelic content and high recombination of MLST genes. Mol. Phylogenet. Evol. 109, 164–179. doi: 10.1016/j.ympev.2016.12.034
Ishmael, N., Hotopp, J. C. D., Ioannidis, P., Biber, S., Sakomoto, J., Siozios, S., et al. (2009). Extensive genomic diversity of closely related Wolbachia strains. Microbiology 155, 2211–2222. doi: 10.1099/mic.0.027581-0
Jiggins, F. M., Bentley, J. K., Majerus, M. E. N., and Hurst, G. D. D. (2001). How many species are infected with Wolbachia? Cryptic sex ratio distorters revealed to be common by intensive sampling. Proc. R. Soc. Lond. B 268, 1123–1126. doi: 10.1098/rspb.2001.1632
Johannesen, J. (2017). Tracing the history and ecological context of Wolbachia double infection in a specialist host (Urophora cardui) – parasitoid (Eurytoma serratulae) system. Ecol. Evol. 7, 986–996. doi: 10.1002/ece3.2713
Kent, B. N., and Bordenstein, S. R. (2010). Phage WO of Wolbachia: lambda of the endosymbiont world. Trends Microbiol. 18, 173–181. doi: 10.1016/j.tim.2009.12.011
Kent, B. N., Funkhouser, L. J., Setia, S., and Bordenstein, S. R. (2011). Evolutionary genomics of a temperate bacteriophage in an obligate intracellular bacteria (Wolbachia). PLoS One 6:e24984. doi: 10.1371/journal.pone.0024984
Klasson, L., Westberg, J., Sapountzis, P., Näslunda, K., Lutnaes, Y., Darby, A. C., et al. (2009). The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc. Natl. Acad. Sci. U.S.A. 106, 5725–5730. doi: 10.1073/pnas.0810753106
LePage, D. P., Metcalf, J. A., Bordenstein, S. R., On, J., Perlmutter, J. I., Shropshire, J. D., et al. (2017). Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543, 243–247. doi: 10.1038/nature21391
Martin, D., and Rybicki, E. (2000). RDP: detection of recombination amongst aligned sequences. Bioinformatics 16, 562–563. doi: 10.1093/bioinformatics/16.6.562
Martin, D. P., Posada, D., Crandall, K. A., and Williamson, C. (2005). A modified BOOTSCAN algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retrov. 21, 98–102. doi: 10.1089/aid.2005.21.98
Masui, S., Kamoda, S., Sasaki, T., and Ishikawa, H. (2000). Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J. Mol. Evol. 51, 491–497. doi: 10.1007/s002390010112
Masui, S., Kuroiwa, H., Sasaki, T., Inui, M., Kuroiwa, T., and Ishikawa, H. (2001). Bacteriophage WO and virus-like particles in Wolbachia, an endosymbiont of arthropods. Biochem. Biophys. Res. Commun. 283, 1099–1104. doi: 10.1006/bbrc.2001.4906
Metcalf, J. A., and Bordenstein, S. R. (2012). The complexity of virus systems: the case of endosymbionts. Curr. Opin. Microbiol. 15, 546–552. doi: 10.1016/j.mib.2012.04.010
Moran, N. A., and Plague, G. R. (2004). Genomic changes following host restriction in bacteria. Curr. Opin. Genet. Dev. 14, 627–633. doi: 10.1016/j.gde.2004.09.003
Narita, S., Kageyama, D., Hiroki, M., Sanpei, T., Hashimoto, S., Kamito, T., et al. (2011). Wolbachia-induced feminisation newly found in Eurema hecabe, a sibling species of Eurema mandarina (Lepidoptera: Pieridae). Ecol. Entomol. 36, 309–317. doi: 10.1111/j.1365-2311.2011.01274.x
Newton, I. L. G., and Bordenstein, S. R. (2011). Correlations between bacterial ecology and mobile DNA. Curr. Microbiol. 62, 198–208. doi: 10.1007/s00284-010-9693-3
Padidam, M., Sawye, S., and Fauquet, C. M. (1999). Possible emergence of new geminiviruses by frequent recombination. Virology 265, 218–225. doi: 10.1006/viro.1999.0056
Posada, D., and Crandall, K. A. (2001). Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. U.S.A. 98, 13757–13762. doi: 10.1073/pnas.241370698
Salunke, B. K., Salunkhe, R. C., Dhotre, D. P., Walujkar, S. A., Khandagale, A. B., Chaudhari, R., et al. (2012). Determination of Wolbachia diversity in butterflies from Western Ghats, India, by a multigene approach. Appl. Environ. Microbiol. 78, 4458–4467. doi: 10.1128/AEM.07298-11
Sanaei, E., Charlat, S., and Engelstädter, J. (2021). Wolbachia host shifts: routes, mechanisms, constraints and evolutionary consequences. Biol. Rev. 96, 433–453. doi: 10.1111/brv.12663
Shropshire, J. D., and Bordenstein, S. R. (2019). Two-by-one model of cytoplasmic incompatibility: synthetic recapitulation by transgenic expression of cifA and cifB in Drosophila. PLoS Genet. 15:e1008221. doi: 10.1371/journal.pgen.1008221
Sintupachee, S., Milne, J. R., Poonchaisri, S., Baimai, V., and Kittayapong, P. (2006). Closely related Wolbachia strains within the pumpkin arthropod community and the potential for horizontal transmission via the plant. Microb. Ecol. 51, 294–301. doi: 10.1007/s00248-006-9036-x
Smith, J. M. (1992). Analyzing the mosaic structure of genes. J. Mol. Evol. 34, 126–129. doi: 10.1007/BF00182389
Su, C. Y., Zhu, D. H., and Yang, X. H. (2021). Design and testing of effective primers for amplification of the orf7 gene of phage WO associated with Andricus hakonensis. Insects 12:713. doi: 10.3390/insects12080713
Swofford, D. L. (2003). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sunderland, MA: Sinauer Associates.
Tagami, Y., and Miura, K. (2004). Distribution and prevalence of Wolbachia in Japanese populations of Lepidoptera. Insect Mol. Biol. 13, 359–364. doi: 10.1111/j.0962-1075.2004.00492.x
Tanaka, K., Furukawa, S., Nikoh, N., Sasaki, T., and Fukatsu, T. (2009). Complete WO phage sequences reveal their dynamic evolutionary trajectories and putative functional elements required for integrationin to the Wolbachia genome. Appl. Environ. Microbiol. 75, 5676–5686. doi: 10.1128/aem.01172-09
van Nieukerken, E. J., Kaila, L., Kitching, I. J., Kristensen, N. P., Lees, D. C., Minet, J., et al. (2011). Order Lepidoptera Linnaeus, 1758. Zootaxa 3148, 212–221. doi: 10.11646/zootaxa.3148.1.41
Wang, G. H., Sun, B. F., Xiong, T. L., Wang, Y. K., Murfin, K. E., Xiao, J. H., et al. (2016). Bacteriophage WO can mediate horizontal gene transfer in endosymbiotic Wolbachia genomes. Front. Microbiol. 7:1867. doi: 10.3389/fmicb.2016.01867
Wang, M., Peng, Y., Huang, X., Jin, Y., and Peng, Y. (2020). Incidence of Wohlbachia, Cardinium, Spiroplasma and phage WO in different geographical populations of Chilo suppressalis (Lepidoptera: Pyralidae) from China. Entomol. News 129, 230–243. doi: 10.3157/021.129.0302
Wang, N. X., Jia, S. S., Xu, H., Liu, Y., and Huang, D. W. (2016). Multiple horizontal transfers of bacteriophage WO and host Wolbachia in fig wasps in a closed community. Front. Microbiol. 7:136. doi: 10.3389/fmicb.2016.00136
Weinert, L. A., Araujo-Jnr, E. V., Ahmed, M. Z., and Welch, J. J. (2015). The incidence of bacterial endosymbionts in terrestrial arthropods. Proc. R. Soc. Lond. B 282:20150249. doi: 10.1098/rspb.2015.0249
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Werren, J. H., Windsor, D., and Guo, L. R. (1995). Distribution of Wolbachia among neotropical arthropods. Proc. R. Soc. Lond. B 262, 197–204. doi: 10.1098/rspb.1995.0196
Yang, X. H., Zhu, D. H., Liu, Z., Zhao, L., and Su, C. Y. (2013). High levels of multiple infections, recombination and horizontal transmission of Wolbachia in the Andricus mukaigawae (Hymenoptera; Cynipidae) communities. PLoS One 8:e78970. doi: 10.1371/journal.pone.0078970
Zhu, D. H., and Gao, S. (2021). A prevalence survey of Wolbachia in butterflies from southern China. Entomol. Exp. Appl. 169, 1157–1166. doi: 10.1111/eea.13112
Zhu, D. H., He, Y. Y., Fan, Y. S., Ma, M. Y., and Peng, D. L. (2007). Negative evidence of parthenogenesis induction by Wolbachia in a gall wasp species, Dryocosmus kuriphilus. Entomol. Exp. Appl. 124, 279–284. doi: 10.1111/j.1570-7458.2007.00578.x
Zhu, D. H., Su, C. Y., Yang, X. H., and Abe, Y. (2021). A case of intragenic recombination dramatically impacting the phage WO genetic diversity in gall wasps. Front. Microbiol. 12:694115. doi: 10.3389/fmicb.2021.694115
Keywords: phage WO, multiple infections, recombination, horizontal transfer, Wolbachia, butterfly, Lepidoptera
Citation: Gao S, Ren Y-S, Su C-Y and Zhu D-H (2022) High Levels of Multiple Phage WO Infections and Its Evolutionary Dynamics Associated With Wolbachia-Infected Butterflies. Front. Microbiol. 13:865227. doi: 10.3389/fmicb.2022.865227
Received: 29 January 2022; Accepted: 28 March 2022;
Published: 21 April 2022.
Edited by:
Claire Valiente Moro, Université Claude Bernard Lyon 1, FranceReviewed by:
Rhitoban Raychoudhury, Indian Institute of Science Education and Research Mohali, IndiaCopyright © 2022 Gao, Ren, Su and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dao-Hong Zhu, ZGFvaG9uZ3podUB5ZWFoLm5ldA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.