
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 08 March 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.865141
This article is part of the Research TopicInsights in Microbial Physiology and Metabolism: 2021View all 13 articles
Protein homeostasis is fundamental to cell function and survival. It relies on an interconnected network of processes involving protein synthesis, folding, post-translational modification and degradation as well as regulators of these processes. Here we provide an update on the roles, regulation and subcellular localization of the protein homeostasis machinery in the Gram-positive model organism Bacillus subtilis. We discuss emerging ideas and current research gaps in the field that, if tackled, increase our understanding of how Gram-positive bacteria, including several human pathogens, maintain protein homeostasis and cope with stressful conditions that challenge their survival.
Native proteins typically fold into well-defined three-dimensional structures. To function properly, all cells need to contain correctly folded proteins and have mechanisms to prevent accumulation of unneeded or aberrant proteins. The folded state of most proteins is marginally more stable than the unfolded state. Therefore, small changes of environmental conditions may affect the equilibrium between the folded and unfolded state. Protein homeostasis (proteostasis) is crucial to achieve a “healthy” proteome, and refers to the dynamic balance between synthesis, folding, post-translational modification, transport, and degradation of proteins (Figure 1; Powers et al., 2009; Richter et al., 2010; Schramm et al., 2020). The main components of the proteostasis network are the ancient and evolutionary conserved chaperones and proteases, which assist in protein folding and degrade specific protein substrates, respectively (Powers and Balch, 2013; Balchin et al., 2016; Olivares et al., 2016). Surprisingly, even though eukaryotic proteomes are typically much larger and complex and contain more aggregation-prone proteins than those of prokaryotes, no new core chaperones appear to have emerged during billion years of evolution (Rebeaud et al., 2021). Instead, the core chaperones and their relative abundance have remained invariant across the domains of life. Maintaining integrity of a more complex and unstable proteome has been dealt with by increasing cellular chaperone levels, as well as promoting cooperation between them (Rebeaud et al., 2021).
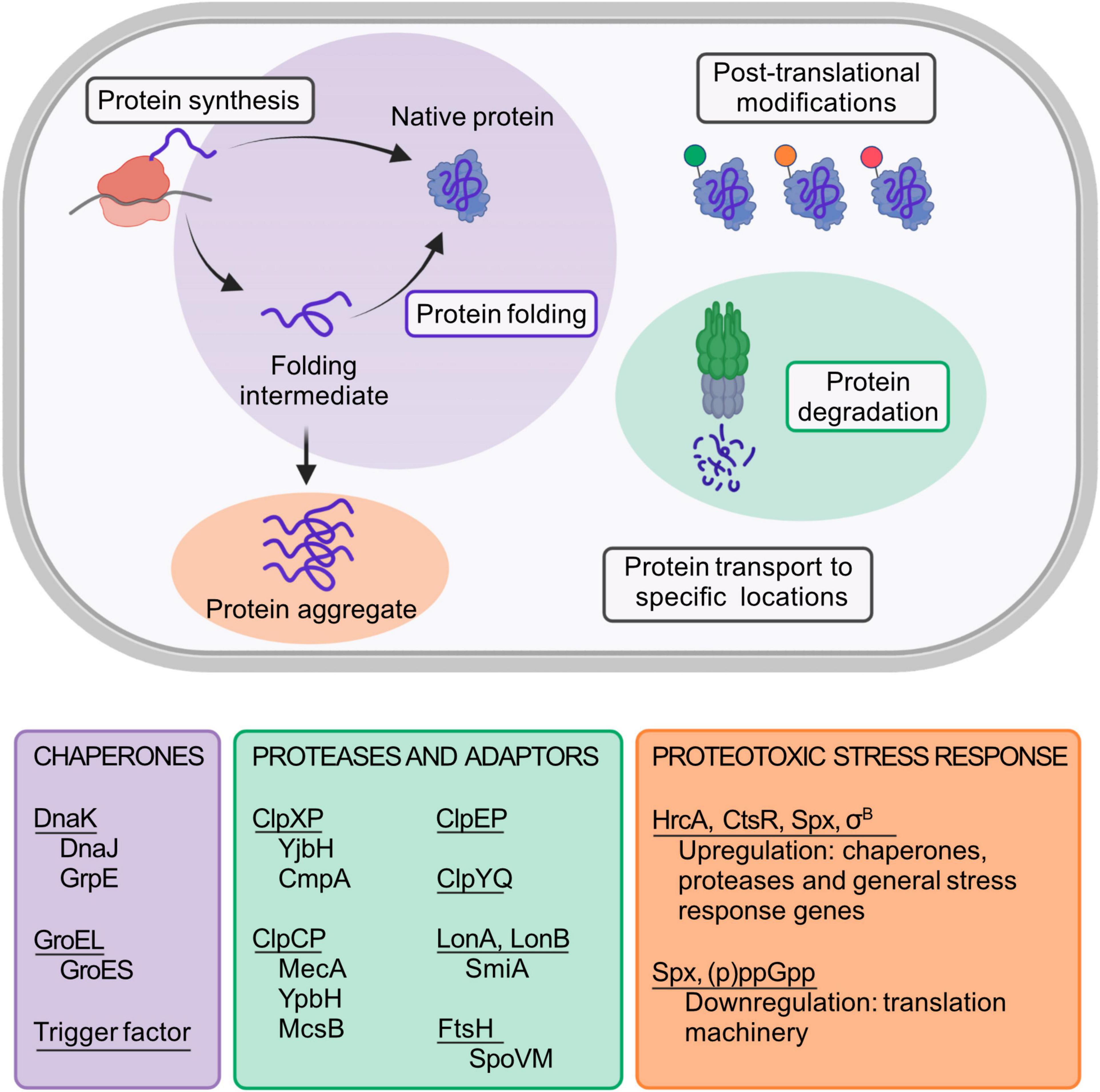
Figure 1. Schematic representation of the B. subtilis proteostasis network and its regulators. Protein folding and degradation are two key cellular processes involved in proteostasis maintenance. While some proteins can spontaneously fold into their functional native state, many others need the assistance of molecular chaperones to do so. Underlined in the purple box are the main B. subtilis chaperones. For its full activity, DnaK requires the co-chaperone DnaJ and the nucleotide exchange factor GrpE. Similarly, GroEL requires the co-chaperonin GroES. Unneeded, misfolded or damaged proteins are eliminated from the cell by proteases (underlined in the green box) and its respective adaptor proteins (shown below each protease complex). In addition to protein folding and degradation, other key processes that affect proteostasis maintenance are protein synthesis, post-translational modifications, and the transport of proteins to specific locations. Failure in proteostasis maintenance often leads to the formation and accumulation of misfolded and aggregated proteins, a condition termed as proteotoxic stress. The main regulators that are known to be involved in the B. subtilis proteotoxic stress response are shown in the orange text box. Created with BioRender.com.
Understanding how cells maintain proteostasis is an important topic to address, not only because proteome integrity is crucial for the correct cellular function, but also because accumulation of protein aggregates – which mainly results from dysregulation of proteostasis – has been linked to aging and to human diseases, such as Parkinson’s and Alzheimer’s, and to defects in growth and survival in prokaryotes (Balchin et al., 2016; Cheng et al., 2018). In addition, the presence of protein aggregates is strongly correlated with dormant antibiotic-resistant cells, called persisters (Leszczynska et al., 2013; Pu et al., 2019; Yu et al., 2019; Dewachter et al., 2021; Huemer et al., 2021). Because of this and other data, disruption of proteostasis has been suggested as an anti-bacterial strategy (Khodaparast et al., 2021).
Most information on chaperones and proteases in bacteria derives from studies in Escherichia coli, but there are many host-specific regulation mechanisms concerning proteostasis to unravel. B. subtilis is adapted to rapid intracellular and environmental fluctuations that challenge the stability of its proteome. Thus, it is a suitable model organism to study not only from the view of fundamental principles of proteostasis, but also regarding proteostasis maintenance in other Gram-positive bacteria, that includes several human pathogens. Here we provide an updated description of the main components of the B. subtilis proteostasis network (Figure 1), and address known and emerging mechanisms for its regulation during proteotoxic stress.
Molecular chaperones are central to proteostasis by ensuring that proteins are correctly folded, and preventing protein misfolding and aggregation (Mogk et al., 2011; Balchin et al., 2016). The ancient and evolutionary conserved DnaK (Hsp70), GroEL (Hsp60), and trigger factor (TF) are three important abundant cytosolic chaperones in B. subtilis (Moliere and Turgay, 2009).
DnaK functions as a monomer and consists of an N-terminal ATPase domain and a C-terminal peptide-binding domain composed of a β-sandwich and an α-helical lid (Zhu et al., 1996; Perales-Calvo et al., 2018). Together with its co-chaperone DnaJ (Hsp40) and the nucleotide exchange factor GrpE, and through ATP hydrolysis, the α-helical lid closes over the β-sandwich, allowing tight binding of unfolded substrates (Liberek et al., 1991; Zhu et al., 1996). DnaK typically recognizes exposed hydrophobic peptide segments (∼5–7 residues) of client proteins that are prone to aggregate during folding (Rudiger et al., 1997; Mogk et al., 1999; Calloni et al., 2012); substrate binding and release cycles decrease the folding rate, and prevent non-native protein species from folding prematurely in a misfolded state or from aggregating (Szabo et al., 1994). Interestingly, some bacteria like E. coli contain more than one Hsp70 and Hsp40 homolog. For instance, apart from DnaK, E. coli harbors two additional Hsp70 proteins – Hsc66 (Seaton and Vickery, 1994) and Hsc62 (Yoshimune et al., 1998) – and five additional DnaJ-like proteins such as Hsc20 and CbpA (Ueguchi et al., 1994; Lelivelt and Kawula, 1995; Clarke et al., 1996; Itoh et al., 1999; Yoshimune et al., 2002). In contrast, B. subtilis appears to have only DnaK and DnaJ. Oligomeric GroEL is composed of two stacked heptameric rings, each forming large cylindrical cavities in which misfolded protein substrates can be enclosed (Langer et al., 1992; Mayhew et al., 1996). The GroES (Hsp10) heptameric co-chaperonin caps GroEL cavities, and through ATP hydrolysis allows complete substrate encapsulation, thus providing a “protected” folding environment (Mayhew et al., 1996). Finally, TF is a ribosome-associated chaperone comprised of three domains adopting an overall elongated shape (Stoller et al., 1995; Zarnt et al., 1997). An N-terminal ribosome-binding domain is followed by a peptidyl-prolyl isomerase domain linked to the C-terminal substrate-binding domain. The substrate-binding domain has two helical arms that form a promiscuous clamp-like structure, providing a shielded environment to nascent polypeptides as translation proceeds, and also slowing the folding rate preventing aggregation (Agashe et al., 2004; Ferbitz et al., 2004; Singhal et al., 2015). The clamp, together with the TF structural flexibility, allows TF to function with a wide range of emerging substrates (Martinez-Hackert and Hendrickson, 2009; Saio et al., 2014). While the monomeric form of TF is bound to the ribosome, its dimeric form exists mainly in the cytosol, and apart from assisting several proteins in their folding, it has anti-aggregation activity (Saio et al., 2018).
In addition to assisting in the folding of cytosolic proteins, DnaK/DnaJ/GrpE (Wild et al., 1992, 1993, 1996), GroEL/GroES (Kusukawa et al., 1989), and TF (Lee and Bernstein, 2002; Genevaux et al., 2004; Ullers et al., 2007; Oh et al., 2011) are also involved in protein secretion by preventing premature folding and aggregation of presecretory proteins in the cytosol. Most proteins are translocated in an unfolded state via the general secretion (Sec) pathway. Typically, the secretion-specific chaperone SecB binds newly synthesized presecretory proteins and targets them for SecA-driven protein translocation. However, SecB is absent in many Gram-positive bacteria, and in B. subtilis CsaA has been suggested to play a similar role since, among other evidence, it interacts with SecA as well as several presecretory proteins (Müller et al., 2000; Linde et al., 2003).
Even though the general mechanisms of action and structures of DnaK, GroEL, and TF chaperones are widely conserved among organisms, their specific roles and their contributions to proteostasis maintenance differ between bacteria. A clear example of such divergence is displayed by the phenotypic differences between E. coli and B. subtilis chaperone-deficient mutants. Although in both organisms groES and groEL are essential genes at all temperatures (Fayet et al., 1989; Commichau et al., 2013), single and double deletions of dnaK and tig (encoding DnaK and TF) give different effects. In E. coli, DnaK is essential for growth at high or low temperature (Paek and Walker, 1987; Bukau and Walker, 1989), and it plays a crucial role under both optimal and proteotoxic stress conditions. Its absence causes cellular defects such as reduced growth rates, dysregulation of the heat-shock genes, and abnormal cell division (Paek and Walker, 1987; Bukau and Walker, 1989, 1990). DnaK is not only needed for the folding of a large number of E. coli “thermolabile” proteins (Mogk et al., 1999), it also regulates the heat shock sigma factor σ32 (Gamer et al., 1992; Liberek et al., 1992).
Deletion of tig in E. coli also leads to cellular defects: it reduces the cell’s tolerance to SDS/EDTA and vancomycin, thereby reducing outer membrane integrity (Oh et al., 2011), and induces the heat shock response (Deuerling et al., 2003). In E. coli, DnaK and TF possess overlapping functions in protein folding, and their shared role seems to be crucial for maintaining proteostasis, even under typical growth temperatures (Deuerling et al., 1999, 2003; Teter et al., 1999; Genevaux et al., 2004; Calloni et al., 2012). This redundancy might explain why a double deletion of dnaK and tig is synthetically lethal at temperatures above 30°C (Deuerling et al., 1999; Teter et al., 1999; Genevaux et al., 2004). Another example of chaperone collaboration is found between Hsp70 and the ATP-dependent Hsp90 chaperone, which in eukaryotes are well known to function together to orchestrate the proteostasis network (Schopf et al., 2017). In bacteria, the function of the Hsp90 homolog HtpG is not as well-characterized [see Wickner et al. (2021) for the latest review], but it has been shown that HtpG and DnaK systems also collaborate during the protein folding process in E. coli (Genest et al., 2011). DnaK-HtpG interaction involves the DnaJ-like protein CbpA (Genest et al., 2015). Unlike in eukaryotes, HtpG is not essential for the growth of many bacteria including E. coli and B. subtilis, and deletion of htpG causes minor growth defects after temperature upshifts in both organisms (Bardwell and Craig, 1988; Thomas and Baneyx, 1998; Versteeg et al., 1999). In addition, recent proteomic studies suggest that HtpG enhances degradation of DnaK/DnaJ client substrates (Fauvet et al., 2021).
While the implications and roles of chaperones in proteostasis are well-characterized in E. coli, far less is known for other bacteria including B. subtilis. In stark contrast to E. coli, the absence of DnaK, TF or both proteins in B. subtilis does not affect cell viability in the 16–52°C temperature range (Schulz et al., 1995; Gothel et al., 1998; Reyes and Yoshikawa, 2002). Apart from a very short study (Reyes and Yoshikawa, 2002), no characterization of the effects of a dnaK tig double deletion in B. subtilis has been published. B. subtilis dnaK tig double mutants are viable below 53°C, suggesting that folding of nascent peptide chains is assisted also by other proteins than DnaK and TF (Reyes and Yoshikawa, 2002).
B. subtilis DnaK and TF are subjected to regulation by phosphorylation. Tyrosine residue 601 in the C-terminal region of DnaK can be phosphorylated by the PtkA kinase and dephosphorylated by the PtpZ phosphatase, influencing its chaperone activity and survival of the cell upon heat-shock (Shi et al., 2016). In the case of TF, phosphorylation of Arg45 by the McsB kinase negatively influences its association with the ribosome (Zhou et al., 2019). Interestingly, spore germination requires Arg45 to be dephosphorylated by the YwlE phosphatase, since this licenses TF to interact with ribosomes and resume translation (Zhou et al., 2019). These observations are in line with the notion that chaperones possess host-specific roles, in addition to their conserved functions.
Unfinished, damaged, misfolded, or unneeded proteins are eliminated from the cell to maintain proteome integrity. In B. subtilis, degradation of most cytoplasmic proteins is performed by the conserved AAA+ family of intracellular proteases (AAA+; ATPases associated with a variety of cellular activities), which recognize, unfold, and degrade specific protein substrates (Sauer and Baker, 2011). B. subtilis has seven AAA+ proteases: ClpCP, ClpEP, ClpXP, ClpYQ, LonA, LonB, and FtsH (Elsholz et al., 2017), whose mechanisms of action have been described in numerous reviews (Sauer et al., 2004; Kirstein et al., 2009; Sauer and Baker, 2011; Olivares et al., 2016; Elsholz et al., 2017). The Clp complexes consist of an AAA+ unfoldase coupled to an ATP-dependent serine protease, whereas LonA, LonB, and FtsH have both unfoldase and protease domains within a single polypeptide (Elsholz et al., 2017).
Proteolysis can be regulated by adaptor proteins, which provide substrate specificity to proteases, usually by interacting with both substrate and protease. Several adaptor proteins have been characterized in B. subtilis, such as the ClpCP adaptor proteins MecA, YpbH, and McsB, and the ClpXP adaptor proteins YjbH and CmpA (Elsholz et al., 2017). The mechanism of adaptors often involves tethering the substrate to the protease to increase the local substrate concentration to facilitate proteolysis (Battesti and Gottesman, 2013). Less common mechanisms of adaptors are also known. For instance, the ClpXP adaptor protein YjbH does not appear to directly interact with ClpX (Chan et al., 2012), but enhances degradation of the stress-responsive regulator Spx by binding and stabilizing it, promoting its recognition by ClpXP (Awad et al., 2019).
AAA+ proteases or their respective adaptors recognize short degradation signals (degrons) located at the N-terminal, internal, or C-terminal position of protein substrates (Kirstein et al., 2009). A degron with relevance in protein homeostasis maintenance is the C-terminal SsrA degradation tag, which is added co-translationally by the transfer-messenger RNA (tmRNA) system to unfinished polypeptides when ribosomes stall (Keiler et al., 1996; Moore and Sauer, 2007). Truncated polypeptides challenge the stability of the proteome, and it is important that they are eliminated from the cell. SsrA-tagged polypeptides are typically degraded by the ClpXP protease complex (Sauer and Baker, 2011), although in E. coli, ClpAP and FtsH proteases can also recognize and degrade SsrA-tagged proteins (Gottesman et al., 1998; Herman et al., 1998). Cryo-EM studies have provided a detailed molecular mechanism of SsrA-tagged substrate recognition by ClpXP. Specific binding of the SsrA degron to ClpX triggers a ClpX conformational change from a “closed-pore” conformation to an “open-pore” conformation, allowing substrate translocation through the channel and subsequent non-specific interactions of the unfolded substrate with inner channel residues (Fei et al., 2020).
Interestingly, a novel ClpXP proteolytic mechanism for degradation of unfinished polypeptides in B. subtilis that is redundant with the SsrA tagging has been uncovered (Lytvynenko et al., 2019). Here, the B. subtilis RqcH recognizes stalled ribosomes and recruits tRNAAla to mark aberrant nascent chains for degradation with C-terminal poly-alanine tails, which are recognized by ClpXP (Lytvynenko et al., 2019). Because the ALAA motif of the SsrA tag and poly-alanine tails are similar, it would be no surprise if their recognition and degradation mechanism would be similar. However, whether the poly-alanine tagged proteins are also degraded by ClpAP and FtsH in B. subtilis remains to be answered.
Another degradation tag of vast importance in B. subtilis proteostasis is the phospho-arginine (pArg) tag introduced by McsB (Trentini et al., 2016). McsB is conserved among Gram-positive bacteria and functions both as an adaptor protein for ClpCP (Kirstein et al., 2007) and as an arginine kinase with a major role in eliminating hundreds of damaged proteins from the cytoplasm, particularly under proteotoxic stress conditions (Elsholz et al., 2012; Trentini et al., 2016). McsB phosphorylates arginine residues, marking proteins for degradation by ClpCP (Trentini et al., 2016). Among the McsB phosphorylated proteins are the protein quality control members CtsR, HrcA, GroEL, TF, ClpC, and ClpP (Schmidt et al., 2014). In the case of the transcriptional repressors CtsR and HrcA, phosphorylation of residues in their DNA-binding domains greatly contributes to induction of the proteotoxic stress response (Kirstein et al., 2005; Fuhrmann et al., 2009; Schmidt et al., 2014). A recent study uncovered the molecular mechanism of McsB targeting (Hajdusits et al., 2021). McsB assemble into octamers, stabilized by auto-phosphorylation to form a molecular chamber-like structure, with the kinase active site buried inside (Hajdusits et al., 2021). McsB octamers are formed upon proteotoxic stress conditions, when McsB levels increase, and possess high selectivity for phosphorylation of unfolded proteins, which are able to access the kinase chamber through a narrow entrance. The phosphorylated proteins are thus targeted for degradation by ClpCP (Hajdusits et al., 2021).
Although it is likely that B. subtilis Lon proteins and FtsH play some role in protein quality control, experimental evidence is lacking. However, roles in degradation of regulatory proteins have been reported (Thi Nguyen and Schumann, 2012; Bradshaw and Losick, 2015; Mukherjee et al., 2015).
B. subtilis ClpC, ClpE, ClpX, and ClpP have been shown to co-localize with heat-induced protein aggregates or PorA inclusion bodies (Kruger et al., 2000; Jurgen et al., 2001; Miethke et al., 2006). In non-stressed cells, without aggregates, ClpP appears in the cytoplasm, while ClpC and ClpX are found both in the cytoplasm and associated with the membrane (Kruger et al., 2000; Jurgen et al., 2001). Three independent publications reported in 2008 that B. subtilis GFP-tagged Clp proteins, such as ClpX and ClpP, form foci with a cell polar localization pattern (Kain et al., 2008; Kirstein et al., 2008; Simmons et al., 2008). The subcellular localization of Lon seems to be developmentally regulated: B. subtilis LonA-GFP associates with the nucleoid under normal growth, and with the forespore during sporulation (Simmons et al., 2008).
It is important to note that used fluorescent protein tags lead to clustering artifacts when fused to homo-oligomers such as Clp proteins, and that, at least in E. coli, Clp proteins are homogenously distributed in the cell (Landgraf et al., 2012). Therefore, the native sub-cellular localization of the B. subtilis proteolytic machinery should be revaluated.
Localization of chaperones seems to be conditional to stress in several bacteria. For example, in E. coli the co-chaperone DnaJ mediates ATP-DnaK binding to protein aggregates (Acebron et al., 2008). Moreover, large heat-induced protein aggregates localize at the cell poles, and such a localization requires DnaK and DnaJ, as well as ATP synthesis and the membrane proton motive force (Rokney et al., 2009). DnaK and the ClpB disaggregase are essential for dissolving polar aggregates (Rokney et al., 2009), but it is not clear whether the polar localization of aggregates in E. coli is energy-dependent, since other studies claim it to be a passive process, driven by the molecular crowding in the nucleoid region (Winkler et al., 2010; Coquel et al., 2013; Gupta et al., 2014; Neeli-Venkata et al., 2016; Oliveira et al., 2016). Large, polar localized, protein aggregates are asymmetrically inherited in E. coli, as division generates cells with aggregates at the old cell poles (Lindner et al., 2008; Winkler et al., 2010). In B. subtilis, protein aggregates have also been shown to locate at cell poles (Kirstein et al., 2008; Runde et al., 2014; Stannek et al., 2014; Hantke et al., 2019; Schafer et al., 2019), but their inheritance after cell division has not been studied.
To our knowledge, chaperone localization in B. subtilis has been only addressed in few studies. B. subtilis GFP-DnaK localizes as multiple discrete foci proximal to the membrane (Meile et al., 2006). In response to short-term ethanol stress, phosphorylated DnaK and GroEL chaperones are recruited to the B. subtilis cytoplasmic membrane (Seydlova et al., 2012).
Collectively, it seems that to cope with proteotoxic stress the cell redirects the protein quality control machinery to sub-cellular areas, containing protein aggregates, such as the cell poles. Interestingly, protein aggregates are typically associated with detrimental effects for cellular fitness (Ross and Poirier, 2004; Lindner et al., 2008; Mortier et al., 2019), but their presence has been reported to pre-adapt lineages to subsequent proteotoxic stress (Govers et al., 2014; Mortier et al., 2019). Such pre-adaptation may arise from the increased levels of protein quality control agents such as proteases and chaperones that co-localize with protein aggregates (Kruger et al., 2000; Jurgen et al., 2001; Acebron et al., 2008; Govers et al., 2014; Mortier et al., 2019).
During proteotoxic stress, response mechanisms are activated which help the bacterium adapt to the new cellular or environmental condition. Of particular importance in B. subtilis are the HrcA and CtsR regulators. HrcA represses transcription of the hrcA-grpE-dnaK-dnaJ-yqeT-yqeU-yqeV and the groES-groEL operons (Schumann, 2016), and thus regulates the synthesis of chaperones. HrcA levels are depleted upon proteotoxic stress by a feedback mechanism involving the GroEL-GroES chaperone complex (Mogk et al., 1997; Schumann, 2016). CtsR, a regulator of protein degradation, represses transcription of the ctsR-mcsA-mcsB-clpC-radA-disA operon, and the clpP and clpE genes (Derre et al., 2000; Kruger et al., 2001; Elsholz et al., 2010). Regulation by CtsR involves a complex regulatory network, where McsB, McsA, and ClpCP play important roles in derepressing the CtsR regulon upon proteotoxic stress conditions [for a review, see Elsholz et al. (2017)].
Another player in the proteotoxic stress response is the Spx protein. Spx was initially characterized as a global regulator of the thiol-specific oxidative stress response (Nakano et al., 2003), controlling ∼144 transcriptional units (Rochat et al., 2012). However, an increasing number of studies have reported the involvement of Spx in the response to other stress conditions, such as heat shock and compounds targeting the cell wall (Runde et al., 2014; Rojas-Tapias and Helmann, 2018; Schafer and Turgay, 2019). The view that Spx is an important regulator of the proteotoxic stress response is becoming established (Rojas-Tapias and Helmann, 2019a,b; Schafer et al., 2019). Spx interacts with the C-terminal domain of the α-subunit (αCTD) of the RNA polymerase (RNAP), activating or repressing target genes in order to cope with the stress (Zuber, 2004; Newberry et al., 2005; Reyes and Zuber, 2008; Lamour et al., 2009; Nakano et al., 2010; Rochat et al., 2012). As revealed by structural studies, redox activated Spx with a disulfide bond between the two cysteine residues (Cys10 and Cys13) interacts both with the αCTD and σA in the holo RNAP, and this complex binds to the −44 position of promoter DNA to enhance transcription activation (Shi et al., 2021). Among the Spx-induced genes are trxA (thioredoxin) and trxB (thioredoxin reductase), as well as the clpX, clpE, and clpC genes, and putatively clpP (Nakano et al., 2003; Rochat et al., 2012). Spx also induces the ctsR operon (Rojas-Tapias and Helmann, 2019a).
Control of the cellular level and activity of Spx involves many layers of regulation that are fine-tuned depending on the type of stress [reviewed in Rojas-Tapias and Helmann (2019b)]. The most important layer of Spx regulation seems to be through ClpXP proteolysis. Efficient degradation of Spx under normal conditions requires the ClpXP adaptor protein YjbH, which aggregates upon proteotoxic stress conditions and causes a decrease in Spx proteolysis (Nakano et al., 2003; Larsson et al., 2007; Garg et al., 2009; Engman and von Wachenfeldt, 2015). By hydrogen-deuterium exchange mass spectrometry it was determined that binding to YjbH decreases the Spx dynamics, reducing the conformational entropy and probably allowing a more efficient recognition of its C-terminal end, needed for ClpXP degradation (Awad et al., 2019). ClpCP and its adaptor McsB are also involved in Spx degradation, although to a lesser extent (Rojas-Tapias and Helmann, 2019a). Interestingly, the Spx paralog MgsR, which is also involved in the oxidative stress response (Reder et al., 2008), has been shown to interact with McsB upon ethanol stress, and McsB enhances MgsR degradation by ClpXP in vivo (Lilge et al., 2020).
Proteotoxic stress conditions also induce the general stress response, which is governed by the alternative sigma factor σB and is one of the most important non-specific stress response mechanisms of B. subtilis. The σB regulon is induced through a signal transduction cascade, involving the RsbV, RsbW, and RsbX regulators, and comprises about 200 genes, defined as class II heat-shock genes (Schumann, 2003; Nannapaneni et al., 2012). Other than heat, the general stress response is triggered by a wide range of stresses (Hecker et al., 2007). Genes included in the σB regulon are, for example, genes that protect against elevated temperatures, such as clpP and clpC, and against oxidative stress, such as thioredoxin (trxA), peroxidase (ohrA), and superoxide dismutase (sodA) (Helmann et al., 2001; Petersohn et al., 2001; Price et al., 2001; Nannapaneni et al., 2012) and regulators (e.g., CtsR and Spx) (Hecker et al., 2007).
Proteotoxic stress and other physiological demands on proteostasis may lead to insufficient protein folding capacity, resulting in accumulation of aberrant proteins. Reducing the rate of translation lowers the protein load, preventing further protein damage, and may help maintenance of proteostasis. Under proteotoxic stress conditions, such as heat or oxidative stress, B. subtilis downregulates transcription of translation-related genes, including ribosomal-protein encoding genes (rplD, rpsC, rplW, and rpsJ), and ribosomal RNA (rRNA) genes (Price et al., 2001; Leichert et al., 2003; Mostertz et al., 2004; Rochat et al., 2012; Schafer et al., 2019). In line with this notion, translation-related proteins have been found in protein aggregates in E. coli (Kwiatkowska et al., 2008; Dewachter et al., 2021). Moreover, the translation rate is reduced in E. coli cells containing a dysfunctional GroEL (Chapman et al., 2006), and protein folding is enhanced by slowing down translation rates in E. coli cells harboring mutant ribosomes (Siller et al., 2010).
Spx, apart from inducing transcription of stress-responsive genes, is also capable of repressing expression of genes for ribosomal proteins and rRNA (Nakano et al., 2003; Rochat et al., 2012; Schafer et al., 2019). Among the Spx downregulated genes are rpoA and rpoC, encoding for RNAP core subunits, and lepA, encoding for elongation factor 4 (EF4/LepA; Schafer et al., 2019), a paralog of the canonical elongation factor EF-G (Evans et al., 2008).
Downregulation of the translation machinery is also observed in cells lacking Spx (Schafer et al., 2019) suggesting that, at least under the stress conditions investigated (heat, oxidative, and cell wall stress), there are additional mechanisms to reduce translation in B. subtilis.
A downregulation of translation is frequently linked to the second messengers of nutrient starvation, ppGpp and pppGpp [collectively referred to as (p)ppGpp], which are synthesized by RelA (Rel in B. subtilis) and mediate the stringent response (Roghanian et al., 2021). Recently, a role of (p)ppGpp in slowing down the translation rate upon proteotoxic stress in B. subtilis has been suggested (Schafer et al., 2020).
McsB, which as mentioned before targets proteins for degradation by ClpCP, also has a potential role in regulating translation upon heat or oxidative stress, since it targets proteins related to translational control (Schmidt et al., 2014).
Inactivation of the B. subtilis methionine synthase MetE could also contribute to limit the translation rate by depleting the biosynthesis of the precursor of the initiation codon formyl-Met (Chi et al., 2011). Indeed, oxidative stress has been shown to inactivate E. coli MetE (Hondorp and Matthews, 2004), and oxidized MetE appears in the aggregate fraction of GroEL-defective mutants cells (Chapman et al., 2006). In response to the oxidative stress agents diamide or sodium hypochlorite, specific cysteine residues of B. subtilis MetE become either S-cysteinylated (Hochgrafe et al., 2007) or S-bacillithiolated, leading to its enzymatic inactivation (Chi et al., 2011). Moreover, MetE is among 108 identified S-thioallylated proteins caused by garlic sulfur compounds, and such compounds were found to induce the HrcA, CtsR, and Spx regulons (Chi et al., 2019).
B. subtilis is tolerant to drastic and rapid environmental changes by having a network of regulators and mechanisms controlling the synthesis, folding, post-translational modifications, sub-cellular localization and clearance of proteins to ensure proteostasis. Several of these processes are controlled differently in E. coli and B. subtilis, reflecting a long history of adaptations of the two model organisms to different niches.
Both authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Work in the laboratory of CW was supported by the Swedish Research Council Grant 2019-05578_3.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank L. Hederstedt for critical reading of the manuscript.
Acebron, S. P., Fernandez-Saiz, V., Taneva, S. G., Moro, F., and Muga, A. (2008). DnaJ recruits DnaK to protein aggregates. J. Biol. Chem. 283, 1381–1390. doi: 10.1074/jbc.M706189200
Agashe, V. R., Guha, S., Chang, H. C., Genevaux, P., Hayer-Hartl, M., Stemp, M., et al. (2004). Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell 117, 199–209. doi: 10.1016/s0092-8674(04)00299-5
Awad, W., Al-Eryani, Y., Ekstrom, S., Logan, D. T., and von Wachenfeldt, C. (2019). Structural basis for YjbH adaptor-mediated recognition of transcription factor Spx. Structure 27, 923–936.e6. doi: 10.1016/j.str.2019.03.009
Balchin, D., Hayer-Hartl, M., and Hartl, F. U. (2016). In vivo aspects of protein folding and quality control. Science 353:aac4354. doi: 10.1126/science.aac4354
Bardwell, J. C., and Craig, E. A. (1988). Ancient heat shock gene is dispensable. J. Bacteriol. 170, 2977–2983. doi: 10.1128/jb.170.7.2977-2983.1988
Battesti, A., and Gottesman, S. (2013). Roles of adaptor proteins in regulation of bacterial proteolysis. Curr. Opin. Microbiol. 16, 140–147. doi: 10.1016/j.mib.2013.01.002
Bradshaw, N., and Losick, R. (2015). Asymmetric division triggers cell-specific gene expression through coupled capture and stabilization of a phosphatase. eLife 4:e08145. doi: 10.7554/eLife.08145
Bukau, B., and Walker, G. C. (1989). Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J. Bacteriol. 171, 2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989
Bukau, B., and Walker, G. C. (1990). Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 9, 4027–4036. doi: 10.1002/j.1460-2075.1990.tb07624.x
Calloni, G., Chen, T., Schermann, S. M., Chang, H. C., Genevaux, P., Agostini, F., et al. (2012). DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 1, 251–264. doi: 10.1016/j.celrep.2011.12.007
Chan, C. M., Garg, S., Lin, A. A., and Zuber, P. (2012). Geobacillus thermodenitrificans YjbH recognizes the C-terminal end of Bacillus subtilis Spx to accelerate Spx proteolysis by ClpXP. Microbiology 158, 1268–1278. doi: 10.1099/mic.0.057661-0
Chapman, E., Farr, G. W., Usaite, R., Furtak, K., Fenton, W. A., Chaudhuri, T. K., et al. (2006). Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc. Natl. Acad. Sci. U.S.A. 103, 15800–15805. doi: 10.1073/pnas.0607534103
Cheng, J., North, B. J., Zhang, T., Dai, X., Tao, K., Guo, J., et al. (2018). The emerging roles of protein homeostasis-governing pathways in Alzheimer’s disease. Aging Cell 17:e12801. doi: 10.1111/acel.12801
Chi, B. K., Gronau, K., Mader, U., Hessling, B., Becher, D., and Antelmann, H. (2011). S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol. Cell Proteomics 10:M111.009506. doi: 10.1074/mcp.M111.009506
Chi, B. K., Huyen, N. T. T., Loi, V. V., Gruhlke, M. C. H., Schaffer, M., Mader, U., et al. (2019). The disulfide stress response and protein S-thioallylation caused by allicin and diallyl polysulfanes in Bacillus subtilis as revealed by transcriptomics and proteomics. Antioxidants 8:605. doi: 10.3390/antiox8120605
Clarke, D. J., Jacq, A., and Holland, I. B. (1996). A novel DnaJ-like protein in Escherichia coli inserts into the cytoplasmic membrane with a type III topology. Mol. Microbiol. 20, 1273–1286. doi: 10.1111/j.1365-2958.1996.tb02646.x
Commichau, F. M., Pietack, N., and Stulke, J. (2013). Essential genes in Bacillus subtilis: a re-evaluation after ten years. Mol. Biosyst. 9, 1068–1075. doi: 10.1039/c3mb25595f
Coquel, A. S., Jacob, J. P., Primet, M., Demarez, A., Dimiccoli, M., Julou, T., et al. (2013). Localization of protein aggregation in Escherichia coli is governed by diffusion and nucleoid macromolecular crowding effect. PLoS Comput. Biol. 9:e1003038. doi: 10.1371/journal.pcbi.1003038
Derre, I., Rapoport, G., and Msadek, T. (2000). The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37 degrees C. Mol. Microbiol. 38, 335–347. doi: 10.1046/j.1365-2958.2000.02124.x
Deuerling, E., Patzelt, H., Vorderwulbecke, S., Rauch, T., Kramer, G., Schaffitzel, E., et al. (2003). Trigger Factor and DnaK possess overlapping substrate pools and binding specificities. Mol. Microbiol. 47, 1317–1328. doi: 10.1046/j.1365-2958.2003.03370.x
Deuerling, E., Schulze-Specking, A., Tomoyasu, T., Mogk, A., and Bukau, B. (1999). Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400, 693–696. doi: 10.1038/23301
Dewachter, L., Bollen, C., Wilmaerts, D., Louwagie, E., Herpels, P., Matthay, P., et al. (2021). The dynamic transition of persistence toward the viable but nonculturable state during stationary phase is driven by protein aggregation. mBio 12:e0070321. doi: 10.1128/mBio.00703-21
Elsholz, A. K., Gerth, U., and Hecker, M. (2010). Regulation of CtsR activity in low GC. Gram+ bacteria. Adv. Microb. Physiol. 57, 119–144. doi: 10.1016/B978-0-12-381045-8.00003-5
Elsholz, A. K., Turgay, K., Michalik, S., Hessling, B., Gronau, K., Oertel, D., et al. (2012). Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 109, 7451–7456. doi: 10.1073/pnas.1117483109
Elsholz, A. K. W., Birk, M. S., Charpentier, E., and Turgay, K. (2017). Functional diversity of AAA+ protease complexes in Bacillus subtilis. Front. Mol. Biosci. 4:44. doi: 10.3389/fmolb.2017.00044
Engman, J., and von Wachenfeldt, C. (2015). Regulated protein aggregation: a mechanism to control the activity of the ClpXP adaptor protein YjbH. Mol. Microbiol. 95, 51–63. doi: 10.1111/mmi.12842
Evans, R. N., Blaha, G., Bailey, S., and Steitz, T. A. (2008). The structure of LepA, the ribosomal back translocase. Proc. Natl. Acad. Sci. U.S.A. 105, 4673–4678. doi: 10.1073/pnas.0801308105
Fauvet, B., Finka, A., Castanie-Cornet, M. P., Cirinesi, A. M., Genevaux, P., Quadroni, M., et al. (2021). Bacterial Hsp90 facilitates the degradation of aggregation-prone Hsp70-Hsp40 substrates. Front. Mol. Biosci. 8:653073. doi: 10.3389/fmolb.2021.653073
Fayet, O., Ziegelhoffer, T., and Georgopoulos, C. (1989). The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J. Bacteriol. 171, 1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989
Fei, X., Bell, T. A., Barkow, S. R., Baker, T. A., and Sauer, R. T. (2020). Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates. eLife 9:e61496. doi: 10.7554/eLife.61496
Ferbitz, L., Maier, T., Patzelt, H., Bukau, B., Deuerling, E., and Ban, N. (2004). Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431, 590–596. doi: 10.1038/nature02899
Fuhrmann, J., Schmidt, A., Spiess, S., Lehner, A., Turgay, K., Mechtler, K., et al. (2009). McsB is a protein arginine kinase that phosphorylates and inhibits the heat-shock regulator CtsR. Science 324, 1323–1327. doi: 10.1126/science.1170088
Gamer, J., Bujard, H., and Bukau, B. (1992). Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor sigma 32. Cell 69, 833–842. doi: 10.1016/0092-8674(92)90294-m
Garg, S. K., Kommineni, S., Henslee, L., Zhang, Y., and Zuber, P. (2009). The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J. Bacteriol. 191, 1268–1277. doi: 10.1128/JB.01289-08
Genest, O., Hoskins, J. R., Camberg, J. L., Doyle, S. M., and Wickner, S. (2011). Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proc. Natl. Acad. Sci. U.S.A. 108, 8206–8211. doi: 10.1073/pnas.1104703108
Genest, O., Hoskins, J. R., Kravats, A. N., Doyle, S. M., and Wickner, S. (2015). Hsp70 and Hsp90 of E. coli directly interact for collaboration in protein remodeling. J. Mol. Biol. 427, 3877–3889. doi: 10.1016/j.jmb.2015.10.010
Genevaux, P., Keppel, F., Schwager, F., Langendijk-Genevaux, P. S., Hartl, F. U., and Georgopoulos, C. (2004). In vivo analysis of the overlapping functions of DnaK and trigger factor. EMBO Rep. 5, 195–200. doi: 10.1038/sj.embor.7400067
Gothel, S. F., Scholz, C., Schmid, F. X., and Marahiel, M. A. (1998). Cyclophilin and trigger factor from Bacillus subtilis catalyze in vitro protein folding and are necessary for viability under starvation conditions. Biochemistry 37, 13392–13399. doi: 10.1021/bi981253w
Gottesman, S., Roche, E., Zhou, Y., and Sauer, R. T. (1998). The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12, 1338–1347. doi: 10.1101/gad.12.9.1338
Govers, S. K., Dutre, P., and Aertsen, A. (2014). In vivo disassembly and reassembly of protein aggregates in Escherichia coli. J. Bacteriol. 196, 2325–2332. doi: 10.1128/JB.01549-14
Gupta, A., Lloyd-Price, J., Neeli-Venkata, R., Oliveira, S. M., and Ribeiro, A. S. (2014). In vivo kinetics of segregation and polar retention of MS2-GFP-RNA complexes in Escherichia coli. Biophys. J. 106, 1928–1937. doi: 10.1016/j.bpj.2014.03.035
Hajdusits, B., Suskiewicz, M. J., Hundt, N., Meinhart, A., Kurzbauer, R., Leodolter, J., et al. (2021). McsB forms a gated kinase chamber to mark aberrant bacterial proteins for degradation. eLife 10:e63505. doi: 10.7554/eLife.63505
Hantke, I., Schafer, H., Janczikowski, A., and Turgay, K. (2019). YocM a small heat shock protein can protect Bacillus subtilis cells during salt stress. Mol. Microbiol. 111, 423–440. doi: 10.1111/mmi.14164
Hecker, M., Pane-Farre, J., and Volker, U. (2007). SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61, 215–236. doi: 10.1146/annurev.micro.61.080706.093445
Helmann, J. D., Wu, M. F., Kobel, P. A., Gamo, F. J., Wilson, M., Morshedi, M. M., et al. (2001). Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183, 7318–7328. doi: 10.1128/JB.183.24.7318-7328.2001
Herman, C., Thevenet, D., Bouloc, P., Walker, G. C., and D’Ari, R. (1998). Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev. 12, 1348–1355. doi: 10.1101/gad.12.9.1348
Hochgrafe, F., Mostertz, J., Pother, D. C., Becher, D., Helmann, J. D., and Hecker, M. (2007). S-cysteinylation is a general mechanism for thiol protection of Bacillus subtilis proteins after oxidative stress. J. Biol. Chem. 282, 25981–25985. doi: 10.1074/jbc.C700105200
Hondorp, E. R., and Matthews, R. G. (2004). Oxidative stress inactivates cobalamin-independent methionine synthase (MetE) in Escherichia coli. PLoS Biol. 2:e336. doi: 10.1371/journal.pbio.0020336
Huemer, M., Mairpady Shambat, S., Bergada-Pijuan, J., Soderholm, S., Boumasmoud, M., Vulin, C., et al. (2021). Molecular reprogramming and phenotype switching in Staphylococcus aureus lead to high antibiotic persistence and affect therapy success. Proc. Natl. Acad. Sci. U.S.A. 118:e2014920118 doi: 10.1073/pnas.2014920118
Itoh, T., Matsuda, H., and Mori, H. (1999). Phylogenetic analysis of the third Hsp70 homolog in Escherichia coli; a novel member of the Hsc66 subfamily and its possible co-chaperone. DNA Res. 6, 299–305. doi: 10.1093/dnares/6.5.299
Jurgen, B., Hanschke, R., Sarvas, M., Hecker, M., and Schweder, T. (2001). Proteome and transcriptome based analysis of Bacillus subtilis cells overproducing an insoluble heterologous protein. Appl. Microbiol. Biotechnol. 55, 326–332. doi: 10.1007/s002530000531
Kain, J., He, G. G., and Losick, R. (2008). Polar localization and compartmentalization of ClpP proteases during growth and sporulation in Bacillus subtilis. J. Bacteriol. 190, 6749–6757. doi: 10.1128/JB.00589-08
Keiler, K. C., Waller, P. R., and Sauer, R. T. (1996). Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993. doi: 10.1126/science.271.5251.990
Khodaparast, L., Wu, G., Khodaparast, L., Schmidt, B. Z., Rousseau, F., and Schymkowitz, J. (2021). Bacterial protein homeostasis disruption as a therapeutic intervention. Front. Mol. Biosci. 8:681855. doi: 10.3389/fmolb.2021.681855
Kirstein, J., Dougan, D. A., Gerth, U., Hecker, M., and Turgay, K. (2007). The tyrosine kinase McsB is a regulated adaptor protein for ClpCP. EMBO J. 26, 2061–2070. doi: 10.1038/sj.emboj.7601655
Kirstein, J., Moliere, N., Dougan, D. A., and Turgay, K. (2009). Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat. Rev. Microbiol. 7, 589–599. doi: 10.1038/nrmicro2185
Kirstein, J., Strahl, H., Moliere, N., Hamoen, L. W., and Turgay, K. (2008). Localization of general and regulatory proteolysis in Bacillus subtilis cells. Mol. Microbiol. 70, 682–694. doi: 10.1111/j.1365-2958.2008.06438.x
Kirstein, J., Zuhlke, D., Gerth, U., Turgay, K., and Hecker, M. (2005). A tyrosine kinase and its activator control the activity of the CtsR heat shock repressor in B. subtilis. EMBO J. 24, 3435–3445. doi: 10.1038/sj.emboj.7600780
Kruger, E., Witt, E., Ohlmeier, S., Hanschke, R., and Hecker, M. (2000). The clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J. Bacteriol. 182, 3259–3265. doi: 10.1128/JB.182.11.3259-3265.2000
Kruger, E., Zuhlke, D., Witt, E., Ludwig, H., and Hecker, M. (2001). Clp-mediated proteolysis in Gram-positive bacteria is autoregulated by the stability of a repressor. EMBO J. 20, 852–863. doi: 10.1093/emboj/20.4.852
Kusukawa, N., Yura, T., Ueguchi, C., Akiyama, Y., and Ito, K. (1989). Effects of mutations in heat-shock genes groES and groEL on protein export in Escherichia coli. EMBO J. 8, 3517–3521. doi: 10.1002/j.1460-2075.1989.tb08517.x
Kwiatkowska, J., Matuszewska, E., Kuczynska-Wisnik, D., and Laskowska, E. (2008). Aggregation of Escherichia coli proteins during stationary phase depends on glucose and oxygen availability. Res. Microbiol. 159, 651–657. doi: 10.1016/j.resmic.2008.09.008
Lamour, V., Westblade, L. F., Campbell, E. A., and Darst, S. A. (2009). Crystal structure of the in vivo-assembled Bacillus subtilis Spx/RNA polymerase alpha subunit C-terminal domain complex. J. Struct. Biol. 168, 352–356. doi: 10.1016/j.jsb.2009.07.001
Landgraf, D., Okumus, B., Chien, P., Baker, T. A., and Paulsson, J. (2012). Segregation of molecules at cell division reveals native protein localization. Nat. Methods 9, 480–482. doi: 10.1038/nmeth.1955
Langer, T., Pfeifer, G., Martin, J., Baumeister, W., and Hartl, F. U. (1992). Chaperonin-mediated protein folding: GroES binds to one end of the GroEL cylinder, which accommodates the protein substrate within its central cavity. EMBO J. 11, 4757–4765. doi: 10.1002/j.1460-2075.1992.tb05581.x
Larsson, J. T., Rogstam, A., and von Wachenfeldt, C. (2007). YjbH is a novel negative effector of the disulphide stress regulator, Spx, in Bacillus subtilis. Mol. Microbiol. 66, 669–684. doi: 10.1111/j.1365-2958.2007.05949.x
Lee, H. C., and Bernstein, H. D. (2002). Trigger factor retards protein export in Escherichia coli. J. Biol. Chem. 277, 43527–43535. doi: 10.1074/jbc.M205950200
Leichert, L. I., Scharf, C., and Hecker, M. (2003). Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185, 1967–1975. doi: 10.1128/JB.185.6.1967-1975.2003
Lelivelt, M. J., and Kawula, T. H. (1995). Hsc66, an Hsp70 homolog in Escherichia coli, is induced by cold shock but not by heat shock. J. Bacteriol. 177, 4900–4907. doi: 10.1128/jb.177.17.4900-4907.1995
Leszczynska, D., Matuszewska, E., Kuczynska-Wisnik, D., Furmanek-Blaszk, B., and Laskowska, E. (2013). The formation of persister cells in stationary-phase cultures of Escherichia coli is associated with the aggregation of endogenous proteins. PLoS One 8:e54737. doi: 10.1371/journal.pone.0054737
Liberek, K., Galitski, T. P., Zylicz, M., and Georgopoulos, C. (1992). The DnaK chaperone modulates the heat shock response of Escherichia coli by binding to the sigma 32 transcription factor. Proc. Natl. Acad. Sci. U.S.A. 89, 3516–3520. doi: 10.1073/pnas.89.8.3516
Liberek, K., Marszalek, J., Ang, D., Georgopoulos, C., and Zylicz, M. (1991). Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. U.S.A. 88, 2874–2878. doi: 10.1073/pnas.88.7.2874
Lilge, L., Reder, A., Tippmann, F., Morgenroth, F., Grohmann, J., Becher, D., et al. (2020). The involvement of the McsB arginine kinase in Clp-dependent degradation of the MgsR regulator in Bacillus subtilis. Front. Microbiol. 11:900. doi: 10.3389/fmicb.2020.00900
Linde, D., Volkmer-Engert, R., Schreiber, S., and Müller, J. P. (2003). Interaction of the Bacillus subtilis chaperone CsaA with the secretory protein YvaY. FEMS Microbiol. Lett. 226, 93–100. doi: 10.1016/S0378-1097(03)00578-0
Lindner, A. B., Madden, R., Demarez, A., Stewart, E. J., and Taddei, F. (2008). Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. U.S.A. 105, 3076–3081. doi: 10.1073/pnas.0708931105
Lytvynenko, I., Paternoga, H., Thrun, A., Balke, A., Muller, T. A., Chiang, C. H., et al. (2019). Alanine tails signal proteolysis in bacterial ribosome-associated quality control. Cell 178, 76–90.e22. doi: 10.1016/j.cell.2019.05.002
Martinez-Hackert, E., and Hendrickson, W. A. (2009). Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell 138, 923–934. doi: 10.1016/j.cell.2009.07.044
Mayhew, M., da Silva, A. C., Martin, J., Erdjument-Bromage, H., Tempst, P., and Hartl, F. U. (1996). Protein folding in the central cavity of the GroEL-GroES chaperonin complex. Nature 379, 420–426. doi: 10.1038/379420a0
Meile, J. C., Wu, L. J., Ehrlich, S. D., Errington, J., and Noirot, P. (2006). Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory. Proteomics 6, 2135–2146. doi: 10.1002/pmic.200500512
Miethke, M., Hecker, M., and Gerth, U. (2006). Involvement of Bacillus subtilis ClpE in CtsR degradation and protein quality control. J. Bacteriol. 188, 4610–4619. doi: 10.1128/JB.00287-06
Mogk, A., Homuth, G., Scholz, C., Kim, L., Schmid, F. X., and Schumann, W. (1997). The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16, 4579–4590. doi: 10.1093/emboj/16.15.4579
Mogk, A., Huber, D., and Bukau, B. (2011). Integrating protein homeostasis strategies in prokaryotes. Cold Spring Harb. Perspect. Biol. 3:a004366. doi: 10.1101/cshperspect.a004366
Mogk, A., Tomoyasu, T., Goloubinoff, P., Rudiger, S., Roder, D., Langen, H., et al. (1999). Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 18, 6934–6949. doi: 10.1093/emboj/18.24.6934
Moliere, N., and Turgay, K. (2009). Chaperone-protease systems in regulation and protein quality control in Bacillus subtilis. Res. Microbiol. 160, 637–644. doi: 10.1016/j.resmic.2009.08.020
Moore, S. D., and Sauer, R. T. (2007). The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 76, 101–124. doi: 10.1146/annurev.biochem.75.103004.142733
Mortier, J., Tadesse, W., Govers, S. K., and Aertsen, A. (2019). Stress-induced protein aggregates shape population heterogeneity in bacteria. Curr. Genet. 65, 865–869. doi: 10.1007/s00294-019-00947-1
Mostertz, J., Scharf, C., Hecker, M., and Homuth, G. (2004). Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150, 497–512. doi: 10.1099/mic.0.26665-0
Mukherjee, S., Bree, A. C., Liu, J., Patrick, J. E., Chien, P., and Kearns, D. B. (2015). Adaptor-mediated Lon proteolysis restricts Bacillus subtilis hyperflagellation. Proc. Natl. Acad. Sci. U.S.A. 112, 250–255. doi: 10.1073/pnas.1417419112
Müller, J. P., Ozegowski, J., Vettermann, S., Swaving, J., Van Wely, K. H., and Driessen, A. J. (2000). Interaction of Bacillus subtilis CsaA with SecA and precursor proteins. Biochem. J. 348, 367–373. doi: 10.1042/bj3480367
Nakano, M. M., Lin, A., Zuber, C. S., Newberry, K. J., Brennan, R. G., and Zuber, P. (2010). Promoter recognition by a complex of Spx and the C-terminal domain of the RNA polymerase alpha subunit. PLoS One 5:e8664. doi: 10.1371/journal.pone.0008664
Nakano, S., Kuster-Schock, E., Grossman, A. D., and Zuber, P. (2003). Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 100, 13603–13608. doi: 10.1073/pnas.2235180100
Nannapaneni, P., Hertwig, F., Depke, M., Hecker, M., Mader, U., Volker, U., et al. (2012). Defining the structure of the general stress regulon of Bacillus subtilis using targeted microarray analysis and random forest classification. Microbiology 158, 696–707. doi: 10.1099/mic.0.055434-0
Neeli-Venkata, R., Martikainen, A., Gupta, A., Goncalves, N., Fonseca, J., and Ribeiro, A. S. (2016). Robustness of the process of nucleoid exclusion of protein aggregates in Escherichia coli. J. Bacteriol. 198, 898–906. doi: 10.1128/JB.00848-15
Newberry, K. J., Nakano, S., Zuber, P., and Brennan, R. G. (2005). Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator. Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 102, 15839–15844. doi: 10.1073/pnas.0506592102
Oh, E., Becker, A. H., Sandikci, A., Huber, D., Chaba, R., Gloge, F., et al. (2011). Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147, 1295–1308. doi: 10.1016/j.cell.2011.10.044
Olivares, A. O., Baker, T. A., and Sauer, R. T. (2016). Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat. Rev. Microbiol. 14, 33–44. doi: 10.1038/nrmicro.2015.4
Oliveira, S. M., Neeli-Venkata, R., Goncalves, N. S., Santinha, J. A., Martins, L., Tran, H., et al. (2016). Increased cytoplasm viscosity hampers aggregate polar segregation in Escherichia coli. Mol. Microbiol. 99, 686–699. doi: 10.1111/mmi.13257
Paek, K. H., and Walker, G. C. (1987). Escherichia coli dnaK null mutants are inviable at high temperature. J. Bacteriol. 169, 283–290. doi: 10.1128/jb.169.1.283-290.1987
Perales-Calvo, J., Giganti, D., Stirnemann, G., and Garcia-Manyes, S. (2018). The force-dependent mechanism of DnaK-mediated mechanical folding. Sci. Adv. 4:eaaq0243. doi: 10.1126/sciadv.aaq0243
Petersohn, A., Brigulla, M., Haas, S., Hoheisel, J. D., Volker, U., and Hecker, M. (2001). Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183, 5617–5631. doi: 10.1128/JB.183.19.5617-5631.2001
Powers, E. T., and Balch, W. E. (2013). Diversity in the origins of proteostasis networks-a driver for protein function in evolution. Nat. Rev. Mol. Cell Biol. 14, 237–248. doi: 10.1038/nrm3542
Powers, E. T., Morimoto, R. I., Dillin, A., Kelly, J. W., and Balch, W. E. (2009). Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78, 959–991. doi: 10.1146/annurev.biochem.052308.114844
Price, C. W., Fawcett, P., Ceremonie, H., Su, N., Murphy, C. K., and Youngman, P. (2001). Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41, 757–774. doi: 10.1046/j.1365-2958.2001.02534.x
Pu, Y., Li, Y., Jin, X., Tian, T., Ma, Q., Zhao, Z., et al. (2019). ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol. Cell 73, 143–156. doi: 10.1016/j.molcel.2018.10.022
Rebeaud, M. E., Mallik, S., Goloubinoff, P., and Tawfik, D. S. (2021). On the evolution of chaperones and cochaperones and the expansion of proteomes across the Tree of Life. Proc. Natl. Acad. Sci. U.S.A. 118:e2020885118. doi: 10.1073/pnas.2020885118
Reder, A., Hoper, D., Weinberg, C., Gerth, U., Fraunholz, M., and Hecker, M. (2008). The Spx paralogue MgsR (YqgZ) controls a subregulon within the general stress response of Bacillus subtilis. Mol. Microbiol. 69, 1104–1120. doi: 10.1111/j.1365-2958.2008.06332.x
Reyes, D. Y., and Yoshikawa, H. (2002). DnaK chaperone machine and trigger factor are only partially required for normal growth of Bacillus subtilis. Biosci. Biotechnol. Biochem. 66, 1583–1586. doi: 10.1271/bbb.66.1583
Reyes, D. Y., and Zuber, P. (2008). Activation of transcription initiation by Spx: formation of transcription complex and identification of a Cis-acting element required for transcriptional activation. Mol. Microbiol. 69, 765–779. doi: 10.1111/j.1365-2958.2008.06330.x
Richter, K., Haslbeck, M., and Buchner, J. (2010). The heat shock response: life on the verge of death. Mol. Cell 40, 253–266. doi: 10.1016/j.molcel.2010.10.006
Rochat, T., Nicolas, P., Delumeau, O., Rabatinova, A., Korelusova, J., Leduc, A., et al. (2012). Genome-wide identification of genes directly regulated by the pleiotropic transcription factor Spx in Bacillus subtilis. Nucleic Acids Res. 40, 9571–9583. doi: 10.1093/nar/gks755
Roghanian, M., Van Nerom, K., Takada, H., Caballero-Montes, J., Tamman, H., Kudrin, P., et al. (2021). (p)ppGpp controls stringent factors by exploiting antagonistic allosteric coupling between catalytic domains. Mol. Cell 81, 3310–3322.e6. doi: 10.1016/j.molcel.2021.07.026
Rojas-Tapias, D. F., and Helmann, J. D. (2018). Induction of the Spx regulon by cell wall stress reveals novel regulatory mechanisms in Bacillus subtilis. Mol. Microbiol. 107, 659–674. doi: 10.1111/mmi.13906
Rojas-Tapias, D. F., and Helmann, J. D. (2019b). Roles and regulation of Spx family transcription factors in Bacillus subtilis and related species. Adv. Microb. Physiol. 75, 279–323. doi: 10.1016/bs.ampbs.2019.05.003
Rojas-Tapias, D. F., and Helmann, J. D. (2019a). Identification of novel Spx regulatory pathways in Bacillus subtilis uncovers a close relationship between the CtsR and Spx regulons. J. Bacteriol. 201, e00151–19. doi: 10.1128/JB.00151-19
Rokney, A., Shagan, M., Kessel, M., Smith, Y., Rosenshine, I., and Oppenheim, A. B. (2009). E. coli transports aggregated proteins to the poles by a specific and energy-dependent process. J. Mol. Biol. 392, 589–601. doi: 10.1016/j.jmb.2009.07.009
Ross, C. A., and Poirier, M. A. (2004). Protein aggregation and neurodegenerative disease. Nat. Med. 10(Suppl.), S10–S17. doi: 10.1038/nm1066
Rudiger, S., Buchberger, A., and Bukau, B. (1997). Interaction of Hsp70 chaperones with substrates. Nat. Struct. Biol. 4, 342–349. doi: 10.1038/nsb0597-342
Runde, S., Moliere, N., Heinz, A., Maisonneuve, E., Janczikowski, A., Elsholz, A. K., et al. (2014). The role of thiol oxidative stress response in heat-induced protein aggregate formation during thermotolerance in Bacillus subtilis. Mol. Microbiol. 91, 1036–1052. doi: 10.1111/mmi.12521
Saio, T., Guan, X., Rossi, P., Economou, A., and Kalodimos, C. G. (2014). Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science 344:1250494. doi: 10.1126/science.1250494
Saio, T., Kawagoe, S., Ishimori, K., and Kalodimos, C. G. (2018). Oligomerization of a molecular chaperone modulates its activity. eLife 7:e35731. doi: 10.7554/eLife.35731
Sauer, R. T., and Baker, T. A. (2011). AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612. doi: 10.1146/annurev-biochem-060408-172623
Sauer, R. T., Bolon, D. N., Burton, B. M., Burton, R. E., Flynn, J. M., Grant, R. A., et al. (2004). Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell 119, 9–18. doi: 10.1016/j.cell.2004.09.020
Schafer, H., Beckert, B., Frese, C. K., Steinchen, W., Nuss, A. M., Beckstette, M., et al. (2020). The alarmones (p)ppGpp are part of the heat shock response of Bacillus subtilis. PLoS Genet. 16:e1008275. doi: 10.1371/journal.pgen.1008275
Schafer, H., Heinz, A., Sudzinova, P., Voss, M., Hantke, I., Krasny, L., et al. (2019). Spx, the central regulator of the heat and oxidative stress response in B. subtilis, can repress transcription of translation-related genes. Mol. Microbiol. 111, 514–533. doi: 10.1111/mmi.14171
Schafer, H., and Turgay, K. (2019). Spx, a versatile regulator of the Bacillus subtilis stress response. Curr. Genet. 65, 871–876. doi: 10.1007/s00294-019-00950-6
Schmidt, A., Trentini, D. B., Spiess, S., Fuhrmann, J., Ammerer, G., Mechtler, K., et al. (2014). Quantitative phosphoproteomics reveals the role of protein arginine phosphorylation in the bacterial stress response. Mol. Cell Proteomics 13, 537–550. doi: 10.1074/mcp.M113.032292
Schopf, F. H., Biebl, M. M., and Buchner, J. (2017). The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 18, 345–360. doi: 10.1038/nrm.2017.20
Schramm, F. D., Schroeder, K., and Jonas, K. (2020). Protein aggregation in bacteria. FEMS Microbiol. Rev. 44, 54–72. doi: 10.1093/femsre/fuz026
Schulz, A., Tzschaschel, B., and Schumann, W. (1995). Isolation and analysis of mutants of the dnaK operon of Bacillus subtilis. Mol. Microbiol. 15, 421–429. doi: 10.1111/j.1365-2958.1995.tb02256.x
Schumann, W. (2003). The Bacillus subtilis heat shock stimulon. Cell Stress Chaperones 8, 207–217. doi: 10.1379/1466-12682003008<0207:tbshss<2.0.co;2
Schumann, W. (2016). Regulation of bacterial heat shock stimulons. Cell Stress Chaperones 21, 959–968. doi: 10.1007/s12192-016-0727-z
Seaton, B. L., and Vickery, L. E. (1994). A gene encoding a DnaK/hsp70 homolog in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 91, 2066–2070. doi: 10.1073/pnas.91.6.2066
Seydlova, G., Halada, P., Fiser, R., Toman, O., Ulrych, A., and Svobodova, J. (2012). DnaK and GroEL chaperones are recruited to the Bacillus subtilis membrane after short-term ethanol stress. J. Appl. Microbiol. 112, 765–774. doi: 10.1111/j.1365-2672.2012.05238.x
Shi, J., Li, F., Wen, A., Yu, L., Wang, L., Wang, F., et al. (2021). Structural basis of transcription activation by the global regulator Spx. Nucleic Acids Res. 49, 10756–10769. doi: 10.1093/nar/gkab790
Shi, L., Ravikumar, V., Derouiche, A., Macek, B., and Mijakovic, I. (2016). Tyrosine 601 of Bacillus subtilis DnaK undergoes phosphorylation and is crucial for chaperone activity and heat shock survival. Front. Microbiol. 7:533. doi: 10.3389/fmicb.2016.00533
Siller, E., DeZwaan, D. C., Anderson, J. F., Freeman, B. C., and Barral, J. M. (2010). Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J. Mol. Biol. 396, 1310–1318. doi: 10.1016/j.jmb.2009.12.042
Simmons, L. A., Grossman, A. D., and Walker, G. C. (2008). Clp and Lon proteases occupy distinct subcellular positions in Bacillus subtilis. J. Bacteriol. 190, 6758–6768. doi: 10.1128/JB.00590-08
Singhal, K., Vreede, J., Mashaghi, A., Tans, S. J., and Bolhuis, P. G. (2015). The trigger factor chaperone encapsulates and stabilizes partial folds of substrate proteins. PLoS Comput. Biol. 11:e1004444. doi: 10.1371/journal.pcbi.1004444
Stannek, L., Gunka, K., Care, R. A., Gerth, U., and Commichau, F. M. (2014). Factors that mediate and prevent degradation of the inactive and unstable GudB protein in Bacillus subtilis. Front. Microbiol. 5:758. doi: 10.3389/fmicb.2014.00758
Stoller, G., Rucknagel, K. P., Nierhaus, K. H., Schmid, F. X., Fischer, G., and Rahfeld, J. U. (1995). A ribosome-associated peptidyl-prolyl cis/trans isomerase identified as the trigger factor. EMBO J. 14, 4939–4948.
Szabo, A., Langer, T., Schroder, H., Flanagan, J., Bukau, B., and Hartl, F. U. (1994). The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK. DnaJ, and GrpE. Proc. Natl. Acad. Sci. U.S.A. 91, 10345–10349. doi: 10.1073/pnas.91.22.10345
Teter, S. A., Houry, W. A., Ang, D., Tradler, T., Rockabrand, D., Fischer, G., et al. (1999). Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97, 755–765. doi: 10.1016/s0092-8674(00)80787-4
Thi Nguyen, H. B., and Schumann, W. (2012). The sporulation control gene spo0M of Bacillus subtilis is a target of the FtsH metalloprotease. Res. Microbiol. 163, 114–118. doi: 10.1016/j.resmic.2011.10.011
Thomas, J. G., and Baneyx, F. (1998). Roles of the Escherichia coli small heat shock proteins IbpA and IbpB in thermal stress management: comparison with ClpA. ClpB, and HtpG In vivo. J. Bacteriol. 180, 5165–5172. doi: 10.1128/JB.180.19.5165-5172.1998
Trentini, D. B., Suskiewicz, M. J., Heuck, A., Kurzbauer, R., Deszcz, L., Mechtler, K., et al. (2016). Arginine phosphorylation marks proteins for degradation by a Clp protease. Nature 539, 48–53. doi: 10.1038/nature20122
Ueguchi, C., Kakeda, M., Yamada, H., and Mizuno, T. (1994). An analogue of the DnaJ molecular chaperone in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 91, 1054–1058. doi: 10.1073/pnas.91.3.1054
Ullers, R. S., Ang, D., Schwager, F., Georgopoulos, C., and Genevaux, P. (2007). Trigger Factor can antagonize both SecB and DnaK/DnaJ chaperone functions in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 104, 3101–3106. doi: 10.1073/pnas.0608232104
Versteeg, S., Mogk, A., and Schumann, W. (1999). The Bacillus subtilis htpG gene is not involved in thermal stress management. Mol. Gen. Genet. 261, 582–588. doi: 10.1007/s004380051004
Wickner, S., Nguyen, T. L., and Genest, O. (2021). The bacterial Hsp90 chaperone: cellular functions and mechanism of action. Annu. Rev. Microbiol. 75, 719–739. doi: 10.1146/annurev-micro-032421-035644
Wild, J., Altman, E., Yura, T., and Gross, C. A. (1992). DnaK and DnaJ heat shock proteins participate in protein export in Escherichia coli. Genes Dev. 6, 1165–1172. doi: 10.1101/gad.6.7.1165
Wild, J., Rossmeissl, P., Walter, W. A., and Gross, C. A. (1996). Involvement of the DnaK-DnaJ-GrpE chaperone team in protein secretion in Escherichia coli. J. Bacteriol. 178, 3608–3613. doi: 10.1128/jb.178.12.3608-3613.1996
Wild, J., Walter, W. A., Gross, C. A., and Altman, E. (1993). Accumulation of secretory protein precursors in Escherichia coli induces the heat shock response. J. Bacteriol. 175, 3992–3997. doi: 10.1128/jb.175.13.3992-3997.1993
Winkler, J., Seybert, A., Konig, L., Pruggnaller, S., Haselmann, U., Sourjik, V., et al. (2010). Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 29, 910–923. doi: 10.1038/emboj.2009.412
Yoshimune, K., Yoshimura, T., and Esaki, N. (1998). Hsc62, a new DnaK homologue of Escherichia coli. Biochem. Biophys. Res. Commun. 250, 115–118. doi: 10.1006/bbrc.1998.9255
Yoshimune, K., Yoshimura, T., Nakayama, T., Nishino, T., and Esaki, N. (2002). Hsc62, Hsc56, and GrpE, the third Hsp70 chaperone system of Escherichia coli. Biochem. Biophys. Res. Commun. 293, 1389–1395. doi: 10.1016/S0006-291X(02)00403-5
Yu, J., Liu, Y., Yin, H., and Chang, Z. (2019). Regrowth-delay body as a bacterial subcellular structure marking multidrug-tolerant persisters. Cell Discov. 5:8. doi: 10.1038/s41421-019-0080-3
Zarnt, T., Tradler, T., Stoller, G., Scholz, C., Schmid, F. X., and Fischer, G. (1997). Modular structure of the trigger factor required for high activity in protein folding. J. Mol. Biol. 271, 827–837. doi: 10.1006/jmbi.1997.1206
Zhou, B., Semanjski, M., Orlovetskie, N., Bhattacharya, S., A.lon, S., Argaman, L., et al. (2019). Arginine dephosphorylation propels spore germination in bacteria. Proc. Natl. Acad. Sci. U.S.A. 116, 14228–14237. doi: 10.1073/pnas.1817742116
Zhu, X., Zhao, X., Burkholder, W. F., Gragerov, A., Ogata, C. M., Gottesman, M. E., et al. (1996). Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614. doi: 10.1126/science.272.5268.1606
Keywords: chaperone, protease, degradation tags, protein quality control, protein aggregation, proteotoxic stress
Citation: Matavacas J and von Wachenfeldt C (2022) Update on the Protein Homeostasis Network in Bacillus subtilis. Front. Microbiol. 13:865141. doi: 10.3389/fmicb.2022.865141
Received: 29 January 2022; Accepted: 15 February 2022;
Published: 08 March 2022.
Edited by:
Katarzyna Potrykus, University of Gdańsk, PolandReviewed by:
Pierre Genevaux, FR3743 Centre de Biologie Intégrative (CBI), FranceCopyright © 2022 Matavacas and von Wachenfeldt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claes von Wachenfeldt, Y2xhZXMudm9uX3dhY2hlbmZlbGR0QGJpb2wubHUuc2U=, orcid.org/0000-0001-8950-3218
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.