- 1Department of Microbiology and Pathogen Biology, Air Force Medical University, Xi’an, China
- 2College of Life Sciences, Northwest University, Xi’an, China
- 3School of Life Sciences, Yan’an University, Yan’an, China
- 4Department of Pediatrics, Tangdu Hospital, Air Force Medical University, Xi’an, China
- 5Graduate School, Chang’an University, Xi’an, China
- 6Department of Immunology and Microbial Disease, Albany Medical College, Albany, NY, United States
Cyclic dimeric adenosine monophosphate (c-di-AMP) is a ubiquitous second messenger of bacteria involved in diverse physiological processes as well as host immune responses. MSMEG_2630 is a c-di-AMP phosphodiesterase (cnpB) of Mycobacterium smegmatis, which is homologous to Mycobacterium tuberculosis Rv2837c. In this study, cnpB-deleted (ΔcnpB), -complemented (ΔcnpB::C), and -overexpressed (ΔcnpB::O) strains of M. smegmatis were constructed to investigate the role of c-di-AMP in regulating mycobacterial physiology and immunogenicity. This study provides more precise evidence that elevated c-di-AMP level resulted in smaller colonies, shorter bacteria length, impaired growth, and inhibition of potassium transporter in M. smegmatis. This is the first study to report that elevated c-di-AMP level could inhibit biofilm formation and induce porphyrin accumulation in M. smegmatis by regulating associated gene expressions, which may have effects on drug resistance and virulence of mycobacterium. Moreover, the cnpB-deleted strain with an elevated c-di-AMP level could induce enhanced Th1 immune responses after M. tuberculosis infection. Further, the pathological changes and the bacteria burden in ΔcnpB group were comparable with the wild-type M. smegmatis group against M. tuberculosis venous infection in the mouse model. Our findings enhanced the understanding of the physiological role of c-di-AMP in mycobacterium, and M. smegmatis cnpB-deleted strain with elevated c-di-AMP level showed the potential for a vaccine against tuberculosis.
Introduction
Mycobacterium tuberculosis is the causative agent of tuberculosis (TB), which is a leading cause of infectious morbidity and mortality with 9.9 million new cases and 1.3–1.5 million deaths in 2020 (WHO, 2021). Currently, the TB burden remains high, mainly due to TB/HIV co-infection and the emergence of drug-resistant M. tuberculosis strains. Live-attenuated Bacille Calmette-Gue’rin (BCG) is the only licensed vaccine against TB. However, the efficiency of BCG varies greatly in protecting against adult pulmonary TB (Mangtani et al., 2014). Deeper insight into the mechanisms regulating physiological and immunological properties of M. tuberculosis is essential for the development of therapeutics and novel or alternative vaccines against TB.
Cyclic di-adenosine monophosphate (c-di-AMP) is a ubiquitous second messenger produced by bacteria and archaea including Bacillus subtilis, Listeria monocytogenes, Streptococcus pneumoniae, and M. tuberculosis (Bai and Bai, 2020; Yin et al., 2020), which regulates diverse cellular processes including bacterial growth, biofilm formation, potassium transport, and virulence (Yin et al., 2020). In bacteria, the c-di-AMP level is controlled by diadenylate cyclase (DisA or DAC) and phosphodiesterase (PDE) (He et al., 2020; Stulke and Kruger, 2020; Zarrella and Bai, 2020). In M. tuberculosis, Rv3586 (DisA) is the sole diadenylate cyclase (Bai et al., 2012), and Rv2837c is a cyclic nucleotide phosphodiesterase (CnpB) for c-di-AMP hydrolysis (Yang et al., 2014). Deletion of cnpB or overexpression of disA could increase c-di-AMP level in M. tuberculosis (Yang et al., 2014; He et al., 2016; Dey et al., 2017), BCG (Zhang et al., 2018; Ning et al., 2019), and M. smegmatis (Tang et al., 2015). It has been proved that c-di-AMP regulates bacterial length in M. tuberculosis and BCG (Yang et al., 2014; Ning et al., 2019), colony morphology in M. smegmatis (Zhang et al., 2013; Tang et al., 2015), potassium transport in S. pneumoniae (Bai et al., 2014), and virulence of M. tuberculosis (Yang et al., 2014; Dey et al., 2017). Furthermore, CnpB degrades nanoRNAs (RNA oligos of ≤ 5 nucleotides) (Postic et al., 2012) and even hydrolyzes c-di-GMP at a lower rate than it dose on c-di-AMP (He et al., 2016). In fact, more phenotypes including those mentioned above have been found in studies of other bacteria such as biofilm, envelope stress, antibiotic resistance, osmoregulation, and virulence (Corrigan et al., 2011; Whiteley et al., 2017; Hu et al., 2020). Thus, it is deduced that more phenotypes controlled by c-di-AMP still need to be discovered in mycobacteria.
Now, c-di-AMP has been considered a key mycobacterial pathogen-associated molecular pattern (PAMP) inducing host innate immune responses, including type I IFN response and autophagy (Woodward et al., 2010; Dey et al., 2015; Devaux et al., 2018; Ning et al., 2019; Singh et al., 2022). Additionally, c-di-AMP as an adjuvant enhanced antigen-induced Th1/Th2/Th17 pattern responses (Ebensen et al., 2011; Ning et al., 2021). It has been noted that cnpB-deleted strain of M. tuberculosis triggered a potent type I IFN response, showing the reduction of bacillary burden in the mouse model, implying virulence attenuation in M. tuberculosis (Yang et al., 2014; Dey et al., 2017). The adaptive immune responses play a major role in the elimination of intracellular mycobacteria. Previous studies showed that disA-overexpressing BCG with extra c-di-AMP as an endogenous adjuvant could induce enhanced immune responses after M. tuberculosis infection in mice (Ning et al., 2019) and provided improved protection compared with BCG against aerosol infection of M. tuberculosis in guinea pig (Dey et al., 2020). Therefore, studying the adaptive immune response induced by recombinant mycobacterium with an elevated c-di-AMP level may provide new methods for the improvement of the tuberculosis vaccine.
Mycobacterium smegmatis is a fast-grown and non-pathogenic mycobacterium and is frequently used as a genetic surrogate to study the physiological processes and gene function of other extremely slow-growing species such as M. tuberculosis (Maarsingh et al., 2019; T et al., 2020). M. smegmatis is also a vaccine vector for a live vaccine against TB. Several recombinant M. smegmatis with different antigens could induce protective immune responses in mice against tuberculosis (Junqueira-Kipnis et al., 2013; Alves Da Silva et al., 2014; Wang et al., 2014; Liu et al., 2015). MSMEG_2630 is homologous with c-di-AMP phosphodiesterase cnpB of M. tuberculosis (Tang et al., 2015). In this study, cnpB-deleted (ΔcnpB), -complemented (ΔcnpB::C), and -overexpressed (ΔcnpB::O) strains of M. smegmatis were constructed to investigate the role of c-di-AMP in regulating mycobacterial physiology and immunogenicity. This study provided more evidence to clarify the role of c-di-AMP on bacterial physiology and immunogenicity and suggested the applicability of ΔcnpB for a vaccine against TB.
Materials and Methods
Bacterial Strains, Plasmids, Antibodies, and Animals
Mycobacterium smegmatis mc2-155 and M. tuberculosis H37Ra were obtained from the National Institutes for Food and Drug Control of China. Mycobacteria were grown in Middlebrook 7H9 broth (BD Difco) supplemented with 0.5% glycerol, 0.05% Tween 80, and 10% oleic acid-albumin-dextrose-catalase (OADC) (BD Difco) at 37°C. E. coli DH5α was grown in the LB medium at 37°C. When required, antibiotics were added as follows: zeocin 100 μg/ml, kanamycin 25 μg/ml, or hygromycin 150 μg/ml. Mycobacteria were plated on Middlebrook 7H10 agar (BD Difco) supplemented with 10% OADC for colony-forming unit (CFU) determination and were grown at 37°C. Recombinant purified proteins of CnpB (MSMEG_2630), DisA (MSMEG_6080), Ag85 (MSMEG_6398), and pyridoxamine 5′-phosphate oxidase (PdxH, MSMEG_5675) and their polyclonal antibody sera were prepared in our laboratory. The strains and plasmids used in this study are listed in Supplementary Table 1. Female BALB/c mice aged 6–8 weeks were purchased from the Animal Center of Air Force Medical University.
Construction and Complementation of Mutants
Mycobacterium smegmatis ΔcnpB mutant was constructed by homologous recombination using a previously reported approach (Van Kessel and Hatfull, 2007). Briefly, upstream and downstream homologous fragments of cnpB were amplified by polymerase chain reaction (PCR) with M. smegmatis mc2-155 genomic DNA as the template. The two flanking DNA fragments were sequentially subcloned into pMSG360zeo at EcoRV/HindIII and XbaI/KpnI restriction sites. The linear recombineering substrates flanked by homologous arms (loxp-zeo-loxp) were generated by EcoRV and KpnI digestion. The linear DNA substrates were electroporated into M. smegmatis cells that carried plasmid of pJV53. Then, cells were plated on 7H10 agar containing 100 μg/ml zeocin. The recombinant clone was verified by PCR and western blot analysis. The plasmid of pJV53 expressing gp60/gp61 was lost spontaneously. Primers used in this study are listed in Supplementary Table 2.
To complement the mutant, the cnpB open reading frame (ORF) was cloned into the single-copy expression vector of PW51 between KpnI/BamHI sites, and this plasmid of PW51-cnpB was transformed into ΔcnpB to complement the mutation (ΔcnpB::C). Meanwhile, cnpB ORF was cloned into the multiple-copy shuttle expression vector of PW54 at HindIII site. The recombinant plasmid of PW54-cnpB was transformed into ΔcnpB to generate cnpB-overexpressed strain (ΔcnpB::O). Transformants were verified by PCR and western blot analysis.
Western Blot Analysis
Mycobacterium smegmatis strains were grown in 15 ml of 7H9 broth supplemented with 0.5% glycerol, 0.05% Tween 80, and 10% OADC at 37°C until OD600 = 1.0. Bacteria were harvested and washed three times with phosphate-buffered saline (PBS). Pellets were resuspended in precooled PBS supplemented with proteinase inhibitor cocktail (Roche) and then lysed by MINI-BEAD BEATER (BioSpec). The lysates were centrifuged at 12,000 rpm for 20 min at 4°C, and the supernatants were collected. Besides, the bacterial culture supernatants were concentrated by vacuum freeze-drying for western blot analysis. The concentrations of proteins were measured by the bicinchoninic acid assay (BCA). Phenotypes of M. smegmatis strains were confirmed by western blot analysis with polyclonal antibodies as the primary antibody, as shown in Supplementary Table 1. PdxH was used as a loading control, and recombinant proteins were used as positive controls.
Determination of c-di-AMP Concentration by High-Performance Liquid Chromatography
Mycobacterium smegmatis strains were grown to OD600 = 1.0 in 7H9 broth with 10% OADC. Intracellular nucleotides were extracted from bacterial pellets according to the reported method (Burhenne and Kaever, 2013). Briefly, bacterial pellets were resuspended in extraction buffer (acetonitrile: methanol: water = 2:2:1), incubated on ice, boiled at 95°C for 10 min, and incubated on ice. The lysates were centrifuged for 10 min at 20,800 × g at 4°C. Supernatants were transferred to a new tube. The extraction procedure was repeated two times. The pooled extractions were lyophilized and dissolved in ddH2O. c-di-AMP levels were determined by high-performance liquid chromatography (HPLC) as previously reported (Ning et al., 2019). Chemically synthesized c-di-AMP (InvivoGen) was used as a standard substance to generate a standard curve. The concentrations of c-di-AMP in M. smegmatis strains (1 g wet weight) were displayed as nanomoles per liter (nM).
Determination of Bacterial Growth and Size
Mycobacterium smegmatis strains (2 × 105CFU) were grown in 15 ml 7H9 broth with 10% OADC or in Sauton’s medium in 50 ml tube under shaking (80 rpm and 180 rpm) or stationary conditions at 37°C. The growth of each strain was determined by measuring OD600 at indicated time points. The bacterial length was determined according to our previous study (Ning et al., 2019). Briefly, bacteria in the late-log phase were collected, washed with PBS, and resuspended in PBS. Bacterial suspensions were dropped on copper grids (200 mesh) for 10 min incubation, and excess bacterial suspension was removed by the filter paper. Bacteria were stained with phosphotungstic acid, and the bacteria were observed by transmission electron microscopy (TEM) (TECNAIG2 Spirit Biotwin). The bacterial length was measured by ImageJ software. The individual cells were manually traced in ImageJ and converted bacterial arbitrary units to actual size according to the scale of 5 μm. Bacterial cells in clumps were omitted during measurement.
Biofilm Formation and Biomass Quantification
Mycobacterium smegmatis strains were grown in a 7H9 medium with 10% OADC to OD600 = 0.8–1.0 at 37°C. Bacteria pellets were resuspended and washed two times by Sauton’s medium in the absence of Tween 80. The cultures were diluted by Sauton’s medium at OD600 = 0.2 and inoculated into 24-well (2 ml/well, for imaging) or 96-well (0.2 ml/well, for quantification) plates. The plates were incubated statically at 37°C for 3–5 days. Biomass was quantified by a crystal violet assay. Briefly, biofilms in the well were washed with ddH2O three times to remove the medium. The biofilms were dried at 37°C and incubated with 200 μl of 1% (w/v) crystal violet for 30 min and washed two times with ddH2O. About 200 μl of 95% ethanol was added to dissolve the biofilm. The OD570 was recorded by a microplate reader. Each experiment was performed in a set of three replicates.
Flow Cytometry Analysis of Long-Term Cultured Bacteria
Mycobacterium smegmatis strains were inoculated in a 7H9 medium and cultured for 3 days (stationary phase) at 80 rpm at 37°C. Subsequently, the cultures were transferred to a 37°C incubator for static culture avoiding light. After incubation at 14 and 60 weeks, bacteria were harvested and washed with PBS. Red autofluorescence of the bacterial cells was monitored by the flow cytometer (BeckmanCoulter EXPO32) in the FL3 channel (excitation 488 nm with a 610LP emission filter). Fresh bacteria (3 days) were used as the control. Flow cytometry data were analyzed using the FlowJo V10.
Absorption and Fluorescence Spectra of Long-Term Cultured Bacterial Supernatant
The 14- and 60-week supernatants of M. smegmatis strains were collected and filtered with a 0.22 μm filter membrane. Absorption spectra of supernatants were recorded by a multifunctional microplate reader (Tecan Spark) from 100 to 1,000 nm. Then, fluorescence measurements were taken with TECAN Spark at the excitation wavelength (λexcitation) of 400 nm.
RNA-seq and qRT-PCR Analysis
Total RNAs were extracted from M. smegmatis by TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. RNA quality was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies) and RNase-free agarose gel electrophoresis. mRNA was enriched by removing rRNA by Ribo-Zero Magnetic Kit (Epicenter) for library construction. RNA sequencing (RNA-seq) was performed using Illumina Novaseq6000 by Gene Denovo Biotechnology Co., Ltd., (Guangzhou, China). The expression levels of differential genes were further verified by real-time quantitative PCR (qRT-PCR). In total, 500 ng total RNA was reverse-transcribed using the HiScript III cDNA Synthesis Kit (Vazyme). The transcriptional levels of target genes were determined by qRT-PCR using SYRB Master Mix (Vazyme). Primers used in the qRT-PCR assay are listed in Supplementary Table 2. The relative transcriptional levels of genes were presented as 2–Δ Δ Ct with M. smegmatis sigA as a reference gene.
Mice Vaccination and Challenge With M. tuberculosis
Female BALB/c mice were anesthetized with an intraperitoneal injection of 50 mg/kg pentobarbital sodium. Mice were immunized with 107CFU M. smegmatis in 100 μl PBS for three times at 2-week intervals through a subcutaneous (s.c) route on the back. An equal volume of PBS was injected into control mice. Then, 4 weeks after immunization, mice were challenged intravenously (i.v) with 5 × 104 CFU M. tuberculosis H37Ra in 200 μl PBS.
ELISA
Whole blood was collected at 4 weeks post-immunization and 8 weeks post-infection. The whole blood was kept at 37°C for 30 min to collect the supernatant as serum. Plates were coated with M. smegmatis total protein extract of mycobacteria and CnpB. The serum was diluted at 1:200 with PBS. Antigen-specific IgG subclasses were determined by indirect enzyme-linked immunosorbent assay (ELISA). For cytokine measurement of IFN-γ, IL-2, and IL-10, splenocyte supernatants were collected after 72 h stimulation with M. smegmatis protein extract (25 μg/ml) as described in our previous study (Ning et al., 2019). Double antibody sandwich ELISA was performed according to the manufacturer’s instructions (Thermo Fisher Scientific).
Splenocytes Proliferation Assay
Spleen single-cell suspensions were prepared as we previously described (Ning et al., 2019). For cells proliferation assay, 1 × 106 cells were seeded in 96-well microplates and stimulated with CnpB (5 μg/ml), M. smegmatis protein extract (25 μg/ml), or M. tuberculosis H37Ra protein extract (25 μg/ml) as indicated. After stimulation at 37°C for 68 h with 5% CO2, MTS reagents (Promega) were added in each well and plates were incubated for another 4 h at 37°C. The absorbance at 490 nm was recorded using a microplate reader. The stimulated index was calculated as described in our previous study (Lu et al., 2018).
Histopathology and CFU Enumeration
For histopathology, the upper lobes of the left lungs were fixed in 10% buffered formalin for 24 h. Then, sections of 5 μm in thickness of tissues were cut into glass slides. Hematoxylin–eosin (H&E) staining for pathohistological analysis was performed by the Department of Histopathology (Air Force Medical University, China). Sections were observed under a light microscope (Olympus). At 8 weeks after M. tuberculosis infection, the lung and the spleen were removed and homogenized in sterile PBS. Homogenates were diluted and spread on 7H10 agar plates with 10% OADC enrichment. Plates were then incubated at 37°C for 3 weeks. The data are represented in Log10CFU (lgCFU).
Ethics Statement
Animal experiments were performed according to the approval and guidance of the Institutional Animal Ethics Center of the Air Force Medical University, and the approval number is 20190213.
Statistical Analysis
Statistical analysis was performed by GraphPad Prism 8.0 software. Statistical significance was determined by Student’s t-test or for multiple comparisons by ANOVA. P < 0.05 were considered statistically significant. Data are shown as means ± SEM or as individual data points, i.e., “*” denotes p < 0.05, “**” denotes p < 0.01, “***” denotes p < 0.001, and “****” denotes p < 0.0001.
Results
Deletion of CnpB Increases c-di-AMP Level in M. smegmatis
Mycobacterium smegmatis CnpB shared 73% identity (247/338 aa) with M. tuberculosis CnpB (Supplementary Figure 1A). In addition, M. smegmatis CnpB carries a highly conserved motif of Asp-His-His (DHH) (Supplementary Figure 1B), which is indispensable for the degradation of c-di-AMP (Yang et al., 2014; Tang et al., 2015). In this study, cnpB mutant (ΔcnpB) of M. smegmatis mc2155 was generated by homologous recombination through Che9c gp60/gp61 recombinase-assisted genome engineering (Figures 1A,B). Meanwhile, complemented (ΔcnpB::C) and overexpressed (ΔcnpB::O) strains of ΔcnpB were generated by different plasmids with the cnpB gene from M. smegmatis. These strains were verified by PCR and western blot analysis (Figures 1B,C). CnpB expression was restored in ΔcnpB::C and ΔcnpB::O (Figure 1C). High-performance liquid chromatography (HPLC) analysis showed that intracellular c-di-AMP concentration was elevated 1.5-folds in ΔcnpB than that of wild-type strain (p < 0.01) (Figure 1D). The level of c-di-AMP in ΔcnpB::C was lower than that of ΔcnpB but was comparable to that of wild-type (Figure 1D). However, the c-di-AMP level of ΔcnpB::O was about a quarter of that in ΔcnpB (Figure 1D), which could be explained that more c-di-AMP was hydrolyzed in M. smegmatis cnpB-overexpressing strain. These results showed that the c-di-AMP level of M. smegmatis could be controlled by the expression level of CnpB.
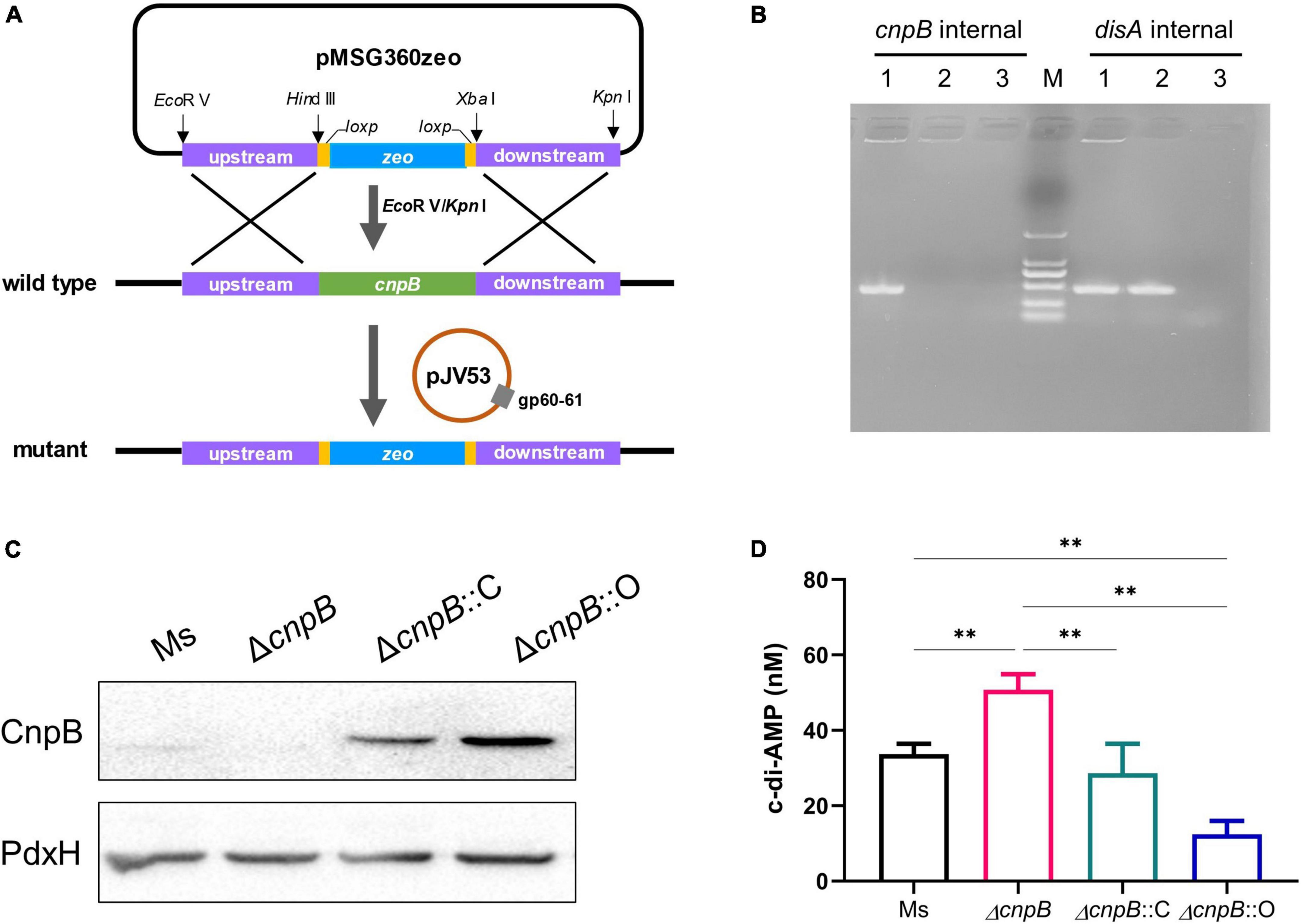
Figure 1. Deletion of cnpB led to elevated bacterial c-di-AMP level. (A) Schematic diagram showing the cnpB knockout strategy in M. smegmatis. (B) PCR verification for the deletion of cnpB in M. smegmatis. disA (MSMEG_6080) was used as a reference gene. The templates were wild-type strain of M. smegmatis (lane 1), cnpB mutant (lane 2), and negative control without template (lane 3), respectively. (C) CnpB expression was determined using western blot with bacterial lysates of wild-type (M. smegmatis, Ms), cnpB mutant (ΔcnpB), cnpB-complemented strain in ΔcnpB (ΔcnpB::C), and cnpB-overexpressed strain in ΔcnpB (ΔcnpB::O), respectively. Pyridoxamine 5′-phosphate oxidase (PdxH, MSMEG_5675) was used as a loading control. (D) The levels of c-di-AMP in each strain were assayed by HPLC. **p < 0.01.
c-di-AMP Controls the Colony Morphology of M. smegmatis
It has been found that increased c-di-AMP level by DisA overexpression in M. smegmatis led to abnormal phenotypes, including cell expansion, cell bulge phenotype, bacterial aggregation, and loss of motility (Zhang and He, 2013; Tang et al., 2015). It was reported that the morphology of CnpB-overexpressed strain was normal as wild-type M. smegmatis (Tang et al., 2015). In our study, the ΔcnpB colonies showed a smaller, smooth, and moist phenotype compared with that of the wild-type (Figure 2A). The colonies of ΔcnpB::O showed a similar appearance but has fewer wrinkles (Figure 2A). Colony diameter measurements showed that ΔcnpB formed smaller colonies with 28% reduction in diameter (p < 0.001) (Figures 2A,B). ΔcnpB::C just partially restored the colony size compared with ΔcnpB (p < 0.05) and smaller than wild-type (p < 0.001) (Figures 2A,B). However, ΔcnpB::O with the lowest c-di-AMP level exhibited a comparable colony size with wild-type, but other appearances seemed to be different from wild-type (Figures 2A,B).
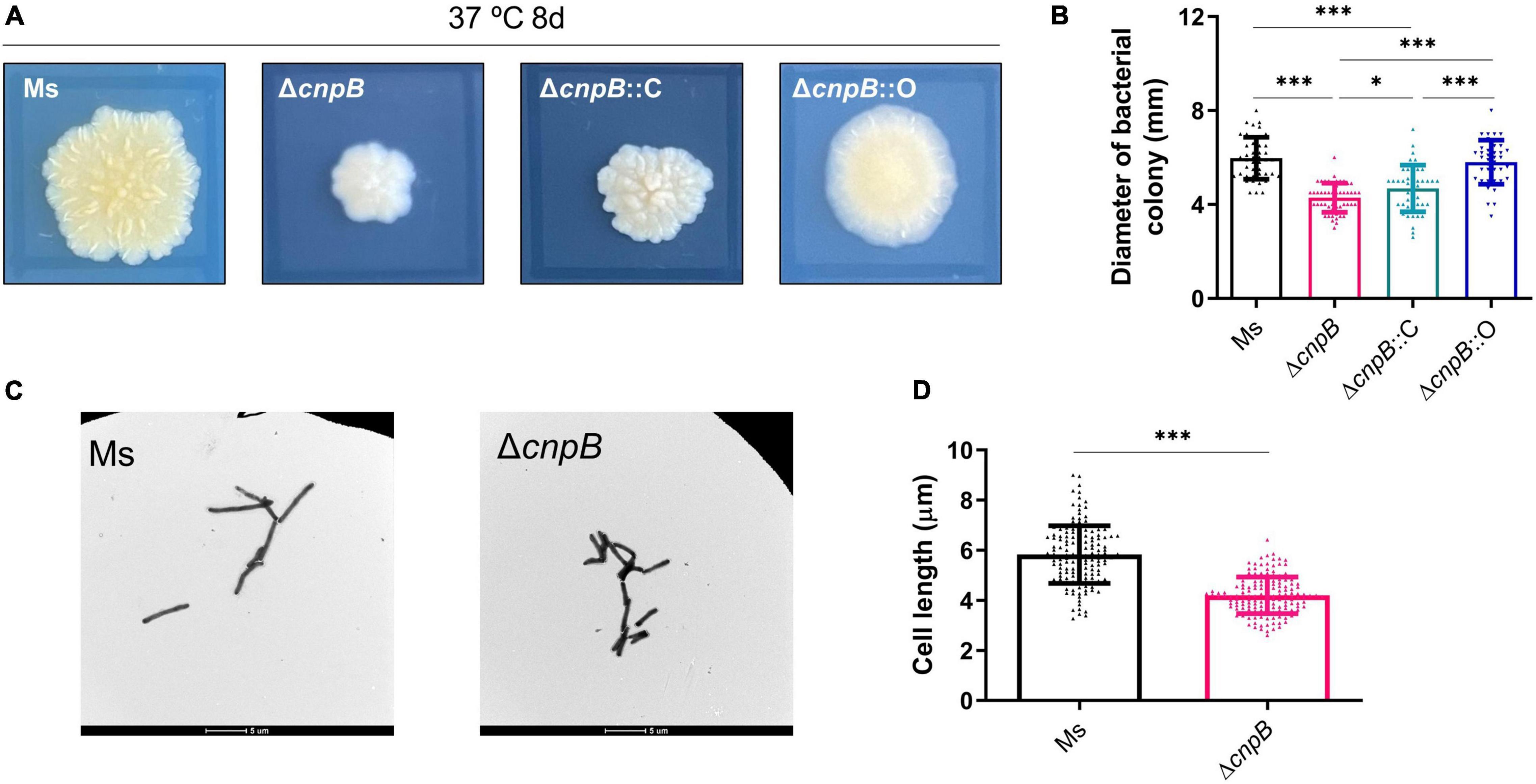
Figure 2. Determination of bacterial colony morphologies and bacterial lengths. (A) Colony morphologies on 7H10 + OADC plates after 8-day culture at 37°C. The black rectangle around each colony is 10 mm × 10 mm. (B) The measurement of colony diameter in panel (A) (40–50 colonies). (C) Observation of the indicated bacterial strains with a transmission electron microscope after culturing in 7H9 + OADC medium for 3 days. (D) The length of bacterial cells in panel (C) was measured (about 150 bacteria). *p < 0.05, ***p < 0.001.
Elevated c-di-AMP level in M. tuberculosis by CnpB deletion or in BCG by DisA overexpression led to a reduction of the single bacterial length (Yang et al., 2014; Ning et al., 2019). We speculated that the smaller bacterial colony is due to the smaller bacterial length. As expected, transmission electron microscope (TEM) observation showed that ΔcnpB (4.16 ± 1.73 μm) exhibited a bacterial length reduction of approximately 28% relative to the wild-type (5.83 ± 1.15 μm) M. smegmatis (Figures 2C,D). These results confirmed previous reports that the c-di-AMP level affected colony morphologies as well as bacterial length in mycobacteria and suggested that other phenotypes were controlled by c-di-AMP.
c-di-AMP Affects M. smegmatis Growth
In M. tuberculosis, deletion of c-di-AMP phosphodiesterase did not affect M. tuberculosis growth by OD600 (Yang et al., 2014), and a similar result was observed in L. monocytogenes (Witte et al., 2013). The growth of wild-type, ΔcnpB, ΔcnpB::C, and ΔcnpB::O was monitored in Middlebrook 7H9 medium and Sauton’s medium. Our data showed that the growth rate of ΔcnpB was slower in the exponential phase in 7H9 determined at both OD600 and CFU detections but reached similar levels at the stationary phase (Figures 3A–D). Meanwhile, ΔcnpB::C and ΔcnpB::O had similar growth rates as that of wild-type in 7H9 broth without the addition of OADC (Figures 3A,B). Similar results were observed when bacteria were grown in 7H9 + OADC (Figures 3C,D). Sauton’s medium is a minimal medium that does not contain exogenous proteins and is commonly used for the preparation of mycobacterial culture filtrates (Venkataswamy et al., 2012). In Sauton’s medium with or without OADC enrichment, ΔcnpB had a significantly reduced growth rate (Figures 3E–H). Both ΔcnpB::C and ΔcnpB::O showed a similar growth rates/CFUs to that of wild-type in 7H9 and Sauton’s medium with OADC (Figures 3E–H). The reduction of ΔcnpB growth was also found under slow shaking conditions (80 rpm) (Supplementary Figures 2A–H). Collectively, elevated c-di-AMP level impaired M. smegmatis growth, especially in media with less complex nutrients.
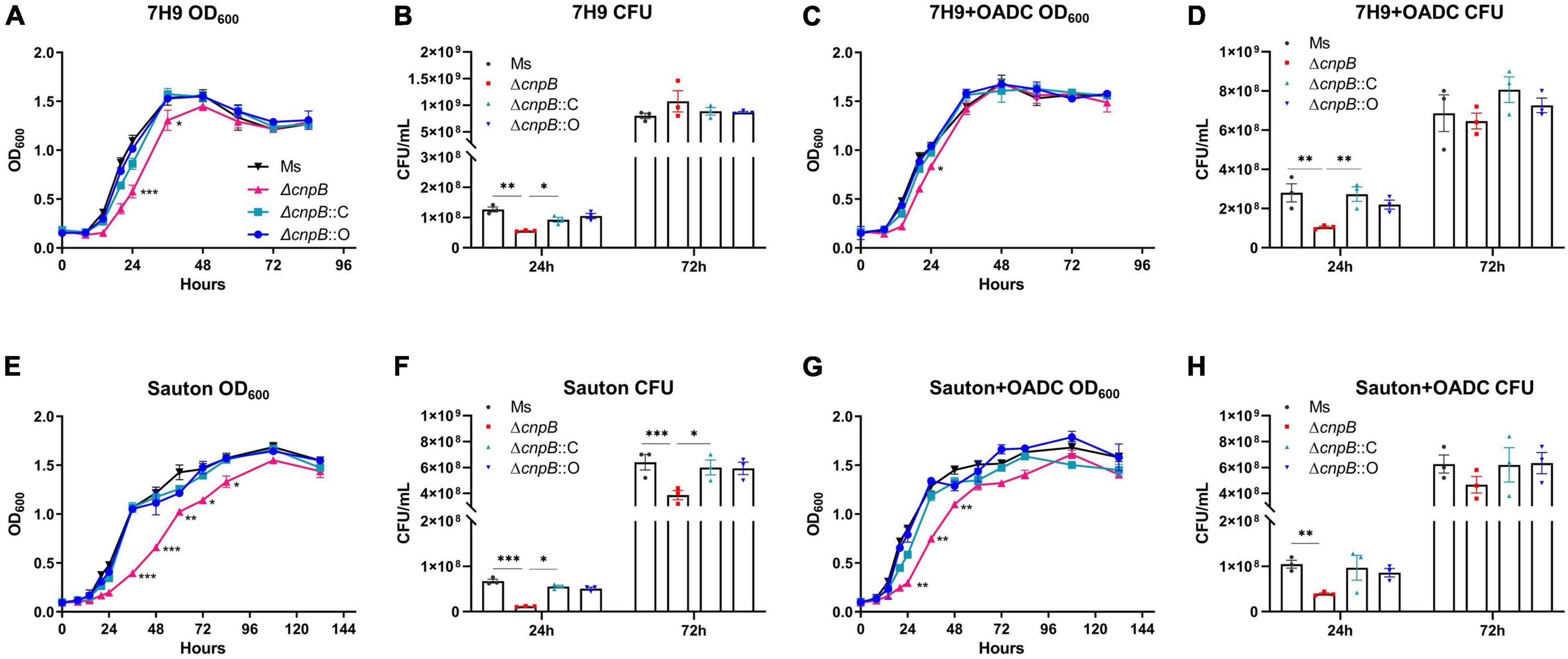
Figure 3. Detection of bacteria growth in liquid media (180 rpm). Each strain was inoculated at 2.5 × 106CFU/ml in media of 7H9 (A), 7H9 + OADC (C), Sauton’s (E), and Sauton’s + OADC (G). Bacteria were monitored at OD600 (A,C,E,G) and corresponding CFUs (B,D,F,H) were numbered at indicated time points, respectively. The growth curves were drawn with the results of three independent experiments with three duplicate cultures. *p < 0.05, **p < 0.01, ***p < 0.001.
Increasing c-di-AMP Level Causes a Reduction in Biofilm Formation in M. smegmatis
It has been reported that smooth or rough/wrinkled colony morphology was associated with the biofilm formation in several bacteria (Friedman and Kolter, 2004; Uhlich et al., 2006; Bharti et al., 2020). Further, the ability of ΔcnpB strain to form biofilm was determined in Sauton’s medium at 48, 72, and 120 h respectively, and biomass was quantified by the crystal violet assay. It was observed that ΔcnpB could not form the biofilm with rough surfaces like M. smegmatis and showed a smooth pellicle with low biomass quantifications by crystal violet assay in 48 and 72 h culture (Figures 4A,B). Though ΔcnpB::C showed a decrease in biofilm formation in 72 h culture, but more biomass quantification was detected in 120 h culture than ΔcnpB and was comparable to the wild-type strain (Figure 4C). To our surprise, ΔcnpB::O with decreased c-di-AMP level showed similar biofilm formation as ΔcnpB::C (Figures 4A–C). Nevertheless, our experimental results demonstrated that c-di-AMP could regulate the biofilm formation of mycobacterium, so the mechanisms related to biofilm regulation should be investigated further.
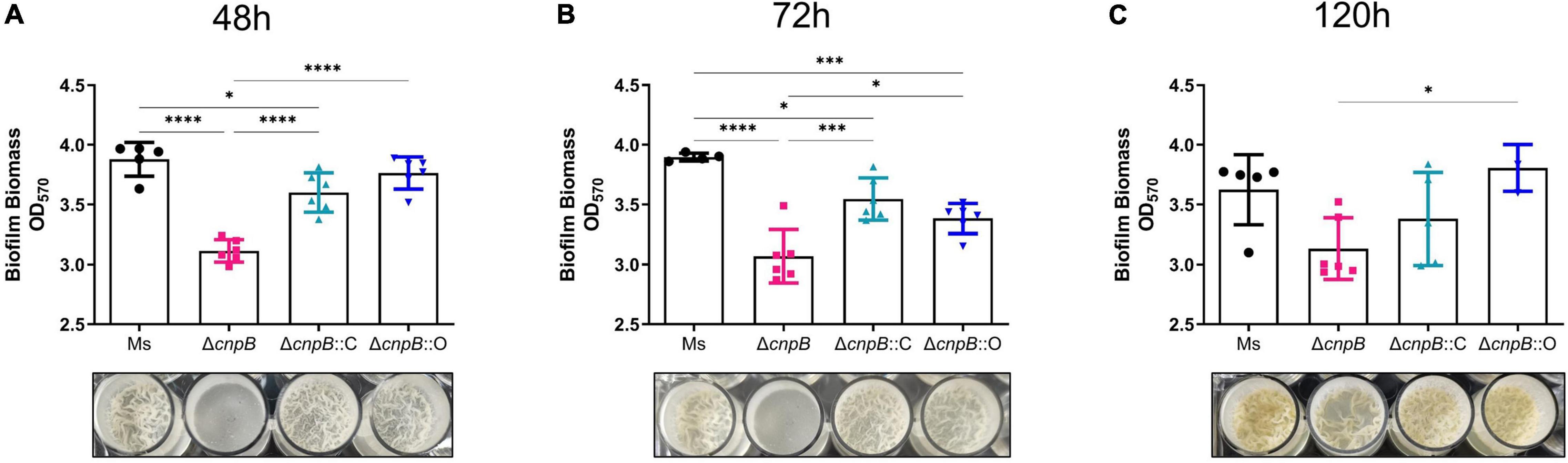
Figure 4. Biofilm formation and biomass quantification. Biomass was quantified by a crystal violet assay at 48 (A), 72 (B), and 120 h (C). The corresponding biofilm images for each strain are shown below each histogram. *p < 0.05, ***p < 0.001, ****p < 0.0001.
c-di-AMP Regulates Porphyrin Metabolism in the Long-Term Culture of M. smegmatis
Unexpectedly, we observed brown coloration in the culture of ΔcnpB, ΔcnpB::C, and ΔcnpB::O over 14 weeks, and supernatants of ΔcnpB::C and ΔcnpB::O showed lighter brown color than that of ΔcnpB (Figures 5A,B). The brown coloration of ΔcnpB::C and ΔcnpB::O supernatants gradually decreases with incubation time and were comparable to wild-type of M. smegmatis in 60-week culture (Figure 5A). After centrifuging, the brown coloration was retained in supernatant rather than bacterial pellets of M. smegmatis strains, suggesting that the unknown brown material substance was water soluble (Figure 5B). In addition, the brown pigment could not be removed by dialysis but be salted out from ΔcnpB supernatant by saturated (NH4)2SO4, which suggested that the brown pigment could bind to protein (Figure 5C and Supplementary Figure 3A). According to the flow cytometry experiments, ΔcnpB cells from long-term (14 and 60 weeks) rather than fresh (3 days) culture revealed red fluorescence (Figure 5D). Then, an absorbance peak at 400 nm was found in ΔcnpB supernatant of 14-week culture recorded by full-wavelength (100–1,000 nm) scanning (Figure 5E) and a lower absorbance peak in that of ΔcnpB::C and ΔcnpB::O (Supplementary Figure 3B). It was observed that M. smegmatis produced a dark brown fluorescent pigment in long-term culture, which was identified as porphyrin by mass spectrometry analysis and 1H-NMR spectra (Nikitushkin et al., 2016). Porphyrin has the typical fluorescence at 400 nm and fluorescence emission maxima at 625 and 675 nm (excitation wavelength, 400 nm) (Gouterman, 1959, 1961; Dolphin, 1978; Nikitushkin et al., 2016). Further, we found the characteristic fluorescence of ΔcnpB supernatant at 625 and 675 nm with an excitation wavelength of 400 nm (Figure 5F and Supplementary Figure 3C). Taken together, our results suggested that the elevated c-di-AMP level may increase porphyrin production in M. smegmatis.
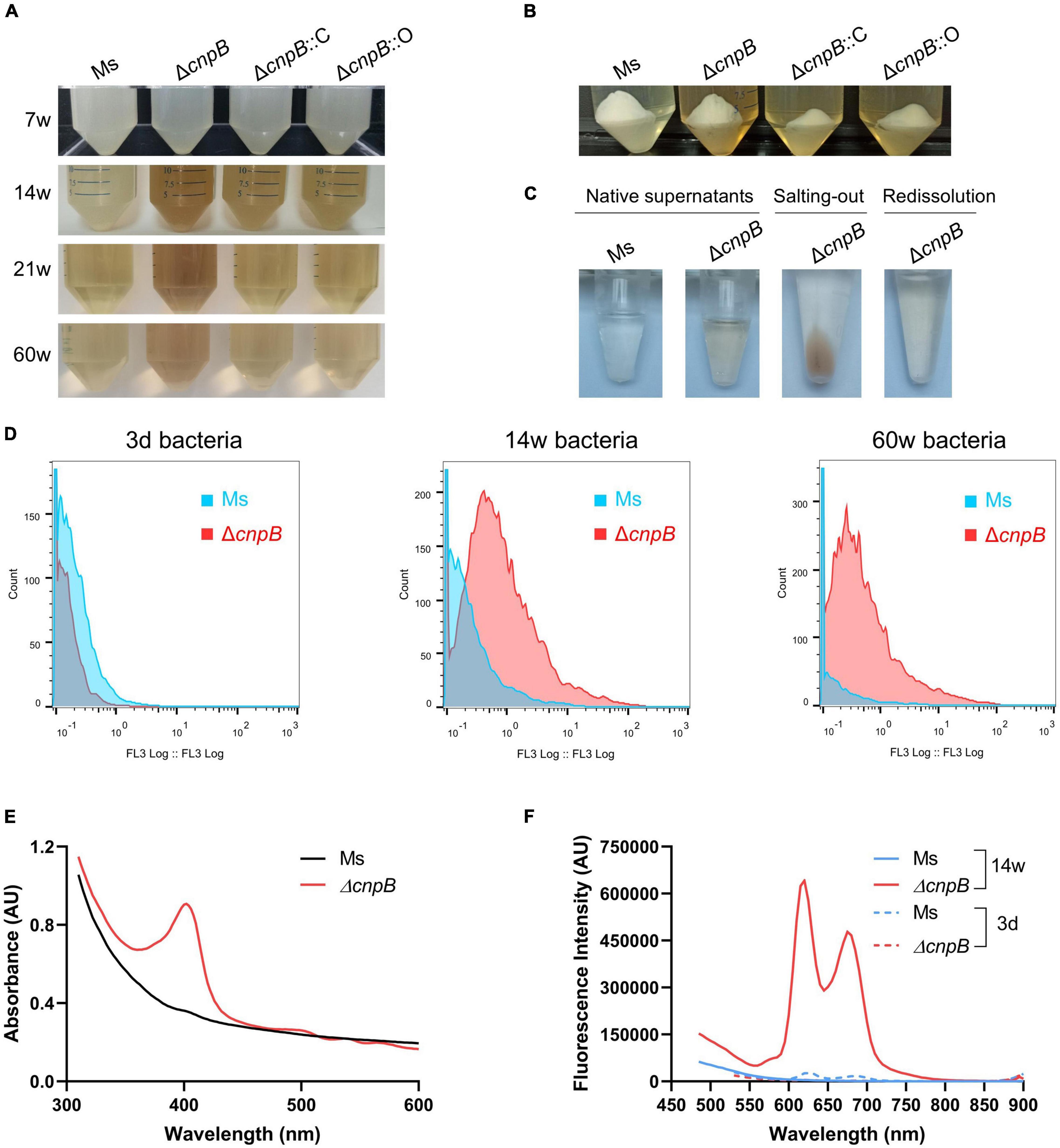
Figure 5. Pigment produced by ΔcnpB in long-term stationary culture. (A) The appearance of each strain was cultured statically for 7, 14, 21 and 60 weeks. Images were captured after gentle mixing. (B) At 14-week incubation, the bacterial pellets and supernatants were separated by centrifugation. (C) Observation of wild-type and ΔcnpB supernatants at 14 weeks. Next, ΔcnpB supernatant was salted-out by saturated ammonium sulfate [(NH4)2SO4] and protein was collected by centrifugation. Then, protein precipitation by salting-out was redissolved by PBS. (D) Autofluorescence of 3 days, 14 weeks, and 60 weeks of bacterial cells were monitored by the flow cytometer (FL3, excitation 488 nm with a 610LP emission filter). (E) Absorption spectra of 14-week supernatants were recorded by full-wavelength (100–1,000 nm) scanning. (F) Fluorescence measurements of 14-week supernatants were analyzed at the excitation wavelength (λexcitation) 400 nm.
c-di-AMP Modulates Broad Gene Expression of M. smegmatis Genes
To further explore gene expression profiles regulated by c-di-AMP, M. smegmatis and ΔcnpB pellets of late logarithmic phase cultures were harvested for RNA-seq analysis. Differentially expressed genes (DEGs) were analyzed at the cutoff of both false discovery rate (FDR) < 0.05 and | log2FC| > 1. Overall, it was found that a total of 2,828 genes were differentially expressed between ΔcnpB and wild-type, including 1,870 upregulated and 958 downregulated compared with wild-type strain. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment showed that several major metabolism-related genes were repressed in ΔcnpB, including nitrogen, alanine, aspartate, and glutamate, microbial metabolism in diverse environments, galactose metabolism, purine, carbon, and so on (Figure 6A). On the other side, elevated c-di-AMP in M. smegmatis induced expressions of genes involved in the biosynthesis of aminoacyl-tRNA, amino acids and so on (Figure 6B). We also noticed that gene expressions of porphyrin and chlorophyll metabolism were also induced in ΔcnpB (Figure 6B).
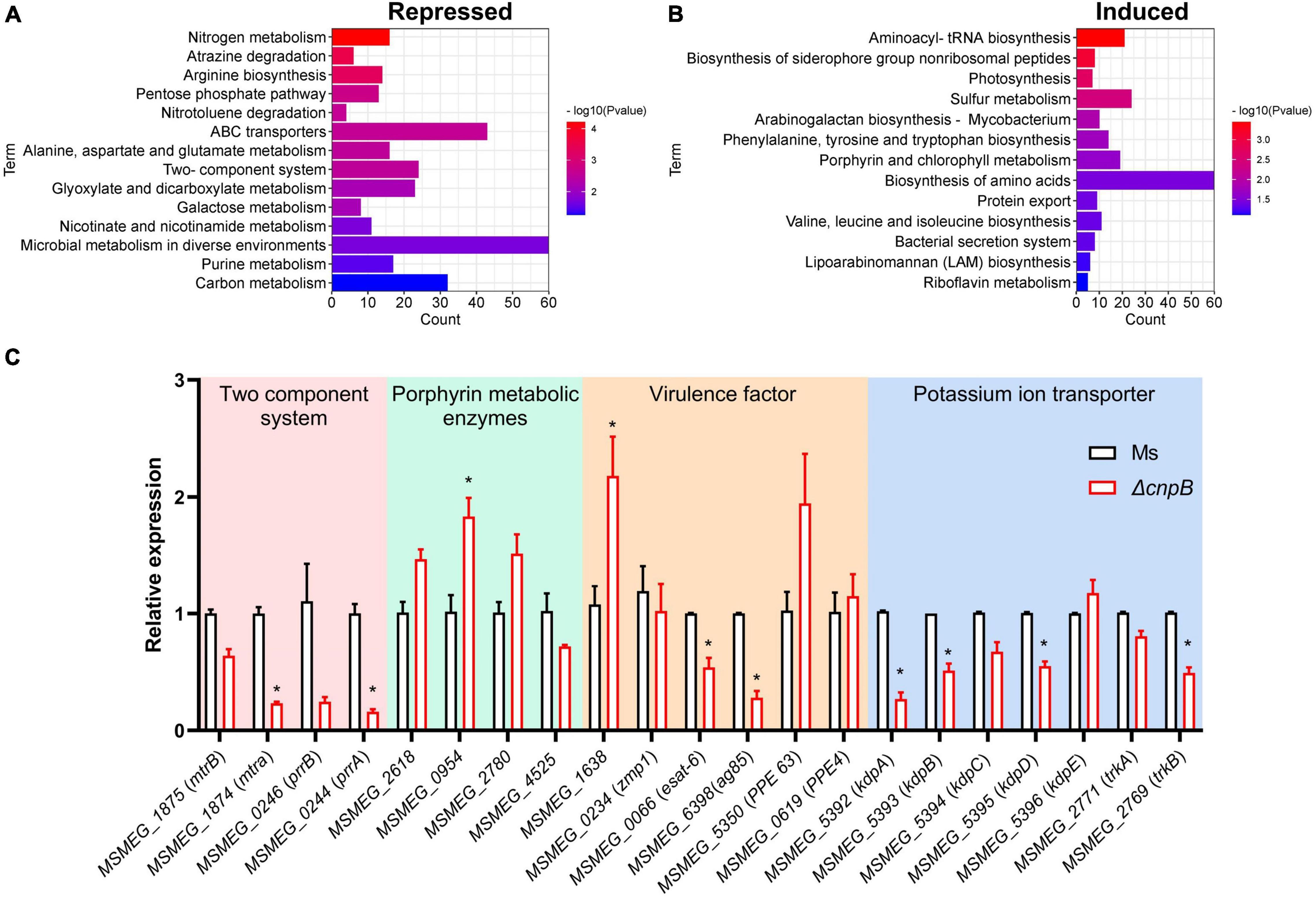
Figure 6. Transcriptional profiles and genes expression of ΔcnpB. Strains were inoculated in 7H9 + OADC medium and cultivated to the late logarithmic stage (3 d) and bacteria pellets were collected for RNA extraction. Differentially expressed genes (DEGs) were screened by RNA-seq with both false discovery rate (FDR) < 0.05 and | log2FC| > 1. KEGG pathway enrichment is associated with DEGs that are repressed (A) or induced (B) in ΔcnpB compared with wild-type M. smegmatis strain. Count means the number of differential gene reads enriched to indicated terms (A,B). (C) The transcription levels of indicated genes were verified by real-time quantitative PCR. *p < 0.05.
Further, qRT-PCR was used for the analysis of gene expressions associated with the phenotypes found above. We had observed that ΔcnpB with high c-di-AMP reduced the biofilm formation in the 7H9 complete medium (Figure 4). The impaired or absent function of the two-component system results in defective biofilm formation in M. smegmatis (Nguyen et al., 2010; Li et al., 2020). Deletion of cnpB with increased c-di-AMP downregulated the genes of two-component systems involved in biofilm formation, such as MSMEG_1875 (mtrB), MSMEG_1874 (mtrA), MSMEG_0246 (prrB), and MSMEG_0244 (prrA) (Figure 6C). ΔcnpB showed porphyrin accumulation in the long-term medium (Figure 5 and Supplementary Figure 3). Elevated c-di-AMP in M. smegmatis significantly upregulated the expression of porphyrin-metabolizing enzymes in 3-day culture, including MSMEG_2618 (uroporphyrin-III C-methyltransferase), MSMEG_0954 (uroporphyrinogen-III synthase), and MSMEG_2780 (uroporphyrinogen decarboxylase) (Figure 6C), and downregulated the expression of MSMEG_4525 (putative oxygen-independent coproporphyrinogen III oxidase) (Figure 6C), which may result in more porphyrin production in ΔcnpB.
It has been reported that c-di-AMP specifically binds to K+ transport proteins such as KdpD and KtrA, TrkA, and TetR family proteins to inhibit potassium transporter function (Bai et al., 2014; Kim et al., 2015; Ali et al., 2017). We found that several potassium ion transport gene transcriptions were downregulated in ΔcnpB, including MSMEG_5392 (kdpA), MSMEG_5393 (kdpB), MSMEG_5394 (kdpC), MSMEG_5395 (kdpD), and MSMEG_2769 (trkB) (Figure 6C). These data suggested that c-di-AMP inhibited the uptake of potassium ions by bacteria not only because of its direct binding to potassium ion transporters but also may be related to the inhibition of transporters transcription.
It has been demonstrated that c-di-AMP modulates bacterial virulence and affects host immune responses (Blander and Barbet, 2018). There were several different protein bands appearing in supernatants between wild-type and ΔcnpB strain but they were not obvious in bacterial lysates (Supplementary Figures 4A,B). qRT-PCR showed that elevated c-di-AMP downregulated the expression of early secreted antigens of MSMEG_0066 (esat-6, esxA) and MSMEG_6398 (ag85, fbpA) in ΔcnpB (Figure 6C), and ΔcnpB seemed to secrete less Ag85 than wild-type M. smegmatis (Supplementary Figure 4C). While host cell death-associated MSMEG_1638 and PE/PPE family proteins of MSMEG_5350 (PPE 63) were upregulated, it was speculated that CnpB deletion might change the immunogenicity of M. smegmatis in vivo.
ΔcnpB Induced Similar Humoral and Cellular Immune Responses in Mice
c-di-AMP could induce innate immune responses and enhance antigen-induced Th1/Th2/Th17 responses as an adjuvant (Ebensen et al., 2011; Devaux et al., 2018; Ning et al., 2021). It has been revealed that M. tuberculosisΔcnpB could trigger host innate immune responses, which could promote the clearance of bacteria (Yang et al., 2014; Dey et al., 2017). We demonstrated that recombinant BCG with elevated c-di-AMP by overexpressing DisA induced enhanced immune responses and provided similar protection as BCG in the same M. tuberculosis intravenous infected mice model (Ning et al., 2019). In this study, humoral and cellular immune responses induced by ΔcnpB were also investigated (Figure 7A). ΔcnpB subcutaneous immunization induced comparable IgG levels with the naïve group (Figure 7B). Splenocyte proliferation assay showed that ΔcnpB immunization induced significant proliferation than the naïve group responding to mycobacterial proteins (p < 0.01) but no more than that of M. smegmatis immunization (Figure 7C). ΔcnpB immunization induced increased Th1 (IFN-γ, IL-2) and Th2 (IL-10) cytokines’ secretion in supernatants of splenocytes restimulated with M. smegmatis proteins than control mice (p < 0.05), but no difference was found between two immunization groups (Figures 7D–F). These results showed that the elevated c-di-AMP level could not enhance the immunogenicity of M. smegmatis as immunized subcutaneously in mice.
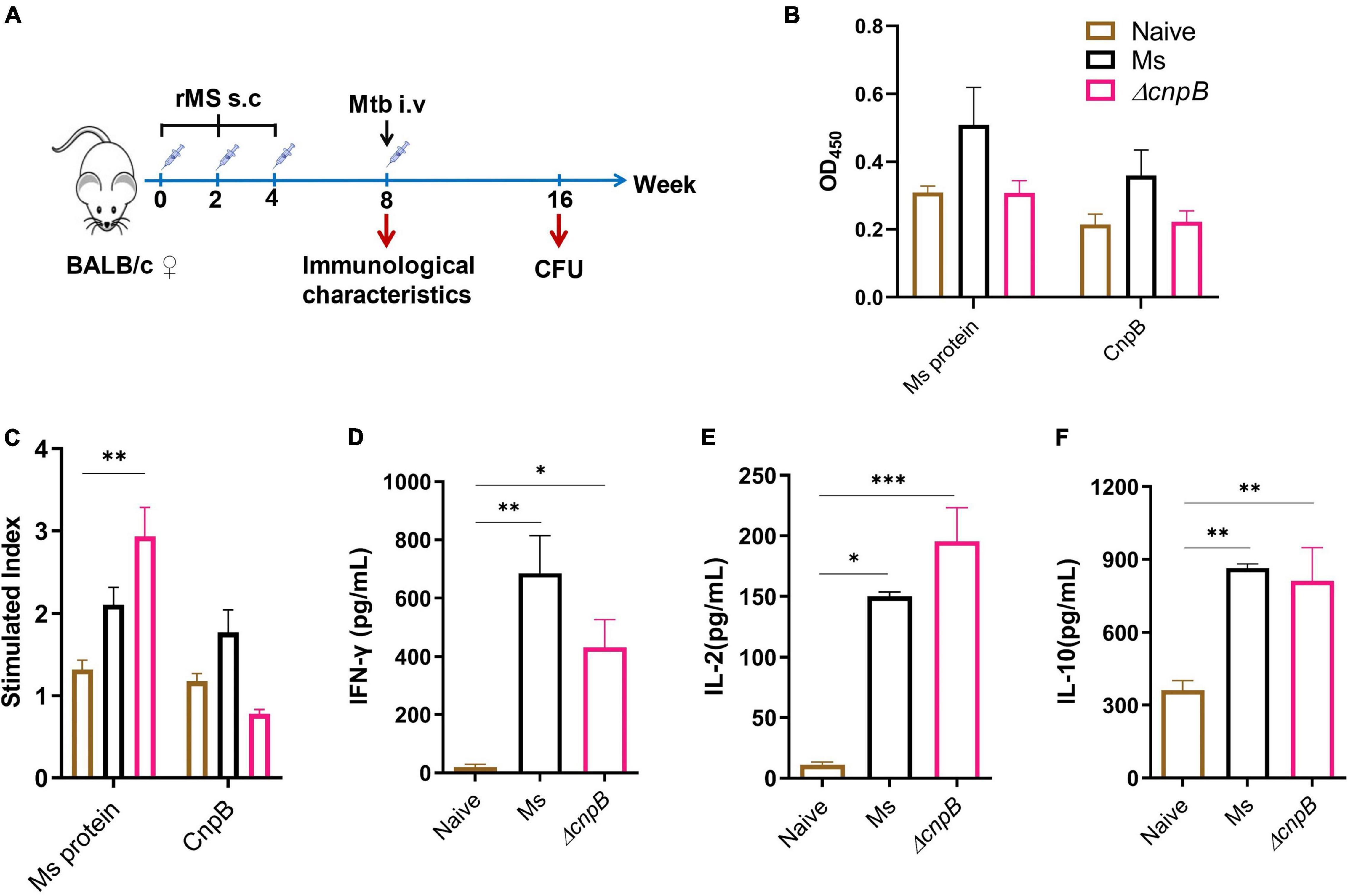
Figure 7. Immune responses induced by ΔcnpB in mice. (A) The immunization and M. tuberculosis infection strategy scheme. Briefly, 6- to 8-week-old female BALB/c mice were immunized subcutaneously (s.c) with PBS (naïve group) and 107 CFU of wild-type M. smegmatis and ΔcnpB strains, respectively. Mice were boosted two times at 2-week intervals. Then, 4 weeks after the last immunization, mice were infected intravenously (i.v) with 5 × 104 CFU of M. tuberculosis H37Ra. (B) Specific IgG levels against M. smegmatis protein and CnpB in sera were assayed by ELISA, and dilutions were 1:200 and 1:400, respectively. (C) Splenocyte proliferation of immunized mice stimulated by M. smegmatis protein extract (25 μg/ml) or CnpB (5 μg/ml) in vitro. (D–F) Splenocytes of mice were stimulated with M. smegmatis protein extract (25 μg/ml) for 72 h in vitro, and cytokine productions of IFN-γ (D), IL-2 (E), and IL-10 (F) in the supernatant were detected using ELISA. *p < 0.05, **p < 0.01, ***p < 0.001.
ΔcnpB Induces Enhanced Immune Responses After M. tuberculosis Infection in Mice
Post 4 weeks of the last immunization, mice were challenged with M. tuberculosis intravenously (Figure 7A). Then, 8 weeks post-infection, ΔcnpB immunized mice showed an increase in IgG level, which were significantly higher than that of challenged mice without immunization (UN), and higher than the Ms group though no significant difference (Figure 8A). After M. tuberculosis infection, both wild-type and ΔcnpB strain-immunized mice produced significantly increased Th1 cytokines (IFN-γ, IL-2) rather than Th2 cytokine (IL-10) compared with unimmunized mice (Figures 8B–D). It was noted that ΔcnpB immunization could induce more IFN-γ and IL-2 secretion in splenocyte supernatant than that of wild-type M. smegmatis (Figures 8B,C). IL-10 secretion showed no differences between groups (Figure 8D). Overall, these results indicated that ΔcnpB immunization promoted significant Th1-type immune responses but not Th2-type immune responses after M. tuberculosis infection in mice.
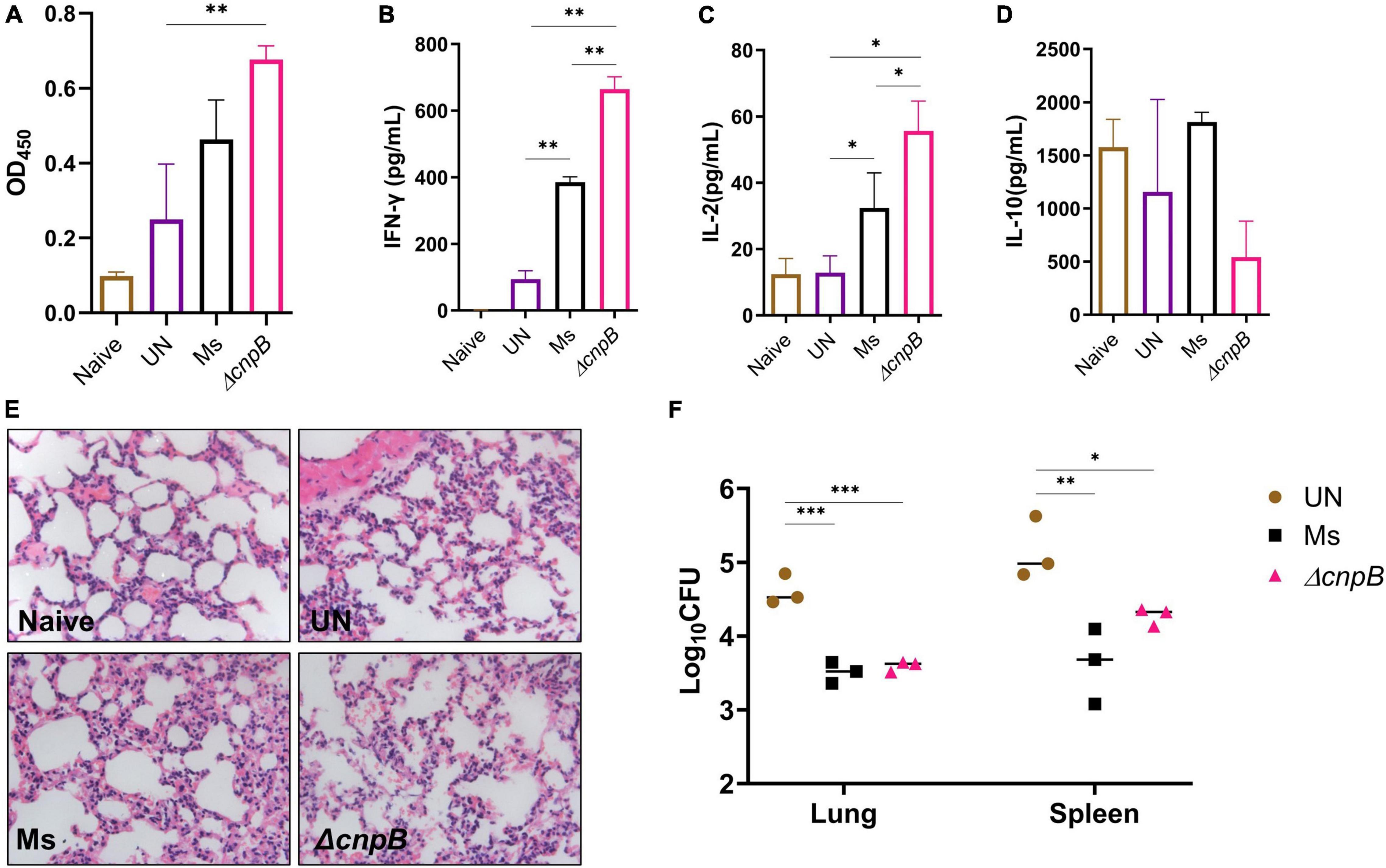
Figure 8. Immune responses induced by ΔcnpB post-M. tuberculosis infection and protection efficiency. (A) Specific IgG level against M. tuberculosis protein in sera (1:200) of immunized mice after M. tuberculosis H37Ra infection. (B–D) Splenocytes of mice were stimulated with H37Ra protein extracts (25 μg/ml) for 72 h in vitro, and cytokine productions of IFN-γ (B), IL-2 (C), and IL-10 (D) in the supernatant were detected using ELISA. (E) H&E-stained lung sections of each group of mice at 8 weeks post-M. tuberculosis infection. Gross pathology changes were observed by microscope (10 × 40). (F) At 8 weeks post-M. tuberculosis infection, bacterial burdens in the mice lungs and spleens were counted by plating on 7H10 plates. *p < 0.05, **p < 0.01, ***p < 0.001.
ΔcnpB Confers Protection Comparable to Wild-Type Strain Against M. tuberculosis Intravenous Infection
ΔcnpB immunization stimulated Th1 instead of Th2 immune responses, which is considered to induce immunopathological damage after M. tuberculosis infection in mice (Da Silva et al., 2015). At 8 weeks post-infection, all infected mice showed similar pathological manifestations of chronic inflammation by H&E staining, including partially damaged alveolar structures, occasional thickening of alveolar mesenchyme, inflammatory cell infiltration, and erythrocyte and histological fluid exudation, but no granuloma or nodules were found in the slide of lungs (Figure 8E).
After mice were infected with M. tuberculosis intravenously, both M. smegmatis strains’ immunization could reduce the bacteria burdens in both lungs (0.87–1.02 lgCFU) and spleens (0.87–1.3 lgCFU) compared to the unimmunized group (p < 0.05) (Figure 8F). There were no statistical differences in bacteria loads of lungs and spleens between M. smegmatis and ΔcnpB-immunized mice (Figure 8F). Overall, ΔcnpB immunization provided similar protections as wild-type M. smegmatis against M. tuberculosis intravenous infection in mice.
Discussion
It has been reported that cnpB deletion did not cause a c-di-AMP level change in M. smegmatis, and overexpressing cnpB led to a drastic decrease in the c-di-AMP level (Tang et al., 2015). In this study, our results showed that cnpB deletion resulted in the accumulation of c-di-AMP at approximately 1.5-folds, and overexpressing cnpB led to decreased c-di-AMP level in M. smegmatis (Figure 1D). Thus, our results confirmed that CnpB as a phosphodiesterase handles c-di-AMP degradation, and the c-di-AMP level could be regulated by the cnpB expression level.
c-di-AMP plays a critical role in several physiological processes and is an indispensable molecule in Bacillus thuringiensis (Zheng et al., 2015). Increasing c-di-AMP levels in bacteria led to smaller colony sizes in Streptomyces venezuelae (Latoscha et al., 2020), as well as in M. smegmatis with DisA overexpression (Zhang and He, 2013; Tang et al., 2015). We found that ΔcnpB with elevated c-di-AMP level forms smaller, whitish, and moist colonies compared with wild-type strain (Figure 2A). Moreover, ΔcnpB showed shorter length (Figure 2D) and slower growth in 7H9 medium (Figure 3 and Supplementary Figure 2), which may be partially attributed to the smaller colonies, as well as reduced optical density (Figure 3 and Supplementary Figure 2). In our previous study, we found that ΔcnpB of M. tuberculosis caused ∼30% reduction in bacterial length (Yang et al., 2014) and a similar reduction in disA-overexpressing BCG (Ning et al., 2019), but did not find obvious changes in colony morphology.
Bacterial biofilms are often associated with chronic infections in human and contribute to the virulence and drug tolerance of M. tuberculosis (Chakraborty et al., 2021). It was reported that inhibition of DisA impedes Enterococcus faecalis biofilm formation (Chen et al., 2018). It was also determined that increased c-di-AMP level by deletion of phosphodiesterase A promoted biofilm formation in Streptococcus mutans (Peng et al., 2016). In this study, we reported that ΔcnpB with elevated c-di-AMP level inhibited biofilm formation (Figure 4). The qRT-PCR analysis identified that biofilm formation genes of two component systems (MtrA/B and PrrA/B) were downregulated and endoglucanase A(MSMEG_6752)was upregulated in ΔcnpB, which supported the observation of the biofilm formation reduction above in the elevated c-di-AMP level strain (Figure 6C, data not shown). Thus, c-di-AMP may regulate biofilm formation through distinct mechanics in different bacteria.
Many mycobacteria produce yellow, orange, or less frequently, salmon pink pigments either in the dark (scotochromogens) or upon exposure to light (photochromogens) (Provvedi et al., 2008; Stockel et al., 2015). These pigments have been characterized as carotenoids, a class of polyterpene lipids whose functions are to act as free radical scavengers and to protect the cells against photodynamic injuries (Provvedi et al., 2008). MSMEG_5609 has been reported as carotenoid oxygenase and is regulated by M. smegmatis MSMEG_1804 (sigF) (Provvedi et al., 2008). In this study, we found that ΔcnpB showed a whitish colony (Figure 2). However, RNA-seq analysis showed that no difference was found in the transcription of MSMEG_5609 and MSMEG_1804 between ΔcnpB and wild-type (data not shown). Thus, the role of c-di-AMP in regulating colony morphology and coloration of mycobacterium remains unknown and needs to be investigated.
Surprisingly, we found that ΔcnpB produced more brown pigment in long-term culture (Figure 5A). By analyzing solubility and properties, fluorescence analysis, full-wavelength scanning, and spectral analysis, the brown pigment accumulation in ΔcnpB supernatant was proved to be a water-soluble porphyrin (Figure 5; Gouterman, 1959, 1961; Dolphin, 1978; Nikitushkin et al., 2016), which was supported by the upregulated porphyrin synthesis-associated genes in ΔcnpB by RNA-seq analysis and qRT-PCR (Figure 6B,C). Ferrous ions could be directly incorporated into various porphyrins for the storage of iron within the bacterial cytoplasm (Ratledge, 2004). The overall process of iron acquisition and its utilization is under very genetic tight control, which contributes to the virulence of mycobacteria as well as the development of tuberculosis (Ratledge, 2004). We detected the iron concentration and found an increase of iron in ΔcnpB::C supernatant of 60-week culture (data not shown), suggesting that c-di-AMP may regulate porphyrin as well as iron metabolism in mycobacterium.
It was reported that the c-di-AMP level was tightly controlled in Bacillus subtilis, and the accumulation of c-di-AMP impaired the growth of B. subtilis, which can be partially suppressed by elevated concentrations of magnesium (Mehne et al., 2013), whereas the lack of c-di-AMP also was detrimental to B. subtilis cell growth (Mehne et al., 2013). In this study, most phenotypes of cnpB-overexpressed strains were similar to those of the wild-type M. smegmatis. It was found that disA-overexpressed strain of BCG showed elevated c-di-AMP and shorter length than ΔcnpB of M. smegmatis but did not find changes in growth and colony morphology (Ning et al., 2019). Thus, we speculated that low c-di-AMP by cnpB overexpression may also be regulated by an unknown compensatory pathway with the unknown mechanism, which will be investigated in our further study.
An increasing number of reports have proved that c-di-AMP regulates diverse bacterial physiological processes (Yin et al., 2020; Yinlan Bai, 2020), as well as host immune responses (Devaux et al., 2018). It was also reported that ΔcnpB of M. tuberculosis was easier to be cleared from the host, which may be the result of a combination of enhanced immune response and strain virulence changes (Yang et al., 2014; Dey et al., 2017). Further, we detected the immunogenicity of ΔcnpB. In fact, ΔcnpB induced similar levels of humoral and cellular immune responses as M. smegmatis did, except significant proliferation of splenocytes against M. smegmatis proteins (Figure 7). Unexpectedly, ΔcnpB induced significant humoral immune responses compared with unimmunized (UN) mice. Moreover, ΔcnpB-immunized mice could produce more Th1 cytokines of IFN-γ and IL-2 rather than Th2 cytokine (IL-10) after M. tuberculosis infection (Figure 8).
Antigen-specific Th1 response is usually considered a marker of protective immune responses against M. tuberculosis infection (Zeng et al., 2018). Compared with wild-type M. smegmatis, it was found that ΔcnpB immunization could induce enhanced Th1 cellular immune responses after M. tuberculosis infection, which could promote the elimination of M. tuberculosis infection. Reports including ours have found that M. smegmatis could be used as a live vaccine vector or a therapeutic vaccine (Junqueira-Kipnis et al., 2013; Alves Da Silva et al., 2014; Wang et al., 2014; Liu et al., 2015). Previously, our and other studies have demonstrated that cnpB-deficient M. tuberculosis exhibited attenuated virulence (Yang et al., 2014; Dey et al., 2017). Therefore, the safety of M. smegmatis ΔcnpB is better in theory than wild tyoe strain, coupled with the stronger immune response induced by ΔcnpB, and it may replace M. smegmatis as a vaccine or vaccine carrier. However, elevated c-di-AMP level did not increase the protective efficiency of M. smegmatis against M. tuberculosis venous infection (Figure 8). Previously, we demonstrated that a recombinant BCG with the elevated c-di-AMP level by overexpressing DisA provided similar protection as BCG in the same mouse model of M. tuberculosis intravenous infection (Ning et al., 2019). It was reported that the same recombinant BCG provided improved protection after aerosol infection of M. tuberculosis in guinea pigs (Dey et al., 2020). Thus, ΔcnpB should be further evaluated for the protection with the aerosol challenge of M. tuberculosis, using more susceptible animals such as guinea pigs.
Taken together, our results provided more evidence that elevated c-di-AMP could affect colony morphology, bacterial length, growth, and potassium transporters of M. smegmatis, consistent with previous research in several bacteria, including other mycobacteria (Corrigan et al., 2011; Du et al., 2014; Chen et al., 2018). Moreover, we reported that elevated c-di-AMP could inhibit biofilm formation and induce porphyrin accumulation, which may affect bacterial drug resistance and virulence, by regulating their associated gene expressions in M. smegmatis. Compared with wild-type M. smegmatis, cnpB-deleted strain with elevated c-di-AMP level could induce enhanced immune responses, especially protective Th1 immune responses after M. tuberculosis intravenous infection in the mouse model, which suggested M. smegmatis with the elevated c-di-AMP level as a vaccine against tuberculosis.
Data Availability Statement
The bulk RNA-seq datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI BioProject—PRJNA808387.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Ethics Center of Air Force Medical University.
Author Contributions
HN, XL, and YX performed the most experiments and analyzed the data. LB, WZ, and LW conducted some animal experiments. JK and YL conducted several immunological assays. YM provided guidance in the study of bacterial physiology. HN and YB wrote the manuscript. YB and GB conceived and designed the research. YB supervised the work. All authors have read and agreed with the data.
Funding
This study was funded by the National Natural Science Foundation (Nos. 81971560, 81371774, and 81671638), the National Major Special Projects of 13th Five-year Plan (No. 2018ZX10302302002004), and the Provincial Natural Science Foundation of Shaanxi Province (No. 2022ZDLSF01-07).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.865045/full#supplementary-material
References
Ali, M. K., Li, X., Tang, Q., Liu, X., Chen, F., Xiao, J., et al. (2017). Regulation of inducible potassium transporter KdpFABC by the KdpD/KdpE two-component system in Mycobacterium smegmatis. Front. Microbiol. 8:570. doi: 10.3389/fmicb.2017.00570
Alves Da Silva, D., Cavalcanti, M. A., Muniz, De Oliveira, F., Trentini, M. M., Junqueira-Kipnis, A. P., et al. (2014). Immunogenicity of a recombinant Mycobacterium smegmatis vaccine expressing the fusion protein CMX in cattle from Goias State. Brazil. J. Vet. Med. Sci. 76, 977–984. doi: 10.1292/jvms.13-0338
Bai, Y., and Bai, G. (2020). “Cyclic di-AMP in Mycobacterium tuberculosis,” in Microbial Cyclic Di-Nucleotide Signaling, ed. N. G. Shan-Ho Chou and T. Vincent (Berlin: Springer), 443–454.
Bai, Y., Yang, J., Zarrella, T. M., Zhang, Y., Metzger, D. W., and Bai, G. (2014). Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J. Bacteriol. 196, 614–623. doi: 10.1128/JB.01041-13
Bai, Y., Yang, J., Zhou, X., Ding, X., Eisele, L. E., and Bai, G. (2012). Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PLoS One 7:e35206. doi: 10.1371/journal.pone.0035206
Bharti, S., Maurya, R. K., Venugopal, U., Singh, R., Akhtar, M. S., and Krishnan, M. Y. (2020). Rv1717 Is a cell wall - associated beta-galactosidase of mycobacterium tuberculosis that is involved in biofilm dispersion. Front. Microbiol. 11:611122. doi: 10.3389/fmicb.2020.611122
Blander, J. M., and Barbet, G. (2018). Exploiting vita-PAMPs in vaccines. Curr. Opin. Pharmacol. 41, 128–136. doi: 10.1016/j.coph.2018.05.012
Burhenne, H., and Kaever, V. (2013). Quantification of cyclic dinucleotides by reversed-phase LC-MS/MS. Methods Mol. Biol. 1016, 27–37. doi: 10.1007/978-1-62703-441-8_3
Chakraborty, P., Bajeli, S., Kaushal, D., Radotra, B. D., and Kumar, A. (2021). Biofilm formation in the lung contributes to virulence and drug tolerance of Mycobacterium tuberculosis. Nat. Commun. 12:1606. doi: 10.1038/s41467-021-21748-6
Chen, L., Li, X., Zhou, X., Zeng, J., Ren, Z., Lei, L., et al. (2018). Inhibition of enterococcus faecalis growth and biofilm formation by molecule targeting Cyclic di-AMP Synthetase Activity. J. Endod. 44, e1381–e1388. doi: 10.1016/j.joen.2018.05.008
Corrigan, R. M., Abbott, J. C., Burhenne, H., Kaever, V., and Grundling, A. (2011). C-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 7:e1002217. doi: 10.1371/journal.ppat.1002217
Da Silva, M. V., Tiburcio, M. G., Machado, J. R., Silva, D. A., Rodrigues, D. B., Rodrigues, V., et al. (2015). Complexity and controversies over the cytokine profiles of t helper cell subpopulations in tuberculosis. J. Immunol. Res. 2015:639107. doi: 10.1155/2015/639107
Devaux, L., Kaminski, P. A., Trieu-Cuot, P., and Firon, A. (2018). Cyclic di-AMP in host-pathogen interactions. Curr. Opin. Microbiol. 41, 21–28. doi: 10.1016/j.mib.2017.11.007
Dey, B., Dey, R. J., Cheung, L. S., Pokkali, S., Guo, H., Lee, J. H., et al. (2015). A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat. Med. 21, 401–406. doi: 10.1038/nm.3813
Dey, R. J., Dey, B., Singh, A. K., Praharaj, M., and Bishai, W. (2020). Bacillus calmette-guerin overexpressing an endogenous stimulator of interferon genes agonist provides enhanced protection against pulmonary tuberculosis. J. Infect. Dis. 221, 1048–1056. doi: 10.1093/infdis/jiz116
Dey, R. J., Dey, B., Zheng, Y., Cheung, L. S., Zhou, J., Sayre, D., et al. (2017). Inhibition of innate immune cytosolic surveillance by an M. tuberculosis phosphodiesterase. Nat. Chem. Biol. 13, 210–217. doi: 10.1038/nchembio.2254
Du, B., Ji, W., An, H., Shi, Y., Huang, Q., Cheng, Y., et al. (2014). Functional analysis of c-di-AMP phosphodiesterase, GdpP, in Streptococcus suis serotype 2. Microbiol. Res. 169, 749–758. doi: 10.1016/j.micres.2014.01.002
Ebensen, T., Libanova, R., Schulze, K., Yevsa, T., Morr, M., and Guzman, C. A. (2011). Bis-(3’,5’)-cyclic dimeric adenosine monophosphate: strong Th1/Th2/Th17 promoting mucosal adjuvant. Vaccine 29, 5210–5220. doi: 10.1016/j.vaccine.2011.05.026
Friedman, L., and Kolter, R. (2004). Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690. doi: 10.1046/j.1365-2958.2003.03877.x
Gouterman, M. (1959). Study of the effects of substitution on the absorption spectra of porphin. J. Chem. Phys. 30, 1139–1161.
He, J., Yin, W., Galperin, M. Y., and Chou, S. H. (2020). Cyclic di-AMP, a second messenger of primary importance: tertiary structures and binding mechanisms. Nucleic Acids Res. 48, 2807–2829. doi: 10.1093/nar/gkaa112
He, Q., Wang, F., Liu, S., Zhu, D., Cong, H., Gao, F., et al. (2016). Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP Phosphodiesterase. J. Biol. Chem. 291, 3668–3681. doi: 10.1074/jbc.M115.699801
Hu, J., Zhang, G., Liang, L., Lei, C., and Sun, X. (2020). Increased excess intracellular Cyclic di-AMP levels impair growth and virulence of bacillus anthracis. J. Bacteriol. 202, e00653–e006619. doi: 10.1128/JB.00653-19
Junqueira-Kipnis, A. P., de Oliveira, F. M., Trentini, M. M., Tiwari, S., Chen, B., Resende, D. P., et al. (2013). Prime-boost with Mycobacterium smegmatis recombinant vaccine improves protection in mice infected with Mycobacterium tuberculosis. PLoS One 8:e78639. doi: 10.1371/journal.pone.0078639
Kim, H., Youn, S. J., Kim, S. O., Ko, J., Lee, J. O., and Choi, B. S. (2015). Structural studies of potassium transport protein KtrA regulator of conductance of K+ (RCK) C domain in complex with cyclic diadenosine monophosphate (c-di-AMP). J. Biol. Chem. 290, 16393–16402. doi: 10.1074/jbc.M115.641340
Latoscha, A., Drexler, D. J., Al-Bassam, M. M., Bandera, A. M., Kaever, V., Findlay, K. C., et al. (2020). C-di-AMP hydrolysis by the phosphodiesterase AtaC promotes differentiation of multicellular bacteria. Proc. Natl. Acad. Sci. U.S.A. 117, 7392–7400. doi: 10.1073/pnas.1917080117
Li, M., Gašparovič, H., Weng, X., Chen, S., Korduláková, J., and Jessen-Trefzer, C. (2020). The two-component locus MSMEG_0244/0246 together with MSMEG_0243 affects biofilm assembly in m. smegmatis correlating with changes in phosphatidylinositol mannosides acylation. Front Microbiol. 11:570606. doi: 10.3389/fmicb.2020.570606
Liu, L., Fu, R., Yuan, X., Shi, C., Wang, S., Lu, X., et al. (2015). Differential immune responses and protective effects in avirulent mycobacterial strains vaccinated BALB/c Mice. Curr. Microbiol. 71, 129–135. doi: 10.1007/s00284-015-0837-3
Lu, Y., Kang, J., Ning, H., Wang, L., Xu, Y., Xue, Y., et al. (2018). Immunological characteristics of Mycobacterium tuberculosis subunit vaccines immunized through different routes. Microb. Pathog. 125, 84–92. doi: 10.1016/j.micpath.2018.09.009
Maarsingh, J. D., Yang, S., Park, J. G., and Haydel, S. E. (2019). Comparative transcriptomics reveals PrrAB-mediated control of metabolic, respiration, energy-generating, and dormancy pathways in Mycobacterium smegmatis. BMC Genomics 20:942. doi: 10.1186/s12864-019-6105-3
Mangtani, P., Abubakar, I., Ariti, C., Beynon, R., Pimpin, L., Fine, P. E., et al. (2014). Protection by BCG vaccine against tuberculosis: a systematic review of randomized controlled trials. Clin. Infect. Dis. 58, 470–480. doi: 10.1093/cid/cit790
Mehne, F. M., Gunka, K., Eilers, H., Herzberg, C., Kaever, V., and Stulke, J. (2013). Cyclic di-AMP homeostasis in bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J. Biol. Chem. 288, 2004–2017. doi: 10.1074/jbc.M112.395491
Nguyen, H. T., Wolff, K. A., Cartabuke, R. H., Ogwang, S., and Nguyen, L. (2010). A lipoprotein modulates activity of the MtrAB two-component system to provide intrinsic multidrug resistance, cytokinetic control and cell wall homeostasis in Mycobacterium. Mol. Microbiol. 76, 348–364. doi: 10.1111/j.1365-2958.2010.07110.x
Nikitushkin, V., Shleeva, M., Zinin, A., Trutneva, K., Ostrovsky, D., and Kaprelyants, A. (2016). The main pigment of the dormant Mycobacterium smegmatis is porphyrin. FEMS Microbiol. Lett. 363:fnw206. doi: 10.1093/femsle/fnw206
Ning, H., Wang, L., Zhou, J., Lu, Y., Kang, J., Ding, T., et al. (2019). Recombinant BCG with bacterial signaling molecule cyclic di-amp as endogenous adjuvant induces elevated immune responses after mycobacterium tuberculosis infection. Front. Immunol. 10:1519. doi: 10.3389/fimmu.2019.01519
Ning, H., Zhang, W., Kang, J., Ding, T., Liang, X., Lu, Y., et al. (2021). Subunit vaccine ESAT-6:c-di-AMP delivered by intranasal route elicits immune responses and protects against mycobacterium tuberculosis infection. Front. Cell Infect. Microbiol. 11:647220. doi: 10.3389/fcimb.2021.647220
Peng, X., Zhang, Y., Bai, G., Zhou, X., and Wu, H. (2016). Cyclic di-AMP mediates biofilm formation. Mol. Microbiol. 99, 945–959. doi: 10.1111/mmi.13277
Postic, G., Danchin, A., and Mechold, U. (2012). Characterization of NrnA homologs from Mycobacterium tuberculosis and Mycoplasma pneumoniae. RNA 18, 155–165. doi: 10.1261/rna.029132.111
Provvedi, R., Kocincova, D., Dona, V., Euphrasie, D., Daffe, M., Etienne, G., et al. (2008). SigF controls carotenoid pigment production and affects transformation efficiency and hydrogen peroxide sensitivity in Mycobacterium smegmatis. J. Bacteriol. 190, 7859–7863. doi: 10.1128/JB.00714-08
Ratledge, C. (2004). Iron, mycobacteria and tuberculosis. Tuberculosis (Edinb). 84, 110–130. doi: 10.1016/j.tube.2003.08.012
Singh, A. K., Praharaj, M., Lombardo, K. A., Yoshida, T., Matoso, A., Baras, A. S., et al. (2022). Re-engineered BCG overexpressing cyclic di-AMP augments trained immunity and exhibits improved efficacy against bladder cancer. Nat. Commun. 13:878. doi: 10.1038/s41467-022-28509-z
Stockel, S., Stanca, A. S., Helbig, J., Rosch, P., and Popp, J. (2015). Raman spectroscopic monitoring of the growth of pigmented and non-pigmented mycobacteria. Anal. Bioanal. Chem. 407, 8919–8923. doi: 10.1007/s00216-015-9031-5
Stulke, J., and Kruger, L. (2020). Cyclic di-AMP signaling in bacteria. Annu. Rev. Microbiol. 74, 159–179. doi: 10.1146/annurev-micro-020518-115943
T, J. A. S., Ranjitha, J., Rajan, A., and Shankar, V. V. (2020). Features of the biochemistry of Mycobacterium smegmatis, as a possible model for Mycobacterium tuberculosis. J. Infect. Public Health 13, 1255–1264. doi: 10.1016/j.jiph.2020.06.023
Tang, Q., Luo, Y., Zheng, C., Yin, K., Ali, M. K., Li, X., et al. (2015). Functional analysis of a c-di-AMP-specific phosphodiesterase MsPDE from Mycobacterium smegmatis. Int. J. Biol. Sci. 11, 813–824. doi: 10.7150/ijbs.11797
Uhlich, G. A., Cooke, P. H., and Solomon, E. B. (2006). Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl. Environ. Microbiol. 72, 2564–2572. doi: 10.1128/AEM.72.4.2564-2572.2006
Van Kessel, J. C., and Hatfull, G. F. (2007). Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152. doi: 10.1038/nmeth996
Venkataswamy, M. M., Goldberg, M. F., Baena, A., Chan, J., Jacobs, W. R. Jr., and Porcelli, S. A. (2012). In vitro culture medium influences the vaccine efficacy of Mycobacterium bovis BCG. Vaccine 30, 1038–1049. doi: 10.1016/j.vaccine.2011.12.044
Wang, P., Wang, L., Zhang, W., Bai, Y., Kang, J., Hao, Y., et al. (2014). Immunotherapeutic efficacy of recombinant Mycobacterium smegmatis expressing Ag85B-ESAT6 fusion protein against persistent tuberculosis infection in mice. Hum. Vaccin Immunother. 10, 150–158. doi: 10.4161/hv.26171
Whiteley, A. T., Garelis, N. E., Peterson, B. N., Choi, P. H., Tong, L., Woodward, J. J., et al. (2017). C-di-AMP modulates Listeria monocytogenes central metabolism to regulate growth, antibiotic resistance and osmoregulation. Mol. Microbiol. 104, 212–233. doi: 10.1111/mmi.13622
WHO (2021). Global Tuberculosis Report [Online]. Available online at: https://www.who.int/publications/i/item/9789240037021. (accessed May 4, 2022).
Witte, C. E., Whiteley, A. T., Burke, T. P., Sauer, J. D., Portnoy, D. A., and Woodward, J. J. (2013). Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. MBio 4, e282–e313. doi: 10.1128/mBio.00282-13
Woodward, J. J., Iavarone, A. T., and Portnoy, D. A. (2010). C-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705. doi: 10.1126/science.1189801
Yang, J., Bai, Y., Zhang, Y., Gabrielle, V. D., Jin, L., and Bai, G. (2014). Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol. Microbiol. 93, 65–79. doi: 10.1111/mmi.12641
Yin, W., Cai, X., Ma, H., Zhu, L., Zhang, Y., Chou, S. H., et al. (2020). A decade of research on the second messenger c-di-AMP. FEMS Microbiol. Rev. 44, 701–724. doi: 10.1093/femsre/fuaa019
Yinlan Bai, G. B. (2020). “Cyclic di-AMP in Mycobacterium tuberculosis,” in Microbial Cyclic Di-Nucleotide Signaling, eds N. G. Shan-Ho Chou and T. Vincent (Berlin: Springer Nature), 443–454.
Zarrella, T. M., and Bai, G. (2020). The many roles of the bacterial second messenger cyclic di-AMP in adapting to stress cues. J. Bacteriol. e348–e420. doi: 10.1128/JB.00348-20
Zeng, G., Zhang, G., and Chen, X. (2018). Th1 cytokines, true functional signatures for protective immunity against TB? Cell Mol. Immunol. 15, 206–215. doi: 10.1038/cmi.2017.113
Zhang, L., and He, Z. G. (2013). Radiation-sensitive gene A (RadA) targets DisA, DNA integrity scanning protein A, to negatively affect cyclic Di-AMP synthesis activity in Mycobacterium smegmatis. J. Biol. Chem. 288, 22426–22436. doi: 10.1074/jbc.M113.464883
Zhang, L., Li, W., and He, Z. G. (2013). DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J. Biol. Chem. 288, 3085–3096. doi: 10.1074/jbc.M112.428110
Zhang, Y., Yang, J., and Bai, G. (2018). Cyclic di-AMP-mediated interaction between Mycobacterium tuberculosis DeltacnpB and macrophages implicates a novel strategy for improving BCG vaccination. Pathog. Dis. 76:fty008. doi: 10.1093/femspd/fty008
Keywords: Mycobacterium smegmatis, M. tuberculosis, c-di-AMP, physiology, immunogenicity
Citation: Ning H, Liang X, Xie Y, Bai L, Zhang W, Wang L, Kang J, Lu Y, Ma Y, Bai G and Bai Y (2022) c-di-AMP Accumulation Regulates Growth, Metabolism, and Immunogenicity of Mycobacterium smegmatis. Front. Microbiol. 13:865045. doi: 10.3389/fmicb.2022.865045
Received: 29 January 2022; Accepted: 14 April 2022;
Published: 24 May 2022.
Edited by:
Michal Letek, Universidad de León, SpainReviewed by:
Herman Sintim, Purdue University, United StatesCara C. Boutte, University of Texas at Arlington, United States
Przemysław Adam Płociński, Institute for Medical Biology (PAN), Poland
Copyright © 2022 Ning, Liang, Xie, Bai, Zhang, Wang, Kang, Lu, Ma, Bai and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangchun Bai, QmFpR0BtYWlsLmFtYy5lZHU=; Yinlan Bai, eWlubGFuYmFpQGZtbXUuZWR1LmNu
†These authors have contributed equally to this work
 Huanhuan Ning
Huanhuan Ning Xuan Liang
Xuan Liang Yanling Xie
Yanling Xie Lu Bai
Lu Bai Wei Zhang4
Wei Zhang4 Lifei Wang
Lifei Wang Jian Kang
Jian Kang Yanzhi Lu
Yanzhi Lu Yanling Ma
Yanling Ma Guangchun Bai
Guangchun Bai Yinlan Bai
Yinlan Bai