
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 May 2022
Sec. Systems Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.863228
This article is part of the Research Topic Probiogenomics of classic and next-generation probiotics View all 6 articles
Lactobacillus species are prominent inhabitants of the human gastrointestinal tract that contribute to maintaining a balanced microbial environment that positively influences host health. These bacterial populations can be altered through use of probiotic supplements or via dietary changes which in turn affect the host health. Utilizing polyphenolic compounds to selectively stimulate the growth of commensal bacteria can have a positive effect on the host through the production of numerous metabolites that are biologically active. Four Lactobacillus strains were grown in the presence of pomegranate (POM) extract. Two strains, namely, L. acidophilus NCFM and L. rhamnosus GG, are commonly used probiotics, while the other two strains, namely, L. crispatus NCK1351 and L. gasseri NCK1342, exhibit probiotic potential. To compare and contrast the impact of POM on the strains' metabolic capacity, we investigated the growth of the strains with and without the presence of POM and identified their carbohydrate utilization and enzyme activity profiles. To further investigate the differences between strains, an untargeted metabolomic approach was utilized to quantitatively and qualitatively define the metabolite profiles of these strains. Several metabolites were produced significantly and/or exclusively in some of the strains, including mevalonate, glutamine, 5-aminoimidazole-4-carboxamide, phenyllactate, and fumarate. The production of numerous discrete compounds illustrates the unique characteristics of and diversity between strains. Unraveling these differences is essential to understand the probiotic function and help inform strain selection for commercial product formulation.
The human gastrointestinal tract (GIT) harbors one of the most densely populated bacterial communities on Earth, which is highly diverse and complex (Holscher, 2017). These microorganisms form a symbiotic relationship with the host as they metabolize undigested food remnants and transform them into compounds that can be used as energy by the host or contribute to host health. Lactobacillus species are prominent microorganisms that help maintain a balanced microbiota within human microbiome habitats such as the oral cavity, gastrointestinal tract, and vaginal tract (Huttenhower et al., 2012). A healthy gut environment can be improved with probiotic supplementation, which is defined by the International Scientific Association for Probiotics and Prebiotics (ISAPP) as “live microorganisms that, when administered in adequate amounts, confer a health benefit to the host”. These effective probiotic strains are transient residents of the host, as they can survive at low pH and high bile concentration environments in the GIT, adhere to intestinal epithelial cells, and inhibit the growth of pathogenic microorganisms (Blum et al., 1999; Likotrafiti and Rhoades, 2016; Somashekaraiah et al., 2019).
In addition to probiotic consumption, the GIT microbiota can be altered through human diets. Prebiotics are substrates that are selectively utilized by host microorganisms to confer a health benefit (Gibson et al., 2017), and traditionally, they consist of non-digestible carbohydrates such as inulin, fructooligosaccharides, galactooligosaccharides, and human milk oligosaccharides. Research in the past decades has shifted to studying plant-derived polyphenolic compounds as potential prebiotics due to their beneficial biological effects. Polyphenols are phytochemicals that are native in plants to protect them from environmental stresses, and these compounds differ considerably in their carbon structure, hydroxylation patterns, and glycosylation and acylation of the heterocyclic rings, which impacts their bioavailability in humans (Scalbert and Williamson, 2000). The polyphenolic compounds that reach the colon to be degraded and depolymerized by microbes result in a variety of 1,3 diphenylpropanes, γ-valerolactones, benzoic acids, phenylacetic acids, phenylpropionic acids, and other aromatic compounds (Justesen et al., 2000; Gonthier et al., 2003; Rios et al., 2003; Urpi-Sarda et al., 2009).
The investigation of the human gut microbiome has progressed over time with advances in high-throughput sequencing technologies to further understand the relationship between the host and microbial population. The most recent emerging field of systems biology research is metabolomics, which provides a comprehensive analysis of all the metabolites involved in general metabolic reactions necessary for the maintenance, growth, and normal functions of a cell (Dettmer et al., 2007). Utilizing metabolomic technology as a tool to study the gut metabolome offers valuable information regarding metabolic pathways, health of the environment, and responses to internal or external perturbations (Bernini et al., 2009; Cambeiro-Perez et al., 2018, 2020).
Metabolomic analysis of Lactobacillus species has gained attention from the food industry due to their use as starter cultures in fermented foods. Studies have observed the metabolite changes during fermentation and analyzed the compounds that contribute to organoleptic properties or bio-preservative effects in fermented vegetables (Park et al., 2016; Zhao et al., 2016), dairy (Piras et al., 2013; Palomo et al., 2014), soy (Namgung et al., 2010; Kim et al., 2012), and beverages (Lee et al., 2009; López-Rituerto et al., 2012). Due to the important role of lactobacilli in human health, researchers have utilized metabolomics in human clinical studies to identify the effects of probiotic treatments (Hong et al., 2011; Ghini et al., 2020). There have been numerous in vivo and in vitro studies to analyze the effects of traditional prebiotics, such as inulin-type fructans (Vandeputte et al., 2017), galactooligosaccharides (Cheng et al., 2018), and xylooligosaccharides (Yang et al., 2019), with probiotics, whereas the use of polyphenolic compounds in combination with probiotics has not been studied as extensively. Pomegranates are rich in polyphenols, specifically punicalagins, punicalins, and ellagic acid, which are part of a group of hydrolyzable tannins designated as ellagitannins (Garcia-Muñoz and Vaillant, 2014). The evidence for pomegranates to have a prebiotic effect still needs to be substantiated by clinical studies; however, in vitro studies have shown to promote the growth of beneficial bacteria and produce smaller metabolites, such as urolithins (Bialonska et al., 2009, 2010). Identifying bacterial fermentation products from various substrates contributes to understanding their beneficial effects on the host. In this study, we used an untargeted metabolomic approach to determine how pomegranate extract influenced the growth and biotransformation abilities of four Lactobacillus strains. We analyzed two commonly used probiotics (L. acidophilus NCFM and L. rhamnosus GG) and two potential probiotics that are found within the human GIT (L. crispatus NCK1351 and L. gasseri NCK1342). This technique allowed for a qualitative measurement of each of the metabolites produced. We aimed to investigate the metabolic profiles of each Lactobacillus strain and compare and contrast the metabolites and associated pathways among them.
The pomegranate extract was composed of POMx spray-dried pomegranate powder from POM Wonderful, Inc., Los Angeles, CA (Heber et al., 2007). The preparation of the POM extract for the bacterial growth curves was followed as described earlier (Henning et al., 2017). POM extract was prepared by dissolving 7 mg/ml of POM extract in ultrapure water and vortexed for 10 min. The stock solution was then centrifuged at 3,226 × g for 10 min, and the supernatant was filtered with a 0.45-μm membrane filter and stored in aliquots at −20°C. Semi-defined media (SDM) was used as the base media (Kimmel and Roberts, 1998) for the growth curves and with added glucose (1% for transfers or 0.5% for growth assay). A previous study reported that 400 μg/ml of POMx stimulated the growth of bifidobacteria and lactobacilli while inhibiting the growth of the Enterobacteriaceae, Clostridia, and Bacteroides fragilis groups (Li et al., 2015). In this study, the stock POM extract was diluted in SDM to 400 μg/ml for the transfers and the growth assays.
The details of the Lactobacillus strains are summarized in Table 1. Lactobacillus acidophilus NCFM, Lacticaseibacillus rhamnosus GG, Lactobacillus crispatus NCK1351, and Lactobacillus gasseri NCK1342 were inoculated from −80°C glycerol stocks into de Man-Rogosa-Sharpe (MRS) broth (Difco Laboratories, Inc., Detroit, MI) and grown overnight in a water bath at 37°C. Overnight cultures were inoculated 1% v/v into SDM (1% glucose, control) and SDM (1% glucose with 400 μg/ml POM extract) and grown overnight in a water bath at 37°C for an additional two transfers. After the second transfer, overnight cultures were then inoculated 1% v/v into SDM (0.5% glucose, control) and SDM (0.5% glucose with 400 μg/ml POM extract) and grown in a water bath at 37°C. Optical density (600 nm) was measured with a Biowave Cell Density Meter CO8000 (Biochrom Ltd, Cambridge, England) for up to 24 h. Simultaneously, at 0, 4, 8, 12, and 24 h cultures were plated on MRS agar medium and incubated anaerobically at 37°C for 48 h before colonies were counted.
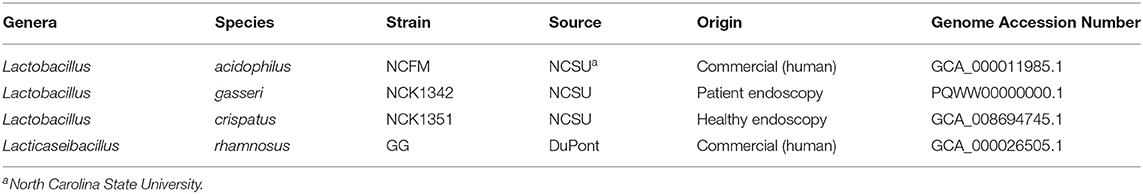
Table 1. Strains used in this study: information regarding Lactobacillus strain type, origin, and accession number.
The carbohydrate utilization and enzyme activity were assayed with Analytical Profile Index (API) tests (Biomerieux, Durham, NC) in duplicate. For the carbohydrate utilization, cultures were grown overnight in MRS broth at 37°C in a water bath; 1 ml of culture was then centrifuged at 8,609 × g for 1 min at room temperature. The cells were washed twice with phosphate-buffered saline (PBS) and then resuspended with 1 ml PBS. Cells (2% v/v) were added to 10 ml PBS, and the absorbance (Abs) at 600 nm was measured. Based on the Abs reading, the volume of the cell suspension needed to achieve ca. Abs 0.05 was calculated. Following the manufacturer's instructions, the calculated cell suspension was added to the API 50 CHL medium and inverted several times. The API tray wells were inoculated with the cell suspension and CHL medium and covered with mineral oil. Trays were incubated at 37°C under ambient atmospheric conditions, and results were recorded at 24 and 48 h.
For the enzyme activity assay, cultures were grown overnight in MRS broth at 37°C in a water bath. Cells were washed in ultrapure distilled water (1:1 v/v) and centrifuged for 5 min at 3,226 × g. Cells achieved a turbidity of 5–6 McFarland, and 65 μl was administered to each cupule. Trays were incubated at 37°C under ambient atmospheric conditions. After incubation, one drop of ZYM A and ZYM B reagents was added to each cupule and placed 10 cm under a light source (1,000 W bulb) for 10 s. Results were recorded 5 min after the 10-s light exposure according to the manufacturer's instructions.
Cultures were grown and transferred as described earlier for the Lactobacillus growth assays. Following two transfers in SDM (1% glucose with 400 μg/ml POM extract), cells were inoculated 1% v/v into 10 ml of SDM (0.5% glucose with 400 μg/ml POM extract) and grown for 16 h at 37°C in a water bath. After 16 h, the cells were centrifuged at 3,226 × g for 10 min, and 200 μl of cell-free supernatant was transferred to labeled cryogenic tubes for immediate storage at −80°C. Controls consisted of SDM media with no POM extract, SDM media with POM extract (400 μg/ml) at 0 h of incubation, and SDM media with POM extract (400 μg/ml) at 16 h of incubation. Quadruplicate cell-free supernatant samples were shipped on dry ice to Metabolon Inc. (Durham, NC) for analysis.
Global metabolite profiles were obtained for each sample by a commercial laboratory (Metabolon, Inc.). Additional standards were purchased and added to the Metabolon library, including 8-O-methyl urolithin A, gallic acid, punicalagin, and punicalin (Sigma-Aldrich, St. Louis, MO). Each sample received was accessioned into the Metabolon Laboratory Information Management System (LIMS) and was assigned by the LIMS a unique identifier that was associated with the original source identifier only. Samples were prepared using the MicroLab STAR® automated system (Hamilton Company, Reno, NE).
Several recovery standards were added prior to the first step in the extraction process for QC purposes. To remove protein, dissociate small molecules bound to protein or trapped in the precipitated protein matrix, and to recover chemically diverse metabolites, proteins were precipitated with methanol under vigorous shaking for 2 min (GenoGrinder 2000, Glen Mills Inc, NJ) followed by centrifugation. The resulting extract was divided into five fractions: two for analysis by two separate reverse-phase (RP)/UPLC-MS/MS methods with positive ion mode electrospray ionization (ESI), one for analysis by RP/UPLC-MS/MS with negative ion mode ESI, one for analysis by HILIC/UPLC-MS/MS with negative ion mode ESI, and one sample was reserved for backup. Samples were placed briefly on a TurboVap® (Zymark) to remove the organic solvent. The sample extracts were stored overnight under nitrogen before preparation for analysis.
All methods utilized a Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo Scientific Q-Exactive high-resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution. The sample extract was dried and then reconstituted in solvents compatible with each of the four methods. Each reconstitution solvent contained a series of standards at fixed concentrations to ensure injection and chromatographic consistency. One aliquot was analyzed using acidic positive ion conditions, chromatographically optimized for more hydrophilic compounds. In this method, the extract was gradient eluted from a C18 column (Waters UPLC BEH C18-2.1 × 100 mm, 1.7 μm) using water and methanol, containing 0.05% perfluoropentanoic acid (PFPA) and 0.1% formic acid (FA). Another aliquot was also analyzed using acidic positive ion conditions; however, it was chromatographically optimized for more hydrophobic compounds. In this method, the extract was gradient eluted from the same aforementioned C18 column using methanol, acetonitrile, water, 0.05% PFPA, and 0.01% FA and was operated at an overall higher organic content. Another aliquot was analyzed using basic negative ion optimized conditions using a separate dedicated C18 column. The basic extracts were gradient eluted from the column using methanol and water, however, with 6.5 mM ammonium bicarbonate, pH 8. The fourth aliquot was analyzed via negative ionization following elution from a HILIC column (Waters UPLC BEH Amide 2.1 × 150 mm, 1.7 μm) using a gradient consisting of water and acetonitrile with 10 mM ammonium formate, pH 10.8. The MS analysis alternated between MS and data-dependent MSn scans using dynamic exclusion. The scan range varied slightly between methods but covered 70–1,000 m/z.
The raw data were extracted, peak-identified, and QC processed using Metabolon's proprietary hardware and software and curated using a variety of procedures to ensure that a high-quality data set was made available for statistical analysis and interpretation of the compounds (Lawton et al., 2008; Dehaven et al., 2010; Evans et al., 2012). The informatics system consisted of four major components, namely, the LIMS, the data extraction and peak-identification software, data processing tools for QC and compound identification, and a collection of information interpretation and visualization tools for use by data analysts. The hardware and software foundations for these informatics components were the LAN backbone and a database server running Oracle 10.2.0.1 Enterprise Edition.
Compounds from the metabolic profiling were semi-quantified using the area under the curve. The data were normalized and scaled to the median value for each compound. Following log transformation and imputation of missing values, if any, a Welch's two-sample t-test of the minimum observed value for each compound was performed in ArrayStudio. The two-sample t-test was used to identify biochemicals that differed significantly (p ≤ 0.05) between the samples and the control media with pomegranate at 16 h, and the resulting values were designated as the fold change values.
Rstudio v1.3. 1093-1 (http://www.rstudio.com/) was used for statistical analysis and graphs. Heat maps were generated with a hierarchical clustering model. The model used “cor()” and “hclust()” functions and a Spearman's rank-order correlation method. Principal component analysis (PCA) and partial least-squares discriminant analysis (PLS_DA) were made using MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/). The boxplots were created by the “geom_boxplot()” and “geom_point()” functions. Apart from growth assays, graphs and boxplots were generated with ggplot2 in Rstudio.
Growth assay was performed in SDM with a concentration of 0.5% w/v glucose, compared with the transfers that contained 1.0% w/v glucose. The decreased sugar content was conducted in order to provide the strains the opportunity to primarily utilize POM as a substrate. The OD 600 nm and CFU/ml counts were measured over 24 h for each strain grown in SDM with and without POM (400 μg/ml). In the case of OD measurements, there was no significant difference (p ≤ 0.05) for L. acidophilus, L. crispatus, and L. gasseri demonstrating POM extract did not have an inhibitory effect on growth. There was a minor but significant decrease in OD 600 nm readings at 4, 6, and 10 h for L. rhamnosus, but by 24 h there was no significant difference between the control and test group (Figure 1). Next, we measured the CFU/ml counts and determined the largest significant difference observed with L. acidophilus where the CFU/ml count increased in the media with POM extract at 24 h (Figure 1). Overall, each strain had similar growth patterns in SDM alone and SDM with POM, demonstrating that these strains are capable of growing in the presence of POM extract and potentially utilize the pomegranate as a substrate.
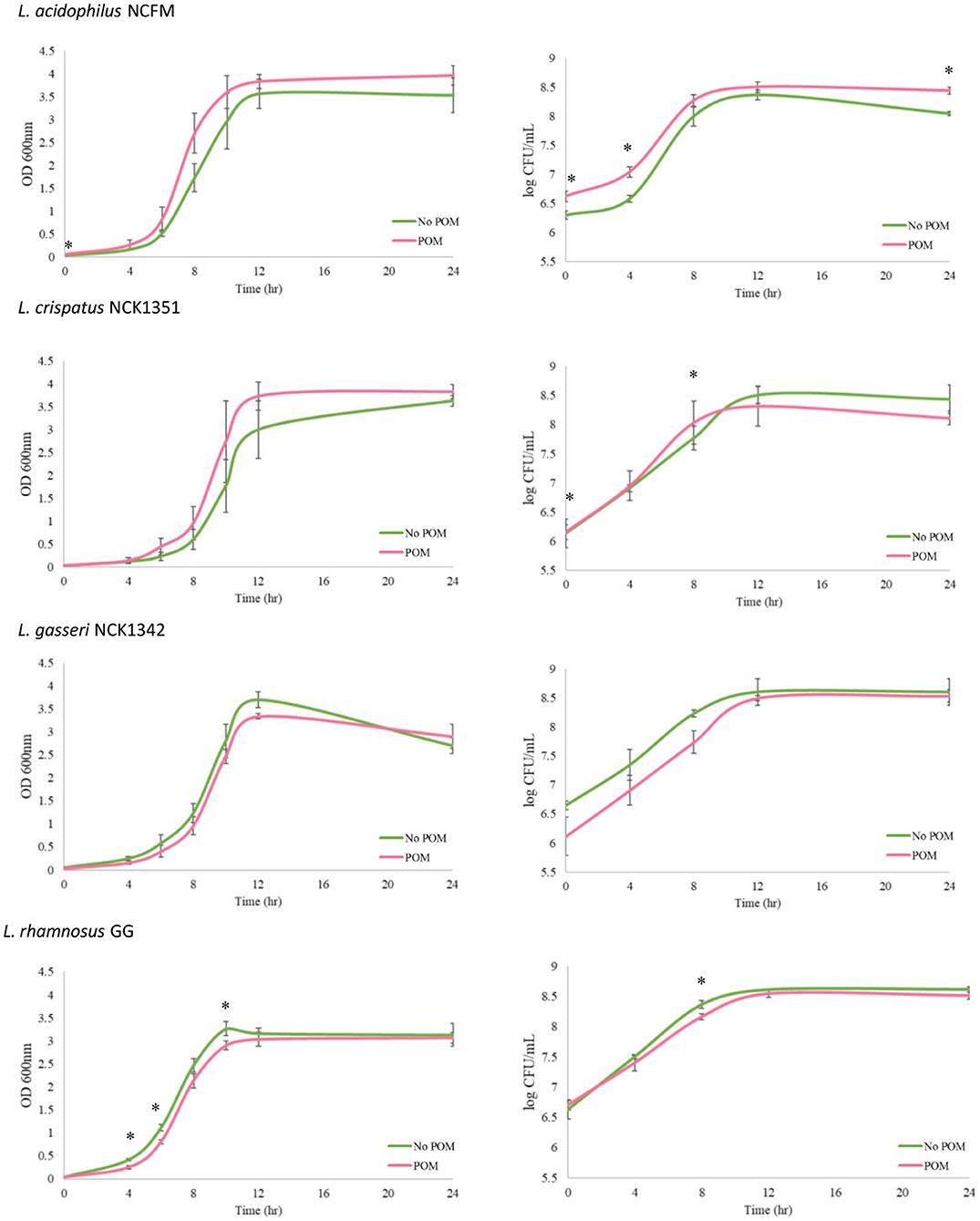
Figure 1. Growth of Lactobacillus strains with and without the POM extract (400 μg/ml). Growth assays were performed over a 24-h period with the L. acidophilus NCFM, L. crispatus NCK1351, L. gasseri NCK1342, and L. rhamnosus GG grown in SDM with 0.5% glucose and no POM extract (green curve, No POM) or in SDM with 0.5% glucose and 400 μg/ml of POM extract (red curve, POM). Graphs on the left-hand side show OD 600 nm values, and log CFU/ml growth curves are shown on the right-hand side. Error bars represent the standard deviation of three biological replicates, and * indicates p ≤ 0.05.
We next determined the carbohydrate utilization and enzymatic activity profile for each of the four strains, namely, L. acidophilus NCFM (Lac), L. crispatus NCK1351 (Lcr), L. gasseri NCK1342 (Lga), and L. rhamnosus GG (Lrh) (Figure 2). Results of the API assays determined that L. rhamnosus fermented the widest range of sugars, including sugar alcohols sorbitol, mannitol, and dulcitol, which were not fermented by the other three strains (Figure 2). One advantageous metabolic feature of lactobacilli is the ability to utilize lactose (Figure 2). In the case of L. rhamnosus GG, this feature is lost due to frameshifts in the antiterminator (lacT) and 6-phospho-β-galactosidase (lacG) genes (Kankainen et al., 2009). The inability of L. rhamnosus GG to ferment lactose was confirmed in the API assay (Figure 2). Additional genetic variations in enzymes or transporters also explain the inability of L. rhamnosus GG to utilize rhamnose, ribose, and maltose (Kankainen et al., 2009). The other three strains showed a similar carbohydrate utilization profile with L. crispatus being the sole strain that utilized both raffinose and starch (Figure 2).
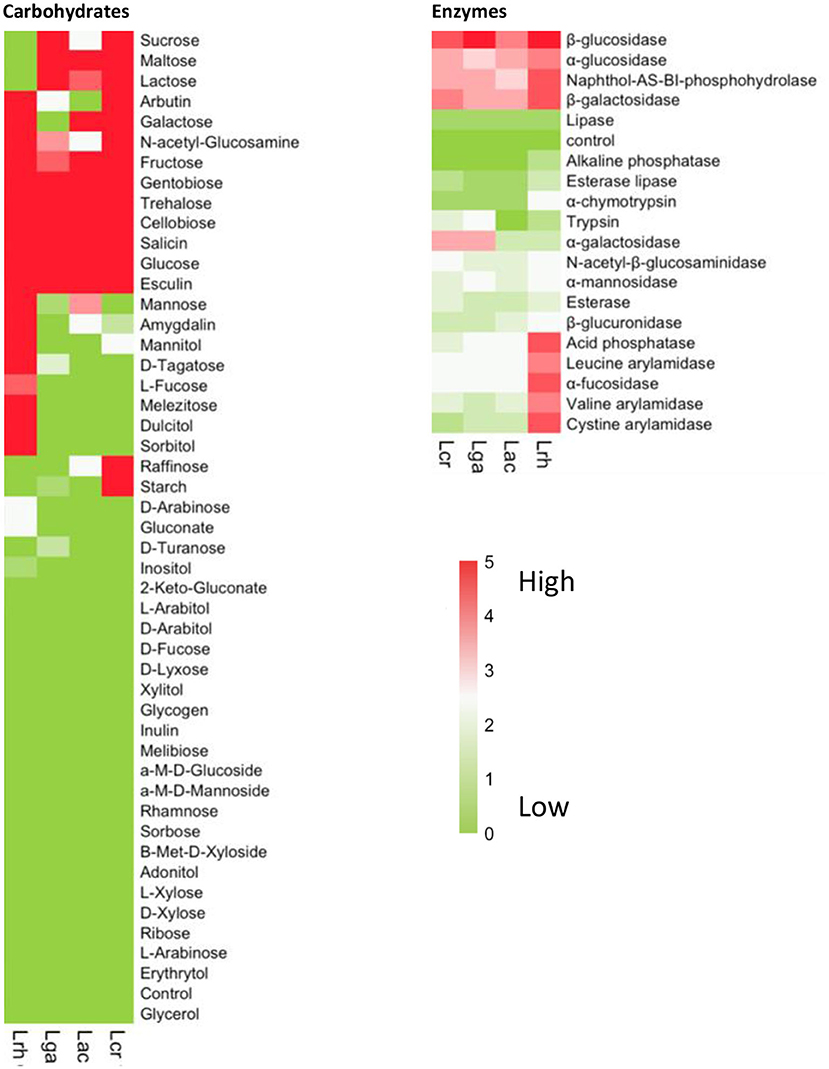
Figure 2. Carbohydrate and enzyme profiles of Lactobacillus strains. Carbohydrate utilization and enzymatic activity were determined using API assays for the four strains, namely, L. acidophilus NCFM (Lac), L. crispatus NCK1351 (Lcr), L. gasseri NCK1342 (Lga), and L. rhamnosus GG (Lrh). The heat maps show the mean values of two biological replicates with the values scored according to the manufacturer's instructions. The values range from 0 to 5 and are assigned to the standard color where zero represents no reaction and 5 represents a reaction of maximum intensity. The relative activity may be estimated from color strength.
Lactobacilli have many glycosyl hydrolases (GHs) that assist them in the metabolism of many types of mono-, di-, and polysaccharides, specifically α- and β-glucosidases and β-galactosidases. All four strains showed activity for these enzymes (Figure 2). Fucosyl-oligosaccharides are natural prebiotics that are found in intestinal mucin and human milk, and α-L-fucosidases have an important role in the cleavage of these compounds and have only been identified in a few Lactobacillus species (Rodríguez-Díaz et al., 2011), including L. rhamnosus GG (Morita et al., 2009). Our data showed that L. rhamnosus GG was the only one of the four strains with high α-L-fucosidases activity (Figure 2). As with the carbohydrate utilization assay, L. rhamnosus exhibited the widest enzymatic activity profile (Figure 2).
To further elucidate which metabolites were produced by the strains when grown in the presence of POM, metabolomic analyses were performed on cell-free supernatants. Untargeted metabolite profiles of the samples from the four Lactobacillus strains grown in the presence of POM identified a total of 552 biochemicals, with 353 compounds confirmed with authentic standards. The number of metabolites that achieved statistical significance (p ≤ 0.05) with respect to the control media with no POM for L. acidophilus, L. crispatus, L. rhamnosus, and L. gasseri was 325, 325, 366, and 274, respectively. Carbohydrates are typically the preferred carbon source for lactobacilli; however, amino acids and other aromatic compounds, such as plant-derived polyphenols, can also serve as a carbon source to support bacterial growth. A hierarchical clustering analysis (HCA) of the fold change values of the entire metabolic profiles in the cell-free supernatants illustrates that L. acidophilus and L. gasseri were the most similar, followed by L. crispatus, and then L. rhamnosus (Figure 3A). Both the POM controls (0 and 16 h) clustered together. The highest number of metabolites detected from the global metabolomic profile were derived from the amino acid metabolic super pathway at 155 metabolites, followed by the nucleotide (48), lipid (41), and carbohydrate (29) super pathways. Hierarchical clustering was then performed on the top 50 significant (p ≤ 0.05) highest detected metabolites compared with the control. Clustering was similar across the four strains (Figure 3A) for the 50 metabolites. To further analyze how the strains differ based on their metabolic profiles, a principal component analysis (PCA) and a partial least-squares discriminant analysis PLS_DA plot of the global metabolite profile values were performed (Figures 3B,C). The PCA and PLS-DA plots of the 552 metabolites detected confirmed the data in the HCA showing L. acidophilus and L. gasseri clustering together, while L. crispatus and L. rhamnosus cluster separately from L. acidophilus and L. gasseri, each indicating differential metabolic profiles for L. crispatus and L. rhamnosus from the other strains.
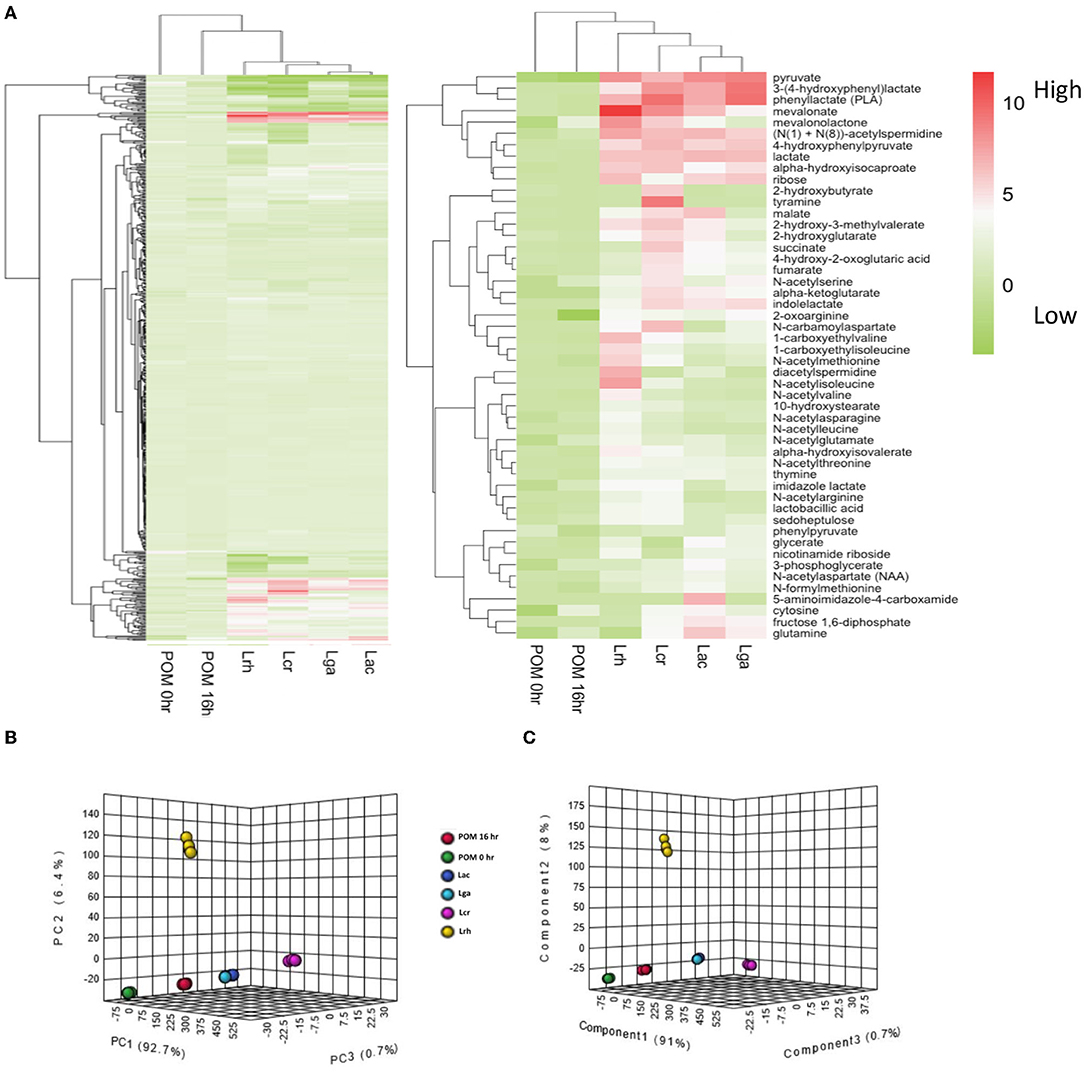
Figure 3. Global metabolomic analysis overview. (A) Hierarchical cluster analysis (HCA) of log2 transformed fold change values of the metabolomic data for the cell-free supernatants of the Lactobacillus strains and controls. The entire metabolite profile (left-hand side) and top 50 significant (p ≤ 0.05) highest produced metabolites (right-hand side) were performed for each strain. The legend shows the log2 transformed fold change values. (B) Principal component analysis (PCA) and (C) partial least-squares discriminant analysis (PLS_DA) of the global metabolite profile for the four Lactobacillus strains and controls. L. acidophilus NCFM (Lac), L. crispatus NCK1351 (Lcr), L. gasseri NCK1342 (Lga), and L. rhamnosus GG (Lrh), SDM media with POM 400 μg/ml at T0 (POM 0 h) and T16 (POM 16 h).
Homofermentative Lactobacillus metabolize monosaccharides through the Embden-Meyerhof pathway (EMP) (Goh and Klaenhammer, 2015), where glucose serves as a carbohydrate energy source that is fermented to two molecules of pyruvate. Pyruvate is often subsequently converted into lactate through NADH-dependent lactate dehydrogenase which helps maintain the redox balance by regenerating NAD+ (Zotta et al., 2017). Surprisingly, L. crispatus had a lower fold change value of pyruvate but a similar fold change value of lactate compared with the other strains. While pyruvate is mainly converted to lactate in Lactobacillus, it is possible that L. crispatus distributed pyruvate among other pathways to generate other products, such as derivatives of branched chain amino acids. Similar to the other strains, L. crispatus has an incomplete TCA cycle, but had a relatively higher fold change value of TCA cycle intermediates. Of the 50 metabolites with the highest fold change values, the amino acid pathway was the super pathway with the greatest number of compounds, followed by the carbohydrate, and then the lipid super pathway. Within the carbohydrate super pathway, pyruvate was the highest metabolite fold change value for all the strains (Figure 4). Interestingly, lactate was the next highest fold change value for all strains except L. rhamnosus, which had ribose as the second highest fold change value. L. rhamnosus is a facultative hetero-fermentative organism that can catabolize pentoses in addition to hexoses (Bintsis, 2018) which may explain why it had a greater fold change value of ribose.
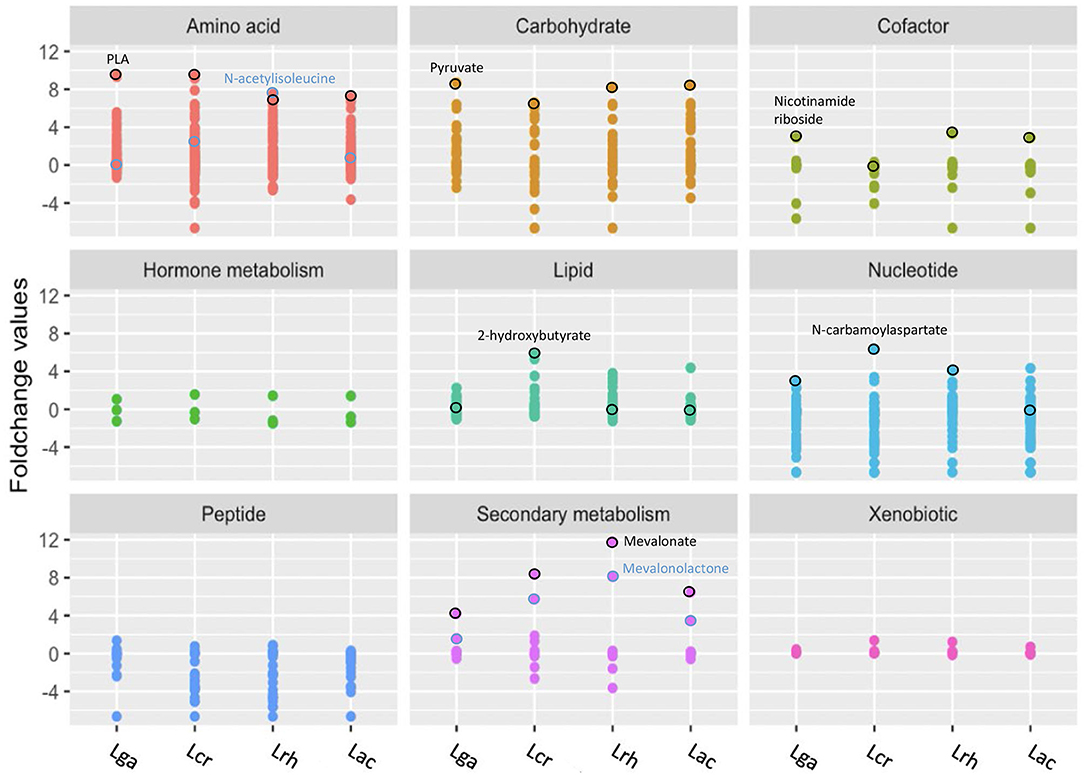
Figure 4. Representation of the global metabolic profile of Lactobacillus strains based on super pathway analysis. Significant (p ≤ 0.05) log2 transformed fold change values for each metabolite were used for mapping to nine biochemical pathways. Distinct colors and grids indicate the overarching biochemical pathway for the detected metabolites. Outlined circles (black and blue) indicate highly detected and unique metabolites for each strain. L. acidophilus NCFM (Lac), L. crispatus NCK1351 (Lcr), L. gasseri NCK1342 (Lga), and L. rhamnosus GG (Lrh).
Phenyllactate (PLA) is a compound derived from the amino acid super pathway and had the greatest increased fold change value by L. gasseri. Phenylpyruvate was identified in significantly smaller quantities than PLA; however, the Kyoto Encyclopedia of Genes and Genomes (KEGG) has no annotated enzymatic activities in lactobacilli to generate these products. An iterative PSI-Blast using fldH (a phenyllactate dehydrogenase from Clostridium sporogenes BL-8) was used to identify potential orthologs. Each species showed a 33–35% identity with potential ortholog, D-2-hydroxyacid dehydrogenase, which suggests they may contain relevant enzymatic capacity. PLA was the highest fold change value metabolite within the amino acid super pathway for all the strains except L. rhamnosus, which had N-acetylisoleucine as the highest fold change value followed by PLA (Figure 4). N-acetylisoleucine was a relatively minimal fold change value in the other strains.
Mevalonate is a secondary metabolite that is derived from the isoprenoid biosynthesis pathway (Kuzuyama and Seto, 2012) and is strongly related with L. rhamnosus. Within the secondary metabolite super pathway, all the strains had the highest fold change value of mevalonate, followed by mevalonolactone.
For the cofactor super pathway, nicotinamide riboside (NR) was the highest fold change value for all the strains except for L. crispatus, which did not have significantly different values than that of the control (Figure 4). Surprisingly, L. crispatus had biotin as the highest fold change value that achieved statistical significance; 2-hydroxybutyrate, a hydroxy fatty acid that falls within the lipid super pathway, had a significantly increased fold change value for L. crispatus but none of the other strains. Interestingly, L. crispatus does not have a complete annotated propanoate metabolism pathway in KEGG, but it does have two L-lactate dehydrogenase enzymes that are necessary for the conversion of 2-oxobutyrate to 2-hydroxybutyrate. N-carbamoylaspartate is a nucleotide derivative from pyrimidine metabolism that was the highest fold change value for all strains except L. acidophilus, which had no significant production of this metabolite and the highest increased fold change value of cytosine (Figure 4).
In order to observe the range of the highlighted metabolites that were unique to each strain, a boxplot was created for selected individual compounds (Figure 5). The values displayed are from the metabolomic data that were normalized and scaled to the median value for each compound. Mevalonate was highly correlated with L. rhamnosus, and this was confirmed from the scaled values as mevalonate was produced in the greatest quantity by L. rhamnosus followed by L. crispatus, L. acidophilus, and L. gasseri (Figure 5). The Lactobacillus strains were able to produce varying amounts of TCA cycle intermediates, including fumarate, succinate, and malate. L. crispatus had the highest fumarate production among the strains which could presumably be attributed to the higher enzymatic activity of fumarate reductase and fumarate hydratase. Glutamine is an α-amino acid that is derived from glutamate within the amino acid superfamily. Glutamine was detected at the highest levels in L. acidophilus, followed by L. gasseri, L. crispatus, and L. rhamnosus. The last illustrated metabolite is also derived from the glutamate family, 5-aminoimidazole-4-carboxamide, and is an intermediate in purine biosynthesis. Interestingly, this compound was only significantly detected in the L. acidophilus sample but in none of the other strains (Figure 5).
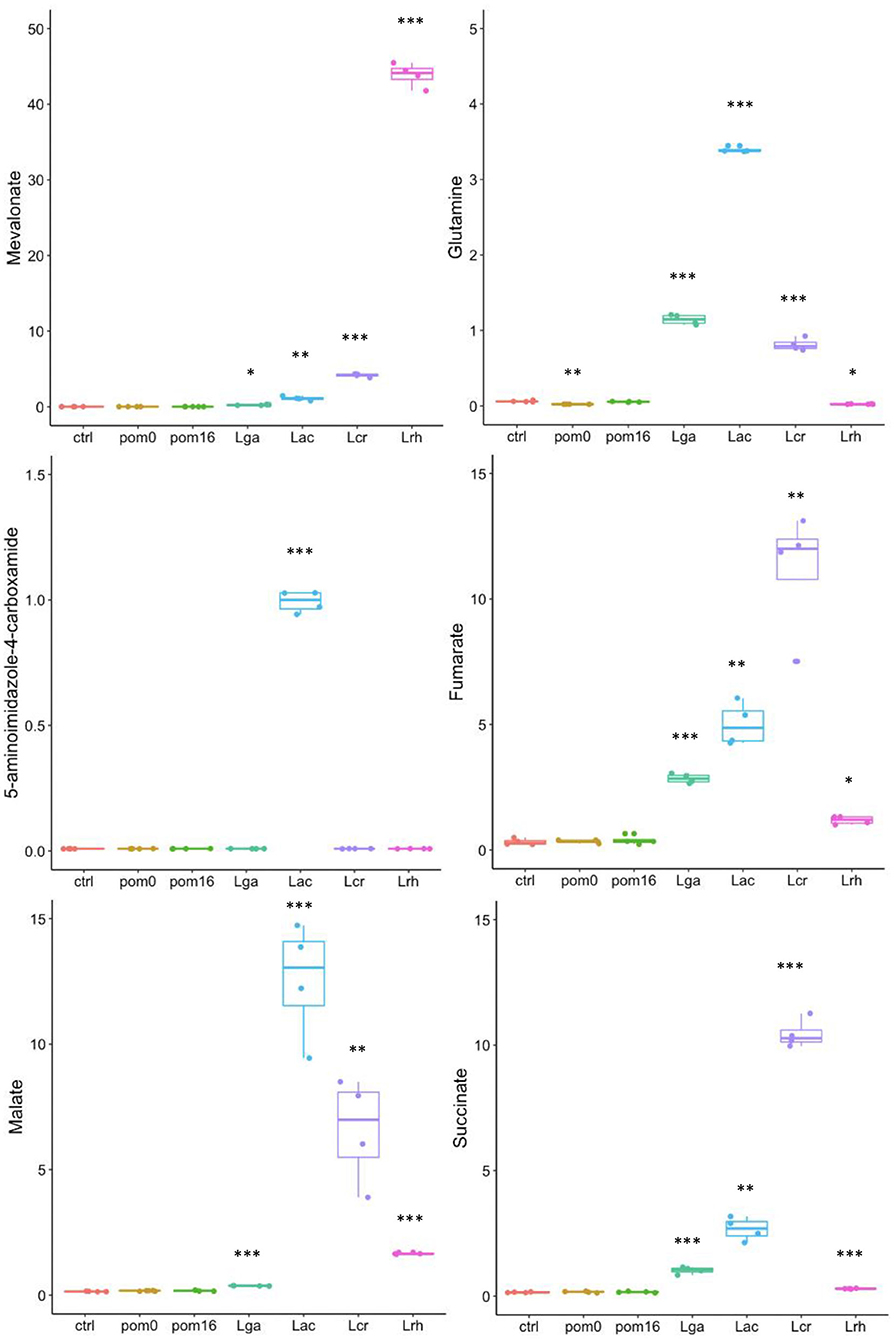
Figure 5. Boxplots of specific metabolites. The data points are from the metabolomic data that were normalized and scaled to the median value for each compound. Boxplots for mevalonate, glutamine, 5-aminoimidazole-4-carboxamide, fumarate, malate, and succinate are shown. L. acidophilus NCFM (Lac), L. crispatus NCK1351 (Lcr), L. gasseri NCK1342 (Lga), and L. rhamnosus GG (Lrh), SDM media alone (ctrl), SDM media with POM 400 μg/ml at T0 (pom0) and T16 (pom16). p-values are indicated as follows: *p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005, and they were determined by comparison to the control.
The metabolomic analysis highlighted several unique metabolites that were associated with a specific strain. To determine whether these differences in metabolite production could be explained by variations in their biochemical pathways, a comparison of the strain's corresponding genes was analyzed. For mevalonate, the genes are split across two main loci. The first locus encodes an acetoacetyl-CoA thiolase (thiL; LBA_RS03260), hydroxymethylglutaryl-CoA synthase (mvaS; LBA_RS03270), and hydroxymethylglutaryl-CoA reductase (mvaA; LBA_RS03265); and the second locus encodes a mevalonate kinase (mvaK; LBA_RS05850), diphosphomevalonate decarboxylase (mvaD; LBA_RS05855), phosphomevalonate kinase (pmvaK; LBA_RS05860), and isopentenyl-diphosphate delta isomerase (fni; LBA_RS05865) (Figure 6A). However, in the case of L. rhamnosus, the mvaK, mvaD, and fni genes are in an operon and the pmvak gene is in its own locus.
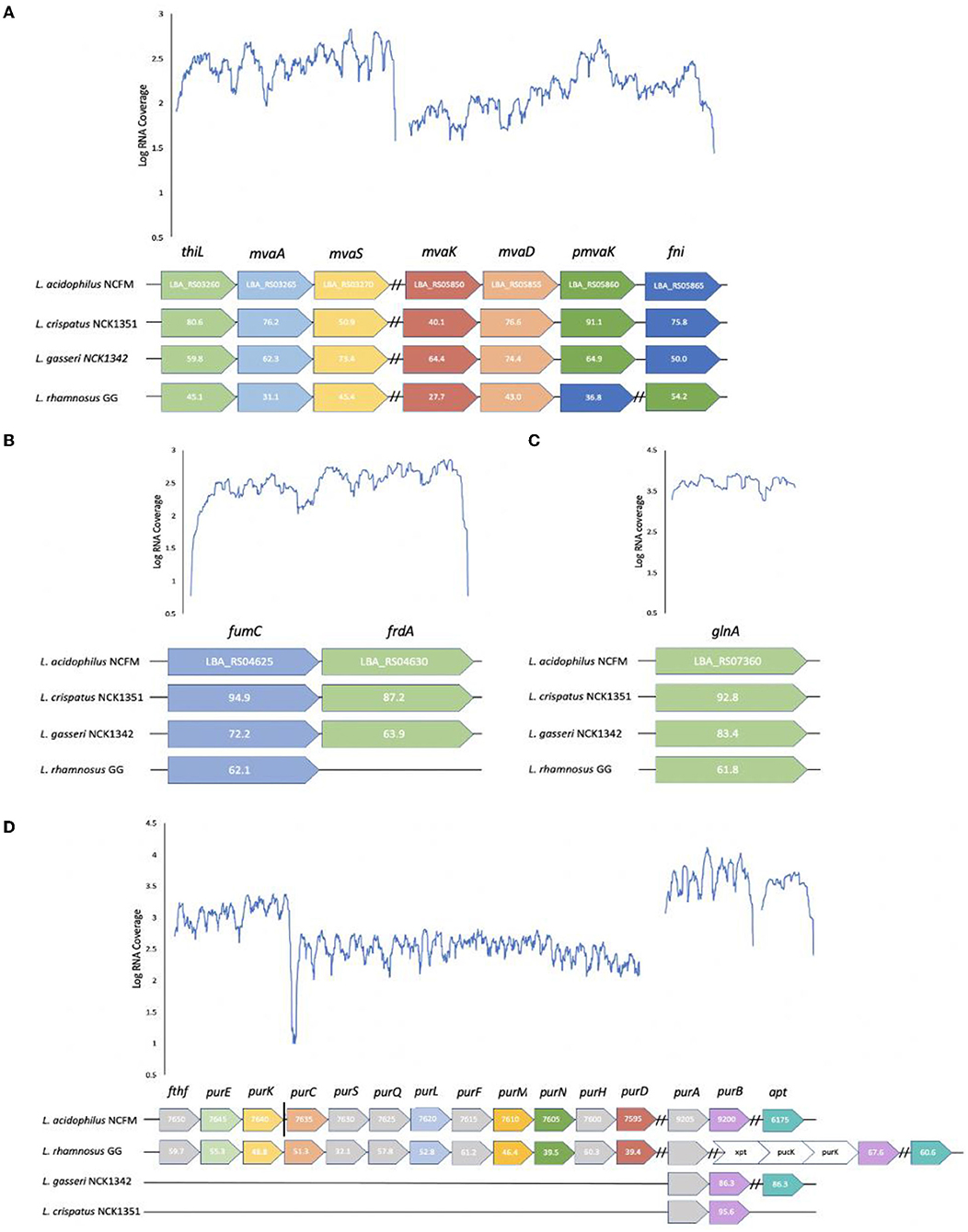
Figure 6. Loci comparison across strains and transcriptional profiles of L. acidophilus NCFM for defined loci in metabolite pathways. Conserved loci and amino acid sequence identities relative to L. acidophilus NCFM are shown in select Lactobacillus strains for (A) mevalonate (B) fumarate (C) glutamine, and (D) 5-aminoimidazole-4-carboxamide. The gray and white arrows represent genes that are not required in the specific metabolic pathway but are located within the operon. The vertical solid black line distinguishes between operons but loci are adjacent to one another on the chromosome. Dashed black lines distinguish between operons on separate parts of the chromosome.
As mentioned previously, fumarate is an intermediate compound in the TCA cycle which is incomplete in lactic acid bacteria (LAB). The active portion of the partial TCA cycle consists of two enzymes that are encoded by a fumarate hydratase (fumC; LBA_RS04630) and a fumarate reductase (frdA; LBA_RS04625) (Figure 6B); however, L. rhamnosus is lacking a fumarate reductase. Fumarate is produced from malate with a fumarate hydratase and from succinate with a fumarate reductase. Not surprisingly, L. rhamnosus produced the smallest amount of succinate and fumarate but had a higher production of malate. Of the three TCA cycle intermediates, L. acidophilus had the highest malate fold change, L. crispatus had the highest succinate fold change, and L. gasseri had the highest fumarate fold change.
The operon involved in the synthesis of glutamine is relatively simple compared with the other metabolites as it only encodes a glutamine synthetase (glnA; LBA_RS07360) in its own locus (Figure 6C). The difference between the strains is reflected in the fold change as L. acidophilus had a fold change value of 63.33, while L. rhamnosus had a value of only 0.42. This could perhaps reflect the potential of L. acidophilus having a more active glutamine synthetase and/or a more highly expressed gene.
The 5-aminoimidazole-4-carboxamide biosynthetic genes are scattered on the chromosome in four transcriptional units for L. acidophilus and L. rhamnosus (Figure 6D). The first locus is composed of formate-tetrahydrofolate ligase (fthf; LBA_RS07650), 5-(carboxyamino) imidazole ribonucleotide mutase (purE; LBA_RS07645), and 5-(carboxyamino) imidazole ribonucleotide synthase (purK; LBA_RS07640). Although fthf is within the operon, it is not required for the production of 5-aminoimidazole-4-carboxamide. The second locus consists of phosphoribosylaminoimidazolesuccinocarboxamide synthase (purC; LBA_RS07635), phosphoribosylformylglycinamidine (pur) synthase subunit purS (LBA_RS07630), pur synthase subunit purQ (LBA_RS07625), pur synthase subunit purL (LBA_RS07620), amidophosphoribosyltransferase (purF; LBA_RS07615), pur cyclo-ligase (purM; LBA_RS07610), phosphoribosylglycinamide formyltransferase (purN; LBA_RS07605), bifunctional AICAR transformylase/IMP cyclohydrolase (purH; LBA_RS07600), and phosphoribosylamine-glycine ligase (purD; LBA_RS07595). The third locus includes adenylosuccinate synthase (purA; LBA_RS09205) and adenylosuccinate lyase (purB; LBA_RS09200), while the fourth locus consists of an adenine phosphoribosyltransferase (aprT; LBA_RS06175) in its own operon. Genes purS, purQ, purF, purH, and purA are not involved in this metabolic pathway. Most lactobacilli are auxotrophic for both purines and pyrimidines and utilize salvage pathways in order to convert the necessary nucleobases or nucleosides to nucleotides (Kilstrup et al., 2005). 5-aminoimidazole-4-carboxamide is an intermediate in the purine metabolism pathway, and the majority of the necessary genes to produce this metabolite are found in L. rhamnosus and L. acidophilus but not in L. gasseri and L. crispatus. The purA and purB genes are conserved in all the strains in their own operon, except for L. rhamnosus which has purA in its own operon and purB in an operon with xpt (xanthine phosphoribosyltransferase), pucK (uric acid permease), and another purK gene.
The human gut microbiome can influence overall host health based on the microbial composition and biochemical function. Lactobacillus species are prominent microorganisms native to the GIT that help maintain a homeostatic environment. Attaining a balanced microbiota can be modulated with the consumption of probiotic bacteria through foods and use of probiotic supplements, which are typically composed of Lactobacillus and Bifidobacterium species. The GIT microbiota can also be positively impacted through dietary changes with the consumption of prebiotic substrates to selectively stimulate the growth of beneficial commensal microorganisms. This study aimed to investigate the metabolic profiles of four Lactobacillus species when grown in the presence of pomegranate extract. Some compounds previously reported as a derived metabolite from ellagitannins, including urolithins and ellagic acid, were not detected.
Evolutionary adaptions to niche environments have enabled Lactobacillus species to specialize and diversify their metabolic capabilities. Lactobacillus species are naturally found within human cavities, including the oral, gastrointestinal, and vaginal cavity. L. acidophilus, L. rhamnosus, L. gasseri, and L. crispatus species are found in all of these human environments (Klaenhammer et al., 2005; Selle and Klaenhammer, 2013; Ghosh et al., 2020). The four strains in this study were investigated to analyze two commonly used probiotics (L. acidophilus NCFM and L. rhamnosus GG) and two potential probiotics (L. crispatus NCK1351 and L. gasseri NCK1342) in combination with a polyphenolic substrate.
Lactobacilli are typically low G+C, facultative anaerobes that are fastidious in nature. From the growth assay results, when grown in SDM and SDM with POM, the strains exhibited similar growth patterns with L. acidophilus demonstrating a higher CFU/ml after 24 h in SDM with POM. These data show that the POM concentration of 400 μg/ml does not produce a toxic environment for these strains. To further differentiate between the strains and confirm their basic saccharolytic and enzymatic capabilities, a carbohydrate utilization and enzyme activity assay was performed. Comparative genome analyses indicate that species that adapt to specific environmental niches tend to have a simplified metabolic capacity, such as L. gasseri, while species from diverse niches, like L. rhamnosus, contain vast regulatory and transport functions (Goh and Klaenhammer, 2009; De Angelis et al., 2016). L. acidophilus species are commonly isolated from acidic environments and are commercially used in a variety of dairy products. L. crispatus and L. gasseri are among six predominant species that belong to the L. acidophilus complex based on similar metabolic and functional properties (Selle et al., 2014). L. rhamnosus is a ubiquitous species that has been isolated from a range of bodily habitats as well as certain fermented dairy products and has also been associated with beer spoilage (Bernardeau et al., 2008). Results from the carbohydrate utilization and enzyme API assays showed L. rhamnosus was able to ferment the widest range of sugars and had the broadest enzyme activity. However, L. rhamnosus GG was not able to utilize lactose, a common substrate forLAB, due to frameshifts in the antiterminator (lacT) and 6-phospho-β-galactosidase (lacG) genes (Kankainen et al., 2009). Glycosyl hydrolases are key enzymes of carbohydrate metabolism that include α- and β-glucosidase, α- and β-galactosidase, β-glucuronidase, α-mannosidase, α-fucosidase, and N-acetyl-β-glucosaminidase from the enzyme activity assay. All of the strains showed strong activity for the α- and β-glucosidases and β-galactosidase, and these results were expected as one of the key features of gastrointestinal lactobacilli is their prominent saccharolytic capabilities. Interestingly, L. rhamnosus was the only strain with α-fucosidase activity and has been reported as one of the only sequenced intestinal lactobacilli with this enzyme (Morita et al., 2009). α-fucosidases cleave biological substrates that contain fucosyl-oligosaccharides into L-fucose compounds, which have been suggested to have protective roles in the gut and systemic infection and inflammation (Pickard and Chervonsky, 2015).
Untargeted metabolomic analysis of cell-free supernatants from the four Lactobacillus strains offered insights into interconnected metabolic pathways and the production of unique metabolites. Phylogenetic analyses of Lactobacillus species have identified key genes acquired through horizontal gene transfer (HGT) and the loss of non-essential genes as the LAB coevolved with their habitats (Makarova et al., 2006; O'sullivan et al., 2009). The complete mevalonate pathway might have been acquired through HGT, and the specific organization of genes in this pathway is conserved in a single operon in most Lactobacillales genomes (Makarova et al., 2006). In these strains, this pattern is observed in L. acidophilus, L. crispatus, and L. gasseri; however, L. rhamnosus has a phosphomevalonate kinase in a separate operon. Lactobacilli are able to utilize various substrates for growth, primarily amino acids, nucleotides, carbohydrates, and lipids. From the HCA (Figure 3A), L. acidophilus and L. gasseri had the most similar metabolite profiles, followed by L. crispatus, and then L. rhamnosus. This is not surprising, as L. gasseri was previously classified as L. acidophilus up until 1980 due to similar phenotypical characteristics, but technological advances with DNA/DNA hybridization proved they are distantly related (Lauer and Kandler, 1980). The top 50 highest fold change value metabolites among the strains were selected and displayed in a heat map (Figure 3A). From these selected metabolites, most of the compounds were derived from the amino acid super pathway, followed by the carbohydrate, and then the lipid super pathway. Most Lactobacillus species are auxotrophic for amino acids and are incapable of synthesizing them de novo. They compensate for this shortcoming with various amino acid and peptide uptake systems and peptidases to effectively utilize exogenous nitrogen sources (Goh and Klaenhammer, 2009). Once LAB acquire amino acids, various enzymes such as aminotransferases and decarboxylases play an important role in these catabolic pathways (Fernández and Zúñiga, 2006). PLA was the highest fold change value metabolite within the amino acid super pathway for all the strains except L. rhamnosus, which had N-acetylisoleucine as the highest produced compound followed by PLA (Figure 4). PLA is converted from phenylpyruvate with a phenyllactate dehydrogenase, which has not been annotated in any of these Lactobacillus species. However, a protein blast with a potential ortholog from C. sporogenes showed that there is potential enzymatic capacity. Many other lactobacilli have the ability to convert phenylalanine to PLA (Valerio et al., 2004), which can be considered a desirable trait. Several studies have reported PLA to have inhibitory effects on several fungal species and bacterial contaminants found in humans and food products, including Listeria monocytogenes, Staphylococcus aureus, and Enterococcus faecalis (Dieuleveux and Guéguen, 1998; Dieuleveux et al., 1998; Lavermicocca et al., 2000).
Glutamine and 5-aminoimidazole-4-carboxamide are derived from glutamate and were produced the most by L. acidophilus. Glutamine synthetase, glnA, catalyzes the formation of L-glutamine by adding ammonia (NH3) to L-glutamate. This reaction plays a crucial role in the regulation of nitrogen metabolism (Siragusa et al., 2014). In addition, GlnA has been reported to be involved in acid stress resistance of L. rhamnosus GG (Koponen et al., 2012). These strains all contain a glnA gene, with L. crispatus having the most sequence similarity to L. acidophilus, followed by L. gasseri and L. rhamnosus. Glutamine is an amino acid found in abundance within the human body that has been shown to have an important role in intestinal physiology (Kim and Kim, 2017). The beneficial role of glutamine in the intestine has been observed by the promotion of enterocyte proliferation, regulation of tight junction proteins, and suppression of proinflammatory signaling pathways (Rhoads et al., 1997; Demarco et al., 2003; Li et al., 2004; Xue et al., 2011).
Purine nucleotides are synthesized de novo and result in the formation of inosine monophosphate (IMP), which then branches into two separate routes that lead to adenosine monophosphate (AMP) or guanosine monophosphate (GMP) (Kilstrup et al., 2005). Prior to IMP formation, 5-aminoimidazole-4-carboxamide can be produced in a 10-step reaction sub-pathway in purine biosynthesis. L. acidophilus and L. rhamnosus encode all of the genes necessary to produce 5-aminoimidazole-4-carboxamide in this specific pathway, while L. gasseri and L. crispatus encode two and one genes, respectively. Although L. gasseri and L. crispatus do not encode all the necessary genes to produce 5-aminoimidazole-4-carboxamide, they can still produce IMP from alternative pathways. Surprisingly, L. gasseri and L. crispatus only contain two purine biosynthesis genes, purA and purB. purB encodes an adenylosuccinate lyase that catalyzes the conversion of succinyl aminoimidazole carboxamide ribotide (SAICAR) into aminoimidazole carboxamide ribotide (AICAR) in the de novo purine synthesis pathway and the conversion of succinyl-AMP (sAMP) to AMP (Singer et al., 2016). Gene purA encodes an adenylosuccinate synthase and performs the reverse reaction of purB by converting sAMP to AMP.
As mentioned earlier, LAB are obligate fermentative organisms, and therefore, they have a partial or incomplete TCA cycle. However, the L. acidophilus, L. gasseri, and L. crispatus genomes encode fumarate reductase and fumarate hydratase that enable the production of fumarate, malate, and succinate. Fumarate reductase is a key enzyme in anaerobic respiration that uses fumarate as the terminal electron acceptor. This is a common property among gram-negative bacteria and some facultative anaerobic gram-positive bacteria (Van Hellemond and Tielens, 1994). L. rhamnosus lacks a fumarate reductase that would inhibit its ability to perform anaerobic respiration via this pathway and reduce its ability to produce fumarate.
Secondary metabolites are organic compounds that are not directly involved in normal growth, development, or reproduction but may confer a selective advantage to bacteria (Craney et al., 2013). Isoprenoids are a diverse group of secondary metabolites that have important biological functions in bacteria such as with electron transport and as cell wall biosynthesis intermediates (Kuzuyama and Seto, 2012). All isoprenoids are synthesized by the condensation of isopentenyl diphosphate (IPP) to dimethylallyl diphosphate (DMAPP). The mevalonate pathway results in the formation of IPP from acetyl-CoA and has been established mainly in mice and Saccharomyces cerevisiae (Kuzuyama and Seto, 2012). Interestingly, most prokaryotes lack the gene homologs of the mevalonate pathway and utilize the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway (Kuzuyama and Seto, 2012). However, all the Lactobacillus strains in this study exhibited the necessary genes for the production of IPP through the mevalonate pathway. Mevalonate was the highest produced secondary metabolite for all strains, followed by mevalonolactone, which is the dehydrated product of mevalonic acid.
The untargeted metabolomic analysis from this study offered insights into the metabolic pathways of several Lactobacillus strains grown in the presence of pomegranate. These species have evolved to adapt to their specific environmental niches, which has altered their metabolic capacities. However, these species are all inhabitants of the human GIT and therefore exhibit some of the same metabolic characteristics. Previous studies have identified specific metabolites derived from pomegranate ellagitannins that may have beneficial effects on the human host, such as urolithins and ellagic acid (Bialonska et al., 2009, 2010; Li et al., 2015). Although these specific compounds were not detected, many other metabolites that are unique to each strain and can contribute to the host health were observed. The production of these metabolites could enhance the probiotic effect of these strains by providing additional health benefits. Understanding specific metabolic processes is useful to mechanistically understand probiotic function in the GIT and is essential in selecting specific probiotic strains in formulation development.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
MC performed the experiments and analyses and wrote the manuscript. SO'F designed the experiments, prepared the samples for metabolomics, and wrote the manuscript. NC designed the experiments and edited the manuscript. RB provided input on the experimental design and edited the manuscript. The data presented were part of MC's MS Thesis (https://repository.lib.ncsu.edu/handle/1840.20/39039). All authors contributed to the article and approved the submitted version.
This work was funded by Elysium Health LLC and the North Carolina Agricultural Foundation. The funder Elysium Health LLC was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the members of the Barrangou Lab, in particular Meichen Pan, for helpful suggestions.
Bernardeau, M., Vernoux, J. P., Henri-Dubernet, S., and Guéguen, M. (2008). Safety assessment of dairy microorganisms: the Lactobacillus genus. Int. J. Food Microbiol. 126, 278–285. doi: 10.1016/j.ijfoodmicro.2007.08.015
Bernini, P., Bertini, I., Luchinat, C., Nepi, S., Saccenti, E., Schäfer, H., et al. (2009). Individual human phenotypes in metabolic space and time. J. Proteome Res. 8, 4264–4271. doi: 10.1021/pr900344m
Bialonska, D., Kasimsetty, S. G., Schrader, K. K., and Ferreira, D. (2009). The effect of pomegranate (Punica granatum L.) byproducts and ellagitannins on the growth of human gut bacteria. J. Agric. Food Chem. 57, 8344–8349. doi: 10.1021/jf901931b
Bialonska, D., Ramnani, P., Kasimsetty, S. G., Muntha, K. R., Gibson, G. R., and Ferreira, D. (2010). The influence of pomegranate by-product and punicalagins on selected groups of human intestinal microbiota. Int. J. Food Microbiol. 140, 175–182. doi: 10.1016/j.ijfoodmicro.2010.03.038
Bintsis, T. (2018). Lactic acid bacteria as starter cultures: an update in their metabolism and genetics. AIMS Microbiol. 4, 665–684. doi: 10.3934/microbiol.2018.4.665
Blum, S., Reniero, R., Schiffrin, E. J., Crittenden, R., Mattila-Sandholm, T., Ouwehand, A. C., et al. (1999). Adhesion studies for probiotics: need for validation and refinement. Trends Food Sci. Technol. 10, 405–410. doi: 10.1016/S0924-2244(00)00028-5
Cambeiro-Perez, N., Hidalgo-Cantabrana, C., Moro-Garcia, M. A., Alonso-Arias, R., Simal-Gandara, J., Sanchez, B., et al. (2018). A metabolomics approach reveals immunomodulatory effects of proteinaceous molecules derived from gut bacteria over human peripheral blood mononuclear cells. Front. Microbiol. 9, 2701. doi: 10.3389/fmicb.2018.02701
Cambeiro-Perez, N., Hidalgo-Cantabrana, C., Moro-Garcia, M. A., Blanco-Miguez, A., Fdez-Riverola, F., Riestra, S., et al. (2020). In silico and functional analyses of immunomodulatory peptides encrypted in the human gut metaproteome. J. Funct. Foods 70, 103969. doi: 10.1016/j.jff.2020.103969
Cheng, W., Lu, J., Lin, W., Wei, X., Li, H., Zhao, X., et al. (2018). Effects of a galacto-oligosaccharide-rich diet on fecal microbiota and metabolite profiles in mice. Food Funct. 9, 1612–1620. doi: 10.1039/C7FO01720K
Craney, A., Ahmed, S., and Nodwell, J. (2013). Towards a new science of secondary metabolism. J. Antibiot. 66, 387–400. doi: 10.1038/ja.2013.25
De Angelis, M., Calasso, M., Cavallo, N., Di Cagno, R., and Gobbetti, M. (2016). Functional proteomics within the genus Lactobacillus. Proteomics 16, 946–962. doi: 10.1002/pmic.201500117
Dehaven, C. D., Evans, A. M., Dai, H., and Lawton, K. A. (2010). Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J. Cheminform. 2, 9. doi: 10.1186/1758-2946-2-9
Demarco, V. G., Li, N., Thomas, J., West, C. M., and Neu, J. (2003). Glutamine and barrier function in cultured Caco-2 epithelial cell monolayers. J. Nutr. 133, 2176–2179. doi: 10.1093/jn/133.7.2176
Dettmer, K., Aronov, P. A., and Hammock, B. D. (2007). Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 26, 51–78. doi: 10.1002/mas.20108
Dieuleveux, V., and Guéguen, M. (1998). Antimicrobial effects of d-3-phenyllactic acid on listeria monocytogenes in TSB-YE medium, milk, and cheese. J. Food Prot. 61, 1281–1285. doi: 10.4315/0362-028X-61.10.1281
Dieuleveux, V., Lemarinier, S., and Guéguen, M. (1998). Antimicrobial spectrum and target site of d-3-phenyllactic acid. Int. J. Food Microbiol. 40, 177–183. doi: 10.1016/S0168-1605(98)00031-2
Evans, A., Mitchell, M., Dai, H., and Dehaven, C. (2012). Categorizing ion -features in liquid chromatography/mass spectrometry metobolomics data. Metabolomics 2. doi: 10.4172/2153-0769.1000110
Fernández, M., and Zúñiga, M. (2006). Amino acid catabolic pathways of lactic acid bacteria. Crit. Rev. Microbiol. 32, 155–183. doi: 10.1080/10408410600880643
Garcia-Muñoz, C., and Vaillant, F. (2014). Metabolic fate of ellagitannins: implications for health, and research perspectives for innovative functional foods. Crit. Rev. Food Sci. Nutr. 54, 1584–1598. doi: 10.1080/10408398.2011.644643
Ghini, V., Tenori, L., Pane, M., Amoruso, A., Marroncini, G., Squarzanti, D. F., et al. (2020). Effects of probiotics administration on human metabolic phenotype. Metabolites 10, 396. doi: 10.3390/metabo10100396
Ghosh, T. S., Arnoux, J., and O'toole, P. W. (2020). Metagenomic analysis reveals distinct patterns of gut lactobacillus prevalence, abundance, and geographical variation in health and disease. Gut Microbes 12, 1–19. doi: 10.1080/19490976.2020.1822729
Gibson, G. R., Hutkins, R., Sanders, M. E., Prescott, S. L., Reimer, R. A., Salminen, S. J., et al. (2017). Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14, 491–502. doi: 10.1038/nrgastro.2017.75
Goh, Y., and Klaenhammer, T. R. (2009). Genomic features of Lactobacillus species. Front. Biosci. 14, 1362–1386. doi: 10.2741/3313
Goh, Y. J., and Klaenhammer, T. R. (2015). Genetic mechanisms of prebiotic oligosaccharide metabolism in probiotic microbes. Annu. Rev. Food Sci. Technol. 6, 137–156. doi: 10.1146/annurev-food-022814-015706
Gonthier, M.-P., Cheynier, V. R., Donovan, J. L., Manach, C., Morand, C., Mila, I., et al. (2003). Microbial aromatic acid metabolites formed in the gut account for a major fraction of the polyphenols excreted in urine of rats fed red wine polyphenols. J. Nutr. 133, 461–467. doi: 10.1093/jn/133.2.461
Heber, D., Seeram, N. P., Wyatt, H., Henning, S. M., Zhang, Y., Ogden, L. G., et al. (2007). Safety and antioxidant activity of a pomegranate ellagitannin-enriched polyphenol dietary supplement in overweight individuals with increased waist size. J. Agric. Food Chem. 55, 10050–10054. doi: 10.1021/jf071689v
Henning, S. M., Summanen, P. H., Lee, R. P., Yang, J., Finegold, S. M., Heber, D., et al. (2017). Pomegranate ellagitannins stimulate the growth of Akkermansia muciniphila in vivo. Anaerobe 43, 56–60. doi: 10.1016/j.anaerobe.2016.12.003
Holscher, H. D. (2017). Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 8, 172–184. doi: 10.1080/19490976.2017.1290756
Hong, Y.-S., Hong, K. S., Park, M.-H., Ahn, Y.-T., Lee, J.-H., Huh, C.-S., et al. (2011). Metabonomic understanding of probiotic effects in humans with irritable bowel syndrome. J. Clin. Gastroenterol. 45, 415–425. doi: 10.1097/MCG.0b013e318207f76c
Huttenhower, C., Gevers, D., Knight, R., Abubucker, S., Badger, J. H., Chinwalla, A. T., et al. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Justesen, U., Arrigoni, E., Larsen, B. R., and Amado, R. (2000). Degradation of flavonoid glycosides and aglycones during in vitro fermentation with human Faecal Flora. LWT Food Sci. Technol. 33, 424–430. doi: 10.1006/fstl.2000.0681
Kankainen, M., Paulin, L., Tynkkynen, S., Von Ossowski, I., Reunanen, J., Partanen, P., et al. (2009). Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc. Natl. Acad. Sci. U. S. A. 106, 17193–17198. doi: 10.1073/pnas.0908876106
Kilstrup, M., Hammer, K., Ruhdal Jensen, P., and Martinussen, J. (2005). Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol. Rev. 29, 555–590. doi: 10.1016/j.fmrre.2005.04.006
Kim, J., John, K. M. M., Kusano, M., Oikawa, A., Saito, K., and Lee, C. H. (2012). GC–TOF-MS- and CE–TOF-MS-based metabolic profiling of cheonggukjang (fast-fermented bean paste) during fermentation and its correlation with metabolic pathways. J. Agric. Food Chem. 60, 9746–9753. doi: 10.1021/jf302833y
Kim, M.-H., and Kim, H. (2017). The roles of glutamine in the intestine and its implication in intestinal diseases. Int. J. Mol. Sci. 18, 1051. doi: 10.3390/ijms18051051
Kimmel, S. A., and Roberts, R. F. (1998). Development of a growth medium suitable for exopolysaccharide production by Lactobacillus delbrueckii ssp. bulgaricus RR. Int. J. Food Microbiol. 40, 87–92. doi: 10.1016/S0168-1605(98)00023-3
Klaenhammer, T. R., Barrangou, R., Buck, B. L., Azcarate-Peril, M. A., and Altermann, E. (2005). Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol. Rev. 29, 393–409. doi: 10.1016/j.fmrre.2005.04.007
Koponen, J., Laakso, K., Koskenniemi, K., Kankainen, M., Savijoki, K., Nyman, T. A., et al. (2012). Effect of acid stress on protein expression and phosphorylation in Lactobacillus rhamnosus GG. J. Proteomics 75, 1357–1374. doi: 10.1016/j.jprot.2011.11.009
Kuzuyama, T., and Seto, H. (2012). Two distinct pathways for essential metabolic precursors for isoprenoid biosynthesis. Proc. Jpn. Acad,. Ser. B. Phys. Biol. Sci. 88, 41–52. doi: 10.2183/pjab.88.41
Lauer, E., and Kandler, O. (1980). Lactobacillus gasseri sp. nov., a new species of the subgenus Thermobacterium. Zentralblatt Bakteriol. 1, 75–78. doi: 10.1016/S0172-5564(80)80019-4
Lavermicocca, P., Valerio, F., Evidente, A., Lazzaroni, S., Corsetti, A., and Gobbetti, M. (2000). Purification and characterization of novel antifungal compounds from the sourdough andlt;emandgt;Lactobacillus plantarumandlt;/emandgt; Strain 21B. Appl. Environ. Microbiol. 66, 4084. doi: 10.1128/AEM.66.9.4084-4090.2000
Lawton, K. A., Berger, A., Mitchell, M., Milgram, K. E., Evans, A. M., Guo, L., et al. (2008). Analysis of the adult human plasma metabolome. Pharmacogenomics 9, 383–397. doi: 10.2217/14622416.9.4.383
Lee, J.-E., Hong, Y.-S., and Lee, C.-H. (2009). Characterization of fermentative behaviors of lactic acid bacteria in grape wines through 1H NMR- and GC-based metabolic profiling. J. Agric. Food Chem. 57, 4810–4817. doi: 10.1021/jf900502a
Li, N., Lewis, P., Samuelson, D., Liboni, K., and Neu, J. (2004). Glutamine regulates Caco-2 cell tight junction proteins. Am. J. Physiol. Gastrointest. Liver Physiol. 287, G726–G733. doi: 10.1152/ajpgi.00012.2004
Li, Z., Summanen, P. H., Komoriya, T., Henning, S. M., Lee, R.-P., Carlson, E., et al. (2015). Pomegranate ellagitannins stimulate growth of gut bacteria in vitro: implications for prebiotic and metabolic effects. Anaerobe 34, 164–168. doi: 10.1016/j.anaerobe.2015.05.012
Likotrafiti, E., and Rhoades, J. (2016). “Chapter 32 - Probiotics, Prebiotics, Synbiotics, and Foodborne Illness,” in Probiotics, Prebiotics, and Synbiotics, eds. R. R. Watson and V. R. Preedy (Cambridge, MA: Academic Press) 469–476. doi: 10.1016/B978-0-12-802189-7.00032-0
López-Rituerto, E., Savorani, F., Avenoza, A., Busto, J. H., Peregrina, J. M., and Engelsen, S. B. (2012). Investigations of La Rioja terroir for wine production using 1H NMR metabolomics. J. Agric. Food Chem. 60, 3452–3461. doi: 10.1021/jf204361d
Makarova, K., Slesarev, A., Wolf, Y., Sorokin, A., Mirkin, B., Koonin, E., et al. (2006). Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103, 15611–15616. doi: 10.1073/pnas.0607117103
Morita, H., Toh, H., Oshima, K., Murakami, M., Taylor, T. D., Igimi, S., et al. (2009). Complete genome sequence of the probiotic Lactobacillus rhamnosus ATCC 53103. J. Bacteriol. 191, 7630–7631. doi: 10.1128/JB.01287-09
Namgung, H.-J., Park, H.-J., Cho, I. H., Choi, H.-K., Kwon, D.-Y., Shim, S.-M., et al. (2010). Metabolite profiling of doenjang, fermented soybean paste, during fermentation. J. Sci. Food Agric. 90, 1926–1935. doi: 10.1002/jsfa.4036
O'sullivan, O., O'callaghan, J., Sangrador-Vegas, A., Mcauliffe, O., Slattery, L., Kaleta, P., et al. (2009). Comparative genomics of lactic acid bacteria reveals a niche-specific gene set. BMC Microbiol. 9, 50. doi: 10.1186/1471-2180-9-50
Palomo, M., Gutiérrez, A. M., Pérez-Conde, M. C., Cámara, C., and Madrid, Y. (2014). Se metallomics during lactic fermentation of Se-enriched yogurt. Food Chem. 164, 371–379. doi: 10.1016/j.foodchem.2014.05.007
Park, S.-E., Yoo, S.-A., Seo, S.-H., Lee, K.-I., Na, C.-S., and Son, H.-S. (2016). GC–MS based metabolomics approach of Kimchi for the understanding of Lactobacillus plantarum fermentation characteristics. LWT Food Sci. Technol. 68, 313–321. doi: 10.1016/j.lwt.2015.12.046
Pickard, J. M., and Chervonsky, A. V. (2015). Intestinal fucose as a mediator of host-microbe symbiosis. J. Immunol. 194, 5588–5593. doi: 10.4049/jimmunol.1500395
Piras, C., Cesare Marincola, F., Savorani, F., Engelsen, S. B., Cosentino, S., Viale, S., et al. (2013). A NMR metabolomics study of the ripening process of the Fiore Sardo cheese produced with autochthonous adjunct cultures. Food Chem. 141, 2137–2147. doi: 10.1016/j.foodchem.2013.04.108
Rhoads, J. M., Argenzio, R. A., Chen, W., Rippe, R. A., Westwick, J. K., Cox, A. D., et al. (1997). L-glutamine stimulates intestinal cell proliferation and activates mitogen-activated protein kinases. Am. J. Physiol. Gastrointest. Liver Physiol. 272, G943–G953. doi: 10.1152/ajpgi.1997.272.5.G943
Rios, L. Y., Gonthier, M.-P., Rémésy, C., Mila, I., Lapierre, C., Lazarus, S. A., et al. (2003). Chocolate intake increases urinary excretion of polyphenol-derived phenolic acids in healthy human subjects. Am. J. Clin. Nutr. 77, 912–918. doi: 10.1093/ajcn/77.4.912
Rodríguez-Díaz, J., Monedero, V., and Yebra, M. J. (2011). Utilization of natural fucosylated oligosaccharides by three novel alpha-L-fucosidases from a probiotic Lactobacillus casei strain. Appl. Environ. Microbiol. 77, 703. doi: 10.1128/AEM.01906-10
Scalbert, A., and Williamson, G. (2000). Dietary intake and bioavailability of polyphenols. J Nutr. 130, 2073S−2085S. doi: 10.1093/jn/130.8.2073S
Selle, K., and Klaenhammer, T. R. (2013). Genomic and phenotypic evidence for probiotic influences of Lactobacillus gasseri on human health. FEMS Microbiol. Rev. 37, 915–935. doi: 10.1111/1574-6976.12021
Selle, K. M., Klaenhammer, T. R., and Russell, W. M. (2014). “LACTOBACILLUS | Lactobacillus acidophilus,” in Encyclopedia of Food Microbiology, 2nd Edn, eds C. A. Batt and M. L. Tortorello (Oxford: Academic Press), 412–417. doi: 10.1016/B978-0-12-384730-0.00179-8
Singer, H. S., Mink, J. W., Gilbert, D. L., and Jankovic, J. (2016). “Chapter 17 - inherited metabolic disorders with associated movement abnormalities,” in Movement Disorders in Childhood, 2nd Edn., eds H. S. Singer, J. W. Mink, D. L. Gilbert, and J. Jankovic (Boston, MA: Academic Press), 337–407. doi: 10.1016/B978-0-12-411573-6.00017-6
Siragusa, S., De Angelis, M., Calasso, M., Campanella, D., Minervini, F., Di Cagno, R., et al. (2014). Fermentation and proteome profiles of Lactobacillus plantarum strains during growth under food-like conditions. J. Proteomics 96, 366–380. doi: 10.1016/j.jprot.2013.11.003
Somashekaraiah, R., Shruthi, B., Deepthi, B. V., and Sreenivasa, M. Y. (2019). probiotic properties of lactic acid bacteria isolated from neera: a naturally fermenting coconut palm nectar. Front. Microbiol. 10, 1382. doi: 10.3389/fmicb.2019.01382
Urpi-Sarda, M., Monagas, M., Khan, N., Lamuela-Raventos, R. M., Santos-Buelga, C., Sacanella, E., et al. (2009). Epicatechin, procyanidins, and phenolic microbial metabolites after cocoa intake in humans and rats. Anal. Bioanal. Chem. 394, 1545–1556. doi: 10.1007/s00216-009-2676-1
Valerio, F., Lavermicocca, P., Pascale, M., and Visconti, A. (2004). Production of phenyllactic acid by lactic acid bacteria: an approach to the selection of strains contributing to food quality and preservation. FEMS Microbiol. Lett. 233, 289–295. doi: 10.1111/j.1574-6968.2004.tb09494.x
Van Hellemond, J. J., and Tielens, A. G. (1994). Expression and functional properties of fumarate reductase. Biochem. J. 304(Pt 2), 321–331. doi: 10.1042/bj3040321
Vandeputte, D., Falony, G., Vieira-Silva, S., Wang, J., Sailer, M., Theis, S., et al. (2017). Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut 66, 1968–1974. doi: 10.1136/gutjnl-2016-313271
Xue, H., Sufit, A. J. D., and Wischmeyer, P. E. (2011). Glutamine therapy improves outcome of in vitro and in vivo experimental colitis models. J. Parenteral Enteral Nutr. 35, 188–197. doi: 10.1177/0148607110381407
Yang, J., Tang, Q., Xu, L., Li, Z., Ma, Y., and Yao, D. (2019). Combining of transcriptome and metabolome analyses for understanding the utilization and metabolic pathways of Xylo-oligosaccharide in Bifidobacterium adolescentis ATCC 15703. Food Sci. Nutr. 7, 3480–3493. doi: 10.1002/fsn3.1194
Zhao, N., Zhang, C., Yang, Q., Guo, Z., Yang, B., Lu, W., et al. (2016). Selection of taste markers related to lactic acid bacteria microflora metabolism for Chinese traditional Paocai: a gas chromatography–mass spectrometry-based metabolomics approach. J. Agric. Food Chem. 64, 2415–2422. doi: 10.1021/acs.jafc.5b05332
Keywords: metabolomics, Lactobacillus, probiotic, pomegranate extract, prebiotic
Citation: Chamberlain M, O'Flaherty S, Cobián N and Barrangou R (2022) Metabolomic Analysis of Lactobacillus acidophilus, L. gasseri, L. crispatus, and Lacticaseibacillus rhamnosus Strains in the Presence of Pomegranate Extract. Front. Microbiol. 13:863228. doi: 10.3389/fmicb.2022.863228
Received: 26 January 2022; Accepted: 06 April 2022;
Published: 18 May 2022.
Edited by:
Eric Guedon, Institut National de recherche pour l'agriculture, l'alimentation et l'environnement (INRAE), FranceReviewed by:
Elena Martínez Carballo, University of Vigo, SpainCopyright © 2022 Chamberlain, O'Flaherty, Cobián and Barrangou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rodolphe Barrangou, cmJhcnJhbkBuc2N1LmVkdQ==; Sarah O'Flaherty, c2pvZmxhaGVAbmNzdS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.