- 1Department of Biological Sciences, Misher College of Arts and Sciences, University of the Sciences in Philadelphia, Philadelphia, PA, United States
- 2Office of the Assistant Secretary for Health, Region 3, U.S. Department of Health and Human Services, Washington, DC, United States
- 3Department of Pharmacology and Physiology, Drexel University College of Medicine, Philadelphia, PA, United States
- 4Department of Infectious Disease and Vaccines, Merck & Co., Inc., Kenilworth, NJ, United States
- 5Center for Neuroimmunology and CNS Therapeutics, Institute of Molecular Medicine and Infectious Diseases, Drexel University of Medicine, Philadelphia, PA, United States
Although HIV-1 replication can be efficiently suppressed to undetectable levels in peripheral blood by combination antiretroviral therapy (cART), lifelong medication is still required in people living with HIV (PLWH). Life expectancies have been extended by cART, but age-related comorbidities have increased which are associated with heavy physiological and economic burdens on PLWH. The obstacle to a functional HIV cure can be ascribed to the formation of latent reservoir establishment at the time of acute infection that persists during cART. Recent studies suggest that some HIV reservoirs are established in the early acute stages of HIV infection within multiple immune cells that are gradually shaped by various host and viral mechanisms and may undergo clonal expansion. Early cART initiation has been shown to reduce the reservoir size in HIV-infected individuals. Memory CD4+ T cell subsets are regarded as the predominant cellular compartment of the HIV reservoir, but monocytes and derivative macrophages or dendritic cells also play a role in the persistent virus infection. HIV latency is regulated at multiple molecular levels in transcriptional and post-transcriptional processes. Epigenetic regulation of the proviral promoter can profoundly regulate the viral transcription. In addition, transcriptional elongation, RNA splicing, and nuclear export pathways are also involved in maintaining HIV latency. Although most proviruses contain large internal deletions, some defective proviruses may induce immune activation by expressing viral proteins or producing replication-defective viral-like particles. In this review article, we discuss the state of the art on mechanisms of virus persistence in the periphery and tissue and summarize interdisciplinary approaches toward a functional HIV cure, including novel capabilities and strategies to measure and eliminate the infected reservoirs and induce immune control.
Introduction
Human Immunodeficiency Virus Type-I (HIV-1) is the infectious agent that causes acquired immunodeficiency syndrome (AIDS) and remains a global concern due to the lack of effective vaccines and curative strategies (No Author, 2006). Despite the ability of combination antiviral therapy (cART) to effectively suppress plasma HIV-1 levels in the majority of people living with HIV (PLWH), HIV-1 persists by the formation of latent reservoirs established very early during the acute phase of HIV infection (Chun et al., 1997; Finzi et al., 1997; Holkmann Olsen et al., 2007; Kousignian et al., 2008; Wong et al., 2019). Furthermore, HIV-infected cells can undergo proliferation and over 50% of latently infected cells observed after prolonged cART are the product of clonal expansion (Ananworanich et al., 2015; Brodin et al., 2016; Yucha et al., 2017; Abrahams et al., 2019; Leite et al., 2019). Upon the cessation of cART, this reservoir can be reactivated and eventually leads to virus rebound in the majority of PLWH (Siliciano et al., 2003). Virus persistence necessitates lifelong cART for the 37.7 million PLWH and is possibly the last unmet challenge toward HIV cure/remission strategies. Persistence depends on multiple factors that are hypothesized to involve complex virus-host interactions. So far, there are two people who have been cured of HIV-1 infection, and one person who is undergoing long-term remission from HIV (Hütter et al., 2009; Allers et al., 2011; Gupta et al., 2019; Peluso et al., 2019). All of them had intercurrent leukemia and underwent a bone marrow transplant from donors who had a homozygous 32 base pair deletion in the CCR5 gene (CCR5-Δ32). Homozygous carriers of Δ32 deletion are largely resistant to HIV-1 infection because the mutation prevents functional expression of CCR5 that is used as a coreceptor for HIV-1 to enter immune cells. This mutation is primarily found in northern Europe because CCR5Δ32 selective pressures were thought to be induced by an intense smallpox epidemic in that region (Klitz et al., 2001; Galvani and Slatkin, 2003). Thus, hematopoietic stem cell transplant is currently the only successful strategy for a cure, but it is high risk and not scalable. Therefore, novel HIV curative strategies are urgently needed. One promising approach to eliminate cells that evade cART is the “kick-and-kill” strategy, where latency reversal agents (LRAs) reactivate the reservoir, allowing for immune cell recognition and elimination of these latently infected cells (Hamer, 2004). In contrast, the “Block-and-Lock” approach aims to induce permanent transcriptional and epigenetic proviral silencing, thereby preventing viral reactivation (Kessing et al., 2017). However, HIV-1 latency is still challenging for functional cure and remission. CRISPR-mediated genome editing is another promising approach to excise and deactivate the integrated HIV-1 DNA (Ebina et al., 2013; Hu et al., 2014). The United States Food and Drug Administration (FDA) recently approved clinical trials with a CRISPR-based strategy for HIV eradication (NCT05144386). Finally, modulation of the immune system has also been considered to address persistent reservoir elimination.
This review article discusses the obstacles to HIV eradication and multiple challenges for a functional cure of persistent HIV-1 infection recently highlighted in our research topic (Sardo et al., 2021).
Human Immunodeficiency Virus Persistence
Cellular Reservoir
The complexity and heterogeneity of HIV-1 reservoirs are caused by the establishment of latent infection into different cell types (Figure 1). HIV-1 primarily infects CD4+ T cells and the differentiation of T cells occurs in a stepwise fashion toward more differentiated cell types. The major cellular reservoirs for HIV-1 during cART reside in the memory CD4+ T cell subsets, especially central memory (TCM) subsets (Chomont et al., 2009; Buzon et al., 2014; Soriano-Sarabia et al., 2014; Ait-Ammar et al., 2020). Although having lower levels of integrated HIV-1 DNA due to their relative resistance to HIV-1 infection, the reactivation rate of latent proviruses in naive CD4+ T cells (TN) by LRAs is higher than in memory subsets (Ostrowski et al., 1999; Chomont et al., 2009; Wightman et al., 2010; Josefsson et al., 2013; Soriano-Sarabia et al., 2014). This suggests that the TN subset might also be an important contributor to viral rebound in PLWH. Latently infected cells can ultimately differentiate into an effector memory subset (TEM), an activated phenotype involved in antigen responses (von Stockenstrom et al., 2015). TEM harbors the largest proportion of intact-replication competent provirus (Hiener et al., 2017) and the largest inducible HIV-1 reservoir in vitro and ex vivo (Kulpa et al., 2019). Gamma delta (γδ) T cells that respond to non-peptide antigens through their distinct T-cell receptors bridge innate and adaptive immunity. The majority of γδ T cells in the blood are comprised of the Vδ2 subset, which also develops a memory phenotype. During HIV-1 infection, Vδ2 T-lymphocytes have been documented to be productively infected and depleted (Ait-Ammar et al., 2020). Vδ2 T cells isolated from HIV-1 infected individuals on prolonged cART contain replication-competent latent HIV-1 proviral DNA (Soriano-Sarabia et al., 2015). Although the memory subset of Vδ2 T cells has not been exclusively studied, peripheral Vδ2 T cells are also suggested to be a potential HIV-1 reservoir and should be targeted in curative approaches. While studies in the cellular reservoir have largely been conducted with the peripheral blood, memory subsets of CD4+ T cells containing HIV-1 sequence in gut-associated lymphoid tissue and lymph nodes (Tissue-resident memory T cells: TRM) were observed to contain 2–4 fold higher levels of HIV-1 DNA than the cells isolated from peripheral blood (Figure 1; Chun et al., 2008).
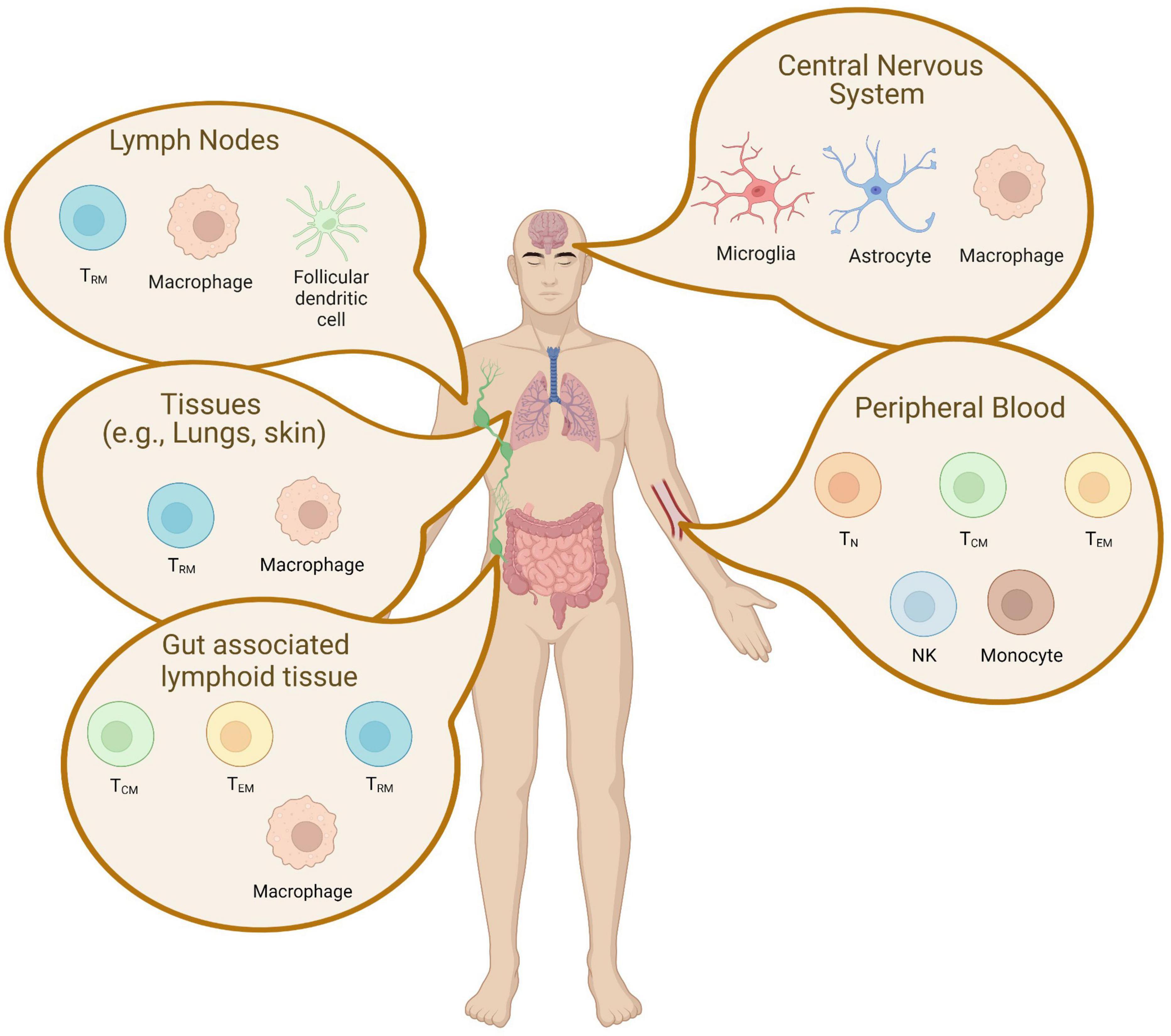
Figure 1. Latent cellular reservoir cells in tissue. A schematic representation of latently infected cells in tissues is depicted. The reservoirs are established in unique cell types and are localized across different tissues. CD4+ memory T cell subsets are found in the peripheral blood, the lymphoid tissue, gut-associated lymphoid tissue, and the central nervous system. This figure was created with BioRender.com.
Great effort to characterize the persistent reservoir cells has been put into identifying markers that could help determine enriched latently HIV-infected cells (Darcis et al., 2019). Some of the markers, including HLA-DR, CD25, CD32, and CD69, are upregulated in the HIV-infected CD4+ T cells (Darcis et al., 2019). On the other hand, CD2 receptor has been found to be expressed on latently infected resting memory CD4+ T cells in virally suppressed individuals (Iglesias-Ussel et al., 2013). The expression of α4β7 integrin has been shown in a T cell subset that is highly susceptible to HIV-1 infection (Ding et al., 2015). Despite the extensive investigation to characterize latently infected CD4+ T cells, a unique and universal marker of the HIV-1 reservoir cells has not been identified yet.
Although CD4+ T cells are the most studied among cellular reservoirs in the peripheral blood, other targets such as cells of the myeloid lineage contribute to persistence. Myeloid cells are thought to be of great importance for the pathogenesis of HIV-1 in the central nervous system (CNS) (Figure 1). As the CNS contains very few CD4+ T cells, CNS reservoirs predominantly include both microglia and perivascular macrophages (Iglesias-Ussel et al., 2013; Ding et al., 2015; Cantero-Pérez et al., 2019; Darcis et al., 2019; Ait-Ammar et al., 2020). Both of these cell types appear to continue to express viral genes despite suppressive cART therapy (Colomer-Lluch et al., 2018; Cantero-Pérez et al., 2019). Like T-cells in the periphery, infected myeloid cells in a tissue can contribute to viral rebound (Gibellini et al., 2008; Ding et al., 2015; Gosselin et al., 2017) and the development of escape mutations (Guihot et al., 2018; Grau-Expósito et al., 2019). Specific to the CNS, these persistently infected myeloid populations are also central drivers of the neuropathologic, behavioral, and cognitive effects collectively known as neuroHIV (Ho et al., 2013; Wightman et al., 2015; Kandathil et al., 2016; Perlman, 2016; Gosselin et al., 2017; Yucha et al., 2017; Darcis et al., 2019; Wong et al., 2019; Zerbato et al., 2019). Myeloid cells have a longer lifespan than lymphocytes and can continue to produce viruses long after initial infection (Perlman, 2016). In addition, myeloid cells are resistant to HIV-induced apoptosis (Morison et al., 2005; Coppé et al., 2010; Perlman, 2016) and myeloid-derived macrophages are resistant against type I IFN inducible innate immunity, and Cytotoxic T-lymphocytes (CTL) mediated immune responses (Cantero-Pérez et al., 2019). This is distinct from infected T cell populations in the periphery (Rangarajan and Weinberg, 2003; Akhtar, 2015) that also become a predominant reservoir.
Laboratory mouse strains play a distinct role in biomedical research due to having many similarities of anatomy and physiology to humans and their ability to be genetically manipulated. However, mice cannot be infected with HIV-1, and thus steps must be taken to modify this usual model organism for use in HIV-1 experimental infections. Humanized mice engrafted with human hematopoietic systems have proven to be versatile experimental models for studying the fundamental aspects of HIV biology (Monod and Jacob, 1961; Rangarajan and Weinberg, 2003; Morison et al., 2005; Coppé et al., 2010; Bailey et al., 2013; Honeycutt et al., 2017). Both viral DNA and RNA were detected in CD4+ T cells and macrophages in bone marrow, liver, and thymus (BLT) transplanted mice (Honeycutt et al., 2017). The latent reservoir formation in either CD4+ T cells or macrophages was previously demonstrated to be independently developed using T-cell-only or myeloid-only humanized mice (Yucha et al., 2017). This suggests that both cell types are capable of establishing a persistent reservoir. HIV-infected macrophages circulate and infiltrate into various tissue compartments including the brain in human myeloid-only mice, suggesting that myeloid cells may seed deep tissue reservoirs. While most human macrophages have a rapid turnover of approximately 1 day in myeloid-only mice, viral rebound could be observed in the infected mice post ART interruption (Honeycutt et al., 2017), suggesting that specialized long-lived macrophages, such as microglia to persistent in the CNS, may contribute to viral reservoir formation over time. Another in vivo model, for instance non-human primates, is needed to further examine the relevance of myeloid-derived cells in HIV-1 reservoir formation.
Mechanisms of Human Immunodeficiency Virus Type-I Latency
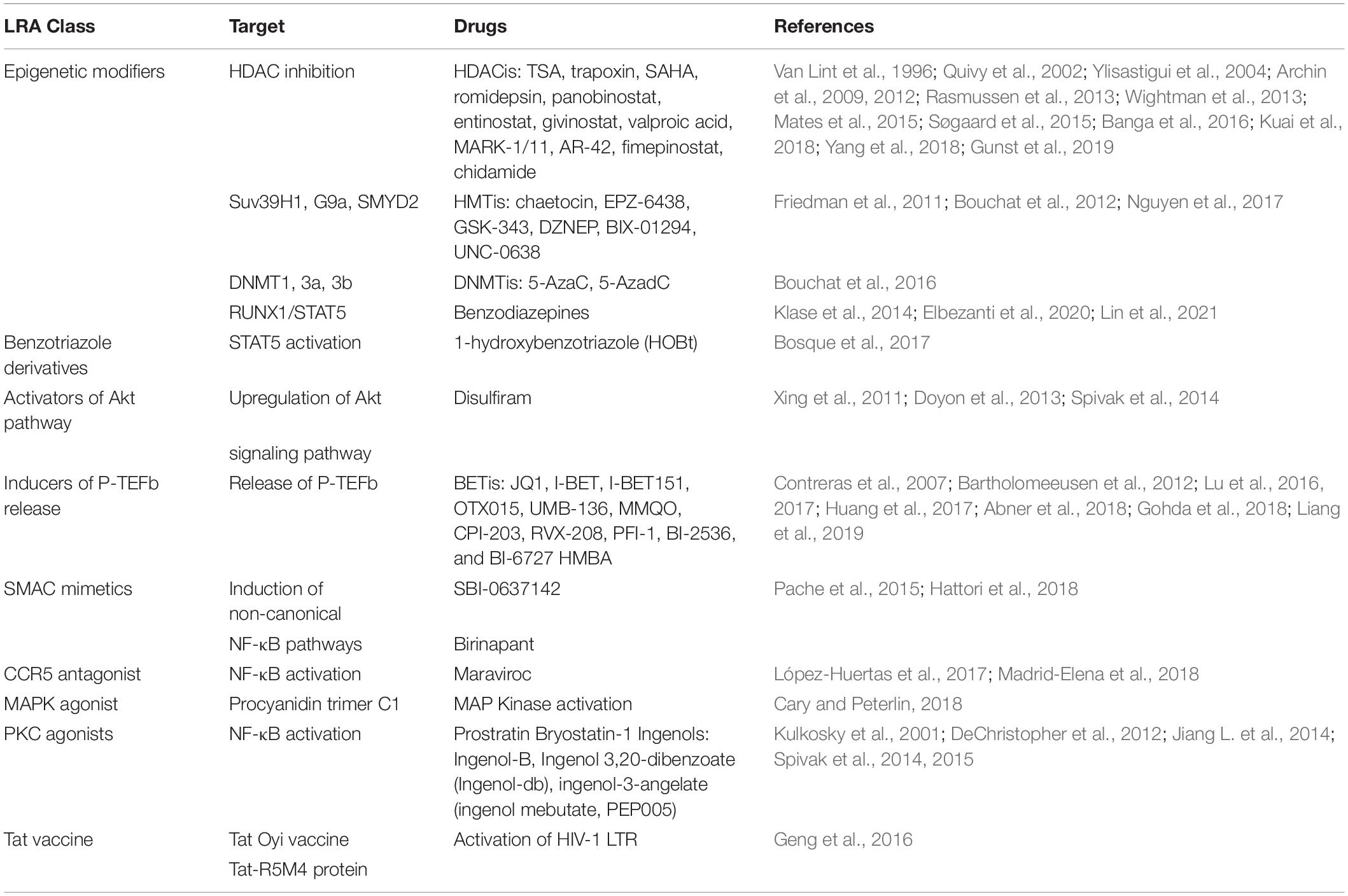
Silent proviruses in infected cells are termed “latent” and are established and maintained, in part, by epigenetic modifications and by transcription factors and signaling molecules that reinforce latency (Van Lint et al., 1996; Triboulet et al., 2007; Sung and Rice, 2009; Mbonye and Karn, 2017). In fact, HIV-1 latency is mediated by both cis- and trans-regulatory mechanisms that prevent transcription factors from accessing the viral promoter region (Figure 2; Hakre et al., 2012; Donahue and Wainberg, 2013; Ruelas and Greene, 2013; Van Lint et al., 2013). Chromatin environments are dominantly cis-regulated, while viral and host transcription factors are trans-regulated in latently infected cells (Besnard et al., 2016). Cis-acting mechanisms depend on the chromatin environment at the virus integration site within the host genome (Figure 2A; Dahabieh et al., 2015). Cis-regulatory elements work through intracellular interactions between different parts of the same molecule, such as promoters, enhancers, and silencers (Goncalves et al., 2012). These elements are located in the vicinity of the genes they regulate and act as binding sites for transcription factors to regulate transcription rates of nearby genes (Davidson, 2021). DNA methylation, histone methylation, acetylation and crotonylation are commonly found in the viral promoter region as cis-regulatory mechanisms in latently infected cells. On the other hand, trans-regulatory factors cooperate with cis-regulatory elements to suppress viral gene expression (McManus et al., 2010; Goncalves et al., 2012; Wang et al., 2019).
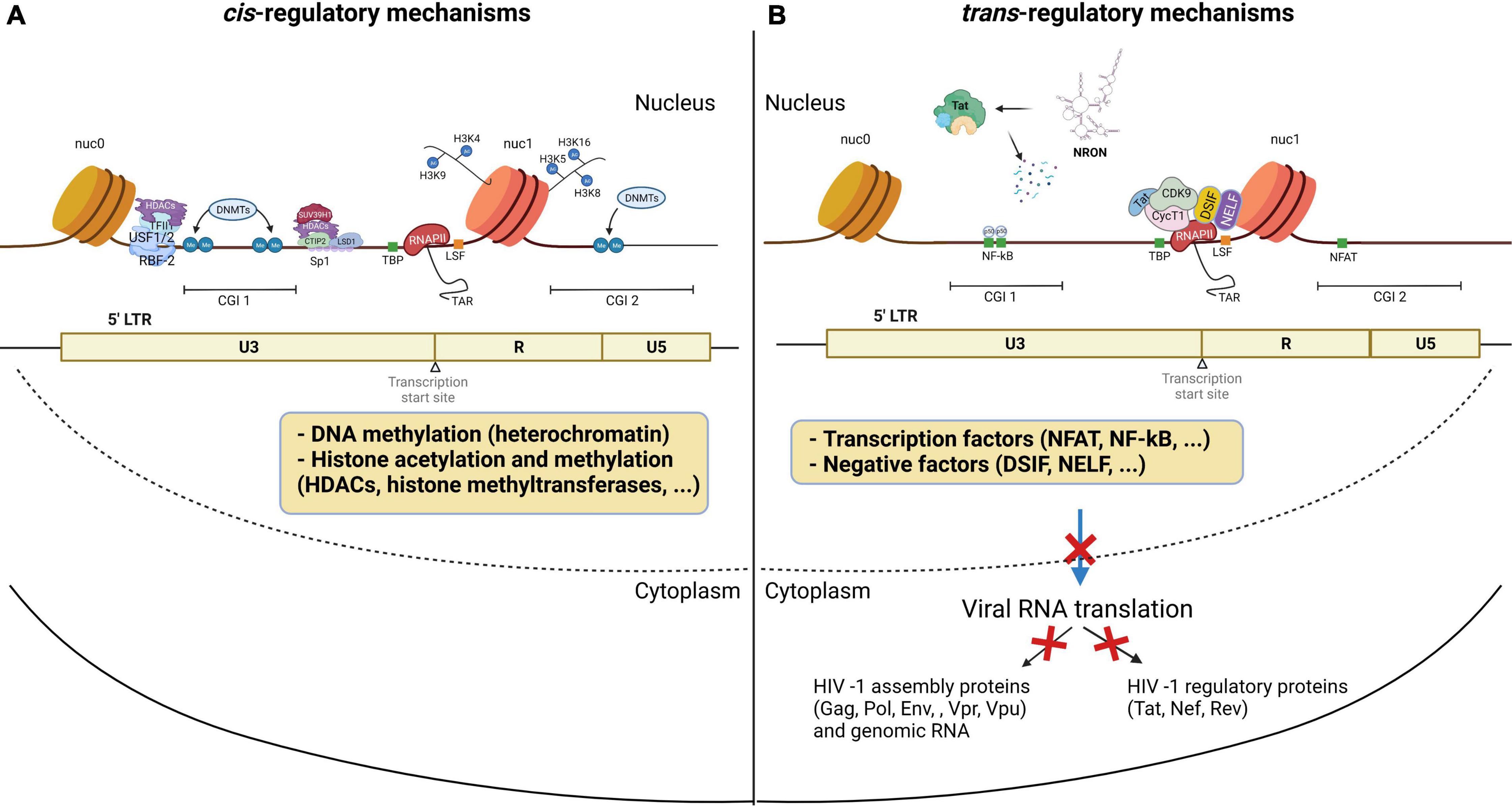
Figure 2. HIV-1 latency mechanisms. Latency is established through (A) cis-regulation elements, and (B) trans-regulation mechanisms predominantly regulate the HIV gene expression in the homeostatically proliferated reservoir cells at the chronic stage. (A) DNA methylation of the CG islands around the HIV-1 transcription start site maintains the HIV-1 promoter in a heterochromatic state to provoke HIV-1 transcriptional repression. Histone acetylation induces structural instability and increases access to transcriptional factors accessibility. (B) Transcription initiation is blocked by the low availability of NF-κB and phosphorylated NFAT at the promoter region. The binding of p50-p50 homodimers to the NF-κB binding site further inhibits transcription initiation. After initiation, RNA Pol II pauses at the promoter region due to the binding of negative elongation factors NELF and DSIF, leading to blocks in transcription elongation. In addition, P-TEFb is sequestered by the 7SK snRNP complex, causing a low expression level of Tat. The lncRNA called NRON degrades HIV-1 Tat to inhibit the P-TEFb formation. miRNAs targeting CyclinT1 regulate viral production and expression by overexpression in resting cells. This figure was created with BioRender.com.
Cellular epigenetic mechanisms to the cis-regulatory proviral regions have been extensively studied in peripheral CD4+ T cells. Clusters of CpG residues termed CG islands around the HIV-1 transcription site may be methylated, which would maintain the HIV-1 promoter in a heterochromatic state to repress HIV-1 transcription (Figure 2A; Blazkova et al., 2009; Kauder et al., 2009; Ruelas and Greene, 2013; Weber et al., 2014; Boltz et al., 2021). Histone acetyltransferases (HATs) are recruited to the viral long terminal repeat (LTR) and lead to increasing H3 and H4 lysine acetylation, specifically at H3K9, H3K4, H4K5, H4K8, and H4K16 residues (Benkirane et al., 1998; Hottiger and Nabel, 1998; Marzio et al., 1998; Col et al., 2001; Lusic, 2003). This acetylation leads to the relaxation of chromatin resulting in increased accessibility of transcription factors to the proviral DNA (Figure 2A; Mariño-Ramírez et al., 2005). Conversely, histone deacetylases (HDAC), remove acetyl groups from histones to promote chromatin condensation, thereby silencing proviruses (Jiang et al., 2007; Marban et al., 2007; Herbein and Wendling, 2010; Seto and Yoshida, 2014). Thus, HDAC inhibitors have been shown to potently reactivate latent cells in vitro (Saayman et al., 2014). In addition, H3 lysine methylations, including H3K4me, H3K36me, and H3K79me, are also involved in HIV DNA transcription and are mediated by histone methyltransferases such as SUV39H1 and EZH2 (du Chéné et al., 2007; Imai et al., 2010; Friedman et al., 2011; Ding et al., 2013; Tchasovnikarova et al., 2015; Li J. et al., 2016). Post-translational modification by lysine crotonylation was found to affect gene expression (Tan et al., 2011; Sabari et al., 2015, 2017). This study showed that histone crotonylation at the LTR through the induction of the enzyme acyl-CoA synthetase short-chain family member 2 (ACSS2) can reactivate latent HIV-1 in vitro and ex vivo (Jiang et al., 2018). Specifically, ACSS2 induction was found to increase histone crotonylation and acetylation at H3K4 while decreasing histone trimethylation at H3K27.
HIV-1 latency also results from the lack of trans-regulatory factors that enable T cell activation. Although many of these mechanisms are still under investigation, some transcription factors have been found to regulate viral gene expression. The positive transcription elongation factor, P-TEFb, is a critical kinase involved in HIV-1 transcription and is hijacked by the HIV-1 trans-activator, Tat (Figure 2B; Burnett et al., 2009). P-TEFb is sequestered by the 7SK small nuclear ribonucleoprotein (snRNP) complex, leading to an initial low expression of Tat (Weinberger et al., 2005). Once Tat is expressed, it associates with P-TEFb at the viral promoter to enhance HIV-1 transcription (Zhu et al., 1997; Yang et al., 2001). P-TEFb consists of CyclinT1 and CDK9 and requires CDK9 phosphorylation to activate the paused RNA Polymerase II (Pol II) (Wei et al., 1998; Ramakrishnan et al., 2009). CDK9 phosphorylation levels are generally lower in the resting CD4+ T cells, which is the dominant compartment of HIV-1 latently infected cells (Budhiraja et al., 2013; Ramakrishnan et al., 2015). A transcription factor, NF-κB, is also involved in the initiation of HIV-1 transcription (Adams et al., 1994; Williams et al., 2006). NF-κB is a host cell master regulator of inflammatory T cells that acts as a transcription factor to initiate HIV transcription in the absence of Tat (West et al., 2001). In resting cells, NF-κB is sequestered in the cytoplasm where it is bound by IκB. The low availability of NF-κB binding at the promoter region causes the change in the chromatin structure surrounding the LTR, leading to the disruption of the recruitment of Poll II and transcriptional initiation (Adams et al., 1994). Moreover, a mature NF-κB subunit, p50, can homodimerize to further suppress the NF-κB mediated transcription by binding to the HIV-1 promoter region and recruiting HDAC1 (Figure 2B; Zhong et al., 2002). Activated cytoplasmic PKC promotes ubiquitination and degradation of IκB to allow for nuclear translocation of NF-κB. The PKC pathway resulting in the activation of NF-κB is one of the most important pathways in HIV reactivation (reviewed in Colin and Van Lint, 2009; McKernan et al., 2012). PKC agonists, such as prostratin (Gulakowski et al., 1997; Williams et al., 2004), bryostatin-1 (Gutiérrez et al., 2016), and ingenol (Pandeló José et al., 2014), are highly effective in inducing latent HIV-1 expression from the viral reservoir through NK-κB signaling (Jiang and Dandekar, 2015). Therefore, PKC inhibitors have been investigated as promising candidates for HIV-1 eradication. In addition to the canonical NF-κB (cNF-κB) pathway, other NF-κB subpathways, including non-canonical (ncNF-κB) signaling, have been reported to regulate NF-κB responsible elements (Sun and Ley, 2008; Sun, 2017; Wong and Jiang, 2021). While cNF-κB signaling leads to RelA/p50 heterodimer binding to the promoter region (Deng et al., 2018), ncNF-κB signaling cleaves p100 into p52, which forms a transcriptional complex with RelB. Activation of ncNF-κB signaling was recently reported to induce HIV expression in both HIV-infected humanized mice and SIV-infected macaques under suppressive treatment (Hennessy et al., 2013; Nixon et al., 2020), indicating that ncNF-κB signaling is also involved in HIV latency.
Even after transcription initiation, RNA Pol II pauses at the promoter region due to the binding of negative elongation factors such as NELF and DSIF, causing RNA Pol II to terminate prematurely (Figure 2B; Yamaguchi et al., 1999). These mechanisms govern the HIV-1 promoter, tangling the LTR and preventing transcription factors from accessing the viral promoter region. Yukl et al. (2018) found that CD4+ T cells from PLWH on cART harbor more blocks to transcriptional elongation, completion, and splicing than transcriptional initiation. This indicates that the repression of HIV-1 transcription and splicing occurs at different stages and is regulated by different mechanisms, overall suggesting that HIV-1 latency is heterogeneous in CD4+ T cells.
Some of the proteins affecting viral transcription have been identified specifically in myeloid cells (Le Douce et al., 2012; Kruize and Kootstra, 2019), and interventions designed to block Tat function have been studied as a means to induce a latent state in macrophages and microglia (Alamer et al., 2020). Epigenetic regulation of HIV transcription does occur in myeloid cells, but studies that have examined changes in epigenetic modifiers at cellular promoters in response to HIV-1 infection in myeloid cells indicate that regulatory mechanisms are distinct from T cells (Wierda et al., 2012; Corley et al., 2016).
Non-coding RNAs (ncRNAs) have also been shown to be involved in gene regulation and mRNA splicing (Ranki et al., 1994), and were found to be involved in HIV-1 latency. Specifically, cellular miRNAs induce RNA silencing and post-transcriptional regulation of gene expression (Bissels et al., 2009). Long non-coding RNAs (lncRNAs) play key roles in RNA splicing and stability (Khalil et al., 2009). miRNAs modulate HIV-1 replication either by directly targeting the HIV-1 mRNA (Hariharan et al., 2005) or host mRNAs that code for transcription factors of HIV-1 (Brass et al., 2008; Zhou et al., 2008; Yeung et al., 2009; Houzet and Jeang, 2011). For instance, miR-17, miR-5p and miR-20a target PCAF (Triboulet et al., 2007), a transcriptional coactivator to interact with Tat and functionally synergize to activate the HIV-1 promoter (Mujtaba et al., 2002), and miR-198, miR-27b, miR-29b, miR-150, miR-223 target CyclinT1 (Sung and Rice, 2009; Chiang et al., 2012; Budhiraja et al., 2013) to regulate viral expression and production. Work by Carpio et al. (2010) and Klase et al. (2007) found that high expression of these miRNAs correlates with less susceptibility to HIV-1 infection in monocytes in comparison to macrophages. A set of mixed miRNAs enriched in monocytes represses the expression of PUR-α, a cofactor of Tat (Shen et al., 2012). The miRNA induced by Tat called miR-217 targets the host protein Sirtuin-1, which deacetylates and inactivates Tat, increasing HIV-1 expression (Zhang et al., 2012). A latency inducible lncRNA represented by NRON is highly expressed in resting CD4+ T cells and is involved in HIV-1 latency by mediating Tat degradation (Figure 2B; Li J. et al., 2016). Interestingly, the knockdown of NRON increases HIV-1 replication by enhancing NFAT activity (Imam et al., 2015). In another study, Li et al. reported that a novel lncRNA AK130181 (also named LOC105747689) was highly expressed in latently infected CD4+ T lymphocytes and inhibited HIV-1 transcription in an NF-κB-dependent manner (Li et al., 2020). Considering these latency mechanisms and other regulations for HIV-1 gene expression (Table 1), HIV-1 latency appears to be established by both cis- and trans-regulatory elements and in a cell lineage-specific manner.
Human Immunodeficiency Virus Integration and Clonal Expansion
Integration is a hallmark of retroviruses and a central step in the HIV replication cycle that enables long-term HIV-1 persistence (Anderson and Maldarelli, 2018). HIV-1 integration into the host genome is generally non-specific with slight preferences toward introns of actively transcribed genes and typically exclude promoter regions (Mack et al., 2003; Han et al., 2004; Ikeda et al., 2007). cART successfully halts HIV replication by targeting various steps in the virus replication cycle, however current therapies do not target HIV-1 transcription or result in direct infected cell killing. The cells that harbor latent HIV-1 proviruses can persist for years despite therapy (Finzi et al., 1997; Wong et al., 1997; Siliciano et al., 2003) and undergo clonal expansion (Maldarelli et al., 2014; Wagner et al., 2014). The landscape of HIV-1 infected cells is shaped over time through cytopathic effects, immune responses, and infected cell proliferation (Anderson et al., 2020; Liu et al., 2020). HIV-1 infected cells can undergo clonal expansion through three distinct mechanisms: (1) homeostatic proliferation, (2) in response to the infected cells cognate antigen, or (3) in some cases as the result of the proviral integration site (Figure 3; Anderson and Maldarelli, 2018; Liu et al., 2020). Each driver of HIV-1 infected cell division can contribute to sustaining the HIV-1 reservoir despite cART. Homeostatic proliferation promotes HIV-1 persistence by allowing latently infected cells to undergo cell division in the absence of viral reactivation or cellular differentiation (Figure 3A; Chomont et al., 2009; Bosque et al., 2011; Musick et al., 2019). This indicates that the homeostatic proliferation of infected cells can sustain and replenish the HIV-1 reservoir while eluding the effects of cART. Antigen-driven clonal expansion occurs when an HIV-1 infected cell recognizes its cognate antigen (Figure 3B; Douek et al., 2002; Mendoza et al., 2020; Simonetti et al., 2021). HIV-1 primarily infects CD4+ T cells, which proliferate in response to antigenic interactions (Douek et al., 2002). The expansion of infected cells driven by antigen interactions is likely reflected in persistent, and/or waxing and waning of clonal populations through chronic and repeat exposures (Wang et al., 2018). Finally, the contribution of integration-site-driven clonal expansion likely plays only a minor role in sustaining the HIV-1 reservoir (Figure 3C) (Coffin et al., 2021). Proviral integration into oncogenes, such as BACH2, MKL2, and STAT5B, occurs rarely and is revealed after years of therapy, indicating the integration site itself may provide a selective advantage for long-term persistence (Maldarelli et al., 2014; Wagner et al., 2014; Cohn et al., 2015). All three mechanisms of HIV-1 infected cell proliferation can drive the persistence and expansion of the HIV-1 reservoir cells and pose major obstacles for curative strategies.
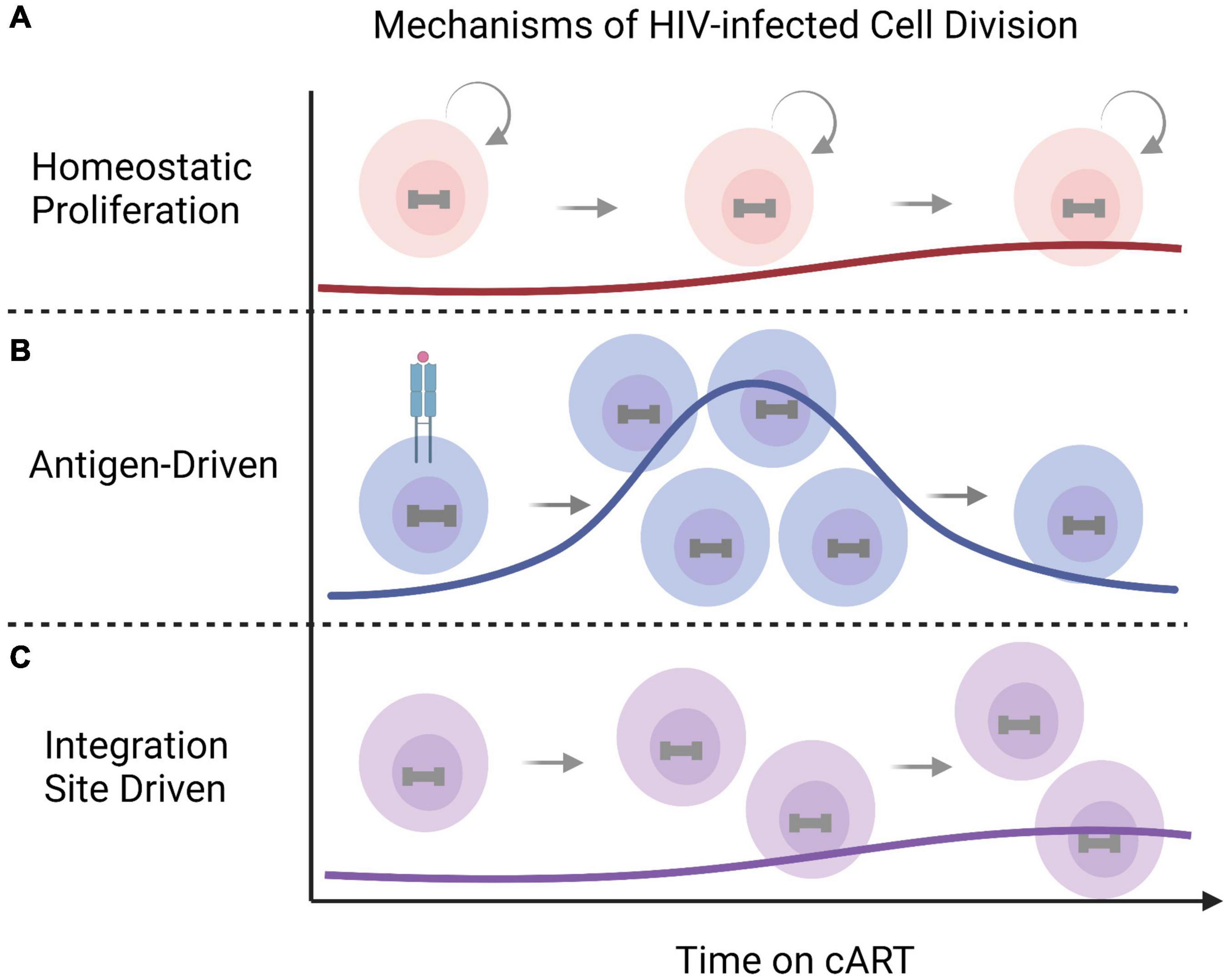
Figure 3. Mechanisms of clonal expansion in HIV-1 infected cells. HIV-infected cells undergo cell division overtime to maintain the HIV reservoir despite antiretroviral therapy. The mechanisms of HIV-1 infected clonal expansion include (A) normal homeostatic proliferation, (B) antigen-driven expansion in response to periodic or persistent cognate antigen exposures, or, in some cases, through (C) integration-site driven expansion (for example BACH2, MKL2, and STAT5B). This figure was created with BioRender.com.
Although the majority of integrated proviruses are defective (Ho et al., 2013; Bruner et al., 2016, 2019; Imamichi et al., 2016), cells harboring intact replication-competent proviruses also undergo clonal expansion to sustain the HIV-1 reservoir despite cART (Simonetti et al., 2016; Bui et al., 2017; Lee et al., 2017; Patro et al., 2019). Once cART is stopped, viremia rebounds from this persistent, replication-competent reservoir to near pre-therapy levels within weeks for most individuals (Joos et al., 2008). It has recently been estimated that over 50% of inducible, replication-competent proviruses within the latent reservoir arose as the result of in vivo proliferation (Lorenzi et al., 2016; Bui et al., 2017; Hosmane et al., 2017). As this is likely an underestimate, these findings indicate that the proliferation of HIV-1 infected cells harboring replication-competent proviruses is one of the major mechanisms that maintains the reservoir and presents a challenge toward an HIV cure. Future curative strategies may aim to directly target cell-killing of HIV-1 infected cells or to blunt infected cell proliferation.
Defective Proviruses
Although HIV-1 rebounds from replication-competent reservoir cells harboring intact proviral DNA upon cART cessation, the majority of proviruses are defective with large internal deletions or lethal mutations and clonally expand in vivo (Coffin and Hughes, 2021). Defective proviruses account for more than 95% of the total proviruses in the peripheral blood isolated from cART-treated PLWH (Siliciano et al., 2003; Eriksson et al., 2013; Crooks et al., 2015). Defective proviruses were initially thought to have little impact on HIV pathogenesis and disease progression due to their inability to produce infectious virions. However, it has recently been discovered that a fraction of defective proviruses are transcriptionally active (Imamichi et al., 2016) and can contribute to chronic immune activation (Pollack et al., 2017). Novel unspliced forms of viral RNAs transcription from defective proviruses can produce HIV-1 Gag and Pol proteins (Imamichi et al., 2020). These observations indicate that defective proviruses are capable of producing virus-like particles (VLPs) or viral proteins, which may become constitutive antigens (Figure 4). Chronic inflammation and persistent immune activation occurs in most individuals on long-term cART despite successful virus suppression and can induce immunological tolerance that enhances autoreactive antibody production, making it difficult to establish protective humoral immunity (van den Dries et al., 2017). VLPs have previously been employed for immunization purposes and demonstrated that immature morphology enhanced immunogenicity of VLPs and strongly induced IFN-γ secretion and antibody production (Álvarez-Fernández et al., 2012; Gonelli et al., 2019). Approximately 20% of the virions harboring intact protease still remain immature morphology after budding (Sarca et al., 2021), which implies that VLPs generated from defective proviruses might contribute to chronic inflammation in PLWH. In addition, proviruses with defective major splice donors or hypermutations induced by APOBEC3 cytidine deaminase enzymes (Izumi et al., 2008; Fukuda et al., 2019) can produce antigens against CTL and constitutively induce their activation (Shankar et al., 2000; Monajemi et al., 2014; Figure 4). It had long been thought that defective proviruses were irrelevant viral DNA sequences. However, defective proviruses capable of transcribing novel unspliced viral RNA can be found in individuals at all stages of virus infection, adding to the complexity of linked chronic immune stimulation in some PLWH.
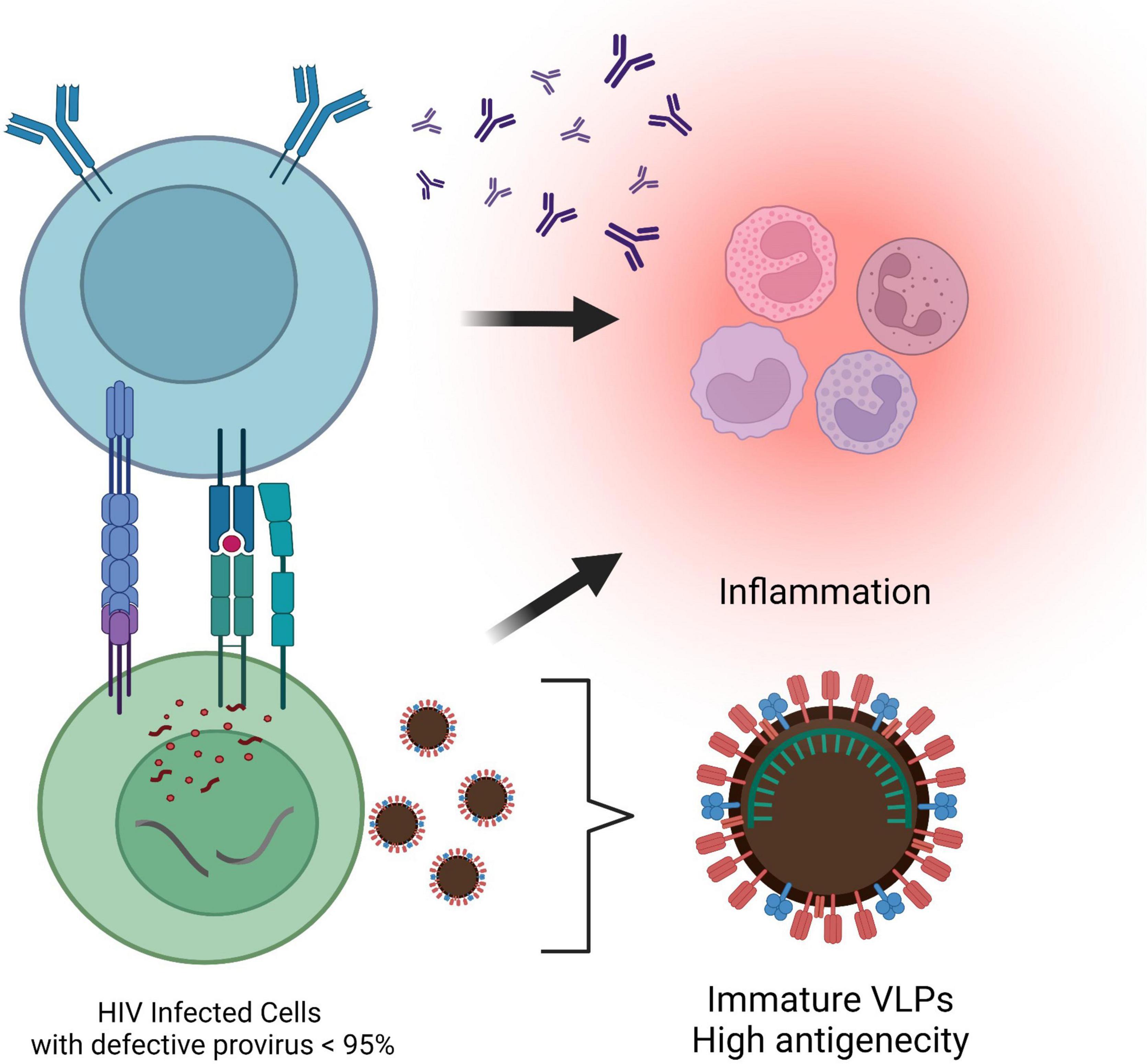
Figure 4. Antigenicity of the defective provirus. Most proviruses are defective with large internal deletions or hypermutations. Viral-like particles are released from novel unspliced viral RNAs transcribed from the defective proviruses. In addition, cytotoxic T lymphocytes are activated by the antigens presented by proviruses with defective major splice donors or hypermutations, which results in leading to chronic inflammation in individuals on cART. This figure was created with BioRender.com.
Cure Strategies
Kick-and-Kill Strategy
One of the potential strategies toward HIV-1 eradication has been named “Kick-and-Kill” therapy. This approach is contingent upon the reactivation of latently infected cells in the presence of cART through the use of LRAs, and subsequent elimination of reactivated HIV-1 infected cells by the immune system or other interventions (Figure 5). Several pathways related to cis- and trans-regulating factors such as histone deacetylase (HDAC), NF-κB signaling, or PKC activation have been targeted to reactivate latently infected cells (Ait-Ammar et al., 2020). Several classes of LRAs have been described, to date (Table 2). Epigenetic modifiers such as histone acetylation/methylation, DNA methylation, or chromatin remodeling affect viral RNA transcription, splicing, or subsequent nuclear export pathway by altering the chromatin structure and DNA accessibility (Darcis et al., 2017; Ait-Ammar et al., 2020). HDAC inhibitors such as vorinostat, panobinostat, and romidepsin have been extensively studied in clinical trials, but the single-use of these inhibitors has not led to the eradication of latently infected cells in vivo (Kim et al., 2018). On the other hand, the combination of HIV-1 specific CD8+ cytotoxic T lymphocytes activation upon the usage of vorinostat showed promise to be effective for purging the latent reservoir cells in ex vivo experiments (Sengupta and Siliciano, 2018).
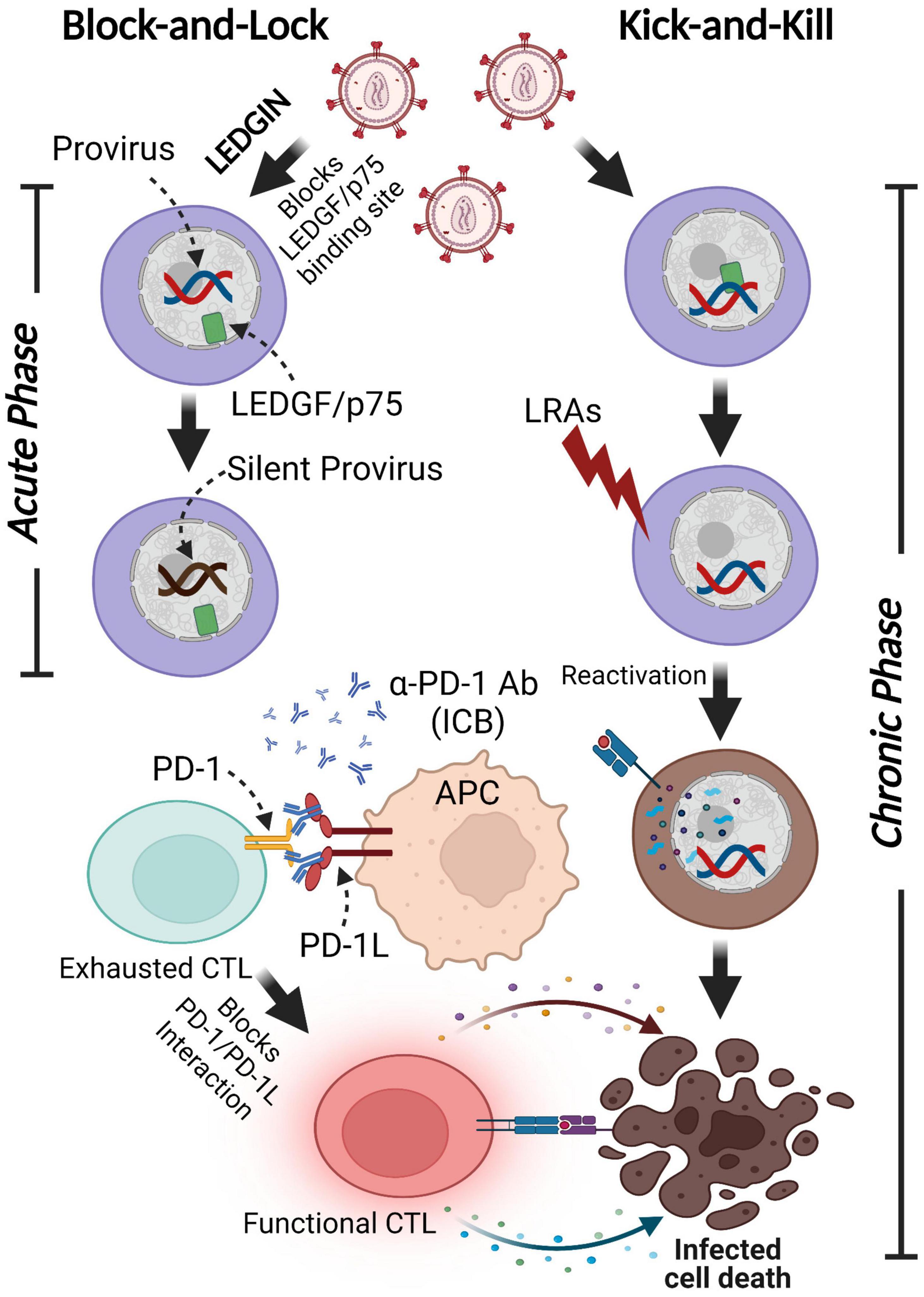
Figure 5. Functional cure strategies: Block-and-Lock and Kick-and-Kill approaches. A minor subset of infected cells harbors latent proviruses following infection. LEDGIN-mediated “Block-and-Lock” functional cure aims to permanently silence the provirus to block viral reactivation in the absence of cART. LEDGINs inhibit the LEDGF/p75-Integrase interaction, resulting in the redirection of integration into transcriptionally silent regions. The “Kick-and-Kill” strategy aims to decrease the size of functional HIV-1 reservoirs by reactivating proviral transcription with LRAs, leading to the elimination of infected cells via immune systems stimulated by ICB. This figure was created with BioRender.com.
PKC agonists may also be a suitable clinical approach for targeting NF-κB signaling in HIV-1 latency reactivation. Bryostatin, targeting NF-κB signaling, has been studied to reactivate latent cells in vivo, but unfortunately, no transcriptional enhancement was observed in HIV-1 latently infected cells in clinical settings (French et al., 2020). On the other hand, Derivatives of ingenol have been gaining interest in the reactivation of HIV-1 expression both in vitro and in vivo through NF-κB signaling (reviewed in Jiang and Dandekar, 2015). Ingenol-3-hexanoate was found to reactivate latent HIV-1 in J-Lat cells at a low concentration, hence lower cellular cytotoxicity (Jiang G. et al., 2014). Ingenol-3-angelate (PEP005), in combination with the P-TEFb agonist, JQ1, can synergistically reactivate latent HIV-1 with 7.5-fold higher effectiveness than PEP005 alone (Jiang et al., 2015). Interestingly, ACSS2-mediated histone crotonylation can also be associated with PEP005 to reactivate latent CD4+ T cells from HIV-infected individuals (Jiang et al., 2018). Ingenol 3,20-dibenzoate was found to activate resting CD4+ T cells from HIV-1 ART-treated aviremic patients and is potentially a marker to measure the reactivation of resting CD4+ T cells from treated PLWH (Spivak et al., 2015).
Recently, a family of IAP inhibitors (IAPi)/the mimetics of second mitochondria-derived activator of caspases (SMACm), such as Debio 1143 and AZD5582, have been proposed as a new class of LRAs via the induction of ncNF-κB signaling (Dashti et al., 2020). Debio 1143 was shown to lead to latent HIV reversal through the degradation of BIRC2/cIAP, a ubiquitin ligase that acts as a repressor of the ncNF-κB pathway in vivo using ART-suppressive BLT humanized mice or ex vivo using resting CD4+ T cells isolated from HIV-infected individuals with cART (Bobardt et al., 2019). AZD5582 was also found to induce SIV-RNA from latency in lymph node tissues of the ART-suppressive SIV-infected rhesus macaques by inhibiting BIRC2/cIAP. While these studies implied a potency of IAPi/SMACm as new LRAs via the novel ncNF-κB signaling pathway (Wong and Jiang, 2021), recent research applying AZD5582 failed to reduce reservoir size in the SHIV model (Nixon et al., 2020), indicating that latency reversal efficacy of IAPi/SMACm monotherapy may not be enough (Wong and Jiang, 2021). Furthermore, the induction of ncNF-κB signaling causes cell death in memory CD4+ T cells where latent HIV provirus is enriched (Wong and Jiang, 2021). Therefore, further studies are required to understand the complexity of both cNF-κB and ncNF-κB signaling pathways to create effective LRAs.
Other agonists targeting the innate immune receptors, TLR7 or TLR9, were also used to reactivate HIV-1 latent cells. The combination of HIV-1 Env-specific broadly neutralizing antibodies (bNAb) PGT121 together with the TLR7 agonist, vesatolimod GS-9620, delayed viral rebound following cART cessation in SIV/HIV chimeric virus (SHIV)-infected rhesus macaques that initiated antiretroviral treatment during acute infection (Bruno and Stoughton, 1984; Gutiérrez et al., 2016). However, the use of GS-9620 in phase I clinical trials with HIV-1 infected individuals on cART (NCT02858401) was discontinued due to adverse effects and an underwhelming impact on the plasma HIV-1 RNA and total DNA in CD4+ T cells. In addition, TLR9 agonist MGN1703 showed a moderate latency-reversing effect in HIV-1 infected participants, but a reduction in the reservoir size was not observed (Vibholm et al., 2017, 2019).
Considering that targeting a single mechanism with LRA monotherapy might not be sufficient to reactivate most latent cells, a combination of several different classes of LRAs targeting heterogenous silencing mechanisms may be required (Ait-Ammar et al., 2020). One study participant treated with MGN1703 showed viral control to undetectable levels upon cART cessation along with robust HIV-1 specific effector memory subset of CD8+ T cells and neutralization antibody production (Schleimann et al., 2019). This suggests that adaptive immunity is essential for the kick-and-kill approach. However, LRAs could impair multiple CD8+ T cell functions (Clutton and Jones, 2018). To date, successful reduction of the reservoir size in vivo has not been observed with LRA monotherapy, but has been seen using romidepsin and immune checkpoint blockade (ICB) nivolumab targeting PD-1 (Chun et al., 1997; Darcis et al., 2017).
As outlined above, the HIV-1 latent reservoir exists in cellular and tissue compartments and likely has multiple maintenance mechanisms. This may require diverse LRAs targeting a wide variety of latent cells, which poses challenges for therapies. Due to a lack of understanding of the various determinants contributing to the heterogeneity in HIV-1 latency, no LRA has been proved successful in clinical trials. Therefore, there is a need to understand the safety and potency of kick-and-kill strategies to develop effective LRA combination with immunostimulants to treat PLWH (Ait-Ammar et al., 2020).
Immunomodulation
As we described before, HIV-1 preferentially infects CD4+ T cells and establishes long-term residency within affected individuals (Douek et al., 2002). In the standard progression of HIV-1 without therapy, normal progressors experience a decline in immune function (Boasso et al., 2009). Within the first 6–8 weeks post-exposure to HIV-1, the host experiences a significant decrease in CD4+ T cells, a concomitant increase in viral load, and a corresponding increase in immune activation (Fauci, 1991; Mellors et al., 1996; Coffin et al., 1997; Boasso et al., 2009) and a decrease in immune function. To control the infection, the host mounts an adaptive immune response via CD8+ T cells (Borrow et al., 1994; Koup et al., 1994). Despite these efforts, HIV persists in a small number of cells that can survive for a prolonged time.
Immune activation and inflammation are persistently driven by the ability of HIV-1 to evade detection and elimination by the immune system, the large-scale depletion of CD4+ T cells that regulate the adaptive immune response to infection, and the inability of cART to eliminate latently infected cells (Hazenberg et al., 2000; Gougeon, 2005; Leonard et al., 2008; Dinoso et al., 2009; Chun et al., 2010; Manel and Littman, 2011). During chronic infection, the host immune system must adapt to find new homeostasis which minimizes tissue damage from persistent inflammation while still maintaining control of infection. To mitigate immunopathology, the immune system has evolved mechanisms to progressively attenuate the immune response during chronic infection. Over time this leads to a state of hypo-responsiveness and eventual immunological tolerance of the virus. T cell exhaustion is one such mechanism in which effector T cells upregulate multiple inhibitory immune checkpoint receptors during chronic infection which results in the hierarchical loss of function (Xiong et al., 2001; Wherry et al., 2003; Saez-Cirion et al., 2007; McLane et al., 2019).
T cell exhaustion has been characterized by the co-expression of multiple inhibitory immune checkpoints such as cytotoxic lymphocyte antigen-4 (CTLA4), programmed cell death-1 (PD-1), T-cell immunoglobulin and mucin-domain-containing 3 (TIM3), T-cell immune receptor with Ig and ITIM domains (TIGIT), and lymphocyte-activation gene 3 (LAG3) on the surface of immune cells (Wherry and Kurachi, 2015). Immune checkpoints receptors, when engaged with their corresponding ligand, negatively regulate T cell activation (Blackburn et al., 2009). T cell exhaustion impacts the ability of CD8+ T cells to suppress viral replication (Mueller et al., 2001; Appay et al., 2002; Wherry et al., 2007; Wherry and Kurachi, 2015).
Recent literature has demonstrated that T cell exhaustion persists despite treatment with cART (Day et al., 2006; Nakanjako et al., 2011; Khoury et al., 2017; de Armas et al., 2019; Macatangay et al., 2020). While cART effectively reduces HIV-1 viral load to undetectable levels and significantly improves life expectancy, it is not a sterilizing cure nor does it fully restore host immune function (Migueles et al., 2009; Deeks, 2011; Arts and Hazuda, 2012; Wilson and Sereti, 2013; Serrano-Villar et al., 2014; Deeks et al., 2015; Perdomo-Celis et al., 2019). Poor immune reconstitution contributes to comorbidities that arise despite long-term treatment with cART, which necessitates an intervention that can restore an exhausted immune system (Baker et al., 2008). One such approach to rejuvenating exhausted T cells is through the use of immunotherapy in which monoclonal antibodies bind to immune checkpoints receptors, block their inhibitory signaling pathways, and consequently boost the immune response to chronic infection or cancer (Śledzińska et al., 2015). There are several FDA-approved cancer immunotherapies that target CTLA-4, PD-1, and PD-L1, which are now being investigated as potential novel strategies to reinvigorate a dysfunctional immune system exhausted by chronic HIV (Abbar et al., 2020; Sahin et al., 2020). The aspirational goal of such studies is to revitalize the immune system in such a way that PLWH could control the virus immunologically, without the need for intensive cART regimens.
The use of immunotherapy for PLWH was initially evaluated in HIV-1 positive cancer patients and it was determined that it was safe and effective for the treatment of cancer in patients with well-controlled viremia (Uldrick et al., 2017). Additional studies determined that the use of immunotherapy in these patients imposed no adverse effects on both CD4+ T cell count and plasma viral load (Cook and Kim, 2019; Abbar et al., 2020; Sahin et al., 2020), while some studies observed that participants experienced grade 3 or higher adverse events with various types of immunotherapy (Sahin et al., 2020). Guihot et al. (2018) demonstrated that treatment with PD-1 immunotherapy resulted in a dramatic decrease in plasma viral RNA and a concomitant increase in HIV-1 specific CD8+ T cells for a single cancer patient living with HIV (Guihot et al., 2018). The possible dual benefits of using immunotherapy to restore the functionality of exhausted CD8+ T cells, reverse HIV-1 latency through activation of viral transcription in CD4+ T cells (Wherry and Kurachi, 2015; Evans et al., 2018; Fromentin et al., 2019; Van der Sluis et al., 2020), and the favorable results of immunotherapy on HIV-1 positive cancer patients have led investigators to evaluate it for the treatment of virally suppressed otherwise healthy PLWH (Gay et al., 2017; Abbar et al., 2020). These studies concluded that participants were able to maintain viral suppression on immunotherapy, restored anti-HIV activity as demonstrated by a further reduction in plasma viral RNA varied (Gay et al., 2017; Abbar et al., 2020).
Immune checkpoint blockade has been utilized as immunotherapy to treat cells exhausted by chronic infection with cancer but these therapies have also been modestly effective at improving anti-tumor immunity (Pardoll, 2012; Shayan et al., 2017). One caveat to utilizing ICB for effective immunotherapy is the co-expression of multiple inhibitory receptors on immune cells (Shayan et al., 2017; Zahavi and Weiner, 2019) which might explain their modest impact at further reducing plasma viremia in otherwise healthy PLWH and warrants the use of combinatorial immunotherapy. Combinatorial therapy using ICB might not be sufficient to reverse the effects of exhaustion and restore the functionality of CD8+ T cells as recent studies have identified epigenetic modifications which may also restrict the effectiveness of ICB.
Epigenetic modifications are known to regulate T cell differentiation and are integral to the formation and heritability of various T cell subsets. These T cell subsets permit both the effective control of infection and also regulate effector function (Scharer et al., 2013; Chen et al., 2018; Henning et al., 2018; Zebley et al., 2020). Recent studies have begun to explore the epigenetic landscape of the exhausted CD8+ T cells and have determined that they are an epigenetically distinct subset of cells (Youngblood et al., 2011; Jadhav et al., 2019; Khan et al., 2019; Calle-Fabregat et al., 2020). Scharer et al. evaluated CD8+ T cell response to acute Lymphocytic Choriomeningitis virus infection from naïve and effector T cells and observed genome-wide DNA methylation of CD8+ T cells following T cell activation which permitted the inheritance of effector functions (Scharer et al., 2013). DNA methylation is a contributing epigenetic mechanism that regulates CD8+ T cell exhaustion (Youngblood et al., 2011, 2013; Scharer et al., 2013; Pauken et al., 2016; Ghoneim et al., 2017; Yates et al., 2021) and has been demonstrated to impact CD8 restoration by ICB (Ahn et al., 2016; Pauken et al., 2016; Ghoneim et al., 2017). It was demonstrated that T cell exhaustion can be categorized into two distinct stages which are delineated by de novo DNA methylation to the PD-1 promoter (Ghoneim et al., 2017). These epigenetic modifications to CD8+ T cells acquired during the effector phase regulate both the formation and heritability of terminally differentiated exhausted CD8+ T cells which preclude restoration by PD-1 blockade (Ahn et al., 2016; Pauken et al., 2016; Ghoneim et al., 2017). Epigenetic targeted therapy could potentially reverse the repressive epigenetic modifications that enforce CD8+ T cell exhaustion which could lead to a novel strategy in the treatment of HIV.
Chimeric antigen receptor (CAR)-T cell therapy is a kind of adaptive immunotherapy that genetically engineers a patient’s T cells to recognize and bind to foreign antigens on the antigen-expressing cells (Maus and Levine, 2016; Hartmann et al., 2017). Patients with hematologic malignancies, such as lymphocytic leukemia, lymphoblastic leukemia, diffuse large B-cell lymphoma, and follicular lymphoma, successfully achieved their treatment plans with CAR-T cell therapy (Porter et al., 2011; Maude et al., 2014; Schuster et al., 2017). Key components of CAR technology are the extracellular single-chain fragment variant derived from the antigen-binding region and the intracellular signaling domains containing CD3ζ, CD28, and 4-1BB (Maher et al., 2002; Imai et al., 2004; Porter et al., 2011; Srivastava and Riddell, 2015). Therefore, CAR can be designed to recognize specific antigens and subsequently induce activation of the immune response against target antigens. CD8+ T cells are collected from HIV-infected individuals and inserted with CAR genes in vitro, whose anti-HIV efficacy was verified, and then autologous HIV-specific CAR-T cells were transplanted into the patients (Qi et al., 2020). Recently, broadly neutralizing antibodies (bNAbs) targeting HIV-1 envelope glycoprotein have been used to construct anti-HIV specific CAR-T cells (Kwong et al., 2013; Ali et al., 2016). Hale et al. showed that CARs engineered with four types of bNAbs (PGT-128, PGT-145, VRC07-523, and 10E8) effectively activate and kill HIV-infected cells. Moreover, the integration of an HIV-1 CAR gene expression cassette into the CCR5 locus via homology-directed repair leads to the suppression of replicating of the virus (Hale et al., 2017). Currently, there are two ongoing clinical trials of CAR-T cell therapy in PLWH under cART (NCT03240328 and NCT03617198) to evaluate CD4-CAR T cells with CCR5 disruption for HIV resistance. Although there are obstacles in CAR-T cell therapy development, such as cell expansion in vivo, off-target effects, and severe cytokine storm (reviewed in Qi et al., 2020), it is worth exploring the potential of CAR-engineered T-cell therapy for an HIV cure.
Although current cART is very effective in targeting HIV pathogenesis, the pervasive nature of this disease requires the continued development of new ways to target viral replication and improve immune function. A focus on understanding the mechanisms of HIV-1 suppression by those with the innate ability to control the virus (Clerici et al., 1992; Kelker et al., 1992; Cao et al., 1995; Huang et al., 1995; Pinto et al., 1995; Rowland-Jones et al., 1995; Fowke et al., 1996; Lefrère et al., 1999; Shacklett, 2006; Dyer et al., 2008; Pereyra et al., 2008; Gonzalo-Gil et al., 2017; Pernas et al., 2018; Lopez-Galindez et al., 2019; Nguyen et al., 2019; Macatangay et al., 2020) has the potential to identify novel ways to improve host immune function, give insight into immune mechanisms that are common to both chronic infection and cancer, and rapidly treat PLWH with FDA approved therapies. As cells harboring reactivated proviruses by LRAs would then be required to be eliminated by CTL, the combination therapy with LRAs and ICBs/CAR-T could achieve a functional HIV-1 cure for chronic HIV infection (Figure 5).
Block-and-Lock Strategy
The inability of the immune system to eradicate latently infected cells, due to the lack of viral protein expression, permits the long-term persistence of HIV-1 infected cells. In contrast to the “Kick-and-Kill” strategy, the “Block-and-Lock” approach aims to promote permanent provirus silencing even after cART cessation. This strategy ultimately seeks to affect both pre-integration and post-integration stages. HIV-1 DNA is preferentially integrated into transcriptionally active sites located near the nuclear pore, where chromatin is decondensed (Demeulemeester et al., 2015). Integration into a transcriptionally inactive site promotes proviral silencing. Lens epithelium-derived growth factor (LEDGF/p75) is a chromatin-binding host protein that supports HIV-1 DNA integration through interactions with the HIV-1 integrase protein (Vranckx et al., 2016). Treatment with a LEDGF inhibitor, LEDGINs, can dramatically redirect HIV-1 DNA integration sites to regions that are resistant to reactivation, potentially leading to a deeply silenced reservoir even after cART cessation (Figure 5; Christ et al., 2012; Kessl et al., 2012; Vranckx et al., 2016). LEDGIN treatment in the case of successful viral integration retargets the transcriptional factors out of active genes, which results in a prolonged latent state. This can lead to the inability of LRAs to reactivate silenced integrated proviruses (Gao et al., 2020). Therefore, LEDGINs might only help to reduce HIV-1 reservoir susceptibility to reactivation early after infection, prior to integration and the seeding of the reservoir.
A post-integration Block-and-Lock strategy aims to permanently suppress HIV-1 transcription to prevent viral reactivation even after successful proviral DNA integration. Post-integration silencing methods target the trans-regulation mechanisms to suppress viral gene expression by inhibiting viral and host transcription factors such as HIV-1 Tat, P-TEFb, and NF-κB. Tat is required for the stimulation of HIV-1 transcriptional elongation by binding an RNA element in the LTR and recruiting several transcription-activating proteins (Roy et al., 1990; Zhu et al., 1997). Didehydro-cortistatin A (dCA), the equipotent analog of cortistatin A, inhibits Tat-mediated transactivation through the interaction with the TAR domain of Tat (Li et al., 2019; Mediouni et al., 2019). Prior studies demonstrated that CDK9 inhibitors block viral transcription by disrupting P-TEFb formation (Pisell et al., 2001; Rice, 2016). In addition, activation of CDK2 that inhibits HIV-1 transcription and activation of the HIV-1 provirus through Tat phosphorylation was also targeted for the “Block-and-Lock” Strategy (Ammosova et al., 2006). Bisacetamide-induced protein (HEXIM-1) and 7SK small nuclear RNA interact and retain P-TEFb away from HIV-1 LTR (Yang et al., 2001; Yik et al., 2003). Bromodomain-containing protein 4 (BRD4) that competes with Tat for the P-TEFb interaction domain to further prevent HIV transcription is another target for permanent silencing (Jang et al., 2005; Bisgrove et al., 2007).
NF-κB is predominantly sequestered in the cytoplasm by IκB and cannot activate HIV-1 transcription in resting latently infected cells (Baeuerle and Henkel, 1994; Baldwin, 1996). Inhibition of NF-κB signaling is also considered as a Block-and-Lock strategy. Latent HIV proviruses were less reactivated by curaxin, a drug also used in immuno-oncology that inhibits NF-κB mediated transcription (Orphanides et al., 1998; Gasparian et al., 2011). This led to the hypothesis that curaxin could induce HIV latency via strengthening NF-κB inhibition.
Tat and NF-κB are required for HIV-1 gene expression, therefore inhibition of these critical components can lead to a post-integration “Block-and-Lock” strategy. The post-integration approach may necessitate PLWH to undergo life-long treatment to permanently suppress viral expression and frequently monitor viremia levels. Therefore, the combination of both methods, “Block-and-Kick-and-Kill” would lead to the functional cure for HIV-1 infection by reducing the reservoir size during acute infection through the redirection of HIV-1 DNA integration sites while subsequently reactivating the residual latently infected cells that are not deeply silenced, and eventually eliminate the reactivated cells by HIV-1 specific immune cells (Figure 5).
Genome Editing Strategy
Genome editing technologies, such as the transcription activator-like nucleases (TALENs), zinc finger nucleases (ZFNs), and clustered regularly interspaced short palindromic repeat (CRISPR)-associated nuclease 9 (Cas9) have been proposed for novel approaches toward cure strategies. The use of Cas9 has recently been investigated with the advantages of precise insertion, deletion, and replacement of target double-strand DNA (dsDNA) (White et al., 2017). Cas9 has quickly become the preferred genome-editing platform for interrogating endogenous gene function in vivo. It was originally found in a bacterial adaptive immune defense system to play a vital role against DNA viruses or plasmids. Cas9 ribonucleoprotein complex consists of two components using endonuclease enzymes with a short-guide RNA (gRNA). Cas9 proteins are a specific class of enzymes that break the target dsDNA identified by gRNA, which is engineered with a particular sequence that guides the Cas9 protein to the target DNA sequence. Cas9 unwinds foreign DNA at sites complementary to the 20 base pair spacer region of the gRNA. If the DNA substrate is complementary to the gRNA, the Cas9 cleaves the invading DNA leading to gene inactivation. According to these unique features of Cas9 enzymatic activity, this system has been used as a genetic engineering technique to modify the genomes of living organisms.
Two people who had been living with HIV have been cured so far, the famous first case of the Berlin patient (Hütter et al., 2009; Allers et al., 2011), the second London Patient (Gupta et al., 2019), and a third individual, known as the Dusseldorf Patient, who is currently experiencing long-term remission (Peluso et al., 2019). These three individuals received a bone marrow transplant from matched donors with a homozygous 32 base pair deletion in the CCR5 gene (CCR5-Δ32) as part of their leukemia treatment. It has been documented that homozygous carriers of Δ32 mutation are largely resistant to R5 tropic HIV-1 infection that is exclusively detected in the transmitted founder viruses during the acute infection because the mutation prevents functional expression of CCR5, a coreceptor used by HIV-1 to enter immune cells (Cocchi et al., 1995; Samson et al., 1996; Biti et al., 1997). In addition, CCR5 gene editing of CD4+ T cells mediated by ZFN has been conducted clinically in HIV-1 infected individuals and was demonstrated to be safe (NCT00842634) (Tebas et al., 2014). Long-term CCR5 disruption in hematopoietic stem cells (HSCs) by the CRISPR/Cas9 system was achieved in a mouse model in 2017 to confer HIV-1 resistance in vivo (Xu et al., 2017). Although the other HIV-1 coreceptor, CXCR4, has also been targeted by Cas9, CXCR4 modified cells showed resistance against HIV-1 infection (Schumann et al., 2015). However, possible adverse side effects after CXCR4 disruption are of great concern as CXCR4 plays a vital role in hematopoietic cell development and thymic differentiation (Samson et al., 1996; Dar et al., 2006). To overcome these challenges, a combination of the Cas9 genome editing system and piggyBac transposase tools enabled the introduction of a point mutation, P191A, in the CXCR4 gene that specifically prohibits HIV-1 infection without disrupting CXCR4 receptor function (Liu et al., 2018). Although it should be noted that the virus rebound had not been observed in spite of the CXCR4-tropic virus existence in the Berlin patient, another case has been reported in a patient with allogeneic transplantation from a CCR5Δ32 donor, where a CXCR4-tropic virus rebounded after cART cessation (Kordelas et al., 2014). In addition, it has also been shown that HIV-1 can infect macrophages in a coreceptor-independent manner, leading to endocytosis of the virus (Gobeil et al., 2012). Therefore, targeting HIV-1 coreceptors by gene editing machinery should be considered carefully for HIV cure strategy.
Cas9 could directly eliminate integrated proviral DNA in vitro by targeting the conserved sequence of the HIV-1 LTR U3 region in a latently infected T cell line, a monocytic cell line, and a microglial cell line (Hu et al., 2014). Recently, the excision of proviruses from latent reservoir cells was demonstrated in vivo in humanized mice by combining a provirus targeting genome editing tool with long-acting slow-effective antiviral therapy (Dash et al., 2019; Figure 6). Looking ahead to the future of CRISPR/Cas9 HIV treatment options, the U.S. Food and Drug Administration (FDA) has recently approved to begin trials testing EBT-101, an in vivo CRISPR/Cas9 gene therapy designed to excise HIV-1 proviral DNA. This is the first time the FDA has given investigational new drug (IND) approval to a CRISPR-based therapy for HIV treatment. Trials will evaluate the safety, tolerability, and efficacy of EBT-101 in healthy individuals living with HIV (NCT05144386).
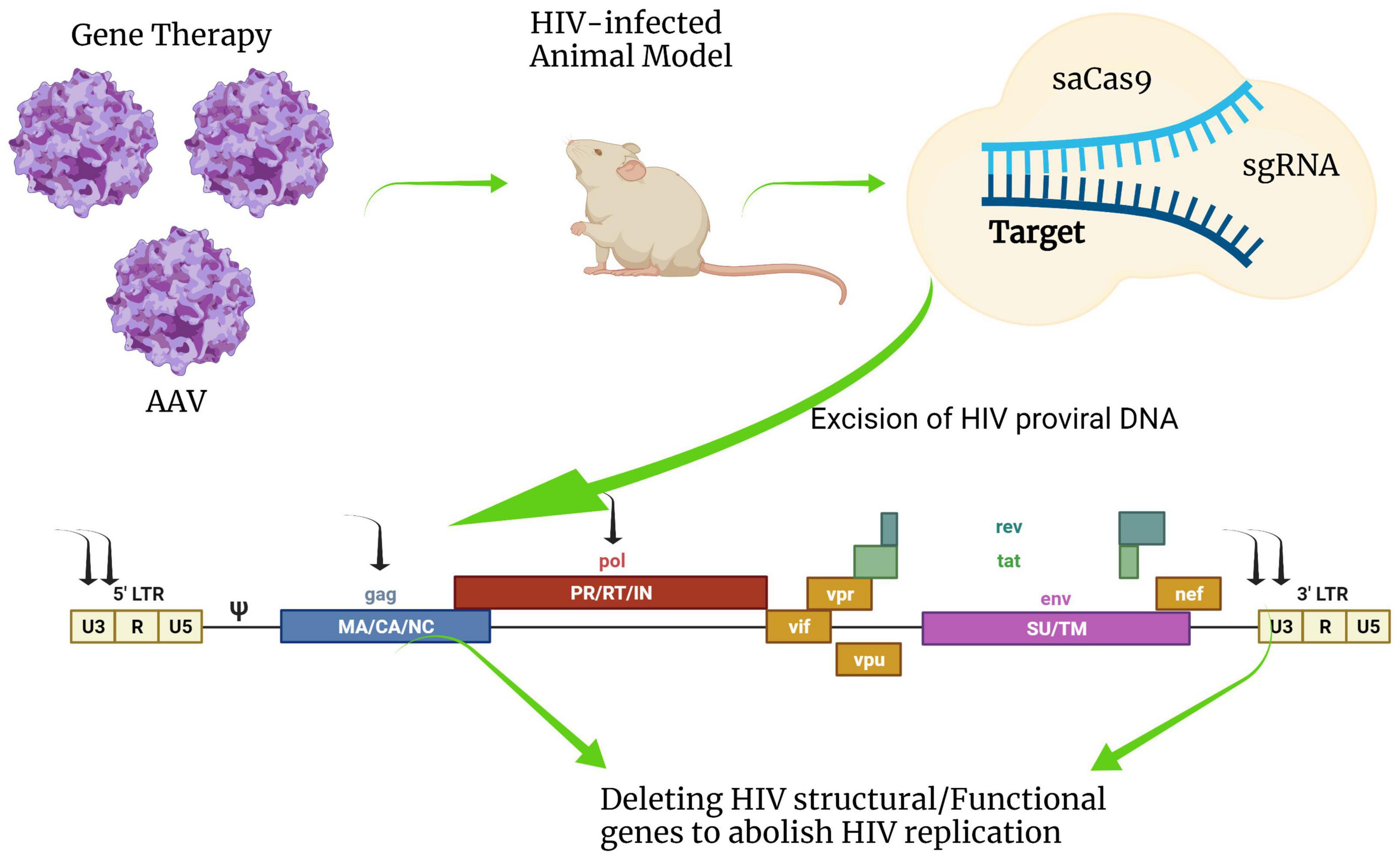
Figure 6. Schematic diagram of HIV-1 provirus DNA deactivation by CRISPR/Cas9 genome editing technology. Cas9 nuclease combined with gRNAs targeting multiple sites in HIV-1 DNA sequences such as 5′-LTR, 3′-LTR, gag, or pol can deactivate integrated viral DNA. Cas9 can be efficiently delivered by an adeno-associated virus vector (AAV) system in vivo. This figure was created with BioRender.com.
Although many classes of viral vectors exist, the adeno-associated virus vector (AAV) has largely been used for delivering genome-editing machinery in vivo (Yin et al., 2017), and in clinical trials (NCT05144386). AAV is thought to be one of the most suitable viral vectors for gene therapy applications and gene transfer in vivo. AAV was approved for a number of human clinical trials in gene augmentation therapies due to its favorable safety profile. One major advantage of using AAV is a very mild immune response and toxicity elicited by AAV in animal models. However, AAV has some disadvantages, such as small cargo capacity, prolonged time for large-scale production, and relatively high cost. AAV has a substantial limitation in small viral genome packing capacity that is generally considered to be less than 5 kb, which is not suitable for large transgenes and is only available for Cas9 derived from smaller orthologs such as Staphylococcus aureus (SaCas9) or Campylobacter jejuni (CjCas9). The gRNA for Cas9 also requires a specific protospacer adjacent motif (PAM) that varies depending on the bacterial species. The most common Cas9 derived from Staphylococcus pyogenes (SpCas9) recognizes NGG directly downstream of the target sequence in the genomic DNA, while the PAM sequence of SaCas9 for optical target requires NNGRRT, limiting the design of specific gRNA target site. The length of the SpCas9 encoding gene is oversized to be packaged in AAV. The AAV-CRISPR system holds the potential to develop therapeutic options, but on the other hand, the development of a novel in vivo Cas9 delivery platform is urgently required to increase its flexibility.
Human Immunodeficiency Virus Type-I Reservoir Detection
Analytic Treatment Interruption
Accurately measuring the latent HIV reservoir is critical to assessing the effectiveness of curative strategies aimed at HIV remission. To date, the only way to definitively evaluate the effectiveness of curative strategies is with an analytic treatment interruption (ATI) in which the individual stops taking cART (Harari et al., 2012). The time that it takes for the viral rebound to occur after treatment cessation can be used to evaluate reservoir reduction. Theoretically, since viral rebound reflects the release of the virus from a stable reservoir, the smaller the size of the reservoir, the longer it takes for the viral rebound to occur (Hill et al., 2016; Li J. Z. et al., 2016). In the hopes to manage adverse effects associated with long-term use of cART, a clinical trial was performed in which HIV-infected individuals were randomly assigned to undergo either continuous suppressive cART or CD4+ count-guided episodic use of cART (NCT00027352) (El-Sadr et al., 2006). This study found that participants who undergo episodic treatment interruptions have significantly higher rates of opportunistic diseases or death from any cause when compared to participants on continuous cART. Moreover, the selection of drug-resistant mutations can occur during repeated treatment interruptions (Martinez-Picado et al., 2002), and rebounding viruses display increased IFNα2 and IFNβ resistance (Gondim et al., 2021). However, in some cases, individuals with high CD4 counts (>500 cells/ul) can safely undergo short CD4+ T cell count guided treatment interruptions without increased risk of morbidity or mortality and without developing drug resistance (Maggiolo et al., 2009; Routy et al., 2012; Ananworanich et al., 2015). While treatment interruption may provide evidence of complete viral eradication, individuals will most likely require long-term monitoring as unpredictable stochastic events can lead to viral rebound months to years later if the latent reservoir is not completely eradicated but only greatly reduced. Furthermore, the individual variability in time to rebound makes it hard to assess the magnitude of reservoir reduction as a result of eradication efforts.
Quantitative Viral Outgrowth Assay
Currently, there is no accurate way to measure the latent reservoir in vivo. The quantitative viral outgrowth assay (QVOA) has been regarded as the “gold standard” for measuring the replication-competent latent reservoir size ex vivo (Finzi et al., 1997; Siliciano et al., 2003). QVOA measures the frequency of resting CD4+ T cells that produce infectious viruses after a single round of maximum global T cell activation. To this end, a large volume of resting CD4+ T cells are isolated from HIV-infected individuals and stimulated with the mitogen phytohemagglutinin (PHA) in the presence of uninfected γ-irradiated allogeneic PBMCs. Then the donor cells are co-cultured with either CD4+ T cells from a healthy donor or a cell line for 2–3 weeks before the infectious virus is measured in the culture supernatant by HIV-1 P24 ELISA (Finzi et al., 1997; Stuelke et al., 2020) or using a quantitative RT-PCR assay (Laird et al., 2013). However, some replication-competent viruses are induced only after multiple rounds of stimulation, indicating that standard QVOA may underestimate the size of the inducible latent reservoir (Abrahams et al., 2019). Furthermore, traditional QVOA is labor intensive and requires large amounts of sample. These limitations have led to the development of other approaches to estimate the size of the latent reservoir.
PCR-Based Methods to Detect Human Immunodeficiency Virus Type-I Proviruses
Standard PCR-based techniques to quantify total HIV DNA are the easiest way to measure HIV-infected cells in PLWH. Total HIV DNA measurements inherently overestimate the size of the HIV reservoir since the majority of proviruses are defective or deleted (Bruner et al., 2016). However, total HIV DNA remains an important biomarker for viral persistence (Avettand-Fènoël et al., 2016), and levels of this marker are associated with viral rebound upon treatment cessation (Yerly et al., 2004; Williams et al., 2014). Recent approaches using droplet digital PCR (ddPCR) employ absolute quantification, which is more accurate than traditional quantitative PCR (qPCR) methods (Strain et al., 2013). Multiplexed ddPCR based assays have been used to track the fraction of deleted proviruses during cART (Anderson and Maldarelli, 2018; Anderson et al., 2020). Still, total HIV DNA measurements are at least two orders of magnitude higher than latent reservoir size measurements by QVOA (Eriksson et al., 2013). Total HIV DNA measurements may be further confounded by unintegrated HIV in either linear or 2-LTR circle forms, as these methods cannot distinguish integrated HIV DNA from non-integrated forms. Efforts to quantify only integrated HIV DNA utilize Alu-PCR (O’Doherty et al., 2002; Brady et al., 2013; De Spiegelaere et al., 2014). Alu-gag-PCR employs an outer forward primer that binds Alu, a repetitive element that is abundant in the human genome, and a reverse primer that is complementary to HIV gag DNA. This method will only amplify HIV proviruses integrated into the host genome and contain the primer target region of gag (O’Doherty et al., 2002; Liszewski et al., 2009). Alu-gag-PCR gives latent reservoir size estimates that are lower than total HIV DNA measurements but are still orders of magnitude higher than QVOA due to the inability of these assays to distinguish between replication-competent and defective proviruses (Eriksson et al., 2013).
Multiple methods have been developed to assess the proportion of intact versus obviously defective or deleted proviruses. Sequencing approaches such as matched integration site and proviral sequencing (MIP-seq) (Einkauf et al., 2019) and multiple-displacement amplification single genome sequencing (MDA-SGS) (Patro et al., 2019) can link full-length proviral sequences with their respective integration sites to infer replication competence as well as clonality. While these assays will provide great insights into the proviral landscape, the costs and labor required may hinder their use in large-scale studies. The intact proviral DNA assay (IPDA) is a high throughput ddPCR assay designed with two sets of primers located in conserved and frequently deleted regions of the viral genome (Bruner et al., 2019). IPDA offers a robust tool to estimate the number of intact proviruses and has been utilized as a surrogate for the latent reservoir (Simonetti et al., 2020). However, PCR failure as a result of primer mismatch due to HIV-1 diversity (Kinloch et al., 2021) and the inability to exclude proviruses that are defective or deleted in other regions (Gaebler et al., 2021) may preclude proper latent reservoir size measurements by IPDA. Other ddPCR strategies that utilize more target regions can further exclude defective proviruses. For example, a triplex digital PCR method (van Snippenberg et al., 2021) and a five-region approach that combines two triplex ddPCR assays (Levy et al., 2021) have been developed to help overcome the mischaracterization of intact proviruses while still utilizing a high throughput and relatively inexpensive digital PCR platform. The recently developed quadruplex PCR with four probes (Q4PCR) assay combines a four-probe qPCR strategy with near full-length sequencing to distinguish between intact and defective proviruses (Gaebler et al., 2019; Cho et al., 2022). In this assay, individual HIV genomes are amplified to near full length and are screened for the presence of 4 HIV targets with qPCR. Wells that are positive for at least two targets are then selected to undergo full-length proviral sequencing (Gaebler et al., 2019). Q4PCR is a lower-throughput assay compared to IPDA but allows for confirmation of proviral intactness. When compared head-to-head, IPDA and Q4PCR measurements correlated with one another but levels of intact proviruses measured with IPDA were approximately 19-fold higher than Q4PCR measurements (Gaebler et al., 2021). These differences in reservoir size estimates are likely from a combination of an overestimation by IPDA due to its inability to exclude defects in other regions of the provirus, and an underestimation by Q4PCR, due to inefficiencies from long-distance PCR. The actual number of intact proviruses may lie in between the measurements from IPDA and Q4PCR. Moreover, not all intact proviruses are replication-competent and ultimately give rise to rebound viremia. The chromatin environment at the proviral integration site as well as defects in transcription and/or translation of the provirus govern its ability to give rise to the infectious virus (Einkauf et al., 2022).
Quantifying Functional Human Immunodeficiency Virus Type-I Proviruses
Strategies to more accurately estimate latent reservoir size aim to quantify replication-competent proviruses. The Tat/rev Induced Limiting Dilution Assay (TILDA) measures the frequency of cells harboring viral genomes that produce tat/rev multiply spliced HIV RNA (msRNA) upon maximum activation of CD4+ T cells (Procopio et al., 2015). The tat/rev msRNA is essential to produce infectious viruses (Pasternak et al., 2008). Many deleted proviruses lack tat and rev genes (Abrahams et al., 2019), and cells that contain tat/rev msRNA likely harbor a replication-competent infectious provirus. However, TILDA may still overestimate the actual size of the HIV reservoir since integrated proviruses that produce msRNA are not guaranteed to be infectious (Procopio et al., 2015). Recent advances in the development of next-generation in situ hybridization (ISH) technologies allow the detection of native viral DNA and RNA markers in histological specimens with greater sensitivity and faster workflow than traditional ISH (Deleage et al., 2016, 2018). These methods, namely DNAScope and RNAScope, can be leveraged to quantify the number of HIV-DNA and RNA-positive cells per gram of tissue (Estes et al., 2017). Uniquely among the methods described in this section, which are based on bulk sampling, tissue imaging approaches bypass the requirement of tissue homogenization and can provide spatial localization of the viral reservoir in tissue compartments. In addition, these allow multiplexing the detection of viral and host biomarkers to phenotype latently and actively infected cells and characterizing the complex heterogeneity of microenvironments that sustain virus persistence. However, these imaging-based approaches remain limited by the scarce accessibility of tissue specimens from clinical trials as opposed to blood and by the two-dimensional analysis of a few representative sections, which is based on the assumption that observations made can be inferred to the entire organ. In this regard, it is encouraging to see the emergence of total body positron emission tomography (PET) scanning (Santangelo et al., 2015; Henrich et al., 2019; Taylor et al., 2021) and whole organ imaging with novel tissue clearing technologies coupled with light-sheet microscopy (Tomer et al., 2014), which may represent new frontiers to detect viral reservoirs and broaden our understanding of virus persistence in tissue. Other approaches to identifying replication-competent reservoirs measure translation competent proviruses (Baxter et al., 2016). A flow-based RNA FISH assay simultaneously measures HIV RNA as well as Gag proteins upon phorbol 12-myristate 13-acetate (PMA) stimulation to identify single cells that are double positive for both cell-associated unspliced HIV gag RNA and its translation product Gag protein (HIVRNA+/Gag+). HIVRNA+/Gag+ cells likely give rise to the infectious virus, enabling this assay to more accurately estimate the latent reservoir size compared to assays that viral RNA transcripts alone (Baxter et al., 2016). The advancement of digital enzyme-linked immunosorbent assay with a single molecule array (Simoa) has enabled the detection of cell-associated HIV gag p24 protein from both peripheral and tissue compartments with sensitivity higher than traditional ELISA (Passaes et al., 2017; Wu et al., 2017, 2021; Stuelke et al., 2020). These assays can be used to quantify steady-state and inducible reservoirs with the advantage of focusing on viruses capable of translating and processing Gag antigens that are relevant for infected cells clearance (Abdel-Mohsen et al., 2020). These further characterize reservoirs independently from their genetic intactness as defective proviruses can produce viral antigens and contribute to chronic immune system stimulation (Imamichi et al., 2020). Overall, rapid and accurate measurements of the true latent reservoir are needed to assess the effectiveness of curative strategies. For now, a combination of the described assays can be used to estimate the size of the latent reservoir, and the true size likely lies somewhere in between QVOA estimates and translation competent measurements.
Summary
Once HIV infection is established, cART cannot eradicate the integrated proviruses due to latency establishment in multiple mechanisms (Table 1). HIV-1 gene transcription and subsequent translation are highly controlled by cis- and trans-regulatory elements and form distinct phenotypes of latent cells (Figure 2). HIV-1 reservoirs are generated in the early acute phase of virus infection with long-lasting treatment-resistant cells that can undergo clonal expansion during cART. Various strategies have been proposed to perturb the latent reservoir. The “Block-and-Lock” strategy aims to permanently silence the latent reservoir using latency-promoting agents such as LEDGINs. LEDGINSs can alter HIV-1 DNA integration sites to deeply silenced regions that are inefficient for reactivation even after cART cessation (Figure 5). While this treatment could be an approach to silence virus expression once and for all, it is not designed to have a profound effect on eradication after reservoir formation. Thus, the “Block-and-Lock” strategy could be effective when used during acute viral infection. The “Kick-and-Kill” strategy is designed to induce the expression of viral antigens by reactivation of latent cells (Figure 5). LRAs target both cis- and trans-mechanisms of the suppressive viral promoter to reactivate the latent cells, leading to the subsequent elimination by immune cells. Since individual LRA studies to date have ineffectively reduced reservoir size in clinical trials, several LRAs with distinct mechanisms of action may be needed to address the heterogeneity in latency.
While CD4+ T cells have been regarded as major cellular reservoir compartment, tissue localized long-lived myeloid cells may also have an essential role in HIV reservoir formation (Figure 1). The memory subsets of latently infected CD4+ T cells containing replication-competent proviral DNA are frequently detected in lymphoid tissue. In addition to the memory T cell subsets in peripheral blood and lymphoid tissue, macrophagic and astrocytic myeloid cells in the CNS pose a unique challenge for virus elimination (Figure 1).
During chronic infection, the immune system is in a state of non-reactivity to HIV-1 antigen caused by constant stimulation by viral antigens presentation or viral-like particles (VLPs) produced by defective proviruses that have large internal deletions or hypermutations (Figure 4).
The immune checkpoint blockade (ICB), which could either activate or inactivate specific immune cells could be useful to purge reactivated cells. The combination of LRAs with ICB has been demonstrated to effectively reduce the reservoir size in vivo, indicating that combinatorial approaches, in which LRAs are used to obtain a more effective shock and ICBs restore HIV-1 specific immune cells to eliminate reactivated reservoirs (Figure 5).
The Food and Drug Administration recently approved a human trial for HIV cure with CRISPR/Cas9 genome editing technology (Figure 6). While the adeno-associated viral vector (AAV) delivery system has several limitations, AAV is a valid option for in vivo Cas9 delivery to deactivate proviruses in tissue-localized reservoir cells in mice models. Cas9 targeting HIV-1 proviral DNA could potentially deactivate intact reservoir cells after the Kick-and-Kill therapy. Taken together, combining appropriate therapies at the stages of HIV acute and chronic infection, could contribute to new cases of cure without resorting to risky and unscalable bone marrow or stem cell transplantation.
Author Contributions
TT, SM, EA, JP, MF, and TI draw figures. EA, LS, ZK, and TI designed the project. ZK and TI contributed to financial assistance. All authors listed wrote the manuscript and contributed to the article and approved the submitted version.
Funding
This project was supported in part by start-up from the University of the Sciences in Philadelphia to TI and from NIDA/NIH (DP2DA044550-01) and PA Department of Health CURE (4100088542) to ZK.
Author Disclaimer
The findings of this article are those of the authors. They do not necessarily reflect the views of the Office of the Assistant Secretary for Health or the U.S. Department of Health and Human Services.
Conflict of Interest
LS is employed by Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, United States.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbar, B., Baron, M., Katlama, C., Marcelin, A.-G., Veyri, M., Autran, B., et al. (2020). Immune checkpoint inhibitors in people living with HIV: what about anti-HIV effects? AIDS 34, 167–175. doi: 10.1097/QAD.0000000000002397
Abdel-Mohsen, M., Richman, D., Siliciano, R. F., Nussenzweig, M. C., Howell, B., Martinez-Picado, J., et al. (2020). Recommendations for measuring HIV reservoir size in cure-directed clinical trials. Nat. Med. 26, 1339–1350. doi: 10.1038/s41591-020-1022-1
Abner, E., Stoszko, M., Zeng, L., Chen, H.-C., Izquierdo-Bouldstridge, A., Konuma, T., et al. (2018). A New Quinoline BRD4 Inhibitor Targets a Distinct Latent HIV-1 Reservoir for Reactivation from Other “Shock” Drugs. J. Virol. 2018:17. doi: 10.1128/JVI.02056-17
Abrahams, M.-R., Joseph, S. B., Garrett, N., Tyers, L., Moeser, M., Archin, N., et al. (2019). The replication-competent HIV-1 latent reservoir is primarily established near the time of therapy initiation. Sci. Transl. Med. 11:eaaw5589. doi: 10.1126/scitranslmed.aaw5589
Adams, M., Sharmeen, L., Kimpton, J., Romeo, J. M., Garcia, J. V., Peterlin, B. M., et al. (1994). Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. 91, 3862–3866. doi: 10.1073/pnas.91.9.3862
Ahn, E., Youngblood, B., Lee, J., Lee, J., Sarkar, S., and Ahmed, R. (2016). Demethylation of the PD-1 Promoter Is Imprinted during the Effector Phase of CD8 T Cell Exhaustion. J. Virol. 90, 8934–8946. doi: 10.1128/JVI.00798-16
Ait-Ammar, A., Kula, A., Darcis, G., Verdikt, R., De Wit, S., Gautier, V., et al. (2020). Current Status of Latency Reversing Agents Facing the Heterogeneity of HIV-1 Cellular and Tissue Reservoirs. Front. Microbiol. 10:3060. doi: 10.3389/fmicb.2019.03060
Akhtar, A. (2015). The Flaws and Human Harms of Animal Experimentation. Camb. Q. Healthc. Ethics 24, 407–419. doi: 10.1017/S0963180115000079
Alamer, E., Zhong, C., Liu, Z., Niu, Q., Long, F., Guo, L., et al. (2020). Epigenetic Suppression of HIV in Myeloid Cells by the BRD4-Selective Small Molecule Modulator ZL0580. J. Virol. 94:19. doi: 10.1128/JVI.01880-19
Ali, A., Kitchen, S. G., Chen, I. S. Y., Ng, H. L., Zack, J. A., and Yang, O. O. (2016). HIV-1-Specific Chimeric Antigen Receptors Based on Broadly Neutralizing Antibodies. J. Virol. 90, 6999–7006. doi: 10.1128/JVI.00805-16
Allers, K., Hütter, G., Hofmann, J., Loddenkemper, C., Rieger, K., Thiel, E., et al. (2011). Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood 117, 2791–2799. doi: 10.1182/blood-2010-09-309591
Álvarez-Fernández, C., Crespo Guardo, A., García-Pérez, J., García, F., Blanco, J., Escribà-García, L., et al. (2012). Generation and characterization of a defective HIV-1 Virus as an immunogen for a therapeutic vaccine. PLoS One 7:e48848. doi: 10.1371/journal.pone.0048848
Ammosova, T., Berro, R., Jerebtsova, M., Jackson, A., Charles, S., Klase, Z., et al. (2006). Phosphorylation of HIV-1 Tat by CDK2 in HIV-1 transcription. Retrovirology 3:78. doi: 10.1186/1742-4690-3-78
Ananworanich, J., Chomont, N., Fletcher, J. L. K., Pinyakorn, S., Schuetz, A., Sereti, I., et al. (2015). Markers of HIV reservoir size and immune activation after treatment in acute HIV infection with and without raltegravir and maraviroc intensification. J. Virus Eradicat. 1, 116–122. doi: 10.1016/S2055-6640(20)30482-9
Anderson, E. M., and Maldarelli, F. (2018). The role of integration and clonal expansion in HIV infection: live long and prosper. Retrovirology 15:71. doi: 10.1186/s12977-018-0448-8
Anderson, E. M., Simonetti, F. R., Gorelick, R. J., Hill, S., Gouzoulis, M. A., Bell, J., et al. (2020). Dynamic Shifts in the HIV Proviral Landscape During Long Term Combination Antiretroviral Therapy: Implications for Persistence and Control of HIV Infections. Viruses 12:E136. doi: 10.3390/v12020136
Appay, V., Papagno, L., Spina, C. A., Hansasuta, P., King, A., Jones, L., et al. (2002). Dynamics of T Cell Responses in HIV Infection. J. Immunol. 168, 3660–3666. doi: 10.4049/jimmunol.168.7.3660
Archin, N. M., Keedy, K. S., Espeseth, A., Dang, H., Hazuda, D. J., and Margolis, D. M. (2009). Expression of latent human immunodeficiency type 1 is induced by novel and selective histone deacetylase inhibitors. AIDS 23, 1799–1806. doi: 10.1097/QAD.0b013e32832ec1dc
Archin, N. M., Liberty, A. L., Kashuba, A. D., Choudhary, S. K., Kuruc, J. D., Crooks, A. M., et al. (2012). Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487, 482–485. doi: 10.1038/nature11286
Arts, E. J., and Hazuda, D. J. (2012). HIV-1 Antiretroviral Drug Therapy. Cold Spring Harb. Perspect. Med. 2, a007161–a007161. doi: 10.1101/cshperspect.a007161
Avettand-Fènoël, V., Hocqueloux, L., Ghosn, J., Cheret, A., Frange, P., Melard, A., et al. (2016). Total HIV-1 DNA, a Marker of Viral Reservoir Dynamics with Clinical Implications. Clin. Microbiol. Rev. 29, 859–880. doi: 10.1128/CMR.00015-16
Baeuerle, P. A., and Henkel, T. (1994). Function and activation of NF-kappa B in the immune system. Annu. Rev. Immunol. 12, 141–179. doi: 10.1146/annurev.iy.12.040194.001041
Bailey, M., Christoforidou, Z., and Lewis, M. C. (2013). The evolutionary basis for differences between the immune systems of man, mouse, pig and ruminants. Vet. Immunol. Immunopathol. 152, 13–19. doi: 10.1016/j.vetimm.2012.09.022
Baker, J. V., Peng, G., Rapkin, J., Abrams, D. I., Silverberg, M. J., MacArthur, R. D., et al. (2008). CD4+ count and risk of non-AIDS diseases following initial treatment for HIV infection. AIDS 22, 841–848. doi: 10.1097/QAD.0b013e3282f7cb76
Baldwin, A. S. (1996). THE NF-κB AND IκB PROTEINS: New Discoveries and Insights. Annu. Rev. Immunol. 14, 649–681. doi: 10.1146/annurev.immunol.14.1.649
Banga, R., Procopio, F. A., Noto, A., Pollakis, G., Cavassini, M., Ohmiti, K., et al. (2016). PD-1+ and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat. Med. 22, 754–761. doi: 10.1038/nm.4113
Bartholomeeusen, K., Xiang, Y., Fujinaga, K., and Peterlin, B. M. (2012). Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J. Biol. Chem. 287, 36609–36616. doi: 10.1074/jbc.M112.410746
Baxter, A. E., Niessl, J., Fromentin, R., Richard, J., Porichis, F., Charlebois, R., et al. (2016). Single-Cell Characterization of Viral Translation-Competent Reservoirs in HIV-Infected Individuals. Cell Host Microbe 20, 368–380. doi: 10.1016/j.chom.2016.07.015
Benkirane, M., Chun, R. F., Xiao, H., Ogryzko, V. V., Howard, B. H., Nakatani, Y., et al. (1998). Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J. Biol. Chem. 273, 24898–24905. doi: 10.1074/jbc.273.38.24898
Besnard, E., Hakre, S., Kampmann, M., Lim, H. W., Hosmane, N. N., Martin, A., et al. (2016). The mTOR Complex Controls HIV Latency. Cell Host Microbe 20, 785–797. doi: 10.1016/j.chom.2016.11.001
Bisgrove, D. A., Mahmoudi, T., Henklein, P., and Verdin, E. (2007). Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. PNAS 104, 13690–13695. doi: 10.1073/pnas.0705053104
Bissels, U., Wild, S., Tomiuk, S., Holste, A., Hafner, M., Tuschl, T., et al. (2009). Absolute quantification of microRNAs by using a universal reference. RNA 15, 2375–2384. doi: 10.1261/rna.1754109
Biti, R., Ffrench, R., Young, J., Bennetts, B., Stewart, G., and Liang, T. (1997). HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat. Med. 3, 252–253. doi: 10.1038/nm0397-252
Blackburn, S. D., Shin, H., Haining, W. N., Zou, T., Workman, C. J., Polley, A., et al. (2009). Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 10, 29–37. doi: 10.1038/ni.1679
Blazkova, J., Trejbalova, K., Gondois-Rey, F., Halfon, P., Philibert, P., Guiguen, A., et al. (2009). CpG Methylation Controls Reactivation of HIV from Latency. PLoS Pathog. 5:e1000554. doi: 10.1371/journal.ppat.1000554
Boasso, A., Shearer, G. M., and Chougnet, C. (2009). Immune dysregulation in human immunodeficiency virus infection: know it, fix it, prevent it? J. Internal Med. 265, 78–96. doi: 10.1111/j.1365-2796.2008.02043.x
Bobardt, M., Kuo, J., Chatterji, U., Chanda, S., Little, S. J., Wiedemann, N., et al. (2019). The inhibitor apoptosis protein antagonist Debio 1143 Is an attractive HIV-1 latency reversal candidate. PLoS One 14:e0211746. doi: 10.1371/journal.pone.0211746
Boehm, D., Jeng, M., Camus, G., Gramatica, A., Schwarzer, R., Johnson, J. R., et al. (2017). SMYD2-Mediated Histone Methylation Contributes to HIV-1 Latency. Cell Host Microbe 21, 569.e–579.e. doi: 10.1016/j.chom.2017.04.011
Boltz, V. F., Ceriani, C., Rausch, J. W., Shao, W., Bale, M. J., Keele, B. F., et al. (2021). CpG Methylation Profiles of HIV-1 Proviral DNA in Individuals on ART. Viruses 13:799. doi: 10.3390/v13050799
Borrow, P., Lewicki, H., Hahn, B. H., Shaw, G. M., and Oldstone, M. B. (1994). Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68, 6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994
Bosque, A., Famiglietti, M., Weyrich, A. S., Goulston, C., and Planelles, V. (2011). Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog. 7:e1002288. doi: 10.1371/journal.ppat.1002288
Bosque, A., Nilson, K. A., Macedo, A. B., Spivak, A. M., Archin, N. M., Van Wagoner, R. M., et al. (2017). Benzotriazoles Reactivate Latent HIV-1 through Inactivation of STAT5 SUMOylation. Cell Rep. 18, 1324–1334. doi: 10.1016/j.celrep.2017.01.022
Bouchat, S., Delacourt, N., Kula, A., Darcis, G., Van Driessche, B., Corazza, F., et al. (2016). Sequential treatment with 5-aza-2’-deoxycytidine and deacetylase inhibitors reactivates HIV-1. EMBO Mol. Med. 8, 117–138. doi: 10.15252/emmm.201505557
Bouchat, S., Gatot, J.-S., Kabeya, K., Cardona, C., Colin, L., Herbein, G., et al. (2012). Histone methyltransferase inhibitors induce HIV-1 recovery in resting CD4+ T cells from HIV-1-infected HAART-treated patients. AIDS 26, 1473–1482. doi: 10.1097/QAD.0b013e32835535f5
Brady, T., Kelly, B. J., Male, F., Roth, S., Bailey, A., Malani, N., et al. (2013). Quantitation of HIV DNA integration: Effects of differential integration site distributions on Alu-PCR assays. J. Virol. Methods 189, 53–57. doi: 10.1016/j.jviromet.2013.01.004
Brass, A. L., Dykxhoorn, D. M., Benita, Y., Yan, N., Engelman, A., Xavier, R. J., et al. (2008). Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926. doi: 10.1126/science.1152725
Brodin, J., Zanini, F., Thebo, L., Lanz, C., Bratt, G., Neher, R. A., et al. (2016). Establishment and stability of the latent HIV-1 DNA reservoir. eLife 5:e18889. doi: 10.7554/eLife.18889
Bruner, K. M., Murray, A. J., Pollack, R. A., Soliman, M. G., Laskey, S. B., Capoferri, A. A., et al. (2016). Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 22, 1043–1049. doi: 10.1038/nm.4156
Bruner, K. M., Wang, Z., Simonetti, F. R., Bender, A. M., Kwon, K. J., Sengupta, S., et al. (2019). A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566, 120–125. doi: 10.1038/s41586-019-0898-8
Bruno, J., and Stoughton, A. M. (1984). Computer-aided communication device for a child with cerebral palsy. Arch. Phys. Med. Rehabil. 65, 603–605.
Budhiraja, S., Famiglietti, M., Bosque, A., Planelles, V., and Rice, A. P. (2013). Cyclin T1 and CDK9 T-Loop Phosphorylation Are Downregulated during Establishment of HIV-1 Latency in Primary Resting Memory CD4 + T Cells. J. Virol. 87, 1211–1220. doi: 10.1128/JVI.02413-12
Bui, J. K., Sobolewski, M. D., Keele, B. F., Spindler, J., Musick, A., Wiegand, A., et al. (2017). Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 13:e1006283. doi: 10.1371/journal.ppat.1006283
Burnett, J. C., Miller-Jensen, K., Shah, P. S., Arkin, A. P., and Schaffer, D. V. (2009). Control of Stochastic Gene Expression by Host Factors at the HIV Promoter. PLoS Pathog. 5:e1000260. doi: 10.1371/journal.ppat.1000260
Buzon, M. J., Sun, H., Li, C., Shaw, A., Seiss, K., Ouyang, Z., et al. (2014). HIV-1 persistence in CD4+ T cells with stem cell–like properties. Nat. Med. 20, 139–142. doi: 10.1038/nm.3445
Calle-Fabregat, C., de la Morante-Palacios, O., and Ballestar, E. (2020). Understanding the Relevance of DNA Methylation Changes in Immune Differentiation and Disease. Genes 11:110. doi: 10.3390/genes11010110
Cantero-Pérez, J., Grau-Expósito, J., Serra-Peinado, C., Rosero, D. A., Luque-Ballesteros, L., Astorga-Gamaza, A., et al. (2019). Resident memory T cells are a cellular reservoir for HIV in the cervical mucosa. Nat. Commun. 10:4739. doi: 10.1038/s41467-019-12732-2
Cao, Y., Qin, L., Zhang, L., Safrit, J., and Ho, D. D. (1995). Virologic and Immunologic Characterization of Long-Term Survivors of Human Immunodeficiency Virus Type 1 Infection. N. Engl. J. Med. 332, 201–208. doi: 10.1056/NEJM199501263320401
Carpio, L., Klase, Z., Coley, W., Guendel, I., Choi, S., Van Duyne, R., et al. (2010). microRNA machinery is an integral component of drug-induced transcription inhibition in HIV-1 infection. J. RNAi Gene Silencing 6, 386–400.
Cary, D. C., and Peterlin, B. M. (2018). Natural Products and HIV/AIDS. AIDS Res. Hum. Retrovir. 34, 31–38. doi: 10.1089/aid.2017.0232
Chávez, L., Kauder, S., and Verdin, E. (2011). In vivo, in vitro, and in silico analysis of methylation of the HIV-1 provirus. Methods 53, 47–53. doi: 10.1016/j.ymeth.2010.05.009
Chen, Y., Zander, R., Khatun, A., Schauder, D. M., and Cui, W. (2018). Transcriptional and Epigenetic Regulation of Effector and Memory CD8 T Cell Differentiation. Front. Immunol. 9:2826. doi: 10.3389/fimmu.2018.02826
Chéné, I., du, Basyuk, E., Lin, Y.-L., Triboulet, R., Knezevich, A., et al. (2007). Suv39H1 and HP1γ are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 26, 424–435. doi: 10.1038/sj.emboj.7601517
Cherrier, T., Le Douce, V., Eilebrecht, S., Riclet, R., Marban, C., Dequiedt, F., et al. (2013). CTIP2 is a negative regulator of P-TEFb. Proc. Natl. Acad. Sci. 110, 12655–12660. doi: 10.1073/pnas.1220136110
Chiang, K., Sung, T.-L., and Rice, A. P. (2012). Regulation of cyclin T1 and HIV-1 Replication by microRNAs in resting CD4+ T lymphocytes. J. Virol. 86, 3244–3252. doi: 10.1128/JVI.05065-11
Cho, A., Gaebler, C., Olveira, T., Ramos, V., Saad, M., Lorenzi, J. C. C., et al. (2022). Longitudinal clonal dynamics of HIV-1 latent reservoirs measured by combination quadruplex polymerase chain reaction and sequencing. PNAS 2022:119. doi: 10.1073/pnas.2117630119
Chomont, N., El-Far, M., Ancuta, P., Trautmann, L., Procopio, F. A., Yassine-Diab, B., et al. (2009). HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 15, 893–900. doi: 10.1038/nm.1972
Christ, F., Shaw, S., Demeulemeester, J., Desimmie, B. A., Marchand, A., Butler, S., et al. (2012). Small-molecule inhibitors of the LEDGF/p75 binding site of integrase block HIV replication and modulate integrase multimerization. Antimicrob. Agents Chemother. 56, 4365–4374. doi: 10.1128/AAC.00717-12
Chun, T., Nickle, D. C., Justement, J. S., Meyers, J. H., Roby, G., Hallahan, C. W., et al. (2008). Persistence of HIV in Gut-Associated Lymphoid Tissue despite Long-Term Antiretroviral Therapy. J. Infect. Dis. 197, 714–720. doi: 10.1086/527324
Chun, T.-W., Justement, J. S., Murray, D., Hallahan, C. W., Maenza, J., Collier, A. C., et al. (2010). Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. AIDS 24, 2803–2808. doi: 10.1097/QAD.0b013e328340a239
Chun, T.-W., Stuyver, L., Mizell, S. B., Ehler, L. A., Mican, J. A. M., Baseler, M., et al. (1997). Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. 94, 13193–13197. doi: 10.1073/pnas.94.24.13193
Clerici, M., Giorgi, J. V., Chou, C.-C., Gudeman, V. K., Zack, J. A., Gupta, P., et al. (1992). Cell-Mediated Immune Response to Human Immunodeficiency Virus (HIV) Type 1 in Seronegative Homosexual Men with Recent Sexual Exposure to HIV-1. J. Infect. Dis. 165, 1012–1019. doi: 10.1093/infdis/165.6.1012
Clutton, G. T., and Jones, R. B. (2018). Diverse Impacts of HIV Latency-Reversing Agents on CD8+ T-Cell Function: Implications for HIV Cure. Front. Immunol. 9:1452. doi: 10.3389/fimmu.2018.01452
Cocchi, F., DeVico, A. L., Garzino-Demo, A., Arya, S. K., Gallo, R. C., and Lusso, P. (1995). Identification of RANTES, MIP-1α, and MIP-1β as the Major HIV-Suppressive Factors Produced by CD8 + T Cells. Science 270, 1811–1815. doi: 10.1126/science.270.5243.1811
Coffin, J. M., and Hughes, S. H. (2021). Clonal Expansion of Infected CD4+ T Cells in People Living with HIV. Viruses 13:2078. doi: 10.3390/v13102078
Coffin, J. M., Bale, M. J., Wells, D., Guo, S., Luke, B., Zerbato, J. M., et al. (2021). Integration in oncogenes plays only a minor role in determining the in vivo distribution of HIV integration sites before or during suppressive antiretroviral therapy. PLoS Pathog. 17:e1009141. doi: 10.1371/journal.ppat.1009141
Coffin, J. M., Hughes, S. H., and Varmus, H. (eds) (1997). Retroviruses. Plainview, NY: Cold Spring Harbor Laboratory Press.
Cohn, L. B., Silva, I. T., Oliveira, T. Y., Rosales, R. A., Parrish, E. H., Learn, G. H., et al. (2015). HIV-1 integration landscape during latent and active infection. Cell 160, 420–432. doi: 10.1016/j.cell.2015.01.020
Col, E., Caron, C., Seigneurin-Berny, D., Gracia, J., Favier, A., and Khochbin, S. (2001). The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator. Tat. J. Biol. Chem. 276, 28179–28184. doi: 10.1074/jbc.M101385200
Colin, L., and Van Lint, C. (2009). Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology 6:111. doi: 10.1186/1742-4690-6-111
Colomer-Lluch, M., Ruiz, A., Moris, A., and Prado, J. G. (2018). Restriction Factors: From Intrinsic Viral Restriction to Shaping Cellular Immunity Against HIV-1. Front. Immunol. 9:2876. doi: 10.3389/fimmu.2018.02876
Contreras, X., Barboric, M., Lenasi, T., and Peterlin, B. M. (2007). HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 3:1459–1469. doi: 10.1371/journal.ppat.0030146
Cook, M. R., and Kim, C. (2019). Safety and Efficacy of Immune Checkpoint Inhibitor Therapy in Patients With HIV Infection and Advanced-Stage Cancer: A Systematic Review. JAMA Oncol. 5:1049. doi: 10.1001/jamaoncol.2018.6737
Coppé, J.-P., Patil, C. K., Rodier, F., Krtolica, A., Beauséjour, C. M., Parrinello, S., et al. (2010). A Human-Like Senescence-Associated Secretory Phenotype Is Conserved in Mouse Cells Dependent on Physiological Oxygen. PLoS One 5:e9188. doi: 10.1371/journal.pone.0009188
Corley, M. J., Dye, C., D’Antoni, M. L., Byron, M. M., Yo, K. L.-A., Lum-Jones, A., et al. (2016). Comparative DNA Methylation Profiling Reveals an Immunoepigenetic Signature of HIV-related Cognitive Impairment. Sci. Rep. 6:33310. doi: 10.1038/srep33310
Crooks, A. M., Bateson, R., Cope, A. B., Dahl, N. P., Griggs, M. K., Kuruc, J. D., et al. (2015). Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies. J. Infect. Dis. 212, 1361–1365. doi: 10.1093/infdis/jiv218
Dahabieh, M., Battivelli, E., and Verdin, E. (2015). Understanding HIV Latency: The Road to an HIV Cure. Annu. Rev. Med. 66, 407–421. doi: 10.1146/annurev-med-092112-152941
Dar, A., Kollet, O., and Lapidot, T. (2006). Mutual, reciprocal SDF-1/CXCR4 interactions between hematopoietic and bone marrow stromal cells regulate human stem cell migration and development in NOD/SCID chimeric mice. Exp. Hematol. 34, 967–975. doi: 10.1016/j.exphem.2006.04.002
Darcis, G., Berkhout, B., and Pasternak, A. O. (2019). The Quest for Cellular Markers of HIV Reservoirs: Any Color You Like. Front. Immunol. 10:2251. doi: 10.3389/fimmu.2019.02251
Darcis, G., Bouchat, S., Kula, A., Van Driessche, B., Delacourt, N., Vanhulle, C., et al. (2017). Reactivation capacity by latency-reversing agents ex vivo correlates with the size of the HIV-1 reservoir. AIDS 31, 181–189. doi: 10.1097/QAD.0000000000001290
Dash, P. K., Kaminski, R., Bella, R., Su, H., Mathews, S., Ahooyi, T. M., et al. (2019). Sequential LASER ART and CRISPR Treatments Eliminate HIV-1 in a Subset of Infected Humanized Mice. Nat. Commun. 10:2753. doi: 10.1038/s41467-019-10366-y
Dashti, A., Waller, C., Mavigner, M., Schoof, N., Bar, K. J., Shaw, G. M., et al. (2020). SMAC Mimetic Plus Triple-Combination Bispecific HIVxCD3 Retargeting Molecules in SHIV.C.CH505-Infected, Antiretroviral Therapy-Suppressed Rhesus Macaques. J. Virol. 94, e793–e720. doi: 10.1128/JVI.00793-20
Davidson, E. (2021). The Regulatory Genome, 1st Edn. Available online at: https://www.elsevier.com/books/the-regulatory-genome/davidson/978-0-12-088563-3 (accessed December 20, 2021)
Day, C. L., Kaufmann, D. E., Kiepiela, P., Brown, J. A., Moodley, E. S., Reddy, S., et al. (2006). PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443, 350–354. doi: 10.1038/nature05115
de Armas, L. R., Pallikkuth, S., Rinaldi, S., Pahwa, R., and Pahwa, S. (2019). Implications of Immune Checkpoint Expression During Aging in HIV-Infected People on Antiretroviral Therapy. AIDS Res. Hum. Retrovir. 35, 1112–1122. doi: 10.1089/aid.2019.0135
De Spiegelaere, W., Malatinkova, E., Lynch, L., Van Nieuwerburgh, F., Messiaen, P., O’Doherty, U., et al. (2014). Quantification of integrated HIV DNA by repetitive-sampling Alu-HIV PCR on the basis of poisson statistics. Clin. Chem. 60, 886–895. doi: 10.1373/clinchem.2013.219378
DeChristopher, B. A., Loy, B. A., Marsden, M. D., Schrier, A. J., Zack, J. A., and Wender, P. A. (2012). Designed, synthetically accessible bryostatin analogues potently induce activation of latent HIV reservoirs in vitro. Nat. Chem. 4, 705–710. doi: 10.1038/nchem.1395
Deeks, S. G. (2011). HIV Infection, Inflammation, Immunosenescence, and Aging. Annu. Rev. Med. 62, 141–155. doi: 10.1146/annurev-med-042909-093756
Deeks, S. G., Overbaugh, J., Phillips, A., and Buchbinder, S. (2015). HIV infection. Nat. Rev. Dis. Primers 1:15035. doi: 10.1038/nrdp.2015.35
Deleage, C., Chan, C. N., Busman-Sahay, K., and Estes, J. D. (2018). Next-generation in situ hybridization approaches to define and quantify HIV and SIV reservoirs in tissue microenvironments. Retrovirology 15:4. doi: 10.1186/s12977-017-0387-9
Deleage, C., Wietgrefe, S. W., Del Prete, G., Morcock, D. R., Hao, X. P., Piatak, M., et al. (2016). Defining HIV and SIV Reservoirs in Lymphoid Tissues. Pathog. Immun. 1, 68–106. doi: 10.20411/pai.v1i1.100
Della Chiara, G., Crotti, A., Liboi, E., Giacca, M., Poli, G., and Lusic, M. (2011). Negative Regulation of HIV-1 Transcription by a Heterodimeric NF-κB1/p50 and C-Terminally Truncated STAT5 Complex. J. Mol. Biol. 410, 933–943. doi: 10.1016/j.jmb.2011.03.044
Demeulemeester, J., De Rijck, J., Gijsbers, R., and Debyser, Z. (2015). Retroviral integration: Site matters: Mechanisms and consequences of retroviral integration site selection. Bioessays 37, 1202–1214. doi: 10.1002/bies.201500051
Deng, L., Zeng, Q., Wang, M., Cheng, A., Jia, R., Chen, S., et al. (2018). Suppression of NF-κB Activity: A Viral Immune Evasion Mechanism. Viruses 10:E409. doi: 10.3390/v10080409
Ding, D., Qu, X., Li, L., Zhou, X., Liu, S., Lin, S., et al. (2013). Involvement of histone methyltransferase GLP in HIV-1 latency through catalysis of H3K9 dimethylation. Virology 440, 182–189. doi: 10.1016/j.virol.2013.02.022
Ding, J., Tasker, C., Lespinasse, P., Dai, J., Fitzgerald-Bocarsly, P., Lu, W., et al. (2015). Integrin α4β7 Expression Increases HIV Susceptibility in Activated Cervical CD4+ T Cells by an HIV Attachment-Independent Mechanism. JAIDS 69, 509–518. doi: 10.1097/QAI.0000000000000676
Dinoso, J. B., Kim, S. Y., Wiegand, A. M., Palmer, S. E., Gange, S. J., Cranmer, L., et al. (2009). Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U S A. 106, 9403–9408. doi: 10.1073/pnas.0903107106
Donahue, D. A., and Wainberg, M. A. (2013). Cellular and molecular mechanisms involved in the establishment of HIV-1 latency. Retrovirology 10:11. doi: 10.1186/1742-4690-10-11
Douek, D. C., Brenchley, J. M., Betts, M. R., Ambrozak, D. R., Hill, B. J., Okamoto, Y., et al. (2002). HIV preferentially infects HIV-specific CD4+ T cells. Nature 417, 95–98. doi: 10.1038/417095a
Doyon, A., Kracht, D., Bayazit, A. K., Deveci, M., Duzova, A., Krmar, R. T., et al. (2013). Carotid Artery Intima-Media Thickness and Distensibility in Children and Adolescents: Reference Values and Role of Body Dimensions. Hypertension 62, 550–556. doi: 10.1161/HYPERTENSIONAHA.113.01297
du Chéné, I., Basyuk, E., Lin, Y.-L., Triboulet, R., Knezevich, A., Chable-Bessia, C., et al. (2007). Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 26, 424–435.
Dyer, W. B., Zaunders, J. J., Yuan, F. F., Wang, B., Learmont, J. C., Geczy, A. F., et al. (2008). Mechanisms of HIV non-progression; robust and sustained CD4+ T-cell proliferative responses to p24 antigen correlate with control of viraemia and lack of disease progression after long-term transfusion-acquired HIV-1 infection. Retrovirology 5:112. doi: 10.1186/1742-4690-5-112
Ebina, H., Misawa, N., Kanemura, Y., and Koyanagi, Y. (2013). Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 3:2510. doi: 10.1038/srep02510
Eilebrecht, S., Le Douce, V., Riclet, R., Targat, B., Hallay, H., Van Driessche, B., et al. (2014). HMGA1 recruits CTIP2-repressed P-TEFb to the HIV-1 and cellular target promoters. Nucleic Acids Res. 42, 4962–4971. doi: 10.1093/nar/gku168
Einkauf, K. B., Lee, G. Q., Gao, C., Sharaf, R., Sun, X., Hua, S., et al. (2019). Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J. Clin. Invest. 129, 988–998. doi: 10.1172/JCI124291
Einkauf, K. B., Osborn, M. R., Gao, C., Sun, W., Sun, X., Lian, X., et al. (2022). Parallel analysis of transcription, integration, and sequence of single HIV-1 proviruses. Cell 185, 266.e–282.e. doi: 10.1016/j.cell.2021.12.011
Elbezanti, W., Lin, A., Schirling, A., Jackson, A., Marshall, M., Duyne, R. V., et al. (2020). Benzodiazepines Drive Alteration of Chromatin at the Integrated HIV-1 LTR. Viruses 12:E191. doi: 10.3390/v12020191
El-Sadr, W. M., Lundgren, J. D., Neaton, J. D., Gordin, F., Abrams, D., Arduino, R. C., et al. (2006). CD4+ Count–Guided Interruption of Antiretroviral Treatment. N. Engl. J. Med. 355, 2283–2296. doi: 10.1056/NEJMoa062360
Eriksson, S., Graf, E. H., Dahl, V., Strain, M. C., Yukl, S. A., Lysenko, E. S., et al. (2013). Comparative Analysis of Measures of Viral Reservoirs in HIV-1 Eradication Studies. PLoS Pathog. 9:e1003174. doi: 10.1371/journal.ppat.1003174
Estes, J. D., Kityo, C., Ssali, F., Swainson, L., Makamdop, K. N., Del Prete, G. Q., et al. (2017). Defining total-body AIDS-virus burden with implications for curative strategies. Nat. Med. 23, 1271–1276. doi: 10.1038/nm.4411
Evans, V. A., van der Sluis, R. M., Solomon, A., Dantanarayana, A., McNeil, C., Garsia, R., et al. (2018). Programmed cell death-1 contributes to the establishment and maintenance of HIV-1 latency. AIDS 32, 1491–1497. doi: 10.1097/QAD.0000000000001849
Fauci, A. S. (1991). Immunopathogenic Mechanisms in Human Immunodeficiency Virus (HIV) Infection. Ann. Intern. Med. 114:678. doi: 10.7326/0003-4819-114-8-678
Finzi, D., Hermankova, M., Pierson, T., Carruth, L. M., Buck, C., Chaisson, R. E., et al. (1997). Identification of a Reservoir for HIV-1 in Patients on Highly Active Antiretroviral Therapy. Science 278, 1295–1300. doi: 10.1126/science.278.5341.1295
Forouzanfar, F., Ali, S., Wallet, C., De Rovere, M., Ducloy, C., El Mekdad, H., et al. (2019). HIV-1 Vpr mediates the depletion of the cellular repressor CTIP2 to counteract viral gene silencing. Sci. Rep. 9:13154. doi: 10.1038/s41598-019-48689-x
Fowke, K. R., Nagelkerke, N. J., Kimani, J., Simonsen, J. N., Anzala, A. O., Bwayo, J. J., et al. (1996). Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet 348, 1347–1351. doi: 10.1016/S0140-6736(95)12269-2
French, A. J., Natesampillai, S., Krogman, A., Correia, C., Peterson, K. L., Alto, A., et al. (2020). Reactivating latent HIV with PKC agonists induces resistance to apoptosis and is associated with phosphorylation and activation of BCL2. PLoS Pathog. 16:e1008906. doi: 10.1371/journal.ppat.1008906
Friedman, J., Cho, W.-K., Chu, C. K., Keedy, K. S., Archin, N. M., Margolis, D. M., et al. (2011). Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J. Virol. 85, 9078–9089. doi: 10.1128/JVI.00836-11
Fromentin, R., DaFonseca, S., Costiniuk, C. T., El-Far, M., Procopio, F. A., Hecht, F. M., et al. (2019). PD-1 blockade potentiates HIV latency reversal ex vivo in CD4+ T cells from ART-suppressed individuals. Nat. Commun. 10:814. doi: 10.1038/s41467-019-08798-7
Fukuda, H., Li, S., Sardo, L., Smith, J. L., Yamashita, K., Sarca, A. D., et al. (2019). Structural Determinants of the APOBEC3G N-Terminal Domain for HIV-1 RNA Association. Front. Cell. Infect. Microbiol. 9:129. doi: 10.3389/fcimb.2019.00129
Gaebler, C., Cetrulo Lorenzi, J. C., Oliveira, T., Nogueira, L., Ramos, V., Lu, C.-L., et al. (2019). Combination of quadruplex qPCR and next-generation sequencing for qualitative and quantitative analysis of the HIV-1 latent reservoir. J. Exp. Med. 216:20190896. doi: 10.1084/jem.20190896
Gaebler, C., Falcinelli, S. D., Stoffel, E., Read, J., Murtagh, R., Oliveira, T. Y., et al. (2021). Sequence Evaluation and Comparative Analysis of Novel Assays for Intact Proviral HIV-1 DNA. J. Virol. 95, e1986–e1920. doi: 10.1128/JVI.01986-20
Galvani, A. P., and Slatkin, M. (2003). Evaluating plague and smallpox as historical selective pressures for the CCR5- 32 HIV-resistance allele. Proc. Natl. Acad. Sci. 100, 15276–15279. doi: 10.1073/pnas.2435085100
Gao, R., Bao, J., Yan, H., Xie, L., Qin, W., Ning, H., et al. (2020). Competition between PAF1 and MLL1/COMPASS confers the opposing function of LEDGF/p75 in HIV latency and proviral reactivation. Sci. Adv. 6:eaaz8411. doi: 10.1126/sciadv.aaz8411
Gasparian, A. V., Burkhart, C. A., Purmal, A. A., Brodsky, L., Pal, M., Saranadasa, M., et al. (2011). Curaxins: Anticancer Compounds That Simultaneously Suppress NF-κB and Activate p53 by Targeting FACT. Sci. Translat. Med. 2011:3002530. doi: 10.1126/scitranslmed.3002530
Gay, C. L., Bosch, R. J., Ritz, J., Hataye, J. M., Aga, E., Tressler, R. L., et al. (2017). Clinical Trial of the Anti-PD-L1 Antibody BMS-936559 in HIV-1 Infected Participants on Suppressive Antiretroviral Therapy. J. Infect. Dis. 215, 1725–1733. doi: 10.1093/infdis/jix191
Geng, Y., Mosyak, L., Kurinov, I., Zuo, H., Sturchler, E., Cheng, T. C., et al. (2016). Structural mechanism of ligand activation in human calcium-sensing receptor. eLife 5:e13662. doi: 10.7554/eLife.13662
Ghoneim, H. E., Fan, Y., Moustaki, A., Abdelsamed, H. A., Dash, P., Dogra, P., et al. (2017). De Novo Epigenetic Programs Inhibit PD-1 Blockade-Mediated T Cell Rejuvenation. Cell 170, 142.e–157.e. doi: 10.1016/j.cell.2017.06.007
Gibellini, D., Borderi, M., De Crignis, E., Cicola, R., Cimatti, L., Vitone, F., et al. (2008). HIV-1 DNA load analysis in peripheral blood lymphocytes and monocytes from naïve and HAART-treated individuals. J. Infect. 56, 219–225. doi: 10.1016/j.jinf.2008.01.001
Gobeil, L.-A., Lodge, R., and Tremblay, M. J. (2012). Differential HIV-1 endocytosis and susceptibility to virus infection in human macrophages correlate with cell activation status. J. Virol. 86, 10399–10407. doi: 10.1128/JVI.01051-12
Gohda, J., Suzuki, K., Liu, K., Xie, X., Takeuchi, H., Inoue, J., et al. (2018). BI-2536 and BI-6727, dual Polo-like kinase/bromodomain inhibitors, effectively reactivate latent HIV-1. Sci. Rep. 8:3521. doi: 10.1038/s41598-018-21942-5
Goncalves, A., Leigh-Brown, S., Thybert, D., Stefflova, K., Turro, E., Flicek, P., et al. (2012). Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome Res. 22, 2376–2384. doi: 10.1101/gr.142281.112
Gondim, M. V. P., Sherrill-Mix, S., Bibollet-Ruche, F., Russell, R. M., Trimboli, S., Smith, A. G., et al. (2021). Heightened resistance to host type 1 interferons characterizes HIV-1 at transmission and after antiretroviral therapy interruption. Sci. Transl. Med. 13:eabd8179. doi: 10.1126/scitranslmed.abd8179
Gonelli, C. A., Khoury, G., Center, R. J., and Purcell, D. F. J. (2019). HIV-1-Based Virus-like Particles that Morphologically Resemble Mature, Infectious HIV-1 Virions. Viruses 11:507. doi: 10.3390/v11060507
Gonzalo-Gil, E., Ikediobi, U., and Sutton, R. E. (2017). Mechanisms of Virologic Control and Clinical Characteristics of HIV+ Elite/Viremic Controllers. Yale J. Biol. Med. 90, 245–259.
Gosselin, A., Wiche Salinas, T. R., Planas, D., Wacleche, V. S., Zhang, Y., Fromentin, R., et al. (2017). HIV persists in CCR6+CD4+ T cells from colon and blood during antiretroviral therapy. AIDS 31, 35–48. doi: 10.1097/QAD.0000000000001309
Gougeon, M.-L. (2005). To kill or be killed: how HIV exhausts the immune system. Cell Death Differ. 12, (Suppl. 1), 845–854. doi: 10.1038/sj.cdd.4401616
Grau-Expósito, J., Luque-Ballesteros, L., Navarro, J., Curran, A., Burgos, J., Ribera, E., et al. (2019). Latency reversal agents affect differently the latent reservoir present in distinct CD4+ T subpopulations. PLoS Pathog. 15:e1007991. doi: 10.1371/journal.ppat.1007991
Guihot, A., Marcelin, A.-G., Massiani, M.-A., Samri, A., Soulié, C., Autran, B., et al. (2018). Drastic decrease of the HIV reservoir in a patient treated with nivolumab for lung cancer. Ann. Oncol. 29, 517–518. doi: 10.1093/annonc/mdx696
Gulakowski, R. J., McMahon, J. B., Buckheit, R. W., Gustafson, K. R., and Boyd, M. R. (1997). Antireplicative and anticytopathic activities of prostratin, a non-tumor-promoting phorbol ester, against human immunodeficiency virus (HIV). Antiviral Res. 33, 87–97. doi: 10.1016/s0166-3542(96)01004-2
Gunst, M., Jarman, K., Yarwood, V., Rokadiya, S., Capsaskis, L., Orcutt, M., et al. (2019). Healthcare access for refugees in Greece: Challenges and opportunities. Health Policy 123, 818–824. doi: 10.1016/j.healthpol.2019.06.003
Gupta, R. K., Abdul-Jawad, S., McCoy, L. E., Mok, H. P., Peppa, D., Salgado, M., et al. (2019). HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature 568, 244–248. doi: 10.1038/s41586-019-1027-4
Gutiérrez, C., Serrano-Villar, S., Madrid-Elena, N., Pérez-Elías, M. J., Martín, M. E., Barbas, C., et al. (2016). Bryostatin-1 for latent virus reactivation in HIV-infected patients on antiretroviral therapy. AIDS 30, 1385–1392. doi: 10.1097/QAD.0000000000001064
Hakre, S., Chavez, L., Shirakawa, K., and Verdin, E. (2012). HIV latency: experimental systems and molecular models. FEMS Microbiol. Rev. 36, 706–716. doi: 10.1111/j.1574-6976.2012.00335.x
Hale, M., Mesojednik, T., Romano Ibarra, G. S., Sahni, J., Bernard, A., Sommer, K., et al. (2017). Engineering HIV-Resistant, Anti-HIV Chimeric Antigen Receptor T Cells. Mol. Ther. 25, 570–579. doi: 10.1016/j.ymthe.2016.12.023
Hamer, D. (2004). Can HIV be Cured? Mechanisms of HIV Persistence and Strategies to Combat It. CHR 2, 99–111. doi: 10.2174/1570162043484915
Han, Y., Lassen, K., Monie, D., Sedaghat, A. R., Shimoji, S., Liu, X., et al. (2004). Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78, 6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004
Harari, A., Rozot, V., Cavassini, M., Bellutti Enders, F., Viganò, S., Castro, E., et al. (2012). NYVAC immunization induces polyfunctional HIV-specific T-cell responses in chronically-infected, ART-treated HIV patients. Eur. J. Immunol. 42:201242696. doi: 10.1002/eji.201242696
Hariharan, M., Scaria, V., Pillai, B., and Brahmachari, S. K. (2005). Targets for human encoded microRNAs in HIV genes. Biochem. Biophys. Res. Commun. 337, 1214–1218. doi: 10.1016/j.bbrc.2005.09.183
Hartmann, J., Schüßler-Lenz, M., Bondanza, A., and Buchholz, C. J. (2017). Clinical development of CAR T cells-challenges and opportunities in translating innovative treatment concepts. EMBO Mol. Med. 9, 1183–1197. doi: 10.15252/emmm.201607485
Hattori, S., Matsuda, K., Tsuchiya, K., Gatanaga, H., Oka, S., Yoshimura, K., et al. (2018). Combination of a Latency-Reversing Agent With a Smac Mimetic Minimizes Secondary HIV-1 Infection in vitro. Front. Microbiol. 9:2022. doi: 10.3389/fmicb.2018.02022
Hazenberg, M. D., Hamann, D., Schuitemaker, H., and Miedema, F. (2000). T cell depletion in HIV-1 infection: how CD4+ T cells go out of stock. Nat. Immunol. 1, 285–289. doi: 10.1038/79724
Hennessy, E. J., Adam, A., Aquila, B. M., Castriotta, L. M., Cook, D., Hattersley, M., et al. (2013). Discovery of a novel class of dimeric Smac mimetics as potent IAP antagonists resulting in a clinical candidate for the treatment of cancer (AZD5582). J. Med. Chem. 56, 9897–9919. doi: 10.1021/jm401075x
Henning, A. N., Roychoudhuri, R., and Restifo, N. P. (2018). Epigenetic control of CD8+ T cell differentiation. Nat. Rev. Immunol. 18, 340–356. doi: 10.1038/nri.2017.146
Henrich, T. J., Hsue, P. Y., and VanBrocklin, H. (2019). Seeing Is Believing: Nuclear Imaging of HIV Persistence. Front. Immunol. 10:2077. doi: 10.3389/fimmu.2019.02077
Herbein, G., and Wendling, D. (2010). Histone deacetylases in viral infections. Clin. Epigenet. 1, 13–24. doi: 10.1007/s13148-010-0003-5
Hiener, B., Horsburgh, B. A., Eden, J.-S., Barton, K., Schlub, T. E., Lee, E., et al. (2017). Identification of Genetically Intact HIV-1 Proviruses in Specific CD4+ T Cells from Effectively Treated Participants. Cell Rep. 21, 813–822. doi: 10.1016/j.celrep.2017.09.081
Hill, A. L., Rosenbloom, D. I. S., Goldstein, E., Hanhauser, E., Kuritzkes, D. R., Siliciano, R. F., et al. (2016). Real-Time Predictions of Reservoir Size and Rebound Time during Antiretroviral Therapy Interruption Trials for HIV. PLoS Pathog. 12:e1005535. doi: 10.1371/journal.ppat.1005535
Ho, Y.-C., Shan, L., Hosmane, N. N., Wang, J., Laskey, S. B., Rosenbloom, D. I. S., et al. (2013). Replication-Competent Noninduced Proviruses in the Latent Reservoir Increase Barrier to HIV-1 Cure. Cell 155, 540–551. doi: 10.1016/j.cell.2013.09.020
Holkmann Olsen, C., Mocroft, A., Kirk, O., Vella, S., Blaxhult, A., Clumeck, N., et al. (2007). Interruption of combination antiretroviral therapy and risk of clinical disease progression to AIDS or death. HIV Med. 8, 96–104. doi: 10.1111/j.1468-1293.2007.00436.x
Honeycutt, J. B., Thayer, W. O., Baker, C. E., Ribeiro, R. M., Lada, S. M., Cao, Y., et al. (2017). HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat. Med. 23, 638–643. doi: 10.1038/nm.4319
Hosmane, N. N., Kwon, K. J., Bruner, K. M., Capoferri, A. A., Beg, S., Rosenbloom, D. I. S., et al. (2017). Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J. Exp. Med. 214, 959–972. doi: 10.1084/jem.20170193
Hottiger, M. O., and Nabel, G. J. (1998). Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J. Virol. 72, 8252–8256. doi: 10.1128/JVI.72.10.8252-8256.1998
Houzet, L., and Jeang, K.-T. (2011). Genome-wide screening using RNA interference to study host factors in viral replication and pathogenesis. Exp. Biol. Med. 236, 962–967. doi: 10.1258/ebm.2010.010272
Hu, W., Kaminski, R., Yang, F., Zhang, Y., Cosentino, L., Li, F., et al. (2014). RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. 111, 11461–11466. doi: 10.1073/pnas.1405186111
Huang, H., Kong, W., Jean, M., Fiches, G., Zhou, D., Hayashi, T., et al. (2019). A CRISPR/Cas9 screen identifies the histone demethylase MINA53 as a novel HIV-1 latency-promoting gene (LPG). Nucleic Acids Res. 47, 7333–7347. doi: 10.1093/nar/gkz493
Huang, H., Liu, S., Jean, M., Simpson, S., Huang, H., Merkley, M., et al. (2017). A Novel Bromodomain Inhibitor Reverses HIV-1 Latency through Specific Binding with BRD4 to Promote Tat and P-TEFb Association. Front. Microbiol. 8:1035. doi: 10.3389/fmicb.2017.01035
Huang, Y., Zhang, L., and Ho, D. D. (1995). Characterization of nef sequences in long-term survivors of human immunodeficiency virus type 1 infection. J. Virol. 1995:1995. doi: 10.1128/jvi.69.1.93-100.1995
Hütter, G., Nowak, D., Mossner, M., Ganepola, S., Müssig, A., Allers, K., et al. (2009). Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 360, 692–698. doi: 10.1056/NEJMoa0802905
Iglesias-Ussel, M., Vandergeeten, C., Marchionni, L., Chomont, N., and Romerio, F. (2013). High levels of CD2 expression identify HIV-1 latently infected resting memory CD4+ T cells in virally suppressed subjects. J. Virol. 87, 9148–9158. doi: 10.1128/JVI.01297-13
Ikeda, T., Shibata, J., Yoshimura, K., Koito, A., and Matsushita, S. (2007). Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J. Infect. Dis. 195, 716–725. doi: 10.1086/510915
Imai, C., Mihara, K., Andreansky, M., Nicholson, I. C., Pui, C.-H., Geiger, T. L., et al. (2004). Chimeric receptors with 4-1BB signaling capacity provoke potent cytotoxicity against acute lymphoblastic leukemia. Leukemia 18, 676–684. doi: 10.1038/sj.leu.2403302
Imai, K., Togami, H., and Okamoto, T. (2010). Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J. Biol. Chem. 285, 16538–16545. doi: 10.1074/jbc.M110.103531
Imam, H., Bano, A. S., Patel, P., Holla, P., and Jameel, S. (2015). The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci. Rep. 5:8639. doi: 10.1038/srep08639
Imamichi, H., Dewar, R. L., Adelsberger, J. W., Rehm, C. A., O’Doherty, U., Paxinos, E. E., et al. (2016). Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. PNAS 113, 8783–8788. doi: 10.1073/pnas.1609057113
Imamichi, H., Smith, M., Adelsberger, J. W., Izumi, T., Scrimieri, F., Sherman, B. T., et al. (2020). Defective HIV-1 proviruses produce viral proteins. PNAS 117, 3704–3710. doi: 10.1073/pnas.1917876117
Izumi, T., Shirakawa, K., and Takaori-Kondo, A. (2008). Cytidine deaminases as a weapon against retroviruses and a new target for antiviral therapy. Mini Rev. Med. Chem. 8, 231–238. doi: 10.2174/138955708783744047
Jadhav, R. R., Im, S. J., Hu, B., Hashimoto, M., Li, P., Lin, J.-X., et al. (2019). Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc. Natl. Acad. Sci. U S A. 116, 14113–14118. doi: 10.1073/pnas.1903520116
Jadlowsky, J. K., Wong, J. Y., Graham, A. C., Dobrowolski, C., Devor, R. L., Adams, M. D., et al. (2014). Negative Elongation Factor Is Required for the Maintenance of Proviral Latency but Does Not Induce Promoter-Proximal Pausing of RNA Polymerase II on the HIV Long Terminal Repeat. Mol. Cell. Biol. 34, 1911–1928. doi: 10.1128/MCB.01013-13
Jang, M. K., Mochizuki, K., Zhou, M., Jeong, H.-S., Brady, J. N., and Ozato, K. (2005). The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19, 523–534. doi: 10.1016/j.molcel.2005.06.027
Jiang, G., and Dandekar, S. (2015). Targeting NF-κB Signaling with Protein Kinase C Agonists As an Emerging Strategy for Combating HIV Latency. AIDS Res. Hum. Retroviruses 31, 4–12. doi: 10.1089/aid.2014.0199
Jiang, G., Espeseth, A., Hazuda, D. J., and Margolis, D. M. (2007). c-Myc and Sp1 Contribute to Proviral Latency by Recruiting Histone Deacetylase 1 to the Human Immunodeficiency Virus Type 1 Promoter. J. Virol. 81, 10914–10923. doi: 10.1128/JVI.01208-07
Jiang, G., Mendes, E. A., Kaiser, P., Sankaran-Walters, S., Tang, Y., Weber, M. G., et al. (2014). Reactivation of HIV latency by a newly modified Ingenol derivative via protein kinase Cδ–NF-κB signaling. AIDS 28, 1555–1566. doi: 10.1097/QAD.0000000000000289
Jiang, G., Mendes, E. A., Kaiser, P., Wong, D. P., Tang, Y., Cai, I., et al. (2015). Synergistic Reactivation of Latent HIV Expression by Ingenol-3-Angelate, PEP005, Targeted NF-kB Signaling in Combination with JQ1 Induced p-TEFb Activation. PLoS Pathog. 11:e1005066. doi: 10.1371/journal.ppat.1005066
Jiang, G., Nguyen, D., Archin, N. M., Yukl, S. A., Méndez-Lagares, G., Tang, Y., et al. (2018). HIV latency is reversed by ACSS2-driven histone crotonylation. J. Clin. Invest. 128, 1190–1198. doi: 10.1172/JCI98071
Jiang, L., Yang, K.-H., Chen, Y., Guan, Q.-L., Zhao, P., Tian, J.-H., et al. (2014). Systematic review and meta-analysis of the effectiveness and safety of extended lymphadenectomy in patients with resectable gastric cancer. Br. J. Surg. 101, 595–604. doi: 10.1002/bjs.9497
Joos, B., Fischer, M., Kuster, H., Pillai, S. K., Wong, J. K., Böni, J., et al. (2008). HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc. Natl. Acad. Sci. U S A. 105, 16725–16730. doi: 10.1073/pnas.0804192105
Josefsson, L., Stockenstrom, S., von, Faria, N. R., Sinclair, E., Bacchetti, P., et al. (2013). The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. PNAS 2013:1308313110. doi: 10.1073/pnas.1308313110
Kandathil, A. J., Sugawara, S., and Balagopal, A. (2016). Are T cells the only HIV-1 reservoir? Retrovirology 13:86. doi: 10.1186/s12977-016-0323-4
Kauder, S. E., Bosque, A., Lindqvist, A., Planelles, V., and Verdin, E. (2009). Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495. doi: 10.1371/journal.ppat.1000495
Kelker, H. C., Seidlin, M., Vogler, M., and Valentine, F. T. (1992). Lymphocytes from Some Long-Term Seronegative Heterosexual Partners of HIV-Infected Individuals Proliferate in Response to HIV Antigens. AIDS Res. Hum. Retroviruses 8, 1355–1359. doi: 10.1089/aid.1992.8.1355
Kessing, C. F., Nixon, C. C., Li, C., Tsai, P., Takata, H., Mousseau, G., et al. (2017). In Vivo Suppression of HIV Rebound by Didehydro-Cortistatin A, a “Block-and-Lock” Strategy for HIV-1 Treatment. Cell Rep. 21, 600–611. doi: 10.1016/j.celrep.2017.09.080
Kessl, J. J., Jena, N., Koh, Y., Taskent-Sezgin, H., Slaughter, A., Feng, L., et al. (2012). Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J. Biol. Chem. 287, 16801–16811. doi: 10.1074/jbc.M112.354373
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. PNAS 106, 11667–11672. doi: 10.1073/pnas.0904715106
Khan, O., Giles, J. R., McDonald, S., Manne, S., Ngiow, S. F., Patel, K. P., et al. (2019). TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218. doi: 10.1038/s41586-019-1325-x
Khoury, A. L., Morey, M. C., Wong, T. C., McNeil, D. L., Humphries, B., Frankey, K., et al. (2017). Diminished physical function in older HIV-infected adults in the Southeastern U.S. despite successful antiretroviral therapy. PLoS One 12:e0179874. doi: 10.1371/journal.pone.0179874
Kim, Y., Anderson, J. L., and Lewin, S. R. (2018). Getting the “Kill” into “Shock and Kill”: Strategies to Eliminate Latent HIV. Cell Host Microbe 23, 14–26. doi: 10.1016/j.chom.2017.12.004
Kinloch, N. N., Ren, Y., Conce Alberto, W. D., Dong, W., Khadka, P., Huang, S. H., et al. (2021). HIV-1 diversity considerations in the application of the Intact Proviral DNA Assay (IPDA). Nat. Commun. 12:165. doi: 10.1038/s41467-020-20442-3
Klase, Z., Kale, P., Winograd, R., Gupta, M. V., Heydarian, M., Berro, R., et al. (2007). HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol. Biol. 8:63. doi: 10.1186/1471-2199-8-63
Klase, Z., Yedavalli, V. S. R. K., Houzet, L., Perkins, M., Maldarelli, F., Brenchley, J., et al. (2014). Activation of HIV-1 from Latent Infection via Synergy of RUNX1 Inhibitor Ro5-3335 and SAHA. PLoS Pathog. 10:e1003997. doi: 10.1371/journal.ppat.1003997
Klitz, W., Brautbar, C., Schito, A. M., Barcellos, L. F., and Oksenberg, J. R. (2001). Evolution of the CCR5 δ32 mutation based on haplotype variation in Jewish and Northern European population samples. Hum. Immunol. 62, 530–538. doi: 10.1016/S0198-8859(01)00239-7
Kordelas, L., Verheyen, J., and Esser, S. (2014). Shift of HIV Tropism in Stem-Cell Transplantation with CCR5 Delta32 Mutation. N. Engl. J. Med. 371, 880–882. doi: 10.1056/NEJMc1405805
Koup, R. A., Safrit, J. T., Cao, Y., Andrews, C. A., McLeod, G., Borkowsky, W., et al. (1994). Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68, 4650–4655. doi: 10.1128/JVI.68.7.4650-4655.1994
Kousignian, I., Abgrall, S., Grabar, S., Mahamat, A., Teicher, E., Rouveix, E., et al. (2008). Maintaining Antiretroviral Therapy Reduces the Risk of AIDS-Defining Events in Patients with Uncontrolled Viral Replication and Profound Immunodeficiency. Clin. Infect. Dis. 46, 296–304. doi: 10.1086/524753
Kruize, Z., and Kootstra, N. A. (2019). The Role of Macrophages in HIV-1 Persistence and Pathogenesis. Front. Microbiol. 10:2828. doi: 10.3389/fmicb.2019.02828
Kuai, Q., Lu, X., Qiao, Z., Wang, R., Wang, Y., Ye, S., et al. (2018). Histone deacetylase inhibitor chidamide promotes reactivation of latent human immunodeficiency virus by introducing histone acetylation. J. Med. Virol. 90, 1478–1485. doi: 10.1002/jmv.25207
Kula, A., Guerra, J., Knezevich, A., Kleva, D., Myers, M. P., and Marcello, A. (2011). Characterization of the HIV-1 RNA associated proteome identifies Matrin 3 as a nuclear cofactor of Rev function. Retrovirology 8:60. doi: 10.1186/1742-4690-8-60
Kulkosky, J., Culnan, D. M., Roman, J., Dornadula, G., Schnell, M., Boyd, M. R., et al. (2001). Prostratin: activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 98, 3006–3015. doi: 10.1182/blood.V98.10.3006
Kulpa, D. A., Talla, A., Brehm, J. H., Ribeiro, S. P., Yuan, S., Bebin-Blackwell, A.-G., et al. (2019). Differentiation into an Effector Memory Phenotype Potentiates HIV-1 Latency Reversal in CD4+ T Cells. J. Virol. 93, e969–e919. doi: 10.1128/JVI.00969-19
Kwong, P. D., Mascola, J. R., and Nabel, G. J. (2013). Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat. Rev. Immunol. 13, 693–701. doi: 10.1038/nri3516
Laird, G. M., Eisele, E. E., Rabi, S. A., Lai, J., Chioma, S., Blankson, J. N., et al. (2013). Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 9:e1003398. doi: 10.1371/journal.ppat.1003398
Le Douce, V., Colin, L., Redel, L., Cherrier, T., Herbein, G., Aunis, D., et al. (2012). LSD1 cooperates with CTIP2 to promote HIV-1 transcriptional silencing. Nucleic Acids Res. 40, 1904–1915. doi: 10.1093/nar/gkr857
Lee, G. Q., Orlova-Fink, N., Einkauf, K., Chowdhury, F. Z., Sun, X., Harrington, S., et al. (2017). Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J. Clin. Invest. 127, 2689–2696. doi: 10.1172/JCI93289
Lefrère, J., Mariotti, M., Morand-Joubert, L., Thauvin, M., and Roudot-Thoraval, F. (1999). Plasma Human Immunodeficiency Virus RNA Below 40 Copies/mL Is Rarein Untreated Persons Even in the First Years of Infection. J. Infect. Dis. 180, 526–529. doi: 10.1086/314906
Leite, T. F., Delatorre, E., Côrtes, F. H., Ferreira, A. C. G., Cardoso, S. W., Grinsztejn, B., et al. (2019). Reduction of HIV-1 Reservoir Size and Diversity After 1 Year of cART Among Brazilian Individuals Starting Treatment During Early Stages of Acute Infection. Front. Microbiol. 10:145. doi: 10.3389/fmicb.2019.00145
Leonard, J. N., Shah, P. S., Burnett, J. C., and Schaffer, D. V. (2008). HIV Evades RNA Interference Directed at TAR by an Indirect Compensatory Mechanism. Cell Host Microbe 4, 484–494. doi: 10.1016/j.chom.2008.09.008
Levy, C. N., Hughes, S. M., Roychoudhury, P., Reeves, D. B., Amstuz, C., Zhu, H., et al. (2021). A highly multiplexed droplet digital PCR assay to measure the intact HIV-1 proviral reservoir. Cell Rep. Med. 2:100243. doi: 10.1016/j.xcrm.2021.100243
Li, C., Mousseau, G., and Valente, S. (2019). Tat inhibition by didehydro-Cortistatin A promotes heterochromatin formation at the HIV-1 long terminal repeat. Epigenetics Chromatin 12:23. doi: 10.1186/s13072-019-0267-8
Li, H., Chi, X., Li, R., Ouyang, J., and Chen, Y. (2020). A Novel lncRNA, AK130181, Contributes to HIV-1 Latency by Regulating Viral Promoter-Driven Gene Expression in Primary CD4+ T Cells. Mol. Ther. Nucleic Acids 20, 754–763. doi: 10.1016/j.omtn.2020.04.011
Li, J., Chen, C., Ma, X., Geng, G., Liu, B., Zhang, Y., et al. (2016). Long noncoding RNA NRON contributes to HIV-1 latency by specifically inducing tat protein degradation. Nat. Commun. 7:11730. doi: 10.1038/ncomms11730
Li, J. Z., Etemad, B., Ahmed, H., Aga, E., Bosch, R. J., Mellors, J. W., et al. (2016). The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. AIDS 30, 343–353. doi: 10.1097/QAD.0000000000000953
Li, Z., Mbonye, U., Feng, Z., Wang, X., Gao, X., Karn, J., et al. (2018). The KAT5-Acetyl-Histone4-Brd4 axis silences HIV-1 transcription and promotes viral latency. PLoS Pathog. 14:e1007012. doi: 10.1371/journal.ppat.1007012
Liang, T., Zhang, X., Lai, F., Lin, J., Zhou, C., Xu, X., et al. (2019). A novel bromodomain inhibitor, CPI-203, serves as an HIV-1 latency-reversing agent by activating positive transcription elongation factor b. Biochem. Pharmacol. 164, 237–251. doi: 10.1016/j.bcp.2019.04.005
Lin, J. E., Asfour, A., Sewell, T. B., Hooe, B., Pryce, P., Earley, C., et al. (2021). Neurological issues in children with COVID-19. Neurosci. Lett. 743:135567. doi: 10.1016/j.neulet.2020.135567
Liszewski, M. K., Yu, J. J., and O’Doherty, U. (2009). Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 47, 254–260. doi: 10.1016/j.ymeth.2009.01.002
Liu, L., Shawki, D., Voulgarakis, A., Kasoar, M., Samset, B. H., Myhre, G., et al. (2018). A PDRMIP Multimodel Study on the Impacts of Regional Aerosol Forcings on Global and Regional Precipitation. J. Clim. 31, 4429–4447. doi: 10.1175/JCLI-D-17-0439.1
Liu, R., Simonetti, F. R., and Ho, Y.-C. (2020). The forces driving clonal expansion of the HIV-1 latent reservoir. Virol. J. 17:4. doi: 10.1186/s12985-019-1276-8
Lopez-Galindez, C., Pernas, M., Casado, C., Olivares, I., and Lorenzo-Redondo, R. (2019). Elite controllers and lessons learned for HIV-1 cure. Curr. Opin. Virol. 38, 31–36. doi: 10.1016/j.coviro.2019.05.010
López-Huertas, M. R., Jiménez-Tormo, L., Madrid-Elena, N., Gutiérrez, C., Rodríguez-Mora, S., Coiras, M., et al. (2017). The CCR5-antagonist Maraviroc reverses HIV-1 latency in vitro alone or in combination with the PKC-agonist Bryostatin-1. Sci. Rep. 7:2385. doi: 10.1038/s41598-017-02634-y
Lorenzi, J. C. C., Cohen, Y. Z., Cohn, L. B., Kreider, E. F., Barton, J. P., Learn, G. H., et al. (2016). Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc. Natl. Acad. Sci. U S A. 113, E7908–E7916. doi: 10.1073/pnas.1617789113
Lu, P., Shen, Y., Yang, H., Wang, Y., Jiang, Z., Yang, X., et al. (2017). BET inhibitors RVX-208 and PFI-1 reactivate HIV-1 from latency. Sci. Rep. 7:16646. doi: 10.1038/s41598-017-16816-1
Lu, Z., Zhang, Q. C., Lee, B., Flynn, R. A., Smith, M. A., Robinson, J. T., et al. (2016). RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 165, 1267–1279. doi: 10.1016/j.cell.2016.04.028
Lusic, M. (2003). Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J. 22, 6550–6561. doi: 10.1093/emboj/cdg631
Macatangay, B. J. C., Gandhi, R. T., Jones, R. B., Mcmahon, D. K., Lalama, C. M., Bosch, R. J., et al. (2020). T cells with high PD-1 expression are associated with lower HIV-specific immune responses despite long-term antiretroviral therapy. AIDS 34, 15–24. doi: 10.1097/QAD.0000000000002406
Mack, K. D., Jin, X., Yu, S., Wei, R., Kapp, L., Green, C., et al. (2003). HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. J. Acquir. Immune Defic. Syndr. 33, 308–320. doi: 10.1097/00126334-200307010-00004
Madrid-Elena, N., García-Bermejo, M. L., Serrano-Villar, S., Díaz-de Santiago, A., Sastre, B., Gutiérrez, C., et al. (2018). Maraviroc Is Associated with Latent HIV-1 Reactivation through NF-κB Activation in Resting CD4 + T Cells from HIV-Infected Individuals on Suppressive Antiretroviral Therapy. J. Virol. 92:17. doi: 10.1128/JVI.01931-17
Maggiolo, F., Airoldi, M., Callegaro, A., Martinelli, C., Dolara, A., Bini, T., et al. (2009). CD4 cell-guided scheduled treatment interruptions in HIV-infected patients with sustained immunologic response to HAART. AIDS 23, 799–807. doi: 10.1097/QAD.0b013e328321b75e
Maher, J., Brentjens, R. J., Gunset, G., Rivière, I., and Sadelain, M. (2002). Human T-lymphocyte cytotoxicity and proliferation directed by a single chimeric TCRζ /CD28 receptor. Nat. Biotechnol. 20, 70–75. doi: 10.1038/nbt0102-70
Maldarelli, F., Wu, X., Su, L., Simonetti, F. R., Shao, W., Hill, S., et al. (2014). HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183. doi: 10.1126/science.1254194
Manel, N., and Littman, D. R. (2011). Hiding in Plain Sight: How HIV Evades Innate Immune Responses. Cell 147, 271–274. doi: 10.1016/j.cell.2011.09.010
Marban, C., Suzanne, S., Dequiedt, F., de Walque, S., Redel, L., Van Lint, C., et al. (2007). Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 26, 412–423. doi: 10.1038/sj.emboj.7601516
Mariño-Ramírez, L., Kann, M. G., Shoemaker, B. A., and Landsman, D. (2005). Histone structure and nucleosome stability. Expert Rev. Proteom. 2, 719–729. doi: 10.1586/14789450.2.5.719
Martinez-Picado, J., Morales-Lopetegi, K., Wrin, T., Prado, J. G., Frost, S. D. W., Petropoulos, C. J., et al. (2002). Selection of drug-resistant HIV-1 mutants in response to repeated structured treatment interruptions. AIDS 16, 895–899. doi: 10.1097/00002030-200204120-00009
Marzio, G., Tyagi, M., Gutierrez, M. I., and Giacca, M. (1998). HIV-1 Tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. PNAS 95, 13519–13524. doi: 10.1073/pnas.95.23.13519
Mates, J. M., de Silva, S., Lustberg, M., Van Deusen, K., Baiocchi, R. A., Wu, L., et al. (2015). A Novel Histone Deacetylase Inhibitor, AR-42, Reactivates HIV-1 from Chronically and Latently Infected CD4+ T-cells. Retrovirology 7, 1–5. doi: 10.4137/RRT.S31632
Maude, S. L., Frey, N., Shaw, P. A., Aplenc, R., Barrett, D. M., Bunin, N. J., et al. (2014). Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 371, 1507–1517. doi: 10.1056/NEJMoa1407222
Maus, M. V., and Levine, B. L. (2016). Chimeric Antigen Receptor T-Cell Therapy for the Community Oncologist. Oncologist 21, 608–617. doi: 10.1634/theoncologist.2015-0421
Mbonye, U., and Karn, J. (2017). The Molecular Basis for Human Immunodeficiency Virus Latency. Annu. Rev. Virol. 4, 261–285. doi: 10.1146/annurev-virology-101416-041646
McKernan, L. N., Momjian, D., and Kulkosky, J. (2012). Protein Kinase C: One Pathway towards the Eradication of Latent HIV-1 Reservoirs. Adv. Virol. 2012:e805347. doi: 10.1155/2012/805347
McLane, L. M., Abdel-Hakeem, M. S., and Wherry, E. J. (2019). CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu. Rev. Immunol. 37, 457–495. doi: 10.1146/annurev-immunol-041015-055318
McManus, C. J., Coolon, J. D., Duff, M. O., Eipper-Mains, J., Graveley, B. R., and Wittkopp, P. J. (2010). Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 20, 816–825. doi: 10.1101/gr.102491.109
Mediouni, S., Chinthalapudi, K., Ekka, M. K., Usui, I., Jablonski, J. A., Clementz, M. A., et al. (2019). Didehydro-Cortistatin A Inhibits HIV-1 by Specifically Binding to the Unstructured Basic Region of Tat. mBio 2019:18. doi: 10.1128/mBio.02662-18
Mellors, J. W., Rinaldo, C. R., Gupta, P., White, R. M., Todd, J. A., and Kingsley, L. A. (1996). Prognosis in HIV-1 Infection Predicted by the Quantity of Virus in Plasma. Science 272, 1167–1170. doi: 10.1126/science.272.5265.1167
Mendoza, P., Jackson, J. R., Oliveira, T. Y., Gaebler, C., Ramos, V., Caskey, M., et al. (2020). Antigen-responsive CD4+ T cell clones contribute to the HIV-1 latent reservoir. J. Exp. Med. 217:e20200051. doi: 10.1084/jem.20200051
Migueles, S. A., Weeks, K. A., Nou, E., Berkley, A. M., Rood, J. E., Osborne, C. M., et al. (2009). Defective Human Immunodeficiency Virus-Specific CD8 + T-Cell Polyfunctionality, Proliferation, and Cytotoxicity Are Not Restored by Antiretroviral Therapy. J. Virol. 83, 11876–11889. doi: 10.1128/JVI.01153-09
Monajemi, M., Woodworth, C. F., Zipperlen, K., Gallant, M., Grant, M. D., and Larijani, M. (2014). Positioning of APOBEC3G/F Mutational Hotspots in the Human Immunodeficiency Virus Genome Favors Reduced Recognition by CD8+ T Cells. PLoS One 9:e93428. doi: 10.1371/journal.pone.0093428
Monod, J., and Jacob, F. (1961). General Conclusions: Teleonomic Mechanisms in Cellular Metabolism, Growth, and Differentiation. Cold Spring Harb. Symposia Quantit. Biol. 26, 389–401. doi: 10.1101/SQB.1961.026.01.048
Morison, I. M., Ramsay, J. P., and Spencer, H. G. (2005). A census of mammalian imprinting. Trends Genet. 21, 457–465. doi: 10.1016/j.tig.2005.06.008
Mueller, Y. M., De Rosa, S. C., Hutton, J. A., Witek, J., Roederer, M., Altman, J. D., et al. (2001). Increased CD95/Fas-Induced Apoptosis of HIV-Specific CD8+ T Cells. Immunity 15, 871–882. doi: 10.1016/S1074-7613(01)00246-1
Mujtaba, S., He, Y., Zeng, L., Farooq, A., Carlson, J. E., Ott, M., et al. (2002). Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol. Cell 9, 575–586. doi: 10.1016/s1097-2765(02)00483-5
Musick, A., Spindler, J., Boritz, E., Pérez, L., Crespo-Vélez, D., Patro, S. C., et al. (2019). HIV Infected T Cells Can Proliferate in vivo Without Inducing Expression of the Integrated Provirus. Front. Microbiol. 10:2204. doi: 10.3389/fmicb.2019.02204
Nakanjako, D., Ssewanyana, I., Mayanja-Kizza, H., Kiragga, A., Colebunders, R., Manabe, Y. C., et al. (2011). High T-cell immune activation and immune exhaustion among individuals with suboptimal CD4 recovery after 4 years of antiretroviral therapy in an African cohort. BMC Infect. Dis. 11:43. doi: 10.1186/1471-2334-11-43
Nguyen, S., Deleage, C., Darko, S., Ransier, A., Truong, D. P., Agarwal, D., et al. (2019). Elite control of HIV is associated with distinct functional and transcriptional signatures in lymphoid tissue CD8 + T cells. Sci. Transl. Med. 11:eaax4077. doi: 10.1126/scitranslmed.aax4077
Nguyen, T., Tagett, R., Diaz, D., and Draghici, S. (2017). A novel approach for data integration and disease subtyping. Genome Res. 27, 2025–2039. doi: 10.1101/gr.215129.116
Nguyen, V. T., Kiss, T., Michels, A. A., and Bensaude, O. (2001). 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414, 322–325. doi: 10.1038/35104581
Nixon, C. C., Mavigner, M., Sampey, G. C., Brooks, A. D., Spagnuolo, R. A., Irlbeck, D. M., et al. (2020). Systemic HIV and SIV latency reversal via non-canonical NF-κB signalling in vivo. Nature 578, 160–165. doi: 10.1038/s41586-020-1951-3
No Author (2006). HHS-CDC News: The Global HIV/AIDS Pandemic, 2006. Ann. Pharmacother. 40, 1708–1708. doi: 10.1345/aph.1N117
O’Doherty, U., Swiggard, W., Jeyakumar, D., McGain, D., and Malim, M. (2002). A Sensitive, Quantitative Assay for Human Immunodeficiency Virus Type 1 Integration. J. Virol. 76, 10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002
Orphanides, G., LeRoy, G., Chang, C. H., Luse, D. S., and Reinberg, D. (1998). FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92, 105–116. doi: 10.1016/s0092-8674(00)80903-4
Ostrowski, M. A., Chun, T. W., Justement, S. J., Motola, I., Spinelli, M. A., Adelsberger, J., et al. (1999). Both memory and CD45RA+/CD62L+ naive CD4(+) T cells are infected in human immunodeficiency virus type 1-infected individuals. J. Virol. 73, 6430–6435. doi: 10.1128/JVI.73.8.6430-6435.1999
Pache, L., Dutra, M. S., Spivak, A. M., Marlett, J. M., Murry, J. P., Hwang, Y., et al. (2015). BIRC2/cIAP1 Is a Negative Regulator of HIV-1 Transcription and Can Be Targeted by Smac Mimetics to Promote Reversal of Viral Latency. Cell Host Microbe 18, 345–353. doi: 10.1016/j.chom.2015.08.009
Pandeló José, D., Bartholomeeusen, K., da Cunha, R. D., Abreu, C. M., Glinski, J., da Costa, T. B. F., et al. (2014). Reactivation of latent HIV-1 by new semi-synthetic ingenol esters. Virology 462–463, 328–339. doi: 10.1016/j.virol.2014.05.033
Pardoll, D. M. (2012). The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264. doi: 10.1038/nrc3239
Passaes, C. P. B., Bruel, T., Decalf, J., David, A., Angin, M., Monceaux, V., et al. (2017). Ultrasensitive HIV-1 p24 Assay Detects Single Infected Cells and Differences in Reservoir Induction by Latency Reversal Agents. J. Virol. 91:16. doi: 10.1128/jvi.02296-16
Pasternak, A. O., Adema, K. W., Bakker, M., Jurriaans, S., Berkhout, B., Cornelissen, M., et al. (2008). Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J. Clin. Microbiol. 46, 2206–2211. doi: 10.1128/JCM.00055-08
Patro, S. C., Brandt, L. D., Bale, M. J., Halvas, E. K., Joseph, K. W., Shao, W., et al. (2019). Combined HIV-1 sequence and integration site analysis informs viral dynamics and allows reconstruction of replicating viral ancestors. Proc. Natl. Acad. Sci. U S A. 116, 25891–25899. doi: 10.1073/pnas.1910334116
Pauken, K. E., Sammons, M. A., Odorizzi, P. M., Manne, S., Godec, J., Khan, O., et al. (2016). Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165. doi: 10.1126/science.aaf2807
Peluso, M. J., Deeks, S. G., and McCune, J. M. (2019). HIV “cure”: A shot in the arm? EBioMed. 42, 3–5. doi: 10.1016/j.ebiom.2019.04.011
Perdomo-Celis, F., Velilla, P. A., Taborda, N. A., and Rugeles, M. T. (2019). An altered cytotoxic program of CD8+ T-cells in HIV-infected patients despite HAART-induced viral suppression. PLoS One 14:e0210540. doi: 10.1371/journal.pone.0210540
Pereyra, F., Addo, M. M., Kaufmann, D. E., Liu, Y., Miura, T., Rathod, A., et al. (2008). Genetic and Immunologic Heterogeneity among Persons Who Control HIV Infection in the Absence of Therapy. J. Infect. Dis. 197, 563–571. doi: 10.1086/526786
Perlman, R. L. (2016). Mouse Models of Human Disease: An Evolutionary Perspective. EMPH 2016:eow014. doi: 10.1093/emph/eow014
Pernas, M., Tarancón-Diez, L., Rodríguez-Gallego, E., Gómez, J., Prado, J. G., Casado, C., et al. (2018). Factors Leading to the Loss of Natural Elite Control of HIV-1 Infection. J. Virol. 92:17. doi: 10.1128/JVI.01805-17
Pinto, L. A., Sullivan, J., Berzofsky, J. A., Clerici, M., Kessler, H. A., Landay, A. L., et al. (1995). ENV-specific cytotoxic T lymphocyte responses in HIV seronegative health care workers occupationally exposed to HIV-contaminated body fluids. J. Clin. Invest. 96, 867–876. doi: 10.1172/JCI118133
Pisell, T. L., Ho, O., Lee, G., and Butera, S. T. (2001). Spectrum of cdk-9 inhibitor activity against HIV-1 replication among various models of chronic and latent infection. Antivir. Chem. Chemother. 12, (Suppl. 1), 33–41.
Pollack, R. A., Jones, R. B., Pertea, M., Bruner, K. M., Martin, A. R., Thomas, A. S., et al. (2017). Defective HIV-1 Proviruses Are Expressed and Can Be Recognized by Cytotoxic T Lymphocytes, which Shape the Proviral Landscape. Cell Host Microbe 21, 494.e–506.e. doi: 10.1016/j.chom.2017.03.008
Porter, D. L., Levine, B. L., Kalos, M., Bagg, A., and June, C. H. (2011). Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 365, 725–733. doi: 10.1056/NEJMoa1103849
Procopio, F. A., Fromentin, R., Kulpa, D. A., Brehm, J. H., Bebin, A.-G., Strain, M. C., et al. (2015). A Novel Assay to Measure the Magnitude of the Inducible Viral Reservoir in HIV-infected Individuals. EBioMedicine 2, 874–883. doi: 10.1016/j.ebiom.2015.06.019
Qi, J., Ding, C., Jiang, X., and Gao, Y. (2020). Advances in Developing CAR T-Cell Therapy for HIV Cure. Front. Immunol. 11:361. doi: 10.3389/fimmu.2020.00361
Quivy, V., Adam, E., Collette, Y., Demonte, D., Chariot, A., Vanhulle, C., et al. (2002). Synergistic Activation of Human Immunodeficiency Virus Type 1 Promoter Activity by NF-κB and Inhibitors of Deacetylases: Potential Perspectives for the Development of Therapeutic Strategies. J. Virol. 2002:2002. doi: 10.1128/JVI.76.21.11091-11103.2002
Ramakrishnan, R., Dow, E. C., and Rice, A. P. (2009). Characterization of Cdk9 T-loop phosphorylation in resting and activated CD4 + T lymphocytes. J. Leukocyte Biol. 86, 1345–1350. doi: 10.1189/jlb.0509309
Ramakrishnan, R., Liu, H., and Rice, A. P. (2015). Short Communication: SAHA (Vorinostat) Induces CDK9 Thr-186 (T-Loop) Phosphorylation in Resting CD4 + T Cells: Implications for Reactivation of Latent HIV. AIDS Res. Hum. Retroviruses 31, 137–141. doi: 10.1089/aid.2013.0288
Rangarajan, A., and Weinberg, R. A. (2003). Comparative biology of mouse versus human cells: modelling human cancer in mice. Nat. Rev. Cancer 3, 952–959. doi: 10.1038/nrc1235
Ranki, A., Lagerstedt, A., Ovod, V., Aavik, E., and Krohn, K. J. E. (1994). Expression kinetics and subcellular localization of HIV-1 regulatory proteins Nef, Tat and Rev in acutely and chronically infected lymphoid cell lines. Arch. Virol. 139, 365–378. doi: 10.1007/BF01310798
Rasmussen, T. A., Søgaard, O. S., Brinkmann, C., Wightman, F., Lewin, S. R., Melchjorsen, J., et al. (2013). Comparison of HDAC inhibitors in clinical development. Hum. Vacc. Immunotherap. 9, 993–1001. doi: 10.4161/hv.23800
Rice, A. P. (2016). Cyclin-dependent kinases as therapeutic targets for HIV-1 infection. Expert Opin. Ther. Targets 20, 1453–1461. doi: 10.1080/14728222.2016.1254619
Routy, J.-P., Boulassel, M., Nicolette, C. A., and Jacobson, J. M. (2012). Assessing risk of a short-term antiretroviral therapy discontinuation as a read-out of viral control in immune-based therapy. J. Med. Virol. 84, 885–889. doi: 10.1002/jmv.23297
Rowland-Jones, S., Sutton, J., Ariyoshi, K., Dong, T., Gotch, F., McAdam, S., et al. (1995). HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat. Med. 1, 59–64. doi: 10.1038/nm0195-59
Roy, S., Delling, U., Chen, C. H., Rosen, C. A., and Sonenberg, N. (1990). A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev. 4, 1365–1373. doi: 10.1101/gad.4.8.1365
Ruelas, D. S., and Greene, W. C. (2013). An Integrated Overview of HIV-1 Latency. Cell 155, 519–529. doi: 10.1016/j.cell.2013.09.044
Saayman, S., Ackley, A., Turner, A.-M. W., Famiglietti, M., Bosque, A., Clemson, M., et al. (2014). An HIV-Encoded Antisense Long Noncoding RNA Epigenetically Regulates Viral Transcription. Mol. Ther. 22, 1164–1175. doi: 10.1038/mt.2014.29
Sabari, B. R., Tang, Z., Huang, H., Yong-Gonzalez, V., Molina, H., Kong, H. E., et al. (2015). Intracellular Crotonyl-CoA Stimulates Transcription through p300-Catalyzed Histone Crotonylation. Mol. Cell 58, 203–215. doi: 10.1016/j.molcel.2015.02.029
Sabari, B. R., Zhang, D., Allis, C. D., and Zhao, Y. (2017). Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 18, 90–101. doi: 10.1038/nrm.2016.140
Saez-Cirion, A., Lacabaratz, C., Lambotte, O., Versmisse, P., Urrutia, A., Boufassa, F., et al. (2007). HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc. Natl. Acad. Sci. 104, 6776–6781. doi: 10.1073/pnas.0611244104
Sahin, I. H., Kane, S. R., Brutcher, E., Guadagno, J., Smith, K. E., Wu, C., et al. (2020). Safety and Efficacy of Immune Checkpoint Inhibitors in Patients With Cancer Living With HIV: A Perspective on Recent Progress and Future Needs. JCO Oncol. Pract. 16, 319–325. doi: 10.1200/JOP.19.00754
Samson, M., Libert, F., Doranz, B. J., Rucker, J., Liesnard, C., Farber, C.-M., et al. (1996). Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382, 722–725. doi: 10.1038/382722a0
Santangelo, P. J., Rogers, K. A., Zurla, C., Blanchard, E. L., Gumber, S., Strait, K., et al. (2015). Whole-body immunoPET reveals active SIV dynamics in viremic and antiretroviral therapy-treated macaques. Nat. Methods 12, 427–432. doi: 10.1038/nmeth.3320
Sarca, A. D., Sardo, L., Fukuda, H., Matsui, H., Shirakawa, K., Horikawa, K., et al. (2021). FRET-Based Detection and Quantification of HIV-1 Virion Maturation. Front. Microbiol. 12:647452. doi: 10.3389/fmicb.2021.647452
Sardo, L., Parolin, C., Yoshida, T., Garzino-Demo, A., and Izumi, T. (2021). Editorial: Novel Insights Into a Functional HIV Cure. Front. Microbiol. 12:3705. doi: 10.3389/fmicb.2021.797570
Sarracino, A., Gharu, L., Kula, A., Pasternak, A. O., Avettand-Fenoel, V., Rouzioux, C., et al. (2018). Posttranscriptional Regulation of HIV-1 Gene Expression during Replication and Reactivation from Latency by Nuclear Matrix Protein MATR3. mBio 9:18. doi: 10.1128/mBio.02158-18
Scharer, C. D., Barwick, B. G., Youngblood, B. A., Ahmed, R., and Boss, J. M. (2013). Global DNA Methylation Remodeling Accompanies CD8 T Cell Effector Function. JI 191, 3419–3429. doi: 10.4049/jimmunol.1301395
Schleimann, M. H., Kobberø, M.-L., Vibholm, L. K., Kjær, K., Giron, L. B., Busman-Sahay, K., et al. (2019). TLR9 agonist MGN1703 enhances B cell differentiation and function in lymph nodes. EBioMedicine 45, 328–340. doi: 10.1016/j.ebiom.2019.07.005
Schumann, K., Lin, S., Boyer, E., Simeonov, D. R., Subramaniam, M., Gate, R. E., et al. (2015). Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. U S A. 112, 10437–10442. doi: 10.1073/pnas.1512503112
Schuster, S. J., Svoboda, J., Chong, E. A., Nasta, S. D., Mato, A. R., and Anak, Ö, et al. (2017). Chimeric Antigen Receptor T Cells in Refractory B-Cell Lymphomas. N. Engl. J. Med. 377, 2545–2554. doi: 10.1056/NEJMoa1708566
Sengupta, S., and Siliciano, R. F. (2018). Targeting the Latent Reservoir for HIV-1. Immunity 48, 872–895. doi: 10.1016/j.immuni.2018.04.030
Serrano-Villar, S., Sainz, T., Lee, S. A., Hunt, P. W., Sinclair, E., Shacklett, B. L., et al. (2014). HIV-Infected Individuals with Low CD4/CD8 Ratio despite Effective Antiretroviral Therapy Exhibit Altered T Cell Subsets, Heightened CD8+ T Cell Activation, and Increased Risk of Non-AIDS Morbidity and Mortality. PLoS Pathog. 10:e1004078. doi: 10.1371/journal.ppat.1004078
Seto, E., and Yoshida, M. (2014). Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 6:a018713. doi: 10.1101/cshperspect.a018713
Shacklett, B. L. (2006). Understanding the “lucky few”: The conundrum of HIV-exposed, seronegative individuals. Curr. HIV/AIDS Rep. 3, 26–31. doi: 10.1007/s11904-006-0005-2
Shankar, P., Russo, M., Harnisch, B., Patterson, M., Skolnik, P., and Lieberman, J. (2000). Impaired function of circulating HIV-specific CD8(+) T cells in chronic human immunodeficiency virus infection. Blood 96, 3094–3101.
Shayan, G., Srivastava, R., Li, J., Schmitt, N., Kane, L. P., and Ferris, R. L. (2017). Adaptive resistance to anti-PD1 therapy by Tim-3 upregulation is mediated by the PI3K-Akt pathway in head and neck cancer. OncoImmunology 6:e1261779. doi: 10.1080/2162402X.2016.1261779
Shen, C.-J., Jia, Y.-H., Tian, R.-R., Ding, M., Zhang, C., and Wang, J.-H. (2012). Translation of Pur-α is targeted by cellular miRNAs to modulate the differentiation-dependent susceptibility of monocytes to HIV-1 infection. FASEB J. 26, 4755–4764. doi: 10.1096/fj.12-209023
Siliciano, J. D., Kajdas, J., Finzi, D., Quinn, T. C., Chadwick, K., Margolick, J. B., et al. (2003). Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9, 727–728. doi: 10.1038/nm880
Simonetti, F. R., Sobolewski, M. D., Fyne, E., Shao, W., Spindler, J., Hattori, J., et al. (2016). Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc. Natl. Acad. Sci. U S A. 113, 1883–1888. doi: 10.1073/pnas.1522675113
Simonetti, F. R., White, J. A., Tumiotto, C., Ritter, K. D., Cai, M., Gandhi, R. T., et al. (2020). Intact proviral DNA assay analysis of large cohorts of people with HIV provides a benchmark for the frequency and composition of persistent proviral DNA. Proc. Natl. Acad. Sci. U S A. 117, 18692–18700. doi: 10.1073/pnas.2006816117
Simonetti, F. R., Zhang, H., Soroosh, G. P., Duan, J., Rhodehouse, K., Hill, A. L., et al. (2021). Antigen-driven clonal selection shapes the persistence of HIV-1-infected CD4+ T cells in vivo. J. Clin. Invest. 131:145254. doi: 10.1172/JCI145254
Śledzińska, A., Menger, L., Bergerhoff, K., Peggs, K. S., and Quezada, S. A. (2015). Negative immune checkpoints on T lymphocytes and their relevance to cancer immunotherapy. Mol. Oncol. 9, 1936–1965. doi: 10.1016/j.molonc.2015.10.008
Søgaard, O. S., Graversen, M. E., Leth, S., Olesen, R., Brinkmann, C. R., Nissen, S. K., et al. (2015). The Depsipeptide Romidepsin Reverses HIV-1 Latency In Vivo. PLoS Pathog. 11:e1005142. doi: 10.1371/journal.ppat.1005142
Soriano-Sarabia, N., Archin, N. M., Bateson, R., Dahl, N. P., Crooks, A. M., Kuruc, J. D., et al. (2015). Peripheral Vγ9Vδ2 T Cells Are a Novel Reservoir of Latent HIV Infection. PLoS Pathog. 11:e1005201. doi: 10.1371/journal.ppat.1005201
Soriano-Sarabia, N., Bateson, R. E., Dahl, N. P., Crooks, A. M., Kuruc, J. D., Margolis, D. M., et al. (2014). Quantitation of Replication-Competent HIV-1 in Populations of Resting CD4 + T Cells. J. Virol. 88, 14070–14077. doi: 10.1128/JVI.01900-14
Spivak, A. M., Andrade, A., Eisele, E., Hoh, R., Bacchetti, P., Bumpus, N. N., et al. (2014). A pilot study assessing the safety and latency-reversing activity of disulfiram in HIV-1-infected adults on antiretroviral therapy. Clin. Infect. Dis. 58, 883–890. doi: 10.1093/cid/cit813
Spivak, A. M., Bosque, A., Balch, A. H., Smyth, D., Martins, L., and Planelles, V. (2015). Ex Vivo Bioactivity and HIV-1 Latency Reversal by Ingenol Dibenzoate and Panobinostat in Resting CD4+ T Cells from Aviremic Patients. Antimicrob. Agents Chemother. 2015:15. doi: 10.1128/AAC.01077-15
Srivastava, S., and Riddell, S. R. (2015). Engineering CAR-T cells: Design concepts. Trends Immunol. 36, 494–502. doi: 10.1016/j.it.2015.06.004
Strain, M. C., Lada, S. M., Luong, T., Rought, S. E., Gianella, S., Terry, V. H., et al. (2013). Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 8:e55943. doi: 10.1371/journal.pone.0055943
Stuelke, E. L., James, K. S., Kirchherr, J. L., Allard, B., Baker, C., Kuruc, J. D., et al. (2020). Measuring the Inducible, Replication-Competent HIV Reservoir Using an Ultra-Sensitive p24 Readout, the Digital ELISA Viral Outgrowth Assay. Front. Immunol. 11:1971. doi: 10.3389/fimmu.2020.01971
Sun, S.-C. (2017). The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 17, 545–558. doi: 10.1038/nri.2017.52
Sun, S.-C., and Ley, S. C. (2008). New insights into NF-κB regulation and function. Trends Immunol. 29, 469–478. doi: 10.1016/j.it.2008.07.003
Sung, T.-L., and Rice, A. P. (2009). miR-198 Inhibits HIV-1 Gene Expression and Replication in Monocytes and Its Mechanism of Action Appears To Involve Repression of Cyclin T1. PLoS Pathog. 5:e1000263. doi: 10.1371/journal.ppat.1000263
Suzuki, K., Hattori, S., Marks, K., Ahlenstiel, C., Maeda, Y., Ishida, T., et al. (2013). Promoter Targeting shRNA Suppresses HIV-1 Infection In vivo Through Transcriptional Gene Silencing. Mol. Therapy Nucleic Acids 2, e137. doi: 10.1038/mtna.2013.64
Tan, M., Luo, H., Lee, S., Jin, F., Yang, J. S., Montellier, E., et al. (2011). Identification of 67 Histone Marks and Histone Lysine Crotonylation as a New Type of Histone Modification. Cell 146, 1016–1028. doi: 10.1016/j.cell.2011.08.008
Taura, M., Song, E., Ho, Y.-C., and Iwasaki, A. (2019). Apobec3A maintains HIV-1 latency through recruitment of epigenetic silencing machinery to the long terminal repeat. Proc. Natl. Acad. Sci. U S A. 116, 2282–2289. doi: 10.1073/pnas.1819386116
Taylor, R. A., Xiao, S., Carias, A. M., McRaven, M. D., Thakkar, D. N., Araínga, M., et al. (2021). PET/CT targeted tissue sampling reveals virus specific dIgA can alter the distribution and localization of HIV after rectal exposure. PLoS Pathog. 17:e1009632. doi: 10.1371/journal.ppat.1009632
Tchasovnikarova, I. A., Timms, R. T., Matheson, N. J., Wals, K., Antrobus, R., Göttgens, B., et al. (2015). Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 348, 1481–1485. doi: 10.1126/science.aaa7227
Tebas, P., Stein, D., Tang, W. W., Frank, I., Wang, S. Q., Lee, G., et al. (2014). Gene Editing of CCR5 in Autologous CD4 T Cells of Persons Infected with HIV. N. Engl. J. Med. 370, 901–910. doi: 10.1056/NEJMoa1300662
Tomer, R., Ye, L., Hsueh, B., and Deisseroth, K. (2014). Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 9, 1682–1697. doi: 10.1038/nprot.2014.123
Triboulet, R., Mari, B., Lin, Y.-L., Chable-Bessia, C., Bennasser, Y., Lebrigand, K., et al. (2007). Suppression of MicroRNA-Silencing Pathway by HIV-1 During Virus Replication. Science 315, 1579–1582. doi: 10.1126/science.1136319
Turrini, F., Marelli, S., Kajaste-Rudnitski, A., Lusic, M., Van Lint, C., Das, A. T., et al. (2015). HIV-1 transcriptional silencing caused by TRIM22 inhibition of Sp1 binding to the viral promoter. Retrovirology 12:104. doi: 10.1186/s12977-015-0230-0
Tyagi, M., and Karn, J. (2007). CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 26, 4985–4995. doi: 10.1038/sj.emboj.7601928
Uldrick, T. S., Ison, G., Rudek, M. A., Noy, A., Schwartz, K., Bruinooge, S., et al. (2017). Modernizing Clinical Trial Eligibility Criteria: Recommendations of the American Society of Clinical Oncology-Friends of Cancer Research HIV Working Group. J. Clin. Oncol. 35, 3774–3780. doi: 10.1200/JCO.2017.73.7338
van den Dries, L., Claassen, M. A. A., Groothuismink, Z. M. A., van Gorp, E., and Boonstra, A. (2017). Immune activation in prolonged cART-suppressed HIV patients is comparable to that of healthy controls. Virology 509, 133–139. doi: 10.1016/j.virol.2017.06.014
Van der Sluis, R. M., Kumar, N. A., Pascoe, R. D., Zerbato, J. M., Evans, V. A., Dantanarayana, A. I., et al. (2020). Combination Immune Checkpoint Blockade to Reverse HIV Latency. JI 204, 1242–1254. doi: 10.4049/jimmunol.1901191
Van Lint, C., Bouchat, S., and Marcello, A. (2013). HIV-1 transcription and latency: an update. Retrovirology 10:67. doi: 10.1186/1742-4690-10-67
Van Lint, C., Emiliani, S., Ott, M., and Verdin, E. (1996). Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15, 1112–1120. doi: 10.1002/j.1460-2075.1996.tb00449.x
van Snippenberg, W., Gleerup, D., Rutsaert, S., Vandekerckhove, L., De Spiegelaere, W., and Trypsteen, W. (2021). Triplex digital PCR assays for the quantification of intact proviral HIV-1 DNA. Methods 2021:S1046202321001328. doi: 10.1016/j.ymeth.2021.05.006
Vibholm, L. K., Konrad, C. V., Schleimann, M. H., Frattari, G., Winckelmann, A., Klastrup, V., et al. (2019). Effects of 24-week Toll-like receptor 9 agonist treatment in HIV type 1+ individuals. AIDS 33, 1315–1325. doi: 10.1097/QAD.0000000000002213
Vibholm, L., Schleimann, M. H., Højen, J. F., Benfield, T., Offersen, R., Rasmussen, K., et al. (2017). Short-Course Toll-Like Receptor 9 Agonist Treatment Impacts Innate Immunity and Plasma Viremia in Individuals With Human Immunodeficiency Virus Infection. Clin. Infect. Dis. 64, 1686–1695. doi: 10.1093/cid/cix201
von Stockenstrom, S., Odevall, L., Lee, E., Sinclair, E., Bacchetti, P., Killian, M., et al. (2015). Longitudinal Genetic Characterization Reveals That Cell Proliferation Maintains a Persistent HIV Type 1 DNA Pool During Effective HIV Therapy. J. Infect. Dis. 212, 596–607. doi: 10.1093/infdis/jiv092
Vranckx, L. S., Demeulemeester, J., Saleh, S., Boll, A., Vansant, G., Schrijvers, R., et al. (2016). LEDGIN-mediated Inhibition of Integrase-LEDGF/p75 Interaction Reduces Reactivation of Residual Latent HIV. EBioMedicine 8, 248–264. doi: 10.1016/j.ebiom.2016.04.039
Wagner, T. A., McLaughlin, S., Garg, K., Cheung, C. Y. K., Larsen, B. B., Styrchak, S., et al. (2014). HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573. doi: 10.1126/science.1256304
Wang, Q., Jia, Y., Wang, Y., Jiang, Z., Zhou, X., Zhang, Z., et al. (2019). Evolution of cis- and trans-regulatory divergence in the chicken genome between two contrasting breeds analyzed using three tissue types at one-day-old. BMC Genomics 20:933. doi: 10.1186/s12864-019-6342-5
Wang, Z., Gurule, E. E., Brennan, T. P., Gerold, J. M., Kwon, K. J., Hosmane, N. N., et al. (2018). Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc. Natl. Acad. Sci. U S A. 115, E2575–E2584. doi: 10.1073/pnas.1720665115
Weber, S., Weiser, B., Kemal, K. S., Burger, H., Ramirez, C. M., Korn, K., et al. (2014). Epigenetic analysis of HIV-1 proviral genomes from infected individuals: Predominance of unmethylated CpG’s. Virology 449, 181–189. doi: 10.1016/j.virol.2013.11.013
Wei, P., Garber, M. E., Fang, S.-M., Fischer, W. H., and Jones, K. A. (1998). A Novel CDK9-Associated C-Type Cyclin Interacts Directly with HIV-1 Tat and Mediates Its High-Affinity, Loop-Specific Binding to TAR RNA. Cell 92, 451–462. doi: 10.1016/S0092-8674(00)80939-3
Weinberger, L. S., Burnett, J. C., Toettcher, J. E., Arkin, A. P., and Schaffer, D. V. (2005). Stochastic Gene Expression in a Lentiviral Positive-Feedback Loop: HIV-1 Tat Fluctuations Drive Phenotypic Diversity. Cell 122, 169–182. doi: 10.1016/j.cell.2005.06.006
West, M. J., Lowe, A. D., and Karn, J. (2001). Activation of Human Immunodeficiency Virus Transcription in T Cells Revisited: NF-κB p65 Stimulates Transcriptional Elongation. J. Virol. 2001:2001. doi: 10.1128/JVI.75.18.8524-8537.2001
Wherry, E. J., and Kurachi, M. (2015). Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15, 486–499. doi: 10.1038/nri3862
Wherry, E. J., Blattman, J. N., Murali-Krishna, K., van der Most, R., and Ahmed, R. (2003). Viral Persistence Alters CD8 T-Cell Immunodominance and Tissue Distribution and Results in Distinct Stages of Functional Impairment. J. Virol. 77, 4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003
Wherry, E. J., Ha, S.-J., Kaech, S. M., Haining, W. N., Sarkar, S., Kalia, V., et al. (2007). Molecular Signature of CD8+ T Cell Exhaustion during Chronic Viral Infection. Immunity 27, 670–684. doi: 10.1016/j.immuni.2007.09.006
White, M. K., Kaminski, R., Young, W., Roehm, P. C., and Khalili, K. (2017). CRISPR Editing Technology in Biological and Biomedical Investigation. J. Cell. Biochem. 118, 3586–3594. doi: 10.1002/jcb.26099
Wierda, R. J., Kuipers, H. F., van Eggermond, M. C. J. A., Benard, A., van Leeuwen, J. C., Carluccio, S., et al. (2012). Epigenetic control of CCR5 transcript levels in immune cells and modulation by small molecules inhibitors. J. Cell. Mol. Med. 16, 1866–1877. doi: 10.1111/j.1582-4934.2011.01482.x
Wightman, F., Lu, H. K., Solomon, A. E., Saleh, S., Harman, A. N., Cunningham, A. L., et al. (2013). Entinostat is a histone deacetylase inhibitor selective for class 1 histone deacetylases and activates HIV production from latently infected primary T cells. AIDS 27, 2853–2862. doi: 10.1097/QAD.0000000000000067
Wightman, F., Solomon, A., Khoury, G., Green, J. A., Gray, L., Gorry, P. R., et al. (2010). Both CD31 + and CD31 – Naive CD4 + T Cells Are Persistent HIV Type 1–Infected Reservoirs in Individuals Receiving Antiretroviral Therapy. J. Infect. Dis. 202, 1738–1748. doi: 10.1086/656721
Wightman, F., Solomon, A., Kumar, S. S., Urriola, N., Gallagher, K., Hiener, B., et al. (2015). Effect of ipilimumab on the HIV reservoir in an HIV-infected individual with metastatic melanoma. AIDS 29, 504–506. doi: 10.1097/QAD.0000000000000562
Williams, J. P., Hurst, J., Stöhr, W., Robinson, N., Brown, H., Fisher, M., et al. (2014). HIV-1 DNA predicts disease progression and post-treatment virological control. eLife 3:e03821. doi: 10.7554/eLife.03821
Williams, S. A., Chen, L.-F., Kwon, H., Fenard, D., Bisgrove, D., Verdin, E., et al. (2004). Prostratin antagonizes HIV latency by activating NF-kappaB. J. Biol. Chem. 279, 42008–42017. doi: 10.1074/jbc.M402124200
Williams, S. A., Chen, L.-F., Kwon, H., Ruiz-Jarabo, C. M., Verdin, E., and Greene, W. C. (2006). NF-κB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 25, 139–149. doi: 10.1038/sj.emboj.7600900
Wilson, E. M. P., and Sereti, I. (2013). Immune restoration after antiretroviral therapy: the pitfalls of hasty or incomplete repairs. Immunol. Rev. 254, 343–354. doi: 10.1111/imr.12064
Wong, J. K., Hezareh, M., Günthard, H. F., Havlir, D. V., Ignacio, C. C., Spina, C. A., et al. (1997). Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295. doi: 10.1126/science.278.5341.1291
Wong, L. M., and Jiang, G. (2021). NF-κB sub-pathways and HIV cure: A revisit. EBioMedicine 63:103159. doi: 10.1016/j.ebiom.2020.103159
Wong, M. E., Jaworowski, A., and Hearps, A. C. (2019). The HIV Reservoir in Monocytes and Macrophages. Front. Immunol. 10:1435. doi: 10.3389/fimmu.2019.01435
Wu, G., Cheney, C., Huang, Q., Hazuda, D. J., Howell, B. J., and Zuck, P. (2021). Improved Detection of HIV Gag p24 Protein Using a Combined Immunoprecipitation and Digital ELISA Method. Front. Microbiol. 12:636703. doi: 10.3389/fmicb.2021.636703
Wu, G., Swanson, M., Talla, A., Graham, D., Strizki, J., Gorman, D., et al. (2017). HDAC inhibition induces HIV-1 protein and enables immune-based clearance following latency reversal. JCI Insight 2:92901. doi: 10.1172/jci.insight.92901
Xing, S., Bullen, C. K., Shroff, N. S., Shan, L., Yang, H.-C., Manucci, J. L., et al. (2011). Disulfiram reactivates latent HIV-1 in a Bcl-2-transduced primary CD4+ T cell model without inducing global T cell activation. J. Virol. 85, 6060–6064. doi: 10.1128/JVI.02033-10
Xiong, Y., Luscher, M. A., Altman, J. D., Hulsey, M., Robinson, H. L., Ostrowski, M., et al. (2001). Simian Immunodeficiency Virus (SIV) Infection of a Rhesus Macaque Induces SIV-Specific CD8 + T Cells with a Defect in Effector Function That Is Reversible on Extended Interleukin-2 Incubation. J. Virol. 75, 3028–3033. doi: 10.1128/JVI.75.6.3028-3033.2001
Xu, L., Yang, H., Gao, Y., Chen, Z., Xie, L., Liu, Y., et al. (2017). CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance In Vivo. Mol. Therapy 25, 1782–1789. doi: 10.1016/j.ymthe.2017.04.027
Yamaguchi, Y., Takagi, T., Wada, T., Yano, K., Furuya, A., Sugimoto, S., et al. (1999). NELF, a Multisubunit Complex Containing RD, Cooperates with DSIF to Repress RNA Polymerase II Elongation. Cell 97, 41–51. doi: 10.1016/S0092-8674(00)80713-8
Yang, C., Dove, M. T., Brazhkin, V. V., and Trachenko, K. (2018). Yang, et al. Reply. Phys. Rev. Lett. 120:219602. doi: 10.1103/PhysRevLett.120.219602
Yang, Z., Zhu, Q., Luo, K., and Zhou, Q. (2001). The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414, 317–322. doi: 10.1038/35104575
Yates, K. B., Tonnerre, P., Martin, G. E., Gerdemann, U., Al Abosy, R., Comstock, D. E., et al. (2021). Epigenetic scars of CD8+ T cell exhaustion persist after cure of chronic infection in humans. Nat. Immunol. 22, 1020–1029. doi: 10.1038/s41590-021-00979-1
Yerly, S., Günthard, H. F., Fagard, C., Joos, B., Perneger, T. V., Hirschel, B., et al. (2004). Proviral HIV-DNA predicts viral rebound and viral setpoint after structured treatment interruptions. AIDS 18, 1951–1953. doi: 10.1097/00002030-200409240-00011
Yeung, M. L., Houzet, L., Yedavalli, V. S. R. K., and Jeang, K.-T. (2009). A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J. Biol. Chem. 284, 19463–19473. doi: 10.1074/jbc.M109.010033
Yik, J. H. N., Chen, R., Nishimura, R., Jennings, J. L., Link, A. J., and Zhou, Q. (2003). Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12, 971–982. doi: 10.1016/s1097-2765(03)00388-5
Yin, C., Zhang, T., Qu, X., Zhang, Y., Putatunda, R., Xiao, X., et al. (2017). In Vivo Excision of HIV-1 Provirus by saCas9 and Multiplex Single-Guide RNAs in Animal Models. Mol. Therapy 25, 1168–1186. doi: 10.1016/j.ymthe.2017.03.012
Ylisastigui, L., Archin, N. M., Lehrman, G., Bosch, R. J., and Margolis, D. M. (2004). Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 18, 1101–1108. doi: 10.1097/00002030-200405210-00003
Youngblood, B., Noto, A., Porichis, F., Akondy, R. S., Ndhlovu, Z. M., Austin, J. W., et al. (2013). Cutting edge: Prolonged exposure to HIV reinforces a poised epigenetic program for PD-1 expression in virus-specific CD8 T cells. J. Immunol. 191, 540–544. doi: 10.4049/jimmunol.1203161
Youngblood, B., Oestreich, K. J., Ha, S.-J., Duraiswamy, J., Akondy, R. S., West, E. E., et al. (2011). Chronic Virus Infection Enforces Demethylation of the Locus that Encodes PD-1 in Antigen-Specific CD8+ T Cells. Immunity 35, 400–412. doi: 10.1016/j.immuni.2011.06.015
Yucha, R. W., Hobbs, K. S., Hanhauser, E., Hogan, L. E., Nieves, W., Ozen, M. O., et al. (2017). High-throughput Characterization of HIV-1 Reservoir Reactivation Using a Single-Cell-in-Droplet PCR Assay. EBioMedicine 20, 217–229. doi: 10.1016/j.ebiom.2017.05.006
Yukl, S. A., Kaiser, P., Kim, P., Telwatte, S., Joshi, S. K., Vu, M., et al. (2018). HIV latency in isolated patient CD4 + T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med. 10:eaa9927. doi: 10.1126/scitranslmed.aap9927
Zahavi, D. J., and Weiner, L. M. (2019). Targeting Multiple Receptors to Increase Checkpoint Blockade Efficacy. Int. J. Mol. Sci. 20:E158. doi: 10.3390/ijms20010158
Zebley, C. C., Gottschalk, S., and Youngblood, B. (2020). Rewriting History: Epigenetic Reprogramming of CD8+ T Cell Differentiation to Enhance Immunotherapy. Trends Immunol. 41, 665–675. doi: 10.1016/j.it.2020.06.008
Zerbato, J. M., McMahon, D. K., Sobolewski, M. D., Mellors, J. W., and Sluis-Cremer, N. (2019). Naive CD4+ T Cells Harbor a Large Inducible Reservoir of Latent, Replication-competent Human Immunodeficiency Virus Type 1. Clin. Infect. Dis. 69, 1919–1925. doi: 10.1093/cid/ciz108
Zhang, H.-S., Wu, T.-C., Sang, W.-W., and Ruan, Z. (2012). MiR-217 is involved in Tat-induced HIV-1 long terminal repeat (LTR) transactivation by down-regulation of SIRT1. Biochim. Biophys. Acta 1823, 1017–1023. doi: 10.1016/j.bbamcr.2012.02.014
Zhang, Z., Nikolai, B. C., Gates, L. A., Jung, S. Y., Siwak, E. B., He, B., et al. (2017). Crosstalk between histone modifications indicates that inhibition of arginine methyltransferase CARM1 activity reverses HIV latency. Nucleic Acids Res. 45, 9348–9360. doi: 10.1093/nar/gkx550
Zhong, H., May, M. J., Jimi, E., and Ghosh, S. (2002). The Phosphorylation Status of Nuclear NF-K B Determines Its Association with CBP/p300 or HDAC-1. Mol. Cell 9, 625–636. doi: 10.1016/S1097-2765(02)00477-X
Zhou, H., Xu, M., Huang, Q., Gates, A. T., Zhang, X. D., Castle, J. C., et al. (2008). Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4, 495–504. doi: 10.1016/j.chom.2008.10.004
Keywords: human immunodeficiency virus (HIV), functional HIV cure, HIV latency, HIV persistence, kick-and-kill strategy, Block-and-Lock strategy, defective proviruses, immunotherapy
Citation: Ta TM, Malik S, Anderson EM, Jones AD, Perchik J, Freylikh M, Sardo L, Klase ZA and Izumi T (2022) Insights Into Persistent HIV-1 Infection and Functional Cure: Novel Capabilities and Strategies. Front. Microbiol. 13:862270. doi: 10.3389/fmicb.2022.862270
Received: 25 January 2022; Accepted: 21 February 2022;
Published: 27 April 2022.
Edited by:
Chunfu Zheng, University of Calgary, CanadaReviewed by:
Shan Cen, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaGuochun Jiang, University of North Carolina at Chapel Hill, United States
Copyright © 2022 Ta, Malik, Anderson, Jones, Perchik, Freylikh, Sardo, Klase and Izumi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taisuke Izumi, dGl6dW1pQHVzY2llbmNlcy5lZHU=
†These authors have contributed equally to this work
 Tram M. Ta
Tram M. Ta Sajjaf Malik
Sajjaf Malik Elizabeth M. Anderson
Elizabeth M. Anderson Amber D. Jones
Amber D. Jones Jocelyn Perchik
Jocelyn Perchik Maryann Freylikh
Maryann Freylikh Luca Sardo
Luca Sardo Zackary A. Klase
Zackary A. Klase Taisuke Izumi
Taisuke Izumi