
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 May 2022
Sec. Extreme Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.862245
This article is part of the Research Topic Thermophilic and Halophilic Extremophiles in Eurasian Environments, Volume II View all 9 articles
Salinization poses great threats to soil fungal communities that would cause the losses of ecosystems services. Soil fungal communities are composed of different functional guilds such as saprotrophic, symbiotrophic, and pathotrophic fungi, and each guild includes many rare taxa and a few abundant taxa. Despite of low abundance, rare taxa may be crucial in determining the responses of entire soil fungal communities to salinization. However, it remains poorly understood how rare taxa mediate the impacts of soil salinization on soil fungal community structure. Here, we took advantage of a salinity gradient in a desert ecosystem ranging from 0.60 to 31.09 g kg−1 that was created by a 12-year saline-water irrigation and assessed how the rare vs. abundant taxa of soil saprotrophic, symbiotrophic, and pathotrophic fungi respond to soil salinization through changes in the community biodiversity and composition. We found that the rare taxa of soil saprotrophic, symbiotrophic, and pathographic fungi were more sensitive to changes in soil salinity compared to the abundant taxa. In addition, the community composition of rare taxa of the saprotrophic and pathotrophic fungi not the symbiotrophic fungi was positively associated with soil salinity change. However, the symbiotrophic fungi showed greater variations in the species richness along the salinity gradient. These findings highlight the importance to differentiate rare taxa in predicting how the biodiversity and functional groups of soil fungal communities respond to soil salinization.
Salinity affects about 1.1 billion hectares of land surface around the world, accounting for 7.4% of global land area (Ivushkin et al., 2019). Soil salinization is becoming a major environmental challenge specifically in drylands due to climate change and/or poor land management, such as saline-water irrigation (Rath and Rousk, 2015). Soil salinization causes osmotic pressure, nutritional imbalance, and ion toxic effects to plants and microorganisms (Mansour, 2000; Munns and Tester, 2008), which threatens ecosystem functions and services (Rath et al., 2016). As one of the most diverse and abundant groups of soil microbiota, soil fungi are important for mitigating the negative impacts of salinization on soil functionality (Ruppel et al., 2013; Frac et al., 2018). For instance, soil fungi use low-quality organic matter in infertile soils (Bardgett, 2005) and show higher resistance to environmental stresses over soil bacteria (Rath et al., 2016; Thiem et al., 2018). Soil fungi display stable network attributes and can maintain high fungal biomass per unit mass of soil organic matter under saline conditions (de Vries et al., 2018; Rath et al., 2019). Therefore, undersatnding the impacts of soil salinization on fungal communities is vital for the conservation of belowground biodiversity in the deteriorating dryland environments.
Soil fungal communities are functionally diverse and can be classified into three major functional guilds based on their trophic modes, such as saprotrophic fungi, pathotrophic fungi, and symbiotrophic fungi (Yang et al., 2016; Frac et al., 2018), which may adapt to soil salinization differently. Saprotrophic fungi decompose litter and transform nutrients and their activities depend on the quality and quantity of soil organic matter (Rath et al., 2019). In contrast, pathotrophic fungi affect disease, pests, and the growth of other organisms (Frac et al., 2018). Symbiotrophic fungi improve plant nutrition by establishing plant-mycorrhizal associations, and their abundance and composition depend on soil nutrient conditions (Moore et al., 2021). Given various functions of soil fungal guilds, treating soil fungal communities a whole without differentiating functional differences may mislead the prediction regarding how the functionality of soil fungal communities responds to salinization. For instance, the relative abundance of saprotrophic fungi increases and that of pathotrophic fungi decreases after ecosystem restoration in saline-alkaline soils (Xu et al., 2021). Although fungal taxonomic groups increase or decrease significantly with salinity changes (Kim et al., 2019; Yang et al., 2020), the responses of functional guilds of soil fungi to salinization are still poorly understood. This hinders our understanding of the mechanisms driving for soil fungal communities in dealing with soil salinization and the cascading effects on ecosystem processes.
Communities of soil fungal guilds are comprised of many rare taxa and a few abundant taxa (Balbuena et al., 2021). Current studies mainly focus on the abundant soil fungi, while little is known about how the rare fungal taxa respond to soil salinization. Although a number of studies report that fungal diversity does not change along salinity gradients (Mohamed and Martiny, 2011; Bharti et al., 2015), increasing evidences show that rare fungal taxa are more sensitive to salinity or other stresses than abundant taxa (Wan et al., 2021). Compared with the abundant taxa, the rare taxa of soil fungal guilds have fewer niches and less clustered phylotypes and can be influenced more by selective pressure and dispersal effects, thus they would be more likely subject to extinction and local environmental variations (Galand et al., 2009; Pascoal et al., 2021; Wan et al., 2021). Evidence from forest and wetland soils show bigger changes in the rare microbial taxa with environmental variations compared to the abundant taxa (Oono et al., 2017; Ji et al., 2020; Wan et al., 2021).
In addition, soil functional fungal guilds have different responses to salinity changes (Xu et al., 2021), which might be induced by the rare taxa. Vanegas et al. (2019) have found that saprotrophic fungi have higher sequence abundance than the symbiotrophic fungi and were expected to be more resistant to salinity changes than the latter. But another study observed that saprotrophic fungi displayed greater variations with salinity changes than symbiotrophic fungi (Bencherif et al., 2015), and that the sequenicng abundance of some rare taxa of saproptrophic fungi was higher than that of the abundant taxa of symbiotrophic fungi (Yang et al., 2019). These findings suggest that the rare taxa of saprotrophic fungi may determine the responses of this functional fungal guild to salinity change. Therefore, we need to elucidate how the rare vs. abundant taxa of functional fungal guilds respond to soil salinization in order to better understand the adaptation mechanisms of soil fungal community in dealing with salinity stress.
In this study, we examined the spatial variability in soil fungal community along a soil salinity gradient ranging from 0.60 to 31.09 g kg−1 to address two questions: (1) How do the diversity and community composition of three fungal guilds respond to soil salinization? (2) Do the rare fungal taxa determine the response of soil fungal communities to salinization? We predict that the diversity of symbiotrophic fungi increases with soil salinity and that of saprotrophic and pathotrophic fungi decrease along the salinity gradient, because symbiotrophic fungi depends more on plants in high saline soils but the growths of sapraotrophic and pathotrophic fungi are inhibited (Bencherif et al., 2015). In addition, symbiotrophic fungi are expected to occupy more niches and sapraotrophic and pathotrophic fungi occupy less niches with increasing soil salinity (Figure 1). Due to the weaker adaption of microbial rare taxa to environmental changes compared to abundant taxa (Montoya-Ciriaco et al., 2020), we predicted that the rare taxa of soil functional fungal guilds are more sensitive to soil salinization than the abundant taxa.
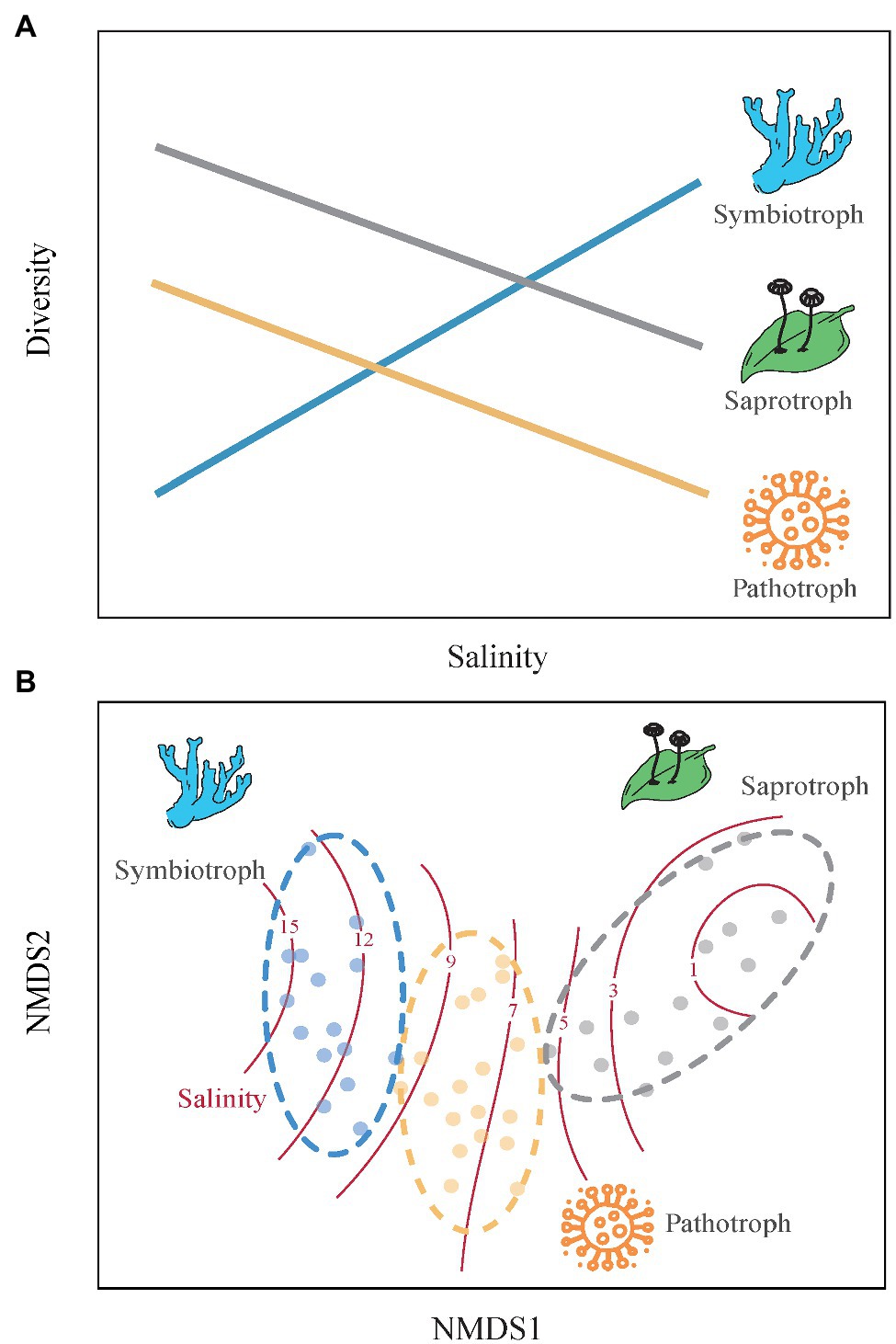
Figure 1. The conceptual framework outlines changes in community structure of three functional guilds of soil fungi along a soil salinity gradient. (A) Alpha diversity. We expect that the alpha-diversity of symbiotrophic fungi would increase with soil salinity because symbiotrophic fungi depend highly on host plant at high-salinity sites (Ren et al., 2016), while that of soil saprotrophic and pathotrophic fungi would decrease with soil salinity because high salinity can hinder the activity of soil fungi that mainly use soil organic matter (Rath et al., 2016). (B) Community composition. Since soil symbiotrophic, sapraotrophic, and pathotrophic fungi have different niches (Frac et al., 2018), we therefore expect that symbiotrophic fungi occur at higher salinity levels, then do pathotrophic and sapraotrophic fungi.
The study sites are in the shrub shelterbelts along the Tarim desert highway, which crosses the Taklamakan desert with a total length of 465 km. The Taklamakan desert has a continental hyper arid climate with mean annual temperature of 2.4°C, mean annual precipitation of 24.6 mm, and mean annual potential evaporation of 3,639 mm. The Taklamakan desert has an average wind speed of 5.6 mph and gale (> 44.7 mph) occurs > 130 days·year−1 (Lei et al., 2008). Soils in the Taklamakan desert are aeolian sandy soils with 87.3% sand, 12.4% silt, and 0.3% clay (Zhang et al., 2016). In 2005, the shrub shelterbelts (72 ~ 78 m in width) were constructed to prevent mobile sand dunes from covering the Tarim desert highway by planting drought and salt tolerant species, such as Haloxylon ammodendron (C. A. Mey.) Bunge., Tamarix chinensis Lour., and Calligonum mongolicum Turcz (Li, 2010). Due to the hyper arid environment of the Taklamakan desert, groundwater has been pumped for drip irrigation to water the shelterbelts from March to October regularly (Zhang et al., 2016). The 12-year drip irrigation by using saline groundwater and high evaporation have resulted in soil salinization (Wang et al., 2012), and H. ammodendron becomes the dominant plant species in the shelterbelts (Li, 2010; Zhang et al., 2016).
The soil salinity gradient was caused by a 12-year saline-water irrigation in the shrub shelterbelts along the Tarim desert highway. The salinity of groundwater used for irrigation decreases from 30.0 to 2.6 g·L−1 from the north to the middle of the Tarim desert highway (Li et al., 2008), and salts accumulate in topsoils along soil profiles due to high evaporation (Supplementary Figure 1). Then, soil salinity genearlly decreases from the north to the middle of the Tarim desert highway and also decreases from surface soils to subsoils. In addition, plants in the shrub shelterbelts are dominated by Haloxylon ammodendron (C. A. Mey.) Bunge., which minimizes the influence of plant variation on fungal guilds and provides great opportunities to investigate the impacts of soil salinization on soil fungi in field. Plants in the shrub shelterbelts were arranged in rows, with a spacing of 2 m between rows and a spacing of 1 m between individuals within each row.
In this study, ten sites in the shelterbelts were selected from the north to the middle of the Tarim desert highway (Supplementary Figure 2). At each site, three spots were randomly selected from locations that were within the two rows of shrubs and about 30-50 cm away from a shrub along the shrub shelterbelts as replications. Soils were collected using soil cores (5 cm in diameter) at the depths of 0–10, 10–20, 20–40, 40–60, 60–80, and 80–100 cm. A total of 180 soil samples were collected from the ten sites in early October 2017. Soils were passed through a 2 mm sieve to remove coarse roots and stones with visible roots picked by hand. A subset of soils for DNA extraction were stored at −20°C, and another subset soils were air-dried to determine soil properties. The details of soil properties along soil depth were listed at Supplementary Figure 1.
Soil salinity, electrical conductivity (EC), and pH were assessed in a suspension with the soil to water ratio of 1:5 (w/v). The sum of Na+, K+, Ca2+, Mg2+, Cl−, and excluded were calculated as soil salinity. The concentration of Na+, K+, Ca2+, and Mg2+ were measured by inductively coupled plasma – optical emission spectrometry (ICP-OES, Optima 6,300 DV, Perkin Elmer, United States). Soil Cl− was measured by the AgNO3 titration method, by ethylenediaminetetraacetic acid (EDTA) titration method, and and by the H2SO4 titration method. Soil pH and EC were measured by a pH and conductivity meter (Mettler Toledo FiveEasy Plus FE38, Greifensee, Switzerland). Soil organic carbon (SOC) was measured by the K2Cr2O7 oxidation method (Nelson and Sommers, 1982). Soil total nitrogen (STN) was measured by an Auto Kjeldahl Unite model (BUCHI K370, Flawil, Switzerland). Soil total phosphorus (STP) was determined by the Mo-Sb colorimetric method (Yuan and Lavkulich, 1995), and soil available P (avaP) was measured using the sodium bicarbonate extraction with Mo-Sb Anti-spectrophotometer method (Olsen, 1954).
Total soil DNA was extracted from 0.25 g of each fresh sample using a PowerSoil DNA Kit (Qiagen, Carlsbad, CA, United States). The final DNA quality and content was measured by NanoDrop (Thermo Fisher Scientific, United States). The fungal internal transcribed spacer 1 (ITS1) regions of nuclear ribosomal RNA genes were amplified using modified ITS5 (Forward primer, ITS5, 5′-GGAAGTAAAAGTCGTAACAAGG-3′; White et al., 1990) and ITS2 (reverse primer, ITS2, 5’-GCTGCGTTCTTCATCGATGC-3′; Smith and Peay, 2014). PCR was conducted at a total volume of 50 μl, containing 25 μl Premix Taq polymerase, 1 μl of each primer (10 mmol·L−1), 60 ng template DNA, and double-distilled H2O to a final reaction volume of 50 μl. The following cycling parameters used were: 94°C for 5 min; 30 cycles at 94°C for 30 s, 52°C for 30 s, and 72°C for 30 s; and a final step at 72°C for 10 min. The PCR products were examined on ethidium bromide-stained 1% agarose gels by electrophoresis and visualized under UV light. Three replicates of the PCR products from one sample were pooled together. The PCR products were purified using the EZNA Gel Extraction Kit (Omega, United States). This DNA mix was then sequenced on Illumina PE250 platform (Illumina, United States).
The sequences were quality-filtered by using Trimmomatic V0.331 and merged by using FLASH V1.2.112 based on the following criteria: the minimum sequence length ≥ 100 bp (excluding barcode and primer sequences); maximum number of primer or barcode mismatches ≤ 2; and minimum mean quality score ≥ 20 in a window of 50 nt; homopolymer length ≤ 10 bp; homopolymer mismatch ratio ≤ 0.1. After quality filtering, 180 samples yielded a total of 1,597,974 sequences, ranging from 37,009 to 157,855 sequences per sample. The sequences were subjected to de novo chimaera detection and clustered into operational taxonomic units (OTUs) at a 97% cutoff sequence similarity by using the USEARCH algorithm. The most abundant sequences were chosen as representative sequences by using UC criteria. OTU sequences were then assigned taxonomies using the RDP classifier method against the UNITE database. All samples yielded a total of 11,289,935 fungal sequences, ranging from 3,699 to 150,718 sequences per sample. Fungal OTU clusters and clusters with <2 reads or < 2 samples were discarded from further processing (Lindahl et al., 2013). Since more sequences per sample will help detect rare taxa, the OTU table in this study was rarefied without replacement to 10,898 reads per sample, which was the 10th smallest number of sequences among all the samples (1 sample from the soil depth of 20–40 cm, 2 samples from 40 to 60 cm, 2 samples from 60 to 80 cm, and 3 samples from 80 to 100 cm) and the smallest number of sequences for the samples from top soil layers. After a standard dilution of fungal sequences set by using the rrarefy command in vegan package (Oksanen et al., 2019), we ultimately obtained 5,422 fungal OTUs. These 5,422 fungal OTUs were blasted against the FUNGuild database (Nguyen et al., 2016). For the fungal OTUs that may have multiple trophic modes, they were classified to fungal functional guilds according to their first trophic mode (Brinkmann et al., 2019). We got 722 saprotrophic fungal OTUs, 742 pathotrophic fungal OTUs, and 177 symbiotrophic fungal OTUs in total. The relative abundance of these three fungal guilds was listed at Supplementary Table 1 and Supplementary Figure 3. Further analyses were conducted for these three fungal guilds. The representative sequences of fungal OTUs were submitted to the European Nucleotide Archive (ENA), which is an archive with a history of 30 years and provides free and unlimited access to the annotated sequences of DNA and RNA, at European Molecular Biology Laboratory - European Bioinformatics Institute (EMBL-EBI) under accession number PRJEB46878.3
One of the goals of this study is to compare the rare with the abundant taxa within each fungal guild. Although there are multiple ways to estimate the diversity of rare and abundant taxa, such as relative abundance, frequency, and weights assignment (Supplementary Table 2), the metrics of alpha diversity and community composition were used to assess the changes of rare and abundant taxa of three fungal guilds along the salinity gradient. Because the relative abundance of symbiotrophic fungi was much lower than that of the other two guilds in the studied ecosystem – Taklamakan desert, using the relative abundance approach (e.g., 0.01% or 0.1% threshold; Liu et al., 2015; Lynch and Neufeld, 2015) would play down the population of abundant symbiotrophic fungi, while using the approach of alpha-diversity metrics can adjust the three fungal guilds to an equal level. The approach of alpha-diversity metrics has been used in many studies defined as a flexible tool to investigate the relative contributions of rare vs. common taxa to changes in community diversity as well as the responses of rare and abundant taxa to environment changes (Gossner et al., 2016; Xu et al., 2020). Besides, we examined the rare and abundant taxa of fungal functional guilds by using the 0.01% threshold method, and found that results analyzed by using these two methods were similar for the saprotrophic and pathogenic fungal guilds (Supplementary Tables 12–15).
As for the approach of alpha-diversity metrics, the estimate of species richness (q = 0) is weighted equally for the rare and abundant species and used to represent the contributions of rare taxa to community diversity. The estimate of the exponential of Shannon entropy (q = 1) is weighted in proportion to species frequency in a community and interpreted as the effective number of species. The estimate of inverse Simpson index (q = 2) gives more weights to abundant taxa than to rare taxa. It is interpreted as the effective number of very abundant species. The inverse Simpson (q = 2) and exponential of Shannon entropy (q = 1) indices are used to represent the contributions of abundant taxa to community diversity. We calculated these alpha-diversity indices by using vegan package 2.5–4 (Oksanen et al., 2019).
where qH is the alpha diversity; S is the number of species in the community; Pi represents the relative abundance of the ith species in the community; q is the order of Hill number. The q values determine the sensitivity of alpha-diversity indices to rare and abundant species (Hill, 1973), such as q = 0 for species richness, q = 1 for the exponential of Shannon entropy, and q = 2 for the inverse Simpson index (Table 1). We also calculated three indices of fungal composition dissimilarity for each fungal guild: Sorensen index, Horn index, and Morisita-Horn index (Jing et al., 2021). The three indices are a function of q values, which are disproportionately sensitive to the rare and abundant species (Chao et al., 2014). Specifically, the estimate of Sorensen index (q = 0) uses binary data (i.e., the rare and abundant species have equal weights) and characterizes the community composition response of rare taxa to soil salinization. The estimate of Horn index (q = 1) uses quantitative data and is weighted in proportion to species frequency. The estimate of Morisita-Horn index (q = 2) favors abundant taxa more than rare taxa. The Morisita-Horn (q = 2) and Horn (q = 1) indices are used to represent the community composition response of abundant taxa to soil salinization (Table 1). We used linear mixed-effects models (LMMs) to examine the effects of soil salinity on the relative abundance and alpha diversity of three fungal guilds. We set soil pH, soil total nitrogen, soil total phosphorus, and available phosphorus as fixed factors to control for the impacts of soil salinity on relative abundance and alpha diversity of fungal guilds. We set soil depth and sampling sites as random effects in the models. We did not use soil organic carbon in the models, because soil organic carbon was highly correlated with soil salinity (r = 0.70, p < 0.01; Supplementary Figure 4). All the factors in the models were subject to forward selection until VIF < 3. We considered soil depth as a random factor because the relative abundance and alpha diversity of three fungal guilds did not significantly differ across soil depths (Supplementary Figure 3) and the interaction between salinity and soil depth showed no significant influence on the relative abundance (Supplementary Table 3) and alpha diversity (Supplementary Tables 4–6) of three fungal guilds. We used both t tests and chi-square tests to examine the significance of variables on the relative abundance of three fungal guilds (Supplementary Table 7). Bivariate regressions and coefficients were used to determine and visualize the effects of soil salinity and soil properties on the relative abundance of fungi (Figure 2). We used both t tests and chi-square tests to examine the significance of soil salinity and properties on fungal alpha diversity (Supplementary Tables 8–10). Bivariate regressions and coefficients were used to determine and visualize the effects of soil salinity (Figure 3) and other soil properties (Figure 4) on fungal alpha diversity.
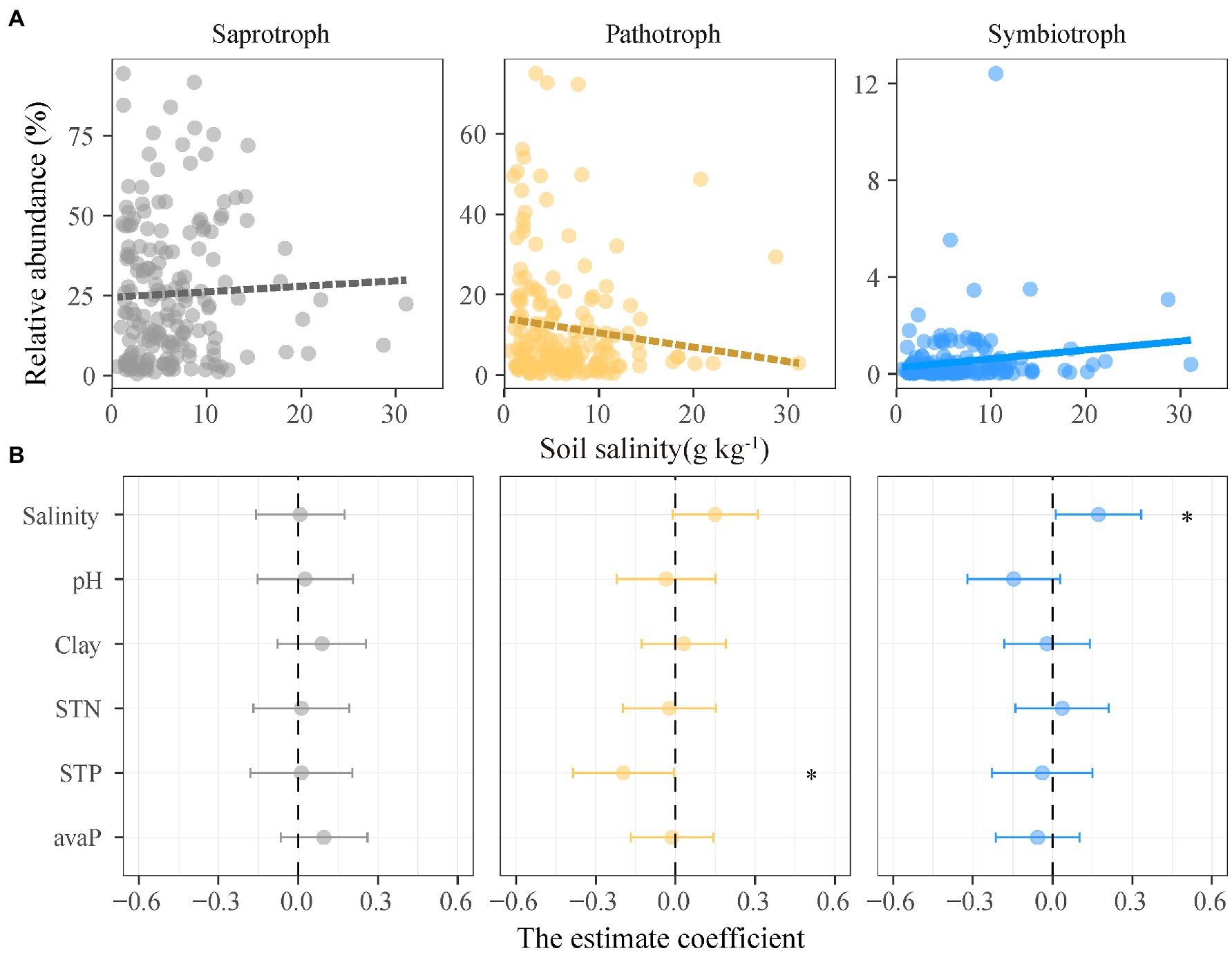
Figure 2. Relationships between the relative abundance of functional guilds of soil fungi and soil salinity and properties. (A) Lines are derived from the linear mixed effects models, and solid and dashed lines denote significant and insignificant regression coefficients, respectively. (B) Points denote regression coefficients and error bars denote 95% confidence intervals. *p < 0.05. Abbreviations: Salinity, soil salinity; STN, soil total nitrogen; Clay, soil clay content; STP, soil total phosphorus; avaP, soil available phosphorus.
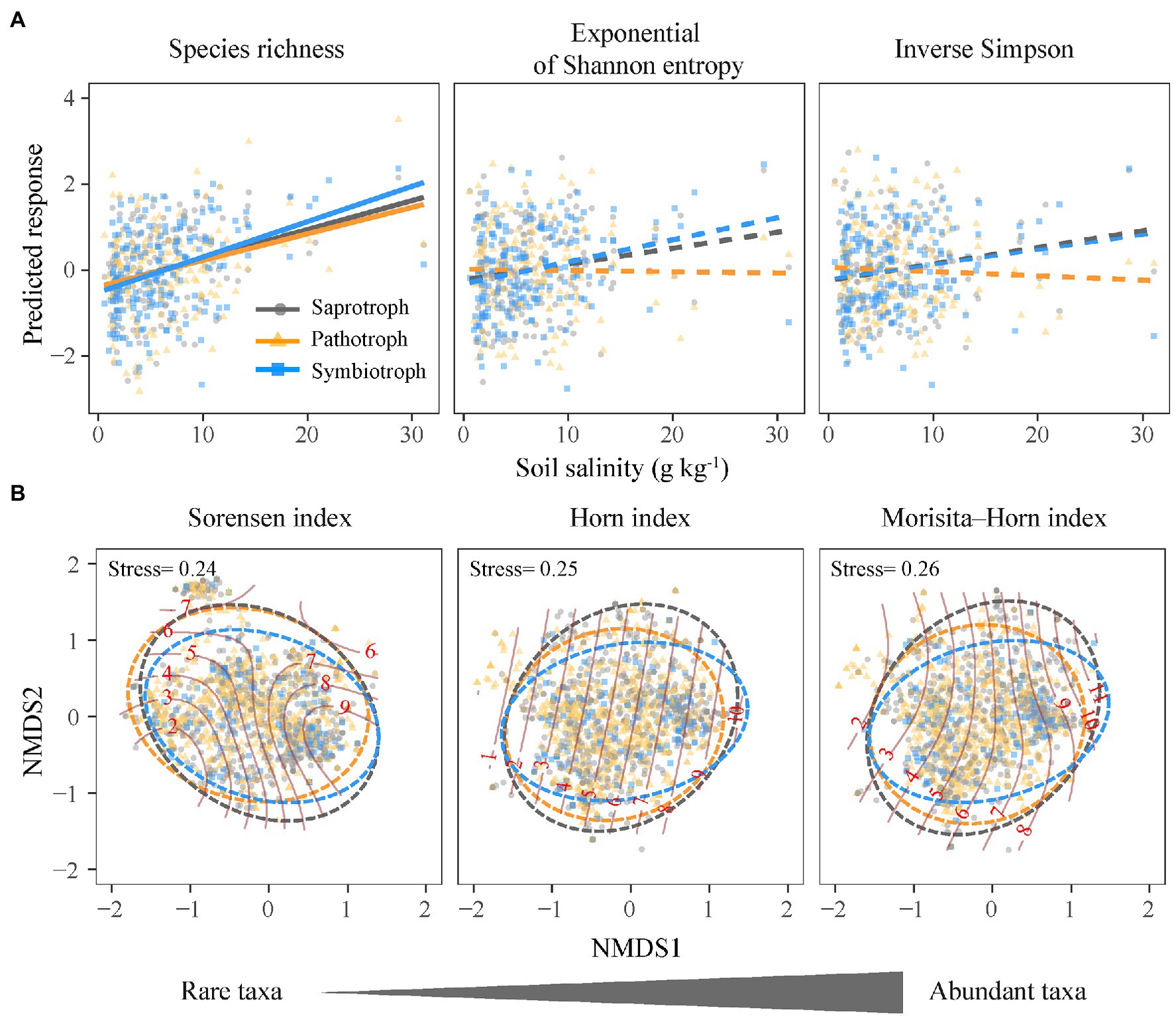
Figure 3. Relationships between the community structure of functional guilds of soil fungi and soil salinity. (A) Alpha diversity; lines are derived from the linear mixed effects models and solid and dashed lines denote significant and insignificant regression coefficients, respectively. (B) Community composition; solid lines denote salinity gradients and dashed lines denote 0.95 confidence ellipses.
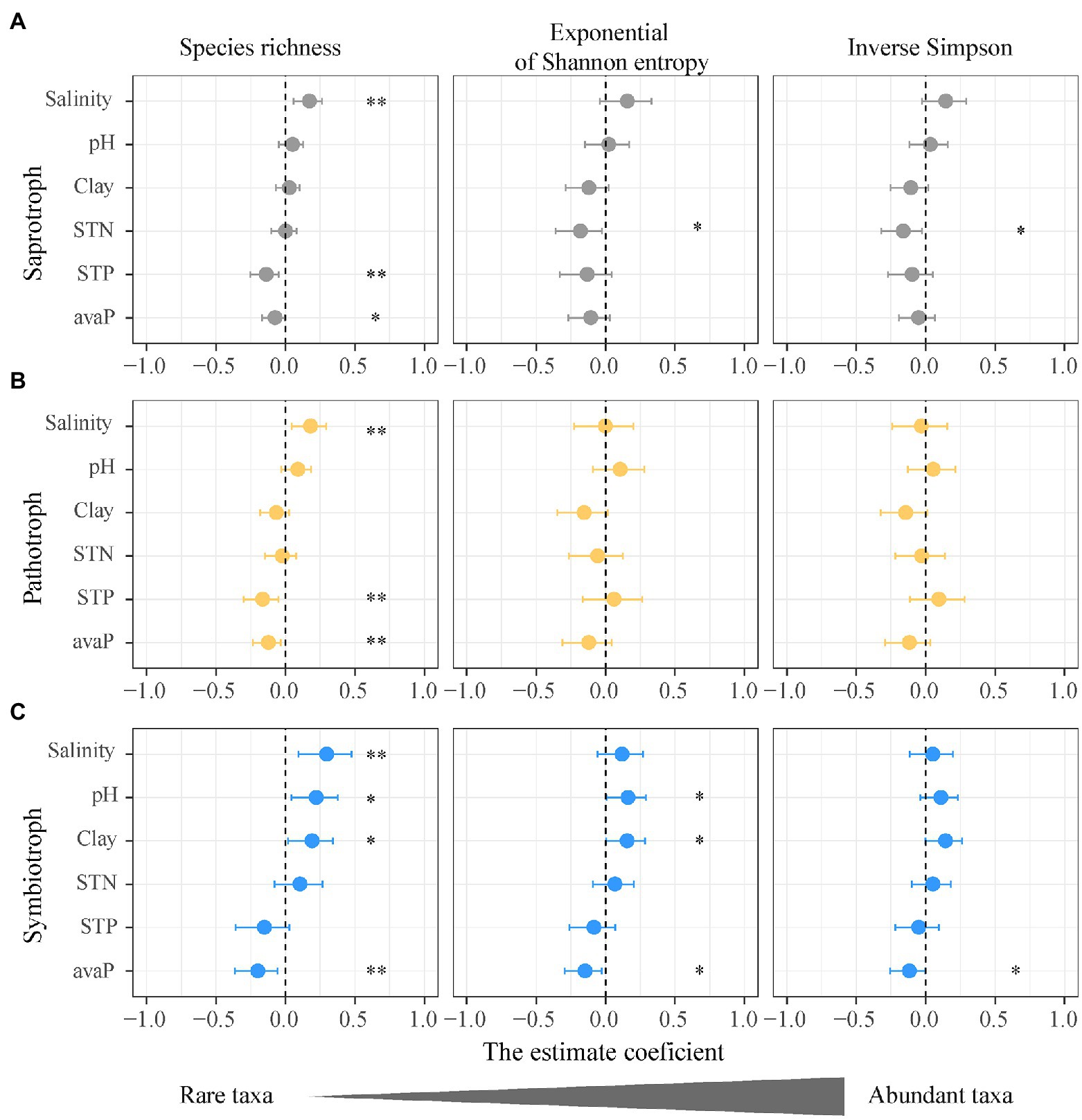
Figure 4. Impacts of soil salinity and properties on the alpha diversity of functional guilds of soil fungi. (A) Saprotrophic fungi; (B) pathotrophic fungi; (C) symbiotrophic fungi. Points denote regression coefficients and error bars denote 95% confidence intervals. *p < 0.05; **p < 0.01. Abbreviations: Salinity, soil salinity; STN, soil total nitrogen; Clay, soil clay content; STP, soil total phosphorus; avaP, soil available phosphorus.
In addition, the Mantel tests (distance approach) and ordination methods (raw-data approach) have been extensively used in studying community composition variation (beta diversity; Tuomisto and Ruokolainen, 2006). In this study, three indices of beta diversity were used to investigate the rare and abundant fungal taxa variations along the salinity gradient. Therefore, the (partial) Mantel tests were used to examine the relationships between soil fungal composition and geographic distance, soil depth, and environmental factors (Supplementary Table 11). In the Mantel tests, we used the Euclidean distances of zero-mean normalized geographic distances, soil depth, and environmental factors as the fixed factors. We used nonmetric multidimensional scaling (NMDS) that was calculated by metaMDSiter and wascores functions in vegan package to visualize the effect of salinity on niche differentiation among three fungal guilds (Figure 3). We used the bivariate partial Mantel coefficients to visualize the effects of changes in geographic distance and soil properties on community dissimilarity (Figure 5).
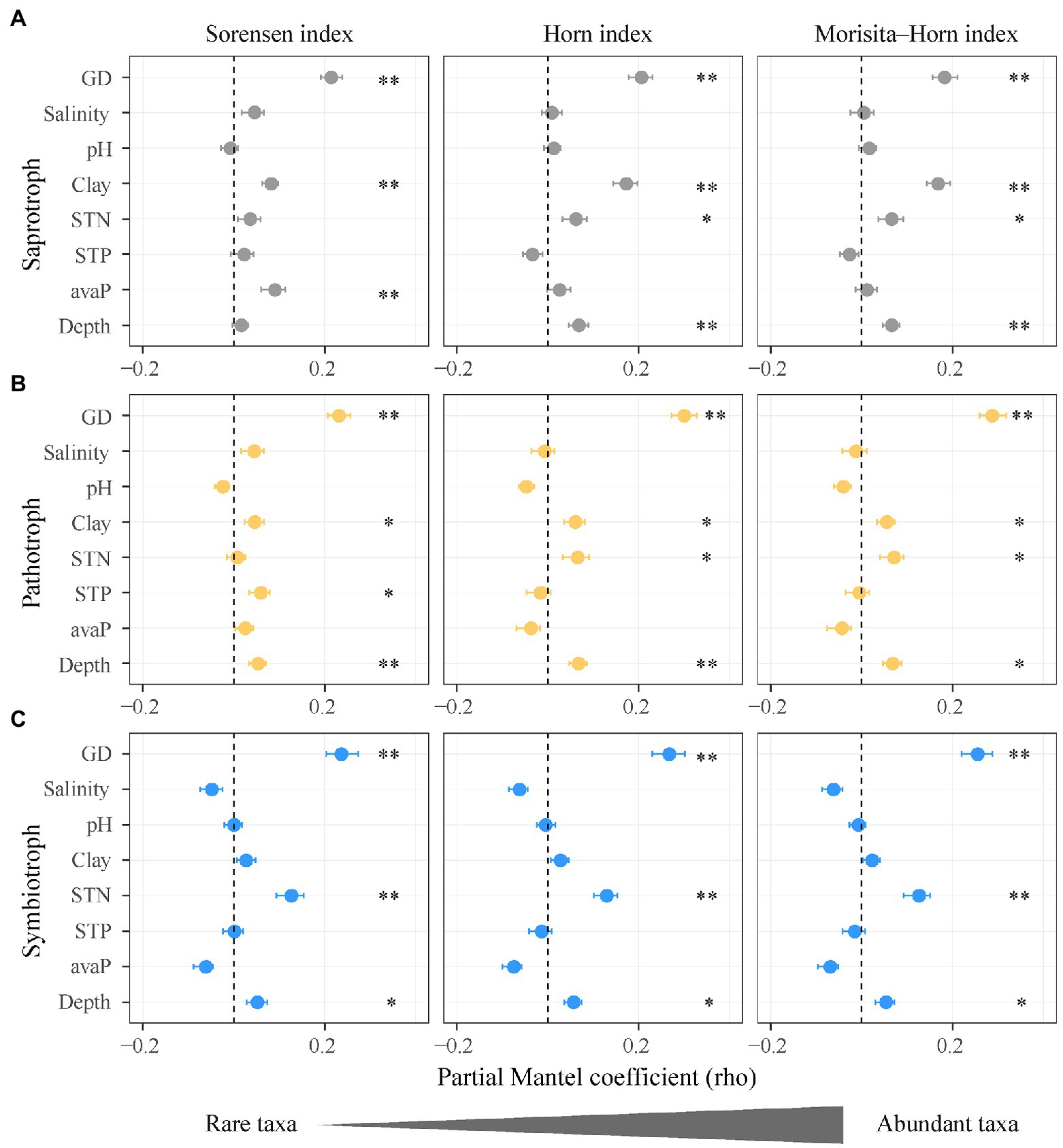
Figure 5. Impacts of soil salinity, geographic distance, and soil properties on the community composition of functional guilds of soil fungi. (A) Saprotrophic fungi; (B) pathotrophic fungi; (C) symbiotrophic fungi. Points denote partial Mantel coefficients and error bars denote 95% confidence intervals. *p < 0.05; **p < 0.01. Abbreviations: GD, geographic distance; Salinity, soil salinity; STN, soil total nitrogen; Clay, soil clay content; STP, soil total phosphorus; avaP, soil available phosphorus; Depth, soil depth.
Saprotrophic, pathotrophic, and symbiotrophic fungal guilds accounted for 25.1, 12.0, and 0.5% of the fungal abundance, respectively (Supplementary Table 1). The unassigned fungal taxa that blasted against FUNGuild database accounted for 62.4% of the fungal abundance. The relative abundance of three fungal guilds showed different patterns along the soil salinity gradient (Figure 2). The relative abundance of symbiotrophic fungi significantly increased with increasing soil salinity, but those of saprotrophic and pathotrophic fungi were not correlated with soil salinity (Figure 2; Supplementary Table 7).
The species richness (q = 0) of saprotrophic, pathotrophic, and symbiotrophic fungi all increased with increasing soil salinity (Figure 3A), and the slope of the standardized regressions between symbiotrophic richness and soil salinity (m = 0.29) was slightly higher than those for saprotrophic (m = 0.16) and pathotrophic richness (m = 0.17; Figure 3A). The species richness (q = 0) of three fungal guilds were influenced by different soil properties, i.e., soil total phosphorus and available phosphorus for the saprotrophic and pathotrophic fungi (Figures 4A,B) and soil pH, clay content and available phosphorus for the symbiotrophic fungi (Figure 4C).
Along the salinity gradient, the community niches of three fungal guilds were not clearly separated (Figure 3B). The community composition of saprotrophic and pathotrophic fungi (e.g., Sorensen index) showed significant Mantel correlation with soil salinity but that of symbiotrophic fungi was not. However, the community composition of three fungal guilds showed no significant partial Mantel correlations with soil salinity (Figure 5; Supplementary Table 11), suggesting the indirect effects of soil salinity on the community composition of saprotrophic and pathotrophic fungi.
The species richness (q = 0) rather than the diversity of abundant taxa (q = 1, 2) for each fungal guild was more susceptible to changes in soil properties (Figure 4). The exponential of Shannon entropy (q = 1) and inverse Simpson diversity (q = 2) of three fungal guilds showed no significant correlations with soil salinity (Figure 3A). Compared with the species richness (q = 0), the inverse Simpson diversities (q = 2) of saprotrophic and pathotrophic fungi showed no correlations with soil total phosphorus and available phosphorus (Figures 4A,B; Supplementary Tables 8 and 9). The soil total phosphorus and available phosphorus were positively correlated soil salinity (Supplementary Figure 4). The inverse Simpson diversity (q = 2) of symbiotrophic fungi showed no significant correlations with soil pH and clay content (Figure 4C; Supplementary Table 10).
The Sorensen index (q = 0) of fungi dissimilarity assigns more weights to rare taxa than the Morisita-Horn index (q = 2). The Sorensen index (q = 0) of saprotrophic fungi dissimilarity showed higher Mantel and partial Mantel coefficients with soil salinity (ρ = 0.098 and ρ = 0.043) than those of the Morisita-Horn index (q = 2; ρ = 0.062 and ρ = 0.003). The Sorensen index (q = 0) of pathotrophic fungi dissimilarity not Morisita-Horn (q = 2) index showed significantly Mantel correlations with soil salinity. Differently, the three indices of symbiotrophic fungi dissimilarity (i.e., Sorensen, Horn, and Morisita-Horn) displayed no Mantel correlation with soil salinity (Figure 5; Supplementary Table 11).
Compared with the Morisita-Horn index (q = 2), the Sorensen index (q = 0) of saprotrophic fungi dissimilarity was associated with soil available phosphorus (ρ = 0.088, p < 0.001) and that of pathotrophic fungi dissimilarity was associated with total phosphorus (ρ = 0.057, p < 0.05). The Sorensen indices (q = 0) of saprotrophic and pathotrophic fungi dissimilarity were not associated with soil total nitrogen (ρ = 0.034, p > 0.05; ρ = 0.005, p > 0.05; Figures 5A,B; Supplementary Table 11). In contrast, the Sorensen, Horn, and Morisita-Horn indices (q = 0, 1, and 2) of symbiotrophic fungi dissimilarity showed similar partial Mantel coefficients with soil total nitrogen (Figure 5C).
The relative abundance of symbiotrophic fungi increased along the salinity gradient, but those of saprotrophic and pathotrophic fungi did not change significantly (Figure 2). Furthermore, the species richness of symbiotrophic fungi showed greater increases to soil salinization than the other two fungal guilds. These results partially supported our prediction that the diversity of symbiotrophic fungi increases with soil salinity and that of saprotrophic and pathotrophic fungi decrease along the salinity gradient. Symbiotrophic fungi can establish mutualistic associations with plants and alleviate the negative impacts of salinization on plants by promoting plant K+ uptake and antioxidant enzymes activity (Akyol et al., 2020). In this study, 59.9% symbiotrophic fungi were arbuscular mycorrhizal fungi. In high saline conditions, symbiotrophic fungi could assist plants in maintaining their productivity (Bencherif et al., 2015). Although the functions of soil fungi were not measured, the response of functional guilds to soil salinization could provide the implications for ecosystem functions. For instance, the diversity of symbiotrophic fungi greatly prompts plant biodiversity, nutrient uptake, and productivity in microcosm (van der Heijden et al., 1998). So, increasing relative abundance and species richness of symbiotrophic fungi would improve the plant uptake of water and mineral nutrients at high salt concentrations. The significance of symbiotrophic fungi to plant growth increases remarkably at higher salt concentrations (Ren et al., 2016). Similarly, the spores and biomass of symbiotrophic fungi increase with increasing salinity in natural sites (Bencherif et al., 2015). More symbiotrophic fungal taxa are associated with plants and plants are less selective in choosing symbiotrophic fungi under stress compared to the unstressed condition (Lin et al., 2021). Increased symbiotrophic fungi (i.e., relative abundance and species richness) were likely to meet the demand of plants on nutrients and offset the adverse impacts of salinity on plant productivity at high salt concentrations.
In contrast, saprotrophic fungi were the most abundant among three functional fungal guilds and their relative abundance is highly resistant to salinity changes (Figure 2), which has been observed in semi-arid mangroves (Vanegas et al., 2019). It is probably because saprotrophic fungi are actively involved in organic matter decomposition and depend highly on organic inputs. Elmajdoub and Marschner (2015) found that repeated organic matter amendments can alleviate the adverse impacts of salinization on the growth of saprotrophic fungi. In this study, the aboveground plant biomass is similar along the salinity gradient (Supplementary Figure 2), resulting in relatively steady organic matter inputs to soil. Similarly, the saprotrophic fungi to microbial biomass ratio slightly increases with salinity in a plant residue addition experiment (Wichern et al., 2006). The resistance of saprotrophic fungi in terms of relative abundance may be confirmed by the functions, such as litter decomposition and nutrient release in the shrub shelterbelts under soil salinization.
Like saprotrophic fungi, the relative abundance of pathotrophic fungi were not correlated with soil salinity, suggesting that pathogenic fungi either adapt or be resistant to salinity changes. Until now, few studies have investigated the response of pathotrophic fungi to soil salinization and little is known about the reasons. Five species of pathotrophic fungi for sugarbeet seedlings exhibited great tolerance to increasing soil salinity (El-Abyad et al., 1988), agreeing with the result of this study.
The species richness of three fungal guilds all increased with increasing soil salinity but in different magnitudes, and the saprotrophic and pathotrophic fungi showed smaller changes than the symbiotrophic fungi (Figure 3A). We speculate that the increasing species richness of saprotrophic and pathotrophic fungi to salinity changes observed here might be attributed to high plasticity of soil fungi under salinity stress (Rath et al., 2019). The plasticity of soil fungi suggests that fungal individuals, over the course of a lifetime, shift trait expression in dealing with changes in the environment (Coleine et al., 2022). High plasticity of soil fungi may imply great stress tolerance but low growth and death rates (Plemenitas et al., 2014). But we are aware that the plasticity of soil fungi should be measured for active fungal communities by using metagenomics or transcriptomics. Thus, advanced techniques should be used to disentangle the mechanisms that drive different responses of fungal functional guilds to salinity stress in future.
Soil organic carbon (SOC) was an important carbon resource for saprotrophic fungi (Bardgett, 2005) and was significantly positively correlated to soil salinity (Supplementary Figure 4). The increment of SOC implies higher soil porosity and water retention capacity (Pathy et al., 2020), providing shelters for soil fungi. Meanwhile, the impacts of geographic distance and soil depth on fungal community dissimilarity imply the stochastic process and dispersal limitation of soil fungi (Wang et al., 2021). The low dispersal rate of soil fungi could be caused by special fungal propagules and the harsh environmental barriers (Zhou and Ning, 2017). Thus, we reason that in studied sites heterogeneous resource at high-salinity soils due to high SOC content could facilitate the migration and occupation of saprotrophic and pathotrophic fungi. In addition, stressful habitats that limit fungal growth may decrease resource competition and increase fungal diversity by suppressing dominating species (Oono et al., 2020). More taxa of saprotrophic fungi would be coexisted with high soil salinity, because the physiological activity of saprotrophic fungi might be inhibited by increasing salinity (Rath et al., 2019). This demonstrates that salinity stress and SOC resource may jointly shape the species richness of saprotrophic fungi under soil salinization.
This study showed that the diversity of the rare fungal taxa (q = 0) not the abundant taxa (q = 1, 2) displayed significant correlations with soil salinity and was more susceptible to other soil properties (Figure 4). The different patterns between the diversities of rare (q = 0) and abundant fungal taxa (q = 1, 2) indicate that the rare taxa of functional guilds are more sensitive to increasing soil salinity than the abundant taxa. Previous studies found that abundant fungi display great resistance to drought stress and ecosystem disturbance (Sun et al., 2017; de Vries et al., 2018). The activity or growth of abundant fungal taxa might be constricted due to high soil salinity or low-quality carbon resource in the desert soils, which promotes the migration rate and diversity of rare fungal taxa (Rath et al., 2016). The growth of a dominant root fungus (the Inula viscosa) was strongly inhibited by soil salinity in vitro assays and the fungal diversity increased at sites with high soil salinity (Macia-Vicente et al., 2012). Oono et al. (2020) indicate that abundant fungal taxa were suppressed in stressful conditions, which promoted the population of rare fungal taxa and increased fungal diversity. The population size of rare fungi would be enlarged with increasing SOC along the salinity gradient, because high SOC content provides more niches for rare fungi. Moreover, the great sensitivity of rare taxa of soil functional fungal guilds to salinity changes could attribute to their high variability, as rare taxa could function as a large reservoir of genetic traits and have redundant functions (Boraks et al., 2020; Pascoal et al., 2021). Therefore, the diversity of rare fungal taxa in this study increased with salinity changes.
This study found that the diversity of rare fungal taxa is more sensitive to soil phosphorus compared to that of abundant fungal taxa. Rare fungal taxa that have phosphorus cycling functions in saline soils can be influenced by soil total phosphorus and available phosphorus. The diversity of rare symbiotrophic fungi (q = 0) was positively correlated with soil pH as well as soil available phosphorus (Figure 4C). Symbiotrophic fungi are associated with plant P and nitrogen nutrition (Ezawa et al., 2002; Lindahl and Tunlid, 2015). In P-poor soils, such as the desert soil with high pH, the mutualistic association between plant and symbiotrophic fungi may be stimulated, causing a high diversity of rare symbiotrophic fungi. Phosphorus uptake of symbiotrophic fungi was found to increase at higher soil pH (Hinsinger, 2001). Unlike the diversities of abundant taxa (q = 1 and 2), the diversity of rare saprotrophic and pathotrophic fungi (q = 0) was negatively correlated with soil total phosphorus and available phosphorus (Figures 4A,B), suggesting that rare taxa may play more important roles in phosphorus cycling than abundant species. The diversity of rare saprotrophic and pathotrophic fungi was positively correlated with soil alkaline phosphatase (Supplementary Table 16).
The community composition of rare saprotrophic and pathotrophic fungi showed higher positive correlations with salinity changes (distance of soil salinity) compared to those of abundant taxa. This result supported the prediction that rare taxa of the functional guilds of soil fungi are more sensitive to salinity changes than the abundant taxa. However, compared with the diversity, the community composition of rare fungal taxa was not significantly correlated with soil salinity partially, suggesting that soil salinity may have indirect effects on community composition of rare fungal taxa. Kim et al. (2019) reported that fungal community composition in the salinity treatment is mainly driven by soil salinity. The impacts of salinity on fungal community composition might be offset by organic matter, as soil salinity and organic carbon were significantly positively correlated (Supplementary Figure 4) and exhibited opposite effects on fungal community composition in a semi-arid mangrovein salt mash (Vanegas et al., 2019).
The community composition of rare saprotrophic and pathotrophic fungi could be indirectly affected by soil salinity through altering soil phosphorus and clay content. Rare taxa of saprotrophic and pathotrophic fungi (i.e., Sorensen index) not the abundant taxa (i.e., Morisita-Horn index) were significantly influenced by changes in soil phosphorus (distance of soil phosphorus; Supplementary Table 11). Similar to this study, the community composition of sapraotrophic and pathotrophic fungi in a natural grassland is mainly driven by soil phosphorus (Delavaux et al., 2021). In current study, soil salinity showed significantly positively correlation with soil phosphorus and clay content and was not correlated with soil total nitrogen, which were controlling factors for the community composition of rare taxa (i.e., Sorensen index). These correlations confirmed that the responses of community composition of saprotrophic and pathotrophic fungi to soil salinization might be induced mainly by rare taxa (i.e., Sorensen index), as abundant taxa have wider variability in abundance under changing resource availability, e.g., soil total nitrogen (Newton and Shade, 2016).
The community composition of rare and abundant symbiotrophic fungi (i.e., Sorensen, Horn, and Morisita-Horn indices) showed no correlations with salinity changes, indicating that salinization had similar stresses on the rare and abundant symbiotrophic fungi. This result partially supported the prediction that the community composition of symbiotrophic fungi is expected to occupy high-salinity niches compared to that of saprotrophic and pathotrophic fungi. The community composition of symbiotrophic fungi was mainly regulated by soil total nitrogen. Symbiotrophic fungi provide mineral nitrogen and phosphorus nutrients to plants in exchange for carbohydrates (Kiers et al., 2011) and enhance nitrogen accumulation in plants with inreasing soil salinity (Zhu et al., 2018), as the accumulation of leaf non-protein nitrogen increase plant resistance to salinity (Mansour, 2000). Given that the leaf N content and N:P ratio of H. ammodendron at high-salinity sites were significantly higher than those at low-salinity sites (Feng et al., in preparation; Supplementary Figure 5), symbiotrophic fungi that provide nitrogen would be promoted with increasing salinity due to plant selection, especially at the high-salinity sites. Furthermore, different symbiotrophic fungi can acquire N from distinctly sources, e.g., ammonium N and nitrate N (Cox et al., 2010) that are influenced by soil total N (Supplementary Figure 6). Therefore, the community composition of symbiotrophic fungi was driven by soil total N under salt stress and resistant to changes in soil salinity.
Soil fungal communities are integral to soil carbon cycling, plant nutrition, and pathogenicity. This study indicates that the rare taxa of soil functional fungal guilds (i.e., saprotrophic, pathotrophic, and symbiotrophic fungi) rather than the abundant taxa would be more sensitive to changes in soil salinity, thus the biodiversity responses of fungal guilds to soil salinization should be reflected by rare taxa. Changes in soil salinity increased the diversity of rare saprotrophic and pathotrophic fungal taxa and indirectly affected their community composition. Compared to saprotrophic and pathotrophic fungi, symbiotrophic fungi increased more in alpha diversity but did not significantly change the community composition with increasing soil salinity. These results advance our understanding of the responses of rare taxa of functional fungal guilds to soil salinization and contribute to soil biodiversity conservation and soil health management regarding fugal functionality in the inland saline environments.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: European Nucleotide Archive (ENA) - PRJEB46878.
LL: methodology, investigation, formal analysis, writing-original draft, and writing-review and editing. XJ: methodology, investigation, formal analysis, review, and editing. ML-B and CS: review and editing. YW: investigation. WF: methodology, investigation, and writing-review and editing. All authors contributed to the article and approved the submitted version.
The financial support for this work is the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences, the National Natural Science Foundation of China (41730638 and U1803342), and K.C. Wong Education Foundation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.862245/full#supplementary-material
Akyol, T., Sato, S., and Turkan, I. (2020). Deploying root microbiome of halophytes to improve salinity tolerance of crops. Plant Biotechnol. Rep. 14, 143–150. doi: 10.1007/s11816-020-00594-w
Balbuena, J. A., Monlleo-Borrull, C., Llopis-Belenguer, C., Blasco-Costa, I., Sarabeev, V. L., and Morand, S. (2021). Fuzzy quantification of common and rare species in ecological communities (FuzzyQ). Methods Ecol. Evol. 12, 1070–1079. doi: 10.1111/2041-210X.13588
Bardgett, R. D. (2005). The Biology of Soil: A Community and Ecosystem Approach. Oxford, UK: Oxford University Press.
Bencherif, K., Boutekrabt, A., Fontaine, J., Laruelle, F., Dalpe, Y., and Sahraoui, A. L. H. (2015). Impact of soil salinity on arbuscular mycorrhizal fungi biodiversity and microflora biomass associated with Tamarix articulata Vahll rhizosphere in arid and semi-arid Algerian areas. Sci. Total Environ. 533, 488–494. doi: 10.1016/j.scitotenv.2015.07.007
Bharti, N., Barnawal, D., Maji, D., and Kalra, A. (2015). Halotolerant PGPRs prevent major shifts in indigenous microbial community structure under salinity stress. Microb. Ecol. 70, 196–208. doi: 10.1007/s00248-014-0557-4
Boraks, A., Plunkett, G. M., Doro, T. M., Alo, F., Sam, C., Tuiwawa, M., et al. (2020). Scale-dependent influences of distance and vegetation on the composition of aboveground and belowground tropical fungal communities. Microb. Ecol. 81, 874–883. doi: 10.1007/s00248-020-01608-4
Brinkmann, N., Schneider, D., Sahner, J., Ballauff, J., Edy, N., Barus, H., et al. (2019). Intensive tropical land use massively shifts soil fungal communities. Sci. Rep. 9:3403. doi: 10.1038/s41598-019-39829-4
Chao, A. N., Chiu, C. H., and Jost, L. (2014). Unifying species diversity, phylogenetic diversity, functional diversity, and related similarity and differentiation measures through Hill numbers. Annu. Rev. Ecol. Evol. Syst. 45, 297–324. doi: 10.1146/annurev-ecolsys-120213-091540
Coleine, C., Stajich, J. E., and Selbmann, L. (2022). Fungi are key players in extreme ecosystems. Trends Ecol. Evol. doi: 10.1016/j.tree.2022.02.002
Cox, F., Barsoum, N., Lilleskov, E. A., and Bidartondo, M. I. (2010). Nitrogen availability is a primary determinant of conifer mycorrhizas across complex environmental gradients. Ecol. Lett. 13, 1103–1113. doi: 10.1111/j.1461-0248.2010.01494.x
Delavaux, C. S., Schemanski, S., House, G. L., Tipton, A. G., Sikes, B., and Bever, J. D. (2021). Root pathogen diversity and composition varies with climate in undisturbed grasslands, but less so in anthropogenically disturbed grasslands. Isme Journal 15, 304–317. doi: 10.1038/s41396-020-00783-z
de Vries, F. T., Griffiths, R. I., Bailey, M., Craig, H., Girlanda, M., Gweon, H. S., et al. (2018). Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 9:3033. doi: 10.1038/s41467-018-05516-7
El-Abyad, M. S., Hindorf, H., and Rizk, M. A. (1988). Impact of salinity stress on soil-borne fungi of sugar beet. I. Pathogenicity implications. Plant Soil 110, 27–32. doi: 10.1007/BF02143535
Elmajdoub, B., and Marschner, P. (2015). Responses of soil microbial activity and biomass to salinity after repeated additions of plant residues. Pedosphere 25, 177–185. doi: 10.1016/S1002-0160(15)60002-9
Ezawa, T., Smith, S. E., and Smith, F. A. (2002). P metabolism and transport in AM fungi. Plant Soil 244, 221–230. doi: 10.1023/A:1020258325010
Frac, M., Hannula, S. E., Belka, M., and Jedryczka, M. (2018). Fungal biodiversity and their role in soil health. Front. Microbiol. 9:707. doi: 10.3389/fmicb.2018.00707
Galand, P. E., Casamayor, E. O., Kirchman, D. L., and Lovejoy, C. (2009). Ecology of the rare microbial biosphere of the Arctic Ocean. Proc. Natl. Acad. Sci. U. S. A. 106, 22427–22432. doi: 10.1073/pnas.0908284106
Gossner, M. M., Lewinsohn, T. M., Kahl, T., Grassein, F., Boch, S., Prati, D., et al. (2016). Land-use intensification causes multitrophic homogenization of grassland communities. Nature 540, 266–269. doi: 10.1038/nature20575
Hill, M. O. (1973). Diversity and evenness: a unifying notation and its consequences. Ecology 54, 427–432. doi: 10.2307/1934352
Hinsinger, P. (2001). Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: A review. Plant Soil 237, 173–195. doi: 10.1023/A:1013351617532
Ivushkin, K., Bartholomeus, H., Bregt, A. K., Pulatov, A., Kempen, B., and de Sousa, L. (2019). Global mapping of soil salinity change. Remote Sens. Environ. 231:111260. doi: 10.1016/j.rse.2019.111260
Ji, M. K., Kong, W. D., Stegen, J., Yue, L. Y., Wang, F., Dong, X. B., et al. (2020). Distinct assembly mechanisms underlie similar biogeographical patterns of rare and abundant bacteria in Tibetan plateau grassland soils. Environ. Microbiol. 22, 2261–2272. doi: 10.1111/1462-2920.14993
Jing, X., Prager, C. M., Borer, E. T., Gotelli, N. J., Gruner, D. S., He, J. S., et al. (2021). Spatial turnover of multiple ecosystem functions is more associated with plant than soil microbial β-diversity. Ecosphere 12:e03644. doi: 10.1002/ecs2
Jost, L. (2007). Partitioning diversity into independent alpha and beta components. Ecology 88, 2427–2439. doi: 10.1890/06-1736.1
Kiers, E. T., Duhamel, M., Beesetty, Y., Mensah, J. A., Franken, O., Verbruggen, E., et al. (2011). Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333, 880–882. doi: 10.1126/science.1208473
Kim, K., Samaddar, S., Chatterjee, P., Krishnamoorthy, R., Jeon, S., and Sa, T. (2019). Structural and functional responses of microbial community with respect to salinity levels in a coastal reclamation land. Appl. Soil Ecol. 137, 96–105. doi: 10.1016/j.apsoil.2019.02.011
Lei, J. Q., Li, S. Y., Fan, D. D., Zhou, H. W., Gu, F., Qiu, Y. Z., et al. (2008). Classification and regionalization of the forming environment of windblown sand disasters along the Tarim Desert Highway. Chin. Sci. Bull. 53, 1–7. doi: 10.1007/s11434-008-6023-2
Li, B. W. (2010). Mechanism of Water-Salt Regulation for the Shelterbelts With Saline Water Irrigation Along the Tarim Desert Highway. China: Beijing Forestry University.
Li, B. W., Xu, X. W., Lei, J. Q., Qiu, Y. Z., Xu, B., Zhou, H. W., et al. (2008). Site type classification for the shelter-forest ecological project along the Tarim Desert highway. Chin. Sci. Bull. 53, 31–40. doi: 10.1007/s11434-008-6003-6
Lin, L. T., Chen, Y., Xu, G. R., Zhang, Y. X., Zhang, S., and Ma, K. M. (2021). Impacts of urbanization undermine nestedness of the plant-arbuscular mycorrhizal fungal network. Front. Microbiol. 12:626671. doi: 10.3389/fmicb.2021.626671
Lindahl, B. D., Nilsson, R. H., Tedersoo, L., Abarenkov, K., Carlsen, T., Kjøller, R., et al. (2013). Fungal community analysis by high-throughput sequencing of amplified markers–a user's guide. New Phytol. 199, 288–299. doi: 10.1111/nph.12243
Lindahl, B. D., and Tunlid, A. (2015). Ectomycorrhizal fungi - potential organic matter decomposers, yet not saprotrophs. New Phytol. 205, 1443–1447. doi: 10.1111/nph.13201
Liu, L., Yang, J., Yu, Z., and Wilkinson, D. M. (2015). The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J. 9, 2068–2077. doi: 10.1038/ismej.2015.29
Lynch, M. D. J., and Neufeld, J. D. (2015). Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 13, 217–229. doi: 10.1038/nrmicro3400
Macia-Vicente, J. G., Ferraro, V., Burruano, S., and Lopez-Llorca, L. V. (2012). Fungal assemblages associated with roots of halophytic and non-halophytic plant species vary differentially along a salinity gradient. Microb. Ecol. 64, 668–679. doi: 10.1007/s00248-012-0066-2
Mansour, M. M. F. (2000). Nitrogen containing compounds and adaptation of plants to salinity stress. Biol. Plant. 43, 491–500. doi: 10.1023/A:1002873531707
Mohamed, D. J., and Martiny, J. B. H. (2011). Patterns of fungal diversity and composition along a salinity gradient. Isme Journal 5, 379–388. doi: 10.1038/ismej.2010.137
Montoya-Ciriaco, N., Gomez-Acata, S., Munoz-Arenas, L. C., Dendooven, L., Estrada-Torres, A., de la Vega-Perez, A. H. D., et al. (2020). Dietary effects on gut microbiota of the mesquite lizard Sceloporus grammicus (Wiegmann, 1828) across different altitudes. Microbiome 8:6. doi: 10.1186/s40168-020-0783-6
Moore, J. A. M., Anthony, M. A., Pec, G. J., Trocha, L. K., Trzebny, A., Geyer, K. M., et al. (2021). Fungal community structure and function shifts with atmospheric nitrogen deposition. Glob. Chang. Biol. 27, 1349–1364. doi: 10.1111/gcb.15444
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nelson, D. W., and Sommers, L. E. (1982). “Total carbon, organic carbon, and organic matter,” in Methods of Soil Analysis. Part 2. Chemical and Microbiological Properties. ed. A. L. Page (American Society of Agronomy, Soil Science Society of America), 539–579.
Newton, R. J., and Shade, A. (2016). Lifestyles of rarity: understanding heterotrophic strategies to inform the ecology of the microbial rare biosphere. Aquat. Microb. Ecol. 78, 51–63. doi: 10.3354/ame01801
Nguyen, N. H., Song, Z. W., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al., (2019). Vegan: community ecology package. R package version 2.5-4. Available at: https://CRAN.R-project.org/package=vegan (Accessed February 4, 2019).
Olsen, S. R. (1954). Estimation of Available Phosphorus in Soils by Extraction With Sodium Bicarbonate. Washington, DC: United States Department of Agriculture.
Oono, R., Black, D., Slessarev, E., Sickler, B., Strom, A., and Apigo, A. (2020). Species diversity of fungal endophytes across a stress gradient for plants. New Phytol. 228, 210–225. doi: 10.1111/nph.16709
Oono, R., Rasmussen, A., and Lefevre, E. (2017). Distance decay relationships in foliar fungal endophytes are driven by rare taxa. Environ. Microbiol. 19, 2794–2805. doi: 10.1111/1462-2920.13799
Pathy, A., Ray, J., and Paramasivan, B. (2020). Biochar amendments and its impact on soil biota for sustainable agriculture. Biochar 2, 287–305. doi: 10.1007/s42773-020-00063-1
Pascoal, F., Costa, R., and Magalhães, C. (2021). The microbial rare biosphere: Current concepts, methods and ecological principles. FEMS Microbiology Ecology 97. doi: 10.1093/femsec/fiaa227
Plemenitas, A., Lenassi, M., Konte, T., Kejzar, A., Zajc, J., Gostincar, C., et al. (2014). Adaptation to high salt concentrations in halotolerant/halophilic fungi: a molecular perspective. Front. Microbiol. 5:199. doi: 10.3389/fmicb.2014.00199
Rath, K. M., Maheshwari, A., Bengtson, P., and Rousk, J. (2016). Comparative toxicities of salts on microbial processes in soil. Appl. Environ. Microbiol. 82, 2012–2020. doi: 10.1128/AEM.04052-15
Rath, K. M., Murphy, D. N., and Rousk, J. (2019). The microbial community size, structure, and process rates along natural gradients of soil salinity. Soil Biol. Biochem. 138:107607. doi: 10.1016/j.soilbio.2019.107607
Rath, K. M., and Rousk, J. (2015). Salt effects on the soil microbial decomposer community and their role in organic carbon cycling: a review. Soil Biol. Biochem. 81, 108–123. doi: 10.1016/j.soilbio.2014.11.001
Ren, C. G., Bai, Y. J., Kong, C. C., Bian, B., and Xie, Z. H. (2016). Synergistic interactions between salt-tolerant rhizobia and arbuscular mycorrhizal fungi on salinity tolerance of sesbania cannabina plants. J. Plant Growth Regul. 35, 1098–1107. doi: 10.1007/s00344-016-9607-0
Ruppel, S., Franken, P., and Witzel, K. (2013). Properties of the halophyte microbiome and their implications for plant salt tolerance. Funct. Plant Biol. 40, 940–951. doi: 10.1071/FP12355
Smith, D. P., and Peay, K. G. (2014). Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS One 9:e90234. doi: 10.1371/journal.pone.0090234
Sun, S., Li, S., Avera, B. N., Strahm, B. D., and Badgley, B. D. (2017). Soil bacterial and fungal communities show distinct recovery patterns during forest ecosystem restoration. Appl. Environ. Microbiol. 83:e00966-17. doi: 10.1128/aem.00966-17
Thiem, D., Golebiewski, M., Hulisz, P., Piernik, A., and Hrynkiewicz, K. (2018). How does salinity shape bacterial and fungal microbiomes of alnus glutinosa roots? Front. Microbiol. 9:651. doi: 10.3389/fmicb.2018.00651
Tuomisto, H., and Ruokolainen, K. (2006). Analyzing or explaining beta diversity? Understanding the targets of different methods of analysis. Ecology 87, 2697–2708. doi: 10.1890/0012-9658(2006)87[2697:AOEBDU]2.0.CO;2
van der Heijden, M. G. A., Klironomos, J. N., Ursic, M., Ursic, M., Moutoglis, P., Streitwolf-Engel, R., et al. (1998). Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396, 69–72. doi: 10.1038/23932
Vanegas, J., Munoz-Garcia, A., Perez-Parra, K. A., Figueroa-Galvis, I., Mestanza, O., and Polania, J. (2019). Effect of salinity on fungal diversity in the rhizosphere of the halophyte Avicennia germinans from a semi-arid mangrove. Fungal Ecol. 42:100855. doi: 10.1016/j.funeco.2019.07.009
Wan, W. J., Gadd, G. M., Yang, Y. Y., Yuan, W. K., Gu, J. D., Ye, L. P., et al. (2021). Environmental adaptation is stronger for abundant rather than rare microorganisms in wetland soils from the Qinghai-Tibet plateau. Mol. Ecol. 30, 2390–2403. doi: 10.1111/mec.15882
Wang, Y. D., Li, S. Y., Xu, X. W., Lei, J. Q., Jin, X. J., Yan, J., et al. (2012). Soil salinization of the windbreak forest belts irrigated with saline water alongside the Tarim Desert highway. Acta Pedol. Sin. 49, 886–891. doi: 10.11766/trxb201103030073
Wang, W. J., Li, J. W., Ye, Z. Q., Wang, J. M., Qu, L. Y., and Zhang, T. H. (2021). Spatial factors and plant attributes influence soil fungal community distribution patterns in the lower reaches of the Heihe River basin, Northwest China. Environ. Microbiol. 23, 2499–2508. doi: 10.1111/1462-2920.15466
White, T., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications. eds. M. Innis, D. Gelfand, and J. T. W. Sninsky (San Diego: Academic Press).
Wichern, J., Wichern, F., and Joergensen, R. G. (2006). Impact of salinity on soil microbial communities and the decomposition of maize in acidic soils. Geoderma 137, 100–108. doi: 10.1016/j.geoderma.2006.08.001
Xu, G., Zhang, Y., Zhang, S., and Ma, K. (2020). Biodiversity associations of soil fauna and plants depend on plant life form and are accounted for by rare taxa along an elevational gradient. Soil Biol. Biochem. 140:107640. doi: 10.1016/j.soilbio.2019.107640
Xu, Y., Zheng, C., Liang, L., Yi, Z. L., and Xue, S. (2021). Quantitative assessment of the potential for soil improvement by planting Miscanthus on saline-alkaline soil and the underlying microbial mechanism. Glob. Change Biol. Bioenergy. 13, 1191–1205. doi: 10.1111/gcbb.12845
Yang, W., Jing, X. Y., Guan, Y. P., Zhai, C., Wang, T., Shi, D. Y., et al. (2019). Response of fungal communities and co-occurrence network patterns to compost amendment in black soil of Northeast China. Front. Microbiol. 10:1562. doi: 10.3389/fmicb.2019.01562
Yang, T., Sun, H. B., Shen, C. C., and Chu, H. Y. (2016). Fungal assemblages in different habitats in an Erman’s birch forest. Front. Microbiol. 7:1368. doi: 10.3389/fmicb.2016.01368
Yang, C., Wang, X. Z., Miao, F. H., Li, Z. Y., Tang, W., and Sun, J. (2020). Assessing the effect of soil salinization on soil microbial respiration and diversities under incubation conditions. Appl. Soil Ecol. 155:103671. doi: 10.1016/j.apsoil.2020.103671
Yuan, G., and Lavkulich, L. M. (1995). Colorimetric determination of phosphorus in citrate-bicarbonate-dithionite extracts of soils. Commun. Soil Sci. Plant Anal. 26, 1979–1988. doi: 10.1080/00103629509369422
Zhang, J. G., Xu, X. W., Li, S. Y., Zhao, Y., Zhang, A. F., Zhang, T. B., et al. (2016). Is the Taklamakan Desert highway shelterbelt sustainable to long-term drip irrigation with high saline groundwater? PLoS One 11:e0164106. doi: 10.1371/journal.pone.0164106
Zhou, J. Z., and Ning, D. L. (2017). Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81:e00002-17. doi: 10.1128/MMBR.00002-17
Keywords: biodiversity, desert, functional guilds, rare biosphere, soil salinization, saprotrophic fungi, symbiotrophic fungi
Citation: Lin L, Jing X, Lucas-Borja ME, Shen C, Wang Y and Feng W (2022) Rare Taxa Drive the Response of Soil Fungal Guilds to Soil Salinization in the Taklamakan Desert. Front. Microbiol. 13:862245. doi: 10.3389/fmicb.2022.862245
Received: 25 January 2022; Accepted: 21 April 2022;
Published: 23 May 2022.
Edited by:
Dilfuza Egamberdieva, Leibniz Center for Agricultural Landscape Research (ZALF), GermanyReviewed by:
Jessica A. Moore, Oak Ridge National Laboratory (DOE), United StatesCopyright © 2022 Lin, Jing, Lucas-Borja, Shen, Wang and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenting Feng, ZmVuZ3dlbnRpbmdAY2Fhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.