- 1Institute for Marine Engineering, Shenzhen International Graduate School, Tsinghua University, Shenzhen, China
- 2Institute of Deep Sea Science and Engineering, Chinese Academy of Sciences, Sanya, China
- 3College of Marine Sciences, University of Chinese Academy of Sciences, Beijing, China
- 4State Key Laboratory of Marine Geology, Tongji University, Shanghai, China
The deep-sea hydrothermal vents (DSHVs) in the Southwest Indian Ridge (SWIR) are formed by specific geological settings. However, the community structure and ecological function of the microbial inhabitants on the sulfide chimneys of active hydrothermal vents remain largely unknown. In this study, our analyses of 16S rRNA gene amplicons and 16S rRNA metagenomic reads showed the dominance of sulfur-oxidizing Ectothiorhodospiraceae, Thiomicrorhabdus, Sulfurimonas, and Sulfurovum on the wall of two active hydrothermal chimneys. Compared with the inactive hydrothermal sediments of SWIR, the active hydrothermal chimneys lacked sulfur-reducing bacteria. The metabolic potentials of the retrieved 82 metagenome-assembled genomes (MAGs) suggest that sulfur oxidation might be conducted by Thiohalomonadales (classified as Ectothiorhodospiraceae based on 16S rRNA gene amplicons), Sulfurovaceae, Hyphomicrobiaceae, Thiotrichaceae, Thiomicrospiraceae, and Rhodobacteraceae. For CO2 fixation, the Calvin-Benson-Bassham and reductive TCA pathways were employed by these bacteria. In Thiohalomonadales MAGs, we revealed putative phytochrome, carotenoid precursor, and squalene synthesis pathways, indicating a possible capacity of Thiohalomonadales in adaptation to dynamics redox conditions and the utilization of red light from the hot hydrothermal chimneys for photolithotrophic growth. This study, therefore, reveals unique microbiomes and their genomic features in the active hydrothermal chimneys of SWIR, which casts light on ecosystem establishment and development in hydrothermal fields and the deep biosphere.
Introduction
Deep-sea hydrothermal vents (DSHVs) are located in tectonically active areas where plate boundaries move at different speeds along mid-ocean ridges. DSHV is an important conduit for the exchange of energy and materials between the Earth’s interior and the ocean. Since the first report in 1977, DSHVs as a deep-sea extreme environment have attracted great concerns about the microbial extremophilic inhabitants with respect to early life form, chemoautotrophy, and adaptation (Corliss et al., 1979; Francheteau et al., 1979; Jannasch and Mottl, 1985; Kelley et al., 2005; Dick, 2019). The mixture of cold, oxic deep-sea water, and highly reducing fluids with high concentrations of hydrogen, sulfide, and methane was ideal for chemoautotrophs and C1 oxidizers (Dick et al., 2013). The complex dynamic habitats have steep thermal and chemical gradients, and different microbial populations are simultaneously involved in many important biogeochemical processes, such as the nitrogen, sulfur, and carbon cycles that also occur in symbionts of megafauna around DSHV (Jannasch and Mottl, 1985; Connelly et al., 2012). Strong dynamics of geochemistry and temperature provide a wide range of habitats for deep-sea microorganisms, and therefore, niche specificity of microbes has been demonstrated in different environmental settings by previous studies (Dick, 2019 and references therein).
Sulfide samples from inactive and active hydrothermal vents are distinct in microbial community structure and ecological function in the Indian Ocean (Zhang et al., 2016; Han et al., 2018; Adam et al., 2020; Hou et al., 2020). Sulfur oxidizing bacteria (SOB) dominate various hydrothermal sediments and flumes and are classified to be aerobes (e.g., SUP05 and Beggiatoa from Gammaproteobacteria), microaerobes (e.g., Sulfurimonas and Sulfurovum from Epsilonbacteraeota), and anaerobes (e.g., Caminibacter and Nautila from Epsilonbacteraeota) (López-García et al., 2003; Dick, 2019; Meier et al., 2019; Hou et al., 2020) with preference to different electron donors. For the microaerobic SOB, the cbb3-type cytochrome c oxidase was involved in adaptation to low oxygen concentrations for respiration (Hou et al., 2020; Dong et al., 2021); however, the mechanism for reducing the damage by high oxygen flux is largely unknown. Considering the highly variable microenvironments adjacent to an active hydrothermal vent, there are perhaps much more SOB species that have evolved to adapt to the varying temperature and redox conditions. A photoautotrophic bacterium has been isolated from a hydrothermal chimney (Beatty et al., 2005). Evidence shows that this bacterial isolate from the Prosthecochloris genus can absorb weak ultra-red light as energy to fix CO2 and oxidize H2S. The abundance and distribution of such photoautotrophic SOB in the dark ocean are still unclear up to date.
Since the first report in the Southwest Indian Ridge (SWIR) more than a decade ago (German et al., 1998), multiple hydrothermal fields have been discovered in the Indian Ocean (Van Dover et al., 2001). Low-temperature hydrothermal activity because of the effect of an ultra-slow spreading speed of tectonic plates was reported in the SWIR (Tao et al., 2012). The discovery of hydrothermal fields in such an ultra-slow spreading ridge provides an unprecedented opportunity to understand microbially mediated biogeochemistry. A serpentinite-hosted hydrothermal site and distinct microbial community resembling those of the Lost City hydrothermal field in the Atlantic Ocean were reported in a magma-poor area of the eastern SWIR (Lecoeuvre et al., 2021). The relatively low temperature and serpentinization of SWIR hydrothermal fields (Zhou and Dick, 2013) had probably shaped the microbial inhabitants in the hydrothermal fields. However, microbial genomics studies on the SWIR hydrothermal areas are still limited. Thiomicrorhabdus has been reported as a dominant SOB in SWIR (Cao et al., 2014), and was also isolated from the other hydrothermal vents (Brazelton and Baross, 2010; Scott et al., 2019). A recent study revealed a large number of Gammaproteobacteria with sulfur oxidation potentials using nitrate and oxygen as electron acceptors in the SWIR inactive hydrothermal sediments (Dong et al., 2021). We employed metagenomics to compare the microbiomes in the active hydrothermal vents with those from inactive ones to reveal microbial community structure and ecological function driven by active “black-smoker” hydrothermal seepage in SWIR. We hereby report a unique microbiome exclusively composed of Thiohalomonadales-dominated SOB, which has not been reported in DSHVs and presumably plays a critical role in the initiation of the deep-sea hydrothermal ecosystem in SWIR.
Materials and Methods
Sample Collection and Mineral Analysis
During the R/V DY35 cruise, the manned submersible “Jiaolong” collected sulfide samples from hydrothermal chimneys by dives no. 96 (49.65°E, 37.78°S; depth: 2,768) and no. 100 (49.65°E, 37.78°S; depth: 2,755 m) in SWIR (Figure 1, Supplementary Table 1, and Supplementary Figure 1). A sulfide chimney sample was obtained from an inactive hydrothermal vent (49.63°E, 37.77°S; depth: 2,789 m) by dive no. 95 at a depth of 2,789 m. The samples were stored in sterile bags and maintained at –80°C until use. The temperature and pH of the hydrothermal fluid were measured in situ with a Miniature Autonomous Plume Recorder (MAPR) (Baker and Milburn, 1997).
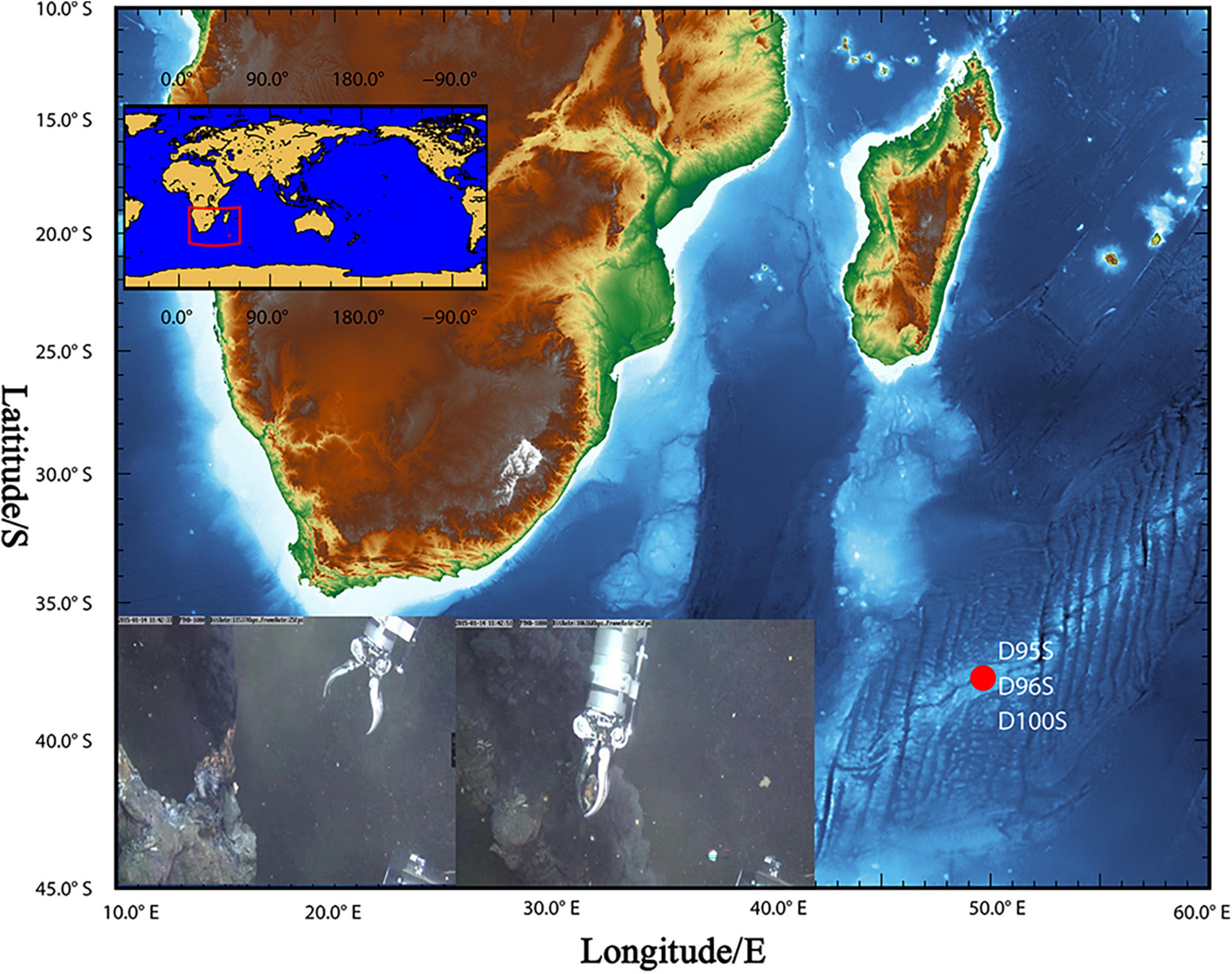
Figure 1. Sampling site at “Longqi” hydrothermal field. During R/V DY35, two chimney rocks (D96S and D100S) were obtained by the “Jiaolong” submersible from active sulfide chimney bodies (inset figures) at “Longqi” hydrothermal field in the Southwest Indian Ocean. A sulfide sample (D95A) was also collected from a nearby inactive chimney by the submersible. The details of the samples are listed in Supplementary Table 1.
An X-ray diffraction (XRD) analysis was carried out to determine the major minerals of the sulfide chimneys using an Empyrean X-ray diffractometer (PANalytical, Malvern, United Kingdom). The detection parameters were Cu Kα radiation at 45 kV and 40 mA; Goniometer PW3050/60 (Theta/Theta); scanning from 3° to 85° with 0.03 step size (°2Th). The mineral components were converted to wt%.
High-Throughput Sequencing and Analyses of 16S rRNA Gene Amplicons
Genomic DNA of sulfide samples was extracted from a 2 g sample using the MO BIO Powersoil DNA isolation kit (Qiagen, Carlsbad, CA, United States). The quality and quantity of the DNA extractions were checked by the Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA, United States). Then, 16S rRNA gene fragments from V3–V4 regions were amplified with a set of universal primers 341F (5′-CCTAYGGGRBGCASCAG-3′) and 802R (5′-TACNVGGGTATCTAATCC-3′) (Wang and Qian, 2009) using 1 ng DNA as a template. PCR conditions of the amplification were performed as follows: initial denaturation at 98°C for 10 s; 28 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 15 s, and extension at 72°C for 30 s; and a final extension at 72°C for 5 min. A PCR replicate was performed. The mixed barcoded 16S rRNA gene amplicons and 100 ng genomic DNA were used for Illumina library preparation using the TruSeq Nano DNA LT kit (Illumina, San Diego, CA, United States), separately, and were sequenced on an Illumina Miseq sequencer to obtain 2 × 300 bp paired-end sequencing reads (Illumina, San Diego, CA, United States).
The sequencing reads of 16S rRNA gene amplicons were checked by FASTQC (v0.11.8)1 and merged by the QIIME 1 workflow (Kuczynski et al., 2011). The adaptors and low-quality reads were trimmed by the NGS QC Toolkit (v2.3.3) (Patel and Jain, 2012). Operational taxonomic units (OTUs) at 97% similarity were selected between the qualified reads by the QIIME 1 workflow (Kuczynski et al., 2011). Taxonomic classification of the OTUs was performed with vsearch against the SILVA database v138 (Quast et al., 2013). A principal component analysis (PCoA) was performed to estimate the similarity between the microbial communities at the family level using vegan in the R package (Dixon, 2003).
Metagenomics Analyses of Microbial Community and Metabolic Potentials
Raw reads of metagenomes were qualified by removing adapters and then filtered by using fastp (version 0.20.0) (Chen et al., 2018) with parameters (-w 16 -q 20 -u 20 -g -c -W 5 -3 -l 50). Low-quality reads (assigned by a quality score < 20 for >20% of the read length), and those shorter than 50 bp and unpaired were removed. Metagenomics reads that were mapped onto sequences in an in-house contaminant database (such as, sequences of human, mouse, and common laboratory contaminant bacterial genomes downloaded from the National Center for Biotechnology Information [NCBI]) (Huang et al., 2019) by Bowtie2 (version.2.4.1) (Langmead and Salzberg, 2012) were discarded.
Furthermore, 16S miTags were extracted from qualified metagenomic reads using rna_hmm3.py (Huang et al., 2019), which employs HMMER (version 3.1b2) to predict ribosomal RNA gene fragments from both forward and reverse metagenomic reads. An in-house python script was used to extract 16S miTags. The 16S miTags were imported into Qiime1 with a setting of –type “SampleData[Sequences]” and dereplicated redundancy to yield representative 16S miTag sequences. The classify-consensus-vsearch command in QIIME v1.9.1 (Kuczynski et al., 2011) was used to classify the representative 16S miTags using the SILVA SSU database v138 as a reference (Quast et al., 2013), as mentioned above.
The qualified reads were assembled using SPAdes (v3.13) (Nurk et al., 2013) with a kmer set of 21, 33, 55, 77, 99, and 127 under the “—careful” mode (metagenome mode). Genome binning from assembled contigs > 2 kbp was performed by running three tools MaxBin (Wu et al., 2016), MetaBAT (Kang et al., 2019), and CONCOCT (Alneberg et al., 2014) using their default settings. Raw genome bins resulting from the three approaches were combined, followed by a selection of the best genome for each genome set using the bin_refinement module in metaWRAP (v1.2) (Uritskiy et al., 2018). During the bin refinement, we applied CheckM_lineage (v1.0.12) (Parks et al., 2015) to evaluate completeness and contamination for each bin. The draft genomes with >50% completeness and <10% contamination were retained for further analysis. Metagenome-assembled genomes (MAGs) were treated with dRep software (Olm et al., 2017) to dereplicate the MAGs by an average nucleotide identity (ANI) threshold of 95% (dereplicate -p 40 -comp 50 -con 10 -pa 0.95 -sa 0.95 -l 10000 -nc 0.30). The taxonomic position of the genomes was identified using genome taxonomy database (GTDB)-tk v0.2.2 (Chaumeil et al., 2020), as well as the calculation of relative evolutionary distance (RED). Coding regions (CDS) for individual MAGs were predicted using Prodigal (version v2.6.3) (Hyatt et al., 2010) with option “-p meta.” Annotation of CDSs was performed using KofamScan (version 1.1.0) (Aramaki et al., 2019) and using Blastp (E value = 1e-5) against the Clusters of Orthologous Gene (COG) and NCBI_nr databases. The results were visualized using the heatmap package of the R platform.
Phylogenetic Tree Construction
For phylogenomic analysis of Thiohalomonadales genomes, 43 conserved proteins were obtained by the CheckM program with default settings (Katoh et al., 2009) and were used for alignment with Mafft (v7.453, setting: –maxiterate 1000-localpair) (Katoh and Standley, 2013), followed by a further optimization with trimAl (v1.4) (Capella-Gutiérrez et al., 2009). A maximum likelihood (ML) tree was reconstructed using IQ-TREE (v1.6.10) (Nguyen et al., 2015) with the “LG + F + R6” model (Price et al., 2009). Bootstrap values were calculated based on 1,000 replicates.
Results
Mineral Composition of the Southwest Indian Ridge Hydrothermal Chimneys
The “Longqi” hydrothermal area is located in the Southwest Indian Ridge. Two sulfide samples, D96S and D100S, were collected from the outer wall of active hydrothermal chimneys on 14 January and 4 February 2015 by the “Jiaolong” submersible (Figure 1 and Supplementary Table 1) in dives nos. 96 and 100, respectively. The chimney rocks were about 10 cm away from the vents. Strong seepage from the hydrothermal vent could be observed. The temperature of the hydrothermal fluid was recorded to be 362 and 365°C for the vents near D96S and D100S, respectively, while the bottom seawater was ∼2.2°C. The pH of the fluid from the two vents was between 3.47 and 3.58. The surface of the D96S sample is gray and has been partially oxidized to grayish brown, while it was light yellow inward with metallic luster (Supplementary Figure 1). There was no megafauna attached to these chimneys. An XRD analysis revealed that D95S and D100S were exclusively composed of sphalerite (ZnS) with a small amount of pyrite (FeS2) (<1%) (Supplementary Figure 2). Oxidized minerals were not present in the XRD results, which is in contrast to the dominance of copper-containing chalcopyrite (CuFeS2) and kusachiite (CuBi2O4) in inactive hydrothermal sediments of SWIR (Cao et al., 2014).
Analysis of Prokaryotic Community Structure
Prokaryotic communities in our chimney samples were examined by sequencing and analysis of 16S rRNA gene amplicons and 16S rRNA gene tags in metagenomic reads (16S miTags), and were then compared with those of the sulfide samples from other SWIR sites (Supplementary Table 1; Dong et al., 2021). There are a total of 30,189 qualified 16S rRNA gene amplicons for the characterization of microbial communities. They were then clustered with 97% similarity into 6,994 OTUs (Supplementary Table 2). Proteobacteria dominated all the SWIR sulfide samples of this study (Figure 2A), except for D95S, in which Bacteroidetes (20%) were the most prevalent phylum. Epsilonbacteraeota was particularly enriched in the chimney samples, and Archaea was not present in these communities. At the family level, Ectothiorhodospiraceae (average 11.9%) and Thiovulaceae (average 6.8%) were the most abundant families in both the 16S rRNA gene amplicon- and miTag-based communities of the D96S sample. Ectothiorhodospiraceae is significantly more abundant in the active chimney samples (D96S and D100S) than in other SWIR hydrothermal sediments using the U-test (p < 10–10). In contrast, D100S contains a high proportion (7.7–19.1%) of Sulfurovaceae, which outcompetes Ectothiorhodospiraceae (∼6.2%) and Thiovulaceae (∼3.2%) as the dominant SOB (Figure 2B). Sulfur reducing bacteria (SRB) represented by Desulfobulbaceae and Thermodesulfovibrionaceae are abundantly present only in D95S and the reference SWIR sulfide samples. Thermodesulfovibrionaceae (Nitrospirae) occupied 6.3% of the community in D95S. The distinct distribution of Ectothiorhodospiraceae and Desulfobulbaceae between active chimney samples (D96S and D100S) and other hydrothermal samples from the SWIR was further illustrated by a chord diagram (Figure 3). The plotting further revealed special enrichment of Thiotrichaceae as the major SOB in inactive hydrothermal sediments, particularly in the S12T1 sample. A PCoA plot separated the microbial communities of D96S and D100S from the references and D95S. For D96S and D100S, the community structures revealed by the 16S rRNA gene amplicon and miTags were consistent with respect to the PCoA clustering (Figure 4).
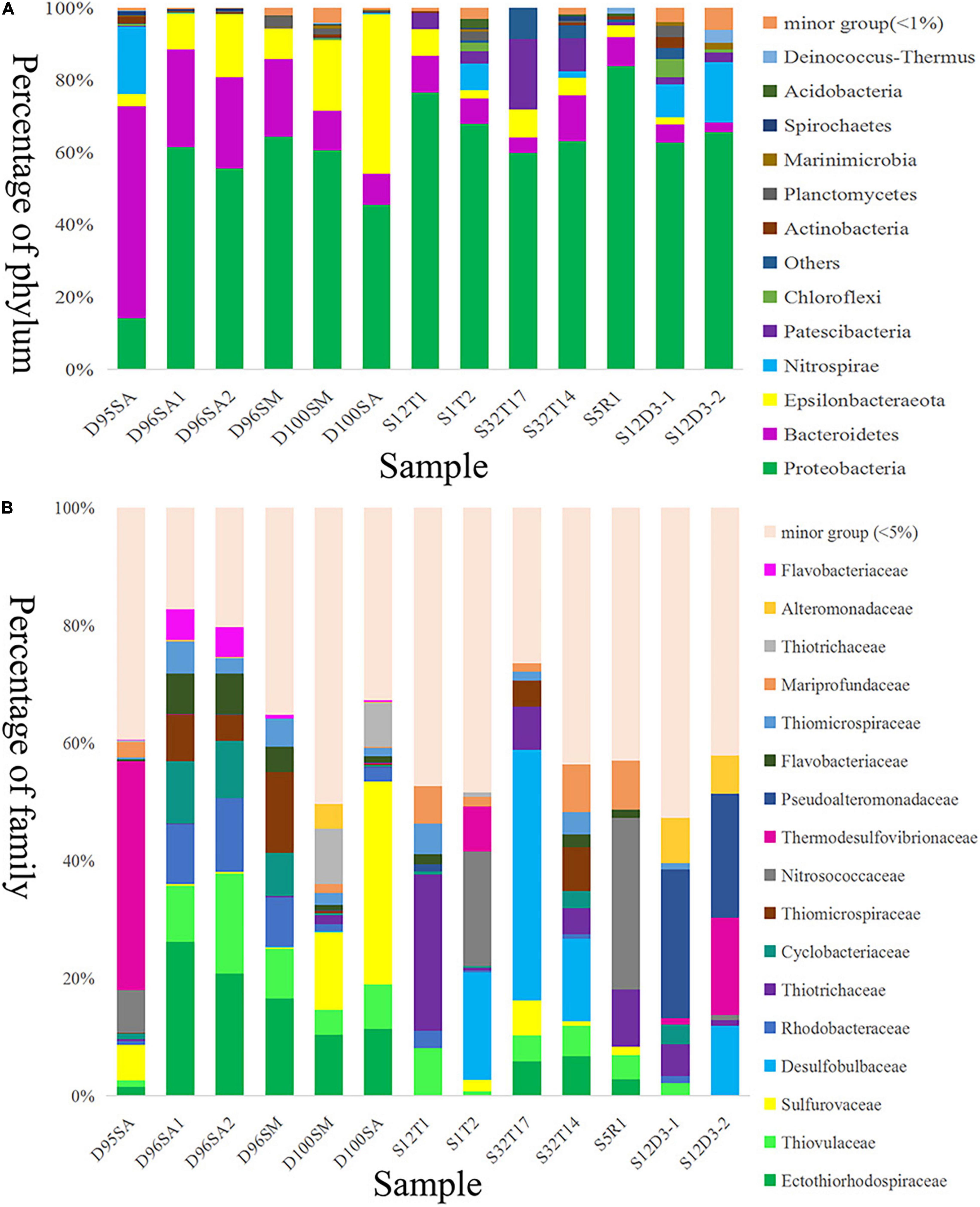
Figure 2. Microbial community structures of hydrothermal samples. Taxonomic classification of operational taxonomic units (OTUs) at 97% similarity was performed by comparing with reference sequences of SILVA database v138. The microbial community structures were shown at phylum (A) and family (B) levels. D100SA, D96SA1, D96SA2, and D95SA refer to 16S rRNA gene amplicons; D100SM and D96SM are 16S miTags. The other samples are referred to Supplementary Table 2.
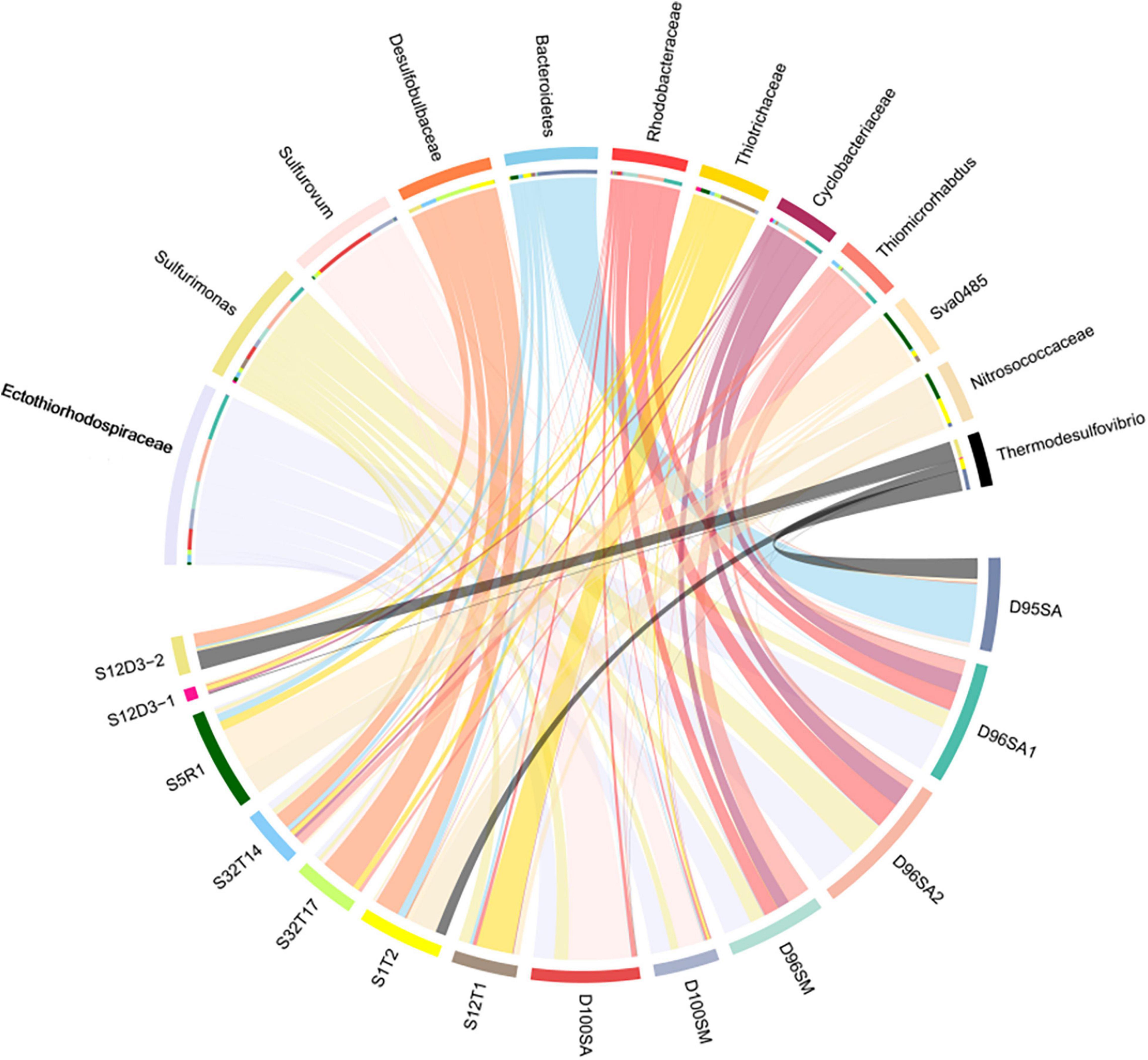
Figure 3. The chord diagram chart showing the distribution of dominant genera. The analysis was performed using the microbial communities at the genus level. D100SA, D96SA1, D96SA2, and D95SA refer to 16S rRNA gene amplicons; D100SM and D96SM are 16S miTags obtained from the metagenomes. The other samples are referred in Supplementary Table 2.
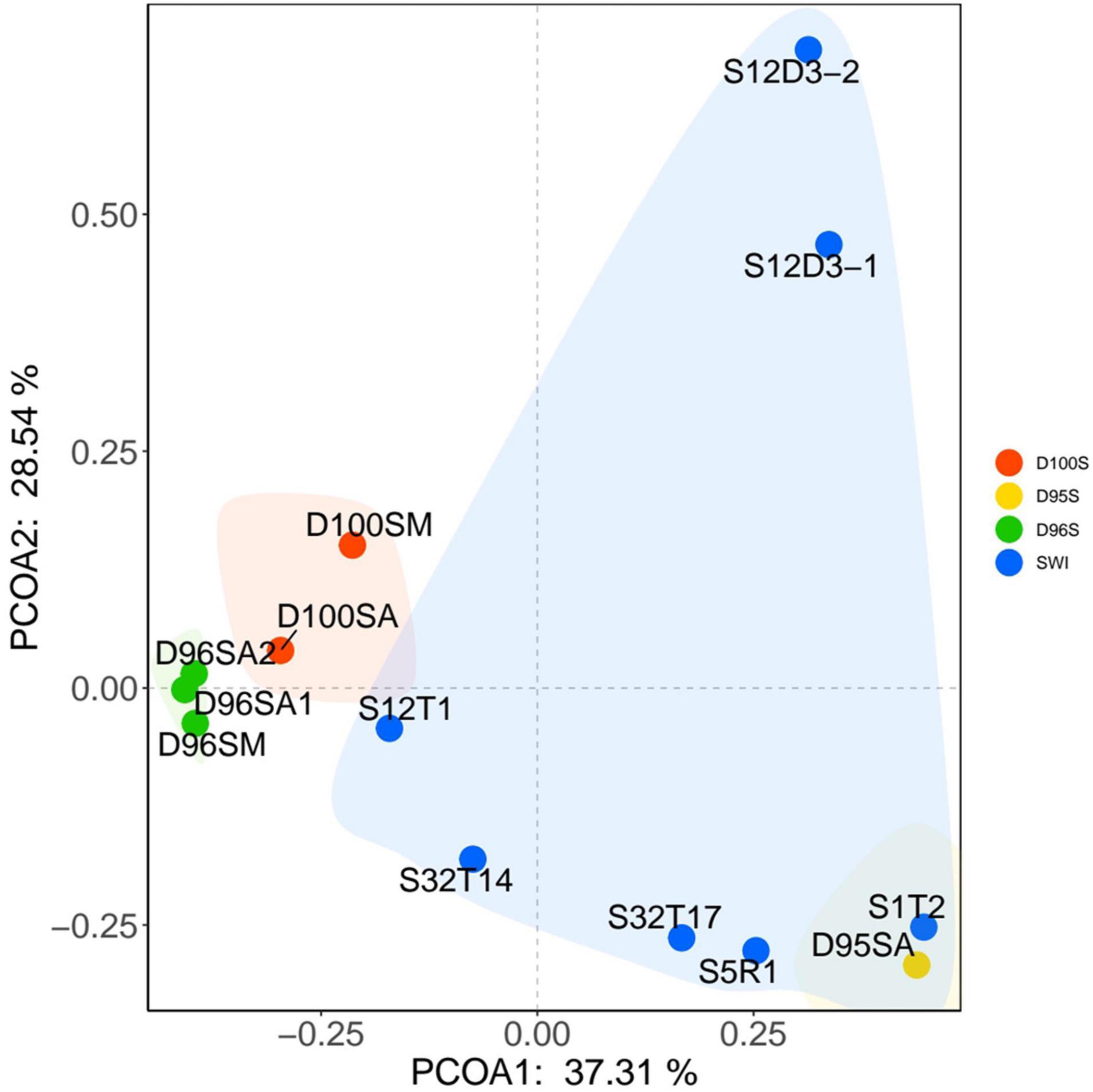
Figure 4. The principal component analysis (PCoA) analysis of microbial communities. The PCoA analysis was performed using the microbial communities at family level. The community structures were based on the classification of 16S miTags (D100SM, D96SM, and the other SWIR reference samples) and 16S rRNA gene amplicons (D96SA1, D96SA2, D100SA, and D95SA) (Supplementary Table 2).
Metagenomics Analysis of Dominant Species Inhabiting Active Hydrothermal Chimney
To understand the ecological role of microbial inhabitants in the active hydrothermal vents, we obtained 12.1 Gb raw Illumina Miseq paired-end sequencing data (2 × 300 bp) and retained 8.3 Gb clean data for subsequent metagenomics work of D96S and D100S (Supplementary Table 3). After assembly and genome binning, a total of 82 MAGs were retrieved from D96S and D100S metagenomic assemblies (Supplementary Table 4), among which 57 MAGs were of good quality (completeness > 80% and contamination < 5%). Taxonomic classification of the MAGs against GTDB indicates that they were affiliated with 12 prokaryotic phyla mainly composed of Proteobacteria (n = 39), Bacteroidota (n = 20), Campylobacterota (n = 7), and Desulfobacterota (n = 5; Supplementary Table 4). The abundant Ectothiorhodospiraceae in D96S and D100S identified by 16S rRNA gene amplicon sequencing were not present in the taxa of the MAGs. By searching the 16S rRNA gene amplicons of Ectothiorhodospiraceae against those extracted from the MAGs (>97% similarity), we reassigned Ectothiorhodospiraceae to the orders Thiohalomonadales SZUA-152 (n = 8) and SZUA-140 (n = 1) of Gammaproteobacteria in GTDB taxonomy. In a phylogenomics tree, these MAGs were clustered with the genomes obtained from hypersaline soda lake sediment, ground water, and other deep-sea hydrothermal vents, such as Mid-Atlantic Ridge, East Pacific Rise, and Lau Basin of Tonga (Figure 5). Three phylogenetic groups were formed with most of our MAGs in Thiohalomonadales SZUA-152, a new order in GTDB.
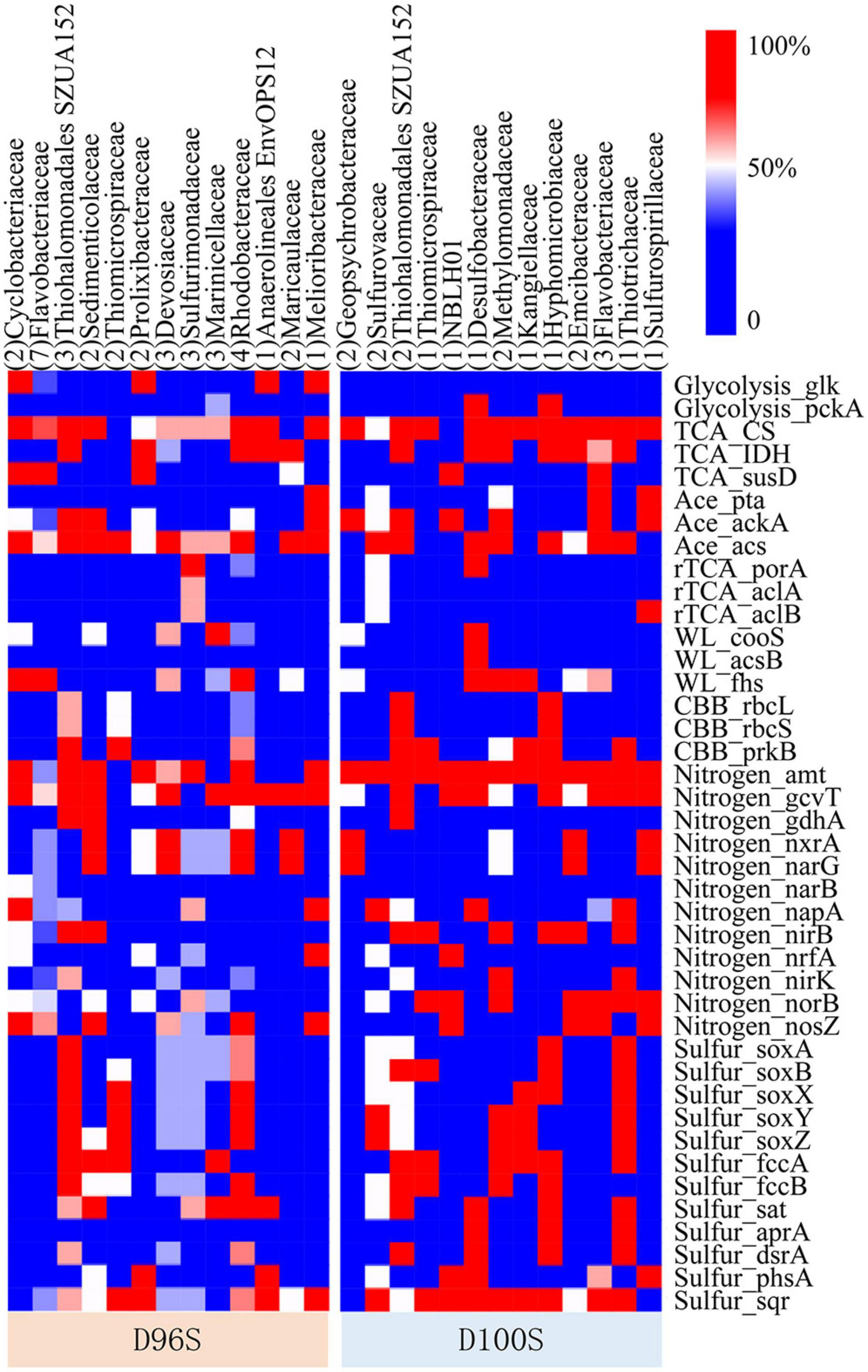
Figure 5. Frequency of functional genes identified in metagenome-assembled genomes (MAGs) belonging to different families. The functional genes of 57 good-quality MAGs were identified by searching against Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The number of MAGs in each genome taxonomy database (GTDB) family was included in the round brackets. The functions of the genes are shown in Supplementary Table 5. The full annotation of KEGG functions is shown in Supplementary Table 6.
On the basis of Kyoto Encyclopedia of Genes and Genomes (KEGG) gene annotation of the 57 good-quality MAGs, the functional genes for carbon, nitrogen, and sulfur metabolisms mediated by the major families of the chimney inhabitants were examined to predict their ecological functions. Hydrothermal vents discharge large amounts of hydrogen, methane, and hydrogen sulfide to deep waters (Jannasch and Mottl, 1985; Kawagucci et al., 2010; Dick, 2019). Thiosulfate oxidizing genes soxABXYZ, and sulfide oxidizing genes dsrAB, fccAB, and sqr were found in ≥50% of the MAGs for Thiohalomonadales (Gammaproteobacteria), Thiomicrospiraceae (Gammaproteobacteria), and Rhodobacteraceae (Alphaproteobacteria) MAGs from D96S; and in Thiohalomonadales, Sulfurovaceae (Campylobacterota), Hyphomicrobiaceae (Alphaproteobacteria), and Thiotrichaceae (Gammaproteobacteria) MAGs from D100S (Figure 6). In the periplasmic space, FccAB catalyzes oxidation of sulfide to elemental sulfur with electrons being transferred to cytochrome c; the SQR enzyme oxidizes sulfide to polysulfide (Schutz et al., 1999). The DsrAB encoded by the SOB carry out sulfide oxidation to sulfite (Anantharaman et al., 2014; Dong et al., 2021). This indicates the capacity of sulfide oxidation to S0, polysulfide or sulfate by these SOB bacteria inhabiting our active chimney samples under different fluxes of electron acceptors. The Rhodobacteraceae MAGs also harbor nitrate reduction genes narG, suggesting the coupling of nitrate reduction and sulfide oxidation. Sulfurimonadaceae and Sulfurovaceae encoded the reverse tricarboxylic acid (rTCA) pathway for autotrophic CO2 fixation. The other SOB, with the exception of Thiotrichaceae, may rely on the Calvin–Benson–Bassham (CBB) pathway for autotrophs, as ribulose-1,5-bisphosphate carboxylase (RuBisCO) genes rbcLS were found in their MAGs. Wood-Ljungdahl pathway as an alternative autotrophic process was only encoded by Desulfobacteraceae MAGs. Acetate metabolism is highly required by the bacterial inhabitant, as evidenced by the prevalence of acetate assimilatory genes pta, acs, and ackA in these MAGs. Genomics data also indicate that Gammaproteobacteria might use hydrogen released from the vents as alternative energy sources for carbon fixation and metabolic activities as indicated by the detection of [NiFe]-hydrogenase genes.
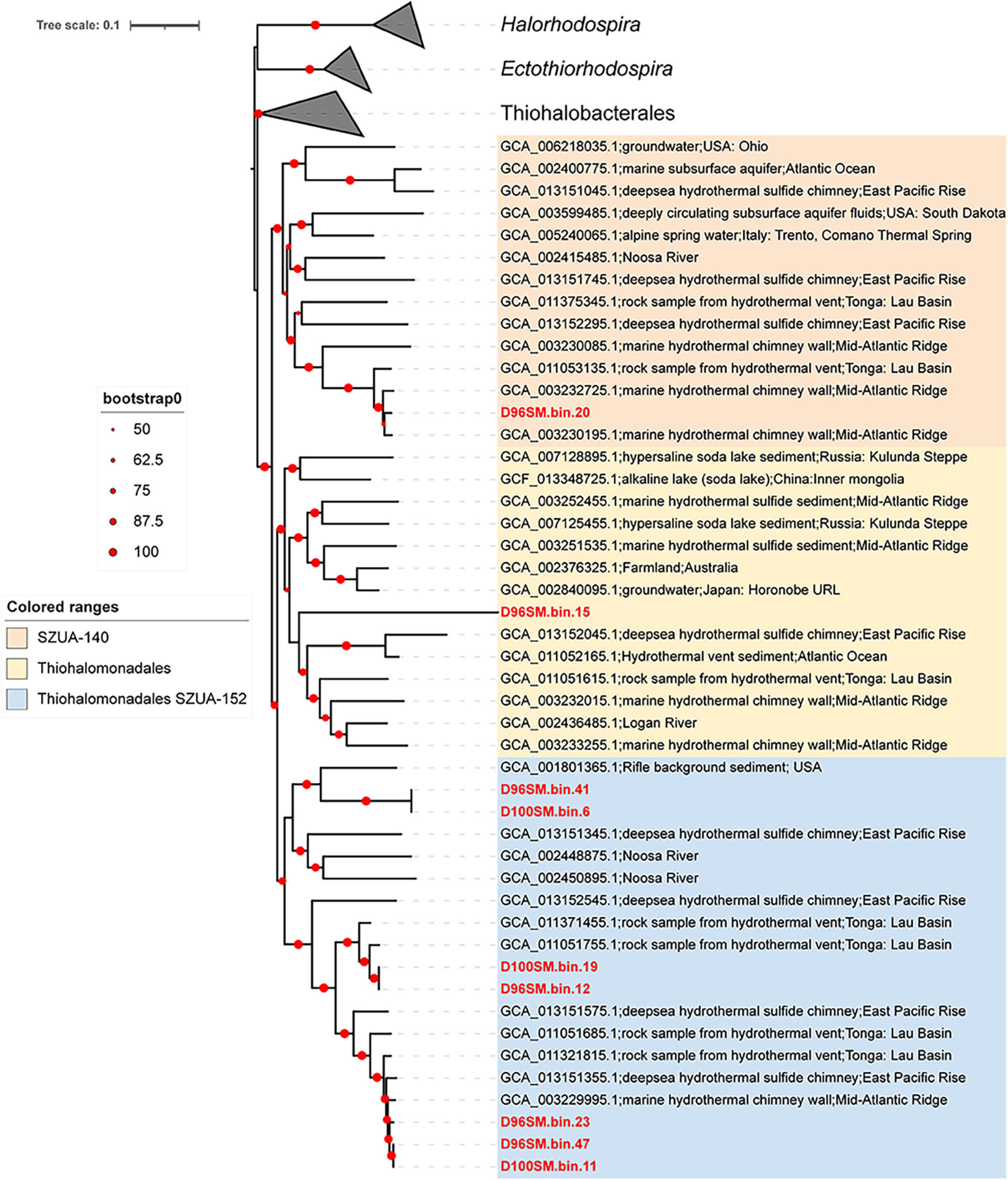
Figure 6. Phylogenomics tree of Thiohalomonadales from the Southwest Indian Ridge (SWIR) hydrothermal chimney samples. The maximum likelihood (ML) phylogenomics tree was constructed using concatenated alignment of 43 conserved proteins extracted from Thiohalomonadales MAGs and reference genomes from GTDB database. The information of the MAGs in red from this study is shown in Supplementary Table 4.
Metabolism Potentials of Thiohalomonadales
We next examined the metabolic potentials and adaptive strategy of the dominant SOB, Thiohalomonadales, from our active hydrothermal vents using the annotation result of their five high-quality MAGs. The Thiohalomonadales MAGs harbor a complete set of sulfur-oxidizing genes, such as soxABXYZ, fccABV, sqr, rdsrAB, and TST, that may yield S0, polysulfide, or sulfate (Figure 7). Nitrate assimilatory reduction may occur in the Thiohalomonadales as the MAGs contain napAB periplasmic nitrate reduction genes and nitrite reduction genes (nirBD) that are involved in ammonia production. Thiohalomonadales might also rely on the CBB pathway for CO2 fixation. Thiohalomonadales MAGs contain cbb3 genes coding for phosphorylation respiration complex IV to overcome low oxygen conditions. Considering the high hydrogen content of the hydrothermal fluids, the microbes on the chimney are expected to be able to utilize hydrogen as an energy source. [NiFe] hydrogenase genes were present in the MAGs, along with hup genes responsible for the uptake and/or export of hydrogen. The Rnf complex as a cross-membrane proton pumping machine for the balance of cytoplasmic pH was encoded by the MAGs (Buckel and Thauer, 2013).
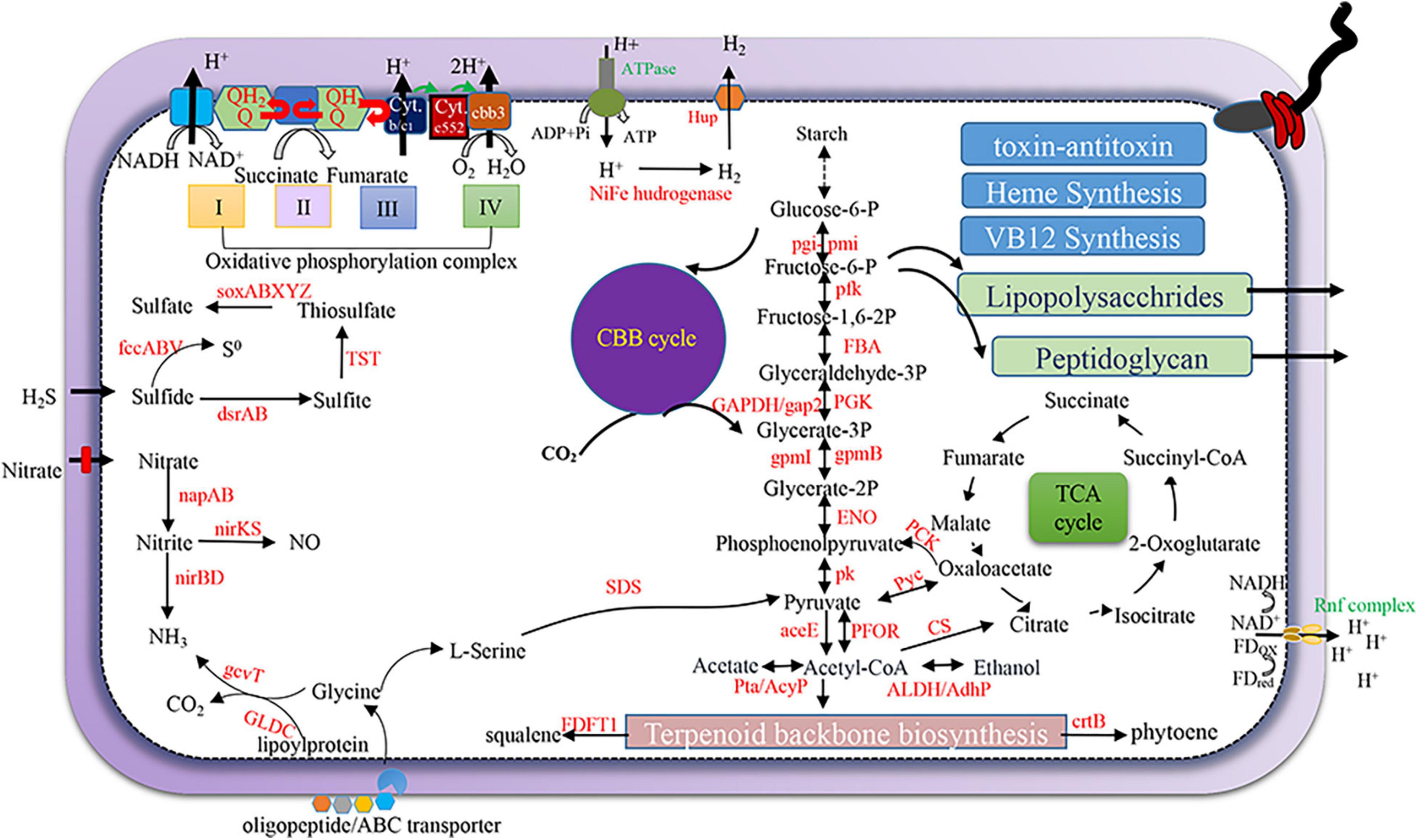
Figure 7. Schematic metabolism of Thiohalomonadales (sulfur oxidizing bacteria) SOB. The metabolic pathways were predicted and depicted based on gene annotation of high-quality MAGs of Thiohalomonadales against KEGG and National Center for Biotechnology Information (NCBI)_nr databases.
We identified all the related genes encoding the enzymes that participate in terpenoid backbone biosynthesis. Using geranylgeranyl diphosphate, Thiohalomonadales might generate phytoene, since the crtB gene encoding 15-cis-phytoene synthase was identified in the MAGs. The CrtB protein of Thiohalomonadales is most similar (82%) to a homolog of Thiotrichaceae from a subseafloor aquifer. Phytoene is a precursor of zeta-carotene and can be catalyzed by phytoene desaturase CtrI for carotene production (Lang et al., 1994). However, ctrI gene is missing from the MAGs, and the capacity of carotene biosynthesis by these SOB is thus questioned. The crtB gene was also present in the MAGs from RBG-16-57-12, Mariprofundaceae, Thiohalomonadales SZUA-140, and Melioribacteraceae. Most of these crtB-bearing MAGs also contain a farnesyl-diphosphate farnesyltransferase coding gene (FDFT1) that functions in squalene synthesis using farnesyl diphosphate.
In the Thiohalomonadales MAGs, a cph2 type bacteriophytochrome coding gene was identified to be 45% similar to a homolog from Pseudomonas aeruginosa. However, the putative bacteriophytochrome is featured with GGDEF and EAL domains but lacks GAF and PHY domains, indicating a discovery of a novel or malfunctional bacteriophytochrome (Gourinchas et al., 2019). The co-factor of bacteriophytochrome is biliverdin with a tetrapyrrolic structure. Heme synthesis pathway and biliverdin-producing heme oxygenase have been identified in the MAGs, which are vital for the potential function of bacteriophytochrome under red and far-red lights for Thiohalomonadales species (Vuillet et al., 2007; Gourinchas et al., 2019).
Discussion
Discovery of Novel Microbiomes in Active Hydrothermal Chimneys
In this study, the “Jiaolong” manned submersible with robotic precision was employed to locate the vent structure of sulfide chimneys in the active SWIR hydrothermal areas. Using these chimney samples, we report the genomes of dominant microbial species and the prevalence of distinct SOB affiliated with Campylobacterota, Gammaproteobacteria, and Alphaproteobacteria inhabiting the sulfide chimney, all of which have been reported in deep-sea cold seeps recently (Li et al., 2021). The microbial community in the active SWIR hydrothermal vents was remarkably different from those from the SWIR inactive hydrothermal sulfide sediments (Cao et al., 2014; Dong et al., 2021) and others DSHVs (Dick, 2019; Hou et al., 2020; Zeng et al., 2021). Culturable Thiohalomonadales SOB, such as Thiohalomonas denitrificans had been isolated from the hydrothermal vents of the Suiyo Seamount in the Pacific Ocean (Mori et al., 2015). In addition, this study demonstrates their distribution in the DSHVs located in the Mid-Atlantic Ridge, East Pacific Rise, and Lau Basin. Given the finding of their relatives in other worldwide sites, the dominance of this order in DSHVs was, however, not reported previously (Flores et al., 2011; Urich et al., 2014; Meier et al., 2017). Dominant SOB differed even between D96S and D100S, owing to different combinations of Thiohalomonadales, Thiomicrorhabdus, Sulfurovum, and Sulfurimonas. This suggests a high diversity of SOB among active hydrothermal vents. Our genomics data predict other potential SOB, such as Hyphomicrobiaceae, Thiotrichaceae (represented by Beggiatoa), and Rhodobacteraceae in our samples, which have been rarely reported in other DSHVs. Considering the highly variable microenvironments adjacent to DSHVs, the composition of SOB inhabitants is predicted to be slightly diversified as reported for D96S and D100S by this study. In the late stages of hydrothermal vents, weak seepage of reducing fluids allows soaking of sulfide with oxic bottom water, which will gradually result in sulfide mineral oxidation and subsequent microbially mediated reduction processes (Li et al., 2017). The microbiomes of D96S and D100S were solely constituted by SOB, which is an indicator of the initial stage of a hydrothermal ecosystem. This has not been noticed in the SWIR and even global DSHVs.
The strong hydrothermal venting probably creates a reducing environment that covers the chimney, prohibiting oxidation of the chimney sulfide in this study. The mineral components in our samples indicate an early stage of the chimney formation at D100S, as there were not any oxidized mineral components. As a result, we could not detect SRB in D100S, while in contrast, prevalent Thermodesulfovibrionaceaea (SRB) was present in D95S. Although the sulfate concentration and nutrients of the samples were not analyzed due to their contact with sea water, we speculate that sulfate from sea water and produced by SOB might probably fuel the SRB in D95S. It seems that Sulfurovaceae, Thiohalomonadales, and Thiovulaceae were distributed differently in the hydrothermal chimneys possibly due to selection of different SOB families by local environmental variants. For example, Sulfurovum is more tolerant to oxygen (Meier et al., 2017) and might prefer a more oxic chimney with lower impact of hydrothermal fluid. In this study, Thiohalomonadales probably employed cbb3 to cope with low oxygen; however, hypoxic exposure may also impair anaerobic and microaerobic microbial inhabitants (Lu and Imlay, 2021). We found that squalene as a bacterial hopanoid was likely synthesized by Thiohalomonadales, which is probably an efficient mechanism to scavenge single oxygen that may damage cell lipid by peroxidation (Kohno et al., 1995). The microbes in D96S and D100S have probably evolved to contain distinct gene profiles for adaptation to environmental changes due to the dynamics of gas and metal fluxes near active hydrothermal vents.
Photolithoautotrophic Potential of Sulfur-Oxidizing Thiohalomonadales
Ectothiorhodospiraceae was previously a family of Chromatiales, but was recently reclassified into a new family of Thiohalomonadales. Members of the Ectothiorhodospiraceae are versatile in metabolisms, including photolithotrophic, photoheterotrophic, chemoheterotrophic, chemolithotrophic, and methylotrophic bacteria using various electron acceptors, such as nitrite, sulfur compounds, and arsenite (Hallberg et al., 2011; Slobodkina et al., 2016). So far, all isolates from hydrothermal fields belonging to this family are sulfur-oxidizing bacteria (Imhoff, 2006; Mori et al., 2015; Slobodkina et al., 2016). In this study, we discovered three phylogenetic groups in Thiohalomonadales belonging to SZUA152, SZUA140, and an unclassified clade. Ectothiorhodospiraceae members are able to capture red/far-red light for anaerobic photosynthesis, by which sulfide is oxidized to elemental sulfur and deposited in periplasm for further oxidation to sulfate (Imhoff, 2006). Bacteriophytochromes are photosensitive proteins employed by bacteria to capture red/far-red light to mediate growth (Vuillet et al., 2007; Gourinchas et al., 2019). At hydrothermal vents with a temperature of ∼350°C, most of the ambient light was detected to fall into near-infrared spectra between 750 and 1,050 nm (Dover et al., 1994). A recent report showed the promoted growth of a bacterial cultivation isolated from the Western Pacific DSHV under stimulation of infrared light at 940 nm (Liu et al., 2021). This is highly supported by a recent report that showed the presence of proteorhodopsin synthesis genes and photoautotrophic microbial groups in some metagenomes of active hydrothermal vents of SWIR (Chen et al., 2022). If the bacteriophytochromes and cofactor biliverdin are functional in Thiohalomonadales, they can probably sense red/near-infrared light from the hydrothermal vents to activate photosynthesis of Thiohalomonadales. In photosynthetic and non-photosynthetic organisms, carotenoids are synthesized to prevent the photooxidative damage (Glaeser and Klug, 2005). In the present study, although the gene coding for the enzyme catabolizing the final step of zeta-carotenoid was not found, there are still perhaps unknown genes responsible for carotenoid synthesis in the genomes. Aside from serving as light sensors, some bacteriophytochromes can be redox sensors to monitor the environmental changes, particularly in the environment approximate to hydrothermal vents where hot reductive fluid meets cold oxic sea water. Whether Thiohalomonadales can synthesize the bona fide bacteriophytochromes for photoautotrophy warrants future experimental efforts using cultivated strains of this order from DSHVs.
Overall, we identified an unusual, uncultivated sulfur-oxidizing bacterial group that is particularly prevalent in active hydrothermal chimneys. Their potential capacity of sulfur oxidization and photoautotrophic lifestyle casts lights on the possible relationship with ancient lineages inhabiting the early earth with similar hydrothermal environments billions of years ago (Lunine, 2006). The finding of this SOB group can probably provide insights into the future evolutionary study of ancient microbial lineages under the climate change in the long history of the Earth.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
H-YB and YW conceived the study and wrote the manuscript. H-GC, P-FZ, and Y-LZ performed the experiments. YW, H-YB, P-FZ, and Y-LZ analyzed the data and summarized the results. J-TL critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Major Scientific and Technological Projects of Hainan Province (ZDKJ2021028).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the cruise members of D/V DY35 and “Jiaolong” submersible pilots for their efforts in sampling. We appreciate the Supercomputing Center of University of Sanya for providing the computation assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.861795/full#supplementary-material
Footnotes
References
Adam, N., Kriete, C., Garbe-Schönberg, D., Gonnella, G., Krause, S., Schippers, A., et al. (2020). Microbial community compositions and geochemistry of sediments with increasing distance to the hydrothermal vent outlet in the Kairei field. Geomicrobiol. J. 37, 242–254. doi: 10.1080/01490451.2019.1694107
Alneberg, J., Bjarnason, B. S., De Bruijn, I., Schirmer, M., Quick, J., Ijaz, U. Z., et al. (2014). Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146.
Anantharaman, K., Duhaime, M. B., Breier, J. A., Wendt, K. A., Toner, B. M., and Dick, G. J. (2014). Sulfur oxidation genes in diverse deep-sea viruses. Science 344, 757–760. doi: 10.1126/science.1252229
Aramaki, T., Blanc-Mathieu, R., Endo, H., Ohkubo, K., Kanehisa, M., Goto, S., et al. (2019). KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252. doi: 10.1093/bioinformatics/btz859
Baker, E. T., and Milburn, H. B. (1997). MAPR: a new instrument for hydrothermal plume mapping. RIDGE Events 8, 23–25.
Beatty, J. T., Overmann, J., Lince, M. T., Manske, A. K., Lang, A. S., Blankenship, R. E., et al. (2005). An obligately photosynthetic bacterial anaerobe from a deep-sea hydrothermal vent. Proc. Natl. Acad. Sci. U.S.A. 102, 9306–9310. doi: 10.1073/pnas.0503674102
Brazelton, W. J., and Baross, J. A. (2010). Metagenomic comparison of two Thiomicrospira lineages inhabiting contrasting deep-aea hydrothermal environments. PLoS One 5:e13530. doi: 10.1371/journal.pone.0013530
Buckel, W., and Thauer, R. K. (2013). Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113. doi: 10.1016/j.bbabio.2012.07.002
Cao, H., Wang, Y., Lee, O. O., Zeng, X., Shao, Z., and Qian, P. Y. (2014). Microbial sulfur cycle in two hydrothermal chimneys on the Southwest Indian Ridge. mBio 5, e980–e913. doi: 10.1128/mBio.00980-13
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chaumeil, P.-A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2020). GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics, [Epub ahead of print]. doi: 10.1093/bioinformatics/btz848
Chen, H., Li, D. H., Jiang, A. J., Li, X. G., Wu, S. J., Chen, J. W., et al. (2022). Metagenomic analysis reveals wide distribution of phototrophic bacteria in hydrothermal vents on the ultraslow-spreading Southwest Indian Ridge. Mar. Life Sci. Technol. doi: 10.1007/s42995-021-00121-y
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Connelly, D. P., Copley, J. T., Murton, B. J., Stansfield, K., Tyler, P. A., German, C. R., et al. (2012). Hydrothermal vent fields and chemosynthetic biota on the world’s deepest seafloor spreading centre. Nat Commun 3:620. doi: 10.1038/ncomms1636
Corliss, J. B., Dymond, J., Gordon, L. I., Edmond, J. M., Von Herzen, R. P., Ballard, R. D., et al. (1979). Submarine thermal sprirngs on the galapagos rift. Science 203, 1073–1083. doi: 10.1126/science.203.4385.1073
Dick, G. J. (2019). The microbiomes of deep-sea hydrothermal vents: distributed globally, shaped locally. Nat. Rev. Microbiol. 17, 271–283. doi: 10.1038/s41579-019-0160-2
Dick, G. J., Anantharaman, K., Baker, B. J., Li, M., Reed, D. C., and Sheik, C. S. (2013). The microbiology of deep-sea hydrothermal vent plumes: ecological and biogeographic linkages to seafloor and water column habitats. Front. Microbiol. 4:124. doi: 10.3389/fmicb.2013.00124
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veget. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Dong, X., Zhang, C., Li, W., Weng, S., Song, W., Li, J., et al. (2021). Functional diversity of microbial communities in inactive seafloor sulfide deposits. FEMS Microbiol. Ecol. 97:fiab108. doi: 10.1093/femsec/fiab108
Dover, C. L. V., Reynolds, G. T., Chave, A. D., and Tyson, J. A. (1994). Light at deep-sea hydrothermal vents. Geophys. Res. Lett. 23, 2049–2052.
Flores, G. E., Campbell, J. H., Kirshtein, J. D., Meneghin, J., Podar, M., Steinberg, J. I., et al. (2011). Microbial community structure of hydrothermal deposits from geochemically different vent fields along the Mid-Atlantic Ridge. Environ. Microbiol. 13, 2158–2171. doi: 10.1111/j.1462-2920.2011.02463.x
Francheteau, J., Needham, H. D., Choukroune, P., Juteau, T., Séguret, M., Ballard, R. D., et al. (1979). Massive deep-sea sulphide ore deposits discovered on the East Pacific Rise. Nature 277, 523–528.
German, C. R., Baker, E. T., Mevel, C., Tamaki, K., and Team, F. S. (1998). Hydrothermal activity along the southwest Indian ridge. Nature 395, 490–493. doi: 10.1038/26730
Glaeser, J., and Klug, G. (2005). Photo-oxidative stress in Rhodobacter sphaeroides: protective role of carotenoids and expression of selected genes. Microbiology 151, 1927–1938. doi: 10.1099/mic.0.27789-0
Gourinchas, G., Etzl, S., and Winkler, A. (2019). Bacteriophytochromes – from informative model systems of phytochrome function to powerful tools in cell biology. Curr. Opin. Struct. Biol. 57, 72–83. doi: 10.1016/j.sbi.2019.02.005
Hallberg, K. B., Hedrich, S., and Johnson, D. B. (2011). Acidiferrobacter thiooxydans, gen. nov. sp. nov.; an acidophilic, thermo-tolerant, facultatively anaerobic iron- and sulfur-oxidizer of the family Ectothiorhodospiraceae. Extremophiles 15, 271–279. doi: 10.1007/s00792-011-0359-2
Han, Y., Gonnella, G., Adam, N., Schippers, A., Burkhardt, L., Kurtz, S., et al. (2018). Hydrothermal chimneys host habitat-specific microbial communities: analogues for studying the possible impact of mining seafloor massive sulfide deposits. Sci. Rep. 8:10386. doi: 10.1038/s41598-018-28613-5
Hou, J., Sievert, S. M., Wang, Y., Seewald, J. S., Natarajan, V. P., Wang, F., et al. (2020). Microbial succession during the transition from active to inactive stages of deep-sea hydrothermal vent sulfide chimneys. Microbiome 8:102. doi: 10.1186/s40168-020-00851-8
Huang, J. M., Baker, B. J., Li, J. T., and Wang, Y. (2019). New microbial lineages capable of carbon fixation and nutrient cycling in deep-sea sediments of the northern South China Sea. Appl. Environ. Microbiol. 85, e523–e519. doi: 10.1128/AEM.00523-19
Hyatt, D., Chen, G.-L., Locascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11:119. doi: 10.1186/1471-2105-11-119
Imhoff, J. F. (2006). “The Family Ectothiorhodospiraceae,” in The Prokaryotes: A Handbook on the Biology of Bacteria Volume 6: Proteobacteria: Gamma Subclass, eds M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (New York: Springer New York), 874–886. doi: 10.1007/0-387-30746-x_32
Jannasch, H. W., and Mottl, M. J. (1985). Geomicrobiology of deep-sea hydrothermal vents. Science 229, 717–725. doi: 10.1126/science.229.4715.717
Kang, D. D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., et al. (2019). MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359.
Katoh, K., Asimenos, G., and Toh, H. (2009). “Multiple alignment of DNA sequences with MAFFT,” in Bioinformatics for DNA Sequence Analysis, Ed. P. David (Berlin: Springer), 39–64. doi: 10.1007/978-1-59745-251-9_3
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kawagucci, S., Toki, T., Ishibashi, J., Takai, K., Ito, M., Oomori, T., et al. (2010). Isotopic variation of molecular hydrogen in 20 degrees-375 degrees C hydrothermal fluids as detected by a new analytical method. J. Geophys. Res. 115:G03021.
Kelley, D. S., Karson, J. A., Fruh-Green, G. L., Yoerger, D. R., Shank, T. M., Butterfield, D. A., et al. (2005). A serpentinite-hosted ecosystem: the Lost City hydrothermal field. Science 307, 1428–1434. doi: 10.1126/science.1102556
Kohno, Y., Egawa, Y., Itoh, S., Nagaoka, S.-I., Takahashi, M., and Mukai, K. (1995). Kinetic study of quenching reaction of singlet oxygen and scavenging reaction of free radical by squalene in n-butanol. Biochim. Biophys. Acta 1256, 52–56. doi: 10.1016/0005-2760(95)00005-w
Kuczynski, J., Stombaugh, J., Walters, W. A., Gonzalez, A., Caporaso, J. G., and Knight, R. (2011). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinform. Chapter 10:10.7.
Lang, H. P., Cogdell, R. J., Gardiner, A. T., and Hunter, C. N. (1994). Early steps in carotenoid biosynthesis: sequences and transcriptional analysis of the crtI and crtB genes of Rhodobacter sphaeroides and overexpression and reactivation of crtI in Escherichia coli and R. sphaeroides. J. Bacteriol. 176, 3859–3869. doi: 10.1128/jb.176.13.3859-3869.1994
Langmead, B., and Salzberg, S. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359.
Lecoeuvre, A., Menez, B., Cannat, M., Chavagnac, V., and Gerard, E. (2021). Microbial ecology of the newly discovered serpentinite-hosted Old City hydrothermal field (southwest Indian ridge). ISME J. 15, 818–832.
Li, J., Cui, J., Yang, Q., Cui, G., Wei, B., Wu, Z., et al. (2017). Oxidative weathering and microbial diversity of an inactive seafloor hydrothermal sulfide chimney. Front. Microbiol. 8:1378. doi: 10.3389/fmicb.2017.01378
Li, W. L., Dong, X., Lu, R., Zhou, Y. L., Zheng, P. F., Feng, D., et al. (2021). Microbial ecology of sulfur cycling near the sulfate-methane transition of deep-sea cold seep sediments. Environ. Microbiol. 23, 6844–6858. doi: 10.1111/1462-2920.15796
Liu, G., Shan, Y., Zheng, R., Liu, R., and Sun, C. (2021). Growth promotion of a deep-sea bacterium by sensing infrared light through a bacteriophytochrome photoreceptor. Environ. Microbiol. 23, 4466–4477. doi: 10.1111/1462-2920.15639
López-García, P., Duperron, S., Philippot, P., Foriel, J., Susini, J., and Moreira, D. (2003). Bacterial diversity in hydrothermal sediment and epsilonproteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ. Microbiol. 5, 961–976. doi: 10.1046/j.1462-2920.2003.00495.x
Lu, Z., and Imlay, J. A. (2021). When anaerobes encounter oxygen: mechanisms of oxygen toxicity, tolerance and defence. Nat. Rev. Microbiol. 19, 774–785. doi: 10.1038/s41579-021-00583-y
Lunine, J. I. (2006). Physical conditions on the early Earth. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1721–1731. doi: 10.1098/rstb.2006.1900
Meier, D. V., Pjevac, P., Bach, W., Hourdez, S., Girguis, P. R., Vidoudez, C., et al. (2017). Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. ISME J. 11, 1545–1558. doi: 10.1038/ismej.2017.37
Meier, D. V., Pjevac, P., Bach, W., Markert, S., Schweder, T., Jamieson, J., et al. (2019). Microbial metal-sulfide oxidation in inactive hydrothermal vent chimneys suggested by metagenomic and metaproteomic analyses. Environ. Microbiol. 21, 682–701. doi: 10.1111/1462-2920.14514
Mori, K., Suzuki, K. I., Yamaguchi, K., Urabe, T., and Hanada, S. (2015). Thiogranum longum gen. nov., sp. nov., an obligately chemolithoautotrophic, sulfur-oxidizing bacterium of the family Ectothiorhodospiraceae isolated from a deep-sea hydrothermal field, and an emended description of the genus Thiohalomonas. Int. J. Syst. Evol. Microbiol. 65, 235–241. doi: 10.1099/ijs.0.070599-0
Nguyen, L.-T., Schmidt, H., Haeseler, A. V., and Minh, B. (2015). IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A. A., Korobeynikov, A., Lapidus, A., et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737.
Olm, M. R., Brown, C. T., Brooks, B., and Banfield, J. F. (2017). dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868.
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genom. Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Patel, R. K., and Jain, M. (2012). NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Schutz, M., Maldener, I., Griesbeck, C., and Hauska, G. (1999). Sulfide-quinone reductase from Rhodobacter capsulatus: requirement for growth, periplasmic localization, and extension of gene sequence analysis. J. Bacteriol. 181, 6516–6523. doi: 10.1128/JB.181.20.6516-6523.1999
Scott, K. M., Leonard, J. M., Boden, R., Chaput, D., Dennison, C., Haller, E., et al. (2019). Diversity in CO(2)-concentrating mechanisms among chemolithoautotrophs from the genera Hydrogenovibrio, Thiomicrorhabdus, and Thiomicrospira, ubiquitous in sulfidic habitats worldwide. Appl. Environ. Microbiol. 85, e2096–e2018. doi: 10.1128/AEM.02096-18
Slobodkina, G. B., Baslerov, R. V., Novikov, A. A., Viryasov, M. B., Bonch-Osmolovskaya, E. A., and Slobodkin, A. I. (2016). Inmirania thermothiophila gen. nov., sp. nov., a thermophilic, facultatively autotrophic, sulfur-oxidizing gammaproteobacterium isolated from a shallow-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 66, 701–706. doi: 10.1099/ijsem.0.000773
Tao, C. H., Lin, J., Guo, S. Q., Chen, Y. J., Wu, G. H., Han, X. Q., et al. (2012). First active hydrothermal vents on an ultraslow-spreading center: southwest Indian Ridge. Geology 40, 47–50. doi: 10.1130/g32389.1
Urich, T., Lanzen, A., Stokke, R., Pedersen, R. B., Bayer, C., Thorseth, I. H., et al. (2014). Microbial community structure and functioning in marine sediments associated with diffuse hydrothermal venting assessed by integrated meta-omics. Environ. Microbiol. 16, 2699–2710. doi: 10.1111/1462-2920.12283
Uritskiy, G. V., Diruggiero, J., and Taylor, J. (2018). MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6:158. doi: 10.1186/s40168-018-0541-1
Van Dover, C. L., Humphris, S. E., Fornari, D., Cavanaugh, C. M., Collier, R., Goffredi, S. K., et al. (2001). Biogeography and ecological setting of Indian Ocean hydrothermal vents. Science 294, 818–823. doi: 10.1126/science.1064574
Vuillet, L., Kojadinovic, M., Zappa, S., Jaubert, M., Adriano, J. M., Fardoux, J., et al. (2007). Evolution of a bacteriophytochrome from light to redox sensor. EMBO J. 26, 3322–3331. doi: 10.1038/sj.emboj.7601770
Wang, Y., and Qian, P.-Y. (2009). Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One 4:e7401. doi: 10.1371/journal.pone.0007401
Wu, Y. W., Simmons, B. A., and Singer, S. W. (2016). MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607.
Zeng, X., Alain, K., and Shao, Z. (2021). Microorganisms from deep-sea hydrothermal vents. Mar. Life Sci. Technol. 3, 204–230.
Zhang, L., Kang, M., Xu, J., Xu, J., Shuai, Y., Zhou, X., et al. (2016). Bacterial and archaeal communities in the deep-sea sediments of inactive hydrothermal vents in the Southwest India Ridge. Sci. Rep. 6:25982. doi: 10.1038/srep25982
Keywords: deep-sea hydrothermal vents, SWIR, thiohalomonadales, metagenomics, red light photosynthesis
Citation: Wang Y, Bi H-Y, Chen H-G, Zheng P-F, Zhou Y-L and Li J-T (2022) Metagenomics Reveals Dominant Unusual Sulfur Oxidizers Inhabiting Active Hydrothermal Chimneys From the Southwest Indian Ridge. Front. Microbiol. 13:861795. doi: 10.3389/fmicb.2022.861795
Received: 25 January 2022; Accepted: 12 April 2022;
Published: 25 May 2022.
Edited by:
Philippe M. Oger, UMR 5240 Microbiologie, Adaptation et Pathogenie (MAP), FranceReviewed by:
Mirjam Perner, GEOMAR Helmholtz Center for Ocean Research Kiel, Helmholtz Association of German Research Centres (HZ), GermanyAnaïs Cario, UMR 5026 Institut de Chimie de la Matière Condensée de Bordeaux (ICMCB), France
Copyright © 2022 Wang, Bi, Chen, Zheng, Zhou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Wang, d2FuZ3lvbmdAc3oudHNpbmdodWEuZWR1LmNu
†These authors have contributed equally to this work
 Yong Wang
Yong Wang Hong-Yu Bi2†
Hong-Yu Bi2† Peng-Fei Zheng
Peng-Fei Zheng Ying-Li Zhou
Ying-Li Zhou Jiang-Tao Li
Jiang-Tao Li