- 1Jeju Branch Office, Honam Regional Center for Disease Control and Prevention, Korea Diseases Control and Prevention Agency, Jeju, South Korea
- 2Division of Infectious Disease Diagnosis Control, Honam Regional Center for Disease Control and Prevention, Korea Diseases Control and Prevention Agency, Gwangju, South Korea
- 3Jeju National Quarantine Station, Korea Diseases Control and Prevention Agency, Jeju, South Korea
The number of coronavirus disease (COVID-19)-positive cases has increased in Jeju Island, Republic of Korea. Identification and monitoring of new mutations in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) are extremely important to fighting the global pandemic. We report a breakout of the B.1.620 lineage, harboring the E484 mutation in the virus spike protein in a general hospital on Jeju Island. A cluster of cases was detected between August 4 and September 10, 2021, involving 20 patients positive for COVID-19 of 286 individuals exposed to the virus, comprising hospital patients, staff, and caregivers. We analyzed the epidemiological characteristics and spike proteins mutation sites using Sanger sequencing and phylogenetic analysis on these 20 patients. By analyzing genomic variance, it was confirmed that 12 of the confirmed patients harbored the SARS-CoV-2 B.1.620 lineage. The breakthrough rate of infection was 2% in fully vaccinated individuals among these patients. Next clade analysis revealed that these SARS-CoV-2 genomes belong to clade 20A. This is the first reported case of SARS-CoV-2 sub-lineage B.1.620, although the B.1.617.2 lineage has prevailed in August and September in Jeju, which has a geographical advantage of being an island. We reaffirm that monitoring the spread of SARS-CoV-2 variants with characteristic features is indispensable for controlling COVID-19 outbreaks.
Introduction
Outbreaks of coronavirus disease (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its various lineages are being reported worldwide (Tegally et al., 2020; Hodcroft et al., 2021). The distinct SARS-CoV-2 lineages may affect transmissibility, diseases severity, and vaccine efficacy (Dudas et al., 2021). Here, we describe an outbreak of the B.1.620 lineage in Jeju Island, Republic of Korea.
The first case of B.1.620 was reported from Lithuania. It has also been reported from Central Africa and several European states, including France, Germany, and Spain (Dudas et al., 2021; Singh et al., 2021). In Republic of Korea, B.1.620 was imported from Kenya and Malawi in March, 2021 (Park et al., 2022). B.1.620 carries 26 mutations and numerous deletions (Dudas et al., 2021) in its genomes, which are different from that of the reference Wuhan Hu-1 strain (Rambaut et al., 2020; O’Toole et al., 2021). In the Pangolin nomenclature, these genomes are assigned to clade 20A (Rambaut et al., 2020; Pirnay et al., 2021). They also carry the D614G mutation, which promotes the infectivity of SARS-CoV-2 by enhancing its interactions with the host angiotensin-converting enzyme 2 (ACE2) receptor via increasing the affinity of its receptor-binding domain (RBD) for ACE2 (Hou et al., 2020; Korber et al., 2020; Yurkovetskiy et al., 2020; Plante et al., 2021). The E484K mutation in the RDB occurs at the periphery of the RDB–ACE2 interface and may introduce new salt bridges with E35/E75 of ACE2 (Lan et al., 2020; Dudas et al., 2021). Therefore, B.1.620 may escape antibody-mediated immunity via interaction with ACE2 (Dudas et al., 2021). A previous study reported that B.1.620 is found 2.4 times in vaccine breakthrough cases compared with its population prevalence (Dudas et al., 2021; Šimaitis, 2021).
B.1.620 was formerly variants under monitoring (VUM) as of July 14, 2021 but was reclassified as formerly monitored variants on November 9, 2021 (WHO, 2021). Formerly monitored variants must meet at least one of the following criteria: (i) no longer spreading at the level of global public health significance, (ii) have been circulating over the long term without any epidemiological concern, or (iii) do not have any concerning properties based on scientific evidence (WHO, 2021).
In this study, we describe the outbreak of the SARS-CoV-2 B.1.620 lineage, the first outbreak in the general hospital of Jeju Island, Republic of Korea. What remains unknown is the influence of B.1.620 lineage on transmission dynamics in 286 individuals, comprising staff, patients, and caregivers, who may or may not be fully vaccinated. Therefore, we describe the epidemiological profile, including vaccination status, type, and symptoms. Over 10% of polymerase chain reaction (PCR)-positive SARS-CoV-2 cases were sequenced and analyzed phylogeny of B.1.620 lineage compared with other SARS-CoV-2 B.1.620 cases in Jeju Island.
Materials and Methods
RNA Isolation and Reverse Transcription (RT)-PCR
COVID-19-positive specimens were provided by the Jeju Special Self-Governing Province Institute of Environment Research. The specimens were handled in a Class II biosafety cabinet (Thermo scientific 1300 series A2) at a biosafety level 2 (BL2) laboratory. RNA was isolated using an RNA extraction kit (Qiagen, Valencia, CA, United States) according to the manufacturer’s instructions. cDNA was synthesized using SuperScript IV First-Strand Synthesis System (Invitrogen, Carlsbad, CA, United States). PCR was conducted using PrimeSTAR GXL DNA Polymerase (Takara, Shiga, Japan) from February to August, 2021. RT-PCR was conducted using a DiaStar 2 × OneStep RT-PCR premix kit (SolGent, Daejeon, South Korea) and a SEQMAX qPCR one-step master mix (Nine Korea, South Korea). Sequences of the primers used for RT-PCR are shown in Table 1. To detect mutations of SARS-CoV-2 spike protein, L71/R75, L76/R79, L80/R84 primer sets were used to conduct RT-PCR, independently. RT-PCR was performed under the following conditions: an initial reverse transcription step at 50°C for 30 min followed by a denaturation step at 95°C for 5 to 15 min. This was followed by 35 cycles of 30 s at 95°C, 30 s at 58°C, 1 min 30 s at 68°C, and a final extension step at 68°C for 7 min.

Table 1. A list of primer sequences used for RT-PCR analysis and sanger sequencing of spike protein.
DNA Sequencing and Analysis
DNA sequencing was performed using a standard protocol (Cosmo Genetech, Seoul, Korea). To analyze the sequences, L71/R75, L76/R79, L80/R84 and inner primers, including L73/R73, L78/R77, L83/R82, were used (Table 1). With sequence data from the COVID-19-positive specimens in Jeju Island, full-length sequences of SARS-CoV-2 spike proteins from different geographical origins were downloaded in FASTA format from GISAID1. The non-coding 3′ and 5′ regions were trimmed using CLC Genomic Workbench 5.0.1 software (CLC bio, Denmark). Multiple sequence alignment was performed using the MAFFT algorithm. Phylogenetic analysis was performed using the maximum likelihood method using Molecular Evolutionary Genetics Analysis version 10.2.5 (MEGA X). Branch support was calculated using bootstrapping, consisting of 1,000 alignments. SARS-CoV-2 B.1.620 lineage genomes from the outbreak cases of the hospital in Jeju Island were classified using Nextclade2. The hCoV-19 genome sequences of B.1.620 strain in Jeju were deposited in GISAID (Table 2).
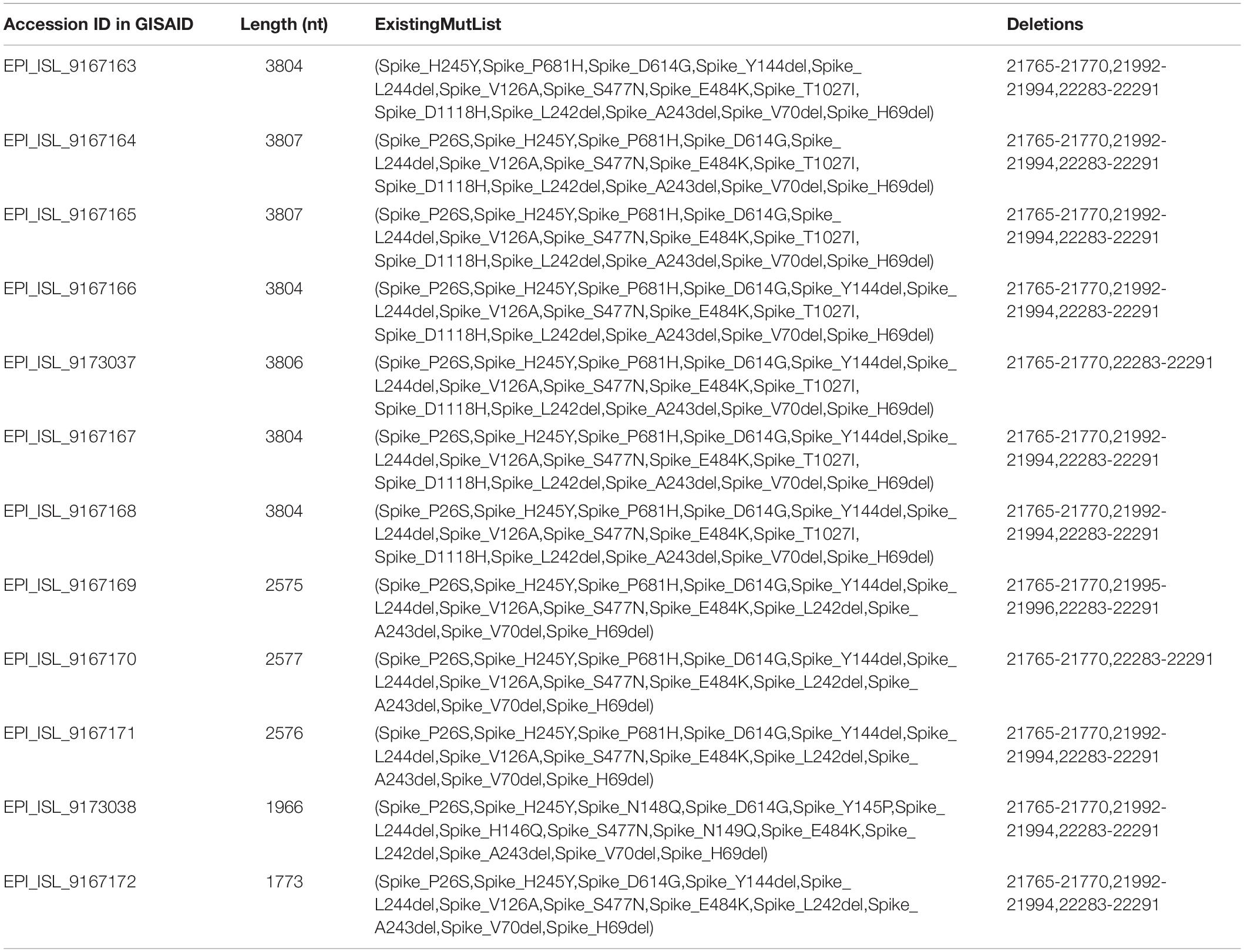
Table 2. Spike protein mutations with their locations identified using 12 sequences of general hospital cases.
Calculation of Infection Attack Rate (IAR)
We calculated the attack rate in 286 individuals of a hospital by vaccination status. We estimated 95% confidence interval (CI) and relative risk (RR), which is the ratio of the attack rates of a disease among vaccinated and not-vaccinated individuals.
Results
Epidemiological Characteristics of the COVID-19 Outbreak Cases
A cluster of cases occurred in the general hospital in Jeju Island between August 4 and September 10, 2021. This cluster included 286 individuals, comprising staff, patients, and caregivers of the hospital. Among them, 20 people, that is, 8 patients, 8 caregivers, and 4 family caregivers, were confirmed to be COVID positive by September 2 (Table 3). Among general hospital individuals exposed to the virus, 12.9% (37) had received their first dose of COVID-19 vaccine, 69.9% (200) were fully vaccinated, and 17.1% (49) were not vaccinated. The vaccinated individuals had received the AstraZeneca, Pfizer-BioNTech, Moderna, or Janssen vaccine. Among the fully vaccinated persons, 60.8% (174) were AstraZeneca vaccine recipients, 7.3% (21) had received the Pfizer-BioNTech vaccine, and 1.7% (5) were Moderna vaccine recipients and were vaccinated 14 days before the outbreak. Therefore, the breakthrough rate of infection was 2% in completing primary COVID-19 vaccination. Of the 20 positive patients, 7 had been vaccinated one time, 6 were completely vaccinated, and 7 had not been vaccinated. The most common symptoms of the positive patients were shiver/chills (20%), cough (10%), sore throat (5%), muscle ache (5%). Thirteen patients did not show any symptoms (Table 3).
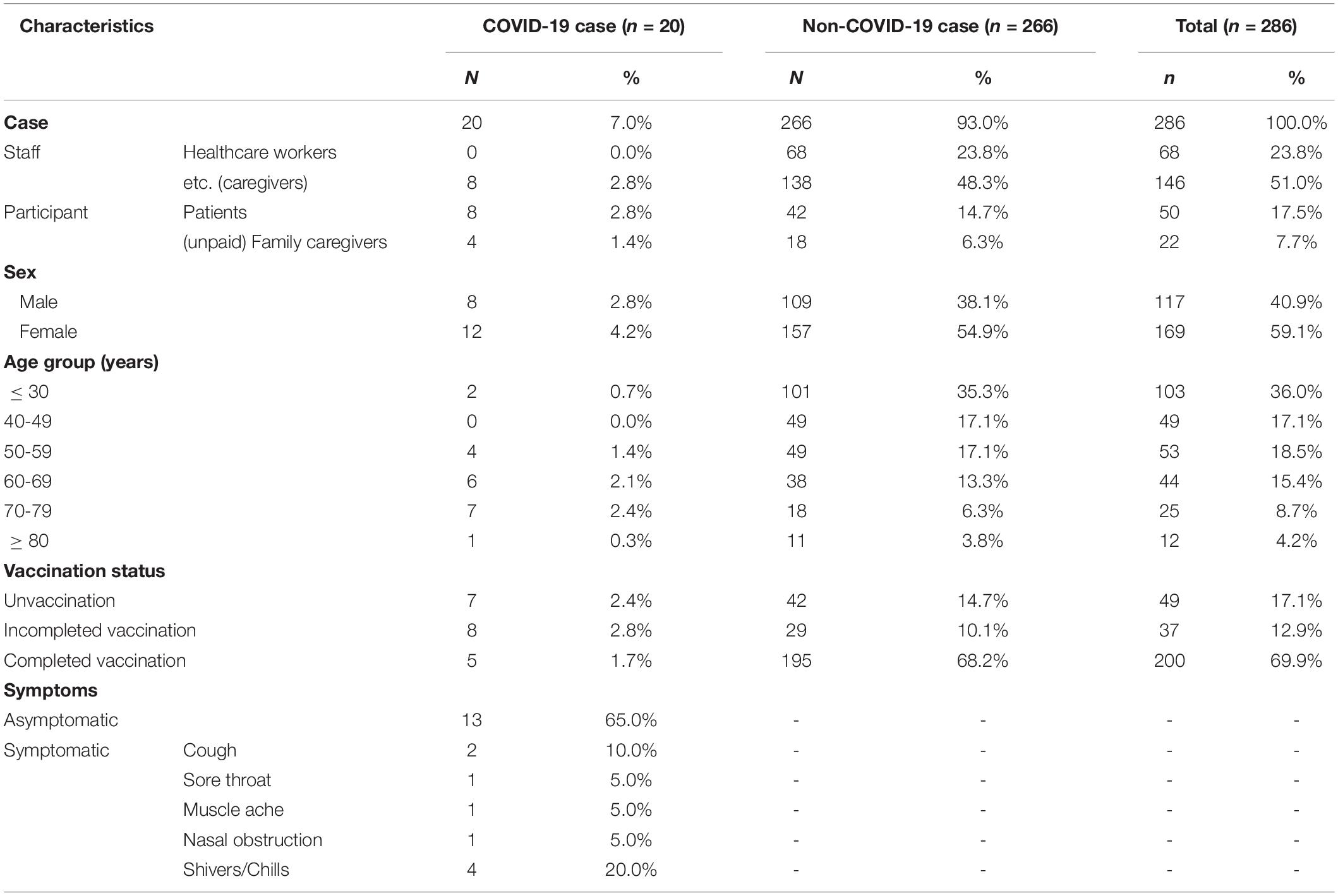
Table 3. Epidemiological characteristics of confirmed cases for the COVID-19 outbreak at the general hospital in Jeju Island, Republic of Korea, in August 2021.
The attack rate was 14.3 and 5.5% in the not-vaccinated and vaccinated individuals, respectively. The relative risk (RR) was 5.5% in vaccinated individuals. The vaccinated individuals were lower than unvaccinated individuals (RR < 1, 95% confidence interval < 0.02). In addition, the total vaccine effectiveness against infection was 96.2%. The vaccine effectiveness was 84.9 and 98.3% in the first dose of COVID-19 vaccine and fully vaccinated individuals, respectively.
Phylogenetic Analysis of SARS-CoV-2 B.1.620 Lineage From the Outbreak Cases
The genome sequences obtained were compared with the reference SARS-CoV-2 isolate, Wuhan Hu-1, genome sequence (accession number: NC_045512.2). The hospital outbreak strains clustered closely around the other B.1.620 lineages in Jeju Island (Figure 1). The group of B.1.620 sequences was closer to the root of the B.1.1.7 lineage (i.e., the alpha variant) than to B.1.351 (the beta variant), P.1 (the gamma variant), and B.1.617.2 (the delta variant). The phylogenetic tree of B.1.620 lineages was composed of the outbreak cases (EPI_ISL_9167163 ∼ EPI_ISL_9167172, EPI_ISL_9173037, and EPI_ISL_9173038) and the other cluster in Jeju Island (EPI_ISL_9175620 ∼ EPI_ISL_9175628). SARS-CoV-2 sequences belonging to the B.1.620 lineages, which were detected in Jeju, were closely related to each other. In addition, we included the B.1.620 lineage, such as hCoV-19/Lithuania/LSMULKKGMMK10C170/2021 (EPI_ISL_1540683), hCoV-19/England/CAMC-13B04C1/2021 (EPI_ISL_1275749), Belgium/Aalst-OLVZ-8042639/2021 (EPI_ISL_1620228), and England/CAMC-139AFAE/2021 (EPI_ISL_1276088). The Nextclade analysis revealed that SARS-CoV-2 genomes from outbreak cases of hospitals in Jeju belong to clade 20A (Supplementary Table 1). Among clade 20A, B.1.620 lineages of Jeju closed with the other B.1.620 lineages, such as hCoV-19/Lithuania/LSMULKKGMMK10C170/2021 (EPI_ISL_1540683), hCoV-19/England/CAMC-13B04C1/2021 (EPI_ISL_1275749), and England/CAMC-139AFAE/2021 (EPI_ISL_1276088) (Figure 1).
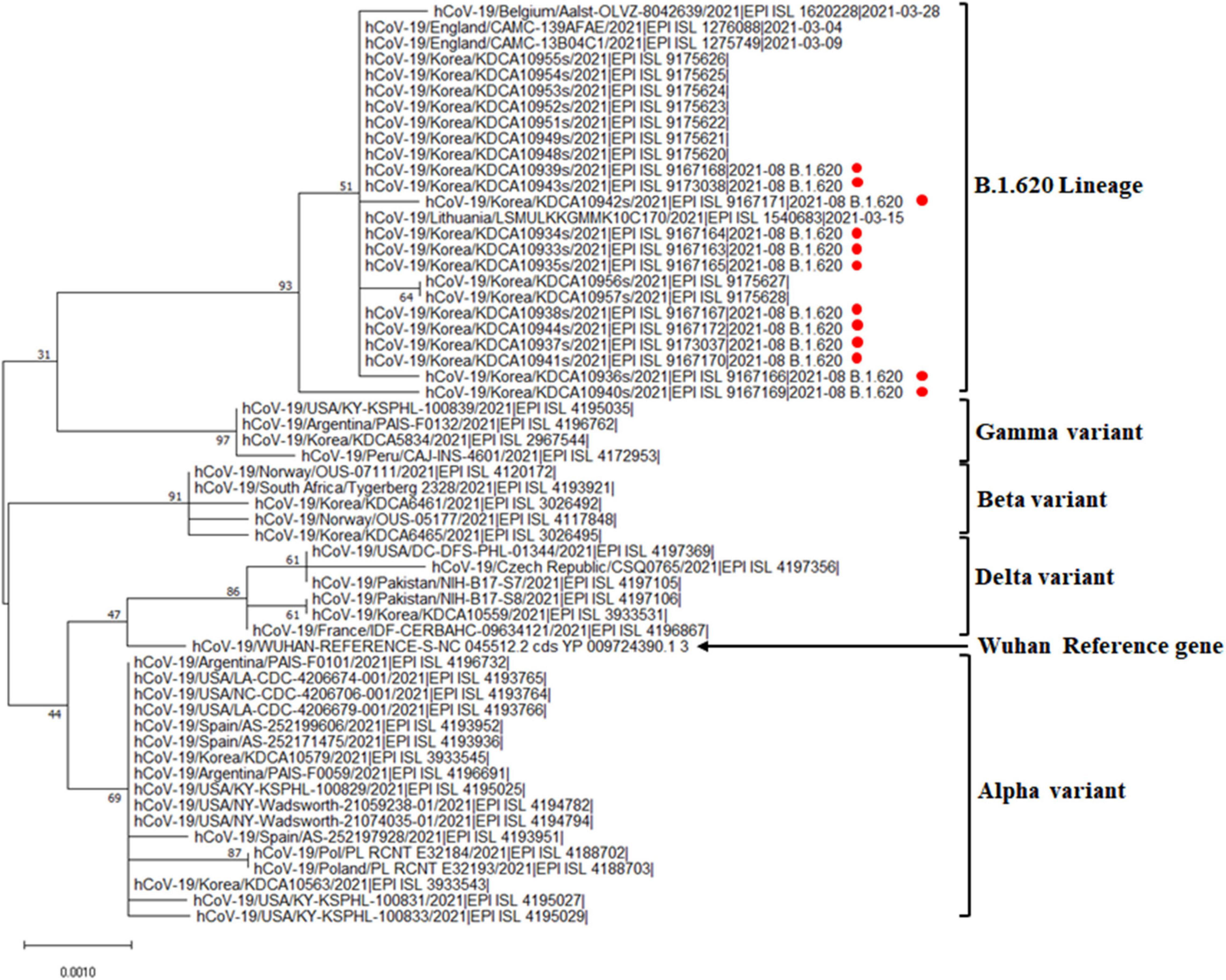
Figure 1. Phylogenetic analysis of the SARS-CoV-2 B.1.620 lineage in the general hospital of Jeju Island. Phylogenetic analysis of alpha, beta, gamma, delta variants, and B.1.620 lineage with outbreak cases in Jeju Island. Red circles indicate the genomes from hospital outbreak cases in Jeju Island.
Conclusion
We described the first outbreak of SARS-CoV-2 lineage B.1.620 in Jeju Island. B.1.620 has been associated with many VOC-like mutations and deletions. Our phylogenetic analysis revealed that the cluster of hospital outbreaks was closed compared with those of other variants. Furthermore, the clustering pattern with branching suggested that this B.1.620 lineage hospital outbreak could indicate community spread with the other B.1.620 lineages in Jeju from April to September 2021.
B.1.620 was introduced to Jeju on April 27, 2021. After introduction, the proportion of B.1.620 increased from April (2.1%) to June (18.9%) (Supplementary Figure 1). However, B.1.617.2 (the delta variant) has become dominant since July (59.3%) to September (96.8%) in Jeju Island (Supplementary Figure 2). Interestingly, the hospital outbreak of the B.1.620 lineage occurred in August and September 2021, although the B.1.617.2 lineage prevailed all over Jeju Island.
Hospitals could be the susceptible place to outbreaks because of highly transmissible variants with less-effective vaccines (Susky et al., 2021). In this study, the outbreak in the hospital of Jeju might contain several epidemiological aspects of learnings. First, most of COVID-19 sub-lineage B.1.620 positive cases were asymptomatic or have only mild symptoms. Secondly, despite vaccination, hospital exposers from the virus may result in transmission of the B.1.620 lineage. Thus, we conclude that continuous monitoring of COVID-19 cases and SARS-CoV-2 variants in Jeju Island is essential for controlling virus transmission and subsequent outbreaks.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.gisaid.org/, EPI_ISL_9167163 – EPI_ISL_9167172; EPI_ISL_9173037; and EPI_ISL_9173038.
Author Contributions
Y-RH contributed to the conception, design, data acquisition, and drafting of the manuscript. E-SS participated in epidemiological surveys. H-JK and E-HH conducted the experiment. J-SP advised the experimental procedure and treatment of specimens. Y-SC conceived the entire study and helped draft the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by a fund (6137-300-210-13) of the Korea Centers for Disease Control and Prevention Agency (KDCA).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.860535/full#supplementary-material
Footnotes
References
Dudas, G., Hong, S. L., Potter, B. I., Calvignac-Spencer, S., Niatou-Singa, F. S., Tombolomako, T. B., et al. (2021). Emergence and spread of SARS-CoV-2 lineage B.1.620 with variant of concern-like mutations and deletions. Nat. Commun. 12:5769. doi: 10.1038/s41467-021-26055-8
Hodcroft, E. B., Zuber, M., Nadeau, S., Vaughan, T. G., Crawford, K. H. D., Althaus, C. L., et al. (2021). Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature 595, 707–712.
Hou, Y. J., Chiba, S., Halfmann, P., Ehre, C., Kuroda, M., Dinnon, K. H., et al. (2020). SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468. doi: 10.1126/science.abe8499
Korber, B., Fischer, W. M., Gnanakaran, S., Yoon, H., Theiler, J., Abfalterer, W., et al. (2020). Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:e819. doi: 10.1016/j.cell.2020.06.043
Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., et al. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220. doi: 10.1038/s41586-020-2180-5
O’Toole, A., Scher, E., Underwood, A., Jackson, B., Hill, V., Mccrone, J. T., et al. (2021). Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 7:veab064. doi: 10.1093/ve/veab064
Park, A. K., Kim, I. H., Man Kim, H., Lee, H., Lee, N. J., Kim, J. A., et al. (2022). SARS-CoV-2 B.1.619 and B.1.620 lineages. South Korea, 2021. Emerg. Infect. Dis. 28, 415–419. doi: 10.3201/eid2802.211653
Pirnay, J. P., Selhorst, P., Hong, S. L., Cochez, C., Potter, B., Maes, P., et al. (2021). Variant analysis of SARS-CoV-2 genomes from belgian military personnel engaged in overseas missions and operations. Viruses 13:1359. doi: 10.3390/v13071359
Plante, J. A., Liu, Y., Liu, J., Xia, H., Johnson, B. A., Lokugamage, K. G., et al. (2021). Author correction: spike mutation D614G alters SARS-CoV-2 fitness. Nature 595:E1. doi: 10.1038/s41586-021-03657-2
Rambaut, A., Holmes, E. C., O’toole, A., Hill, V., Mccrone, J. T., Ruis, C., et al. (2020). A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 5, 1403–1407. doi: 10.1038/s41564-020-0770-5
Šimaitis, A. (2021). A Situation Report to the Lithuanian Government Regarding SARS-CoV-2 [Online]. Available online at: https://lrv.lt/uploads/main/documents/files/20210511%20COVID-19%20situacijos%20ap%C5%BEvalga.pdf (accessed November 18, 2021).
Singh, D. D., Parveen, A., and Yadav, D. K. (2021). SARS-CoV-2: emergence of new variants and effectiveness of vaccines. Front. Cell Infect. Microbiol. 11:777212. doi: 10.3389/fcimb.2021.777212
Susky, E. K., Hota, S., Armstrong, I. E., Mazzulli, T., Kestenberg, S., Casaubon, L. K., et al. (2021). Hospital outbreak of the severe acute respiratory coronavirus virus 2 (SARS-CoV-2) delta variant in partially and fully vaccinated patients and healthcare workers in Toronto, Canada. Infect. Control Hosp. Epidemiol. 28, 1–4. doi: 10.1017/ice.2021.471
Tegally, H., Wilkinson, E., Giovanetti, M., Iranzadeh, A., Fonseca, V., Giandhari, J., et al. (2020). Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv [Preprint] doi: 10.1101/2020
WHO (2021). Tracking SARS-CoV-2 Variants [Online]. Available online at: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed November 10, 2021).
Keywords: COVID-19, SARS-CoV-2, spike protein, Republic of Korea, lineage B.1.620
Citation: Ha Y-R, Shin E-s, Kim H-J, Hyeon E-H, Park J-S and Chung Y-S (2022) Outbreak of Severe Acute Respiratory Syndrome Coronavirus-2 B.1.620 Lineage in the General Hospital of Jeju Island, Republic of Korea. Front. Microbiol. 13:860535. doi: 10.3389/fmicb.2022.860535
Received: 23 January 2022; Accepted: 07 March 2022;
Published: 05 April 2022.
Edited by:
Pragya Dhruv Yadav, ICMR-National Institute of Virology, IndiaReviewed by:
Arif Ansori, Airlangga University, IndonesiaKenji Sadamasu, Tokyo Metropolitan Institute of Public Health, Japan
Copyright © 2022 Ha, Shin, Kim, Hyeon, Park and Chung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoon-Seok Chung, cm9sbHN0b25lOTNAa29yZWEua3I=
 Young-Ran Ha
Young-Ran Ha Een-suk Shin2
Een-suk Shin2 Yoon-Seok Chung
Yoon-Seok Chung