
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 March 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.858857
This article is part of the Research Topic Insights in Evolutionary and Genomic Microbiology: 2021 View all 12 articles
Composite genomic islands (GIs) are useful models for studying GI evolution if they can revert into the previous components. In this study, CGI48—a 48,135-bp native composite GI that carries GI21, whose homologies specifically integrated in the conserved yicC gene—were identified in Shewanella putrefaciens CN-32. CGI48 was integrated into the tRNATrp gene, which is a conserved gene locus for the integration of genomic islands in Shewanella. Upon expressing integrase and excisionase, CGI48 and GI21 are excised from chromosomes via site-specific recombination. The shorter attachment sites of GI21 facilitated the capture of GI21 into CGI48. Moreover, GI21 encodes a functional HipAB toxin–antitoxin system, thus contributing to the maintenance of CGI48 in the host bacteria. This study provides new insights into GI evolution by performing the excision process of the inserting GI and improves our understanding of the maintenance mechanisms of composite GI.
Genomic islands (GIs) are discrete DNA segments acquired by horizontal transfer, and they always differ among closely related strains. GIs vary in size from a few to several kilobase pairs and have a mosaic structure that evolves by gene acquisition and loss (Bellanger et al., 2014). Horizontal transfer of GIs can be advantageous for the host, influencing traits, such as pathogenicity, symbiosis, metabolism, phage resistance, and fitness (Dobrindt et al., 2004; Bellanger et al., 2014). Therefore, an understanding of GI evolution is critical for understanding the acquisition of these important adaptive traits.
Composite GI formation is a special type of GI evolution in which one mobile genetic element (MGE) is inserted within another or into the attachment sites of a resident GI (tandem accretion; Bellanger et al., 2014). Many composite GIs have been found through genome comparison (Bellanger et al., 2014), such as the SGI1 variant SGI1-B2 from Proteus mirabilis (Lei et al., 2015), ICESt1 and CIME302 elements of Streptococcus thermophilus (Burrus et al., 2000), and ICE6013 from Staphylococcus aureus (Smyth and Robinson, 2009). The native composite GIs have likely undergone some complicated recombination events; therefore, it is difficult to reconstruct their precise evolutionary history. To date, the formation processes of a few native composite GIs have been determined, such as the tripartite integrative and conjugative element (ICE) assembled through recombination from two GIs with integrases and one ICE without an integrase in Mesorhizobium ciceri (Haskett et al., 2016), the tandem structure of GIprfC inserting in the integration site for SXT/R391 ICEs in Pseudoalteromonas sp. SCSIO 11900 in our previous study (Wang et al., 2017). Native composite islands that can replicate their evolutionary processes under laboratory conditions would be especially useful for improving our understanding of GI evolution. Interestingly, how composite GIs maintain structural stability should also be explored.
Toxin–antitoxin (TA) systems were originally discovered on conjugative plasmids and participated in their stable maintenance in host bacteria (Ogura and Hiraga, 1983; Roberts et al., 1994). The TA system consists of two neighboring genes, encoding a stable toxin killing the cell or inhibiting cell growth and an unstable antitoxin that masks its toxicity (Wang et al., 2021). A proposed mechanism post-segregationally killing (PSK) was established based on the differential stability of the antitoxin and toxin components. In PSK, plasmid-loss cells do not survive, so the plasmid is maintained in the population (Jurenas et al., 2022). Currently, TA systems have also been found to be ubiquitous in bacterial chromosomes and have been suggested to contribute to the maintenance of integrative MGEs. For example, the MosAT system promotes the maintenance of SXT family ICE carried by some Vibrio cholerae strains (Wozniak and Waldor, 2009); the ParESO/CopASO system stabilizes prophage CP4So in Shewanella oneidensis (Yao et al., 2018). Whether the TA system also participates in the maintenance of composite GI is unknown. In this study, a new composite island CGI48 was identified and characterized from Shewanella putrefaciens CN32 using genome comparison and excision assay. It evolved by inserting a 21-kb genomic island GI21 into the internal region of CGI48. We further show that GI21 carries a functional HipAB toxin–antitoxin system and contributes to the maintenance of CGI48 in the bacterial host.
The bacterial strains and plasmids used in this study are listed in Table 1. Shewanella was grown in LB medium at 30°C. Escherichia coli WM3064 was grown in LB medium containing 0.3 mM 2,6-diamino-pimelic acid (DAP) at 37°C. Chloramphenicol (Cm; 30 μg ml−1), kanamycin (50 μg ml−1), and ampicillin (100 μg ml−1) were used in E. coli, and chloramphenicol (10 μg ml−1) was used in Shewanella. Isopropyl-β-D-thiogalactopyranoside (IPTG) was used as an inducer.
The primers used in this study are listed in Table 2. The encoding regions of xis21, xisPO1, xisANA3, int48, and int2894 were amplified from the original bacterial host and cloned into the EcoRI and BamHI sites of pHGECm using T4 ligase, generating pXis21, pXisPO1, pXisANA3, and pInt48. The encoding regions of hipA, hipB, and hipAB were amplified from CN32 and inserted into the SalI and PstI sits of pCA24N, generating pHipA, pHipB, and pHipAB. The promoter and encoding region of hipAB was amplified from CN32 and inserted into pMD19-T, generating pMD19-T-hipAB. To construct the lacZ reporter plasmid pHGI01-Pint, the reporter region of the integrase gene Sputcn32_2900 was amplified with the primer pair pHGI01-Pint-F/−R and fused with the lacZ gene in pHGI01. Then, the integrative plasmid pHGI01-Pint was transferred into CN32 and ΔhipAB by conjugation and integrated into the promoter region of Sputcn32_2900, generating CN32 Pint::lacZ and ΔhipAB Pint::lacZ. The primer sets mob-F/int-R and Int-F/lacZ-R were used to confirm the construct. To construct pHGR01-PhipA, the promoter of hipAB was amplified with primers pHGR01-PhipA-F/−R from CN32, and inserted into the promoterless-lacZ reporter plasmid pHGR01.
The deletion mutant ΔhipAB was constructed based on pK18mobsacB-Cm as described previously (Wang et al., 2015). Briefly, the upstream and downstream regions of hipAB were amplified from CN32 using the primers listed in Table 2 and inserted into pK18mobsacB-Cm using T4 ligase, producing pK18Cm-hipAB. Then, pK18Cm-hipAB was introduced into CN32 by conjugation. After mating, cells were spread on LB plates containing Cm to screen the single crossover mutant in which pK18Cm-hipAB had integrated into the CN32 genome. The mutant was then grown on LB medium without antibiotics for 8 h. To select mutants in which the second recombination had occurred, the culture was diluted, spread on LB medium containing 10% sucrose, and grown at 30°C for 24–36 h. Single colonies were transferred onto LB- and LB-containing Cm plates simultaneously, and colonies sensitive to Cm were collected and confirmed by PCR followed by DNA sequencing.
The plasmids in this study were transferred from E. coli WM3064 into Shewanella strains by conjugation assays as described previously (Wang et al., 2015). Briefly, equal amounts of donor and recipient cells were mixed and dropped onto LB medium containing DAP. The plates were incubated at 30°C for 6–8 h, and cells were collected from the lawn and streaked on LB medium with antibiotics to select for transconjugants.
Specific β-galactosidase activity was determined by monitoring the absorbance at 420 nm using the Miller assay (Miller, 1972). To determine the promoter activity of hipAB under overexpression of HipB and HipB-HipA, plasmids pHipB or pHipAB were transformed into the E. coli host carrying the reporter plasmid pHGR01-PhipA. Overnight cultures were diluted 1:100 in LB with Kan and Cm and induced with 0.1 mM IPTG at an OD600 of 1.0. After induction for 2 h, cells were collected to determine the β-galactosidase activity.
For GI21, GISspANA3, GISpuPO1, and CGI48, attB/gyrB indicated the excision rate of the target GIs after excision. We conducted real-time quantitative PCR (qPCR) assays to quantify the attB of these GIs as previously reported methods (Burrus and Waldor, 2003; Wang et al., 2017). The primers used for the qPCR assays are listed in Table 2, and chromosomal gyrB was used as the reference gene. To test the regulation of Xis and Int on the excision of GI21, GISspANA3, GISpuPO1, and CGI48, pXis21-, pXisPO1-, pXisANA3-, and pInt48-containing strains were induced with 1.0 mM IPTG for 6 h at an OD600 of 0.8–1.
Both CGI48 and GI21 are non-replicable, and loss of CGI48 and GI21 only occurs after their excision. Therefore, to visualize the loss of CGI48 and GI21, the wild-type and ΔhipAB strains carrying pXis21 or pInt48 were induced with 1 mM IPTG for 6 h to overproduce Xis21 (to induce GI21 excision) or Int48 (to induce CGI48 excision). Then, the cells were plated on LB plates containing X-gal to calculate the numbers of white colonies (losing CGI48 or GI21) plus blue colonies, and the white colonies were also confirmed by PCR assay.
The contribution of HipA/HipB TA system to plasmid stability was tested as described previously (Yao et al., 2015). Overnight cultures of E. coli BW25113 containing plasmid pHipAB or empty vector pCA24N were grown in LB medium with Cm. Then, the preculture was used to inoculate 3 ml LB without antibiotics. Every 12 h of growth, bacterial suspensions were diluted 1,000-fold in 3 ml fresh LB medium. The cultures were serially diluted in 10-fold dilution steps from 0 to 108 h, and 10 μl was dropped on LB plates with or without Cm. The colony-forming unit (CFU) assay was conducted every 12 h for 108 h, and the number of CFUs was determined. Each experiment was performed in triplicate with two independent cultures.
Comparing the genome sequence of S. putrefaciens CN32 with the related strain S. putrefaciens W3-18-1, a large region within 3,340,000–3,400,000 of CN32 was absent in the same gene locus (1,160,000–1,170,000) of W3-18-1 (Figure 1A), suggesting that this region was acquired horizontally. Moreover, the internal sequence within 3,360,000–3,380,000 of this large region showed high homology with another region 335,000–360,000 of W3-18-1 (Figure 1B). These results suggested that the region within 3,340,000–3,400,000 of CN32 is a putative composite genomic island. It is 48 kb in length; thus, it is designated CGI48 hereafter (Table 3). Further analysis showed that region 3,360,000–3,380,000 of CN32 contains a 21 kb genomic island (designated GI21), which exhibits sequence identity with genomic islands integrated in the conserved yicC gene, such as GISpuPO1 in S. putrefaciens W3-18-1, GISspANA3 in Shewanella sp. ANA-3, and GIPspSM9913 in Pseudoalteromonas sp. SM9913 (Figure 2A). GI21 exhibits 99% sequence identity with the two ends of GISpuPO1 in W3-18-1. The left region of GI21 contains an integrase and an excisionase gene next to the left attachment site (attL21), and the right region contains a putative hipA-hipB toxin–antitoxin pair next to the right attachment site (attR21). The middle region contains 12 genes encoding a restriction–modification system and hypothetical proteins (Figure 2A).
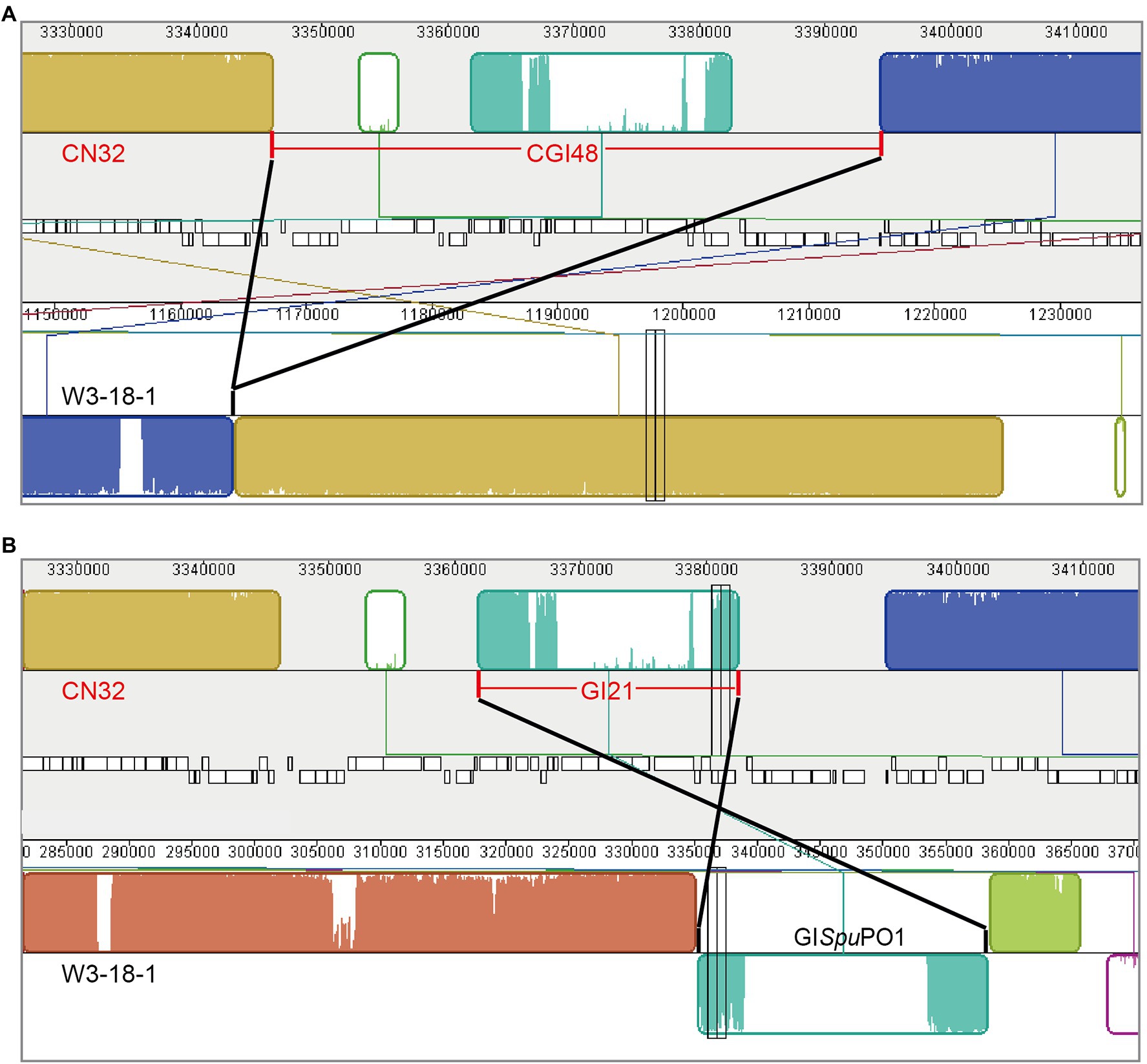
Figure 1. Schematic view of the composite island CGI48 in the CN32 genome. CGI48 (A) and its component genomic island GI21 (B) were identified by comparing the genome sequence of S. putrefaciens CN32 (CP000681) with S. putrefaciens W3-18-1 (CP000503) with Mauve.
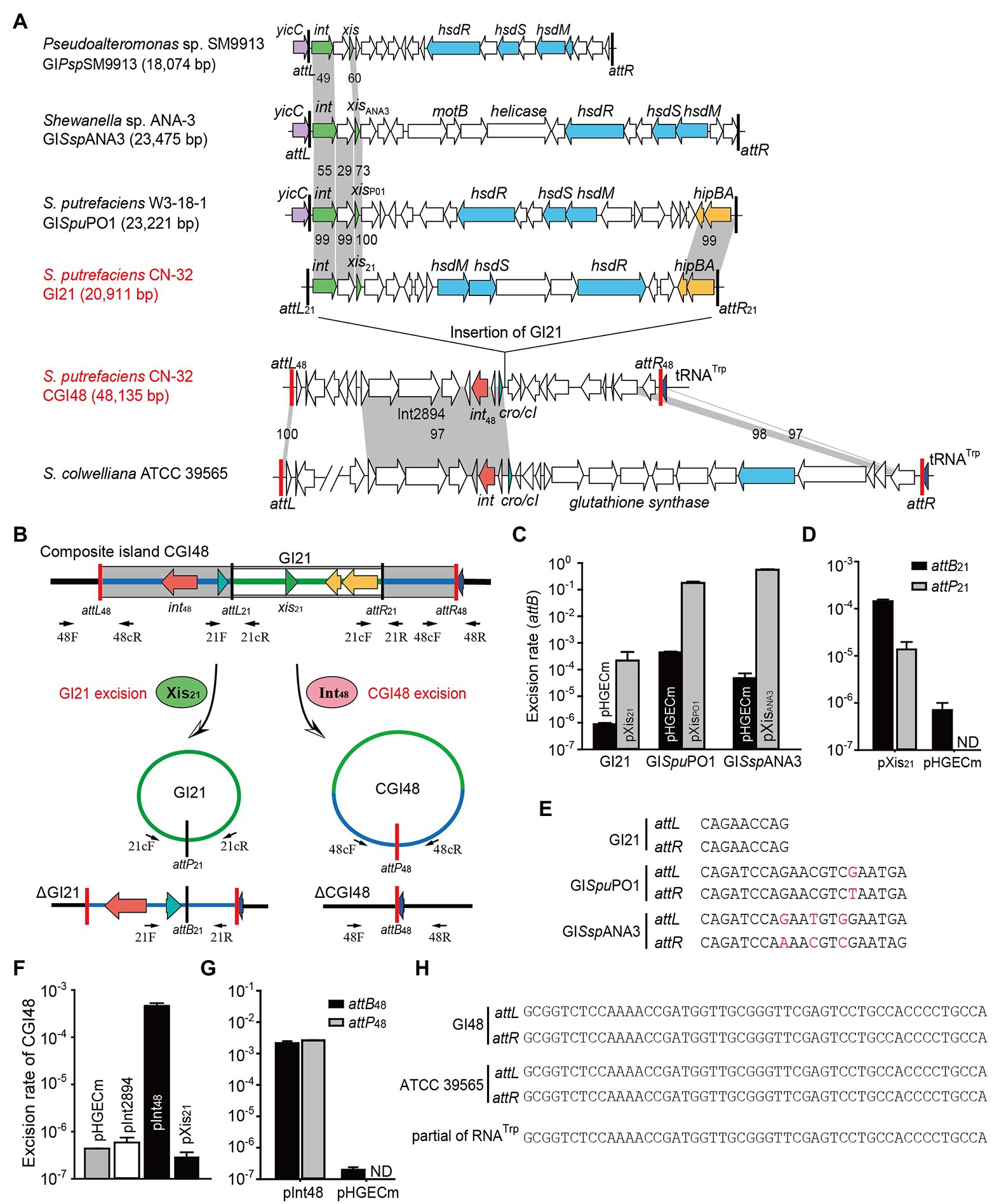
Figure 2. The composite island CGI48 and its component GI21 can be excised from the CN32 genome. (A) Sequence analysis of CGI48 with the related genomic islands. Open reading frames with putative functions are shown in different colors. The attL and attR attachment sites of CGI48 and GI21 are shown in red and black, respectively. The sequence of the GI integrated into tRNATrp of ATCC 39565 genome was in L876DRAFT_scaffold 00018.18_C (82,217–85,620 bp) to scaffold 00033.33_C (1,478–38,068 bp). (B) Schematic of the excision of CGI48 and GI21. (C) The excision rate of GI21, GISpuPO1, and GISspANA3 was quantified when cognate excisionase was overexpressed. (D) Comparison of the excision rate (attB21) and circular form of GI21 (attP21) in CN32 when Xis21 was overexpressed. ND indicates not detected. (E) Sequence comparison of the attachments of GI21, GISpuPO1, and GISspANA3. (F) The excision rate of CGI48 when Int48 was overexpressed. (G) Comparison of the excision rate (attB48) and circular form of CGI48 (attP48) in CN32 when Int48 was overexpressed. ND indicates not detected. (H) Sequence comparison of the attachment sites of CGI48 and GI in ATCC 39565 compared with the 3′ end of RNATrp in Shewanella.
Excision of GI followed by formation of circular forms of GI is prequisite for its horizontal transfer. Integrase is essential for GI excision and integration, and some GIs also encode recombination directionality factors (or excisionases Xis) directing the reaction toward excision (Lewis and Hatfull, 2001). We wondered whether GI21 can be excised from the CGI48 genome by recombining the attachment attL21 and attR21, and producing attB21 and attP21 sites (Figure 2B). Quantitative PCR (qPCR) was used to quantify the excision rate by measuring the percentage of cells in the culture containing attB21, which is only present after GI21 excision. In this assay, the amount of attB21 sites is compared to the amount of the reference gene gyrB, which is used to quantify the total number of cells in the culture. Excisionase Sputcn32_2902 (Xis21) was induced in strain CN32 with 1 mM IPTG for 6 h. Additionally, the excisionases XisPO1 and XisANA3 were also overexpressed in W3-18-1 and ANA-3 as a control. The results showed that Xis21 mediated GI21 excision, resulting in a 440-fold increase in the excision rate of GI21 and reaching (3.8 ± 0.3) × 10−4. However, the excision rate of GISpuPO1 and GISspANA3 reached 17.9%–55.6% when XisPO1 and XisANA3 were overexpressed, which was much higher than that of GI21 (Figure 2C). qPCR was also used to quantified the circular form of GI21 by measuring attP21, which is present after GI21 is circularized or replicated after excision. The number of attP21 is less than attB21, suggesting that GI21 is non-replicable in wild-type CN32 or expressing Xis21 (Figure 2D). PCR sequencing showed that the attachment sites of GISpuPO1 and GISspANA3 were 21 bp in length, and the attachment sites of GI21 were 9 bp (Figure 2E). In CGI48, GI21 was integrated in the untranslated region between Sputcn32_2899 and Sputcn32_2917, which encoded a predicted transcriptional regulator of the Cro/CI family and a conserved hypothetical protein, respectively (Figure 2A). The excision and integration of GI21 did not cause any sequence changes in the neighboring genes. The results suggested that GI21 can be excised from CN32 by site-specific recombination of attL21 and attR21, and the shorter attachment sites may greatly limit the recombination efficiency.
We then evaluated the excision of CGI48 (Figure 2B), and the integrase genes Sputcn32_2894 and Sputcn32_2897 were cloned into pHGECm for their overexpression. Overproduction of Sputcn32_2897 (named Int48) resulted in a 1.070-fold increase in the excision rate of CGI48 and reached (4.7 ± 0.6) × 10−4, and Sputcn32_2894 did not affect the excision of CGI48 (Figure 2F). Quantification of attP48 indicated that CGI48 is non-replicable in wild-type CN32 or expressing Int48 (Figure 2G). Sequence analysis showed that CGI48 was integrated in the 5′ end of tRNATrp, a conserved integration locus of GIs, such as the GI in S. colwelliana ATCC 39565 (Figure 2A). PCR sequencing confirmed that CGI48 and GI in S. colwelliana ATCC 39565 shared 100% identical attachment sites of 50 bp in length, and the excision and integration did not cause sequence changes in tRNATrp (Figure 2H). Phylogenetic tree analysis of Int21 and Int48 revealed that GI21 homologs are widely distributed in Shewanella, Pseudoalteromonas, Halomonas, and Vibrio strains (Figure 3A), and CGI48 homologs are widely distributed in Shewanella, Pseudomonas, Halomonas, and Photobacterium strains (Figure 3B). Collectively, CGI48 and the component GI21 can be excised from the CN32 genome, suggesting that CGI48 is an active composite island in host bacteria.
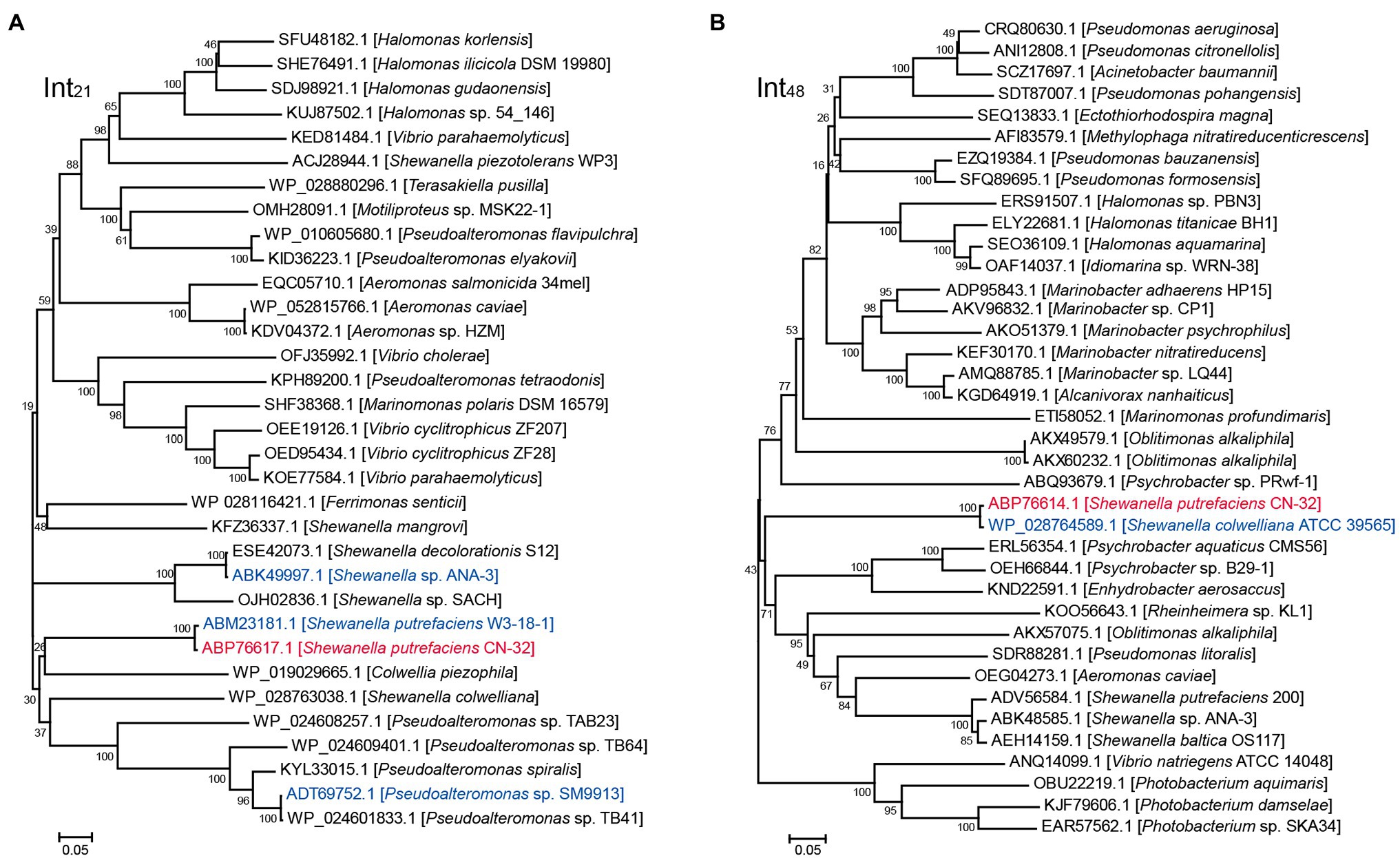
Figure 3. Phylogenetic analysis of Int21 and Int48 homologs. (A) Neighbor-joining phylogenetic tree of 32 homologs of Int21, and (B) 37 homologs of Int48 based on amino acid (aa) sequences. Proteins with homology to Int21 and Int48 were selected by BLASTp at the NCBI server. CN32 was indicated in red, and other genomic islands shown in Figure 2A were indicated in blue.
In GI21, two neighboring genes that are only 4 bp apart, Sputcn32_2916 and Sputcn32_2915, were identified as a putative hipA-hipB TA pair. In HipA/HipB TA system characterized in E. coli K-12, HipAK-12 toxin functions as a serine/threonine protein kinase that inhibits cell growth, and HipBK-12 antitoxin encoded by the gene upstream to hipA blocks its effects (Germain et al., 2013). Here, the putative hipA-hipB TA pair in GI21 has a genetic architecture reversed to that of hipB-HipA in E. coli K-12 (Figure 4A). Sputcn32_2916 encodes a HipA domain protein that is 448 aa in length, and it has 40% identity and 6% coverage with HipAK-12. Sputcn32_2915 encodes a XRE family transcriptional regulator of 152 aa that contains a Helix-turn-helix (HTH) domain in the C-terminal and has 33% amino acid sequence identity and 23% coverage with HipBK-12 (Figure 4B). To determine whether Sputcn32_2916 and Sputcn32_2915 constitute a functional TA pair, open reading frames of the two genes were cloned into plasmid pCA24N to obtain pHipA and pHipB, respectively. Expression of hipA or hipB was induced in E. coli BW25113 with 0.5 mM IPTG. Cell growth (turbidity) and cell viability (CFU ml−1) were measured for 8 h. Overproducing HipA in BW25113 cells led to growth inhibition (Figures 4C–E). To further assess whether HipB can block the toxicity of HipA, we cloned the coding regions of hipA and hipB into plasmid pCA24N to construct pHipAB. Coexpression of hipA and hipB via plasmid pHipAB in BW25113 cells showed that HipB could partially neutralize the toxic effect of HipA (Figures 4C–E); this may result from the too high load of toxins driven by the strong lac promoter on the high copy number plasmid pCA24N. Then, we cloned hipA-hipB with its native promoter into pMD19-T to generate pMD19-T-hipAB. The strain BW25113/pMD19-T-hipAB exhibited similar cell viability with that of BW25113/pMD19-T, suggesting that HipB could fully neutralize the toxic effect of HipA under the native promoter (Figure 4F). Taken together, HipA and HipB in GI21 form a TA pair in which HipA is a potent toxin and HipB is the cognate antitoxin.
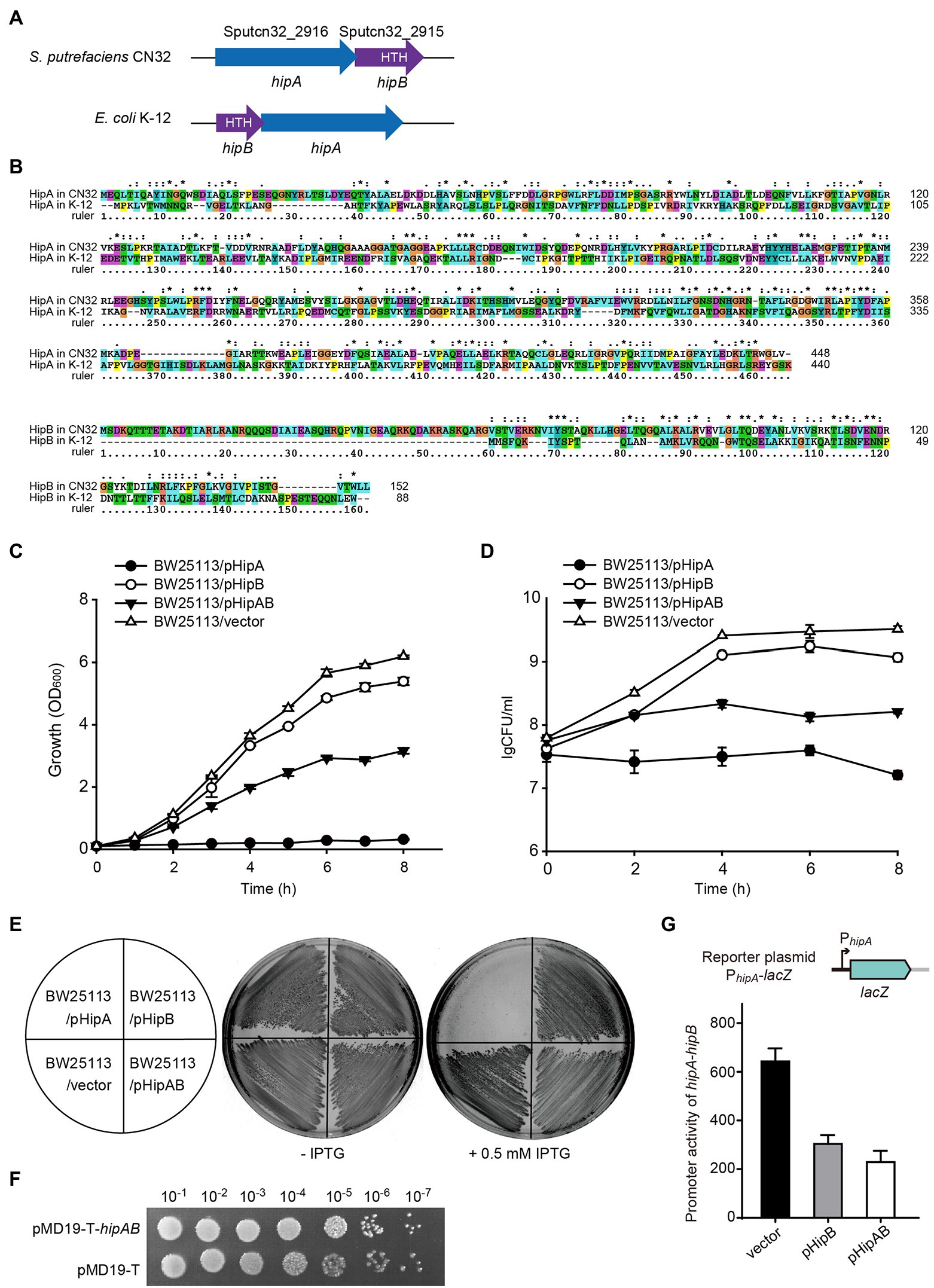
Figure 4. HipA and HipB in GI21 constitute a Toxin–antitoxin (TA) pair. (A) Comparison of the hipA-hipB operon in GI21 and hipB-hipA operon in E. coli K-12. (B) Sequence alignment was carried out using ClustalX to compare the amino acid sequence identity of HipA/HipB in S. putrefaciens CN32 and E. coli K-12. Cell growth (C) and cell viability (D) of cells overexpressing hipA, hipB, and hipA-hipB via pCA24N-based plasmids in E. coli BW25113. (E) Growth of BW25113 cells overexpressing hipA, hipB, and hipA-hipB via pCA24N-based plasmids on LB plates with and without 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). (F) CFU of strain BW25113 containing pMD19-T-hipAB or empty vector pMD19-T on LB plates with ampicillin. (G)The activity of the hipA-hipB promoter in GI21 was measured by overexpressing hipB or hipA-hipB.
In HipAK-12/HipBK-12, the antitoxin HipBK-12 or the TA complex bind DNA and autoregulate the transcription of the TA operon (Black et al., 1994). Similar to HipBK-12, HipB in CN32 also contains a HTH domain, thus we wondered whether HipB in GI21 can regulate the hipA-hipB operon. Using the plasmid by fusing lacZ with the hipA-hipB promoter as the reporter, we found that overproduction of HipB exhibited 2.1 ± 0.1-fold decrease in the promoter activity compared to empty vector. Moreover, overproduction of HipA/HipB complex via pHipAB showed a 2.9 ± 0.4-fold decrease in the promoter activity (Figure 4G). These results suggested that GI21-encoded HipB and the HipA/HipB complex can repress the TA operon.
To test whether the HipA/HipB TA system affects the excision of CGI48, we deleted the hipAB region in CN32. qPCR assays showed no significant difference in the excision rate of CGI48 in the hipAB deletion mutant compared to wild-type CN32 (Figure 5A). As reported in our previous study, the TA system in prophage CP4So in S. oneidensis stabilizes CP4So after its excision (Yao et al., 2018). We wondered whether GI21-encoding HipA/HipB played a role in the maintenance of CGI48 after its excision. A blue–white reporter screening assay was designed to detect the loss of GI21 and CGI48 after their excision. In brief, the lacZ gene was fused with the promoter of the integrase gene Sputcn32_2900 to generate a Pint::lacZ fusion and cloned into the integrative plasmid pHGI01, generating pHGI01-Pint. The constructed plasmid was site specifically integrated into GI21 in CN32. Blue colonies indicated the presence of GI21 in CN32, irrespective of whether it was integrated in the host chromosome or existed in a circular form after GI21 or CGI48 was excised. White colonies indicated a complete loss of GI21 from CGI48 or a complete loss of CGI48 from the CN32 genome (Figure 5B). To activate the excision of GI21 and CGI48, Xis21 and Int48 were induced with 1 mM IPTG for 6 h, and cells were then plated on X-gal plates to detect GI21- and CGI48-free cells using the reporter plasmid (Figure 5C). No loss of GI21 was detected in wild-type CN32, and 0.39% of GI21-free cells were exhibited in the hipAB deletion mutant when Xis21 was overexpressed. Similarly, no loss of CGI48 was detected in wild-type CN32, and 0.82% of CGI48-free cells was exhibited in the hipAB deletion mutant when Int48 was overexpressed (Figure 5D). Then, two white colonies (indicated with blue arrows) from the Xis21-induced plates and two (indicated with blue arrows) from the Int48-induced plates were randomly selected to confirm the loss of GI21 and CGI48 (Figure 5E) by PCR. In addition, we also test the contribution of GI21-encoded HipA/HipB on plasmid stability. As shown in Figure 5F, plasmid pCA24N was completely lost from E. coli BW25113 after 72 h, while pHipAB which contains hipAB in pCA24N was stably maintained in E. coli after 108 h of culturing. Altogether, these results thus demonstrate that HipA/HipB not only stabilizes GI21 and CGI48 but also provides plasmid stabilization.
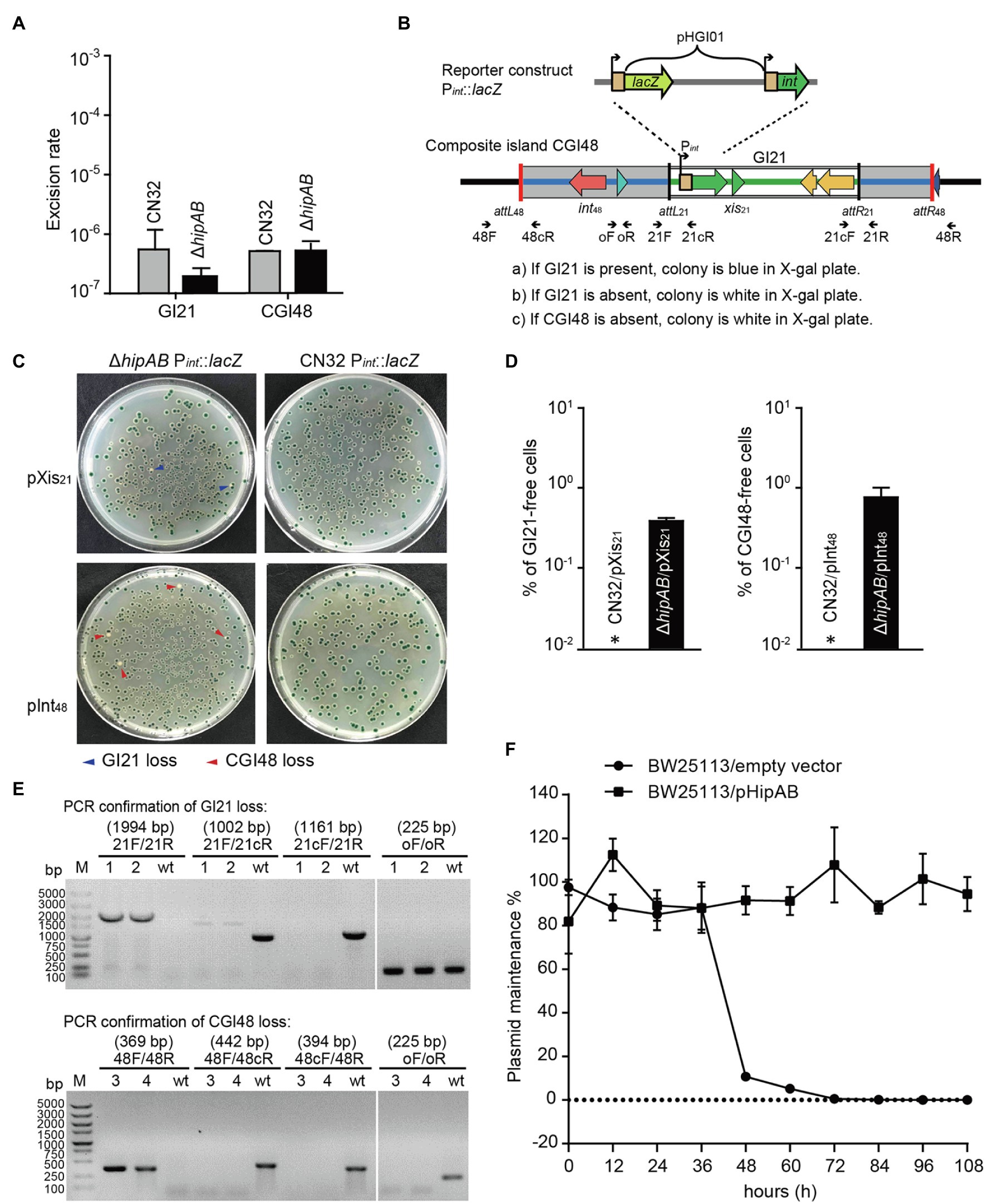
Figure 5. GI21-encoded HipAB promotes the maintenance of CGI48. (A) The excision rate of GI21 and CGI48 in the CN32 wild-type and ΔhipAB mutant strains. (B) Schematic of the lacZ reporter constructs in the CN32 wild-type and ΔhipAB strains. (C) Observation of GI21 loss when Xis21 is overexpressed (upper plates) and of CGI48 loss when Int48 is overexpressed (lower plates) on X-gal plates using the lacZ reporter system. (D) % of GI21-free cells (left panel) and % CGI48-free cells (right panel) were quantified by counting five plates, a representative image as shown in (C). Asterisks indicate that the frequency of GI21 and CGI48 loss was below the limit of detection of the assays (<1 × 10−5). (E) Confirmation of GI21 (upper panel) and CGI48 (lower panel) loss by PCR using the indicated primers in (B). 1 and 2 indicate the DNA templates extracted from the colonies with blue arrows in (C); 3 and 4 indicate the DNA templates extracted from the colonies with red arrows in (C); wt indicates the DNA templates from wild-type CN32 used as a control. Lane M indicates DNA Marker DL5k. The expected product sizes are indicated at the top of the primer sets. (F) GI21-encoded HipAB confers plasmid stability in E. coli. E. coli BW25113 harboring plasmids pHipAB and empty vector pCA24N were used in this assay. Three independent cultures were conducted, and the data are shown as means ± SDs.
In this study, a new composite island, CGI48, was detected in the genome of S. putrefaciens CN-32. CGI48 harbors genes encoding adaptive traits, such as antibiotics and restriction–modification systems. CGI48 evolved by inserting a genomic island, GI21, showing high identity with GIs integrated in the yicC locus. Because the conserved yicC locus is intact and available in CN32 genome, GI21 might integrated into CN32 accompanied by the composite CGI48. Another possibility is that GI21 is integrated into the secondary attachment site within CGI48 genome after horizontal gene transfer. Many genomic islands preferentially integrated into a primary attachment site in the bacterial genome. Studies on the ICEs, ICEBs1 found that ICEBs1 can also integrate into secondary attachment site, especially when the primary site is absent. However, the excision of ICEBs1 from secondary sites is greatly reduced compared to the primary site, limiting the dissemination of ICEBs1 (Menard and Grossman, 2013). In vitro assays showed that the efficiency of integrase-mediated site-specific recombination is related to the length of the attachment site, and the reduction of the core attachment site produced a dramatically decrease in the recombination activity (Ghosh et al., 2003). Thus, we speculated that the shorter attachment sites flanking GI21 may limit its excision and stabilize the composite structure. Some composite GIs are also found to be stabilized by truncated attachment sites or integrases (Bellanger et al., 2014). In this study, we also found that a functional TA system maintain the stability of the composite GI. All these mechanisms explain the complexity and diversity of GIs.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, CP000503; https://www.ncbi.nlm.nih.gov/genbank/, CP000681.
XW and PW conceptualized and designed the project. YZ, WW, JY, XW, DL, and PW did the investigation, data curation, and analysis. YZ, XW, DL, and PW wrote, reviewed, and edited the original draft. All authors contributed to the article and approved the submitted version.
This work was supported by the Guangdong Major Project of Basic and Applied Basic Research (2019B030302004), the Natural Science Foundation of Guangdong Province (2019A1515011912), the Science and Technology Planning Project of Guangzhou (202002030493), Hainan Provincial Joint Project of Sanya Yazhou Bay Science and Technology City (320LH047), the Youth Innovation Promotion Association CAS (2021345 to PW), the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou; GML2019ZD0407), the Natural Science Foundation of Hebei Province (C2019205044), Research Fund of Hebei Normal University (L2016Z03), and Science and Technology Research Project of Hebei University (ZD2018070).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. doi: 10.1038/msb4100050
Bellanger, X., Payot, S., Leblond-Bourget, N., and Guedon, G. (2014). Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol. Rev. 38, 720–760. doi: 10.1111/1574-6976.12058
Black, D. S., Irwin, B., and Moyed, H. S. (1994). Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 176, 4081–4091. doi: 10.1128/jb.176.13.4081-4091.1994
Burrus, V., Roussel, Y., Decaris, B., and Guedon, G. (2000). Characterization of a novel integrative element, ICESt1, in the lactic acid bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 66, 1749–1753. doi: 10.1128/AEM.66.4.1749-1753.2000
Burrus, V., and Waldor, M. K. (2003). Control of SXT integration and excision. J. Bacteriol. 185, 5045–5054. doi: 10.1128/JB.185.17.5045-5054.2003
Caro-Quintero, A., Deng, J., Auchtung, J., Brettar, I., Hofle, M. G., Klappenbach, J., et al. (2011). Unprecedented levels of horizontal gene transfer among spatially co-occurring Shewanella bacteria from the Baltic Sea. ISME J. 5, 131–140. doi: 10.1038/ismej.2010.93
Dehio, C., and Meyer, M. (1997). Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179, 538–540. doi: 10.1128/jb.179.2.538-540.1997
Dobrindt, U., Hochhut, B., Hentschel, U., and Hacker, J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424. doi: 10.1038/nrmicro884
Fu, H., Jin, M., Ju, L., Mao, Y., and Gao, H. (2014). Evidence for function overlapping of CymA and the cytochrome bc1 complex in the Shewanella oneidensis nitrate and nitrite respiration. Environ. Microbiol. 16, 3181–3195. doi: 10.1111/1462-2920.12457
Germain, E., Castro-Roa, D., Zenkin, N., and Gerdes, K. (2013). Molecular mechanism of bacterial persistence by HipA. Mol. Cell 52, 248–254. doi: 10.1016/j.molcel.2013.08.045
Ghosh, P., Kim, A. I., and Hatfull, G. F. (2003). The orientation of mycobacteriophage Bxb1 integration is solely dependent on the central dinucleotide of attP and attB. Mol. Cell 12, 1101–1111. doi: 10.1016/s1097-2765(03)00444-1
Haskett, T. L., Terpolilli, J. J., Bekuma, A., O’hara, G. W., Sullivan, J. T., Wang, P., et al. (2016). Assembly and transfer of tripartite integrative and conjugative genetic elements. Proc. Natl. Acad. Sci. U. S. A. 113, 12268–12273. doi: 10.1073/pnas.1613358113
Jurenas, D., Fraikin, N., Goormaghtigh, F., and Van Melderen, L. (2022). Biology and evolution of bacterial toxin-antitoxin systems. Nat. Rev. Microbiol. doi: 10.1038/s41579-021-00661-1 [Epub ahead of print].
Kitagawa, M., Ara, T., Arifuzzaman, M., Ioka-Nakamichi, T., Inamoto, E., Toyonaga, H., et al. (2005). Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12, 291–299. doi: 10.1093/dnares/dsi012
Lei, C. W., Zhang, A. Y., Liu, B. H., Wang, H. N., Yang, L. Q., Guan, Z. B., et al. (2015). Two novel salmonella genomic island 1 variants in Proteus mirabilis isolates from swine farms in China. Antimicrob. Agents Chemother. 59, 4336–4338. doi: 10.1128/AAC.00120-15
Lewis, J. A., and Hatfull, G. F. (2001). Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and cox proteins. Nucleic Acids Res. 29, 2205–2216. doi: 10.1093/nar/29.11.2205
Menard, K. L., and Grossman, A. D. (2013). Selective pressures to maintain attachment site specificity of integrative and conjugative elements. PLoS Genet. 9:e1003623. doi: 10.1371/journal.pgen.1003623
Miller, J.H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press
Ogura, T., and Hiraga, S. (1983). Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. U. S. A. 80, 4784–4788. doi: 10.1073/pnas.80.15.4784
Roberts, R. C., Strom, A. R., and Helinski, D. R. (1994). The parDE operon of the broad-host-range plasmid RK2 specifies growth inhibition associated with plasmid loss. J. Mol. Biol. 237, 35–51. doi: 10.1006/jmbi.1994.1207
Smyth, D. S., and Robinson, D. A. (2009). Integrative and sequence characteristics of a novel genetic element, ICE6013, in Staphylococcus aureus. J. Bacteriol. 191, 5964–5975. doi: 10.1128/JB.00352-09
Wang, X., Yao, J., Sun, Y. C., and Wood, T. K. (2021). Type VII toxin/antitoxin classification system for antitoxins that enzymatically neutralize toxins. Trends Microbiol. 29, 388–393. doi: 10.1016/j.tim.2020.12.001
Wang, P., Yu, Z., Li, B., Cai, X., Zeng, Z., Chen, X., et al. (2015). Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas. Microb. Cell Factories 14:11. doi: 10.1186/s12934-015-0194-8
Wang, P., Zeng, Z., Wang, W., Wen, Z., Li, J., and Wang, X. (2017). Dissemination and loss of a biofilm-related genomic island in marine Pseudoalteromonas mediated by integrative and conjugative elements. Environ. Microbiol. 19, 4620–4637. doi: 10.1111/1462-2920.13925
Wozniak, R. A., and Waldor, M. K. (2009). A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 5:e1000439. doi: 10.1371/journal.pgen.1000439
Yao, J., Guo, Y., Wang, P., Zeng, Z., Li, B., Tang, K., et al. (2018). Type II toxin/antitoxin system ParESO/CopASO stabilizes prophage CP4So in Shewanella oneidensis. Environ. Microbiol. 20, 1224–1239. doi: 10.1111/1462-2920.14068
Keywords: Shewanella putrefaciens, mobile genetic element, stability, genomic island, toxin–antitoxin
Citation: Zhao Y, Wang W, Yao J, Wang X, Liu D and Wang P (2022) The HipAB Toxin–Antitoxin System Stabilizes a Composite Genomic Island in Shewanella putrefaciens CN-32. Front. Microbiol. 13:858857. doi: 10.3389/fmicb.2022.858857
Received: 20 January 2022; Accepted: 24 February 2022;
Published: 21 March 2022.
Edited by:
Daniel Yero, Universidad Autónoma de Barcelona, SpainReviewed by:
Alberto J. Martín-Rodríguez, Karolinska Institutet (KI), SwedenCopyright © 2022 Zhao, Wang, Yao, Wang, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong Liu, cHF3MTIzNEAxNjMuY29t, orcid.org/0000-0003-1546-8215; Pengxia Wang, d2FuZ3Blbmd4aWFAc2NzaW8uYWMuY24=,orcid.org/0000-0002-1267-6118
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.