
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 May 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.858799
This article is part of the Research Topic Antimicrobial Resistance in Foodborne Pathogens: ecology, epidemiology, and mechanisms View all 12 articles
 Xiangyun Wu1
Xiangyun Wu1 Jiayi Liu1
Jiayi Liu1 Jiawei Feng1
Jiawei Feng1 Muhammad Abu Bakr Shabbir2
Muhammad Abu Bakr Shabbir2 Yali Feng1
Yali Feng1 Rui Guo1
Rui Guo1 Meifang Zhou1
Meifang Zhou1 Sulin Hou1
Sulin Hou1 Guiqiang Wang3
Guiqiang Wang3 Haihong Hao1,4
Haihong Hao1,4 Guyue Cheng1,4
Guyue Cheng1,4 Yulian Wang1,4*
Yulian Wang1,4*Klebsiella pneumoniae (K. pneumoniae) is an opportunistic pathogen, which causes serious infections in humans and animals. To investigate the antimicrobial resistance pattern and virulence profile of K. pneumoniae, a total of 887 samples were collected from both the healthy and mastitis cows and the bedding, feed, feces, air, drinking water, spraying water, washing water, and milk cup swabs from five dairy farms in Hubei, China, during 2019 and 2020. K. pneumoniae was isolated and identified using PCR of the khe and 16S rDNA sequencing. A genotypic characterization was performed for K. pneumoniae isolates using wzi typing and multilocus sequence typing (MLST). Antimicrobial resistances were confirmed using broth microdilution against 17 antimicrobial agents and resistance and virulence genes were determined by PCR. The prevalence of K. pneumoniae was 26.94% (239/887) distributed in 101 wzi allele types (199/239, 83.26%) and 100 sequence types (STs) (209/239, 87.45%), including 5 new wzi allele type and 25 new STs. Phylogenetic analysis showed that K. pneumoniae isolated from milk, nipple swab, feed, and feces is classified in the same clone complex. By comparing with the PubMLST database, at least 67 STs have the risk of spreading in different species and regions. Interestingly, 60 STs have been isolated from humans. The isolates were highly sensitive to meropenem and colistin, but resistant to ampicillin (100%), sulfisoxazole (94.56%), cephalothin (47.28%), streptomycin (30.13%), and so on. Noteworthy, multidrug-resistant (MDR) rate was found to be 43.93% in this study. By PCR, 30 of 68 antimicrobial resistance (AMR) genes were identified; the prevalence rate of blaTEM, blaSHV, strA, strB, aadA1, and aac(6′)-Ib-cr was more than 50%. Eleven CTX-M-producing K. pneumoniae were found. The detection rate of fimH, mrkD, uge, wabG, entB, iutA, iroN, and ureA was over 85%. This study reinforces the epidemiological importance of K. pneumoniae in food-producing animals in Hubei. The emergence and spread of environmental MDR K. pneumoniae may pose a potential threat to food safety and public health.
Cow mastitis is considered to be one of the most common and frequent diseases in dairy herds. This disease only affects the estrus and pregnancy of dairy cows, resulting in the decline of milk production and quality, but also increases treatment costs and causes high economic losses in dairy industries worldwide (Käppeli et al., 2019). The global economic losses per year due to mastitis amount to $35 billion and for the United States dairy industry $2 billion per year (Sathiyabarathi et al., 2016; Li et al., 2019). Besides, mastitis can pose a threat to human and animal health via the transfer of antimicrobial resistance bacteria and food poisoning (Li et al., 2019). Klebsiella pneumoniae (K. pneumoniae) is one of the main environmental pathogens causing mastitis, as well as a pathogen of zoonotic conditions that can cause a series of serious infections such as respiratory tract infections, urinary tract infections, soft-tissue infections, and bloodstream infections (Lee et al., 2017; Massé et al., 2020).
Virulence factors play an important role in the pathogenic mechanism of K. pneumoniae. Capsular, iron carriers, pili, and lipopolysaccharide (LPS) have been widely demonstrated to be involved in the adhesion, invasion, and growth of K. pneumoniae (Shon et al., 2014; Cheng et al., 2020). Capsular can prevent K. pneumoniae from being recognized by the host immune system through some immune escape mechanisms such as antiphagocytosis, inhibition of early inflammatory response, neutralization of antimicrobial peptides to reduce the body’s immune response, and inhibition of dendritic cell maturation (Paczosa and Mecsas, 2016). These bacteria can absorb the iron of the host via four siderophores such as aerobactin, salmochelin, enterobactin and yersiniabactin for metabolism and enhance the virulence to cause infection (Wang G. et al., 2020). Two common types of fimbriae are found in K. pneumoniae: type 1 (fim) and type 3 (mrk) fimbriae. Type 1 fimbriae can enhance virulence by adhering to mucosal or epithelial surfaces and type 3 fimbriae adhere to the cell surface and promote biofilm formation (Schroll et al., 2010). Lipid A, a component of lipopolysaccharide, reduces the inflammatory response during K. pneumoniae infection and prevents the bactericidal effect of cationic antimicrobial peptides. O antigen is the outermost subunit of LPS, which eliminates the lysis of bacteria by the complement membrane attack complex (Paczosa and Mecsas, 2016).
Previous studies have shown that mucoviscosity-associated gene (magA), uridine diphosphate galactose 4 epimerase encoding gene (uge), regulator of the mucoid phenotype (rmpA), iron uptake system gene (kfu, aerobactin), and K1 and K2 capsule serotypes are important virulence genes in invasive K. pneumoniae strains causing mastitis (Osman et al., 2014; Massé et al., 2020). According to virulence characteristics, K. pneumoniae can be divided into classic Klebsiella pneumoniae (cKP) and hypervirulence Klebsiella pneumoniae (hvKP). Most K. pneumoniae infections in the world are cKP infections, but cKP can evolve into hvKP by obtaining virulence factors (such as plasmids) (Wyres et al., 2020). Studies have shown that the genes peg-344, iroB, iucA, prmpA, and prmpA2 can distinguish cKP and hvKP with high accuracy (Harada and Doia, 2018; Russo et al., 2018). Some scholars have found that K1 and K2 capsule serotype is closely related to high virulence (Follador et al., 2016; Guo et al., 2017). Since hvKP was first detected in Taiwan in 1986, the number of clinical infection cases of the bacteria has gradually increased and Wuhan was the city with the highest hvKP prevalence (73.9%) reported among 230 K. pneumoniae isolates from 10 cities in China (Liu et al., 1986, 2014; Zhang et al., 2016). More worryingly, with the inappropriate use of antibiotics, multidrug resistant (MDR) has been increased all over the world that is considered as a public health threat. Several recent investigations reported the emergence of multidrug-resistant bacterial pathogens from different origins, including humans, birds, cattle, and fish that increase the need for new potent and safe antimicrobial agents (Algammal et al., 2020a,b, 2021). Epidemics and outbreaks of multidrug-resistant K. pneumoniae have also been reported around the world (Gu et al., 2018; Alanezi et al., 2022; Arteaga-Livias et al., 2022). The worldwide spread of extended-spectrum β-lactamase (ESBL)-producing K. pneumoniae remains a critical concern for therapies against multidrug-resistant bacteria (Chong et al., 2018). “Superbug” derived from the combination of virulence and resistance of K. pneumoniae will bring great challenges to clinical anti-infective treatment. In addition, hundreds of mobile antimicrobial resistance (AMR) genes have been found in K. pneumoniae (Navon-Venezia et al., 2017). K. pneumoniae is considered to be a major transporter of resistance genes from environmental sources to clinically important bacteria and some isolates can carry acquired AMR genes or plasmid to move between environmental, human, and animal (Wyres and Holt, 2018).
As the most important measures for disease prevention and control, finding the source of infection and cutting off the transmission route are to block the transmission of pathogens from infected animals/people to susceptible animals/people (Chen et al., 2021). Genotyping technology is mainly used to understand the transmission route of pathogens and trace the source of infection. Multilocus sequence typing (MLST) is widely used because of its good typing ability, comparability, high reproducibility, high throughput, and convenient data sharing (Pérez-Losada et al., 2013). The sequence types (STs) of K. pneumoniae reported to be important and prevalent globally include ST11, ST15, ST23, ST258, ST395, ST512, and so on (Zhang et al., 2019; Gato et al., 2020; Zhou et al., 2020; Di Pilato et al., 2021). Currently, the host and regional epidemiological characteristics of K. pneumoniae in various regions are still unclear and surveillance data on resistance and virulence of K. pneumoniae in the cow are very scarce. This study aimed to investigate the prevalence, antimicrobial resistance pattern, resistance genes, and virulence genes of K. pneumoniae isolated from dairy cows in Hubei, China. K. pneumoniae from milk and environmental samples was genotyped using PCR to determine genetic diversity and explore potential reservoirs and transmission.
Samples were collected from five commercial dairy farms in Hubei during 2019 and 2020. The number of lactating cows in farms A, B, C, D, and E are 202, 300, 400, 1,200, and 287, respectively. Five farms participated in monthly Dairy Herd Improvement (DHI). Lactating cows in five farms were milked 2 times/day in milking parlors. The clinical symptoms of cows with clinical mastitis (CM) are elevated body temperature, redness, and pain in the udder. The judgment of subclinical mastitis (SCM) refers to the Chinese agricultural industry standard NY/T 2692-2015 (National standards of the people’s Republic of China, 2015) [the number of somatic cell count (SCC) is > 500,000 cells/ml and no visible pathological changes]. Daily care of cows in five farms was provided by the veterinarian; the farms routinely used different antibacterial agents such as ceftiofur, amoxicillin/clavulanic acid, lincomycin, florfenicol, kanamycin, and gentamicin for regular prophylactic and treatment protocols. The sample collection includes animal samples (nipple milk, nipple skin swabs, anal swabs from healthy cows, CM cows, and SCM cows) and environmental samples (bedding, feed, feces, air, drinking water, spraying water, washing water, and milk cup swabs). Lactating cows of parity 3rd to 4th were selected for sample collection. Briefly, milk samples were collected from lactating cows by the milkers after premilking disinfection and the first 3 streams of milk were discarded. Commercial liquid delivery mediums (Haibo Biotech, Qingdao, China) were used to collect nipple skin swabs, anal swabs, and milk cup swabs. All the nipples of each cow were rubbed on the skin with swabs 4 to 5 times from the root to the end of teat after milking. For the anal swab, the swab was inserted into the anus of the cow and rotated for 2 to 3 circles. The swab of the milk cup is collected in milking parlors (5 per farm). Feed, bedding, and feces samples were stored in sterile sampling bags and air samples were collected by natural sedimentation method [suspended sterile sampling tube with 15 ml Trypticase Soy Broth (Haibo Biotech, Qingdao, China) on the column in the barn for 15 to 30 min]. Three sampling points were evenly selected in the barn and the interval between sampling points was not less than 5 m and then mix the samples with equal weight. All the samples were stored at 4°C and transported to the laboratory for bacterial culturing and identification within 24 h. SCC of milk sample was analyzed by DHI Center of Livestock and Poultry Breeding Centre in Hubei.
All the samples were inoculated in 5 ml Trypticase Soy Broth (Haibo Biotech, Qingdao, China) and incubated at 37°C for 18 to 24 h. A loopful of broth culture was streaked onto Columbia Blood Agar and MacConkey Inositol Adonitol Carbenicillin (MIAC) agar (Haibo Biotech, Qingdao, China) and incubated at 37°C for 12 to 18 h. A single pink, slimy, and non-hemolytic suspected colony was picked and purified on MIAC agar (Gao et al., 2010). Purified bacteria were selected for biochemical identification and PCR testing. Gram staining was performed with the Gram Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Biochemical identification tubes (Haibo Biotech, Qingdao, China) were used for motility tests, indole tests, urease tests, oxidase experiments, and citrate utilization tests (Massé et al., 2020). Further identification and confirmation of the isolates were carried out using PCR of khe and 16s rDNA gene (He et al., 2016; Cheng et al., 2021). Positive clones were sequenced by the Tsingke Biotechnology Corporation Ltd. (Wuhan, China). K. pneumoniae ATCC 700603 (khe positive) and Escherichia coli ATCC 25922 (khe negative) were used as control strains.
Genomic DNA of K. pneumoniae was extracted by boiling method (Kuang, 2015). 3 ml suspended plaque samples were incubated for 15 to 20 min at 99°C. Subsequently, the supernatants were immediately transferred to −20°C for 15 min and centrifuged at 12,000 rpm/min for 15 min at 4°C. The supernatants were diluted by sterile water (1:10) and kept at −20°C for PCR testing.
Capsular polysaccharide serotypes were determined by wzi gene sequencing (Brisse et al., 2013). MLST detection method refers to the K. pneumoniae MLST online database1. The seven housekeeping genes (gapA, infB, mdh, phoE, pgi, rpoB, and tonB) were amplified by PCR. All the positive products were sequenced by Sanger and sequences were submitted to the website2 to get the wzi allele types and the subtypes of each housekeeping gene of MLST. Submit the allele profile to the MLST website3 to get ST. wzi alleles and STs that had not been previously described were submitted to the curator of the PubMLST database and were assigned new designations. Sanger sequencing was completed by Tsingke Biotechnology Corporation Ltd. (Wuhan, China).
The ST phylogeny tree (Tamura 3-parameter model) was constructed using the maximum likelihood method in the MEGA-X software and was modified visually with interactive tree of life (iTOL)4. The phylogenetic tree is tested by the bootstrap method and the number of tests is 1,000 times. The information of 726 K. pneumoniae isolates from different countries and 724 K. pneumoniae isolates from different hosts was downloaded from the PubMLST database (the same 100 STs as in this study). Minimum spanning tree (MST) was constructed by Bionumerics 8.1.
All the 239 K. pneumoniae were evaluated for the presence of 23 known virulence genes, including transporter (peg344), fimbriae synthesis-related gene (fimH, mrkD), lipopolysaccharide-related gene (uge, wabG), capsular polysaccharide synthesis and synthesis regulation-related gene (wcaG, crmpA, prmpA, prmpA2, and magA), iron uptake system (iroB, iroN, aerobactin, iutA, irp2, iucA, ybtA, kfu, and entB), urease-related gene (allS, ureA), tellurite resistance gene (terB), and hemolysin (hly) by PCR (Supplementary Table 1).
The susceptibility of K. pneumoniae isolates to 17 antimicrobials, which are commonly used on dairy farms and become clinically important broad-spectrum antimicrobials, was determined using the microbroth dilution method recommended by the Clinical and Laboratory Standardization Institute [Clinical and Laboratory Standards Institute [CLSI] (2021)], including penicillins: ampicillin (AMP); β-lactam/β-lactamase-inhibitor combinations: amoxicillin/clavulanic acid (AMC); cephalosporins: cephalothin (CEP), ceftiofur (EFT), and ceftriaxone (CRO); carbapenems: meropenem (MEM); aminoglycosides: streptomycin (STR), kanamycin (KAN), and gentamicin (GEN); tetracyclines: tetracycline (TET) and doxycycline (DOX); amphenicols: florfenicol (FFC) and polymyxins: colistin (CT), rifamycins: rifaximin (RFX), fluoroquinolones: ciprofloxacin (CIP); and sulfonamides: sulfisoxazole (SOX) and sulfamethoxazole/trimethoprim (SXT). Results were interpreted according to the CLSI M100-Ed31 and VET01S-Ed5 standards [Clinical and Laboratory Standards Institute [CLSI] (2020, 2021)]. No CLSI interpretative criteria for ceftiophene and streptomycin are currently available and the resistance breakpoint of ceftiophene and streptomycin refers to ceftiofur and kanamycin, respectively. The criteria for resistance of rifaximin were analyzed by Ecofinder software recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (Turnidge et al., 2006). The phenotypic resistance patterns are categorized into MDR, extensively drug-resistant (XDR), and pandrug-resistant (PDR) as previously described by Magiorakos et al. (2012). The quality control strain is K. pneumoniae ATCC 700603 and Escherichia coli ATCC 25922.
Antimicrobial resistance genes that confer resistance to β-lactams (blaCTX–M–2, blaCTX–M–10, blaCTX–M–14, blaCTX–M–15, blaSHV, blaTEM, and blaOXA–1), carbapenems (IMP, VIM, blaOXA–48, blaOXA–181, NDM, and KPC), AmpCs (MOX, CIT, DHA, ACC, EBC, and FOX), aminoglycosides [armA, rmtA, rmtB, rmtC, rmtD, rmtE, npmA, aadA1, aadA2, aadB, aacC1, aacC2, aac(3)-IV, aacA4, aphA1, aphA2, aphA6, strA, and strB], plasmid-mediated quinolone resistance (PMQR) genes [aac(6′)-Ib-cr, qnrA, qnrB, qnrC, qnrD, and qnrS], tetracyclines [tet(A), tet(B), tet(C), tet(D), tet(E), and tet(G)], sulfonamides (sul1, sul2, and sul3), macrolides (mefA, ereA, ereB, ermB, mphA, and mphB), polymyxins (mcr-1 to mcr-5), and phenicols (floR, cmlA, and Cat1) were investigated by PCR (Supplementary Table 2). For each positive gene, some PCR products were selected and sent to Quintara Biotechnology Corporation Ltd. (Wuhan, China) for Sanger sequencing for further confirmation.
All of ciprofloxacin-resistant K. pneumoniae were selected to determine the DNA sequence of quinolone resistance-determining region (QRDR) genes (gyrA, gyrB, parC, and parE) (Horii et al., 2008; Pitondo-Silva et al., 2015; Cheng et al., 2018). Three ciprofloxacin-intermediate and 7 ciprofloxacin-sensitive isolates were randomly selected as controls. Genomic DNA was extracted by the Universal Genomic DNA Kit of ComWin Biotechnology Corporation Ltd. (Beijing, China). PCR products were purified and sequenced in both the directions by Quintara Biotechnology Corporation Ltd. (Wuhan, China). The nucleotide sequences and the deduced amino acids were compared with that of K. pneumoniae ATCC 13883 (GenBank JOOW00000000.1) using the Bioedit (Azargun et al., 2019).
SPSS statistics version 26 and GraphPad Prism version 8.0.1 software were used for statistical analysis and the chi-squared test (χ2) was used to compare the statistical significance between the different groups. p < 0.05 means significant difference and p < 0.01 indicates extremely significant difference. Correlations were assessed by calculating the Spearman’s rank correlation coefficient (r). |r| ≥ 0.8 means high correlation, 0.5 ≤ | r| < 0.8 means moderate correlation, 0.3 ≤ | r| < 0.5 means low correlation, and | r| < 0.3 means no correlation.
Klebsiella pneumoniae isolates grew well on MIAC agar due to resistance to carbenicillin and gave characteristic pink colonies as a result of inositol and adonitol fermentation. On Columbia Blood Agar, the colonies are not hemolytic and tend to be viscous and stringy. K. pneumoniae was observed as Gram-negative bacillus under a light microscope. Besides, it showed the characteristics of non-motility, indole negativity, production of urease, negative oxidase testing, and utilization of citrate.
A total of 239 K. pneumoniae (26.94%) were identified in 887 samples through biochemical identification, khe amplification, and 16s rDNA sequencing (Figure 1 and Supplementary Table 3). The samples collected in farm C (114/338, 33.73%) showed a significantly higher K. pneumoniae prevalence than farm A (18/109, 16.51%), farm B (37/179, 20.67%), and farm E (48/189, 25.40%). The K. pneumoniae prevalence in farm D (22/72, 30.56%) was also significantly higher than farm A (18/109, 16.51%) (p < 0.05). There was no significant difference in the K. pneumoniae prevalence of nipple milk, skin swabs, and anal swabs between healthy and mastitis cows (p > 0.05). Besides, K. pneumoniae was found in a variety of environmental sources, such as milk cup swab, feed, bedding, feces, air, drinking water, and spray water and the separation rate is between 15.79 and 47.83%.
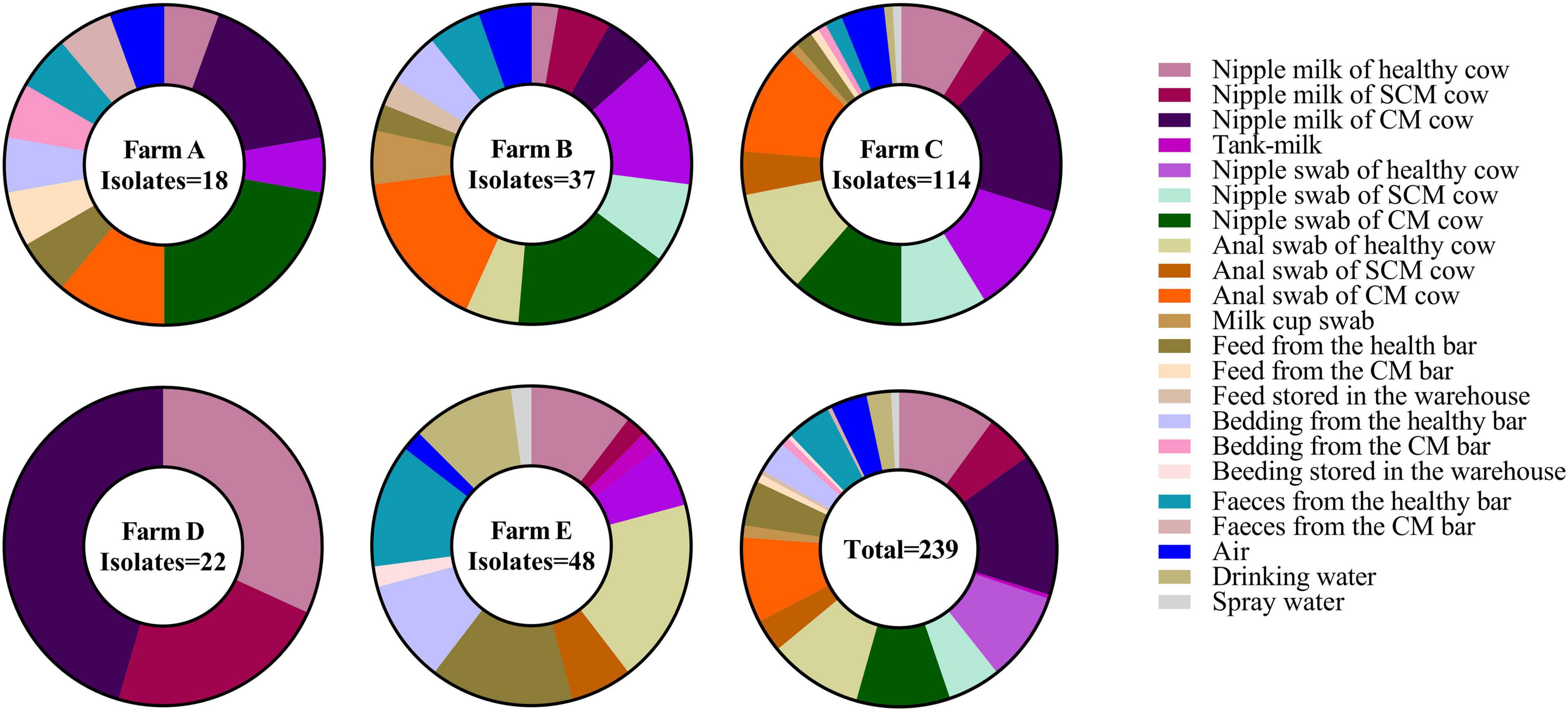
Figure 1. The percentage of Klebsiella pneumoniae (K. pneumoniae) in different sample types in five farms.
Based on the wzi gene sequences, 239 isolates were classified as 101 different wzi allele types (Figure 2). wzi150-KL163KL27KL46 (40/239, 16.74%) accounted for the most and wzi706, wzi727, wzi728, wzi729, and wzi730 were the new wzi allele. The wzi genotypes of 40 strains were unknown, which might be new alleles. K1 and K2 capsule serotypes commonly found in hvKP were not detected in this study. The results of MLST on these strains showed that 100 different STs were obtained (Figure 2), among which ST2854 (30/239, 12.55%) accounted for the largest proportion, followed by ST5960 (15/239, 6.28%) and ST5915 (13/239, 5.44%) and ST5367, ST5369, ST5370, ST5372, ST5855, ST5874, ST5875, ST5876, ST5914, ST5915, ST5917, ST5918, ST5919, ST5920, ST5947, ST5949, ST5951, ST5952, ST5953, ST5954, ST5955, ST5957, ST5959, ST5960, and ST5961 were the new STs. There are still 30 strains of unknown ST, which might be a new allele. We did not find STs such as ST11 and ST23 that have been reported to have a global epidemic of highly toxic in humans. According to the ST phylogeny tree of 239 isolates (Figure 2), K. pneumoniae isolated from five farms in Hubei has a rich variety of serotypes and genotypes. Strains of the same serotype may have differences in the genome and strains of the same genotype may also have different serotypes, such as ST2584 and ST5370 are wzi150, isolates of ST5960 have at least 10 different wzi types.
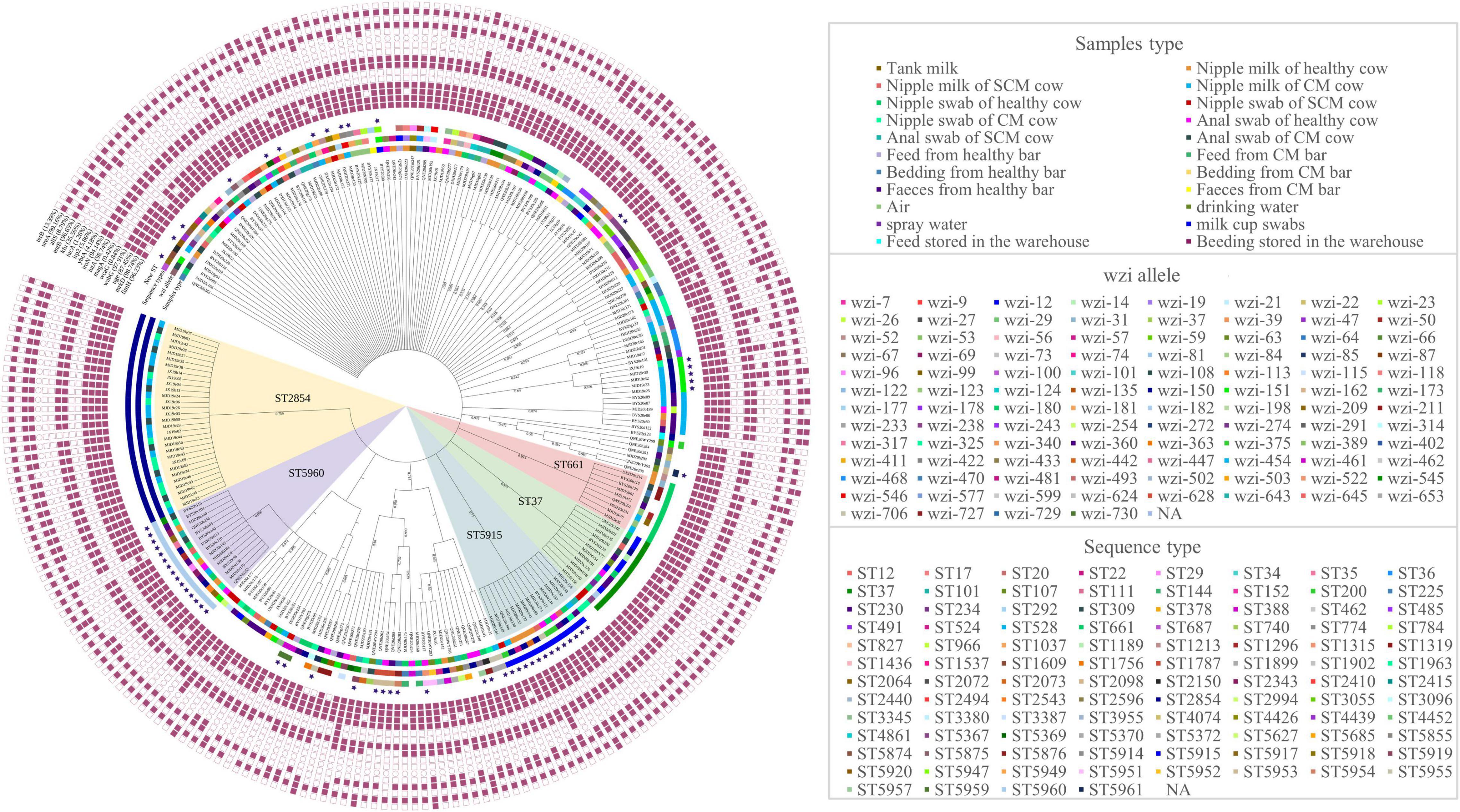
Figure 2. Sequence type (ST) phylogenetic tree, wzi allele, and detection of virulence of 239 K. pneumoniae. The blue stars represent the new STs. PaleVioletRed squares represent the presence of virulence genes and white squares represent the absence of virulence genes, respectively. NA: No allele number assigned.
The MST of 100 different STs showed that there was diversity in the population structure of STs in this study (Figure 3). The founder ST20 and ST17 and ST22 with single locus variants (SLVs) comprise clonal complex 20 (CC20); the founder ST200 and ST292, ST966, ST1537, and ST5627 with SLVs comprise CC200. K. pneumoniae isolated from milk, nipple swab, feed, and feces is classified in the same clone complex, which indicated that the contamination of milk may have a great correlation with the hygiene of nipple surface and environment.
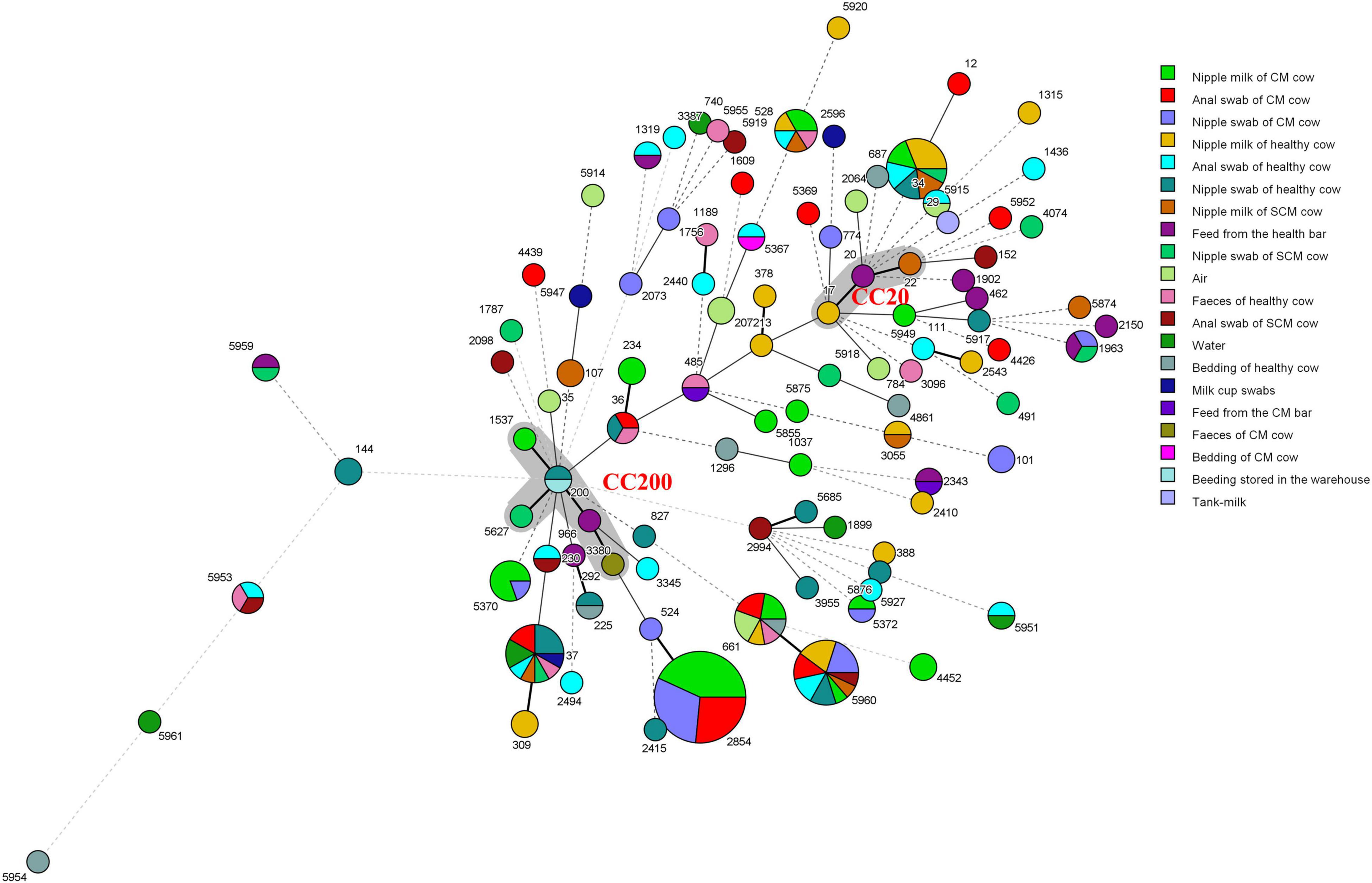
Figure 3. Minimum spanning tree (MST) of 239 K. pneumoniae. The gray area represents a clonal complex and the number represents ST. Each ST is grouped by different sample types. One allele difference is represented by a bold line, two allele differences are represented by a straight line, and three or more allele differences are represented by a dotted line.
Compared with the PubMLST database, we found that 58 STs were isolated in other countries (Figure 4A). These genotypes are mainly from Italian isolates; ST17, ST20, ST29, ST34, ST36, ST37, and ST101 have a wide range of the region. Furthermore, the host sources of 67 STs were not only cows, but also humans, pigs, chickens, dogs, cats, horses, rhizosphere, environment, and so on. According to Figure 4B, there are 66 STs known to come from multiple hosts, of which 60 STs had existed in humans.
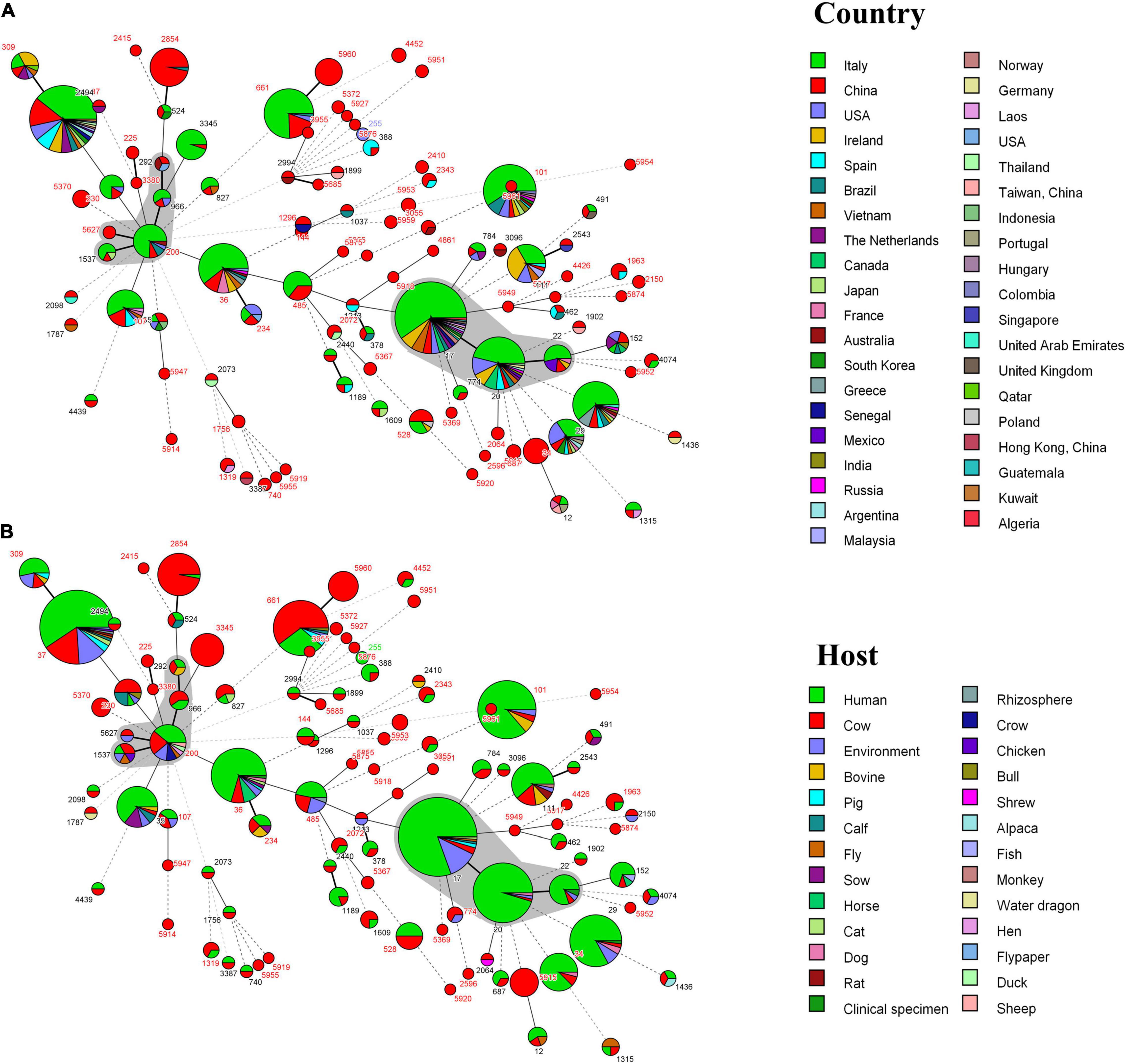
Figure 4. (A) MST of 239 K. pneumoniae in this study and 722 isolates with the same 104 ST from the PubMLST database. Each ST is grouped by different countries. (B) MST of 239 K. pneumoniae in this study and 720 isolates with the same 104 ST from the PubMLST database. Each ST is grouped by different hosts. The gray area represents a clonal complex. One allele difference is represented by a bold line, two allele differences are represented by a straight line, and three or more allele differences are represented by a dotted line.
Sixteen of 23 virulence genes were positively detected by PCR (Figure 2). The detection rate of fimbriae synthesis-related gene (fimH, mrkD), lipopolysaccharide-related gene (uge, wabG), iron uptake system (entB, iutA, and iroN), and urease-related gene (ureA) was over 85% and others were between 0.42 and 35.56%. Notably, there are 3 isolates [ST34 (n = 2), ST2410 (n = 1)] carrying iucA (Figure 2), which are related to high virulence. Besides, 99.16% (237/239) K. pneumoniae carried at least 5 virulence genes and three isolates were positive for 12 virulence genes (Figure 2). There are differences in the virulence genes carried by strains of the same serotype or ST; no significant correlation has been found between the virulence genes and the serotypes or genotypes in this study.
The results of the susceptibility test to 17 antimicrobials are shown in Figure 5 and Supplementary Table 4. K. pneumoniae isolates were highly sensitive to meropenem (99.16%) and colistin (99.58%). However, K. pneumoniae was completely resistant to ampicillin and highly resistant to sulfisoxazole (94.56%); the resistance rate to cephalothin, streptomycin, gentamicin, tetracycline, doxycycline, florfenicol, and sulfamethoxazole/trimethoprim was between 20.08 and 47.28%. It is worth noting that 43.93% (105/239) of the isolates were MDR strains and neither XDR nor PDR strains were found in this study (Figure 6A). The resistance rate of K. pneumoniae to ceftriaxone, streptomycin, and gentamicin in mastitis milk (nipple milk of SCM and CM cows) is significantly higher than that in healthy milk (p < 0.05) (Figure 6B). Ninety-three resistance profiles have been identified and the dominant ones were AMP-SOX (57/239, 23.85%), followed by AMP-CEP-SOX (26/239, 10.88%), AMP-STR-GEN-SOX (11/239, 4.66%), and AMP-CEP-STR-GEN-SOX (11/239, 4.66%) (Supplementary Figure 1).
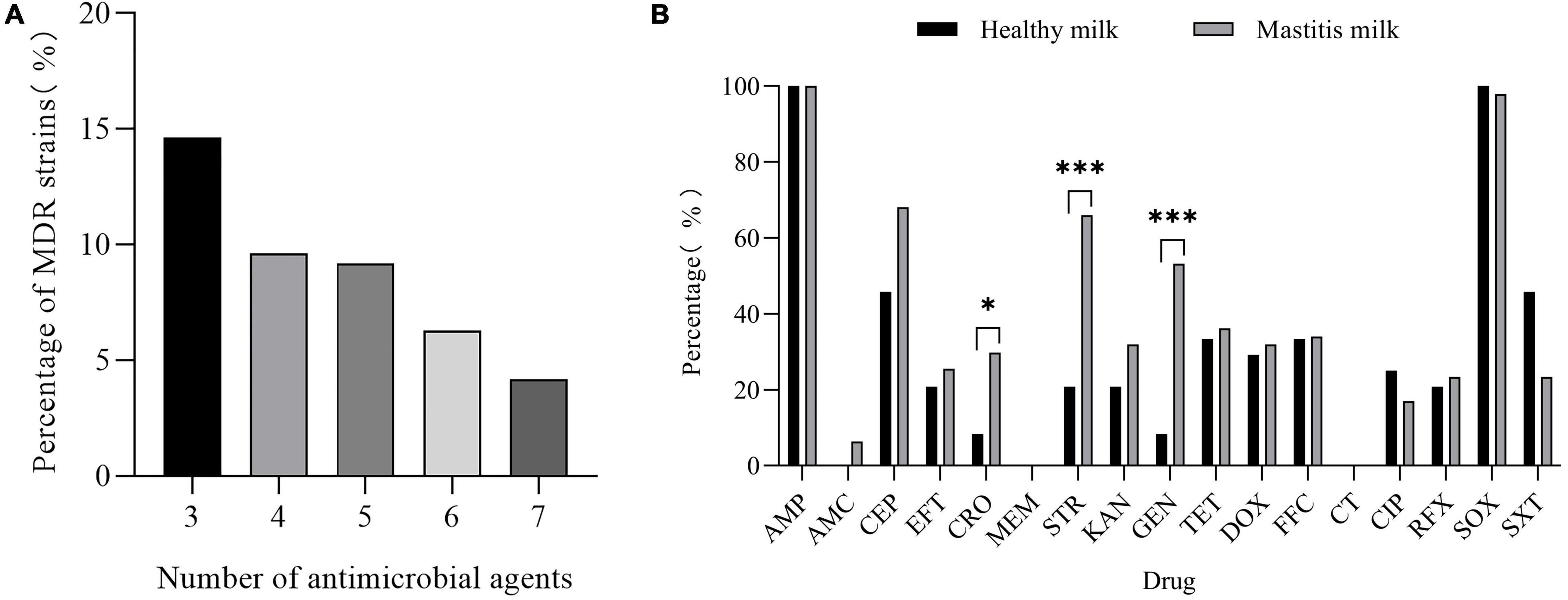
Figure 6. (A) Proportion of multidrug-resistant (MDR) K. pneumoniae resistant to different amounts of antibiotics. (B) Resistance rate of healthy milk and mastitis milk [milk from CM and subclinical mastitis (SCM) cows] to 17 antimicrobials. “*” indicates p < 0.05 and “*” indicates p < 0.001.
Thirty of 68 AMR genes were identified by PCR (Figure 7). The detection rate of blaTEM, blaSHV, strA, strB, aadA1, and aac(6′)-Ib-cr was more than 50% and others were between 0.42 and 38.08%. Eleven CTX-M-producing K. pneumoniae were found. Among the isolates resistant to β-lactam or aminoglycoside antibiotics, more than 90% of isolates have detected at least one β-lactam resistance gene or aminoglycoside resistance gene; among the isolates resistant to TET, DOX, FFC, CIP, and SXT, more than 60% of isolates have detected at least one resistance gene corresponding to the resistance phenotype (Supplementary Figure 1 and Supplementary Table 5). The correlation analysis performed between different phenotypic antibiotic resistance and the antibiotic resistance genes is shown in Figure 8. Among β-lactam antibiotic phenotypes and antibiotic resistance genes, the obtained results revealed low positive correlations between EFT, CRO, and blaCTX–M–15 (r = 0.492–0.474) and AMC and blaOXA–1 (r = 0.590). Moreover, a low positive correlation was observed between CEP and EFT (r = 0.405). Among aminoglycosides, a high positive correlation was found between GEN and aacC2 (r = 0.823); moderate positive correlation was observed between STR and GEN (r = 0.606), STR and strB (r = 0.501), and KAN and aphA1 (r = 0.568); low positive correlation was observed between STR and aacC2 (r = 0.488) and KAN and aacA4 (r = 0.428). Among tetracyclines, moderate positive correlations were observed between TET, DOX, and tetA (r = 0.580–0.686). Among sulfonamides and amphenicols, low positive correlation was observed between sul1 and sul2 (r = 0.482), sul2 and sul3 (r = 0.303), FFC, floR, and cmlA (r = 0.316–0.457). No significant association was found between fluoroquinolone resistance phenotype and PMQR genes. DNA sequencing of QRDR of gyrA, gyrB, parC, and parE demonstrated that no amino acid changes were found in these genes whether K. pneumoniae isolates were resistant, moderately resistant, or sensitive to ciprofloxacin and silent mutations found are shown in Supplementary Table 6.
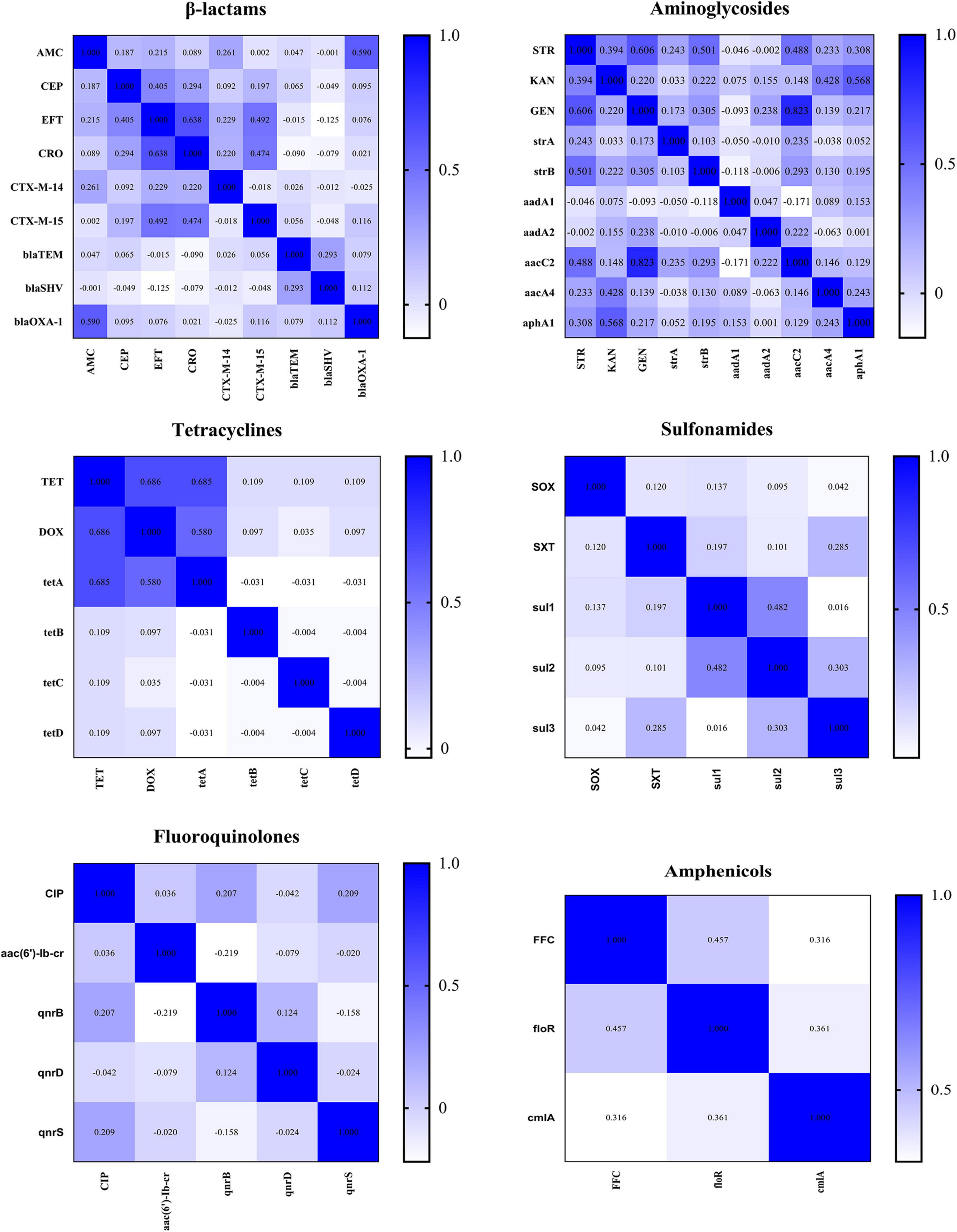
Figure 8. Correlations between different classes of antimicrobials and resistance genes. The intensity of the color indicates the numerical value of the correlation coefficient (r).
Klebsiella pneumoniae is an important opportunistic pathogen that has attracted global attention due to the difficult clinical cure of mastitis caused by K. pneumoniae, the low effectiveness of antibiotic treatment, and the lack of advancement in preventive measures (Yang et al., 2019b). The prevalence and transmission of multidrug-resistant and hypervirulent K. pneumoniae have brought unexpected harm and loss to humans and animals. At present, most of the study on K. pneumoniae is in the field of human medicine, while the study data in the field of mastitis is very scarce. This study showed the prevalence and molecular characteristics of virulence, resistance, capsular serotyping, and genotype of K. pneumoniae in five cattle farms in Hubei, which will help to track the infection trend of K. pneumoniae and emphasized the importance of environmental sanitation in the breeding process. The prevalence of K. pneumoniae varied among the five farms involved in this study, which may be related to stocking density and farm environment. The K. pneumoniae isolation rate in milk from CM cows was 25.36% (35/138), which was similar to the study by Yang et al. (2021). However, we found that there was no significant difference in the K. pneumoniae prevalence of nipple milk, skin swabs, and anal swabs between healthy and mastitis cows, which was different from the previous study (Koovapra et al., 2016). This may be due to that most of the CM cows are in the period of treatment or recovery. The results also confirmed that K. pneumoniae, as an environmental pathogen, is also ubiquitous in the environment (Wareth and Neubauer, 2021).
At present, the hvKP was reported to be most common in K. pneumoniae with ST11, serotype K1 or K2 (Wang et al., 2021). No strains of these serotypes and sequence types were found in this study, but K54 (wzi66, wzi115, n = 2) and K57 (wzi57, n = 1) were found, which may also be related to high virulence (Zhong et al., 2020). The genetic diversity of K. pneumoniae is very rich and its serotypes and sequence types are not in a one-to-one correspondence, which is similar to previous studies (Benulič et al., 2020; Lepuschitz et al., 2020). Many studies confirmed that the occurrence of K. pneumoniae is related to the environment of farms and K. pneumoniae can be spread through contaminated feed, feces, drinking water, and so on (Wareth and Neubauer, 2021; Zhao et al., 2021). Similar results were also observed in our MST analysis. K. pneumoniae isolated from milk, nipple swab, feed, and feces is closely related and 58 STs exist in different countries and 66 STs have been found in different hosts. Strains of these genotypes have potential harm of cross-species and regional transmission, which cannot be ignored. Enhancing the sanitation of the breeding environment and disinfection of the cattle stalls may be an effective ways to prevent mastitis.
The pathogenicity of K. pneumoniae is inseparable from the role of virulence factors. In this study, peg-344, iroB, prmpA, and prmpA2 related to high virulence were not found, but 3 strains were detected carrying the iucA; further study is needed to verify the virulence, such as animal experiments. In addition, PCR detection results showed that fimbriae-related genes (fimH, mrkD), iron uptake system (iutA, iroN, and entB), urease-related genes (ureA), and the lipopolysaccharide-related genes (uge, wabG) were widely distributed in the isolates, which were similar to the previous results (Zhang et al., 2018; Yang et al., 2019a). These results can preliminarily elucidate the pathogenic mechanism of the K. pneumoniae that may include: synthesizing fimbriae to adhere to the surface of host cells or form biofilms for virulence, secreting siderophores to absorb iron in the host for metabolism to enhance virulence, evading serum killing of phagocytes and suppress host immunity by utilizing capsular polysaccharides, and lipopolysaccharide aggregates into complexes on the surface of the K. pneumoniae so that the bacteria can escape or resist the killing of the host’s innate immunity (Schroll et al., 2010; Li et al., 2014; Liu et al., 2019).
Antibiotic resistance has always been a key and difficult issue of global concern. The results of antibiotic susceptibility test showed that K. pneumoniae isolates were highly sensitive to meropenem and colistin. It is mainly because carbapenems are strictly forbidden to be used in animals; the Chinese government has officially banned polymyxin as an animal growth promoter on 30 April 2017 (Wang Y. et al., 2020; Yang et al., 2021). However, K. pneumoniae has different degrees of resistance to other antibiotics and has a complex antimicrobial spectrum. As previously reported, K. pneumoniae shows intrinsic resistance to ampicillin (Fu et al., 2007). The rate of MDR (43.93%) and resistance to SOX (94.56%) are higher than that of the study by Zhang et al. (2020). β-lactam and aminoglycoside antibiotics have long been used to treat mastitis in five farms, which may explain why the resistance rate of isolates in mastitis milk to CEP, STR, and GEN was significantly higher than that of healthy milk. The development of antibiotic resistance is inseparable from the existence and spread of resistance genes. Our results show that blaTEM and blaSHV are present in mostly K. pneumoniae and blaCTX–M–15 is the main CTX-M gene detected, which is similar to the results of previous studies (Timofte et al., 2014; Carvalho et al., 2021). These extended-spectrum β-lactamase (ESBL) resistance genes can lead to resistance by hydrolysis of penicillins and cephalosporins (Algammal et al., 2020a). Correlation analysis showed that the resistance of K. pneumoniae to AMC, EFT, and CRO may be caused by hydrolysis mediated by blaOXA–1 and blaCTX–M–15. A variety of aminoglycoside-modifying enzyme genes (aadA1, aacc2, aacA4, and aphA1) were detected in the isolates. These enzymes can modify the active antimicrobial drugs that enter the cell and make them inactive (Belaynehe et al., 2017). The resistance of STR is mainly mediated by strA and strB and correlation analysis shows that the resistance of the STR of K. pneumoniae in this study may be more related to strB. It has been reported that tetA and floR/cmlA mediate the efflux of tetracycline and florfenicol, respectively, which can pump the drug out of the cell and reduce the intracellular drug concentration to generate resistance (Acosta-Pérez et al., 2015; Wang et al., 2018; Graesbøll et al., 2019). Notably, tetA and floR are the most frequently observed AMR genes in K. pneumoniae resistant to tetracycline and amide alcohols, respectively, which are consistent with the study by Nobrega et al. (2021). Although the three dihydrofolate synthase genes (sul1, sul2, and sul3) were detected to varying degrees, no significant correlation was found between them and SOX and SXT. Other resistance mechanisms, such as the expression of dihydrofolate reductase gene (dfr) and permeability barriers, are the causes of sulfonamide resistance (Skold, 2001). K. pneumoniae can be against quinolones through several mechanisms, including mutations in quinolone resistance-determining regions, plasmid-mediated quinolone resistance (PMQR), increased activity of efflux pumps, and decreased cellular permeability. Among them, mutation of QRDR is the main mechanism mediating quinolone resistance, especially gyrA and parC (Kareem et al., 2021). However, no amino acid changes were found in this study, which is different from the study by Saiful Anuar et al. (2013) and Fu et al. (2008), but similar to the study by Kim et al. (2018). Although the PMQR gene was detected to varying degrees in the isolates, its correlation with the ciprofloxacin resistance phenotype was not significant. We speculate that the activity of efflux pumps and the reduction of cell permeability may be the main reasons for mediating K. pneumoniae resistance to ciprofloxacin, but further verification is needed.
The occurrence of mastitis may be closely related to environmental hygiene. Virulence factors such as fimbriae, iron uptake, and lipopolysaccharide may play important roles in the pathogenesis of K. pneumoniae. The MDR of K. pneumoniae is a serious public health problem that still needs to be paid attention to, especially, the high resistance caused by the frequent use of β-lactams, aminoglycosides, and sulfonamides. The resistance of some antibiotics is attributed to the existence of resistance genes and other mechanisms such as efflux pumps and decreased permeability may be involved in the resistance of K. pneumoniae to sulfonamides and fluoroquinolones. Multiple sequence types of K. pneumoniae have the risk of cross-species and regional transmission. It is recommended to strengthen and regularly conduct surveillance, antibiotic resistance investigation, and traceability study on this strain. Further studies will be required to clarify whether the resistance and virulence characteristics of these isolates are affected by some movable genetic elements (such as plasmids) and whether they pose a risk of transmission.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
YW conceived the study and directed the study. XW, JL, JF, YF, RG, SH, and MZ performed the experiments. GW performed the somatic cell count of milk. XW performed the data analysis and wrote the manuscript. YW, HH, GC, and MS revised the manuscript. All authors have read and approved the final version of the manuscript.
This study was supported by the National Program for Quality and Safety Risk Assessment of Agricultural Products of China (GJFP2019027).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the Institut Pasteur teams for the curation and maintenance of BIGSdb-Pasteur databases at http://bigsdb.pasteur.fr/.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.858799/full#supplementary-material
Acosta-Pérez, G., Ibáñez-Cervantes, G., Bello-López, J. M., Hernández, J. M., Hernández-Montañez, Z., Giono-Cerezo, S., et al. (2015). Structural diversity of class 1 integrons in multiresistant strains of Escherichia coli isolated from patients in a hospital in Mexico city. Curr. Microbiol. 71, 501–508. doi: 10.1007/s00284-015-0876-9
Alanezi, G., Almulhem, A., Aldriwesh, M., and Bawazeer, M. (2022). A triple antimicrobial regimen for multidrug-resistant Klebsiella pneumonia in a neonatal intensive care unit outbreak: a case series. J Infect Public Health. 15, 138–141. doi: 10.1016/j.jiph.2021.10.008
Algammal, A. M., Hashem, H. R., Alfifi, K. J., Hetta, H. F., Sheraba, N. S., Ramadan, H., et al. (2021). atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci. Rep. 11:9476. doi: 10.1038/s41598-021-88861-w
Algammal, A. M., Hetta, H. F., Batiha, G. E., Hozzein, W. N., El Kazzaz, W. M., Hashem, H. R., et al. (2020a). Virulence-determinants and antibiotic-resistance genes of MDR-E. coli isolated from secondary infections following FMD-outbreak in cattle. Sci. Rep. 10:19779. doi: 10.1038/s41598-020-75914-9
Algammal, A. M., Mabrok, M., Sivaramasamy, E., Youssef, F. M., Atwa, M. H., El-kholy, A. W., et al. (2020b). Emerging MDR-Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and blaTEM, blaCTX-M, and tetA antibiotic-resistance genes. Sci. Rep. 10:15961. doi: 10.1038/s41598-020-72264-4
Arteaga-Livias, K., Pinzas-Acosta, K., Perez-Abad, L., Panduro-Correa, V., Rabaan, A. A., Pecho-Silva, S., et al. (2022). A multidrug-resistant Klebsiella pneumoniae outbreak in a Peruvian hospital: another threat from the COVID-19 pandemic. Infect Control Hosp. Epidemiol. 43, 267–268. doi: 10.1017/ice.2020.1401
Azargun, R., Soroush Barhaghi, M. H., Samadi Kafil, H., Ahangar Oskouee, M., Sadeghi, V., Memar, M. Y., et al. (2019). Frequency of DNA gyrase and topoisomerase IV mutations and plasmid-mediated quinolone resistance genes among Escherichia coli and Klebsiella pneumoniae isolated from urinary tract infections in Azerbaijan, Iran. J. Glob Antimicrob Resist. 17, 39–43. doi: 10.1016/j.jgar.2018.11.003
Belaynehe, K. M., Shin, S. W., Hong-Tae, P., and Yoo, H. S. (2017). Occurrence of aminoglycoside-modifying enzymes among isolates of Escherichia coli exhibiting high levels of aminoglycoside resistance isolated from Korean cattle farms. FEMS Microbiol. Lett. 364:fnx129. doi: 10.1093/femsle/fnx129
Benulič, K., Pirš, M., Couto, N., Chlebowicz, M., Rossen, J. W. A., Zorec, T. M., et al. (2020). Whole genome sequencing characterization of Slovenian carbapenem-resistant Klebsiella pneumoniae, including OXA-48 and NDM-1 producing outbreak isolates. PLoS One. 15:e0231503. doi: 10.1371/journal.pone.0231503
Brisse, S., Passet, V., Haugaard, A. B., Babosan, A., Kassis-Chikhani, N., Struve, C., et al. (2013). Wzi gene sequencing, a rapid method for determination of capsulartype for Klebsiella strains. J. Clin. Microbiol. 51, 4073–4078. doi: 10.1128/JCM.01924-13
Carvalho, I., Carvalho, J. A., Martínez-Álvarez, S., Sadi, M., Capita, R., Alonso-Calleja, C., et al. (2021). Characterization of ESBL-producing Escherichia coli and Klebsiella pneumoniae isolated from clinical samples in a Northern Portuguese hospital: Predominance of CTX-M-15 and high genetic diversity. Microorganisms 9:1914. doi: 10.3390/microorganisms9091914
Chen, F., Liu, Y., Ya, B., He, J., Leng, T., Sun, Y., et al. (2021). Latest effective measures to combat COVID-19: a review. Iran. J. Public Health. 50, 640–648. doi: 10.18502/ijph.v50i4.5989
Cheng, F., Li, Z., Lan, S., Liu, W., Li, X., Zhou, Z., et al. (2018). Characterization of Klebsiella pneumoniae associated with cattle infections in southwest China using multi-locus sequence typing (MLST), antibiotic resistance and virulence-associated gene profile analysis. Braz J. Microbiol. 49, 93–100. doi: 10.1016/j.bjm.2018.06.004
Cheng, J., Zhang, J., Han, B., Barkema, H. W., Cobo, E. R., Kastelic, J. P., et al. (2020). Klebsiella pneumoniae isolated from bovine mastitis is cytopathogenic for bovine mammary epithelial cells. J. Dairy Sci. 103, 3493–3504. doi: 10.3168/jds.2019-17458
Cheng, J., Zhou, M., Nobrega, D. B., Barkema, H. W., Xu, S., Li, M., et al. (2021). Genetic diversity and molecular epidemiology of outbreaks of Klebsiella pneumoniae mastitis on two large Chinese dairy farms. J. Dairy Sci. 104, 762–775. doi: 10.3168/jds.2020-19325
Chong, Y., Shimoda, S., and Shimono, N. (2018). Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol. 61, 185–188. doi: 10.1016/j.meegid.2018.04.005
Clinical and Laboratory Standards Institute [CLSI] (2020). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th Edn. Available online at: http://www.emeraldinsight.com (accessed November 3, 2020).
Clinical and Laboratory Standards Institute [CLSI] (2021). Performance Standards for Antimicrobial Susceptibility Testing 31st Edn. Available online at: http://www.emeraldinsight.com (accessed March 26, 2021).
Di Pilato, V., Errico, G., Monaco, M., Giani, T., Del Grosso, M., Antonelli, A., et al. (2021). The changing epidemiology of carbapenemase-producing Klebsiella pneumoniae in Italy: toward polyclonal evolution with emergence of high-risk lineages. J. Antimicrob Chemother. 76, 355–361. doi: 10.1093/jac/dkaa431
Follador, R., Heinz, E., Wyres, K. L., Ellington, M. J., Kowarik, M., Holt, K. E., et al. (2016). The diversity of Klebsiella pneumoniae surface polysaccharides. Microb Genom. 2:e000073. doi: 10.1099/mgen.0.000073
Fu, Y., Guo, L., Xu, Y., Zhang, W., Gu, J., Xu, J., et al. (2008). Alteration of GyrA amino acid required for ciprofloxacin resistance in Klebsiella pneumoniae isolates in China. Antimicrob Agents Chemother. 52, 2980–2983. doi: 10.1128/AAC.00151-08
Fu, Y., Zhang, F., Zhang, W., Chen, X., Zhao, Y., Ma, J., et al. (2007). Differential expression of blaSHV related to susceptibility to ampicillin in Klebsiella pneumoniae. Int. J. Antimicrob. Ag. 29, 344–347. doi: 10.1016/j.ijantimicag.2006.10.015
Gao, H., Gao, Q. L., Zhang, X., Guan, C., Luo, M. H., Zhang, H. B., et al. (2010). Improved medium for detection of Klebsiella in powdered milk. J. Food Saf. 30, 12–23. doi: 10.1111/j.1745-4565.2009.00186.x
Gato, E., Vázquez-Ucha, J. C., Rumbo-Feal, S., Álvarez-Fraga, L., Vallejo, J. A., Martínez-Guitián, M., et al. (2020). Kpi, a chaperone-usher pili system associated with the worldwide-disseminated high-risk clone Klebsiella pneumoniae ST-15. Proc. Natl. Acad. Sci. U.S.A. 117, 17249–17259. doi: 10.1073/pnas.1921393117
Graesbøll, K., Larsen, I., Clasen, J., Birkegård, A. C., Nielsen, J. P., Christiansen, L. E., et al. (2019). Effect of tetracycline treatment regimens on antibiotic resistance gene selection over time in nursery pigs. BMC Microbiol. 19:269. doi: 10.1186/s12866-019-1619-z
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
Guo, Y., Wang, S., Zhan, L., Jin, Y., Duan, J., Hao, Z., et al. (2017). Microbiological and clinical characteristics of hypermucoviscous Klebsiella pneumoniae isolates associated with invasive infections in China. Front Cell Infect Microbiol. 7:24. doi: 10.3389/fcimb.2017.00024
Harada, S., and Doia, Y. (2018). Hypervirulent Klebsiella pneumoniae: A call for consensus definition and international collaboration. J. Clin. Microbiol. 56:e00959-18. doi: 10.1128/JCM.00959-18
He, Y., Guo, X., Xiang, S., Li, J., Li, X., Xiang, H., et al. (2016). Comparative analyses of phenotypic methods and 16S rRNA, khe, rpoB genes sequencing for identification of clinical isolates of Klebsiella pneumoniae. Antonie Van Leeuwenhoek. 109, 1029–1040. doi: 10.1007/s10482-016-0702-9
Horii, T., Osaki, M., and Muramatsu, H. (2008). Fluoroquinolone resistance in clinical isolates of Klebsiella oxytoca. Chemotherapy. 54, 323–327. doi: 10.1159/000151266
Käppeli, N., Morach, M., Zurfluh, K., Corti, S., Nüesch-Inderbinen, M., and Stephan, R. (2019). Sequence types and antimicrobial resistance profiles of Streptococcus uberis isolated from bovine mastitis. Front Vet Sci. 6:234. doi: 10.3389/fvets.2019.00234
Kareem, S. M., Al-Kadmy, I., Kazaal, S. S., Mohammed, A. A., Aziz, S. N., Makharita, R. R., et al. (2021). Detection of gyrA and parC mutations and prevalence of plasmid-mediated quinolone resistance genes in Klebsiella pneumoniae. Infect Drug Resist. 14, 555–563. doi: 10.2147/IDR.S275852
Kim, S. Y., Jhun, B. W., Moon, S. M., Shin, S. H., Jeon, K., Jung Kwon, O., et al. (2018). Mutations in gyrA and gyrB in moxifloxacin-resistant mycobacterium avium complex and mycobacterium abscessus complex clinical isolates. Antimicrob Agents Chemother. 62:e00527-18. doi: 10.1128/AAC.00527-18
Koovapra, S., Bandyopadhyay, S., Das, G., Bhattacharyya, D., Banerjee, J., Mahanti, A., et al. (2016). Molecular signature of extended spectrum β-lactamase producing Klebsiella pneumoniae isolated from bovine milk in eastern and north-eastern India. Infect Genet Evol. 44, 395–402. doi: 10.1016/j.meegid.2016.07.032
Kuang, X. (2015). Prevalence and Molecular Characteristic of Resistance and Virulence of Three Different Livestock Pathogen Bacteria in Central China. Wuhan: HuaZhong Agricultural University.
Lee, C. R., Lee, J. H., Park, K. S., Jeon, J. H., Kim, Y. B., Cha, C. J., et al. (2017). Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology. hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 7:483. doi: 10.3389/fcimb.2017.00483
Lepuschitz, S., Hauser, K., Schriebl, A., Schlagenhaufen, C., Stöger, A., Chakeri, A., et al. (2020). Fecal Klebsiella pneumoniae carriage is intermittent and of high clonal diversity. Front Microbiol. 11:581081. doi: 10.3389/fmicb.2020.581081
Li, B., Zhao, Y., Liu, C., Chen, Z., and Zhou, D. (2014). Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 9, 1071–1081. doi: 10.2217/fmb.14.48
Li, L., Chen, X., and Chen, Z. (2019). Identification of key candidate genes in dairy cow in response to Escherichia coli mastitis by bioinformatical analysis. Front. Genet. 10:1251. doi: 10.3389/fgene.2019.01251
Liu, Y. C., Cheng, D. L., and Lin, C. L. (1986). Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med. 146, 1913–1916. doi: 10.1001/archinte.146.10.1913
Liu, Y. M., Li, B. B., Zhang, Y. Y., Zhang, W., Shen, H., Li, H., et al. (2014). Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland china. Antimicrob. Agents Ch. 58, 5379–5385. doi: 10.1128/AAC.02523-14
Liu, Z., Gu, Y., Li, X., Liu, Y., Ye, Y., Guan, S., et al. (2019). Identification and characterization of NDM-1-producing hypervirulent (hypermucoviscous) Klebsiella pneumoniae in China. Ann. Lab. Med. 39, 167–175. doi: 10.3343/alm.2019.39.2.167
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Massé, J., Dufour, S., and Archambault, M. (2020). Characterization of Klebsiella isolates obtained from clinical mastitis cases in dairy cattle. J. Dairy Sci. 103, 3392–3400. doi: 10.3168/jds.2019-17324
National standards of the people’s Republic of China. (2015). Rapid Diagnostic Technology For Subclinical Mastitis In Dairy Cow. Available online at: http://down.foodmate.net/standard/yulan.php?itemid=89398
Navon-Venezia, S., Kondratyeva, K., and Carattoli, A. (2017). Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275. doi: 10.1093/femsre/fux013
Nobrega, D. B., Calarga, A. P., Nascimento, L. C., Chande, V. C., de Lima, E. M., Langoni, H., et al. (2021). Molecular characterization of antimicrobial resistance in Klebsiella pneumoniae isolated from Brazilian dairy herds. J. Dairy Sci. 104, 7210–7224. doi: 10.3168/jds.2020-19569
Osman, K. M., Hassan, H. M., Orabi, A., and Abdelhafez, A. S. T. (2014). Phenotypic, antimicrobial susceptibility profile and virulence factors of Klebsiella pneumoniae isolated from buffalo and cow mastitic milk. Pathog. Glob. Health. 108, 191–199. doi: 10.1179/2047773214Y.0000000141
Paczosa, M. K., and Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. R. 80, 629–661. doi: 10.1128/MMBR.00078-15
Pérez-Losada, M., Cabezas, P., Castro-Nallar, E., and Crandall, K. A. (2013). Pathogen typing in the genomics era: MLST and the future of molecular epidemiology. Infect Genet Evol. 16, 38–53. doi: 10.1016/j.meegid.2013.01.009
Pitondo-Silva, A., Martins, V. V., Silva, C. F., and Da Stehling, E. G. (2015). Conjugation between quinolone-susceptible bacteria can generate mutations in the quinolone resistance-determining region, inducing quinolone resistance. Int. J. Antimicrob Agents. 45, 119–123. doi: 10.1016/j.ijantimicag.2014.07.018
Russo, T. A., Olson, R., Fang, C. T., Stoesser, N., Miller, M., MacDonald, U., et al. (2018). Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J. Clin. Microbiol. 56:e00776-18. doi: 10.1128/JCM.00776-18
Saiful Anuar, A. S., Mohd Yusof, M. Y., and Tay, S. T. (2013). Prevalence of plasmid-mediated qnr determinants and gyrase alteration in Klebsiella pneumoniae isolated from a university teaching hospital in Malaysia. Eur. Rev. Med. Pharmacol. Sci. 17, 1744–1747.
Sathiyabarathi, M., Jeyakumar, S., Manimaran, A., Jayaprakash, G., Pushpadass, H. A., Sivaram, M., et al. (2016). Infrared thermography: a potential noninvasive tool to monitor udder health status in dairy cows. Vet. World 9, 1075–1081. doi: 10.14202/vetworld.2016.1075-1081
Schroll, C., Barken, K. B., Krogfelt, K. A., and Struve, C. (2010). Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 10:179. doi: 10.1186/1471-2180-10-179
Shon, A. S., Bajwa, R. P. S., and Russo, T. A. (2014). Hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Virulence. 4, 107–118. doi: 10.4161/viru.22718
Skold, O. (2001). Resistance to trimethoprim and sulfonamides. Vet. Res. 32, 261–273. doi: 10.1051/vetres:2001123
Timofte, D., Maciuca, I. E., Evans, N. J., Williams, H., Wattret, A., Fick, J. C., et al. (2014). Detection and molecular characterization of Escherichia coli CTX-M-15 and Klebsiella pneumoniae SHV-12 β-Lactamases from bovine mastitis isolates in the United Kingdom. Antimicrob. Agents Ch. 58, 789–794. doi: 10.1128/AAC.00752-13
Turnidge, J., Kahlmeter, G., and Kronvall, G. (2006). Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin. Microbiol. Infect. 12, 418–425. doi: 10.1111/j.1469-0691.2006.01377.x
Wang, G., Zhao, G., Chao, X., Xie, L., and Wang, H. (2020). The characteristic of virulence, biofilm and antibiotic resistance of Klebsiella pneumoniae. Int. J. Env. Res. Pub. He. 17:6278. doi: 10.3390/ijerph17176278
Wang, T., Lin, J., Chang, J., Hiaso, Y., Wang, C., Chiu, S. K., et al. (2021). Virulence among different types of hypervirulent Klebsiella pneumoniae with multi-locus sequence type (MLST)-11, Serotype K1 or K2 strains. Gut Pathog. 13:40. doi: 10.1186/s13099-021-00439-z
Wang, Y., Li, X., Chen, C., Zhang, J., and Wang, G. (2018). Detection of floR gene and active efflux mechanism of Escherichia coli in Ningxia. China. Microb. Pathog. 117, 310–314. doi: 10.1016/j.micpath.2018.02.042
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020). Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect Dis. 20, 1161–1171. doi: 10.1016/S1473-3099(20)30149-3
Wareth, G., and Neubauer, H. (2021). The animal-foods-environment interface of Klebsiella pneumoniae in Germany: an observational study on pathogenicity, resistance development and the current situation. Vet Res. 52:16. doi: 10.1186/s13567-020-00875-w
Wyres, K. L., and Holt, K. E. (2018). Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 45, 131–139. doi: 10.1016/j.mib.2018.04.004
Wyres, K. L., Lam, M. M. C., and Holt, K. E. (2020). Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 18, 344–359. doi: 10.1038/s41579-019-0315-1
Yang, F., Deng, B., Liao, W., Wang, P., Chen, P., and Wei, J. (2019a). High rate of multiresistant Klebsiella pneumoniae from human and animal origin. Infect Drug Resist. 12, 2729–2737. doi: 10.2147/IDR.S219155
Yang, Y., Higgins, C. H., Rehman, I., Galvao, K. N., Brito, I. L., Bicalho, M. L., et al. (2019b). Genomic diversity, virulence, and antimicrobial resistance of Klebsiella pneumoniae Strains from Cows and Humans. Appl. Environ. Microbiol. 85:e02654-18. doi: 10.1128/AEM.02654-18
Yang, Y., Peng, Y., Jiang, J., Gong, Z., Zhu, H., Wang, K., et al. (2021). Isolation and characterization of multidrug-resistant Klebsiella pneumoniae from raw cow milk in Jiangsu and Shandong provinces. China. Transbound Emerg. Dis. 68, 1033–1039. doi: 10.1111/tbed.13787
Zhang, S., Yang, G., Ye, Q., Wu, Q., Zhang, J., and Huang, Y. (2018). Phenotypic and genotypic characterization of Klebsiella pneumoniae isolated from retail foods in China. Front Microbiol. 9:289. doi: 10.3389/fmicb.2018.00289
Zhang, S., Zhang, X., Wu, Q., Zheng, X., Dong, G., Fang, R., et al. (2019). Clinical, microbiological, and molecular epidemiological characteristics of Klebsiella pneumoniae-induced pyogenic liver abscess in southeastern China. Antimicrob Resist Infect Control. 8:166. doi: 10.1186/s13756-019-0615-2
Zhang, X. F., Wang, L., Hu, B., Zhang, A. H., Zheng, J. H., Lu, J. J., et al. (2020). Detection rate, antimicrobial resistance and molecular types of Klebsiella pneumoniae from stool samples of outpatients with diarrhea-syndrome in Tai’an. Chin J Epidemiol. 41, 423–428. doi: 10.3760/cma.j.issn.0254-6450.2020.03.027
Zhang, Y., Zhao, C., Wang, Q., Wang, X., Chen, H., Li, H., et al. (2016). High prevalence of hypervirulent Klebsiella pneumoniae infection in china: Geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob. Agents Ch. 60, 6115–6120. doi: 10.1128/AAC.01127-16
Zhao, Q., Berglund, B., Zou, H., Zhou, Z., Xia, H., Zhao, L., et al. (2021). Dissemination of blaNDM-5 via IncX3 plasmids in carbapenem-resistant Enterobacteriaceae among humans and in the environment in an intensive vegetable cultivation area in eastern China. Environ. Pollut. 273:116370. doi: 10.1016/j.envpol.2020.116370
Zhong, X. S., Li, Y. Z., Ge, J., Xiao, G., Mo, Y., Wen, Y. Q., et al. (2020). Comparisons of microbiological characteristics and antibiotic resistance of Klebsiella pneumoniae isolates from urban rodents, shrews, and healthy people. BMC Microbiol. 20:12. doi: 10.1186/s12866-020-1702-5
Keywords: Klebsiella pneumoniae, cow mastitis, antimicrobial resistance, virulence gene, wzi gene sequencing, MLST
Citation: Wu X, Liu J, Feng J, Shabbir MAB, Feng Y, Guo R, Zhou M, Hou S, Wang G, Hao H, Cheng G and Wang Y (2022) Epidemiology, Environmental Risks, Virulence, and Resistance Determinants of Klebsiella pneumoniae From Dairy Cows in Hubei, China. Front. Microbiol. 13:858799. doi: 10.3389/fmicb.2022.858799
Received: 20 January 2022; Accepted: 08 April 2022;
Published: 04 May 2022.
Edited by:
Bao-Tao Liu, Qingdao Agricultural University, ChinaReviewed by:
Chang-Wei Lei, Sichuan University, ChinaCopyright © 2022 Wu, Liu, Feng, Shabbir, Feng, Guo, Zhou, Hou, Wang, Hao, Cheng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yulian Wang, d2FuZ3l1bGlhbkBtYWlsLmh6YXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.