
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 March 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.858559
 Wen-Liang Yin1,2,3
Wen-Liang Yin1,2,3 Zhen-Yu Xie1,2,3,4*
Zhen-Yu Xie1,2,3,4* Yan-Hua Zeng1,2
Yan-Hua Zeng1,2 Ju Zhang1,2,3
Ju Zhang1,2,3 Hao Long1,2
Hao Long1,2 Wei Ren1,2,3,4
Wei Ren1,2,3,4 Xiang Zhang1,2,3,4
Xiang Zhang1,2,3,4 Xiao-Ni Cai1,2,3,4
Xiao-Ni Cai1,2,3,4 Ai-You Huang1,2,3,4
Ai-You Huang1,2,3,4
The stringent response mediated by the signal molecule (p)ppGpp is involved in response to multiple environmental stresses and control of various physiological processes. Studies have revealed that (p)ppGpp strongly affects the formation and maintenance of several bacterial biofilms. However, the specific regulatory roles of (p)ppGpp in biofilms, especially in the expression of genes related to cell motility and exopolysaccharides (EPSs) production, remain poorly understood. We recently reported two (p)ppGpp synthetase genes relA and spoT from the epizootic pathogen Vibrio alginolyticus. Herein, we found that the (p)ppGpp synthetase genes of V. alginolyticus contributed to biofilm formation at low cell density and biofilm detachment at high cell density, respectively, in polystyrene microtiter plates. Quantitative reverse transcription PCR (qRT-PCR) analysis revealed that the expression levels of both EPSs and motility associated genes were consistent with the development of biofilms. Besides, the (p)ppGpp synthetase gene spoT was found to be closely involved in the regulation of flagellum, smooth/translucent colony morphology and spotty pellicle at the air-liquid interface. Interestingly, pleiotropic phenotypes of ΔrelAΔspoT were similar to that of the rpoN (σ54) deletion mutant. Meanwhile, the absence of (p)ppGpp synthetase genes significantly reduced the expression levels of rpoN at low cell density, suggesting that (p)ppGpp may mediate the formation via positively affecting the alternative sigma factor RpoN. These findings allow us to propose (p)ppGpp as a crucial regulator for biofilm development in V. alginolyticus, in view of the regulatory roles of relA and spoT in cell motility and EPSs production.
Vibrio alginolyticus is a halophilic, facultative anaerobic, and Gram-negative opportunistic pathogen that inhabits coastal waters and estuaries. V. alginolyticus has been reported as a notorious causative agent of vibriosis in many marine animals, including fish, shellfish, and shrimp (Lee et al., 1996; Gómez-León et al., 2005; Kahla-Nakbi et al., 2006; Austin, 2010). Besides, it can cause ear infections, gastroenteritis and septicemia in humans through wounds or ingestion of contaminated food (Mustapha et al., 2013; Jacobs Slifka et al., 2017). The robust response abilities to environmental stresses and stringent control of virulence genes are thought to be essential for the pathogenicity of this bacterium (Wang et al., 2007; Rui et al., 2008).
Biofilm formation is one of the most effective strategies for pathogenic Vibrio spp. to confront harsh environmental conditions (Faruque et al., 2006; Croxatto et al., 2007; Hall and Mah, 2017). Initially, flagella and other motility factors accelerate bacterial attachment to the surface and initiate microcolony formation (O’Toole and Kolter, 1998; Utada et al., 2014). Exopolysaccharides (EPSs), the most prevalent component of extracellular matrix, are produced to promote the development of mature biofilms (Yildiz and Visick, 2009; Teschler et al., 2015) and responsible for the transition of colony morphology between opaque/rugose (Op) and translucent/smooth (Tr) (Chen et al., 2009). EPSs production depends on transcription of the Vibrio polysaccharide synthesis (vps) genes (Yildiz and Schoolnik, 1999). Deletion of vpsM or other vps genes lead to a reduction in biofilm formation and EPSs production in V. cholera, indicating vps genes have important roles in biofilm formation (Fong et al., 2010). A complex intracellular regulatory network, including transcriptional activators, alternative sigma factors and other factors, could directly regulate the vps genes (Zhu et al., 2002; Teschler et al., 2015). VpsT, a member of the UhpA (FixJ) family of transcriptional regulators, is found to be required for vps genes expression and development of rugose colonial morphology in V. cholerae O1 El Tor (Casper-Lindley and Yildiz, 2004; Beyhan et al., 2007). The alternative sigma factor-54 (RpoN) has been reported to be strongly involved in affecting vps genes expression of Vibrio spp. (Yildiz and Visick, 2009; Cheng et al., 2018). Interestingly, RpoN could indirectly promote the vps gene expression at low cell density in V. cholera (Herzog et al., 2019). Finally, bacteria stimulated by nutrient stress or extracellular signals would downregulate vps genes, degradate polysaccharides, and reorientate in direction to escape from the biofilm matrix mesh (Fong and Yildiz, 2007; Pratt et al., 2009; Hay and Zhu, 2015; Bridges et al., 2020).
The stringent response mediated by the bacterial alarmones pppGpp and ppGpp [collectively termed (p)ppGpp] is considered as an important adaptive response to stressful conditions, whether living in the environment or in the host (Srivatsan and Wang, 2008). In many Gram-negative bacteria, such as V. alginolyticus, the metabolism of (p)ppGpp is controlled by two conserved enzymes: monofunctional synthetase RelA specialized for the synthesis of (p)ppGpp and bifunctional synthetase/hydrolase SpoT (Magnusson et al., 2005; Gaca et al., 2015). As a global regulator, (p)ppGpp is able to activate or repress the transcription of many genes by directly regulating RNA polymerase in cooperation with the RNA polymerase-binding transcription factor DksA or indirectly interacting with σ-factors (Dalebroux and Swanson, 2012). It has been reported that (p)ppGpp synthetases are involved in the regulation of biofilm formation in several bacterial species (Sugisaki et al., 2013; Azriel et al., 2016; Díaz-Salazar et al., 2017; Colomer-Winter et al., 2019). The lack of (p)ppGpp synthetase genes in most pathogens showed reduced biofilm formation (Renier et al., 2011; He et al., 2012; Sugisaki et al., 2013), while some (p)ppGpp-deletion mutants could form significantly enhanced biofilms compared with the wild type, such as Actinobacillus pleuropneumoniae (Li et al., 2015) and Pseudomonas putida KT2440 (Liu et al., 2017). This means that (p)ppGpp synthetase genes play significant regulatory roles in biofilm formation among bacterial species.
Although a close link between (p)ppGpp synthetase genes and biofilm formation has been found, the regulation mechanism of (p)ppGpp synthetase genes on biofilm formation remains unclear. In this manuscript, we constructed the (p)ppGpp-synthetase deletion mutants (ΔrelA, ΔrelAΔspoT) and observed the alteration of biofilm formation, EPSs production, colonial morphology, and flagellum by comparison with the wild type (WT) and complemented strains (ΔrelA-pRelA, ΔrelAΔspoT-pSpoT). Finally, we elaborated the potential regulatory role of (p)ppGpp synthetase genes in biofilm formation by analyzing these phenotypes and the expression levels of several key genes related to biofilm formation and motility. These findings help us better understand the complex regulatory network of biofilm formation in V. alginolyticus.
Vibrio alginolyticus HN08155 and its derivative strains were cultured at 30°C and 180 rpm in 2216E medium consisting of 5 g/L tryptone, 1 g/L yeast extract, and 0.01 g/L FePO4. Escherichia coli β2163 was grown at 37°C and 180 rpm in standard Luria-Bertani (LB) medium containing 0.3 mM diaminopimelic acid (DAP) (Luo et al., 2015). Antibiotics were used at the following concentrations: 100 μg/mL ampicillin (Amp) was used for the growth of Vibrio spp., 10 μg/mL chloramphenicol (Cm) was additionally added to complemented strains for integrated plasmids. All reagents were purchased from Solarbio (Beijing, China).
All strains and plasmids used are described in Supplementary Table 1. The (p)ppGpp synthetase gene mutants of V. alginolyticus HN08155 were generated by allelic exchange (Milton et al., 1996). This method has been described in detail in our previous work (Yin et al., 2021), and the oligonucleotides used in these procedures are listed in Supplementary Table 2. In brief, the sucrose-sensitive suicide plasmid pDM4-relA containing the fusion product of a 542 bp upstream fragment and a 542 bp downstream fragment of relA coding sequence was introduced into E. coli β2163 cells by heat shock. Subsequently, the recombinant plasmid pDM4-relA was integrated into wild type through conjugation, and the relA internal coding sequence was deleted after homologous recombination occurred on both homologous arms. Single crossover mutants with plasmids integrated into specific chromosomal loci were obtained on 2216E agar plates supplemented with Amp and Cm. The second crossover mutants were screened on 2216E agar plates supplemented with Amp and 10% sucrose. The in-frame deletion mutant ΔrelA was confirmed by PCR and DNA sequencing. For generation of the double deletion strain ΔrelAΔspoT, the suicide plasmid pDM4-spoT containing the fusion product of a 563 bp upstream fragment and a 500 bp downstream fragment of spoT coding sequence was transferred into the single deletion mutant ΔrelA, and the spoT internal coding sequence was deleted after double cross-over recombination. High cellular (p)ppGpp level in strains would inhibit cell growth, due to the lack of hydrolysis activity mediated by SpoT to degrade (p)ppGpp synthesized by RelA, so we failed to construct a spoT single mutant (Xiao et al., 1991). In order to construct the complemented plasmids, the open reading frame of relA and the linear segment pACYC184 (without tetracycline resistance but with chloramphenicol resistance gene) were amplified, respectively. The open reading frame of relA was cloned into the linear segment of pACYC184 by using ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). The opening reading frame of tetracycline resistance gene was replaced by that of relA gene but with its native promoter to produce the pACYC184-relA plasmids expressing relA gene. And then, the resulting pACYC184-relA plasmid was transformed into the ΔrelA mutant to generate the complemented strain with AmpR and CmR, designated ΔrelA-pRelA. Similarly, the pACYC184-spoT plasmid was transformed into the ΔrelAΔspoT mutant to generate the complemented strain with AmpR and CmR, designated ΔrelAΔspoT-pSpoT.
The biofilm assay was performed following a previously described protocol with modification (O’Toole and Kolter, 1998). After overnight incubation, the cultures were adjusted to an OD600 of 0.6, diluted 1:100 into 50 mL fresh LB medium without antibiotics. Each 200 μL sample was taken from the diluted medium into the 96-well polystyrene microplate and cultivated statically at 30°C. After culturing for 2, 4, 6, 8, 10, 12, 14, 18, 24, and 36 h respectively, bacterial cells were removed, rinsed once with phosphate buffer saline (PBS) and 220 μL of 0.1% crystal violet was added to stain biofilm for 30 min. The residual stains were then removed, and the stained biofilms were washed twice with PBS. Finally, the amount of biofilm was determined at 570 nm using a microplate reader (Epoch2, BioTeK) after dissolving crystal violet with 200 μL of 95% ethanol for 1 h. Three separate experiments were performed with biological triplicates each. To exclude growth effects on biofilm formation, the cell density was measured at 600 nm, and biofilm formation was normalized by dividing total biofilm by the cell density.
In spotty pellicle assay, 5 mL above diluted medium was added in each glass tube and incubated without shaking for 16 h at 30°C. For colonial morphology assay, the cultures were adjusted to an OD600 of 0.6, and collected by centrifuging at 12,000 rpm for 2 min. After removing the supernatant, bacteria are resuspended in 10 μL LB medium. Subsequently, 10 μL mixed medium is spotted on a trypticase soy sheep blood agar plate (Huankai Microbial, Guangdong, China) and cultivated statically at 30°C for 24 h, after which the colonial morphology was observed and photographed.
The modified phenol-sulfuric acid method was used to determine the polysaccharides production (Liu et al., 2021). In the beginning, 8 mg glucose was dissolved into the 100 mL volumetric flask, and then 0, 0.2, 0.4, 0.6, 0.8, and 1 mL glucose solution were added to each centrifuge tube, respectively. Double-distilled water was added to the centrifuge tube containing less than 1 mL glucose solution. Subsequently, 1 mL phenol solution and 5 mL concentrated sulfuric acid were added into each tube, after 30 min of reaction, the absorbance at 490 nm was respectively measured to draw the standard glucose curve. Subsequently, 1 mL phenol solution and 5 mL concentrated sulfuric acid were added to the tube, after 30 min of reaction, the absorbance at 490 nm was respectively measured to draw the standard glucose curve. Each overnight culture was adjusted to an OD600 of 0.6, diluted 1:100 into 50 mL LB medium and 6 mL broth was incubated to the 6-well polystyrene microplate statically at room temperature for 6 and 24 h, respectively. 5 mL bacterial culture was collected and centrifuged at 5,000 rpm and 4°C for 5 min, and 1 mL supernatant was collected and transferred into a new tube. Afterward, the supernatant, phenol solution (9%, v/v) and concentrated sulfuric acid were added to the tube sequentially in the ratio of 1:1:5 (v:v:v). After 30 min of reaction, the absorbance at 490 nm was measured, and the amount of polysaccharides was further calculated according to the above standard glucose curve.
Vibrio containing Amp antibiotic (OD600 = 0.6) were fixed in 2.5% glutaric dialdehyde, and then bacterial flagella were observed by using the transmission electron microscope (TEM, JEM-2100, Japan). To evaluate swarming ability, fresh cultures of V. alginolyticus strains containing antibiotics (OD600 = 0.6) were centrifuged, precipitated, and resuspended in 10 μL 2216E medium, and then 1 μL culture was spotted on 0.9% (w/v) 2216E agar plates at room temperature.
The WT and mutants were cultured in LB medium containing Amp overnight, and then diluted 1:100 into fresh medium and grown to the exponential phase (OD600 = 0.6) and stationary phase (OD600 = 1.2). RNA was extracted with a Bacteria Total RNA Extraction Kit (Promega, Madison, WI, United States), and then HiScript II Q RT SuperMix (+gDNA wiper) (Vazyme, Nanjing, China) was used for reverse transcription after determining the RNA quality by Nanophotometer NP80 (IMPLEN, München, Germany). The final cDNA samples were analyzed by Quantitative Reverse Transcription PCR (qRT-PCR) using ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China). Primers used in the qRT-PCR analysis are listed in Table 1. Expression of the genes encoding helix-turn-helix transcriptional regulator VpsT, polysaccharides biosynthesis protein VpsM and VpsH, chemotaxis protein CheR, flagellar hook protein FlgE, flagellar basal body L-ring protein FliD, and alternative sigma transcription factor RpoN was determined in triplicate. The 16S rRNA gene was used as an endogenous control for normalization of expression values. The 2–ΔΔCt method was used to quantify and compare each gene expression (Livak and Schmittgen, 2001).
Three separate experiments were performed with biological triplicates each, and results were presented as mean ± standard deviation (SD; n = 3). Analysis of statistical differences was conducted on GraphPad Prism version 7.01 for Windows (GraphPad Software, SanDiego, CA, United States) through one-way analysis of variance (ANOVA) with Dunnett’s post-test. Differences were considered significant by calculated p value (*p ≤ 0.05, **p ≤ 0.01), and p value ≥ 0.05 was regarded as no significant.
The amount of biofilm formation of WT, ΔrelA, ΔrelAΔspoT, ΔrelA-pRelA, and ΔrelAΔspoT-pSpoT was evaluated by staining with crystal violet. Afterward, biofilm formation was normalized by dividing total biofilm by the cell density (Supplementary Figure 1) to exclude growth effects (Figure 1A). Overall, the trend of biofilm formation in WT was to continuously increase at low cell density and then reduce at high cell density. The increasing amount of biofilm formation reached a peak at 6 h (OD570/OD600 = 3.61), and kept a high level for a period of time until 10 h. Interestingly, the second small peak of biofilm formation appeared at 12 h (OD570/OD600 = 1.04) and decreased again to a low level. The amount of biofilm formation in ΔrelA was significantly lower than that of the WT at low cell density (Figure 1B, p ≤ 0.01). However, it maintained a high level (OD570/OD600 = 2.61–3.36) during 8–14 h and then began to decompose slowly until close to the level of WT (Figure 1C). In terms of the biofilm formation in ΔrelAΔspoT, its absorbance value was significantly lower than that of WT at low cell density (Figure 1B, p ≤ 0.05). Surprisingly, the amount of biofilm formation reached its peak at 8 h (OD570/OD600 = 4.66), but the biofilm did not decompose within 36 h, even maintained at a high level (Figure 1C). The biofilm formation of ΔrelA-pRelA and ΔrelAΔspoT-pSpoT were similar to WT and ΔrelA, respectively. An obvious crystal violet-stained circular ring appeared at the air-liquid interface (data not shown), suggesting that bacteria might tend to accumulate in oxygen-rich areas. However, there was additional biofilm staining at the bottom of the microtiter plates in ΔrelAΔspoT at high cell density (8–36 h), exhibiting robust bacterial self-aggregation ability. These results revealed that the deletion of two (p)ppGpp synthetase genes led to postponed biofilm formation at low cell density, and the absence of relA and spoT resulted in delayed and failed biofilm disassembly, respectively.
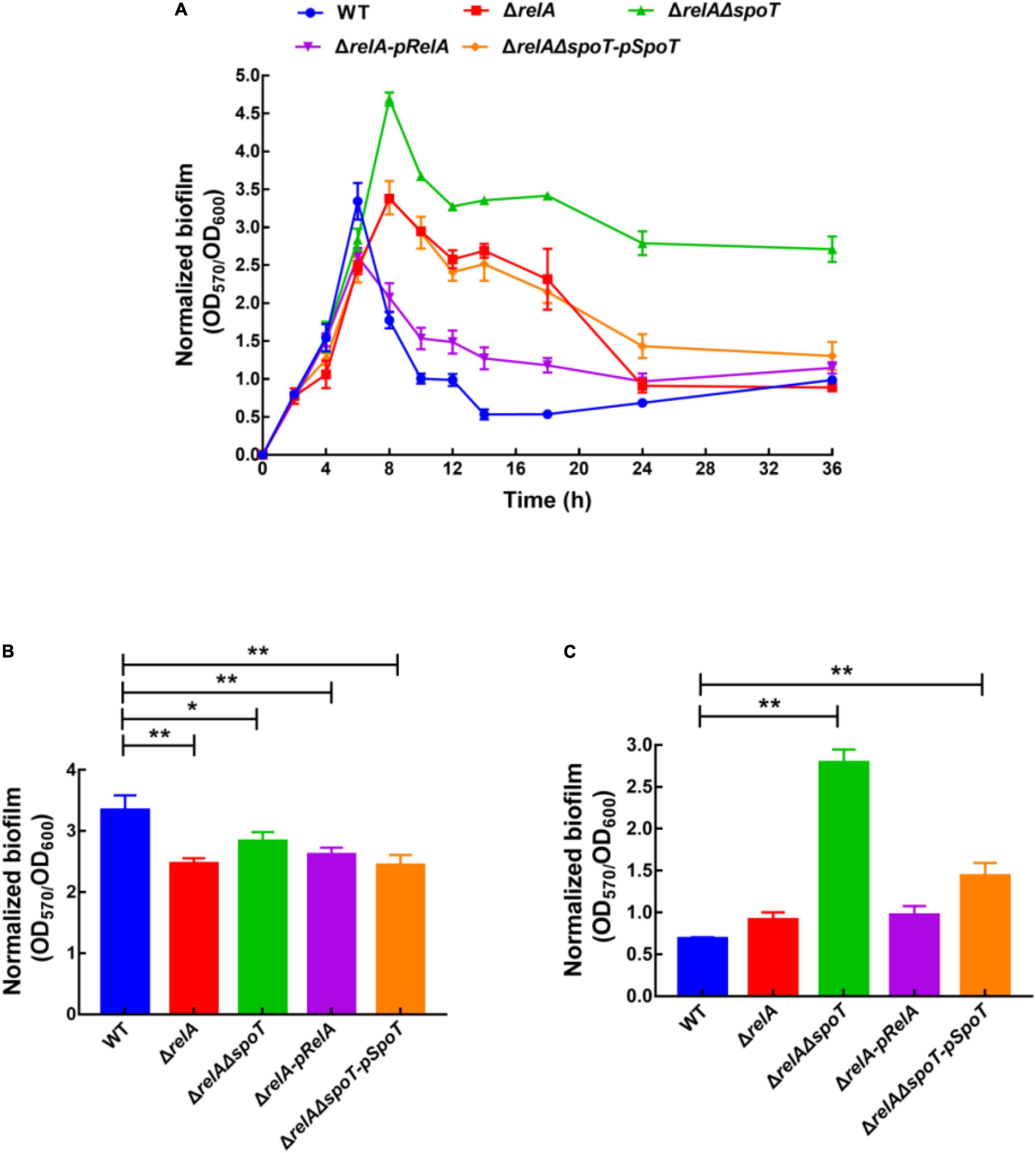
Figure 1. Normalized biofilm formation (total amount of biofilm/growth) of WT, ΔrelA, ΔrelAΔspoT, ΔrelA-pRelA, and ΔrelAΔspoT-pSpoT in 96-well polystyrene microtiter plates at different cell densities (A). The normalized biofilm differences between WT and ΔrelA, ΔrelAΔspoT, ΔrelA-pRelA, and ΔrelAΔspoT-pSpoT were shown at 6 (B) and 24 h (C), respectively. Three separate experiments were performed with biological triplicates each, and results were presented as mean ± SD (n = 3). * indicates p ≤ 0.05, and ** indicates p ≤ 0.01.
It has been reported that the formation of spotty pellicle strongly depends on oxygen level (Hare et al., 1981). After static incubation at 30°C for 16 h in the glass tube, the liquid solutions of WT, ΔrelA, and ΔrelAΔspoT-pSpoT (OD600 = 0.8–1.0) were more turbid than that of ΔrelAΔspoT (OD600 = 0.6–0.8), without apparent bacterial flocculation and transparent air-liquid surface layer (Figure 2A). However, a spotty, unattached floating and thicker pellicle of ΔrelAΔspoT was observed at the air-liquid surface layer (Figure 2A). This result implied that spoT might participate in sensing oxygen and regulation of spotty pellicle.
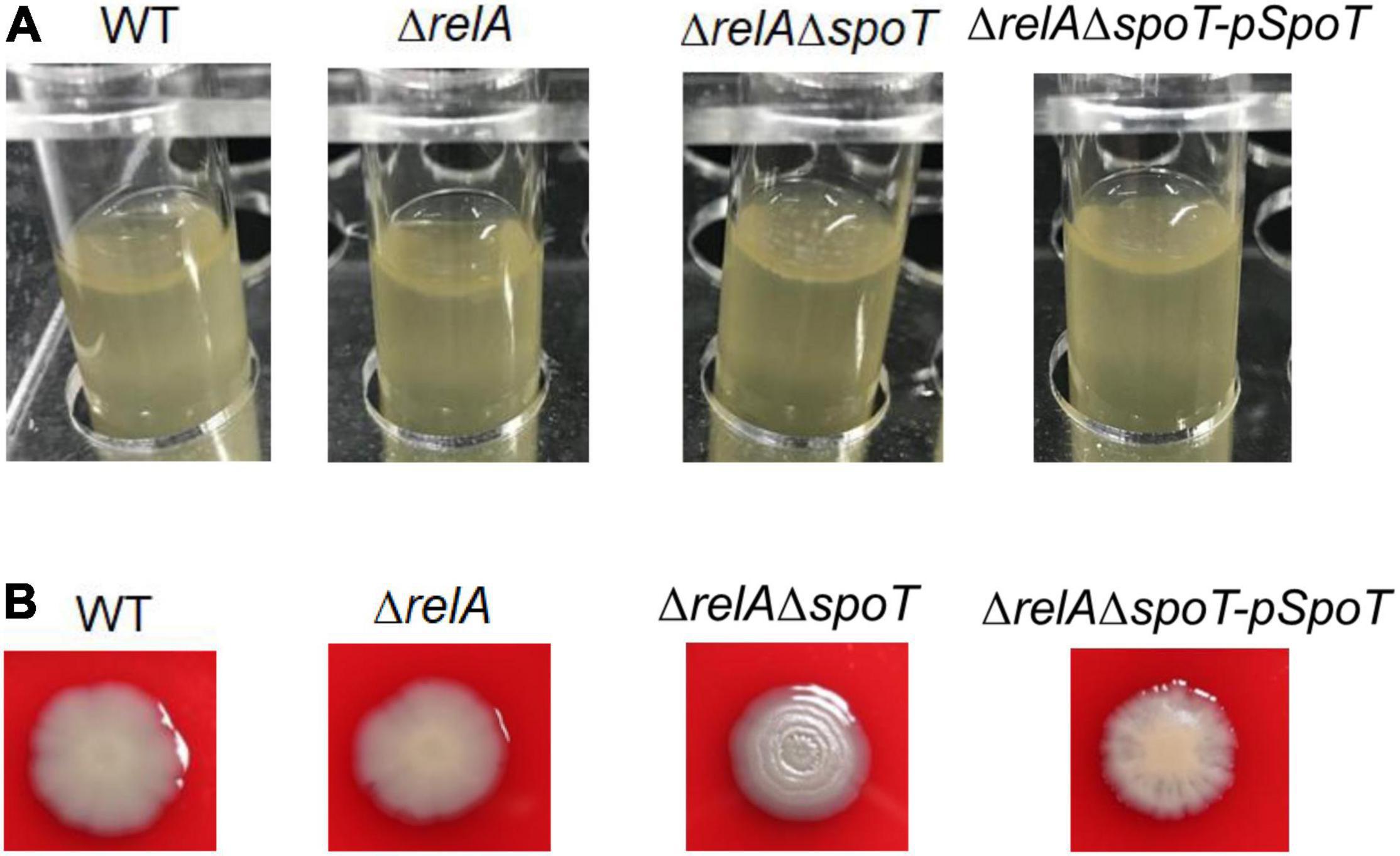
Figure 2. The effects of relA and spoT on spotty pellicle and colony morphology. (A) Air-liquid interface pellicle formed by WT, ΔrelA, ΔrelAΔspoT, and ΔrelAΔspoT-pSpoT in the glass tube for 16 h at 30°C without shaking. (B) Colony morphologies of WT, its derivative and complemented strains. Three colonies of each strain were grown on a trypticase soy sheep blood agar plate for 24 h at 30°C. Three separate experiments were performed with biological triplicates each, and a representative image was displayed.
A surprising but expected phenomenon was discovered in the hemolysis test when we determined whether the hemolytic ability of V. alginolyticus HN08155 could be affected by the absence of (p)ppGpp synthetase genes. In the beginning, there was no obvious difference in colony morphology among all strains (data not shown), whereas the colony morphology of ΔrelAΔspoT began to wrinkle gradually during cultivation. As shown in Figure 2B, WT and ΔrelA grew into gray-white, smooth/translucent, and neat-edged colonies after 24 h of cultivation on trypticase soy sheep blood agar plate. In contrast, the colony morphology of ΔrelAΔspoT showed lackluster and surface rugose, and ΔrelAΔspoT-pSpoT exhibited slightly wrinkled colony morphology. This finding indicated that spoT rather than relA played a variable role in controlling colony phase variation of V. alginolyticus: the lack of spoT resulted in the transformation of gray-white and smooth/translucent colonies into surface wrinkled colonies, while the presence of spoT inhibited the formation of opaque/rugose colonies.
Exopolysaccharides are an indispensable component of biofilm formation in V. alginolyticus (Chen et al., 2009). Based on our previous experiments, 6 and 24 h were chosen as the representatives of the low cell density and high cell density, respectively (Zhang et al., 2021). Our results indicated that the EPSs content of WT was the highest at low cell density (Figure 3A), but it gradually decreased with the disruption of (p)ppGpp synthetase genes, which was almost 3- and 5-fold of that in ΔrelA and ΔrelAΔspoT, respectively (Figure 3B). Complementation of the relA or spoT gene into mutant strains ΔrelA-pRelA and ΔrelAΔspoT-pSpoT respectively increased the synthesis of EPSs, although these contents were relatively lower in comparison with that in WT (p ≤ 0.01). At high cell density, WT had the lowest EPSs content, followed by ΔrelA, while ΔrelAΔspoT had the highest EPSs content. Notably, the content of EPSs in ΔrelAΔspoT was almost 1.5 times that of WT and ΔrelA. The EPSs content in ΔrelA was almost the same as that in WT (p ≥ 0.05), and supplementation of the spoT gene in ΔrelAΔspoT could reduce the secretion of EPSs to some extent. Overall, both relA and spoT favored the synthesis of EPSs at low cell density, and only spoT helped the degradation of it at high cell density.
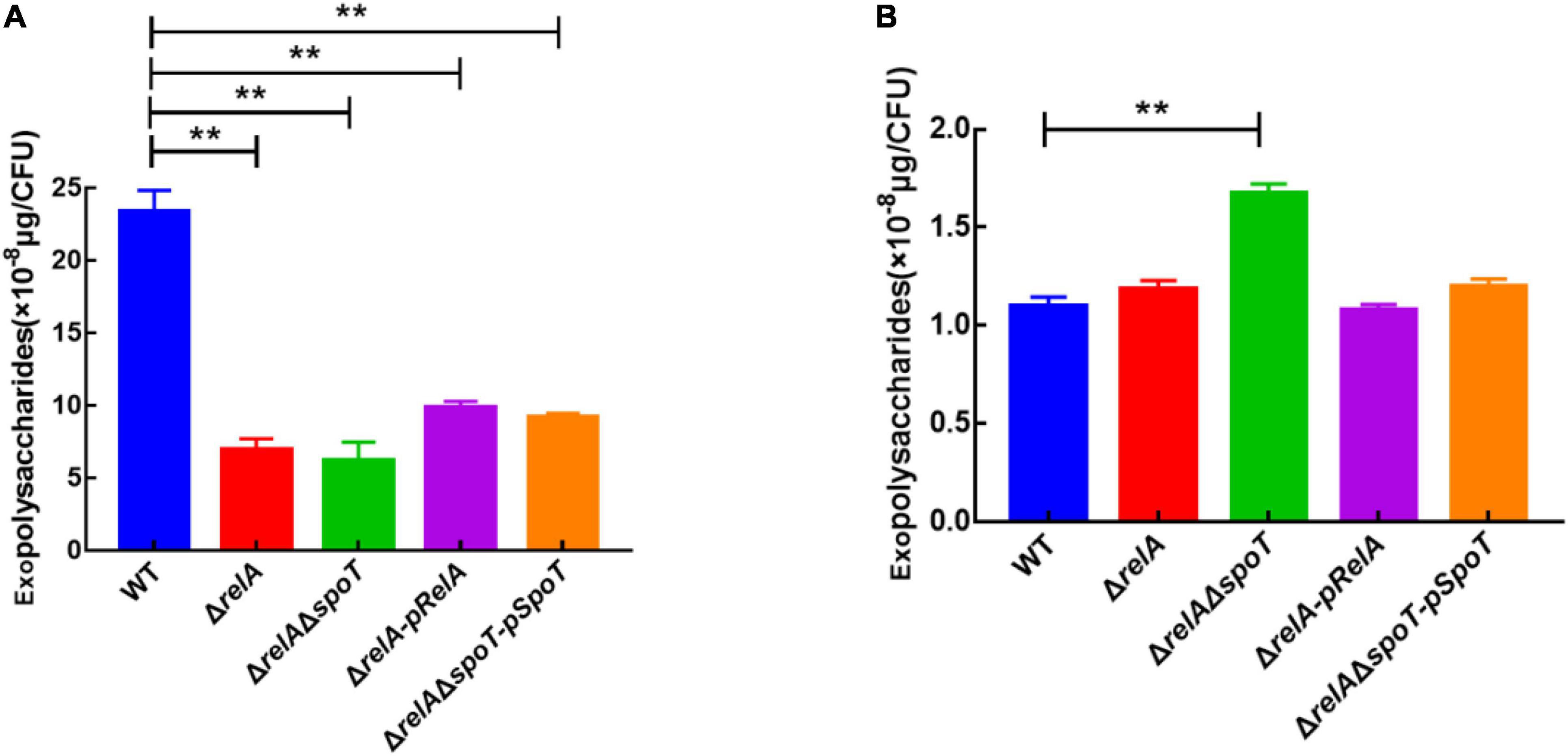
Figure 3. The effects of relA and spoT on EPSs production. The content of EPSs was measured by phenol-sulfuric acid method in WT, ΔrelA, ΔrelAΔspoT, ΔrelA-pRelA, and ΔrelAΔspoT-pSpoT at 6 (A) and 24 h (B), respectively. Three separate experiments were performed with biological triplicates each, and results were presented as mean ± SD (n = 3). ** indicates p ≤ 0.01.
Transmission electron microscope observation showed that V. alginolyticus HN08155 was short and rod-shaped, with a single curved polar flagellum that was 2–3 times longer than that of the bacterium (Figure 4A). The flagellum of ΔrelA and ΔrelAΔspoT-pSpoT was identical to WT (Figures 4B,D), but the lack of spoT resulted in the loss of flagellum (Figure 4C). There was no doubt that spoT played a critical role in synthesizing flagellum. Previous study revealed that the swimming ability of ΔrelAΔspoT decreased sharply (Yin et al., 2021). Here we further investigated whether the absence of spoT would affect its swarming ability. After 12 h of continuous culture on 0.9% 2216E agar plates at room temperature, the swarming halo diameter of the WT, ΔrelA, and ΔrelAΔspoT-pSpoT averaged 6.80 ± 0.3, 7.65 ± 0.5, and 5.75 ± 0.4 mm, respectively, while those of ΔrelAΔspoT averaged 2.10 ± 0.8 mm only (Figures 5A,B). The diameters of the swarming rings of ΔrelAΔspoT were significantly smaller than that of the WT, ΔrelA and complemented strains (Figure 5B, p ≤ 0.01), demonstrating that the swarming ability of ΔrelAΔspoT was sharply diminished. Taken together, the disruption of spoT in V. alginolyticus not only negatively affected the production of flagellum, but also significantly impaired its swarming ability.
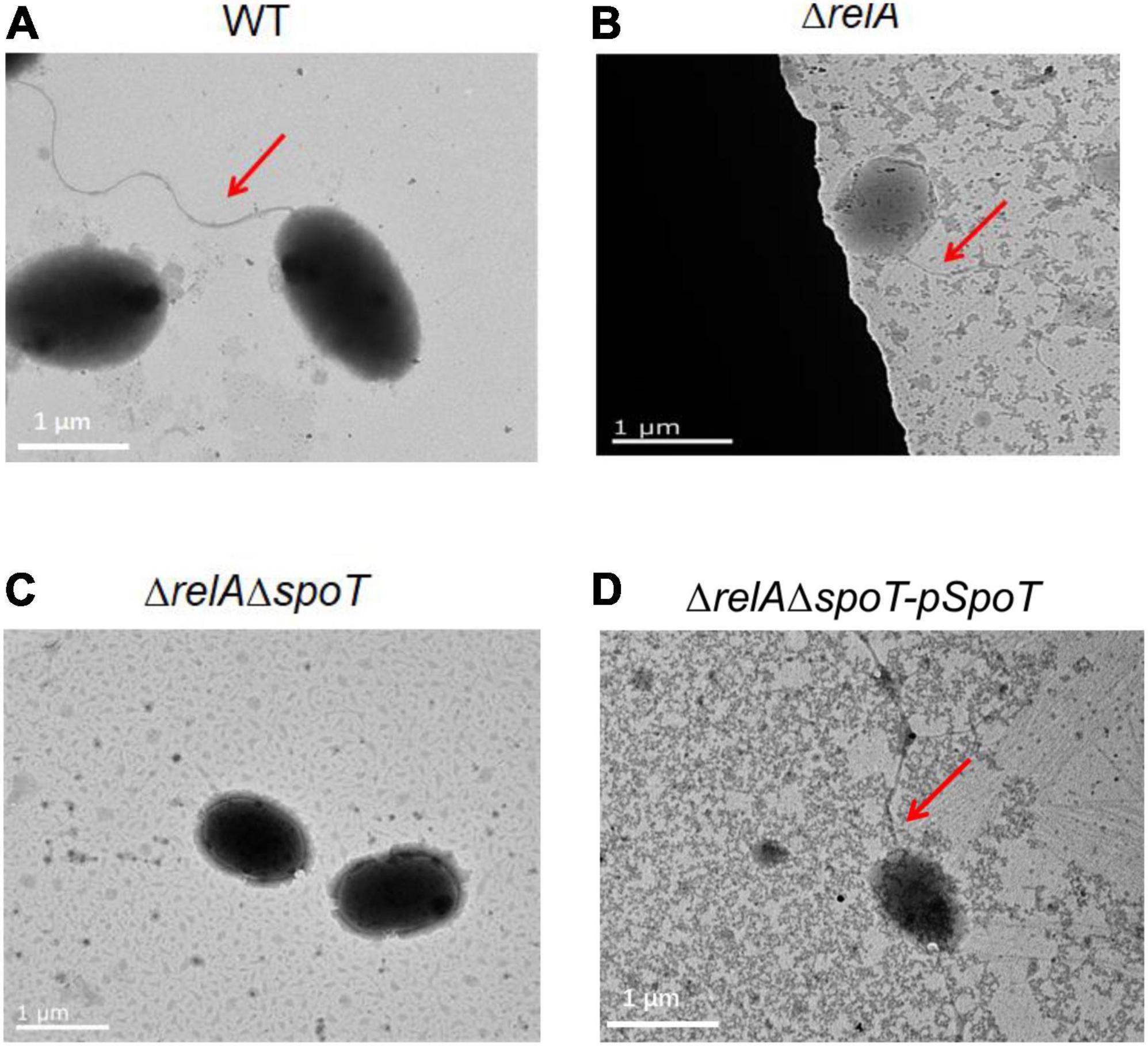
Figure 4. The effects of relA and spoT on the synthesis of flagellum. Transmission electron microscopic visualization of flagellum in WT (A), ΔrelA (B), ΔrelAΔspoT (C) and ΔrelAΔspoT -pSpoT (D). Red arrow indicates the flagellum and scale bar = 1 μm for all panels.
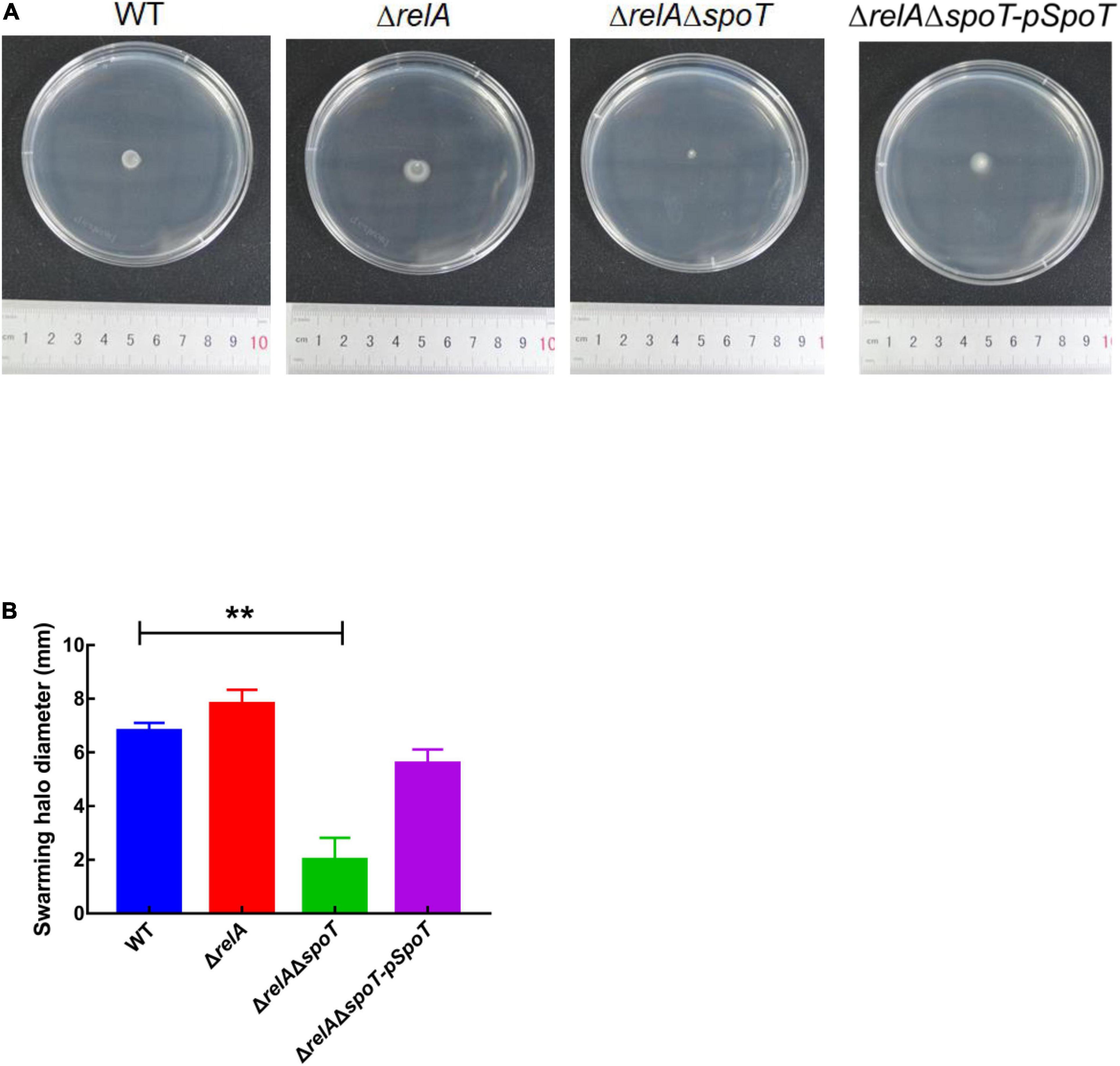
Figure 5. The effects of relA and spoT on the swarming ability. (A,B) The diameter of swarming rings of WT, ΔrelA, ΔrelAΔspoT, and ΔrelAΔspoT-pSpoT was recorded after 12 h of continuous culture on 0.9% 2216E agar plates at room temperature. Results were presented as mean ± SD (n = 3). ** indicates p ≤ 0.01. Three separate experiments were performed with biological triplicates each, and a representative image was displayed.
Although the content of extracellular polysaccharides can be determined by the phenol-sulfuric acid method by which the product concentration is qualified by observing the absorbance value (Liu et al., 2021). However, we could not rule out the interference of the residual nucleic acid and other sugars in the samples. Considering that vps-related genes affect the synthesis and transport of bacterial polysaccharides (Casper-Lindley and Yildiz, 2004; Chang et al., 2010), and cheR, flgH, and fliD are closely related to bacterial flagella synthesis and motility (Echazarreta and Klose, 2019; Liu et al., 2020), the expression levels of some functional genes were further determined by qRT-PCR. As shown in Figures 6A–C, at low cell density, the expression level of vpsH in WT was 1.92- and 6.14-fold higher than that of ΔrelA and ΔrelAΔspoT, respectively. Conversely, at high cell density, the transcription of vpsH in ΔrelAΔspoT was 5- and 3.5-fold higher than that of WT and ΔrelA, respectively. Similarly, the expression levels of the other EPSs-related genes, vpsT and vpsM, in WT and ΔrelA showed a trend of first increasing and then decreasing, which was similar to the trend of biofilm formation and EPSs production in the culture medium under the same conditions. As for motility-related genes, the expression level of cheR and flgE in ΔrelA was significantly lower than that of WT, whereas the expression levels of flgH in ΔrelA were not significantly different from that of WT (Figure 6D). The transcriptional profiles of these motility-related genes in ΔrelAΔspoT exhibited lower levels compared with WT, which was consistent with the results of the swarming ability test. These results will further help us understand the effects of (p)ppGpp synthetase gene-mediated alteration in EPSs and motility on biofilms.
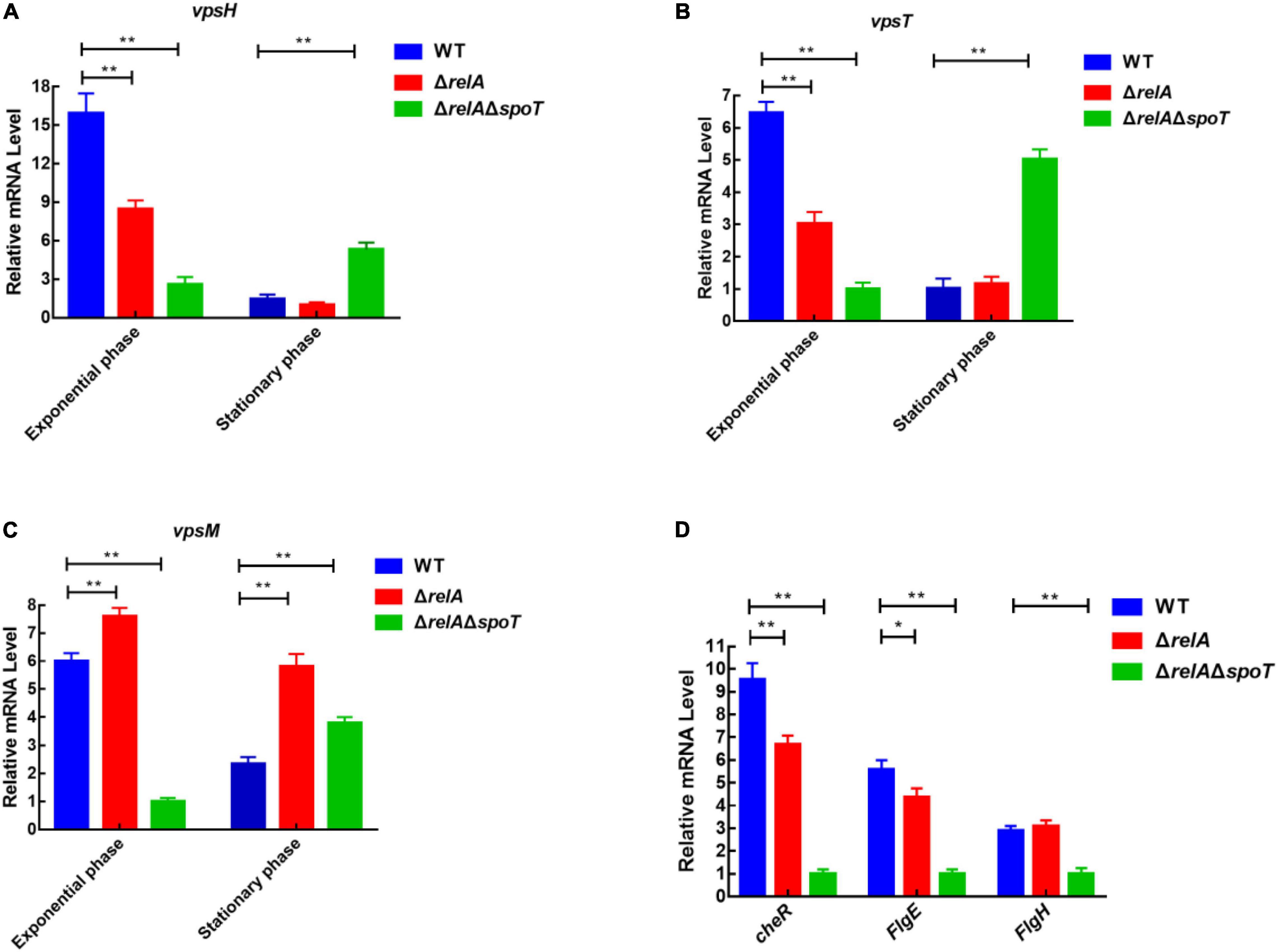
Figure 6. qRT-PCR analysis of transcriptional levels in WT, ΔrelA, and ΔrelAΔspoT. (A–C) The relative mRNA levels of EPSs-related genes were detected at low and high cell densities, respectively. (D) The relative mRNA levels of motility-related genes were detected when bacterial cultures entered the exponential phase (OD600 = 0.6). The results are normalized to the control gene 16S rRNA using the 2–ΔΔCt method. Results are shown as the means ± SD (n = 3). The blue columns, red columns, and green columns represent WT, ΔrelA, and ΔrelAΔspoT, respectively. * indicates p ≤ 0.05, and ** indicates p ≤ 0.01.
Previous study has shown that the deletion of rpoN leads to failure of biofilm detachment, loss of flagellum, p roduction of spotty pellicle, transparent surface rugose colonies, and decreased swimming motility (Zhang et al., 2021). To explore the relationship between (p)ppGpp synthetase genes and rpoN, we measured the expression level of rpoN at low and high cell densities. At low cell density, the expression levels of rpoN in ΔrelA was significantly higher than that in WT, but that in ΔrelAΔspoT was significantly reduced by almost 3-fold compared with WT (Figure 7). At high cell density, the transcription levels of rpoN in ΔrelA and ΔrelAΔspoT was lower than that of WT even if the difference was not significant. It was likely that (p)ppGpp synthetase genes had no obvious effect on rpoN at high cell density. These results implied that the deletion of (p)ppGpp synthetase genes had negative effects on the expression of rpoN at low cell density.
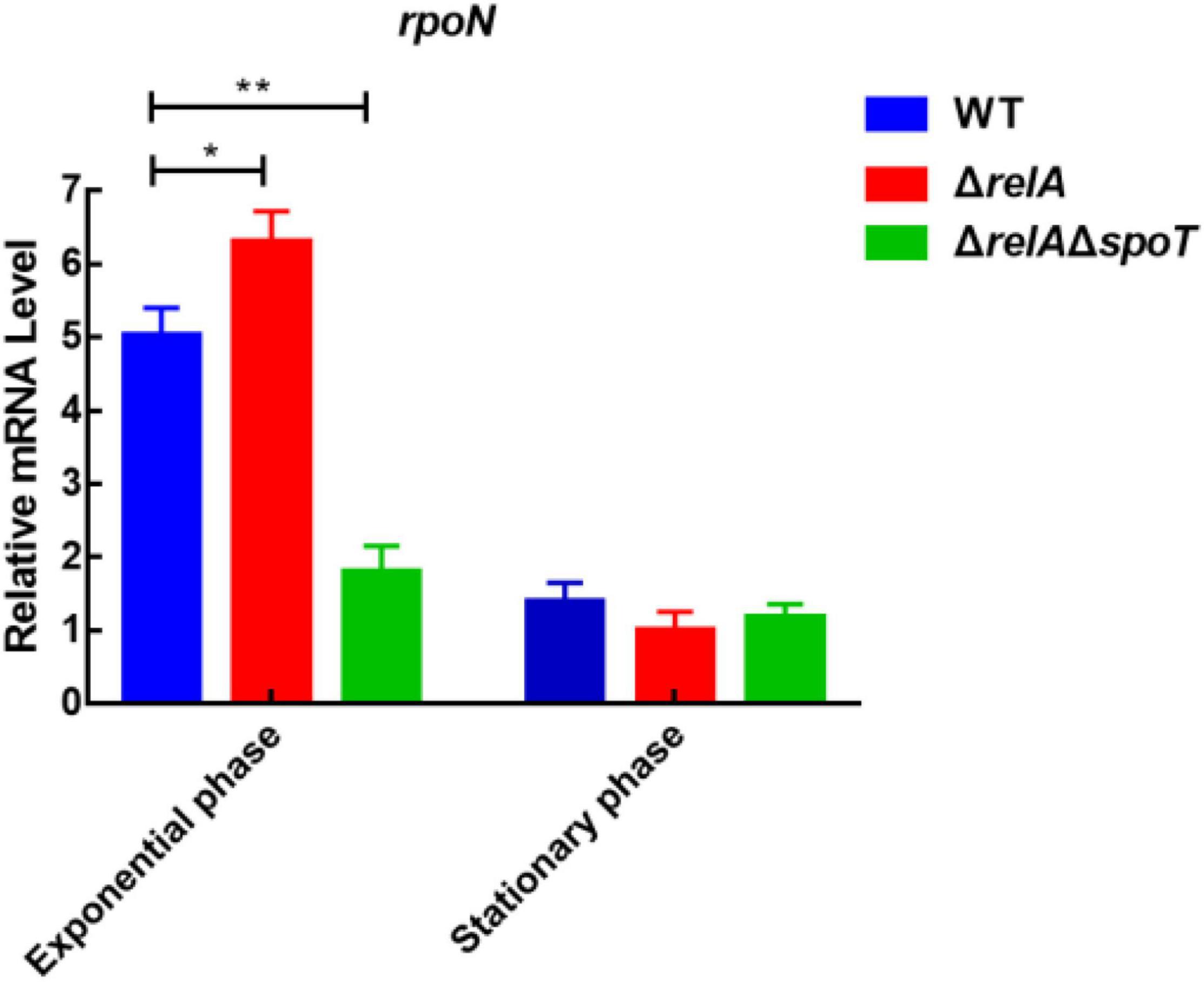
Figure 7. The relative mRNA levels of rpoN was detected by qRT-PCR in WT, ΔrelA, and ΔrelAΔspoT at low and high densities, respectively. The results are normalized to the control gene 16S rRNA using the 2–ΔΔCt method. Results are shown as the means ± SD (n = 3). The blue columns, red columns, and green columns represent WT, ΔrelA, and ΔrelAΔspoT, respectively. * indicates p ≤ 0.05, and ** indicates p ≤ 0.01.
To date, many internal factors and external stimuli are known to regulate biofilm formation (Martin-Rodriguez and Romling, 2017; Ranieri et al., 2018; Bridges et al., 2020). Recently, the roles of nucleotide-based second messenger (p)ppGpp in biofilm regulation have attracted more and more attention (Díaz-Salazar et al., 2017; Colomer-Winter et al., 2019; Salzer et al., 2020). However, little information is available on the (p)ppGpp-mediated regulation of biofilm formation at different cell densities. In this study, we provide evidence by polystyrene plate assays that the (p)ppGpp synthetase genes, relA and spoT, favor biofilm formation at low cell density and biofilm detachment at high cell density, respectively, in V. alginolyticus.
The initial adhesion mediated by flagella is essential for biofilm formation (Utada et al., 2014). Our study indicated that the lack of (p)ppGpp synthetase gene spoT rather than relA led to loss of flagella in V. alginolyticus. Meanwhile, we found that the biofilm of WT and other strains formed more rapidly than that of ΔrelAΔspoT within 6 h, which may depend on the flagella-mediated aggregation. This result was similar to that of Vibrio cholerae O139 mutant strain, which was deficient in flagella synthesis and failed to form an obvious biofilm stained by crystal violet (Watnick et al., 2001). Nevertheless, ΔrelAΔspoT could form biofilm, which might be due to other motility-related mechanisms such as type IV pili (Klausen et al., 2003). Flagella or other motility-related organelles also play an indispensable role in the biofilm detachment (Bridges et al., 2020). Interestingly, both ΔrelA and ΔrelAΔspoT showed a higher level of biofilm formation in comparison with WT between 8 and 18 h, then the biofilm in ΔrelA instead of in ΔrelAΔspoT began to decompose, implying that flagella in ΔrelA could play an important role in promoting biofilm detachment. Considering that bacterial motility, including swimming and swarming, is beneficial to the biofilm development (Bridges et al., 2020). We thus speculated the motility ability might be negatively affected in ΔrelAΔspoT. As expected, the swarming motility of WT and ΔrelA was not affected (p ≥ 0.05), while that of ΔrelAΔspoT significantly decreased relative to WT (p ≤ 0.01). qRT-PCR analysis further demonstrated that only the lack of spoT in V. alginolyticus significantly decreased the expression level of cheR, flgH, and flgE. In summary, the flagella synthesis and motility ability mediated by spoT would contribute to biofilm formation at low cell density and is necessary for the decomposition of biofilms at high cell density.
The most prevalent extracellular matrix component in biofilms is EPSs (Teschler et al., 2015). To better understand the mechanism of biofilm formation, we first measured the content of EPSs at low and high cell densities. Our results showed that the EPSs content of WT was higher than those of other strains at low cell density, whereas lower than those of ΔrelAΔspoT at high cell density. Both relA and spoT promoted EPSs production at low cell density and only spoT inhibited EPSs at high cell density. Similarly, the biofilm in WT was the highest at low cell density and the lowest at high cell density compared with that of other strains. Therefore, we propose that the content of EPSs is positively correlated with the amount of biofilm formation at low and high cell densities in V. alginolyticus. qRT-PCR analysis further showed that two (p)ppGpp synthetase genes relA and spoT increased the expression level of vpsT and vpsH at low cell density, whereas only spoT negatively regulated them at high cell density. Unlike vpsH and vpsT, only spoT contributed to the expression level of vpsM at low cell density, and two (p)ppGpp synthetase genes had negative effects on the expression of vpsM at high cell density. Our results indicated that (p)ppGpp synthetase genes could positively regulate transcription of some EPSs-related genes at low cell density, but negatively regulate expression level of them at high cell density. Besides, ΔrelAΔspoT was discovered to change the colony morphology from smoothness to rugosity, and form a spotty pellicle at the air-liquid interface. Yip et al. (2006) showed that induction of polysaccharide biosynthetic genes resulted in wrinkled colonies, pellicle formation and matrix production in Vibrio fischeri. We speculated that the higher expression levels of vpsT, vpsM, and vpsH in ΔrelAΔspoT than that in WT and ΔrelA were key elements to produce these biofilm phenotypes. In addition, the degradation of the biofilm in ΔrelA was delayed for a long time (8–18 h) might be explained, at least in part, by the higher expression of vpsM in ΔrelA than that in WT at high cell density.
Many studies have found that (p)ppGpp is closely related to the expression of rpoS and biofilm formation including in Pseudomonas aeruginosa and Pseudomonas putida KT2440 (van Delden et al., 2001; Liu et al., 2017). Previous study revealed that rpoN of V. alginolyticus played essential roles in controlling biofilm mediated by flagellum and EPSs (Zhang et al., 2021). In this study, we focused on the alteration of rpoN and rpoS expression in (p)ppGpp-deficient mutants. Although no obvious regular changes mediated by rpoS was observed (data not shown), the expression level of rpoN in ΔrelAΔspoT was lower than that in WT at low cell density, suggesting that (p)ppGpp synthetase genes could promote the biofilm formation by increasing the transcription level of rpoN. But unexpectedly, (p)ppGpp likely does not decompose the biofilm via rpoN-related pathways at high cell density (Figure 8). Lange et al. (1995) found that ppGpp played an important role in transcriptional elongation of rpoS. We speculated (p)ppGpp might have a similar effect on rpoN. In addition, the reason why ΔrelA had a higher gene expression level of rpoN than that of WT, but kept a notable decreased biofilm was that low concentration of (p)ppGpp might not be beneficial to the accumulation and stability of the alternative sigma factors in cell (Bougdour and Gottesman, 2007).
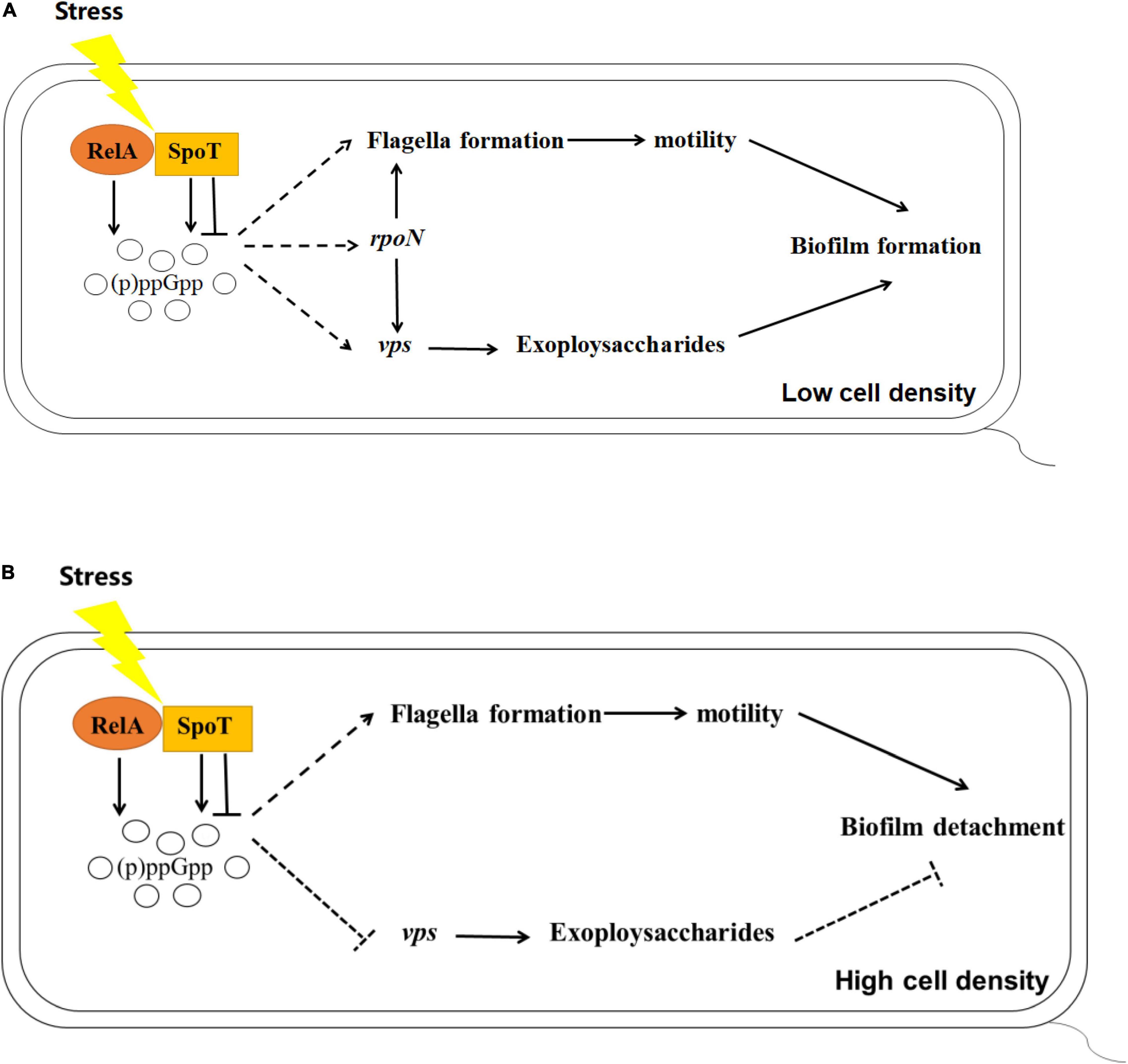
Figure 8. Schematics outlining the putative regulation of biofilm mediated by (p)ppGpp in V. alginolyticus at low (A) or high (B) cell density, respectively. Pathways have been mentioned by previous reports are depicted with the solid lines, while the inferences of this study are represented with the dashed lines. Arrows indicate promotion and horizontal symbols mean inhibition.
In summary, our results revealed the impact of (p)ppGpp-mediated regulation on biofilm, including biofilm formation in multi-well plates, the spotty pellicle at air-liquid interface and colony morphology. Combined with the results of cell motility, EPSs production, and the expression of related functional genes, we found that (p)ppGpp synthetase genes may facilitate biofilm formation at low cell density and biofilm detachment at high cell density through flagella-mediated motility, EPSs production and rpoN-regulated pathways.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
WY and ZX designed the study and analyzed the data. WY, ZX, and YZ wrote and revised the manuscript. WY, JZ, WR, HL, XZ, XC, and AH performed the experiments. All authors contributed to the article and approved the submitted version.
This research was supported by the Key Research and Development Project of Hainan Province (ZDYF2020095), National Natural Science Foundation of China (31660744 and 32060835), and National “13th Five-Year Plan” Marine Economic Innovation Development Demonstration City Project (HHCL201813 and HHCL201802).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Na Zhang and Shun Zhang from Hainan University for technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.858559/full#supplementary-material
Austin, B. (2010). Vibrios as causal agents of zoonoses. Vet. Microbiol. 140, 310–317. doi: 10.1016/j.vetmic.2009.03.015
Azriel, S., Goren, A., Rahav, G., and Gal-Mor, O. (2016). The stringent response regulator DksA is required for Salmonella enterica serovar Typhimurium growth in minimal medium, motility, biofilm formation, and intestinal colonization. Infect. Immun. 84, 375–384. doi: 10.1128/iai.01135-15
Beyhan, S., Bilecen, K., Salama, S. R., Casper-Lindley, C., and Yildiz, F. H. (2007). Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J. Bacteriol. 189, 388–402. doi: 10.1128/JB.00981-06
Bougdour, A., and Gottesman, S. (2007). ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc. Natl. Acad. Sci. U.S.A. 104, 12896–12901. doi: 10.1073/pnas.0705561104
Bridges, A. A., Fei, C., and Bassler, B. L. (2020). Identification of signaling pathways, matrix-digestion enzymes, and motility components controlling Vibrio cholerae biofilm dispersal. Proc. Natl. Acad. Sci. U.S.A. 117, 32639–32647. doi: 10.1073/pnas.2021166117
Casper-Lindley, C., and Yildiz, F. H. (2004). VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186, 1574–1578. doi: 10.1128/JB.186.5.1574-1578.2004
Chang, C., Jing-Jing, Z., Chun-Hua, R., and Chao-Qun, H. (2010). Deletion of valR, a homolog of Vibrio harveyiś luxR generates an intermediate colony phenotype between opaque/rugose and translucent/smooth in Vibrio alginolyticus. Biofouling 26, 595–601. doi: 10.1080/08927014.2010.499511
Chen, C., Xie, J., and Hu, C. Q. (2009). Phenotypic and genetic differences between opaque and transparent colonies of Vibrio alginolyticus. Biofouling 25, 525–531. doi: 10.1080/08927010902964578
Cheng, A. T., Zamorano-Sanchez, D., Teschler, J. K., Wu, D., and Yildiz, F. H. (2018). NtrC adds a new node to the complex regulatory network of biofilm formation and vps expression in Vibrio cholerae. J. Bacteriol. 200:e00025-18. doi: 10.1128/JB.00025-18
Colomer-Winter, C., Flores-Mireles, A. L., Kundra, S., Hultgren, S. J., and Lemos, J. A. (2019). (p)ppGpp and CodY promote Enterococcus faecalis virulence in a murine model of catheter-associated urinary tract infection. mSphere 4:e00392-19. doi: 10.1128/mSphere.00392-19
Croxatto, A., Lauritz, J., Chen, C., and Milton, D. L. (2007). Vibrio anguillarum colonization of rainbow trout integument requires a DNA locus involved in exopolysaccharide transport and biosynthesis. Environ. Microbiol. 9, 370–382. doi: 10.1111/j.1462-2920.2006.01147.x
Dalebroux, Z. D., and Swanson, M. S. (2012). ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10, 203–212. doi: 10.1038/nrmicro2720
Díaz-Salazar, C., Calero, P., Espinosa-Portero, R., Jiménez-Fernández, A., Wirebrand, L., Velasco-Domínguez, M. G., et al. (2017). The stringent response promotes biofilm dispersal in Pseudomonas putida. Sci. Rep. 7:18055. doi: 10.1038/s41598-017-18518-0
Echazarreta, M. A., and Klose, K. E. (2019). Vibrio flagellar synthesis. Front. Cell. Infect. Microbiol. 9:131. doi: 10.3389/fcimb.2019.00131
Faruque, S. M., Biswas, K., Udden, S. M., Ahmad, Q. S., Sack, D. A., Nair, G. B., et al. (2006). Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. U.S.A. 103, 6350–6355. doi: 10.1073/pnas.0601277103
Fong, J. C. N., Syed, K. A., Klose, K. E., and Yildiz, F. H. (2010). Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholera pathogenesis. Microbiology 156, 2757–2769. doi: 10.1099/mic.0.040196-0
Fong, J. C. N., and Yildiz, F. H. (2007). The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J. Bacteriol. 189, 2319–2330. doi: 10.1128/JB.01569-06
Gaca, A. O., Colomer-Winter, C., and Lemos, J. A. (2015). Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J. Bacteriol. 197, 1146–1156. doi: 10.1128/JB.02577-14
Gómez-León, J., Villamil, L., Lemos, M. L., Novoa, B., and Figueras, A. (2005). Isolation of Vibrio alginolyticus and Vibrio splendidus from aquacultured carpet shell clam (Ruditapes decussatus) larvae associated with mass mortalities. Appl. Environ. Microbiol. 71, 98–104. doi: 10.1128/AEM.71.1.98-104.2005
Hall, C. W., and Mah, T. F. (2017). Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS. Microbiol. Rev. 41, 276–301. doi: 10.1093/femsre/fux010
Hare, P., Long, S., Robb, F. T., and Woods, D. R. (1981). Regulation of exoprotease production by temperature and oxygen in Vibrio alginolyticus. Arch. Microbiol. 130, 276–280. doi: 10.1007/BF00425940
Hay, A. J., and Zhu, J. (2015). Host intestinal signal-promoted biofilm dispersal induces Vibrio cholerae colonization. Infect. Immun. 83, 317–323. doi: 10.1128/IAI.02617-14
He, H., Cooper, J. N., Mishra, A., and Raskin, D. M. (2012). Stringent response regulation of biofilm formation in Vibrio cholerae. J. Bacteriol. 194, 2962–2972. doi: 10.1128/jb.00014-12
Herzog, R., Peschek, N., Fröhlich, K. S., Schumacher, K., and Papenfort, K. (2019). Three autoinducer molecules act in concert to control virulence gene expression in Vibrio cholerae. Nucleic Acids Res. 47, 3171–3183. doi: 10.1093/nar/gky1320
Jacobs Slifka, K. M., Newton, A. E., and Mahon, B. E. (2017). Vibrio alginolyticus infections in the USA, 1988-2012. Epidemiol. Infect. 145, 1491–1499. doi: 10.1017/S0950268817000140
Kahla-Nakbi, A. B., Chaieb, K., Besbes, A., Zmantar, T., and Bakhrouf, A. (2006). Virulence and enterobacterial repetitive intergenic consensus PCR of Vibrio alginolyticus strains isolated from Tunisian cultured gilthead sea bream and sea bass outbreaks. Vet. Microbiol. 117, 321–327. doi: 10.1016/j.vetmic.2006.06.012
Klausen, M., Heydorn, A., Ragas, P., Lambertsen, L., Aaes-Jørgensen, A., Molin, S., et al. (2003). Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48, 1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x
Lange, R., Fischer, D., and Hengge-Aronis, R. (1995). Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the sigma S subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 177, 4676–4680. doi: 10.1128/jb.177.16.4676-4680.1995
Lee, K. K., Yu, S. R., Yang, T. I., Liu, P. C., and Chen, F. R. (1996). Isolation and characterization of Vibrio alginolyticus isolated from diseased kuruma prawn, Penaeus japonicus. Lett. Appl. Microbiol. 22, 111–114. doi: 10.1111/j.1472-765x.1996.tb01121.x
Li, G., Xie, F., Zhang, Y., Bossé, J. T., Langford, P. R., and Wang, C. (2015). Role of (p)ppGpp in viability and biofilm formation of Actinobacillus pleuropneumoniae S8. PLoS One 10:e0141501. doi: 10.1371/journal.pone.0141501
Liu, H., Xiao, M., Zuo, J., He, X., Lu, P., Li, Y., et al. (2021). Vanillic acid combats Vibrio alginolyticus by cell membrane damage and biofilm reduction. J. Fish. Dis. 44, 1799–1809. doi: 10.1111/jfd.13498
Liu, H., Xiao, Y., Nie, H., Huang, Q., and Chen, W. (2017). Influence of (p)ppGpp on biofilm regulation in Pseudomonas putida KT2440. Microbiol. Res. 204, 1–8. doi: 10.1016/j.micres.2017.07.003
Liu, J., Yu, M., Chatnaparat, T., Lee, J. H., Tian, Y., Hu, B., et al. (2020). Comparative transcriptomic analysis of global gene expression mediated by (p) ppGpp reveals common regulatory networks in Pseudomonas syringae. BMC Genomics 21:296. doi: 10.1186/s12864-020-6701-2
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, P., He, X., Liu, Q., and Hu, C. (2015). Developing universal genetic tools for rapid and efficient deletion mutation in Vibrio species based on suicide T-vectors carrying a novel counterselectable Marker, vmi480. PLoS One 10:e0144465. doi: 10.1371/journal.pone.0144465
Magnusson, L. U., Farewell, A., and Nyström, T. (2005). ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13, 236–242. doi: 10.1016/j.tim.2005.03.008
Martin-Rodriguez, A. J., and Romling, U. (2017). Nucleotide second messenger signaling as a target for the control of bacterial biofilm formation. Curr. Top. Med. Chem. 17, 1928–1944. doi: 10.2174/1568026617666170105144424
Milton, D. L., O’Toole, R., Horstedt, P., and Wolf-Watz, H. (1996). Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178, 1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996
Mustapha, S., Mustapha, E. M., and Nozha, C. (2013). Vibrio Alginolyticus: an emerging pathogen of foodborne diseases. Int. J. Sci. Tech. 2, 302–309.
O’Toole, G. A., and Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
Pratt, J. T., McDonough, E., and Camilli, A. (2009). PhoB regulates motility, biofilms, and cyclic di-GMP in Vibrio cholerae. J. Bacteriol. 191, 6632–6642. doi: 10.1128/JB.00708-09
Ranieri, M. R., Whitchurch, C. B., and Burrows, L. L. (2018). Mechanisms of biofilm stimulation by subinhibitory concentrations of antimicrobials. Curr. Opin. Microbiol. 45, 164–169. doi: 10.1016/j.mib.2018.07.006
Renier, S., Hébraud, M., and Desvaux, M. (2011). Molecular biology of surface colonization by Listeria monocytogenes: an additional facet of an opportunistic Gram-positive foodborne pathogen. Environ. Microbiol. 13, 835–850. doi: 10.1111/j.1462-2920.2010.02378.x
Rui, H., Liu, Q., Ma, Y., Wang, Q., and Zhang, Y. (2008). Roles of LuxR in regulating extracellular alkaline serine protease A, extracellular polysaccharide and mobility of Vibrio alginolyticus. FEMS. Microbiol. Lett. 285, 155–162. doi: 10.1111/j.1574-6968.2008.01185.x
Salzer, A., Keinhörster, D., Kästle, C., Kästle, B., and Wolz, C. (2020). Small alarmone synthetases RelP and RelQ of Staphylococcus aureus are involved in biofilm formation and maintenance under cell wall stress conditions. Front. Microbiol. 11:575882. doi: 10.3389/fmicb.2020.575882
Srivatsan, A., and Wang, J. D. (2008). Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 11, 100–105. doi: 10.1016/j.mib.2008.02.001
Sugisaki, K., Hanawa, T., Yonezawa, H., Osaki, T., Fukutomi, T., Kawakami, H., et al. (2013). Role of (p)ppGpp in biofilm formation and expression of filamentous structures in Bordetella pertussis. Microbiology 159, 1379–1389. doi: 10.1099/mic.0.066597-0
Teschler, J. K., Zamorano-Sánchez, D., Utada, A. S., Warner, C. J., Wong, G. C., Linington, R. G., et al. (2015). Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol. 13, 255–268. doi: 10.1038/nrmicro3433
Utada, A. S., Bennett, R. R., Fong, J. C. N., Gibiansky, M. L., Yildiz, F. H., Golestanian, R., et al. (2014). Vibrio cholerae use pili and flagella synergistically to effect motility switching and conditional surface attachment. Nat. Commun. 5:4913. doi: 10.1038/ncomms5913
van Delden, C., Comte, R., and Bally, A. M. (2001). Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 183, 5376–5384. doi: 10.1128/JB.183.18.5376-5384.2001
Wang, Q., Liu, Q., Ma, Y., Zhou, L., and Zhang, Y. (2007). Isolation, sequencing and characterization of cluster genes involved in the biosynthesis and utilization of the siderophore of marine fish pathogen Vibrio alginolyticus. Arch. Microbiol. 188, 433–439. doi: 10.1007/s00203-007-0261-6
Watnick, P. I., Lauriano, C. M., Klose, K. E., Croal, L., and Kolter, R. (2001). The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39, 223–235. doi: 10.1046/j.1365-2958.2001.02195.x
Xiao, H., Kalman, M., Ikehara, K., Zemel, S., Glaser, G., and Cashel, M. (1991). Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266, 5980–5990. doi: 10.1016/S0021-9258(19)67694-5
Yildiz, F. H., and Schoolnik, G. K. (1999). Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 96, 4028–4033. doi: 10.1073/pnas.96.7.4028
Yildiz, F. H., and Visick, K. L. (2009). Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17, 109–118. doi: 10.1016/j.tim.2008.12.004
Yin, W. L., Zhang, N., Xu, H., Gong, X. X., Long, H., Ren, W., et al. (2021). Stress adaptation and virulence in Vibrio alginolyticus is mediated by two (p)ppGpp synthetase genes, relA and spoT. Microbiol. Res. 253:126883. doi: 10.1016/j.micres.2021.126883
Yip, E. S., Geszvain, K., DeLoney-Marino, C. R., and Visick, K. L. (2006). The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol. 62, 1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x
Zhang, N., Zhang, S., Ren, W., Gong, X. X., Long, H., Zhang, X., et al. (2021). Roles of rpoN in biofilm formation of Vibrio alginolyticus HN08155 at different cell densities. Microbiol. Res. 247:126728. doi: 10.1016/j.micres.2021.126728
Keywords: Vibrio alginolyticus, stringent response, (p)ppGpp, biofilm formation, motility, EPSs, rpoN
Citation: Yin W-L, Xie Z-Y, Zeng Y-H, Zhang J, Long H, Ren W, Zhang X, Cai X-N and Huang A-Y (2022) Two (p)ppGpp Synthetase Genes, relA and spoT, Are Involved in Regulating Cell Motility, Exopolysaccharides Production, and Biofilm Formation of Vibrio alginolyticus. Front. Microbiol. 13:858559. doi: 10.3389/fmicb.2022.858559
Received: 20 January 2022; Accepted: 28 February 2022;
Published: 29 March 2022.
Edited by:
Haike Antelmann, Freie Universität Berlin, GermanyReviewed by:
Christiane Wolz, University of Tübingen, GermanyCopyright © 2022 Yin, Xie, Zeng, Zhang, Long, Ren, Zhang, Cai and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen-Yu Xie, eGllenlzY3V0YUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.