- 1CAS Key Laboratory of Tropical Marine Bio-Resources and Ecology, Guangdong Key Laboratory of Marine Materia Medica, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 2College of Earth and Planetary Sciences, University of Chinese Academy of Sciences, Beijing, China
- 3Guangxi Zhuang Yao Medicine Center of Engineering and Technology, Guangxi University of Chinese Medicine, Nanning, China
- 4State Key Laboratory for Chemistry and Molecular Engineering of Medicinal Resources, School of Chemistry and Pharmaceutical Sciences, Guangxi Normal University, Guilin, China
- 5Guangdong Eco-Engineering Polytechnic, Guangzhou, China
- 6Guangdong Ocean Association, Guangzhou, China
- 7Wuya College of Innovation, Shenyang Pharmaceutical University, Shenyang, China
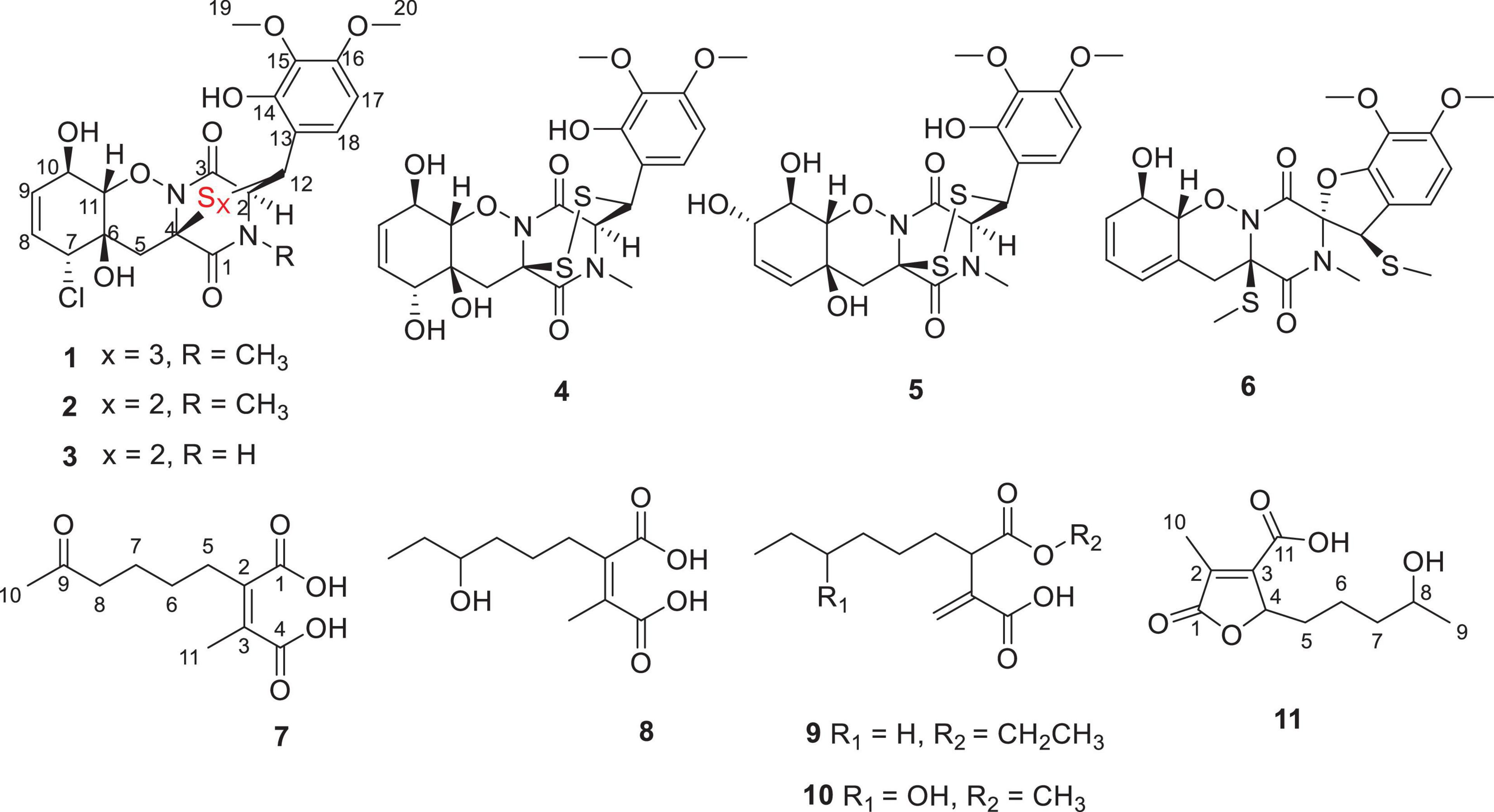
A new trithiodiketopiperazine derivative, adametizine C (1), and five new alkane derivatives (7–11), were isolated from the mangrove sediment–derived fungus Penicillium ludwigii SCSIO 41408, together with five known dithiodiketopiperazine derivatives (2–6). Their structures were elucidated on the basis of spectroscopic analysis, and the absolute configuration of 1 was determined by X-ray crystallographic analysis. In a variety of bioactivity screening, 1–5 exhibited some selective antifungal or antibacterial activities. Compounds 1–3 showed cytotoxicity against prostate cancer cell line 22Rv1 with half maximal inhibitory concentration (IC50) values of 13.0–13.9 μM; moreover, 3 showed obvious activity against another prostate cancer PC-3 cells with an IC50 value of 5.1 μM. Further experiments revealed that 3 could significantly reduce PC-3 cells colony formation and induce apoptosis in a dose-dependent manner. Several compounds also exhibited obvious inhibitory activities of lipopolysaccharide–induced nuclear factor-κB with IC50 values range from 8.2 to 21.5 μM, and 1, 5, and 9 were further evaluated for their effects on receptor activator of NF-κB ligand (RANKL)-induced osteoclastogenesis. Adametizine C (1), with the strongest inhibitory activity against RANKL-induced osteoclast differentiation in bone marrow macrophage cells with 10 μM, was suggested to be the promising lead compound for the treatment of osteoclast-related diseases.
Introduction
Mangroves, a special ecosystem characterized by high salinity, muddy or sandy soil, and low pH, as well as partly anoxic and periodically soaked by the tides, play an important role in tropical and subtropical coastal ecosystems. Mangroves nourish various microorganisms due to their complex ecosystem. The variety and complexity of the mangrove soil environment leads to the diversity of soil microorganisms (He et al., 2019). Mangrove soil or sediment-derived microbes play an essential role in maintaining the biosphere balance and are also a prolific source of structurally unique and novel bioactive secondary metabolites (Li et al., 2022). Mainly derived from marine fungi, thiodiketopiperazines have been recently reported to have a broad range of significant biological activities, such as brine shrimp lethality (Liu et al., 2015a), antibacterial (Liu et al., 2015a), antifungal (Liu et al., 2015b), cytotoxic (Yurchenko et al., 2016), and C-terminal inhibitor (Dai et al., 2019) activities. During our ongoing search for novel bioactive secondary metabolites from mangrove fungi (Luo et al., 2018; Luo et al., 2019; Chen et al., 2021a,b; Cai et al., 2021), a new trithiodiketopiperazine, five new alkane derivatives (7–11), and five dithiodiketopiperazine derivatives (2–6) (Figure 1) were isolated from the mangrove sediment–derived fungus Penicillium ludwigii SCSIO 41408. We performed screening for antibacterial and antifungal activities, cytotoxicity against anti-prostate cancer cells, and inhibitory activities of lipopolysaccharide (LPS)–induced nuclear factor-κB (NF-κB) activation. NF-κB exhibited an important role in receptor activator of NF-κB ligand (RANKL)-induced osteoclast differentiation (Hong et al., 2020; Tan et al., 2020; Zhou et al., 2020). Our further research found that Adametizine C (1) was suggested to be the promising lead compound for the treatment of osteoclast-related diseases.
Materials and Methods
General Experimental Procedures
Optical rotations were measured on a PerkinElmer MPC 500 (Waltham) polarimeter. The UV, IR, and CD spectra were recorded on a Shimadzu UV-2600 PC spectrometer (Shimadzu), an IR Affinity-1 spectrometer (Shimadzu), and a Chirascan circular dichroism spectrometer (Applied Photophysics), respectively. NMR spectra were recorded on a Bruker Avance spectrometer (Bruker) operating at 500 and 700 MHz for 1H NMR and 125 and 175 MHz for 13C NMR that used tetramethylsilane as an internal standard. High resolution electrospray ionization mass spectroscopy (HRESIMS) spectra were acquired on a Bruker miXis TOF-QII mass spectrometer (Bruker). Column chromatography was performed over silica gel (200–300 mesh) (Qingdao Marine Chemical Factory). Spots were detected on TLC (Qingdao Marine Chemical Factory) under 254-nm UV light. All solvents employed were analytical grade (Tianjin Fuyu Chemical and Industry Factory). Semipreparative HPLC was performed using an octadecylsilyl (ODS) column (YMC-pack ODS-A, YMC Co., Ltd., 10 mm × 250 mm, 5 μm). Artificial sea salt was a commercial product (Guangzhou Haili Aquarium Technology Company).
Fungal Material
The fungal strain Penicillium ludwigii SCSIO 41408 was isolated from a mangrove sediment sample, collected in the Hongsha River estuary to South China Sea, in Sanya city, Hainan Island. The fungus was identified according to the internally transcribed spacer (ITS) region sequence data of the rDNA, and the sequence was deposited in GenBank with the accession number OL823084. The strain was stored on Muller Hinton broth agar (malt extract, 15 g; sea salt, 10 g; agar, 15 g; H2O, 1 L; pH 7.4–7.8) at 4°C and deposited in the CAS Key Laboratory of Tropical Marine Bioresources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China.
Fermentation, Extraction, and Isolation
The strain Penicillium ludwigii SCSIO 41408 was cultured in the seed medium (malt extract, 15 g; sea salt, 10 g; H2O, 1 L; pH 7.4–7.8) for 4 days at 28°C on a rotating shaker (180 rpm). A large scale of fermentation was incubated statically at 26°C for 60 days in 1 L × 65 conical flasks with a rice medium (each flack contains 180 g of rice, 3 g of sea salt, 200 ml of H2O). The fermented cultures were overlaid and extracted with EtOAc three times to afford a brown extract (346.6 g).
The EtOAc crude extract was chromatographed over a silica gel column eluted with PE/CH2Cl2 (0–100%, v/v) and CH2Cl2/MeOH (0–100%, v/v) in a gradient to yield fourteen fractions (Frs. 1–14). Fr. 2 (0.9 g) was separated by semipreparative HPLC (65% MeCN/H2O, 2 ml/min) to afford 9 (3.4 mg, tR = 17.0 min) and Fr. 2-1. Fr. 2-1 was separated again by semipreparative HPLC (40% MeCN/H2O, 2.7 ml/min) to afford 7 (6.7 mg, tR = 16.1 min) and 8 (7.5 mg, 14.5 min). Fr. 4 (6.4 g) was separated by semipreparative HPLC (48% MeCN/H2O, 3 ml/min) to afford 6 (2.1 mg, tR = 17.3 min). Fr. 8 (8.5 g) was purified by semipreparative HPLC (65% MeOH/H2O, 2 ml/min) to afford 2 (5.2 mg, tR = 10.1 min). Fr.10 was divided into four subfractions by ODS silica gel chromatography eluting with MeOH/H2O (10–100%). Fr. 10 was divided into four subfractions by Sephadex LH-20. Fr.10-1 was purified by semipreparative HPLC (35% MeCN/H2O, 2 ml/min) to afford 11 (10.2 mg, tR = 8.1 min) and 1 (6.0 mg, tR = 27.1 min). Fr. 10-4 was purified by semipreparative HPLC (55% MeOH/H2O, 3 ml/min) to afford 3 (6.0 mg, tR = 7.5 min). Fr. 13 was divided into four subfractions by ODS silica gel chromatography eluting with MeOH/H2O (10–100%). Fr. 13-1 was further purified by semipreparative HPLC (45% MeOH/H2O, 2.7 ml/min) to afford 4 (26.0 mg, tR = 12.4 min), 5 (5.7 mg, tR = 14.1 min), and 10 (16.4 mg, tR = 21.9 min).
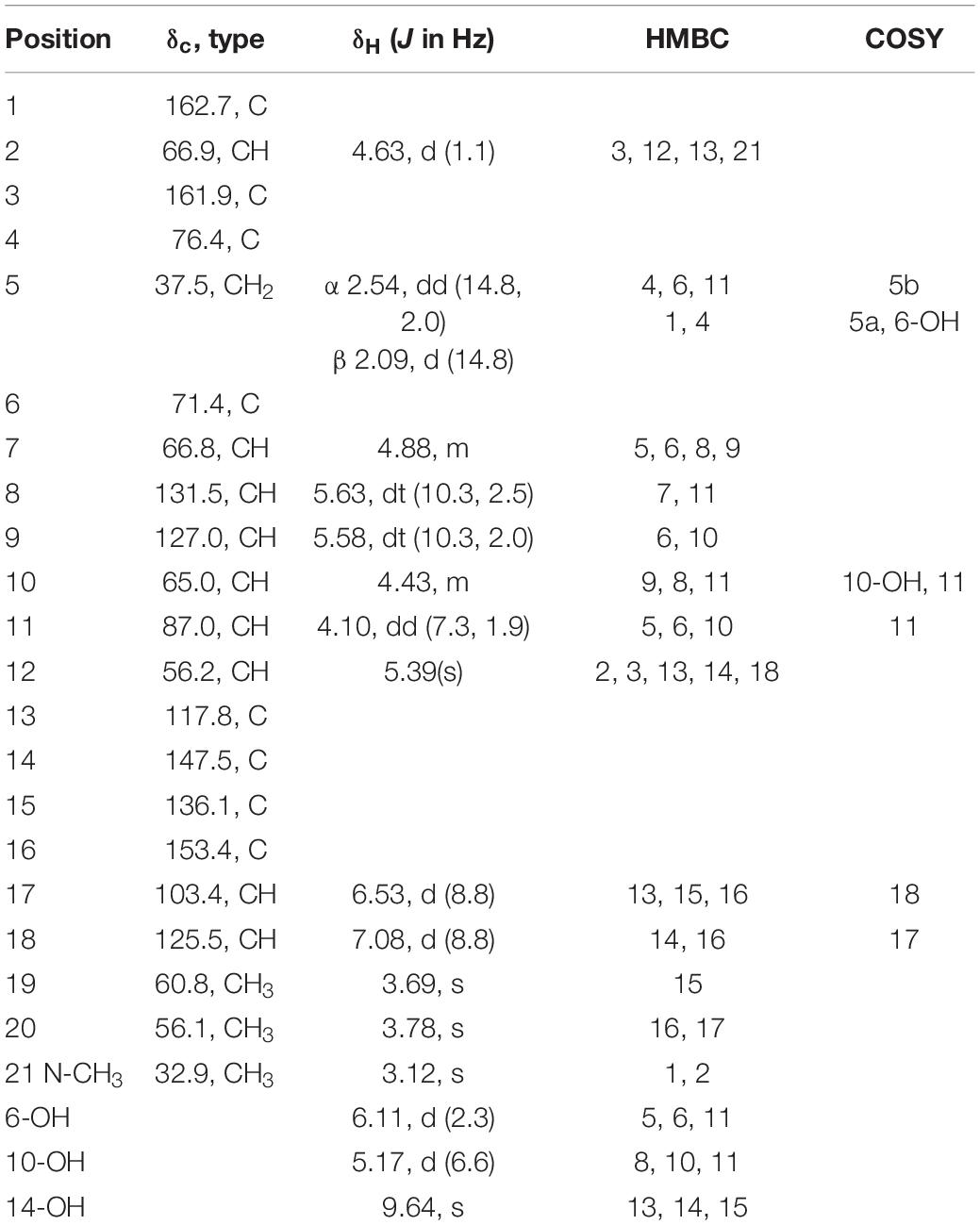
Adametizine C (1): colorless needles; [α]25 D, −128.5 (c 0.1, MeOH); UV (MeOH) λmax (log ε), 205 (3.52) nm; ECD (0.36 mM, MeOH) λmax (Δε), 203 (−10.98), 242 (−9.06), and 293(+1.33); IRνmax, 3,352, 2,945, 2,835, 1,670, 1,506, 1,431, 1,250, 1,204, 1,096, 1,018, 831, 795, 677, 601, and 557 cm–1; 1H and 13C NMR, data see Table 1; HRESIMS at m/z 563.0381 [M + H]+ (calculated for C21H24ClN2O8S3, 563.0378).
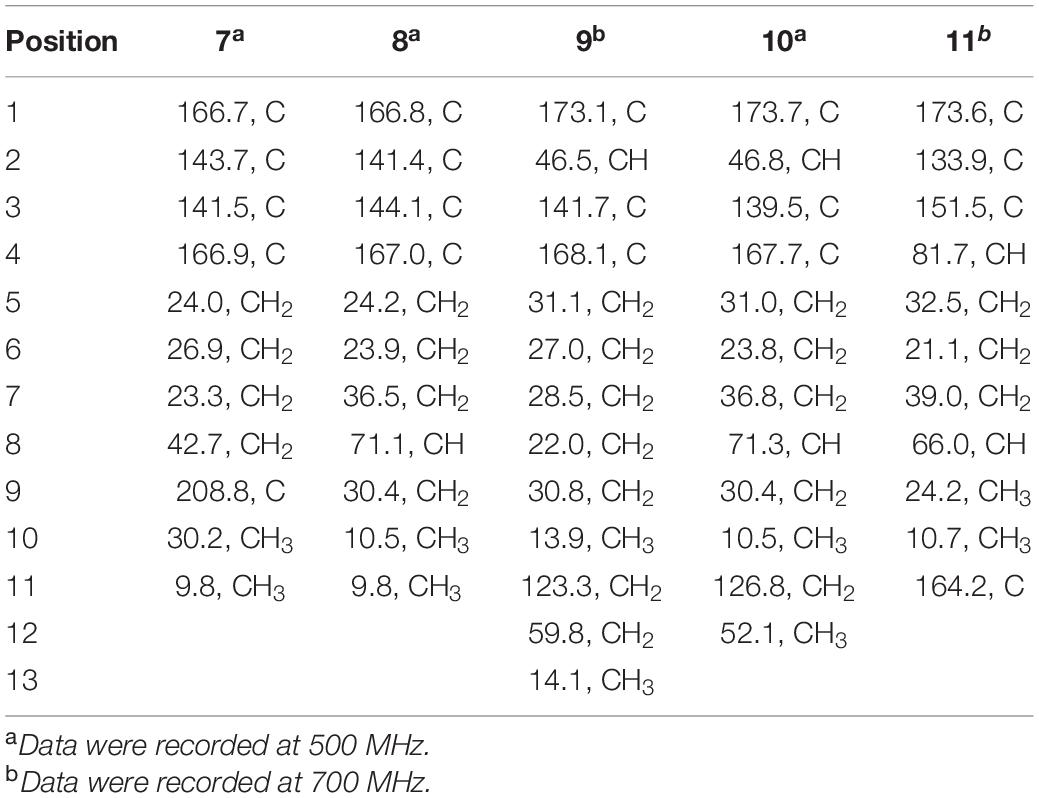
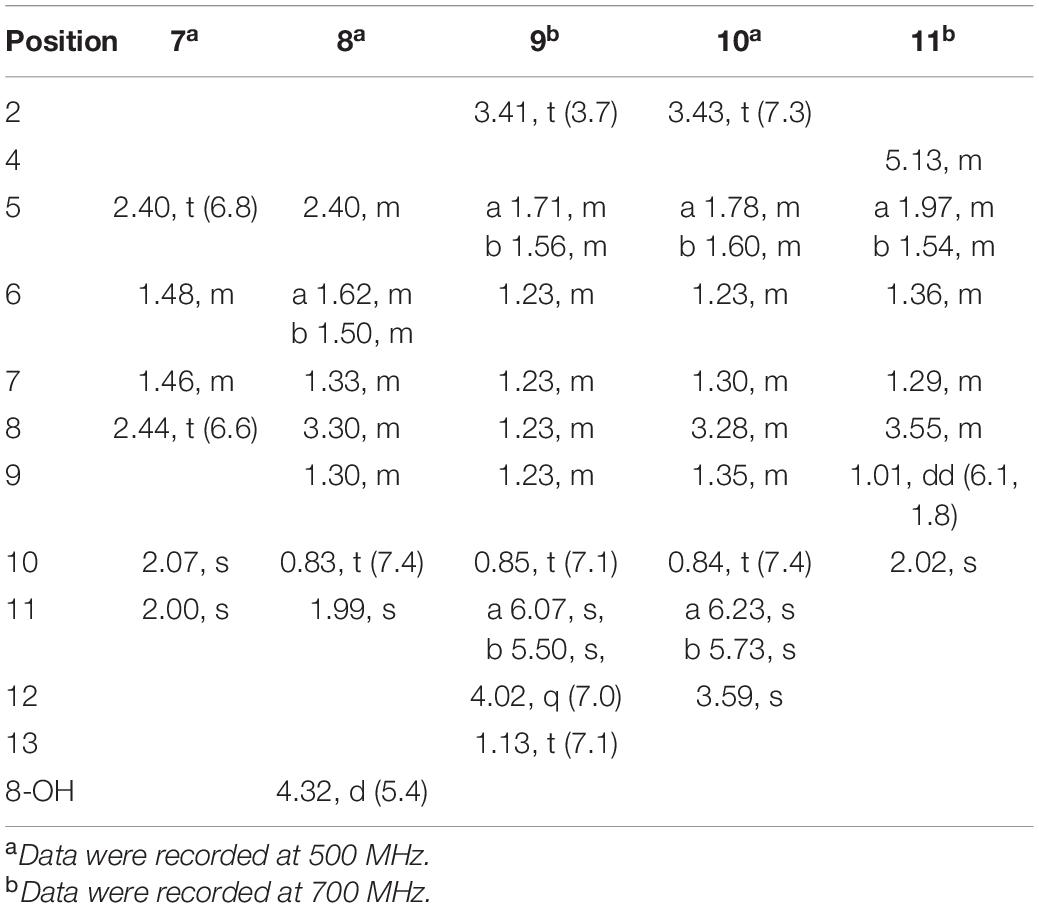
2-methyl-3-(5-oxohexyl) maleic acid (7): colorless oil; UV (MeOH) λmax (log ε), 250 (3.95) and 207 (3.69) nm; IRνmax, 2,938, 2,866, 1,759, 1,712, 1,362, 1,273, 916, and 735 cm–1; 1H and 13C NMR data, see Tables 2, 3; HRESIMS at m/z 227.0927 [M-H]– (calculated for C11H15O5, 227.0925).
2-(4-hydroxyhexyl)-3-methylmaleic acid (8): colorless oil; [α]25 D, −0.7 (c 0.1, MeOH); ECD (2.46 mM, MeOH) λmax (Δε), 203 (−2.49) and 213 (+2.27); UV (MeOH) λmax (log ε), 250 (3.25), 206 (3.65) nm; IRνmax, 3,390, 2,943, 1,763, 1,682, 1,435, 1,283, 1,202, 1,136, 1,026, 800, and 721 cm–1; 1H and 13C NMR data, see Tables 2, 3; HRESIMS at m/z 229.1082 [M-H]– (calculated for C11H17O5, 229.1081).
3-(ethoxycarbonyl)-2-methylenenonanoic acid (9): brown oil; [α]25 D, +5.9 (c 0.1, MeOH); ECD (3.83 mM, MeOH) λmax (Δε) 200 (−10.62), 214 (+0.68), and 227 (−2.64); UV (MeOH) λmax (log ε), 204 (3.86) nm; IRνmax, 2,955, 2,928, 2,857, 1,732, 1,715, 1,202, 1,153, 1,113, 1,036, 953, and 835 cm–1; 1H and 13C NMR data, see Tables 2, 3; HRESIMS at m/z 243.1594 [M + H]+ (calculated for C13H23O4, 243.1591).
7-hydroxy-3-(methoxycarbonyl)-2-methylenenonanoic acid (10): colorless oil; [α], +1.9 (c 0.1, MeOH); ECD (2.05 mM, MeOH) λmax (Δε), 200 (−3.34), 201 (+4.57), 205 (+2.35), and 211 (−0.98); UV (MeOH) λmax (log ε), 205 (2.93) nm; IRνmax, 3,447, 2,936, 2,866, 1,717, 1,628, 1,456, 1,435, 1,204, 1,155, 1,024, 9,54.8, and 824 cm–1; 1H and 13C NMR data, see Tables 2, 3; HRESIMS at m/z 245.1388 [M + H]+ (calculated for C12H21O5, 245.1384).
2-(4-hydroxypentyl)-4-methyl-5-oxo-2,5-dihydrofuran-3-carbo xylic acid (11): brown oil; [α], +2.6 (c 0.1, MeOH); ECD (4.39 mM, MeOH) λmax (Δε), 200 (−10.62), 214 (+0.68), and 227 (−2.64); UV (MeOH) λmax (log ε), 225 (3.85) nm; IRνmax, 3,414, 2,932, 1,744, 1,715, 1,337, 1,231, 1,117, 1,024, 947, 764, and 721 cm–1; HRESIMS at m/z 229.1076 [M + H]+ (calculated for C11H17O5, 229.1072).
X-Ray Crystal Structure Analysis
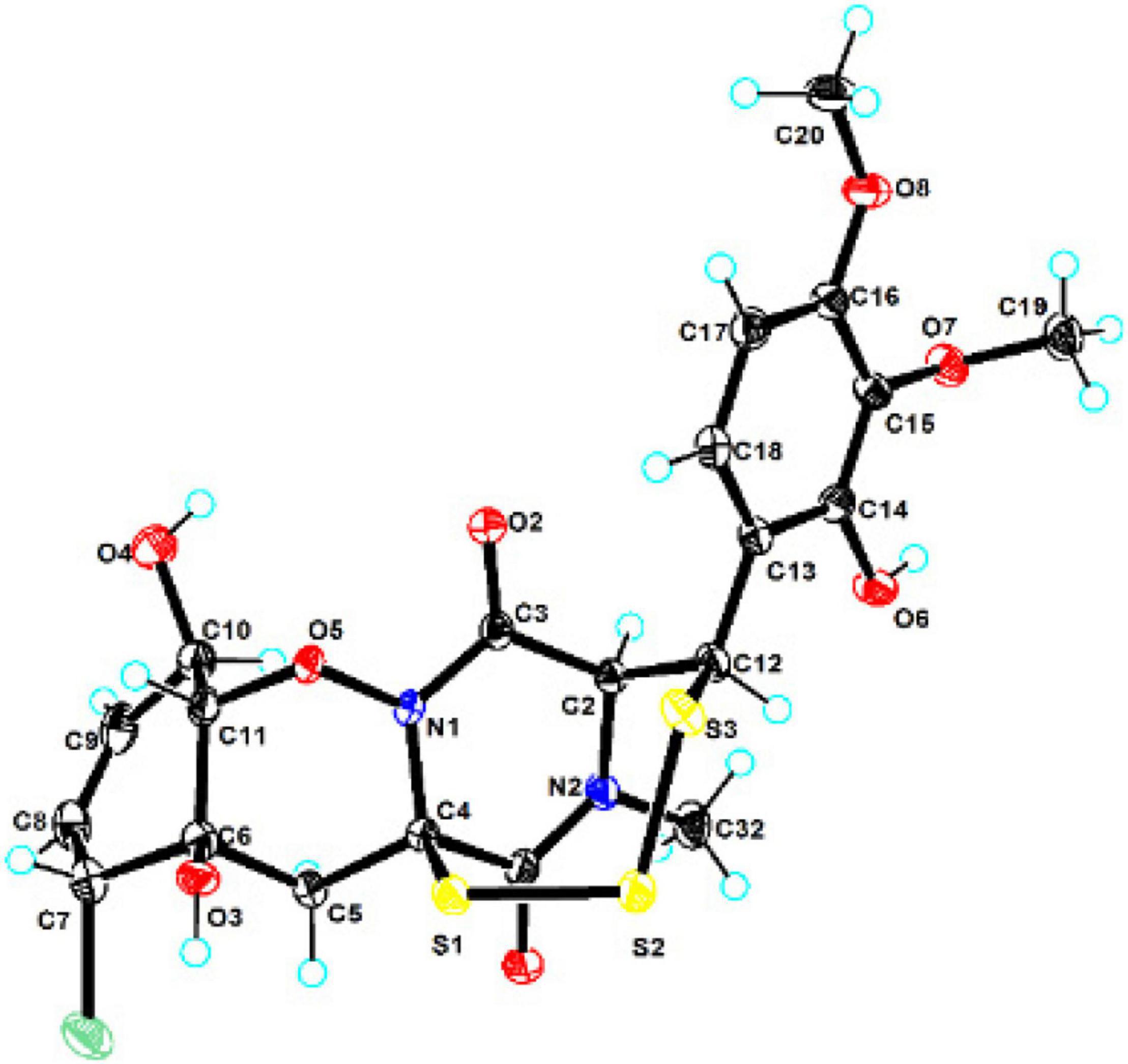
The crystallographic data of compound 1 obtained in MeOH were collected with a Rigaku XtaLAB PRO single-crystal diffractometer using Cu Kα radiation (λ = 1.54184). Briefly, their X-ray crystal structure was solved using SHELXS97, expanded by difference Fourier techniques, and refined by full-matrix least-squares calculation finally. The non-hydrogen atoms were refined anisotropically, and hydrogen atoms were fixed at calculated positions. The crystallographic data of compound 1 have been deposited in the Cambridge Crystallographic Data Centre.
Crystal Data for adametizine C (1): C21H31ClN2O12S3, Mr = 635.11, crystal size 0.1 mm × 0.08 mm × 0.06 mm, orthorhombic, a = 9.16490 (10) Å, b = 11.25800 (10) Å, c = 27.3339 (2) Å, α = β = γ = 90°, V = 2407.9 (4) Å3, Z = 4, T = 100.00 (10) K, Space group P212121, μ = 3.837 mm–1, ρcalc = 1.496 g/cm3, 14,343 reflections measured (6.468° ≤ 2Θ ≤ 148.482°), 5,546 unique (Rint = 0.0275, Rsigma = 0.0325). The final R1 values were 0.0287 [I > 2σ(I)]. The final wR (F2) values were 0.0758 [I > 2σ(I)]. The final R1 values were 0.0299 (all data). The final wR (F2) values were 0.0764 (all data). The goodness of fit on F2 was 1.049. The Flack parameter is 0.002 (6) (CDCC 2130918).
Antibacterial Activity Assay
The antimicrobial activities against five bacteria (Erysipelothrix rhusiopathiae WH13013, Streptococcus suis SC19, Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, and Staphylococcus aureus ATCC 25923) and four fungi (Botrytis cinerea, Septoria nodorum Berk., Fusarium graminearum Schw., and Rhizoctonia solani Kühn) were evaluated using the methods described previously (Wan et al., 2014). Cephalosporin and cycloheximide were used as positive controls against bacteria and fungi, respectively.
NF-κB Bioassay
The inhibitory activities of LPS-induced NF-κB activation in RAW264.7 cells were evaluated as detected by luciferase reporter gene assay as described previously (Tan et al., 2020). In brief, the RAW264.7 cells stably transfected with a luciferase reporter gene were plated in 96-well plates and then pretreated with tested compounds (20 μM) and BAY11-7082 (NF-κB inhibitor as positive control, 5 μM, Sigma-Aldrich) for 30 min, followed by LPS stimulation (5 μg/ml) for 8 h. Cells were harvested, and luciferase activities of the triplicate tests were measured by the luciferase assay system (Promega, Madison, WI, United States). For further study of compounds 1, 5, and 9 on osteoclastogenesis, bone marrow macrophage cells (BMMCs) were added with macrophage-stimulating factor (50 ng/ml) and RANKL (100 ng/ml) stimulation at 5 μM concentrations for 3 days. Then, the cells were fixed and stained for TRAP activity, and the images were photographed by using an inverted microscope (Nikon, Japan). Data are expressed as the mean ± SD and analyzed using GraphPad Prism 7.0 software (San Diego, CA, United States). Statistical differences among groups were performed using one-way analysis of variance with Bonferroni post hoc test. A p-value of < 0.05 was considered statistically significant.
Cytotoxicity Bioassay
Cell viability was analyzed by 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide (MTT) assay as previous described (Wang et al., 2021). In brief, cells were seeded in 96-well plate at a density of 5 × 103 per well overnight and treated with compounds for demand time. OD570 values were detected using a Hybrid Multi-Mode Reader (Synergy H1, BioTek). The experiment was repeated three times independently.
Plate Clone Formation Assay
PC-3 cells were seeded in six-well plate at a density of 1,000 cell per well overnight, and then, cells were treated with dimethyl sulfoxide (DMSO) (0.1%, v/v), docetaxel (1 μM), and compound 3 (1.25, 2.5, 5, and 10 μM), respectively, for demand time. The cell clone colonies were formed after treating for 2 weeks, and cells were fixed with 4% formaldehyde for 30 min, washed with phosphate buffer saline (PBS) buffer, and then stained with crystal violet stain solution for 30 min. The dye solution was removed, and the cells were washed with PBS buffer again. Cell colonies were recorded and analyzed by the colony count analysis system (GelCount, Oxford Optronix). The experiment was repeated three times independently.
Apoptosis Assay
PC-3 cells were seeded in six-well plate at a density of 2.0 × 105 cell per well overnight and treated with DMSO (0.1%, v/v), docetaxel (1 μM), and compound 3 (1.25, 2.5, 5, and 10 μM), respectively, for 48 h. Then, cells were collected and stained with annexin V–fluoresceine isothiocyanate (FITC) and propidium iodide (PI) solution following the manufacturer’s manual (BMS500FI-300, Thermo Fisher Scientific). The apoptotic rate of PC-3 cells was examined and analyzed by flow cytometer (NovoCyte, Agilent). Each experiment was repeated three times independently.
Results and Discussion
Structural Elucidation
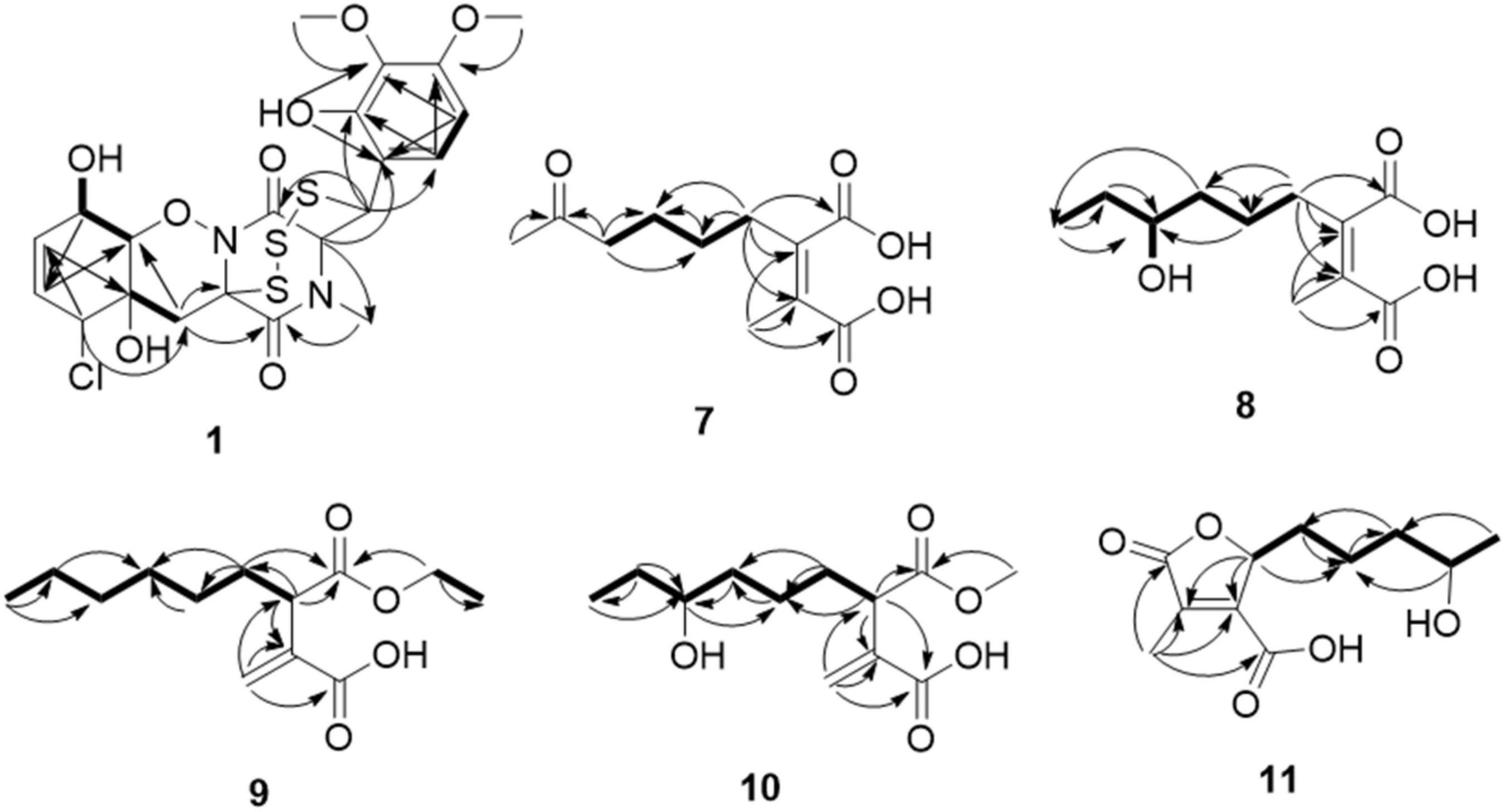
Compound 1 was obtained as colorless needles and had the molecular formula C21H23ClN2O8S3 with 11 degrees of unsaturation as established by HRESIMS. The 1H-NMR (Table 1) and HSQC experiment of 1 showed the typical pattern of an thiodiketopiperazines skeleton with three exchangeable protons, assigned to 14-OH (δH 9.64, s), 10-OH (δH 5.17, d, J = 6.6 Hz), and 6-OH (δH 6.11, d, J = 2.3 Hz), two aromatic methine protons [H-17 (δH 6.53, d, J = 8.8 Hz) and H-18 (δH 7.08, d, J = 8.8 Hz)], two olefinic protons [H-8 (δH 5.63, dt, J = 10.3, 2.5 Hz) and H-9 (δH 5.58, dt, J = 10.3, 2.0 Hz)], five methines [H-2 (δH 4.63, d, J = 1.1 Hz), H-7 (δH 4.88, m), H-10 (δH 4.43, m), H-11 (δH 4.10, dd, J = 7.3, 1.9 Hz), and H-12 (δH 5.39, s)], one methylene [H2-5α (δH 2.54, dd, J = 14.8, 2.0 Hz) and H2-5β (δH 2.09, d, J = 14.8 Hz)], one N-methyl [H3-21 (δH 3.12, s)], and two O-methyl [H3-19 (δH 3.69, s) and H3-20 (δH 3.78, s)]. Besides the above 13 corresponding hydrogen-bearing carbons, eight non-protonated (with six sp2 and two sp3) carbon atoms remained in the 13C NMR spectrum. Detailed analysis of the above NMR data and 2D NMR correlations (Figure 2 and Table 1) resulted in the elucidation of the planar structure of 1. Upon slow evaporation of the solvent MeOH, which was achieved by storing the sample in a refrigerator for 4 weeks, single crystals of adequate quality of 1 were obtained, making an X-ray diffraction study possible that could unequivocally confirm the chemical structure of 1. The absolute configuration was determined on the basis of measuring the anomalous dispersion effects by collecting Friedel pair reflections in the X-ray diffraction experiment (Figure 3).
Compound 7 was isolated as colorless oil, and its molecular formula was determined as C11H16O5 with four degrees of unsaturation, as a deprotonated ion peak at m/z 227.0927 [M-H]– in the HRESIMS spectrum. The NMR data of 7 (Tables 2, 3) indicated the presence of a ketone group (δC 208.8), two carboxylic groups (δC 166.7, 166.9), one double bond (δC 143.7, 141.5), two methyl group (δC 30.2, 9.8), and four aliphatic methylene groups. Through inspection of the 1H-1H COSY spectrum, we easily established a long spin system that started from H2-5 (δH 2.40, t) and terminated at H2-8 (δH 2.44, t) (Figure 2). In the HMBC spectrum, the H2-5 showed correlations to the double bond (C-2/C-3), as well as to carboxylic group (C-1, δC 166.7), indicating that C-5 and C-1 were attached to the allylic carbon (C-2). The HMBC correlations from H3-11 to double bond (C-3/C-2), as well as to C-4, indicating that C-11 and C-4 were attached to the allylic carbon (C-2). C-9 connected to C-10 and C-8 was substantiated by the HMBC correlations from H2-8 to C-9 and from H3-10 to C-9 and C-8. Therefore, 7 was established as 2-methyl-3-(5-oxohexyl) maleic acid.
Compound 8 exhibited UV maximum absorption at 206 and 250 nm similar to that of 7, indicating that they shared a similar chromophore. The HRESIMS data of 8 determined the molecular formula C11H18O5, with two hydrogen atoms more than 7. Compared to the NMR data, 8 contained one less carbonyl group at C-9, but one more hydroxyl group at C-8 than 7. Thus, 8 was determined as 2-(4-hydroxyhexyl)-3-methylmaleic acid.
Compound 9 showed a prominent peak at m/z 243.1594 [M + H]+ in the HRESIMS spectrum, corresponding to the molecular formula C13H23O4. Analysis of the NMR data (Tables 2, 3) of 9 revealed that it was also structurally related to 7. Compound 9 established a long chain from C-5 to C-10 through COSY spectrum (Figure 2). The position of the double bond is at C-3/C-11 and an ethyl group attached to the ester group (C1). Thus, compound 9 was determined as 3-(ethoxycarbonyl)-2-methylenenonanoic acid.
Compound 10, a colorless oil, was found to have the molecular formula C12H21O5 on the basis of HRESIMS data. In addition, 10 exhibited a similar UV maximum absorption as 9 at 206 nm, indicating that they have similar chromophores. The NMR spectrum of 10 (Tables 2, 3) is similar to that of 9, with one more oxygen atom and one methylene group. The 1H-1H COSY correlations (Figure 2) of H2-7/H-8 and H-8/H2-9, as well as HMBC correlations from H-8 to C-6 and from C-8 to H3-10, H2-9, and H2-7 indicated that the position of the oxymethine (δH 3.28; δC 71.3). Thus, 10 was elucidated as 7-hydroxy-3-(methoxycarbonyl)-2-methylenenonanoic acid.
Compound 11 was obtained as brown oil. HREIMS ion peak at m/z 229.1076 [M + H]+ gave the molecular formula C11H17O5, suggesting four degrees of unsaturation. The 13C NMR spectrum (Table 2) revealed two carbonyl carbons (δC 173.6 and 164.2), together with a fully substituted double bond (δC 133.9 and 151.5). The 1H–1H COSY spectrum (Figure 2) revealed a spin system consistent with an n-pentyl chain (from C-5 to C-9). An oxymethine of C-8 (δH 3.55; δC 66.0) had HMBC correlations extending to C-6 (δC 21.1). The HMBC correlations from δH 5.13 (1H, m, H-4) to C-2 (δC 133.9) and C-3 (δC 151.5) constructed an α, β-unsaturated five-member lactone ring. In addition, a methyl group substituted at C-2 was supported by the HMBC correlations of C-10 (δH 2.02, δC 10.7) to C-1, C-2, and C-3, and a carboxyl substituted at C-3 was supported by the correlations of H-10 to C-11. Finally, compound 11 was identified and named 2-(4-hydroxypentyl)-4-methyl-5-oxo-2,5-dihydrofuran-3-carboxylic acid.
The optical rotations of compounds 8–11 were close to zero, and these compounds showed little cotton effect in CD spectroscopy, suggesting them to be racemic mixtures (Supplementary Material). The α-hydro-carbon and C-8 with hydroxyl groups led to chiral centers. It had been reported in the literature that the alkane derivatives were found as enantiomers (Akone et al., 2014).
In addition, the known thiodiketopiperazine derivatives were elucidated as adametizine A (2) (Liu et al., 2015a), DC1149B (3) (Yamazaki et al., 2015), outovirin B (4) (Kajula et al., 2016), pretrichodermamide E (5) (Yurchenko et al., 2016), and peniciadametizine A (6) (Liu et al., 2015b), respectively, by comparing their physicochemical properties and spectroscopic data with the reported literature values.
Bioassays of Compounds
All obtained compounds were evaluated for their antibacterial and antifungal activities. Compounds 1–5 exhibited weak antibacterial activities against Erysipelothrix rhusiopathiae WH13013 and Streptococcus suis SC19, with the minimal inhibitory concentration (MIC) values of 50–100 μg/ml. Compound 2 also exhibited activity against fungi Botrytis cinerea and Septoria nodorum Berk., with the MIC values of 25 μg/ml. However, all isolated compounds showed no activities against the other three bacteria (Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, and Staphylococcus aureus ATCC 25923) and two fungi (Fusarium graminearum Schw. and Rhizoctonia solani Kühn). Cephalosporin and cycloheximide were used as the positive controls in the antibacterial (MIC values of 0.78 μg/ml) and antifungal (MIC values of 6.25 μg/ml) tests, respectively (Table 4).

Table 4. Antibacterial, antifungal, cytotoxic, and anti-inflammatory activities of the obtained compounds.
Two human prostate cancer cell lines, PC-3 (androgen receptor negative) and 22Rv1 (androgen receptor positive), were used in the cytotoxicity tests. Compounds 1–3 exhibited cytotoxicity against 22Rv1 cells with half maximal inhibitory concentration (IC50) values of 13.9, 13.0, and 13.6 μM, respectively, whereas 1 and 3 showed activities against PC-3 cells with IC50 values of 44.0 and 5.1 μM, respectively. Compound 3 was further evaluated for its anti-tumor effect by plate clone formation assay and flow cytometry on PC-3 cells. The results showed that 3 reduced PC-3 cells colony formation (Figures 4A,C) and induced cell apoptosis in a dose-dependent manner (Figures 4B,D).
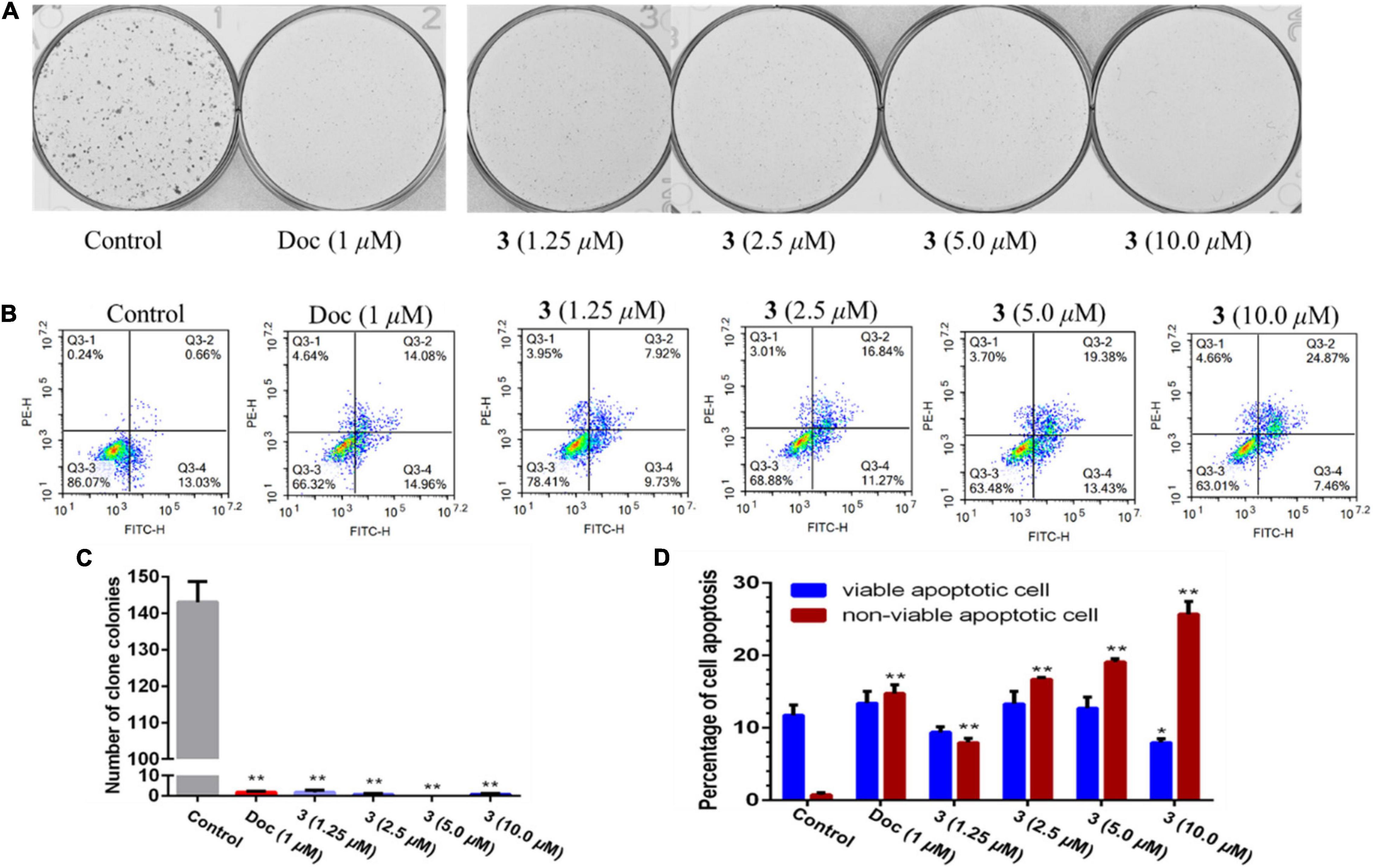
Figure 4. Compound 3 reduced PC-3 cells colony formation (A,C) and induced apoptosis (B,D). All results were presented as mean ± SD. Statistical significance was determined with one-way ANOVA. *P < 0.05 and **P < 0.01 were considered statistically significant.
Compounds 1–11 were screened for their inhibitory activities of LPS-induced NF-κB activation in RAW264.7 cells. Compounds 1, 2, 5, 9, and 11 exhibited obvious inhibitory activities against LPS-induced NF-κB with IC50 values of 8.2, 15.1, 12.6, 10.7, and 21.5 μM, respectively. Moreover, in the further study for evaluation with their effects on RANKL-induced osteoclastogenesis, 1, 5, and 9 could suppress the RANKL-induced osteoclast differentiation in BMMCs obviously, with the concentration of 10 μM. The new trithiodiketopiperazine derivative adametizine C (1) showed the strongest activity, relatively (Figure 5). Consequently, it is revealed that these compounds could be the promising osteoclast differentiation inhibitors for the treatment of osteoclast-related diseases.
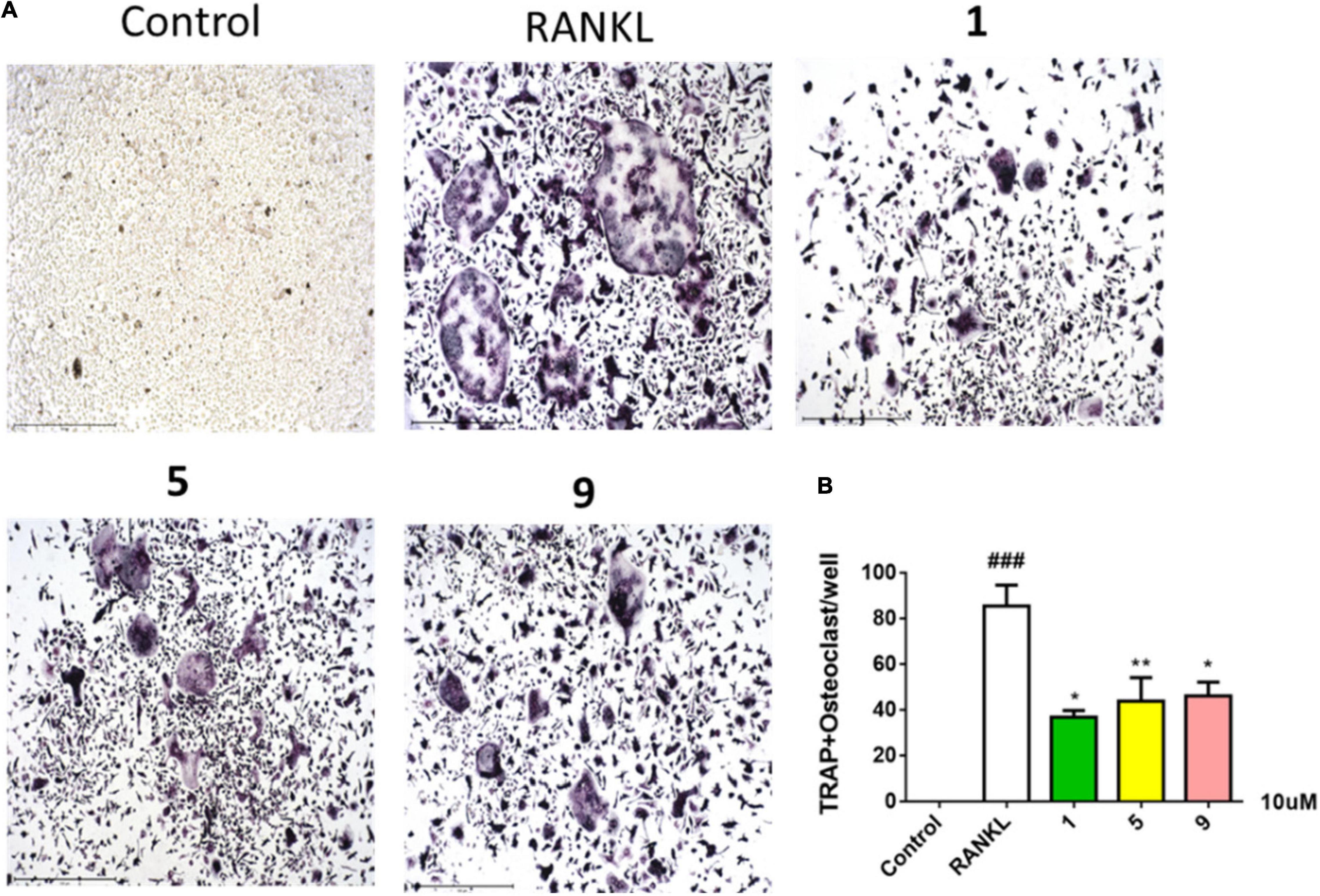
Figure 5. Compounds 1, 5, and 9 suppressed RANKL-induced osteoclast differentiation. Representative images of osteoclasts from BMMCs treated with 1, 5, and 9 (10 μM) for 3 days, tartrate-resistant acidic phosphatase (TRAP)–positive multinucleated cells were regarded as osteoclasts (A) and quantified (B). All experiments were performed at least three times. The data are presented as the mean ± SD of representative experiments. ###p < 0.001 vs. control group; *p < 0.05 and **p < 0.01 vs. RANKL group.
In addition, we also found that the thiodiketopiperazine derivatives exhibited various activities reported in the literatures. For example, pretrichodermide A was active against Mycobacterium tuberculosis (Seephonkai et al., 2006); Adametizine A (2) was found to be active against a variety of bacteria (Liu et al., 2015a); Outovirin C was active against fungus Botrytis cinerea (Kajula et al., 2016). Gliovirin showed inhibitory effects on the expression of cytokines [tumor necrosis factor (TNF)-α and interleukin-2 (IL-2)] and pro-inflammatory enzymes [cyclooxygenase-2 (COX-2) and inducible nitric oxide synthase (INOS)] in T cells and monocytes/macrophages (Rether et al., 2007).
Conclusion
In summary, chemical investigation of the mangrove sediment–derived fungus Penicillium ludwigii SCSIO41408 led to the isolation of a new trithiodiketopiperazine, five new alkane derivatives (7–11), and five dithiodiketopiperazine derivatives (2–6). In a variety of bioactivity screening, 1–5 exhibited some selective antifungal or antibacterial activities; 1–3 showed cytotoxicity against prostate cancer cell line 22Rv1or PC-3 cells; moreover, 3 could significantly reduce PC-3 cells’ colony formation and induce apoptosis in a dose-dependent manner. Several compounds also exhibited obvious inhibitory activities of LPS-induced NF-κB, and 1, 5, and 9 were suppressed RANKL-induced osteoclast differentiation in BMMCs at 10 μM. Adametizine C (1), with the strongest inhibitory activity against RANKL-induced osteoclast differentiation, was suggested to be the promising lead compound for the treatment of osteoclast-related diseases.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JC, BP, YL, and XZ contributed to the conception and design of the study. JC performed the experiments, analyzed data, and wrote the manuscript. XZ reviewed and revised the manuscript. XW and ZY performed the cytotoxicity against 22Rv1 and PC-3 cells. YT did the inhibitory activities of LPS-induced NF-κB activation. All authors contributed to manuscript revision and reviewed and approved the submitted version.
Funding
This work was supported by grants from the Guangdong Local Innovation Team Program (2019BT02Y262), the Marine Economy Development Project of Guangdong Province [GDNRC (2021)52], National Natural Science Foundation of China (U20A20101, 81973235), K. C. Wong Education Foundation (GJTD-2020-12), and Liao Ning Revitalization Talents Program (XLYC1802037).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to the analytical facilities (Xiao, Zheng, Sun, Zhang, and Ma) in SCSIO.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.857041/full#supplementary-material
References
Akone, S. H., Rahn, S., Henrich, B., Daletos, G., Vardamides, J. C., Nkengfack, A. E., et al. (2014). 2-Pentenedioic acid derivatives from a soil-derived fungus Gongronella butleri. Phytochem. Lett. 10, 184–188. doi: 10.1016/j.phytol.2014.09.001
Cai, J., Chen, C. M., Tan, Y. H., Chen, W. H., Luo, X. W., Luo, L. X., et al. (2021). Bioactive polyketide and diketopiperazine derivatives from the mangrove-sediment-derived fungus. Aspergill. Mol. 26:4851. doi: 10.3390/molecules26164851
Chen, C. M., Chen, W. H., Pang, X. Y., Liao, S. R., Wang, J. F., Lin, X. P., et al. (2021a). Pyrrolyl 4-quinolone alkaloids from the mangrove endophytic fungus Penicillium steckii SCSIO 41025: Chiral resolution, configurational assignment, and enzyme inhibitory activities. Phytochemistry 186:112730. doi: 10.1016/j.phytochem.2021.112730
Chen, C. M., Chen, W. H., Tao, H. M., Yang, B., Zhou, X. F., Luo, X. W., et al. (2021b). Diversified polyketides and nitrogenous compounds from the mangrove endophytic fungus Penicillium steckii SCSIO 41025. Chin. J. Chem. 39, 2132–2140. doi: 10.1002/cjoc.202100226
Dai, J. J., Chen, A., Zhu, M. L., Qi, X., Tang, W., Liu, M., et al. (2019). Penicisulfuranol A, a novel C-terminal inhibitor disrupting molecular chaperone function of Hsp90 independent of ATP binding domain. Biochem. Pharmacol. 163, 404–415. doi: 10.1016/j.bcp.2019.03.012
He, F., Li, X. B., Yu, J. H., Zhang, X. Y., Nong, X. H., Chen, G. Y., et al. (2019). Secondary metabolites from the mangrove sediment-derived fungus Penicillium pinophilum SCAU037. Fitoterapia 136:104177. doi: 10.1016/j.fitote.2019.104177
Hong, G. J., Zhou, L., Han, X. R., Sun, P., Chen, Z. Q., He, W., et al. (2020). Asiatic acid inhibits OVX-induced osteoporosis and osteoclastogenesis via regulating RANKL-mediated NF-κB and NFATC1 signaling pathways. Front. Pharmacol. 11:331. doi: 10.3389/fphar.2020.00331
Kajula, M., Ward, J. M., Turpeinen, A., Tejesvi, M. V., Hokkanen, J., Tolonen, A., et al. (2016). Bridged epipolythiodiketopiperazines from Penicillium raciborskii, an endophytic fungus of Rhododendron tomentosum harmaja. J. Nat. Prod. 79, 685–690. doi: 10.1021/np500822k
Li, K. L., Chen, S. Q., Pang, X. Y., Cai, J., Zhang, X. Y., Liu, Y. H., et al. (2022). Natural products from mangrove sediments-derived microbes: Structural diversity, bioactivities, biosynthesis, and total synthesis. Eur. J. Med. Chem. 2022:117. doi: 10.1016/j.ejmech.2022.114117
Liu, Y., Li, X. M., Meng, L. H., Jiang, W. L., Xu, G. M., Huang, C. G., et al. (2015a). Bisthiodiketopiperazines and acorane sesquiterpenes produced by the marine-derived fungus Penicillium adametzioides AS-53 on different culture media. J. Nat. Prod. 78, 1294–1299. doi: 10.1021/acs.jnatprod.5b00102
Liu, Y., Mándi, A., Li, X. M., Meng, L. H., Kurtán, T., and Wang, B. G. (2015b). Peniciadametizine A, a dithiodiketopiperazine with a unique spiro [furan-2,7’-pyrazino [1,2-b][1,2]oxazine] skeleton, and a related analogue, peniciadametizine B, from the marine sponge-derived fungus Penicillium adametzioides. Mar. Drugs 13, 3640–3652. doi: 10.3390/md13063640
Luo, X. W., Chen, C. M., Tao, H. M., Lin, X. P., Yang, B., Zhou, X. F., et al. (2019). Structurally diverse diketopiperazine alkaloids from the marine-derived fungus Aspergillus Versicolor SCSIO 41016. Org. Chem. Front. 6, 736–740. doi: 10.1039/C8QO01147H
Luo, X. W., Lin, X. P., Tao, H. M., Wang, J. F., Li, J., Yang, B., et al. (2018). Isochromophilones A–F, cytotoxic chloroazaphilones from the marine mangrove endophytic fungus Diaporthe sp. SCSIO 41011. J. Nat. Prod. 81, 934–941. doi: 10.1021/acs.jnatprod.7b01053
Rether, J., Serwe, A., Anke, T., and Erkel, J. (2007). Inhibition of inducible tumor necrosis factor-α expression by the fungal epipolythiodiketopiperazine gliovirin. Biol. Chem. 388, 627–637. doi: 10.1515/BC.2007.066
Seephonkai, P., Kongsaeree, S., Prabpai, S., Isaka, M., and Thebtaranonth, Y. (2006). Transformation of an irregularly bridged epidithiodiketopiperazine to trichodermamide A. Org. Lett. 8, 3073–3075. doi: 10.1021/ol061046l
Tan, Y. H., Dang, W. D., Yang, Y. Y., Ke, M. H., Zou, B. H., Luo, X. W., et al. (2020). A marine fungus-derived nitrobenzoyl sesquiterpenoid suppresses receptor activator of NF-κB ligand-induced osteoclastogenesis and inflammatory bone destruction. Brit. J. Pharmacol. 177, 4242–4260. doi: 10.1111/bph.15179
Wan, Z. Y., Fang, W., Shi, L. Q., Wang, K. M., Zhang, Y. N., and Zhang, Z. G. (2014). Novonestmycins A and B, two new 32-membered bioactive macrolides from Streptomyces phytohabitans HBERC-20821. J. Antibiot. 68, 185–190. doi: 10.1038/ja.2014.123
Wang, X., Zhu, J., Yan, H., Shi, M., Zheng, Q., Wang, Y., et al. (2021). Kaempferol inhibits benign prostatic hyperplasia by resisting the action of androgen. Eur. J. Pharmacol. 907:174251. doi: 10.1016/j.ejphar.2021.174251
Yamazaki, H., Takahashi, O., Murakami, K., and Namikoshi, M. (2015). Induced production of a new unprecedented epitrithiodiketopiperazine, chlorotrithiobrevamide, by a culture of the marine-derived trichoderma cf. brevicompactum with dimethyl sulfoxide. Tetrahed. Lett. 56, 6262–6265. doi: 10.1016/j.tetlet.2015.09.113
Yurchenko, A. N., Smetanina, O. F., Ivanets, E. V., Kalinovsky, A. I., and Dyshlovoy, S. A. (2016). Pretrichodermamides D–F from a marine algicolous fungus Penicillium sp. KMM 4672. Mar. Drugs 14, 122. doi: 10.3390/md14070122
Zhou, Y., Wang, C. W., Si, J. Y., Wang, B. X., Zhang, D. H., Ding, D., et al. (2020). Melatonin up-regulates bone marrow mesenchymal stem cells osteogenic action but suppresses their mediated osteoclastogenesis via MT2 -inactivated NF-κB pathway. Brit. J. Pharmacol. 177, 2106–2122. doi: 10.1111/bph.14972
Keywords: mangrove-sediment-derived fungus, penicillium ludwigii, thiodiketopiperazines, PC-3, NF-κB, osteoclast differentiation
Citation: Cai J, Wang X, Yang Z, Tan Y, Peng B, Liu Y and Zhou X (2022) Thiodiketopiperazines and Alkane Derivatives Produced by the Mangrove Sediment–Derived Fungus Penicillium ludwigii SCSIO 41408. Front. Microbiol. 13:857041. doi: 10.3389/fmicb.2022.857041
Received: 18 January 2022; Accepted: 16 February 2022;
Published: 28 March 2022.
Edited by:
Xian-Wen Yang, Third Institute of Oceanography, Ministry of Natural Resources, ChinaReviewed by:
Chang-Lun Shao, Ocean University of China, ChinaKandasamy Saravanakumar, Kangwon National University, South Korea
Copyright © 2022 Cai, Wang, Yang, Tan, Peng, Liu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanhui Tan, dHloNTMzQDEyNi5jb20=; Bo Peng, cGVuZ2JvQGdpZy5hYy5jbg==; Xuefeng Zhou, eGZ6aG91QHNjc2lvLmFjLmNu
 Jian Cai1,2
Jian Cai1,2 Xueni Wang
Xueni Wang Yonghong Liu
Yonghong Liu Xuefeng Zhou
Xuefeng Zhou