- Faculty of Engineering, University of Nottingham, Nottingham, United Kingdom
[FeFe]-hydrogenases are complex metalloenzymes, key to microbial energy metabolism in numerous organisms. During anaerobic metabolism, they dissipate excess reducing equivalents by using protons from water as terminal electron acceptors, leading to hydrogen production. This reaction is coupled to reoxidation of specific redox partners [ferredoxins, NAD(P)H or cytochrome c3], that can be used either individually or simultaneously (via flavin-based electron bifurcation). [FeFe]-hydrogenases also serve additional physiological functions such as H2 uptake (oxidation), H2 sensing, and CO2 fixation. This broad functional spectrum is enabled by a modular architecture and vast genetic diversity, which is not fully explored and understood. This Mini Review summarises recent advancements in identifying and characterising novel [FeFe]-hydrogenases, which has led to expanding our understanding of their multiple roles in metabolism and functional mechanisms. For example, while numerous well-known [FeFe]-hydrogenases are irreversibly damaged by oxygen, some newly discovered enzymes display intrinsic tolerance. These findings demonstrate that oxygen sensitivity varies between different [FeFe]-hydrogenases: in some cases, protection requires the presence of exogenous compounds such as carbon monoxide or sulphide, while in other cases it is a spontaneous built-in mechanism that relies on a reversible conformational change. Overall, it emerges that additional research is needed to characterise new [FeFe]-hydrogenases as this will reveal further details on the physiology and mechanisms of these enzymes that will enable potential impactful applications.
Introduction
Microbial hydrogen metabolism is thought to have appeared in the very early days of life on Earth, before oxygen began to accumulate in the atmosphere 2.4 billion years ago (Lyons et al., 2014). It has existed ever since, and it still plays a key role in numerous environments such as soil, wetlands, marine sediments, freshwaters, oceans, geothermal springs, and animal guts (Boyd et al., 2010; Greening et al., 2016; Piche-Choquette and Constant, 2019).
Hydrogenases are specialised metalloenzymes essential to microbial hydrogen metabolism. They are classified in three classes based on the metals found at the active site: [FeFe]-hydrogenases, [NiFe]-hydrogenases, and [Fe]-hydrogenases (also known as Hmd) (Vignais and Billoud, 2007).
[FeFe]-hydrogenases are found in the genome of numerous microorganisms, both Prokaryotes and Eukaryotes but not in Archaea (Peters et al., 2015). Their enzymatic features depend on a biologically unusual iron sulphur centre, named H-cluster, composed of a cubane [4Fe4S]-subcluster linked to a [FeFe]-subcluster via a conserved cysteine residue. Concerted proton and electron transfer steps lead to H2 production, via a mechanism that is under debate (Haumann and Stripp, 2018; Birrell et al., 2021). The ability of [FeFe]-hydrogenases to catalyse reversible H2 production at high turnover rates and low overpotential has put them under the spotlight for potential exploitation in devices to produce green hydrogen (Morra et al., 2017; Evans et al., 2019; Brown and King, 2020). [FeFe]-hydrogenases have also inspired the synthesis of artificial catalysts that mimic their natural properties (Ahmed and Dey, 2019; Karayilan et al., 2019). Synthetic biology has also explored the potential of exploiting [FeFe]-hydrogenases in vivo, to improve or to instal H2 production in several microbial hosts (Klein et al., 2010; Kelly et al., 2015; Noone et al., 2017; Kanygin et al., 2020; Wegelius et al., 2021).
This Mini Review will focus on [FeFe]-hydrogenases and highlight how recent research is revolutionising our understanding of these enzymes.
[FeFe]-Hydrogenases Diversity: A Poorly Explored Space
In the post-genomic and multi-omics era, thousands of putative [FeFe]-hydrogenase sequences can be retrieved from public databases. The common factor to all [FeFe]-hydrogenases is the H-domain, a ∼40 kDa (350 amino acids) protein domain that hosts the H-cluster. In addition to sequence variability within this core domain, [FeFe]-hydrogenases display a highly modular genetic organisation, featuring several additional domains and subunits that lead to both monomeric and multi-subunit enzyme complexes. Research in this field has identified massive diversity and several classification schemes have been proposed (Meyer, 2007; Vignais and Billoud, 2007; Calusinska et al., 2010; Winkler et al., 2013). In addition to studying the hydrogenase gene phylogeny, recent studies have also included the analysis of flanking genes. Poudel et al. (2016) compiled 714 sequences and proposed three groups: (G1) monomeric HydA; (G2) trimeric HydABC; (G3) tetrameric HydABCD. Greening et al. (2016) curated 1,222 sequences and proposed a classification into three groups (Figure 1): (A) prototypical and bifurcating; (B) putative ancestral; (C) putative sensory; by analysing variations in the domain organisation and probable quaternary structure, group A can be further split into four subtypes.
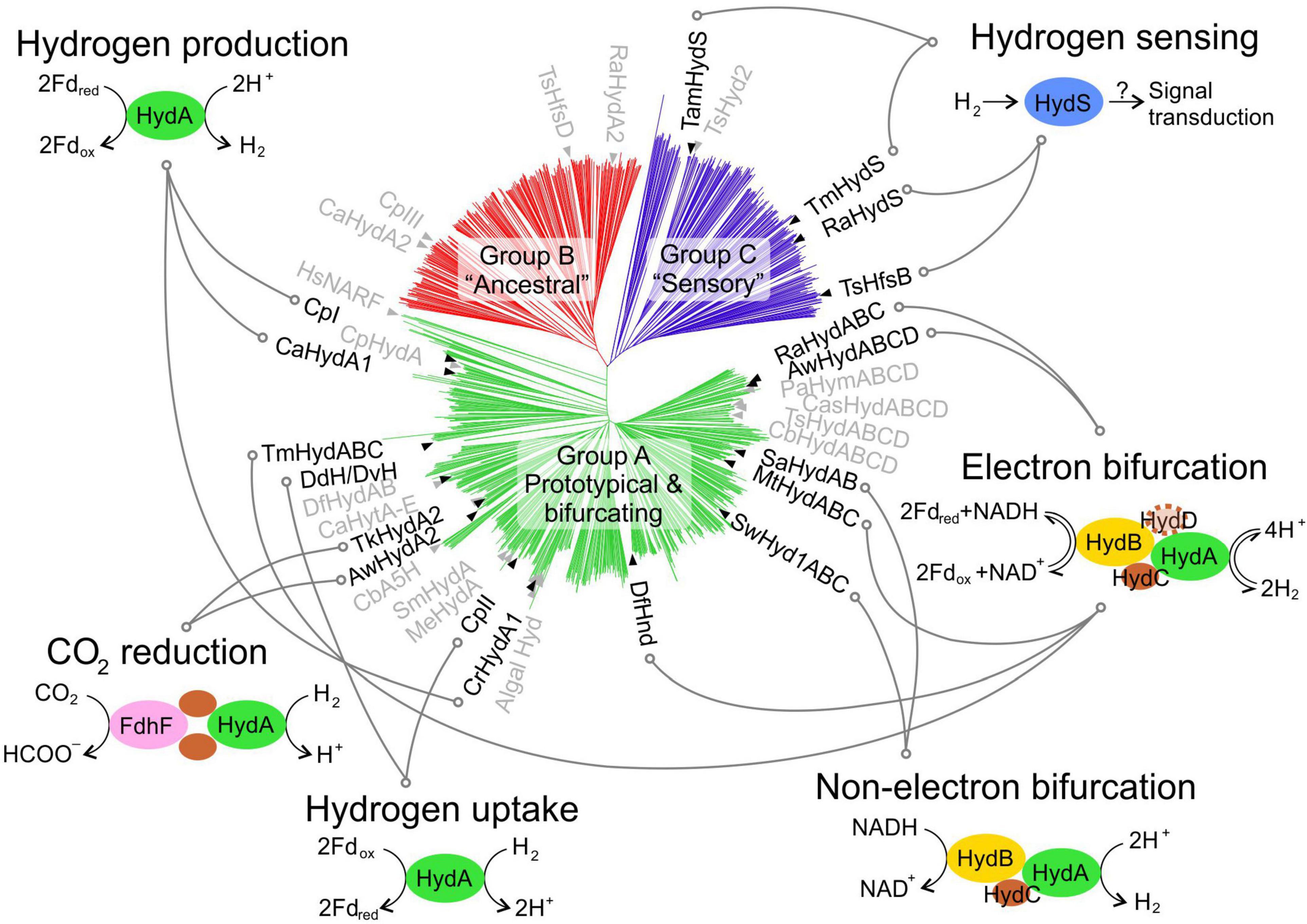
Figure 1. [FeFe]-hydrogenases phylogeny and known functions. A phylogenetic tree shows the phylogeny of [FeFe]-hydrogenase sequences from public databases, as previously proposed (Greening et al., 2016). Enzymes that have been experimentally characterised are indicated on the tree to show their relative position. The proposed physiological function of each enzyme is also presented, where known. A full list of enzymes is available in Supplementary Table 1, including details on the enzyme identifier/acronym used here. Hyd, hydrogenase subunit; FdhF, formate dehydrogenase subunit; Fdrex/ox, reduced/oxidised ferredoxin; NADH/NAD+, reduced/oxidised nicotinamide adenine dinucleotide.
However, only three model enzymes have been experimentally characterised to a high level of detail using various techniques, and they all belong to group A (subtype A1). These are: CpI (from the anaerobic nitrogen-fixing bacterium Clostridium pasteurianum) (Nakos and Mortenson, 1971; Peters et al., 1998; Therien et al., 2017); DvH (from the sulphate-reducing bacterium Desulfovibrio vulgaris) (Legall et al., 1971; Nicolet et al., 1999; Pohorelic et al., 2002); and CrHydA1 (from the eukaryotic green alga Chlamydomonas reinhardtii) (Happe and Naber, 1993; Happe and Kaminski, 2002; Mulder et al., 2010). DvH and DdH (from Desulfovibrio desulfuricans) sequences have been claimed to be identical (Nicolet et al., 1999), even if a closer inspection at genomes denotes numerous differences. Despite this, the nomenclature has been used interchangeably, and biophysical characterisation has been carried out on DvH sequence under the DdH name (Nicolet et al., 1999; Birrell et al., 2016; Rodriguez-Macia et al., 2020).
From the late 1990s, research has expanded to more [FeFe]-hydrogenases thanks to important technical advances, such as recombinant overexpression (Girbal et al., 2005; King et al., 2006; Kuchenreuther et al., 2010) and production of semi-synthetic hydrogenases (Berggren et al., 2013; Esselborn et al., 2013). Currently, approximately 40 [FeFe]-hydrogenases have been studied experimentally (Figure 1 and Supplementary Table 1). A simple comparison of these numbers with the count of putative [FeFe]-hydrogenases makes it evident that only very little is currently known about these enzymes’ diversity. Furthermore, the level of characterisation is highly variable, and in most cases only a very limited amount of information is available (Supplementary Table 1).
Beyond Hydrogen Production: Multiple Functional Roles
[FeFe]-hydrogenases are well-known for their prototypical role in hydrogen production (Figure 1): the enzyme acts as a sink for reducing equivalents, allowing for dissipation of excess reducing power from energy metabolism. This role is well-characterised in some clostridial hydrogenases, such as CpI (Therien et al., 2017), and in algal hydrogenases, such as CrHydA1 (Happe and Kaminski, 2002). Other hydrogenases with very high sequence similarity are believed to acts in a similar way both within green algae (Florin et al., 2001; Winkler et al., 2002; Kamp et al., 2008; Meuser et al., 2011; Cornish et al., 2015) and Clostridia (Demuez et al., 2007; Morra et al., 2016b). However, H2-producing hydrogenases do not cluster together in the phylogenetic tree, demonstrating that the function cannot simply be predicted from the sequence alone, as previously noted (Greening et al., 2016).
In addition to this flagship role, several new functions have been identified. During hydrogen uptake (Figure 1), the enzyme oxidises hydrogen and transfers the low potential electrons to a suitable cellular acceptor. This function has been proposed for relatively few [FeFe]-hydrogenases, such as the cytoplasmic CpII (Therien et al., 2017) and periplasmic DdH/DvH (Pohorelic et al., 2002).
Reactions of additional complexity are catalysed by multi-subunit [FeFe]-hydrogenases. AwHydA2 (from Acetobacterium woodii) (Schuchmann and Muller, 2013) and TkHydA2 (from Thermoanaerobacter kivui) (Schwarz et al., 2018) have been shown to form stable heterotetrameric complexes with two FeS subunits and a formate dehydrogenase (FdhF) subunit. These enzymes have been named HDCR (Hydrogen-Dependent Carbon dioxide Reductase) after their unprecedented ability to catalyse direct CO2 reduction using H2 as the sole reducing agent, in a single and efficient step (Schuchmann et al., 2018; Leo et al., 2021).
Following the discovery of TmHydABC (from Thermotoga maritima) (Schut and Adams, 2009), numerous other heterotrimeric and heterotetrameric [FeFe]-hydrogenases have been shown to perform flavin-based electron bifurcation (FBEB). These enzymes couple the thermodynamically favourable oxidation of ferredoxin to the unfavourable oxidation of NADH leading to H2 production. The coupling is synergistic and provides a clear physiological advantage over “conventional” hydrogen production from ferredoxin only, as it allows simultaneous reoxidation of both NADH and ferredoxin from glycolysis, thus facilitating ATP production in the absence of aerobic respiration (Buckel and Thauer, 2018; Peters et al., 2018; Schuchmann et al., 2018).
Two [FeFe]-hydrogenases SwHyd1ABC (from Syntrophomonas wolfei) (Losey et al., 2017) and SaHydAB (from Syntrophus aciditrophicus) (Losey et al., 2020) have been discovered to catalyse H2 production from NADH without the requirement of reduced ferredoxin. Despite overall sequence and predicted structural similarity to FBEB enzymes, closer inspection of the flavin-containing HydB subunit revealed differences that may explain the absence of synergistic bifurcation (Losey et al., 2020).
A sensory role has been proposed for a number of group C [FeFe]-hydrogenases, such as TmHydS (Chongdar et al., 2018) and TamHydS (Land et al., 2020). This function has been proposed based the presence of a PAS domain, that is known to take part in signal transduction of other proteins. Furthermore, the lack of a conserved cysteine near the H-cluster slows down the turnover rate, thus making unlikely an active metabolic role. Additional evidence supporting a sensory role has been reported for RaHydS (from Ruminococcus albus) (Zheng et al., 2014) and TsHfsB (from Thermoanaerobacterium saccharolyticum) (Shaw et al., 2009). However, given the little direct physiological evidence for such role, this assignment is considered putative.
Group B is currently much less characterised and a tentative denomination as “ancestral” has been proposed, but due to lack of evidence it is difficult to discuss further on their function. Some data are available for CpIII, showing that the gene is transcribed (Therien et al., 2017) and that the enzyme is active and displays a different catalytic bias to both CpI and CpII (Artz et al., 2020).
It is also notable that proteins with sequence similarity to [FeFe]-hydrogenases can be found in higher eukaryotes, including humans (HsNARF, Figure 1), where they lost H2-linked functionality and evolved to other roles (Barton and Worman, 1999; Ding et al., 2020).
The impressive variety of functional roles for [FeFe]-hydrogenases is not surprising given the large number of organisms that rely on them, and their evolutionary history that allowed adaptation to numerous ecological niches. However, such diversity does not only occur across different organisms, but also within them: several species possess multiple [FeFe]-hydrogenase genes annotated in the genome, often in addition to other H2-activating enzymes such as [NiFe]-hydrogenases and nitrogenases (Calusinska et al., 2010; Baffert et al., 2019). The apparent redundancy of hydrogen-related enzymes may provide advantages by quickly adapting the metabolism in response to the environmental changes. However, a comprehensive investigation on multiple hydrogenases within an organism is currently missing.
Thermoanaerobacterium saccharolyticum has four putative [FeFe]-hydrogenases and a putative [NiFe]-hydrogenase genes. A systematic knockout study revealed that the hfs genes encoding for TsHfsB and TsHfsD [FeFe]-hydrogenases are essential for H2 production, while deletion of the other genes had no effect on this function. Moreover, deletion of hfs genes downregulated the expression of all the other genes (hyd and ech), suggesting a regulatory or sensory role for TsHfsB. Consequently, group B TsHfsD may be the main enzyme for H2 production in this organism (Shaw et al., 2009).
Within green algae, it is common to find two closely related [FeFe]-hydrogenase genes that are likely originating from gene duplication (Meuser et al., 2011). For example, Chlamydomonas reinhardtii encodes for CrHydA1 and CrHydA2, whose expression profile is very similar (Forestier et al., 2003). The two isoenzymes differ for their affinity for the ferredoxin PetF and for catalytic bias, suggesting different functional roles (Engelbrecht et al., 2021).
The well-known solvent producer Clostridium acetobutylicum has two [FeFe]-hydrogenases (CaHydA1 and CaHydA2), a [NiFe]-hydrogenase and nitrogenase. It has been suggested that CaHydA1 is the main enzyme for hydrogen production during acidogenesis (Demuez et al., 2007), but more recently a ΔCaHydA1 mutant strain has shown abundant H2 production (Du et al., 2021) suggesting a significant contribution from other enzymes. On the other hand, the [NiFe]-hydrogenase has been shown to play a key role in hydrogen cycling during solventogenesis (i.e., H2 reoxidation to provide reducing power for the acid-to-solvent conversion) (Germane et al., 2018).
In the case of Clostridium pasteurianum (three [FeFe]-hydrogenases, a [NiFe]-hydrogenase and nitrogenase), it has been proposed that CpI is the key H2 producer under non-nitrogen-fixing conditions (Adams, 1990), while CpII and the [NiFe]-hydrogenase would act as H2 uptake enzymes during nitrogen fixation. This would allow for reducing equivalents to be recovered from the highly uncoupled reaction of nitrogenase, ultimately improving the efficiency of this energy-consuming process (Therien et al., 2017).
Gene expression studies of multiple [FeFe]-hydrogenases in Clostridium butyricum, Clostridium beijerinckii, and Clostridium perfringens have shown that all genes are transcribed and regulation is actively occurring, suggesting different functional roles that have not yet been determined (Morra et al., 2014; Calusinska et al., 2015; Arizzi et al., 2021).
Oxygen Sensitivity: Not an Insurmountable Problem
For long time it has been assumed that [FeFe]-hydrogenases were extremely sensitive to O2, with their catalytic activity disappearing irreversibly and very quickly, in contrast to [NiFe]-hydrogenases that often display reversible inhibition or complete tolerance (Goldet et al., 2009; Lautier et al., 2011; Kubas et al., 2017). While this is certainly true for some of the most studied model [FeFe]-hydrogenases, the characterisation of novel enzymes has recently revealed that the phenomenon of oxygen sensitivity is highly variable across the class, and several examples of oxygen-stable enzymes exist (Figure 2).
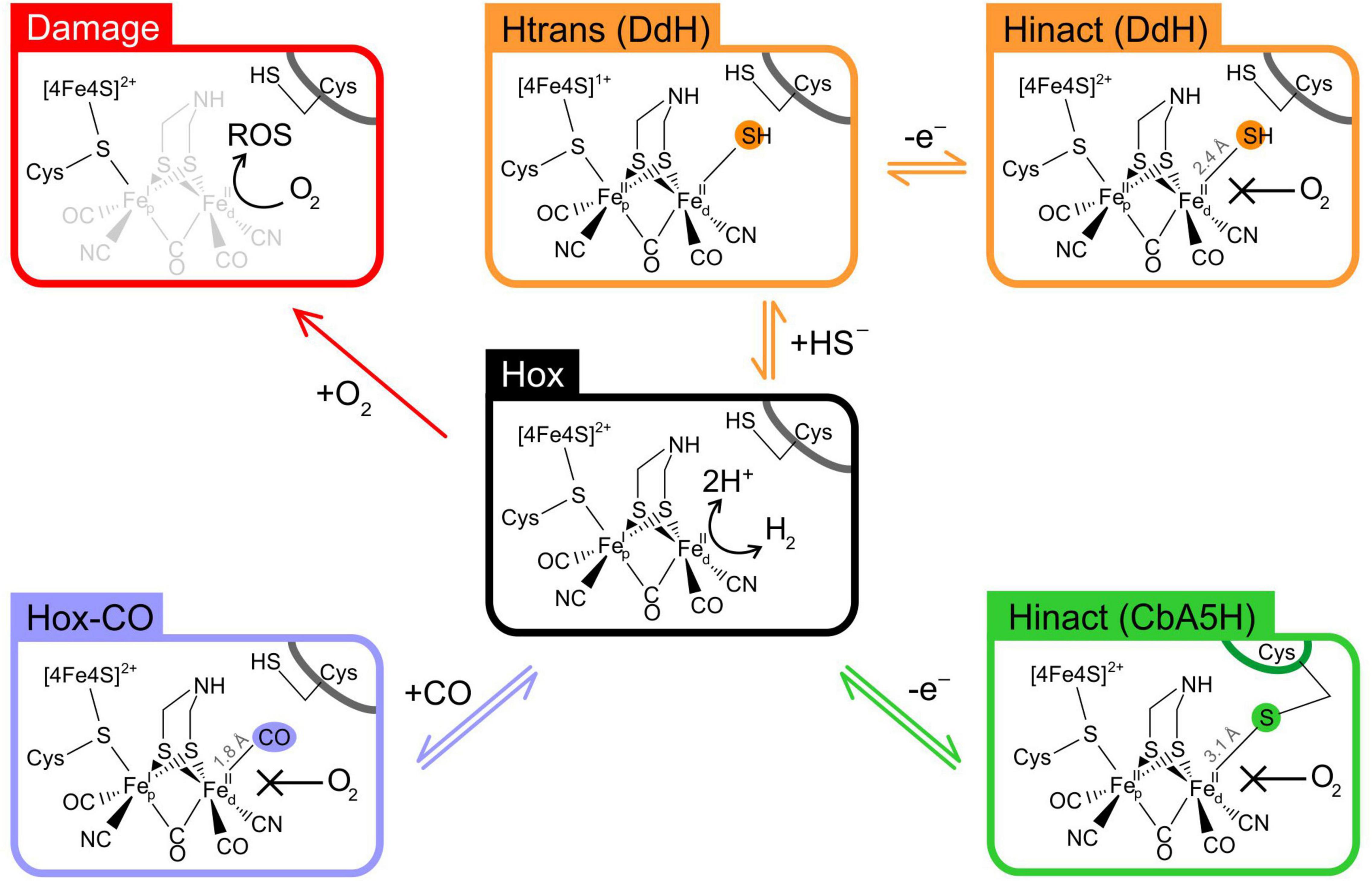
Figure 2. Oxygen tolerance strategies in [FeFe]-hydrogenases. Schematic representation of the H-cluster in the oxidised active state Hox (centre). In the absence of any exogenous protectant, numerous [FeFe]-hydrogenases undergo irreversible inactivation due to H-cluster damage with loss of Fe atoms (red pathway); carbon monoxide acts as a protective agent due to its ability to form Hox-CO, by binding reversibly to the H-cluster at the same site as O2 (purple pathway); in DdH, a similar mechanism occurs when sulphide binds to the H-cluster forming Hinact, via the Htrans intermediate (orange pathway); in CbA5H, a conformational change in the protein structure allows for a conserved cysteine to directly bind to the H-cluster, forming Hinact (green pathway). Fep, proximal iron atom; Fed, distal iron atom; Cys, cysteine residue.
Studies on model enzymes CrHydA1 and CpI have shown that the H-cluster suffers severe and irreversible structural damage when exposed to oxygen, resulting in loss of several Fe atoms and their non-protein ligands (Stripp et al., 2009; Swanson et al., 2015; Esselborn et al., 2019). The exact degradation mechanism is still under debate, but there is agreement that this requires O2 binding to the distal iron atom (Fed) followed by electron and proton transfer, leading to the formation of reactive oxygen species (ROS) that in turn would cause the actual damage (Figure 2). Protection from oxygen can occur in the presence of carbon monoxide via competition, as CO is able to bind to Fed faster than oxygen, forming Hox-CO (Lemon and Peters, 1999; Goldet et al., 2009).
Oxygen sensitivity of [FeFe]-hydrogenases is an obvious practical limitation to working with such enzymes and raises concerns over their prospective exploitation for potential applications (Ghirardi, 2015; Karayilan et al., 2019). As such, several attempts have been made to improve oxygen tolerance of model [FeFe]-hydrogenases, adopting both rational and random mutagenesis approaches but the improvements reported are limited (Lautier et al., 2011; Bingham et al., 2012; King et al., 2014).
More progress has been made by looking at other [FeFe]-hydrogenases. For example, it has been known for decades that DdH and DvH can be purified under air as an inactive state (named Hinact) that can be reactivated by a reductive treatment (Pierik et al., 1998; Roseboom et al., 2006). However, the first attempts to generate Hinact in vitro were unsuccessful, and the exact protection mechanism has been elusive for years. Only recently it was shown that DdH requires the addition of exogenous sulphide to form Hinact, via the intermediate species Htrans. It has been demonstrated that Hinact in DdH (Figure 2) is an overoxidized H-cluster with sulphide bound to Fed, thus preventing O2 binding by direct competition and protecting the enzyme from damage. Given the importance of sulphur metabolism in Desulfovibrio, it has been suggested that this mechanism may play a physiological role in vivo. Interestingly, sulphide dependent Hinact formation is not an exclusive feature of DdH, as it occurs in CrHydA1 as well; however, this mechanism is not effective on CpI, highlighting variability between different [FeFe]-hydrogenases (Rodriguez-Macia et al., 2018; Rodriguez-Macia et al., 2020).
Also recently, it has been shown that CbA5H from C. beijerinckii is able to form Hinact in a fully reversible manner; the enzyme can be inactivated and reactivated multiple times without any loss of activity (Morra et al., 2016a). Further characterisation showed that Hinact formation in CbA5H does not require exogenous sulphide and spectro-electrochemical titrations showed that the Hox/Hinact transition in CbA5H is a 1-electron process that occurs at an unusually low potential, without forming Htrans (Corrigan et al., 2020). Xray crystallography has recently confirmed that Hinact in CbA5H is indeed independent of exogenous sulphide, as a sulphur atom from a conserved cysteine can bind directly Fed, following a conformational change within the enzyme (Figure 2). This is particularly fascinating since the immediate surroundings of the H-cluster in CbA5H are conserved and do not differ from other [FeFe]-hydrogenases. Hence, it has been proposed that the protection mechanism depends on three non-conserved amino acids, situated far away from the H-cluster, that allow for the local conformational change to occur. This finding highlights the complexity of the interplay between the H-cluster and the protein residues as a fundamental part of [FeFe]-hydrogenase function, challenging the common assumption of [FeFe]-hydrogenases being a rigid scaffold that simply hosts a metal cluster responsible for their peculiar features (Winkler et al., 2021).
Protection from oxygen is not limited to few fortunate cases, as it has been shown that two other [FeFe]-hydrogenases are likely to display similar protection mechanisms: DfHnd, the heterotetrameric bifurcating [FeFe]-hydrogenase from Desulfovibrio fructosovorans forms Hinact and is protected from oxygen damage (Kpebe et al., 2018); also CpIII can form a species whose spectroscopy is reminiscent of Hinact (Artz et al., 2020).
Overall, recent research has highlighted that oxygen sensitivity/tolerance in [FeFe]-hydrogenases is much more complex than previously assumed. Different enzymes display completely different reactivity toward oxygen, with a growing number of them being able to tolerate it. It emerges that long term protection from oxygen damage in [FeFe]-hydrogenases invariably requires a nucleophilic species (CO, H2S or a cysteine thiol) to directly bind Fed thus competing for O2 binding (Figure 2). While carbon monoxide protection seems to be a universal feature of all hydrogenases, sulphur-based protection appears to vary significantly between different enzymes. This clearly highlights that the protein environment plays a crucial role in determining the enzyme’s fate when exposed to O2, but the exact determinants for such a diversity are not comprehensively understood. Also, the physiological implications of oxygen tolerance in [FeFe]-hydrogenases have not been fully addressed yet.
Conclusion and Future Perspectives
Recent characterisation of novel [FeFe]-hydrogenases has highlighted a broad functional spectrum that exceeds their prototypical role in hydrogen production. Involvement in hydrogen sensing, electron bifurcation and CO2 reduction demonstrate how diverse [FeFe]-hydrogenase functions are, and how crucial they are to support cellular metabolism under different conditions. It has also become clear that several [FeFe]-hydrogenases have evolved strategies to cope with oxygen, making them oxygen tolerant.
Despite the major advancements summarised here, the current understanding of [FeFe]-hydrogenase diversity is still limited because few enzymes have been characterised in detail so far. Studying novel [FeFe]-hydrogenases and populating the phylogenetic tree with experimental evidence from this unexplored space will be essential to improve our understanding of [FeFe]-hydrogenase function.
Research in this field has shown that relying mainly on few model enzymes and extrapolating information, in the assumption that they represent a vast enzyme class, inevitably leads to severe biases. Providing accurate predictions purely based on primary sequence data and phylogenetic positioning will always be a risky task but it is reasonable to expect that expanding our knowledge to additional enzymes will make comparisons between closely related sequences more reliable. Improving our understanding of [FeFe]-hydrogenase function will provide numerous benefits, for example when interpreting-omics data, as this task heavily relies on previous information being available and correctly annotated in databases. Also, the availability of additional enzymes will inevitably expand the portfolio of [FeFe]-hydrogenases with desirable features for a given application, either in vitro in a technological device, or in vivo within an engineered organism, or as an inspirational example for artificial catalysts, thus increasing the chances for success in future applications.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
SM gratefully acknowledges support from the University of Nottingham via the Nottingham Research Fellowship scheme.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.853626/full#supplementary-material
References
Adams, M. W. W. (1990). The structure and mechanism of iron-hydrogenases. Biochim. Biophys. Acta 1020, 115–145. doi: 10.1016/0005-2728(90)90044-5
Ahmed, M. E., and Dey, A. (2019). Recent developments in bioinspired modelling of NiFe - and FeFe -hydrogenases. Curr. Opin. Electrochem. 15, 155–164. doi: 10.1021/acs.accounts.0c00315
Arizzi, M., Morra, S., Gilardi, G., Pugliese, M., Gullino, M. L., and Valetti, F. (2021). Improving sustainable hydrogen production from green waste: FeFe -hydrogenases quantitative gene expression RT-qPCR analysis in presence of autochthonous consortia. Biotechnol. Biofuels 14:17. doi: 10.1186/s13068-021-02028-3
Artz, J. H., Zadvornyy, O. A., Mulder, D. W., Keable, S. M., Cohen, A. E., Ratzloff, M. W., et al. (2020). Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases. J. Am. Chem. Soc. 142, 1227–1235. doi: 10.1021/jacs.9b08756
Baffert, C., Kpebe, A., Avilan, L., and Brugna, M. (2019). Hydrogenases and H-2 metabolism in sulfate-reducing bacteria of the Desulfovibrio genus. Adv. Microbial. Physiol. 74, 143–189. doi: 10.1016/bs.ampbs.2019.03.001
Barton, R. M., and Worman, H. J. (1999). Prenylated prelamin A interacts with Narf, a novel nuclear protein. J. Biol. Chem. 274, 30008–30018.
Berggren, G., Adamska, A., Lambertz, C., Simmons, T. R., Esselborn, J., Atta, M., et al. (2013). Biomimetic assembly and activation of FeFe -hydrogenases. Nature 499:66. doi: 10.1038/nature12239
Bingham, A. S., Smith, P. R., and Swartz, J. R. (2012). Evolution of an FeFe hydrogenase with decreased oxygen sensitivity. Int. J. Hydrogen Energy 37, 2965–2976. doi: 10.1016/j.ijhydene.2011.02.048
Birrell, J. A., Rodriguez-Macia, P., Reijerse, E. J., Martini, M. A., and Lubitz, W. (2021). The catalytic cycle of FeFe hydrogenase: A tale of two sites. Coordinat. Chem. Rev. 449:214191. doi: 10.1016/j.ccr.2021.214191
Birrell, J. A., Wrede, K., Pawlak, K., Rodriguez-Macia, P., Rudiger, O., Reijerse, E. J., et al. (2016). Artificial Maturation of the Highly Active Heterodimeric FeFe Hydrogenase from Desulfovibrio desulfuricans ATCC 7757. Israel J. Chem. 56, 852–863. doi: 10.1002/ijch.201600035
Boyd, E. S., Hamilton, T. L., Spear, J. R., Lavin, M., and Peters, J. W. (2010). FeFe -hydrogenase in Yellowstone National Park: evidence for dispersal limitation and phylogenetic niche conservatism. Isme J. 4, 1485–1495. doi: 10.1038/ismej.2010.76
Brown, K. A., and King, P. W. (2020). Coupling biology to synthetic nanomaterials for semi-artificial photosynthesis. Photosynth. Res. 143, 193–203. doi: 10.1007/s11120-019-00670-5
Buckel, W., and Thauer, R. K. (2018). Flavin-Based Electron Bifurcation, Ferredoxin, Flavodoxin, and Anaerobic Respiration With Protons (Ech) or NAD(+) (Rnf) as Electron Acceptors: A Historical Review. Front. Microbiol. 9:401. doi: 10.3389/fmicb.2018.00401
Calusinska, M., Hamilton, C., Monsieurs, P., Mathy, G., Leys, N., Franck, F., et al. (2015). Genome-wide transcriptional analysis suggests hydrogenase- and nitrogenase-mediated hydrogen production in Clostridium butyricum CWBI 1009. Biotechnol. Biofuels 8:27. doi: 10.1186/s13068-015-0203-5
Calusinska, M., Happe, T., Joris, B., and Wilmotte, A. (2010). The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiol. Sgm 156, 1575–1588. doi: 10.1099/mic.0.032771-0
Chongdar, N., Birrell, J. A., Pawlak, K., Sommer, C., Reijerse, E. J., Rudiger, O., et al. (2018). Unique Spectroscopic Properties of the H-Cluster in a Putative Sensory FeFe Hydrogenase. J. Am. Chem. Soc. 140, 1057–1068. doi: 10.1021/jacs.7b11287
Cornish, A. J., Green, R., Gartner, K., Mason, S., and Hegg, E. L. (2015). Characterization of Hydrogen Metabolism in the Multicellular Green Alga Volvox carteri. PLos One 10:e0125324. doi: 10.1371/journal.pone.0125324
Corrigan, P. S., Tirsch, J. L., and Silakov, A. (2020). Investigation of the Unusual Ability of the FeFe Hydrogenase from Clostridium beijerinckii to Access an O-2-Protected State. J. Am. Chem. Soc. 142, 12409–12419. doi: 10.1021/jacs.0c04964
Demuez, M., Cournac, L., Guerrini, O., Soucaille, P., and Girbal, L. (2007). Complete activity profile of Clostridium acetobutylicum FeFe -hydrogenase and kinetic parameters for endogenous redox partners. FEMS Microbiol. Lett. 275, 113–121. doi: 10.1111/j.1574-6968.2007.00868.x
Ding, D., Valdivia, A. O., and Bhattacharya, S. K. (2020). Nuclear prelamin a recognition factor and iron dysregulation in multiple sclerosis. Metabolic Brain Dis. 35, 275–282. doi: 10.1007/s11011-019-00515-z
Du, G. Q., Che, J., Wu, Y. D., Wang, Z. Z., Jiang, Z. Y., Ji, F., et al. (2021). Disruption of hydrogenase gene for enhancing butanol selectivity and production in Clostridium acetobutylicum. Biochem. Engin. J. 5:171.
Engelbrecht, V., Liedtke, K., Rutz, A., Yadav, S., Gunzel, A., and Happe, T. (2021). One isoform for one task? The second hydrogenase of Chlamydomonas reinhardtii prefers hydrogen uptake. Int. J. Hydrogen Energy 46, 7165–7175. doi: 10.1016/j.ijhydene.2020.11.231
Esselborn, J., Kertess, L., Apfel, U. P., Hofmann, E., and Happe, T. (2019). Loss of Specific Active-Site Iron Atoms in Oxygen-Exposed FeFe -Hydrogenase Determined by Detailed X-ray Structure Analyses. J. Am. Chem. Soc. 141, 17721–17728. doi: 10.1021/jacs.9b07808
Esselborn, J., Lambertz, C., Adamska-Venkatesh, A., Simmons, T., Berggren, G., Nothl, J., et al. (2013). Spontaneous activation of FeFe -hydrogenases by an inorganic 2Fe active site mimic. Nat. Chem. Biol. 9, 607–609. doi: 10.1038/nchembio.1311
Evans, R. M., Siritanaratkul, B., Megarity, C. F., Pandey, K., Esterle, T. F., Badiani, S., et al. (2019). The value of enzymes in solar fuels research - efficient electrocatalysts through evolution. Chem. Soc. Rev. 48, 2039–2052. doi: 10.1039/c8cs00546j
Florin, L., Tsokoglou, A., and Happe, T. (2001). A novel type of iron hydrogenase in the green alga Scenedesmus obliquus is linked to the photosynthetic electron transport chain. J. Biol. Chem. 276, 6125–6132. doi: 10.1074/jbc.M008470200
Forestier, M., King, P., Zhang, L. P., Posewitz, M., Schwarzer, S., Happe, T., et al. (2003). Expression of two Fe -hydrogenases in Chlamydomonas reinhardtii under anaerobic conditions. Europ. J. Biochem. 270, 2750–2758. doi: 10.1046/j.1432-1033.2003.03656
Germane, K. L., Liu, S. C., Gerlach, E. S., Savage, A. M., Renberg, R. L., Zu, T. N. K., et al. (2018). Hydrogen-Cycling during Solventogenesis in Clostridium acetobutylicum American Type Culture Collection (ATCC) 824 Requires the NiFe -Hydrogenase for Energy Conservation. Ferment. Basel 4:16.
Ghirardi, M. L. (2015). Implementation of photobiological H-2 production: the O-2 sensitivity of hydrogenases. Photosynth. Res. 125, 383–393. doi: 10.1002/mbo3.37
Girbal, L., Von Abendroth, G., Winkler, M., Benton, P. M. C., Meynial-Salles, I., Croux, C., et al. (2005). Homologous and heterologous overexpression in Clostridium acetobutylicum and characterization of purified clostridial and algal Fe-only hydrogenases with high specific activities. Appl. Environ. Microbiol. 71, 2777–2781. doi: 10.1128/AEM.71.5.2777-2781.2005
Goldet, G., Brandmayr, C., Stripp, S. T., Happe, T., Cavazza, C., Fontecilla-Camps, J. C., et al. (2009). Electrochemical Kinetic Investigations of the Reactions of FeFe -Hydrogenases with Carbon Monoxide and Oxygen: Comparing the Importance of Gas Tunnels and Active-Site Electronic/Redox Effects. J. Am. Chem. Soc. 131, 14979–14989. doi: 10.1021/ja905388j
Greening, C., Biswas, A., Carere, C. R., Jackson, C. J., Taylor, M. C., Stott, M. B., et al. (2016). Genomic and metagenomic surveys of hydrogenase distribution indicate H-2 is a widely utilised energy source for microbial growth and survival. ISME J. 10, 761–777. doi: 10.1038/ismej.2015.153
Happe, T., and Kaminski, A. (2002). Differential regulation of the Fe-hydrogenase during anaerobic adaptation in the green alga Chlamydomonas reinhardtii. Europ. J. Biochem. 269, 1022–1032. doi: 10.1046/j.0014-2956.2001.02743.x
Happe, T., and Naber, J. D. (1993). Isolation, characterization and N-terminal amino acid sequence of hydrogenase from the green alga Chlamydomonas reinhardtii. Europ. J. Biochem. 214, 475–481. doi: 10.1111/j.1432-1033.1993.tb17944.x
Haumann, M., and Stripp, S. T. (2018). The Molecular Proceedings of Biological Hydrogen Turnover. Accounts Chem. Res. 51, 1755–1763. doi: 10.1021/acs.accounts.8b00109
Kamp, C., Silakov, A., Winkler, M., Reijerse, E. J., Lubitz, W., and Happe, T. (2008). Isolation and first EPR characterization of the FeFe -hydrogenases from green algae. Biochim. Biophys. Acta Bioenerget. 1777, 410–416. doi: 10.1016/j.bbabio.2008.02.002
Kanygin, A., Milrad, Y., Thummala, C., Reifschneider, K., Baker, P., Marco, P., et al. (2020). Rewiring photosynthesis: a photosystem I-hydrogenase chimera that makes H-2 in vivo. Energy Environ. Sci. 13, 2903–2914.
Karayilan, M., Brezinski, W. P., Clary, K. E., Lichtenberger, D. L., Glass, R. S., and Pyun, J. (2019). Catalytic Metallopolymers from 2Fe-2S Clusters: Artificial Metalloenzymes for Hydrogen Production. Angew. Chem. Int. Edition 58, 7537–7550. doi: 10.1002/anie.201813776
Kelly, C. L., Pinske, C., Murphy, B. J., Parkin, A., Armstrong, F., Palmer, T., et al. (2015). Integration of an [FeFe]-hydrogenase into the anaerobic metabolism of Escherichia coli. Biotechnol. Rep. 8, 94–104.
King, P., Ghirardi, M. L., and Seibert, M. (2014). Oxygen-resistant hydrogenases and methods for designing and making same. USA Patent Appl. 8:958.
King, P. W., Posewitz, M. C., Ghirardi, M. L., and Seibert, M. (2006). Functional studies of FeFe hydrogenase maturation in an Escherichia coli biosynthetic system. J. Bacteriol. 188, 2163–2172.
Klein, M., Ansorge-Schumacher, M. B., Fritsch, M., and Hartmeier, W. (2010). Influence of hydrogenase overexpression on hydrogen production of Clostridium acetobutylicum DSM 792. Enzyme Microbial Technol. 46, 384–390.
Kpebe, A., Benvenuti, M., Guendon, C., Rebai, A., Fernandez, V., Le Laz, S., et al. (2018). A new mechanistic model for an O-2-protected electron-bifurcating hydrogenase, Hnd from Desulfovibrio fructosovorans. Biochim. Biophys. Acta Bioenerget. 1859, 1302–1312. doi: 10.1016/j.bbabio.2018.09.364
Kubas, A., Orain, C., De Sancho, D., Saujet, L., Sensi, M., Gauquelin, C., et al. (2017). Mechanism of O-2 diffusion and reduction in FeFe hydrogenases. Nat. Chem. 9, 88–95. doi: 10.1038/nchem.2592
Kuchenreuther, J. M., Grady-Smith, C. S., Bingham, A. S., George, S. J., Cramer, S. P., and Swartz, J. R. (2010). High-Yield Expression of Heterologous FeFe Hydrogenases in Escherichia coli. PLoS One 5:e15491. doi: 10.1371/journal.pone.0015491
Land, H., Sekretareva, A., Huang, P., Redman, H. J., Nemeth, B., Polidori, N., et al. (2020). Characterization of a putative sensory FeFe -hydrogenase provides new insight into the role of the active site architecture. Chem. Sci. 11, 12789–12801. doi: 10.1039/d0sc03319g
Lautier, T., Ezanno, P., Baffert, C., Fourmond, V., Cournac, L., Fontecilla-Camps, J. C., et al. (2011). The quest for a functional substrate access tunnel in FeFe hydrogenase. Faraday Dis. 148, 385–407. doi: 10.1039/c004099c
Legall, J., Dervartanian, D. V., Spilker, E., Lee, J. P., and Peck, H. D. (1971). Evidence for the involvement of non-heme iron in the active site of hydrogenase from Desulfovibrio vulgaris. Biochim. Biophys. Acta 234, 526–30.
Lemon, B. J., and Peters, J. W. (1999). Binding of exogenously added carbon monoxide at the active site of the iron-only hydrogenase (CpI) from Clostridium pasteurianum. Biochemistry 38, 12969–12973. doi: 10.1021/bi9913193
Leo, F., Schwarz, F. M., Schuchmann, K., and Muller, V. (2021). Capture of carbon dioxide and hydrogen by engineered Escherichia coli: hydrogen-dependent CO2 reduction to formate. Appl. Microbiol. Biotechnol. 105, 5861–5872. doi: 10.1007/s00253-021-11463-z
Losey, N. A., Mus, F., Peters, J. W., Le, H. M., and Mcinerney, M. J. (2017). Syntrophomonas wolfei Uses an NADH-Dependent, Ferredoxin-Independent FeFe -Hydrogenase To Reoxidize NADH. Appl. Environ. Microbiol. 83, e01335–17. doi: 10.1128/AEM.01335-17
Losey, N. A., Poudel, S., Boyd, E. S., and Mcinerney, M. J. (2020). The Beta Subunit of Non-bifurcating NADH-Dependent FeFe -Hydrogenases Differs From Those of Multimeric Electron-Bifurcating FeFe -Hydrogenases. Front. Microbiol. 11:14. doi: 10.3389/fmicb.2020.01109
Lyons, T. W., Reinhard, C. T., and Planavsky, N. J. (2014). The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315. doi: 10.1038/nature13068
Meuser, J. E., Boyd, E. S., Ananyev, G., Karns, D., Radakovits, R., Murthy, U. M. N., et al. (2011). Evolutionary significance of an algal gene encoding an FeFe -hydrogenase with F-domain homology and hydrogenase activity in Chlorella variabilis NC64A. Planta 234, 829–843. doi: 10.1007/s00425-011-1431-y
Meyer, J. (2007). FeFe hydrogenases and their evolution: a genomic perspective. Cell. Mole. Life Sci. 64, 1063–1084. doi: 10.1007/s00018-007-6477-4
Morra, S., Arizzi, M., Allegra, P., La Licata, B., Sagnelli, F., Zitella, P., et al. (2014). Expression of different types of FeFe -hydrogenase genes in bacteria isolated from a population of a bio-hydrogen pilot-scale plant. Int. J. Hydrog. Energ. 39, 9018–9027.
Morra, S., Arizzi, M., Valetti, F., and Gilardi, G. (2016a). Oxygen Stability in the New FeFe -Hydrogenase from Clostridium beijerinckii SM10 (CbA5H). Biochemistry 55, 5897–5900. doi: 10.1021/acs.biochem.6b00780
Morra, S., Mongili, B., Maurelli, S., Gilardi, G., and Valetti, F. (2016b). Isolation and characterization of a new FeFe -hydrogenase from Clostridium perfringens. Biotechnol. Appl. Biochem. 63, 305–311. doi: 10.1002/bab.1382
Morra, S., Valetti, F., and Gilardi, G. (2017). FeFe -hydrogenases as biocatalysts in bio-hydrogen production. Rendiconti Lincei Scienze Fisiche E Naturali 28, 183–194.
Mulder, D. W., Boyd, E. S., Sarma, R., Lange, R. K., Endrizzi, J. A., Broderick, J. B., et al. (2010). Stepwise FeFe -hydrogenase H-cluster assembly revealed in the structure of HydA(Delta EFG). Nature 465, 248–U143. doi: 10.1038/nature08993
Nakos, G., and Mortenson, L. (1971). Purification and properties of hydrogenase, an iron sulfur protein, from Clostridium pasteurianum W5. Biochim. Biophys. Acta 227:576. doi: 10.1016/0005-2744(71)90008-8
Nicolet, Y., Piras, C., Legrand, P., Hatchikian, C. E., and Fontecilla-Camps, J. C. (1999). Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. Struct. Folding Design 7, 13–23. doi: 10.1016/s0969-2126(99)80005-7
Noone, S., Ratcliff, K., Davis, R., Subramanian, V., Meuser, J., Posewitz, M. C., et al. (2017). Expression of a clostridial FeFe -hydrogenase in Chlamydomonas reinhardtii prolongs photo-production of hydrogen from water splitting. Algal Res. Biomass Biofuels Bioprod. 22, 116–121.
Peters, J. W., Beratan, D. N., Bothner, B., Dyer, R. B., Harwood, C. S., Heiden, Z. M., et al. (2018). A new era for electron bifurcation. Curr. Opin. Chem. Biol. 47, 32–38. doi: 10.1016/j.cbpa.2018.07.026
Peters, J. W., Lanzilotta, W. N., Lemon, B. J., and Seefeldt, L. C. (1998). X-ray crystal structure of the Fe-only hydrogenase (Cpl) from Clostridium pasteurianum to 1.8 angstrom resolution. Science 282, 1853–1858. doi: 10.1126/science.282.5395.1853
Peters, J. W., Schut, G. J., Boyd, E. S., Mulder, D. W., Shepard, E. M., Broderick, J. B., et al. (2015). FeFe - and NiFe -hydrogenase diversity, mechanism, and maturation. Biochim. Biophys. Acta Mole. Cell Res. 1853, 1350–1369. doi: 10.1016/j.bbamcr.2014.11.021
Piche-Choquette, S., and Constant, P. (2019). Molecular Hydrogen, a Neglected Key Driver of Soil Biogeochemical Processes. Appl. Environ. Microbiol. 85, e02418–18. doi: 10.1128/AEM.02418-18
Pierik, A. J., Hulstein, M., Hagen, W. R., and Albracht, S. P. J. (1998). A low-spin iron with CN and CO as intrinsic ligands forms the core of the active site in Fe -hydrogenases. Europ. J. Biochem. 258, 572–578. doi: 10.1046/j.1432-1327.1998.2580572.x
Pohorelic, B. K. J., Voordouw, J. K., Lojou, E., Dolla, A., Harder, J., and Voordouw, G. (2002). Effects of deletion of genes encoding Fe-only hydrogenase of Desulfovibrio vulgaris Hildenborough on hydrogen and lactate metabolism. J. Bacteriol. 184, 679–686. doi: 10.1128/JB.184.3.679-686.2002
Poudel, S., Tokmina-Lukaszewska, M., Colman, D. R., Refai, M., Schut, G. J., King, P. W., et al. (2016). Unification of FeFe -hydrogenases into three structural and functional groups. Biochim. Biophys. Acta General Subj. 1860, 1910–1921. doi: 10.1016/j.bbagen.2016.05.034
Rodriguez-Macia, P., Galle, L. M., Bjornsson, R., Lorent, C., Zebger, I., Yoda, Y., et al. (2020). Caught in the H-inact: Crystal Structure and Spectroscopy Reveal a Sulfur Bound to the Active Site of an O-2-stable State of FeFe Hydrogenase. Angewandte Chem. Int. Edition 59, 16786–16794. doi: 10.1002/anie.202005208
Rodriguez-Macia, P., Reijerse, E. J., Van Gastel, M., Debeer, S., Lubitz, W., Rudiger, O., et al. (2018). Sulfide Protects FeFe Hydrogenases From O-2. J. Am. Chem. Soc. 140, 9346–9350.
Roseboom, W., De Lacey, A. L., Fernandez, V. M., Hatchikian, E. C., and Albracht, S. P. J. (2006). The active site of the FeFe -hydrogenase from Desulfovibrio desulfuricans. II. Redox properties, light sensitivity and CO-ligand exchange as observed by infrared spectroscopy. J. Biol. Inorg. Chem. 11, 102–118. doi: 10.1007/s00775-005-0040-2
Schuchmann, K., Chowdhury, N. P., and Muller, V. (2018). Complex Multimeric FeFe Hydrogenases: Biochemistry, Physiology and New Opportunities for the Hydrogen Economy. Front. Microbiol. 9:2911. doi: 10.3389/fmicb.2018.02911
Schuchmann, K., and Muller, V. (2013). Direct and Reversible Hydrogenation of CO2 to Formate by a Bacterial Carbon Dioxide Reductase. Science 342, 1382–1385. doi: 10.1126/science.1244758
Schut, G. J., and Adams, M. W. W. (2009). The Iron-Hydrogenase of Thermotoga maritima Utilizes Ferredoxin and NADH Synergistically: a New Perspective on Anaerobic Hydrogen Production. J. Bacteriol. 191, 4451–4457. doi: 10.1128/JB.01582-08
Schwarz, F. M., Schuchmann, K., and Muller, V. (2018). Hydrogenation of CO2 at ambient pressure catalyzed by a highly active thermostable biocatalyst. Biotechnol. Biofuels 11:11. doi: 10.1186/s13068-018-1236-3
Shaw, A. J., Hogsett, D. A., and Lynd, L. R. (2009). Identification of the FeFe -Hydrogenase Responsible for Hydrogen Generation in Thermoanaerobacterium saccharolyticum and Demonstration of Increased Ethanol Yield via Hydrogenase Knockout. J. Bacteriol. 191, 6457–6464. doi: 10.1128/JB.00497-09
Stripp, S. T., Goldet, G., Brandmayr, C., Sanganas, O., Vincent, K. A., Haumann, M., et al. (2009). How oxygen attacks FeFe hydrogenases from photosynthetic organisms. Proc. Natl. Acad. Sci. U.S.A. 106, 17331–17336. doi: 10.1073/pnas.0905343106
Swanson, K. D., Ratzloff, M. W., Mulder, D. W., Artz, J. H., Ghose, S., Hoffman, A., et al. (2015). FeFe -Hydrogenase Oxygen Inactivation Is Initiated at the H Cluster 2Fe Subcluster. J. Am. Chem. Soc. 137, 1809–1816. doi: 10.1021/ja510169s
Therien, J. B., Artz, J. H., Poudel, S., Hamilton, T. L., Liu, Z. F., Noone, S. M., et al. (2017). The Physiological Functions and Structural Determinants of Catalytic Bias in the FeFe -Hydrogenases CpI and CpII of Clostridium pasteurianum Strain W5. Front. Microbiol. 8:11. doi: 10.3389/fmicb.2017.01305
Vignais, P. M., and Billoud, B. (2007). Occurrence, classification, and biological function of hydrogenases: An overview. Chem. Rev. 107, 4206–4272. doi: 10.1021/cr050196r
Wegelius, A., Land, H., Berggren, G., and Lindblad, P. (2021). Semisynthetic FeFe -hydrogenase with stable expression and H-2 production capacity in a photosynthetic microbe. Cell Rep. Phys. Sci. 2:100376. doi: 10.1016/j.xcrp.2021.100376
Winkler, M., Duan, J. F., Rutz, A., Felbek, C., Scholtysek, L., Lampret, O., et al. (2021). A safety cap protects hydrogenase from oxygen attack. Nat. Commun. 12:756. doi: 10.1038/s41467-020-20861-2
Winkler, M., Esselborn, J., and Happe, T. (2013). Molecular basis of FeFe -hydrogenase function An insight into the complex interplay between protein and catalytic cofactor. Biochim. Biophys. Acta Bioenerg. 1827, 974–985. doi: 10.1016/j.bbabio.2013.03.004
Winkler, M., Heil, B., and Happe, T. (2002). Isolation and molecular characterization of the Fe -hydrogenase from the unicellular green alga Chlorella fusca. Biochim. Biophys. Acta Gene Struct. Exp. 1576, 330–334. doi: 10.1016/s0167-4781(02)00239-7
Zheng, Y. N., Kahnt, J., Kwon, I. H., Mackie, R. I., and Thauer, R. K. (2014). Hydrogen Formation and Its Regulation in Ruminococcus albus: Involvement of an Electron-Bifurcating FeFe -Hydrogenase, of a Non-Electron-Bifurcating FeFe -Hydrogenase, and of a Putative Hydrogen-Sensing FeFe -Hydrogenase. J. Bacteriol. 196, 3840–3852. doi: 10.1128/JB.02070-14
Keywords: hydrogenase, energy metabolism, oxygen sensitivity, H-cluster, metalloenzymes
Citation: Morra S (2022) Fantastic [FeFe]-Hydrogenases and Where to Find Them. Front. Microbiol. 13:853626. doi: 10.3389/fmicb.2022.853626
Received: 12 January 2022; Accepted: 10 February 2022;
Published: 02 March 2022.
Edited by:
Stefan Frielingsdorf, Technical University of Berlin, GermanyReviewed by:
Eric Stephen Boyd, Montana State University, United StatesMyriam Brugna, Aix-Marseille Université, France
Copyright © 2022 Morra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simone Morra, c2ltb25lLm1vcnJhQG5vdHRpbmdoYW0uYWMudWs=
 Simone Morra
Simone Morra